
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Plant Sci., 08 February 2024
Sec. Plant Physiology
Volume 15 - 2024 | https://doi.org/10.3389/fpls.2024.1343928
This article is part of the Research TopicMolecular and Genetic Mechanisms of Plant Architecture RegulationView all 8 articles
 Mehmood Jan1,2,3†
Mehmood Jan1,2,3† Sajid Muhammad4†
Sajid Muhammad4† Weicai Jin3,5
Weicai Jin3,5 Wenhao Zhong3
Wenhao Zhong3 Shaolong Zhang1,5
Shaolong Zhang1,5 Yanjie Lin3
Yanjie Lin3 Yueni Zhou3
Yueni Zhou3 Jinlong Liu3
Jinlong Liu3 Haifeng Liu3,5
Haifeng Liu3,5 Raheel Munir4
Raheel Munir4 Qiang Yue2
Qiang Yue2 Muhammad Afzal1,2,3*
Muhammad Afzal1,2,3* Guoping Wang1,5*
Guoping Wang1,5*Root architecture is an important agronomic trait that plays an essential role in water uptake, soil compactions, nutrient recycling, plant–microbe interactions, and hormone-mediated signaling pathways. Recently, significant advancements have been made in understanding how the complex interactions of phytohormones regulate the dynamic organization of root architecture in crops. Moreover, phytohormones, particularly auxin, act as internal regulators of root development in soil, starting from the early organogenesis to the formation of root hair (RH) through diverse signaling mechanisms. However, a considerable gap remains in understanding the hormonal cross-talk during various developmental stages of roots. This review examines the dynamic aspects of phytohormone signaling, cross-talk mechanisms, and the activation of transcription factors (TFs) throughout various developmental stages of the root life cycle. Understanding these developmental processes, together with hormonal signaling and molecular engineering in crops, can improve our knowledge of root development under various environmental conditions.
Plant architecture is considered the most important trait that affects stress resistance and yield (Guo et al., 2020a). Although the above-ground parts (green parts) of plants participate vigorously in photosynthesis and transpiration (Sharma et al., 2021), reduction in plant height, leaf width, and length, branching pattern, and even the angles of the leaves have a significant effect on light capturing, photosynthesis, and, finally, the plant’s development and growth (Berry and Argueso, 2022). Moreover, the changes in these traits can directly affect the aptitude of the crops to grow in high-density planting (HDP) systems and their mechanical harvesting at the end (Huot et al., 2014; Berry and Argueso, 2022). For example, in maize, the sparsely and upright-branched phenotype is better for HDP (Tian et al., 2019); however, in rice, the semi-dwarf phenotype overcomes lodging (Lo et al., 2017; Wang et al., 2017).
Similarly, the hidden parts of the plants (roots) control the wide-ranging processes that are essential for the regular development and growth of plants, such as water acquisition, nutrient uptake from soil and transfer to other parts, and soil anchorage, and also for mechanical support (Li et al., 2018). Additionally, roots also form a site for the symbiotic relationship of plant–microbe interaction, such as nitrogen fixation (Afzal et al., 2022). Root architecture (RA) optimization mainly depends upon the plant species, environmental conditions, and the demand from the green parts of the plants (Hodge et al., 2009). Therefore, modifications in RA, such as reduced length of primary roots (PRs), changes in structure and numbers of the lateral roots (LRs), and root hairs (RHs) preclude crops from regularly obtaining water and the uptake of vital nutrients from the soil for normal development. In the current era, the main challenge for crop breeders in modern agricultural practices is producing an ideal plant with improved plant architecture suitable for more yield under various environmental conditions.
Owing to the sessile condition of plants, understanding the RA would be helpful for improving yield and optimization for agricultural land. The usage of newly developed technologies, such as three-dimensional (3-D) root imaging, advances in digital photography, fluorescence-based imaging systems, and X-ray tomography, and using transparent soil have assisted scientists in understanding the complex system of RA in different plant species (Tötzke et al., 2019). Similarly, with advancements in the new techniques of modern molecular biology, the identification of the important gene and signaling pathways in plant species has become a reality (Šimášková et al., 2015).
In addition, plant hormones regulate numerous metabolic pathways to facilitate plant adoption in harsh environments. In different plant species, many mutants of hormonal biosynthesis and signaling have been identified and used to investigate the importance of hormones in plant architecture under different environmental conditions (Iqbal, 2014; EL Sabagh et al., 2022). For instance, gibberellin is documented to be shown as a regulator effect on root development. The study on gibberellin-deficient Arabidopsis mutants showed reduced root elongation, which was connected with lower production of gibberellin in roots (Willige et al., 2007). Correspondingly, numerous studies demonstrated the cross-talk between phytohormones such as auxin and gibberellic acid (GA) (Niu et al., 2013). Auxins have been noted to modulate the expression of GA synthesis-related genes in several plant species (Frigerio et al., 2006), while GA is involved in increasing the polar-auxin transport in plants (Björklund et al., 2007). Similarly, auxin signaling was noted in the regulation of the RGA (repressor of ga1-3), thus indorsing GA signaling, and root development (Fu and Harberd, 2003). Moreover, the exogenous application of GA was reported as an RH promotor.
Plant RA is influenced by a complex interaction of regulatory networks that involve phytohormones, transcription factors (TFs), and miRNAs (Tang and Chu, 2017; Li et al., 2018). In addition, phytohormone signaling pathways and their cross-talk govern the developmental programs of whether to begin, continue, or maybe arrest. Therefore, this review aims to elucidate the potential role of phytohormone-regulated TFs, which results in root development and their possible interaction for ideal RA. Gaining insights into these developmental processes, together with engineering their expression and activity of related factors, can enhance our understanding of creating smart crops under various environmental conditions.
The three-dimensional structure of the plant root is known as the root system architecture (RSA), which consists of PRs, the numbers and patterns of LRs, adventitious roots (ARs), and RHs (Figure 1A). The RSA in different plant species exhibits high differences in the morphological traits, which help plants adapt to competitive environmental conditions. The cross-talk among phytohormones includes intricate and complex molecular mechanisms that regulate RA in plants (Figure 2). Furthermore, two different kinds of root systems exist in plants tap roots and fibrous roots, which are defined by their branching patterns.
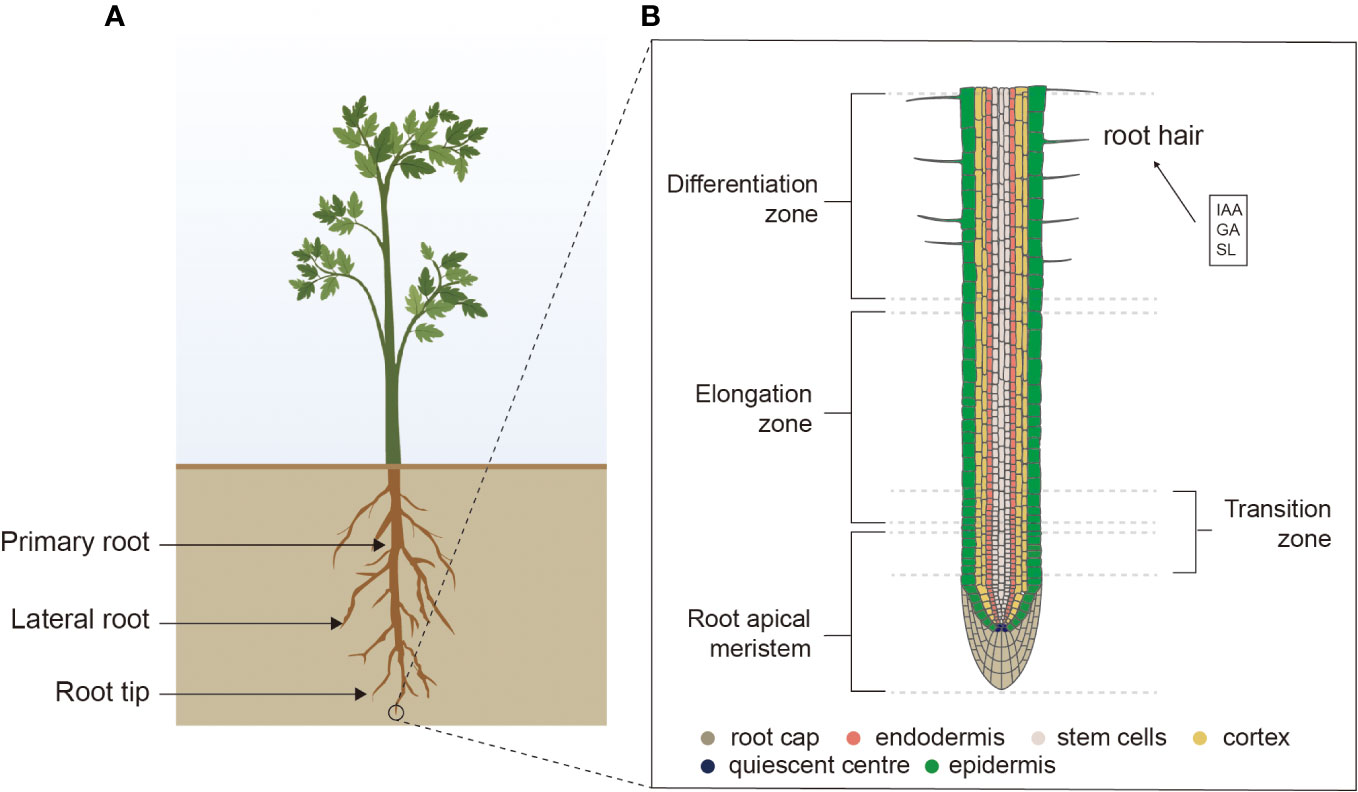
Figure 1 The tap root system. (A) RSA of dicots representing different parts of roots. (B) Root tip showing root cap, endodermis, stem cells, cortex, quiescent center, epidermis, root apical meristem, elongation zone, transition zone, differentiation zone, and RHs. RSA, root system architecture; RHs, root hairs.
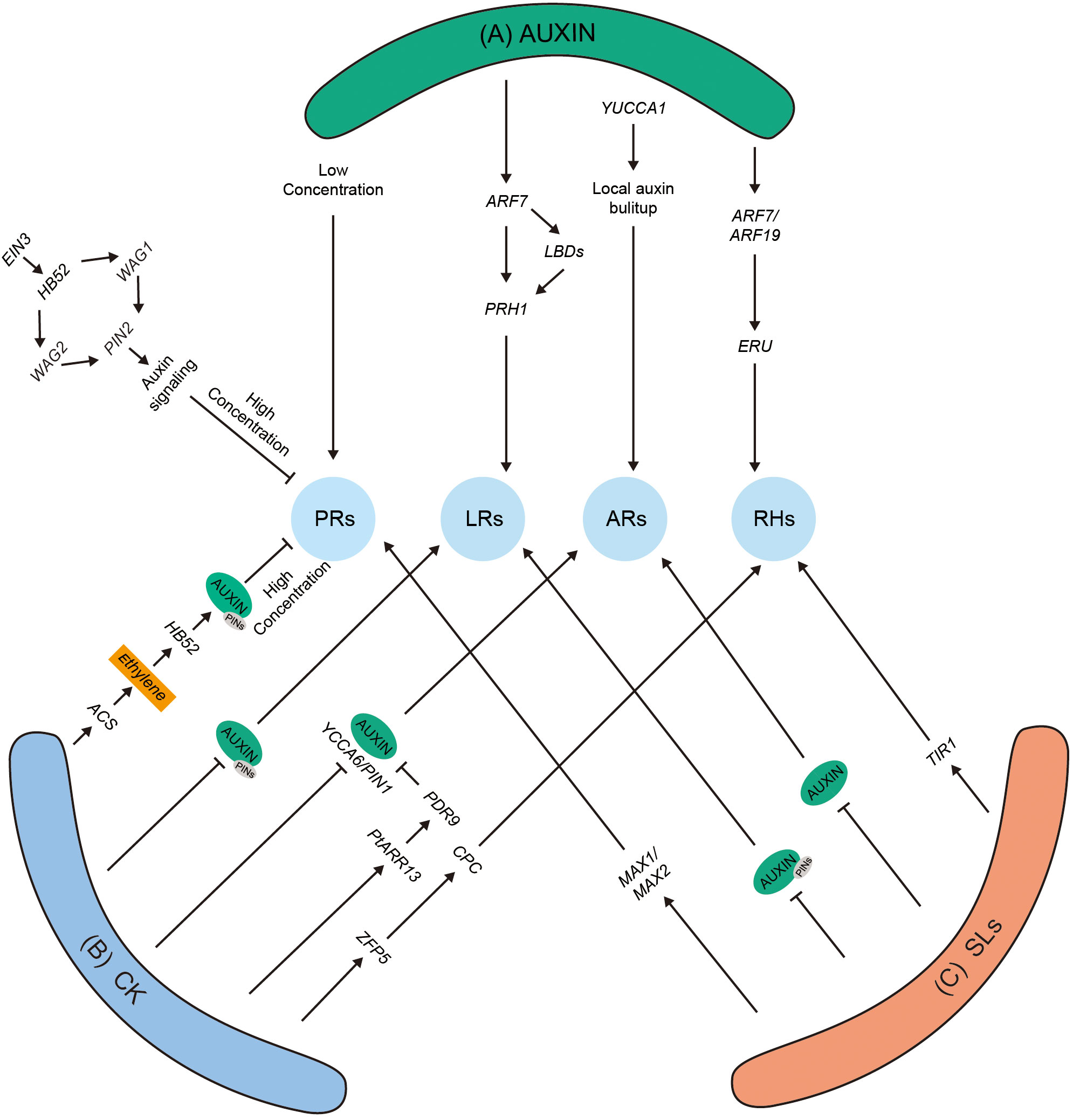
Figure 2 The role of auxin and its hormonal cross-talk with other phytohormones. (A) High auxin level negatively affects the PR length. HB52 binds to the promotor of WAG1, WAG2, and PIN2 and positively modulates their transcriptional levels. As a result, more auxin is transported to EZ. Auxin promotes LR development by inducing RPH1 expression dependent on ARF7 and LBDs. The overexpression of YUCCA or exogenously applied auxin promotes ARs in Arabidopsis. Auxins positively promote RHs by ERU-dependent (ERULUS is a core regulator for RHs growth) pathway. (B) CK positively regulates ACS gene, which produces more ethylene, and ethylene stabilizes PIN2, resulting in the modulation of more auxin in the roots, which further negatively regulates PRs. Application of CK negatively modulates LRs with disturbance of auxin levels viaPIN expression. The application of CK regulates auxin levels in ARs by negatively modulating YUCCA, ARR13, and PIN genes. CK-induced RHs via CPC-mediated complex. (C) Exogenous application of SLs promotes PRs through max-dependent pathway. SL modulates LRs and ARs by altering auxin levels in roots in Arabidopsis. Under low Pi environments, SLs promote the expression of TIR gene, increasing auxin levels and promoting RHs. The local auxin accumulation in RSA promotes PR, LR, AR, and RH growth and development. ACS, ACC synthase 2; ARF, auxin response factor; ARs, adventitious roots; CK, cytokinins; EZ, elongation zone; EIN3, ethylene-insensitive3; PRs, primary roots; LRs, lateral roots; HB52, homeobox protein 52; LBD, lateral organ boundaries domain; PIN, pin formed; PDR, pleiotropic drug resistance transporter; WAG, wavy root growth; RPH1, leaf rust resistance gene; TIR, transport inhibitor response 1; RHs, root Hairs; zfp, zinc finger protein 5.
In RSA, the PRs constitute the fundamental component that originates from the embryonic meristematic tissues close to the early globular embryo’s base and are the first to emerge. These PRs exhibit distinct developmental regions in RA (Svolacchia et al., 2020). At the root apical meristem, all the cells originate from stem cells or precursors within the stem cell niche (SCN), which is located in the tip. After that, daughter cells undergo differential rates of division and proportions, delineating a zone known as the division zone (DZ) (Figure 1B). Upon exiting the cell division cycle, these cells elongate and generate the zone of elongation (EZ) (Svolacchia et al., 2020). Subsequently, these cells differentiate, acquiring characteristics of specific tissues, based on their radical position, leading to the differentiation zone’s constitution. In RSA, the transition zone (TZ) assists as a developmental boundary between the division and differentiation of the cells. Furthermore, each of the developmental zones corresponds to a classified distribution of phytohormones, peptides, mRNA, and transcription factors. Notably, the functions of cytokinin and auxin are prominent in SCN and help in the facilitation of cell differentiation. The auxin gradient was noted higher in SCN and gradually decreased toward the TZ (Galinha et al., 2007).
LRs contribute to enhanced soil anchorage, promote efficient nutrient absorption and water uptake, and foster symbiotic relationships with microorganisms (Du and Scheres, 2018; Motte et al., 2019). The development of LRs mainly directed by the auxin activity, response maxima, and downstream signaling pathways govern the pattering of LRs in angiosperms. LR formations take place within the single-layered pericycle tissues located in the xylem poles and are known as xylem pole pericycle cells. Through continuous cell elongation and anticlinal division, xylem pole pericycle cells switch from the meristem to auxin-rich zones (Jansen et al., 2012). In addition, transcriptomic studies of LR initiations pointed out some common pathways in maize and Arabidopsis, which may suggest that conserved genes are involved in the LR initiation in angiosperms (Jansen et al., 2013).
AR roots are the roots that ascend from any other point than the radicle (embryonic roots) or the root axis of the plants; some can ascend from the stem tissues (Steffens and Rasmussen, 2016). These can manifest as crown roots, arising from the underground nodes, or the brace roots, which develop from the nodes that are above the ground as well as seminal roots and stem roots. Various root categories serve different structural and absorptive roles in plants. PRs and seminal roots are pivotal during the initial phases of plant growth, whereas brace roots become significant in the later stage of monocot RA and provide structure support to the plants (Steffens and Rasmussen, 2016).
RHs are elongated, tip-growing structures from the epidermis of roots, playing an important role in increasing the surface area of roots for enhancing nutrient and water absorption (Miguel et al., 2015). Additionally, they also promote root adhesion to the surrounding rhizosphere and facilitate soil, plant, and microbial interactions. Several studies have demonstrated the compromised fitness of plants with RH mutants or loss of function when they were subjected to harsh environmental conditions in soil (Schoenaers et al., 2018). Thus, the dynamic development of RH offers an opportunity for agricultural exploitation, enhancing nutrient and water acquisition in diverse soil environments. The initiation of RH development commences with positional signaling originating from the roots’ cortical cells. Overall, the above evidence suggests that environmental conditions and phytohormones in various developmental stages and tissues of RA significantly regulate root development.
Previous studies have demonstrated a strong relationship between root development and auxin; therefore, it was named root-forming hormone (Zhu et al., 2019; Moreno-Risueno et al., 2010). Auxin showed a regulatory effect on almost every phase of root development and growth (Figure 2A). PR is stimulated in maize, Arabidopsis, tomato, and rice under low auxin concentrations (Figure 2A). In comparison, inhibited growth was observed at high concentrations through the TRANSPORT INHIBITOR RESPONSE 1 (TIR1) receptor, mediating signaling by an unknown non-transcriptional mechanism (Fendrych et al., 2018). It has been noted that free IAA/Aux can positively regulate root development; in contrast, ubiquitinated Aux/IAA proteins can negatively regulate root development. Compared to other signals in root development, auxins are considered the most essential hormone that instructs root meristem and organogenesis (Fendrych et al., 2018). It was shown that auxin is produced through a tryptophan-dependent pathway in roots, in which tryptophan converts into indole acetic acid (IAA). TRYPTOPHAN AMINOTRANSFERASE RELATED 2 (TAR2) protein and its homologous WEAK ETHYLENE INSENSITIVE 8 (WEI8) are the two key enzymes that are involved in producing and maintaining the auxin homeostasis in roots. Mutants of WEI8/TAR2 showed lower auxin concentrations and a loss of the meristem zone in plant roots (Brumos et al., 2018). Additionally, the roots deficient with auxin levels could not be recovered with shoot-derived auxins through a grafting experiment (Brumos et al., 2018).
In Arabidopsis, a gene Nitrilase 1 (NIT1) coding for nitrilase 1 enzyme modulates the auxin synthesis, and the overexpression of NIT1 is responsible for shorter PR because of changes in IAA and IAN levels. A study on rice showed that FPF1-like protein (OsFPFL4) regulates the auxin levels and root growth. Overexpression of OsFPFL4 resulted in shorter PRs because of the high endogenous accumulation of auxin and also altered the expression level of auxin’s biosynthesis and transporter-related genes (Guo et al., 2020b). Additionally, a gene MADS-box Transcription Factor (OsMADS25) activates the biosynthesis and transport of auxin in rice, modulating root development and growth (Zhang et al., 2018). Moreover, the mutant lines of (rice auxin influx carrier) OsAUX3-1 and OsAUX3-2 resulted in shorter PRs by changing the concentration of auxin in rice roots (Nan et al., 2014). Furthermore, in rice, the OsWOX4 (WUSHEL-related homeobox protein) has been proven to be responsible for root elongation by transcriptional activation of OsAux1 gene (Chen et al., 2020). The soybean gene GmYUC2a is considered an important regulator gene for root development and growth by regulating auxin biosynthesis (Wang et al., 2019). The auxin biosynthesis in roots can be regulated by other alternative mechanisms, such as exogenously applied chemicals and protein phosphorylation (Nikonorova et al., 2021). A study on exogenously applied cyclic guanosine 3′,5′-monophosphate (cGMP) provided evidence that PR growth in Arabidopsis was modulated due to IAA/AUX protein degradation (Nan et al., 2014). The phosphoproteome studies by Nikonorova et al. (2021) identified some new growth regulators/inhibitors, such as MAP kinases and some receptor-like kinases, by applying different concentrations of auxins and suggesting that cell wall modification, H+-ATPases, cell wall sensing receptors, and auxin concentration are responsible for the elongation of the cell root tips. It has been previously reported in many studies that auxin interacts with other phytohormones to govern the development of roots (Figure 2). The gene homeobox protein (HB52) has been shown to regulate the cross-talk between auxin and ethylene and modulate PR growth. HB52 acts as the downstream of ethylene-insensitive3 (EIN3) and also modulates the expression level of auxin transporters like PIN2 and WAVY ROOT GROWTH (WAG1 and WAG2) by binding with their promoters. The genes WAG1 and WAG2 phosphorylate PIN2, which gathers in the apical side of the root’s cells and results in more auxins being transported to the TZ, and because of the higher accumulation of auxins, the PRs were noted to be shorter (Figure 2A) (Mao et al., 2016). In addition to auxin’s transporters, the ethylene signaling gene ETHYLENE RESPONSE FACTOR (ERF1) was noted to regulate the auxin biosynthesis genes (YUCCAs) and ANTHRANILATE SYNTHASE alfa 1 (ASA1) during root development (Mao et al., 2016).
Few previous studies already discussed the interaction of cytokinin (CK) with auxin in modulating the cell architecture in the initial stages of root growth and development. In Arabidopsis, a newly identified AKH3 transmembrane receptor belonging to the CK family, in conjunction with type B ARABIDOPSIS RESPONSE REGULATOR (ARR1 and 2), plays a vital role in the redistribution of auxins in the root’s meristem (Ioio et al., 2008a). They also provided evidence that changes in the auxin distribution are due to the modulation in the transcript level of PIN genes via the activation of auxin repressor genes like SHORT HYPOCOTYL2 (SHY2) and IAA3, resulting in cell differentiation. Another study showed that CK modulates the transcription of PIN gene through the involvement of CYTOKININS RESPONSE FACTORS (CRFI), which degrades SHY2, resulting in the upregulation of PIN genes (Ioio et al., 2008b). Therefore, CKs are important for the homeostatic modulation of SHY2 factors and the root architecture and development. The octuplet mutant of arrs (3, 4, 5, 6, 7, 8, 9, and 15) showed smaller meristem size as compared to control plants and was highly sensitive to NPA inhibitor, which targets the auxin transporters (Zhang et al., 2011). Furthermore, when CK was applied externally to arr mutants, the reduced PIN4-GFP was noted in the root cap (Zhang et al., 2011).
Auxin-induced LR development has been widely studied in Arabidopsis and tomato by modulating their auxin signaling and biosynthesis genes; however, few studies claimed that a lower concentration of auxin (25 nM) reduced LR growth when applied externally (Banda et al., 2019). A canonical downstream target (LBD16/ASYMMETRIC LEAVES2-LIKE18; ASL18), that is, the conventional downstream target of (SOLITARY ROOT (SLR)/IAA14-ARF19-ARF7) auxin signaling module, modulates the growth and development of LRs, presumably through the transcriptional modulation via its associated LBD/ASLs, resulting in the asymmetric cell division in the initiations of LRs and primordium development (Goh et al., 2012). Some reports have documented that (ATP-BINDING CASSETTE) ABCB transporters can play a significant role in the emergence of LRs, and delayed emergence of LRs was observed in the mutants of ABCB21 (Jenness et al., 2019). Similarly, these studies also suggest that ABCB21 is also responsible for the modulation of auxin distribution and plays a role in LR formation. Based on earlier studies on Arabidopsis, it was noted that LR formation requires ARF7, and also LATERAL ORGAN BOUNDARIES DOMAIN (LBD), for normal development and growth. Recently, it was revealed that LR development in the Arabidopsis is modulated by PRH1 through endogenous auxin concentration and the auxin signaling pathway. Furthermore, PRH1 expression is induced by auxin, and reduced LRs were noted in PRH1 mutant lines. Moreover, PRH1 is shown as a direct transcriptional target of LBDs and ARF7, thus playing a positive role in cell wall loosening because it may able to regulate EXPANSIN (EXP) gene expression (Figure 2A) (Zhang et al., 2020).
Both LRs and ARs mostly use the same pathways; however, there are differences in some regulator processes. For example, ARF7 and ARF19 are responsible for regulating LRs and activating LBD genes, whereas for ARs, the TF WOX11 overtakes the functions of ARFs. In procambium and its adjacent cells, WOX11 responds to the wound-induced auxins and activates the downstream LBD 16 and 29, together with its homologous protein WOX12, regulating the fate transition from procambium to founder cells of the roots (Liu et al., 2014). Similarly, few researchers suggested that ARF17 and 19 are responsible for the regulation of ARs in hypocotyls, which describes the exact molecular mechanism for AR and LR development (Wilmoth et al., 2005). Moreover, the overexpression line 35S:YUCCA1 resulted in higher numbers of ARs as compared to WT, which was consistent with 100 nM exogenously applied IAA, which can increase ARs in Arabidopsis (Figure 2A). Few studies have clearly defined the role of auxin transporters during AR development (Table 1). The AT-binding cassette ABCB19 and IBA have been linked to inducing AR formation. It has been suggested by a recent study that AR development is mediated by ARF8, ARF17, and ARF6, which further modulates the transcriptional levels of GH3.3, 3.5, and 3.6; it has been documented that ARF6 and 8 are the two positive modulators, while ARF17 acts as a negative regulator (Gutierrez et al., 2012). In addition, these three genes control JA homeostasis and lead to an increase in JA-lle, which activates the JA.lle/COL1 signaling pathway and negatively modulates ARs in Arabidopsis. In conclusion, auxin modulates AR initiation by the activation of ARF8 and ARF7, which results in AR ambitions through the COL1 signaling pathway (Gutierrez et al., 2012).
Another essential role of auxin in the RSA is the development and growth of RHs (Hossain et al., 2015). A new study on trichoblasts explained that the expression level of (ROOT HAIR DEFECTIVE 6 LIKE 4) RSL4 is controlled by ARF5, ARF7, ARF8, and ARF19 by binding with its promoters. In addition, it was also noted that auxins control the modulation of cell wall receptor protein ERULUS (the member of Catharanthus roseus receptor Like-Kinase-1-Like) in an ARF19- and ARF7-dependent manner (Figure 2A) (Schoemaers et al., 2018). The overexpression of ARF1–4 and ARF16 negatively modulated the growth of RHs and suggested the different roles of ARFs in RHs. Furthermore, ethylene and auxin were demonstrated to regulate RH growth and development synergistically (Bruex et al., 2012). Taken together, auxin biosynthesis, transport, and concentration are important RSA.
CK is another phytohormone that plays a vital role in forming PRs. Exogenous application or high endogenous concentration of CKs inhibits proper root development by negatively affecting the cell division in the apical meristem of roots (Tiwari and Bhatia, 2020). Similarly, CK signaling mutants of type A ARR and type B ARR showed a strong impact on PR length in some monocots and dicots (Gao et al., 2014; Márquez et al., 2019). Furthermore, single or double CK-deficient mutants showed an increased root phenotype (Werner et al., 2003; Higuchi et al., 2004).
Cytokinin–auxin interaction regulates plant growth in numerous plant species, i.e., rice, tomato, and maize (Kurepa et al., 2018). CK performs a crucial role in the modulation of cell differentiation by enhancing the positive effects of auxins on cell division (Ioio et al., 2007; Ioio et al., 2008a. Through the ARR1/AKH3 pathway, CK activates the transcription of auxin/IAA and SHY2/IAA3 genes within vascular tissues of the TZ (Ioio et al., 2008b). SHY2 negatively influences PIN expression and changes the auxin transport to roots (Ioio et al., 2008a). In addition, the overexpression of SHY2-2 resulted in impaired root growth upon exogenous CK application, and loss of function resulted in higher root length, which is closely similar to arr1 mutants. Furthermore, the PIN expression was downregulated in shy2-2 mutant lines, providing a relationship among SHY2, PINs, and root development (Ioio et al., 2008a). Moreover, CK induces ethylene biosynthesis by stabilizing the ACS gene; ethylene through HB52, WAG1, and WAG2 stabilizes PIN2 gene and inhibits PR growth due to elevated auxin levels (Figure 2B) (Qin et al., 2019a). In summary, these findings suggested that the plants require minimum CK and auxin levels for normal development of PRs.
Concurrently, CK negatively regulates LR formation in plants. Increased levels of endogenous level or exogenously applied CK inhibit the LRs in rice and Arabidopsis (Debi et al., 2005; Li et al., 2006). In comparison to the mutant plants of the AKH and type B ARR, the gain of function displayed enhanced LR formation (Werner and Schmülling, 2009; Del Bianco et al., 2013). Similarly in Arabidopsis, some type A and type B ARR mutants displayed reduced LR numbers as compared to the control (To et al., 2004). It was noted that the reduction of LRs through CK was because of modulation in auxin transport and the disturbance of PIN1 localization (Mason et al., 2005). Exogenously applied CK rapidly regulated PIN1 gene expression, further clarifying CK’s role in modulating auxin transport (Figure 2B) (Laplaze et al., 2007). In addition, a recent report identified a new TRANSPORTER OF IBA (TOB1) gene, which is highly expressed in lateral root caps and showed repose to CK. TOB1 is responsible for transporting IBA in the cytoplasm of the cell, which converts into active form IAA, and the mutant of tob1 increased the production of LR number (Michniewicz et al., 2019).
Numerous studies have explored the interaction between CK and auxin in modulating the development and growth of ARs in many species (Table 1). Additionally, in Arabidopsis, CK showed negative influences on PIN (like Aux1), LAX3, and YUCCA6 genes, resulting in the alteration of auxin flow and repression of ARs (Figure 2B) (Li, 2021). In Populus tremula, PtRR13 modulates CK signaling and results in AR repression. PtRR13 modulates the expression of PLEIOTROPIC DRUG RESISTANCE TRANSPORTER (PDR9), which is responsible for the auxin’s efflux pump, which shows further impact on the vascular tissue formation in AR rooting, thus confirming the role of CK and auxin cross-talk (Figure 2B) (Ramirez-Carvajal et al., 2009). Furthermore, PtRR13 also inhibits AR development and growth by negatively modulating two ethylene-responsive TINY-like transcription factors, bridging the gap between CK and ethylene (Ramirez-Carvajal et al., 2009). In addition, AR rooting culture for Medicago truncatula CK downregulated the expression of PLETHORA (PLT2 and PLT1) while upregulating the expression level of (AINTEGUMENTA-like) AIL1, WOX4, and AINTEGUMENTA (ANT) and the SHOOT MERISTEMLESS (STM) transcriptional factors. In poplar, WOX genes like Pewor11b or Pewox11a enhanced AR formation, with wox11 acting as a mediator between CK and auxin by downregulating type B CK response regulators (Kitomi et al., 2011; Xu et al., 2015; Zhao et al., 2009). These findings elucidate the interaction of CK and auxin pathways in AR development.
Similarly, a recent article suggested the positive role of CK in root hair elongation (Takatsuka et al., 2023). It noted that RH elongation involves type A ARRs, ARR12 and ARR1; ARR1 can directly induce the transcriptional level of RSL4, which plays an important role in CK-dependent RH development (Takatsuka et al., 2023). Exogenously applied CK promoted RH elongation, whereas the overexpression of cytokinin oxidase 2 (CKX2 code for CK degrading) displayed shorter RH formation (Zhang et al., 2016). In Arabidopsis, some TFs regulate RH growth and development, and mutants of zinc finger 5 (zfp5) displayed fewer root hairs than WT. The modulation in the expression level of ZFP5 was increased by the application of 6-benzylaminopurine, which is known as RH inducer. ZFP5 has the ability to induce the transcriptional level of MYB proteins, CAPRICE (CPC), which binds to their promotors that are involved in the RH patterning. Furthermore, this complex inhibits the negative modulators of root hairs like GLARBAI (GL2), as shown in Figure 2B (Wada et al., 2002).
The roots in plants are considered the central part of strigolactone (SL) biosynthesis (Dun et al., 2009). Numerous studies suggested that SL interferes with the auxin’s transporters to regulate root growth and development (Xie et al., 2010, Rasmussen et al., 2012; Xie et al., 2015). In different plant species, several mutants of SL biosynthesis (max4 and max3) and signaling (D14 and D3) have been identified (Claassens and Hills, 2018). SL promotes PRs and AR growth, but a negative effect has been noted in LR growth (Figure 2C) (Kumar et al., 2015; Sun et al., 2016). At lower concentrations, GR-24 has been shown as an LR growth and development enhancer, while higher concentrations (2.5 μM) reduced LR growth in a few species (Jain et al., 2007; Shinohara et al., 2013).
In Arabidopsis, the PR length was reduced because of smaller numbers of cells in the meristem zone of the roots in the mutant lines as compared to the wild type (Ruyter-Spira et al., 2011). Under specific conditions, the biosynthesis mutant of SL, max1 (cytochrome P450), and max4 (carotenoid cleavage dioxygenase), max3, and d14 were observed with shorter PR lengths as compared to WT. Exogenous application of GR-24 recovered PRs in the max4 mutant line, while signaling mutants showed no significant change in roots (Ruyter-Spira et al., 2011). Similarly, PR recovery was noted in SL-deficient mutants in tomato, rice, and M. truncatula after the application of GR-24, while no change or non-significant changes were noted in WT or signaling mutants (Koltai et al., 2010; Sun et al., 2014; De Cuyper et al., 2015).
Exogenously applied GR-24 treatment appears to inhibit LRs in numerous plant species (Arite et al., 2012; Marzec et al., 2016; and De Cuyper et al., 2015). A signaling mutant of SL (hvd14.d) in Hordeum vulgare L. was noted with higher lateral root density, while the change in the root length was non-significant as compared to the wild type (Marzec et al., 2016). It was observed in Lotus japonicus L., in which the SL-biosynthesis mutant line carotenoid cleavage dioxygenase 7 (LjCCD7) showed more LRs than WT (Liu et al., 2013). SL may affect LRs by changing the concentration of auxin in roots. The application of GR-24 affects the PIN auxin efflux in roots, which results in reduced PIN-GFP intensity in LRs (Figure 2C) (Koltai et al., 2010; Ruyter-Spira et al., 2011).
In addition, both SL-deficient and signaling mutants in pea and Arabidopsis resulted in more ARs as compared to WT, which shows that SL interferes with AR formation (Figure 2C) (Srivastava et al., 2018). To confirm the interaction between auxin and SL, Rasmussen et al. examined the SL response to an over-producing line of auxin 35:YUCCA1 and demonstrated that the over-producing line increased AR formation as compared to WT, which is in line with the property of auxin, which promotes ARs. In contrast, the 35:YUCCA1 line treated with exogenous application of SL reduced AR formation by up to 50%. In the case of the max3 mutant treated with IAA, IBA produced more ARs as compared to the control. Similarly, ARs were decreased in pea and Arabidopsis mutants, and further investigation of CYCLIN B1 confirmed that SL decreased the number of ARs by interfering in the cell division of founder’s cells (Rasmussen et al., 2012; Brewer et al., 2013). Interestingly, in rice, SL was shown to positively regulate ARs (Arite et al., 2012; Sun et al., 2014). Furthermore, SL mutants with biosynthesis and signaling in rice showed fewer ARs in the seedling and tillering stage (maturation stage). Treatment with exogenously applied GR-24 recovers the ARs in the (d10) biosynthesis mutant while remaining unchanged in the signaling mutant (Sun et al., 2015). In addition, maize (Zea mays) SL-deficient mutant zmccd8 resulted in shorter nodal ARs as compared to the control plants, which shows the regulator role of SL in AR development (Guan et al., 2012). Altogether, these consequences suggest that SLs can negatively regulate the AR development in eudicot plants like Arabidopsis. In contrast, increased number and elongation of ARs were noted in grasses like maize and rice plants. Auxin signaling is important for normal AR development, and SL regulates the auxin levels in the pericycle and modulates AR initiation (Rasmussen et al., 2012).
Exogenously applied GR-24 was noted to increase RHs in tomatoes and Arabidopsis (Koltai et al., 2011; Kapulnik et al., 2011). It was demonstrated that SLs and ethylene modulate root hair elongation. Furthermore, the treatment of SLs increased the expression of ACC synthase 2 (ACS2) gene and activated the acs enzyme, suggesting a shared regulatory pathway for RH development. Additionally, SLs were noted to regulate RH elongations through an auxin-dependent pathway (Kapulnik et al., 2011; Kohlen et al., 2012). A study has shown that phosphate deficiency promotes SL accumulation and induces the regulation of auxin receptor gene TIR1 in the WT plants but not in the max4-deficient mutant, signifying that under specific conditions, both SL and auxins affect the root hair density (Figure 2C). In summary, SLs are proposed to modulate auxin transport and regulate RSA.
This review emphasizes the complexity of plant phytohormones in RSA and highlights hormonal effects depending on specific combinations or individual actions. This study explores cross-talk between various hormones like auxin–SL and auxin–CK during different parts of RSA, indicating that different hormones coincide in regulating different processes in plants. Indeed, ample evidence demonstrates that the response of phytohormone changes during various stages of organ development, potentially due to diverse cellular and tissue environments. The configuration of root morphology is influenced by a highly complex network of phytohormones, highlighting the importance of comprehending key transcriptional and post-transcriptional pathways, along with their associated genes. Mapping these genes could offer insights into hormonal regulation and developmental outcomes. More systemic investigations and computational models are crucially needed to understand the hormonal cross-talk for root development under realistic field conditions. These investigations will not only help to consolidate the knowledge about how roots perceive internal and external signals and translate them into cellular responses but will also enable breeders to create predictive models to pinpoint essential regulators and integrators for governing RSA during various environmental conditions.
MJ: Conceptualization, Investigation, Methodology, Project administration, Software, Writing – original draft, Writing – review & editing, Validation. MS: Conceptualization, Data curation, Investigation, Methodology, Writing – original draft, Writing – review & editing, Project administration, Resources, Visualization. WJ: Conceptualization, Data curation, Investigation, Methodology, Software, Writing – review & editing. WZ: Conceptualization, Formal Analysis, Investigation, Resources, Visualization, Writing – original draft. SZ: Data curation, Formal analysis, Resources, Software, Visualization, Writing – original draft. YL: Conceptualization, Data curation, Funding acquisition, Investigation, Project administration, Resources, Validation, Writing – original draft. YZ: Conceptualization, Data curation, Formal analysis, Writing – original draft. JL: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Project administration, Software, Writing – review & editing. HL: Conceptualization, Data curation, Software, Supervision, Writing – original draft. RM: Conceptualization, Data curation, Software, Validation, Writing – original draft. QY: Formal analysis, Project administration, Software, Supervision, Writing – original draft. MA: Methodology, Project administration, Visualization, Writing – original draft, Writing – review & editing. GW: Conceptualization, Data curation, Funding acquisition, Project administration, Resources, Supervision, Validation, Visualization, Writing – original draft.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study was supported by the Heyuan division of Guangdong Laboratory for Lingnan Modern Agriculture Project (DT20220017) and Guangzhou Science and Technology Plan Project (2023B03J1356 and 2023B03J1271).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Afzal, M., Tang, C., Yu, M., Muhammad, N., Zhao, H., Xu, J. (2022). Water regime is important to determine cadmium toxicity on rice growth and rhizospheric nitrifier communities in contaminated paddy soils. Plant Soil 472 (1-2), 609–628. doi: 10.1007/s11104-021-05272-6
Ai, G., Huang, R., Zhang, D., Li, M., Li, G., Li, W., et al. (2023). SlGH3. 15, a member of the GH3 gene family, regulates lateral root development and gravitropism response by modulating auxin homeostasis in tomato. Plant Sci. 330, 111638. doi: 10.1016/j.plantsci.2023.111638
Arite, T., Kameoka, H., Kyozuka, ,. J. (2012). Strigolactone positively controls crown root elongation in rice. J. Plant Growth Regul. 31, 165–172. doi: 10.1007/s00344-011-9228-6
Arshad, M., Feyissa, B. A., Amyot, L., Aung, B., Hannoufa, A. (2017). MicroRNA156 improves drought stress tolerance in alfalfa (Medicago sativa) by silencing SPL13. Plant Sci. 258, 122–136. doi: 10.1016/j.plantsci.2017.01.018
Banda, J., Bellande, K., von Wangenheim, D., Goh, T., Guyomarc’h, S., Laplaze, L., et al. (2019). Lateral root formation in Arabidopsis: a well-ordered LRexit. Trends Plant Sci. 24 (9), 826–839. doi: 10.1016/j.tplants.2019.06.015
Berry, H. M., Argueso, C. T. (2022). More than growth: Phytohormone-regulated transcription factors controlling plant immunity, plant development and plant architecture. Curr. Opin. Plant Biol. 70, 102309. doi: 10.1016/j.pbi.2022.102309
Björklund, S., Antti, H., Uddestrand, I., Moritz, T., Sundberg, B. (2007). Cross-talk between gibberellin and auxin in development of Populus wood: gibberellin stimulates polar auxin transport and has a common transcriptome with auxin. Plant J. 52 (3), 499–511. doi: 10.1111/j.1365-313X.2007.03250.x
Brewer, P. B., Koltai, H., Beveridge, C. A. (2013). Diverse roles of strigolactones in plant development. Mol. Plant 6 (1), 18–28. doi: 10.1093/mp/sss130
Bruex, A., Kainkaryam, R. M., Wieckowski, Y., Kang, Y. H., Bernhardt, C., Xia, Y., et al. (2012). A gene regulatory network for root epidermis cell differentiation in Arabidopsis. PloS Genet. 8 (1), e1002446. doi: 10.1371/journal.pgen.1002446
Brumos, J., Robles, L. M., Yun, J., Vu, T. C., Jackson, S., Alonso, J. M., et al. (2018). Local auxin biosynthesis is a key regulator of plant development. Dev. Cell 47 (3), 306–318.e305. doi: 10.1016/j.devcel.2018.09.022
Chen, R., Xu, N., Yu, B., Wu, Q., Li, X., Wang, G., et al. (2020). The WUSCHEL-related homeobox transcription factor OsWOX4 controls the primary root elongation by activating OsAUX1 in rice. Plant Sci. 298, 110575. doi: 10.1016/j.plantsci.2020.110575
Debi, B. R., Taketa, S., Ichii, ,. M. (2005). Cytokinin inhibits lateral root initiation but stimulates lateral root elongation in rice (Oryza sativa). J. Plant Physiol. 162 (5), 507–515. doi: 10.1016/j.jplph.2004.08.007
De Cuyper, C., Fromentin, J., Yocgo, R. E., De Keyser, A., Guillotin, B., Kunert, K., et al. (2015). From lateral root density to nodule number, the strigolactone analogue. GR24 shapes the root architecture of Medicago truncatula. J. Exp. Bot. 66 (1), 137–146. doi: 10.1093/jxb/eru404
Del Bianco, M., Giustini, L., Sabatini, S. (2013). Spatiotemporal changes in the role of cytokinin during root development. New Phytol. 199 (2), 324–338. doi: 10.1111/nph.12338
Du, Y., Scheres, B. (2018). Lateral root formation and the multiple roles of auxin. J. Exp. Bot. 69 (2), 155–167. doi: 10.1093/jxb/erx223
Dun, E. A., Brewer, P. B., Beveridge, ,. C. A. (2009). Strigolactones: discovery of the elusive shoot branching hormone. Trends Plant Sci. 14 (7), 364–372. doi: 10.1016/j.tplants.2009.04.003
EL Sabagh, A., Islam, M. S., Hossain, A., Iqbal, M. A., Mubeen, M., Waleed, M., et al. (2022). Phytohormones as growth regulators during abiotic stress tolerance in plants. Front. Agron. 4, 765068. doi: 10.3389/fagro.2022.765068
Fendrych, M., Akhmanova, M., Merrin, J., Glanc, M., Hagihara, S., Takahashi, K., et al. (2018). Rapid and reversible root growth inhibition by TIR1 auxin signalling. Nat. Plants 4 (7), 453–459. doi: 10.1038/s41477-018-0190-1
Frigerio, M., Alabadí, D., Pérez-Gómez, J., García-Cárcel, L., Phillips, A. L., Hedden, P., et al. (2006). Transcriptional regulation of gibberellin metabolism genes by auxin signaling in Arabidopsis. Plant Physiol. 142 (2), 553–563. doi: 10.1104/pp.106.084871
Fu, X., Harberd, N. P. (2003). Auxin promotes Arabidopsis root growth by modulating gibberellin response. Nature 421 (6924), 740–743. doi: 10.1038/nature01387
Galinha, C., Hofhuis, H., Luijten, M., Willemsen, V., Blilou, I., Heidstra, R., et al. (2007). PLETHORA proteins as dose-dependent master regulators of Arabidopsis root development. Nature 449 (7165), 1053–1057. doi: 10.1038/nature06206
Gao, S., Fang, J., Xu, F., Wang, W., Sun, X., Chu, J., et al. (2014). CYTOKININ OXIDASE/DEHYDROGENASE4 integrates cytokinin and auxin signaling to control rice crown root formation. Plant Physiol. 165 (3), 1035–1046. doi: 10.1104/pp.114.238584
Goh, T., Joi, S., Mimura, T., Fukaki, H. (2012). The establishment of asymmetry in Arabidopsis lateral root founder cells is regulated by LBD16/ASL18 and related LBD/ASL proteins. Development 139 (5), 883–893. doi: 10.1242/dev.071928
Guan, J., Koch, K., Suzuki, M., Wu, S., Latshaw, S., Petruff, T., et al. (2012). Diverse roles of strigolactone signaling in maize architecture and the uncoupling of a branching-specific subnetwork. Plant Physiol 160, 1303–1317. doi: 10.1104/pp.112.204503
Guo, W., Chen, L., Herrera-Estrella, L., Cao, D., Tran, L.-S. P (2020a). Altering plant architecture to improve performance and resistance. Trends Plant Sci. 25 (11), 1154–1170. doi: 10.1016/j.tplants.2020.05.009
Guo, Y., Wu, Q., Xie, Z., Yu, B., Zeng, R., Min, Q., et al. (2020b). OsFPFL4 is involved in the root and flower development by affecting auxin levels and ROS accumulation in rice (Oryza sativa). Rice 13, 1–15. doi: 10.1186/s12284-019-0364-0
Gutierrez, L., Mongelard, G., Floková, K., Păcurar, D. I., Novák, O., Staswick, P., et al. (2012). Auxin controls Arabidopsis adventitious root initiation by regulating jasmonic acid homeostasis. Plant Cell 24 (6), 2515–2527. doi: 10.1105/tpc.112.099119
Higuchi, M., Pischke, M. S., Mähönen, A. P., Miyawaki, K., Hashimoto, Y., Seki, M., et al. (2004). In planta functions of the Arabidopsis cytokinin receptor family. Proc. Natl. Acad. Sci. 101 (23), 8821–8826. doi: 10.1073/pnas.0402887101
Hodge, A., Berta, G., Doussan, C., Merchan, F., Crespi, M. (2009). Plant root growth, architecture and function. Plant and Soil 321, 153–187. doi: 10.1007/s11104-009-9929-9
Hossain, M. S., Joshi, T., Stacey, G. (2015). System approaches to study root hairs as a single cell plant model: current status and future perspectives. Front. Plant Sci. 6, 36. doi: 10.3389/fpls.2015.00363
Huot, B., Yao, J., Montgomery, B. L., He, S. Y. (2014). Growth–defense tradeoffs in plants: a balancing act to optimize fitness. Mol. Plant 7 (8), 1267–1287. doi: 10.1093/mp/ssu049
Ioio, R. D., Linhares, F. S., Sabatini, S. (2008a). Emerging role of cytokinin as a regulator of cellular 5differentiation. Curr. Opin. Plant Biol. 11 (1), 23–27. doi: 10.1016/j.pbi.2007.10.006
Ioio, R. D., Linhares, F. S., Scacchi, E., Casamitjana-Martinez, E., Heidstra, R., Costantino, P., et al. (2007). Cytokinins determine Arabidopsis root-meristem size by controlling cell differentiation. Curr. Biol. 17 (8), 678–682. doi: 10.1016/j.cub.2007.02.047
Ioio, R. D., Nakamura, K., Moubayidin, L., Perilli, S., Taniguchi, M., Morita, M. T., et al (2008b). A genetic framework for the control of cell division and differentiation in the root meristem. Science 322 (5906), 1380–1384. doi: 10.1126/science.1164147
Iqbal, M. A. (2014). Improving the growth and yield of Canola (Brassica napus L.) with seed treatment and foliar sprays of Brassica (Brassica naups L.) and moringa (Moringa olifera L.) leaf extracts. American-Eurasian J. Agric. Environ. Sci. 14 (10), 1067–1073. doi: 10.5829/idosi.aejaes.2014.14.10.12429
Irving, T. B., Chakraborty, S., Maia, L. G. S., Knaack, S., Conde, D., Schmidt, H. W., et al. (2022). An LCO-responsive homolog of NODULE INCEPTION positively regulates lateral root formation in Populus sp. Plant Physiol. 190 (3), 1699–1714. doi: 10.1093/plphys/kiac356
Jain, A., Poling, M. D., Karthikeyan, A. S., Blakeslee, J. J., Peer, W. A., Titapiwatanakun, B., et al. (2007). Differential effects of sucrose and auxin on localized phosphate deficiency-induced modulation of different traits of root system architecture in Arabidopsis. Plant Physiol. 144 (1), 232–247. doi: 10.1104/pp.106.092130
Jansen, L., Hollunder, J., Roberts, I., Forestan, C., Fonteyne, P., Van Quickenborne, C., et al. (2013). Comparative transcriptomics as a tool for the identification of root branching genes in maize. Plant Biotechnol. J 11 (9), 1092–1102. doi: 10.1111/pbi.12104
Jansen, L., Roberts, I., De Rycke, R., Beeckman, T. (2012). Phloem-associated auxin response maxima determine radial positioning of lateral roots in maize. Philos. Trans. R. Soc. B: Biol. Sci. 367 (1595), 1525–1533. doi: 10.1098/rstb.2011.0239
Jenness, M. K., Carraro, N., Pritchard, C. A., Murphy, A. S. (2019). The Arabidopsis ATP-BINDING CASSETTE transporter ABCB21 regulates auxin levels in cotyledons, the root pericycle, and leaves. Front. Plant Sci. 10, 806. doi: 10.3389/fpls.2019.00806
Jiang, D., Chen, W., Dong, J., Li, J., Yang, F., Wu, Z., et al. (2018). Overexpression of miR164b-resistant OsNAC2 improves plant architecture and grain yield in rice. J. Exp. Bot. 69 (7), 1533–1543. doi: 10.1093/jxb/ery017
Kapulnik, Y., Delaux, P.-M., Resnick, N., Mayzlish-Gati, E., Wininger, S., Bhattacharya, C., et al. (2011). Strigolactones affect lateral root formation and root-hair elongation in Arabidopsis. Planta 62 (8), 2915–2924. doi: 10.1007/s00425-010-1310-y
Khandal, H., Gupta, S. K., Dwivedi, V., Mandal, D., Sharma, N. K., Vishwakarma, N. K., et al. (2020). Root-specific expression of chickpea cytokinin oxidase/dehydrogenase 6 leads to enhanced root growth, drought tolerance and yield without compromising nodulation. Plant Biotechnol. J. 18 (11), 2225–2240. doi: 10.1111/pbi.13378
Kitomi, Y., Ito, H., Hobo, T., Aya, K., Kitano, H., Inukai, ,. Y. (2011). The auxin responsive AP2/ERF transcription factor CROWN ROOTLESS5 is involved in crown root initiation in rice through the induction of OsRR1, a type-A response regulator of cytokinin signaling. Plant J. 67 (3), 472–484. doi: 10.1111/j.1365-313X.2011.04610.x
Kohlen, W., Charnikhova, T., Lammers, M., Pollina, T., Tóth, P., Haider, I., et al. (2012). The tomato CAROTENOID CLEAVAGE DIOXYGENASE 8 (S l CCD 8) regulates rhizosphere signaling, plant architecture and affects reproductive development through strigolactone biosynthesis. New Phytol. 196 (2), 535–547. doi: 10.1111/j.1469-8137.2012.04265.x
Koltai, H., Dor, E., Hershenhorn, J., Joel, D. M., Weininger, S., Lekalla, S., et al. (2010). Strigolactones’ effect on root growth and root-hair elongation may be mediated by auxin-efflux carriers. J. Plant Growth Regul. 29, 129–136. doi: 10.1007/s00344-009-9122-7
Koltai, H. (2011). Strigolactones are regulators of root development. New Phytologist 130 (3), 545–549. doi: 10.1111/j.1469-8137.2011.03678.x
Kumar, M., Pandya-Kumar, N., Kapulnik, Y., Koltai, H. (2015). Strigolactone signaling in root development and phosphate starvation. Plant Signaling Behav. 10 (7), e1045174. doi: 10.1080/15592324.2015.1045174
Kurepa, J., Shull, T. E., Karunadasa, S. S., Smalle, J. A. (2018). Modulation of auxin and cytokinin responses by early steps of the phenylpropanoid pathway. BMC Plant Biol. 18, 1–15. doi: 10.1186/s12870-018-1477-0
Laplaze, L., Benkova, E., Casimiro, I., Maes, L., Vanneste, S., Swarup, R., et al. (2007). Cytokinins act directly on lateral root founder cells to inhibit root initiation. Plant Cell 19 (12), 3889–3900. doi: 10.1105/tpc.107.055863
Li, S.-W. (2021). Molecular bases for the regulation of adventitious root generation in plants. Front. Plant Sci. 12, 614072. doi: 10.3389/fpls.2021.614072
Li, X. J., Chen, X. J., Guo, X., Yin, L. L., Ahammed, G. J., Xu, C. J., et al. (2016). DWARF overexpression induces alteration in phytohormone homeostasis, development, architecture and carotenoid accumulation in tomato. Plant Biotechnol. J. 14 (3), 1021–1033. doi: 10.1111/pbi.12474
Li, K., Liang, Y., Xing, L., Mao, J., Liu, Z., Dong, F., et al. (2018). Transcriptome analysis reveals multiple hormones, wounding and sugar signaling pathways mediate adventitious root formation in apple rootstock. Int. J. Mol. Sci. 19 (8), 2201. doi: 10.3390/ijms19082201
Li, X., Mo, X., Shou, H., Wu, P. (2006). Cytokinin-mediated cell cycling arrest of pericycle founder cells in lateral root initiation of Arabidopsis. Plant Cell Physiol. 47 (8), 1112–1123. doi: 10.1093/pcp/pcj082
Lin, Q., Gong, J., Zhang, Z., Meng, Z., Wang, J., Wang, S., et al. (2023). The Arabidopsis thaliana trehalose-6-phosphate phosphatase gene AtTPPI regulates primary root growth and lateral root elongation. Front. Plant Sci. 13, 1088278. doi: 10.3389/fpls.2022.1088278
Liu, J., Novero, M., Charnikhova, T., Ferrandino, A., Schubert, A., Ruyter-Spira, C., et al. (2013). Carotenoid cleavage dioxygenase 7 modulates plant growth, reproduction, senescence, and determinate nodulation in the model legume Lotus japonicus. J. Exp. Bot. 64 (7), 1967–1981.
Liu, J., Sheng, L., Xu, Y., Li, J., Yang, Z., Huang, H., et al. (2014). WOX11 and 12 are involved in the first-step cell fate transition during de novo root organogenesis in Arabidopsis. Plant Cell 26 (3), 1081–1093. doi: 10.1105/tpc.114.122887
Lo, S. F., Ho, T. H. D., Liu, Y. L., Jiang, M. J., Hsieh, K. T., Chen, K. T., et al. (2017). Ectopic expression of specific GA 2 oxidase mutants promotes yield and stress tolerance in rice. Plant Biotechnol. J. 15 (7), 850–864. doi: 10.1111/pbi.12681
Luo, X., Xu, J., Zheng, C., Yang, Y., Wang, L., Zhang, R., et al. (2023). Abscisic acid inhibits primary root growth by impairing ABI4-mediated cell cycle and auxin biosynthesis. Plant Physiol. 191 (1), 265–279. doi: 10.1093/plphys/kiac407
Mao, C., He, J., Liu, L., Deng, Q., Yao, X., Liu, C., et al. (2020). OsNAC2 integrates auxin and cytokinin pathways to modulate rice root development. Plant Biotechnol. J. 18 (2), 429–442. doi: 10.1111/pbi.13209
Mao, H., Li, S., Wang, Z., Cheng, X., Li, F., Mei, F., et al. (2020). Regulatory changes in TaSNAC8-6A are associated with drought tolerance in wheat seedlings. Plant Biotechnol. J. 18 (4), 1078–1092. doi: 10.1111/pbi.13277
Mao, J.-L., Miao, Z.-Q., Wang, Z., Yu, L.-H., Cai, X.-T., Xiang, C.-B. (2016). Arabidopsis ERF1 mediates cross-talk between ethylene and auxin biosynthesis during primary root elongation by regulating ASA1 expression. PloS Genet. 12 (1), e1005760. doi: 10.1371/journal.pgen.1005760
Márquez, G., Alarcón, M. V., Salguero, J. (2019). Cytokinin inhibits lateral root development at the earliest stages of lateral root primordium initiation in maize primary root. J. Plant Growth Regul. 38 (1), 83–92. doi: 10.1007/s00344-018-9811-1
Marzec, M., Gruszka, D., Tylec, P., Szarejko, I. (2016). Identification and functional analysis of the HvD14 gene involved in strigolactone signaling in Hordeum vulgare. Physiologia Plantarum 158 (3), 341–355. doi: 10.1111/ppl.12460
Mason, M. G., Mathews, D. E., Argyros, D. A., Maxwell, B. B., Kieber, J. J., Alonso, J. M., et al. (2005). Multiple type-B response regulators mediate cytokinin signal transduction in Arabidopsis. Plant Cell 17 (11), 3007–3018. doi: 10.1105/tpc.105.035451
Michniewicz, M., Ho, C. H., Enders, T. A., Floro, E., Damodaran, S., Gunther, L. K., et al. (2019). TRANSPORTER OF IBA1 links auxin and cytokinin toinfluence root architecture. Dev. Cell 50, 599–609.e4. doi: 10.1016/j.devcel.2019.06.010
Miguel, M. A., Postma, J. A., Lynch, J. P. (2015). Phene synergism between root hair length and basal root growth angle for phosphorus acquisition. Plant Physiol. 167 (4), 1430–1439. doi: 10.1104/pp.15.00145
Moreno-Risueno, M. A., Van Norman, J. M., Moreno, A., Zhang, J., Ahnert, S. E., Benfey, P. N. (2010). Oscillating gene expression determines competence for periodic Arabidopsis root branching. Science 329 (5997), 1306–1311. doi: 10.1126/science.1191937
Motte, H., Vanneste, S., Beeckman, T. (2019). Molecular and environmental regulation of root development. Annu. Rev. Plant Biol. 70, 465–488. doi: 10.1146/annurev-arplant-050718-100423
Nakagami, S., Aoyama, T., Sato, Y., Kajiwara, T., Ishida, T., Sawa, S. (2023). CLE3 and its homologs share overlapping functions in the modulation of lateral root formation through CLV1 and BAM1 in Arabidopsis thaliana. Plant J. 113 (6), 1176–1191. doi: 10.1111/tpj.16103
Nan, W., Wang, X., Yang, L., Hu, Y., Wei, Y., Liang, X., et al. (2014). Cyclic GMP is involved in auxin signalling during Arabidopsis root growth and development. J. Exp. Bot. 65 (6), 1571–1583. doi: 10.1093/jxb/eru019
Nikonorova, N., Murphy, E., Fonseca de Lima, C. F., Zhu, S., van de Cotte, B., Vu, L. D., et al. (2021). Auxin-triggered changes in the Arabidopsis root tip (phospho) proteome reveal novel root growth regulators. bioRxiv, 1–47. doi: 10.1101/2021.03.04.433936
Niu, S., Li, Z., Yuan, H., Fang, P., Chen, X., Li, W. (2013). Proper gibberellin localization in vascular tissue is required to regulate adventitious root development in tobacco. J. Exp. Bot. 64 (11), 3411–3424.
Ogura, T., Goeschl, C., Filiault, D., Mirea, M., Slovak, R., Wolhrab, B., et al. (2019). Root system depth in Arabidopsis is shaped by EXOCYST70A3 via the dynamic modulation of auxin transport. Cell 178 (2), 400–412. e416. doi: 10.1016/j.cell.2019.06.021
Peng, Y., Jiang, S., Wang, J., Xu, X., Gong, X., Jin, W., et al. (2023). Control of lateral root initiation by DA3 in Arabidopsis. Cell Rep. 42 (1), 111913. doi: 10.1016/j.celrep.2022.111913
Qin, H., He, L., Huang, R. (2019a). The coordination of ethylene and other hormones in primary root development. Front. Plant Sci. 10. doi: 10.3389/fpls.2019.00874
Qin, H., Wang, J., Chen, X., Wang, F., Peng, P., Zhou, Y., et al. (2019b). Rice Os DOF 15 contributes to ethylene-inhibited primary root elongation under salt stress. New Phytol. 223 (2), 798–813. doi: 10.1111/nph.15824
Ramirez-Carvajal, G. A., Morse, A. M., Dervinis, C., Davis, J.M. (2009). The cytokinin type-B response regulator PtRR13 is a negative regulator of adventitious root development in Populus. Plant Physiol. 150, 759–771. doi: 10.1104/pp.109.137505
Rasmussen, A., Mason, M. G., De Cuyper, C., Brewer, P. B., Herold, S., Agusti, J., et al. (2012). Strigolactones suppress adventitious rooting in Arabidopsis and pea. Plant Physiol. 158 (4), 1976–1987. doi: 10.1104/pp.111.187104
Ruyter-Spira, C., Kohlen, W., Charnikhova, T., van Zeijl, A., van Bezouwen, L., De Ruijter, N., et al. (2011). Physiological effects of the synthetic strigolactone analog GR24 on root system architecture in Arabidopsis: another belowground role for strigolactones? Plant Physiol. 155 (2), 721–734. doi: 10.1104/pp.110.166645
Schoenaers, S., Balcerowicz, D., Breen, G., Hill, K., Zdanio, M., Mouille, G., et al. (2018). The auxin-regulated CrRLK1L kinase ERULUS controls cell wall composition during root hair tip growth. Curr. Biol. 28 (5), 722–732. e726. doi: 10.1016/j.cub.2018.01.050
Sharma, M., Singh, D., Saksena, H. B., Sharma, M., Tiwari, A., Awasthi, P., et al. (2021). Understanding the intricate web of phytohormone signalling in modulating root system architecture. Int. J. Mol. Sci. 22 (11), 5508. doi: 10.3390/ijms22115508
Shibata, M., Favero, D. S., Takebayashi, R., Takebayashi, A., Kawamura, A., Rymen, B., et al. (2022). Trihelix transcription factors GTL1 and DF1 prevent aberrant root hair formation in an excess nutrient condition. New Phytol. 235 (4), 1426–1441. doi: 10.1111/nph.18255
Shinohara, N., Taylor, C., Leyser, O. (2013). Strigolactone can promote or inhibit shoot branching by triggering rapid depletion of the auxin efflux protein PIN1 from the plasma membrane. PloS Biol. 11 (1), e1001474. doi: 10.1371/journal.pbio.1001474
Šimášková, M., O’Brien, J. A., Khan, M., Van Noorden, G., Ötvös, K., Vieten, A., et al. (2015). Cytokinin response factors regulate PIN- FORMED auxin transporters. Nat. Commun. 6 (1), 8717. doi: 10.1038/ncomms9717
Singh, S., Yadav, S., Singh, A., Mahima, M., Singh, A., Gautam, V., et al. (2020). Auxin signaling modulates LATERAL ROOT PRIMORDIUM 1 (LRP 1) expression during lateral root development in Arabidopsis. Plant J. 101 (1), 87–100. doi: 10.1111/tpj.14520
Srivastava, S., Upadhyay, M. K., Srivastava, A. K., Abdelrahman, M., Suprasanna, P., Tran, L. P. (2018). Cellular and subcellular phosphate transport machinery in plants. Int. J. Mol. Sci. 19, 1914. doi: 10.3390/ijms19071914
Steffens, B., Rasmussen, A. (2016). The physiology of adventitious roots. Plant Physiol. 170 (2), 603–631 617. doi: 10.1104/pp.15.01360
Sun, C., Li, D., Gao, Z., Gao, L., Shang, L., Wang, M., et al. (2022). OsRLR4 binds to the OsAUX1 promoter to negatively regulate primary root development in rice. J. Integr. Plant Biol. 64 (1), 118–134. doi: 10.1111/jipb.13183
Sun, H., Tao, J., Gu, P., Xu, G., Zhang, Y. (2016). The role of strigolactones in root development. Plant Signaling Behav. 11 (1), e1110662. doi: 10.1080/15592324.2015.1110662
Sun, H., Tao, J., Hou, M., Huang, S., Chen, S., Liang, Z., et al. (2015). A strigolactone signal is required for adventitious root formation in rice. Ann. Bot. 115 (7), 642 1155–1162. doi: 10.1093/aob/mcv052
Sun, H., Tao, J., Liu, S., Huang, S., Chen, S., Xie, X., et al. (2014). Strigolactones are involved in phosphate-and nitrate-deficiency-induced root development and auxin transport in rice. J. Exp. Bot. 65 (22), 6735–6746. doi: 10.1093/jxb/eru029
Svolacchia, N., Salvi, E., Sabatini, S. (2020). Arabidopsis primary root growth: let it grow, can't hold it back anymore! Curr. Opin. Plant Biol. 57, 133–141. doi: 10.1016/j.pbi.2020.08.005
Takatsuka, H., Sasaki, A., Takahashi, N., Shibata, M., Sugimoto, K., Tanaka, M., et al. (2023). Cytokinin signaling promotes root hair growth by directly regulating RSL4 expression. J. Exp. Bot. 74 (12), 3579-3594. doi: 10.1093/jxb/erad091
Tang, J., Chu, C. (2017). MicroRNAs in crop improvement: fine-tuners for complex traits. Nat. Plants 3 (7), 1–11. doi: 10.1038/nplants.2017.77
Tian, J., Wang, C., Xia, J., Wu, L., Xu, G., Wu, W., et al. (2019). Teosinte ligule allele narrows plant architecture and enhances high-density maize yields. Science 6454), 658–664. doi: 10.1126/science.aax5482
Tiwari, M., Bhatia, S. (2020). Expression profiling of miRNAs indicates crosstalk between phytohormonal response and rhizobial infection in chickpea. J. Plant Biochem. Biotechnol. 29, 380–394. doi: 10.1007/s13562-019-00545-9
To, J. P., Haberer, G., Ferreira, F. J., Deruere, J., Mason, M. G., Schaller, G. E., et al. (2004). Type-A Arabidopsis response regulators are partially redundant negative regulators of cytokinin signaling. Plant Cell 16 (3), 658–671. doi: 10.1105/tpc.018978
Tötzke, C., Kardjilov, N., Lenoir, N., Manke, I., Oswald, S. E., Tengattini, A. (2019). What comes NeXT?–high-speed neutron tomography at ILL. Optics Express 27 (20), 28640–28648. doi: 10.1364/OE.27.028640
Veerappa, R., Slocum, R. D., Siegenthaler, A., Wang, J., Clark, G., Roux, S. J. (2019). Ectopic expression of a pea apyrase enhances root system architecture and drought survival in Arabidopsis and soybean. Plant Cell Environ. 42 (1), 337–353. doi: 10.1111/pce.13425
Wada, T., Kurata, T., Tominaga, R., Koshino-Kimura, Y., Tachibana, T., Goto, K., et al. (2002). Role of a positive regulator of root hair development, CAPRICE, in Arabidopsis root epidermal cell differentiation. Development 129 (23), 5409–5419. doi: 10.1242/dev.00111
Wang, H., Hu, Z., Huang, K., Han, Y., Zhao, A., Han, H., et al. (2018). Three genomes differentially contribute to the seedling lateral root number in allohexaploid wheat: evidence from phenotype evolution and gene expression. Plant J. 95 (6), 976–987. doi: 10.1111/tpj.14005
Wang, J., Wang, R., Mao, X., Li, L., Chang, X., Zhang, X., et al. (2019). TaARF4 genes are linked to root growth and plant height in wheat. Ann. Bot. 124 (6), 903–915. doi: 10.1093/aob/mcy218
Wang, Y., Yang, W., Zuo, Y., Zhu, L., Hastwell, A. H., Chen, L., et al. (2019). GmYUC2a mediates auxin biosynthesis during root development and nodulation in soybean. J. Exp. Bot. 70 (12), 3165–3176. doi: 10.1093/jxb/erz144
Wang, J., Yu, H., Xiong, G., Lu, Z., Jiao, Y., Meng, X., et al. (2017). Tissue-specific ubiquitination by IPA1 INTERACTING PROTEIN1 modulates IPA1 protein levels to regulate plant architecture in rice. Plant Cell 29 (4), 697–707. doi: 10.1105/tpc.16.00879
Werner, T. S., Motyka, V., Laucou, V., Smets, R., Van Onckelen, H., Schmülling, T. (2003). Cytokinin-deficient transgenic Arabidopsis plants show multiple developmental alterations indicating opposite functions of cytokinins in the regulation of shoot and root meristem activity. Plant Cell 15 (11), 2532–2550. doi: 10.1105/tpc.014928
Werner, T., Schmülling, T. (2009). Cytokinin action in plant development. Curr. Opin. Plant Biol. 12 (5), 527–538. doi: 10.1016/j.pbi.2009.07.002
Willige, B. C., Ghosh, S., Nill, C., Zourelidou, M., Dohmann, E. M., Maier, A., et al. (2007). The DELLA domain of GA INSENSITIVE mediates the interaction with the GA INSENSITIVE DWARF1A gibberellin receptor of Arabidopsis. Plant Cell 19 (4), 1209–1220. doi: 10.1105/tpc.107.051441
Wilmoth, J. C., Wang, S., Tiwari, S. B., Joshi, A. D., Hagen, G., Guilfoyle, T. J., et al. (2005). NPH4/ARF7 and ARF19 promote leaf expansion and auxin-induced lateral root formation. Plant J. 43 (1), 118–130. doi: 10.1111/j.1365-313X.2005.02432.x
Xie, X., Yoneyama, K., Kisugi, T., Nomura, T., Akiyama, K., Asami, T., et al. (2015). Strigolactones are transported from roots to shoots, although not through the xylem. J. Pesticide Sci. 40 (4), 214–216. doi: 10.1584/jpestics.D15-045
Xie, X., Yoneyama, K., Yoneyama, ,. K. (2010). The strigolactone story. Annu. Rev. Phytopathol. 48, 93–117. doi: 10.1146/annurev-phyto-073009-114453
Xu, M., Xie, W., Huang, M. (2015). Two WUSCHEL-related HOMEOBOX genes, PeWOX11a and PeWOX11b, are involved in adventitious root formation of poplar. Physiol. Plant 155, 446–456. doi: 10.1111/ppl.12349
Zhang, S., Huang, L., Yan, A., Liu, Y., Liu, B., Yu, C., et al. (2016). Multiple phytohormones promote root hair elongation by regulating a similar set of genes in the root epidermis in Arabidopsis. J. Exp. Bot. 67, (22) 6363–6372.
Zhang, W., To, J. P., Cheng, C. Y., Eric Schaller, G., Kieber, J. J. (2011). Type-A response regulators are required for proper root apical meristem function through post-transcriptional regulation of PIN auxin efflux carriers. Plant J. 68 (1), 1–10.
Zhang, G., Xu, N., Chen, H., Wang, G., Huang, J. (2018). OsMADS25 regulates root system development via auxin signalling in rice. Plant J. 95 (6), 1004–1022.
Zhang, F., Tao, W., Sun, R., Wang, J., Li, C., Kong, X., et al. (2020). PRH1 mediates ARF7-707 LBD dependent auxin signaling to regulate lateral root development in Arabidopsis thaliana. PloS Genetics 16 (2), e1008044. doi: 10.1371/journal.pgen.1008044
Zhao, Y., Hu, Y., Dai, M., Huang, L., Zhou, D.-X. (2009). The WUSCHEL-related homeobox gene WOX11 is required to activate shoot-borne crown root development in rice. Plant Cell 21 (3), 736–748.
Zhao, J., Zheng, L., Wei, J., Wang, Y., Chen, J., Zhou, Y., et al. (2022). The soybean PLATZ transcription factor GmPLATZ17 suppresses drought tolerance by interfering with stress-associated gene regulation of GmDREB5. Crop J. 10 (4), 1014–1025. doi: 10.1016/j.cj.2022.03.009
Keywords: auxin crosstalk, auxin-CK, auxin-SL interaction, phytohormones, root architecture, root development
Citation: Jan M, Muhammad S, Jin W, Zhong W, Zhang S, Lin Y, Zhou Y, Liu J, Liu H, Munir R, Yue Q, Afzal M and Wang G (2024) Modulating root system architecture: cross-talk between auxin and phytohormones. Front. Plant Sci. 15:1343928. doi: 10.3389/fpls.2024.1343928
Received: 24 November 2023; Accepted: 08 January 2024;
Published: 08 February 2024.
Edited by:
Lei Huang, Purdue University, United StatesReviewed by:
Faisal Islam, Jiangsu University, ChinaCopyright © 2024 Jan, Muhammad, Jin, Zhong, Zhang, Lin, Zhou, Liu, Liu, Munir, Yue, Afzal and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Muhammad Afzal, bWFmemFsQHNjYXUuZWR1LmNu; Guoping Wang, Z3B3YW5nQHNjYXUuZWR1LmNu
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.