
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 28 February 2024
Sec. Plant Pathogen Interactions
Volume 15 - 2024 | https://doi.org/10.3389/fpls.2024.1357141
This article is part of the Research Topic Plant Parasitic Nematode–Host Interactions: Mechanisms and Exploitative Management Strategies View all 11 articles
 Zhiwen Li1
Zhiwen Li1 Honghong Wang2
Honghong Wang2 Yuqing Cao1
Yuqing Cao1 Xiaoling Shan1
Xiaoling Shan1 Xiaoxian He1
Xiaoxian He1 Qiuling Huang1
Qiuling Huang1 Kan Zhuo1,3
Kan Zhuo1,3 Jinling Liao1,2*
Jinling Liao1,2* Borong Lin1,3*
Borong Lin1,3*The migratory endoparasitic phytonematodes Bursaphelenchus xylophilus is the causal agent of pine wilt disease and causes significant economic damage to pine forests in China. Effectors play a key role in the successful parasitism of plants by phytonematodes. In this study, 210 genes obtained by transcriptomics analyses were found to be upregulated in B. xylophilus infecting Pinus massoniana that were not functionally annotated nor reported previously in B. xylophilus infecting P. thunbergii. Among these differentially expressed genes, a novel effector, BxICD1, that could induce cell death in the extracellular space of Nicotiana benthamiana was identified. BxICD1 was upregulated in the early stages of infection, as shown by RT-qPCR analyses. In situ hybridization analysis showed that BxICD1 was expressed in the esophageal gland of nematodes. The yeast signal sequence trap system indicated that BxICD1 possessed an N-terminal signal peptide with secretion functionality. Using an Agrobacterium-mediated transient expression system, it was demonstrated that the cell death-inducing activity of BxICD1 was dependent on N. benthamiana brassinosteroid-insensitive 1-associated kinase 1 (NbBAK1). Finally, BxICD1 contributed to B. xylophilus virulence and migration in host pine trees, as demonstrated by RNAi silencing assays. These findings indicate that BxICD1 both induces plant cell death and also contributes to nematode virulence and migration in P. massonian.
The pine wood nematode (PWN), Bursaphelenchus xylophilus, is a migratory endoparasitic nematode. The nematodes enter the trees through the wounds formed by Monochamus beetles feeding (Edwards and Linit, 1992), then colonize the tree’s xylem and cortex resin canals, obstructing water and nutrient flow. This ultimately leads to tree wilting and death (Futai, 2013), termed pine wilt disease (PWD). PWD has significant economic and ecological impacts on pine forests in Asia and Europe (Kikuchi et al., 2011). Currently, the methods for preventing and controlling PWD primarily involve directly cutting and incinerating infested trees, fumigating logs with pesticides, applying pesticides to manage Monochamus beetles, or injecting nematicides against B. xylophilus (Cui et al., 2014). However, these strategies possess certain drawbacks, including environmental pollution and significant costs. To formulate novel and eco-friendly strategies for PWD management, it is necessary to understand the mechanisms of PWN parasitism.
When plant-parasitic nematodes (PPNs) infect host plants, they secrete numerous proteins into the host. These proteins, termed effectors, play crucial roles in successful nematode parasitism, and understanding their functions is crucial to uncovering the molecular mechanisms underlying parasitism (Haegeman et al., 2012; Vieira and Gleason, 2019). Many previous studies have shown that PPN effectors mainly function in plant cell wall degradation, feeding site formation, and the suppression of host immunity (Popeijus et al., 2000; Kikuchi et al., 2006; Kudla et al., 2007; Vanholme et al., 2007; Chen et al., 2017; Verma et al., 2018; Chen et al., 2021; Song et al., 2021). Most studies have focused on sedentary endoparasitic nematodes, such as root-knot nematodes and cyst nematodes. Recently, some effectors from B. xylophilus have been characterized, and effectors with plant cell wall-degradation or host immunity suppression functions have been discovered (Kikuchi et al., 2006; Wen et al., 2021, Wen et al., 2022; Hu et al., 2022a; Qiu et al., 2023). Intriguingly, a number of effectors from B. xylophilus were found to induce host cell death. Of these, BxCDP1 can trigger plant cell death (PCD) to induce Pinus thunbergii resistance against nematodes (Hu et al., 2020; Hu et al., 2022b). Several other effectors, such as BxSapB1, BxSapB2, and BxSapB3, while capable of inducing plant cell death, also function as a virulence factor, during B. xylophilus infection (Huang et al., 2019; Hu et al., 2019; Zhao et al., 2020). This phenomenon is relatively common in effectors from fungi and oomycetes, such as the Fusarium graminearum effector Fg62 (Wang et al., 2023), the Phytophthora infestans effector Pi23226 (Lee et al., 2023), etc., which can induce host cell death and promote pathogen parasitism.
Masson pine (Pinus massoniana) is a pioneer species for afforestation timber and oleoresin production, widely distributed in South China. However, it is highly sensitive to B. xylophilus (Xue et al., 2013; Liu et al., 2022a). Despite this, the roles of effectors during B. xylophilus parasitism in P. massoniana are poorly understood. In this study, transcriptomes of B. xylophilus feeding on Pestalotiopsis sp. and infecting P. massoniana were analyzed by RNA sequencing (RNA-seq). Differentially expressed genes of B. xylophilus were identified in these two conditions, which were analyzed to identify candidate effectors involved in B. xylophilus parasitism in P. massoniana parasitism. Among the candidate effectors, a novel effector (named BxICD1) that was expressed in the esophageal gland of B. xylophilus and upregulated during nematode parasitism in P. massoniana was identified. Here, we showed that the effector BxICD1 not only induces PCD, but also promotes nematode virulence and migration in P. massoniana.
B. xylophilus was isolated from a wilted Masson pine in Guangdong province, China, purified using a single fertilized adult female, and cultured in the fungus Pestalotiopsis sp. grown on potato dextrose agar (PDA) medium at 25°C under 16 h light/8 h dark (16/8 LD) conditions for 6 days.
Approximately 3-year-old P. massoniana seedlings were obtained from Yangshan County (24°17′1.6″N, 112°33′42.4″E), Qingyuan City, Guangdong province, China, and grown at temperatures ranging from 25°C to 30°C. Nicotiana benthamiana LAB were grown in a glasshouse at 25°C with a relative humidity of 60% under 16/8 LD conditions.
B. xylophilus was inoculated into a small, approximately 1 cm, wound in P. massoniana stems. After 12 h, mixed-life-stage nematodes were isolated from the pine stems using a Baermann funnel technique (Hu et al., 2019), and approximately 20,000 nematodes were used for RNA extraction. The same number of B. xylophilus nematodes were also isolated from Pestalotiopsis sp. grown on PDA plates for RNA extraction. Each treatment consisted of two replicates. Total RNA was extracted using the RNAprep Pure Micro Kit (DP420, TianGen Biotech), and first-strand cDNA was synthesized from 1 µg of total RNA using a TransScript One-Step gDNA Removal and the cDNA Synthesis SuperMix kits (AT311, TransGen Biotech, Beijing, China) according to the manufacturer’s instruction.
Sequencing libraries were constructed using the NEBNext Ultra RNA Library Prep Kit (E7530, NEB, USA) according to the manufacturer’s instructions. RNA integrity was determined by the RNA Nano 6000 Assay Kit of the Bioanalyzer 5400 system (Agilent Technologies, CA, USA). Total RNA was used as input material for the RNA library preparations. The library preparations were sequenced on an Illumina Novaseq platform, generating 6.49-6.82 GB of data of 150-base long paired reads per sample (Modi et al., 2021). The raw RNA-Seq data in this study are available through the National Center for Biotechnology Information under the accession number PRJNA1027064.
Paired-end reads were aligned to the reference genome (https://parasite.wormbase.org/Bursaphelenchus_xylophilus_prjea64437/Info/Index) using Hisat2 v2.0.5 (Kim et al., 2019). The reads mapped to each gene were measured using FeatureCounts v1.5.0-p3 (Liao et al., 2014). The FPKM values of each gene were calculated based on the length of the gene and the reads count mapped to this gene (Trapnell et al., 2010).
Differential expression analysis was performed using the DESeq2 R package (1.20.0). The resulting P-values were adjusted using the Benjamini and Hochberg’s approach for controlling the false discovery rate (Benjamini and Hochberg, 1995). Genes identified by DESeq with an adjusted P-value<0.05 and |Log2(fold change)|>1 were considered differentially expressed genes.
Real-time quantitative PCR (RT-qPCR) was performed using Green qPCR SuperMix (AQ101, TransGen Biotech, Beijing, China) on a Thermal Cycler Dice Real Time System (Takara, Beijing, China). The RT-qPCR reaction mixture consisted of 1.0 μL cDNA, 0.4 μL of each of the 10 pM primers, 10 μL 2 × Green qPCR SuperMix, and 8.2 μL RNase-free ddH2O. The ubiquitin-conjugating enzyme gene BxUBI2 (GenBank accession number CAD5208953.1) and NbEF1α (Heese et al., 2007) were used as endogenous controls for gene expression normalization in the different species. All the gene-specific primers used in this study are listed in Supplementary Table 1. The RT-qPCR cycling conditions were as follows: 94°C for 30 sec, followed by 40 cycles of 94°C for 10 s, 55°C annealing for 15 s, 72°C elongation for 10 s. The relative changes in gene expression were determined using the 2−ΔΔCT method (Livak and Schmittgen, 2001). Three independent experiments were conducted.
Putative secreted proteins encoded by differentially expressed genes (DEGs) were predicted using both SignalP 5.0 (https://services.healthtech.dtu.dk/service.-php?SignalP-5.0) and TMHMM-2.0 (https://services.healthtech.dtu.dk/service.php?TMHMM-2.0) for the presence of an N-terminal signal peptide (SP) and for the absence of a transmembrane domain, respectively. These steps were implemented as a ‘secreted protein prediction’ workflow (Petitot et al., 2016). Protein functional domains were predicted by EGGNOG-MAPPER (http://eggnog-mapper.embl.de/). The phylogenetic tree was constructed using the NJ method and visualized using MEGA-X (Kumar et al., 2018).
The full-length cDNA sequences of 10 candidate genes were amplified by using the corresponding gene-specific primers. The PCR reaction was performed in a total volume of 50 μL containing 1.0 U KOD-FX (KFX-101, Toyobo, Osaka, Japan), 25 μL 2 × PCR Buffer, 10 μL 2 mM dNTPs, 1.5 μL of each of the 10 pM primers, 200 ng cDNA, and sterilized distilled water up to 50 μL. The PCR cycling conditions were as follows: 95°C for 5 min, followed by 30 cycles of 98°C for 10 s, annealing at 55°C for 30 s, elongation at 68°C for 30 s, and final elongation at 68°C for 5 min.
The CaMV35S driven β-glucuronidase (GUS) gene of the pCAMBIA1305.1 vector was removed and replaced with the eGFP gene to generate the binary vector pCambia1305.1ΔGUS and pCambia1305.1:eGFP. To construct the overexpression vector, the 10 candidate genes mentioned above, with or without the native SP sequence, were amplified and ligated to the linearized pCambia1305.1:eGFP digested by NcoI and PmlI (FD0574 and FD0364, Thermo Fisher Scientific, MA, USA) to generate pCambia1305.1:gene:eGFP, following the instructions of the ClonExpress II One Step Cloning Kit (C112-01, Vazyme, Nanjing, China). BxICD1:Flag was amplified and ligated to the linearized pCambia1305.1ΔGUS to generate pCambia1305.1:BxICD1:Flag. For pSUC2, vectors were digested by EcoRI and XhoI (FD0274 and FD0694, Thermo Fisher Scientific, MA, USA) in the appropriate conditions. The SP sequences of BxICD1 and Avr1b were linked to linearized pSUC2 to generate pSUC2:SPBxICD1 and pSUC2:SPAvr1b following the instructions of the ClonExpress II One Step Cloning Kit.
Approximately 10,000 mixed-life-stage nematodes were collected from pine stems. The primer pairs ISH-BxICD1-F/ISH-BxICD1-R were employed to synthesize digoxygenin (DIG)-labeled sense and antisense DNA probes based on the BxICD1 fragments of 96–281 bp using a PCR DIG Probe Synthesis Kit (1636090910, Roche Applied Science, Rotkreuz, Switzerland). The primer pair ISH-BxICD2-F/ISH-BxICD2-R was employed to synthesize sense and antisense DNA probes based on the BxICD2 fragments of 103-311 bp in size. The in situ hybridization and staining of the nematodes was performed as described previously (De Boer et al., 1998). Nematodes were then examined using a Nikon ECLIPSE Ni microscope (Nikon, Tokyo, Japan).
The cell death assay was performed as described previously (Chen et al., 2017). Briefly, the pCambia1305.1:gene:eGFP constructs mentioned above were individually introduced into Agrobacterium tumefaciens GV3101. Then the transformed bacteria were suspended in a buffer containing 10 mM 2-(N-morpholino) ethanesulfonic acid (MES) (pH 5.5) and 200 μM acetosyringone until a 600 nm absorbance (OD600) of 0.6 was reached. Then, the A. tumefaciens cell suspensions were infiltrated into 4-week-old N. benthamiana leaves. After 3 days, the cell-death phenotype was observed and photographed. eGFP and the P. infestans elicitor gene INF1, cloned individually into the pCambia1305.1 vector, were used as the negative and positive control, respectively. The infiltration assay was performed three times, and five different plants with two inoculated leaves were used in each assay.
The secretion function of the BxICD1 SP was verified using the yeast signal sequence trap system described previously (Yin et al., 2018). Briefly, the SP sequence of BxICD1 was cloned into the pSUC2 vector containing the invertase gene but lacking Methionine (Met) and SP sequence to generate pSUC2:SPBxICD1-Invertase. Additionally, the SP of Phytophthora sojae RXLR effector Avr1b has been demonstrated to have secretion function in yeast (Dou et al., 2008) and was also cloned into pSUC2 to generate pSUC2:SPAvr1b-Invertase as a positive control. pSUC2:SPBxICD1-Invertase and pSUC2:SPAvr1b-Invertase were transformed into the yeast strain YTK12 by the lithium acetate method (Gietz and Schiestl, 2007; Yin et al., 2018). The yeast strains YTK12 and YTK12 carrying the pSUC2 empty vector were used as negative controls. YTK12 and YTK12 containing pSUC2-derived plasmids were grown on a CMD-W medium (0.67% yeast N base without amino acids, 0.075% W dropout supplement, 2% sucrose, 0.1% glucose, and 2% agar) to detect the expression of pSUC2-derived plasmids. The enzymatic activity of invertase was confirmed by the reduction of the dye 2,3,5-triphenyltetrazolium chloride (TTC) to the insoluble red-colored 1,3,5-triphenylformazan (TPF).
The subcellular localization assays were performed as described previously (Chen et al., 2017). Briefly, BxICD1 sequences with or without the native SP were cloned into the pCambia1305.1 vector to generate BxICD1:eGFP and BxICD1Δsp:eGFP, respectively. eGFP alone was used as the control. The constructs were used for the transformation of 4-week-old N. benthamiana leaves by agroinfiltration. Then the N. benthamiana plants were cultured for 3 days at 25°C under 16/8 LD. N. benthamiana leaves expressing the constructs were infiltrated with 30% glycerol for 20 min to induce plasmolysis for the observation of extracellular localization (Chen et al., 2021). The fluorescence was observed using an SP5 Leica confocal microscope (Nikon, Tokyo, Japan).
Western blotting was performed to verify the production of intact BxICD1:eGFP and BxICD1Δsp:eGFP fusion proteins in N. benthamiana, as described previously (Chen et al., 2021). Briefly, the total proteins from N. benthamiana leaves were extracted using RIPA lysis buffer. The proteins were denatured, separated on an SDS-PAGE gel, and transferred to a nitrocellulose membrane (PALL, Washington, NY, United States). After blocking with 5% (w/v) non-fat milk for 2 h at room temperature, the membranes were incubated with a primary mouse anti-GFP antibody (1:5000 dilution) (HT801, TransGen Biotech, Beijing, China) in a blocking solution for 2 h. Then membranes were incubated with an anti-mouse horseradish peroxidase-conjugated secondary antibody at a 1:5000 dilution (HS201, TransGen Biotech, Beijing, China). Proteins were visualized using the Immobilon Western Chemiluminescent system with Pierce ECL Western Blotting Substrate (Thermo Fisher Scientific, MA, USA). Ponceau S staining was used to assess equal loading (Goldman et al., 2016).
Electrolyte leakage assays were performed in N. benthamiana, as described previously (Yu et al., 2012). The A. tumefaciens harboring BxICD1:eGFP, BxICD1Δsp:eGFP, eGFP and INF1 were infiltrated into N. benthamiana leaves as described above. 36 h after agroinfiltration, five leaf discs (9 mm diameter) were floated on 5 mL distilled water for each sample and shaken at 25°C for 3 h. Then the conductivity of the solution was measured with a conductivity meter (DDS-307A, Shanghai INESA Scientific Instrument) to obtain the value A. In addition, the conductivity of the solution containing the leaf discs was measured after boiling for 30 min to obtain the value B. Ion leakage was calculated according to the formula (value A/value B) × 100%. The experiment was performed three times. Western blotting was performed to detect protein expression as described above.
Virus-induced gene silencing targeting NbBAK1 and NbSOBIR1 in N. benthamiana was performed as described previously (Lin et al., 2016). Briefly, the vectors pTRV1, pTRV2:NbBAK1, pTRV2:NbSOBIR1, and pTRV2:eGFP were individually transformed into A. tumefaciens GV3101. A. tumefaciens GV3101 harboring pTRV1 was mixed in a 1:1 ratio with those harboring pTRV2:NbBAK1, pTRV2:NbSOBIR1 or pTRV2:eGFP, respectively, and infiltrated into N. benthamiana leaves. The silencing efficiency of NbBAK1 and NbSOBIR1 was validated by RT-qPCR. RT-qPCR was performed as described above. The infiltration experiment was performed thrice and three different plants, with three inoculated leaves in each, were used in each assay.
The BxICD1 gene was silenced using in vitro RNAi, as described previously (Liu et al., 2022b). Briefly, the T7-promoter sequence was introduced in the sense and antisense direction of a 186-bp BxICD1 fragment using the primer pairs RNAi-T7BxICD1-F/RNAi-BxICD1-R and RNAi-BxICD1-F/RNAi-T7BxICD1-R. The PCR was performed as described above. The obtained PCR products were used to synthesize double-stranded RNA (dsRNA) using the Thermo T7 Transcription Kit (TSK-101, Toyobo, Shanghai, China) according to the manufacturer’s instructions. The same approach was used to synthesize dsRNA for eGFP using the primer pairs RNAi-T7eGFP-F/RNAi-eGFP-R and RNAi-eGFP-F/RNAi-T7eGFP-R. Nematodes were then inoculated in a dsRNA and a non-dsRNA solution at 25°C for 24 h. The treated nematodes were washed with ddH2O three times to remove external dsRNA. Subsequently, approximately 5,000 nematodes were collected for RNA extraction. The remaining nematodes were used for nematode virulence, migration, and reproduction analysis. The extent of BxICD1 silencing was assessed using RT-qPCR. RNA extraction and RT-qPCR were performed as described above, except for the different primer pairs that were used in the RT-qPCR.
Each 3-year-old P. massoniana seedling was inoculated with 1,000 B. xylophilus treated with a BxICD1 dsRNA, eGFP dsRNA, and non-dsRNA solution. Nine to ten pine trees were inoculated with each treatment. The experiment was repeated 3 times. Following the classification by Hu et al. (2022a), the morbidity degree of the P. massoniana seedlings was categorized into five different grades: 0, all needles were green; I, a few needles have turned yellow; II, approximately half of the needles have turned yellow or brown; III, most of the needles turned brown; and IV, the entire seedling was withered. The following formulas were used to calculate morbidity and the disease severity index.
The migration ability of B. xylophilus in P. massoniana was determined as described previously (Kusumoto et al., 2014). Briefly, approximately 800 nematodes treated with BxICD1 dsRNA, eGFP dsRNA, and non-dsRNA solutions were used to inoculate 3-year-old P. massoniana seedlings. 12 h after inoculation, the pine stem segments 1-2, 2-3, 3-4 and 4-5 cm below the inoculation point were used for nematode extraction using the Baermann funnel method. Nematodes isolated from these stem segments were counted. The experiment was repeated twice with four biological replicates each time. Statistically significant differences between treatments and controls were determined by Duncan’s multiple-range test.
The reproduction and feeding rate of B. xylophilus in fungi was assessed as described previously (Hu et al., 2022a). Approximately 100 nematodes treated with BxICD1 dsRNA, eGFP dsRNA, and non-dsRNA solutions were inoculated to PDA plates covered with Pestalotiopsis sp. After culturing at 25°C for 8 d, the nematodes were collected from each PDA plate using the Baermann funnel method and counted to calculate their reproduction. The amount of Pestalotiopsis sp. remaining on PDA plates was measured to determine the feeding rate of B. xylophilus. Six PDA plates were used for each treatment, and the experiment was repeated three times. Statistically significant differences between treatments and controls were determined by Duncan’s multiple-range test.
RNA was extracted from B. xylophilus during the mycetophagous and phytophagous stages to construct respective RNA-Seq libraries. In total, 181,144,358 raw reads were obtained from all samples. After filtering out low-quality sequences, 177,125,722 clean reads were retained. The GC content of individual libraries was between 48%-49%. On average, 92% of the clean reads mapped to the B. xylophilus reference genome (Supplementary Table 2). The high genome coverage of our RNA-Seq data indicated that the transcriptome data were reliable for the further bioinformatics analyses.
Differential expression analysis showed that 1,233 genes were differentially expressed between the mycetophagous and phytophagous stages of B. xylophilus. Compared with the mycetophagous stage, 765 genes were significantly up-regulated, and 468 genes were significantly down-regulated in the phytophagous stage (Figure 1A). To validate the identified DEGs based on the RNA-seq data, seven genes were selected and validated by RT-qPCR. Similar expression levels were observed for all seven genes in both RNA-seq and RT-qPCR datasets, further confirming the reliability of the RNA-seq transcriptome data (Figure 1B).
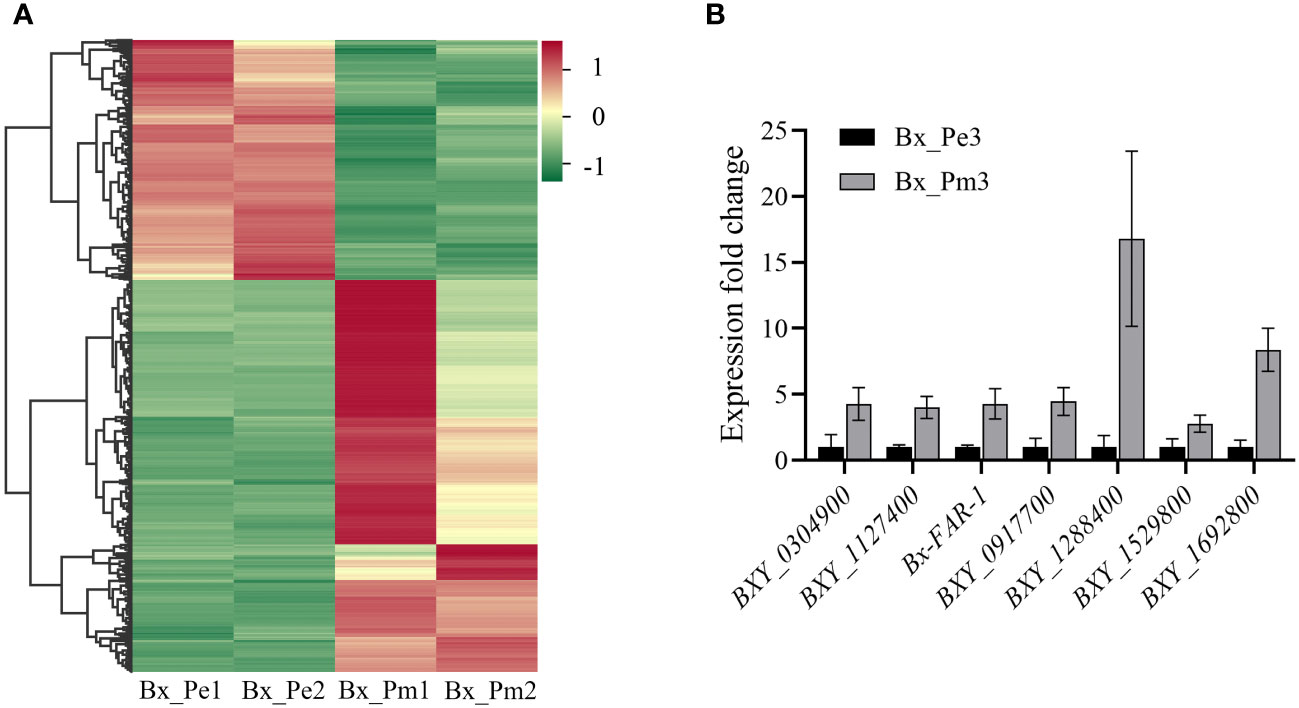
Figure 1 Differentially expressed genes at the mycetophagous and phytophagous stages of Bursaphelenchus xylophilus. (A) Heatmap of differentially expressed genes at the mycetophagous and phytophagous stages. (B) Validation of differentially expressed genes (DEGs). Validation of seven DEGs identified in the RNA-seq dataset by quantitative reverse transcription PCR. The relative expression levels were calculated using the 2−ΔΔCt method; data represent shown as the mean ± standard deviation (SD) of three replicated; the experiments were performed twice with similar results. Bx_Pe1, Bx_Pe2, and Bx_Pe3 correspond to the three B. xylophilus samples from the mycetophagous stage; Bx_Pm1, Bx_Pm2, and Bx_Pm3 correspond to the three B. xylophilus samples from the phytophagous stage.
Among the 765 up-regulated genes, 394 encoded proteins contained an SP and lacked transmembrane domains, as predicted by SignalP 5.0 and TMHMM-2.0. Of these, 210 genes had no functional annotations and were not identified in the transcriptome analysis of B. xylophilus infecting Pinus thunbergii. 31 of these 210 genes were further considered to be candidate genes encoding for effectors (Supplementary Table 3) based on the following criteria: Log2(fold change) > 3 and FPKM > 10 with regards to their differential expression, and a gene open reading frame (ORF) is smaller than 600 bp. From this set, 10 genes were randomly selected and were transiently expressed in N. benthamiana. The results showed that the genes BXY_0304900 and BXY_1398900 can induce cell death (ICD) in the presence of the native SP. They were named BxICD1 and BxICD2, respectively (Figure 2).
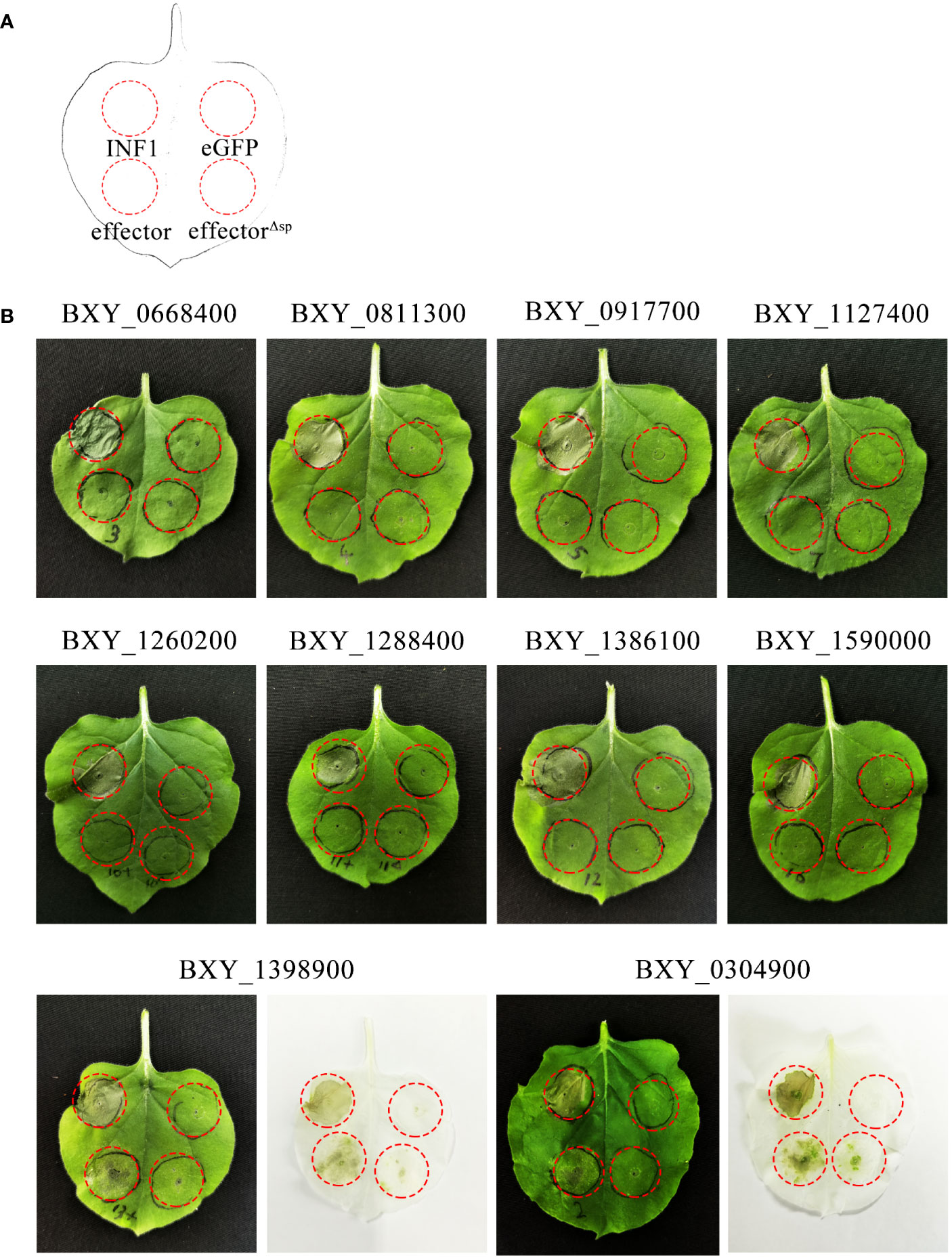
Figure 2 Screening of candidate effectors from Bursaphelenchus xylophilus capable of inducing plant cell death. (A) Schematic diagram of the location of Agrobacterium tumefaciens-injected tobacco leaves. INF1 is Phytophthora infestans elicitin, a known apoplastic protein that induces cell death in Nicotiana benthamiana, used as a positive control. eGFP, an enhanced green fluorescent protein that cannot induce cell death, was used as a negative control. “Effector” represents candidate effectors with the native signal peptide; “effectorΔsp” represents candidate effectors without the native signal peptide. (B) Ten candidate effectors were expressed in N. benthamiana leaves. The infiltration assay was performed thrice, and five different plants with two inoculated leaves were used in each assay. Similar results were obtained from all experiments.
The nematode tissue localization of BxICD1 and BxICD2 in the nematodes was further determined by in situ hybridization. Hybridization signals were detected in the esophageal gland cells with the DIG-labeled antisense cDNA probe of BxICD1 and in the intestinal terminus with the antisense cDNA probe of BxICD2. No hybridization signals were detected in B. xylophilus tissues with the sense cDNA probe of BxICD1 and BxICD2 (Figure 3A, Supplementary Figure 1). The results indicated that BxICD1 should be a nematode effector because it was expressed in the typical secretory organs of nematodes. Therefore, we selected the effector BxICD1 for further analysis.
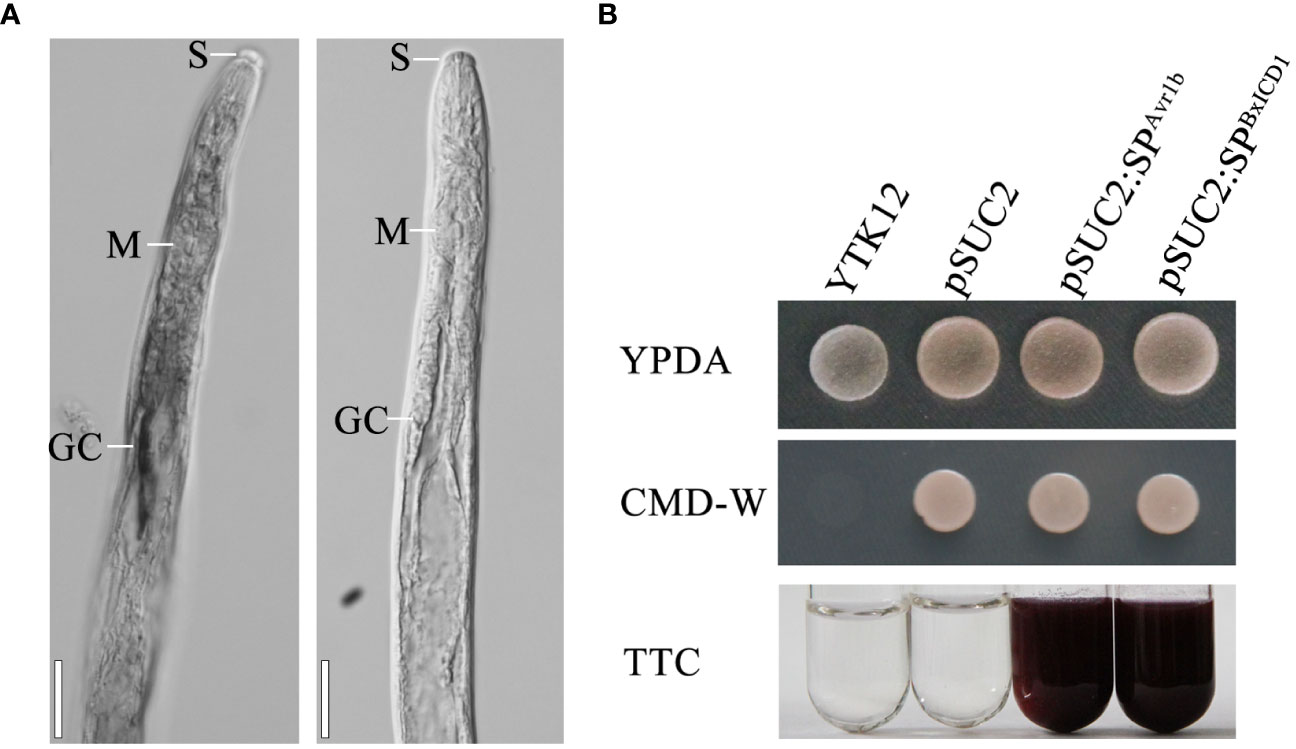
Figure 3 Localization of BxICD1 mRNA and secretion function analysis of BxICD1 signal peptide. (A) Localization of BxICD1 mRNA in esophageal gland cells of Bursaphelenchus xylophilus by in-situ hybridization. Fixed nematodes were hybridized with digoxigenin-labeled antisense (left) and sense (right) cDNA probes from BxICD1. S, stylet; M, median bulb; GC, esophageal gland. Scale bars = 50 µm (B) Secretion function analysis of the BxICD1 signal peptide. The predicted signal peptide of BxICD1 was cloned into the yeast vector pSUC2 to generate pSUC2:SPBxICD1-invertase constructs, and pSUC2: SPAvr1b-Invertase was used as a positive control. YTK12 can grow on YPDA plates. CMD-W media was used to ensure the expression of pSUC2-derived plasmids. Invertase can reduce 2,3,5-Triphenyltetrazolium Chloride (TTC) to insoluble red-colored 1,3,5-Triphenylformazan (TPF).
The BxICD1 gene has a 396-bp ORF, which encodes a 131-amino acid protein. The protein possesses a 19-amino acid SP, as predicted by the SignalP program, and has no putative transmembrane domain according to TMHMM. It also shows similarities to the proteins of B. xylophilus by a protein BLAST search, including CAD5219553 (62.3% identity), CAD5219557 (44.6% identity), CAD5219551 (40.2% identity) etc., while no sequences were matched to other organisms. A neighbor-joining (NJ) tree was constructed to examine the phylogenetic relationships among these proteins, with BxICD1 and CAD5219553 being located in the same branch (Supplementary Figure 2B).
To determine whether the SP of BxICD1 is functional, the predicted SP was cloned into the yeast vector pSUC2. The growth of yeast YTK12 yeast carrying pSUC2, pSUC2:SPBxICD1 and pSUC2:SPAvr1b constructs on CMD-W demonstrated the successful transformation with the plasmids. The yeast strain YTK12 carrying pSUC2:SPBxICD1-Invertase or the positive control pSUC2:SPAvr1b-Invertase could reduce TTC to the red-colored TPF. By contrast, no color change was observed in the YTK12 strain used as a negative control, and YTK12 carried the empty pSUC2 vector (Figure 3B). These results suggested that BxICD1 carries a functional secretory SP.
Based on the above results, BxICD1 could induce PCD in the presence of the native SP, therefore the subcellular localization of BxICD1 was determined. Three constructs, BxICD1:eGFP, BxICD1Δsp:eGFP and eGFP were generated and transiently expressed in N. benthamiana leaves. Based on the results, the BxICD1:eGFP fusion protein was accumulated in the edges of the tobacco cell, while BxICD1Δsp:eGFP and eGFP were localized in the cytoplasm (Figure 4A). Plasmolysis was further performed to distinguish the plasma membrane from the apoplast. After the plasmolysis treatment, BxICD1:eGFP fusion proteins were observed in the extracellular space, while BxICD1Δsp:eGFP and eGFP were still localized in the intracellular space (Figure 4B). Western blot analysis using a GFP antibody revealed bands at approximately 42, 40 and 27 kDa, corresponding to BxICD1:eGFP, BxICD1Δsp:eGFP, and eGFP, respectively (Figure 4C), indicating that BxICD1:eGFP and BxICD1Δsp:eGFP fusion proteins were expressed intact. At the same time, BxICD1:eGFP could induce tobacco cell death, while BxICD1Δsp:eGFP and eGFP could not (Figure 4D). In addition, the electrolyte leakage induced by INF1 or BxICD1:eGFP was significantly greater than that induced by BxICD1Δsp:eGFP or eGFP (Figure 4E). These results suggested that the presence of BxICD1 in N. benthamiana apoplasts triggers cell death.
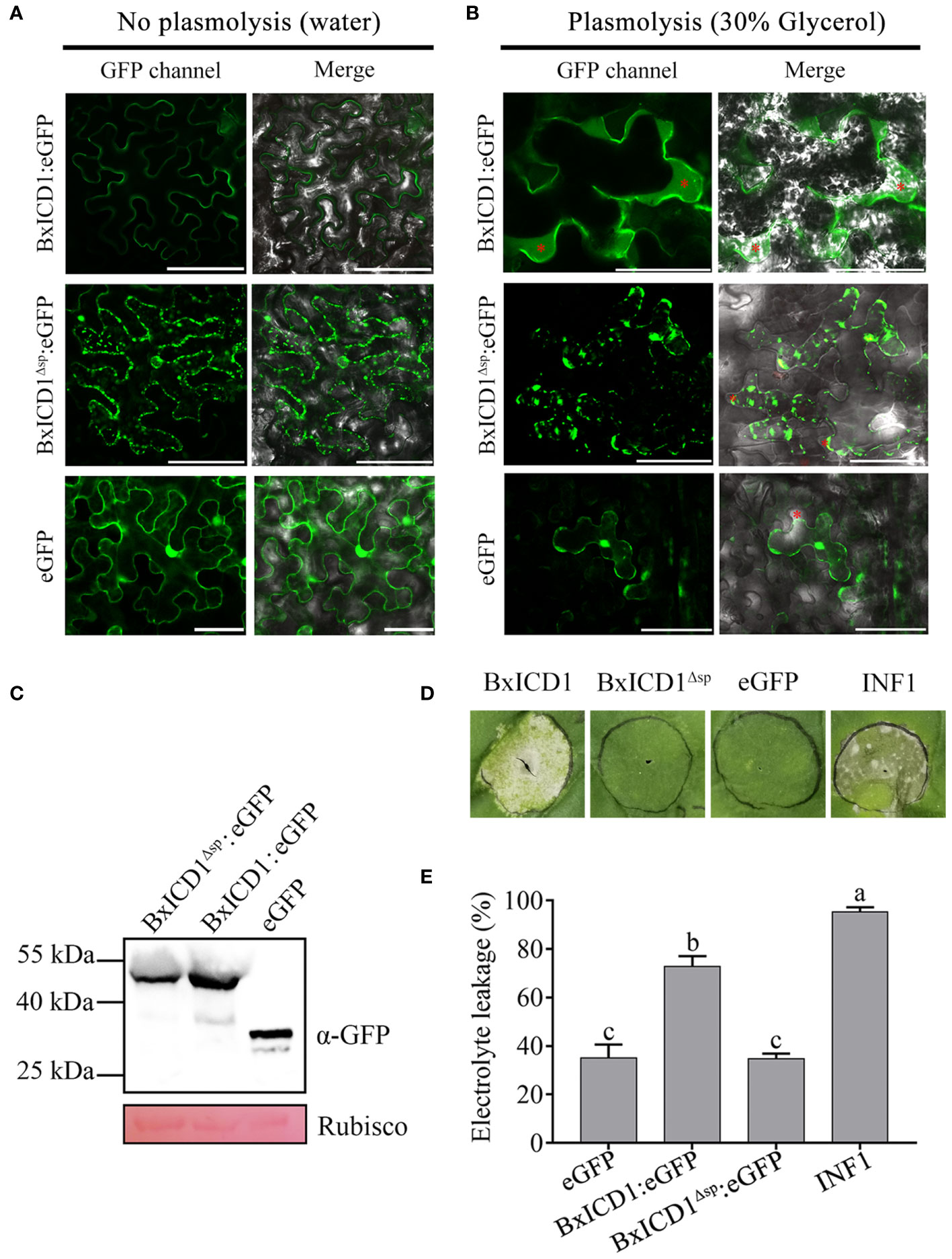
Figure 4 Apoplastic localization of BxICD1 is required for the induction of cell death in Nicotiana benthamiana. (A) Agrobacterium strain GV3101 carrying the fusion constructs BxICD1:eGFP, BxICD1Δsp:eGFP and the eGFP control were infiltrated in Nicotiana benthamiana leaves for transient expression analysis. (B) N. benthamiana leaves expressing BxICD1:eGFP, BxICD1:eGFPΔsp and eGFP were treated with 30% glycerol for plasmolysis. GFP signals were observed at 3 days after infiltration. The red asterisks indicate the apoplast region. Scale bars=50 µm. (C) BxICD1-triggered cell death in N. benthamiana. Agrobacterium strain GV3101 carrying the constructs BxICD1:eGFP, BxICD1Δsp:eGFP, eGFP, and INF1 were infiltrated in N. benthamiana leaves for transient expression analysis. The leaf phenotypes were observed after 3 days. (D) Quantification of cell death by measuring electrolyte leakage in N. benthamiana leaves at 3 days post-infiltration with constructs encoding the indicated proteins. Data represent the mean of three repeats ± SD. Three independent experiments were performed with similar results, with three technical replicates for each reaction. (E) Western blot analysis was performed to confirmed the expression of proteins in N. benthamiana. Ponceau S staining of RuBisCO was used as indicate the protein loading control.
Many studies have demonstrated that BAK1 and SOBIR1 are indispensable for cell death triggered by many apoplastic effectors (Liebrand et al., 2014). To determine whether NbBAK1 and NbSOBIR1 are involved in BxICD1-triggered cell death, they were knocked down using virus-induced gene silencing (VIGS) in N. benthamiana. Two weeks after viral inoculation, N. benthamiana plants were agroinfiltrated with the pCAMBIA1305.1:BxICD1:Flag, pCAMBIA1305.1:INF1, and pCAMBIA1305.1:eGFP : Flag-expressing constructs, respectively. Based on the results, INF1 failed to induce cell death in NbBAK1- or NbSOBIR1-silenced plants. Notably, although BxICD1 was unable to induce cell death in NbBAK1-silenced plants, it retained the capacity to induce cell death in NbSOBIR1-silenced plants (Figure 5A). Neither eGFP nor the EV, used as negative controls, triggered cell death in pTRV2:NbBAK1-, pTRV2:NbSOBIR1- and pTRV2:eGFP- infiltrated plants (Figure 5A). Western blot analysis was used to confirm BxICD1 protein expression in NbBAK1-, NbSOBIR1- and eGFP-silenced plants (Figure 5B), and RT-qPCR confirmed the successful silencing of NbBAK1 and NbSOBIR1 in plants (Figures 5C, D). Taken together, these results indicated that BxICD1-triggered cell death is dependent on NbBAK1.
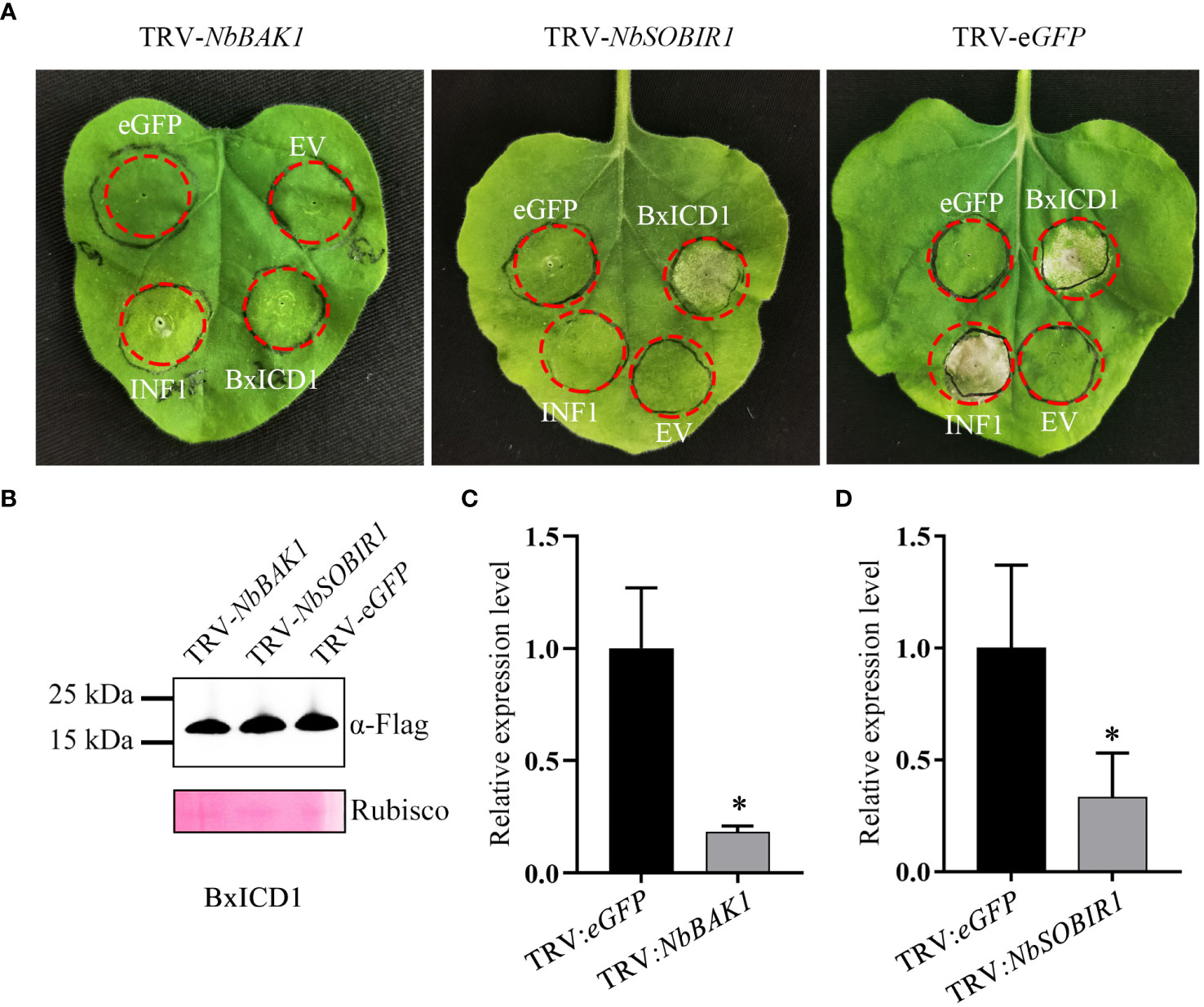
Figure 5 BxICD1-triggered cell death depends on NbBAK1. (A) NbBAK1, but not NbSOBIR1, is required for BxICD1-triggered cell death in Nicotiana benthamiana. N. benthamiana plants were subjected to VIGS by inoculation with TRV constructs (TRV:eGFP, TRV : NbBAK1, or TRV : NbSOBIR1). Three weeks after inoculation, BxICD1, INF1, the empty vector (EV), and eGFP were transiently expressed in the gene-silenced leaves and photographed after 3 d. The infiltration experiment was performed thrice, with three different plants with three inoculated leaves were used in each assay. All experiments were performed with similar results. (B) Western blot analysis confirmed the expression of BxICD1 protein in gene-silenced plants. Ponceau S staining of RuBisCO was used as indicate the protein loading control. (C, D) The relative expression levels of NbBAK1 and NbSOBIR1 in gene-silenced plant were determined by RT-qPCR. Data represent the mean of three replicates ± SD. Three independent experiments were performed with similar results. Asterisks indicate significant differences (P<0.05).
To assess the role of BxICD1 in nematode parasitism, an RNAi assay was performed by soaking B. xylophilus in BxICD1 dsRNA. The transcript levels of BxICD1 were significantly decreased after incubation with the BxICD1 dsRNA, compared to the treatments with eGFP dsRNA and non-dsRNA solution (Figure 6A), demonstrating that RNAi was successful.
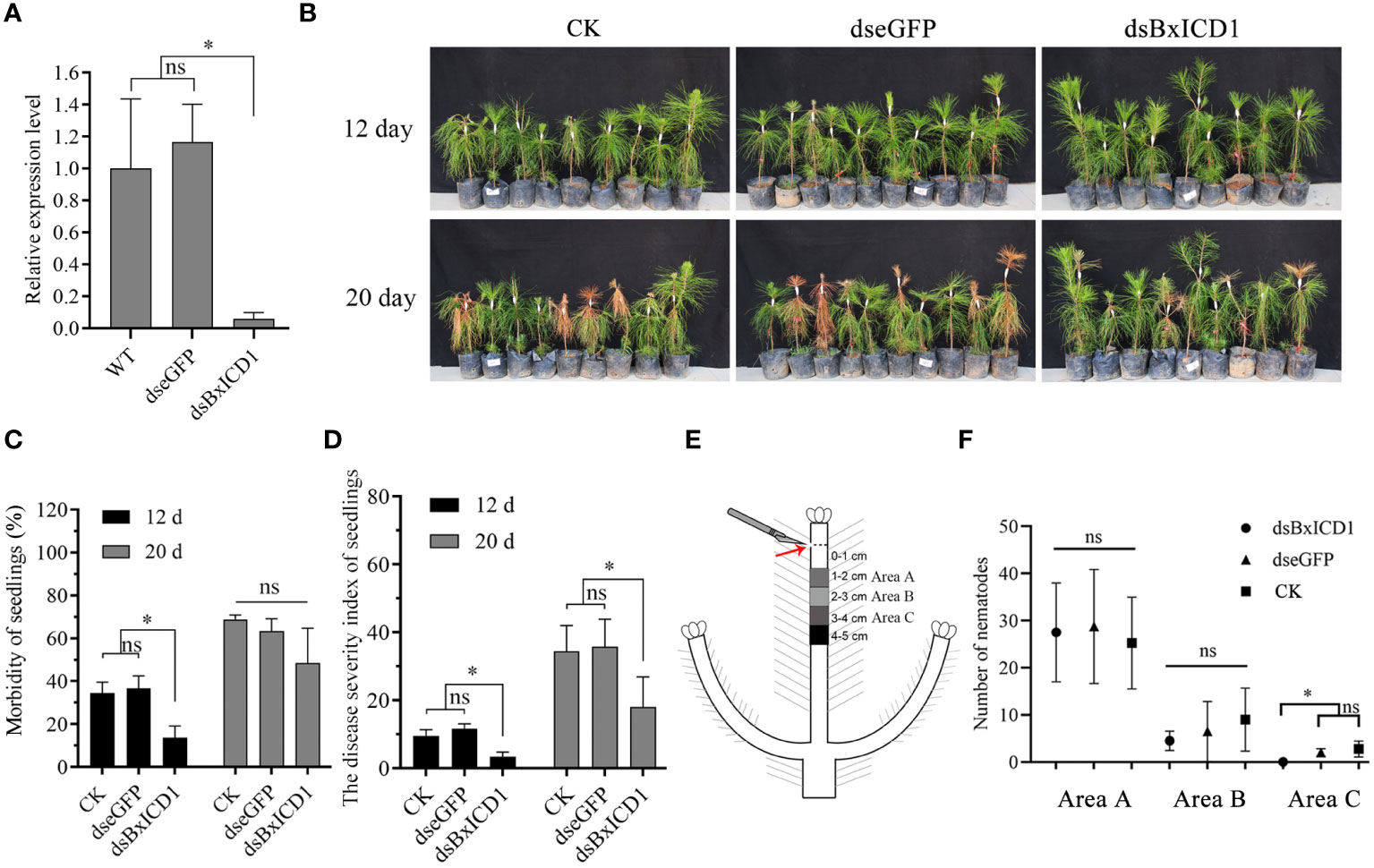
Figure 6 BxICD1 contributes to the virulence and migration of Bursaphelenchus xylophilus. (A) BxICD1 silencing efficiency after treatment with BxICD1 dsRNA in B. xylophilus. Asterisks indicate significant differences (P<0.05). ns indicates no significant differences (P≥0.05). (B) Representative photographs of Pinus massoniana seedlings at 12 days and 20 days post-inoculation. dseGFP, dsBxICD1 and CK correspond to nematodes inoculated in eGFP-, BxICD1- and non-dsRNA solution, respectively. (C) The morbidity of Pinus massoniana seedlings inoculated with dseGFP-, dsBxICD1-, and non-dsRNA solution -treated nematodes. (D) The disease severity index of P. massoniana seedlings inoculated with dseGFP-, dsBxICD1-, and non-dsRNA solution-treated nematodes. (E) Schematic depiction of nematode inoculation and sampling. (F) The number of nematodes in Area A, Area B, and Area C wood sections under three different treatments. “ns” indicates no sigificant difference (p≥0.05).
Subsequently, inoculation experiments were performed using B. xylophilus that underwent different pre-treatment. Compared with the eGFP dsRNA-treated and H2O-treated nematodes at 12 dpi, BxICD1 dsRNA-treated nematodes caused to a significantly lower morbidity and disease severity index (DSI) in P. massoniana. The morbidity was calculated at 36.7%, 34.4%, and 13.7% after inoculation with eGFP dsRNA-, H2O- and BxICD1 dsRNA-treated nematodes, respectively (Figure 6B), while corresponding DSI was 11.7, 9.5, and 3.4 (Figure 6C). At 20 dpi, the DSI of P. massoniana inoculated with BxICD1 dsRNA-treated nematodes was still significantly lower compared with P. massoniana inoculated with eGFP dsRNA and non-dsRNA solution-treated nematodes. However, no significant differences in morbidity were observed (Figures 6C, D). Moreover, at 12 h post-inoculation, B. xylophilus was isolated from pine stem segments at different distances from the inoculation site. No significant differences were observed in the number of BxICD1 dsRNA, eGFP dsRNA, and the non-dsRNA solution-treated nematodes at pine stem segments 1-2 and 2-3 cm below the inoculation site. However, the number of BxICD1 dsRNA-treated nematodes was significantly lower compared with eGFP dsRNA and non-dsRNA solution-treated nematodes isolated from pine stem segments 3-4 cm below the inoculation site. No nematodes were isolated from pine stem segments 4-5 cm below the inoculation point in all treatments (Figures 6E, F).
In addition, BxICD1 dsRNA, eGFP dsRNA, and the non-dsRNA solution-treated nematodes were cultured on a PDA medium covered with Pestalotiopsis sp. After eight days, no significant differences were observed in the amount of nematodes and fungi mycelium in different treatments (Supplementary Figure 3). The above results showed that BxICD1 affected B. xylophilus parasitism and migration in P. massoniana, but did not affect the feeding rate and reproduction of B. xylophilus in fungi.
Effectors are the key factors affecting PPNs virulence and parasitism against hosts (Goverse and Smant, 2014). In recent years, a growing number of PPN candidate effectors have been identified by transcriptome sequencing. More than 100 candidate effectors from B. xylophilus infecting P. thunbergii have been obtained, while a few have been functionally characterized (Tsai et al., 2016; Hu et al., 2019). Interestingly, it was reported that different proteins were detected in the secretomes of B. xylophilus during the infection of different pine hosts (Silva et al., 2021). Accordingly, we screened effectors from B. xylophilus infecting P. massoniana for the first time. As result, 394 candidate effectors were identified through transcriptome data and bioinformatics analysis. Among them, 210 were different from the candidate effectors of B. xylophilus infesting P. thunbergii and were not annotated in the HMMER homology database, which might be pioneers effectors against P. massoniana. One of them, named BxICD1, was found to be significantly upregulated in the early parasitic stages of B. xylophilus and specifically present in the esophageal gland cell, a classical effector secretion organ of nematodes (Mitchum et al., 2013). Furthermore, the signal peptide of BxICD1 was confirmed to be functional based on yeast system assays. These data suggest that BxICD1 is a novel effector of B. xylophilus.
In this study, we found that BxICD1 could induce PCD in the extracellular space of N. benthamiana, which was dependent on NbBAK1. As is known, PCD has different effects on different nutritional pathogens with different lifestyles and feeding strategies. Necrotrophic pathogens need to kill cells and tissues of host plants, and then absorb nutrients. Therefore, PCD facilitates the parasitism of necrotrophic pathogens (Laluk and Mengiste, 2010). However, biotrophic pathogens must acquire nutrients directly from living plant cells and tissues without immediately killing host cells or tissues. Thus, PCD is often detrimental to biotrophic pathogens (Gebrie, 2016). Additionally, hemibiotrophs are biotrophic in the early stage and necrotrophic in the late stage of parasitism. Therefore, PCD inhibits the infection by hemibiotrophs in the early parasitism stage, while it promotes hemibiotroph colonization and facilitates pathogen transition from a biotrophic to a necrotrophic state in the late parasitism stage (Jia et al., 2000; Orbach et al., 2000; Qutob et al., 2002). PPNs are biotrophic pathogens, and it is generally considered that PCD negatively affects nematode parasitism. For example, the potato protein Gpa2 recognizes the Globodera pallida effector Gp-Rbp-1 and triggers PCD, which inhibits nematode parasitism (Sacco et al., 2009). A hypersensitive response-like reaction was observed in the Meloidogyne graminicola-resistant rice plants but not in the M. graminicola-susceptible rice plants under nematode infection (Cabasan et al., 2014). To survive, PPNs secrete effectors to suppress PCD. In recent years, many PPN effectors with PCD-inhibiting properties have been identified, especially from sedentary endoparasitic phytonematodes, e.g., root-knot nematodes and cyst nematodes (Postma et al., 2012; Chen et al., 2017; Song et al., 2021; Chen et al., 2022). In the migratory endoparasitic phytonematode B. xylophilus, six effectors were also found to inhibit PCD (Hu et al., 2021; Wen et al., 2021, Wen et al., 2022; Zhang et al., 2022; Hu et al., 2022a; Qiu et al., 2023). Interestingly, five effectors from B. xylophilus were confirmed to have the ability to activate PCD. One of them not only induced PCD but also inhibited nematode parasitism. However, BxSapB1, BxSapB2, and BxSapB3, induced PCD but at the same time also enhanced nematode virulence (Huang et al., 2019; Hu et al., 2019, Hu et al., 2020; Zhao et al., 2020; Shinya et al., 2021). In the present study, we found that BxICD1 also promots parasitism of B. xylophilus based on RNAi gene silencing assays. This seemingly paradoxical phenomenon was also observed in effectors from oomycetes and fungi, such as PsXEG1 (Ma et al., 2015) from Phytophthora sojae, Avh238 (Yang et al., 2017) from P. essential, PlAvh142 (Situ et al., 2020) from Peronophythora litchi and Fg12 (Yang et al., 2020) from F. graminearum. Two hypotheses have been proposed to explain this phenomenon. First, the accumulation of effectors is insufficient to induce PCD under natural conditions. Second, other interacting effectors or proteins could suppress PCD activated by effectors during the successful parasitism of pathogens (Ma et al., 2015; Yang et al., 2017; Situ et al., 2020).
This study demonstrated that BxICD1 contributed to nematode migration in P. massoniana, but was not involved and did not affect nematode feeding and reproduction in fungi based on the RNAi assays results. It has been reported that pine trees develop resistance by inducing pine resin secretion and lignification in the resin canals. Lignin biosynthesis in cell walls can effectively inhibits the migration of B. xylophilus (Kusumoto et al., 2014). In pine trees, highly virulent B. xylophilus strains can rapidly destroy the resin-secreting parenchyma cells of cortical and xylem resin canals before cell wall lignification (Jin and Ye, 2007), while weakly virulent strains do not have such capacity. Taken together, the effector BxICD1 has the capacity to induce PCD, resulting in the destruction of parenchyma cells in resin canals, which is beneficial for B. xylophilus virulence and migration.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: BioProject, PRJNA1027064.
The manuscript presents research on animals that do not require ethical approval for their study.
ZL: Writing – original draft, Writing – review & editing, Data curation, Formal analysis, Investigation, Methodology, Software. HW: Writing – original draft, Methodology, Writing – review & editing, Formal analysis, Investigation. YC: Writing – original draft, Formal analysis, Investigation. XS: Writing – original draft, Formal analysis, Investigation. XH: Writing – original draft, Formal analysis, Investigation. QH: Investigation, Writing – original draft, Formal analysis. KZ: Methodology, Writing – review & editing, Writing – original draft. JL: Writing – original draft, Writing – review & editing, Methodology. BL: Writing – review & editing, Formal analysis, Investigation, Methodology.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the 2023 Special Fund for Affairs of Natural Resources—Project of Ecological Construction of Forestry, (Guangdong Forestry Finance [2022] No.8) and Dynamic Monitoring of Forest Nematode Diversity in Guangdong Province (Guangdong Resources and Environment Financial [2020] No.28).
We acknowledge Guanghui Kong (South China Agricultural University, China) for providing the plasmid pSUC2 and yeast strain YTK12, and Lili Hu (Guangdong Academy of Forestry) for providing Pinus massoniana seedlings.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2024.1357141/full#supplementary-material
Supplementary Figure 1 | Localization of BxICD2 mRNA in the intestine terminus of Bursaphelenchus xylophilus by in-situ hybridization. DIG, digoxygenin; Scale bars = 50 µm.
Supplementary Figure 2 | Multiple sequence alignment and phylogenetic analyses of BxICD1 (BXY_0304900). (A) Multiple sequence alignment of the BxICD1 protein with other homologous sequences from Bursaphelenchus xylophilus. (B) The phylogenetic tree of BxICD1 protein homologs.
Supplementary Figure 3 | BxICD1 does not affect reproduction or fungal feeding rate in Bursaphelenchus xylophilus (A) The propagation rate of B. xylophilus after culture on Pestalotiopsis spp. for 8 days. Data represent the means, and the error bars represent ± standard deviation of six biological replicates. (B) The fungal feeding rate of B. xylophilus grown on Pestalotiopsis spp. Two independent experiments were performed with similar results. dseGFP, dsBxICD1 and CK indicate nematodes inoculated in eGFP-, BxICD1- and non-dsRNA solution, respectively.
Benjamini, Y., Hochberg, Y. (1995). Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc Series. B Stat. Methodol. 57, 289–300. doi: 10.1111/j.2517-6161.1995.tb02031.x
Cabasan, M. T. N., Kumar, A., Bellafiore, S., De Waele, D. (2014). Histopathology of the rice root-knot nematode, Meloidogyne graminicola, on Oryza sativa and O. glaberrima. Nematology 16, 73–81. doi: 10.1163/15685411-00002746
Chen, J., Chen, S., Xu, C., Yang, H., Achom, M., Wang, X. (2022). A key virulence effector from cyst nematodes targets host autophagy to promote nematode parasitism. New Phytol. 237, 1374–1390. doi: 10.1111/nph.18609
Chen, J., Li, Z., Lin, B., Liao, J., Zhuo, K. (2021). A Meloidogyne graminicola pectate lyase is involved in virulence and activation of host defense responses. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.651627
Chen, J., Lin, B., Huang, Q., Hu, L., Zhuo, K., Liao, J. (2017). A novel Meloidogyne graminicola effector, MgGPP, is secreted into host cells and undergoes glycosylation in concert with proteolysis to suppress plant defenses and promote parasitism. PLoS Pathog. 13, e1006301. doi: 10.1371/journal.ppat.1006301
Cui, H., Jin, H., Liu, Q., Yan, Z., Ding, L., Qin, B. (2014). Nematicidal metabolites from roots of Stellera chamaejasme against Bursaphelenchus xylophilus and Bursaphelenchus mucronatus. Pest Manage. Sci. 70, 827–835. doi: 10.1002/ps.3625
De Boer, J. M., Yan, Y., Smant, G., Davis, E. L., Baum, T. J. (1998). In-situ hybridization to messenger RNA in Heterodera glycines. J. Nematol. 30, 309–312.
Dou, D., Kale, S. D., Wang, X., Jiang, R. H., Bruce, N. A., Arredondo, F. D., et al. (2008). RXLR-mediated entry of Phytophthora sojae effector Avr1b into soybean cells does not require pathogen-encoded machinery. Plant Cell 20, 1930–1947. doi: 10.1105/tpc.107.056093
Edwards, O. R., Linit, M. J. (1992). Transmission of Bursaphelenchus xylophilus through oviposition wounds of Monochamm carolinensis (Coleoptera: Cerambycidae). J. Nematol. 24, 133–139.
Futai, K. (2013). Pine wood nematode, Bursaphelenchus xylophilus. Annu. Rev. Phytopathol. 51, 61–83. doi: 10.1146/annurev-phyto-081211-172910
Gebrie, S. A. (2016). Biotrophic fungi infection and plant defense mechanism. J. Plant Pathol. Microbiol. 7, 2. doi: 10.4172/2157-7471.1000378
Gietz, R. D., Schiestl, R. H. (2007). High-efficiency yeast transformation using the LiAc/SS carrier DNA/PEG method. Nat. Protoc. 2, 38–41. doi: 10.1038/nprot.2007.15
Goldman, A., Harper, S., Speicher, D. W. (2016). Detection of proteins on blot membranes. Curr. Protoc. Protein Sci. 86, 10.8.1–10.8.11. doi: 10.1002/cpps.15
Goverse, A., Smant, G. (2014). The activation and suppression of plant innate immunity by parasitic nematodes. Annu. Rev. Phytopathol. 52, 243–265. doi: 10.1146/annurev-phyto-102313-050118
Haegeman, A., Mantelin, S., Jones, J. T., Gheysen, G. (2012). Functional roles of effectors of plant-parasitic nematodes. Gene 492, 19–31. doi: 10.1016/j.gene.2011.10.040
Heese, A., Hann, D. R., Gimenez-Ibanez, S., Jones, A. M., He, K., Li, J., et al. (2007). The receptor-like kinase SERK3/BAK1 is a central regulator of innate immunity in plants. Proc. Natl. Acad. Sci. U.S.A. 104, 12217–12222. doi: 10.1073/pnas.0705306104
Hu, L. J., Wu, X. Q., Ding, X. L., Ye, J. R. (2021). Comparative transcriptomic analysis of candidate effectors to explore the infection and survival strategy of Bursaphelenchus xylophilus during different interaction stages with pine trees. BMC Plant Biol. 21, 224. doi: 10.1186/s12870-021-02993-9
Hu, L. J., Wu, X. Q., Li, H. Y., Wang, Y. C., Huang, X., Wang, Y., et al. (2020). BxCDP1 from the pine wood nematode Bursaphelenchus xylophilus is recognized as a novel molecular pattern. Mol. Plant Pathol. 21, 923–935. doi: 10.1111/mpp.12939
Hu, L. J., Wu, X. Q., Li, H. Y., Zhao, Q., Wang, Y. C., Ye, J. R. (2019). An Effector, BxSapB1, induces cell death and contributes to virulence in the pine wood nematode Bursaphelenchus xylophilus. Mol. Plant Microbe Interact. 32, 452–463. doi: 10.1094/MPMI-10-18-0275-R
Hu, L. J., Wu, X. Q., Wen, T. Y., Qiu, Y. J., Rui, L., Zhang, Y., et al. (2022a). A Bursaphelenchus xylophilus effector, BxSCD3, suppresses plant defense and contributes to virulence. Int. J. Mol. Sci. 23, 6417. doi: 10.3390/ijms23126417
Hu, L. J., Wu, X. Q., Wen, T. Y., Ye, J. R., Qiu, Y. J., Rui, L., et al. (2022b). The key molecular pattern BxCDP1 of Bursaphelenchus xylophilus induces plant immunity and enhances plant defense response via two small peptide regions. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.937473
Huang, X., Hu, L. J., Wu, X. Q. (2019). Identification of a novel effector BxSapB3 that enhances the virulence of pine wood nematode Bursaphelenchus xylophilus. Acta Biochim. Biophys. Sin. 51, 1071–1078. doi: 10.1093/abbs/gmz100
Jia, Y., McAdams, S. A., Bryan, G. T., Hershey, H. P., Valent, B. (2000). Direct interaction of resistance gene and avirulence gene products confers rice blast resistance. EMBO J. 19, 4004–4014. doi: 10.1093/emboj/19.15.4004
Jin, G., Ye, J. R. (2007). Histopathological study on the nematode in seedling of Pinus thunbergii. J. Nanjing Forestry Uni. 50, 115–120. doi: 10.3969/j.jssn.1000-2006.2007.04.026
Kikuchi, T., Cotton, J. A., Dalzell, J. J., Hasegawa, K., Kanzaki, N., McVeigh, P., et al. (2011). Genomic insights into the origin of parasitism in the emerging plant pathogen Bursaphelenchus xylophilus. PLoS Pathog. 7, e1002219. doi: 10.1371/journal.ppat.1002219
Kikuchi, T., Shibuya, H., Aikawa, T., Jones, J. T. (2006). Cloning and characterization of pectate lyases expressed in the esophageal gland of the pine wood nematode Bursaphelenchus xylophilus. Mol. Plant Microbe Interact. 19, 280–287. doi: 10.1094/MPMI-19-0280
Kim, D., Paggi, J. M., Park, C., Bennett, C., Salzberg, S. L. (2019). Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 37, 907–915. doi: 10.1038/s41587-019-0201-4
Kudla, U., MILAC, A. L., Qin, L., Overmars, H., Roze, E., Holterman, M., et al. (2007). Structural and functional characterization of a novel, host penetration-related pectate lyase from the potato cyst nematode Globodera rostochiensis. Mol. Plant Pathol. 8, 293–305. doi: 10.1111/j.1364-3703.2007.00394.x
Kumar, S., Stecher, G., Li, M., Knyaz, C., Tamura, K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547. doi: 10.1093/molbev/msy096
Kusumoto, D., Yonemichi, T., Inoue, H., Hirao, T., Watanabe, A., Yamada, T. (2014). Comparison of histological responses and tissue damage expansion between resistant and susceptible Pinus thunbergii infected with pine wood nematode Bursaphelenchus xylophilus. J. For. Res. 19, 285–294. doi: 10.1007/s10310-013-0417-y
Laluk, K., Mengiste, T. (2010). Necrotroph attacks on plants: wanton destruction or covert extortion? arabidopsis Book 8, e0136. doi: 10.1199/tab.0136
Lee, S., Kim, J., Kim, M. S., Min, C. W., Kim, S. T., Choi, S. B., et al. (2023). The Phytophthora nucleolar effector Pi23226 targets host ribosome biogenesis to induce necrotrophic cell death. Plant Commun. 4, 100606. doi: 10.1016/j.xplc.2023.100606
Liao, Y., Smyth, G. K., Shi, W. (2014). featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930. doi: 10.1093/bioinformatics/btt656
Liebrand, T. W., van den Burg, H. A., Joosten, M. H. (2014). Two for all: receptor-associated kinases SOBIR1 and BAK1. Trends Plant Sci. 19, 123–132. doi: 10.1016/j.tplants.2013.10.003
Lin, B. R., Zhuo, K., Chen, J. S., Hu, L. L., Sun, L. H., Wang, X. H., et al. (2016). A novel nematode effector suppresses plant immunity by activating host reactive oxygen species-scavenging system. New Phytol. 209, 1159–1173. doi: 10.1111/nph.13701
Liu, B., Xie, Y., Yin, H., Zhou, Z., Liu, Q. (2022a). Identification and defensive characterization of PmCYP720B11v2 from Pinus massoniana. Int. J. Mol. Sci. 23, 6640. doi: 10.3390/ijms23126640
Liu, X., Zhou, X., Zhou, L., Hu, J., Guo, K. (2022b). Application of RNA interference in the pinewood nematode, bursaphelenchus xylophilus. J. Vis. Exp. 181, e63645. doi: 10.3791/63645
Livak, K. J., Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2–ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Ma, Z., Song, T., Zhu, L., Ye, W., Wang, Y., Shao, Y., et al. (2015). A Phytophthora sojae glycoside hydrolase 12 protein is a major virulence factor during soybean infection and is recognized as a PAMP. Plant Cell 27, 2057–2072. doi: 10.1105/tpc.15.00390
Mitchum, M. G., Hussey, R. S., Baum, T. J., Wang, X., Elling, A. A., Wubben, M., et al. (2013). Nematode effector proteins: an emerging paradigm of parasitism. New Phytol. 199, 879–894. doi: 10.1111/nph.12323
Modi, A., Vai, S., Caramelli, D., Lari, M. (2021). The illumina sequencing protocol and the novaSeq 6000 system. Methods Mol. Biol. 2242, 15–42. doi: 10.1007/978-1-0716-1099-2_2
Orbach, M. J., Farrall, L., Sweigard, J. A., Chumley, F. G., Valent, B. (2000). A telomeric avirulence gene determines efficacy for the rice blast resistance gene Pi-ta. Plant Cell 12, 2019–2032. doi: 10.1105/tpc.12.11.2019
Petitot, A. S., Dereeper, A., Agbessi, M., Silva, C., Guy, J., Ardisson, M., et al. (2016). Dual RNA-seq reveals Meloidogyne graminicola transcriptome and candidate effectors during the interaction with rice plants. Mol. Plant Pathol. 17, 860–874. doi: 10.1111/mpp.12334
Popeijus, H., Overmars, H., Jones, J., Blok, V., Goverse, A., Helder, J., et al. (2000). Degradation of plant cell walls by a nematode. Nature 406, 36–37. doi: 10.1038/35017641
Postma, W. J., Slootweg, E. J., Rehman, S., Finkers-Tomczak, A., Tytgat, T. O., van Gelderen, K., et al. (2012). The effector SPRYSEC-19 of Globodera rostochiensis suppresses CC-NB-LRR-mediated disease resistance in plants. Plant Physiol. 160, 944–954. doi: 10.1104/pp.112.200188
Qiu, Y. J., Wu, X. Q., Wen, T. Y., Hu, L. J., Rui, L., Zhang, Y., et al. (2023). The Bursaphelenchus xylophilus candidate effector BxLip-3 targets the class I chitinases to suppress immunity in pine. Mol. Plant Pathol. 24, 1033–1046. doi: 10.1111/mpp.13334
Qutob, D., Kamoun, S., Gijzen, M. (2002). Expression of a Phytophthora sojae necrosis-inducing protein occurs during transition from biotrophy to necrotrophy. Plant J. 32, 361–373. doi: 10.1046/j.1365-313X.2002.01439.x
Sacco, M. A., Koropacka, K., Grenier, E., Jaubert, M. J., Blanchard, A., Goverse, A., et al. (2009). The cyst nematode SPRYSEC protein RBP-1 elicits Gpa2-and RanGAP2-dependent plant cell death. PLoS Pathog. 5, e1000564. doi: 10.1371/journal.ppat.1000564
Shinya, R., Kirino, H., Morisaka, H., Takeuchi-Kaneko, Y., Futai, K., Ueda, M. (2021). Comparative secretome and functional analyses reveal glycoside hydrolase family 30 and cysteine peptidase as virulence determinants in the pinewood nematode Bursaphelenchus xylophilus. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.640459
Silva, H., Anjo, S. I., Manadas, B., Abrantes, I., Fonseca, L., Cardoso, J. M. (2021). Comparative analysis of Bursaphelenchus xylophilus secretome under Pinus pinaster and P. pinea stimuli. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.668064
Situ, J., Jiang, L., Fan, X., Yang, W., Li, W., Xi, P., et al. (2020). An RXLR effector PlAvh142 from Peronophythora litchii triggers plant cell death and contributes to virulence. Mol. Plant Pathol. 21, 415–428. doi: 10.1111/mpp.12905
Song, H., Lin, B., Huang, Q., Sun, L., Chen, J., Hu, L., et al. (2021). The Meloidogyne graminicola effector MgMO289 targets a novel copper metallochaperone to suppress immunity in rice. J. Exp. Bot. 72, 5638–5655. doi: 10.1093/jxb/erab208
Trapnell, C., Williams, B. A., Pertea, G., Mortazavi, A., Kwan, G., Van Baren, M. J., et al. (2010). Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28, 511–515. doi: 10.1038/nbt.1621
Tsai, I. J., Tanaka, R., Kanzaki, N., Akiba, M., Yokoi, T., Espada, M., et al. (2016). Transcriptional and morphological changes in the transition from mycetophagous to phytophagous phase in the plant-parasitic nematode Bursaphelenchus xylophilus. Mol. Plant Pathol. 17, 77–83. doi: 10.1111/mpp.12261
Vanholme, B., Van Thuyne, W., Vanhouteghem, K., De Meutter, J. A. N., Cannoot, B., Gheysen, G. (2007). Molecular characterization and functional importance of pectate lyase secreted by the cyst nematode Heterodera schachtii. Mol. Plant Pathol. 8, 267–278. doi: 10.1111/j.1364-3703.2007.00392.x
Verma, A., Lee, C., Morriss, S., Odu, F., Kenning, C., Rizzo, N., et al. (2018). The novel cyst nematode effector protein 30D08 targets host nuclear functions to alter gene expression in feeding sites. New Phytol. 219, 697–713. doi: 10.1111/nph.15179
Vieira, P., Gleason, C. (2019). Plant-parasitic nematode effectors—insights into their diversity and new tools for their identification. Curr. Opin. Plant Biol. 50, 37–43. doi: 10.1016/j.pbi.2019.02.007
Wang, S., Yang, S., Dai, K., Zheng, W., Zhang, X., Yang, B., et al. (2023). The effector Fg62 contributes to Fusarium graminearum virulence and induces plant cell death. Phytopathol. Res. 5, 12. doi: 10.1186/s42483-023-00167-z
Wen, T. Y., Wu, X. Q., Hu, L. J., Qiu, Y. J., Rui, L., Zhang, Y., et al. (2021). A novel pine wood nematode effector, BxSCD1, suppresses plant immunity and interacts with an ethylene-forming enzyme in pine. Mol. Plant Pathol. 22, 1399–1412. doi: 10.1111/mpp.13121
Wen, T. Y., Wu, X. Q., Ye, J. R., Qiu, Y. J., Rui, L., Zhang, Y. (2022). A Bursaphelenchus xylophilus pathogenic protein Bx-FAR-1, as potential control target, mediates the jasmonic acid pathway in pines. Pest Manage. Sci. 78, 1870–1880. doi: 10.1002/ps.6805
Xue, L., Lie, G., Lu, G., Shao, Y. (2013). Allometric scaling among tree components in Pinus massoniana stands with different sites. Ecol. Res. 28, 327–333. doi: 10.1007/s11284-012-1021-x
Yang, B., Wang, Q., Jing, M., Guo, B., Wu, J., Wang, H., et al. (2017). Distinct regions of the Phytophthora essential effector Avh238 determine its function in cell death activation and plant immunity suppression. New Phytol. 214, 361–375. doi: 10.1111/nph.14430
Yang, B., Wang, Y., Tian, M., Dai, K., Zheng, W., Liu, Z., et al. (2020). Fg12 ribonuclease secretion contributes to Fusarium graminearum virulence and induces plant cell death. J. Integr. Plant Biol. 63, 365–377. doi: 10.1111/jipb.12997
Yin, W., Wang, Y., Chen, T., Lin, Y., Luo, C. (2018). Functional evaluation of the signal peptides of secreted proteins. Bio Protoc. 8, e2839. doi: 10.21769/BioProtoc.2839
Yu, X., Tang, J., Wang, Q., Ye, W., Tao, K., Duan, S., et al. (2012). The RxLR effector Avh241 from Phytophthora sojae requires plasma membrane localization to induce plant cell death. New Phytol. 196, 247–260. doi: 10.1111/j.1469-8137.2012.04241.x
Zhang, Y., Wen, T. Y., Wu, X. Q., Hu, L. J., Qiu, Y. J., Rui, L. (2022). The Bursaphelenchus xylophilus effector BxML1 targets the cyclophilin protein (CyP) to promote parasitism and virulence in pine. BMC Plant Biol. 22, 216. doi: 10.1186/s12870-022-03567-z
Keywords: Bursaphelenchus xylophilus, transcriptome, Pinus massoniana, effector, plant cell death activation
Citation: Li Z, Wang H, Cao Y, Shan X, He X, Huang Q, Zhuo K, Liao J and Lin B (2024) A Bursaphelenchus xylophilus effector BxICD1 inducing plant cell death, concurrently contributes to nematode virulence and migration. Front. Plant Sci. 15:1357141. doi: 10.3389/fpls.2024.1357141
Received: 17 December 2023; Accepted: 16 February 2024;
Published: 28 February 2024.
Edited by:
Andressa Machado, Agronema, BrazilReviewed by:
Jun Su, Fujian Agriculture and Forestry University, ChinaCopyright © 2024 Li, Wang, Cao, Shan, He, Huang, Zhuo, Liao and Lin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Borong Lin, Qm9yb25nbGluQHNjYXUuZWR1LmNu; Jinling Liao, amxsaWFvQHNjYXUuZWR1LmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.