- 1Laboratory of Plant Protection and Biotechnology, Intercollegiate Faculty of Biotechnology, University of Gdansk and Medical University of Gdansk, Gdansk, Poland
- 2Department of Pharmaceutical Microbiology, Faculty of Pharmacy, Medical University of Gdansk, Gdansk, Poland
- 3DNA Sequencing & Synthesis Facility, Institute of Biochemistry & Biophysics, Polish Academy of Sciences, Warsaw, Poland
- 4Department of Pharmaceutical Biochemistry, Faculty of Pharmacy, Medical University of Gdansk, Gdansk, Poland
Introduction: Bacteria of genus Pectobacterium, encompassing economically significant pathogens affecting various plants, includes the species P. betavasculorum, initially associated with beetroot infection. However, its host range is much broader. It causes diseases of sunflower, potato, tomato, carrots, sweet potato, radish, squash, cucumber, and chrysanthemum. To explain this phenomenon, a comprehensive pathogenomic and phenomic characterisation of P. betavasculorum species was performed.
Methods: Genomes of P. betavasculorum strains isolated from potato, sunflower, and artichoke were sequenced and compared with those from sugar beet isolates. Metabolic profiling and pathogenomic analyses were conducted to assess virulence determinants and adaptation potential. Pathogenicity assays were performed on potato tubers and chicory leaves to confirm in silico predictions of disease symptoms. Phenotypic assays were also conducted to assess the strains ability to synthesise homoserine lactones and siderophores.
Results: The genome size ranged from 4.675 to 4.931 kbp, and GC % was between 51.0% and 51.2%. The pangenome of P. betavasculorum is open and comprises, on average, 4,220 gene families. Of these, 83% of genes are the core genome, and 2% of the entire pangenome are unique genes. Strains isolated from sugar beet have a smaller pangenome size and a higher number of unique genes than those from other plants. Interestingly, genomes of strains from artichoke and sunflower share 391 common CDS that are not present in the genomes of other strains from sugar beet or potato. Those strains have only one unique gene. All strains could use numerous sugars as building materials and energy sources and possessed a high repertoire of virulence determinants in the genomes. P. betavasculorum strains were able to cause disease symptoms on potato tubers and chicory leaves. They were also able to synthesise homoserine lactones and siderophores.
Discussion: The findings underscore the adaptability of P. betavasculorum to diverse hosts and environments. Strains adapted to plants with high sugar content in tissues have a different composition of fatty acids in membranes and a different mechanism of replenishing nitrogen in case of deficiency of this compound than strains derived from other plant species. Extensive phenomics and genomic analyses performed in this study have shown that P. betavasculorum species is an agronomically relevant pathogen.
1 Introduction
Pectobacterium, necrotrophic phytopathogenic rods, are a common causative agent of soft rot and blackleg diseases in agricultural and ornamental plants. They possess a wide range of plant cell-wall-degrading enzymes, which are their main virulence factor (Toth et al., 2021). Economic losses caused by these bacteria classified them in the 10th position out of the top 10 most important bacterial phytopathogens in the world (Mansfield et al., 2012).
Among Pectobacterium species, P. betavasculorum is considered a causative agent of vascular necrosis in sugar beets (Thomson, 1981; Rastgou et al., 2022). Bacteria from this species were first isolated from sugar beets with soft rot symptoms in the USA in 1972 (Thomson et al., 1977). In 1981, they were delineated as a new subspecies of Erwinia carotovora subsp. betavasculorum (Thomson, 1981). Previously, this species was thought to have a narrow host range restricted to sugar beet. However, further studies show that, in addition to beets, P. betavasculorum species can infect artichokes, carrots, cucumber, potatoes, sweet potatoes, radishes, sunflowers, squash, tomato, zucchini, and chrysanthemum (Whitney and Duffus, 1986; Harveson et al., 2009; Khan and Siddiqui 2020; Saleh et al., 1996).
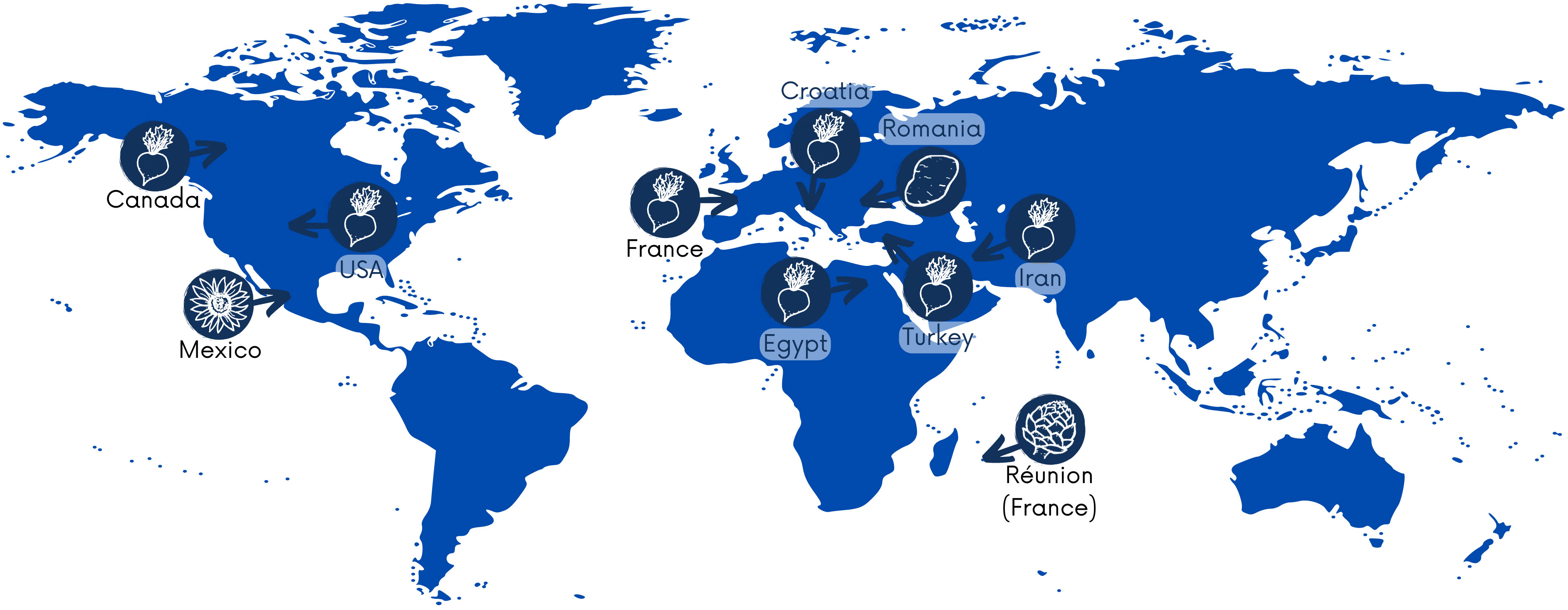
P. betavasculorum causes severe losses in sugar crops globally, and these bacteria have been reported on four continents, including Asia, Africa, Europe, and North America. According to the Centre for Agriculture and Bioscience International Organization (CABI) https://www.cabi.org/, as of 31 March 2021, the occurrence of P. betavasculorum had been reported in nine countries, such as the USA (Thomson, 1981), Egypt (Saleh et al., 1996), the French island La Reunion and Mexico (Gardan et al., 2003), Croatia (Ðermić, 2010), Iran (Rezaei and Taghavi, 2010; Nedaienia and Fassihiani, 2011; Baghaee-Ravari et al., 2011; Rezaee Danesh et al. 2021), Canada (Gilbert et al., 2015), Turkey (Ozturk et al., 2019), and France and Romania (Portier et al., 2020) (Figure 1). They are also present in Russia. In 2022, gene sequences from five P. betavasculorum strains isolated in Russia have been deposited in the GenBank database under accession numbers (OP593502–OP593506).
The industrial production of crops like potatoes, sugar beets, and sunflowers is essential for agriculture. According to the Food and Agriculture Organization (FAO) data, in 2021, a total of 376 million tons of potatoes were produced worldwide, and the total world production of sugar beets was 252.9 million tons. (FAOSTAT September 2023). These crops are vital for human consumption and play a crucial role in the food market. They are also extensively used in the biotechnological industry. For example, sugar beets, sucrose, potatoes, starch, and the waste from their production are used to produce biodegradable polymers (including the most popular polylactide, PLA), bioethanol, hydrogen, or biogas (Tomaszewska et al., 2018). Sunflowers have been recognized as functional foods or nutraceuticals, feedstock to produce biodiesel and bioplastics (Geneau-Sbartaï, 2008), and edible membranes, which serve as a packaging material in the food industry (Petraru and Amariei, 2022).
This intricate web of dependencies may be easily disrupted in case of a sudden decrease in crop yield caused by the instability of the Earth’s climate due to global warming. Plant development and crop yield depend on weather conditions during the entire vegetation period. From 1990 to 2020, there has been an increase of more than 0.5°C in average global temperature and more than 1.5°C in Europe, Asia, and Africa (FAOSTAT September 2023). This continuous increase in temperature results in water limitation, increasing soil salinity, and reduced nutrient availability, which negatively affect plant growth and reduce their resistance to pathogens such as viruses, nematodes, fungi, and bacteria. Microorganisms with increased resistance to draught conditions especially pose a significant threat to weakened plants.
Despite widespread occurrence, little is still known about P. betavasculorum species. One reason may be that, so far, no rapid detection and identification methods have been developed. Unfortunately, the PCR method commonly used to detect bacteria of the genus Pectobacterium using primers developed by Darrasse et al. (1994) is not useful in the case of P. betavasculorum. The primers were designed based on the pelY gene sequence, which is not present in the genomes of P. betavasculorum. The identification of this species is based on PCR-RFLP (Toth et al., 2001; Waleron et al., 2002), AFLP (Avrova et al., 2002), sequencing analyses of the 16S rRNA gene (Kwon et al., 1997; Hauben et al., 1998), and housekeeping genes (Gardan et al., 2003; Ma et al., 2007; Waleron et al., 2018; Portier et al., 2020). The difficulty of identification undoubtedly results in major underestimation of this pathogen’s significance and global prevalence.
By November 2023, in the GenBank database, only two genomic sequences are available for P. betavasculorum strains, NCPPB2793 (JQHM00000000) and NCPPB2795T (JQHL00000000), both isolated from sugar beet in the USA. It further emphasizes the need for thorough research in order to improve our understanding of this little-studied plant pathogen. Therefore, in the present study, we undertook genomic sequencing of four strains, CFBP1520 isolated from a sunflower in Mexico, SF142.2 originating from artichoke from the French island of La Reunion, CFBP3291 isolated from a potato in Romania, and strain Ecb168, derived from sugar beet in the USA.
The aim of this study was the comprehensive genetic and phenotypic characterization of the P. betavasculorum strains, which causes diseases of various plants that are an important part of the food market and are also used as raw materials in biotechnology.
The polyphasic analysis was performed to gain the broadest possible understanding of the biology of this underestimated Pectobacterium species. Extensive phenomic and genomic analyses were applied to verify their adaptive capabilities.
A special emphasis was placed on testing their potential to infect plants with high sugar or lipid content. It is worth highlighting that P. betavasculorum has a broad host range and may infect other plants than those from which it was originally isolated. It might be important for agriculture, especially in the era of progressive climate change, as such bacteria may be naturally more resistant to drought conditions and readily infect weakened plants. It may, in turn, lead to substantial losses in the crop industry. This is a significant concern, as no effective preventive and eradication methods exist for this pathogen. Thus, these bacteria may easily spread to new niches due to the intensive international trade of seeds, horticultural and ornamental plants, and plant-originated foods.
2 Methods
2.1 Bacterial strains
The 14 P. betavasculorum strains used in this study are listed in Table 1, and the reference Pectobacterium strains used for comparison are specified in Supplementary Table S1. The strains of various Pectobacterium species and Chromobacterium violaceum CV026 are a part of the Pectobacterium culture collection at the Intercollegiate Faculty of Biotechnology, Gdansk, Poland. Pectobacterium strains were maintained on the Crystal Violet Pectate (CVP) medium (Hélias et al. 2012), and Chromobacterium violaceum CV026 was cultivated on the Mueller–Hinton (MH) medium at 28°C. All strains were stored in 40% glycerol at −80°C.
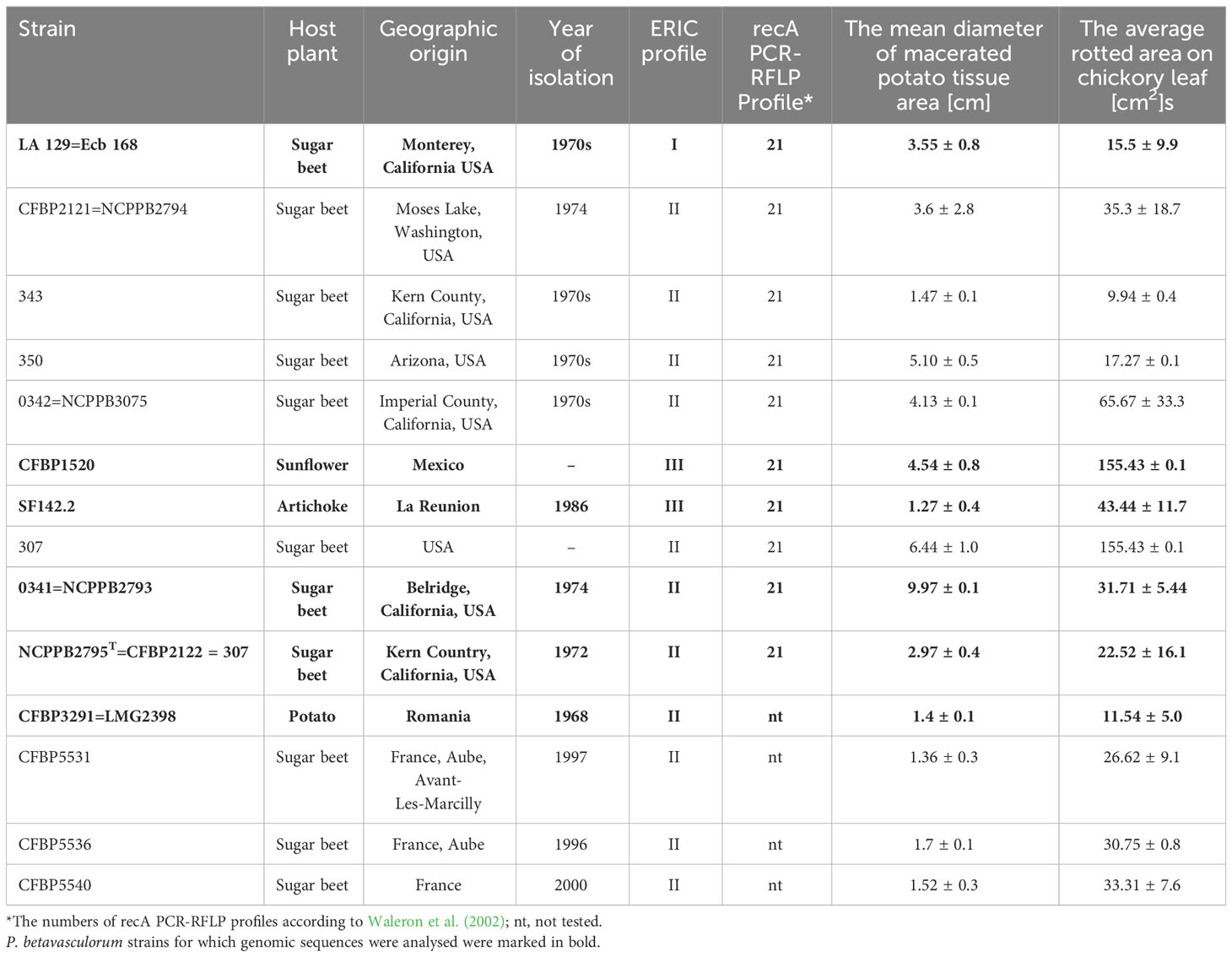
Table 1 Host plant, geographical origin, year of isolation, ERIC profile, recA PCR-RFLP profile, and pathogenicity of the studied P. betavasculorum strains.
2.2 Phenotypic characterization
2.2.1 Biochemical and physiological tests
Biochemical tests were performed with the BIOLOG GENIII plates (Biolog Inc., Hayward, CA, USA) according to the manufacturer’s instructions, using inoculation fluid A. Briefly, overnight cultures of bacteria on MHII plates were resuspended in 0.85% saline solution. A volume of 100 μl of bacterial suspension was added into each of the microplate wells. The change in color of the wells was evaluated with the naked eye. The reference or type strains of Pectobacterium species listed in Supplementary Table S1 were used for comparison. Additionally, the production of acid from α-methyl glucoside or reducing substances from sucrose, utilization of citrate, dulcitol and dextrose as the only carbon source, fermentation of 10 sugars, inulin, mannitol and sorbitol and production of 17 enzymes using the DIATABS™, Diagnostic Tablets (RoscoDiagnostica A/S, Taastrup, Denmark) were assessed.
2.2.2 Growth assays, adaptation to various environmental conditions, and metabolic activity measurements
The growth of P. betavasculorum strains in a microtiter plate was determined by absorbance (OD) measurement at 600 nm each hour for 72 h of incubation at 28°C using a microplate reader InfiniteM200Pro (Tecan, Männedorf, Switzerland).
The assays were performed in 96-well titration plates. The 200-μL of tryptic soy broth (TSB) medium with various pH, salinity, and PEG concentrations was inoculated with 5 μL of bacterial suspension with an optical density of 0.5 McF. The plates were incubated with shaking at 28°C. The absorbance readings at 600 nm were made after 0, 6, 24, and 48 h of incubation, using the Infinite M200 Pro (Tecan, Männedorf, Switzerland). The experiments with two replicates were performed twice. The effect of temperature on the growth of P. betavasculorum strains was investigated in the TSB medium. Plates were incubated with shaking at 15°C, 20°C, 28°C, and 37°C. The effect of pH on bacterial growth was studied in a TSB medium under pH values of 4, 5, 6, 7, 8, 9, 10, and 11, respectively. The ability to grow in various salinity conditions was conducted in TSB medium supplemented with 0 g L−1, 10 g L−1, 20 g L−1, 30 g L−1, 40 g L−1, 50 g L−1, 60 g L−1, 70 g L−1 NaCl, and 80 g L−1. The tolerance for limited water availability was estimated in TSB medium supplemented with 0.0 g L−1, 50.0 g L−1, 75.0 g L−1, 100.0 g L−1, 200.0 g L−1, 300.0 g L−1, 400.0 g L−1, and 500.0 g L−1 of polyethylene glycol (PEG).
The metabolic activity of P. betavasculorum strains cultivated in media with different sugars and plant extracts was assessed by the polypyridyl complex of Ru(II) method as described in (Jońca et al., 2021). The following media were used: M63 medium supplemented with 0.5% solutions of four polyols, namely, arabitol, erythritol, sorbitol, and xylitol,; eight monosaccharides, namely, ribose, arabinose, xylose, glucose, fructose, fucose, mannose, rhamnose, nine disaccharides, sucrose, lactose, maltose, isomaltose, melibiose, cellobiose, turanose, trehalose, pallatinose; one trisaccharide–raphinose; three polymers and starch such as (consisting of numerous glucose units), inulin (composed mainly of fructose units—fructans), pectin (pectic polysaccharide, rich in galacturonic acid), and taurine (non-protein amino acid). Moreover, M63 medium supplemented with 10% of extracts obtained from plants such as sugar beet, fodder beet, calla lily, blackberry, pitaya, ground cherry, black nightshade, orchid, viviparous, and Arabidopsis thaliana were used for the experiments. Plant extracts were prepared according to the procedure previously described by Jońca et al. (2021). Absorbance was read at 600 nm and fluorescence at 480 nm, each hour for 72 h of incubation at 28°C using a microplate reader InfiniteM200Pro (Tecan, Männedorf, Switzerland). The experiment was performed with three replicate samples.
2.2.3 Antibiotic susceptibility assay
The antibiotic susceptibility of P. betavasculorum strains was tested by a standard disk diffusion method. Antibiotic disks containing ampicillin 10 µg, erythromycin 15 µg, gentamicin 250 µg, kanamycin 30 µg, streptomycin 10 µg, and tetracycline 15 µg (Biomaxima, Gdansk, Poland) were used. Briefly, 100 µL of bacterial suspension was spread on a Mueller–Hinton (MH) medium with an optical density of 0.5 McF. After applying the antibiotic disks, the plates were incubated for 24 h at 28°C. Then, the zone of bacterial growth inhibition around the antibiotic disk was assessed. Moreover, the production of beta-lactamases and carbapenemases was determined with the application of chromogenic medium, ESBL (extended-spectrum beta-lactamases), and KPC (Klebsiella pneumoniae carbapenemase) plates (Graso Biotech, Owidz, Poland) as described previously (Smoktunowicz et al., 2022).
2.2.4 Plant tissue maceration assays
Pathogenicity of the P. betavasculorum strains was assessed for chicory leaves and potato tubers. Tests were performed as described previously (Lee et al., 2013; Smoktunowicz et al., 2022). Briefly, plant leaves and tuber slices were surface sterilized by soaking in 5% (v/v) sodium hypochlorite (NaOCl) and then rinsed thrice with distilled water. Bacterial cultures were grown on CVP medium for 48 h and then harvested and resuspended in Ringer solution to an approximate cell density of 108 CFU mL−1. A small incision was made on each chicory leaf using a pipette tip, and 25 μL of each bacterial suspension was inoculated into the damaged section. Pipette tips containing 50 μL of each bacterial suspension were driven into the flesh of the surface disinfected potato slices. Plants were placed in sterilized plastic boxes with sufficient moisture and stored at 28°C for several days. Plants inoculated with Ringer solutions were used as a negative control. The experiments were repeated three times, each with three replicate samples.
2.2.5 PCWDEs, N-acyl homoserine lactone siderophore and auxin production
The activity of PCWDEs was assessed as described previously (Smoktunowicz et al., 2022; Andro et al., 1984; Miller, 1972; Tindall et al., 2014; Himpsl and Mobley, 2019). Briefly, for polygalacturonase assay, isolates were incubated on a solid M63 medium (Miller, 1972) containing polygalacturonic acid. After 48 h incubation at 28°C, plates were stained by flooding with 10% (w/v) copper acetate, which forms a blue complex with the polymer, leaving clear haloes around colonies that produce pectolytic enzymes. Cellulase activity was assessed on a medium containing 0.1% (w/v) carboxymethylcellulose (CMC). After 48 h of incubation at 30°C, the plates were stained with an aqueous 0.1% (w/v) Congo red solution for 1 h at room temperature and washed with 1 M NaCl. Cellulase-producing colonies formed “halo” zones. Protease, lipase, and oligo-1,6-glucosidase activities were assessed on skimmed milk, egg yolk agar, and starch agar, respectively (Tindall et al., 2014). Siderophore production was assessed with chrome azurol S (CAS) agar plate assay (Himpsl and Mobley, 2019). The diameter of “halo” zones around bacterial colonies was measured in each case. Experiments were performed in two repetitions, and the means were calculated for each strain. Gelatinase production was assayed with the standard gelatin stab method (Tindall et al., 2014). Liquefaction of the medium was assessed after 1 week of incubation. Malonate utilization as a sole carbon source was evaluated using the malonate broth (Tindall et al., 2014). A shift in the pH indicator color from green to dark violet indicated malonate utilization.
The capability of the strains to produce N-acyl homoserine lactone (AHL) was assessed using Chromobacterium violaceum CV026 as a biosensor, as described by Ravn et al. (2001). Briefly, tested strains were streaked on the LA plates and then incubated overnight at 28°C. After that time, C. violaceum CV2026 was streaked on the plates in parallel. Plates were then once again incubated at 30°C overnight. After that time, the plates were evaluated for the purple violacein pigment that is produced by C. violaceum in the presence of AHLs.
The auxin production by P. betavasculorum strains was assessed by confirming with a colorimetric assay using the Salkowski reagent method (Gang et al., 2019). Strains were cultivated overnight at 30°C in TSB medium supplemented with 0.5 g of tryptophan. After centrifugation, 100 µL of supernatant and 100 µL of Salkowski reagent were mixed on a 96-well plate incubated for 30 min in the dark. The color intensity was measured spectrophotometrically at a wavelength of 536 nm. A non-inoculated medium was used as a control. An Escherichia coli strain ATCC8339 was used as a positive control, while ΔtnaA deletion mutant, which lost l-tryptophan degradation activity (Shimazaki et al., 2012), was used as a non-producing auxins control.
2.3 Fatty acids analysis
Total lipids were extracted from bacteria suspension in Ringer solution, using a mixture of chloroform and methanol (2:1 v/v). Lipid extracts were subjected to hydrolysis by 0.5 M KOH. After hydrolysis and washing with water/n-hexane, the n-hexane phase was evaporated to dryness under a stream of nitrogen. 19-Methyl eicosanoate was used as an internal standard (IS). Free fatty acids (unesterified FAs) were methylated with 10% boron trifluoride reagent in methanol. The fatty acid methyl esters (FAMEs) were analyzed with gas chromatography/electron ionization mass spectrometry (GC-EI-MS). FAMEs were separated in a 30 m×0.25 mm i.d. RTX-MS-5 capillary column (film thickness, 0.25 μm). The column temperature was programmed from 60°C to 300°C at a rate of 4°C/min. The injector temperature and ion source was 300°C and 200°C, respectively. The carrier gas was helium, and the electron energy will be 70 eV.
A standard mix of fatty acids methyl esters (Supelco ® 37 Component FAME Mix. 1890. cis/trans) were used for the identification of the major fatty acids based on retention time. Individual FAs were expressed as percentages of the total FA’s present in the chromatogram.
2.4 Genetic characterization
2.4.1 Genomic DNA preparation
Strains of P. betavasculorum were stored in 20% glycerol at −80°C. For the preparation of genomic DNA, bacteria were first grown for 48 h on CVP medium (Hélias et al. 2012) at 28°C. A volume of 10 ml of the liquid TSB medium (bioMerieux, Marcy-l’Étoile, France) was inoculated with a single colony from the CVP plate and grown overnight at 28°C with shaking. Obtained cultures were centrifuged for cell harvest for 20 min at 3,200g at 4°C and resuspended into TE buffer (10 mM Tris–HCl, pH 7.5, 1 mM EDTA). The DNA was extracted according to the cetyltrimethylammonium bromide (CTAB) protocol (www.jgi.doe.gov, accessed on 5 August 2023). The quantity and quality of the DNA were assessed by the spectrophotometric analysis with a microplate reader InfiniteM200Pro (Tecan, Männedorf, Switzerland) and the 1% agarose gel electrophoresis.
2.4.2 DNA fingerprinting analyses
A total genomic DNA fingerprinting was performed by the repetitive element PCR fingerprinting (rep-PCR) using enterobacterial repetitive intergenic consensus (ERIC) primers (Versalovic et al., 1991). Analysis was performed with BioNumerics V6.6 (http://www.applied-maths.com). The neighbor-joining cladogram was created with a band-based Jaccard coefficient.
2.4.3 Multi-locus sequence analysis
The sequences of 13 housekeeping genes (acnA, 2,692 bp; gapA, 1,005 bp; gyrA, 2,649 bp; gyrB, 2,418 bp; icdA, 1,254 bp; mdh, 940 bp; mtlD, 1,149 bp; pgi, 1,644 bp; proA, 1,259 bp; recA, 1,074 bp; recN, 1,662 bp; rpoA, 990 bp; and rpoS, 1,514 bp) commonly used for multi-locus sequence analysis of Pectobacterium (Ma et al., 2007; Waleron et al., 2018), were extracted from genomes of six P. betavasculorum strains (listed in Table 1). Their sequences were merged and aligned with sequences retrieved from genomes of the type strains of each of Pectobacterium species that are available at the GenBank database with using the MUSCLE algorithm with the default settings in Geneious Prime (www.geneious.com).
Phylogenetic analyses were performed on the concatenated data set (20,770 bp) with the Maximum-Likelihood method and the General Time Reversible as the best nucleotide substitution model selected using jModeltest 2.1.9 software. Bootstrapping was executed with 1,000 replications. The gene sequences of D. solani IFB0099 (JXRS00000000) were used as an outgroup.
2.4.4 Genome sequencing, assembly, and annotation
The genomes of two P. betavasculorum strains, CFBP1520 and SF142.2, were sequenced using the Illumina MiSeq technology (Table 2). Quality and adapter trimming of the raw reads was performed using Trimmomatic v0.32 (http://www.usadellab.org/cms/?page=trimmomatic, accessed on 5 August 2023); kmer length and distribution were analyzed using KmerGenie v1.6949 (http://kmergenie.bx.psu.edu/, accessed on 5 August 2023). De novo assembling was performed using SPAdes v3.10.1 (http://cab.spbu.ru/software/spades/, accessed on 5 August 2023), Velvet v1.2.10 (https://www.mybiosoftware.com/velvet-1-1-07-sequence-assembler-short-reads.html, accessed on 5 August 2023), and Ray v2.3.1 (http://denovoassembler.sourceforge.net/, accessed on 5 August 2023). The contigs were integrated using CISA v1.3 (http://sb.nhri.org.tw/CISA/en/CISA, accessed on 5 August 2023) software and scaffolded with SSPACE v3.0 (https://github.com/nsoranzo/sspace_basic, accessed on 5 August 2023). The final assembly was evaluated using Quast v4.5 (http://quast.sourceforge.net/quast, accessed on 5 August 2023) software. Another two strains, CFBP3291 and Ecb168, were sequenced in a hybrid mode using the Illumina HiSeq 2000 and Oxford Nanopore platforms (Table 2). An Illumina-compatible paired-end sequencing library was constructed using the NEB Ultra II FS Preparation Kit (New England Biolabs, Beverly, USA) according to the manufacturer’s instructions. The library was sequenced using an Illumina MiSeq platform (Illumina, San Diego, CA, USA) with 2 × 300 paired-end reads using v3 600-cycle sequencing kit. Sequence quality metrics were assessed using FASTQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/), and quality was trimmed using fastp (Chen et al., 2018). Prior to long-read library preparation, genomic DNA was sheared into ~30 kb fragments using 26G needle, followed by size selection using a Short Read Eliminator kit (Circulomics, Baltimore, MD, USA). Five micrograms of the recovered DNA was taken for 1D library construction using SQK-LSK109 kit, and 0.8 µg of the final library was loaded into R9.4.1 flow cell and sequenced on GridION sequencer (Oxford Nanopore Technologies, Oxford, UK). Raw nanopore data was basecalled using Guppy v5.0.7 in super accuracy mode (Oxford Nanopore Technologies, Oxford, UK). After quality filtering using NanoFilt (De Coster et al., 2018) and residual adapter removal using Porechop (https://github.com/rrwick/Porechop), the obtained dataset was quality checked using NanoPlot (De Coster et al., 2018). Long nanopore reads were then assembled in hybrid mode using Unicycler (Wick et al., 2017). The remaining ambiguities in the genome assemblies were verified by the PCR amplification of DNA fragments, followed by Sanger sequencing with an ABI3730xl Genetic Analyzer (Life Technologies, Carlsbad, CA, USA) using BigDye Terminator Mix v. 3.1 chemistry (Life Technologies, Carlsbad, CA, USA). All of the possible sequence errors and miss-assemblies were further manually corrected using Seqman software (DNAStar, Madison, WI, USA) to obtain the complete nucleotide sequence of bacterial genomes). novo assembling was performed using Flye v 2.9.1 (https://github.com/fenderglass/Flye, accessed on 5 August 2023) and Unicycler v 0.5.0 (https://github.com/rrwick/Unicycler, accessed on 5 August 2023). The final assembly was evaluated using Quast v4.5 (http://quast.sourceforge.net/quast, accessed on 5 August 2023) software.
Annotation was performed by the NCBI PGAPpipeline (https://www.ncbi.nlm.nih.gov/genome/annotation_prok/, accessed on 5 August 2023) and RAST (Rapid Annotation using Subsystem Technology) (http://rast.nmpdr.org/, accessed on 5 August 2023). The genomic sequence was additionally annotated with BlastKOALA (KEGG Orthology And LinksAnnotation) (https://www.kegg.jp/blastkoala/, accessed on 5 August 2023).
2.4.5 Genomic analyses
Comparative genomics was performed with the use of Pathway Tools Software v27.0 (http://bioinformatics.ai.sri.com/ptools/, accessed on 14 August 2023), Bacterial Pangenome Analysis (BPGA) pipeline (Chaudhari et al., 2016) (https://iicb.res.in/bpga/, accessed on 13 September 2023), and a GET_HOMOLOGUES software package (Contreras-Moreira and Vinuesa, 2013). The obtained results were subjected to the BLAST analysis against the UniProt protein database (https://www.uniprot.org/, accessed on 13 September 2023). The entire set of genes identified in all P. betavasculorum genomes was recognized as a pangenome. The core of the pangenome consists of the gene families that were present on all six analyzed genomes. While the accessory genome contains gene families that were present in at least two up to five of six analyzed P. betavasculorum genomes. The unique genes were only found in a single genome.
The visual comparison of genome homology was done with BRIG (BLAST Ring Image Generator) (http://sourceforge.net/projects/brig, accessed on 5 August 2023) with the default settings (Alikhan et al., 2011). The genomes of P. betavasculorum NCPPB2795T (JQHM00000000) and P. betavasculorum NCPPB2793 (JQHL00000000) have been included for genomic comparison.
The phylogenomic analysis based on the core proteins sequence comparison was done using PhyloPhlAn computational pipeline (https://huttenhower.sph.harvard.edu/phylophlan, accessed on 4 August 2023). The core protein analysis was performed using the sequences of other Pectobacterium members available on GenBank and the phylophlan library of 400 conserved proteins of Prokaryotes and Archea. The sequence of Duffyella gerundensis AR (CP073262) was used as an outgroup. The bootstrap consensus was inferred from 1,000 replicates. To establish the relationship between P. betavasculorum strains isolated from different plant species, the ML tree based on 3,573 core genes per genome (3,588,885 bp per genome) have been created with using the EDGAR platform for comparative genomics (https://edgar3.computational.bio.uni-giessen.de/cgi-bin/edgar_login.cgi). The calculation of the average nucleotide identity (ANI), the average amino-acid identity (AAI), and the Pairwise Percentage of Conserved Proteins (POCP) was performed using the EDGAR pipeline (Dieckmann et al. 2021). To evaluate the assignment of the studied strains to species, in silico and the average nucleotide identity (ANI) were calculated between the genomes of P. betavasculorum and the genomes of the other type or reference strains of the members of Pectobacteriaceae family with using g JSpeciesWS: a web server (Richter et al., 2016).
The presence of ICE and IME elements in the P. betavasculorum genomes was investigated using ICEfinder, a web-based tool (https://bioinfo-mml.sjtu.edu.cn/ICEfinder/ICEfinder.html, accessed on 20 September 2023).
The biosynthetic gene clusters encoding secondary metabolites related to the synthesis of phytotoxins, antibiotics, and antimicrobial compounds were predicted using antiSMASH 4.0 (Blin et al., 2023) (https://antismash.secondarymetabolites.org/#!/about, accessed on 20 September 2023).
2.5 Statistical analyses
Statistical significance of the pathogenicity tests and phenotypic assays was assessed using independent Student’s t-tests and one-way analysis of variance (ANOVA). A post-hoc analysis was performed using Student’s t-test with a base mean as a reference. Statistical significance was assumed if p-value < 0.05. The statistical analysis was done using R software for Windows version 4.1.2 (R Foundation, Vienna, Austria) and the following packages: ComplexHeatmap (Gu et al., 2016), ggplot2 (Wickham, 2009), and ggpubr (Kassambara, 2023).
3 Results
3.1 Phenotypic characterization
P. betavasculorum strains are Gram-negative, motile, non-sporulating, and facultative anaerobic bacteria that belong to the order Enterobacteriales within the class of Gammaproteobacteria. Cells are rod shaped with a length of approximately 2 μm in the exponential growth phase. P. betavasculorum strains produce pits on a crystal violet pectate medium (Supplementary Figure S1). All studied P. betavasculorum strains were able to macerate potato tuber and chicory leaf tissue efficiently (Table 1).
All of the 14 tested P. betavasculorum strains (Table 1) could perform glucose, maltose, mannose, rhamnose, raffinose, sucrose, d-xylose, and mannitol fermentation. Additionally, all tolerated salinity was up to 4% NaCl. What is more, all strains exhibited enzyme activity of α- and β-glucosidase, α- and β-galactosidase, β-glucuronidase, ornithine decarboxylase, arginine dihydrolase, tryptophan deaminase, and urease, but not lysine decarboxylase and β-xylosidase. They do not produce acid from α-methyl glucoside, lactose, trehalose, adonitol, sorbitol, or reducing substances from sucrose. In addition, all strains were sensitive to ampicillin, tetracycline, gentamicin, kanamycin, and streptomycin but resistant to erythromycin. None of the tested strains exhibited resistance to beta-lactamases or carbapenems (Supplementary Figure S2, Supplementary Table S1).
Metabolic profiling showed that all tested P. betavasculorum strains could use as sole carbon sources the following compounds: dextrin, α-D-glucose, D-galactose (except type strain NCPPB2795T), gentiobiose, D-fructose, D-maltose, D-mannose, L-rhamnose, D-cellobiose, D-mannitol, myoinositol, glycerol, D-sorbitol, D-glucose-6-PO4, sucrose, D-trehalose, D-turanose, β-methyl-D-glucoside, D-salicin, N-acetyl-D-glucosamine, D-aspartic acid, L-aspartic acid, L-glutamic acid, D-gluconic acid, L-galactonic acid lactone, acetic acid, bromosuccinic acid, formic acid, L-lactic acid, L-malic acid, mucic acid, D-saccharic acid, and pectin. However, they were not able to utilize stachyose, D-raffinose, α-D-Lactose, D-melibiose, D-Fucose, L-Fucose, and D-arabitol, D-serine, L-histidine, L-pyroglutamic acid, glucuronamide, quinic acid, p-hydroxyphenyl acetic acid, citric acid, D-malic acid, γ-aminobutyric acid, N-acetyl-β-D-mannosamine, N-acetyl-D-galactosamine, N-acetyl neuraminic acid, 3-methyl glucose, inosine, D-serine, gelatin, L-arginine, D-lactic acid methyl ester, D-malic acid, Tween 40, β-hydroxy-D l-butyric acid, α-keto butyric acid, propionic acid, sodium bromide. In addition, all P. betavasculorum strains could grow at pH 6, 1% sodium chloride and 4% sodium chloride (except strain NCPPB2793), and 1% sodium lactate, and were resistant to troleandomycin, rifamycin SV, lincomycin, niaproof 4, vancomycin, tetrazolium violet, and tetrazolium blue. Only two strains, NCPPB 2794 and NCPPB 3075, did not grow at pH 5 and in 4% NaCl, while the rest did. All tested strains were sensitive to fusidic acid, D-serine, minocycline, lincomycin, nalidixic acid, guanidine HCl, lithium chloride, and potassium tellurite (Supplementary Table S2).
Analysis of biochemical properties showed that P. betavasculorum strains CFBP3291, CFBP1520, and SF142.2 isolated from potato, sunflower, and artichoke differ in metabolic activity from sugar beet strains, as they cannot utilize 3-methyl-glucose. What is more, the strains CFBP1520 and SF142.2 also cannot utilize lactose and raffinose. Unlike sugar beet strains, they could use α-hydroxybutyric acid as their only carbon source. In addition, only these two strains are resistant to aztreonam and nalidixic acid. It should be noted that sugar beet strains differ from one another. For example, strain Ecb168, like the sunflower and artichoke strains, does not use lactose and raffinose. Unlike all other strains, this strain did not use inosine, glycyl-L-proline, or L-serine.
In contrast, the type strain, P. betavasculorum NCPPB2795T, can use propionic acid but does not use D-galactose and is resistant to minocycline. The strain NCPPB2793 was the only one capable of growing with the presence of Tween40, whereas strains NCPPB2794, NCPPB3075, and Ecb168 could not grow in the medium containing 4% NaCl. Moreover, three French strains, CFBP5540, CFBP5536, and CFBP5531, could utilize fucose as a carbon source (Figure 2, Supplementary Table S2).

Figure 2 Phenotypic characters that differentiate ten P. betavasculorum strains, IFB5271, CF142.2. CFBP3291, Ecb168, CFBP5531, CFBP5536, CFBP5540, NCPPB2795T, NCPPB2793, NCPPB3075, and other closely related species: P. atrosepticum ICMP1526T, P. parmentieri SCC3193, P. peruviense IFB5232T, P. polonicum DPMP315T, P. punjabense SS95T, P. wasabiae CFBP 3304T, and P. zantedeschiae 9MT. Navy color means a positive reaction, blue means a weak activity, and white means an adverse reaction.
The metabolic profiling results of P. betavasculorum strains were extended by simultaneous measurements of bacterial growth and metabolism on minimal medium M63 supplemented with 0.5% solutions of various polyols, sugars, polymers, or 10% plant extracts. All strains could use numerous sugars as a building material and energy source. The strains grew efficiently on minimal medium M63 supplemented with 0.5% solutions of ribose, xylose, glucose, trehalose, lactose, maltose, or sorbitol while growing half as fast on media supplemented with cellobiose, palatinose, raffinose, rhamnose, ribose, melibiose, and isomaltose. Feeble growth was observed when 0.5% inulin and turanose were used as a source of organic compounds. No growth ability was observed on media containing 0.5% arabitol, erythritol, xylitol, and taurine (Supplementary Figure S3). In addition, P. betavasculorum strains can grow on minimal medium containing, as the only carbon source, extracts of various plant species, such as sugar beet, fodder beet, carrot, physalis, dragon fruit, Arabidopsis thaliana, and potato tubers. Moreover, poor growth was observed in the case of extracts from blueberry and dragon fruits. None of the strains tested showed the ability to grow in the presence of extracts from ornamental plants like the Calla lily, orchids, viviparous, or black nightshade (Supplementary Figure S4).
All tested strains can synthesize signal molecules (acyl-homoserine lactones), siderophores, and plant hormones, which were confirmed using various assays (Supplementary Figures S5-S7).
A total of 14 tested P. betavasculorum strains were able to macerate plant tissue at laboratory conditions. The soft rot disease symptoms were observed on potato tuber slices and chicory leaves inoculated by wounding (Table 1).
Bacteria of the species P. betavasculorum are highly adaptable to environmental conditions. P. betavasculorum strains grew most effectively in the 20°C–28°C temperature range. Reducing the incubation temperature to 15°C resulted in an average of twofold poorer bacterial growth, and the observed difference was statistically significant while increasing the incubation temperature to 37°C resulted in a two- to fivefold decrease in bacterial growth (Supplementary Figure S8A). All P. betavasculorum strains grow well in salinity (0%–7%) with optimum at 1%–2% NaCl concentration (Supplementary Figure S8B). Tested bacteria grow efficiently at pH from 5 up to 11 (Supplementary Figure S8C) and tolerate the reduced availability of water up to 10% of PEG concentration in the growth medium (Supplementary Figure S8D). Furthermore, P. betavasculorum strains are able to grow and were metabolically active in media with different sucrose concentrations from 0% to 20% (Supplementary Figure S9).
3.2 Fatty acid methyl ester profiling
Fatty acid methyl ester (FAME) profiling was performed for four selected strains, two isolated from sugar beet (Ecb168. NCPPB2795T) and two from other plants (SF142. CFBP1520). Strain CFBP3291 isolated from potato was not included, as it is more similar phenotypically and genetically to sugar beet strains. P. betavasculorum strains demonstrate the highest amounts of seven fatty acids: palmitic (16:0), palmitoleic acids (16:1), oleic (18:1), pentadecylic acid (15:0), stearic acids (18:0), followed by margaric (17:0) and linoleic acid (18:2n-6) (Table 3).
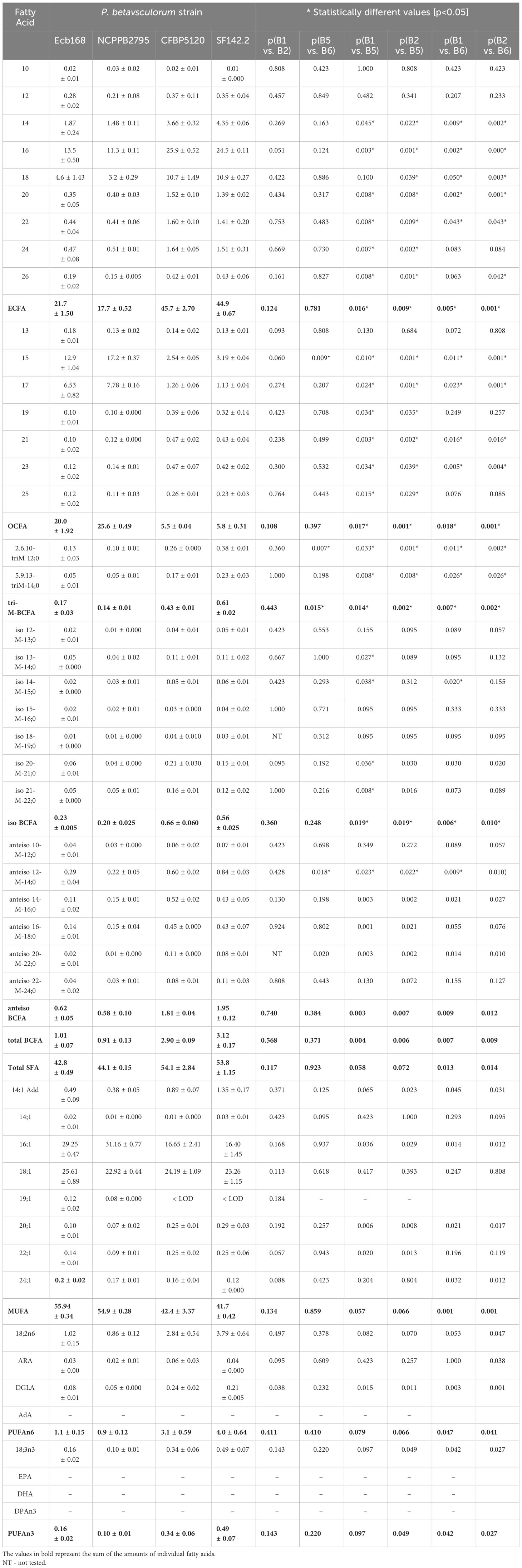
Table 3 The total fatty acid composition of P. betavasculorum strains NCPPB2795T, Ecb168, CFBP1520, and SF142.2.
All tested P. betavasculorum strains demonstrate the same fatty acid composition but differ in their proportions. Strains isolated from sugar beets have more (56%–57%) unsaturated fatty acids (UFAs) represented mostly by monounsaturated fatty acid (MUFA) than saturated (SFA) (43%–44%). The opposite proportions were observed for strains isolated from sunflower and artichoke: 46% of unsaturated and 54% of saturated fatty acids (Figure 3A, Table 3). Polyunsaturated fatty acids (PUFAs) were more abundant in strains from sunflower and artichoke (Figure 3D, Table 3). Strains from sugar beets had significantly fewer branched fatty acids (BCFAs) than strains from sunflower and artichoke (Figure 3C, Table 3). In addition, the sugar beet strains had two to three times fewer even-chain fatty acids (ECFAs) while four to five times more odd-chain fatty acids (OCFA) than strains that came from other host plants (Figure 3B, Table 3). The differences in fatty acid composition observed between strains isolated from sugar beet and other plants were statistically significant (Table 3, Supplementary Figure S10).
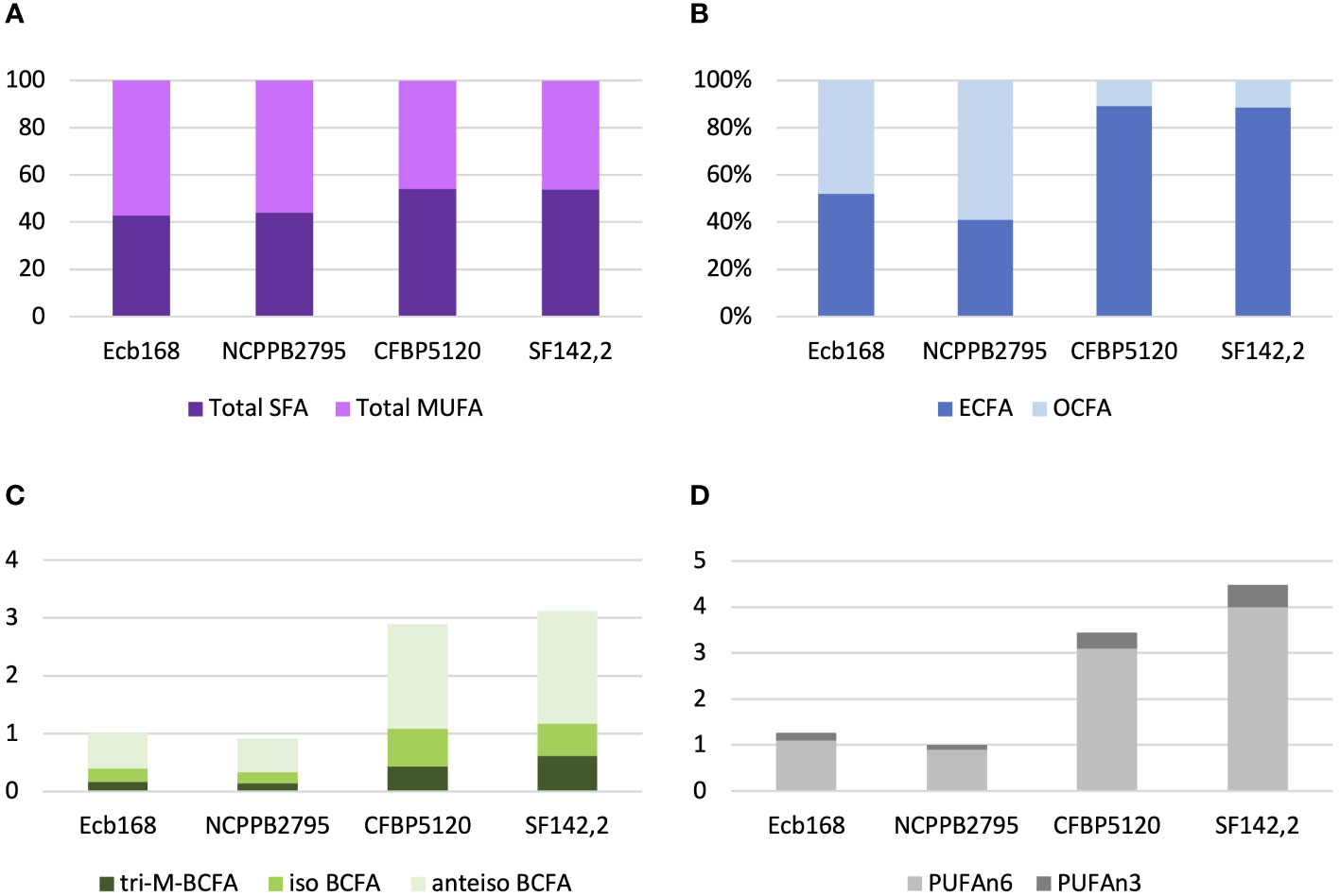
Figure 3 The proportions of the different fatty acid groups present in strains of P. betavasculorum isolated from a different host plant. (A) The ratio of total saturated fatty acids (SFAs) to monounsaturated fatty acids (MUFAs), (B) the proportion of even-chain fatty acids (ECFAs) to odd-chain fatty acids (OCFA), (C) the content (%) of polyunsaturated fatty acids (PUFAs) n6 and n3, and (D) the content (%) of various branched fatty acids (BCFAs). P. betavasculorum strains isolated from sugar beet, Ecb168 and NCPPB2795T, P. betavasculorum strain CFBP1520 isolated from sunflower, and P. betavasculorum strain SF142.2 isolated from the artichoke.
3.3 Genotypic characterization
3.3.1 Fingerprinting with ERIC-PCR
The genetic diversity of 14 P. betavasculorum strains originating from four different plant species and three different continents was assessed by the ERIC-PCR method and multi-locus sequence analysis.
Three ERIC profiles were discriminated against (Table 1, Figure 4). For strains CFBP1520 and SF142.2 isolated from sunflower and artichoke, the same fingerprint pattern was observed; however, it was distinct from those strains derived from sugar beet and potato. Furthermore, the sugar beet strains showed genetic diversity as two patterns were distinguished. Strain Ecb168 differed from the other strains isolated from this host plant.
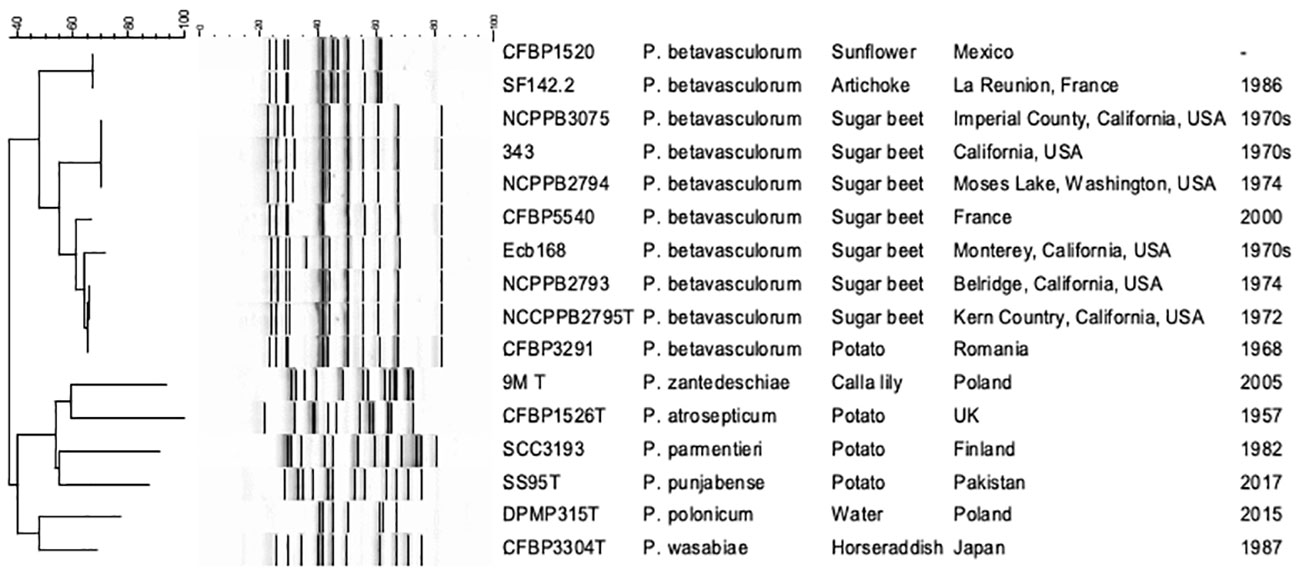
Figure 4 The electrophoretic patterns obtained after REP-PCR with ERIC primers. 1, P. betavasculorum Ecb168; 2, P. betavasculorum NCPPB2795T; 3, P. betavasculorum NCPPB2794; 4, P. betavasculorum 343; 5, P. betavasculorum 350; 6, P. betavasculorum NCPPB3075; 7, P. betavasculorum NCPPB2793; 8, P. betavasculorum 307; 9, P. betavasculorum CFBP5540; 12, P. betavasculorum CFBP3291; 13, P. betavasculorum CFBP1520; 14, P. betavasculorum SF142.2; 15, P. zantedeschiae 9MT; 16, P. atrosepticum ICMP1526T; 17, P. parmentieri SCC3193; 18, P. wasabiae CFBP 3304T; 19, P. punjabense SS95T; 20, P. polonicum DPMP315T. The neighbor joining cladogram was created with band base Jaccard coefficient.
3.3.2 Multi-locus sequence analysis
Multi-locus sequence analysis (MLSA) analysis was performed for seven P. betavasculorum strains, namely, Ecb168, NCPPB2795T, NCPPB2794, CFBP3291, CFBP1520, SF142.2, and NCPPB3075, that were selected based on ERIC fingerprinting profiles. A total of 13 housekeeping genes, namely, acnA, gapA, gyrA, gyrB, icdA, mdh, mtlD, pgi, proA, recA, recN, rpoA, and rpoS, were merged. Next, the Maximum Likelihood phylogenetic analysis on concatenated gene sequences (20,770 bp) was performed to evaluate the evolutionary relationships among P. betavasculorum strains and other species within the Pectobacterium genus. The generated tree has shown the presence of two major clusters. The species P. betavasculorum belongs to the first of these and is most closely related to P. zantedeschiae, P. atrosepticum, and P. peruviense, while P. polonicum, P. punjabense, P. parmentieri, and P. wasabiae constitute their sister clade. All P. betavasculorum strains form two subgroups that form one clearly separated branch.
The second clade included all the other species of the genus Pectobacterium described so far except for P. fontis and P. cacticidum. These two species, in turn, are grouped separately (Supplementary Figure S11). Moreover, the ML tree showed that P. betavasculorum strains are separated into two groups. Strain CFBP3291 isolated from potato groups together with two sugar beet strains, NCPPB2793 and NCPPB2795T. At the same time, sugar beet strain Ecb168 is more related to strains CFBP150 and SF142.2 that originated from sunflower and artichoke, respectively.
3.3.3 Phylogenomics
The phylogenomic analysis based on core proteins showed similar results as MLSA and fingerprinting methods. All P. betavasculorum strains form one group (marked in coral pink) that is separated from other Pectobacterium species. P. betavasculorum is more closely related with species P. peruviense, P. atrosepticum, and P. zantedeschiae and more distantly with P. polonicum, P. punjabense, P. parmentieri, and P. wasabiae (Figure 5).
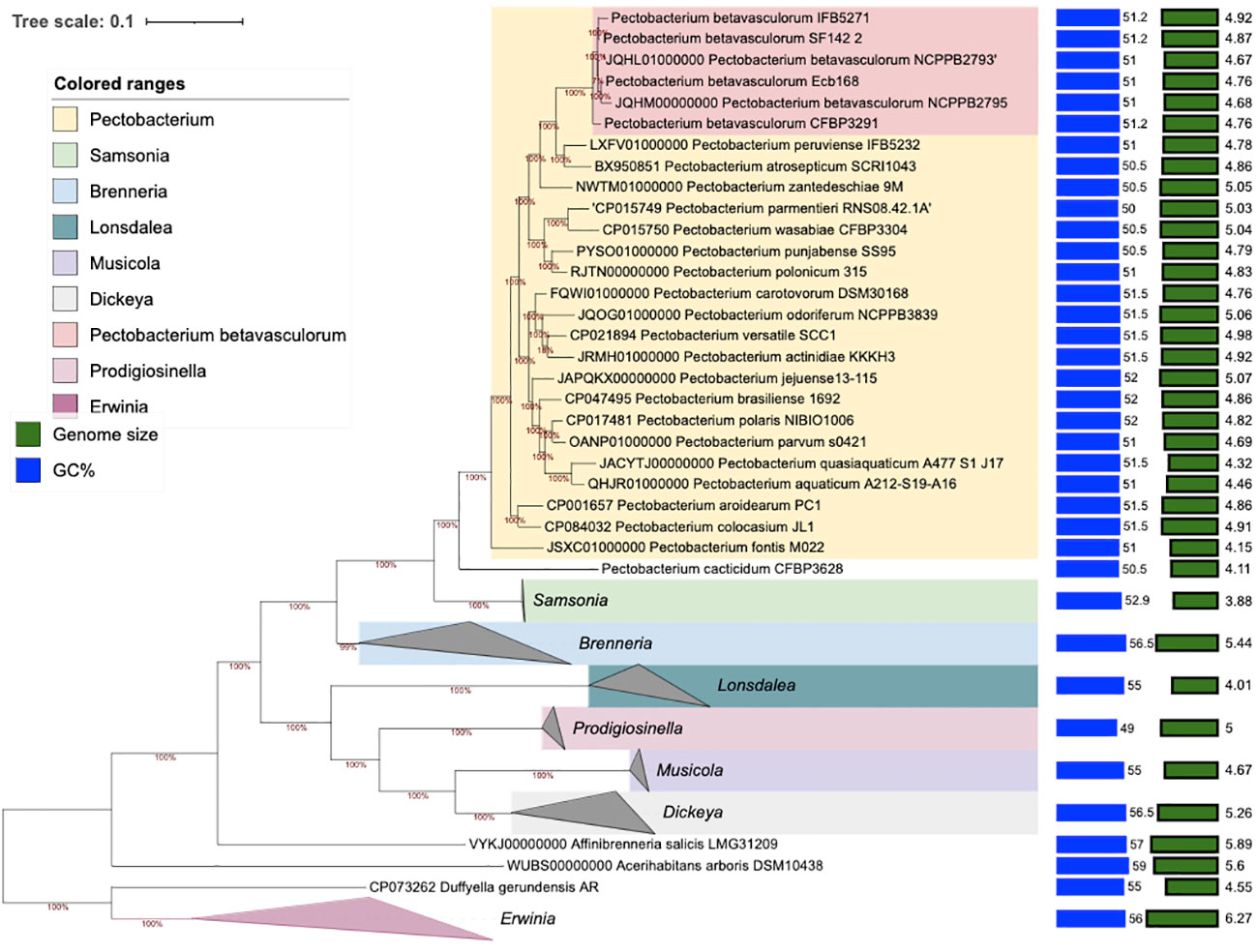
Figure 5 The phylogenomic analysis of P. betavasculorum strains based on the 400 most conserved universal proteins. The Maximum Likelihood tree was constructed using PhyloPhlAn computational pipeline (https://huttenhower.sph.harvard.edu/phylophlan, accessed on 7 August 2023).
The ML tree for six genomes, built out of a core of 3,573 genes per genome (3,588,885 bp per genome) to establish a relationship between P. betavasculorum strains isolated from different plant species, was created. Strains CFBP150 and SF142.2 isolated from sunflower and artichoke are most similar to each other. Strain Ecb168 is most closely related to type strain NCPPB2795T, while strain CFBP3291 from potato is more related to strain NCBBP2793 from sugar beet (Supplementary Figure S12A). Additional genomic analyses, Percent of Conserved Proteins (POCP), Average Nucleotide Identity (ANI), and Average Amino acid Identity (AAI) were performed to resolve this dilemma also. According to POCP analysis, strains CFBP1520 and SF142.2 derived from sunflower and artichoke are most similar to each other and are a bit distant from strains from sugar beet NCPPB2793, NCPPB2795T, and Ecb168, and strain CFBP3291 from potato (Supplementary Figure S12B). In contrary to ANI, AAI analyses showed that strains CFBP150 and SF142.2 isolated from sunflower and artichoke are more similar to two strains isolated from sugar beet, Ecb168 and NCPPB2793, and all together are a bit distant from strains NCPPB2795T from sugar beet and CFBP3291 derived from potato, which are most similar to each other (Supplementary Figures S12C, D).
3.3.4 Genome sequencing and analysis
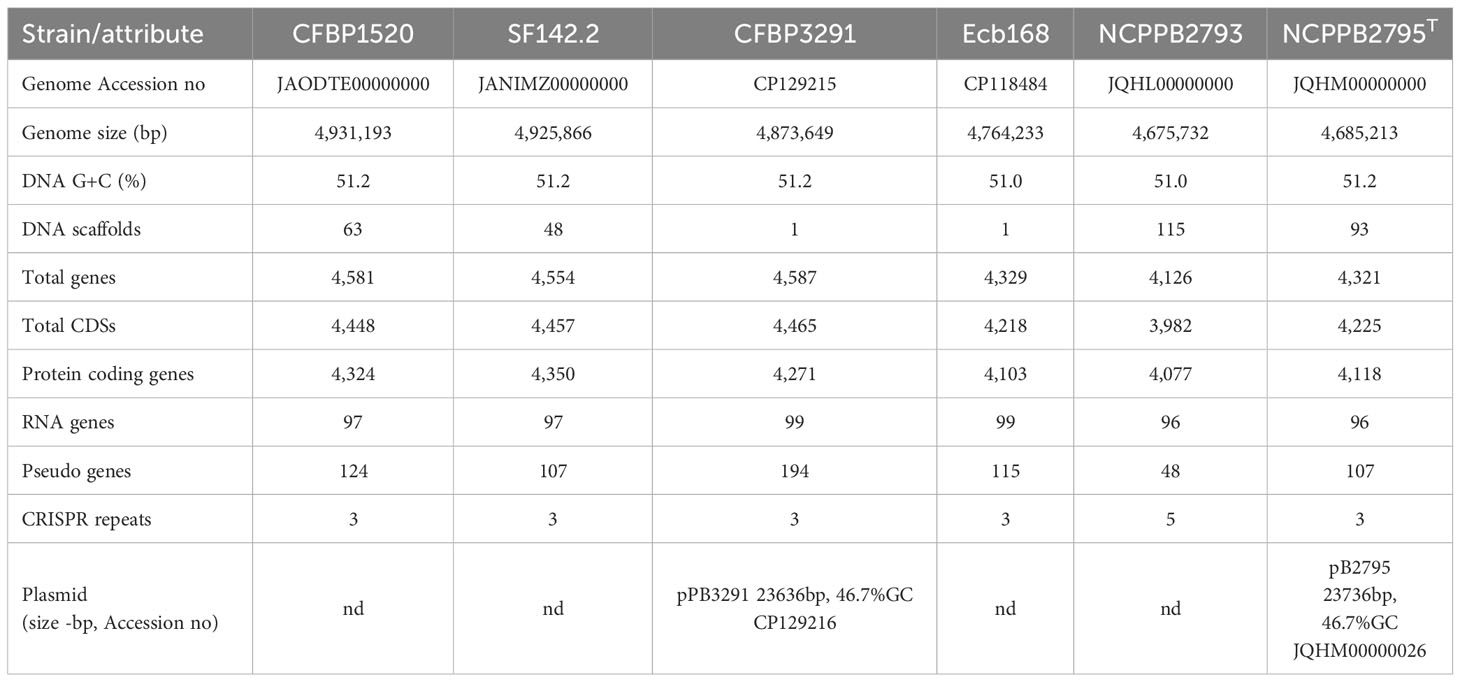
The genomes of four P. betavasculorum strains, CFBP1520 originated from sunflower in Mexico, SF142.2 isolated from artichoke on the French island La Reunion, CFBP3291 from potato grown in Romania, and Ecb168 from sugar beet in the USA, were sequenced, assembled, annotated, and deposited in GenBank under the following accession numbers CP118484, CP129215, JANIMZ000000000, and JARDVT000000000, respectively. The final genome sequences of strains Ecb168 and CFBP3291 are circulated and closed, while sequences of strains SF142.2 and CFBP1520 contain 48 or 63 scaffolds representing one chromosome each. The genome size ranged from 4.76 kBP to 4.93 kBP, and GC % was between 51.0% and 51.2% (Table 2).
Among the six P. betavasculorum isolates, only two strains, CFBP3291 and CBPPB2795T, harbor 23,763 bp long plasmid that is similar in 94.27% (100% coverage) with 21,742 bp long plasmid IncFII(pCRY) (NC005814) harboring a cryptic type of IVA secretory system (T4ASS) from Yersinia pestis biovar Microtus str. 91001 (Song et al., 2004). Plasmids pPB3291 (CP129216) and pPB2795 (JQHM00000026) from P. betavasculorum strains CFBP3291 and CBPPB2795T, respectively, are identical in 100%. Likewise, plasmid pCRY and both plasmids from P. betavasculorum strains contain 10 VirB proteins from T4SS (VirB1, TrbC/VirB2, ATPase VirB3, minor pilin of type IV secretion complex VirB5, VirB6, VirB8, VirB9, VirB10, and VirB11) and VirD, sequences for plasmid mobilization relaxosome protein MobB and MobC, RepA protein, chromosome partitioning protein ParA, sequences for dopa decarboxylase, conjugal transfer protein, Cag pathogenicity island protein Cag12, transcriptional antiterminator NusG, transcriptional regulator, EexN lipoprotein, conjugal transfer protein TrbL, and membrane protein. Moreover, the contig 4 corresponding to plasmid pPB3291 contains a thermonuclease, type II toxin-antitoxin system RelE/ParE, while scaffold 26, corresponding to plasmid pPB2795, contains additionally TriB protein ATP-binding protein, toxin-antitoxin system, toxin component HigB, micrococcal nuclease-like protein, and seven hypothetical proteins. The plasmids of P. betavasculorum strains are similar in 97.84% with plasmid pRHBSTW-00515_4 from Klebsiella pneumoniae strain RHBSTW-00515 (CP056428) isolated from livestock host (AbuOun et al., 2021, Microb Genom.) and in 94.8% (92% coverage) with numerous plasmids from clinical isolates of Citrobacter freundii (CP110909, CP110906, and CP110893).
The newly obtained genomic sequences were compared with genomes of two strains, NCPPB2793 and NCPPB2795T, isolated from sugar beet in the USA, which sequences are available in the GenBank under accession numbers JQHL00000000 and JQHM00000000, respectively. Genomes of strains isolated from sugar beet were slightly smaller (~4.7 kBP) than those of strains originating from other host plant species (~4.9 kBP). Consequently, this has resulted in a lower number of genes encoding proteins in the case of three strains isolated from sugar beet, NCPPB2793 (4,126), NCPPB2795T (4,321), and Ecb168 (4,329), than for strains isolated from sunflower (4,581), artichoke (4,554), and potato (4,587). However, it should be noted that four out of six of the analyzed genomes are not closed. Summary information is presented in Table 2.
3.3.4.1 Functional genome annotation
The RASTtk ModelSEED database was used to identify the metabolic pathways in P. betavasculorum using the Kyoto Encyclopedia of Gene and Genomes (KEGG). The subsystem category distribution of P. betavasculorum genomes annotated at RASTtk showed that approximately 30% of functioning features were present in ~350 subsystems (Table 4). Of these, the most abundant were features involved in amino acid and derivative subsystems 269–298 (14.7%) and metabolism of carbohydrate 308–358 (14.8%), protein 200–230 (12%), cofactors, vitamins, prosthetic group, and pigments 155–167 (8%), and features involved in membrane transport 123–148 (7.5%).
We observed differences between P. betavasculorum strains in the number of genes involved in nitrogen and aromatic compounds metabolism. Strains isolated from sugar beet and potato do not have nif operon encoding genes responsible for atmospheric nitrogen fixation and genes responsible for salicylate and gentisate catabolism that are present in two strains originated from sunflower and artichoke (Table 4).
Furthermore, to determine differences in functions of each gene of P. betavasculorum genome, we analyzed our data with KEGG and COG; 7157/038 functional categories (including not annotated sequences) with P. betavasculorum strain Ecb168 as a reference were found in the selected contigs, distributed in core dispensable and singleton genes. Analyzing all annotated KEGG/COG genes, 0.33%/0.28% have inconsistent category information.
According to the cluster of orthologous genes (COG) database, 86.5% of genes were annotated, and 41% of them were involved in metabolism processes, 27% were engaged in information processing and storage, 12% in cellular processing and signaling, while 20% of genes belonged to the poorly characterized category. In the cases of all P. betavasculorum genomes, the two s subgroups among the metabolism category contain genes annotated to the metabolism and transport of amino acids and carbohydrates; they contain almost 9% and 8% of genes, respectively (Figure 6, Supplementary Table S3).
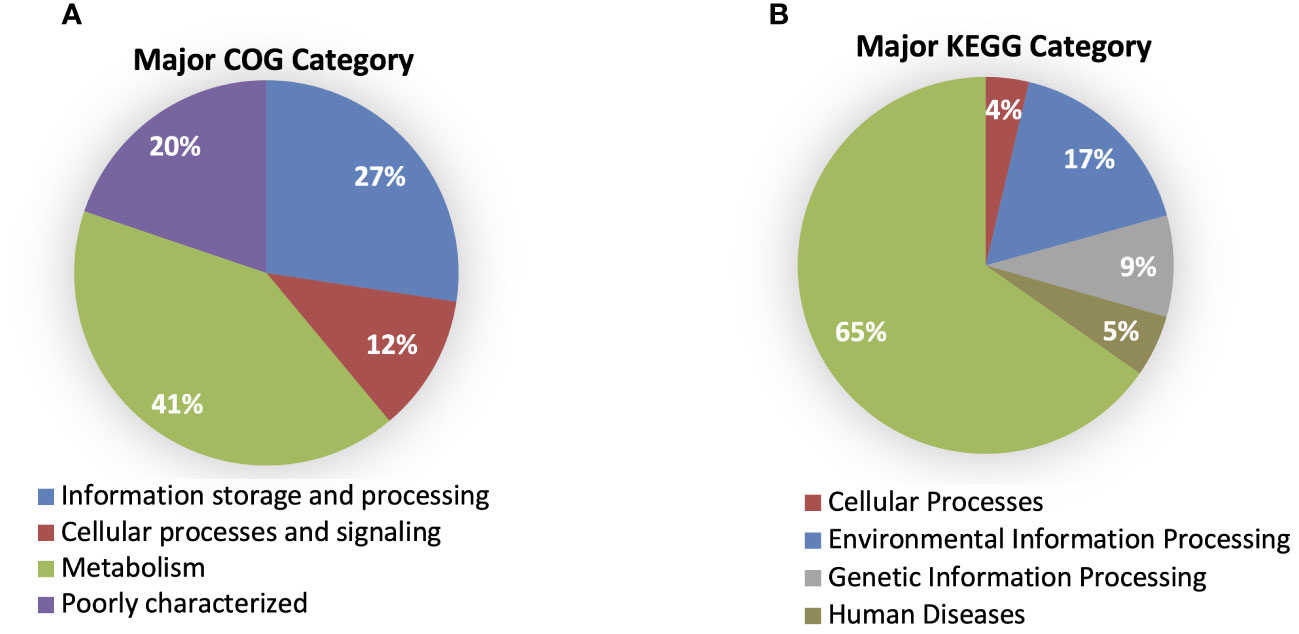
Figure 6 The % of genes annotated to major (A) COG and (B) KEGG categories for P. betavasculorum genomes.
Using the KEGG database, 73% of genes have been annotated; among them, 65% of genes were annotated to metabolic processes, 17% to environmental information processing, 9% to genetic information processing, 5% were classified to human diseases category, and 4% of genes were connected to cellular processes. In all of the six analyzed genomes of P. betavasculorum, the same number of genes classified into each KEGG category was observed. Two categories, the signaling and cellular processes and genetic information processing, were the most prominent groups and contained 16% (916–929) and 11% of genes (612–640), respectively. The third largest group was a category of genes whose function was related to processing environmental information (450–462 genes, 8%). Approximately 4.5% of the genes each fell into the category of cellular processes and metabolism of carbohydrates and proteins (Figure 6, Supplementary Table S3). A visual comparison of genome homology done using BRIG (BLAST Ring Image Generator) showed that most regions within the analyzed genomes were conserved between genomes of P. betavasculorum strains. However, there were apparent differences between them (Figure 7).
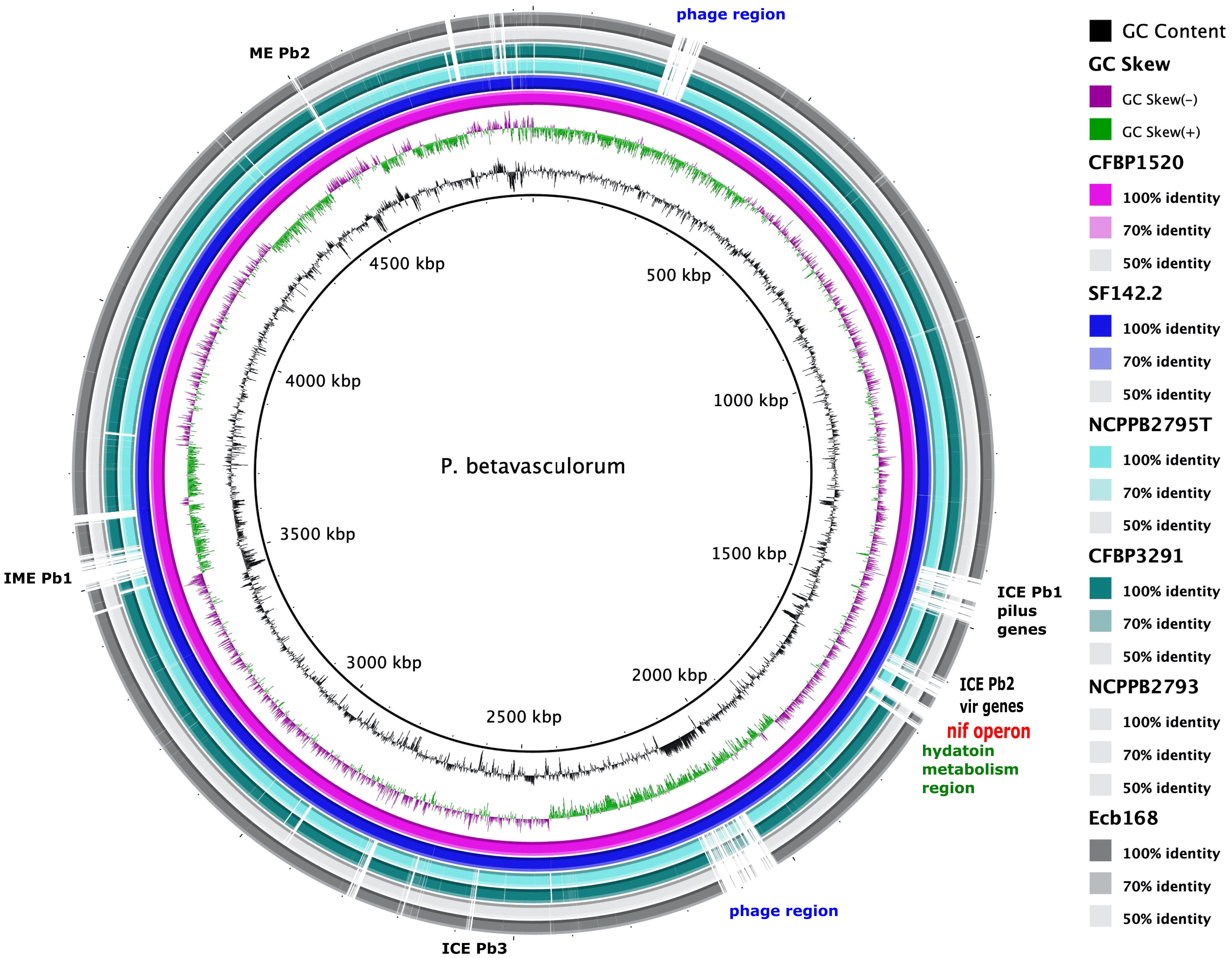
Figure 7 A graphical comparison of the whole genome sequences of P. betavasculorum strains: CFBP3291, Ecb168; NCPPB2795T, CFBP1520, and SF142.2. The circular map was created using BLAST Ring Image Generator software (Alikhan et al., 2011). The genome of strain P. betavasculorum CFBP3628 type strain was used as a reference. First rings correspond to the GC content and GC skew, respectively. Each of the depicted rings refers to one P. betavasculorum genome according to the listed coloration. White regions mark dissimilarities.
3.3.5 Pangenomics
The pangenomic analysis revealed that the pangenome of P. betavasculorum is open and comprises, on average, 4,361 gene families. Of these, 87% of genes are the core genome, and the unique genes are only 2% of the entire pangenome of this species (Figure 8).
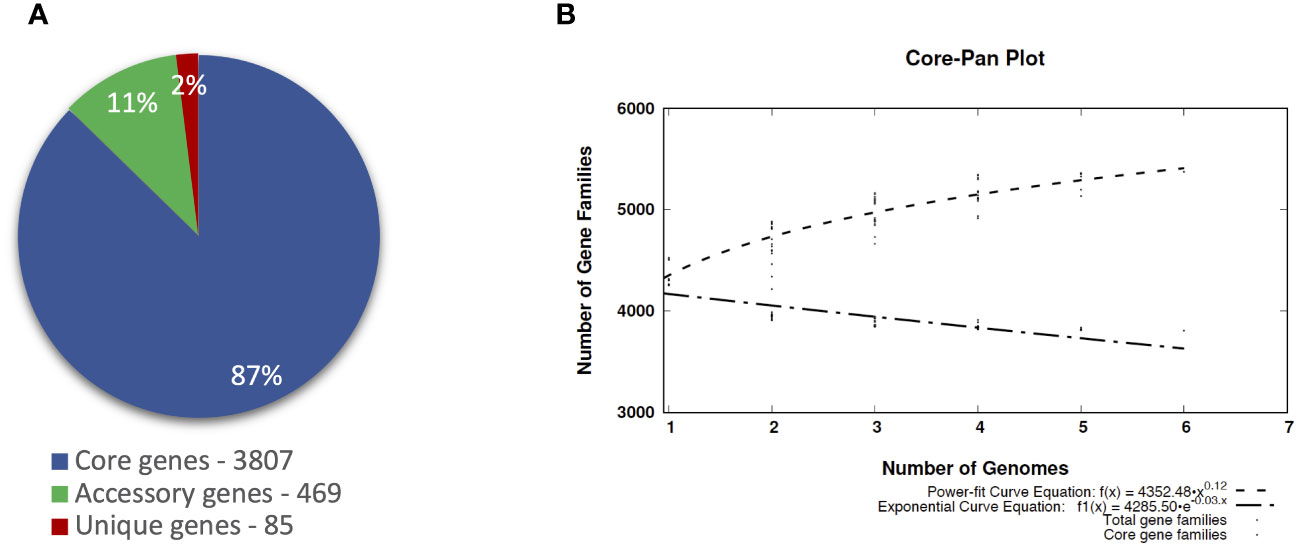
Figure 8 The pangenome profile of P. betavasculorum species calculated with BPGA software Chaudhari et al., 2016). (A) Abundancy of the core, accessory, and unique pangenome fractions within the pangenome of P. betavasculorum. (B) The number of distinct gene families referring to the pangenome size (dashed line; power-fit curve equation: f(x) = 4,224.574 · x0.14) in addition to the number of core gene families (dash-dotted line; exponential curve equation: f1(x) = 4272.24 · e−0.004x) are plotted against the number of genomes included.
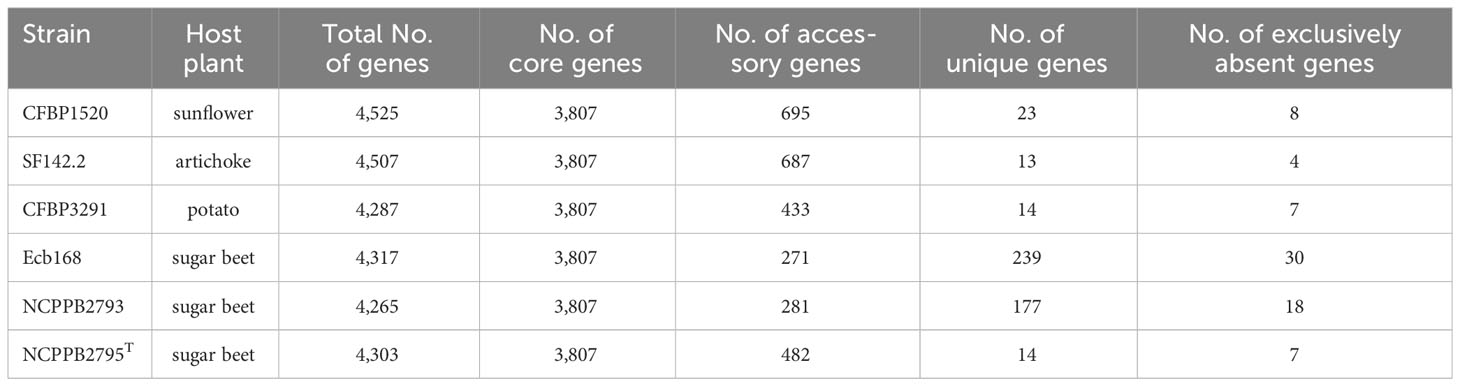
The largest number of genes, 4,525, was recorded for the strain CFBP1520 from sunflower and the smallest, 4,265, for the strain NCPPB2793 isolated from sugar beet. Interestingly, the number of genes of strains CFBP1520 and SF142.2 originating from sunflower and artichoke, respectively, were bigger than those observed for strains NCPPB2793, NCPPB2795T Ecb168, and CFBP3291 isolated from sugar beet and potato (Table 5). The complete genomes of these bacteria share 3,807 gene families, which represent over 80% of CDS in each genome. The highest unique CDS number was observed in the case of two strains isolated from sugar beet Ecb168 and NCPPB2793 (239 and 177, respectively), which was at least 10 times more than the unique CDS detected in genomes of four strains. In the case of strains SF142.2 NCPPB2795T and CFBP3291, all unique CDS are annotated as hypothetical proteins. For the strain Ecb168, there were 239 unique CDS. The majority of them, 181 (70%), are coding hypothetical proteins. The CDS that are unique for this strain are genes encoding phage-related proteins, and three different type II toxin-antitoxin systems, RelB/DinJ, YafO/YafM, and HicA/B, VirB family type IV secretion system proteins. Among 177 CDS unique for strain NCPPB2793, 65 encoded hypothetical proteins, six CDS of membrane proteins, three genes for integrases, three encoding flavodoxin, one gene encoding flavodoxin/nitric oxide synthase, and two genes for aldehyde oxidase, transposases, DNA primases, transcriptional regulators, cold-shock proteins, non-ribosomal peptide synthetase module, LysR family transcriptional regulators, and type IV secretion protein Rhs, and 83 single genes listed in Supplementary Table S4.
The independent pangenomic analysis was performed with the implementation of the GET_HOMOLOGUES software package (Contreras-Moreira and Vinuesa, 2013), which allows the calculation of core genome with the application of three different methods (COG, BBDH, and OrthoMCL) and the discrimination of intersections between strains of different origins. (Supplementary Figure S13).
The genomes of three P. betavasculorum strains, Ecb168, NCPPB2793, and NCPPB2795T isolated from sugar beet, shared only one common and unique gene that was not present in the genomes of strains isolated from other vegetables. Unfortunately, the function of this protein is not known.
Among five genes that were unique for three strains CFBP3291, CFBP1520, and SF142.2 originated from plants other than sugar beet, there were two hypothetical proteins, AlpA family transcriptional regulator, phage-related DUF4222 domain-containing protein and zinc-ribbon domain-containing protein.
Interestingly, genomes of two strains, CFBP1520 and SF142.2, isolated from plants whose tissues do not contain high sugar concentration (artichoke and sunflower) share 391 common CDS that are not present in other strains from sugar beet or potato. Many of them, 138 (35%), are hypothetical proteins. Interestingly, only these two strains harbored genes nifA, nifB, nifD, knife, nifH, nifL, nifM, nifN, nifQ, nifT, nifU, nifW, nifX, and nifZ that formed nif operon and are involved in nitrogen fixation (Figure 7, Supplementary Figure S14). Moreover, they shared two different type II TA systems, YdaS/YdaT and yafN/yafO, type IV TA system AbiEJ/AbiG, three restriction endonucleases including Eco47II-like restriction-modification (R-M) system type II, genes related to prophages, seven integrases, proteins Ync and Ynd being a part of conjugal type IV macromolecular transfer systems secretion system, outer membrane ferripyoverdine receptor from Ton and Tol transport systems, DNA-cytosine methyltransferase (EC 2.1.1.37), and two enzymes responsible for salicylate catabolism, salicylate esterase (SalE), and fumarylacetoacetate hydrolase (Fhf), and transporters and transcription regulators (Supplementary Table S4).
Four strains, CFBP3291, Ecb168, NCPPB2793, and NCPPB2795T, derived from plants with high sugar content (potato and sugar beet) share only 31 unique CDS. Among them are genetic determinants of enzymes responsible for histidine degradation [forminoglutamic iminohydrolase (EC 3.5.3.13), histidine ammonia-lyase (EC 4.3.1.3), histidine utilization repressor, imidazolonepropionase (EC 3.5.2.7), N-formylglutamate deformylase (EC 3.5.1.68), urocanate hydratase (EC 4.2.1.49)] (Supplementary Figure S15), ABC transporters (substrate-binding protein and permease-cluster 13, osmolytes), stresses-induced protein Ves (HutD), formiminoglutamic iminohydrolase (EC 3.5.3.13), and genes for phage-related proteins and 10 hypothetical proteins with unknown functions.
Moreover, comparative analysis allows us to identify 162 genes that are unique for two strains CFBP3291 and NCPPB2795T containing plasmid. Among them, 71 are coding hypothetical proteins and genes of phage-related proteins, genes mcrBC and mrr encoding type IV RM system, and genes hsdM, hsdR, and hsdS coding complete RM system type I. They shared two different type II TA systems, yafN/yafO and relE/rarE, and the solitary gene of yeeU encoding antitoxin to YeeV, and the genes of flagellin lysine methyltransferase FliB, cag pathogenicity island Cag12 family protein, and aromatic l-amino acid decarboxylase. On the contrary, four strains without plasmid have only 32 unique genes mainly encoding hypothetical proteins, phage-related proteins, integrases, relaxase, and transcriptional regulators (Supplementary Table S4).
Functional annotation of genes within the core genome, accessory, and unique genes according to both COG and KEGG databases showed that most functions focused on metabolic processes (Figure 9). However, twice as few genes determining metabolic processes belong to the accessory part of the genome and four times fewer among unique genes. The opposite trend can be observed in the case of genes related to environmental information processing and information storage processing. In the core genome, the four COGs categories are the most numerous: [J] Translation ribosomal structure and biogenesis, [P] Inorganic ion transport and metabolism, [H] Coenzyme transport and metabolism, and [F] Nucleotide transport and metabolism. In the accessory part of the genome, the most abundant is the [Q] Secondary metabolites biosynthesis transport and catabolism category, while among the unique genes, the largest are two categories, [U] Intracellular trafficking secretion and vesicular transport and [V] Defense mechanisms (Figure 10). Using KEGG database, over 70% of genes from each category belonged to the core genome. The same KEGG sub-category related to carbohydrate metabolism is the largest in core, accessory, and unique part of P. betavasculorum genomes. The second largest subcategory in the core genome gathers genes related to membrane transport; in the accessory part of the genome, it was the amino acid metabolism subcategory, while within the unique genes, replication and repair category (Supplementary Table S3).
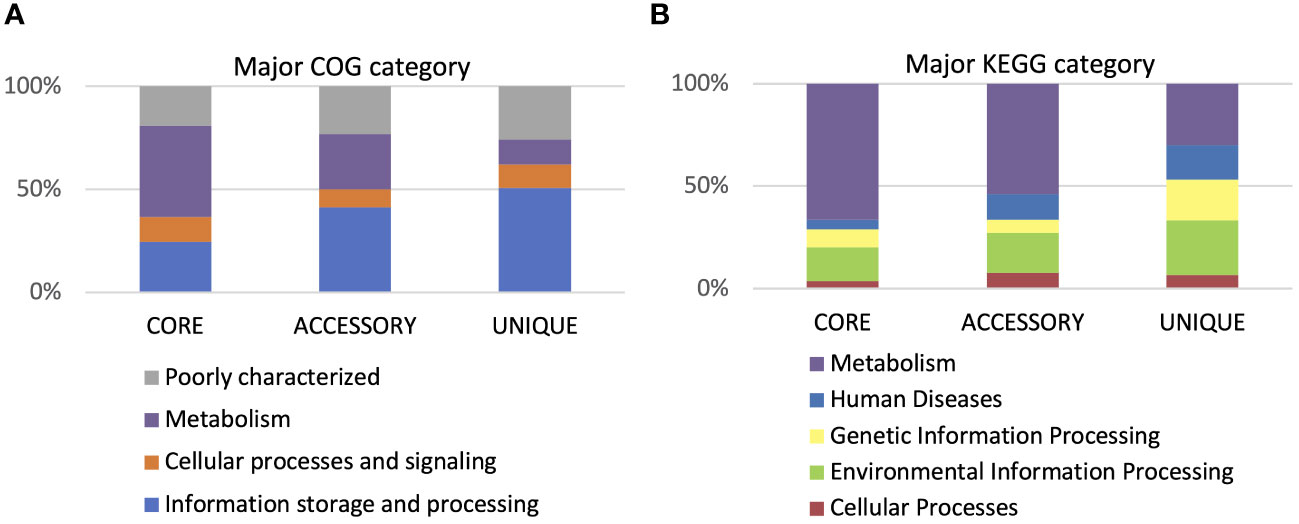
Figure 9 Gene annotation by COG and KEGG categories. (A) Major COG category and (B) major KEGG category.

Figure 10 Gene annotation by COG and KEGG sub-categories. (A) Major COG sub-category and (B) major KEGG sub-category.
3.3.6 Comparative genomics
Comparative genomics analyses were also performed to further identify distinctive traits between the P. betavasculorum and other Pectobacterium species for which genomes are available in the GenBank. Using the function-based comparison tool under RAST [29], 59 genes were predicted to be specific for P. betavasculorum (Table 6, Supplementary Table S2). Among them, we found 25 genes encoding hypothetical proteins, three genes encoding ABC transporters, two genes of transposases of insertion sequence (IS), two genes encoding major facilitator family (MSF) transporters, four encoding transcriptional regulators from LysR and GntR families, efflux transporter periplasmic adaptor subunit (hlyD), hemagglutinin repeat family protein, thiol: disulfide intercharge protein, or two phage-related proteins. Moreover, unique CDs encode proteins involved in amino acid synthesis and transport. Interestingly, P. betavasculorum is the only species of the genus Pectobacterium with genes encoding isopentyl transferase, allantoin permease, urea carboxylase and allophanate hydrolase gene cluster, genetic determinants of acetoin and butanediol biosynthesis, or genes responsible for sulfoquinovose metabolism (Supplementary Figure S16).
3.3.7 Pathogenomics
The genomic comparison of P. betavasculorum strains revealed the presence of 106 genes responsible for the synthesis of various virulence factors like plant cell wall-degrading enzymes (51) and toxins (25), and 106 genes encoding proteins budling six secretion systems (T1SS–T6SS) and outer membrane vesicles (known as T0SS) (Table 7).
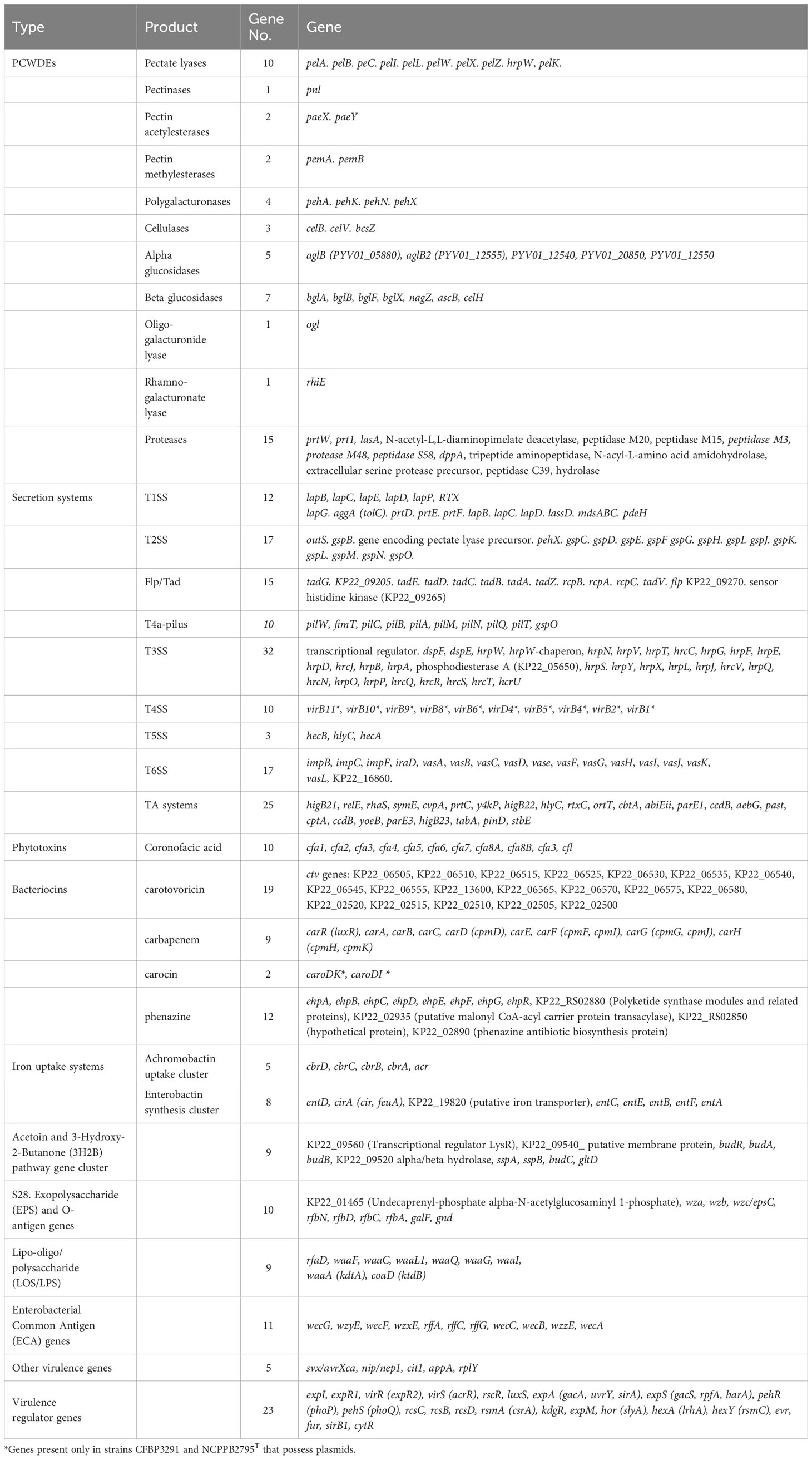
Table 7 List of genes encoding virulence factors and virulence regulators detected in the in P. betavasculorum genomes.
3.3.7.1 PCWDEs
A total of 40 known or putatively related genes encoding pectinases, cellulases, and proteinases were identified (Table 7). The genomes of P. betavasculorum, like other Pectobacterium species, contain 10 genes: pelA, pelB, pelC, pelZ, pelI, pelK, encoding pectate lyases, pelW—pectate disaccharide lyase, including pelX—exopolygalacturonate lyase, hrpW that encodes type III effector HrpW, hairpin with pectate lyase domain, and pectate lyase L precursor pelL. However, strains of P. betavasculorum, as the only species of the genus Pectobacterium, lack the pelY gene encoding a periplasmic pectate lyase. In addition, the following genes were detected: pnl for a pectin lyase; pemA and pemB for pectinesterase; paeX and paeY for pectin acetylesterase; pehX, pehN, pehA, and pehK for polygalacturonases; ogl for an oligogalacturonide lyase; and rhiE for a rhamnogalacturonate lyase.
Three genes in P. betavasculorum are involved in cellulose degradation, including glycoside hydrolase celB(celS), and two endoglucanase-encoding genes, celV and bcsZ. Moreover, five beta-glucosidase-encoding genes bglA, bglB, bglD, nagZ, and celH, and five an alpha-glucosidase-encoding genes were detected. An operon of nine genes encoding cellulose synthetase, including bcsC, bcsA, bcsB, bcsG, bcsE, bcsZ, bcsQ, bcsF, and bcsR, was also identified in all of six analyzed P. betavasculorum genomes. Moreover, 21 genes encoding proteases or peptidases were detected.
3.3.7.2 Secretion systems
Six types of secretion systems (TSS) were detected in the genomes of analyzed six P. betavasculorum strains. In total, 12 genes were involved in the secretion system Type I (T1SS), 17 in Type II (T2SS), 32 in Type III (T3SS), 10 in Type IV (T4SS), three in Type V (T5SS), and 17 in Type VI (T6SS) secretion systems (Table 7).
P. betavasculorum strains and other Pectobacterium species possess genes related to T1SS, prtDEF, lssBEF, lapBCDEG, aggA, and pdeH, while genes encoding the hasDEF are missing in their genomes. The complete T2SS Out secretion system and the twin-arginine translocation (Tat) protein system are present in the analyzed genomes of P. betavasculorum strains. The T3SS gene cluster in P. betavasculorum strains consists of hypersensitive response and pathogenicity conserved genes (hcr)/hypersensitive response and pathogenicity or hairpin proteins (hrp), genes encoding DspE (disease-specific effector protein E), DspF (disease-specific chaperone protein F), HrpW- and HrpK-T3SS effector proteins, and chaperon proteins. Interestingly, the type IV secretion system consisting of 12 vir genes was detected only in the case of two strains CFBP3291 and NCPPB2795T, as it was located on the plasmid that is harbored by these strains. The complete set of genes (pilABCDTQMNOPVW) encoding type IV pilus machinery was present in all of P. betavasculorum genomes. The type Vb secretion system (T5SS) consisting of two genes, hecB and hecA, encoding hemolysin activation protein and hemolysin/hemagglutinin-like protein, were found in P. betavasculorum genomes. In the case of the T6SS, only the core part of this injectosome, delivering toxic effector proteins into bacterial or host cells, was present. The 17 genes encode the outer membrane lipoprotein (VasD), inner membrane proteins (ImpL/VasK and ImpK/VasF), ATPase (ClpV), and regulatory proteins or structure proteins (ImpB, ImpC, TssE, ImpG, ImpH, ImpI, ImpJ, VasH, VasI, VasJ, and VasL). While the gene encoding hypothetical proteinKP22_16860, gene vgrG (valine–glycine repeat protein G), and hcp2 gene that encodes extracellular structural components of the secretion machine of P. betavasculorum strains were absent in their genomes. In addition, genes encoding Sigma-fimbriae uncharacterized paralogous subunit sF-SU1, chaperone protein, usher protein, and tip adhesin forming T7SS (Chaperone/Usher pathway, CU) were identified.
3.3.7.3 Conjugative and mobile elements
The genomes of P. betavasculorum strains were screened for the presence of integrative conjugative elements (ICEs) and integrative mobile elements (IMEs) that are known to affect the pathogenicity and lifestyle of phytopathogens (de Assis et al., 2022). In total, four different ICE elements harboring genetic determinant of T4SS and six IME elements were detected. Integrative conjugative elements were present in all of P. betavasculorum genomes except strain Ecb168, which harbor four different IME elements only, whereas strain SF142.2 has only ICEs. The ICEAPb1 element encoding type IV secretion pilus proteins was present in the genomes of four strains, CFBP1520, SF142.2, CFBP3291, and NPPB2795T, while T4SS-like element ICEPb2 harboring vir genes was detected in four strains CFBP1520, SF142.2, NPPB2793, and NPPB2795T. Of the six IMEs detected, only IMEPb1 was unique to one P. betavasculorum strain CFBP1520; the others were present in two or three of the analyzed genomes (Supplementary Table S6).
3.3.7.4 Prophages
Between 6 and 10 prophage regions were detected in all P. betavasculorum genomes analyzed (Supplementary Table S7). Most genomes differed in phage regions. The exceptions were strains CFBP1520 and SF142.2, in which 10 phage regions each were detected in their genomes, nine of which were the same. Interestingly, the phage BcepMu (NC_005882) from Burkholderia cenocepacia was detected in all analyzed genomes. The prophage region in strain CFBP3291 encodes type II toxin-antitoxin system CcdA/CcdB.
3.3.7.5 Toxins and bacteriocins
Coronofacic acid (cfa) biosynthetic cluster consisting of 10 genes (cfa1-8A and B, and cfl), the carotovoricin (ctv) biosynthetic cluster, the carbapenem (carRABCDEFGH) biosynthesis cluster, and phenazine antibiotic biosynthesis gene cluster (ehpABCDEFGR) were detected in all of the six analyzed P. betavasculorum genomes. The carocin biosynthesis cluster consisting of two genes caroDK encoding bacteriocin killing protein (carocin D) and caroDI bacteriocin immunity protein was present in the genomes of two strains CFBP3291 and NCPPB2795T only (Table 7). Those two strains possess plasmid; however, carocin genes were located on the chromosome.
The phytotoxin operon of biosynthesis of coronofacic acid (cfa) is located in the P. betavasculorum in the genomic island, like in the other species, P. atrosepticum, P. brasiliense, P. peruviense, and P. actinidiae (Panda et al., 2016; Arizala and Arif, 2019).
The detected genes of bacteriocins, carotovoricin, phenazine, and carocins are not unique for P. betavasculorum and are also produced by other Pectobacterium species (Marquez-Villavicencio et al., 2011). Carotovoricin is a phage tail-like bacteriocin encoded by a cluster of genes (ctv) located in the genetic island containing prophage remnant genes. In P. betavasculorum, the cluster of genes ctv was identified by the Phaster platform as a region similar to prophage BcepMu from Burkholderia cenocepacia (NC_005882) (Supplementary Figure S5). Such genomic island was described earlier in other Pectobacterium species, P. atrosepticum SCRI1043 and P. carotovorum (Nguyen et al., 2002; Yamada et al., 2006) or P. brasiliense 1692 (Shyntum et al. 2019). Carbapenem is an antibiotic produced by many bacteria including various Pectobacterium species (Marquez-Villavicencio et al., 2011). Synthesis of this bacteriocin is regulated by quorum-sensing molecules N-(3-oxohexanoyl)-L-homoserine (OHHL) and occurs simultaneously with the production of PCWD enzymes (Põllumaa et al., 2012). Oxygenic conditions suppress it (Shyntum et al. 2019). Carocin D colicin-like bacteriocin with DNase activity is also produced by P. carotovorum (Roh et al., 2010).
3.3.7.6 Iron acquisition systems
Two distinct gene clusters involved in iron uptake were detected in all of the P. betavasculorum genomes. The Achromobactin uptake cluster consists of five genes, cbrABCD and acr, while the enterobactin synthesis cluster is built from eight genes, entAFBECD, cirA, and gene putative encoding iron transporter (Table 7). The ability to chelate iron was confirmed for all strains by using CAS plate assay (Supplementary Figure S5). P. betavasculorum strains do not pose genes encoding other iron uptake systems, like ferric citrate uptake cluster (fecIRABCDE), heme acquisition cluster (hasRADEF), hemin storage cluster (hmsHFRS), and ferredoxin uptake cluster (fusBACD), which were identified in the genomes of other Pectobacterium species (Arizala and Arif, 2019).
3.3.7.7 The production of auxin and acetoin and 2,3-butanediol, other virulence genes, and virulence regulators
The indole-3-acetic acid (IAA) and 3-hydroxy-2-butanone (3H2B) pathways gene clusters and genes encoding avirulence protein (svx/avrXca) necrosis inducing virulence protein/necrosis and ethylene-inducing peptide 1 (nip/nep1), citrate transporter (cit1), glucose-1-phosphatase (appA), ribosomal protein RplY (rplY) were present in all of P. betavasculorum genomes. What is more, 12 virulence regulators like the two-component PhoP-PhoQ system, RcsCBD phosphorelay system, Gac/Rsm system, the quorum-sensing N-acyl homoserine lactone (AHL) including ExpI, a LuxI-type AHL synthase, two AHL receptors ExpR1 and ExpR2, two-component transcriptional response virulence regulator (evr), ferric uptake regulation protein FUR, sirohydrochlorin ferrochelatase (sirB1), transcriptional regulators KdgR and ExpM (regulator of RpoS), transcriptional regulator SlyA, and transcriptional (co)regulator CytR were detected (Table 7). The production of IAA, acetoin, and 3-hydroxy-2-butanone (3H2B) was confirmed experimentally (Supplementary Figures S2, S7, and Supplementary Table S1). We detected the same set of genes as (Arizala and Arif, 2019) in earlier analyses, which were performed for only two P. betavasculorum genomes (JQHM00000000 and JQHL00000000) that were available in the GenBank. Similarly, like Arizala and Arif, 2019, in none of the six P. betavasculorum genomes did we detect the presence of other virulence factors, such as metal-dependent isothiocyanate hydrolases (saxA1, saxA2, and saxA3) or Erwinia virulence factor Evf (Table 7).
3.3.7.8 Polymer synthesis
The newly sequenced genomes of P. betavasculorum strains had the same repertoire of genetic determinants responsible for biopolymer synthesis, as it was described for strains NCPPB2793 and NCPPB25T by Arizala and Arif (2019). Regardless of the origin of P. betavasculorum strains, in all genomes, the following gene clusters encoding bacterial cellulose synthase complex (bcsCABGEZQFR), capsular polysaccharide (cps) biosynthesis, Enterobacterial Common Antigen (ECA), exopolysaccharide (EPS), and O-antigen biosynthesis and lipo-oligo/polysaccharide (LOS/LPS) were detected (Table 7).
3.3.7.9 Production of secondary metabolites
In the genomes of all five strains of P. betavasculorum, clusters of genes for the biosynthesis of 12 secondary metabolites such as coronophoric acid, 1.6-phenazinedimethanol, amonabactin P 750, linear peptides containing azol(in)e, O-antigens, siderophores, homoserine lactones, beta-lactones containing a protease inhibitor, β-lactams, bicornutin A, and rhizomide were detected.
Xenoamicin A biosynthetic gene cluster was unique for strains IFB5271 and SF142.2 that did not originate from sugar beets. Gene clusters responsible for synthesizing Le-pyrrolopyrazines were a characteristic for strain IFB5271 from sunflower, while gacamide A and holrhizin were unique for strain SF142.2 from artichoke. Strains from sugar beets have different compositions of gene clusters predicted to encode non-ribosomal peptides. For type strains NCPPB2795T, these were teixobactin, syringomycin, and N-myristoyl-D-asparagine/cis-7-tetradecenoyl-D-asparagine/(R)-N1-[(S)-5-oxohexan-2-yl]-2-tetradecanamidosuccinamide. Strain NCPPB2793 were nunapeptin/nunamycin, microsclerodermin, and thanamycin (Supplementary Table S8).
4 Discussion
The currently ongoing climate changes have the most significant impact in agriculture. Rising land temperatures, decreased water availability, and increased soil salinity related to, among others, the use of excessive amounts of plant protection products or fertilizers have a negative impact on plant growth and development. At the same time, intensive international trade of plant materials, such as seeds, tubers, bulbs, ornamentals, herbs, or plant food, results in the intensive spread of plant-associated bacteria, including pathogens from the Pectobacterium genus (Smoktunowicz et al., 2022). Climate warming makes it easier for phytopathogens to acclimatize to the new climate zone and/or settle in a new host. In order to effectively detect and monitor the spread of phytopathogens, a comprehensive genetic and phenotypic characterization and the broadest possible knowledge about their biology, adaptive abilities, and the possibility of changing hosts are necessary.
Therefore, in this study, the research was undertaken to understand the biology of P. betavasculorum, which causes diseases of various plants that are an important part of the food market and are also used as raw materials in biotechnology. The polyphasic analysis was performed to verify whether strains of P. betavasculorum are adapted for the colonization of plants with high sugar or lipid content.
The comprehensive phenotypic characteristic based on 130 phenotypic features (Figure 2; Supplementary Tables S1, S2) of14 strains isolated from sugar beet, potato, sunflower, and artichoke isolated in Europe, Africa, and Central America showed that P. betavasculorum strains differ from other Pectobacterium species, and strains CFBP1520 and SF142.2 from sunflower and artichoke have nearly identical biochemical properties. These observations agreed with previous studies of Thomson (1981) and Gardan et al. (2003) except the fact that strains isolated from sugar beet were phenotypically diverse and could not be differentiated from strains isolated from other plants. Strain NCPPB2795T was more similar to strains CFBP1520 and SF142.2 from sunflower and artichoke than with other sugar beet strains. Strain CFBP3291 from potato was more similar to strains isolated from sugar beet. In contrast, strain Ecb168 differed significantly from all P. betavasculorum strains, formed smaller colonies, and was characterized by a slower growth rate.
The results of metabolic profiling of P. betavasculorum strains showed very high metabolic plasticity of these bacteria, which is a fundamental ability that allows them to adapt their metabolic status to specific requirements during growth in response to various stimuli or in response to stress conditions. P. betavasculorum strains showed the ability to use various polyols, sugars, and polymers as a building material and energy source (Supplementary Figure S3) and plant extracts (Supplementary Figure S4) regardless of the host plant from which the bacteria were isolated.
All tested strains are characterized by the same high adaptability to changing environmental conditions, showing effective growth in the temperature range from 15°C to 37°C, salinity gradient of 0%–6%, pH from 2 to 11, and in conditions of limited access to water. A similar ability to grow under osmotic stress caused by high sucrose concentration (up to 20%) is demonstrated by strains isolated from sugar beet, sunflower, and artichoke (Supplementary Figure S9). Those results indicate that due to their high adaptability, P. betavasculorum may be able to spread to various climatic zones, especially in times of ongoing climate change. In addition, due to their ability to use various nutrients, P. betavasculorum strains are able to colonize various plant hosts. It has been shown that the ability to grow in a minimal medium containing 10% tissue extracts from eight plant species as the only source of organic compounds (Supplementary Figure S4). The tissues of most of the tested plant species contain significant amounts of sugars. However, it was not observed that strains from sugar beet were characterized by a faster growth rate than strains isolated from other plants. All tested strains showed weaker growth in the presence of blueberry and pitaya extracts. This fact can be correlated with the high content of flavonoids, including anthocyanins, known for their antibacterial properties (Cisowska et al., 2011), in the tissue of these plants. However, it was not observed that the SF142.2 strain isolated from artichoke, also containing large amounts of flavonoids, had a more remarkable ability to grow in the presence of blueberry and pitaya extracts than strains from sugar beet or sunflower (Supplementary Figure S4).
In order to compare the pathogenicity of the tested strains, we used potato tubers and chicory leaves, as those tests are most commonly performed on those plants, which allows comparison of their virulence with other Pectobacterium species. All tested P. betavasculorum strains showed the ability to macerate potato tubers and chicory leaves in laboratory conditions, in tests in which the plants were inoculated by wounding. No differences were observed in the maceration ability of plant species depending on the plant species from which the bacteria were isolated. Strains isolated from sugar beet or potato did not cause more intense symptoms of soft rot in tests on potato tubers than strains from sunflower or artichoke. Like the artichoke strain, it did not rot the chicory leaves more severely (Table 1). The obtained results are not surprising as the comparative analyses of genomes revealed that all analyzed strains had the same set of genes encoding PCWD enzymes. However, it should be highlighted that even though P. betavasculorum strains do not possess the pelY gene, they do not differ in terms of virulence from strains of other Pectobacterium species as was shown in our earlier work (Waleron et al., 2014).
The phenotypic analysis, metabolic profiling, and pathogenicity tests of P. betvasculorum strains did not show that the strains isolated from plants characterized by high or low sugar or lipid content differed much in terms of biochemical properties, virulence, or adaptability.
However, bacteria can adapt to various environmental conditions also by modifying the lipid composition of their membranes. Unsaturated fatty acids and a decrease in the fatty acids chain length increase the resistance of microorganisms to lower temperatures, especially anteiso-branching (Suutari and Laakso, 1994). Physiological tests showed that all P. betavasculorum strains could grow well at temperatures between 15°C and 28°C. The results of FAME analysis showed that all analyzed P. betavasculorum strains demonstrate the same fatty acid composition but differ in their proportions. The lack of change in the fatty acid composition may be because, for FAME analysis, all strains were cultured in the same rich medium at the same 28°C temperature, which is optimal for the growth of P. betavasculorum. Surprisingly, the differences in the proportions of fatty acids were observed between P. betavasculorum strains, and they can be connected with differences in the optimal growth temperatures of the host plants from which the bacteria were isolated. Bacterial strains isolated from sugar beets are characterized by higher amounts of UFAs than strains from sunflower and artichoke (Figure 3A), which correlates with the optimal growth temperatures of the host plants. Sugar beets, which are grown mainly in the northern hemisphere, show more significant growth and yield more abundantly at lower temperatures than in temperatures that are the best for growing sunflower or artichoke. The increase in UFAs during growth at cold temperatures is observed. Increased relative abundance of UFAs increases membrane fluidity at cold temperatures (Diomande et al., 2014; de Carvalho and Caramujo, 2018; Français et al., 2021). Increased proportions of UFA as growth temperature decreases have been reported for E. coli (Marr and Ingraham, 1962) and Vibrio spp (Bhakoo and Herbert, 1979). On the contrary, higher amounts of SFA than UFA were recorded for P. betavasculorum strains isolated from sunflower and artichoke originating from warm climates (Figure 3A, Table 3). Likewise, it was noticed that, in the case of E. coli, when the growth temperature was elevated from 15°C to 43°C, the ratio of saturated to unsaturated fatty acids in the membrane also increased (Sinensky, 1971; 1974). Similar observations apply to MUFA, which were observed in greater amounts in P. betavasculorum strains isolated from sugar beet than in strains from sunflower and artichoke (Figure 3A, Table 3). This observation is consistent with previous studies that reported higher MUFA proportions at cold than at optimal temperatures in B. cereus (Kaneda, 1972; Haque and Russell, 2004; de Sarrau et al., 2012; Chazarreta Cifré et al., 2013).
The total amounts of OCFA and MUFA are higher in sugar beet strains than in strains from other plants (Figure 3B, Table 3). Higher sucrose content might favor the fermentation process, leading to a decrease in the pH value. Bacteria to adapt to acidic conditions increase the OCFA and MUFA contents and increase membrane fluidity, as was observed in the case of E. coli (Gianotti et al., 2009). Moreover, monounsaturated fatty acids and odd-chain long fatty acids can help maintain the microbial membrane’s fluidity in anaerobic conditions (Fulco, 1983; Jenkins, 1993).
The high sucrose content available for P. betavasculorum strains in sugar beet tissue might result in such adaptation as lower BCFA content. It was observed that cellulolytic bacteria increase membrane fluidity by increasing BCFA in cellulose’s presence. An increase in BCFA was observed after changing from a medium rich in simple sugars to one containing cellulose. The increase in membrane fluidity allows the translocation of large polymers and the secretion of cellulolytic enzymes outside the cell to hydrolyse cellulose (Vlaeminck et al., 2005; Mitchell et al., 2023). It can be hypothesized that P. betavasculorum isolated from sugar beet with substantial amounts of readily available sucrose contain less BCFA than strains isolated from artichoke and sunflower, which are forced to hydrolyze polymers as a result of less availability of simpler compounds that do not require hydrolysis. It should be noted, however, that all strains tested, including those from sugar beet, showed cellulolytic activity and possessed the same number (three) of genes encoding cellulases.
P. betavasculorum strains Ecb168 and NCPPB2795T, isolated from sugar beet, had a higher proportion of unsaturated fatty acids than strains SF142.2 and CFBP1520 isolated from artichoke and sunflower, respectively. On the other hand, strains SF142.2 and CFBP1520 had more polyunsaturated and branched fatty acids, which could compensate for the lower amounts of unsaturated fatty acids. P. betavasculorum strains, in general, also possessed the highest amounts of fatty acids of low chain length and palmitic (16:0) and palmitoleic acids (16:1). What is more, unsaturated long-chain fatty acids increase the permeability of the membranes and cell resistance to osmotic stress (Montgomerie et al., 1973). Therefore, the higher proportions of unsaturated fatty acids in Ecb168 and NCPPB2795T strains could help them adapt to high concentration of sucrose found in sugar beetroot, which may be as high as 20% (Getz, 2000).
Finally, it can also be hypothesized that the differences in the amounts of SFA and BCFA observed between P. betavasculorum strains may result from the adaptation of these bacteria to use nutrients available in the host plant’s tissues for FA synthesis. Strains adapted to grow in plant tissues with a high sugar content modulate the FA composition in membranes more through the synthesis of SFA because sugars are substrates needed for their synthesis (Salvador et al., 1993), while the primers for BCFA synthesis are branched amino acids (Kaneda, 1991; Kaiser et al., 2016; Dullius et al., 2020), which is why they are more abundant in isolated strains of artichoke or sunflower than in strains derived from sugar beet.
The genetic characterization of P. betavasculorum showed that analyzed strains are genetically diverse, and three different fingerprinting profiles were observed. Strains CFBP152 from sunflower and SF142.2 from artichoke are almost identical and differ from the sugar beet strains. No correlations between geographic origin and year of isolation could be determined (Table 1, Supplementary Figure S12A). However, further genetic characterization of P. betavasculorum strains agree with the results of phenotypic analyses and shows that strains from sugar beet are not separated from strains originating from other plant species. It should be noted that observed phylogenetic relationships between P. betavasculorum strains depend on the genomic method used. Based on MLSA (Supplementary Figure S11), core protein-based phylogeny (Figure 5) and POCP (Supplementary Figure S12B) strain CFBP3291 from potato is more related to strains from sugar beet NCPPB2793, NCPPB2795, and Ecb168 and all together are a bit distant from strains CFBP1520 and SF142.2 derived from sunflower and artichoke that are most similar to each other. In contrast, ANI (Supplementary Figure S12C) and AAI (Supplementary Figure S12D) methods show that NCPPB2795 and CFBP3291 strains are more distant from other P. betavasculorum strains.
Pangenomic analyses allowed the detection of common and unique features for P. betavasculorum strains isolated from different plants. The sugar beet and potato strains do not have the nif operon that enabled fixation of atmospheric nitrogen but are equipped with the hut genes encoding enzymes allowing for histidine degradation. In the case of strains originating from the artichoke and sunflower, it is the opposite. The ability to degrade histidine is common among bacteria, and the enzymes and genes of Hut pathway are conserved. The complete catabolism of histidine to glutamate, ammonia, and either formate or formamide allows the use of histidine as a carbon and nitrogen source. Moreover, intermediates of the pathway might serve as signaling molecules that affect bacterial virulence or biofilm formation (Bender, 2012). In the presence of high sugar content, as it is in sugar beet or potato tuber tissues, the catabolic repression inhibits the histidine degradation, but under nitrogen starvation, the repression is abolished, and then, histidine degradation provides both the carbon and nitrogen necessary for bacteria growth. In the case of strains that lack hut genes but possess the nif operon, nitrogen can be obtained by its fixation from the atmosphere. P. betavasculorum strains isolated from plants with high sugar content (sugar beet and potato) lacked genes of atmospheric nitrogen fixation. It is common for sugar beet microbiome and is correlated with field fertilization (Wolfgang et al., 2023). Moreover, it was shown that NifA is a regulator that represses auxin synthesis. NifA mutant exhibited higher pathogenicity and root infection (Bellés-Sancho et al., 2022). It can be, therefore, a factor that increases the virulence of P. betavasculorum strains without nif operon genes on sugar beet than P. betavasculorum strains derived from other host plants. The presence or absence of hut genes can be related to the histidine content in plant tissue. The red beets, potato tubers, carrot, chicory, or eggplant contain 0.02–0.04 g of histidine per 100 g of plant tissue. A total of 100 g of sugar beet and potato starch have 0.3 g and 0.17 g of histidine, while in artichoke and sunflower seeds, this amino acid was not detected (https://fitaudit.com/categories/fvs/histidine). However, free histidine was present in sunflower leaves but was absent in the bound amino acids (Roy et al., 2013).
The interchangeable occurrence of nif and hut genes in P. betavasculorum strains correlated with the host plant from which they were isolated can be considered an adaptation of the bacteria to the host plants, enabling optimal use of the nutrients available in their host tissues. However, to confirm this hypothesis, further in-depth research should be conducted on the ability of P. betavasculorum to colonize and cause disease symptoms in various host plants.
Comparative genomics of P. betavasculorum with other Pectobacterium species revealed several unusual features of this species that might have an influence on their environmental faintness.
In the genomes of P. betavasculorum, the genetic determinants of enzymes allowing for the sulfoquinovose utilization were identified. Sulfoquinovose is a product of hydrolysis of plant sulfolipids, which are present in plant membranes. The enzyme sulfoquinovose isomerase carries out the isomerization of sulfoquinovose to 6-deoxy-6-sulfofructose (Liu et al., 2023). In the sulfur deficiency conditions, which are typical for intensively cultivated soils, the presence of sulfur-scavenging enzymes may be advantageous (Roy et al., 2003).
Another important features unique for P. betavasculorum are cytokines, which are hormones promoting plant growth (Wei et al., 2023). P. betavasculorum strains possess isopentenyl transferase (IPT) enzyme gene in their genomes and several other genes with IPT activity, such as tRNA-IPTs and adenylate IPTs, which can produce various forms of cytokines, including the most active form, trans-Zeatin. Cytokines produced by bacteria may promote plant growth and enhance their resistance under stress conditions (Wei et al., 2023; Pereira et al., 2024). However, some phytopathogenic bacteria increase cytokine production at the infection site, which lengthens their survival in plant tissues (Wei et al., 2023). Therefore, cytokine production may contribute to enhanced virulence.
What is more, P. betavasculorum strains pose allantoin permease, or urea carboxylase and allophanate hydrolase gene cluster, which enable it to extract nitrogen from purines under anaerobic conditions in nitrogen starvation (Izaguirre-Mayoral et al., 2018).
The results of pathogenomic analyses correspond well with the phenotypic characterization of the P. betavasculorum strains. All strains have the same wide repertoire of virulence factors and an almost identical set of genes responsible for the production of PCWDEs, toxins, bacteriocins, siderophores, plant growth hormones, and volatile compounds influencing the interaction with plants, and are equipped with a set of seven secretion systems.
It is worth noting that the genes unique to P. betavasculorum selected based on comparative genomics can be used to develop a method for detecting and identifying this species, which is not currently available. In consequence, the number of P. betavasculorum strains available in international collections is very limited. Strains isolated from sugar beet predominate. As a result, it was only possible to use three strains from other plants in our study. When more strains become available, further studies are needed.
It is important, as no methods of protection or effective eradication of Pectobacterium exist. An effective detection method could prevent the spread of this pathogen and limit losses in beetroot, sunflower, and potato crops. Pectinolytic bacteria can spread rapidly. For example, Dickeya solani and P. brasilense were first observed on ornamental plants and vegetables grown in greenhouses in colder climates before spreading to the field crops (Sławiak et al., 2009; van der Wolf et al., 2014; Waleron et al., 2015; Jonkheer et al., 2021; Toth et al., 2021).
Summarizing the results of the analyses carried out within this research, it can be stated that species P. betavasculorum is a group of bacteria that specialize to colonize and infect particular plants. However, thanks to their metabolic plasticity, bacteria can easily change the host plant and adapt to changing environmental conditions, and thus, they are able to spread on different climatic regions. Therefore, the species P. betavasculorum should be considered an important pathogen in agriculture, especially in the era of progressive climate change and intensification of international trade in plants, especially on the food market.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author contributions
MB-B: Writing – original draft, Formal analysis, Investigation, Visualization. MS: Formal analysis, Investigation, Writing – original draft. DH: Formal analysis, Investigation, Visualization, Writing – original draft. JJ: Formal analysis, Investigation, Methodology, Resources, Validation, Visualization, Writing – original draft. MMW: Data curation, Formal analysis, Investigation, Validation, Visualization, Writing – original draft. JG: Data curation, Formal analysis, Investigation, Writing – original draft. AM: Formal analysis, Investigation, Visualization, Writing – original draft. TS: Methodology, Writing – original draft. KW: Conceptualization, Data curation, Methodology, Supervision, Writing – original draft, Writing – review & editing. MW: Conceptualization, Data curation, Formal Analysis, Funding acquisition, Methodology, Project administration, Resources, Supervision, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. The study was funded by the Polish National Science Centre grants OPUS9-2015/17/B/NZ9/01730 and OPUS18-2019/35/B/NZ9/01973 and Task D801-23, Plant Protection and Biotechnology Laboratory research capacity funds. MS was supported by project POWR.03.02.00-00-I035/16-00 co-financed by the European Union through the European Social Fund under the Operational Programme Knowledge Education Development 2014–2020.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2024.1352318/full#supplementary-material
References
AbuOun, M, Jones, H, Stubberfield, E, Gilson, D, Shaw, L. P., Hubbard, ATM, et al (2021). A genomic epidemiological study shows that prevalence of antimicrobial resistance in Enterobacterales is associated with the livestock host, as well as antimicrobial usage. Microb. Genom. 7(10), 000630. doi: 10.1099/mgen.0.000630
Alikhan, N. F., Petty, N. K., Ben Zakour, N. L., Beatson, S. A. (2011). BLAST Ring Image Generator (BRIG): Simple prokaryote genome comparisons. BMC Genomics. 12, 1–10. doi: 10.1186/1471-2164-12-402/TABLES/3
Andro, T., Chambost, J. P., Kotoujansky, A., Cattaneo, J., Bertheau, Y., Barras, F., et al (1984). Mutants of Erwinia chrysanthemi defective in secretion of pectinase and cellulase. J. Bacteriol. 160, 1199–1203. doi: 10.1128/JB.160.3.1199-1203.1984
Arizala, D., Arif, M. (2019). Genome-wide analyses revealed remarkable heterogeneity in pathogenicity determinants, antimicrobial compounds, and CRISPR-Cas systems of complex phytopathogenic genus pectobacterium. Pathogens 8, 247. doi: 10.3390/pathogens8040247
Avrova, A. O., Hyman, L. J., Toth, R. L., Toth, I. K. (2002). Application of amplified fragment length polymorphism fingerprinting for taxonomy and identification of the soft rot bacteria Erwinia carotovora and Erwinia chrysanthemi. Appl. Environ. Microbiol. 68, 1499–1508. doi: 10.1128/AEM.68.4.1499-1508.2002
Baghaee-Ravari, S., Rahimian, H., Shams-bakhsh, M., Lopez-Solanilla, E., Antúnez-Lamas, M., Rodriguez, P. (2011). Characterization of Pectobacterium species from Iran using biochemical and molecular methods. Eur. J. Plant Pathol. 129, 413–425. doi: 10.1007/s10658-010-9704-z
Bellés-Sancho, P., Liu, Y., Heiniger, B., von Salis, E., Eberl, L., Ahrens, C. H., et al. (2022). A novel function of the key nitrogen-fixation activator NifA in beta-rhizobia: Repression of bacterial auxin synthesis during symbiosis. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.991548
Bender, R. A. (2012). Regulation of the histidine utilization (hut) system in bacteria. Microbiol. Mol. Biol. Rev. 76, 565–584. doi: 10.1128/MMBR.00014-12
Bhakoo, M., Herbert, R. A. (1979). The effects of temperature on the fatty acid and phospholipid composition of four obligately psychrophilicVibrio Spp. Arch. Microbiol. 121, 121–127. doi: 10.1007/BF00689975
Blin, K., Shaw, S., Augustijn, H. E., Reitz, Z. L., Biermann, F., Alanjary, M., et al (2023). A’ntiSMASH 7.0: new and improved predictions for detection, regulation, chemical structures and visualisation. Nucleic Acids Res. 51(), W46–W50. doi: 10.1093/NAR/GKAD344
Chaudhari, N. M., Gupta, V. K., Dutta, C. (2016). BPGA- an ultra-fast pan-genome analysis pipeline. Sci. Rep. 13 6, 24373. doi: 10.1038/srep24373
Chazarreta Cifré, L., Alemany, M., de Mendoza, D., Altabe, S. (2013). Exploring the biosynthesis of unsaturated fatty acids in bacillus cereus ATCC 14579 and functional characterization of novel acyl-lipid desaturases. Appl. Environ. Microbiol. 79, 6271–6279. doi: 10.1128/AEM.01761-13
Chen, S., Zhou, Y., Chen, Y., Gu, J. (2018). fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics, 34(17), i884–i890. doi: 10.1093/bioinformatics/bty560
Cisowska, A., Wojnicz, D., Hendrich, A. B. (2011). Anthocyanins as antimicrobial agents of natural plant origin. Natural Product Commun. 6, 1934578X1100600. doi: 10.1177/1934578X1100600136
Contreras-Moreira, B., Vinuesa, P. (2013). GET_HOMOLOGUES, a versatile software package for scalable and robust microbial pangenome analysis. Appl. Environ. Microbiol. 79, 7696–701. doi: 10.1128/AEM.02411-1
Darrasse, A., Priou, S., Kotoujansky, A., Bertheau, Y. (1994). PCR and restriction fragment length polymorphism of a pel gene as a tool to identify Erwinia carotovora in relation to potato diseases. Appl. Environ. Microbiol. 60 (5), 1437–43. doi: 10.1128/aem.60.5.1437-1443.1994
de Assis, J.C.S., Gonçalves, O.S., Fernandes, A.S., de Queiroz, M. V., Bazzolli, D. M.S., Santana, M. F., et al. (2022). Genomic analysis reveals the role of integrative and conjugative elements in plant pathogenic bacteria. Mobile DNA 13, 19. doi: 10.1186/s13100-022-00275-1
de Carvalho, C., Caramujo, M. (2018). The various roles of fatty acids. Molecules 23, 2583. doi: 10.3390/molecules23102583
De Coster, W., D'Hert, S., Schultz, D. T., Cruts, M., Van Broeckhoven, C. (2018). NanoPack: visualizing and processing long-read sequencing data. Bioinformatics 34 (15), 2666–2669. doi: 10.1093/bioinformatics/bty149
de Sarrau, B., Clavel, T., Clerté, C., Carlin, F., Giniès, C., Nguyen-The, C. (2012). Influence of Anaerobiosis and low temperature on bacillus cereus growth, metabolism, and membrane properties. Appl. Environ. Microbiol. 78, 1715–1723. doi: 10.1128/AEM.06410-11
Dieckmann, M. A., Beyvers, S., Nkouamedjo-Fankep, R. C., Hanel, P. H. G., Jelonek, L., Blom, J., et al (2021). EDGAR3.0: comparative genomics and phylogenomics on a scalable infrastructure. Nucleic Acids Res. 49(W1), W185–W192. doi: 10.1093/nar/gkab341
Ðermić, E (2010). Bacterial diseases of sugarbeet. (Bakterioze šećerne repe.). Glasilo Biljne Zaštite. 10 (3), 191–193. Available at: http://www.hdbz.hr.
Diomandé, S. E., Chamot, S., Antolinos, V., Vasai, F., Guinebretière, M. -H., Bornard, I., et al. (2014). The CasKR Two-Component System Is Required for the Growth of Mesophilic and Psychrotolerant Bacillus cereus Strains at Low Temperatures. Appl. Environ. Microbiol. 80, 2493–2503. doi: 10.1128/AEM.00090-14
Dullius, A., Fassina, P., Giroldi, M., Goettert, M. I., de Souza, C. F. V. (2020). A biotechnological approach for the production of branched chain amino acid containing bioactive peptides to improve human health: A review. Food research international, 131, 109002. doi: 10.1016/j.foodres.2020.109002"10.1016/j.foodres.2020.109002
Français, M., Bott, R., Dargaignaratz, C., Giniès, C., Carlin, F., Broussolle, V., et al. (2021). Short-chain and unsaturated fatty acids increase sequentially from the lag phase during cold growth of Bacillus cereus. Front. Microbiol. 12. doi: 10.3389/fmicb.2021.694757
Fulco, A. J. (1983). Fatty acid metabolism in bacteria. Prog. Lipid Res. 22, 133–160. doi: 10.1016/0163-7827(83)90005-X
Gang, S., Sharma, S., Saraf, M., Buck, M., Schumacher, J. (2019). Analysis of indole-3-acetic acid (IAA) production in Klebsiellaby LC-MS/MS and the Salkowski method. Bio Protoc. 9, e3230. doi: 10.21769/BioProtoc.3230
Gardan, L., Gouy, C., Christen, R., Samson, R. (2003). Elevation of three subspecies of Pectobacterium carotovorum to species level: Pectobacterium atrosepticum sp. nov., Pectobacterium betavasculorum sp. nov. and Pectobacterium wasabiae sp. nov. Int. J. Syst. Evol. Microbiol. 53, 381–391. doi: 10.1099/ijs.0.02423-0
Geneau-Sbartaï, C., Leyris, J., Silvestre, F., Rigal, L. (2008). Sunflower cake as a natural composite: composition and plastic properties. J Agric Food Chem. 56, 11198–208. doi: 10.1021/jf8011536
Getz, H. P. (2000). Sucrose accumulation and synthesis in sugar beet, Developments in Crop Science Editor(s): Gupta, A.K., Kaur, N., (Elsevier), 26, 55–77. doi: 10.1016/S0378-519X(00)80004-5
Gianotti, A., Iucci, L., Guerzoni, E., Lanciotti, R. (2009). Effect of acidic conditions on fatty acid composition and membrane fluidity ofEscherichia coli strains isolated from Crescenza cheese. Ann. Microbiol. 59, 603–610. doi: 10.1007/BF03175152
Gilbert, G., Caron, J., Shallow, N., Breton, A.-M., Dumont, K. (2015). Maladies diagnostiques sur les echantillons de cultures commerciales en 2014 au laboratoire de diagnostic en phytoprotection du MAPAQ. Canadian Plant Disease Survey. 95, 49–67.
Haque, M. A., Russell, N. J. (2004). Strains of Bacillus cereus vary in the phenotypic adaptation of their membrane lipid composition in response to low water activity, reduced temperature and growth in rice starch. Microbiology 150, 1397–1404. doi: 10.1099/mic.0.26767-0
Gu, Z., Eils, R., Schlesner, M. (2016). Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 32, 2847–2849. doi: 10.1093/BIOINFORMATICS/BTW313
Harveson, R. M., Hanson, L. E., Hein, G. L. (2009)Compendium of beet diseases and pests. In: Compendium of beet diseases and pests. Available online at: https://www.cabdirect.org/cabdirect/abstract/20093183793 (Accessed December 7, 2023).
Hauben, L., Moore, E. R., Vauterin, L., Steenackers, M., Mergaert, J., Verdonck, L., et al. (1998). Phylogenetic position of phytopathogens within the Enterobacteriaceae. Syst. Appl. Microbiol. 21, 384–397. doi: 10.1016/S0723-2020(98)80048-9
Hélias, V., Hamon, P., Huchet, E., Wolf, J. V. D., Andrivon, D. (2012). Two new effective semiselective crystal violet pectate media for isolation of Pectobacterium and Dickeya. Plant Pathol. 61, 339–345. doi: 10.1111/j.1365-3059.2011.02508.x
Himpsl, S. D., Mobley, H. L. T. (2019). Siderophore detection using chrome Azurol S and cross-feeding assays. Methods Mol. Biol. 2021, 97–108. doi: 10.1007/978-1-4939-9601-8_10/COVER
Jenkins, T. C. (1993). Lipid metabolism in the rumen. J. Dairy Sci. 76, 3851–3863. doi: 10.3168/jds.S0022-0302(93)77727-9
Izaguirre-Mayoral, M.L., Lazarovits, G., Baral, B. (2018). Ureide metabolism in plant-associated bacteria: purine plant-bacteria interactive scenarios under nitrogen deficiency. Plant Soil 428, 1–34. doi: 10.1007/s11104-018-3674-x
Jońca, J., Stachowska, A., Chylewska, A., Turecka, K., Waleron, K., Waleron, M. (2021). Practical considerations in the application of a polypyridyl complex of Ru(II) in physiological and biochemical studies of Pectobacterium spp. and other bacteria. Eur. J. Plant Pathol. 159, 371–383. doi: 10.1007/s10658-020-02168-6
Jonkheer, E. M., Brankovics, B., Houwers, I. M., van der Wolf, J. M., Bonants, P. J. M., Vreeburg, R. A. M., et al. (2021). The Pectobacterium pangenome, with a focus on Pectobacterium brasiliense, shows a robust core and extensive exchange of genes from a shared gene pool. BMC Genomics 22, 265. doi: 10.1186/s12864-021-07583-5
Kaneda, T. (1972). Positional preference of fatty-acids in phospholipids of Bacillus cereus and its relation to growth temperature. Biochim. Biophys. Acta 280, 297–305. doi: 10.1016/00052760(72)90097-5
Kaiser, J. C., Sen, S., Sinha, A., Wilkinson, B. J., Heinrichs, D. E. (2016). The role of two branched-chain amino acid transporters in Staphylococcus aureus growth, membrane fatty acid composition and virulence. Mol. Microbiol. 102, 850–864. doi: 10.1111/mmi.13495
Kassambara, A. (2023). Ggpubr: “ggplot2” Based publication ready plots. Available at: https://cran.r-project.org/web/packages/ggpubr/index.html
Kaneda, T. (1991). Iso- and anteiso-fatty acids in bacteria: biosynthesis, function, and taxonomic significance. Microbiol. Rev. 55, 288–302. doi: 10.1128/mr.55.2.288-302.1991
Khan, M. R., Siddiqui, Z. A. (2020). Effects of Meloidogyne incognita, Pectobacterium betavasculorum and Rhizoctonia solani interactions on growth, physiological and biochemical changes of beetroot. Israel J. Plant Sci. 67, 194–211. doi: 10.1163/22238980-bja10015
Kwon, S.-W., Go, S.-J., Kang, H.-W., Ryu, J.-C., Jo, J.-K. (1997). Phylogenetic analysis of Erwinia species based on 16S rRNA gene sequences†. Int. J. Systematic Evolutionary Microbiol. 47, 1061–1067. doi: 10.1099/00207713-47-4-1061
Lee, D. H., Kim, J. B., Kim, M., Roh, E., Jung, K., Choi, M., et al (2013). Microbiota on spoiled vegetables and their characterization. J. Food Prot. 76, 1350–1358. doi: 10.4315/0362-028X.JFP-12-439
Liu, L., Chen, X., Ye, J., Ma, X., Han, Y., He, Y., et al. (2023). Sulfoquinovose is a widespread organosulfur substrate for Roseobacter clade bacteria in the ocean. ISME J. 17, 393–405. doi: 10.1038/s41396-022-01353-1
Ma, B., Hibbing, M. E., Kim, H.-S., Reedy, R. M., Yedidia, I., Breuer, J., et al. (2007). Host range and molecular phylogenies of the soft rot enterobacterial genera Pectobacterium and Dickeya. Phytopathology® 97, 1150–1163. doi: 10.1094/PHYTO-97-9-1150
Mansfield, J., Genin, S., Magori, S., Citovsky, V., Sriariyanum, M., Ronald, P., et al. (2012). Top 10 plant pathogenic bacteria in molecular plant pathology. Mol. Plant Pathol. 13, 614–629. doi: 10.1111/j.1364-3703.2012.00804.x
Marquez-Villavicencio, M., del, P., Weber, B., Witherell, R. A., Willis, D. K., Charkowski, A. O. (2011). The 3-hydroxy-2-butanone pathway is required for pectobacterium carotovorum pathogenesis. PLoS One 6, e22974. doi: 10.1371/journal.pone.0022974
Marr, A. G., Ingraham, J. L. (1962). EFFECT OF TEMPERATURE ON THE COMPOSITION OF FATTY ACIDS IN ESCHERICHIA COLI. J. Bacteriol 84, 1260–1267. doi: 10.1128/jb.84.6.1260-1267.1962
Miller, J. (1972). Experiments in molecular genetics, Cold Spring Harbor Laboratory (Cold Spring Harbor, NY, United States).
Mitchell, K. E., Lee, C., Socha, M. T., Kleinschmit, D. H., Firkins, J. L. (2023). Supplementing branched-chain volatile fatty acids in dual-flow cultures varying in dietary forage and corn oil concentrations. III: Protein metabolism and incorporation into bacterial protein. J. Dairy Sci. 106, 7566–7577. doi: 10.3168/jds.2022-23193
Montgomerie, J. Z., Kalmanson, G. M., Guze, L. B. (1973). Fatty Acid Composition of L-Forms of Streptococcus faecalis Cultured at Different Osmolalities. J. Bacteriology 115, 73–75. doi: 10.1128/jb.115.1.73-75.1973
Nedaienia, R., Fassihiani, A. (2011)Host range and distribution of pectobacterium betavasculorum ¸ the causal agent of bacterial vascular necrosis and root rot of sugarbeet in fars province *. In: Iranian Journal of Plant Pathology. Available online at: https://www.semanticscholar.org/paper/HOST-RANGE-AND-DISTRIBUTION-OF-Pectobacterium-%C2%B8-THE-Nedaienia-Fassihiani/1c3fecff0d0d3f4212c684f2ef0efeab66fbadf8 (Accessed December 7, 2023).
Nguyen, H. A., Kaneko, J., Kamio, Y. (2002). Temperature-dependent production of carotovoricin Er and pectin lyase in phytopathogenic erwinia carotovora subsp. carotovora Er. Bioscience biotechnology Biochem. 66, 444–447. doi: 10.1271/bbb.66.444
Ozturk, M., Eroglu, Z., Soylu, S. (2019). First report of Pectobacterium betavasculorum associated with bacterial vascular necrosis and root rot disease of sugar beet in Turkey. New Dis. Rep. 39, 20–20. doi: 10.5197/j.2044-0588.2019.039.020
Panda, P., Vanga, B. R., Lu, A., Fiers, M., Fineran, P. C., Butler, R., et al. (2016). Pectobacterium atrosepticum and Pectobacterium carotovorum Harbor Distinct, Independently Acquired Integrative and Conjugative Elements Encoding Coronafacic Acid that Enhance Virulence on Potato Stems. Front. Microbiol. 7. doi: 10.3389/fmicb.2016.00397
Pereira, W. J., Boyd, J., Conde, D., Triozzi, P. M., Balmant, K. M., Dervinis, C., et al. (2024). The single-cell transcriptome program of nodule development cellular lineages in Medicago truncatula. Cell Rep. 43 (2), 113747. doi: 10.1016/j.celrep.2024.113747
Petraru, A., Amariei, S. (2022). Sunflower iilcake as a potential source for the development of edible membranes. Membranes. 12, 789. doi: 10.3390/membranes12080789
Põllumaa, L., Alamäe, T., Mäe, A. (2012). Quorum sensing and expression of virulence in pectobacteria. Sensors (Basel) 12, 3327–3349. doi: 10.3390/s120303327
Portier, P., Pédron, J., Taghouti, G., Dutrieux, C., Barny, M.-A. (2020). Updated taxonomy of pectobacterium genus in the CIRM-CFBP bacterial collection: when newly described species reveal “Old” Endemic population. Microorganisms 8, 1441. doi: 10.3390/microorganisms8091441
Rastgou, M., Rezaee Danesh, Y., Ercisli, S., Sayyed, R. Z., El Enshasy, H. A., Dailin, D. J. (2022). The effect of some wild grown plant extracts and essential oils on Pectobacterium betavasculorum: the causative agent of bacterial soft rot and vascular wilt of sugar beet. Plants (Basel) 11, 1155. doi: 10.3390/plants11091155
Ravn, L., Christensen, A. B., Molin, S., Givskov, M., Gram, L. (2001). Methods for detecting acylated homoserine lactones produced by Gram-negative bacteria and their application in studies of AHL-production kinetics. J. Microbiol. Methods 44, 239–251. doi: 10.1016/S0167-7012(01)00217-2
Rezaee Danesh, Y., Rastgou, M., Kariman, K. (2021). Phenotypic and molecular characterization of the bacterial causal agents of sugar beet soft rot and vascular necrosis disease in Southeastern Iran. Int. J. Agric. Tech. 17, 2487–2502.
Rezaei, R., Taghavi, S. M. (2010). Phenotypic and genotypic characterization of Iranian soft rot bacterial isolates from different hosts. Phytopathologia Mediterranea. 49, 194–204. Available at: https://www.proquest.com/scholarly-journals/phenotypic-genotypic-characteristics-iranian-soft/docview/865320856/se-2.
Richter, M., Rosselló-Móra, R., Glöckner, O. F., Peplies, J. (2016). JSpeciesWS: a web server for prokaryotic species circumscription based on pairwise genome comparison. Bioinformatics. 32, 929–31. doi: 10.1093/bioinformatics/btv681
Roh, E., Park, T.-H., Kim, M., Lee, S., Ryu, S., Oh, C.-S., et al. (2010). Characterization of a New Bacteriocin, Carocin D, from Pectobacterium carotovorum subsp. carotovorum Pcc21. Appl. Environ. Microbiol. 76, 7541–7549. doi: 10.1128/AEM.03103-09
Roy, A. B., Hewlins, M. J. E., Ellis, A. J., Harwood, J. L., White, G. F. (2003). Glycolytic breakdown of Sulfoquinovose in bacteria: a missing link in the sulfur cycle. Appl. Environ. Microbiol. 69, 6434–6441. doi: 10.1128/AEM.69.11.6434-6441.2003
Roy, N., Laskar, S., Barik, A. (2013). Amino acids through developmental stages of sunflower leaves. Acta Botanica Croatica 72, 23–33. doi: 10.2478/v10184-012-0009-5
Saleh, O. I., Huang, P.-Y., Huang, J.-S. (1996). Bacterial vascular necrosis and root rot disease of sugar beet in Egypt. J. Phytopathol. 144, 225–230. doi: 10.1111/j.1439-0434.1996.tb01520.x
Salvador, V., Cherbut, C., Barry, J. L., Bertrand, D., Bonnet, C., Delort-Laval, J. (1993). Sugar composition of dietary fibre and short-chain fatty acid production during in vitro fermentation by human bacteria. Br. J. Nutr. 70, 189–197. doi: 10.1079/BJN19930116
Shimazaki, J., Furukawa, S., Ogihara, H., Morinaga, Y. (2012). l-Tryptophan prevents Escherichia coli biofilm formation and triggers biofilm degradation. Biochem. Biophys. Res. Commun. 419, 715–718. doi: 10.1016/j.bbrc.2012.02.085
Shyntum, D. Y., Nkomo, N. P., Shingange, N. L., Gricia, A. R., Bellieny-Rabelo, D., Moleleki, L. N. (2019). The Impact of Type VI Secretion System, Bacteriocins and Antibiotics on Bacterial Competition of Pectobacterium carotovorum subsp. brasiliense and the Regulation of Carbapenem Biosynthesis by Iron and the Ferric-Uptake Regulator. Front. Microbiol. 10. doi: 10.3389/fmicb.2019.02379
Sinensky, M. (1971). Temperature control of phospholipid biosynthesis in Escherichia coli. J. Bacteriology 106, 449–455. doi: 10.1128/jb.106.2.449-455.1971
Sinensky, M. (1974). Homeoviscous adaptation—A homeostatic process that regulates the viscosity of membrane lipids in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 71, 522–525. doi: 10.1073/pnas.71.2.522
Sławiak, M., Łojkowska, E., Van der Wolf, J. M. (2009). First report of bacterial soft rot on potato caused by Dickeya sp.(syn. Erwinia chrysanthemi) in Poland. Plant Pathol. 58, 794. doi: 10.1111/j.1365-3059.2009.02028.x
Smoktunowicz, M., Jonca, J., Stachowska, A., May, M., Waleron, M. M., Waleron, M., et al. (2022). The international trade of ware vegetables and orna-mental plants—An underestimated risk of accelerated spreading of phytopathogenic bacteria in the era of globalisation and ongoing climatic changes. Pathogens 11, 728. doi: 10.3390/pathogens11070728
Song, Y., Tong, Z., Wang, J., Wang, L., Guo, Z., Han, Y., et al. (2004). Complete genome sequence of Yersinia pestis strain 91001, an isolate avirulent to humans. DNA Res. 11(3), 179–97. doi: 10.1093/dnares/11.3.179
Suutari, M., Laakso, S. (1994). Microbial fatty acids and thermal adaptation. Crit. Rev. Microbiol. 20, 285–328. doi: 10.3109/10408419409113560
Thomson, S. V. (1981). Sugar and root yield of sugar beets as affected by bacterial vascular necrosis and rot, nitrogen fertilization, and plant spacing. Phytopathology 71, 605. doi: 10.1094/Phyto-71-605
Thomson, S. V., Schroth, M. N., Hills, F. J., Whitney, E. D., Hildebrand, D. C. (1977). Bacterial vascular necrosis and rot of sugarbeet: general description and etiology. Phytopathology 67, 1183–1189. doi: 10.1094/Phyto-67-1183
Tindall, B. J., Sikorski, J., Smibert, R. A., Krieg, N. R. (2014). Phenotypic characterization and the principles of comparative systematics. Methods Gen. Mol. Microbiol., 15, 330–393. doi: 10.1128/9781555817497.CH15
Tomaszewska, J., Bielinski, D., Binczarski, M., Berlowska, J., Dziugan, P., Piotrowski, J., et al. (2018). Products of sugar beet processing as raw materials for chemicals and biodegradable polymers. RSC Adv. 8, 3161–3177. doi: 10.1039/c7ra12782k
Toth, I. K., Avrova, A. O., Hyman, L. J. (2001). Rapid identification and differentiation of the soft rot erwinias by 16S-23S intergenic transcribed spacer-PCR and restriction fragment length polymorphism analyses. Appl. Environ. Microbiol. 67, 4070–4076. doi: 10.1128/AEM.67.9.4070-4076.2001
Toth, I. K., Barny, M., Brurberg, M. B., Condemine, G., Czajkowski, R., Elphinstone, J. G., et al. (2021). “Pectobacterium and dickeya: environment to disease development,” in Plant Diseases Caused by Dickeya and Pectobacterium Species. Eds. Gijsegem, F.V., Wolf, J.M.v. d., Toth, I. K. (Springer International Publishing, Cham), 39–84. doi: 10.1007/978-3-030-61459-1_3
van der Wolf, J. M., Nijhuis, E. H., Kowalewska, M. J., Saddler, G. S., Parkinson, N., Elphinstone, J. G., et al. (2014). Dickeya solani sp. nov., a pectinolytic plant-pathogenic bacterium isolated from potato (Solanum tuberosum). Int. J. Systematic Evolutionary Microbiol. 64, 768–774. doi: 10.1099/ijs.0.052944-0
Versalovic, J., Koeuth, T., Lupski, J. R. (1991). Distribution of repetitive DNA sequences in eubacteria and application to fingerprinting of bacterial genomes. Nucleic Acids Res. 19, 6823–6831. doi: 10.1093/nar/19.24.6823
Vlaeminck, B., Fievez, V., van Laar, H., Demeyer, D. (2005). Rumen odd and branched chain fatty acids in relation to in vitro rumen volatile fatty acid productions and dietary characteristics of incubated substrates. J. Anim. Physiol. Anim. Nutr. 88, 401–411. doi: 10.1111/j.1439-0396.2004.00497.x
Waleron, M., Misztak, A., Waleron, M., Franczuk, M., Wielgomas, B., Waleron, K. (2018). Transfer of Pectobacterium carotovorum subsp. carotovorum strains isolated from potatoes grown at high altitudes to Pectobacterium Peruviense sp. nov. Systematic Appl. Microbiol. 41, 85–93. doi: 10.1016/j.syapm.2017.11.005
Waleron, M., Waleron, K., Lojkowska, E. (2014). Characterization of Pectobacterium carotovorum subsp. Odoriferum Causing Soft Rot of Stored Vegetables. Eur. J. Plant Pathol. 139, 457–469. doi: 10.1007/s10658-014-0403-z
Waleron, M., Waleron, K., Lojkowska, E. (2015). First Report of Pectobacterium carotovorum subsp. Brasiliense Causing Soft Rot on Potato and Other Vegetables in Poland. Plant Dis. 99, 1271. doi: 10.1094/PDIS-02-15-0180-PDN
Waleron, M., Waleron, K., Podhajska, A. J., Łojkowska, E. (2002). Genotyping of bacteria belonging to the former Erwinia genus by PCR-RFLP analysis of a recA gene fragment. Microbiology 148, 583–595. doi: 10.1099/00221287-148-2-583
Wei, X., Moreno-Hagelsieb, G., Glick, B. R., Doxey, A. C. (2023). Comparative analysis of adenylate isopentenyl transferase genes in plant growth-promoting bacteria and plant pathogenic bacteria. Heliyon. 9, e13955. doi: 10.1016/j.heliyon.2023.e13955
Whitney, E. D., Duffus, J. E. (1986). Compendium of beet diseases and insects. In: Compendium of beet diseases and insects. Available online at: https://www.cabdirect.org/cabdirect/abstract/19860538155 (Accessed December 7, 2023).
Wick, R. R., Judd, L. M., Gorrie, C. L., Holt, K. E. (2017). Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 13 (6), e1005595. doi: 10.1371/journal.pcbi.1005595
Wickham, H. (2009). Ggplot2: Elegant Graphics for Data Analysis. (Springer). doi: 10.1007/978-3-319-24277-4_5
Wolfgang, A., Temme, N., Tilcher, R., Berg, G. (2023). Understanding the sugar beet holobiont for sustainable agriculture. Front. Microbiol. 14. doi: 10.3389/fmicb.2023.1151052
Yamada, K., Hirota, M., Niimi, Y., Nguyen, H. A., Takahara, Y., Kamio, Y., et al. (2006). Nucleotide Sequences and Organization of the Genes for Carotovoricin (Ctv) from Erwinia carotovora Indicate That Ctv Evolved from the Same Ancestor as Salmonella typhi Prophage. Biosci. Biotechnol. Biochem. 70, 2236–2247. doi: 10.1271/bbb.60177
Keywords: Pectobacterium betavasculorum, genomics, pangenomics, pathogenomics, fatty acid composition, host specificity, adaptation capacity, sugar beet
Citation: Borowska-Beszta M, Smoktunowicz M, Horoszkiewicz D, Jonca J, Waleron MM, Gawor J, Mika A, Sledzinski T, Waleron K and Waleron M (2024) Comparative genomics, pangenomics, and phenomic studies of Pectobacterium betavasculorum strains isolated from sugar beet, potato, sunflower, and artichoke: insights into pathogenicity, virulence determinants, and adaptation to the host plant. Front. Plant Sci. 15:1352318. doi: 10.3389/fpls.2024.1352318
Received: 07 December 2023; Accepted: 26 February 2024;
Published: 21 March 2024.
Edited by:
Stephen Ernest Strelkov, University of Alberta, CanadaReviewed by:
Ömür Baysal, Mugla University, TürkiyeSandra Marcela Velasco Cuervo, University of Alberta, Canada
Copyright © 2024 Borowska-Beszta, Smoktunowicz, Horoszkiewicz, Jonca, Waleron, Gawor, Mika, Sledzinski, Waleron and Waleron. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Krzysztof Waleron, a3J6eXN6dG9mLndhbGVyb25AZ3VtZWQuZWR1LnBs; Malgorzata Waleron, bWFsZ29yemF0YS53YWxlcm9uQGJpb3RlY2gudWcuZWR1LnBs
 Maria Borowska-Beszta
Maria Borowska-Beszta Magdalena Smoktunowicz
Magdalena Smoktunowicz Daria Horoszkiewicz
Daria Horoszkiewicz Joanna Jonca
Joanna Jonca Michal Mateusz Waleron
Michal Mateusz Waleron Jan Gawor
Jan Gawor Adriana Mika
Adriana Mika Tomasz Sledzinski
Tomasz Sledzinski Krzysztof Waleron
Krzysztof Waleron Malgorzata Waleron
Malgorzata Waleron