
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 05 April 2024
Sec. Plant Bioinformatics
Volume 15 - 2024 | https://doi.org/10.3389/fpls.2024.1379750
This article is part of the Research TopicGenetically Deciphering the Adaptive Growth and Development of Photosynthetic Organisms: A Holistic Omics ApproachView all 7 articles
Sucrose content is one of the important factors to determine longan fruit flavor quality. To gain deep insight of molecular mechanism on sucrose accumulation in longan, we conducted comparative transcriptomic analysis between low sucrose content longan cultivar ‘Qingkebaoyuan’ and high sucrose content cultivar ‘Songfengben’. A total of 12,350 unique differentially expressed genes (DEGs) were detected across various development stages and different varieties, including hexokinase (HK) and sucrose-phosphate synthase (SPS), which are intricately linked to soluble sugar accumulation and metabolism. Weighted gene co-expression network analysis (WGCNA) identified magenta module, including DlSPS gene, was significantly positively correlated with sucrose content. Furthermore, transient expression unveiled DlSPS gene play crucial role in sucrose accumulation. Moreover, 5 transcription factors (MYB, ERF, bHLH, C2H2, and NAC) were potentially involved in DlSPS regulation. Our findings provide clues for sucrose metabolism, and lay the foundation for longan breeding in the future.
Longan (Dimocarpus longan Lour.) is a typical tropical and subtropical fruit tree which from the Sapindaceae family (Wang et al., 2022). It is widely cultivated in numerous countries, including China, Australia, East Malaysia, and Sri Lanka. Notably, China boasts the largest planting area and highest yield of longan (Lithanatudom et al., 2017). This fruit is rich in nutritional and medical values, earning the nickname of a “treasure among fruits” (Huang et al., 2021). With the enhancement of people’s living standards, evolving consumption patterns, and the globalization of the product market, consumers are increasingly discerning about fruit flavors. Sugar content plays a significant role in determining fruit flavor and quality.
During photosynthesis, sugars are key products of carbon and energy utilization. Within plants, they not only serve as a carbon source but also function as signaling molecules that respond to environmental signals, developmental cues, and metabolic information (Lastdrager et al., 2014). Furthermore, sweetness, an essential component of fruit organoleptic quality, has a direct impact on consumers’ preferences for fresh fruit (Fang et al., 2020a; Fang et al., 2020b). The intensity of sweetness depends on the sugar composition and content as different types of soluble sugars possess varying degrees of sweetness relative to one another (Ma et al., 2015).
In plant, sugars are synthesized in leaves (the source) and transported to sink cells using sugar transporters or plasmodesmata from sieve element-companion cell (SE-CC). These sugars are then stored in vacuoles through the action of sugar transporters located on the vacuole membrane (Chen et al., 2015; Chen et al., 2021; Zhu et al., 2021). Notably, sucrose serves as the primary carbohydrate shuttled from photosynthetically active source tissues to heterotrophic sink tissues, playing a pivotal role in the resource allocation system (Ruan, 2014; Hedrich et al., 2015; Chen, 2022). The accumulation of sucrose in fruits is determined by its transportation and metabolism. Multiple key enzymes were involved in sucrose metabolism, including sucrose phosphate synthase (SPS), sucrose synthase (SS), and invertase (INV) (Ren and Zhang, 2013). SPS converts uridine diphosphate-glucose (UDPG) and fructose-6 phosphate (F6P) into sucrose (Ma et al., 2020). Overexpression of a SPS gene from Arabidopsis family in tobacco resulted in significantly higher sucrose concentrations in the storage sinks compared to WT plants (Park et al., 2008). The loss of the two SPS genes in Arabidopsis thaliana confined sucrose synthesis (Volkert et al., 2014). SuSy, which catalyzes reversible reactions between sucrose and UDPG, participated in the sucrose degradation pathway (Stein and Granot, 2019). A single nucleotide polymorphism (SNP) variation in the SoSUS gene has been found to be significantly associated with sugar-related traits (Khanbo et al., 2023). Overexpression of PtSS3 alters fructose, glucose, and sucrose levels in Populus tomentosa stems and leaves (Li et al., 2020). INV, including acid invertases (AINV) and neutral invertases (NINV), catalyzes sucrose into fructose and glucose (Chen et al., 2021). Inhibition of NtNINV10 gene expression decreased glucose and fructose levels in tobacco leaves (Cheng et al., 2023). Additionally, hexokinase (HK) and fructokinase (FK) play important roles in glucose metabolism. Overexpression of Prunus HXK3 in Arabidopsis significantly decreased soluble sugars content compared to wild-type plants (Pérez-Díaz et al., 2021). Fruits from transgenic apple trees that overexpressed MdFRK2 exhibited significantly elevated levels of fructose compared to wild-type (Su et al., 2023).
Transcription factors (TFs) play important roles in fruit sugar accumulation by influencing the expression levels of sugar metabolism and transport genes (Li et al., 2020; Yue et al., 2020; Wang et al., 2021; Gao et al., 2024; Zhang et al., 2024). In citrus, CitZAT5 adjusts the hexose proportion by regulating the expression of two sugar transport genes (Fang et al., 2023). In apples, MdAREB1.1/1.2 regulate sugar concentration by binding to the promoter of MdTST1/2, thereby altering its expression (Zhu et al., 2023). In pears, PuPRE6 and PuMYB12 act as antagonistic complexes to regulate the transcription of PuSUT4-like and sucrose accumulation (Gao et al., 2024), while PbrbZIP15 promotes soluble sugar accumulation by activating PbrXylA1 transcription during development (Jia et al., 2023).
To investigate the potential molecular regulation mechanism of sucrose metabolism and accumulation in longan, a comparative transcriptomic analysis was performed in high sucrose content cultivar ‘Songfengben’ and low sucrose content cultivar ‘Qingkebaoyuan’ during fruit development. Combined DEG and WGCNA analysis, several key genes involved in sugar accumulation were uncovered, such as sucrose phosphate synthase (DlSPS) and hexokinase (DlHK). Transient overexpressing the DlSPS gene in strawberry fruit increased sucrose accumulation. The findings shed light on the regulatory networks involved in sucrose accumulation during fruit development in longan, which will provide a scientific basis for breeding for this trait.
Two longan cultivars with contrasting sucrose contents, namely, ‘Songfengben’ (high sucrose accumulating type, SFB) and ‘Qingkebaoyuan’ (low sucrose accumulating type, QKBY), were used in this study for RNA-seq. The cultivars were collected at Fruit Research Institute, Fujian Academy of Agricultural Sciences, Fuzhou, China. Fruits of uniform size and free from visible defects were collected from three trees, with ten fruits from one tree were regarded as one replication, at 60, 90, and 120 days after flowering (DAF), respectively. After removing the peel and seed coat, the pulp was immediately preserved in liquid nitrogen and stored at −80°C until further analysis. The ‘Hongyan’ strawberry cultivar used for transient transformation were grown in a greenhouse with 16 h/8 h light conditions at 22°C.
Sugar content was measured using high performance liquid chromatography (HPLC) according to our reported protocol (Fang et al., 2020a). In brief, 0.5 g of fruit sample powder was dissolved in 6 mL deionized water and ultrasonic treatment for 15 min. After centrifugation at 5,000 rpm for 15 min at 4°C, the supernatant was collected and then filtered through a 0.22 μm Sep-Pak filter (ANPEL, Shanghai, China). Sugar content was detected using a 1260 HPLC system (Agilent, USA) with refractive index detector (RID). The mobile phase was deionized water and elution flow rate was 0.5 mL/min.
The RNAprep Pure PlantPlus Kit (Tiangen, Beijing, China) was used to extract RNA in accordance with the manufacturer’s instructions. Subsequently, the quality of the extracted RNA was assessed using both a NanoDrop 2000 spectrophotometer (Thermo, USA) and an Agilent 2100 Bioanalyzer (Agilent, USA). For library preparation, 1 μg of RNA from each sample was used as the starting material. The NEBNext Ultra™ RNA Library Prep Kit for Illumina (NEB, USA) was then utilized to construct the sequencing librariesaccording to the manufacturer’s guidelines. Sequencing was carried out on the Illumina NovaSeq 6000 platform, yielding paired-end RNA-seq reads. Raw data underwent preprocessing to eliminate adapter sequences, reads with poly-N sequences, and those of poor quality. The clean reads were then aligned to the ‘Honghezi’ longan genome (Lin et al., 2017) using HISAT2 with standard settings (Kim et al., 2015). The expression levels of genes were calculated using fragments per kilobase of transcript per millionmapped reads (FPKM). The differentially expressed genes (DEGs) analysis was performed using the DEseq2 R package, with an absolute log2 (fold change) ≥1 and false discovery rate ≤ 0.05 (Love et al., 2014). Both Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses were facilitated by BMKcloud (https://www.biocloud.net/).
The WGCNA was conducted using the WGCNA plug-in (https://github.com/ShawnWx2019/WGCNA-shinyApp). The modules were constructed using the automatic network build function with the following parameters: power=22 and minModuleSize=30. Module-trait associations were considered statistically significant at p<0.05. Genes that were significantly associated with traits were identified within the modules, and their relationships were analyzed. The co-expression network was mapped using Cytoscape V3.8.2 software.
The qRT-PCR analysis were performed on the LightCycler 96 Real-Time PCR Systems (Roche, USA) with the TB Green Premix Ex Taq II (supplied by Takara in Dalian, China) and the primer sequences listed in Supplementary Table S1. Actin was chosen as the reference gene (Jue et al., 2018). The relative gene expression levels were calculated using the 2-ΔΔCt method (Livak and Schmittgen, 2001).
The coding region of DlSPS gene was amplified from the cDNA of ‘SFB’ fruit and inserted into the site of pSAK277 vector between Hind III and XbaI enzyme sites using ClonExpress® IIOne Step Cloning Kit (Vazyme, China). Primer sequences are listed in Supplementary Table S1. The recombinant constructs pSAK277-DlSPS and the empty vector pSAK277 were individually transferred into Agrobacterium tumefaciens EHA105 with the freeze-thaw method. Transient overexpressed in strawberry fruits was conducted as previously described (Pi et al., 2019).
Values are means ± standard deviation (SD) of three biological replicates. The significant differences between mean values was determined by the Student’s t test (*p<0.05 and **p<0.01) calculated using SPSS 21.0 (IBM, USA).
The soluble sugar component and content in two cultivars was mainly carried out at 60, 90, and 120 DAF. Throughout the developmental process, the sucrose content increased most rapidly and becomes the dominant soluble sugar at mature stage (Figure 1). Notably, ‘SFB’ fruit exhibits a significantly higher sucrose content compared to ‘QKBY’ fruit at 120 DAF. However, no significant differences are observed at other developmental stages (Figure 1A). The glucose and fructose contents showed a slight increase from 60 DAF to 90 DAF, followed by a slight decrease from 90 DAF to 120 DAF. Differences in glucose content between the two cultivars are significant at three stages, while significant differences in fructose content are observed at 60 DAF and 120 DAF (Figures 1B, C). Based on the results, we infer that the higher sugar content and sweeter taste of ‘SFB’ longan fruit can be attributed to the higher accumulation of sucrose.
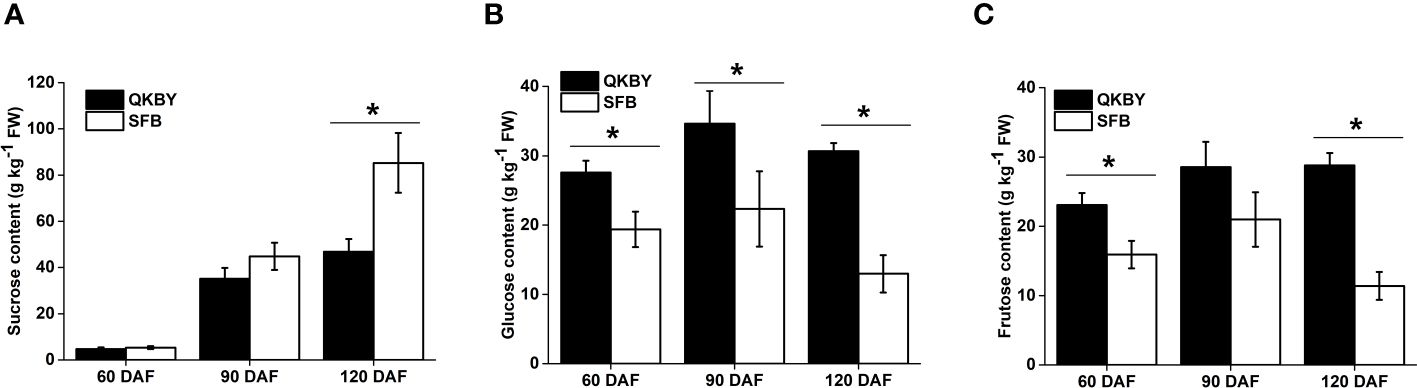
Figure 1 Changes in sugar contents in two longan cultivars during fruit development. (A) Sucrose; (B) Glucose; (C) Fructose. Small star above the bars indicate significant differences (* p ≤ 0.05).
To study the transcriptional regulation of soluble sugar accumulation in longan, RNA-seq was conducted on two cultivars, ‘SFB’ and ‘QKBY’, during three critical developmental stages. An overview of the sequencing quality is provided in Supplementary Table S2. After removing low-quality reads, the number of clean reads per library ranged from 20,418,767 to 81,405,449, with 72.57%-92.28% of the clean reads successfully mapped to the longan genome. The percentage of Q30 bases ranged from 94.60% to 95.63%, indicating the sequencing data was high quality. The GC content of the clean reads varied between 44.19% and 46.14%.
In total, 33,655 genes, including 4,960 novel genes, were identified across all fruit samples tested. For the annotation of these novel unigenes, we conducted a comprehensive search in public databases, including NR, Swiss-Prot, eggNOG, COG, GO, Pfam, KOG, and KEGG. The majority of the novel unigenes were annotated in the NR protein database (2539), followed by eggNOG (2204), Swiss-Prot (1465), KOG (1448), GO (1369), Pfam (1216), KEGG (669), and COG (363) (Supplementary Table S3).
The gene expression levels among different fruit samples were assessed base on the FPKM value. Genes with FPKM values≥1.0 were considered expressed genes. Among the 33,655 identified genes, 17,055, 16,355, and 16,107 genes were expressed at 60 DAF, 90 DAF, and 120 DAF, respectively (Figure 2A). Of these genes, 14,015 were expressed throughout fruit development, while 1,458, 301, and 1,269 were specifically expressed at 60 DAF, 90 DAF, and 120 DAF, respectively (Figure 2B). A principal component analysis (PCA) of the expressed genes clustered them into six groups (Figure 2C). We further classified the expressed genes into five different groups based on their expression levels: extremely low-expressed genes with FPKM values less than 10, low-expressed genes with 10-30 FPKM, medium-expressed genes with 30-100 FPKM, and highly expressed genes with FPKM values above 100 (Figure 2D). Upon comparing the qRT-PCR evaluation of ten genes against the transcriptomic FPKM datasets, the expression patterns were similar to the RNA-Seq data (Supplementary Figure S1).
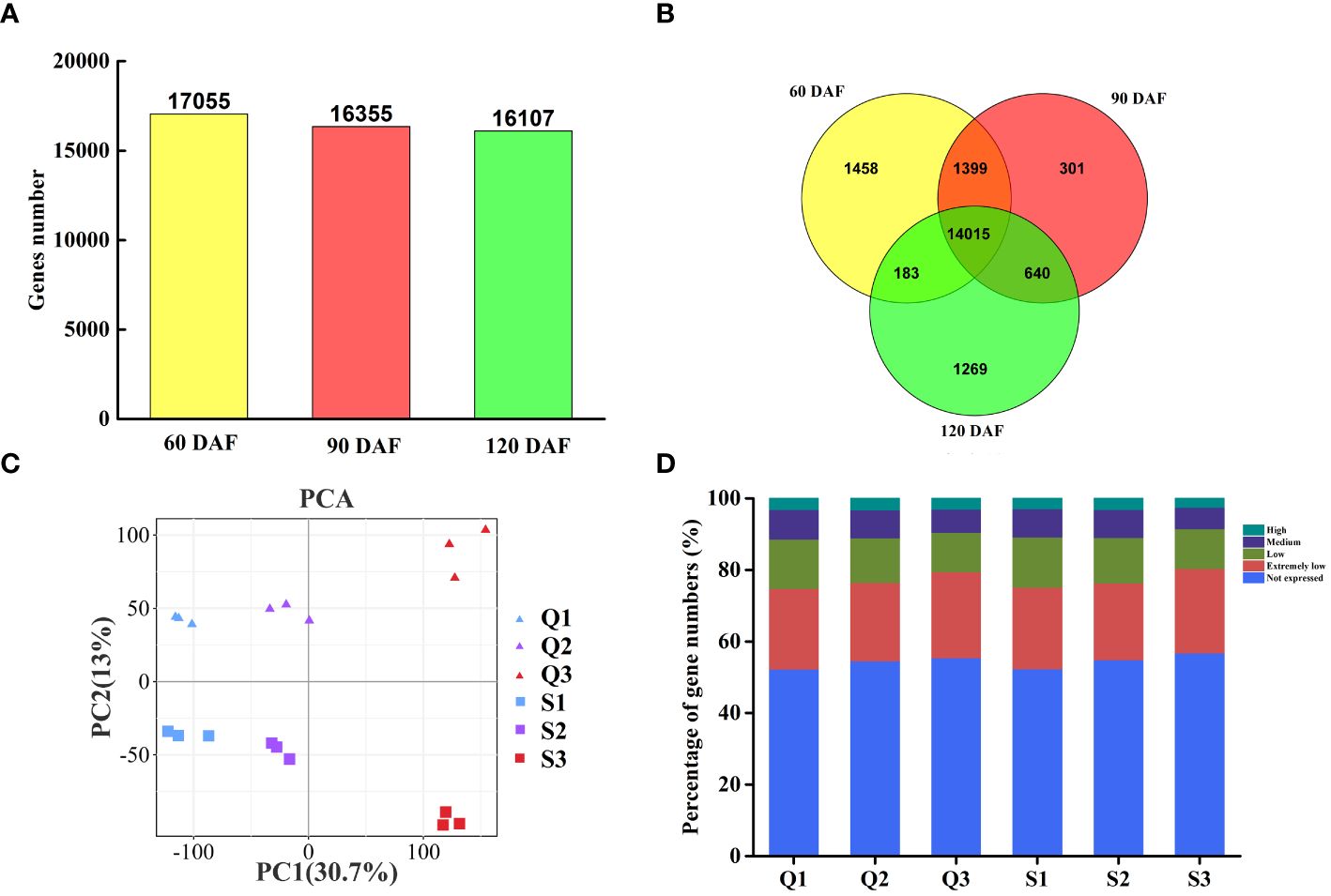
Figure 2 Gene expression and correlation between the transcriptomes of three representative stages each of two longan cultivars. (A) The proportion of expressed genes in two longan cultivars of three typical stages. (B) Venn diagram of expressed genes in two longan cultivars of three typical stages. (C) Principal component analysis (PCA) plot showing clustering of transcriptomes of three representative stages in ‘Songfengben’ and ‘Qingkebaoyuan’. (D) Percentage of gene numbers in different categories according to their expression levels in each sample based on the FPKM values. Q1: QKBY at 60 DAF; Q2: QKBY at 90 DAF; Q3: QKBY at 120 DAF; S1: SFB at 60 DAF; S2: SFB at 90 DAF; S3: SFB at 120 DAF.
To explore the expression-profile differences between two cultivars, the DEGs analysis was conducted. In ‘SFB’, 7,831 and 3,248 genes were found to be differentially expressed in S3 compared to S1 and S2 (Figure 3A). Of these, 3,512 genes were differentially expressed in S2 compared to S1 (Figure 3A). Specifically, 334 genes were up-regulated and 605 genes were down-regulated in S3/S1, S3/S2, and S2/S1 (Figures 3B, C). Similarly, in ‘QKBY’, 7,382 and 3,945 genes were differentially expressed in Q3 compared to Q1 and Q2 (Figure 3D). Among these, 2,987 genes were differentially expressed in Q2 compared to Q1 (Figure 3D). Notably, 346 genes were up-regulated and 347 genes were down-regulated in Q3/Q1, Q3/Q2, and Q2/Q1 (Figures 3E, F). When comparing ‘QKBY’ and ‘SFB’ at the same fruit development time points, 1,894 (Q1 vs. S1), 1,986 (Q2 vs. S2), and 2,667 (Q3 vs. S3) differentially expressed genes (DEGs) were identified (Figure 3G). After excluding redundant genes, a total of 12,350 unique DEGs were detected across various development stages and different longan varieties.
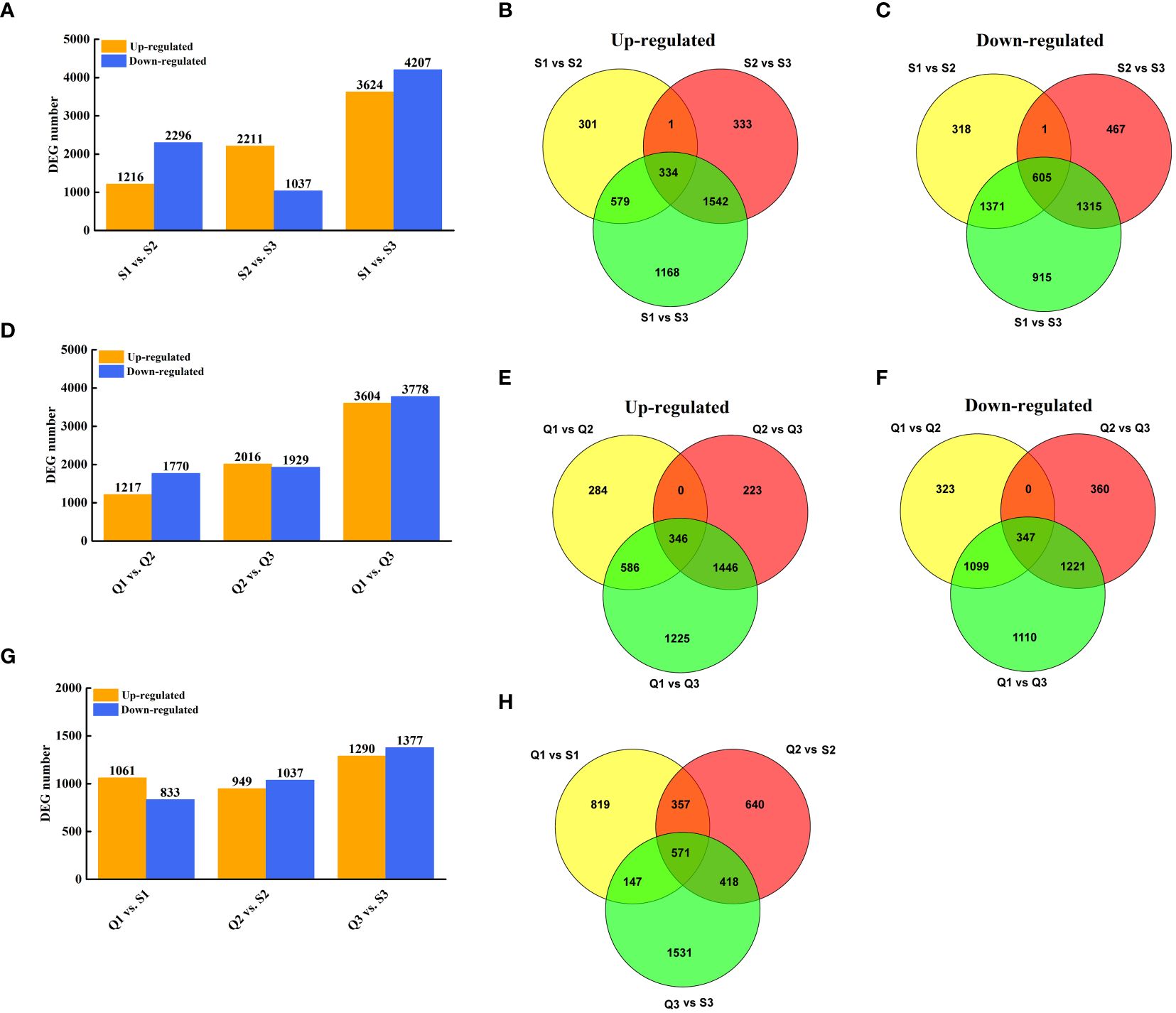
Figure 3 Identification of DEGs at three representative stages each of two longan cultivars. (A) The number of DEGs between two stages in ‘SFB’. (B) Venn diagram of the number of upregulated DEGs during fruit development in ‘SFB’. (C) Venn diagram of the number of downregulated DEGs during fruit development in ‘SFB’. (D) The number of DEGs between two stages in ‘QKBY’. (E) Venn diagram of the number of upregulated DEGs during fruit development in ‘QKBY’. (F) Venn diagram of the number of downregulated DEGs during fruit development in ‘QKBY’. (G) The number of DEGs between two cultivars at same stage. (H) Venn diagram of the number of DEGs between two cultivars at same stage. Q1: QKBY at 60 DAF; Q2: QKBY at 90 DAF; Q3: QKBY at 120 DAF; S1: SFB at 60 DAF; S2: SFB at 90 DAF; S3: SFB at 120 DAF.
To gain insight into the functional properties of the DEGs, a comprehensive GO and KEGG analysis was conducted. The DEGs were categorized into three broad domains: molecular functions, cellular components, and biological processes. Within the cellular component category, DEGs were highly enriched in specific subcategories including the anchored component of the plasma membrane, the plant-type cell wall, the chloroplast envelope, the plasma membrane, and the chloroplast thylakoid membrane. In the biological process category, DEGs were further subclassified into a range of processes including response to light stimulus, regulation of cell size, cell proliferation, chlorophyll biosynthesis process, and polysaccharide biosynthetic process. Notably, within the molecular function category, the sole enriched function was ATP binding (Figure 4A).
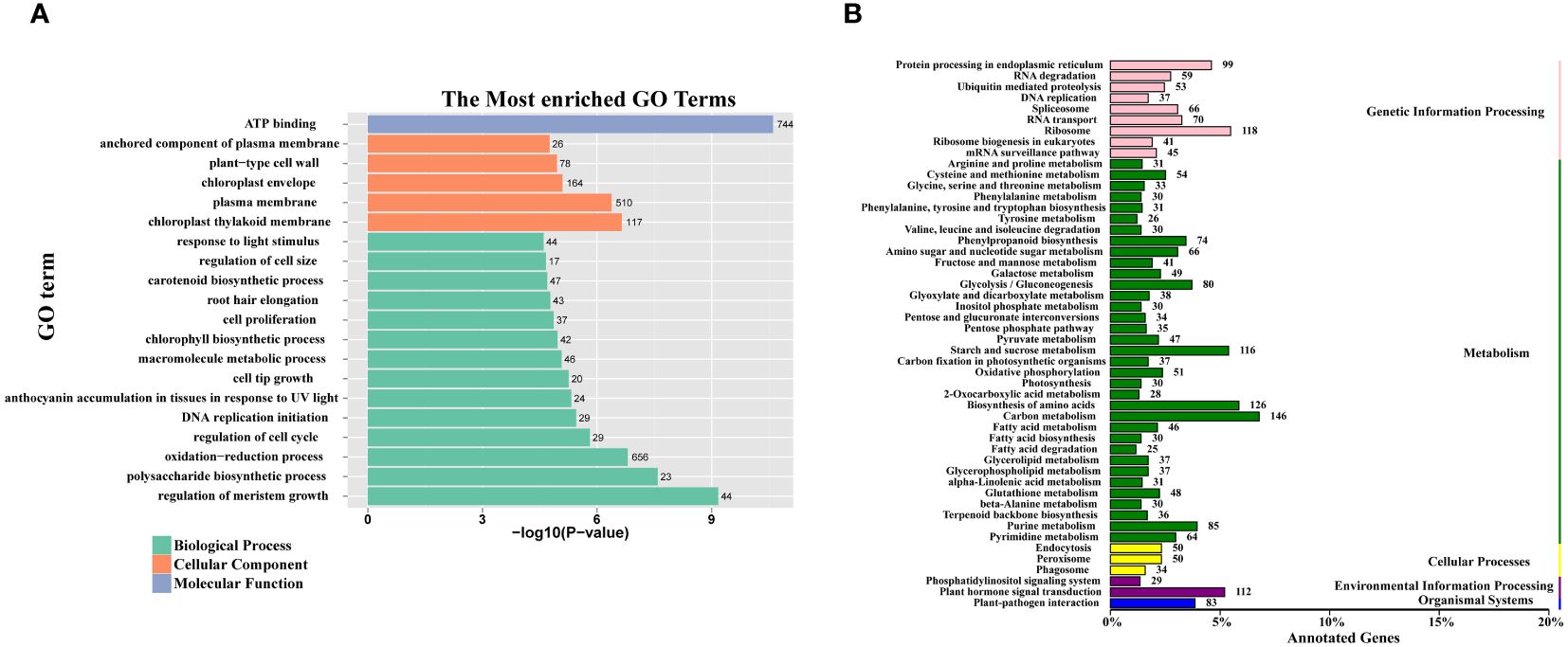
Figure 4 GO enrichment and KEGG class of all DEGs. (A) The 20 most significantly GO terms of DEGs. The numerals beside the histogram indicate the number of DEGs. (B) The KEGG class of DEGs. The numerals beside the histogram indicate the number of DEGs involved in the pathway.
The KEGG pathway classification analysis revealed that the DEGs were significantly associated with several key metabolic pathways. These included fructose and mannose metabolism, starch and sucrose metabolism, glucosinolate biosynthesis, and glycolysis/gluconeogenesis (Figure 4B).
As shown in Figure 1, the sucrose content in ‘SFB’ fruit only exhibit significantly higher than that in ‘QKBY’ at 120 DAF. Therefore, we focused on the DEGs between the two varieties at this stage. Venn analysis revealed that 1,531 genes, which show differential expression specifically at 120 DAF, have been designated as 120 DAF-preferential (Figure 3G). To identify the key regulatory pathways involved in longan sucrose accumulation, we conducted KEGG pathway analysis on the 120 DAF stage-preferential genes. The results indicate that 11 annotated unigenes related to sucrose metabolism (starch and sucrose metabolism) are involved, including genes encoding pectinesterase, beta-glucosidase, endo-(1, 4)-beta-D-glucanase, 1, 4-alpha-glucan-branching, nudix hydrolase, sucrose-phosphate synthase, galacturonosyltransferase, phosphoglucomutase, and UDP-glucuronic acid decarboxylase (Supplementary Figure S2 and Supplementary Table S4). Additionally, the glucose content showed significant differences between ‘SFB’ and ‘QKBY’ during fruit development. A total of 571 common DEGs were identified (Figure 3H). KEGG pathway analysis revealed that 5 annotated unigenes related to glucose metabolism (Glucosinolate biosynthesis and Glycolysis/Gluconeogenesis) are involved, including genes encoding UDP-glycosyltransferase, alcohol dehydrogenase, ATP-dependent 6-phosphofructokinase, pyruvate kinase, and hexokinase (Supplementary Figure S3 and Supplementary Table S5).
Moreover, the fructose content in ‘SFB’ and ‘QKBY’ fruits exhibits significant variations at 60 DAF and 120 DAF. Notably, 718 common DEGs were identified in these two stages. KEGG pathway analysis of these genes led to the identification of two genes involved in fructose metabolism (Fructose and mannose metabolism), including one encoding ATP-dependent 6-phosphofructokinase and another encoding hexokinase (Supplementary Figure S4 and Supplementary Table S6).
We conducted WGCNA to identify the co-expressed gene modules significantly associated with the content of sucrose, glucose, and fructose. The result identified 10 distinct modules, with the number of genes per module ranging from 134 (magenta) to 4,793 (turquoise) (Supplementary Figure S5 and Supplementary Table S7). Notably, the red (r=0.9, p=0.00000028) and magenta (r=0.74, p=0.00047) module was significantly positively correlated with sucrose content. The yellow module was significantly positively correlated with glucose (r=0.86, p=0.000004) and fructose (r=0.83, p=0.000018) content (Figure 5A). To further explore key genes involved in sugar accumulation, venn plot was used to reveal the overlapped filtered genes of significantly modules genes and DEGs. As shown in Figure 5B, one gene (DlSPS) was identified to be involved in sucrose content. Furthermore, one gene (DlHK) and two genes (DlADH and DlHK) were involved in glucose and fructose content, respectively (Supplementary Figure S6).
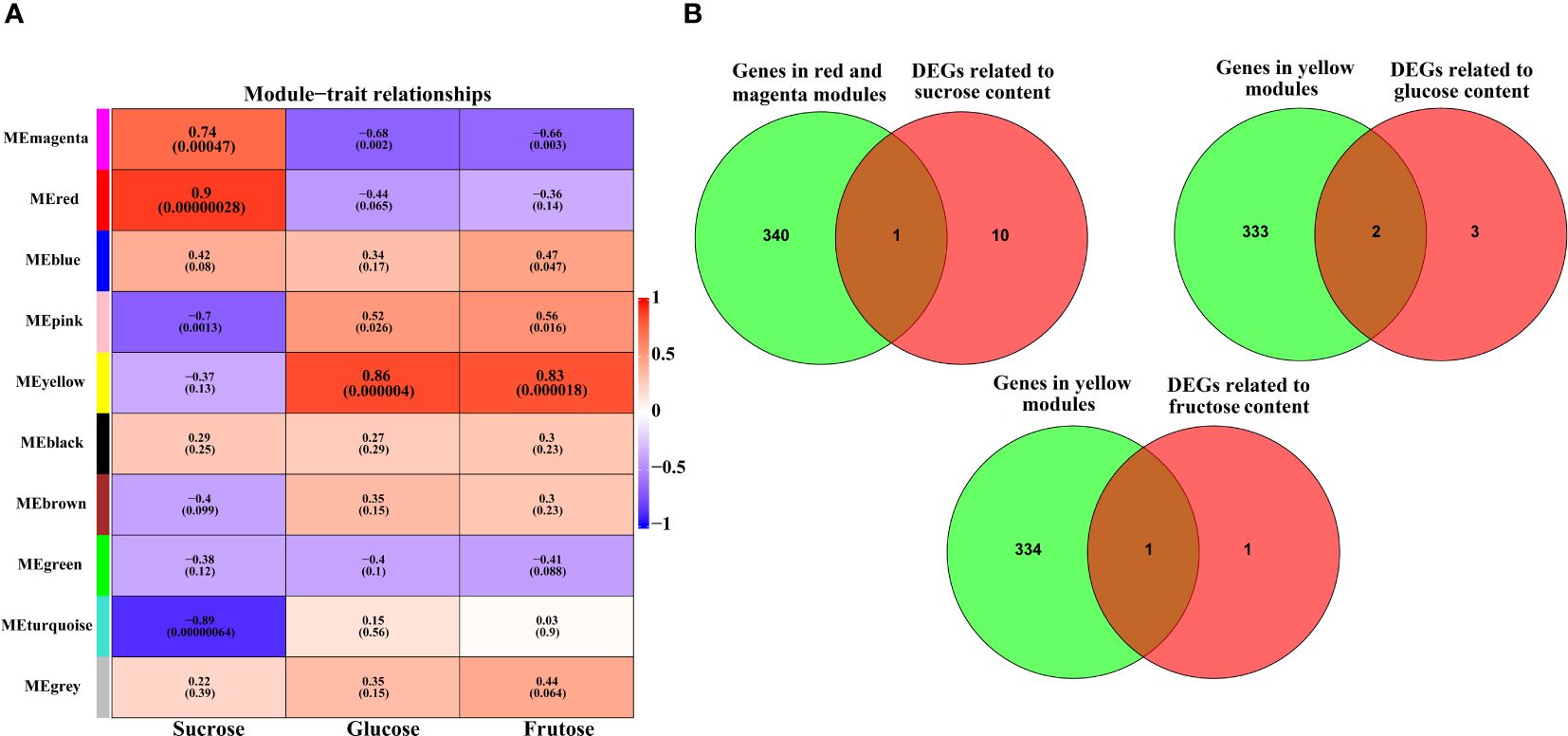
Figure 5 Identification of key genes involved in sugar accumulation. (A) C The heatmap of module−sugar relationships. Each row represents a module indicated by different colors. The color key from blue to red represents correlation values from−1 to 1. (B) Venn diagram of the key genes in each module with DEGs related to sugar metabolism.
To examine the possible role of DlSPS in fruit sucrose accumulation, we transiently transferred pSAK277-DlSPS vector into immature white fruit of strawberry with empty pSAK277 vector as control. The qRT-PCR analysis showed that DlSPS was significantly higher expressed in transgenic fruit than that in the control (Figure 6A). The concentrations of sucrose and total sugar in the DlSPS-overexpression were significantly higher than those in the control. However, the glucose and fructose showed no significant difference between fruits overexpressing DlSPS or empty vector (Figure 6B).
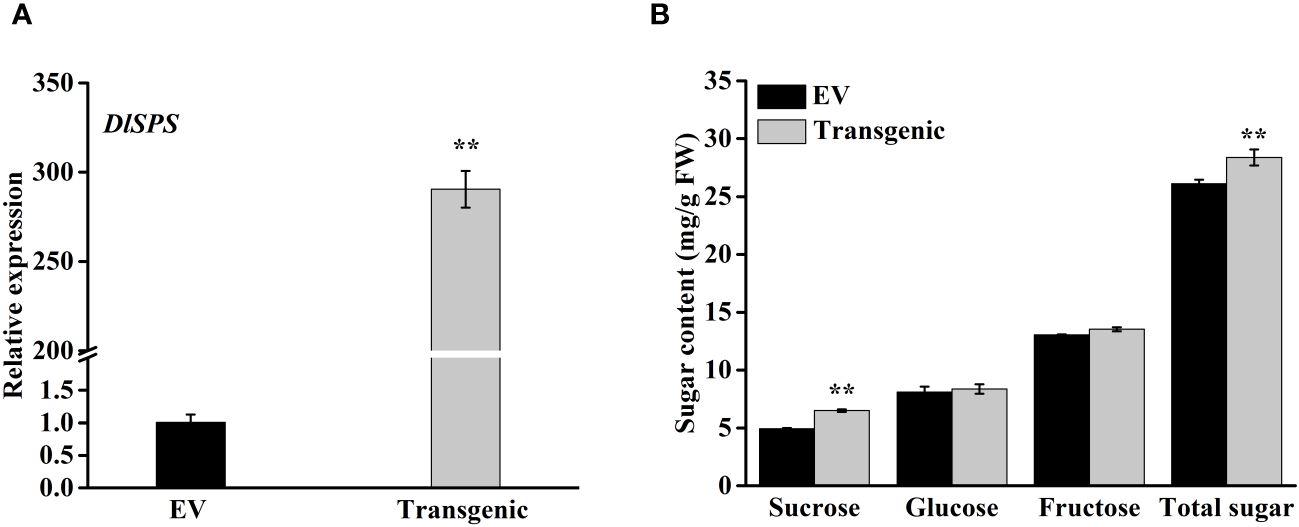
Figure 6 Overexpression of DlSPS in strawberry fruit. (A) Relative expression of DlSPS in strawberry fruits infiltrated with DlSPS and the entry vector, respectively. (B) Sugar content in control and transgenic strawberry fruits, respectively. Small star above the bars indicate significant differences (** p ≤ 0.01).
To identify the key transcription factors regulating sugar content in longan, we first identified the transcription factors present in the magenta, red and yellow modules. The results revealed that a total of 30 transcription factors exist in these three modules, including bHLH (3), bZIP (1), ERF (4), MYB (4), NAC (3), and WRKY (1) (Figure 7A). As shown in Figure 5A, DlSPS is key gene regulating sucrose synthesis in longan fruit. Therefore, we investigated the potential regulation pathway between TFs and DlSPS. Notably, five TF genes that were categorized in the magenta module (Figure 7B) had similar expression trends to DlSPS. Furthermore, many binding sites of this five TFs on the promoters of DlSPS genes were also identified (Supplementary Table S8).
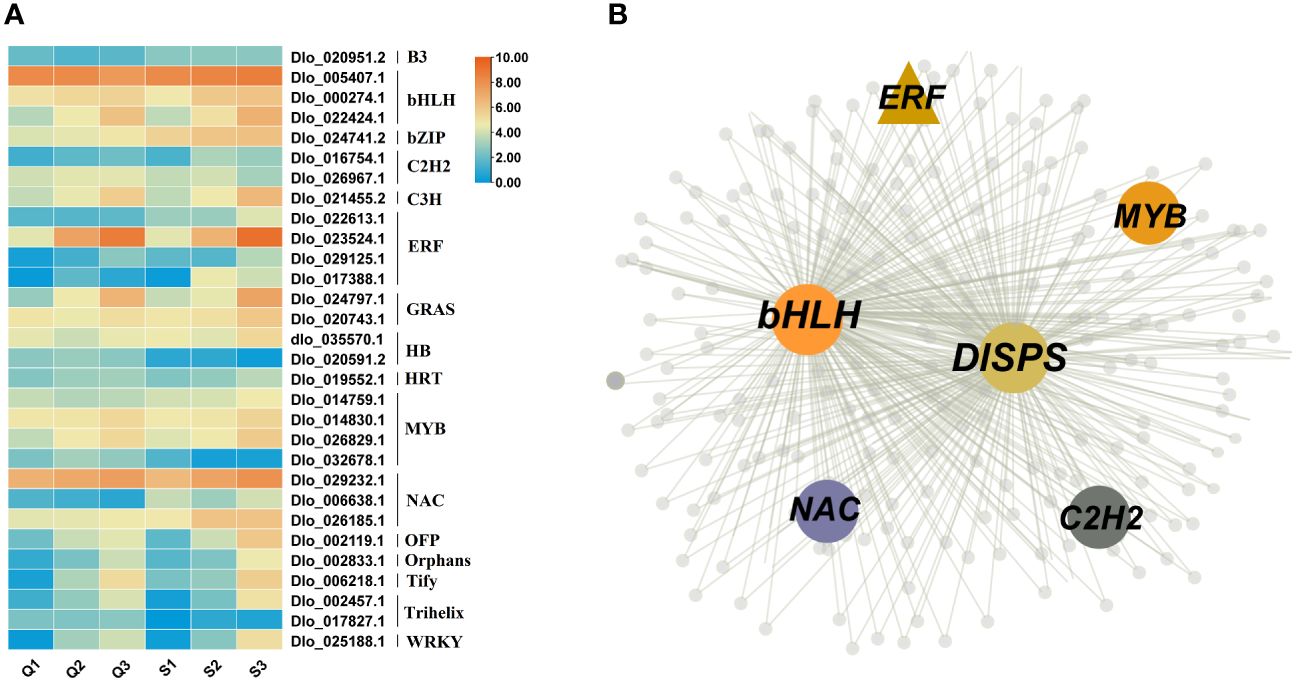
Figure 7 Identification of transcription factors Involved in soluble sugars accumulation. (A) Expression profiles of 30 transcription factors in magenta, red and yellow modules. Q1: QKBY at 60 DAF; Q2: QKBY at 90 DAF; Q3: QKBY at 120 DAF; S1: SFB at 60 DAF; S2: SFB at 90 DAF; S3: SFB at 120 DAF. (B) The co-expression network contained DlSPS genes for the magenta module.
Sweetness, which depends on the soluble sugar components and content, plays crucial role in determining fruit flavor and quality in longan. Various components of soluble sugar have been observed in plants, such as sucrose, fructose, glucose, raffinose, and galactose (Kotha et al., 2020; Zheng et al., 2022). In this study, we found that the main soluble sugars in longan fruit were sucrose, glucose, and fructose, which consistent with previous reports in other fruits such as apples (Ma et al., 2015) and peaches (Vimolmangkang et al., 2016). However, each soluble sugar component showed a distinct accumulation pattern, for instance, the sucrose content increased markedly during fruit development and exhibited significant differences between ‘SFB’ and ‘QKBY’ at the mature stage. While, the sucrose content was notably lower than previously research which conducted on the same two varieties in Guangdong province (Luo et al., 2019). This discrepancy may be caused by different weather and cultivation conditions, such as rainfall, light exposure, temperature, and fertilization. Furthermore, we observed a slight increase in glucose and fructose contents from 60 DAF to 90 DAF, followed by a slight decrease from 90 DAF to 120 DAF. At the mature stage, the contents of glucose and fructose were significantly lower than those of sucrose, consistent with previous research demonstrating that sucrose is the primary component of soluble sugar in longan.
The sugar accumulation during fruit development is regulated by a variety of biological processes, such as sugar metabolism and transportation (Chen et al., 2021). In previous studies, many important enzymes and transporters have been identified in longan, such as soluble acid invertases (SAI), vacuolar glucose transporters family (VGTs), and sugars will eventually be exported transporters (SWEET) (Luo et al., 2019; Fang et al., 2020a, 2022).
To gain a deeper understanding of the molecular mechanisms of sugar accumulation in longan, a comparative transcriptomic analysis was conducted and identified several potential genes involved in sugar metabolism, including SPS and HK. SPS is a key enzyme in sucrose metabolism. In melon, the activity of SPS is positively correlated with sucrose accumulation (Gao et al., 2024; Wang et al., 2023). In sugarcane, overexpression the SoSPS1 gene increased sucrose content in leaves (Anur et al., 2020). In this study, we found expression level of one SPS gene and sucrose content were significant differences between two cultivars at 120 DAF, suggesting that the SPS gene may be involved in sucrose metabolism in longan fruit. Furthermore, transient overexpression of the DlSPS gene enhances sucrose and total sugar content in strawberry fruit, demonstrating DlSPS gene is a strong candidate for the regulation of sugar accumulation in longan fruit. Hexokinase (HK), a multifunctional protein that serves as a sugar sensor for glucose signaling and a catalyst for glycolysis, has also been implicated in sugar metabolism. Overexpression of the PbHXK1 gene in tomato significantly reduced sugar content in leaves (Zhao et al., 2019). In this study, one HK gene showed significant expression levels between two cultivars during fruit development, indicating its potential function in longan soluble sugar metabolism.
Several studies have demonstrated that transcription factors play crucial roles in fruit sugar accumulation. For instance, in apple, MdbHLH3 modulates the soluble sugar content by activating phosphofructokinase gene expression (Yu et al., 2022), while MdMYB305 interacts with sugar-related genes, including MdCWI1, MdVGT3, and MdTMT2, thereby enhancing their activities and ultimately increasing the sugar content in fruits (Zhang et al., 2023). PuWRKY31 bound to the PuSWEET15 promoter and induced its transcription to increase sucrose content in pear (Li et al., 2020). In present study, three modules, including 30 transcription factors (TFs), were significantly positively correlated with soluble sugar content. Our RNA-seq results revealed that DlSPS expression was significantly different between the ‘SFB’ and ‘QKBY’ fruits (Supplementary Table S4). The transient overexpression of the DlSPS gene in strawberry fruit validated its positive role in regulating sucrose accumulation. Previous studies revealed various TFs participated in sucrose synthesis via binding to promoters of SPS to alter its expression level. For instance, CdWRKY2 is a positive regulator in cold stress by targeting CdSPS1 promoters and activating its expression to mediate sucrose biosynthesis in bermudagrass (Huang et al., 2022). CmMYB44 repressed the transcriptional activation of CmSPS1, leading to lower sucrose content in melon (Gao et al., 2024). In present study, we found that five TFs exist in the same module as DlSPS gene and further analysis indicated thatseveral binding sites of this five TFs were found on the promoters of DlSPS genes, suggesting that these TFs might be involved in regulating sucrose pathways via interaction with DlSPS genes, thus affecting sucrose accumulation in longan. Taken together, these results partially elucidate the sucrose accumulation regulation process in longan and provide a theoretical foundation for fruit quality improvement. Obviously, more in-depth works needed to be carried out to identify our hypothesis.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
YL: Writing – original draft, Investigation, Methodology. RR: Investigation, Methodology, Writing – original draft. RP: Investigation, Writing – original draft. YB: Investigation, Writing – original draft. TX: Investigation, Writing – original draft. LZ: Writing – original draft, Project administration, Writing – review & editing. TF: Project administration, Writing – original draft, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This research was funded by the National Natural Science Foundation of China (32102343) and the construction of the plateau discipline of Fujian province (102/71201801101).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2024.1379750/full#supplementary-material
Anur, R. M., Mufithah, N., Sawitri, W. D., Sakakibara, H., Sugiharto, B. (2020). Overexpression of sucrose phosphate synthase enhanced sucrose content and biomass production in transgenic sugarcane. Plants (Basel). 9, 200. doi: 10.3390/plants9020200
Chen, L. Q., Cheung, L. S., Feng, L., Tanner, W., Frommer, W. B. (2015). Transport of sugars. Annu. Rev. Biochem. 84, 865–894. doi: 10.1146/annurev-biochem-060614-033904
Chen, L. Q. (2022). Improved understanding of sugar transport in various plants. Int. J. Mol. Sci. 23, 10260. doi: 10.3390/ijms231810260
Chen, L., Zheng, F., Feng, Z., Li, Y., Ma, M., Wang, G., et al. (2021). A vacuolar invertase csvi2 regulates sucrose metabolism and increases drought tolerance in Cucumis sativus L. Int. J. Mol. Sci. 23, 176. doi: 10.3390/ijms23010176
Chen, T., Zhang, Z., Li, B., Qin, G., Tian, S. (2021). Molecular basis for optimizing sugar metabolism and transport during fruit development. aBIOTECH 2, 330–340. doi: 10.1007/s42994-021-00061-2
Cheng, L., Jin, J., He, X., Luo, Z., Wang, Z., Yang, J., et al. (2023). Genome-wide identification and analysis of the invertase gene family in tobacco (Nicotiana tabacum) reveals NtNINV10 participating the sugar metabolism. Front. Plant Sci. 14. doi: 10.3389/fpls.2023.1164296
Fang, T., Cai, Y., Yang, Q., Ogutu, C. O., Liao, L., Han, Y. (2020a). Analysis of sorbitol content variation in wild and cultivated apples. J. Sci. Food Agric. 100, 139–144. doi: 10.1002/jsfa.10005
Fang, T., Peng, Y., Rao, Y., Li, S., Zeng, L. (2020b). Genome-wide identification and expression analysis of sugar transporter (ST) gene family in Longan (Dimocarpus longan L.). Plants (Basel). 9, 342. doi: 10.3390/plants9030342
Fang, T., Rao, Y., Wang, M., Li, Y., Liu, Y., Xiong, P., et al. (2022). Characterization of the SWEET gene family in Longan (Dimocarpus longan) and the role of DlSWEET1 in cold tolerance. Int. J. Mol. Sci. 23, 8914. doi: 10.3390/ijms23168914
Fang, H., Shi, Y., Liu, S., Jin, R., Sun, J., Grierson, D., et al. (2023). The transcription factor CitZAT5 modifies sugar accumulation and hexose proportion in citrus fruit. Plant Physiol. 192, 1858–1876. doi: 10.1093/plphys/kiad156
Gao, S., Yin, M., Xu, M., Zhang, H., Li, S., Han, Y., et al. (2024). Transcription factors PuPRE6/PuMYB12 and histone deacetylase PuHDAC9-like regulate sucrose levels in pear. Plant Physiol. 194 (3), 1577–1592. doi: 10.1093/plphys/kiad628
Hedrich, R., Sauer, N., Neuhaus, H. E. (2015). Sugar transport across the plant vacuolar membrane: nature and regulation of carrier proteins. Curr. Opin. Plant Biol. 25, 63–70. doi: 10.1016/j.pbi.2015.04.008
Huang, X., Cao, L., Fan, J., Ma, G., Chen, L. (2022). CdWRKY2-mediated sucrose biosynthesis and CBF-signalling pathways coordinately contribute to cold tolerance in Bermudagrass. Plant Biotechnol. J. 20, 660–675. doi: 10.1111/pbi.13745
Huang, S., Han, D., Wang, J., Guo, D., Li, J. (2021). Floral induction of Longan (Dimocarpus longan) by potassium chlorate: application, mechanism, and future perspectives. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.670587
Jia, L., Zhang, X., Zhang, Z., Luo, W., Nambeesan, S. U., Li, Q., et al. (2024). PbrbZIP15 promotes sugar accumulation in pear via activating the transcription of the glucose isomerase gene PbrXylA1. Plant J. 117 (5), 1392–1412. doi: 10.1111/tpj.16569
Jue, D., Sang, X., Liu, L., Shu, B., Wang, Y., Xie, J., et al. (2018). The ubiquitin-conjugating enzyme gene family in longan (Dimocarpus longan lour.): Genome-wide identification and gene expression during flower induction and abiotic stress responses. Molecules 23, 662. doi: 10.3390/molecules23030662
Khanbo, S., Somyong, S., Phetchawang, P., Wirojsirasak, W., Ukoskit, K., Klomsa-Ard, P., et al. (2023). A SNP variation in the Sucrose synthase (SoSUS) gene associated with sugar-related traits in sugarcane. Peer J. 11, e16667. doi: 10.7717/peerj.16667
Kim, D., Langmead, B., Salzberg, S. (2015). HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360. doi: 10.1038/nmeth.3317
Kotha, R. R., Finley, J. W., Luthria, D. L. (2020). Determination of soluble mono, di, and oligosaccharide content in 23 dry beans (Phaseolus vulgaris L.). J. Agric. Food Chem. 68, 6412–6419. doi: 10.1021/acs.jafc.0c00713
Lastdrager, J., Hanson, J., Smeekens, S. (2014). Sugar signals and the control of plant growth and development. J. Exp. Bot. 65, 799–807. doi: 10.1093/jxb/ert474
Li, J., Gao, K., Lei, B., Zhou, J., Guo, T., An, X. (2020). Altered sucrose metabolism and plant growth in transgenic Populus tomentosa with altered sucrose synthase PtSS3. Transgenic Res. 29, 125–134. doi: 10.1007/s11248-019-00184-9
Li, X. Y., Guo, W., Li, J. C., Yue, P. T., Bu, H. D., Jiang, J., et al. (2020). Histone acetylation at the promoter for the transcription factor PuWRKY31 affects sucrose accumulation in pear fruit. Plant Physiol. 182, 2035–2046. doi: 10.1104/pp.20.00002
Lin, Y., Min, J., Lai, R., Wu, Z., Chen, Y., Yu, L., et al. (2017). Genome-wide sequencing of longan (Dimocarpus longan Lour.) provides insights into molecular basis of its polyphenol-rich characteristics. Gigascience 6, 1–14. doi: 10.1093/gigascience/gix023
Lithanatudom, S. K., Chaowasku, T., Nantarat, N., Jaroenkit, T., Smith, D. R., Lithanatudom, P. (2017). A first phylogeny of the genus Dimocarpus and suggestions for revision of some taxa based on molecular and morphological evidence. Sci. Rep. 7, 6716. doi: 10.1038/s41598-017-07045-7
Livak, K. J., Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the2–ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Love, M. I., Huber, W., Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550. doi: 10.1186/s13059-014-0550-8
Luo, T., Shuai, L., Liao, L., Li, J., Duan, Z., Guo, X., et al. (2019). Soluble acid invertases act as key factors influencing the sucrose/hexose ratio and sugar receding in Longan (Dimocarpus longan Lour.) Pulp. J. Agric. Food Chem. 67, 352–363. doi: 10.1021/acs.jafc.8b05243
Ma, B., Chen, J., Zheng, H., Fang, T., Ogutu, C., Li, S., et al. (2015). Comparative assessment of sugar and Malic acid composition in cultivated and wild apples. Food Chem. 172, 86–91. doi: 10.1016/j.foodchem.2014.09.032
Ma, P., Zhang, X., Chen, L., Zhao, Q., Zhang, Q., Hua, X., et al. (2020). Comparative analysis of sucrose phosphate synthase (SPS) gene family between Saccharum officinarum and Saccharum spontaneum. BMC Plant Biol. 20, 422. doi: 10.1186/s12870-020-02599-7
Park, J. Y., Canam, T., Kang, K. Y., Mansfield, S. D. (2008). Over-expression of an Arabidopsis family a sucrose phosphate synthase (SPS) gene alters plant growth and fibre development. Transgenic Res. 17, 181–192. doi: 10.1007/s11248-007-9090-2
Pérez-Díaz, J., Batista-Silva, W., Almada, R., Medeiros, D. B., Arrivault, S., Correa, F., et al. (2021). Prunus Hexokinase 3 genes alter primary C-metabolism and promote drought and salt stress tolerance in Arabidopsis transgenic plants. Sci. Rep. 11, 7098. doi: 10.1038/s41598-021-86535-1
Pi, M., Gao, Q., Kang, C. (2019). Transient expression assay in strawberry fruits. Bio-Protoc. 9, 11. doi: 10.21769/BioProtoc.3249
Ren, X., Zhang, J. (2013). Research progresses on the key enzymes involved in sucrose metabolism in maize. Carbohydr Res. 368, 29–34. doi: 10.1016/j.carres.2012.10.016
Ruan, Y. L. (2014). Sucrose metabolism: gateway to diverse carbon use and sugar signaling. Annu. Rev. Plant Biol. 65, 33–67. doi: 10.1146/annurev-arplant-050213-040251
Stein, O., Granot, D. (2019). An overview of sucrose synthases in plants. Front. Plant Sci. 10. doi: 10.3389/fpls.2019.00095
Su, J., Jiao, T., Liu, X., Zhu, L., Ma, B., Ma, F., et al. (2023). Calcyclin-binding protein-promoted degradation of MdFRUCTOKINASE2 regulates sugar homeostasis in apple. Plant Physiol. 191, 1052–1065. doi: 10.1093/plphys/kiac549
Vimolmangkang, S., Zheng, H., Peng, Q., Jiang, Q., Wang, H., Fang, T., et al. (2016). Assessment of sugar components and genes involved in the regulation of sucrose accumulation in peach fruit. J. Agric. Food Chem. 64, 6723–6729. doi: 10.1021/acs.jafc.6b02159
Volkert, K., Debast, S., Voll, L. M., Voll, H., Schießl, I., Hofmann, J., et al. (2014). Loss of the two major leaf isoforms of sucrose-phosphate synthase in Arabidopsis thaliana limits sucrose synthesis and nocturnal starch degradation but does not alter carbon partitioning during photosynthesis. J. Exp. Bot. 65, 5217–5229. doi: 10.1093/jxb/eru282
Wang, C., Jiang, H., Gao, G., Yang, F., Guan, J., Qi, H. (2023). CmMYB44 might interact with CmAPS2-2 to regulate starch metabolism in oriental melon fruit. Plant Physiol. Biochem. 196, 361–369. doi: 10.1016/j.plaphy.2023.01.047
Wang, J., Li, J., Li, Z., Liu, B., Zhang, L., Guo, D., et al. (2022). Genomic insights into longan evolution from a chromosome-level genome assembly and population genomics of longan accessions. Hortic. Res. 9, uhac021. doi: 10.1093/hr/uhac021
Wang, J. F., Wang, Y. P., Zhang, J., Ren, Y., Li, M. Y., Tian, S. W., et al. (2021). The NAC transcription factor ClNAC68 positively regulates sugar content and seed development in watermelon by repressing ClINV and ClGH3.6. Hortic. Res. 8, 214. doi: 10.1038/s41438-021-00649-1
Yu, J. Q., Gu, K. D., Zhang, L. L., Sun, C. H., Zhang, Q. Y., Wang, J. H., et al. (2022). MdbHLH3 modulates apple soluble sugar content by activating phosphofructokinase gene expression. J. Integr. Plant Biol. 64, 884–900. doi: 10.1111/jipb.13236
Yue, P. T., Lu, Q., Liu, Z., Lv, T. X., Li, X. Y., Bu, H. D., et al. (2020). Auxin-activated MdARF5 induces the expression of ethylene biosynthetic genes to initiate apple fruit ripening. New Phytol. 226, 1781–1795. doi: 10.1111/nph.16500
Zhang, R. X., Liu, Y., Zhang, X., Chen, X., Sun, J., Zhao, Y., et al. (2024). Two adjacent NAC transcription factors regulate fruit maturity date and flavor in peach. New Phytol. 241, 632–649. doi: 10.1111/nph.19372
Zhang, S., Wang, H., Wang, T., Liu, W., Zhang, J., Fang, H., et al. (2023). MdMYB305-MdbHLH33-MdMYB10 regulates sugar and anthocyanin balance in red-fleshed apple fruits. Plant J. 113, 1062–1079. doi: 10.1111/tpj.16100
Zhao, B., Qi, K., Yi, X., Chen, G., Liu, X., Qi, X., et al. (2019). Identification of hexokinase family members in pear (Pyrus × bretschneideri) and functional exploration of PbHXK1 in modulating sugar content and plant growth. Gene 711, 143932. doi: 10.1016/j.gene.2019.06.022
Zheng, X., Yuan, Y., Huang, B., Hu, X., Tang, Y., Xu, X., et al. (2022). Control of fruit softening and Ascorbic acid accumulation by manipulation of SlIMP3 in tomato. Plant Biotechnol. J. 20, 1213–1225. doi: 10.1111/pbi.13804
Zhu, L., Li, Y., Wang, C., Wang, Z., Cao, W., Su, J., et al. (2023). The SnRK2.3-AREB1-TST1/2 cascade activated by cytosolic glucose regulates sugar accumulation across tonoplasts in apple and tomato. Nat. Plants. 9, 951–964. doi: 10.1038/s41477-023-01443-8
Keywords: longan, sucrose, sucrose-phosphate synthase, interact network, transcription factor
Citation: Li Y, Ren R, Pan R, Bao Y, Xie T, Zeng L and Fang T (2024) Comparative transcriptome analysis identifies candidate genes related to sucrose accumulation in longan (Dimocarpus longan Lour.) pulp. Front. Plant Sci. 15:1379750. doi: 10.3389/fpls.2024.1379750
Received: 31 January 2024; Accepted: 22 March 2024;
Published: 05 April 2024.
Edited by:
Dongli He, Hubei University, ChinaReviewed by:
Alexandria Harkey, Wake Forest University, United StatesCopyright © 2024 Li, Ren, Pan, Bao, Xie, Zeng and Fang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ting Fang, ZmFuZ3RpbmdAZmFmdS5lZHUuY24=; Lihui Zeng, bGh6ZW5nQGZhZnUuZWR1LmNu
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.