
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 02 April 2024
Sec. Functional and Applied Plant Genomics
Volume 15 - 2024 | https://doi.org/10.3389/fpls.2024.1356922
Among the bioactive compounds, lipid-soluble tanshinone is present in Salvia miltiorrhiza, a medicinal plant species. While it is known that ethephon has the ability to inhibit the tanshinones biosynthesis in the S. miltiorrhiza hairy root, however the underlying regulatory mechanism remains obscure. In this study, using the transcriptome dataset of the S. miltiorrhiza hairy root induced by ethephon, an ethylene-responsive transcriptional factor EIN3-like 1 (SmEIL1) was identified. The SmEIL1 protein was found to be localized in the nuclei, and confirmed by the transient transformation observed in tobacco leaves. The overexpression of SmEIL1 was able to inhibit the tanshinones accumulation to a large degree, as well as down-regulate tanshinones biosynthetic genes including SmGGPPS1, SmHMGR1, SmHMGS1, SmCPS1, SmKSL1 and SmCYP76AH1. These are well recognized participants in the tanshinones biosynthesis pathway. Further investigation on the SmEIL1 was observed to inhibit the transcription of the CPS1 gene by the Dual-Luciferase (Dual-LUC) and yeast one-hybrid (Y1H) assays. The data in this work will be of value regarding the involvement of EILs in regulating the biosynthesis of tanshinones and lay the foundation for the metabolic engineering of bioactive ingredients in S. miltiorrhiza.
Salvia miltiorrhiza, commonly known as Danshen, is a medicinal plant of the family Lamiaceae (Jia et al., 2019). Historically, the dried roots of Danshen were popular in the treatment of diseases related to the cardiovascular, digestive and cerebrovascular systems. They are also recorded as being able to perform pharmacological actions (Hu et al., 2014; Shi et al., 2019). Several Danshen products, such as the Danshen injection, continue to find wide usage in clinical practice. Given the wide application of S. miltiorrhiza, the study of medicinal compounds biosynthesis is important to ensure its production to meet clinical needs. From previous studies it appears that the principal active components of S. miltiorrhiza can be distinguished as either lipid-soluble or water-soluble components in nature (Liu et al., 2022). The lipid-soluble components include the tanshinones namely, tanshinone I (TA-I), tanshinone IIA (TA-II), dihydrotanshinone (DT), and cryptotanshinone (CT); the water-soluble components include the phenolic acids namely, salvianolic acid A (Sal A), salvianolic acid B (Sal B), caffeic acid (CA) and rosmarinic acid (RA) (Ma et al., 2015; Mao et al., 2020). Over the recent years, growing attention has been focused on the compounds above, mainly on ways to improve the tanshinones yield in S. miltiorrhiza. Tanshinone is confirmed to accumulate mainly in the S. miltiorrhiza roots, and three distinct stages can be observed in its biosynthesis pathway. First, the common terpenoid precursors, which include isopentenyl diphosphate (IPP) and dimethylallyl diphosphate (DMAPP) produced by two distinct processes, namely the mevalonate (MVA) pathway in cytosol and 2-C-Methyl-D-erythritol-4-phosphate (MEP) pathway in the plastids (Rohmer et al., 1993; Lange et al., 2000; Miziorko, 2011). Second, three known synthases are evident namely, copalyl diphosphate synthase 1 (CPS1), kaurene synthase-like 1 (KSL1), and miltiradiene oxidase (CYP76AH1); incidentally, several yet-to-be-identified enzymes are present, which form the skeleton of the tanshinones. Finally, through post-synthesis modifications namely, oxidation, methylation, decarboxylation, or cyclization, diverse tanshinones can be generated (Jiang et al., 2023). Data from several studies confirmed that the overexpression or downregulation of one or two synthase genes was able to promote the tanshinones biosynthesis process (Wang and Wu, 2010; Huang et al., 2019). The active ingredient present in medicinal plants can be enhanced by metabolic engineering strategy. This process requires a deep understanding of the biosynthesis pathway of the active ingredients, which includes the biosynthetic genes and the regulatory factors (Yu et al., 2020; Chen et al., 2022).
In plants, the transcription factors (TFs) were validated to regulate the processes of anti-stress and metabolic engineering (Zhou and Memelink, 2016; Sun et al., 2018; Yan et al., 2021). The EIN3/EIL1 is one member of the family of EIL transcription factors (Dolgikh et al., 2019), and it significantly affects plant growth and development, as well as the responses to biotic and abiotic stressors (Zhong et al., 2009; Zhu et al., 2022; Xu et al., 2023). The Arabidopsis thaliana genome has six EIN3/EIL1 members, while five are present in Nicotiana tabacum, six in Oryza sativa (rice), and six in S. miltiorrhiza (Solano et al., 1998; Zhang et al., 2018). Such types of TF possess many special structural features: 1. The N-terminal DNA binding domain has a highly conservative and unique folding structure. 2. The portion of the transmembrane amino acid shows acidic amino acid portions, five small basic amino acid clusters (BD I-V), and proline-abundant sections. 3. Coil structure (Wang et al., 2012). The coil structure is principally the region where the proteins interact and take part in DNA binding (Chao et al., 1997). In the EIL family, a rich alkaline amino acid is found to be close to the N-terminal helix motif of α spiral part, similar to the DNA binding domain observed in the bZIP family. The acidic amino acid portion, as well as the glutamine abundant and proline rich activation domains are regarded as the transcriptional activation domains (Kong et al., 2018). The presence of these motifs in the EIN3/EIL protein implies the possibility of their function in transcriptional regulation.
While ethylene is the simplest of the olefin gases, it is also the first gas molecule confirmed to have hormonal action and significantly promote the accumulation of the secondary metabolites in plants (Bleecker and Kende, 2000). In fact, EIN3/EIL1 is considered as one of the important regulatory factors, bearing a close relationship to ethylene signaling (Guo and Ecker, 2004). The EIN3 promotes the lengthening of the root hair through the direct activation of the RHD6-LIKE4 (RSL4) gene (Feng et al., 2017). Besides, the homeotic genes CpEIN3a and CpNAC2 of the EIN3 family in papaya, influence the carotenoid biosynthesis by the direct transcription and activation of the expression of CpPDS4 and CpCHY-b genes, which are linked to the biosynthesis of the carotenoid during the stage of ripening of the fruit (Fu et al., 2017). In apple, however, the homeotic gene MdEIL1 of the EIN3 family directly binds to the MdMYB1 promoter and transcriptionally activates its expression, showing an effect on the biosynthesis of anthocyanidin in apple fruit (An et al., 2018). However, the precise transcriptional mechanism in the ethylene-induced tanshinones biosynthesis continues to remain cryptic.
In this work, a new EIN3/EIL1 transcription factor SmEIL1 was isolated and identified in S. miltiorrhiza. This study closely examined the role played by SmEIL1 in the tanshinones biosynthesis induced by ethylene in S. miltiorrhiza, and gained insight into the regulatory network. Overall, our findings thus provide new insights into the underlying molecular mechanisms involved in the ethylene-induced tanshinones biosynthesis in S. miltiorrhiza.
The S. miltiorrhiza plantlets were raised by adopting either the standard greenhouse conditions or the Murashige and Skoog (MS) plant growth medium. The temperature was maintained at 25°C, following a 16-h day and 8-h night cycle. The hairy roots of the S. miltiorrhiza were subcultured in 250 mL Erlenmeyer flasks, to which were added 100 mL 1/2 MS liquid medium. They were then placed in a shaker at 110 rpm/min in the dark at 25°C. The Nicotiana benthamiana were planted in the soil and raised in pots, maintaining the identical conditions as for the S. miltiorrhiza. Finally, RNA isolation was conducted using tissue samples drawn from the main root, lateral root, fibrous root, stem, petiole, young leaf, mature leaf, and flower tissues of a single one-year-old S. miltiorrhiza plant. Hairy roots of S. miltiorrhiza were expanded and cultured in 1/2 MS liquid medium with 70 μM ethephon treatment for about 50 days.
Using the local datasets, identification of all the EIL families in S. miltiorrihiza was done. The SmEIL1, of the EIL family, was cloned by employing a homology-based cloning method, the details of which were explained earlier (Shi et al., 2016a). The primer pair utilized in the gene cloning was cited in the Supplementary Table 1. The BLAST-Protein (BLASTP) analysis of the SmEIL1 was accomplished using the non-redundant (NR) protein sequence database (www.ncbi.nlm.nih.gov). Then four amino acid sequence highly homologous to SmEIL1 alignment was performed using DNAMAN software. Alignment sequences includes Arabidopsis thaliana AtEIL1 (accession number AT2G27050), Arabidopsis thaliana AtEIN3 (AT3G20770), Lithospermum erythrorhizon LeEIL1 (ACP56697.1) and Malus domestica MdEIL1 (ADE41153.1).Then, based on the amino acid sequences, a phylogenetic tree was built up using the neighbor-joining method and employing the MEGA 6.0 software. The bootstrap method was used to assess the reliability of each node in the tree and 1,000 replicates were done (Tamura et al., 2013).
To verify the subcellular localization profile of SmEIL1, the open reading frame (ORF) of SmEIL1 was cloned and constructed into the pHB-GFP vector driven by the CaMV35S promoter to form the SmEIL1-GFP fusion protein (Supplementary Figure 5). An empty pHB-GFP vector was used as the negative control. The fusion vector and the control vector were introduced into the Agrobacterium rhizogenes strain GV3101 for transient transformation, respectively. In order to observe the nuclei, a solution of 4’, 6-diamidino-2-phenylindole dihydrochloride (DAPI) at a concentration of 10 μg/mL was introduced into N. benthamiana leaves via syringe injection, 3-h of growth prior to microscopic examination. Then, the fluorescence signal was observed by laser confocal microscope (Zeiss, Germany) under the excitation of the 405 nm and 488 nm laser. The experiments were repeated with at least three biological replicates (Shi et al., 2016a).
Quantitative real-time PCR (qRT-PCR) detection was done adopting the method used by Shi et al., 2016b. Different samples were collected from eight tissues including the main root, lateral root, fibrous root, stem, petiole, young leaf, mature leaf, and flower within a single one-year-old plant. Hairy roots treated with ethylene at 0, 1, 6, 12, and 24 h time points, were harvested and then frozen in liquid nitrogen. For the internal control, the SmActin gene was used. The primer sequences for the real-time PCR detection are listed in the Supplementary Table 1. Using the comparative Ct method, quantification of the gene expression levels was conducted (Shi et al., 2016b).
The open reading frame (ORF) of SmEIL1 was placed in the double restriction insertion site of BamHI and SpeI of the pHB vector and controlled by the CaMV35S promoter and NOS terminator (Supplementary Figure 5). To accomplish the SmEIL1 knock-out (KO) vector construction, analysis of the potential gene editing sites of the SmEIL1 gene sequence was done using the Optimized CRISPR Design (http://crispr.dbcls.jp/). Next, synthesis of a pair of complementary oligos was done. This was then ligated to the CRISPR/Cas9 system expression protein to enable the sgRNA and hSpCas9 to combine. Hence, the hSpCas9 was driven by the CaMV35S promoter, and the SmEIL1 sgRNA was driven by the AtU6 promoter (Supplementary Figure 6). The expression cassette was then inserted into the linearized plant expression vector pCAMBIA1300 in order to infect the S. miltiorrhiza leaves and thus produce the hairy roots. For the control, the pCAMBIA1300 empty vector without the sgRNA sequence was used (Supplementary Figure 6). All the constructs were finally introduced into the Agrobacterium strain C58C1, which were then transformed into the S. miltiorrhiza, and the transgenic hairy root lines were generated as explained earlier on Huang et al., 2019.
After continuous culture in 1/2 MS liquid medium for 60 days, each hairy root line was harvested and freeze dried for 24 h. Using high-performance liquid chromatography (HPLC), all the metabolites were quantified as described earlier (Huang et al., 2019; Sun et al., 2019; Huang et al., 2020). Next, the dried roots were finely powdered. Samples of 200 mg were then extracted with 16 mL of methanol/dichloromethane (3:1, v/v), sonicated for 1 h, and centrifuged at room temperature of 25°C. Post centrifugation, the supernatant was transferred into a distillation flask. The supernatant was centrifuged once more, poured into a distillation flask and spun dry at 50°C. Next, 2 mL of methanol was added into the distillation flask to dissolve the material present there. After one more centrifugation at 6,500 × g for 10 min, the samples were drawn out with a 1 mL sterile syringe and filtered separately through 0.22 µM organic membranes (Jinten, China). Finally, the resulting filtrate collected was prepared for HPLC detection (Huang et al., 2019).
Dual-LUC assays were done as cited earlier (Sun et al., 2019). The promoter portions of the candidate genes (~2000 bp) were inserted into a pGreen0800-LUC vector driving a firefly LUC reporter gene; the 35S promoter controlled the Renilla luciferase (REN) gene. Later, the recombinant vectors were transferred into A. tumefaciens (GV3101) along with a helper plasmid (pSoup-P19) encoding a cosuppressor. The test was performed as reported earlier (Sun et al., 2019). While the pHB-GFP construct was used as the negative control, and the REN gene was utilized as the internal control.
The yeast one-hybrid (Y1H) assay was done adopting the technique reported by Sun et al (Sun et al., 2019). The full-length coding sequence of the SmEIL1 gene was constructed in combination with the pB42AD vector. Two EBS-box sequences in the promoter (5’-ATGAATCCT-3’ and 5’-TCCATGCA-3’) were inserted separately into the pLacZ2u plasmid in triplicate. The recombinants produced through this procedure were co-transformed into the EGY48a yeast strain. The transformants were cultivated on the SD/-Ura/-Trp medium for 48 h, and the positive binding activity was evident as a blue color on the SD/-Ura/-Trp medium with 5-bromo-4-chloro-3-indolyl-b-D-galactopyranoside (X-Gal) for 24 h. The empty vectors of pB42CE and pLacZ2u functioned as the negative controls.
Each of the findings indicate the average value of three independently conducted experiments. In this study, all the experiments were performed in triplicate. The contents and degrees of gene expression involved in the biosynthesis of tanshinones and phenolic acids are given as the mean value ± SD. SPSS 16.0 software was employed to conduct the single-sample t-test and One-Way ANOVA; the P-value < 0.05 were regarded as statistically significant.
Ethylene is a gaseous hormone which exerts a significant effect on the plant in terms of growth, development, and stress responses (Bleecker and Kende, 2000). Initially, HPLC detection was used to examine the content of tanshinones and phenolic acid in the S. miltiorrhiza hairy roots treated with ethephon. Within 24-h of treatment, the total tanshinones content present in the S. miltiorrhiza hairy roots revealed a remarkable drop in comparison to the control group (0-h of treatment). All four tanshinone compounds achieved their lowest levels, post treatment of 12 hours duration. The dihydrotanshinone (DT) content particularly registered a 90% drop when compared to the control group, while the tanshinone I (TA-I) content exhibited a 4.4-fold reduction, and the cryptotanshinone (CT) level dropped by 77% in comparison to the control (Figure 1A). However, with the ethylene treatment from 1- to 24-h, the accumulation of phenolic acids did not reveal a very regulative variation (Figure 1B). The results cited above reiterate that ethylene signaling can inhibit the accumulation of tanshinones in the S. miltiorrhiza hairy roots.
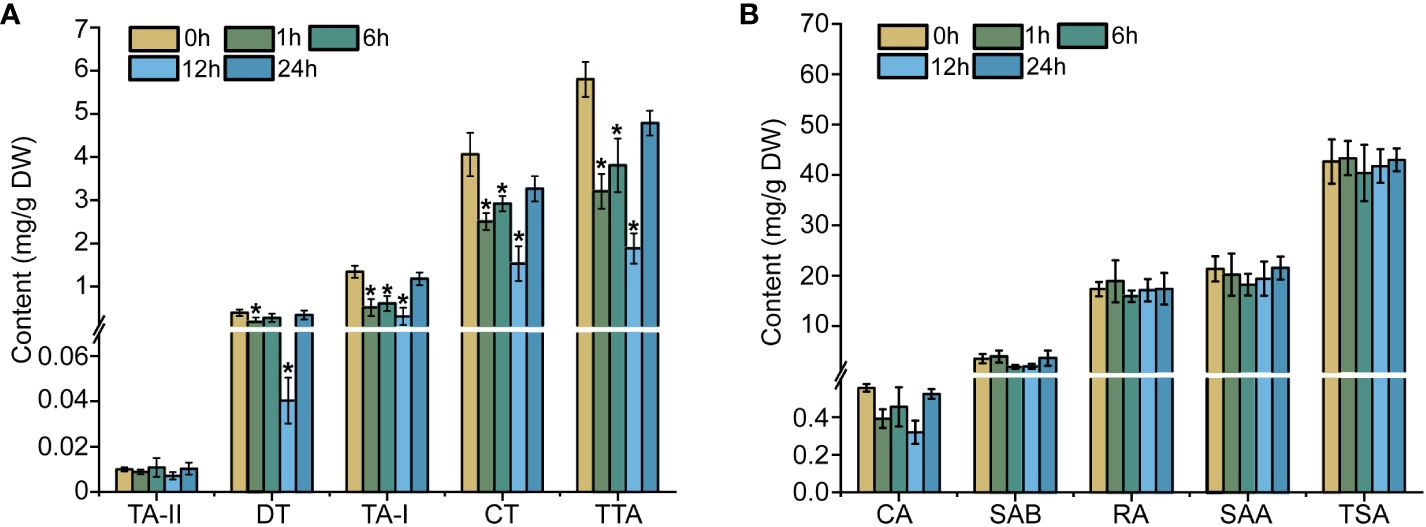
Figure 1 Effects of ethylene on tanshinones (A) and phenolic acids (B) in hairy roots. The error line indicates the error for three biological replicates; t-test was done for significant differences (*P< 0.05).
To investigate whether the SmEIL1 was induced by ethylene, the SmEIL1 expression post the exogenous ethephon treatment in the S. miltiorrhiza hairy roots was examined by qRT-PCR at different time intervals from 0- to 24-h. The ethephon was then found to significantly upregulate the expression of SmEIL1, and its transcript level rapidly peaked after the 1 h ethephon treatment (Figure 2A). The expression levels of the principal genes that participated in the biosynthesis pathway of the tanshinones in the hairy roots after ethephon treatment were quantified using qRT-PCR detection. The eight genes involved in the tanshinones biosynthesis including copalyl diphosphate synthase 1 (CPS1), 1-deoxy-D-xylulose 5-phosphate reductoisomerase (DXR1), 1-deoxy-D-xylulose 5-phosphate synthase 1 (DXS2), geranylgeranyl pyrophosphate synthase (GGPPS), kaurene synthase-like 1 (KSL), miltiradiene oxidase (CYP76AH1), 3-hydroxy-3-methylglutaryl-coenzyme A reductase (HMGR) and 3-hydroxy-3-methylglutaryl coenzyme A synthase (HMGS) genes, exhibited visible variations with the treatment of ethephon. Of these, the CPS1, GGPPS, and CYP76AH1 genes showed a maximum of 4-, 5.3-, and 3-fold reductions, respectively, when compared with the control (Figure 2B). This finding suggests that ethylene signaling can inhibit the expression of the three pivotal genes (CPS1, GGPPS, and CYP76AH1) involved in the tanshinones biosynthesis process.
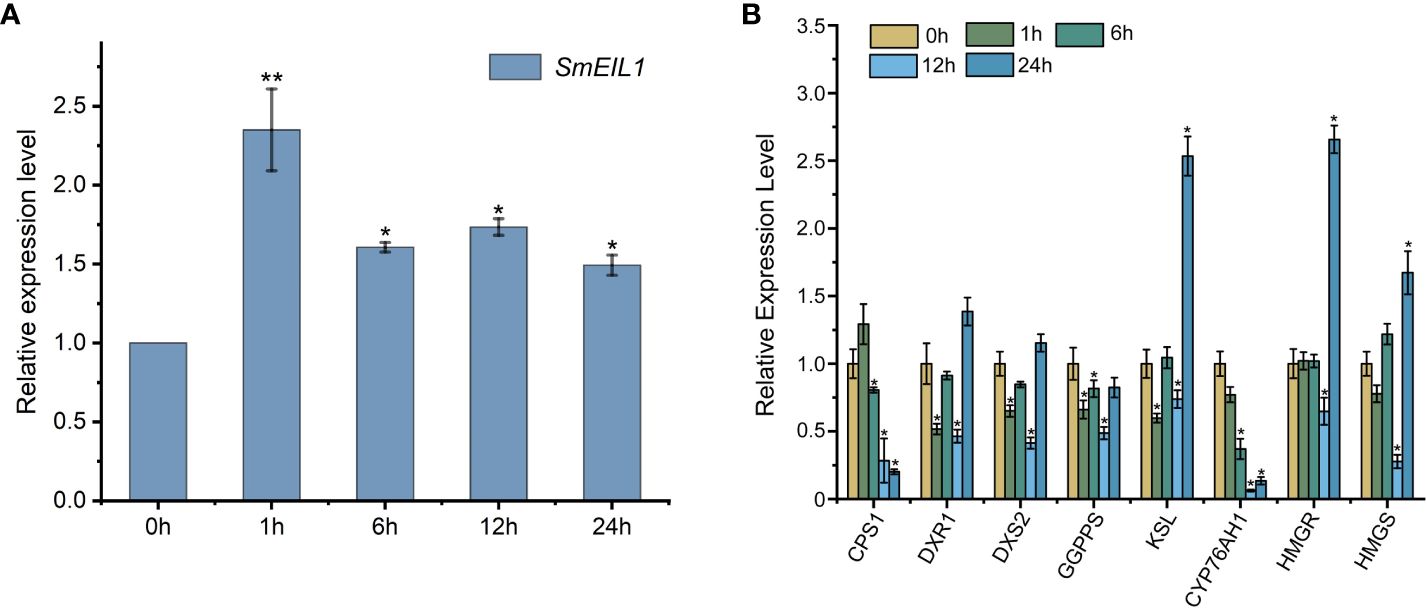
Figure 2 Expression profiles of SmEIL1 gene and the genes involved in the tanshinones biosynthesis pathway in response to the treatment with ethylene. (A) The expression profiles of SmEIL1 gene after the induction of ethylene. (B) Expression profiles of genes in the tanshinones biosynthesis pathway after treatment with ethylene. Fold changes of the relative gene expression level were all normalized to the 0 h treatment. The error line indicates the error for three biological replicates; t-test was done for significant differences (**P< 0.01; *P< 0.05).
It is a known fact that the EIL1 gene is assumed to be the core regulator in ethylene signaling (Meng et al., 2018, 2018). In the study cited above, ethylene signaling has been verified as an inhibitor of tanshinones accumulation in the S. miltiorrhiza hairy roots. Therefore, cloning the EIL1 gene and studying its molecular mechanism as a regulator in the biosynthesis of tanshinones is a worthwhile endeavor. Using the data of the S. miltiorrhiza transcriptome, the entire cDNA sequence of the SmEIL1 gene in S. miltiorrhiza was firstly spliced, using the sequence of the AtEIL1 (AT2G27050) gene in Arabidopsis as the reference. Soon after, the SmEIL1 gene was isolated via homology-based cloning; the gene was found to be 1848-bp long and encoded 616 amino acids (Figure 3A). The SmEIL1 protein is composed of several conserved motifs, such as the acidic amino acid portion (AD), alkaline amino acid part (BDI-V), and proline abundant (PR) domains (Figure 3C). The EIN3/EIL1 members of the family Arabidopsis, Lithospermum erythrorhizon LeEIL1, Malus domestica MdEIL1 and SmEIL1 were used to construct a phylogenetic tree. From the results, it was evident the closest evolutionary relationship existed between SmEIL1 and LeEIL1 (Figure 3B). LeEIL1 acts as a positive regulator of purslane synthesis in the hairy roots of comfrey Lithospermum erythrorhizon and promotes purslane accumulation (Fang et al., 2016). Hence, this suggests that the SmEIL1 gene plays a similar role in S. miltiorrhiza.
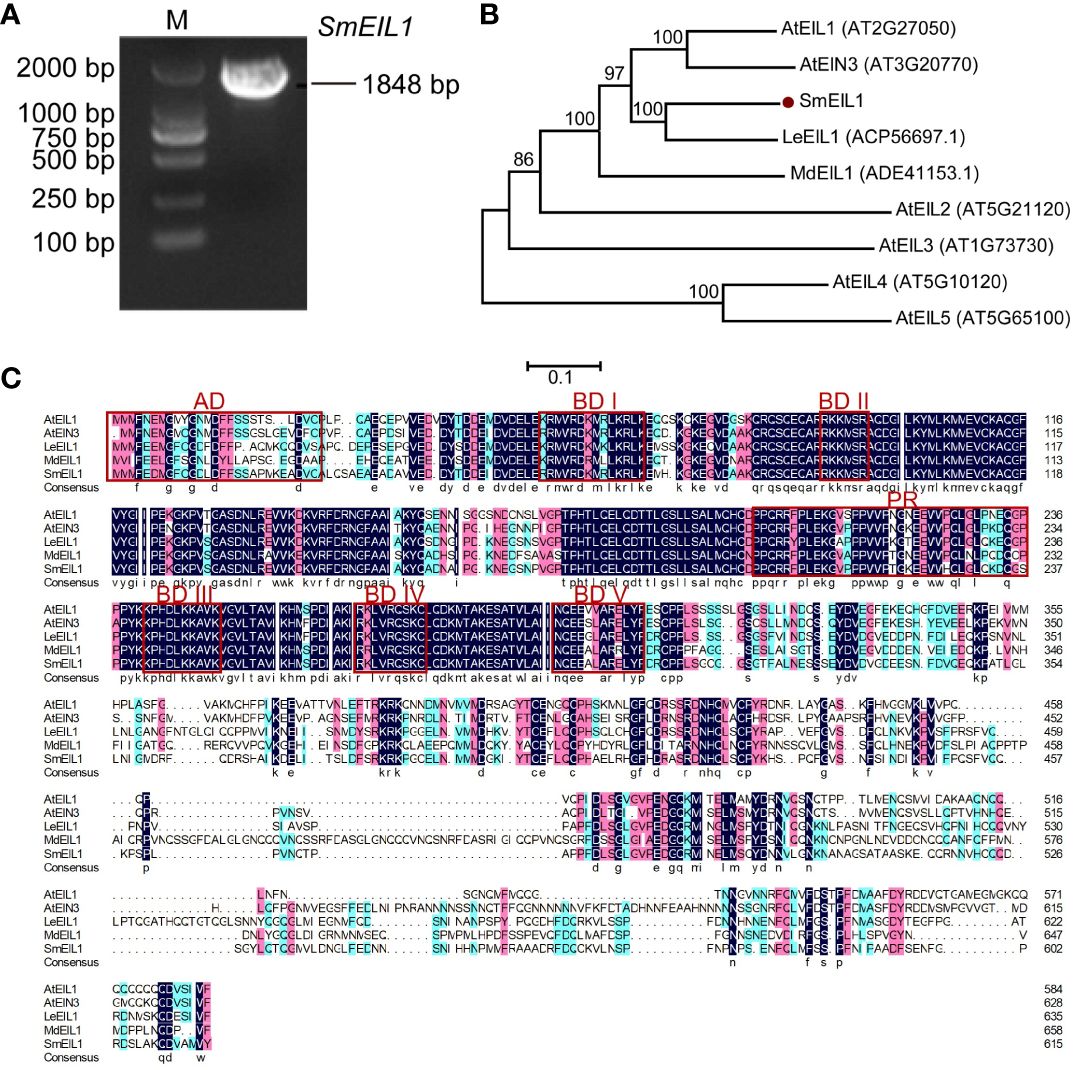
Figure 3 Characterization and subcellular localization of SmEIL1. (A) Cloning of SmEIL1 gene. (B) Phylogenetic tree used in analyzing SmEIL1 and other EIL protein members in plants by neighbor-joining method, with MEGA 6 software. The numbers on the nodes indicate the bootstrap values post 1000 replicates. (C) Multiple protein sequences alignment between SmEIL1 and AtEILs transcription factors (AD, acidic amino acid regions; PR, Proline-rich regions; BD I-V, Basic amino acid regions I-V). The frame represents conserved domains.
An examination of the SmEIL1 gene expression profiles in different tissues was conducted using qRT-PCR detection. The branch roots and young leaves exhibited the highest degree of expression, while the main roots showed the lowest degree of expression (Figure 4A). Later, the subcellular localization of the SmEIL1 was investigated in the tobacco cells via the transient expression of SmEIL1 combined with GFP. Vigorous fluorescence was evident only in the nuclei containing the construct of the 35S::SmEIL1-GFP recombinant; the control recombinant of 35S::GFP, however, displayed fluorescence throughout the whole cell (Figure 4B), which implies that the SmEIL1 protein is localized in the nuclei in S. miltiorrhiza.
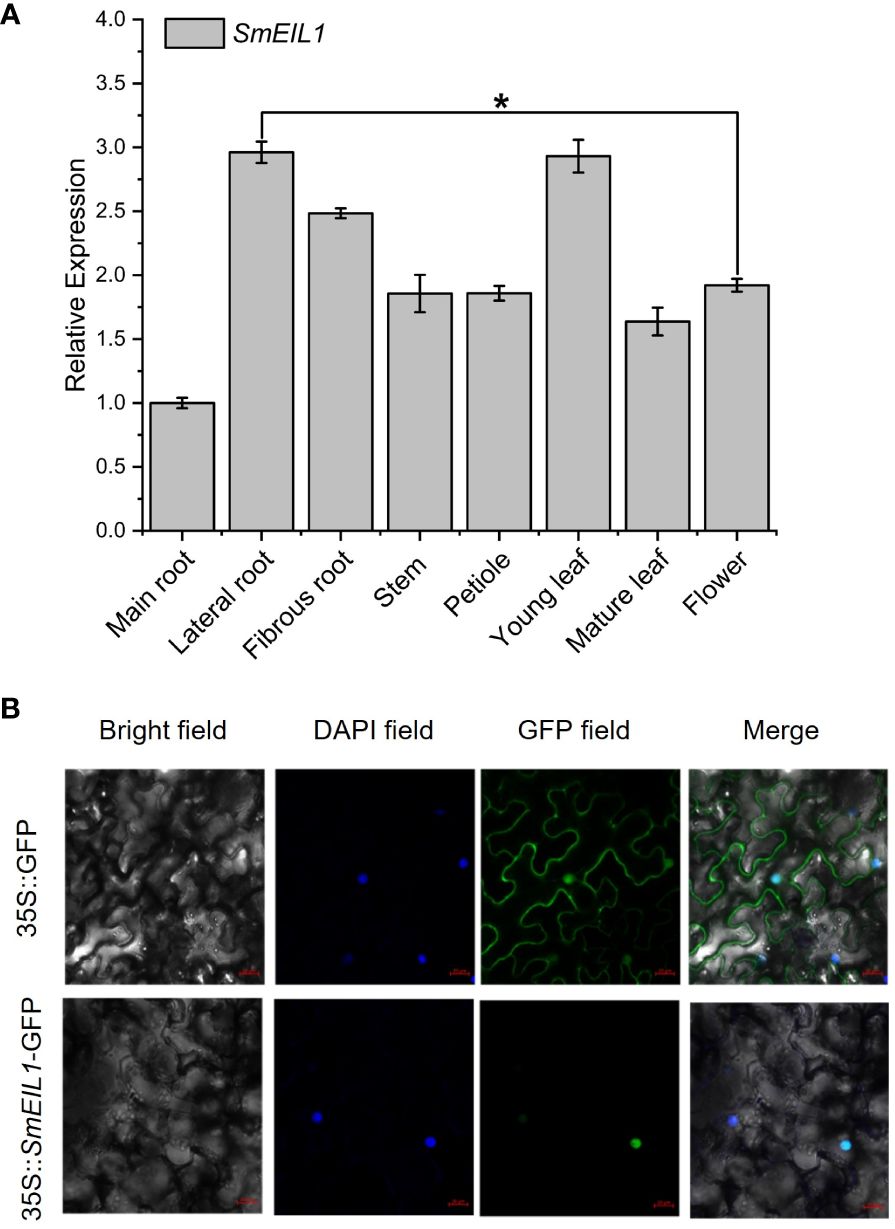
Figure 4 Tissue expression and subcellular location analysis of SmEIL1. (A) Expression patterns of the SmEIL1 gene in different tissues. Fold changes in the relative gene expression level of all the other tissues are normalized to the main root. (B) Subcellular localization of SmEIL1 in leaf epidermal cells of N. benthamiana. DAPI was the positive control. (The error line indicates the error for three biological replicates.) (*P<0.05).
On further investigation of whether SmEIL1 can influence the tanshinones biosynthesis in the S. miltiorrhiza hairy roots, an examination was done of the over-expression (OE) and knock-out (KO) of the SmEIL1 gene in the transgenic hairy root lines, independent of each other, through genomic PCR detection (Supplementary Figure 1); after this, DNA sequencing was introduced to select the positive hairy root lines (Supplementary Figure 3) and qRT-PCR assays (Supplementary Figures 3, 4) were done as well. For further analysis, a selection was made of four independent OE lines (OE-1, OE-2, OE-3, and OE-4), which showed the greatest levels of expression, and four KO lines (KO-1, KO-2, KO-3, and KO-4) with the lowest expression levels. On comparison with the control lines, the four SmEIL1 OE lines exhibited higher SmEIL1 transcript levels (30 to 70-fold rise) (Supplementary Figure 3). Besides, the SmHMGR1, SmHMGS1, SmGGPPS1, SmCPS1, SmKSL1 and SmCYP76AH1 were examined to be remarkably downregulated in the four OE lines (Figure 5A). On the contrary, SmHMGR1, SmHMGS1, SmCPS1 and SmGGPPS1 were found to show a significant upregulation in all the four KO lines (Figure 5B). From these findings it become clear that the SmEIL1 may act as a negative regulatory factor to participate in the tanshinones biosynthesis process.
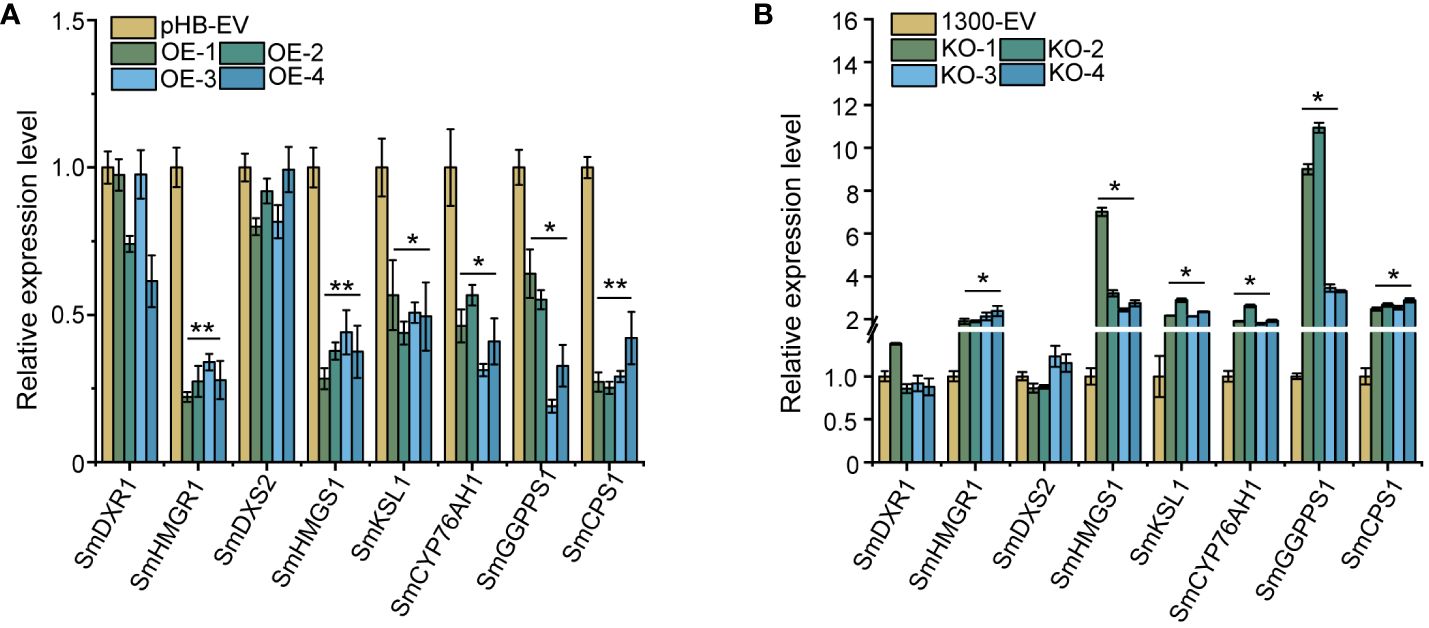
Figure 5 Gene expression levels of the main enzyme genes in tanshinones biosynthesis pathway in overexpression (A) and knock-out (B) hairy roots. The error line indicates the error for three biological replicates; t-test was done for significant differences (**P< 0.01; *P< 0.05).
In line with the gene expression profiles cited above, four tanshinones in the hairy root lines of 45-day-old, namely tanshinone I (TA-I), tanshinone II (TA-II), cryptotanshinone (CT), dihydrotanshinone (DT) and total tanshinones (TTA) were identified through HPLC detection. The TTA contents all exhibited a significant reduction in the four OE lines (OE-1, OE-2, OE-3, and OE-4); of these, the OE-1 line showed the lowest TTA concentration, dropping to 74% less than the control. Consistently, the TTA content in the four KO lines (KO-1, KO-2, KO-3, and KO-4) all showed significant elevation in the range from 2- to 4.66 times more than the control (Figure 6).
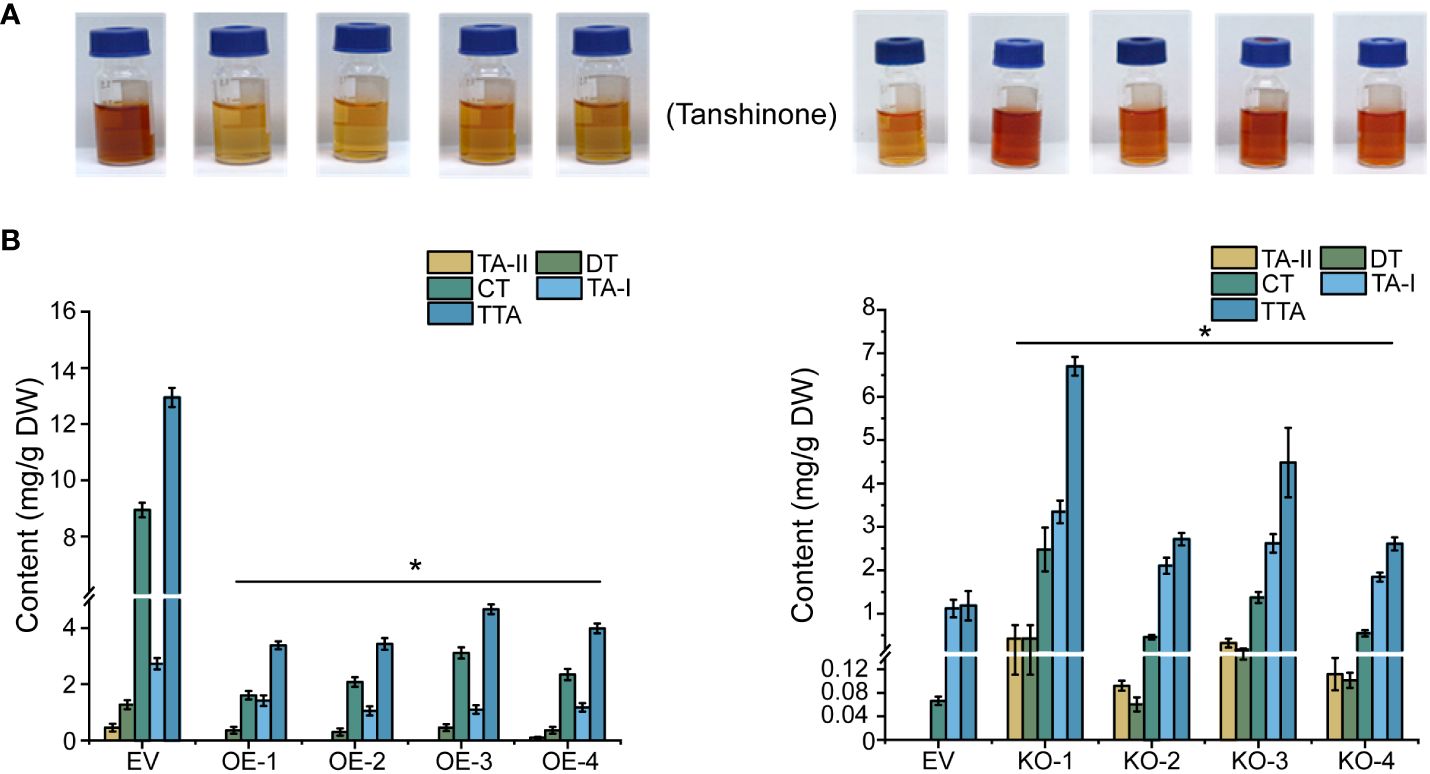
Figure 6 Extraction and determination of tanshinones in transgenic hairy roots. (A) Tanshinones extract from overexpression and knock-out hairy roots. (B) Determination of tanshinones content in overexpressed and knock-out hairy roots. The error line indicates the error for three biological replicates; t-test was done for significant differences (*P< 0.05).
Dual-Luc and Y1H assays were performed to clarify the potential molecular mechanism controlled by SmEIL1, and confirmed the downstream target genes which participated in the tanshinones biosynthesis pathway. The Dual-Luc assay confirm that SmEIL1 inhibits the biosynthesis of tanshinones, most probably by the inhibition of the transcription of the SmHMGR1 and SmCPS1 genes (Figure 7A). The specific binding element (EBS-box, A (C/T) G (A/T) A (C/T) CT) for the SmEIL1 protein is present in the promoter portion of the SmHMGR1 and SmCPS1 genes. The Y1H assay reveal that the SmEIL1 is unable to bind with the EBS-box element in the promoter region of the SmHMGR1 gene; however, it can bind with the EBS-box element in the promoter portion of the SmCPS1 gene. From these results it becomes clear that the SmCPS1 may be a direct target for the SmEIL1 to regulate the tanshinones biosynthesis in a negative manner.
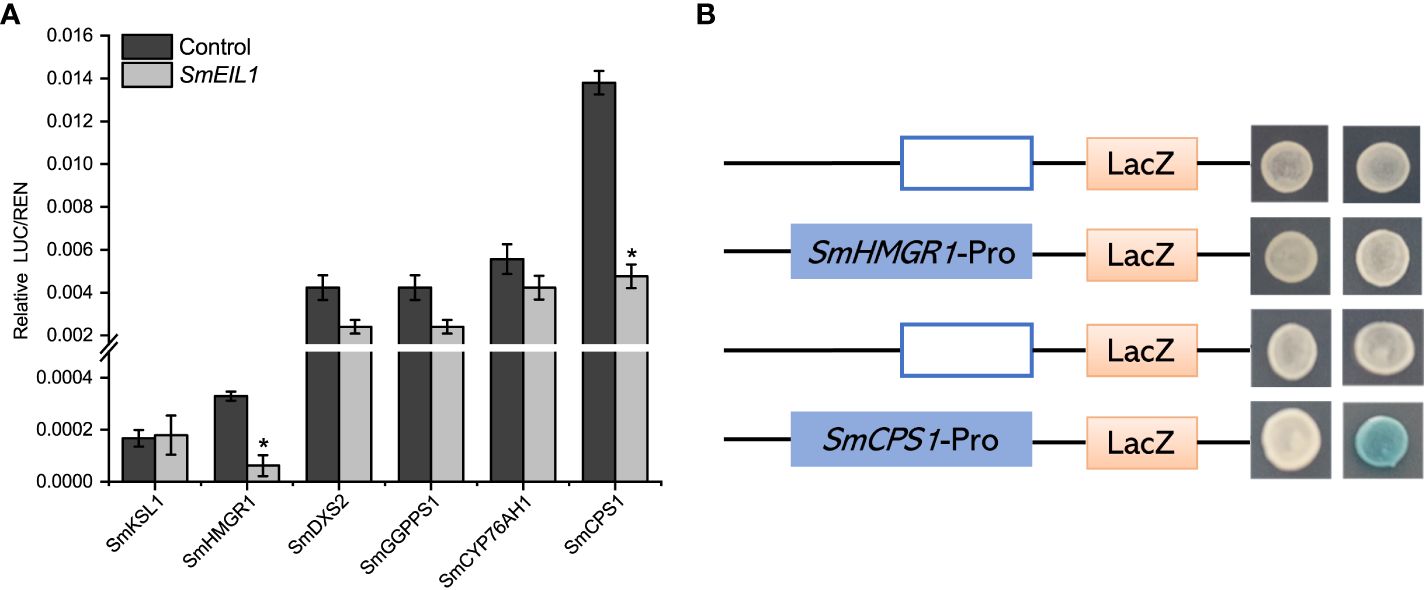
Figure 7 SmEIL1 inhibits the SmCPS1 expression. (A) Dual-Luc results of SmEIL1 on key enzyme promoters in tanshinones synthesis pathway. The error line indicates the error for three biological replicates; t-test was done for significant differences (*P< 0.05). (B)Y1H assay. (AD: pB42AD vector.).
Ethylene (ET) is a crucial phytohormone which modulates the plant secondary metabolites, growth, and adaptation to the environment (Heredia and Cisneros-Zevallos, 2009; Chen et al., 2012; Cheng et al., 2018; Ke et al., 2018; Li et al., 2018). In the signaling pathway of plant, EIN3/EIL1 gene was thought to act as a core integration node between ET and other signals (Shi et al., 2012; Liang et al., 2013; Liu et al., 2017; Bai et al., 2020). With the mediation of EIN3/EIL1 gene, ethylene can promote the accumulation of phenolic antioxidants in Daucus carota (Heredia and Cisneros-Zevallos, 2009), the production of shikonin and its derivatives in Lithospermum erythrorhizon (Fang et al., 2016),anthocyanin biosynthesis in apple (An et al., 2018), and the biosynthesis of capsaicinoids in Capsicum (Wen et al., 2022). ET-stabilized transcription factors EIN3 and EIL1 were validated to interact with multiple transcription factors to synergistically regulate anthocyanin accumulation, trichome formation, and defense against insect attack (Song et al., 2022). In the present work, with the treatment of ethephon in S. miltiorrhiza hairy roots, we revealed that the production of tanshinones including DT, CT, TA-I, and TTA were rapidly decreased at the induction time point of 1 h. Although the contents of DT, TA-I, CT and TTA recovered after 24 h of treatment, their contents were still lower than the control. This phenomenon may be caused by the fact that S. miltiorrhiza hairy roots in response to ET is usually short-term and rapid (Li et al., 2024). No visible variations were noted for the CA and SAB accumulation; however, the trend of variations in the RA, SAA and TSA biosynthesis showed irregularities (Figure 1). Hence, it pushes us to explore the molecular mechanism of how SmEIL1 mediate the ET signaling to regulate the biosynthesis of tanshinones in S. miltiorrhiza.
Based on the S. miltiorrhiza transcriptome collected in this study, the whole ORF sequence of SmEIL1 was firstly assembled using the AtEIL1 gene in A. thaliana as the reference sequence, and then it was cloned. Six members of EIL families were found in the S. miltiorrhiza transcriptome. In fact, six members are present in A. thaliana (Chao et al., 1997), four members in S. lycopersicum (Tieman et al., 2001; Yokotani et al., 2003), and five members in N. tabacum (Kosugi and Ohashi, 2000; Rieu et al., 2003). Obviously, EIL is a gene family and may conduct multiple functions by different gene members. In A. thaliana, the EIN3, EIL1 and EIL2 genes were observed to play a part in the ethylene signaling to regulate plant growth (Houben et al., 2022). The LeEIL1 gene, in Lithospermum erythrorhizon, was validated to participate in regulating the biosynthesis of shikonin, whereas MdEIL1 gene in Malus domestica was identified to promote the accumulation of anthocyanin (An et al., 2018). Through the construction of evolutionary tree, the SmEIL1 protein was found to fall into the same subgroup along with LeEIL1, implying that the SmEIL1 might exert a regulatory influence on the secondary metabolites triggered by the ethylene signal in S. miltiorrhiza.
Through examining the expression profile of SmEIL1 gene in diverse tissues, it was found that the lateral and fibrous roots, and young leaf showed a degree of expression much higher than the other tissues like those of the flower, mature leaf, petiole, stem, and main root (Figure 4A). While the leaf is an important vegetative organ which is able to receive the ethylene signals directly, the lateral and fibrous roots are thought to be the principal tissues for the accumulation of medicinal substances in S. miltiorrhiza (Zhou et al., 2021). Moreover, the relative expression of SmEIL1 increased significantly in hairy roots treated with ethylene, reaching a peak after 1 h of treatment (Figure 2A). The tissue expression and induced expression profile of SmEIL1 gene is similar to the SmERF1b-like gene, which shows a significant response to exogenous ethylene supply and exhibits the highest expression level in leaves after 1 h of induction in S. miltiorrhiza (Li et al., 2024). Soon after, the gain- and loss-of-function assays revealed that the SmEIL1 was able to negatively regulate the ethylene-mediated accumulation of tanshinones in the S. miltiorrhiza hairy root lines (Figures 5, 6). In fact, the total tanshinones concentration in the SmEIL1 knock-out transgenic hairy root line was at maximum 4.66 times as that of the non-transgenic line (Figure 5B). The EBS-box (A (C/T) G (A/T) A (C/T) CT) element is considered essential in the binding by the EIL1 protein (Yamasaki et al., 2005), found in the promoter sequence of the SmKSL1, SmHMGR1, SmDXS2, SmGGPPS1, SmCYP76AH1, and SmCPS1 genes. Later, the Dual-LUC and Y1H assays performed revealed that the SmEIL1 directly activated its expression of the SmCPS1 gene (Figure 7). The qRT-PCR assay revealed that the overexpression of SmEIL1 in the four transgenic hairy roots not only inhibited the transcript level of its target gene SmCPS1, but also decreased the expression of three genes in the tanshinones biosynthesis pathway (Figure 5). The overexpression of the SmERF1L1 encourages the expression of the DXS2, DXR, HMGS, CPS1, and KSL1; however, the DXR alone is the target of the SmERF1L1 (Huang et al., 2019). In the transgenic hairy roots, the overexpression of the SmMYB1 activated the expression of the PAL1, C4H1, 4CL1, TAT1, HPPR1, and RAS1 genes; however, the CYP98A14 alone was confirmed as the target of the SmMYB1 (Zhou et al., 2021). From these findings, it seems likely that the ectopic expression of certain transcriptional genes can regulate the degree of expression of their target genes as well as that of the other genes present in the same biosynthesis pathway.
The present work, to the best of our knowledge, is the first report on the identification of the tanshinones biosynthesis gene, acting as the direct target of the SmEIL1 protein, the ethylene signaling gene in S. miltiorrhiza. On considering the observations cited above, a model was proposed for the role of SmEIL1 as a negative regulator inhibiting tanshinones accumulation (Figure 8). On being exposed to triggering by the ethylene, the SmEIL1 downregulates the transcription of the CPS1 gene by binding with the EBS-box elements present in the promoter of the CPS1 gene. This caused an inhibition of the tanshinones accumulation in the S. miltiorrhiza hairy roots (Figure 8).
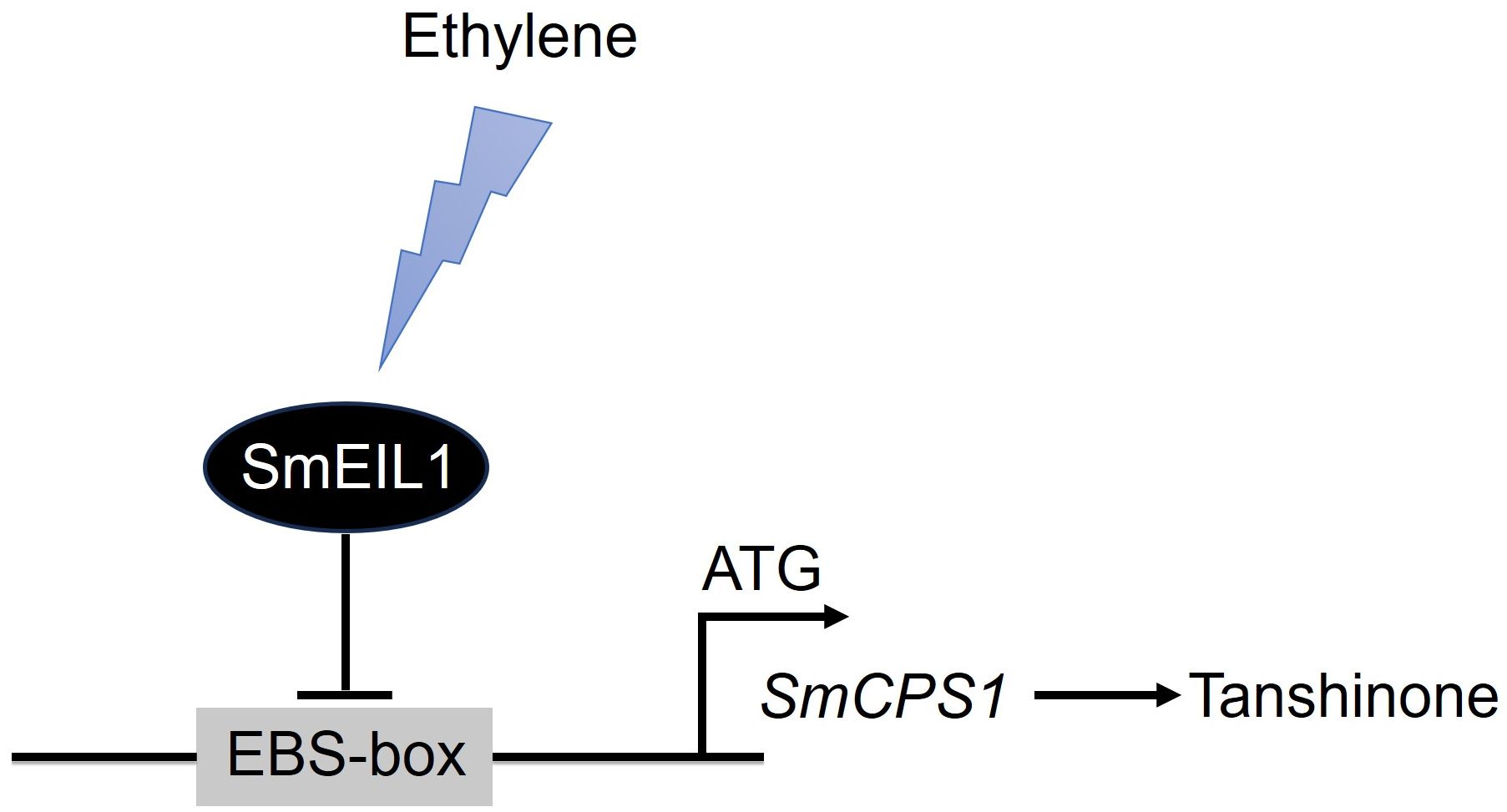
Figure 8 A model proposed for the role of SmEIL1 in modulating tanshinones biosynthesis in S. miltiorrhiza.
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
XL: Writing – original draft, Writing – review & editing. MX: Writing – original draft, Writing – review & editing. KZ: Writing – original draft. SH: Writing – original draft. LL: Writing – review & editing. LW: Writing – original draft. WZ: Writing – original draft, Writing – review & editing. GK: Writing – original draft, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by ‘Sannongjiufang’ Research Joint Project of Zhejiang Province (2023SNJF029), National Natural Science Foundation of China (82373979, 82204553), the Key Scientific and Technological Grant of Zhejiang for Breeding New Agricultural Varieties (2021C02074-3), National ‘Ten-thousand Talents Program’ for Leading Talents of Science and Technology Innovation in China, National Young Qihuang Scholars Training Program, Zhejiang Provincial Natural Science Foundation of China (LZ24H280002), and Research Projects of Zhejiang Chinese Medical University (2021JKZKTS011A, 2021JKZDZC06, 2020010 and YB20023).
We appreciate the great experimental support from the Public Platform of Pharmaceutical Research Center, Academy of Chinese Medical Science, Zhejiang Chinese Medical University.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2024.1356922/full#supplementary-material
An, J. P., Wang, X. F., Li, Y. Y., Song, L. Q., Zhao, L. L., You, C. X., et al. (2018). EIN3-LIKE1, MYB1, and ETHYLENE RESPONSE FACTOR3 act in a regulatory loop that synergistically modulates ethylene biosynthesis and anthocyanin accumulation. Plant Physiol. 178, 808–823. doi: 10.1104/pp.18.00068
Bai, Z., Wu, J., Huang, W., Jiao, J., Zhang, C., Hou, Z., et al. (2020). The ethylene response factor SmERF8 regulates the expression of SmKSL1 and is involved in tanshinone biosynthesis in Saliva miltiorrhiza hairy roots. J. Plant Physiol. 244, 153006. doi: 10.1016/j.jplph.2019.153006
Bleecker, A. B., Kende, H. (2000). Ethylene: a gaseous signal molecule in plants. Annu. Rev. Cell Dev. Biol. 16, 1–18. doi: 10.1146/annurev.cellbio.16.1.1
Chao, Q., Rothenberg, M., Solano, R., Roman, G., Terzaghi, W., Ecker, J. R. (1997). Activation of the ethylene gas response pathway in Arabidopsis by the nuclear protein ETHYLENE-INSENSITIVE3 and related proteins. Cell 89, 1133–1144. doi: 10.1016/s0092-8674(00)80300-1
Chen, J., Shi, D. Y., Liu, S. L., Zhong, L. (2012). Tanshinone IIA induces growth inhibition and apoptosis in gastric cancer in vitro and in vivo. Oncol. Rep. 27, 523–528. doi: 10.3892/or.2011.1524
Chen, Y., Wang, Y., Guo, J., Yang, J., Zhang, X., Wang, Z., et al. (2022). Integrated transcriptomics and proteomics to reveal regulation mechanism and evolution of SmWRKY61 on tanshinone biosynthesis in Salvia miltiorrhiza and Salvia castanea. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.820582
Cheng, J., Song, N., Wu, J. (2018). A patatin-like protein synergistically regulated by jasmonate and ethylene signaling pathways plays a negative role in Nicotiana attenuata resistance to Alternaria alternata. Plant Divers. 41, 7–12. doi: 10.1016/j.pld.2018.12.001
Dolgikh, V. A., Pukhovaya, E. M., Zemlyanskaya, E. V. (2019). Shaping ethylene response: the role of EIN3/EIL1 transcription factors. Front. Plant Sci. 10. doi: 10.3389/fpls.2019.01030
Fang, R., Zou, A., Zhao, H., Wu, F., Zhu, Y., Zhao, H., et al. (2016). Transgenic studies reveal the positive role of LeEIL-1 in regulating shikonin biosynthesis in Lithospermum erythrorhizon hairy roots. BMC Plant Biol. 16, 121. doi: 10.1186/s12870-016-0812-6
Feng, Y., Xu, P., Li, B., Li, P., Wen, X., An, F., et al. (2017). Ethylene promotes root hair growth through coordinated EIN3/EIL1 and RHD6/RSL1 activity in Arabidopsis. Proc. Natl. Acad. Sci. U. S. A. 114, 13834–13839. doi: 10.1073/pnas.1711723115
Fu, C. C., Han, Y. C., Kuang, J. F., Chen, J. Y., Lu, W. J. (2017). Papaya CpEIN3a and CpNAC2 co-operatively regulate carotenoid biosynthesis-related genes CpPDS2/4, CpLCY-e and CpCHY-b during fruit ripening. Plant Cell Physiol. 58, 2155–2165. doi: 10.1093/pcp/pcx149
Guo, H., Ecker, J. R. (2004). The ethylene signaling pathway: new insights. Curr. Opin. Plant Biol. 7, 40–49. doi: 10.1016/j.pbi.2003.11.011
Heredia, J. B., Cisneros-Zevallos, L. (2009). The effects of exogenous ethylene and methyl jasmonate on the accumulation of phenolic antioxidants in selected whole and wounded fresh produce. Food Chem. 115, 1500–1508. doi: 10.1016/j.foodchem
Houben, M., Vaughan-Hirsch, J., Mou, W., Van de Poel, B. (2022). Ethylene Insensitive 3-Like 2 is a Brassicaceae-specific transcriptional regulator involved in fine-tuning ethylene responses in Arabidopsis thaliana. J. Exp. Bot. 73, 4793–4805. doi: 10.1093/jxb/erac198
Hu, T., To, K. K., Wang, L., Zhang, L., Lu, L., Shen, J., et al. (2014). Reversal of P-glycoprotein (P-gp) mediated multidrug resistance in colon cancer cells by cryptotanshinone and dihydrotanshinone of Salvia miltiorrhiza. Phytomedicine 21, 1264–1272. doi: 10.1016/j.phymed.2014.06.013
Huang, P., Dong, Z., Guo, P., Zhang, X., Qiu, Y., Li, B., et al. (2020). Salicylic acid suppresses apical hook formation via NPR1-mediated repression of EIN3 and EIL1 in Arabidopsis. Plant Cell. 32, 612–629. doi: 10.1105/tpc.19.00658
Huang, Q., Sun, M., Yuan, T., Wang, Y., Shi, M., Lu, S., et al. (2019). The AP2/ERF transcription factor SmERF1L1 regulates the biosynthesis of tanshinones and phenolic acids in Salvia miltiorrhiza. Food Chem. 274, 368–375. doi: 10.1016/j.foodchem.2018.08.119
Jia, Q., Zhu, R., Tian, Y., Chen, B., Li, R., Li, L., et al. (2019). Salvia miltiorrhiza in diabetes: a review of its pharmacology, phytochemistry, and safety. Phytomedicine 58, 152871. doi: 10.1016/j.phymed.2019.152871
Jiang, J. S., Gu, Q., Feng, Z. M., Yuan, X., Zhang, X., Zhang, P. C., et al. (2023). The tanshinones from the plant of Salvia miltiorrhiza. Phytochemistry 210, 113673. doi: 10.1016/j.phytochem.2023.113673
Ke, S. W., Chen, G. H., Chen, C. T., Tzen, J. T. C., Yang, C. Y. (2018). Ethylene signaling modulates contents of catechin and ability of antioxidant in Camellia sinensis. Bot. Stud. 59, 11. doi: 10.1186/s40529-018-0226-x
Kong, X., Li, C., Zhang, F., Yu, Q., Gao, S., Zhang, M., et al. (2018). Ethylene promotes cadmium-induced root growth inhibition through EIN3 controlled XTH33 and LSU1 expression in Arabidopsis. Plant Cell Environ. 41, 2449–2462. doi: 10.1111/pce.13361
Kosugi, S., Ohashi, Y. (2000). Cloning and DNA-binding properties of a tobacco Ethylene-Insensitive3 (EIN3) homolog. Nucleic Acids Res. 28, 960–967. doi: 10.1093/nar/28.4.960
Lange, B. M., Rujan, T., Martin, W., Croteau, R. (2000). Isoprenoid biosynthesis: the evolution of two ancient and distinct pathways across genomes. Proc. Natl. Acad. Sci. U. S. A. 97, 13172–13177. doi: 10.1073/pnas.240454797
Li, D., Liu, Y., Chen, G., Yan, Y., Bai, Z. (2024). The SmERF1b-like regulates tanshinone biosynthesis in Salvia miltiorrhiza hairy root. AoB. Plants. 16, 1–7. doi: 10.1093/aobpla/plad086
Li, W., Nishiyama, R., Watanabe, Y., Ha, C., Kojima, M., An, P., et al. (2018). Effects of overproduced ethylene on the contents of other phytohormones and expression of their key biosynthetic genes. Plant Physiol. Biochem. 128, 170–177. doi: 10.1016/j.plaphy.2018.05.013
Liang, Z., Ma, Y., Xu, T., Cui, B., Liu, Y., Guo, Z., et al. (2013). Effects of abscisic acid, gibberellin, ethylene and their interactions on production of phenolic acids in salvia miltiorrhiza bunge hairy roots. PloS One. 8, e72806. doi: 10.1371/journal.pone.0072806
Liu, S., Wang, Y., Shi, M., Maoz, I., Gao, X., Sun, M., et al. (2022). SmbHLH60 and SmMYC2 antagonistically regulate phenolic acids and anthocyanins biosynthesis in Salvia miltiorrhiza. J. Adv. Res. 42, 205–219. doi: 10.1016/j.jare.2022.02.005
Liu, X., Liu, R., Li, Y., Shen, X., Zhong, S., Shi, H. (2017). EIN3 and PIF3 form an interdependent module that represses chloroplast development in buried seedlings. Plant Cell. 29, 3051–3067. doi: 10.1105/tpc.17.00508
Ma, X. H., Ma, Y., Tang, J. F., He, Y. L., Liu, Y. C., Ma, X. J., et al. (2015). The biosynthetic pathways of tanshinones and phenolic acids in Salvia miltiorrhiza. Molecules. 20, 16235–16254. doi: 10.3390/molecules200916235
Mao, Y., Ma, Y., Chen, T., Ma, X., Xu, Y., Bu, J., et al. (2020). Functional integration of two CYP450 genes involved in biosynthesis of tanshinones for improved diterpenoid production by synthetic biology. ACS Synth. Biol. 9, 1763–1770. doi: 10.1021/acssynbio.0c00136
Meng, L. S., Xu, M. K., Wan, W., Yu, F., Li, C., Wang, J. Y., et al. (2018). Sucrose signaling regulates anthocyanin biosynthesis through a MAPK cascade in Arabidopsis thaliana. Genetics. 210, 607–619. doi: 10.1534/genetics.118.301470
Miziorko, H. M. (2011). Enzymes of the mevalonate pathway of isoprenoid biosynthesis. Arch. Biochem. Biophys. 505, 131–143. doi: 10.1016/j.abb.2010.09.028
Rieu, I., Mariani, C., Weterings, K. (2003). Expression analysis of five tobacco EIN3 family members in relation to tissue-specific ethylene responses. J. Exp. Bot. 54, 2239–2244. doi: 10.1093/jxb/erg240
Rohmer, M., Knani, M., Simonin, P., Sutter, B., Sahm, H. (1993). Isoprenoid biosynthesis in bacteria: a novel pathway for the early steps leading to isopentenyl diphosphate. Biochem. J. 295, 517–524. doi: 10.1042/bj2950517
Shi, M., Huang, F., Deng, C., Wang, Y., Kai, G. (2019). Bioactivities, biosynthesis and biotechnological production of phenolic acids in Salvia miltiorrhiza. Crit. Rev. Food Sci. Nutr. 59, 953–964. doi: 10.1080/10408398.2018.1474170
Shi, M., Luo, X., Ju, G., Li, L., Huang, S., Zhang, T., et al. (2016a). Enhanced diterpene tanshinone accumulation and bioactivity of transgenic Salvia miltiorrhiza hairy roots by pathway engineering. J. Agric. Food Chem. 64, 2523–2530. doi: 10.1021/acs.jafc.5b04697
Shi, Y., Tian, S., Hou, L., Huang, X., Zhang, X., Guo, H., et al. (2012). Ethylene signaling negatively regulates freezing tolerance by repressing expression of CBF and type-A ARR genes in Arabidopsis. Plant Cell. 24, 2578–2595. doi: 10.1105/tpc.112.098640
Shi, M., Zhou, W., Zhang, J., Huang, S., Wang, H., Kai, G. (2016b). Methyl jasmonate induction of tanshinone biosynthesis in Salvia miltiorrhiza hairy roots is mediated by JASMONATE ZIM-DOMAIN repressor proteins. Sci. Rep. 6, 20919. doi: 10.1038/srep20919
Solano, R., Stepanova, A., Chao, Q., Ecker, J. R. (1998). Nuclear events in ethylene signaling: a transcriptional cascade mediated by ETHYLENE-INSENSITIVE3 and ETHYLENE-RESPONSE-FACTOR1. Genes Dev. 12, 3703–3714. doi: 10.1101/gad.12.23.3703
Song, S., Liu, B., Song, J., Pang, S., Song, T., Gao, S., et al (2022). A molecular framework for signaling crosstalk between jasmonate and ethylene in anthocyanin biosynthesis, trichome development, and defenses against insect herbivores in Arabidopsis. J. Integr. Plant Biol. 64, 1770–1788. doi: 10.1111/jipb.13319
Sun, M., Shi, M., Wang, Y., Huang, Q., Yuan, T., Wang, Q., et al. (2019). The biosynthesis of phenolic acids is positively regulated by the JA-responsive transcription factor ERF115 in Salvia miltiorrhiza. J. Exp. Bot. 70, 243–254. doi: 10.1093/jxb/ery349
Sun, X., Wang, Y., Sui, N. (2018). Transcriptional regulation of bHLH during plant response to stress. Biochem. Biophys. Res. Commun. 503, 397–401. doi: 10.1016/j.bbrc.2018.07
Tamura, K., Stecher, G., Peterson, D., Filipski, A., Kumar, S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. doi: 10.1093/molbev/mst197
Tieman, D. M., Ciardi, J. A., Taylor, M. G., Klee, H. J. (2001). Members of the tomato LeEIL (EIN3-like) gene family are functionally redundant and regulate ethylene responses throughout plant development. Plant J. 26, 47–58. doi: 10.1046/j.1365-313x.2001.01006.x
Wang, Y., Wan, L., Zhang, L., Zhang, Z., Zhang, H., Quan, R., et al. (2012). An ethylene response factor OsWR1 responsive to drought stress transcriptionally activates wax synthesis related genes and increases wax production in rice. Plant Mol. Biol. 78, 275–288. doi: 10.1007/s11103-011-9861-2
Wang, J. W., Wu, J. Y. (2010). Tanshinone biosynthesis in Salvia miltiorrhiza and production in plant tissue cultures. Appl. Microbiol. Biotechnol. 88, 437–449. doi: 10.1007/s00253-010-2797-7
Wen, J., Lv, J., Zhao, K., Zhang, X., Li, Z., Zhang, H., et al. (2022). Ethylene-inducible AP2/ERF transcription factor involved in the capsaicinoid biosynthesis in Capsicum. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.832669
Xu, Y., Huo, L., Zhao, K., Li, Y., Zhao, X., Wang, H., et al. (2023). Salicylic acid delays pear fruit senescence by playing an antagonistic role toward ethylene, auxin, and glucose in regulating the expression of PpEIN3a. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.1096645
Yan, H., Pei, X., Zhang, H., Li, X., Zhang, X., Zhao, M., et al. (2021). MYB-mediated regulation of anthocyanin biosynthesis. Int. J. Mol. Sci. 22, 3103. doi: 10.3390/ijms22063103
Yamasaki, K., Kigawa, T., Inoue, M., Yamasaki, T., Yabuki, T., Aoki, M., et al. (2005). Solution structure of the major DNA-binding domain of Arabidopsis thaliana ethylene-insensitive3-like3. J. Mol. Biol. 348, 253–264. doi: 10.1016/j.jmb.2005.02.065
Yokotani, N., Tamura, S., Nakano, R., Inaba, A., Kubo, Y. (2003). Characterization of a novel tomato EIN3-like gene (LeEIL4). J. Exp. Bot. 54, 2775–2776. doi: 10.1093/jxb/erg308
Yu, H., Jiang, M., Xing, B., Liang, L., Zhang, B., Liang, Z. (2020). Systematic analysis of Kelch repeat F-box (KFB) protein gene family and identification of phenolic acid regulation members in Salvia miltiorrhiza Bunge. Genes (Basel). 11, 557. doi: 10.3390/genes11050557
Zhang, J., Xu, H., Wang, N., Jiang, S., Fang, H., Zhang, Z., et al. (2018). The ethylene response factor MdERF1B regulates anthocyanin and proanthocyanidin biosynthesis in apple. Plant Mol. Biol. 98, 205–218. doi: 10.1007/s11103-018-0770-5
Zhong, S., Zhao, M., Shi, T., Shi, H., An, F., Zhao, Q., et al. (2009). EIN3/EIL1 cooperate with PIF1 to prevent photo-oxidation and to promote greening of Arabidopsis seedlings. Proc. Natl. Acad. Sci. U. S. A. 106, 21431–21436. doi: 10.1073/pnas.0907670106
Zhou, M., Memelink, J. (2016). Jasmonate-responsive transcription factors regulating plant secondary metabolism. Biotechnol. Adv. 34, 441–449. doi: 10.1016/j.bioteChadv.2016.02.004
Zhou, W., Shi, M., Deng, C., Lu, S., Huang, F., Wang, Y., et al. (2021). The methyl jasmonate-responsive transcription factor SmMYB1 promotes phenolic acid biosynthesis in Salvia miltiorrhiza. Hortic. Res. 8, 10. doi: 10.1038/s41438-020-00443-5
Keywords: Salvia miltiorrhiza, ethylene, SmEIL1 transcription factor, tanshinone, regulatory mechanism
Citation: Li X, Xu M, Zhou K, Hao S, Li L, Wang L, Zhou W and Kai G (2024) SmEIL1 transcription factor inhibits tanshinone accumulation in response to ethylene signaling in Salvia miltiorrhiza. Front. Plant Sci. 15:1356922. doi: 10.3389/fpls.2024.1356922
Received: 16 December 2023; Accepted: 18 March 2024;
Published: 02 April 2024.
Edited by:
Xiaozeng Yang, Beijing Academy of Agricultural and Forestry Sciences, ChinaReviewed by:
Shi Qiu, Shanghai University of Traditional Chinese Medicine, ChinaCopyright © 2024 Li, Xu, Zhou, Hao, Li, Wang, Zhou and Kai. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wei Zhou, emhvdXdlaTE5ODEwNTAxQDE2My5jb20=; Guoyin Kai, Z3VveWlua2FpMUAxMjYuY29t
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.