- 1School of Marine Biology and Fisheries, State Key Laboratory of Marine Resource Utilization in South China Sea, Hainan University, Haikou, Hainan, China
- 2Institute of Hydrobiology, Chinese Academy of Sciences, Wuhan, Hubei, China
Microorganisms are important members of seagrass bed ecosystems and play a crucial role in maintaining the health of seagrasses and the ecological functions of the ecosystem. In this study, we systematically quantified the assembly processes of microbial communities in fragmented seagrass beds and examined their correlation with environmental factors. Concurrently, we explored the relative contributions of species replacement and richness differences to the taxonomic and functional β-diversity of microbial communities, investigated the potential interrelation between these components, and assessed the explanatory power of environmental factors. The results suggest that stochastic processes dominate community assembly. Taxonomic β-diversity differences are governed by species replacement, while for functional β-diversity, the contribution of richness differences slightly outweighs that of replacement processes. A weak but significant correlation (p < 0.05) exists between the two components of β-diversity in taxonomy and functionality, with almost no observed significant correlation with environmental factors. This implies significant differences in taxonomy, but functional convergence and redundancy within microbial communities. Environmental factors are insufficient to explain the β-diversity differences. In conclusion, the assembly of microbial communities in fragmented seagrass beds is governed by stochastic processes. The patterns of taxonomic and functional β-diversity provide new insights and evidence for a better understanding of these stochastic assembly rules. This has important implications for the conservation and management of fragmented seagrass beds.
1 Introduction
Assembly mechanisms are closely linked to changes in community structure, diversity, and the ecological functions they support (Fukami, 2015). This phenomenon has been extensively studied in various ecosystems, such as soil microbes (Wang et al., 2022; Cao et al., 2023), bacterioplankton (Dai et al., 2017), intertidal communities (Loke and Chisholm, 2023), and plants (Peter and Harrington, 2018; Tanunchai et al., 2023). Assembly processes are influenced by various environmental factors, with community turnover cycles limiting exploration. Microorganisms, which are often the most abundant group within systems, play a crucial role in material and energy cycling (Tarquinio et al., 2019; Lemay et al., 2021; Nguyen et al., 2022). Due to their short life cycles, rapid community turnover, and sensitivity to the environment, they are excellent subjects for investigating community assemblage (Vogel et al., 2021a, Vogel et al., 2021b). Seagrass bed ecosystems are known for their highly diverse habitats and host abundant microbial communities, providing habitats for various marine organisms and numerous ecosystem services (Yarnall et al., 2022; Han et al., 2023). These characteristics make seagrass beds an ideal environment for studying microbial community assembly mechanisms (Santos et al., 2016; Du et al., 2020). However, due to factors like climate change, pollution influx, and commercial fishing, seagrass beds in China are rapidly declining (Santos et al., 2016; Xu et al., 2022), resulting in significant losses of ecosystem services they provide (Xiao et al., 2020; Li et al., 2022). Despite ongoing attention and research on related issues, our comprehension of microbial community assembly in fragmented seagrass beds remains limited (Du et al., 2020; Hu et al., 2021), constraining our ability to develop applications for seagrass bed conservation and restoration.
The widely embraced null model method, which quantifies community assembly using beta-mean nearest taxon distance (β-NTI) based on phylogenetic distance and species abundance (Stegen et al., 2012), is designed to elucidate spatial alterations in phylogenetic β-diversity (Zhou and Ning, 2017). Assembly processes, which encompass deterministic and stochastic components, provide insights into variations in community structure composition and diversity dynamics (Prosser et al., 2007). Currently, the latter has garnered significant attention and research focus in seagrass habitats (Crump et al., 2018; Liu et al., 2022; Kardish and Stachowicz, 2023). However, investigations into community assembly, particularly concerning microbial communities, remain relatively limited (Leibold et al., 2017; Zhou and Ning, 2017; Fahrig, 2019). Understanding changes in microbial communities is crucial for seagrass growth (Milbrandt et al., 2008), health (Vandenkoornhuyse et al., 2015), and the ecological functions they provide (Liu et al., 2017; Zhang et al., 2023). Additionally, related research indicates that microbial communities are seldom uniformly distributed in the environment and their spatial variation is primarily dependent on diffusion (Vincent et al., 2021). However, it is currently unknown how microbial function varies as a result.
β-diversity refers to the variations in species composition between different communities, delineated by the processes of species replacement and richness differences. Methods for partitioning β-diversity aim to discern the roles of these processes in total β-diversity, investigating their collective impact on species distribution patterns across diverse spatiotemporal dimensions (Baselga, 2010; Podani and Schmera, 2011). Both taxonomic and functional β-diversity can be quantified between communities, offering valuable insights into the rules governing micro-community assembly (Luo et al., 2023). However, analysis of taxonomic data is inherently limited as it does not provide information about the ecological roles of species (McGill et al., 2006). The use of functional data aids in obtaining a more nuanced comprehension of biodiversity patterns and processes (Swenson et al., 2012; Villéger et al., 2012), which is a well-established application in related research (Swenson et al., 2011; Cardoso et al., 2014).
Since the introduction of β-diversity patterns (Williams, 1996; Lennon et al., 2001), the diversity patterns of plant and animal species have been extensively studied, resulting in various hypotheses (Shen et al., 2020). More recently, attention has also been given to microbial communities, particularly in soils (Li et al., 2020), with fewer investigations in aquatic environments (Sun et al., 2024). However, there is still a lack of understanding regarding fragmented seagrass bed habitats. These patches may resemble individual islands, but their dynamics are significantly influenced by the ebb and flow of seawater. Therefore, classical island ecology theories, such as the area-diversity theory, may not be directly applicable to seagrass bed patches (Chisholm et al., 2016). Additionally, some studies suggest that ecological and evolutionary processes, rather than island biogeography theories, primarily govern species assembly on islands (HilleRisLambers et al., 2012; Mittelbach and Schemske, 2015). Research on microbial communities in seawater indicates that species replacement significantly contributes to β-diversity patterns (Lyu et al., 2023).
This work randomly selected seagrass patches of varying sizes, inculding Enhalus acoroides and Thalassia hemprichi. Microorganisms attached to the leaves and those present in the surrounding seawater, were systematically sampled. The study analyzed the patterns of β-diversity, considering both taxonomic and functional aspects, and used null models to investigate the mechanisms of microbial community assembly in fragmented seagrass beds. The hypotheses are as follows: (1) The phylogenetic relationships of metapopulation provide a more comprehensive background for community assembly within fragmented seagrass bed habitats. Stochastic processes predominantly shape the constitution of local species pools, thereby influencing on community assembly. (2) Replacement processes are the primary driver of β-diversity differences in microbial communities. (3) Microbial functions play a significant role in community assembly through mechanisms that involve environmental adaptation and niche differentiation. Despite taxonomic differences, functional convergence suggests functional redundancy.
2 Materials and methods
2.1 Sampling
Microbial samples were collected in July 2022 from a fragmented seagrass bed area located between Nanhai Village and Baozhi Village (19.4741°N – 19.4784°N, 110.8096°E – 110.8192°E) in Wenchang City, within the South China Sea. A total of 43 seagrass patches were selected, with 27 patches dominated by Enhalus acoroides, 13 patches dominated by Thalassia hemprichii, and 3 patches co-dominated by both seagrasses. Each patch had designated sampling points at the center and the edge, where microbial samples were collected from seagrass leaf and the surrounding seawater (Figure 1A). Specifically, microbial samples from the seawater and seagrass leaves were collected at the center and edge of E. acoroides patches (including patches with both seagrasses). For T. hemprichii patches, also including 3 patches with both seagrasses, microbial samples were collected from seagrass leaves at the center and edge. In total, we obtained 60 seawater microbial samples, 60 microbial samples attached to E. acoroides, and 32 microbial samples attached to T. hemprichii, with half at the center and half at the edge of patches. Environmental variables, such as total nitrogen (TN), total phosphorus (TP), and phosphate (PO43-), as well as patch characteristics, such as seagrass biomass, patch area, and isolation, were measured. Both environmental variables and patch characteristics are classified as environmental factors.
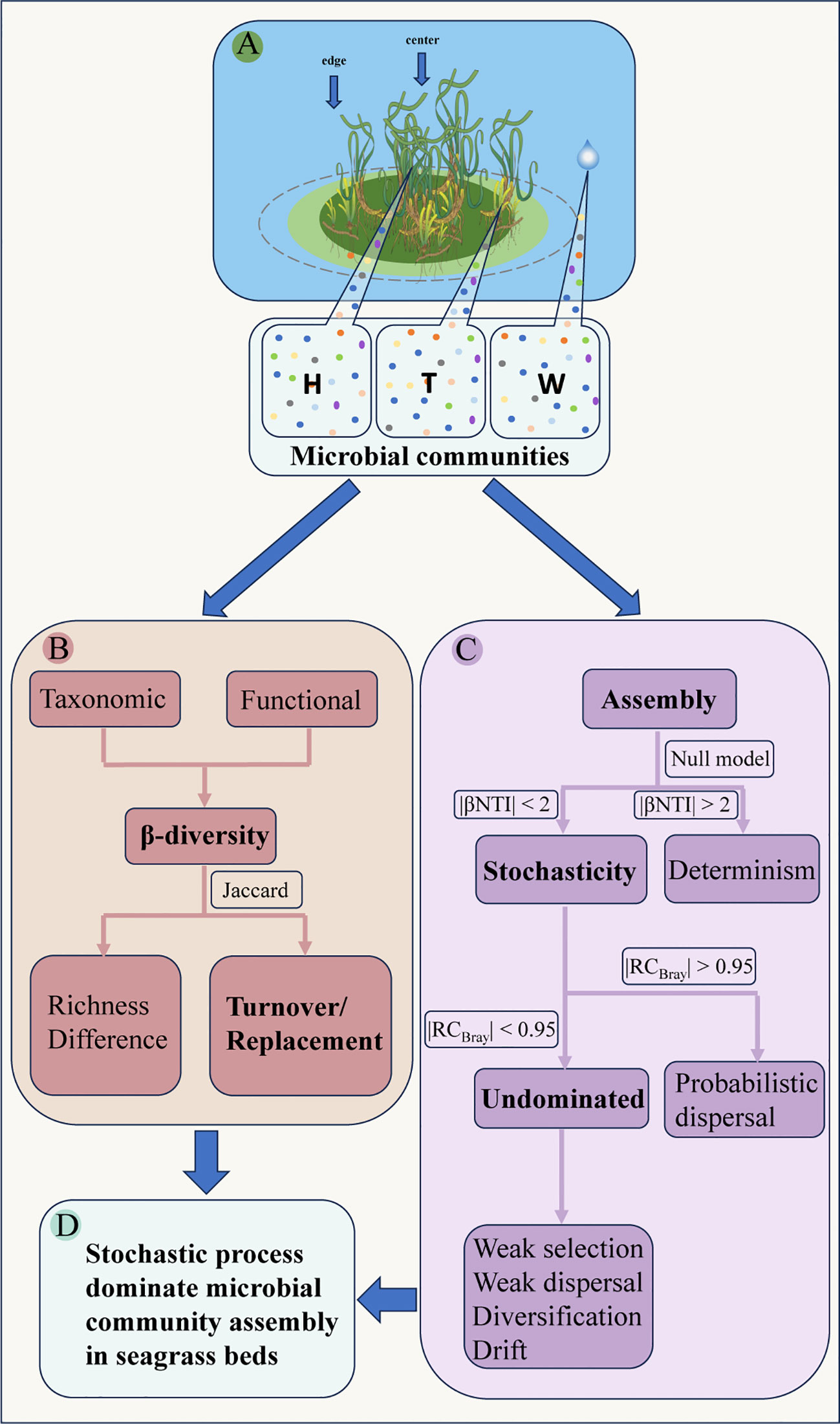
Figure 1 Schematic diagram of microbial community assembly and β-diversity patterns in seagrass beds. (A, conceptual diagram of sampling; B, anaylsis of β-diversity; C, anaylsis of assembly process; D, the main conclusion; W, surrounding seawater; H, E. acoroides; T, T. hemprichii).
Microbial samples were filtered onto 0.22 μm diameter filter membranes, with seawater initially filtered through a 5 μm before further filtration to 0.22 μm, with a volume of 1 L. Microorganisms from seagrass leaves were gently brushed into distilled water and subsequently filtered onto the filter membranes. The membranes were stored at -80°C until genomic DNA extraction. Another 1 L seawater sample was collected and transported promptly to the laboratory for measuring TN, TP, and PO43- concentrations, using spectrophotometer and molybdenum blue methods (Murphy and Riley, 1962). Seagrass biomass, including both aboveground and belowground parts, was weighted at wet and dry conditions. Patch morphological assessments and patch areas measurements were conducted using unmanned aerial vehicles (DJI Mini 4 Pro) and GPS markers.
2.2 DNA sequencing
Microbial DNA was extracted from the filter membranes using a Thermo genomic DNA extraction kit. A biological company was contracted for high-throughput sequencing services. The OD260/280 values of the DNA samples were within the range of 1.8 to 2.0. The concentration, determined using a Nanodrop 2000 spectrophotometer, exceeded 50 μg/μL, with a total amount surpassing 2 μg. The DNA samples mentioned above were diluted to 1 ng/μL with sterile water and used as a template for PCR amplification of the 16S rRNA target gene. High-fidelity enzymes and primers with barcode tags were used for the amplification. After amplification, PCR products of equal volumes were mixed based on their concentration, and 2% agarose gel electrophoresis was conducted for detection. Subsequently, the target bands were purified using gel extraction kits from QIAGEN. Finally, the raw sequencing data was obtained though sequencing with the Hiseq2500 platform and library preparation using Illumina kits.
The processes of assembly, quality control, and filtering are necessary to convert data into valid data. Subsequently, the Uparse software (Edgar, 2013) was used with a sequence similarity threshold of 97% (Schloss and Handelsman, 2005) to obtain operational taxonomic units (OTUs). After taxonomic annotation based on representative sequences and normalization of OTUs abundance information (Green et al., 2017) based on the minimum sequence count, a total of 4320 OTUs were obtained from seawater, 6739 from E. acoroides, and 3157 from T. hemprichii, respectively. The taxa, classified at the Family level by OTUs, were used for data analysis in this study. All sequences utilized in this study are publicly accessible at the NCBI Sequence Read Archive (http://www.ncbi.nlm.nih.gov/Traces/sra) under accession PRJNA1048314.
2.3 Analysis of microbial community assembly
The microbial community assembly was quantified using a well-accepted null model (Chase et al., 2011) that evaluates the deterministic and stochastic processes based on the abundance of microbial taxonomic groups and their phylogenetic evolutionary distances. This analysis involved 999 randomizations. The phylogenetic relatedness of taxa within the community was evaluated using nearest taxon index (NTI) and its calculation was performed using the “picante” package (Webb et al., 2008). A homogeneous assembly is indicated by NTI values >2 or mean NTI > 0 (at the regional level), while heterogeneous assembly is suggested by NTI values < -2 or mean NTI < 0 (at the regional level) (Kembel, 2009; Stegen et al., 2013).
The β-NTI (Webb et al., 2008; Kembel et al., 2011) and RCBray (Stegen et al., 2013) values were used to quantify deterministic and stochastic processes. These values were calculated using the “pNST” function in the “NST” package. Assembly of community is considered deterministic if |β-NTI| > 2, while stochastic processes dominate assembly if |β-NTI| < 2 (Kembel et al., 2011). The RCBray value provides a more precise understanding of stochastic processes. If |RCBray| is greater than 0.95, it is likely that community assembly is dominated by probabilistic dispersal. On the other hand, if |RCBray| is less than 0.95, community assembly can be attributed to undominated processes such as weak selection, weak dispersal, diversification, and drift (Stegen et al., 2015) (Figure 1C). Stegen et al. (2013) developed a statistical framework to describe the ecological processes of community assembly (Figure 1C).
To explore the impact of environmental factors on community assembly, the mantel test based on matrix data was employed to observe the correlation between the two. Euclidean distances for both environmental variables and patch characteristics were calculated using the “dist” function of the “vegan” R package, resulting in matrix data. The significance threshold was set at p < 0.05. The patch isolation was calculated based on the coordinates between sampling sites using the “distm” function in the “geosphere” R package.
2.4 Analysis of microbial community β-diversity patterns
A β-diversity partitioning approach based on the Jaccard dissimilarity index (Podani and Schmera, 2011; Carvalho et al., 2012; Cardoso et al., 2014) was used to calculate three paired β-diversity metrics for both microbial taxonomy and ecological functional groups (referred to as “functional”): total diversity (Total), turnover or replacement (Repl), and richness differences (RichDiff) (Figure 1B). Ternary plots were utilized to illustrate the varying contributions of different metrics to β-diversity differences. The differences between taxonomic and functional were compared for each diversity metric, and their correlation was explored using the mantel test. Additionally, the mantel test was used to indicate the relationship between the β-diversity metrics and environmental factors.
The β-diversity analysis in microbial taxonomy employed the data obtained from the classification of OTUs at the Family level, which yielded 416 taxa in seawater, 464 taxa in E. acoroides, and 364 taxa in T. hemprichii. Ecological function groups of microbial communities in seagrass beds were obtained by comparing OTUs classification tables with the functional annotation of prokaryotic taxa (FAPROTAX) (http://www.zoology.ubc.ca/louca/FAPROTAX/) using Python scripts. These functional groups effectively explained and predicted the functions related to the cycling of elements such as carbon (C), nitrogen (N), phosphorus (P), and sulfur (S) (Louca et al., 2016; Kumar et al., 2018; Picazo et al., 2021). A total of 57 ecological functional groups were identified from microbial communities of seawater and E. acoroides, and 48 in T. hemprichii. These data were used for β-diversity analysis in microbial functionality.
The analyses were conducted using the R software (Team, 2023). The β-diversity calculation used the “adespatial” package, the mantel test used the “vegan” package, and functional annotations were based on the Linux system and accomplished using the “FAPROTAX” package.
3 Results
3.1 Stochasticity, undominated process specifically, determines microbial community assembly
The average NTI values of microbial communities in both seagrass species are higher than those in the surrounding seawater, and both are greater than zero (Supplementary Figure S1). This suggests a homogeneous assembly of communities in seagrass beds and a closer phylogenetic relationship in the microbial communities attached to seagrasses. The assembly of microbial communities in seagrass beds is primarily governed by stochastic processes, accounting for over 75%, as evidenced by β-NTI values (Supplementary Figure S2; Figure 2A). The contribution of deterministic processes in seawater and T. hemprichii patches is higher at the edge than at the center, while the opposite is observed in E. acoroides patches (Figure 2A). Further identification revealed that determinism is primarily attributed to heterogeneous selection, whereas stochasticity is mainly attributed to undominated processes, including weak selection, weak dispersal, diversification, and drift (Figure 2B).

Figure 2 The proportion of different assembly processes of microbial communities in seagrass beds (A, the proportion of deterministic and stochastic processes; B, the proportion of components of deterministic and stochastic processes; W, surrounding seawater; H, E. acoroides; T, T. hemprichii; C, center of the patch; E, edge of the patch).
Mantel test results (Supplementary Table S1) indicate a significant correlation between the assembly processes of microbial communities (β-NTI) at the centers (p < 0.05) of E. acoroides patches with seagrass biomass and a partially significant correlation (p < 0.08) at the edges. At the centers of T. hemprichii patches, community assembly is partially correlated with TN and isolation, while at the edges, it is significantly correlated with PO43- (Supplementary Table S1). We analyzed whether there were patterns of variation in dominant taxa, defined as taxa with abundances exceeding 1% across all sampling sites, within the community in fragmented seagrass bed patches (Supplementary Figure S3). The results showed no discernible patterns, suggesting that dominant taxa do not dictate the assembly of microbial communities in fragmented seagrass beds.
3.2 Replacement is responsible for the β-diversity difference of microbial communities
The analysis of β-diversity partition indicates that the differences in microbial community composition are primarily driven by species replacement, which contributes significantly more to β-diversity than richness differences (Figure 3; Table 1). Furthermore, no significant differences were observed between seawater and the two seagrass species, nor between the center and edge of patches. In seawater microbial communities, species replacement processes accounted for an average of 73.96%, while richness differences processes accounted for 26.04% (Table 1). In the microbial communities attached to E. acoroides, species replacement processes were 73.42% and richness differences processes were 26.58%. For the microbial communities attached to T. hemprichii, species replacement processes accounted for 71.65% and richness differences processes accounted for 28.35% (Table 1). The mantel test revealed that the species replacement processes of most communities did not exhibit significant correlation with environmental factors (Supplementary Table S2). Specifically, the replacement process was significantly and positively correlated with patch area, isolation, and PO43- in the central community of E. acoroides patches. TP and TN were significantly and positively correlated with the replacement process at the edge of T. hemprichii patches (Supplementary Table S2).
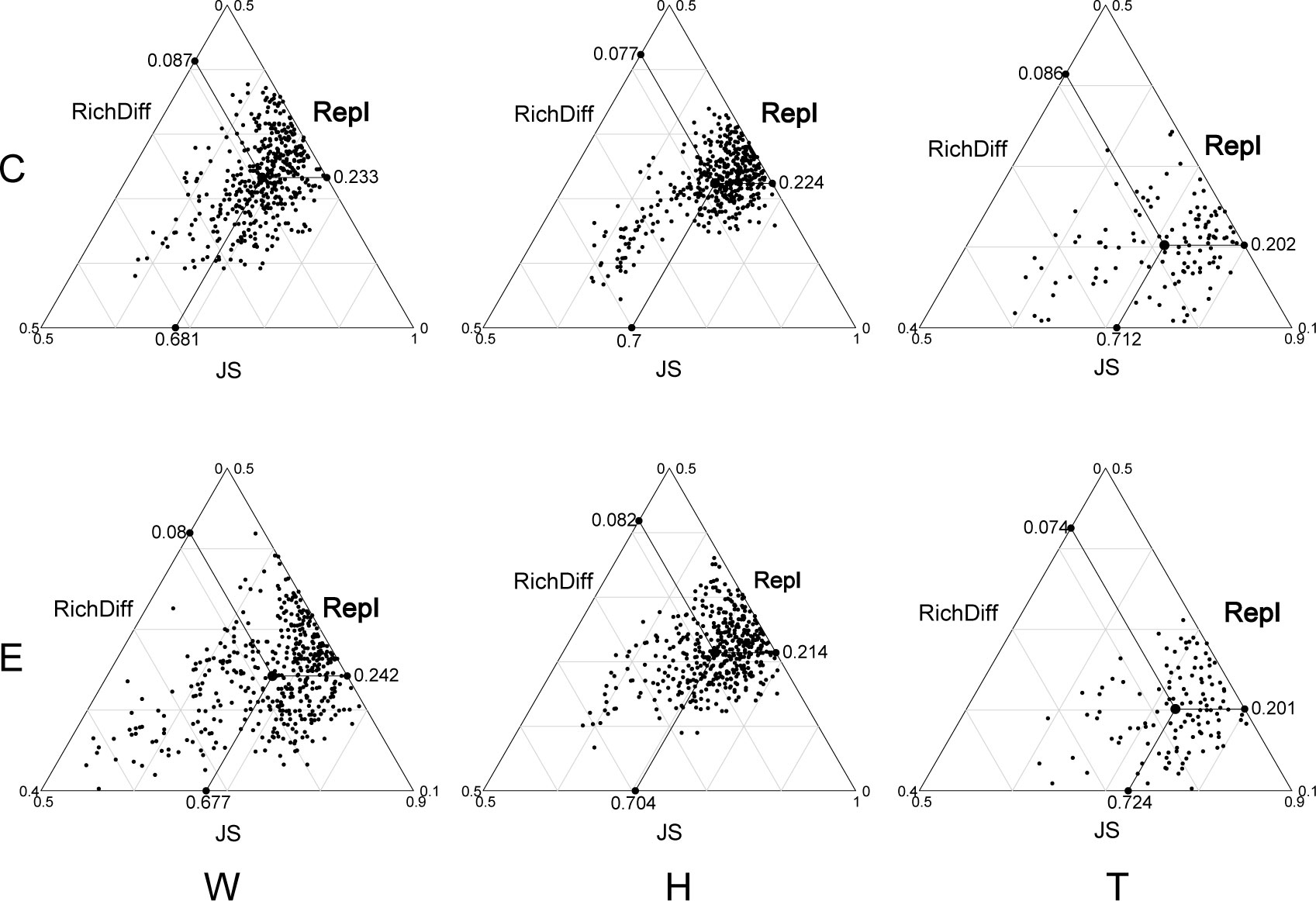
Figure 3 Triangular plots of taxonomic β-diversity comparisons (using Jaccard dissimilarity index) in seagrass beds among all sites (Its position is determined by a triplet of values from the JS (Jaccard Similarity), Repl (replacement) and RichDiff (richness difference) matrices. The mean values of JS, Repl and RichDiff are shown. Each point represents a pair of sites with 435 pairs sites in the W and H, 120 pairs sites in the T. W, surrounding seawater; H, E. acoroides; T, T. hemprichii; C, center of the patch; E, edge of the patch.).
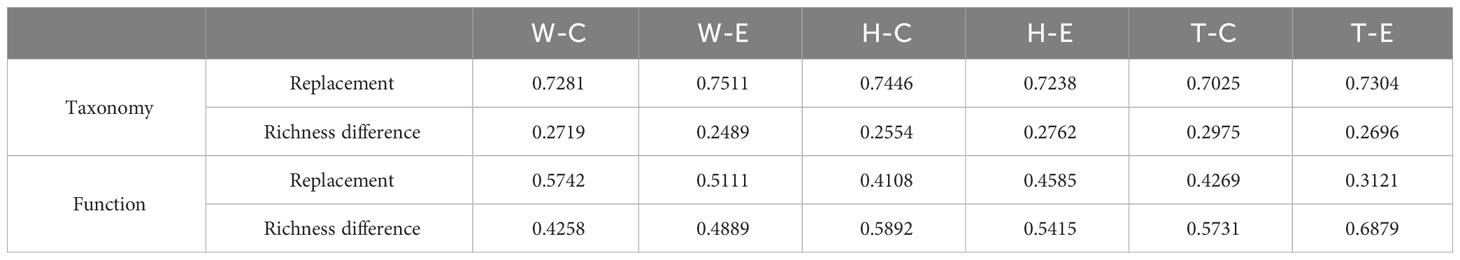
Table 1 The contribution of replacement and richness differences to β-diversity differences (W, surrounding seawater; H, E. acoroides; T, T. hemprichii; C, center of the patch; E, edge of the patch).
Among the top 15 most abundant ecological function groups, the majority are associated with the C cycle (Supplementary Figure S4). However, their relative abundance varies. Chemoheterotrophy and aerobic_chemoheterotrophy are most abundant in the microbial communities of seawater and T. hemprichii, whereas the opposite is true for E. acoroides (Supplementary Figure S4). In contrast to differences in community composition, microbial functional differences in seawater are mainly contributed by replacement processes, whereas in seagrass richness differences play a predominant role (Figure 4; Table 1). Specifically (Table 1), in seawater microbial communities, replacement processes contribute an average of 54.27%, while richness difference processes contribute 45.73%. For the communities attached to E. acoroides, replacement processes contribute an average of 43.47% and richness differences processes contribute 56.53%. In the microbial communities attached to T. hemprichii, replacement processes were 37%, and richness differences processes contributed 63%. Replacement processes of most communities were not significantly correlated with environmental factors, except for a significant positive correlation with PO43- and patch isolation in seawater at the edge of patches (Supplementary Table S3). There was no significant correlation between all richness difference processes and environmental factors (Supplementary Table S4).
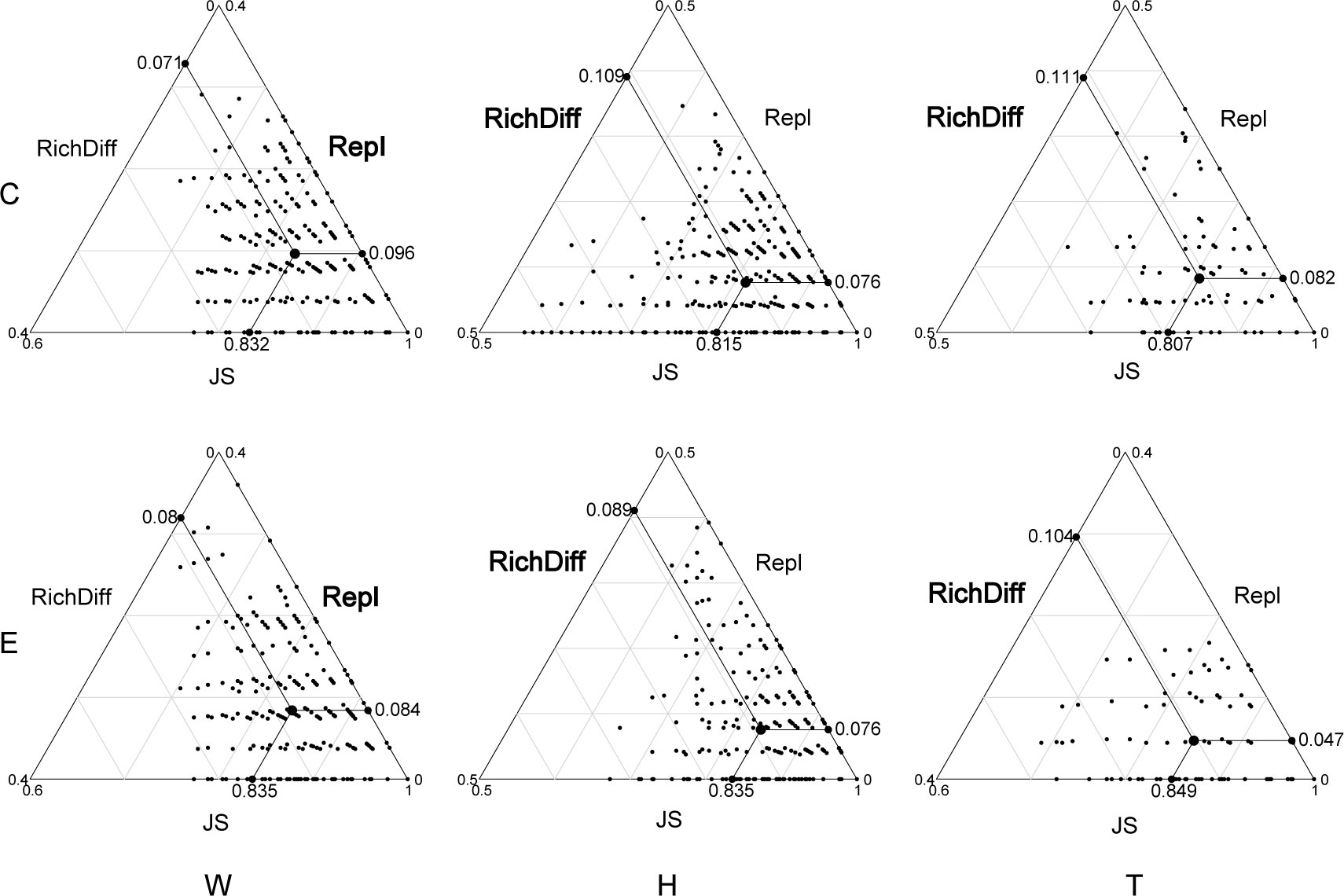
Figure 4 Triangular plots of functional β-diversity comparisons (using Jaccard dissimilarity index) for microbes in seagrass beds among all sites. (Its position is determined by a triplet of values from the JS (Jaccard Similarity), Repl (replacement) and RichDiff (richness difference) matrices. The mean values of JS, Repl and RichDiff are shown. Each point represents a pair of sites with 435 pairs sites in the W and H, 120 pairs sites in the T. W, surrounding seawater; H, E. acoroides; T, T. hemprichii; C, center of the patch; E, edge of the patch.).
A comparison was conducted between taxonomy and functionality for the three components of β-diversity: total β-diversity, replacement, and richness differences. The results indicate that both total β-diversity and replacement exhibit higher values in taxonomy than in functionality, while richness differences are relatively similar between the two (Figure 5). Mantel analyses were performed to examine the relationship between taxonomic and functional β-diversity. Significant positive correlations were found for total β-diversity in seawater, including both the center and edge. Only at the edge of E. acoroides patches, total β-diversity exhibited a significant positive correlation. At the center and edge of T. hemprichii patches, significant positive correlations of total β-diversity and richness difference were observed. However, replacement processes existed a partially significant positive correlation at the center (Table 2).
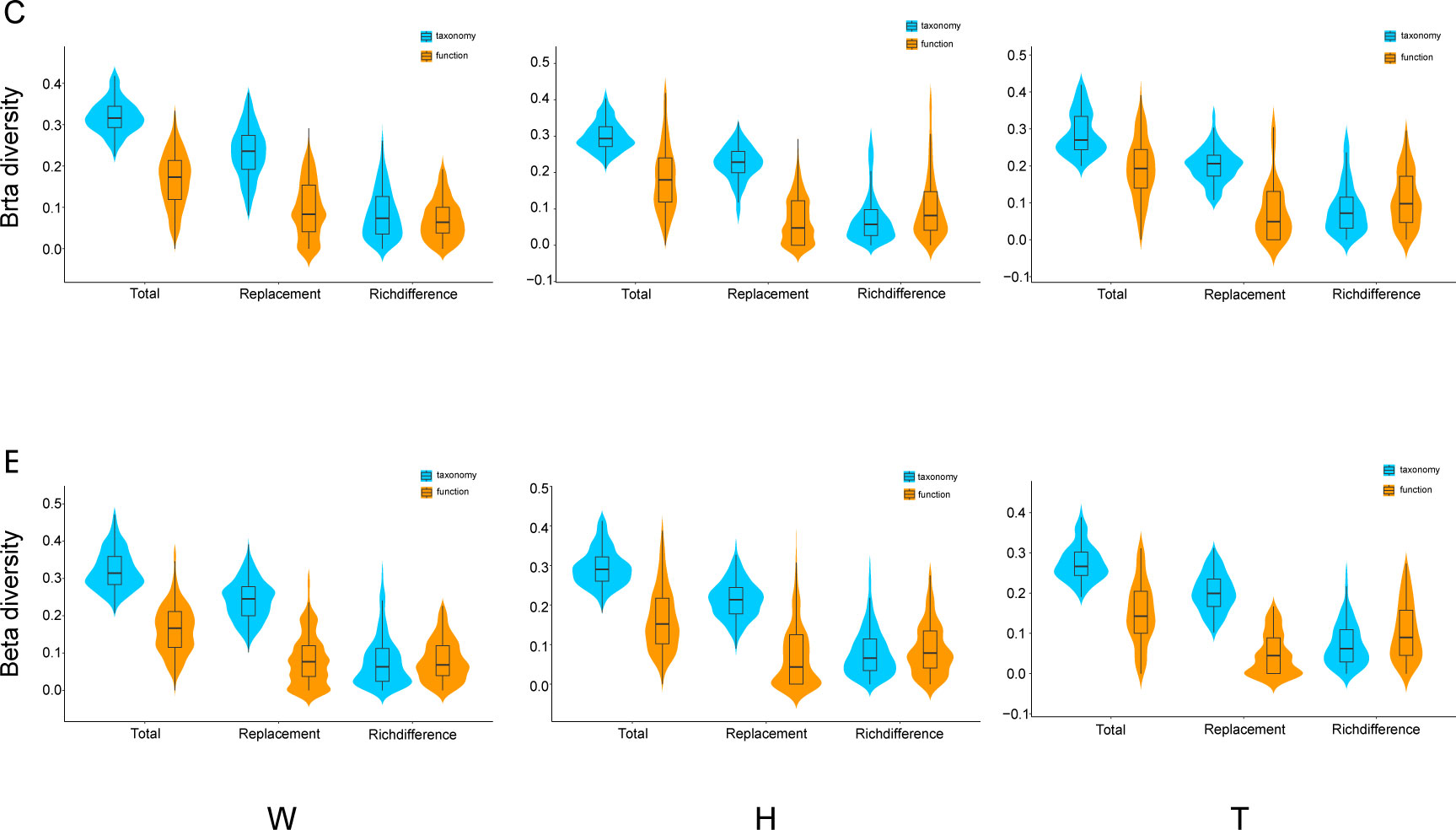
Figure 5 Comparisons of β-diversity components (total, replacement and richness difference) for taxonomy and ecological function groups of microorganisms in seagrass beds (W, surrounding seawater; H, E. acoroides; T, T. hemprichii; C, center of the patch; E, edge of the patch.).
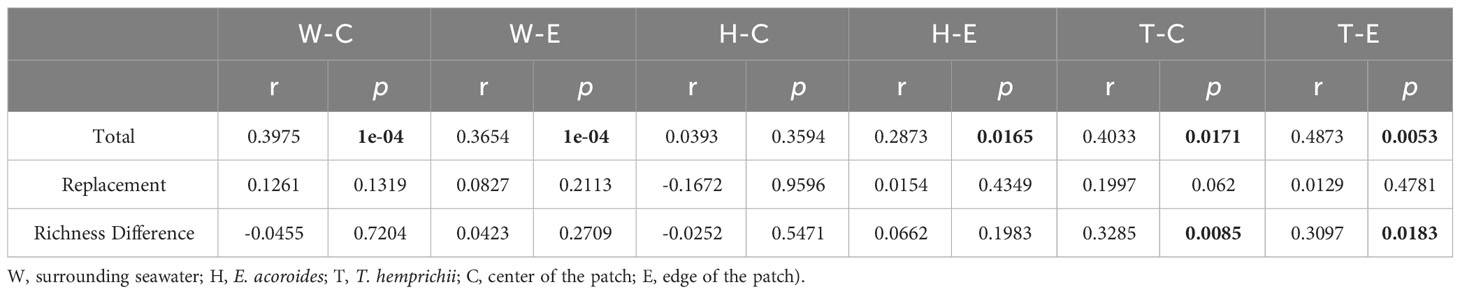
Table 2 Mantel test between the β-diversity components (total, replacement and richness difference) and taxonomy, ecological function groups of microorganisms in seagrass beds (Bold numbers indicates significant correlation, p < 0.05.
4 Discussion
While it is widely recognized that both deterministic and stochastic processes impact community assembly (Ofiţeru et al., 2010; Chase and Myers, 2011; Zhou and Ning, 2017), there is an ongoing debate regarding their relative importance in governing community variations (Zhou et al., 2013; Vellend et al., 2014). Notably, the significance of stochastic processes has historically been understated in this discourse (Zhou and Ning, 2017; Li and Gao, 2023). The study highlights the significant role of stochastic processes in microbial community assembly in fragmented seagrass beds, in contrast to traditional island ecology perspectives. Additionally, the study reveals differences in the patterns of functional and taxonomic β-diversity, including the relative contributions of replacement and richness difference processes and their responses to environmental factors. This aids in a better understanding and elucidation of these stochastic rules. On the basis of the dominant factor of stochastic assembly, perhaps focusing on community function in future management and restoration is an ingenious perspective.
4.1 The stochastic assembly processes in microbial communities
The NTI results reflect the phylogenetic clustering relationships within communities. An average NTI across multiple communities at the regional level demonstrates the homogeneous assembly of microbial communities across the region, rather than relying on a single NTI. In other words, the phylogenetic relationships of metapopulation provide background information for the microbial community assembly in seagrass beds. According to Stegen et al. (2013), undominant processes in stochasticity, such as weak selection, weak dispersal, diversification, and drift, are responsible for the assembly of microbial communities. The mantel results indicate a lack of significant correlation between β-NTI and environmental factors, supporting this notion. The findings suggest that community assembly in seagrass bed patches is minimally explained by environmental variables and patch characteristics, which is consistent with previous research on microbes (Ramette and Tiedje, 2007; Zhou et al., 2008; Graham et al., 2016).
Generally, diversification and dispersal are often considered components of stochastic processes, especially in microbial communities (Chase and Myers, 2011), as microbial dispersal is typically passive (Nemergut et al., 2013) and passive dispersal is regarded as random (Vellend et al., 2014). Drift refers to the random changes in the relative abundance of different species within a community over time, driven by inherent stochastic processes such as birth and death (Vellend, 2010; Nemergut et al., 2013). It is undeniably stochastic (Vellend et al., 2014). The challenge lies in the inability to directly measure these stochastic processes in community ecology. Furthermore, in microbial communities, drift is likely associated with functional redundancy and plays a crucial role in shaping community structure (Ofiţeru et al., 2010; Zhou et al., 2013), as the common functional redundancy in microbial communities can induce drift by increasing neutrality (Allison and Martiny, 2008; Delgado-Baquerizo et al., 2016). It is clear that there are dynamic exchanges between microorganisms attached to seagrass and the species pool in the surrounding seawater (Fahimipour et al., 2017). Seagrasses prevent or selectively filter microbial colonization for various reasons. Combining our results, it can be inferred that stochastic processes have a significant impact on the assembly of attached microbial communities by predominantly shaping the formation of local species pools.
4.2 Microbial community assembly from a β-diversity patterns perspective
Understanding the contributions of the two components of β-diversity is crucial for comprehending how communities change, and provides a new perspective on community assembly (Carvalho et al., 2012). From a taxonomic standpoint, differences in microbial community β-diversity are mainly driven by species replacement. From a functional perspective, replacement processes contribute slightly more than richness differences in seawater microbial communities, which is contrary to the observed pattern in communities attached to seagrasses. These findings align with related research on seawater microbial communities (Sun et al., 2024). Furthermore, we observed that the proportion of dominant taxa did not exhibit consistent patterns with increasing or decreasing patch area of seagrass beds. This statement contrasts with the area-diversity theory in island ecology, which suggests that species diversity is proportional to island size (Li et al., 2020). The area-diversity theory is not applicable to fragmented seagrass bed patches; instead, replacement processes dominate the β-diversity differences in microbial communities. Previous studies suggest that species dispersal may cause species turnover among communities (Swenson et al., 2011). Dispersal is common in microbial communities (Wu et al., 2021).
Taxonomically, β-diversity is primarily influenced by replacement processes, while the contribution of richness differences to functional β-diversity slightly surpasses that of replacement. This implies a significant convergence of functional traits among communities (Carvalho et al., 2020), reinforcing the concept of heightened functional redundancy within microbial communities. The taxonomic dissimilarity and functional convergence suggest turnover among microbial taxa with similar functions (Wu et al., 2021). There is a weak yet significant correlation between taxonomic and function-based β-diversity components, indicating that these two facets provide complementary ecological information (Devictor et al., 2010). This pattern has been observed across diverse biological groups, including fish, planktonic plants, and zooplankton, as well as various habitats, encompassing different types of water bodies (Gianuca et al., 2018; Wu et al., 2018; Hill et al., 2019). Furthermore, there is limited correlation between β-diversity components from both taxonomic and functional perspectives and environmental factors, including environmental variables and patch characteristics. These findings suggest that environmental factors play a minimal role in driving replacement processes within microbial communities. The collective findings demonstrate that the β-diversity patterns provide a new perspective and reference for better understanding the stochastic assembly rules of microbial communities in fragmented seagrass beds.
Research indicates a complex and intimate interaction between plants and their associated microorganisms, whether in the rhizosphere microbiome (Gabriel Nuto et al., 2023; Tang et al., 2023) or the phyllosphere microbiome (Lehnen et al., 2016; Ettinger et al., 2017). Seagrasses are no exception, as changes in the structure and diversity of attached microbial communities, along with their responses to the environment, significantly impact the health, growth, and functions of seagrasses (Chen et al., 2022; O’Connor et al., 2022). For instance, alterations in rhizosphere microbial communities play a pivotal role in the successful transplantation of seagrasses (Christiaen et al., 2013). Undoubtedly, the ecosystem services provided by seagrass beds, such as high productivity and a robust carbon sequestration capacity (Fourqurean et al., 2012; Han et al., 2023), predominantly rely on seagrasses. Therefore, delving into community assembly mechanisms is crucial for understanding of the intricate relationship between microbial communities and seagrasses. This knowledge can guide the development of targeted management strategies to protect and restore seagrass bed ecosystems.
5 Conclusion
The assembly processes of microbial communities attached to seagrass play a crucial role in the growth and health of seagrasses and, consequently, the stability, diversity, and multifunctionality of seagrass bed ecosystems. We emphasize the dominance of stochastic rules, particularly undominant processes such as weak selection, weak dispersal, diversification, and drift, in the assembly of these communities (Figure 1D). The taxonomic-based differences in β-diversity are driven by replacement processes, whereas in functional β-diversity, the contribution of richness differences slightly surpasses that of replacement processes. Although there are significant taxonomic distinctions among microbial communities in seagrass beds, functional convergence and redundancy are observed. Environmental factors, indicative of deterministic processes, provide minimal explanation for the patterns of β-diversity. In summary, stochastic processes govern the assembly of microbial communities in fragmented seagrass beds. The taxonomic and functional β-diversity patterns provide a new perspective and support for better comprehending these stochastic assembly rules. This study provides foundational evidence for the mechanisms of microbial community assembly in fragmented seagrass beds, offering novel insights for the conservation and management of seagrass beds.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics statement
The manuscript presents research on animals that do not require ethical approval for their study.
Author contributions
XN: Conceptualization, Formal analysis, Writing – original draft, Visualization. WR: Formal analysis, Methodology, Writing – review & editing. CX: Data curation, Methodology, Writing – review & editing. RW: Data curation, Methodology, Writing – review & editing. JZ: Formal analysis, Methodology, Writing – review & editing. HW: Conceptualization, Data curation, Funding acquisition, Methodology, Project administration, Supervision, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This research was supported by the National Natural Science Foundation of China (Grant No. 32001151 and 42377469) and Hainan University Start-up Funding for Scientific Research (KYQD (ZR)-23, 086) and Hainan University Start-up Funding for Scientific Research (KYQD (ZR)-23, 087).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2024.1367773/full#supplementary-material
References
Allison, S. D., Martiny, J. B. H. (2008). Resistance, resilience, and redundancy in microbial communities. Proc. Natl. Acad. Sci. 105, 11512–11519. doi: 10.1073/pnas.0801925105
Baselga, A. (2010). Partitioning the turnover and nestedness components of beta diversity. Global Ecol. Biogeography 19, 134–143. doi: 10.1111/j.1466-8238.2009.00490.x
Cao, J., Zhang, Y., Dai, G., Cui, K., Wu, X., Qin, F., et al. (2023). The long-acting herbicide mesosulfuron-methyl inhibits soil microbial community assembly mediating nitrogen cycling. J. Hazard Mater 443, 130293. doi: 10.1016/j.jhazmat.2022.130293
Cardoso, P., Rigal, F., Carvalho, J. C., Fortelius, M., Borges, P. A. V., Podani, J., et al. (2014). Partitioning taxon, phylogenetic and functional beta diversity into replacement and richness difference components. J. Biogeography 41, 749–761. doi: 10.1111/jbi.12239
Carvalho, J. C., Cardoso, P., Gomes, P. (2012). Determining the relative roles of species replacement and species richness differences in generating beta-diversity patterns. Global Ecol. Biogeography 21, 760–771. doi: 10.1111/j.1466-8238.2011.00694.x
Carvalho, J. C., Malumbres-Olarte, J., Arnedo, M. A., Crespo, L. C., Domenech, M., Cardoso, P. (2020). Taxonomic divergence and functional convergence in Iberian spider forest communities: Insights from beta diversity partitioning. J. Biogeography 47, 288–300. doi: 10.1111/jbi.13722
Chase, J. M., Kraft, N. J. B., Smith, K. G., Vellend, M., Inouye, B. D. (2011). Using null models to disentangle variation in community dissimilarity from variation in α-diversity. Ecosphere 2(2):art24. doi: 10.1890/ES10-00117.1
Chase, J. M., Myers, J. A. (2011). Disentangling the importance of ecological niches from stochastic processes across scales. Philos. Trans. R Soc. Lond B Biol. Sci. 366, 2351–2363. doi: 10.1098/rstb.2011.0063
Chen, J., Zang, Y., Yang, Z., Qu, T., Sun, T., Liang, S., et al. (2022). Composition and functional diversity of epiphytic bacterial and fungal communities on marine macrophytes in an intertidal zone. Front. Microbiol. 13. doi: 10.3389/fmicb.2022.839465
Chisholm, R. A., Fung, T., Chimalakonda, D., O'dwyer, J. P. (2016). Maintenance of biodiversity on islands. Proc. R. Soc. B: Biol. Sci. 283, 20160102. doi: 10.1098/rspb.2016.0102
Christiaen, B., Mcdonald, A., Cebrian, J., Ortmann, A. C. (2013). Response of the microbial community to environmental change during seagrass transplantation. Aquat. Bot. 109, 31–38. doi: 10.1016/j.aquabot.2013.03.008
Crump, B. C., Wojahn, J. M., Tomas, F., Mueller, R. S. (2018). Metatranscriptomics and amplicon sequencing reveal mutualisms in seagrass microbiomes. Front. Microbiol. 9. doi: 10.3389/fmicb.2018.00388
Dai, W., Zhang, J., Tu, Q., Deng, Y., Qiu, Q., Xiong, J. (2017). Bacterioplankton assembly and interspecies interaction indicating increasing coastal eutrophication. Chemosphere 177, 317–325. doi: 10.1016/j.chemosphere.2017.03.034
Delgado-Baquerizo, M., Giaramida, L., Reich, P. B., Khachane, A. N., Hamonts, K., Edwards, C., et al. (2016). Lack of functional redundancy in the relationship between microbial diversity and ecosystem functioning. J. Ecol. 104, 936–946. doi: 10.1111/1365-2745.12585
Devictor, V., Mouillot, D., Meynard, C., Jiguet, F., Thuiller, W., Mouquet, N. (2010). Spatial mismatch and congruence between taxonomic, phylogenetic and functional diversity: the need for integrative conservation strategies in a changing world. Ecol. Lett. 13, 1030–1040. doi: 10.1111/j.1461-0248.2010.01493.x
Du, J., Hu, W., Nagelkerken, I., Sangsawang, L., Loh, K. H., Ooi, J. L.-S., et al. (2020). Seagrass meadows provide multiple benefits to adjacent coral reefs through various microhabitat functions. Ecosystem Health Sustainability 6, 1812433. doi: 10.1080/20964129.2020.1812433
Edgar, R. C. (2013). UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 10, 996–998. doi: 10.1038/nmeth.2604
Ettinger, C. L., Voerman, S. E., Lang, J. M., Stachowicz, J. J., Eisen, J. A. (2017). Microbial communities in sediment from Zostera marina patches, but not the Z. marina leaf or root microbiomes, vary in relation to distance from patch edge. PeerJ 5, e3246. doi: 10.7717/peerj.3246
Fahimipour, A. K., Kardish, M. R., Lang, J. M., Green, J. L., Eisen, J. A., Stachowicz, J. J. (2017). Global-scale structure of the eelgrass microbiome. Appl. Environ. Microbiol. 83 (12), e03391-16. doi: 10.1128/AEM.03391-16
Fahrig, L. (2019). Habitat fragmentation: A long and tangled tale. Global Ecol. Biogeography 28, 33–41. doi: 10.1111/geb.12839
Fourqurean, J. W., Duarte, C. M., Kennedy, H., Marbà, N., Holmer, M., Mateo, M. A., et al. (2012). Seagrass ecosystems as a globally significant carbon stock. Nat. Geosci. 5, 505–509. doi: 10.1038/ngeo1477
Fukami, T. (2015). Historical contingency in community assembly: integrating niches, species pools, and priority effects. Annu. Rev. Ecology Evolution Systematics 46, 1–23. doi: 10.1146/annurev-ecolsys-110411-160340
Gabriel Nuto, N., Pedro Avelino Maia De, A., Hermano Melo, Q., Arthur Prudncio De Arajo, P., Margareth Da Silva, C., Daniel, G., et al. (2023). Bridging soil biogeochemistry and microbial communities (archaea and bacteria) in tropical seagrass meadows. Front. Mar. Sci. 10. doi: 10.3389/fmars.2023.1190497
Gianuca, A. T., Engelen, J., Brans, K. I., Hanashiro, F. T. T., Vanhamel, M., Van Den Berg, E. M., et al. (2018). Taxonomic, functional and phylogenetic metacommunity ecology of cladoceran zooplankton along urbanization gradients. Ecography 41, 183–194. doi: 10.1111/ecog.02926
Graham, E. B., Knelman, J. E., Schindlbacher, A., Siciliano, S., Breulmann, M., Yannarell, A., et al. (2016). Microbes as engines of ecosystem function: when does community structure enhance predictions of ecosystem processes? Front. Microbiol. 7. doi: 10.3389/fmicb.2016.00214
Green, S. J., Wen, C., Wu, L., Qin, Y., Van Nostrand, J. D., Ning, D., et al. (2017). Evaluation of the reproducibility of amplicon sequencing with Illumina MiSeq platform. PloS One 12 (4), e0176716. doi: 10.1371/journal.pone.0176716
Han, Q., Qiu, C., Zeng, W., Chen, S., Zhao, M., Shi, Y., et al. (2023). Sediment carbon sequestration and driving factors in seagrass beds from Hainan island and the Xisha islands. Processes 11, 456. doi: 10.3390/pr11020456
Hill, M. J., Heino, J., White, J. C., Ryves, D. B., Wood, P. J. (2019). Environmental factors are primary determinants of different facets of pond macroinvertebrate alpha and beta diversity in a human-modified landscape. Biol. Conserv. 237, 348–357. doi: 10.1016/j.biocon.2019.07.015
HilleRisLambers, J., Adler, P. B., Harpole, W. S., Levine, J. M., Mayfield, M. M. (2012). Rethinking community assembly through the lens of coexistence theory. Annu. Rev. Ecology Evolution Systematics 43, 227–248. doi: 10.1146/annurev-ecolsys-110411-160411
Hu, W., Zhang, D., Chen, B., Liu, X., Ye, X., Jiang, Q., et al. (2021). Mapping the seagrass conservation and restoration priorities: coupling habitat suitability and anthropogenic pressures. Ecol. Indic. 129, 107960. doi: 10.1016/j.ecolind.2021.107960
Kardish, M. R., Stachowicz, J. J. (2023). Local environment drives rapid shifts in composition and phylogenetic clustering of seagrass microbiomes. Sci. Rep. 13, 3673. doi: 10.1038/s41598-023-30194-x
Kembel, S. W. (2009). Disentangling niche and neutral influences on community assembly: assessing the performance of community phylogenetic structure tests. Ecol. Lett. 12, 949–960. doi: 10.1111/j.1461-0248.2009.01354.x
Kembel, S. W., Eisen, J. A., Pollard, K. S., Green, J. L. (2011). The phylogenetic diversity of metagenomes. PloS One 6, e23214. doi: 10.1371/journal.pone.0023214
Kumar, A., Ng, D. H. P., Wu, Y., Cao, B. (2018). Microbial community composition and putative biogeochemical functions in the sediment and water of tropical granite quarry lakes. Microbial Ecol. 77, 1–11. doi: 10.1007/s00248-018-1204-2
Lehnen, N., Marchant, H. K., Schwedt, A., Milucka, J., Lott, C., Weber, M., et al. (2016). High rates of microbial dinitrogen fixation and sulfate reduction associated with the Mediterranean seagrass Posidonia oceanica. Systematic Appl. Microbiol. 39, 476–483. doi: 10.1016/j.syapm.2016.08.004
Leibold, M. A., Chase, J. M., Ernest, S. K. M. (2017). Community assembly and the functioning of ecosystems: how metacommunity processes alter ecosystems attributes. Ecology 98, 909–919. doi: 10.1002/ecy.1697
Lemay, M. A., Chen, M. Y., Mazel, F., Hind, K. R., Starko, S., Keeling, P. J., et al. (2021). Morphological complexity affects the diversity of marine microbiomes. ISME J. 15, 1372–1386. doi: 10.1038/s41396-020-00856-z
Lennon, J. J., Koleff, P., Greenwood, J. J. D., Gaston, K. J. (2001). The geographical structure of British bird distributions: diversity, spatial turnover and scale. J. Anim. Ecol. 70, 966–979. doi: 10.1046/j.0021-8790.2001.00563.x
Li, Q., Jin, R., Ye, Z., Gu, J., Dan, L., He, J., et al. (2022). Mapping seagrass meadows in coastal. Geocarto Int. 37, 12602–12617. doi: 10.1080/10106049.2022.2070672
Li, S.-P., Wang, P., Chen, Y., Wilson, M. C., Yang, X., Ma, C., et al. (2020). Island biogeography of soil bacteria and fungi: similar patterns, but different mechanisms. ISME J. 14, 1886–1896. doi: 10.1038/s41396-020-0657-8
Li, T. Q., Gao, J. Y. (2023). Attribution of dispersal limitation can better explain the assembly patterns of plant microbiota. Front. Plant Sci. 14. doi: 10.3389/fpls.2023.1168760
Liu, S., Jiang, Z., Zhang, J., Wu, Y., Huang, X., Macreadie, P. I. (2017). Sediment microbes mediate the impact of nutrient loading on blue carbon sequestration by mixed seagrass meadows. Sci. Total Environ. 599-600, 1479–1484. doi: 10.1016/j.scitotenv.2017.05.129
Liu, S., Trevathan-Tackett, S. M., Jiang, Z., Cui, L., Wu, Y., Zhang, X., et al. (2022). Nutrient loading decreases blue carbon by mediating fungi activities within seagrass meadows. Environ. Res. 212, 113280. doi: 10.1016/j.envres.2022.113280
Loke, L. H. L., Chisholm, R. A. (2023). Unveiling the transition from niche to dispersal assembly in ecology. Nature 618, 537–542. doi: 10.1038/s41586-023-06161-x
Louca, S., Parfrey, L. W., Doebeli, M. (2016). Decoupling function and taxonomy in the global ocean microbiome. Science 353, 1272–1277. doi: 10.1126/science.aaf4507
Luo, W., Wang, Y., Cahill, J. F., Luan, F., Zhong, Y., Li, Y., et al. (2023). Root-centric β diversity reveals functional homogeneity while phylogenetic heterogeneity in a subtropical forest. Ecology 105 (1), e4189. doi: 10.1002/ecy.4189
Lyu, Y., Zhang, J., Chen, Y., Li, Q., Ke, Z., Zhang, S., et al. (2023). Distinct diversity patterns and assembly mechanisms of prokaryotic microbial sub-community in the water column of deep-sea cold seeps. J. Environ. Manage. 348, 119240. doi: 10.1016/j.jenvman.2023.119240
McGill, B. J., Enquist, B. J., Weiher, E., Westoby, M. (2006). Rebuilding community ecology from functional traits. Trends Ecol. Evol. 21, 178–185. doi: 10.1016/j.tree.2006.02.002
Milbrandt, E. C., Greenawalt-Boswell, J., Sokoloff, P. D. (2008). Short-term indicators of seagrass transplant stress in response to sediment bacterial community disruption. Botanica Marina 51, 103–111. doi: 10.1515/BOT.2008.020
Mittelbach, G. G., Schemske, D. W. (2015). Ecological and evolutionary perspectives on community assembly. Trends Ecol. Evol. 30, 241–247. doi: 10.1016/j.tree.2015.02.008
Murphy, J., Riley, J. P. (1962). A modified single solution method for the determination of phosphate in natural waters. Anal. Chim. Acta 27, 678–681. doi: 10.1016/S0003-2670(00)88444-5
Nemergut, D. R., Schmidt, S. K., Fukami, T., O'neill, S. P., Bilinski, T. M., Stanish, L. F., et al. (2013). Patterns and processes of microbial community assembly. Microbiol. Mol. Biol. Rev. 77, 342–356. doi: 10.1128/MMBR.00051-12
Nguyen, X. V., Phan, T. T. H., Cao, V. L., Nhat, N. T. N., Nguyen, T. H., Nguyen, X. T., et al. (2022). Current advances in seagrass research: a review from Viet Nam. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.991865
O’Connor, M., Griffiths, G., Smith, R., Hessing-Lewis, M., Davis, K., Forbes, C., et al. (2022). A reciprocal transplant experiment sheds new light on a classic marine seagrass-algal symbiosis and suggests influence of epiphytic symbiont on seagrass microbiota. Aquat. Bot. 179, 103511. doi: 10.1016/j.aquabot.2022.103511
Ofiţeru, I. D., Lunn, M., Curtis, T. P., Wells, G. F., Criddle, C. S., Francis, C. A., et al. (2010). Combined niche and neutral effects in a microbial wastewater treatment community. Proc. Natl. Acad. Sci. 107, 15345–15350. doi: 10.1073/pnas.1000604107
Peter, D. H., Harrington, T. B. (2018). Effects of forest harvesting, logging debris, and herbicides on the composition, diversity and assembly of a western Washington, USA plant community. For. Ecol. Manage. 417, 18–30. doi: 10.1016/j.foreco.2018.01.045
Picazo, A., Villaescusa, J. A., Rochera, C., Miralles-Lorenzo, J., Quesada, A., Camacho, A. (2021). Functional metabolic diversity of bacterioplankton in maritime Antarctic lakes. Microorganisms 9 (10), 2077. doi: 10.3390/microorganisms9102077
Podani, J., Schmera, D. (2011). A new conceptual and methodological framework for exploring and explaining pattern in presence – absence data. Oikos 120, 1625–1638. doi: 10.1111/j.1600-0706.2011.19451.x
Prosser, J. I., Bohannan, B. J. M., Curtis, T. P., Ellis, R. J., Firestone, M. K., Freckleton, R. P., et al. (2007). The role of ecological theory in microbial ecology. Nat. Rev. Microbiol. 5, 384–392. doi: 10.1038/nrmicro1643
Ramette, A., Tiedje, J. M. (2007). Multiscale responses of microbial life to spatial distance and environmental heterogeneity in a patchy ecosystem. Proc. Natl. Acad. Sci. 104, 2761–2766. doi: 10.1073/pnas.0610671104
Santos, R. O., Lirman, D., Pittman, S. J. (2016). Long-term spatial dynamics in vegetated seascapes: fragmentation and habitat loss in a human-impacted subtropical lagoon. Mar. Ecol. 37, 200–214. doi: 10.1111/maec.12259
Schloss, P. D., Handelsman, J. (2005). ). Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Appl. Environ. Microbiol. 71, 1501–1506. doi: 10.1128/AEM.71.3.1501-1506.2005
Shen, C., Gunina, A., Luo, Y., Wang, J., He, J. Z., Kuzyakov, Y., et al. (2020). Contrasting patterns and drivers of soil bacterial and fungal diversity across a mountain gradient. Environ. Microbiol. 22, 3287–3301. doi: 10.1111/1462-2920.15090
Stegen, J. C., Lin, X., Fredrickson, J. K., Chen, X., Kennedy, D. W., Murray, C. J., et al. (2013). Quantifying community assembly processes and identifying features that impose them. ISME J. 7, 2069–2079. doi: 10.1038/ismej.2013.93
Stegen, J. C., Lin, X., Fredrickson, J. K., Konopka, A. E. (2015). Estimating and mapping ecological processes influencing microbial community assembly. Front. Microbiol. 6, 370. doi: 10.3389/fmicb.2015.00370
Stegen, J. C., Lin, X., Konopka, A. E., Fredrickson, J. K. (2012). Stochastic and deterministic assembly processes in subsurface microbial communities. ISME J. 6, 1653–1664. doi: 10.1038/ismej.2012.22
Sun, Y., Li, H., Zhang, J., Wang, H., Cui, X., Gao, X., et al. (2024). Assembly mechanisms of microbial communities in plastisphere related to species taxonomic types and habitat niches. Mar. Pollut. Bull. 198, 115894. doi: 10.1016/j.marpolbul.2023.115894
Swenson, N. G., Anglada-Cordero, P., Barone, J. A. (2011). Deterministic tropical tree community turnover: evidence from patterns of functional beta diversity along an elevational gradient. Proc. R. Soc. B: Biol. Sci. 278, 877–884. doi: 10.1098/rspb.2010.1369
Swenson, N. G., Stegen, J. C., Davies, S. J., Erickson, D. L., Forero-Montaña, J., Hurlbert, A. H., et al. (2012). Temporal turnover in the composition of tropical tree communities: functional determinism and phylogenetic stochasticity. Ecology 93, 490–499. doi: 10.1890/11-1180.1
Tang, L. Y., Zhan, L., Han, Y. A., Wang, Z. R., Dong, L., Zhang, Z. (2023). Microbial community assembly and functional profiles along the soil-oot continuum of salt-tolerant Suaeda glauca and Suaeda salsa. Front. Plant Sci. 14. doi: 10.3389/fpls.2023.1301117
Tanunchai, B., Ji, L., Schroeter, S. A., Wahdan, S. F. M., Thongsuk, K., Hilke, I., et al. (2023). Tree mycorrhizal type regulates leaf and needle microbial communities, affects microbial assembly and co-occurrence network patterns, and influences litter decomposition rates in temperate forest. Front. Plant Sci. 14. doi: 10.3389/fpls.2023.1239600
Tarquinio, F., Hyndes, G. A., Laverock, B., Koenders, A., Säwström, C. (2019). The seagrass holobiont: understanding seagrass-bacteria interactions and their role in seagrass ecosystem functioning. FEMS Microbiol. Lett. 366 (6), fnz057. doi: 10.1093/femsle/fnz057
Team, R. C. (2023). A language and environment for statistical computing (Vienna, Austria: R foundation for statistical computing).
Vandenkoornhuyse, P., Quaiser, A., Duhamel, M., Le Van, A., Dufresne, A. (2015). The importance of the microbiome of the plant holobiont. New Phytol. 206, 1196–1206. doi: 10.1111/nph.13312
Vellend, M. (2010). Conceptual synthesis in community ecology. Q Rev. Biol. 85, 183–206. doi: 10.1086/652373
Vellend, M., Srivastava, D. S., Anderson, K. M., Brown, C. D., Jankowski, J. E., Kleynhans, E. J., et al. (2014). Assessing the relative importance of neutral stochasticity in ecological communities. Oikos 123, 1420–1430. doi: 10.1111/oik.01493
Villéger, S., Miranda, J. R., Hernandez, D. F., Mouillot, D. (2012). Low functional β-Diversity despite high taxonomic β-Diversity among tropical estuarine fish communities. PloS One 7, e40679. doi: 10.1371/journal.pone.0040679
Vincent, S. G. T., Jennerjahn, T., Ramasamy, K. (2021). “Chapter 3 - Environmental variables and factors regulating microbial structure and functions,” in Microbial communities in coastal sediments. Eds. Vincent, S. G. T., Jennerjahn, T., Ramasamy, K. (Elsevier), 79–117. doi: 10.1016/B978-0-12-815165-5.00003-0
Vogel, M. A., Mason, O. U., Miller, T. E. (2021a). Composition of seagrass phyllosphere microbial communities suggests rapid environmental regulation of community structure. FEMS Microbiol. Ecol. 97 (3), fiab013. doi: 10.1093/femsec/fiab013
Vogel, M. A., Mason, O. U., Miller, T. E. (2021b). Environmental stressors alter the composition of seagrass phyllosphere microbial communities. Climate Change Ecol. 2, 100042. doi: 10.1016/j.ecochg.2021.100042
Wang, X. D., Li, Y., Yan, Z. Q., Hao, Y. B., Kang, E. Z., Zhang, X. D., et al. (2022). The divergent vertical pattern and assembly of soil bacterial and fungal communities in response to short-term warming in an alpine peatland. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.986034
Webb, C. O., Ackerly, D. D., Kembel, S. W. (2008). Phylocom: software for the analysis of phylogenetic community structure and trait evolution. Bioinformatics 24, 2098–2100. doi: 10.1093/bioinformatics/btn358
Williams, P. H. (1996). Mapping variations in the strength and breadth of biogeographic transition zones using species turnover. Proc. R. Soc. B: Biol. Sci. 263, 579–588. doi: 10.1098/rspb.1996.0087
Wu, N., Qu, Y., Guse, B., Makarevičiūtė, K., To, S., Riis, T., et al. (2018). Hydrological and environmental variables outperform spatial factors in structuring species, trait composition, and beta diversity of pelagic algae. Ecol. Evol. 8, 2947–2961. doi: 10.1002/ece3.3903
Wu, N., Zhou, S., Zhang, M., Peng, W., Guo, K., Qu, X., et al. (2021). Spatial and local environmental factors outweigh geo-climatic gradients in structuring taxonomically and trait-based β-diversity of benthic algae. J. Biogeography 48, 1842–1857. doi: 10.1111/jbi.14108
Xiao, X., Huang, Y., Holmer, M. (2020). Current trends in seagrass research in China, (2010-2019). Aquat. Bot. 166, 103266. doi: 10.1016/j.aquabot.2020.103266
Xu, C., Su, G., Zhao, K., Xu, X., Li, Z., Hu, Q., et al. (2022). Current status of greenhouse gas emissions from aquaculture in China. Water Biol. Secur. 1, 100041. doi: 10.1016/j.watbs.2022.100041
Yarnall, A. H., Byers, J. E., Yeager, L. A., Fodrie, F. J. (2022). Comparing edge and fragmentation effects within seagrass communities: A meta-analysis. Ecology 103, e3603. doi: 10.1002/ecy.3603
Zhang, J., Yang, Q., Yue, W., Yang, B., Zhou, W., Chen, L., et al. (2023). Seagrass Thalassia hemprichii and associated bacteria co-response to the synergistic stress of ocean warming and ocean acidification. Environ. Res. 236, 116658. doi: 10.1016/j.envres.2023.116658
Zhou, J., Kang, S., SChadt, C. W., Garten, C. T. (2008). Spatial scaling of functional gene diversity across various microbial taxa. Proc. Natl. Acad. Sci. 105, 7768–7773. doi: 10.1073/pnas.0709016105
Zhou, J., Liu, W., Deng, Y., Jiang, Y.-H., Xue, K., He, Z., et al. (2013). Stochastic assembly leads to alternative communities with distinct functions in a bioreactor microbial community. mBio 4 (2), e00584-12. doi: 10.1128/mbio.00584-00512
Keywords: stochastic assembly, β-diversity, replacement, attached microbial communities, seagrass bed, habitat fragmentation
Citation: Niu X, Ren W, Xu C, Wang R, Zhang J and Wang H (2024) Taxonomic and functional β-diversity patterns reveal stochastic assembly rules in microbial communities of seagrass beds. Front. Plant Sci. 15:1367773. doi: 10.3389/fpls.2024.1367773
Received: 09 January 2024; Accepted: 14 February 2024;
Published: 28 February 2024.
Edited by:
Qiang Yang, German Centre for Integrative Biodiversity Research (iDiv), GermanyReviewed by:
Yun Li, Chinese Academy of Sciences (CAS), ChinaJing Chen, Wuhan Polytechnic University, China
Copyright © 2024 Niu, Ren, Xu, Wang, Zhang and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Huan Wang, d2FuZ2h1YW5AaGFpbmFudS5lZHUuY24=
 Xiaofeng Niu
Xiaofeng Niu Wenjing Ren
Wenjing Ren Congjun Xu2
Congjun Xu2 Jingwei Zhang
Jingwei Zhang Huan Wang
Huan Wang