
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 09 February 2024
Sec. Plant Abiotic Stress
Volume 15 - 2024 | https://doi.org/10.3389/fpls.2024.1353365
This article is part of the Research Topic Surviving and Thriving: How Crops Perceive and Respond to Temperature Stress View all 13 articles
 Wei Wei1
Wei Wei1 Jisheng Ju1
Jisheng Ju1 Xueli Zhang1
Xueli Zhang1 Pingjie Ling1
Pingjie Ling1 Jin Luo1
Jin Luo1 Ying Li1
Ying Li1 Wenjuan Xu1
Wenjuan Xu1 Junji Su1,2*
Junji Su1,2* Xianliang Zhang2,3*
Xianliang Zhang2,3* Caixiang Wang1*
Caixiang Wang1*Introduction: Abiotic stress during growth readily reduces cotton crop yield. The different survival tactics of plants include the activation of numerous stress response genes, such as BREVIS RADIX (BRX).
Methods: In this study, the BRX gene family of upland cotton was identified and analyzed by bioinformatics method, three salt-tolerant and cold-resistant GhBRX genes were screened. The expression of GhBRX.1, GhBRX.2 and GhBRXL4.3 in upland cotton was silenced by virus-induced gene silencing (VIGS) technique. The physiological and biochemical indexes of plants and the expression of related stress-response genes were detected before and after gene silencing. The effects of GhBRX.1, GhBRX.2 and GhBRXL4.3 on salt and cold resistance of upland cotton were further verified.
Results and discussion: We discovered 12, 6, and 6 BRX genes in Gossypium hirsutum, Gossypium raimondii and Gossypium arboreum, respectively. Chromosomal localization indicated that the retention and loss of GhBRX genes on homologous chromosomes did not have a clear preference for the subgenomes. Collinearity analysis suggested that segmental duplications were the main force for BRX gene amplification. The upland cotton genes GhBRX.1, GhBRX.2 and GhBRXL4.3 are highly expressed in roots, and GhBRXL4.3 is also strongly expressed in the pistil. Transcriptome data and qRT‒PCR validation showed that abiotic stress strongly induced GhBRX.1, GhBRX.2 and GhBRXL4.3. Under salt stress and low-temperature stress conditions, the activities of superoxide dismutase (SOD), peroxidase (POD) and catalase (CAT) and the content of soluble sugar and chlorophyll decreased in GhBRX.1-, GhBRX.2- and GhBRXL4.3-silenced cotton plants compared with those in the control (TRV: 00). Moreover, GhBRX.1-, GhBRX.2- and GhBRXL4.3-silenced cotton plants exhibited greater malondialdehyde (MDA) levels than did the control plants. Moreover, the expression of stress marker genes (GhSOS1, GhSOS2, GhNHX1, GhCIPK6, GhBIN2, GhSnRK2.6, GhHDT4D, GhCBF1 and GhPP2C) decreased significantly in the three target genes of silenced plants following exposure to stress. These results imply that the GhBRX.1, GhBRX.2 and GhBRXL4.3 genes may be regulators of salt stress and low-temperature stress responses in upland cotton.
Cotton is an annual herbaceous plant of the Malvaceae family that is not only used to produce natural textile fibers but is also one of the major cash crops and a significant source of protein and seed oil worldwide (John and Crow, 1992). Despite the economic importance of cotton, various environmental factors, including both biotic and abiotic stresses, pose a great threat to cotton production (Potters et al., 2007). Due to global climate change, drought, salinity, extreme temperature, waterlogging, heavy metals, hypoxia and other major abiotic stresses will impede the development and growth of plants and affect crop yield and quality and sustainable agricultural development (Hasanuzzaman et al., 2020). To increase their chances of survival, plants have developed numerous defense strategies and mechanisms to handle a range of challenging circumstances (Westerheide et al., 2012; Van Zelm et al., 2020; Nian et al., 2021). In plant defense mechanisms, many stress-response genes help plants tolerate the adverse effects of various stressors by regulating their transcriptome levels.
The highly conserved BREVIS RADIX (BRX) family of plant-specific genes is found in all higher plants for which data are known but not in animals or single-celled organisms (Mouchel et al., 2004). For the first time, BRX was isolated from Arabidopsis thaliana UK-1 plants with a short root phenotype using map-based cloning (Mouchel et al., 2004). There are five BRX genes in Arabidopsis: BRX, BRXL1 to BRXL4. Although the sequences of BRX are highly conserved, the functions of the five genes in the Arabidopsis BRX family are largely nonredundant (Briggs et al., 2006). BRX proteins have four highly conserved domains, two of which are short N-terminal domains and two of which are BRX domains (Briggs et al., 2006; Beuchat et al., 2010a; Liu et al., 2010). These include a 9–10 amino acid region at the N-terminus that is thought to contain palmitoylation signals and is crucial for the membrane localization of BRX (Rowe et al., 2019). The adjacent domains are a 25-amino acid N-terminal domain with a KDMA motif and two BRX domains with 55 amino acid extensions consisting of tandem repeats (Koh et al., 2021). The BRX domain may represent a new protein–protein interaction domain, which is the first indication of the biological function of the BRXL protein (Briggs et al., 2006). Adding one BRX domain to the corresponding BRXN140 fragment partially restored functionality (Briggs et al., 2006). However, the addition of two BRX domains to the corresponding BRXN140 fragment, as with full-length BRX, significantly alleviated the brx root growth phenotype and elicited hypocotyl function to acquire the phenotype (Briggs et al., 2006; Scacchi et al., 2009). The conserved N-terminal domain of BRX family proteins may play only secondary functional roles (Briggs et al., 2006). However, according to Ka/Ks analysis, the diversity of BRX family genes may be caused by the variable N-terminal region, which could be the cause of the nonredundant functions of most AtBRX family genes (Briggs et al., 2006; Beuchat et al., 2010a).
BRX is a growth regulator needed for root growth that regulates cell proliferation and elongation in root growth areas (Mouchel et al., 2006). Auxin substantially stimulates BRX expression, while brassinosteroid (BR) marginally inhibits expression (Mouchel et al., 2006). This finding suggests that BRX forms a feedback loop between BR and auxin, which maintains brassinosteroid thresholds and controls the root response to auxin, while auxin completes the cycle by controlling BRX expression (Mouchel et al., 2006). The BRX functional allele (brx-2) is highly sensitive to ABA-mediated root growth inhibition, and it has also been shown to be insensitive to cytokinin-induced lateral root initiation inhibition, indicating crosstalk between BR and cytokinin (Bari and Jones, 2009; Scacchi et al., 2009). Therefore, BRX could be a key node in the interconnection of auxin, BR, ABA, and cytokinin signaling during root development. (John and Crow, 1992; Scacchi et al., 2009). The BRX protein is associated with the plasma membrane but is translocated to the nucleus after auxin treatment to regulate gene expression (Scacchi et al., 2009). The BRX gene family has been studied and identified in a variety of plant species (Li et al., 2009; Liu et al., 2010; Zhang Y. et al., 2021; Tiwari et al., 2023). BRX is involved in the longitudinal and radial expansion of hypocotyls and roots, the development of embryos and leaves, and the asymmetric division of stomatal lineage cells in Arabidopsis, and unlike their partial or nonredundant roles in roots, BRX genes play redundant roles in stomatal development (Rowe et al., 2019). The brx mutants also exhibited significant reductions in cotyledon and leaf growth, and deletion of the BRX functional allele (brx-2) resulted in a decrease in rosette area in comparison to that of Col-0, but the quantity of leaves remained the same (Rodrigues et al., 2009). In contrast, plants with functionally acquired BRX exhibit elongated hypocotyls and epicotyl leaves (Scacchi et al., 2009; Gill and Tuteja, 2010). In rice, compared with those of WT plants, OsBRXL4-overexpressing transgenic plants had significantly longer roots and greater sensitivity to auxin under normal growth conditions (Liu et al., 2010). These results suggest that OsBRXL4 may regulate primary root growth via auxin signaling (Liu et al., 2010). The optimal tillering angle is essential for an ideal plant structure, and the molecular mechanism controlling the tillering angle in rice will improve our ability to rationally change the structure of rice plants, thereby increasing grain yield. OsBRX regulates auxin transport to control the tiller angle of rice plants. (Scacchi et al., 2009; Liu et al., 2010). Overexpression of three different BrBRX genes in Brassica rapa significantly increased the number of rosette leaves, decreased the rosette area and increased the petiole length in transgenic plants (Zhang Y. et al., 2021). TaBRXL1 is generally expressed in all analyzed tissues except flag leaves, and the expression levels of TaBRXL2, TaBRXL3 and TaBRXL4 are significantly increased under auxin treatment, indicating that TaBRX family genes may contribute to functional diversity (Tiwari et al., 2023).
Studies have shown that the expression of BRXs is differentially induced by different types of abiotic stress (Liu et al., 2010; Tiwari et al., 2023). In rice, OsBRXL1 and OsBRXL4 respond to drought, salt and cold stress; OsBRXL3 responds to drought and salt stress; OsBRXL2 and OsBRXL5 respond only to cold stress; and the expression of these five OsBRXL genes is upregulated under drought and salt stress and downregulated under low-temperature stress (Liu et al., 2010). TaBRXL1 was found to be involved primarily in developmental processes, whereas TaBRXL2 was highly regulated by development, hormones, and other abiotic stimuli (Tiwari et al., 2023). In addition to TaBRXL2, the other TaBRX genes were significantly downregulated under drought conditions in common wheat. Under osmotic stress (200 mM mannitol), TaBRXL2, TaBRXL3 and TaBRXL4 were upregulated (Tiwari et al., 2023). In summary, the BRX gene family plays an important role in enhancing plant tolerance to abiotic stress.
Cotton is a valuable economic crop that provides “oil, fiber, feed, and medicine.” Yield losses readily occur due to abiotic stress during the growth development process of cotton. Thus, it is important to screen and apply key genes in cotton that respond well to abiotic stress and improve cotton stress resistance through biological breeding. To date, the relationship between the GhBRX gene and abiotic stress has not been studied. The whole genomes of 12 GhBRX genes were discovered in this work. Its evolutionary model, physical and chemical properties, chromosomal location, gene structure, cis-acting elements and expression pattern were comprehensively analyzed. Using the virus-induced gene silencing (VIGS) approach, we further elucidated the biological function of the GhBRX.1, GhBRX.2 and GhBRXL4.3 genes in response to salt and cold stress.
To identify cotton BRX gene families, we used CottonFGD (http://www.cottonfgd.org/) and Arabidopsis thaliana genome sequence data from TAIR (http://www.arabidopsis.org) for five AtBRX protein sequences from Pfam (https://pfam.xfam.org/), which was subsequently used to download the PF08381 hidden Markov model (HMM) version 3.0 (El-Gebali et al., 2019). Then, we used HMMER 3.0 software with default parameter settings (http://www.HMMER.org/) to obtain the BRX gene, for which the E value was <1e-5 (Zhu et al., 2017). We used Pfam (https://www.ebi.ac.uk/Tools/pfa/pfamscan/) and SMART (https://smart.embl.de/) to further evaluate the results of our genes for confirmation. Finally, we manually confirmed the identified BRX genes. The BRX protein sequence was predicted through ExPASy (https://us.expasy.org/tools/protparam.html) to predict the molecular weight (MW), theoretical isoelectric point (pI), etc. In addition, Wolfpsort (https://www.wolfpsort.hgc.jp/) was used to predict the subcellular localization of the cotton BRX protein.
The ClustalW program (version 2.0) was used to align the full-length amino acid sequences of the BRX-encoded Gossypium hirsutum (Gh), Gossypium arboreum (Ga), Gossypium raimondii (Gr), Arabidopsis thaliana (At), Brassica rapa (Br), Oryza sativa (Os), and Triticum aestivum (Ta). The alignment was then manually modified in MEGA 7.0. Subsequently, we constructed a neighbor joining (NJ) tree with 1000 bootstrap repetitions using MEGA 7.0’s Poisson substitution model with default parameters (Kumar et al., 2016). The Interactive Tree of Life (iTOL) tool was utilized to enhance the visualization of the phylogenetic tree (http://itol.embl.de/).
To find conserved protein motifs, we utilized the motif elicitation (multiple EM for motif elicitation) website (http://meme-suite.org/) (Bailey et al., 2009). A conservative motif map was generated using TBtools software (Chen et al., 2018). The upland cotton CDS and genome sequence and NWK file from phylogenetic tree analysis were used to map gene structure through the Server (Gene Structure Display Server, GSDS) program (http://gsds.cbi.pku.edu.cn/).
GFF3 files extracted from the CottonFGD database were used to determine chromosome locations. The GhBRX gene was located on the chromosome using TBtools software (Chen et al., 2020). During collinear analysis, to compare the GhBRX protein sequences, the Basic Local Alignment Search Tool (BLAST) was utilized, and the cutoff E value was < 10-5. The MCScanX tool of the TBtools software was subsequently used to evaluate the above BLASTP results, extract collinear pairs of GhBRX family proteins, and construct a collinearity map of the GhBRX family using TBtools (Chen et al., 2020). Ka/Ks values of the GhBRX gene were determined by using TBtools (Chen et al., 2020). TBtools was subsequently used to construct interspecific collinearity maps of G. hirsutum, G. raimondii and G. arboretum (Chen et al., 2020).
We obtained an upstream sequence of our genes spanning 2 kb from the translation start site from CottonFGD (https://cottonfgd.net/) to identify cis-regulatory regions in BRX genes. Then, we predicted the cis-regulatory elements in the promoter region of the GhBRX genes using the PlantCARE website (http://bioinformatics.psb.ugent.be/beg).
To verify the GhBRX gene expression profile in upland cotton organizations, RNA-seq data from Zhejiang University (ZJU) (http://cotton.zju.edu.cn/) were downloaded to determine the GhBRX gene organization and response to salt, drought, cold and heat stresses(Zhang et al., 2015). Heatmaps of 12 GhBRX genes were generated using TBtools.
Healthy plants of the new upland cotton variety XinshiK25 were selected and treated with 15% PEG (PEG-6000), 250 mmol/L NaCl, 12°C and 42°C, respectively. The leaves were removed every 3 h and treated for 24 h. RNA and reverse transcription cDNA were extracted using a kit produced by Tiangen Biochemical Technology (Beijing) Co., Ltd. GhBRX gene primers were constructed using NCBI Prime-BLAST (primer design tool, https://www.ncbi.nlm.nih.gov/tools/primer-blast/index.cgi?LINK_LOC=BlastHome). Supplementary Table S1 shows the primers used. Real-time fluorescence quantitative PCR was performed (SYBR Green, FP209, Tiangen, China) according to the instructions for the thermal cycling process. AY305733 is an internal control gene that employs the 2-ΔΔCT technique (Livak and Schmittgen, 2001). The comparative expression values of GhActin and GhBRXs were calculated, and the relative expression levels of three independent biological replicates and technical replicates were averaged.
The coding sequences of the GhBRX.1, GhBRX.2 and GhBRXL4.3 genes were downloaded from CottonFGD, and specific primers (Supplementary Table S2) were designed. Using the online tool NCBI Prime-BLAST, specific primers for gene silencing and related stress response gene fluorescence were designed (Supplementary Table S3). Specific cDNA sequences of GhBRX.1, GhBRX.2 and GhBRXL4.3 were amplified, constructed into Figure vectors, and subsequently introduced into Agrobacterium strain GV3101. Then, the cotyledons of two 8-day-old XinshiK25 plants were grown at 25°C for 24 h in the dark and injected with the vector (Gao et al., 2013). RNA was collected from cotton leaves that had been silenced at the four-leaf stage, and RT–qPCR was used to assess the silencing efficacy. RT–qPCR was used to test the expression of nine stress-responsive genes, GhSOS1, GhSOS2, GhNHX1, GhCIPK6, GhBIN2, GhSnRK2.6, GhHDT4D, GhCBF1 and GhPP2C, associated with salt stress and cold stress in control and silenced plants.
The activities of peroxidase (POD), catalase (CAT), and superoxide dismutase (SOD), three antioxidant enzymes that are important to plants under abiotic stress conditions, were evaluated. SOD activity was determined by tracking the suppression of the photochemical reduction of nitroblue tetrazole (Giannopolitis and Ries, 1977). POD activity was evaluated using ortho-methoxyphenol (guaiacol) as a substrate (Castro et al., 2017). POD can oxidize guaiacol to o-4-methoxyphenol, which can be detected via spectrophotometry at 470 nm. CAT activity is determined by UV absorption (Beers and Sizer, 1952), and the malondialdehyde (MDA) content is a marker of lipid peroxidation (Liu et al., 2018). Chlorophyll was extracted with 95% ethanol and measured spectrophotometrically at 665 nm and 649 nm (Lichtenthaler and Wellburn, 1983).
The amino acid sequences of the BRX proteins found in Arabidopsis and rice were used as query sequences. As shown in Supplementary Table S4, the presence of 12, 6, and 6 BRX genes was confirmed in G. hirsutum, G. raimondii and G. arboreum. The number of BRX genes in allotetraploid cotton was twice as high as that in the two diploid cotton lines, suggesting that the BRX gene family experienced expansion during evolution in Gossypium spp. while maintaining their unique genetic makeup. The predicted protein sequences were used to calculate the number of amino acids, MW, and pI. The 24 BRX proteins that were found had amino acid ranges of 342–405, protein pIs that varied from 5.73 to 8.59, and MWs that varied from 38.58 to 45.59 kDa. Bioinformatics analysis revealed that 24 BRX proteins were predicted to locate in the nucleus. All features and chromosomal locations of the identified BRXs are shown in Additional file Supplementary Table S4.
Using 57 BRX protein sequences obtained from G. hirsutum, G. raimondii, G. arboreum, B. rapa, A. thaliana, T. aestivum, and O. sativa, we constructed a phylogenetic tree based on multiple alignment analyses using the neighbor–joining (NJ) method to examine the phylogenetic relationships among the BRX family genes (Figure 1). Based on bootstrap values (=1,000), the 57 BRX proteins were found to precluster into two main groups (Group I and Group II). Group I was defined by AtBRX (At1G31880) and had no BRX members from the three cotton species. Group II was divided into three subgroups: II-1, II-2 and II-3. All the OsBRX genes clustered in group I and subgroup II-1, and all three Gossypium spp. species BRX genes clustered in subgroups II-2 and II-3. As shown in Figure 1, mutually homologous BRX family genes clustered together, with one copy in diploid cotton species and two copies in heterotetraploid cotton species in almost every direct homologous group. The results of the clustering analysis provide additional evidence that the upland cotton species heterotetraploid is the product of an ancestral cross between G. raimondii and G. arboreum, two diploid cotton species, and doubles in number.
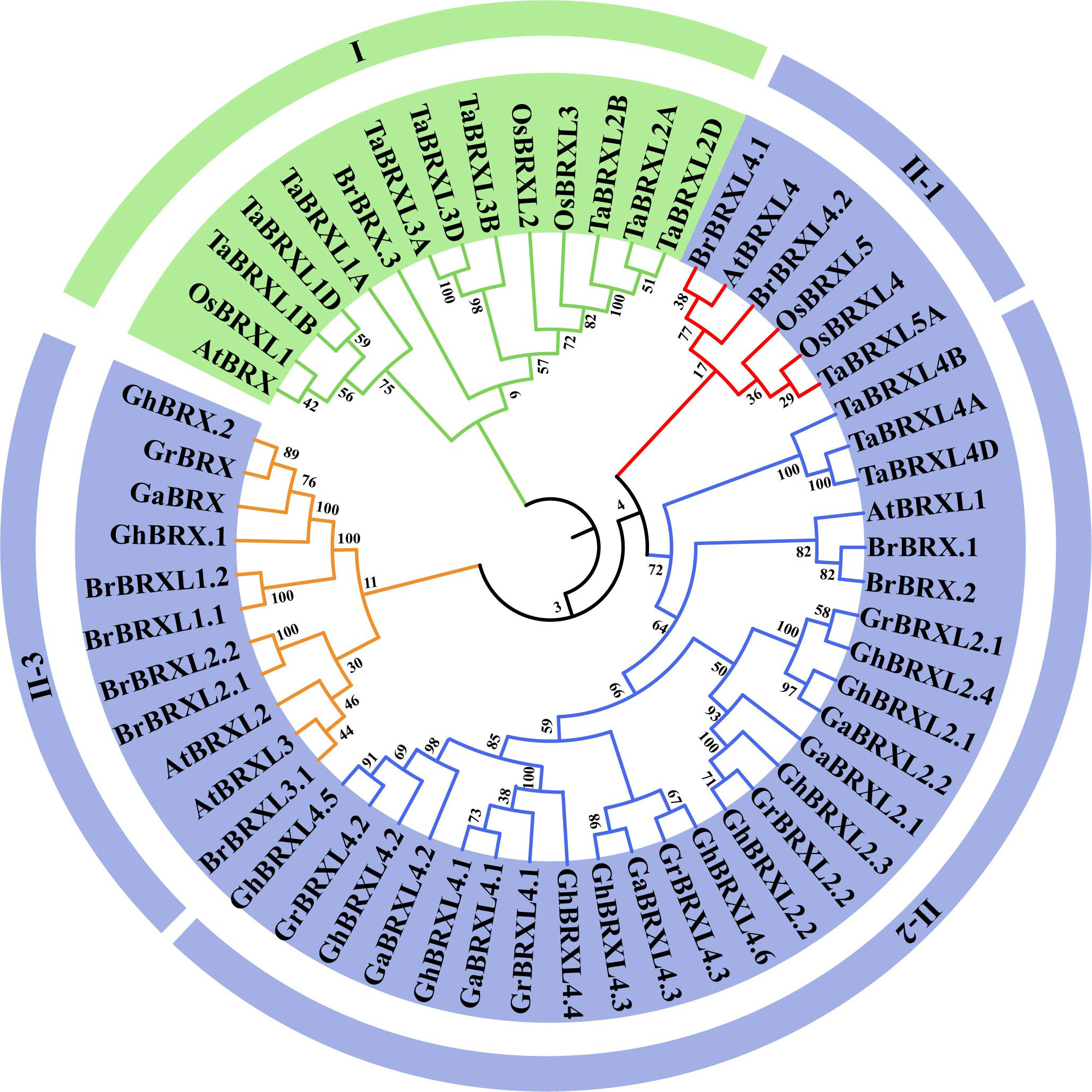
Figure 1 Phylogenetic relationships of BRX proteins in seven plant species. Gossypium hirsutum (Gh), Gossypium raimondii (Gr), Gossypium arboreum (Ga), Brassica rapa (Br), Arabidopsis thaliana (At), Tariticum aestivum (Ta), and Oryza sativa (Os). At each node, the bootstrap values were displayed.
We analyzed the structural features of GhBRX using the GSDS program and the exon and intron structures and conserved structural domains using the MEME tool. The GhBRX genes exhibited a comparatively high level of structural similarity according to the exon/intron structure analysis (Supplementary Figure S1A). All the GhBRX genes contained five exons. Most of the homologous genes had similar gene lengths, with GhBRXL4.3 being the longest. The results suggest that exon structure is associated with phylogenetic relationships, further supporting the structural classification of the GhBRX gene family. Using MEME software, the functional regions of the GhBRX proteins were divided into five different motifs (Supplementary Figure S1B). Motifs 1, 2, 3 and 5 (BRX motifs) were widely distributed among all GhBRX family members, with only GhBRX.1 and GhBRX.2 without motif 4; these two genes also had the shortest lengths. The motif compositions of GhBRX proteins in a given branch were strikingly similar, indicating that these proteins may early every play comparable role.
Gene duplication events in G. hirsutum were explored with TBtools to elucidate the amplification patterns and determine the homologous locus linkages of the GhBRX gene family members between the At and Dt subgenomes (Figure 2). One tandem duplicate gene was identified on chromosome A05. In addition, the GhBRX gene family contained 20 segmentally duplicated genes (Figure 3). These data suggest that segmental duplications are important in the evolution of GhBRX gene families, indicating the dominance of segmental repeats over tandem repeats in the evolution of the GhBRX gene family (Supplementary Table S5). Furthermore, to gain a deeper understanding of the homologous gene functions and evolutionary relationships of the BRX genes, the outcomes of the genome symbiosis study between upland cotton and two other species of cotton were examined (Supplementary Figure S2). In conclusion, the current findings suggest that BRX genes may undergo certain genomic rearrangements during polyploidy. The nonsynonymous substitution (Ka), synonymous substitution (Ks), and Ka/Ks ratio were estimated to better understand the evolutionary constraints regulating the functional divergence of the GhBRX gene family (Supplementary Table S6). The Ka/Ks ratio of all duplicate GhBRX gene pairs was less than 1, indicating that selective pressure may have been applied to the GhBRX family genes throughout their evolutionary history.
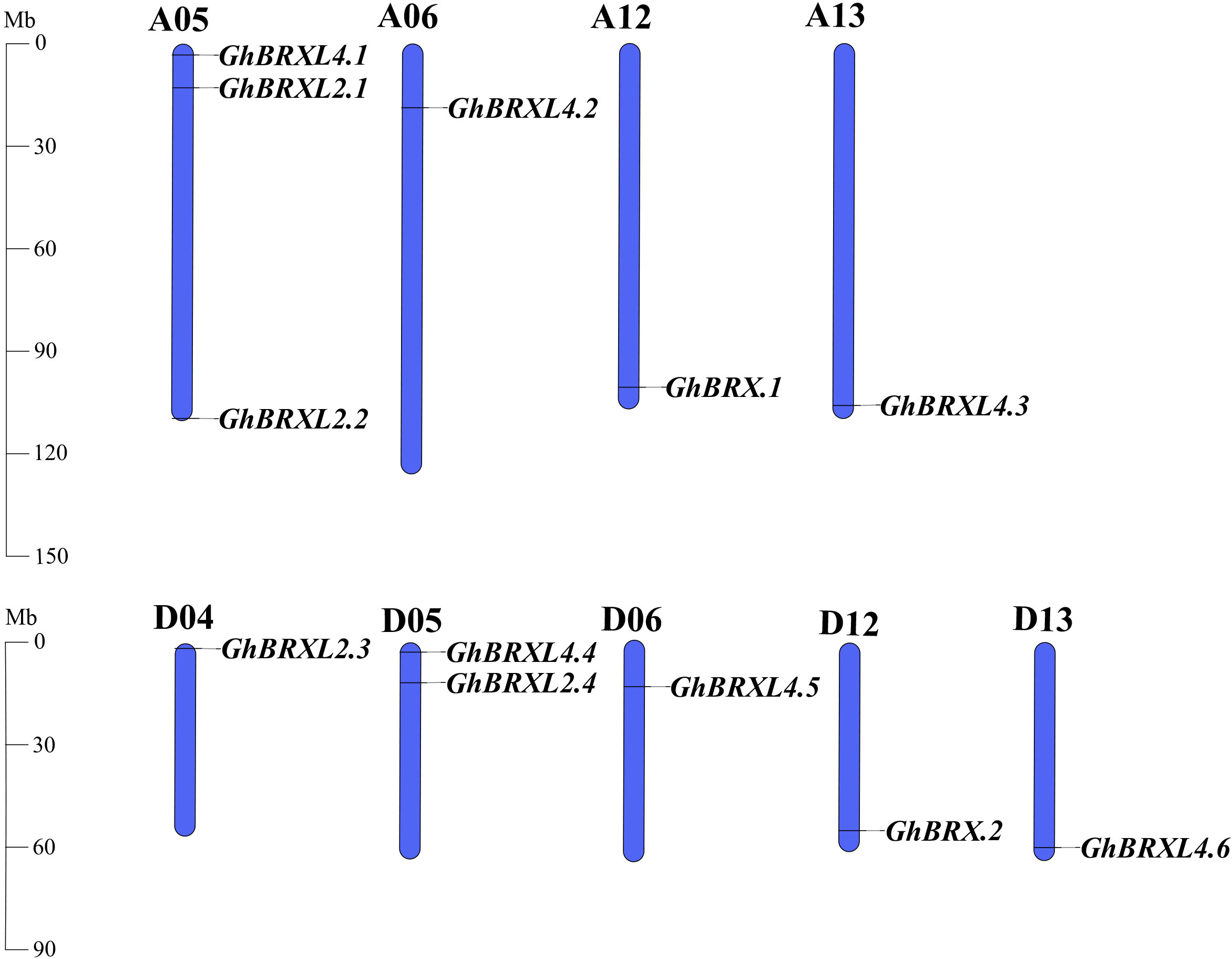
Figure 2 GhBRX locations on Gossypium hirsutum chromosomes. The blue bars represent the At and Dt subgenomes of G. hirsutum. The subgenomes’ matching gene names were showned on the right side of each chromosome.
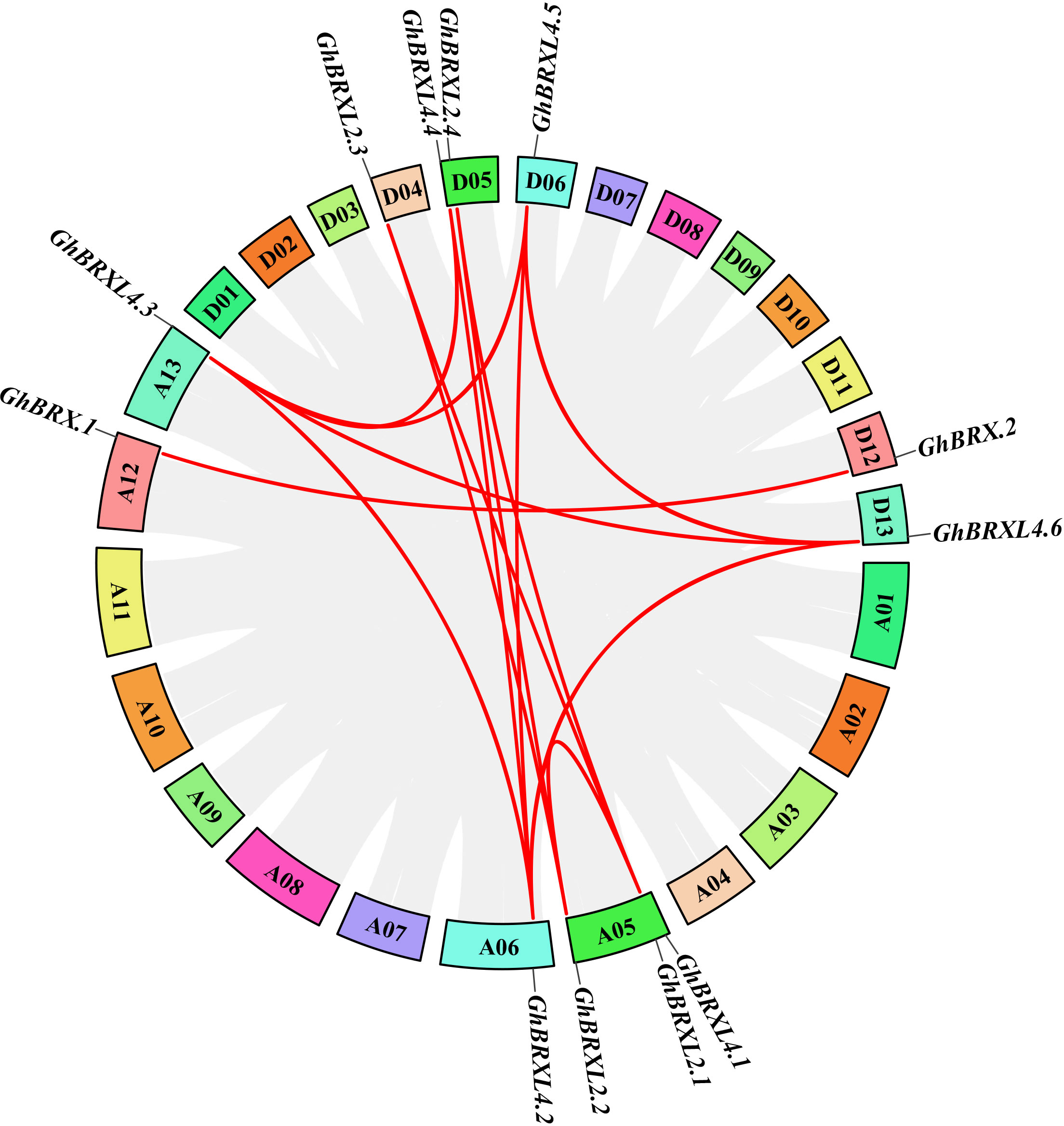
Figure 3 Duplication of GhBRX genes. Duplications of GhBRX genes on the chromosome of G. hirsutum; the red lines represented gene pairs of GhBRX. The rectangles represented chromosomes.
We examined and characterized the cis-acting region in the 2 kb promoter sequence of GhBRXs to further investigate the putative regulatory roles of the BRX gene in response to abiotic stressors. We discovered 38 different types of cis-regulatory elements that are involved in tissue-specific expression, stress responses, phytohormone responses, and light responses (Figure 4). Five elements associated with tissue-specific expression were identified, namely, the RY element, O2 site, CAT box, GCN4 motif and HD-Zip 1. The cis-regulatory element associated with meristematic tissue expression (CAT-box) was only present in the promoter regions of the homologs GhBRXL2.1 and GhBRXL2.4, and the seed-specific regulatory element (RY-element) is specific to GhBRXL2.4. Six elements associated with stress responsiveness were identified, and these cis-acting elements are involved in defense and stress responsiveness, drought, low temperature, anaerobic and wounding responses. Abiotic stress response elements, such as the drought response element (MBS) and low temperature response element (LTR), were present in variable amounts in the homologous genes GhBRX.1, GhBRX.2, GhBRXL4.2 and GhBRXL4.5. The cis-acting regulatory elements necessary for anaerobic induction (ARE) were more abundant in most GhBRX genes. In addition, four hormone-related elements were identified, namely, the gibberellin response element (GARE motif-containing element, P-box and TATC motif-containing element), the abscisic acid response element (ABRE), the salicylic acid response element (TCA-element) and the methyl jasmonate (MeJA) response element (CGTCA motif-containing element and TGACG motif-containing element). Among them, ABRE and MeJA response elements were relatively abundant in most GhBRX genes. Light-responsive cis-elements, including Box 4, G-box, and TCT-motif, were present in all the GhBRX promoters. Among the light-responsive cis-acting regulatory elements, Box 4 and G-box were relatively more common. Taken together, these findings imply that GhBRX genes might be crucial for defense-related signaling, phytohormone responses, and abiotic stress responses.
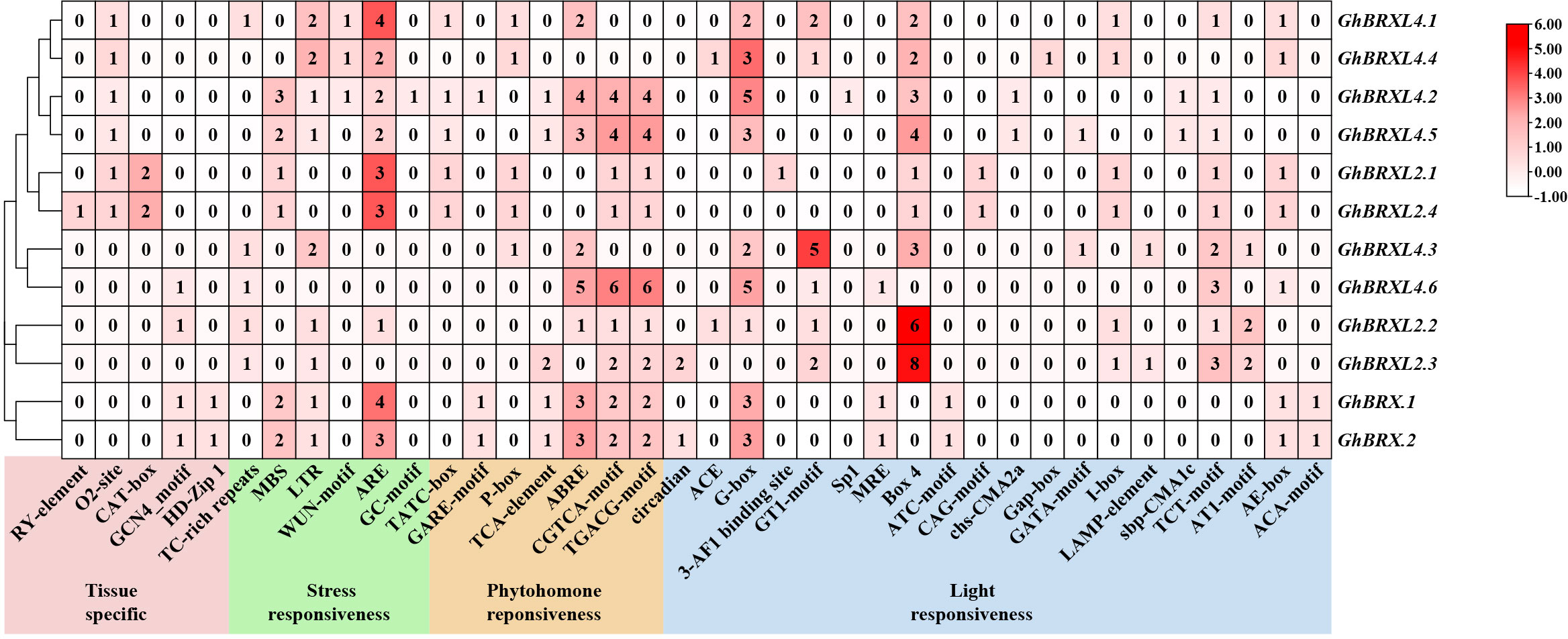
Figure 4 Cis-regulatory element prediction outcomes in the promoter regions of GhBRX gene family members. Shades and digits in the boxes denoted the number of cis-elements.
To determine the purpose of the GhBRX gene, we analyzed the expression profile data in the cotton functional database. Tissue-specific expression analysis indicated that the GhBRX.1, GhBRX.2, GhBRXL2.3, GhBRXL4.1 and GhBRXL4.4 genes were expressed mainly in roots; GhBRXL2.2, GhBRXL4.2, and GhBRXL4.5 were expressed mainly in ovules; and GhBRXL2.4, GhBRXL4.3 and GhBRXL4.6 were expressed highly in fibers, pistils, and flowers, respectively (Figure 5A). According to the expression analysis, the response of five genes, namely, GhBRX.1, GhBRXL2.1, GhBRXL2.4, GhBRXL4.3 and GhBRXL4.6, to abiotic stress significantly increased under salt treatment (Figure 5B). Under PEG stress, GhBRX.2, GhBRXL4.2, GhBRXL4.3, GhBRXL4.5 and GhBRXL4.6 were also significantly upregulated (Figure 5B). Under heat stress treatment, four genes were upregulated, namely, GhBRXL4.1, GhBRXL4.2, GhBRXL4.3 and GhBRXL4.6 (Figure 5C). Under cold stress treatment, GhBRX.1 and GhBRXL4.6 were upregulated, while GhBRXL2.1 and GhBRXL2.4 were downregulated (Figure 5C).
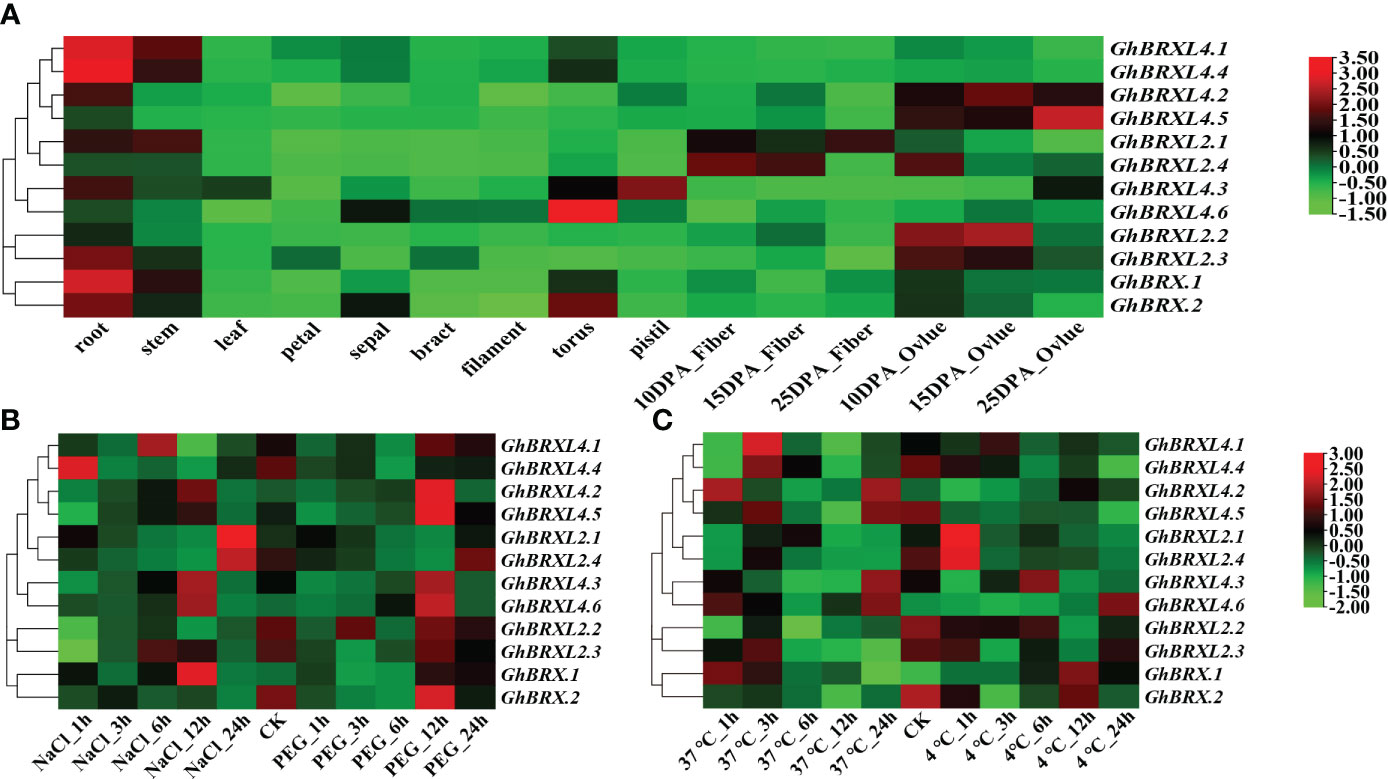
Figure 5 GhBRX gene expression patterns in distinct tissues and under four abiotic stimuli in upland cotton. (A) Expression patterns of 12 GhBRXs in various tissues. (B) Salt and PEG stress expression patterns of 12 GhBRXs. (C) Expression patterns of 12 GhBRXs under heat and cold stress.
To further determine whether the level of GhBRX gene family expression was related to abiotic stress, we investigated the expression levels of 12 GhBRX genes via qRT–PCR. Seedlings leaves were sampled at five different stress periods (0, 1, 3, 6, 12 and 24 h) to analyze the expression of GhBRX genes under different abiotic stress conditions, including salt, drought, cold and heat stress (250 mM NaCl, 15% PEG, 12°C and 42°C). Under four different stresses (PEG, NaCl, cold, and heat), all the tested genes responded to at least one stress (Figure 6). Following a three-hour salt stress treatment, the expression levels of GhBRX.1, GhBRX.2, GhBRXL2.4, GhBRXL4.1, and GhBRXL4.4 increased when the stress duration was extended. GhBRXL4.5 and GhBRXL4.6 exhibited increasing and then decreasing trends, respectively. After 3 h of drought stress treatment, the expression of GhBRX.1, GhBRX.2, GhBRXL2.2, GhBRXL2.3, GhBRXL2.4 and GhBRXL4.3 first increased and then decreased, and that of GhBRXL2.1, GhBRXL4.1, GhBRXL4.2, GhBRXL4.4, GhBRXL4.5 and GhBRXL4.6 first decreased and then increased. GhBRX.1, GhBRX.2, GhBRXL2.2, GhBRXL2.3, GhBRXL4.1, GhBRXL4.2, GhBRXL4.3, GhBRXL4.4, GhBRXL4.5 and GhBRXL4.6 were subjected to high-temperature stress for 3 h, after which the expression levels increased with increasing duration of stress. Similarly, the expression of GhBRX.1, GhBRX.2, GhBRXL4.2, GhBRXL4.3 and GhBRXL4.5 significantly increased after 24 h of low-temperature stress treatment compared with that at 0 h. In summary, GhBRX.1, GhBRX.2 and GhBRXL4.3, which are highly responsive to all four kinds of stress, were selected as stress candidate genes.
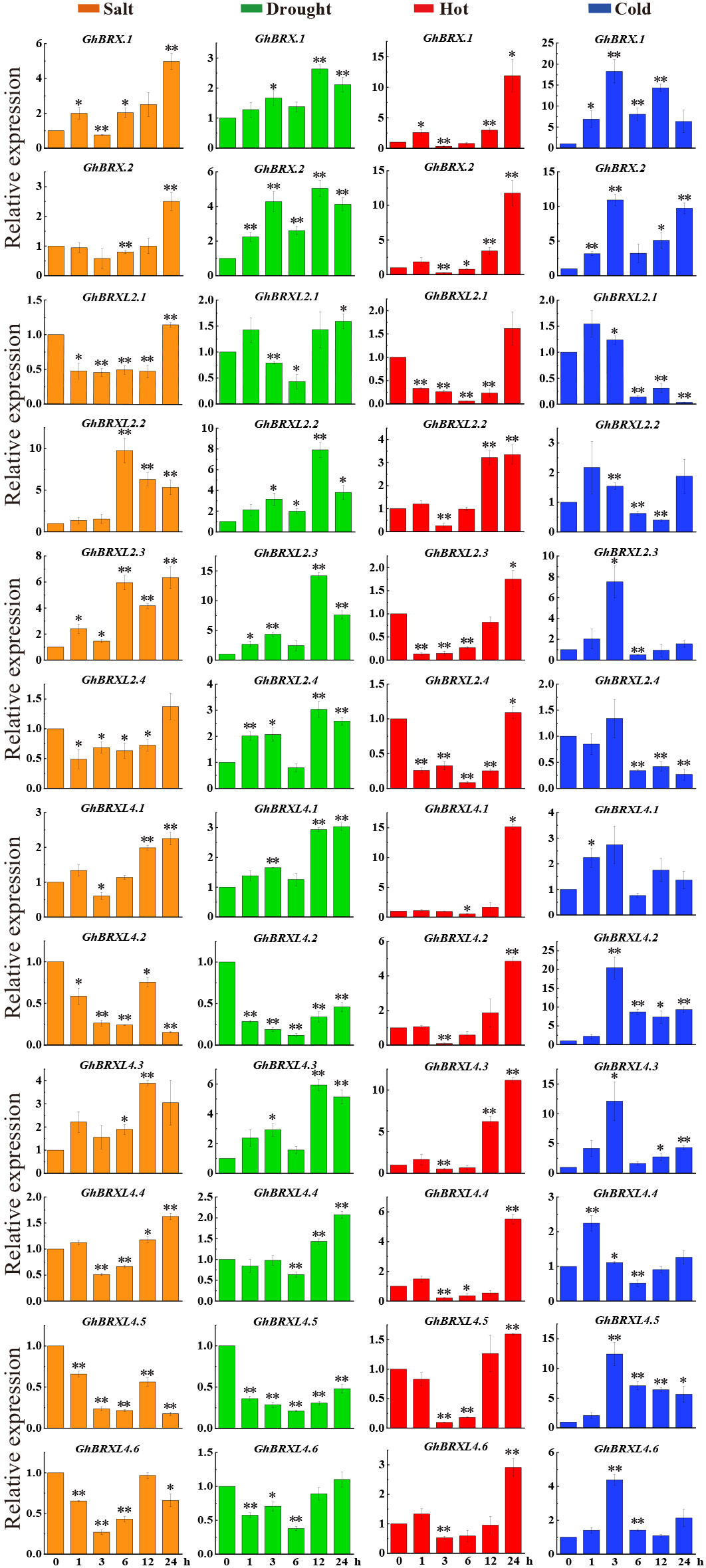
Figure 6 Relative GhBRXs expression levels in response to heat, cold, drought, and salt treatments. The standard deviations of the three biological replicates were represented by error bars. Orange denoted salt stress, green denoted drought stress, red denoted heat stress, and blue denoted cold stress. Asterisks were used to indicate a significant difference in expression compared to the control value (* P <0.05; * * P <0.01).
We selected the GhBRX.1, GhBRX.2, and GhBRXL4.3 genes for further investigation based on the transcriptome and qRT–PCR results. It is assumed that the GhBRX.1, GhBRX.2 and GhBRXL4.3 genes are potentially important in the regulation of the stress response. To test our hypothesis, we used the VIGS method to knock down the GhBRX.1, GhBRX.2 and GhBRXL4.3 genes by constructing the vectors TRV: GhBRX.1, TRV: GhBRX.2 and TRV: GhBRXL4.3, respectively. TRV: CLA served as a positive control (Figures 7A, B). Ten days after VIGS, when albino plants were observed in the positive control group, the expression levels in the leaves of the VIGS-silenced and control plants were determined via qRT–PCR. The qRT–PCR results showed that GhBRX.1, GhBRX.2 and GhBRXL4.3 were effectively repressed (Figure 7C). Silenced plants (TRV: GhBRX.1, TRV: GhBRX.2, TRV: GhBRXL4.3) and control plants (TRV: 00) were treated under different abiotic stress conditions, and the silenced plants showed more severe wilting after two weeks of salt and cold stress treatment (Figure 7D). Compared with those in control plants (TRV: 00), the expression levels of GhBRX.1, GhBRX.2 and GhBRXL4.3 were significantly lower after 0 days (CK) and 8 days (salt and low temperature) in the silenced plants (TRV: GhBRX.1, TRV: GhBRX.2, TRV: GhBRXL4.3) (Figure 7E). Taken together, these findings showed that cotton tolerance to cold and salt stress was decreased by silencing GhBRX.1, GhBRX.2, and GhBRXL4.3.
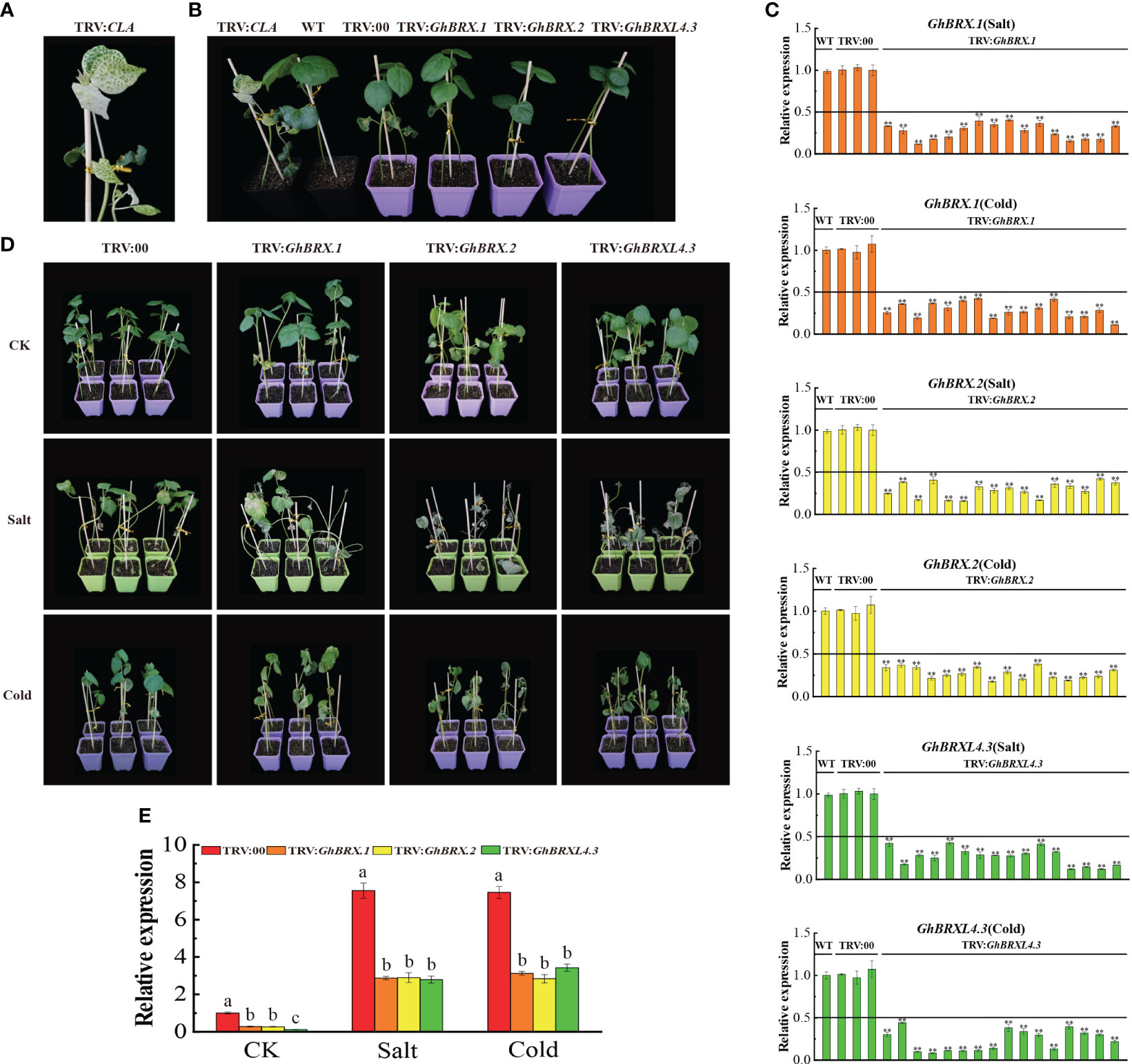
Figure 7 Silencing of GhBRX.1, GhBRX.2 and GhBRXL4.3 affects tolerance to salt and cold stress in upland cotton plants. (A) Positive control. (B) Representative VIGS images (TRV: CLA, WT, TRV: 00, TRV: GhBRX.1, TRV: GhBRX.2, and TRV: GhBRXL4.3). (C) The silencing efficiency of the WT, TRV: 00, TRV: GhBRX.1, TRV: GhBRX.2 and TRV: GhBRXL4.3 plants were tested for salt and cold stress, and the standard deviation determined from three separate experiments were represented by the error lines. (D) Phenotypes of the target gene-silenced plants in comparison to those of the control plants growing under normal growth conditions (CK) and under stress conditions (250 mmol/L NaCl, 12°C). (E) RT–qPCR analysis of changes in the expression levels of the GhBRX.1, GhBRX.2 and GhBRXL4.3 genes in target gene-silenced plants before and after stress treatment. “TRV: 00” represented plants carrying the empty vector control; “TRV: GhBRX.1, TRV: GhBRX.2, and TRV: GhBRXL4.3” represented plants with GhBRX.1, GhBRX.2 and GhBRXL4.3 silenced, respectively. The error line represented the standard deviation calculated from three independent experiments. Asterisks indicated a t test for statistically significant differences (* * P <0.01). Significant changes between control and gene-silenced plants were indicated by different letters (ANOVAs; P<0.05).
Plants with silenced target genes were less resistant to salt and cold stress. To investigate the impact of salt and cold stress on the silenced plants, we determined the activities of the reactive oxygen species (ROS) scavenger enzymes SOD, POD, and CAT and the MDA, soluble sugar and chlorophyll contents in the leaves before and 8 days after salt or cold stress. Under normal growth conditions, there was no significant difference in physiological parameters between the silenced plants (TRV: GhBRX.1, TRV: GhBRX.2, TRV: GhBRXL4.3) and the control plants (TRV:00). After 8 days of salt and cold stress, the SOD, POD and CAT activities of the silenced plants (TRV: GhBRX.1, TRV: GhBRX.2, and TRV: GhBRXL4.3) were significantly lower than those of the control plants (TRV: 00) (Figures 8A–C), indicating that the VIGS-silenced plants suffered extensive oxidative damage. The contents of soluble sugars and chlorophyll in the silenced plants were significantly lower than those in the control plants, while the content of MDA in the silenced plants was significantly greater than that in the control plants (Figures 8D–F), indicating that the resistance of the silenced plants decreased under adverse conditions and that the degree of adverse damage increased. The results showed that the silenced plants were very sensitive to salt stress and cold stress and that silencing the GhBRX.1, GhBRX.2 and GhBRXL4.3 genes significantly reduced their ability to tolerate salt stress and cold stress.
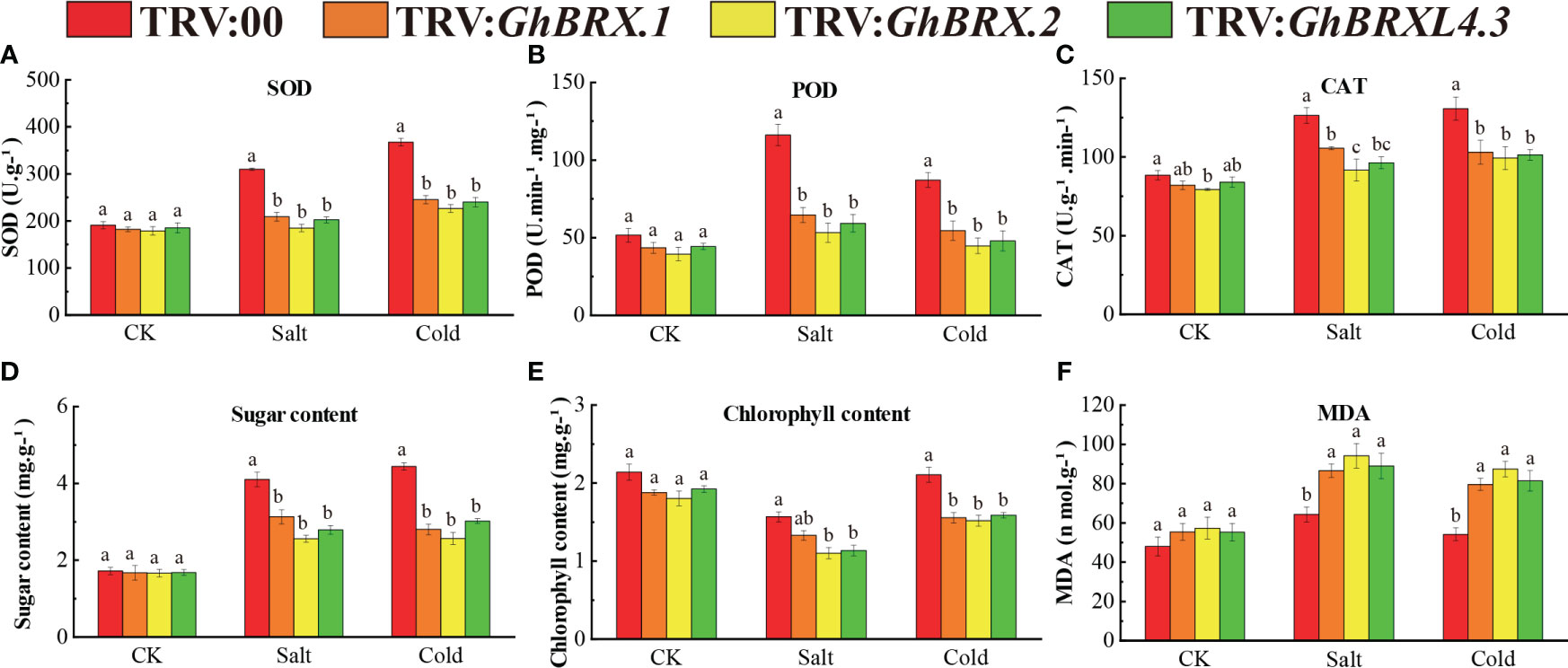
Figure 8 Determination of antioxidant enzyme activities and soluble sugar, chlorophyll and MDA concentrations in GhBRX.1-, GhBRX.2- and GhBRXL4.3-silenced plants and control plants under abiotic stress: (A) SOD activity; (B) POD activity; (C) CAT activity; (D) soluble sugar content; (E) chlorophyll activity; (F) MDA concentration. The standard deviation of three biological replicates were shown by error bars. Significant changes between control and gene-silenced plants were indicated by different letters (ANOVA; P < 0.05).
The nine genes associated with tolerance to salt stress or low-temperature stress were selected for analysis of the response characteristics of the control and targeted gene-silenced plants under salt and cold conditions. The nine genes included GhSOS1 (Na+/H+ antiporter salt overly sensitive 1), GhSOS2 (salt overly sensitive 2), GhNHX1 (Na+/H+ antiporter), GhCIPK6 (Ser/Thr protein kinase 6), GhBIN2 (glycogen synthase kinase 3 (GSK3)-like kinase), GhSnRK2.6 (SnRK2 protein kinase), GhHDT4D (a member of the HD2 subfamily of histone deacetylases), GhCBF1 (C-repeat binding factor) and GhPP2C (protein phosphatase 2C). The expression levels of these nine genes were high in the leaves of the control plants but were significantly lower in the targeted gene-silenced plants under salt or cold stress (Figure 9). The downregulated expression of these genes indicated that the plants were very sensitive to salt and cold stress and had a significantly reduced ability to tolerate various abiotic stress factors, resulting in greater oxidative damage.
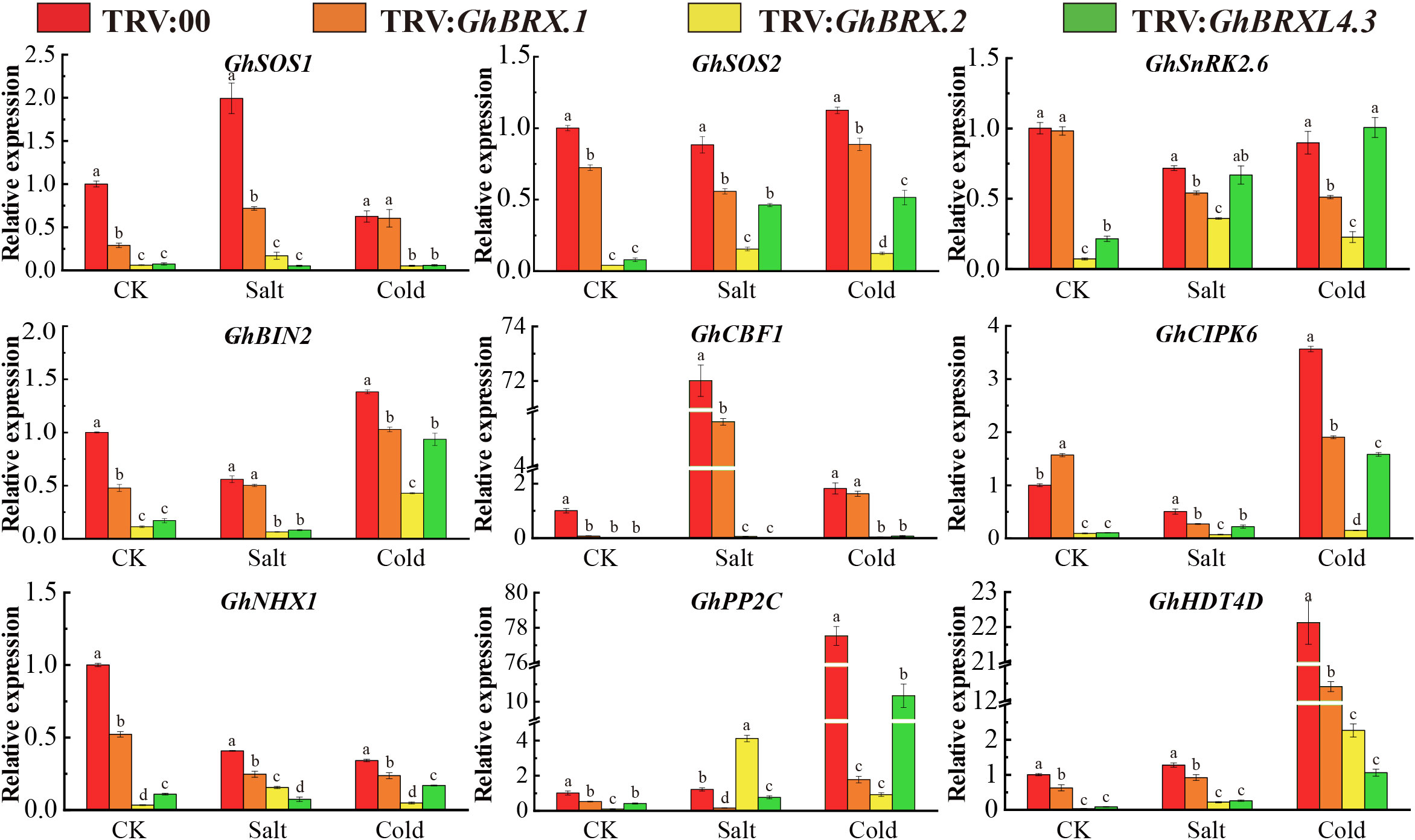
Figure 9 Expression of stress-responsive genes associated with salt stress tolerance or cold stress tolerance in control and silenced plants before and after stress. The error line represents the standard deviation calculated from three independent experiments. Significant changes between control and gene-silenced plants were indicated by different letters (ANOVA; P < 0.05).
Drought, high temperature, salinity, and cold are all environmental stressors that harm plant growth and output (Suzuki et al., 2014), change the internal balance within plants, and affect all biological and physiological activities within plants (Aasamaa and Sõber, 2011). Therefore, it is crucial to develop new varieties with enhanced performance and maintain and improve cotton production (Kuppu et al., 2013; Kirungu et al., 2019; Hajihashemi et al., 2020; Li et al., 2021). BRX is involved in the control of plant embryonic development, root and bud growth, tiller angle and stomatal development (Scacchi et al., 2009; Beuchat et al., 2010b; Liu et al., 2010; Li et al., 2019; Rowe et al., 2019; Zhang Y. et al., 2021; Tiwari et al., 2023). Further studies have shown that BRX regulates crosstalk between signaling pathways of various phytohormones, including BRs, auxin, ABA and cytokinin (Mouchel et al., 2006; Li et al., 2009; Rodrigues et al., 2009). The BRX gene family in rice may be implicated in BR and auxin signaling, and BRX genes respond differently to abiotic stress and may play a role in the abiotic stress response (Liu et al., 2010). TaBRXL1 is involved primarily in developmental processes, whereas TaBRXL2 is extensively regulated by hormones, development, and other abiotic stressors (Tiwari et al., 2023).
Although BRX genes have been found throughout the genomes of many plant species, only rice (Liu et al., 2010) and wheat (Tiwari et al., 2023) have been found to contain evidence of their possible roles in stress adaptation. The BRX gene has not been identified in cotton, and its function in cotton has rarely been studied. By using the AtBRX protein as a query, we identified 12, 6 and 6 BRX gene family members in G. hirsutum, G. raimondii and G. arboreum, respectively. There are 5 BRX genes in Arabidopsis and rice, 10 in B. rapa, and 13 in T. aestivum (Briggs et al., 2006; Liu et al., 2010; Zhang Y. et al., 2021; Tiwari et al., 2023). The monocots wheat and rice clustered together with stronger homology. Among the dicotyledonous plants, the three cotton species exhibited greater similarity and formed a cluster. The allotetraploid cotton species G. hirsutum is the product of the doubling of the ancestral cross between two diploid cotton species, G. raimondii and G. arboreum, according to a phylogenetic tree study. Research has demonstrated that sequence similarities across genes belonging to the same taxonomic category can result in similar activities (Nan et al., 2021). The gene structure of the GhBRX genes is largely conserved between orthologous genes. Homologs of GhBRX, GhBRX2 and GhBRXL4 are predicted to have exons similar to those reported for AtBRX, AtBRXL2 and AtBRXL4. The GhBRX proteins have similar MEME motifs. A few motifs have varying copy numbers in different proteins, and some motifs share two or more proteins, which could account for the functional discrepancies described among BRX family proteins.
The primary mechanism of gene family expansion is gene duplication. Segment and tandem repeats are thought to be the primary drivers of gene family expansion in plants (Cannon et al., 2004; Flagel and Wendel, 2009). Twenty segmentally duplicated genes and one tandemly duplicated gene were identified in the GhBRX gene family. Therefore, our study points to segmental duplication as the primary cause of the increase in GhBRXs. One technique for researching gene evolution and relationships is to analyze the collinearity of various species (Yu et al., 2020). The results of intergenomic symbiosis analysis between upland cotton and the other two cotton species were compared to acquire better knowledge of the homologous gene functions and evolutionary linkages of the genes. The results indicate that BRX gene duplication events and chromosomal rearrangements may be conserved in cotton given the numbers of G. raimondii and G. arboreum. Analysis revealed the same number of direct homologous gene pairs between G. hirsutum and G. raimondii and G. arboreum, indicating high conservation of BRX genes in cotton. To study the differences after gene duplication, the Ka and Ks values of the replicated GhBRX genes in upland cotton were calculated. The results showed that the Ka/Ks ratio of all the duplicate GhBRX gene pairs was <1, indicating that the GhBRX gene family experienced selection pressure during evolution.
Identification and characterization of cis-regulatory DNA sequences in response to coordinated developmental and environmental cues are critical for plant biology (Schmitz et al., 2022). We isolated the upstream promoter segments of the candidate genes and examined the distribution of cis-acting elements in the promoter region of GhBRX to gain additional insight into the potential role of GhBRX in upland cotton under various environmental conditions. In the present study, 38 cis-acting elements (those involved in stress responsiveness, tissue specificity, plant hormone responsiveness, and photoresponsiveness) were confirmed within the promoters of the GhBRXs. Photoresponsive elements are widely found in plants; for example, AT-rich G-boxes, GT1, Box 4, and I-boxes are commonly present in photoinducible promoters (Lam and Chua, 1989; Gilmartin et al., 1992; Foster et al., 1994). The upland cotton genes GhBRX.1, GhBRX.2, and GhBRXL4.3 were highly expressed in roots, and GhBRXL4.3 was also strongly expressed in pistils. These findings were based on the prediction of cis-elements in GhBRX genes and RNAseq expression data. The GhBRX gene promoter contains cis-acting regions linked to the abiotic stress response, including MBS (drought-induced MYB junction) and LTR (cis-acting element of cold responsiveness), suggesting that the regulation of drought and cold stress in cotton may be mediated by GhBRX genes. The presence of cis-elements in the promoter regions of genes provides clues to the spatiotemporal and hormonal regulation of genes and their response to different environmental stresses. We studied the expression of GhBRXs in various cotton leaves under normal and abiotic stress conditions to better understand the use of GhBRXs in cotton growth and abiotic stress resistance. The transcriptomic data of GhBRX genes in leaves under abiotic stress were obtained from the RNA-seq data of Zhejiang University, and most of the GhBRX genes responded to at least one stressor. In addition, to confirm the prior transcriptome findings, GhBRX transcript levels under abiotic stress were evaluated using quantitative RT–PCR. Quantitative RT–PCR analysis revealed that the expression of the GhBRX.1, GhBRX.2 and GhBRXL4.3 genes was significantly upregulated 24 h after the four stress treatments. GhBRXL2.2, GhBRXL2.3 and GhBRXL4.1 were significantly elevated under salt and drought stress, while GhBRXL4.2 was significantly elevated under both heat and cold stress. These findings suggested that these genes may play a significant biological role in enhancing upland cotton tolerance to abiotic stress.
Since genes were significantly upregulated under all four stress treatments, to further explore the role of the GhBRX.1, GhBRX.2 and GhBRXL4.3 genes in abiotic stress regulation in upland cotton, we constructed a VIGS vector for further functional analysis. We treated the silenced plants with abiotic stress (salt, drought, high temperature and low temperature) and found that the silenced plants were more sensitive to salt stress and low-temperature stress, and the silenced plants exhibited a more obvious phenotype and water loss phenotype. These findings may indicate that the GhBRX.1, GhBRX.2 and GhBRXL4.3 genes may play significant biological roles in enhancing tolerance to salt and cold stress in cotton plants. To investigate the stress-related mechanisms of these three genes, we analyzed the physiological and biochemical indicators, including ROS scavenger enzyme (SOD, POD, CAT) activities and MDA, soluble sugar and chlorophyll contents, of silenced plants and control plants before and after stress. The induction of salt stress and cold stress leads to the overproduction of ROS and other oxygen radicals, leading to oxidative destruction of plant cell structure and their components and eventually plant death; antioxidant defense systems work together to control uncontrolled oxidative cascades and protect plant cells from oxidative damage by removing ROS (Gill and Tuteja, 2010; Malhan et al., 2015). Therefore, the removal of excess ROS is a key process for plant protection against salt stress and cold stress (Zhang et al., 2011; Ullah et al., 2018). Essential ROS scavenging enzymes include SOD, POD, and CAT, whose activities increase in plants exposed to cold and salt stress (Salih et al. 2024). The results showed that SOD, POD, and CAT enzyme activities increased significantly after stress due to the overproduction of plant ROS in upland cotton, and the ability to eliminate ROS was significantly reduced after the silencing of GhBRX.1, GhBRX.2 and GhBRXL4.3. Therefore, after stress, the SOD, POD and CAT activities of the GhBRX.1-, GhBRX.2- and GhBRXL4.3-silenced plants significantly decreased compared with those of the control plants. The MDA concentration is a crucial indicator of the body’s ability to respond to antioxidants and can also be used to infer the extent of cell damage (Yu et al., 2016). The soluble sugar content can reflect not only the growth status of crops but also their quality (Jiang et al., 2020). After stress, the degree of oxidative damage to the cotton plants increased, the MDA content of the silenced plants increased, and the soluble sugar and chlorophyll contents decreased, indicating that the VIGS-mediated silencing of these plants increased cell damage and decreased quality. This further revealed the important role of the proteins encoded by the GhBRX.1, GhBRX.2 and GhBRXL4.3 genes in enhancing salt tolerance and low-temperature tolerance in cotton.
Finally, we examined the expression levels of nine known stress response genes, GhSOS1, GhSOS2, GhNHX1, GhCIPK6, GhBIN2, GhSnRK2.6, GhHDT4D, GhCBF1 and GhPP2C, in the leaf tissue of VIGS and control plants (TRV:00) under salt and cold stress conditions. Under salt and cold stress conditions, most stress-related genes exhibited considerable downregulation. The SOS1 and SOS2 genes can improve the salt tolerance of transgenic plants (Liu et al., 2000; Yue et al., 2012). Before and after salt stress, the expression of both GhSOS1 and GhSOS2, including GhBRX.2 and GhBRXL4.3, was downregulated, and the two gene-silenced plants were the most significantly downregulated. The SnRK2.6 and CBF1 genes play important roles in improving salt and cold tolerance in plants under stress (Novillo et al., 2007; Song et al., 2016; Wang et al., 2018; Yan et al., 2023). PCaP2 can activate the CBF and SnRK2 transcriptional networks and play an important role in cold stress tolerance (Wang et al., 2018). When subjected to salt and cold stress, although GhSnRK2.6 was not significantly expressed in the GhBRXL4.3 gene-silenced plants, both GhSnRK2.6 and GhCBF1 were downregulated in the VIGS-treated plants. BIN2 interacts with and phosphorylates the CBF EXPRESSION1 inducer (ICE1) to inhibit SOS2 kinase activity and further inhibit the salt stress response, thus negatively regulating salt stress and low-temperature stress (Ye et al., 2019). Similarly, compared with that in the absence of stress, GhBIN2 expression in silenced plants was broadly upregulated. The CIPK6, NHX1, PP2C and HDT4D genes play important roles in salt stress tolerance and cold stress and can be used to regulate growth and development and improve crop tolerance to salt and low-temperature stress (Teakle et al., 2010; Chen et al., 2013; Dubrovina et al., 2015; Imran et al., 2020; Zhang Y. et al., 2021; Zhu et al., 2022; Wu et al., 2023). Compared with those in control plants, except for in GhBRX.2 gene-silenced plants, the expression of the CIPK6, NHX1, PP2C and HDT4D genes was significantly lower in VIGS-treated plants. The expression of stress-related genes in VIGS–generated cotton leaves revealed that silencing GhBRX.1, GhBRX.2, and GhBRXL4.3 affected the expression of genes involved in the cotton stress response under salt and cold stress conditions, suggesting that BRX genes play an important role in upland cotton tolerance to salt and cold stress.
In summary, the cotton genome encodes 24 highly conserved BRX genes. The BRX genes in upland cotton have similar gene structures. Multiple cis-acting regions linked with hormonal or abiotic stress responses can be found in the GhBRX promoter sequence. qRT–PCR data also showed that different abiotic stresses could induce GhBRX expression. Further functional characterization of GhBRX.1, GhBRX.2 and GhBRXL4.3 by VIGS indicated that silencing of the GhBRX.1, GhBRX.2 and GhBRXL4.3 genes may weaken the response of cotton to salt and low-temperature stress. This work could lead to additional research on the function of GhBRXs in the cotton response and resistance to abiotic stress.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
WW: Writing – original draft, Writing – review & editing, Data curation, Formal Analysis, Investigation, Validation, Visualization, Conceptualization, Methodology, Project administration, Software, Supervision. JJ: Validation, Writing – review & editing, Software. XuZ: Writing – review & editing. PL: Writing – review & editing, Validation. JL: Writing – review & editing, Validation. YL: Writing – review & editing, Validation. WX: Writing – review & editing, Validation. JS: Writing – review & editing, Funding acquisition, Resources, Supervision. XiZ: Writing – review & editing, Funding acquisition, Resources, Supervision. CW: Writing – review & editing, Formal Analysis, Funding acquisition, Resources, Supervision.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This research was funded by the State Key Laboratory of Aridland Crop Science, Gansu Agricultural University (GSCS-2019-10); the Natural Science Foundation of Xinjiang Uygur Autonomous Region (Project Nos. 2022D01E103 and 2023D01A015); and the Project for Postdoctoral and High-level Flexible Talents of Xinjiang Uygur Autonomous Region (Grant. No: RSSQ00066509); Changji Prefecture “Two Districts” Science and Technology Development Plan Project: (No: 2023LQG04); Major Science and Technology Program of Changji Hui Autonomous Prefecture (Grant. No: 2021Z01-01); Central Leading Local Science and Technology Development Fund Project of Xinjiang Uygur Autonomous Region (No: ZYYD2023C06).
The author is especially special thanks to Qifeng Ma, Institute of Cotton Research of CAAS, for the TRV vectors and Yonglin Yang, Shihezi Academy of Agricultural Sciences, for the cotton seeds of XinshiK25.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as potential conflicts of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2024.1353365/full#supplementary-material
Aasamaa, K., Sõber, A. (2011). Stomatal sensitivities to changes in leaf water potential, air humidity, CO2 concentration and light intensity, and the effect of abscisic acid on the sensitivities in six temperate deciduous tree species. Environ. Exp. Bot. 71 (1), 72–78. doi: 10.1016/j.envexpbot.2010.10.013
Bailey, T. L., Boden, M., Buske, F. A., Frith, M., Grant, C. E., Clementi, L., et al. (2009). MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 37 (suppl_2), W202–W208. doi: 10.1093/nar/gkp335
Bari, R., Jones, J. D. (2009). Role of plant hormones in plant defence responses. Plant Mol. Biol. 69, 473–488. doi: 10.1007/s11103-008-9435-0
Beers, R. F., Sizer, I. W. (1952). A spectrophotometric method for measuring the breakdown of hydrogen peroxide by catalase. J. Biol. Chem. 195 (1), 133–140. doi: 10.1016/S0021-9258(19)50881-X
Beuchat, J., Li, S., Ragni, L., Shindo, C., Kohn, M. H., Hardtke, C. S. (2010a). A hyperactive quantitative trait locus allele of Arabidopsis BRX contributes to natural variation in root growth vigor. Proc. Natl. Acad. Sci. U.S.A. 107 (18), 8475–8480. doi: 10.1073/pnas.0913207107
Beuchat, J., Scacchi, E., Tarkowska, D., Ragni, L., Strnad, M., Hardtke, C. S. (2010b). BRX promotes Arabidopsis shoot growth. New Phytol. 188 (1), 23–29. doi: 10.1111/j.1469-8137.2010.03387.x
Briggs, G. C., Mouchel, C. F., Hardtke, C. S. (2006). Characterization of the plant-specific BREVIS RADIX gene family reveals limited genetic redundancy despite high sequence conservation. Plant Physiol. 140 (4), 1306–1316. doi: 10.1104/pp.105.075382
Cannon, S. B., Mitra, A., Baumgarten, A., Young, N. D., May, G. (2004). The roles of segmental and tandem gene duplication in the evolution of large gene families in. Arabidopsis thaliana. BMC Plant Biol. 4 (1), 1–21. doi: 10.1186/1471-2229-4-10
Castro, D., Contreras, L. M., Kurz, L., Wilkesman, J. (2017). “Detection of guaiacol peroxidase on electrophoretic gels,” in Zymography: methods mol. Biol., vol. 1626. (New York, NY: Humana Press). doi: 10.1007/978-1-4939-7111-4_18
Chen, C., Chen, H., He, Y., Xia, R. (2018). TBtools, a toolkit for biologists integrating various biological data handling tools with a user-friendly interface. BioRxiv 289660 (10.1101), 289660. doi: 10.1101/289660
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: an integrative toolkit developed for interactive analyses of big biological data. Plant Mol. 13 (8), 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chen, L., Wang, Q. Q., Zhou, L., Ren, F., Li, D. D., Li, X. B. (2013). Arabidopsis CBL-interacting protein kinase (CIPK6) is involved in plant response to salt/osmotic stress and ABA. Mol. Biol. Rep. 40, 4759–4767. doi: 10.1007/s11033-013-2572-9
Dubrovina, A. S., Kiselev, K. V., Khristenko, V. S., Aleynova, O. A. (2015). VaCPK20, a calcium-dependent protein kinase gene of wild grapevine Vitis amurensis Rupr., mediates cold and drought stress tolerance. J. Plant Physiol. 185, 1–12. doi: 10.1016/j.jplph.2015.05.020
El-Gebali, S., Mistry, J., Bateman, A., Eddy, S. R., Luciani, A., Potter, S. C., et al. (2019). The Pfam protein families database in 2019. Nucleic Acids Res. 47 (D1), D427–D432. doi: 10.1093/nar/gky995
Flagel, L. E., Wendel, J. F. (2009). Gene duplication and evolutionary novelty in plants. New Phytol. 183 (3), 557–564. doi: 10.1111/j.1469-8137.2009.02923.x
Foster, R., Izawa, T., Chua, N. H. (1994). Plant bZIP proteins gather at ACGT elements. FASEB J. 8 (2), 192–200. doi: 10.1096/fasebj.8.2.8119490
Gao, W., Long, L., Zhu, L. F., Xu, L., Gao, W. H., Sun, L. Q., et al. (2013). Proteomic and virus-induced gene silencing (VIGS) analyses reveal that gossypol, brassinosteroids, and jasmonic acid contribute to the resistance of cotton to. Verticillium dahliae. Mol. Cell Proteomics 12 (12), 3690–3703. doi: 10.1074/mcp.M113.031013
Giannopolitis, C. N., Ries, S. K. (1977). Superoxide dismutases: II. Purification and quantitative relationship with water-soluble protein in seedlings. Plant Physiol. 59 (2), 315–318. doi: 10.1104/pp.59.2.315
Gill, S. S., Tuteja, N. (2010). Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol. Biochem. 48 (12), 909–930. doi: 10.1073/pnas.95.12.6705
Gilmartin, P. M., Memelink, J., Hiratsuka, K., Kay, S. A., Chua, N. H. (1992). Characterization of a gene encoding a DNA binding protein with specificity for a light-responsive element. Plant Cell 4 (7), 839–849. doi: 10.1105/tpc.4.7.839
Hajihashemi, S., Brestic, M., Landi, M., Skalicky, M. (2020). Resistance of Fritillaria imperialis to freezing stress through gene expression, osmotic adjustment and antioxidants. Sci. Rep. 10 (1), 10427. doi: 10.1038/s41598-020-63006-7
Hasanuzzaman, M., Bhuyan, M. B., Zulfiqar, F., Raza, A., Mohsin, S. M., Mahmud, J. A., et al. (2020). Reactive oxygen species and antioxidant defense in plants under abiotic stress: Revisiting the crucial role of a universal defense regulator. Antioxidants 9 (8), 681. doi: 10.3390/antiox9080681
Imran, M., Shafiq, S., Naeem, M. K., Widemann, E., Munir, M. Z., Jensen, K. B., et al. (2020). Histone deacetylase (HDAC) gene family in allotetraploid cotton and its diploid progenitors: In silico identification, molecular characterization, and gene expression analysis under multiple abiotic stresses, DNA damage and phytohormone treatments. Int. J. Mol. Sci. 21 (1), 321. doi: 10.3390/ijms21010321
Jiang, W., Li, N., Zhang, D., Meinhardt, L., Cao, B., Li, Y., et al. (2020). Elevated temperature and drought stress significantly affect fruit quality and activity of anthocyanin-related enzymes in jujube (Ziziphus jujuba Mill. cv. ‘Lingwuchangzao’). PloS One 15 (11), e0241491. doi: 10.1371/journal.pone.0241491
John, M. E., Crow, L. J. (1992). Gene expression in cotton (Gossypium hirsutum L.) fiber: cloning of the mRNAs. Proc. Natl. Acad. Sci. U.S.A. 89 (13), 5769–5773. doi: 10.1073/pnas.89.13.5769
Kirungu, J. N., Magwanga, R. O., Lu, P., Cai, X., Zhou, Z., Wang, X., et al. (2019). Functional characterization of Gh_A08G1120 (GH3.5) gene reveal their significant role in enhancing drought and salt stress tolerance in cotton. BMC Genom. 20, 1–17. doi: 10.1186/s12863-019-0756-6
Koh, S. W., Marhava, P., Rana, S., Graf, A., Moret, B., Bassukas, A. E., et al. (2021). Mapping and engineering of auxin-induced plasma membrane dissociation in BRX family proteins. Plant Cell 33 (6), 1945–1960. doi: 10.1093/plcell/koab076
Kumar, S., Stecher, G., Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33 (7), 1870–1874. doi: 10.1093/molbev/msw054
Kuppu, S., Mishra, N., Hu, R., Sun, L., Zhu, X., Shen, G., et al. (2013). Water-deficit inducible expression of a cytokinin biosynthetic gene IPT improves drought tolerance in cotton. PloS One 8 (5), e64190. doi: 10.1371/journal.pone.0064190
Lam, E., Chua, N. H. (1989). ASF-2: a factor that binds to the cauliflower mosaic virus 35S promoter and a conserved GATA motif in Cab promoters. Plant Cell 1 (12), 1147–1156. doi: 10.1105/tpc.1.12.1147
Li, J., Mo, X., Wang, J., Chen, N., Fan, H., Dai, C., et al. (2009). BREVIS RADIX is involved in cytokinin-mediated inhibition of lateral root initiation in Arabidopsis. Planta 229, 593–603. doi: 10.1007/s00425-008-0854-6
Li, X. T., Feng, X. Y., Zeng, Z., Liu, Y., Shao, Z. Q. (2021). Comparative analysis of HSF genes from Secale cereale and its Triticeae relatives reveal ancient and recent gene expansions. Front. Genet. 12. doi: 10.3389/fgene.2021.801218
Li, Z., Liang, Y., Yuan, Y., Wang, L., Meng, X., Xiong, G., et al. (2019). OsBRXL4 regulates shoot gravitropism and rice tiller angle through affecting LAZY1 nuclear localization. Plant Mol. 12 (8), 1143–1156. doi: 10.1016/j.molp.2019.05.014
Lichtenthaler, H. K., Wellburn, A. R. (1983). Determinations of total carotenoids and chlorophylls a and b of leaf extracts in different solvents. Biochem. Soc Trans. 11, 591–592. doi: 10.1042/BST0110591
Liu, H., Jiang, W., Cao, J., Ma, L. (2018). A combination of 1-methylcyclopropene treatment and intermittent warming alleviates chilling injury and affects phenolics and antioxidant activity of peach fruit during storage. Sci. Hortic. 229, 175–181. doi: 10.1016/j.scienta.2017.11.010
Liu, J., Ishitani, M., Halfter, U., Kim, C. S., Zhu, J. K. (2000). The Arabidopsis thaliana SOS2 gene encodes a protein kinase that is required for salt tolerance. Proc. Natl. Acad. Sci. U.S.A. 97 (7), 3730–3734. doi: 10.1073/pnas.97.7.3730
Liu, J., Liang, D., Song, Y., Xiong, L. (2010). Systematic identification and expression analysis of BREVIS RADIX-like homologous genes in rice. Plant Sci. 178 (2), 183–191. doi: 10.1016/j.plantsci.2009.11.009
Livak, K. J., Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2– ΔΔCT method. Methods 25 (4), 402–408. doi: 10.1006/meth.2001.1262
Malhan, D., Bhatia, S., Yadav, R. K. (2015). Genome wide gene expression analyses of Arabidopsis shoot stem cell niche cell populations. Plant Signal. Behav. 10 (4), e1011937. doi: 10.1080/15592324.2015.1011937
Mouchel, C. F., Briggs, G. C., Hardtke, C. S. (2004). Natural genetic variation in Arabidopsis identifies BREVIS RADIX, a novel regulator of cell proliferation and elongation in the root. Genes Dev. 18 (6), 700–714. doi: 10.1101/gad.1187704
Mouchel, C. F., Osmont, K. S., Hardtke, C. S. (2006). BRX mediates feedback between brassinosteroid levels and auxin signalling in root growth. Nature 443 (7110), 458–461. doi: 10.1038/nature05130
Nan, H., Lin, Y., Wang, X., Gao, L. (2021). Comprehensive genomic analysis and expression profiling of cysteine-rich polycomb-like transcription factor gene family in tea tree. Hortic. Plant J. 7 (5), 469–478. doi: 10.1016/j.hpj.2021.03.001
Nian, L., Zhang, X., Yi, X., Liu, X., Nu, A., Yang, Y., et al. (2021). Genome-wide identification of ABA receptor PYL/RCAR gene family and their response to cold stress in Medicago sativa L. Physiol. Mol. Biol. Plants 27, 1979–1995. doi: 10.1007/s12298-021-01066-3
Novillo, F., Medina, J., Salinas, J. (2007). Arabidopsis CBF1 and CBF3 have a different function than CBF2 in cold acclimation and define different gene classes in the CBF regulon. Proc. Natl. Acad. Sci. U.S.A. 104 (52), 21002–21007. doi: 10.1073/pnas.0705639105
Potters, G., Pasternak, T. P., Guisez, Y., Palme, K. J., Jansen, M. A. (2007). Stress-induced morphogenic responses: growing out of trouble? Trends Plant Sci. 12 (3), 98–105. doi: 10.1016/j.tplants.2007.01.004
Rodrigues, A., Santiago, J., Rubio, S., Saez, A., Osmont, K. S., Gadea, J., et al. (2009). The short-rooted phenotype of the brevis radix mutant partly reflects root abscisic acid hypersensitivity. Plant Physiol. 149 (4), 1917–1928. doi: 10.1104/pp.108.133819
Rowe, M. H., Dong, J., Weimer, A. K., Bergmann, D. C. (2019). A plant-specific polarity module establishes cell fate asymmetry in the Arabidopsis stomatal lineage. BioRxiv, 614636. doi: 10.1101/614636
Salih, H., Bai, W., Liang, W., Yang, R., Zhao, M., Muhammd, S. M. (2024). ROS scavenging enzyme-encoding genes play important roles in the desert moss Syntrichia caninervis response to extreme cold and desiccation stresses. Int. J. Biol. Macromol. 254, 127778. doi: 10.1016/j.ijbiomac.2023.127778
Scacchi, E., Osmont, K. S., Beuchat, J., Salinas, P., Navarrete-Gómez, M., Trigueros, M., et al. (2009). Dynamic, auxin-responsive plasma membrane-to-nucleus movement of Arabidopsis BRX. Development 136 (12), 2059–2067. doi: 10.1242/dev.035444
Schmitz, R. J., Grotewold, E., Stam, M. (2022). Cis-regulatory sequences in plants: Their importance, discovery, and future challenges. Plant Cell 34 (2), 718–741. doi: 10.1093/plcell/koab281
Song, X., Yu, X., Hori, C., Demura, T., Ohtani, M., Zhuge, Q. (2016). Heterologous overexpression of poplar SnRK2 genes enhanced salt stress tolerance in Arabidopsis thaliana. Front. Plant Sci. 7. doi: 10.3389/fpls.2016.00612
Suzuki, N., Rivero, R. M., Shulaev, V., Blumwald, E., Mittler, R. (2014). Abiotic and biotic stress combinations. New Phytol. 203 (1), 32–43. doi: 10.1111/nph.12797
Teakle, N. L., Amtmann, A., Real, D., Colmer, T. D. (2010). Lotus tenuis tolerates combined salinity and waterlogging: maintaining O2 transport to roots and expression of an NHX1-like gene contribute to regulation of Na+ transport. Physiol. Plant 139 (4), 358–374. doi: 10.1111/j.1399-3054.2010.01373.x
Tiwari, S., Muthusamy, S. K., Roy, P., Dalal, M. (2023). Genome wide analysis of BREVIS RADIX gene family from wheat (Triticum aestivum): A conserved gene family differentially regulated by hormones and abiotic stresses. Int. J. Biol. Macromol. 232, 123081. doi: 10.1016/j.ijbiomac.2022.12.300
Ullah, A., Sun, H., Yang, X., Zhang, X. (2018). A novel cotton WRKY gene, GhWRKY6-like, improves salt tolerance by activating the ABA signaling pathway and scavenging of reactive oxygen species. Physiol. Plant 162 (4), 439–454. doi: 10.1111/ppl.12651
Van Zelm, E., Zhang, Y., Testerink, C. (2020). Salt tolerance mechanisms of plants. Annu. Rev. Plant Biol. 71, 403–433. doi: 10.1146/annurev-arplant-050718-100005
Wang, X., Wang, L., Wang, Y., Liu, H., Hu, D., Zhang, N., et al. (2018). Arabidopsis PCaP2 plays an important role in chilling tolerance and ABA response by activating CBF-and SnRK2-mediated transcriptional regulatory network. Front. Plant Sci. 9. doi: 10.3389/fpls.2018.00215
Westerheide, S. D., Raynes, R., Powell, C., Xue, B., N Uversky, V. (2012). HSF transcription factor family, heat shock response, and protein intrinsic disorder. Curr. Protein Pept. Sci. 13 (1), 86–103. doi: 10.2174/138920312799277956
Wu, Z., Luo, L., Wan, Y., Liu, F. (2023). Genome-wide characterization of the PP2C gene family in peanut (Arachis hypogaea L.) and the identification of candidate genes involved in salinity-stress response. Front. Plant Sci. 14. doi: 10.3389/fpls.2023.1093913
Yan, J., Liu, Y., Yan, J., Liu, Z., Lou, H., Wu, J. (2023). The salt-activated CBF1/CBF2/CBF3-GALS1 module fine-tunes galactan-induced salt hypersensitivity in Arabidopsis. J. Integr. Plant Biol. 65 (8), 1904–1917. doi: 10.1111/jipb.13501
Ye, K., Li, H., Ding, Y., Shi, Y., Song, C., Gong, Z., et al. (2019). BRASSINOSTEROID-INSENSITIVE2 negatively regulates the stability of transcription factor ICE1 in response to cold stress in Arabidopsis. Plant Cell 31 (11), 2682–2696. doi: 10.1105/tpc.19.00058
Yu, J., Xie, Q., Li, C., Dong, Y., Zhu, S., Chen, J. (2020). Comprehensive characterization and gene expression patterns of LBD gene family in Gossypium. Planta 251 (4), 1–16. doi: 10.1007/s00425-020-03364-8
Yu, X., Liu, Y., Wang, S., Tao, Y., Wang, Z., Shu, Y., et al. (2016). CarNAC4, a NAC-type chickpea transcription factor conferring enhanced drought and salt stress tolerances in Arabidopsis. Plant Cell Rep. 35, 613–627. doi: 10.1007/s00299-015-1907-5
Yue, Y., Zhang, M., Zhang, J., Duan, L., Li, Z. (2012). SOS1 gene overexpression increased salt tolerance in transgenic tobacco by maintaining a higher K+/Na+ ratio. J. Plant Physiol. 169 (3), 255–261. doi: 10.1016/j.jplph.2011.10.007
Zhang, B., Chen, N., Peng, X., Shen, S. (2021). Identification of the PP2C gene family in paper mulberry (Broussonetia papyrifera) and its roles in the regulation mechanism of the response to cold stress. Biotechnol. Lett. 43, 1089–1102. doi: 10.1007/s10529-021-03110-4
Zhang, T., Hu, Y., Jiang, W., Fang, L., Guan, X., Chen, J., et al. (2015). Sequencing of allotetraploid cotton (Gossypium hirsutum L. acc. TM-1) provides a resource for fiber improvement. Nat. Biotechnol. 33 (5), 531–537. doi: 10.1038/nbt.3207
Zhang, Y., Liang, J., Cai, X., Chen, H., Wu, J., Lin, R., et al. (2021). Divergence of three BRX homoeologs in Brassica rapa and its effect on leaf morphology. Hortic. Res. 8, 68. doi: 10.1038/s41438-021-00504-3
Zhang, Y. J., Yang, J. S., Guo, S. J., Meng, J. J., Zhang, Y. L., Wan, S. B., et al. (2011). Over-expression of the Arabidopsis CBF1 gene improves resistance of tomato leaves to low temperature under low irradiance. Plant Biol. (Stuttg) 13 (2), 362–367. doi: 10.1111/j.1438-8677.2010.00365.x
Zhu, L., Fang, H., Lian, Z., Zhang, J., Li, X., Shi, J., et al. (2022). Genome-wide investigation and expression analysis of the Nitraria sibirica Pall. CIPK gene family. Int. J. Mol. Sci. 23 (19), 11599. doi: 10.3390/ijms231911599
Keywords: BREVIS RADIX, salt stress, cold stress, virus-induced gene silencing (VIGS), upland cotton
Citation: Wei W, Ju J, Zhang X, Ling P, Luo J, Li Y, Xu W, Su J, Zhang X and Wang C (2024) GhBRX.1, GhBRX.2, and GhBRX4.3 improve resistance to salt and cold stress in upland cotton. Front. Plant Sci. 15:1353365. doi: 10.3389/fpls.2024.1353365
Received: 10 December 2023; Accepted: 23 January 2024;
Published: 09 February 2024.
Edited by:
Yi Wang, Chinese Academy of Sciences (CAS), ChinaReviewed by:
Qian Tong, Changzhi Medical College, ChinaCopyright © 2024 Wei, Ju, Zhang, Ling, Luo, Li, Xu, Su, Zhang and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Junji Su, c3VqakBnc2F1LmVkdS5jbg==; Xianliang Zhang, emhhbmd4aWFubGlhbmdAY2Fhcy5jbg==; Caixiang Wang, d2FuZ2NhaXhAZ3NhdS5lZHUuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.