- 1Institute of Floriculture, Liaoning Academy of Agriculture Sciences, Shenyang, Liaoning, China
- 2Liaoning Provincial Key Laboratory of Floriculture, Shenyang, Liaoning, China
Currently, soil heavy metal contamination is a severe issue, particularly with Cd pollution. The metal tolerance protein (MTP) proteins, as plant divalent cation transporters, play a crucial role in the transport and tolerance of heavy metals in plants. This study conducted comprehensive identification and characterization of the MTP gene family in the tulip. A total of 11 TgMTP genes were identified and phylogenetically classified into three subfamilies. Conserved motif and gene structure analyses unveiled commonalities and variations among subfamily members. Expression profiling demonstrated several TgMTPs were markedly upregulated under Cd exposure, including the TgMTP7.1. Heterologous expression in yeast validated that TgMTP7.1 could ameliorate Cd sensitivity and enhance its tolerance. These results provide primary insights into the MTP gene family in tulip. Phylogenetic relationships and functional analyses establish a framework for elucidating the transporters and molecular mechanisms governing Cd accumulation and distribution in tulip. Key TgMTPs identified, exemplified by TgMTP7.1, may illuminate molecular breeding efforts aimed at developing Cd-tolerant cultivars for the remediation of soil Cd contamination.
1 Introduction
Tulip (Tulipa gesneriana) is one of the most important ornamental geophytes, cultivated worldwide for its spectacularly colored flowers. As the third largest cut flower after rose and chrysanthemum, the global production of tulip has significant market value (Byczyńska et al., 2019). However, with industrial development, heavy metal pollution has become an increasing concern, ornamental plants grown along roadsides are often subjected to heavy metal stress, tulip also belongs to ornamental plants (Dodangeh et al., 2018). There are significant differences in the accumulation of cadmium and lead between different genotypes of plants or different organs in tulipa (Alexander et al., 2006; Osmani et al., 2018). Excessive accumulation of these non-essential metals could lead to inhibited plant growth, reduced bulb yield and quality through oxidative damage, genotoxicity and interference with nutrient homeostasis (Djebali et al., 2005; Liu et al., 2008).
To cope with metal stress, plants have evolved a complex metal homeostatic network consisting of various membrane transporters and chelators that tightly regulate the uptake, efflux, sequestration, trafficking and detoxification of metal ions (Hall and Williams, 2003). The cation diffusion facilitator (CDF) family, also known as metal tolerance proteins (MTPs) in plants, plays pivotal roles in this network by mediating efflux or sequestration of diverse metal ions across biomembranes (Gustin et al., 2011). There are three subfamilies of MTPs in plants, contain Mn-CDF, Fe/Zn-CDF, and Zn-CDF, and they were classified into seven groups in Arabidopsis (Montanini et al., 2007; Gustin et al., 2011). Plant MTPs function as H+/metal-ion antiporters driven by proton gradients. They contribute to metal tolerance by sequestration of non-essential toxic metals like Cd2+ and Ni2+ into vacuoles. Their transport of essential metals like Zn2+, Mn2+, Fe2+ and Co2+ also facilitates normal growth and development (Paulsen and Saier, 1997; Haney et al., 2005). ShMTP1 conferred Mn2+ tolerance through internal sequestration (Delhaize et al., 2003). The ZAT gene of Arabidopsis confers increased Zn2+ tolerance when overexpressed in plants (Van der Zaal et al., 1999).
The MTP gene family has been investigated in several plant species, including Arabidopsis thaliana (Van der Zaal et al., 1999), rice (Ram et al., 2019), grape (Shirazi et al., 2019) and soybean (El-Sappah et al., 2023), common bean (Yilmaz et al., 2023) and tomato (El-Sappah et al., 2021). And many MTPs function have been identification, such as the heterologous expression of CsMTP8.2 in A. thaliana could decrease the accumulation of Mn in plants (Zhang et al., 2020), the heterologous expression GmMTP8.1 restored growth of Mn-hypersensitive yeast mutant Δpmr1 (Li et al., 2021). Meanwhile, it is noteworthy that phylogenetic analyses have classified plant MTPs into three major subfamilies, but substrate specificities are not absolute, as some MTPs transport multiple metals (Montanini et al., 2007). MTP genes also show differential tissue-specific and metal-responsive expression patterns (Hassinen, 2009). Overexpression of particular MTPs has been found to increase tolerance to Zn, Co, Ni, Fe and Cd (Montanini et al., 2007; Chen et al., 2013; Menguer et al., 2013). In tobacco, under heavy metal toxicity, the NtMTP gene exhibited distinct responses in tobacco leaves and root systems. NtMTP8.1, NtMTP8.4 and NtMTP11.1 displayed Mn transport capability in yeast cells (Liu et al., 2019). In rice, OsMTP11 may play an important role in Mn and other heavy metal transport in rice (Zhang and Liu, 2017). In Eucalyptus grandis, The up-regulation of EgMTP6, EgMTP5 and EgMTP11.1 upon excessive exposure to Cd2+ and Cu2+ may account for the metal translocation from root to leaf (Shirazi et al., 2023). In Populus trichocarpa, PtrMTP8.1, PtrMTP9 and PtrMTP10.4 displayed Mn transport capability in yeast cells, while PtrMTP6 was able to transport Co, Fe and Mn (Gao et al., 2020).
Given the importance of MTP transporters in plant metal homeostasis, characterizing the MTP gene family in tulip will provide critical insights into the mechanistic basis of heavy metal accumulation and distribution in this species. Genome-wide identification of MTPs in tulip may also present valuable targets for developing cultivars tolerant to metal stresses. Moreover, as an ornamental plant, comprehensive identification of MTPs in tulip will facilitate its application in restoring heavy metal polluted soils, like other ornamental plants (Dodangeh et al., 2018). However, genome-wide analysis and functional characterization of MTPs has not yet been performed in tulip.
In this study, we identified and comprehensively analyzed the MTP gene family in tulip through Iso-seq mining and phylogenetic comparisons with A. thaliana and rice MTPs. Gene and protein structures were determined, along with conserved motifs and phylogenetic relationships. Expression profiles of TgMTPs were examined using RNA-seq data. Several MTPs showing metal responsiveness were selected for functional analyses via heterologous expression in yeast. Yeast complementation assays tested Cd transport capabilities. This multifaceted investigation of the MTP gene family provides the first insights into the molecular basis of heavy metal accumulation and tolerance mechanisms in tulip. This study is expected to enhance the understanding of TgMTPs’ functions and establish a solid foundation for scrutinizing the roles and mechanisms of TgMTP proteins.
2 Methods and materials
2.1 Identification of TgMTP gene family members in tulip
Based on the full-length transcriptome assembly results, the CDS sequence, protein sequence, and annotation file of the genome were obtained (bioproject_accession: PRJNA1028662). The Pfam numbers PF16916 and PF01545 of MTP were obtained from the InterPro (https://www.ebi.ac.uk/interpro/ database to download its Hidden Markov Model (HMM) file. The HMM file of MTP was used as input, and hmmsearch (v3.3.2) software (Johnson et al., 2010) was used to search the tulip protein sequence file with E-value < 0.05.
Secondly, the sequence files of MTP proteins in rice and A. thaliana were downloaded from Phytozome (https://phytozome-next.jgi.doe.gov/) database as submission files. Blastp (v2.13.0) (Kelley et al., 2004) was used to search for potential MTP proteins in tulip protein files, and the results of hmmsearch and blastp were combined to obtain candidate gene ID. The protein sequences of candidate genes were extracted and submitted to Pfam, CDD (https://www.ncbi.nlm.nih.gov/Structure/cdd/cdd.shtml) and SMART (http://smart.embl-heidelberg.de) database for domain confirmation to obtain the determined TgMTP protein.
2.2 Analysis of TgMTPs’ physical and chemical properties
By comparing with A. thaliana and rice MTP family members and naming TgMTP family members, the physicochemical properties of TgMTP family members were analyzed by Expasy (https://web.expasy.org/protparam/) software. Cell-PLoc-2 (http://www.csbio.sjtu.edu.cn/bioinf/Cell-PLoc-2/) software was used to predict the subcellular localization of TgMTP.
2.3 Conserved motifs and gene structure analysis of TgMTP protein
All MTP protein sequences of tulips were used as input files, and the conserved motifs of TgMTP family were analyzed by MEME software (version: v5.0.5, http://meme-suite.org/) (Bailey et al., 2006). The parameter motif search number is 10, the minimum motif amino acid length is 6, and the maximum motif amino acid length is 100. The R package gggenomes (https://thackl.github.io/gggenomes/) was used to visually analyze the conserved motif and domain of TgMTP, and TBtools was used to visualize the CDS and UTR regions. The signal peptide of TgMTP protein was predicted by SignalP (v5.0) software (https://services.healthtech.dtu.dk/service.php?SignalP-5.0).
2.4 Ka/Ks analysis of TgMTP gene family members
Ka/Ks analysis, a prevalent and crucial tool in bioinformatics, plays a significant role in examining the evolutionary dynamics of nucleic acid molecules. Within the realm of genetics, the Ka/Ks ratio signifies the balance between the nonsynonymous substitution rate (Ka) and the synonymous substitution rate (Ks) in two protein-coding genes. This metric serves as an indicator of selective pressure acting on these genes, using KaKs _ Calculator (v2.0) (Wang et al., 2010) software for analysis.
2.5 Phylogenetic analysis of TgMTP gene family members
The MTP family protein sequences of A. thaliana, rice, tomato and soybean were downloaded from the Phytozome database, and the phylogenetic tree was drawn using MEGA (v10) software (Kumar et al., 2018). Clustal W (Thompson et al., 1994) was selected as the comparison method, and the phylogenetic tree was constructed by Neighbor-joining method. The model was p-distance, the missing data method was Partial deletion, the cutoff was 50%, and Bootstrap was set to 1000. We use Evolview3 (https://www.evolgenius.info/evolview/) to beautify the tree.
2.6 Protein structure analysis and GO annotation of TgMTP
The protein secondary structure of TgMTP was predicted by sopma (https://npsa-prabi.ibcp.fr/cgi-bin/npsa_automat.pl?page=/NPSA/npsa_sopma.html) website, and the protein tertiary structure of TgMTP was predicted by SwissModel (https://swissmodel.expasy.org/). The clusterProfiler (Yu et al., 2012) in R was used to analyze the GO (Gene Ontology) annotation of TgMTP.
2.7 Regulation network analysis of miRNA- TgMTP gene family members
The psRNATarget online tool (https://www.zhaolab.org/psRNATarget/) was use to analysis the miRNA target gene network of TgMTP gene family. Additionally, since miRNA research in tulip remains scarce, we selected the model plant A. thaliana, which has the most extensive miRNA databases, to analyze the miRNAs regulating TgMTP genes. Cytoscape (Smoot et al., 2011) was used to visualize the interaction networks.
2.8 qRT-PCR analysis
Tulip cultivar “Dow Jones” was exposed at 5°C for 4 weeks and then plant them in the soil (growing media 4.0, Shangdao Co., Ltd, Jinan, China). After treatment, untreated tulip bulbs were used as CK group (0 d), and tulip bulbs were washed and placed in the medium containing different concentrations of metal ions (for FeSO4: 1 mM; for MnSO4: 5 mM; for ZnSO4: 1 mM and for CdCl2: 5 mM) in Hoagland nutrient solution. They were then cultivated in a conventional water-fertilizer incubator in soil with a 25°C/21°C day-night temperature and 16 h day-1 photoperiod. Bulbs subjected to these conditions were sampled at 1, 3, 5, and 7 days and total RNA was extracted using the BioRun Plant Total RNA Extraction Kit (Biorun Co., Ltd, Wuhan, China). cDNA was synthesized, followed by qRT-PCR to measure relative expression. Relative gene expression was calculated using the 2-ΔΔCt method. Graphpad prism 9 software (https://www.graphpad.com/features) was used for plotting and statistical analysis. Primers were designed at NCBI and are listed in Supplementary Table 1.
2.9 Heterologous expression of TgMTP7.1 in yeast
The heterogeneic expression of TgMTP7.1 in yeast BY4741 and ycf1Δ were used to investigate whether TgMTP7.1 can improve the Cd tolerance of yeast. The ORF of TgMTP7.1 was amplified from tulip cDNA and inserted into the yeast expression vector pYES2 under the control of a galactose-inducible promoter. The resulting pYES2-TgMTP7.1 and empty vector pYES2 were transformed into yeast cells. Transgenic yeast strains were cultured in synthetic complete (SC) medium lacking uracil with 2% (w/v) galactose to induce gene expression. For Cd tolerance assays, the exponentially growing yeast cultures were normalized to an OD600 of 0.2, and serial dilutions (100, 101, 102, 103) were prepared (Liu et al., 2019) 2 μL of each dilution was spotted onto SC-Ura agar medium containing 5 mM CdCl2. Plates were incubated at 30°C for 3-5 days before comparing growth differences. To ascertain metal tolerance, the growth of yeast (as measured by the OD600 value) was quantified at various intervals, employing methodologies delineated by Peng (Peng et al., 2018).
3 Results
3.1 Identification of TgMTP gene family members
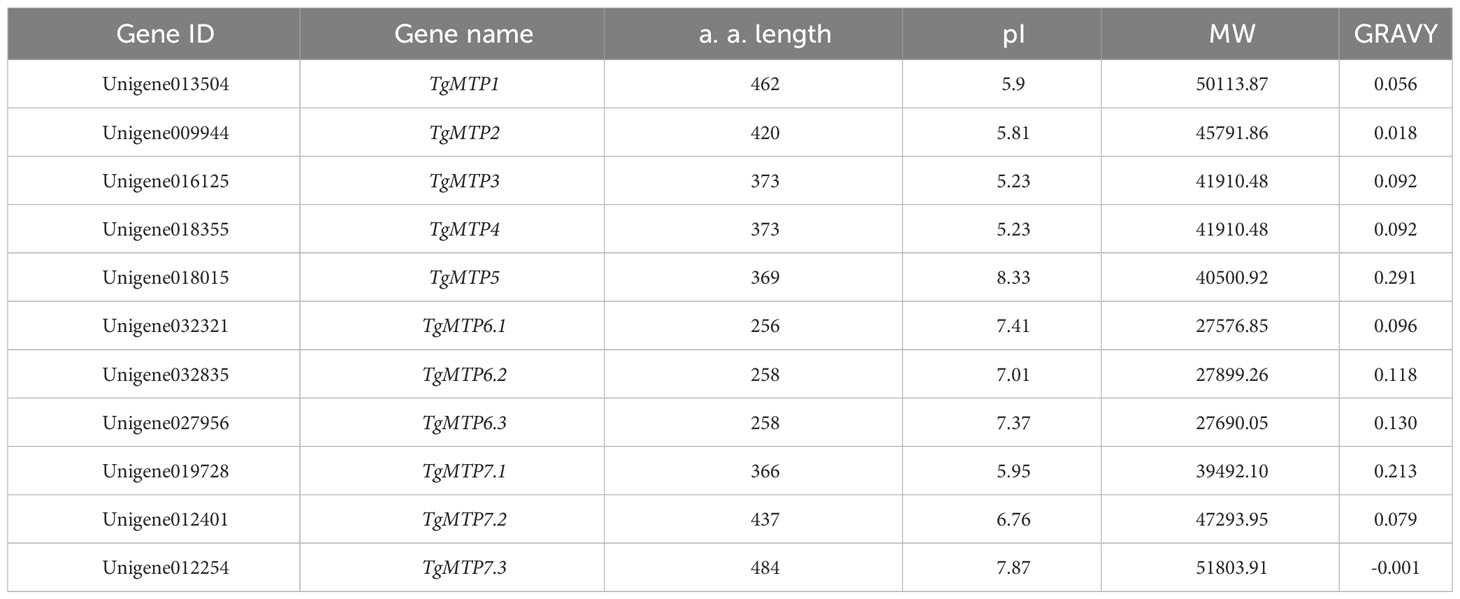
Based on homologous sequence alignment and conserved domain analysis, a total of 11 TgMTP gene family members were identified in this study, named as TgMTP1, TgMTP2, TgMTP3, TgMTP4, TgMTP5, TgMTP6.1, TgMTP6.2, TgMTP6.3, TgMTP7.1, TgMTP7.2 and TgMTP7.3 (Table 1). The amino acid lengths (a. a. length) of the 11 TgMTP proteins ranged from 256 aa (TgMTP6.1) to 484 aa (TgMTP7.3). The theoretical isoelectric points (pI) and molecular weights (MW) of TgMTP proteins were 5.23 (TgMTP3 and TgMTP4) - 8.33 (TgMTP5) and 27576.85 Da (TgMTP6.1) - 51803.91 Da (TgMTP7.3), respectively. The grand average of hydropathicity (GRAVY) values distributed from -0.001 (TgMTP7.3) to 0.291 (TgMTP5). Except for TgMTP7.3 with a GRAVY of -0.001, all other members had GRAVY values greater than 0, indicating that the TgMTP family proteins are mostly hydrophobic, with only one member TgMTP7.3 being hydrophilic.
3.2 Phylogenetic relationships of TgMTP protein
The phylogenetic relationships of TgMTPs with MTP members from soybean, tomato, rice and A. thaliana were displayed in the phylogenetic tree (Figure 1). The 63 MTP proteins were clustered into three major clades, designated as Mn-CDF, Fe/Zn-CDF and Zn-CDF subfamilies. The Mn-CDF clade contained TgMTP3 and TgMTP4. The Fe/Zn-CDF subfamily included 6 TgMTP members (TgMTP6.1-6.3, TgMTP7.1-7.3). TgMTP1, TgMTP2 and TgMTP5 belonged to the Zn-CDF group. Each clade also incorporated the homologous MTPs from the other plant species. The phylogenetic analysis revealed that the 11 TgMTPs were distributed among the 3 subfamilies together with respective orthologs. This suggests that the divergence of TgMTPs into different subfamilies preceded the speciation of tulip, soybean, tomato, rice and A. thaliana. The results provide insights into the evolutionary relationships and putative functional specificities of TgMTP transporters based on their classification into Mn-CDF, Fe/Zn-CDF and Zn-CDF subfamilies.
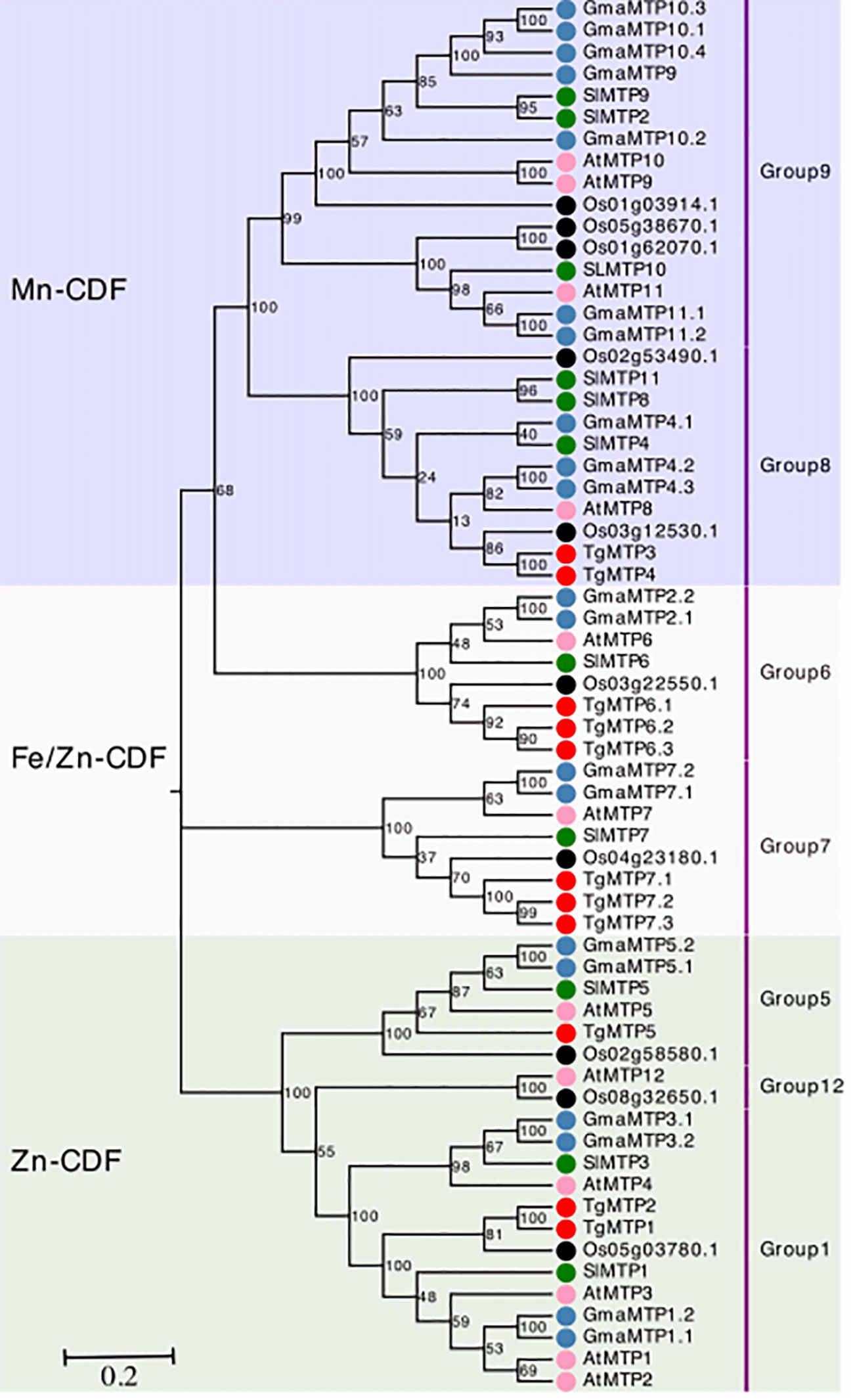
Figure 1 Phylogenetic relationships of TgMTPs with soybean (Gm), tomato (Sl), rice (Os) and A. thaliana (At) MTPs. The three clades were marked by different color. The MTP proteins are represented by solid circles with different colors denoting different plant species. Each group of MTP was annotated on right by text.
3.3 Structure and subcellular localization of MTP proteins
The predicted secondary structure composition of the 11 TgMTP proteins is summarized in Figure 2A. Four types of secondary structure elements were analyzed, including alpha helix, beta turn, random coil and extended strand. Among the TgMTPs, TgMTP3 and TgMTP4 contained the highest percentages of alpha helices at 63.27%, while TgMTP5 was enriched in extended strands at 12.47%. TgMTP7.1 exhibited the lowest content of random coils at 21.04%, whereas TgMTP7.3 showed the highest random coil percentage of 31.20%. The remaining TgMTPs had moderate compositions of the four secondary structures. In general, alpha helix appeared to be the predominant element in most TgMTPs, accounting for 38.96%-63.27% of their sequences. However, the proportions of alpha helices, beta turns, random coils and extended strands varied among different TgMTP members. These compositional divergences in secondary structures may contribute to functional specificities of the TgMTP transporters.
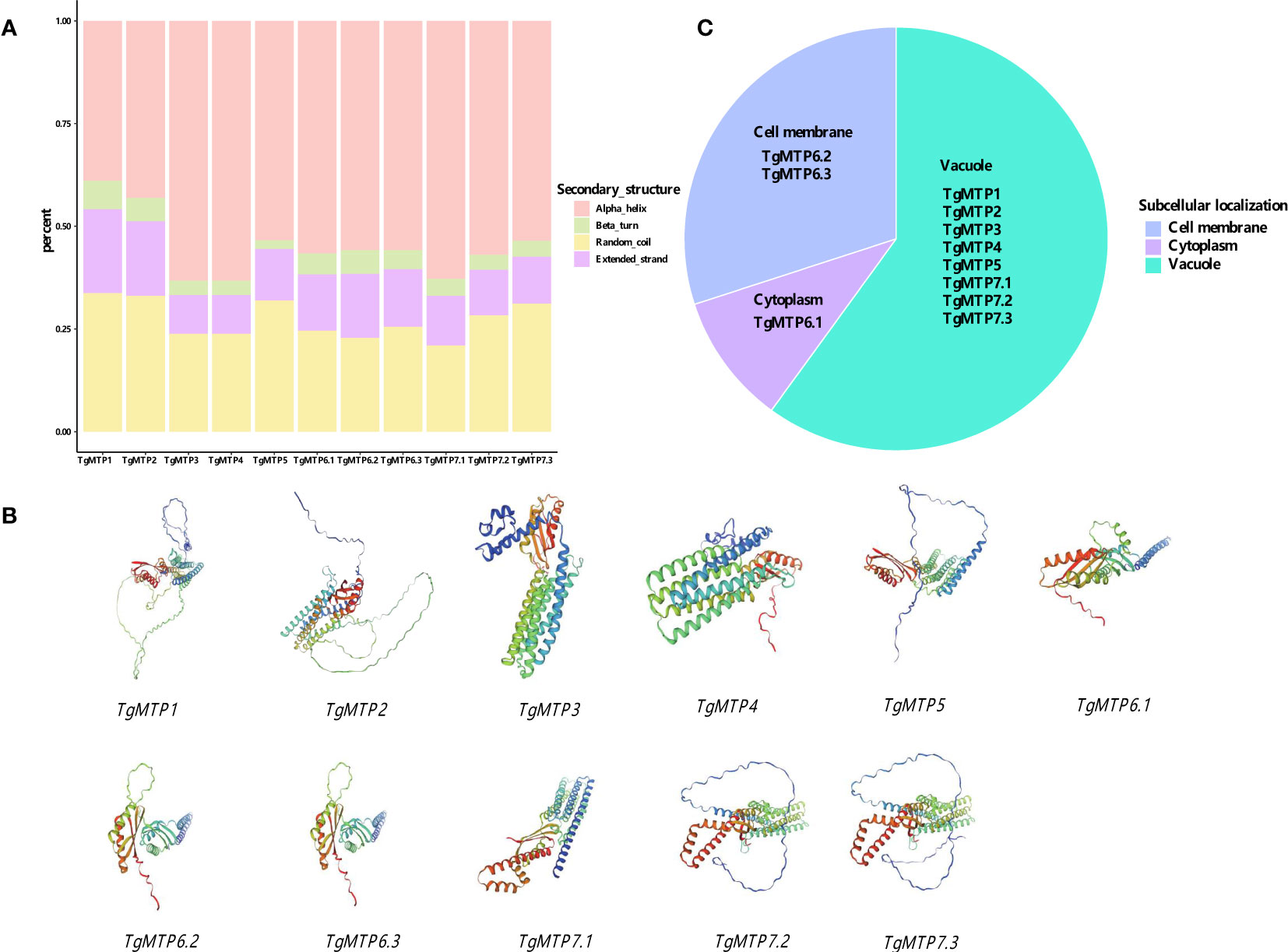
Figure 2 Structure and subcellular localization of MTP proteins. (A) The secondary structure composition of the 11 TgMTP proteins. (B) The predicted tertiary structures of the 11 TgMTP proteins. (C) The subcellular localizations of the 11 TgMTP proteins.
The predicted tertiary structures of the 11 TgMTP proteins are presented in Figure 2B. Homology modeling was performed using SWISS-MODEL with the best-match templates. The tertiary structures of TgMTPs displayed the typical architecture of MTP/CDF transporters, consisting of 6 transmembrane helices (TM1-6) and connecting loops.
The subcellular localizations of the 11 TgMTP proteins were predicted using Cell-PLoc-2 (Figure 2C). The results showed TgMTP1, TgMTP2, TgMTP3, TgMTP4, TgMTP5, TgMTP7.1, TgMTP7.2 and TgMTP7.3 were localized to the vacuolar membrane. TgMTP6.2 and TgMTP6.3 were predicted to target the plasma membrane. Whereas TgMTP6.1 was identified as a cytoplasmic protein.
3.3 Conserved motif, domains and CDS of TgMTP genes
The phylogenetic relationship of TgMTP genes were shown in Figure 3A, the phylogenetic tree illustrates a high correspondence between evolutionary relationships and conserved motif as well as domain architectures. To better understand the diversity and similarity of TgMTP genes, conserved motifs in TgMTP proteins were analyzed using the MEME software (Figure 3B). The results revealed 10 conserved motifs. TgMTP5 contained the fewest motifs with only motif 4 among all TgMTP family members. Motif 1 was only present in TgMTP members belonging to the Mn-CDF subfamily and TgMTP6.1, 6.2, 6.3 of the Fe/Zn-CDF subfamily. Motif 2 existed in all TgMTPs of the Fe/Zn-CDF and Zn-CDF subfamilies but not in the Mn-CDF subfamily. Motif 3 was specific to the Fe/Zn-CDF subfamily members. Motif 7 was only found in TgMTP6.1, 6.2 and 6.3 of the Fe/Zn-CDF subfamily. Motif 4 and 6 were absent only in TgMTP6.1, 6.2 and 6.3. Motif 5 was unique to TgMTP7.1, 7.2 and 7.3 of the Fe/Zn-CDF subfamily. Motifs 9 and 10 were present in all TgMTP members except the Mn-CDF subfamily. The sequences of all motifs are shown in Supplementary Figure 1.
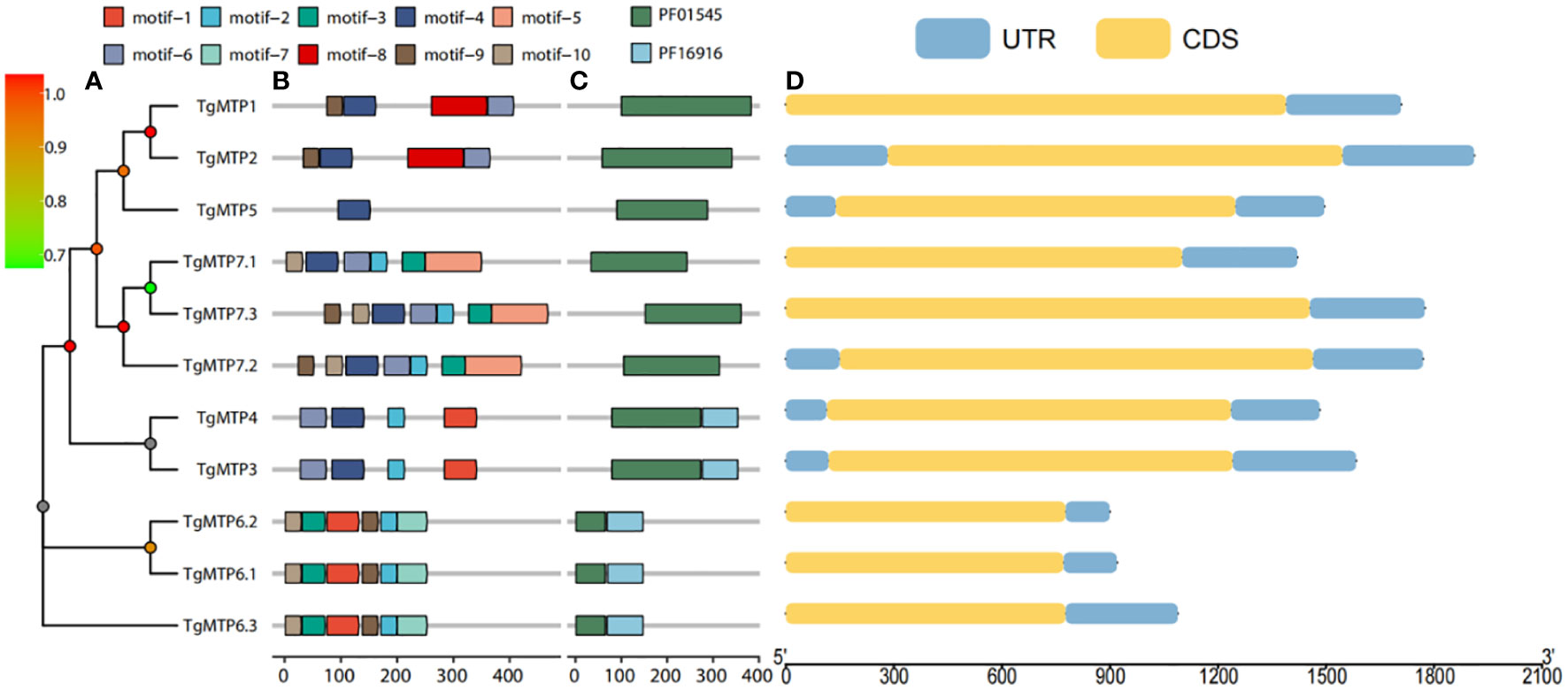
Figure 3 Phylogenetic tree (A), conserved motifs (B) and conserved domains (C) of TgMTP proteins, and CDS (D) of TgMTP genes.
Figure 3C displays two major protein domains (PF16916 and PF01545) in the TgMTP family. PF16916 only existed in the Mn-CDF clade and TgMTP6.1-6.3, while PF01545 distributed across all members.
Gene structure analysis (Figure 3D) showed that TgMTP members of the same clade had higher consistency in CDS length, such as TgMTP6.1, TgMTP6.2 and TgMTP6.3. Moreover, as the identification was based on full-length transcriptome, all members displayed only one CDS region and UTR region, without intron region.
3.4 Ka/Ks analysis of TgMTP genes
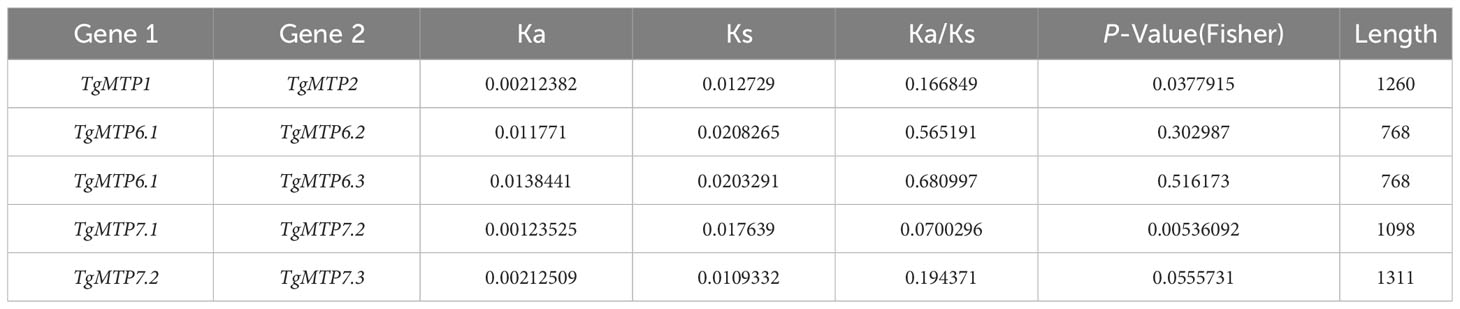
The Ka/Ks analysis revealed varied selection pressures among TgMTP gene pairs (Table 2). The Ka/Ks ratio of TgMTP1 and TgMTP2 was 0.166849, significantly lower than 1, indicating purifying selection between these two members. TgMTP6.1 and TgMTP6.2 had a Ka/Ks ratio of 0.565191, while TgMTP6.1 and TgMTP6.3 showed a Ka/Ks ratio of 0.680997, slightly higher than 0.5, suggesting relatively weak positive selection. Notably, an extremely low Ka/Ks value of 0.0700296 was observed between TgMTP7.1 and TgMTP7.2, evidencing robust purifying selection. TgMTP7.2 and TgMTP7.3 also exhibited a Ka/Ks ratio of 0.194371, lower than 1, implying purifying selection as well. Collectively, most TgMTP gene pairs were under various extents of purifying selection, reflecting functional differentiation and constraints among family members. However, a relaxation of selective pressure was observed between TgMTP6.1/6.2 and TgMTP6.1/6.3. Overall, this analysis provided insights into the molecular evolution patterns and selection forces shaping diversity and conservation within the TgMTP gene family.
3.5 GO annotation of TgMTP genes
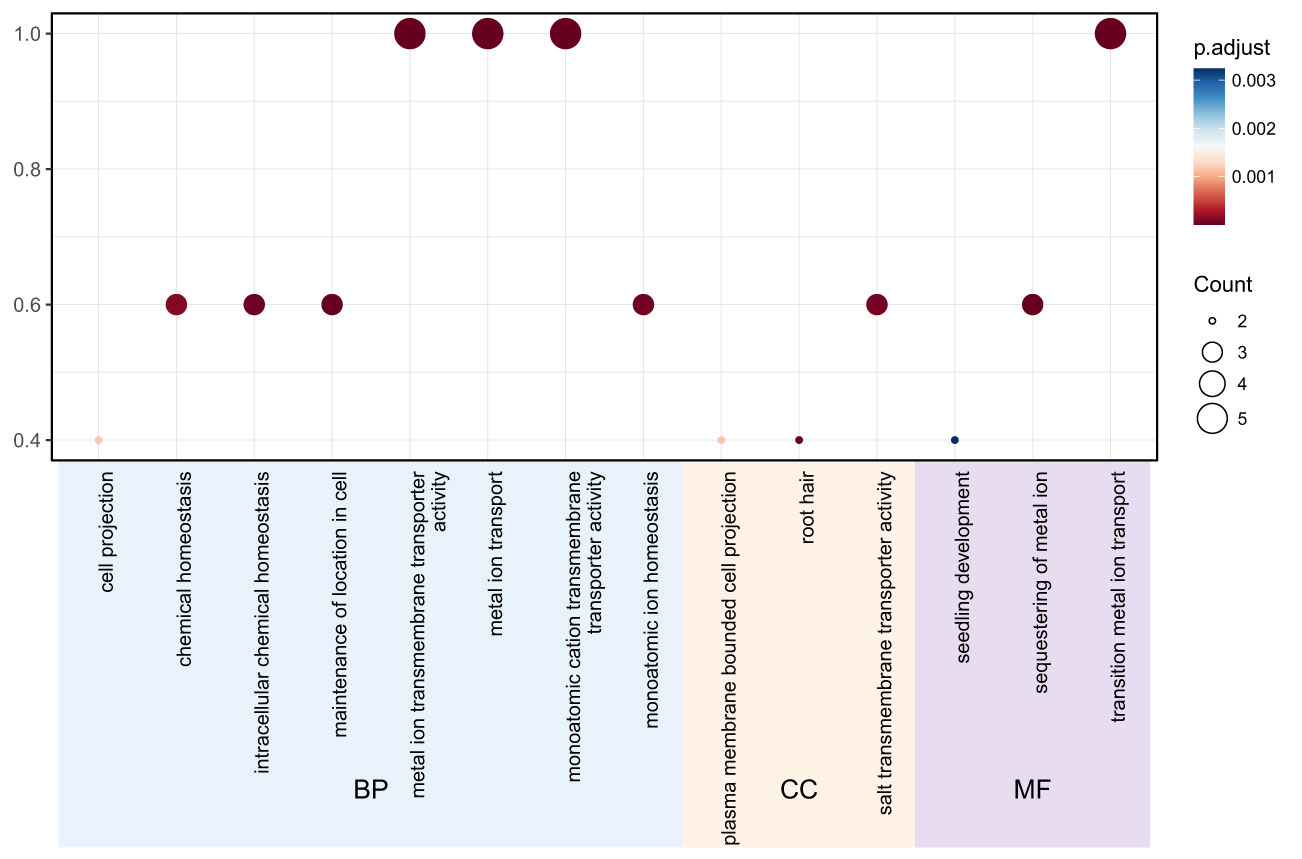
Gene ontology (GO) analysis was performed to predict the functional roles of the 11 TgMTP members. As shown in Figure 4, the significantly enriched GO terms could be categorized into biological process (BP), cellular component (CC) and molecular function (MF). For BP, the major terms included metal ion transport, metal ion homeostasis, transition metal ion transport and sequestering of metal ion. Other notable terms were intracellular chemical homeostasis, chemical homeostasis and seedling development. In the CC category, key enriched terms were plasma membrane bounded cell projection and root hair. The predominant MF terms were metal ion transmembrane transporter activity, monoatomic cation transmembrane transporter activity and monoatomic ion homeostasis. These results indicate that the TgMTP family is associated with metal transport, homeostasis and sequestration, consistent with their predicted roles as metal cation transporters. The GO terms related to root and plasma membrane localization further suggest their functional importance in roots for metal acquisition and compartmentalization. This GO analysis provides an overview of putative biological roles of the TgMTP transporters and will inform further investigations into their metal transport capabilities and physiological functions in tulip.
3.6 miRNA regulation network for TgMTP genes
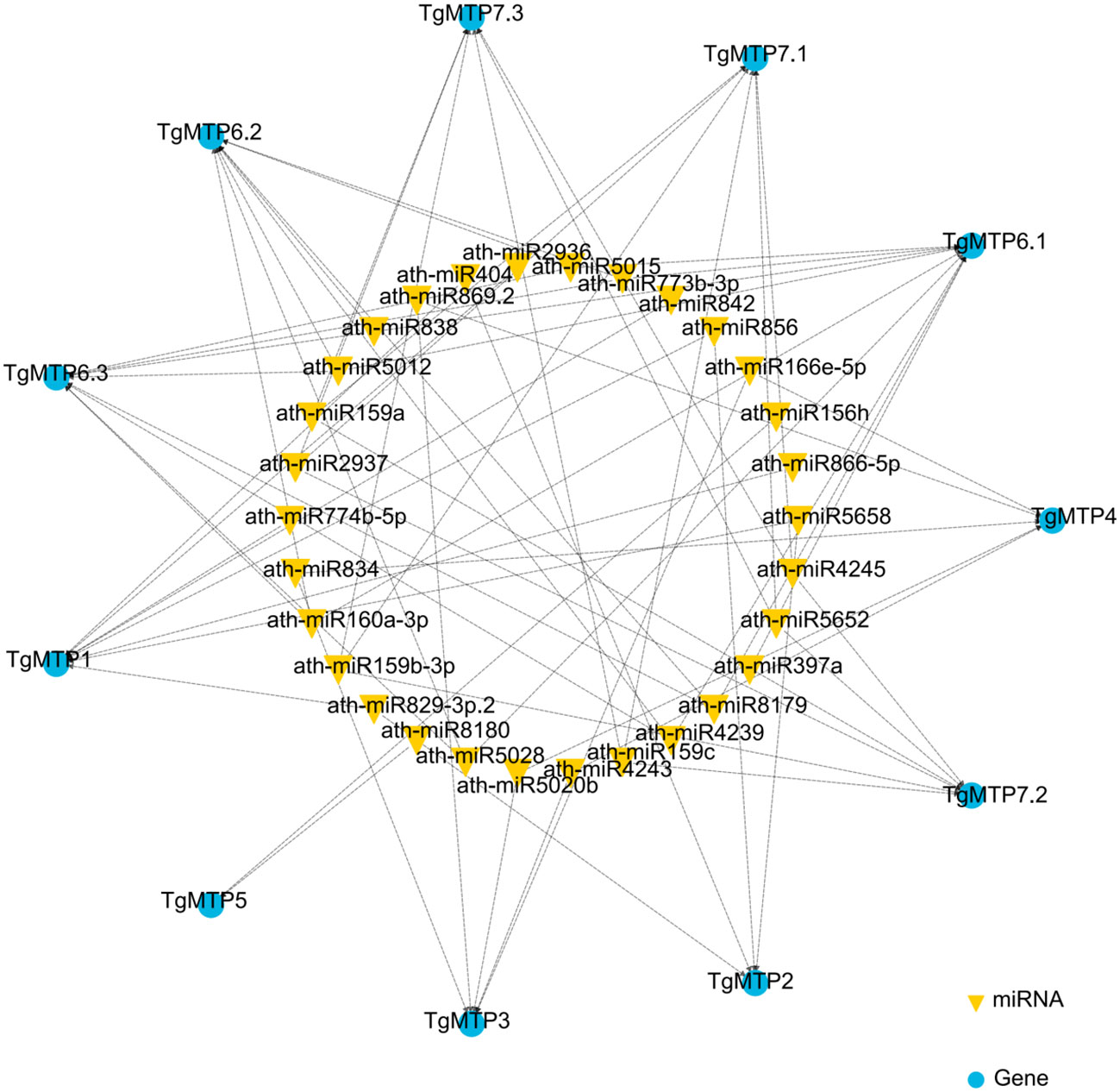
We predicted the potential miRNA binding sites of TgMTPs using psRNATarget (Dai and Zhao, 2011) In total, 30 miRNAs were identified that may regulate TgMTPs (Figure 5; Supplementary Table 1). Most miRNAs had multiple TgMTP targets, such as miR159a, which could target 3 TgMTP genes (Figure 5 and Supplementary Table 1). In contrast, some TgMTPs could be targeted by several miRNAs. For instance, TgMTP7.1 could be targeted by 6 miRNAs, including ath-miR159a, ath-miR159b-3p and ath-miR159c (Figure 5; Supplementary Table 1). The relationships between TgMTPs and miRNAs warrant further investigation.
3.7 qRT-PCR analysis of TgMTP genes under metal treatment
The relative expression profiles of 11 TgMTP genes in response to Fe2+, Mn2+, Cd2+ and Zn2+ treatments are shown in Figure 6. The transcript levels were examined by qRT-PCR at 0, 1, 3, 5 and 7 days after metal exposure. Significant variations in expression were observed among TgMTP genes by metal treatments over time. Overall, most TgMTPs were markedly induced by four metal (except TgMTP4 under Cd treatment), especially at later time points for most TgMTP genes. For instance, TgMTP7.1 displayed over 30-fold upregulation after Cd treatment for 1, 3, 5 and 7 days compared to 0 day. Substantial stimulation of transcript levels was also evident for TgMTP3, TgMTP6.1 and TgMTP6.3 following Cd exposure. For Fe2+ treatment, 72.7% (8/11) of the TgMTP genes exhibited over 10-fold upregulation in expression. Similarly, greater than 10-fold increased expression was observed for 54.5% (6/11) and 81.8% (9/11) of the genes under Zn2+ and Mn2+ exposure, respectively. These results demonstrate that most TgMTPs are responsive to heavy metal stresses. However, the percentage of TgMTP genes that could be markedly induced (with relative expression elevated by over 10-fold) by Cd2+ treatment was 45.4% (5/11). The data reveals the potential roles of these TgMTP genes in metal tolerance and accumulation in tulip. Further functional analyses of the responsive transporters such as TgMTP7.1 may provide insights into heavy metal detoxification mechanisms in this species.
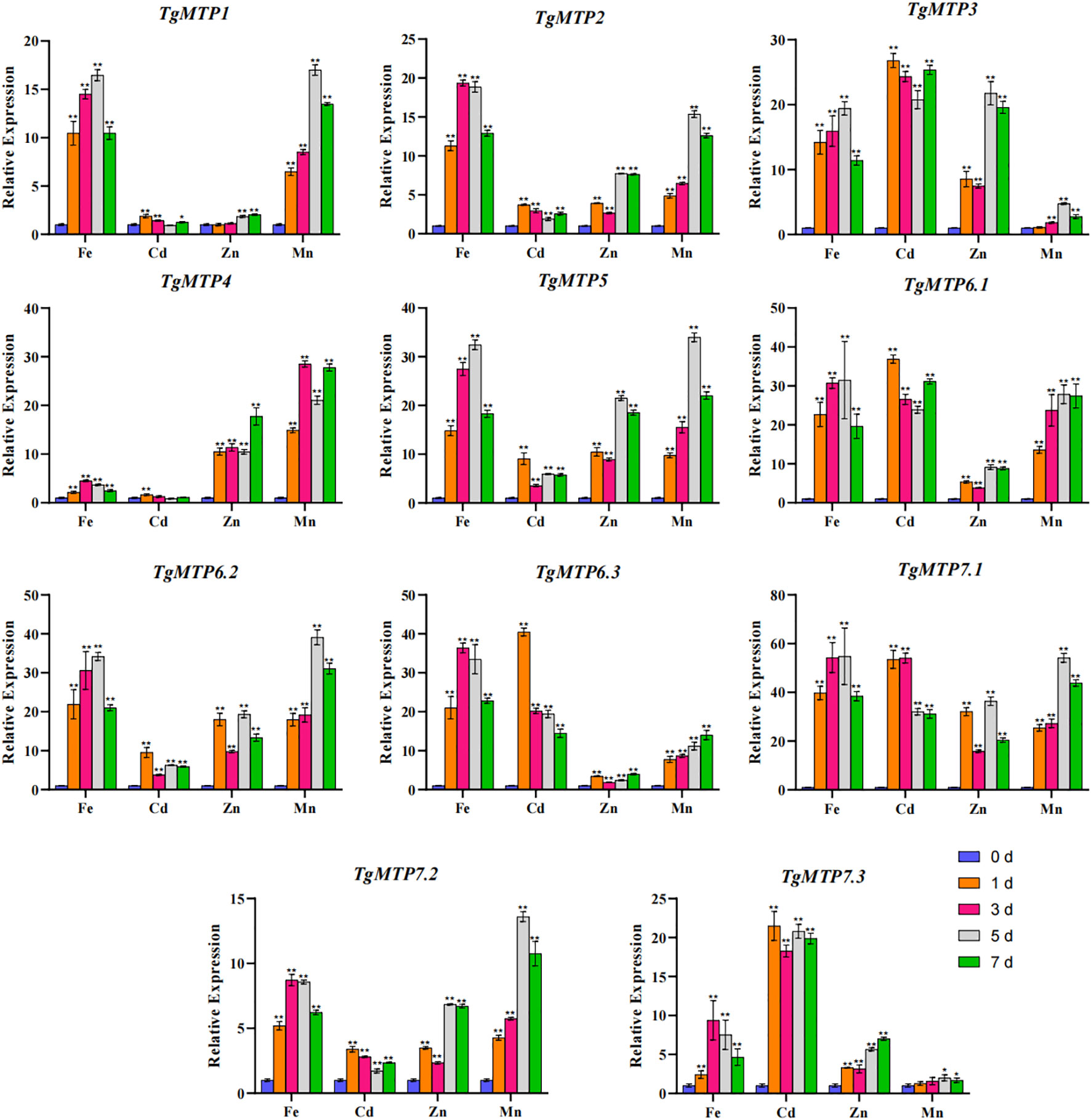
Figure 6 Relative expression level of TgMTP genes under different metal treatment. Two-way ANOVA analysis was used to compare the expression level.Data represent means ± SD, n = 3. * means p<0.05, ** means p<0.001.
3.8 Heterologous expression of TgMTP7.1
The TgMTP7.1 gene was selected for functional validation as it exhibited the most pronounced upregulation upon Cd treatment when compared with other TgMTP genes. The TgMTP7.1 was heterologous expressed in the parental strain BY4741 and a yeast mutants that are highly sensitive to Cd (ycf1Δ), respectively. As shown in Figure 7, yeast cells transformed with empty vector pYES2 displayed severely inhibited growth on the medium containing 5 mM CdCl2, whether in plates (Figure 7A) or liquid medium (Figure 7B). In contrast, ycf1Δ expressing pYES2-TgMTP7.1 exhibited significantly enhanced tolerance to Cd toxicity. At the 102 dilution factor, the survival rate of the pYES2 transformants (ycf1Δ+pYES2) was 0, and at the 103 dilution factor, the survival rate of the pYES2 transformants (BY4741+pYES2) was 0. Whereas yeast cells with TgMTP7.1 (ycf1Δ+pYES2-TgMTP7.1) grew readily at dilutions of 101 and 102, even the dilutions of 103, the ycf1Δ+pYES2-TgMTP7.1 transformants showed a survival. These results demonstrate that TgMTP7.1 expression could effectively rescue the Cd sensitivity of yeast cells. The increased Cd tolerance conferred by TgMTP7.1 heterologous expression indicates it possesses Cd transport activity and likely functions in vacuolar sequestration of Cd in plants.
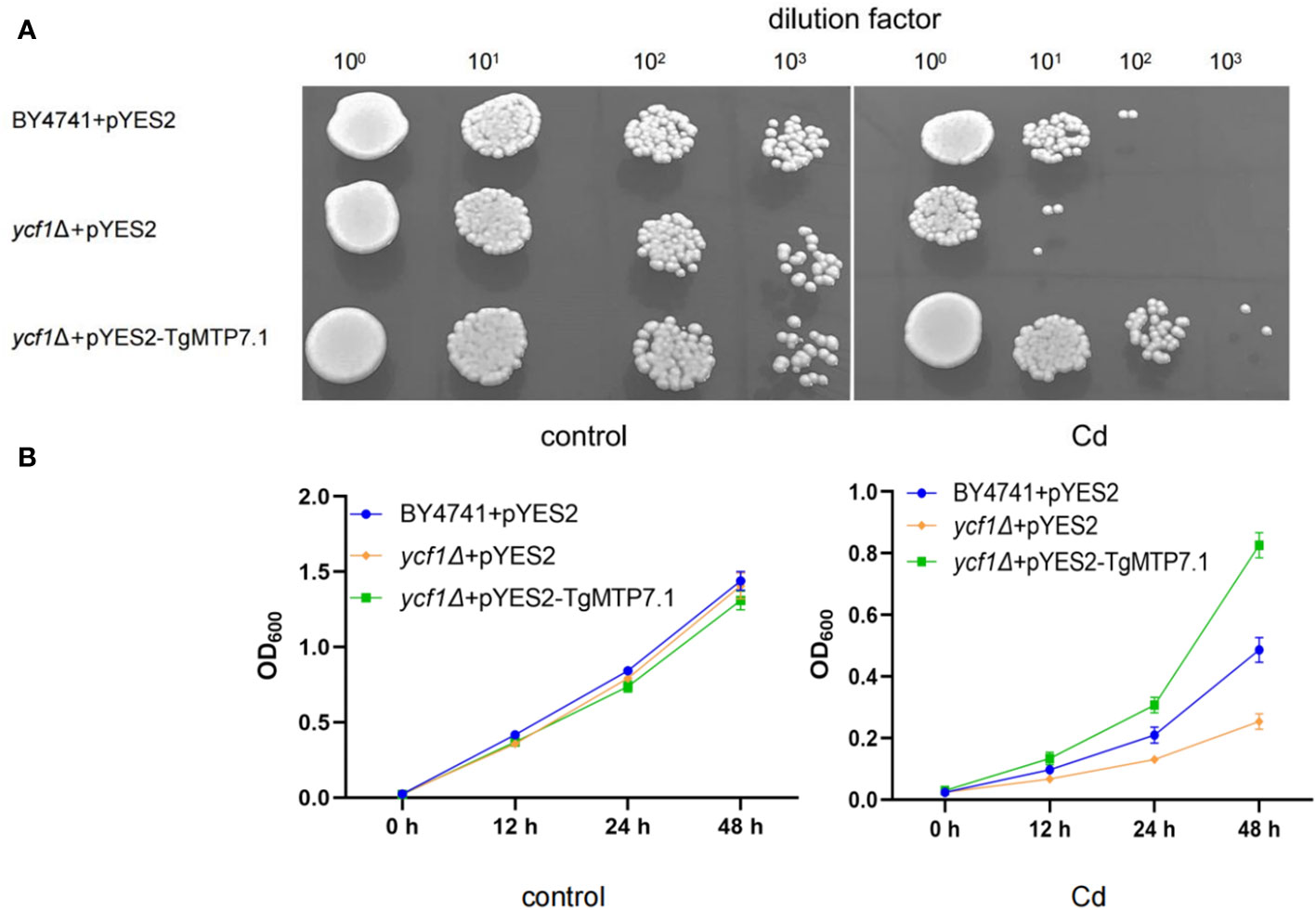
Figure 7 Sensitivity to Cd in yeast. (A) Complementation of yeast mutants on solid medium containing heavy metals. Yeast wild-type strain BY4741 was transformed with the empty vector pYES2, Cadmium-sensitive yeast mutants were transformed with either the same empty vector pYES2 or the pYES2 vector harboring the TgMTP7.1 expression cassette. Transformants were cultured to mid-logarithmic phase at OD600 of 0.2 before preparation of 10-fold serial dilutions spanning four orders of magnitude. Subsequently, 2 μL of each diluted sample was pipetted onto medium plates supplemented with 5 mM CdCl2 to assess growth phenotypes. (B) Sensitivity to Cd when grown in liquid medium contain CdCl2, error bar = mean ± SD, n=3.
4 Discussion
Mn, Zn, and Fe are trace elements critical for proper growth, development, function, and sustainment of plants. Notwithstanding their significance in many biochemical processes, superfluous quantities of these elements can be toxic to plants, eliciting decreases in agricultural yield and quality (Ghori et al., 2019). Meanwhile, other non-essential elements, including cadmium (Cd) can likewise be absorbed and elicit toxicity even at very low concentrations (Clemens, 2001). Cd exposure elicits toxicity symptoms in plants such as growth inhibition, declines in photosynthesis and pigmentation, cellular damage from oxidative stress, and antagonistic effects on the uptake of essential nutrients (Mishra et al., 2006; Mobin and Khan, 2007; Wang et al., 2009).
Concomitant with the progression of industrialization, heavy metals (particularly Cd) are amassing abundantly in the environment. These elements ingress plants and other organisms through the food chain, eliciting toxicity symptoms even at diminished concentrations. This phenomenon poses a substantial hazard to ecosystems and may also imperil human wellbeing (El-Sappah et al., 2012; Elrys et al., 2018). Ornamental plants, not intended for human consumption, demonstrate a robust potential for the remediation of heavy metal-contaminated soils, given their ability to thrive in such environments (Khan et al., 2021). The MTP gene, which functions in the transport of metal ions within plants, plays a crucial role in the resistance of plants to heavy metals (Ricachenevsky et al., 2013). However, research on the MTP gene family in tulips, as one of the significant ornamental plants, has not yet been initiated.
In this study, we performed a comprehensive genome-wide identification and characterization of the MTP gene family in tulip. A total of 11 TgMTP members were identified based on conserved MTP/CDF domains and phylogenetic analysis. The nomenclature and classification of TgMTPs were assigned according to sequence homology with A. thaliana and rice MTPs (Mäser et al, 2001). As the first investigation of MTPs in tulip, we comprehensively studied the genomic organization, evolutionary relationships, structural features and expression patterns of TgMTP.
Phylogenetic analysis clustered the 11 TgMTPs into three distinct subfamilies (Table 1), together with MTP orthologs from soybean, tomato, rice and A. thaliana (Figure 1) (Montanini et al., 2007; Gustin et al., 2011; Migocka, 2015). This suggests that the divergence of plant MTPs into different subfamilies preceded the speciation of monocot and dicot plants. In tomatoes, 11 MTP proteins are identified, six of which may play a significant role in the response to heavy metal stress (El-Sappah et al., 2021). Similarly, this study has also identified five TgMTP genes that exhibit a strong response to Cd (Figure 6). The phylogenetic classifications of TgMTPs provide a foundation to investigate their functional specificities.
Conserved motif analysis revealed shared motifs between TgMTPs of the same subfamily, implying similar structure-function relationships. Distinct motifs among subfamilies may contribute to functional divergences. The predominant α-helical transmembrane topology is consistent with their predicted roles as membrane transporters (El-Sappah et al., 2021). Differences in secondary structure composition and length of hydrophilic loops connecting the TMs could impact substrate access, transport kinetics and metal selectivity (El-Sappah et al., 2021). The two duplicated pseudosymmetrical domains (TMs 1-3 and TMs 2-4-5-6) form the translocation pathway and mediate alternating access of metals. Further mutational and structural analyses are required to elucidate the structure-function relationships of TgMTPs.
Varied subcellular localizations of TgMTPs were predicted, including vacuolar membrane, plasma membrane and cytoplasm (Figure 2). Similar localizations have been confirmed for MTPs in A. thaliana, soybean and rice (Delhaize et al., 2003; Yuan et al., 2012; Zhang et al., 2020). The vacuolar TgMTPs likely sequester toxic metals like Cd into vacuoles. Plasma membrane TgMTPs may regulate metal uptake and efflux. Whereas cytoplasmic members could be involved in intracellular metal transfer or storage. Determining the precise localizations and tissue expression patterns of TgMTPs will clarify their physiological roles in tulip. The construction of miRNA-gene regulatory networks may also engender avenues for investigating the mechanisms of action of TgMTPs, particularly miRNA159 (targets TgMTP7.1) which has been implicated in abiotic stress tolerance in sugarcane (Patade and Suprasanna, 2010) and rice (Mohsenifard et al., 2017).
Most MTP genes exhibited metal-responsive expression changes, especially induction by Cd (Sasaki et al., 2014). The considerable stimulation of predominant TgMTPs like TgMTP7.1 under Cd implies their importance for metal tolerance and sequestration in tulip (Figure 6). Further functional analyses of these metal and responsive transporters will uncover their contribution to low Cd traits in tulip.
Notably, heterologous expression of TgMTP7.1 could rescue the Cd sensitivity of yeast cells. Enhanced Cd tolerance indicates TgMTP7.1 possesses Cd transport activity in plants, likely for vacuolar sequestration (Delhaize et al., 2003). Characterization of TgMTP7.1 and other responsive TgMTPs will help elucidate the mechanistic basis of heavy metal homeostasis and distribution in tulip. Engineering key TgMTPs may generate tulip cultivars with reduced heavy metal accumulation or heavy metal tolerance to address and mitigate soil cadmium contamination.
Tulip is an economically important ornamental geophyte cultivated worldwide, with bulbs prone to accumulating toxic heavy metals that pose potential toxicity risks (Byczyńska et al., 2019). In wheat, by genetic diversity could be exploited for developing low-metal wheat cultivars through molecular breeding approaches (Zaid et al., 2018). Our study represents the first genome-wide analysis of MTP transporters that underlie heavy metal accumulation in tulip. The results lay an important foundation for elucidating the transporters and molecular mechanisms governing metal distribution and detoxification in this species.
Notably, several Cd-responsive TgMTPs were identified as high priority candidates for functional analyses. Due to the substantial threat that Cd poses to human health (Fowler, 2009). In furture, overexpression or knockdown/knockout of these key transporters in tulip would help establish their importance for vacuolar sequestration and shoot translocation of Cd. Comparing Cd contents and oxidative damage between transgenic and wild-type tulip plants under Cd exposure could reveal impacts on Cd tolerance and accumulation phenotypes. Furthermore, introducing mutations into specific TgMTPs by genome editing may help determine structural domains or residues critical for substrate recognition and transport kinetics. This will play a crucial role in elucidating the mechanisms by which MTP proteins contribute to Cd tolerance or transport in tulips. Concurrently, it offers significant insights for the cultivation of tulip varieties with superior heavy metal remediation capabilities.
In addition to genetic manipulation in tulip itself, heterologous expression also provides a rapid screening method for identifying MTP genes with heavy metal tolerance or transport capabilities. Cd-sensitive yeast could be complemented by key TgMTPs to assess their ability to restore Cd tolerance. Expressing TgMTPs in A. thaliana metal hyperaccumulator or hypertolerance mutants would test whether they can alter natural variation in Cd accumulation or sensitivity. Analyzing fluctuations in root/shoot metal contents and mapping TgMTP expression patterns in the heterologous hosts could provide deeper insights into transporter roles in metal distribution between tissues and subcellular compartments (Verbruggen et al., 2009).
In summary, this comprehensive study revealed important molecular and functional characterisics of the TgMTP gene family as putative key determinants of heavy metal accumulation in tulip. Our results lay a foundation for further elucidating TgMTP transport capabilities, expression regulation and physiological roles. Genetic manipulation in heterologous expression systems present promising approaches to establish structure-function relations and contributions of specific TgMTPs to Cd homeostasis. Key TgMTPs identified also present potential targets for engineering or breeding low Cd cultivars to reduce metal toxicity risks and enable sustainable production. Further deciphering TgMTP-mediated metal transport and detoxification mechanisms will provide deeper insights into heavy metal accumulation traits in geophytes.
5 Conclusions
In this study, leveraging Iso-seq data, 11 MTP genes were identified from tulips and categorized into three subfamilies based on phylogenetic relationships. The MTP proteins within the same subfamily exhibit structural similarities, yet they show differential relative expression in response to metal stress. The identification of the key Cd-responsive gene TgMTP7.1, along with its heterologous expression in yeast, preliminarily elucidates the function of TgMTP7.1. This provides vital genetic material for in-depth future research into the functionality of MTP genes. Moreover, this study offers valuable insights into using ornamental plants, specifically tulips, for the phytoremediation of heavy metal-contaminated soils.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Author contributions
JL: Data curation, Writing – original draft. GX: Software, Writing – original draft. YZ: Data curation, Writing – original draft. HZ: Software, Writing – original draft. TW: Data curation, Writing – original draft. ZT: Conceptualization, Writing – review & editing. LQ: Conceptualization, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This research was funded by the Program of China Agriculture Research System(CARS-23-G57), Subject Construction Program of Liaoning Academy of Agricultural Sciences (2022DD051608) and Presidential Foundation of Liaoning Academy of Agricultural Sciences (2023QN2403).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2024.1346255/full#supplementary-material
References
Alexander, P., Alloway, B., Dourado, A. (2006). Genotypic variations in the accumulation of Cd, Cu, Pb and Zn exhibited by six commonly grown vegetables. Environ. pollut. 144, 736–745. doi: 10.1016/j.envpol.2006.03.001.
Bailey, T. L., Williams, N., Misleh, C., Li, W. W. (2006). MEME: discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 34, W369–W373. doi: 10.1093/nar/gkl198.
Byczyńska, A., Zawadzińska, A., Salachna, P. (2019). Silver nanoparticles preplant bulb soaking affects tulip production. Acta Agriculturae Scandinavica Section B—Soil Plant Sci. 69, 250–256. doi: 10.1080/09064710.2018.1545863
Chen, Z., Fujii, Y., Yamaji, N., Masuda, S., Takemoto, Y., Kamiya, T., et al. (2013). Mn tolerance in rice is mediated by MTP8. 1, a member of the cation diffusion facilitator family. J. Exp. Bot. 64, 4375–4387. doi: 10.1093/jxb/ert243.
Clemens, S. (2001). Molecular mechanisms of plant metal tolerance and homeostasis. Planta 212, 475–486. doi: 10.1007/s004250000458.
Dai, X., Zhao, P. X. (2011). psRNATarget: a plant small RNA target analysis server. Nucleic Acids Res. 39, W155–W159. doi: 10.1093/nar/gkr319.
Delhaize, E., Kataoka, T., Hebb, D. M., White, R. G., Ryan, P. R. (2003). Genes encoding proteins of the cation diffusion facilitator family that confer manganese tolerance. Plant Cell 15, 1131–1142. doi: 10.1105/tpc.009134.
Djebali, W., Zarrouk, M., Brouquisse, R., El Kahoui, S., Limam, F., Ghorbel, M., et al. (2005). Ultrastructure and lipid alterations induced by cadmium in tomato (Lycopersicon esculentum) chloroplast membranes. Plant Biol. 7, 358–368. doi: 10.1055/s-2005-837696.
Dodangeh, H., Rahimi, G., Fallah, M., Ebrahimi, E. (2018). Investigation of heavy metal uptake by three types of ornamental plants as affected by application of organic and chemical fertilizers in contaminated soils. Environ. Earth Sci. 77, 1–15. doi: 10.1007/s12665-018-7620-2.
Elrys, A. S., Merwad, A.-R. M., Abdo, A. I., Abdel-Fatah, M. K., Desoky, E.-S. M. (2018). Does the application of silicon and Moringa seed extract reduce heavy metals toxicity in potato tubers treated with phosphate fertilizers? Environ. Sci. pollut. Res. 25, 16776–16787. doi: 10.1007/s11356-018-1823-7.
El-Sappah, A. H., Abbas, M., Rather, S. A., Wani, S. H., Soaud, N., Noor, Z., et al. (2023). Genome-wide identification and expression analysis of metal tolerance protein (MTP) gene family in soybean (Glycine max) under heavy metal stress. Mol. Biol. Rep. 50, 2975–2990. doi: 10.1007/s11033-022-08100-x.
El-Sappah, A. H., Elrys, A. S., Desoky, E.-S. M., Zhao, X., Bingwen, W., El-Sappah, H. H., et al. (2021). Comprehensive genome wide identification and expression analysis of MTP gene family in tomato (Solanum lycopersicum) under multiple heavy metal stress. Saudi J. Biol. Sci. 28, 6946–6956. doi: 10.1016/j.sjbs.2021.07.073.
El-Sappah, A., Shawky, A., Sayed-Ahmad, M., Youssef, M. (2012). Nile tilapia as bio indicator to estimate the contamination of water using SDS-PAGE and RAPDPCR techniques. Egypt J. Genet. Cytol 41, 209–227. doi: 10.21608/ejgc.2012.10536.
Fowler, B. A. (2009). Monitoring of human populations for early markers of cadmium toxicity: a review. Toxicol. Appl. Pharmacol. 238, 294–300. doi: 10.1016/j.taap.2009.05.004.
Gao, Y., Yang, F., Liu, J., Xie, W., Zhang, L., Chen, Z., et al. (2020). Genome-wide identification of metal tolerance protein genes in Populus trichocarpa and their roles in response to various heavy metal stresses. Int. J. Mol. Sci. 21, 1680. doi: 10.3390/ijms21051680.
Ghori, N.-H., Ghori, T., Hayat, M., Imadi, S., Gul, A., Altay, V., et al. (2019). Heavy metal stress and responses in plants. Int. J. Environ. Sci. Technol. 16, 1807–1828. doi: 10.1007/s13762-019-02215-8.
Gustin, J. L., Zanis, M. J., Salt, D. E. (2011). Structure and evolution of the plant cation diffusion facilitator family of ion transporters. BMC evolutionary Biol. 11, 1–13. doi: 10.1186/1471-2148-11-76.
Hall, J.Á., Williams, L. E. (2003). Transition metal transporters in plants. J. Exp. Bot. 54, 2601–2613. doi: 10.1093/jxb/erg303.
Haney, C. J., Grass, G., Franke, S., Rensing, C. (2005). New developments in the understanding of the cation diffusion facilitator family. J. Ind. Microbiol. Biotechnol. 32, 215–226. doi: 10.1007/s10295-005-0224-3.
Hassinen, V. H. (2009). Search for metal-responsive genes in plants (Kuopio: University of Kuopio, Department of Environmental Science).
Johnson, L. S., Eddy, S. R., Portugaly, E. (2010). Hidden Markov model speed heuristic and iterative HMM search procedure. BMC Bioinf. 11, 1–8. doi: 10.1186/1471-2105-11-431.
Kelley, B. P., Yuan, B., Lewitter, F., Sharan, R., Stockwell, B. R., Ideker, T. (2004). PathBLAST: a tool for alignment of protein interaction networks. Nucleic Acids Res. 32, W83–W88. doi: 10.1093/nar/gkh411.
Khan, A. H. A., Kiyani, A., Mirza, C. R., Butt, T. A., Barros, R., Ali, B., et al. (2021). Ornamental plants for the phytoremediation of heavy metals: Present knowledge and future perspectives. Environ. Res. 195, 110780. doi: 10.1016/j.envres.2021.110780.
Kumar, S., Stecher, G., Li, M., Knyaz, C., Tamura, K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547. doi: 10.1093/molbev/msy096.
Li, J., Dong, R., Jia, Y., Huang, J., Zou, X., An, N., et al. (2021). Characterization of metal tolerance proteins and functional analysis of GmMTP8. 1 involved in manganese tolerance in soybean. Front. Plant Sci. 12, 683813. doi: 10.3389/fpls.2021.683813
Liu, J., Gao, Y., Tang, Y., Wang, D., Chen, X., Yao, Y., et al. (2019). Genome-wide identification, comprehensive gene feature, evolution, and expression analysis of plant metal tolerance proteins in tobacco under heavy metal toxicity. Front. Genet. 10, 345. doi: 10.3389/fgene.2019.00345.
Liu, K. L., Shen, L., Wang, J. Q., Sheng, J. P. (2008). Rapid inactivation of chloroplastic ascorbate peroxidase is responsible for oxidative modification to Rubisco in tomato (Lycopersicon esculentum) under cadmium stress. J. Integr. Plant Biol. 50, 415–426. doi: 10.1111/j.1744-7909.2007.00621.x.
Mäser, P., Thomine, S., Schroeder, J. I., Ward, J. M., Hirschi, K., Sze, H., et al. (2001). Phylogenetic relationships within cation transporter families of Arabidopsis. Plant Physiol. 126, 1646–1667. doi: 10.1104/pp.126.4.1646.
Menguer, P. K., Farthing, E., Peaston, K. A., Ricachenevsky, F. K., Fett, J. P., Williams, L. E. (2013). Functional analysis of the rice vacuolar zinc transporter OsMTP1. J. Exp. Bot. 64, 2871–2883. doi: 10.1093/jxb/ert136.
Migocka, M. (2015). Copper-transporting ATPases: The evolutionarily conserved machineries for balancing copper in living systems. IUBMB Life 67, 737–745. doi: 10.1002/iub.1437.
Mishra, S., Srivastava, S., Tripathi, R., Govindarajan, R., Kuriakose, S., Prasad, M. (2006). Phytochelatin synthesis and response of antioxidants during cadmium stress in Bacopa monnieri L◊. Plant Physiol. Biochem. 44, 25–37. doi: 10.1016/j.plaphy.2006.01.007.
Mobin, M., Khan, N. A. (2007). Photosynthetic activity, pigment composition and antioxidative response of two mustard (Brassica juncea) cultivars differing in photosynthetic capacity subjected to cadmium stress. J. Plant Physiol. 164, 601–610. doi: 10.1016/j.jplph.2006.03.003.
Mohsenifard, E., Ghabooli, M., Mehri, N., Bakhshi, B. (2017). Regulation of miR159 and miR396 mediated by Piriformospora indica confer drought tolerance in rice. J. Plant Mol. Breed. 5, 10–18. doi: 10.22058/JPMB.2017.60864.1129
Montanini, B., Blaudez, D., Jeandroz, S., Sanders, D., Chalot, M. (2007). Phylogenetic and functional analysis of the Cation Diffusion Facilitator (CDF) family: improved signature and prediction of substrate specificity. BMC Genomics 8, 1–16. doi: 10.1186/1471-2164-8-107.
Osmani, M., Tuna, M., Elezaj, I. R. (2018). Concentration of some metals in soil and plant organs and their biochemical profiles in Tulipa luanica, T. kosovarica and T. albanica native plant species. Physiol. Mol. Biol. Plants 24, 1117–1126. doi: 10.1007/s12298-018-0539-y.
Patade, V. Y., Suprasanna, P. (2010). Short-term salt and PEG stresses regulate expression of MicroRNA, miR159 in sugarcane leaves. J. Crop Sci. Biotechnol. 13, 177–182. doi: 10.1007/s12892-010-0019-6.
Paulsen, I., Saier, J. (1997). MH. A novel family of ubiquitous heavy metal ion transport proteins. J. membrane Biol. 156, 99–103. doi: 10.1007/s002329900192.
Peng, F., Wang, C., Cheng, Y., Kang, H., Fan, X., Sha, L., et al. (2018). Cloning and characterization of tpNRAMP3, a metal transporter from polish wheat (Triticum polonicum L.). Front. Plant Sci. 20, 9:1354. doi: 10.3389/fpls.2018.01354
Ram, H., Kaur, A., Gandass, N., Singh, S., Deshmukh, R., Sonah, H., et al. (2019). Molecular characterization and expression dynamics of MTP genes under various spatio-temporal stages and metal stress conditions in rice. PloS One 14, e0217360. doi: 10.1371/journal.pone.0217360.
Ricachenevsky, F. K., Menguer, P. K., Sperotto, R. A., Williams, L. E., Fett, J. P. (2013). Roles of plant metal tolerance proteins (MTP) in metal storage and potential use in biofortification strategies. Front. Plant Sci. 4, 144. doi: 10.3389/fpls.2013.00144.
Sasaki, A., Yamaji, N., Ma, J. F. (2014). Overexpression of OsHMA3 enhances Cd tolerance and expression of Zn transporter genes in rice. J. Exp. Bot. 65, 6013–6021. doi: 10.1093/jxb/eru340.
Shirazi, Z., Abedi, A., Kordrostami, M., Burritt, D. J., Hossain, M. A. (2019). Genome-wide identification and characterization of the metal tolerance protein (MTP) family in grape (Vitis vinifera L.). 3 Biotech. 9, 1–17. doi: 10.1007/s13205-019-1728-2.
Shirazi, Z., Khakdan, F., Rafiei, F., Balalami, M. Y., Ranjbar, M. (2023). Genome-wide identification and expression profile analysis of metal tolerance protein gene family in Eucalyptus grandis under metal stresses. BMC Plant Biol. 23, 240. doi: 10.1186/s12870-023-04240-9.
Smoot, M. E., Ono, K., Ruscheinski, J., Wang, P.-L., Ideker, T. (2011). Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics 27, 431–432. doi: 10.1093/bioinformatics/btq675.
Thompson, J. D., Higgins, D. G., Gibson, T. J. (1994). CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680. doi: 10.1093/nar/22.22.4673.
Van der Zaal, B. J., Neuteboom, L. W., Pinas, J. E., Chardonnens, A. N., Schat, H., Verkleij, J. A., et al. (1999). Overexpression of a novel Arabidopsis gene related to putative zinc-transporter genes from animals can lead to enhanced zinc resistance and accumulation. Plant Physiol. 119, 1047–1056. doi: 10.1104/pp.119.3.1047.
Verbruggen, N., Hermans, C., Schat, H. (2009). Molecular mechanisms of metal hyperaccumulation in plants. New Phytol. 181, 759–776. doi: 10.1111/j.1469-8137.2008.02748.x.
Wang, C., Sun, Q., Wang, L. (2009). Cadmium toxicity and phytochelatin production in a rooted-submerged macrophyte Vallisneria spiralis exposed to low concentrations of cadmium. Environ. Toxicology: Int. J. 24, 271–278. doi: 10.1002/tox.20429.
Wang, D., Zhang, Y., Zhang, Z., Zhu, J., Yu, J. (2010). KaKs_Calculator 2.0: a toolkit incorporating gamma-series methods and sliding window strategies. Genomics Proteomics Bioinf. 8, 77–80. doi: 10.1016/S1672-0229(10)60008-3.
Yilmaz, H., Özer, G., Baloch, F. S., Çiftçi, V., Chung, Y. S., Sun, H.-J. (2023). Genome-wide identification and expression analysis of MTP (Metal ion transport proteins) genes in the common bean. Plants 12, 3218. doi: 10.3390/plants12183218.
Yu, G., Wang, L.-G., Han, Y., He, Q.-Y. (2012). clusterProfiler: an R package for comparing biological themes among gene clusters. Omics: J. Integr. Biol. 16, 284–287. doi: 10.1089/omi.2011.0118.
Yuan, L., Yang, S., Liu, B., Zhang, M., Wu, K. (2012). Molecular characterization of a rice metal tolerance protein, OsMTP1. Plant Cell Rep. 31, 67–79. doi: 10.1007/s00299-011-1140-9.
Zaid, I. U., Zheng, X., Li, X. (2018). Breeding low-cadmium wheat: Progress and perspectives. Agronomy 8, 249. doi: 10.3390/agronomy8110249.
Zhang, X., Li, Q., Xu, W., Zhao, H., Guo, F., Wang, P., et al. (2020). Identification of MTP gene family in tea plant (Camellia sinensis L.) and characterization of CsMTP8. 2 in manganese toxicity. Ecotoxicology Environ. Saf. 202, 110904. doi: 10.1016/j.ecoenv.2020.110904
Keywords: Tulipa gesneriana, heavy metal accumulation, metal tolerance proteins (MTPs), phylogenetic analysis, gene expression regulation
Citation: Lu J, Xing G, Zhang Y, Zhang H, Wu T, Tian Z and Qu L (2024) Genome-wide identification, expression and function analysis of the MTP gene family in tulip (Tulipa gesneriana). Front. Plant Sci. 15:1346255. doi: 10.3389/fpls.2024.1346255
Received: 01 December 2023; Accepted: 05 February 2024;
Published: 19 February 2024.
Edited by:
Mehmet Cengiz Baloglu, Kastamonu University, TürkiyeReviewed by:
Emre Aksoy, Middle East Technical University, TürkiyeAnuj Kumar, Dalhousie University, Canada
Copyright © 2024 Lu, Xing, Zhang, Zhang, Wu, Tian and Qu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lianwei Qu, cXVsaWFud2VpMjAyM0AxNjMuY29t
 Jiaojiao Lu
Jiaojiao Lu Guimei Xing
Guimei Xing Yanqiu Zhang1
Yanqiu Zhang1 Lianwei Qu
Lianwei Qu