
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Plant Sci. , 23 February 2024
Sec. Plant Pathogen Interactions
Volume 15 - 2024 | https://doi.org/10.3389/fpls.2024.1355178
This article is part of the Research Topic Wheat Disease Resistance: Diagnosis, Germplasm Mining, and Molecular Breeding View all 15 articles
Systemic acquired resistance (SAR) is an inducible disease resistance phenomenon in plant species, providing plants with broad-spectrum resistance to secondary pathogen infections beyond the initial infection site. In Arabidopsis, SAR can be triggered by direct pathogen infection or treatment with the phytohormone salicylic acid (SA), as well as its analogues 2,6-dichloroisonicotinic acid (INA) and benzothiadiazole (BTH). The SA receptor non-expressor of pathogenesis-related protein gene 1 (NPR1) protein serves as a key regulator in controlling SAR signaling transduction. Similarly, in common wheat (Triticum aestivum), pathogen infection or treatment with the SA analogue BTH can induce broad-spectrum resistance to powdery mildew, leaf rust, Fusarium head blight, and other diseases. However, unlike SAR in the model plant Arabidopsis or rice, SAR-like responses in wheat exhibit unique features and regulatory pathways. The acquired resistance (AR) induced by the model pathogen Pseudomonas syringae pv. tomato strain DC3000 is regulated by NPR1, but its effects are limited to the adjacent region of the same leaf and not systemic. On the other hand, the systemic immunity (SI) triggered by Xanthomonas translucens pv. cerealis (Xtc) or Pseudomonas syringae pv. japonica (Psj) is not controlled by NPR1 or SA, but rather closely associated with jasmonate (JA), abscisic acid (ABA), and several transcription factors. Furthermore, the BTH-induced resistance (BIR) partially depends on NPR1 activation, leading to a broader and stronger plant defense response. This paper provides a systematic review of the research progress on SAR in wheat, emphasizes the key regulatory role of NPR1 in wheat SAR, and summarizes the potential of pathogenesis-related protein (PR) genes in genetically modifying wheat to enhance broad-spectrum disease resistance. This review lays an important foundation for further analyzing the molecular mechanism of SAR and genetically improving broad-spectrum disease resistance in wheat.
As early as 1901, researchers found that plants infected with pathogens develop higher levels of resistance to secondary infections (Görlach et al., 1996). Over the next 30 years, several descriptive studies were conducted, which collectively indicated the presence of systemic immunity in plants. Among them, the concept of SAR was widely accepted, referring to the ability of plants to develop broad-spectrum resistance against secondary pathogens during the response to primary pathogen infection (Görlach et al., 1996). Tobacco mosaic virus and its Solanaceae hosts were used in the early study of SAR (Görlach et al., 1996). With the deepening of research, SAR has been demonstrated to exist widely in various plants, and effectively combat viral, bacterial, and fungal diseases, and induce disease resistance responses with long-term and systematic nature (Zhang et al., 1999).
Usually caused by local infection of pathogenic bacteria, SAR is an important part of plant disease resistance system, and is an inducible broad-spectrum immunity of plant resistance to pathogenic bacteria, lacks specificity toward the initial infection (Görlach et al., 1996). When a non-lethal pathogen causes localized programmed cell death, it triggers the accumulation of the plant hormone SA and the expression of PR genes, thereby protecting the rest of the plant from secondary infections for weeks to months. SAR can even be transmitted to offspring through epigenetic regulation (Fu and Dong, 2013).
In the model plant Arabidopsis thaliana, infection with pathogenic microorganisms or treatment with SA and its analogs INA and BTH can induce SAR response, associated with the transcriptional activation of PR genes (Fu and Dong, 2013). Specifically, the SA receptor protein NPR1 in Arabidopsis is a key transcriptional regulatory factor for SAR. Upon infection with a primary pathogen or treatment with SA analogs, the intracellular SA level significantly increases, and NPR1 protein undergoes redox modification to translocate from cytoplasm to nucleus. After phosphorylation, it forms a complex with TGA transcription factors, promoting the expression of various PR genes and enhancing the plant resistance to secondary pathogens (Delaney et al., 1995; Mou et al., 2003). The endogenous level of SA in plant defense responses is largely dependent on the intensity of the hypersensitive response (HR) induced by the pathogen (Mou et al., 2003).
In Arabidopsis, the homologous proteins of NPR1, NPR3 and NPR4, participate in E3 ligase-mediated degradation of NPR1 in an SA-dependent manner (Fu et al., 2012). When the SA level is low, proteases bind to NPR4 and degrade NPR1. However, during pathogen infection, when the SA level increases, it competitively binds to NPR4. This binding leads to the accumulation of NPR1 and activates the NPR1-mediated defense response. When plants develop HR as part of the defense response, SA levels reach very high levels in the plant, and in this scenario, SA binds to NPR3 to promote its interaction with NPR1, ultimately leading to the transformation of NPR1 (Fu et al., 2012; Moreau et al., 2012). Recently, the crystal structure of the NPR1 protein in Arabidopsis has been resolved, revealing its existence as a “bird-shaped” dimer. NPR1-induced defense gene reprogramming in response to various biotic and abiotic stresses may involve not only TGA homodimers but also heterologous transcription activators, repressors, and lipid metabolites (Kumar et al., 2022).
In rice, infection with Pseudomonas syringae or treatment with BTH can induce SAR-like response against the rice blast fungus and is associated with the transcriptional activation of PR genes (Smith and Métraux, 1991; Schweizer et al., 1999) However, compared to Arabidopsis, rice has relatively higher endogenous levels of SA, which do not increase upon pathogen infection (Silverman et al., 1995; Chen et al., 1997). The protein interaction between rice NPR1 homolog (rNH1) and TGA transcription factors is conserved (Chern et al., 2001). Overexpression of the Arabidopsis AtNPR1 gene in rice significantly enhances the broad-spectrum resistance of plant to various pathogens, including Xanthomonas oryzae pv. oryzae (Xoo), Magnaporthe oryzae, and Cochliobolus miyabeanus (Quilis et al., 2008; Xu et al., 2017). Further studies indicate that the overexpression of the rNH1 gene in rice not only enhances resistance to Xoo but also increases sensitivity to light and BTH treatment (Chern et al., 2005; Xu et al., 2017). Additionally, the WRKY transcription factor OsWRKY45 is a key regulator of the SA/BTH signaling pathway independent of rNH1 in rice (Shimono et al., 2007; Nakayama et al., 2013).
In Triticeae crops of wheat and barley, both primary pathogen infection and BTH treatment can induce broad-spectrum resistance against diseases like powdery mildew, rust, and Fusarium head blight. However, SAR in Triticeae crops such as wheat and barley, when induced by pathogen infection or BTH treatment, displays distinct characteristics and regulatory pathways compared to model plants like Arabidopsis and rice. As a result, three different “SAR-like responses” have been observed in wheat and barley: acquired resistance (AR), systemic immunity (SI), and BTH-induced resistance (BIR) (Wang et al., 2018) (Table 1, Figure 1).
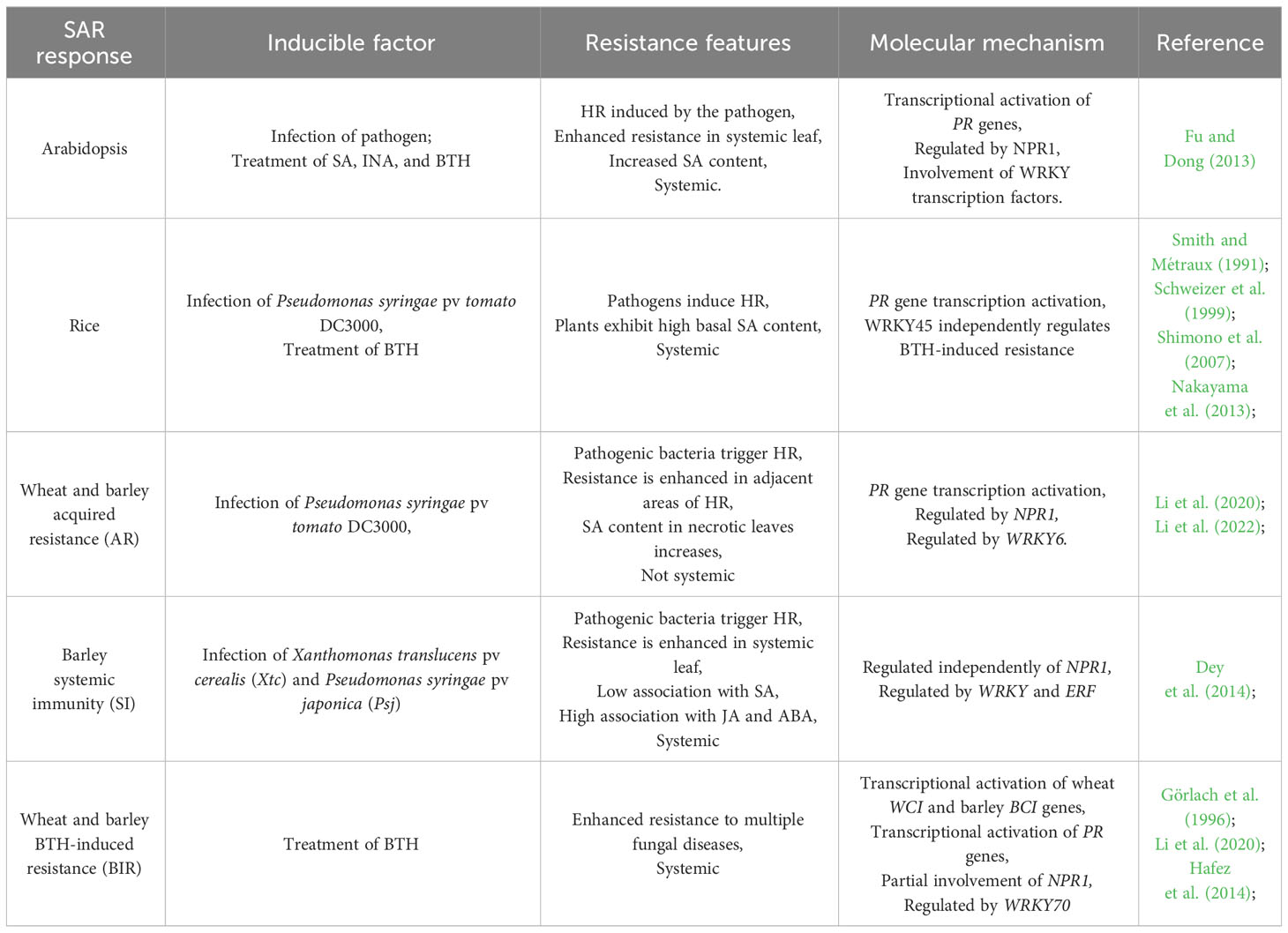
Table 1 Features and molecular mechanism of SAR response in Arabidopsis, rice, and Triticeae crops of wheat and barley.
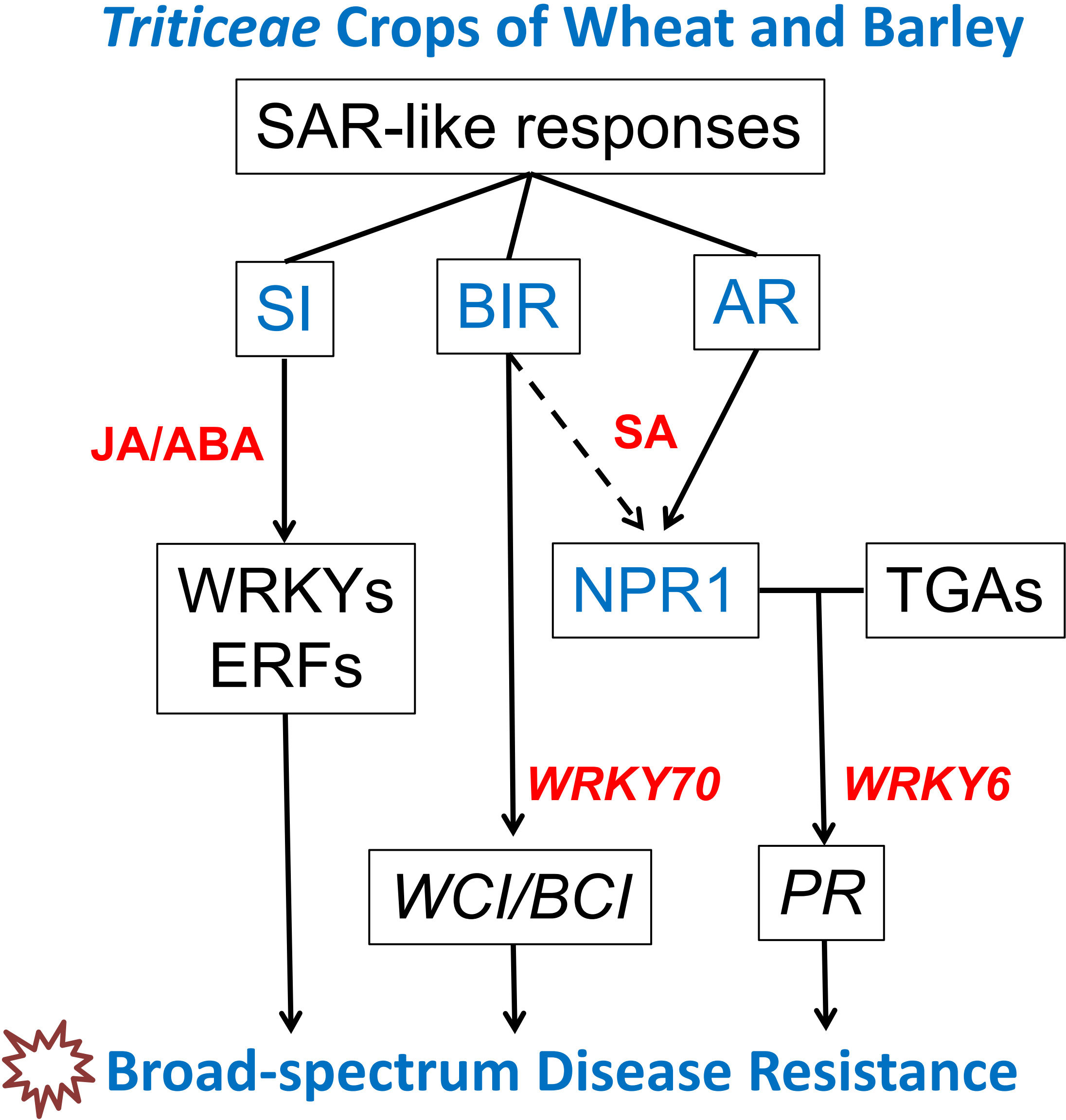
Figure 1 Regulatory network of SAR-like responses in Triticeae crops of wheat and barley. This network encompasses three SAR-like responses: acquired resistance (AR), systemic immunity (SI), and benzothiadiazole-induced resistance (BIR). Recent studies utilizing transcriptome sequencing and wheat transgenic lines have contributed to the prediction of this regulatory network. Understanding the molecular mechanisms that underlie SAR-like responses provides valuable gene resources for enhancing the broad-spectrum resistance of wheat to diseases such as stripe/leaf rust, powdery mildew, Fusarium head blight, Fusarium crown rot, sharp eyespot, and others.
In Triticeae crops of wheat and barley, AR like the SAR in model plant can be observed in the neighboring area after injection with Pseudomonas syringae pv. tomato strain DC3000. This AR response enhances resistance against the secondary pathogen M. oryzae by upregulating PR genes (Colebrook et al., 2012). However, unlike SAR in Arabidopsis, the AR response in barley does not extend to other leaves, meaning it lacks systemic properties. Inducing AR response in wheat by injecting P. syringae DC3000 significantly enhances resistance to the highly virulent Puccinia tritici pathotype THTT (Gao et al., 2018). Overexpression of the wheat NPR1 gene in barley (wNPR1-OE) significantly enhances resistance to the M. oryzae strain Guy11, while gene silencing of the barley HvNPR1 gene (HvNPR1-Kd) significantly reduces AR response to Guy11, indicating that the NPR1 gene directly regulates the AR response in barley. Using quantitative real-time polymerase chain reaction (qRT-PCR), it was found that the induction levels of barley PR genes, including HvPR1b, HvPR2, HvPR3_Chit2b, and HvPR5, were closely correlated with the expression levels of NPR1 transgene in wNPR1-OE and HvNPR1-Kd barley transgenic plants (Wang et al., 2016). Further transcriptomic analysis revealed that the expression of HvPR1, HvPR2, HvPR3, HvPR5, HvPR9, and HvPR13 genes was significantly induced in the AR response to DC3000 in barley, and the induction patterns were positively correlated with the expression levels of the NPR1 gene, suggesting that these genes are regulated by the NPR1 gene in the AR response (Gao et al., 2018).
By transcriptome sequencing, it was found that barley transcription factor gene HvWRKY6 may act as a regulatory factor for AR in wheat (Li et al., 2020). The AR response was induced by injecting DC3000, and the resistance level of wheat transgenic lines overexpressing HvWRKY6 (HvWRKY6-OE) to M. oryzae strain P131 was significantly improved. The expression levels of TaPR1a, TaPR2 and TaPR4b genes in HvWRKY6-OE were significantly increased, indicating that HvWRKY6 gene plays an important regulatory role in AR response. HvWRKY6-OE have shown enhanced resistance to P. tritici pathotype THTT, Puccinia striiformis f. sp. tritici pathotype CYR32, Fusarium crown rot, and sharp eyespot to varying degrees (Li et al., 2020; Li et al., 2022). Transcriptome sequencing analysis showed that HvWRKY6 gene activated wheat defense response in a pathogen-independent manner. HvWRKY6 partially activates the SAR-associated transcripts, including calcium-associated disease resistance pathways and part of the effector-triggered immunity (ETI) (Li et al., 2022). HvWRKY6-mediated resistance response is related to the activation of SA pathway and the inhibition of ABA and JA pathways (Li et al., 2022).
Injecting Xanthomonas translucens pv. cerealis (Xtc) or Pseudomonas syringae pv. japonica (Psj) into barley leaves can induce SI against Xtc infection in other systemic healthy leaves (Dey et al., 2014). However, unlike SAR in Arabidopsis, the SI process in barley is not induced by SA or BTH, but is closely associated with jasmonic acid (JA) and abscisic acid (ABA). Further studies have shown that the SI process in barley is regulated by several WRKY and ERF transcription factors but has a relatively low correlation with the NPR1 gene.
BTH has been successfully developed as a commercial plant resistance inducer and applied in agricultural production. In Arabidopsis and tobacco, induction of resistance and PR gene expression by INA/BTH has been observed (Uknes et al., 1992; Lawton et al., 1995). It is likely that SA/INA/BTH induce SAR through the same signal transduction pathway (Görlach et al., 1996). Treatment of wheat with BTH induces systemic BIR against multiple diseases such as powdery mildew, leaf rust, and Fusarium head blight. However, most PR genes are insensitive to SA and BTH, while another class of BTH-induced genes, such as the wheat chemical induced (WCI) gene and the barley chemical induced (BCI) gene, may play important roles in the BIR (Beßer et al., 2000; Hafez et al., 2014). Transcriptome sequencing showed that the barley transcription factor gene HvWRKY70 may act as a regulatory factor of BIR in wheat. BTH-induced SAR reaction showed that several PR genes and BCI genes in wheat transgenic lines overexpressing HvWRKY70 (HvWRKY70-OE) were significantly up-regulated by BTH, indicating that HvWRKY70 gene was involved in the regulation of BIR. In addition, HvWRKY70-OE showed increased resistance to P. striiformis f. sp. tritici pathotype CYR32 and Blumeria graminis f. sp. tritici pathotype E20 (Li et al., 2020).
NPR1, as a SA receptor protein, possesses three conserved domains: the N-terminal broad-complex, tramtrack, and brica-brac/poxvirus, zinc finger (BTB/POZ) domain, the central ankyrin-repeat (ANK) domain, and the C-terminal NPR1/NIM1-like domain. The BTB/POZ domain is a potential target for the E3 ubiquitin degradation pathway (Petroski and Deshaies, 2005), while the ANK domain is mainly involved in the interaction with TGA transcription factors (Cao et al., 1997; Sedgwick and Smerdon, 1999). The NPR1/NIM1-like domain participates in SA binding along with the BTB/POZ domain (Wu et al., 2012).
NIMIN is a protein family first discovered in Arabidopsis and plays a crucial role in regulating the NPR1-mediated immune signaling pathway. It finely regulates the dynamic defense process against diseases. Hermann et al. reported that during different stages of SAR, different NIMIN-NPR1 complexes are formed to promote the activation of defense genes. In normal plant growth, the binding of NIMIN3 to NPR1 prevents excessive activation of immune responses (Hermann et al., 2013). However, upon pathogen attack, NIMIN2 and NIMIN1 sequentially bind to NPR1, promoting the binding with SA and the expression of PR genes, thereby enhancing plant immune responses (Maier et al., 2011). NIMIN proteins regulate the dynamic balance of NPR1 and contribute to the rapid upregulation of defense genes, ensuring successful resistance against invading pathogens (Hermann et al., 2013). TEOSINTE BRANCHED 1, CYCLOIDEA, PCF (TCP) transcription factors are also essential participants in the SA signaling pathway. NPR1 interacts with nuclear TCP transcription factors TCP8, TCP14 and TCP15, promoting SAR function. TCP15 directly binds to the TCP binding site in the PR5 promoter, enhancing PR5 expression, contributing to ETI, and playing an important role in SAR (Li et al., 2018). NPR1 has also been reported to interact with cyclin-dependent kinase 8 (CDK8) and WRKY18 in Arabidopsis, and SA can promote their interaction. CDK8 facilitates the expression of NPR1 and PR1 genes by recruiting RNA polymerase II to their promoter and coding regions. NPR1, in turn, recruits CDK8, promoting its own and target gene expression and contributing to the establishment of plant immunity (Chen et al., 2019).
Both fungal pathogen Colletotrichum fructicola effector protein CfEC12 and apple MdNPR1 interact with NIM1-interacting (MdNIMIN2) protein. CfEC12 competes with MdNIMIN2 in binding to the 13-63 amino acid position of MdNPR1, thereby suppressing the expression of downstream PR genes and immune responses (Shang et al., 2023). NPR1 plays a key role in limiting co-infection of TuMV, a member of the largest RNA virus genus in plant, and this resistance is counteracted by viral RNA-dependent RNA polymerase nuclear inclusion body B (NIb). NIb interacts with NPR1 and targets its SUMO interacting motif 3 (SIM3); NIb interferes with NPR1-SIM3 interaction and subsequent SUMoization; NIb also affects the SUMO-dependent phosphorylation of NPR1; targeting NPR1-SIM3 is a conserved ability of NIBs from different descendant viruses. Potyvirus uses NIb to suppress NPR1-mediated resistance by disrupting NPR1 SUMO (Liu et al., 2023a). The RXLR effector protein RXLR48 in Phytophthora capsica interacts with NPR1 and inhibits plant defense. RxLR48 promotes nuclear localization of NPR1 and inhibits its proteasome-mediated degradation, suggesting that RxLR48 inhibits SA signaling by targeting the transcriptional regulator NPR1 (Li et al., 2019). Using yeast two-hybrid screening, a virulent effector protein, PNPi, that directly targets the NPR1 protein was identified in wheat rust, inhibiting the interaction between NPR1 and TGA2, and suppressing wheat SAR (Wang et al., 2016). There were 19 PNPi-like secreted proteins with RlpA-like double-psi beta-barrel (DPBB_1) conserved structure in wheat leaf rust. Twelve PNPi-like effector protein genes were cloned and their interaction with wheat NPR1 protein was verified by yeast two-hybrid system. Among them, four PNPi-like effector proteins could interact with wheat NPR1 protein (Zhao et al., 2020).
In the Triticeae crops of wheat and barley, significant progress has been made in the study of NPR1 gene (Table 2). For example, overexpression of Arabidopsis AtNPR1 or oat ScNPR1 in common wheat significantly enhances plant resistance against Fusarium graminearum (Gao et al., 2013; Yu et al., 2017). The interaction between NPR1 homologous proteins and TGA transcription factor homologous proteins is highly conserved in wheat and rice, suggesting that wheat NPR1 homologous proteins have conserved disease resistance functions (Cantu et al., 2013). Bioinformatics analysis revealed the presence of nine NPR1 homologous genes (TaNPR1) in wheat, with six members located on the homoeologous group 3 chromosomes named TaG3NPR1, and three members located on the homoeologous group 7 chromosomes named TaG7NPR1. TaG3NPR regulates the expression of PR genes in the SA signaling pathway. Additionally, a novel fusion pattern of NPR1 protein, as NPR1 fused with nucleotide-binding adaptor shared by APAF-1, R proteins, and CED-4 (NB-ARC-NPR1) was discovered on the 7A chromosome in common wheat (Ta7ANPR1). Under biotic stress conditions, the Ta7ANPR1 gene simultaneously transcribes two mRNAs, one encoding an NB-ARC protein and the other encoding an NB-ARC-NPR1 fusion protein. The Ta7ANPR1 gene negatively regulates plant defense responses against wheat rust through the NB-ARC-NPR1 fusion protein (Wang et al., 2020b). Calcineurin B-like interacting protein kinases (CIPKs) have been shown to be essential for biological stress tolerance in plant-pathogen interactions. A CIPK homolog, TaCIPK10, was identified and cloned from wheat. TaCIPK10 physically interacts and phosphorylates with AtNPR3/4 homologous TaNH2 to regulate wheat resistance to P. striiformis f. sp. tritici (Liu et al., 2019a).
Furthermore, it has been reported that about 40 NPR1 homologous genes or NPR family coding genes can be identified in Triticum aestivum, Triticum urartu, Triticum dicoccoides, and Aegilops tauschii. NPR1 homologous genes exhibit good collinearity in common wheat and its close relatives. Based on RNA-seq data, TaNPR1 homologous genes exhibit different tissue-specific expression patterns, and TaNPR1-A/B/D, TaNPR3-A/B/D, and TaNPR4-A/B/D are significantly induced under biotic stress conditions (Liu et al., 2019b). Through gene expression profile analysis, three NPR1 homologous genes named TaNPR1, TaNPR2, and TaNPR3 were cloned from wheat near-isogenic lines resistant to Fusarium head blight (Yang et al., 2013). Among them, TaNPR1 and TaNPR3 showed significant upregulation in response to Fusarium graminearum, suggesting their involvement in wheat defense against Fusarium head blight. Association analysis using a natural population consisting of 178 winter wheat genotypes revealed that two NPR genes located on the 2AL and 2DL chromosomes of common wheat were associated with resistance to Fusarium head blight (Diethelm et al., 2014). Both SA and the biocontrol agent Trichoderma induce the expression of the wheat NPR1 gene and enhance plant resistance against powdery mildew (Ahangar et al., 2017).
PR proteins are a class of water-soluble proteins produced by plants in response to pathogen invasion or non-biological stimuli. When subjected to biotic or abiotic stress, the expression levels of PR genes rapidly increase and are often used as markers of plant immune activation. Numerous studies have shown that PR proteins play important roles in plant disease resistance and SAR (Hamamouch et al., 2011). To date, a total of 18 PR gene families have been identified in various plant species in response to different pathogens (van Loon et al., 2006; Ferreira et al., 2007; Sels et al., 2008; Wang et al., 2018). In addition, PR gene has been widely used as indicator gene for wheat resistance response to monitor the intensity of resistance response (Wu et al., 2019).
The PR1 protein family, as the earliest identified defense-related proteins, has been extensively studied for its disease resistance mechanisms. PR1 homologous genes (PR1a and PR1b) have been successfully cloned in wheat and barley and induced to express by a variety of pathogens (Molina et al., 1999; Gao et al., 2015). These genes are considered as key downstream regulatory genes of NPR1 in SAR-like responses in Triticeae crops of wheat and barley (Wang et al., 2016). RNA-seq transcriptome sequencing was performed on resistant wheat line carrying leaf rust resistance gene TcLr19 and susceptible wheat variety “Chinese Spring” inoculated with the P. tritici pathotype PHNT, and seven SA-induced TaPR1 genes associated with plant disease resistance were identified. qRT-PCR results showed that among these TaPR1 genes, TaPR1-4 had the largest induction effect by the infection of leaf rust (Wang et al., 2022a). The TdPR1.2 gene identified from Trticum turgidum also plays a vital role in enhancing plant resistance to abiotic stress (Ghorbel et al., 2021). The expression and genetic polymorphisms of PR1, PR2, PR4, PR9 and PR10 in sixteen Egyptian wheat genotypes were analyzed to clarify the expression mechanism of PR genes during stripe rust infection (Esmail et al., 2020).
The PR1 protein plays a pivotal role as an interaction hub in the extracellular space. TaPR1a protein in wheat interacts with lipid transfer protein TaLTP3 (PR14) in the extracellular matrix, and overexpression of TaLTP3 in wheat transgenic line can specifically activate the transcription of TaPR1a gene, as well as multiple plant hormone pathways including SA, JA, and auxin, providing new insights into the synergistic mechanism of PR proteins (Zhao et al., 2021). TaPR1-4 interacts with thaumatin-like protein TaPR5/TaTLP1 through the αIV helix and participates in the defense process against wheat leaf rust through the CAPE1 motif (Wang et al., 2022a).
Research has shown that wheat TaPR1 protein can directly interact with the fungal toxin ToxA produced by the tan spot pathogen (Pyrenophora tritici-repentis) and mediate the induction of necrotic reactions in susceptible wheat (Lu et al., 2014). The effector SnTox3 from Parastagonospora nodorum elicits a strong necrotic response in susceptible wheat and also interacts with wheat TaPR-1, SnTox3 prevented CAPE1 from being released from TaPR1 in vitro, SnTox3 independently induced necrosis through Snn3 recognition, and inhibited host defense through direct interaction with the TaPR1 protein (Sung et al., 2021). The wheat transgenic lines overexpressing TaPR1a gene (TaPR1a-OE) showed increased resistance to both wheat leaf rust and stripe rust. By targeting TaPR1a protein in the extracellular space, wheat rust effector protein PNPi inhibits plant disease resistance and is conducive to rust infection (Bi et al., 2020).
Significant efforts have been dedicated to the genome-wide identification of PR1 genes in wheat. Twelve PR-1 genes encoding the CAP superfamily domain were identified in the genome of the Triticum turgidum subspecies. Phylogenetic analysis showed that PR1 gene could be divided into three groups according to variations in conserved domain. Most TdPR1 proteins present an N-terminal signaling peptide. The expression profile analysis showed that the PR-1 gene family was organism specific and could be induced by different abiotic stresses (Zribi et al., 2023). Bioinformatics tools and RNA sequencing discovered 86 potential wheat TaPR1 genes, and TaPR1 genes were involved in SA signaling pathway, MAPK signaling pathway, and phenylalanine metabolism in response to infection of P. striiformis f. sp. tritici pathotype CYR34. One particular gene, TaPR1-7, was found to be associated with resistance to P. striiformis f. sp. tritici in a biparental wheat population (Liu et al., 2023b). TaPR1, as the representative of wheat SAR downstream defense related protein genes, holds significant potential and is deserving of further exploration and investigation.
PR2 and PR3 genes encode β-1,3-endoglucanases and chitinase proteins, respectively. Molecular docking analysis of β-1,3-endoglucanases and chitinase proteins revealed key amino acid residues involved in ligand binding and important interactions, which may play an important role in plant defense against fungal pathogens (Numan et al., 2021). The PR4 family features a Barwin domain at the C-terminus, which endows the host plant with disease resistance. A total of four PR-4 genes were identified from the genome of the Qingke (Hordeum vulgare L. var. nudum) by HMM analysis. Expression profile analysis confirmed that PR-4 was involved in the defense response to drought, cold, and powdery mildew infection, and the transcription of two barley PR4 genes were differentially regulated by MeJA and SA (Wang et al., 2022b).
Wheat thaumatin-like protein (TaTLP/TaPR5) are secreted into the apoplastic space, and when stimulated by biological or abiotic stresses, their expression levels increase rapidly, and they show antifungal activity in various plant species, which is an important component of plant SAR and a sign of plant disease resistance (Han et al., 2023). Wheat TaTLP1 is involved in the resistance to leaf rust. TaPR1 and TaTLP1 also have direct protein interaction in the extracellular space, positively regulating wheat resistance to leaf rust in a reactive oxygen species (ROS)-dependent and direct germicidal manner (Wang et al., 2022a). Leaf rust effector protein Pt_21 directly targets wheat TaTLP1 and inhibits host defense response by inhibiting the antifungal activity of TaTLP1 (Wang et al., 2023).
Currently, other PR genes in barley and wheat have been rarely reported. However, considering the research potential and application prospects of PR genes in other crops, they hold great promise for genetic improvement of wheat disease resistance. Drawing from information on PR genes reported in other plant species, we conducted a preliminary prediction and classification of 18 PR gene families in wheat (Table 3).
The SAR-like responses observed in Triticeae crops of wheat and barley (AR, SI, and BIR) exhibit significant differences compared to those in Arabidopsis and rice. In recent years, studies on the NPR1 homologous genes in wheat have provided preliminary clues to understanding the molecular mechanisms underlying these differences. The key regulatory factors and downstream functional proteins in SAR, including the SA receptor proteins NPR3/4, WRKY transcription factors, and PR proteins, still require further exploration. Uncovering the key nodal genes involved in SAR-like responses in wheat, as well as the co-regulated downstream genes involved in these biological processes, will provide important genetic resources for broad-spectrum disease resistance improvement in wheat. Additionally, with the continuous advancement of genomics and the widespread application of gene editing technologies, knockout of key negative regulatory genes involved in SAR-like responses in wheat can generate innovative disease-resistant germplasm resources, demonstrating significant research prospects and application potential.
SZ: Data curation, Investigation, Visualization, Writing – original draft, Software. ML: Data curation, Investigation, Software, Visualization, Writing – original draft. XR: Data curation, Investigation, Writing – original draft. CW: Data curation, Investigation, Writing – original draft. XS: Formal analysis, Funding acquisition, Supervision, Writing – review & editing. MS: Formal analysis, Funding acquisition, Supervision, Writing – review & editing. XY: Formal analysis, Supervision, Writing – review & editing. XW: Conceptualization, Funding acquisition, Project administration, Supervision, Writing – review & editing, Formal analysis.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the National Key Research and Development Program of China (2023YFD1201002), Provincial Natural Science Foundation of Hebei for Outstanding Young Scientists (C2022204010), Provincial Natural Science Foundation of Hebei (C2021204008 and C2021204010), Local Science and Technology Development Fund Projects Guided by the Central Government (236Z6501G and 236Z6302G), Provincial Natural Science Foundation of Hebei for Excellent Young Scientists (C2023204188), the State Key Laboratory of North China Crop Improvement and Regulation (NCCIR2021ZZ-17), and S&T Program of Hebei (23567601H).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Aglas, L., Soh, W. T., Kraiem, A., Wenger, M., Brandstetter, H., Ferreira, F. (2020). Ligand binding of PR-10 proteins with a particular focus on the bet v 1 allergen family. Curr. Allergy Asthm. Rep. 20, 1–11. doi: 10.1007/s11882-020-00918-4
Ahangar, L., Ranjbar, G. A., Babaeizad, V., Najafi Zarrini, H., Biabani, A. (2017). Assay of NPR1 gene expression in wheat under powdery mildew stress. J. Crop Prot. 6, 157–166.
Beßer, K., Jarosch, B., Langen, G., Kogel, K. H. (2000). Expression analysis of genes induced in barley after chemical activation reveals distinct disease resistance pathways. Mol. Plant Pathol. 1, 277–286. doi: 10.1046/j.1364-3703.2000.00031.x
Bi, W., Zhao, S., Zhao, J., Su, J., Yu, X., Liu, D., et al. (2020). Rust effector PNPi interacting with wheat TaPR1a attenuates plant defense response. Phytopathol. Res. 2, 1–14. doi: 10.1186/s42483-020-00075-6
Blein, J. P., Coutos-Thévenot, P., Marion, D., Ponchet, M. (2002). From elicitins to lipid-transfer proteins: a new insight in cell signalling involved in plant defence mechanisms. Trends Plant Sci. 7, 293–296. doi: 10.1016/S1360-1385(02)02284-7
Cantu, D., Yang, B., Ruan, R., Li, K., Menzo, V., Fu, D., et al. (2013). Comparative analysis of protein-protein interactions in the defense response of rice and wheat. BMC Genomics 14, 1–12. doi: 10.1186/1471-2164-14-166
Cao, H., Glazebrook, J., Clarke, J. D., Volko, S., Dong, X. (1997). The Arabidopsis NPR1 gene that controls systemic acquired resistance encodes a novel protein containing ankyrin repeats. Cell 88, 57–63. doi: 10.1016/S0092-8674(00)81858-9
Chen, J., Mohan, R., Zhang, Y., Li, M., Chen, H., Palmer, I. A., et al. (2019). NPR1 promotes its own and target gene expression in plant defense by recruiting CDK8. Plant Physiol. 181, 289–304. doi: 10.1104/pp.19.00124
Chen, Z., Iyer, S., Caplan, A., Klessig, D. F., Fan, B. (1997). Differential accumulation of salicylic acid and salicylic acid-sensitive catalase in different rice tissues. Plant Physiol. 114, 193–201. doi: 10.1104/pp.114.1.193
Chern, M., Fitzgerald, H. A., Canlas, P. E., Navarre, D. A., Ronald, P. C. (2005). Overexpression of a rice NPR1 homolog leads to constitutive activation of defense response and hypersensitivity to light. Mol. Plant-Microbe Interact. 18, 511–520. doi: 10.1094/MPMI-18-0511
Chern, M. S., Fitzgerald, H. A., Yadav, R. C., Canlas, P. E., Dong, X., Ronald, P. C. (2001). Evidence for a disease-resistance pathway in rice similar to the NPR1-mediated signaling pathway in Arabidopsis. Plant J. 27, 101–113. doi: 10.1046/j.1365-313x.2001.01070.x
Christensen, A. B., Cho, B. H., Næsby, M., Gregersen, P. L., Brandt, J., Madriz-Ordeñana, K., et al. (2002). The molecular characterization of two barley proteins establishes the novel PR-17 family of pathogenesis-related proteins. Mol. Plant Pathol. 3, 135–144. doi: 10.1046/j.1364-3703.2002.00105.x
Colebrook, E. H., Creissen, G., McGrann, G. R. D., Dreos, R., Lamb, C., Boyd, L. A. (2012). Broad-spectrum acquired resistance in barley induced by the Pseudomonas pathosystem shares transcriptional components with Arabidopsis systemic acquired resistance. Mol. Plant-Microbe Interact. 25, 658–667. doi: 10.1094/MPMI-09-11-0246
Custers, J. H., Harrison, S. J., Sela-Buurlage, M. B., Van Deventer, E., Lageweg, W., Howe, P. W., et al. (2004). Isolation and characterisation of a class of carbohydrate oxidases from higher plants, with a role in active defence. Plant J. 39, 147–160. doi: 10.1111/j.1365-313X.2004.02117.x
Delaney, T. P., Friedrich, L., Ryals, J. A. (1995). Arabidopsis signal transduction mutant defective in chemically and biologically induced disease resistance. Proc. Natl. Acad. Sci. U.S.A. 92, 6602–6606. doi: 10.1073/pnas.92.14.6602
Dey, S., Wenig, M., Langen, G., Sharma, S., Kugler, K. G., Knappe, C., et al. (2014). Bacteria-triggered systemic immunity in barley is associated with WRKY and ETHYLENE RESPONSIVE FACTORs but not with salicylic acid. Plant Physiol. 166, 2133–2151. doi: 10.1104/pp.114.249276
Diethelm, M., Schmolke, M., Groth, J., Friedt, W., Schweizer, G., Hartl, L. (2014). Association of allelic variation in two NPR1-like genes with Fusarium head blight resistance in wheat. Mol. Breed. 34, 31–43. doi: 10.1007/s11032-013-0010-2
Epple, P., Apel, K., Bohlmann, H. (1995). An Arabidopsis thaliana thionin gene is inducible via a signal transduction pathway different from that for pathogenesis-related proteins. Plant Physiol. 109, 813. doi: 10.1104/pp.109.3.813
Esmail, S. M., Aboulila, A. A., Abd El-Moneim, D. (2020). Variation in several pathogenesis-related (PR) protein genes in wheat (Triticum aestivum) involved in defense against Puccinia striiformis f. sp. tritici. Physiol. Mol. Plant Pathol. 112, 101545. doi: 10.1016/j.pmpp.2020.101545
Ferreira, R. B., Monteiro, S. A. R. A., Freitas, R., Santos, C. N., Chen, Z., Batista, L. M., et al. (2007). The role of plant defence proteins in fungal pathogenesis. Mol. Plant Pathol. 8, 677–700. doi: 10.1111/j.1364-3703.2007.00419.x
Fu, Z. Q., Dong, X. (2013). Systemic acquired resistance: turning local infection into global defense. Annu. Rev. Plant Biol. 64, 839–863. doi: 10.1146/annurev-arplant-042811-105606
Fu, Z. Q., Yan, S., Saleh, A., Wang, W., Ruble, J., Oka, N., et al. (2012). NPR3 and NPR4 are receptors for the immune signal salicylic acid in plants. Nature 486, 228–232. doi: 10.1038/nature11162
Gaddour, K., Vicentecarbajosa, J., Lara, P., Isabellamoneda, I., Díaz, I., Carbonero, P. (2001). A constitutive cystatin-encoding gene from barley (Icy) responds differentially to abiotic stimuli. Plant Mol. Biol. 45, 599–608. doi: 10.1023/A:1010697204686
Gao, J., Bi, W., Li, H., Wu, J., Yu, X., Liu, D., et al. (2018). WRKY transcription factors associated with NPR1-mediated acquired resistance in barley are potential resources to improve wheat resistance to Puccinia triticina. Front. Plant Sci. 9, 1486. doi: 10.3389/fpls.2018.01486
Gao, C. S., Kou, X. J., Li, H. P., Zhang, J. B., Saad, A. S. I., Liao, Y. C. (2013). Inverse effects of Arabidopsis NPR1 gene on Fusarium seedling blight and Fusarium head blight in transgenic wheat. Plant Pathol. 62, 383–392. doi: 10.1111/j.1365-3059.2012.02656.x
Gao, L., Wang, S., Li, X. Y., Wei, X. J., Zhang, Y. J., Wang, H. Y., et al. (2015). Expression and functional analysis of a pathogenesis-related protein 1 gene, TcLr19PR1, involved in wheat resistance against leaf rust fungus. Plant Mol. Biol. Rep. 33, 797–805. doi: 10.1007/s11105-014-0790-5
Ghorbel, M., Zribi, I., Haddaji, N., Besbes, M., Bouali, N., Brini, F. (2021). The wheat pathogenesis related protein (TdPR1. 2) ensures contrasting behaviors to E. coli transformant cells under stress conditions. Adv. Microbiol. 11, 453–468. doi: 10.4236/aim.2021.119034
Giżyńska, M., Witkowska, J., Karpowicz, P., Rostankowski, R., Chocron, E. S., Pickering, A. M., et al. (2018). Proline-and arginine-rich peptides as flexible allosteric modulators of human proteasome activity. J. Med. Chem. 62, 359–370. doi: 10.1021/acs.jmedchem.8b01025
Görlach, J., Volrath, S., Knauf-Beiter, G., Hengy, G., Beckhove, U., Kogel, K. H., et al. (1996). Benzothiadiazole, a novel class of inducers of systemic acquired resistance, activates gene expression and disease resistance in wheat. Plant Cell. 8, 629–643. doi: 10.1105/tpc.8.4.629
Gregersen, P. L., Thordal-Christensen, H., Förster, H., Collinge, D. B. (1997). Differential gene transcript accumulation in barley leaf epidermis and mesophyll in response to attack by Blumeria graminis f. sp. hordei (syn. Erysiphe graminis f. sp. hordei). Physiol. Mol. Plant Pathol. 51, 85–97. doi: 10.1006/pmpp.1997.0108
Hafez, Y. M., Soliman, N. K., Saber, M. M., Imbabi, I. A., Abd-Elaziz, A. S. (2014). Induced resistance against Puccinia triticina, the causal agent of wheat leaf rust by chemical inducers. Egypt. J. Biol. Pest Control 24, 173.
Hamamouch, N., Li, C., Seo, P. J., PARK, C. M., Davis, E. L. (2011). Expression of Arabidopsis pathogenesis-related genes during nematode infection. Mol. Plant Pathol. 12, 355–364. doi: 10.1111/j.1364-3703.2010.00675.x
Han, Z., Guang, D., Schneiter, R., Tian, C. (2023). The function of plant PR1 and other members of the CAP protein superfamily in plant–pathogen interactions. Mol. Plant Pathol. 24, 651–668. doi: 10.1111/mpp.13320
Hermann, M., Maier, F., Masroor, A., Hirth, S., Pfitzner, A. J., Pfitzner, U. M. (2013). The arabidopsis NIMIN proteins affect NPR1 differentially. Front. Plant Sci. 4. doi: 10.3389/fpls.2013.00088
Jordá, L., Conejero, V., Vera, P. (2000). Characterization of P69E and P69F, two differentially regulated genes encoding new members of the subtilisin-like proteinase family from tomato plants. Plant Physiol. 122, 67–74. doi: 10.1104/pp.122.1.67
Kumar, S., Zavaliev, R., Wu, Q., Zhou, Y., Cheng, J., Dillard, L., et al. (2022). Structural basis of NPR1 in activating plant immunity. Nature 605, 561–566. doi: 10.1038/s41586-022-04699-w
Lawton, K., Weymann, K., Friedrich, L., Vernooij, B., Uknes, S., Ryals, J. (1995). Systemic acquired resistance in Arabidopsis requires salicylic acid but not ethylene. Mol. Plant-Microbe Interact. 8, 863–870. doi: 10.1094/MPMI-8-0863
Li, M., Chen, H., Chen, J., Chang, M., Palmer, I. A., Gassmann, W., et al. (2018). TCP transcription factors interact with NPR1 and contribute redundantly to systemic acquired resistance. Front. Plant Sci. 9, 1153. doi: 10.3389/fpls.2018.01153
Li, Q., Chen, Y., Wang, J., Zou, F., Jia, Y., Shen, D., et al. (2019). A Phytophthora capsica virulence effector associates with NPR1 and suppresses plant immune responses. Phytopathol. Res. 1, 1–11. doi: 10.1186/s42483-019-0013-y
Li, H., Wu, J., Shang, X., Geng, M., Gao, J., Zhao, S., et al. (2020). WRKY transcription factors shared by BTH-induced resistance and NPR1-mediated acquired resistance improve broad-spectrum disease resistance in wheat. Mol. Plant-Microbe Interact. 33, 433–443. doi: 10.1094/MPMI-09-19-0257-R
Li, M., Zhao, S., Yang, J., Ren, Y., Su, J., Zhao, J., et al. (2022). Exogenous expression of barley HvWRKY6 in wheat improves broad-spectrum resistance to leaf rust, Fusarium crown rot, and sharp eyespot. Int. J. Biol. Macromolecules 218, 1002–1012. doi: 10.1016/j.ijbiomac.2022.07.138
Liu, P., Guo, J., Zhang, R., Zhao, J., Liu, C., Qi, T., et al. (2019a). TaCIPK10 interacts with and phosphorylates TaNH2 to activate wheat defense responses to stripe rust. Plant Biotechnol. J. 17, 956–968. doi: 10.1111/pbi.13031
Liu, X., Liu, Z., Niu, X., Xu, Q., Yang, L. (2019b). Genome-wide identification and analysis of the NPR1-like gene family in bread wheat and its relatives. Int. J. Mol. Sci. 20, 5974. doi: 10.3390/ijms20235974
Liu, R., Lu, J., Xing, J., Xue, L., Wu, Y., Zhang, L. (2023b). Characterization and functional analyses of wheat TaPR1 genes in response to stripe rust fungal infection. Sci. Rep. 13, 3362. doi: 10.1038/s41598-023-30456-8
Liu, J., Wu, X., Fang, Y., Liu, Y., Bello, E. O., Li, Y., et al. (2023a). A plant RNA virus inhibits NPR1 sumoylation and subverts NPR1-mediated plant immunity. Nat. Commun. 14, 3580. doi: 10.1038/s41467-023-39254-2
Lu, S., Faris, J. D., Sherwood, R., Friesen, T. L., Edwards, M. C. (2014). A dimeric PR-1-type pathogenesis-related protein interacts with ToxA and potentially mediates ToxA-induced necrosis in sensitive wheat. Mol. Plant Pathol. 15, 650–663. doi: 10.1111/mpp.12122
Maier, F., Zwicker, S., Hueckelhoven, A., Meissner, M., Funk, J., Pfitzner, A. J., et al. (2011). NONEXPRESSOR OF PATHOGENESIS-RELATED PROTEINS1 (NPR1) and some NPR1-related proteins are sensitive to salicylic acid. Mol. Plant Pathol. 12, 73–91. doi: 10.1111/j.1364-3703.2010.00653.x
Molina, A., Görlach, J., Volrath, S., Ryals, J. (1999). Wheat genes encoding two types of PR-1 proteins are pathogen inducible, but do not respond to activators of systemic acquired resistance. Mol. Plant-Microbe Interact. 12, 53–58. doi: 10.1094/MPMI.1999.12.1.53
Moreau, M., Tian, M., Klessig, D. F. (2012). Salicylic acid binds NPR3 and NPR4 to regulate NPR1-dependent defense responses. Cell Res. 22, 1631–1633. doi: 10.1038/cr.2012.100
Mou, Z., Fan, W., Dong, X. (2003). Inducers of plant systemic acquired resistance regulate NPR1 function through redox changes. Cell 113, 935–944. doi: 10.1016/S0092-8674(03)00429-X
Nakayama, A., Fukushima, S., Goto, S., Matsushita, A., Shimono, M., Sugano, S., et al. (2013). Genome-wide identification of WRKY45-regulated genes that mediate benzothiadiazole-induced defense responses in rice. BMC Plant Biol. 13, 1–11. doi: 10.1186/1471-2229-13-150
Numan, M., Bukhari, S. A., Rehman, M. U., Mustafa, G., Sadia, B. (2021). Phylogenetic analyses, protein modeling and active site prediction of two pathogenesis related (PR2 and PR3) genes from bread wheat. PloS One 16, e0257392. doi: 10.1371/journal.pone.0257392
Passardi, F., Penel, C., Dunand, C. (2004). Performing the paradoxical: how plant peroxidases modify the cell wall. Trends Plant Sci. 9, 534–540. doi: 10.1016/j.tplants.2004.09.002
Petroski, M. D., Deshaies, R. J. (2005). Function and regulation of cullin–RING ubiquitin ligases. Nat. Rev. Mol. Cell Biol. 6, 9–20. doi: 10.1038/nrm1547
Quilis, J., Peñas, G., Messeguer, J., Brugidou, C., Segundo, B. S. (2008). The Arabidopsis AtNPR1 inversely modulates defense responses against fungal, bacterial, or viral pathogens while conferring hypersensitivity to abiotic stresses in transgenic rice. Mol. Plant-Microbe Interact. 21, 1215–1231. doi: 10.1094/MPMI-21-9-1215
Schweizer, P., Schlagenhauf, E., Schaffrath, U., Dudler, R. (1999). Different patterns of host genes are induced in rice by Pseudomonas syringae, a biological inducer of resistance, and the chemical inducer benzothiadiazole (BTH). Eur. J. Plant Pathol. 105, 659–665. doi: 10.1023/A:1008791223608
Sedgwick, S. G., Smerdon, S. J. (1999). The ankyrin repeat: a diversity of interactions on a common structural framework. Trends Biochem. Sci. 24, 311–316. doi: 10.1016/S0968-0004(99)01426-7
Sels, J., Mathys, J., De Coninck, B. M., Cammue, B. P., De Bolle, M. F. (2008). Plant pathogenesis-related (PR) proteins: a focus on PR peptides. Plant Physiol. Biochem. 46, 941–950. doi: 10.1016/j.plaphy.2008.06.011
Shang, S., Liu, G., Zhang, S., Liang, X., Zhang, R., Sun, G. (2023). A fungal CFEM-containing effector targets NPR1 regulator NIMIN2 to suppress plant immunity. Plant Biotechnol. J. 22, 82–97. doi: 10.1111/pbi.14166
Shimono, M., Sugano, S., Nakayama, A., Jiang, C. J., Ono, K., Toki, S., et al. (2007). Rice WRKY45 plays a crucial role in benzothiadiazole-inducible blast resistance. Plant Cell 19, 2064–2076. doi: 10.1105/tpc.106.046250
Silverman, P., Seskar, M., Kanter, D., Schweizer, P., Métraux, J. P., Raskin, I. (1995). Salicylic acid in rice (biosynthesis, conjugation, and possible role). Plant Physiol. 108, 633–639. doi: 10.1104/pp.108.2.633
Smith, J. A., Métraux, J. P. (1991). Pseudomonas syringae pv. syringae induces systemic resistance to Pyricularia oryzae in rice. Physiol. Mol. Plant Pathol. 39, 451–461. doi: 10.1016/0885-5765(91)90011-6
Sung, Y. C., Outram, M. A., Breen, S., Wang, C., Dagvadorj, B., Winterberg, B., et al. (2021). PR1-mediated defence via C-terminal peptide release is targeted by a fungal pathogen effector. New Phytol. 229, 3467–3480. doi: 10.1111/nph.17128
Terras, F. R., Eggermont, K., Kovaleva, V., Raikhel, N. V., Osborn, R. W., Kester, A., et al. (1995). Small cysteine-rich antifungal proteins from radish: Their role in host defense. Plant Cell 7, 573. doi: 10.2307/3870116
Uknes, S., Mauch-Mani, B., Moyer, M., Potter, S., Williams, S., Dincher, S., et al. (1992). Acquired resistance in arabidopsis. Plant Cell 4, 645–656. doi: 10.1105/tpc.4.6.645
van Loon, L. C., Rep, M., Pieterse, C. M. (2006). Significance of inducible defense-related proteins in infected plants. Annu. Rev. Phytopathol. 44, 135–162. doi: 10.1146/annurev.phyto.44.070505.143425
Wang, X., Bi, W. S., Gao, J., Yu, X., Wang, H., Liu, D. (2018). Systemic acquired resistance, NPR1, and pathogenesis-related genes in wheat and barley. J. Integr. Agricult. 17, 60345–60347. doi: 10.1016/S2095-3119(17)61852-5
Wang, L., Lu, H., Zhan, J., Shang, Q., Wang, L., Yin, W., et al. (2022b). Pathogenesis-related protein-4 (PR-4) gene family in Qingke (Hordeum vulgare L. var. nudum): Genome-wide identification, structural analysis and expression profile under stresses. Mol. Biol. Rep. 49, 9397–9408. doi: 10.1007/s11033-022-07794-3
Wang, F., Shen, S., Zhao, C., Cui, Z., Meng, L., Wu, W., et al. (2022a). TaPR1 interacts with TaTLP1 via the αIV helix to be involved in wheat defense to Puccinia triticina through the CAPE1 motif. Front. Plant Sci. 13, 874654. doi: 10.3389/fpls.2022.874654
Wang, X., Yang, B., Li, K., Kang, Z., Cantu, D., Dubcovsky, J. (2016). A conserved Puccinia striiformis protein interacts with wheat NPR1 and reduces induction of pathogenesis-related genes in response to pathogens. Mol. Plant-Microbe Interact. 29, 977–989. doi: 10.1094/MPMI-10-16-0207-R
Wang, F., Yuan, S., Wu, W., Yang, Y., Cui, Z., Wang, H., et al. (2020a). TaTLP1 interacts with TaPR1 to contribute to wheat defense responses to leaf rust fungus. PloS Genet. 16, e1008713. doi: 10.1371/journal.pgen.1008713
Wang, X., Zhang, H., Nyamesorto, B., Luo, Y., Mu, X., Wang, F., et al. (2020b). A new mode of NPR1 action via an NB-ARC–NPR1 fusion protein negatively regulates the defence response in wheat to stem rust pathogen. New Phytol. 228, 959–972. doi: 10.1111/nph.16748
Wang, F., Shen, S., Cui, Z., Yuan, S., Qu, P., Jia, H., et al. (2023). Puccinia triticina effector protein Pt_21 interacts with wheat thaumatin-like protein TaTLP1 to inhibit its antifungal activity and suppress wheat apoplast immunity. Crop J. 11, 1431–1440. doi: 10.1016/j.cj.2023.04.006
Wu, J., Gao, J., Bi, W., Zhao, J., Yu, X., Li, Z., et al. (2019). Genome-wide expression profiling of genes associated with the Lr47-mediated wheat resistance to leaf rust (Puccinia triticina). Int. J. Mol. Sci. 20, 4498. doi: 10.3390/ijms20184498
Wu, Y., Zhang, D., Chu, J. Y., Boyle, P., Wang, Y., Brindle, I. D., et al. (2012). The Arabidopsis NPR1 protein is a receptor for the plant defense hormone salicylic acid. Cell Rep. 1, 639–647. doi: 10.1016/j.celrep.2012.05.008
Xu, G., Yuan, M., Ai, C., Liu, L., Zhuang, E., Karapetyan, S., et al. (2017). uORF-mediated translation allows engineered plant disease resistance without fitness costs. Nature 545, 491–494. doi: 10.1038/nature22372
Yang, Z., Ma, X., Wu, S., Wang, H., Sun, X., Ji, X., et al. (2013). Cloning of NPR1-like genes and their response to Fusarium graminearum infection in wheat. Acta Agronom. Sin. 39, 1775–1782. doi: 10.3724/SP.J.1006.2013.01775
Yu, G., Zhang, X., Yao, J., Zhou, M., Ma, H. (2017). Resistance against Fusarium Head Blight in transgenic wheat plants expressing the ScNPR1 gene. J. Phytopathol. 165, 223–231. doi: 10.1111/jph.12553
Zhang, Y., Fan, W., Kinkema, M., Li, X., Dong, X. (1999). Interaction of NPR1 with basic leucine zipper protein transcription factors that bind sequences required for salicylic acid induction of the PR-1 gene. Proc. Natl. Acad. Sci. U.S.A. 96, 6523–6528. doi: 10.1073/pnas.96.11.6523
Zhang, W. J., Pedersen, C., Kwaaitaal, M., Gregersen, P. L., Mørch, S. M., Hanisch, S., et al. (2012). Interaction of barley powdery mildew effector candidate CSEP0055 with the defence protein PR17c. Mol. Plant Pathol. 13, 1110–1119. doi: 10.1111/j.1364-3703.2012.00820.x
Zhao, J., Bi, W., Zhao, S., Su, J., Li, M., Ma, L., et al. (2021). Wheat apoplast-localized lipid transfer protein TaLTP3 enhances defense responses against Puccinia triticina. Front. Plant Sci. 12, 771806. doi: 10.3389/fpls.2021.771806
Zhao, S., Shang, X., Bi, W., Yu, X., Liu, D., Kang, Z., et al. (2020). Genome-wide identification of effector candidates with conserved motifs from the wheat leaf rust fungus Puccinia triticina. Front. Microbiol. 11, 1188. doi: 10.3389/fmicb.2020.01188
Zhou, F., Zhang, Z., Gregersen, P. L., Mikkelsen, J. D., de Neergaard, E., Collinge, D. B., et al. (1998). Molecular characterization of the oxalate oxidase involved in the response of barley to the powdery mildew fungus. Plant Physiol. 117, 33–41. doi: 10.1104/pp.117.1.33
Keywords: wheat, systemic acquired resistance, genetic improvement, NPR1, PR genes
Citation: Zhao S, Li M, Ren X, Wang C, Sun X, Sun M, Yu X and Wang X (2024) Enhancement of broad-spectrum disease resistance in wheat through key genes involved in systemic acquired resistance. Front. Plant Sci. 15:1355178. doi: 10.3389/fpls.2024.1355178
Received: 13 December 2023; Accepted: 22 January 2024;
Published: 23 February 2024.
Edited by:
Xinli Zhou, Southwest University of Science and Technology, ChinaReviewed by:
Huamin Chen, Chinese Academy of Agricultural Sciences, ChinaCopyright © 2024 Zhao, Li, Ren, Wang, Sun, Sun, Yu and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaodong Wang, emhid3hkQGhlYmF1LmVkdS5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.