- 1Department of Pesticide Science, State Key Laboratory for Conservation and Utilization of Bio-Resource in Yunnan, College of Plant Protection, Yunnan Agricultural University, Kunming, Yunnan, China
- 2Sanya Institute of China Agricultural University, Sanya, Hainan, China
- 3Hainan Seed Industry Laboratory, Sanya, Hainan, China
- 4Department of Plant Pathology, Beijing Key Laboratory of Seed Disease Testing and Control, China Agricultural University, Beijing, China
- 5MOA Key Lab of Pest Monitoring and Green Management, China Agricultural University, Beijing, China
Aglaia odorata, native to Guangdong, Guangxi, and Hainan provinces in China, has long been utilized as an herbal remedy in ancient China. In this study, we assembled and annotated the complete mitochondrial genome (mitogenome) of A. odorata, which spans a total length of 537,321 bp. Conformation of the A. odorata recombination was verified through PCR experiments and Sanger sequencing. We identified and annotated 35 protein-coding genes (PCGs), 22 tRNA genes, and 3 rRNA genes within the mitogenome. Analysis of repeated elements revealed the presence of 192 SSRs, 29 pairs of tandem repeats, and 333 pairs of dispersed repeats in the A. odorata mitogenome. Additionally, we analyzed codon usage and mitochondrial plastid DNAs (MTPTs). Twelve MTPTs between the plastome and mitogenome of A. odorata were identified, with a combined length of 2,501 bp, accounting for 0.47% of the mitogenome. Furthermore, 359 high-confidence C to U RNA editing sites were predicted on PCGs, and four selected RNA editing sites were specially examined to verify the creation of start and/or stop codons. Extensive genomic rearrangement was observed between A. odorata and related mitogenomes. Phylogenetic analysis based on mitochondrial PCGs were conducted to elucidate the evolutionary relationships between A. odorata and other angiosperms.
Introduction
Aglaia odorata Lour. belongs to the Aglaia genus within the Meliaceae family. It is native to Guangdong, Guangxi and Hainan provinces in China, as well as various Southeast Asian countries (Zhang et al., 2020b). This plant is commonly found in sparsely forested areas and shrubbery within low-altitude mountain regions (Liu et al., 2014). In ancient China, A. odorata has traditionally been employed as an herbal remedy for treating heart diseases, bruises, traumatic injuries, and fever (Yang et al., 2022). Modern pharmacological studies indicate that A. odorata exhibits anti-cancer, anti-inflammatory, antibacterial, and antiviral activities (Inad et al., 2001). Despite its economic value, there has been limited research on it, and the genomic information of this species is still lacking at present. So far, in various databases such as the GenBank database, only the chloroplast genome resource is available (accession number: NC_048994.1).
Mitochondria play a pivotal role in synthesizing and converting energy for diverse cellular physiological processes, rendering them essential for plant growth and development (Ye et al., 2017). They transform biomass energy into chemical energy via phosphorylation and participate in cellular processes such as cell division, differentiation, and apoptosis (Kroemer and Reed, 2000; van Loo et al., 2002; Bonora et al., 2014). Mitochondria stand as distinctive cellular structures separate from the nucleus, housing their own genome. This genetic material is inherited in a haploid, asexual, and maternal fashion (Cheng et al., 2021). Following the endosymbiotic theory, mitochondria’s origins trace back to the mutualistic relationship between alpha-bacteria and archaea-derived host cells, eventually evolved into integral organelles within eukaryotic cells (Roger et al., 2017). While mitogenomes, like to plastidial genomes, are maternally inherited and encompass a smaller gene set, significant evolutionary distinctions exist between these two genomes. Compared to the mitogenomes, the plastidial genomes are relatively compact and remarkably conserved. Plant mitogenomes display significant size variation, spanning from 60 kb to over 11 Mb among different species, a considerably broader range than what is observed in plastid genomes (Sloan et al., 2012; Skippington et al., 2015). Higher plant mitogenomes exhibit linear, circular, complex branching and reticular structures, whereas the majority of plant plastidial genomes have a circular structure (Cheng et al., 2017; Kozik et al., 2019; Fischer et al., 2022). Plant mitogenomes tend to exhibit higher mutation rates in comparison to nuclear genomes, a consequence attributed to the absence of robust DNA repair systems (Gualberto et al., 2014; Morley et al., 2019). This higher mutation rate contributes to rearrangements, duplications, and the generation of subgenomic configurations within the mitogenome. Furthermore, certain plant mitogenomes have assimilated genes through horizontal gene transfer from external organisms. This occurrence is especially prevalent in higher plants, where they have incorporated several plastid sequences from neighboring chloroplasts. This evolutionary process has transpired over an extended timeframe and is likely ongoing (Choi and Park, 2021; Garcia et al., 2021; Lin et al., 2022). Currently, the availability of mitogenome resources for the plants is limited.
In this study, we assembled and annotated the mitogenome and plastidial genome of A. odorata, analyzed the codon usage, repeated elements, and mitochondrial plastid DNAs (MTPTs). We also analyzed the RNA editing sites in mitochondrial PCGs. Lastly, we inferred the phylogenetic relationships of A. odorata and other angiosperms based on mitochondrial PCGs. As the first reported mitogenome within genus Aglaia, this study provides valuable reference for mitogenome analysis in Aglaia species. Additionally, it offers important insights into RNA editing, mitochondrial genome evolution, genome rearrangement, and phylogenetics of angiosperms. Furthermore, we also provide reliable genomic resources for studying the organelle genomes of Meliaceae plants.
Materials and methods
Plant sampling, DNA extracting and sequencing
The fresh leaves of A. odorata were collected in Sanya, Hainan, China. These specimens have been deposited in our lab (Seed Health Centre of China Agricultural University, Sanya Institute of China Agricultural University and Yunnan Agricultural University). Genomic DNA was extracted using the Tiangen Biotech DNA kit (Beijing). For library construction, we utilized the NEBNext® library building kit with an insert size of 350 bp. The constructed DNA library was sequenced on the NovaSeq 6000 platform at Benagen (Wuhan, China). To ensure data quality, we applied Trimmomatic (Bolger et al., 2014) to remove low-quality sequences, including those with a quality value (Q) of less than or equal to 5, which accounted for more than 50% of the total bases, as well as sequences containing more than 10% “N” bases. Furthermore, the plant sample used for Illumina sequencing was also subjected to Oxford Nanopore sequencing based on PromethION devices. Purified DNA was prepared for long-read sequencing following the protocol outlined in the SQK-LSK109 genomic sequencing kit (ONT, Oxford, UK).
RNA extracting and sequencing
For long non-coding RNA (lncRNA) extraction, total RNA was isolated from fresh A. odorata leaves using a high-quality RNA extraction kit (TRIzol® Reagent, Thermo Fisher Scientific, Waltham, MA, USA), following the manufacturer’s instructions. The extracted RNA was reverse-transcribed into cDNA using random primers, and rRNA was subsequently removed. The processed cDNA was fragmented and constructed into a library with an average length of 500 bp. The integrity and concentration of the RNA were assessed using Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA) and NanoDrop spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA). The enriched lncRNA was then used to construct a cDNA library employing a protocol compatible with lncRNA sequencing. The library was subsequently sequenced using an Illumina HiSeq platform. Quality control measures were implemented to filter out low-quality sequences, and bioinformatics analyses were performed on the resulting data to identify and characterize lncRNAs.
Organelle genome assembly
For plastome assembly, we utilized GetOrganelle v1.7.4.1 with the following parameters: ‘-R 15 -k 21,45,65,85,105 -F embplant_pt’ to assemble the Illumina short-reads (Jin et al., 2020). GetOrganelle generated two complete plastome sequences, and we selected the one where the SSC region aligns in the same direction as Arabidopsis thaliana (NC_000932.1). Subsequently, we performed de novo assembly for long-reads. The long-reads were polished using Canu (Koren et al., 2017) and then assembled using PMAT assembler (Bi et al., 2024a) with the default parameters. BLASTn (Chen et al., 2015) was employed to identify the draft mitogenome from the assembled sequences. Six mitochondrial contigs were successfully identified. Considering the low accuracy of long-reads from Oxford Nanopore sequencing, we further utilize assembled mitochondrial sequences as the reference sequence. We establish an index using BWA (Li and Durbin, 2009). Subsequently, ‘bwa mem’ was utilized to obtain reads successfully mapped to the reference sequence. Indexing and mapping of the long-reads were performed using minimap2 (Li, 2018), with specific parameters ‘minimap2 -d’ for index creation and ‘minimap2 -ax map-ont -t 8 –secondary=no’ for mapping long-reads. Finally, we performed hybrid assembly using Unicycler (Wick et al., 2017) by combining Illumina short-reads and Nanopore long reads. The mapped Illumina short-reads were initially assembled using SPAdes (Bankevich et al., 2012), and then the Nanopore long-reads were employed to resolve repetitive sequence regions in the assembly, using minimap2 (Li, 2018). After multiple iterations and adjustments, we determined the optimal kmer value of 89. The resulting GFA format files generated by Unicycler were visualized using Bandage (Wick et al., 2015). Ultimately, Unicycler generated a complete circular genome. Notably, as these contigs were assembled based on Illumina short-reads, no additional polishing steps were necessary.
Verification of the mitogenome structure
In our study, we employed PCR experiments to investigate the structure of A. odorata. Specifically, we designed eight specific primers to verify the accuracy of assembly generated by PMAT assembler. The primer design was conducted using the Primer designing tool on NCBI (https://www.ncbi.nlm.nih.gov/tools/primer-blast/) with default parameters. The primer sequences used for PCR reactions are listed in Supplementary Table 1. Subsequently, DNA was extracted, and the amplifications were performed using a Pro-Flex PCR system (Applied Biosystems, Waltham, MA, USA). The PCR reaction volume was 25 µL, comprising 2 µL of template DNA, 0.5 µL of forward primer, 0.5 µL of reverse primer, 12.5 µL of 2 × Taq PCR Master Mix, and 9.5 µL of ddH2O. The amplification conditions consisted of an initial denaturation at 94 °C for 5 min, followed by 30 cycles of denaturation at 94 °C for 30 s, annealing at 58 °C for 30 s, extension at 72 °C for 60 s, and a final extension step at 72 °C for 5 min. The PCR amplicons were visualized using 1% agarose gel electrophoresis. Subsequently, the single bright bands were excised and sent to Sangon Biotech (Shanghai, China) Co., Ltd. for Sanger sequencing.
Mitogenome and plastidial genome annotation
The plastome of our A. odorata was annotated using CPGAVAS2 (Shi et al., 2019). The plastome of published A. odorata (NC_048994.1) was used as the reference genome. The annotation results were further verified using CPGView (Liu et al., 2023) to ensure accurate gene annotations. We utilized IPMGA (http://www.1kmpg.cn/ipmga/) to annotate the assembled mitogenome of A. odorata. We selected a database of mitochondrial genes of angiosperms on IPMGA. IPMGA generates annotated files in the standard GenBank format. The tRNA annotations were performed using tRNAscan-SE (Lowe and Eddy, 1997) while rRNA annotations were obtained through BLASTn (Chen et al., 2015). To ensure accuracy, manual edits were made to the annotations using Apollo (Lewis et al., 2002). Finally, the genome map was generated using OGDRAW (v1.3.1) (Alverson et al., 2010).
Repetitive elements
The long tandem repeats were detected by Tandem Repeats Finder (TRF, https://tandem.bu.edu/trf/trf.html) with the default parameters (Benson, 1999). The simple sequence repeats (SSRs) of the assembled mitogenome were identified using the online website MISA (https://webblast.ipk-gatersleben.de/misa/), the parameters of the minimum numbers of mono-, di-, tri-, tetra-, penta-, and hexanucleotides were set as 10, 5, 4, 3, 3, and 3, respectively. Additionally, forward, reverse, palindromic, and complementary repeat sequences were identified using REPuter (https://bibiserv.cebitec.uni-bielefeld.de/reputer/) with the following settings: hamming distance of three and minimal repeat size of 30 bp, and e-value is limited to less than 1e-5. The visualization of the repetitive elements was done using the Circos package (Zhang et al., 2013).
Codon usage of mitochondrial genes
We employed PhyloSuite software (v1.2.2) (Zhang et al., 2020a) to parse the GenBank format file of the A. odorata mitogenome, extracting the protein-coding genes (PCGs). Subsequently, we conducted an analysis of the codon usage in mitochondrial PCGs using Mega 7.0 software (Kumar et al., 2016), which involved the calculation of Relative Synonymous Codon Usage (RSCU) values. An RSCU value of 1 signifies a neutral preference for codon utilization, whereas an RSCU value exceeding 1 indicates a relatively higher frequency of codon usage.
Identification of the mitochondrial plastid sequences
To identify the mitochondrial plastid DNAs (MTPTs), we compare the plastome and mitogenome DNAs of A. odorata by using BLASTn (Chen et al., 2015) program with the following parameters: -evalue 1e-5, -word_size 9, -gapopen 5, - gapextend 2, -reward 2, -penalty -3. The BLASTn results were visualized using Circos package (Zhang et al., 2013). The identified MTPTs were also annotated by using GeSeq.
Analysis of RNA editing sites
We employed a two-step approach for predicting RNA editing sites. Initially, lncRNA-Seq reads were mapped to the coding sequences (CDS) of each protein-coding gene (PCG) using BWA software (Li and Durbin, 2009) with default parameters. Subsequently, we utilized REDItools (Picardi and Pesole, 2013) to predict RNA editing sites based on the mapping results. The prediction criteria were established as follows: coverage exceeding 30, frequency equal to or greater than 0.1, and p-value equal to or greater than 0.05. Afterward, the Illumina short-reads of DNA were aligned to the CDS of each PCG using BWA software with default parameters. Genomic SNPs were predicted using BCFtools (Li, 2011) based on the mapping outcomes, with thresholds set at coverage greater than 30 and frequency less than or equal to 0.1. These heterogeneous sites of natural variation need to be excluded from RNA editing sites. Finally, after excluding SNP sites, the remaining sites identified in the lncRNA-seq mapping will be considered as genuine RNA editing sites.
To confirm the accuracy of the predicted RNA editing sites, we further designed experiments to validate these four specific sites. The primers were designed on both sides of the editing sites (Supplementary Table 2), and amplification was performed using genomic DNA (gDNA) and cDNA obtained from RNA reverse transcription using random primers as templates. The reaction conditions for amplification are described above. The amplified products were subsequently subjected to Sanger sequencing. Finally, by comparing the sequences of the products obtained from gDNA and cDNA, we determined the occurrence of RNA editing events.
Collinear analysis
For the collinear analysis with A. odorata, we selected five closely related species: Citrus unshiu (NC_057142.1), Citrus maxima (NC_057143.1), Citrus sinensis (NC_037463.1), Toona ciliata (NC_065060.1) and Toona sinensis (NC_065061.1). We identified collinear blocks based on sequence similarity using the BLASTn program with the following parameters: -evalue 1e-5, -word_size 9, -gapopen 5, - gapextend 2, -reward 2, -penalty -3. Only collinear blocks longer than 1 kb were retained for downstream analysis. To visualize the collinear relationships, we generated a multiple synteny plot using TBtools (Chen et al., 2023).
Phylogenetic analysis
We retrieved a total of 31 mitogenomes, including two outgroups (Stylosanthes capitata and Glycine max), from the GenBank database. These mitogenomes were used to construct a phylogenetic tree with A. odorata. Firstly, PhyloSuite (v.1.2.2) (Zhang et al., 2020a) was employed to identify and extract orthologous protein-coding genes (PCGs) across the analyzed species. The nucleotide sequences corresponding to these PCGs were then aligned using MAFFT (v7.471) (Katoh and Standley, 2013). Subsequently, the aligned sequences were concatenated to generate the input for phylogenetic tree construction. The maximum likelihood (ML) method was implemented using IQ-TREE (version 2.1.4-beta) (Minh et al., 2020) with the parameters “–alrt 1000 -B 1000”. Using the Bayesian Information Criterion (BIC) for model selection, the results indicate that the best-fit model is GTR+F+R2. The bootstrap analysis was performed with 1,000 replicates. Finally, the resulting phylogenetic tree was visualized and edited using the online tool ITOL (Letunic and Bork, 2019).
Results
Genomic structure of the A. odorata mitogenome
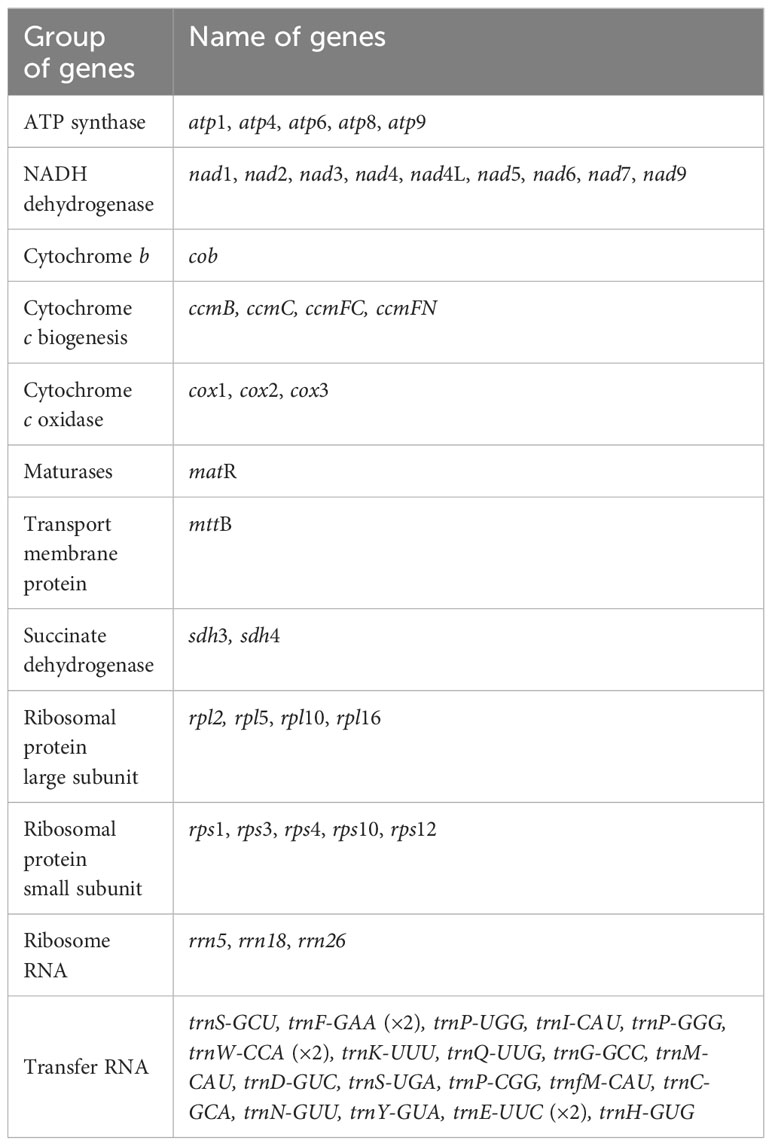
The assembly is composed of six distinct nodes and eight edges (paths), visually depicted in Figure 1A. Each of these nodes signifies an assembled contig, demonstrating a region of overlap along the linkages. It’s worth noting that contig5 and contig6 exhibited distinct characteristics suggestive of potential repetitive sequences. These two repeat sequences each showcased four distinct paths (designated as p1–p4 and p5–p8, Figure 1B). To confirm the presence of these paths within the A. odorata mitogenome, we conducted PCR experiments. The four primer pairs (F1 + R1, F2 + R1, F3 + R3, F3 + R2) were employed to validate the repeated sequence within contig5 and the paths, while the remaining pairs (F4 + R4, F5 + R5, F6 + R4, F7 + R7) were utilized to confirm the repetitive sequence within contig6 and the paths. The PCR products exhibited conformity with the anticipated outcome (Supplementary Figure 1). The results of these PCR experiment not only validated the accuracy of the assembly and the eight paths, but also helped us to propose four possible genomic configurations. Configuration 1 presents a master circular structure, incorporating all six contigs (Figure 1C). In configuration 2, contig2, contig3, contig4, contig5, and contig6 collectively form a circular arrangement, and contig1 and contig6 form a smaller circular (Figure 1D). Similarly, configuration 3 showcases a circular arrangement encompassing all six contigs, while the contig3 was inverted compare to configuration 1 (Figure 1E). Lastly, configuration 4 is similar to configuration 2, with the inversion of contig3 (Figure 1F). Our PCR experiments show the possibility of multiple configurations, and here we use configuration 1, a master circle represent the complete mitogenome, for subsequent analysis.
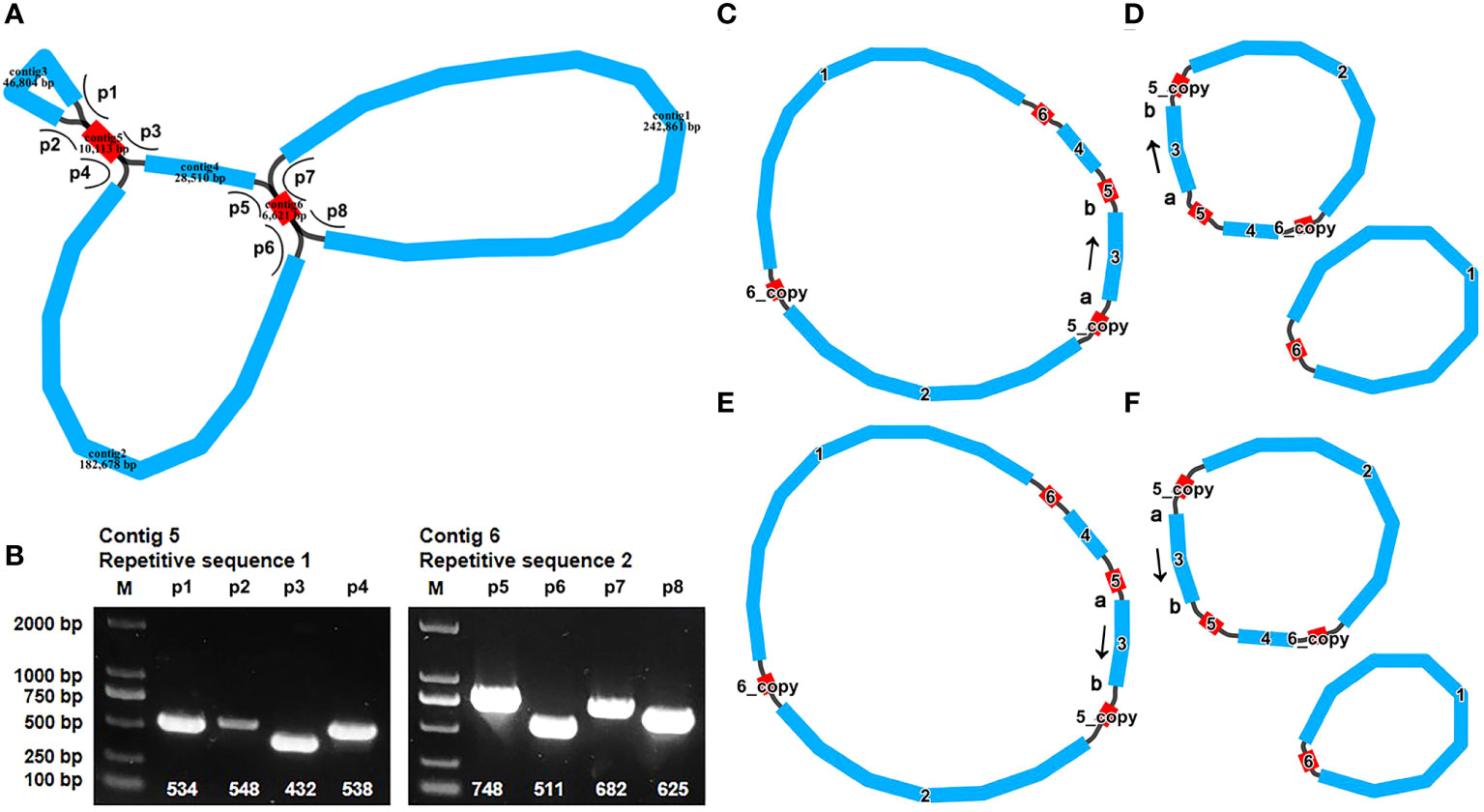
Figure 1 The graphic assembly and verification of A odorata mitogenome. (A). The graphic mitogenome consists of six contigs with different lengths, and they connected to each other. The length of the six contigs are 242,861 bp, 182,678 bp, 46,804 bp, 28,510 bp, 10,113 bp and 6,621 bp, respectively. Contig 5 and contig6 are the repetitive sequence. (B). represents the electropherogram of eight paths. (C-F) represent four conformations mediated by two pairs of repeat sequences, respectively. The arrows in panel (C-F) indicates the direction of sequence 3. These four conformations can dynamically change between them.
Gene content of the A. odorata mitogenome
The mitogenome maps of A. odorata was visually presents in Figure 2A. The total length of A. odorata mitogenome is 534,321 bp, and consists of 35 distinct protein-coding genes (PCGs) (Table 1), including five ATP synthase genes (atp1, atp4, atp6, atp8, and atp9), four cytochrome c biogenesis genes (ccmB, ccmC, ccmFC, and ccmFN), nine NADH dehydrogenase genes (nad1, nad2, nad3, nad4, nad4L, nad5, nad6, nad7, and nad9), three cytochrome c oxidase genes (cox1, cox2, and cox3), one transport membrane protein gene (mttB), one maturases gene (matR), and one cytochrome b gene (cob), four large subunits of ribosomal proteins (rpl2, rpl5, rpl10, and rpl16), five small subunits of ribosomal proteins (rps1, rps3, rps4, rps10 and rps12), as well as two succinate dehydrogenases (sdh3 and sdh4).
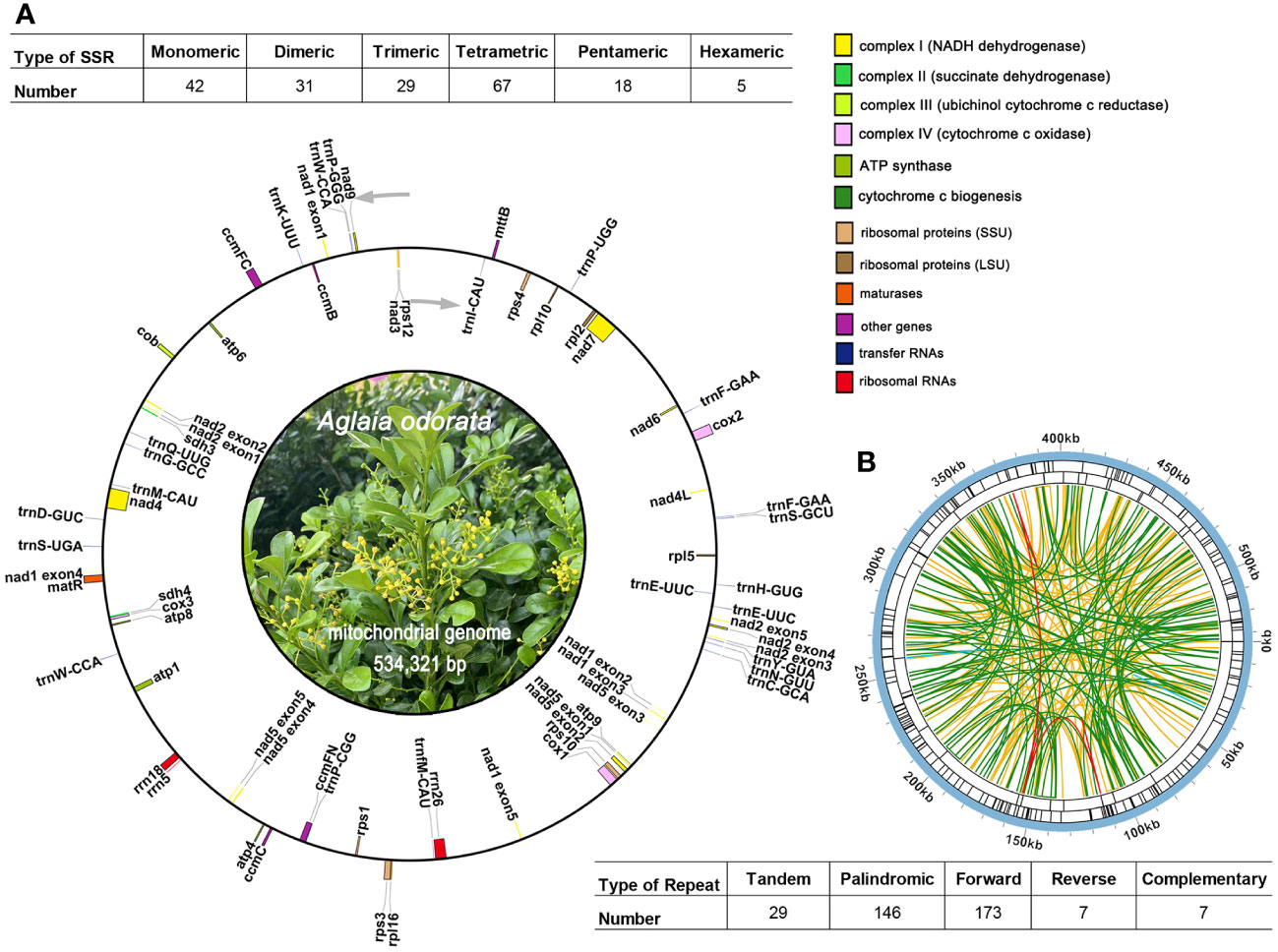
Figure 2 The mitogenome map of A odorata. (A). The figure shows the master circle of A odorata mitogenome. Genes transcript clockwise or counter-clockwise strands are drawn on the upper or lower of the circles, respectively. Genes belonging to different functional groups are color-coded. The table showing the number of SSRs of each type, with tetrameric being the most and Hexametric being the least. (B). The identified dispersed repeats (≥ 30bp). The green ribbons represent the forward repeats, the yellow ribbons represent the palindromic repeats, the red ribbons represent the complementary repeats and the blue ribbons represent the reverse repeats. The detailed information about dispersed repeats can be found in Table S6.
In the A. odorata mitogenome, a total of 22 tRNA genes have been annotated, with 19 being unique. Among these, 11 tRNA genes are mitochondrial native. Furthermore, our investigation has revealed 7 tRNA genes originating from the plastid: trnN-GUU, trnH-GUG, trnM-CAU, trnD-GUC, trnW-CCA, trnP-UGG, and trnI-CAU. Notably, our exploration has led us to the identification of a tRNA gene with bacterial origins, trnC-GCA, exhibiting a remarkable level of sequence homology with previously documented genes (Kitazaki et al., 2011). The remaining tRNA genes, devoid of sequence homology with known organelle tRNA genes, are unknown about their origin (Li, 2011; Rice et al., 2013). Furthermore, we have successfully pinpointed three distinct rRNA genes within the A. odorata mitogenome, namely rrn5, rrn18, and rrn26. The precise positions of each gene can be referenced in Supplementary Table 3. Among the entirety of the genes that have been annotated, 10 PCGs encompass introns (Supplementary Table 3). To elaborate, the genes ccmFC, cox2, cox1, trnP-CGG, and rps10 each feature one intron, whereas nad4 encompasses two introns. On the other hand, the genes nad1, nad2, nad5, and nad7 encompass four introns each.
Repetitive elements
Microsatellites, also known as simple sequence repeats (SSRs), typically consist of tandem sequences with a length of up to 6 base pairs in eukaryotic genomes. Within the mitogenome of A. odorata, a comprehensive total of 192 SSRs has been meticulously identified (as shown in Supplementary Table 4). Among this array of SSRs, tetrameric repeats stand out as the most prominent, encompassing 34.90% (67) of the overall count. This pattern is subsequently trailed by monomeric repeats (42), dimeric repeats (31), trimeric repeats (29), pentameric repeats (18), and hexametric repeats (5) (Figure 2A). And we have detected 29 long tandem repeat elements (Supplementary Table 5).
In addition, we have meticulously identified a total of 333 pairs of dispersed repeats within the A. odorata mitogenome, each with lengths equal to or exceeding 30 base pairs. This collection encompasses 173 pairs of forward repeats, 146 pairs of palindromic repeats, and 7 pairs of reverse repeats and complementary repeats, as outlined in Supplementary Table 6. Most of these repeat elements exhibit a length of less than 200 bp, and it’s evident that the number of dispersed repeats surpasses that of both SSRs and tandem repeats. The only two long dispersed repeats identified in the A. odorata mitogenome is contig5 and contig6 (10,113 bp palindromic repeat element and 6,621 bp repeat element). Cumulatively, the extent of these dispersed repeats encompasses 30,754 bp, constituting 5.76% of the entire A. odorata mitogenome. Furthermore, we provide a visual representation employing the Circos (v1.120) package (Krzywinski et al., 2009) to depict the dispersed repeats of the A. odorata mitogenome and counted the number of repetitive sequences in Figure 2B. These dispersed repeats are distributed in various regions of the mitogenome, effectively increasing the size of the genome.
Codon usage analysis
We conducted an analysis of the codon usage within the PCGs. The comprehensive codon usage data for all PCGs is presented in Supplementary Table 7. As shown in Figure 3A, revealing a discernible preference for specific codons among mitochondrial protein-coding genes. Notably, the RSCU values for the start codons AUG (Met) and UGG (Trp) both equate to 1. Furthermore, the RSCU values for the termination codons UGA (End), UAA (End), and UAG (End) are recorded as 1.20, 1.02, and 0.78, respectively. In terms of specific codons, GCU (Ala), UAA (End), CAU (His), CCU (Pro), and UAU (Tyr) emerge as the four most frequently employed codons within A. odorata. Conversely, GCG (Ala), UAG (End), CAC (His), and UAC (Tyr) are identified as the four least utilized codons. A visual representation in Figure 3A underscores the prevalence of arginine (Arg), leucine (Leu), and serine (Ser) codons, while methionine (Met) and tryptophan (Trp) codons exhibit relatively lower occurrence rates.
Figure 3 (A) The codon usage and RSCU value of A odorata PCGs. Codon families are shown on the x-axis. RSCU values are the number of times a particular codon is observed relative to the number of times that codon would be expected for uniform synonymous codon usage. (B) Schematic representation of the distribution of MTPTs between the mitogenome and the plastome of A odorata. The MTPTs on the chloroplast IR regions were counted only once. The location of each MTPT has been marked on the Figure. (C) Characteristics of the RNA editing sites identified in mitochondrial PCGs of A odorata. The ordinate shows the number of RNA editing sites identified in PCGs, the abscissa shows the name of PCGs identified in the mitogenome of A odorata. (D) The figure shows the results of PCR experiments on cDNA and gDNA of four genes. The experimental results are as expected. gDNA amplification length of rpl10-412 gene is longer because of the presence of intron in the gene. (E). The figure shows the results of Sanger sequencing experiments on cDNA and gDNA of four genes, and it shows that RNA editing is not universally present in all cases.
Characteristic of mitochondrial plastid DNAs
In our study, we conducted an annotation of the plastidial genome of A. odorata and performed a comprehensive comparison with its mitogenome. Employing the BLASTn program, we successfully identified a total of 12 instances of homologous sequences, we considered they might be potential MTPTs occurring between these two organelle genomes. These 12 MTPTs collectively span a length of 2,501 bp, contributing to 0.47% of the mitogenome’s total size (Supplementary Table 8). Among these MTPTs, MTPT12 stands out as the longest, spanning 1,122 bp, while MTPT1 emerges as the briefest, encompassing a mere 29 bp. Subsequently, our efforts focused on annotating these MTPTs, revealing a revelation: every MTPT encompassed plastidial genes or gene fragments. As depicted in Figure 3B and Supplementary Table 8, MTPT12 harbors a set of plastid genes, primarily associated with the photosystem II protein complex. It included psbJ, psbL, psbF, and psbE. Moreover, our analysis unearthed various gene fragments that resulted from the process of plastid migration. These fragments encompassed genes such as petG, ndhD, psbC, and atpH. It is plausible that these gene fragments underwent sequence loss during migration.
Analysis of RNA editing sites
We have successfully discerned a total of 427 high-confidence C to U RNA editing sites across 32 mitochondrial protein-coding genes (Supplementary Table 9). These editing sites are supported by our lncRNA data with an average depth of nearly 5100 times. The RNA editing sites for each gene are visually represented in Figure 3C. Within this set of mitochondrial genes, ccmB boasts the highest number of RNA editing sites at 45, closely followed by mttB with 41, positioning them as the foremost two genes in terms of RNA editing occurrences. Conversely, genes like atp1, atp6 and rpl5 possess the least number of editing sites, with only a solitary C to U edit detected for each. Based on our findings, we have identified C to U RNA editing events in three genes, which lead to the creation of premature stop codons. These genes are ccmFC, atp9 and rps10 (where CGA transitions to UGA). Notably, RNA editing plays a role in the formation of start codons as well, such as gene nad4L, have their start codons generated through RNA editing, converting ACG codon to AUG.
To evaluate the precision of this prediction, we employed PCR amplification and Sanger sequencing as a means of substantiating the occurrence of RNA editing. The four primer sequences can be found in Supplementary Table 2. Among these, every RNA site underwent successful verification, namely nad4L-2, atp9-223, ccmFC-1324, and rps10-412 (as depicted in Figure 3D). And all the validated sites encompassed C to U substitutions (Figure 3E), ccmFC-1324 is an exception, probably because of the lower frequency of editing, but the peak plot shows a hybrid peak of base U (uracil). Notably, the genomic DNA (gDNA) of rps10 has a longer PCR product because our primers span its introns, and the banding here also confirms that the DNA was sufficiently removed in the cDNA experimental group. Moreover, it’s noteworthy that each of these sites involved non-synonymous substitutions, as detailed in Supplementary Table 9. These PCR experiment firmly attesting to the dependability of the anticipated RNA editing sites.
Collinear and phylogenetic analysis
To delve into the rearrangements and conserved sequence blocks within the mitogenomes, we utilized the BLASTn program to pinpoint homologous collinear blocks. Illustrated in Figure 4A, each ribbon connecting two neighboring mitogenomes signifies a remarkably homologous collinear block or sequence. When comparing A. odorata and T. ciliata, we observed one large adjacent collinear block. Furthermore, we uncovered three collinear blocks exceeding 10 kb in length between these two mitogenomes. However, in the case of comparing A. odorata with C. sinensis, no extensive collinear blocks were detected, with the longest identified block measuring only 7.5 kb. Overall, while longer collinear blocks tend to be present between closely related species, the mitogenomes demonstrated limited collinearity, with several regions lacking homology. These findings underscore the prevalence of extensive genomic rearrangements between A. odorata and its related mitogenomes, suggesting that the genomic structure of the mitogenomes is not conserved.
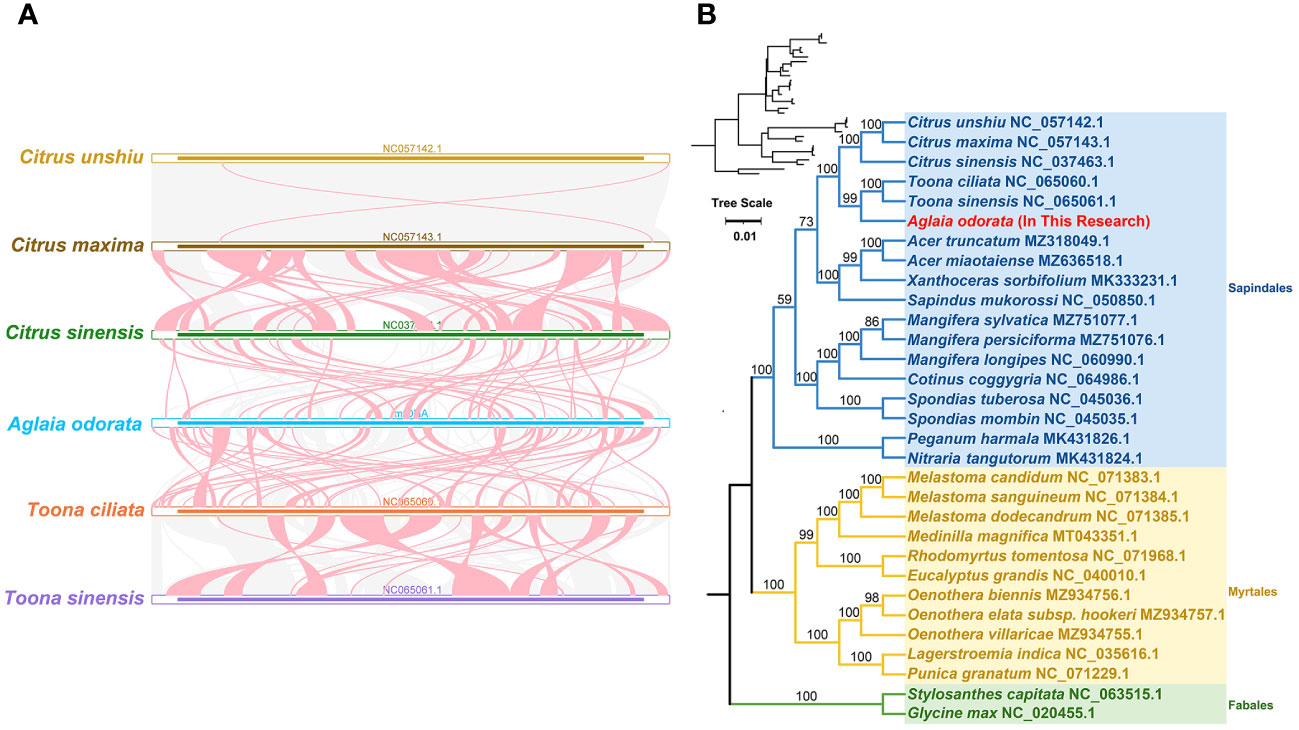
Figure 4 (A) Collinear analysis of A odorata mitogenome and its related species. The colorful bars indicated the mitogenomes, and the ribbons showed the homologous sequences between the adjacent species. The blue ribbons indicate regions with homology and the red ribbons indicate where the inversion occurred. The homologous blocks less than 0.5 kb in length are not remaining, and regions that fail to have a homologous block indicate that they are unique to the species. (B) The phylogenetic relationships of A odorata and another 30 species based on conserved mitochondrial genes. The tree was constructed based on the nucleotide sequences of conserved mitochondrial protein-coding genes (PCGs). We used Maximum Likelihood (ML) method to reconstruct the phylogenetic tree. The ML topology is indicated with ML bootstrap support values. T. capitata and G max were used as outgroups. The species list and its accession numbers that used in phylogenetic analysis are shown in Table S10.
In addition, we carried out a phylogenetic analysis utilizing 31 mitogenomes of related species, with T. capitata and G. max serving as outgroups for reference. The detailed list of species and their corresponding GenBank accessions utilized for this analysis is available in Supplementary Table 10. By aligning and concatenating the shared protein-coding genes (PCGs), we created the matrix data for analysis. The outcome of our phylogenetic investigation yielded a maximum likelihood (ML) tree that exhibits robust support along the primary basal branches (refer to Figure 4B). In terms of evolutionary relationships, A. odorata exhibits its closest affinity with T. ciliata, both species falling under the Sapindales order. While the overall structure of the phylogenetic tree broadly aligns with the APG IV system (The Angiosperm Phylogeny et al., 2016) at the order level, it’s noteworthy that within the Sapindales order, a couple of nodes lack bootstrap support. This suggests that deriving phylogenetic inferences solely from mitochondrial PCGs might have limitations in accurately resolving lower taxonomic categories.
Discussion
Our study successfully assembled the complete mitogenome of A. odorata, with a total length of 534,321 bp. To accomplish this task, we employed a hybrid assembly approach, combining both Illumina short-reads and Oxford Nanopore long-reads. In contrast to the stability observed in plant plastomes, plant mitogenomes have undergone significant transformations throughout evolution, resulting in complex structures (Bi et al., 2022; Han et al., 2022; Ma et al., 2022; Bi et al., 2024b). Numerous researchers have delved into the intricate structural variations within plant mitogenomes (Chevigny et al., 2020), leading to the development of various tools for decoding these dynamically evolving genomes (He et al., 2023; Shan et al., 2023). Some studies propose that the diversity within mitogenomes may arise from long repeat-mediated recombination (Wang et al., 2023a). Within the mitogenome of A. odorata, our analysis unveiled two pairs of lengthy repetitive sequences, measuring 10,113 bp and 6,621 bp, respectively. This prolific presence of repeats points towards their potential significance not only in genome reconfiguration but also in influencing genome size dynamics. Remarkably, these two pairs of long repetitive sequences each possibly facilitated two conformations, and up to four potential configurations can be created. This highlights that the mitogenome structure of A. odorata is not static but rather dynamically varies among these four conformations. The phenomenon of long repeat-mediated recombination is not unique to A. odorata; it has also been observed in other plant species such as Mimulus guttatus (Mower et al., 2012), Scutellaria tsinyunensis (Li et al., 2021), Photinia serratifolia (Wang et al., 2023a), and Ginkgo biloba (Guo et al., 2016). Furthermore, extensive research has demonstrated that short dispersed repeats contribute to mitogenome recombination in various plant species, including Nymphaea colorata (Dong et al., 2018), Silene latifolia (Sloan et al., 2012), and Ginkgo biloba (Guo et al., 2016).
Horizontal gene transfer (HGT) among organellar genomes and the nuclear genome is a common phenomenon that plays a pivotal role in plant evolution (Sprinzl and Vassilenko, 2005). The genome structure and evolutionary dynamics of plant mitogenomes render them particularly prone to acquiring and assimilating foreign DNA (Christensen, 2013). In contrast to plastid DNA, plant mitogenomes exhibit a greater propensity to accept and incorporate foreign genetic material, a phenomenon frequently observed. In the case of A. odorata, we have observed some sequences transferred from the plastidial genome to the mitogenome (Supplementary Table 8, Figure 3B). These MTPTs are believed to hold substantial implications for eukaryotic evolution, fostering genetic diversity. Among the MTPTs identified in the A. odorata mitogenome, MTPT12 stands out as the longest, spanning 1,122 base pairs. However, in comparison to the mitogenomes of other published species, A. odorata displays fewer MTPTs, and they tend to be of shorter lengths. For instance, in Suaeda glauca, MTPTs covering 26.87 kb constitute 5.18% of its mitogenome. Some studies have unveiled a notable degree of diversity in MTPTs among various species. Previous research has revealed that tRNAs within plant mitochondria have diverse origins. A portion of these tRNAs is inherited from the ancestral mitochondria, while another part is acquired from chloroplasts through HGT (Sprinzl and Vassilenko, 2005). By leveraging sequence similarities and existing findings, we successfully identified specific tRNA genes in the A. odorata mitogenome that originated from the plastid and were transferred to the mitochondria (Richardson et al., 2013). Within the A. odorata mitogenome, we identified the following tRNA genes as potential acquisitions from the plastid: trnN-GUU, trnH-GUG, trnM-CAU, trnD-GUC, trnW-CCA, trnP-UGG, and trnI-CAU. Over the course of evolutionary timescales, these MTPTs have led to the incorporation of functional tRNAs, as evidenced by their widespread conservation across angiosperms (Chaw et al., 2008). For instance, trnH-GUG and trnM-CAU were early additions to the mitogenome and remain functional (Joyce and Gray, 1989). However, during the transfer of DNA fragments from chloroplasts to the mitogenome, some PCGs are often carried along and tend to become nonfunctional pseudogenes. This phenomenon also occurs within the mitogenome of A. odorata (Supplementary Table Table 8).
Plant mitochondrial RNA editing is a biological phenomenon where specific nucleotide positions within the mitochondrial RNA sequence undergo base mutations catalyzed by mitochondrial RNA editing enzymes (Wang et al., 2019; Liu et al., 2020; Wang et al., 2023b). These RNA editing processes can convert C to U or U to C in the RNA sequence (Gerke et al., 2020), playing a pivotal role in mitochondrial gene expression and function (Sosso et al., 2012). This is because many of these RNA editing events can result in changes in RNA sequences, leading to variations in the final protein products (Edera et al., 2018). And RNA editing of mitochondrial genes is believed to be an important factor in regulating plant cytoplasmic inheritance-related traits (Liu et al., 2013). In our predictions, we observed that most RNA editing sites occur at the first or second positions of the triplet codon, a pattern similar to that seen in many other plants (Grewe et al., 2014; Kovar et al., 2018). Furthermore, as depicted in Figure 3E, the frequency of editing varies greatly at different sites. For example, in gene ccmFC-1324, the frequency of RNA editing is lower than that of non-editing events. Furthermore, in our RNA editing experiments, we have successfully confirmed the occurrence of RNA editing events that generate stop codons in gene ccmFC, rps10 and atp9. The emergence of RNA editing events that yielded start codons also been found in gene nad4L. This new start and stop codons are typically generated to encode proteins that exhibit greater conservation and homology with corresponding proteins in other species, thereby enhancing gene expression within the mitochondria. In future cases, the annotation of these genes should fully account for the influence of RNA editing events, otherwise the wrong coding sequence will be obtained.
Conclusion
In our study, we have accomplished the successful assembly of the mitogenome of A. odorata, revealing a circular genome structure. We conducted thorough analyses to explore its gene content, repetitive elements, codon usage, MTPTs, and RNA editing sites, along with making phylogenetic inferences. To the best of our knowledge, this represents the first comprehensive description of a complete mitogenome within A. odorata. Our findings illuminate previously uncharted aspects of the evolutionary dynamics of mitochondrial genes, providing valuable insights into the evolutionary history of mitogenomes.
Data availability statement
The mitogenome sequence is available in nucleotide database of GenBank (https://www.ncbi.nlm.nih.gov/nucleotide/) with accession numbers: OR680716.1 (plastome) and OR680718.1 (mitogenome). The sequencing reads used for mitogenome assembly in this study have been released on the NCBI with those accession numbers: PRJNA1031331 (BioProject); SAMN37933744 (BioSample) and SRR26513849, SRR26513847 and SRR26513848 (SRA).
Author contributions
ZH: Data curation, Formal analysis, Funding acquisition, Methodology, Software, Writing – original draft, Writing – review & editing. ZZ: Data curation, Formal analysis, Methodology, Writing – review & editing. JZ: Investigation, Resources, Software, Writing – original draft. XC: Methodology, Resources, Software, Writing – original draft. JL: Supervision, Validation, Writing – review & editing. LL: Investigation, Supervision, Writing – review & editing. YL: Formal analysis, Funding acquisition, Validation, Visualization, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. The research was supported by The Open Fund of State Key Laboratory for Conservation and Utilization of Bio-Resources in Yunnan, Grant No: gzkf2022007, Sanya Institute of China Agricultural University (SYND-2022-11), and the Young Talent Project of Yunnan Revitalization Talent Support Program (XDYC-QNRC-2022-0719) and Hainan Seed Industry Laboratory (Grant No: B23C10004).
Acknowledgments
I sincerely thank the experimental personnel and bioinformatics analysis at MitoRun research group participated in this project.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2024.1362045/full#supplementary-material
Abbreviations
PCR, Polymerase chain reaction; SSR, Simple sequence repeat; ML, Maximum-likelihood; BI, Bayesian inferences; NCBI, National Center for Biotechnology Information; BLAST, Basic Local Alignment Search Tool; PCGs, Protein-coding gene sequences; MTPT, Mitochondrial plastid; DNA lncRNA, Long non-coding RMA.
References
Alverson, A. J., Wei, X., Rice, D. W., Stern, D. B., Barry, K., Palmer, J. D. (2010). Insights into the evolution of mitochondrial genome size from complete sequences of Citrullus lanatus and Cucurbita pepo (Cucurbitaceae). Mol. Biol. Evol. 27, 1436–1448. doi: 10.1093/molbev/msq029
Bankevich, A., Nurk, S., Antipov, D., Gurevich, A. A., Dvorkin, M., Kulikov, A. S., et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477. doi: 10.1089/cmb.2012.0021
Benson, G. (1999). Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 27, 573–580. doi: 10.1093/nar/27.2.573
Bi, C., Qu, Y., Hou, J., Wu, K., Ye, N., Yin, T. (2022). Deciphering the multi-chromosomal mitochondrial genome of populus simonii. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.914635
Bi, C., Shen, F., Han, F., Qu, Y., Hou, J., Xu, K., et al. (2024a). PMAT: an efficient plant mitogenome assembly toolkit using low coverage HiFi sequencing data. Horticult. Res., uhae023. doi: 10.1093/hr/uhae023
Bi, C., Sun, N., Han, F., Xu, K., Yang, Y., Ferguson, D. K. (2024b). The first mitogenome of Lauraceae (Cinnamomum chekiangense). Plant Diversity 46, 144–148. doi: 10.1016/j.pld.2023.11.001
Bolger, A. M., Lohse, M., Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Bonora, M., De Marchi, E., Patergnani, S., Suski, J. M., Celsi, F., Bononi, A., et al. (2014). Tumor necrosis factor-alpha impairs oligodendroglial differentiation through a mitochondria-dependent process. Cell Death Differ. 21, 1198–1208. doi: 10.1038/cdd.2014.35
Chaw, S. M., Shih, A. C., Wang, D., Wu, Y. W., Liu, S. M., Chou, T. Y. (2008). The mitochondrial genome of the gymnosperm Cycas taitungensis contains a novel family of short interspersed elements, Bpu sequences, and abundant RNA editing sites. Mol. Biol. Evol. 25, 603–615. doi: 10.1093/molbev/msn009
Chen, C., Wu, Y., Li, J., Wang, X., Zeng, Z., Xu, J., et al. (2023). TBtools-II: A “One for all, all for one” Bioinformatics platform for biological big-data mining. Mol. Plant. 16, 1733–1742. doi: 10.1016/j.molp.2023.09.010
Chen, Y., Ye, W., Zhang, Y., Xu, Y. (2015). High speed BLASTN: an accelerated MegaBLAST search tool. Nucleic Acids Res. 43, 7762–7768. doi: 10.1093/nar/gkv784
Cheng, Y., He, X., Priyadarshani, S., Wang, Y., Ye, L., Shi, C., et al. (2021). Assembly and comparative analysis of the complete mitochondrial genome of Suaeda glauca. BMC Genomics 22, 167. doi: 10.1186/s12864-021-07490-9
Cheng, N., Lo, Y. S., Ansari, M. I., Ho, K. C., Jeng, S. T., Lin, N. S., et al. (2017). Correlation between mtDNA complexity and mtDNA replication mode in developing cotyledon mitochondria during mung bean seed germination. New Phytol. 213, 751–763. doi: 10.1111/nph.14158
Chevigny, N., Schatz-Daas, D., Lotfi, F., Gualberto, J. M. (2020). DNA repair and the stability of the plant mitochondrial genome. Int. J. Mol. Sci. 21, 1–24. doi: 10.3390/ijms21010328
Choi, K. S., Park, S. (2021). Complete plastid and mitochondrial genomes of aeginetia indica reveal intracellular gene transfer (IGT), horizontal gene transfer (HGT), and cytoplasmic male sterility (CMS). Int. J. Mol. Sci. 22, 1–12. doi: 10.3390/ijms22116143
Christensen, A. C. (2013). Plant mitochondrial genome evolution can be explained by DNA repair mechanisms. Genome Biol. Evol. 5, 1079–1086. doi: 10.1093/gbe/evt069
Dong, S., Zhao, C., Chen, F., Liu, Y., Zhang, S., Wu, H., et al. (2018). The complete mitochondrial genome of the early flowering plant Nymphaea colorata is highly repetitive with low recombination. BMC Genomics 19, 614. doi: 10.1186/s12864-018-4991-4
Edera, A. A., Gandini, C. L., Sanchez-Puerta, M. V. (2018). Towards a comprehensive picture of C-to-U RNA editing sites in angiosperm mitochondria. Plant Mol. Biol. 97, 215–231. doi: 10.1007/s11103-018-0734-9
Fischer, A., Dotzek, J., Walther, D., Greiner, S. (2022). Graph-based models of the Oenothera mitochondrial genome capture the enormous complexity of higher plant mitochondrial DNA organization. NAR Genom. Bioinform. 4, lqac027. doi: 10.1093/nargab/lqac027
Garcia, L. E., Edera, A. A., Palmer, J. D., Sato, H., Sanchez-Puerta, M. V. (2021). Horizontal gene transfers dominate the functional mitochondrial gene space of a holoparasitic plant. New Phytol. 229, 1701–1714. doi: 10.1111/nph.16926
Gerke, P., Szovenyi, P., Neubauer, A., Lenz, H., Gutmann, B., McDowell, R., et al. (2020). Towards a plant model for enigmatic U-to-C RNA editing: the organelle genomes, transcriptomes, editomes and candidate RNA editing factors in the hornwort Anthoceros agrestis. New Phytol. 225, 1974–1992. doi: 10.1111/nph.16297
Grewe, F., Edger, P. P., Keren, I., Sultan, L., Pires, J. C., Ostersetzer-Biran, O., et al. (2014). Comparative analysis of 11 Brassicales mitochondrial genomes and the mitochondrial transcriptome of Brassica oleracea. Mitochondrion 19 Pt B, 135–143. doi: 10.1016/j.mito.2014.05.008
Gualberto, J. M., Mileshina, D., Wallet, C., Niazi, A. K., Weber-Lotfi, F., Dietrich, A. (2014). The plant mitochondrial genome: dynamics and maintenance. Biochimie 100, 107–120. doi: 10.1016/j.biochi.2013.09.016
Guo, W., Grewe, F., Fan, W., Young, G. J., Knoop, V., Palmer, J. D., et al. (2016). Ginkgo and welwitschia mitogenomes reveal extreme contrasts in gymnosperm mitochondrial evolution. Mol. Biol. Evol. 33, 1448–1460. doi: 10.1093/molbev/msw024
Han, F., Qu, Y., Chen, Y., Xu, L., Bi, C. (2022). Assembly and comparative analysis of the complete mitochondrial genome of Salix wilsonii using PacBio HiFi sequencing. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.1031769
He, W., Xiang, K., Chen, C., Wang, J., Wu, Z. (2023). Master graph: an essential integrated assembly model for the plant mitogenome based on a graph-based framework. Brief Bioinform. 24, 1–13. doi: 10.1093/bib/bbac522
Inad, A., Nishino, H., Kuchide, M., Takayasu, J., Mukainaka, T., Nobukuni, Y., et al. (2001). Cancer chemopreventive activity of odorine and odorinol from Aglaia odorata. Biol. Pharm. Bull. 24, 1282–1285. doi: 10.1248/bpb.24.1282
Jin, J. J., Yu, W. B., Yang, J. B., Song, Y., dePamphilis, C. W., Yi, T. S., et al. (2020). GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21, 241. doi: 10.1186/s13059-020-02154-5
Joyce, P. B., Gray, M. W. (1989). Chloroplast-like transfer RNA genes expressed in wheat mitochondria. Nucleic Acids Res. 17, 5461–5476. doi: 10.1093/nar/17.14.5461
Katoh, K., Standley, D. M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. doi: 10.1093/molbev/mst010
Kitazaki, K., Kubo, T., Kagami, H., Matsumoto, T., Fujita, A., Matsuhira, H., et al. (2011). A horizontally transferred tRNA(Cys) gene in the sugar beet mitochondrial genome: evidence that the gene is present in diverse angiosperms and its transcript is aminoacylated. Plant J. 68, 262–272. doi: 10.1111/j.1365-313X.2011.04684.x
Koren, S., Walenz, B. P., Berlin, K., Miller, J. R., Bergman, N. H., Phillippy, A. M. (2017). Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 27, 722–736. doi: 10.1101/gr.215087.116
Kovar, L., Nageswara-Rao, M., Ortega-Rodriguez, S., Dugas, D. V., Straub, S., Cronn, R., et al. (2018). PacBio-based mitochondrial genome assembly of leucaena trichandra (Leguminosae) and an intrageneric assessment of mitochondrial RNA editing. Genome Biol. Evol. 10, 2501–2517. doi: 10.1093/gbe/evy179
Kozik, A., Rowan, B. A., Lavelle, D., Berke, L., Schranz, M. E., Michelmore, R. W., et al. (2019). The alternative reality of plant mitochondrial DNA: One ring does not rule them all. PloS Genet. 15, e1008373. doi: 10.1371/journal.pgen.1008373
Kroemer, G., Reed, J. C. (2000). Mitochondrial control of cell death. Nat. Med. 6, 513–519. doi: 10.1038/74994
Krzywinski, M., Schein, J., Birol, I., Connors, J., Gascoyne, R., Horsman, D., et al. (2009). Circos: an information aesthetic for comparative genomics. Genome Res. 19, 1639–1645. doi: 10.1101/gr.092759.109
Kumar, S., Stecher, G., Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Letunic, I., Bork, P. (2019). Interactive Tree Of Life (iTOL) v4: recent updates and new developments. Nucleic Acids Res. 47, W256–W259. doi: 10.1093/nar/gkz239
Lewis, S. E., Searle, S. M., Harris, N., Gibson, M., Lyer, V., Richter, J., et al. (2002). Apollo: a sequence annotation editor. Genome Biol. 3, RESEARCH0082. doi: 10.1186/gb-2002-3-12-research0082
Li, H. (2011). A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 27, 2987–2993. doi: 10.1093/bioinformatics/btr509
Li, H. (2018). Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics 34, 3094–3100. doi: 10.1093/bioinformatics/bty191
Li, H., Durbin, R. (2009). Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760. doi: 10.1093/bioinformatics/btp324
Li, J., Xu, Y., Shan, Y., Pei, X., Yong, S., Liu, C., et al. (2021). Assembly of the complete mitochondrial genome of an endemic plant, Scutellaria tsinyunensis, revealed the existence of two conformations generated by a repeat-mediated recombination. Planta 254, 36. doi: 10.1007/s00425-021-03684-3
Lin, Q., Banerjee, A., Stefanovic, S. (2022). Mitochondrial phylogenomics of Cuscuta (Convolvulaceae) reveals a potentially functional horizontal gene transfer from the host. Genome Biol. Evol. 14, 1–11. doi: 10.1093/gbe/evac091
Liu, R., Cao, S. K., Sayyed, A., Yang, H. H., Zhao, J., Wang, X., et al. (2020). The DYW-subgroup pentatricopeptide repeat protein PPR27 interacts with ZmMORF1 to facilitate mitochondrial RNA editing and seed development in maize. J. Exp. Bot. 71, 5495–5505. doi: 10.1093/jxb/eraa273
Liu, S., Ni, Y., Li, J., Zhang, X., Yang, H., Chen, H., et al. (2023). CPGView: A package for visualizing detailed chloroplast genome structures. Mol. Ecol. Resour. 23, 694–704. doi: 10.1111/1755-0998.13729
Liu, Y. J., Xiu, Z. H., Meeley, R., Tan, B. C. (2013). Empty pericarp5 encodes a pentatricopeptide repeat protein that is required for mitochondrial RNA editing and seed development in maize. Plant Cell 25, 868–883. doi: 10.1105/tpc.112.106781
Liu, S., Yang, W., Liu, S. B., Wang, H., Guo, Z. K., Zeng, Y. B., et al. (2014). A new dolabellane diterpenoid and a sesquilignan from Aglaia odorata var. microphyllina. Nat. Prod Commun. 9, 7–8. doi: 10.1177/1934578X1400900103
Lowe, T. M., Eddy, S. R. (1997). tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25, 955–964. doi: 10.1093/nar/25.5.955
Ma, Q., Wang, Y., Li, S., Wen, J., Zhu, L., Yan, K., et al. (2022). Assembly and comparative analysis of the first complete mitochondrial genome of Acer truncatum Bunge: a woody oil-tree species producing nervonic acid. BMC Plant Biol. 22, 29. doi: 10.1186/s12870-021-03416-5
Minh, B. Q., Schmidt, H. A., Chernomor, O., Schrempf, D., Woodhams, M. D., von Haeseler, A., et al. (2020). IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 37, 1530–1534. doi: 10.1093/molbev/msaa015
Morley, S. A., Ahmad, N., Nielsen, B. L. (2019). Plant organelle genome replication. Plants (Basel) 8, 1–18. doi: 10.3390/plants8100358
Mower, J. P., Case, A. L., Floro, E. R., Willis, J. H. (2012). Evidence against equimolarity of large repeat arrangements and a predominant master circle structure of the mitochondrial genome from a monkeyflower (Mimulus guttatus) lineage with cryptic CMS. Genome Biol. Evol. 4, 670–686. doi: 10.1093/gbe/evs042
Picardi, E., Pesole, G. (2013). REDItools: high-throughput RNA editing detection made easy. Bioinformatics 29, 1813–1814. doi: 10.1093/bioinformatics/btt287
Rice, D. W., Alverson, A. J., Richardson, A. O., Young, G. J., Sanchez-Puerta, M. V., Munzinger, J., et al. (2013). Horizontal transfer of entire genomes via mitochondrial fusion in the angiosperm Amborella. Science 342, 1468–1473. doi: 10.1126/science.1246275
Richardson, A. O., Rice, D. W., Young, G. J., Alverson, A. J., Palmer, J. D. (2013). The “fossilized” mitochondrial genome of Liriodendron tulipifera: ancestral gene content and order, ancestral editing sites, and extraordinarily low mutation rate. BMC Biol. 11, 29. doi: 10.1186/1741-7007-11-29
Roger, A. J., Munoz-Gomez, S. A., Kamikawa, R. (2017). The origin and diversification of mitochondria. Curr. Biol. 27, R1177–R1192. doi: 10.1016/j.cub.2017.09.015
Shan, Y., Li, J., Zhang, X., Yu, J. (2023). The complete mitochondrial genome of Amorphophallus albus and development of molecular markers for five Amorphophallus species based on mitochondrial DNA. Front. Plant Sci. 14. doi: 10.3389/fpls.2023.1180417
Shi, L., Chen, H., Jiang, M., Wang, L., Wu, X., Huang, L., et al. (2019). CPGAVAS2, an integrated plastome sequence annotator and analyzer. Nucleic Acids Res. 47, W65–w73. doi: 10.1093/nar/gkz345
Skippington, E., Barkman, T. J., Rice, D. W., Palmer, J. D. (2015). Miniaturized mitogenome of the parasitic plant Viscum scurruloideum is extremely divergent and dynamic and has lost all nad genes. Proc. Natl. Acad. Sci. U.S.A. 112, E3515–E3524. doi: 10.1073/pnas.1504491112
Sloan, D. B., Alverson, A. J., Chuckalovcak, J. P., Wu, M., McCauley, D. E., Palmer, J. D., et al. (2012). Rapid evolution of enormous, multichromosomal genomes in flowering plant mitochondria with exceptionally high mutation rates. PloS Biol. 10, e1001241. doi: 10.1371/journal.pbio.1001241
Sosso, D., Mbelo, S., Vernoud, V., Gendrot, G., Dedieu, A., Chambrier, P., et al. (2012). PPR2263, a DYW-Subgroup Pentatricopeptide repeat protein, is required for mitochondrial nad5 and cob transcript editing, mitochondrion biogenesis, and maize growth. Plant Cell 24, 676–691. doi: 10.1105/tpc.111.091074
Sprinzl, M., Vassilenko, K. S. (2005). Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res. 33, D139–D140. doi: 10.1093/nar/gki012
The Angiosperm Phylogeny, G., Chase, M. W., Christenhusz, M. J. M., Fay, M. F., Byng, J. W., Judd, W. S., et al. (2016). An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG IV. Botanic. J. Linn. Soc. 181, 1–20. doi: 10.1111/boj.12385
van Loo, G., Saelens, X., van Gurp, M., MacFarlane, M., Martin, S. J., Vandenabeele, P. (2002). The role of mitochondrial factors in apoptosis: a Russian roulette with more than one bullet. Cell Death Differ. 9, 1031–1042. doi: 10.1038/sj.cdd.4401088
Wang, Y., Chen, S., Chen, J., Chen, C., Lin, X., Peng, H., et al. (2023a). Characterization and phylogenetic analysis of the complete mitochondrial genome sequence of Photinia serratifolia. Sci. Rep. 13, 770. doi: 10.1038/s41598-022-24327-x
Wang, Y., Li, H., Huang, Z. Q., Ma, B., Yang, Y. Z., Xiu, Z. H., et al. (2023b). Maize PPR-E proteins mediate RNA C-to-U editing in mitochondria by recruiting the trans deaminase PCW1. Plant Cell 35, 529–551. doi: 10.1093/plcell/koac298
Wang, Y., Liu, X. Y., Yang, Y. Z., Huang, J., Sun, F., Lin, J., et al. (2019). Empty Pericarp21 encodes a novel PPR-DYW protein that is required for mitochondrial RNA editing at multiple sites, complexes I and V biogenesis, and seed development in maize. PloS Genet. 15, e1008305. doi: 10.1371/journal.pgen.1008305
Wick, R. R., Judd, L. M., Gorrie, C. L., Holt, K. E. (2017). Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PloS Comput. Biol. 13, e1005595. doi: 10.1371/journal.pcbi.1005595
Wick, R. R., Schultz, M. B., Zobel, J., Holt, K. E. (2015). Bandage: interactive visualization of de novo genome assemblies. Bioinformatics 31, 3350–3352. doi: 10.1093/bioinformatics/btv383
Yang, X., Yu, Y., Wu, P., Liu, J., Li, Y., Tao, L., et al. (2022). Phenolic and bisamide derivatives from Aglaia odorata and their biological activities. Nat. Prod Res., 1–12. doi: 10.1080/14786419.2022.2162514
Ye, N., Wang, X., Li, J., Bi, C., Xu, Y., Wu, D., et al. (2017). Assembly and comparative analysis of complete mitochondrial genome sequence of an economic plant Salix suchowensis. PeerJ 5, e3148. doi: 10.7717/peerj.3148
Zhang, D., Gao, F., Jakovlic, I., Zou, H., Zhang, J., Li, W. X., et al. (2020a). PhyloSuite: An integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol. Ecol. Resour. 20, 348–355. doi: 10.1111/1755-0998.13096
Zhang, J., Li, Y., Wang, Y. (2020b). The complete chloroplast genome sequence of Aglaia odorata. Mitochondrial. DNA B Resour. 5, 472–473. doi: 10.1080/23802359.2019.1704649
Keywords: Aglaia odorata, mitochondrial genome, recombination, MTPT, RNA editing
Citation: Hao Z, Zhang Z, Zhang J, Cui X, Li J, Luo L and Li Y (2024) The complete mitochondrial genome of Aglaia odorata, insights into its genomic structure and RNA editing sites. Front. Plant Sci. 15:1362045. doi: 10.3389/fpls.2024.1362045
Received: 27 December 2023; Accepted: 21 February 2024;
Published: 06 March 2024.
Edited by:
Gregory Thyssen, Agricultural Research Service (USDA), United StatesReviewed by:
Hoang Dang Khoa Do, Independent researcherChangwei Bi, Nanjing Forestry University, China
Copyright © 2024 Hao, Zhang, Zhang, Cui, Li, Luo and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Laixin Luo, bHVvbGFpeGluQGNhdS5lZHUuY24=; Yingbin Li, bGl5YjMwQDE2My5jb20=
†These authors have contributed equally to this work
 Zhigang Hao
Zhigang Hao Zhiping Zhang
Zhiping Zhang Jinan Zhang2
Jinan Zhang2 Jianqiang Li
Jianqiang Li Laixin Luo
Laixin Luo Yingbin Li
Yingbin Li