- 1Department of Agronomy, Faculty of Agriculture at Kamphaeng Saen, Kasetsart University, Nakhon Pathom, Thailand
- 2Department of Agronomy, Faculty of Agriculture, Khon Kaen University, Khon Kaen, Thailand
- 3Plant Breeding Research Center for Sustainable Agriculture, Faculty of Agriculture, Khon Kaen University, Khon Kaen, Thailand
- 4Department of Agronomy, Iowa State University, Ames, IA, United States
- 5National Center for Genetic Engineering and Biotechnology (BIOTEC), National Science and Technology Development Agency (NSTDA), Pathum Thani, Thailand
- 6Rice Science Center, Kasetsart University, Nakhon Pathom, Thailand
Doubled haploid (DH) technology becomes more routinely applied in maize hybrid breeding. However, some issues in haploid induction and identification persist, requiring resolution to optimize DH production. Our objective was to implement simultaneous marker-assisted selection (MAS) for qhir1 (MTL/ZmPLA1/NLD) and qhir8 (ZmDMP) using TaqMan assay in F2 generation of four BHI306-derived tropical × temperate inducer families. We also aimed to assess their haploid induction rate (HIR) in the F3 generation as a phenotypic response to MAS. We highlighted remarkable increases in HIR of each inducer family. Genotypes carrying qhir1 and qhir8 exhibited 1 – 3-fold higher haploid frequency than those carrying only qhir1. Additionally, the qhir1 marker was employed for verifying putative haploid seedlings at 7 days after planting. Flow cytometric analysis served as the gold standard test to assess the accuracy of the R1-nj and the qhir1 marker. The qhir1 marker showed high accuracy and may be integrated in multiple haploid identifications at early seedling stage succeeding pre-haploid sorting via R1-nj marker.
Introduction
Maize is one of the most important cereal crops in the world as food, feed, and fuel (Prasanna, 2012). The success of hybrid maize breeding relies on robust pipelines of germplasm, genetics, phenotyping, and selection processes (Cooper et al., 2014). Traditionally, the breeding process for the market release of a new cultivar extended over a decade, until the advent of doubled haploid (DH) technology (Chaikam et al., 2019). A notable advantage of DH technology is associated with the substantial reduction of breeding cycles required to develop fully homozygous lines within just two generations (Geiger and Gordillo, 2009). Haploids can be produced in vitro or in vivo. The in vitro method requires laboratory procedures, where gametophytic tissues such as microspores and egg cells are used to produce paternal and maternal haploids, respectively. However, this method gains low success rates due to the high levels of genotype dependency (Jacquier et al., 2021). The in vivo method involves four main steps: (1) haploid induction, (2) haploid identification, (3) haploid genome doubling, and (4) self-pollination of haploid plants to obtain DH0 seeds (Chaikam et al., 2019). For maternal haploid induction, haploid inducers are used as male parents to pollinate source germplasm for haploid induction. Efficient DH line production depends on the availability of inducer genotypes with high induction ability.
In 2012, a QTL study involving four populations, all sharing the inbred inducer UH400 as common parent, identified 8 QTL. Notably, qhir1 and qhir8 emerged as two major QTL located on chromosomes 1 and 9, explaining 66% and 20% of the genetic variance, respectively (Prigge et al., 2012). The qhir1 region in bin 1.04 plays pivotal roles in triggering haploid induction, gametophytic segregation distortion, and embryo abortion (Barret et al., 2008; Prigge et al., 2012; Xu et al., 2013). Mutation of the gene MTL/ZmPLA1/NLD in qhir1 has been shown to generate an average haploid induction rate (HIR) up to 6.7% (Gilles et al., 2017; Kelliher et al., 2017; Liu et al., 2017). However, qhir1 is not sufficient for commercial productions of DH lines. To fully leverage this technology, the average HIR of modern haploid inducers should surpass 10% (Hu et al., 2016). Zhong et al. (2019) discovered a novel gene named ZmDMP underlying QTL qhir8. A mutation of ZmDMP markedly enhances haploid induction, resulting in a 2–3-fold increase in HIR. It is important to note that both MTL/ZmPLA1/NLD and ZmDMP act synergistically, suggesting the potential for a substantial 5–6-fold increase in the HIR when both mutations are present (Zhong et al., 2019). Marker assisted selection (MAS) for qhir1 has been applied to improve the HIR of maternal haploid inducers in different maize backgrounds. For instance, Chaikam et al. (2018) were able to obtain promising second-generation Tropically Adapted Inducer Lines (2GTAILs) with an average HIR of 13.1%, a 48.9% improvement over TAILs. Liu et al. (2022) developed an elite oil haploid inducer, CHOI4, with an averaged HIR of 15.8%, a 58.0% increase compared to CAU2, the founder parent of CHOI4. While these results are promising, further enhancements could be achieved through MAS for two loci, qhir1 and qhir8. Considering that HIR is a polygenic trait, selection of a single locus may not be sufficient to obtain inducers with optimum HIR (Dong et al., 2014). Nevertheless, limited evidence exists to illustrate the feasibility of MAS for both loci simultaneously in breeding haploid inducers for high HIR.
Haploids are commonly identified via the R1-navajo (R1-nj), a dominant monogenic biomarker (Nanda and Chase, 1966) integrated in haploid inducers. This marker distinguishes progeny seeds derived from haploid induction based on anthocyanin expression in different parts of the kernel. Haploid kernels show a purple crown in the endosperm but a colorless scutellum in the embryo, while diploids express both purple endosperm and embryo (Dermail et al., 2021). Despite practical and non-destructive features, the effectiveness of R1-nj expressions may be constrained by the presence of dominant C1 anthocyanin inhibitors (Chaikam et al., 2015), naturally occurring anthocyanins in donor germplasm (Chaikam et al., 2016), morpho-physiological kernel properties (Prigge et al., 2011; Trentin et al., 2022), and environments (Sintanaparadee et al., 2022; Dermail et al., 2023; Thawarorit et al., 2023). These factors contribute to high misclassification rates (MCRs), hindering selection gains on HIR and emphasizing the need for alternative markers for haploid selection. While simple sequence repeat (SSR) has been successfully used in maize haploid identification (Qiu et al., 2014; Dong et al., 2018; Li et al., 2021), the practical use of SNP markers for that purpose is still lacking. Since most paternal chromosomes of inducers are excluded from haploid embryonic cells within a week after pollination, the haploid individuals carry only maternal chromosomes from the donor germplasm (Zhao et al., 2013). Codominant SNP markers can differentiate between homozygotes (donor female) and heterozygotes (F1 diploids). Considering remarkable allelic variation for qhir1 and qhir8 haploid inducers versus non-inducer genotypes, there is an encouraging prospect of applying these loci for haploid identification using TaqMan probes. Kelliher et al. (2017) employed TaqMan assays for qhir1, proposing that haploids carry zero copies of the mtl allele and two copies of the maternal MTL allele, whereas diploids carry one copy of the mtl allele and one copy of the MTL allele.
Our study aimed to utilize the qhir1 and qhir8 loci in marker-assisted selection, with a dual focus on breeding haploid inducers for high HIR and accurately identifying true haploids in maize. We hypothesized that (i) inducer genotypes carrying qhir1 and qhir8 should demonstrate a higher capacity to induce haploids compared to those carrying qhir1 alone and (ii) molecular markers using TaqMan assay are more reliable than the R1-nj marker when validated with flow cytometry. This study will provide an insight into the advantages of molecular assays, especially TaqMan probes, to accelerate the improvements of haploid inducers underpinning HIR. Additionally, it seeks to enhance the accuracy of identifying true haploids at early seedling stage.
Materials and methods
Breeding scheme, haploid induction, and HIR evaluation
A temperate inbred inducer, BHI306, and four tropical inducer families (K8, K11, KHI49, and KHI54) were selected as founder parents. The BHI306 genotype, an RWS/RWK-76-derived haploid inducer, has 10–15% of HIR, carries both qhir1 and qhir8 loci (Supplementary Table S1 and Supplementary Figure S1), kernel anthocyanin R1-nj and red root Pl-1 selectable markers. BHI306 was developed by the DH Facility of Iowa State University (DHF-ISU) (https://www.doubledhaploid.biotech.iastate.edu/). Four genotypes, K8, K11, KHI49, and KHI54 belong to qhir1−/qhir8− group (Supplementary Table S1 and Supplementary Figure S1), Stock-6-derived haploid inducers, had low HIRs (<6.0%) but possess favorable tropical adaptations. These genotypes were developed by the Plant Breeding Research Center for Sustainable Agriculture of Khon Kaen University in Thailand (Dermail et al., 2021; Sintanaparadee et al., 2022; Thawarorit et al., 2023). A 1 × 4 factorial mating scheme was performed by assigning BHI306 as a male and four tropical inducers as females to establish four tropical × temperate inducer base populations including K8/BHI306, K11/BHI306, KHI49/BHI306, and KHI54/BHI306. In the F2 generation, approximately 100 F2 seedlings per inducer population underwent random marker-assisted selection (MAS) for qhir1 and qhir8. Plants carrying qhir1 only and both qhir1 and qhir8 were labeled as qhir1+/qhir8− and qhir1+/qhir8+ genotypes, respectively. These targeted genotypes were subsequently transplanted into the field and self-pollinated to obtain F3 seeds. At that generation, we did not perform haploid induction. Thus, there was no preliminary information regarding the actual HIR. At the F3 generation, repeated genotyping of each individual plant and phenotyping on actual HIR were performed in each population (Supplementary Table S1).
Maternal haploid induction was performed to evaluate HIRs. A commercial hybrid Pacific789 (P789), developed by Pacific Seeds, Thailand, was used as a donor female. This genotype is resistant to tropical diseases, high-yielding, and large-seeded with flat embryos, facilitating haploid selection based on the R1-nj marker at the seed stage. Each F3 inducer plant in each qhir genotype and family was used to pollinate four donor ears to minimize the errors due to unstable inducer pollen. Shoot bagging and detasseling of donor plants were routinely performed to prevent pollen contamination.
Haploid seed was selected via the R1-nj marker at the seed stage. Haploids showed a purple crown endosperm but a colorless embryo, while diploids expressed purple colorations on both crown endosperm and embryo (Nanda and Chase, 1966; Dermail et al., 2023). The HIR was calculated as the frequency of haploid seeds per induction cross, as follows:
where seed set represents the total seed number of haploid seeds, diploid seeds, and the seeds without the R1-nj marker.
About 10 putative haploid seeds per genotype in each inducer family were sampled for further true haploid confirmation through molecular assays.
Marker development
Two TaqMan® markers (qhir1 and qhir8) for two targeted genes namely MATRILINEAL (MTL/ZmPLA1/NLD) and ZmDMP, respectively, were constructed (Figure 1). The marker for the MTL gene (GRMZM2G471240) was developed at 4 bp (CGAG) insertion in the 4th exon of the gene that led to premature stop codon (Gilles et al., 2017; Kelliher et al., 2017; Liu et al., 2017). The ZmDMP gene (GRMZM2G465053) was developed at single nucleotide substitution from T to C at 131 bp on coding sequence that led to amino acid change from methionine to threonine (Zhong et al., 2019).
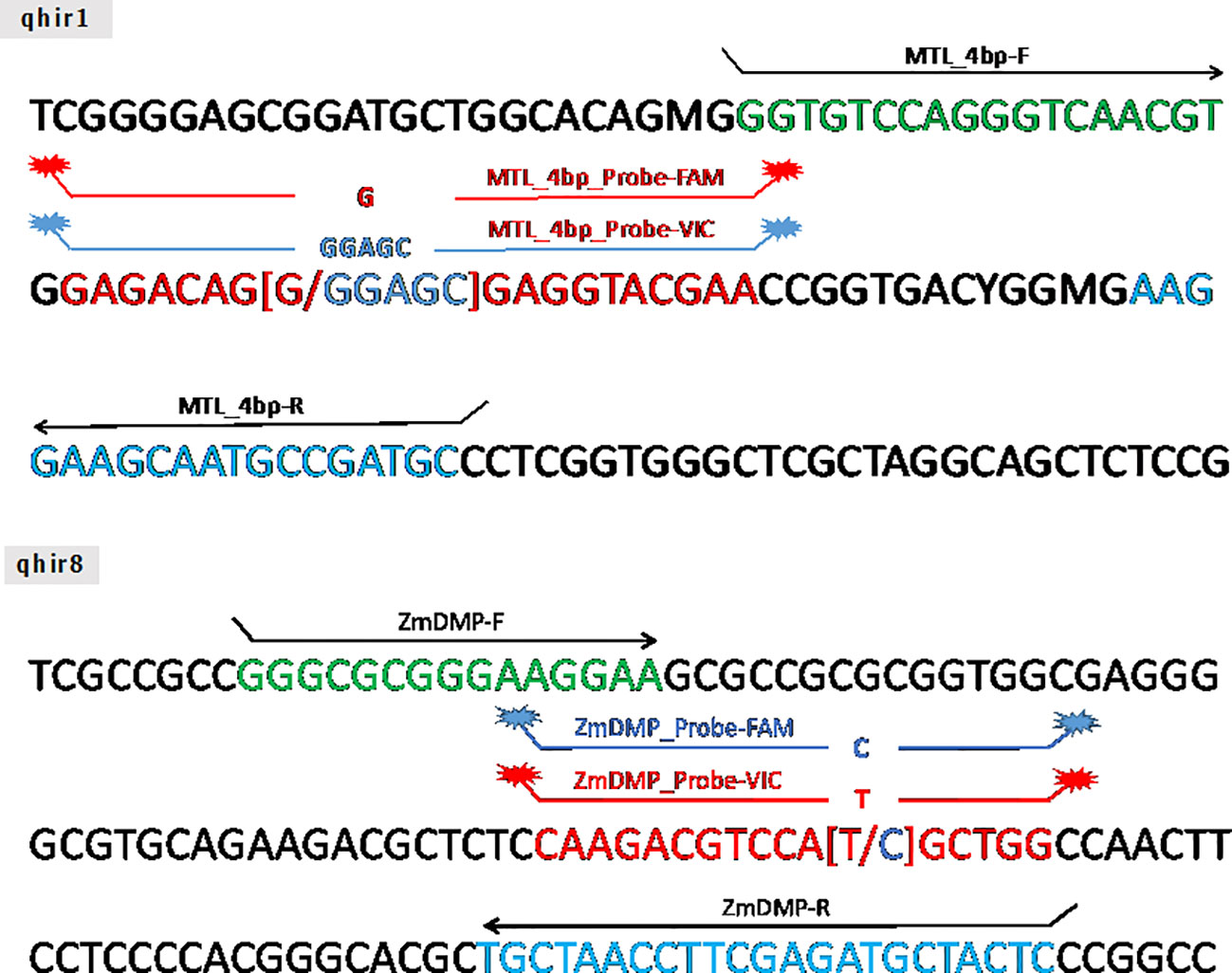
Figure 1 The schematic of TaqMan® probe design on MATRILINEAL gene (GRMZM2G471240) and ZmDMP gene (GRMZM2G465053).
Genotyping and DNA extraction
High-quality genomic DNA (gDNA) was isolated from maize leaves at 14 days after germination using the DNeasy® Plant Mini Kit (QIAGEN, Germany). Genotyping for qhir1 and qhir8 markers was carried out with ready-to-order TaqMan assays (Thermo Fisher Scientific, Watham, MA USA) (Figure 1). In the amplification process, 20 ng gDNA was utilized. For the PCR reaction, the total volume was 5 µl composed of 2 µl of template DNA, 1.5 µl of 2X TaqMan® Gene Expression Master Mix (Thermo Fisher Scientific, Watham, MA USA), 0.0375 µl of TaqMan assay, and 1.4625 µl of dH2O. The PCR cycling conditions were set at 95°C for 5 min, followed by 36 cycles at 94°C for 30 s, 60°C for 1 min, and 60°C for 2 min. For the PCR product, the amplicons were melted at 60°C using QuantStudio 6 Real-Time PCR Systems (Thermo Fisher Scientific, Watham, MA USA) for 30 s to detect single nucleotide polymorphism (SNP).
Flow cytometry analysis
Three subsets of populations derived from induction crosses between female donor P789 and three male inducers, BHI306, KHI49/BHI306, and KHI54/BHI306, were used for haploid validation via flow cytometry analysis. The number of samples was 24, derived from false positives previously assumed as putative haploids based on the R1-nj marker but eventually true diploids regarding the qhir1 marker. Those 24 samples composed of 1 putative haploid of P789/BHI306, 10 putative haploids of P789/(KHI49/BHI306), and 13 putative haploids of P789/(KHI54/BHI306). The FC analysis on those 24 samples served as the gold standard classification method to verify if qhir1 marker is reliable to determine the true haploids. The FC graph of each sample can be found in the Supplementary Figure S3, and the result of FC analysis corresponding to the qhir1 marker assay can be found in Table 1.
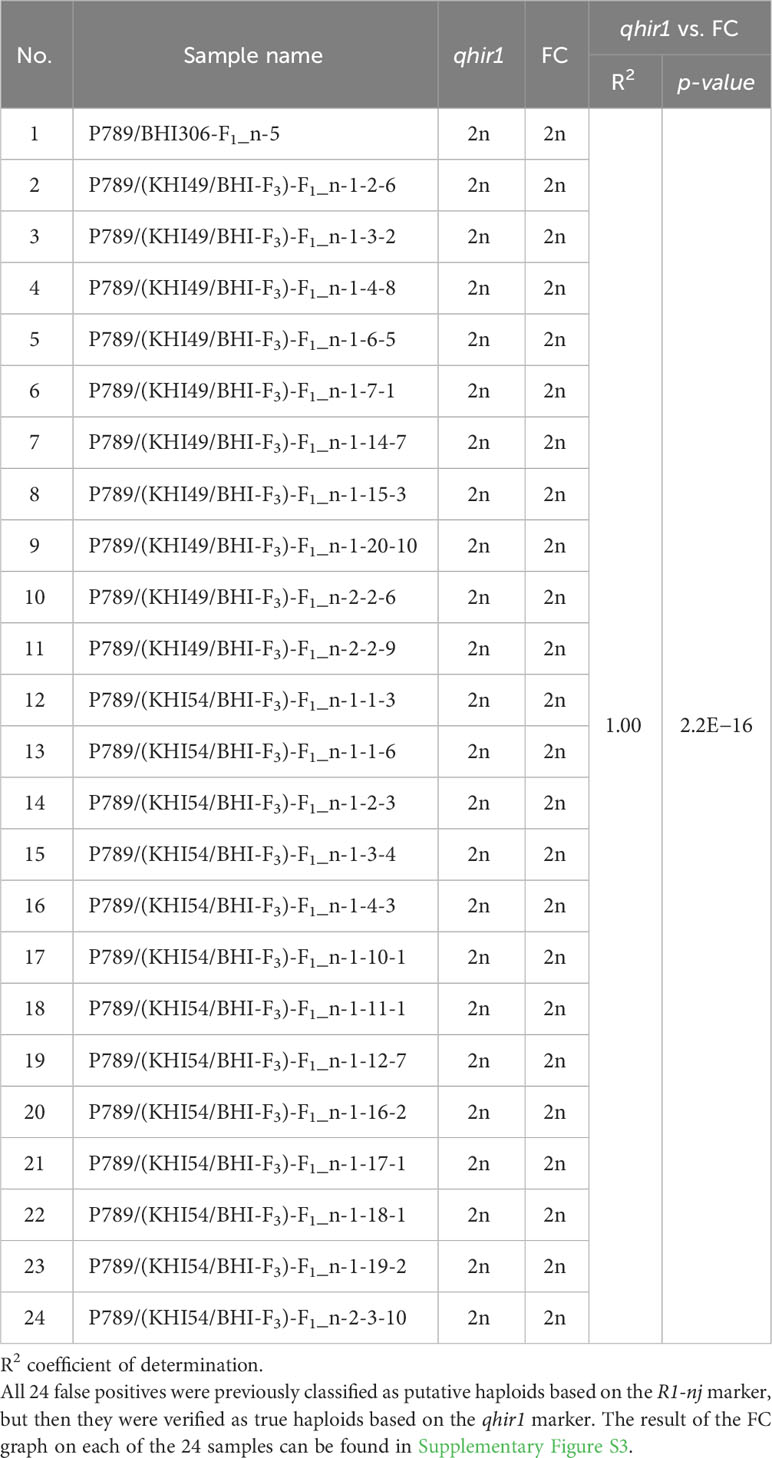
Table 1 Haploid validation via qhir1 marker and flow cytometry (FC) analysis of 24 false positives derived from subsets of F1 induction crosses between female donor P789 and three male inducers BHI306, KHI49/BHI306, and KHI54/BHI306.
Two maize leaves at 14 days after germination were cut about 3 cm in length (50-100 mg fresh weight) and placed into a plastic petri dish on ice. Then, 1.5 ml of LB01 buffer (15 mM Tris, 2 mM Na2EDTA, 0.5 mM spermine.4HCl, 80 mM KCl, 20 mM NaCl, 0.1% (v/v) Triton X-100, pH 7.5) (Doležel, 1997) was added, and the leaves were chopped in this buffer using a razor blade to facilitate the release of the nuclei (Pfosser et al., 1995). After that, 500 ul of the cell solution was transferred into a 1.5 ml tube. Propidium Iodide 1 mg/mlI-stained nuclei and RNaseA were then added to the solution. The BD Accuri™ C6 Plus flow cytometer (BD Biosciences, USA) was employed for measurement. The ploidy status of each sample can be determined by the fluorescence intensity of stained cell nuclei isolated from plant tissue. The peak value (G1) of haploid is commonly set to half of the diploid reference (Supplementary Figure S3).
Statistical analysis
A total of 237 inducer plants were evaluated for HIR performance including K8/BHI306 (32 plants), K11/BHI306 (54 plants), KHI49/BHI306 (52 plants), and KHI54/BHI306 (99 plants). Each induced donor ear was represented as a technical replicate, resulting in four replications for each inducer plant within each genotype and family. The HIR for each genotype was calculated as the mean HIR across these four replications. The data were subjected to the unpaired samples t-test with 95% confidence interval (CI), Tukey’s Honestly Significant Difference (HSD) Test at 5%, and linear regression analysis.
Results
Haploid inducer breeding via marker-assisted selection for qhir1 and qhir8
The median HIR of qhir1+/qhir8+ genotypes was significantly (P<0.01) higher than that of the qhir1+/qhir8− genotypes within each F3 inducer family (Figure 2A). Across the four families, the average HIR for the qhir1+/qhir8+ genotype ranged from 3.85 to 9.48%, while the average HIR for the qhir1+/qhir8− genotype was significantly (P<0.01) lower, ranging from 1.18 to 4.89% (Figure 2B; Table 2). This suggests that inducer genotypes fixed for both targeted loci for HIR, qhir1 and qhir8, have remarkable abilities to induce haploids, showing an increase of 3–5% or 1–3-fold higher than inducer genotypes fixed for qhir1 only.
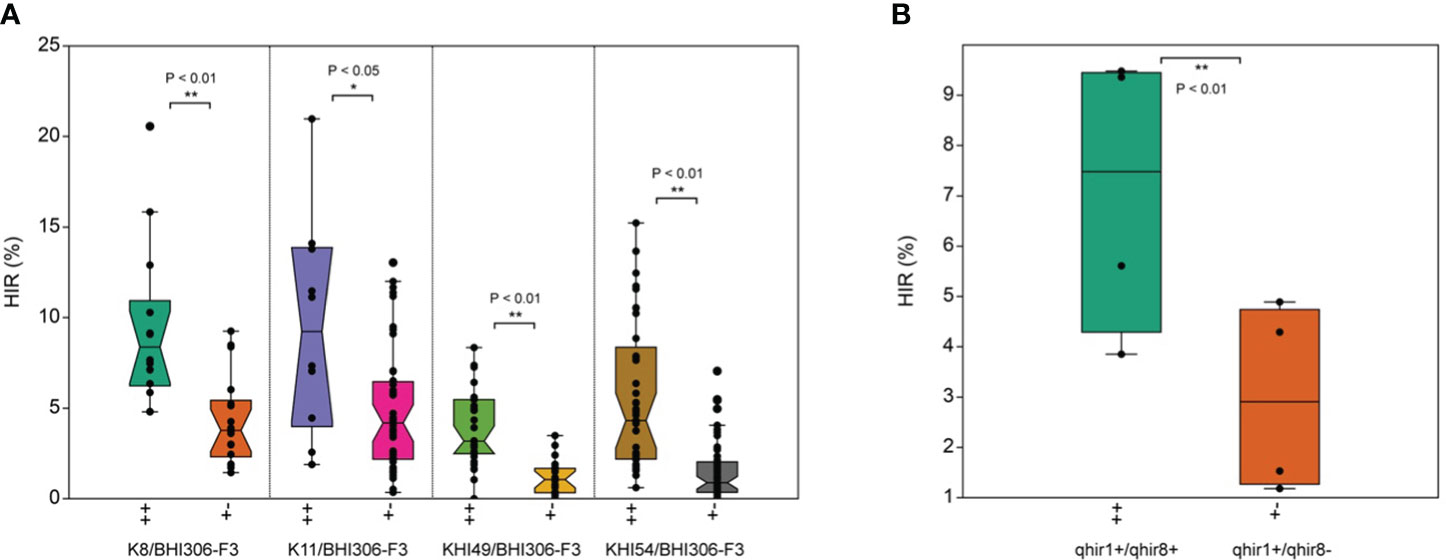
Figure 2 Haploid induction rate (HIR) in F3 inducer families with qhir1+/qhir8+ (++) and qhir1+/qhir8− (+−) genotypes involving (A) four different inducer parents; and (B) averaged means over four respective inducer populations. *, and ** data significant through paired samples t-test at P<0.05 and P<0.01, respectively.
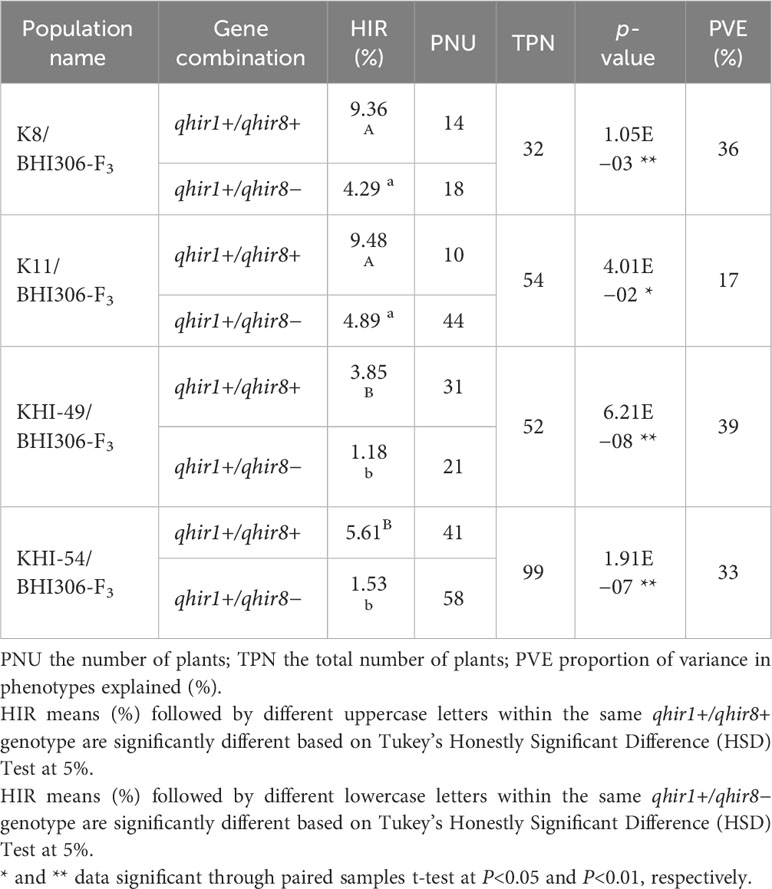
Table 2 The mean comparison between qhir1+/qhir8+ and qhir1+/qhir8− genotypes of each F3 family on haploid induction rate (HIR, %).
The proportion of phenotypic variation explained (PVE) across inducer families ranged from 17% to 39% (Table 2). These values, within acceptable ranges, indicated that MAS for two loci was effective in identifying haploid inducers with high HIR. We also found that the HIR between families within the same qhir1+/qhir8+ genotype was significantly different (Table 2). For instance, families K8/BHI306 and K11/BHI306 demonstrated a significantly higher HIR than families KHI-49/BHI306 and KHI-54/BHI306. The evidence of low %PVE (<50%) (Table 2), outliers, and overlapping values between two inducer groups on HIR (Figure 2), suggests the potential existence of other minor QTL influencing HIR.
Haploid validation via qhir1 marker and flow cytometry analysis
Marker-assisted selection (MAS) for qhir1 was applied to validate putative haploids and diploids derived from the R1-nj marker system as a preliminary haploid identification among the F1 progenies of induction crosses. Both parents, BHI306 and P789, were included as positive and negative controls for qhir1, respectively (Figure 3). Through the TaqMan assay, all samples of P789, the female donor, were found to be homozygous for qhir1− (G/G), while all samples of BHI306, the male inducer, were homozygous qhir1+ (GGAGC/GGAGC). The sample progenies were then distributed into two pools according to haplotypes: (1) the diploid class, heterozygous for qhir1 (G/GGAGC) and (2) the haploid class, homozygous for qhir1− (G/G), which was grouped with the donor female P789 (Figure 3B, Table 3). Similar results for other populations can be seen in Supplementary Table S2 and Supplementary Figure S2. A few numbers of false positives were found in putative haploid populations derived from induction crosses, accounting for 1, 10, and 13 samples in populations P789/BHI306-F1, P789/(KHI49/BHI306-F3)-F1, and P789/(KHI54/BHI306-F3)-F1, respectively (Table 3). The reliability of the qhir1 for haploid determination was further validated by flow cytometric analysis. We found that the result of FC analysis among 24 false positives (Supplementary Figure S3) corresponded to the qhir1 marker, as indicated by R2 = 1.00 (Table 1). This implies that the qhir1 marker using the TaqMan assay was effective to identify true haploids, indicated by a 0-false positive rate, which could thus serve as an alternative gold standard test compared to flow cytometry in future. It also suggests that a single SNP marker, like qhir1, is ultimately sufficient for haploid identification to reduce the cost of genotyping.
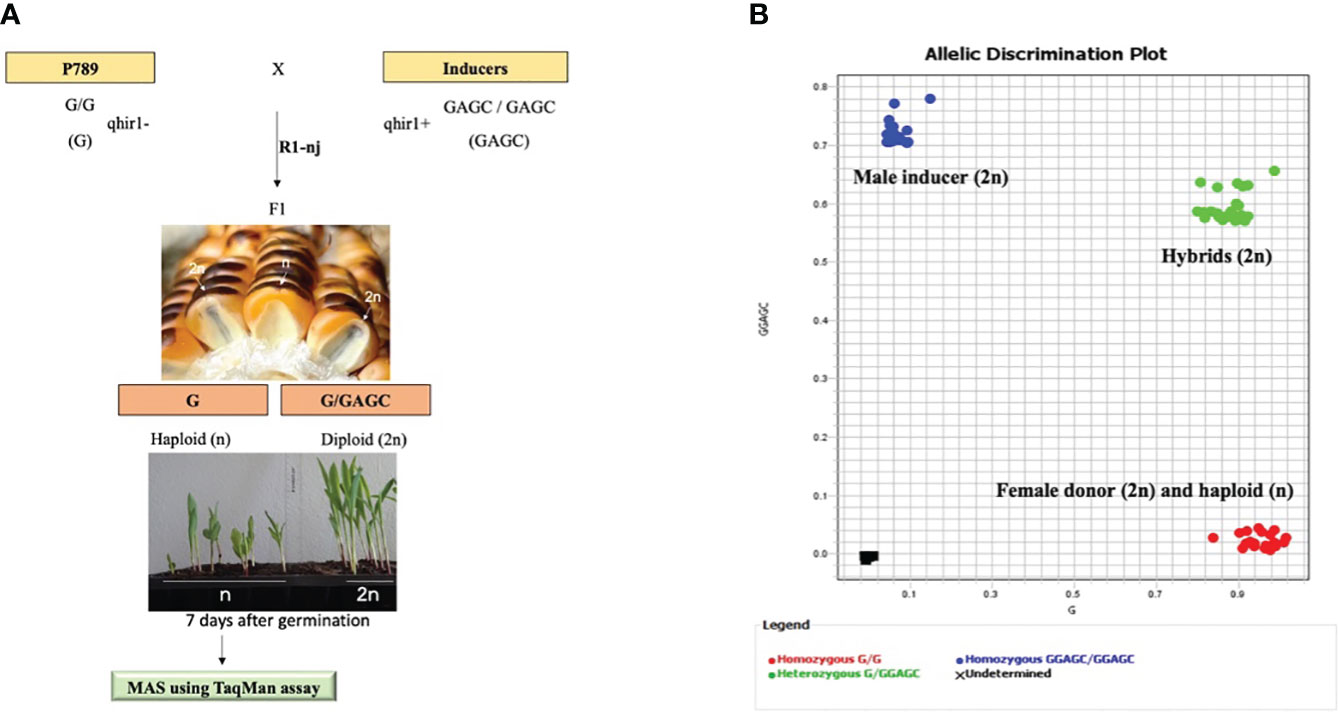
Figure 3 Workflow for haploid identification through the R1-nj and haploid validation by MAS using qhir1 TaqMan assay (A) and the allelic distribution plot from TaqMan assay (B). The diploid and haploid classes are represented by the green and red dots, respectively. Genotype BHI306 was used as male inducer, whereas genotype P789 was used as female donor. The SNP graphs for other populations can be found in Supplementary Figure S2.
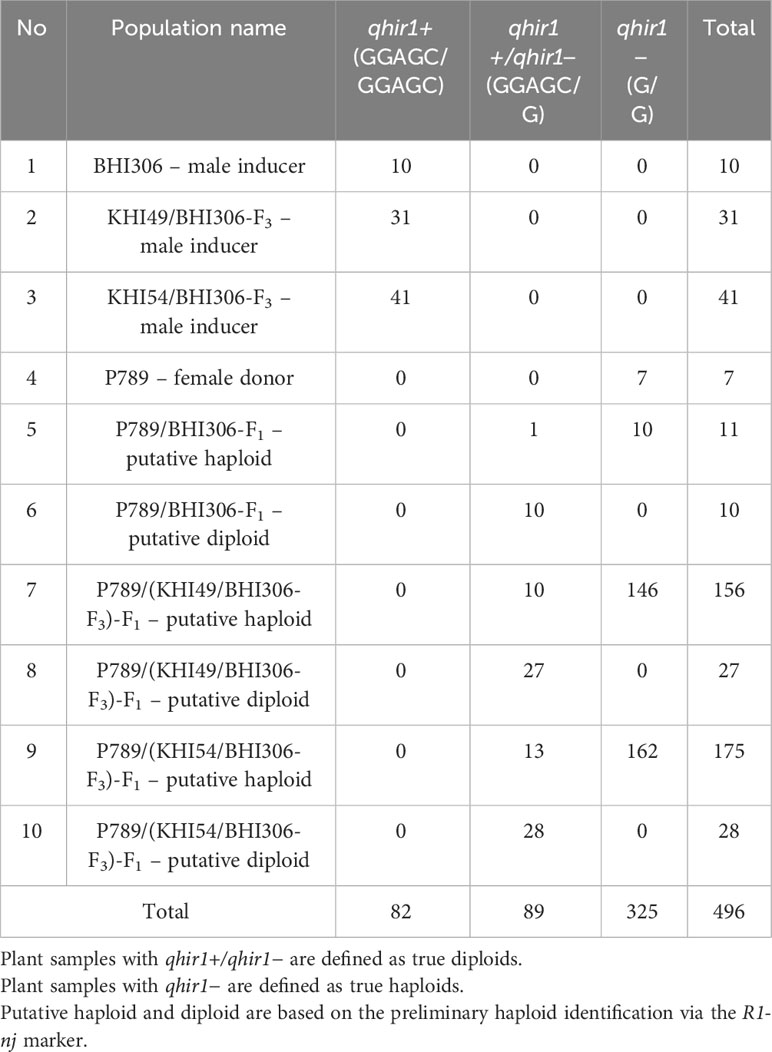
Table 3 Ploidy identification (haploid vs. diploid) via TaqMan assay for qhir1 in a sub-set population of induction crosses between a male inducer BHI306 and a female donor P789.
Discussion
Marker-assisted selection (MAS) may accelerate breeding programs by indirectly selecting target traits using molecular markers tightly linked to underlying genes (Xu and Crouch, 2008). Plant breeders can benefit from this approach especially when targeted traits pose challenges for improvement through traditional phenotypic selections. Technical issues such as resource intensiveness and genetic properties like low heritability, complex inheritance, and presence of recessive alleles make phenotypic selection difficult (Koebner, 2004; Collard et al., 2005; Xu et al., 2005). It is suitable for our breeding objectives to accelerate the rates of haploid induction (HIR) possessing multiple recessive alleles and QTL (Prigge et al., 2012) and prone to the environments of haploid induction (Kebede et al., 2011; De La Fuente et al., 2018; Sintanaparadee et al., 2022).
The effectiveness of MAS for qhir1 has been reported in the breeding high-oil inducers (Dong et al., 2014) and the development of CIMMYT second-generation Tropically Adapted Inducer Lines (CIM2GTAILs) (Chaikam et al., 2018). Trentin et al. (2020) suggested a stratified MAS approach, initially targeting the mtl allele or qhir1 in a large F2 population and later for zmdmp allele or qhir8 in F3 plants carrying the mtl allele or qhir1. In our study, we validated the efficacy of simultaneous MAS for qhir1 and qhir8 in F2 segregating populations, leading to enhanced HIR in F3 genotypes by 1–3-fold. We also noticed that the genotype of qhir1−/qhir8+ and heterozygous qhir1/qhir8+ showed lower HIR than genotypes with qhir1+ (data not shown). Our findings align with Zhong et al. (2019), who identified a novel mutation in the ZmDMP gene in the CAUHOI (qhir1+) genotype and demonstrated its impact on HIR. They found that the genotype with qhir1+/qhir8+ exhibited inflating HIR by 5–6-fold compared to qhir1+/qhir8−. The implementation of MAS in the early generations proves beneficial by significantly reducing the number of F3 plants that need evaluation for actual HIR through resource-intensive haploid induction and haploid selection. Chen et al. (2020) also reported the effectiveness of simultaneous MAS for qhir1 and qhir8, resulting in a substantial increase in HIR by 3–14% and the elimination of approximately 90% of low-HIR genotypes.
Our study did not include inducer families with qhir8 only because we aimed to investigate the synergistic effects between qhir1 and qhir8 on HIR. Previous studies have reported that qhir8 alone resulted in poor or even null HIR. For instance, Chen et al. (2020) reported that the HIR of the plants with qhir8 only ranged from 0.70% to 1.04%, which was significantly lower than either those that carried a heterozygous qhir1 allele or those that carried a homozygous qhir1, with HIRs of 3.77% to 5.27% and 10.02% to 14.42%, respectively.
Previous studies reported six minor QTL (qhir2, qhir3, qhir4, qhir5, qhir6, and qhir7) (Prigge et al., 2012) and a novel gene, ZmPLD3 (Li et al., 2021). Mutations of the ZmPLD3 gene resulted in a haploid induction rate (HIR) comparable to that of the homozygous recessive MTL gene. This mutation showed synergistic effects rather than functional redundancy in tripling HIR in the presence of the homozygous recessive MTL gene. Later in 2022, Meng and colleagues manipulated the Stock6-derived inducer lines by overexpressing maize CENH3 fused with different fluorescent protein tags and found that the engineered Stock6-derived lines showed a noticeable increase in the maternal HIR up to 16.3%, which was increased by ~6.1% than Stock6-derived lines control (Meng et al., 2022). Hu et al. (2016) found two minor QTL responsible for HIR expression, namely qhir11 and qhir12, which are closely linked to the major QTL qhir1. While the qhir11 was not diagnostic for differentiating inducers and non-inducers, the qhir12 had a haplotype allele common to all inducer lines observed but not found in all non-inducers studied. In addition, they noticed that the qhir12 region was related to three candidate genes involved in DNA or amino acid binding. (Nair et al., 2017) performed a genome wide association study (GWAS) and identified more than 20 SNPs associated with HIR in two different association mapping panels. A recent genome-wide association study (GWAS) involving 159 haploid inducers has confirmed the polygenic nature of HIR and identified a major gene near MTL, a significant QTL on chromosome 10, and other minor QTL on six of the ten chromosomes (Trentin et al., 2023a). It is conceivable that these QTL, or even undiscovered ones, may be present in our inducer genotypes, highlighting the need for further investigations to discover novel QTL conferring HIR. Drawing insights from Prigge et al. (2012), this endeavor is feasible, as the number of QTL and the magnitude of QTL effects for HIR can vary across populations and generations.
This present study serves as a continuation of the haploid inducer breeding program, focusing on achieving high HIR and local adaptation to the tropical savanna in Thailand. In our previous reports, relying solely on phenotypic selection in the breeding strategy did not yield promising haploid inducers with satisfactory HIR, i.e., below 6% in two families KHI49 and KHI54 (Dermail et al., 2021) and two populations K8 and K11 (Thawarorit et al., 2023). The incorporation of genetic recombination with BHI306, an elite inducer stock carrying favorable alleles for HIR, and the implementation of precise selections such as MAS for simultaneous loci have now enabled us to obtain promising inducer genotypes. Notably, some individual plants within qhir1+/qhir8+ genotypes in families K8/BHI306 and K11/BHI306 demonstrated HIRs exceeding 20%, surpassing both founder parents (Supplementary Table S1).
The significant variations for HIR among families within the same qhir1+/qhir8+ genotype (Table 2) imply the importance of the genetic background of founder parents to establish those inducer families. We noticed that families KHI-49/BHI306 and KHI-54/BHI306 had significantly lower abilities to induce haploids than families K8/BHI306 and K11/BHI306. Although the four females (KHI-49, KHI-54, K8, and K11) derived from the same haploid inducer, Stock-6, they experienced different selection schemes. Regarding the pedigree information, the females KHI-49 and KHI-54 had experienced long-term selections, including six for non-HIR related traits and the following three for R1-nj kernel marker. Some favorable alleles responsible for HIR may be lost during selections since Chaikam et al. (2019) argued that non-inducer pollen showed selection advantages over inducer pollen. In contrast, the females K8 and K11 only experienced one selection cycle among F2 populations derived from crosses between Stock-6 haploid inducer and non-inducer waxy maize germplasm. We assumed that the proportion of HIR-related favorable alleles in the K8 and K11 genotypes was higher than in KHI-49 and KHI-54.
Although per se on HIR can be altered by different testing environments and donor germplasm (Kebede et al., 2011; Prigge et al., 2011; De La Fuente et al., 2018; Sintanaparadee et al., 2022), our current finding suggests the presence of transgressive segregants in F3 families. We recommend further phenotypic evaluations in inducer families with qhir1+/qhir8+ genotypes, focusing on traits related to the ideotype of haploid inducers, such as plant height, ear height, flowering behaviors, tassel and pollen attributes, and seed set. This assessment will help determine the feasibility of those genotypes in haploid induction stage, whether in induction nurseries or isolation fields (Trentin et al., 2020; Trentin et al., 2023b). Considering the polygenic nature governing HIR and those mentioned agronomic traits, genomic selection approach can be applied to simultaneously identify promising parents to generate progenies with favorable performance on targeted traits prior to field evaluation. Almeida et al. (2020) implemented genomic prediction for cross prediction and parental selection in a haploid inducer breeding program with varying levels of accuracy depending on traits evaluated and suggested that HIR and agronomic traits can be improved simultaneously.
In our study, we proved that MAS for qhir1 is effective to confirm the true-to-type of haploids. The induced progenies were clustered into two pools according to haplotypes. This allelic clustering can be explained by two hypotheses: (1) single fertilization occurs when only the egg or the central cell is fertilized, resulting in kernels with haploid embryos (Sarkar and Coe, 1966) and (2) selective elimination of inducer genomes from embryonic cells (Zhao et al., 2013). Acknowledging the small sample sizes used, Linnet (1999) suggested that the minimum sample size for optimizing the regression analysis should fall withing the range of 40 to 100 samples. Therefore, conducting further replicated trials with a larger sample size is encouraged before fully realizing the potential of this approach in haploid identification in maize. As a practical proposal, molecular markers could be employed to verify R1-nj-based putative haploids at the early seedling stage, not exceeding seven days after planting (DAP). This timeline aligns with the common practice of haploid genome doubling using colchicine at 10-12 DAP (Vanous et al., 2017). To prevent the risk of R1-nj marker misclassification, an additional phenotypic marker, the red root phenotype at seedling stage from Pl-1 gene, was used. This phenotype results from light-independent anthocyanin production, although exposure to light conditions can induce anthocyanin pigmentation for some genotypes (Coe, 1994). Moreover, oil content was used as a screening criterion for haploid and diploid using nuclear magnetic resonance (NMR) (Wang et al., 2016). The success of this method required high-oil haploid inducers (Liu et al., 2022). Preventing high false positives through molecular assays can help to reduce the DH line production costs, because false positives can be discarded prior to haploid genome doubling (Baleroni et al., 2021).
Conclusions
Our study revealed that implementing marker-assisted selection (MAS) for qhir1 and qhir8 at an early generation (F3) substantially enhanced the haploid induction rate (HIR) of tropical × temperate haploid inducer families. On average, the HIRs of families homozygous for both qhir1+ and qhir8+ were 1–3-fold higher than those homozygous for qhir1+ only. The qhir1 marker, utilizing the TaqMan assay, effectively distinguished diploid/haploid progenies at the early seedling stage (≤7 DAP) with high accuracy (100%), as validated by flow cytometric analysis. We propose the integration of MAS to expedite the breeding of haploid inducers for high HIR, complementing the use of the R1-nj marker for the identification of true haploids.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author contributions
KK: Writing – original draft, Conceptualization, Data curation, Formal analysis, Methodology. AD: Writing – original draft, Conceptualization, Methodology, Writing – review & editing. KS: Writing – review & editing, Conceptualization, Funding acquisition, Methodology, Supervision. TL: Supervision, Writing – review & editing. SW: Supervision, Writing – review & editing. BT: Supervision, Writing – review & editing. WP: Supervision, Writing – review & editing. TT: Supervision, Writing – review & editing. VR: Conceptualization, Methodology, Supervision, Writing – review & editing. SA: Writing – review & editing, Supervision.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. The National Science and Technology Development Agency (NSTDA) (Grant No. P-20-52286, P-21-50610 and P-23-51489). Also, the National Science, Research and Innovation Fund, Thailand Science Research and innovation (TSRI).
Acknowledgments
The authors would like to thank the Plant Breeding Research Center for Sustainable Agriculture, Faculty of Agriculture, Khon Kaen University, Thailand, for providing plant materials and research facilities. As well as the High-Quality Research Graduate Development Cooperation Project between Kasetsart University and the National Science and Technology Development Agency (NSTDA) for providing the scholarship.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2024.1337463/full#supplementary-material
References
Almeida, V. C., Trentin, H. U., Frei, U. K., Lübberstedt, T. (2020). Genomic prediction of maternal haploid induction rate in maize. Plant Genome. 13, e20014. doi: 10.1002/tpg2.20014
Baleroni, A. G., Ré, F., Pelozo, A., Kamphorst, S. H., Carneiro, J. W. P., Rossi, R. M., et al. (2021). Identification of haploids and diploids in maize using seedling traits and flow cytometry. Crop Breed. Appl. Biotechnol. 21, e38422145. doi: 10.1590/1984-70332021v21n4a54
Barret, P., Brinkmann, M., Beckert, M. A. (2008). Major locus expressed in the male gametophyte with incomplete penetrance is responsible for in situ gynogenesis in maize. Theor. Appl. Genet. 117, 581–594. doi: 10.1007/s00122-008-0803-6
Chaikam, V., Martinez, L., Melchinger, A. E., Schipprack, W., Boddupalli, P. M. (2016). Development and validation of red root marker-based haploid inducers in maize. Crop Sci. 56, 1678–1688. doi: 10.2135/cropsci2015.10.0653
Chaikam, V., Molenaar, W., Melchinger, A. E., Boddupalli, P. M. (2019). Doubled haploid technology for line development in maize: technical advances and prospects. Theor. Appl. Genet. 132, 3227–3243. doi: 10.1007/s00122-019-03433-x
Chaikam, V., Nair, S. K., Babu, R., Martinez, L., Tejomurtula, J., Boddupalli, P. M. (2015). Analysis of effectiveness of R1-nj anthocyanin marker for in vivo haploid identification in maize and molecular markers for predicting the inhibition of R1-nj expression. Theor. Appl. Genet. 128, 159–171. doi: 10.1007/s00122-014-2419-3
Chaikam, V., Nair, S. K., Martinez, L., Lopez, L. A., Utz, H. F., Melchinger, A. E., et al. (2018). Marker-assisted breeding of improved maternal haploid inducers in maize for the tropical/subtropical regions. Front. Plant Sci. 9. doi: 10.3389/fpls.2018.01527
Chen, C., Xiao, Z., Zhang, J., Li, W., Li, J., Liu, C., et al. (2020). Development of in vivo haploid inducer lines for screening haploid immature embryos in maize. Plants. 9, 739. doi: 10.3390/plants9060739
Coe, E. H. (1994). ““Anthocyanin genetics,”,” in The maize handbook (Springer, New York, NY, USA), 279–281.
Collard, B. C. Y., Jahufer, M. Z. Z., Brouwer, J. B., Pang, E. C. K. (2005). An introduction to markers, quantitative trait loci (QTL) mapping and marker-assisted selection for crop improvement: the basic concepts. Euphytica 142, 169–196. doi: 10.1007/s10681-005-1681-5
Cooper, M., Gho, C., Leafgren, R., et al. (2014). Breeding drought-tolerant maize hybrids for the US corn-belt: discovery to product. J. Exp. Bot. 65, 6191–6204. doi: 10.1093/jxb/eru064
De La Fuente, G. N., Frei, U. K., Trampe, B., et al. (2018). A diallel analysis of a maize donor population response to in vivo maternal haploid induction: I. Inducibility. Crop Sci. 58, 1830–1837. doi: 10.2135/cropsci2017.05.0285
Dermail, A., Chankaew, S., Lertrat, K., Lübberstedt, T., Suriharn, K. (2021). Selection gain of maize haploid inducers for the tropical savanna environments. Plants. 10, 2812. doi: 10.3390/plants10122812
Dermail, A., Lübberstedt, T., Suwarno, W. B., Chankaew, S., Lertrat, K., Ruanjaichon, V., et al. (2023). Combining ability of tropical× temperate maize inducers for haploid induction rate, R1-nj seed set, and agronomic traits. Front. Plant Sci. 14. doi: 10.3389/fpls.2023.1154905
Doležel, J. (1997). Application of flow cytometry for the study of plants genomes. J. Appl. Genet. 38, 285–302.
Dong, L., Li, L., Liu, C., Liu, C., Geng, S., Li, X., et al. (2018). Genome editing and double-fluorescence proteins enable robust maternal haploid induction and identification in maize. Mol. Plant 11, 1214–1217. doi: 10.1016/j.molp.2018.06.011
Dong, X., Xu, X., Li, L., Liu, C., Tian, X., Li, W., et al. (2014). Marker-assisted selection and evaluation of high oil in vivo haploid inducers in maize. Mol. Breed 34, 1147–1158. doi: 10.1007/s11032-014-0106-3
Gilles, L. M., Khaled, A., Laffaire, J. B., Chaignon, S., Gendrot, G., Laplaige, J., et al. (2017). Loss of pollen-specific phospholipase NOT LIKE DAD triggers gynogenesis in maize. EMBO J. 36, 707–717. doi: 10.15252/embj.201796603
Hu, H., Schrag, T. A., Peis, R., Unterseer, S., Schipprack, W., Chen, S., et al. (2016). The genetic basis of haploid induction in maize identified with a novel genome-wide association method. Genetics. 202, 267–1276. doi: 10.1534/genetics.115.184234
Jacquier, N. M. A., Gilles, L. M., Martinant, J. P., Rogowsky, P. M., Widiez, T. (2021). Maize in planta haploid inducer lines: A cornerstone for doubled haploid technology. Doubled Haploid Technol. 2, 25–48. doi: 10.1007/978-1-0716-1335-1_2
Kebede, A. Z., Dhillon, B. S., Schipprack, W., Araus, J. L., Bänziger, M., Semagn, K., et al. (2011). Effect of source germplasm and season on the in vivo haploid induction rate in tropical maize. Euphytica. 180, 219–226. doi: 10.1007/s10681-011-0376-3
Kelliher, T., Starr, D., Richbourg, L., Chintamanani, S., Delzer, B., Nuccio, M. L., et al. (2017). MATRILINEAL, a sperm-specific phospholipase, triggers maize haploid induction. Nature. 542, 105–109. doi: 10.1038/nature20827
Koebner, R. M. (2004). Marker assisted selection in the cereals: the dream and the reality. Cereal Genomics, 317–329. doi: 10.1007/1-4020-2359-6_10
Li, Y., Lin, Z., Yue, Y., et al. (2021). Loss-of-function alleles of ZmPLD3 cause haploid induction in maize. Nat. Plants. 7, 1579–1588. doi: 10.1038/s41477-021-01037-2
Linnet, K. (1999). Necessary sample size for method comparison studies based on regression analysis. Clin. Chem. 45, 882–894. doi: 10.1093/clinchem/45.6.882
Liu, C., Li, J., Chen, M., Li, W., Zhong, Y., Dong, X., et al. (2022). Development of high-oil maize haploid inducer with a novel phenotyping strategy. Crop J. 10, 524–531. doi: 10.1016/j.cj.2021.07.009
Liu, C., Li, X., Meng, D., Zhong, Y., Chen, C., Dong, X., et al. (2017). A 4-bp insertion at ZmPLA1 encoding a putative phospholipase A generates haploid induction in maize. Mol. Plant 10, 520–522. doi: 10.1016/j.molp.2017.01.011
Meng, D., Luo, H., Dong, Z., Huang, W., Liu, F., Li, F., et al. (2022). Overexpression of modified CENH3 in Maize Stock6-derived inducer lines can effectively improve maternal haploid induction rates. Front. Plant Sci. 13, 892055. doi: 10.3389/fpls.2022.892055
Nair, S. K., Molenaar, W., Melchinger, A. E., Boddupalli, P. M., Martinez, L., Lopez, L. A., et al. (2017). Dissection of a major QTL qhir1 conferring maternal haploid induction ability in maize. Theor. Appl. Genet. 130, 1113–1122. doi: 10.1016/j.cj.2019.09.008
Nanda, D. K., Chase, S. S. (1966). An embryo marker for detecting monoploids of maize (Zea mays L.). Crop Sci. 6, 213–215. doi: 10.2135/cropsci1966.0011183X000600020036x
Pfosser, M., Amon, A., Lelley, T., Heberlebors, E. (1995). Evaluation of sensitivity of flow-cytometry in detecting aneuploidy in wheat using disomic and ditelosomic wheat-rye addition lines. Cytometry 21, 387–393. doi: 10.1002/cyto.990210412
Prasanna, B. M. (2012). Diversity in global maize germplasm: Characterization and utilization. J. Biosci. 37, 843–855. doi: 10.1007/s12038-012-9227-1
Prigge, V., Sánchez, C., Dhillon, B. S., Schipprack, W., Araus, J. L., Bänziger, M., et al. (2011). Doubled haploids in tropical maize: I. Effects of inducers and source germplasm on in vivo haploid induction rates. Crop Sci. 51, 1498–1506. doi: 10.2135/cropsci2010.10.0568
Prigge, V., Xu, X., Li, L., Babu, R., Chen, S., Atlin, G. N., et al. (2012). New insights into the genetics of in vivo induction of maternal haploids, the backbone of doubled haploid technology in maize. Genetics. 190, 781–793. doi: 10.1534/genetics.111.133066
Qiu, F., Liang, Y., Li, Y., Liu, Y., Wang, L., Zheng, Y. (2014). Morphological, cellular and molecular evidences of chromosome random elimination in vivo upon haploid induction in maize. Curr. Plant Biol. 1, 83–90. doi: 10.1016/j.cpb.2014.04.001
Sarkar, K. R., Coe, E. H. (1966). A genetic analysis of the origin of maternal haploids in maize. Genetics. 54, 453–464. doi: 10.1093/genetics/54.2.453
Sintanaparadee, P., Dermail, A., Lübberstedt, T., Lertrat, K., Chankaew, S., Ruanjaichon, V., et al. (2022). Seasonal variation of tropical savanna altered agronomic adaptation of Stock-6-derived inducer lines. Plants. 11, 2902. doi: 10.3390/plants11212902
Thawarorit, A., Dermail, A., Lertrat, K., Chankaew, S., Suriharn, K. (2023). Stratified haploid identification system through the R1-nj kernel and reduced seedling vigor in tropical maize germplasm. Biodiversitas. 24, 4262–4268. doi: 10.13057/biodiv/d240807
Trentin, H. U., Batîru, G., Frei, U. K., Dutta, S., Lübberstedt, T. (2022). Investigating the effect of the interaction of maize inducer and donor backgrounds on haploid induction rates. Plants. 11, 1527. doi: 10.3390/plants11121527
Trentin, H. U., Frei, U. K., Lübberstedt, T. (2020). Breeding maize maternal haploid inducers. Plants. 9, 614. doi: 10.3390/plants9050614
Trentin, H. U., Krause, M. D., Zunjare, R. U., Almeida, V. C., Peterlini, E., Rotarenco, V., et al. (2023a). Genetic basis of maize maternal haploid induction beyond MATRILINEAL and ZmDMP. Front. Plant Sci. 14. doi: 10.3389/fpls.2023.1218042
Trentin, H. U., Yavuz, R., Dermail, A., Frei, U. K., Dutta., S., Lübberstedt, T. (2023b). A comparison between inbred and hybrid maize haploid inducers. Plants. 12, 1095. doi: 10.3390/plants12051095
Vanous, K., Vanous, A., Frei, U. K., Lübberstedt, T. (2017). Generation of maize (Zea mays) doubled haploids via traditional methods. Curr. Protoc. Plant Biol. 2, 147–157. doi: 10.1002/cppb.20050
Wang, H., Liu, J., Xu, X., Huang, Q., Chen, S., Yang, P., et al. (2016). Fully-automated high-throughput NMR system for screening of haploid kernels of maize (corn) by measurement of oil content. PloS One 11, e0159444. doi: 10.1371/journal.pone.0159444
Xu, X., Li, L., Dong, X., Jin, W., Melchinger, A. E., Chen, S. (2013). Gametophytic and zygotic selection leads to segregation distortion through in vivo induction of a maternal haploid in maize. J. Exp. Bot. 64, 1083–1096. doi: 10.1093/jxb/ers393
Xu, Y., Crouch, J. H. (2008). Marker-assisted selection in plant breeding: From publications to practice. Crop Sci. 48, 391–407. doi: 10.2135/cropsci2007.04.0191
Xu, Y., McCouch, S. R., Zhang, Q. (2005). How can we use genomics to improve cereals with rice as a reference genome? Plant Mol. Biol. 59, 7–26. doi: 10.1007/s11103-004-4681-2
Zhao, X., Xu, X., Xie, H., Chen, S., Jin, W. (2013). Fertilization and uniparental chromosome elimination during crosses with maize haploid inducers. Plant Physiol. 163, 721–731. doi: 10.1104/pp.113.223982
Keywords: hybrid breeding, doubled haploid, haploid induction, haploid identification, molecular assay
Citation: Khammona K, Dermail A, Suriharn K, Lübberstedt T, Wanchana S, Thunnom B, Poncheewin W, Toojinda T, Ruanjaichon V and Arikit S (2024) Accelerating haploid induction rate and haploid validation through marker-assisted selection for qhir1 and qhir8 in maize. Front. Plant Sci. 15:1337463. doi: 10.3389/fpls.2024.1337463
Received: 13 November 2023; Accepted: 19 February 2024;
Published: 05 March 2024.
Edited by:
Ting Peng, Henan Agricultural University, ChinaReviewed by:
Zenglu Li, University of Georgia, United StatesAnilkumar C, National Rice Research Institute (ICAR), India
Copyright © 2024 Khammona, Dermail, Suriharn, Lübberstedt, Wanchana, Thunnom, Poncheewin, Toojinda, Ruanjaichon and Arikit. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Siwaret Arikit, c2l3YXJldC5hQGt1LnRo; Vinitchan Ruanjaichon, dmluaXRjaGFuLnJ1YUBiaW90ZWMub3IudGg=
†These authors have contributed equally to this work
 Kanogporn Khammona
Kanogporn Khammona Abil Dermail
Abil Dermail Khundej Suriharn
Khundej Suriharn Thomas Lübberstedt
Thomas Lübberstedt Samart Wanchana
Samart Wanchana Burin Thunnom
Burin Thunnom Wasin Poncheewin
Wasin Poncheewin Theerayut Toojinda
Theerayut Toojinda Vinitchan Ruanjaichon
Vinitchan Ruanjaichon Siwaret Arikit
Siwaret Arikit