- Department of Biology, University of Fribourg, Fribourg, Switzerland
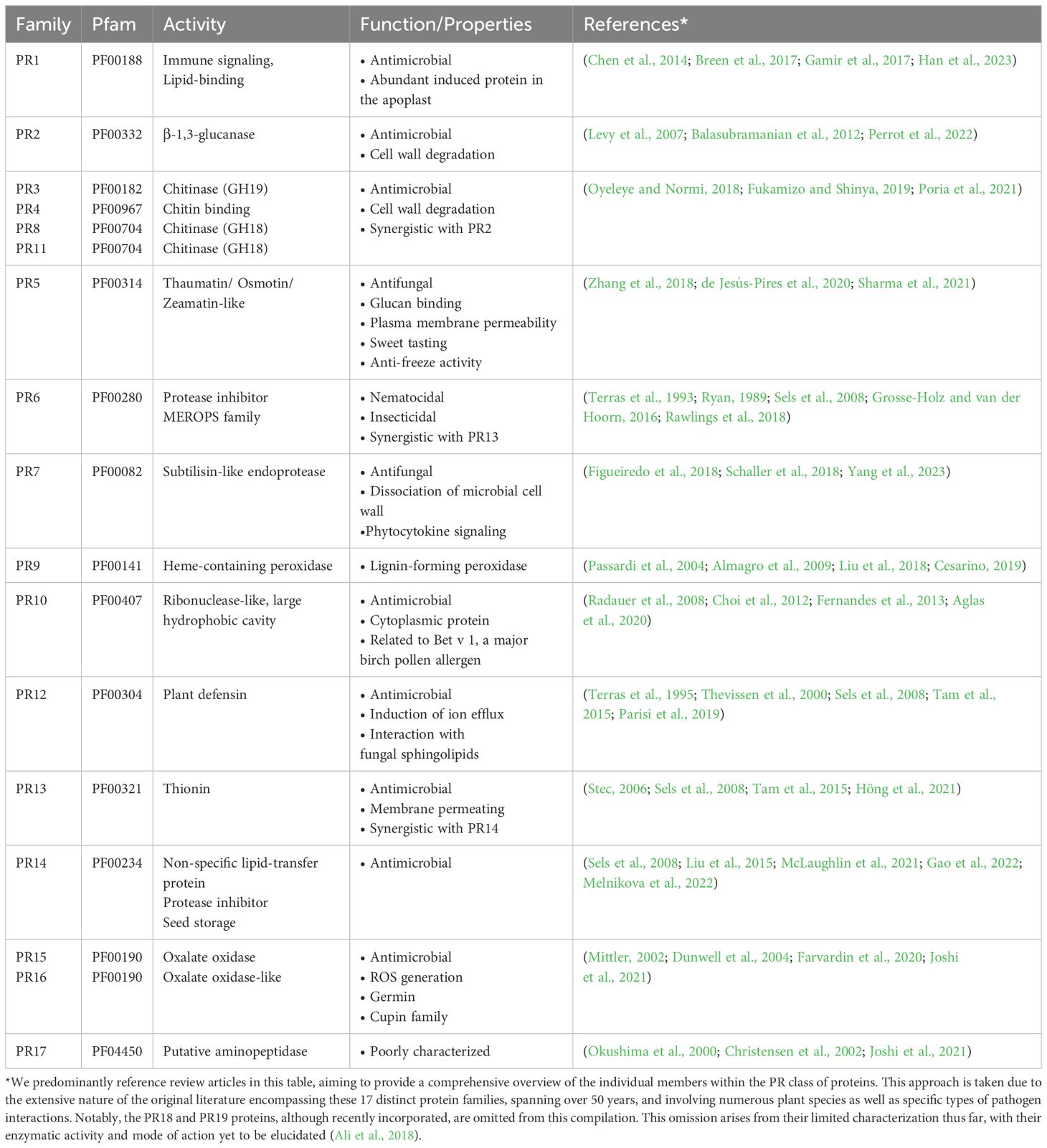
Plants respond to pathogen exposure by activating the expression of a group of defense-related proteins known as Pathogenesis-Related (PR) proteins, initially discovered in the 1970s. These PR proteins are categorized into 17 distinct families, denoted as PR1-PR17. Predominantly secreted, most of these proteins execute their defensive roles within the apoplastic space. Several PR proteins possess well-defined enzymatic functions, such as β-glucanase (PR2), chitinases (PR3, 4, 8, 11), proteinase (PR7), or RNase (PR10). Enhanced resistance against pathogens is observed upon PR protein overexpression, while their downregulation renders plants more susceptible to pathogen infections. Many of these proteins exhibit antimicrobial activity in vitro, and due to their compact size, some are classified as antimicrobial peptides. Recent research has unveiled that phytopathogens, including nematodes, fungi, and phytophthora, employ analogous proteins to bolster their virulence and suppress plant immunity. This raises a fundamental question: how can these conserved proteins act as antimicrobial agents when produced by the host plant but simultaneously suppress plant immunity when generated by the pathogen? In this hypothesis, we investigate PR proteins produced by pathogens, which we term “PR-like proteins,” and explore potential mechanisms by which this class of virulence factors operate. Preliminary data suggests that these proteins may form complexes with the host’s own PR proteins, thereby interfering with their defense-related functions. This analysis sheds light on the intriguing interplay between plant and pathogen-derived PR-like proteins, providing fresh insights into the intricate mechanisms governing plant-pathogen interactions.
Introduction
Plants are constantly challenged by various organisms, including fungi, oomycetes, bacteria, and viruses, which can compromise the plant’s fitness and survival (Teixeira et al., 2019). Plant pathogens affect forest plantations and most staple crops, decreasing productivity worldwide and severely compromising food security (Fones et al., 2020). The situation is expected to get worse, given the current rate of growth of the human population, the effect of climate change, the prevalence of monocultures, and the rise in pathogen resistance (Singh et al., 2023).
To combat the incursion of pathogens, plants have developed an intricate defense strategy comprising both inherent and inducible mechanisms (Jones and Dangl, 2006; Han, 2019). Constitutive defenses, operating as the foremost line of protection, encompass features like cutin, waxes, robust lignin deposition on cell walls, and the synthesis of antimicrobial small molecules, such as phytoanticipins (Li et al., 2020). The inducible defense mechanisms can be broadly categorized into two main types: Pathogen-associated molecular pattern (PAMP)-triggered immunity (PTI) and effector-triggered immunity (ETI). Furthermore, plants can develop systemic acquired resistance (SAR), a sophisticated response that fortifies defense throughout the plant following localized pathogen attack (Zhou and Zhang, 2020; Tanaka and Heil, 2021; Ngou et al., 2022).
Pathogenesis-related proteins
Amidst the spectrum of plant defense mechanisms, PR proteins stand as a prominent line of primary defense. These proteins are categorized into various families, denoted as PR1 to PR17 and beyond, based on their unique structural and functional characteristics (van Loon and van Kammen, 1970). Typically, PR proteins are induced in response to pathogen invasion and complement the action of small organic defense compounds that primarily serve to fend off herbivores but also exhibit antimicrobial activities (Westrick et al., 2021).
The discovery of PR proteins traces back to pioneering studies in the 1970s, where their robust induction in response to tobacco mosaic virus infection was first observed (Gianinazzi et al., 1970; van Loon and van Kammen, 1970). Subsequent research extended this finding to various plant species facing diverse pathogens, including oomycetes, fungi, bacteria, viruses, viroids, nematodes, and insect pests (van Loon et al., 1987; Stintzi et al., 1993; Van Loon and Van Strien, 1999; Edreva, 2005; van Loon et al., 2006; Jain and Khurana, 2018; Zribi et al., 2021). The transcripts encoding PR proteins show rapid accumulation following PTI and ETI, with their expression often regulated by the signaling molecule salicylic acid (SA). Notably, PR1 proteins are distinguished as crucial molecular markers for heightened plant defense due to the induction of SAR (Vlot et al., 2009).
PR proteins exhibit distinct biochemical properties, such as low molecular weight (ranging from 6 to 43 kDa), extractability and stability at low pH (below 3, a condition under which most other proteins denature), thermostability, and resistance to proteases (Van Loon and Van Strien, 1999). They are found throughout various plant organs, with leaves being particularly rich in these proteins, where they can constitute up to 5-10% of total leaf proteins. The PR1 family, for example, can comprise 1-2% of total leaf proteins (Van Loon and Van Strien, 1999). In plants, multiple genes usually represent each PR protein family, enabling the synthesis of diverse protein isoforms. For example, Arabidopsis thaliana has 22 genes encoding PR1 homologs, and rice contains 39 PR1-type genes (Mitsuhara et al., 2008). Some of these PR1 genes are constitutively expressed in roots or floral tissues, implying roles in plant development. This wide distribution of defense-related proteins across monocots and dicots underscores their multifaceted functions beyond defense (van Loon et al., 2006).
PR proteins can be categorized based on their isoelectric points, with acidic variants primarily induced upon immune activation and secreted to the apoplast. In contrast, those with a basic isoelectric point are often involved in developmental processes, showing limited induction upon pathogen infection, and typically localizing intracellularly, particularly in vacuoles (Farvardin et al., 2020; Zribi et al., 2021). Certain PR proteins also respond to various abiotic stressors like wounding, dehydration, salt, or cold stress, while others possess anti-freeze activity, reflecting their roles under adverse environmental conditions (Griffith and Yaish, 2004; Islam et al., 2023). Importantly, several PR proteins present in pollen, fruits, and vegetables can trigger allergic reactions in humans, making them significant contributors to plant allergens (Arora et al., 2020).
Over the past five decades, extensive research has been dedicated to characterizing individual PR proteins, elucidating their basic enzymatic activities, and establishing their direct role in defense against microbial pathogens (van Loon et al., 2006; Ferreira et al., 2007; Ali et al., 2018; Dos Santos and Franco, 2023) (see Table 1). For instance, PR1 proteins exhibit lipid-binding activity and inhibit the growth of sterol auxotrophic oomycetes (Gamir et al., 2017; Han et al., 2023). PR1 proteins also harbor a C-terminal peptide known as CAP-derived peptide 1 (CAPE1), which, when cleaved from the full-length PR1 protein, stimulates plant immune defense (Chen et al., 2014; Breen et al., 2017; Chen et al., 2023). PR2 proteins share sequence homology with β-1,3-glucanases and can hydrolyze β-1,3-glucans, which are present in the cell walls of microbes, generating oligomers that serve as elicitors. PR3, PR8, and PR11 exhibit chitinase activity, often synergizing with PR2, and PR4 binds chitin, a key component of fungal cell walls (Levy et al., 2007; Balasubramanian et al., 2012; Perrot et al., 2022). PR5 encompasses thaumatin-like proteins (TLPs) that exert antimicrobial activity by rapidly permeabilizing microbial plasma membranes (Zhang et al., 2018b; de Jesús-Pires et al., 2020; Sharma et al., 2021). PR6 encodes a protease inhibitor and shows synergy with thionins (PR13) (Ryan, 1989; Terras et al., 1993; Sels et al., 2008; Grosse-Holz and van der Hoorn, 2016; Rawlings et al., 2018). PR7 encodes a subtilisin-like endoprotease, believed to attack and degrade microbial cell wall proteins. However, these proteolytic enzymes are also important for peptide signaling, for example, by releasing serine rich endogenous peptides (SCOOPs) in Brassicacea, which are then perceived by the leucine-rich repeat receptor kinase male discovery 1-interacting receptor-like kinase 2 (MIK2) to elicit immunity (Yang et al., 2023). PR9 exhibits heme-dependent peroxidase activity, crucial for lignification, wound healing, and oxidative degradation of phenolic compounds (Passardi et al., 2004; Almagro et al., 2009; Liu et al., 2018; Cesarino, 2019). PR10 proteins are members of the major latex-like family and have been reported to possess ribonuclease activity, but this might be attributed to copurifying RNase contaminations (Fernandes et al., 2013; Aglas et al., 2020; Longsaward et al., 2023). PR10 has a hydrophobic cavity capable of binding various lipids, including steroids and fatty acids (Radauer et al., 2008). Intriguingly, PR10 members are localized in the cytoplasm, but secreted into the apoplastic space when complexed with and activated by leucine-rich repeat protein 1 (LRR1) (Choi et al., 2012).
PR12 comprises plant defensins, small proteins with antimicrobial activity but an uncharacterized mode of action (Terras et al., 1995; Thevissen et al., 2000; Sels et al., 2008; Tam et al., 2015; Parisi et al., 2019). PR6, PR12, PR13, and PR14, due to their low molecular weight and antimicrobial activity, are classified as antimicrobial peptides (Sels et al., 2008). PR13 belongs to the class of thionins, small, basic, and cysteine-rich peptides that, like PR12 peptides, cause the permeabilization of microbial cell membranes. PR13 exhibits synergistic antimicrobial activity with PR14 (Stec, 2006; Sels et al., 2008; Tam et al., 2015; Höng et al., 2021). PR14 proteins can transfer phospholipids between membranes in vitro and, due to their low substrate specificity, are known as non-specific lipid transfer proteins (ns-LTPs) (Sels et al., 2008; Liu et al., 2015; Gao et al., 2022; Melnikova et al., 2022). PR15 and PR16, oxalate oxidase and oxalate oxidase-like proteins, contribute to the generation of apoplastic reactive oxygen species (ROS), initiating signal transduction cascades and activating plant defense mechanisms (Mittler, 2002; Dunwell et al., 2004; Farvardin et al., 2020; Joshi et al., 2021). Lastly, PR17, the least understood class, is postulated to possess aminopeptidase activity (Okushima et al., 2000; Christensen et al., 2002; Joshi et al., 2021).
Numerous PR proteins display antimicrobial activity in vitro, and their overexpression in plants enhances resistance to various pathogens across diverse plant species (Alexander et al., 1993; Niderman et al., 1995; Epple et al., 1997; Anand et al., 2004; van Loon et al., 2006; Ferreira et al., 2007; Sels et al., 2008; Dos Santos and Franco, 2023). Conversely, silencing the expression of PR1 or PR5 renders plants more susceptible to pathogens (Riviere et al., 2008; Zhang et al., 2018b). Despite their antimicrobial activity, the precise functions of many PR proteins in defense responses remain incompletely understood, extending beyond direct pathogen inhibition to encompass roles in cell wall reinforcement, scavenging of ROS, and modulation of defense signaling pathways (van Loon et al., 2006; Islam et al., 2023). Given the protective effects conferred by the induction and accumulation of PR proteins, their overexpression, and heterologous expression are currently explored as strategies to establish stress-tolerant plants (Ali et al., 2018; Boccardo et al., 2019; Islam et al., 2023).
Pathogenesis-related-like proteins produced by pathogens
While PR proteins are typically produced by plants in response to pathogen infection as part of their defense mechanism, recent findings have unveiled a fascinating twist: pathogens themselves synthesize pathogenesis-related-like proteins, which we will refer to as PR-like proteins, that play crucial roles in promoting pathogen virulence (Han et al., 2023). Unlike the induction and secretion of antimicrobial proteins by the host plant upon pathogen attack, which are well studied, the precise function and contribution of PR-like proteins to pathogen virulence remain enigmatic.
Among PR-like proteins, the PR1-like family is perhaps the most extensively characterized. PR1 proteins belong to a large protein superfamily, also known as CAP proteins (CRISP/Ag5/PR1) or SCPs (sperm coating proteins) and are related to VALs/VAPs (venom allergen-like proteins made by nematodes) (Gibbs et al., 2008; Cantacessi and Gasser, 2012; Wilbers et al., 2018; Han et al., 2023). Recent research has unveiled PR1-like proteins from various pathogenic nematodes and fungi as novel virulence factors. For example, PR1-like proteins from hemibiotrophic Fusarium species, including Fpr1 from Fusarium oxysporum, FgPR1L-4 from Fusarium graminearum, as well as FvSCP1 from Fusarium verticillioides, have all been shown to enhance fungal virulence in their respective host plants (Prados-Rosales et al., 2012; Lu and Edwards, 2018; Zhang et al., 2018a). More recently, a family of three highly related PR1-like proteins was identified in the necrotrophic fungal pathogens Cytospora chrysosperma and Valsa mali, causal agents of canker disease in poplar and apple, respectively. Deletion of CcCAP1 in C. chrysosperma reduced fungal virulence and increased sensitivity to ROS, highlighting its importance (Han et al., 2021). Additionally, two of the three V. mali PR1-like proteins, VmPR1a and VmPR1c, were found to be essential for pathogen virulence (Wang et al., 2021). Recent host-induced gene silencing experiments further demonstrated that three out of six PR1-like proteins from the wheat stripe rust fungus Puccinia striiformis f. sp. tritici are necessary for fungal virulence (Zhao et al., 2023).
Furthermore, in susceptible tomato plants, GrVAP1 secreted by the potato cyst nematode (Globodera rostochiensis) is required for successful infection (Lozano-Torres et al., 2014). However, in other cultivars, GrVAP1 interacts with the tomato papain-like cysteine protease Rcr3, activating the membrane-localized immune receptor Cf-2, thereby inducing the host’s immune response (Lozano-Torres et al., 2012). Similarly, a PR1-like protein from Phytophthora sojae has been found to trigger an immune response in Nicotiana benthamiana, dependent on its recognition by the leucine-rich repeat receptor-like protein (LRR-RLP) RCAP1. This recognition involves the shared immune coreceptors BAK1 and SOBIR1 and leads to increased plant resistance against Phytophthora (Gust and Felix, 2014; Liebrand et al., 2014; Jiang et al., 2023). PsCAP1, the Phytophthora PR1-like protein, contains an N-terminal PAN domain and exhibits immune-stimulatory activities such as triggering ROS bursts, activating mitogen-activated protein kinase (MAPK), and inducing cell death. Importantly, these activities are mediated by the PAN domain, which is distinct from the CAP domain found in canonical PR1 proteins. The PAN domain has been proposed to facilitate protein-protein and protein-carbohydrate interactions, but its precise role in plant-microbe interactions remains a subject of study (Jiang et al., 2023). This PAN domain containing PsCAP1 protein is conserved among phytopathogenic oomycetes but absent in the genomes of plants, diatoms, bacteria, or fungi (Jiang et al., 2023).
Interestingly, heterologous expression of PR1-like proteins from pathogens, such as GrVAP1 from G. rostochiensis or CcCAP1 from C. chrysosperma, in host plants suppresses the plant’s PTI response. Expression of CcCAP1 in tobacco inhibits pathogen-induced induction of PR1 and PR4 and the expression of GrVAP1 selectively suppresses the activation of the programmed cell death by surface-localized immune receptors (Lozano-Torres et al., 2014; Han et al., 2021). These observations suggest that these proteins possess potent immune modulatory activity, rendering plants hypersensitive to various unrelated pathogens (Lozano-Torres et al., 2014; Han et al., 2021).
The corn smut Ustilago maydis UmPR1-like protein has recently been shown to sense plant-derived phenolic compounds to eliciting hyphal-like growth to guide fungal invasion in plants. In addition, secretion of UmPR1-like promotes fungal virulence by hijacking a plant cysteine protease to release a UmCAPE-like signaling peptide from UmPR1-like and suppress plant immunity (Lin et al., 2023).
These PR1-like proteins from pathogens appear to function in ways similar to plant hormones produced by pathogenic and symbiotic fungi. They may have dual roles: (i) perturbing plant processes, either positively or negatively, to promote invasion and nutrient uptake by the pathogens; and (ii) serving as signals for the fungi to engage in appropriate developmental and physiological processes adapted to their environment (Chanclud and Morel, 2016).
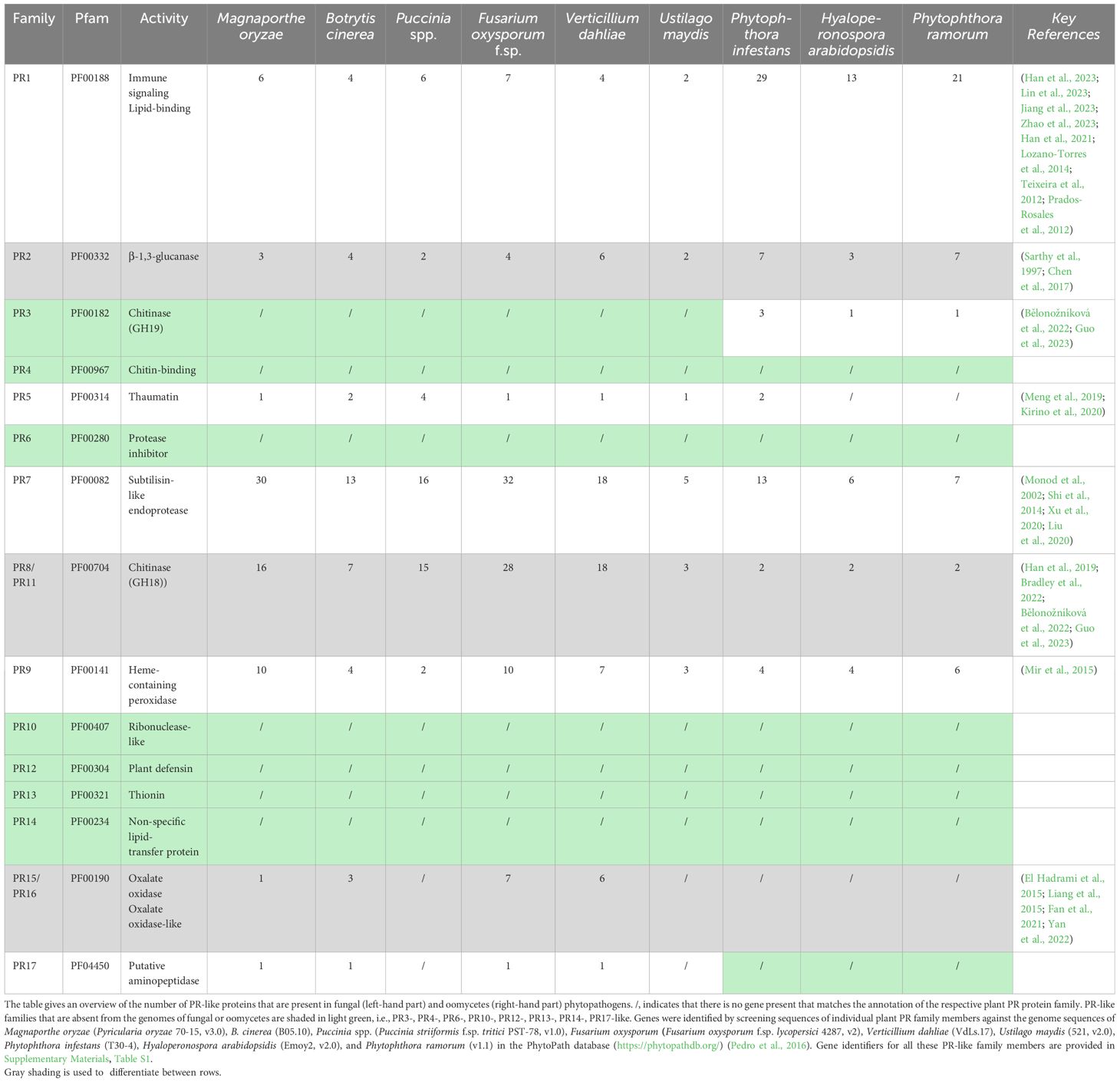
Building upon these insights into PR1-like proteins, we explored the genomes of various plant pathogens for the presence of other PR-like genes. Remarkably, we found that not only PR1-like genes are prevalent in phytopathogen genomes but that many other PR-like protein family members are also present. Except for PR4 (chitinase), PR6 (protease inhibitor), PR10 (ribonuclease-like), PR12 (plant defensin), PR13 (thionin), and PR14 (non-specific lipid transfer protein), multiple copies of genes encoding PR-like proteins are frequently identified in the genomes of model phytopathogens, particularly in those of fungi and oomycetes (Dean et al., 2012; Kamoun et al., 2015). Notable examples include the rice blast fungus Magnaporthe oryzae (Tan et al., 2023), the gray mold fungus Botrytis cinerea (Bi et al., 2023), the rust fungus Puccinia spp (Avasthi et al., 2023), the soil-borne ascomycete Fusarium oxysporum (Srinivas et al., 2019), the causative agent of wilt disease Verticillium dahliae (Klosterman et al., 2009), the corn smut fungus Ustilago maydis (Yu et al., 2023), as well as the oomycetes Phytophthora infestans (Whisson et al., 2016), which cause late blight disease on potato and tomato, the downy mildew causing Hyaloperonospora arabidopsidis (Coates and Beynon, 2010), and the sudden oak death disease causing Phytophthora ramorum (Grünwald et al., 2008) (see Table 2; Supplementary Materials Table S1). These intriguing observations suggests that the phenomena described for PR1-like proteins likely extend to other PR-like protein families as well. Consequently, some of the key questions that arise are: What functions do these PR-like proteins serve in phytopathogens? Do their deletions impact pathogen virulence? Can their heterologous expression in plants render them more susceptible to a broader spectrum of pathogens? What are the mechanisms of action employed by pathogen-produced PR-like proteins?
The role of PR-like proteins in pathogen vegetative growth, virulence, or their function as effectors modulating the host’s immune response are not well characterized, except for the cell wall remodeling enzymes including β-1,3-glucanases, such as Bgl2 in Candida albicans, and chitinases, which have established roles in filamentous growth, conidial germination, or haustorium establishment (Sarthy et al., 1997; Chen et al., 2017; Han et al., 2019; Guo et al., 2023). However, these cell wall remodeling enzymes may function primarily as morphogenetic factors rather than classical effectors, even though chitin can induce strong PTI, and its immune modulation involves processes such as shielding through lectin binding or deacetylation to chitosan (Gong et al., 2020). Interestingly, PR8 and PR11 chitinases belong the glycosyl hydrolase family 18 (GH18), a bacterial type of endochitinase, which are widely distributed in almost all organisms including plant pathogens (Bradley et al., 2022) (Table 2). PR3, on the other hand, belongs to the glycosyl hydrolase family 19 (GH19), which are mostly found in plants, and possess a specific chitin binding domain, which is absent in the bacterial type of enzyme (Henrissat and Bairoch, 1993). Members of this family are thought to be produced as part of a defense response against fungal pathogens. The overall structures and catalytic domains of these two classes of chitinase differ greatly. The GH19 family chitinases have an α-helix-rich lysozyme-like domain characterized by a deep cleft, whereas GH18 chitinases are characterized by a catalytic region that consists of a triosephosphate isomerase (TIM) (β/α)8-barrel domain (Oyeleye and Normi, 2018; Fukamizo and Shinya, 2019; Poria et al., 2021) (Figure 1). Interestingly, oomycetes express the GH19 plant type of chitinase as well, whereas most of the fungal pathogens do not (Klinter et al., 2019). This is particularly intriguing given that the cell wall of oomycetes is primarily composed of cellulose and β-glucans rather than chitin (Mélida et al., 2013; Wanke et al., 2021). These GH19 family chitinases in oomycetes have likely been acquired by horizontal gene transfer and been proposed to be important for the degradation of complex carbohydrates present in fungal cell walls during mycoparasitism (Liang et al., 2020; Bělonožníková et al., 2022).
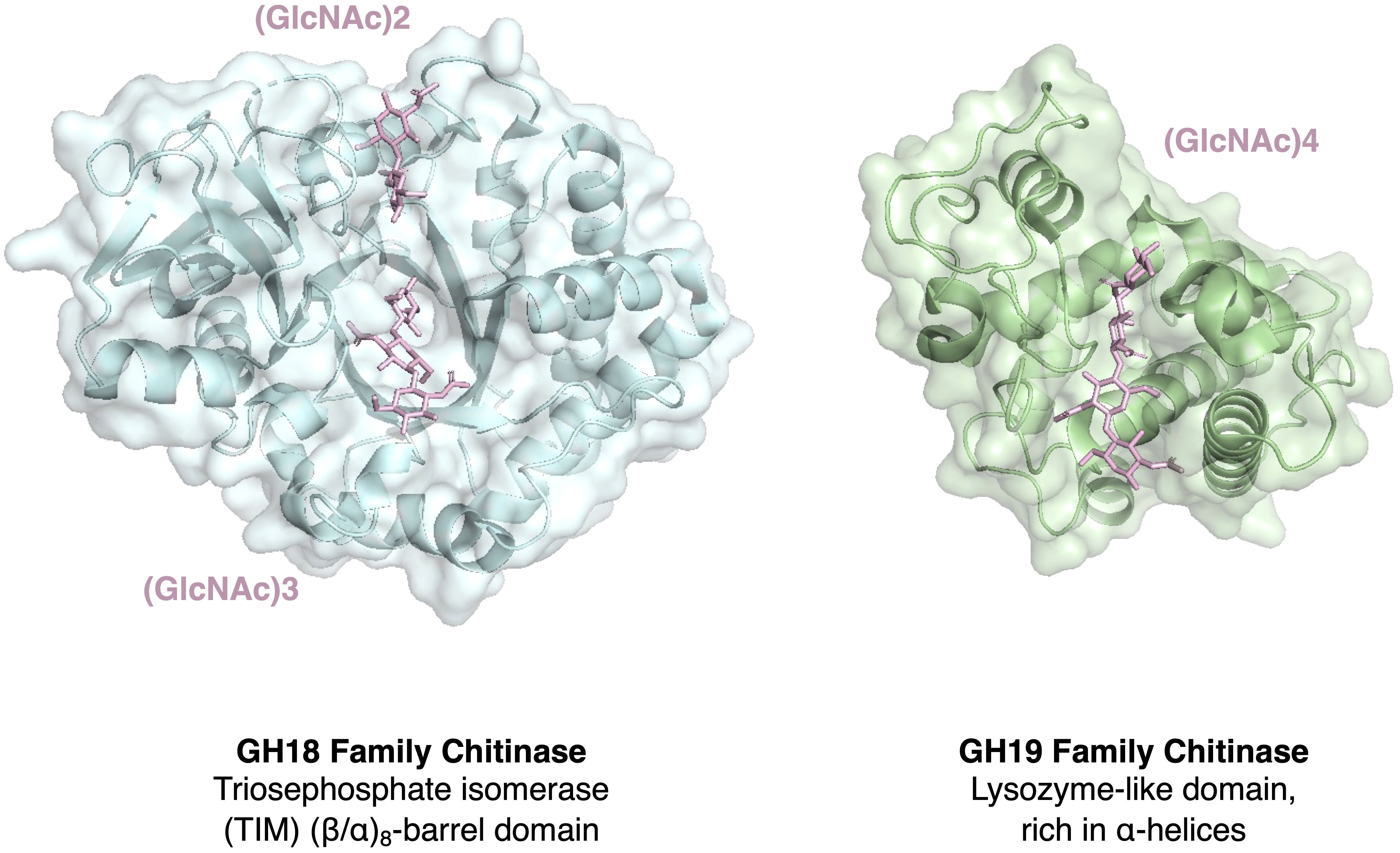
Figure 1 Comparison of the structure of chitinases belonging to two major families. Structural comparison of the ubiquitous chitinases belonging to the glycoside hydrolase family 18 (GH18) and the more plant-specific GH19 family. PR8 and PR11 are members of the bacterial/fungal GH18 family whereas PR3 is a member of the GH19 family and has structural similarity to some lysozymes (Monzingo et al., 1996). Shown are the structures of the triosephosphate isomerase (TIM) (β/α)8-barrel domain-containing GH18 family chitinase from Cycas revoluta in complex with the chitin dimer (GlcNAc)2 and chitin trimer (GlcNAc)3 (PDB 4MNK), and the α-helix-rich lysozyme-like GH19 family chitinase from Bryum coronatum in complex with the chitin tetramer (GlcNAc)4 (PDB 4IJ4; Ohnuma et al., 2014).
On the other hand, silencing of the PR5-like thaumatin-like protein from the pine wood nematode Bursaphelenchus xylophilus has been shown to reduce the pathogen’s reproduction and pathogenicity. When expressed in tobacco, it induces a robust cell death response (Meng et al., 2019; Kirino et al., 2020; Meng et al., 2022). Thaumatin-like proteins have been reported to bind β-1,3-glucans, exhibit endo-β-1,3-glucanase activity, inhibit α-amylase, or permeabilize cell membranes, yet their precise antimicrobial mechanisms remain ambiguous (Roberts and Selitrennikoff, 1990; Abada et al., 1996; Trudel et al., 1998; Grenier et al., 1999; Koiwa et al., 1999; Franco et al., 2002; Menu-Bouaouiche et al., 2003; de Jesús-Pires et al., 2020; Sharma et al., 2021). Thaumatin-like proteins are found in fungi, nematodes, and insects but are absent in vertebrates (Brandazza et al., 2004; Sakamoto et al., 2006; Belaish et al., 2008; Meng et al., 2019; de Jesús-Pires et al., 2020; Kirino et al., 2020).
Discussion
Several studies have highlighted the interactions between plant and fungal PR1 and PR1-like proteins, shedding light on their potential roles in modulating the host’s immune response. Notably, some of these proteins form homodimers, exemplified by the wheat protein TaPR1-5, Fpr1 from the soilborne fungal pathogen F. oxysporum, and S. cerevisiae Pry1 (Prados-Rosales et al., 2012; Lu et al., 2013; Darwiche et al., 2016). Furthermore, it has been shown that the dimeric form of TaPR1-5 is a specific target of ToxA, a host-selective virulence factor secreted by the causal agent of wheat tan spot disease, Pyrenophora tritici-repentis and the leaf/glume blotch fungus Stagonospora nodorum (Sn) (Lu et al., 2014). The binding of SnToxA to TaPR1-5 appears to compromise the immune-protective function of PR1 in wheat, thereby promoting necrosis (Ciuffetti et al., 2010).
Intriguingly, a second effector protein, SnTox3, secreted by S. nodorum, interacts with a broader range of wheat PR1 isoforms than SnToxA. SnTox3 effectively inhibits the release of CAPE1, thus suppressing the plant’s immune defense mechanisms (Breen et al., 2016; Sung et al., 2021). These findings suggest a multifaceted strategy by phytopathogens to subvert the host’s immune response, utilizing distinct effectors to target different components of the plant’s defense system.
Beyond the interactions with pathogenic effectors, PR1 has been shown to form heteromeric complexes with other PR proteins, particularly PR5 and PR14. The thaumatin-like PR5 is secreted into the apoplastic space and rapidly accumulates in response to various stressors, both biotic and abiotic (Hakim et al., 2018; Zhang et al., 2018b). Notably, wheat PR5 (TaTLP1) directly interacts with TaPR1, and the antimicrobial activity of the resulting heteromeric complex surpasses that of either PR5 or PR1-4 alone. This synergy suggests that these proteins act cooperatively to enhance the plant’s defense against invading pathogens (Wang et al., 2020, 2022).
On the other hand, PR14 belongs to the ns-LTP family. These extracellular ns-LTPs are known to bind to and transfer lipids between membranes in vitro. In vivo, they may serve as lipid sensors or sequester lipids to modulate their potential signaling functions (Missaoui et al., 2022). Wheat PR14 (TaLTP3) associates with TaPR1 in the apoplast, and plants overexpressing both proteins activate multiple signaling cascades, including the SA, jasmonic acid, and auxin pathways, and they exhibit enhanced production of ROS during the defense response. This interaction, together with the fact that purified PR14 exhibits antimicrobial activity in vitro, highlights the role of PR14 in reinforcing plant immunity (McLaughlin et al., 2021; Zhao et al., 2021).
These reported protein interactions suggest that the association between PR1-like proteins and their various PR family members may function to modulate plant-pathogen interactions. Given that PR1-like proteins from certain pathogens, such as the nematode G. rostochiensis (GrVAP1) or the fungal pathogen C. chrysosperma (CcCAP1), have been shown to reduce host immunity when expressed in plants (Lozano-Torres et al., 2014; Han et al., 2021), these PR1-like proteins may interfere with the immune-stimulating actions of endogenous PR1 proteins, similarly to the effectors SnToxA or SnTox3. This interference may occur through the disruption of protein complexes between plant’s own PR1 and PR5 and/or PR14 or by impeding the CAPE1-mediated signaling of PR1 (Breen et al., 2017; Han et al., 2023).
Consistent with the potential mode of action of PR-like proteins, in silico docking experiments suggest that PR1-like proteins from pathogens can indeed form protein complexes with plant endogenous PR5 and PR14, thus potentially undermining the host’s PR-based defense mechanisms (Figure 2). The prediction of protein complexes is emerging as a new powerful tool to identify potential microbial effectors. A recent bioinformatics screen using AlphaFold-Multimer, for example, has identified PR7, a subtilisin-like endoprotease PR7 (also known as P69 subtilase), as an effector hub targeted by different microbial kingdoms (Homma et al., 2023). This discovery lends further support to the notion that PR-like proteins may play pivotal roles in manipulating the plant’s immune response through the formation of cross-kingdom heteromeric protein complexes (Figure 3).
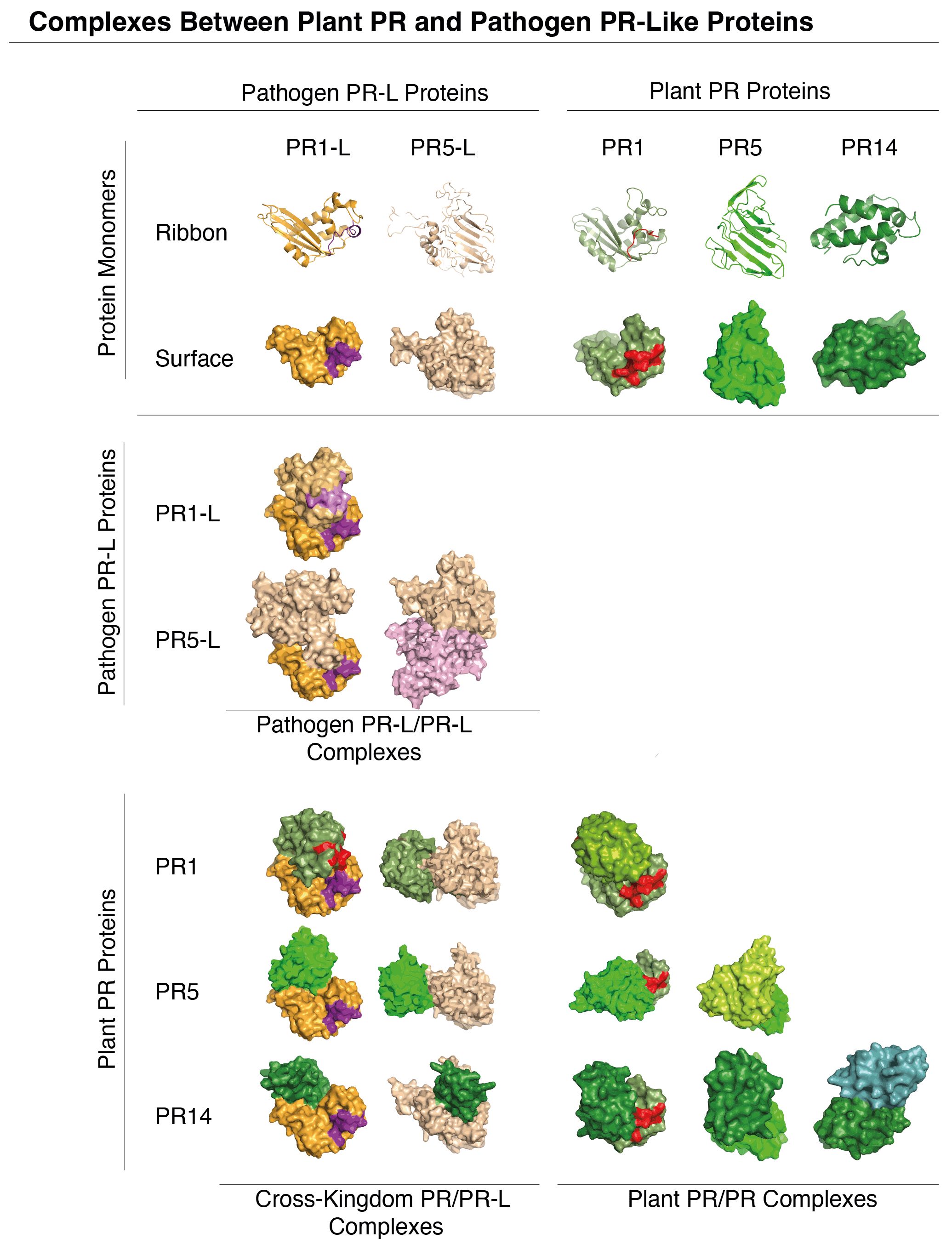
Figure 2 Molecular docking of phytopathogen PR1-like and PR5-like proteins with plant PR1, PR5 and PR14. Predicted molecular interactions between PR1-like and PR5-like proteins (orange) from phytopathogens and plant PR1, PR5, and PR14 (green) are visualized in the matrix. Notably, PR-like (PR-L) proteins from phytopathogens can interact both with themselves, and with host plant PR proteins. The docking simulations of PR1-like and PR5-like proteins from Botrytis cinerea with plant PR1, PR5, and PR14 (Arabidopsis thaliana) were performed using UCSF Chimera (Pettersen et al., 2004). The pathogen’s CAPE-like and the plant’s CAPE immune stimulatory peptides in the C-terminal end of PR1 and PR1-like are indicated in violet and red, respectively. Structures of monomeric proteins shown in the top two rows are represented both by ribbon diagrams and as space-filling models.
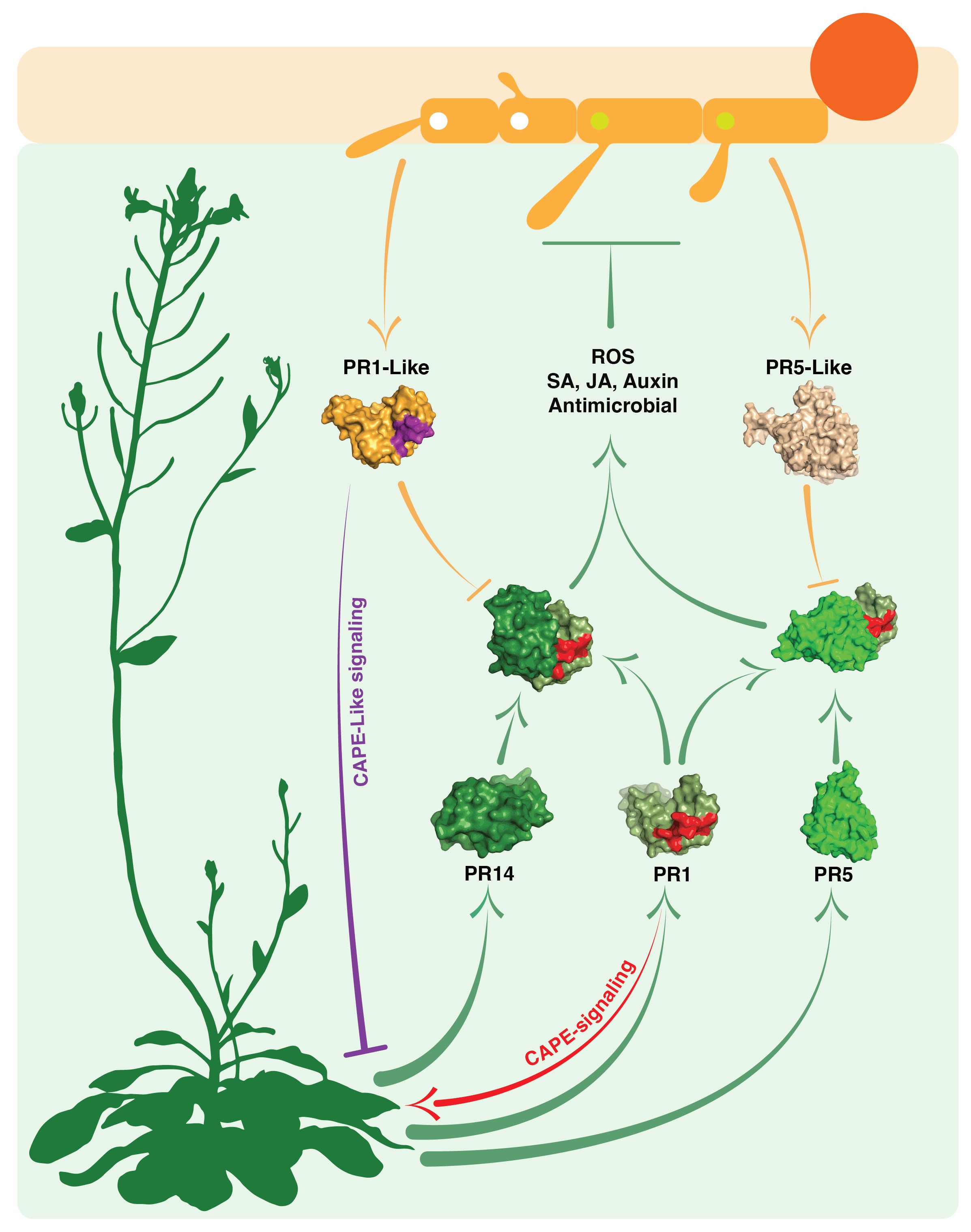
Figure 3 Interactions between plant defense-related PR proteins and phytopathogen PR-like proteins. This schematic diagram illustrates the complex interactions occurring in the apoplastic space of host plants (light green shaded space). It showcases the interplay between the host plant’s own PR1, PR5, and PR14 proteins (depicted by green arrows and green space-filling models) and the PR1-like and PR5-like proteins secreted by invading phytopathogens (depicted by orange arrows and orange space-filling models). The light green shading represents the host’s apoplastic space. The figure displays the structures of heteromeric complexes formed by plant PR1 with PR5 or PR14 proteins. The formation of these complexes contributes to immune stimulatory processes (involving SA, salicylic acid; JA, jasmonic acid; and auxin) as well as antimicrobial responses (including ROS production and direct antimicrobial activity). Importantly, the figure also suggests that these interactions can be disrupted by pathogen-derived PR-like proteins. Furthermore, it highlights CAPE immune stimulatory signaling by plant PR1 (indicated by the red arrow) and CAPE-like inhibitory signaling mediated by the pathogen’s PR1-like protein (indicated by the violet blunt arrow).
In conclusion, accumulating evidence suggests that phytopathogens have evolved strategies involving PR-like proteins to subdue their host’s immune response. These proteins appear to have coopted elements of the plant’s innate defense mechanisms, our hypothesis is that these proteins potentially interfere with the formation of protein complexes involving PR1, PR5, and PR14 within the apoplastic space and with key signaling pathways mediated by the CAPE peptide of PR1. While these findings offer valuable insights, it is important to emphasize that further experimental validation is necessary to establish the exact mechanisms underlying the interactions between PR and PR-like proteins and their impact on plant immunity.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Author contributions
ZH: Conceptualization, Data curation, Funding acquisition, Investigation, Validation, Visualization, Writing – original draft, Writing – review & editing. RS: Conceptualization, Funding acquisition, Project administration, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the Swiss National Science Foundation (SNF, 320030-227838, and 310030_207870).
Acknowledgments
We would like to thank Stéphanie Cottier for helpful comments on the manuscript, Jiri Stribny for expert assistance with protein docking, and Matteo Schneiter for expert support with figure illustrations.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2024.1368467/full#supplementary-material
References
Abada, L. R., D’Urzo, M. P., Liua, D., Narasimhan, M. L., Reuveni, M., Zhua, J. K., et al. (1996). Antifungal activity of tobacco osmotin has specificity and involves plasma membrane permeabilization. Plant Sci. 118, 11–23. doi: 10.1016/0168-9452(96)04420-2
Aglas, L., Soh, W. T., Kraiem, A., Wenger, M., Brandstetter, H., Ferreira, F. (2020). Ligand binding of PR-10 proteins with a particular focus on the Bet v 1 allergen family. Curr. Allergy Asthma Rep. 20, 25. doi: 10.1007/s11882-020-00918-4
Alexander, D., Goodman, R. M., Gut-Rella, M., Glascock, C., Weymann, K., Friedrich, L., et al. (1993). Increased tolerance to two oomycete pathogens in transgenic tobacco expressing pathogenesis-related protein 1a. Proc. Natl. Acad. Sci. U.S.A. 90, 7327–7331. doi: 10.1073/pnas.90.15.7327
Ali, S., Ganai, B. A., Kamili, A. N., Bhat, A. A., Mir, Z. A., Bhat, J. A., et al. (2018). Pathogenesis-related proteins and peptides as promising tools for engineering plants with multiple stress tolerance. Microbiol. Res. 212-213, 29–37. doi: 10.1016/j.micres.2018.04.008
Almagro, L., Gómez Ros, L. V., Belchi-Navarro, S., Bru, R., Ros Barceló, A., Pedreño, M. A. (2009). Class III peroxidases in plant defence reactions. J. Exp. Bot. 60, 377–390. doi: 10.1093/jxb/ern277
Anand, A., Lei, Z., Sumner, L. W., Mysore, K. S., Arakane, Y., Bockus, W. W., et al. (2004). Apoplastic extracts from a transgenic wheat line exhibiting lesion-mimic phenotype have multiple pathogenesis-related proteins that are antifungal. Mol. Plant Microbe Interact. 17, 1306–1317. doi: 10.1094/MPMI.2004.17.12.1306
Arora, R., Kumar, A., Singh, I. K., Singh, A. (2020). Pathogenesis related proteins: a defensin for plants but an allergen for humans. Int. J. Biol. Macromol 157, 659–672. doi: 10.1016/j.ijbiomac.2019.11.223
Avasthi, S., Gautam, A. K., Niranjan, M., Verma, R. K., Karunarathna, S. C., Kumar, A., et al. (2023). Insights into diversity, distribution, and systematics of rust genus puccinia. J. Fungi (Basel) 9, 639. doi: 10.3390/jof9060639
Balasubramanian, V., Vashisht, D., Cletus, J., Sakthivel, N. (2012). Plant β-1,3-glucanases: their biological functions and transgenic expression against phytopathogenic fungi. Biotechnol. Lett. 34, 1983–1990. doi: 10.1007/s10529-012-1012-6
Belaish, R., Sharon, H., Levdansky, E., Greenstein, S., Shadkchan, Y., Osherov, N. (2008). The Aspergillus nidulans cetA and calA genes are involved in conidial germination and cell wall morphogenesis. Fungal Genet. Biol. 45, 232–242. doi: 10.1016/j.fgb.2007.07.005
Bělonožníková, K., Hýsková, V., Chmelík, J., Kavan, D., Čeřovská, N., Ryšlavá, H. (2022). Pythium oligandrum in plant protection and growth promotion: Secretion of hydrolytic enzymes, elicitors and tryptamine as auxin precursor. Microbiol. Res. 258, 126976. doi: 10.1016/j.micres.2022.126976
Bi, K., Liang, Y., Mengiste, T., Sharon, A. (2023). Killing softly: a roadmap of Botrytis cinerea pathogenicity. Trends Plant Sci. 28, 211–222. doi: 10.1016/j.tplants.2022.08.024
Boccardo, N. A., Segretin, M. E., Hernandez, I., Mirkin, F. G., Chacón, O., Lopez, Y., et al. (2019). Expression of pathogenesis-related proteins in transplastomic tobacco plants confers resistance to filamentous pathogens under field trials. Sci. Rep. 9, 2791. doi: 10.1038/s41598-019-39568-6
Bradley, E. L., Ökmen, B., Doehlemann, G., Henrissat, B., Bradshaw, R. E., Mesarich, C. H. (2022). Secreted glycoside hydrolase proteins as effectors and invasion patterns of plant-associated fungi and oomycetes. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.853106
Brandazza, A., Angeli, S., Tegoni, M., Cambillau, C., Pelosi, P. (2004). Plant stress proteins of the thaumatin-like family discovered in animals. FEBS Lett. 572, 3–7. doi: 10.1016/j.febslet.2004.07.003
Breen, S., Williams, S. J., Outram, M., Kobe, B., Solomon, P. S. (2017). Emerging insights into the functions of pathogenesis-related protein 1. Trends Plant Sci. 22, 871–879. doi: 10.1016/j.tplants.2017.06.013
Breen, S., Williams, S. J., Winterberg, B., Kobe, B., Solomon, P. S. (2016). Wheat PR-1 proteins are targeted by necrotrophic pathogen effector proteins. Plant J. 88, 13–25. doi: 10.1111/tpj.13228
Cantacessi, C., Gasser, R. B. (2012). SCP/TAPS proteins in helminths–where to from now? Mol. Cell Probes 26, 54–59. doi: 10.1016/j.mcp.2011.10.001
Cesarino, I. (2019). Structural features and regulation of lignin deposited upon biotic and abiotic stresses. Curr. Opin. Biotechnol. 56, 209–214. doi: 10.1016/j.copbio.2018.12.012
Chanclud, E., Morel, J. B. (2016). Plant hormones: a fungal point of view. Mol. Plant Pathol. 17, 1289–1297. doi: 10.1111/mpp.12393
Chen, Y. L., Lee, C. Y., Cheng, K. T., Chang, W. H., Huang, R. N., Nam, H. G., et al. (2014). Quantitative peptidomics study reveals that a wound-induced peptide from PR-1 regulates immune signaling in tomato. Plant Cell 26, 4135–4148. doi: 10.1105/tpc.114.131185
Chen, Y. L., Lin, F. W., Cheng, K. T., Chang, C. H., Hung, S. C., Efferth, T., et al. (2023). XCP1 cleaves pathogenesis-related protein 1 into CAPE9 for systemic immunity in Arabidopsis. Nat. Commun. 14, 4697. doi: 10.1038/s41467-023-40406-7
Chen, X., Zhang, R., Takada, A., Iwatani, S., Oka, C., Kitamoto, T., et al. (2017). The role of Bgl2p in the transition to filamentous cells during biofilm formation by. Candida albicans. Mycoses 60, 96–103. doi: 10.1111/myc.12554
Choi, D. S., Hwang, I. S., Hwang, B. K. (2012). Requirement of the cytosolic interaction between PATHOGENESIS-RELATED PROTEIN10 and LEUCINE-RICH REPEAT PROTEIN1 for cell death and defense signaling in pepper. Plant Cell 24, 1675–1690. doi: 10.1105/tpc.112.095869
Christensen, A. B., Cho, B. H., Næsby, M., Gregersen, P. L., Brandt, J., Madriz-Ordeñana, K., et al. (2002). The molecular characterization of two barley proteins establishes the novel PR-17 family of pathogenesis-related proteins. Mol. Plant Pathol. 3, 135–144. doi: 10.1046/j.1364-3703.2002.00105.x
Ciuffetti, L. M., Manning, V. A., Pandelova, I., Betts, M. F., Martinez, J. P. (2010). Host-selective toxins, Ptr ToxA and Ptr ToxB, as necrotrophic effectors in the Pyrenophora tritici-repentis-wheat interaction. New Phytol. 187, 911–919. doi: 10.1111/j.1469-8137.2010.03362.x
Coates, M. E., Beynon, J. L. (2010). Hyaloperonospora Arabidopsidis as a pathogen model. Annu. Rev. Phytopathol. 48, 329–345. doi: 10.1146/annurev-phyto-080508-094422
Darwiche, R., Kelleher, A., Hudspeth, E. M., Schneiter, R., Asojo, O. A. (2016). Structural and functional characterization of the CAP domain of pathogen-related yeast 1 (Pry1) protein. Sci. Rep. 6, 28838. doi: 10.1038/srep28838
Dean, R., Van Kan, J. A., Pretorius, Z. A., Hammond-Kosack, K. E., Di Pietro, A., Spanu, P. D., et al. (2012). The Top 10 fungal pathogens in molecular plant pathology. Mol. Plant Pathol. 13, 414–430. doi: 10.1111/j.1364-3703.2011.00783.x
de Jesús-Pires, C., Ferreira-Neto, J. R. C., Pacifico Bezerra-Neto, J., Kido, E. A., de Oliveira Silva, R. L., Pandolfi, V., et al. (2020). Plant thaumatin-like proteins: Function, evolution and biotechnological applications. Curr. Protein Pept. Sci. 21, 36–51. doi: 10.2174/1389203720666190318164905
Dos Santos, C., Franco, O. L. (2023). Pathogenesis-related proteins (PRs) with enzyme activity activating plant defense responses. Plants (Basel) 12, 2226. doi: 10.3390/plants12112226
Dunwell, J. M., Purvis, A., Khuri, S. (2004). Cupins: the most functionally diverse protein superfamily. Phytochemistry 65, 7–17. doi: 10.1016/j.phytochem.2003.08.016
Edreva, A. (2005). Pathogenesis-related proteins: research progress in the last 15 years. Gen. Appl. Plant Physiol. 31, 105–124.
El Hadrami, A., Islam, M. R., Adam, L. R., Daayf, F. (2015). A cupin domain-containing protein with a quercetinase activity (VdQase) regulates Verticillium dahliae’s pathogenicity and contributes to counteracting host defenses. Front. Plant Sci. 6. doi: 10.3389/fpls.2015.00440
Epple, P., Apel, K., Bohlmann, H. (1997). Overexpression of an endogenous thionin enhances resistance of Arabidopsis against. Fusarium oxysporum. Plant Cell 9, 509–520. doi: 10.1105/tpc.9.4.509
Fan, H., Yang, W., Nie, J., Lin, C., Wu, J., Wu, D., et al. (2021). Characterization of a secretory YML079-like cupin protein that contributes to sclerotinia sclerotiorum pathogenicity. Microorganisms 9, 2519. doi: 10.3390/microorganisms9122519
Farvardin, A., González-Hernández, A. I., Llorens, E., García-Agustín, P., Scalschi, L., Vicedo, B. (2020). The apoplast: a key player in plant survival. Antioxidants (Basel) 9, 604. doi: 10.3390/antiox9070604
Fernandes, H., Michalska, K., Sikorski, M., Jaskolski, M. (2013). Structural and functional aspects of PR-10 proteins. FEBS J. 280, 1169–1199. doi: 10.1111/febs.12114
Ferreira, R. B., Monteiro, S., Freitas, R., Santos, C. N., Chen, Z., Batista, L. M., et al. (2007). The role of plant defence proteins in fungal pathogenesis. Mol. Plant Pathol. 8, 677–700. doi: 10.1111/j.1364-3703.2007.00419.x
Figueiredo, J., Sousa Silva, M., Figueiredo, A. (2018). Subtilisin-like proteases in plant defence: the past, the present and beyond. Mol. Plant Pathol. 19, 1017–1028. doi: 10.1111/mpp.12567
Fones, H. N., Bebber, D. P., Chaloner, T. M., Kay, W. T., Steinberg, G., Gurr, S. J. (2020). Threats to global food security from emerging fungal and oomycete crop pathogens. Nat. Food 1, 332–342. doi: 10.1038/s43016-020-0075-0
Franco, O. L., Rigden, D. J., Melo, F. R., Grossi-De-Sá, M. F. (2002). Plant alpha-amylase inhibitors and their interaction with insect alpha-amylases. Eur. J. Biochem. 269, 397–412. doi: 10.1046/j.0014-2956.2001.02656.x
Fukamizo, T., Shinya, S. (2019). Chitin/chitosan-active enzymes involved in plant-microbe interactions. Adv. Exp. Med. Biol. 1142, 253–272. doi: 10.1007/978-981-13-7318-3_12
Gamir, J., Darwiche, R., Van’t Hof, P., Choudhary, V., Stumpe, M., Schneiter, R., et al. (2017). The sterol-binding activity of PATHOGENESIS-RELATED PROTEIN 1 reveals the mode of action of an antimicrobial protein. Plant J. 89, 502–509. doi: 10.1111/tpj.13398
Gao, H., Ma, K., Ji, G., Pan, L., Zhou, Q. (2022). Lipid transfer proteins involved in plant-pathogen interactions and their molecular mechanisms. Mol. Plant Pathol. 23, 1815–1829. doi: 10.1111/mpp.13264
Gianinazzi, S., Martin, C., Vallee, J. C. (1970). Hypersensitivity to viruses, temperature and soluble proteins in Nicotiana xanthi n.c. appearance of new macromolecules at the repression of viral synthesis. C R Acad. Sci. Hebd Seances Acad. Sci. D 270, 2383–2386.
Gibbs, G. M., Roelants, K., O’Bryan, M. K. (2008). The CAP superfamily: cysteine-rich secretory proteins, antigen 5, and pathogenesis-related 1 proteins–roles in reproduction, cancer, and immune defense. Endocr. Rev. 29, 865–897. doi: 10.1210/er.2008-0032
Gong, B. Q., Wang, F. Z., Li, J. F. (2020). Hide-and-seek: Chitin-triggered plant immunity and fungal counterstrategies. Trends Plant Sci. 25, 805–816. doi: 10.1016/j.tplants.2020.03.006
Grenier, J., Potvin, C., Trudel, J., Asselin, A. (1999). Some thaumatin-like proteins hydrolyse polymeric beta-1,3-glucans. Plant J. 19, 473–480. doi: 10.1046/j.1365-313x.1999.00551.x
Griffith, M., Yaish, M. W. (2004). Antifreeze proteins in overwintering plants: a tale of two activities. Trends Plant Sci. 9, 399–405. doi: 10.1016/j.tplants.2004.06.007
Grosse-Holz, F. M., van der Hoorn, R. A. (2016). Juggling jobs: roles and mechanisms of multifunctional protease inhibitors in plants. New Phytol. 210, 794–807. doi: 10.1111/nph.13839
Grünwald, N. J., Goss, E. M., Press, C. M. (2008). Phytophthora ramorum: a pathogen with a remarkably wide host range causing sudden oak death on oaks and ramorum blight on woody ornamentals. Mol. Plant Pathol. 9, 729–740. doi: 10.1111/j.1364-3703.2008.00500.x
Guo, J., Mou, Y., Li, Y., Yang, Q., Wang, X., Lin, H., et al. (2023). Silencing a chitinase gene, PstChia1, reduces virulence of Puccinia striiformis f. sp. tritici. Int. J. Mol. Sci. 24, 8215. doi: 10.3390/ijms24098215
Gust, A. A., Felix, G. (2014). Receptor like proteins associate with SOBIR1-type of adaptors to form bimolecular receptor kinases. Curr. Opin. Plant Biol. 21, 104–111. doi: 10.1016/j.pbi.2014.07.007
Hakim, U. A., Hussain, A., Shaban, M., Khan, A. H., Alariqi, M., Gul, S., et al. (2018). Osmotin: a plant defense tool against biotic and abiotic stresses. Plant Physiol. Biochem. 123, 149–159. doi: 10.1016/j.plaphy.2017.12.012
Han, G. Z. (2019). Origin and evolution of the plant immune system. New Phytol. 222, 70–83. doi: 10.1111/nph.15596
Han, Y., Song, L., Peng, C., Liu, X., Liu, L., Zhang, Y., et al. (2019). A Magnaporthe chitinase interacts with a rice jacalin-related lectin to promote host colonization. Plant Physiol. 179, 1416–1430. doi: 10.1104/pp.18.01594
Han, Z., Xiong, D., Schneiter, R., Tian, C. (2023). The function of plant PR1 and other members of the CAP protein superfamily in plant-pathogen interactions. Mol. Plant Pathol. 24, 651–668. doi: 10.1111/mpp.13320
Han, Z., Xiong, D., Xu, Z., Liu, T., Tian, C. (2021). The Cytospora chrysosperma virulence effector CcCAP1 mainly localizes to the plant nucleus to suppress plant immune responses. mSphere 6, e00883–e00820. doi: 10.1128/msphere.00883-20
Henrissat, B., Bairoch, A. (1993). New families in the classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem. J. 293, 781–788. doi: 10.1042/bj2930781
Homma, F., Huang, J., van der Hoorn, R. A. L. (2023). AlphaFold-Multimer predicts cross-kingdom interactions at the plant-pathogen interface. Nat. Commun. 14, 1–13. doi: 10.1038/s41467-023-41721-9
Höng, K., Austerlitz, T., Bohlmann, T., Bohlmann, H. (2021). The thionin family of antimicrobial peptides. PloS One 16, e0254549. doi: 10.1371/journal.pone.0254549
Islam, M. M., El-Sappah, A. H., Ali, H. M., Zandi, P., Huang, Q., Soaud, S. A., et al. (2023). Pathogenesis-related proteins (PRs) countering environmental stress in plants: a review. South Afr. J. Bot. 160, 414–427. doi: 10.1016/j.sajb.2023.07.003
Jain, D., Khurana, J. P. (2018). “Role of pathogenesis-Related (PR) proteins in plant microbes defence mechanism,” in Molecular aspects of plant-pathogen interactions. Eds. Singh, A., Singh, I. K. (Springer Nature, Singapore), 265–281.
Jiang, H., Xia, Y., Zhang, S., Zhang, Z., Feng, H., Zhang, Q., et al. (2023). The CAP superfamily protein PsCAP1 secreted by Phytophthora triggers immune responses in Nicotiana benthamiana through a leucine-rich repeat receptor-like protein. New Phytol. 240, 784–801. doi: 10.1111/nph.19194
Jones, J. D., Dangl, J. L. (2006). The plant immune system. Nature 444, 323–329. doi: 10.1038/nature05286
Joshi, V., Joshi, N., Vyas, A., Jadhav, S. K. (2021). Pathogenesis-related proteins: Role in plant defense. Biocontrol Agents Secondary Metabolites, 573–590. doi: 10.1016/B978-0-12-822919-4.00025-9
Kamoun, S., Furzer, O., Jones, J. D., Judelson, H. S., Ali, G. S., Dalio, R. J., et al. (2015). The Top 10 oomycete pathogens in molecular plant pathology. Mol. Plant Pathol. 16, 413–434. doi: 10.1111/mpp.12190
Kirino, H., Yoshimoto, K., Shinya, R. (2020). Thaumatin-like proteins and a cysteine protease inhibitor secreted by the pine wood nematode Bursaphelenchus xylophilus induce cell death in Nicotiana benthamiana. PloS One 15, e0241613. doi: 10.1371/journal.pone.0241613
Klinter, S., Bulone, V., Arvestad, L. (2019). Diversity and evolution of chitin synthases in oomycetes (Straminipila: Oomycota). Mol. Phylogenet Evol. 139, 106558. doi: 10.1016/j.ympev.2019.106558
Klosterman, S. J., Atallah, Z. K., Vallad, G. E., Subbarao, K. V. (2009). Diversity, pathogenicity, and management of verticillium species. Annu. Rev. Phytopathol. 47, 39–62. doi: 10.1146/annurev-phyto-080508-081748
Koiwa, H., Kato, H., Nakatsu, T., Oda, J., Yamada, Y., Sato, F. (1999). Crystal structure of tobacco PR-5d protein at 1.8 A resolution reveals a conserved acidic cleft structure in antifungal thaumatin-like proteins. J. Mol. Biol. 286, 1137–1145. doi: 10.1006/jmbi.1998.2540
Levy, A., Guenoune-Gelbart, D., Epel, B. L. (2007). Beta-1,3-Glucanases: Plasmodesmal gate keepers for intercellular communication. Plant Signal Behav. 2, 404–407. doi: 10.4161/psb.2.5.4334
Li, P., Lu, Y. J., Chen, H., Day, B. (2020). The lifecycle of the plant immune system. CRC Crit. Rev. Plant Sci. 39, 72–100. doi: 10.1080/07352689.2020.1757829
Liang, D., Andersen, C. B., Vetukuri, R. R., Dou, D., Grenville-Briggs, L. J. (2020). Horizontal Gene Transfer and Tandem Duplication Shape the Unique CAZyme Complement of the Mycoparasitic Oomycetes Pythium oligandrum and Pythium periplocum. Front. Microbiol. 11. doi: 10.3389/fmicb.2020.581698
Liang, X., Moomaw, E. W., Rollins, J. A. (2015). Fungal oxalate decarboxylase activity contributes to Sclerotinia sclerotiorum early infection by affecting both compound appressoria development and function. Mol. Plant Pathol. 16, 825–836. doi: 10.1111/mpp.12239
Liebrand, T. W., van den Burg, H. A., Joosten, M. H. (2014). Two for all: receptor-associated kinases SOBIR1 and BAK1. Trends Plant Sci. 19, 123–132. doi: 10.1016/j.tplants.2013.10.003
Lin, Y. H., Xu, M. Y., Hsu, C. C., Damei, F. A., Lee, H. C., Tsai, W. L., et al. (2023). Ustilago maydis PR-1-like protein has evolved two distinct domains for dual virulence activities. Nat. Commun. 14, 5755. doi: 10.1038/s41467-023-41459-4
Liu, Q., Luo, L., Zheng, L. (2018). Lignins: Biosynthesis and biological functions in plants. Int. J. Mol. Sci. 19, 335. doi: 10.3390/ijms19020335
Liu, X., Xie, J., Fu, Y., Jiang, D., Chen, T., Cheng, J. (2020). The subtilisin-like protease bcser2 affects the sclerotial formation, conidiation and virulence of botrytis cinerea. Int. J. Mol. Sci. 21, 603. doi: 10.3390/ijms21020603
Liu, F., Zhang, X., Lu, C., Zeng, X., Li, Y., Fu, D., et al. (2015). Non-specific lipid transfer proteins in plants: presenting new advances and an integrated functional analysis. J. Exp. Bot. 66, 5663–5681. doi: 10.1093/jxb/erv313
Longsaward, R., Sanguankiattichai, N., Viboonjun, U., van der Hoorn, R. A. L. (2023). Letter to the editor: cautionary note on ribonuclease activity of recombinant PR-10 proteins. Plant Cell Physiol. 64, 847–849. doi: 10.1093/pcp/pcad062
Lozano-Torres, J. L., Wilbers, R. H., Gawronski, P., Boshoven, J. C., Finkers-Tomczak, A., Cordewener, J. H., et al. (2012). Dual disease resistance mediated by the immune receptor Cf-2 in tomato requires a common virulence target of a fungus and a nematode. Proc. Natl. Acad. Sci. U.S.A. 109, 10119–10124. doi: 10.1073/pnas.1202867109
Lozano-Torres, J. L., Wilbers, R. H., Warmerdam, S., Finkers-Tomczak, A., Diaz-Granados, A., van Schaik, C. C., et al. (2014). Apoplastic venom allergen-like proteins of cyst nematodes modulate the activation of basal plant innate immunity by cell surface receptors. PloS Pathog. 10, e1004569. doi: 10.1371/journal.ppat.1004569
Lu, S., Edwards, M. C. (2018). Molecular characterization and functional analysis of PR-1-like proteins identified from the wheat head blight fungus. Fusarium graminearum. Phytopathol. 108, 510–520. doi: 10.1094/PHYTO-08-17-0268-R
Lu, S., Faris, J. D., Sherwood, R., Edwards, M. C. (2013). Dimerization and protease resistance: new insight into the function of PR-1. J. Plant Physiol. 170, 105–110. doi: 10.1016/j.jplph.2012.08.006
Lu, S., Faris, J. D., Sherwood, R., Friesen, T. L., Edwards, M. C. (2014). A dimeric PR-1-type pathogenesis-related protein interacts with ToxA and potentially mediates ToxA-induced necrosis in sensitive wheat. Mol. Plant Pathol. 15, 650–663. doi: 10.1111/mpp.12122
McLaughlin, J. E., Darwish, N. I., Garcia-Sanchez, J., Tyagi, N., Trick, H. N., McCormick, S., et al. (2021). A lipid transfer protein has antifungal and antioxidant activity and suppresses fusarium head blight disease and DON accumulation in transgenic wheat. Phytopathology 111, 671–683. doi: 10.1094/PHYTO-04-20-0153-R
Mélida, H., Sandoval-Sierra, J. V., Diéguez-Uribeondo, J., Bulone, V. (2013). Analyses of extracellular carbohydrates in oomycetes unveil the existence of three different cell wall types. Eukaryot Cell 12, 194–203. doi: 10.1128/EC.00288-12
Melnikova, D. N., Finkina, E. I., Bogdanov, I. V., Tagaev, A. A., Ovchinnikova, T. V. (2022). Features and possible applications of plant lipid-binding and transfer proteins. Membranes (Basel) 13, 2. doi: 10.3390/membranes13010002
Meng, F., Li, Y., Liu, Z., Feng, Y., Wang, X., Zhang, X. (2022). Expression of the thaumatin-like protein-1 gene (Bx-tlp-1) from pine wood nematode Bursaphelenchus xylophilus affects terpene metabolism in pine trees. Phytopathology 112, 888–897. doi: 10.1094/PHYTO-07-21-0289-R
Meng, F., Li, Y., Wang, X., Feng, Y., Liu, Z., Zhang, W., et al. (2019). Thaumatin-like protein-1 gene (Bx-tlp-1) is associated with the pathogenicity of Bursaphelenchus xylophilus. Phytopathology 109, 1949–1956. doi: 10.1094/PHYTO-03-19-0082-R
Menu-Bouaouiche, L., Vriet, C., Peumans, W. J., Barre, A., Van Damme, E. J., Rougé, P. (2003). A molecular basis for the endo-beta 1,3-glucanase activity of the thaumatin-like proteins from edible fruits. Biochimie 85, 123–131. doi: 10.1016/s0300-9084(03)00058-0
Mir, A. A., Park, S. Y., Abu Sadat, M., Kim, S., Choi, J., Jeon, J., et al. (2015). Systematic characterization of the peroxidase gene family provides new insights into fungal pathogenicity in. Magnaporthe oryzae. Sci. Rep. 5, 11831. doi: 10.1038/srep11831
Missaoui, K., Gonzalez-Klein, Z., Pazos-Castro, D., Hernandez-Ramirez, G., Garrido-Arandia, M., Brini, F., et al. (2022). Plant non-specific lipid transfer proteins: an overview. Plant Physiol. Biochem. 171, 115–127. doi: 10.1016/j.plaphy.2021.12.026
Mitsuhara, I., Iwai, T., Seo, S., Yanagawa, Y., Kawahigasi, H., Hirose, S., et al. (2008). Characteristic expression of twelve rice PR1 family genes in response to pathogen infection, wounding, and defense-related signal compounds (121/180). Mol. Genet. Genomics 279, 415–427. doi: 10.1007/s00438-008-0322-9
Mittler, R. (2002). Oxidative stress, antioxidants and stress tolerance. Trends Plant Sci. 7, 405–410. doi: 10.1016/s1360-1385(02)02312-9
Monod, M., Capoccia, S., Léchenne, B., Zaugg, C., Holdom, M., Jousson, O. (2002). Secreted proteases from pathogenic fungi. Int. J. Med. Microbiol. 292, 405–419. doi: 10.1078/1438-4221-00223
Monzingo, A. F., Marcotte, E. M., Hart, P. J., Robertus, J. D. (1996). Chitinases, chitosanases, and lysozymes can be divided into procaryotic and eucaryotic families sharing a conserved core. Nat. Struct. Biol. 3, 133–140. doi: 10.1038/nsb0296-133
Ngou, B. P. M., Ding, P., Jones, J. D. G. (2022). Thirty years of resistance: Zig-zag through the plant immune system. Plant Cell 34, 1447–1478. doi: 10.1093/plcell/koac041
Niderman, T., Genetet, I., Bruyere, T., Gees, R., Stintzi, A., Legrand, M., et al. (1995). Pathogenesis-related PR-1 proteins are antifungal. Isolation and characterization of three 14-kilodalton proteins of tomato and of a basic PR-1 of tobacco with inhibitory activity against. Phytophthora infestans. Plant Physiol. 108, 17–27. doi: 10.1104/pp.108.1.17
Ohnuma, T., Umemoto, N., Nagata, T., Shinya, S., Numata, T., Taira, T., et al. (2014). Crystal structure of a “loopless” GH19 chitinase in complex with chitin tetrasaccharide spanning the catalytic center. Biochim. Biophys. Acta 1844, 793–802. doi: 10.1016/j.bbapap.2014.02.013
Okushima, Y., Koizumi, N., Kusano, T., Sano, H. (2000). Secreted proteins of tobacco cultured BY2 cells: identification of a new member of pathogenesis-related proteins. Plant Mol. Biol. 42, 479–488. doi: 10.1023/a:1006393326985
Oyeleye, A., Normi, Y. M. (2018). Chitinase: diversity, limitations, and trends in engineering for suitable applications. Biosci. Rep. 38, BSR2018032300. doi: 10.1042/BSR20180323
Parisi, K., Shafee, T. M. A., Quimbar, P., van der Weerden, N. L., Bleackley, M. R., Anderson, M. A. (2019). The evolution, function and mechanisms of action for plant defensins. Semin. Cell Dev. Biol. 88, 107–118. doi: 10.1016/j.semcdb.2018.02.004
Passardi, F., Penel, C., Dunand, C. (2004). Performing the paradoxical: how plant peroxidases modify the cell wall. Trends Plant Sci. 9, 534–540. doi: 10.1016/j.tplants.2004.09.002
Pedro, H., Maheswari, U., Urban, M., Irvine, A. G., Cuzick, A., McDowall, M. D., et al. (2016). PhytoPath: an integrative resource for plant pathogen genomics. Nucleic Acids Res. 44, D688–D693. doi: 10.1093/nar/gkv1052
Perrot, T., Pauly, M., Ramírez, V. (2022). Emerging roles of β-glucanases in plant development and adaptative responses. Plants (Basel) 11, 1119. doi: 10.3390/plants11091119
Pettersen, E. F., Goddard, T. D., Huang, C. C., Couch, G. S., Greenblatt, D. M., Meng, E. C., et al. (2004). UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612. doi: 10.1002/jcc.20084
Poria, V., Rana, A., Kumari, A., Grewal, J., Pranaw, K., Singh, S. (2021). Current perspectives on chitinolytic enzymes and their agro-industrial applications. Biol. (Basel) 10, 1319. doi: 10.3390/biology10121319
Prados-Rosales, R. C., Roldan-Rodriguez, R., Serena, C., Lopez-Berges, M. S., Guarro, J., Martinez-del-Pozo, A., et al. (2012). A PR-1-like protein of Fusarium oxysporum functions in virulence on mammalian hosts. J. Biol. Chem. 287, 21970–21979. doi: 10.1074/jbc.M112.364034
Radauer, C., Lackner, P., Breiteneder, H. (2008). The Bet v 1 fold: an ancient, versatile scaffold for binding of large, hydrophobic ligands. BMC Evol. Biol. 8, 286. doi: 10.1186/1471-2148-8-286
Rawlings, N. D., Barrett, A. J., Thomas, P. D., Huang, X., Bateman, A., Finn, R. D. (2018). The MEROPS database of proteolytic enzymes, their substrates and inhibitors in 2017 and a comparison with peptidases in the PANTHER database. Nucleic Acids Res. 46, D624–D632. doi: 10.1093/nar/gkx1134
Riviere, M. P., Marais, A., Ponchet, M., Willats, W., Galiana, E. (2008). Silencing of acidic pathogenesis-related PR-1 genes increases extracellular beta-(1-3)-glucanase activity at the onset of tobacco defence reactions. J. Exp. Bot. 59, 1225–1239. doi: 10.1093/jxb/ern044
Roberts, W. K., Selitrennikoff, C. P. (1990). Zeamatin, an antifungal protein from maize with membrane-permeabilizing activity. J. Gen. Microbiolol. 136, 1771–1778. doi: 10.1099/00221287-136-9-1771
Ryan, C. A. (1989). Proteinase inhibitor gene families: strategies for transformation to improve plant defenses against herbivores. Bioessays 10, 20–24. doi: 10.1002/bies.950100106
Sakamoto, Y., Watanabe, H., Nagai, M., Nakade, K., Takahashi, M., Sato, T. (2006). Lentinula edodes tlg1 encodes a thaumatin-like protein that is involved in lentinan degradation and fruiting body senescence. Plant Physiol. 141, 793–801. doi: 10.1104/pp.106.076679
Sarthy, A. V., McGonigal, T., Coen, M., Frost, D. J., Meulbroek, J. A., Goldman, R. C. (1997). Phenotype in Candida albicans of a disruption of the BGL2 gene encoding a 1,3-beta-glucosyltransferase. Microbiol. (Reading) 143, 367–376. doi: 10.1099/00221287-143-2-367
Schaller, A., Stintzi, A., Rivas, S., Serrano, I., Chichkova, N. V., Vartapetian, A. B., et al. (2018). From structure to function–a family portrait of plant subtilases. New Phytol. 218, 901–915. doi: 10.1111/nph.14582
Sels, J., Mathys, J., De Coninck, B. M., Cammue, B. P., De Bolle, M. F. (2008). Plant pathogenesis-related (PR) proteins: a focus on PR peptides. Plant Physiol. Biochem. 46, 941–950. doi: 10.1016/j.plaphy.2008.06.011
Sharma, A., Sharma, H., Rajput, R., Pandey, A., Upadhyay, S. K. (2021). Molecular characterization revealed the role of thaumatin-like proteins of bread wheat in stress response. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.807448
Shi, L., Li, R., Liao, S., Bai, L., Lu, Q., Chen, B. (2014). Prb1, a subtilisin-like protease, is required for virulence and phenotypical traits in the chestnut blight fungus. FEMS Microbiol. Lett. 359, 26–33. doi: 10.1111/1574-6968.12547
Singh, B. K., Delgado-Baquerizo, M., Egidi, E., Guirado, E., Leach, J. E., Liu, H., et al. (2023). Climate change impacts on plant pathogens, food security and paths forward. Nat. Rev. Microbiol. 21, 640–656. doi: 10.1038/s41579-023-00900-7
Srinivas, C., Nirmala Devi, D., Narasimha Murthy, K., Mohan, C. D., Lakshmeesha, T. R., Singh, B., et al. (2019). Fusarium oxysporum f. sp. lycopersici causal agent of vascular wilt disease of tomato: Biology to diversity- A review. Saudi J. Biol. Sci. 26, 1315–1324. doi: 10.1016/j.sjbs.2019.06.002
Stec, B. (2006). Plant thionins–the structural perspective. Cell Mol. Life Sci. 63, 1370–1385. doi: 10.1007/s00018-005-5574-5
Stintzi, A., Heitz, T., Prasad, V., Wiedemann-Merdinoglu, S., Kauffmann, S., Geoffroy, P., et al. (1993). Plant ‘pathogenesis-related’ proteins and their role in defense against pathogens. Biochimie 75, 687–706. doi: 10.1016/0300-9084(93)90100-7
Sung, Y. C., Outram, M. A., Breen, S., Wang, C., Dagvadorj, B., Winterberg, B., et al. (2021). PR1-mediated defence via C-terminal peptide release is targeted by a fungal pathogen effector. New Phytol. 229, 3467–3480. doi: 10.1111/nph.17128
Tam, J. P., Wang, S., Wong, K. H., Tan, W. L. (2015). Antimicrobial peptides from plants. Pharm. (Basel) 8, 711–757. doi: 10.3390/ph8040711
Tan, J., Zhao, H., Li, J., Gong, Y., Li, X. (2023). The devastating rice blast airborne pathogen magnaporthe oryzae-A review on genes studied with mutant analysis. Pathogens 12, 379. doi: 10.3390/pathogens12030379
Tanaka, K., Heil, M. (2021). Damage-associated molecular patterns (DAMPs) in plant innate immunity: applying the danger model and evolutionary perspectives. Annu. Rev. Phytopathol. 59, 53–75. doi: 10.1146/annurev-phyto-082718-100146
Teixeira, P. J. P., Colaianni, N. R., Fitzpatrick, C. R., Dangl, J. L. (2019). Beyond pathogens: microbiota interactions with the plant immune system. Curr. Opin. Microbiol. 49, 7–17. doi: 10.1016/j.mib.2019.08.003
Teixeira, P. J., Thomazella, D. P., Vidal, R. O., do Prado, P. F., Reis, O., Baroni, R. M., et al. (2012). The fungal pathogen Moniliophthora perniciosa has genes similar to plant PR-1 that are highly expressed during its interaction with cacao. PloS One 7, e45929. doi: 10.1371/journal.pone.0045929
Terras, F. R., Eggermont, K., Kovaleva, V., Raikhel, N. V., Osborn, R. W., Kester, A., et al. (1995). Small cysteine-rich antifungal proteins from radish: their role in host defense. Plant Cell 7, 573–588. doi: 10.1105/tpc.7.5.573
Terras, F., Schoofs, H., Thevissen, K., Osborn, R. W., Vanderleyden, J., Cammue, B., et al. (1993). Synergistic enhancement of the antifungal activity of wheat and barley thionins by radish and oilseed rape 2S albumins and by barley trypsin inhibitors. Plant Physiol. 103, 1311–1319. doi: 10.1104/pp.103.4.1311
Thevissen, K., Cammue, B. P., Lemaire, K., Winderickx, J., Dickson, R. C., Lester, R. L., et al. (2000). A gene encoding a sphingolipid biosynthesis enzyme determines the sensitivity of Saccharomyces cerevisiae to an antifungal plant defensin from dahlia (Dahlia merckii). Proc. Natl. Acad. Sci. U.S.A. 97, 9531–9536. doi: 10.1073/pnas.160077797
Trudel, J., Grenier, J., Potvin, C., Asselin, A. (1998). Several thaumatin-like proteins bind to beta-1,3-glucans. Plant Physiol. 118, 1431–1438. doi: 10.1104/pp.118.4.1431
van Loon, L. C., Gerritsen, Y. A. M., Ritter, C. E. (1987). Identification, purification, and characterization of pathogenesis-related proteins from virus-infected Samsun NN tobacco leaves. Plant Mol. Biol. 9, 593–609. doi: 10.1007/BF00020536
van Loon, L. C., Rep, M., Pieterse, C. M. (2006). Significance of inducible defense-related proteins in infected plants. Annu. Rev. Phytopathol. 44, 135–162. doi: 10.1146/annurev.phyto.44.070505.143425
van Loon, L. C., van Kammen, A. (1970). Polyacrylamide disc electrophoresis of the soluble leaf proteins from Nicotiana tabacum var. “Samsun” and “Samsun NN”. II. Changes in protein constitution after infection with tobacco mosaic virus. Virology 40, 190–211. doi: 10.1016/0042-6822(70)90395-8
Van Loon, L. C., Van Strien, E. A. (1999). The families of pathogenesis-related proteins, their activities, and comparative analysis of PR-1 type proteins. Physiol. Mol. Plant Pathol. 55, 85–97. doi: 10.1006/pmpp.1999.0213
Vlot, A. C., Dempsey, D. A., Klessig, D. F. (2009). Salicylic acid, a multifaceted hormone to combat disease. Annu. Rev. Phytopathol. 47, 177–206. doi: 10.1146/annurev.phyto.050908.135202
Wang, F., Shen, S., Zhao, C., Cui, Z., Meng, L., Wu, W., et al. (2022). TaPR1 interacts with TaTLP1 via the αIV helix to be involved in wheat defense to Puccinia triticina through the CAPE1 motif. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.874654
Wang, C., Yin, Z., Nie, J., Lin, Y., Huang, L. (2021). Identification and virulence analysis of CAP superfamily genes in Valsa Mali. Scientia Agricultura Sin. 54, 3440–3450. doi: 10.3864/j.issn.0578-1752.2021.16.007
Wang, F., Yuan, S., Wu, W., Yang, Y., Cui, Z., Wang, H., et al. (2020). TaTLP1 interacts with TaPR1 to contribute to wheat defense responses to leaf rust fungus. PloS Genet. 16, e1008713. doi: 10.1371/journal.pgen.1008713
Wanke, A., Malisic, M., Wawra, S., Zuccaro, A. (2021). Unraveling the sugar code: the role of microbial extracellular glycans in plant-microbe interactions. J. Exp. Bot. 72, 15–35. doi: 10.1093/jxb/eraa414
Westrick, N. M., Smith, D. L., Kabbage, M. (2021). Disarming the host: detoxification of plant defense compounds during fungal necrotrophy. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.651716
Whisson, S. C., Boevink, P. C., Wang, S., Birch, P. R. (2016). The cell biology of late blight disease. Curr. Opin. Microbiol. 34, 127–135. doi: 10.1016/j.mib.2016.09.002
Wilbers, R. H. P., Schneiter, R., Holterman, M. H. M., Drurey, C., Smant, G., Asojo, O. A., et al. (2018). Secreted venom allergen-like proteins of helminths: Conserved modulators of host responses in animals and plants. PloS Pathog. 14, e1007300. doi: 10.1371/journal.ppat.1007300
Xu, L., Wang, H., Zhang, C., Wang, J., Chen, A., Chen, Y., et al. (2020). System-wide characterization of subtilases reveals that subtilisin-like protease FgPrb1 of Fusarium graminearum regulates fungal development and virulence. Fungal Genet. Biol. 144, 103449. doi: 10.1016/j.fgb.2020.103449
Yan, T., Zhou, X., Li, J., Li, G., Zhao, Y., Wang, H., et al. (2022). FoCupin1, a Cupin_1 domain-containing protein, is necessary for the virulence of Fusarium oxysporum f. sp. cubense tropical race 4. Front. Microbiol. 13. doi: 10.3389/fmicb.2022.1001540
Yang, H., Kim, X., Skłenar, J., Aubourg, S., Sancho-Andrés, G., Stahl, E., et al. (2023). Subtilase-mediated biogenesis of the expanded family of SERINE RICH ENDOGENOUS PEPTIDES. Nat. Plants 9, 2085–2094. doi: 10.1038/s41477-023-01583-x
Yu, C., Qi, J., Han, H., Wang, P., Liu, C. (2023). Progress in pathogenesis research of Ustilago maydis, and the metabolites involved along with their biosynthesis. Mol. Plant Pathol. 24, 495–509. doi: 10.1111/mpp.13307
Zhang, H., Mukherjee, M., Kim, J. E., Yu, W., Shim, W. B. (2018a). Fsr1, a striatin homologue, forms an endomembrane-associated complex that regulates virulence in the maize pathogen. Fusarium verticillioides. Mol. Plant Pathol. 19, 812–826. doi: 10.1111/mpp.12562
Zhang, J., Wang, F., Liang, F., Zhang, Y., Ma, L., Wang, H., et al. (2018b). Functional analysis of a pathogenesis-related thaumatin-like protein gene TaLr35PR5 from wheat induced by leaf rust fungus. BMC Plant Biol. 18, 76. doi: 10.1186/s12870-018-1297-2
Zhao, J., Bi, W., Zhao, S., Su, J., Li, M., Ma, L., et al. (2021). Wheat apoplast-localized lipid transfer protein TaLTP3 enhances defense responses against. Puccinia triticina. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.771806
Zhao, M., Zhang, Y., Guo, H., Gan, P., Cai, M., Kang, Z., et al. (2023). Identification and functional analysis of CAP genes from the wheat stripe rust fungus Puccinia striiformis f. sp. tritici. J. Fungi (Basel) 9, 734. doi: 10.3390/jof9070734
Zhou, J. M., Zhang, Y. (2020). Plant immunity: Danger perception and signaling. Cell 181, 978–989. doi: 10.1016/j.cell.2020.04.028
Keywords: plant immunity, fungal pathogens, secretion, apoplast, virulence, immune signaling, sperm coating proteins (SCPs), venom allergen-like proteins (VALs/VAPs)
Citation: Han Z and Schneiter R (2024) Dual functionality of pathogenesis-related proteins: defensive role in plants versus immunosuppressive role in pathogens. Front. Plant Sci. 15:1368467. doi: 10.3389/fpls.2024.1368467
Received: 10 January 2024; Accepted: 03 July 2024;
Published: 02 August 2024.
Edited by:
Mario Serrano, National Autonomous University of Mexico, MexicoReviewed by:
Xiao-Ren Chen, Yangzhou University, ChinaArtemio Mendoza-Mendoza, Lincoln University, New Zealand
Copyright © 2024 Han and Schneiter. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Roger Schneiter, Um9nZXIuU2NobmVpdGVyQFVuaUZyLmNo
 Zhu Han
Zhu Han Roger Schneiter
Roger Schneiter