- 1The Engineering Research Institute of Agriculture and Forestry, Ludong University, Yantai, China
- 2State Key Laboratory of Tree Genetics and Breeding, Research Institute of Forestry, Chinese Academy of Forestry, Beijing, China
- 3State Key Laboratory of Subtropical Silviculture, College of Forestry and Biotechnology, Zhejiang A&F University, Hangzhou, China
Introduction: Hybrid poplars are industrial trees in China. An understanding of the molecular mechanism underlying wood formation in hybrid poplars is necessary for molecular breeding. Although the division and differentiation of vascular cambial cells is important for secondary growth and wood formation, the regulation of this process is largely unclear.
Methods: In this study, mPagGRF15 OE and PagGRF15-SRDX transgenic poplars were generated to investigate the function of PagGRF15. RNA-seq and qRT-PCR were conducted to analyze genome-wide gene expression, while ChIP‒seq and ChIP-PCR were used to identified the downstream genes regulated by PagGRF15.
Results and discussion: We report that PagGRF15 from hybrid poplar (Populus alba × P. glandulosa), a growth-regulating factor, plays a critical role in the regulation of vascular cambium activity. PagGRF15 was expressed predominantly in the cambial zone of vascular tissue. Overexpression of mPagGRF15 (the mutated version of GRF15 in the miR396 target sequence) in Populus led to decreased plant height and internode number. Further stem cross sections showed that the mPagGRF15 OE plants exhibited significant changes in vascular pattern with an increase in xylem and a reduction in phloem. In addition, cambium cell files were decreased in the mPagGRF15 OE plants. However, dominant suppression of the downstream genes of PagGRF15 using PagGRF15-SRDX showed an opposite phenotype. Based on the RNA-seq and ChIP-seq results, combining qRT-PCR and ChIP-PCR analysis, candidate genes, such as WOX4b, PXY and GID1.3, were obtained and found to be mainly involved in cambial activity and xylem differentiation. Accordingly, we speculated that PagGRF15 functions as a positive regulator mediating xylem differentiation by repressing the expression of the WOX4a and PXY genes to set the pace of cambial activity. In contrast, PagGRF15 mediated the GA signaling pathway by upregulating GID1.3 expression to stimulate xylem differentiation. This study provides valuable information for further studies on vascular cambium differentiation mechanisms and genetic improvement of the specific gravity of wood in hybrid poplars.
1 Introduction
As one of the fastest-growing industrial trees, hybrid poplars are suitable for cultivation (Sannigrahi et al., 2010). Their widespread distribution also makes them important contributors to ecological diversity. Annotating the poplar genome has provided a way to breed new clones optimized for timber production (Sannigrahi et al., 2010) and has made poplars model plants for tree molecular biology and biotechnology (Brunner et al., 2004). Changes in the cambium during growth, water availability, insect and bacterial attacks, gravitropic effects, and other environmental influences are key energy-related characteristics of lignocellulosic feedstock and can be enhanced by genetic modifications (Sannigrahi et al., 2010). The specific gravity of wood and its lignin and cellulose contents have been proposed as prime targets for genetic modification (Dinus, 2001; Sannigrahi et al., 2010).
Cell proliferation of the vascular cambium generates wood (Smetana et al., 2019). The vascular cambium is a lateral meristem responsible for the production of secondary xylem (wood) inward and secondary phloem outward during secondary growth (Włoch et al., 2023). The differentiation of vascular cambium to xylem and phloem occurs underlying a series of highly ordered and complicated developmental process, which involves initiation, patterning and direction of vascular tissue differentiation (Johnsson and Fischer, 2016). Thus, the pace of the division and differentiation of vascular cambial cells is a rate-limiting step for wood formation. However, the underlying molecular mechanism that regulates vascular cambium expansion and xylem differentiation is not well understood.
Growth-regulating factors (GRFs) are plant-specific transcription factors that are involved in various developmental events and mainly function in organ size control (Hepworth and Lenhard, 2014; Kim, 2019). GRFs contain conserved QLQ and WRC domains in their N-terminal regions, which were the most distinctive characteristics of the family members in plant species (Kuijt et al., 2014). The QLQ domain mediates protein-protein interaction, whereas the WRC domain is necessary for DNA-binding (Omidbakhshfard et al., 2015). Currently, the GRF family consists of 9 genes in Arabidopsis thaliana (Kim et al., 2003) and 19 genes in Populus trichocarpa (Cao et al., 2016). Most GRFs are the target of miR396, and they work with miR396 to regulate the development of leaves, roots, stems, flowers and seeds (Kim and Tsukaya, 2015; Omidbakhshfard et al., 2015; Kim, 2019). Notably, the function of GRFs in meristem development has increasingly attracted attention of the researchers. Previous studies showed that grf1/2/3/4 displays a shoot meristemless phenotype (Kim and Lee, 2006), but the root apical meristem (RAM) size was enlarged in grf1/2/3 Arabidopsis, and the miR396-GRF-PLETHORA (PLT) module was required for the transition of stem cells into transit-amplifying cells to establish the boundary between the stem cell niche and the transit-amplifying region (Rodriguez et al., 2015). GRF directly binds to the promotor of KNOX, a key regulator involved in meristem activity (Kuijt et al., 2014). In addition, GRF-interacting factor 1 (GIF1) can interact with GRFs to regulate meristem determinacy in maize (Zhang et al., 2018). These results suggest that GRFs play an essential role in apical meristem activity by interacting with other regulators. In hybrid poplar 84K (Populus alba × P. glandulosa), we previously reported that PagGRF12a interacted with PagGIF1b to control PagXND1a expression and thereby inhibited xylem development (Wang et al., 2021). Recently, a novel module for GRF15 regulating secondary vascular development was reported in P. tomentosa (Wang et al., 2023). The PtoTCP20-miR396d-PtoGRF15 module mediated vascular development during the transition from primary to secondary vascular development. PtoTCP20 activated the transcription of miR396d to downregulate PtoGRF15 expression. 35S:PtoGRF15 plants exhibited delayed secondary growth, indicating that GRF15 was a negative regulator of secondary vascular development in P. tomentosa (Wang et al., 2023).
In a previous study, we reported that PagGRF15 is highly expressed in leaves and can modulate leaf development by regulating expansin genes (Zhou et al., 2019). Overexpression of PagGRF15 caused significantly enlarged leaves and increased palisade cell size in mPagGRF15 OE plants, whereas PagGRF15-SRDX plants exhibited significantly decreased leaf size and palisade cell area. PagGRF15-SRDX plants have opposite effects to the overexpressing lines, because expression of genes downstream of PagGRF15 would be repressed by SRDX, an active transcriptional repression domain. Moreover, PagGRF15 exhibits the highest expression in stems among PagGRFs (Zhou et al., 2019), implying that PagGRF15 may play a role in the regulation of vascular cambium activity, which is worth further investigation. In this study, we investigated the role of PagGRF15 in stem development. Overexpression of PagGRF15 in Populus resulted in increased xylem production and decreased phloem tissue growth, while PagGRF15-dominant repressed transgenic plants demonstrated the opposite phenotype. The genes involved in vascular cambium differentiation are targeted by PagGRF15. These findings increase the understanding of vascular differentiation mechanisms in hybrid poplar 84K and provide information to support improvements in hybrid poplar cultivation and the specific gravity of wood by genetic modification.
2 Materials and methods
2.1 Plant materials
Hybrid poplar 84K was used for gene cloning and genetic transformation. mPagGRF15OE plants, PagGRF15pro::GUS plants and PagGRF15 SRDX plants were obtained as previously reported (Zhou et al., 2019). All plants were cultivated in a phytotron with a light and dark cycle of 16 h and 8 h at 22°C-25°C.
2.2 GUS staining
GUS staining was performed as previously reported (Zhou et al., 2019). Briefly, 1-month-old PagGRF15pro:GUS plants were incubated in 90% acetone (v/v) for at least 2 h on ice and then incubated in GUS staining solution (50 mM sodium phosphate, 2 mM potassium ferricyanide, 2 mM potassium ferrocyanide, 0.2% Triton X-100, and 1 mM X-Gluc) for at least 2 h at 37°C. Following staining, plants were cleared with 95% ethanol and preserved in 75% ethanol and photographed. Sections were obtained using a vibratome (VT1000S; Leica, Wetzlar, Germany) at a thickness of 30 μm and photographed using an Axio Imager A1 microscope (Zeiss).
2.3 RNA isolation and qRT-PCR
For RNA-seq, samples were collected by scraping the juicy materials from the surface of debarked bark of two-year-old CK and PagGRF15 transgenic plants grown in the experimental field. Then, total RNA was isolated using the RNeasy Plant Mini Kit (QIAGEN). Six independent cDNA libraries of CK- and mPagGRF15-overexpressing plants were sequenced using an Illumina Solexa sequencing platform. Then, RNA-seq reads were aligned to the P. trichocarpa genome (Phytozome 10.0). Genes with |log2fold change (FC)| > 1 and false discovery rate (FDR) ≤ 0.05 in all three biological replicates were designated as differentially expressed. For qRT-PCR, actin7 (Potri.001G309500, Tang et al., 2019) was used as the internal control.
2.4 Microscopy
Different internodes of CK and PagGRF15 transgenic plants were fixed in 4% paraformaldehyde (prepared with 1×PBS solution, 0.137 M NaCl, 0.0027 M KCl, 0.008 M Na2HPO4, 0.002 M KH2PO4, pH 7.2–7.4), dehydrated in a graded ethanol series (30%, 50%, 70%, 90% and 100%), exchanged in a graded Spurr’s resin (ERL 4221:D.E.R. 736:NSA:DMAE=4.10:1.43:5.90:0.10, SPI-CHEM™):ethanol series (1:3, 1:1 and 3:1), and polymerized in 100% Spurr’s resin at 60°C. Then, the samples were sectioned using a microtome (Leica EM UC7) with a thickness of 4 μm, stained with 0.05% (w/v) toluidine blue and visualized using a microscope (OLYMPUS BX51, Olympus).
2.5 RNA-seq analysis
For RNA-seq, samples were collected by scraping the juicy materials from the surface of debarked bark of two-year-old CK and PagGRF15 transgenic plants grown in the experimental field. Then, total RNA was isolated using the RNeasy Plant Mini Kit (QIAGEN). Six independent cDNA libraries of CK- and mPagGRF15-overexpressing plants were sequenced using an Illumina Solexa sequencing platform. Then, RNA-seq reads were aligned to the P. trichocarpa genome (Phytozome 10.0). Genes with |log2fold change (FC)| > 1 and false discovery rate (FDR) ≤ 0.05 in all three biological replicates were designated as differentially expressed (BioProject ID: PRJNA693036).
2.6 ChIP-seq and ChIP-qPCR
ChIP assays were performed as described previously (Li et al., 2014). Briefly, 2.5 g stems of 1-month-old 35S:GFP-PagGRF15 plants were collected and ground to a fine powder in liquid nitrogen. The powder was transferred to 20 ml cross-linking buffer for a total of 30 min under vacuum at room temperature. 2.5 ml of 2 M glycine was added to quench the cross-linking reaction, and the solution centrifuged at 1,800g for 10 min. The pellet was resuspended in 30 ml buffer 1 (0.4 M sucrose, 10 mM Tris-HCl (pH 8.0), 5 mM β-mercaptoethanol, 1 mM PMSF, 1 µg/ml pepstatin A and 1 µg/ml leupeptin) and the mixture was gradually passed through three layers of Miracloth into a new, ice-cold 50-ml conical tube, and the Miracloth squeezed to collect all of the liquid. The solution was centrifuged at 1,800g for 10 min at 4°C, the supernatant discarded, and the pellet resuspended in 2 ml buffer 2 (0.25 M sucrose, 10 mM Tris-HCl (pH 8.0), 5 mM β-mercaptoethanol, 10 mM MgCl2, 1% (vol/vol) Triton X-100, 1 mM PMSF, 1 µg/ml pepstatin A and 1 µg/ml leupeptin) and transferred to a 2.0-ml microcentrifuge tube. After centrifugation at 16,000g for 10 min at 4°C, the pellet was resuspended in 400 µl of buffer 3 (1.7 M sucrose, 10 mM Tris–HCl (pH 8.0), 5 mM β-mercaptoethanol, 2 mM MgCl2, 0.15% (vol/vol) Triton X-100, 1 mM PMSF, 1 µg/ml pepstatin A and 1 µg/ml leupeptin). After centrifugation at 16,000g for 10 min at 4°C, the pellet was resuspended in 400 µl of buffer 3 once more, then centrifuged at 16,000g for 1 h at 4°C and the supernatant removed. The pellet (chromatin) was resuspended in 400 µl of lysis buffer (50 mM Tris-HCl (pH 8.0), 10 mM EDTA, 1% (wt/vol) SDS, 1 mM PMSF, 1 µg/ml pepstatin A and 1 µg/ml leupeptin) and sonicated. Then, the sample was centrifuged for chromatin isolation. Chromatin extracted from 35S:GFP-PagGRF15 plants was immunoprecipitated with anti-GFP antibody (No. 11814460001; Roche) with a Plant ChIP-seq kit (Diagenode). Following decrosslinking, isolation, and purification of the immunoprecipitated DNA, libraries were constructed and subjected to sequencing on the Illumina HiSeq 2000 platform. ChIP-seq reads were aligned to the Populus trichocarpa genome (Phytozome 10.0). Peaks were identified as significantly enriched (P < 0.05) in each of the ChIP-seq libraries compared with input DNA. Three independent biological replicates of the ChIP assay were performed. For ChIP-qPCR, SYBR Premix Ex Taq (TaKaRa) was used to quantify DNA targets immunoprecipitated by anti-GFP antibodies relative to input DNA with three biological replicates each with four technical replicates. The primers used are listed in Supplementary Table 5. Actin7 (POPTR_0019s02630.1, Li et al., 2014) was used as the internal control, and the fold enrichment of the DNA target was relative to the input sample.
3 Results
3.1 Expression pattern of PagGRF15 during vascular development
In a previous study, we reported that PagGRF15 was the most highly expressed PagGRF in stem tissue, with a higher expression in young stems than in mature stems (Zhou et al., 2019). GUS staining assays revealed strong signals in young stems (Supplementary Figure 1B), consistent with the expression of GRF15 from the apex to the 5th internode detected through joint PacBio Iso-Seq and RNA-seq analysis (Supplementary Figure 1; Chao et al., 2019). To examine the expression pattern of PagGRF15 in detail, we analyzed cross sections of 1-month-old PagGRF15pro:GUS transgenic seedlings. Cross sections of the 2nd to 5th internodes revealed strong signals in the vascular tissue and were emphasized in the cambial zone (Figure 1). qRT-PCR results also confirmed the expression of PagGRF15 in vascular tissue (Supplementary Figure 1C). These results suggest that PagGRF15 plays a crucial role in vascular development, likely through the regulation of cambium activity.
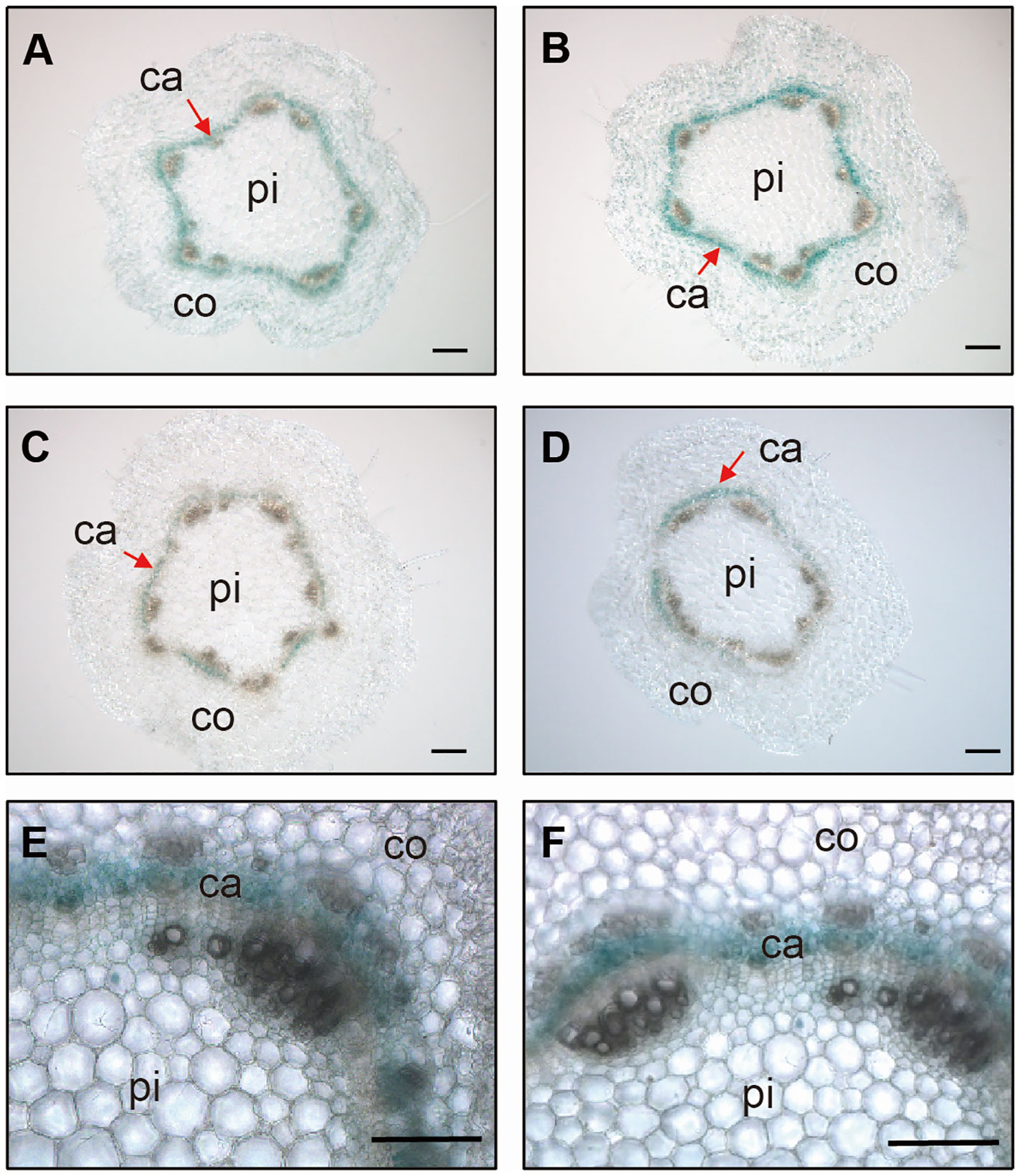
Figure 1 Expression patterns of GRF15 in stems. (A-D) Stem sections of the 2nd (A), 3rd (B), 4th (C) and 5th (D) internodes from 4-week-old PagGRF15pro:GUS transgenic plants. Bars = 100 μm. (E, F) Further cross sections of the 2nd (E) and 3rd (F) internodes showed that GUS signals were concentrated in the vascular cambium. Bars = 100 μm. Ca, cambium; pi, pith; co, cortex.
3.2 Altered PagGRF15 expression led to the alteration of secondary vascular development
We have reported that the height of mPagGRF15 OE plants was decreased, while PagGRF15-SRDX grew slightly higher than the nontransgenic control (CK) plants (Zhou et al., 2019). Statistically, 4-month-old mPagGRF15 OE transgenic plants demonstrated an approximately 16.5% decrease in plant height, while PagGRF15-SRDX plants displayed an approximately 10% increase (Figures 2A, B). The basal diameter, however, was not significantly altered in either PagGRF15 transgenic plant (Figure 2C). We also counted the internode number and measured the internode length of all transgenic lines. The internode number was decreased by 20% in mPagGRF15 OE transgenic plants but increased by 20% in PagGRF15-SRDX plants. The internode length showed no obvious alteration in mPagGRF15 OE transgenic plants but decreased in PagGRF15-SRDX plants compared with CK plants (Figure 2D). Interestingly, the internode length showed no significant difference in mPagGRF15 OE plants but decreased in PagGRF15-SRDX plants (Figure 2D). These results indicated that PagGRF15 could affect stem development by regulating the differentiation and elongation of internodes.
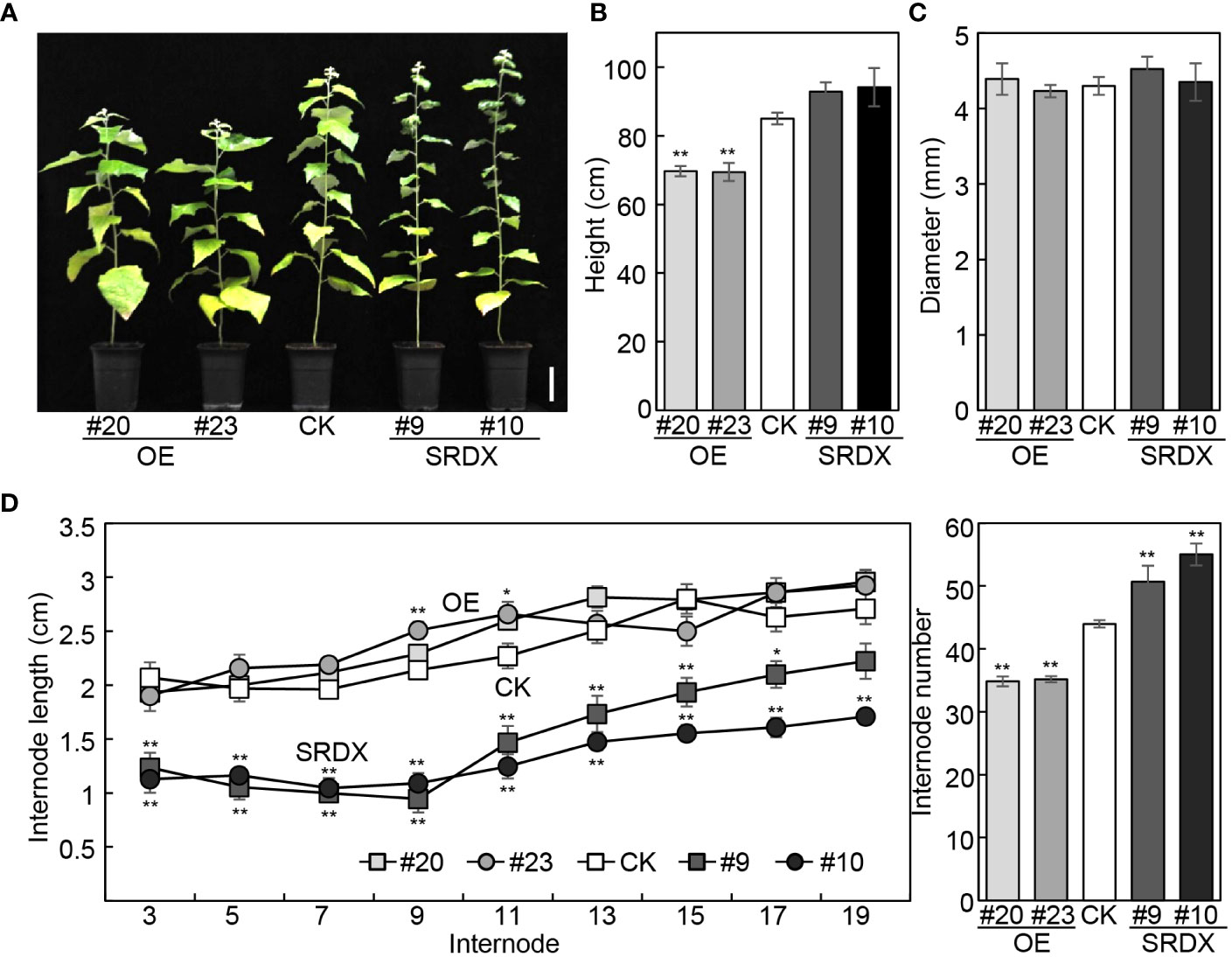
Figure 2 Growth characteristics of mPagGRF15 overexpression (OE) and PagGRF15 dominant repression (SRDX) transgenic plants. (A) Photograph of 4-month-old mPagGRF15 OE and PagGRF15-SRDX transgenic plants. Bar = 10 cm. (B, C) Plant height (B) and basal diameter (C) of 4-month-old mPagGRF15 OE and PagGRF15-SRDX transgenic plants. (D) Internode length and internode number of 4-month-old mPagGRF15 OE and internode number of 4-month-old mPagGRF15 OE and PagGRF15-SRDX transgenic plants. Data are presented as the means ± SDs (n = 6). Asterisks indicate significant differences (Dunnett test) compared to CK. CK, the control line. *, P ≤ 0.05; **, P ≤ 0.01.
We then assessed the vascular tissues in PagGRF15 transgenic plants in comparison with CK plants. Cross sections of the 1st to 6th internodes of CK and PagGRF15 transgenic plants were obtained, and the vascular cambium ring was not significantly different between PagGRF15 transgenic and CK plants (Supplementary Figure 2). Subsequently, cross sections of 4-month-old PagGRF15 transgenic and CK plants were analyzed. Cross sections of the 7th, 9th and 23rd internodes of CK and PagGRF15 transgenic plants were obtained, and secondary xylem developed faster in mPagGRF15 OE plants but slower in PagGRF15-SRDX plants than in CK plants (Figures 3 and Supplementary Figure 3). Statistical data showed that the width of the secondary xylem from mPagGRF15 OE transgenic plants was increased compared with that of CK plants, with an approximately 55% increase in the 9th internode (Figures 3A, B). In contrast, PagGRF15-SRDX plants exhibited a narrower xylem zone, with an average of a 28% decrease in the 9th internode (Figures 3A, B). On the other hand, the distance from phloem fiber to xylem in the 9th internode decreased by approximately 28% on average in the mPagGRF15 OE plants but increased by 17% in the PagGRF15-SRDX plants (Figure 3C). The number of layers of vascular cambium in the 9th internode of 4-month-old seedlings was significantly increased in mPagGRF15 OE, and decreased in PagGRF15-SRDX as compared with CK plants (Figure 3D). These results suggested that PagGRF15 mediated secondary vascular development by affecting cambial activity to set the pace of xylem and phloem differentiation.
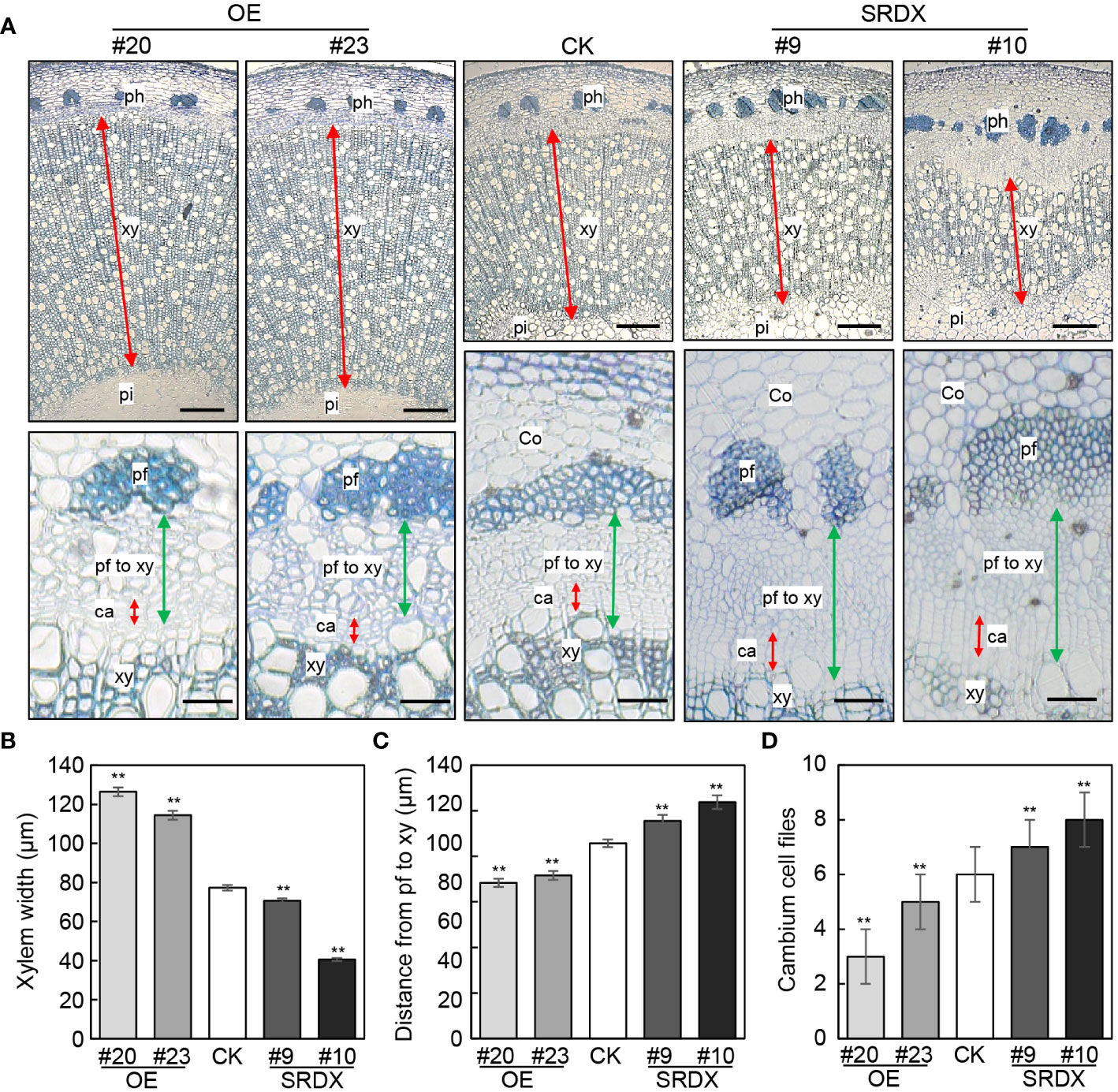
Figure 3 Stem development of mPagGRF15 OE and PagGRF15-SRDX transgenic plants. (A) Cross sections of the 9th internode from 4-month-old mPagGRF15 OE and PagGRF15-SRDX plants. Co, cortex; ph, phloem; pf, phloem fiber; xy, xylem; ca, cambium. Bar = 200 µm in the upper panel and bar = 20 µm in the lower panel. (B-D) Xylem width, phloem width and cambium cell files of the 9th internode from 4-month-old mPagGRF15 OE and PagGRF15-SRDX transgenic plants, respectively. Data are presented as the means ± SDs. 3 separate plants and 20 biological replicates were used for the statistical analyses in (B-D). Asterisks indicate significant differences (t test) compared to CK. CK, the control line. **, P ≤ 0.01.
3.3 The potential downstream genes of PagGRF15
For determination of how PagGRF15 regulates vascular cambial differentiation, genome-wide gene expression in the cambial zone of mPagGRF15 OE plants was profiled by RNA-seq. Analysis of transcript abundance showed that 2013 genes were upregulated and 1487 genes were downregulated in the PagGRF15 OE transgenic plants (Supplementary Table 1). Gene Ontology (GO) enrichment analysis indicated that the differentially expressed genes (DEGs) were associated with xylem development, glucuronoxylan metabolic process, and xylem biosynthetic process (Supplementary Figure 4), indicating the regulatory role of PagGRF15 in vascular cambial differentiation. To further identify the downstream genes regulated by PagGRF15, we performed chromatin immunoprecipitation followed by high-throughput DNA sequencing (ChIP-seq) using GFP-mPagGRF15 OE plants (Supplementary Figure 5). Illumina libraries were sequenced for three independent ChIP-seq biological replicates for GFP antibody libraries, and the DNA ‘input’ libraries were used as a control for ChIP-seq peak calling. A total of 18261 enriched peaks belonging to 13660 genes were identified (Supplementary Table 2). Next, the above DEGs found in the transcriptional profile were screened in ChIP-seq data, and 1241 overlapping genes were found (Supplementary Table 3). In particular, 81 of these genes were previously reported to be involved in vascular development (Supplementary Table 4, Ye and Zhong, 2015; Zhong and Ye, 2015). For instance, GID1.3 (Mauriat and Moritz, 2009) is involved in regulating xylem differentiation, while WhOX4b (Kucukoglu et al., 2017) and PXY (Etchells and Turner, 2010) are marker genes for cambial cell activity.
To validate the RNA-seq and ChIP-seq results, we performed qRT-PCR and ChIP-qPCR to quantify the above marker genes involved in cambial activity and xylem differentiation. Consistent with the RNA-seq results, the expression of WOX4b, PXY, and APL was downregulated, while the expression of GID1.3, ERF6, JAS1, and MYC2 was upregulated in mPagGRF15 OE plants in qRT-PCR detection (Figures 4A and Supplementary Figure 6). Consistent with the ChIP-seq results, ChIP-qPCR showed more than 2-fold, 3-fold, and 50-fold enrichment in binding over the input control for WOX4b, PXY, and GID1.3, respectively (Figure 4B), suggesting that these genes could be directly regulated by PagGRF15.
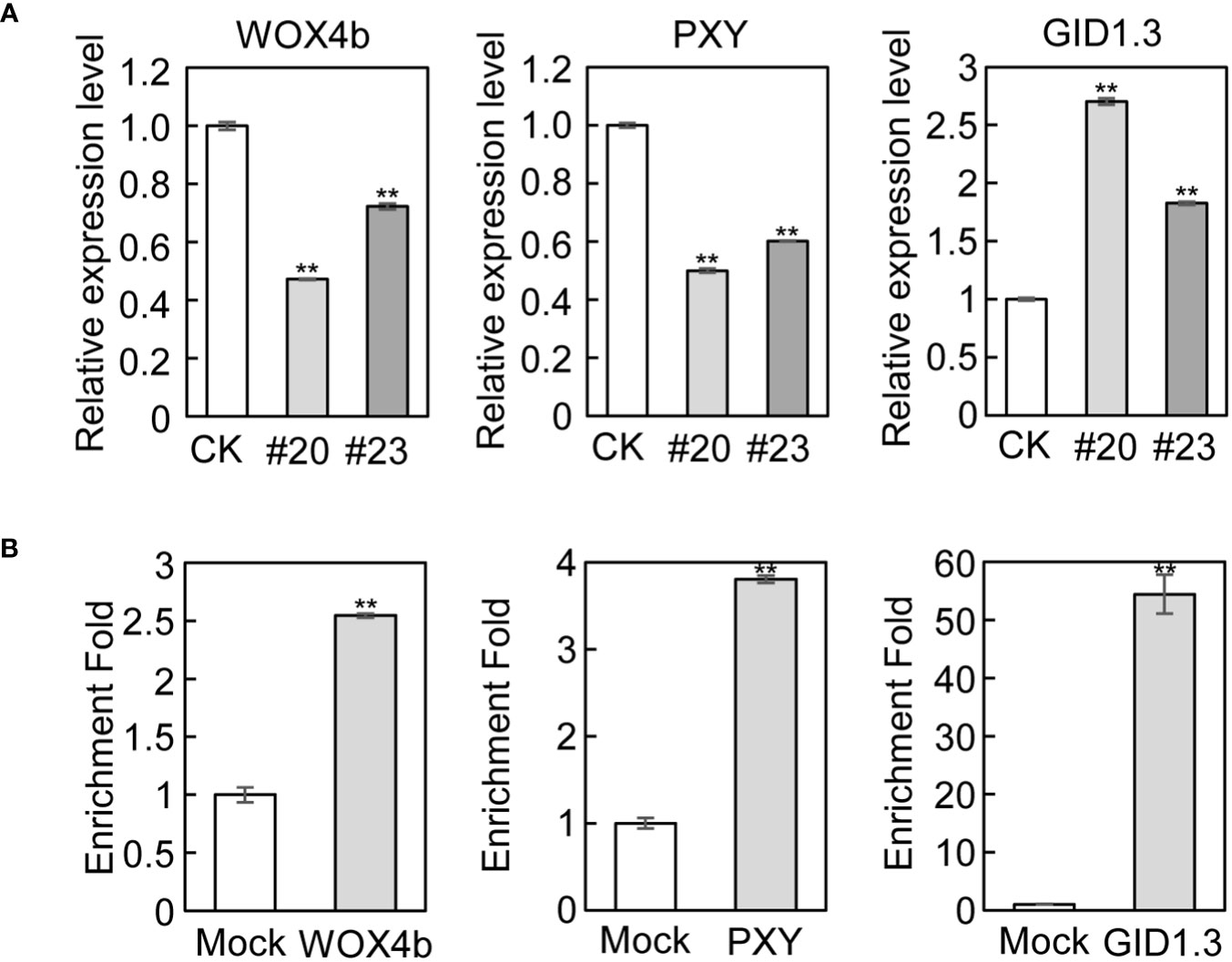
Figure 4 Genes related to vascular development in mPagGRF15 OE plants. (A) qRT-PCR detection of the expression of WOX4b, PXY and GID1.3 in mPagGRF15 OE plants. (B) ChIP-qPCR verification of WOX4b, PXY and GID1.3 regulated by PagGRF15. Data are presented as the means ± SDs. 3 biological and technical replicates were used for the statistical analyses. Asterisks indicate significant differences (t test) compared to CK. CK, the control line. **, P ≤ 0.01.
4 Discussion
GRFs are reported to be involved in many developmental events, including stem development (Kim, 2019). Overexpressing OsGRF1 in Arabidopsis completely abolished stem elongation (van der Knaap et al., 2000). Similarly, overexpression of ZmGRF10 and ZmGRF1R (miR396a-resistant version of GRF1) in maize both led to a reduction in plant height (Wu et al., 2014; Nelissen et al., 2015). However, overexpressing OsGRF3 and OsGRF10 in rice could increase tiller length, while downregulation of the expression of GRF resulted in visibly shorter internodes (Kuijt et al., 2014). In hybrid poplar 84K, we reported that plants overexpressing PagGRF12a exhibited no significant alteration in plant height (Wang et al., 2021). In addition to P. tomentosa, 35S:PtoGRF15 plants also showed no significant difference in height (Wang et al., 2023). Here, we found that overexpression of PagGRF15 could lead to a decrease in plant height. Thus, different GRFs may play different roles in stem elongation regulation, and PagGRF15 is a negative regulator of plant height in hybrid poplar 84K. Secondary growth is another aspect of stem development in woody plants. Poplar trees overexpressing PagGRF12a as well as PtoGRF15 showed delayed secondary growth and xylem production reduction (Wang et al., 2021, 2023), indicating the negative regulatory role of GRF in vascular development. We also found that abnormal expression of PagGRF15 could affect vascular tissue development. Plants with PagGRF15 overexpression exhibited increased xylem production, whereas PagGRF15-SRDX plants showed decreased xylem production. Interestingly, the plant height decreased while the secondary xylem width increased in mPagGRF15 OE plants, while PagGRF15-SRDX plants demonstrated the opposite phenotype. Therefore, PagGRF15 functions as an activator that regulates not only leaf size but also secondary xylem growth in woody plants.
Previous reports have shown that GRF may have a role in meristem activity (Kim and Lee, 2006; Rodriguez et al., 2015; Zhang et al., 2018), and gene function in the meristem always also has a role in vascular cambium activity (Groover, 2005). In secondary growth, xylem and phloem are produced by cambium activity during wood formation (Spicer and Groover, 2010). Here, we showed that PagGRF15 is highly expressed in vascular cambium and that its overexpression resulted in fewer cell files in the cambial zone and more cell files in the secondary xylem zone, while PagGRF15-SRDX plants had the opposite phenotype, indicating that PagGRF15 could affect secondary xylem development by reducing cambial cell proliferation but accelerating xylem differentiation. AtGRF5 was reported to accelerate cell proliferation activities and reduce cell expansion (an indication of differentiation in plants) to affect the movement of the cell cycle arrest front (AF) during leaf development (Rodriguez et al., 2010; Vercruyssen et al., 2015). In poplar, we reported that PagGRF12a inhibits xylem development by upregulating PagXND1a expression (Wang et al., 2021). It was reported that XND1 affects xylem formation by modulating the differentiation of cambial cells to xylem cells (Zhao et al., 2008; Tang et al., 2018; Grant et al., 2010; Zhao et al., 2020). Recently, in P. tomentosa, PtoGRF15 was found to delay secondary vascular development by decreasing the number of layers of xylem cells between primary vascular bundles during the transition from primary to secondary vascular development (Wang et al., 2023). Moreover, PtoTCP20 integrated with the miR396d-PtoGRF15 regulatory module and thereby formed the PtoTCP20-miR396d-PtoGRF15 signaling cascade that functions in secondary vascular development (Wang et al., 2023). In hybrid polar 84K, we showed that PagGRF15 was localized in the vascular cambium zone, which contributed to the development of secondary tissues, indicating the potential roles of PagGRF15 in vascular development during wood formation. Overexpression of PagGRF15 caused an enlarged xylem width, a decreased amount of phloem and a significant reduction in cambium dividing cells, whereas PagGRF15-SRDX plants produced a decreased amount of xylem, an enlarged phloem width and an increased number of cambium dividing cells, resulting in no significant changes in the stem diameter of PagGRF15 transgenic plants compared with CK plants. These results suggested that PagGRF15 in hybrid poplar acted as a positive regulator of xylem development by affecting cambial activity to set the pace of xylem and phloem. Taken together, the results indicated that GRF15 participates in more complex regulatory loops in secondary vascular tissue growth.
WOX4 is the target of PXY, and both are marker genes for cambial activity (Etchells et al., 2013, 2015; Kucukoglu et al., 2017). PXY is a cambium-specific leucine-rich repeat receptor-like kinase that acts as a receptor of the peptide ligands CLE41/CLE44/TDIF, which stimulates WOX4 transcription factor activity and regulates the proliferation of cambial cells and vascular tissue patterning in stems by interaction with CLE peptides (Sugimoto et al., 2022). WOX4 plays a dominant role in regulating the activity of the vascular cambium (Kucukoglu et al., 2017). In Arabidopsis, AtWOX4 is necessary for stimulating the proliferation of procambium/cambium stem cells, but not required for repressing their commitment to become xylem cells (Hirakawa et al., 2010), whereas PttWOX4 not only promotes cambial proliferation, but also enhances the differentiation of secondary xylem in poplar (Kucukoglu et al., 2017). Repressed PttWOX4a expression could result in reduced vascular cambium width (Kucukoglu et al., 2017), while overexpression of PttPXY could result in increased cambial cell division (Etchells et al., 2015). In P. tomentosa, PtoTCP has reported to interact with PtoWOX4a to control vascular cambium proliferation and promote xylem cell differentiation by activating PtoWND6 transcription (Hou et al., 2020). Recently, the genetic regulatory network underlying WOX4-mediated wood formation at the post-transcriptional level has been elucidated (Tang et al., 2022). PagWOX4 RNAi transgenic plants exhibited severely inhibited secondary growth with a reduced proportion of xylem and significantly reduced layers of cambial cells. Genetic analysis showed that PagDA1 functions as a repressor to destabilize PagWOX4 mediated by PagDA2 to affect cambium development (Tang et al., 2022). As shown in this study, the expression of PagWOX4b and PagPXY was downregulated in mPagGRF15 OE plants and resulted in decreased cambial cell files. In addition, PagGRF15 can directly regulate PagWOX4b and PagPXY, as revealed by ChIP-seq and ChIP-qRT analysis. Therefore, GRF15 may be involved in the PXY-WOX pathway to set the pace of cambial activity. Interestingly, mPagGRF15 OE plants exhibited a significantly increased proportion of xylem, suggesting that there may be another regulatory network underlying PagGRF15-mediated wood formation.
GID is a soluble GA receptor that functions as a key mediator of GA response pathways (Mauriat and Moritz, 2009). The GA/GID1 complex binds to DELLA and leads to the degradation of DELLA to derepress GA signaling (Mauriat and Moritz, 2009). In hybrid aspen, the pith/xylem ratio was lower in PttGID1.3-overexpressing plants than in wild-type plants, indicating that overexpression of PttGID1.3 stimulates xylem differentiation (Mauriat and Moritz, 2009). The xylem width was significantly increased in mPagGRF15 OE plants compared to CK plants (Figures 3 and Supplementary Figure 4). Thus, the upregulated PagGID1.3 could be responsible for the enlarged xylem observed in mPagGRF15 OE plants.
5 Conclusions
Hybrid poplars are among the fastest-growing industrial trees in China. P. alba × P. glandulosa is a hybrid poplar species of economic and ecological importance. The specific gravity of wood has been proposed as a prime target for genetic modification. Accordingly, functional analysis of PagGRF15 from P. alba × P. glandulosa was performed to improve the understanding of the molecular mechanisms of wood formation. Overexpression of PagGRF15 in hybrid poplar positively promoted xylem differentiation during plant secondary growth. RNA-seq and ChIP-seq results combining qRT-PCR and ChIP-PCR analysis revealed that PagGRF15 may be involved in setting the pace of cambial activity by downregulating WOX4b and PXY expression and mediating the GA signaling pathway by accelerating PagGID1.3 expression to stimulate xylem differentiation. The results provide valuable information for further studies on the wood formation mechanisms of hybrid poplar with the aim of improving the specific gravity of wood.
Data availability statement
The original contributions presented in the study are publicly available. This data can be found here: https://www.ncbi.nlm.nih.gov/bioproject/; PRJNA693036.
Author contributions
HZ: Data curation, Formal analysis, Funding acquisition, Investigation, Validation, Writing – original draft, Writing – review & editing. XS: Conceptualization, Formal analysis, Funding acquisition, Methodology, Project administration, Visualization, Writing – original draft, Writing – review & editing. ML: Conceptualization, Project administration, Supervision, Writing – review & editing, Methodology.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the National Natural Science Foundation of China (Grant no. 32001330, 31570676).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2024.1343312/full#supplementary-material
References
Brunner, A. M., Busov, V. B., Strauss, S. H. (2004). Poplar genome sequence: functional genomics in an ecologically dominant plant species. Trends Plant Sci. 9, 49–56. doi: 10.1016/j.tplants.2003.11.006
Cao, Y., Han, Y., Jin, Q., Lin, Y., Cai, Y. (2016). Comparative genomic analysis of the GRF genes in Chinese pear (Pyrus bretschneideri Rehd), poplar (Populous), grape (Vitis vinifera), Arabidopsis and rice (Oryza sativa). Front. Plant Sci. 7. doi: 10.3389/fpls.2016.01750
Chao, Q., Gao, Z. F., Zhang, D., Zhao, B. G., Dong, F. Q., Fu, C. X., et al. (2019). The developmental dynamics of the Populus stem transcriptome. Plant Biotechnol. J. 17, 206–219. doi: 10.1111/pbi.12958
Dinus, R. J. (2001). Genetic improvement of poplar feedstock quality for ethanol production. Appl. Biochem. Biotechnol. 91, 23–34. doi: 10.1007/978-1-4612-0217-2_2
Etchells, J. P., Mishra, L. S., Kumar, M., Campbell, L., Turner, S. R. (2015). Wood formation in trees is increased by manipulating PXY-regulated cell division. Curr. Biol. 25, 1050–1055. doi: 10.1016/j.cub.2015.02.023
Etchells, J. P., Provost, C. M., Mishra, L., Turner, S. R. (2013). WOX4 and WOX14 act downstream of the PXY receptor kinase to regulate plant vascular proliferation independently of any role in vascular organisation. Development 140, 2224–2234. doi: 10.1242/dev.091314
Etchells, J. P., Turner, S. R. (2010). The PXY-CLE41 receptor ligand pair defines a multifunctional pathway that controls the rate and orientation of vascular cell division. Development 137, 767–774. doi: 10.1242/dev.044941
Grant, E. H., Fujino, T., Beers, E. p., Brunner, A. M. (2010). Characterization of NAC domain transcription factors implicated in control of vascular cell differentiation in Arabidopsis and Populus. Planta 232, 337–352. doi: 10.1007/s00425-010-1181-2
Groover, A. T. (2005). What genes make a tree a tree? Trends Plant Sci. 10, 210–214. doi: 10.1016/j.tplants.2005.03.001
Hepworth, J., Lenhard, M. (2014). Regulation of plant lateral-organ growth by modulating cell number and size. Curr. Opin. Plant Biol. 17, 36–42. doi: 10.1016/j.pbi.2013.11.005
Hirakawa, Y., Kondo, Y., Fukuda, H. (2010). TDIF peptide signaling regulates vascular stem cell proliferation via the WOX4 homeobox gene in Arabidopsis. Plant Cell 22, 2618–2629. doi: 10.1105/tpc.110.076083
Hou, J., Xu, H., Fan, D., Ran, L., Li, J., Wu, S., et al. (2020). MiR319a-targeted PtoTCP20 regulates secondary growth via interactions with PtoWOX4 and PtoWND6 in Populus tomentosa. New Phytol. 228, 1354–1368. doi: 10.1016/j.pbi.2013.11.005
Johnsson, C., Fischer, U. (2016). Cambial stem cells and their niche. Plant Sci. 252, 239–245. doi: 10.1016/j.plantsci.2016.08.002
Kim, J. H. (2019). Biological roles and an evolutionary sketch of the GRF-GIF transcriptional complex in plants. BMB Rep. 52, 227–238. doi: 10.5483/BMBRep.2019.52.4.051
Kim, J. H., Chi, D., Kenda, H. (2003). The AtGRF family of putative transcription factors is involved in leaf and cotyledon growth in Arabidopsis. Plant J. 36, 94–104. doi: 10.1046/j.1365-313X.2003.01862.x
Kim, J. H., Lee, B. H. (2006). GROWTH-REGULATING FACTOR4 of Arabidopsis thaliana is required for development of leaves, cotyledons, and shoot apical meristem. J. Plant Biol. 49, 463–468. doi: 10.1007/BF03031127
Kim, J. H., Tsukaya, H. (2015). Regulation of plant growth and development by the GROWTH-REGULATING FACTOR and GRF-INTERACTING FACTOR duo. J. Exp. Bot. 66, 6093–6107. doi: 10.1093/jxb/erv349
Kucukoglu, M., Nilsson, J., Zheng, B., Chaabouni, S., Nilsson, O. (2017). WUSCHEL-RELATED HOMEOBOX4 (WOX4)-like genes regulate cambial cell division activity and secondary growth in Populus trees. New Phytol. 215, 642–657. doi: 10.1111/nph.14631
Kuijt, S. J., Greco, R., Agalou, A., Shao, J., ‘t Hoen, C. C., Overnäs, E., et al. (2014). Interaction between the GROWTH-REGULATING FACTOR and KNOTTED1-LIKE HOMEOBOX families of transcription factors. Plant Physiol. 164, 1952–1966. doi: 10.1104/pp.113.222836
Li, W., Lin, Y. C., Li, Q., Shi, R., Lin, C. Y., Chen, H., et al. (2014). A robust chromatin immunoprecipitation protocol for studying transcription factor-DNA interactions and histone modifications in wood-forming tissue. Nat. Protoc. 9, 2180–2193. doi: 10.1038/nprot.2014.146
Mauriat, M., Moritz, T. (2009). Analyses of GA20ox- and GID1-over-expressing aspen suggest that gibberellins play two distinct roles in wood formation. Plant J. 58, 989–1003. doi: 10.1111/j.1365-313X.2009.03836.x
Nelissen, H., Eeckhout, D., Demuynck, K., Persiau, G., Walton, A., van Bel, M., et al. (2015). Dynamic changes in ANGUSTIFOLIA3 complex composition reveal a growth regulatory mechanism in the maize leaf. Plant Cell 27, 1605–1619. doi: 10.1105/tpc.15.00269
Omidbakhshfard, M. A., Proost, S., Fujikura, U., Mueller-Roeber, B. (2015). Growth-regulating factors (GRFs): a small transcription factor family with important functions in plant biology. Mol. Plant 8, 998–1010. doi: 10.1016/j.molp.2015.01.013
Rodriguez, R. E., Ercoli, M. F., Debernardi, J. M., Breakfield, N. W., Mecchia, M. A., Sabatini, M., et al. (2015). MicroRNA miR396 regulates the switch between stem cells and transit-amplifying cells in Arabidopsis roots. Plant Cell 27, 3354–3366. doi: 10.1105/tpc.15.00452
Rodriguez, R. E., Mecchia, M. A., Debernardi, J. M., Schommer, C., Weigel, D., Palatnik, J. F. (2010). Control of cell proliferation in Arabidopsis thaliana by microRNA miR396. Development 137, 103–112. doi: 10.1242/dev.043067
Sannigrahi, P., Ragauskas, A. J., Tuskan, G. A. (2010). Poplar as a feedstock for biofuels: a review of compositional characteristics. Biofuel Bioprod. Bior. 4, 209–226. doi: 10.1002/bbb.206
Smetana, O., Mäkilä, R., Lyu, M., Amiryousefi, A., Sánchez Rodríguez, F., Wu, M. F., et al. (2019). High levels of auxin signalling define the stem-cell organizer of the vascular cambium. Nature 565, 485–489. doi: 10.1038/s41586-018-0837-0
Spicer, R., Groover, A. (2010). Evolution of development of vascular cambia and secondary growth. New Phytol. 186, 577–592. doi: 10.1111/j.1469-8137.2010.03236.x
Sugimoto, H., Tanaka, T., Muramoto, N., Kitagawa-Yogo, R., Mitsukawa, N. (2022). Transcription factor NTL9 negatively regulates Arabidopsis vascular cambium development during stem secondary growth. Plant Physiol. 190, 1731–1746. doi: 10.1093/plphys/kiac368
Tang, F., Chu, L., Shu, W., He, X., Wang, L., Lu, M. (2019). Selection and validation of reference genes for quantitative expression analysis of miRNAs and mRNAs in Poplar. Plant Methods 15, 1–15. doi: 10.1186/s13007-019-0420-1
Tang, N., Shahzad, Z., Lonjon, F., Loudet, O., Vailleau, F., Maurel, C. (2018). Natural variation at XND1 impacts root hydraulics and trade-off for stress responses in Arabidopsis. Nat. Commun. 9, 3884. doi: 10.1038/s41467-018-06430-8
Tang, X., Wang, C., Chai, G., Wang, D., Xu, H., Liu, Y., et al. (2022). Ubiquitinated DA1 negatively regulates vascular cambium activity through modulating the stability of WOX4 in Populus. Plant Cell 34, 3364–3382. doi: 10.1093/plcell/koac178
van der Knaap, E., Kim, J. H., Kende, H. (2000). A novel gibberellin-induced gene from rice and its potential regulatory role in stem growth. Plant Physiol. 122, 695–704. doi: 10.1104/pp.122.3.695
Vercruyssen, L., Tognetti, V. B., Gonzalez, N., Van Dingenen, J., De Milde, L., Bielach, A., et al. (2015). GROWTH REGULATING FACTOR5 stimulates Arabidopsis chloroplast division, photosynthesis, and leaf longevity. Plant Physiol. 167, 817–832. doi: 10.1104/pp.114.256180
Wang, L., Hou, J., Xu, H., Zhang, Y., Huang, R., Wang, D., et al. (2023). The PtoTCP20-miR396d-PtoGRF15 module regulates secondary vascular development in Populus. Plant Commun. 4, 100494. doi: 10.1016/j.xplc.2022.100494
Wang, J., Zhou, H., Zhao, Y., Jiang, C., Li, J., Tang, F., et al. (2021). PagGRF12a interacts with PagGIF1b to regulate secondary xylem development through modulating PagXND1a expression in Populus alba× P. glandulosa. J. Integr. Plant Biol. 63, 1683–1694. doi: 10.1111/jipb.13102
Włoch, W., Iqbal, M., Jura-Morawiec, J. (2023). Calculating the growth of vascular cambium in woody plants as the cylindrical surface. Bot. Rev. 89, 237–249. doi: 10.1007/s12229-023-09291-z
Wu, L., Zhang, D., Xue, M., Qian, J., He, Y., Wang, S. (2014). Overexpression of the maize GRF10, an endogenous truncated growth-regulating factor protein, leads to reduction in leaf size and plant height. J. Integr. Plant Biol. 56, 1053–1063. doi: 10.1111/jipb.12220
Ye, Z. H., Zhong, R. (2015). Molecular control of wood formation in trees. J. Exp. Bot. 66, 4119–4131. doi: 10.1093/jxb/erv081
Zhang, D., Sun, W., Singh, R., Zheng, Y., Cao, Z., Li, M., et al. (2018). GRF-interacting factor 1 regulates shoot architecture and meristem determinacy in maize. Plant Cell 30, 360–374. doi: 10.1105/tpc.17.00791
Zhao, C., Avci, U., Grant, E. H., Haigler, C. H., Beers, E. P. (2008). XND1, a member of the NAC domain family in Arabidopsis thaliana, negatively regulates lignocellulose synthesis and programmed cell death in xylem. Plant J. 53, 425–436. doi: 10.1111/j.1365-313X.2007.03350.x
Zhao, Y., Song, X., Zhou, H., Wei, K., Jiang, C., Wang, J., et al. (2020). KNAT2/6b, a class I KNOX gene, impedes xylem differentiation by regulating NAC domain transcription factors in poplar. New Phytol. 225, 1531–1544. doi: 10.1111/nph.16036
Zhong, R., Ye, Z. H. (2015). Secondary cell walls: biosynthesis, patterned deposition and transcriptional regulation. Plant Cell Physiol. 56, 195–214. doi: 10.1093/pcp/pcu140
Keywords: hybrid poplar, PagGRF15, vascular cambium, division, xylem differentiation
Citation: Zhou H, Song X and Lu M-Z (2024) Growth-regulating factor 15-mediated vascular cambium differentiation positively regulates wood formation in hybrid poplar (Populus alba × P. glandulosa). Front. Plant Sci. 15:1343312. doi: 10.3389/fpls.2024.1343312
Received: 23 November 2023; Accepted: 01 February 2024;
Published: 15 February 2024.
Edited by:
Sergio Tombesi, Catholic University of the Sacred Heart, ItalyReviewed by:
David Collings, Australian National University, AustraliaMiyuki T. Nakata, Nara Institute of Science and Technology (NAIST), Japan
Copyright © 2024 Zhou, Song and Lu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Houjun Zhou, aGp6aG91QGxkdS5lZHUuY24=; Xueqin Song, eHFzb25nQGNhZi5hYy5jbg==
 Houjun Zhou
Houjun Zhou Xueqin Song
Xueqin Song Meng-Zhu Lu
Meng-Zhu Lu