- 1Key Laboratory of Sugarcane Biology and Genetic Breeding, Ministry of Agriculture and Rural Affairs, National Engineering Research Center for Sugarcane, College of Agriculture, Fujian Agriculture and Forestry University, Fuzhou, Fujian, China
- 2Key Laboratory of Ministry of Education for Genetics, Breeding and Multiple Utilization of Crops, College of Crop Science, Fujian Agriculture and Forestry University, Fuzhou, China
- 3Fujian Key Laboratory of Subtropical Plant Physiology and Biochemistry, Fujian Institute of Subtropical Botany, Xiamen, Fujian, China
Introduction: Remorins (REMs) are plant-specific membrane-associated proteins that play important roles in plant–pathogen interactions and environmental adaptations. Group I REMs are extensively involved in virus infection. However, little is known about the REM gene family in sugarcane (Saccharum spp. hyrid), the most important sugar and energy crop around world.
Methods: Comparative genomics were employed to analyze the REM gene family in Saccharum spontaneum. Transcriptomics or RT-qPCR were used to analyze their expression files in different development stages or tissues under different treatments. Yeast two hybrid, bimolecular fluorescence complementation and co-immunoprecipitation assays were applied to investigate the protein interaction.
Results: In this study, 65 REMs were identified from Saccharum spontaneum genome and classified into six groups based on phylogenetic tree analysis. These REMs contain multiple cis-elements associated with growth, development, hormone and stress response. Expression profiling revealed that among different SsREMs with variable expression levels in different developmental stages or different tissues. A pair of alleles, ScREM1.5e-1/-2, were isolated from the sugarcane cultivar ROC22. ScREM1.5e-1/-2 were highly expressed in leaves, with the former expressed at significantly higher levels than the latter. Their expression was induced by treatment with H2O2, ABA, ethylene, brassinosteroid, SA or MeJA, and varied upon Sugarcane mosaic virus (SCMV) infection. ScREM1.5e-1 was localized to the plasma membrane (PM), while ScREM1.5e-2 was localized to the cytoplasm or nucleus. ScREM1.5e-1/-2 can self-interact and interact with each other, and interact with VPgs from SCMV, Sorghum mosaic virus, or Sugarcane streak mosaic virus. The interactions with VPgs relocated ScREM1.5e-1 from the PM to the cytoplasm.
Discussion: These results reveal the origin, distribution and evolution of the REM gene family in sugarcane and may shed light on engineering sugarcane resistance against sugarcane mosaic pathogens.
1 Introduction
Membrane rafts are ordered liquid nanodomains formed by lateral aggregation of sterols, sphingolipids and specific proteins on the cell plasma membrane (PM) and are important platforms for signal transduction (Mongrand et al., 2010; Otero et al., 2016; Perraki et al., 2018). Remorins (REMs), first discovered in tomato and potato, are a class of membrane raft-associated proteins unique to land plants (Farmer et al., 1989; Jacinto et al., 1993; Reymond et al., 1996). The REM protein consists of a highly variable N-terminal domain (PF03766) and a conserved C-terminal domain (PF03763) containing a coiled-coil motif and an anchor (RemCA) (Raffaele et al., 2007; Perraki et al., 2012; Yue et al., 2014; Badawi et al., 2019; Wang et al., 2022; Li et al., 2023). Although REM contains no transmembrane domain, the C-terminal coiled-coil motif and RemCA can distribute REM to the PM (Perraki et al., 2014). In addition, the palmitoylation of cysteine residues in the C-terminal domain and the phosphorylation of serine residues in the N-terminal domain contribute to the PM localization of REM proteins (Marín and Ott, 2012; Marín et al., 2012; Konrad et al., 2014; Fu et al., 2018; Gouguet et al., 2021; Legrand et al., 2023). Based on their structural characteristics, REM proteins are divided into 6 main groups (Raffaele et al., 2007). REM gene families have been found in many plant species, including poplar (Populus trichocarpa), Arabidopsis (Arabidopsis thaliana), tomato (Solanum lycopersicum), rice (Oryza sativa), wheat (Triticum aestivum), maize (Zea mays) and foxtail millet (Setaria italica) (Raffaele et al., 2007; Badawi et al., 2019; Wang et al., 2022; Li et al., 2023). Sugarcane (Saccharum spp. hybrid) is the most important sugar and energy crop in China and worldwide, but the REM gene family has not been reported for this plant.
REMs are involved in plant growth and development and in responding to biotic or abiotic stresses (Bray, 2002; Reddy et al., 2002; Nohzadeh Malakshah et al., 2007; Checker and Khurana, 2013; Li et al., 2014; Cai et al., 2018; Fu et al., 2018; Cheng et al., 2020). Overexpression of remorin GSD1 (from Group 6) in rice increased callose accumulation in the plasmodesmata and inhibited the transport of assimilates to grains (Gui et al., 2014). Overexpression of SlREM1 in tomato stimulated fruit ripening and promoted ethylene production and lycopene accumulation (Cai et al., 2018). The literature has shown that REMs respond to biological stresses such as infection by viruses (Raffaele et al., 2009; Jarsch and Ott, 2011; Perraki et al., 2012; Wu et al., 2013; Perraki et al., 2014; Fu et al., 2018; Perraki et al., 2018; Sasaki et al., 2018), bacteria (Lefebvre et al., 2010; Tóth et al., 2012; Bozkurt et al., 2014; Liang et al., 2018) and fungi (Son et al., 2015; Jamann et al., 2016; Vilakazi et al., 2017); abiotic stresses such as drought, salt and low temperature (Nohzadeh Malakshah et al., 2007; Li et al., 2012; Checker and Khurana, 2013; Yue et al., 2014; Badawi et al., 2019; Zhang et al., 2020; Gouguet et al., 2021; Wang et al., 2022); and hormones (Li et al., 2014; Gui et al., 2016; Badawi et al., 2019; Huang et al., 2019; Gouguet et al., 2021; Wang et al., 2022). However, Group 1 REMs are involved mainly in the response to virus infection. For example, OsREM1.4 of rice or NbREM1 of Nicotiana benthamiana interact with the movement protein NSvc4 of Rice stripe virus (RSV), and overexpression of OsREM1.4 or NbREM1 inhibited RSV infection (Fu et al., 2018). Overexpression of tomato StREM1.3 inhibited the 30 kD movement protein of Tobacco mosaic virus (TMV) and the intercellular movement of Potato virus Y (PVY) HC-Pro (Perraki et al., 2014). In tomato, StREM1.3 interacts with TGBp1, a movement protein of Potato virus X (PVX), and overexpression of StREM1.3 impairs PVX infection by gating the plasmodesmata (Rajamäki and Valkonen, 2009; Perraki et al., 2012, 2014). REMs can oligomerize into a scaffolding structure and cooperate with flotillins to stabilize large-scale membrane conformations and the actin cytoskeleton (Liang et al., 2018; Su et al., 2023). The oligomerization of REMs also reduced plasma membrane fluidity and plasmodesmata permeability to impair CMV infection (Huang et al., 2019). In Arabidopsis, overexpression of REM1.2 or REM1.3 inhibited TuMV infection (Cheng et al., 2020).
Sugarcane mosaic disease seriously threatens sugarcane production. The main causative agents are Sugarcane mosaic virus (SCMV; Potyvirus), Sorghum mosaic virus (SrMV, Potyvirus) and Sugarcane streak mosaic virus (SCSMV; Poacevirus), all belonging to the Potyviridae family (Xu et al., 2008; Dong et al., 2017; Yao et al., 2017; Akbar et al., 2020, 2021; Hincapie et al., 2021). These 3 pathogens are single-stranded positive RNA viruses with a genome length of approximately 10 kb that encode 2 polyproteins, which are eventually hydrolyzed into 11 mature proteins: P1, HC-Pro, P3, P3N-PIPO, 6K1, CI, 6K2, NIa-Pro, VPg, NIb and CP (Ward and Shukla, 1991; Riechmann et al., 1992; Hall et al., 1998; Urcuqui-Inchima et al., 2001; Xu et al., 2010; Li et al., 2011; Olspert et al., 2015; Wang, 2015; Olspert et al., 2016; Cheng et al., 2017; Cui et al., 2017). Among these viral proteins, VPg is involved in viral replication, translation, movement and counteraction against host RNAi through interactions with other viral proteins and host proteins (Cotton et al., 2009; Huang et al., 2010; Cheng and Wang, 2017; Li and Wang, 2018; Cheng et al., 2020; Zhai et al., 2021). For example, TuMV utilizes VPg to interact with SGS3, REM1.2 or NbNdhM to mediate the degradation of SGS3 and REM1.2 via autophagy, the 26S ubiquitin proteasome system, or the perinuclear chloroplast aggregation of NbNdhM, respectively, to promote infection (Cheng and Wang, 2017; Cheng et al., 2020; Zhai et al., 2021). However, whether sugarcane Group 1 REMs interact with the VPgs of these three viruses (SCMV, SrMV and SCSMV) has not been determined.
In this study, members of the REM family were identified in sugarcane for the first time, and their phylogeny, chromosomal localization, physical and chemical properties, gene structure, conserved mods, cis-acting elements of promoters, and collinearity within and between species were analyzed. The expression of REM gene family members in different tissues and across leaf gradients was subsequently analyzed. Because Group 1 REMs mainly respond to viral infection, we cloned a pair of REM1 alleles from the sugarcane cultivar ROC22 and designated them ScREM1.5e-1/-2 based on phylogenetic tree analysis. The expression levels and subcellular localization of REM1 alleles in different sugarcane tissues were analyzed, and the expression patterns of REM1 alleles in response to SCMV infection were analyzed. The interactions between ScREM1.5e-1/-2 and the VPg proteins of SCMV, SCSMV, and SrMV were investigated, and whether ScREM1.5e-1 or ScREM1.5e-2 self-interact or interact with each other was evaluated. Finally, the expression levels of ScREM1.5e-1/-2 under NaCl, PEG, H2O2, ABA, ETH, BR, SA and MeJA stress were analyzed. The aim of this study was to lay a foundation for the functional study of the REM genes in sugarcane and to provide potential molecular targets for engineering sugarcane resistance against sugarcane mosaic pathogens.
2 Materials and methods
2.1 Plant materials and methods
Tissue-cultured sugarcane cultivar ROC22 plantlets were cultured in a greenhouse with a 14/10-h light/dark cycle at 28°C and were individually inoculated with SCMV at the 5–6 leaf stage, as previously described (Zhang et al., 2019; Yang et al., 2021). The mock-inoculated ROC22 plantlets with 0.01 M phosphate buffer (pH 7.0) were used as the negative control. The inoculated or mock-inoculated leaves were sampled at 0 h, 6 h, 12 h, 24 h, 48 h, 5 d, 8 d and 14 d post-inoculation. For abiotic treatments, ROC22 plantlets of were grown in water for one week and then transferred to conical tubes for the following eight different treatments with a 16 h light/8 h dark cycle at 28°C. The plantlets were treated with 250 mM NaCl, 25% PEG (polyethylene glycol), 10 mM H2O2 (Su et al., 2014), 100 µM ABA, 400 mg/L ethylene solution (Chen et al., 2019) (Coolaber, Beijing, China), 25 mg/L BR (Wang et al., 2020), 5 mM SA, or 25 µM MeJA (methyl jasmonate) in 0.1% (v/v) ethanol, 0.05% (v/v) Tween-20, and were incubated for 0 h, 6 h, 12 h, 24 h and 48 h. After the treatments, three sugarcane plantlets per time point were sampled, frozen in liquid nitrogen immediately and kept at −80°C before use. Tissue samples from the leaves, roots and stem piths of 10-month-old healthy plants of ROC22 were also collected.
2.2 Identification, chromosomal localization and duplication events of the REM gene family
The genome sequence of Saccharum spontaneum was obtained from published genomic information (http://www.life.illinois.edu/ming/downloads/Spontaneum_genome) (Zhang et al., 2018). A Hidden Markov Model (HMM) of REM (Remorin_N: PF03766; Remorin_C: PF03763) was obtained from the Pfam database (http://pfam.xfam.org/) (Mistry et al., 2021) and was used to identify the REM genes in S. spontaneum. The gff3 files for S. spontaneum were downloaded (Zhang et al., 2018) to acquire the length and location information for the REM genes, then TBtools software was used to visualize the chromosomal locations of the REM genes. The chromosome color was set to black with the remaining parameters set to the default values. MCScanX software was used to analyze the replication pattern of the REM gene family with the parameters set to the default values (Wang et al., 2012).
2.3 Phylogenetic and physicochemical properties and subcellular localization
The phylogenetic tree of REM was constructed by using MEGA6 version with the neighbor−joining method and 1000 bootstrap replicates. The physicochemical properties of the REM protein, including the number of encoded amino acids, molecular weight, isoelectric point, instability index, aliphatic index and grand average of hydropathicity, were predicted by the ProtParam tool of ExPASy (https://web.expasy.org/protparam/) (Gasteiger et al., 2003). The Cell-PLoc 2.0 (http://www.csbio.sjtu.edu.cn/bioinf/Cell-PLoc-2/) website was used to predict the subcellular localization of REM proteins (Chou and Shen, 2008).
2.4 Motif and gene structure analysis
The S. spontaneum REM gene family motif information was acquired from the MEME website (https://meme-suite.org/meme/doc/meme.html) with the parameter was set to show 17 conserved motifs and the rest set as the default values (Bailey et al., 2009). The exon−intron structure information of the REM gene family was acquired from the gff3 files of S. spontaneum. The visualization of conserved motifs or gene structure of the REM gene family was conducted using TBtools software (Chen et al., 2020).
2.5 Synteny and homologous gene pairs
MCScanX software was used to analyze the synteny of the REM gene family with the parameters set to the default values (Wang et al., 2012). TBtools software was used to generate a synteny map within the genomes of S. spontaneum, sorghum (Sorghum bicolor), maize (Zea mays), wheat (Triticum aestivum), barley (Hordeum vulgare), and rice (Oryza sativa). The genome information of sorghum, maize, wheat, barley, and rice was acquired from Ensembl Plants (https://plants.ensembl.org/index.html). TBtools software was used to calculate the values of nonsynonymous (Ka)/synonymous (Ks) between homologous gene pairs based on the synteny relationship (Chen et al., 2020).
2.6 Cis-element analysis of the REM genes
The upstream 2,000 bp promoter sequences of the REM gene family members were acquired from the databases of S. spontaneum genome. The prediction of the cis-elements in the REM gene family was conducted through the PlantCARE online website (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) (Lescot et al., 2002), while the visualization of the cis-elements was conducted with TBtools software (Chen et al., 2020).
2.7 Expression profiles of REM gene families in various tissues
Sugarcane tissue-specific expression data included leaf and stem data at the seedling stage (35 d), early maturity stage (9 months) and maturity stage (12 months). Materials were taken from SES2086 sugarcane plants for 11 d. Leaves were divided into 15 segments from the base to the tip (a total of 15 cm); each segment was 1 cm long, and RNA was extracted from each leaf segment. For the leaf segment model, three representative regions of each material were taken. All SsREMs FPKMs (fragments per kilobase of transcript per million segments mapped) were used to construct heatmaps and perform cluster analysis through TBtools.
2.8 Total RNA extraction, cDNA first-strand synthesis and real-time quantitative PCR
TRIzol was used to isolate total RNA (Connolly et al., 2006). The Primer-Script® RT−PCR Kit (TaKaRa Biotechnology, Dalian, China) was used to synthesize first-strand cDNA. For the real-time quantitative PCR (RT-qPCR) analysis, three technical replicates were completed for each sample with 25S rRNA and eEF-1a were used as internal references (Iskandar et al., 2004; Guo et al., 2014; Ling et al., 2014). The specificity of each primer pair were confirmed by the melting curve analysis. The relative gene expression was calculated by using the 2−ΔΔCt method in triplicate and presented as the means ± standard error.
2.9 Expression pattern of ScREM1.5e-1/-2 in sugarcane
The ChamQ Universal SYBR qPCR Master Mix (Vazyme, Shanghai, China) was used to determine the expression patterns of ScREM1.5e-1/-2 by RT-qPCR with the sugarcane cDNA using as the templates, and 25S rRNA and eEF-1a were used as internal references (Iskandar et al., 2004; Guo et al., 2014; Ling et al., 2014) (Supplementary Table 1). The primers ScREM1.5e-1/-2-qPCR (Supplementary Table 1) were designed by the Primer Premier 5.0 software. RT−qPCR assays were conducted on the An ABI 7500 Real-time PCR System (Applied Biosystems, Foster City, CA, USA). The relative gene expression was calculated by using the 2−ΔΔCt method in triplicate and presented as the means ± standard error.
2.10 Plasmid construction
The primers used for plasmid generation are listed in Supplementary Table 1. For the Yeast two hybrid (Y2H) assays, the Y2H vectors and all DNA fragments were individually ligated via Sfi I sites. SCMV-/SCSMV-/SrMV-VPg or ScREM1.5e-1/-2 were individually cloned into the bait vector pGBKT7, and ScREM1.5e-1/-2 were individually cloned into the prey vector pGADT7. For the transient protein expression and bimolecular fluorescence complementation (BiFC) assays, all plasmids were constructed using Gateway technology as previously described (Cheng et al., 2020). All the plasmids generated in the present study were verified by sequencing.
2.11 Protein interactions as determined by Y2H, BiFC and Co-immunoprecipitation assays
For the Y2H assay, the Matchmaker Gold Yeast Two-Hybrid System (Clontech, Mountain View, CA, USA) was used according to the manufacturer’s instructions. The prey vector pGADT7 and bait vector pGBKT7 harboring the genes to be tested were cotransformed into the yeast (Saccharomyces cerevisiae) strain AH109. The yeast transformants were spread on SD/-Trp/-Leu agar plates and incubated at 30°C for 2–3 d. Colonies grown on SD/-Trp/-Leu agar plates were suspended in SD/-Trp/-Leu liquid medium to an optical density of 0.6 at 600 nm (OD600). A 10× dilution series of 5 µL aliquots of yeast transformants were spotted onto SD/-Trp/-Leu or SD/-Trp/-Leu/-His/-Ade agar plates supplemented with 5-bromo-4-chloro-3-indolyl-α-D-galactopyranoside (X-α-Gal). Then the plates were incubated at 30 °C for 2–3 d. The yeast AH109 cotransformed with pGADT7-T and pGBKT7-53, which interact in Y2H assays, were used as positive controls, and the yeast cotransformed with pGADT7-T and pGBKT7-Lam, which do not interact, were used as negative controls.
For the BiFC assays, two YFP fusion constructs were cotransformed into the Agrobacterium tumefaciens GV3101, then cultured to 0.6 at OD600. Equal volumes of each culture were mixed and infiltrated into the epidermal cells of N. benthamiana leaves using a needleless syringe. The agroinfiltrated plants were cultured under normal growth conditions for 48 to 72 h.
For Co-IP assays, total proteins were extracted as previously described (Vijayapalani et al., 2012) and then centrifuged at 16,000 × g for 10 min at 4°C for three times. The supernatants were incubated with anti-mCherry Nanobody Magarose Beads (Shenzhen KangTi Life Technology, Shenzhen, China) overnight at 4°C with gentle shaking. Then the Magarose beads were centrifuged at 500 × g for 3 min at 4°C and washed five times with IP buffer. Western blotting was conducted by using mCherry antibodies as described above.
2.12 Transient expression
The constructs harboring the genes to be tested were individually transformed into Agrobacterium, which were then agroinfiltrated into N. benthamiana leaves using needleless syringes. The GV3101 transformants were cultured overnight in Luria–Bertani medium supplemented with the appropriate antibiotics and collected by centrifugation, and were resuspended in 10 mM MgCl2 supplemented with 100 mM acetosyringone (Sigma, St. Louis, MO, USA). After incubation at room temperature for 2-3 h, the culture was diluted to an OD600 of 0.2-0.5 and agroinfiltrated into the leaves of N. benthamiana plants. The agroinfiltrated plants were maintained under normal growth conditions for 48 to 72 h.
2.13 Confocal microscopy
Leica SP8 X inverted confocal microscope with an argon laser (Leica, Wetzlar, Germany) was used to image the agroinfiltrated leaf sections at room temperature. YFP was excited at 514 nm, and the emission was captured at 530–590 nm. The RFP was excited at 561 nm, and the emission was captured at 588–648 nm. Images were captured digitally and processed using Leica Application Suite Advanced Fluorescence Lite software (Leica Microsystems). ImageJ (http://rsbweb.nih.gov/ij/) was used to quantify the fluorescence intensity.
3 Results
3.1 Identification, phylogenetic analysis, and gene structure analysis of the SsREM gene family
A total of 65 SsREM genes were identified in the S. spontaneum genome (Zhang et al., 2018) based on the N-terminal (Pfam ID PF03766) and C-terminal (Pfam ID PF03763) regions of the REM proteins. To investigate the evolutionary relationships of the REM gene family, a phylogenetic tree was constructed to associate the SsREMs with those of five other monocotyledonous plants (Zea mays, Setaria italica, Triticum aestivum, Oryza sativa, and Allium cepa), ten dicotyledonous plants (Arabidopsis thaliana, Solanum tuberosum, Solanum lycopersicum, Populus trichocarpa, Persea Americana, Nicotiana tabacum, Nuphar advena, Medicago truncatula, Mesembryanthemum crystallinum and Amborella trichopoda) and three gymnosperms (Pinus taeda, Pinus pinaster and Picea sitchensis) (Raffaele et al., 2007; Badawi et al., 2019; Wang et al., 2022) (Supplementary Table 2). The 65 SsREM proteins were classified into 6 groups (Figure 1), namely, Group 0.1, Group 0.2, Group 1, Group 4, Group 5, and Group 6, which contained 2, 7, 18, 8, 10, and 20 members, respectively (Figure 1; Supplementary Figure 1A; Supplementary Table 2). These results are similar to those of previous reports (Raffaele et al., 2007; Badawi et al., 2019; Wang et al., 2022).
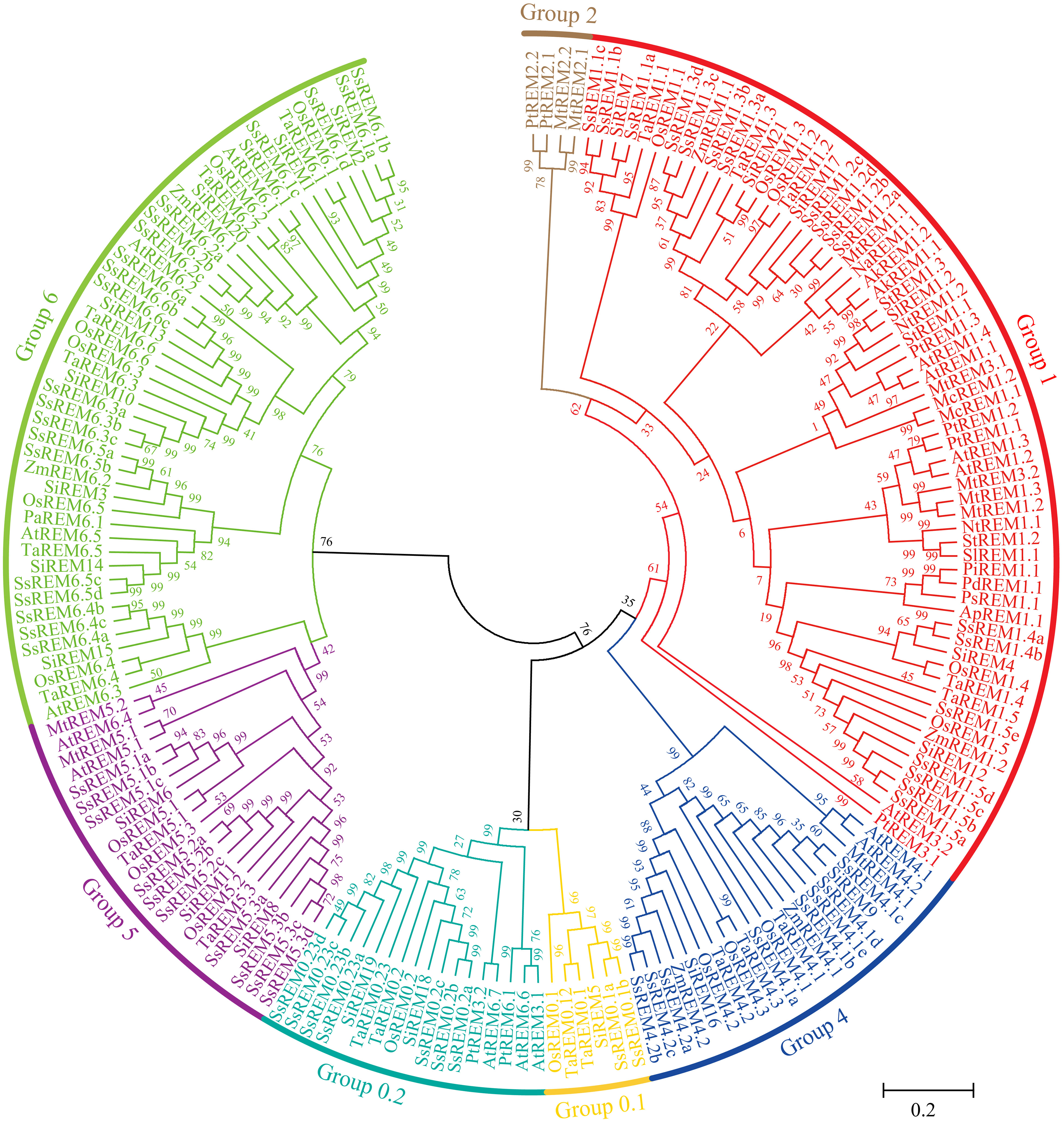
Figure 1 Phylogenetic tree analysis of Remorins (REMs) proteins. The phylogenetic tress was constructed using the maximum likelihood method with 1000 bootstrap replicates. The REMs sequences are from the databases of 5 monocotyledons (Oryza sativa, Zea mays, Triticum aestivum, Setaria italic, Allium cepa, and Saccharum spontaneum), 10 dicotyledons (Arabidopsis thaliana, Solanum tuberosum, Solanum lycopersicon, Populus trichocarpa, Persea Americana, Nicotiana tabacum, Nuphar advena, Medicago truncatula, Mesembryanthemum crystallinum and Amborella trichopoda) and 3 gymnosperm plants (Pinus tadea, Pinus pinaster and Picea sitchensis). The S. spontaneum remorins were grouped into six distinct groups and annotated with different colors.
The online software MEME (http://meme-suite.org/tools/meme) was used to investigate the motifs contained in the SsREM proteins, and a total of 20 conserved motifs were identified (Supplementary Figure 1B). With the exceptions of SsREM1.1c, SsREM1.3a, SsREM1.3b, SsREM4.1a, SsREM5.3a/b/c/d, SsREM6.4a/b/c and SsREM6.5c/d, the SsREM proteins contain Motif 1, Motif 3 and Motif 7; Group 0.1 and Group 5 do not contain Motif 2. Moreover, SsREM0.1a, SsREM1.1b/c, SsREM1.3a/b, and SsREM5.3a contain only three motifs, and Group 0.2, Group 5 and Group 6 only contain Motif 8, Motif 9, Motif 15 and Motif 19. SsREM0.1a and SsREM1.1b contain Motif 1, Motif 3 and Motif 7. SsREM1.1c and SsREM1.3a/b contain Motif 1, Motif 2 and Motif 7. SsREM5.3a contains Motif 1, Motif 4 and Motif 6.
The exon−intron distribution patterns of the SsREM genes were also investigated via TBtools (Supplementary Figure 1C). The results showed that the number of exons in the SsREM genes varied from 1 to 9, and the exons also varied within the same group. For example, in Group 1, SsREM1.2a had only 1 exon, while SsREM1.1a had 6 exons. Most of the genes contained 4 to 6 exons, which is similar to the findings of previous reports (Badawi et al., 2019).
3.2 Analysis of SsREM gene synteny, duplication, selection pressure, chromosomal location and physicochemical properties
To investigate the evolutionary mechanisms of the SsREM gene family, the synteny of the SsREM gene family within S. spontaneum itself was analyzed. As shown in Figure 2, there were 60 pairs of homologous SsREM genes in the genome of S. spontaneum. There were 2, 27, 3, 11 and 17 pairs of homologous SsREM genes in Group 0.2, Group 1, Group 4, Group 5 and Group 6, respectively. However, Group 0.1 contained no gene pairs. To obtain additional information on the evolution of the REM gene family, synteny analysis was conducted on S. spontaneum compared with sorghum, maize, rice, barley, and wheat. The results revealed 37 pairs of homologous genes between S. spontaneum and sorghum, 40 pairs between S. spontaneum and maize, 33 pairs between S. spontaneum and rice, 15 pairs between S. spontaneum and barley, and 69 pairs between S. spontaneum and wheat (Figure 3; Supplementary Table 3).
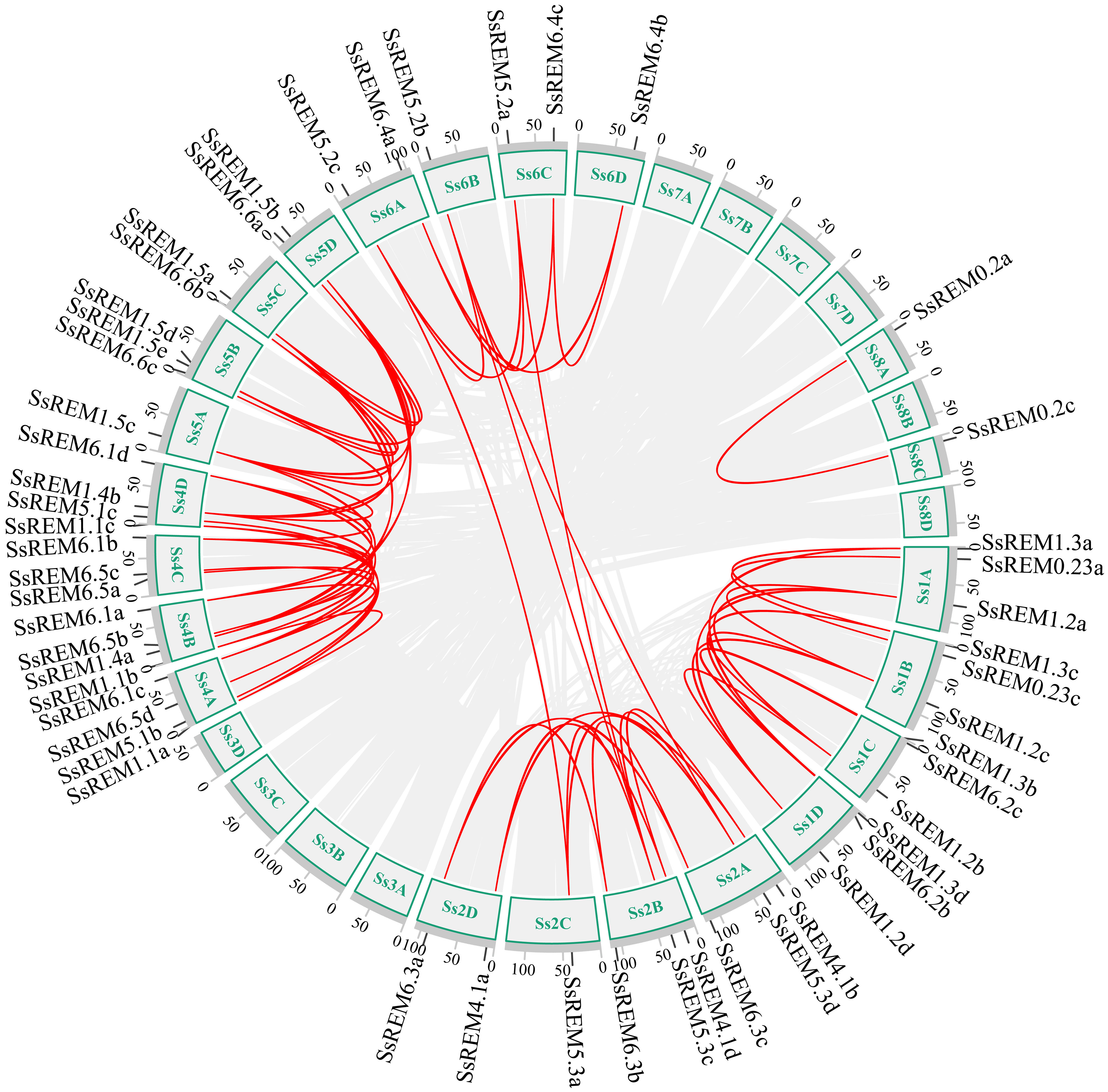
Figure 2 Interchromosomal collinearity relationship analysis of the REM gene family from Saccharum spontaneum. The red lines represent replicated REM gene pairs.
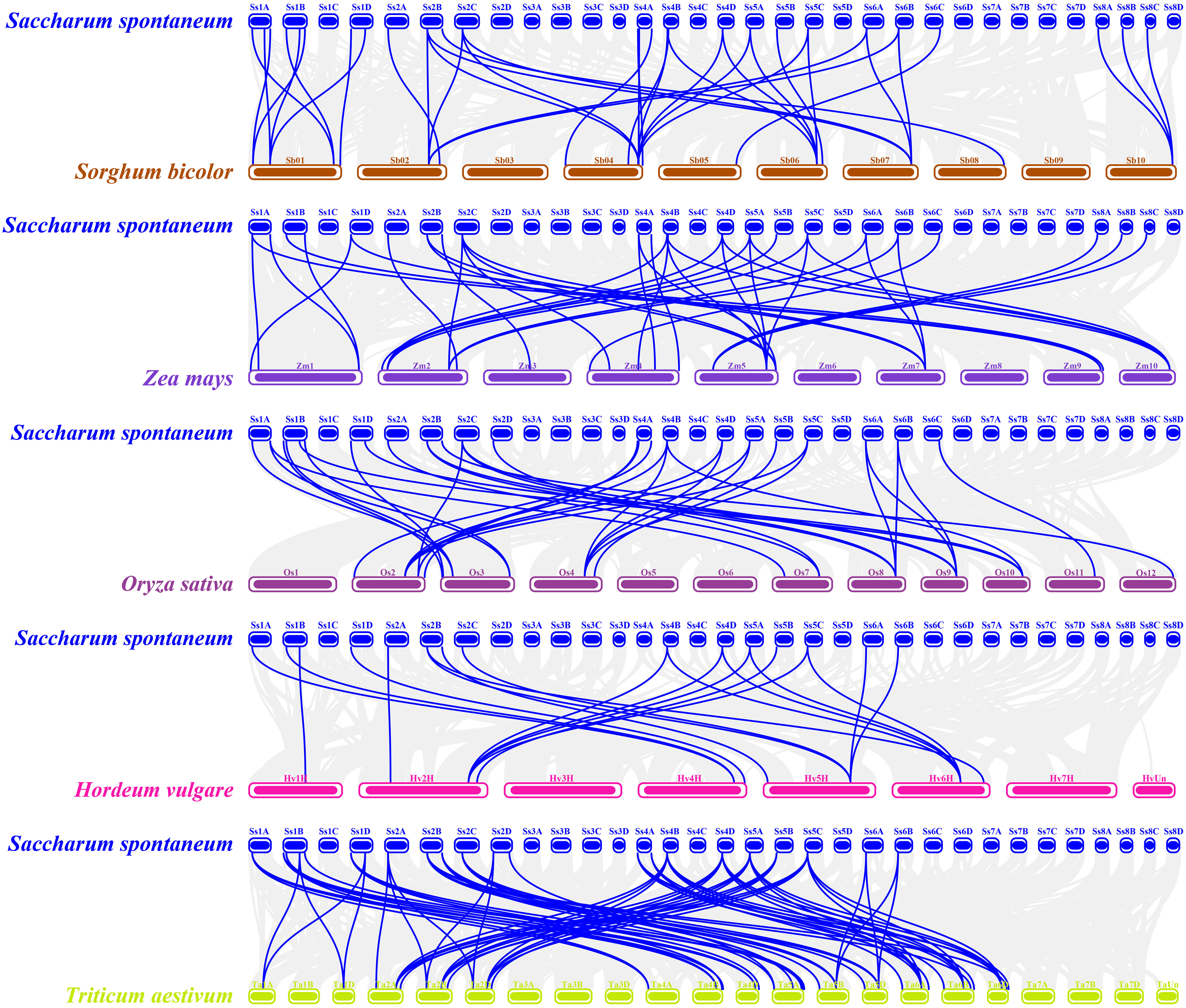
Figure 3 Synteny analysis of REM genes between Saccharum spontaneum and other five Poaceae species. The gray lines in the background represent collinear blocks in the genomes of millet and the other plant species, i.e., Sorghum bicolor, Zea mays, Oryza sativa, Hordeum vulgare and Triticum aestivum, respectively. The blue lines represent the SsREM homologous gene pairs.
The gene duplication types or events were identified by MCScanX software. As shown in Supplementary Table 4, dispersed, tandem and whole-genome duplications (WGD)/segmental duplications were present in the SsREM genes. The main duplication types for the SsREM genes were WGD/segmental (25/65, 38%) and dispersed (22/65, 34%). The Ka/Ks values of the SsREM gene pairs were investigated to evaluate the evolutionary selection pressure on the SsREM gene family. As shown in Supplementary Table 3, the Ka/Ks ratios of all the gene pairs were < 1, indicating that the homologous genes among S. spontaneum, sorghum, maize, rice, barley and wheat had undergone strong purifying selection.
Chromosomal mapping revealed that the SsREM genes were unevenly distributed on the 23 chromosomes (Supplementary Figure 2; Supplementary Table 5), with one on chromosomes 2C, 5A, 6B, 6D, 8A, 8B and 8C (Supplementary Figure 2); two on chromosomes 5C, 5D, 6A and 6C (Supplementary Figure 2); three on chromosomes 1C, 1D, 2B and 5B (Supplementary Figure 2); four on chromosomes 2A, 2D, 4A, 4B and 4C (Supplementary Figure 2); and five, six and seven on chromosomes 4D, 1B and 1A, respectively (Supplementary Figure 2; Supplementary Table 5).
The physicochemical properties and subcellular localization of the 65 SsREM proteins were calculated using the ProtParam tool in ExPASy (Supplementary Table 6). The 65 SsREM proteins encoded proteins ranging from 99 to 939 amino acids, with molecular weights ranging from 11.28 kDa to 101.46 kDa. Among them, 53 SsREM proteins were basic proteins, and 12 SsREM proteins were acidic proteins. The instability coefficient indicated that the 65 SsREM proteins were unstable, with the single exception of SsREM4.1b, which was stable. The average hydrophobicity index indicated that all 65 SsREM proteins were hydrophilic. Subcellular localization prediction via Cell-PLoc 2.0 software (online site) revealed that 62 SsREM proteins localize to the plasma membrane, whereas only three SsREM proteins (SsREM6.2a, SsREM6.5b, and SsREM6.6c) localize to the nucleus (Supplementary Table 6).
3.3 Cis-element analysis of the SsREM gene family
According to the genome of S. spontaneum, the cis-regulatory elements within the first 2,000 bp upstream of 65 SsREMs were predicted. A total of 17 cis-regulatory elements were screened; these elements are involved in the responses to stress, growth and development, and hormones (Figure 4 and Supplementary Table 7). More than 80% of the SsREM genes contained elements related to anaerobic induction (ARE, 81.5%), methyl jasmonate (TGACG and CGTCA motifs, 86.2%), ABA (ABRE, 83.1%) and light responsiveness (G-box, 84.6%). Among the elements involved in growth and development, the ACE element is present in SsREM1.5e, SsREM5.1e and SsREM6.5a/b; the motif I element is present in SsREM5.1a/b, SsREM6.3b/c and SsREM6.6c, SsREM0.2c; and the cell cycle regulation element (MSA-like) is present in SsREM4.1c, SsREM5.1a/b/c and SsREM6.1a/d. Moreover, only SsREM4.1c and SsREM6.3b/c contain the TATC box. However, SsREM0.1b, SsREM1.3d, SsREM1.4a and SsREM5.2b contain 4 cis-elements (SsREM0.1b: TGACG-motif, LTR, TCA-element and GCN4-motif; SsREM1.3d: MBS, ARE, TCA-element and TC-rich repeats; SsREM1.4a: ARE, ABRE, G-box and GCN4-motif; SsREM5.2b: TGACG-motif, ABRE, G-box and TC-rich repeats) (Figure 4; Supplementary Table 7).
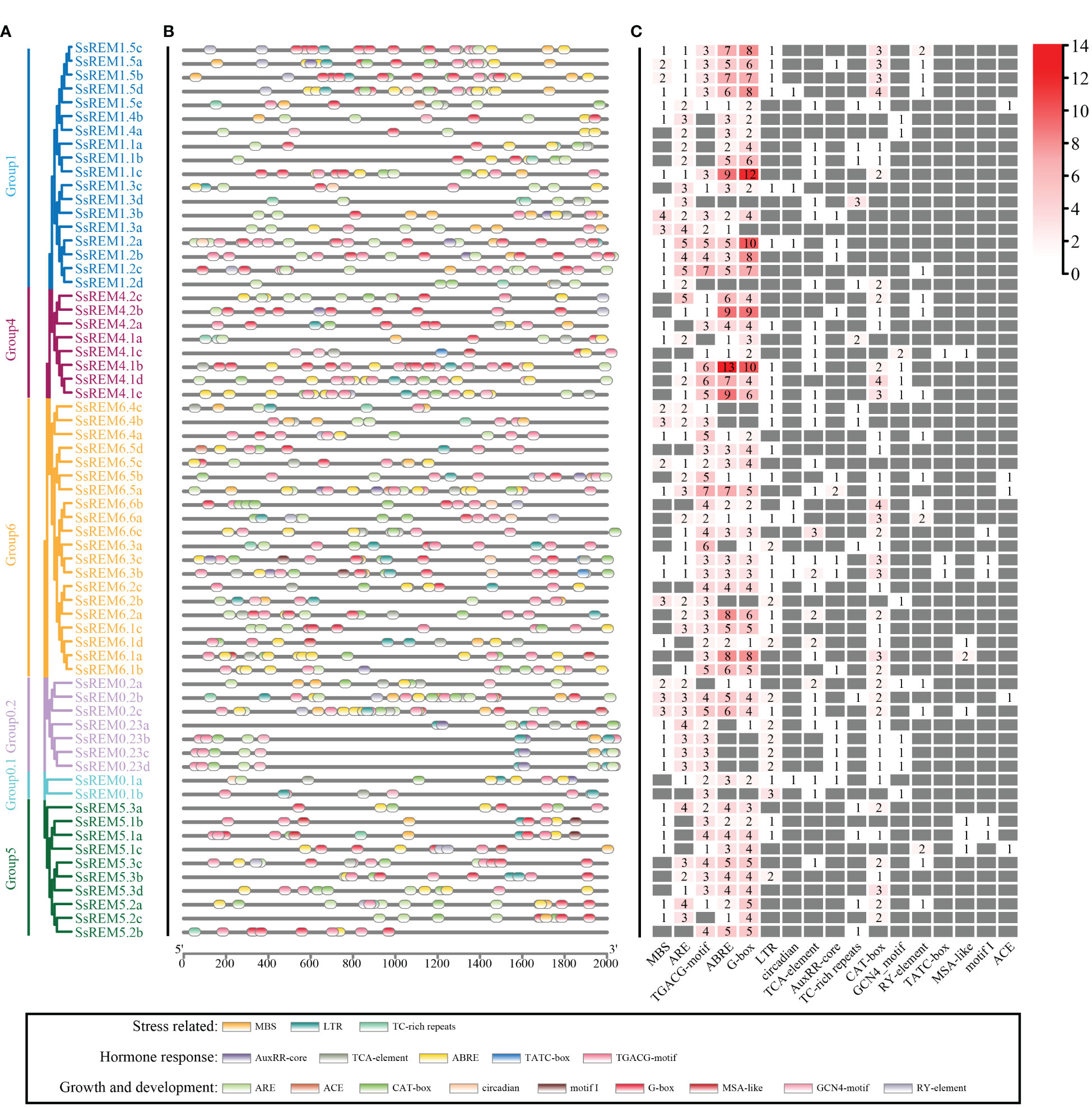
Figure 4 Cis-elements analysis of the REM gene family in Saccharum spontaneum. (A) The phylogenetic tree of SsREM from S. spontaneum. (B) Distribution of cis-elements in the promoter regions of SsREM genes family. (C) Number of cis-elements in the promoter regions of SsREM genes family. Different colored boxes represent different cis-elements.
3.4 Expression patterns of SsREM genes in sugarcane
Transcriptomic data were used to explore the expression patterns of SsREM genes in different tissues and developmental stages of S. spontaneum (Li et al., 2021). The expression levels of the SsREM genes in Group 1 and Group 4 were higher than those in Group 0.1, Group 0.2, Group 5 and Group 6. Moreover, the SsREM genes of Group 0.1, Group 0.2, Group 5 and Group 6 were expressed at very low levels or were undetectable in the premature stage and mature stage (Figure 5A). Notably, the SsREM genes in Group 1 (SsREM1.5ea/b/c/d/e) and Group 4 (SsREM4.1a/b/c/d/e) were expressed at very high levels in the stems during the premature or mature stage (Figure 5A).
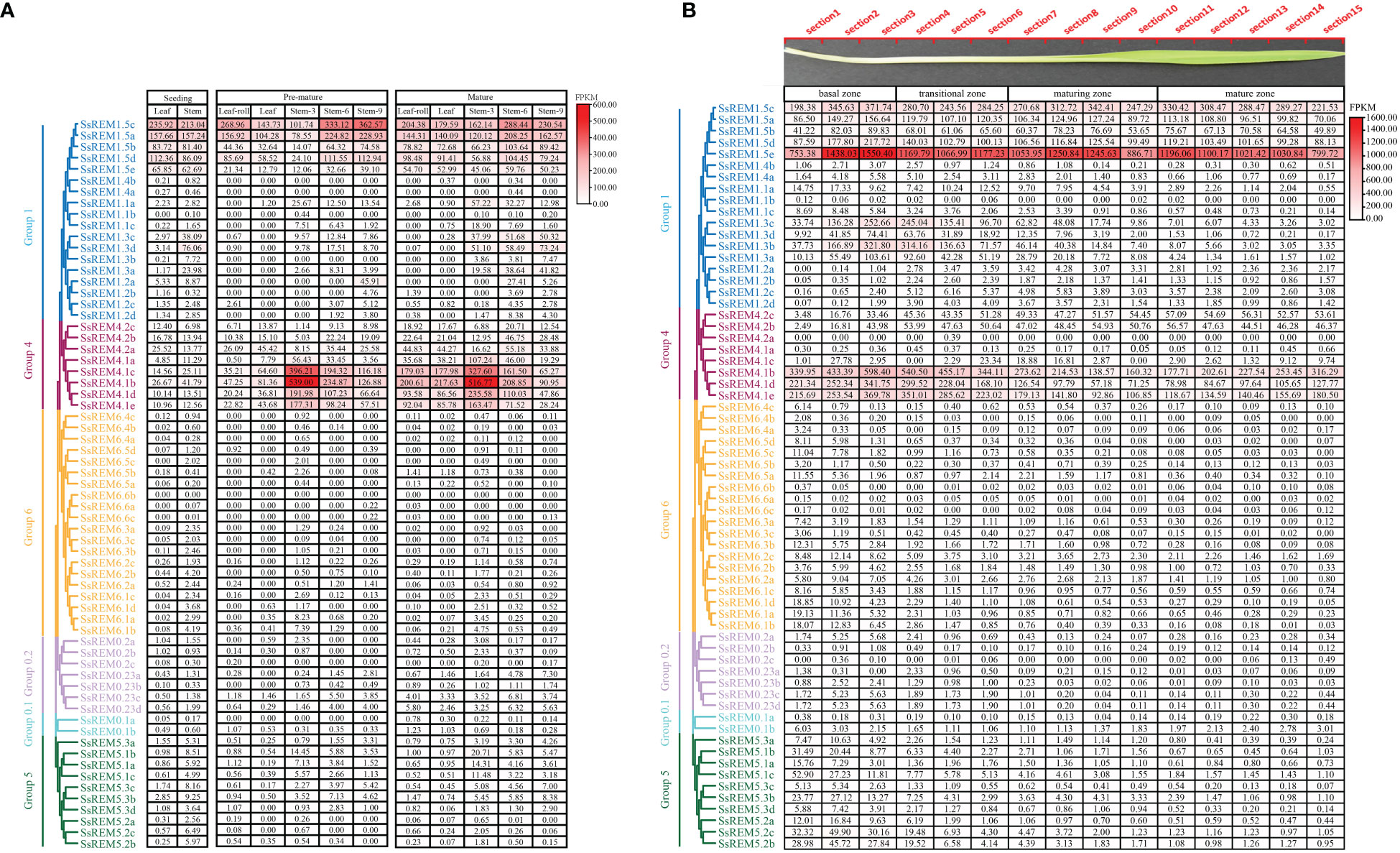
Figure 5 The expression patterns of Remorin (SsREM) genes family in different tissues and across leaf gradients. (A) The expression patterns of SsREM gene family based on FPKM in different tissues of different stages in Saccharum spontaneum. (B) The expression patterns of SsREM genes based on FPKM across leaf gradients in Saccharum spontaneum.
To further investigate the functions of the SsREM genes in the photosynthetic tissues of S. spontaneum, we analyzed the transcriptomes of the leaves along a continuous developmental gradient as described in previous studies (Li et al., 2021). The results showed that the expression levels of the SsREM genes were very high in Group 1 and Group 4 but very low in the other groups. Five genes (SsREM1.5c/e and SsREM4.1b/d/e) exhibited very high expression levels, suggesting that these genes play a key role in leaf development in S. spontaneum. SsREM1.5e presented the highest expression levels across the 15 sections of the leaf, whereas SsREM4.2a exhibited very low or undetectable expression levels (Figure 5B). The results indicate that the SsREM genes may function differently at various developmental stages, thereby affecting biological processes in different tissues.
3.5 Cloning of the ScREM1.5e-1/-2 genes
Two Group 1 REM alleles were cloned from the sugarcane cultivar ROC22 and designated ScREM1.5e-1 and ScREM1.5e-2, respectively, based on phylogenetic tree analysis with the 65 SsREM genes (Supplementary Figure 3). The open reading frame (ORF) of ScREM1.5e-1 (OR805348) is 555 bp in length and encodes 184 amino acids. While, the ORF of ScREM1.5e-2 (OR805349) is 420 bp in length and encodes 139 amino acids. The genomic structures of ScREM1.5e-1 or ScREM1.5e-2 are similar to that of Group I REM in other plants, with five exons and four introns (Figure 6). The difference between ScREM1.5e-1 or ScREM1.5e-2 at the genomic level is that the fourth intron of ScREM1.5e-1 is 29 bp longer than that of ScREM1.5e-2 (Figure 6). The fourth intron of ScREM1.5e-1 is 239 bp in length with the first 2 nucleic acids at the 5´ terminus are GT and the last 3 nucleic acids at the 3´ terminus are TAG. As GT and AG are conserved splicing sites, the fourth intron of ScREM1.5e-1 could be successfully be spliced, resulting a typical REM protein, ScREM1.5e-1, which contains conserved Remorin_N domain (PF03766, 17-68), Remorin_C domain (PF03763, 71-177) domain (in which contains a coiled-coil domain) (Figure 6). A conserved C-terminal anchor (RemCA) was also found at the C-terminal (155-182) of ScREM1.5e-1 by sequence alignment with the well-studied StREM1.3 or AtREM1.4 (Perraki et al., 2012; Raffaele et al., 2013) (Figure 6). However, for the ScREM1.5e-2, the splicing sites AG at the end of the fourth intron mutated to AA, resulting failure in splicing. Whereas, the splicing occurred at the A143G144 of the fourth intron, resulting in retainment of the 66 bp of the fourth intron in the ORF of ScREM1.5e-2 and a meaningful termination codon TAA (Figure 6). Thus, ScREM1.5e-2 protein losses whole RemCA and partial Remorin_C or coiled-coil domain (Figure 6).
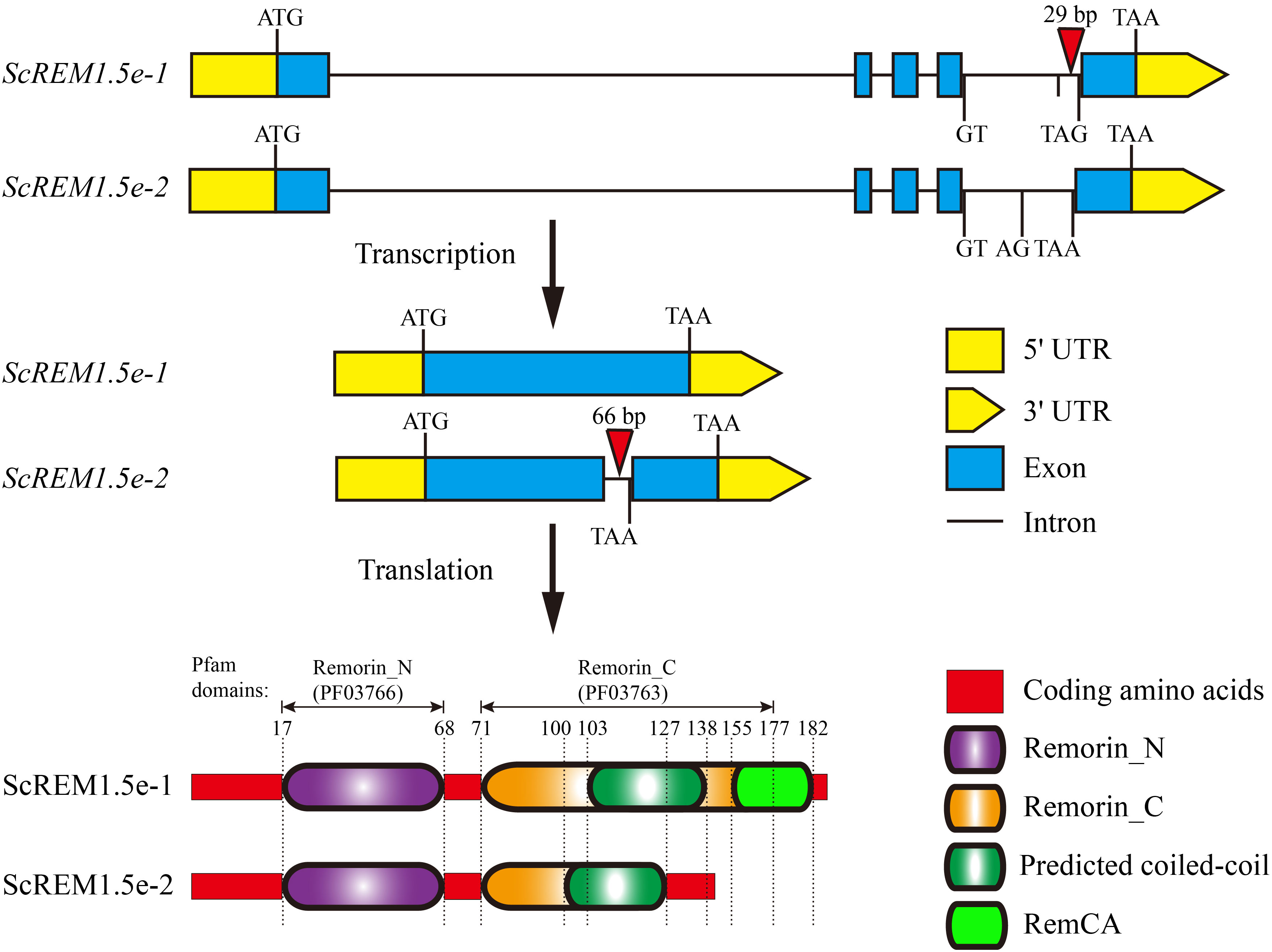
Figure 6 The diagram schematics of gene and protein structures of ScREM1.5e-1 and ScREM1.5e-2. The inverted triangle indicates the insertion of nucleic acids.
3.6 Expression patterns of ScREM1.5e-1/-2 in different tissues or upon stress
The expression patterns of ScREM1.5e-1/-2 in different tissues of the sugarcane cultivar ROC22 were determined via RT−qPCR analysis. Low expression levels of ScREM1.5e-1 were observed in the root and 8th internode. However, a greater expression level (> 6-fold greater than that in the roots) of ScREM1.5e-1 was found in the leaf rolls and 1st leaves than in the 8th internode and roots (Figure 7). In addition, the 8th internode presented the lowest ScREM1.5e-1 expression level (Figure 7). However, the lowest expression level of ScREM1.5e-2 was found in the roots, and the expression levels were significantly greater in the leaves (> 10-fold higher than those in the roots) and internodes (Figure 7). These results suggest that ScREM1.5e-1/-2 may have different functions in the same tissues.
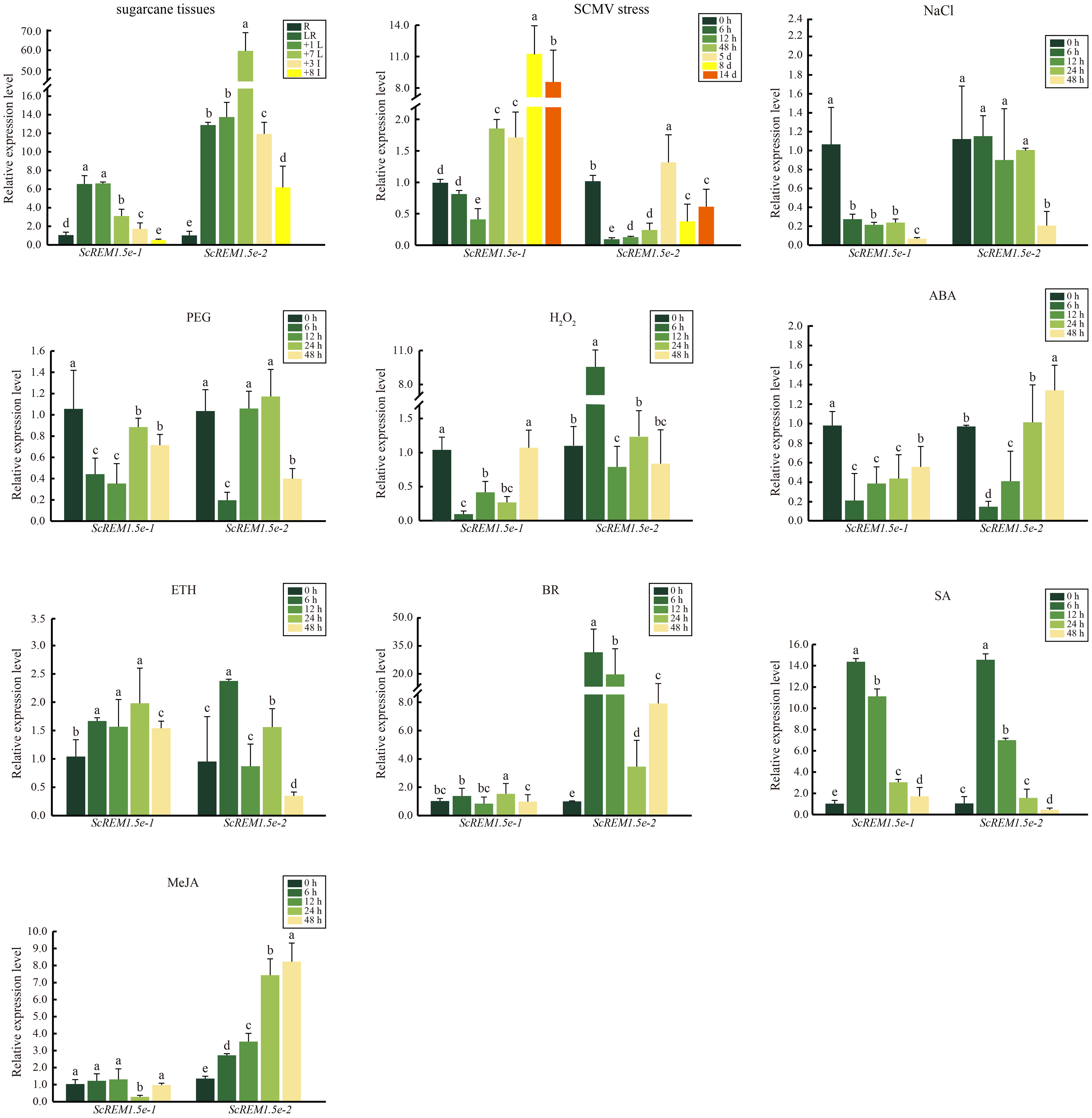
Figure 7 The expression profiles of ScREM1.5e-1 and ScREM1.5e-2 in different tissues of sugarcane cultivar ROC22, or under the challenge by Sugarcane mosaic virus (SCMV) or other treatments. For the study of the expression patterns of ScREM1.5e-1 and ScREM1.5e-2, the sugarcane cultivar ROC22 plants about 120-day-old were used. LR: leaf roll; +1 L: 1st leaf; +7 L: 7st leaf; +3 I: the 3rd internode; +8 I: the 8th internode and R: root. For the expression profiles of ScREM1.5e-1 and ScREM1.5e-2 genes under the challenge by SCMV, leaves of ROC22 plantlets were inoculated with SCMV and sampled at different time points. Plants mock inoculated with 0.01 mM phosphate buffer (pH 7.0) were used as the negative controls. The Y axes indicates the relative expression of ScREM1.5e-1/-2. The X axes indicates the time point of material collection. Error bars indicate SD (n = 3), a, b, c, d and e indicate significance at the corresponding time points, Student’s t-test, P < 0.05. Results were representative of three independent experiments. Simulated plant stresses conditions, including 250 mM NaCl, 25% PEG, 10 mM H2O2, 100 µM ABA, 400 mg/L ETH, 25 mg/L BR, 5 mM SA or 25 µM MeJA, for 0 h, 12 h, 24 h and 48 h. Error bars indicate SD (n = 3), a, b, c, d and e indicate significance at the corresponding time points, Student’s t-test, P < 0.05. Results were representative of three independent experiments.
As the Group 1 remorin are extensively involved in the response to pathogen infection, we investigated the transcription profiles of ScREM1.5e-1/-2 in ROC22 plantlets challenged by SCMV using RT−qPCR. The results showed that the expression levels of ScREM1.5e-1 were significantly downregulated 12 h after SCMV inoculation and subsequently upregulated more than 8-fold at 8 or 14 d (Figure 7). However, the expression of ScREM1.5e-2 was significantly downregulated at 12 h after SCMV inoculation, then significantly (but not more than 1.5-fold) upregulated at 5 d, then downregulated to levels significantly lower than those in the controls (Figure 7).
As many cis-elements involved in plant responses to stress and hormones were found in the promoters of Group 1 remorin, we investigated the expression patterns of ScREM1.5e-1/-2 in the sugarcane cultivar ROC22 under various abiotic stresses and hormonal treatments by RT−qPCR. Generally, the expression patterns of ScREM1.5e-1/-2 were similar under treatment with 25% PEG, 100 µM ABA or 5 mM SA but were different under treatment with 250 mM NaCl, 10 mM H2O2, 400 mg/L ETH, 25 mg/L BR or 25 µM MeJA (Figure 7). In addition, ScREM1.5e-1/-2 was downregulated at 6 h and then upregulated to the level of the control or slightly lower than the control level under 25% PEG and 100 µM ABA. However, under 5 mM SA, the expression levels of ScREM1.5e-1/-2 at 6 h and 12 h were 6- to14-fold greater than those in the controls. 250 mM NaCl treatment significantly suppressed the expression of ScREM1.5e-1 but had no impact on the expression of ScREM1.5e-2 except at 48 h. Treatment with 10 mM H2O2 also significantly suppressed the expression of ScREM1.5e-1 but significantly promoted the expression of ScREM1.5e-2 at 6 h. In particular, 25 mg/L BR or 25 µM MeJA did not impact the expression levels of ScREM1.5e-1 but significantly upregulated the expression of ScREM1.5e-2. In the 400 mg/L ETH treatment, ScREM1.5e-1 had a greater expression level than the control at 6 h, 12 h, 24 h, and 48 h. Inversely, ScREM1.5e-1 had a higher expression level than the control only at 6 h.
3.7 Interaction of ScREM1.5e-1/-2 with SCMV-/SCSMV-/SrMV-VPg
In our previous study, we found that the VPg of TuMV interacts with AtREM1.2/1.3 to facilitate virus infection, and this interaction relocalizes AtREM1.2/1.3 from the plasma membrane to the cytoplasm (Cheng et al., 2020). To test the possible influence of SCMV-/SCSMV-/SrMV-VPg on the subcellular localization of ScREM1.5e-1/-2, we first investigated the subcellular localization of ScREM1.5e-1/-2 or SCMV-/SCSMV-/SrMV-VPg in the epidermal cells of N. benthamiana leaves. The recombinant plasmids YFP-ScREM1.5e-1/-2 and mCherry-AtREM1.2, HDEL-RFP or H2B-RFP were subsequently transformed into N. benthamiana leaves by agroinfiltration as previously described (Cheng et al., 2020). Confocal observation was conducted at 48 hpi. The YFP-ScREM1.5e-1 fluorescence was strongly distributed on the plasma membrane and merged with the red fluorescence of RFP-AtREM1.2, a PM marker, indicating the PM localization of ScREM1.5e-1 (Supplementary Figure 4A). However, the fluorescent signal of YFP-ScREM1.5e-2 merged with the fluorescent signal of HDEL-RFP, a cytoplasmic marker, or H2B-RFP, a nuclear marker (Supplementary Figure 4A), indicating the cytoplasmic or nuclear localization of ScREM1.5e-2. For SCMV-/SCSMV-/SrMV-VPg-YFP, the fluorescent signals merged with those of HDEL-RFP or H2B-RFP, indicating the cytoplasmic and nuclear localization of SCMV-/SCSMV-/SrMV-VP, respectively (Supplementary Figures 4B, C). All the localization assay results were further confirmed by fluorescence intensity measurements (Supplementary Figure 4).
To test the interaction of ScREM1.5e-1/-2 with SCMV, SCSMV, and SrMV-VPg, Y2H assays based on GAL4 and BiFC assays were conducted. For the Y2H assays, pGBKT7-SCMV-/SCSMV-/SrMV-VPg was individually cotransformed with pGADT7-ScREM1.5e-1/-2 into the yeast strain AH109. The combination of pGBKT7-53 and pGADT7-T was used as a positive control, while the combination of pGBKT7-Lam and pGADT7-T was used as a negative control. The results showed that the yeast cells cotransformed with the combination of pGBKT7-SCMV-/SCSMV-/SrMV-VPg with pGADT7-ScREM1.5e-1/-2 produced blue colonies on SD/-Trp/-Leu and SD/-Trp/-Leu/-His/-Ade culture medium supplemented with X-α-Gal as the positive control (Figure 8A), indicating an interaction between SCMV-/SCSMV-/SrMV-VPg and ScREM1.5e-1/-2, while no colonies developed for the negative control (Figure 8A).
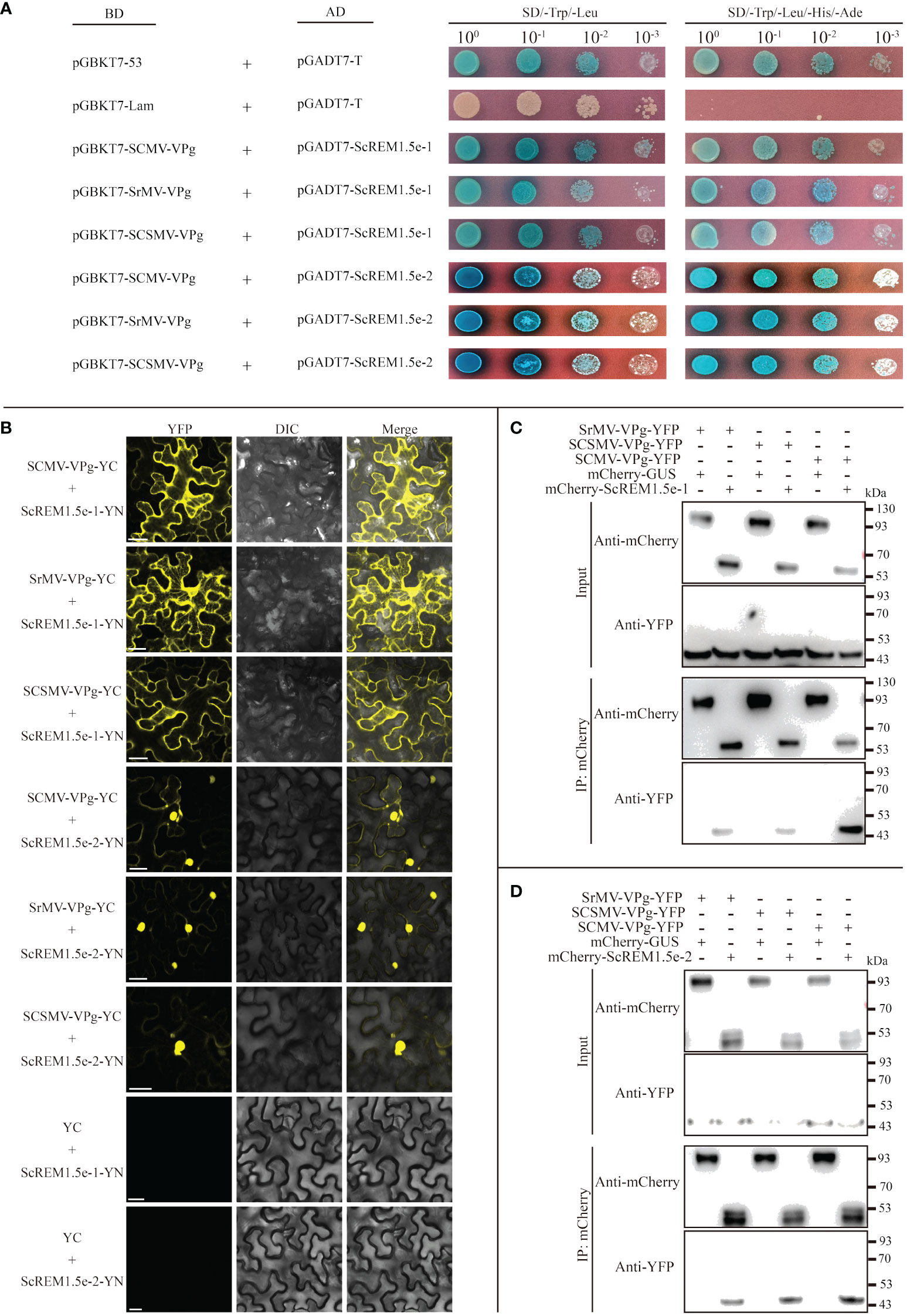
Figure 8 Interaction of ScREM1.5e-1or ScREM1.5e-2 with SCMV-/SCSMV-/SrMV-VPg. (A) Y2H assays. pGADT7-ScREM1.5e-1 or pGADT7-ScREM1.5e-2 were individually pairwise co-transformed with the vector pGBKT7-SCMV-VPg, pGBKT7-SCSMV-VPg, or pGBKT7-SrMV-VPg into the yeast AH109 cells in a 10 × dilution series of 10-µL aliquots, which were then plated on a non-selective medium SD/-Leu/-Trp or quadruple dropout medium SD/-Leu/-Trp/-His/-Ade supplemented with X-α-Gal. Yeast cells co-transformed with pGBKT7-53 and pGADT7-T were used as a positive control, pGBKT7-Lam and pGADT7-T were used as a negative control. (B) BiFC assays. Agrobacteria harboring YC/YN fusion proteins were individually pairwise co-infiltrated into N. benthamiana leaves. The leaf epidermal cells pairwise co-transformed with ScREM1.5e-1-YN or ScREM1.5e-12-YN plus YC were used as negative controls. The images were captured at 48 h post infiltration. Bars = 25 µm. (C, D) Co-immunoprecipitation (co-IP) assays to confirm the interaction of ScREM1.5e-1 or ScREM1.5e-2 with SCMV-/SCSMV-/SrMV-VPg, respectively. Total proteins were extracted from N. benthamiana leaves expressing the mCherry-ScREM1.5e-1/-2, mCherry-GUS or SCMV-/SCSMV-/SrMV-VPg-YFP construct. The immune complexes were immobilized on anti-mCherry magnetic beads, and the co-precipitation of SCMV-/SCSMV-/SrMV-VPg were examined by western blotting using antibodies against YFP or mCherry.
For the BiFC assays, the fusion construct SCMV-/SCSMV-/SrMV-VPg-YC was individually cotransformed with ScREM1.5e-1/-2-YN into the Agrobacterium strain GV3101 and then agroinfiltrated into N. benthamiana leaves. Confocal observation was performed at 48 hpi. The results showed that SCMV-VPg, SCSMV-VPg and SrMV-VPg interact with ScREM1.5e-1/-2 individually; SCMV-VPg, SSMV-VPg and SrMV-VPg interact with ScREM1.5e-1 mainly in the cytoplasm, whereas the interaction with ScREM1.5e-2 mainly occurs in the nucleus (Figure 8B). As expected, the negative controls ScREM1.5e-1/-2-YN and YC emitted no fluorescence signals (Figure 8B).
To further confirm the interaction of SCMV-/SCSMV-/SrMV-VPg with ScREM1.5e-1/-2, Co-IP assays were performed. We found that SCMV-/SCSMV-/SrMV-VPg could be individually pulled down by ScREM1.5e-1/2 but not GUS (Figures 8C, D). Collectively, these results demonstrated the interaction of ScREM1.5e-1/-2 with SCMV-/SCSMV-/SrMV-VPg both in vitro and in vivo (Figure 8).
3.8 Self-interaction of and interaction between ScREM1.5e-1/-2
As previous studies have shown that REMs can form homo or hetero-oligomers (Keinath et al., 2010; Martinez et al., 2019; Cai et al., 2020; Ma et al., 2022; Su et al., 2023), we performed Y2H, BiFC and Co-IP assays to evaluate the self-interaction of ScREM1.5e-1/-2 or the interaction between ScREM1.5e-1 and ScREM1.5e-2. For the Y2H assays, the combinations of pGBKT7-ScREM1.5e-1 plus pGADT7-ScREM1.5e-1, pGBKT7-ScREM1.5e-1 plus pGADT7-ScREM1.5e-2, and pGBKT7-ScREM1.5e-2 plus pGADT7-ScREM1.5e-2 were individually cotransformed into the yeast strain AH109. Like yeast cells cotransformed with the positive control plasmids pGBKT7-53 and pGADT7-T, yeast cells cotransformed with the combination of pGBKT7-ScREM1.5e-1 and pGADT7-ScREM1.5e-1, pGBKT7-ScREM1.5e-1 and pGADT7-ScREM1.5e-2, pGBKT7-ScREM1.5e-2 and pGADT7-ScREM1.5e-2 produced blue colonies on SD/-Trp/-Leu and SD/-Trp/-Leu/-His/-Ade culture medium supplemented with X-α-Gal, indicating interactions between ScREM1.5e-1 and ScREM1.5e-2 and self-interactions (Figure 9A), and the negative control cotransformed with plasmids pGBKT7-Lam and pGADT7-T did not produce blue colonies (Figure 9A). For the BiFC assays, the fusion construct ScREM1.5e-1/-2-YN/-YC was individually cotransformed with ScREM1.5e-1/-2-YN/-YC into the Agrobacterium strain GV3101 and then agroinfiltrated into N. benthamiana leaves. At 48 hpi, yellow fluorescence corresponding to YFP was observed via confocal microscopy. The results confirmed the ScREM1.5e-1/-2 interaction and their self-interactions (Figure 9B). As expected, the negative controls ScREM1.5e-1/-2-YN/-YC and YC/YN emitted no fluorescence signals (Figure 9B).
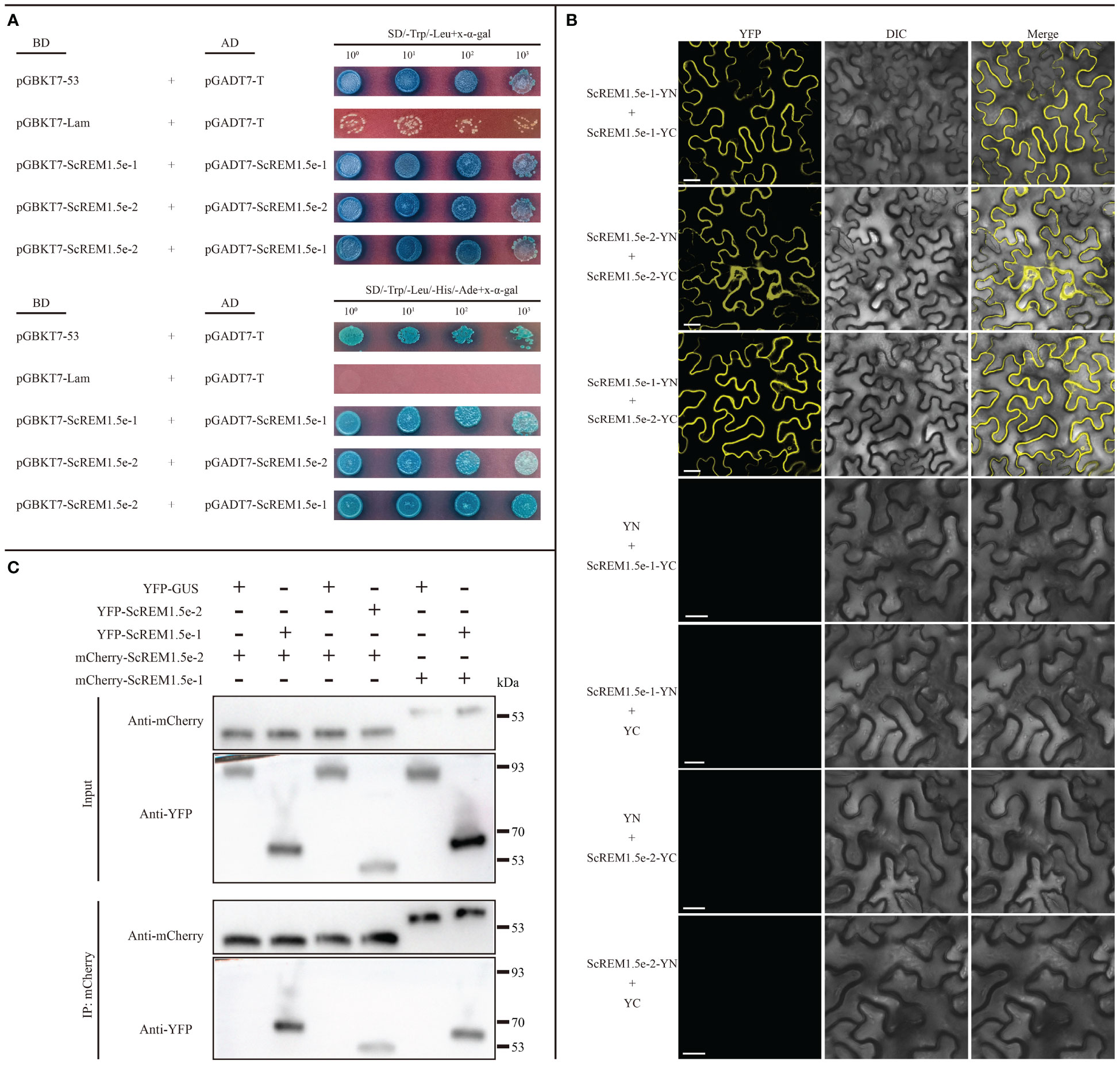
Figure 9 Selfinteraction of or interaction between ScREM1.5e-1 and ScREM1.5e-2. (A) Y2H assays. The pGADT7-ScREM1.5e-1 or pGADT7-ScREM1.5e-2 were infused into the prey vector pGADT7 or pGBKT7, then pairwise co-transformed into the yeast AH109 cells in a 10× dilution series of 10-µL aliquots, which were then plated on a non-selective medium SD/-Leu/-Trp or quadruple dropout medium SD/-Leu/-Trp/-His/-Ade supplemented with X-α-Gal. Yeast cells co-transformed with pGBKT7-53 and pGADT7-T were used as a positive control, while the yeast cells co-transformed with pGBKT7-Lam and pGADT7-T were used as a negative control. (B) BiFC assays. Agrobacteria harboring YC/YN fusion proteins were co-infiltrated into N. benthamiana leaves, respectively. The leaf epidermal cells pairwise co-transformed with YN plus ScREM1.5e-1-YC, ScREM1.5e-1-YN plus YC, YN plus ScREM1.5e-2-YC, or ScREM1.5e-1-YN-plus YC were used as negative controls. The images were captured at 48 h post agroinfiltration. Bars = 25 µm. (C) Co-IP assays to confirm the interaction of ScREM1.5e-1 with ScREM1.5e-2 or the self-interaction of ScREM1.5e-1 or ScREM1.5e-2. Total proteins were extracted from N. benthamiana leaves individually expressing the YFP-ScREM1.5e-1, YFP-ScREM1.5e-2, YFP-GUS, mCherry-ScREM1.5e-1, or mCherry-ScREM1.5e-2 construct. The immune complexes were immobilized on anti-mCherry magnetic beads, and the co-precipitation of ScREM1.5e-1 or ScREM1.5e-2 were examined by western blotting using antibodies against YFP or mCherry.
To further confirm the interaction between ScREM1.5e-1/-2 and the self-interactions, we conducted Co-IP assays. The results showed that ScREM1.5e-1/-2 could be pulled down by ScREM1.5e-1/-2 but not by GUS (Figure 9C). Collectively, these results demonstrate the interaction between ScREM1.5e-1 and ScREM1.5e-2 and the self-interactions of each protein both in vitro and in vivo.
4 Discussion
In view of the important role of REMs in plant growth and development and in response to biological and abiotic stresses (Raffaele et al., 2007; Jarsch and Ott, 2011; Gouguet et al., 2021), the number of studies on the REM gene family at the whole-genome level has gradually increased in recent years (Raffaele et al., 2007; Jamann et al., 2016; Badawi et al., 2019; Wang et al., 2022; Li et al., 2023). In the present study, 65 REM genes were identified from S. spontaneum (Figure 1). There are different numbers of REMs in different monocotyledonous crops, e.g., 19 in rice, 20 in wheat, 21 in foxtail millet, 21 in sorghum, and 33 in maize (Raffaele et al., 2007; Badawi et al., 2019; Wang et al., 2022; Li et al., 2023). The numbers of REMs are generally comparable across species. For Panicoideae crops, the number of REMs in foxtail millet is comparable to that in sorghum, while maize has a greater number of REMs, which might be due to the tetraploidization of the maize genome 5-12 million years ago (Schnable et al., 2011). However, the number of REMs in S. spontaneum is 3 times that in sorghum, which is speculated to be related to two whole-genome duplication events in sugarcane (Zhang et al., 2018, 2022). The expansion of gene families is always accompanied by gene duplication. The present study showed that the expansion of the REM gene family in S. spontaneum occurred mainly through WGD/segmentation and dispersed replication, which is consistent with the findings of previous studies (Wang et al., 2018; Zhang et al., 2018). In the present study, the S. spontaneum REMs were classified into 6 groups, in which Group 0.2 may exist only in monocotyledonous plants, while Group 4 may be differentiated in monocots and dicots (Figure 1), which is consistent with the findings of previous studies (Raffaele et al., 2007; Badawi et al., 2019).
REM gene families are extensively involved in plant growth and development and exhibit diverse expression patterns (Huang et al., 2019; Li et al., 2023). In this study, the transcriptomic data showed that Group 1 REMs (SsREM1.5a/b/c/d/e) and Group 4 REMs (SsREM4.1a/b/c/d/e) were highly expressed in internodes and leaves (Figure 5). However, the expression levels of ScREM1.5e-1/-2 were significantly greater in leaves than in internodes or roots, with the expression level of ScREM1.5e-2 being much greater than that of ScREM1.5e-1 in the sugarcane cultivar ROC22 (Figure 7), indicating the different roles of these REM alleles in sugarcane growth and development.
During the growth and development process, crops are subjected to various biotic and abiotic stresses, which seriously affect production and yield (Zhu, 2016; Gong et al., 2020; Ding and Yang, 2022; Waadt et al., 2022). Therefore, crops adopt complex mechanisms to cope with stress, among which transcriptional reprogramming is one of the most important means (Kumar, 2014; Jones, 2016; Carvalhais et al., 2017; Cheng et al., 2018; Xie et al., 2018; Wu et al., 2021; Shang et al., 2023). Transcriptomic studies have shown that REMs respond to drought, high salt concentrations, low temperature, ABA, BR, SA and MeJA (Bray, 2002; Reddy et al., 2002; Nohzadeh Malakshah et al., 2007; Checker and Khurana, 2013; Hu et al., 2013; Yue et al., 2014; Gui et al., 2016; Kong et al., 2016; Badawi et al., 2019; Huang et al., 2019; Wang et al., 2022). For example, the abscisic acid (ABA)-responsive DREB-binding protein (SiARDP) from foxtail millet can regulate the expression of SiREM6 (Li et al., 2014). The tolerance of mutant Arabidopsis plants overexpressing SiREM6 to high salt and low temperature improved with the exogenous application of ABA (Checker and Khurana, 2013; Li et al., 2014; Yue et al., 2014). ABA activates the transcription factor OsbZIP23 to upregulate the expression of OsREM4.1 in rice, whereas OsREM4.1 negatively regulates the BR signaling pathway (Gui et al., 2016). Overexpression of mulberry (Morus indica) MiREM confers tolerance to drought and salt stresses in mutant Arabidopsis plants (Checker and Khurana, 2013). The expression of the Antarctic hairgrass (Deschampsia antarctica) transcription factor DaCBF7 was induced by drought, low temperature, and salinity; however, overexpression of DaCBF7 in rice plants only conferred cold tolerance (Byun et al., 2015). Further investigation revealed that REMs were upregulated, and putative CRT/DRE or low-temperature responsive elements were found in their promoter regions (Byun et al., 2015). Analysis of the REM families of Arabidopsis, foxtail millet, rice, wheat and tomato showed that their upstream promoter regions contain a large number of responsive elements related to growth, development, hormones and stress (Raffaele et al., 2007; Yue et al., 2014; Badawi et al., 2019; Wang et al., 2022; Li et al., 2023). Different REMs possess different responsive elements in their upstream promoter regions, thereby exhibiting different expression profiles in some situations. For example, most wheat REMs respond to ABA induction, but only Group 4 REMs are strongly induced under cold stress (Badawi et al., 2019). Our analysis of cis-acting elements in the promoter regions of SsREMs demonstrated that a large number of elements are involved in the regulation of growth and development and in responses to stress and hormones (Figure 4). Therefore, we speculate that the REMs of S. spontaneum might be widely involved in these biological processes and exhibit different expression patterns, as indicated in Figure 4. The upstream promoter regions of SsREM1.5e contain MBS, ARE, TGACG-motif, ABRE, G-box, TCA-element, TC-rich repeat, CATbox, and ACE elements (Figure 4), resulting in different expression profiles of ScREM1.5e-1/-2 under different treatments (Figure 7). The present study showed that both ScREM1.5e-1 and ScREM1.5e-2 responded to PEG or H2O2 treatment; however, ScREM1.5e-1 was downregulated under NaCl treatment, whereas ScREM1.5e-2 was not (Figure 7). These results indicate that different REMs might play different roles in response to abiotic stress in sugarcane, and further investigation is needed.
Our previous study showed that TuMV employs P3N-PIPO to recruit PCaP1 to degrade actin filaments in close proximity to or within plasmodesmata to promote TuMV cell-to-cell movement. However, REM interacts with PCaP1 to interfere with the binding of PCaP1 to actin filaments. As a counteractive response, TuMV employs VPg to interact with and mediate REM degradation through autophagy and the 26S proteasome pathway to establish systemic infection (Cheng et al., 2020). Our previous study also confirmed the interaction of SCMV P3N-PIPO with ScPCaP1 (Cheng et al., 2017). In this study, we found that SCMV infection upregulated the expression of ScREM1.5e-1 and ScREM1.5e-2, and the former was significantly greater than the latter (Figure 7). In general, SCMV infection significantly upregulated the expression of ScREM1.5e-1 2 d post-inoculation but downregulated the expression of ScREM1.5e-2 (Figure 7). It is worth investigating whether ScREM1.5e-1 or ScREM1.5e-2 can interfere with the interaction of ScPCaP1 with actin filaments. The C-terminal anchor RemCA plays key role in the PM localization of REMs (Perraki et al., 2012). In the present study, the subcellular localization assays revealed that ScREM1.5e-1 was localized to the plasma membrane, while ScREM1.5e-2 was localized to the cytoplasm and nucleus (Supplementary Figure 4), possibly due to the loss of RemCA (Figure 6). Therefore, we speculate that C-terminal anchors are critical for the localization of REM protein plasma membrane. VPg plays an important role in the translation of the potyvirus genome. In addition, ScREM1.5e-1 interacts with the VPgs of SCMV, SCSMV, and SrMV in the plasma membrane and cytoplasm; however, ScREM1.5e-2 interacts with these proteins in the nucleus (Figure 8B) and might differentially impact the function of these proteins. In our previous study, we predicted in silico that SCMV, SCSMV, and SrMV-VPg all contain domains that interact with ATG8 (Yang et al., 2020). However, further studies are needed to investigate whether the interaction of ScREM1.5e-1 or ScREM1.5e-2 with all the VPgs interferes with the interaction of SCMV-/SCSMV-/SrMV-VPg with SceIF4Es, thereby impairing the translation of viral genomes or mediating ScREM1.5e-1 or ScREM1.5e-2 degradation via the autophagy pathway or the 26S proteasome pathway. REMs can oligomerize to homologous or heterologous dimers or trimers, thereby participating in the initial immune response of plants (Keinath et al., 2010; Cai et al., 2020; Ma et al., 2022; Su et al., 2023). In the present study, we found that ScREM1.5e-1 and ScREM1.5e-2 self-interact and interact with each other (Figure 9), indicating that they can oligomerize. We speculated that oligomerized REMs may also reduce plasma membrane fluidity and plasmodesmata permeability, thereby inhibiting SCMV infection, reminiscent of REMs inhibiting CMV infection (Huang et al., 2019). It would be interesting to further investigate the biological roles of hetero-oligomers of ScREM1.5e-1 with ScREM1.5e-2 because the interaction of ScREM1.5e-1 with ScREM1.5e-2 occurs on the plasma membrane and changes the cytoplasmic localization of ScREM1.5e-2 (Figure 9B), suggesting that the biological function of ScREM1.5e-2 is affected to some extent.
5 Conclusion
In this study, 65 REM genes were identified in S. spontaneum. SsREM gene duplication events were mainly dispersed and characterized as WGD or segmental. The upstream promoter regions of the SsREM family contain multiple cis-acting elements associated with stress, growth, and hormonal responses, indicating that the SsREM family may be involved in the response to various stresses, growth and development. SsREMs were constitutively expressed in different sugarcane tissues, as indicated by the RNA-seq database. In addition, a pair of alleles, ScREM1.5e-1 and ScREM1.5e-2, was cloned from the sugarcane cultivar ROC22. In addition, ScREM1.5e-1 and ScREM1.5e-2 were highly expressed in leaves, while the expression of ScREM1.5e-2 was significantly higher than that of ScREM1.5e-1 in the internodes. Exogenous application of ABA, ETH, SA or SCMV infection upregulated the expression of ScREM1.5e-1 and ScREM1.5e-2. ScREM1.5e-1 was localized to the PM, while ScREM1.5e-2 was localized to the nucleus and cytoplasm. The interaction of ScREM1.5e-1 or ScREM1.5e-2 with the VPgs of SCMV, SCSMV, or SrMV was individually confirmed by Y2H, BiFC, and co-IP assays. The homo- or hetero-oligomerization of ScREM1.5e-1 with ScREM1.5e-2 was also confirmed. The present study sheds light on the biological functions of ScREM1.5e-1 and ScREM1.5e-2 and will be valuable for engineering sugarcane resistance to sugarcane mosaic disease.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
Author contributions
ZY: Data curation, Writing – original draft, Formal Analysis, Investigation, Visualization. GC: Data curation, Formal Analysis, Investigation, Visualization, Writing – original draft, Funding acquisition. QY: Data curation, Formal Analysis, Investigation, Writing – original draft. KZ: Data curation, Formal Analysis, Investigation, Writing – original draft. WJ: Data curation, Formal Analysis, Investigation, Writing – original draft. TL: Data curation, Formal Analysis, Investigation, Writing – original draft. HZ: Data curation, Formal Analysis, Investigation, Writing – original draft. HS: Data curation, Formal Analysis, Writing – original draft. GH: Writing – original draft, Resources. FW: Resources, Conceptualization, Formal Analysis, Writing – review & editing. YG: Conceptualization, Formal Analysis, Resources, Writing – review & editing, Supervision. JX: Conceptualization, Supervision, Writing – review & editing, Data curation, Funding acquisition, Project administration, Writing – original draft.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the National Natural Science Foundation of China (31971991) and the Scientific Research Foundation of Graduate School of Fujian Agriculture and Forestry University (YB2020002).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2024.1365995/full#supplementary-material
Supplementary Figure 1 | Phylogenetic relationships, conserved motifs and structure analysis of the Remorin (SsREM) genes family. (A) The phylogenetic tree of Remorin proteins in Saccharum spontaneum. (B) Conserved motifs analysis of the Remorin genes family. (C) Gene structure analysis of the Remorin genes family. The groupings of clusters are shown in different colors. Conserved motifs of Remorin proteins are indicated by differently colored boxes. Offwhite boxes represent UTR (untranslated region), dark blue boxes represent exon, black line represent intron.
Supplementary Figure 2 | Chromosomal locations of Remorin (SsREM) genes in the Saccharum spontaneum genome. The distributions of the 65 Remorin genes were determined according to the scaffold number and are shown in red. The numbers on the top indicate each chromosome number and the genome.
Supplementary Figure 3 | Evolutionary tree of the two genes of sugarcane cultivar ROC22 with 65 SsREM genes. The two genes of sugarcane cultivar ROC22 were highlighted by the “leaves” in red, indicated that it belongs to REM1.5e branch.
Supplementary Figure 4 | Subcellular localization of ScREM1.5e-1/-2 or SCMV-/SCSMV-/SrMV-VPg in the epidermal cells of Nicotiana benthamiana. (A) Subcellular localization of YFP-tagged ScREM1.5e-1/-2 in N. benthamiana leaf epidermal cells. Bars = 20 μm. (B, C) Subcellular localization of YFP-tagged SCMV-/SCSMV-/SrMV-VPg in N. benthamiana leaf epidermal cells. The fifth column shows overlapping fluorescence spectra analysis of YFP and RFP signals marked in red dashed line. Bars = 20 μm.
References
Akbar, S., Wei, Y., Yuan, Y., Khan, M. T., Qin, L., Powell, C. A., et al. (2020). Gene expression profiling of reactive oxygen species (ROS) and antioxidant defense system following Sugarcane mosaic virus (SCMV) infection. BMC Plant Biol. 20, 532. doi: 10.1186/s12870-020-02737-1
Akbar, S., Yao, W., Yu, K., Qin, L., Ruan, M., Powell, C. A., et al. (2021). Photosynthetic characterization and expression profiles of sugarcane infected by Sugarcane mosaic virus (SCMV). Photosynth. Res. 150, 279–294. doi: 10.1007/s11120-019-00706-w
Badawi, M. A., Agharbaoui, Z., Zayed, M., Li, Q., Byrns, B., Zou, J., et al. (2019). Genome-wide identification and characterization of the wheat remorin (TaREM) family during cold acclimation. Plant Genome 12, 10.3835/plantgenome2018.06.0040. doi: 10.3835/plantgenome2018.06.0040
Bailey, T. L., Boden, M., Buske, F. A., Frith, M., Grant, C. E., Clementi, L., et al. (2009). MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 37, W202–W208. doi: 10.1093/nar/gkp335
Bozkurt, T. O., Richardson, A., Dagdas, Y. F., Mongrand, S., Kamoun, S., Raffaele, S. (2014). The plant membrane-associated REMORIN1.3 accumulates in discrete perihaustorial domains and enhances susceptibility to Phytophthora infestans. Plant Physiol. 165, 1005–1018. doi: 10.1104/pp.114.235804
Bray, E. A. (2002). Abscisic acid regulation of gene expression during water-deficit stress in the era of the Arabidopsis genome. Plant Cell Environ. 25, 153–161. doi: 10.1046/j.1365-3040.2002.00746.x
Byun, M. Y., Lee, J., Cui, L. H., Kang, Y., Oh, T. K., Park, H., et al. (2015). Constitutive expression of DaCBF7, an Antarctic vascular plant Deschampsia Antarctica CBF homolog, resulted in improved cold tolerance in transgenic rice plants. Plant Sci. 236, 61–74. doi: 10.1016/j.plantsci.2015.03.020
Cai, J., Chen, T., Wang, Y., Qin, G., Tian, S. (2020). SlREM1 triggers cell death by activating an oxidative burst and other regulators. Plant Physiol. 183, 717–732. doi: 10.1104/pp.20.00120
Cai, J., Qin, G., Chen, T., Tian, S. (2018). The mode of action of remorin1 in regulating fruit ripening at transcriptional and post-transcriptional levels. New Phytol. 219, 1406–1420. doi: 10.1111/nph.15264
Carvalhais, L. C., Schenk, P. M., Dennis, P. G. (2017). Jasmonic acid signalling and the plant holobiont. Curr. Opin. Microbiol. 37, 42–47. doi: 10.1016/j.mib.2017.03.009
Checker, V. G., Khurana, P. (2013). Molecular and functional characterization of mulberry EST encoding remorin (MiREM) involved in abiotic stress. Plant Cell Rep. 32, 1729–1741. doi: 10.1007/s00299-013-1483-5
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chen, Z., Qin, C., Wang, M., Liao, F., Liao, Q., Liu, X., et al. (2019). Ethylene-mediated improvement in sucrose accumulation in ripening sugarcane involves increased sink strength. BMC Plant Biol. 19, 285. doi: 10.1186/s12870-019-1882-z
Cheng, G., Dong, M., Xu, Q., Peng, L., Yang, Z., Wei, T., et al. (2017). Dissecting the Molecular Mechanism of the Subcellular Localization and Cell-to-cell Movement of the Sugarcane mosaic virus P3N-PIPO. Sci. Rep. 7, 9868. doi: 10.1038/s41598-017-10497-6
Cheng, X., Wang, A. (2017). The potyvirus silencing suppressor protein VPg mediates degradation of SGS3 via ubiquitination and autophagy pathways. J. Virol. 91. doi: 10.1128/JVI.01478-16
Cheng, C., Wang, Y., Chai, F., Li, S., Xin, H., Liang, Z. (2018). Genome-wide identification and characterization of the 14-3-3 family in Vitis vinifera L. during berry development and cold- and heat-stress response. BMC Genomics 19, 579. doi: 10.1186/s12864-018-4955-8
Cheng, G., Yang, Z., Zhang, H., Zhang, J., Xu, J. (2020). Remorin interacting with PCaP1 impairs Turnip Mosaic Virus intercellular movement but is antagonized by VPg. New Phytol. 225, 2122–2139. doi: 10.1111/nph.16285
Chou, K. C., Shen, H. B. (2008). Cell-PLoc: a package of Web servers for predicting subcellular localization of proteins in various organisms. Nat. Protoc. 3, 153–162. doi: 10.1038/nprot.2007.494
Connolly, M. A., Clausen, P. A., Lazar, J. G. (2006). Preparation of RNA from plant tissue using trizol. CSH Protoc. 2006, 4105. doi: 10.1101/pdb.prot4105
Cotton, S., Grangeon, R., Thivierge, K., Mathieu, I., Ide, C., Wei, T., et al. (2009). Turnip mosaic virus RNA replication complex vesicles are mobile, align with microfilaments, and are each derived from a single viral genome. J. Virol. 83, 10460–10471. doi: 10.1128/JVI.00819-09
Cui, X., Yaghmaiean, H., Wu, G., Wu, X., Chen, X., Thorn, G., et al. (2017). The C-terminal region of the Turnip mosaic virus P3 protein is essential for viral infection via targeting P3 to the viral replication complex. Virology 510, 147–155. doi: 10.1016/j.virol.2017.07.016
Ding, Y., Yang, S. (2022). Surviving and thriving: How plants perceive and respond to temperature stress. Dev. Cell 57, 947–958. doi: 10.1016/j.devcel.2022.03.010
Dong, M., Cheng, G., Peng, L., Xu, Q., Yang, Y., Xu, J. (2017). Transcriptome analysis of sugarcane response to the infection by sugarcane steak mosaic virus (SCSMV). Trop. Plant Biol. 10, 45–55. doi: 10.1007/s12042-016-9183-2
Farmer, E. E., Pearce, G., Ryan, C. A. (1989). In vitro phosphorylation of plant plasma membrane proteins in response to the proteinase inhibitor inducing factor. Proc. Natl. Acad. Sci. U.S.A. 86, 1539–1542. doi: 10.1073/pnas.86.5.1539
Fu, S., Xu, Y., Li, C., Li, Y., Wu, J., Zhou, X. (2018). Rice stripe virus interferes with S-acylation of remorin and induces its autophagic degradation to facilitate virus infection. Mol. Plant 11, 269–287. doi: 10.1016/j.molp.2017.11.011
Gasteiger, E., Gattiker, A., Hoogland, C., Ivanyi, I., Appel, R. D., Bairoch, A. (2003). ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 31, 3784–3788. doi: 10.1093/nar/gkg563
Gong, Z., Xiong, L., Shi, H., Yang, S., Herrera-Estrella, L. R., Xu, G., et al. (2020). Plant abiotic stress response and nutrient use efficiency. Sci. China Life Sci. 63, 635–674. doi: 10.1007/s11427-020-1683-x
Gouguet, P., Gronnier, J., Legrand, A., Perraki, A., Jolivet, M. D., Deroubaix, A. F., et al. (2021). Connecting the dots: from nanodomains to physiological functions of REMORINs. Plant Physiol. 185, 632–649. doi: 10.1093/plphys/kiaa063
Gui, J., Liu, C., Shen, J., Li, L. (2014). Grain setting defect1, encoding a remorin protein, affects the grain setting in rice through regulating plasmodesmatal conductance. Plant Physiol. 166, 1463–1478. doi: 10.1104/pp.114.246769
Gui, J., Zheng, S., Liu, C., Shen, J., Li, J., Li, L. (2016). OsREM4.1 interacts with OsSERK1 to coordinate the interlinking between abscisic acid and brassinosteroid signaling in rice. Dev. Cell 38, 201–213. doi: 10.1016/j.devcel.2016.06.011
Guo, J., Ling, H., Wu, Q., Xu, L., Que, Y. (2014). The choice of reference genes for assessing gene expression in sugarcane under salinity and drought stresses. Sci. Rep. 4, 7042. doi: 10.1038/srep07042
Hall, J. S., Adams, B., Parsons, T. J., French, R., Lane, L. C., Jensen, S. G. (1998). Molecular cloning, sequencing, and phylogenetic relationships of a new potyvirus: sugarcane streak mosaic virus, and a reevaluation of the classification of the potyviridae. Mol. Phylogenet. Evol. 10, 323–332. doi: 10.1006/mpev.1998.0535
Hincapie, M., Sood, S., Mollov, D., Odero, D. C., Grisham, M., Rott, P. (2021). Eight species of poaceae are hosting different genetic and pathogenic strains of sugarcane mosaic virus in the everglades agricultural area. Phytopathology 111, 1862–1869. doi: 10.1094/PHYTO-11-20-0489-R
Hu, Y., Jiang, L., Wang, F., Yu, D. (2013). Jasmonate regulates the inducer of cbf expression-C-repeat binding factor/DRE binding factor1 cascade and freezing tolerance in Arabidopsis. Plant Cell 25, 2907–2924. doi: 10.1105/tpc.113.112631
Huang, D., Sun, Y., Ma, Z., Ke, M., Cui, Y., Chen, Z., et al. (2019). Salicylic acid-mediated plasmodesmal closure via Remorin-dependent lipid organization. Proc. Natl. Acad. Sci. U.S.A. 116, 21274–21284. doi: 10.1073/pnas.1911892116
Huang, T. S., Wei, T., Laliberté, J. F., Wang, A. (2010). A host RNA helicase-like protein, AtRH8, interacts with the potyviral genome-linked protein, VPg, associates with the virus accumulation complex, and is essential for infection. Plant Physiol. 152, 255–266. doi: 10.1104/pp.109.147983
Iskandar, H. M., Simpson, R. S., Casu, R. E., Bonnett, G. D., Maclean, D. J., Manners, J. M. (2004). Comparison of reference genes for quantitative real-time polymerase chain reaction analysis of gene expression. Plant Mol. Biol. Rep. 22, 325–337. doi: 10.1007/BF02772676.
Jacinto, T., Farmer, E. E., Ryan, C. A. (1993). Purification of potato leaf plasma membrane protein pp34, a protein phosphorylated in response to oligogalacturonide signals for defense and development. Plant Physiol. 103, 1393–1397. doi: 10.1104/pp.103.4.1393
Jamann, T. M., Luo, X., Morales, L., Kolkman, J. M., Chung, C. L., Nelson, R. J. (2016). A remorin gene is implicated in quantitative disease resistance in maize. Theor. Appl. Genet. 129, 591–602. doi: 10.1007/s00122-015-2650-6
Jarsch, I. K., Ott, T. (2011). Perspectives on remorin proteins, membrane rafts, and their role during plant-microbe interactions. Mol. Plant Microbe Interact. 24, 7–12. doi: 10.1094/MPMI-07-10-0166
Jones, A. M. (2016). A new look at stress: abscisic acid patterns and dynamics at high-resolution. New Phytol. 210, 38–44. doi: 10.1111/nph.13552
Keinath, N. F., Kierszniowska, S., Lorek, J., Bourdais, G., Kessler, S. A., Shimosato-Asano, H., et al. (2010). PAMP (pathogen-associated molecular pattern)-induced changes in plasma membrane compartmentalization reveal novel components of plant immunity. J. Biol. Chem. 285, 39140–39149. doi: 10.1074/jbc.M110.160531
Kong, C.-Y., Luo, Y.-P., Duan, T.-T., Xue, Z., Gao, X.-D., Zhao, X., et al. (2016). Potato remorin gene StREMa4 cloning and its spatiotemporal expression pattern under Ralstonia solanacearum and plant hormones treatment. Phytoparasitica 44, 575–584. doi: 10.1007/s12600-016-0536-z
Konrad, S. S., Popp, C., Stratil, T. F., Jarsch, I. K., Thallmair, V., Folgmann, J., et al. (2014). S-acylation anchors remorin proteins to the plasma membrane but does not primarily determine their localization in membrane microdomains. New Phytol. 203, 758–769. doi: 10.1111/nph.12867
Kumar, D. (2014). Salicylic acid signaling in disease resistance. Plant Sci. 228, 127–134. doi: 10.1016/j.plantsci.2014.04.014
Lefebvre, B., Timmers, T., Mbengue, M., Moreau, S., Hervé, C., Tóth, K., et al. (2010). A remorin protein interacts with symbiotic receptors and regulates bacterial infection. Proc. Natl. Acad. Sci. U.S.A. 107, 2343–2348. doi: 10.1073/pnas.0913320107
Legrand, A., G-Cava, D., Jolivet, M. D., Decossas, M., Lambert, O., Bayle, V., et al. (2023). Structural determinants of REMORIN nanodomain formation in anionic membranes. Biophys. J. 122, 2192–2202. doi: 10.1016/j.bpj.2022.12.035
Lescot, M., Déhais, P., Thijs, G., Marchal, K., Moreau, Y., Van de Peer, Y., et al. (2002). PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 30, 325–327. doi: 10.1093/nar/30.1.325
Li, W., He, Z., Li, S., Huang, Y., Zhang, Z., Jiang, D., et al. (2011). Molecular characterization of a new strain of sugarcane streak mosaic virus (SCSMV). Arch. Virol. 156, 2101–2104. doi: 10.1007/s00705-011-1090-0
Li, F., Wang, A. (2018). RNA decay is an antiviral defense in plants that is counteracted by viral RNA silencing suppressors. PloS Pathog. 14, e1007228. doi: 10.1371/journal.ppat.1007228
Li, Z., Wang, G., Liu, X., Wang, Z., Zhang, M., Zhang, J. (2021). Genome-wide identification and expression profiling of DREB genes in Saccharum spontaneum. BMC Genomics 22, 456. doi: 10.1186/s12864-021-07799-5
Li, H., Wang, X., Zhuo, Y., Chen, S., Lin, J., Ma, H., et al. (2023). Molecular characterization and expression analysis of the remorin genes in tomato (Solanum lycopersicum L.). Front. Plant Sci. 14. doi: 10.3389/fpls.2023.1175153
Li, C., Yue, J., Wu, X., Xu, C., Yu, J. (2014). An ABA-responsive DRE-binding protein gene from Setaria italica, SiARDP, the target gene of SiAREB, plays a critical role under drought stress. J. Exp. Bot. 65, 5415–5427. doi: 10.1093/jxb/eru302
Li, B., Zhang, C., Cao, B., Qin, G., Wang, W., Tian, S. (2012). Brassinolide enhances cold stress tolerance of fruit by regulating plasma membrane proteins and lipids. Amino Acids 43, 2469–2480. doi: 10.1007/s00726-012-1327-6
Liang, P., Stratil, T. F., Popp, C., Marín, M., Folgmann, J., Mysore, K. S., et al. (2018). Symbiotic root infections in Medicago truncatula require remorin-mediated receptor stabilization in membrane nanodomains. Proc. Natl. Acad. Sci. U.S.A. 115, 5289–5294. doi: 10.1073/pnas.1721868115
Ling, H., Wu, Q., Guo, J., Xu, L., Que, Y. (2014). Comprehensive selection of reference genes for gene expression normalization in sugarcane by real time quantitative rt-PCR. PloS One 9, e97469. doi: 10.1371/journal.pone.0097469
Ma, Z., Sun, Y., Zhu, X., Yang, L., Chen, X., Miao, Y. (2022). Membrane nanodomains modulate formin condensation for actin remodeling in Arabidopsis innate immune responses. Plant Cell 34, 374–394. doi: 10.1093/plcell/koab261
Marín, M., Ott, T. (2012). Phosphorylation of intrinsically disordered regions in remorin proteins. Front. Plant Sci. 3. doi: 10.3389/fpls.2012.00086
Marín, M., Thallmair, V., Ott, T. (2012). The intrinsically disordered N-terminal region of AtREM1.3 remorin protein mediates protein-protein interactions. J. Biol. Chem. 287, 39982–39991. doi: 10.1074/jbc.M112.414292
Martinez, D., Legrand, A., Gronnier, J., Decossas, M., Gouguet, P., Lambert, O., et al. (2019). Coiled-coil oligomerization controls localization of the plasma membrane REMORINs. J. Struct. Biol. 206, 12–19. doi: 10.1016/j.jsb.2018.02.003
Mistry, J., Chuguransky, S., Williams, L., Qureshi, M., Salazar, G. A., Sonnhammer, E. L. L., et al. (2021). Pfam: The protein families database in 2021. Nucleic Acids Res. 49, D412–d419. doi: 10.1093/nar/gkaa913
Mongrand, S., Stanislas, T., Bayer, E. M., Lherminier, J., Simon-Plas, F. (2010). Membrane rafts in plant cells. Trends Plant Sci. 15, 656–663. doi: 10.1016/j.tplants.2010.09.003
Nohzadeh Malakshah, S., Habibi Rezaei, M., Heidari, M., Salekdeh, G. H. (2007). Proteomics reveals new salt responsive proteins associated with rice plasma membrane. Biosci. Biotechnol. Biochem. 71, 2144–2154. doi: 10.1271/bbb.70027
Olspert, A., Carr, J. P., Firth, A. E. (2016). Mutational analysis of the Potyviridae transcriptional slippage site utilized for expression of the P3N-PIPO and P1N-PISPO proteins. Nucleic Acids Res. 44, 7618–7629. doi: 10.1093/nar/gkw441
Olspert, A., Chung, B. Y., Atkins, J. F., Carr, J. P., Firth, A. E. (2015). Transcriptional slippage in the positive-sense RNA virus family Potyviridae. EMBO Rep. 16, 995–1004. doi: 10.15252/embr.201540509
Otero, S., Helariutta, Y., Benitez-Alfonso, Y. (2016). Symplastic communication in organ formation and tissue patterning. Curr. Opin. Plant Biol. 29, 21–28. doi: 10.1016/j.pbi.2015.10.007
Perraki, A., Binaghi, M., Mecchia, M. A., Gronnier, J., German-Retana, S., Mongrand, S., et al. (2014). StRemorin1.3 hampers Potato virus X TGBp1 ability to increase plasmodesmata permeability, but does not interfere with its silencing suppressor activity. FEBS Lett. 588, 1699–1705. doi: 10.1016/j.febslet.2014.03.014
Perraki, A., Cacas, J. L., Crowet, J. M., Lins, L., Castroviejo, M., German-Retana, S., et al. (2012). Plasma membrane localization of Solanum tuberosum remorin from group 1, homolog 3 is mediated by conformational changes in a novel C-terminal anchor and required for the restriction of potato virus X movement]. Plant Physiol. 160, 624–637. doi: 10.1104/pp.112.200519
Perraki, A., Gronnier, J., Gouguet, P., Boudsocq, M., Deroubaix, A. F., Simon, V., et al. (2018). REM1.3’s phospho-status defines its plasma membrane nanodomain organization and activity in restricting PVX cell-to-cell movement. PloS Pathog. 14, e1007378. doi: 10.1371/journal.ppat.1007378
Raffaele, S., Bayer, E., Lafarge, D., Cluzet, S., German Retana, S., Boubekeur, T., et al. (2009). Remorin, a solanaceae protein resident in membrane rafts and plasmodesmata, impairs potato virus X movement. Plant Cell 21, 1541–1555. doi: 10.1105/tpc.108.064279
Raffaele, S., Mongrand, S., Gamas, P., Niebel, A., Ott, T. (2007). Genome-wide annotation of remorins, a plant-specific protein family: evolutionary and functional perspectives. Plant Physiol. 145, 593–600. doi: 10.1104/pp.107.108639
Raffaele, S., Perraki, A., Mongrand, S. (2013). The Remorin C-terminal Anchor was shaped by convergent evolution among membrane binding domains. Plant Signal Behav. 8, e23207. doi: 10.4161/psb.23207
Rajamäki, M. L., Valkonen, J. P. (2009). Control of nuclear and nucleolar localization of nuclear inclusion protein a of picorna-like Potato virus A in Nicotiana species. Plant Cell 21, 2485–2502. doi: 10.1105/tpc.108.064147
Reddy, A. R., Ramakrishna, W., Sekhar, A. C., Ithal, N., Babu, P. R., Bonaldo, M. F., et al. (2002). Novel genes are enriched in normalized cDNA libraries from drought-stressed seedlings of rice (Oryza sativa L. subsp. indica cv. Nagina 22). Genome 45, 204–211. doi: 10.1139/g01-114
Reymond, P., Kunz, B., Paul-Pletzer, K., Grimm, R., Eckerskorn, C., Farmer, E. E. (1996). Cloning of a cDNA encoding a plasma membrane-associated, uronide binding phosphoprotein with physical properties similar to viral movement proteins. Plant Cell 8, 2265–2276. doi: 10.1105/tpc.8.12.2265
Riechmann, J. L., Laín, S., García, J. A. (1992). Highlights and prospects of potyvirus molecular biology. J. Gen. Virol. 73, 1–16. doi: 10.1099/0022-1317-73-1-1
Sasaki, N., Takashima, E., Nyunoya, H. (2018). Altered subcellular localization of a tobacco membrane raft-associated remorin protein by tobamovirus infection and transient expression of viral replication and movement proteins. Front. Plant Sci. 9. doi: 10.3389/fpls.2018.00619
Schnable, J. C., Springer, N. M., Freeling, M. (2011). Differentiation of the maize subgenomes by genome dominance and both ancient and ongoing gene loss. Proc. Natl. Acad. Sci. U.S.A. 108, 4069–4074. doi: 10.1073/pnas.1101368108
Shang, H., Fang, L., Qin, L., Jiang, H., Duan, Z., Zhang, H., et al. (2023). Genome-wide identification of the class III peroxidase gene family of sugarcane and its expression profiles under stresses. Front. Plant Sci. 14. doi: 10.3389/fpls.2023.1101665
Son, S., Oh, C. J., Bae, J.-H., Lee, H., An, C. S. (2015). GmREM1.1 and GmREM2.1, which encode the remorin proteins in soybean, have distinct roles during root nodule development. J. Plant Biol. 58, 17–25. doi: 10.1007/s12374-014-0273-0
Su, Y., Guo, J., Ling, H., Chen, S., Wang, S., Xu, L., et al. (2014). Isolation of a novel peroxisomal catalase gene from sugarcane, which is responsive to biotic and abiotic stresses. PloS One 9, e84426. doi: 10.1371/journal.pone.0084426
Su, C., Rodriguez-Franco, M., Lace, B., Nebel, N., Hernandez-Reyes, C., Liang, P., et al. (2023). Stabilization of membrane topologies by proteinaceous remorin scaffolds. Nat. Commun. 14, 323. doi: 10.1038/s41467-023-35976-5
Tóth, K., Stratil, T. F., Madsen, E. B., Ye, J., Popp, C., Antolín-Llovera, M., et al. (2012). Functional domain analysis of the Remorin protein LjSYMREM1 in Lotus japonicus. PloS One 7, e30817. doi: 10.1371/journal.pone.0030817
Urcuqui-Inchima, S., Haenni, A. L., Bernardi, F. (2001). Potyvirus proteins: a wealth of functions. Virus Res. 74, 157–175. doi: 10.1016/S0168-1702(01)00220-9
Vijayapalani, P., Maeshima, M., Nagasaki-Takekuchi, N., Miller, W. A. (2012). Interaction of the trans-frame potyvirus protein P3N-PIPO with host protein PCaP1 facilitates potyvirus movement. PloS Pathog. 8, e1002639. doi: 10.1371/journal.ppat.1002639
Vilakazi, C. S., Dubery, I. A., Piater, L. A. (2017). Identification of lipopolysaccharide-interacting plasma membrane-type proteins in Arabidopsis thaliana. Plant Physiol. Biochem. 111, 155–165. doi: 10.1016/j.plaphy.2016.11.025
Waadt, R., Seller, C. A., Hsu, P. K., Takahashi, Y., Munemasa, S., Schroeder, J. I. (2022). Plant hormone regulation of abiotic stress responses. Nat. Rev. Mol. Cell Biol. 23, 680–694. doi: 10.1038/s41580-022-00479-6
Wang, A. (2015). Dissecting the molecular network of virus-plant interactions: the complex roles of host factors. Annu. Rev. Phytopathol. 53, 45–66. doi: 10.1146/annurev-phyto-080614-120001
Wang, Y., Li, J., Li, M., Li, Y., Zhao, Z., Li, C., et al. (2022). Genome-wide characterization of remorin genes in terms of their evolution and expression in response to hormone signals and abiotic stresses in foxtail millet (Setaria italica). Diversity 14, 711. doi: 10.3390/d14090711.
Wang, P., Moore, B. M., Panchy, N. L., Meng, F., Lehti-Shiu, M. D., Shiu, S. H. (2018). Factors influencing gene family size variation among related species in a plant family, solanaceae. Genome Biol. Evol. 10, 2596–2613. doi: 10.1093/gbe/evy193
Wang, Y., Tang, H., Debarry, J. D., Tan, X., Li, J., Wang, X., et al. (2012). MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 40, e49. doi: 10.1093/nar/gkr1293
Wang, L., Yan, Y., Lu, W., Lu, D. (2020). Application of Exogenous Phytohormones at Silking Stage Improve Grain Quality under Post-Silking Drought Stress in Waxy Maize. Plants (Basel) 10, 48. doi: 10.3390/plants10010048
Ward, C. W., Shukla, D. D. (1991). Taxonomy of potyviruses: current problems and some solutions. Intervirology 32, 269–296. doi: 10.1159/000150211
Wu, X., Lai, Y., Rao, S., Lv, L., Ji, M., Han, K., et al. (2021). Genome-wide identification reveals that Nicotiana benthamiana hypersensitive response (HR)-like lesion inducing protein 4 (NbHRLI4) mediates cell death and salicylic acid-dependent defense responses to turnip mosaic virus. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.627315
Wu, L., Wang, S., Chen, X., Wang, X., Wu, L., Zu, X., et al. (2013). Proteomic and phytohormone analysis of the response of maize (Zea mays L.) seedlings to sugarcane mosaic virus. PloS One 8, e70295. doi: 10.1371/journal.pone.0070295
Xie, T., Chen, C., Li, C., Liu, J., Liu, C., He, Y. (2018). Genome-wide investigation of WRKY gene family in pineapple: evolution and expression profiles during development and stress. BMC Genomics 19, 490. doi: 10.1186/s12864-018-4880-x
Xu, D. L., Park, J. W., Mirkov, T. E., Zhou, G. H. (2008). Viruses causing mosaic disease in sugarcane and their genetic diversity in southern China. Arch. Virol. 153, 1031–1039. doi: 10.1007/s00705-008-0072-3
Xu, D. L., Zhou, G. H., Xie, Y. J., Mock, R., Li, R. (2010). Complete nucleotide sequence and taxonomy of Sugarcane streak mosaic virus, member of a novel genus in the family Potyviridae. Virus Genes 40, 432–439. doi: 10.1007/s11262-010-0457-8
Yang, Z., Dong, M., Cheng, G., Liu, S., Zhang, H., Shang, H., et al. (2021). Selective interaction of sugarcane eIF4E with VPgs from sugarcane mosaic pathogens. Viruses 13, 518. doi: 10.3390/v13030518
Yang, M., Ismayil, A., Liu, Y. (2020). Autophagy in plant-virus interactions. Annu. Rev. Virol. 7, 403–419. doi: 10.1146/annurev-virology-010220-054709
Yao, W., Ruan, M., Qin, L., Yang, C., Chen, R., Chen, B., et al. (2017). Field performance of transgenic sugarcane lines resistant to sugarcane mosaic virus. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.00104
Yue, J., Li, C., Liu, Y., Yu, J. (2014). A remorin gene SiREM6, the target gene of SiARDP, from foxtail millet (Setaria italica) promotes high salt tolerance in transgenic Arabidopsis. PloS One 9, e100772. doi: 10.1371/journal.pone.0100772
Zhai, Y., Yuan, Q., Qiu, S., Li, S., Li, M., Zheng, H., et al. (2021). Turnip mosaic virus impairs perinuclear chloroplast clustering to facilitate viral infection. Plant Cell Environ. 44, 3681–3699. doi: 10.1111/pce.14157
Zhang, H., Cheng, G., Yang, Z., Wang, T., Xu, J. (2019). Identification of sugarcane host factors interacting with the 6K2 protein of the sugarcane mosaic virus. Int. J. Mol. Sci. 20, 3867. doi: 10.3390/ijms20163867
Zhang, H., Deng, C., Wu, X., Yao, J., Zhang, Y., Zhang, Y., et al. (2020). Populus euphratica remorin 6.5 activates plasma membrane H+-ATPases to mediate salt tolerance. Tree Physiol. 40, 731–745. doi: 10.1093/treephys/tpaa022
Zhang, Q., Qi, Y., Pan, H., Tang, H., Wang, G., Hua, X., et al. (2022). Genomic insights into the recent chromosome reduction of autopolyploid sugarcane Saccharum spontaneum. Nat. Genet. 54, 885–896. doi: 10.1038/s41588-022-01084-1
Zhang, J., Zhang, X., Tang, H., Zhang, Q., Hua, X., Ma, X., et al. (2018). Allele-defined genome of the autopolyploid sugarcane Saccharum spontaneum L. Nat. Genet. 50, 1565–1573. doi: 10.1038/s41588-018-0237-2
Keywords: sugarcane, Remorin, gene family, virus, VPg
Citation: Yang Z, Cheng G, Yu Q, Jiao W, Zeng K, Luo T, Zhang H, Shang H, Huang G, Wang F, Guo Y and Xu J (2024) Identification and characterization of the Remorin gene family in Saccharum and the involvement of ScREM1.5e-1/-2 in SCMV infection on sugarcane. Front. Plant Sci. 15:1365995. doi: 10.3389/fpls.2024.1365995
Received: 05 January 2024; Accepted: 08 February 2024;
Published: 23 February 2024.
Edited by:
Harold Meijer, Wageningen University and Research, NetherlandsReviewed by:
Katarzyna Otulak-Kozieł, Warsaw University of Life Sciences, PolandYinglang Wan, Hainan University, China
Copyright © 2024 Yang, Cheng, Yu, Jiao, Zeng, Luo, Zhang, Shang, Huang, Wang, Guo and Xu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jingsheng Xu, eHVqaW5nc2hlbmdAMTI2LmNvbQ==; Fengji Wang, d2FuZ2ZlbmdqaUBmYWZ1LmVkdS5jbg==; Ying Guo, YmlieWd5bGNAMTYzLmNvbQ==
†These authors have contributed equally to this work
 Zongtao Yang
Zongtao Yang Guangyuan Cheng
Guangyuan Cheng Quanxin Yu1
Quanxin Yu1 Heyang Shang
Heyang Shang Jingsheng Xu
Jingsheng Xu