
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 24 July 2024
Sec. Plant Genetics, Epigenetics and Chromosome Biology
Volume 15 - 2024 | https://doi.org/10.3389/fpls.2024.1364086
 Mingli Shen1,2
Mingli Shen1,2 Kai Zhao1,2*
Kai Zhao1,2* Xianmei Luo1,2
Xianmei Luo1,2 Lingling Guo1
Lingling Guo1 Zhirui Ma1
Zhirui Ma1 Lei Wen1
Lei Wen1 Siqing Lin1
Siqing Lin1 Yingxuan Lin1
Yingxuan Lin1 Hongyan Sun1
Hongyan Sun1 Sagheer Ahmad3
Sagheer Ahmad3As an upright tree with multifunctional economic application, Machilus pauhoi is an excellent choice in modern forestry from Lauraceae. The growth characteristics is of great significance for its molecular breeding and improvement. However, there still lack the information of WUSCHEL-related homeobox (WOX) and Auxin response factor (ARF) gene family, which were reported as specific transcription factors in plant growth as well as auxin signaling. Here, a total of sixteen MpWOX and twenty-one MpARF genes were identified from the genome of M. pauhoi. Though member of WOX conserved in the Lauraceae, MpWOX and MpARF genes were unevenly distributed on 12 chromosomes as a result of region duplication. These genes presented 45 and 142 miRNA editing sites, respectively, reflecting a potential post-transcriptional restrain. Overall, MpWOX4, MpWOX13a, MpWOX13b, MpARF6b, MpARF6c, and MpARF19a were highly co-expressed in the vascular cambium, forming a working mode as WOX-ARF complex. MpWOXs contains typical AuxRR-core and TGA-element cis-acting regulatory elements in this auxin signaling linkage. In addition, under IAA and NPA treatments, MpARF2a and MpWOX1a was highly sensitive to IAA response, showing significant changes after 6 hours of treatment. And MpWOX1a was significantly inhibited by NPA treatment. Through all these solid analysis, our findings provide a genetic foundation to growth mechanism analysis and further molecular designing breeding in Machilus pauhoi.
Plants produce shoot apical meristem (SAM) tissues during development, which contain a population of stem cells. These stem cells have the ability to produce new cells, ensuring the continuous formation of new tissues and organs as the plant develops (Bowman and Eshed, 2000; Barton, 2010). SAM is crucial for plant growth and development in the above-ground portion of the plant. The regulatory network responsible for its formation and development is highly complex, influenced by various external environmental factors and internal regulatory factors. The genes CUC1 and CUC2 (Aida et al., 1997), SHOOT MERISTEMLESS (Barton and Poethig, 1993) and WUSCHEL (WUS) (Mayer et al., 1998) have been proven to be involved in the regulatory network. The WUS gene is specifically required for maintaining the structural and functional integrity of shoot and floral meristems, which are crucial for central meristem identity (Laux et al., 1996).The WUSCHEL-related homeobox (WOX) gene family is a plant-specific class of transcription factors that belongs to a subclass of the homeobox (HOX) superfamily (Alvarez et al., 2018). Based on their phylogenetic relationships, plant WOX proteins can be classified into three categories: the WUS clade, the intermediate clade and the ancient clade (van der Graaff et al., 2009). The ancient clade includes conserved WOX genes from algae to angiosperms. The intermediate clade consists of members from ferns to angiosperms, and the modern clade is exclusive to seed plants. This classification reflects the evolutionary time when WOX genes appeared in plants (Lian et al., 2014).WOX genes are characterized by short chains of amino acids (60-66 residues) folded into a helix-loop-helix-turn-helix structure known as the Homeodomain (HD), which is responsible for DNA-binding (Gehring et al., 1990; Gehring et al., 1994; Gu et al., 2020). In addition to the HD structural domain, WOX proteins contain a unique WUS-box motif (TLXLFP) that is conserved in the WOX gene and is essential for WUS activity (Ikeda et al., 2009). The WUS-box is specific to the WUS clade members and functions as an activator and contains a C-terminal EAR domain that involves transcriptional repression. EAR-motif interacts with TOPLESS (TPL)/TPL-related (TPR) corepressor to repress the transcription of auxin-responsive genes (Szemenyei et al., 2008; van der Graaff et al., 2009). WUS (WUSCHEL) was the first gene in the WOX family to be identified. This gene is essential for maintaining the structural and functional integrity of stem and inflorescence meristems in A. thaliana (Laux et al., 1996). The WUS protein performs dual function, acting both as a transcriptional repressor and as an activator involved in the regulation of AGMOUS (AG) expression in A. thaliana (Ikeda et al., 2009). AtWUS transcription factors regulate the differentiation of apical stem cells by controlling auxin hormone signaling and response pathways through the regulation of histone acetylation (Ma et al., 2019). The WUS gene also affects the formation of tiller buds in rice. Xia et al. discovered that the loss of OsWUS function led to decreased tillering and increased apical dominance (Xia et al., 2020). Researchers have recently identified and analyzed the WOX gene families of several other plants. For example, 13, 18, 18, 14 and 31 WOX members are identified in Ginkgo biloba (Nardmann et al., 2009), Populus (Liu et al., 2014b), tea (Wang et al., 2019), Pinus pinaster (Alvarez et al., 2018) and maize (Zhang et al., 2010), respectively. WOX performs specific functions in key developmental processes, including embryonic patterning, stem cell maintenance, organogenesis, floral development, and hormone signaling (Haecker et al., 2004; Cheng et al., 2014; Costanzo et al., 2014; Dolzblasz et al., 2016). WOX4 has been demonstrated to be associated with cambium formation in A. thaliana (Suer et al., 2011) and Populus (Kucukoglu et al., 2017). WOX5 acts downstream of SHORTROOT (SHR)/SCARECROW (SCR) genes in maintaining the stem cell identity (homeostasis) of the root apical meristem (RAM) quiescent center in A. thaliana (Stahl et al., 2009). PtrWUSa/b and PtrWOX13a/b/c are expressed in the vascular cambium and differentiating xylem cells in poplar (Haghighat et al., 2024). PtoWOX5a is participating in the development of adventitious roots in poplar (Li et al., 2018b) and the GhWOX13 gene affects the development of cotton fiber (He et al., 2019). WOX14 promotes the differentiation and lignification of vascular cells in the inflorescence stems of A. thaliana (Denis et al., 2017). However, there are limited studies on the regulatory functions of this gene family in forest trees, particularly in Lauraceae.
The primary form of auxin in plants is indole-3-acetic acid (IAA). It regulates the growth, division, and specific differentiation of cells, participating in the growth and development of various plant parts (Ljung, 2013). The highly conserved nuclear auxin signal transduction pathway is composed of the TIR1/AFB-Aux/IAA auxin co-receptors, the transcriptional co-repressor TOPLESS (TPL), and the AUXIN RESPONSE FACTORS (ARFs) (Galli et al., 2018). ARFs bind with specificity to TGTCTC auxin response elements (AuxRE) in promoters of these genes and function in combination with Aux/IAA (auxin/indole acetic acid) repressors, which dimerize with ARF activators in an auxin-regulated manner (Guilfoyle and Hagen, 2007). The ARF protein has a specific structure and can perform distinct functions, enabling it to participate in multiple signal transduction pathways and other regulatory processes. ARF typically contain three domains: the N-terminal B3-like DNA-binding domain (DBD), the transcriptional regulatory ARF domain of the middle region (MR), and the C-terminal dimerization domain (CTD) (Tiwari et al., 2003; Li et al., 2016). The DBD domain directly and specifically binds to the AuxRE of plant auxin-responsive genes, such as GH3 and SAUR. ARF domains can be categorized as activation domains (AD) or repression domains (RD) based on their functions. The CTD domain is capable of forming dimeric interactions between ARF-ARF or ARF-Aux/IAA to regulate the auxin response (Guilfoyle and Hagen, 2007). The stems of plants play a crucial role in providing support and transporting nutrients, and their growth and development are significantly influenced by auxin. PoptrARF2.1, PoptrARF2.2, PoptrARF3.1, PoptrARF3.4, PoptrARF6.2, and PoptrARF6.3 were found to regulate the growth and development of phloem and xylem (Kalluri et al., 2007) and ARF7 as a molecular bridge of GA and auxin signaling pathways to regulate cambial development in poplar (Hu et al., 2022). PoptrARF5 plays a key role in the development of secondary xylem (Johnson and Douglas, 2007). Moreover, PtoARF5 is able to drive the PtoIAA9-dependent cellular behaviors for secondary xylem differentiation in poplar (Xu et al., 2019). EgrARF of Eucalyptus grandis are also expressed in all parts, with the highest expression of EgrARF3 and EgrARF4 in the stem and phloem. EgrARF5 is highly expressed in both xylem and phloem, while EgrARF10 and EgrARF19A are highly expressed in the vascular cambium (Yu et al., 2014).
It has been demonstrated that WOX11 and 12 are direct target genes for growth hormones in de novo organogenesis (Liu et al., 2014a). In addition, WOX9 is predicted to be a downstream target gene of the MP/BDL (ARF5)-dependent auxin signaling pathway (Haecker et al., 2004). Moreover, ARF5 promotes xylem production mainly through the direct activation of xylem-related genes and repression of WOX4 (Brackmann et al., 2018). Meanwhile, intermediate branches of related WOX genes (IC-WOXs) and class A auxin response factors (A-ARFs) form various protein complexes to activate three distinct root types in A. thaliana (Zhang et al., 2023b). A study on the molecular mechanism of leaf flattening to form broad leaves in A. thaliana, has is found that redundant abaxial-rich ARF repressors can inhibit the expression of WOX1 and PRS through direct DNA-binding (Guan et al., 2017). The aforementioned studies demonstrate a complex regulatory relationship between the WOX genes and ARF.
Machilus pauhoi is an evergreen broad-leaved tree of the genus Machilus in the family Lauraceae, which is characteristics by its strong budding ability, adaptability and versatility. With a straight stem and rapid growth, it is a fast-growing species with high economic value among broad-leaved trees (Chunhui et al., 2019). The rapid and high growth of forest trees is attributed to the vigorous top advantage of the plant. Recently, there have been numerous studies on the agronomic traits and physical characteristics of M. pauhoi (QuanLin et al., 2002; Quanlin et al., 2008; Leilei et al., 2016; Man et al., 2016; Pan et al., 2016; Yan et al., 2018; Yu-xing et al., 2018). However, there is no available report on the MpWOX and MpARF gene currently. Therefore, in this study, we identified and analyzed the WOX and ARF gene family in the M. pauhoi genome, including phylogenetic tree, gene collinearity, gene structure and expression pattern analysis under treatments with IAA and NPA. The aim of this study was to provide valuable information for further investigation of the functions of the WOX and ARF gene families and their interactions in the growth and development of M. pauhoi.
A total of sixteen MpWOX and twenty-one MpARF genes were identified through HMM search and BLAST. The MpWOX and MpARF proteins were named according to their homology with A. thaliana WOX and ARF proteins. The analysis of the physicochemical properties of MpWOX and MpARF revealed significant differences among members of different gene families (Supplementary Table 2). The molecular weights of MpWOX ranged from 19864.29 to 37964.66 Da and the isoelectric points ranged from 5.76 to 9.19. The molecular weights of MpARF ranged from 41288.75 to 131242.66 Da, and the isoelectric points ranged from 5.5 to 8.54. The grand average of hydropathicity (GRAVY) index of all proteins were negative indicating that MpWOX and MpARF had good hydrophilicity. The aliphatic index was greater than 40, showing their thermostability. Subcellular prediction showed that MpWOXs and MpARFs were all localized in the nucleus, this provided a possible interaction and linkage common space.
To gain a deeper understanding of the evolutionary relationships of proteins in various species, particularly in Lauraceae, phylogenetic tree was constructed for 99 WOX and 163 ARF proteins in 7 species. The evolutionary tree was clearly divided into two main branches, WOX and ARF. The 99 WOX proteins could be divided into three branches: the WUS clade (WC), intermediate clade (IC) and ancient clade (AC). The WC contained the most WOX genes (58). The number of MpWOX proteins varied greatly in different branches (Figure 1A). The ancient clade contained members of the MpWOX13a and MpWOX13b. The intermediate clade contained 4 MpWOX members. The remaining MpWOXs were found in the WUS clade. According to the phylogenetic tree, 163 ARF proteins can be classified into 4 classes (I-IV). The ARF proteins in the seven species were primarily distributed in class I and class II. Class II contained the most ARF proteins (58). The MpARF proteins contained eight members in class II, seven members in class I, and only three members in both class III and IV (Figure 1A). Polyploidy in plant evolution is one of the important driving forces behind gene family expansion and the diversification of gene functions (Fernández-Mazuecos and Glover, 2017). There were numerous multi-copy genes in both WOX and ARF gene families of M. pauhoi, particularly MpWOX2, MpARF2, MpARF19 and MpARF6, each of which has three copies in the M. pauhoi genome.
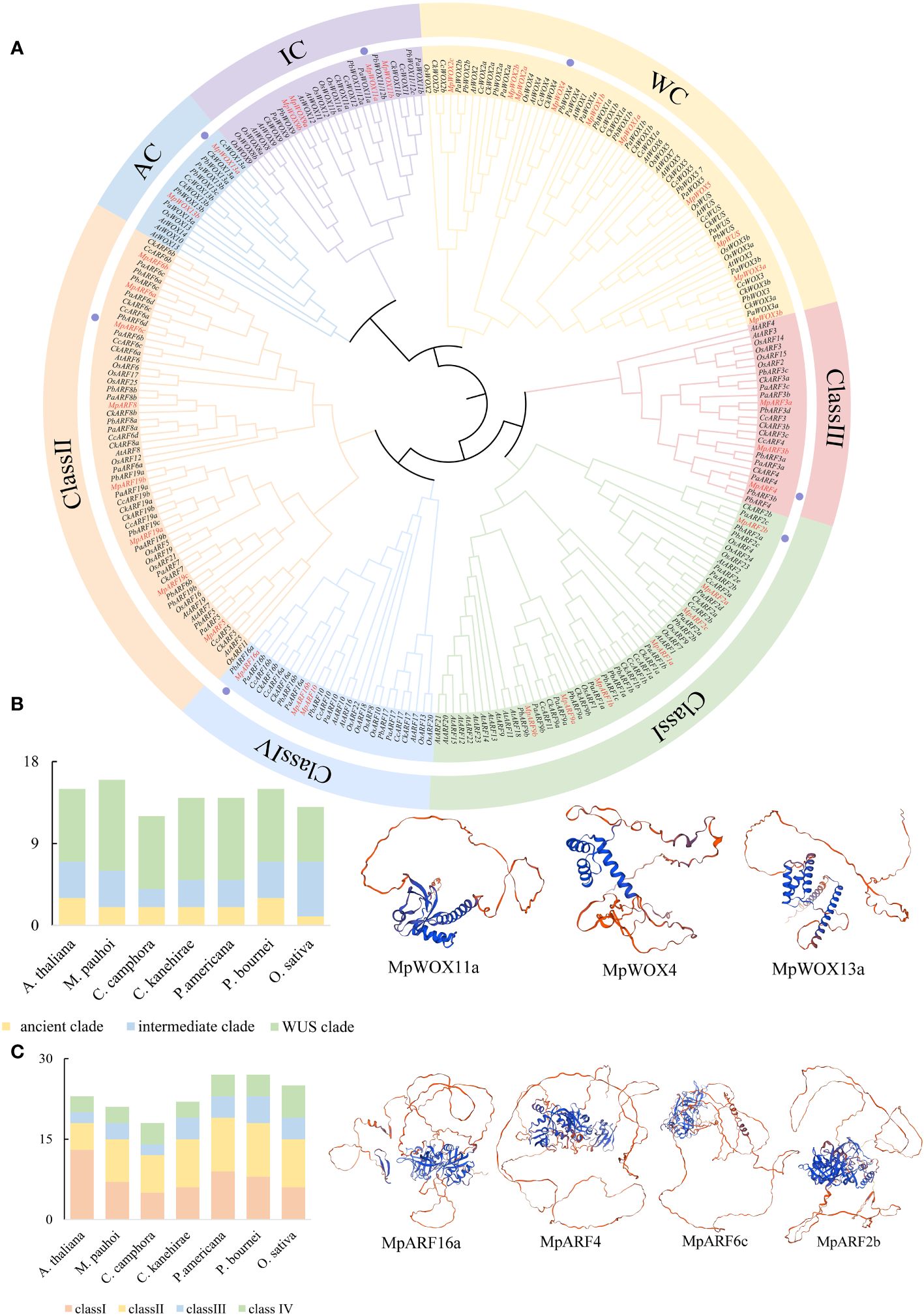
Figure 1 Phylogenetic analysis of WOX and ARF. (A) The evolutionary tree contains a total of 99 WOX and 163 ARF genes. The 99 WOX proteins can be divided into three different branches. The WC is WUS clade. IC, which stands for intermediate clade. AC is an ancient clade. There are 163 ARF proteins divided into 4 classes (I-IV). Different branches are marked with various colors. (B) Number of WOX gene members on different evolutionary branches in seven species. (C) Number of ARF gene members on different evolutionary branches in seven species. The tree contains sequences from Machilus pauhoi (Mp), Cinnamomum kanehirae (Ck), Cinnamomum camphora (Cc), Persea americana (Pa), Phoebe bournei (Pb), Arabidopsis thaliana (At) and Oryza sativa (Os). The WOX and ARF proteins in M. pauhoi are marked with red. In addition, the tertiary structures of some proteins are displayed, and proteins with tertiary structures are labeled with purple dots in part A.
In addition, the number of WOX genes varied from twelve to sixteen in different species (Figure 1B). The number of genes in different subfamilies showed a similar trend, with the WUS clade having the highest number of genes, the intermediate clade having the next highest and the ancient clade having the lowest numbers among all species. This trend in the number of WOX genes was particularly evident in Lauraceae. The number of ARF genes varied from 18 to 27 in different species (Figure 1C). The number of ARF genes fluctuated greatly in Lauraceae plants. P. americana and P. bournei have the same number of ARF genes (27), while C. camphora contains only 18 ARF genes. Compared to A. thaliana, the number of class II members tended to increase in the five species of Lauraceae. In contrast, the number of class II members tended to decrease in Lauraceae, possibly due to the presence of functional redundancy in some genes.
The prediction and analysis showed that MpWOX and MpARF proteins of the same subfamily tended to have similar secondary structures, including alpha-helix, beta-turn, and random coil distributions (Supplementary Table 4; Supplementary Figure 1), as well as tertiary structures (Supplementary Figure 2). The secondary structures of proteins in different subfamilies were quite different, which was mainly reflected in the large fluctuation of alpha helix content (Supplementary Table 4).
The conservative motif showed that MpWOX contained ten distinct motifs. Motif 2 and motif 1 were commonly found in the MpWOX proteins. Motif 10 was specifically present in the ancient clade of the MpWOX. Interestingly, motif 7 and motif 4 only existed in the intermediate clade. Motif 3, motif 8, motif 2, motif 1 and motif 5 were present in all MpARF proteins. Motif 14 was specifically present in the three MpARF proteins of class I, while all three genes of class I were missing motif 6. The results of protein structure analysis showed that members with similar phylogenetic relationships also had similar intron/exon structures. The number of introns varied between one and three in the MpWOX genes. The numbers of introns in the MpARF gene were highly variable, ranging from four (MpARF16b) to eighteen (MpARF2c). Long introns were found in both MpWOX and MpARF genes, such as MpWOX13a, MpWOX2c, MpARF6c, MpARF2a. The presence of longer introns may be due to the insertion of transposons within the genes (Figures 2A, B).
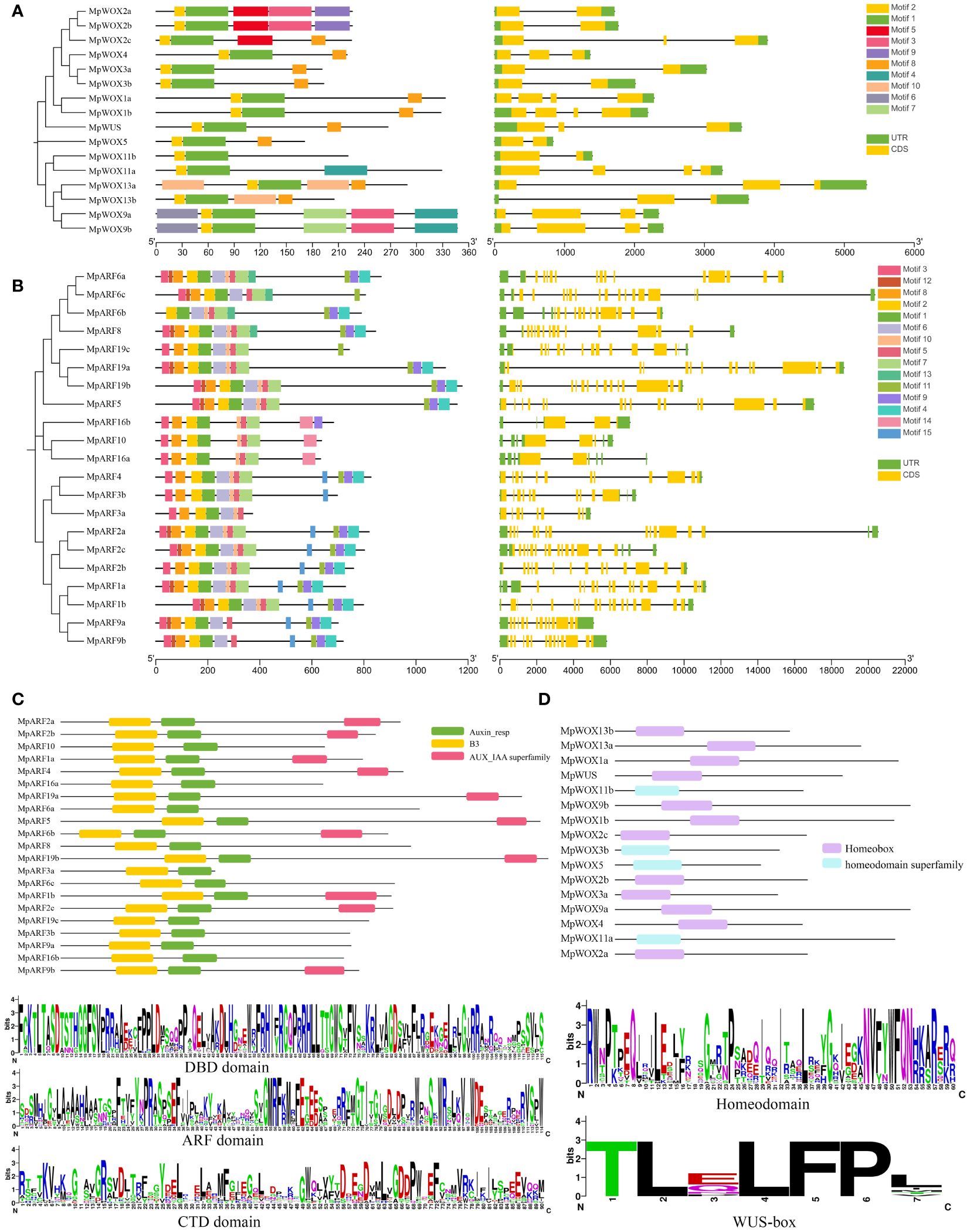
Figure 2 Characteristic of MpWOX and MpARF proteins. (A, B) denotes conserved domain and gene structure analysis of MpWOX and MpARF, respectively. (C) Conserved structural domains of the MpARF. Sequence logo of DBD domain, ARF domain, and CTD domain. (D) Conserved structural domains of the MpWOX. Sequence logo of homeodomain and the WUS-box specific to the WC member.
Analysis of the conserved protein sequences of AtWOXs and MpWOXs revealed that 31 amino acid sequences contained a conserved homeodomain with an average length of 60 aa. Further multi-sequence alignment of the amino acids in the WOX homeodomain revealed that the AtWOX and MpWOX homeodomains were highly conserved. The WUS-box (TLXLFP) is an important structure for the functioning of WC members. The WUS-box was observed in all ten WC members of M. pauhoi (Figure 2D; Supplementary Figure 4). The Batch CD analysis found that all MpARFs contained Auxin_resp and B3 structural domains, which further confirms the accuracy of the identified MpARF genes. Additionally, there were ten MpARFs that did not contain the AUX/IAA superfamily structural domain, and these genes were mainly found in class I and class II. We conducted a multiple sequence comparison among them and the results showed that all MpARFs contained DBD and ARF domains. The CTD structure of certain MpARFs were missing to varying degrees. The conserved structural domains of each gene had a relatively similar sequence composition to one another (Figure 2C; Supplementary Figure 3).
All MpWOX and MpARF genes were mapped to chromosomes based on the gene coordinate annotation data. The 16 MpWOX genes were unevenly distributed on 8 out of the 12 chromosomes and located mainly at the ends of each chromosome (Figure 3A). No MpWOX genes were located on chromosomes 5, 9, 11, and 12. Chromosome 2 boasted the highest count of MpWOX genes, totaling five. Collinearity analysis showed that there were three pairs of segmental duplication genes in MpWOX. Two of them were involved genes from WUS clades, one was involved a homologous gene from intermediate clades, and none was involved a homologous gene from ancient clades (Supplementary Table 5). The 21 MpARF genes were unevenly distributed on 11 out of the 12 chromosomes of M. pauhoi. Chromosome 5 was the sole chromosome that lacked any MpARF genes. Meanwhile, a total of 10 pairs of segmental duplication genes were identified among the 21 MpARF genes, spanning classes I-IV. These co-linear gene pairs predominantly consisted of genes that exhibit multi-copying phenomenon. Notably, chromosome 4 was involved in the most segmental duplication events, with a total of six duplication pairs of the MpARF gene localized on this chromosome. In addition, tandem duplication genes were not present in either the MpWOX genes or the MpARF genes. Furthermore, the collinearity between M. pauhoi and A. thaliana was analyzed (Figure 4B). A total of 15 pairs of collinear relationships between A. thaliana and M. pauhoi, which harbored six pairs of homologous genes for MpWOX and nine pairs of homologous genes for MpARF, respectively (Figure 3B).
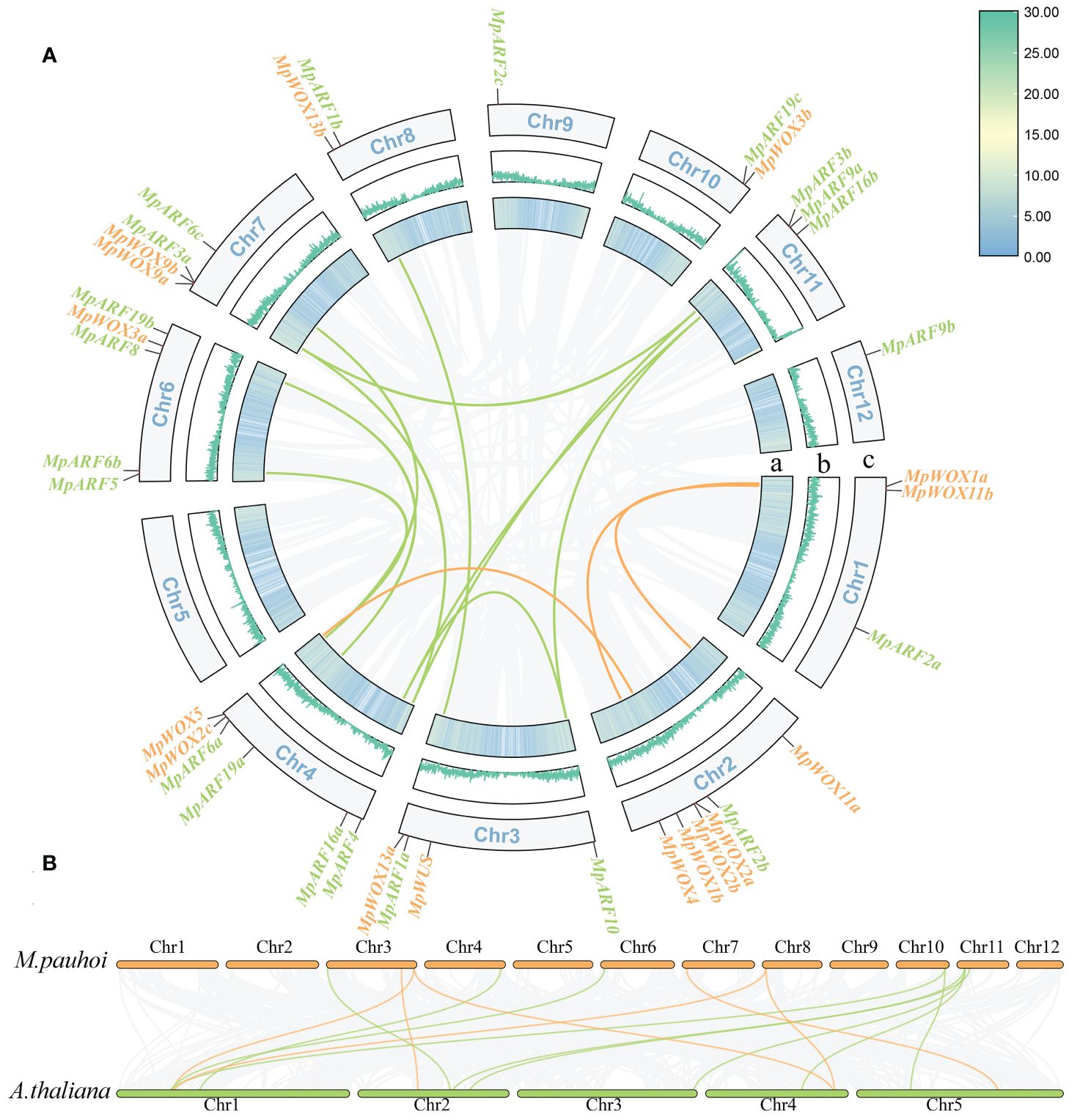
Figure 3 Intragenomic collinearity map of MpWOX and MpARF. (A) Intragenomic collinearity map of MpWOX and MpARF. a: The colored lines indicate gene density. b: The width of the blue lines represents the gene density. c: Chr1-Chr12 represent 12 chromosomes. Green and orange lines represent collinear pairs between ARF and WOX genes, respectively. Gray lines indicate all synteny blocks in the genome. (B) Synteny analysis of WOX and ARF genes between M. pauhoi and A.thaliana. Green and orange lines represent collinear pairs between ARF and WOX genes, respectively. Gray lines indicate all synteny blocks in the genome.
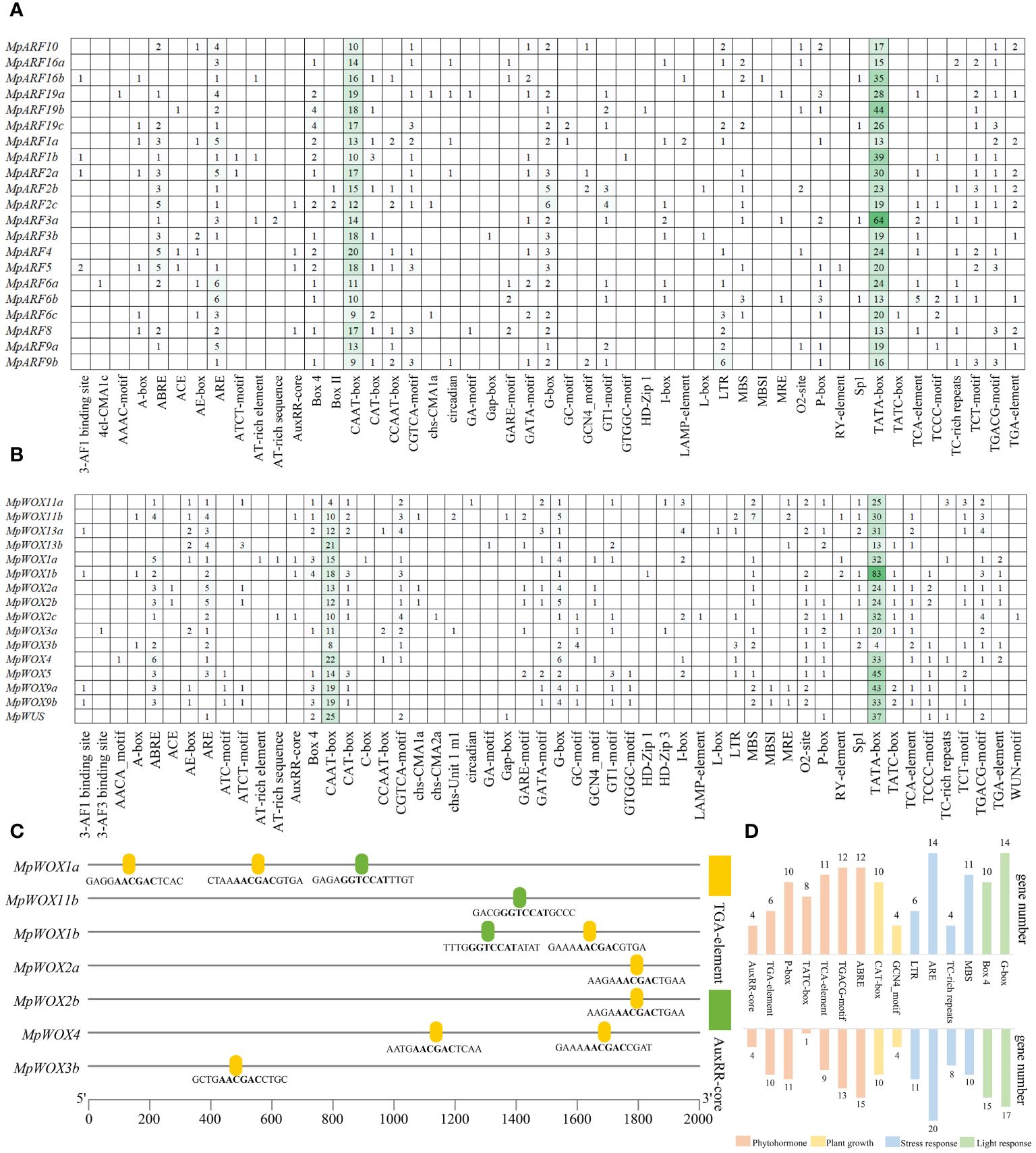
Figure 4 Cis-regulatory element in MpWOXs and MpARFs. (A, B) represent the number and type of cis-regulatory element in MpWOX and MpARF, respectively. (C) Structure and sequence of AuxRR-core and TGA-element in MpWOX promoter. (D) Quantitative statistics of major cis-acting elements in genes.
1206 and 1304 cis-acting regulatory elements were predicted in the promoters of MpWOXs and MpARFs, respectively (Figures 4A, B). Most of these different cis-regulatory elements were involved in plant hormonal regulation, abiotic stress, plant growth, and light response. For MpWOXs, apart from the CAAT-box and TATA-box, the most prevalent regulatory element was the G-box, which is a universal regulatory element in plant responses to environmental stimuli and is widely present in the promoters of light-regulated genes. ABREs (regulating of abscisic acid) and AREs (anaerobic induction) were widely present in MpWOXs. Some auxin-responsive functional elements such as TGA-element (AACGAC) and AuxRR-core (GGTCCAT) exist in seven MpWOX genes (Figure 4C). MpARFs contained regulatory elements for auxin, abscisic acid, gibberellin, salicylic acid, and MeJA. In addition, it contained numerous light-responsive elements, such as G-box, Box4, and GATA-motif (Figure 4D). In conclusion, all the predicted results indicated that MpWOX and MpARF may play important regulatory roles in the responses or regulation by the environment, phytohormones, and abiotic stresses.
Plant miRNA is a group of 21-nucleotides that belongs to endogenous non-coding single-stranded RNA. To gain a better understanding of the role of miRNAs in the post-transcriptional regulation of MpWOXs and MpARFs, the miRNA editing sites were predicted based on the psRNATarget database. 45 and 142 miRNA editing sites were predicted in 16 MpWOX and 21 MpARF genes, respectively (Figure 5; Supplementary Table 6). Among them, there were no miRNA editing sites in MpWOX3a. However, there were more miRNA editing sites in MpWUS, MpWOX9a, MpWOX9b, MpWOX11a. MpARF9a contains the most miRNA-targeting sites (20). MpARF19c contained only one miRNA editing site. At the same time, these predicted miRNA editing sites were mainly distributed in the upstream region of the genes.
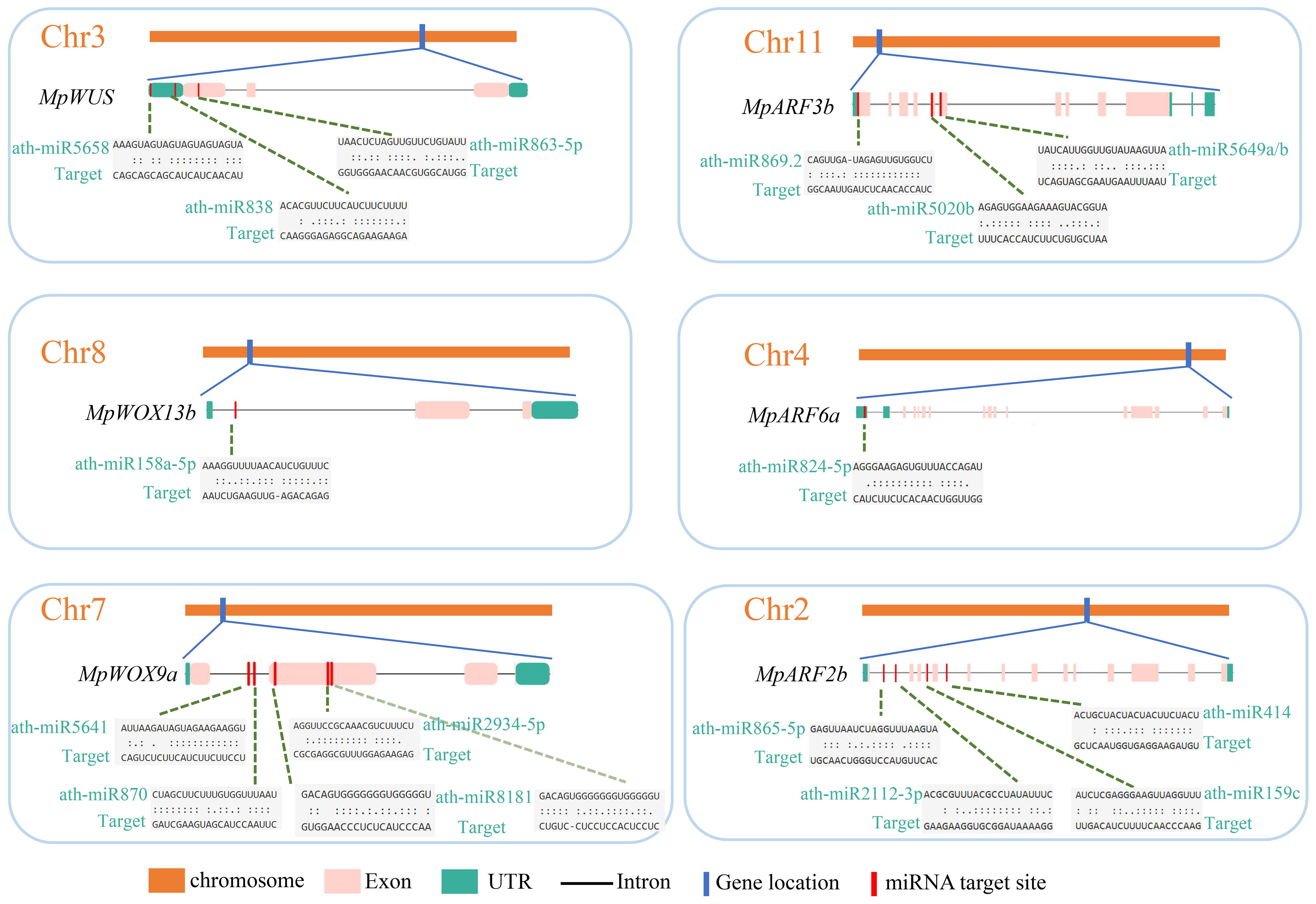
Figure 5 The predicted target sites of miRNA of MpWOX and MpARF gene family. Left and right represent the distribution of miRNA editing sites in MpWOX and MpARF genes, respectively. Detailed miRNA editing information for each gene can be found in Supplementary Table 6.
Based on the available transcriptome data, we analyzed the expression of MpWOX and MpARF genes in leaf, stem, phloem and vascular cambium (Figures 6C, D). There were significant differences in the expression of all MpWOX genes across different tissues. MpWOX4, MpWOX13a and MpWOX13b were expressed by higher levels in all tissues compared with other members of the WOX gene family. This suggested that these three genes may act as important regulatory roles in the growth and development of these four tissues. All MpWOX genes exhibited similar expression profiles in two distinct lineages. In particular, the expression of MpWOX4 gene increased sharply during the transition from vascular cambium 2 to vascular cambium 3. The change of expression level may be related to the thickening of plant growth during the later stages of growth. Compared to the MpWOX genes, all the MpARF genes, except MpARF5, MpARF2b, and MpARF16b, exhibited higher expression in various tissues. Among them, MpARF6c, MpARF19a, MpARF8, MpARF2c, and MpARF4 showed higher expression in phloem, indicating that these genes may play an important role in phloem formation. In addition, MpARF6b, MpARF6c, and MpARF19a were significantly expressed in the vascular cambium, suggesting that they may perform essential activities in this tissue. Similarly to MpWOXs, the MpARF genes showed similar expression patterns in different lineages (Figure 6A). Based on the expression trend of genes, aside from genes in cluster 4 and cluster 5, the remaining genes were mainly clustered in vascular cambium across various periods in different lineages (Figure 6B).
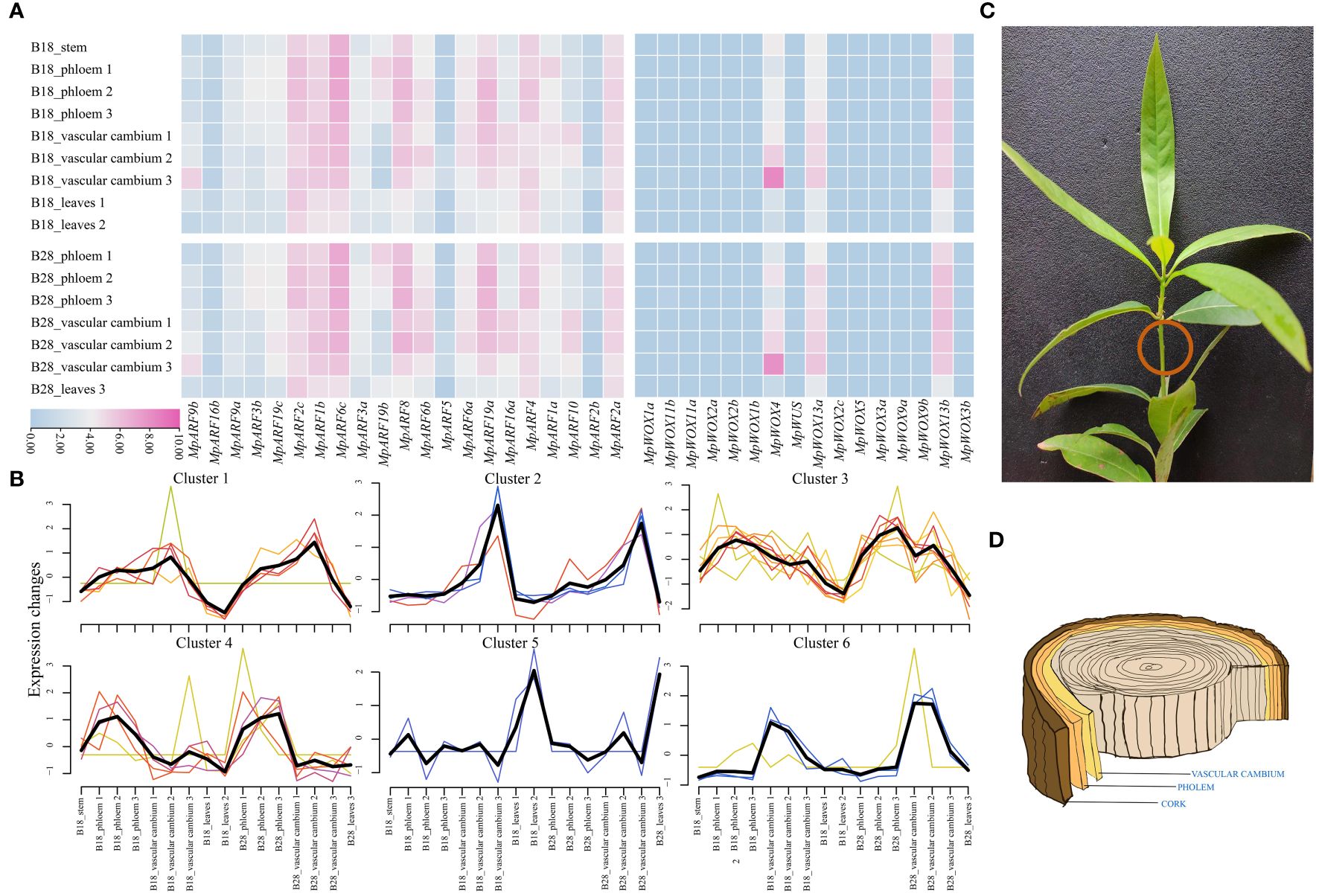
Figure 6 Expression patterns of MpWOX and MpARF genes in M. pauhoi. (A) The transcriptome data of M. pauhoi were obtained from two different lineage, B18 and B28. Among them, B18 lineage plant materials from Ganzhou, Jiangxi province, and B28 lineage plant materials from Jiande, Zhejiang province. Phloem 1: 1 year after planting; Vascular cambium 1: 1 year after planting; Leaves 1: 1 year after planting; Phloem 2: 2 years after planting; Vascular cambium 2: 2 years after planting; Leaves 2: 2 years after planting; Phloem 3: 3 years after planting; Vascular cambium 3: 3 years after planting. FPKM values are logarithmically transformed using a base of 2 for plotting the heatmap. (B) Gene expression trends of the MpWOX and MpARF gene. The black line represents the central trend change, and the other color lines represent the trend change of different genes. (C) M. pauhoi plants. (D) Schematic diagram of the phloem and vascular cambium inside the stem of M. pauhoi.
To further understand the biological function and MpWOXs and MpARFs regulatory networks, the protein-protein interaction (PPI) of MpWOXs and MpARFs were predicted and constructed based on the interaction information of A. thaliana in STRING database. This interactome map consisted of the essential auxin signaling transduction components mediated by WOX-ARF proteins (Figure 7). TPL/TPR corepressors were first described as direct interactors of the Arabidopsis homeodomain transcription factor WUSCHEL (Kieffer et al., 2006). In auxin resting state, AUX/IAA repressors binded auxin response factors (ARFs) and repress their transcription by recruiting the TOPLESS and TOPLESS-related co-repressors (TPL/TPRs). In the presence of auxin, AUX/IAAs binded to TIR1/AFB receptors, they were quickly ubiquitinated and degraded, subsequently releasing the repression of auxin-responsive genes (Mockaitis and Estelle, 2008). Four WOX and nine ARF proteins directly interacted with TPL in the interaction network. There were also interactions between TIR1 and three WOX proteins. In conclusion, based on the predicted results of the interaction network, WOX proteins may be a potential regulatory role of ARFs in plant auxin signaling.
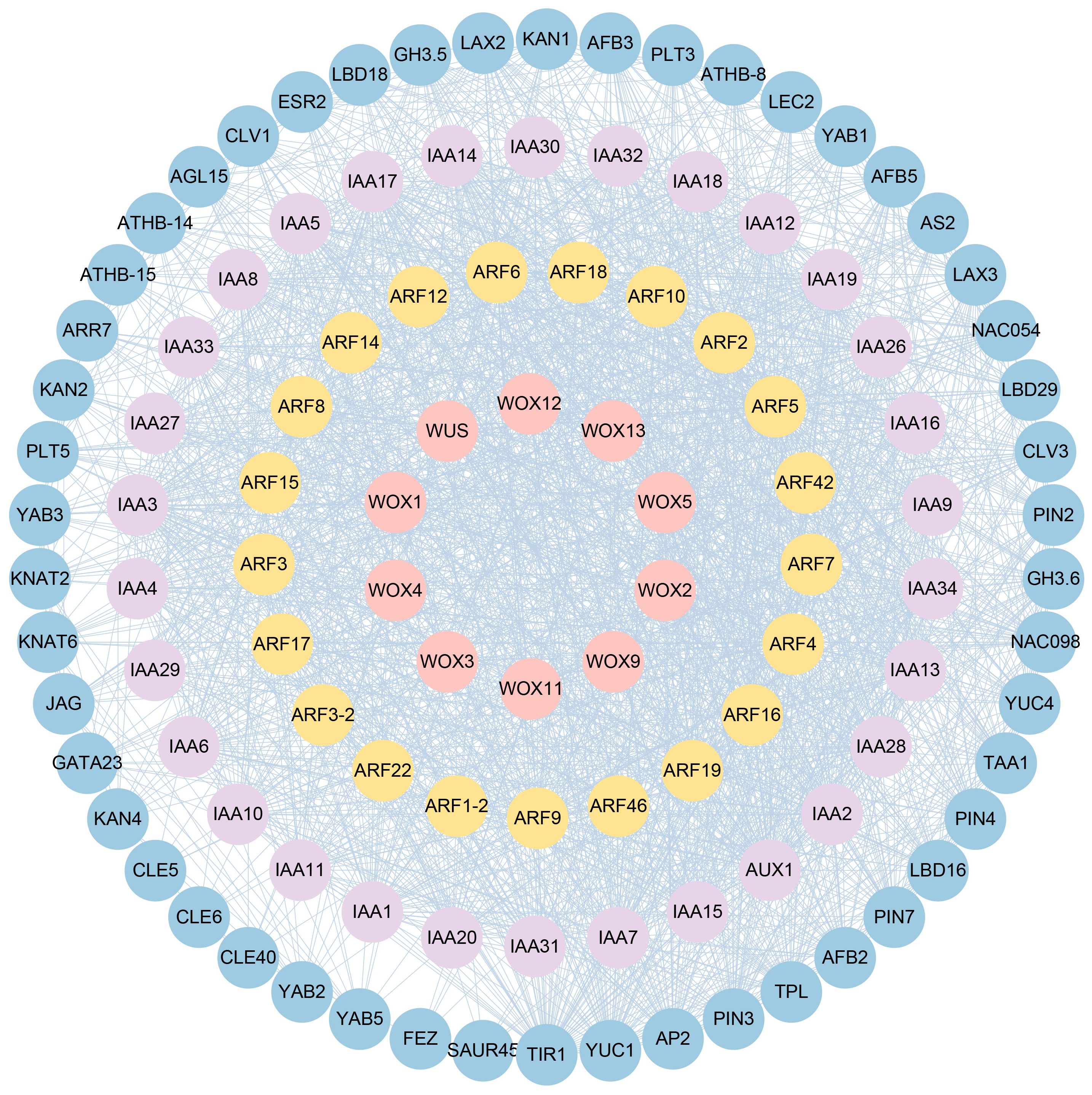
Figure 7 WOX-ARF protein interaction network. Interaction networks between the MpWOX and MpARF are predicted based on the homolog to A. thaliana.
Based on the prediction of cis-regulatory element and PPI, there was a potential interaction between MpWOX and auxin. We treated 1-year-old M. pauhoi with exogenous hormones and analyzed their leaves using RT-qPCR. The results indicated that the expression of all MpARF genes, as well as MpWOX genes, except for MpWOX4, were strongly induced under IAA treatment (Figure 8A). The MpARF genes exhibited a more consistent trend in expression, with fluctuations of varying degrees following a significant increase in expression upon induction for 6 hours. The expression levels of MpWOX13a and MpWOX13b increased with the prolongation of treatment time. The expression levels of MpWUS and MpWOX1a were strongly induced after 6 hours of treatment, and showed a decreasing trend. For NPA treatment, all genes showed down-regulated expression after 6h treatment except MpWUS and MpWOX4. MpWUS, MpWOX13a and MpARF6c had no significant change across all time points. The expressions of MpWOX1a, MpWOX13b, MpARF1b, MpARF2a, and MpARF16a were inhibited at all time points (Figure 8B). Based on the correlation network, there were mainly positive correlations between MpWOX and MpARF genes, except for MpWOX4 (Figure 8C).
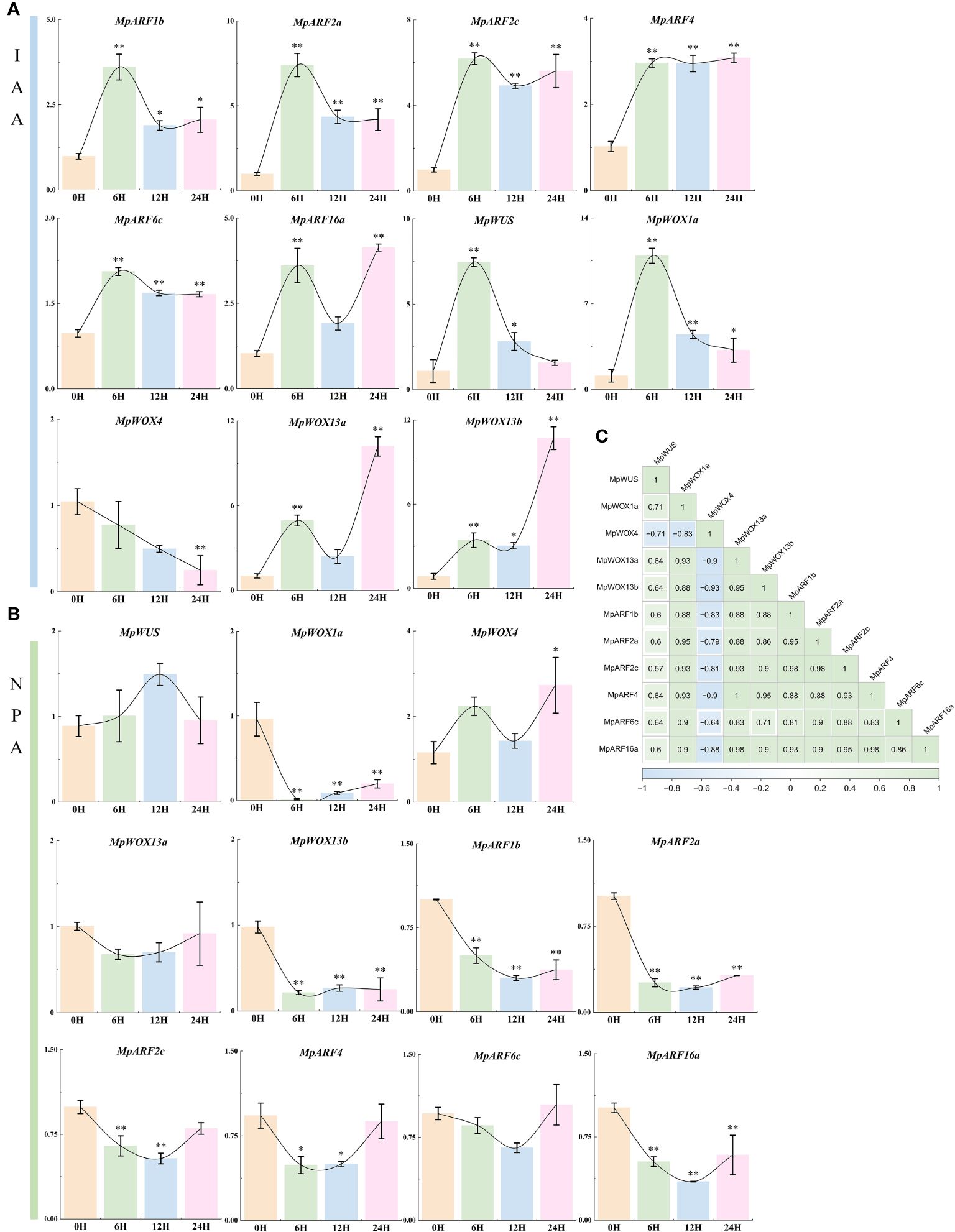
Figure 8 The relative expression level of MpWOX and MpARF under hormone treatment. The data is the average ± standard deviation of the repetition of the three organisms. * and * * stand for p < 0.05 and p < 0.01 respectively, by One-way ANOVAs. (A) The relative expression level of MpWOX and MpARF gene under IAA treatment. (B) The relative expression level of MpWOX and MpARF gene under NPA treatment. (C) Correlation Heatmap.
The WOX gene is a plant-specific TF and plays a crucial role in stem cell formation, maintenance, differentiation, and organogenesis (Dolzblasz et al., 2016). ARFs play a crucial role in the auxin signaling pathway by regulating their own expression and influencing the transcriptional activity of downstream target genes. They achieve this by binding to the response elements in the promoters of auxin responsive genes (Liu et al., 2015). Although the WOX and ARF gene families have been extensively studied in some model plants, these gene families have been less studied in woody plants, especially in Lauraceae. Earlier studies have also demonstrated that ARF-WOX forms a complex in plants to regulate plant growth and development. In this study, AtWOXs and AtARFs from Arabidopsis were utilized to identify WOXs and ARFs proteins in five Lauraceae plants, revealing a range of 12 to 16 WOXs and 18 to 27 ARFs. The quantities differed less from those of A. thaliana [15 AtWOX (Haecker et al., 2004), 23 AtARF (Guilfoyle and Hagen, 2001)] and rice [13 OsWOX (Zhang et al., 2010), 25 OsARF (Wang et al., 2007)], suggesting that there may not have been a large-scale gene duplication event in these two gene families in Lauraceae. Members of the MpARF and MpWOX subfamilies exhibit differences in molecular weight, isoelectric point, and gene structure. These variances are commonly observed in other plants (Yang et al., 2022; Chen et al., 2023). Referring to the classification system of A. thaliana WOX and ARF genes, WOXs and ARFs can be categorized into different evolutionary branches, with variations in the number of genes among these branches. Deletions of A. thaliana homologous genes were present in both MpWOXs and MpARFs and these deletions may be attributed to their functional substitutability (Feng et al., 2021). Members of the WUS branch accounted for a large proportion of the species were counted, indicating that each member of this branch had a specific function in plant growth. At the same time, the large number of WUS clades might be attributed to gene duplication during evolution, particularly in seed plants, to ensure the normal growth of plants (Tang et al., 2017).
The homeodomain is conserved across the WOX gene family in different species and maintains the functional integrity of the WOX gene (Kanchan and Sembi, 2020). Our results show that the HD structure is present in all MpWOX proteins. It has also been demonstrated that the typical WUS-box is limited to the WUS clade. Additionally, the WUS-box interacts with TOPLESS-type co-repressors (WUS, WOX1, WOX5) to mediate gene repression through histone deacetylation (Zhang et al., 2014; Pi et al., 2015). All members of the WUS clade have two amino acid motifs T-L at the beginning of the WUS-box, whereas non-WUS members of the WOX family show variations at this position (van der Graaff et al., 2009). In general, ARF proteins contain three domains: DNA-binding domain (DBD), middle region (MR), and C-terminal domain (CTD). All MpARFs have a typical B3 domain required for efficient binding of AuxRE and ARF domain. However, some MpARFs lack the CTD structure, which may not participate in the auxin signal transduction pathway in plants and perform independent functions (Rui-E et al., 2011). For MpARFs containing DNA-binding domain (DBD) and AUX/IAA domains, we found that these proteins were highly conserved for residues crucial for DNA and IAA binding. These highly conserved structures ensure these proteins accurately perform regulatory functions. The collinearity analysis results revealed the presence of intraspecific collinear homologous genes in both MpWOX and MpARF genes, suggesting the occurrence of gene duplication events in M. pauhoi during evolution, with segmental duplication being the primary mode. As a result of this intragenic segmental duplication event, the MpWOX and MpARF gene family members turn to be multi-copy.
The miRNA complements and connects with target gene mRNA to repress gene expression or cleave mRNA, thereby achieving negative regulation of the target gene. The regulatory role of miRNA on the expression of certain genes has been discovered and confirmed in A. thaliana (Wu et al., 2006). In our study, we find that genes with a higher number of miRNA editing sites exhibit lower expression levels in various tissues. In addition, among the MpARF genes, some genes lacking the complete CTD structural domains, such as MpARF10, MpARF16a, MpARF3a, and MpARF19c, had relatively low expression levels. Comparison of additional conserved structural domains revealed that these genes had a low number of motifs, and the absence of certain motifs may have impacted their normal regulatory functions. It has been demonstrated that WOX4 is located downstream of ARF7, which can directly regulate WOX4 expression and promote the activity of cambium stem cells in poplar (Hu et al., 2022). Previous studies have confirmed the existence of a complex regulatory network between WOX and ARF (Liu et al., 2014a). Some of the MpWOX genes contain auxin response elements, suggesting that these genes may be potentially regulated by ARF genes. In MpWOX genes, while canonical auxin response elements (AuxREs) are absent, they do contain AuxRR-core and TGA-elements. RhARF18 was able to directly bind the AuxRR cis-element in the RhAG promoter and suppress its transcription activity in Rosa hybrida (Chen et al., 2021). Meanwhile, ARF7 and ARF19 are able to bind to the auxin-response elements of the PHR1 promoter in vitro and vivo (Huang et al., 2018). WOX4 and WOX14 can regulate the division activity of cambium cells (Etchells et al., 2013). In our study, MpWOX4 was highly expressed in the vascular cambium, which was consistent with previous research. During secondary development of A. thaliana, the ARF5 is activated by auxin, regulating vascular proliferation early in development but inhibiting vascular cambium expansion later in secondary development (Li et al., 2018a). ARF5 also promotes PIN1 expression in the pro-cambium and increases the number of vascular cambium cells (Wenzel et al., 2007; Smetana et al., 2019). MpARF6b, MpARF6c, and MpARF19a exhibited higher transcription levels in the vascular cambium, suggesting that these genes play crucial roles in the expansion of M. pauhoi cambium cells. Based on the expression trends of genes at different stages, MpWOX and MpARF genes exhibit similar expression patterns, showing high expression during various developmental stages of vascular cambium. Furthermore, the prediction results of the protein interaction network confirmed the presence of an interaction between the ARF-WOX genes. Based on the analysis results and existing research, we believe there exists a linkage-based expression regulatory pattern between WOX and ARF in M. pauhoi, especially playing a crucial role in regulating vascular cambium development. However, interactions between these genes require further experimentation, such as Yeast Two-Hybrid (Y2H) and Bimolecular Fluorescence Complementation (BiFC).
In the auxin signaling pathway, the Aux/IAA-TIR1-ARF signaling cascade regulates the transcription of auxin, facilitating adjustments in auxin concentration to indicate alterations in the transcriptional activity of numerous genes (Paponov et al., 2008). In addition to the Aux/IAA-TIR1-ARF signaling pathway, ARF is also involved in other signaling pathways and regulated by other transcription factors, miRNAs, and ta-siRNAs (Yan-Lin et al., 2017). Based on the prediction from the STRING database, the protein interaction network is primarily regulated by Aux/IAA, TIR1, and ARF genes. It is also suggested that the Aux/IAA-TIR1-ARF signaling pathway plays a crucial role in auxin signal regulation. In Phoebe bournei (Zhang et al., 2023a), the PbWUS gene was strongly induced by IAA treatment, which supports our conclusion. However, the expression levels of PbWOX13a and PbWOX13b decreased under IAA treatment, which differed from our results. This discrepancy may be attributed to variations in species and treatments. In addition, MpWOX1a exhibited a greater increase in expression compared to several other MpWOX genes following IAA treatment, likely due to the presence of two auxin responsive binding elements, TGA-element and AuxRR-core, in MpWOX1a.There are numerous reports on the response of the ARF gene to auxin. The transcripts AtARF4, AtARF5, AtARF16, AtARF19, OsARF1, and OsARF23 exhibited slightly increased expression after auxin treatment (Nagpal et al., 2005; Okushima et al., 2005; Garcia et al., 2006; Wang et al., 2007). The expression of MpARF increased after auxin treatment, consistent with previous studies.
The genome data of M. pauhoi used in this study came from local research group of Fujian Normal University. We utilized HMMER 3.3.2, based on the Hidden Markov model files of WOX (PF00046) and ARF (PF06507) from the Pfam (Pfam: Home page (xfam.org), accessed on 15 October 2023), to search for potential WOX and ARF genes in the M. pauhoi genome. The potential gene family sequences in M. pauhoi were obtained by using BLAST to compare them with the WOX and ARF protein sequences of A. thaliana from the PlantTFDB database (PlantTFDB - Plant Transcription Factor Database @ CBI, PKU (gao-lab.org), accessed on 15 October 2023). Finally, the results obtained from HMM search and BLAST(2.5.0) (Altschul et al., 1990) comparison were intersected to obtain the candidate sequences for the final gene family. The candidate proteins were verified to contain the WOX, B3 and Auxin-resp domain using NCBI-CDD (Conserved Domains Database (CDD) and Resources (nih.gov), accessed on 15 October 2023) with the default parameters. Naming of MpWOX and MpARF genes is based on their homology to AtWOX and AtARF genes. The WOX and ARF gene families of Cinnamomum kanehirae (Chaw et al., 2019), Phoebe bournei (Han et al., 2022; Zhang et al., 2023a), Persea americana (Nath et al., 2022) and Cinnamomum camphora (Shen et al., 2022) were screened and identified using the same method. The physicochemical properties of the gene family were analyzed by ExPASy (http://web.expasy.org/protparam). DeepLoc 2.0 (DeepLoc 2.0 - DTU Health Tech - Bioinformatic Services, accessed on 23 November 2023) was used to predict protein subcellular localization.
The gene sequences of Oryza sativa were obtained from the PlantTFDB database. The gene ID and gene name are shown in Supplementary Table 3. Phylogenetic trees were constructed using MEGAX (Kumar et al., 2018) (Maximum Likelihood, Bootstrap 1000) and visualized using iTOL (iTOL: Interactive Tree Of Life (embl.de), accessed on 2 November 2023). Gene annotation information was used to map genes on chromosomes. MCScanX (Wang et al., 2012) was used to identify collinearity blocks in M. pauhoi. Chromosome localization and collinear results in members of the WOX and ARF gene family are visualized using TBtools.
The conserved motifs of the gene family proteins were analyzed using MEME (Bailey and Elkan, 1994) (Introduction - MEME Suite (meme-suite.org), accessed on 3 November 2023) (with the following parameter settings: the number of motifs: 10, motif width: 6-100). Phylogenetic relationships, conserved motifs and exon and intron structures of the WOXs and ARFs were visualized using TBtools (Chen et al., 2020). Multiple sequence comparison maps and logo maps of conserved structures of the M. pauhoi and A. thaliana WOX gene families were drawn using ESPript (https://espript.ibcp.fr/ESPript/cgi-bin/ESPript.cgi) and WebLogo (WebLogo - Create Sequence Logos (berkeley.edu), accessed on 3 November 2023), respectively. SOPMA (https://npsa-prabi.ibcp.fr/cgi-bin/npsa_automat.pl?page=/NPSA/npsa_sopma.html, accessed on 3 November 2023) was used for protein secondary structure prediction. The tertiary structure of protein was predicted using SWISS-MODEL (Waterhouse et al., 2018).
The genome sequence and gene annotation information files were added to the TBtools GFF3 Sequence Extractor submenu. The upstream bases were set to 2000, and the 2000 bp nucleotide sequence upstream of the MpWOX and MpARF genes were used as the promoter sequence for each MpWOX and MpARF gene. The promoter cis-regulatory elements of MpWOX and MpARF genes were predicted using PlantCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/, accessed on 1 November 2023). Excel was used to count cis-elements implicated in significant biological processes such as phytohormone signaling, stress response, site-binding, light responsiveness, and plant growth. The cis-regulatory elements of the MpWOX and MpARF gene were visualized in Excel.
The miRNA and target of MpWOX and MpARF gene were analyzed using psRNATarget (psRNATarget: A Plant Small RNA Target Analysis Server (2017 Update) (zhaolab.org), accessed on 16 November 2023). The transcriptome data used for the study were obtained from our group. The transcriptome data of M. pauhoi were obtained from two different lineages, B18 and B28. Among them, B18 lineage plant materials were obtained from Ganzhou, Jiangxi province, and B28 lineage plant materials were provided by Jiande, Zhejiang province. Firstly, all the data were quality controlled and filtered using fastp. The obtained data were aligned to the reference genome using hista2 and the FPKM values of all MpWOX and MpARF genes were calculated using the FPKM function in the edgeR package. Finally, TBtools was used to visualize the results and generate the expression heatmap of MpWOX and MpARF. FPKM data Logarithm, set base = 2. Using STRING (https://cn.string-db.org/, accessed on 16 November 2023) protein interaction data, we predicted and constructed protein interaction network.
Plants were grown at Fujian Normal University. We sprayed exogenous hormones to normal growing M. pauhoi plants. The hormones consisted of 3-Indoleacetic acid (IAA, 100 μM) and N-1-Naphthylphthalamic acid (NPA, 100 μM). The surface of the plants was sprayed with the atomizer until the leaves are completely wet but without any condensed droplets. Plant leaves were collected in 2.5 mL sterile and enzyme-free cryopreservation tubes before, 6h, 12h and 24h after hormone treatment, respectively, and rapidly frozen in liquid nitrogen. Total RNA was extracted using Plant RNA Kit R6827 and quantified by measuring A260/280 nm and A260/A230 nm values. The obtained RNA was then reverse transcribed into cDNA using Hifair® III 1st Strand cDNA Synthesis SuperMix. Primer sequence information for the MpWOX and MpARF gene were listed in Supplementary Table 1. The RT-qPCR was performed using the Mpactin gene as an internal reference gene, and the experiment was set up with three biological replicates. The results were used to calculate the relative gene expression levels using the 2−ΔΔCt method.
In this study, we identified two gene families related to meristematic tissue growth in five Lauraceae plants, with WOX ranging from 12 to 16 and ARF from 18 to 27. The upstream region of the MpWOX4 promoter contains two auxin response elements (TGA-elements) and shows a similar expression pattern in the vascular cambium as most MpARF genes. MpWOX4 serve as a conserved gene in the WOX-ARF linkage regulating development in the vascular cambium development.
In addition, MpWUS, MpWOX1a, and MpWOX13 was strongly induced by IAA treatment. However, the expression of MpWOX1a and MpWOX13b was significantly suppressed by NPA treatment. The MpARF genes were induced to varying degrees by IAA. Furthermore, there existed a stable linkage between MpWOX and MpARF genes. Our results will help further investigate the function of MpWOX and MpARF genes in vascular tissue system, thus providing an important foundation for the cultivation of precious forest resources by genome editing and further synthetic biology.
The original M. pauhoi genome and RNA-seq data described in this article have been deposited to NGDC (https://ngdc.cncb.ac.cn/gsub/submit/gsa/list) under the bioproject: PRJCA021595 (CRA013677 and CRA013720). All data generated or analyzed during this study are included in this published article and are also available from the corresponding author on reasonable request.
MS: Data curation, Investigation, Software, Writing – original draft. KZ: Conceptualization, Funding acquisition, Resources, Supervision, Writing – original draft, Writing – review & editing. XL: Data curation, Formal analysis, Writing – original draft. LG: Resources, Software, Writing – original draft. ZM: Resources, Software, Writing – original draft. LW: Resources, Software, Writing – original draft. SL: Resources, Writing – original draft. YL: Resources, Writing – original draft. HS: Resources, Writing – original draft. SA: Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. The research was supported by Forestry Bureau Project of Fujian Province of China (2022FKJ12), the National Natural Science Foundation of China (No. 31901353), the Natural Science Foundation of Fujian province (2022J01639), Science and technology special correspondent project of Fujian province, the Science and Technology Program of Fujian province (2020N0009) and the scientific research innovation program “Xiyuanjiang River Scholarship” from College of Life Sciences, Fujian Normal University.
We would like to thank the editor and reviewers for their helpful comments on the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2024.1364086/full#supplementary-material
Aida, M., Ishida, T., Fukaki, H., Fujisawa, H., Tasaka, M. (1997). Genes involved in organ separation in Arabidopsis: an analysis of the cup-shaped cotyledon mutant. Plant Cell 9, 841–857. doi: 10.1105/tpc.9.6.841
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., Lipman, D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. doi: 10.1016/S0022-2836(05)80360-2
Alvarez, J. M., Bueno, N., Cañas, R. A., Avila, C., Cánovas, F. M., Ordás, R. J. (2018). Analysis of the WUSCHEL-RELATED HOMEOBOX gene family in Pinus pinaster: new insights into the gene family evolution. Plant Physiol. Biochem. 123, 304–318. doi: 10.1016/j.plaphy.2017.12.031
Bailey, T. L., Elkan, C. (1994). Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf Intell. Syst. Mol. Biol. 2, 28–36.
Barton, M. K. (2010). Twenty years on: The inner workings of the shoot apical meristem, a developmental dynamo. Dev. Biol. 341, 95–113. doi: 10.1016/j.ydbio.2009.11.029
Barton, M. K., Poethig, R. S. (1993). Formation of the shoot apical meristem in Arabidopsis thaliana: an analysis of development in the wild type and in the shoot meristemless mutant. Development. 119, 823–831. doi: 10.1242/dev.119.3.823
Bowman, J. L., Eshed, Y. (2000). Formation and maintenance of the shoot apical meristem. Trends Plant Sci. 5, 110–115. doi: 10.1016/S1360-1385(00)01569-7
Brackmann, K., Qi, J., Gebert, M., Jouannet, V., Schlamp, T., Grünwald, K., et al. (2018). Spatial specificity of auxin responses coordinates wood formation. Nat. Commun. 9, 875. doi: 10.1038/s41467-018-03256-2
Chaw, S.-M., Liu, Y.-C., Wu, Y.-W., Wang, H.-Y., Lin, C.-Y. I., Wu, C.-S., et al. (2019). Stout camphor tree genome fills gaps in understanding of flowering plant genome evolution. Nat. Plants 5, 63–73. doi: 10.1038/s41477-018-0337-0
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chen, F., Zhang, J., Ha, X., Ma, H. (2023). Genome-wide identification and expression analysis of the Auxin-Response factor (ARF) gene family in Medicago sativa under abiotic stress. BMC Genomics 24, 498. doi: 10.1186/s12864-023-09610-z
Chen, J., Li, Y., Li, Y., Li, Y., Wang, Y., Jiang, C., et al. (2021). AUXIN RESPONSE FACTOR 18–HISTONE DEACETYLASE 6 module regulates floral organ identity in rose (Rosa hybrida). Plant Physiol. 186, 1074–1087. doi: 10.1093/plphys/kiab130
Cheng, S., Huang, Y., Zhu, N., Zhao, Y. (2014). The rice WUSCHEL-related homeobox genes are involved in reproductive organ development, hormone signaling and abiotic stress response. Gene 549, 266–274. doi: 10.1016/j.gene.2014.08.003
Chunhui, L., Min, Y., Lu, Z., Xiang, L., Zishan, C., Mengyun, C., et al. (2019). Study on the relationship between main wood anatomical traits and wood properties rapid assessment indexes of Machilus pauhiol. J. Forestry Eng. 4, 46–53. doi: 10.13360/j.issn.2096-1359.2019.05.007
Costanzo, E., Trehin, C., Vandenbussche, M. (2014). The role of WOX genes in flower development. Ann. Bot. 114, 1545–1553. doi: 10.1093/aob/mcu123
Denis, E., Kbiri, N., Mary, V., Claisse, G., Conde e Silva, N., Kreis, M., et al. (2017). WOX14 promotes bioactive gibberellin synthesis and vascular cell differentiation in Arabidopsis. Plant J. 90, 560–572. doi: 10.1111/tpj.13513
Dolzblasz, A., Nardmann, J., Clerici, E., Causier, B., van der Graaff, E., Chen, J., et al. (2016). Stem cell regulation by arabidopsis WOX genes. Mol. Plant. 9, 1028–1039. doi: 10.1016/j.molp.2016.04.007
Etchells, P., Provost, C., Mishra, L., Turner, S. (2013). WOX4 and WOX14 act downstream of the PXY receptor kinase to regulate plant vascular proliferation independently of any role in vascular organisation. Development 140, 2224–3224. doi: 10.1242/dev.091314
Feng, C., Shuaiyu, Z., Gao, P., Wang, Z. (2021). In silico identification, characterization expression profile of WUSCHEL-Related Homeobox (WOX) gene family in two species of kiwifruit. PeerJ. 9, e12348. doi: 10.7717/peerj.12348
Fernández-Mazuecos, M., Glover, B. J. (2017). The evo-devo of plant speciation. Nat. Ecol. Evol. 1, 0110. doi: 10.1038/s41559-017-0110
Galli, M., Khakhar, A., Lu, Z., Chen, Z., Sen, S., Joshi, T., et al. (2018). The DNA binding landscape of the maize AUXIN RESPONSE FACTOR family. Nat. Commun. 9, 4526. doi: 10.1038/s41467-018-06977-6
Garcia, D., Collier, S. A., Byrne, M. E., Martienssen, R. A. (2006). Specification of leaf polarity in Arabidopsis via the trans-acting siRNA pathway. Curr. Biol. 16, 933–938. doi: 10.1016/j.cub.2006.03.064
Gehring, W. J., Müller, M., Affolter, M., Percival-Smith, A., Billeter, M., Qian, Y. Q., et al. (1990). The structure of the homeodomain and its functional implications. Trends Genet. 6, 323–329. doi: 10.1016/0168-9525(90)90253-3
Gehring, W. J., Qian, Y. Q., Billeter, M., Furukubo-Tokunaga, K., Schier, A. F., Resendez-Perez, D., et al. (1994). Homeodomain-DNA recognition. Cell. 78, 211–223. doi: 10.1016/0092-8674(94)90292-5
Gu, R., Song, X., Liu, X., Yan, L., Zhou, Z., Zhang, X. (2020). Genome-wide analysis of CsWOX transcription factor gene family in cucumber (Cucumis sativus L.). Sci. Rep. 10, 6216. doi: 10.1038/s41598-020-63197-z
Guan, C., Wu, B., Yu, T., Wang, Q., Krogan, N. T., Liu, X., et al. (2017). Spatial auxin signaling controls leaf flattening in arabidopsis. Curr. Biol. 27, 2940–2950.e4. doi: 10.1016/j.cub.2017.08.042
Guilfoyle, T. J., Hagen, G. (2001). Auxin response factors. J. Plant Growth Regul. 20, 281–291. doi: 10.1007/s003440010026
Guilfoyle, T. J., Hagen, G. (2007). Auxin response factors. Curr. Opin. Plant Biol. 10, 453–460. doi: 10.1016/j.pbi.2007.08.014
Haecker, A., Groß-Hardt, R., Geiges, B., Sarkar, A., Breuninger, H., Herrmann, M., et al. (2004). Expression dynamics of WOX genes mark cell fate decisions during early embryonic patterning in Arabidopsis thaliana. Development. 131, 657–668. doi: 10.1242/dev.00963
Haghighat, M., Zhong, R., Ye, Z.-H. (2024). WUSCHEL-RELATED HOMEOBOX genes are crucial for normal vascular organization and wood formation in poplar. Plant Sci. 346, 112138. doi: 10.1016/j.plantsci.2024.112138
Han, X., Zhang, J., Han, S., Chong, S. L., Meng, G., Song, M., et al. (2022). The chromosome-scale genome of Phoebe bournei reveals contrasting fates of terpene synthase (TPS)-a and TPS-b subfamilies. Plant Commun. 3, 100410. doi: 10.1016/j.xplc.2022.100410
He, P., Zhang, Y., Liu, H., Yuan, Y., Wang, C., Yu, J., et al. (2019). Comprehensive analysis of WOX genes uncovers that WOX13 is involved in phytohormone-mediated fiber development in cotton. BMC Plant Biol. 19, 312. doi: 10.1186/s12870-019-1892-x
Hu, J., Su, H., Cao, H., Wei, H., Fu, X., Jiang, X., et al. (2022). AUXIN RESPONSE FACTOR7 integrates gibberellin and auxin signaling via interactions between DELLA and AUX/IAA proteins to regulate cambial activity in poplar. Plant Cell. 34, 2688–2707. doi: 10.1093/plcell/koac107
Huang, K.-L., Ma, G.-J., Zhang, M.-L., Xiong, H., Wu, H., Zhao, C.-Z., et al. (2018). The ARF7 and ARF19 transcription factors positively regulate PHOSPHATE STARVATION RESPONSE1 in arabidopsis roots. Plant Physiol. 178, 413–427. doi: 10.1104/pp.17.01713
Ikeda, M., Mitsuda, N., Ohme-Takagi, M. (2009). Arabidopsis WUSCHEL is a bifunctional transcription factor that acts as a repressor in stem cell regulation and as an activator in floral patterning. Plant Cell. 21, 3493–3505. doi: 10.1105/tpc.109.069997
Johnson, L. A., Douglas, C. J. (2007). Populus trichocarpa MONOPTEROS/AUXIN RESPONSE FACTOR5 (ARF5) genes: comparative structure, sub-functionalization, and Populus-Arabidopsis microsynteny. Can. J. Bot. 85, 1058–1070. doi: 10.1139/B07-065
Kalluri, U. C., DiFazio, S. P., Brunner, A. M., Tuskan, G. A. (2007). Genome-wide analysis of Aux/IAA and ARF gene families in Populus trichocarpa. BMC Plant Biol. 7, 59. doi: 10.1186/1471-2229-7-59
Kanchan, M., Sembi, J. (2020). Comparative transcriptomic analyses of four Phalaenopsis species to identify and characterize the WUSCHEL -related homeobox (WOX) gene family. Biotechnologia. 101, 309–322. doi: 10.5114/bta.2020.100423
Kieffer, M., Stern, Y., Cook, H., Clerici, E., Maulbetsch, C., Laux, T., et al. (2006). Analysis of the transcription factor WUSCHEL and its functional homologue in antirrhinum reveals a potential mechanism for their roles in meristem maintenance. Plant Cell. 18, 560–573. doi: 10.1105/tpc.105.039107
Kucukoglu, M., Nilsson, J., Zheng, B., Chaabouni, S., Nilsson, O. (2017). WUSCHEL-RELATED HOMEOBOX4 (WOX4)-like genes regulate cambial cell division activity and secondary growth in Populus trees. New Phytol. 215, 642–657. doi: 10.1111/nph.14631
Kumar, S., Stecher, G., Li, M., Knyaz, C., Tamura, K., MEGA, X. (2018). Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Laux, T., Mayer, K. F. X., Berger, J., Jürgens, G. (1996). The WUSCHEL gene is required for shoot and floral meristem integrity in Arabidopsis. Development. 122 1, 87–96. doi: 10.1242/dev.122.1.87
Leilei, Z., Quanlin, Z., Dongliang, C., Ling, F., Jing, L., Yonghong, W., et al. (2016). Biomass relative growth rate of Machilus pauhoi in relation to leaf carbon, nitrogen, and phosphorus stoichiometry properties. Acta Ecologica Sin. 36, 2607–2613. doi: 10.5846/stxb201501160136
Li, X., Tao, S., Wei, S., Ming, M., Huang, X., Zhang, S., et al. (2018a). The mining and evolutionary investigation of AP2/ERF genes in pear (Pyrus). BMC Plant Biol. 18, 46. doi: 10.1186/s12870-018-1265-x
Li, S.-B., Xie, Z.-Z., Hu, C.-G., Zhang, J.-Z. (2016). A review of auxin response factors (ARFs) in plants. Front. Plant Sci. 7, 47. doi: 10.3389/fpls.2016.00047
Li, J., Zhang, J., Jia, H., Liu, B., Sun, P., Hu, J., et al. (2018b). The WUSCHEL-related homeobox 5a (PtoWOX5a) is involved in adventitious root development in poplar. Tree Physiol. 38, 139–153. doi: 10.1093/treephys/tpx118
Lian, G., Ding, Z., Wang, Q., Zhang, D., Xu, J. (2014). Origins and evolution of WUSCHEL-related homeobox protein family in plant kingdom. Sci. World J. 2014, 534140. doi: 10.1155/2014/534140
Liu, J., Sheng, L., Xu, Y., Li, J., Yang, Z., Huang, H., et al. (2014a). WOX11 and 12 Are Involved in the First-Step Cell Fate Transition during de Novo Root Organogenesis in Arabidopsis. Plant Cell 26, 1081–1093. doi: 10.1105/tpc.114.122887
Liu, B., Wang, L., Zhang, J., li, J., Zheng, H., Chen, J., et al. (2014b). WUSCHEL-related Homeobox genes in Populus tomentosa: Diversified expression patterns and a functional similarity in adventitious root formation. BMC Genomics. 15, 296. doi: 10.1186/1471-2164-15-296
Liu, K., Yuan, C., Li, H., Lin, W., Yang, Y., Shen, C., et al. (2015). Genome-wide identification and characterization of auxin response factor (ARF) family genes related to flower and fruit development in papaya (Carica papaya L.). BMC Genomics. 16, 901. doi: 10.1186/s12864-015-2182-0
Ljung, K. (2013). Auxin metabolism and homeostasis during plant development. Development. 140, 943–950. doi: 10.1242/dev.086363
Ma, Y., Miotk, A., Šutiković, Z., Ermakova, O., Wenzl, C., Medzihradszky, A., et al. (2019). WUSCHEL acts as an auxin response rheostat to maintain apical stem cells in Arabidopsis. Nat. Commun. 10, 5093. doi: 10.1038/s41467-019-13074-9
Man, L., Bingjie, J., Quanlin, Z., Yuzhu, M., Hongdian, L., Bingqiao, G., et al. (2016). Effect of nitrogen and phosphorus fertilization on leaf N and P stoichiometriccharacteristics of Machilus pauhoi seedlings. Chin. J. Appl. Environ. Biol. 22, 285–291. doi: 10.3724/SP.J.1145.2015.06022
Mayer, K. F. X., Schoof, H., Haecker, A., Lenhard, M., Jürgens, G., Laux, T. (1998). Role of WUSCHEL in regulating stem cell fate in the arabidopsis shoot meristem. Cell. 95, 805–815. doi: 10.1016/S0092-8674(00)81703-1
Mockaitis, K., Estelle, M. (2008). Auxin receptors and plant development: A new signaling paradigm. Annu. Rev. Cell Dev. Biol. 24, 55–80. doi: 10.1146/annurev.cellbio.23.090506.123214
Nagpal, P., Ellis, C. M., Weber, H., Ploense, S. E., Barkawi, L. S., Guilfoyle, T. J., et al. (2005). Auxin response factors ARF6 and ARF8 promote jasmonic acid production and flower maturation. Development. 132, 4107–4118. doi: 10.1242/dev.01955
Nardmann, J., Reisewitz, P., Werr, W. (2009). Discrete shoot and root stem cell-promoting WUS/WOX5 functions are an evolutionary innovation of angiosperms. Mol. Biol. Evol. 26, 1745–1755. doi: 10.1093/molbev/msp084
Nath, O., Fletcher, S. J., Hayward, A., Shaw, L. M., Masouleh, A. K., Furtado, A., et al. (2022). A haplotype resolved chromosomal level avocado genome allows analysis of novel avocado genes. Horticult. Res. 9, uhac157. doi: 10.1093/hr/uhac157
Okushima, Y., Overvoorde, P. J., Arima, K., Alonso, J. M., Chan, A., Chang, C., et al. (2005). Functional genomic analysis of the AUXIN RESPONSE FACTOR gene family members in Arabidopsis thaliana: unique and overlapping functions of ARF7 and ARF19. Plant Cell. 17, 444–463. doi: 10.1105/tpc.104.028316
Pan, P., Quanlin, Z., Dongliang, C., Chaobin, X., Zhongrui, Z. (2016). The effect of nitrogen and phosphorus foliage spray on the growth of young free growing Machilus pauhoi plantations. Chin. J. Appl. Environ. Biol. 22, 831–838. doi: 10.3724/SP.J.1145.2016.04048
Paponov, I. A., Paponov, M., Teale, W., Menges, M., Chakrabortee, S., Murray, J. A. H., et al. (2008). Comprehensive transcriptome analysis of auxin responses in Arabidopsis. Mol. Plant. 1, 321–337. doi: 10.1093/mp/ssm021
Pi, L., Aichinger, E., van der Graaff, E., Llavata-Peris, C. I., Weijers, D., Hennig, L., et al. (2015). Organizer-derived WOX5 signal maintains root columella stem cells through chromatin-mediated repression of CDF4 expression. Dev. Cell. 33, 576–588. doi: 10.1016/j.devcel.2015.04.024
Quanlin, Z., Songzhu, H., Lizhong, H., Chengcai, T. (2008). Analysis of primary light response parameters of machilus pauhoi from different provenances. Scientia Silvae Sinicae. 44, 118–123. doi: 10.11707/j.1001-7488.20080720
QuanLin, Z., SongZhu, H., ZhiQiang, H., JianHua, C., ShuiQing, X. (2002). Growth characters and effects of ecological factors on Machilus pauhoi. Scientia Silvae Sinicae. 38, 165–168. doi: 10.11707/j.1001-7488.20020228
Rui-E, L., Chang-Gui, H., Yu-Qiang, S. (2011). Advances in plant auxin response factors. Plant Physiol. J. 47, 669–679. doi: 10.13592/j.cnki.ppj.2011.07.010
Shen, T., Qi, H., Luan, X., Xu, W., Yu, F., Zhong, Y., et al. (2022). The chromosome-level genome sequence of the camphor tree provides insights into Lauraceae evolution and terpene biosynthesis. Plant Biotechnol. J. 20, 244–246. doi: 10.1111/pbi.13749
Smetana, O., Mäkilä, R., Lyu, M., Amiryousefi, A., Sánchez Rodríguez, F., Wu, M.-F., et al. (2019). High levels of auxin signalling define the stem-cell organizer of the vascular cambium. Nature. 565, 485–489. doi: 10.1038/s41586-018-0837-0
Stahl, Y., Wink, R. H., Ingram, G. C., Simon, R. (2009). A signaling module controlling the stem cell niche in arabidopsis root meristems. Curr. Biol. 19, 909–914. doi: 10.1016/j.cub.2009.03.060
Suer, S., Agusti, J., Sanchez, P., Schwarz, M., Greb, T. (2011). WOX4 imparts auxin responsiveness to cambium cells in arabidopsis. Plant Cell. 23, 3247–3259. doi: 10.1105/tpc.111.087874
Szemenyei, H., Hannon, M., Long, J. A. (2008). TOPLESS mediates auxin-dependent transcriptional repression during arabidopsis embryogenesis. Science. 319, 1384–1386. doi: 10.1126/science.1151461
Tang, F., Chen, N., Zhao, M., Wang, Y., He, R., Peng, X., et al. (2017). Identification and functional divergence analysis of WOX gene family in paper mulberry. Int. J. Mol. Sci. 18, 1782. doi: 10.3390/ijms18081782
Tiwari, S. B., Hagen, G., Guilfoyle, T. (2003). The roles of auxin response factor domains in auxin-responsive transcription. Plant Cell. 15, 533–543. doi: 10.1105/tpc.008417
van der Graaff, E., Laux, T., Rensing, S. A. (2009). The WUS homeobox-containing (WOX) protein family. Genome Biol. 10, 248. doi: 10.1186/gb-2009-10-12-248
Wang, P., Guo, Y., Chen, X., Zheng, Y., Sun, Y., Yang, J., et al. (2019). Genome-wide identification of WOX genes and their expression patterns under different hormone and abiotic stress treatments in tea plant (Camellia sinensis). Trees (Berl West). 33, 1129–1142. doi: 10.1007/s00468-019-01847-0
Wang, D., Pei, K., Fu, Y., Sun, Z., Li, S., Liu, H., et al. (2007). Genome-wide analysis of the auxin response factors (ARF) gene family in rice (Oryza sativa). Gene. 394, 13–24. doi: 10.1016/j.gene.2007.01.006
Wang, Y., Tang, H., DeBarry, J. D., Tan, X., Li, J., Wang, X., et al. (2012). MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 40, e49. doi: 10.1093/nar/gkr1293
Waterhouse, A., Bertoni, M., Bienert, S., Studer, G., Tauriello, G., Gumienny, R., et al. (2018). SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 46, W296–W303. doi: 10.1093/nar/gky427
Wenzel, C. L., Schuetz, M., Yu, Q., Mattsson, J. (2007). Dynamics of MONOPTEROS and PIN-FORMED1 expression during leaf vein pattern formation in Arabidopsis thaliana. Plant J. 49, 387–398. doi: 10.1111/j.1365-313X.2006.02977.x
Wu, M.-F., Tian, Q., Reed, J. W. (2006). Arabidopsis microRNA167 controls patterns of ARF6 and ARF8 expression, and regulates both female and male reproduction. Development. 133, 4211–4218. doi: 10.1242/dev.02602
Xia, T., Chen, H., Dong, S., Ma, Z., Ren, H., Zhu, X., et al. (2020). OsWUS promotes tiller bud growth by establishing weak apical dominance in rice. Plant J. 104, 1635–1647. doi: 10.1111/tpj.15026
Xu, C., Shen, Y., He, F., Fu, X., Yu, H., Lu, W., et al. (2019). Auxin-mediated Aux/IAA-ARF-HB signaling cascade regulates secondary xylem development in Populus. New Phytol. 222, 752–767. doi: 10.1111/nph.15658
Yan, W., Quanlin, Z., Chaobin, X., Zhongrui, Z., Dongliang, C. (2018). Effect of adding a combination of nitrogen and phosphorus on fine root morphology and soil microbes of Machilus pauhoi seedling. Acta Ecologica Sin. 38, 2271–2278. doi: 10.5846/stxb201704240735
Yang, T., Gao, T., Wang, C., Wang, X., Chen, C., Tian, M., et al. (2022). In silico genome wide identification and expression analysis of the WUSCHEL-related homeobox gene family in Medicago sativa. Genomics Inform. 20, e19. doi: 10.5808/gi.22013
Yan-Lin, L., Zhi-Hong, G., Juan, S., Wan-Xu, W., Ting, S. (2017). Auxin response factor (ARF) and its functions in plant growth and development. Plant Physiol. J. 53, 1842–1858. doi: 10.13592/j.cnki.ppj.2017.0271
Yu, H., Soler, M., Mila, I., San Clemente, H., Savelli, B., Dunand, C., et al. (2014). Genome-wide characterization and expression profiling of the AUXIN RESPONSE FACTOR (ARF) gene family in eucalyptus grandis. PloS One. 9, e108906. doi: 10.1371/journal.pone.0108906
Yu-xing, Z., Quan-lin, Z., Ya-ling, Y., Hua, Y., Wen-ting, Z., Jia-jing, C., et al. (2018). Short-term effects of nitrogen and water treatments on fine root order morphology of Machilus pauhoi seedlings. Chin. J. Appl. Ecol. 29, 2323–2329. doi: 10.13287/j.1001-9332.201807.014
Zhang, M., Chen, X., Lou, X., Zhang, Y., Han, X., Yang, Q., et al. (2023a). Identification of WUSCHEL-related homeobox (WOX) gene family members and determination of their expression profiles during somatic embryogenesis in Phoebe bournei. Forestry Res. 3, 5. doi: 10.48130/FR-2023-0005
Zhang, T., Ge, Y., Cai, G., Pan, X., Xu, L. (2023b). WOX-ARF modules initiate different types of roots. Cell Rep. 42, 112966. doi: 10.1016/j.celrep.2023.112966
Zhang, F., Wang, Y., Li, G., Tang, Y., Kramer, E. M., Tadege, M. (2014). STENOFOLIA Recruits TOPLESS to Repress ASYMMETRIC LEAVES2 at the Leaf Margin and Promote Leaf Blade Outgrowth in Medicago truncatula. Plant Cell. 26, 650–664. doi: 10.1105/tpc.113.121947
Keywords: Lauraceae, WOX-ARF linkage, miRNA binding sites, IAA response, development
Citation: Shen M, Zhao K, Luo X, Guo L, Ma Z, Wen L, Lin S, Lin Y, Sun H and Ahmad S (2024) Genome mining of WOX-ARF gene linkage in Machilus pauhoi underpinned cambial activity associated with IAA induction. Front. Plant Sci. 15:1364086. doi: 10.3389/fpls.2024.1364086
Received: 01 January 2024; Accepted: 09 July 2024;
Published: 24 July 2024.
Edited by:
Chun-Ping Yu, Academia Sinica, TaiwanReviewed by:
Khalid Abdallah Hussein, Assiut University, EgyptCopyright © 2024 Shen, Zhao, Luo, Guo, Ma, Wen, Lin, Lin, Sun and Ahmad. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kai Zhao, emhhb2thaUBmam51LmVkdS5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.