- College of Plant Protection, Yangzhou University, Yangzhou, China
The gene encoding 9-cis-epoxycarotenoid dioxygenase 3 (NCED3) functions in abscisic acid (ABA) biosynthesis, plant growth and development, and tolerance to adverse temperatures, drought and saline conditions. In this study, three rice lines were used to explore the function of OsNCED3, these included an OsNCED3-overexpressing line (OsNCED3-OE), a knockdown line (osnced3-RNAi) and wild-type rice (WT). These rice lines were infested with the brown plant hopper (BPH; Nilaparvata lugens) and examined for physiological and biochemical changes, hormone content, and defense gene expression. The results showed that OsNCED3 activated rice defense mechanisms, which led to an increased defense enzyme activity of superoxide dismutase, peroxidase, and polyphenol oxidase. The overexpression of OsNCED3 decreased the number of planthoppers and reduced oviposition and BPH hatching rates. Furthermore, the overexpression of OsNCED3 increased the concentrations of jasmonic acid, jasmonyl-isoleucine and ABA relative to WT rice and the osnced3-RNAi line. These results indicate that OsNCED3 improved the stress tolerance in rice and support a role for both jasmonates and ABA as defense compounds in the rice-BPH interaction.
Highlights
● The brown planthopper (BPH, Nilaparvata lugens) is one of the most important pests of rice in China and causes damage by ingesting phloem sap. OsNCED3 is a key gene in abscisic acid (ABA) synthesis and functions in drought resistance. Interestingly, it is not known if OsNCED3 overexpression can increase resistance to BPH or whether drought resistance is correlated with pest resistance. In this study, the role of OsNCED3 in BPH resistance was evaluated in rice by conducting physiological and biochemical assays, monitoring changes in hormones, and evaluating expression of defense genes. Our results show that the defense gene OsNCED3 is induced by BPH feeding and correlates with improved plant resistance to BPH in 48 h. Our findings also support a role for several plant hormones as defense compounds in the rice-BPH interaction. Although the control of BPH is still based on chemical methods, the results of this study indicate that modulation of endogenous genes in rice may also be utilized to lessen yield loss, which would be beneficial for the environment due to the reduced use of chemicals.
1 Introduction
The phytohormone abscisic acid (ABA) is a sesquiterpenoid with a C15 backbone (Nambara and Marion-Poll, 2005). ABA was initially identified as a compound that accelerated abscission (abbreviated as “abscission II”) in cotton and induced dormancy (abbreviated as “dormin”) in the leaves of Firmiana simplex. Further analyses showed that dormin has the same structure as abscission II, and both compounds are considered analogues of ABA (Taishi et al., 2011). The physiological effects of ABA were first detected in the early 1950s (Addicott and Lyon, 1969) when researchers isolated acidic fractions from plant extracts that had growth inhibitory effects on oat germinal sheaths. These extracts were classified as members of the β-inhibitor complex, and their function was consistent with what we know about the physiological effects of ABA (Taishi et al., 2011). Abiotic stressors have huge impacts on agricultural productivity and induce the production of many compounds that function in stress tolerance. ABA has numerous functions at the cellular level including the induction of the dehydration-responsive element-binding (DREB) transcription factors. DREB proteins regulate the expression of stress-induced genes by binding to DRE/CRT cis-elements in the promoter region to improve drought and salinity tolerance (Li et al., 2014). In response to abiotic stressors, mutations in the gene encoding serine-threonine protein kinase (Open Stomata1, OST1) rendered Arabidopsis thaliana guard cells insensitive to ABA, which kept the stomata open (Assmann, 2003). ABA also plays an important role in temperature stress and can reduce the risk of frost damage in plants. In tobacco plants exposed to a short period of heat stress, increased ABA concentrations reduced the cellular damage caused by high temperatures (Itai et al., 1978). ABA can also increase tolerance to biotic stressors; for example, ABA improved the resistance of rice to fungi, bacteria and the brown planthopper (BPH; Nilaparvata lugens) (Dunn et al., 1990; Robert-Seilaniantz et al., 2007; Fan et al., 2009; De Vleesschauwer et al., 2010; Liu et al., 2014; AbuQamar et al., 2017; Liu et al., 2017; Camisón et al., 2019; Darma et al., 2019; Boba et al., 2020). ABA has important roles in regulating plant growth, inhibiting seed germination, and promoting leaf senescence (Hao et al., 2009; Liang et al., 2014). Furthermore, ABA reduces water consumption by regulating stomatal closure, decreasing transpiration (Christmann et al., 2007) and regulating the activity of water channel proteins (Parent et al., 2009). ABA increased hydraulic conductivity and promotes water uptake by roots when soil begins to harden and dry out (Hose et al., 2000), which influences root and shoot growth (Hao et al., 2009). Thus, ABA has crucial roles in plant growth, development and stress tolerance.
In higher plants, 9-cis-epoxycarotenoid dioxygenase (NCED) is a key enzyme in ABA biosynthesis and is involved in the rate-limiting step of ABA biosynthesis. NCED is induced earlier than other ABA synthase genes and is considered a pivotal step in ABA synthesis (Qin and Zeevaart, 1999). In rice, the OsNCED gene family is comprised of five genes, OsNCED1–5 (Zhu et al., 2009), and these have different roles in plant growth and response to external stressors. OsNCED1 is primarily expressed in rice leaves but is significantly suppressed during water stress; whereas OsNCED2 is more highly expressed in rice seeds as compared to the other four OsNCED genes (Zhu et al., 2009; Ye et al., 2011). OsNCED3 is highly induced during water stress (Ye et al., 2011) and regulates the accumulation of the dehydrin and late embryogenesis abundant (LEA) proteins (Hundertmark and Hincha, 2008; Xiang et al., 2008). OsNCED3 and OsNCED4 exhibit an overlapping expression pattern in rice seeds, which suggests a level of functional redundancy and common control of ABA biosynthesis in rice seeds in response to salinity stress (Hwang et al., 2018). In response to rice black-stripe dwarf virus, the expression of OsNCED4 and OsNCED5 increased in rice as the duration of viral infection became more prolonged, which suggests a role for both genes in the regulation of ABA synthesis during viral infection (Ni et al., 2015).
ABA is synthesized via oxidative cleavage of epoxy-carotenoids. In maize, xanthophyll epoxygenase catalyzes the epoxidation of zeaxanthin and antioxidant xanthophylls to form purple xanthophylls and neoxanthophylls. These products are isomerized to produce 9-cis-isomers that are cleaved by NCED to form xanthotoxin; the latter is converted to ABA by short-chain dehydrogenase/reductase and aldehyde oxidase 3 (Wen, 2019).
OsNCED3 is systematically expressed in various tissues of rice and is induced by NaCl, PEG and H2O2, which supports a role for OsNCED3 in abiotic stress tolerance (Huang et al., 2018). When MhNCED3 from Malus hupehensis was expressed in Arabidopsis thaliana exposed to chlorine stress, the growth and development of transgenic A. thaliana improved, and plants exhibited an increase in ABA content and a decrease in transpiration (Zhang et al., 2015). Hwang et al. (2010) reported that heterologous expression of NCED3 in Arabidopsis increased ABA levels (Hwang et al., 2010). NtNCED3–2 is one of the NCED genes in tobacco, and NtNCED3–2 knockout plants had reduced levels of diterpenes, photosynthetic pigments, and phytohormones. Furthermore, knockdown of NtNCED3–2 resulted in decreased expression of genes in the isoprenoid metabolic pathway as compared to wild-type plants, resulting in reduced photosynthetic capacity (Yang et al., 2018). In rice plants, drought stress significantly induced OsNCED3 expression, which was down-regulated when watering was resumed. Transgenic lines overexpressing OsNCED3 after drought stress had higher ABA levels (Xu et al., 2018a). Southern hybridization experiments in Lycium barbarum showed that NCED was present in low copy numbers, and NCED expression gene was synchronized with the accumulation of endogenous ABA after salt and dehydration stress (Lu et al., 2013). Our previous work found that OsNCED3 had a positive role in defense against the brown planthopper through transcriptome profiling but more details not showed (Sun et al., 2022). In this study, we used OsNCED3 overexpression, RNA interference and wild-type (WT) rice to investigate the role of OsNCED3 in conferring resistance to the brown planthopper (BPH, Nilaparvata lugens). Resistance was evaluated by examining physiological and biochemical parameters, changes in hormone content, and defense gene expression. The results provide a foundation for analyzing ABA function in the regulation of BPH resistance.
2 Materials and methods
2.1 Plant and insect materials
The wild-type rice variety used in the experiment was Zhonghua 11(ZH11), and both OsNCED3 overexpression (OE-5) and silencing (RNAi-5) rice seeds were provided by the School of Life Sciences, South China Agricultural University and the phenotype was shown in Xu et al. paper (Xu et al., 2018b). The test rice was grown normally in the test field at 28~36 °C in summer. The BPH populations were collected from the China Rice Research Institute (Hangzhou, China) and kept in the greenhouse of the Ecological Laboratory under the following conditions: the temperature was (26 ± 2) °C, the humidity was maintained at 65%~75%, and the photoperiod was controlled at 16 L:8 D. Green house cultured BPH were transferred to experimental field, propagated for 3 additional generations and subsequently used for all the experiments (Sun et al., 2022).
2.2 RNA extraction and quantitative RT-PCR
Total RNA was isolated by FastPure® Universal Plant Total RNA Isolation Kit (Vazyme, Nanjing, China). A 2 mL sample of first-strand cDNA was analyzed in each 20μL reaction by qRT-PCR. All the tests were performed in three replicates. qRT-PCR was carried out using SYBR Select Mas-ter Mix (TaKaRa Biotech, Osaka, Japan) under the following reaction program: qRT-PCR was performed in a 20 μL reaction volume containing 10 μL of SYBR GreenPCR Master Mix, 2 μL of cDNA template (100 ng), and 1 μL each of forward and reverse primer. Each PCR was performed in a total volume of 20 µL following the manufacturer’s protocol. The expression level was calculated using the ΔΔCt (threshold cycle) method (Livak and Schmittgen, 2001). Three biological replicates were used per sample, and the expression level of each gene was normalized to that of the reference gene OsActin1.
2.3 Bioinformatics analysis of OsNCED3 gene in rice
The amino acid sequence of the CDS region of the rice gene OsNCED3 was obtained from NCBI (https://www.ncbi.nlm.nih.gov/), and the amino acid similarity was searched through the BLASTp program (https://blast.ncbi.nlm.nih.gov/Blast.cgi). Based on the search results, 21 NCED protein amino acid sequences from 21 species containing complete coding for aminoacids in protein (CDS) in Genbank were selected for phylogenetic analysis with OsNCED3, and a phylogenetic tree was constructed with Mega 7, and 5 amino acid sequences with higher similarity were selected and compared with OsNCED3 by DNAMAN. The five amino acid sequences with high similarity were selected and compared with OsNCED3 using DNAMAN; the tertiary structure of the protein was predicted using SWISSMODEL (https://swissmodel.expasy.org/); and the possible functional cooperating proteins of the protein were predicted using String (https://string-db.org).
2.4 Phenotypic differences among OE, RNAi and WT rice
The growth status of the three types of rice, such as differences in plant height, root length, was observed again 30 days after rice germination. ZH11, OsNCED3-OE and osnced3-RNAi lines (n=10 per genotype) were selected.
2.5 Treatment of OsNCED3 transgenic rice against BPH
10 plants (ZH11, OsNCED3-OE and osnced3-RNAi lines) were taken when the rice at the age of 4-leaf stage, and each line was set up with 10 replications, BPH infestation with 30 3rd-instar nymphs per plant, and the insects were picked up after starved treatment for 1 h. Cultivation was under normal environment and observations were made and samples of plant sheath were taken at four-time intervals: 0, 6, 12 and 24 h. All treatments were cultured in normal environment, and the fertilizer and water were managed properly.
2.6 Determination of rice injury levels and functional plant loss index after BPH feeding
ZH11, OsNCED3-OE and osnced3-RNAi lines (n=10 per genotype) were selected. Plants were infected as previously described (Sun et al., 2022). Briefly a flexible cylinder (5 cm diameter, 12 cm high) made from a polyvinyl chloride (PVC) sheet was inserted into the soil along the rim of the cup. BPH infestation was done by transferring 30 number of 3rd-instar nymphs per plant, to the cylinder followed by its sealing with a gauze. The injury level of rice in each plastic cup was checked at seven days. After determining injury levels, rice plants were cut into pieces, then washed, dried at 110°C for 20 min, and then dried to constant weight at 60°C. Dry weights were measured with a precision electronic balance, and the functional plant loss index (FPLI) was calculated.
2.7 Determination of insect resistant substances in rice
For the determination of flavonoid content, the method of Wang et al. (2005) was referred (Wang et al., 2005): the absorbance at the wavelength of 510 nm was measured with UV spectrophotometer, and the standard curve was made with rutin standard, and then the content of flavonoids in each sample was calculated.
For the determination of soluble sugar content, the method of Chen et al. (2017) was referred (Shi et al., 2018): the absorbance at 630 nm was measured by UV spectrophotometer, while the standard curve was made with glucose standard solution (100 μg/ml) to calculate the content of soluble sugar in each sample.
For the determination of oxalic acid content, reference was made to the method of Zhan et al. (2006) (Zhan et al., 2006): the absorbance was measured at 400 nm with a UV spectrophotometer, and a standard curve was made with oxalic acid standard to calculate the content of oxalic acid in each sample.
For the determination of free amino acid content, the method of Wang et al. (2007) was referred (Wang, 2007): the absorbance was measured at the wavelength of 570 nm with an ultraviolet spectrophotometer, while the standard curve was made with glutamic acid standard to calculate the content of free amino acid in each sample.
2.8 Extraction and assay of defense-related enzyme activities
For the determination of superoxide dismutase (SOD) activity, the method of Chen et al. (2017) was referred (Livak and Schmittgen, 2001): the absorbance at 560 nm was measured by UV spectrophotometer under the condition of avoiding light.
For the determination of peroxidase (POD) activity, the method of Han et al. (2018) was referred (Han, 2018): the absorbance was measured at 470 nm using a UV spectrophotometer. The data were recorded with a change of 0.01 in A470 per 1 min as a peroxidase activity unit U.
Catalase (CAT) activity was determined by referring the method of Lu et al. (2013) (Lu, 2014): the data were recorded with a change of 0.01 in A240 every 1 min as a catalase activity unit U.
For the determination of polyphenol oxidase (PPO) activity, the method of Soliva et al. (2000) was referred (Soliva et al., 2000): the data were recorded with a change of 0.01 in A410 per 1 min as one unit of peroxidase activity U. The data were recorded with a change of 0.01 in A410 per 1 min.
2.9 Determination of hormone content
The hormone content was determined according to the method of Zhang et al. (2017) (Zhang et al., 2017): the sample was analyzed using an HPLC-MS. HPLC-MS was used to determine the content of each hormone.
2.10 Statistical analysis
The statistical significance of differences between treatments was determined by analysis of variance (ANOVA; Systat Inc.) followed by Duncan´s multiple range test for multiple comparisons. For ANOVA, data were analyzed directly if normally distributed; data that were not normally distributed were transformed to ensure homogeneity of variances among different groups. Data were denoted as mean± SE, and analyzed using SPSS 11.0 software (SPSS).
3 Results
3.1 Multiple sequence alignment and phylogenetic analysis
The predicted amino acid sequence of OsNCED3 was obtained by searching databases at the National Center for Biotechnology Information (NCBI). OsNCED3 from Oryza sativa showed 92% similarity with ObNCED1 in the wild rice, O. brachyantha. Multiple sequence alignments of OsNCED3, ObNCED1, BdNCED3 (Brachypodium distachyon), SbNCED1 (Sorghum bicolor), SiNCED1 (Setaria italica), and ZmNCED1 (Zea mays) were conducted with DNAman (https://www.lynnon.com/) (Figure 1A). These sequences contained both highly and relatively-conserved amino acid residues. The amino acid sequences of 29 NCED proteins from different plant species were downloaded from NCBI, and MEGA 7.0 was used for multiple sequence alignment and construction of a phylogenetic tree (Figure 1B). Phylogenetic analysis showed that the dendogram could be divided into branches representing dicots and monocots. The NCEDs in monocots such as rice, B. distachyon, millet and sorghum are represented in a branch that differed from the dicots grouped in the other branch (e.g., Cucurbita pepo, Cucumis melo, and Malus domestica). The NCEDs in graminaceous plants were closely related to plants in Chenopodiaceae and Papaveraceae. The tertiary structure of OsNCED3 was predicted using SWISS-MODEL (https://swissmodel.expasy.org/), which revealed that OsNCED3 contained 12 α-helices and 36 β-folds (Figure 1C). Proteins that potentially interact with OsNCED3 were identified with String (https://string-db.org/). Ten possible interacting proteins were predicted (Figure 1D), and the highest and lowest scores were 0.941 and 0.613, respectively. Potential interacting proteins included OSJ_13064 containing ketoreductase and fatty acid synthase domains.
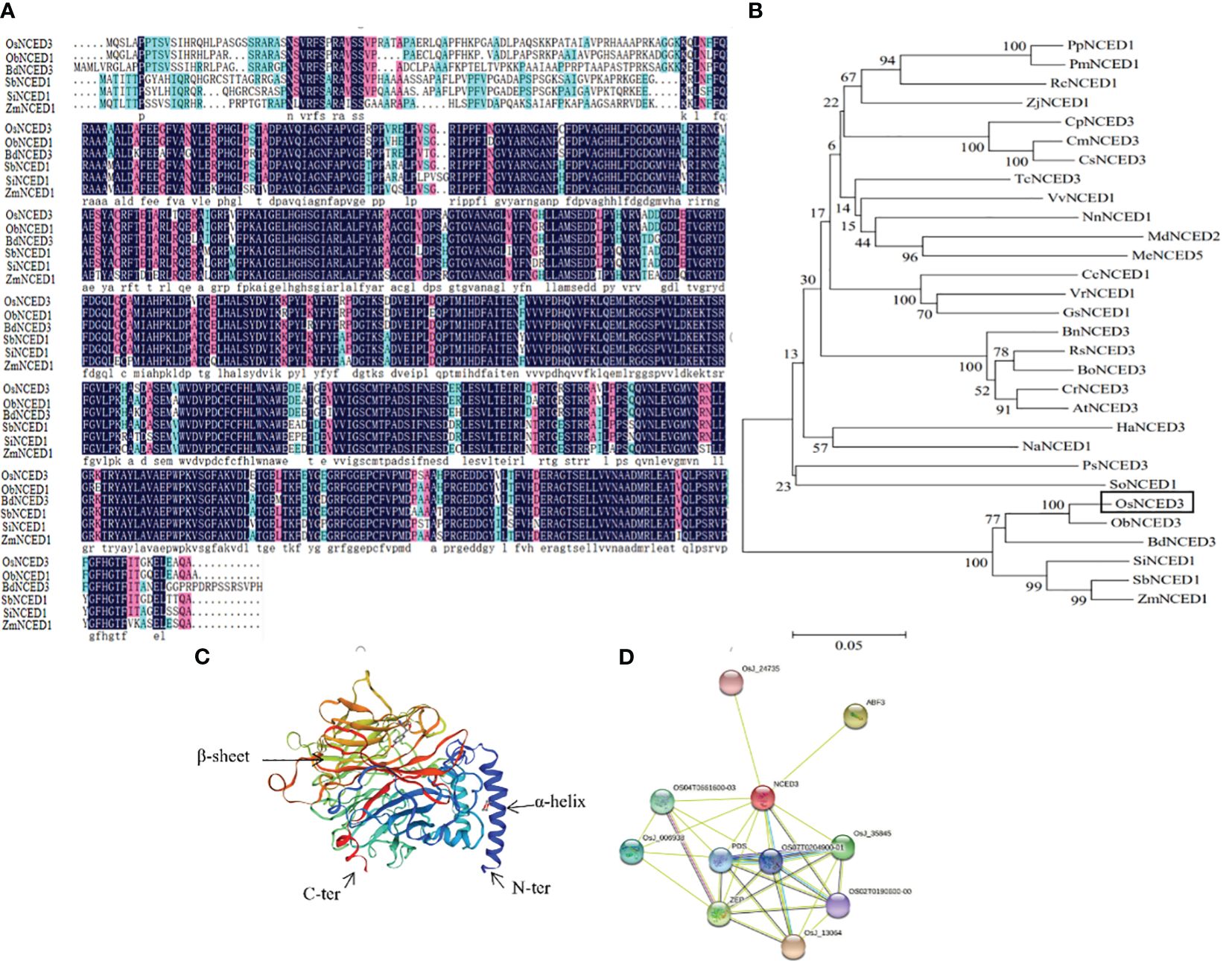
Figure 1 Bioinformatics analysis of OsNCED3 gene in rice. (A) comparison of deduced protein sequence encoded by OsNCED3. Black part represents highly conserved residues; red represents conservative substitution; light blue represents semi conservative substitution. OsNCED3 (Oryza sativa); ObNCED1 (Oryza brachyantha); SbNCED1 (Sorghum bicolor); ZmNCED1 (Zea mays); BdNCED3 (Brachypodium distachyon); SiNCED1 (Setaria italica). (B) phylogenetic tree of amino acid sequence of OsNCED gene from different sources; OsNCED3 (Oryza sativa); ObNCED1 (Oryza brachyantha); SbNCED1 (Sorghum bicolor); ZmNCED1 (Zea mays); BdNCED3 (Brachypodium distachyon); SiNCED1 (Setaria italica); RsNCED3 (Raphanus sativus); PpNCED1 (Prunus persica); RcNCED1 (Rosa chinensis); CrNCED3 (Capsella rubella); AtNCED3 (Arabidopsis thaliana); BnNCED3 (Brassica napus); BoNCED3 (Brassica oleracea); VvNCED1 (Vitis vinifera); NnNCED1 (Nelumbo nucifera); CmNCED3 (Cucumis melo); PmNCED1 (Prunus mume); CpNCED3 (Cucurbita pepo); MdNCED2 (Malus domestica); CsNCED3 (Cucumis sativus); HaNCED3 (Helianthus annuus); VrNCED1 (Vigna radiata); PsNCED3 (Papaver somniferum); NaNCED1 (Nicotiana attenuata); ZjNCED1 (Ziziphus jujuba); TcNCED3 (Theobroma cacao); MeNCED5 (Manihot esculenta); GsNCED1 (Glycine soja); CcNCED1 (Cajanus cajan); SoNCED1 (Spinacia oleracea). (C) Predicted 3D structure of OsNCED3 using PyMOL software. (D) interacting proteins of OsNCED3 using the STRING database.
3.2 Phenotypic differences among OE, RNAi and WT rice lines
The phenotypes of wild-type (WT) rice ZH11, the overexpressing line OsNCED3-OE and the knockdown osnced3-RNAi were compared in 30-d-old seedlings (Figure 2A). There were no significant differences in plant height (Figure 2B), root length (Figure 2C), or fresh and dry weights (Figures 2D, E) when osnced3-RNAi rice was compared with WT. However, OsNCED3-OE plants were shorter with reduced root development and lower fresh and dry weights as compared with osnced3-RNAi and the WT, indicating that OsNCED3 is involved in rice growth and development.
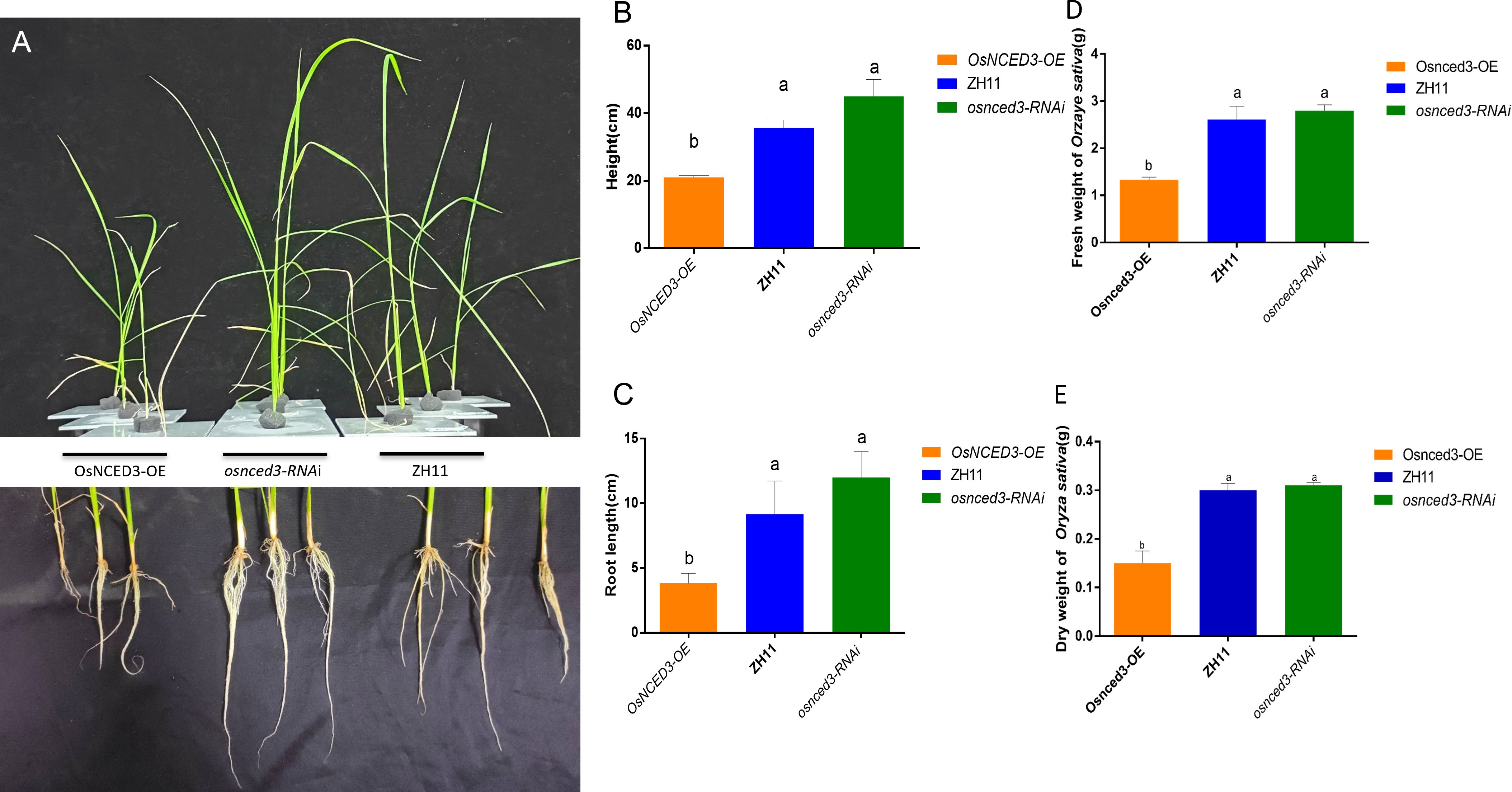
Figure 2 Growth genotypes of OsNCED3-OE, osnced3-RNAi and WT rice plants. (A) Phenotypes of plant height and root length of indicated genotypes(n=10); (B) comparison of plant height; (C) comparison of root length; (D) comparison of fresh weight of plants; (E) comparison of dry weight of plants. The data are mean ± SE. Bars with different letters show significant different at P< 0.05 by PLSD test.
3.3 Interactions and validation of insect resistance in transgenic rice
There were significant differences in average injury level and function plant loss index when BPH fed on OsNCED3-OE, osnced3-RNAi and WT rice (Figures 3A, B). It indicated that BPH cause more serious damage on osnced3-RNAi, BPH clearly preferred to feed on the osnced3-RNAi line, and BPH populations on the OsNCED3-OE line were significantly lower than those on the WT and osnced3-RNAi lines (Figure 3C). BPH that fed on the OsNCED3-OE line had lower larval survival rates (Figure 3D). and lower numbers of eggs per plant than osnced3-RNAi and WT rice (Figure 3E). Furthermore, expression levels of OsNCED3 in the OsNCED3-OE line continued to increase when BPH was allowed to feed (Figure 3F), whereas no changes in expression levels were detected during BPH feeding in the osnced3-RNAi line. Overall, these results indicate that BPH feeding was higher on osnced3-RNAi rice as compared to the OsNCED3-OE line.
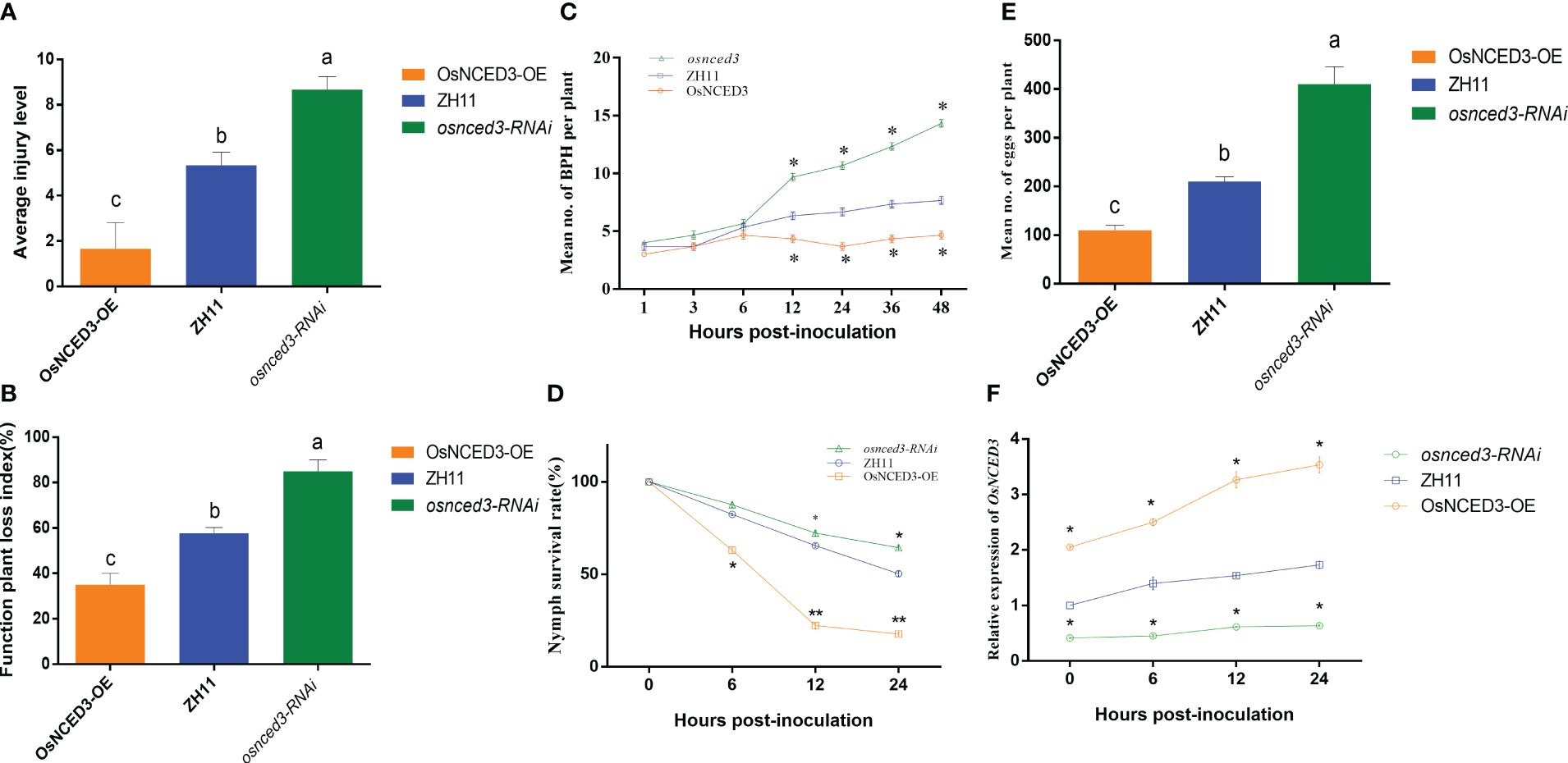
Figure 3 OsNCED3 positively regulates BPH resistance in rice (A). (B) Average injury levels, and FPLI after BPH feeding on ZH11, OsNCED3-OE, osnced3-RNAi line. BPH (n=30) were allowed to feed on WT, and OE rice for 7 d, and injury, and FPLI values were then obtained; (C) statistical analysis of number of BPH in plant after feeding on ZH11, OsNCED3-OE, osnced3-RNAi line; (D) statistical analysis of nymph survival rates; (E) statistical analysis of number of eggs per plant; (F) qRT-PCR analysis of OsNCED3 transcripts in ZH11, OsNCED3-OE, osnced3-RNAi line after BPH infestation. The data are mean ± SE. Bars with different letters show significant different at P< 0.05 by PLSD test.
3.4 The content of antibiotic-resistant substances in rice increased with the infestation of brown planthopper
Flavonoid content in the OsNCED3-OE line increased rapidly after 12 h of BPH feeding and was 33.45% higher than levels in the WT (Figure 4A). the flavonoid content at 24 h was significantly lower (32.84%) in the osnced3-RNAi line as compared to the WT. The soluble sugar content was significantly higher in the OsNCED3-OE line as compared to the WT in the 6–24 h time period after exposure to BPH (Figure 4B). In contrast, the soluble sugar content in the OsNCED3-OE line was significantly lower than the osnced3-RNAi line at 0–12 h, but this difference disappeared at 24 h. The results showed that the soluble sugar content of the BPH treatment was significantly lower than that of WT (Figure 4B). Significant differences in oxalic acid content were observed after 12 h of BPH exposure, and levels were significantly higher in the OsNCED3-OE line as compared to the osnced3-RNAi and WT lines (Figure 4C). The oxalic acid content in the OsNCED3-OE line was 44.39% higher than the WT at 12 h. The osnced3-RNAi line had significantly reduced oxalic acid content (down 28.56%) as compared to the WT at 24 h after BPH exposure. There were no significant differences in the oxalic acid content of the three lines at 0 and 6 h. Furthermore, no significant differences were observed in the free amino acid content of the osnced3-RNAi and WT rice lines at 0–24 h of BPH feeding (Figure 4D). In contrast, the free amino acid content in the OsNCED3-OE line was significantly lower than levels in the WT and osnced3-RNAi lines after 6–24 h of BPH feeding.
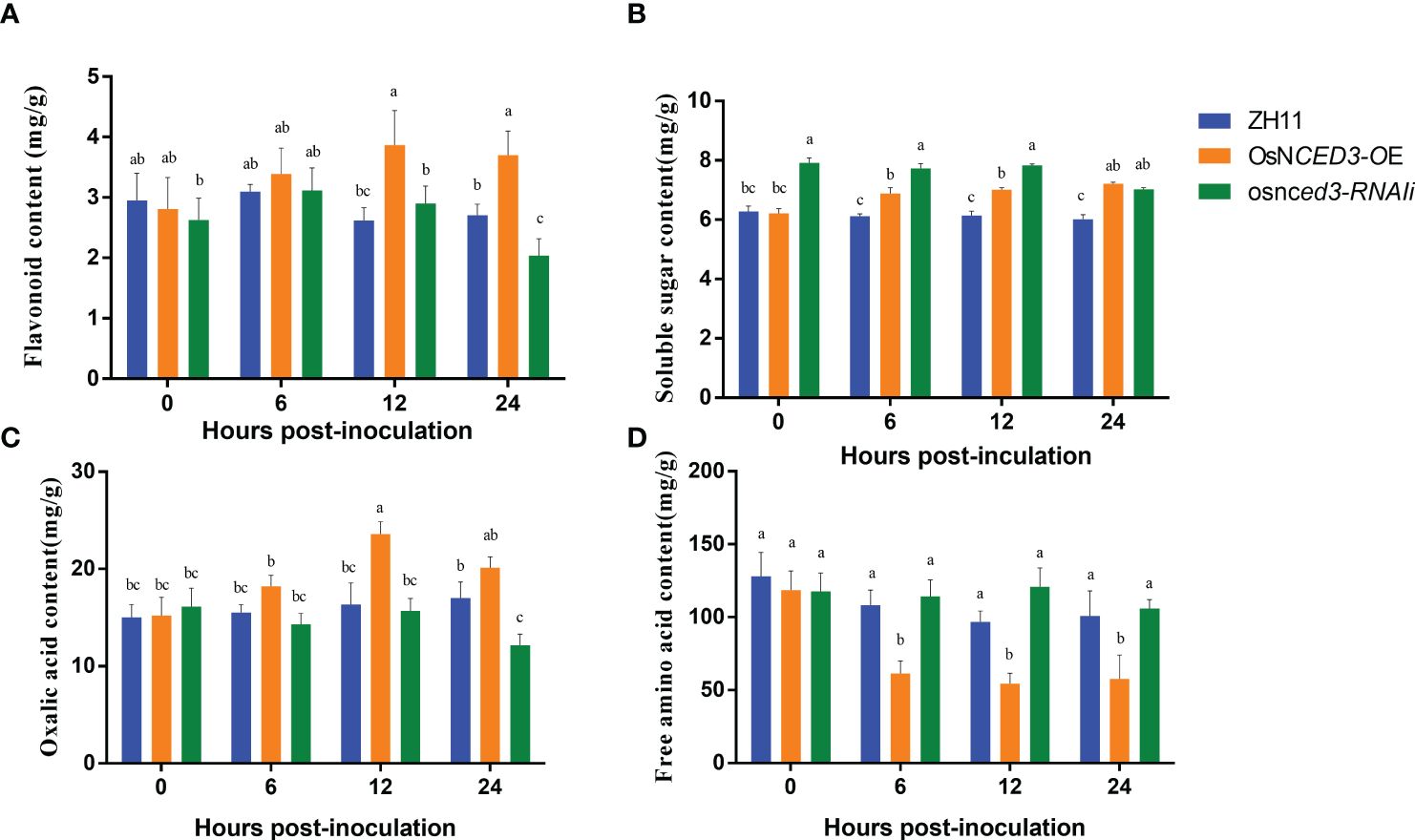
Figure 4 Changes of resistance substances in OsNCED3 transgenic rice after BPH feeding. (A) the results of flavonoid content; (B) the results of soluble sugar content; (C) the results of oxalic acid content; (D) the results of free amino acid content. The data are mean ± SE. Bars with different letters show significant different at P< 0.05 by PLSD test.
3.5 The activities of defense enzymes in rice increased with the infection of brown planthopper
SOD activity in the OsNCED3-OE line was significantly higher than the WT and osnced3-RNAi lines from 0 to 24 h after BPH feeding (Figure 5A). which indicated that OsNCED3 overexpression caused a significant increase in SOD levels. POD activity in the OsNCED3-OE line was significantly higher than levels in the WT and osnced3-RNAi lines from 0–12 h, but this difference was not apparent at 12 or 24 h (Figure 5B). PPO activity in the OsNCED3-OE, osnced3-RNAi and WT rice lines was not significantly different at 0 or 6 h; however, PPO activity began to increase at 12 h after BPH feeding (Figure 5C). PPO activity in the OsNCED3-OE line was significantly higher than the osnced3-RNAi line at 12 and 24 h after BPH feeding. CAT activity in the OsNCED3-OE line was consistently higher than in the osnced3-RNAi and WT rice (Figure 5D). CAT activity showed a rapid increase from 0–6 h in the OsNCED3-OE line and remained high throughout the sampling times.
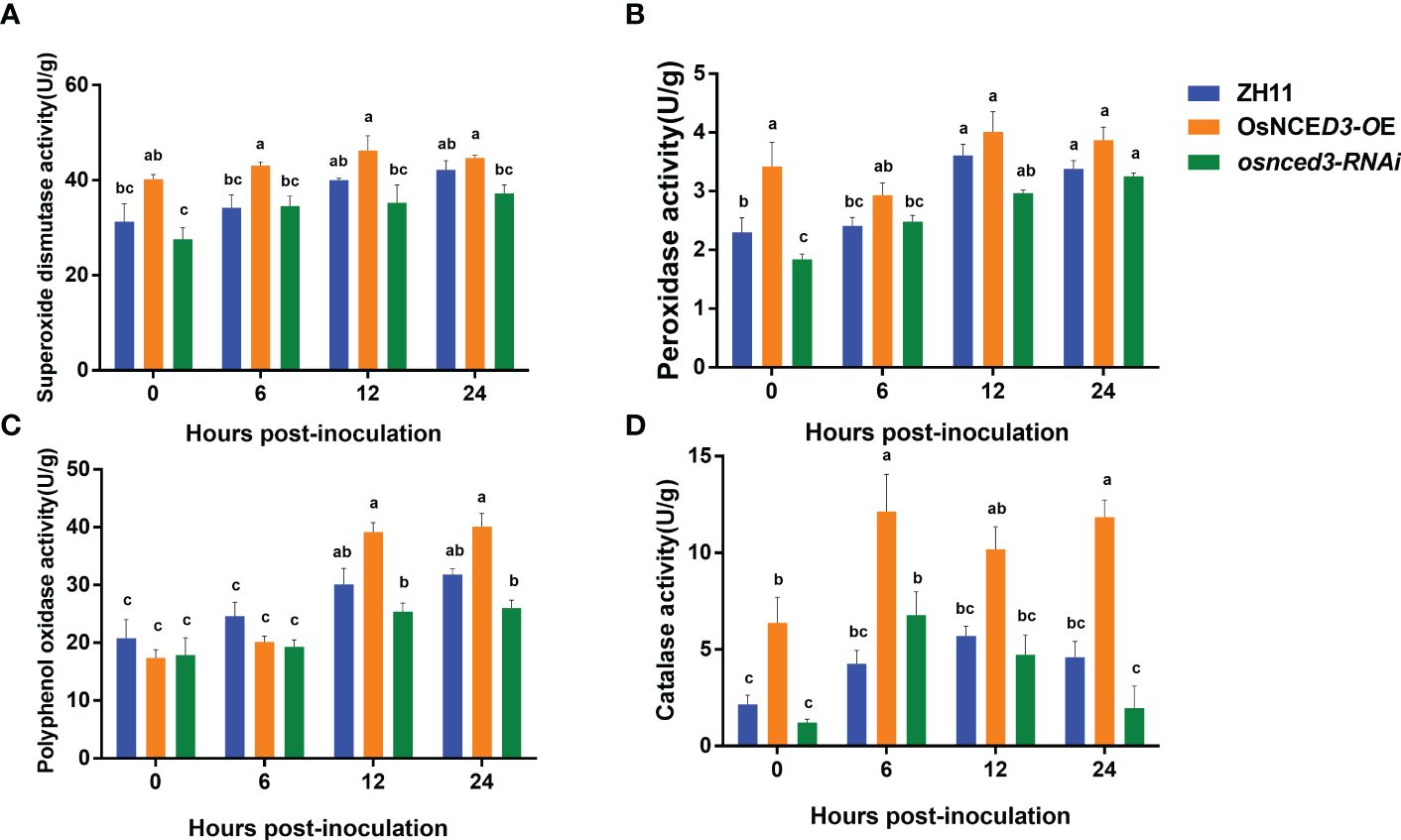
Figure 5 Changes of defense enzyme activity of OsNCED3 transgenic rice after BPH feeding. (A) the results of superoxide dismutase activity; (B) the results of peroxidase activity; (C) the results of polyphenol oxidase activity; (D) the results of Catalase activity. The data are mean ± SE. Bars with different letters show significant different at P< 0.05 by PLSD test.
3.6 The hormones ABA and JA in transgenic rice plants were significantly increased under brown planthopper infestation
There were no significant differences in SA content in OsNCED3-OE, osnced3-RNAi and WT rice at 0, 12 and 24 h of BPH feeding. The SA content in WT plants was significantly higher than levels in OsNCED3-OE and osnced3-RNAi lines at 6 h (Figure 6A). The JA content in the OsNCED3-OE line was significantly higher than the WT and osnced3-RNAi plants at all sampling times (Figure 6B). The JA content in the OsNCED3-OE line was highest at 6 h after BPH feeding; afterwards, the JA content decreased but remained significantly higher than levels in the WT and osnced3-RNAi plants. The ABA content was significantly higher in OsNCED3-OE rice at 6 and 12 h after BPH infestation (Figure 6C). ABA levels stabilized in the OsNCED3-OE line at 12 and 24 h after BPH exposure. In the osnced3-RNAi line, there was a transient increase in the ABA content at 6 h, which decreased at 12 and 24 h; these results indicated that osnced3-RNAi modulated ABA levels in the early stages of BPH infestation (Figure 6C). The JA-Ile content in the OsNCED3-OE line was significantly elevated as compared with the WT and osnced3-RNAi lines at 6–12 h of inoculation (Figure 6D); however this difference disappeared at 24 h. These results indicate that JA-Ile levels rapidly accumulate after BPH infestation, which is similar to results observed with JA (Figure 6B). There were no significant differences in JA-Ile levels in osnced3-RNAi and WT rice. In summary, most of the hormones monitored in this study showed significant changes early after BPH infestation, and these levels declined as the infestation time increased.
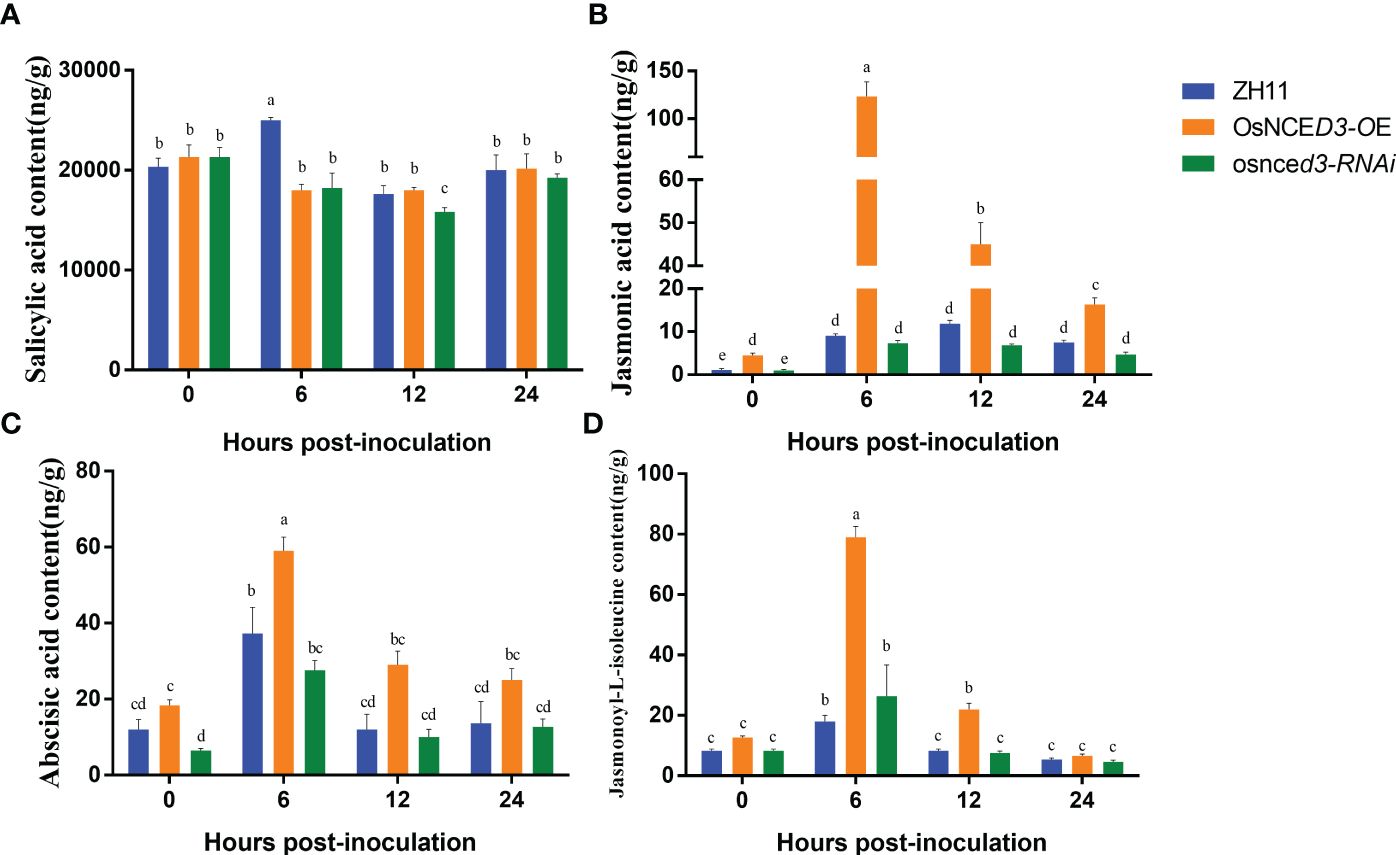
Figure 6 Determination of hormone content in OsNCED3 transgenic rice. (A) salicylic acid content; (B) jasmonic acid content; (C) abscisic acid content; (D) jasmonoyl-L-isoleucine content. The data are mean ± SE. Bars with different letters show significant different at P< 0.05 by PLSD test.
3.7 The expression of defense-related genes in transgenic rice plants was significantly increased under brown planthopper infestation
OsAOS1 (LOC_Os03g55800) and OsMYC2 (LOC_Os10g42430) are involved in JA biosynthesis and regulation, respectively. Both OsAOS1 and OsMYC2 were significantly upregulated in the OsNCED3-OE line as compared to WT rice after BPH feeding (Figure 7). In contrast, OsAOS1 and OsMYC2 expression were significantly reduced in osnced3-RNAi rice as compared to the WT. OsJAZ1 (LOC_Os04g55920) is a transcriptional repressor of JA, and its expression was significantly elevated in osnced3-RNAi rice; this suggests that OsNCED3 is involved in both JA synthesis and signaling. OsABA8ox3 (LOC_Os09g28390) and OsPYL9 (LOC_Os06g36670) are ABA catabolism and ABA receptor genes, respectively. There was no significant change in OsABA8ox3 expression in OsNCED3-OE rice as compared to the WT after BPH feeding, but the expression of osnced3-RNAi decreased significantly. OsPYL9 expression in OsNCED3-OE rice was significantly higher than expression in the osnced3-RNAi and WT lines. Rice BPH resistance genes OsbZIP23 (LOC_Os02g52780), Osbph6 (LOC_Os04g35210), and OsKSL4 (LOC_Os04g10060) were all induced and expressed at higher levels in OsNCED3-OE plants as compared to WT and the osnced3-RNAi line. In summary, our results indicate that OsNCED3 is involved in the expression of genes that confer insect resistance.
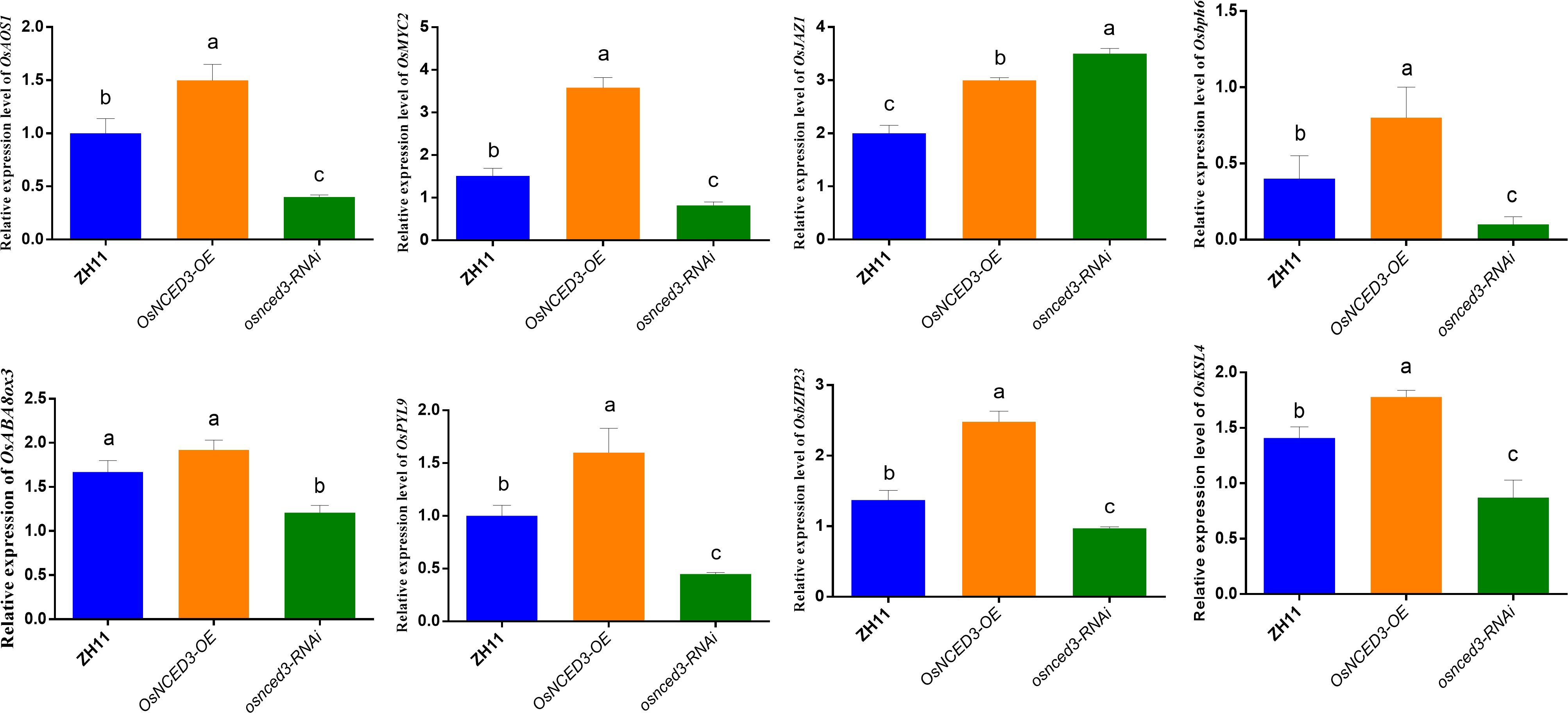
Figure 7 Changes of key gene expression in OsNCED3 transgenic rice after BPH feeding. The relative expression level of OsAOS1(LOC_Os03g55800). The relative expression of OsMYC2 (Jasmonic acid transcription factor, LOC_Os10g42430). The relative expression level of OsJAZ1(LOC_Os04g55920). The relative expression level of Osbph6(LOC_Os04g35210). The relative expression of OsABA8ox3 (LOC_Os09g28390). The relative expression level of OsPYL9(LOC_Os06g36670). The relative expression of OsbZIP23 (LOC_Os02g52780). The relative expression of OsKSL4 (kaurene synthase-like 4, LOC_Os04g10060). The data are mean ± SE. Bars with different letters show significant different at P< 0.05 by PLSD test.
4 Discussion
Plant insect tolerance is the ability of plants to tolerate adversity and compensate for it by relying on factors such as their own growth as well as reproductive capacity when subjected to harsh biotic stresses similar to those of insect-sensitive species (Yang, 2005). Our findings suggest that BPH preferred to feed on osnced3-RNAi rice as compared to WT and OsNCED3-OE rice. The overexpression of OsNCED3 reduced BPH-induced damage and improved insect tolerance. These results suggest that OsNCED3 expression is induced by BPH feeding and correlates with improved insect tolerance in a very short amount of time.
The overexpression and silencing of OsNCED3 in rice resulted in multiple changes in the concentrations of flavonoids, soluble sugars, oxalic acid, and amino acids in the OsNCED3-OE and osnced3-RNAi rice lines. We observed a rapid increase in flavonoid content in OsNCED3-OE rice that was significantly higher than levels in the knockdown line, osnced3-RNAi. Plants produce a number of polyphenolic compounds including flavonoids (Ballester et al., 2010; Wang et al., 2011), which function as antioxidants that scavenge free radicals in plants. Furthermore, flavonoids have key roles in antimicrobial activity and stress tolerance (Heim et al., 2002; Chen et al., 2007). Soluble sugars provide energy for growth and development and have signaling functions in plants (Takahashi et al., 2003; Kato-Noguchi et al., 2010; Boriboonkaset et al., 2013). There are two opinions on the roles of soluble sugar in plant resistance to insects; for example, one view is that higher levels of soluble sugars improve plant resistance to insects. For example, Ji et al. (2006) studied oviposition and feeding selectivity of the tobacco whitefly, Bemisia tabaci, on different cucumber varieties. Higher concentrations of soluble sugars in cucumber reduced whitefly numbers and development (Ji, 2006). He et al. (2017) also reported that peppers with higher resistance would have higher resistance to whitefly (Jing et al., 2017). After Solanum lycopersicum was treated with exogenous MeJA, Zhang et al. (2009) analyzed the nutrients, secondary metabolites, and defense enzymes in tomato leaves infested with whiteflies. The content of soluble sugars and proteins were reduced and resistance to whiteflies improved, leading the authors to speculated that exogenous MeJA improved plant resistance by reducing nutrients such as soluble sugars (Zhang et al., 2020). Our data shows that soluble sugars weaken plant resistance to plant hopper. Oxalic acid is another defensive compound that has an important role in plant resistance to abiotic and biotic stressors (Tian et al., 2009). For example, kiwifruit sprayed with oxalic acid had increased levels of defense enzymes and were more resistant to Penicillium causativeum (Zhu et al., 2016). Melons (Cucumis melo) treated with oxalic acid were more resistant to pink mold rot caused by Trichothecium roseum, and oxalic acid treatment elevated the activity of defense enzymes such as POD and PPO (Deng et al., 2015). Free amino acids function in the metabolic homeostasis of plants and also play an important role in plant resistance (Gao, 2016). Zeng et al. (1992) reported that the free amino acid content in BPH-resistant varieties of rice was lower than in susceptible cultivars (Zeng et al., 1992).
SOD, POD and CAT are important antioxidant enzymes that scavenge free radicals in plants and protect cells from oxidative damage. SOD scavenges free radicals and converts them to O2 and hydrogen peroxide, which reduces cellular damage. CAT protects plant cells by utilizing hydrogen peroxide as a substrate and converting free radicals to oxygen and water, thus eliminating the toxicity of H2O2 (Xing et al., 2015; Hu, 2020). In addition to metabolizing hydrogen peroxide, POD catalyzes the polymerization of phenolics into lignin, which strengthens the plant cell wall and provides a level of protection from invading insects and pathogens (Tian et al., 2001; Jia et al., 2004). PPO catalyzes the conversion of phenolic compounds into highly reactive quinones and provides precursors for defense compounds such as lignin and phytochelatins (Zhang et al., 2020). In this study, we observed higher levels of SOD, POD and CAT in the OsNCED3-OE rice line as compared to osnced3-RNAi and WT rice; this was especially true for CAT activity. PPO activity increased rapidly beginning at 12 h and levels were significantly higher than those in the osnced3-RNAi and WT lines.
Our results definitively show that defense compounds such as flavonoids and oxalic acid were highest in the OsNCED3-OE line, and the increase was more obvious beginning 12 h after BPH exposure. The soluble sugar content was comparable in all three rice lines. The levels of the antioxidant enzymes SOD, POD and CAT were higher in the OsNCED3-OE line than the WT beginning at 0 h. In contrast, PPO activity began to rise at 12 h after BPH exposure. The average level of damage and the index of functional loss were exclusively discussed in Sun’s article for overexpression lines, while no mention was made regarding RNAi interference line, this study demonstrates that the OsNCED3-OE line exhibits higher insect resistance compared to the osnced3-RNAi line (Sun et al., 2022). Sun’s EPG analysis reveals that the feeding of BPH is inhibited by the OsNCED3-OE line. The data presented by Sun aligns with our results, indicating the involvement of OsNCED3 in rice’s mechanism of insect resistance (Sun et al., 2022). Collectively, these results also indicate that levels of defense compounds were highest in the OsNCED3-OE line, which is also is more resistant to BPH than WT and osnced3-RNAi rice.
SA has been extensively studied in plants since its original isolation from willow bark in 1828. SA has an important role in tolerance to abiotic (e.g., cold and salt stress) and biotic stressors (diseases and insects) (Kang et al., 2007; Palma et al., 2009; Hussain et al., 2014; Ueda et al., 2019; Cheng et al., 2020). JA-Ile is the bioactive form of JA, and induces the expression of defense-related genes to protect the plant from biotic stress (Wang, 2018). Although no significant changes were observed in SA content in the three rice lines (Figure 6A), both JA and JA-Ile levels increased rapidly in the OsNCED3-OE line beginning at 6 h after BPH feeding. Beginning at 0 h, the ABA content in OsNCED3-OE rice was higher than the levels in osnced3-RNAi rice, suggesting a role for OsNCED3 in regulating ABA synthesis. At 6–12 h after BPH feeding, the ABA content in all three rice lines increased and then declined rapidly, which was similar to the increase and decrease in JA and JA-Ile levels (Figure 6). At 24 h of BPH feeding, the ABA content was comparable in all three rice lines. Based on the existing research in our group, we hypothesized that there was a synergistic interaction between ABA and JA in plants after BPH feeding.
Allene oxide synthase (AOS1) is the second enzyme in the JA biosynthesis pathway, whereas MYC2 is a transcription factor that positively regulates JA biosynthesis. Vos et al. (2013) found that exogenous ABA induced MYC2 expression in Arabidopsis, which resulted in a JA-mediated defense response. The ABA synthesis loss-of-function mutant aba2–1 impeded the transcription of the cabbage worm Pieris rapae post-feeding expression of resistance genes regulated by the transcription factor MYC2 (Vos et al., 2013). JAZ (Jasmonate ZIM-domain) is a transcriptional repressor of JA signaling that inhibits JA-activated responses by repressing MYC2. JAZ proteins are dependent on the JA signaling pathway to negatively regulate plant defense against biotic and abiotic stressors (Browse, 2009; Fu et al., 2017). OsKSL4, a biosynthesis gene for the diterpene phytocannabinoid ryanodiolactone, acts as a positive regulator of the defense response in rice (Xu et al., 2007; Yoshida et al., 2017). OsABA8ox3 is an ABA catabolism-related gene in rice, and OsPYL9 (pyrabactin resistance 9-like) is an ABA receptor. OsPYL positively regulates the ABA response when seed germination occurs, whereas overexpression of OsPYL9 can significantly improve drought and cold tolerance in rice (Tian et al., 2015). OsbZIP23 belongs to the rice bZIP transcription factor family, which promotes in plant resistance (Mellacheruvu et al., 2016). Osbph6 (brown plant hopper resistance 6) confers broad-spectrum resistance to BPH (Guo et al., 2018). RT-qPCR after BPH feeding showed that the expression of 8 genes was higher in OsNCED3-OE rice than in the WT and osnced3-RNAi lines. The expression patterns of OsAOS1, OsMYC2, and OsJAZ1 correlated with changes in JA hormone content, indicating that JA biosynthesis was promoted during BPH feeding. Expression of the ABA catabolism gene OsABA8ox3 in OsNCED3-OE rice was not significantly different from expression in WT rice (Figure 7). However, expression of OsPYL9 in OsNCED3-OE was significantly higher than the WT and osnced3-RNAi lines. Based on the results obtained for OsKSL4, OsbZIP23 and Osbph6, we concluded that the resistance of rice lines overexpressing OsNCED3 gene was enhanced and the expressed genes identified in Sun et al. (2022)’s article were found to align with the transcriptome data, and their expression patterns exhibited consistency, suggesting a positive impact on enhancing rice resistance against insects (Sun et al., 2022).
5 Conclusions
The resistance of three rice lines (OsNCED3-OE, osnced3-RNAi, and WT) to BPH and the role of OsNCED3 in BPH tolerance was examined by monitoring physiological and biochemical parameters, changes in hormone content, and defense gene expression. The results indicate that OsNCED3 expression is induced by BPH feeding and correlates with improved insect tolerance in a very short amount of time. The overexpression of OsNCED3 reduced BPH-induced damage and improved insect tolerance. BPH clearly preferred to feed on osnced3-RNAi vs. the OsNCED3-OE and WT lines, and BPH populations on the OsNCED3-OE line were significantly lower than those on the WT and osnced3-RNAi. BPH that fed on the osnced3-RNAi line also had lower larval survival rates and reduced numbers of eggs per plant as compared to osnced3-RNAi and WT rice. one graphical summary model showed in Figure 8.
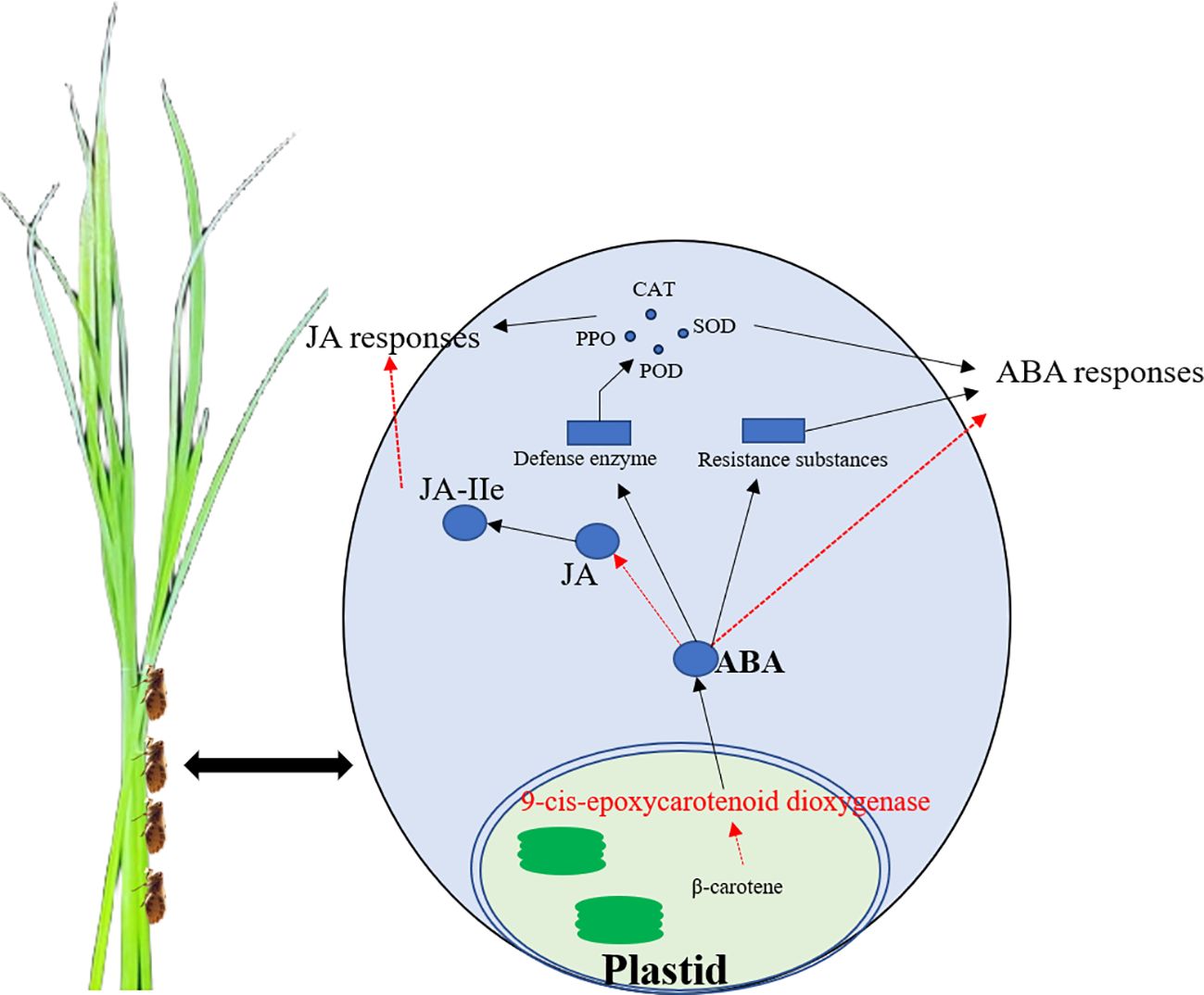
Figure 8 The schematic model of OsNCED3 regulated the complex crosstalk between ABA and JA signaling pathway after BPH feeding. OsNCED3 increased the expression of defensed genes, defense enzyme activity and resistance substances content to response biotic stress.
Our results indicate that levels of defense compounds (flavonoids and OA) were highest in the OsNCED3-OE line, which was more resistant to BPH than WT and osnced3-RNAi rice. Our findings also show that OsNCED3 activated rice defense mechanisms, which led to increases in the defense enzymes superoxide dismutase, peroxidase, and polyphenol oxidase. JA, JA-Ile and ABA concentrations rapidly accumulated in the OsNCED3-OE line after BPH infestation, and these levels declined as the infestation time increased. The findings outlined in this study indicate that modulation of endogenous genes in rice may be a valid management tactic for reducing yield loss, which is beneficial for the environment due to the reduced use of chemical agents.
Data availability statement
The original contributions presented in the study are included in the article/supplementary materials. Further inquiries can be directed to the corresponding authors.
Author contributions
JtL: Data curation, Formal analysis, Investigation, Methodology, Project administration, Resources, Software, Validation, Visualization, Writing – original draft, Writing – review & editing. HL: Investigation, Formal analysis, Software, Writing – original draft. XL: Software, Supervision, Visualization, Writing – original draft. WW: Investigation, Formal analysis, Writing – original draft. XL: Investigation, Conceptualization, Formal analysis, Methodology, Writing – original draft. LC: Conceptualization, Writing – original draft. YW: Conceptualization, Methodology, Project administration, Software, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing. JlL: Writing – review & editing, Conceptualization, Data curation, Investigation, Methodology, Project administration, Resources, Supervision, Validation, Visualization.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This research was funded by The National Natural Science Foundation of China grant numbers 32172426 and 31871978.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
AbuQamar, S., Moustafa, K., Tran, L. S. (2017). Mechanisms and strategies of plant defense against Botrytis cinerea. Crit. Rev. Biotechnol. 37, 262–274. doi: 10.1080/07388551.2016.1271767
Addicott, F. T., Lyon, J. L. (1969). Physiology of abscisic acid and related substances. Annu. Rev. Plant Physiol. 20), 139–164. doi: 10.1146/annurev.pp.20.060169.001035
Assmann, S. M. (2003). Open Stomata1 opens the door to ABA signaling in Arabidopsis guard cells. Trends Plant Sci. 8, 151–153. doi: 10.1016/S1360-1385(03)00052-9
Ballester, A. R., Molthoff, J., de Vos, R., Hekkert, B. T., Orzaez, D., Fernández-Moreno, J. P., et al. (2010). Biochemical and molecular analysis of pink tomatoes: deregulated expression of the gene encoding transcription factor SlMYB12 leads to pink tomato fruit color. Plant Physiol. 152, 71–84. doi: 10.1104/pp.109.147322
Boba, A., Kostyn, K., Kozak, B., Wojtasik, W., Preisner, M., Prescha, A., et al. (2020). Fusarium oxysporum infection activates the plastidial branch of the terpenoid biosynthesis pathway in flax, leading to increased ABA synthesis. Planta 251, 50. doi: 10.1007/s00425-020-03339-9
Boriboonkaset, T., Theerawitaya, C., Yamada, N., Pichakum, A., Supaibulwatana, K., Cha-Um, S., et al. (2013). Regulation of some carbohydrate metabolism-related genes, starch and soluble sugar contents, photosynthetic activities and yield attributes of two contrasting rice genotypes subjected to salt stress. Protoplasma 250, 1157–1167. doi: 10.1007/s00709-013-0496-9
Browse, J. (2009). Jasmonate passes muster: a receptor and targets for the defense hormone. Annu. Rev. Plant Biol. 60, 183–205. doi: 10.1146/annurev.arplant.043008.092007
Camisón, Á., Martín, M.Á., Sánchez-Bel, P., Flors, V., Alcaide, F., Morcuende, D., et al. (2019). Hormone and secondary metabolite profiling in chestnut during susceptible and resistant interactions with Phytophthora cinnamomi. J. Plant Physiol. 241, 153030. doi: 10.1016/j.jplph.2019.153030
Chen, Y., Liu, K., Zhang, G. L., Tan, W. Z. (2007). Progress on the biological activity and chemical composition of the invasive weed, Pelargonium huangdianense. Weed Sci. 04), 1–3. doi: 10.19588/j.issn.1003-935x.2007.04.001
Cheng, T., Yao, X. Z., Wu, C. Y., Zhang, W., Wei, He. (2020). Endophytic Bacillus megaterium triggers salicylic acid-dependent resistance and improves the rhizosphere bacterial community to mitigate rice spikelet rot disease. Appl. Soil Ecol. 156, 103710. doi: 10.1016/j.apsoil.2020.103710
Christmann, A., Weiler, E. W., Steudle, E., Grill, E. (2007). A hydraulic signal in root-to-shoot signalling of water shortage. Plant J. 52, 167–174. doi: 10.1111/j.1365-313X.2007.03234.x
Darma, R., Lutz, A., Elliott, C. E., Idnurm, A. (2019). Identification of a gene cluster for the synthesis of the plant hormone abscisic acid in the plant pathogen Leptosphaeria maculans. Fungal Genet. Biol. 130, 62–71. doi: 10.1016/j.fgb.2019.04.015
De Vleesschauwer, D., Yang, Y., Cruz, C. V., Höfte, M. (2010). Abscisic acid-induced resistance against the brown spot pathogen Cochliobolus miyabeanus in rice involves MAP kinase-mediated repression of ethylene signaling. Plant Physiol. 152, 2036–2052. doi: 10.1104/pp.109.152702
Deng, J. J., Bi, Y., Zhang, Z. K., Xie, D. F. (2015). Postharvest oxalic acid treatment induces resistance against pink rot by priming in muskmelon (Cucumis melo) fruit. Postharvest Biol. Tec. 106, 53–61. doi: 10.1016/j.postharvbio.2015.04.005
Dunn, R. M., Hedden, P., Bailey, J. A. (1990). A physiologically-induced resistance of Phaseolus vulgaris to a compatible race of Colletotrichum lindemuthianum is associated with increases in ABA content. Physiol. Mol. Plant Pathol. 36, 0885–5765. doi: 10.1016/0885-5765(90)90063-4
Fan, J., Hill, L., Crooks, C., Doerner, P., Lamb, C. (2009). Abscisic acid has a key role in modulating diverse plant-pathogen interactions. Plant Physiol. 150, 1750–1761. doi: 10.1104/pp.109.137943
Fu, J., Wu, H., Ma, S., Xiang, D., Liu, R., Xiong, L. (2017). OsJAZ1 attenuates drought resistance by regulating JA and ABA signaling in rice. Front. Plant Sci. 8, 2108. doi: 10.3389/fpls.2017.02108
Gao, X., Xiao, H., Zhang, Z., Zheng, N. (2016). Determination of free amino acids in fragrans leaves by using high performance liquid chromatography.. Earth and environment. 44 (01), 103–109. doi: 10.14050/j.cnki.1672-9250.2016.01.014
Guo, J., Xu, C., Wu, D., Zhao, Y., Qiu, Y., Wang, X., et al. (2018). Bph6 encodes an exocyst-localized protein and confers broad resistance to planthoppers in rice. Nat. Genet. 50, 297–306. doi: 10.1038/s41588-018-0039-6
Han, S. N. (2018). Screening and identification of biocontrol bacteria inducing rice resistance to rice blast and preliminary study on the mechanism. Shenyang Agric. University., 11–77.
Hao, G. G., Sun, Z. F., Zhang, L. Q., Du, K. M. (2009). Research overview of the plant resistance to adverse environment by using abscisic acid. Chin Agricul Sci Bull. 25, 212–215. doi: 10.11924/j.issn.1000-6850.2009-0827
Heim, K. E., Tagliaferro, A. R., Bobilya, D. J. (2002). Flavonoid antioxidants: chemistry, metabolism and structure-activity relationships. J. Nutr. Biochem. 13, 572–584. doi: 10.1016/S0955-2863(02)00208-5
Hose, E., Steudle, E., Hartung, W. (2000). Abscisic acid and hydraulic conductivity of maize roots: a study using cell- and root-pressure probes. Planta 211, 874–882. doi: 10.1007/s004250000412
Hu, T. (2020). Effects of low temperature on the physiological characteristics of rice root system and its gene expression. Shenyang Agric. University. 02–52. doi: 10.27327/d.cnki.gshnu.2019.000066
Huang, Y., Guo, Y., Liu, Y., Zhang, F., Wang, Z., Wang, H., et al. (2018). 9-cis-Epoxycarotenoid dioxygenase 3 regulates plant growth and enhances multi-abiotic stress tolerance in rice. Front. Plant Sci. 9, 162. doi: 10.3389/fpls.2018.00162
Hundertmark, M., Hincha, D. K. (2008). LEA (late embryogenesis abundant) proteins and their encoding genes in Arabidopsis thaliana. BMC Genomics 9, 118. doi: 10.1186/1471-2164-9-118
Hussain, B., War, A., Sharma, H. (2014). Jasmonic and salicylic acid-induced resistance in sorghum against the stem borer Chilo partellus. Phytoparasitica. 42), 99–108. doi: 10.1007/s12600-013-0343-8
Hwang, S. G., Chen, H. C., Huang, W. Y., Chun, Y. C., Shii, C. T., Cheng, W. H. (2010). Ectopic expression of rice OsNCED3 in Arabidopsis increases ABA level and alters leaf morphology. Plant Sci. 178, 12–22. doi: 10.1016/j.plantsci.2009.09.014
Hwang, S. G., Lee, C. Y., Tseng, C. S. (2018). Heterologous expression of rice 9-cis-epoxycarotenoid dioxygenase 4 (OsNCED4) in Arabidopsis confers sugar oversensitivity and drought tolerance. Bot. Stud. 59, 2. doi: 10.1186/s40529-018-0219-9
Itai, C., Benzioni, A., Munz, S. (1978). Heat stress: Effects of abscisic acid and kinetin on response and recovery of tobacco leaves. Plant Cell Physiol. 19, 453–459. doi: 10.1093/oxfordjournals.pcp.a075614
Ji, X. Z. (2006). Oviposition and feeding selection and mechanism of cigarette louse on different cucumber varieties. China Agric. University. 06–52.
Jia, Z., Song, Z. W., Jin, Z. Y., Wang, L. (2004). Effects of hawthorn leaf mite damage on POD of begonia leaves. Northwest J. Botany. 11, 2136–2139.
Jing, H., Zhou, F. C., Su, H. H., Zhao, B., Shao, Y. D., Yang, A. M., et al. (2017). Resistance studies of chili pepper varieties to sooty mold. J. Appl. Entomol. 54, 629–638.
Kang, G. Z., Wang, Z. X., Xia, K. F., Sun, G. C. (2007). Protection of ultrastructure in chilling-stressed banana leaves by salicylic acid. J. Zhejiang Univ Sci. B. 8, 277–282. doi: 10.1631/jzus.2007.B0277
Kato-Noguchi, H., Yasuda, Y., Sasaki, R. (2010). Soluble sugar availability of aerobically germinated barley, oat and rice coleoptiles in anoxia. J. Plant Physiol. 167, 1571–1576. doi: 10.1016/j.jplph.2010.06.017
Li, C., Yue, J., Wu, X., Xu, C., Yu, J. (2014). An ABA-responsive DRE-binding protein gene from Setaria italica, SiARDP, the target gene of SiAREB, plays a critical role under drought stress. J. Exp. Bot. 65, 5415–5427. doi: 10.1093/jxb/eru302
Liang, C., Wang, Y., Zhu, Y., Tang, J., Hu, B., Liu, L., et al. (2014). OsNAP connects abscisic acid and leaf senescence by fine-tuning abscisic acid biosynthesis and directly targeting senescence-associated genes in rice. Proc. Natl. Acad. Sci. U.S.A. 111, 10013–10018. doi: 10.1073/pnas.1321568111
Liu, J., Du, H., Ding, X., Zhou, Y., Xie, P., Wu, J. (2017). Mechanisms of callose deposition in rice regulated by exogenous abscisic acid and its involvement in rice resistance to Nilaparvata lugens Stål (Hemiptera: Delphacidae). Pest Manag Sci. 73, 2559–2568. doi: 10.1002/ps.4655
Liu, J. L., Chen, X., Zhang, H. M., Yang, X., Wong, A. (2014). Effects of exogenous plant growth regulator abscisic acid-induced resistance in rice on the expression of vitellogenin mRNA in Nilaparvata lugens (Hemiptera: Delphacidae) adult females. J. Insect Sci. 14, 213. doi: 10.1093/jisesa/ieu075
Livak, K. J., Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Lu, J. (2014). Preliminary study on the interactions between Rhizopus oryzae and rice. Huazhong Agric. University. 02–58.
Lu, P., Tian, Y. S., Wang, M. X., Li, S., Zhao, J. Y. (2013). Cloning and expression analysis of NCED, a key enzyme gene for abscisic acid biosynthesis in Lycium barbarum. Plant Genet. Resour. 14 (02), 303–310. doi: 10.13430/j.cnki.jpgr.2013.02.006
Mellacheruvu, S., Tamirisa, S., Vudem, D. R., Khareedu, V. R. (2016). Pigeonpea hybrid-proline-rich protein (CcHyPRP) confers biotic and abiotic stress tolerance in transgenic rice. Front. Plant Sci. 6, 1167. doi: 10.3389/fpls.2015.01167
Nambara, E., Marion-Poll, A. (2005). Abscisic acid biosynthesis and catabolism. Annu. Rev. Plant Biol. 56, 165–185. doi: 10.1146/annurev.arplant.56.032604.144046
Ni, H. P., Xu, Q. F., Lan, Y., Chen, Q. Q., Zhang, J. F., Zhou, Y. J. (2015). Effect of RBSDV infestation on the expression of ABA metabolism-related genes in rice. China. Rice. Sci. 29, 319–326. doi: 10.3969/j.issn.1001-7216.2015.03.012
Palma, F., Lluch, C., Iribarne, C., García-Garrido, J., Garcia, N. T. (2009). Combined effect of salicylic acid and salinity on some antioxidant activities, oxidative stress and metabolite accumulation in Phaseolus vulgaris. Plant Growth Regul. 58, 307–316. doi: 10.1007/s10725-009-9380-1
Parent, B., Hachez, C., Redondo, E., Simonneau, T., Chaumont, F., Tardieu, F. (2009). Drought and abscisic acid effects on aquaporin content translate into changes in hydraulic conductivity and leaf growth rate: a trans-scale approach. Plant Physiol. 149, 2000–2012. doi: 10.1104/pp.108.130682
Qin, X., Zeevaart, J. A. (1999). The 9-cis-epoxycarotenoid cleavage reaction is the key regulatory step of abscisic acid biosynthesis in water-stressed bean. Proc. Natl. Acad. Sci. U.S.A. 96, 15354–15361. doi: 10.1073/pnas.96.26.15354
Robert-Seilaniantz, A., Navarro, L., Bari, R., Jones, J. D. (2007). Pathological hormone imbalances. Curr. Opin. Plant Biol. 10, 372–379. doi: 10.1016/j.pbi.2007.06.003
Shi, Y. C., Liu, J. Y., Wang, X. Y., Chen, Q. Y., Wang, X. Y., Liu, W. Q. (2017). Enhancing resistance to TMV of tobacco by exogenous trehalose and preliminary analysis of its mechanism. Acta Tabacaria Sinica. 23 (3). doi: 10.16472/j.chinatobacco.2016.378
Soliva, R. C., Elez, P., Sebastián, M., Olga, M. (2000). Evaluation of browning effect on avocado purée preserved by combined methods-Science Direct. Innov. Food Sci. Emerg. Technol. 1, 261–268. doi: 10.1016/S1466-8564(00)00033-3
Sun, L., Li, J., Liu, Y., Noman, A., Chen, L., Liu, J. (2022). Transcriptome profiling in rice reveals a positive role for OsNCED3 in defense against the brown planthopper, Nilaparvata lugens. BMC Genomics 23, 634. doi: 10.1186/s12864-022-08846-5
Taishi, U., Takashi, H., Takashi, K., Shinozaki, K. (2011). The regulatory networks of plant responses to abscisic acid. Adv. Bot. Res. 57, 201–248. doi: 10.1016/B978-0-12-387692-8.00006-0
Takahashi, F., Sato-Nara, K., Kobayashi, K., Suzuki, M., Suzuki, H. (2003). Sugar-induced adventitious roots in Arabidopsis seedlings. J. Plant Res. 116, 83–91. doi: 10.1007/s10265-002-0074-2
Tian, G. Z., Li, H. F., Qiu, W. F. (2001). Progress of plant peroxidase. Wuhan Botanical Res. 04), 332–344.
Tian, H., Wang, L., Duan, M. Y., Li, G. X. (2009). Research progress on oxalic acid metabolism and regulation in plants. Mod. Agric. Sci. Technol. 08), 212–213.
Tian, X., Wang, Z., Li, X., Lv, T., Liu, H., Wang, L., et al. (2015). Characterization and functional analysis of pyrabactin resistance-like abscisic acid receptor family in rice. Rice (N Y). 8, 28. doi: 10.1186/s12284-015-0061-6
Ueda, H., Kugimiya, S., Tabata, J., Kitamoto, H., Mitsuhara, I. (2019). Accumulation of salicylic acid in tomato plant under biological stress affects oviposition preference of Bemisia tabaci. J. Plant Interact. 14, 73–78. doi: 10.1080/17429145.2018.1550220
Vos, I. A., Verhage, A., Schuurink, R. C., Watt, L. G., Pieterse, C. M., Van Wees, S. C. (2013). Onset of herbivore-induced resistance in systemic tissue primed for jasmonate-dependent defenses is activated by abscisic acid. Front. Plant Sci. 4, 539. doi: 10.3389/fpls.2013.00539
Wang, F. (2007). Selective pesticide-induced rice leaf borer reemergence and varietal resistance. Yangzhou University., 06–62.
Wang, J. N. (2018). Molecular and biochemical mechanisms of OsRCI-1 regulating induced resistance to brown planthopper and white-backed planthopper in rice. Zhejiang Agric. Forestry University., 07–62.
Wang, W. J., Li, X. Y., Zu, Y. G. (2005). Dynamic feature of flavonoids content in different organs of larch (Larix gmelinii). J. Forestry. Res. 16, 89–92. doi: 10.1007/BF02857896
Wang, Y., Chen, S., Yu, O. (2011). Metabolic engineering of flavonoids in plants and microorganisms. Appl. Microbiol. Biotechnol. 91, 949–956. doi: 10.1007/s00253-011-3449-2
Wen, A. N. (2019). Research on the function and genetic diversity of OsNCED gene in rice low temperature tolerance. Hunan Normal University., 12–73.
Xiang, Y., Tang, N., Du, H., Ye, H., Xiong, L. (2008). Characterization of OsbZIP23 as a key player of the basic leucine zipper transcription factor family for conferring abscisic acid sensitivity and salinity and drought tolerance in rice. Plant Physiol. 148, 1938–1952. doi: 10.1104/pp.108.128199
Xing, S. L., Yao, Y. L., Xu, L., Hu, X. W., Liu, Y. (2015). Cloning and bioinformatics analysis of land rice catalase gene (OsCATB-lh). Guangdong Agric. Science. 42, 106–110. doi: 10.16768/j.issn.1004-874x.2015.19.018
Xu, X. Z., Wang, T., Wan, W., Li, S. H., Zhu, G. H. (2018a). Rice ABA biosynthesis gene OsNCED3 responds to drought stress. J. Crops. 44, 24–31. doi: 10.3724/SP.J.1006.2018.00024
Xu, X. Z., Wang, T., Wang., W., Li, S. H., Zhu, G. H. (2018b). ABA biosynthesis gene OsNCED3 confers drought stress tolerance in rice. Acta Agron. Sin. 44, 24–31. doi: 10.3724/SP.J.1006.2018.00024
Xu, M., Wilderman, P. R., Morrone, D., Xu, J., Roy, A., Margis-Pinheiro, M., et al. (2007). Functional characterization of the rice kaurene synthase-like gene family. Phytochemistry 68, 312–326. doi: 10.1016/j.phytochem.2006.10.016
Yang, S. J. (2005). Experimental study on the resistance of hybrid progeny of medicinal wild rice to rice leaf borer. J. Agric. Sci. Technol. 33, 34–35.
Yang, Y. X., Zhou, Q. Q., Xu, J. Y., Li, Q. C., Zhang, S. T. (2018). RNA interference of NtNCED3 reduces drought tolerance and impairs plant growth through feedback regulation of isoprenoids in Nicotiana tabacum. Environ. Exp. Bot. 155), 332–344. doi: 10.1016/j.envexpbot.2018.07.016
Ye, N., Zhu, G., Liu, Y., Li, Y., Zhang, J. (2011). ABA controls H2O2 accumulation through the induction of OsCATB in rice leaves under water stress. Plant Cell Physiol. 52, 689–698. doi: 10.1093/pcp/pcr028
Yoshida, Y., Miyamoto, K., Yamane, H., Nishizawa, Y., Minami, E., Nojiri, H., et al. (2017). OsTGAP1 is responsible for JA-inducible diterpenoid phytoalexin biosynthesis in rice roots with biological impacts on allelopathic interaction. Physiol. Plant 161, 532–544. doi: 10.1111/ppl.12638
Zeng, L., Wu, R. Z., Feng, C., Li, J. Q. (1992). Relationship between free amino acid content and resistance to brown rice louse in rice varieties. J. South China Agric. University. 04), 69–76.
Zhan, H. J., Zhou, C. Z., Zhang, L., Gong, Y. Z. (2006). Determination of oxalic acid content in beer. Brewing Sci. Technology. 04), 102–104. doi: 10.13746/j.njkj.2006.04.038
Zhang, X. X., Hong, B., Jing, L. L., Cao, S., Jia, Y. X. (2020). Exogenous methyl jasmonate induces insect resistance in tomato against sooty mold. J. Ecol. 39, 3651–3657. doi: 10.13292/j.1000-4890.202011.026
Zhang, J., Luo, T., Wang, W., Cao, T., Li, R., Lou, Y. (2017). Silencing OsSLR1 enhances the resistance of rice to the brown planthopper Nilaparvata lugens. Plant Cell Environ. 40, 2147–2159. doi: 10.1111/pce.13012
Zhang, H. X., Shi, W. K., Guo, R., Zhang, Y. J., Guo, H. B. (2020). Protein screening and validation of danshen polyphenol oxidase interactions. J. Tradit. Chin. Med. 45, 2523–2532. doi: 10.19540/j.cnki.cjcmm.20200329.103
Zhang, W. W., Yang, H. Q., You, S. Z., Fan, S. L., Ran, K. (2015). MhNCED3, a gene encoding 9-cis-epoxycarotenoid dioxygenase in Malus hupehensis Rehd., enhances plant tolerance to Cl- stress by reducing Cl- accumulation. Plant Physiol. Biochem. 89, 85–91. doi: 10.1016/j.plaphy.2015.02.012
Zhu, G., Ye, N., Zhang, J. (2009). Glucose-induced delay of seed germination in rice is mediated by the suppression of ABA catabolism rather than an enhancement of ABA biosynthesis. Plant Cell Physiol. 50, 644–651. doi: 10.1093/pcp/pcp022
Keywords: OsNCED3, Nilaparvata lugens, abscisic acid (ABA), jasmonic acid (JA), rice
Citation: Li J, Liu H, Lv X, Wang W, Liang X, Chen L, Wang Y and Liu J (2024) A key ABA biosynthetic gene OsNCED3 is a positive regulator in resistance to Nilaparvata lugens in Oryza sativa. Front. Plant Sci. 15:1359315. doi: 10.3389/fpls.2024.1359315
Received: 21 December 2023; Accepted: 07 June 2024;
Published: 26 June 2024.
Edited by:
Manu Agarwal, University of Delhi, IndiaReviewed by:
Girdhar Kumar Pandey, University of Delhi, IndiaAnuradha Singh, Michigan State University, United States
Copyright © 2024 Li, Liu, Lv, Wang, Liang, Chen, Wang and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jinglan Liu, bGl1amxAeXp1LmVkdS5jbg==; Yiping Wang, eXB3YW5nMDFAeXp1LmVkdS5jbg==
 Jitong Li
Jitong Li Hao Liu
Hao Liu Xinyi Lv
Xinyi Lv Wenjuan Wang
Wenjuan Wang Xinyan Liang
Xinyan Liang Lin Chen
Lin Chen Yiping Wang
Yiping Wang Jinglan Liu
Jinglan Liu