- 1Department of Molecular, Cellular and Developmental Biology, Faculty of Arts and Sciences, Yale University, New Haven, CT, United States
- 2Flowering Lab, School of Biological Sciences, University of Auckland, Auckland, New Zealand
- 3Mt Albert Research Centre, The New Zealand Institute for Plant and Food Research Limited, Auckland, New Zealand
Optimized flowering time is an important trait that ensures successful plant adaptation and crop productivity. SOC1-like genes encode MADS transcription factors, which are known to play important roles in flowering control in many plants. This includes the best-characterized eudicot model Arabidopsis thaliana (Arabidopsis), where SOC1 promotes flowering and functions as a floral integrator gene integrating signals from different flowering-time regulatory pathways. Medicago truncatula (Medicago) is a temperate reference legume with strong genomic and genetic resources used to study flowering pathways in legumes. Interestingly, despite responding to similar floral-inductive cues of extended cold (vernalization) followed by warm long days (VLD), such as in winter annual Arabidopsis, Medicago lacks FLC and CO which are key regulators of flowering in Arabidopsis. Unlike Arabidopsis with one SOC1 gene, multiple gene duplication events have given rise to three MtSOC1 paralogs within the Medicago genus in legumes: one Fabaceae group A SOC1 gene, MtSOC1a, and two tandemly repeated Fabaceae group B SOC1 genes, MtSOC1b and MtSOC1c. Previously, we showed that MtSOC1a has unique functions in floral promotion in Medicago. The Mtsoc1a Tnt1 retroelement insertion single mutant showed moderately delayed flowering in long- and short-day photoperiods, with and without prior vernalization, compared to the wild-type. In contrast, Mtsoc1b Tnt1 single mutants did not have altered flowering time or flower development, indicating that it was redundant in an otherwise wild-type background. Here, we describe the generation of Mtsoc1a Mtsoc1b Mtsoc1c triple mutant lines using CRISPR-Cas9 gene editing. We studied two independent triple mutant lines that segregated plants that did not flower and were bushy under floral inductive VLD. Genotyping indicated that these non-flowering plants were homozygous for the predicted strong mutant alleles of the three MtSOC1 genes. Gene expression analyses using RNA-seq and RT-qPCR indicated that these plants remained vegetative. Overall, the non-flowering triple mutants were dramatically different from the single Mtsoc1a mutant and the Arabidopsis soc1 mutant; implicating multiple MtSOC1 genes in critical overlapping roles in the transition to flowering in Medicago.
Introduction
Optimal timing of flowering is a major adaptive trait in plants and a key determinant of productivity in crops, including legumes (Graham and Vance, 2003; Jung and Muller, 2009; Tadege et al., 2015; Weller and Ortega, 2015; Foyer et al., 2016). Medicago truncatula (Medicago) is a temperate legume with powerful genetic and genomic resources for investigating the molecular pathways underlying flowering (Benlloch et al., 2006; Tadege et al., 2009; Young et al., 2011; Putterill et al., 2013; Meng et al., 2017; Cañas and Beltrán, 2018; Weller and Macknight, 2018; Jaudal et al., 2020a). Interestingly, Medicago and Arabidopsis thaliana (Arabidopsis), share similarities and striking differences in their flowering time regulation. Multiple signalling pathways control Arabidopsis flowering (Kim et al., 2009; Fornara et al., 2010; Srikanth and Schmid, 2011; Andrés and Coupland, 2012). Both winter annual Arabidopsis and Medicago are induced to flower by extended winter cold (vernalization, V) followed by warm, long day (LD) photoperiods (VLD) (Clarkson and Russell, 1975). However, Medicago lacks the well-known regulators of these processes, the repressor FLOWERING LOCUS C (FLC), and the activator CONSTANS (CO) (Putterill et al., 2013; Wong et al., 2014; Weller and Macknight, 2018; Jaudal et al., 2020a).
Nevertheless, progress has been made in identifying Medicago flowering-time regulators using forward and reverse genetics. These include the LD activators MtPHYTOCHROME A (Jaudal et al., 2020b), MtFE (Thomson et al., 2021), and MtPHYTOCHROMOBILIN SYNTHASE (Perez-Santangelo et al., 2022), and repressors MtCYCLING DOF FACTORS genes (Zhang et al., 2019). The polycomb repressive complex 2 component MtVERNALISATION2 represses Medicago flowering until after vernalization (Jaudal et al., 2016). MtINHIBITOR OF GROWTH 2 (MtING2) promoted flowering particularly in response to VLD (Jaudal et al., 2022). The floral integrator genes include the FLOWERING LOCUS T (FT)-like gene MtFTa1 (Laurie et al., 2011; Yeoh et al., 2013; Jaudal et al., 2020a; Zhang et al., 2021b) and the FLOWERING LOCUS D (FD)-like gene MtFDa. MtFDa interacts with MtFTa1 and stimulates the transition to flowering, as has been observed for Arabidopsis FT and FD (Srikanth and Schmid, 2011; Collani et al., 2019; Cheng et al., 2021; Zhang et al., 2021b; Zhu et al., 2021). Mtfta1 Mtfda double mutants have a bushy phenotype and fail to transition to flowering, indicating complementary critical functions for these genes in flowering time (Laurie et al., 2011; Cheng et al., 2021). This is unlike Arabidopsis, where the ft-10 twin sister of ft (tsf-1) fd-3 triple mutants showed only delayed flowering (Romera-Branchat et al., 2020). Other likely players include multiple paralogs of genes encoding MADS transcription factors, such as the three MtSUPPRESSOR OF OVEREXPRESSION OF CO1 (SOC1) (MtSOC1a, b, c), three MtFUL genes (MtFULa, b, c), three MtSHORT VEGETATIVE PHASE (MtSVP1, 2, c), and additional FT/TERMINAL FLOWER 1 (TFL1) genes including MtFTa2, MtFTb1, MtFTb2, and MtTFL1a, c (Laurie et al., 2011; Jaudal et al., 2014, 2015; Sussmilch et al., 2017; Cheng et al., 2018; Fudge et al., 2018; Jaudal et al., 2018; Thomson et al., 2019; Jaudal et al., 2022).
SOC1 functions as an important floral integrator gene in Arabidopsis, integrating signals from different flowering time pathways, including the LD photoperiod, gibberellin (GA), ambient temperature, and age pathways (Melzer et al., 2008; Gregis et al., 2009; Liu et al., 2009; Lee and Lee, 2010; Balanzà et al., 2014; Romera-Branchat et al., 2020). SOC1 binds to and controls the expression of numerous flowering regulators, including SVP, APETALA (AP2)-like repressor genes, and floral homeotic genes, and represses its own expression (Immink et al., 2012; Tao et al., 2012). SOC1 homologues regulate flowering or phenotypes, such as dormancy, in other plants (Lee and Lee, 2010; Immink et al., 2012; Tao et al., 2012; Voogd et al., 2015; Jaudal et al., 2018). Medicago has three MtSOC1 genes that encode proteins with ~66% amino acid identity with SOC1. These include the Fabaceae group A SOC1 gene, MtSOC1a, on chromosome 7, and two Fabaceae group B SOC1 genes, MtSOC1b and MtSOC1c, adjacent to each other on chromosome 8 (Fudge et al., 2018; Jaudal et al., 2018). The latter shares 93% amino acid identity and is ~66% identical to MtSOC1a. MtSOC1b and MtSOC1c duplications have only been observed in the genus Medicago, suggesting that it is relatively recent (Fudge et al., 2018; Jaudal et al., 2018). The three MtSOC1 genes partially complemented the delayed flowering of the Arabidopsis soc1 mutant (Fudge et al., 2018) and MtSOC1a overexpression strongly accelerated flowering in some wild-type (WT) Arabidopsis transgenic plants (Jaudal et al., 2018). In Medicago, the three MtSOC1 genes showed elevated expression in the shoot apex in response to floral inductive signals of VLD (Fudge et al., 2018; Jaudal et al., 2018). In addition, their expression was reduced in the late-flowering Mtfta1 mutant but elevated in transgenic plants overexpressing MtFTa1. This indicates that their expression relies partly on functional MtFTa1, as observed for the SOC1-mediated LD promotion of flowering in Arabidopsis (Fudge et al., 2018; Jaudal et al., 2018). Recently, a role for MtFDa in promoting MtSOC1 gene expression was reported, with the three MtSOC1 genes showing reduced expression in the Mtfda late-flowering mutant (Cheng et al., 2021, Zhang et al., 2021b).
In our previous study, we showed that MtSOC1a promotes flowering in Medicago, and that overexpression of MtSOC1a partially rescued the late flowering phenotype of the Mtsoc1a Tnt1 insertion mutant (Jaudal et al., 2018). However, Mtsoc1b Tnt1 insertion mutants had no flowering time phenotype, suggesting that MtSOC1b function may be redundant in regulating flowering in the WT (Fudge et al., 2018; Jaudal et al., 2018). Mtsoc1c Tnt1 mutants have not been previously identified; however, overexpression of MtSOC1c in WT R108 accelerated Medicago flowering under LD conditions, indicating that it is likely involved in floral induction (Fudge et al., 2018; Jaudal et al., 2018; Yuan et al., 2023).
To analyze the combined functions of the three MtSOC1 genes, we generated Mtsoc1a Mtsoc1b Mtsoc1c triple mutant lines using CRISPR/Cas9 gene editing. Strikingly, two independent triple mutant lines segregated plants that did not flower and were bushy, even after 5 months, compared with WT plants that flowered at ~3–4 weeks in VLD conditions. RNA-seq and RT-qPCR were carried out to analyze the molecular basis of these phenotypes and indicated that the non-flowering mutant plants remained vegetative, implicating a combined critical role for multiple MtSOC1 genes in promoting the transition to flowering.
Materials and methods
Plant materials, growth conditions, and phenotyping
The wild-type (WT) Medicago plants R108_C3 (R108) (Trinh et al., 1998) and the Tnt1 insertion Mtsoc1a (NF1705) single mutant in the R108 background have been previously reported (Jaudal et al., 2018). The gene-edited Mtsoc1a Mtsoc1b Mtsoc1c triple mutant lines, named Mtsoc1-1 and Mtsoc1-2, were generated in this study in the WT background, as described below. For the typical growth of Medicago plants under floral-inductive vernalized long-day conditions (VLD), seeds were first scarified between two pieces of sandpaper (grade P600), sterilized in a chlorine solution (Millipore, USA) for 5 min–10 min, and germinated overnight at 15°C in the dark. For the vernalization treatment, germinated seeds were placed on moist filter paper and stored at 4°C in the dark for 3 weeks. Germinated seeds with or without vernalization were planted in seed-raising mix (Daltons Limited, NZ) and placed on rockwool mats subirrigated with hydroponics (Gibeaut et al., 1997, without Na2O3Si). Seedlings were transplanted after 11–14 days to 2 L pots of soil mix consisting of nine parts of potting mix (Daltons Limited, NZ), three parts of vermiculite (Pacific Growers Supplies Limited, NZ), one part of number 2 sand (Daltons Limited, NZ). Plants were grown in controlled rooms under long days (LD, 16 h light/8 h dark) at 22°C with fluorescent lights at ~160 μmol m−2s−1. Plants were watered with tap water and hydroponic medium.
Flowering time was scored in days after planting and the number of nodes on the primary axis at the time the first floral bud was observed on the plant. The length of the primary shoot axis (main stem) was measured from the monofoliate leaf node to the uppermost shoot apical bud and the number of nodes was counted. The longest secondary axis that branched off from the primary shoot axis was also identified, and its length and number of nodes were measured. The flowering time and shoot axis measurements are shown as box plots. Statistical significance was determined using the Wilcoxon test (P-value with Bonferroni correction: *P ≤0.05, **P ≤0.01, ***P ≤0.001, ****P ≤0.0001).
General bioinformatics
Medicago gene sequences MtSOC1a (Medtr7g075870), MtSOC1b (Medtr8g033250), and MtSOC1c (Medtr8g033220) were obtained from the ‘Jemalong A17’ accession in the M. truncatula Genome Database (Mt4.0v2) http://blast.jcvi.org/Medicago-Blast/index.cgi) (Tang et al., 2014). The WT R108 genome assembly (v1.0, http://blast.jcvi.org/Medicago-Blast/index.cgi) (Moll et al., 2017) was used for the primer design and gene editing. Primers were designed using the Geneious software package (≥v8.0) (Kearse et al., 2012) using the Primer3 plugin (Untergasser et al., 2012).
Generation of CRISPR-Cas9 gene-edited Mtsoc1 triple mutant lines
Gene editing was used to generate the Mtsoc1a Mtsoc1b Mtsoc1c triple mutant lines. Two triple mutant lines were studied further and were named Mtsoc1-1 and Mtsoc1-2. Seven single guide RNAs (guides) were designed using the Geneious Prime software (version 2019.1.1) (Kearse et al., 2012) to target the coding sequences of MtSOC1a, MtSOC1b, and MtSOC1c in Medicago WT A17 (Table 1; Supplementary Table 1). Two guides (guide 1 and guide 2) targeted exons 5 and 7 of MtSOC1a, which encode part of the K-domain and C-terminal domain of MtSOC1a, respectively. Two guides (guides 3 and 4) targeted MtSOC1b in exons 3 and 7, which encode a part of the K-domain and C-terminal domain of MtSOC1b, respectively. Four guides (guide 3, guide 5, guide 6, and guide 7) targeted exons 3, 4, 5, and 7 of MtSOC1c, respectively. These exons encode part of the K-domain (guides 3, 5, and 6) or the C-terminal domain (guide 7) of MtSOC1c. The guide sequences are listed in Supplementary Table 1. The guides were identified based on N(17)VVR(NGG)H target selection criteria and activity scoring by Doench et al. (2014), and checked for the absence of off-target sites in the Medicago R108 genome (v1.0). One construct consisting of a polycistronic pre-tRNA-sgRNA-scaffold cassette (Xie et al., 2015) placed downstream of the Medicago U6 promoter with all seven guides and HindIII restriction sites added on both ends was commercially synthesized by GenScript (USA). The construct was inserted into the pCBSG041 plasmid vector backbone with Cas9 driven by the constitutive CAMV 35S promoter, as previously described (Jaudal et al., 2022). The plasmid was transformed into Agrobacterium tumefaciens strain EHA105 to transform Medicago WT R108 leaf tissue. Independent T0 transformant plants were selected using phosphinothricin (PPT) and regenerated as described previously (Cosson et al., 2006; Jaudal et al., 2018). Young regenerant plants were also sprayed with the Basta herbicide (Bayer, Germany) to select transgenic plants. T0 mutant plants were self-crossed to produce segregating T1 and T2 progenies for analysis. MtSOC1a, MtSOC1b, and MtSOC1c were genotyped for gene editing by polymerase chain reaction (PCR) using the primers listed in Supplementary Table 1. The identity of the edits in MtSOC1 genes was confirmed by DNA sequencing of the PCR products (Macrogen, South Korea).
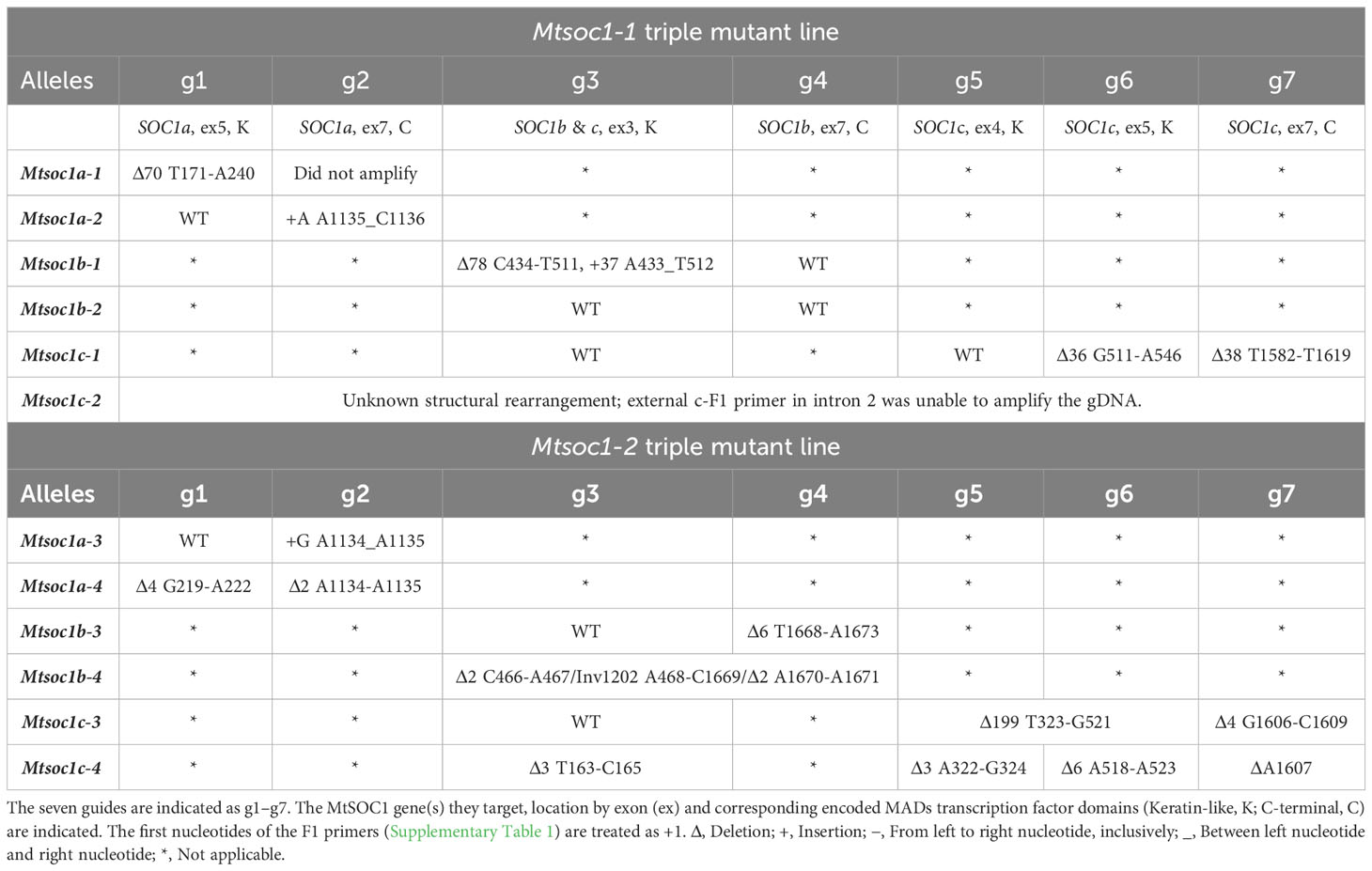
Table 1 Summary of CRISPR-Cas9 gene edits in Mtsoc1a, Mtsoc1b, and Mtsoc1c alleles segregating in Mtsoc1-1 and Mtsoc1-2 triple mutant lines.
Plant tissue harvesting and RNA extraction
For RNA-seq of a single Mtsoc1a mutant, apex samples were harvested from 15-day old Mtsoc1a and WT plants. The upper portion of three primary stems and their shoot apices, from three plants were pooled to make up one biological replicate, with three biological replicates harvested for each genotype. For real time reverse transcription quantitative PCR (RT-qPCR) and RNA-seq on the Mtsoc1a Mtsoc1b Mtsoc1c triple mutant line Mtsoc1-2, shoot apices from non-flowering Mtsoc1-2 plants and WT plants were harvested. Three primary shoot apices from three Mtsoc1-2 plants were harvested at 83 days (after phenotyping) and pooled to make up one biological replicate, with three biological replicates harvested. WT shoot apex samples for RT-qPCR comparisons with Mtsoc1-2 plants were harvested on day 14. Three biological replicates were harvested, each consisting of three primary apices. Shoot apices (apex) (15 mg–100 mg), were harvested from plants grown under VLD conditions at zeitgeber time 4 (4 h after dawn). Tissues were snap-frozen with metal beads and homogenized using a Geno/Grinder (New Jersey, USA) into powder in liquid nitrogen. Total RNA was extracted using an RNeasy Plant Mini Kit (Qiagen, Germany) following the manufacturer’s protocol. The quantity and quality of the RNA were checked using a Bioanalyzer 2100 (Agilent Technologies, USA).
Gene expression analysis by RT-qPCR
The WT and Mtsoc1-2 triple mutant apex RNA samples were treated with DNase (TURBO DNA-free Kit; Invitrogen) before being subjected to cDNA synthesis and RT-qPCR, as previously described (Laurie et al., 2011; Zhang et al., 2019). Primer sequences used for RT-qPCR are listed in Supplementary Table 1. Relative gene expression was calculated based on the comparative Ct method (Livak and Schmittgen, 2001), with modifications (Bookout and Mangelsdorf, 2003), using the formula 2−ΔCT, where ΔCT was obtained by normalizing the gene of interest to the reference gene, PROTEIN PHOSPHATASE 2A (PP2A, Medtr6g084690). Statistical significance was calculated using the t-test, assuming unequal variance (p ≤0.05).
RNA-seq and bioinformatic analysis
Shoot apex RNA samples of the Mtsoc1a single mutant and WT were sent to Novogene (Hong Kong) and six strand-specific mRNA libraries with polyA enrichment were prepared and subjected to Illumina NovaSeq 6000, 150 bp paired-end sequencing. Similarly, three strand-specific mRNA libraries with polyA enrichment were prepared from the shoot apex RNA of the non-flowering plants of the Mtsoc1-2 mutant line. The FASTQ file read quality was evaluated, and the Fastp version (0.21) was used for trimming (Chen et al., 2018). When quality was below the PHRED score of 20, reads were trimmed from the 3’end, and reads <36bp in length were excluded. The remaining reads were mapped against the Mt4.0v2 transcriptome (Young et al., 2011; Tang et al., 2014) using Salmon (v0.8.2) (Patro et al., 2017). The resulting count tables were imported into R (R Core Team, 2018) using the tximport package (v1.12.0) (Soneson et al., 2015). DESeq2 (v1.24.0) (Love et al., 2014) was used for normalization and differential expression analyses. Differentially expressed transcripts were filtered using a cut-off adjusted p-value ≤0.05 and log2 fold-change ≥1 or ≤−1. To identify candidate direct target genes of MtSOC1s, we used Blastx (Altschul et al., 1990) against Arabidopsis SOC1 bound and regulated genes (Immink et al., 2012). All the genes with an e-value <e−100 were selected, and we removed the genes that were not expressed or weakly expressed, which gave a list of 253 transcripts (Supplementary Table 4). Raw RNA-seq data were available from GEO (GSE247931).
Gene ontology enrichment analysis
ShinyGO v0.77 (Ge et al., 2020) was used for gene ontology enrichment analysis of the differentially expressed genes that are significantly over-represented in biological processes, cellular components, and molecular functions. Medicago genes (MedtrA17_4.0) and a false discovery rate p-value cut-off 0.05 was used.
Results
CRISPR-Cas9 gene editing to generate Mtsoc1a Mtsoc1b Mtsoc1c triple mutants
To investigate the combined function of the three MtSOC1 genes MtSOC1a, MtSOC1b, and MtSOC1c in Medicago development and flowering, we created a CRISPR-Cas9 gene editing vector with seven guides to generate triple mutant plants. These targeted the exons that encode a part of the MADS transcription factor conserved K domain or the variable C domain (Supplementary Table 1, Table 1) (Lai et al., 2021). Of the seven guides, two (guides 1 and 2) specifically targeted MtSOC1a, one (guide 3) targeted both MtSOC1b and MtSOC1c, one (guide 4) targeted MtSOC1b and three (guides 5–7) targeted MtSOC1c, (Supplementary Table 1, Table 1). WT R108 Medicago leaf explants were transformed using Agrobacterium-mediated gene transfer, and T0 transgenic plants were regenerated via somatic embryogenesis. The transformants were genotyped to identify plants with Mtsoc1a, Mtsoc1b, and Mtsoc1c mutations. This was followed by further analyses to segregate T1 and T2 progeny generations. Two independent Mtsoc1a Mtsoc1b Mtsoc1c triple mutant lines were analyzed. These lines were named Mtsoc1-1 and Mtsoc1-2, respectively (Table 1).
The Mtsoc1 triple mutant lines segregate plants that are non-flowering
The flowering time phenotypes of plants from the two triple mutant lines Mtsoc1-1 and Mtsoc1-2 were compared with those of WT plants under VLD. These lines segregated non-flowering plants (Figures 1A–D, 2A–C; Supplementary Figures 1A, B; Supplementary Table 2). Strikingly, the latter plants did not flower even when grown for 5 months or more, compared to WT plants that flowered much earlier at ~1 month of age. Mtsoc1-1 T1 plants segregated three plants with a non-flowering phenotype (Figures 1A, B, D). The remaining 17 siblings generally showed delayed flowering, ranging from 28 to 42 days, with increased node numbers on the primary axis (Figures 1A, B), similar to the Mtsoc1a Tnt1 single mutant (Jaudal et al., 2018). One plant with the non-flowering phenotype was again observed in a separate sowing of Mtsoc1-1 T1 plants, and five non-flowering plants were observed out of the 20 plants in the progeny T2 line tested (Figure 1C; Supplementary Figures 1A, B). Genotyping by PCR and DNA sequencing indicated 100% co-segregation between the non-flowering phenotype and homozygosity of the predicted strong deleterious mutant alleles Mtsoc1a-1, Mtsoc1b-1, and Mtsoc1c-1, with large deletions at one or more guide target sites in each gene (Table 1; Supplementary Figure 1; Supplementary Table 2). Genotyping and segregation analyses also indicated that the specific Mtsoc1b and Mtsoc1c alleles were inherited together, as predicted for genes adjacent to each other on the same chromosome. Thus, the Mtsoc1b-1 and Mtsoc1c-1 alleles were tightly linked, as were Mtsoc1b-2 (wild-type allele) and Mtsoc1c-2 (Table 1; Supplementary Table 2).
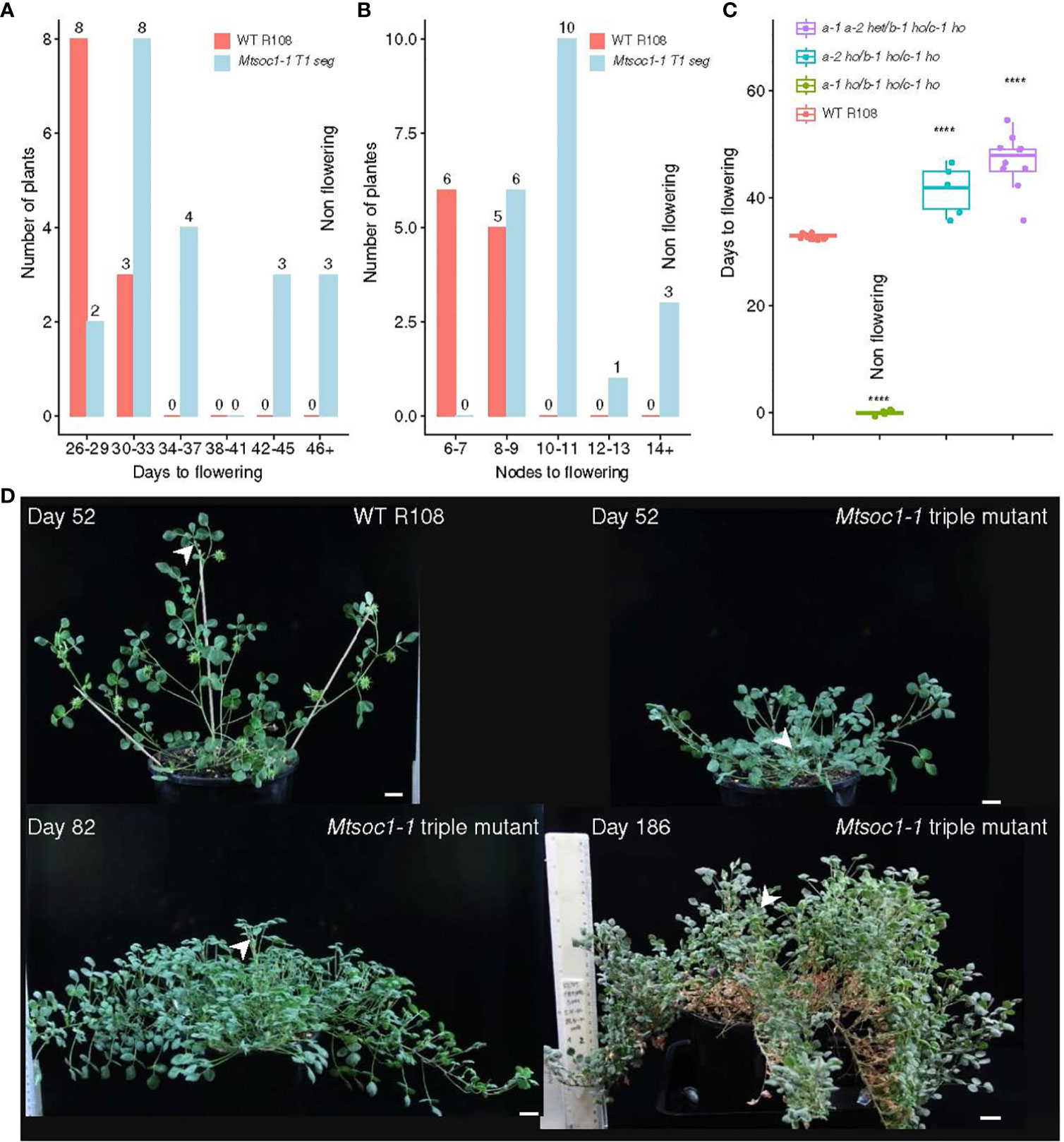
Figure 1 The Mtsoc1a Mtsoc1b Mtsoc1c triple mutant line Mtsoc1-1 segregates plants that do not flower. (A, B) Distribution graphs in days (A) and node numbers on the primary axis (B) of the first flower of WT R108 (WT) and Mtsoc1-1 T1 segregating line (R545) under VLD. The sample sizes are shown above each bar (C) Box plots showing the number of days to first flower of the WT and Mtsoc1-1 T2 segregating lines (R760 and R761) under VLD. The homozygous (ho) and heterozygous (het) genotypes are shown. Statistical significance was determined using the Wilcoxon test (P-value with Bonferroni correction: ****P ≤0.0001). (D) Photographs of WT and non-flowering Mtsoc1-1 triple mutants (R656-6, right top and left bottom, and R545-2, right bottom) under VLD. The white stealth arrows indicate the apex of the primary axis. Scale is 2 cm.
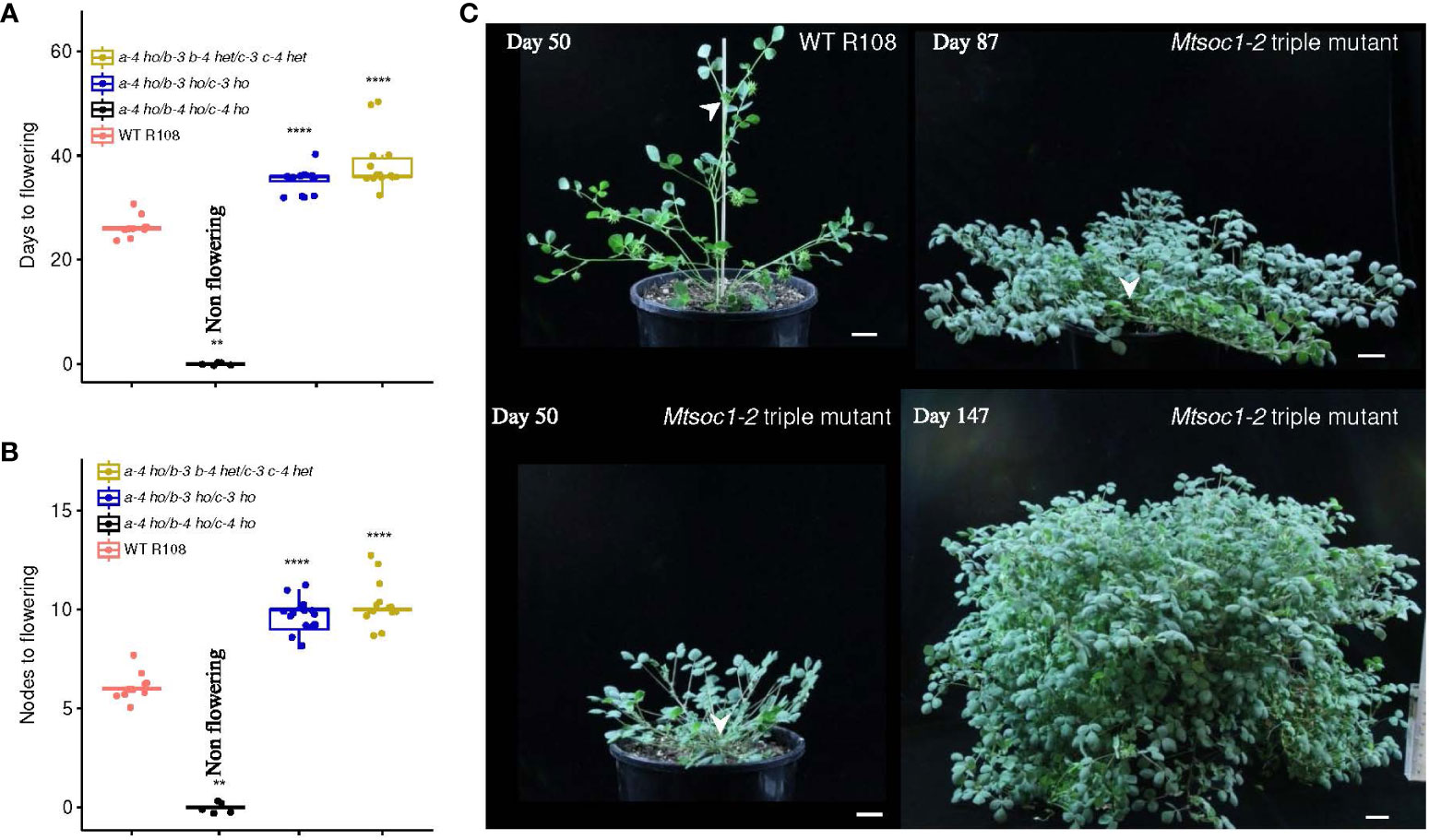
Figure 2 The triple mutant line Mtsoc1-2 segregates non-flowering plants. (A, B) Box plots showing the number of days (A) and node number on the primary axis (B) to the first flower of the WT and Mtsoc1-2 T2 segregating line (R647) under VLD. Homozygous (ho) and heterozygous (het) genotypes are shown. Statistical significance was determined using the Wilcoxon test (P-value with Bonferroni correction: ****P ≤0.0001). (C) Photographs of the WT and non-flowering Mtsoc1-2 triple mutant (R647-26) taken on days 50, 87, and 147 under VLD. White stealth arrows indicate the apices of the primary axis. Scale is 2 cm.
In the second triple mutant line, Mtsoc1-2, five non-flowering T2 plants were observed in a total of 35 plants grown (Figures 2A-C; Supplementary Table 2). Genotyping indicated that the non-flowering plants were homozygous for the predicted strongly deleterious mutant alleles Mtsoc1a-4, Mtsoc1b-4, and Mtsoc1c-4, with deletions at multiple guide targets and a large inversion in the case of Mtsoc1b-4 (Table 1). There was 100% co-segregation between the presence of homozygous Mtsoc1a-4, Mtsoc1b-4, and Mtsoc1c-4 alleles and the non-flowering phenotype (Figures 2A-C; Supplementary Table 2).
Non-flowering triple mutants remain vegetative with a bushy aerial architecture and a short primary axis
We also scored the aerial architectural phenotype of plants at different ages segregated in the two triple mutant lines (Mtsoc1-1, Figures 3A-F; Supplementary Figures 1C–F; Mtsoc1-2, Figures 3G-L; Supplementary Figures 2A–D). The non-flowering plants had a strikingly short primary axis compared with the WT plants in the VLD (Figures 1D, 2C, 3A, E, G, K; Supplementary Figure 1C, 2A). These plants were homozygous for Mtsoc1a-1 Mtsoc1b-1 Mtsoc1c-1 alleles in the Mtsoc1-1 line or Mtsoc1a-4 Mtsoc1b-4 Mtsoc1c-4 in the Mtsoc1-2 line. However, the number of nodes on the primary axis was similar to or greater than that of the WT in these plants (Figures 3B, E, I, K; Supplementary Figures 1E, 2C). This indicates that the reduced height of these mutants was not due to slower plant development, as measured by the production of nodes. In contrast, the length of the longest secondary axis was generally similar between the WT and triple mutants (Figures 3C, F, H, L; Supplementary Figure 1D), except in one experiment (Supplementary Figure 2B). However, there was a significantly increase in the node number on the longest secondary axis in the mutant plants at all time points compared to the WT (Figures 3D, F, J, L; Supplementary Figures 1F, 2D). This indicated a change in the aerial architecture of the mutant plants, with increased node density relative to the WT on the secondary axes.
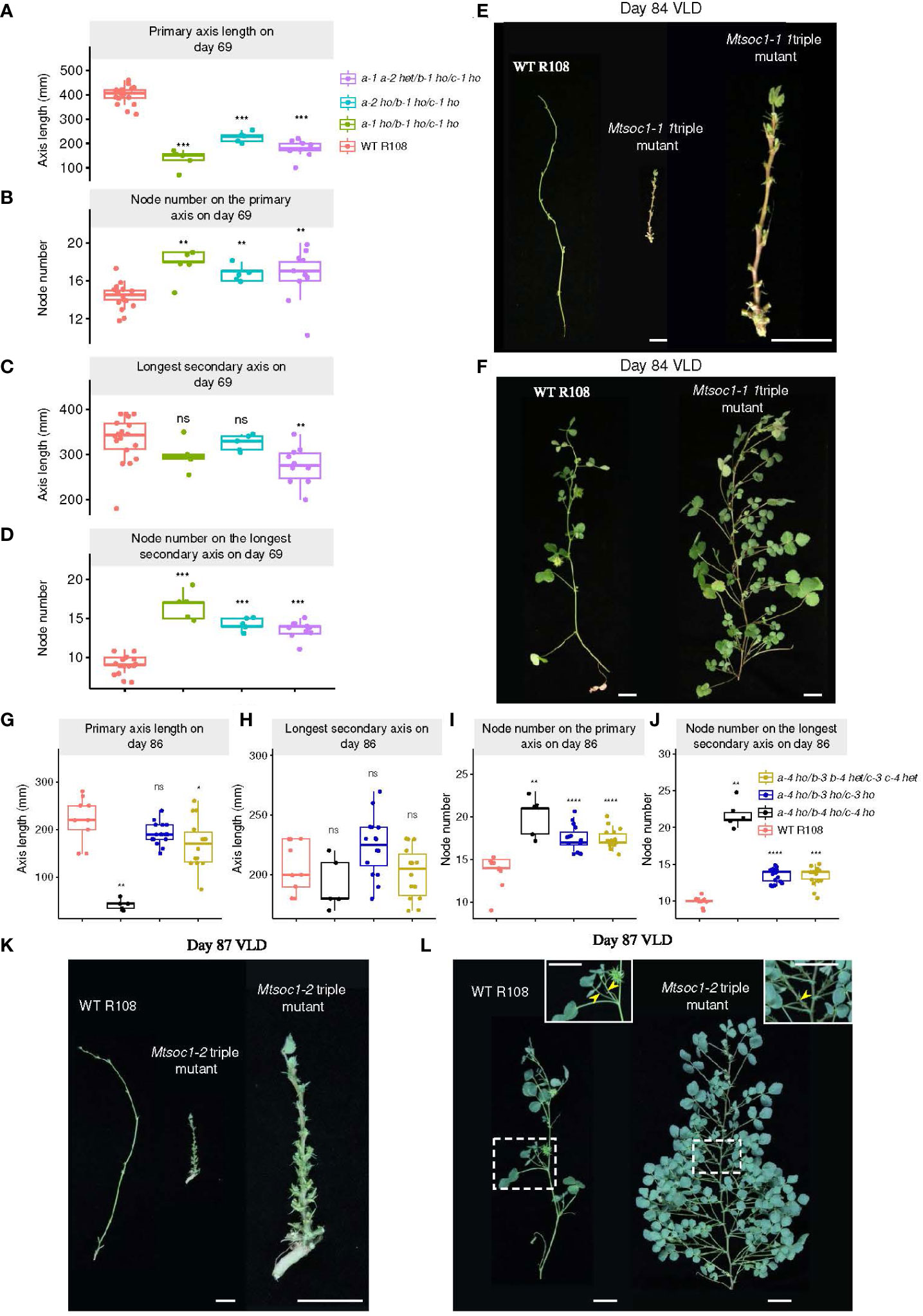
Figure 3 Aerial architecture of plants segregating in triple mutant lines Mtsoc1-1 and Mtsoc1-2. (A–D) Aerial architecture of the WT and Mtsoc1-1 T2 lines (R760 and R761) under VLD, shown as boxplots of primary axis length for each plant (A), Boxplots of number on the primary axis for each plant (B), Boxplots of the longest secondary axis length for each plant (C), Boxplots of the node number on the longest secondary axis for each plant (D). Homozygous (ho) and heterozygous (het) genotypes are shown. (E) Left: Photographs of primary axes on day 84 under VLD of the WT and non-flowering Mtsoc1-1 triple mutant (R760-11), with leaves and branches removed. Right: Close-up view of the Mtsoc1-1 triple mutant primary axis on day 84 under VLD. (F) Photographs of the longest secondary axes on day 84 under VLD of the WT (left) and Mtsoc1-1 triple mutant (R760-11, right). (G–J) Aerial architecture of the WT and Mtsoc1-2 T2 segregating line (R647) under VLD, shown as a boxplot of the primary axis length for each plant (G), boxplot of the longest secondary axis length for each plant (H), boxplot of the node number on the primary axis for each plant (I), and boxplot of the node number on the longest secondary axis for each plant (J). (K) Left: Photographs of primary axes on day 87 under VLD of the WT and non-flowering Mtsoc1-2 triple mutant (R647-18), with leaves and branches removed. Right: Close-up view of the Mtsoc1-2 triple mutant primary axis on day 87 under VLD. (L) Photographs of the longest secondary axes on day 87 under VLD of the WT (left) and Mtsoc1-2 triple mutant (R647-18, right). Photographs on the top right are close-ups of the regions in dashed rectangles. Yellow stealth arrows indicate the lateral structures. Scale is 2 cm. Statistical significance was determined using a Wilcoxon test (P-value with Bonferroni correction: *P ≤0.05, **P ≤0.01, ***P ≤0.001, ****P ≤0.0001). ns, not significant.
The non-flowering Mtsoc1 triple mutant plants exhibited a bushy phenotype correlated with increased growth of the lateral branch in the leaf axils and increased node density, as noted above, compared with the WT (Figures 1D, 2C, 3F, L). However, these triple mutants produced only one lateral structure in the leaf axils (Figure 3L), which is consistent with the remaining vegetative (Cheng et al., 2021). This contrasts with reproductive WT plants, which produce two structures: a lateral branch and a compound flower in the leaf axils (Figure 3L) (Cheng et al., 2021).
Gene expression analyses indicate that the Mtsoc1 triple mutants remain vegetative
To investigate the molecular basis of the Mtsoc1 triple mutant phenotypes, we first compared shoot apex gene expression in non-flowering triple mutants with the Mtsoc1a single mutant and WT by RNA-seq. Shoot apical samples were taken from 15-day old Mtsoc1a and WT plants and from 83-day-old non-flowering Mtsoc1-2 triple mutants. We obtained 129 and 447 upregulated genes and 46 and 1,258 downregulated genes in the single and triple mutants, respectively (Figure 4A, Supplementary Table 3). Consistent with their dramatically different mutant phenotypes (Jaudal et al., 2018), only a few genes (14) were affected in the same way in the two genotypes (13 upregulated and 1 downregulated). We then made a Gene ontology enrichment analysis for the genes up- and down in the two different mutants. We observed that the upregulated genes in the triple mutant were enriched for pathways related to photosynthesis, which is consistent with the vegetative phenotype (Zhang et al., 2021a), and genes that were downregulated were involved in biotic and abiotic stress and hormone-related genes (Supplementary Table 3).
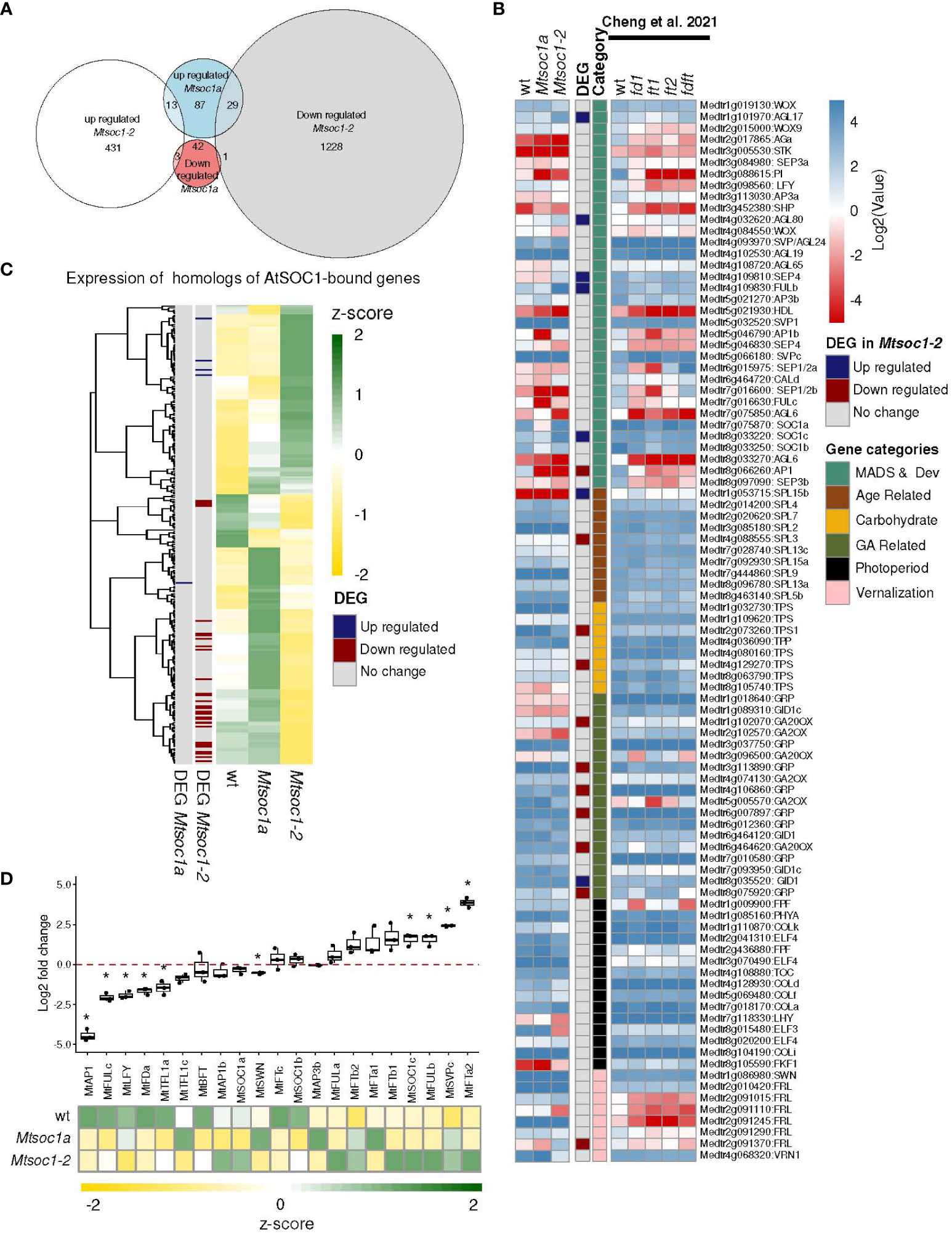
Figure 4 Gene expression in the non-flowering Mtsoc1-2 triple mutants was consistent with their vegetative phenotypes. (A) Euler diagrams showing the number of upregulated and downregulated genes (two-fold change) in the shoot apex of the Mtsoc1-2 triple mutant (83 days old) and the Mtsoc1a Tnt1 single mutant (15 days old) compared to WT (15 days old) (Padj <0.05). (B) Expression of Medicago homologs of Arabidopsis genes bound and regulated by AtSOC1 in ChIP-seq from Immink et al. (2012), shown as z-scores extracted from the TPM values. (C) Heat map of Mtsoc1-2 triple mutant (83 days old) and the Mtsoc1a Tnt1 single mutant (15 days old) and vegetative WT (15 days old) (left, heatmap log2(TPM) is shown), and mutants Mtfda (30 days old), Mtfta1 (30 days old, ft1, 63 days old, ft2), MtfdaMttfta1 (63 days old), and flowering WT (30 days old) from Cheng et al. (2021) (right part of the heatmap (log2(FKPM) is shown). The 93 genes shown are a list extracted from Cheng et al. (2021) of the selected candidate flowering genes. The DEG in Mtsoc1-2 genes were upregulated or downregulated with a fold change of 2 and padj ≤0.05. (D) Upper panel: RT-qPCR analysis of candidate flowering genes at the apex of the Mtsoc1-2 triple mutant and WT. Graph showing the relative expression of candidate flowering genes in the apex samples of the Mtsoc1-2 triple mutant (day 83) and the WT (day 14) under VLD. Relative gene expression was calculated using the formula 2−ΔCT, where ΔCT was obtained by normalizing the gene of interest to the reference gene, MtPP2A, and presenting the log2 fold change in the mutant relative to WT. the box plot shows three biological replicates. Asterisks indicate significant differences between the mutant and WT strains using the t-test, assuming unequal variance (p ≤0.05). Lower panel: Expression shown as z-scores extracted from the TPM values from RNA-seq.
Then, to compare the regulation by Arabidopsis SOC1, we asked if the expression of homologs of genes that were bound and regulated by SOC1 was also altered in the Medicago mutants in RNA-seq (Figure 4B). We used Blastx to identify the Medicago homologs of a list of AtSOC1-bound and regulated genes identified by Immink et al. (2012) (Supplementary Table 4). Strikingly, this indicated that over half of the homologs of the AtSOC1-bound and -regulated genes had altered expression in the non-flowering Mtsoc1-2 triple mutants (Figure 4B). These included two AP2/B3 domain candidate flowering repressors, MtTEMPRANILLO (MtTEM)1 and MtTEM2, which were both significantly downregulated in the triple mutant (Supplementary Table 4).
Next, we analyzed the expression of a list of candidate Medicago flowering genes identified by Cheng et al. (2021). In Figure 4C, the triple mutant shoot apex (83 days old) was compared with 15-day old Mtsoc1a and WT plants, with no visible floral buds (Supplementary Table 5). Data from Cheng et al. (2021) on Mtfta1 Mtfda double and single mutants and a flowering WT are also shown. Consistent with the vegetative phenotype, MADS genes, including the inflorescence meristem identity gene MtAP1 and the B class floral organ identity gene MtPISTILLATA (PI), were downregulated in the Mtsoc1 triple and Mtfta1 Mtfda mutants. Inflorescence identity genes MtLEAFY (LFY) and MtFULc (Cheng et al., 2018) were reduced compared to WT, as were other developmentally important genes required for flowering, such as the WUSCHEL homolog HEADLESS (HDL) (Meng et al., 2018).
Finally, we measured the expression of 21 candidate flowering regulator genes in Mtsoc1-2 triple mutants (83 days old) and WT shoot apices (14 days old) RT-qPCR (Figure 4D, Supplementary Table 5). We found that 10 genes were significantly differentially expressed (six upregulated and four downregulated) in the triple mutant compared to the WT in RT-qPCR. These included four key genes that function in the development of the Medicago compound inflorescence; MtTFL1a is involved in conferring primary inflorescence (I1) identity, MtFULc in secondary inflorescence (I2) identity, MtAP1 in floral meristem identity, and MtLFY is a common primordia determination that gives rise to petals, stamens, and carpels (Benlloch et al., 2015; Cheng et al., 2018). All four genes showed elevated expression levels during the transition to flowering in Medicago (Cheng et al., 2018). Consistent with the vegetative phenotype, MtAP1, MtFULc, MtLFY, and MtTFL1a levels were lower in the triple mutant than in the WT. The flowering time and I2 inflorescence meristem identity gene, MtFDa, were also downregulated. Four genes, including the likely flowering repressor MtSVPc and candidate FT-like flowering repressor MtFTa2 (Jaudal et al., 2022), were upregulated in the triple mutant. RNA-seq analysis of the triple Mtsoc1 mutant yielded similar results.
Discussion
Analysis of Mtsoc1a Mtsoc1b Mtsoc1c triple mutants indicates combined critical roles of the three MtSOC1 genes in Medicago flowering in floral inductive VLD
SOC1-like genes are known to play important roles in flowering control in many plants, but soc1 mutations have not previously been associated with non-flowering phenotypes. For example, in soybeans, mutations in one or both duplicated GmSOC1 A-class genes, GmSOC1a and GmSOC1b, led to delayed flowering and increased node number, with GmSOC1a having stronger effects, whereas the double mutants showed additive effects on shoot architecture and greater delay in flowering (Kou et al., 2022). Importantly, naturally occurring GmSOC1a alleles contribute to optimal latitudinal adaptation in soybean (Kou et al., 2022). Within the Medicago genus in legumes, multiple gene duplication events have given rise to three MtSOC1 paralogs: one Fabaceae A group MtSOC1a gene and two B-group genes, MtSOC1b and MtSOC1c. Here, we describe the generation of Mtsoc1a Mtsoc1b Mtsoc1c triple mutant plants by CRISPR-Cas9 gene editing using guides predominantly targeting the exons encoding the K-domain or C-terminal domain. Two independent triple mutant lines, Mtsoc1-1 and Mtsoc1-2, were studied. These lines segregated non-flowering plants in floral inductive VLD. These non-flowering plants were homozygous for the strong mutant alleles of Mtsoc1a, Mtsoc1b, and Mtsoc1c. Phenotyping and gene expression analyses indicated that these non-flowering plants remained vegetative, because they did not transition to flowering. The triple mutant non-flowering phenotype was thus dramatically different from the single Mtsoc1a Tnt1 mutant, which had a moderate delay to flowering of ~10 days compared with the WT in VLD (Jaudal et al., 2018). These results imply a combined critical role of multiple MtSOC1 genes in the transition to flowering in Medicago.
MtSOC1 genes and primary axis elongation
The non-flowering triple mutants had shorter primary axes than the WT (Figures 3E, K), consistent with previous studies implicating MtSOC1a in primary axis elongation. The Mtsoc1a Tnt1 single mutant had a shorter primary axis than the WT, while overexpression of MtSOC1a caused increased primary axis elongation compared with the WT (Jaudal et al., 2018). Interestingly, in the Mtsoc1-1 line, the primary axis was also shorter in plants homozygous for the strong Mtsoc1b-1 and Mtsoc1c-1 alleles but heterozygous or homozygous for the predicted weaker Mtsoc1a-2 allele (Figure 3A; Supplementary Figures 1C, 2A; Table 1). Mtsoc1a-2 is predicted to affect the C-terminal domain, which is highly variable in sequence between different MADs transcription factors (Lai et al., 2021) and has a weak effect on flowering in the triple mutant (Figure 1C, Supplementary Figures 1A, B). Therefore, the Mtsoc1b-1 and Mtsoc1c-1 alleles likely contribute strongly to the short primary axis phenotype of the triple mutant, which correlates with the strong expression of MtSOC1b and MtSOC1c in primary axis stems (Fudge et al., 2018).
The non-flowering Mtsoc1 triple mutants appear to remain vegetative
The non-flowering triple mutants were also bushy in appearance because of the increased node density observed on the secondary axes, and increased growth of the lateral branch in their leaf axils (Figure 3). They only produce one lateral structure, a lateral branch in their leaf axils (Figure 3L) indicating that they remain vegetative in the VLD (Cheng et al., 2021). The non-flowering triple mutant phenotype thus appears like the Mtfta1 Mtfda double mutant that failed to transition to flowering (Cheng et al., 2021). Similar to the triple mutants, Mtfta1Mtfda double mutants remained vegetative with a bushy phenotype, producing only one lateral branch in the leaf axils. In contrast, the non-flowering Mtfulc mutant does transition to flowering but cannot produce flowers. Instead, it produces two lateral branches in the leaf axils due to the aberrant elevation of MtTFL1a in the secondary inflorescence meristem (Cheng et al., 2018).
Gene expression analysis also supported the idea that the triple mutants remained vegetative. Four key genes that promote the development of Medicago compound inflorescence, MtAP1, MtTFL1a, MtLFY, and MtFULc, were downregulated in the apex of the triple mutant, consistent with the remaining vegetative cells (Figure 4D) (Cheng et al., 2018, 2021). MtFDa was also downregulated at the apex of the triple mutant. The candidate floral repressors MtFTa2 and MtSVPc were upregulated in the triple mutant (Figure 4D), consistent with their elevation in the late-flowering Mting2 mutant (Jaudal et al., 2022), indicating that they are likely to play a repressive role in Medicago flowering. MtFULb and MtSOC1c levels were elevated in the triple mutant, suggesting their misregulation in the shoot apex. Other MADS-box genes that are likely to promote floral meristem and floral organ identity and/or development were also downregulated in the triple mutant shoot apex, consistent with the non-flowering phenotype (Supplementary Table 3). These include genes homologous to SEP, AGL6, B function genes PI and AP3, AG-like C function genes, and a predicted D function gene, MtSEEDSTICK-like (STK).
Perspectives
While the three MtSOC1 genes are likely to function predominantly downstream of MtFTa1 and MtFDa in promoting the vegetative-to-reproductive transition (Fudge et al., 2018; Jaudal et al., 2018; Cheng et al., 2021; Zhang et al., 2021b), we found that MtFDa gene expression was also reduced in the Mtsoc1a single mutant and the triple mutant. This may indicate negative feedback regulation of MtSOC1s on MtFDa, consistent with the multiple interactions observed between SOC1 and flowering regulators in Arabidopsis (Immink et al., 2012; Tao et al., 2012). It is also possible that MtSOC1 genes function in both floral induction and I1 and/or I2 inflorescence development, as has been reported for MtFDa (Cheng et al., 2021; Zhang et al., 2021b). One mechanism by which MtFDa regulates inflorescence development is via stimulation of MtFULc expression in I2, which in turn represses MtTFL1a in I2, enabling floral meristem formation via activation of MtPIM (Cheng et al., 2021; Zhang et al., 2021b). The MtSOC1a transcript was detected in I1 and I2 primordia via in situ hybridization in the WT (Jaudal et al., 2018). In addition, a sharp elevation of MtSOC1b transcripts in the shoot apex was detected at flowering by RT-qPCR, later than the increase in expression observed for MtSOC1a and MtSOC1c (Jaudal et al., 2018). This indicates potential additional roles for MtSOC1s downstream of floral induction that remain to be uncovered. Taken together, our phenotyping and global transcriptomic analyses implicated multiple MtSOC1 genes in overlapping and complementary functions, which are essential for the vegetative-to-floral transition in VLD and the development of normal aerial architecture, unlike Arabidopsis SOC1 (Lee and Lee, 2010) or MtSOC1a alone (Jaudal et al., 2018). Ultimately, the non-flowering phenotype may be used to improved forage and biofuel productivity in temperate legumes (Jung and Muller, 2009; Tadege et al., 2015; Wolabu et al., 2023).
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author contributions
AP: Conceptualization, Data curation, Formal analysis, Visualization, Writing – original draft, Writing – review & editing. MZ: Investigation, Writing – review & editing. YP: Data curation, Investigation, Project administration, Writing – original draft, Writing – review & editing. FT: Data curation, Investigation, Writing – review & editing. MJ: Conceptualization, Methodology, Writing – review & editing. LZ: Investigation, Methodology, Writing – review & editing. JW: Funding acquisition, Writing – review & editing. JP: Conceptualization, Funding acquisition, Project administration, Resources, Supervision, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was financially supported by a grant from the New Zealand Marsden Fund (www.royalsociety.org.nz/programmes/funds/marsden/) (contract 17-UOA-075 awarded to JP and used to support MJ and YP).
Acknowledgments
We thank Matthew Mayo Smith for help with DNA sequence analysis of gene edits and critical reading of the manuscript, Erika Varkonyi-Gasic for insightful feedback on the manuscript and Nathan Deed for glasshouse support.
Conflict of interest
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2024.1357924/full#supplementary-material
Supplementary Table 1 | List of primers and guides.
Supplementary Table 2 | Lists of plant genotypes and flowering time.
Supplementary Table 3 | RNA-seq analysis for Mtsoc1a and Mtsoc1-2 triple mutant and Gene Ontology analysis.
Supplementary Table 4 | Expression of Medicago homologs of AtSOC1-bound and regulated genes listed in Immink et al. (2012).
Supplementary Table 5 | Expression of candidate flowering genes listed in Cheng et al. (2021) and genes used in RT-qPCR.
References
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., Lipman, D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. doi: 10.1016/s0022-2836(05)80360-2
Andrés, F., Coupland, G. (2012). The genetic basis of flowering responses to seasonal cues. Nat. Rev. Genet. 13, 627–639. doi: 10.1038/nrg3291
Balanzà, V., Martínez-Fernández, I., Ferrándiz, C. (2014). Sequential action of FRUITFULL as a modulator of the activity of the floral regulators SVP and SOC1. J. Exp. Bot. 65, 1193–1203. doi: 10.1093/jxb/ert482
Benlloch, R., Berbel, A., Ali, L., Gohari, G., Millán, T., Madueño, F. (2015). Genetic control of inflorescence architecture in legumes. Front. Plant Sci. 6. doi: 10.3389/fpls.2015.00543
Benlloch, R., d’Erfurth, I., Ferrandiz, C., Cosson, V., Beltran, J. P., Canas, L. A., et al. (2006). Isolation of mtpim proves Tnt1 a useful reverse genetics tool in Medicago truncatula and uncovers new aspects of AP1-like functions in legumes. Plant Physiol. 142, 972–983. doi: 10.1104/pp.106.083543
Bookout, A. L., Mangelsdorf, D. J. (2003). Quantitative real-time PCR protocol for analysis of nuclear receptor signaling pathways. Nucl. Receptor Signaling 1, e012. doi: 10.1621/nrs.01012
Cañas, L. A., Beltrán, J. P. (2018). “Model Legumes: Functional Genomics Tools in Medicago truncatula,” in Functional Genomics in Medicago truncatula. Methods in Molecular Biology, vol. 1822 . Eds. Cañas, L., Beltrán, J. (Humana Press, New York, NY). doi: 10.1007/978-1-4939-8633-0_2
Chen, S., Zhou, Y., Chen, Y., Gu, J. (2018). fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, i884–i890. doi: 10.1093/bioinformatics/bty560
Cheng, X., Li, G., Krom, N., Tang, Y., Wen, J. (2021). Genetic regulation of flowering time and inflorescence architecture by MtFDa and MtFTa1 in Medicago truncatula. Plant Physiol. 185, 161–178. doi: 10.1093/plphys/kiaa005
Cheng, X., Li, G., Tang, Y., Wen, J. (2018). Dissection of genetic regulation of compound inflorescence development in Medicago truncatula. Development 145. doi: 10.1242/dev.158766
Clarkson, N. M., Russell, J. S. (1975). Flowering responses to vernalisation and photoperiod in annual medics (Medicago spp). Aust. J. Agric. Res. 26, 831–838. doi: 10.1071/AR9750831
Collani, S., Neumann, M., Yant, L., Schmid, M. (2019). FT modulates genome-wide DNA-binding of the bZIP transcription factor FD. Plant Physiol. 180, 367–380. doi: 10.1104/pp.18.01505
Cosson, V., Durand, P., Erfurth, I., Kondorosi, A., Ratet, P. (2006). Medicago truncatula transformation using leaf explants. Methods Mol. Biol. 343, 115–127. doi: 10.1385/1-59745-130-4:115
Doench, J. G., Hartenian, E., Graham, D. B., Tothova, Z., Hegde, M., Smith, I., et al. (2014). Rational design of highly active sgRNAs for CRISPR-Cas9–mediated gene inactivation. Nat. Biotechnol. 32, 1262–1267. doi: 10.1038/nbt.3026
Fornara, F., de Montaigu, A., Coupland, G. (2010). SnapShot: control of flowering in arabidopsis. Cell 141, 550. doi: 10.1016/j.cell.2010.04.024
Foyer, C. H., Lam, H. M., Nguyen, H. T., Siddique, K. H., Varshney, R. K., Colmer, T. D., et al. (2016). Neglecting legumes has compromised human health and sustainable food production. Nat. Plants 2, 16112. doi: 10.1038/nplants.2016.112
Fudge, J. B., Lee, R. H., Laurie, R. E., Mysore, K. S., Wen, J., Weller, J. L., et al. (2018). Medicago truncatula SOC1 genes are up-regulated by environmental cues that promote flowering. Front. Plant Sci. 9. doi: 10.3389/fpls.2018.00496
Ge, S. X., Jung, D., Yao, R. (2020). ShinyGO: a graphical gene-set enrichment tool for animals and plants. Bioinformatics 36, 2628–2629. doi: 10.1093/bioinformatics/btz931
Gibeaut, D. M., Hulett, J., Cramer, G. R., Seemann, J. R. (1997). Maximal biomass of Arabidopsis thaliana using a simple, low-maintenance hydroponic method and favorable environmental conditions. Plant Physiol. 115, 317–319. doi: 10.1104/pp.115.2.317
Graham, P., Vance, C. (2003). Legumes: importance and constraints to greater use. Plant Physiol. 131, 872–877. doi: 10.1104/pp.017004
Gregis, V., Sessa, A., Dorca-Fornell, C., Kater, M. M. (2009). The Arabidopsis floral meristem identity genes AP1, AGL24 and SVP directly repress class B and C floral homeotic genes. Plant J. 60, 626–637. doi: 10.1111/j.1365-313X.2009.03985.x
Immink, R. G. H., Posé, D., Ferrario, S., Ott, F., Kaufmann, K., Valentim, F. L., et al. (2012). Characterization of SOC1’s central role in flowering by the identification of its upstream and downstream regulators. Plant Physiol. 160, 433–449. doi: 10.1104/pp.112.202614
Jaudal, M., Mayo-Smith, M., Poulet, A., Whibley, A., Peng, Y., Zhang, L., et al. (2022). MtING2 encodes an ING domain PHD finger protein which affects Medicago growth, flowering, global patterns of H3K4me3, and gene expression. Plant J. 112, 1029–1050. doi: 10.1111/tpj.15994
Jaudal, M., Monash, J., Zhang, L., Wen, J., Mysore, K. S., Macknight, R., et al. (2014). Overexpression of Medicago SVP genes causes floral defects and delayed flowering in Arabidopsis but only affects floral development in Medicago. J. Exp. Bot. 65, 429–442. doi: 10.1093/jxb/ert384
Jaudal, M., Thomson, G., Zhang, L., Che, C., Wen, J., Mysore, K. S., et al. (2020a). “Forward and reverse screens to identify genes that control vernalization and flowering time in Medicago truncatula.,” in The Model Legume Medicago truncatula. Ed. de Bruijn, F. (Wiley/Blackwell, New York, USA), 189–196.
Jaudal, M., Wen, J., Mysore, K. S., Putterill, J. (2020b). Medicago PHYA promotes flowering, primary stem elongation and expression of flowering time genes in long days. BMC Plant Biol. 20, 329. doi: 10.1186/s12870-020-02540-y
Jaudal, M., Zhang, L., Che, C., Hurley, D. G., Thomson, G., Wen, J., et al. (2016). MtVRN2 is a Polycomb VRN2-like gene which represses the transition to flowering in the model legume Medicago truncatula. Plant J. 86, 145–160. doi: 10.1111/tpj.13156
Jaudal, M., Zhang, L., Che, C., Li, G., Tang, Y., Wen, J., et al. (2018). A SOC1-like gene MtSOC1a promotes flowering and primary stem elongation in Medicago. J. Exp. Bot. 69, 4867–4880. doi: 10.1093/jxb/ery284
Jaudal, M., Zhang, L., Che, C., Putterill, J. (2015). Three Medicago MtFUL genes have distinct and overlapping expression patterns during vegetative and reproductive development and 35S:MtFULb accelerates flowering and causes a terminal flower phenotype in Arabidopsis. Front. Genet. 5. doi: 10.3389/fgene.2015.00050
Jung, C., Muller, A. E. (2009). Flowering time control and applications in plant breeding. Trends Plant Sci. 14, 563–573. doi: 10.1016/j.tplants.2009.07.005
Kearse, M., Moir, R., Wilson, A., Stones-Havas, S., Cheung, M., Sturrock, S., et al. (2012). Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28, 1647–1649. doi: 10.1093/bioinformatics/bts199
Kim, D. H., Doyle, M. R., Sung, S., Amasino, R. M. (2009). Vernalization: winter and the timing of flowering in plants. Annu. Rev. Cell Dev. Biol. 25, 277–299. doi: 10.1146/annurev.cellbio.042308.113411
Kou, K., Yang, H., Li, H., Fang, C., Chen, L., Yue, L., et al. (2022). A functionally divergent SOC1 homolog improves soybean yield and latitudinal adaptation. Curr. Biol. 32, 1728–1742. doi: 10.1016/j.cub.2022.02.046
Lai, X., Vega-Léon, R., Hugouvieux, V., Blanc-Mathieu, R., van der Wal, F., Lucas, J., et al. (2021). The intervening domain is required for DNA-binding and functional identity of plant MADS transcription factors. Nat. Commun. 12, 4760. doi: 10.1038/s41467-021-24978-w
Laurie, R. E., Diwadkar, P., Jaudal, M., Zhang, L., Hecht, V., Wen, J., et al. (2011). The Medicago FLOWERING LOCUS T homolog, MtFTa1, is a key regulator of flowering time. Plant Physiol. 156, 2207–2224. doi: 10.1104/pp.111.180182
Lee, J., Lee, I. (2010). Regulation and function of SOC1, a flowering pathway integrator. Journal of Experimental Botany 61, 2247–2254.
Liu, C., Xi, W., Shen, L., Tan, C., Yu, H. (2009). Regulation of floral patterning by flowering time genes. Dev. Cell 16, 711–722. doi: 10.1016/j.devcel.2009.03.011
Livak, K. J., Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Love, M. I., Huber, W., Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550. doi: 10.1186/s13059-014-0550-8
Melzer, S., Lens, F., Gennen, J., Vanneste, S., Rohde, A., Beeckman, T. (2008). Flowering-time genes modulate meristem determinacy and growth form in Arabidopsis thaliana. Nat. Genet. 40, 1489–1492. doi: 10.1038/ng.253
Meng, Y., Hou, Y., Wang, H., Ji, R., Liu, B. (2017). Targeted mutagenesis by CRISPR/Cas9 system in the model legume Medicago truncatula. Plant Cell Rep. 36, 371–374. doi: 10.1007/s00299-016-2069-9
Meng, Y., Liu, H., Wang, H., Liu, Y., Zhu, B., Wang, Z., et al. (2018). HEADLESS, a WUSCHEL homolog, uncovers novel aspects of shoot meristem regulation and leaf blade development in Medicago truncatula. J. Exp. Bot. 70, 149–163. doi: 10.1093/jxb/ery346
Moll, K. M., Zhou, P., Ramaraj, T., Fajardo, D., Devitt, N. P., Sadowsky, M. J., et al. (2017). Strategies for optimizing BioNano and Dovetail explored through a second reference quality assembly for the legume model, Medicago truncatula. BMC Genomics 18, 1–16. doi: 10.1186/s12864-017-3971-4
Patro, R., Duggal, G., Love, M. I., Irizarry, R. A., Kingsford, C. (2017). Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 14, 417–419. doi: 10.1038/nmeth.4197
Perez-Santangelo, S., Napier, N., Robson, F., Weller, J. L., Bond, D. M., Macknight, R. C. (2022). A Point Mutation in Phytochromobilin synthase Alters the Circadian Clock and Photoperiodic Flowering of Medicago truncatula. Plants 11, 239. doi: 10.3390/plants11030239
Putterill, J., Zhang, L., Yeoh, C., Balcerowicz, M., Jaudal, M., Varkonyi Gasic, E. (2013). FT genes and regulation of flowering in the legume Medicago truncatula. Funct. Plant Biol. 40, 1199–1207. doi: 10.1071/FP13087
R Core Team (2018). R: A language and environment for statistical computing. R Foundation for statistical computing (Vienna, Austria) Available at: https://www.R-project.org/.
Romera-Branchat, M., Severing, E., Pocard, C., Ohr, H., Vincent, C., Née, G., et al. (2020). Functional divergence of the arabidopsis florigen-interacting bZIP transcription factors FD and FDP. Cell Rep. 31, 107717. doi: 10.1016/j.celrep.2020.107717
Soneson, C., Love, M. I., Robinson, M. D. (2015). Differential analyses for RNA-seq: transcript-level estimates improve gene-level inferences. F1000Res 4, 1521. doi: 10.12688/f1000research.7563.2
Srikanth, A., Schmid, M. (2011). Regulation of flowering time: all roads lead to Rome. Cell. Mol. Life Sci. 68, 2013–2037. doi: 10.1007/s00018-011-0673-y
Sussmilch, F. C., Hecht, V., Vander Schoor, J. K., Weller, J. L. (2017). Identification of the SHORT VEGETATIVE PHASE (SVP)-like MADS-box genes in pea (Pisum sativum L.). Plant Gene 12, 72–79. doi: 10.1016/j.plgene.2017.08.003
Tadege, M., Chen, F., Murray, J., Wen, J., Ratet, P., Udvardi, M. K., et al. (2015). Control of vegetative to reproductive phase transition improves biomass yield and simultaneously reduces lignin content in medicago truncatula. Bioenergy Res. 8, 857–867. doi: 10.1007/s12155-014-9565-y
Tadege, M., Wang, T. L., Wen, J. Q., Ratet, P., Mysore, K. S. (2009). Mutagenesis and beyond! Tools for understanding legume biology. Plant Physiol. 151, 978–984. doi: 10.1104/pp.109.144097
Tang, H., Krishnakumar, V., Bidwell, S., Rosen, B., Chan, A., Zhou, S., et al. (2014). An improved genome release (version Mt4.0) for the model legume Medicago truncatula. BMC Genomics 15, 1–14. doi: 10.1186/1471-2164-15-312
Tao, Z., Shen, L., Liu, C., Liu, L., Yan, Y., Yu, H. (2012). Genome-wide identification of SOC1 and SVP targets during the floral transition in Arabidopsis. Plant J. 70, 549–561. doi: 10.1111/j.1365-313X.2012.04919.x
Thomson, G., Taylor, J., Putterill, J. (2019). The transcriptomic response to a short day to long day shift in leaves of the reference legume Medicago truncatula. PeerJ 7, e6626. doi: 10.7717/peerj.6626
Thomson, G., Zhang, L., Wen, J., Mysore, K. S., Putterill, J. (2021). The candidate photoperiod gene mtFE promotes growth and flowering in medicago truncatula. Front. Plant Sci. 12, 455. doi: 10.3389/fpls.2021.634091
Trinh, T. H., Ratet, P., Kondorosi, E., Durand, P., Kamaté, K., Bauer, P., et al. (1998). Rapid and efficient transformation of diploid Medicago truncatula and Medicago sativa ssp. falcata lines improved in somatic embryogenesis. Plant Cell Rep. 17, 345–355. doi: 10.1007/s002990050405
Untergasser, A., Cutcutache, I., Koressaar, T., Ye, J., Faircloth, B. C., Remm, M., et al. (2012). Primer3—new capabilities and interfaces. Nucleic Acids Research 40, e115
Voogd, C., Wang, T., Varkonyi-Gasic, E. (2015). Functional and expression analyses of kiwifruit SOC1-like genes suggest that they may not have a role in the transition to flowering but may affect the duration of dormancy. J. Exp. Bot. 66, 4699–4710. doi: 10.1093/jxb/erv234
Weller, J. L., Macknight, R. C. (2018). Functional genomics and flowering time in Medicago truncatula: an overview. In: Cañas, L., Beltrán, J. (eds) Functional Genomics in Medicago truncatula. Methods in Molecular Biology, vol 1822. New York, NY: Humana Press. doi: 10.1007/978-1-4939-8633-0_17
Weller, J. L., Ortega, R. (2015). Genetic control of flowering time in legumes. Front. Plant Sci. 6. doi: 10.3389/fpls.2015.00207
Wolabu, T. W., Mahmood, K., Jerez, I. T., Cong, L., Yun, J., Udvardi, M., et al. (2023). Multiplex CRISPR/Cas9-mediated mutagenesis of alfalfa FLOWERING LOCUS Ta1 (MsFTa1) leads to delayed flowering time with improved forage biomass yield and quality. Plant Biotechnol. J. 21, 1383–1392. doi: 10.1111/pbi.14042
Wong, A. C. S., Hecht, V. F. G., Picard, K., Diwadkar, P., Laurie, R. E., Wen, J., et al. (2014). Isolation and functional analysis of CONSTANS-LIKE genes suggests that a central role for CONSTANS in flowering time control is not evolutionarily conserved in Medicago truncatula. Front. Plant Sci. 5. doi: 10.3389/fpls.2014.00486
Xie, K., Minkenberg, B., Yang, Y. (2015). Boosting CRISPR/Cas9 multiplex editing capability with the endogenous tRNA-processing system. Proceedings of the National Academy of Sciences 112 (11), 3570–3575.
Yeoh, C. C., Balcerowicz, M., Zhang, L., Jaudal, M., Brocard, L., Ratet, P., et al. (2013). Fine mapping links the FTa1 flowering time regulator to the dominant spring1 locus in medicago. PloS One 8, e53467. doi: 10.1371/journal.pone.0053467
Young, N. D., Debellé, F., Oldroyd, G. E., Geurts, R., Cannon, S. B., Udvardi, M. K., et al. (2011). The Medicago genome provides insight into the evolution of rhizobial symbioses. Nature 480, 520–524. doi: 10.1038/nature10625
Yuan, J., Long, H., Qiu, F., Wang, Y., Zhang, M., Chao, Y., et al. (2023). MADS-box protein MtSOC1c regulates flowering and seed development in Medicago truncatula. Ind. Crops Prod 193. doi: 10.1016/j.indcrop.2022.116125
Zhang, T. Q., Chen, Y., Wang, J. W. (2021a). A single-cell analysis of the Arabidopsis vegetative shoot apex. Dev. Cell 56, 1056–1074 e1058. doi: 10.1016/j.devcel.2021.02.021
Zhang, L., Jiang, A., Thomson, G., Kerr-Phillips, M., Phan, C., Krueger, T., et al. (2019). Overexpression of Medicago MtCDFd1_1 causes delayed flowering in Medicago via repression of MtFTa1 but not MtCO-like genes. Front. Plant Sci. 10, 1148. doi: 10.3389/fpls.2019.01148
Zhang, P., Liu, H., Mysore, K. S., Wen, J., Meng, Y., Lin, H., et al. (2021b). MtFDa is essential for flowering control and inflorescence development in Medicago truncatula. J. Plant Physiol. 260, 153412. doi: 10.1016/j.jplph.2021.153412
Keywords: MtSOC1a, MtSOC1b, MtSOC1c, legume, Medicago, flowering time, gene editing, CRISPR-Cas9
Citation: Poulet A, Zhao M, Peng Y, Tham F, Jaudal M, Zhang L, van Wolfswinkel JC and Putterill J (2024) Gene-edited Mtsoc1 triple mutant Medicago plants do not flower. Front. Plant Sci. 15:1357924. doi: 10.3389/fpls.2024.1357924
Received: 19 December 2023; Accepted: 02 February 2024;
Published: 26 February 2024.
Edited by:
Liqin Li, Huzhou Central Hospital, ChinaReviewed by:
Fang Yang, Huazhong Agricultural University, ChinaMd Mostofa Uddin Helal, Shanxi Agricultural University, China
Copyright © 2024 Poulet, Zhao, Peng, Tham, Jaudal, Zhang, van Wolfswinkel and Putterill. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Joanna Putterill, ai5wdXR0ZXJpbGxAYXVja2xhbmQuYWMubno=
†These authors share first authorship
 Axel Poulet
Axel Poulet Min Zhao
Min Zhao Yongyan Peng
Yongyan Peng FangFei Tham
FangFei Tham Mauren Jaudal
Mauren Jaudal Lulu Zhang
Lulu Zhang Josien C. van Wolfswinkel
Josien C. van Wolfswinkel Joanna Putterill
Joanna Putterill