- 1College of Life Sciences, State Key Laboratory of Crop Genetics & Germplasm Enhancement and Utilization, Nanjing Agricultural University, Nanjing, China
- 2Sanya Institute of Nanjing Agricultural University, Nanjing Agricultural University, Sanya, China
Pectin methylesterase (PME), a family of enzymes that catalyze the demethylation of pectin, influences seed germination. Phytohormone abscisic acid (ABA) inhibits seed germination. However, little is known about the function of PMEs in response to ABA-mediated seed germination. In this study, we found the role of PME31 in response to ABA-mediated inhibition of seed germination. The expression of PME31 is prominent in the embryo and is repressed by ABA treatment. Phenotype analysis showed that disruption of PME31 increases ABA-mediated inhibition of seed germination, whereas overexpression of PME31 attenuates this effect. Further study found that ABI5, an ABA signaling bZIP transcription factor, is identified as an upstream regulator of PME31. Genetic analysis showed that PME31 functions downstream of ABI5 in ABA-mediated seed germination. Detailed studies showed that ABI5 directly binds to the PME31 promoter and inhibits its expression. In the plants, PME31 expression is reduced by ABI5 in ABA-mediated seed germination. Taken together, PME31 is transcriptionally inhibited by ABI5 and negatively regulates ABA-mediated seed germination inhibition. These findings shed new light on the mechanisms of PMEs in response to ABA-mediated seed germination.
Introduction
Seed germination is a vital step in plant growth and development and impacts the crop yield. The timing and rate of seed germination are controlled by a variety of internal and external factors (Graeber et al., 2012; Mei et al., 2023). Abscisic acid (ABA) and gibberellin acid (GA) are the primary internal factors that regulate the transition from dormancy to germination (Singh and Datta, 2023). ABA promotes seed dormancy and represses seed germination. GA is essential for the release of dormancy and for the initiation of germination. They antagonistically modulate seed germination, which is at least in part regulated by the balance and sensitivity to ABA and GA (Shu et al., 2013; Loades et al., 2023). In adverse environmental conditions, such as drought, high salinity, and extreme temperatures, ABA dominates the dormant state. The mechanisms of ABA-mediated seed germination inhibition have been investigated (Graeber et al., 2012; Tuan et al., 2018; Sharma et al., 2023). For instance, ABA reduces the water potential on the seed surface and water uptake to hinder seed germination (Berry and Bewley, 1992; Grainge et al., 2022). ABA also represses cell division and regulates the ion transport of seeds in seed germination (Liu et al., 2022). Besides, ABA modulates gene expression and regulates the activity of enzymes, such as amylases and protein kinases, to control seed germination (Xue et al., 2021; Li et al., 2022; Loades et al., 2023).
ABA initiates the cell response by activating downstream signaling genes. Several components have been identified in ABA signaling pathway, including PYRABACTIN RESISTANCE (PYR1)/PYR1-LIKE (PYL)/REGULATORY COMPONENT OF ABSCISIC ACID RECEPTOR (RCAR), SNF1-related protein kinase 2s (SnRK2s), protein phosphatases 2C (PP2Cs), ABSCISIC ACID INSENSITIVE3 (ABI3), ABSCISIC ACID INSENSITIVE4 (ABI4), and ABSCISIC ACID INSENSITIVE5 (ABI5) (Yan et al., 2020; Zhao et al., 2020). Among these, bZIP transcription factor ABI5 performs a crucial part in ABA-mediated seed germination (Lopez-Molina et al., 2001; Liu and Stone, 2010; Rushton et al., 2012; Nie et al., 2022). The abi5 mutants showed insensitivity to ABA in seed germination, while ABI5 overexpressors exhibited hypersensitivity (Lopez-Molina et al., 2001), suggesting that ABI5 positively regulated ABA-mediated seed germination inhibition. Studies showed that ABI5 directly regulated downstream target genes by binding to the G-box (ACGT) motif (Bi et al., 2017; Zhao et al., 2020). Bi et al. (2017) discovered that ABI5 was bound to the G-box of CAT1 promoter and activated CAT1 expression during seed germination. ABI5 also modulated the expression of ABA receptor genes (PYL11 and PYL12) by directly binding to their promoters in ABA-mediated seed germination (Zhao et al., 2020). A key negative regulator of GA signaling DELLA protein RGL2 was directly bound to the ABI5 promoter to promote its expression (Liu et al., 2016). Furthermore, ABI5 interacted with AFP, KEG, and MIEL1 in regulating seed germination (Liu and Stone, 2010; Nie et al., 2022). These studies demonstrated that ABI5 modulated seed germination in various manner.
Pectin methylesterase (PME) catalyzes the demethylation of pectin and alters cell wall structure and characteristics. The PME gene family is a large and diverse group with at least 66 members in Arabidopsis thaliana (Weber et al., 2013). Approximately 75% of Arabidopsis PME genes had tissue- and stress-specific expression profiles. The PME expression pattern showed the diversity of their involvement in cell wall modification throughout the development as well as stress responses (Pelloux et al., 2007; Huang et al., 2017). PMEs influence plant growth and development by changing the biomechanical features of the cell wall (Hongo et al., 2012; Zúñiga-Sánchez et al., 2014; Huang et al., 2017; Zhang et al., 2023). For instance, PME35 reduced the degree of methylation in pectin and affected the mechanical characteristics of the stem cell wall (Hongo et al., 2012). Mutation of PME3 influenced cell wall structure and zinc ion absorption (Weber et al., 2013). PMEs also play an important role in response to abiotic stresses. PME34 improved plant heat tolerance by promoting stomatal opening, and increased water evaporation and temperature regulation capabilities under high-temperature conditions (Huang et al., 2017). PME31 positively regulated salt stress tolerance (Yan et al., 2018). Brassinosteroids (BRs) modulated total PME activity in Arabidopsis by regulating PME41 expression under chilling stress (Qu et al., 2011). Seed germination is intimately connected to the degree of pectin methylesterification. The demethylesterification of pectin promoted seed coat rupture and endosperm release (Müller et al., 2013). Zúñiga-Sánchez et al. (2014) found that the DUF642 gene BIIDXI improved PME activity to enhance Arabidopsis seed germination performance. In addition, PME activity was inhibited when garden cress seeds were treated with ABA (Scheler et al., 2015), indicating that ABA influenced PME activity in seed germination. However, the role of ABA and PME genes in seed germination is still unknown.
Here, we showed the role of PME31 in ABA-mediated seed germination inhibition. PME31 negatively modulated ABA-mediated seed germination inhibition. We further identified the upstream regulator of PME31, an ABA-signaling bZIP transcription factor ABI5. We found that PME31 expression was inhibited by ABI5 in ABA-mediated seed germination. Our findings shed light on the mechanisms of PMEs in response to ABA-mediated seed germination.
Materials and methods
Plant material and growth conditions
The Arabidopsis Biological Resource Center (ABRC) provided Arabidopsis thaliana L. Heynh. Ecotype Columbia (Col-0), Landsberg erecta (Ler-0), pme31-1 (SALK_074820), pme31-2 (CS25163), abi3-1 (CS24), abi4-1 (CS8104) and abi5-1 (CS8105). The abi5-1 pme31-1 double mutant was created through a genetic cross between pme31-1 and abi5-1. The identified primers were listed in Supplementary Table S1. Seeds were planted in 1/2 MS medium and grew in a light incubator at 22°C/20°C (day/night), with a photoperiod of 16 h/8 h (day/night) and photosynthetic active radiation of 200 µmol m–2 s–1. Seeds were stratified at 4°C in darkness for various times (0, 12, 24, 36, 48 h) to detect PME31 gene expression in seed germination. Samples were collected and promptly preserved in liquid nitrogen for analysis.
Generation of transgenic plants
To generate PME31-overexpressing plants, the coding sequence of PME31 with specific primers (Supplementary Table S1) was amplified and inserted into the pEarleyGate 101 vector from Earley et al. (2006). To investigate PME31 tissue-specific expression, the PME31 promoter sequence (2142 bp) was cloned into the pMD19-T vector (Takara, Japan) and inserted into the pGWB3 vector from Nakagawa et al. (2007) (Supplementary Table S1). The reporter GUS gene was driven by the PME31 promoter (proPME31:GUS). The recombinant vectors were transformed into Agrobacterium tumefaciens strain GV3101. Transgenic plants were created by floral dip transformation (Clough and Bent, 1998). Positive plants were screened on 1/2 MS medium with 50 μg mL-1 Basta. Homozygous T3 transgenic plants were selected and collected for further analysis. ABI5 overexpressors were obtained from the Arashare platform. ABI5 was overexpressed by a β-estradiol-inducible promoter (Coego et al., 2014). ABI4 overexpressor was obtained from Yan et al. (2020).
Histochemical staining for β-glucuronidase (GUS) activity
The peeled seeds, rosette leaves, and roots of the proPME31:GUS transgenic plants were placed in GUS staining solution (10 mg mL X-Gluc, 0.1 M Na2HPO4-NaH2PO4 pH 7.0, 0.1% Triton X-100, 8 mM β-mercaptoethanol, 0.5 mM K3[Fe(CN)6], 0.5 mM K4[Fe(CN)6]) and vacuum-infiltrated for 30 min. The samples were stained at 37°C overnight and decolorized with 75% ethanol. The images were taken using a stereomicroscope (Optec SZ680, China).
Real-time quantitative PCR (RT-qPCR) analyses
Total RNA was extracted from plant materials using TRIzol Reagent (CoWin Biotech, China). RNAs were then reverse-transcribed into cDNA by the 5×All-In-One MasterMix Kit (Abm Inc., China). The cDNA was amplified using the CFX96 Touch Real-Time PCR System (Bio-Rad, USA). The reaction system was 20 μL, containing 10μL AceQ qPCR SYBR Green Master Mix (Vazyme, China), 1.0 μL of each primer, 100 ng cDNA. The amplification conditions were as follows: 95 ° for 10 min; 95 ° for 15 s; 60 ° for 1 min, 40 Cycles. The internal reference was Actin2. The details of the specific primers were listed in Supplementary Table S1. The relative expression of each gene was calculated using the 2-ΔΔCt method (Livak and Schmittgen, 2001).
Determination of PME activity
PME activity was determined using a PME activity assay kit (Thermo Fisher Scientific, USA). Samples were extracted from seeds with PBS buffer (0.01 M, pH7.4). The protein samples were centrifuged at 12,000g for 20 min and collected supernatants. Protein concentration was measured by Bradford assays (Bradford, 1976). Samples were measured according to the manufacturer’s instructions. After the reaction, the absorbance value was measured at 450nm. The PME activity was calculated using the standard curve.
Assays for seed germination and cotyledon greening
Assays for seed germination and cotyledon greening were performed as previously described (Yan et al., 2020). Each transgenic plant had more than 100 seeds collected simultaneously. After sterilization, seeds were sown on 1/2 MS medium either with or without 0.5 μM ABA. Once the radicle had emerged from the testa, the seed germination rate was recorded every 12 h. After 72 h, the cotyledon greening rate was measured. To investigate PME31 in response to ABA at the seedling stage, five-day-old seedlings were transferred to 1/2 MS medium with or without 10 µM ABA and then kept in an incubator under normal conditions for 14 d. Time to 50% germination and the percentage of maximum germination were recorded.
Yeast-one-hybrid assays (Y1H)
Y1H assays were performed as previously described (Zhou et al., 2020; Xiang et al., 2021a). The pLacZi-proPME31 and pB42AD-ABI5 constructs were transformed into yeast strain EGY48. Transformants were screened on SD/–Trp–Ura plates and cultured in SD/–Trp–Ura/Gal/Raf/X-Gal (80 μg mL−1) plates for color development. The β-galactosidase activity was detected using a β-Galactosidase Assay Kit (Beyotime, Shanghai, China). The β-galactosidase activity was calculated as previously described (Xiang et al., 2021a). The pLacZi and pB42AD vectors were used as the negative control.
Electrophoretic mobility shift assays (EMSA)
EMSA was performed as previously described (Xiang et al., 2021b). The coding sequence of ABI5 was inserted into the pET-30a vector (Novagen, USA). His-ABI5 proteins were expressed in Escherichia coli Rosetta (DE3) for 6 h at 24°C following being induced with 0.5 mM isopropyl-1-thio-β-D-galactopyranoside (IPTG). The recombined proteins were purified using the MagnetHis™ protein purification system (Promega, USA) and eluted with elution buffer (100 mM HEPES, 500 mM imidazole, pH7.5). The probe contained the ttaCACGTag sequence (−945 to −954 bp) and was labeled using an EMSA Probe Biotin Labeling Kit (Beyotime, Shanghai, China). EMSAs were performed using a Chemiluminescent EMSA Kit (Beyotime, Shanghai, China). The unlabeled probe (Comp) and the mutated probe (mComp) were used as competitors (Supplementary Table S1).
Luciferase assays
The PME31 promoter was inserted into the p1381-LUC vector from Xiang et al. (2021a) (Supplementary Table S1). The coding sequence of ABI5 was recombined into the pCAMBIA1300-221-3×flag vector (generated by insertion of a 35S promoter and a Flag tag into the pCAMBIA1300 vector) from Xiang et al. (2021a) (Supplementary Table S1). The recombinant plasmids were transformed into Agrobacterium tumefaciens strain GV3101. The proPME31:LUC-35S:REN reporter was infiltrated into tobacco (Nicotiana benthamiana) leaves. After infiltration for 72 h, the LUC signals were imaged using a Tanon 5200 multi-chemiluminescent imaging system (Tanon, China). The tobacco leaves were collected and frozen in liquid nitrogen. The luciferase activities were detected using a Dual Luciferase Reporter Gene Assay Kit (Yeasen, Shanghai, China). The LUC/REN ratio was used to calculate the relative promoter activity.
Accession numbers
The genes in this study have the following IDs: PME31, AT3G29090; ABI5, AT2G36270; Actin2, AT3G18780; ABI3, AT3G24650; ABI4, AT2G40220.
Results
The expression pattern of PME31 during seed germination
To investigate the tissue-specific expression of PME31, transgenic plants with PME31 promoter-driven GUS reporters were generated. Different tissues from various development stages were collected and stained by GUS histochemical staining. The peeled seeds were obtained from Arabidopsis seed at 12 h imbibition. The Arabidopsis seed embryo exhibited a blue color. When Arabidopsis grew to 10 days old, the rosette leaves and roots were stained by GUS and showed that the leaf vasculature and primary root displayed a blue color (Figure 1A). The results suggested PME31 was expressed in embryo, leaf vasculature, and primary root in Arabidopsis.
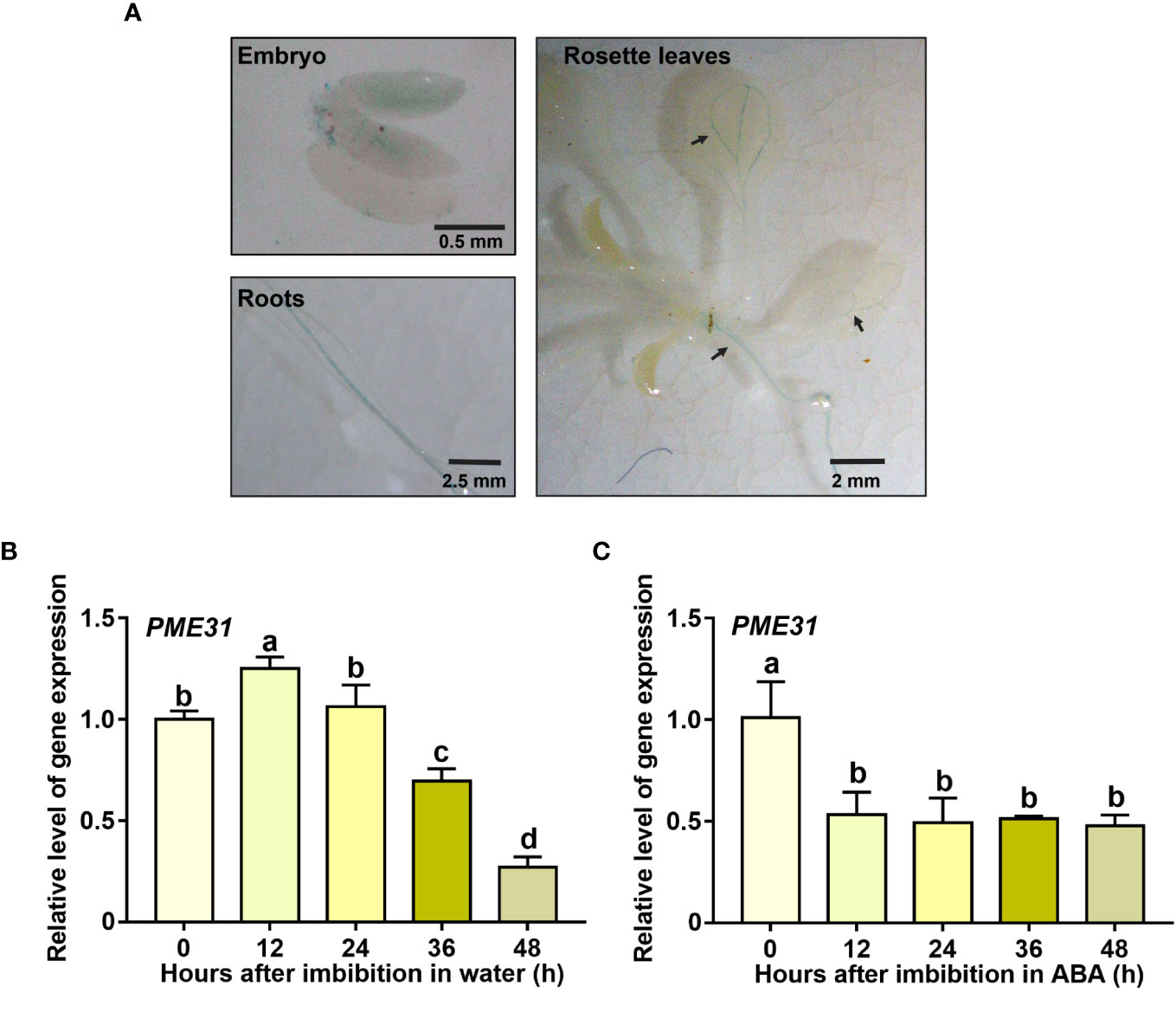
Figure 1 The expression pattern of PME31 in different tissue and seed germination. (A) Tissue-specific expression analysis of PME31 using GUS histochemical staining. The peeled seeds were obtained from Arabidopsis seed at 12 h imbibition. The rosette leaves and roots were obtained from 10-day-old Arabidopsis seedlings on 1/2 MS medium. The tissues were stained in GUS staining solution and vacuum-infiltrated for 30 min. The samples were stained at 37°C overnight and decolorized with 75% ethanol. The images were taken. (B) The expression of PME31 during seed germination. Seeds were imbibed at 25°C in darkness for various times (0, 12, 24, 36, 48 d). (C) The expression of PME31 in the presence of ABA during seed germination. Seeds were imbibed at 25°C in darkness and treated with 0.5 μM ABA. The gene expression of PME31 was determined using RT-qPCR. Actin2 was used as the internal reference. Data are means (± SD) of three biological replicates. Different letters indicate significant differences at P < 0.05 according to two-way ANOVA (Tukey’s multiple comparison test).
We next determined the expression pattern of PME31 during seed germination. After imbibition, the PME31 expression had a significant accumulation at 12 h imbibition and then decreased at 36 h and 48 h imbibition compared with dry seeds (Figure 1B). It suggested that PME31 have an impact on seed germination. Previous studies showed that seed germination is tightly controlled by ABA (Zhao et al., 2020). To investigate whether PME31 is involved in ABA-mediated seed germination, seeds were treated with ABA. The PME31 expression decreased in ABA treatment for 12 h and was maintained until 48 h. The results showed that PME31 expression was downregulated under ABA treatment (Figure 1C).
Disruption of PME31 promotes ABA-mediated inhibition of seed germination
To investigate whether PME31 is involved in ABA-mediated seed germination, we screened two pme31 mutants by PCR identification. The insertion site of the pme31-1 mutant was in exon, while that of the pme31-2 mutant was in 5’-UTR. RT-qPCR analysis showed that the expression of PME31 decreased in the pme31-2 mutant (Supplementary Figure S1). Thus, PME31 was knocked out in the pme31-1 mutant and knocked down in the pme31-2 mutant. Under normal conditions, the pme31-1 and pme31-2 mutants had similar cotyledon greening rates and germination rates compared with the Col-0. However, the cotyledon greening rate and germination rate of pme31-1 and pme31-2 mutants were significantly lower than that of Col-0 under ABA treatment (Figures 2A–C). Moreover, the time to 50% germination of pme31-1 and pme31-2 mutants was higher compared with the Col-0 under ABA treatment (Figure 2D). The percentage of maximum germination had no significant difference in the Col-0, pme31-1, and pme31-2 mutants (Figure 2E). The results suggested that disruption of PME31 increased sensitivity to ABA during seed germination.
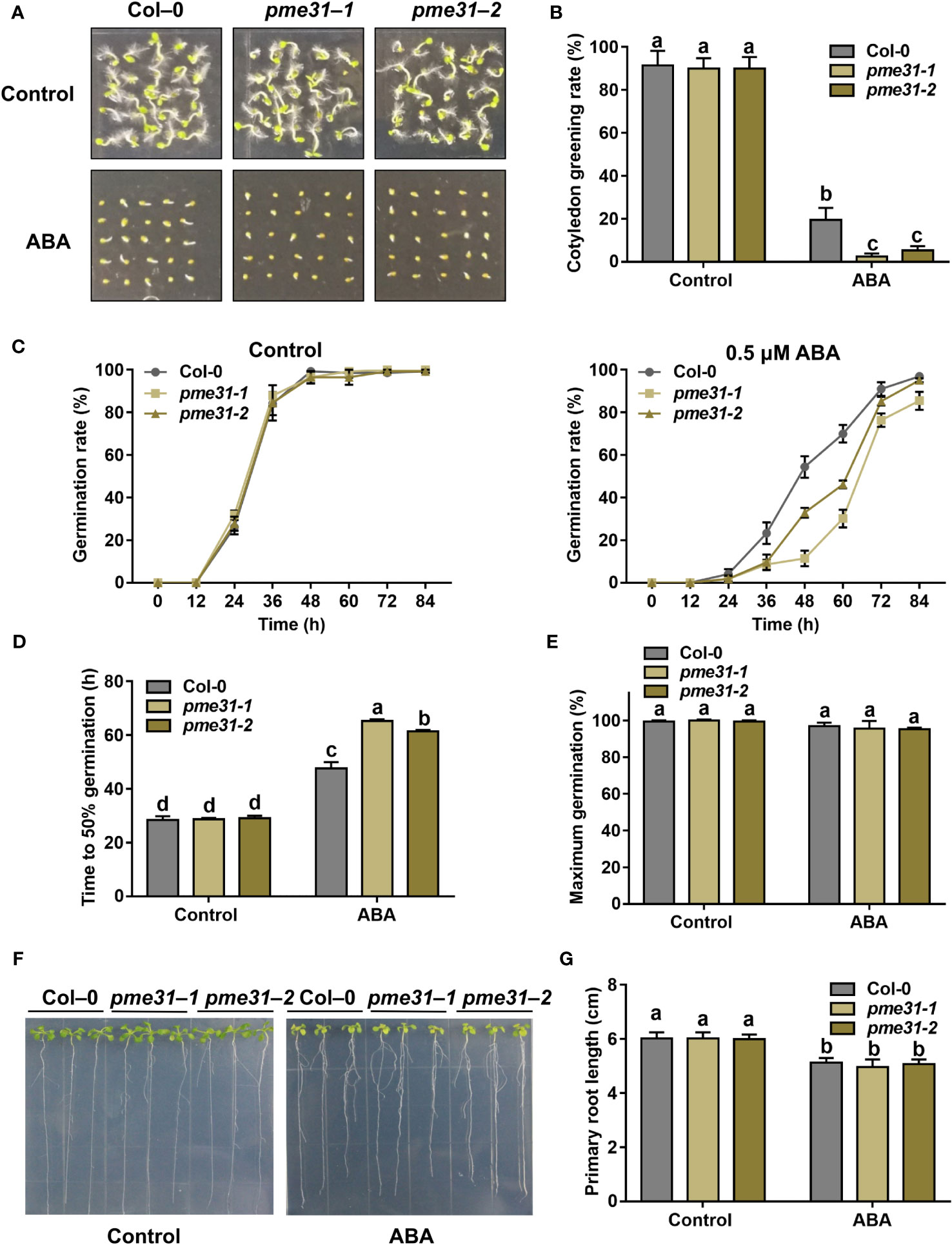
Figure 2 Disruption of PME31 improves ABA-mediated inhibition of seed germination. (A) Phenotype of seed germination in Col-0 and pme31 mutants treated with ABA. Seeds were sown on 1/2 MS medium with or without 0.5 µM ABA, and photographs were taken at 48 h of seed germination. (B) Green cotyledons rate was recorded after 72 h. (C) Germination rate of Col-0 and pme31 mutants. Seeds were sown on 1/2 MS medium with or without 0.5 μM ABA. The seed germination rate was recorded every 12 h. (D) Time to 50% gemination of Col-0 and pme31 mutants. (E) The percentage of maximum germination of Col-0 and pme31 mutants. Seeds were sown on 1/2 MS medium with 0.5 μM ABA. (F) Phenotype of seedlings in Col-0 and pme31 mutants treated with ABA. Five-day-old seedlings were selected and transferred to 1/2 MS medium with or without 10 µM ABA and placed in an incubator with normal conditions for 14 d. (G) Primary root length was recorded after growing for 14 d. Data are means (± SD) of three biological replicates. Different letters indicate significant differences at P < 0.05 according to two-way ANOVA (Tukey’s multiple comparison test).
To further test whether PME31 respond to ABA in the seedling stage, five-day-old seedlings were treated with ABA. Under normal conditions, the growth phenotype of pme31-1 and pme31-2 seedlings was not significantly different from Col-0. When supplemented with 10 µM ABA, there was also no significant difference in Col-0, pme31-1, and pme31-2 seedlings (Figures 2F, G), indicating that the PME31 mutation did not affect sensitivity to ABA in Arabidopsis seedlings. The above results suggested that disruption of PME31 improves ABA-mediated inhibition of seed germination.
Overexpression of PME31 attenuates ABA-mediated inhibition of seed germination
To further elucidate the role of PME31 in ABA-mediated seed germination, we created transgenic plants overexpressing PME31 under the control of the 35S promoter (Supplementary Figure S2A). RT-qPCR analysis showed that transgenic plants overexpressing PME31 (OE-PME31#3 and OE-PME31#4) had higher PME31 expression (Supplementary Figure S2B). Under normal conditions, PME31 overexpressors showed a similar cotyledon greening rate and germination rate as Col-0. Under ABA treatment conditions, the cotyledon greening rate and germination rate in PME31 overexpressors were significantly higher than that in Col-0 (Figures 3A–C). The time to 50% germination of PME31 overexpressors was lower compared with the Col-0 under ABA treatment (Figure 2D). The percentage of maximum germination had no significant difference in the Col-0 and PME31 overexpressors (Figure 2E). It indicated that PME31 overexpression reduced ABA-mediated inhibition of seed germination.
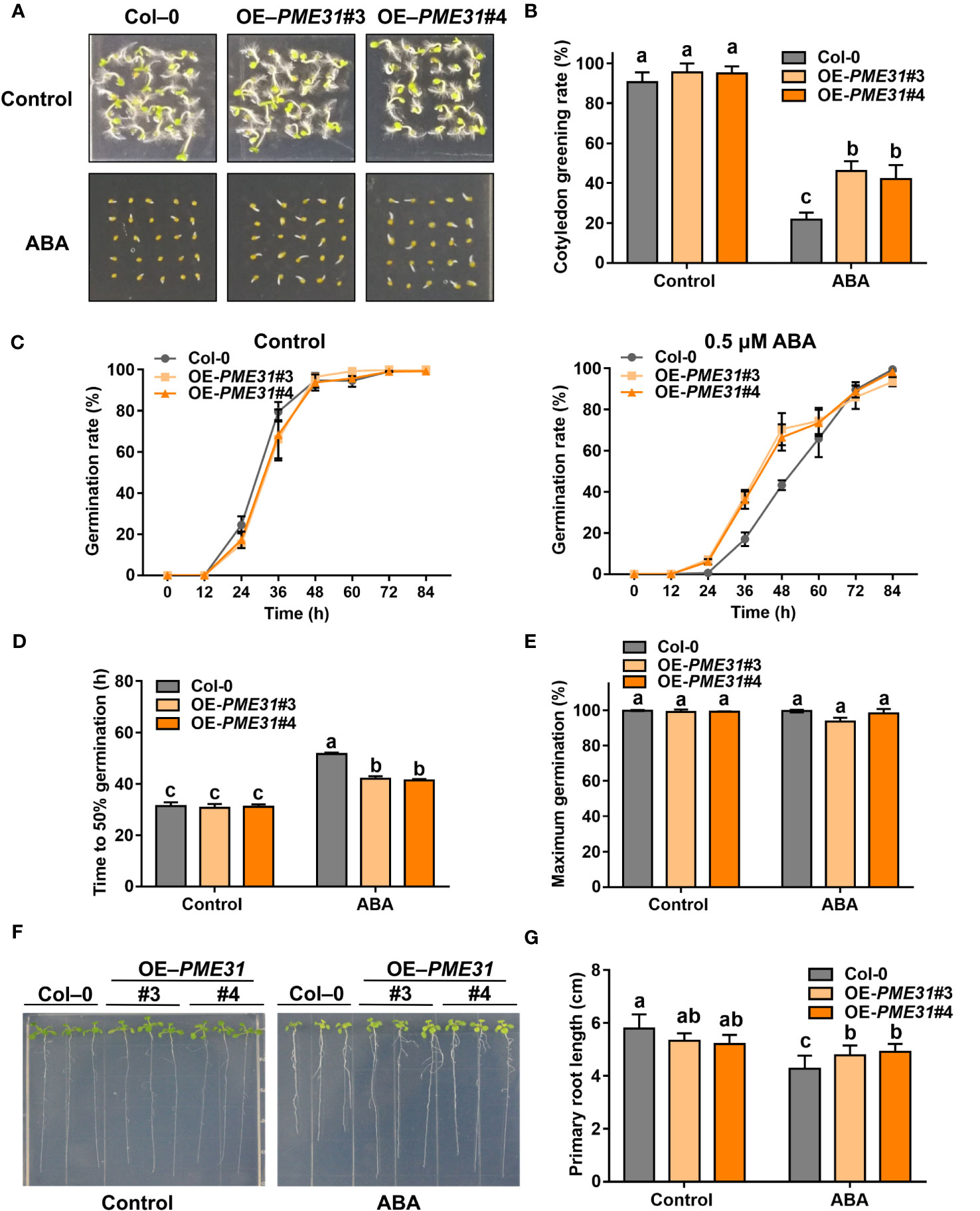
Figure 3 PME31 overexpression reduces ABA-mediated inhibition of seed germination. (A) Phenotype of seed germination in Col-0 and PME31 overexpressors treated with ABA. Seeds were sown on 1/2 MS medium with or without 0.5 µM ABA, and photographs were taken at 48 h of seed germination. (B) Green cotyledons rate was recorded after 72 h. (C) Germination rate of Col-0 and PME31 overexpressors mutants. Seeds were sown on 1/2 MS medium with or without 0.5 μM ABA. The seed germination rate was recorded every 12 h. (D) Time to 50% gemination of Col-0 and PME31 overexpressors. (E) The percentage of maximum germination of Col-0 and PME31 overexpressors. Seeds were sown on 1/2 MS medium with 0.5 μM ABA. (F) Phenotype of seedlings in Col-0 and PME31 overexpressors treated with ABA. Five-day-old seedlings were selected and transferred to 1/2 MS medium with or without 10 µM ABA and placed in an incubator with normal conditions for 14 d. (G) Primary root length was recorded after growing for 14 d. Data are means (± SD) of three biological replicates. Different letters indicate significant differences at P < 0.05 according to two-way ANOVA (Tukey’s multiple comparison test).
We then investigated the effect of PME31 overexpression on Arabidopsis seedlings treated with ABA. Under normal conditions, the primary root length of OE-PME31#3 and OE-PME31#4 was similar to that of Col-0, but with the addition of 10 µM ABA, the primary root length of OE-PME31#3 and OE-PME31#4 was longer than that of the Col-0 (Figures 3F, G). Those results showed that PME31 overexpression reduces ABA-mediated inhibition of seed germination.
ABI5 directly binds to the PME31 promoter and inhibits its expression
To identify upstream regulators of PME31, the PME31 promoter was used as bait in a yeast-one-hybrid (Y1H) screen. The results showed that a bZIP transcription factor ABI5 was identified. ABI5 plays a vital role in seed germination, early seedling growth, and abiotic stresses in the presence of ABA (Skubacz et al., 2016). To investigate whether PME31 was regulated by other ABI transcription factors, we analyzed the PME31 expression in seeds of abi3-1, abi4-1 mutants, and ABI4 overexpressor and found that the PME31 expression had no significant difference in those mutants (Supplementary Figure S4).
We then performed Y1H assays to test if ABI5 directly binds to the PME31 promoter. As shown in Figure 4A, the color turned blue in EGY48 yeast cells transformed with pB42AD-ABI5 and pLacZi-proPME31 constructs, but not in yeast cells with other constructs. β-Galactosidase assays showed that yeast cells transformed with pB42AD-ABI5 and pLacZi-proPME31 had higher β-galactosidase activity compared with yeast cells transformed with other constructs (Figure 4B), suggesting that ABI5 directly binds to the PME31 promoter.
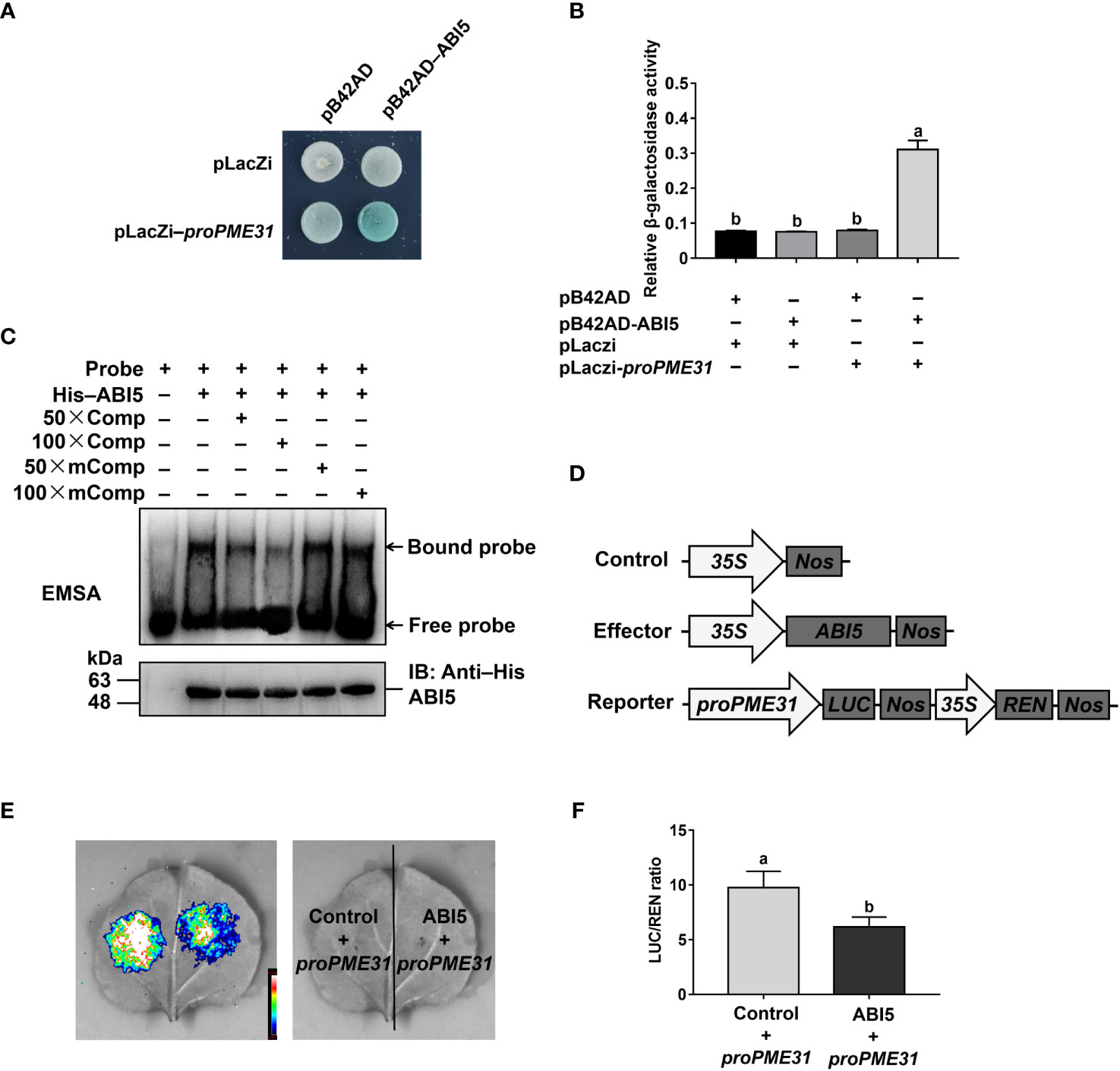
Figure 4 ABI5 directly binds to the PME31 promoter and represses its expression. (A) Yeast-one-hybrid assays showing ABI5 directly binding to the promoter of PME31. The pLacZi-proPME31 constructs were transformed with pB42AD-ABI5 into yeast strain EGY48. Transformants were screened on SD/–Trp–Ura plates and cultured on SD/–Trp–Ura/Gal/Raf/X-Gal (80 μg mL−1) for blue color development at 30°C for 3 d. The pLacZi and pB42AD vectors were used as the negative control. (B) β-Galactosidase activity in (A). (C) Electrophoretic mobility shift assays showing ABI5 directly binding to the promoter of PME31. The 50-bp oligonucleotide containing the ttaCACGTag sequence (−945 to −954 bp) was synthesized and labeled as a probe. The 50× and 100× unlabeled probes (Comp) and 100× mutated probes (mComp) were used as competitors. (D) Schematic diagram of the effector and reporter constructs. The coding sequence of ABI5 was inserted into the pCAMBIA1300-221-3×flag vector to generate an effector. The PME31 promoter was fused with LUC to generate a reporter. (E) Luciferase assays showing ABI5 acted as a transcriptional repressor of PME31. The proPME31:LUC-35S:REN reporter was infiltrated into tobacco leaves. The LUC signals were imaged. (F) LUC and REN activities. The relative activity of the promoter was calculated by the LUC/REN ratio. Data are means (± SD) of three biological replicates. Different letters indicate significant differences at P < 0.05 according to two-way ANOVA (Tukey’s multiple comparison test).
We next search ABI5 binding elements in the promoters of PME31 by using the Plant Promoter Analysis Navigator (PlantPAN; http://PlantPAN2.itps.ncku.edu.tw) (Chow et al., 2016) and found that the putative binding sequence TTACACGTAG located in −945 to −954 bp. We performed EMSAs to verify whether ABI5 directly binds to the TTACACGTAG sequence of the PME31 promoter. The 50-bp oligonucleotide containing the TTACACGTAG sequence (−945 to −954 bp) was synthesized and labeled as a probe. When the His-ABI5 protein was incubated with the labeled probe, the protein-DNA complex with a slower migration speed was observed. When the His-ABI5 protein was incubated with the unlabeled probe, the protein-DNA binding was suppressed, but not with the mutated probe (Figure 4C). These results showed that ABI5 directly binds to the TTACACGTAG sequence of the PME31 promoter.
To test if ABI5 was an activator or a repressor of PME31, we then performed luciferase assays. The effector was generated by the coding sequence of ABI5 inserted into the pCAMBIA1300-221-3×flag vector. The reporter was generated by the PME31 promoter fusing with LUC (Figure 4D). Tobacco leaves were infiltrated with the proPME31:LUC-35S:REN reporter and the 35S:ABI5 effector. The results showed that ABI5 inhibited LUC activity under the control of the PME31 promoter (Figures 4E, F). Overall, those results indicated that ABI5 directly binds to the PME31 promoter and acts as a transcriptional repressor of the PME31 gene.
PME31 acts genetically downstream of ABI5 in ABA-mediated seed germination
To dissect the genetic relationship between PME31 and ABI5, we crossed pme31-1 with abi5-1 and analyzed seed germination in Col-0, pme31-1, abi5-1, and abi5-1 pme31-1 mutants under ABA treatment. Under normal conditions, the cotyledon greening rate and germination rate had no significant difference in pme31-1, abi5-1, and abi5-1 pme31-1 compared with the Col-0. Under ABA treatment conditions, the cotyledon greening rate and germination rate of abi5-1 were higher than that of the Col-0. It indicated abi5-1 was insensitive to ABA-mediated inhibition of seed germination. The cotyledon greening rate and germination rate of pme31-1 were lower than that of Col-0 and abi5-1, while that of abi5-1 pme31-1 were intermediate between pme31-1 and abi5-1 (Figures 5A–C). The time to 50% germination of abi5-1 pme31-1 mutant was more than that of abi5-1 and less than that of pme31-1 (Figure 5D). The percentage of maximum germination had no significant difference in those mutants (Figure 5E). Taken together, those results showed that PME31 acts genetically downstream of ABI5 in ABA-mediated seed germination.
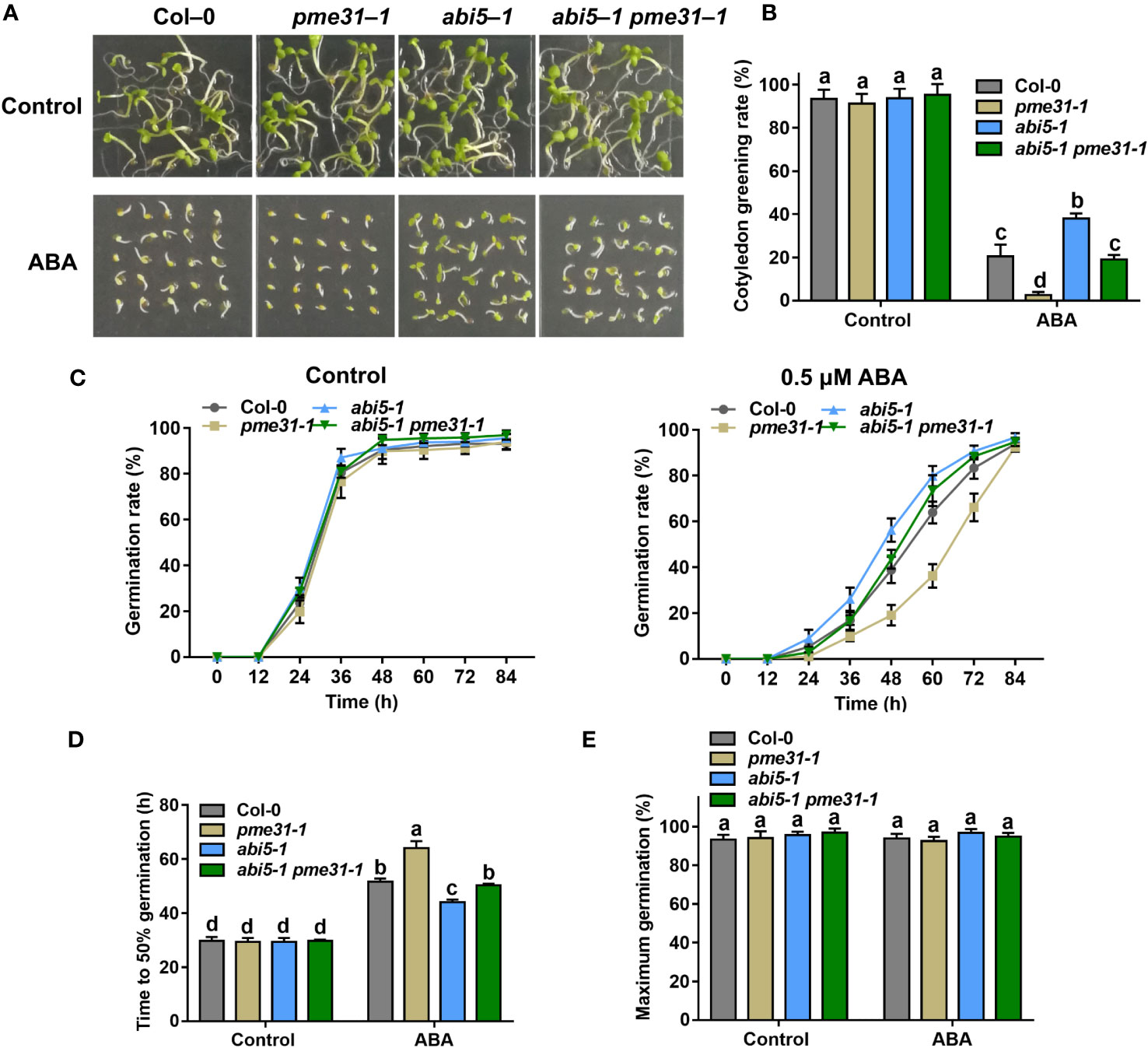
Figure 5 PME31 acts genetically downstream of ABI5 in ABA-mediated inhibition of seed germination. (A) Phenotype of seed germination in Col-0, pme31-1, abi5-1, abi5-1 pme31-1 mutants treated with ABA. Seeds were sown on 1/2 MS medium with or without 0.5 µM ABA, and photographs were taken at 48 h of seed germination. (B) Green cotyledons rate was recorded after 72 h. (C) Germination rate of Col-0, pme31-1, abi5-1, abi5-1 pme31-1 mutants. Seeds were sown on 1/2 MS medium with or without exogenous ABA (0.5 μM). The seed germination rate was recorded every 12 h. (D) Time to 50% gemination of Col-0, pme31-1, abi5-1, abi5-1 pme31-1 mutants. (E) The percentage of maximum germination of Col-0, pme31-1, abi5-1, abi5-1 pme31-1 mutants. Seeds were sown on 1/2 MS medium with 0.5 μM ABA. Data are means (± SD) of three biological replicates. Different letters indicate significant differences at P < 0.05 according to two-way ANOVA (Tukey’s multiple comparison test).
PME31 expression is reduced by ABI5 in ABA-mediated seed germination
To further explore the effect of ABI5 on PME31 expression in the plants, we examined the PME31 expression in the abi5-1 mutant and ABI5 overexpressors during seed germination. Under normal conditions, PME31 expression in the abi5-1 mutant was significantly lower than in the wild-type. PME31 expression increased significantly in the abi5-1 mutant after 12 h imbibition. After 12 h imbibition, the PME31 expression had a significant increase in the abi5-1 mutant (Figure 6A). We then examined the PME31 expression in ABI5 overexpressors. ABI5 overexpression was driven by a β-estradiol-inducible promoter. The ABI5 expression was significantly increased in the presence of β-estradiol treatment (Figure 6B). After 12 h imbibition with β-estradiol induction, the PME31 expression was significantly reduced in ABI5 overexpressors compared with the Col-0 (Figure 6C). Therefore, ABI5 decreases PME31 expression in seed germination.
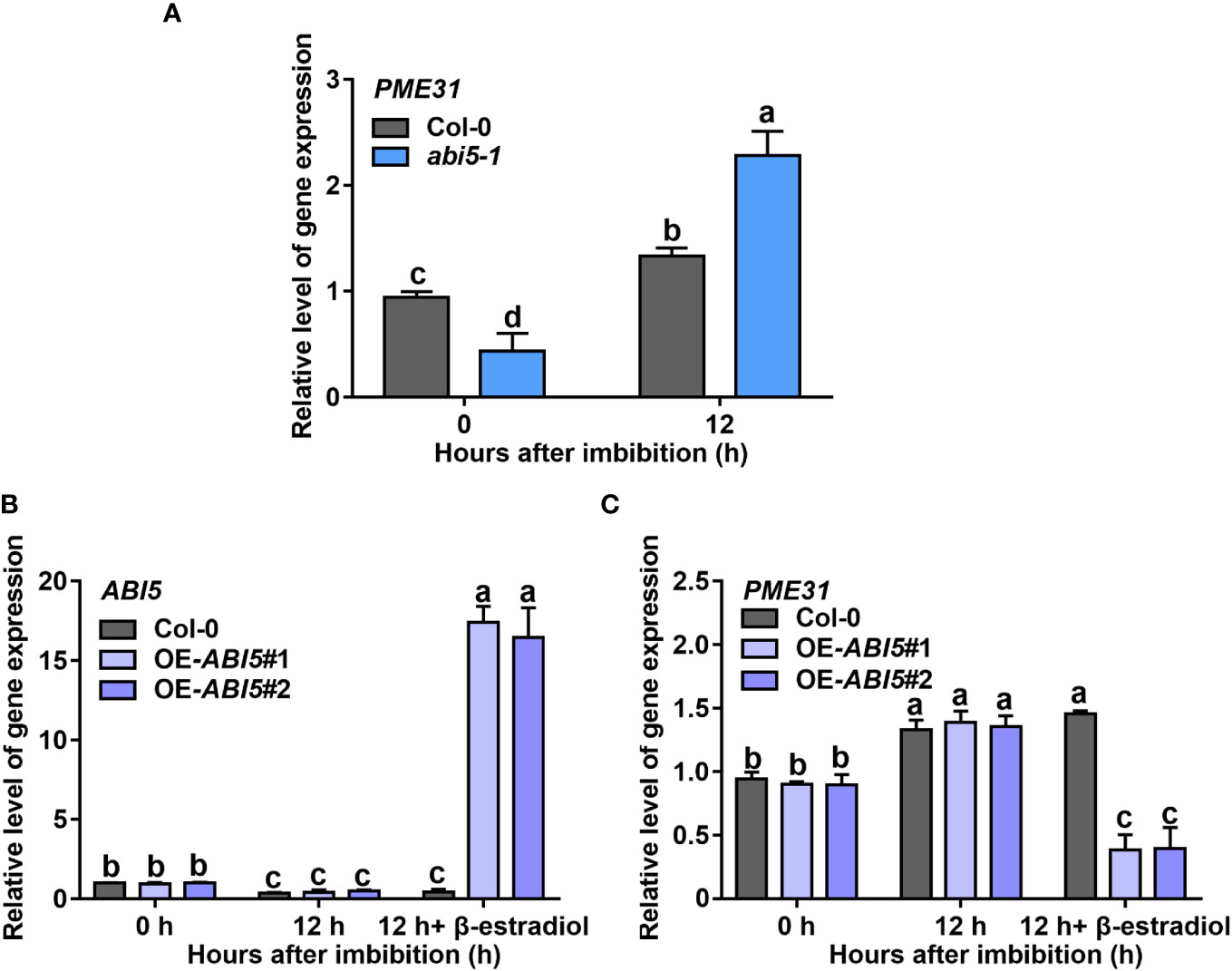
Figure 6 ABI5 decreases PME31 expression in ABA-mediated seed germination (A) The expression of PME31 in the abi5-1 mutant. (B) The expression of ABI5 in ABI5 overexpressors. (C) The expression of PME31 in ABI5 overexpressors. Seeds were imbibed at 25°C in darkness for 12 h. The expression of ABI5 was induced with or without 10 μM β-estradiol. Actin2 was used as the internal reference. Data are means (± SD) of three biological replicates. Different letters indicate significant differences at P < 0.05 according to two-way ANOVA (Tukey’s multiple comparison test).
Discussion
Endospermic seeds, such as Arabidopsis, tobacco, and garden cress, often germinate in a two-step process: after the initial phase of water uptake by the dry seeds (imbibition), testa rupture (TR) occurs and is subsequently followed by endosperm rupture (ER) (Müller et al., 2006; Linkies et al., 2010). Cell walls are highly dynamic structures that provide mechanical support to plant cells. Hemicelluloses bind and crosslink with pectin in cell walls to generate a methylesterified hydrated matrix. Modifications of pectin are important to the process of TR and ER. PMEs regulate the methylesterification status of pectins by removing the methyl group of pectin. It results in an anionically charged matrix and alters the structure properties of the cell wall (Müller et al., 2013; Chandrasekaran et al., 2022). In garden cress, exogenous PME treatment increased testa permeability and promoted TR (Scheler et al., 2015). Salanenka et al. (2009) found that the activity of PMEs was associated with endosperm weakening in cucumber seeds, suggesting PMEs were closely related to seed germination. PME genes were more than 66 members in Arabidopsis. Most PME genes displayed tissue- and stress-specific expression profiles in plant development and stress responses (Pelloux et al., 2007; Weber et al., 2013; Huang et al., 2017; Tang et al., 2023). For instance, Weber et al. (2013) found that the root cell size in the pme3 mutant seedlings was smaller in the presence of Zn2+. PbrPME44 greatly inhibited pollen tube growth and alleviated the increase in methyl-esterified pectin levels caused by PbrS–RNase (Tang et al., 2023). Previous studies showed that PME activity was close to seed germination. The increased PME activity improved the demethylesterification of pectin to facilitate the release of endosperm and promote seed germination (Müller et al., 2013). Scheler et al. (2015) found that ABA suppressed PME activity during seed germination. However, the mechanism of PMEs in response to ABA-mediated seed germination remains unknown. Studies showed that disruption of PME31 reduced the release of seed coat mucilage and salt stress tolerance (Yan et al., 2018; Zhang et al., 2023), but the role of PME31 in seed germination is not clear. In this study, we found that the PME31 expression is prominent in the embryo and was downregulated in the presence of ABA (Figure 1). Phenotype analysis showed that PME31 negatively regulated ABA-mediated seed germination inhibition (Figures 2 and 3). We further identified an upstream regulator of PME31, ABI5, which is an ABA signaling bZIP transcription factor (Figure 4A). PME31 expression was downregulated in the abi5-1 mutant, while it had a significant increase in the abi5-1 mutant after 12 h imbibition. In seeds of abi3-1, abi4-1 mutants, and ABI4 overexpressor, the PME31 expression had no significant difference (Figure 6A; Supplementary Figure S4). It suggested that PME31 expression is mainly influenced by ABI5.
ABA inhibits seed germination and hinders post-germination growth at early developmental stages in plants. Studies have shown that ABI5 negatively regulated ABA-mediated seed germination (Lopez-Molina et al., 2001; Bi et al., 2017; Zhao et al., 2020). ABI5 interacted with a variety of factors, including functional proteins, transcription factors, and enzymes, to regulate seed germination. For instance, the circadian clock proteins PSEUDO-RESPONSE REGULATOR5 (PRR5) collaborated with ABI5 to increase ABA signaling and inhibit seed germination (Yang et al., 2021). In addition, ABA relieved the interaction of C-type Cyclin1;1 (CycC1;1) and inhibition of ABI5, which activated ABI5 activity in the ABA responses and inhibited seed germination (Guo et al., 2022). ABI5 also was bound to the target genes in ABA-mediated seed germination. ABI5 could bind to the promoters of ABI3, CAT1, and PYR/PYL/RCAR genes to influence their gene expression during seed germination (Lopez‐Molina et al., 2002; Bi et al., 2017; Zhao et al., 2020). However, whether ABI5 transcriptionally regulates PMEs expression is not clear. We found that ABI5 was directly bound to the PME31 promoter and suppressed its expression (Figure 4). Meanwhile, genetic analysis revealed that ABI5 acted upstream of PME31 in ABA-mediated seed germination (Figure 5). Furthermore, ABI5 reduced PME31 expression in plant seed germination (Figure 6). Consequently, it suggested that ABI5 was directly bound and downregulated PME31 expression in ABA-mediated seed germination.
The stability of ABI5 is strictly regulated as an important regulator of ABA-responsive genes during seed germination (Lopez-Molina et al., 2001; Nie et al., 2022). ABA elevated the stability of the ABI5 protein, but 26S proteasomes quickly broke down the protein when ABA was removed (Lopez-Molina et al., 2001). Numerous factors regulated the degradation of ABI5. For instance, ABI five binding protein (AFP) promoted ABI5 protein degradation (Lopez-Molina et al., 2003). ABA facilitated ABI5 accumulation by inducing the ubiquitination and proteasomal degradation of KEG (Liu and Stone, 2010). Moreover, the ubiquitin E3 ligase MIEL1 interacted with and ubiquitinated ABI5 to facilitate its degradation in the process of seed germination (Nie et al., 2022). It has been proved that as seeds germinate, the amount of ABA and the level of ABI5 protein gradually drop (Price et al., 2003; Zhao et al., 2020). We therefore hypothesized that as seed germination proceeds, ABA content and ABI5 protein level decrease, and the reduced ABI5 alleviates its transcriptional repression of PME31, thereby responding to the ABA-mediated seed germination (Figure 7).
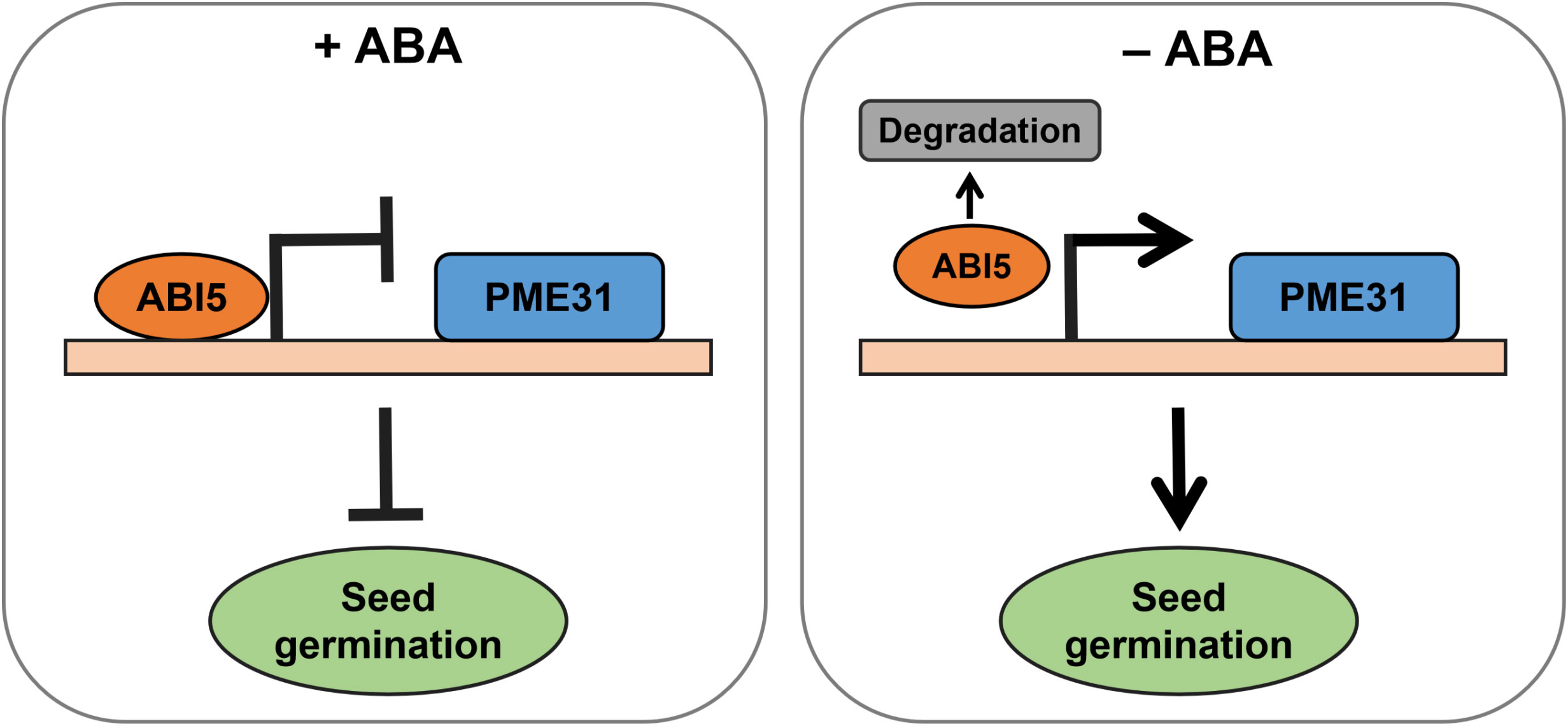
Figure 7 A working model for transcriptional repression of PME31 by ABI5 in ABA-mediated seed germination As seed germination proceeded, ABA content and ABI5 protein level reduced, and the reduced ABI5 alleviated its transcriptional repression of PME31, thereby responding to the ABA-mediated seed germination.
In conclusion, PME31 is transcriptionally repressed by ABI5 to negatively regulate ABA-mediated seed germination inhibition. Our findings shed light on the mechanisms that PME31 is in response to ABA-mediated seed germination.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Files, further inquiries can be directed to the corresponding author/s.
Author contributions
YX: Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Software, Writing – original draft, Writing – review & editing. CZ: Data curation, Investigation, Writing – original draft, Writing – review & editing. QL: Data curation, Investigation, Writing – original draft, Writing – review & editing. YN: Data curation, Methodology, Writing – original draft, Writing – review & editing. YP: Data curation, Investigation, Methodology, Writing – original draft. GL: Data curation, Investigation, Methodology, Writing – original draft. YC: Data curation, Investigation, Methodology, Writing – original draft. AZ: Funding acquisition, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study was supported by National Natural Science Foundation of China (32201707), the Fundamental Research Funds for the Central Universities (KYQN2023025), Natural Science Foundation of Jiangsu Province (BK20220999), China Postdoctoral Science Foundation (2021M701739, 2023T160323), Jiangsu Funding Program for Excellent Postdoctoral Talent (2022ZB330), Open Competition Mechanism to Select the Best Candidates Fund of Jiangsu Province (JBGS[2021]012), Key Research and Development Program of Ningxia Hui Autonomous Region (2023BCF01009), and the Achievement Transformation Fund Project of Hainan Research Institute of Nanjing Agricultural University (NAUSY-CG-YB07).
Acknowledgments
We gratefully acknowledge Professor Jingwei Yan from Zhejiang Agriculture and Forestry University for providing ABI4 overexpressor.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2024.1336689/full#supplementary-material
References
Berry, T., Bewley, J. D. (1992). A role for the surrounding fruit tissues in preventing the germination of tomato (Lycopersicon esculentum) seeds. Plant Physiol. 100, 951–957. doi: 10.1104/pp.100.2.951
Bi, C., Ma, Y., Wu, Z., Yu, Y. T., Liang, S., Lu, K., et al. (2017). Arabidopsis ABI5 plays a role in regulating ROS homeostasis by activating CATALASE 1 transcription in seed germination. Plant Mol. Biol. 94, 197–213. doi: 10.1007/s11103-017-0603-y
Bradford, M. M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254. doi: 10.1016/0003-2697(76)90527-3
Chandrasekaran, U., Zhao, X., Luo, X., Wei, S., Shu, K. (2022). Endosperm weakening: The gateway to a seed’s new life. Plant Physiol. Bioch. 178, 31–39. doi: 10.1016/j.plaphy.2022.02.016
Chow, C. N., Zheng, H. Q., Wu, N. Y., Chien, C. H., Huang, H. D., Lee, T. Y., et al. (2016). PlantPAN 2.0: an update of plant promoter analysis navigator for reconstructing transcriptional regulatory networks in plants. Nucleic Acids Res. 44, 1154–1160. doi: 10.1093/nar/gkv1035
Clough, S. J., Bent, A. F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. doi: 10.1046/j.1365-313x.1998.00343.x
Coego, A., Brizuela, E., Castillejo, P., Ruíz, S., Koncz, C., del Pozo, J. C., et al. (2014). The TRANSPLANTA collection of Arabidopsis lines: a resource for functional analysis of transcription factors based on their conditional overexpression. Plant J. 77, 944–953. doi: 10.1111/tpj.12443
Earley, K. W., Haag, J. R., Pontes, O., Opper, K., Juehne, T., Song, K., et al. (2006). Gateway-compatible vectors for plant functional genomics and proteomics. Plant J. 45, 616–629. doi: 10.1111/j.1365-313X.2005.02617.x
Graeber, K. A. I., Nakabayashi, K., Miatton, E., Leubner-Metzger, G., Soppe, W. J. J. (2012). Molecular mechanisms of seed dormancy. Plant Cell Environ. 35, 1769–1786. doi: 10.1111/j.1365-3040.2012.02542.x
Grainge, G., Nakabayashi, K., Steinbrecher, T., Kennedy, S., Ren, J., Iza, F., et al. (2022). Molecular mechanisms of seed dormancy release by gas plasma-activated water technology. J. Exp. Bot. 73, 4065–4078. doi: 10.1093/jxb/erac150
Guo, J. X., Song, R. F., Lu, K. K., Zhang, Y., Chen, H. H., Zuo, J. X., et al. (2022). CycC1;1 negatively modulates ABA signaling by interacting with and inhibiting ABI5 during seed germination. Plant Physiol. 190, 2812–2827. doi: 10.1093/plphys/kiac456
Hongo, S., Sato, K., Yokoyama, R., Nishitani, K. (2012). Demethylesterification of the primary wall by PECTIN METHYLESTERASE35 provides mechanical support to the Arabidopsis stem. Plant Cell 24, 2624–2634. doi: 10.1105/tpc.112.099325
Huang, Y. C., Wu, H. C., Wang, Y. D., Liu, C. H., Lin, C. C., Luo, D. L., et al. (2017). PECTIN METHYLESTERASE34 contributes to heat tolerance through its role in promoting stomatal movement. Plant Physiol. 174, 748–763. doi: 10.1104/pp.17.00335
Li, H., Li, X., Wang, G., Zhang, J., Wang, G. (2022). Analysis of gene expression in early seed germination of rice: landscape and genetic regulation. BMC Plant Biol. 22, 70. doi: 10.1186/s12870-022-03458-3
Linkies, A., Graeber, K., Knight, C., Leubner-Metzger, G. (2010). The evolution of seeds. New Phytol. 186, 817–831. doi: 10.1111/j.1469-8137.2010.03249.x
Liu, H., Stone, S. L. (2010). Abscisic acid increases Arabidopsis ABI5 transcription factor levels by promoting KEG E3 ligase self-ubiquitination and proteasomal degradation. Plant Cell 22, 2630–2641. doi: 10.1105/tpc.110.076075
Liu, X., Hu, P., Huang, M., Tang, Y., Li, Y., Li, L., et al. (2016). The NF-YC-RGL2 module integrates GA and ABA signalling to regulate seed germination in Arabidopsis. Nat. Commun. 7, 12768. doi: 10.1038/ncomms12768
Liu, Z., Ma, C., Hou, L., Wu, X., Wang, D., Zhang, L., et al. (2022). Exogenous SA affects rice seed germination under salt stress by regulating Na+/K+ balance and endogenous GAs and ABA homeostasis. Int. J. Mol. Sci. 23, 3293. doi: 10.3390/ijms23063293
Livak, K. J., Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2–ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Loades, E., Pérez, M., Turečková, V., Tarkowská, D., Strnad, M., Seville, A., et al. (2023). Distinct hormonal and morphological control of dormancy and germination in Chenopodium album dimorphic seeds. Front. Plant Sci. 14. doi: 10.3389/fpls.2023.1156794
Lopez-Molina, L., Mongrand, S., Kinoshita, N., Chua, N. H. (2003). AFP is a novel negative regulator of ABA signaling that promotes ABI5 protein degradation. Genes Dev. 17, 410–418. doi: 10.1101/gad.1055803
Lopez-Molina, L., Mongrand, S., McLachlin, D. T., Chait, B. T., Chua, N. H. (2002). ABI5 acts downstream of ABI3 to execute an ABA-dependent growth arrest during germination. Plant J. 32, 317–328. doi: 10.1046/j.1365-313X.2002.01430.x
Lopez-Molina, L., Mongrand, S., Nam-Hai, C. (2001). A postgermination developmental arrest checkpoint is mediated by abscisic acid and requires the ABI5 transcription factor in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 98, 4782–4787. doi: 10.1073/pnas/081594298
Mei, S., Zhang, M., Ye, J., Du, J., Jiang, Y., Hu, Y. (2023). Auxin contributes to jasmonate-mediated regulation of abscisic acid signaling during seed germination in Arabidopsis. Plant Cell 35, 1110–1133. doi: 10.1093/plcell/koac362
Müller, K., Levesque-Tremblay, G., Bartels, S., Weitbrecht, K., Wormit, A., Usadel, B., et al. (2013). Demethylesterification of cell wall pectins in Arabidopsis plays a role in seed germination. Plant Physiol. 161, 305–316. doi: 10.1104/pp.112.205724
Müller, K., Tintelnot, S., Leubner-Metzger, G. (2006). Endosperm-limited Brassicaceae seed germination: abscisic acid inhibits embryo-induced endosperm weakening of Lepidium sativum (cress) and endosperm rupture of cress and Arabidopsis thaliana. Plant Cell Physiol. 47, 864–877. doi: 10.1093/pcp/pcj059
Nakagawa, T., Kurose, T., Hino, T., Tanaka, K., Kawamukai, M., Niwa, Y., et al. (2007). Development of series of gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J. Biosci. Bioeng. 104, 34–41. doi: 10.1263/jbb.104.34
Nie, K., Zhao, H., Wang, X., Niu, Y., Zhou, H., Zheng, Y. (2022). The MIEL1-ABI5/MYB30 regulatory module fine tunes abscisic acid signaling during seed germination. J. Integr. Plant Biol. 64, 930–941. doi: 10.1111/jipb.13234
Pelloux, J., Rusterucci, C., Mellerowicz, E. (2007). New insights into pectin methylesterase structure and function. Trends Plant Sci. 12, 267–277. doi: 10.1016/j.tplants.2007.04.001
Price, J., Li, T. C., Kang, S. G., Na, J. K., Jang, J. C. (2003). Mechanisms of glucose signaling during germination of Arabidopsis. Plant Physiol. 132, 1424–1438. doi: 10.1104/pp.103.020347
Qu, T., Liu, R., Wang, W., An, L., Chen, T., Liu, G., et al. (2011). Brassinosteroids regulate pectin methylesterase activity and AtPME41 expression in Arabidopsis under chilling stress. Cryobiology 63, 111–117. doi: 10.1016/j.cryobiol.2011.07.003
Rushton, D. L., Tripathi, P., Rabara, R. C., Lin, J., Ringler, P., Boken, A. K., et al. (2012). WRKY transcription factors: key components in abscisic acid signalling. Plant Biotechnol. J. 10, 2–11. doi: 10.1111/j.1467-7652.2011.00634.x
Salanenka, Y. A., Goffinet, M. C., Taylor, A. G. (2009). Structure and histochemistry of the micropylar and chalazal regions of the perisperm-endosperm envelope of cucumber seeds associated with solute permeability and germination. J. Am. Soc Hortic. Sci. 134, 479–487. doi: 10.21273/JASHS.134.4.479
Scheler, C., Weitbrecht, K., Pearce, S. P., Hampstead, A., Büttner-Mainik, A., Lee, K. J. D., et al. (2015). Promotion of testa rupture during garden cress germination involves seed compartment-specific expression and activity of pectin methylesterases. Plant Physiol. 167, 200–215. doi: 10.1104/pp.114.247429
Sharma, E., Majee, M., Penfield, S. (2023). Seed germination variability: why do genetically identical seeds not germinate at the same time? J. Exp. Bot. 74, 3462–3475. doi: 10.1093/jxb/erad101
Shu, K., Zhang, H., Wang, S., Chen, M., Wu, Y., Tang, S., et al. (2013). ABI4 regulates primary seed dormancy by regulating the biogenesis of abscisic acid and gibberellins in Arabidopsis. PloS Genet. 9, e1003577. doi: 10.1371/journal.pgen.1003577
Singh, D., Datta, S. (2023). BBX30/miP1b and BBX31/miP1a form a positive feedback loop with ABI5 to regulate ABA-mediated postgermination seedling growth arrest. New Phytol. 238, 1908–1923. doi: 10.1111/nph.18866
Skubacz, A., Daszkowska-Golec, A., Szarejko, I. (2016). The role and regulation of ABI5 (ABA-Insensitive 5) in plant development, abiotic stress responses and phytohormone crosstalk. Front. Plant Sci. 71884. doi: 10.3389/fpls.2016.01884
Tang, C., Wang, P., Zhu, X., Qi, K., Xie, Z., Zhang, H., et al. (2023). Acetylation of inorganic pyrophosphatase by S-RNase signaling induces pollen tube tip swelling by repressing pectin methylesterase. Plant Cell 35, 3544–3565. doi: 10.1093/plcell/koad162
Tuan, P. A., Kumar, R., Rehal, P. K., Toora, P. K., Ayele, B. T. (2018). Molecular mechanisms underlying abscisic acid/gibberellin balance in the control of seed dormancy and germination in cereals. Front. Plant Sci. 9. doi: 10.3389/fpls.2018.00668
Weber, M., Deinlein, U., Fischer, S., Rogowski, M., Geimer, S., Tenhaken, R., et al. (2013). A mutation in the Arabidopsis thaliana cell wall biosynthesis gene pectin methylesterase 3 as well as its aberrant expression cause hypersensitivity specifically to Zn. Plant J. 76, 151–164. doi: 10.1111/tpj.12279
Xiang, Y., Bian, X., Wei, T., Yan, J., Sun, X., Han, T., et al. (2021a). ZmMPK5 phosphorylates ZmNAC49 to enhance oxidative stress tolerance in maize. New Phytol. 232, 2400–2417. doi: 10.1111/nph.17761
Xiang, Y., Sun, X., Bian, X., Wei, T., Han, T., Yan, J., et al. (2021b). The transcription factor ZmNAC49 reduces stomatal density and improves drought tolerance in maize. J. Exp. Bot. 72, 1399–1410. doi: 10.1093/jxb/eraa507
Xue, X., Du, S., Jiao, F., Xi, M., Wang, A., Xu, H., et al. (2021). The regulatory network behind maize seed germination: Effects of temperature, water, phytohormones, and nutrients. Crop J. 9, 718–724. doi: 10.1016/j.cj.2020.11.005
Yan, J., Fang, L., Yang, L., He, H., Huang, Y., Liu, Y., et al. (2020). Abscisic acid positively regulates L-arabinose metabolism to inhibit seed germination through ABSCISIC ACID INSENSITIVE4-mediated transcriptional promotions of MUR4 in Arabidopsis thaliana. New Phytol. 225, 823–834. doi: 10.1111/nph.16149
Yan, J., He, H., Fang, L., Zhang, A. (2018). Pectin methylesterase31 positively regulates salt stress tolerance in Arabidopsis. Biochem. Biophys. Res. Commun. 496, 497–501. doi: 10.1016/j.bbrc.2018.01.025
Yang, M., Han, X., Yang, J., Jiang, Y., Hu, Y. (2021). The Arabidopsis circadian clock protein PRR5 interacts with and stimulates ABI5 to modulate abscisic acid signaling during seed germination. Plant Cell 33, 3022–3041. doi: 10.1093/plcell/koab168
Zhang, X., Guo, H., Xiao, C., Yan, Z., Ning, N., Chen, G., et al. (2023). PECTIN METHYLESTERASE INHIBITOR18 functions in stomatal dynamics and stomatal dimension. Plant Physiol. 192, 1603–1620. doi: 10.1093/plphys/kiad145
Zhao, H., Nie, K., Zhou, H., Yan, X., Zhan, Q., Zheng, Y., et al. (2020). ABI5 modulates seed germination via feedback regulation of the expression of the PYR/PYL/RCAR ABA receptor genes. New Phytol. 228, 596–608. doi: 10.1111/nph.16713
Zhou, C., Lin, Q., Lan, J., Zhang, T., Liu, X., Miao, R., et al. (2020). WRKY transcription factor OsWRKY29 represses seed dormancy in rice by weakening abscisic acid response. Front. Plant Sci. 11, 691. doi: 10.3389/fpls.2020.00691
Zúñiga-Sánchez, E., Soriano, D., Martínez-Barajas, E., Orozco-Segovia, A., Alicia, G. D. (2014). BIIDXI, the At4g32460 DUF642 gene, is involved in pectin methyl esterase regulation during Arabidopsis thaliana seed germination and plant development. BMC Plant Biol. 14, 338. doi: 10.1186/s12870-014-0338-8
Keywords: abscisic acid, seed germination, PME31, ABI5, transcription repression
Citation: Xiang Y, Zhao C, Li Q, Niu Y, Pan Y, Li G, Cheng Y and Zhang A (2024) Pectin methylesterase 31 is transcriptionally repressed by ABI5 to negatively regulate ABA-mediated inhibition of seed germination. Front. Plant Sci. 15:1336689. doi: 10.3389/fpls.2024.1336689
Received: 11 November 2023; Accepted: 18 January 2024;
Published: 02 February 2024.
Edited by:
Muhammad Awais Farooq, Hebei Agricultural University, ChinaReviewed by:
Xianhai Zhao, Brookhaven National Laboratory (DOE), United StatesRaquel Iglesias Fernández, Polytechnic University of Madrid, Spain
Copyright © 2024 Xiang, Zhao, Li, Niu, Pan, Li, Cheng and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Aying Zhang, YXl6aGFuZ0BuamF1LmVkdS5jbg==
 Yang Xiang
Yang Xiang Chongyang Zhao1
Chongyang Zhao1 Aying Zhang
Aying Zhang