
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 02 April 2024
Sec. Plant Systematics and Evolution
Volume 15 - 2024 | https://doi.org/10.3389/fpls.2024.1367917
Aster yaoshanensis sp. nov., a new species of the family Asteraceae is here described and illustrated. The species is presently known only from rock crevices of mountain valleys in Dayaoshan National Nature Reserve, Guangxi autonomous region, China. Phylogenetic analyses based on ITS sequences and complete plastome data have shown that this new species is a member of genus Aster with high support. Morphologically, it mostly resembles A. jishouensis, but it can be easily distinguished from the latter by bract indumentum (glabrous except margin ciliate vs. villous especially on veins abaxially, glabrous adaxially) and color (green vs. purple), shorter corolla (3.2–3.5 mm vs. 4.5–5.3 mm), bract stalk (obvious, ca.1.2 mm vs. sessile), and different distribution (Guangxi vs. Hunan). The detailed description, distribution map, and photos are provided. This study further elucidates the species identification, phylogeny and characteristic evolution of Aster.
The genus Aster with about 152 species is one of the largest genera in tribe Astereae (Asteraceae) (Nesom, 1994; Noyes and Rieseberg, 1999; Nesom and Robinson, 2007; Brouillet et al., 2009; Chen et al., 2011; Fu et al., 2016). So far, 123 species of Aster have been identified from different regions of China (Chen et al., 2011; Xiong et al., 2019). To revise the genus Aster, we have conducted extensive collections and observations in the field in Eurasia. In Guangxi province of China, Asteraceae is represented by 81 genera and 233 species, and the genus Aster includes 15 species (Wei, 2018).
On 3 November 2020, Dr. Pan Li (Zhejiang University) collected some specimens of a species belonging to genus Aster in Dayaoshan National Nature Reserve, Jinxiu county, Laibin city, Guangxi province, China. Then, in 12 November 2020, Dr. Zhixi Fu revisited and collected the distinctive species in the same location. Through field investigation and morphological study, we confirmed that this species does not match any previously published description from southwest China to central Himalaya. Further molecular analyses revealed significant differences between this species and its relatives. The results allow us to infer that these newly collected specimens from Guangxi belong to a new species.
High rates of morphological divergence are a challenge to morphology-based classification, because some of this variation originated by parallel evolution whereas other characters are apomorphic (Wei et al., 2013). In previous studies, the reconstruction of the ancestral state of morphological characteristics has confirmed the hypothesis on the origin and evolution of the morphological diversity of some species in Asteraceae (Norbert et al., 2017). There are still insufficient studies on the character evolution of Aster, as in other Asteraceae genera with many species and a complex taxonomy (Wagensommer et al., 2016). Based on the ancestral reconstruction, we analyzed some morphological traits of the new species and its relatives in Aster.
In the context of the ongoing taxonomic revision of the genus Aster, this finding allows us to more precisely define its morphological characters, complete plastome characters, phylogenetic position, taxonomic affinities, and ancestral trait. Therefore, the present work aims (1) to provide a complete morphological record of the new species and distribution map; (2) to examine the phylogenetic affinities of A. yaoshanensis and its related species using the complete plastome sequences and nuclear ribosomal DNA internal transcribed spacer (ITS) sequences; and (3) to provide hypotheses for the evolution of morphological characters of A. yaoshanensis and by integrating available information, to provide an adequate taxonomic treatment for this new species.
Our taxon sampling strategy was designed to include Aster and relatives genera based on the recent classification (Li et al., 2012; Fu et al., 2016). We downloaded nrDNA ITS sequence matrix of 71 species from other molecular phylogenetic studies (Li et al., 2012; Zhang et al., 2015a; Zhang et al., 2019b), representing 17 genera and major clades of Aster and its relatives. Meanwhile, we downloaded the complete plastome sequence matrix of 17 species, representing the genus Aster and its relatives.
One fresh leave for each specimen was dried in silica gel (the list of samples is shown in Table 1). Total genomic DNA was extracted using the Plant Genomic DNA Kit (Tiangen Biotech, Beijing, China) following the CTAB DNA extraction protocol (Doyle and Doyle, 1987) and the manufacturer’s instructions. In this study, to determine the systematic position of the new species, we used nuclear ribosomal DNA ITS sequences for phylogenetic inference. The spacer region of the nuclear ribosomal repeat (nrDNA ITS) was separately amplified with standard PCR following the method described in Zhang et al. (2015, 2019). The new species, A. yaoshanensis was sequenced, and the GenBank accession numbers and sample information of the ITS sequences used in this study are shown in Table 2 (accession number of A. yaoshanensis: OL461705.1). Voucher specimens were deposited at the herbarium of the Sichuan Normal University (SCNU).
In this study, we also used the complete plastome sequence of A. yaoshanensis for phylogenetic inference. We followed the CTAB DNA extraction protocol (Doyle and Doyle, 1987) and the manufacturer’s instructions, to extract total genomic DNA of A. yaoshanensis, using the Plant Genomic DNA Kit (Tiangen Biotech, Beijing, China). Then, the NanoDrop 2000 Spectrophotometer and Qubit 4 Fluorometer (Thermo Fisher Scientific, Wilmington, DE, USA) were used to test quality and quantity of the total genomic DNA. We constructed DNA libraries using Illumina Paired-End DNA Library Kit (Illumina Inc., San Diego, CA, USA) in the NovaSeq 6000 platform with a paired-end reading length of 150 bp (NovoGene Inc., Beijing, China).
The complete plastome of A. yaoshanensis was assembled by SPAdes3.15.1 (Bankevich et al., 2012) with default parameters. The circular plastome was obtained by using Bandage (Wick et al., 2015). Subsequently, the results were annotated using PGA (Qu et al., 2019). The annotation results were checked using Geneious R11 (Kearse et al., 2012). The plastome map was drawn on the OGDRAW (https://chlorobox.mpimp-golm.mpg.de/OGDraw.html). The tRNA sequences were also confirmed by tRNAscan-SE v2.0.7 (Lowe and Eddy, 1997), on the platform Geseq (https://chlorobox.mpimp-golm.mpg.de/geseq.html). The genome sequence of A. yaoshanensis has been deposited in GenBank (accession number: ON120847.1). We performed nucleobase content and genes analysis using the platform JSHYClound (www.jshycloud.net).
Phylogenetic analyses of the ITS data set (Table 2) were performed using maximum likelihood (ML) and Bayesian inference (BI) analysis using RAxML (Stamatakis et al., 2008) and MrBayes (Ronquist et al., 2012), respectively, on the CIPRES science gateway portal (https://www.phylo.org/portal2/, Miller et al., 2010). Prior to ML and BI analyses, the best fit model selected was GTR+T+G according to the Akaike information criterion (AIC) implemented in ModelTest (Posada and Crandall, 1998). RAxML was conducted with the fast bootstrap option, using 1,000 replicates, bootstrap value (BS) of 80%–100% were interpreted as strong support, 60%–80% as moderate. In BI analysis, four Markov chain Monte Carlo chains were run for 7,000,000 generations each, and were sampled every 1,000 generations, starting with a random tree, and Bayesian posterior probabilities (PP) were calculated for the majority consensus tree of all sampled trees after discarding 25% of trees sampled. Bayesian PP more than 0.95 were considered to be strong support, from 0.9 to 0.95 to be moderate. Finally, the trees were visualized using FigTree (Rambaut, 2012). The species Chrysanthemum indicum L. and Dendranthema indicum (L.) Des Moul. were selected as the outgroup following previous works (Panero and Funk, 2002, 2008; Panero et al., 2014).
To investigate the phylogenetic positions of A. yaoshanensis within the Aster genus, 17 complete plastome sequences were downloaded from the NCBI database (Table 3). Phylogenetic analyses of the complete plastome data set were performed using ML in RAxML on the CIPRES platform (https://www.phylo.org/portal2/). We used ModelTest to predict the best fit model. The fast bootstrap option and 1,000 replicates were used in RAxML. Finally, the trees were visualized using FigTree.
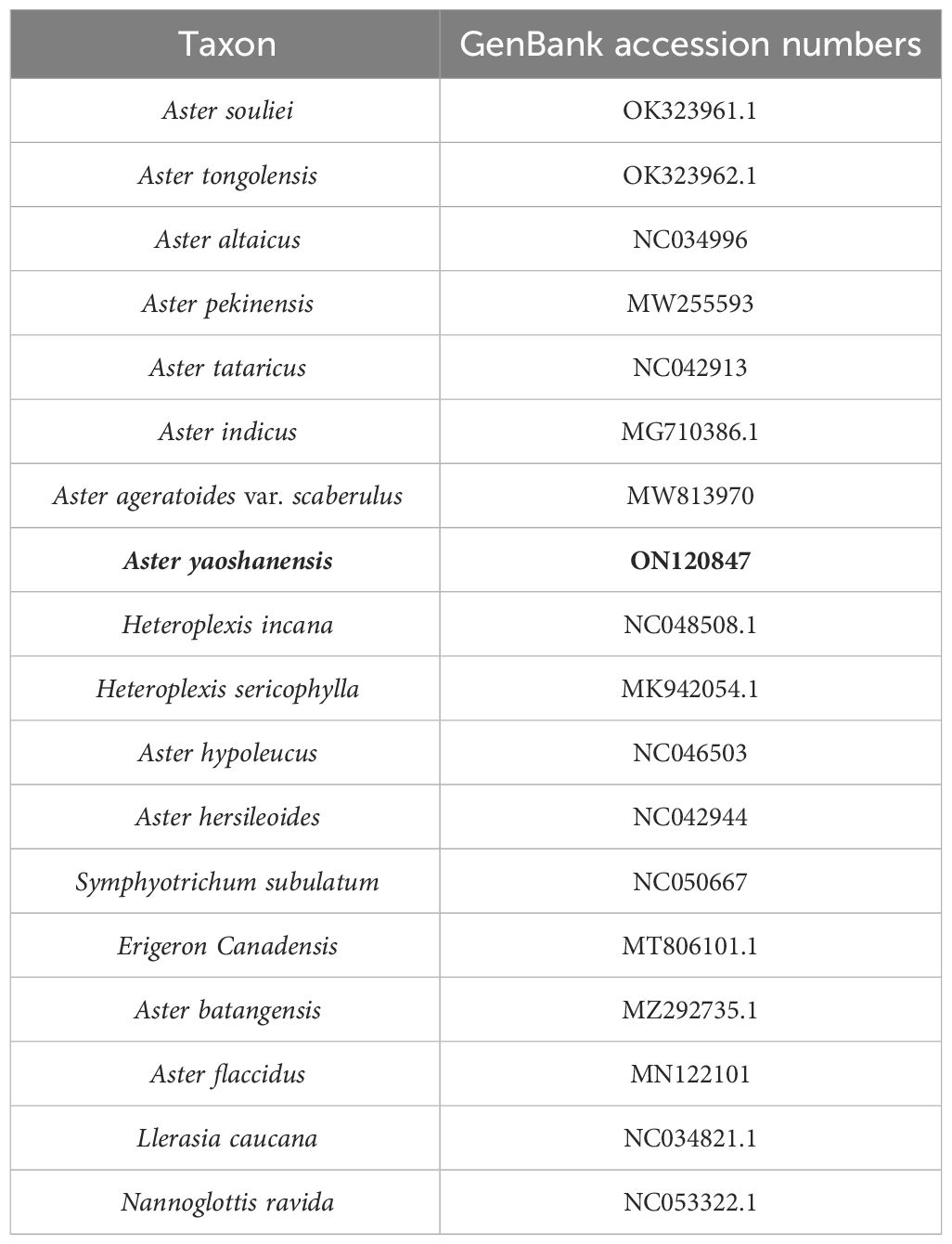
Table 3 Taxa sampled and their GenBank accession numbers for the whole plastome sequences used in this study.
Based on the topology of the ITS sequence for ML analysis, in Mesquite v3.81 (Maddison, 2008), we used the “Trace character history” option to analyze the ancestral morphological characters in the genus Aster. An ML approach using Markov k-state 1 parameter model (Mk1; Lewis, 2001) was used to reconstruct character evolution. The data employed for the reconstruction of the evolution of morphological characters were obtained from literatures (Li et al., 2012; Norbert et al., 2017), and our own observations of morphological character variation using herbarium specimens held at SCNU and PE (Thiers, 2024), and CVH (https://www.cvh.ac.cn/). The codes associated with these morphological features include (A) Inflorescence type: listed in order of 0–7, (B) the length of pappus: (0) none pappus, (1) short pappus, (2) long pappus, (3) longer pappus; (C) habit (0) dwarf herb, (1) tall herb, (2) subshrub, (3) shrub. A list of morphological characters and their character state coding used for the ancestral state reconstruction is detailed in Table 4. The differences of eight types of inflorescence can be seen in the statement at the end of Table 4. Based on the codes, we reconstructed the evolution of three morphological characters for genera of Aster and its relatives.
With the information collected through fieldwork, the conservation status of the new species was evaluated according to the IUCN, 2024 categories and criteria.
In this study, the structure of plastome of A. yaoshanensis is highly conserved, and it has standard quadripartite structure, including a large single-copy (LSC), a small single-copy (SSC), and a pair of IRs (IRa and IRb). The size o f A. yaoshanensis is 152,384bp, and GC content is 37.37%. Overall, the plastome have 133 genes, including 87 protein-coding genes, 38 tRNA, and 8 rRNA, of which 115 were unique and 18 were duplicated in the IR regions (Table 5). The arrangements of these 133 genes were totally collinear. There are two introns of three genes (rps12, ycf3, clpP), and single intron of 16 genes (ndhA, ndhB, petB, petD, atpF, rbcL, rpl16, rpl2, rps16, rpoC1, trnA-UGC, trnG, trnI-GAU, trnK-UUU, trnL-UAA, trnV-UAC). The gene rps12 was trans-spliced and the genes ndhD, psbL were RNA editing (Table 5). These results were performed in the plastome map (Figure 1).
The total length of ITS sequence alignment with gaps was 650 bp. Analysis of the data using ML and BI methods obtained similar trees with high ML Bootstrap (BS) and BI posterior probability (PP). Phylogeny analysis showed that A. yaoshanensis is positioned in the Aster clade, and it formed a clade with A. homochlamydeus Hand.-Mazz and A. handelii Onno (ML BS ≥ 90% and BI PP ≥ 0.90) (Figure 2).
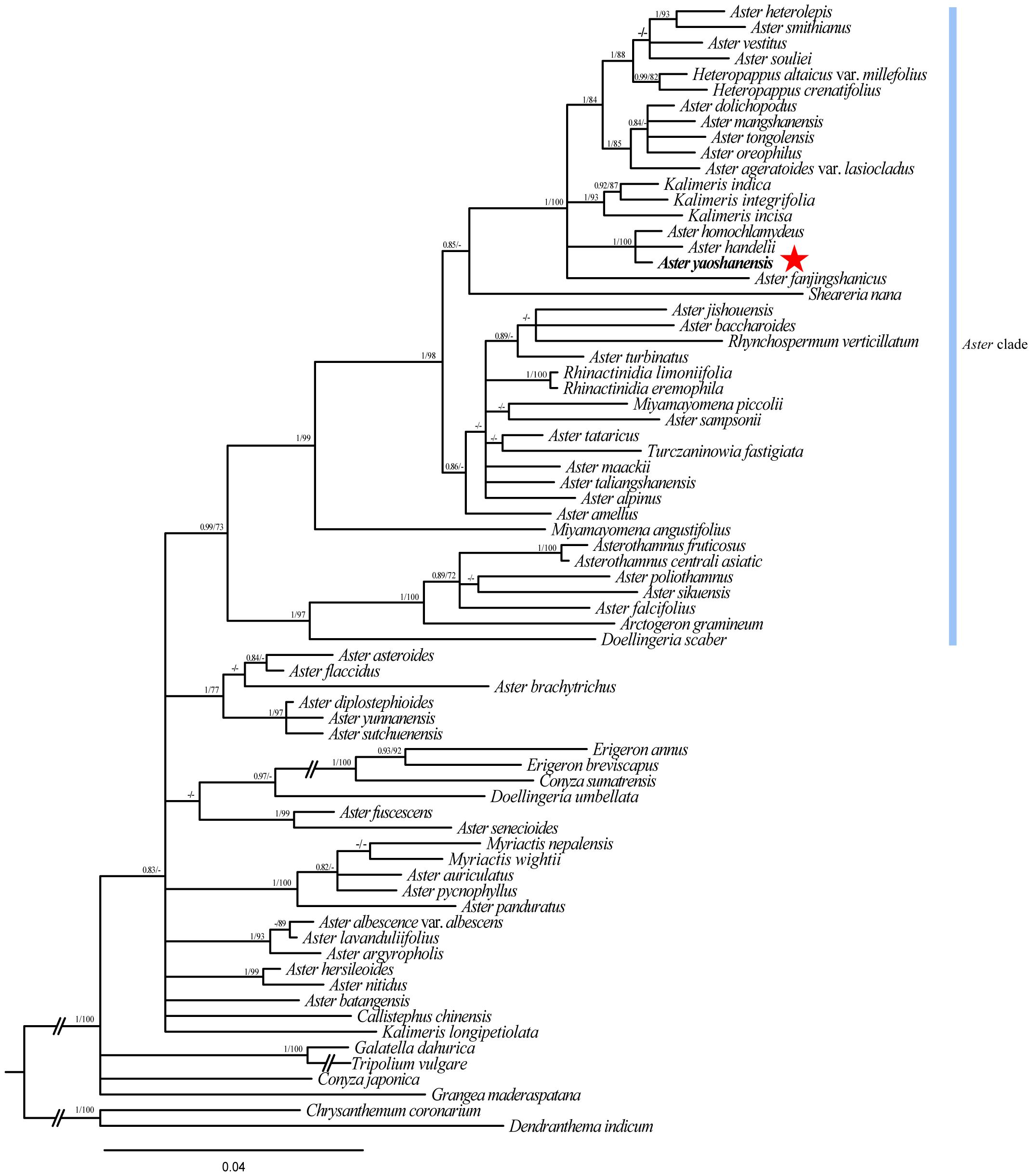
Figure 2 Cladogram of the Beyasian inference (BI) phylogenetic tree of Aster. Phylogenetic tree based on nrDNA data (ITS), showing the position of A. yaoshanensis (in red star). Values above branch represent Bayesian posterior probabilities (PP) and bootstrap values (BS), respectively; bootstrap support values >0.80 (BI) or >60% (ML) are shown. Blue represents Aster clade.
Based on phylogenetic analysis, A. yaoshanensis is positioned in the Aster clade, and it is the sister species to A. ageratoides var. scaberulus (Miq.) Y. Ling. (BS = 100) (Figure 3).
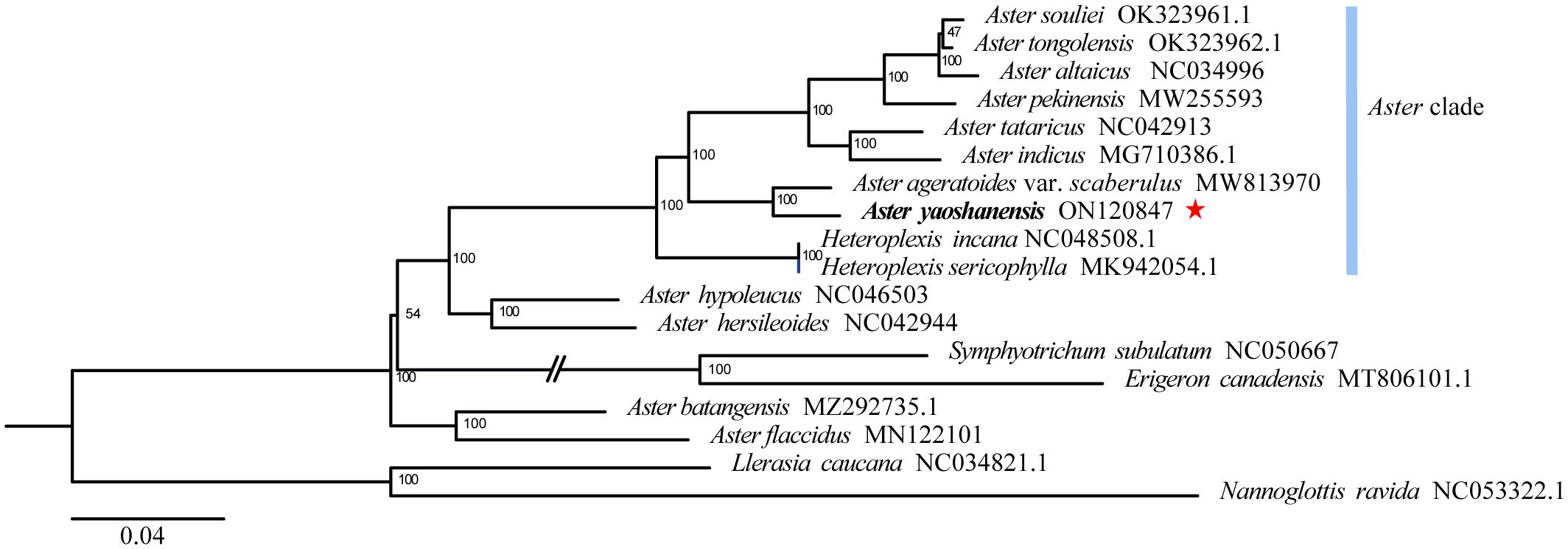
Figure 3 Maximum likelihood (ML) phylogenetic tree based on complete plastome sequences, showing the position of A. yaoshanensis (in red star). Blue represent Aster clade.
Considering the similarity between new species and its related species in Aster, we obtained the morphological characteristics of the latter from specimens and literature, and made morphological comparisons between these species (Table 6). The new species is most similar to Aster jishouensis W.P. Li & S.X. Liu (Figure 4). It is mainly distinguished from the latter by its erect (versus oblique ascensional) stem, deep purple (vs. green) basal leaves, 4–5 seriate (vs. 5–7seriate) phyllaries, numerous (vs. 1–4) capitula. In addition, the results of ancestral trait reconstruction were shown in Figures 5–7.

Figure 4 Isotype of A. jishouensis W.P. Li and S.X. Liu (voucher, W.P. Li & G.X. Chen 0776301 (barcode PE 00935961).
Aster yaoshanensis K. Qin, Z.X. Fu & P. Li, sp. nov. (Figures 8, 9)

Figure 8 Holotype of A. yaoshanensis sp. nov. K. Qin, Z.X. Fu and P. Li. (Upper part) (voucher, Z.X. Fu 5501, SCNU).

Figure 9 Holotype of A. yaoshanensis sp. nov. K. Qin, Z.X. Fu and P. Li. (Lower part) (voucher, Z.X. Fu 5501, SCNU).
Type (Holotype). China, Guangxi, Laibin city, Jinxiu county, Dayaoshan Mountain National Nature Reserve, ca. 1100 m a.s.l., 24.1988°N, 110.2692°E, limestone, 12 November 2020, Zhixi Fu FZX5501 (SCNU).
Description. Perennial herbs, 90–130 cm tall, basally woody, rhizomatous. Stems erect, unbranched, finely striate, smooth and glabrous with many basal leaves and some cauline leaves. Basal leaves rather thick, ovate-lanceolate, apex acute to slightly obtuse, petiole narrowly winged, 3–3.5 cm × 1–1.5 cm, margin undulant serrate, abruptly mucronate, adaxially green, strigillose; abaxially amaranth, the vein strigillose. Cauline leaves, slightly reduced downward, adaxially green, densely strigose; abaxially subglabrous, midvein prominent abaxially, lowest leaves withered by anthesis, lower to upper leaves sessile or shortly petiole, the uppermost very small, blade narrowly lanceolate to oblanceolate, 2–14 cm × 1–4 cm, margin entire to undulant serrate (teeth mucronulate). Flowering branches attach to the leaf-axils, capitula numerous, in terminal or axillary radialized, 1.5–2 cm in diam; peduncles 2–8 mm, smooth and glabrous, bracts ovate, margin entire. Involucres campanulate, 6–7 mm × 2–3 mm, phyllaries 4-5 seriate, unequal, narrowly green apically, abaxially moderately to densely strigose, margin scarious, apex acute to obtuse, ciliate, outer phyllaries ovate, 1.5–2 × ca. 1 mm, middle phyllaries lanceolate-oblong, 2–2.5 × ca. 1 mm, inner phyllaries approximately oblong, 3–3.5 × 1–1.5 mm. Ray florets 8-18, white, lamina 6–7 mm × 1–1.5 mm, glabrous, eglandular; disk florets yellowish-green, 5–6 mm, tube base flared, lobes capreolary, tip recurved to cannular. Achenes long-obovoid, compressed, ca. 1.8–2.2 mm, 2-3-ribbed, sparsely strigillose. Pappus, bristles 2.5–3.5 mm. (Figures 10, 11).

Figure 10 A. yaoshanensis K. Qin, Z.X. Fu and P. Li (voucher, Z.X. Fu 5501, SCNU): (A) Habit; (B) Inflorescences, showing capitula terminal on lateral branches.

Figure 11 A. yaoshanensis (voucher, Z. X. Fu 5501, SCNU): (A) Habitat; (B) Capitulum, lateral view, showing the involucre. (C) Abaxial surface of leaf blade; (D) Adaxial surface of leaf blade. Scale Bar: 2 cm.
Etymology. The specific epithet refers the famous Dayaoshan Mountain in Jinxiu county, Laibin city, Guangxi autonomous region, China.
Distribution and habitat. The new species, A. yaoshanensis is mainly distributed at an altitude of about 1000 m in Dayaoshan Mountain, Guangxi (Figure 12), and mainly growing on limestone, slopes, forest margins, flooded lands, along streams, wet places.
Phenology. Flowering and fruiting from September to November.
Conservation status. According to the IUCN, 2024 categories and criteria, newly described species with a restricted distribution area are often considered in a threat category (CR, EN, VU) (Zhang et al., 2019; Blasco et al., 2021; Wagensommer and Venanzoni, 2021; Ibáñez et al., 2022), but more exploration in the occurring area of the new species is necessary for defining the distribution, population size, threats, etc., before being able to assess the conservation status of A. yaoshanensis. The new species is currently only found in rock crevices of mountain valleys in Dayaoshan National Nature Reserve. It is a narrowly distributed species with no more than 30 individuals found. It is very possible that the species is found in other localities with a more comprehensive collecting programme. Therefore, it would be best to assess the conservation status of the species as DD (Data Deficient).
Additional specimens examined (Paratypes). CHINA. Guangxi: Jinxiu county, Dayaoshan Mountain National Nature Reserve, alt. 1100m a.s.l., limestone, 12 November 2020, FZX5505 (SCNU), FZX5508 (SCNU), FZX5509 (SCNU) FZX5510 (SCNU), and FZX5511 (SCNU).
In our study, we successfully assembled the complete plastome of A. yaoshanensis collected from Guangxi for the first time. Based on the analysis of the plastome map, we found that the structure, gene location, size and gene content of A. yaoshanensis are highly conserved, which is very similar to the plastome structure of most reported Aster plants, such as A. ageratoides (Feng et al., 2021; Zhang et al., 2021; Wang and Liu, 2023). The plastomes of these Aster have a standard quadripartite structure, including two inverted repeat regions (IRs), an LSC region and an SSC region, and the general GC content was low in the whole sequence. This indicates the close relationship between A. yaoshanensis and Aster.
In order to further determine the evolutionary position of A. yaoshanensis in Aster, we conducted phylogenetic analysis of ITS sequences of Aster and its related genera (Figure 2). Heteropappus, Kalimeris, Sheareria, Rhynchospermum, Rhinactinidia, Miyamayomena, Turczaninowia, Doellingeria, and other Aster taxa were deeply nested within the core Aster clade; this is consistent with previous studies (Li et al., 2012). In the Aster clade, A. yaoshanensis formed a compact clade with A. homochlamydeus and A. handelii (PP = 1.00, BS = 100), and the clade was sister to the clades comprising some Aster taxa, Heteropappus and Kalimeris. The result further proved that A. yaoshanensis is closely related to Aster. In addition, A. jishouensis and A. yaoshanensis were apparently embedded in different clades, indicating that the two species are not closely related.
Phylogenetic analysis of 17 plastomes within Aster and its related genera revealed that A. yaoshanensis is sister to A. ageratoides, and they form a powerful clade (BS = 100 in Figure 3), which was sister clade to the clade comprising with six Aster species (BS = 100). At the same time, Heteroplexis is not monophyletic and embedded within the Aster clade, which is sister clade to above eight Aster species (including A. yaoshanensis, BS = 100). This is consistent with Duan’s findings (Duan et al., 2022). Therefore, the Aster clade had strong support, and A. yaoshanensis should be merged into Aster.
Pappus length, inflorescence, and habit were often considered to be important diagnostic characters in previous taxonomic treatments of Aster and its related genera (Li et al., 2012; Norbert et al., 2017).
Pappus length. In Asteraceae, sepals usually degenerate to pappus to help the dispersal of fruits or seeds, and the pappus of Aster plants are varied in morphology (Chen et al., 2011; Norbert et al., 2017). Therefore, it is one of the important features in the classification of Aster and its related genera (Chen, 1986; Li et al., 2012). Based on the length of pappus, these plants are divide into four types (I–IV): none pappus, short pappus, long pappus, and longer pappus. The result indicated that type IV with a probability of 98% was the common ancestral trait (Figure 5). A. yaoshanensis have longer pappus, belonging to the typical type IV.
Inflorescence. In genus Aster, the inflorescences belong to capitula (Chen et al., 2011), and the evolution of inflorescence is diverse (Li et al., 2012). Based on the number of capitulum and the way of inflorescences branching, Aster and its related genera could be divided into seven types (I–VII), and the legend can be seen in Figure 6 for details. The type of inflorescence is one of the important identification morphological characters of A. yaoshanensis as a new species, we define it as Type VII. Compared with A. handelii (I) and A. homochlamydeus (III) of the same subclade, A. yaoshanensis has more numbers of capitula, which mostly grows in the branch ends and leaf axils. A. jishouensis (VI) is very similar to A. yaoshanensis, but inflorescences have only 1–4 capitula. In addition, the ancestral reconstruction result showed that type I evolved independently at least 10 times in this taxa, type II originated from type III, and the evolutionary transformation may have occurred more than once.
Habit. In our analysis, we divided the habit of Aster and its relatives into dwarf herb, tall herb, subshrub, and shrub (Chen et al., 2011; Norbert et al., 2017; Jabbour et al., 2018). Phylogenetic analysis confirmed that tall herbs were the most probable ancestral feature of these plants (96%, Figure 7). Like A. jishouensis, A. yaoshanensis belongs to the tall herb, which may be the result of the evolution of the two plants to adapt to similar habitats. In addition, we found that dwarf herbs evolved independently at least nine times, subshrubs four times, and shrubs only twice.
Based on the multi-methods analysis, A. yaoshanensis is indicated as a new species. We assembled and analyzed the whole plastome analysis and understood the structural characteristics of the whole plastome. Phylogenetic analyses of ITS sequence with 71 species of Aster and its related genera and the whole plastome with 17 species of Aster further revealed the evolutionary relationship of A. yaoshanensis. The results showed that A. yaoshanensis is located in the Aster clade. Results of morphological comparison and ancestral traits reconstruction showed that A. yaoshanensis and its some morphological related Aster spp., especially A. jishouensis, are similar, but there are some significant differences. For example, the former has more capitula.
The results of this study can provide references for future studies on species identification, phylogeny and characteristic evolution of Aster. At the same time, the analysis of phylogeny, morphological comparison and ancestral reconstruction in this study still have some limitations. In future studies, we need to expand the sample size and obtain more morphological data to increase the availability of data and more comprehensively analyze and discuss the phylogenetic and evolutionary characteristics of Aster.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, OL461705.1 https://www.ncbi.nlm.nih.gov/, ON120847.1.
XZ: Conceptualization, Writing – original draft. KQ: Investigation, Writing – review & editing. TL: Data curation, Methodology, Writing – review & editing. TQ: Data curation, Methodology, Writing – review & editing. JL: Data curation, Methodology, Writing – review & editing. GZ: Conceptualization, Writing – original draft. BL: Conceptualization, Writing – original draft. PL: Supervision, Writing – review & editing. ZF: Investigation, Supervision, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study was financially supported by the National Natural Science Foundation of China (No. 32000158, 31970225), the National Science and Technology Fundamental Resources Investigation Program of China (No. 2021XJKK0702), the Foundation of Sustainable Development Research Center of Resources and Environment of Western Sichuan, Sichuan Normal University (No. 2020CXZYHJZX03), Key Laboratory of Chemistry in Ethnic Medicinal Resources (Yunnan Minzu University), State Ethnic Affairs Commission & Ministry of Education (No. MZY2301) and Laboratory equipment research projects, Sichuan Normal University (No. SYJS20220014).
The authors thank the curator and relevant staff of the PE herbarium who granted us access to their collections and photos and Dr. Caifei Zhang (Wuhan Botanical Garden, the Chinese Academy of Sciences), Mr. Meng Wei, and Dr. Haihua Hu (Institute of Botany, the Chinese Academy of Sciences), for their critical comments and great help. We also thank Dr. Jiangtao Xiao (Sichuan Normal University) for drawing distribution map and reviewer for constructive criticism to the original manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Bankevich, A., Nurk, S., Antipov, D., Gurevich, A. A., Dvorkin, M., Kulikov, A. S., et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477. doi: 10.1089/cmb.2012.0021
Blasco, F. A., Rubite, R. R., Cortes, J. C., Alejandro, G. J. D. (2021). Begonia lanuzaensis (sect. Petermannia, Begoniaceae) a new species from Surigao del Sur, Mindanao Island, Philippines. Phytotaxa 523, 203–207. doi: 10.11646/phytotaxa.523.3.1
Brouillet, L., Lowrey, T. K., Urbatsch, L., Karaman-Castro, V., Sancho, G., Wagstaff, S., et al. (2009). “Astereae,” in Systematics, evolution and biogeography of the Compositae. Eds. Funk, V. A., Susanna, A., Stuessy, T., Bayer, R. (IAPT, Vienna), 449–490.
Chen, Y. L. (1986). Systematic notes on the genus Miyamayomena Kitam. (Compositae). Bull. Botanical Res. 6, 37–46.
Chen, Y. L., Brouillet, L., Semple, J. C. (2011). “Aster L,” in Flora of China, vol. 20-21 . Eds. Wu, Z. Y., Raven, P. H., Hong, D. Y. (Science Press, Beijing), 574–632.
Doyle, J. J., Doyle, J. L. (1987). A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochemical Bulletin. 19, 11–15.
Duan, N., Deng, L., Zhang, Y., Shi, Y., Liu, B. (2022). Comparative and phylogenetic analysis based on chloroplast genome of Heteroplexis (Compositae), a protected rare genus. BMC Plant Biol. 22, 605. doi: 10.1186/s12870-022-04000-1
Feng, J. Y., Wu, Y. Z., Wang, R. R., Xiao, X. F., Wang, R. H., Qi, Z. C., et al. (2021). The complete chloroplast genome of balsam aster (Aster ageratoides Turcz. var. scaberulus (Miq.) Ling., Asteraceae). Mitochondrial DNA Part B. 6, 2464–2465. doi: 10.1080/23802359.2021.1955030
Fu, Z. X., Jiao, B. H., Nie, B., Zhang, G. J., Gao, T. G. (2016). A comprehensive generic-level phylogeny of the sunflower family: Implications for the systematics of Chinese Asteraceae. J. Syst. Evol. 54, 416–437. doi: 10.1111/jse.12216
Ibáñez, S. T., Muñoz-Schick, M., Scherson, R. A., Moreira-Muñoz, A. (2022). A new species of Diplostephium (Asteraceae, Astereae) from the Atacama Desert, Chile. PhytoKeys 215, 51–63. doi: 10.3897/phytokeys.215.89175
IUCN (2024) Guidelines for using the IUCN Red List categories and criteria. Version 16. Prepared by the Standards and Petitions Committee. Available online at: https://www.iucnredlist.org/resources/redlistguidelines (Accessed March 1, 2024).
Jabbour, F., Gaudeul, M., Lambourdière, J., Ramstein, G., Hassanin, A., Labat, J. N., et al. (2018). Phylogeny, biogeography and character evolution in the tribe Desmodieae (Fabaceae: Papilionoideae), with special emphasis on the New Caledonian endemic genera. Mol. Phylogenet. Evol. 118, 108–121. doi: 10.1016/j.ympev.2017.09.017
Kearse, M., Moir, R., Wilson, A., Stones-Havas, S., Cheung, M., Sturrock, S., et al. (2012). Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28, 1647–1649. doi: 10.1093/bioinformatics/bts199
Lewis, P. O. (2001). A likelihood approach to estimating phylogeny from discrete morphological character data. Syst. Biol. 50, 913–925. doi: 10.1080/106351501753462876
Li, W. P., Yang, F. S., Jivkova, T., Yin, G. S. (2012). Phylogenetic relationships and generic delimitation of Eurasian Aster (Asteraceae: Astereae) inferred from ITS, ETS and trnL-F sequence data. Ann. Bot. 109, 1341–1357. doi: 10.1093/aob/mcs054
Lowe, T. M., Eddy, S. R. (1997). tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25, 955–964. doi: 10.1093/nar/25.5.0955
Maddison, W. P. (2008) Mesquite: A modular system for evolutionary analysis. Version 3.81. Available online at: http://www.mesquiteproject.org/.
Miller, M. A., Pfeiffer, W., Schwartz, T. (2010). Creating the CIPRES science gateway for inference of large phylogenetic trees. 2010 Gateway Computing Environments Workshop (GCE) IEEE. 1–8. doi: 10.1109/GCE.2010.5676129
Nesom, G. L. (1994). Review of the taxonomy of Aster sensu lato (Asteraceae: Astereae), emphasizing the New World species. Phytologia 77, 141–297.
Nesom, G. L., Robinson, H. (2007). “Astereae,” in The Families and Genera of Vascular Plants, vol. 8 . Eds. Kadereit, J. W., Jeffrey, C. (Springer, Germany, Berlin), 284–342.
Norbert, K., Mercè, G. C., Ronny, S., Christoph, O., Rob, S., Antony, M., et al. (2017). Systematics of Libinhania, a new endemic genus of Gnaphalieae (Asteraceae) from the Socotra archipelago (Yemen), inferred from plastid, low-copy nuclear and nuclear ribosomal DNA loci. Bot. J. Linn. Soc 183, 373–412. doi: 10.1093/botlinnean/bow013
Noyes, R. D., Rieseberg, L. H. (1999). ITS sequence data support a single origin for North American Astereae (Asteraceae) and reflect deep geographic divisions in Aster s.l. Am. J. Bot. 86, 398–412. doi: 10.2307/2656761
Panero, J. L., Freire, S. E., Ariza Espinar, L., Crozier, B. S., Barboza, G. E., Cantero, J. J. (2014). Resolution of deep nodes yields an improved backbone phylogeny and a new basal lineage to study early evolution of Asteraceae. Mol. Phylogenet. Evol. 80, 43–53. doi: 10.1016/j.ympev.2014.07.012
Panero, J. L., Funk, V. A. (2002). Toward a phylogenetic subfamilial classification for the Compositae (Asteraceae). Proc. Biol. Soc Wash. 115, 909–922.
Panero, J. L., Funk, V. A. (2008). The value of sampling anomalous taxa in phylogenetic studies: major clades of the Asteraceae revealed. Mol. Phylogenet. Evol. 47, 757–782. doi: 10.1016/j.ympev.2008.02.011
Posada, D., Crandall, K. A. (1998). MODELTEST: testing the model of DNA substitution. Bioinformatics 14, 817–818. doi: 10.1093/bioinformatics/14.9.817
Qu, X. J., Moore, M. J., Li, D. Z., Yi, T. S. (2019). PGA: a software package for rapid, accurate, and flexible batch annotation of plastomes. Plant Methods 15, 50. doi: 10.1186/s13007-019-0435-7
Rambaut, A. (2012) FigTree v 1.4.0. Available online at: http://tree.bio.ed.ac.uk/software/Figtree/.
Ronquist, F., Teslenko, M., van der Mark, P., Ayres, D. L., Darling, A., Höhna, S., et al. (2012). MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61, 539–542. doi: 10.1093/sysbio/sys029
Stamatakis, A., Hoover, P., Rougemont, J. (2008). A rapid bootstrap algorithm for the RAxML Web servers. Syst. Biol. 57, 758–771. doi: 10.1080/10635150802429642
Thiers, B. (2024) Index Herbariorum: A global directory of public herbaria and associated staff. New York Botanical Garden’s Virtual Herbarium. Available online at: https://sweetgum.nybg.org/science/ih/ (Accessed March 1, 2024).
Wagensommer, R. P., Perrino, E. V., Albano, A., Medagli, P., Passalacqua, N. G. (2016). Lectotypification of four Lacaita’s names in the genus Centaurea (Asteraceae). Phytotaxa 269, 54–58. doi: 10.11646/phytotaxa.269.1.7
Wagensommer, R. P., Venanzoni, R. (2021). Geranium lucarinii sp. nov. and re-evaluation of G. kikianum (Geraniaceae). Phytotaxa 489, 252–262. doi: 10.11646/phytotaxa.489.3.2
Wang, X. Y., Liu, Y. (2023). The complete chloroplast genome of Aster altaicus Willd. (Asteraceae: Aster) and phylogenetic analysis. Mitochondrial DNA Part B. 8, 819–822. doi: 10.1080/23802359.2023.2238358
Wei, Y. G. (2018). “Aster L,” in The Distribution and Conservation Status of Native Plants in Guangxi, China. Ed. Wei, Y. G. (China Forestry Publishing House, Beijing), 493–494.
Wei, Y. G., Wen, F., Fu, L. F., Wang, W. T. (2013). Three new species of Elatostema J.R. Forst & G. Forst. (Urticaceae) in limestone caves from Guangxi and Guizhou, China. Bangl. J. Plant Taxon. 20, 1–8. doi: 10.3329/bjpt.v20i1.15458
Wick, R. R., Schultz, M. B., Zobel, J., Holt, K. E. (2015). Bandage: interactive visualization of de novo genome assemblies. Bioinformatics 31, 3350–3352. doi: 10.1093/bioinformatics/btv383
Xiong, Y. C., Li, Z., Xiao, J. W., Li, W. P. (2019). Aster saxicola sp. nov. (Asteraceae, Astereae), a new species from Guizhou, China: evidence from morphological, molecular and cytological studies. Nord. J. Bot. 37, 1–2. doi: 10.1111/njb.02364
Zhang, G. J., Hu, H. H., Gao, T. G., Gilbert, M. G., Jin, X. F. (2019). Convergent origin of the narrowly lanceolate leaf in the genus Aster-with special reference to an unexpected discovery of a new Aster species from East China. Peer J. 7, e6288. doi: 10.7717/peerj.6288
Zhang, G. J., Hu, H. H., Zhang, C. F., Tian, X. J., Peng, H., Gao, T. G. (2015). Inaccessible biodiversity on limestone cliffs: Aster tianmenshanensis (Asteraceae), a new critically endangered species from China. PloS One 10, e0134895. doi: 10.1371/journal.pone.0134895
Keywords: Asteraceae, Aster, Guangxi, new species, phylogeny
Citation: Zheng X, Qin K, Li T, Qu T, Luo J, Zhang G, Li B, Li P and Fu Z (2024) A new species, Aster yaoshanensis (Asteracae, Astereae), from Guangxi (China), based on morphology and molecular phylogenetic data. Front. Plant Sci. 15:1367917. doi: 10.3389/fpls.2024.1367917
Received: 09 January 2024; Accepted: 15 March 2024;
Published: 02 April 2024.
Edited by:
Robert Philipp Wagensommer, Free University of Bozen-Bolzano, ItalyReviewed by:
Enrico Vito Perrino, International Centre for Advanced Mediterranean Agronomic Studies, ItalyCopyright © 2024 Zheng, Qin, Li, Qu, Luo, Zhang, Li, Li and Fu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Pan Li, cGFubGlfemp1QDEyNi5jb20=; Zhixi Fu, ZnV6eDIwMTdAc2ljbnUuZWR1LmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.