- 1Department of Extension Plant Sciences, New Mexico State University, Las Cruces, NM, United States
- 2Seed Biotechnology Center, University of California, Davis, Davis, CA, United States
Introduction: Domestic production of pepper (Capsicum spp.) is shrinking while demand within the US is growing. Lack of availability and cost of labor often present an obstacle for domestic producers both practically and economically. As a result, switching to harvesting peppers mechanically is anticipated as a key strategy to help domestic producers compete in the international market. Mechanical harvest efficiency can be improved through breeding. One important trait that mechanical harvest compatible material should have is an easy destemming trait: low force separation of the pedicel and calyx from the fruit.
Methods: To detect the genetic sources underlying a novel easy destemming trait for the purpose of future breeding efforts in New Mexico pod-type green chile, we performed QTL analysis on three F2:F3 populations, coming from three New Mexico pod-type varieties: ‘NuMex Odyssey,’ ‘NuMex Iliad,’ and ‘NuMex Joe E. Parker,’ each crossed with a parent with an easy destemming trait: MUC14. Genotyping was done through genotyping by sequencing (GBS) and phenotyping was done for destemming and fruit trait measurements. Correlations between measurements were found through the R package hmisc and QTL analysis was done through R/qtl.
Results: A strong relationship was seen between destemming and aspects of fruit morphology, particularly, destemming force and fruit width (Pearson’s correlation coefficient r=0.75). Major QTLs for destemming and fruit size were discovered. Of these, the largest destemming force QTLs for all populations (PVE=34.5-69.9%) were on chromosome 10, and in two populations QTLs for destemming force were found on chromosome 3 (Percent Variance Explained (PVE)=10.7-18.8%). Fruit size-related QTLs in all populations colocalized in these same areas on chromosomes 3 and 10.
Discussion: This suggests that fruit shape may be genetically linked to destemming, and breeders interested in selecting for easy destemming pepper will also have to pay attention to fruit size and shape.
1 Introduction
In New Mexico, chile pepper (Capsicum spp.) is an economically and culturally important crop. The state is a major hot pepper producer, accounting for 82.8%, 80.2%, and 83.7% of the total US chile pepper harvest reported for 2020, 2021, and 2022, respectively (USDA and NASS, 2023). It is also the origin of a type of pepper that bears the state’s name: New Mexico pod–type pepper (this pod shape is sometimes also called Anaheim pepper). Long, smooth, and mild to moderately hot, New Mexico pod–type peppers were first developed and released by the horticulturalist Fabian Garcia at New Mexico State University in the early 20th century (Coon et al., 2008). Today, New Mexico pod–type peppers are an integral part of “Mexican” cuisine in the American southwest, with many varieties (New Mexico 6-4, NuMex Big Jim, and NuMex Joe E. Parker) being commonly grown both within New Mexico and in nearby states (Bosland and Walker, 2014).
Both red (ripe) and green (un-ripe) chile are grown and consumed, but, whereas the red chile crop is mostly mechanically harvested, the green chile crop is still entirely hand-harvested: a task that presents challenges for producers within the US (Wall et al., 2003; Walker and Funk, 2014). An aging workforce, changing immigration laws, and labor reforms affect labor availability, making it uncertain that there will be a workforce available at harvest time. In addition, it is estimated that manual harvesting is responsible for up to 50% of the cost of domestic chile production (Hawkes et al., 2008). Because of this, US pepper production is not increasing alongside demand. Instead, the total acreage of chile pepper grown within the US is shrinking, whereas imports from countries with cheaper and more readily available labor are increasing (Gandonou and Waliczek, 2013; Lillywhite and Tso, 2021). As a result, the implementation of mechanical harvesting for green chile is anticipated as a key strategy to help domestic pepper producers compete in an international market.
Field trials indicate that successful mechanical harvesting in green chile requires a systems approach because the choice of harvester, field management techniques, and pepper varieties impact the efficacy of mechanical harvesting (Funk et al., 2011). For the delicate green chile crop, which deteriorates quickly if broken, the harvester chosen should be relatively gentle. Currently, the most promising harvester in this respect is an inclined double helix machine produced by Yung-Etgar (Bet-Lehem-Haglilit, Israel) (Funk and Walker, 2010). The height and growth habit of plants also affect mechanical harvesting, so field management decisions about plant spacing and transplanting vs. direct seeding that impact plant structure are relevant to the system as well (Havlik, 2021). Finally, access to plant material that is bred to have traits suitable for mechanical harvesting is critical.
There are certain plant and fruit traits that are important for mechanical harvest pepper varieties. To be compatible with the double helix harvester, plants should have upright architecture with a tall, single major stem, lack of low-lying lateral branches (sometimes called basal branches), and larger basal stem diameters (Joukhadar et al., 2018). Fruit traits like uniform ripening, thicker fruit walls (pericarp thickness), and easy to remove pedicels (easy destemming) would also be conducive to an ideal result: mechanically harvested, uniformly mature, stemless, and intact green chile.
Efforts to develop mechanical harvest–amenable plant material are at the early stages. Recently, a New Mexico pod–type variety called ‘NuMex Odyssey’ that was bred to have a main single stem and increased height to fruit set, ideal plant architecture traits for green chile mechanical harvest, was released (Walker et al., 2021). This is currently the only commercially available green chile New Mexico pod–type variety that has been developed with mechanical harvest in mind. With regard to the genetics of stem removal, a quantitative trait locus (QTL) study was recently conducted for two populations derived from a semi-domesticated pepper with a novel easy destemming trait called UCD-14, revealing several QTLs with a large effect (Hill et al., 2023). Studies such as this can be a step toward breeding through marker-assisted selection, an expedited breeding method in which individuals of a population are screened and selected on the basis of the presence of genes that confer traits of interest.
Considering this, to aid in the development of New Mexico pod–type green chile varieties that are amenable to mechanical harvest, we conducted a QTL study for three populations that came from crosses between a New Mexico pod–type variety and a semi-domesticated landrace with a novel easy destemming trait.
2 Materials and methods
2.1 Population descriptions
Three intercross populations were created, each from two parent reciprocal crosses where one parent was a green chile New Mexico pod–type variety, and the other parent was a variety with an easy destemming trait (MUC14). The three New Mexico pod–type parents were ‘NuMex Iliad,’ ‘NuMex Odyssey,’ and ‘NuMex Joe E. Parker.’ Of these, two were breeding lines developed at New Mexico State University, ‘NuMex Iliad’ and ‘NuMex Odyssey,’ to be compatible with mechanical harvesting in terms of their plant architecture. The other, ‘NuMex Joe E. Parker,’ was a line that had very few destemmed fruits harvested in mechanical harvest trials. The parent crossed with each New Mexico pod–type is MUC14. This is a line derived from a cross between a semi-domesticated Mexican landrace with small fruit and a novel destemming trait (UCD-14) and Maor that was selected for low destemming force and performance in mechanical harvest. For all three populations, the F1 and F2 generations were grown in a greenhouse at Fabian Garcia Science Center, Las Cruces, NM. The F3 generation was grown at the NMSU Agricultural Science Center in Los Lunas, NM. The F3 families were direct-seeded in five-foot plots, with two replicates per family. We aimed to collect 40 mature green fruits per plot, and we collected an average of 38 fruits from six plants per plot. The field was planted in April and harvested in September 2021.
2.2 Phenotyping
Five measurements were taken from each fruit collected: fruit width, fruit length, pericarp thickness, destemming score, and destemming force. The height was measured at the tallest length of the fruit, and the width was measured across the fruit body below the shoulder. Fruits were cut at the width measurement, and the pericarp thickness was measured with calipers. A representation of overall fruit size, fruit size index, was recorded as fruit width times fruit length. To measure the destemming force, a Tohnichi torque gauge (Tohnichi America Corporation, Buffalo Grove, IL, United States) with a custom metal fork attachment that enables the removal of a pedicel with a twisting motion, meant to resemble the motion of the harvester, was used (Hill et al., 2023). At the same time, a subjective rating of destemming efficiency, called “destemming score,” was taken, where the measurer rated the quality of the fruit after destemming. The scale was from 1 to 5, where 1 meant a perfectly destemmed fruit, 2 to 4 meant the stem was removed, but with increasing damage to the fruit, and 5 meant there was a failure to destem. For each measurement, an average was taken for all the fruit measurements per plot, and the average of the two replicate plots was used as the final phenotype representing the F2 parent.
2.3 DNA extraction
Young leaf tissue was taken from the parents (‘NuMex Joe E. Parker,’ ‘NuMex Odyssey,’ ‘NuMex Iliad,’ and MUC14) and F2 individuals from each population and collected into 2-mL screw cap tubes with 2.5-mm Zirconia/Silica beads and stored at −80°C until extraction. Before homogenization with a Precellys 24 Homogenizer (Bertin instruments Paris, France), buffer AP1 (QIAGEN, Germany) was added to the tubes. The QIAGEN DNeasy Plant Mini Kit was used to extract DNA, and the concentration and absorbance ratios were checked with a Nanodrop 2000 Spectrophotometer (Thermo Fisher Scientific, Waltham, MA US). RNA contamination was checked using gel electrophoresis.
2.4 Genotyping by sequencing
Libraries were constructed using the protocol described by Elshire et al. (2011), with some modifications. Instead of using ApeKI, the restriction enzyme MlyI was used (Hill et al., 2023). Sequencing was done on an Illumina HS4000 in pools of 96 at the UC Davis Genome Center (Davis, California).
2.5 SNP calling using BCF and VCFtools
Demultiplexed files were downloaded onto the NMSU computer Kraken (Penguin Relion XE1112 384 GiB). The reads were trimmed for quality, and adaptor sequences were removed using Cutadapt v4.1 (Martin, 2011). The trimmed reads were processed into a variant call format (VCF) file following the guidelines of the GB-eaSy pipeline (Wickland et al., 2017). The paired-end reads were aligned to the UCD10X genome (Hulse-Kemp et al., 2018) in parallel (Tange, 2018) using the Burrows Wheeler Aligner (BWA) (Li and Durbin, 2009). Packages BCFtools (Li, 2011) and VCFtools (Danecek et al., 2011) were used to call SNPs and filter for read depth, respectively. Insertions and deletions were ignored, and Single Nucleotide Polymorphism (SNPs) were called and then filtered to leave only SNPs with a minimum average read depth of 10.
2.6 Imputation and phasing using TASSEL
Each population was loaded into TASSEL (Bradbury et al., 2007), filtered by table sites with site min count = 80 (markers must have information for at least 80 individuals to be kept, 89.9%–97.5% of the complete populations), and then filtered by table taxa with min proportion of sites present = 0.3 (taxa must have 30% or more of marker information). After the filtering, remaining marker sites with missing data were imputed using the Linkage Disequilibrium k-Nearest Neighbors Imputation (LD KNNi) option with nearest neighbors = 5. The resulting genotype table was then transformed into ABH format using the ABH genotype option in TASSEL, which phases offspring genotypes given the genotypes of the parents.
2.7 QTL mapping with R/qtl
Genotype information from the TASSEL generated ABH files were read into R and combined with their matching phenotype information using the readCross function from R/qtl (v1.60; Broman et al., 2003). A series of filtering steps through R/qtl were also done to each pepper cross object. Duplicate markers were dropped, markers were filtered for segregation distortion, and duplicate F2 members and F2 members with unusually high numbers of crossover events were dropped. The genetic maps were also created using R/qtl, following the guidelines of Broman (2012) with an exception: instead of using functions formLinkageGroups and orderMarkers to put markers together on linkage groups and order them according to recombination fractions, markers were separated into linkage groups (chromosomes) and ordered along chromosomes according to the marker position on the physical map using the reference genome UCD v1.0 (Hulse-Kemp et al., 2018). This information was readily available because the markers were aligned to a reference genome. If a map showed a gap on a chromosome larger than 40 cM, then the markers were identified using iplotMap from the package qtlcharts (v0.16; Broman, 2015), and markers nearby those gaps were dropped. For each trait, a multiple-QTL model was found using stepwiseqtl, with the maximum number of QTLs set to the number of peaks seen in the summary plot of the logarithm of the odds (LOD) scores across the genome. This model was fit to the data using fitqtl, which reported LOD scores, p-value of chi-squared test, and percent variance explained, for both the model and individual QTL comprising the model. Additive and dominant effects were estimated using the effectscan function from R/qtl, and locations of QTLs were visualized using LinkageMapView (v2.1.2; Ouellette et al., 2018).
2.8 Statistical analysis
Correlations between phenotypes from all F3 families were found through the R package hmisc (v5.1-0; Harrell, 2023) and plotted with the R package corrplot (v0.92; Wei et al., 2021) in RStudio version 4.0.3 (R Core Team, 2020). Heritability estimates were done for the F3 families of each population using genotypes and phenotypes through the R package sommer (v3.4.2; Covarrubias-Pazaran, 2016) in RStudio. Statistics from detected QTLs were reported by the stepwiseqtl function in R/qtl (v1.60; Broman et al., 2003).
3 Results
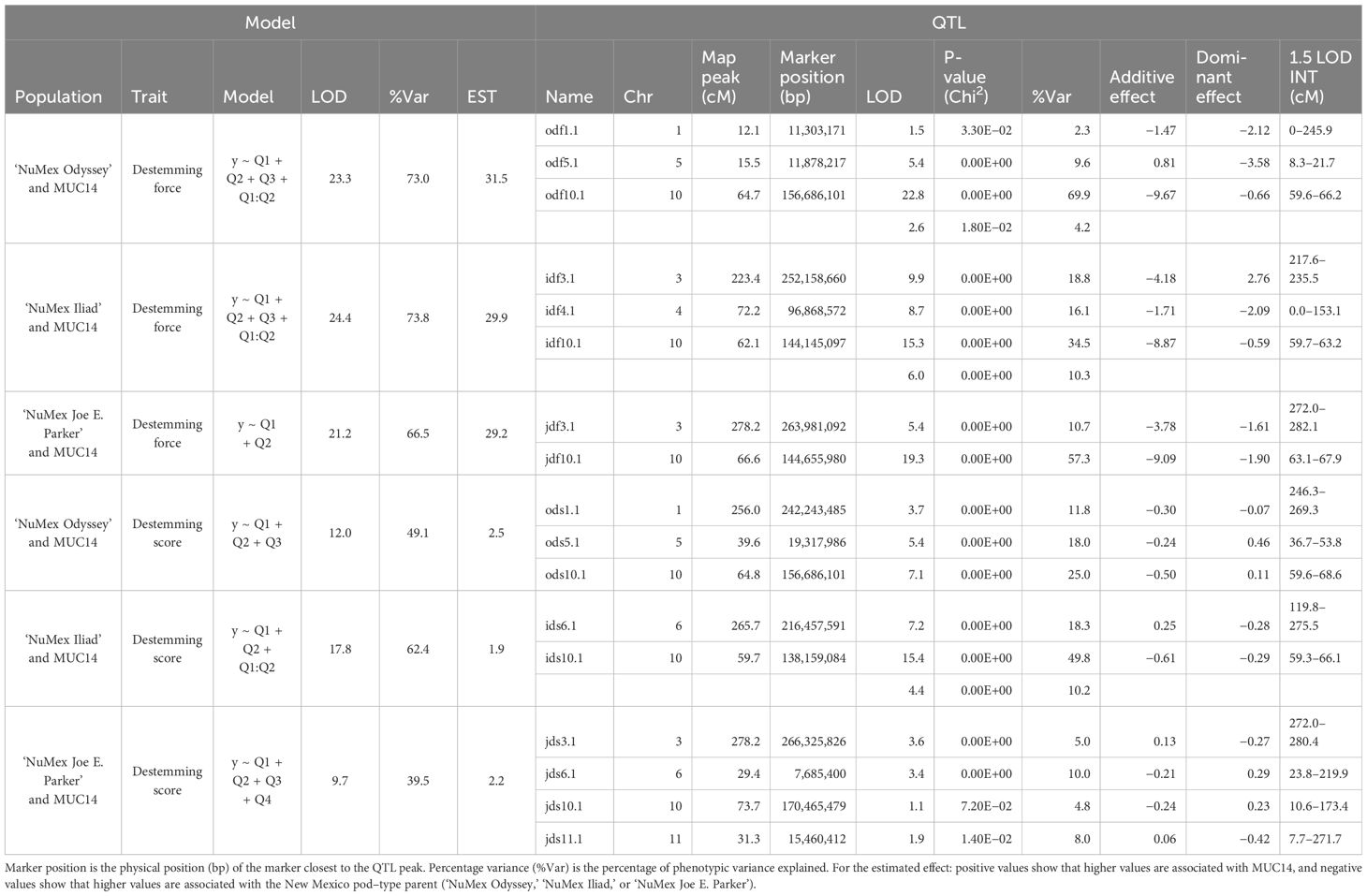
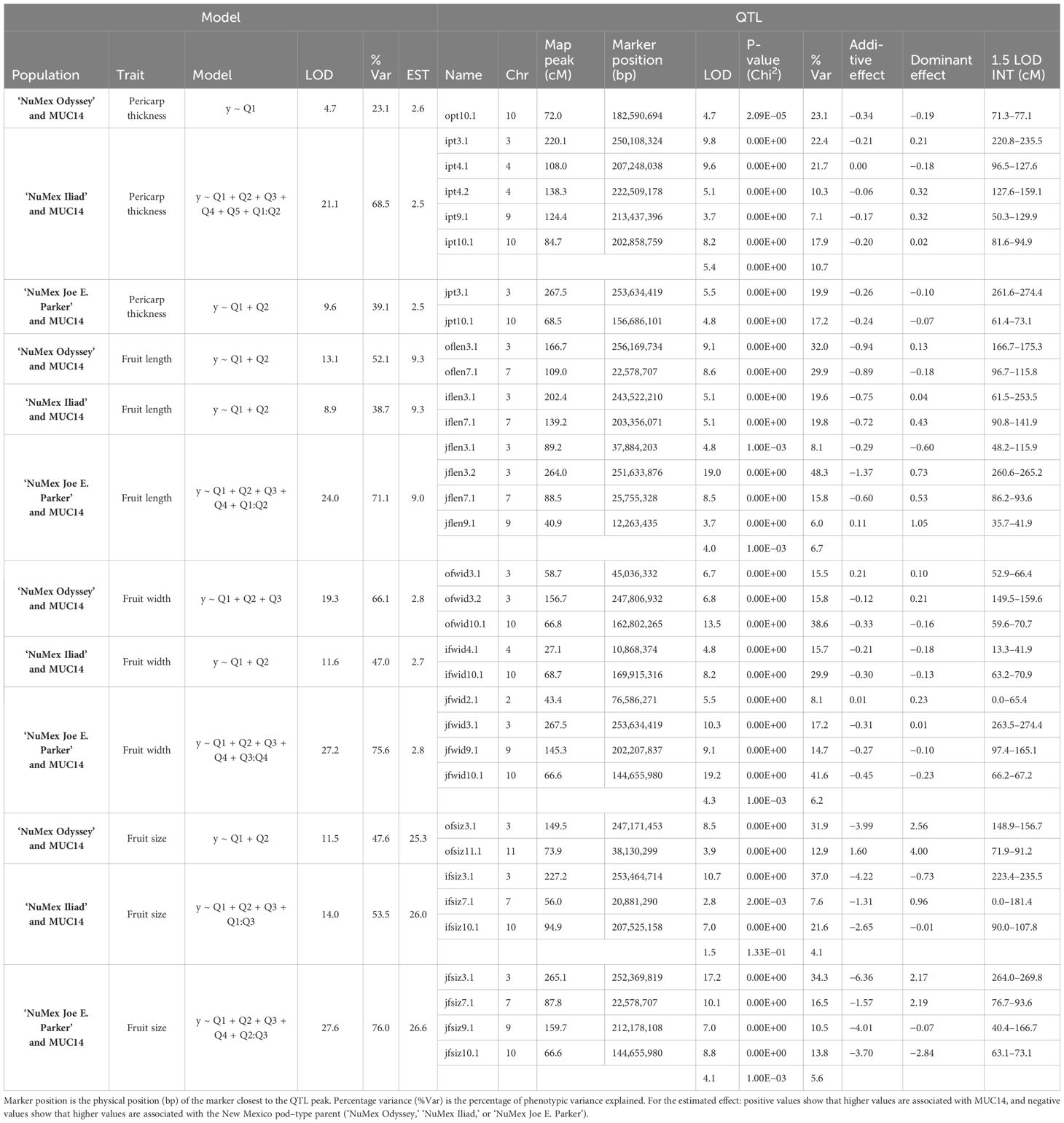
Three F2 populations were created from one of three New Mexico pod–type parents, and a common other parent: a novel easy destemming parent derived from UCD-14, MUC14. For brevity in this section, they are referred to as Odyssey for the population from ‘NuMex Odyssey’ and MUC14, Iliad for the population from ‘NuMex Iliad’ and MUC14, and Joe E. Parker for the population from ‘NuMex Joe E. Parker’ and MUC14. Each population was created from reciprocal crosses between the parents. The F2 members of each population were grown in a greenhouse, their tissue was collected, DNA was extracted, and genotyping by sequencing (GBS) was carried out. The F2:F3 families of all the populations were grown out, and their fruit collected and measured. With phenotypic means of the F3 families representing the F2 parent genotype (Table 1), QTL analysis in R/qtl was done for the measurements destemming score, destemming force, fruit size index, fruit length, fruit width, and pericarp thickness. A handful of QTLs were found for each population, with a few QTLs having consensus between populations.
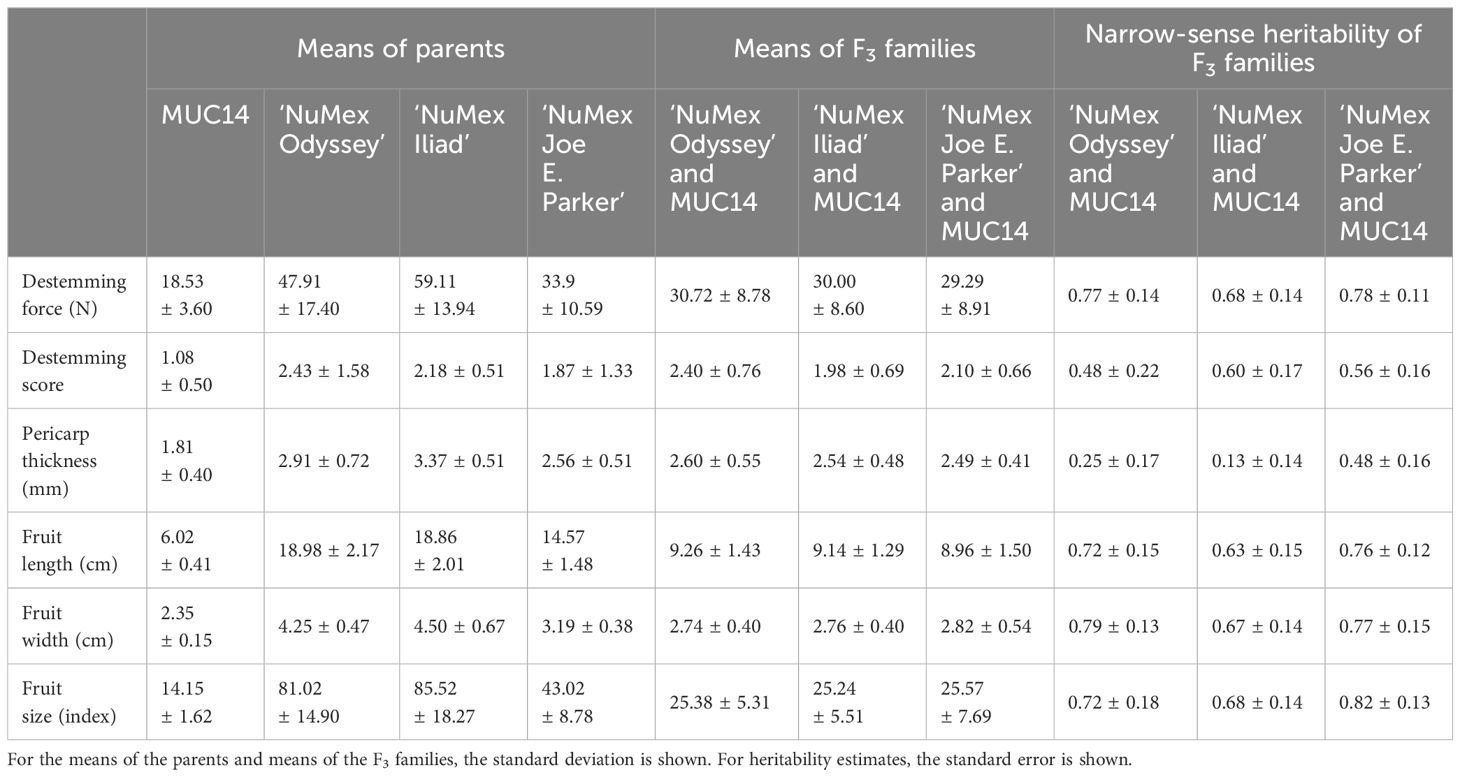
Table 1 Phenotypic means of parents and means and narrow-sense heritability estimates of F3 populations.
3.1 GBS results
For each population, 107 DNA samples from the F2 generation were sent for GBS. The average value of Q30 for the sequences of the populations were 90.5, 91.0, and 91.6 for the Odyssey, Iliad, and Joe E. Parker populations, respectively, showing good sequence quality that was fairly consistent overall. After creating the variant calling files and filtering for depth, the Odyssey population had 3,384,423 markers, the Iliad population had 2,790,049 markers, and the Joe E. Parker population had 4,903,549 markers. After the filtering steps in TASSEL and in R/qtl described previously, the total number of markers decreased to 1,464, 2,613, and 8,254, respectively. The number of F2:F3 individuals in the phenotypic analysis was also reduced due to some plants not surviving to adulthood. There were 82 individuals in the Odyssey population, 84 in the Iliad population, and 89 individuals used in the Joe E. Parker population.
3.2 Genetic map construction and QTL analysis in R/qtl
For the Odyssey population, the map was constructed, and linkage analysis was done with 82 individuals and 1,464 markers (Table 2). The average chromosome length was 191.0 cM, with the largest being chromosome 1 with 287.1 cM and with the smallest being chromosome 10 with 93.1 cM. The map of the Iliad population was constructed with 93 individuals, and QTL linkage analysis was done with 84 individuals and 2,613 markers. The average chromosome length was 181.1 cM, with the largest being chromosome 6 with 292.9 cM and with the smallest being chromosome 5 with 126.6 cM. For the Joe E. Parker population, the map was built, and linkage analysis was done with 89 individuals and 8,254 markers. The average chromosome length was 238.4 cM, with the largest being chromosome 1 at 345.5 cM and with the smallest being chromosome 5 with 140.2 cM. All three populations had 12 linkage groups corresponding to the 12 chromosomes of Capsicum annuum (Table 2).
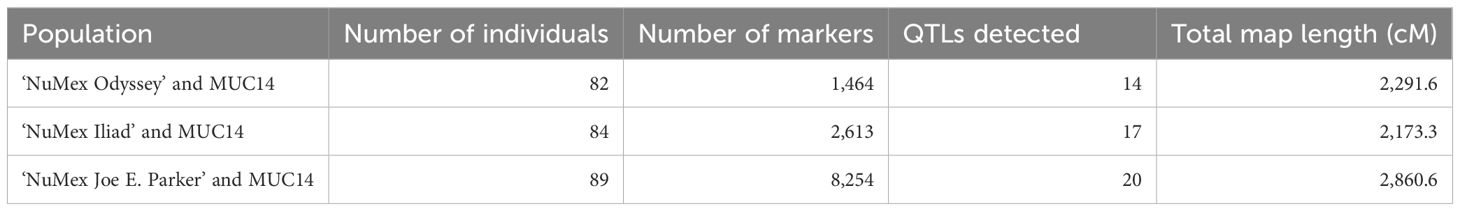
Table 2 Number of individuals and markers in each population used for QTL analyses and number of QTLs discovered.
For all populations, QTL analysis was done on six traits. In total, there were five measurements collected, which all relate to traits that are important for breeding peppers compatible with mechanical harvesting. Two of these were measurements related specifically to the removal of the stem, and the other three were related to the size of the fruit. Another measurement, a fruit size index, was created by multiplying fruit width by fruit length. A variety of fruit sizes was seen among F3 families (Figure 1).

Figure 1 Fruit shape diversity seen in a selection of fruits from F3 families that came from all three populations created from MUC14 and a New Mexico pod–type parent. From these, some common fruit shapes observed were (A) oblong (most resembling MUC14), (B) oblong with narrow diameter at calyx attachment, (C) elongated, (D) conical, (E) rectangular, and (F) triangular (most resembling NM pod-type pepper) (USDA, 2015).
For destemming traits, QTLs were found. For destemming force, all three populations had major QTLs on chromosome 10 and two of the populations, Iliad and Joe E. Parker, identified additional major QTLs (PVE > 10%) (Collard et al., 2005) on chromosome 3 (Table 3). Similarly, for all three populations, QTLs on chromosome 10 were found for destemming score. In the Odyssey population, the marker at the peak of QTLs for destemming force and destemming score was the same. For the Odyssey and Iliad populations, QTLs on chromosome 5 were found for the destemming score phenotype. Other QTLs on chromosomes 1, 4, 6, 11, and 12 were identified for destemming score or force, but none were shared between populations except for the QTL on chromosome 6, which may be shared between the Iliad and Joe E. Parker populations (Table 3).
For chile fruit traits, QTLs were found. For pericarp thickness, QTLs on chromosome 10 were found for all three populations nearby QTLs for destemming force and score. The Iliad and Joe E. Parker populations also shared major QTLs for pericarp thickness on chromosome 3. Three QTLs, two on chromosome 4 and one on chromosome 9 were found just in the Iliad population. For fruit size, QTLs were found in all populations on chromosomes 3 and Iliad and Joe E. Parker populations had QTLs on chromosomes 7, and 10. For fruit length, major QTLs on chromosomes 3 and 7 were found for all three populations. Fruit size and length QTLs were found on chromosome 7. These showed up at two positions: around 20 Mbp and 200 Mbp. The Odyssey population had a QTL around 20 Mbp for fruit length, whereas the population from Iliad had a QTL around 200 Mbp for fruit length and around 20 Mbp for fruit size. In the population from Joe E. Parker, a QTL at 20 Mbp was found for both fruit length and fruit size. An additional minor QTL on chromosome 9 was found only in the Joe E. Parker population. For fruit width, QTLs on chromosomes 10 and 3 were found for all populations with some population specific QTLs showing up on chromosomes 2, 4, and 9 (Table 4).
Several QTLs for different traits were found nearby one another on the chromosomes. Fruit length, width, size, pericarp thickness, destemming score, and destemming force QTL peaks were seen around the same area on chromosome 3 (Figure 2). Markers closest to QTL peaks around this area on chromosome 3 were clustered around 243–263 Mbp. For the Iliad and Joe E. Parker populations, size QTL peaks were around 20 Mbp on chromosome 7. Similarly, length QTL peaks for the Odyssey and Joe E. Parker populations are seen close together around 20 Mbp on chromosome 7. Width and size QTL peaks from the Joe E. Parker population and pericarp thickness from the Iliad population were clustered around 213 Mbp on chromosome 9. Strikingly, QTLs for destemming force, destemming score, fruit width, fruit size, and pericarp thickness were scattered along the lower half of chromosome 10 (Figure 2). When the phenotype was plotted by genotype, the general pattern seen was that the more alleles at QTLs on chromosomes 3 and 10 an F2 individual had from MUC14, the lesser destemming force was and the less wide the fruit was in the F3 family (Figure 3 and Figure 4).

Figure 2 Physical map of QTLs reported in this study from the three biparental populations ‘NuMex Iliad’ and MUC14 (red), ‘NuMex Odyssey’ and MUC14 (blue), and ‘NuMex Joe E. Parker’ and MUC14 (yellow). QTLs previously reported from Hill et al. (2023) (purple), and genes of interest related to fruit size or abscission (black) are also shown. The location of the QTL is plotted approximately in the center of its confidence interval. Genes are plotted at their starting location. Chromosome length is according to the UCD v1.0 genome. Chromosomes on which no QTLs were found in this study were excluded (chromosomes 8 and 12).
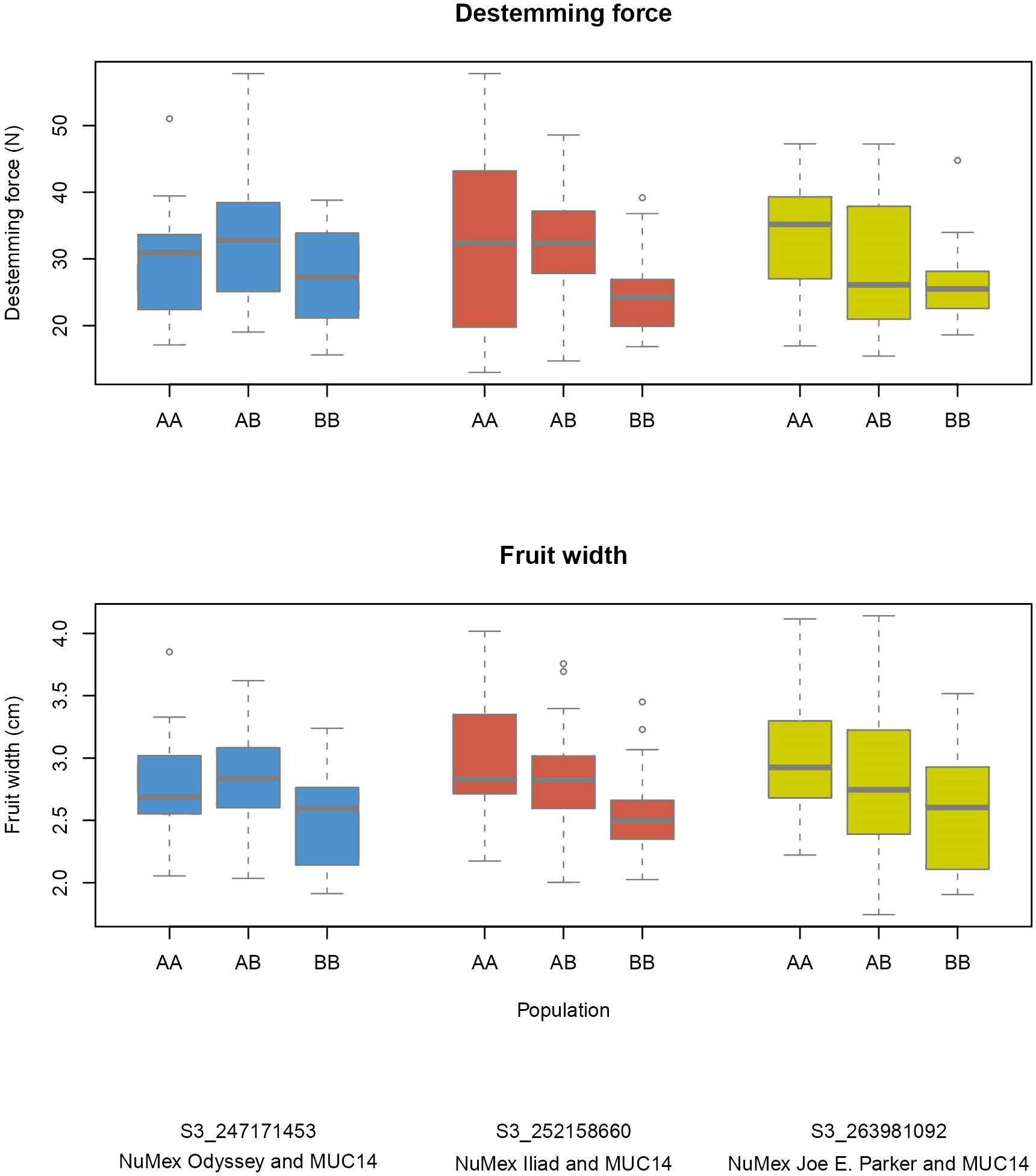
Figure 3 Phenotype by genotype boxplots for QTLs of destemming force and fruit width on chromosome 3. Labeled as AA is the genotype of the New Mexico pod–type parent (NuMex Odyssey, NuMex Iliad, and NuMex Joe E. Parker), and BB is the genotype of MUC14.
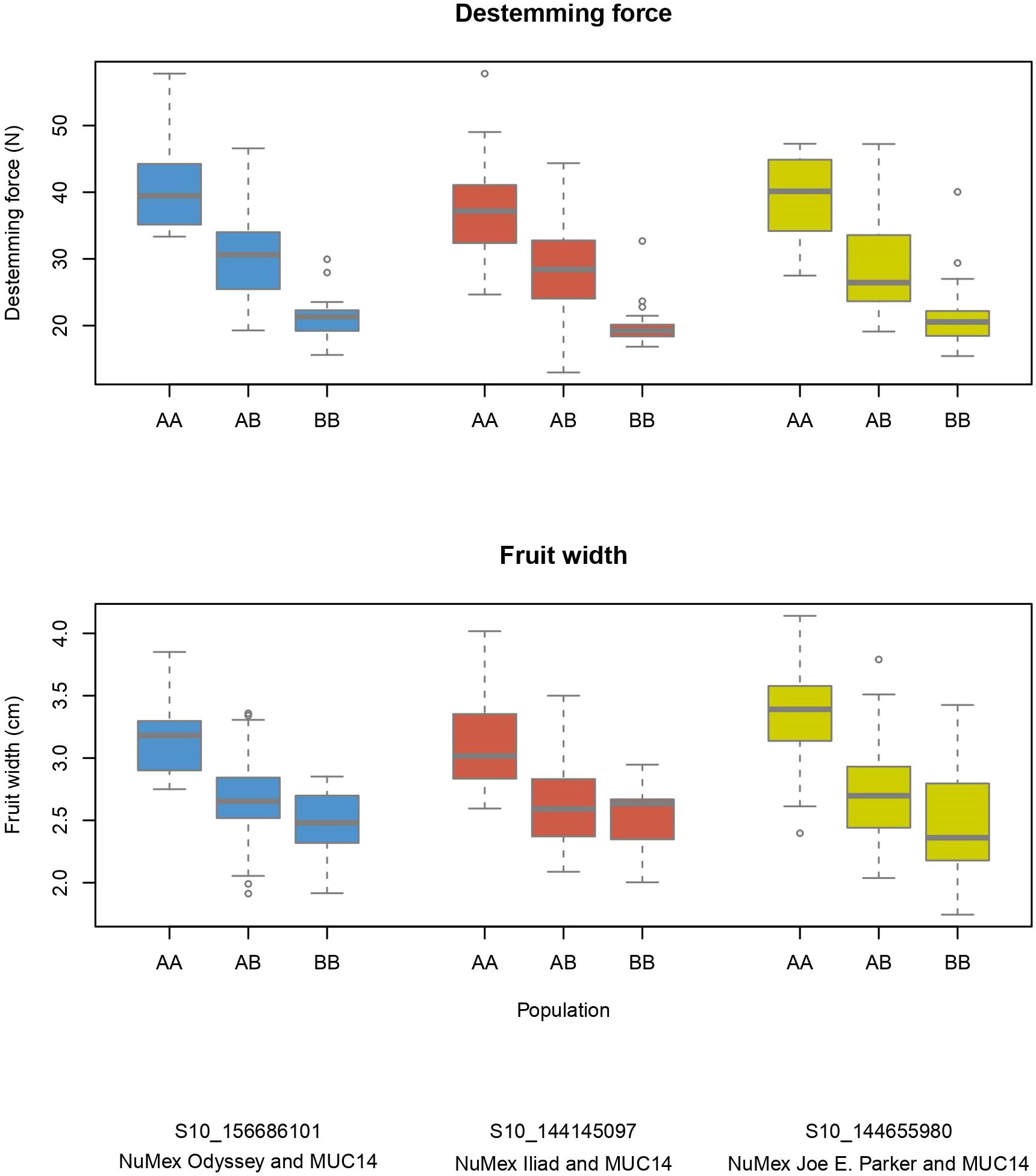
Figure 4 Phenotype by genotype boxplots for QTLs of destemming force and fruit width on chromosome 10. Labeled as AA is the genotype of the New Mexico pod–type parent (NuMex Odyssey, NuMex Iliad, and NuMex Joe E. Parker), and BB is the genotype of MUC14.
3.3 Fruit trait correlations using hmisc
Pearson’s correlation coefficients and their p-values were found between each pair of phenotypes. The phenotype pair with the strongest relationship, which was also significant was between destemming force and fruit width (r = 0.75, p = 0.00e+00), followed by pericarp thickness and destemming force (r = 0.56, p = 0.00e+00) and pericarp thickness and fruit width (r = 0.52, p = 0.00e+00). Pairs with a moderately positive relationship were between destemming score and destemming force (r = 0.37, p = 2.09e−10), destemming score and fruit width (r = 0.24, p = 4.69e−05), and fruit width and fruit length (r = 0.17, p = 3.39e−03). One pair, fruit length and destemming score, had a negative relationship (r = −0.21, p = 3.04e−04). The pairs fruit length and destemming force, fruit length and pericarp thickness, and destemming score and pericarp thickness did not have significant correlation coefficients (p-values are 5.24e−01, 2.59e−01, and 3.81e−01, respectively) (Figure 5).
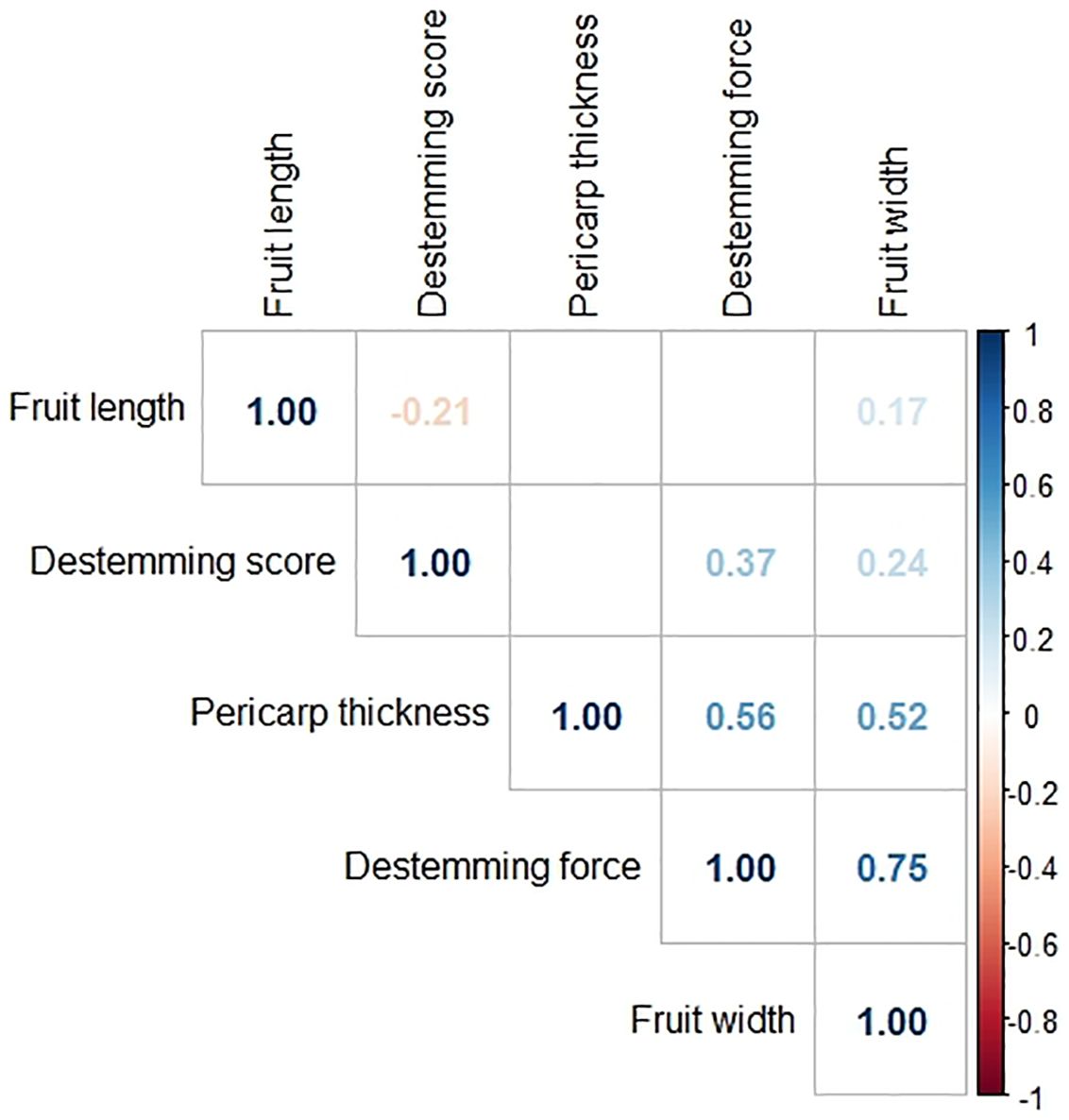
Figure 5 Correlation matrix among phenotypes of the F3 family means for all populations. Pearson’s correlation coefficients are shown for trait pairs with significant correlations (p-value threshold=0.05).
4 Discussion
Implementation of mechanical harvesting for green chile is a goal of the industry in the United States. Crucial for this is the development of varieties with traits that facilitate the process of mechanical harvesting. One such important trait is easy destemming: because the green chile pepper’s woody pedicel is inedible, it is usually removed by hand in the field. For mechanical harvesting, varieties in which pedicels can be removed completely with little force applied would be ideal. This study sought to trace the genetic source of a novel easy destemming trait in crosses between a semi-domesticated UCD-14 derived pepper, MUC14, and several New Mexico pod–type peppers. All three biparental F2 populations were genotyped using GBS, F3 families were grown, and destemming and fruit measurements were taken from harvested F3 fruit. For the three populations studied, MUC14 was crossed to ‘NuMex Odyssey,’ ‘NuMex Iliad,’ and ‘NuMex Joe E. Parker,’ and QTL analyses were performed, discovering 14, 17, and 20 QTLs, respectively, for six traits. For several of these QTLs, consensus between populations, as well as overlap between QTLs for different traits, was observed.
One other study has searched for destemming force QTLs in chile pepper. Studying populations that came from the easy destemming Mexican landrace UCD-14, Hill et al. (2023) reported destemming force QTLs on chromosomes 2, 3, 4, 9, 10, and 11. In the present study, QTLs were found close to theirs on chromosomes 3, 4, and 10. In this study on chromosome 3, fruit length, fruit width, fruit size, pericarp thickness, destemming force, and destemming score QTLs were detected nearby where they found pull force and force QTLs (Figure 2). In this region, they point out a candidate gene, Mitogen-activated protein kinase kinase 5 (MKK5), that is involved in the abscission signal transduction pathway in Arabidopsis thaliana (Cho et al., 2008). This is notable as they suggest easy destemming of UCD-14 could come from the activation of an abscission zone at the pedicel/fruit boundary. On one end of chromosome 4, a pericarp thickness QTL in this study and destemming force and rate QTLs in theirs were found nearby one another. On the other end of chromosome 4, a fruit width QTL in this study and a destemming rate QTL in theirs were nearby one another. These are close to the genes CST and SERK2, which are also related to abscission (Burr et al., 2011; Meng et al., 2016) (Figure 2). Finally, on chromosome 10, we found destemming force, destemming score, fruit width, fruit size, and pericarp thickness QTLs nearby where they found destemming force, rate, and pull rate QTLs (Figure 2).
In addition to destemming traits, this study measured fruit size. In pepper, there are many reports of QTLs for fruit size traits being found clustered together on chromosome 3 (Hill et al., 2017; Lozada et al., 2021). In an early QTL study that covered fruit traits in pepper, fruit diameter, fruit length, fruit shape, and pericarp thickness were all found to have QTLs in an overlapping region on chromosome 3 (Chaim et al., 2001). For an interspecific cross between C. annuum and C. frutescens, QTLs for the fruit traits maximum width, maximum height, width mid area, and fruit area, were found together in a region on their map between 54.8 cM and 100 cM on chromosome 3 (Yarnes et al., 2013). A double haploid C. annuum study found QTLs for fruit length, fruit diameter, and fruit shape at the same marker on chromosome 3 (Mimura et al., 2012). For a C. annuum/C. annuum cross, QTLs for fruit traits—fruit length, fruit diameter, and fruit shape—were found concentrated on chromosome 3, mostly in the range between 107 Mbp and 252 Mbp (Han et al., 2016). Similarly, fruit longitude and fruit diameter in a population derived from the blocky sweet pepper Maor and pungent disease resistant CM334 (both C. annuum) found QTLs for these traits within a region on chromosome 3 between 227 Mbp and 247 Mbp in the CM334 genome (Chunthawodtiporn et al., 2018). These two latter studies both have a physical range on the genome where regions of QTLs on chromosome 3 from this study overlap (Supplementary Table S1).
A couple of genes have been proposed to underlie the previously discovered fruit size QTLs on pepper chromosome 3. A SNP marker at 183,386,147 bp in the CM334 genome was found within a gene, Longifolia 1-like, that was homologous to lng1 (LONGIFOLIA1) and lng2 (LONGIFOLIA2), genes that encode TONNEAU1-recruiting motif (TRM) proteins, which are known to promote cell elongation in the model organism Arabidopsis thaliana (Zhang et al., 2020). In pepper, this location was related to fruit shape, determining whether the fruit is more circular or oval shaped (Colonna et al., 2019). Although our chromosome 3 QTLs do not overlap with this region, the predicted gene Longifolia 2 (LOC107862335) sitting at 252,383,854–252,389,858 bp in the UCD v1.0 genome, is nearby our QTLs. Another gene proposed to regulate cell division is CaARF2-i2, which is on chromosome 3 at 244,772,375–250,606,405 bp in the CM334 genome (Hill et al., 2017).
There is also precedence for fruit size QTLs in pepper on chromosome 10. In one study, fruit shape, fruit diameter, and pericarp thickness QTLs were all reported around a QTL called fs10.1 on chromosome 10 responsible for fruit shape (Chaim et al., 2001, Chaim et al., 2003). Recently, a multi-environment GWAS done for a C. annuum collection found FShI-P10.1, a fruit shape index QTL, and FLe-P10.1, a fruit length QTL, on chromosome 10 in all locations that they studied (McLeod et al., 2023). For fruit size traits measured through tomato analyzer—transverse perimeter and transverse area—QTLs were found on chromosome 10 for RILs from a cross between two C. annuum parents (Chunthawodtiporn, et al., 2018). Fruit width and diameter QTLs on chromosome 10 were also reported by Eggink et al. (2013) and Mimura et al. (2012). All of these, except for Chaim et al. (2003), reported their QTL for this area as accounting for about 10% of variance for their respective trait. In this study, when fruit size is broken down into its components, fruit length and width, QTLs on chromosome 10 are only found for fruit width in all three populations, accounting for a much greater percent variance explained (29%–47%). This is more similar to results from Chaim et al. (2003) who reported percent variance explained for fruit shape of this QTL to be 30%–44% (Supplementary Table S2).
An early mutagenesis study hinted that the gene underlying fs10.1 was likely creating variation in overall fruit size through a change in cell shape (Borovsky and Paran, 2011). Later, this research group identified through bulked segregant analysis using RNA sequencing of F2 ovary tissue that the gene underlying fs10.1 is an ortholog of the tomato suppressor of ovate family proteins (OFPs), SlOFP20, in pepper named CaOFP20 (Borovsky et al., 2022). When CaOFP20 was silenced, knockdown fruits appeared elongated compared to control fruits, suggesting that CaOFP20 acts to repress fruit elongation. OFPs have been implicated in fruit shape in land plants like Arabidopsis (Wang et al., 2011) and tomato (Liu et al., 2002) and have been identified in shaping pepper fruit size/weight before (Tsaballa et al., 2011). OFPs are thought to confer this effect in part though interactions with TRM proteins, which impacts the formation of the pre-prophase band during cell division (Snouffer et al., 2020). CaOFP20 (LOC107845902) is found on the UCD v1.0 genome on chromosome 10 at 147,584,627–147,586,245 bp. This is nearby major fruit width, destemming force, and destemming score related QTLs found close together on chromosome 10 found for all three populations in this study. Moreover, in a genome-wide analysis, Luo et al. (2022) searched for OFPs throughout the Capsicum annuum genomes of Zunla-1, CM334, and Chiltepin, finding 8, 5, and 7 OFP genes (respectively) residing on a distal end of chromosome 10. The OFPs “CaOFP15-21” in their study were found in tandem repeats on chromosome 10 and showed similar expression in different tissues. The redundancy of OFPs on chromosome 10 in pepper may explain the span of positions for fruit size–related QTLs in this region.
Breeding for easy destemming may be complicated by the connection between destemming force and fruit size. Fruit size and shape are a fundamental part of the appeal of pepper varieties, so selecting for traits that are closely linked to genetic sources of fruit variability can be a challenge (Naegele et al., 2014). In this study, significant positive correlations were seen between destemming force and the fruit traits pericarp thickness and fruit width (Figure 5). Additionally, the largest QTLs for destemming force were found at the two high-density QTL regions for fruit traits on chromosomes 3 and 10 (Figure 2). Plotting destemming force and fruit width measurements alongside one another at the markers closest to QTL peaks on chromosomes 3 and 10 displays this relationship, as similar patterns of changing phenotype according to genotype can be seen for both traits (Figure 3 and Figure 4). This could be the result of pleiotropy or linkage and may complicate breeding for easy destemming while maintaining a New Mexico pod–type.
5 Conclusion
Easy destemming in chile pepper appears to be intertwined with fruit size. In particular, the region on chromosome 10 between 138 Mbp and 211 Mbp holds QTLs, which account for a large percentage variance for both fruit width and destemming force. This same region is occupied by OFPs, a gene family that is known to mediate fruit shape in several species, including tomato, and the gene CaOFP20 in pepper is documented as having a dramatic effect on pepper fruit elongation when knocked down. It is thought that OFPs modify fruit shape through interactions with TRMs. Two out of three populations had destemming force QTLs, and all three populations had fruit size QTLs, on chromosome 3 around 247–264 Mbp. In this region, there is a TRM called Longifolia 2, a homolog of a gene in Arabidopsis that mediates cell elongation. These areas on chromosomes 3 and 10 have been implicated in a separate destemming force study in California.
It is possible that genes influencing fruit shape are nearby other genes that are contributing to the easy destemming trait. For example, in the region of interest on chromosome 3, there is a homolog of a gene (EVR), which can contribute to floral organ abscission in Arabidopsis (Leslie et al., 2010). At this point, it is not clear whether the fruit shape genes implicated in this study are solely responsible for easy destemming or if it is connected to contributing genes through linkage. Either way, future breeding efforts toward easy destemming in pepper will require consideration of fruit size and shape.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ebi.ac.uk/eva/, PRJEB70773.
Author contributions
FO: Data curation, Formal analysis, Investigation, Visualization, Writing – original draft, Writing – review & editing. TH: Conceptualization, Funding acquisition, Writing – review & editing. AD: Conceptualization, Funding acquisition, Writing – review & editing. AG-L: Investigation, Writing – review & editing. SW: Funding acquisition, Resources, Supervision, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was funded by USDA/NIFA award 2021-67013-33942 and New Mexico State University Agriculture Experiment Stations.
Acknowledgments
The authors wish to acknowledge the NMSU Agriculture Experiment Stations, the NM Chile Association and NM Chile Commission for their support. The sequencing was carried out by the Genome and Biomedical Sciences Facility at UC Davis supported by NIH Shared Instrumentation Grant 1S10OD010786-01.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2024.1357986/full#supplementary-material
Supplementary Table 1 | Previously reported chile pepper fruit size QTLs on chromosome 3.
Supplementary Table 2 | Previously reported chile pepper fruit size QTLs on chromosome 10.
References
Borovsky, Y., Paran, I. (2011). Characterization of fs10. 1, a major QTL controlling fruit elongation in Capsicum. Theor. Appl. Genet. 123, 657–665. doi: 10.1007/s00122-011-1615-7
Borovsky, Y., Raz, A., Doron-Faigenboim, A., Zemach, H., Karavani, E., Paran, I. (2022). Pepper fruit elongation is controlled by Capsicum annuum ovate family protein 20. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.815589
Bosland, P. W., Walker, S. (2014). Growing Chiles in New Mexico (New Mexico State University Guide H-230).
Bradbury, P. J., Zhang, Z., Kroon, D. E., Casstevens, T. M., Ramdoss, Y., Buckler, E. S. (2007). TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics 23, 2633–2635. doi: 10.1093/bioinformatics/btm308
Broman, K. W. (2012). A brief tour of R/QTL. Available online at: https://rqtl.org/tutorials/rqtltour.pdf.
Broman, K. W. (2015). R/qtlcharts: Interactive graphics for quantitative trait locus mapping. Genetics 99, 359–361. doi: 10.1534/genetics.114.172742
Broman, K. W., Wu, H., Sen, Ś., Churchill, G. A. (2003). R/qtl: QTL mapping in experimental crosses. Bioinformatics 19, 889–890. doi: 10.1093/bioinformatics/btg112
Burr, C. A., Leslie, M. E., Orlowski, S. K., Chen, I., Wright, C. E., Daniels, M. J., et al. (2011). Cast away, a membrane-associated receptor-like kinase, inhibits organ abscission in Arabidopsis. Plant Physiol. 156, 1837–1850. doi: 10.1104/pp.111.175224
Chaim, A., Borovsky, Y., De Jong, W., Paran, I. (2003). Linkage of the A locus for the presence of anthocyanin and fs10. 1, a major fruit-shape QTL in pepper. Theor. Appl. Genet. 106, 889–894. doi: 10.1007/s00122-002-1132-9
Chaim, A. B., Paran, I., Grube, R. C., Jahn, M., Van Wijk, R., Peleman, J. (2001). QTL mapping of fruit-related traits in pepper (Capsicum annuum). Theor. Appl. Genet. 102, 1016. doi: 10.1007/s001220000461
Cho, S. K., Larue, C. T., Chevalier, D., Wang, H., Jinn, T.-L., Zhang, S., et al. (2008). Regulation of floral organ abscission in Arabidopsis thaliana. Proc. Natl. Acad. Sci. 105, 15629–15634. doi: 10.1073/pnas.0805539105
Chunthawodtiporn, J., Hill, T., Stoffel, K., Van Deynze, A. (2018). Quantitative trait loci controlling fruit size and other horticultural traits in bell pepper (Capsicum annuum). Plant Genome 11, 160125. doi: 10.3835/plantgenome2016.12.0125
Collard, B. C., Jahufer, M. Z., Brouwer, J. B., Pang, E. C. (2005). An introduction to markers, quantitative trait loci (QTL) mapping and marker-assisted selection for Crop Improvement: The basic concepts. Euphytica 142, 169–196. doi: 10.1007/s10681-005-1681-5
Colonna, V., D’Agostino, N., Garrison, E., Albrechtsen, A., Meisner, J., Facchiano, A., et al. (2019). Genomic diversity and novel genome-wide association with fruit morphology in Capsicum, from 746k polymorphic sites. Sci. Rep. 9, 1–14. doi: 10.1038/s41598-019-46136-5
Coon, D., Votava, E., Bosland, P. W. (2008). The Chile cultivars of New Mexico State University released from 1913 to 2008 (New Mexico State University Research Report 763).
Covarrubias-Pazaran, G. (2016). Genome-assisted prediction of quantitative traits using the R package sommer. PLoS One 11, e0156744. doi: 10.1371/journal.pone.0156744
Danecek, P., Auton, A., Abecasis, G., Albers, C. A., Banks, E., DePristo, M. A., et al. (2011). The variant call format and VCFtools. Bioinformatics 27, 2156–2158. doi: 10.1093/bioinformatics/btr330
Eggink, P. M., Tikunov, Y., Maliepaard, C., Haanstra, J. P., de Rooij, H., Vogelaar, A., et al. (2013). Capturing flavors from Capsicum baccatum by introgression in Sweet Pepper. Theor. Appl. Genet. 127, 373–390. doi: 10.1007/s00122-013-2225-3
Elshire, R. J., Glaubitz, J. C., Sun, Q., Poland, J. A., Kawamoto, K., Buckler, E. S., et al. (2011). A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species. PLoS One 6, e19379. doi: 10.1371/journal.pone.0019379
Funk, P. A., Walker, S. J. (2010). Evaluation of five green Chile cultivars utilizing five different harvest mechanisms. Appl. Eng. Agric. 26, 955–964. doi: 10.13031/2013.35906
Funk, P. A., Walker, S. J., Herbon, R. P. (2011). A systems approach to Chile harvest mechanization. Int. J. Veg. Sci. 17, 296–309. doi: 10.1080/19315260.2010.549167
Gandonou, J. M., Waliczek, T. M. (2013). An analysis of the recent trends in US Chile pepper production, consumption and imports. J. Food Agr. Environ. 11, 361–367. doi: 10.5555/20133112965
Han, K., Jeong, H.-J., Yang, H.-B., Kang, S.-M., Kwon, J.-K., Kim, S., et al. (2016). An ultra-high-density bin map facilitates high-throughput qtl mapping of horticultural traits in pepper (Capsicum annuum). DNA Res. 23, 81–91. doi: 10.1093/dnares/dsv038
Harrell, F. E. (2023). R HMISC package (Hmisc). Available at: https://hbiostat.org/r/hmisc/.
Havlik, C. (2021). Investigating field Protocols for Mechanized Harvest and Season Extension to Expand Chile (Capsicum annuum L.) Production in New Mexico. New Mexico State University, Las Cruces (NM.
Hawkes, J., Libbin, J. D., Jones, B. A. (2008). Chile production in New Mexico and northern Mexico. J. Am. Soc Farm Manage 83–92. doi: 10.22004/ag.econ.189883
Hill, T., Cassibba, V., Joukhadar, I., Tonnessen, B., Havlik, C., Ortega, F., et al. (2023). Genetics of destemming in pepper: A step towards mechanical harvesting. Front. Genet. 14. doi: 10.3389/fgene.2023.1114832
Hill, T. A., Chunthawodtiporn, J., Ashrafi, H., Stoffel, K., Weir, A., Van Deynze, A. (2017). Regions underlying population structure and the genomics of organ size determination in Capsicum annuum. Plant Genome 10. doi: 10.3835/plantgenome2017.03.0026
Hulse-Kemp, A. M., Maheshwari, S., Stoffel, K., Hill, T. A., Jaffe, D., Williams, S. R., et al. (2018). Reference Quality Assembly of the 3.5-GB genome of Capsicum annuum from a single linked-read library. Hortic. Res. 5. doi: 10.1038/s41438-017-0011-0
Joukhadar, I. S., Walker, S. J., Funk, P. A. (2018). Comparative mechanical harvest efficiency of six New Mexico pod–type green Chile pepper cultivars. Horttechnology 28, 310–318. doi: 10.21273/HORTTECH03999-18
Leslie, M. E., Lewis, M. W., Youn, J.-Y., Daniels, M. J., Liljegren, S. J. (2010). The Evershed receptor-like kinase modulates floral organ shedding inarabidopsis. Development 137, 467–476. doi: 10.1242/dev.041335
Li, H. (2011). A statistical framework for SNP Calling, mutation discovery, association mapping and Population Genetical Parameter Estimation from Sequencing Data. Bioinformatics 27, 2987–2993. doi: 10.1093/bioinformatics/btr509
Li, H., Durbin, R. (2009). Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25, 1754–1760. doi: 10.1093/bioinformatics/btp324
Lillywhite, J., Tso, S. (2021). Consumers within the spicy pepper supply chain. Agronomy 11, 2040. doi: 10.3390/agronomy11102040
Liu, J., Van Eck, J., Cong, B., Tanksley, S. D. (2002). A new class of regulatory genes underlying the cause of pear-shaped tomato fruit. Proc. Natl. Acad. Sci. 99, 13302–13306. doi: 10.1371/journal.pone.0023896
Lozada, D. N., Whelpley, M., Acuña-Galindo, A. (2021). Genetic architecture of Chile pepper (Capsicum spp.) QTLome revealed using Meta-QTL analysis. Horticulturae 7, 227. doi: 10.3390/horticulturae7080227
Luo, Y., Yang, S., Luo, X., Li, J., Li, T., Tang, X., et al. (2022). Genome-wide analysis of OFP gene family in pepper (Capsicum annuum L.). Front. Genet. 13. doi: 10.3389/fgene.2022.941954
Martin, M. (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 17, 10–12. doi: 10.14806/ej.17.1.200
McLeod, L., Barchi, L., Tumino, G., Tripodi, P., Salinier, J., Gros, C., et al. (2023). Multi-Environment association study highlights candidate genes for robust agronomic quantitative trait loci in a novel worldwide capsicum core collection. Plant J. 116, 1508–1528. doi: 10.1111/tpj.16425
Meng, X., Zhou, J., Tang, J., Li, B., de Oliveira, M. V. V., Chai, J., et al. (2016). Ligand-induced receptor-like kinase complex regulates floral organ abscission in Arabidopsis. Cell Rep. 14, 1330–1338. doi: 10.1016/j.celrep.2016.01.023
Mimura, Y., Inoue, T., Minamiyama, Y., Kubo, N. (2012). An SSR-based genetic map of pepper (Capsicum annuum L.) serves as an anchor for the alignment of major pepper maps. Breed. Sci. 62, 93–98. doi: 10.1270/jsbbs.62.93
Naegele, R. P., Ashrafi, H., Hill, T. A., Chin-Wo, S. R., Van Deynze, A. E., Hausbeck, M. K. (2014). QTL mapping of fruit rot resistance to the plant pathogen phytophthora capsici in a recombinant inbred line capsicum annuum population. Phytopathology® 104, 479–483. doi: 10.1094/PHYTO-05-13-0143-R
Ouellette, L. A., Reid, R. W., Blanchard, S. G., Brouwer, C. R. (2018). LinkageMapView—rendering high-resolution linkage and QTL maps. Bioinformatics 34, 306–307. doi: 10.1093/bioinformatics/btx576
R Core Team (2020). R: A language and environment for statistical computing. (Vienna, Austria:R Foundation for Statistical Computing). Available at: https://www.R-project.org/.
Snouffer, A., Kraus, C., van der Knaap, E. (2020). The shape of things to come: ovate family proteins regulate plant organ shape. Curr. Opin. Plant Biol. 53, 98–105. doi: 10.1016/j.pbi.2019.10.005
Tsaballa, A., Pasentsis, K., Darzentas, N., Tsaftaris, A. S. (2011). Multiple evidence for the role of an ovate-like gene in determining fruit shape in pepper. BMC Plant Biol. 11:1–14. doi: 10.1186/1471-2229-11-46
USDA (2015). ST 470-56 Objective description of variety pepper (Capsicum spp.) Exhibit C (Washington, DC: Plant Variety Protection Office).
USDA, NASS (2023). Vegetables Annual Summary. Available online at: https://downloads.usda.library.cornell.edu/usda-esmis/files/02870v86p/hq37x121v/4b29ck28c/vegean23.pdf.
Walker, S. J., Funk, P. A. (2014). Mechanizing Chile peppers: Challenges and advances in transitioning harvest of New Mexico’s signature crop. HortTechnology 24, 281–284. doi: 10.21273/HORTTECH.24.3.281
Walker, S. J., Funk, P., Joukhadar, I., Place, T., Havlik, C., Tonnessen, B. (2021). ‘NuMex Odyssey’, a New MEXICO–type Green CHILE pepper for mechanical harvest. HortScience 56, 1605–1607. doi: 10.21273/HORTSCI15793-21
Wall, M. M., Walker, S. J., Wall, A. D., Hughs, E., Phillips, R. (2003). Yield and quality of machine harvested red Chile peppers. HortTechnology 13, 296–302. doi: 10.21273/HORTTECH.13.2.0296
Wang, S., Chang, Y., Guo, J., Zeng, Q., Ellis, B. E., Chen, J. G. (2011). Arabidopsis ovate family proteins, a novel transcriptional repressor family, control multiple aspects of plant growth and development. PLoS One 6, e23896. doi: 10.1371/journal.pone.0023896
Wei, T., Simko, V., Levy, M., Xie, Y., Jin, Y., Zemla, J., et al. (2021). corrplot package. An introduction to corrplot package. Available online at: https://cran.r-project.org/web/packages/corrplot/vignettes/corrplot-intro.html.
Wickland, D. P., Battu, G., Hudson, K. A., Diers, B. W., Hudson, M. E. (2017). A comparison of genotyping-by-sequencing analysis methods on low-coverage crop datasets shows advantages of a new workflow, GB-eaSy. BMC Bioinform. 18, 1–12. doi: 10.1186/s12859-017-2000-6
Yarnes, S. C., Ashrafi, H., Reyes-Chin-Wo, S., Hill, T. A., Stoffel, K. M., Van Deynze, A. (2013). Identification of QTLs for capsaicinoids, fruit quality, and plant architecture-related traits in an interspecific Capsicum RIL population. Genome 56, 61–74. doi: 10.1139/gen-2012-0083
Keywords: Capsicum annuum, pepper, Chile pepper, destemming, fruit size, mechanical harvest
Citation: Ortega F, Hill T, Van Deynze A, Garcia-Llanos A and Walker S (2024) Identification of QTLs involved in destemming and fruit quality for mechanical harvesting of New Mexico pod–type green chile. Front. Plant Sci. 15:1357986. doi: 10.3389/fpls.2024.1357986
Received: 19 December 2023; Accepted: 12 June 2024;
Published: 01 July 2024.
Edited by:
Ahmed H. El-Sappah, Zagazig University, EgyptCopyright © 2024 Ortega, Hill, Van Deynze, Garcia-Llanos and Walker. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Stephanie Walker, c3dhbGtlckBubXN1LmVkdQ==; Franchesca Ortega, ZnJhbmNoZXNAbm1zdS5lZHU=
 Franchesca Ortega
Franchesca Ortega Theresa Hill
Theresa Hill Allen Van Deynze
Allen Van Deynze Armando Garcia-Llanos2
Armando Garcia-Llanos2 Stephanie Walker
Stephanie Walker