- 1Institute of Vegetables, Henan Academy of Agricultural Sciences, Graduate T&R Base of Zhengzhou University, Zhengzhou, Henan, China
- 2School of Agricultural Sciences, Zhengzhou University, Zhengzhou, Henan, China
- 3Department of Genetics and Plant Breeding, Bangladesh Agricultural University, Mymensingh, Bangladesh
Clubroot disease poses a significant threat to Brassica crops, necessitating ongoing updates on resistance gene sources. In F2 segregants of the clubroot-resistant inbred line BrT18-6-4-3 and susceptible DH line Y510, the genetic analysis identified a single dominant gene responsible for clubroot resistance. Through bulk segregant sequencing analysis and kompetitive allele-specific polymerase chain reaction assays, CRA8.1.6 was mapped within 110 kb (12,255–12,365 Mb) between markers L-CR11 and L-CR12 on chromosome A08. We identified B raA08g015220.3.5C as the candidate gene of CRA8.1.6. Upon comparison with the sequence of disease-resistant material BrT18-6-4-3, we found 249 single-nucleotide polymorphisms, seven insertions, six deletions, and a long terminal repeat (LTR) retrotransposon (5,310 bp) at 909 bp of the first intron. However, the LTR retrotransposon was absent in the coding sequence of the susceptible DH line Y510. Given the presence of a non-functional LTR insertion in other materials, it showed that the LTR insertion might not be associated with susceptibility. Sequence alignment analysis revealed that the fourth exon of the susceptible line harbored two deletions and an insertion, resulting in a frameshift mutation at 8,551 bp, leading to translation termination at the leucine-rich repeat domain’s C-terminal in susceptible material. Sequence alignment of the CDS revealed a 99.4% similarity to Crr1a, which indicate that CRA8.1.6 is likely an allele of the Crr1a gene. Two functional markers, CRA08-InDel and CRA08-KASP1, have been developed for marker-assisted selection in CR turnip cultivars. Our findings could facilitate the development of clubroot-resistance turnip cultivars through marker-assisted selection.
1 Introduction
Turnip (Brassica rapa L. spp. rapifera) is a vegetable belonging to the genus Brassica. It originated in Afghanistan, Pakistan, Transcaucasia (part of Asia), and the Mediterranean. The Asian turnip variety is predominantly distributed in China and western Japan (Pu et al., 2018). Clubroot disease is caused by Plasmodiophora brassicae (P. brassicae) and has been reported in numerous countries (Chai et al., 2014). It affects plants in the Brassicaceae family, such as Chinese cabbage, turnips, radish, cauliflower, and mustard (Howard et al., 2010). P. brassicae first infects root hairs by free primary spores and then releases secondary zoospores to invade the cortex (Kageyama and Asano, 2009). This results in gradual swelling of the roots in affected plants, hindering nutrient absorption and ultimately leading to wilting and death of the entire plant. Dormant spores of P. brassicae can remain active in the soil for up to 20 years, posing a long-term threat to cruciferous plants (Dixon, 2009). Clubroot diseases tend to exacerbate annually, making control challenging through chemical, biological, and agricultural means. From an ecological standpoint, developing resistant varieties through breeding is a promising solution (Piao et al., 2009). Zhang et al. (2024) successfully identified a CR gene, facilitating CR-resistant breeding in B. oleracea.
Single-nucleotide polymorphisms (SNPs) represent a prevalent form of DNA variation across the genome, offering advantages such as high throughput and seamless integration. Consequently, they find wide applications in various fields, such as disease treatment, drug development, and plant breeding. The kompetitive allele-specific polymerase chain reaction (PCR) (KASP) technique is a precise allele-specific PCR method capable of accurately identifying SNPs by matching terminal primer bases. KASP technology is renowned for its time efficiency, reduced error rates, and cost-effectiveness in genotyping, providing flexibility for genotyping multiple samples with minimal SNP loci (Gouda et al., 2021).
Significant progress has been made in developing clubroot-resistant genotypes. European fodder turnips (Brassica rapa L. spp. rapifera) (AA, 2n = 20) ‘ECD01-04,’ ‘Gelria R,’ ‘Siloga,’ ‘Debra,’ and ‘Milan White’ (Eckholm, 1993; Elke et al., 2009) have emerged as a widely utilized resistant source, successfully integrated into CR breeding programs in Chinese cabbage and canola (Piao et al., 2009). To date, 36 clubroot-resistant loci have been identified across Brassica species, including B. rapa, B. oleracea, B. napus, and B. nigra (Hasan et al., 2021; Pang et al., 2022). Among these loci, the majority are found on chromosomes A03 and A08. Chromosome A03 harbors 19 resistance loci, encompassing CRa, CRb, CRd, CRq, CRk, Rcr1, Rcr2, Rcr4, Rcr5, pbBa3.1, pbBa3.2, pbBa3.3, CR6b, CRbkato, BraA.CRa, BraA.CRc, BraA3PSX.CRa/bkato1.1, BraA3PSX.CRa/bkato1.2, and Crr3 (Etsuo et al., 1998; Hirai et al., 2004; Piao et al., 2004; Ueno et al., 2012; Chen et al., 2013; Chu et al., 2014; Sun et al., 2016; Yu et al., 2017; Pang et al., 2018; Huang et al., 2019). On chromosome A08, nine resistance loci are identified, namely, CRs, Crr1, Rcr3, Rcr9, PbBa8.1, qBrCR38-2, RCr9wa, PbBrA08Banglim, and BraACRb (Hirani et al., 2018; Laila et al., 2019; Md. Masud et al., 2020; Yu et al., 2022). Additionally, chromosome A01 hosts three loci, namely, CR6a, Crr2, and PbBa1.1 (Suwabe et al., 2006). Chromosome A02 carries two loci, namely, CRc and Rcr8 (Yu et al., 2017), whereas resistance loci CrrA5, Crr4, and qBrCR38-1 are located on chromosomes A05, A06, and A07, respectively (Suwabe et al., 2006; Nguyen et al., 2018; Zhu et al., 2019). The collection of resistance loci appears to be extensive; however, only Crr1a, CRa, and their alleles have been cloned (Ueno et al., 2012; Hatakeyama et al., 2013; Yang et al., 2022). These cloned genes have been found to contain important components of effector-induced immunity, TIR-NBS-LRR [Toll/interleukin-1 (IL-1) receptor–like nucleotide binding site, leucine-rich repeat] protein domain family (Mariana et al., 2002).
CRA8.1.6 was finely mapped and cloned from the F2 population, resulting from a cross between the resistant inbred line BrT18-6-4-3 and the susceptible DH line Y510. Our investigation aimed to elucidate the mechanisms underlying the loss of resistance by analyzing the structure of candidate genes in the susceptible line. Gene sequence alignment revealed a frameshift mutation in the susceptible line Y510, resulting in translation termination at 8,551 bp. This mutation altered the C-terminal leucine-rich repeat (LRR) domain, leading to a loss of resistance. The disease index survey demonstrated a significantly lower disease index in overexpression transgenic Arabidopsis than in wild-type Arabidopsis. The results of the target gene expression revealed a significant increase in the relative expression of CRA8.1.6 in the T2 generation of Arabidopsis compared to the wild-type. This indicates that the conferred CRA8.1.6 gene is indeed a clubroot-resistant gene. Functional markers, CRA08-InDel and CRA08-KASP1, linked to CRA8.1.6 were successfully developed and validated. These markers represent valuable resources for marker-assisted selection in breeding CR cultivars against P. brassicae.
2 Materials and methods
2.1 Plant materials
In the current study, F1 and F2 populations were developed by crossing the clubroot-resistant turnip inbred line BrT18-6-4-3 (P1) with the susceptible Chinese cabbage DH line Y510 (P2) followed by the selfing of F1. Parental lines, F1, and F2 populations, were inoculated with P. brassicae race four isolate ‘XY-2’ (Yuan et al., 2017). The F2 population was cultivated between September 2021 and November 2021, and the inheritance pattern of the resistant gene was determined using a Chi-square test (χ2). Furthermore, 26 disease-resistant and 29 susceptible lines were used to analyze mutations in the candidate gene (Supplementary Table 1). All materials used in this study were provided by the Institute of Vegetables, Henan Academy of Agricultural Sciences.
2.2 Estimation of disease index
The disease index was assessed by inoculating the plants on the 20th day after sowing using a root irrigation method with a concentration of 107 spores/mL of P. brassicae suspension (Luo et al., 2014). AKIMEKI and ECD05 were utilized for disease index estimation as controls for disease resistance and susceptibility, respectively (Matsumoto et al., 2012). The inoculated plants were maintained at 22°C ± 2°C and 16-h/8-h (light/dark) photoperiod (Yuan et al., 2021). Samples were collected and immediately frozen in liquid nitrogen and then stored at −80°C for subsequent use. Root samples from resistant and susceptible materials were collected at different time points: 0 days after inoculation (DAI; indicating no infection), 3 DAI (representing cortical infection), 9 DAI (indicating early onset of disease), and 20 DAI (indicating late onset of disease). One-centimeter-long root segments were cut from the junction of the rhizome and fixed with Formalin-Aceto-Alcohol (FAA) solution (containing 5 mL of acetic acid + 5 mL of formalin + 50 mL of 95% alcohol + 35 mL of sterile water) for 24 h. After fixation with FAA, the roots were rinsed with sterile water, dried with absorbent paper, and stained with 0.5% fluorescent pink for 3 h. The infected roots were observed under a light microscope (Li et al., 2022).
2.3 Bulked segregant analysis by resequencing
DNA was extracted from resistant and susceptible parents, along with 30 highly resistant (DI = 0; R-pool) and susceptible F2 plants (DI = 5–7; S-pool) (Supplementary Table 1) following the cetyl trimethyl ammonium bromide method. Bulked segregant analysis (BSA) sequencing (BSA-seq) was performed on four DNA pools (Huang et al., 2017), and sequencing was conducted using the Illumina HiSeq platform, with analysis carried out using NovoGene (http://www.novogene.com). The raw reads underwent quality control trimming to remove low-quality paired reads with the adapter and retained clean reads. The clean reads from the four DNA bulks were combined using BWA software (V0.7.17) and compared with B. rapa (Chiifu-401) genome version 3.0 (http://brassicadb.org/brad/index.phpM) (Kathiresan et al., 2014). SAMtools software (V1.3.1) was employed to identify SNPs and insertion/deletion (InDel) variations between the R and S pools. The ΔSNP index for all genomic positions in the R and S libraries was calculated (Supplementary Table 2) using a sliding window analysis with a window width of 1 Mb and a sliding window step size of 10 kb.
2.4 Marker analysis and linkage map construction
Sequences of the genes were downloaded from the Brassica database (http://brassicadb.org/brad/downloadOverview.php), and the SNPs of candidate genes were validated. Thirty-five pairs of primers for KASP markers were designed using DNAMAN software (V6.0) to verify the parents and F2 population (Supplementary Table 3), and, ultimately, 21 pairs of polymorphic primers were selected (Xiang et al., 2007).
The KASP markers were utilized for genotyping the F2 population consisting of 98 individuals. Linked loci were identified using JoinMap4.0 to form linkage groups (Voorrips, 2002), and a genetic linkage map was constructed according to the Kosambi mapping function (Kosambi, 1944).
An additional large F2 population of 1,489 individuals was planted for recombination selection, from which 438 susceptible individuals were selected and used to map candidate genes.
2.5 Cloning and sequencing of the CRA8.1.6 gene
DNA and cDNA sequences of CRA8.1.6 candidate genes were cloned from the parents using Phanta Super-Fidelity DNA polymerase Mix (Vazyme, Nanjing, China) (Supplementary Table 4). Standard PCR was conducted with 30-s denaturation at 98°C followed by 35 cycles of 98°C for 10 s, 58°C for 5 s, and 72°C for 60 s.
PCR products were sequenced by Sangon Biotech Co., Ltd. (Zhengzhou, China) and analyzed using DNAMAN software (V6.0). The complete coding sequence (CDS) of the CRA8.1.6 gene from BrT18-6-4-3 and Y510 has been submitted to GenBank under the accession number AB605024.1. Protein’s physical and chemical properties were determined with Expasy (http://web.expasy.org/protparam/).
2.6 Quantitative real-time PCR and semi-quantitative RT-PCR analyses
Total RNA was extracted from root samples collected at 0 DAI, 3 DAI, 9 DAI, and 20 DAI of BrT18-6-4-3 and Y510 lines using the RNA Prep Pure Plant Kit (Beijing Day, China) (Wei et al., 2021). Single-stranded cDNA was synthesized using Trans-script after a two-step removal of genomic DNA (gDNA) (Trans, Beijing, China).
The quantitative real-time PCR (qRT-PCR) was performed in a Roche Light Cycler 480-II system (Roche Applied Sciences, Beijing, China). GAPDH was used as an internal reference gene to calculate relative expression levels using the 2−ΔΔCt method (Livak and Schmittgen, 2001). B. rapa sequence information was utilized to design RT-PCR primers. Standard PCR was conducted by denaturing for 5 min at 94°C, followed by 30 s at 94°C, 30 s at 55°C, and 60 s at 72°C for 25 cycles. PCR products were visualized after electrophoresis on a 1% agarose gel (Supplementary Table 4).
2.7 Vector construction and transformation
Specific full-length primers were designed to incorporate restriction sites and protective bases. This was achieved using the plasmid DNA of the target gene as a template. The PCR amplification procedure was conducted as follows: 95°C for 5 min; 30 cycles of 95°C for 30 s, 50°C for 45 s, and 72°C for 258 s; followed by 72°C for 10 min; and 16°C for 30 min.
The purified PCR product was digested with pBWA (V) HS expression vector (Bio Run, Hu Bei, China), and the digested product was further purified and ligated with the vector using T4 ligase to obtain the recombinant plasmid pBWA (V) HS-BrT18-6. The ligation product was then transformed into competent DH5α cells and PCR-screened positive clones. After sequencing and verification, Arabidopsis thaliana was transformed using dip method (Hamann et al., 2002). The resulting T0 transgenic A. thaliana was identified (Pang et al., 2022), and positive plants were selected for future planting to obtain T2 generation seeds.
2.8 Inoculation of overexpression transgenic A. thaliana and identification of candidate gene
The T2 generation seeds of A. thaliana were sown on a medium containing hygromycin to select homozygous strains based on positive reactions on specific media (Tang, 2007). T2 generation A. thaliana seeds were planted (Suwabe et al., 2006). The P. brassicae strain used in this study was obtained from a clubroot-infected Chinese cabbage field (B. rapa) in Xinye County, Henan Province, China (113.97°E, 35.05°N), and the Williams system was used to confirm the P. brassicae strain as race 4 (Yuan et al., 2017).
Twenty-five-day-old transgenic and wild-type A. thaliana were inoculated with P. brassicae. Similarly, another set was inoculated with fresh water without any P. brassicae spores, which served as the control group. After 30 days of inoculation, the wild-type and transgenic A. thaliana were examined in control and experimental groups. The frequency of clubroot disease and expression of candidate genes were observed (Keita et al., 2012).
3 Results
3.1 Phenotypic and genetic analyses and cytological observation
After 30 days of infection with P. brassicae, all F1 individuals resisted clubroot disease. We inoculated two fractions of the F2 population with P. brassicae to account for the nature of disease-resistance gene action. In a small fraction of the F2 population, 341 plants were resistant and 132 were susceptible, whereas, in a large fraction, 1,134 plants were resistant and 335 were susceptible. Both F2 populations exhibited a 3:1 segregation, indicating Mendelian genetics for a single dominant gene (Table 1).
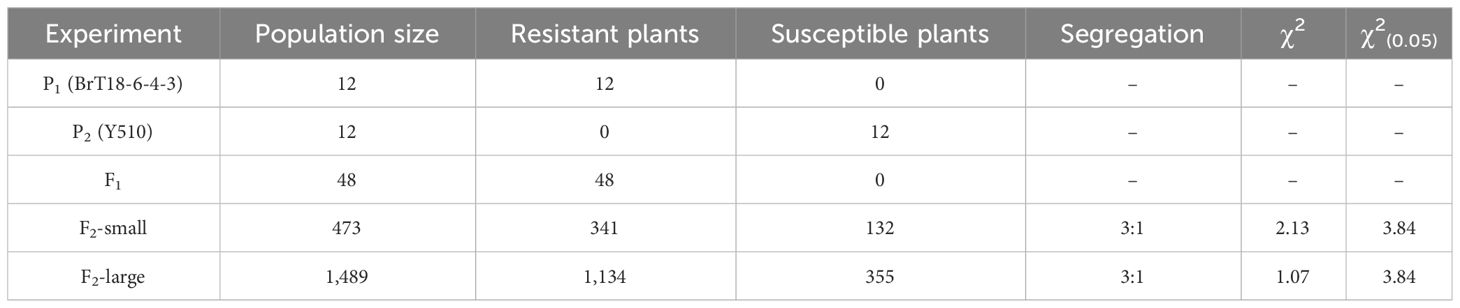
Table 1 Genetic analysis of clubroot resistance and susceptibility in F2 populations of the cross between BrT18-6-4-3 and Y510.
Microscopic examination revealed abundant spores attached to root hairs in all test plants at 1 DAI, indicating sufficient spores to initiate root infection. However, no cortical infection was detected in the resistant line (BrT18-6-4-3) at 1 DAI, 2 DAI, and 3 DAI or in the control group. In contrast, root hair infection was observed in the susceptible line (Y510) at 1 DAI, with cortical infection and free spores in epidermal cells observed at 3 DAI. No such symptoms were observed in the susceptible lines of the control group.
At 9 DAI, the roots of the resistant line appeared normal, whereas the roots of the susceptible line were slightly swollen. Numerous spores were found in the swollen root cortical cells of the susceptible line. In contrast, only a few spores were observed in the cortical cells of the resistant line (Figure 1), indicating cortical infection occurred between 3 DAI and 9 DAI.
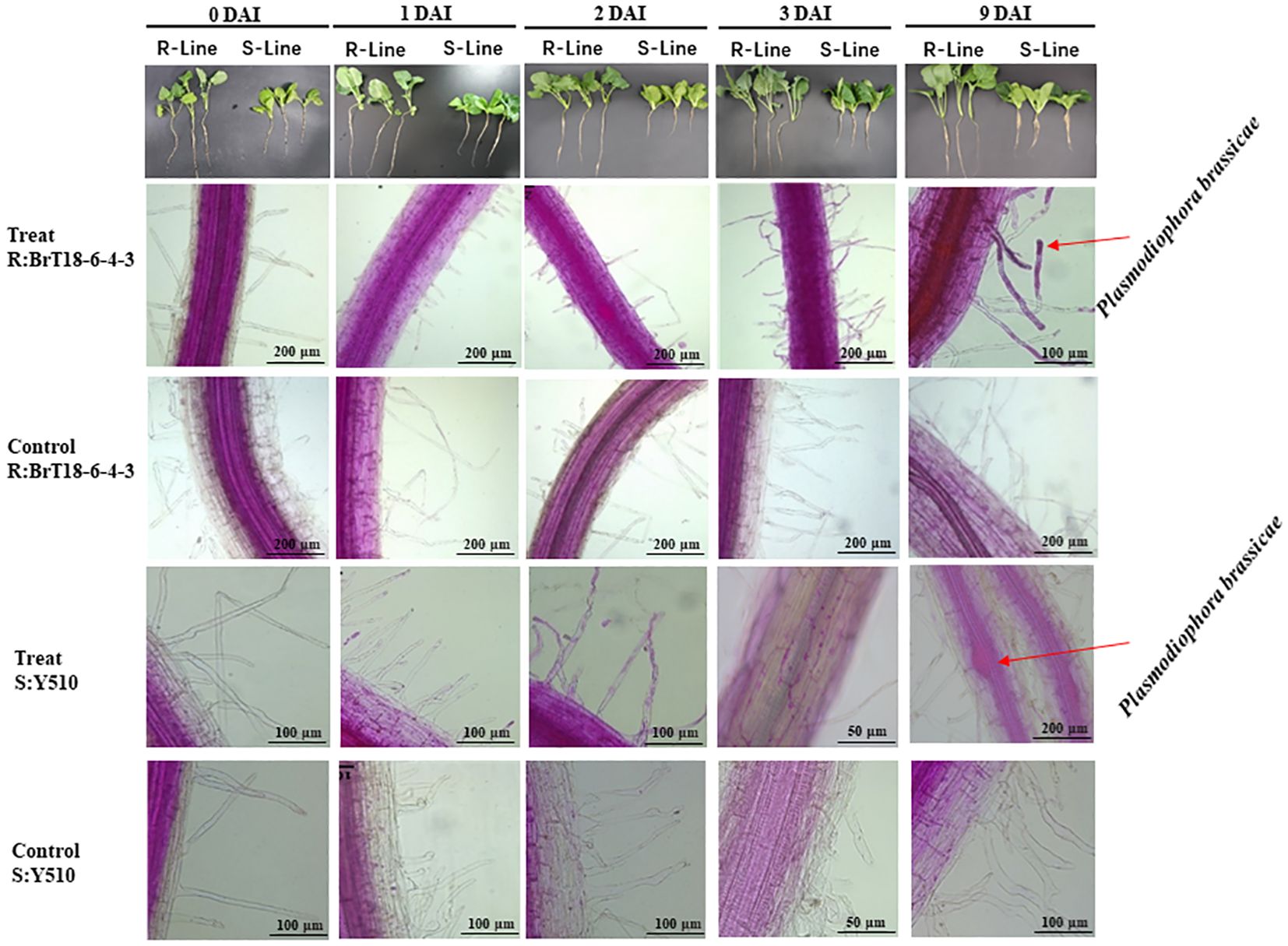
Figure 1 Comparison of root infection status between R-line (BrT18-6-4-3) and S-line (Y510) of control and P. brassicae inoculated groups. BrT18-6-4-3 initiated root hair infection at 1 DAI, and few spores were present in cortical cells at 9 DAI. Y510 showed root hair infection at 1 DAI and cortical infection at 3 DAI to 9 DAI, with many spores in cortical cells at 9 DAI. Bar = 200 μm, 100 μm, and 50 μm.
3.2 Mapping of CRA8.1.6 QTL
BSA-seq analysis generated 36,416,118 and 38,958,860 clean reads from the R and S pools, respectively (Supplementary Table 2). These reads were aligned to the B. rapa genome, identifying 218,150 SNPs and 53,040 InDels between the R and S pools. △ (SNP index) for each locus was calculated using sliding window analysis. Subsequently, a new quantitative trait locus (QTL) called CRA8.1.6 was identified on chromosome A08, likely between 11.04 Mb and 16.10 Mb (Supplementary Figure 1).
Thirty-five KASP markers were created on the basis of SNP variants within the candidate interval and polymorphism between the resistant (BrT18-6-4-3) and susceptible (Y510) lines, with 21 markers selected for genotyping of 98 F2 plants to develop markers for chromosome A08. Within the reported CRA8.1.6 QTL, one recombinant individual was found each for markers L-CR10 and L-CA01 with a genetic distance of 0.9 cM and 1.2 cM, respectively (Figure 2A). The order of markers on the genetic map was consistent with the physical map.
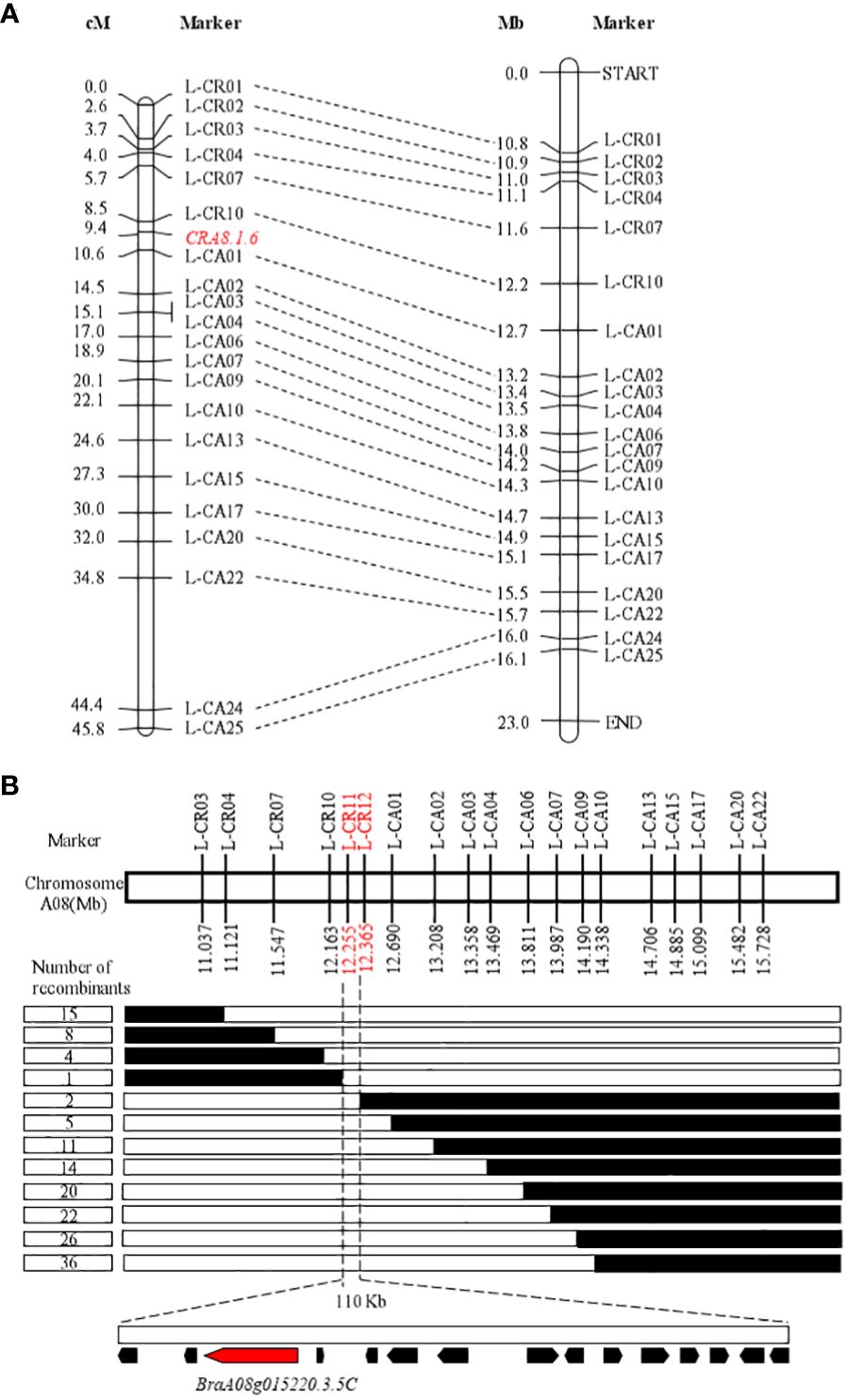
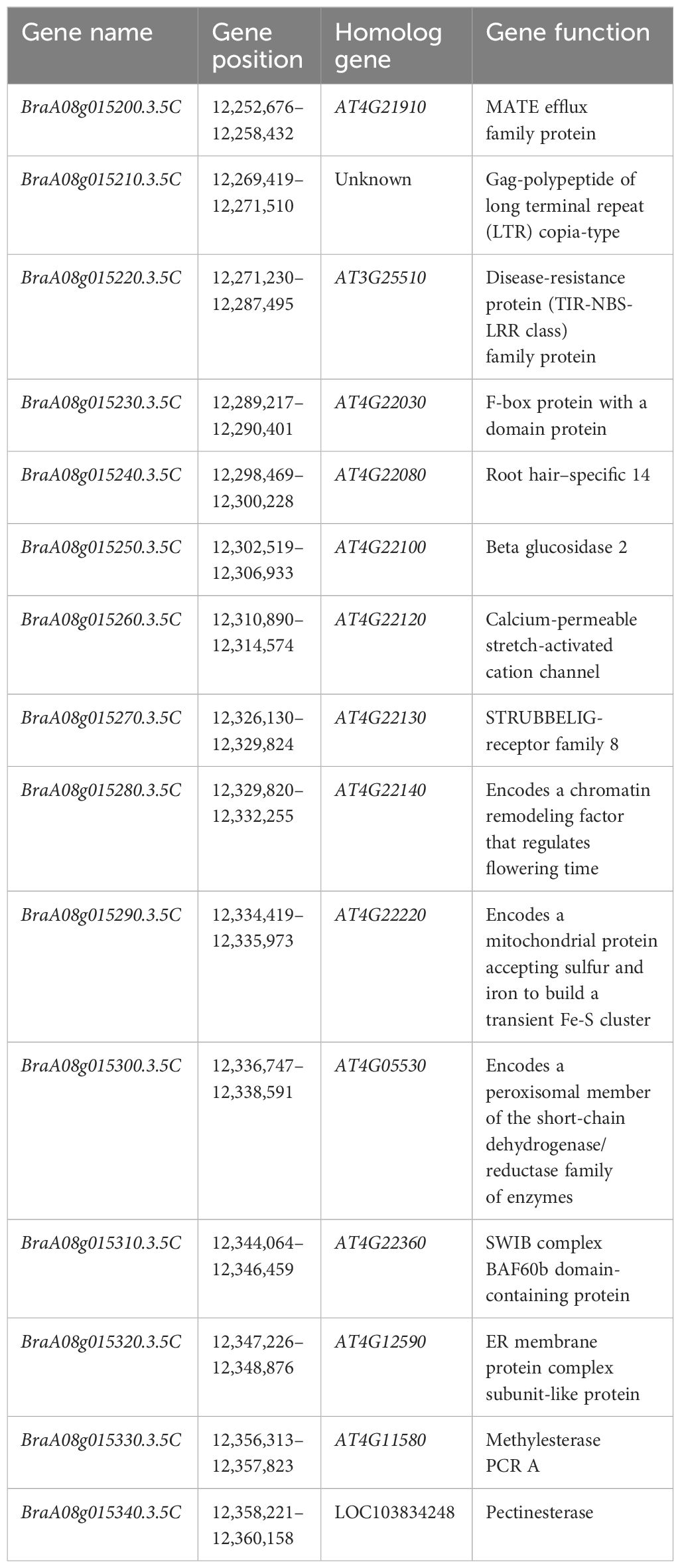
Figure 2 Initial and fine mapping of the CRA8.1.6 in Chinese Turnip. (A) Preliminary mapping of the area CRA8.1.6. The left side depicts the CRA8.1.6 genetic map, with marker distance indicated in cM. The corresponding physical map is also shown on the right, with units in megabases (Mb). (B) The CRA8.1.6 gene was delineated between markers L-CR11 and L-CR12, spanning an estimated length of 110 kb. Fifteen genes were annotated on the basis of the reference genome sequence (Chiifu-401-42). Each genotype is represented, with white indicating the homozygote and black representing the heterozygote. The number of recombinants is displayed in the boxes to the left of the map.
To refine the CRA8.1.6 QTL, 438 susceptible homozygous F2 individuals were screened for polymorphisms between markers L-CR03 and L-CA22, identifying 36 recombinants. All 51 recombinants were further genotyped using markers L-CR10 and L-CA01, identifying nine recombinants. Five KASP markers were developed, two of which showed polymorphism. Ultimately, a candidate gene for the CRA8.1.6 QTL was determined at a position (12,255–12,365 Mb) on chromosome A08 marked by markers L-CR11 and L-CR12 (Figure 2B).
3.3 Candidate gene analysis
Detailed mapping of the CRA8.1.6 QTL region revealed 15 genes, with BraA08g015220.3.5C being the sole gene encoding the TIR-NBS-LRR domains (Table 2). We utilized qRT-PCR to examine the expression of the hypothesized candidate gene. Among the 15 genes, only BraA08g015220.3.5C exhibited differential expression between the parental lines (Figure 3A). The expression of the BraA08g015220.3.5C gene was analyzed across various periods. The BraA08g015220.3.5C gene exhibited a four-fold increase in expression in the inoculated resistant line (BrT18-6-4-3) compared to the non-inoculated resistant line and both the inoculated and non-inoculated susceptible (Y510) lines at 3 DAI. Similarly, at 9 DAI, there was a two-fold increase in expression in the inoculated resistant line compared to the other lines (Figure 3B). The trend of gene expression analyzed by RT-PCR was consistent with that of qPCR (Figure 3C). Hence, the BraA08g015220.3.5C gene was identified as a potential candidate gene responsible for imparting resistance against clubroot disease.
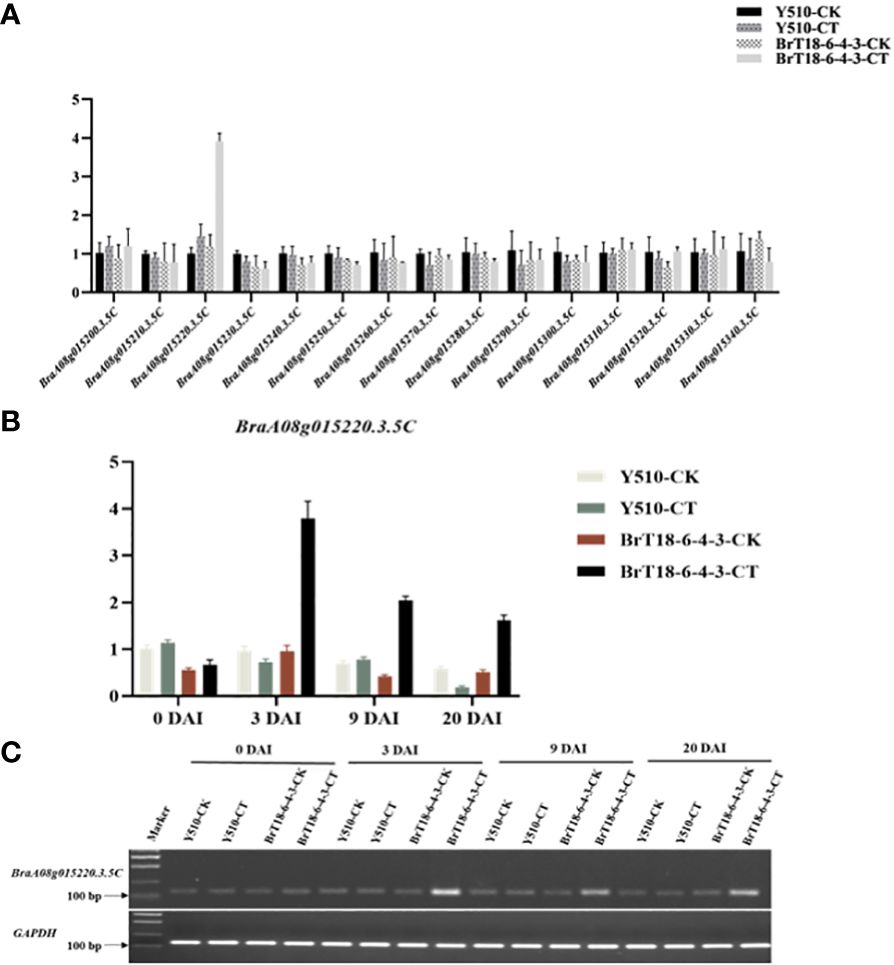
Figure 3 Gene expression of two parents in experimental and control groups at different periods. (A) qRT-PCR analysis of 15 candidate genes. (B) The expression of BraA08g015220.3.5c across different periods. (C) Semi-quantitative RT-PCR of CRA8.1.6 (BraA08g015220.3.5c), with GAPDH served as the internal control.
3.4 Sequence variation of candidate genes between resistant and susceptible lines
PCR products of the BraA08g015220.3.5C gene were utilized to determine gDNA and CDS. Specific primers amplify disease-resistant (BrT18-6-4-3) and susceptible (Y510) lines. In the resistant line, the genome and CDS sequences of BraA08g015220.3.5C were 4295 bp and 3675 bp, respectively. Conversely, in the susceptible parent, the genome and CDS sequences were 9586 bp and 3644 bp, respectively (Supplementary Figure 2). Conserved domain analysis unveiled several domains within the BraA08g015220.3.5C gene, including a Toll and IL-1 domain (TIR, amino acids 70–235), a nucleotide binding site (NBS-ARC, amino acids 260–484), and an LRR (amino acids 701–908). Compared to disease-resistant and susceptible materials, we identified 249 SNPs, seven insertions, and six deletions in the susceptible line. Among these mutations, the insertion of the long terminal repeat (LTR) retrotransposon does not influence the CRA8.1.6 function, as it does not alter the CDS sequence (Figure 4A, Supplementary Figure 2). The sequencing results revealed six non-synonymous mutations that did not influence the resistance of the wild, because these mutations were contained in both resistant and susceptible materials (Supplementary Figures 3, 4). Furthermore, sequence alignment analysis revealed that the fourth exon of the susceptible line harbored two deletions and an insertion, leading to premature termination of the C-terminus and destruction of the LRR domain (Figure 4B). This premature termination consequently led to the loss of CRA8.1.6 function, rendering Y510 susceptible to clubroot disease.
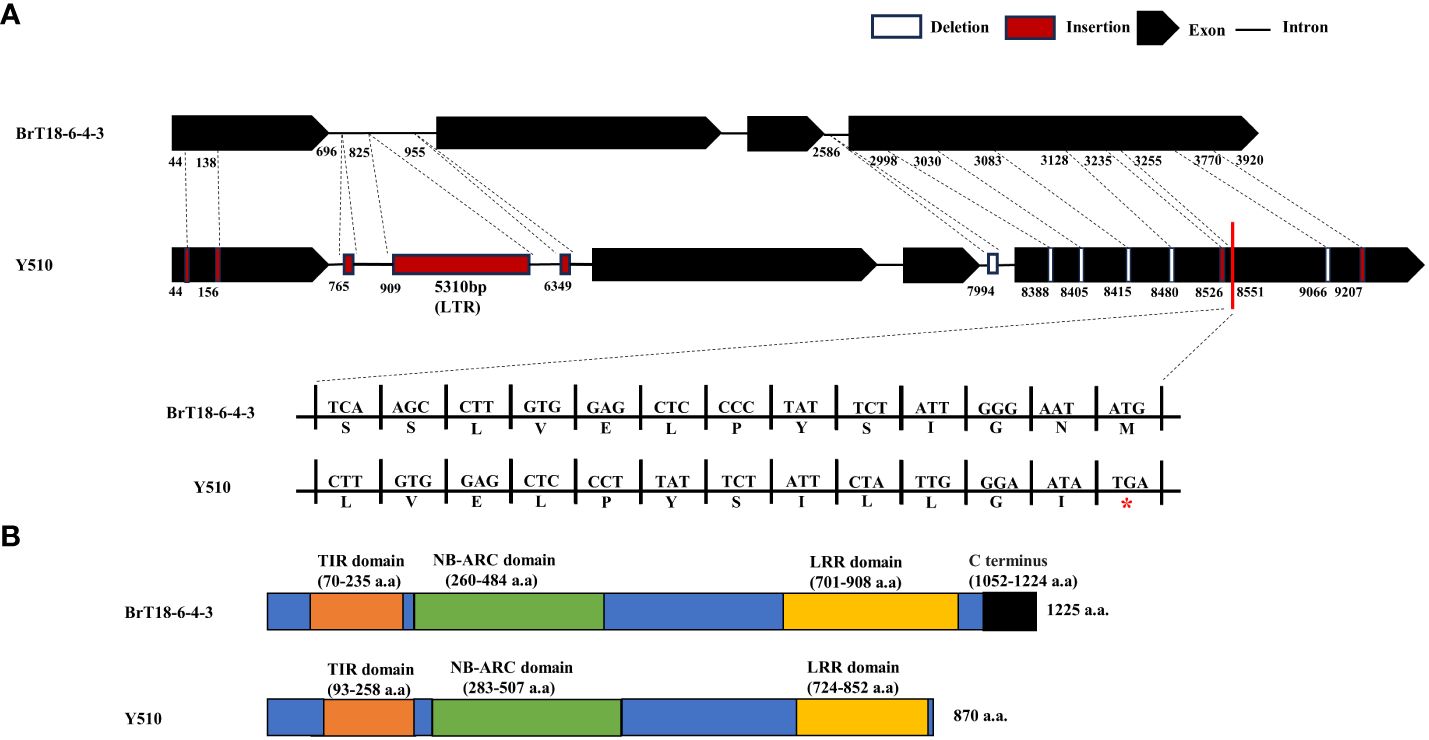
Figure 4 CRA8.1.6 gene structure, amino acid analysis, and protein structure prediction. (A) CRA8.1.6 comprises four exons and three introns. Gene sequences in the disease-susceptible material exhibited 249 SNPs, seven insertions, and six deletions, including 5,310 bp insertions at 909 bp. INS, insertion; DEL, deletion. (B) A frameshift mutation caused premature termination at 870 amino acids of exon four, disrupting the LRR domain in the C-terminal region.
Using the sequence variation observed in the fourth exon between the parental lines, we designed upstream and downstream primers for 2,855 bp and 3,395 bp, respectively. Functional markers, including CRA08-InDeL (Supplementary Table 4), were also developed to distinguish individuals of BrT18-6-4-3, Y510, and F2 populations. The amplified fragment size was 541 bp in the resistant materials and 447 bp in the susceptible materials. The results indicated complete concordance between the phenotypes of resistant, susceptible, and heterozygous F2 plants with their respective genotypes. This suggests that CRA08-InDel is a functional marker capable of accurately distinguishing clubroot-resistance phenotypes within segregating populations (Figure 5A). Within a natural population, the genotypes of disease-resistant materials consistently matched their phenotypes. However, the identification of susceptible materials was not sufficiently consistent. Hence, we sequenced materials that did not match this marker and identified a trait-associated SNP at 3,209 bp. Subsequently, we developed the KASP functional marker CRA08-KASP1 (Supplementary Table 4). Upon screening F2 populations with this marker, we observed that the expected amplified products for resistant, susceptible, and heterozygous F2 individuals perfectly matched the anticipated results. This suggests that the marker CRA08-KASP1 segregated in the F2 population with a clubroot-resistant phenotype (Figure 5B). The CRA08-KASP1 markers were also screened for 24 disease-resistant and 26 disease-susceptible materials. Among these, the phenotype and genotype of all 23 resistant materials were matched entirely, whereas the phenotype and genotype of 22 susceptible materials were matched. However, only five materials (DH40, BrT127, BrT47-1-1-1, BrT133, and BrT71-1-1-4-3) did not correspond with the genotype (Figure 5C). In our future research, we will clone the genes of these five non-matching materials identified by the CRA08-KASP1 marker to explore other variations that might cause clubroot disease.
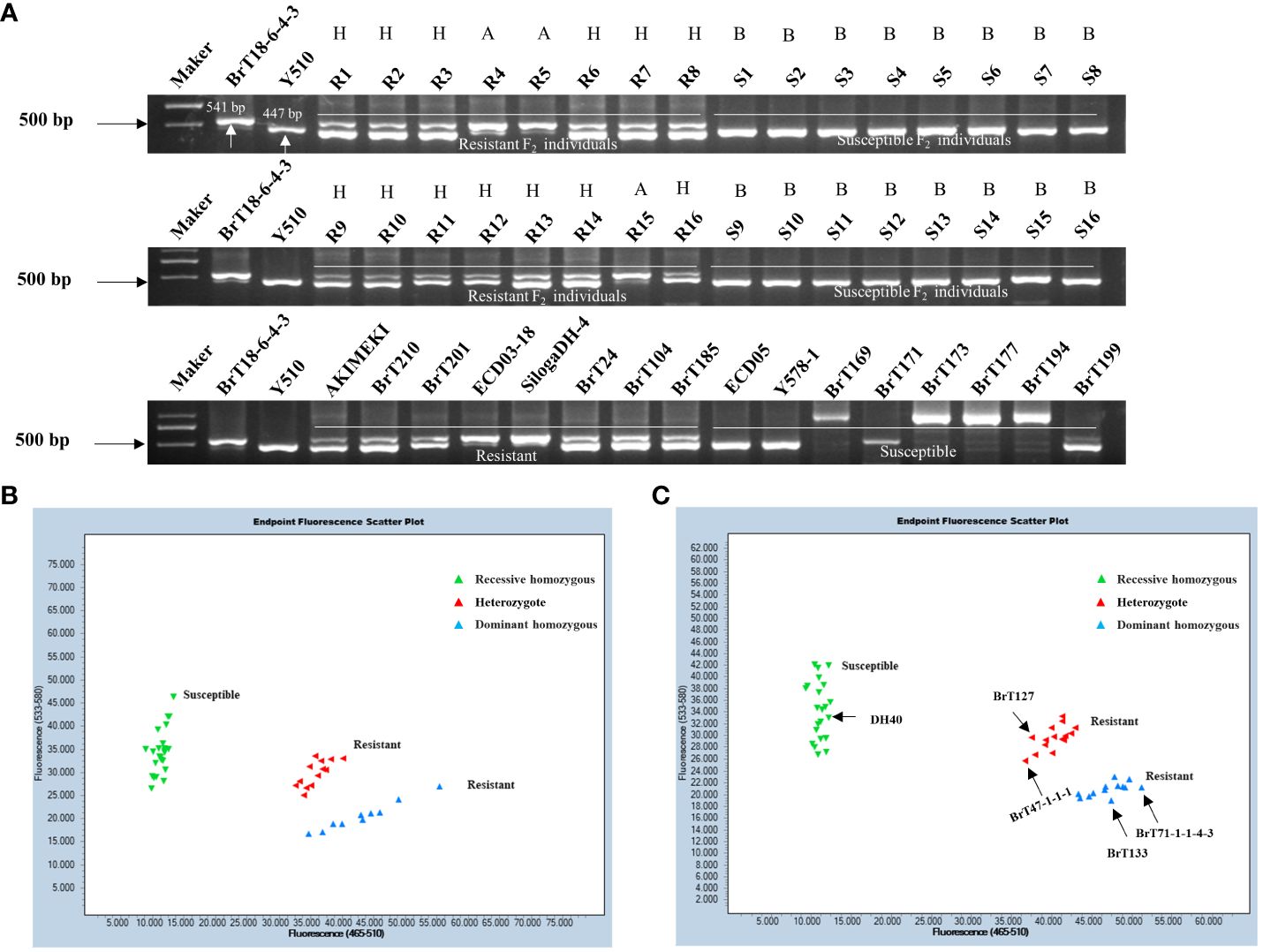
Figure 5 Validation of functional markers. (A) Validation of the CRA08-Indel marker in F2 individuals and natural populations. (B) Validation of the CRA08-KASP1 marker in F2 individuals. (C) Validation of the CRA08-KASP1 marker in 24 disease-resistant (AKIMEKI, ECD03-18, ECD04-1, ECD04-15, SilogaDH-4, SilogaDH-5, BrT22, BrT24, BrT25, BrT30-2-1-3, BrT81, BrT104, BrT114DH-1, BrT114DH-2, BrT114DH-3, BrT185, BrT201, BrT210, BrT238, BrT242, BrT243, WJ2-5-1, and DH40) and 26 disease-susceptible materials (ECD05, R16, BrT121, BrT127, BrT47-1-1-1, BrT130, BrT131, BrT133, BrT165, BrT169, BrT171, BrT71-1-1-4-3, BrT177, BrT193, BrT194, BrT199, BrT221, BrT222, BrT223, BrT224, BrT225, BrT226, BrT228, BrT229, BrT232, and Y578-1). Note: A, dominant homozygous; B, recessive homozygous; and H, heterozygote.
3.5 Construction of an overexpression vector and the development of transgenic Arabidopsis
PCR was conducted using specific full-length primers to amplify the target gene from the subclone vector, which has a length of 4,000 bp. The correct cloning vector was digested using XhoI + EcoRI. Subsequently, the exogenous fragment was ligated into the pBWA (V) HS vector using T4 DNA ligase to obtain the recombinant plasmid (Supplementary Figure 4). The recombinant plasmid was confirmed to be correct, and the overexpression vector was successfully constructed. Overexpression Arabidopsis lines were identified in hygromycin-resistant medium, and homozygous lines were obtained through consecutive selfing over two generations. DNA extracted from T1 transgenic A. thaliana was identified (Supplementary Figure 5). The recombinant plasmid was the positive control, whereas water was the negative control. The PCR bands observed in the eight transgenic plants were consistent with the bands of the positive control. Consequently, eight T2 lines (CR-1, CR-2, CR-3, CR-6, CR-7, CR-8, CR-11, and CR-12) were identified as overexpressing the target gene (Figures 6A, B).
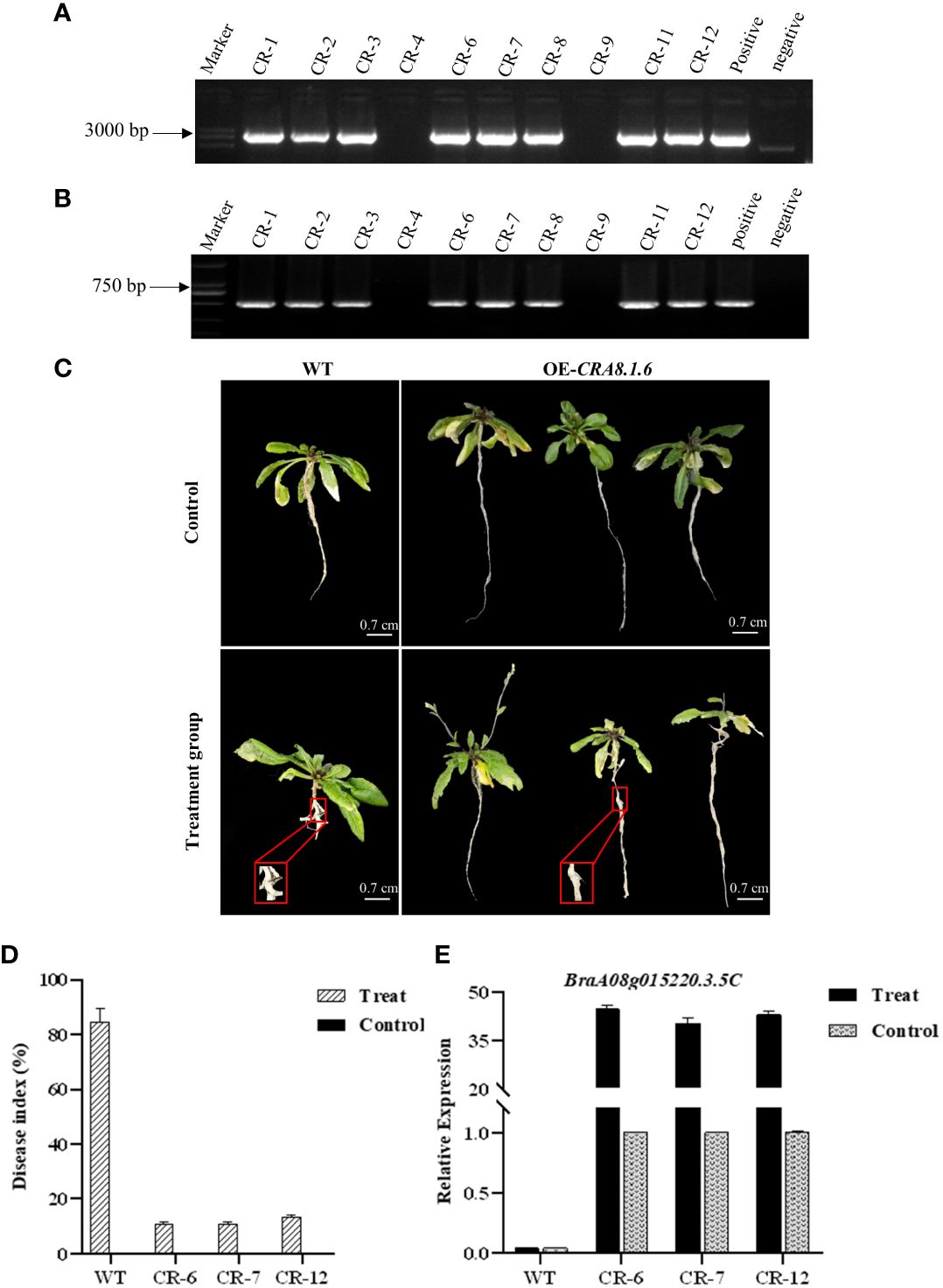
Figure 6 Detection map of genomic primers and hygromycin primers of T2 transgenic Arabidopsis lines, phenotypic characteristics of overexpressed Arabidopsis thaliana, disease index investigation, and qPCR analysis. (A) Primer CRA8-full-F/R; (B) primer Hyg-F/R. CR1-12: T1 lines; positive control: recombinant plasmid; and negative control: H2O. (C) Disease incidence in wild-type roots and overexpression transgenic Arabidopsis following infection with P. brassicae. WT, wild-type Arabidopsis; OE-CRA8.1.6, overexpression Arabidopsis. Bar = 0.7 cm, 0.7 cm, 0.7 cm, and 0.7 cm. (D) Disease index comparison between wild-type and overexpression transgenic Arabidopsis CR-6, CR-7, and CR-12. (E) Analysis of candidate gene expression levels in wild-type and overexpression transgenic Arabidopsis CR-6, CR-7, and CR-12.
3.6 Identification of transgenic Arabidopsis with target gene expression
Transgenic lines CR-6, CR-7, and CR-12, along with wild-type Arabidopsis, were chosen for clubroot phenotypic identification. One set of 25-day-old Arabidopsis plants was inoculated with P. brassicae spores, whereas the other set was inoculated with water only as an experimental control. The experiments were performed with three biological replicates.
After 30 days of inoculation, the control wild-type and transgenic Arabidopsis plants displayed no disease symptoms. In the experimental group, within the overexpression line, CR-6, 22, 5, and 7 out of the 34 plants displayed disease grades 0, 1, and 3, respectively, resulting in a disease index of 10.92%. For the CR-7 line, 19, 5, and 6 out of the 30 plants exhibited disease grades 0, 1, and 3, respectively, with a disease index of 10.95%. Regarding the CR-12 overexpression line, 20, 3, and 9 out of the 32 plants showed disease grades 0, 1, and 3, respectively, yielding a disease index of 13.39% (Supplementary Table 6). In wild-type Arabidopsis, the frequency of disease stages in CR-6, CR-7, and CR-12 was 5 or 7, with disease rates of 85.71%, 89.01%, and 79.59%, respectively (Figure 6C).
The disease index in the overexpression transgenic Arabidopsis was significantly lower than that of the wild-type Arabidopsis in the experimental group (Figure 6D). RNA was extracted from transgenic Arabidopsis plants and reverse transcribed into cDNA to analyze the expression of candidate genes. Criterion 1 was used to evaluate the expression level of the candidate genes in transgenic Arabidopsis based on their expression levels in the control group. The relative expression of the candidate gene in the wild A. thaliana from the experimental and control groups was only 0.04. In contrast, gene expression levels were over 40 in transgenic lines, significantly higher than those in the control (Figure 6E).
4 Discussion
This study analyzed the clubroot resistance of the inbred line BrT18-6-4-3 and the susceptible DH line Y510. Microscopic observation and qRT-PCR verification of gene expression led us to speculate that the stages between 3 DAI and 9 DAI are crucial for cortical infection and clubroot formation in turnip. The infection cycle of P. brassicae typically involves primary infection of the root epidermis followed by secondary infection of cortex tissue. Root hair and cortex infections are thought to occur in host and non-host organisms. We proceeded to morphologically characterize the polymorphic developmental structures of P. brassicae during the primary infection in root hairs and epidermal cells, a process that concluded within 7 days of inoculation (Liu et al., 2020a). In this study, we observed that root hair infection occurred in parents at 1 DAI, and swelling of the root of the susceptible parent commenced at 9 DAI. The cortical cells of the susceptible parent harbored numerous spores, whereas those of the resistant parent contained only a few spores. We hypothesize that cortical infection is inhibited in the resistant parent, which is consistent with the study by Liu et al. (2020b).
The CRA8.1.6 QTL was successfully mapped to a physical interval between 12.255 Mb and 12.365 Mb. Functional annotation of the genes within the QTL suggests that the BraA08g015220.3.5C gene is a potential candidate for clubroot resistance. Analysis of gene sequence alignment between lines BrT18-6-4-3 and Y510 revealed 249 SNPs, seven insertions, and six deletions. Among these variations, an LTR retrotransposon (5,310 bp) was identified as an insertion at 909 bp within the first intron. This LTR retrotransposon was not detected in the resistant line; however, another non-functional LTR insertion was detected in our other materials. Therefore, we concluded that LTR insertion may not be responsible for susceptibility.
Additionally, amino acid sequence alignment indicated six non-synonymous SNPs in the TIR domain but had no effect on clubroot resistance. Because these six non-synonymous mutations are present in both resistant and susceptible materials of natural populations. The frameshift mutation that we identified led to early translation termination at 8,551 bp in the susceptible line Y510. This premature termination in the C-terminal LRR domain resulted in a complete loss of function, leading to the loss of clubroot resistance. Recently, the clubroot-resistant genes Crr1a, CRa, and their alleles have been cloned (Ueno et al., 2012; Hatakeyama et al., 2013, Hatakeyama et al., 2017, Hatakeyama et al., 2022). All of these genes encode nucleotide-binding LRR receptors and feature a TIR domain at the N-terminus. Wang et al. (2023) identified a broad-spectrum clubroot-resistance gene, WeiTsing, in A. thaliana, which was induced in the pericycle to hinder the colonization of P. brassicae in the stele. Moreover, WeiTsing’s channel activity is essential for increasing [Ca2+]cyt and enhancing plant defense. WeiTsing is situated in the endoplasmic reticulum and functions as a calcium-permeable cation-selective channel. However, no WeiTsing homologs have been discovered in B. rapa or B. oleracea (Ochoa et al., 2023). Consequently, current breeding efforts for clubroot resistance in Brassica primarily rely on NBS-LRR genes. Hatakeyama et al. (2013) discovered an LTR retrotransposon inserted into the first exon of the susceptible Crr1aA9709 allele, with a similar result observed in Chiifu-401. However, this insertion is not prevalent in CR-resistant Chinese cabbage varieties. Two susceptible CR alleles lacking 172 amino acids in the C-terminal region were identified in A. thaliana. A chimeric Crr1a transgene restored resistance in susceptible A. thaliana (Hatakeyama et al., 2022), suggesting that susceptibility is attributed to the absence of the C-terminus.
The candidate gene of CRA8.1.6 QTL was located in the same region as previously reported CR genes: Crr1G004, Crr1aKinami90-a, Crr1aKiko85-a, and Crr1aHiroki-b. Gene sequencing revealed a candidate CRA8.1.6 with a genomic length of 4,295 bp and a CDS length of 3,675 bp in the resistant line BrT18-6-4-3. In contrast, in the susceptible line Y510, the genome sequence was 9,586 bp, with a CDS length of 3,644 bp. Sequence alignment of the CDS revealed a 99.4% similarity to Crr1G004 (Supplementary Table 5, Supplementary Figure 4). We hypothesize that CRA8.1.6 is likely an allele of the Crr1G004 gene.
The RPS4, RPP1, and RPP5 genes in tobacco belonging to the TIR-NB-LRR class R genes are functionally impaired by TIR domain deletion or point mutation. In this study, we aligned the TIR region of the susceptible lines Y510, Crr1aKiko85_a, and Crr1aHiroki_b with the disease-resistant lines Crr1G004 and Crr1aKinami90_a. Six non-synonymous SNPs were identified through amino acid sequence alignment and confirmed to be unrelated to the trait in natural populations. In the susceptible Y510 line, we observed premature translation termination at 8,551 bp and alterations in the LRR domain. The underlying cause could be the insertion and deletion of large fragments in the LRR region, resulting in structural damage to this domain. This mutation results in losing resistance in clubroot (Hatakeyama et al., 2022). We developed two variant-based functional markers, CRA08-InDel and CRA08-KASP1, compatible with genotypes and phenotypes. They showed >90% concordance with the clubroot-resistant phenotype.
5 Conclusion
The fine localization distance of CRA8.1.6 was 110 kb. BraA08g0152203.5C is likely the candidate gene for CRA8.1.6, encoding a TIR-NBS-LRR protein. Compared to disease-resistant and susceptible materials, we identified 249 SNPs, seven insertions, and six deletions in CRA8.1.6 (BraA08g015220.3.5C). We discovered premature translation termination at 8,551 bp, leading to the loss of the LRR domain at the C-terminus and ultimately resulting in the loss of clubroot resistance. Furthermore, we developed and validated two functional markers for CRA8.1.6. This accomplishment represents a significant advancement in molecular research on turnip clubroot resistance.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
Author contributions
XW: Writing – original draft, Writing – review & editing. SX: Writing – original draft, Writing – review & editing. YZ: Data curation, Writing – review & editing. LZ: Data curation, Writing – review & editing. UN: Writing – review & editing. SY: Methodology, Data curation, Writing – review & editing. HS: Project administration, Software, Supervision, Writing – review & editing. WZ: Resources, Writing – review & editing. ZW: Investigation, Writing – review & editing. BT: Formal Analysis, Writing – review & editing. FW: Investigation, Writing – review & editing. YY: Conceptualization, Supervision, Writing – review & editing. XZ: Supervision, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the National Science Foundation of China (Grant No. 31801874, 31872945), Science and Technology R & D Joint Fund of Henan Province (232301420024), Young Elite Scientists Sponsorship Program by Henan Association for Science and Technology (Grant No. 2023HYTP022) and Sci-Tech Innovation Team of Henan Academy Agricultural Sciences (2023TD06).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2024.1355090/full#supplementary-material
References
Chai, A. L., X.-w., X., Shi, Y. X., Li, B. J. (2014). Research status of clubroot (Plasmodiophora brassicae) on cruciferous crops in China. Can. J. Plant Pathology. 36, 142–153. doi: 10.1080/07060661.2013.868829
Chen, J., Jing, J., Zhan, Z., Zhang, T., Zhang, C., Piao, Z. (2013). Identification of novel QTLs for isolate-specific partial resistance to Plasmodiophora brassicae in Brassica rapa. PloS One 8, e85307. doi: 10.1371/journal.pone.0085307
Chu, M., Song, T., Falk, K. C., Zhang, X., Liu, X., Chang, A., et al. (2014). Fine mapping of Rcr1 and analyses of its effect on transcriptome patterns during infection by Plasmodiophora brassicae. BMC Genomics 15, 1166. doi: 10.1186/1471-2164-15-1166
Dixon, G. R. (2009). The occurrence and economic impact of plasmodiophora brassicae and clubroot disease. J. Plant Growth Regulation. 28, 194–202. doi: 10.1007/s00344-009-9090-y
Eckholm, E. (1993). Studies on breeding of clubroot resistance in cole Cruciferae crops. Bull. Natl. Res. Inst Veg Ornam Plants Tea Jpn Ser. A 7.
Elke, D., Martin, F., Enrico, G. A. L., Katsunori, H., Masashi, H. (2009). Status and perspectives of clubroot resistance breeding in crucifer crops. J. Plant Growth Regul. 28, 265–281. doi: 10.1007/s00344-009-9100-0
Etsuo, M., Chika, Y., Michio, O., Motohisa, T. (1998). Linkage analysis of RFLP markers for clubroot resistance and pigmentation in Chinese cabbage (Brassica rapa ssp. pekinensis). Euphytica 104, 79–86. doi: 10.1023/A:1018370418201
Gouda, A. C., Warburton, M. L., Djedatin, G. L., Kpeki, S. B., Wambugu, P. W., Gnikoua, K., et al. (2021). Development and validation of diagnostic SNP markers for quality control genotyping in a collection of four rice (Oryza) species. Sci. Rep. 11, 18617. doi: 10.1038/s41598-021-97689-3
Hamann, T., Benkova, E., Bäurle, I., Kientz, M., Jürgens, G. (2002). The Arabidopsis BODENLOS gene encodes an auxin response protein inhibiting MONOPTEROS-mediated embryo patterning. Genes Dev. 16, 1610–1615. doi: 10.1101/gad.229402
Hasan, J., Megha, S., Rahman, H. (2021). Clubroot in Brassica: recent advances in genomics, breeding, and disease management. Genome. 64, 735–760. doi: 10.1139/gen-2020-0089
Hatakeyama, K., Niwa, T., Kato, T., Ohara, T., Kakizaki, T., Matsumoto, S. (2017). The tandem repeated organization of NB-LRR genes in the clubroot-resistant CRb locus in Brassica rapa L. Mol. Genet. genomics: MGG. 292, 397–405. doi: 10.1007/s00438-016-1281-1
Hatakeyama, K., Suwabe, K., Tomita, R. N., Kato, T., Nunome, T., Fukuoka, H., et al. (2013). Identification and characterization of Crr1a, a gene for resistance to clubroot disease (Plasmodiophora brassicae Woronin) in Brassica rapa L. PloS One 8, e54745. doi: 10.1371/journal.pone.0054745
Hatakeyama, K., Yuzawa, S., Tonosaki, K., Takahata, Y., Matsumoto, S. (2022). Allelic variation of a clubroot resistance gene (Crr1a) in Japanese cultivars of Chinese cabbage (Brassica rapa L). Breed. Science. 72, 115–123. doi: 10.1270/jsbbs.21040
Hirai, M., Harada, T., Kubo, N., Tsukada, M., Suwabe, K., Matsumoto, S. (2004). A novel locus for clubroot resistance in Brassica rapa and its linkage markers. TAG. 108, 639–643. doi: 10.1007/s00122-003-1475-x
Hirani, A. H., Gao, F., Liu, J., Fu, G., Wu, C., McVetty, P. B. E., et al. (2018). Combinations of independent dominant loci conferring clubroot resistance in all four Turnip Accessions (Brassica rapa) from the European clubroot differential set. Front. Plant science. 9, 1628. doi: 10.3389/fpls.2018.01628
Howard, R. J., Strelkov, S. E., Harding, M. W. (2010). Clubroot of cruciferous crops - new perspectives on an old disease. Can. J. Plant Pathology: Rev. Can. phytopathologie (1), 32. doi: 10.1080/07060661003621761
Huang, Z., Peng, G., Gossen, B. D., Yu, F. (2019). Fine mapping of a clubroot resistance gene from turnip using SNP markers identified from bulked segregant RNA-Seq. Mol. Breeding. 39, 131. doi: 10.1007/s11032-019-1038-8
Huang, Z., Peng, G., Liu, X., Deora, A., Falk, K. C., Gossen, B. D., et al. (2017). Fine mapping of a clubroot resistance gene in Chinese Cabbage using SNP markers identified from Bulked Segregant RNA Sequencing. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.01448
Kageyama, K., Asano, T. (2009). Life cycle of Plasmodiophora brassicae. J. Plant Growth Regulation. 28, 203–211. doi: 10.1007/s00344-009-9101-z
Kathiresan, N., Temanni, M. R., Al-Ali, R. (2014). Performance improvement of BWA MEM algorithm using data-parallel with concurrent parallelization. Int. Conf. Parallel Distributed Grid Computing. 406–411. doi: 10.1109/PDGC.2014.7030780
Keita, S., Go, S., Tsukasa, N., Katsunori, H., Yasuhiko, M., Hiroyuki, F., et al. (2012). Microstructure of a Brassica rapa genome segment homoeologous to the resistance gene cluster on Arabidopsis chromosome 4. Breed. Science. 62, 170–177. doi: 10.1270/jsbbs.62.170
Kosambi, D. D. (1944). The estimation of map distance from recombination values. Ann. Eugen. 12, 172–175. doi: 10.1111/j.1469-1809.1943.tb02321.x
Laila, R., Park, J.-I., Robin, A. H. K., Natarajan, S., Vijayakumar, H., Shirasawa, K., et al. (2019). Mapping of a novel clubroot resistance QTL using ddRAD-seq in Chinese cabbage (Brassica rapa L). BMC Plant Biol. 19, 13. doi: 10.1186/s12870-018-1615-8
Li, S., Yang, N., Chen, L. (2022). Paraffin section observation of flower bud differentiation of Chimonanthus praecox in Kunming and comparison of the differentiation processes in different regions, China. Hortic. Plant J. 8, 221–229. doi: 10.1016/j.hpj.2021.11.001
Liu, L., Qin, L., Cheng, X., Zhang, Y., Wei, Y. (2020a). Comparing the infection biology of Plasmodiophora brassicae in clubroot susceptible and resistant hosts and non-hosts. Front. Microbiol. 11, 507036. doi: 10.3389/fmicb.2020.507036
Liu, L., Qin, L., Zhou, Z., Hendriks, W., Liu, S., Wei, Y. (2020b). Refining the life cycle of Plasmodiophora brassicae. Phytopathology. 110, 1704–1712. doi: 10.1094/PHYTO-02-20-0029-R
Livak, K. J., Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods: A Companion to Methods Enzymology 25 (4), 402–408. doi: 10.1006/meth.2001.1262
Luo, H., Chen, G., Liu, C., Yun, H. (2014). An improved culture solution technique for Plasmodiophora brassicae infection and the dynamic infection in the root hair. Australas. Plant Pathology. 43, 53–60. doi: 10.1007/s13313-013-0240-0
Mariana, M.-P., Meyers, B. C., Michelmore, R. W., Gaut, B. S. (2002). Patterns of positive selection in the complete NBS-LRR gene family of Arabidopsis thaliana. Genome Res. 12, 1305–1315. doi: 10.1101/gr.159402
Matsumoto, S., Hatakeyama, K., Takashita, S., Miyazaki, T., Kondo, T. (2012). Clubroot and verticillium-resistant Chinese cabbage F1 cultivar, ‘Akimeki’, developed by DNA marker-assisted selection. Agriculture forestry fisheries Technol. Res. J. 35, 1–7.
Md. Masud, K., Abdulsalam, D., Zhang, Y., Chen, Q., Peng, G., Stephen, E. S., et al. (2020). Two clubroot-resistance genes, Rcr3 and Rcr9wa, mapped in Brassica rapa using Bulk Segregant RNA Sequencing. Int. J. Mol. Sci. 21, 5033. doi: 10.3390/ijms21145033
Nguyen, M. L., Monakhos, G. F., Komakhin, R. A., Monakhos, S. G. (2018). The new clubroot resistance locus is located on Chromosome A05 in Chinese Cabbage (Brassica rapa L). Russian J. Genet. 54, 296–304. doi: 10.1134/S1022795418030080
Ochoa, J. C., Mukhopadhyay, S., Bieluszewski, T., Jędryczka, M., Malinowski, R., Truman, W. (2023). Natural variation in Arabidopsis responses to Plasmodiophora brassicae reveals an essential role for resistance to Plasmodiophora brasssicae 1 (RPB1). Plant journal: Cell Mol. Biol. 6, 511079. doi: 10.1111/tpj.16438
Pang, W., Fu, P., Li, X., Zhan, Z., Yu, S., Piao, Z. (2018). Identification and mapping of the clubroot resistance gene CRd in Chinese Cabbage (Brassica rapa ssp. pekinensis). Front. Plant science. 9, 653. doi: 10.3389/fpls.2018.00653
Pang, W., Zhang, X., Ma, Y., Wang, Y., Zhan, Z., Piao, Z. (2022). Fine mapping and candidate gene analysis of CRA3.7 conferring clubroot resistance in Brassica rapa. TAG. 135, 4541–4548. doi: 10.1007/s00122-022-04237-2
Piao, Z., Ramchiary, N., Lim, Y. P. (2009). Genetics of clubroot resistance in Brassica species. J. Plant Growth Regulation. 28, 252–264. doi: 10.1007/s00344-009-9093-8
Piao, Z. Y., Deng, Y. Q., Choi, S. R., Park, Y. J., Lim, Y. P. (2004). SCAR and CAPS mapping of CRb, a gene conferring resistance to Plasmodiophora brassicae in Chinese cabbage (Brassica rapa ssp. pekinensis). TAG. 108, 1458–1465. doi: 10.1007/s00122-003-1577-5
Pu, Y., Yang, D., Yin, X., Wang, Q., Chen, Q., Yang, Y., et al. (2018). Genome-wide analysis indicates diverse physiological roles of the turnip (Brassica rapa var. rapa) oligopeptide transporters gene family. Plant Diversity. 40, 57–67. doi: 10.1016/j.pld.2018.03.001
Sun, Z., Anfeng, M. A., Fei, C., Dandan, C., Pingping, R., Jianwei, G. (2016). Rapid detection of Crr3 gene against clubroot disease by PCR and selection of corresponding germplasm in Chinese Cabbage(Brassica pekinensis L). Northern Horticulture. (22), 120–123. doi: 10.11937/bfyy.201622030
Suwabe, K., Tsukazaki, H., Iketani, H., Hatakeyama, K., Kondo, M., Fujimura, M., et al. (2006). Simple sequence repeat-based comparative genomics between Brassica rapa and Arabidopsis thaliana: the genetic origin of clubroot resistance. Genetics. 173, 309–319. doi: 10.1534/genetics.104.038968
Tang, W. (2007). Genetic analysis of several transgenic plant progenies. Anhui Agric. Sci. (31), 9867–9868. doi: 10.13989/j.cnki.0517-6611.2007.31.020
Ueno, H., Matsumoto, E., Aruga, D., Kitagawa, S., Matsumura, H., Hayashida, N. (2012). Molecular characterization of the CRa gene conferring clubroot resistance in Brassica rapa. Plant Mol. Biol. Reporter. 80, 621–629. doi: 10.1007/s11103-012-9971-5
Voorrips, R. E. (2002). MapChart: software for the graphical presentation of linkage maps and QTLs. J. Hered. 93, 77–78. doi: 10.1093/jhered/93.1.77
Wang, W., Qin, L., Zhang, W., Tang, L., Zhang, C., Dong, X., et al. (2023). WeiTsing, a pericycle-expressed ion channel, safeguards the stele to confer clubroot resistance. Cell 186, 2656–2671.e2618. doi: 10.1016/j.cell.2023.05.023
Wei, X., Zhang, Y., Zhao, Y., Xie, Z., Hossain, M. R., Yang, S., et al. (2021). Root transcriptome and metabolome profiling reveal key phytohormone-related genes and pathways involved clubroot resistance in Brassica rapa L. Front. Plant Science. 12, 759623. doi: 10.3389/fpls.2021.759623
Xiang, Y., Huang, X., Wang, T., Zhang, Y., Liu, Q., Hussey, P. J., et al. (2007). ACTIN BINDING PROTEIN 29 from Lilium pollen plays an important role in dynamic actin remodeling. Plant Cell. 19, 1930–1946. doi: 10.1105/tpc.106.048413
Yang, Z., Jiang, Y., Gong, J., Li, Q., Dun, B., Liu, D., et al. (2022). R gene triplication confers European fodder turnip with improved clubroot resistance. Plant Biotechnol. J. 20, 1502–1517. doi: 10.1111/pbi.13827
Yu, F., Zhang, X., Peng, G., Falk, K. C., Strelkov, S. E., Gossen, B. D. (2017). Genotyping-by-sequencing reveals three QTL for clubroot resistance to six pathotypes of Plasmodiophora brassicae in Brassica rapa. Sci. Rep. 7, 4516. doi: 10.1038/s41598-017-04903-2
Yu, F., Zhang, Y., Wang, J., Chen, Q., Karim, M. M., Gossen, B. D., et al. (2022). Identification of two major QTLs in Brassica napus lines with introgressed clubroot resistance from Turnip Cultivar ECD01. Front. Plant Science. 12, 785989. doi: 10.3389/fpls.2021.785989
Yuan, Y., Qin, L., Su, H., Yang, S., Wei, X., Wang, Z., et al. (2021). Transcriptome and coexpression network analyses reveal hub genes in Chinese Cabbage (Brassica rapa L. ssp. pekinensis) during different stages of Plasmodiophora brassicae Infection. Front. Plant Science. 12, 650252. doi: 10.3389/fpls.2021.650252
Yuan, Y., Zhao, Y., Wei, X., Yao, Q., Jiang, W., Wang, Z., et al. (2017). Pathotype identification of Plasmodiophora brassicae woron. collected from Chinese Cabbage in Henan Province. J. Henan Agric. Sci. 46 (7), 71–76. doi: 10.15933/j.cnki.1004-3268
Zhang, X., Han, F., Li, Z., Wen, Z., Cheng, W., Shan, X., et al. (2024). Map-based cloning and functional analysis of a major quantitative trait locus, BolC.Pb9.1, controlling clubroot resistance in a wild Brassica relative (Brassica macrocarpa). Theor. Appl. Genet. 137 (2), 41. doi: 10.1007/s00122-024-04543-x
Keywords: turnip, clubroot, fine mapping, C-terminal, CRA08-InDel
Citation: Wei X, Xiao S, Zhao Y, Zhang L, Nath UK, Yang S, Su H, Zhang W, Wang Z, Tian B, Wei F, Yuan Y and Zhang X (2024) Fine mapping and candidate gene analysis of CRA8.1.6, which confers clubroot resistance in turnip (Brassica rapa ssp. rapa). Front. Plant Sci. 15:1355090. doi: 10.3389/fpls.2024.1355090
Received: 13 December 2023; Accepted: 21 March 2024;
Published: 17 May 2024.
Edited by:
Xiangshu Dong, Yunnan University, ChinaReviewed by:
Wenxing Pang, Shenyang Agricultural University, ChinaZhansheng Li, Chinese Academy of Agricultural Sciences, China
Chunyu Zhang, Huazhong Agricultural University, China
Copyright © 2024 Wei, Xiao, Zhao, Zhang, Nath, Yang, Su, Zhang, Wang, Tian, Wei, Yuan and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yuxiang Yuan, eXV4aWFuZ3l1YW4xMjZAMTI2LmNvbQ==; Xiaowei Zhang, eGlhb3dlaTU3MzdAMTYzLmNvbQ==
†These authors have contributed equally to this work
 Xiaochun Wei
Xiaochun Wei Shixiong Xiao1,2†
Shixiong Xiao1,2† Ujjal Kumar Nath
Ujjal Kumar Nath Shuangjuan Yang
Shuangjuan Yang Henan Su
Henan Su Fang Wei
Fang Wei Yuxiang Yuan
Yuxiang Yuan Xiaowei Zhang
Xiaowei Zhang