- 1School of Biological Science, Jining Medical University, Rizhao, Shandong, China
- 2Agricultural Science and Engineering School, Liaocheng University, Liaocheng, Shandong, China
Nitrogen is an essential macronutrient for plant growth and development. Nitrate is the major form of nitrogen acquired by most crops and also serves as a vital signaling molecule. Nitrate is absorbed from the soil into root cells usually by the low-affinity NRT1 NO3- transporters and high-affinity NRT2 NO3- transporters, with NRT2s serving to absorb NO3- under NO3–limiting conditions. Seven NRT2 members have been identified in Arabidopsis, and they have been shown to be involved in various biological processes. In this review, we summarize the spatiotemporal expression patterns, localization, and biotic and abiotic responses of these transporters with a focus on recent advances in the current understanding of the functions of the seven AtNRT2 genes. This review offers beneficial insight into the mechanisms by which plants adapt to changing environmental conditions and provides a theoretical basis for crop research in the near future.
1 Introduction
Plants rely on nitrogen (N) as an essential macronutrient that is vital for their growth and productivity. Nitrate (NO3-) is the most abundant source of inorganic nitrogen taken up by most terrestrial plant species (Crawford and Forde, 2002). Kinetic criteria have been used to characterize such nitrate uptake as being mediated by three distinct systems, including a low-affinity transport system (LATS) as well as inducible and constitutive high-affinity transport systems (iHATS and cHATS) (Glass et al., 1995; Crawford and Glass, 1998; Forde, 2000). The cHATS and iHATS systems are generally active at NO3- concentrations in the 10-250 μM range, whereas LATS activity is only apparent when these concentrations exceed 250 μM. The NO3- affinity of the iHATS (Km: 13-79 μM) is lower than that of the cHATS (Km: 6-20 μM) (Forde and Clarkson, 1999), iHATS capacity for the uptake of NO3- greatly exceeds that of cHATS. For example, analyses of iHATS activity in response to induction with a NO3- concentration of 100 μM yielded a Vmax that was roughly 25-fold higher than that for the cHATS (Siddiqi et al., 1990). Members of the NRT1 protein family serve as low-affinity NO3- transporters with Km values in the mM range, while members of the NRT2 protein family function as high-affinity transporters with Km values in the μM range (Wang et al., 2012; O'Brien et al., 2016).The Aspergillus nidulans crnA gene was the first high-affinity NO3- transporter cloned from a eukaryotic species, and mutations in this gene can confer chlorate (ClO3-) resistance while resulting in the partial impairment of NO3- uptake (Brownlee and Arst, 1983; Unkles et al., 1991). The uptake of NO2- and NO3- in Chlamydomonas reinhardtii was subsequently determined to be under the control of three genes related to crnA, including CrNRT2.1, CrNRT2.2, and CrNRT2.3 (Quesada et al., 1994; Galván et al., 1996; Quesada et al., 1998). More recently, researchers have identified high-affinity nitrate transporters from a wide range of plants including Arabidopsis thaliana, Hordeum vulgare, and Nicotiana plumbaginifolia (Quesada et al., 1997; Filleur and Daniel-Vedele, 1999; Zhuo et al., 1999; Vidmar et al., 2000). In the case of Arabidopsis, RT-PCR analyses performed using degenerate primers led to the initial cloning of the two related AtNRT2.1 and AtNRT2.2 transporter genes (Zhuo et al., 1999). With the completion of the Arabidopsis genome project, a total of seven Arabidopsis NRT2 family members were identified in this model species. The roots are the predominant site of expression for all AtNRT2 genes other than AtNRT2.7, which is expressed at the highest levels in seeds (Orsel et al., 2002; Okamoto et al., 2003; Chopin et al., 2007a). The responsivity of different NRT2 genes to changes in nitrate availability following N starvation varies markedly. For example, NO3- exposure strongly induced the expression of NRT2.1 and NRT2.2, while NRT2.4 was modestly upregulated, NRT2.5 expression was repressed, and the expression of NRT2.3, NRT2.6, and NRT2.7 was unaffected by the available nitrate supply (Okamoto et al., 2003). In addition to their roles as mediators of high-affinity NO3- influx, these NRT2 family members have been reported to play key roles in an array of biological processes involved in regulating processes such as N starvation, root architecture, seed development, cadmium uptake, plant-microbe interactions, systemic nitrate signaling, and the maintenance of appropriate nitrogen and carbon homeostasis (Table 1). This review offers an overview of information that is currently known regarding the molecular mechanisms and functions associated with plant members of the NRT2 gene family.
2 High-affinity nitrate absorption
To date, seven NRT2 proteins have been identified in Arabidopsis thaliana (Glass et al., 2001). Of these, high-affinity root NO3- influx is only mediated by NRT2.1, NRT2.2, NRT2.4, and NRT2.5, each of which exhibits distinct context-dependent contributions to this absorptive process (Filleur et al., 2001; Kiba et al., 2012; Lezhneva et al., 2014).
2.1 AtNRT2.1 and AtNRT2.2
The ability of AtNRT2.1 and AtNRT2.2 to function in an iHATS is supported by the fact that the Atnrt2 T-DNA mutant, in which both of these genes are disrupted, exhibited a reduction in high-affinity nitrate uptake (Cerezo et al., 2001; Filleur et al., 2001). Notably, AtNRT2.1 transcript levels are strictly correlated with high-affinity uptake of nitrogen when nitrate is supplied to plants that were initially nitrate-deprived (Okamoto et al., 2003). Further studies have explored the iHATS, cHATS, and LATS systems in Atnrt2.1 mutant plants that were cultivated for 4 weeks in a 1 mM NH4NO3 solution followed by a 7-day nitrogen deprivation period in order to deplete nitrogen reserves (Li et al., 2007). Upon initial exposure of these plants to 100 μM 13NO3-, cHATS flux is first observed. Moreover, after the 1-week nitrogen deprivation period, plants were treated for 6 h with 1 mM KNO3 followed by exposure to 100 μM 13NO3-, with the resultant flux representing the combination of iHATS and cHATS flux. These two flux measurements can be used to estimate iHATS activity based on the difference between the two. These sample plants were also utilized to assess LATS influx after a 6-hour induction period with 1 mM KNO3 and subsequent exposure to 10 mM 13NO3-. These analyses revealed a ~72% reduction in iHATS activity in Atnrt2.1 mutants without any corresponding change in cHATS or LATS flux (Li et al., 2007). Similarly, a 19% drop in iHATS flux was observed in Atnrt2.2 mutants, whereas cHATS and LATS fluxes remained intact (Li et al., 2007). AtNRT2.2 expression levels are significantly lower than AtNRT2.1 levels (Zhuo et al., 1999; Orsel et al., 2002; Okamoto et al., 2003), but these levels were ~3-fold higher in Atnrt2.1 mutant plants, indicating that AtNRT2.2 overexpression may partially compensate for the loss of Atnrt2.1. Consistent with such a mechanism, nrt2.1 nrt2.2 double mutants exhibit more dramatic iHATS and cHATS fluxes by ~80% and 30%, respectively, relative to nrt2.1 and nrt2.2 single mutants. These data emphasize the importance of NRT2.1 as the key driver of iHATS activity, whereas NRT2.2 exhibits a smaller compensatory role in this context (Li et al., 2007).
AtNRT2.1 expression is primarily evident in roots (Orsel et al., 2002), and it primarily localizes to the plasma membrane of root epidermal and cortical cells, consistent with this being the primary site of nitrate uptake (Wirth et al., 2007; Chopin et al., 2007b). NRT2.1 protein level changes reportedly differ from corresponding shifts in the mRNA expression of NRT2.1 (Wirth et al., 2007), and 35S::NRT2.1 transformants constitutively overexpressing NRT2.1 still exhibited reductions in HATS activity (Laugier et al., 2012), consistent with mechanisms responsible for post-translationally regulating NRT2.1. Phosphoproteomic analyses indicated that NRT2.1 is subject to phosphorylation, with the degree of its phosphorylation shifting as a function of the availability of nitrate (Engelsberger and Schulze, 2012; Menz et al., 2016). NRT2.1 reportedly harbors four phosphorylation sites as confirmed through high-accuracy mass spectrometry-based efforts to detect phosphopeptides (Engelsberger and Schulze, 2012; Menz et al., 2016; Jacquot et al., 2020). The Ser28 phosphorylation of NRT2.1 is evident in plants subject to N starvation, but dephosphorylation occurs rapidly when nitrate becomes available (Engelsberger and Schulze, 2012). Consistent with this observation, other studies have confirmed the stabilization and enhanced Ser28 phosphorylation of NRT2.1 under conditions of nitrate limitation. To explore the role of Ser28 phosphorylation, researchers established transgenic NRT2.1S28E and NRT2.1S28A plants that respectively mimic the phosphorylates and dephosphorylated forms of this protein (Zou et al., 2020). The Ser28 alanine substitution was associated with NRT2.1 destabilization, and NRT2.1S28A overexpression under conditions of limited nitrate availability failed to rescue defective nrt2 mutant plant phonotypes. In contrast, greater levels of the NRT2.1S28E isoform enhanced protein stability and were sufficient to restore nrt2 mutant phonotypes when cultivated in the presence of low nitrate levels (Zou et al., 2020). NRT2.1 Ser28 phosphorylation thus plays a key role in regulating NRT2.1 stability. Jacquot et al. further determined that the C-terminal portion of NRT2.1 (aa 494-513) is essential for the appropriate function of this protein, as demonstrated using transgenic nrt2.1-2 mutant plants expressing truncated NRT2.1 isoforms (NRT2.1ΔC494-530 and NRT2.1ΔC514-530) (Jacquot et al., 2020). While the pNRT2.1::NRT2.1ΔC494-530 transgene was unable to restore HAST activity and growth to wild-type levels, the pNRT2.1::NRT2.1ΔC514-530 transgene was able to do so. Through mass spectrometry-based phosphopeptide detection efforts, the authors were able to identify the Ser501 phosphorylation site within this region of the protein, and the phonotypes of phosphomimetic S501D transgenic plants were comparable to those of nrt2 mutants. Higher levels of Ser501 phosphorylation were observed under cultivation on 1 mM NO3- followed by transfer for 4 h onto 10 mM NH4NO3, consistent with a reduction in the influx of nitrate evident in wild-type plants (Jacquot et al., 2020). Ser501 phosphorylation is thus capable of inactivating the activity of NRT2.1. Notably, this Ser501 phosphorylation site is highly conserved across plant species, emphasizing the key role that it plays as a regulator of NRT2.1 functionality (Jacquot et al., 2020). This protein has also been shown to harbor N-terminal Ser11 and C-terminal Thr521 phosphorylation sites (Menz et al., 2016), although additional research will be necessary to clarify their functions.
2.2 AtNRT2.4 and AtNRT2.5
AtNRT2.4 is a high-affinity nitrate transporter as demonstrated by its expression in plants and heterologous expression in Xenopus laevis oocytes. When nrt2.1 nrt2.2 double mutant plants exhibiting impaired high-affinity uptake of nitrate were transformed with NRT2.4 cDNA under the control of the root-specific RolD promoter (Fraisier et al., 2000), NRT2.4 overexpression was associated with a pronounced increase in 15NO3- uptake relative to non-transformed nrt2.1 ntr2.2 double mutant plants under conditions of low nitrate availability (0.2 mM NO3-), supporting the ability of NRT2.4 to regulate the high-affinity uptake of NO3 (Kiba et al., 2012). To further confirm its ability to function in this regulatory context, Xenopus oocytes were injected for 3 days with NRT2.4 mRNA or with water as a vehicle control, followed by exposure for 16 h to 0.2 mM Na15NO3. Subsequent analyses of the accumulation of 15N within oocytes revealed that those oocytes injected with the NRT2.4 mRNA-injected oocytes took up significantly more NO3- than water-injected controls (Kiba et al., 2012).
AtNRT2.4 levels in plant roots were lower than those of AtNRT2.1 at baseline, but it is strongly upregulated in response to N deprivation. When growing plants on complete N medium for 7 days followed by N starvation for 5 days, wild-type plants exhibited maximal NRT2.4 expression. Significantly decreased 15NO3- uptake relative to wild-type was detected in nrt2.4 null mutants supplied with extremely low concentrations of 15NO3- (0.025 or 0.01 mM), whereas no differences between the two were apparent when the available concentration of 15NO3- was higher (0.2, 0.5, or 6 mM). This highlights a role for NRT2.4 as a mediator of very-high-affinity NO3- uptake. Much like NRT2.4, the transformation of nrt2.1 ntr2.2 double mutants with NRT2.5 under the control of the RolD promoter resulted in a pronounced increase in the influx of 15NO3- in roots as compared to non-transformed double mutants in the presence of 0.2 mM NO3- conditions, consistent with the ability of NRT2.5 to serve as a NO3- transporter (Lezhneva et al., 2014). In contrast to nrt2.4 mutants for which no alterations in 15NO3- influx were evident relative to wild-type plants, nrt2.5 mutants exhibited significantly reduced high-affinity 15NO3- influx in the presence of 0.2 mM NO3- (Lezhneva et al., 2014). NRT2.5 therefore functions as a high-affinity transporter of nitrate.
2.3 Two-component high-affinity nitrate transporters
Besides transcriptional regulation, posttranscriptional events also can influence NRT2 protein activity and/or abundance, strongly influencing HATS functionality. Early studies demonstrated that the functionality of the C. reinhardtii was dependent on two gene products. The genes that encode these two proteins, CrNRT2 and CrNAR2, are present within a single cluster of nitrate-regulated genes, and mutant plants with deletions in this region of the genome exhibit dramatically lower levels of high-affinity nitrate uptake that were only restored by the transformation of these plants with constructs encoding CrNAR2 and either CrNRT2.1 or CrNRT2.2, whereas none of these constructs alone were sufficient (Quesada et al., 1994). Studies of Xenopus oocytes provided further confirmation of the existence of this two-component high-affinity nitrate transport system, as the injection of mRNAs CrNAR2 or CrNRT2.1 alone failed to induce nitrate currents, whereas high levels of nitrate uptake were evident when both were co-injected with one another (Zhou et al., 2000a). Similar findings were also detected in barley such that only the co-injection of Xenopus oocytes with the HvNRT2.1 and HvNAR2.3 mRNAs encoding homologous barley proteins was sufficient to enhance nitrate transport (Tong et al., 2005; Ishikawa et al., 2009).
Through subsequent research efforts, researchers determined that Arabidopsis also encodes a two-component high-affinity nitrate transport system. Okamoto et al. (2006) searched for genes homologous to the NAR2 sequences from C. reinhardtii, ultimately leading to the identification of the AtNRT3.1 and AtNRT3.2 genes (Okamoto et al., 2006; Feng et al., 2011a), the former of which was expressed at much higher levels than the latter. Strong AtNRT3.1 upregulation was evident when the roots of plants that had been N starved were treated for 3 or 6 h with 1 mM KNO3, whereas only limited upregulation of AtNRT3.2 was evident at the 6 h time point (Okamoto et al., 2006). Relative to wild-type plants, Atnrt3.1 mutants exhibited a significant reduction in root nitrate influx under conditions of low 13NO3- availability (10 - 150 μM), consistent with a role for this gene product as a regulator of NO3- HATS activity. A ~70% reduction in iHATS activity was reported for Atntr2.1 mutants (Filleur et al., 2001; Li et al., 2007), whereas this reduction was upwards of 95% when AtNRT3.1 was mutated (Okamoto et al., 2006; Orsel et al., 2006). Oocyte injection experiments in which the NRT2.1 or NRT3.1 mRNAs were individually injected or co-injected revealed that significant uptake of 15NO3- was only apparent following the co-injection of both genes (Orsel et al., 2006). Arabidopsis HATS activity is thus dependent on both the AtNRT2.1 and AtNRT3.1 genes, in line with the phenotypes observed in C. reinhardtii (Quesada et al., 1994; Zhou et al., 2000a). These data suggest that while NRT3.1 is dispensable for the regulation of NRT2.1 transcription, it can serve as a facilitator of the transport activity of the NRT2.1 protein, potentially through a mechanism mediated by direct interactions. Additional yeast split-ubiquitin system assays indicated that the NRT2.1 and NRT3.1 proteins are capable of interacting with one another (Orsel et al., 2006), and this interaction localizes to the plasma membrane (Yong et al., 2010). Consistently, an absence of NRT2.1 plasma membrane localization was evident in nrt3.1 mutants (Wirth et al., 2007). Further confirming this result, Yong et al. conducted western blotting experiments in which they used anti-NRT2.1 to detect a 150-kDa oligomeric polypeptide extracted from the root membrane fraction, and this fraction was further resolved, revealing it to be composed of NRT2.1 (48 kDa) and myc-tagged NRT3.1 (26 kDa). This, coupled with the absence of this 150-kDa complex in nrt2.1 or nrt3.1 mutants, suggests that a tetrameric complex composed of two NRT2.1 subunits and two NRT3.1 subunits may be responsible for high-affinity nitrate uptake activity (Yong et al., 2010). With the exception of AtNRT2.7, which was identified as a tonoplast transporter (Chopin et al., 2007a), all NRT2 family members were shown to be capable of engaging in strong interactions with NRT3.1 in bimolecular fluorescence complementation and yeast two-hybrid experiments (Kotur et al., 2012). In Xenopus oocytes, different NRT2 mRNAs were injected alone or in combination with NRT3.1 to evaluate the effects on nitrate uptake. These experiments revealed that NRT3.1 and NRT2 co-injections were associated with greater 15NO3- uptake, with this effect being particularly pronounced for NRT3.1 coinjection with NRT2.1/NRT2.5, which yielded respective increases in nitrate uptake of 532% and 334%, as compared to only slight increases when co-injected with NRT2.3/NRT2.4 (Kotur et al., 2012). Much like NRT2.1, NRT2.5 was also capable of forming a 150-kDa tetrameric complex with NRT3.1 at the plasma membrane to facilitate the high-affinity uptake of nitrate (Yong et al., 2010; Kotur and Glass, 2015). The existence of two-component NRT2/NAR2 nitrate uptake machinery has also been confirmed in plants including barley (Tong et al., 2005; Ishikawa et al., 2009), rice (Yan et al., 2011), wheat (Taulemesse et al., 2015), maize (Pii et al., 2016; Liu et al., 2020), and chrysanthemum (Gu et al., 2016). This system is not universal, however, as the NRT2.1 homolog in A. nidulans, crnA, did not require any corresponding NAR2 activity in Xenopus oocytes to facilitate nitrate current generation (Zhou et al., 2000b). These differences may be related to the longer crnA central loop and the lack of any homolog of NAR2 in A. nidulans (Yong et al., 2010). Strikingly, all NRT2 proteins other than AtNRT2.1 were capable of mediating small levels of nitrate flux following the injection of the individual encoding mRNA sequences into Xenopus oocytes, with this being most apparent for NRT2.4 and NRT2.7 (Chopin et al., 2007a; Kiba et al., 2012; Kotur et al., 2012). Additional research focused on the specific mechanisms whereby NRT2 family proteins mediate high-affinity nitrate transport is thus warranted.
3 N starvation and remobilization
Under conditions of N deficiency, the NO3- that is stored in plants can undergo remobilization and phloem-mediated transport (Wang et al., 2012). Marked increases in NRT2.4 and NRT2.5 expression are evident in response to N deprivation, with NRT2.5 being induced at much higher levels than NART2.4 in shoots and roots. Both of these genes are expressed in shoot vascular tissue in N-starved plants (Kiba et al., 2012; Lezhneva et al., 2014). In experiment in which plants were growth with access to normal N levels for 6 weeks followed by a 4-week period of N starvation, a 45% reduction in leaf phloem exudate NO3- concentrations was observed in nrt2.4 mutants relative to wild-type plants, without any corresponding change in nrt2.5 mutants, and an even stronger reduction in nrt2.4 nrt2.5 double mutants such that these exudate levels were just 20% of those observed in wild-type plants. This phenotype was restricted to phloem exudate NO3- levels, as none of these mutants exhibited changes in whole leaf NO3- concentrations or phloem exudate amino acid content, demonstrating specific roes for NRT2.4 and NRT2.5 in the remobilization of nitrate within shoots in response to N starvation (Kiba et al., 2012; Lezhneva et al., 2014).
A summary of the various contributions of different NRT2 family members to specific phases of the processes of nitrate uptake and allocation is presented in Figure 1 and Table 1. In Arabidopsis, NRT2.1, NRT2.2, NRT2.4, and NRT2.5 all serve as mediators of high-affinity nitrate uptake, although the functions of the latter two of these proteins are only evident in the context of N starvation. After an extended starvation interval, NRT2.5 expression levels are increased such that it serves as the primary high-affinity uptake transporter protein. NRT2.1, NRT2.4, and NRT2.5 also exhibit differences in their spatial expression profiles, with NRT2.1 expression primarily being evident in older portions of primary roots (Nazoa et al., 2003), whereas NRT2.4 is most prominently expressed in younger portions of the primary roots and distal areas of lateral roots (Kiba et al., 2012), and NRT2.5 is expressed in the root hair regions of both primary and lateral roots (Figure 1). Future research is warranted to clarify the degree to which nitrate affinity differs among these transporters, given that higher-affinity transporters may be important to allow plants to better deal with the stress associated with extended N starvation in soil with poor fertility. NRT2.4 and NRT2.5 expression are also evident in the phloem of the major and minor shoot veins, influencing shoot phloem nitrate levels under certain conditions or in the context of specific mutations. NRT2.7 expression is primarily evident during seed development in the tonoplast, wherein it serves to regulate seed nitrate levels.
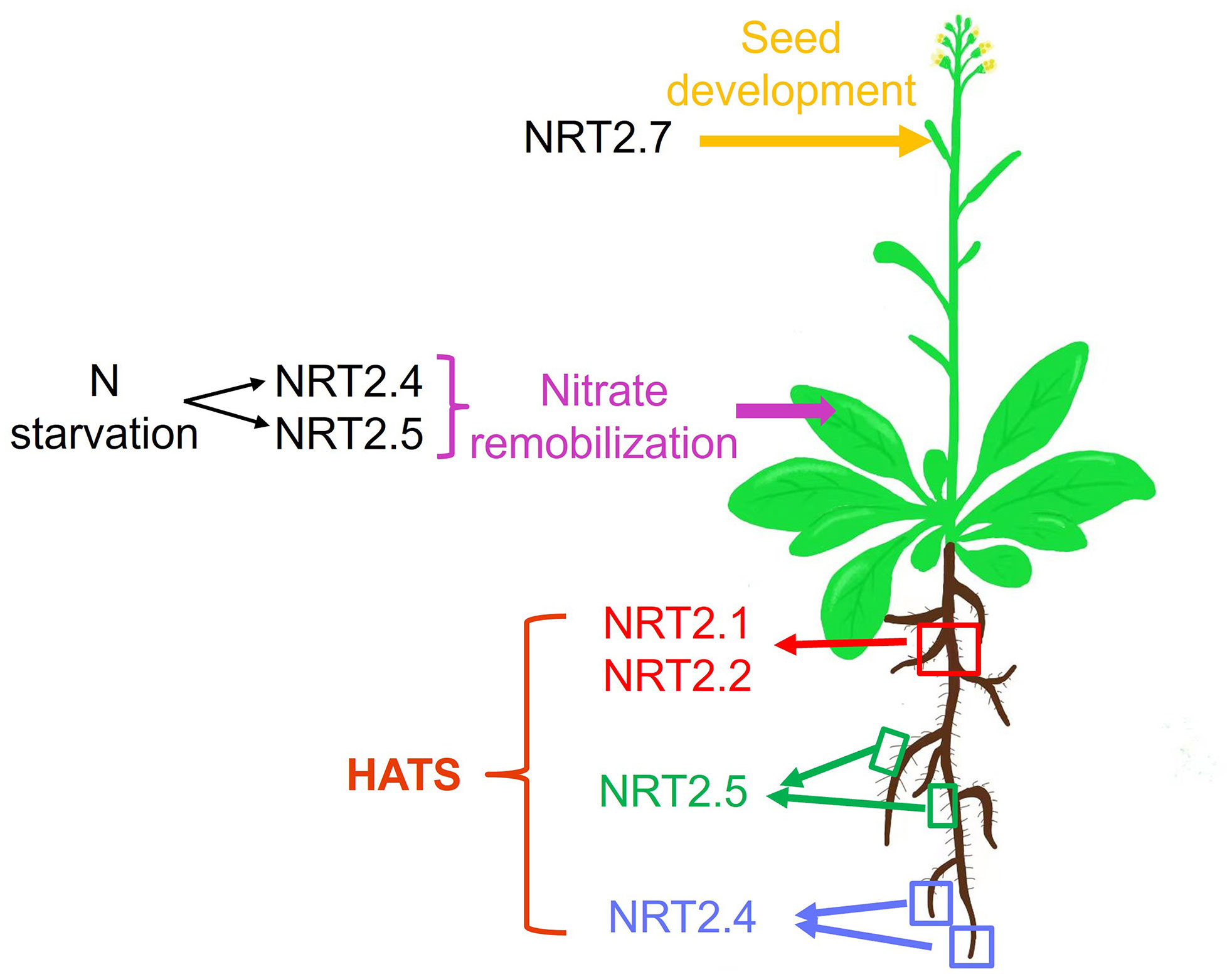
Figure 1 The physiological roles played by Arabidopsis NRT2 nitrate transporters, from the uptake of soil nitrate to its remobilization among leaves and the function of these transporters during seed development. HATS: high-affinity transport system; red square indicates the older parts of both primary roots and lateral roots; green squares indicate root hair of both primary roots and lateral roots; blue squares indicate the younger parts of both primary roots and lateral roots.
4 Plant growth and development
4.1 Shoot growth
Members of the NRT2 family are vital to the activity of the NO3- HATS (Cerezo et al., 2001; Filleur et al., 2001; Li et al., 2007), and they thereby strongly influence plant growth when cultivating plants in the context of NO3–poor solution (Orsel et al., 2004; Li et al., 2007; Kiba et al., 2012; Lezhneva et al., 2014). Indeed, nrt2.1 nrt2.2 double mutant plants exhibit markedly reduced shoot growth as compared to wild-type plants, with their leaves turning a pink color (Li et al., 2007). The growth of Arabidopsis plants for 4 weeks in hydroponic tanks in a 1 mM NH4NO3 solution with subsequent nitrogen starvation for 1 week resulted in significantly lower shoot-to-root ratios and shoot weight for nrt2.1 mutants as compared with wild-type plants, while no corresponding changes were evident for nrt2.2 single mutants, and even greater reductions in both parameters were evident for nrt2.1 nrt2.2 double mutants relative to plants in which only nrt2.1 was mutated (Li et al., 2007). This supports a model wherein both of these genes serve as important regulators of growth, albeit with NRT2.1 playing the most important role in this context. While no significant differences in nrt2.4 single mutant plant fresh weight were detected when compared to wild-type controls cultivated in the presence of low (0.05 or 0.5 mM) KNO3 levels, the triple nrt2.1 nrt2.2 nrt2.4 mutation was associated with even greater reductions in biomass productivity relative to nrt2.1 nrt2.2 mutants, particularly at the lower tested KNO3 level. This may be attributable to the ability of NRT2.4 to mediate the high-affinity transport of nitrate only when its concentrations are very low (Kiba et al., 2012). In a similar vein, nrt2.5 mutants and wild-type Arabidopsis exhibited comparable shoot biomass, while a 10% drop in shoot fresh weight was detected for triple nrt2.1 nrt2.2 nrt2.5 mutant plants, and this decline was even more marked if nrt2.4 was also mutated (Lezhneva et al., 2014). Together, these data provide clear evidence for the essential roles that NRT2.1, NRT2.2, NRT2.4, and NRT2.5 play in supporting plant growth when N levels are limited. This is consistent with the fact that the expression of these four genes is largely restricted to the roots, wherein they facilitate root NO3- influx (Filleur and Daniel-Vedele, 1999; Orsel et al., 2002; Okamoto et al., 2003; Vidal et al., 2020). Rightfully so, mutation of the partner protein NRT3.1 resulted in the poor shoot growth when plant grown on plates containing 250 mM NO3- as the sole nitrogen source (Okamoto et al., 2006). Shoot growth deficiencies in these mutants may thus be attributable to the long-distance effects of NO3-, indicative of the shifts in whole-plant N distributions.
4.2 Root system architecture
In order to contend with shifting soil N source availability under changing environmental conditions, plants have evolved a range of adaptive strategies that include root system architecture plasticity (Robinson, 1994; Zhang and Forde, 2000). Both nitrate and sugar can induce the expression of NRT2.1 (Lejay et al., 1999; Cerezo et al., 2001). Relative to the use of standard growth medium, cultivating seedlings on media with a high C/N ratio results in the significant repression of lateral root initiation compared to a standard growth medium (Malamy and Ryan, 2001), whereas this repression was not evident in nrt2.1 mutant plants (Little et al., 2005), supporting a role for NRT2.1 in this repressive mechanism. Under nitrate-free conditions, such repression of lateral root initiation was still evident, simulating defective transport activity and thus revealing that this impairment of lateral root initiation was not nitrogen uptake-dependent (Remans et al., 2006b). NRT2.1 may thus serve as a sensor or signal transducer for nitrate involved in a signaling pathway that ultimately represses lateral root initiation. Relative to wild-type plants, those harboring nrt2.1 mutations also presented with lower levels of lateral root growth following transfer from nitrate-rich to nitrate-poor medium (10 to 0.5 mM) (Remans et al., 2006b; Li et al., 2007), with this reduction being even more pronounced in nrt2.1 nrt2.2 double mutants consistent with both of these genes serving as important factors involved in the regulation of lateral root growth (Li et al., 2007). NRT2.1 thus appears to help coordinate the development of lateral roots when NO3- availability is limited.
Nitrate treatment can reportedly enhance the initiation and emergence of lateral roots (Vidal et al., 2010, 2013), with both of these processes being impaired for nrt2.1 nrt2.2 mutant plants relative to wild-type controls cultivated in the presence of 1 mM ammonium for 2 weeks in a hydroponic system followed by a 3-day treatment with 5 mM KNO3. Strikingly, tga1 tga4 and nrt2.1 nrt2.2 plants presented with similar lateral root initiation phenotypes (Alvarez et al., 2014). In subsequent analyses, the TGA1/TGA4 transcription factors were identified as direct regulators of NRT2.1/NRT2.2 (Alvarez et al., 2014), supporting their ability to regulate the development of lateral roots at least in part through the control of NRT2.1 and NRT2.2 expression.
A recent forward genetic screening effort additionally established NRT2.1 as a key regulator of primary root elongation under limited NO3- stress conditions, as evidenced by the significant increase in primary root length for nrt2.1 mutant seedlings cultivated in the presence of 0.05 mM NO3- relative to wild-type controls. The root tips of these nrt2.1 seedlings also exhibited higher levels of the key root growth regulator auxin as compared to wild-type root tips in the presence of low nitrate concentrations. However, nrt2.1 pin7 double mutants exhibited root tips comparable to those of wild-type plants and shorter than those of nrt2.1 plants, consistent with the ability of PIN7, which is an auxin efflux carrier, to function downstream of NRT2.1 as a regulator of the growth of roots in the presence of limited NO3- availability. A series of assays confirmed the ability of PIN7 and NRT2.1 to physically interact with one another when NO3- levels are low, thereby suppressing the PIN7-mediated acropetal efflux of auxin, thus slowing the elongation of primary roots. Together these results support a model in which NRT2.1 is capable of influencing root growth activity through interactions with the PIN7-mediated auxin transport machinery when levels of available NO3- are low (Wang et al., 2023).
4.3 Seed development and germination
There are three primary stages to the process of Arabidopsis seed development. After initial morphogenesis, a maturation phase occurs that entails the production of N and C storage compounds in the form of seed storage proteins (Heath et al., 1986; Baud et al., 2002). NPF2.12/NRT1.6 localizes to the plasma membrane and is vital for early embryonic development (Almagro et al., 2008), with mutations in this gene reducing rates of nitrate accumulation within mature seeds while enhancing seed abortion rates. NPF2.12 expression was only detectable in funiculus and silique vascular tissues, with upregulation immediately following pollination. This suggests a role for NPF2.12 in the delivery of nitrate from maternal tissues to nascent embryos (Almagro et al., 2008). In contrast, NRT2.7 is primarily active within mature seeds. The homology of NRT2.7 is markedly lower relative to other NRT2 family members, sharing just 55% similarity with NRT2.1, for example (Orsel et al., 2002; Chopin et al., 2007a). Unlike most other members of this gene family, it is also primarily expressed in developing seeds rather than in roots, with its upregulation being particularly pronounced as seeds undergo dessication (Orsel et al., 2002; Okamoto et al., 2003; Chopin et al., 2007a). While oocyte-based experiments have confirmed that NRT2.7 can function as a nitrate transporter, it has no role in the direct uptake of soil nitrate via the roots, nor does it impact the distribution of nitrate in plant vegetative organs (Chopin et al., 2007a). Subcellular localization analyses have demonstrated that NRT2.7 primarily localizes to the tonoplast surrounding the vacuoles. Studies of the effects of nrt2.7 mutations on seeds have been conducted with the mutant nrt2.7-1 (Col-8 background) and nrt2.7-2 (Ws background) plant lines. Both exhibit similar seed weights to those of wild-type plants, but reduced seed nitrate levels under nonlimiting N conditions. Nitrate has also been posited to serve as a signal that can trigger seeds to break dormancy and begin germination (Alboresi et al., 2005; Chopin et al., 2007a). In line with such a model, when the same batches of freshly harvested seeds were sown on water-containing medium, both nrt2.7 mutants exhibited germination delays relative to wild-type controls within 2 days. While nrt2.7-2 mutants exhibited lower rates of germination throughout a 7-day analytical period relative to the control Ws line, no apparent difference in germination was evident between the Col-8 and nrt2.7-1 mutant lines from days 3-7 post-sowing (Chopin et al., 2007a). Col seeds and foliar tissues exhibited higher nitrate storage capabilities relative to those of Ws plants, suggesting that Col plants are better able to tolerate N deprivation (Chopin et al., 2007a; North et al., 2009). Differences in such tolerance among plant ecotypes may thus account for varying seed germination phenotypes. Overall, these data highlight a key role for NRT2.7 in seed nitrate concentration and germination.
David et al. performed further characterization of nrt2.7-2 mutants exhibiting a distinctive phenotype consisting of a seed coat that was a plane-brown color in contrast to that of wild-type Ws (David et al., 2014). Seed coloration is generally related to flavonoid oxidation levels (Pourcel et al., 2005; Lepiniec et al., 2006; Routaboul et al., 2012), and additional analysis indicated that these nrt2.7-2 mutant seeds accumulated higher levels of soluble proanthocyanidins (PAs) that could undergo oxidation in the testa with seed dessication (David et al., 2014). This seed PA accumulation was apparently unrelated to fluctuations in seed NO3- content, in line with the observation that npf2.12 and clca mutant seeds did not exhibit any change in color or PA content despite the reduction in NO3- levels therein (Almagro et al., 2008; Monachello et al., 2009). These data support a specific link between the accumulation of PA in seeds and the function of NRT2.7, rather than linking it to NO3- accumulation. Lower NO3- levels and higher concentrations of soluble PAs were also apparent in nrt2.7-2 mutant seeds relative to Ws, resembling tt10 mutant phenotypes (David et al., 2014). The TRANSPARENT TESTA 10 (TT10) protein serves as a laccase candidate enzyme that facilitates the oxidative polymerization of PAs and other flavonoids (Pourcel et al., 2005). No studies to date, however, have revealed any ability of NRT2.7 to influence the enzymatic activity of TT10, and additional research aimed at clarifying the activity of TT10 will be vital to understanding the mechanisms that ultimately result in the higher levels of soluble Pas within nrt2.7-2 seeds. These findings thus reveal a central role for NRT2.7 as a regulator of the accumulation and oxidation of PAs within seeds. While NRT1 family proteins have been shown to serve as transporters for non-nitrate molecules (Léran et al., 2014), whether NRT2 proteins can function in a similar manner remains poorly understood, and additional research will be vital to test this hypothesis.
5 AtNRT2.1, AtNRT2.5, and AtNRT2.6 are influence plant-microbe interactions
Plant nutritional status can strongly shape the ability of these plants to defend against pathogens such as Pseudomonas syringae (Long et al., 2000; Modolo et al., 2005, 2006). Relative to wild-type controls, nrt2.1 and nrt2.1 nrt2.2 mutant plants exhibit a reduction in susceptibility to P. syringae pv tomato DC3000 (Pst) (Camañes et al., 2012). Under infection conditions, nrt2.1 exhibited more robust and more rapid SA-dependent defense priming, which was a key mechanism responsible for enhanced Pst resistance (Zimmerli et al., 2000; Conrath et al., 2006; Jung et al., 2009). These nrt2.1 mutants were also partially deficient in their ability to detect coronatine, a bacterial effector important in the context of infection (Brooks et al., 2004; Melotto et al., 2008; Camañes et al., 2012). These decreases in nrt2.1 susceptibility to Pst may thus stem from both coronatine insensitivity and improved SA priming. The inoculation of plants with the phytopathogen Erwinia amylovora also resulted in an increase in the expression of NRT2.6, with plants expressing lower NRT2.6 levels exhibiting greater pathogen susceptibility as a consequence of impaired reactive oxygen species production, although these nrt2.6 mutants did not exhibit any apparent nitrate-associated phenotypes (Dechorgnat et al., 2012). Together, these data suggest that members of the NRT2 family can serve as sensors for a range of environmental stimuli, thereby coordinating abiotic and biotic stress responses in addition to shaping the ability of plants to respond to nutritional cues.
The plant growth-promoting rhizobacterium (PGPR) strain Phyllobacterium brassicacearum STM196 has been reported to promote the growth of Arabidopsis and to overcome lateral root developmental inhibition under conditions of high nitrate availability (Mantelin et al., 2006). Notable increases in NRT2.5 and NRT2.6 expression have been observed in plants exposed to STM196, but nrt2.5 and nrt2.6 mutants failed to exhibit such STM196-induced growth (Kechid et al., 2013), indicating that these two genes encode proteins that can influence the outcomes of beneficial biotic interactions.
6 Biological processes by AtNRT2.1
6.1 AtNRT2.1 controls cadmium uptake
Supplying plants with NO3- has been shown to result in higher concentrations of Cd and more pronounced Cd toxicity in exposed plants (Mao et al., 2014; Yang et al., 2015; Cheng et al., 2020). Moreover, nitrate transporters NPF6.3/NRT1.1, NPF7.3/NRT1.5, and NPF7.2/NRT1.8 are responsive to Cd stress conditions in Arabidopsis, regulating the accumulation of Cd under conditions of both high and normal NO3- availability (Li et al., 2010; Chen et al., 2012; Mao et al., 2014; Wang et al., 2018). Further studies have indicated that Cd can suppress the expression of key HATS-related genes including NRT2.1, NRT2.2, and NRT2.4, thereby suppressing the uptake and accumulation of nitrate in roots when nitrate levels are low, which results in a corresponding reduction in root Cd uptake (Guan et al., 2021). This suggests that efforts to control nitrate transporter activity may provide a means of abrogating Cd accumulation when growing crops in soil contaminated with this heavy metal.
6.2 AtNRT2.1 influences light-responsive carbon and nitrogen metabolism
As reported previously, root transport systems are generally regulated by shoot photosynthetic activity, especially in the context of the uptake of NO3- (Delhon et al., 1995; Forde, 2002). NO3- uptake is under the control of downwardly transported sugars, CO2, carboxylic acids, and other photosynthates (Delhon et al., 1996). Root NRT2.1 expression has been demonstrated to be both sugar- and light-inducible (Lejay et al., 1999). However, sugar analog treatment or analyses of sugar-sensing mutant plants revealed no changes in sugar-induced NRT2.1 induction, suggesting that this process occurs through a mechanism distinct from the primary mechanisms that have been documented to facilitate plant sugar sensing (Lejay et al., 2003). Mutants lacking the expression of hexokinase (HXK), in contrast, exhibited an absence of sugar-induced NRT2.1 expression, consistent with the metabolic activity downstream of HXK being key to this regulatory process, rather than sugar itself (Lejay et al., 2003). HXK catalyzes a reaction that produces glucose-6-P (G6P), and treating roots with glycerol to reduce G6P concentrations can strongly impair normal NRT2.1 upregulation following the dark/light transition (Lejay et al., 2008). However, the treatment of plants with 6-aminonicotinamide (6-AN), which can potently inhibit phosphor-gluconate dehydrogenase activity and impair OPPP, the near total absence of sugar-induced NRT2.1 expression was evident despite no corresponding change in G6P levels relative to sucrose treatment. This suggests that NRT2.1 upregulation in response to C signals is associated with OPPP activity rather than being directly induced by G6P (Lejay et al., 2008). Relative to wild-type plants, gin2-1 mutant plants with defective sugar responses exhibited impaired NRT2.1 upregulation in response to Glc, while treatment with the OPPP intermediates shikimate and pyruvate was sufficient to restore this defect (de Jong et al., 2014). Sugar-induced NRT2.1 expression is thus dependent on the OPPP pathway.
C and N acquisition rates are regulated in a tightly coupled manner (Matt et al., 2010; Nunes-Nesi et al., 2010), and light serves an important regulatory role for both of these processes (Lillo, 2008). Wild-type Arabidopsis seedlings with shoots and roots respectively exposed to light and dark conditions [S(L)/R(D)] exhibited significant increases in both primary root length and NO3- uptake as compared to wild-type seedlings placed under the opposite conditions [S(D)/R(L)], supporting a model wherein light-induced shoot-to-root signaling activity can favor nitrate uptake and the growth of roots (Chen X. et al., 2016). Strikingly, mutations in the HY5 gene encoding a photomorphogenic bZIP transcription factor were capable of eliminating this nitrate uptake and root growth induced by shoot illumination, and further hypocotyl graft chimera-based studies codified HY5 as a shoot-root phloem-mobile signal (Chen X. et al., 2016). The nrt2.1 mutant plants also exhibited reduced levels of NO3- uptake in response to shoot illumination, with HY5 derived from shoot tissue promoting the autoactivation of HY5 in the roots, thereby promoting NO3- uptake in the roots via NRT2.1 activation (Chen X. et al., 2016). NRT2.1 promoter binding by HY5 can be enhanced when sucrose availability, with HY5 regulating its fixation and translocation (Chen X. et al., 2016). Mobile HY5 thus serves as an important regulator of NRT2.1 in the context of illumination-responsive N and C metabolism.
6.3 AtNRT2.1 involves in systemic nitrate signaling mechanisms
Soil nitrate distributions are generally heterogeneous. To adapt to this inconsistent availability, plants have evolved intricate systemic responses whereby stimuli that are perceived at the local level can be communicated to distant organs. For example, in plants grown in split-root plates for which half of their root system was nitrate-deprived while the other half was in a nitrate-rich environment, more pronounced proliferation of roots on the nitrate-rich side was evident relative to plants cultivated under homogenously nitrate-rich conditions, with this response being dependent on shoot nitrate accumulation (Remans et al., 2006a; Ruffel et al., 2011; Vidal et al., 2020). The transcription factor gene TEOSINTE BRANCHED1/CYCLOIDEA/PROLIFERATING CELL FACTOR1-20 (TCP20) mutations in these split-root experiments were sufficient to impair the preferential growth of lateral roots on the nitrate-rich site (Guan et al., 2014). Additional analyses demonstrated the ability of TCP20 to interact directly with HOMOLOG OF BRASSINOSTEROID ENHANCED EXPRESSION2 INTERACTING WITH IBH1 (HBI1), a bHLH transcription factor, with nitrate starvation enhancing this interaction. The resultant HBI1-TCP20 complex was then capable of directly binding the C-terminally encoded peptide (CEP) promoters, inducing their expression in a cooperative fashion (Chu et al., 2021). The resultant CEPs were secreted from roots and functioned as ascending signals of N starvation that were detected by the LRR-RK receptor (CEPR) in shoots, thereby triggering shoot-derived descending CEPD1 (CEP downstream 1) and CEPD2 peptide production. These peptides, in turn, triggered the upregulation of NRT2.1, NRT3.1, and other nitrate-related genes in the roots, thereby inducing higher levels of nitrate uptake and root proliferation in areas rich in nitrates (Tabata et al., 2014; Ohkubo et al., 2017). The CEPD-like 2 (CEPDL2) peptide was also induced by low nitrate concentrations and shoot N deprivation in a CEPR-independent fashion, whereupon it functions as a leaf-derived systemic signal that controls the root-mediated uptake of nitrate by regulating the expression of key genes including NRT2.1, NRT3.1, and NRT2.4 (Ota et al., 2020). The RNAPII complex component IWS1 is also capable of suppressing the expression of NRT2.1 under high levels of nitrate availability by enhancing the H3K27me3 of the chromatin region encoding this gene (Girin et al., 2010; Widiez et al., 2011). A range of post-translational mechanisms also shape the activity of NRT2.1 in response to systemic N-related signaling activity. For example, the CEPD1/2- and CEPDL2-mediated signals are capable of promoting the upregulation of CEPH, a root-specific PP2C phosphatase that can dephosphorylate NRT2.1 Ser501 to activate the high-affinity uptake of nitrate in Arabidopsis (Ohkubo et al., 2021). At the systemic level, both transcriptional and post-translational switches govern the activity of NRT2.1 to maintain N homeostasis, particularly under conditions of limited soil N availability and/or increased shoot N demands (Figure 2).
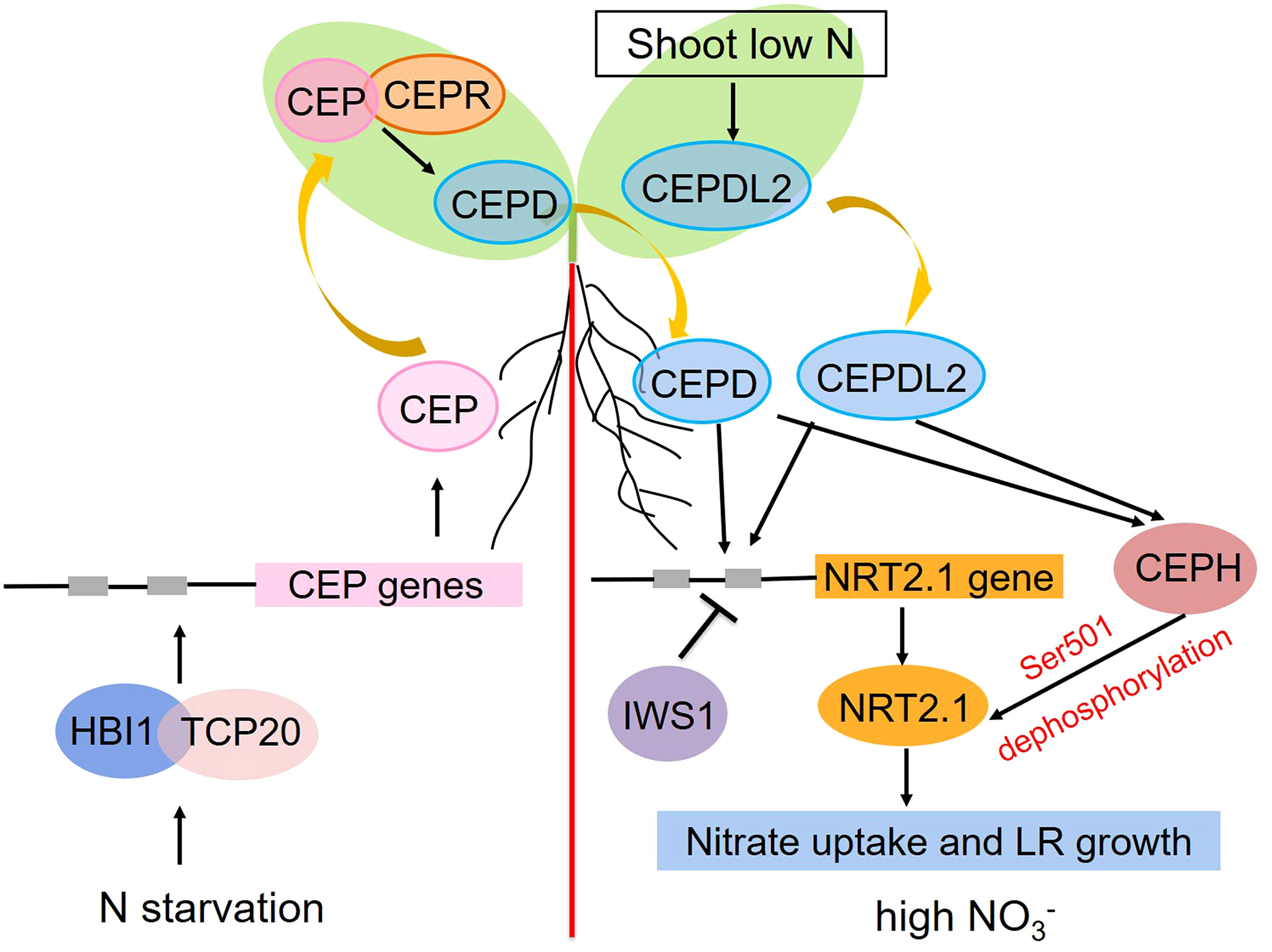
Figure 2 Schematic overview of the roles that AtNRT2.1 plays in the systemic nitrate signaling pathway under heterogeneous conditions. Positive and negative regulation are respectively represented with arrows and blunted lines.
7 Conclusions
Since the first identification of crnA as the founding member of the NRT2 gene family, countless studies have sought to clarify the characteristics and functions of different NRT2 proteins in specific plant species. This review offers an overview of the spatiotemporal expression patterns, localization, biotic and abiotic responses, and functional roles of the seven AtNRT2 genes (Table 1). While some progress has been made on this front to date, much remains to be done. For example, how AtNRT2.3 and AtNRT2.6 contribute to nitrate-related regulatory processes remains uncertain, and it remains uncertain as to whether NRT2 proteins can serve as transporters for molecules other than nitrate. Similarly, the status of NRT2.1 as a potential nitrate sensor warrants further investigation. As such, additional studies will be vital to fully clarify how plants are able to sense and respond to changing environmental conditions in order to provide a sound basis for crop-focused research.
Arabidopsis-based findings can be extended to other economically important plant species, highlighting valuable targets for efforts to enhance crop yields in harsh or otherwise unfavorable environments. In recent years, significant progress has been made in the research of NRT2 genes in crops. In rice, overexpression of OsNRT2.1 increased the transcription level of auxin transporter genes OsPIN1a/b/c and OsPIN2 and which in turn promoted total root length under 0.5 mM NO3- conditions (Naz et al., 2019). Knockout of OsNRT2.4 inhibited lateral root length and number under 0.25 Mm and 2.5 mM NO3- conditions (Wei et al., 2018). In addition, knockdown of OsNAR2.1 suppressed lateral root formation under low NO3- conditions (Song et al., 2020). These results suggest that OsNRT2.1/NRT2.4/NAR2.1 play a critical role in controlling root development. Moreover, transgenic lines overexpressing OsNRT2.1/2.2 could improve nitrogen use efficiency (NUE) and grain yield in rice (Chen J. et al., 2016; Hu et al., 2023). Transgenic lines co-overexpressing OsNAR2.1 and OsNRT2.3a increased agronomic nitrogen use efficiency (Chen et al., 2020). Meanwhile, Fan’s group reported that overexpression of OsNRT2.3b could also enhance NUE and rice grain yield in the field (Fan et al., 2016). In wheat, transcription factor TaNAC2-5A could directly bind to the promoters of TaNRT2.1-6B, TaNRT2.5-3B, TaNPF7.1-6D, and TaGS2-2A and activate their expression to affect NO3- transport and assimilation, and ultimately increased grain yield and NUE (Li et al., 2020; Gao et al., 2022). Furthermore, new information have been obtained about unexpected peculiar roles played by some NRT2 genes. The NRT2 transporter family has at least 4 members in Lotus japonicus (Criscuolo et al., 2012), 5 in Oryza sativa (Feng et al., 2011b) and 49 in the Wheat genome (Deng et al., 2023). In Lotus japonicus, high NO3- treatment attenuated nodulation, but the effect of nitrate was suppressed by the LjNRT2.1 mutation (Misawa et al., 2022). Of note, the nodulation phenotypes of the Ljnlp1 and the Ljnlp4 mutants are similar to those of the Ljnrt2.1 mutants under high NO3- conditions (Nishida et al., 2021). Further investigation suggested that NODULE INCEPTION (NIN)-LIKE PROTEIN1 (LjNLP1) could directly bind to the LjNRT2.1 promoter and activate its transcript and subsequently promote nitrate uptake/transport process, which ultimately affected nuclear localization of LjNLP4 and subsequent regulation of the expression of nodulation-related genes (Misawa et al., 2022). Phylogenic analysis revealed LjNRT2.4 to be a close relative of AtNRT2.7 which was the most diverged of all the NRT2 sequence (Valkov et al., 2020). Unlike AtNRT2.7 which expressed mainly in seeds and the protein localized to vacuolar membrane (Chopin et al., 2007a), LjNRT2.4 was expressed in root and nitrogen-fixing nodule vascular tissues and localized at the plasma membrane. Mutation of LjNRT2.4 caused much more severe N2-fixation related phenotypes in nodulated plants grown under hydroponic conditions (Valkov et al., 2020). In rice, all OsNRT2 members except OsNRT2.4 which shares the highest value of amino acid identity with AtNRT2.7 need OsNAR2.1 for root NO3- acquisition in response to both low- and high- nitrate supply (Wei et al., 2018). Interestingly, unlike other NRT2s which function as the high-affinity NO3- transporter, OsNRT2.4 is a dual-affinity NO3- transporter (Wei et al., 2018). These findings are just the tip of the iceberg, more endeavors are needed to decipher the mechanism of NRT2 family, improve NUE in crops, eliminate the pollution from N as field fertilizer, and maintain nutrient homeostasis.
Author contributions
NX: Writing – original draft. LC: Writing – original draft. YK: Writing – original draft. GC: Writing – review & editing. LZ: Writing – review & editing. FL: Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This research was funded by the National Natural Science Foundation of China (No. 32000194), the Shandong Provincial University Youth Innovation Team, China (No. 2022KJ102), the Shandong Provincial Natural Science Foundation (No. ZR201702210017), the Open Project Program of State Key Laboratory of Crop Biology (No. 2021KF04), the Education Science Planning Project for the Subject of Innovation Capability of Shandong Province (No. 2022CYB210).
Acknowledgments
The authors acknowledge the support of many fundings.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Alboresi, A., Gestin, C., Leydecker, M. T., Bedu, M., Meyer, C., Truong, H. N. (2005). Nitrate, a signal relieving seed dormancy in Arabidopsis. Plant Cell Environ. 28, 500–512. doi: 10.1111/j.1365-3040.2005.01292.x
Almagro, A., Lin, S. H., Tsay, Y. F. (2008). Characterization of the Arabidopsis nitrate transporter NRT1.6 reveals a role of nitrate in early embryo development. Plant Cell 20, 3289–3299. doi: 10.1105/tpc.107.056788
Alvarez, J. M., Riveras, E., Vidal, E. A., Gras, D. E., Contreras-López, O., Tamayo, K. P., et al. (2014). Systems approach identifies TGA1 and TGA4 transcription factors as important regulatory components of the nitrate response of Arabidopsis thaliana roots. Plant J. 80, 1–13. doi: 10.1111/tpj.12618
Baud, S., Boutin, J. P., Miquel, M., Lepiniec, L. C., Rochat, C. (2002). An integrated overview of seed development in Arabidopsis thaliana ecotype WS. Plant Physiol. Biochem. 40, 151–160. doi: 10.1016/S0981-9428(01)01350-X
Brooks, D. M., Hernández-Guzmán, G., Kloek, A. P., Alarcón-Chaidez, F., Sreedharan, A., Rangaswamy, V., et al. (2004). Identification and characterization of a well-defined series of coronatine biosynthetic mutants of Pseudomonas syringae pv. tomato DC3000. Mol. Plant Microbe Interact. 17, 162–174. doi: 10.1094/MPMI.2004.17.2.162
Brownlee, A. G., Arst, H. N. (1983). Nitrate uptake in Aspergillus nidulans and involvement of the third gene of the nitrate assimilation gene cluster. J. Bacteriol. 155, 1138–1146. doi: 10.1128/jb.155.3.1138-1146.1983
Camañes, G., Pastor, V., Cerezo, M., García-Andrade, J., Vicedo, B., García-Agustín, P., et al. (2012). A deletion in NRT2.1 attenuates Pseudomonas syringae-induced hormonal perturbation, resulting in primed plant defenses. Plant Physiol. 158, 1054–1066. doi: 10.1104/pp.111.184424
Cerezo, M., Tillard, P., Filleur, S., Munos, S., Daniel-Vedele, F., Gojon, A. (2001). Major alterations of the regulation of root NO3- uptake are associated with the mutation of Nrt2.1 and Nrt2.2 genes in Arabidopsis. Plant Physiol. 127, 262–271. doi: 10.1104/pp.127.1.262
Chen, J., Liu, X., Liu, S., Fan, X., Zhao, L., Song, M., et al. (2020). Co-overexpression of OsNAR2.1 and OsNRT2.3a increased agronomic nitrogen use efficiency in transgenic rice plants. Front. Plant Sci. 11. doi: 10.3389/fpls.2020.01245
Chen, C. Z., Lv, X. F., Li, J. Y., Yi, H. Y., Gong, J. M. (2012). Arabidopsis NRT1.5 is another essential component in the regulation of nitrate reallocation and stress tolerance. Plant Physiol. 159, 1582–1590. doi: 10.1104/pp.112.199257
Chen, J., Yong, Z., Tan, Y., Min, Z., Fan, X. (2016). Agronomic nitrogen-use efficiency of rice can be increased by driving OsNRT2.1 expression with the OsNAR2.1 promoter. Plant Biotechnol. J. 14, 1705–1715. doi: 10.1111/pbi.12531
Chen, X., Yao, Q., Gao, X., Jiang, C., Harberd, N. P., Fu, X. (2016). Shoot-to-root mobile transcription factor HY5 coordinates plant carbon and nitrogen acquisition. Curr. Biol. 26, 640–646. doi: 10.1016/j.cub.2015.12.066
Cheng, Y., Bao, Y., Chen, X., Yao, Q., Wang, C., Chai, S., et al. (2020). Different nitrogen forms differentially affect Cd uptake and accumulation in dwarf Polish wheat (Triticum polonicum L.) seedlings. J. Hazard. Mater. 400, 123209. doi: 10.1016/j.jhazmat.2020.123209
Chopin, F., Orsel, M., Dorbe, M. F., Chardon, F., Truong, H. N., Miller, A. J., et al. (2007a). The Arabidopsis ATNRT2.7 nitrate transporter controls nitrate content in seeds. Plant Cell 19, 1590–1602. doi: 10.1105/tpc.107.050542
Chopin, F., Wirth, J., Dorbe, M. F., Lejay, L., Krapp, A., Gojon, A., et al. (2007b). The Arabidopsis nitrate transporter AtNRT2.1 is targeted to the root plasma membrane. Plant Physiol. Biochem. 45, 630–635. doi: 10.1016/j.plaphy.2007.04.007
Chu, X., Li, M., Zhang, S., Fan, M., Han, C., Xiang, F., et al. (2021). HBI1-TCP20 interaction positively regulates the CEPs-mediated systemic nitrate acquisition. J. Integr. Plant Biol. 63, 902–912. doi: 10.1111/jipb.13035
Conrath, U., Beckers, G. J., Flors, V., García-Agustín, P., Jakab, G., Mauch, F., et al. (2006). Priming: Getting ready for battle. Mol. Plant Microbe Interact. 19, 1062–1071. doi: 10.1094/MPMI-19-1062
Crawford, N. M., Forde, B. G. (2002). Molecular and developmental biology of inorganic nitrogen nutrition. Arabidopsis Book 1, e0011. doi: 10.1199/tab.0011
Crawford, N. M., Glass, A. D. M. (1998). Molecular and physiological aspects of nitrate uptake in plants. Trends Plant Sci. 3, 389–395. doi: 10.1016/S1360-1385(98)01311-9
Criscuolo, G., Valkov, V. T., Parlati, A., Alves, L. M., Chiurazzi, M. (2012). Molecular characterization of the Lotus japonicus NRT1(PTR) and NRT2 families. Plant Cell Environ. 35, 1567–1581. doi: 10.1111/j.1365-3040.2012.02510.x
David, L. C., Dechorgnat, J., Berquin, P., Routaboul, J. M., Debeaujon, I., Daniel-Vedele, F., et al. (2014). Proanthocyanidin oxidation of Arabidopsis seeds is altered in mutant of the high-affinity nitrate transporter NRT2.7. J. Exp. Bot. 65, 885–893. doi: 10.1093/jxb/ert481
Dechorgnat, J., Patrit, O., Krapp, A., Fagard, M., Daniel-Vedele, F. (2012). Characterization of the Nrt2.6 gene in Arabidopsis thaliana: A link with plant response to biotic and abiotic stress. PloS One 7, e42491. doi: 10.1371/journal.pone.0042491
de Jong, F., Thodey, K., Lejay, L. V., Bevan, M. W. (2014). Glucose elevates NITRATE TRANSPORTER2.1 protein levels and nitrate transport activity independently of its HEXOKINASE1-mediated stimulation of NITRATE TRANSPORTER2.1 expression. Plant Physiol. 164, 308–320. doi: 10.1104/pp.113.230599
Delhon, P., Gojon, A., Tilard, P., Passama, L. (1995). Diurnal regulation of NO3- uptake in soybean plants I. Changes in NO3- influx, efflux, and N utilization in the plant during the day/night cycle. J. Exp. Bot. 46, 1585–1594. doi: 10.1093/jxb/46.10.1585
Delhon, P., Gojon, A., Tilard, P., Passama, L. (1996). Diurnal regulation of NO3- uptake in soybean plants. III. Implication of the Dijkshoorn-Ben Zioni model in relation with the diurnal changes in NO3- assimilation. J. Exp. Bot. 47, 885–892. doi: 10.1093/jxb/47.7.885
Deng, Q. Y., Luo, J. T., Zheng., J. M., Tan, W. F., Pu, Z. J., Wang, F. (2023). Genome-wide systematic characterization of the NRT2 gene family and its expression profile in wheat (Triticum aestivum L.) during plant growth and in response to nitrate deficiency. BMC. Plant Biol. 23, 353. doi: 10.1186/s12870-023-04333-5
Engelsberger, W. R., Schulze, W. X. (2012). Nitrate and ammonium lead to distinct global dynamic phosphorylation patterns when resupplied to nitrogen-starved Arabidopsis seedlings. Plant J. 69, 978–995. doi: 10.1111/j.1365-313X.2011.04848.x
Fan, X., Tang, Z., Tan, Y., Zhang, Y., Luo, B., Yang, M., et al. (2016). Overexpression of a pH-sensitive nitrate transporter in rice increases crop yields. Proc. Natl. Acad. Sci. U. S. A. 114, 7118–7123. doi: 10.1073/pnas.1525184113
Feng, H., Fan, X., Fan, X., Liu, X., Miller, A. J., Xu, G. (2011a). Multiple roles of nitrate transport accessory protein NAR2 in plants. Plant Signal. Behav. 6, 1286–1289. doi: 10.4161/psb.6.9.16377
Feng, H., Yan, M., Fan, X., Li, B., Shen, Q., Miller, A. J., et al. (2011b). Spatial expression and regulation of rice high-affinity nitrate transporters bynitrogen and carbon status. J. Exp. Bot. 62, 2319–2332. doi: 10.1093/jxb/erq403
Filleur, S., Daniel-Vedele, F. (1999). Expression analysis of a high-affinity nitrate transporter isolated from Arabidopsis thaliana by differential display. Planta 207, 461–469. doi: 10.1007/s004250050505
Filleur, S., Dorbe, M. F., Cerezo, M., Orsel, M., Granier, F., Gojon, A., et al. (2001). An arabidopsis T-DNA mutant affected in Nrt2 genes is impaired in nitrate uptake. FEBS Lett. 489, 220–224. doi: 10.1016/S0014-5793(01)02096-8
Forde, B. G. (2000). Nitrate transporters in plants: Structure, function and regulation. Biochim. Biophys. Acta 1465, 219–235. doi: 10.1016/S0005-2736(00)00140-1
Forde, B. G. (2002). Local and long-range signaling pathways regulating plant responses to nitrate. Annu. Rev. Plant Biol. 53, 203–224. doi: 10.1146/annurev.arplant.53.100301.135256
Forde, B. G., Clarkson, D. T. (1999). Nitrate and ammonium nutrition of plants: Physiological and molecular perspectives. Adv. Bot. Res. 30, 1–90. doi: 10.1016/S0065-2296(08)60226-8
Fraisier, V., Gojon, A., Tillard, P., Daniel-Vedele, F. (2000). Constitutive expression of a putative high-affinity nitrate transporter in Nicotiana plumbaginifolia: Evidence for post-transcriptional regulation by a reduced nitrogen source. Plant J. 23, 489–496. doi: 10.1046/j.1365-313x.2000.00813.x
Galván, A., Quesada, A., Fernández, E. (1996). Nitrate and nitrate are transported by different specific transport systems and by a bispecific transporter in Chlamydomonas reinhardtii. J. Biol. Chem. 271, 2088–2092. doi: 10.1074/jbc.271.4.2088
Gao, Y., Qi, S., Wang, Y. (2022). Nitrate signaling and use efficiency in crops. Plant Commun. 3, 100353. doi: 10.1016/j.xplc.2022.100353
Girin, T., El-Kafafi, E. S., Widiez, T., Erban, A., Hubberten, H. M., Kopka, J., et al. (2010). Identification of Arabidopsis mutants impaired in the systemic regulation of root nitrate uptake by the nitrogen status of the plant. Plant Physiol. 153, 1250–1260. doi: 10.1104/pp.110.157354
Glass, A. D. M., Brito, D. T., Kaiser, B. N., Kronzucker, H. J., Kumar, A., Okamoto, M., et al. (2001). Nitrogen transport in plants, with an emphasis on the regulation of fluxes to match plant demand. J. Plant Nutr. Soil Sci. 164, 199–207. doi: 10.1002/1522-2624(200104)164:2<199::AID-JPLN199>3.0.CO;2-K
Glass, A. D. M., Siddiqi, M. Y. (1995). “Nitrogen absorption by plant roots,” in Nitrogen Nutrition in Higher Plants. Ed. Srivastava, H. S., Singh, R. P. (New Delhi, India: Associated Publishing Co.), pp. 21–56.
Gu, C., Song, A., Zhang, X., Wang, H., Li, T., Chen, Y., et al. (2016). Cloning of chrysanthemum high-affinity nitrate transporter family (CmNRT2) and characterization of CmNRT2.1. Sci. Rep. 6, 23462. doi: 10.1038/srep23462
Guan, M., Chen, M., Cao, Z. (2021). NRT2.1, a major contributor to cadmium uptake controlled by high-affinity nitrate transporters. Ecotoxicol. Environ. Saf. 218, 112269. doi: 10.1016/j.ecoenv.2021.112269
Guan, P., Wang, R., Nacry, P., Breton, G., Kay, S. A., Pruneda-Paz, J. L., et al. (2014). Nitrate foraging by Arabidopsis roots is mediated by the transcription factor TCP20 through the systemic signaling pathway. Proc. Natl. Acad. Sci. U. S. A. 111, 15267–15272. doi: 10.1073/pnas.1411375111
Heath, J. D., Weldon, R., Monnot, C., Meinke, D. W. (1986). Analysis of storage proteins in normal and aborted seeds from embryo-lethal mutants of Arabidopsis thaliana. Planta 169, 304–312. doi: 10.1007/BF00392124
Hu, Z., Guo, Y., Ying, S., Tang, Y., Niu, J., Wang, T., et al. (2023). OsCBL1 modulates rice nitrogen use efficiency via negative regulation of OsNRT2.2 by OsCCA1. BMC Plant Biol. 23, 502. doi: 10.1186/s12870-023-04520-4
Ishikawa, S., Ito, Y., Sato, Y., Fukaya, Y., Takahashi, M., Morikawa, H., et al. (2009). Two-component high-affinity nitrate transport system in barley: Membrane localization, protein expression in roots and a direct protein-protein interaction. Plant Biotechnol. 26, 197–205. doi: 10.5511/plantbiotechnology.26.197
Jacquot, A., Chaput, V., Mauries, A., Li, Z., Tillard, P., Fizames, C., et al. (2020). NRT2.1 C-terminus phosphorylation prevents root high affinity nitrate uptake activity in Arabidopsis thaliana. New Phytol. 228, 1038–1054. doi: 10.1111/nph.16710
Jung, H. W., Tschaplinski, T. J., Wang, L., Glazebrook, J., Greenberg, J. T. (2009). Priming in systemic plant immunity. Science 324, 89–91. doi: 10.1126/science.1170025
Kechid, M., Desbrosses, G., Rokhsi, W., Varoquaux, F., Djekoun, A., Touraine, B. (2013). The NRT2.5 and NRT2.6 genes are involved in growth promotion of Arabidopsis by the plant growth-promoting rhizobacterium (PGPR) strain Phyllobacterium brassicacearum STM196. New Phytol. 198, 514–524. doi: 10.1111/nph.12158
Kiba, T., Feria-Bourrellier, A. B., Lafouge, F., Lezhneva, L., Boutet-Mercey, S., Orsel, M., et al. (2012). The Arabidopsis nitrate transporter NRT2.4 plays a double role in roots and shoots of nitrogen-starved plants. Plant Cell 24, 245–258. doi: 10.1105/tpc.111.092221
Kotur, Z., Glass, A. D. (2015). A 150 kDa plasma membrane complex of AtNRT2.5 and AtNAR2.1 is the major contributor to constitutive high-affinity nitrate influx in Arabidopsis thaliana. Plant Cell Environ. 38, 1490–1502. doi: 10.1111/pce.12496
Kotur, Z., Mackenzie, N., Ramesh, S., Tyerman, S., Kaiser, B., Glass, A. (2012). Nitrate transport capacity of the Arabidopsis thaliana NRT2 family members and their interactions with AtNAR2.1. New Phytol. 194, 724–731. doi: 10.1111/j.1469-8137.2012.04094.x
Laugier, E., Bouguyon, E., Mauriès, A., Tillard, P., Gojon, A., Lejay, L. (2012). Regulation of high-affinity nitrate uptake in roots of Arabidopsis depends predominantly on posttranscriptional control of the NRT2.1/NAR2.1 transport system. Plant Physiol. 158, 1067–1078. doi: 10.1104/pp.111.188532
Lejay, L., Gansel, X., Cerezo, M., Tillard, P., Müller, C., Krapp, A., et al. (2003). Regulation of root ion transporters by photosynthesis: Functional importance and relation with hexokinase. Plant Cell 15, 2218–2232. doi: 10.1105/tpc.013516
Lejay, L., Tillard, P., Lepetit, M., Olive, D. F., Filleur, S., Daniel-Vedele, F., et al. (1999). Molecular and functional regulation of two NO3- uptake systems by N- and C-status of Arabidopsis plants. Plant J. 18, 509–519. doi: 10.1046/j.1365-313X.1999.00480.x
Lejay, L., Wirth, J., Pervent, M., Cross, J. M., Tillard, P., Gojon, A. (2008). Oxidative pentose phosphate pathway-dependent sugar sensing as a mechanism for regulation of root ion transporters by photosynthesis. Plant Physiol. 146, 2036–2053. doi: 10.1104/pp.107.114710
Lepiniec, L., Debeaujon, I., Routaboul, J. M., Baudry, A., Pourcel, L., Nesi, N., et al. (2006). Genetics and biochemistry of seed flavonoids. Annu. Rev. Plant Biol. 57, 405–430. doi: 10.1146/annurev.arplant.57.032905.105252
Léran, S., Varala, K., Boyer, J. C., Chiurazzi, M., Crawford, N., Daniel-Vedele, F., et al. (2014). A unified nomenclature of NITRATE TRANSPORTER 1/PEPTIDE TRANSPORTER family members in plants. Trends Plant Sci. 19, 5–9. doi: 10.1016/j.tplants.2013.08.008
Lezhneva, L., Kiba, T., Feria-Bourrellier, A. B., Lafouge, F., Boutet-Mercey, S., Zoufan, P., et al. (2014). The Arabidopsis nitrate transporter NRT2.5 plays a role in nitrate acquisition and remobilization in nitrogen-starved plants. Plant J. 80, 230–241. doi: 10.1111/tpj.12626
Li, J. Y., Fu, Y. L., Pike, S. M., Bao, J., Tian, W., Zhang, Y., et al. (2010). The Arabidopsis nitrate transporter NRT1.8 functions in nitrate removal from the xylem sap and mediates cadmium tolerance. Plant Cell 22, 1633–1646. doi: 10.1105/tpc.110.075242
Li, W., He, X., Chen, Y., Jing, Y., Shen, C., Yang, J., et al. (2020). A wheat transcription factor positively sets seed vigor by regulating the grain nitrate signal. New Phytol. 225, 1667–1680. doi: 10.1111/nph.16234
Li, W., Wang, Y., Okamoto, M., Crawford, N. M., Siddiqi, M. Y., Glass, A. D. (2007). Dissection of the AtNRT2.1:AtNRT2.2 inducible high-affinity nitrate transporter gene cluster. Plant Physiol. 143, 425–433. doi: 10.1104/pp.106.091223
Lillo, C. (2008). Signaling cascades integrating light-enhanced nitrate metabolism. Biochem. J. 415, 11–19. doi: 10.1042/BJ20081115
Little, D. Y., Rao, H., Oliva, S., Daniel-Vedele, F., Krapp, A., Malamy, J. E. (2005). The putative high-affinity nitrate transporter NRT2.1 represses lateral root initiation in response to nutritional cues. Proc. Natl. Acad. Sci. U. S. A. 102, 13693–13698. doi: 10.1073/pnas.0504219102
Liu, R., Jia, T., Cui, B., Song, J. (2020). The expression patterns and putative function of nitrate transporter 2.5 in plants. Plant Signal. Behav. 15, 1815980. doi: 10.1080/15592324.2020.1815980
Long, D. H., Lee, F. N., TeBeest, D. O. (2000). Effect of nitrogen fertilization on disease progress of rice blast on susceptible and resistant cultivars. Plant Dis. 84, 403–409. doi: 10.1094/PDIS.2000.84.4.403
Malamy, J. E., Ryan, K. S. (2001). Environmental regulation of lateral root initiation in Arabidopsis. Plant Physiol. 127, 899–909. doi: 10.1104/pp.010406
Mantelin, S., Desbrosses, G., Larcher, M., Tranbarger, T. J., Cleyet-Marel, J. C., Touraine, B. (2006). Nitrate-dependent control of root architecture and N nutrition are altered by a plant growth-promoting Phyllobacterium sp. Planta 223, 591–603. doi: 10.1007/s00425-005-0106-y
Mao, Q. Q., Guan, M. Y., Lu, K. X., Du, S. T., Fan, S. K., Ye, Y. Q., et al. (2014). Inhibition of nitrate transporter 1.1-controlled nitrate uptake reduces cadmium uptake in Arabidopsis. Plant Physiol. 166, 934–944. doi: 10.1104/pp.114.243766
Matt, P., Geiger, M., Walch-Liu, P., Engels, C., Krapp, A., Stitt, M. (2010). Elevated carbon dioxide increases nitrate uptake and nitrate reductase activity when tobacco is growing on nitrate, but increases ammonium uptake and inhibits nitrate reductase activity when tobacco is growing on ammonium nitrate. Plant Cell Environ. 24, 1119–1137. doi: 10.1046/j.1365-3040.2001.00771.x
Melotto, M., Underwood, W., He, S. Y. (2008). Role of stomata in plant innate immunity and foliar bacterial diseases. Annu. Rev. Phytopathol. 46, 101–122. doi: 10.1146/annurev.phyto.121107.104959
Menz, J., Li, Z., Schulze, W. X., Ludewig, U. (2016). Early nitrogen-deprivation responses in Arabidopsis roots reveal distinct differences on transcriptome and (phospho-) proteome levels between nitrate and ammonium nutrition. Plant J. 88, 717–734. doi: 10.1111/tpj.13272
Misawa, F., Ito, M., Nosaki, S., Nishida, H., Watanabe, H., Suzuki, T., et al. (2022). Nitrate transport via NRT2.1 mediates NIN-LIKE PROTEIN-dependent suppression of root nodulation in Lotus japonicus. Plant Cell 34, 1844–1862. doi: 10.1093/plcell/koac046
Modolo, L. V., Augusto, O., Almeida, I. M., Magalhaes, J. R., Salgado, I. (2005). Nitrite as the major source of nitric oxide production by Arabidopsis thaliana in response to Pseudomonas syringae. FEBS Lett. 579, 3814–3820. doi: 10.1016/j.febslet.2005.05.078
Modolo, L. V., Augusto, O., Almeida, I. M. G., Pinto-Maglio, C. A. F., Oliveira, H. C., Seligman, K., et al. (2006). Decreased arginine and nitrite levels in nitrate reductase-deficient Arabidopsis thaliana plants impair nitric oxide synthesis and the hypersensitive response to Pseudomonas syringae. Plant Sci. 171, 34–40. doi: 10.1016/j.plantsci.2006.02.010
Monachello, D., Allot, M., Oliva, S., Krapp, A., Daniel-Vedele, F., Barbier-Brygoo, H., et al. (2009). Two anion transporters AtClCa and AtClCe fulfil interconnecting but not redundant roles in nitrate assimilation pathways. New Phytol. 183, 88–94. doi: 10.1111/j.1469-8137.2009.02837.x
Naz, M., Luo, B., Guo, X., Li, B., Chen, J., Fan, X. (2019). Overexpression of nitrate transporter OsNRT2.1 enhances nitrate-dependent root elongation. Genes 10, 290. doi: 10.3390/genes10040290
Nazoa, P., Vidmar, J. J., Tranbarger, T. J., Mouline, K., Damiani, I., Tillard, P., et al. (2003). Regulation of the nitrate transporter gene AtNRT2.1 in Arabidopsis thaliana: Responses to nitrate, amino acids and developmental stage. Plant Mol. Biol. 52, 689–703. doi: 10.1023/A:1024899808018
Nishida, H., Nosaki, S., Suzuki, T., Ito, M., Miyakawa, T., Nomoto, M., et al. (2021). Different DNA-binding specificities of NLP and NIN transcription factors underlie nitrate-induced control of root nodulation. Plant Cell 33, 2340–2359. doi: 10.1093/plcell/koab103
North, K. A., Ehlting, B., Koprivova, A., Rennenberg, H., Kopriva, S. (2009). Natural variation in Arabidopsis adaptation to growth at low nitrogen conditions. Plant Physiol. Biochem. 47, 912–918. doi: 10.1016/j.plaphy.2009.06.009
Nunes-Nesi, A., Fernie, A. R., Stitt, M. (2010). Metabolic and signaling aspects underpinning the regulation of plant carbon nitrogen interactions. Mol. Plant 3, 973–996. doi: 10.1093/mp/ssq049
O'Brien, J. A., Vega, A., Bouguyon, E., Krouk, G., Gojon, A., Coruzzi, G., et al. (2016). Nitrate transport, sensing, and responses in plants. Mol. Plant 9, 837–856. doi: 10.1016/j.molp.2016.05.004
Ohkubo, Y., Kuwata, K., Matsubayashi, Y. (2021). A type 2C protein phosphatase activates high-affinity nitrate uptake by dephosphorylating NRT2.1. Nat. Plants 7, 310–316. doi: 10.1038/s41477-021-00870-9
Ohkubo, Y., Tanaka, M., Tabata, R., Ogawa-Ohnishi, M., Matsubayashi, Y. (2017). Shoot-to-root mobile polypeptides involved in systemic regulation of nitrogen acquisition. Nat. Plants 3, 17029. doi: 10.1038/nplants.2017.29
Okamoto, M., Kumar, A., Li, W., Wang, Y., Siddiqi, M. Y., Crawford, N. M., et al. (2006). High-affinity nitrate transport in roots of Arabidopsis depends on expression of the NAR2-like gene AtNRT3.1. Plant Physiol. 140, 1036–1046. doi: 10.1104/pp.105.074385
Okamoto, M., Vidmar, J. J., Glass, A. D. M. (2003). Regulation of NRT1 and NRT2 gene families of Arabidopsis thaliana: Responses to nitrate provision. Plant Cell Physiol. 44, 304–317. doi: 10.1093/pcp/pcg036
Orsel, M., Chopin, F., Leleu, O., Smith, S., Krapp, A., Daniel-Vedele, F., et al. (2006). Characterization of a two-component high-affinity nitrate uptake system in Arabidopsis. Physiology and protein-protein interaction. Plant Physiol. 142, 1304–1317. doi: 10.1104/pp.106.085209
Orsel, M., Eulenburg, K., Krapp, A., Daniel-Vedele, F. (2004). Disruption of the nitrate transporter genes AtNRT2.1 and AtNRT2.2 restricts growth at low external nitrate concentration. Planta 219, 714–721. doi: 10.1007/s00425-004-1266-x
Orsel, M., Krapp, A., Daniel-Vedele, F. (2002). Analysis of the NRT2 nitrate transporter family in Arabidopsis. Structure and gene expression. Plant Physiol. 129, 886–896. doi: 10.1104/pp.005280
Ota, R., Ohkubo, Y., Yamashita, Y., Ogawa-Ohnishi, M., Matsubayashi, Y. (2020). Shoot-to-root mobile CEPD-like 2 integrates shoot nitrogen status to systemically regulate nitrate uptake in Arabidopsis. Nat. Commun. 11, 641. doi: 10.1038/s41467-020-14440-8
Pii, Y., Alessandrini, M., Dall'Osto, L., Guardini, K., Prinsi, B., Espen, L., et al. (2016). Time-resolved investigation of molecular components involved in the induction of [Formula: see text] high affinity transport system in maize roots. Front. Plant Sci. 7. doi: 10.3389/fpls.2016.01657
Pourcel, L., Routaboul, J. M., Kerhoas, L., Caboche, M., Lepiniec, L., Debeaujon, I. (2005). TRANSPARENT TESTA10 encodes a laccase-like enzyme involved in oxidative polymerization of flavonoids in Arabidopsis seed coat. Plant Cell 17, 2966–2980. doi: 10.1105/tpc.105.035154
Quesada, A., Galván, A., Fernández, E. (1994). Identification of nitrate transporter genes in Chlamydomonas reinhardtii. Plant J. 5, 407–419. doi: 10.1111/j.1365-313X.1994.00407.x
Quesada, A., Hidalgo, J., Fernández, E. (1998). Three Nrt2 genes are differentially regulated in Chlamydomonas reinhardtii. Mol. Gen. Genet. 258, 373–377. doi: 10.1007/s004380050743
Quesada, A., Krapp, A., Trueman, L. J., Daniel-Vedele, F., Fernández, E., Forde, B. G., et al. (1997). PCR-identification of a Nicotiana plumbaginifolia cDNA homologous to the high-affinity nitrate transporters of the crnA family. Plant Mol. Biol. 34, 265–274. doi: 10.1023/A:1005872816881
Remans, T., Nacry, P., Pervent, M., Filleur, S., Diatloff, E., Mounier, E., et al. (2006a). The Arabidopsis NRT1.1 transporter participates in the signaling pathway triggering root colonization of nitrate-rich patches. Proc. Natl. Acad. Sci. U. S. A. 103, 19206–19211. doi: 10.1073/pnas.0605275103
Remans, T., Nacry, P., Pervent, M., Girin, T., Tillard, P., Lepetit, M., et al. (2006b). A central role for the nitrate transporter NRT2.1 in the integrated morphological and physiological responses of the root system to nitrogen limitation in Arabidopsis. Plant Physiol. 140, 909–921. doi: 10.1104/pp.105.075721
Robinson, D. (1994). The responses of plants to non-uniform supplies of nutrients. New Phytol. 127, 635–674. doi: 10.1111/j.1469-8137.1994.tb02969.x
Routaboul, J. M., Dubos, C., Beck, G., Marquis, C., Bidzinski, P., Loudet, O., et al. (2012). Metabolite profiling and quantitative genetics of natural variation for flavonoids in Arabidopsis. J. Exp. Bot. 63, 3749–3764. doi: 10.1093/jxb/ers067
Ruffel, S., Krouk, G., Ristova, D., Shasha, D., Birnbaum, K. D., Coruzzi, G. M. (2011). Nitrogen economics of root foraging: transitive closure of the nitrate-cytokinin relay and distinct systemic signaling for N supply vs. demand. Proc. Natl. Acad. Sci. U. S. A. 108, 18524–18529. doi: 10.1073/pnas.1108684108
Siddiqi, M. Y., Glass, A. D., Ruth, T. J., Rufty, T. W. (1990). Studies of the uptake of nitrate in barley: I. kinetics of 13NO3- influx. Plant Physiol. 93, 1426–1432. doi: 10.1104/pp.93.4.1426
Song, M., Fan, X., Chen, J., Qu, H., Luo, L., Xu, G. (2020). OsNAR2.1 interaction with OsNIT1 and OsNIT2 functions in root-growth responses to nitrate and ammonium. Plant Physiol. 183, 289–303. doi: 10.1104/pp.19.01364
Tabata, R., Sumida, K., Yoshii, T., Ohyama, K., Shinohara, H., Matsubayashi, Y. (2014). Perception of root-derived peptides by shoot LRR-RKs mediates systemic N-demand signaling. Science 346, 343–346. doi: 10.1126/science.1257800
Taulemesse, F., Le Gouis, J., Gouache, D., Gibon, Y., Allard, V. (2015). Post-flowering nitrate uptake in wheat is controlled by N status at flowering, with a putative major role of root nitrate transporter NRT2.1. PloS One 10, e0120291. doi: 10.1371/journal.pone.0120291
Tong, Y., Zhou, J. J., Li, Z., Miller, A. J. (2005). A two-component high-affinity nitrate uptake system in barley. Plant J. 41, 442–450. doi: 10.1111/j.1365-313X.2004.02310.x
Unkles, S. E., Hawker, K. L., Grieve, C., Campbell, E. I., Montague, P., Kinghorn, J. R. (1991). crnA encodes a nitrate transporter in Aspergillus nidulans. Proc. Natl. Acad. Sci. U. S. A. 88, 204–208. doi: 10.1073/pnas.88.1.204
Valkov, V. T., Sol, S., Rogato, A., Chiurazzi, M. (2020). The functional characterization of LjNRT2.4 indicates a novel, positive role of nitrate for an efficient nodule N2-fixation activity. New Phytol. 228, 682–696. doi: 10.1111/nph.16728
Vidal, E. A., Alvarez, J. M., Araus, V., Riveras, E., Brooks, M. D., Krouk, G., et al. (2020). Nitrate in 2020: Thirty years from transport to signaling networks. Plant Cell 32, 2094–2119. doi: 10.1105/tpc.19.00748
Vidal, E. A., Araus, V., Lu, C., Parry, G., Green, P. J., Coruzzi, G. M., et al. (2010). Nitrate-responsive miR393/AFB3 regulatory module controls root system architecture in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U. S. A. 107, 4477–4482. doi: 10.1073/pnas.0909571107
Vidal, E. A., Moyano, T. C., Riveras, E., Contreras-López, O., Gutiérrez, R. A. (2013). Systems approaches map regulatory networks downstream of the auxin receptor AFB3 in the nitrate response of Arabidopsis thaliana roots. Proc. Natl. Acad. Sci. U. S. A. 110, 12840–12845. doi: 10.1073/pnas.1310937110
Vidmar, J. J., Zhuo, D., Siddiqi, M. Y., Glass, A. D. (2000). Isolation and characterization of HvNRT2.3 and HvNRT2.4, cDNAs encoding high-affinity nitrate transporters from roots of barley. Plant Physiol. 122, 783–792. doi: 10.1104/pp.122.3.783
Wang, Y. Y., Hsu, P. K., Tsay, Y. F. (2012). Uptake, allocation and signaling of nitrate. Trends Plant Sci. 17, 458–467. doi: 10.1016/j.tplants.2012.04.006
Wang, T., Hua, Y., Chen, M., Zhang, J., Guan, C., Zhang, Z. (2018). Mechanism enhancing arabidopsis resistance to cadmium: The role of NRT1.5 and proton pump. Front. Plant Sci. 9. doi: 10.3389/fpls.2018.01892
Wang, Y., Yuan, Z., Wang, J., Xiao, H., Wan, L., Li, L., et al. (2023). The nitrate transporter NRT2.1 directly antagonizes PIN7-mediated auxin transport for root growth adaptation. Proc. Natl. Acad. Sci. U. S. A. 120, e2221313120. doi: 10.1073/pnas.2221313120
Wei, J., Zheng, Y., Feng, H., Qu, H., Fan, X., Yamaji, N., et al. (2018). OsNRT2.4 encodes a dual-affinity nitrate transporter and functions in nitrate-regulated root growth and nitrate distribution in rice. J. Exp. Bot. 69, 1095–1107. doi: 10.1093/jxb/erx486
Widiez, T., El Kafafi, E. S., Girin, T., Berr, A., Ruffel, S., Krouk, G., et al. (2011). High nitrogen insensitive 9 (HNI9)-mediated systemic repression of root NO3- uptake is associated with changes in histone methylation. Proc. Natl. Acad. Sci. U. S. A. 108, 13329–13334. doi: 10.1073/pnas.1017863108
Wirth, J., Chopin, F., Santoni, V., Viennois, G., Tillard, P., Krapp, A., et al. (2007). Regulation of root nitrate uptake at the NRT2.1 protein level in Arabidopsis thaliana. J. Biol. Chem. 282, 23541–23552. doi: 10.1074/jbc.M700901200
Yan, M., Fan, X., Feng, H., Miller, A. J., Shen, Q., Xu, G. (2011). Rice OsNAR2.1 interacts with OsNRT2.1, OsNRT2.2 and OsNRT2.3a nitrate transporters to provide uptake over high and low concentration ranges. Plant Cell Environ. 34, 1360–1372. doi: 10.1111/j.1365-3040.2011.02335.x
Yang, Y., Xiong, J., Chen, R., Fu, G., Chen, T. T., Tao, L. (2015). Excessive Nitrate Enhances Cadmium (Cd) Uptake by Up-regulating the expression of OsIRT1 in rice (Oryza sativa). Environ. Exp. Bot. 122, 141–149. doi: 10.1016/j.envexpbot.2015.10.001
Yong, Z., Kotur, Z., Glass, A. D. (2010). Characterization of an intact two-component high-affinity nitrate transporter from Arabidopsis roots. Plant J. 63, 739–748. doi: 10.1111/tpj.2010.63.issue-5
Zhang, H., Forde, B. G. (2000). Regulation of Arabidopsis root development by nitrate availability. J. Exp. Bot. 51, 51–59. doi: 10.1093/jxb/51.342.51
Zhou, J. J., Fernández, E., Galván, A., Miller, A. J. (2000a). A high affinity nitrate transport system from Chlamydomonas requires two gene products. FEBS Lett. 466, 225–227. doi: 10.1016/S0014-5793(00)01085-1
Zhou, J. J., Trueman, L. J., Boorer, K. J., Theodoulou, F. L., Forde, B. G., Miller, A. J. (2000b). A high affinity fungal nitrate carrier with two transport mechanisms. J. Biol. Chem. 275, 39894–39899. doi: 10.1074/jbc.M004610200
Zhuo, D., Okamoto, M., Vidmar, J. J., Glass, A. D. (1999). Regulation of a putative high-affinity nitrate transporter (Nrt2;1At) in roots of Arabidopsis thaliana. Plant J. 17, 563–568. doi: 10.1046/j.1365-313X.1999.00396.x
Zimmerli, L., Jakab, G., Metraux, J. P., Mauch-Mani, B. (2000). Potentiation of pathogen-specific defense mechanisms in Arabidopsis by beta -aminobutyric acid. Proc. Natl. Acad. Sci. U. S. A. 97, 12920–12925. doi: 10.1073/pnas.230416897
Keywords: Arabidopsis, high-affinity nitrate transport system (HATS), NRT2s, C-N homeostasis, plant-microbe interactions, systemic nitrate signaling
Citation: Xu N, Cheng L, Kong Y, Chen G, Zhao L and Liu F (2024) Functional analyses of the NRT2 family of nitrate transporters in Arabidopsis. Front. Plant Sci. 15:1351998. doi: 10.3389/fpls.2024.1351998
Received: 07 December 2023; Accepted: 06 February 2024;
Published: 04 March 2024.
Edited by:
Néstor Fernández Del-Saz, University of the Balearic Islands, SpainReviewed by:
Maurizio Chiurazzi, National Research Council (CNR), ItalyMeng Xu, Chinese Academy of Agricultural Sciences, China
Copyright © 2024 Xu, Cheng, Kong, Chen, Zhao and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Fei Liu, bGl1ZmVpMjgwNDI5QDE2My5jb20=
 Na Xu
Na Xu Li Cheng1
Li Cheng1 Fei Liu
Fei Liu