- 1College of Life Sciences, Gannan Normal University, Ganzhou, China
- 2China-USA Citrus Huanglongbing Joint Laboratory, National Navel Orange Engineering Research Center, College of Life Sciences, Gannan Normal University, Ganzhou, Jiangxi, China
Introduction: Heavy metal-associated isoprenylated plant proteins (HIPPs) play vital roles in maintaining heavy metal balance and responding to both biotic and abiotic stresses in vascular plants. However, the role of HIPPs in the response to Huanglongbing (HLB), a harmful disease of citrus caused by the phloem-colonizing bacterium Candidatus Liberibacter asiaticus (CLas), has not been examined.
Methods and results: In this study, a total of 26 HIPP genes were identified in Citrus sinensis, and they were grouped into 5 clades. The CsHIPP genes are distributed on 8 chromosomes and exhibited considerable synteny with HIPPs found in Arabidopsis thaliana. Additionally, we analyzed the gene structure, conserved motifs and domains of the CsHIPPs. Various cis-acting elements related to plant hormones and stress responses were identified in the promoters of CsHIPPs. Public transcriptome data and RT-qPCR analysis showed that the expression level of CsHIPP03 was significantly reduced in samples infected by CLas and Xanthomonas citri ssp. citri (Xcc). Furthermore, silencing the homologous gene of CsHIPP03 in Nicotiana benthamiana increased the disease resistance of plants to bacteria.
Discussion: Our results provide a basis for functional studies of HIPP gene family in C. sinensis, highlighting their functions in bacterial resistance, and improve our understanding to the susceptibility mechanism of HLB.
Introduction
Heavy metal-associated isoprenylated plant proteins (HIPPs) are metal-binding metallochaperones consisting of one or two heavy metal-associated (HMA) domains and an isoprenylation motif (Barr et al., 2023). Moreover, glycine-rich or proline-rich regions are commonly found between the HMA domains and the isoprenylation motif (Suzuki et al., 2002; Barth et al., 2009). Genome-wide identification of HIPP members indicated various quantities of HIPPs in plants, as there were 45 members in Arabidopsis thaliana, 59 in Oryza sativa, 74 in Populus trichocarpa, 51 in Setaria italica, 5 in Selaginella moellendorffii, and 13 in Fagopyrum tataricum (de Abreu-Neto et al., 2013; Ye et al., 2022). Moreover, in Triticeae species, 114 HIPPs were identified in Triticum aestivum, 33 in Triticum urartu, 40 in Aegilops tauschii, 58 in Triticum dicoccoides, and 33 in Hordeum vulgare (Zhang et al., 2020c). The HIPPs were categorized into five clades through phylogenetic analysis, and segmental duplication played a significant role in the proliferation of the HIPP gene family (Khan et al., 2019; Zhang et al., 2020c).
The HMA domain contained a CXXC motif that was highly conserved, along with a βαββαβ-fold structure. This structure is responsible for binding Cu2+, Cd2+, or Zn2+ and is involved in the regulation of heavy metal homeostasis and detoxification (Dykema et al., 1999; Robinson and Winge, 2010). The C-terminus of the HIPPs usually includes a CaaX motif, which is crucial for its biological functions, including protein–membrane and protein–protein interactions (de Abreu-Neto et al., 2013). Research on HIPPs focuses on their connection to both abiotic and biotic stresses (Guo et al., 2021). AtCdI19 in A. thaliana showed induction in response to Cd2+, Hg2+, Fe2+, and Cu2+ stress treatments, and its overexpression resulted in increased Cd tolerance in transgenic Arabidopsis (Suzuki et al., 2002). The expression of A. thaliana HIPP20, HIPP22, HIPP26, and HIPP27 in yeast resulted in the enhancement of Cd resistance in the Cd-sensitive yeast strain ycf1. Nevertheless, the A. thaliana hipp20/21/22 triple mutant exhibited heightened sensitivity to Cd (Tehseen et al., 2010). The heterologous expression of VvHIPP21 from Vitis vinifera in A. thaliana resulted in decreased resistance to cold and drought in plants (Zheng et al., 2023). Furthermore, A. thaliana HIPP5, HIPP6, HIPP7, and HIPP34 were implicated in plant endoplasmic reticulum-associated degradation (Guo et al., 2021). Various HIPP genes were targeted by pathogen effectors and served as susceptibility genes (Cowan et al., 2018; Radakovic et al., 2018). For example, the potato mop-top virus (PMTV) movement protein TGB1 interacted with NbHIPP26 to activate drought stress response genes and facilitate virus long-distance movement (Cowan et al., 2018). Knocking out of AtHIPP27 in A. thaliana plants resulted in decreased susceptibility to beet cyst nematode (Heterodera schachtii) infection (Radakovic et al., 2018). Additionally, overexpression of TaHIPP1 increased plant susceptibility to Puccinia striiformis f. sp. Tritici, while knocking down TaHIPP1 expression enhanced wheat resistance (Zhang et al., 2015).
Susceptibility genes (S) are crucial for the pathogenicity and virulence of pathogens. It is believed that the products of S genes may be utilized by pathogens for host recognition, infiltration, acquisition of nutrients, proliferation, and transmission, or to suppress the host immune response (Langner et al., 2018; Zaidi et al., 2018). There were many S genes in different plants, like elF4E in A. thaliana (Pyott et al., 2016), SlMlo1 in tomato (Nekrasov et al., 2017), DIPM-1 in apple (Malnoy et al., 2016), TaEDR1 in wheat (Zhang et al., 2017), OsSWEET14 in rice (Li et al., 2012), and CsLOB1 in citrus (Peng et al., 2017). Inactivating the S genes can disrupt the baseline susceptibility of hosts and result in disease resistance (Xu et al., 2019). A deleterious mutation in the elF4E gene locus of A. thaliana results in plants that are completely resistant to Turnip mosaic virus (Pyott et al., 2016). Rice resistance to bacterial blight was enhanced when the expression of OsSWEET14 was suppressed (Li et al., 2012). By utilizing CRISPR/Cas9 technology, grapefruit and sweet orange were able to develop resistance to canker disease by editing the promoter and coding region of the citrus susceptibility gene CsLOB1 (Peng et al., 2017).
Citrus huanglongbing (HLB) is a highly damaging disease that has led to a substantial decrease in citrus production on a global scale (Bové, 2006). In field, the Asian citrus psyllid (Diaphorina citri) transmit CLas to citrus while feeding on sap. Subsequently, CLas established itself in the phloem sieve element, ultimately causing disease symptoms (da Graca et al., 2016). As the citrus industry rapidly develops and the scale of citrus trade continues to expand, CLas and Asian citrus psyllid have spread globally (Wang, 2019). There are no effective strategies for curing HLB at the moment, and all citrus cultivars could potentially be affected by the HLB-associated bacterium CLas (Hu et al., 2021). It is difficult to uncover the pathogenic mechanism of CLas because it cannot be cultured in vitro (Pagliaccia et al., 2017). The CLas has a genome size of approximately 1.23 Mb and does not have type III and type IV secretion systems. However, it does have a complete type I secretion system and the general secretory pathway (GSP) (Duan et al., 2009). The GSP is crucial for transporting bacterial proteins, while the Sec-dependent effectors (SDEs) play a significant role in the infection of plants by pathogens (Liu et al., 2021). Researches have indicated that the immune responses of citrus can be influenced by several SDEs of CLas (Clark et al., 2018; Zhang et al., 2020b, 2023; Shi et al., 2023a, Shi et al., 2023b).
Previously, we had identified CsHIPP03 (namely, HIPP7) as a potential interactor of CLas core effector SDE34 in yeast two-hybrid (Y2H) screening (Hu et al., 2023). However, a thorough analysis of the HIPP gene family in citrus has not been documented. In this study, we identified 26 HIPP genes and systematically investigated the putative functions of HIPP genes in C. sinensis. Their detailed phylogenetic relationships, gene structures, subcellular localization, and expression profiles under CLas infection were investigated. Additionally, we performed a functional analysis of CsHIPP03 in Nicotiana benthamiana. Our research has established a foundation for further examination of the role of HIPP genes in CLas-citrus interaction.
Materials and methods
Plant materials
The A. thaliana and N. benthamiana plants were cultivated in an artificial climate culture chamber under specific conditions (16/8 h day/night cycle, 23°C ± 2°C temperature, and 50%–60% relative humidity). The leaves and branches selected for gene cloning and expression analysis were obtained from healthy and CLas-infected C. sinensis cv. Newhall navel trees in Ganzhou, China. These trees were 3 years old and had Citrange rootstock.
Identification of the HIPPs in C. sinensis
The protein sequences of C. sinensis (version 3.0) were obtained from the Citrus Pan-genome to Breeding Database (CPBD) (http://citrus.hzau.edu.cn/) (Liu et al., 2022). The protein sequences of C. sinensis with the HMA domain (PF00403.28) were searched using HMMER web server (Finn et al., 2011). Afterward, the candidate sequences were sent to the NCBI CDD database to search for conserved domains, and any sequences that included the HMA domain were retained (Marchler-Bauer et al., 2017). Subsequently, the reserved sequences that were predicted containing the isoprenylation motif CaaX using PrePS were recognized as HIPP proteins (Maurer-Stroh and Eisenhaber, 2005).
Phylogenetic analysis
The protein sequences of HIPPs in A. thaliana were generated from a previous published study (de Abreu-Neto et al., 2013). The protein sequence of HIPPs in A. thaliana and C. sinensis was aligned using MAFFT (Katoh et al., 2019). Optimized partitioning scheme and evolutionary models were determined by PartitionFinder2 (Lanfear et al., 2017). Maximum likelihood phylogenetic analysis was inferred with IQ-TREE (Nguyen et al., 2015).
Chromosomal distribution, synteny analysis, and exon–intron structural analysis
Chromosomal distribution of the CsHIPP genes was visualized using TBtools (Chen et al., 2020) with the GFF3 file downloaded from CPBD (Liu et al., 2022). The collinearity relationship of HIPP genes from A. thaliana and C. sinensis was conducted using MCScanX and BLASTP methods integrated in TBtools (Wang et al., 2012; Chen et al., 2020). Moreover, the exon–intron structure of HIPP genes were determined using the “Visualize Gene Structure” plug-in of TBtools (Chen et al., 2020).
Conserved motifs and domain analysis
The conserved motif of HIPP proteins was assessed using MEME suite online (Bailey et al., 2015). Then, conserved domains of HIPP proteins were determined using “Batch CD-Search” tools in the NCBI CDD database (Marchler-Bauer et al., 2017).
RNA-seq analysis
Released RNA-seq data related with HLB and citrus canker of C. sinensis in SRA database were downloaded and reanalyzed locally as mentioned previously with few modifications (Pertea et al., 2016). In brief, selected RNA-seq data were downloaded using the SRA Toolkit from the SRA database. Raw RNA-seq data were trimmed with Trimmomatic 0.36.0 (Bolger et al., 2014) and then transcript quantification was performed using the HISAT2-StringTie-ballgown RNA-seq pipeline (Fu et al., 2019). The reference genome of C. sinensis (version 3.0) for RNA-seq analysis was download from CPBD (Liu et al., 2022). The expression data of HIPPs were extracted and normalized using the Z-score method for heat map construction with the Pheatmap package (Kolde and Kolde, 2015). The BioSamples accessions of RNA-seq data used in this study are listed in Supplementary Table 1.
RT-qPCR analysis
Total RNA of plant samples was extracted using the EasyPure® Plant RNA Kit (TransGen Biotech) according to the manufacturer’s protocol. First-strand cDNA was synthesized with EasyScript® All-in-One First-Strand cDNA Synthesis SuperMix for qPCR (One-Step gDNA Removal) (TransGen Biotech) and diluted 20 times for RT-qPCR using specific primers (Supplementary Table 2). Reactions were performed in a volume of 20 μL with 1× TransStart® Tip Green qPCR SuperMix, using forward and reverse primers at concentrations of 0.2 μM. The PCR cycling began with an initial activation step at 98°C for 10 min, then proceeded with 40 cycles of 95°C for 15 s and 60°C for 40 s. All cDNA samples were run in triplicate. The C. sinensis glyceraldehyde-3-phosphate dehydrogenase (GAPDH) gene (NCBI Reference Sequence: XM_006468885.2) and N. benthamiana actin gene (NCBI Reference Sequence: AY179605.1) were used as endogenous controls. Relative expression levels were calculated using the 2−ΔΔCT method (Livak and Schmittgen, 2001).
Cis-acting elements analysis
The 2,000-bp upstream genomic sequences of CsHIPPs were obtained from the reference genome of C. sinensis (version 3.0) (Liu et al., 2022). The cis-acting elements in the promoter regions of CsHIPPs were in silico analysis with the PlantCARE database (Lescot et al., 2002).
Vector construction
The plasmids were digested using the corresponding restriction enzymes. The gene fragments with homologous arms to the plasmids were amplified and subjected to homologous recombination with the linearized plasmids using the pEASY®-T&B Simple Cloning Kit (TransGen Biotech, China). Subsequently, the recombinant plasmid was transformed into Escherichia coli (TOP10) chemically competent cells. Positive clones were identified by PCR and then confirmed by sequencing (Tsingke). The plasmids, restriction sites, and primers used in this study are listed in Supplementary Table 2, with details as follows. For the promoter activity assay, the promoter sequences of CsHIPP03 were cloned and inserted into the plant binary vector pCAMBIA1380-GUS digested by Sbf I and Xba I to drive expression of β-glucuronidase (GUS). For subcellular localization, the coding sequences of CsHIPP03, CsHIPP10, CsHIPP13, CsHIPP19, CsHIPP20, CsHIPP22, CsHIPP23, and CsHIPP26, which do not contain stop codons, were cloned and inserted into the plant binary vector pCAMBIA2300-GFP digested by Sac I and Xba I. For Y2H assays, the coding sequences of NbHIPP3.1 (Niben101Scf07236g00011), NbHIPP3.2 (Niben101Scf06423g01011), and NbHIPP3.3 (Niben101Scf26202g00005) from N. benthamiana were amplified and inserted into the prey vector pGADT7 digested with EcoR I and BamH I. For virus-induced gene silencing (VIGS) assays, 300-bp fragments of NbHIPP3.1, NbHIPP3.2, and NbHIPP3.3 were cloned and inserted into vector pTRV2 digested with EcoR I and BamH I.
Genetic transformation of A. thaliana and citrange hairy roots
PHIPP03-GUS and PHIPPM-GUS were transformed into A. thaliana through floral dipping method (Clough and Bent, 1998). In addition, these vectors were transformed into Agrobacterium rhizogenes (K599), and the Carrizo citrange hairy roots were transformed by the A. rhizogenes-mediated method as described previously (Ma et al., 2022). The transformed hairy roots were identified by GFP fluorescence observation.
GUS staining
The transgenic A. thaliana and citrange hairy roots were used for GUS staining and observed using an optical microscope. In brief, plant tissue samples were submerged in GUS staining buffer (Coolaber, China) and stained for a period of 8 h. Subsequently, samples were decolorized by repeated immersion in 70% alcohol three times prior to photography.
Subcellular localization
A. tumefaciens strain GV3101 (pMP90) cells containing binary vectors were cultured overnight in Luria Bertani liquid medium with 20 μg mL−1 rifampicin and 50 μg mL−1 kanamycin and resuspended in MES solution (10 mM MgCl2, 10 mM MES, pH 5.6, and 200 mM acetosyringone). The cultures were diluted to OD600 of 0.4. The diluted A. tumefaciens suspension was used to infiltrate the leaves of N. benthamiana, which were then incubated in darkness at 22°C for 2 days. Subsequently, the fluorescence signals were observed using a Leica Sp8 confocal microscope (Huang et al., 2019).
Yeast two-hybrid assays
BD-SDE34 and AD-NbHIPPs were co-transformed into yeast Y2Hgold strain, and the resultant diploid yeasts were cultured on synthetic dropout medium (SD)/-Leu/-Trp (DDO) and SD/-Leu/-Trp/-His (TDO) agar plates supplemented with 40 μg/mL X-a-gal to monitor the interaction. Images were captured 3 days post-plating.
TRV-based VIGS in N. benthamiana
A. tumefaciens strain GV3101 (pMP90) cultures carrying the TRV-RNA1 plasmid and TRV-RNA2 cassettes were grown overnight, collected by centrifugation at 3,500 rpm for 5 min, and then resuspended in MES solution to a final OD600 of 0.5. The TRV-RNA2 and TRV-RNA1 solutions were mixed in equal volumes and then infiltrated into the abaxial side of two N. benthamiana leaves on seedlings at the four-leaf stage. Three weeks post-infection, RT-qPCR was carried out to detect the VIGS efficiency. Pseudomonas syringae pv. tomato DC3000 mutant ΔhopQ1-1 were inoculated at 1.0 × 104 colony-forming units (CFU) mL−1 for virulence assays (Wei et al., 2007). In order to assess bacterial growth, the 1-cm2 leaf discs from N. benthamiana were ground in 1 ml of sterile water, and then serial dilutions were spotted onto KB medium. CFU were counted 2 days after incubation at 28°C (Ramos et al., 2021).
Statistical analysis
The statistical significance of the results was assessed by conducting a two-sided, unpaired Student’s t-test using SPSS 25.0. The significance levels were indicated as follows: * for p < 0.05, ** for p < 0.01, and *** for p < 0.001.
Results
Identification and characterization of the HIPP members in C. sinensis
After hmmsearch using the HMM profile of HMA (PF00403.28) and confirmation using the NCBI CDD database, 26 genes encoding HIPP proteins containing the HMA domain and isoprenylation motif were identified in the genome of C. sinensis. The protein lengths of CsHIPPs ranged from 101 to 554 amino acids, with the molecular weight ranging from 11.43 to 59.91 kDa. The isoelectric point of CsHIPPs ranged from 4.96 to 10.16 and the grand average of hydropathy value varied from −1.215 to −0.118 (Supplementary Table 3).
Phylogenetic analysis of the HIPP members
To reveal the phylogeny of HIPP proteins, the 26 CsHIPPs and 45 AtHIPPs were phylogenetically analyzed and a Maximum likelihood tree was built using IQ-TREE (Figure 1). HIPP members were divided into five clades based on the topology structure of the phylogenetic tree. The minimum clade, clade 4, consisted of four HIPP proteins, namely, CsHIPP09, AtHIPP17, AtHIPP18, and AtHIPP19, while the maximum clade, clade 2, contained 25 HIPP proteins, namely, 11 CsHIPPs and 14 AtHIPPs. In detail, clade 1 contained 6 CsHIPPs and 8 AtHIPPs, clade 3 contained 4 CsHIPPs and 7 AtHIPPs, and clade 5 contained 4 CsHIPPs and 13 AtHIPPs (Figure 1).
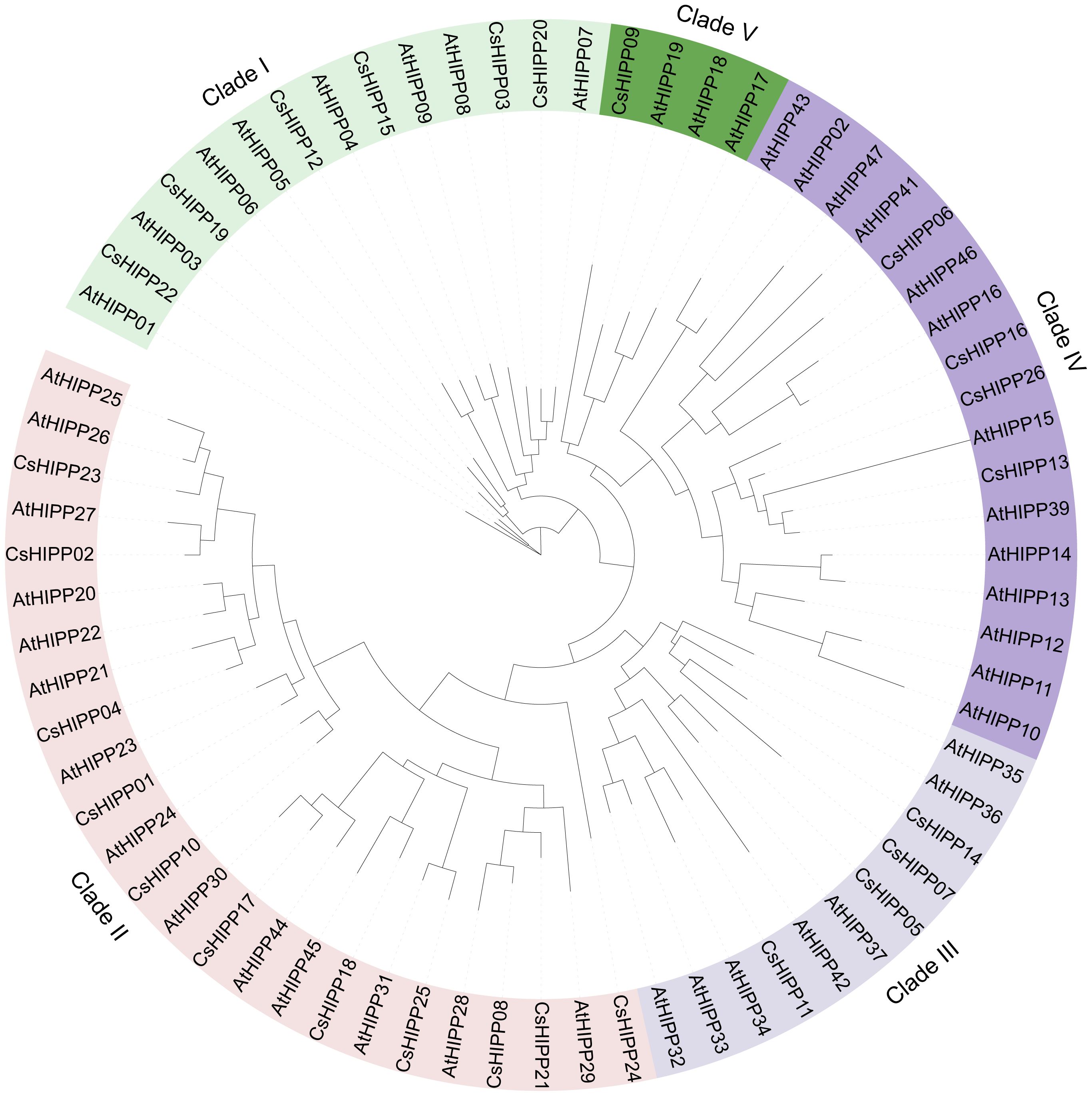
Figure 1 The phylogenetic analysis of the HIPPs from Citrus sinensis and Arabidopsis thialina. The phylogenetic tree contains 71 HIPPs, which were classified into five clades, namely, 26 HIPPs from C. sinensis and 45 HIPPs from A. thialina. Each clade’s label is positioned adjacent to the respective protein identifiers.
Chromosomal distribution and synteny analysis of the HIPP members
According to the gene location annotation information, the chromosomal distribution of CsHIPPs was performed. As shown in Figure 2, the HIPP members distributed separately on eight out of nine chromosomes of C. sinensis except chromosome 4. Chromosome 2 contained seven CsHIPPs, chromosome 5 contained six CsHIPPs, chromosome 9 contained three CsHIPPs, and there are two CsHIPPs distributed on each of the other remaining chromosomes (Figure 2A). Moreover, the collinearity relationships between the CsHIPPs and AtHIPPs were investigated. Twenty-one collinearity events occurred. Particularly, 14 CsHIPPs had collinearity relationships with 19 AtHIPPs (Figure 2B).
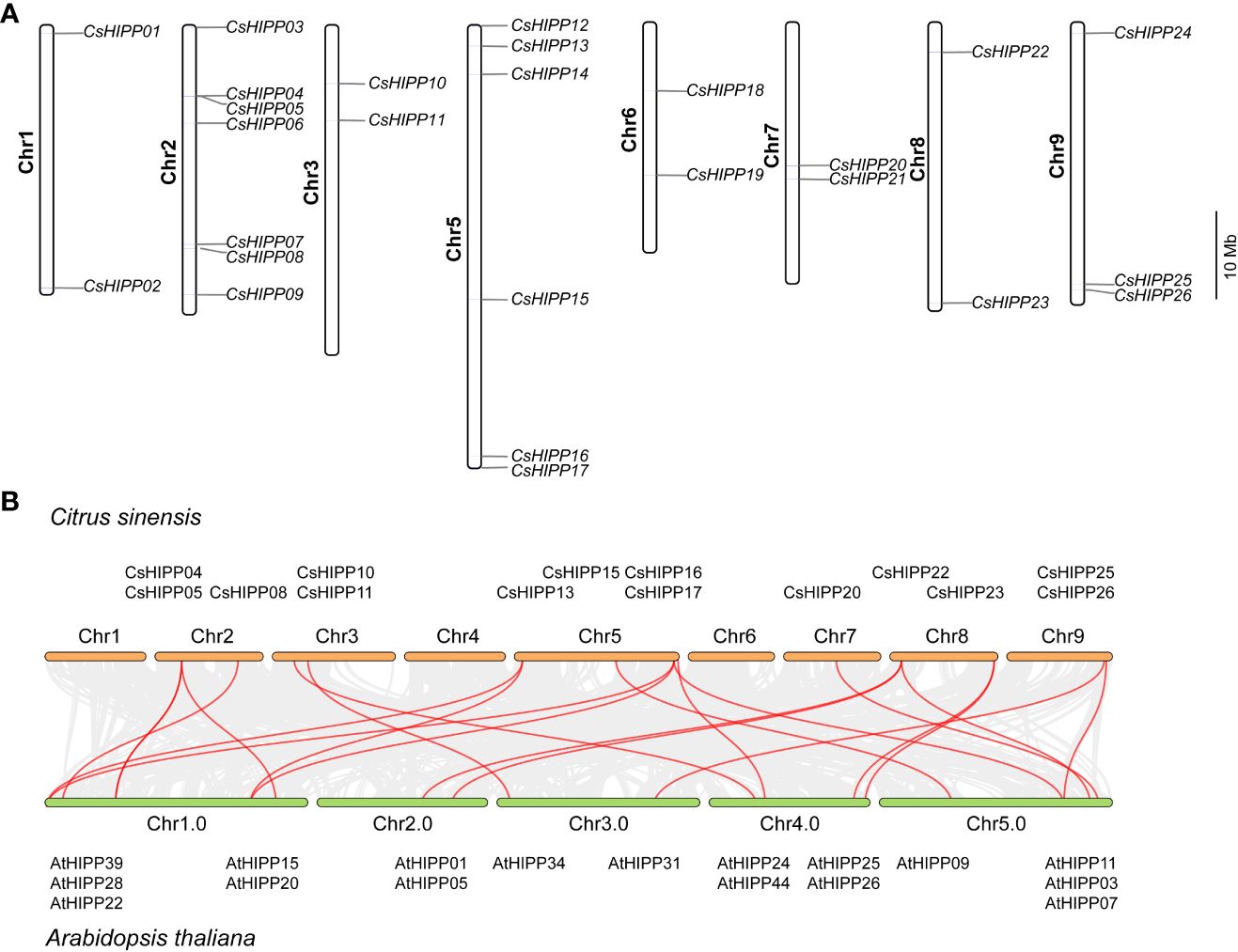
Figure 2 Chromosomal distribution of the CsHIPP genes and the collinearity relationships of CsHIPPs and AtHIPPs. (A) Chromosomal distribution of the HIPPs from C. sinensis. The identities of the chromosomes are marked on the left side of each chromosome, while the HIPP gene names are shown to the right of each chromosome. The scale on the right is in megabases (Mb). (B) The collinearity relationships between the CsHIPP genes and AtHIPP genes. Red lines indicate the existence of collinear relationship between the two, with a total of 21 collinear events.
Gene structure analysis of CsHIPPs
The gene structures of CsHIPPs were explored to understand the connections between gene structure and gene function (Figure 3). The number of exons in CsHIPPs ranged from 2 to 5. Thirteen CsHIPPs contained three exons, which is the primary gene structure of CsHIPPs, and five CsHIPPs had five exons, four CsHIPPs had four exons, and four CsHIPPs had two exons (Figure 3B). The phylogenetic relationships of CsHIPPs (Figure 3A) were consistent with their gene structures (Figure 3B).
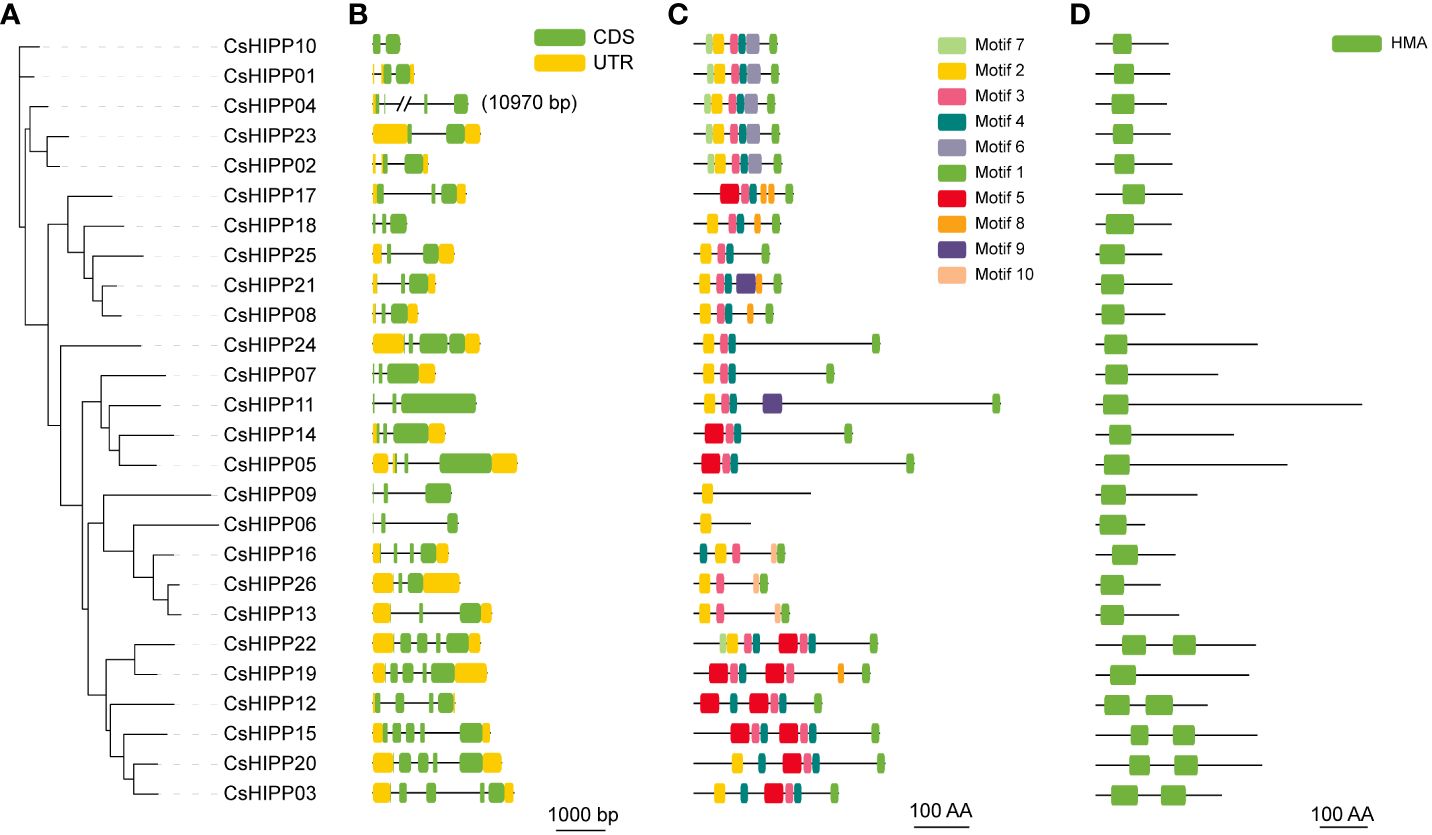
Figure 3 The gene structure, motif, and conserved domain analysis of CsHIPPs. (A) Phylogenetic tree of CsHIPPs. (B) The exons/introns structure of CsHIPP genes (green indicates CDSs, yellow indicates upstream/downstream sequences, and black line indicates introns). (C) Conservation motif analysis of CsHIPPs. (D) HMA conservative domain of CsHIPPs.
Conserved motifs and domains analysis of the HIPP members in C. sinensis
A total of 10 conserved motifs and one conserved domain were identified from CsHIPPs (Figure 3C). Similar to the clusters in Figure 1, CsHIPPs were divided into five clades (Figure 3A, Supplementary Table 3). All clade I members contained motifs 1, 3, 4 and 5. Clade II and III members shared motifs 1, 3, and 4. In addition, clades IV and V shared motif 2. Most of the CsHIPPs had more than four motifs except CsHIPP06 and CsHIPP09, which contained only one motif 4. Moreover, motif 9 presents only in CsHIPP11 and CsHIPP21 (Figure 3C). All CsHIPPs shared a conserved domain “HMA”. However, Clade I members contained two HMA domains except CsHIPP19, and the other clade members contained only one HMA domain (Figure 3D).
Response of CsHIPPs to HLB and citrus canker by transcriptome analysis
To understand the dynamic expression profiles of CsHIPPs in response to HLB and citrus canker, the two major diseases in citrus industry, the expression quantity of CsHIPPs was generated by reanalyzing related RNA-seq data in the SRA database. In the progress of CLas infection, the expression tendency of CsHIPPs varied irregularly. However, three genes (e.g., CsHIPP03) downregulated when CLas-infected samples in contrast to CLas-free samples at the 26- and 46-week time points. In addition, nine CsHIPPs upregulated in CLas-infected samples at the 46-week time point (Figure 4A). In response to citrus canker, eight CsHIPPs (e.g., CsHIPP01) upregulated after citrus bacterial canker infected for 24 and 48 h. Thirteen CsHIPPs (e.g., CsHIPP03) downregulated when citrus bacterial canker infected for 24 h while nine CsHIPPs (e.g., CsHIPP10) downregulated for 48 h (Figure 4B). Interestingly, two CsHIPPs, CsHIPP03 and CsHIPP10, showed downregulation in response to the infection of CLas and citrus bacterial canker (Figure 4).
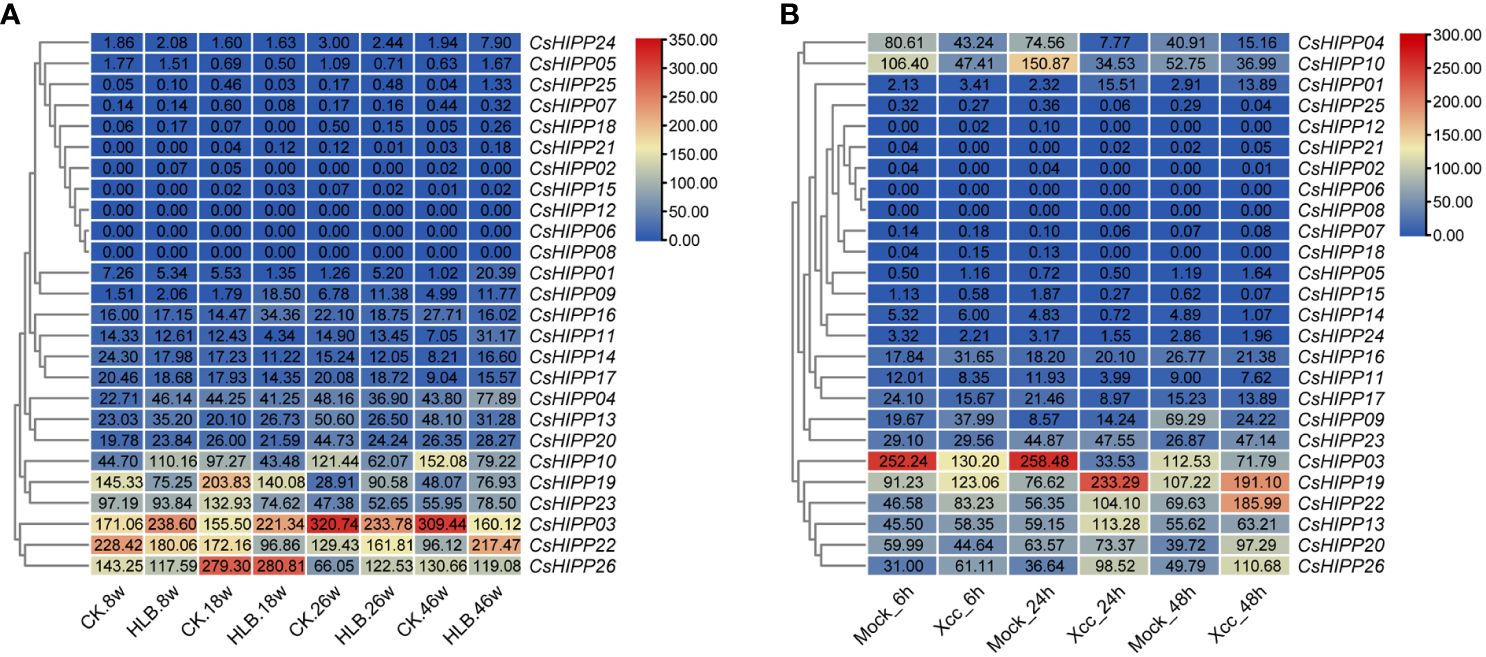
Figure 4 Expression profiles of the CsHIPP genes in different periods after infection by Candidatus Liberobacter asiaticus (CLas) (A) and Xanthomonas citri ssp. citri (Xcc) (B). “CK” and “Mock” represent control samples, while “HLB” signifies CLas-infected samples and “Xcc” refers to samples infected with Xcc. The durations post-CLas infection were 8 weeks, 18 weeks, 26 weeks, and 46 weeks, denoted as 8w, 18w, 26w, and 46w, respectively. The durations post-Xcc infection were 6 h, 24 h, and 48 h, denoted as 6h, 24h, and 48h, respectively. The numbers within the rectangle represent the transcripts per million value for each gene in each sample.
Gene expression of CsHIPPs in response to CLas infection
Many studies have shown that the HIPPs play an important role in the phytopathogenic–bacterial interaction system. To further validate the expression changes of CsHIPPs under the induction of HLB, we selected eight genes, namely, CsHIPP03, CsHIPP10, CsHIPP13, CsHIPP19, CsHIPP20, CsHIPP22, CsHIPP23, and CsHIPP26, for expression pattern analysis. The expression levels were detected by RT-qPCR in the midrib tissues of CLas-free and -infected C. sinensis cv. Newhall. Results showed that after CLas infection, the expression of CsHIPP03 was significantly downregulated in young and mature tissues (Figure 5A). Furthermore, with the exception of CsHIPP22 showing no significant difference in expression level, the other six genes exhibited upregulation during HLB infection (Figure 5).
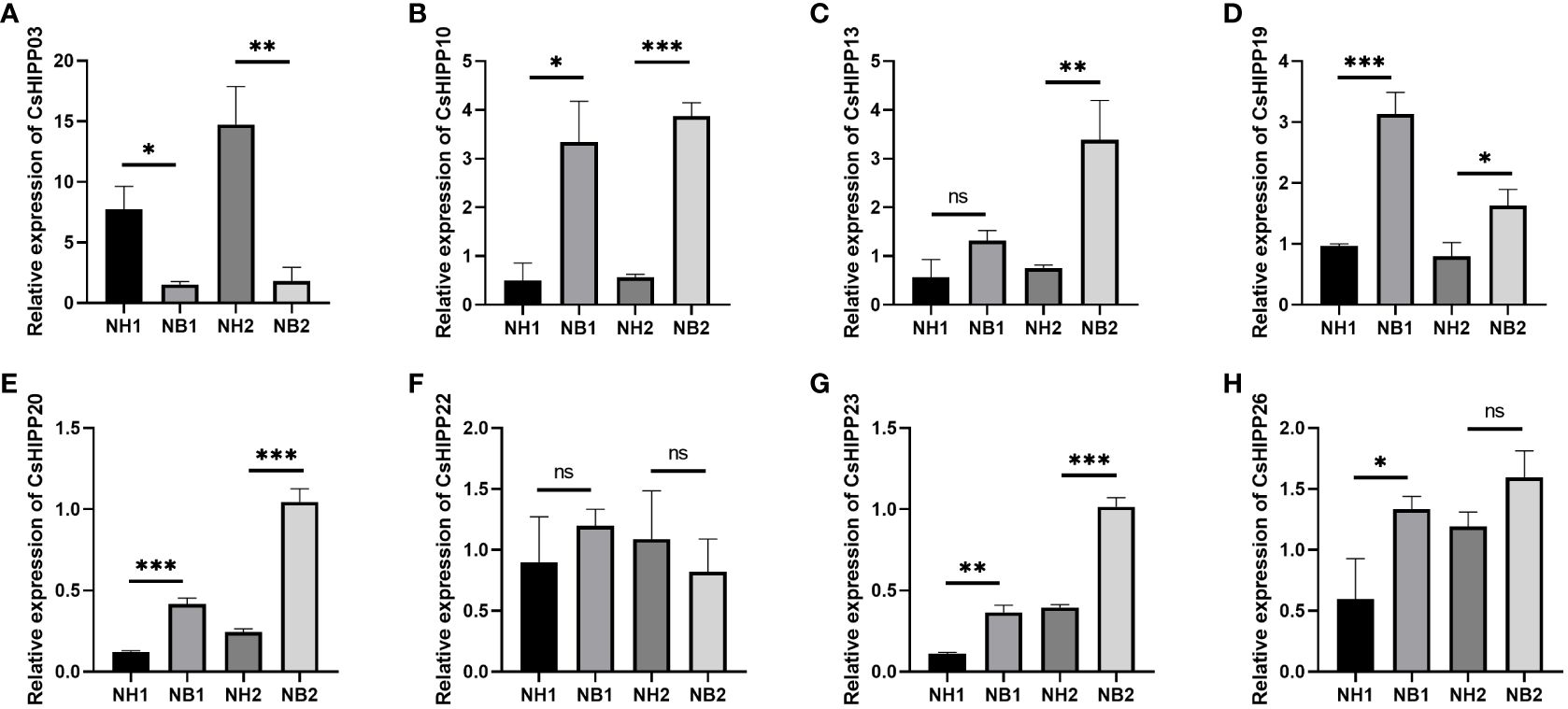
Figure 5 RT-qPCR analysis of eight CsHIPP genes in healthy and huanglongbing-positive C. sinensis. Relative expression of (A) CsHIPP03, (B) CsHIPP10, (C) CsHIPP13, (D) CsHIPP19, (E) CsHIPP20, (F) CsHIPP22, (G) CsHIPP23, and (H) CsHIPP26. Asterisks indicate that the corresponding genes were distinctly up- or downregulated in different samples by t-test (*p < 0.05, **p < 0.01, ***p < 0.001). The citrus GAPDH gene was used as internal reference, three biological replicates were adopted, and the relative expression of CsHIPP genes was calculated by the 2−ΔΔCT method. NB: Newhell CLas infection, NH: Newhall CLas free, 1: young leaves, 2: mature leaves. ns, not significant.
Cis-acting elements distribution in the promoter region of CsHIPPs
Promoters are pivotal in regulating gene expression, whereas cis-acting elements in the promoter region served as binding sites for associated transcription factors (Hernandez-Garcia and Finer, 2014). To understand the dynamic expression patterns of CsHIPPs when citrus was infected with CLas and Xanthomonas citri ssp. citri (Xcc), the cis-acting elements distributed on the upstream genomic region of CsHIPPs were searched and those related with phytohormone, abiotic, and biotic stress responsiveness were reserved. As shown in Supplementary Figure 1, abscisic acid responsiveness, auxin responsiveness, defense and stress responsiveness, DRE, gibberellin responsiveness, low-temperature responsiveness, MeJA responsiveness, and salicylic acid responsiveness dispersedly distributed on the promoter region of CsHIPPs. Twenty-two CsHIPPs contained the abscisic acid responsiveness elements, and CsHIPP10, which encompassed six, had the most abscisic acid responsiveness elements. In addition, 22 CsHIPPs had the MeJA responsiveness elements. Moreover, 16 CsHIPPs contained the gibberellin responsiveness elements. Fourteen CsHIPPs contained the salicylic acid responsiveness elements. Twelve CsHIPPs contained the auxin responsiveness elements. Eight CsHIPPs contained the low-temperature responsiveness elements. Seven CsHIPPs contained the defense and stress responsiveness elements. It is worth noting that only CsHIPP03 and CsHIPP23 contained the DRE elements (Supplementary Figure 1). In short, CsHIPPs could response to various phytohormone, abiotic, and biotic stress while each CsHIPP had its unique cis-acting element array mode.
Promoter activity of CsHIPP03
It has been reported that HIPP26 from A. thaliana and N. benthamiana is highly expressed in plant vascular tissues (Barth et al., 2009; Cowan et al., 2018). Since the phloem is the specific colonization site for CLas, we were interested in that whether CsHIPP03, a target of CLas (Hu et al., 2023), is expressed in vascular tissues of citrus. To assess the promoter activity of CsHIPP03, a 2,287-bp sequence upstream of the gene was cloned into the vector pCAMBIA1380-GUS to construct PHIPP03-GUS. Additionally, a mutated sequence including 149-bp gaps and 11 mismatches was identified during sequencing, which was named PHIPPM-GUS. Both PHIPP03-GUS and PHIPPM-GUS constructs were transformed into A. thaliana and citrange hairy roots to investigate their activity. GUS staining of different developmental stages and tissues showed that both promoters were active in the leaves and roots of A. thaliana seedlings, with higher expression in leaf veins. Furthermore, they were significantly induced by wound treatment in mature A. thaliana plants (Figure 6A). In transgenic citrange, GUS staining revealed that blue staining was observed in the cortex, columnar sheath, and phloem of the hairy roots, but not in the xylem. This was different from the 35s promoter (Figure 6B).
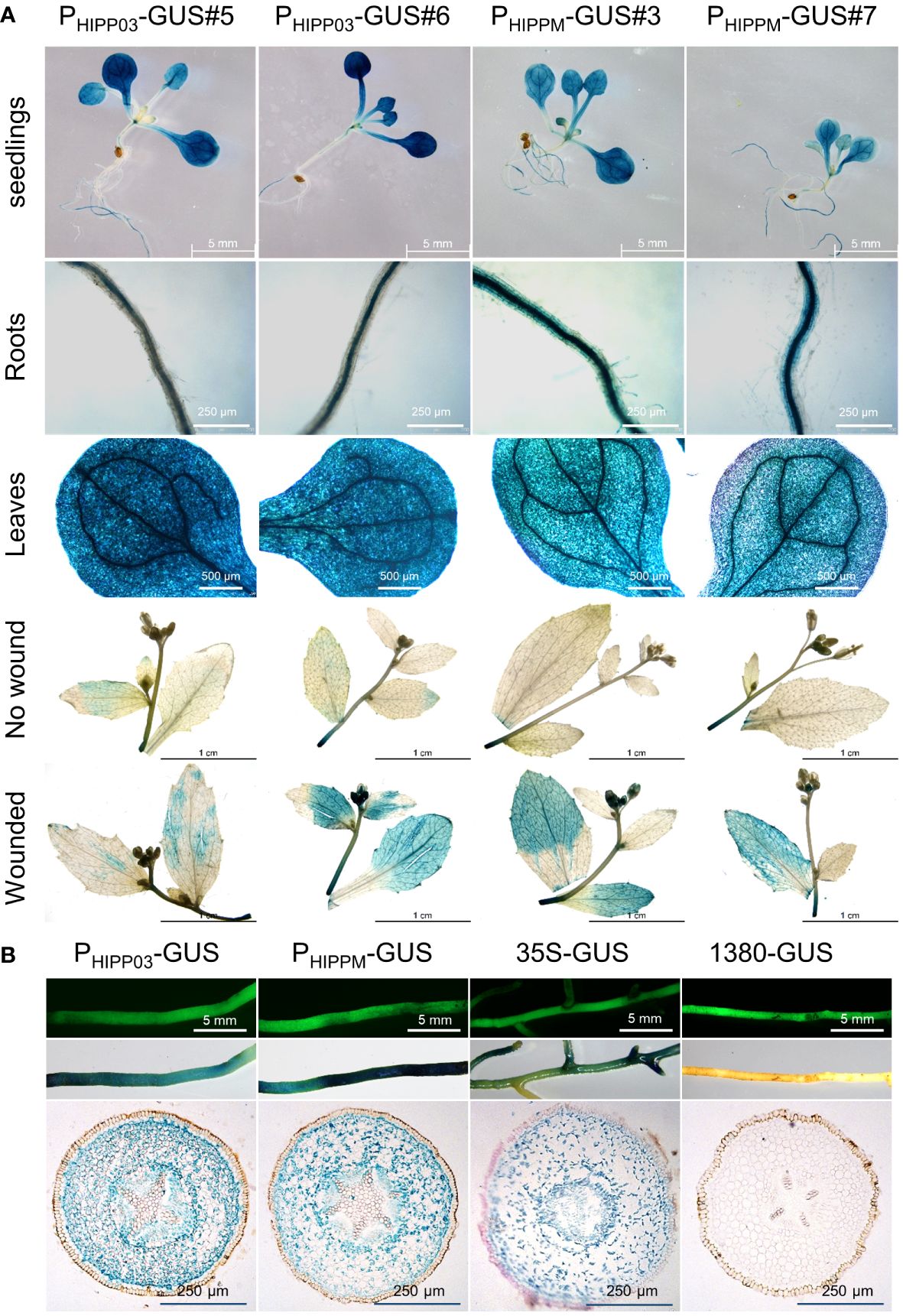
Figure 6 CsHIPP03 is induced by wound and expressed in different tissues. (A) GUS staining of seedlings and mature plants with or without wound of PHIPP03-GUS and PHIPPM-GUS transgenic A. thaliana. (B) GUS staining and GFP fluorescence observation of transgenic citrange hairy roots. GFP fluorescence indicates that the root has been successfully transformed. PHIPP03 represents the promoter that matches the reference genome sequence and PHIPPM-GUS represents a mutat with gaps and mismatches.
Subcellular localization analysis of CsHIPPs
To understand the subcellular localization of CsHIPPs, we selected eight CsHIPPs (namely, CsHIPP03, CsHIPP10, CsHIPP13, CsHIPP19, CsHIPP20, CsHIPP22, CsHIPP23, and CsHIPP26) for further analysis. The results showed that all CsHIPPs, with the exception of CsHIPP23, were found in the plasma membrane (PM), although they exhibited varying levels of fluorescence intensity (Figure 7). The fluorescence intensity of CsHIPP10, CsHIPP19, CsHIPP22, and CsHIPP23 was stronger in the nucleolus than others. CsHIPP23 showed many spot-like fluorescence signals in the nucleus. In addition, CsHIPP19 and CsHIPP22 are located in the nucleus and microtubule, and there are dotted signals at the junction of microtubules. Interestingly, we found that overexpression of CsHIPP26 would stimulate plant necrosis in N. benthamiana (Supplementary Figure 2).
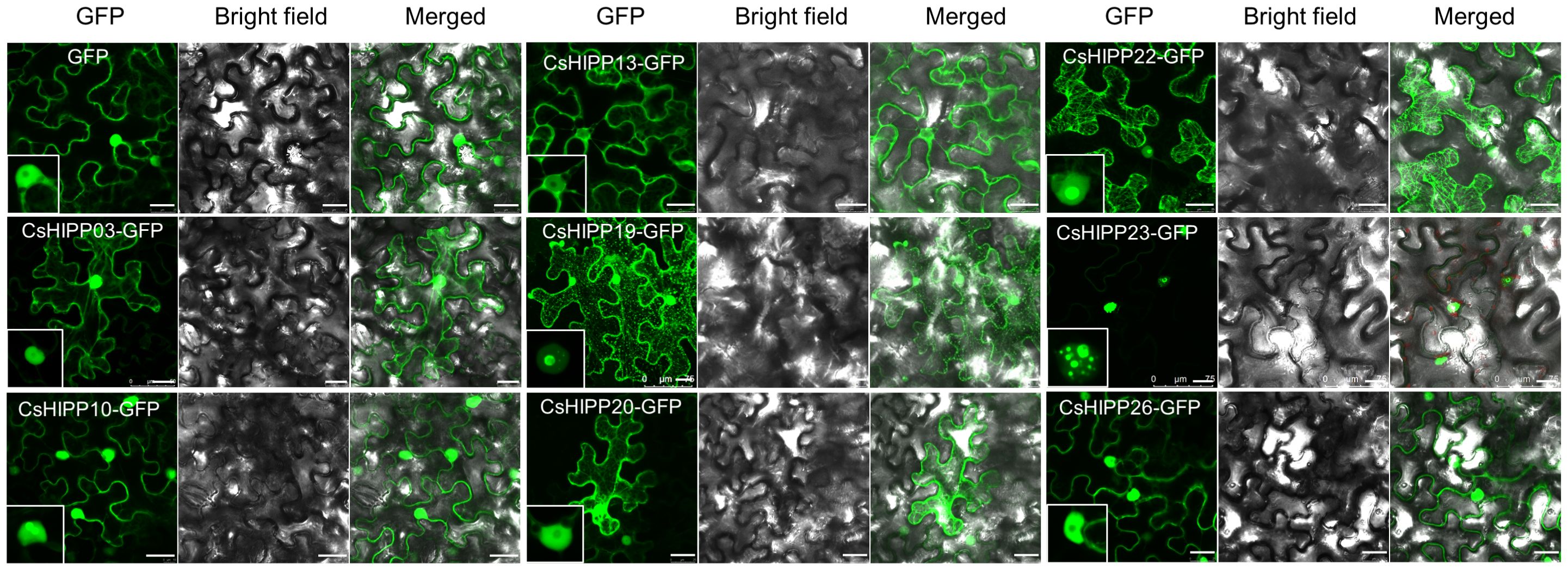
Figure 7 Subcellular localization of CsHIPPs in Nicotiana benthamiana. Subcellular localization of CsHIPP03-GFP, CsHIPP10-GFP, CsHIPP13-GFP, CsHIPP19-GFP, CsHIPP20-GFP, CsHIPP22-GFP, CsHIPP23-GFP, and CsHIPP26-GFP. GFP was used as a control. The insert images display the nucleus. Bar = 25 μm.
Silencing of NbHIPP3 enhances the N. benthamiana resistance to P. syringae
To verify whether CsHIPP03, the target of SDE34 (Hu et al., 2023), is a negative immune regulator, a tobacco rattle virus-based system was used to knock down the expression of NbHIPP3 in N. benthamiana. Through sequence alignment, we identified three homologous genes of CsHIPP03 in N. benthamiana, named NbHIPP3.1, NbHIPP3.2, and NbHIPP3.3 (Supplementary Figure 3). Y2H assay indicated that SDE34 interacted with NbHIPP3.2 and NbHIPP3.3 (Figure 8A). Subsequently, we designed a 300-bp silencing sequence for each homologous gene, and these fragments were cloned into TRV2 in an antisense orientation respectively, and we conducted VIGS experiments to determine the role of NbHIPP3s in the bacterial resistance of N. benthamiana. VIGS results showed that expression of the NbHIPP3s was reduced in inoculated plants compared with the control (TRV-GFP) (Figure 8B). Among them, the silencing efficiency of NbHIPP3.1 and NbHIPP3.2 is as high as 90% (Figures 8C, D), and NbHIPP3.3 also has a silencing efficiency of approximately 60% (Figure 8E). Next, the VIGS plants were inoculated with the P. syringae pv. tomato DC3000 mutant ΔhopQ1-1, which can cause disease in N. benthamiana (Wei et al., 2007). The symptoms and the level of bacterial colonization were determined 2 days later. The results showed that compared with the control group (TRV-GFP), silencing NbHIPP3s significantly reduced the bacterial colonization (Figure 8F). In conclusion, silencing homologous genes of CsHIPP03 in N. benthamiana enhanced the disease resistance.
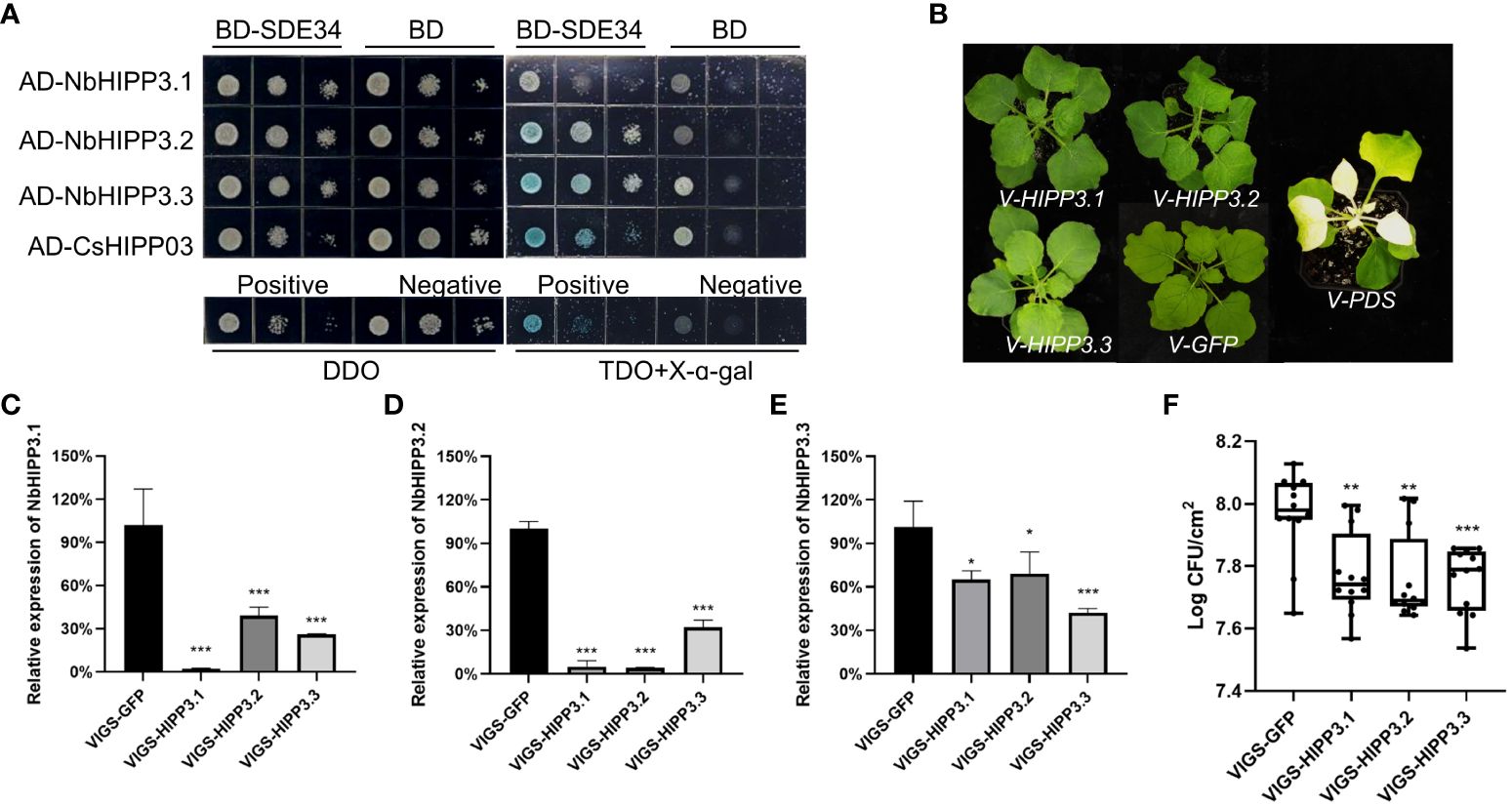
Figure 8 Silencing of NbHIPP3 genes inhibits the colonization of Pto DC3000ΔhopQ1-1 in Nicotiana benthamiana leaves. (A) SDE34 interacted with NbHIPP3.1, NbHIPP3.2, and NbHIPP3.3 by yeast two-hybrid. DDO indicates synthetic dropout medium (SD)/-Leu/-Trp and TDO indicates SD/-Leu/-Trp/-His. Positive control, pGBKT7-53 + pGADT7-T; Negative control, pGBKT7-Lam + pGADT7-T. (B) The growth phenotype of N. benthamiana after virus induced silencing of NbHIPP3.1, NbHIPP3.2, and NbHIPP3.3, with GFP and PDS as control. Relative expression of the (C) NbHIPP3.1, (D) NbHIPP3.2, and (E) NbHIPP3.3 after gene silencing by VIGS. There were six biological replicates for each experimental group, with TRV-GFP as a control. Total RNAs were extracted from the upper leaves 20 dpi and were used for RT-qPCR. The N. benthamiana actin gene was used as endogenous controls. Asterisks indicate significant downregulation of genes by t-test (*p < 0.05, **p < 0.01, ***p < 0.001). (F) The colonization of Pto DC3000ΔhopQ1-1 was lower in NbHIPP3.1, NbHIPP3.2, and NbHIPP3.3 gene-silenced N. benthamiana leaves. This experiment were repeated three times with consistent results. * denotes p < 0.05, ** denotes p < 0.01, and *** denotes p < 0.001.
Discussion
HIPPs are key proteins involved in widespread biological processes such as transport of metallic ions, cold and drought stresses, and plant–pathogen interactions (de Abreu-Neto et al., 2013). Genome-wide identification and characterization analysis of the HIPP gene family have been implemented in A. thaliana, O. sativa, P. trichocarpa, S. italica, S. moellendorffii, F. tataricum, T. aestivum, T. urartu, A. tauschii, T. dicoccoides, and H. vulgare (de Abreu-Neto et al., 2013; Zhang et al., 2020c; Ye et al., 2022). In the present study, we performed a systematic analysis of the HIPP gene family in C. sinensis. A total of 26 HIPP members were identified in the genome of C. sinensis, and the number is less than HIPPs of other plants like 45 for Arabidopsis, 59 for rice, 74 for poplar, and 51 for millet (de Abreu-Neto et al., 2013). The difference could be on account of gene duplication events during polyploidization or whole genome duplication. The 26 HIPPs members in C. sinensis were divided into five clusters when compared to the phylogenetic analysis of HIPPs in A. thaliana, which is consistent with the cluster analysis in O. sativa (Khan et al., 2019). The protein conserved domains are responsible for biological functions of proteins in cells. Two types of proteins with the HMA domain have been discovered in plants: heavy metal-associated plant proteins (HPPs) and HIPPs (Barth et al., 2009; Tehseen et al., 2010). The difference is that HIPPs contain an isoprenylation motif on the C-terminus end. All identified C. sinensis HIPPs have one or two HMA domains and an isoprenylation motif. Interestingly, Clade I HIPPs of C. sinensis contain two HMA domains except for CsHIPP19 while the other HIPPs contain only one HMA domain. Similarly, the same cluster of HIPPs have a similar distribution of motifs.
Even though HIPPs have a significant impact on the metabolic regulation of metallic ions, we are paying close attention to their roles in citrus–bacterial interactions. The public transcriptome data in the NCBI SRA database related with HLB- or Xcc-infected samples were used for the dynamic expression analysis of CsHIPPs (Chin et al., 2021; Tang et al., 2021). CsHIPP09 and CsHIPP16 were significantly induced when C. sinensis was infected by CLas for 18 weeks. Alternatively, CsHIPP01, CsHIPP04, CsHIPP11, CsHIPP21, CsHIPP24, and CsHIPP25 were significantly induced while CsHIPP03, CsHIPP10, and CsHIPP16 were downregulated at 46 weeks. As for Xcc-infected samples, CsHIPP21, CsHIPP19, and CsHIPP26 were significantly upregulated at 24 and 48 h. Conversely, CsHIPP03, CsHIPP04, CsHIPP14, CsHIPP15, CsHIPP24, and CsHIPP25 were downregulated at 24 and 48 h. CsHIPP03 were downregulated during the process of HLB and Xcc infection. In consideration of CsHIPP03 as a target of the effector from CLas, the function of CsHIPP03 in the interaction between citrus and CLas needs to be understood. VIGS assays were used to silence the CsGAPCs analogues in N. benthamiana, resulting in increased resistance to Phytophthora capsici infection. This finding supports the postulated role of CsGAPCs in citrus as a potential suppressor of immunity, suggesting a function that is conserved across disparate species (Shi et al., 2023b). Since the effector protein SDE34 of CLas can interact with CsHIPP03 (Hu et al., 2023), we speculate that CsHIPP03 plays an important role in the citrus–HLB interaction system. Furthermore, VIGS demonstrated that the knockdown of NbHIPP3 increased the resistance of plants to bacteria, suggesting that CsHIPP03 may be a susceptibility gene for HLB in citrus. However, further research is needed to confirm its biological function.
HIPP proteins were localized on the PM, including AtFP6 from Arabidopsis (Gao et al., 2009), NbHIPP26 from N. benthamiana (Cowan et al., 2018), OsHIPP29 from O. sativa (Zhang et al., 2020a), and HIPP1-V from H. villosa (Zhang et al., 2020c). The isoprenylation motif is an integral part of the HIPP protein structure, and any mutations in this motif may lead to changes in the protein’s localization. For instance, the AtIPT3 farnesylation directed its localization to the cytoplasm and nucleus, whereas the nonfarnesylated protein was found in the plastids (Galichet et al., 2008). Similarly, wild-type CdI19 was localized to the PM, whereas the farnesylation motif mutant was found in the entire cytoplasm (Suzuki et al., 2002). In our study, apart from CsHIPP23, all HIPP proteins from C. sinensis were localized on the PM. Notably, CsHIPP22 was also found to be localized on microtubules, like the co-localization result of TGB1 and NbHIPP26. However, NbHIPP26 is localized in the PM, nucleus, and motile vesicles (Cowan et al., 2018). In addition, during the cloning of CsHIPP22, a conserved sequence containing an intron was obtained, but the test results showed that the intron sequence did not affect its subcellular localization in N. benthamiana. Protein localization is related to function. The richness of the subcellular localization of citrus HIPP family indicates its functional diversity, but its specific function is still unknown. The HMA domain is known to be involved in heavy metal transport and maintenance of homeostasis, and the isoprenylation motif has a direct impact on the function of HIPPs. For instance, HIPP1-V, a positive regulator of powdery mildew resistance in H. villosa, was observed to be rapidly induced by Blumeria graminis f. sp. tritics (Bgt). Transiently or stably heterologous overexpression of HIPP1‐V in wheat was found to suppress the haustorium formation and enhance powdery mildew resistance, with isoprenylation being critical for PM localization, interaction with E3‐ligase CMPG1‐V, and function in powdery mildew resistance (Wang et al., 2023).
Conclusion
In this study, we identified 26 CsHIPPs in C. sinensis genome and performed a systematic analysis of the CsHIPP gene family, including phylogenetic relationship, chromosome location, conserved motifs and domain, cis-acting elements, and expression pattern analysis. RT-qPCR analysis showed that the expression levels of six among the eight CsHIPPs were increased, and only the expression level of CsHIPP03 gene was decreased. Further research has shown that silencing the expression of NbHIPP3, homologous genes of CsHIPP03, increased the resistance of N. benthamiana to P. syringae. The comprehensive analysis of the CsHIPPs at a genome-wide level will lay the groundwork for functional examinations of CsHIPPs. Furthermore, exploring the functions of HIPPs will aid in gaining insight into their biological roles.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author contributions
GH: Funding acquisition, Investigation, Methodology, Software, Writing – original draft, Writing – review & editing. YH: Investigation, Methodology, Software, Writing – original draft. FuL: Funding acquisition, Methodology, Writing – original draft. XZ: Investigation, Software, Writing – original draft. XW: Investigation, Software, Writing – original draft. FeL: Investigation, Software, Writing – original draft. RL: Conceptualization, Funding acquisition, Project administration, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the National Natural Science Foundation of China [grant numbers 32160621 and 32260659], the Innovation Program of short-term Leader of Jiangxi Province [grant number jxsq2020102129], and the Innovation Fund Designated for Graduate Students of Jiangxi Province [grant number YC2023-S850].
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2024.1369883/full#supplementary-material
References
Bailey, T. L., Johnson, J., Grant, C. E., Noble, W. S. (2015). The MEME suite. Nucleic Acids Res. 43, 39–49. doi: 10.1093/nar/gkv416
Barr, Z. K., Werner, T., Tilsner, J. (2023). Heavy metal-associated isoprenylated plant proteins (HIPPs) at plasmodesmata: exploring the link between localization and function. Plants 12, 3015. doi: 10.3390/plants12163015
Barth, O., Vogt, S., Uhlemann, R., Zschiesche, W., Humbeck, K. (2009). Stress induced and nuclear localized HIPP26 from Arabidopsis thaliana interacts via its heavy metal associated domain with the drought stress related zinc finger transcription factor ATHB29. Plant Mol. Biol. 69, 213–226. doi: 10.1007/s11103-008-9419-0
Bolger, A. M., Lohse, M., Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Bové, J. M. (2006). Huanglongbing: a destructive, newly-emerging, century-old disease of citrus. J. Plant Pathol. 88, 7–37. doi: 10.4454/jpp.v88i1.828
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Xia, R. (2020). TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chin, E. L., Ramsey, J., Saha, S., Mishchuk, D., Chavez, J., Howe, K., et al. (2021). Multi-omics Comparison Reveals Landscape of Citrus limon and Citrus sinensis Response to ‘Candidatus Liberibacter asiaticus’. PhytoFrontiers™ 1, 76–84. doi: 10.1094/PHYTOFR-09-20-0018-R
Clark, K., Franco, J. Y., Schwizer, S., Pang, Z., Hawara, E., Liebrand, T. W. H., et al. (2018). An effector from the Huanglongbing-associated pathogen targets citrus proteases. Nat. Commun. 9, 1718. doi: 10.1038/s41467-018-04140-9
Clough, S. J., Bent, A. F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. doi: 10.1046/j.1365-313x.1998.00343.x
Cowan, G. H., Roberts, A. G., Jones, S., Kumar, P., Kalyandurg, P. B., Gil, J. F., et al. (2018). Potato mop-top virus co-opts the stress sensor HIPP26 for long-distance movement. Plant Physiol. 176, 2052–2070. doi: 10.1104/pp.17.01698
da Graca, J. V., Douhan, G. W., Halbert, S. E., Keremane, M. L., Lee, R. F., Vidalakis, G., et al. (2016). Huanglongbing: An overview of a complex pathosystem ravaging the world's citrus. J. Integr. Plant Biol. 58, 373–387. doi: 10.1111/jipb.12437
de Abreu-Neto, J. B., Turchetto-Zolet, A. C., De Oliveira, L. F. V., Bodanese Zanettini, M. H., Margis-Pinheiro, M. (2013). Heavy metal-associated isoprenylated plant protein (HIPP): characterization of a family of proteins exclusive to plants. FEBS J. 280, 1604–1616. doi: 10.1111/febs.12159
Duan, Y., Zhou, L., Hall, D. G., Li, W., Doddapaneni, H., Lin, H., et al. (2009). Complete genome sequence of citrus Huanglongbing bacterium, ‘Candidatus liberibacter asiaticus’ Obtained through metagenomics. Mol. Plant-Microbe Interactions® 22, 1011–1020. doi: 10.1094/MPMI-22-8-1011
Dykema, P. E., Sipes, P. R., Marie, A., Biermann, B. J., Crowell, D. N., Randall, S. K. (1999). A new class of proteins capable of binding transition metals. Plant Mol. Biol. 41, 139–150. doi: 10.1023/A:1006367609556
Finn, R. D., Clements, J., Eddy, S. R. (2011). HMMER web server: interactive sequence similarity searching. Nucleic Acids Res. 39, W29–W37. doi: 10.1093/nar/gkr367
Fu, B., Yu, X., Tong, J., Pang, M., Zhou, Y., Liu, Q., et al. (2019). Comparative transcriptomic analysis of hypothalamus-pituitary-liver axis in bighead carp (Hypophthalmichthys nobilis) with differential growth rate. BMC Genomics 20, 328. doi: 10.1186/s12864-019-5691-4
Galichet, A., Hoyerova, K., Kaminek, M., Gruissem, W. (2008). Farnesylation directs AtIPT3 subcellular localization and modulates cytokinin biosynthesis in Arabidopsis. Plant Physiol. 146, 1155–1164. doi: 10.1104/pp.107.107425
Gao, W., Xiao, S., Li, H. Y., Tsao, S. W., Chye, M. L. (2009). Arabidopsis thaliana acyl-CoA-binding protein ACBP2 interacts with heavy-metal-binding farnesylated protein AtFP6. New Phytol. 181, 89–102. doi: 10.1111/j.1469-8137.2008.02631.x
Guo, T., Weber, H., Niemann, M. C. E., Theisl, L., Leonte, G., Novák, O., et al. (2021). Arabidopsis HIPP proteins regulate endoplasmic reticulum-associated degradation of CKX proteins and cytokinin responses. Mol. Plant 14, 1918–1934. doi: 10.1016/j.molp.2021.07.015
Hernandez-Garcia, C. M., Finer, J. J. (2014). Identification and validation of promoters and cis-acting regulatory elements. Plant Sci. 217-218, 109–119. doi: 10.1016/j.plantsci.2013.12.007
Hu, B., Rao, M. J., Deng, X., Pandey, S. S., Hendrich, C., Ding, F., et al. (2021). Molecular signatures between citrus and Candidatus Liberibacter asiaticus. PloS Pathog. 17, e1010071. doi: 10.1371/journal.ppat.1010071
Hu, Y., Zeng, L., Li, Z., Li, F., Huang, Y., Li, R., et al. (2023). Construction of yeast two-hybrid library and screening the host targets of the effector SDE34 from Candidatus Liberibacter asiaticus. J. Agric. Biotechnol. 31, 2589–2598. doi: 10.3969/j.issn.1674-7968.2023.12.013
Huang, G., Liu, Z., Gu, B., Zhao, H., Jia, J., Fan, G., et al. (2019). An RXLR effector secreted by Phytophthora parasitica is a virulence factor and triggers cell death in various plants. Mol. Plant Pathol. 20, 356–371. doi: 10.1111/mpp.12760
Katoh, K., Rozewicki, J., Yamada, K. D. (2019). MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief Bioinform. 20, 1160–1166. doi: 10.1093/bib/bbx108
Khan, I. U., Rono, J. K., Zhang, B. Q., Liu, X. S., Wang, M. Q., Wang, L. L., et al. (2019). Identification of novel rice (Oryza sativa) HPP and HIPP genes tolerant to heavy metal toxicity. Ecotoxicol. Environ. Saf. 175, 8–18. doi: 10.1016/j.ecoenv.2019.03.040
Lanfear, R., Frandsen, P. B., Wright, A. M., Senfeld, T., Calcott, B. (2017). PartitionFinder 2: new methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Mol. Biol. Evol. 34, 772–773. doi: 10.1093/molbev/msw260
Langner, T., Kamoun, S., Belhaj, K. (2018). CRISPR crops: plant genome editing toward disease resistance. Annu. Rev. Phytopathol. 56, 479–512. doi: 10.1146/annurev-phyto-080417-050158
Lescot, M., Déhais, P., Thijs, G., Marchal, K., Moreau, Y., Van De Peer, Y., et al. (2002). PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 30, 325–327. doi: 10.1093/nar/30.1.325
Li, T., Liu, B., Spalding, M. H., Weeks, D. P., Yang, B. (2012). High-efficiency TALEN-based gene editing produces disease-resistant rice. Nat. Biotechnol. 30, 390–392. doi: 10.1038/nbt.2199
Liu, L., Wang, Z., Li, J., Wang, Y., Yuan, J., Zhan, J., et al. (2021). Verticillium dahliae secreted protein Vd424Y is required for full virulence, targets the nucleus of plant cells, and induces cell death. Mol. Plant Pathol. 22, 1109–1120. doi: 10.1111/mpp.13100
Liu, H., Wang, X., Liu, S., Huang, Y., Guo, Y.-X., Xie, W.-Z., et al. (2022). Citrus Pan-Genome to Breeding Database (CPBD): A comprehensive genome database for citrus breeding. Mol. Plant 15, 1503–1505. doi: 10.1016/j.molp.2022.08.006
Livak, K. J., Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2– ΔΔCT method. methods 25, 402–408. doi: 10.1006/meth.2001.1262
Ma, H., Meng, X., Xu, K., Li, M., Gmitter, F. G., Jr., Liu, N., et al. (2022). Highly efficient hairy root genetic transformation and applications in citrus. Front. Plant Sci. 13, 1039094. doi: 10.3389/fpls.2022.1039094
Malnoy, M., Viola, R., Jung, M.-H., Koo, O.-J., Kim, S., Kim, J.-S., et al. (2016). DNA-free genetically edited grapevine and apple protoplast using CRISPR/Cas9 ribonucleoproteins. Front. Plant Sci. 7, 1904. doi: 10.3389/fpls.2016.01904
Marchler-Bauer, A., Bo, Y., Han, L., He, J., Lanczycki, C. J., Lu, S., et al. (2017). CDD/SPARCLE: functional classification of proteins via subfamily domain architectures. Nucleic Acids Res. 45, D200–d203. doi: 10.1093/nar/gkw1129
Maurer-Stroh, S., Eisenhaber, F. (2005). Refinement and prediction of protein prenylation motifs. Genome Biol. 6, R55. doi: 10.1186/gb-2005-6-6-r55
Nekrasov, V., Wang, C., Win, J., Lanz, C., Weigel, D., Kamoun, S. (2017). Rapid generation of a transgene-free powdery mildew resistant tomato by genome deletion. Sci. Rep. 7, 1–6. doi: 10.1038/s41598-017-00578-x
Nguyen, L. T., Schmidt, H. A., Von Haeseler, A., Minh, B. Q. (2015). IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274. doi: 10.1093/molbev/msu300
Pagliaccia, D., Shi, J., Pang, Z., Hawara, E., Clark, K., Thapa, S. P., et al. (2017). A pathogen secreted protein as a detection marker for citrus huanglongbing. Front. Microbiol. 8, 2041. doi: 10.3389/fmicb.2017.02041
Peng, A., Chen, S., Lei, T., Xu, L., He, Y., Wu, L., et al. (2017). Engineering canker-resistant plants through CRISPR/Cas9-targeted editing of the susceptibility gene CsLOB1 promoter in citrus. Plant Biotechnol. J. 15, 1509–1519. doi: 10.1111/pbi.12733
Pertea, M., Kim, D., Pertea, G. M., Leek, J. T., Salzberg, S. L. (2016). Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 11, 1650–1667. doi: 10.1038/nprot.2016.095
Pyott, D. E., Sheehan, E., Molnar, A. (2016). Engineering of CRISPR/Cas9-mediated potyvirus resistance in transgene-free Arabidopsis plants. Mol. Plant Pathol. 17, 1276–1288. doi: 10.1111/mpp.12417
Radakovic, Z., Anjam, M., Escobar, E., Chopra, D., Cabrera, J., Silva, A., et al. (2018). Arabidopsis HIPP27 is a host susceptibility gene for the beet cyst nematode Heterodera schachtii. Mol. Plant Pathol. 19, 1917–1298. doi: 10.1111/mpp.12668
Ramos, R. N., Martin, G. B., Pombo, M. A., Rosli, H. G. (2021). WRKY22 and WRKY25 transcription factors are positive regulators of defense responses in Nicotiana benthamiana. Plant Mol. Biol. 105, 65–82. doi: 10.1007/s11103-020-01069-w
Robinson, N. J., Winge, D. R. (2010). Copper metallochaperones. Annu. Rev. Biochem. 79, 537–562. doi: 10.1146/annurev-biochem-030409-143539
Shi, J., Gong, Y., Shi, H., Ma, X., Zhu, Y., Yang, F., et al. (2023b). ‘Candidatus Liberibacter asiaticus’ secretory protein SDE3 inhibits host autophagy to promote Huanglongbing disease in citrus. Autophagy 19, 2558–2574. doi: 10.1080/15548627.2023.2213040
Shi, H., Yang, Z., Huang, J., Wu, H., Fu, S., Li, W., et al. (2023a). An effector of ‘Candidatus Liberibacter asiaticus’ manipulates autophagy to promote bacterial infection. J. Exp. Bot. 74 (15), 4670–4684. doi: 10.1093/jxb/erad176
Suzuki, N., Yamaguchi, Y., Koizumi, N., Sano, H. (2002). Functional characterization of a heavy metal binding protein CdI19 from Arabidopsis. Plant J. 32, 165–173. doi: 10.1046/j.1365-313X.2002.01412.x
Tang, X., Wang, X., Huang, Y., Ma, L., Jiang, X., Rao, M. J., et al. (2021). Natural variations of TFIIAγ gene and LOB1 promoter contribute to citrus canker disease resistance in Atalantia buxifolia. PloS Genet. 17, e1009316. doi: 10.1371/journal.pgen.1009316
Tehseen, M., Cairns, N., Sherson, S., Cobbett, C. S. (2010). Metallochaperone-like genes in Arabidopsis thaliana. Metallomics 2, 556–564. doi: 10.1039/c003484c
Wang, N. (2019). The citrus huanglongbing crisis and potential solutions. Mol. Plant 12, 607–609. doi: 10.1016/j.molp.2019.03.008
Wang, Y., Tang, H., Debarry, J. D., Tan, X., Li, J., Wang, X., et al. (2012). MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 40, e49. doi: 10.1093/nar/gkr1293
Wang, Z., Zhang, H., Li, Y., Chen, Y., Tang, X., Zhao, J., et al. (2023). Isoprenylation modification is required for HIPP1-mediated powdery mildew resistance in wheat. Plant Cell Environ. 46, 288–305. doi: 10.1111/pce.14479
Wei, C. F., Kvitko, B. H., Shimizu, R., Crabill, E., Alfano, J. R., Lin, N. C., et al. (2007). A Pseudomonas syringae pv. tomato DC3000 mutant lacking the type III effector HopQ1-1 is able to cause disease in the model plant Nicotiana benthamiana. Plant J. 51, 32–46. doi: 10.1111/j.1365-313X.2007.03126.x
Xu, Z., Xu, X., Gong, Q., Li, Z., Li, Y., Wang, S., et al. (2019). Engineering broad-spectrum bacterial blight resistance by simultaneously disrupting variable TALE-binding elements of multiple susceptibility genes in rice. Mol. Plant 12, 1434–1446. doi: 10.1016/j.molp.2019.08.006
Ye, X., Liu, C., Yan, H., Wan, Y., Wu, Q., Wu, X., et al. (2022). Genome-wide identification and transcriptome analysis of the heavy metal-associated (HMA) gene family in Tartary buckwheat and their regulatory roles under cadmium stress. Gene 847, 146884. doi: 10.1016/j.gene.2022.146884
Zaidi, S. S., Mukhtar, M. S., Mansoor, S. (2018). Genome editing: targeting susceptibility genes for plant disease resistance. Trends Biotechnol. 36, 898–906. doi: 10.1016/j.tibtech.2018.04.005
Zhang, Y., Bai, Y., Wu, G., Zou, S., Chen, Y., Gao, C., et al. (2017). Simultaneous modification of three homoeologs of Ta EDR 1 by genome editing enhances powdery mildew resistance in wheat. Plant J. 91, 714–724. doi: 10.1111/tpj.13599
Zhang, C., Du, P., Yan, H., Zhu, Z., Wang, X., Li, W. (2020b). A Sec-dependent secretory protein of the Huanglongbing-associated pathogen suppresses hypersensitive cell death in Nicotiana benthamiana. Front. Microbiol. 11, 594669. doi: 10.3389/fmicb.2020.594669
Zhang, X., Feng, H., Feng, C., Xu, H., Huang, X., Wang, Q., et al. (2015). Isolation and characterisation of cDNA encoding a wheat heavy metal-associated isoprenylated protein involved in stress responses. Plant Biol. (Stuttg) 17, 1176–1186. doi: 10.1111/plb.12344
Zhang, B. Q., Liu, X. S., Feng, S. J., Zhao, Y. N., Wang, L. L., Rono, J. K., et al. (2020a). Developing a cadmium resistant rice genotype with OsHIPP29 locus for limiting cadmium accumulation in the paddy crop. Chemosphere 247, 125958. doi: 10.1016/j.chemosphere.2020.125958
Zhang, S., Wang, X., He, J., Zhang, S., Zhao, T., Fu, S., et al. (2023). A Sec-dependent effector, CLIBASIA_04425, contributes to virulence in 'Candidatus Liberibater asiaticus'. Front. Plant Sci. 14, 1224736. doi: 10.3389/fpls.2023.1224736
Zhang, H., Zhang, X., Liu, J., Niu, Y., Chen, Y., Hao, Y., et al. (2020c). Characterization of the heavy-metal-associated isoprenylated plant protein (HIPP) gene family from Triticeae species. Int. J. Mol. Sci. 21, 6191. doi: 10.3390/ijms21176191
Keywords: Citrus sinensis, heavy metal-associated isoprenylated plant proteins, phylogenetic analysis, susceptible gene, huanglongbing
Citation: Huang G, Hu Y, Li F, Zuo X, Wang X, Li F and Li R (2024) Genome-wide characterization of heavy metal-associated isoprenylated plant protein gene family from Citrus sinensis in response to huanglongbing. Front. Plant Sci. 15:1369883. doi: 10.3389/fpls.2024.1369883
Received: 13 January 2024; Accepted: 12 March 2024;
Published: 27 March 2024.
Edited by:
Jian Chen, Jiangsu University, ChinaReviewed by:
Yasser Nehela, University of Florida, United StatesGuilherme Souza Prado, Instituto Agronômico de Campinas (IAC), Brazil
Copyright © 2024 Huang, Hu, Li, Zuo, Wang, Li and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ruimin Li, bGlydWltaW5AZ25udS5lZHUuY24=
 Guiyan Huang
Guiyan Huang Yanan Hu
Yanan Hu Fuxuan Li1
Fuxuan Li1 Ruimin Li
Ruimin Li