- 1Área de Genética, Facultad de Ciencias, Universidad de Málaga, Málaga, Spain
- 2Instituto de Hortofruticultura Subtropical y Mediterránea “La Mayora”, Consejo Superior de Investigaciones Científicas (IHSM-UMA-CSIC), Málaga, Spain
- 3Institute for Multidisciplinary Research in Applied Biology, Universidad Pública de Navarra (UPNA), Edificio de Agrobiotecnología, Mutilva Baja, Spain
GacS/GacA is a widely distributed two-component system playing an essential role as a key global regulator, although its characterization in phytopathogenic bacteria has been deeply biased, being intensively studied in pathogens of herbaceous plants but barely investigated in pathogens of woody hosts. P. savastanoi pv. savastanoi (Psv) is characterized by inducing tumours in the stem and branches of olive trees. In this work, the model strain Psv NCPPB 3335 and a mutant derivative with a complete deletion of gene gacA were subjected to RNA-Seq analyses in a minimum medium and a medium mimicking in planta conditions, accompanied by RT-qPCR analyses of selected genes and phenotypic assays. These experiments indicated that GacA participates in the regulation of at least 2152 genes in strain NCPPB 3335, representing 37.9 % of the annotated CDSs. GacA also controls the expression of diverse rsm genes, and modulates diverse phenotypes, including motility and resistance to oxidative stresses. As occurs with other P. syringae pathovars of herbaceous plants, GacA regulates the expression of the type III secretion system and cognate effectors. In addition, GacA also regulates the expression of WHOP genes, specifically encoded in P. syringe strains isolated from woody hosts, and genes for the biosynthesis of phytohormones. A gacA mutant of NCPPB 3335 showed increased virulence, producing large immature tumours with high bacterial populations, but showed a significantly reduced competitiveness in planta. Our results further extend the role of the global regulator GacA in the virulence and fitness of a P. syringae pathogen of woody hosts.
Introduction
Two-component regulatory systems (TCSs) play a fundamental role in bacterial detection of extracellular signals and transduction of this information, causing physiological responses that facilitate adaptation to a changing environment (Stock et al., 2000; Gao et al., 2007; Papon and Stock, 2019). TCSs are very common and conserved in bacteria (Wuichet et al., 2010), being the GacS/GacA system one of the most intensively studied. GacS/GacA homologues are widely distributed in Gram-negative bacteria, controlling the expression of numerous genes involved in robust growth as well as a wide range of virulence factors in both animal and plant pathogenic bacteria (Heeb and Haas, 2001; Gooderham and Hancock, 2009; Gauthier et al., 2010; Song et al., 2023). Phytopathogenic bacteria rely on the expression of pathogenicity and virulence factors to cause disease in host plants, and activation of many of them depends on the GacS/GacA pathway (Hrabak and Willis, 1992; Kitten et al., 1998; Chancey et al., 1999; Ferreiro and Gallegos, 2021). However, this system is characterized by displaying a remarkable variability in the pool of regulated genes across different genera, species and pathovars of bacterial plant pathogens.
In Pseudomonas species, the GacS/GacA system is initiated by GacS, a histidine kinase located at the plasma membrane that promotes its own phosphorylation after recognition of a yet unknown stimulus. Next, the phosphoryl group is transferred to the cytoplasmic response regulator GacA, which triggers the transcription of a variable number of small non-coding RNAs (sRNAs), among which rsmX, rsmY and rsmZ are the most abundant in Pseudomonas and the most intensively investigated (Lapouge et al., 2008; Sobrero and Valverde, 2020). These sRNA molecules modulate the activity of the RsmA family of proteins, which are global posttranscriptional regulators of gene expression in bacteria within the regulatory cascade Gac-Rsm (Sobrero and Valverde, 2020). The RsmA protein has been widely referred to as CsrA, among other names; here, we will adhere to the proposed uniform nomenclature (Sobrero and Valverde, 2020) and use only the Rsm abbreviation for RsmA and homologues. Additionally, in pseudomonads and few other bacteria, the sensor kinases LadS and RetS act as positive or negative regulators, respectively, of the activity of GacS (Sobrero and Valverde, 2020). For the interested reader, several excellent recent reviews include complete descriptions of the Gac-Rsm system (Sobrero and Valverde, 2020; Ferreiro et al., 2021; Song et al., 2023).
The Gac-Rsm system regulates a plethora of processes and phenotypes including the intracellular carbon flux, motility and biofilm formation, pathogenicity and virulence, and biocontrol, among others (Lapouge et al., 2008; Ferreiro and Gallegos, 2021). It has been generally assumed that the regulatory activity of GacA is largely or entirely mediated transcriptionally by direct regulation of the sRNAs and, through the activity of these sRNAs, post-transcriptionally via the Rsm system (Brencic et al., 2009; Zere et al., 2015). However, in P. aeruginosa GacA can bind to its own promoter, suggesting autoregulation of its expression, and it can co-regulate the expression of certain genes in collaboration with additional transcription factors, suggesting a complex regulatory network involving transcriptional, posttranscriptional, and translational mechanisms (Huang et al., 2019).
P. syringae sensu lato is one of the most important plant-pathogenic bacteria, displaying an outstanding capacity to infect hundreds of species of herbaceous plants, shrubs, and trees (Baltrus et al., 2011; Dillon et al., 2019). In this bacterium, GacS/GacA modulates the expression of many diverse virulence factors and prominently that of the type III secretion system (T3SS), likely the most important pathogenicity determinant of P. syringae. The T3SS, encoded by the hrp/hrc genes, allows bacterial translocation of effector proteins (T3Es) into the host cytoplasm (Alfano and Collmer, 1997; Lindgren, 1997; He, 1998; Hutcheson et al., 2001). However, regulation of the T3SS and virulence by GacS/GacA seems to be strain dependent.
The characterization of the GacS/GacA system in P. syringae appears to be deeply biased, being intensively studied in pathogens of herbaceous plants (Hrabak and Willis, 1992; Marutani et al., 2008; Vargas et al., 2013; Ramírez-Zapata et al., 2020) and only reported for a single strain pathogenic to woody hosts, P. syringae pv. actinidiae strain A18. A gacA mutant of strain A18 showed complete abolition of phaseolotoxin production, and reduction of several virulence-related phenotypes, such as bacterial motility, biofilm formation, production of exopolysaccharide, and HR induction in tobacco plants (Zhang et al., 2018).
P. savastanoi pv. savastanoi (Psv) is characterized by inducing hyperplastic growths in the stem and branches of olive trees (Ramos et al., 2012). The pathogenicity of Psv depends on the coordination and expression of several virulence factors (Matas et al., 2012), most notably: production of cytokinins and indole-3-acetic acid (Rodríguez-Moreno et al., 2008; Aragón et al., 2014; Añorga et al., 2020), a Na+/Ca2+ exchanger (Moretti et al., 2019), quorum sensing (Caballo-Ponce et al., 2018), production of cyclic-di-GMP (Aragón et al., 2015; Martínez-Gil and Ramos, 2018), translocation of type III effector proteins (T3Es) through the T3SS (Pérez-Martínez et al., 2010; Castañeda-Ojeda et al., 2017a; Castañeda-Ojeda et al., 2017b) or the WHOP genomic region (Caballo-Ponce et al., 2017a), related with degradation of phenolic compounds. Psv has become a well-studied model for the mechanisms of pathogenicity in woody hosts and the induction of plant tumours. Here, we aimed to characterize the role of GacA in the pathogenicity/virulence of Psv. For this purpose, we constructed a gacA deletion mutant of strain Psv NCPPB 3335 and characterized, by RNA-Seq, the GacA regulon using two different culture media: a standard succinate medium (SSM) and a Hrp inducing medium (HIM), which simulates the apoplastic conditions. Results from this study correlate changes in transcript abundance with diverse virulence-related phenotypes in this bacterial pathogen of woody hosts.
Experimental procedures
Bacterial strain, plasmids, and growth conditions
Bacterial strains, plasmids and primers used in this study are summarized in Supplementary Tables S1–S3, respectively. P. savastanoi strains were grown at 28 °C using lysogeny broth (LB) (Lennox, 1955) without glucose and containing 0.5 % NaCl, King’s B (KB) (King et al., 1954), SSM (Meyer and Abdallah, 1978), super optimal broth (SOB) (Hanahan, 1983) or HIM (Huynh et al., 1989) culture media. Escherichia coli strains were grown using LB at 37 °C. When required, media were supplemented with appropriate antibiotics at the following final concentrations. For P. savastanoi: ampicillin (Ap) (400 μg/mL), gentamicin (Gm) (10 μg/mL), kanamycin (Km) (7 μg/mL), nitrofurantoin (25 μg/mL), and cycloheximide (100 μg/mL). For E. coli: Ap (100 μg/mL), Gm (10 μg/mL) and Km (50 μg/mL).
Construction of P. savastanoi mutants and complemented strains
Gene gacA was removed from Psv NCPPB 3335 using plasmid pgacA-Km, containing the DNA immediately flanking gene gacA in both sides (approximately 1.2 kb on each side) separated by an nptII (Km)-resistance gene (Supplementary Tables S2, S6), and following previous procedures (Pérez-Martínez et al., 2007; Matas et al., 2014). Afterwards, the Km gene was removed using the pFLP2 plasmid (Supplementary Table S2).
For construction of the complemented strains, the complete coding sequences of gacA and uvrC, together with their predicted ribosomal binding sites, were amplified from Psv NCPPB 3335 by PCR. After verifying the correct sequence of the resulting amplicons, those fragments were individually cloned under the control of Plac in pBBR1MCS-5 (GmR) and pBBR1MCS-2 (KmR), respectively (Supplementary Table S2).
Genomic organization and gene expression of the gacA-uvrC operon in Psv NCPPB 3335
Promoters were predicted with BPROM (Solovyev and Salamov, 2011), a bacterial sigma 70 promoter recognition program (http://www.softberry.com/).
5’ Rapid amplification of complementary DNA (cDNA) ends (RACE) was used to establish the transcription start point and genomic boundary of uvrC in Psv NCPPB 3335. Transcripts were obtained from bacterial cultures grown in HIM after 6 hours of induction. 5´ RACE strategy was performed using the 5’/3’ RACE Kit 2nd Generation (Roche Applied Science, Mannheim, Germany). According to the manufacturer’s instructions, three different primers at 357 (SP1), 214 (SP2) and 142 (SP3) base pairs (bp) distance from the ATG of uvrC were used for amplification (Supplementary Table S3).
RNA-Seq analysis
To purify RNA, pre-inocula of 20 mL of NCPPB 3335 and Psv-ΔgacA were grown overnight in both KB and SSM media at 28 °C. Bacterial cells from each medium were diluted in fresh KB or SSM to a final optical density at 600 nm (OD600) of 0.1, to prepare three biological replicates of 110 mL for each medium and grown at 28 °C with gentle shaking to a final OD600 of 0.5 (approximately 5x107 CFUs/mL). Next, bacterial cells from these SSM cultures were collected, each biological replicate resuspended in 12 mL of fresh SSM and divided in twelve 1 mL samples that were frozen in liquid nitrogen and stored at -80 °C. Liquid cultures from each biological replicate grown on KB were pelleted, washed twice with one volume of sterile 10 mM MgCl2, and each resuspended in 110 mL of HIM and incubated at 28 °C for 6 hours. After this, bacterial cells were collected, and each biological replicate resuspended in 12 mL of HIM and divided in twelve 1 mL samples. After a centrifugation step, the pellets were flash-frozen in liquid nitrogen and stored at -80 °C. RNA extractions were performed with the RNAeasy Mini Kit (Qiagen; Hilden, Germany) and following the manufacturer’s instructions. Subsequently, the RNA was cleaned up from traces of genomic DNA using the TURBO DNA-free kit (Invitrogen Corp, CA, USA) following the manufacturer’s instructions. For ribosomal RNA degradation, the Illumina Ribo-Zero Plus rRNA Depletion kit Bacteria (Illumina; CA, USA) was used. The resulting RNA was quantified by spectrophotometry, its integrity assessed by agarose gel electrophoresis, and its quality evaluated on an Agilent Bioanalyzer 2100 using a Pico 6000 RNA bioanalyzer chip (Agilent Technologies, Santa Clara, CA, USA). For genotyping and sequencing, the two biological replicates for each medium with the best quality of purified RNA were used. The TruSeq Stranded mRNA kit (Illumina; CA, USA) was used for genotyping and the Illumina NextSeq550 kit (Illumina; CA, USA) was used for sequencing.
Reads obtained from the RNA-Seq analysis were processed in collaboration with the Ultra sequencing Service of the University of Málaga and the Andalusian Bioinformatics Platform, using a workflow of previously described software packages (Moreno-Pérez et al., 2021). SeqTrimNext (v.2.0.60) was used to clean up and process the sequences, and the FastQC software for quality control; then, sequences were aligned to the reference whole chromosome genome (accession no. CP008742.1) and the three native plasmids of Psv NCPPB 3335 (pPsv48A, FR820585.2; pPsv48B, FR820586.1; pPsv48C, FR820587.2) with the Bowtie2 tool (Langmead and Salzberg, 2012). Transcript abundance, measured in fragments per kilobase per million mapped reads (FPKM), and differential gene expression were calculated using Tuxedo together with the Cufflinks suit of tools (Trapnell et al., 2010; Ghosh and Chan, 2016). CuffDiff was used to identify differentially expressed genes (DEGs). Only those genes with an adjusted P-value (q-value) < 0.05 and a log2 fold change ≤ -0.5 or ≥ 0.5 were considered significant. Graphical representation of the differential expression results was carried out using the CummeRbund package in R (Goff et al., 2012).
RT-qPCR assays
For real-time quantitative PCR (RT-qPCR), DNA-free total RNA samples described in the previous section were used. cDNA synthesis was carried out using random hexamers included in the iScriptTM cDNA synthesis kit (BioRAD, CA, USA) and 1 μg of DNA-free total RNA as template. RT-qPCR primers were designed with Primer3Plus (Untergasser et al., 2007) and Bacon Designer Free (Thornton and Basu, 2015) (Supplementary Table S3). Efficiency curve analysis to confirm the amplification specificity of primer pairs was carried out as described (Vargas et al., 2011). The relative transcript abundance was calculated using the ΔΔ cycle-threshold (Ct) method (Livak and Schmittgen, 2001). Transcriptional data was then normalized according to the expression of the housekeeping gene gyrA by calculating the difference between the relative expression of the analysed gene and that of the gyrA gene (ΔΔCt = Ct studied gene – Ct gyrA). Fold change values were calculated as 2-ΔΔCt (Pfaffl, 2004; Rotenberg et al., 2006). RT-qPCR analyses were carried out in triplicate with three biological replicates. Change rate is represented by the Psv-ΔgacA expression divided by the wild-type strain expression.
Bioinformatics
Identification and comparison of homologous rsm genes in Psv NCPPB 3335 was carried out by blastp and blastn, using the Geneious 8.1.9 program (Kearse et al., 2012), and the previously described rsm sequences in 1448A (Ramírez-Zapata et al., 2020), and Pto DC3000 (Chatterjee et al., 2003; Ferreiro et al., 2018). Sequence alignments were performed using Multalin (http://multalin.toulouse.inra.fr/multalin/) (Corpet, 1988) and Needle servers (https://www.ebi.ac.uk/Tools/psa/emboss_needle/) (Madeira et al., 2022). Additionally, the different rsm transcripts were identified using the IGV program by aligning the original.fna sequence with the.gtf and bowtie.bam files extracted from the RNA-Seq analysis.
Transmission electron microscopy (TEM)
For transmission electron microscopy (TEM) analysis, Psv strains were grown at 28 °C for 48 h on LB. Then, bacteria were collected, washed three times with 10 mM MgCl2, and resuspended in SSM medium. After incubation at 28 °C overnight, bacterial cultures were resuspended in SSM to an OD600 of 0.1 and incubated with shaking at 28 °C until they reached an OD600 of 0.6. Grids were deposited over 30 μL of bacterial suspensions and incubated for 1 h at room temperature. Subsequently, grids were treated with 4 % paraformaldehyde for 10 minutes, washed with water for 5 minutes, negatively stained with 1 % uranyl acetate for 30 seconds and washed once with water for 30 seconds. The grids were allowed to dry for 24 h and imaged on a FEI Tecnai G2 20 TWIN TEM at an accelerating voltage of 80 kV. Images were acquired using TIA FEI Imaging Software v.4.14 (Berlanga-Clavero et al., 2022).
Swimming motility assay
Psv strains were grown at 28 °C for 48 h on KB in the dark. Cells were resuspended in KB to an OD600 of 1 (approximately 1x109 CFUs/mL). Two microliters of the bacterial suspension were spotted onto soft KB agar (KB with 0.3 % agar) and plates were incubated for 72 h at 25 °C and 60 % relative humidity in the dark (Antúnez-Lamas et al., 2009). The surface area of swimming colonies was photographed and quantified using the area selection tool of the image software FiJi (Schindelin et al., 2012).
Hydrogen peroxide tolerance assay
Bacterial suspensions were prepared to an OD600 of 1 (approximately 1x109 CFUs/mL) and 100 μL plated on SSM using a Drigalski plating loop. Thirteen-millimetre diameter Whatman paper discs were wetted with 100 μL of 1.5 % H2O2 and allowed to dry for 15 minutes. The discs were placed in the centre of each plate and incubated at 28 °C for 48 h before measuring the diameter of the growth inhibition haloes.
Detection of reactive oxygen species inside live cells
To determine the intrinsic oxidative stress of the cells, the Psv strains were grown overnight in 20 mL of LB at 28 °C. Bacterial cells were diluted 1:10 and disaggregated using a 23-gauge needle. Samples of 100 μL each were incubated with 0.2 μL of 2.5 μM CellROX™ Green Reagent (Invitrogen, Corp, CA, USA) for 30 minutes at 37 °C with shaking. Cells were washed three times with Phosphate Buffered Saline (PBS) and then thoroughly resuspended by passing through a needle (Cámara-Almirón et al., 2020). Five microliters of each sample were dropped on a slide and observed using a Leica SP5 confocal microscope equipped with a HCX PL APO lambda blue 63.0x1.40 OIL UV objective. Images were processed using the Leica Application Suite Advance Fluorescence v.2.7.3.9723 (LCS Lite, Leica Microsystems) and the FIJI/ImageJ software. The laser settings, scanning speed, photomultiplier detector gain, and pinhole aperture were constant for the five images per strain acquired for each of the three experiments performed, and were as described (Berlanga-Clavero et al., 2022).
Hypersensitivity reaction assay
Two-month-old Nicotiana tabacum var. Newdel plants were used for the HR assays. Plants were grown with a photoperiod of 16 h of light, 8 h of darkness at 26 °C. Bacterial suspensions were prepared in 10 mM MgCl2 with low population density, 1x106 CFUs/mL. Then, a small wound was made on the abaxial side of the leaf with a hypodermic needle and the bacterial suspension was introduced into the periplasmic space using a 1 mL blunt syringe. The development of symptoms was observed 2 days and 3 days post inoculation (dpi). Images were taken with a high-resolution digital camera (Nikon DXM 1200, Nikon Corp).
Plant bioassays
Pathogenicity was tested on Olea europaea plants, derived from seeds of cv. Arbequina germinated in vitro, grown at 60 % humidity and 26 °C. For inoculation, bacterial suspensions were prepared in 10 mM MgCl2 to a final OD600 of 0.1 (approximately 1x107 CFUs/mL) and two wounds made in the stem of the plant with a scalpel were each inoculated with 5 μL. Four plants were used per strain in each of the three replicate assays. During the 54-day trial, the volume of tumours in each of the inoculated wounds was measured every week. At the end of the trial, symptoms were photographed with a high-resolution digital camera (Nikon DXM 1200). Bacteria were recovered from tumours by grinding them using a mortar and pestle containing 1 mL of sterile 10 mM MgCl2. Serial dilutions were plated onto LB supplemented with nitrofurantoin, cycloheximide and the corresponding antibiotic, as described (Penyalver et al., 2006; Pérez-Martínez et al., 2007; Matas et al., 2012).
Competitive index (CI) assays were performed on in vitro olive plants obtained from a germinated seed of O. europaea cv. Arbequina and grown on DKW medium (Driver and Kuniyuki, 1984) supplemented differently in each of the three culture periods (proliferation, channelling, and maintenance). Plants were inoculated with bacterial suspensions in 10 mM MgCl2 in a 1:1 ratio (wild-type strain: mutant strain). A single wound per plant was inoculated with approximately 5x103 CFUs of each strain, using six plants per experiment and repeating the experiment three times (a total of 18 inoculated wounds). After 30 dpi under 25 °C, 50-60 % humidity and 16 h light photoperiod, symptoms were visualized using a stereo microscope (Leica MZ FLIII; Leica Microsystems, Wetzlar, Germany). Bacteria were recovered from tumours as above, and serial dilutions were seeded on LB and LB+Km for colony counting after 2 days at 28 °C. The number of CFUs of the mutant strain was obtained from the colonies that grew on the Km plates, while the number of wild-type bacteria was obtained by subtracting the number of mutant bacteria from the total number of bacteria grown on LB plates (Macho et al., 2007; Macho et al., 2016). The CI is defined as the mutant to wild-type ratio in the output sample divided by the mutant to wild-type ratio in the input (inoculum) sample (Freter et al., 1981; Taylor et al., 1987), the input ratio being close to 1.
Statistics and reproducibility
Statistical analyses were performed using GraphPad Prism version 9. p-values < 0.05 were considered significantly different. Asterisks indicate the level of statistical significance: *p < 0.05, **p < 0.01, ***p < 0.001 and ****p < 0.0001. When needed, data were normalized to the corresponding internal standard of the samples. All experiments were repeated at least three independent times, with similar results.
Results
Genomic conservation of GacS/GacA downstream regulatory elements in Psv NCPPB 3335
Pseudomonas species contain two to seven Rsm gene homologues in their genome, with different levels of sequence identity (Sobrero and Valverde, 2020). Genomic analysis of Psv NCPPB 3335 revealed four Rsm homologues (RsmA, RsmC, RsmE and RsmH1) and seven different sRNA genes (rsmX1, rsmX2, rsmX3, rsmX4, rsmX5, rsmY and rsmZ) (Table 1). The Rsm deduced products and the sRNA genes showed a high sequence identity with their respective homologues in Pto DC3000 (Ferreiro et al., 2018) and Pph 1448A (Ramírez-Zapata et al., 2020) (Table 1). Therefore, Psv NCPPB 3335 contains a similar, but distinct complement of Rsm homologues and sRNA genes to other P. syringae strains, which should allow for the full functionality of the GacS/GacA regulatory system.
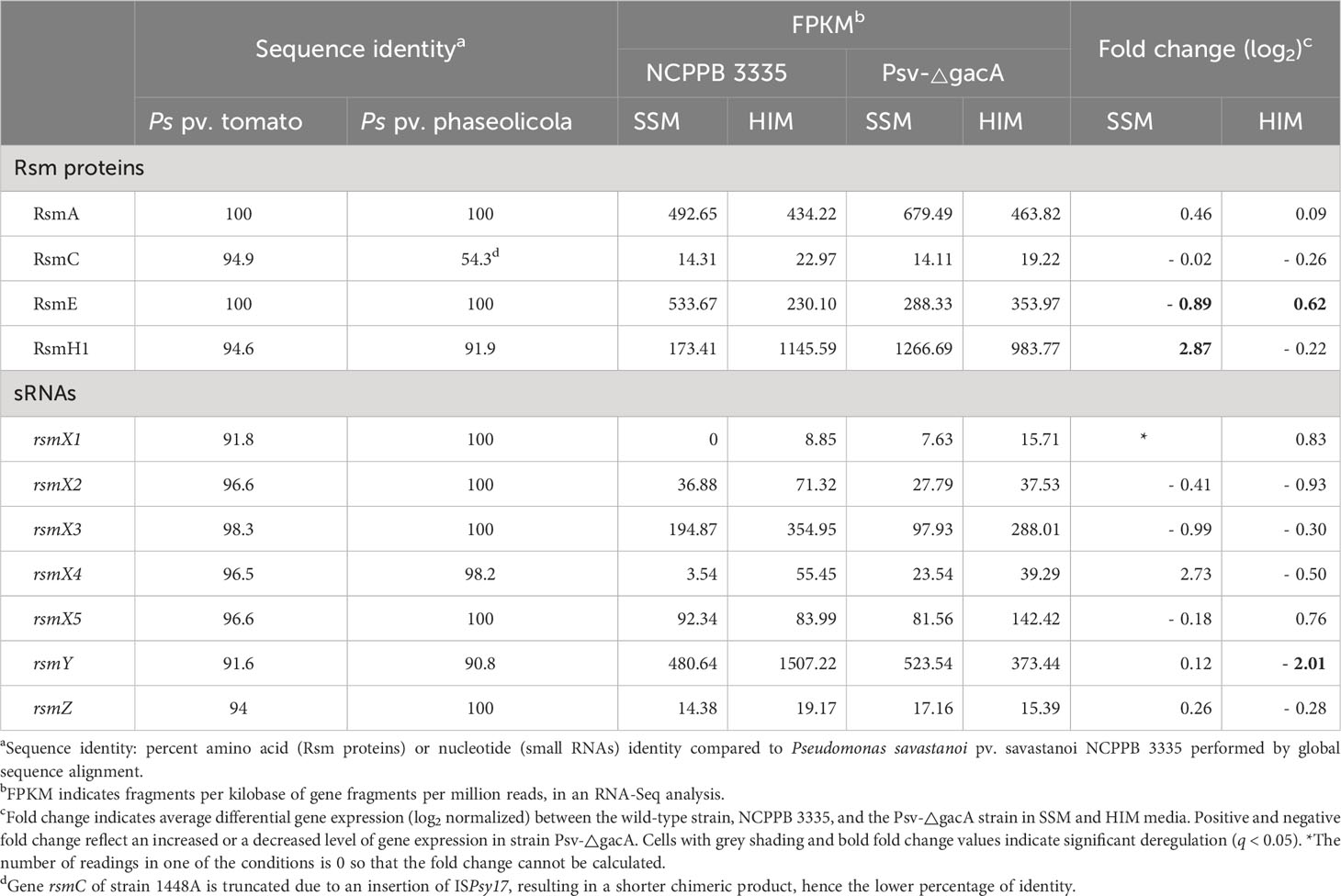
Table 1 Characterization of GacS/GacA transcriptional regulation systems in Pseudomonas savastanoi pv. savastanoi NCPPB 3335.
RNA-Seq analysis
Total RNA was purified from cultures of Psv NCPPB 3335 and its ΔgacA mutant grown in SSM, a minimal medium, and HIM medium, which simulates conditions within the plant apoplast and causes the induction of the hrp genes (Huynh et al., 1989). Following previous analyses of the role of GacA in P. syringae (Chatterjee et al., 2003; Marutani et al., 2008; Ortiz-Martín et al., 2010; Ferreiro et al., 2018), cells were collected during the log phase (OD600 of 0.5) in order to better capture the early induction of virulence genes. To compare transcripts abundance, two biological replicates per strain and media were subjected to RNA Illumina sequencing.
In total, 143.2 and 131.0 million high-quality reads (clean reads) were obtained for the SSM and HIM samples, respectively (Supplementary Tables S4, S5). By iterative sequence alignment, an average of 99.40 % (SSM) and 99.44 % (HIM) of clean reads were mapped to the Psv NCPPB 3335 genome. Out of the 5508 genes annotated in the GenBank record of the Psv NCPPB 3335 chromosome, a total of 5472 genes (99.35 % of the total) were covered by Illumina sequencing in SSM for strain Psv-ΔgacA (Supplementary Table S6). In HIM medium, Illumina sequencing covered 99.44 % (5477 genes) and 99.44 % (5476 genes) of the genes in strains NCPPB 3335 and Psv-ΔgacA, respectively (Supplementary Table S6). Furthermore, in both media and strains, the Illumina RNA sequencing also covered the 68, 53 and 51 genes annotated in the GenBank records of plasmids pPsv48A, pPsv48B and pPsv48C, respectively (Supplementary Table S6).
Genomic context of gene gacA in P. savastanoi pv. savastanoi NCPPB 3335
O’Malley and Anderson (2021) showed that deletion of the gacA gene in Pto DC3000 did not affect the expression of the downstream genes uvrC and psgA, which is relevant because of the regulatory activity associated to gene uvrC (Humann et al., 2009; O’Malley et al., 2019). This is in contrast with the significant downregulation of uvrC observed in our RNA-Seq analysis of strain Psv-ΔgacA (Table 2). We therefore sought to analyse the transcriptional organization of these genes in strain NCPPB 3335.
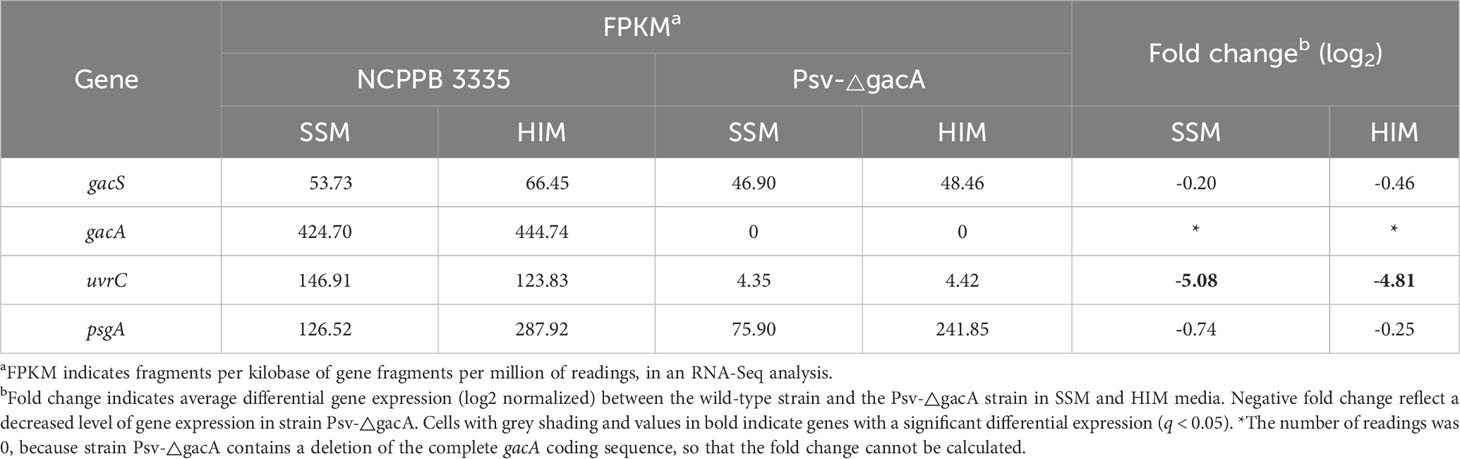
Table 2 RNA-Seq analysis expression values of genes gacS and gacA, and genes downstream of gacA, from Pseudomonas savastanoi pv. savastanoi NCPPB 3335 in SSM and HIM media.
An RT-qPCR analysis of cells grown in SSM and HIM media showed a significantly reduced expression of uvrC in strain Psv-ΔgacA, in agreement with the RNA-Seq data, whereas the expression of gene psgA was comparable to that of the wild-type strain (Figure 1A). Using BPROM, we identified several -35 and -10 regulatory elements typical of Pribnow-type promoters in the gacA DNA region (Figure 1B). A 5´ RACE assay with strain NCPPB 3335 showed that the uvrC transcription start site (+1) was located 193 bp upstream from its predicted start codon (Supplementary Figure S1A). This result confirms that the coding sequence of gacA includes a functional promoter for uvrC, but upstream of the -35 and -10 regions predicted bioinformatically. However, in strain Psv-ΔgacA we did not observe a complete absence of transcripts covering gene uvrC (Table 2, Supplementary Figure S1B), likely suggesting a low level of uvrC transcription from the promoter upstream of gene gacA (Figure 1B).
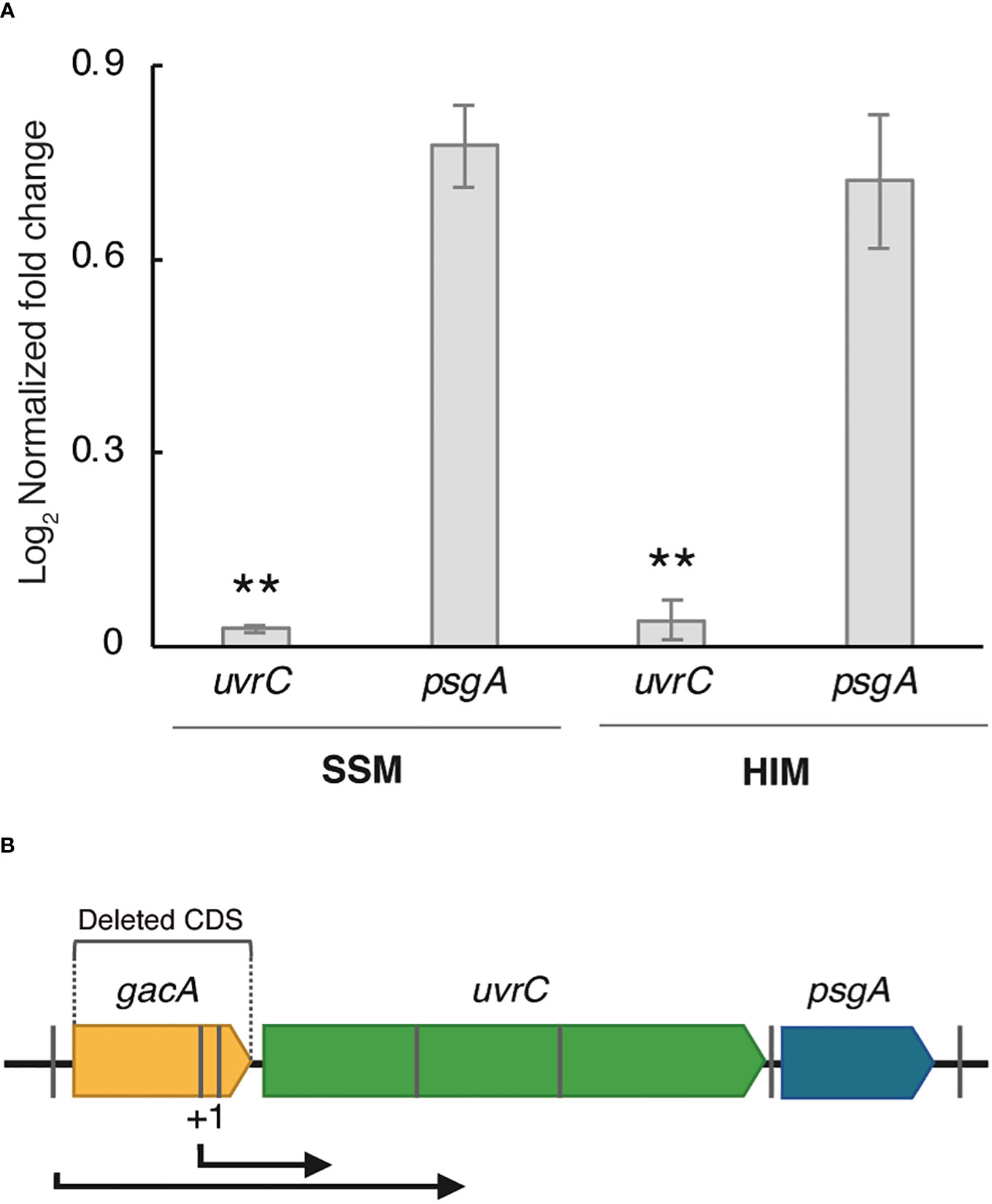
Figure 1 Characterization of the genomic context of gene gacA. (A) Relative expression of genes uvrC and psgA in SSM and HIM media by RT-qPCR. Bars represent the average expression values of strain Psv-ΔgacA relative to those of NCPPB 3335, both of which were previously normalized to the constitutive expression of gene gyrA. Error bars represent the standard deviation. Asterisks (**) indicate significant differences (Student’s t-test, p < 0.01). (B) Relative position of genes gacA, uvrC and psgA in the genome of strain NCPPB 3335. Grey bars indicate the approximate location of relevant -35/-10 promoters predicted by BPROM. The short and long bent arrows indicate transcription from, respectively, the transcription start site identified by 5’ RACE (+1) and a predicted promoter, as supported by RNA-Seq.
Nevertheless, the altered transcription of gene uvrC that is associated to certain mutations in gacA was shown to confound the assessment of the actual regulatory role of GacA (O’Malley and Anderson, 2021). Therefore, and considering the irregular expression of uvrC observed in strain Psv-ΔgacA, we decided to construct derivatives of this strain complemented separately with genes uvrC (Psv::uvrC) and gacA (Psv::gacA) in order to perform the following phenotypical assays.
Characterization of the GacA regulon
Distribution of the normalized FPKM values did not show significant differences between the biological replicates for any combination of strain and medium (Supplementary Figure S2), suggesting that no technical bias was introduced during library construction or sequencing. The normalized expression levels of wild type and mutant strains were compared to detect DEGs, which were selected considering a fold change of ± 0.5 and a statistical value of q < 0.05 (Figures 2A, B). Strain Psv-ΔgacA showed a total of 1020 (SSM) and 1239 (HIM) DEGs (Figures 2C, D), with 654 genes downregulated and 366 genes upregulated in SSM, and 623 genes downregulated, and 616 genes upregulated in HIM medium.
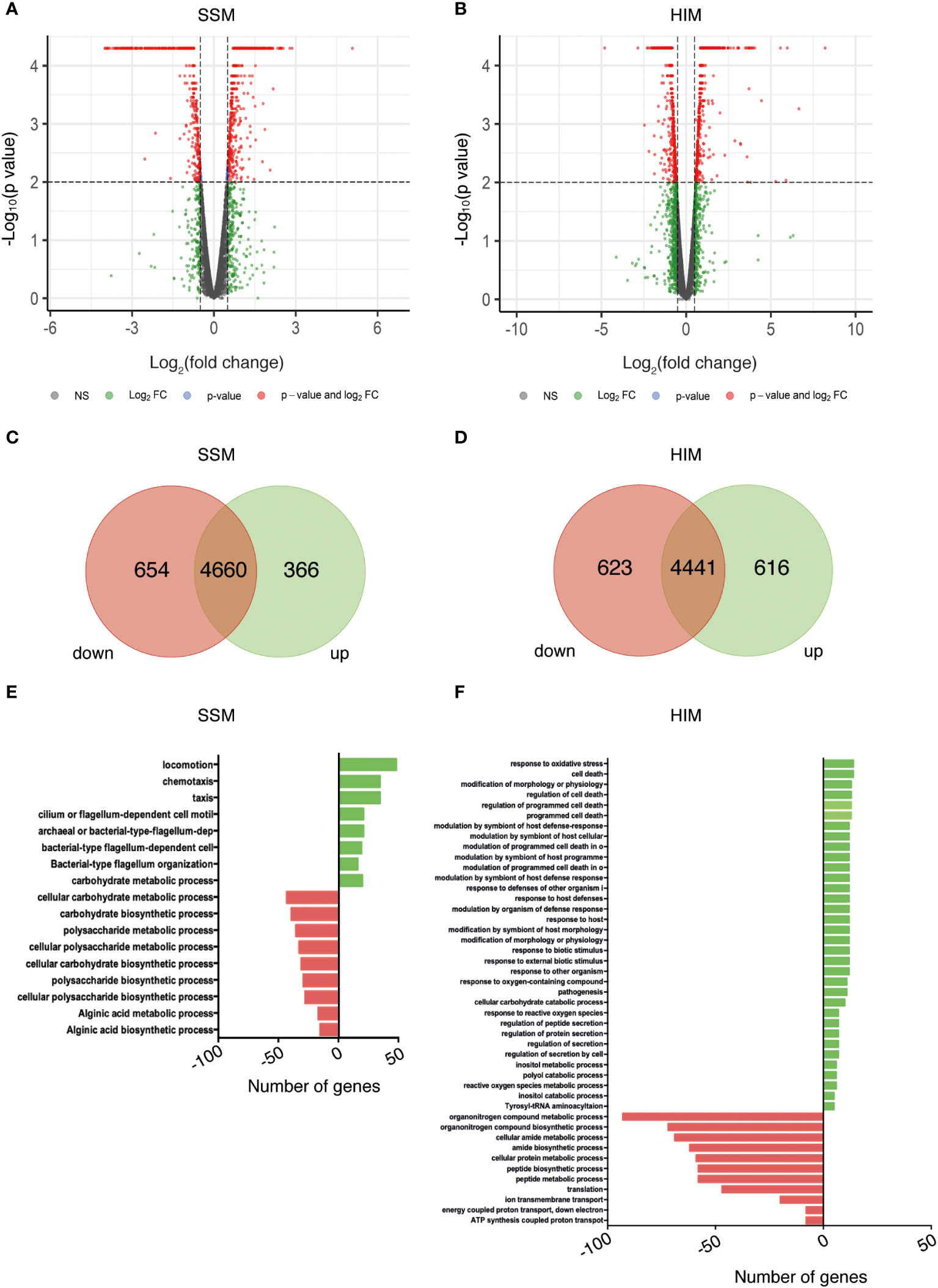
Figure 2 Identification of GacA-dependent genes in P. savastanoi pv. savastanoi NCPPB 3335 by RNA-Seq in SSM (A, C, E) and HIM (B, D, F) media. (A, B) Volcano plots showing DEGs in strain Psv-△gacA with respect to strain NCPPB 3335. Red dots represent significant DEGs (q < 0.05). (C, D) Venn diagrams of significant DEGs. (E, F) Functional categorisation of DEGs. In panels (C-F), downregulated and upregulated DEGs are shown to the left (in red) and to the right (in green), respectively.
As could be expected, the growing conditions determined specific changes in transcript abundance (Figure 2). Thus, genes upregulated in Psv-ΔgacA in SSM medium were mainly related to bacterial motility (Figure 2E), but were classified in the functional categories of pathogenesis, host response and programmed cell death in HIM medium, which favours expression of virulence-related genes under the assayed conditions (Figure 2F). Likewise, most genes downregulated in SSM were related to the metabolism of carbohydrates (Figure 2E), whereas in HIM they belonged to the categories of metabolism of nitrogen and proton transport (Figure 2F).
In fact, there appears to be a minor overlap of the pool of genes deregulated in media SSM and HIM. On the one hand, only seven genes showed the same type of differential expression in both media (Supplementary Figure S3; Supplementary Table S7). Among these genes is uvrC, which is located immediately downstream of gene gacA and showed the highest downregulation (Supplementary Table S7). On the other hand, we also found opposing regulation for a limited number of genes. Specifically, 58 genes were upregulated in HIM but downregulated in SSM, while 42 genes were downregulated in HIM but upregulated in SSM (Supplementary Figure S3). The functions that exhibited the most notable changes in expression depending on the growth medium were related to metabolic processes or locomotion dependent on flagellum activity. These 100 genes are of considerable interest because they may represent genes that are expressed in free-living conditions (SSM) but are repressed during the interaction with the plant (HIM) and vice versa.
Lack of GacA deregulates the expression of genes required for motility in P. savastanoi pv. savastanoi
Previous studies reported the involvement of the GacS/GacA system in the motility of Pto DC3000 (Chatterjee et al., 2003; Ferreiro et al., 2018; Ferreiro et al., 2021). The 42 genes for flagellar biosynthesis in diverse Pseudomonas, including NCPPB 3335, are grouped in nine different operons plus one monocistronic gene, fliK (Figure 3A) (McCarter, 2006). Our transcriptomic analysis in SSM medium showed that 37 out of the 42 genes were significantly upregulated in strain Psv-ΔgacA (Figures 3A, B). In agreement with this, genes fliC, fliE, fliM, flhA and flhF showed a significantly higher expression in strain Psv-ΔgacA than in strain NCPPB 3335 in an RT-qPCR analysis of cells cultivated in SSM (Figure 3C). This could suggest that the GacS/GacA system might directly or indirectly regulate motility in Psv by regulating flagellar biogenesis.
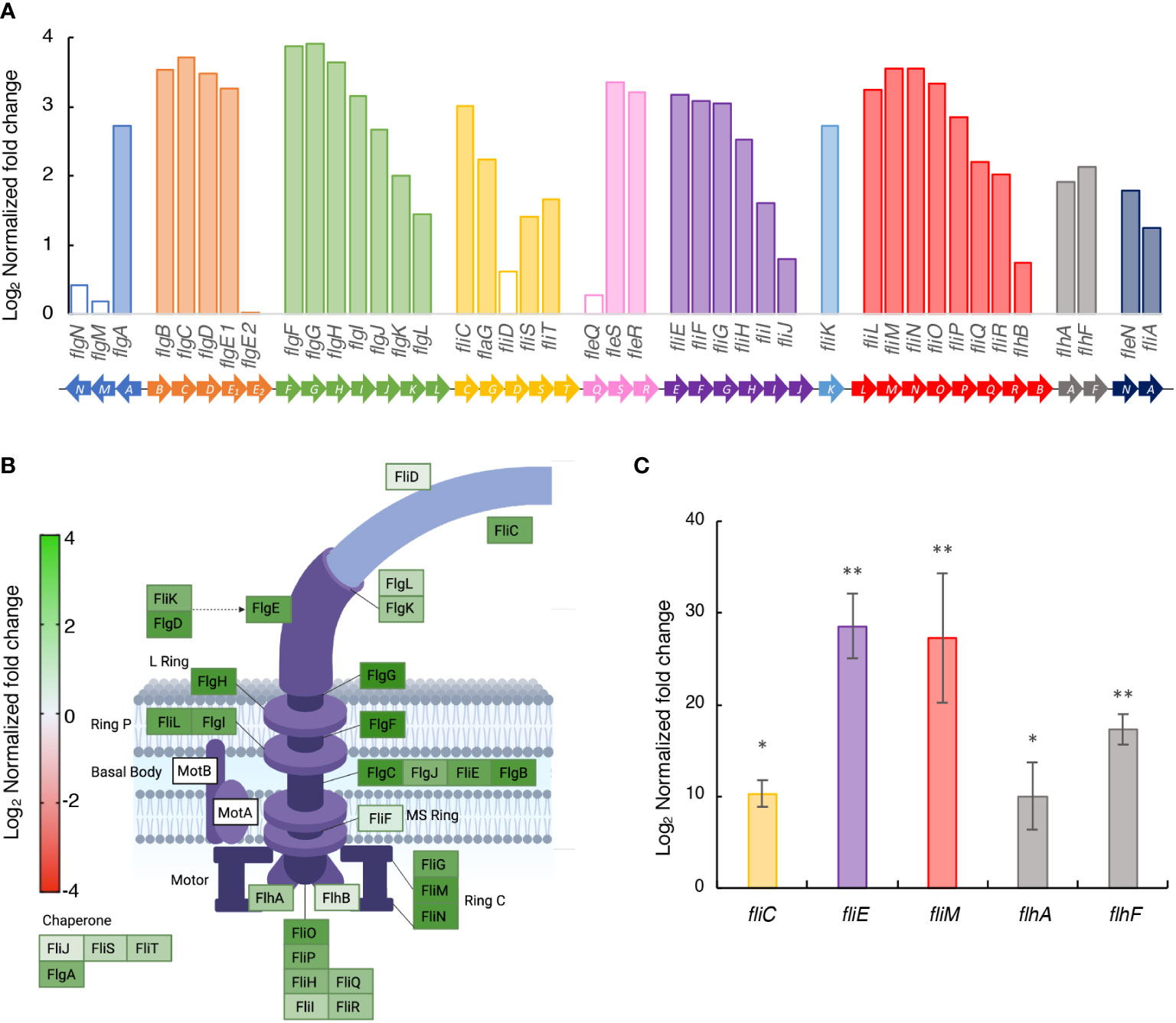
Figure 3 Upregulation in strain Psv-ΔgacA of genes involved in the biosynthesis of flagella in SSM medium. (A) Bars represents the average log2 of the fold change (q < 0.05) in the RNA-Seq analysis of strain Psv-△gacA relative to NCPPB 3335, with empty bars indicating nonsignificant different values. Genes of the same colour are part of the same operon. (B) Genes involved in flagellum structure. The colour gradation for each gene is proportional to the log2 of the fold change in strain Psv-△gacA relative to NCPPB 3335. Figure created with Biorender. (C) RT-qPCR analysis of selected genes upregulated in strain Psv-ΔgacA. Bars represent the average expression values of strain Psv-ΔgacA relative to those of NCPPB 3335, both of which were previously normalized to the constitutive expression of gene gyrA. Error bars represent the standard deviation and asterisks indicate significant differences (Student’s t-test; *p < 0.05, **p < 0.01).
Due to the significant number of deregulated genes encoding structural proteins of the bacterial flagella, we decided to analyse the morphology and motility of strain Psv-ΔgacA. Transmission electron microscopy (Figure 4A) showed a characteristic rod-shaped morphology with several amphitrichous flagella for the wild-type strain, whereas no regular flagella could be seen in cells from strain Psv-ΔgacA, which showed an abnormal conical morphology at their poles (Figure 4A).
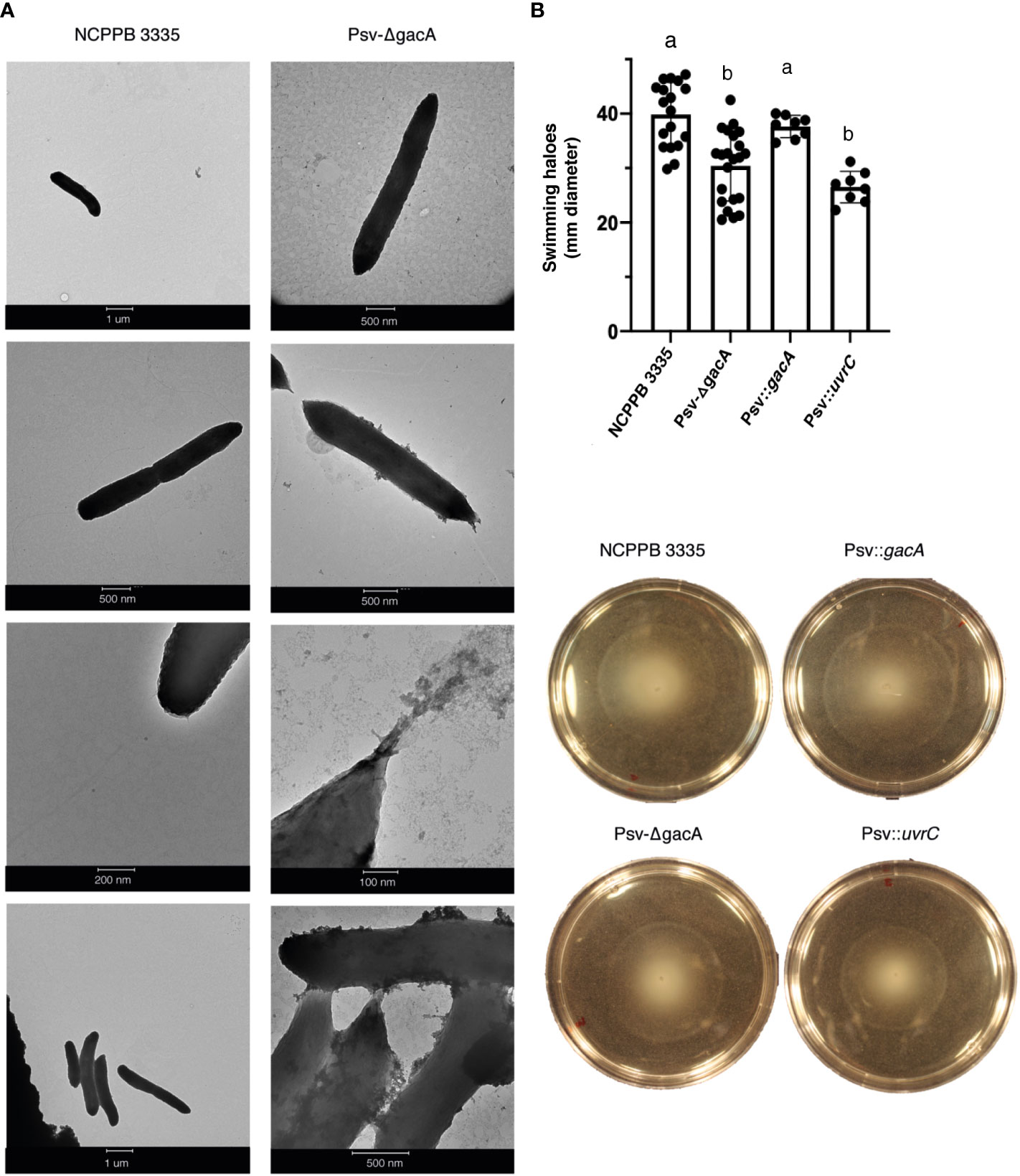
Figure 4 Role of GacA in motility of P. savastanoi pv. savastanoi NCPPB 3335. (A) Transmission electron microscopy (TEM) images of strains NCPPB 3335 and Psv-ΔgacA grown in SSM medium. (B) Swimming motility assay on LB plates. Bars indicate the average diameter of haloes, measured after 48 hours, with standard deviation, and black dots indicate the value of each repetition. Different letters indicate significant differences after Student´s t-test (p < 0.05). The four plates under the graph show representative plates for each of the strains, showing the swimming haloes. Psv::gacA and Psv::uvrC indicate the Psv-ΔgacA mutant complemented with genes gacA and uvrC, respectively.
We then tested the swimming capacity of strain Psv-ΔgacA in comparison with the wild-type strain and the gacA and uvrC complemented strains (Figure 4B). After 72 h, both strain Psv-ΔgacA and the complemented strain Psv::uvrC exhibited a significantly reduced swimming halo when compared to strain NCPPB 3335 or the complemented strain Psv::gacA (Figure 4B). These results indicate that the overexpression of flagellar genes observed in strain Psv-ΔgacA does not correlate with an increased motility or a higher abundance of flagella in the experimental conditions analysed. Additionally, the complete absence of flagella in strain Psv-ΔgacA suggest that they are not essential for swimming motility in Psv NCPPB 3335, as occurs with other bacteria (McCarren and Brahamsha, 2009; Wadhwa and Berg, 2022).
GacA participates in the oxidative stress alleviation in Psv NCCPB 3335
Excessive levels of reactive oxygen species (ROS) or oxidants can cause cellular damage and disrupt cellular signalling processes. Therefore, a critical cell defence mechanism against oxidative stress involves the regulation of genes maintaining ROS homeostasis (Ray et al., 2012). In this sense, previous studies have shown that the GacS/GacA system enhances resistance to H2O2 and participates in the regulation of diverse genes involved in resistance to oxidative stress in biocontrol strains of Pseudomonas (Heeb et al., 2005; Hassan et al., 2010; Kim et al., 2014).
The RNA-Seq analysis showed a differential expression of several genes involved in the oxidative stress response, although the expression patterns were dependent on the growth medium (Supplementary Table S8). In cells of strain Psv-ΔgacA incubated in HIM, all the genes encoding the different subunits of the ATP synthase complex, as well as five out of 13 genes encoding the NADH oxidoreductase complex, were significantly downregulated (Supplementary Table S8). In contrast, 19 genes encoding catalases, peroxidases, aquaporins and reductases showed significantly higher expression levels, likely because of a differential expression of transcriptional regulator encoding genes (Supplementary Table S8). This pattern of regulation is essentially reversed in the ΔgacA mutant grown in SSM medium, where many of the genes that were downregulated in HIM medium were either upregulated or did not show a differential expression.
To evaluate the role of GacA on the response of Psv to an external oxidative stress, we measured growth inhibition around a filter paper disc soaked in H2O2. After 48 hours, both strains Psv-ΔgacA and Psv::uvrC showed a reduced growth inhibition halo compared to strain NCPPB 3335 (Figure 5A). Notably, this phenotype was reversed in strain Psv::gacA, complemented with the gacA gene. Additionally, the oxidative stress state of the different mutants and the wild-type strain was measured using the cell permeable dye provided with the CellROX™ Green Reagent (Thermofisher). This dye penetrates through the plasma membrane and remains in a non-fluorescent version in a reduced state but emits fluorescence upon oxidation by ROS. The emission of green fluorescence detected by confocal microscopy was significantly reduced for both strains Psv-ΔgacA and Psv::uvrC, suggesting a lower abundance of ROS in their cells when compared with strains NCPPB 3335 or Psv::gacA (Figures 5B, C). These results would indicate that the activity of GacA not only contributes to a decreased tolerance to H2O2, but also to a reduction in the amount of intracellular ROS in free-living bacterial cells not exposed to oxidative stresses.
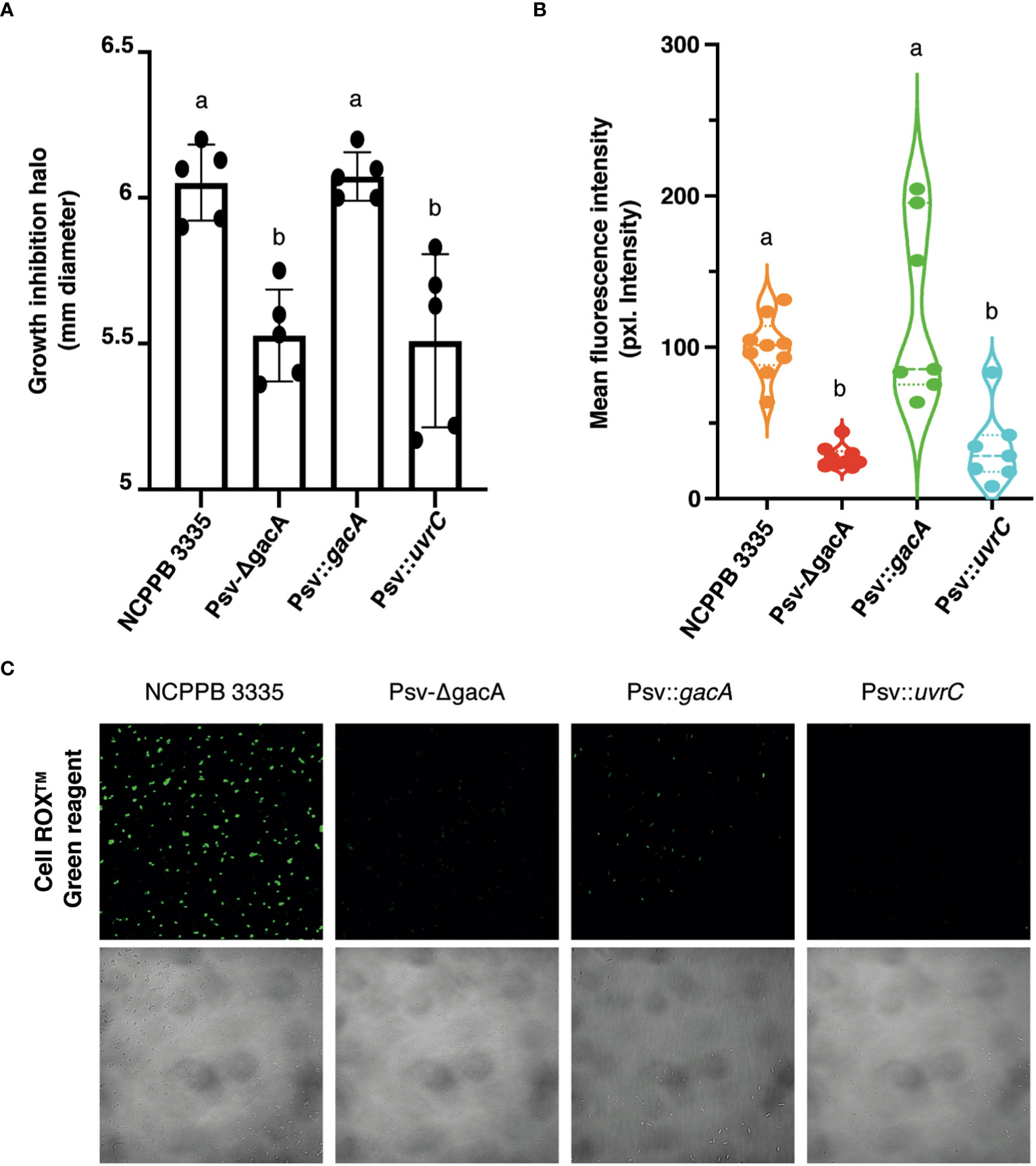
Figure 5 Role of GacA in tolerance to oxidative stress. (A) Bacterial growth sensitivity assay of the wild-type, mutant, and complemented strains to H2O2. Bars are average growth inhibition haloes with standard deviation. (B) Level of cell fluorescence, in intensity of pixels, in cultures after being exposed to CellROX™ Green Reagent; fluorescence is proportional to the amount of reactive oxygen species in live cells. (C) Representative image of confocal microscopy analysis of intrinsic oxidative stress detection in living cells using the CellROX™ Green reagent. In panels (A, B), dots represent values of individual repetitions and letters indicate significant differences (Student´s t-test (p < 0.05). Description of strains is as in Figure 4.
Expression of the type III secretion system is upregulated in a Psv gacA mutant
The T3SS is one of the most important pathogenicity factors in plant pathogenic bacteria, including Psv NCPBB 3335 (Pérez-Martínez et al., 2010), facilitating the delivery of T3Es into the host cell cytoplasm (Hueck, 1998; Göhre and Robatzek, 2008). However, the influence of the GacS/GacA pathway over the T3SS can vary among different phytopathogenic Pseudomonas strains (Valentini et al., 2018; O’Malley et al., 2019). Our RNA-Seq data in HIM, revealed that 25 out of the 29 genes from the hrp/hrc cluster were significantly upregulated in strain Psv-ΔgacA compared to the wild-type strain (Figure 6A). Additionally, nearly two thirds of the T3E genes were also upregulated in the ΔgacA mutant exclusively in HIM medium (Supplementary Table S9), suggesting that GacA also participates, directly or indirectly, in ensuring the correct level of transcription of effector genes during pathogenesis. Furthermore, RT-qPCR analyses showed that the expression levels of two T3SS regulatory genes (hrpA and hrpL) and two T3Es genes (hopAZ1 and avrRpm2) were significantly increased in Psv-ΔgacA compared to the wild-type strain (Figure 6B).
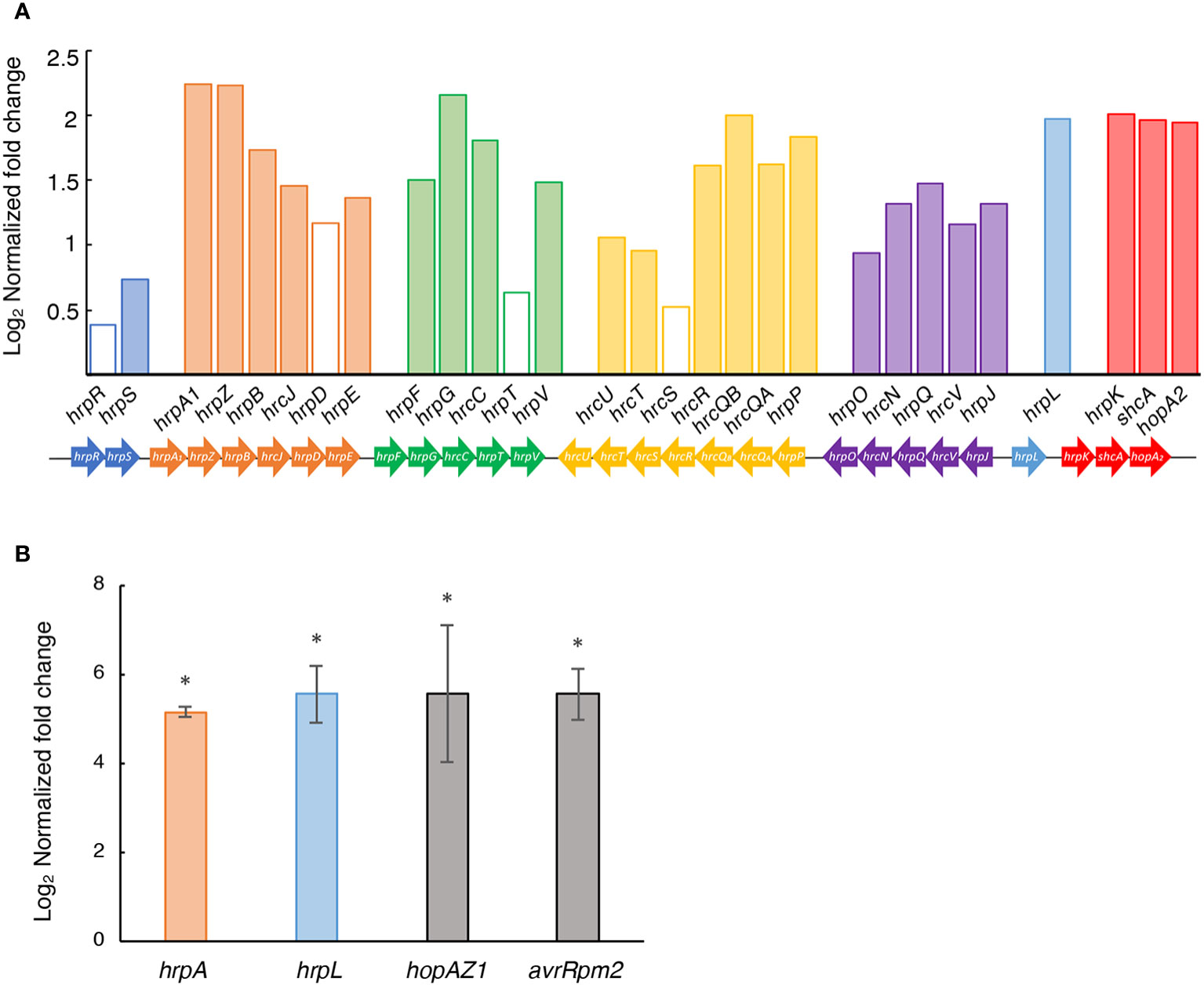
Figure 6 Role of GacA in the expression of the type III secretion system in HIM medium. (A) Bars represent the average log2 of the fold change (q < 0.05) in the RNA-Seq analysis of strain Psv-△gacA relative to NCPPB 3335, with empty bars indicating nonsignificant different values. Genes of the same colour are part of the same operon. (B) RT-qPCR analysis of selected genes upregulated in strain Psv-ΔgacA. Bars represent the average expression values of strain Psv-ΔgacA relative to those of NCPPB 3335, both of which were previously normalized to the constitutive expression of gene gyrA. Error bars represent the standard deviation and asterisks indicate significant differences (Student’s t-test; p < 0.05).
Since genes for the biosynthesis and delivery of T3Es are overexpressed in strain Psv-ΔgacA, we evaluated its ability to induce an HR in tobacco leaves. In line with this overexpression, after 48 h of infiltration, strain Psv-ΔgacA showed a more intense HR than that observed for the wild-type strain or the gacA complemented strain (Figure 7). The Psv-ΔgacA strain complemented with gene uvrC consistently displayed an intermediate phenotype (Figure 7), which was difficult to interpret. It is possible that this is the consequence of a partial complementation in our experimental conditions, as occurs with other systems (e.g., Añorga et al., 2020; Ramírez-Zapata et al., 2020), or a possible contribution of uvrC to the regulation of the secretion process.

Figure 7 Effect of genes gacA and uvrC on the induction of the hypersensitive response (HR). Nicotiana tabacum var. Newdel leaves were infiltrated into the abaxial side with a total of 105 CFUs of the indicated strains, and the response recorded 48 hours post-inoculation. Negative control, infiltrated leaves with 10 mM MgCl2; Psv::gacA and Psv::uvrC indicate the Psv-ΔgacA mutant complemented with genes gacA and uvrC, respectively.
GacA modulates virulence and fitness of Psv NCPPB 3335 in olive plants
Besides the genes presented above, several other genes involved in virulence were overexpressed in the RNA-Seq analysis of strain Psv-ΔgacA (Supplementary Table S10). Among these, five genes located within the ant and cat operons of the WHOP region (Caballo-Ponce et al., 2017b), a genomic island involved in the degradation of aromatic compounds, were significantly upregulated in HIM medium. Additionally, genes involved in the biosynthesis of the phytohormones indole-3-acetic acid (iaaH1 and iaaM1) and cytokinins (ptz) were also significantly upregulated in SSM medium.
To assess the possible impact on virulence of the lack of gene gacA, we carried out virulence assays in olive seedlings under controlled conditions. In these conditions, we measured virulence on the basis of tumour volume and total population within the tumours, as has been done previously (Rodríguez-Moreno et al., 2008; Añorga et al., 2020; Moreno-Pérez et al., 2021). After 54 dpi, tumours induced by strain Psv-ΔgacA and the complemented strain Psv::uvrC were noticeably bigger than tumours induced by strains NCPPB 3335 and Psv::gacA (Figure 8A). As expected, the average tumour volume was significantly higher for strain Psv-ΔgacA and its uvrC-complemented derivative (Figure 8B), while they reached significantly higher cell densities within the tumours than the wild-type and the gacA-complemented strains (Figure 8C). Nevertheless, relative bacterial populations were not significantly different for strains NCPPB 3335 and Psv-gacA, reaching 6.1x105 and 7.4x105 CFU per gram of tumour tissue, respectively. Therefore, the higher virulence of the gacA mutant resulted in larger tumours that, because of a higher volume, supported larger total bacterial populations. However, these tumours are not representative of typical disease symptoms because tumours induced by the wild-type strain showed a more compact and necrotized tissue, whereas those induced by strain Psv-ΔgacA had a softer and more greenish aspect and showed significant parts of the tissue occupied by aggregations of dedifferentiated cells (Figure 8D).
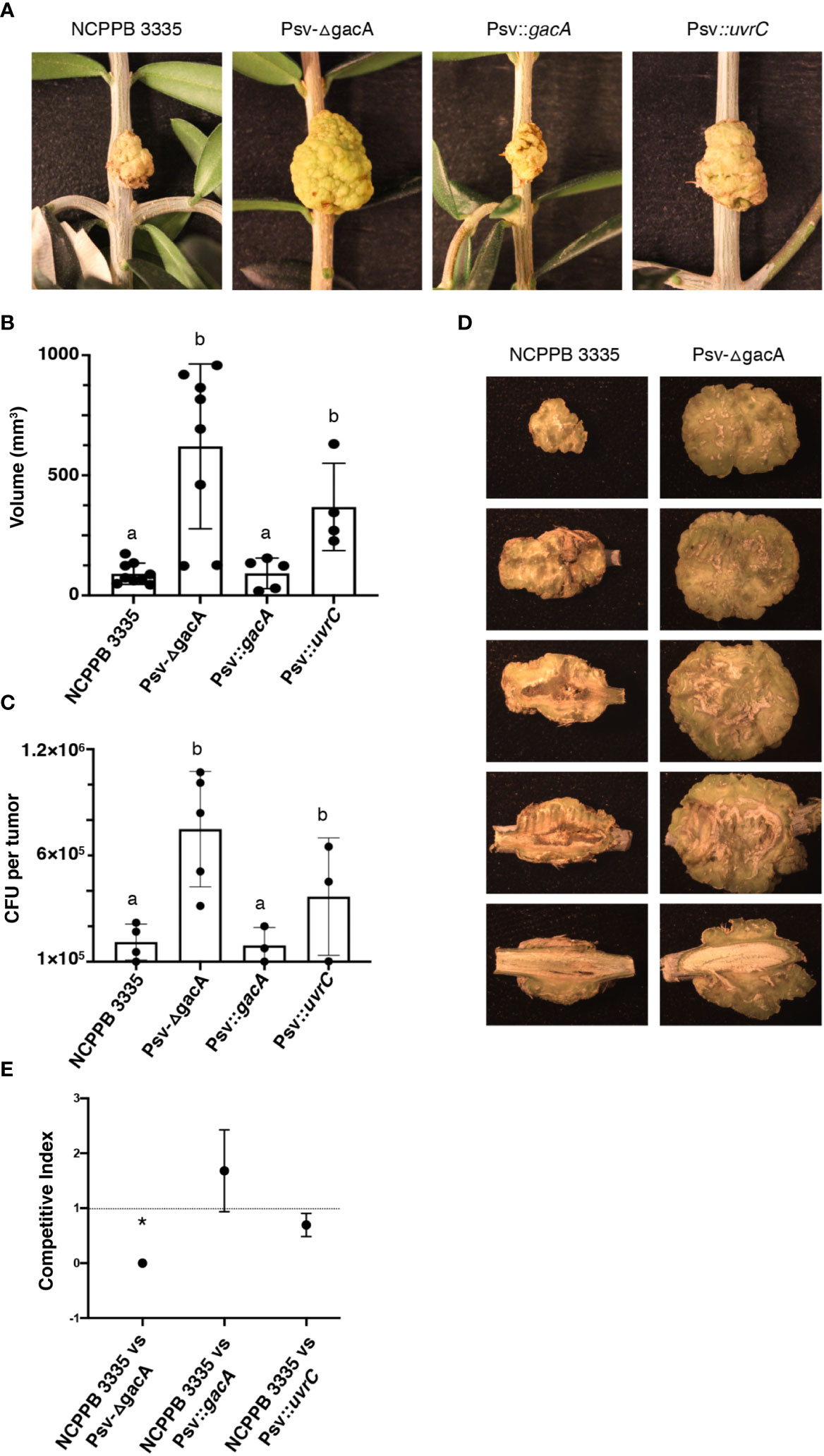
Figure 8 Effect of genes gacA and uvrC on the virulence of P. savastanoi pv. savastanoi NCPPB 3335 in olive plants. (A) Symptoms generated in olive plants at 54 days post-inoculation (dpi). Negative control inoculated with 10 mM MgCl2. (B, C) Bars represent average volume of tumours (B) and average total bacterial populations per tumour (C) at 54 dpi, with dots indicating values of individual samples and error bars representing standard error. Different letters indicate means that are significantly different using ANOVA followed by Bonferroni t-test (p < 0.05). (D) Serial longitudinal sections of a representative tumour per strain; dedifferentiated tissue, apparent as greenish areas where active cell division is occurring, is predominating in tumours induced by strain Psv-ΔgacA. (E) Competitive index for mixed inoculations in micropropagated olive plants; points are the mean from three independent experiments with error bars indicating the standard error. The asterisk indicates a value significantly different from one using Student´s t-test with a threshold of p = 0.05. Description of strains is as in Figure 4.
To investigate competitiveness, micropropagated olive plants were inoculated with a total of 5x103 CFUs of a 1:1 ratio mixture of the wild-type strain and either 1) strain Psv-ΔgacA, 2) strain Psv-ΔgacA complemented with gene gacA (Psv::gacA), or 3) strain Psv-ΔgacA complemented with gene uvrC (Psv::uvrC). The analysis of the bacterial populations of tumours showed no significant difference in competitiveness between the wild-type strain and either one of the two complemented strains (Figure 8E). However, population counts of strain Psv-ΔgacA were significantly lower than those of the wild-type strain, indicating a lower competitiveness of strain Psv-ΔgacA. This indicates that genes gacA and uvrC act redundantly to increase the competitive fitness of the bacterium.
In summary, these results indicate that GacA is necessary for the development of typical disease symptoms, characterized by tumours with increased cavity formation and necrotic tissue. Conversely, the absence of this gene increases virulence, inducing significantly larger tumours with increased bacterial populations, but reduces the competitive advantage of the strain in planta.
Discussion
The Gac-Rsm regulatory pathway has been widely studied in Pseudomonas (Marutani et al., 2008; Ortiz-Martín et al., 2010; Anderson et al., 2017; Zhang et al., 2018; Ramírez-Zapata et al., 2020; Zhang et al., 2020). GacA transcriptionally targets small rsm RNAs, which control the activity of post-transcriptional regulatory proteins of the Csr/Rsm family (Sobrero and Valverde, 2020). Species of Pseudomonas contain a variable number of rsm sRNAs, typically seven in the analysed strains of P. syringae sensu lato, and from two to seven functional csr/rsm protein genes (Sobrero and Valverde, 2020). In this work, we identified seven rsm sRNAs and four different Rsm/Csr gene homologues (rsmA, rsmC, rsmE and rsmH1) in the genome of Psv NCPPB 3335 (Table 1). RsmA and RsmE from Psv NCPPB 3335 showed the highest identity (100 %) with their respective homologues in Pto DC3000 and Pph 1448A (Table 1), suggesting a highly conserved and relevant function for these two proteins in bacteria belonging to P. syringae sensu lato. However, despite the high conservation of all these molecules, their regulatory roles are strain dependent, as has been shown for the regulation of the T3SS in P. syringae (Chatterjee et al., 2003; Marutani et al., 2008; O’Malley et al., 2020; Ramírez-Zapata et al., 2020; O’Malley and Anderson, 2021); and the production of iron-chelating siderophores in P. fluorescens (Cheng et al., 2013; Yu et al., 2014). These regulatory differences might reflect differential affinities of sRNAs to different RsmA homologues and/or distinct expression patterns (Sonnleitner and Haas, 2011; Ge et al., 2019). In fact, in Pph 1448A, GacA regulates the expression of six of the seven sRNAs but does not alter expression of genes encoding Rsm proteins (Ramírez-Zapata et al., 2020). However, in Psv NCPPB 3335, GacA modulated the expression of RsmE and RsmH1 and two sRNAs under the tested conditions (Table 1).
In many bacterial species, the gacA gene is recurrently located upstream of the uvrC gene (Goodier and Ahmer, 2001; Heeb and Haas, 2001), coding for subunit UvrC of the nucleotide excision repair endonuclease UvrABC, which catalyses removal of very diverse chemical and structural DNA lesions. Additionally, uvrY (the gacA homologue in E. coli) and uvrC appear to form a single transcriptional unit in E. coli K12 (Moolenaar et al., 1987), whereas gacA mutants of P. fluorescens, P. aeruginosa, Salmonella typhimurium and Erwinia carotovora showed a significant UV tolerance reduction, suggesting that both genes could also be co-transcribed in these strains (Laville et al., 1992; Reimmann et al., 1997; Ahmer et al., 1998; Eriksson et al., 1998). A 5´ RACE analysis revealed that the 5´-UTR of the uvrC transcripts in the wild-type strain, Psv NCPPB 3335, was located within the gacA CDS (Figures 1B, Supplementary Figure S1A). Therefore, it was unexpected that the RNA-Seq data for strain Psv-ΔgacA showed the existence of a minor amount of complete uvrC transcripts as part of the total RNA samples (Supplementary Figure S1B). These transcripts likely originate from a functional promoter preceding gene gacA. Additionally, RT-qPCR analyses showed a significantly reduced, but still detectable, transcription of uvrC in strain Psv-ΔgacA (Figure 1A). These results suggest that gacA and uvrC form a co-transcriptional unit in Psv NCPPB 3335, as occurs in other bacteria, but that uvrC can still be transcribed from an alternative promoter located within the gacA CDS. This situation also likely occurs in many other bacteria. For instance, uvrC can be transcribed from three functional promoters in E. coli, although only the distal one, located more than 1 kb upstream of the uvrC start codon and likely preceding gacA, leads to an efficient synthesis of the UvrC product (Sharma et al., 1984). Additionally, polar gacA mutations in Pto DC3000 and Pph 1448A reduced, but did not abolish, transcription of uvrC (O’Malley et al., 2019; Ramírez-Zapata et al., 2020). Nevertheless, reduction of the uvrC expression due to mutations on gacA might confound the assessment of the GacA regulon because of the demonstrated changes in diverse phenotypes and gene expression patterns in uvrC mutant backgrounds in other bacteria (Humann et al., 2009; O’Malley et al., 2019). It is unclear how the lack of UvrC could determine widespread transcriptional changes, although it may be due to reductions in DNA repair activities or a failure to adequately respond to cellular stresses (Humann et al., 2009; O’Malley et al., 2019). However, we are confident in our identification of phenotypes regulated by GacA because we routinely used as controls derivative strains of Psv-ΔgacA complemented with either the gacA or the uvrC gene.
Many phytopathogenic bacteria combine epiphytic and endophytic lifestyles during their host colonization process, for which bacterial motility is crucial to enable colonization of different niches by microorganisms. For instance, flagella formation, as well as swimming, swarming, and twitching movements, are negatively regulated by GacS/GacA in the biocontrol strains P. fluorescens F113 (Navazo et al., 2009) and P. chlororaphis O6 (Kim et al., 2014). P. fluorescens strains carrying a gacS deletion produce higher swimming haloes (Martínez-Granero et al., 2006), however, this phenotype disappears when RsmA and RsmE are overexpressed in this mutant (Martínez-Granero et al., 2012). Additionally, GacA is required by Pto DC3000 for swarming and swimming motility (Chatterjee et al., 2003; Vargas et al., 2013), as well as for full virulence on leaf surfaces (O’Malley et al., 2020). In this sense, our RNA-Seq analysis showed that 37 out of 42 genes encoding the flagella machinery were significantly upregulated in strain Psv-ΔgacA grown in SSM (Figure 3), but not in HIM medium. However, in spite of the higher expression of flagella genes, Psv-ΔgacA showed smaller swimming haloes than the wild-type strain NCPPB 3335 or the Psv::gacA complemented strains (Figure 4B). Intriguingly, the mutant strain Psv-ΔgacA was unable to polymerize regular flagella, showing an abnormal cone-shaped morphology at the cell poles (Figure 4A). Since swimming motility depends on a correct flagella disposition, the reduced motility haloes (Figure 4B) would likely be caused by the inability of strain Psv-ΔgacA to assemble regular flagella (Figure 4A). Reduced swimming capacities and altered colony morphologies have also been reported in GacA mutants of P. aeruginosa and P. fluorescens (Goodier and Ahmer, 2001). Furthermore, similar phenotypes have also been observed in animal pathogenic bacteria carrying mutations in gene csrA, i.e., Legionella pneumophila (Molofsky and Swanson, 2003), Bacillus subtilis (Yakhnin et al., 2007) or Borrelia burgdorferi (Sanjuan et al., 2009). All these results together demonstrate a connection between the Gac-Rsm regulatory pathway and bacterial motility and morphology in gamma proteobacteria (Goodier and Ahmer, 2001; O’Malley and Anderson, 2021; Song et al., 2023). Nevertheless, it remains to be elucidated whether the effect of gacA in the expression of flagellar genes takes place at the transcriptional or post-transcriptional levels. For instance, LetA, the GacA homologue of L. pneumophila, regulates expression of flagellar genes in an sRNA-independent manner (Sahr et al., 2009). Proteomics analyses would likely contribute to address this issue in Psv.
One of the earliest plant defence mechanisms is the generation of reactive oxygen species (ROS), contributing, together with the acidic pH in the apoplast, to cause oxidative stress (Sharma et al., 2012). In turn, GacS/GacA participates in the regulation of genes involved in the metabolism of ROS. For instance, ROS-related genes are upregulated in GacS/GacA mutants of various P. fluorescens and P. protegens strains (Whistler et al., 1998; Hassan et al., 2010; Cheng et al., 2013). In P. fluorescens SBW25, the sod gene cluster, including a super-oxide dismutase (sodA) and a fumarate hydratase (fumC1), was among the most differentially regulated locus in a gacS mutant (Cheng et al., 2013). Likewise, diverse genes related to the redox status of the cell also appear to be regulated by the GacS/GacA system in Psv NCPPB 3335, depending strongly on the growth medium (Supplementary Table S8). In particular, the soxR gene, which encodes a negative LysR family transcriptional regulator with a critical role in oxidative stress responses in numerous bacteria (Flores-Cruz and Allen, 2011; Toledo et al., 2011; Burbank and Roper, 2014), was inversely deregulated in Psv-ΔgacA during growth in SSM (upregulated) and HIM (downregulated) (Supplementary Table S8). Therefore, this suggests that GacA could indirectly regulate the expression of genes related to the oxidative stress response through the transcriptional regulatory activity of SoxR. Genes encoding the NADH-quinone oxidoreductase complex also show a similar inverse deregulation in SSM and HIM media (Supplementary Table S8). Considering that HIM media simulates the oxidative environment found for bacterial cells during plant invasion, a reduced activity of the NADH-quinone oxidoreductase complex could avoid the generation of additional ROS within the bacterial cells. Nevertheless, we cannot establish a clear relationship between gene expression mediated by GacA in NCPPB 3335 and its resistance to oxidative stresses. Thus, GacA mediates repression of various catalases, peroxidases, or reductases genes in HIM medium (Supplementary Table S8), although we would have expected that these genes would be highly expressed natively because HIM mimics the oxidative conditions found in the plant apoplast. Additionally, we identified only a few genes potentially involved in resistance to oxidative stresses that were upregulated in Psv-ΔgacA, even though this strain showed a higher tolerance to hydrogen peroxide in in vitro assays (Figure 5A) and a higher capacity to reduce the accumulation of ROS inside the bacterial cells than the wild-type strain (Figures 5B, C). Finally, we did not observe a significant deregulation of other genes that are involved in tolerance and detoxification of ROS, such as the sod locus or genes pqiB and rubB, previously identified in Psv NCPPB 3335 as required for survival in olive plant tissues (Matas et al., 2012). Therefore, in Psv NCPPB 3335 GacA participates, directly or indirectly, in the regulation of genes involved in maintaining the redox status of the cell and the tolerance to oxidative stresses, but further research is necessary to identify the genes involved in these functions and the regulatory level at which GacA exerts its action.
Induction of T3SS genes in P. syringae sensu lato cultures requires incubation in the synthetic HIM medium used in this study (Huynh et al., 1989), which mimics apoplast conditions. Incubation of low cell density cultures in this medium allowed the evaluation of the role of the Gac-Rsm system in P. syringae pathogens of bean, tomato, and tobacco plants (Chatterjee et al., 2003; Marutani et al., 2008; Ortiz-Martín et al., 2010; Ferreiro et al., 2018). The role of GacS/GacA in regulation of the T3SS, one of the most relevant pathogenicity factors in phytopathogenic bacteria (Alfano and Collmer, 2004), is variable among P. syringae strains (Hirano et al., 1997; Chatterjee et al., 2003; Marutani et al., 2008; Ortiz-Martín et al., 2010; Vargas et al., 2013). GacA negatively regulates T3SS genes in Pto DC3000 and Pph 1448A (Chatterjee et al., 2003; Ortiz-Martín et al., 2010). Similarly, GacA negatively regulates both the T3SS and effector genes in NCPPB 3335 (Figure 6, Supplementary Table S9) and the strain with a mutated gacA gene produced a more intense and expanded HR reaction in tobacco (Figure 7), unlike in strains DC3000 and 1448A. Remarkably, overexpression of the T3SS and T3Es in Psv-ΔgacA also resulted in increased virulence, characterized by significantly larger tumour volume and bacterial populations for inoculations of strain Psv-ΔgacA. However, the tumours showed an immature appearance and a greenish colour, with no apparent tissue necrosis, indicating an altered infection process (Figures 8A, C). We do not have a satisfactory explanation for the larger tumours produced by the gacA mutant, because genes involved in the biosynthesis of phytohormones are not significantly differentially expressed in medium HIM (Supplementary Table S10). It is possible, however, that the overexpression of gene idi (Supplementary Table S10) could contribute to an imbalance of phytohormones impacting the development of tumours, as shown before with mutants in this gene (Añorga et al., 2020). Despite the higher population of cells reached in planta by Psv-ΔgacA, this strain showed a significantly reduced competitiveness. Therefore, our assays suggest that the ability of the gacA mutant to grow in the plant tissue is apparently not impaired when infecting the plant by itself. However, the infection process is likely affected because it cannot colonize the tissue as efficiently as the wild-type strain. These results underscore the variability of the GacA regulon in diverse strains, as a gacA mutant of Pto DC3000 was not affected in virulence in Arabidopsis whereas that of Pph 1448A showed a moderately reduced virulence on bean (Phaseolus vulgaris).
In summary, in this work we analysed the GacA regulon of Psv using an RNA-Seq analysis in a standard minimal growth medium, SSM, and a hrp-inducing medium, HIM. We identified DEGs in both growth media, suggesting differences in the regulation of metabolic pathways depending on environmental conditions. Most of the genes significantly upregulated in Psv-ΔgacA in SSM medium were related to bacterial motility. Conversely, the most relevant genes significantly upregulated in HIM medium were those involved in the redox balance and tolerance to oxidative stress, as well as those encoding the T3SS and T3Es. The phenotypic analyses shown here demonstrate that GacA participates, directly or indirectly, in the regulation of relevant physiological processes in Psv NCPPB 3335, including motility, resistance to oxidative stress, virulence, and fitness during invasion and colonization of plant tissue. Further proteomics analysis would be required to determine the level at which GacA exerts its function in this bacterial pathogen of woody hosts.
Data availability statement
The datasets presented in this study can be found online at the NCBI, accession number GSE254022.
Author contributions
CL-B: Data curation, Formal analysis, Investigation, Methodology, Software, Writing – original draft. JM: Conceptualization, Data curation, Formal analysis, Methodology, Supervision, Writing – original draft, Writing – review & editing, Funding acquisition. MM-G: Investigation, Methodology, Writing – original draft. CR: Conceptualization, Data curation, Formal analysis, Funding acquisition, Methodology, Project administration, Supervision, Writing – original draft. Writing – review & editing. LR-M: Conceptualization, Data curation, Formal analysis, Funding acquisition, Methodology, Project administration, Supervision, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This research was supported by project grants PID2020-115177RB-C21 and PID2020-115177RB-C22 financed by the Spanish Ministry of Science and Innovation (MCIN)/Agencia Estatal de Investigación (AEI)/10.13039/501100011033/ and by the European Regional Development Fund (ERDF) “A way to make Europe”. CL-B was supported by the FPI2018-084276 predoctoral grant. Open access was partially funded by grant QUAL21 012 IHSM, Consejería de Universidad, Investigación e Innovación, Junta de Andalucía.
Acknowledgments
We are grateful to P. García-Vallejo and I. Imbroda for their excellent technical assistance. We thank the Servicios Centrales de Apoyo a la Investigación (SCAI) of Málaga University for the technical support. We also thank T.H. Osinga for critical reading of the manuscript and for advice on English usage.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2024.1347982/full#supplementary-material
References
Ahmer, B. M., Van Reeuwijk, J., Timmers, C. D., Valentine, P. J., Heffron, F. (1998). Salmonella typhimurium encodes an SdiA homolog, a putative quorum sensor of the LuxR family, that regulates genes on the virulence plasmid. J. Bacteriology 180 (5), 1185–1193. doi: 10.1128/JB.180.5.1185-1193.1998
Alfano, J. R., Collmer, A. (1997). The type III (Hrp) secretion pathway of plant pathogenic bacteria: trafficking harpins, Avr proteins, and death. J. Bacteriology 179 (18), 5655–5662. doi: 10.1128/jb.179.18.5655-5662.1997
Alfano, J. R., Collmer, A. (2004). Type III secretion system effector proteins: double agents in bacterial disease and plant defense. Annu. Rev. Phytopathol. 42, 385–414. doi: 10.1146/annurev.phyto.42.040103.110731
Anderson, A. J., Kang, B. R., Kim, Y. C. (2017). The Gac/Rsm signaling pathway of a biocontrol bacterium, Pseudomonas chlororaphis O6. Res. Plant Dis. 23 (3), 212–227. doi: 10.5423/RPD.2017.23.3.212
Añorga, M., Pintado, A., Ramos, C., De Diego, N., Ugena, L., Novák, O., et al. (2020). Genes ptz and idi, coding for cytokinin biosynthesis enzymes, are essential for tumorigenesis and in planta growth by P. syringae pv. savastanoi NCPPB 3335. Front. Plant Sci. 11, 1294. doi: 10.3389/fpls.2020.01294
Antúnez-Lamas, M., Cabrera-Ordonez, E., Lopez-Solanilla, E., Raposo, R., Trelles-Salazar, O., Rodriguez-Moreno, A., et al. (2009). Role of motility and chemotaxis in the pathogenesis of Dickeya dadantii 3937 (ex Erwinia chrysanthemi 3937). Microbiology 155 (2), 434–442. doi: 10.1099/mic.0.022244-0
Aragón, I. M., Pérez-Martínez, I., Moreno-Pérez, A., Cerezo, M., Ramos, C. (2014). New insights into the role of indole-3-acetic acid in the virulence of Pseudomonas savastanoi pv. savastanoi. FEMS Microbiol. Lett. 356 (2), 184–192. doi: 10.1111/1574-6968.12413
Aragón, I. M., Pérez-Mendoza, D., Moscoso, J. A., Faure, E., Guery, B., Gallegos, M. T., et al. (2015). Diguanylate cyclase DgcP is involved in plant and human Pseudomonas spp. infections. Environ. Microbiol. 17 (11), 4332–4351. doi: 10.1111/1462-2920.12856
Baltrus, D. A., Nishimura, M. T., Romanchuk, A., Chang, J. H., Mukhtar, M. S., Cherkis, K., et al. (2011). Dynamic evolution of pathogenicity revealed by sequencing and comparative genomics of 19 Pseudomonas syringae isolates. PloS Pathog. 7 (7), e1002132. doi: 10.1371/journal.ppat.1002132
Berlanga-Clavero, M. V., Molina-Santiago, C., Caraballo-Rodríguez, A. M., Petras, D., Díaz-Martínez, L., Pérez-García, A., et al. (2022). Bacillus subtilis biofilm matrix components target seed oil bodies to promote growth and anti-fungal resistance in melon. Nat. Microbiol. 7 (7), 1001–1015. doi: 10.1038/s41564-022-01134-8
Brencic, A., McFarland, K. A., McManus, H. R., Castang, S., Mogno, I., Dove, S. L., et al. (2009). The GacS/GacA signal transduction system of Pseudomonas aeruginosa acts exclusively through its control over the transcription of the RsmY and RsmZ regulatory small RNAs. Mol. Microbiol. 73, 434–445. doi: 10.1111/j.1365-2958.2009.06782.x
Burbank, L., Roper, M. C. (2014). OxyR and SoxR modulate the inducible oxidative stress response and are implicated during different stages of infection for the bacterial phytopathogen Pantoea stewartii subsp. stewartii. Mol. Plant-Microbe Interact. 27 (5), 479–490. doi: 10.1094/MPMI-11-13-0348-R
Caballo-Ponce, E., Meng, X., Uzelac, G., Halliday, N., Cámara, M., Licastro, D., et al. (2018). Quorum sensing in Pseudomonas savastanoi pv. savastanoi and Erwinia toletana: role in virulence and interspecies interactions in the olive knot. Appl. Environ. Microbiol. 84 (18), e00950–e00918. doi: 10.1128/AEM.00950-18
Caballo-Ponce, E., Murillo, J., Martínez-Gil, M., Moreno-Pérez, A., Pintado, A., Ramos, C. (2017a). Knots untie: molecular determinants involved in knot formation induced by Pseudomonas savastanoi in woody hosts. Front. Plant Sci. 8, 1089. doi: 10.3389/fpls.2017.01089
Caballo-Ponce, E., Van Dillewijn, P., Wittich, R. M., Ramos, C. (2017b). WHOP, a genomic region associated with woody hosts in the Pseudomonas syringae complex contributes to the virulence and fitness of Pseudomonas savastanoi pv. savastanoi in olive plants. Mol. Plant-Microbe Interact. 30 (2), 113–126. doi: 10.1094/MPMI-11-16-0233-R
Cámara-Almirón, J., Navarro, Y., Díaz-Martínez, L., Magno-Pérez-Bryan, M. C., Molina-Santiago, C., Pearson, J. R., et al. (2020). Dual functionality of the amyloid protein TasA in Bacillus physiology and fitness on the phylloplane. Nat. Commun. 11 (1), 1859. doi: 10.1038/s41467-020-15758-z
Castañeda-Ojeda, M. P., López-Solanilla, E., Ramos, C. (2017b). Differential modulation of plant immune responses by diverse members of the Pseudomonas savastanoi pv. savastanoi HopAF type III effector family. Mol. Plant Pathol. 18 (5), 625–634. doi: 10.1111/mpp.12420
Castañeda-Ojeda, M. P., Moreno-Pérez, A., Ramos, C., López-Solanilla, E. (2017a). Suppression of plant immune responses by the Pseudomonas savastanoi pv. savastanoi NCPPB 3335 type III effector tyrosine phosphatases HopAO1 and HopAO2. Front. Plant Sci. 8, 680. doi: 10.3389/fpls.2017.00680
Chancey, S. T., Wood, D. W., Pierson, L. S., III (1999). Two-component transcriptional regulation of N-acyl-homoserine lactone production in Pseudomonas aureofaciens. Appl. Environ. Microbiol. 65 (6), 2294–2299. doi: 10.1128/AEM.65.6.2294-2299.1999
Chatterjee, A., Cui, Y., Yang, H., Collmer, A., Alfano, J. R., Chatterjee, A. K. (2003). GacA, the response regulator of a two-component system, acts as a master regulator in Pseudomonas syringae pv. tomato DC3000 by controlling regulatory RNA, transcriptional activators, and alternate sigma factors. Mol. Plant-Microbe Interact. 16 (12), 1106–1117. doi: 10.1094/mpmi.2003.16.12.1106
Cheng, X., de Bruijn, I., van der Voort, M., Loper, J. E., Raaijmakers, J. M. (2013). The Gac regulon of Pseudomonas fluorescens SBW 25. Environ. Microbiol. Rep. 5 (4), 608–619. doi: 10.1111/1758-2229.12061
Corpet, F. (1988). Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 16 (22), 10881–10890. doi: 10.1093/nar/16.22.10881
Dillon, M. M., Almeida, R. N., Laflamme, B., Martel, A., Weir, B. S., Desveaux, D., et al. (2019). Molecular evolution of Pseudomonas syringae type III secreted effector proteins. Front. Plant Sci. 10, 418. doi: 10.3389/fpls.2019.00418
Driver, J. A., Kuniyuki, A. H. (1984). In vitro propagation of Paradox walnut rootstock. HortScience 19 (4), 507–509. doi: 10.21273/HORTSCI.19.4.507
Eriksson, A. R., Andersson, R. A., Pirhonen, M., Palva, E. T. (1998). Two-component regulators involved in the global control of virulence in Erwinia carotovora subsp. carotovora. Mol. Plant-Microbe Interact. 11 (8), 743–752. doi: 10.1094/MPMI.1998.11.8.743
Ferreiro, M. D., Behrmann, L. V., Corral, A., Nogales, J., Gallegos, M. T. (2021). Exploring the expression and functionality of the rsm sRNAs in Pseudomonas syringae pv. tomato DC3000. RNA Biol. 18 (11), 1818–1833. doi: 10.1080/15476286.2020.1871217
Ferreiro, M. D., Gallegos, M. T. (2021). Distinctive features of the Gac-Rsm pathway in plant-associated Pseudomonas. Environ. Microbiol. 23 (10), 5670–5689. doi: 10.1111/1462-2920.15558
Ferreiro, M. D., Nogales, J., Farias, G. A., Olmedilla, A., Sanjuán, J., Gallegos, M. T. (2018). Multiple CsrA proteins control key virulence traits in Pseudomonas syringae pv. tomato DC3000. Mol. Plant-Microbe Interact. 31 (5), 525–536. doi: 10.1094/MPMI-09-17-0232-R
Flores-Cruz, Z., Allen, C. (2011). Necessity of OxyR for the hydrogen peroxide stress response and full virulence in Ralstonia solanacearum. Appl. Environ. Microbiol. 77 (18), 6426–6432. doi: 10.1128/AEM.05813-11
Freter, R., Allweiss, B., O’brien, P. C., Halstead, S. A., Macsai, M. S. (1981). Role of chemotaxis in the association of motile bacteria with intestinal mucosa: in vitro studies. Infection Immun. 34 (1), 241–249. doi: 10.1128/iai.34.1.241-249.1981
Gao, R., Mack, T. R., Stock, A. M. (2007). Bacterial response regulators: versatile regulatory strategies from common domains. Trends Biochem. Sci. 32 (5), 225–234. doi: 10.1016/j.tibs.2007.03.002
Gauthier, J. D., Jones, M. K., Thiaville, P., Joseph, J. L., Swain, R. A., Krediet, C. J., et al. (2010). Role of GacA in virulence of Vibrio vulnificus. Microbiology 156 (12), 3722–3733. doi: 10.1099/mic.0.043422-0
Ge, Y., Lee, J. H., Liu, J., Yang, H. W., Tian, Y., Hu, B., et al. (2019). Homologues of the RNA binding protein RsmA in Pseudomonas syringae pv. tomato DC3000 exhibit distinct binding affinities with non-coding small RNAs and have distinct roles in virulence. Mol. Plant Pathol. 20, 1217–1236. doi: 10.1111/mpp.12823
Ghosh, S., Chan, C. K. K. (2016). “Analysis of RNA-Seq data using TopHat and Cufflinks,” in Plant Bioinformatics: Methods and Protocols (New York, NY, USA; Humana Press), 339–361.
Goff, L. A., Trapnell, C., Kelley, D. (2012). CummeRbund: visualization and exploration of Cufflinks high-throughput sequencing data. R package version (Cambridge, MA, USA: Computer Science & Artificial Intelligence Laboratory), Vol. 2.
Göhre, V., Robatzek, S. (2008). Breaking the barriers: microbial effector molecules subvert plant immunity. Annu. Rev. Phytopathol. 46, 189–215. doi: 10.1146/annurev.phyto.46.120407.110050
Gooderham, W. J., Hancock, R. E. (2009). Regulation of virulence and antibiotic resistance by two-component regulatory systems in Pseudomonas aeruginosa. FEMS. Microbiol. Rev. 33 (2), 279–294. doi: 10.1111/j.1574-6976.2008.00135.x
Goodier, R. I., Ahmer, B. M. (2001). SirA orthologs affect both motility and virulence. J. Bacteriology 183 (7), 2249–2258. doi: 10.1128/JB.183.7.2249-2258.2001
Hanahan, D. (1983). Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166 (4), 557–580. doi: 10.1016/S0022-2836(83)80284-8
Hassan, K. A., Johnson, A., Shaffer, B. T., Ren, Q., Kidarsa, T. A., Elbourne, L. D., et al. (2010). Inactivation of the GacA response regulator in Pseudomonas fluorescens Pf-5 has far-reaching transcriptomic consequences. Environ. Microbiol. 12 (4), 899–915. doi: 10.1111/j.1462-2920.2009.02134.x
He, S. Y. (1998). Type III protein secretion systems in plant and animal pathogenic bacteria. Annu. Rev. Phytopathol. 36 (1), 363–392. doi: 10.1146/annurev.phyto.36.1.363
Heeb, S., Haas, D. (2001). Regulatory roles of the GacS/GacA two-component system in plant-associated and other gram-negative bacteria. Mol. Plant-Microbe Interact. 14 (12), 1351–1363. doi: 10.1094/MPMI.2001.14.12.1351
Heeb, S., Valverde, C., Gigot-Bonnefoy, C., Haas, D. (2005). Role of the stress sigma factor RpoS in GacA/RsmA-controlled secondary metabolism and resistance to oxidative stress in Pseudomonas fluorescens CHA0. FEMS Microbiol. Lett. 243 (1), 251–258. doi: 10.1016/j.femsle.2004.12.008
Hirano, S. S., Ostertag, E. M., Savage, S. A., Baker, L. S., Willis, D. K., Upper, C. D. (1997). Contribution of the regulatory gene lemA to field fitness of Pseudomonas syringae pv. syringae. Appl. Environ. Microbiol. 63 (11), 4304–4312. doi: 10.1128/aem.63.11.4304-4312.1997
Hrabak, E. M., Willis, D. K. (1992). The lemA gene required for pathogenicity of Pseudomonas syringae pv. syringae on bean is a member of a family of two-component regulators. J. Bacteriology 174 (9), 3011–3020. doi: 10.1128/jb.174.9.3011-3020.1992
Huang, H., Shao, X., Xie, Y., Wang, T., Zhang, Y., Wang, X., et al. (2019). An integrated genomic regulatory network of virulence-related transcriptional factors in. Pseudomonas aeruginosa. Nat. Commun. 10 (1), 2931. doi: 10.1038/s41467-019-10778-w
Hueck, C. J. (1998). Type III protein secretion systems in bacterial pathogens of animals and plants. Microbiol. Mol. Biol. Rev. 62 (2), 379–433. doi: 10.1128/MMBR.62.2.379-433.1998
Humann, J. L., Ziemkiewicz, H. T., Yurgel, S. N., Kahn, M. L. (2009). Regulatory and DNA repair genes contribute to the desiccation resistance of Sinorhizobium meliloti Rm1021. Appl. Environ. Microbiol. 75, 446–453. doi: 10.1128/AEM.02207-08
Hutcheson, S. W., Bretz, J., Sussan, T., Jin, S., Pak, K. (2001). Enhancer-binding proteins HrpR and HrpS interact to regulate hrp-encoded type III protein secretion in Pseudomonas syringae strains. J. Bacteriology 183 (19), 5589–5598. doi: 10.1128/JB.183.19.5589-5598.2001
Huynh, T. V., Dahlbeck, D., Staskawicz, B. J. (1989). Bacterial blight of soybean: regulation of a pathogen gene determining host cultivar specificity. Science 245 (4924), 1374–1377. doi: 10.1126/science.278128
Kearse, M., Moir, R., Wilson, A., Stones-Havas, S., Cheung, M., Sturrock, S., et al. (2012). Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28 (12), 1647–1649. doi: 10.1093/bioinformatics/bts199
Kim, C. H., Kim, Y. H., Anderson, A. J., Kim, Y. C. (2014). Proteomic analysis of a global regulator GacS sensor kinase in the rhizobacterium, Pseudomonas chlororaphis O6. Plant Pathol. J. 30 (2), 220. doi: 10.5423/PPJ.NT.02.2014.0012
King, E. O., Ward, M. K., Raney, D. E. (1954). Two simple media for the demonstration of pyocyanin and fluorescin. J. Lab. Clin. Med. 44 (2), 301–307. doi: 10.5555/uri:pii:002221435490222X
Kitten, T., Kinscherf, T. G., McEvoy, J. L., Willis, D. K. (1998). A newly identified regulator is required for virulence and toxin production in Pseudomonas syringae. Mol. Microbiol. 28 (5), 917–929. doi: 10.1046/j.1365-2958.1998.00842.x
Langmead, B., Salzberg, S. L. (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9 (4), 357–359. doi: 10.1038/nmeth.1923
Lapouge, K., Schubert, M., Allain, F. H. T., Haas, D. (2008). Gac/Rsm signal transduction pathway of γ-proteobacteria: from RNA recognition to regulation of social behaviour. Mol. Microbiol. 67 (2), 241–253. doi: 10.1111/j.1365-2958.2007.06042.x
Laville, J., Voisard, C., Keel, C., Maurhofer, M., Defago, G., Haas, D. (1992). Global control in Pseudomonas fluorescens mediating antibiotic synthesis and suppression of black root rot of tobacco. Proc. Natl. Acad. Sci. 89 (5), 1562–1566. doi: 10.1073/pnas.89.5.1562
Lennox, E. S. (1955). Transduction of linked genetic characters of the host by bacteriophage P1. Virology 1, 190–206. doi: 10.1016/0042-6822(55)90016-7
Lindgren, P. B. (1997). The role of hrp genes during plant-bacterial interactions. Annu. Rev. Phytopathol. 35 (1), 129–152. doi: 10.1146/annurev.phyto.35.1.129
Livak, K. J., Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2– ΔΔCT method. Methods 25 (4), 402–408. doi: 10.1006/meth.2001.1262
Macho, A. P., Rufián, J. S., Ruiz-Albert, J., Beuzón, C. R. (2016). Competitive index: mixed infection-based virulence assays for genetic analysis in Pseudomonas syringae-plant interactions. Plant Signal Transduction: Methods Protoc., 209–217. doi: 10.1007/978-1-4939-3115-6_17
Macho, A. P., Zumaquero, A., Ortiz-Martín, I., Beuzon, C. R. (2007). Competitive index in mixed infections: a sensitive and accurate assay for the genetic analysis of Pseudomonas syringae–plant interactions. Mol. Plant Pathol. 8 (4), 437–450. doi: 10.1111/j.1364-3703.2007.00404.x
Madeira, F., Pearce, M., Tivey, A. R. N., Basutkar, P., Lee, J., Edbali, O., et al. (2022). Search and sequence analysis tools services from EMBL-EBI. Nucleic Acids Res. 50, 276–279. doi: 10.1093/nar/gkac240
Martínez-Gil, M., Ramos, C. (2018). Role of cyclic di-GMP in the bacterial virulence and evasion of the plant immunity. Curr. Issues Mol. Biol. 25 (1), 199–222. doi: 10.21775/cimb.025.199
Martínez-Granero, F., Navazo, A., Barahona, E., Redondo-Nieto, M., Rivilla, R., Martín, M. (2012). The Gac-Rsm and SadB signal transduction pathways converge on AlgU to downregulate motility in Pseudomonas fluorescens. PloS One 7 (2), e31765. doi: 10.1371/journal.pone.0031765
Martínez-Granero, F., Rivilla, R., Martín, M. (2006). Rhizosphere selection of highly motile phenotypic variants of Pseudomonas fluorescens with enhanced competitive colonization ability. Appl. Environ. Microbiol. 72 (5), 3429–3434. doi: 10.1128/AEM.72.5.3429-3434.2006
Marutani, M., Taguchi, F., Ogawa, Y., Hossain, M. M., Inagaki, Y., Toyoda, K., et al. (2008). Gac two-component system in Pseudomonas syringae pv. tabaci is required for virulence but not for hypersensitive reaction. Mol. Genet. Genomics 279, 313–322. doi: 10.1007/s00438-007-0309-y
Matas, I. M., Castañeda-Ojeda, M. P., Aragón, I. M., Antúnez-Lamas, M., Murillo, J., Rodríguez-Palenzuela, P., et al. (2014). Translocation and functional analysis of Pseudomonas savastanoi pv. savastanoi NCPPB 3335 type III secretion system effectors reveals two novel effector families of the Pseudomonas syringae complex. Mol. Plant-Microbe Interact. 27 (5), 424–436. doi: 10.1094/MPMI-07-13-0206-R
Matas, I. M., Lambertsen, L., Rodríguez-Moreno, L., Ramos, C. (2012). Identification of novel virulence genes and metabolic pathways required for full fitness of Pseudomonas savastanoi pv. savastanoi in olive (Olea europaea) knots. New Phytol. 196 (4), 1182–1196. doi: 10.1111/j.1469-8137.2012.04357.x
McCarren, J., Brahamsha, B. (2009). Swimming motility mutants of marine Synechococcus affected in production and localization of the S-layer protein SwmA. J. Bacteriology 191 (3), 1111–1114. doi: 10.1128/JB.01401-08
McCarter, L. L. (2006). Regulation of flagella. Curr. Opin. Microbiol. 9 (2), 180–186. doi: 10.1016/j.mib.2006.02.001
Meyer, J. A., Abdallah, M. A. (1978). The fluorescent pigment of Pseudomonas fluorescens: biosynthesis, purification and physicochemical properties. Microbiology 107 (2), 319–328. doi: 10.1099/00221287-107-2-319
Molofsky, A. B., Swanson, M. S. (2003). Legionella pneumophila CsrA is a pivotal repressor of transmission traits and activator of replication. Mol. Microbiol. 50 (2), 445–461. doi: 10.1046/j.1365-2958.2003.03706.x
Moolenaar, G. F., van Sluis, C. A., Backendorf, C., De Putte, P. V. (1987). Regulation of the Escherichia coli excision repair gene uvrC. Overlap between the uvrC structural gene and the region coding for a 24 kD protein. Nucleic Acids Res. 15 (10), 4273–4289. doi: 10.1093/nar/15.10.4273
Moreno-Pérez, A., Ramos, C., Rodríguez-Moreno, L. (2021). HrpL regulon of bacterial pathogen of woody host Pseudomonas savastanoi pv. savastanoi NCPPB 3335. Microorganisms 9 (7), 1447. doi: 10.3390/microorganisms9071447
Moretti, C., Trabalza, S., Granieri, L., Caballo-Ponce, E., Devescovi, G., Del Pino, A. M., et al. (2019). A Na+/Ca2+ exchanger of the olive pathogen Pseudomonas savastanoi pv. savastanoi is critical for its virulence. Mol. Plant Pathol. 20 (5), 716–730. doi: 10.1111/mpp.12787
Navazo, A., Barahona, E., Redondo-Nieto, M., Martínez-Granero, F., Rivilla, R., Martín, M. (2009). Three independent signalling pathways repress motility in Pseudomonas fluorescens F113. Microbial Biotechnol. 2 (4), 489–498. doi: 10.1111/j.1751-7915.2009.00103.x
O’Malley, M. R., Anderson, J. C. (2021). Regulation of the Pseudomonas syringae type III secretion system by host environment signals. Microorganisms 9 (6), 1227. doi: 10.3390/microorganisms9061227
O’Malley, M. R., Chien, C. F., Peck, S. C., Lin, N. C., Anderson, J. C. (2020). A revised model for the role of GacS/GacA in regulating type III secretion by Pseudomonas syringae pv. tomato DC3000. Mol. Plant Pathol. 21 (1), 139–144. doi: 10.1111/mpp.12876
O’Malley, M. R., Weisberg, A. J., Chang, J. H., Anderson, J. C. (2019). Re-evaluation of a Tn5::gacA mutant of Pseudomonas syringae pv. tomato DC3000 uncovers roles for uvrC and anmK in promoting virulence. PloS One 14 (10), e0223637. doi: 10.1371/journal.pone.0223637
Ortiz-Martín, I., Thwaites, R., Macho, A. P., Mansfield, J. W., Beuzón, C. R. (2010). Positive regulation of the Hrp type III secretion system in Pseudomonas syringae pv. phaseolicola. Mol. Plant-Microbe Interact. 23, 665–681. doi: 10.1094/MPMI-23-5-0665
Papon, N., Stock, A. M. (2019). Two-component systems. Curr. Biol. 29 (15), 724–725. doi: 10.1016/j.cub.2019.06.010
Penyalver, R., García, A., Ferrer, A., Bertolini, E., Quesada, J. M., Salcedo, C. I., et al. (2006). Factors affecting Pseudomonas savastanoi pv. savastanoi plant inoculations and their use for evaluation of olive cultivar susceptibility. Phytopathology 96 (3), 313–319. doi: 10.1094/PHYTO-96-0313
Pérez-Martínez, I., Rodríguez-Moreno, L., Lambertsen, L., Matas, I. M., Murillo, J., Tegli, S., et al. (2010). Fate of a Pseudomonas savastanoi pv. savastanoi type III secretion system mutant in olive plants (Olea europaea L.). Appl. Environ. Microbiol. 76 (11), 3611–3619. doi: 10.1128/AEM.00133-10
Pérez-Martínez, I., Rodriguez-Moreno, L., Matas, I. M., Ramos, C. (2007). Strain selection and improvement of gene transfer for genetic manipulation of Pseudomonas savastanoi isolated from olive knots. Res. Microbiol. 158 (1), 60–69. doi: 10.1016/j.resmic.2006.09.008
Ramírez-Zapata, D., Ramos, C., Aguilera, S., Bardaji, L., Martínez-Gil, M., Murillo, J. (2020). Two homologues of the global regulator Csr/Rsm redundantly control phaseolotoxin biosynthesis and virulence in the plant pathogen Pseudomonas amygdali pv. phaseolicola 1448A. Microorganisms 8 (10), 1536. doi: 10.3390/microorganisms8101536
Ramos, C., Matas, I. M., Bardaji, L., Aragón, I. M., Murillo, J. (2012). Pseudomonas savastanoi pv. savastanoi: some like it knot. Mol. Plant Pathol. 13 (9), 998–1009. doi: 10.1111/j.1364-3703.2012.00816.x
Ray, P. D., Huang, B. W., Tsuji, Y. (2012). Reactive oxygen species (ROS) homeostasis and redox regulation in cellular signaling. Cell. Signalling 24 (5), 981–990. doi: 10.1016/j.cellsig.2012.01.008
Reimmann, C., Beyeler, M., Latifi, A., Winteler, H., Foglino, M., Lazdunski, A., et al. (1997). The global activator GacA of Pseudomonas aeruginosa PAO positively controls the production of the autoinducer N-butyryl-homoserine lactone and the formation of the virulence factors pyocyanin, cyanide, and lipase. Mol. Microbiol. 24 (2), 309–319. doi: 10.1046/j.1365-2958.1997.3291701.x
Rodríguez-Moreno, L., Barceló-Muñoz, A., Ramos, C. (2008). In vitro analysis of the interaction of Pseudomonas savastanoi pv. savastanoi and nerii with micropropagated olive plants. Phytopathology 98 (7), 815–822. doi: 10.1094/PHYTO-98-7-0815
Rotenberg, D., Thompson, T. S., German, T. L., Willis, D. K. (2006). Methods for effective real-time RT-PCR analysis of virus-induced gene silencing. J. Virological Methods 138 (1-2), 49–59. doi: 10.1016/j.jviromet.2006.07.017
Sahr, T., Bruggemann, H., Jules, M., Lomma, M., Albert-Weissenberger, C., Cazalet, C., et al. (2009). Two small ncRNAs jointly govern virulence and transmission in Legionella pneumophila. Mol. Microbiol. 72 (3), 741–762. doi: 10.1111/j.1365-2958.2009.06677.x
Sanjuan, E., Esteve-Gassent, M. D., Maruskova, M., Seshu, J. (2009). Overexpression of CsrA (BB0184) alters the morphology and antigen profiles of Borrelia burgdorferi. Infection Immun. 77 (11), 5149–5162. doi: 10.1128/IAI.00673-09
Schindelin, J., Arganda-Carreras, I., Frise, E., Kaynig, V., Longair, M., Pietzsch, T., et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat. Methods 9 (7), 676–682. doi: 10.1038/nmeth.2019
Sharma, P., Jha, A. B., Dubey, R. S., Pessarakli, M. (2012). Reactive oxygen species, oxidative damage, and antioxidative defense mechanism in plants under stressful conditions. J. Bot. 2012, 217037. doi: 10.1155/2012/217037
Sharma, S., Stark, T., Moses, R. E. (1984). Distal regulatory functions for the uvrC of E. coli. Nucleic Acids Res. 12 (13), 5341–5354. doi: 10.1093/nar/12.13.5341
Sobrero, P. M., Valverde, C. (2020). Comparative genomics and evolutionary analysis of RNA-binding proteins of the CsrA family in the genus Pseudomonas. Front. Mol. Biosci. 7, 127. doi: 10.3389/fmolb.2020.00127
Solovyev, A., Salamov, V. S. A. (2011). Automatic annotation of microbial genomes and metagenomic sequences. Metagenomics Its Appl. Agriculture Biomedicine Environ. Stud., 61–78.
Song, H., Li, Y., Wang, Y. (2023). Two-component system GacS/GacA, a global response regulator of bacterial physiological behaviors. Eng. Microbiol. 3 (1), 100051. doi: 10.1016/j.engmic.2022.100051
Sonnleitner, E., Haas, D. (2011). Small RNAs as regulators of primary and secondary metabolism in Pseudomonas species. Appl. Microbioly Biotechnoly 91, 63–79. doi: 10.1007/s00253-011-3332-1
Stock, A. M., Robinson, V. L., Goudreau, P. N. (2000). Two-component signal transduction. Annu. Rev. Biochem. 69 (1), 183–215. doi: 10.1146/annurev.biochem.69.1.183
Taylor, R. K., Miller, V. L., Furlong, D. B., Mekalanos, J. J. (1987). Use of phoA gene fusions to identify a pilus colonization factor coordinately regulated with cholera toxin. Proc. Natl. Acad. Sci. 84 (9), 2833–2837. doi: 10.1073/pnas.84.9.2833
Thornton, B., Basu, C. (2015). Rapid and simple method of qPCR primer design. PCR primer design. Methods Mol. Biol., 173–179. doi: 10.1007/978-1-4939-2365-6_13
Toledo, M. A. S. D., Schneider, D. R., Azzoni, A. R., Favaro, M. T. D. P., Pelloso, A. C., Santos, C. A., et al. (2011). Characterization of an oxidative stress response regulator, homologous to Escherichia coli OxyR, from the phytopathogen Xylella fastidiosa. Protein Expression Purification 75 (2), 204–210. doi: 10.1016/j.pep.2010.10.004
Trapnell, C., Williams, B., Pertea, G., Mortazavi, A., Kwan, G., Van Baren, M. J., et al. (2010). Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28, 511–515. doi: 10.1038/nbt.1621
Untergasser, A., Nijveen, H., Rao, X., Bisseling, T., Geurts, R., Jack, A. M. (2007). Leunissen: Primer3Plus, an enhanced web interface to Primer3. Nucleic Acids Res. 35, 71–74. doi: 10.1093/nar/gkm306
Valentini, M., Gonzalez, D., Mavridou, D. A., Filloux, A. (2018). Lifestyle transitions and adaptive pathogenesis of Pseudomonas aeruginosa. Curr. Opin. Microbiol. 41, 15–20. doi: 10.1016/j.mib.2017.11.006
Vargas, P., Farias, G. A., Nogales, J., Prada, H., Carvajal, V., Barón, M., et al. (2013). Plant flavonoids target Pseudomonas syringae pv. tomato DC 3000 flagella and type III secretion system. Environ. Microbiol. Rep. 5 (6), 841–850. doi: 10.1111/1758-2229.12086
Vargas, P., Felipe, A., Michán, C., Gallegos, M. T. (2011). Induction of Pseudomonas syringae pv. tomato DC3000 MexAB-OprM multidrug efflux pump by flavonoids is mediated by the repressor PmeR. Mol. Plant-Microbe Interact. 24 (10), 1207–1219. doi: 10.1094/MPMI-03-11-0077
Wadhwa, N., Berg, H. C. (2022). Bacterial motility: machinery and mechanisms. Nat. Rev. Microbiol. 20 (3), 161–173. doi: 10.1038/s41579-021-00626-4
Whistler, C. A., Corbell, N. A., Sarniguet, A., Ream, W., Loper, J. E. (1998). The two-component regulators GacS and GacA influence accumulation of the stationary-phase sigma factor ςS and the stress response in Pseudomonas fluorescens Pf-5. J. Bacteriology 180 (24), 6635–6641. doi: 10.1128/JB.180.24.6635-6641.1998
Wuichet, K., Cantwell, B. J., Zhulin, I. B. (2010). Evolution and phyletic distribution of two-component signal transduction systems. Curr. Opin. Microbiol. 13 (2), 219–225. doi: 10.1016/j.mib.2009.12.011
Yakhnin, H., Pandit, P., Petty, T. J., Baker, C. S., Romeo, T., Babitzke, P. (2007). CsrA of Bacillus subtilis regulates translation initiation of the gene encoding the flagellin protein (hag) by blocking ribosome binding. Mol. Microbiol. 64 (6), 1605–1620. doi: 10.1111/j.1365-2958.2007.05765.x
Yu, X., Chen, M., Jiang, Z., Hu, Y., Xie, Z. (2014). The two-component regulators GacS and GacA positively regulate a nonfluorescent siderophore through the Gac/Rsm signaling cascade in high-siderophore-yielding Pseudomonas sp. strain HYS. J. Bacteriology 196 (18), 3259–3270. doi: 10.1128/JB.01756-14
Zere, T. R., Vakulskas, C. A., Leng, Y., Pannuri, A., Potts, A. H., Dias, R., et al. (2015). Genomic targets and features of BarA-UvrY (-SirA) signal transduction systems. PloS One 10, e0145035. doi: 10.1371/journal.pone.0145035
Zhang, L., Shi, Y., Wu, Z., Tan, G. (2018). Characterization of response regulator GacA involved in phaseolotoxin production, hypersensitive response and cellular processes in Pseudomonas syringae pv. actinidiae A18. Physiol. Mol. Plant Pathol. 103, 137–142. doi: 10.1016/j.pmpp.2018.07.001
Keywords: Pseudomonas savastanoi, Pseudomonas syringae, GacS/GacA two component system, RNA-seq analysis, Gac-Rsm system, woody host
Citation: Lavado-Benito C, Murillo J, Martínez-Gil M, Ramos C and Rodríguez-Moreno L (2024) GacA reduces virulence and increases competitiveness in planta in the tumorigenic olive pathogen Pseudomonas savastanoi pv. savastanoi. Front. Plant Sci. 15:1347982. doi: 10.3389/fpls.2024.1347982
Received: 01 December 2023; Accepted: 08 January 2024;
Published: 05 February 2024.
Edited by:
Andrea Chini, Spanish National Research Council (CSIC), SpainReviewed by:
Claudio Valverde, National University of Quilmes, ArgentinaJacob George Malone, John Innes Centre, United Kingdom
Copyright © 2024 Lavado-Benito, Murillo, Martínez-Gil, Ramos and Rodríguez-Moreno. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Cayo Ramos, Y3JyQHVtYS5lcw==; Luis Rodríguez-Moreno, bGdyb2RyaWd1ZXpAdW1hLmVz
 Carla Lavado-Benito
Carla Lavado-Benito Jesús Murillo
Jesús Murillo Marta Martínez-Gil1
Marta Martínez-Gil1 Cayo Ramos
Cayo Ramos Luis Rodríguez-Moreno
Luis Rodríguez-Moreno