- 1State Key Laboratory for Quality Ensurance and Sustainable Use of Dao-di Herbs, Institute of Medicinal Plant Development, Chinese Academy of Medical Sciences & Peking Union Medical College, Beijing, China
- 2Engineering Research Center of Chinese Medicine Resource, Ministry of Education, Institute of Medicinal Plant Development, Chinese Academy of Medical Sciences & Peking Union Medical College, Beijing, China
- 3College of Pharmaceutical Sciences, Chengdu Medical College, Chengdu, China
Prunella vulgaris is an important material for Chinese medicines with rosmarinic acid (RA) as its index component. Based on the chromosome-level genome assembly we obtained recently, 51 RA biosynthesis-related genes were identified. Sequence feature, gene expression pattern and phylogenetic relationship analyses showed that 17 of them could be involved in RA biosynthesis. In vitro enzymatic assay showed that PvRAS3 catalyzed the condensation of p-coumaroyl-CoA and caffeoyl-CoA with pHPL and DHPL. Its affinity toward p-coumaroyl-CoA was higher than caffeoyl-CoA. PvRAS4 catalyzed the condensation of p-coumaroyl-CoA with pHPL and DHPL. Its affinity toward p-coumaroyl-CoA was lower than PvRAS3. UPLC and LC-MS/MS analyses showed the existence of RA, 4-coumaroyl-3’,4’-dihydroxyphenyllactic acid, 4-coumaroyl-4’-hydroxyphenyllactic acid and caffeoyl-4’-hydroxyphenyllactic acid in P. vulgaris. Generation and analysis of pvras3 homozygous mutants showed significant decrease of RA, 4-coumaroyl-3’,4’-dihydroxyphenyllactic acid, 4-coumaroyl-4’-hydroxyphenyllactic acid and caffeoyl-4’-hydroxyphenyllactic acid and significant increase of DHPL and pHPL. It suggests that PvRAS3 is the main enzyme catalyzing the condensation of acyl donors and acceptors during RA biosynthesis. The role of PvRAS4 appears minor. The results provide significant information for quality control of P. vulgaris medicinal materials.
Introduction
Prunella vulgaris L. is a perennial medicinal plant of Lamiaceae, which is widely distributed in Asia, North America, Europe and North Africa (National Pharmacopoeia Committee, 2020; Hu et al., 2023). The whole plants and spikes of P. vulgaris are commonly used to treat thyroiditis, mastitis, tuberculosis, infectious hepatitis and hypertension in East Asia, the Middle East, and Europe (Tang et al., 2023). In addition, P. vulgaris spikes are used as the main raw materials of functional herbal tea in the southern provinces of China. Its fresh leaves are used as seasonal vegetables in southeastern China. The whole plants are often used as urban landscape plants for urban greening (Chen et al., 2019). The demand for P. vulgaris in the production of Chinese patented medicines and functional herbal tea is approximately 60 million kilograms per year (Chen et al., 2022; Li et al., 2022).
P. vulgaris is rich in polyphenols, of which rosmarinic acid (RA) is an index component in evaluating the quality of P. vulgaris medicinal materials and Chinese patented medicines. As the main polyphenol component produced in P. vulgaris, RA has a variety of pharmacological activities, such as antioxidant, anti-inflammatory, anti-tumor, anti-allergy, anti-depression, and anti-anxiety (Taguchi et al., 2017). It also has unique pharmacological effects in improving sleep, neurological prevention, reducing testicular injury and inhibiting elastin degradation (Kwon et al., 2017), and has obvious inhibitory effect on liver tumor cells, lung tumor cells and stomach tumor cells (Radziejewska et al., 2018). In addition, RA is easily absorbed and no toxic side effects on blood cells, kidney, and liver (Noguchi-Shinohara et al., 2015).
RA is a depside condensed from two single phenolic acids (Scarpati and Oriente, 1958). One of them is derived from the general phenylpropanoid pathway. It serves as the acyl donor during condensation. The other one is come from the tyrosine-derived pathway. It serves as the acyl acceptor (Figure 1). RA is present in some hornworts, ferns, and multiple taxa of flowering plants and its biosynthetic pathways are probably evolved independently in differently species (Petersen et al., 2009; Levsh et al., 2019). Analysis of RA biosynthesis in Coleus blumei, Phacelia campanularia and Salvia miltiorrhiza showed the existence of three proposed RA biosynthetic routes in different plants (Figure 1), which include the biosynthesis of 4-coumaroyl-4’-hydroxyphenyllactic acid from p-coumaroyl-CoA and p-hydroxyphenyllactic acid (pHPL) in C. blumei and P. campanularia (route 1), the biosynthesis of 4-coumaroyl-3’,4’-dihydroxyphenyllactic acid from p-coumaroyl-CoA and 3,4-dihydroxyphenyllactic acid (danshensu, DHPL) in S. miltiorrhiza (route 2), and the biosynthesis of caffeoyl-4’-hydroxyphenyllactic acid from caffeoyl-CoA and pHPL in S. miltiorrhiza (route 3) (Eberle et al., 2009; Petersen et al., 2009; Di et al., 2013; Levsh et al., 2019; Liu et al., 2019; Lu, 2021). The biosynthetic routes of RA in P. vulgaris are largely unknown.
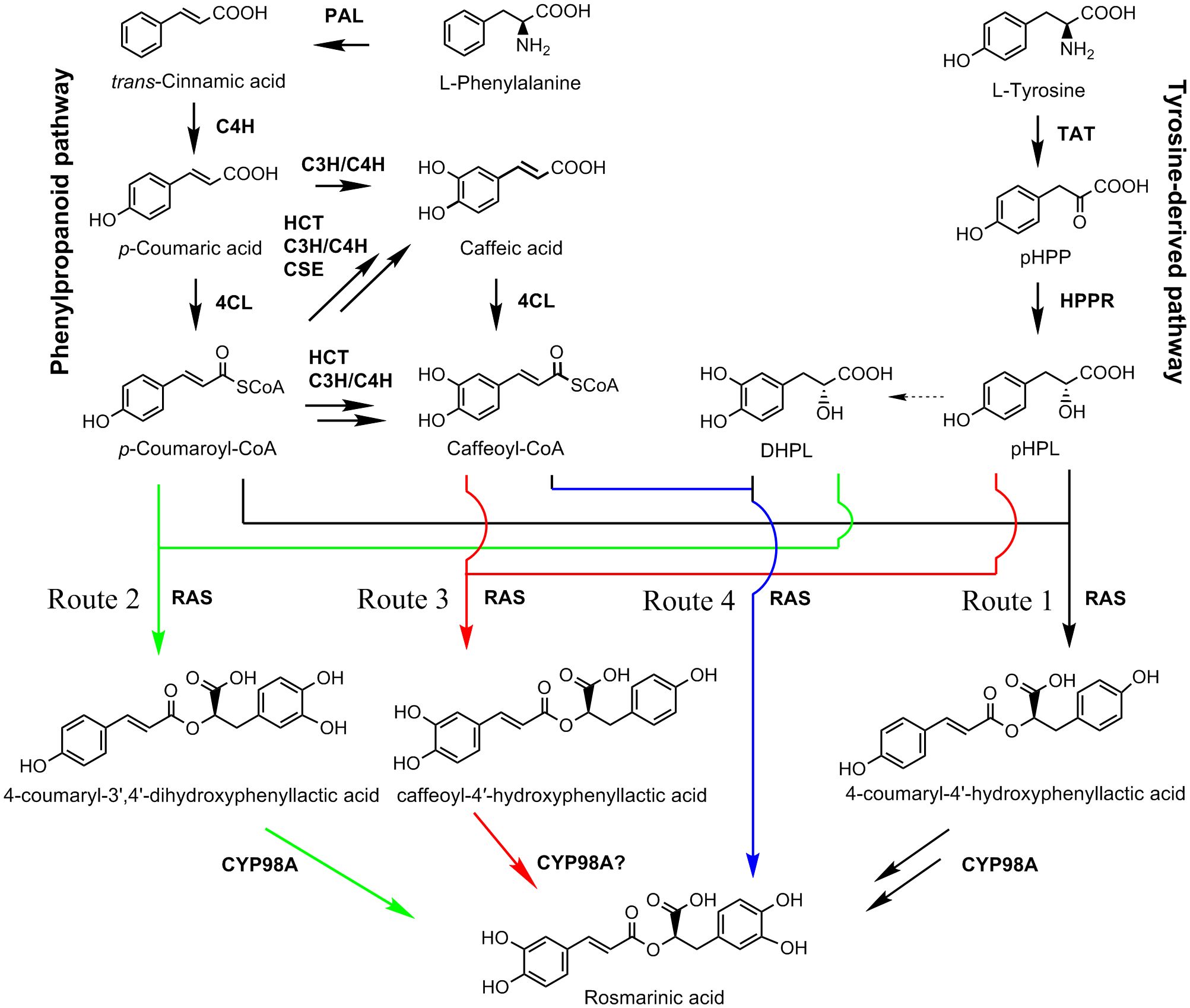
Figure 1 The proposed RA biosynthetic pathways (Updated from Lu, 2021). Solid arrow represents single biosynthetic step. Two arrows represent two or more steps. Dashed arrow indicates the enzyme involved in the reaction is unknown. Four proposed biosynthetic routes of RA are shown in black, green, red, and blue, respectively. Route 1 was found in C. blumei and P. campanularia (Eberle et al., 2009; Petersen et al., 2009; Levsh et al., 2019). Routes 2 and 3 were found in S. miltiorrhiza (Di et al., 2013; Liu et al., 2019). Route 4 was proposed in this study. 4CL, 4-coumaroyl CoA ligase; C3H, p-coumaroyl shikimate 3’-hydroxylase/coumarate 3-hydroxylase; C4H, cinnamate 4-hydroxylase; CSE, caffeoyl shikimate esterase; DHPL, 3,4-dihydroxyphenyllactate (danshensu); HCT, p-hydroxycinnamoyl-CoA: shikimate p-hydroxycinnamoyltransferase; HPPR, p-hydroxyphenylpyruvate reductase; PAL, phenylalanine ammonia lyase; pHPL, p-hydroxyphenyllactic acid; pHPP, p-hydroxyphenylpyruvic acid; RAS, RA synthase; TAT, tyrosine aminotransferase.
Recently, we sequenced and assembled the genome of P. vulgaris, which provide a solid foundation for analyzing RA biosynthetic routes in P. vulgaris (Zhang et al., 2024). In this study, a total of 51 P. vulgaris genes belonging to seven RA biosynthesis-related gene families were systematically studied through genome-wide identification, feature analysis, expression analysis, and phylogenetic analysis. Among them, seventeen were identified as candidate genes for RA biosynthesis. In vitro enzymatic assay of PvRAS3 and PvRAS4, in vivo phenolic acid compound determination and PvRAS3 transgenic analysis showed that PvRAS3 was the main enzyme catalyzing the condensation of acyl donors and acceptors during RA biosynthesis, whereas PvRAS4 played a minor role.
Materials and methods
Plant materials and growth conditions
A wild and whole genome sequenced Prunella vulgaris L. line, named Bangshan-XKC, was transplanted from Bangshan village, Shunchang county of Fujian Province of China and grown in a greenhouse at the Institute of Medicinal Plant Development in Beijing of China. Shoots were cut from the plant and surface-sterilized using 75% ethanol for 1 min and 5% sodium hypochlorite for 20 min. Subsequently, the shoots were rinsed three times with sterile water and inserted into MS medium supplemented with 30 g L-1 sucrose with pH value adjusted to 5.8. After two weeks, the resulting sterile plantlets were transferred to a fresh MS medium. To induce rooting, the apical and axillary buds were cut and placed on 1/2 MS medium containing 0.1 mg L-1 indole-3-butyric acid (IBA). The sterile plantlets were sub-cultivated in a tissue culture room on 1/2 MS medium supplemented with 30 g L-1 sucrose under a 16/8 h light/dark photoperiod at 25°C.
Sequence retrieval and gene prediction
The deduced amino acid sequences of RA biosynthesis-related genes from S. miltiorrhiza were downloaded from NCBI GenBank (https://www.ncbi.nlm.nih.gov/protein) (Wang et al., 2015). BLAST analysis of the downloaded proteins against the chromosome-level assembly of P. vulgaris was carried out using the tBLASTn algorithm (Altschul et al., 1997; Zhang et al., 2024). An E-value cut-off of 10-5 was applied to the homologue recognition. Gene models were predicted from the retrieved P. vulgaris genomic DNA sequences based on the downloaded S. miltiorrhiza genes and through BLASTx analysis of retrieved sequences against the NR database using the default parameters (https://blast.ncbi.nlm.nih.gov/Blast.cgi). The predicted gene models were further examined and corrected manually through BLASTn analysis against P. vulgaris transcriptome sequencing data (Altschul et al., 1997; Zhang et al., 2024).
Gene and protein feature analysis
The theoretical isoelectric point (pI) and molecular weight (Mw) were calculated using the Compute pI/MW tool on the ExPASy server (https://web.expasy.org/compute_pi/). Protein subcellular localization was predicted using Plant-mPLoc version 2.0 (http://www.csbio.sjtu.edu.cn/bioinf/plant-multi/#). The number of transmembrane regions was predicted using DeepTMHMM version 1.0.24 (https://dtu.biolib.com/DeepTMHMM). Distribution of genes on the chromosomes of P. vulgaris was visualized using TBtools (Chen et al., 2020). Intron/exon structures were predicted using GSDS2.0 on the Gene Structure Display Server (http://gsds.gao-lab.org/).
Phylogenetic analysis
RA biosynthesis-related protein sequences from various plant species were downloaded from NCBI GenBank (https://www.ncbi.nlm.nih.gov/protein). Sequence alignment was carried out using the ClustalW algorithms in MEGA version 7.0.26 (Kumar et al., 2016). Neighbor-joining trees were constructed for amino acid sequences using MEGA versopm 7.0.26 with default parameters (Kumar et al., 2016). The number of bootstrap replications was 1000.
Quantitative real-time PCR analysis of gene expression
Total RNA was extracted from roots, stems, leaves and spikes using the EASYspin Plus Complex Plant RNA kit (Aidlab, China) as described previously (Cui et al., 2022). Genomic DNA contamination was eliminated by treating with RNase-free DNase (Aidlab, China). RNA integrity was evaluated on a 1% argarose gel. RNA quantity was determined using a NanoDrop 2000C Spectrophotometer (Thermo Scientific, USA). RNA was reverse-transcribed into single-stranded cDNA using Superscript III Reverse Transcriptase (Invitrogen, USA). qRT-PCR was carried out using TB Green Premix Ex Taq II (Takara, Japan) on a Bio-Rad CFX96 Real-Time system. Primers used for qRT-PCR were designed using Primer Premier 5 (Lalitha, 2000) and are shown in Supplementary Table 1. Gene amplification efficiency of each primer pair was evaluated using the standard curves. Primer pairs with an appropriate PCR amplification efficiency were used for subsequent analysis (Supplementary Figure 1). PveIF-2 was selected as the reference gene as described before (Zheng et al., 2022). The specificity of amplification was assessed by dissociation curve analysis. Relative abundance of transcripts was determined using the 2-ΔΔCt method. Standard deviations were calculated from three biological replicates and three PCR replicates per biological replicates.
Analysis of gene expression using RNA-seq data
Gene expression was analyzed using the published transcriptome data from roots, stems, leaves, and spikes (Zhang et al., 2024). Salmon software (v1.10.3) (Sahraeian et al., 2017) was used to quantify the level of gene expression. Heat maps were constructed using the TBtools software (Chen et al., 2020).
PvRAS3 and PvRAS4 gene cloning and expression vector construction
Total RNA extracted from leaves of P. vulgaris was reverse-transcribed into cDNA using the SuperScript III First-Strand Synthesis System for RT-PCR (Invitrogen, USA). PvRAS3 and PvRAS4 were amplified by nested PCR using cDNA from P. vulgaris leaves as the template. The nesting and nested primers used for PCR are listed in Supplementary Table 1. PCR products were inserted into pGEX-4T-1 and verified by Sanger sequencing.
Heterologous expression of PvRAS3 and PvRAS4 proteins in E. coli
The pGEX-4T-1 vector with PvRAS3 or PvRAS4 was introduced into E. coli strain BL21 (DE3). Heterologous expression of PvRAS3 and PvRAS4 proteins were induced with 0.5 mmol L-1 IPTG at 16 °C for 20–24 h. Cells were collected through centrifugation at 6,000 rpm for 10 min at 4 °C. After resuspension in 10 mM PBS buffer (pH 7.2), the cells were sonicated on ice. Purification of soluble proteins was carried out using the PurKine™ GST-Tag Protein Purification kit (Glutathione) (Abbkine, China). Concentration of the purified proteins was determined using the BCA Protein Assay kit (Takara Biomedical Technology, Beijing).
In vitro enzymatic activity assay of PvRAS3 and PvRAS4 recombinant proteins
The enzymatic activity assay was carried out in a 500 µl reaction system comprising 100 µg purified proteins, 1 mM caffeoyl-CoA or p-coumaroyl-CoA as the acyl donors, 1 mM pHPL or DHPL as the acyl receptors. The reactions were incubated at 25 °C for 60 min and terminated by adding 10 µl of 10 M acetic acid. Controls were carried out using total proteins from E. coli transformed with the empty pGEX-4T-1 vector. Reaction products were collected and analyzed using ACQUITY UPLC system (Waters, Milford, MA, USA). MS/MS data were recorded on a Xevo G2-XS Q-ToF Mass Spectrometer (Waters, Milford, MA, USA) coupled to a Waters Acquity I-Class UPLC system (Waters, Milford, MA, USA). MS/MS analyses were conducted in negative-ion mode. The samples were separated on an ACQUITY UPLC BEH C18 column (1.7 μm, 100×2.1 mm) at 25°C. The mobile phase A was 0.1% (v/v) formic acid-acetonitrile. The mobile phase B was 0.1% (v/v) formic acid in water. The flow rate was 0.3 mL min−1. The mobile phases changed with the following gradient: 0–6 min, 5% A and 95% B; 6–8 min, 20% A and 80% B; 8–14 min, 21% A and 79% B; 14–18 min, 95% A and 5% B. MS was analyzed using electrospray ionization (ESI) at negative ion mode. MS-MS data were analyzed using the MssLynx V4.1 software (Waters) as described previously (Pan et al., 2023).
Kinetic analysis of PvRAS3 and PvRAS4 recombinant proteins
Kinetic analysis of PvRAS3 and PvRAS4 was carried out in a 200 μL reaction system consisting of Tris-HCl buffer (100 mM Tris-HCl, pH 7.0, 2 mM DTT, 4 mM MgCl2, 10% glycerol), 100 μg recombinant protein, and different concentrations of substrates. The reactions were incubated at 25 °C for 30 min and terminated by adding 10 µl of 10 M acetic acid. The reaction products were analyzed using UPLC system as described as in vitro enzymatic activity assay of recombinant proteins. Enzyme activity was determined by measuring the variation of substrate contents. To determine kinetic parameters, PvRAS3 or PvRAS4 was incubated with different concentrations of acyl donor and acyl acceptor. The saturation concentration of one substrate was set at 2 mM, while the concentration of another substrate was varied at different levels, including 10 μM, 50 μM, 100 μM, 250 μM, 350 μM, 500 μM, and 1000 μM, respectively. The kinetic constants of the donor substrates were calculated based on contents of the product. The kinetic constants of the acceptor substrates were determined through monitoring the consumption of the acceptor substrates. Enzyme assays were performed in triplicate at each concentration of substrate. Vmax and Km values were calculated using Origin 8.0 software with nonlinear regression analysis.
UPLC and LC-MS/MS analyses of phenolic acids
Roots, stems, leaves and spikes of two-year-old P. vulgaris were ground in liquid nitrogen. The ground samples (0.5 g) were dissolved in 10 ml of 80% ethanol and sonicated for 60 min. The extracts were collected by centrifugation and filtered using a 0.22 μm filter (Merk Millipore, USA). UPLC and LC-MS/MS analyses were performed using the ACQUITY UPLC I-Class system (Waters) as described as in vitro enzymatic activity assay of recombinant proteins. Three biological and three technological replicates were carried out for analysis of each tissue.
Generation and analysis of pvras3 mutants
Pvras3 mutants of P. vulgaris hairy roots were generated using the CRISPR/Cas9 system described previously (Wang et al., 2022a). Briefly, PCR amplification was carried out using two pairs of primers containing two dividual guide RNAs (sgRNAs) sequences of PvRAS3. pDT1T2 vector was used as a template. The products were purified, digested with Bsa I, and ligated into the binary vector pHEE401E. The resulting constructs were transferred into Agrobacterium strain ATCC15834.
Leaf discs from thirty-day-old sterile plantlets were cultivated on 1/2 MS medium in dark for two days, immersed for 10 min in the suspension of Agrobacterium cells with or without the constructs, and co-cultivated on MS medium for 2 days. The leaf discs were then transferred onto 1/2 MS medium supplemented with 30 mg L-1 of hygromycin and 400 mg L-1 of cefotaxime for generation of hairy roots. Leaf discs were subcultured every two weeks. Hairy roots generated were transferred to 1/2 MS medium supplemented with 200 mg L-1 of cefotaxime and cultivated for about two weeks. Newly generated hairy roots were then transferred to 1/2 MS medium supplemented with 100 mg L-1 of cefotaxime and cultivated for about two weeks. Finally, newly generated hairy roots from medium with 100 mg L-1 of cefotaxime were transferred to 1/2 MS medium without cefotaxime and cultivated for two weeks. Root tips with 3-4 cm in length were cut, transferred to 100 ml of 1/2 MS medium in 250 ml-flasks, and cultivated at 25°C in dark with 100 rpm shaking.
To analyze the mutations of PvRAS3 in transgenic hairy roots, genomic DNA was extracted. DNA fragments around the target site were PCR-amplified using gene-specific primers, Mut-F: GTCGTTTGCTCCCTTACAAAT, and Mut-R: GATCGAAGTGAAGGAGTCGACG. PCR products were sequenced using the primer Mut-F. Hairy roots generated from leaf discs through inoculation with Agrobactrium without the constructs were used as a control. UPLC analysis of chemical compounds was performed using the ACQUITY UPLC I-Class system (Waters). Three biological and three technological replicates were carried out for analysis of each transgenic hairy root line.
Results and discussion
Genome-wide identification of genes associated with RA biosynthesis in P. vulgaris
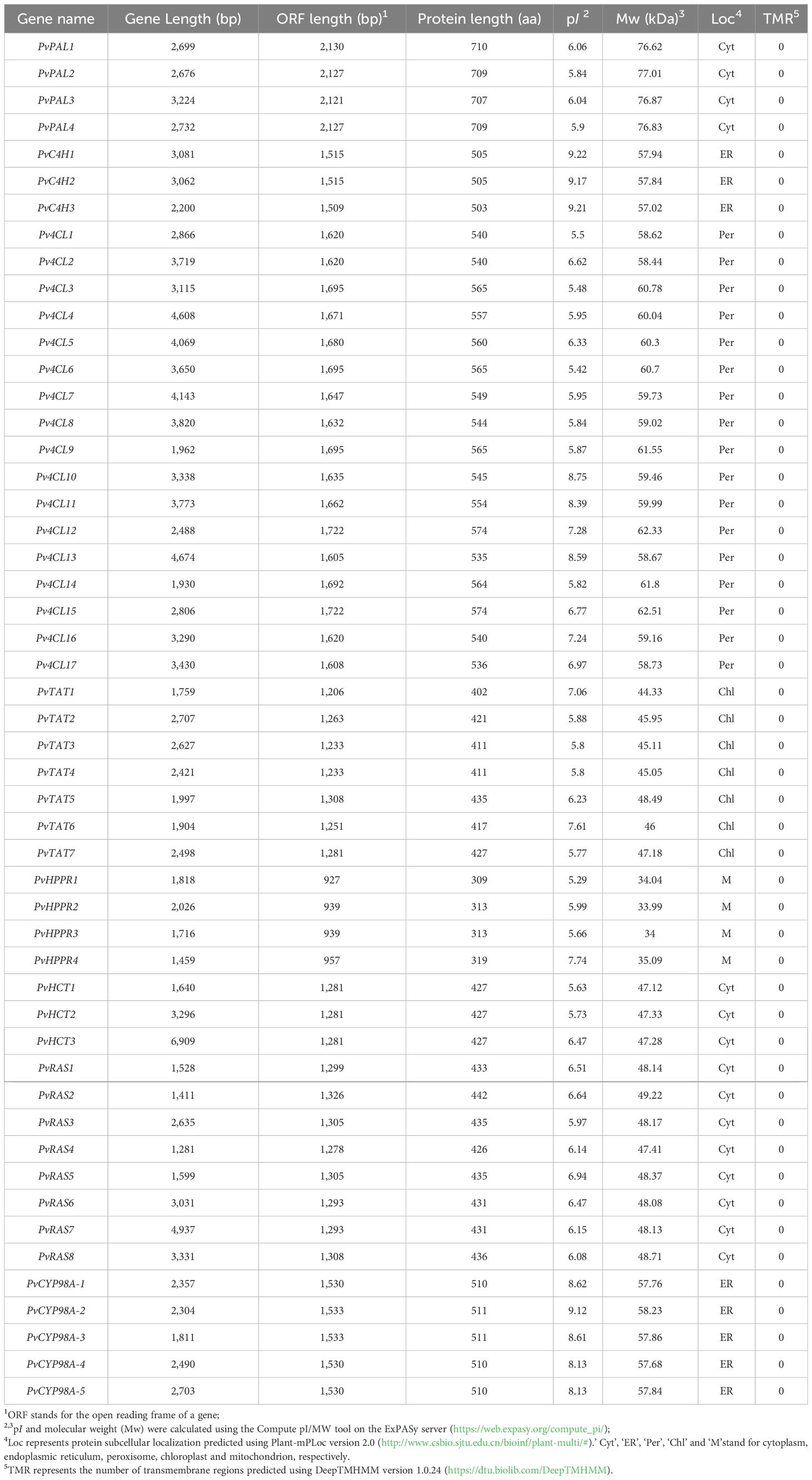
RA is synthesized through the general phenylpropanoid pathway and the tyrosine-derived pathway, involving at least nine enzymes encoded by seven gene families (Figure 1) (Deng and Lu, 2017; Lu, 2021). In order to identify P. vulgaris genes involved in RA biosynthesis, tblastn analysis of the deduced protein sequences of RA biosynthesis-related genes in Salvia miltiorrhiza against the whole-genome assembly of P. vulgaris (2n=28) was carried out (Altschul et al., 1997; Wang et al., 2015; Zhang et al., 2024). It resulted in the identification of 51 full-length candidate genes, including four putative PvPALs, three putative PvC4Hs, seventeen putative Pv4CLs, seven putative PvTATs, four putative PvHPPRs, three putative PvHCTs, eight putative PvRASs, and five putative PvCYP98As (Table 1). These genes were designated as PvPAL1–PvPAL4, PvC4H1–PvC4H3, Pv4CL1–Pv4CL17, PvTAT1–PvTAT7, PvHPPR1–PvHPPR4, PvHCT1–PvHCT3, PvRAS1–PvRAS8, and PvCYP98A-1–PvCYP98A-5, respectively. Among them, PvPAL1 and PvC4H1 showed 99% identities at the amino acid level with the reported PvPAL (KJ010815) and PvC4H (KJ010816), respectively (Kim et al., 2014). Pv4CL3 showed 97% identities at the amino acid level with the reported Pv4CL1 (KJ010814), whereas Pv4CL8 showed 99% amino acid identities with the reported 5’ truncated Pv4CL2 (KJ010817) (Kim et al., 2014). PvTAT3, PvRAS3 and PvCYP98A-1 showed 99% identities at the amino acid level with the reported PvTAT (M053278), PvRAS (KM053280) and PvCYP98A101 (AJW87635), respectively (Ru et al., 2017a; Ru et al., 2017b). PvHPPR3 was identical to the sequence under the GenBank accession number KM053279. The other 43 genes have not been reported previously.
The identified genes are distributed on the 14 chromosomes of the whole genome assembly of P. vulgaris (Supplementary Figure 2) (Zhang et al., 2024). The deduced proteins have length varying from 309 to 709 amino acid residues, pI varying from 5.29 to 9.22, and molecular weight varying from 34.00 to 77.01 (Table 1). All of them do not contain transmembrane regions and were predicted to be localized in the cytoplasm, endoplasmic reticulum, peroxisome, chloroplast and mitochondrion, respectively (Table 1). The predicted localization of PvPALs, PvHCTs and PvRASs in the cytoplasm is consistent with the experimental results from tobacco, S. miltiorrhiza (Achnine et al., 2004; Di et al., 2013). The predicted localization of PvC4Hs and PvCTP98As in the endoplasmic reticulum is consistent with the experimental results from Populus, S. miltiorrhiza, and P. vulgaris (Ro et al., 2001; Di et al., 2013; Ru et al., 2017b). Pv4CLs, PvTATs and PvHPPRs were predicted to be localized in the peroxisome, chloroplast and mitochondrion, respectively. However, Peucedanum praeruptorum 4CL, P. vulgaris TAT and S. miltiorrhiza HPPR were previously found to be located in the cytoplasm (Liu et al., 2017; Wang et al., 2017; Ru et al., 2017a). Thus, the actual subcellular localization of Pv4CLs, PvTATs and PvHPPRs remain to be experimentally analyzed.
Characterization and expression analysis of genes involved in the general phenylpropanoid pathway
The general phenylpropanoid pathway involves three enzymes, including phenylalanine ammonia lyase (PAL), cinnamate 4-hydroxylase (C4H), and 4-coumaroyl CoA ligase (4CL) (Figure 1). PAL catalyzes the conversion of L-phenylalanine to trans-cinnamic acid through deamination of L-phenylalamine. It is the first reaction of the general phenylpropanoid pathway and a rate limiting step mediating the influx from primary metabolism into the general phenylpropanoid pathway (Raes et al., 2003). In a plant, PAL is usually encoded by a small gene family. For instance, there are two PAL genes in tobacco, three in S. miltiorrhiza, and four in Arabidopsis (Raes et al., 2003; Achnine et al., 2004; Wang et al., 2015). Genome-wide analysis showed that there were four putative PvPAL genes in P. vulgaris (Table 1), all of which contained an intron and had similar gene structures (Figure 2A). qRT-PCR analysis showed that the four PvPAL genes were differentially expressed (Figure 2B). RNA-seq analysis showed that PvPAL1 and PvPAL2 were expressed relatively higher than the other two PvPALs (Figure 3A). The expression patterns revealed between qRT-PCR and RNA-seq were largely consistent for PvPALs and other genes analyzed hereinafter. However, discrepancy was also observed, which could be results from the difference of detection technologies, plant tissues, data analysis method, or other unknown factors. Analysis of the deduced PvPAL proteins showed that all of them contained the conserved “GTITASGDLVPLSYIA” motif involved in substrate binding and catalytic activity and the conserved “FL” residues impartent for substrate specificity (Poppe and Rétey, 2005; Watts et al., 2006; Ma et al., 2013). In addition, other three conserved catalytic active sites, including “GLALVNG”, “NDN” and “HNQD”, were also found (Supplementary Figure 3) (He et al., 2020). It indicates that all of the four identified PvPALs have catalytic activity. Phylogenetic analysis of PALs from P. vulgaris, S. miltiorrhiza, Arabidospsis, Populus trichocarpa and various other plant showed that PvPAL1, PvPAL2 and PvPAL4 were grouped with SmPAL1, SmPAL3 and MoPAL involved in RA biosynthesis (Figure 2C) (Weitzel and Petersen, 2010; Song and Wang, 2011; Hou et al., 2013). Taken together with previous results for PvPAL1 (Kim et al., 2014), the presence of RA in roots, stems, leaves and flowers (Kim et al., 2014), and the results from gene expression analysis, we speculated that PvPAL1 and PvPAL2 could be the main PvPALs for RA biosynthesis.
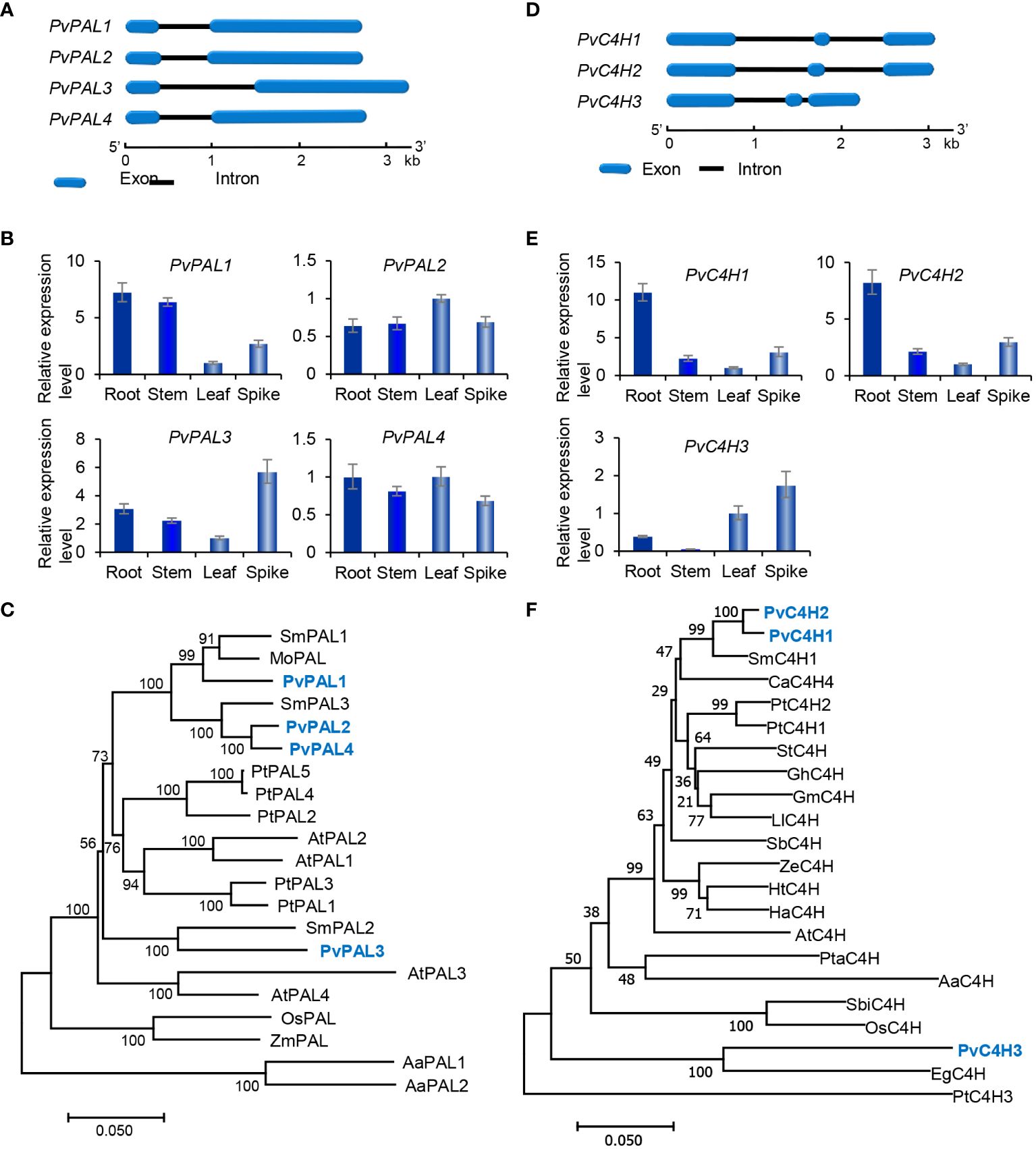
Figure 2 Gene structures, expression patterns and phylogenetic analysis of PvPAL and PvC4H genes and their deduced proteins. (A, D) The intron-exon structures of PvPAL (A) and PvC4H (D) genes. (B, E) Fold changes of PvPAL (B) and PvC4H (E) gene expression in roots, stems, leaves and spikes of P. vulgaris plants. The expression level in leaves was arbitrarily set to 1, respectively. (C) Phylogenetic analysis of PAL proteins. The rooted Neighbor-Joining tree was constructed using the MEGA program (version 7.0) with default parameters. AaPAL1 (QPI70499.1) and AaPAL2 (SPO49995.1) from Anthoceros agrestis were used as outgroup. Ingroup consists of four PvPALs and the PALs from S. miltiorrhiza (Sm), Arabidopsis (At), Populus trichocarpa (Pt), Melissa officinalis (Mo), and maize (Zm) (Supplementary Table 2). (F) Phylogenetic analysis of C4H proteins. C4Hs included are three PvC4Hs and the C4Hs from S. miltiorrhiza (Sm), Coffea Arabica (Ca), P. trichocarpa (Pt), Solanum tuberosum (St), Gossypium hirsutum (Gh), Glycine max (Gm), Leucaena leucocephala (Ll), Scutellaria baicalensis (Sb), Zinnia elegans (Ze), Helianthus tuberosus (Ht), Helichrysum aureonitens (Ha), Arabidopsis (At), Pinus taeda (Pt), A. agrestis (Aa), Sorghum bicolor (Sb), rice (Os), and Erythranthe guttata (Eg) (Supplementary Table 2).
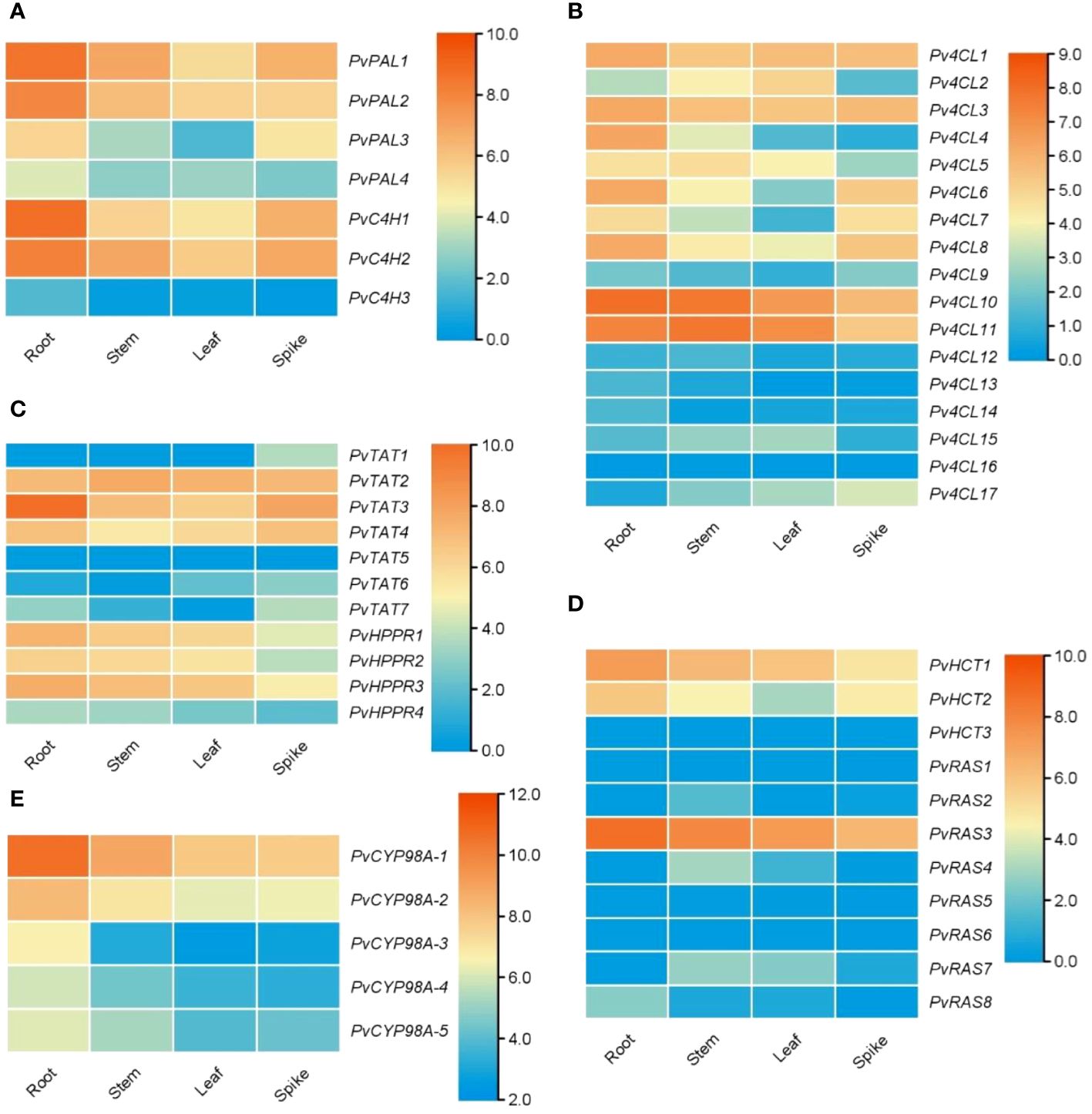
Figure 3 Expression of RA biosynthesis-related genes in roots, stems, leaves and spikes of P. vulgaris. (A–E) Hierarchical clustering of the expression levels of RA biosynthesis-related genes analyzed using RNA-seq clean data.
C4H catalyzes the hydroxylation of trans-cinnamic acid to p-coumaric acid (Figure 1). It is encoded by the members of CYP73A gene subfamily. Genome-wide analysis showed that there were three putative PvC4H genes in P. pulgaris. All of them contained two introns (Figure 2D). PvC4H1 and PvC4H2 showed similar expression patterns with the highest expression in roots, followed by spikes, stems and leaves (Figures 2E, 3A). High expression of PvC4H1 and PvC4H2 is consistent with high content of RA in roots, stems, leaves and flowers (Kim et al., 2014). The expression of PvC4H3 was very low in four tissues analyzed (Figure 3A). Analysis of the deduced proteins showed that all of the three PvC4Hs contained five conserved P450 motifs, including the proline-rich motif “PPGP”, the oxygen binding motif “AAIETT”, the “ETLR” motif, the “PERF” motif, and the heme-binding motif “FGVGRRSCPG” (Supplementary Figure 4) (Khatri et al., 2023). Phylogenetic analysis of C4Hs from P. vulgaris, S. miltiorrhiza, Arabidospsis, P. trichocarpa and various other plants showed that PvC4H1 and PvC4H2 were grouped with SmC4H1 involved in RA biosynthesis (Figure 2F), indicating the involvement of PvC4H1 and PvC4H2 in RA biosynthesis (Xiao et al., 2011; Kim et al., 2014; Wang et al., 2015).
4CL is the third and the last enzyme of the general phenylpropanoid pathway. It catalyzes the thioesterification of p-coumaric acid (Figure 1). The product, p-coumaroyl-CoA, can be funneled into downstream branch pathways for lingnins, flavonoids, coumarins, lignans, and RA (Deng and Lu, 2017). 4CL is encoded by a multiple gene family. For instance, there are seventeen Pt4CLs in P. trichocarpa, ten Sm4CLs in S. miltiorrhiza, and thirteen At4CLs and At4CL-likes in Arabidopsis (Raes et al., 2003; Shi et al., 2010; Wang et al., 2015). Genome-wide analysis showed that there were seventeen putative Pv4CL genes with 3–5 introns in P. pulgaris (Figure 4A). Gene expression analysis showed that the seventeen Pv4CLs had differential expression patterns (Figure 4B). RNA-seq analysis showed that the levels of Pv4CL1, Pv4CL3, Pv4CL8, Pv4CL10 and Pv4CL11 were relatively high, whereas the levels of Pv4CL9 and Pv4CL12–Pv4CL17 were very low in the four tissues analyzed (Figure 3B).
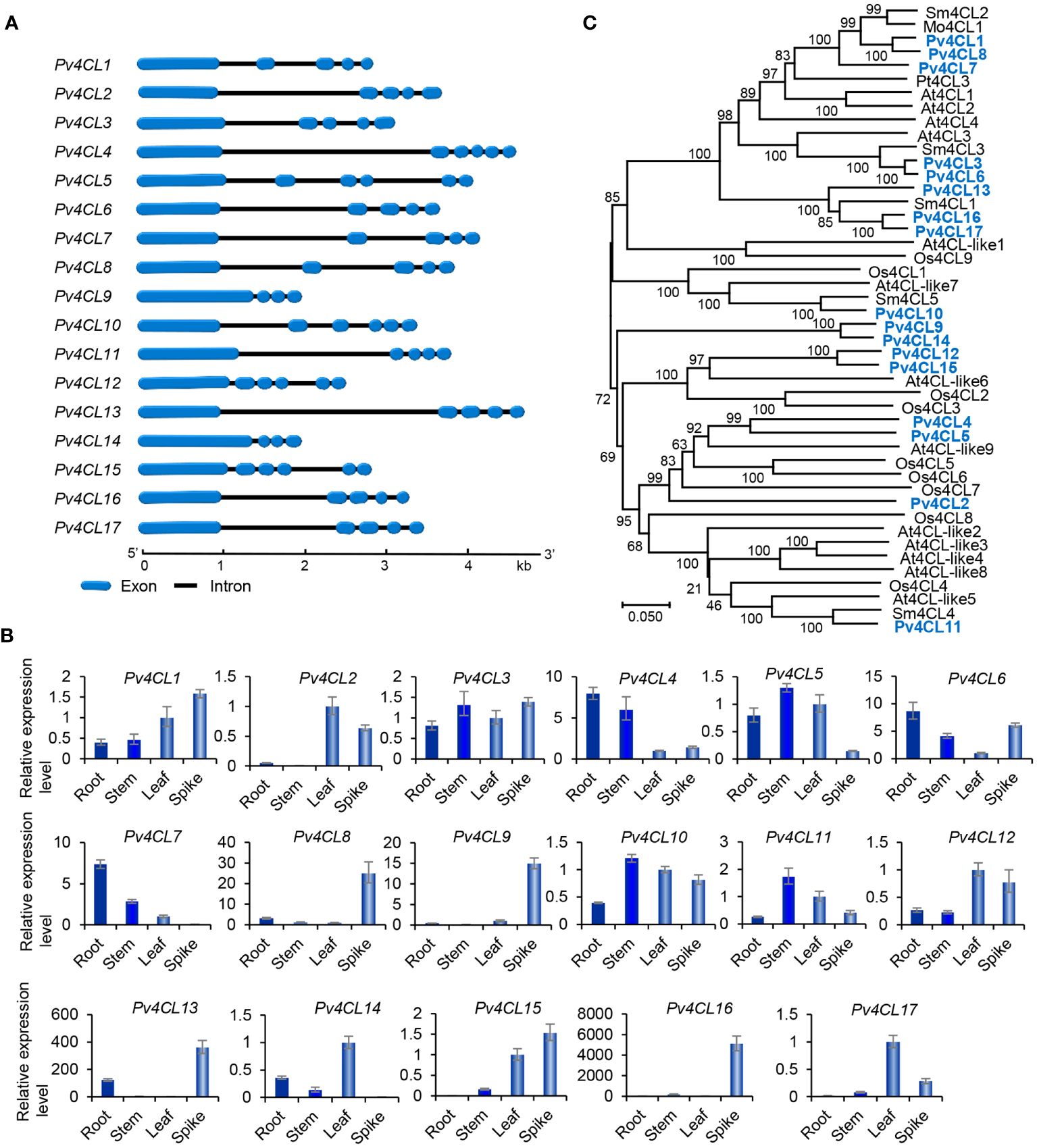
Figure 4 Gene structures, expression patterns and phylogenetic analysis of Pv4CL genes and their deduced proteins. (A) The intron-exon structures of Pv4CL genes. (B) Fold changes of Pv4CL gene expression in roots, stems, leaves and spikes of P. vulgaris plants. The expression level in leaves was arbitrarily set to 1, respectively. (C) Phylogenetic analysis of 4CL proteins. The unrooted Neighbor-Joining tree was constructed using the MEGA program (version 7.0) with default parameters. 4CLs included are seventeen Pv4CLs and other 4CLs from S. miltiorrhiza (Sm), Arabidopsis (At), rice (Os), M. officinalis (Mo), and P. trichocarpa (Pt) (Supplementary Table 2).
It is generally known that the 4CL proteins contain two conserved motifs, including Box I with the representative sequence “SSGTTGLPKGV” and Box II with the representative sequence “GEICIRG” (Uhlmann and Ebel, 1993). Box I is conserved in adenylate-forming enzymes and involved in adenosine monophosphate (AMP)-binding. Box II is conserved in 4CL and related to the spatial conformation of the enzyme (Wang et al., 2022b). Sequence alignment of the seventeen Pv4CL proteins showed that Pv4CLs also had the two conserved motifs. However, their sequences were divergent (Supplementary Figure 5). It indicates that the identified seventeen Pv4CLs could be functionally diverse. Phylogenetic analysis of 4CLs from P. vulgaris, S. miltiorrhiza, Arabidospsis, rice and various other plants showed that Pv4CL1, Pv4CL3, Pv4CL6, Pv4CL7 and Pv4CL8 were grouped with Mo4CL1, Sm4CL2 and Sm4CL3 involved in RA biosynthesis (Figure 4C) (Zhao et al., 2006; Weitzel and Petersen, 2010; Wang et al., 2015). These Pv4CLs could be associated with RA biosynthesis in P. vulgaris (Kim et al., 2014).
Characterization and expression analysis of genes involved in the tyrosine-derived pathway
The tyrosine-derived pathway involves two enzymes, including tyrosine aminotransferase (TAT) and p-hydroxyphenylpyruvate reductase (HPPR) (Figure 1). TAT catalyzes the conversion of L-tyrosine to pHPP, a precursor for the biosynthesis of various secondary metabolites, such as plastoquinone, tocopheros, benzylisoquinoline alkaloids, and RA. In plants, TAT is encoded by multiple gene family with three members in S. miltiorrhiza, four in apple, and eight in Arabidopsis (Wang et al., 2015, 2018). Through genone-wide mining, we identified seven putative PvTAT genes that had five or six introns (Figure 5A). Among them, PvTAT2–PvTAT3 showed relatively high expression (Figures 3C, 5B). The expression of other four PvTATs, including PvTAT1 and PvTAT5–PvTAT7, was mainly in spikes and the level was very low in the tissues analyzed, in comparison with PvTAT2–PvTAT3 (Figures 3C, 5B). Sequence alignment of the seven PvTAT proteins showed that all of them contained the conserved Motif 1 for aminotransferase family-I pyridoxal phosphate binding site and Motif 2 with the highly conserved residue Arg (Supplementary Figure 6) (Lu et al., 2013; Wang et al., 2018). Phylogenetic analysis of seven PvTATs and TATs from S. miltiorrhiza, Arabidopsis and other plants showed that PvTAT3 and PvTAT4 were clustered with SmTAT1 and PfTAT involved in RA biosynthesis (Figure 5C) (Xiao et al., 2011; Lu et al., 2013). PvTAT3 was previously shown to participate in the biosynthesis of RA in P. vulgaris and its high expression in four tissues analyzed is consistent with the accumulation of RA (Figure 5B) (Kim et al., 2014; Ru et al., 2017a). PvTAT4 could be a novel PvTAT playing a redundant role with PvTAT3 in RA biosynthesis.
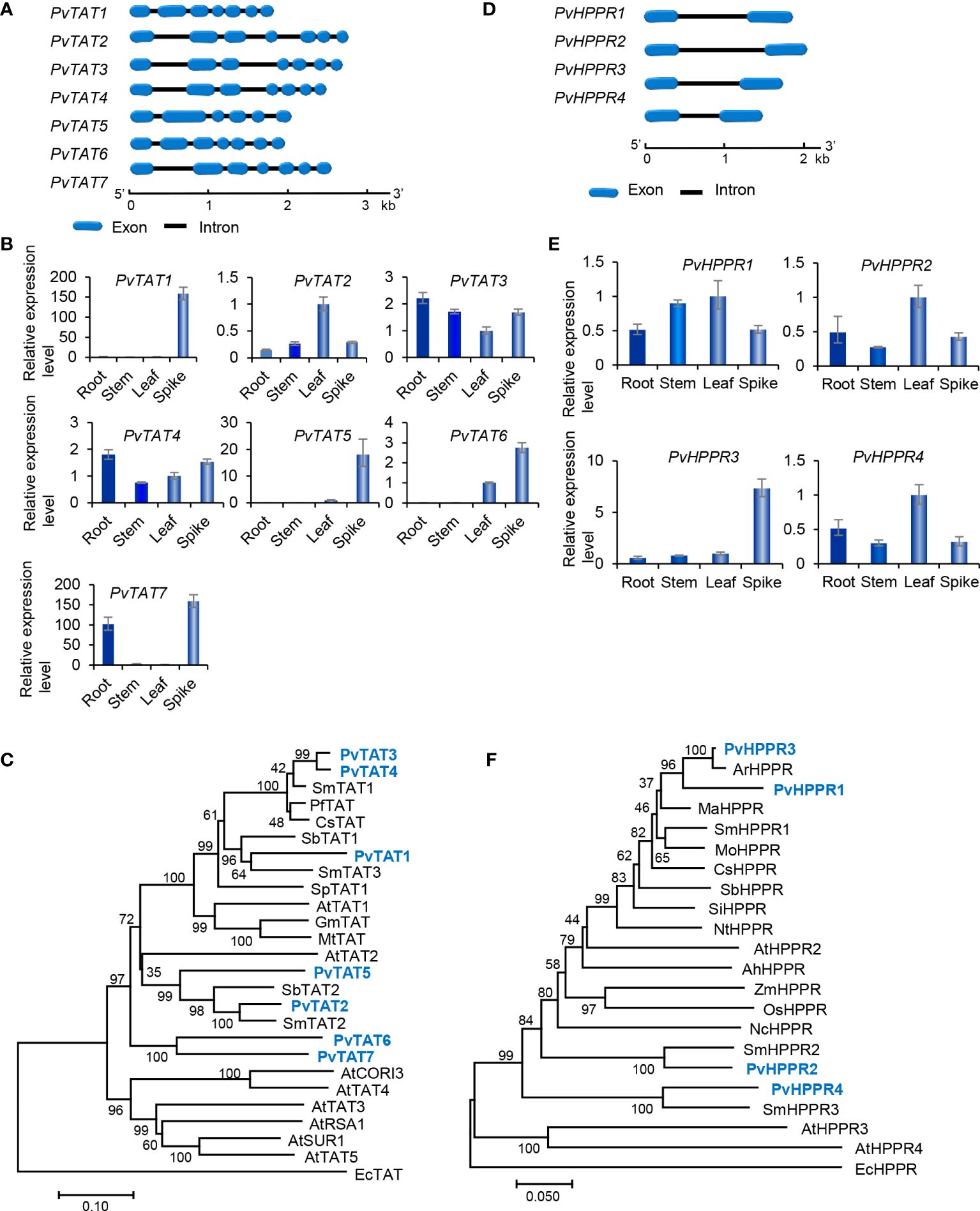
Figure 5 Gene structures, expression patterns and phylogenetic analysis of PvTAT and PvHPPR genes and their deduced proteins. (A, D) The intron-exon structures of PvTAT (A) and PvHPPR (D) genes. (B, E) Fold changes of PvTAT (B) and PvHPPR (E) gene expression in roots, stems, leaves and spikes of P. vulgaris plants. The expression level in leaves was arbitrarily set to 1, respectively. (C) Phylogenetic analysis of TAT proteins. The rooted Neighbor-Joining tree was constructed using the MEGA program (version 7.0) with default parameters. EcTAT (NP_418478.1) from Escherichia coli was used as outgroup. Ingroup TATs include PvTAT1–PvTAT7 and others from S. miltiorrhiza (Sm), Arabidopsis (At), Perilla frutescens (Pf), Coleus scutellarioides (Cs), S. baicalensis (Sb), Solanum pennellii (Sp), G. max (Gm), and Medicago truncatula (Mt) (Supplementary Table 2). (F) Phylogenetic analysis of HPPR proteins. The rooted Neighbor-Joining tree was constructed using the MEGA program (version 7.0) with default parameters. EcHPPR (WP_000811015.1) from Escherichia coli was used as outgroup. Ingroup HPPRs include PvHPPR1–PvHPPR4 and others from S. miltiorrhiza (Sm), Arabidopsis (At), Agastache rugosa (Ar), Mentha aquatica (Ma), M. officinalis (Mo), C. scutellarioides (Cs), S. baicalensis (Sb), Sesamum indicum (Si), Nicotiana tomentosiformis (Nt), Arachis hypogaea (Ah), Zea mays (Zm), Oryza sativa (Os), and Nymphaea colorata (Nc) (Supplementary Table 2).
HPPR, belonging to the family of D-isomer-specific 2-hydroxyacid dehydrogenases, is the other enzyme involved in the tyrosine-derived pathway. It catalyzes the conversion of pHPP to pHPL (Kim et al., 2004) (Figure 1). HPPR is encoded by a small gene family, such ast there are three SmHPPR genes in S. miltiorrhiza and four in Arabidopsis (Wang et al., 2015; Xu et al., 2018). Its involvement in RA biosynthesis has been verified through functional analysis of MoHPPR from Melissa offcianalis (Mansouri and Mohammadi, 2021), CsHPPR from Coleus scutellarioides (Kim et al., 2004), and SmHPPR1 from S. miltiorrhiza (Xiao et al., 2011; Wang et al., 2017). Genome-wide analysis showed that there were four PvHPPR genes in P. vulgaris, all of which contained one intron and had similar structures (Figure 5D). All of them showed differential expression patterns, and the overall expression level of PvHPPR1–PvHPPR3 was higher than PvHPPR4 (Figures 3C,5E). Amino acid sequence alignment showed that all four PvHPPR proteins contained the NAD(P)H binding motif with the representative sequence “GLGRIG” and the putative myristylation site with the representative sequences “GTVETR” and “GNLEA” (Supplementary Figure 7) (Wang et al., 2017). Phylogenetic analysis of four PvHPPRs and HPPRs from S. miltiorrhiza, Arabidopsis and other plants showed that PvHPPR1 and PvHPPR3 were grouped with SmHPPR1, MoHPPR and CsHPPR involved in RA biosynthesis (Figure 5F). It suggests that PvHPPR1 and PvHPPR3 could be involved in the biosynthesis of RA in P. vulgaris. The function of PvHPPR2 and PvHPPR4 remain to be elucidated.
Characterization and expression analysis of genes involved in downstream of the RA biosynthetic pathway
The downstream of RA biosynthetic pathway involves four known enzymes, including p-hydroxycinnamoyl-CoA: shikimate p-hydroxycinnamoyltransferase (HCT), rosmarinic acid synthase (RAS), p-coumaroyl shikimate 3’-hydroxylase/coumarate 3-hydroxylase (C3H), and the enzyme catalyzing the final step of RA biosynthetic pathway (Figure 1). Enzymes involved in DHPL biosynthesis are currently unknown (Figure 1). Among the four known enzymes, HCT catalyzes the coupling of p-coumaroyl-CoA with shikimate to form p-coumaroyl shikimic acid. It also catalyzes the reverse reaction converting caffeoyl shikimate ester to caffeoyl-CoA (Hoffmann et al., 2003). Differently, RAS catalyzes the coupling of p-coumaroyl-CoA/caffeoyl-CoA and pHPL/DHPL to form 4-coumaroyl-4’-hydroxyphenyllactic acid, 4-coumaroyl-3’,4’-dihydroxyphenyllactic acid, and/or caffeoyl-4’-hydroxyphenyllactic acid in different plant species (Figure 1) (Eberle et al., 2009; Petersen et al., 2009; Di et al., 2013; Levsh et al., 2019; Liu et al., 2019; Lu, 2021). Both HCT and RAS belong to the BAHD acyltransferase family and are known as CoA-ester-dependent BAHD hydroxycinnamoyltransferases (Petersen et al., 2009).
Genome-wide analysis showed that there were three PvHCT and eight PvRAS genes in P. vulgaris (Table 1). Except that PvRAS4 had no intron and PvRAS8 had two introns, other three PvHCTs and six PvRASs contained an intron and shared similar gene structures (Figure 6A). qRT-PCR analysis showed that the three PvHCTs showed differential expression (Figure 6B). RNA-seq analysis showed that PvHCT1 had the highest expression, followed by PvHCT2 (Figure 3D). The expression of PvHCT3 was very low (Figure 3D). It indicates that, among the three PvHCTs, PvHCT1 could be most likely to be involved in RA biosynthesis in P. vulgaris. Among the eight PvRASs, PvRAS3 showed the highest expression and was highly expressed in spikes, followed by stems, leaves, and roots (Figures 3D, 6B). The expression of other seven PvRASs was relatively low in the tissues analyzed (Figure 3D).
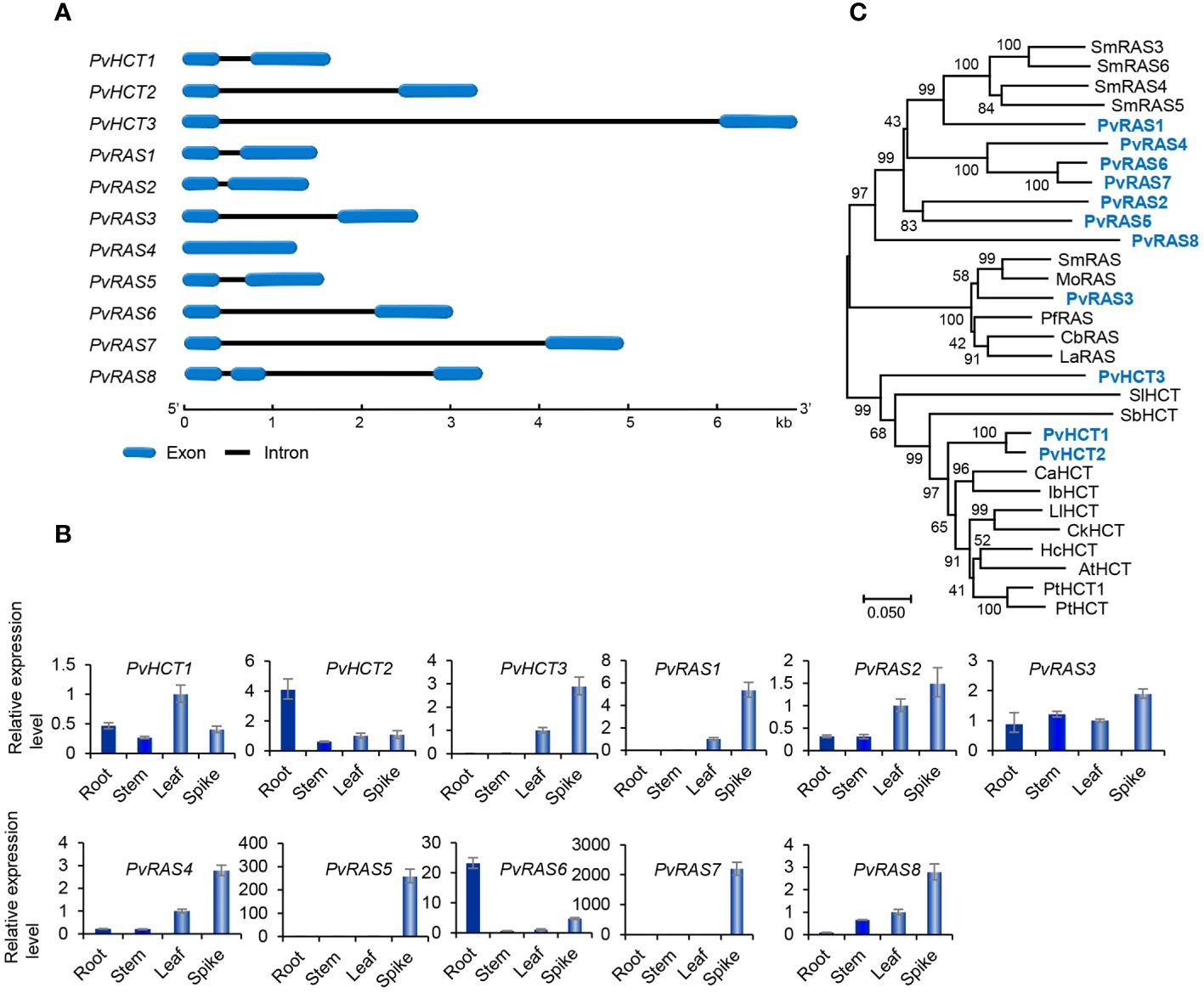
Figure 6 Gene structures, expression patterns and phylogenetic analysis of PvHCT and PvRAS genes and their deduced proteins. (A) The intron-exon structures of three PvHCT and eight PvRAS genes. (B) Fold changes of PvHCT and PvRAS gene expression in roots, stems, leaves and spikes of P. vulgaris plants. The expression level in leaves was arbitrarily set to 1, respectively. (C) Phylogenetic analysis of PvHCT and PvRAS proteins. The unrooted Neighbor-Joining tree was constructed using the MEGA program (version 7.0) with default parameters. Proteins included are PvHCT1–PvHCT3, PvRAS1–PvRAS8, and the HCTs and RASs from S. miltiorrhiza (Sm), M. officinalis (Mo), P. frutescens (Pf), C. blumei (Cb), L. angustifolia (La), Solanum lycopersicum (Sl), S. bicolor (Sb), C. arabica (Ca), Ipomoea batatas (Ib), L. leucocephala (Ll), Caragana korshinskii (Ck), Hibiscus cannabinus (Hc), Arabidopsis (At), P. trichocarpa (Pt) (Supplementary Table 2).
Sequence alignment of PvHCT and PvRAS proteins showed that all of them contained the conserved “HXXXD” and “DFGWG” motifs (Supplementary Figure 8) (Berger et al., 2006). Phylogenetic analysis of three PvHCTs, eight PvRASs, and HCTs and RASs from various other plants showed that RASs and HCTs separated into two clades (Figure 6C). All HCTs were clustered in one clade, whereas all RASs were clustered in the other one. In addition, the RAS clade could be divided into two sub-clades. PvRAS3 was clustered with functionally known RASs from C. blumei, Melissa officinalis, Lavandula Angustifolia and S. miltiorrhiza in a sub-clade (Berger et al., 2006; Landmann et al., 2011; Sander and Petersen, 2011; Weitzel and Petersen, 2011; Di et al., 2013; Zhou et al., 2018; Fu et al., 2020). Taken together with the high expression of PvRAS3 gene (Figure 6B), the results suggest the involvement of PvRAS3 in RA biosynthesis. The other RAS sub-clade included PvRAS1, PvRAS2, PvRAS4–PvRAS8, and four putative SmRASs (Figure 5C). The function of these RASs is currently unknown. Among them, the expression of PvRAS2, PvRAS4 and PvRAS8 showed similar patterns with RA distribution (Kim et al., 2014) (Figure 6B). It indicates that these PvRASs could also be associated with RA biosynthesis.
C3H is the other enzyme involved in downstream of the RA biosynthetic pathway (Figure 1). It catalyzes the hydroxylation of p-coumaroyl shikimic acid, a shikimate ester of p-coumarate generated from p-coumaroyl-CoA and shikimate under the catalysis of HCT, into caffeoyl shikimic acid, a shikimate ester of caffeic acid (Schoch et al., 2001). C3H is a cytochrome P450 encoded by members of the CYP98 gene family (Schoch et al., 2001; Franke et al., 2002; Ralph et al., 2006). Similarily, the enzyme involved in the final step of RA biosynthetic pathway is also a cytochrome P450 encoded by members of the CYP98 gene family. It introduces the hydroxyl group(s) to the products of RAS (Eberle et al., 2009; Di et al., 2013; Levsh et al., 2019; Liu et al., 2019; Fu et al., 2020) (Figure 1).
Genome-wide analysis showed that there were five PvCYP98 genes in P. vulgaris (Table 1). All of them contain two introns and share similar gene structures (Figure 7A). They showed differential expression patterns and PvCYP98A-1 and PvCYP98A-2 had relatively high expression among the five PvCYP98s (Figures 3E, 7B). Similar to PvC4Hs, the other family of cytochrome P450 proteins involved in RA biosynthesis (Supplementary Figure 4), PvCYP98 proteins also contain the five conserved P450 motifs, including “PPGP”, “(A/G)(A/G)X(D/E)T(T/S)”, “EXLR”, “PERF”, and “FGXGRRXCXG” (Supplementary Figure 9) (Eberle et al., 2009; Khatri et al., 2023). Phylogenetic analysis showed that PvCYP98A-4 and PvCYP98A-5 were clustered with the functionally known C3Hs (Figure 7C), such as AtC3H1 (AtCYP98A3) and PtC3H3 (Schoch et al., 2001; Chen et al., 2011). PvCYP98A-1 and PvCYP98A-2 were clustered with CsCYP98A14 and SmCYP98A78 involved in the last step of RA biosynthetic pathway (Figure 7C) (Eberle et al., 2009; Di et al., 2013; Liu et al., 2019; Fu et al., 2020). PvCYP98A-3 was clustered with the putative Sesamum indicum SiC3H and P. campanularia PcCYP98A113 involved in the last step of RA biosynthetic pathway (Figure 6C) (Anterola et al., 2002; Levsh et al., 2019). Taken together with the expression patterns of PvCYP98As (Figures 3E, 7B) and the distribution patterns of RAs in P. vulgaris (Kim et al., 2014), the results indicated that PvCYP98A-1 and PvCYP98A-2 could be involved in the hydroxylation of RAS products, PvCYP98A-4 and PvCYP98A-5 could be PvC3Hs, whereas the function of PvCYP98A-3 remained to be analyzed.
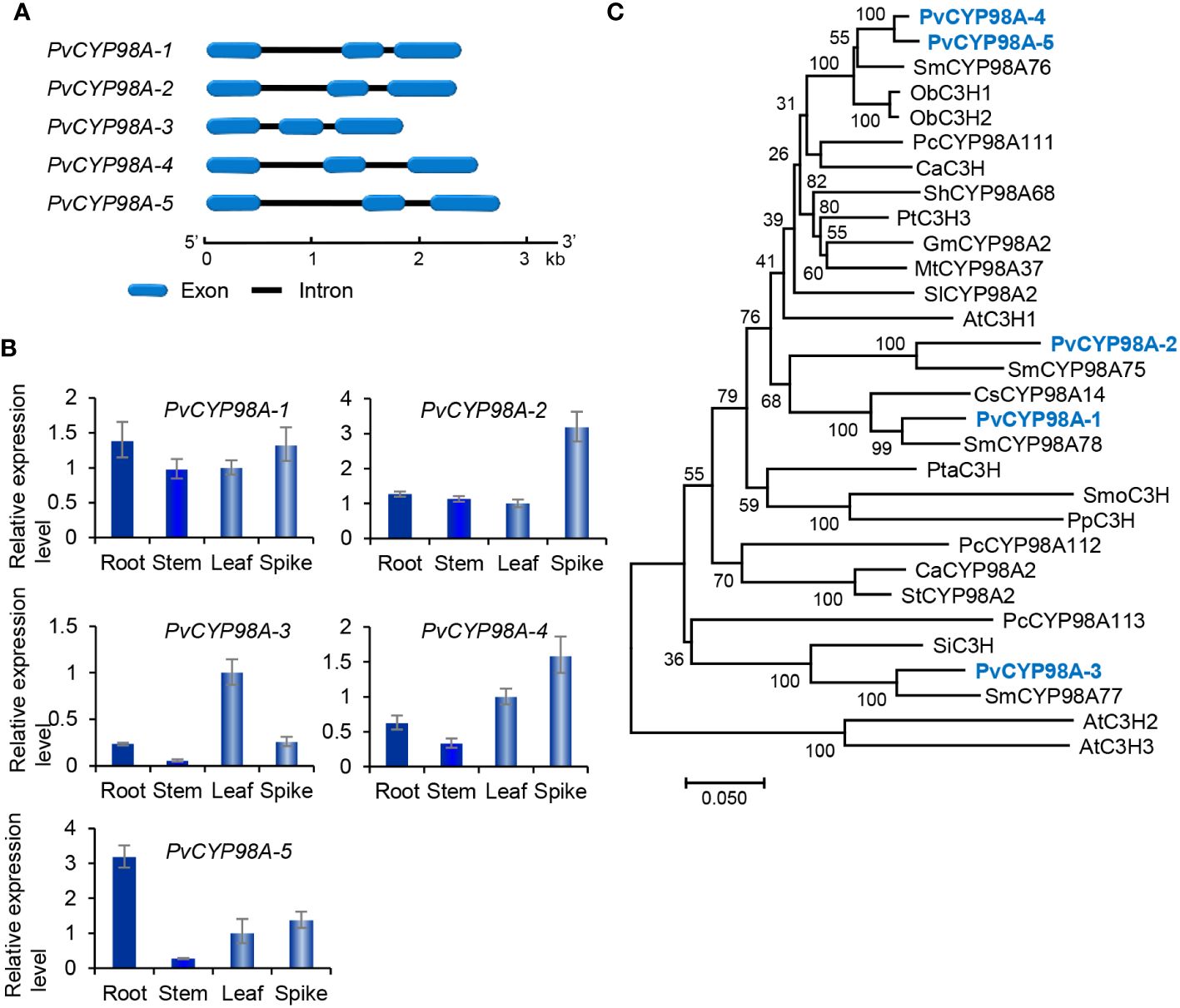
Figure 7 Gene structures, expression patterns and phylogenetic analysis of PvCYP98 genes and their deduced proteins. (A) The intron-exon structures of five PvCYP98 genes. (B) Fold changes of PvCYP98 gene expression in roots, stems, leaves and spikes of P. vulgaris plants. The expression level in leaves was arbitrarily set to 1, respectively. (C) Phylogenetic analysis of PvCYP98 proteins. The unrooted Neighbor-Joining tree was constructed using the MEGA program (version 7.0) with default parameters. Proteins included are PvCYP98A-1–PvCYP98A-5 and the C3Hs and CYP98As from Arabidopsis (At), S. miltiorrhiza (Sm), Ocimum basilicum (Ob), P. campanularia (Pc), C. arabica (Ca), Sinopodophyllum hexandrum (Sh), P. trichocarpa (Pt), (G) max (Gm), M. truncatula (Mt), S. lycopersicum (Sl), C. scutellarioides (Cs), P. taeda (Pta), Selaginella moellendorffii (Smo), Physcomitrium patens (Pp), Capsicum annuum (Ca), S. tuberosum (St), and S. indicum (Si) (Supplementary Table 2).
PvRAS3 and PvRAS4 were involved in RA biosynthesis in vitro
Based on gene expression patterns, phylogenetic relationships and the significance of RAS in RA biosynthesis, PvRAS3 and PvRAS4 were selected for functional analysis using experimental approaches. Among them, PvRAS3 showed the highest expression among the eight PvRAS genes identified and was clustered with functionally known RASs in the phylogenetic tree constructed (Figures 6B, C). PvRAS4 was one of the three PvRAS genes with expression patterns similar to RA distribution patterns (Kim et al., 2014) (Figure 6B). PvRAS3 and PvRAS4 cDNAs were cloned and introduced into E. coli competent cells. Recombinant proteins were induced and purified (Supplementary Figure 10). To assay the activity of recombinant PvRAS3 enzyme, p-coumaroyl-CoA or caffeoyl-CoA was used as the acyl donor, and pHPL or DHPL was used as the acyl acceptor. Negative controls were performed with E. coli BL21 (DE3) cells transformed with the empty pGEX-4T-1 vector (Supplementary Figure 11). LC-MS/MS analysis showed that PvRAS3 could catalyze the condensation of acyl donors and acceptors to generate four compounds, respectively (Figures 8A–D). Compound 1, generated through the condensation of caffeoyl-CoA and DHPL, was identified as RA based on UPLC and LC-MS/MS analyses (Figures 8A, E) and previous publication (Landmann et al., 2011; Di et al., 2013). Compounds 2 (Figure 8B) and 3 (Figure 8C) had the formula C18H16O7 according to the MS spectra in negative mode [M-H]– (m/z = 343) (Figures 8F, G). They corresponded to an ester of caffeoyl-CoA and DHPL (caffeoyl-4’-hydroxyphenyllactic acid) or p-coumaroyl-CoA and pHPL (4-coumaroyl-3’,4’-dihydroxyphenyllactic acid), respectively, as described previously (Landmann et al., 2011). Compound 4 was determined to be the ester of p-coumaroyl-CoA and pHPL based on the negative ion spectra (m/z = 327, C18H15O6) (Figures 8D, H) and previous publication (Landmann et al., 2011).
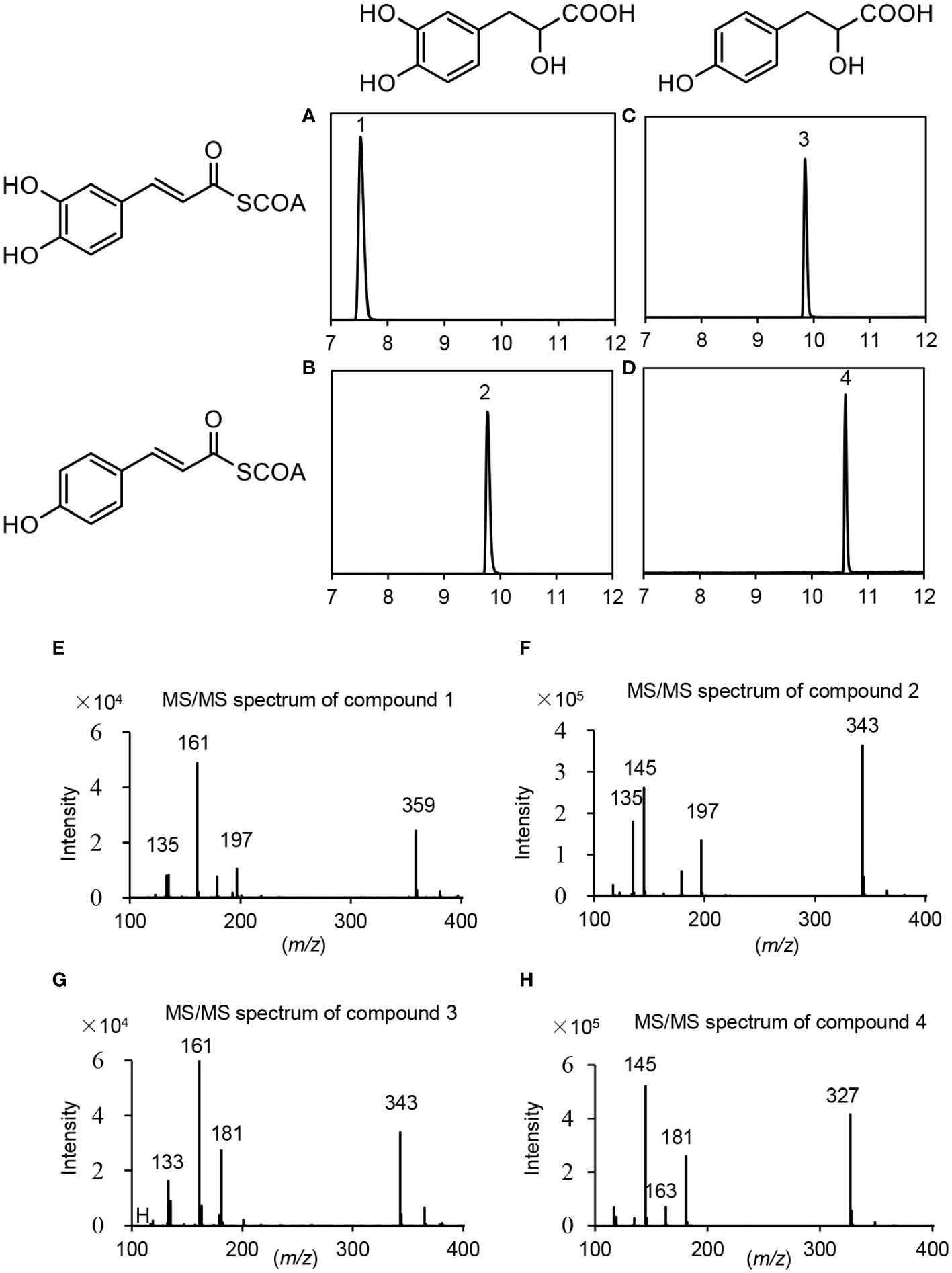
Figure 8 UPLC and LC-MS/MS detection of in vitro reaction products catalyzed by PvRAS3. (A–D) UPLC chromatograms of the reaction products catalyzed by PvRAS3 using caffeoyl-CoA and DHPL (A), p-coumaroyl-CoA and DHPL (B), caffeoyl-CoA and pHPL (C), and p-coumaroyl-CoA and pHPL (D) as acyl donors and acceptors, respectively. (E–H) MS/MS spectra of compounds 1, 2, 3, and 4.
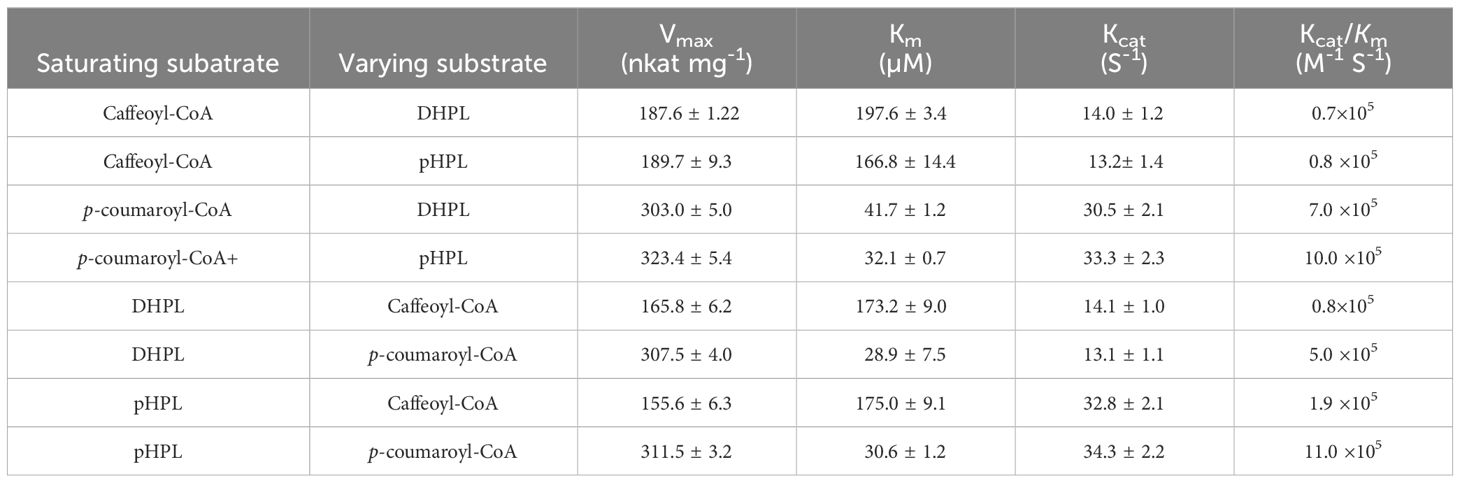
For kinetic analysis of PvRAS3, p-coumaroyl-CoA or caffeoyl-CoA was used as the acyl donor and pHPL or DHPL was used as the acyl acceptor. To test the acceptor specificity, the concentration of the donor substrates remained saturated, while the levels of the acceptor substrates were varied (Supplementary Figure 12). When caffeoyl-CoA was used as the donor, the Km values of PvRAS3 with DHPL and pHPL were 197.6 and 166.8 μM, respectively (Table 2). Using p-coumaroyl-CoA as the donor, the Km values of DHPL and pHPL were 41.7 and 32.1 μM, respectively. The results suggested that the Km values of DHPL and pHPL with p-coumaroyl-CoA were lower than those with caffeoyl-CoA. To assess the donor affinity, the concentrations of the acceptors (DHPL and pHPL) were kept constant while the levels of caffeoyl-CoA and p-coumaroyl-CoA were varied. The results showed that the Km value of PvRAS3 for Caffeoyl-CoA was approximately 5.7-fold higher than that for p-coumaroyl-CoA (Table 2). Overall, PvRAS3 exhibited a high affinity toward DHPL and pHPL when p-coumaroyl-CoA was used as the acyl donor.
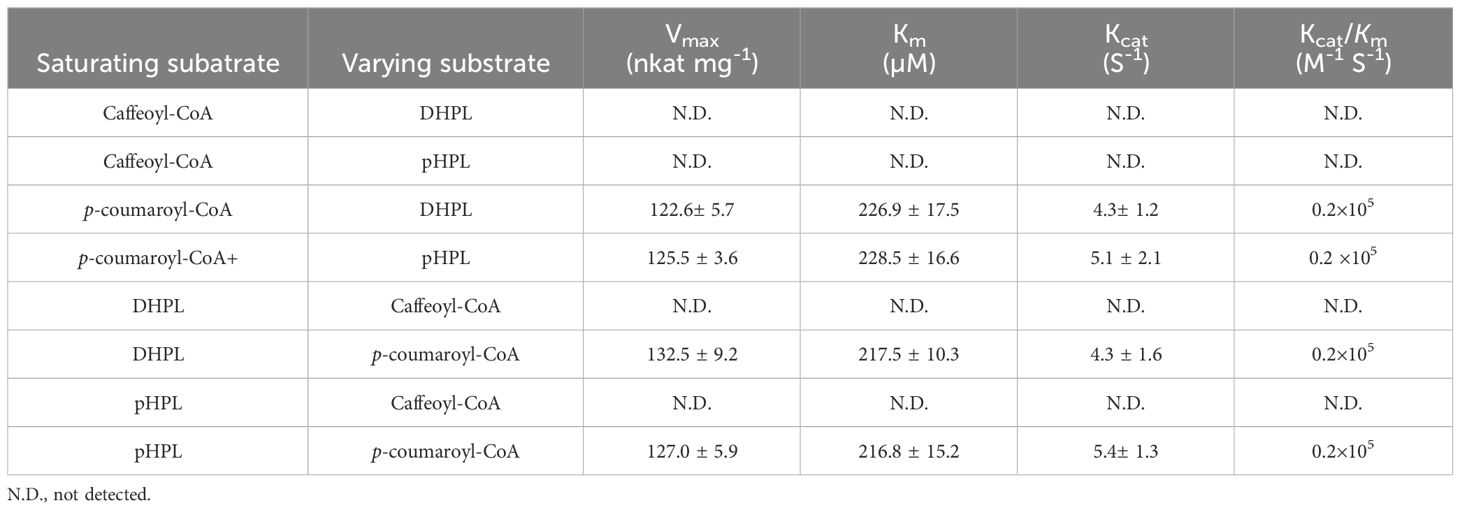
Similarly, p-coumaroyl-CoA, caffeoyl-CoA, pHPL and DHPL were also used as substrates for the analysis of PvRAS4 (Supplementary Figure 13). LC-MS/MS analysis showed that products could be detected when p-coumaroyl-CoA was used as acyl donor and pHPL or DHPL were used as acyl acceptor (Figures 9B, D). Based on MS spectra, the products were identical to compounds 2 and 4 catalyzed by PvRAS3 (Figures 9E, F). The Kcat/Km values of PvRAS4 for p-coumaroyl-CoA and pHPL or DHPL were smaller than those of PvRAS3 (Table 3). No product was found when caffeoyl-CoA was used as acyl donor (Figures 9A, B). The results indicated that PvRAS4 could use p-coumaroyl-CoA, but not caffeoyl-CoA, as acyl donor. However, its affinity toward p-coumaroyl-CoA was lower than PvRAS3.
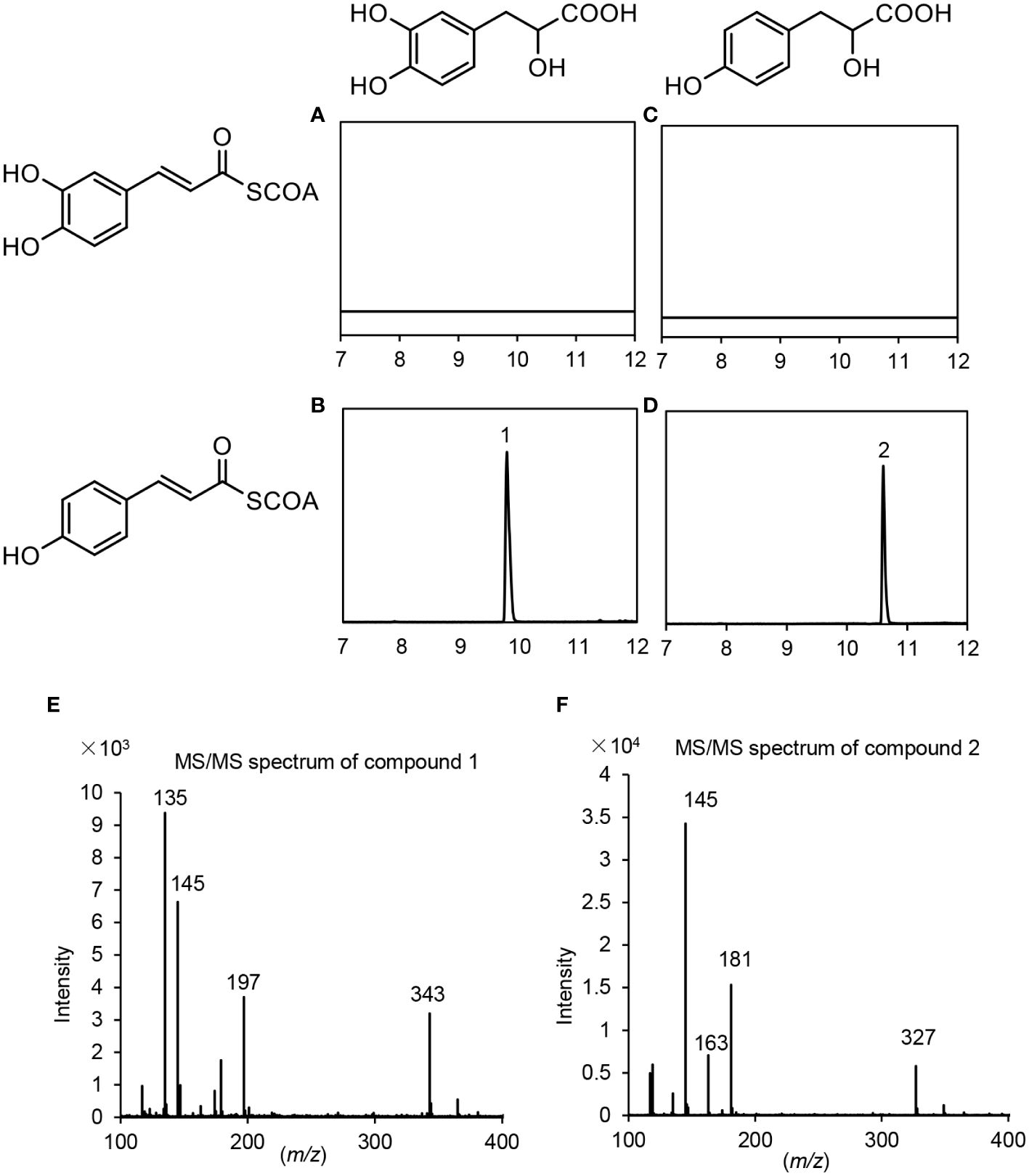
Figure 9 UPLC and LC-MS/MS detection of in vitro reaction products catalyzed by PvRAS4. (A–D) UPLC chromatograms of the reaction products catalyzed by PvRAS4 using caffeoyl-CoA and DHPL (A), p-coumaroyl-CoA and DHPL (B), caffeoyl-CoA and pHPL (C), and p-coumaroyl-CoA and pHPL (D) as acyl donors and acceptors, respectively. (E, F) MS/MS spectra of compounds 1 and 2.
Existence of RA, 4-coumaroyl-3’,4’-dihydroxyphenyllactic acid, 4-coumaroyl-4’-hydroxyphenyllactic acid and caffeoyl-4’-hydroxyphenyllactic acid in P. vulgaris plants
Enzyme activity assay showed that PvRAS3 could condense p-coumaroyl-CoA and caffeoyl-CoA with pHPL and DHPL in vitro. To analyze whether the products exist in P. vulgaris plants, phenolic acid compounds were extracted from roots, stems, leaves and flowers and analyzed using UPLC and LC-MS/MS as described (Kim et al., 2014). The results showed that all of the four products, including RA, 4-coumaroyl-3’,4’-dihydroxyphenyllactic acid, 4-coumaroyl-4’-hydroxyphenyllactic acid and caffeoyl-4’-hydroxyphenyllactic acid, could be detected in the tissues analyzed (Figure 10; Supplementary Figure 14). RA was highly accumulated at the level of mg g-1 fresh weight (FW) with the highest level of 6.1 mg g-1 FW in flowers and the lowest level of 3.1 mg g-1 FW in stems (Figure 10A). The contents of 4-coumaroyl-4’-hydroxyphenyllactic acid ranged from 51.9 µg g-1 FW in roots to 140.3 µg g-1 FW in leaves (Figure 10B). The contents of 4-coumaroyl-4’-hydroxyphenyllactic acid were relatively low, which ranged from 32.9 µg g-1 FW in roots to 93.5 µg g-1 FW in flowers (Figure 10C). The contents of 4-coumaroyl-3’,4’-dihydroxyphenyllactic acid ranged from 133.3 µg g-1 FW in roots to 368.0 µg g-1 FW in flowers (Figure 10D). The contents of RA and 4-coumaroyl-3’,4’-dihydroxyphenyllactic acid in P. vulgaris were higher but comparable to those in lavender flowers, which were 2 mg g-1 FW and 150 µg g-1 FW, respectively (Landmann et al., 2011). To our best knowledge, this is the first report to detect 4-coumaroyl-4’-hydroxyphenyllactic acid and caffeoyl-4’-hydroxyphenyllactic acid in plants. In addition, we also analyzed the contents of 3,4-dihydroxyphenyllactic acid and 4-hydroxyphenyllactic acid. Among them, 3,4-dihydroxyphenyllactic acid was highly accumulated in roots, leaves and flowers (Figure 10E), whereas the content of 4-hydroxyphenyllactic acid was relatively higher in leaves and flowers than roots and stems (Figure 10F). The results suggest that both the substrates and products of the four reactions catalyzed by PvRAS3 and/or PvRAS4 in vitro exist in P. vulgaris plants.
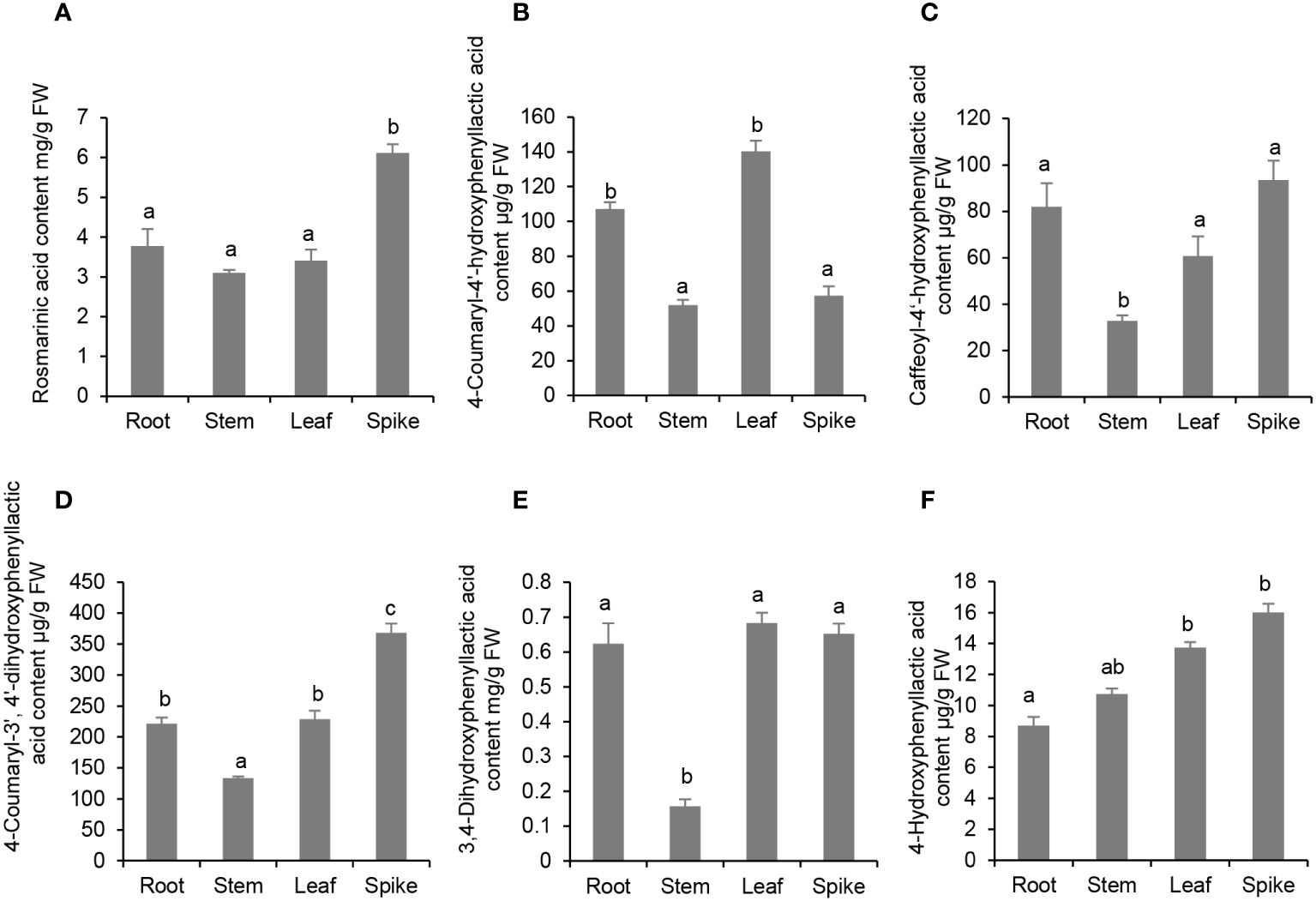
Figure 10 Analysis of phenolic acid contents in roots, stems, leaves and spikes of P. vulgaris. (A) Rosmarinic acid; (B) 4-Coumaroyl-4’-hydroxyphenyllactic acid; (C) Caffeoyl-4’-hydroxyphenyllactic acid; (D) 4-Coumaroyl-3’, 4’-dihydroxyphenyllactic acid; (E) 3,4-Dihydroxyphenyllactic acid; (F) 4-Hydroxyphenyllactic acid. Error bars represent standard deviations of mean value from three biological replicates. ANOVA (analysis of variance) was calculated using SPSS. P < 0.05 was considered statistically significant and was represented by different letters above the bars. P ≥ 0.05 was considered statistically non-significant and was represented by same letters above the bars.
CRISPR/Cas9-mediated functional analysis of PvRAS3 in P. vulgaris hairy roots
Gene expression profiling and In vitro enzyme activity assay showed that PvRAS3 could be the main RAS catalyzing RA biosynthesis in P. vulgaris. To gain further insight into the involvement of PvRAS3 in RA biosynthesis, we designed two guide RNAs (gRNAs) targeting the first coding exon of PvRAS3 for the CRISPR/Cas9 gene-editing tool. Five lines of transgenic hairy roots with the same insertion and deletion patterns in PvRAS3 gene were obtained (Figure 11A). It indicated that these transgenic lines were homozygous mutants of PvRAS3, hereinafter referred to as pvras3-1, pvras3-2, pvras3-3, pvras3-4 and pvras3-5, respectively. Analysis of RA, 4-coumaroyl-3’,4’-dihydroxyphenyllactic acid, 4-coumaroyl-4’-hydroxyphenyllactic acid and caffeoyl-4’-hydroxyphenyllactic acid showed that the contents of these compounds decreased significantly in pvras3 mutants with the contents of RA and caffeoyl-4’-hydroxyphenyllactic acid almost below the detection limit (Figures 11B, E–G). On the contrary, the contents of DHPL and pHPL increased significantly in the mutants (Figures 10C, D). It confirmed the catalytical function of PvRAS3 and its significance in RA biosynthesis.
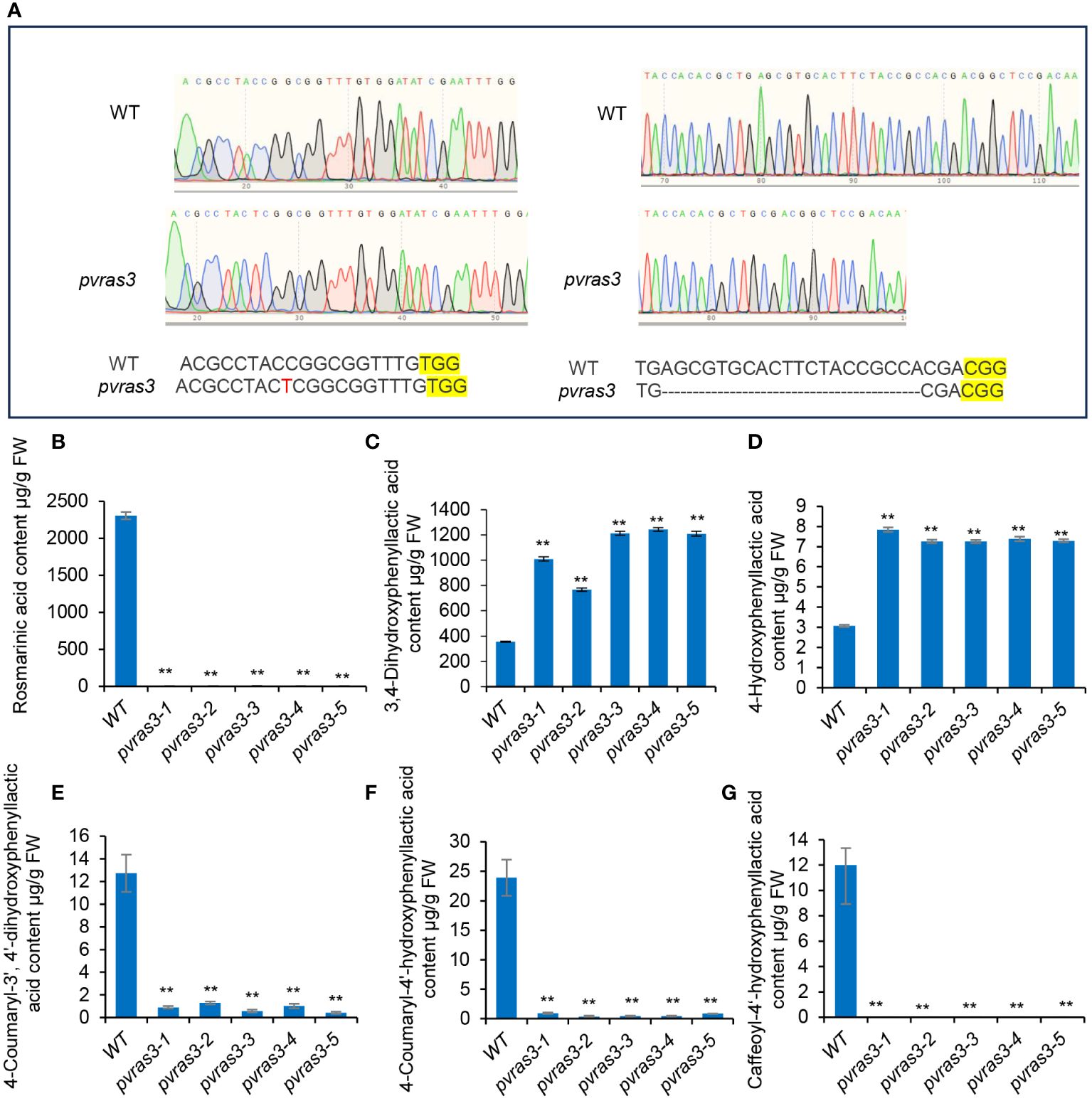
Figure 11 Analysis of phenolic acid contents in hairy roots of wild type and pvras3 mutants. (A) The sgRNA-edited nucleotide sequences in pvras3 mutants. WT, wild type. (B–G) Contents of rosmarinic acid (B), 3,4-dihydroxyphenyllactic acid (C), 4-hydroxyphenyllactic acid (D), 4-coumaroyl-3’,4’-dihydroxyphenyllactic acid (E), 4-coumaroyl-4’-hydroxyphenyllactic acid (F) and caffeoyl-4’-hydroxyphenyllactic acid (G) in wild type (WT) and five pvras3 mutant lines. P < 0.05 (*) and P < 0.01 (**) were considered statistically significant and highly significant, respectively.
Conclusions
P. vulgaris is a species of the Lamiaceae family with significant medicinal value. RA is one of the major bioactive components in P. vulgaris medicinal materials. Through genome-wide analysis, a total of 51 P. vulgaris genes belonging to seven RA biosynthesis-related gene families were identified. Subsequent gene and protein feature analysis, gene expression analysis and phylogenetic relationship analysis showed that seventeen of them, including PvPAL1, PvPAL2, PvC4H1, PvC4H2, Pv4CL1, Pv4CL3, Pv4CL6, Pv4CL7, Pv4CL8, PvTAT3, PvTAT4, PvHPPR1, PvHPPR3, PvRAS3, PvRAS4, PvCYP98A-1 and PvCYP98A-2, could be involved in RA biosynthesis. In vitro enzymatic assay showed that both of PvRAS3 and PvRAS4 were involved in RA biosynthesis. PvRAS3 could catalyze the condensation of p-coumaroyl-CoA and caffeoyl-CoA with pHPL and DHPL. The affinity of PvRAS3 toward p-coumaroyl-CoA was higher than caffeoyl-CoA. PvRAS4 only catalyzed the condensation of p-coumaroyl-CoA with pHPL and DHPL. The affinity of PvRAS4 toward p-coumaroyl-CoA was lower than PvRAS3. These results were consistent with in vivo phenolic acid compound determination and PvRAS3 transgenic analysis. UPLC analysis of phenolic acid compounds showed the existence of RA, 4-coumaroyl-3’,4’-dihydroxyphenyllactic acid, 4-coumaroyl-4’-hydroxyphenyllactic acid and caffeoyl-4’-hydroxyphenyllactic acid in roots, stems, leaves and flowers of P. vulgaris. Generation of pvras3 homozygous mutants through CRISPR/Cas9 technology and subsequent chemical compound analysis showed that the contents of RA, 4-coumaroyl-3’,4’-dihydroxyphenyllactic acid, 4-coumaroyl-4’-hydroxyphenyllactic acid and caffeoyl-4’-hydroxyphenyllactic acid decreased significantly, whereas the contents of DHPL and pHPL increased significantly in pvras3 mutants. These results indicate the existence of four possible RA biosynthetic routes in P. vulgaris, which remains to be further confirmed through the analysis of PvCYP98A genes (Figure 1). Among them, routes 1 and 2 could be the main routes. PvRAS3 was the main enzyme catalyzing the condensation of acyl donors and acyl acceptors during RA biosynthesis in P. vulgaris. PvRAS4 could play a minor role. Further functional analysis of other fourteen candidate genes, particularly PvCYP98A-1 and PvCYP98A-2, may provide a more complete picture of RA biosynthetic pathway.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
Author contributions
CY: Data curation, Writing – original draft. CL: Data curation, Methodology, Validation, Writing – review & editing. MJ: Data curation, Writing – review & editing. YX: Data curation, Writing – review & editing. SZ: Writing – review & editing. XH: Writing – review & editing. YC: Writing – review & editing. SL: Methodology, Software, Supervision, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the Chinese Academy of Medical Sciences Medical and Health Science and Technology Innovation Project [grant No. 2022-I2M-2-001], the National Natural Science Foundation of China [grant No. 31500263], and the Sichuan Science and Technology Program [grant No. 19YJ0368].
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2024.1374912/full#supplementary-material
References
Achnine, L., Blancaflor, E. B., Rasmussen, S., Dixon, R. A. (2004). Colocalization of L-phenylalanine ammonia-lyase and cinnamate 4-hydroxylase for metabolic channeling in phenylpropanoid biosynthesis. Plant Cell 16, 3098–3109. doi: 10.1105/tpc.104.024406
Altschul, S. F., Madden, T. L., Schaffer, A. A., Zhang, J., Zhang, Z., Miller, W., et al. (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402. doi: 10.1093/nar/25.17.3389
Anterola, A. M., Jeon, J. H., Davin, L. B., Lewis, N. G. (2002). Transcriptional control of monolignol biosynthesis in Pinus taeda: factors affecting monolignol ratios and carbon allocation in phenylpropanoid metabolism. J. Biol. Chem. 277, 18272–18280. doi: 10.1074/jbc.M112051200
Berger, A., Meinhard, J., Petersen, M. (2006). Rosmarinic acid synthase is a new member of the superfamily of BAHD acyltransferases. Planta 224, 1503–1510. doi: 10.1007/s00425-006-0393-y
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: an integrative toolkit developed for interactive analyses of bigbiological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chen, H. C., Li, Q., Shuford, C. M., Liu, J., Muddiman, D. C., Sederoff, R. R., et al. (2011). Membrane protein complexes catalyze both 4- and 3-hydroxylation of cinnamic acid derivatives in monolignol biosynthesis. Proc. Natl. Acad. Sci. United States America 108, 21253–21258. doi: 10.1073/pnas
Chen, Y., Zhang, X., Guo, Q., Cao, L., Qin, Q., Li, C., et al. (2019). Plant morphology, physiological characteristics, accumulation of secondary metabolites and antioxidant activities of Prunella vulgaris L. under UV solar exclusion. Biol. Res. 52, 17. doi: 10.1186/s40659-019-0225-8
Chen, Y., Li, Z., Guo, Q., Wang, C., Cao, L., Tang, H., et al. (2022). Identification of key genes related to flowering by transcriptome of flowering and nonflowering Prunella vulgaris L. Biol. Plantarum 66, 1–13. doi: 10.32615/bp.2021.056
Cui, X., Meng, F., Pan, X., Qiu, X., Zhang, S., Li, C., et al. (2022). Chromosome-level genome assembly of Aristolochia contorta provides insights into the biosynthesis of benzylisoquinoline alkaloids and aristolochic acids. Horticulture Res. 9, uhac005. doi: 10.1093/hr/uhac005
Deng, Y., Lu, S. (2017). Biosynthesis and regulation of phenylpropanoids in plants. Crit. Rev. Plant Sci. 36, 257–290. doi: 10.1080/07352689.2017.1402852
Di, P., Zhang, L., Chen, J., Tan, H., Xiao, Y., Dong, X., et al. (2013). ¹³C tracer reveals phenolic acids biosynthesis in hairy root cultures of Salvia miltiorrhiza. ACS Chem. Biol. 8, 1537–1548. doi: 10.1021/cb3006962
Eberle, D., Ullmann, P., Werck-Reichhart, D., Petersen, M. (2009). cDNA cloning and functional characterisation of CYP98A14 and NADPH: cytochrome P450 reductase from Coleus blumei involved in rosmarinic acid biosynthesis. Plant Mol. Biol. 69, 239–253. doi: 10.1007/s11103-008-9420-7
Franke, R., Hemm, M. R., Denault, J. W., Ruegger, M. O., Humphreys, J. M., Chapple, C. (2002). Changes in secondary metabolism and deposition of an unusual lignin in the ref8 mutant of Arabidopsis. Plant J. 30, 47–59. doi: 10.1046/j.1365-313X.2002.01267.x
Fu, R., Shi, M., Deng, C., Zhang, Y., Zhang, X., Wang, Y., et al. (2020). Improved phenolic acid content and bioactivities of Salvia miltiorrhiza hairy roots by genetic manipulation of RAS and CYP98A14. Food Chem. 331, 127365. doi: 10.1016/j.foodchem.2020.127365
He, Y., Zhong, X., Jiang, X., Cong, H., Sun, H., Qiao, F. (2020). Characterisation, expression and functional analysis of PAL gene family in Cephalotaxus hainanensis. Plant Physiol. Biochem. 156, 461–470. doi: 10.1016/j.plaphy.2020.09.030
Hoffmann, L., Maury, S., Martz, F., Geoffroy, P., Legrand, M. (2003). Purification, cloning, and properties of an acyltransferase controlling shikimate and quinate ester intermediates in phenylpropanoid metabolism. J. Biol. Chem. 278, 95–103. doi: 10.1074/jbc.M209362200
Hou, X., Shao, F., Ma, Y., Lu, S. (2013). The phenylalanine ammonia-lyase gene family in Salvia miltiorrhiza: genome-wide characterization, molecular cloning and expression analysis. Mol. Biol. Rep. 40, 4301–4310. doi: 10.1007/s11033-013-2517-3
Hu, J., Yan, C., Li, S., Tang, H., Chen, Y. (2023). Comparative physiological responses and transcriptome analysis revealing the metabolic regulatory mechanism of Prunella vulgaris L. induced by exogenous application of hydrogen peroxide. Ind. Crops Products 192, 116065. doi: 10.1016/j.indcrop.2022.116065
Khatri, P., Chen, L., Rajcan, I., Dhaubhadel, S. (2023). Functional characterization of cinnamate 4-hydroxylase gene family in soybean (Glycine max). PloS One 18, e0285698. doi: 10.1371/journal.pone.0285698
Kim, K. H., Janiak, V., Petersen, M. (2004). Purification, cloning and functional expression of hydroxyphenylpyruvate reductase involved in rosmarinic acid biosynthesis in cell cultures of Coleus blumei. Plant Mol. Biol. 54, 311–323. doi: 10.1023/B:PLAN.0000036367.03056.b2
Kim, Y. B., Shin, Y., Tuan, P. A., Li, X., Park, Y., Park, N. I., et al. (2014). Molecular cloning and characterization of genes involved in rosmarinic acid biosynthesis from Prunella vulgaris. Biol. Pharm. Bull. 37, 1221–1227. doi: 10.1248/bpb.b14-00139
Kumar, S., Stecher, G., Tamura, K. (2016). MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Kwon, Y. O., Hong, J. T., Oh, K. W. (2017). Rosmarinic acid potentiates pentobarbital-induced sleep behaviors and non-rapid eye movement (NREM) sleep through the activation of GABAA-ergic systems. Biomolecules Ther. 25, 105–111. doi: 10.4062/biomolther.2016.035
Lalitha, S. (2000). Primer premier 5. Biotech. Software Internet Rep. 1, 270–272. doi: 10.1089/152791600459894
Landmann, C., Hücherig, S., Fink, B., Hoffmann, T., Dittlein, D., Coiner, H. A., et al. (2011). Substrate promiscuity of a rosmarinic acid synthase from lavender (Lavandula angustifolia L.). Planta 234, 305–320. doi: 10.1007/s00425-011-1400-5
Levsh, O., Pluskal, T., Carballo, V., Mitchell, A. J., Weng, J. K. (2019). Independent evolution of rosmarinic acid biosynthesis in two sister families under the Lamiids clade of flowering plants. J. Biol. Chem. 294, 15193–15205. doi: 10.1074/jbc.RA119.010454
Li, Z., Hu, J., Tang, H., Cao, L., Guo, Q., Wang, C., et al. (2022). Temperature and photoperiod change the flowering process in Prunella vulgaris by inducing changes in morphology, endogenous hormones, carbon/nitrogen metabolites and gene expression. J. Am. Soc. Hortic. Sci. 147, 73–81. doi: 10.21273/JASHS05144-21
Liu, Q., Yao, L., Xu, Y., Cheng, H., Wang, W., Liu, Z., et al. (2019). In vitro evaluation of hydroxycinnamoyl CoA: quinate hydroxycinnamoyl transferase expression and regulation in Taraxacum antungense in relation to 5-caffeoylquinic acid production. Phytochemistry 162, 148–156. doi: 10.1016/j.phytochem.2019.02.014
Liu, T., Yao, R., Zhao, Y., Xu, S., Huang, C., Luo, J., et al. (2017). Cloning, functional characterization and site-directed mutagenesis of 4-coumarate: Coenzyme A ligase (4CL) involved in coumarin biosynthesis in Peucedanum praeruptorum Dunn. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.00004
Lu, S. (2021). Biosynthesis and regulatory mechanisms of bioactive compounds in Salvia miltiorrhiza, a model system for medicinal plant biology. Crit. Rev. Plant Sci. 40, 243–283. doi: 10.1080/07352689.2021.1935719
Lu, X., Hao, L., Wang, F., Huang, C., Wu, S. (2013). Molecular cloning and overexpression of the tyrosine aminotransferase (TAT) gene leads to increased rosmarinic acid yield in Perilla frutescens. Plant Cell Tissue Organ Culture 115, 69–83. doi: 10.1007/s11240-013-0341-z
Ma, W. L., Wu, M., Wu, Y., Ren, Z., Zhong, Y. (2013). Cloning and characterization of a phenylalanine ammonia-lyase gene from Rhus chinensis. Plant Cell Rep. 32, 1179–1190. doi: 10.1007/s00299-013-1413-6
Mansouri, M., Mohammadi, F. (2021). Transcriptome analysis to identify key genes involved in terpenoid and rosmarinic acid biosynthesis in lemon balm (Melissa officinalis). Gene 773, 145417. doi: 10.1016/j.gene.2021.145417
National Pharmacopoeia Committee (2020). Pharmacopoeia of the people’s Republic of China (part I) (Beijing: China Medical Science and Technology Press).
Noguchi-Shinohara, M., Ono, K., Hamaguchi, T., Iwasa, K., Nagai, T., Kobayashi, S., et al. (2015). Pharmacokinetics, safety and tolerability of Melissa officinalis extract which contained rosmarinic acid in healthy individuals: a randomized controlled trial. PloS One 10, e0126422. doi: 10.1371/journal.pone.0126422
Pan, X., Chang, Y., Li, C., Qiu, X., Cui, X., Meng, F., et al. (2023). Chromosome-level genome assembly of Salvia miltiorrhiza with orange roots uncovers the role of Sm2OGD3 in catalyzing 15,16-dehydrogenation of tanshinones. Horticulture Res. 10, uhad069. doi: 10.1093/hr/uhad069
Petersen, M., Abdullah, Y., Benner, J., Eberle, D., Gehlen, K., Hücherig, S., et al. (2009). Evolution of rosmarinic acid biosynthesis. Phytochemistry 70, 1663–1679. doi: 10.1016/j.phytochem.2009.05.010
Poppe, L., Rétey, J. (2005). Friedel-crafts-type mechanism for the enzymatic elimination of ammonia from histidine and phenylalanine. Angewandte Chemie (Internattional Ed. English) 44, 3668–3688. doi: 10.1002/anie.200461377
Radziejewska, I., Supruniuk, K., Nazaruk, J., Karna, E., Poplawska, B., Bielawska, A., et al. (2018). Rosmarinic acid influences collagen, MMPs, TIMPs, glycosylation and MUC1 in CRL-1739 gastric cancer cell line. Biomedicine Pharmacotherapy 107, 397–407. doi: 10.1016/j.biopha.2018.07.123
Raes, J., Rohde, A., Christensen, J. H., Van de Peer, Y., Boerjan, W. (2003). Genome-wide characterization of the lignification toolbox in Arabidopsis. Plant Physiol. 133, 1051–1071. doi: 10.1104/pp.103.026484
Ralph, J., Akiyama, T., Kim, H., Lu, F., Schatz, P. F., Marita, J. M., et al. (2006). Effects of coumarate 3-hydroxylase down-regulation on lignin structure. J. Biol. Chem. 281, 8843–8853. doi: 10.1074/jbc.M511598200
Ro, D. K., Mah, N., Ellis, B. E., Douglas, C. J. (2001). Functional characterization and subcellular localization of poplar (Populus trichocarpa x Populus deltoides) cinnamate 4-hydroxylase. Plant Physiol. 126, 317–329. doi: 10.1104/pp.126.1.317
Ru, M., Wang, K., Bai, Z., Peng, L., He, S., Pei, T., et al. (2017b). Molecular cloning and characterisation of two enzymes involved in the rosmarinic acid biosynthesis pathway of Prunella vulgaris L. Plant Cell Tissue Organ Culture 128, 381–390. doi: 10.1007/s11240-016-1117-z
Ru, M., Wang, K., Bai, Z., Peng, L., He, S., Wang, Y., et al. (2017a). A tyrosine aminotransferase involved in rosmarinic acid biosynthesis in Prunella vulgaris L. Sci. Rep. 7, 4892. doi: 10.1038/s41598-017-05290-4
Sahraeian, S. M. E., Mohiyuddin, M., Sebra, R., Tilgner, H., Afshar, P. T., Au, K. F., et al. (2017). Gaining comprehensive biological insight into the transcriptome by performing a broad-spectrum RNA-seq analysis. Nat. Communication 8, 59. doi: 10.1038/s41467-017-00050-4
Sander, M., Petersen, M. (2011). Distinct substrate specificities and unusual substrate flexibilities of two hydroxycinnamoyltransferases, rosmarinic acid synthase and hydroxycinnamoyl-CoA: shikimate hydroxycinnamoyl-transferase, from Coleus blumei Benth. Planta 233, 1157–1171. doi: 10.1007/s00425-011-1367-2
Scarpati, M., Oriente, G. (1958). Isolation and constitution of rosmarinic acid from Rosmarinus officinalis. La Ricerca Scientifica 28, 2329–2333.
Schoch, G., Goepfert, S., Morant, M., Hehn, A., Meyer, D., Ullmann, P., et al. (2001). CYP98A3 from Arabidopsis thaliana is a 3′-hydroxylase of phenolic esters, a missing link in the phenylpropanoid pathway. J. Biol. Chem. 276, 36566–36574. doi: 10.1074/jbc.M104047200
Shi, R., Sun, Y. H., Li, Q., Heber, S., Sederoff, R., Chiang, V. L. (2010). Towards a systems approach for lignin biosynthesis in Populus trichocarpa: transcript abundance and specificity of the monolignol biosynthetic genes. Plant Cell Physiol. 51, 144–163. doi: 10.1093/pcp/pcp175
Song, J., Wang, Z. (2011). RNAi-mediated suppression of the phenylalanine ammonia-lyase gene in Salvia miltiorrhiza causes abnormal phenotypes and a reduction in rosmarinic acid biosynthesis. J. Plant Res. 124, 183–192. doi: 10.1007/s10265-010-0350-5
Taguchi, R., Hatayama, K., Takahashi, T., Hayashi, T., Sato, Y., Sato, D., et al. (2017). Structure-activity relations of rosmarinic acid derivatives for the amyloid β aggregation inhibition and antioxidant properties. Eur. J. Medicinal Chem. 138, 1066–1075. doi: 10.1016/j.ejmech.2017.07.026
Tang, H., Hu, J., Zhao, M., Cao, L., Chen, Y. (2023). Comparative study of the physiological responses, secondary metabolites, and gene expression of medicinal plant Prunella vulgaris L. treated with exogenous methyl jasmonate and salicylic acid. Acta Physiologiae Plantarum 45, 20. doi: 10.1007/s11738-022-03498-0
Uhlmann, A., Ebel, J. (1993). Molecular cloning and expression of 4-coumarate-Coenzyme A ligase, an enzyme involved in the resistance response of soybean (Glycine max L.) against pathogen attack. Plant Physiol. 102, 1147–1156. doi: 10.1104/pp.102.4.1147
Wang, G. Q., Chen, J. F., Yi, B., Tan, H. X., Zhang, L., Chen, W. S. (2017). HPPR encodes the hydroxyphenylpyruvate reductase required for the biosynthesis of hydrophilic phenolic acids in Salvia miltiorrhiza. Chin. J. Natural Medicines 15, 917–927. doi: 10.1016/S1875-5364(18)30008-6
Wang, H., Dong, Q., Duan, D., Zhao, S., Li, M., van Nocker, S., et al. (2018). Comprehensive genomic analysis of the TYROSINE AMINOTRANSFERASE (TAT) genes in apple (Malus domestica) allows the identification of MdTAT2 conferring tolerance to drought and osmotic stresses in plants. Plant Physiol. Biochem. 133, 81–91. doi: 10.1016/j.plaphy.2018.10.033
Wang, Y., Guo, L., Zhao, Y., Zhao, X., Yuan, Z. (2022b). Systematic analysis and expression profiles of the 4-coumarate: CoA ligase (4CL) gene family in Pomegranate (Punica granatum L.). Int. J. Mol. Sci. 23, 3509. doi: 10.3390/ijms23073509
Wang, B., Sun, W., Li, Q., Li, Y., Luo, H., Song, J., et al. (2015). Genome-wide identification of phenolic acid biosynthetic genes in Salvia miltiorrhiza. Planta 241, 711–725. doi: 10.1007/s00425-014-2212-1
Wang, C., Yang, J., Song, P., Zhang, W., Lu, Q., Yu, Q., et al. (2022a). FIONA1 is an RNA N6-methyladenosine methyltransferase affecting Arabidopsis photomorphogenesis and flowering. Genome Biol. 23, 40. doi: 10.1186/s13059-022-02612-2
Watts, K. T., Mijts, B. N., Lee, P. C., Manning, A. J., Schmidt-Dannert, C. (2006). Discovery of a substrate selectivity switch in tyrosine ammonia-lyase, a member of the aromatic amino acid lyase family. Chem. Biol. 13, 1317–1326. doi: 10.1016/j.chembiol.2006.10.008
Weitzel, C., Petersen, M. (2010). Enzymes of phenylpropanoid metabolism in the important medicinal plant Melissa officinalis L. Planta 232, 731–742. doi: 10.1007/s00425-010-1206-x
Weitzel, C., Petersen, M. (2011). Cloning and characterisation of rosmarinic acid synthase from Melissa officinalis L. Phytochemistry 72, 572–578. doi: 10.1016/j.phytochem.2011.01.039
Xiao, Y., Zhang, L., Gao, S., Saechao, S., Di, P., Chen, J., et al. (2011). The c4h, tat, hppr and hppd genes prompted engineering of rosmarinic acid biosynthetic pathway in Salvia miltiorrhiza hairy root cultures. PloS One 6, e29713. doi: 10.1371/journal.pone.0029713
Xu, J. J., Fang, X., Li, C. Y., Zhao, Q., Martin, C., Chen, X. Y., et al. (2018). Characterization of Arabidopsis thaliana hydroxyphenylpyruvate reductases in the tyrosine conversion pathway. Front. Plant Sci. 9. doi: 10.3389/fpls.2018.01305
Zhang, S., Meng, F., Pan, X., Qiu, X., Li, C., Lu, S. (2024). Chromosome-level genome assembly of Prunella vulgaris L. provides insights into pentacyclic triterpenoid biosynthesis. Plant J. doi: 10.1111/tpj.16629
Zhao, S. J., Hu, Z. B., Liu, D., Leung, F. C. (2006). Two divergent members of 4-coumarate: coenzyme A ligase from Salvia miltiorrhiza Bunge: cDNA cloning and functional study. J. Integr. Plant Biol. 48, 1355–1364. doi: 10.1111/j.1744-7909.2006.00302.x
Zheng, H., Zhao, H., Zhang, X., Liang, Z., He, Q. (2022). Systematic identification and validation of suitable reference genes for the normalization of gene expression in Prunella vulgaris under different organs and spike development stages. Genes 13, 1947. doi: 10.3390/genes13111947
Keywords: biosynthetic pathway, CRISPR/Cas9, in vitro enzymatic activity assay, Prunella vulgaris, PvRAS3, PvRAS4, rosmarinic acid, rosmarinic acid synthase
Citation: Yan C, Li C, Jiang M, Xu Y, Zhang S, Hu X, Chen Y and Lu S (2024) Systematic characterization of gene families and functional analysis of PvRAS3 and PvRAS4 involved in rosmarinic acid biosynthesis in Prunella vulgaris. Front. Plant Sci. 15:1374912. doi: 10.3389/fpls.2024.1374912
Received: 23 January 2024; Accepted: 15 April 2024;
Published: 01 May 2024.
Edited by:
Naoki Kitaoka, Hokkaido University, JapanReviewed by:
Takafumi Shimizu, Research Institute of Innovative Technology for the Earth (RITE), JapanSibongile Mafu, University of Massachusetts Amherst, United States
Copyright © 2024 Yan, Li, Jiang, Xu, Zhang, Hu, Chen and Lu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shanfa Lu, c2ZsdUBpbXBsYWQuYWMuY24=; Yuhang Chen, Y2hlbnl1aGFuZzIyMUAxMjYuY29t
†These authors have contributed equally to this work
 Chao Yan1,2,3†
Chao Yan1,2,3† Caili Li
Caili Li Yuhang Chen
Yuhang Chen Shanfa Lu
Shanfa Lu