- 1College of Life Science and Technology, Tarim University, Alar, Xinjiang, China
- 2National-Local Joint Engineering Laboratory of High Efficiency and Superior Quality Cultivation and Fruit Deep Processing Technology on Characteristic Fruit Trees, Alar, Xinjiang, China
- 3National Key Laboratory for Germplasm Innovation & Utilization of Horticultural Crops, Huazhong Agricultural University, Wuhan, Hubei, China
- 4College of Horticulture and Forestry, Tarim University, Alar, Xinjiang, China
Hexokinase (HXK) plays a crucial role in plants, catalyzing the phosphorylation of hexose substances, which is one of the key steps in sugar metabolism and energy production. While HXK genes have been well-studied in model plants, the evolutionary and functional characteristics of HXK gene family in jujube is unknow. In this study, the HXK gene family members were identified by bioinformatics methods, the key members regulating glucose metabolism were identified by transcriptome data, and finally the function of the key genes was verified by instantaneous and stable genetic transformation. Our results showed that seven HXK genes were identified in the jujube genome, all of which were predict located in the chloroplast and contain Hexokinase-1 (PF00349) and Hexokinase-2 (PF03727) conserved domains. Most of HXK proteins were transmembrane protein with stable, lipid-soluble, hydrophilic. The secondary structure of ZjHXK proteins main α-helix, and contains two distinct tertiary structure. All ZjHXK genes contain nine exons and eight introns. Predictions of cis-regulatory elements indicate that the promoter region of ZjHXK contains a large number of MeJA responsive elements. Finally, combined with the analysis of the relationship between the expression and glucose metabolism, found that ZjHXK5 and ZjHXK6 may the key genes regulating sugar metabolism. Transient overexpression of ZjHXK5 and ZjHXK6 on jujube, or allogeneic overexpression of ZjHXK5 and ZjHXK6 on tomato would significantly reduce the content of total sugar and various sugar components. Transient silencing of ZjHXK5 and ZjHXK6 genes results in a significant increase in sucrose and total sugar content. Interestingly, the expression of ZjHXK5 and ZjHXK6 were also affected by methyl jasmonate.
Introduction
Ziziphus jujuba Mill. is origin from China and widely distributed in temperate and subtropical regions of the Northern Hemisphere (Huang, 2020). Jujube fruits are rich in nutrition, especially the ascorbic acid, amino acid and sugar (Liu et al., 2020; Liao et al., 2023). The sugar content in ripe fresh jujube fruits is up to 25%~40%, which is a traditional fruit with the same origin as medicine and food (Li et al., 2007; Chen and Tsim, 2020). The rich germplasm resources and the variation of sugar content make jujube an important species for studying sugar metabolism.
In higher plants, sugar can not only be used as an energy source and structural component, but also participate in the growth and development process as a transmission signal (Jang and Sheen, 1994). Sugar metabolism is regulated by several enzymes, among which hexokinase (HXK) is closely related to glucose metabolism and as a hexose sensor, HXK can senses the levels of hexoses and phosphorylation states. Then through signal transduction pathways, HXK convey information to the cell nucleus, this process playing a crucial role in sugar sensing and signaling (Moore et al., 2003; Rolland et al., 2006). Importantly, HXK participates in sucrose-induced signal transduction and regulates the expression of genes related to sucrose metabolism and starch synthesis (Xiao et al., 2000; Polit and Ciereszko, 2009). For example, reduced StHXK1 activity leads to accumulation of glucose and starch, and decreased sucrose levels (Veramendi et al., 2002). Hexokinase-like (HKL) is an isoenzyme of HXK, and although the function of HLK is unknown, defective phosphorylation of AtHKL1 can negatively regulate plant growth (Karve et al., 2008). Recent studies also indicate that HXK effect root development (Granot et al., 2014), leaf senescence (Swartzberg et al., 2011), regulates photosynthesis (Roth et al., 2019), pollen germination (Karni and Aloni, 2002), and sugar content during fruit development (Ji et al., 2023).
Currently, HXK genes have been isolated from many plants, including pear (Zhao et al., 2019), apple (Zhu et al., 2021), and peach (Xu et al., 2021), existing in the form of gene families in plants (Karve et al., 2010). Six and ten HXK gene family members have been identified in Arabidopsis thaliana (Karve et al., 2008) and rice (Cho et al., 2006) respectively. Different HXK members showed tissue-specific expression. For example, AtHXK1 was expressed in all organs, while AtHXK3 showed higher expression in roots and siliques (Karve et al., 2008).
In addition, the sugar content and HXK expression were induced by abiotic stress, such as temperature and methyl jasmonate (Sami et al., 2016; Saddhe et al., 2021). In model plants, the structure and function of HXK have been well understood. However, the identification and there few reports on the functional analysis of HXK gene in jujube. Therefore, the aim of this study was to identify the members of the ZjHXK gene family at the whole genome level, and to clarify the function of key genes regulating glucose metabolism and their response to abiotic stress.
Materials and methods
Materials and treatments
Different developmental stages of cv ‘Fucuimi’ (higher sugar content) and cv ‘Jing 39’ (lower sugar content) fruit were harvested, including the young fruit stage (YF), early white maturation stage (EF), white maturation stage (WM), half-red fruit stage (HR), and full-red fruit stage (FR). Nine cv ‘Fucuimi’ and nine cv ‘Jing 39’ were cultivated at the Tarim University Jujube Germplasm Resource Repository (40°54′N, 81°30′E), with uniform cultivation conditions. Three plants were a biological replicate, and each plant collected 15 fruits from four directions (east, south, west, and north). The collected samples were treated with liquid nitrogen at once, and stored at -80°C until use. The Beijing Novogene Biotech Co., Ltd. performed sequencing with HiSeq2500, the winter jujube genome (PRJNA251714) (Liu et al., 2014) was used as reference genome. Transcriptome data of different winter jujube tissues were downloaded from the NCBI database (PRJNA260241).
‘Jing39’ jujube callus tissues were used as the experimental material for different concentration MeJA treatment, including control (0 mmol/L), T1 (0.05 mmol/L), T2 (0.10 mmol/L), T3 (0.20 mmol/L) and T4 (0.30 mmol/L). Each treatment was repeated three times, and the callus was treated in 25°C darkness for 10 d, and then frozen in liquid nitrogen.
Identification of ZjHXK
Firstly, the jujube genome (PRJNA251714) was downloaded from National Center for Biotechnology Information database, and then the protein sequence of AtHXK in Arabidopsis was used as a probe to blast in jujube proteins to obtain ZjHXK candidate sequence (Liu et al., 2014). The ZjHXK candidate sequences that contain the Hexokinase-1 (PF00349) and Hexokinase-2 (PF03727) domain were considered the ZjHXK gene family members by the Pfam database (http://pfam.xfam.org/).
Bioinformatics analysis
TMHMM Serverv. 2. R2.0 (http://www.cbs.dtu.dk/services/TMHMM) and ProtParam (https://web.expasy.org/protparam/) predicted ZjHXK physicochemical properties. WoLF PSORT (https://wolfpsort.hgc.jp/) predicted the subcellular localization of ZjHXK. NPSA (https://npsa-prabi.ibcp.fr/cgi-bin/npsa_automat.pl?page=npsa_sopma.html) and Swiss model (https://swissmodel.expasy.org/) predicted the secondary structure and tertiary structure of ZjHXK, respectively. Signalp 6.0 (https://services.healthtech.dtu.dk/services/SignalP-6.0/) and Netphos 3.1 (https://services.healthtech.dtu.dk/services/NetPhos-3.1/) predicted signal peptide, phosphorylation and glycosylation sites, respectively. The TBtools-II (Chen et al., 2023) was used to extract genome, promoter and CDS sequences, and the gene structure display by GSDS (http://gsds.gao-lab.org/index.php) (Hu et al., 2015). PlantCARE (http://bioinformatics.psb.ugent.Be/webtools/PlantCARE/HTML/) was used for predicted the cis regulatory elements. MEME (https://meme-suite.org/meme/tools/meme) (Bailey et al., 2006) predicted conservative motifs with 10 ordinals and 5-50 aa length. The ClustalW method in MEGA11.0 software was used to blast, then constructed phylogenetic tree with the Neighbor-Joining method, the bootstrap validation value was set to 1,000. The phylogenetic tree was beautified by Evolview (https://evolgenius.info//evolview-v2). TBtools-II (Chen et al., 2023) was used to conduct multicollinearity analysis among gene family members.
Cloning and vector construction of ZjHXK5 and ZjHXK6
Specific primers for the full-length coding sequences (CDS) of ZjHXK5 (GenBank accession number: XM_048465612.1) and ZjHXK6 (GenBank accession number: XM_016018332.3) genes were designed using Primer5.0 software. The primer sequences were as follows: For ZjHXK5 CDS, the forward primer was ZjHXK5-CDS-F: ATGGGGAAATTGGCGGTGGGTG, and the reverse primer was ZjHXK5-CDS-R: TTAAGACTCTTCTACCTCTAAG. For ZjHXK6 CDS, the forward primer was ZjHXK6-CDS-F: ATGGGGAGGGTGGTGGTGGGAG, and the reverse primer was ZjHXK6-CDS-R: CTATCCACACTGTTGGATGTTG. Total RNA from mature ‘Jing39’ fruit at the ripe stage was extracted using the TIANGEN RNAprep Pure Polysaccharide and Polyphenolics Plant Total RNA Extraction Kit (Tiangen Biochemical Technology Co., LTD., Beijing, China). Subsequently, cDNA was synthesized by the HiScript II Q RT SuperMix for qPCR (+gDNA wiper) kit (Novozymes Biotechnology Co., Ltd., Nanjing, China). Gene cloning and LR reaction refer to the previous method (Wasaya et al., 2023) to construct CDS sequences of ZjHXK5 and ZjHXK6 into pK7WG2D vector. Tsingke Biotech Co., Ltd. (Wuhan, China) performed the sequencing. The sequencing confirmed cloned plasmids were extracted using the SIMGEN Rapid Plasmid Mini Kit (Xinjing biological reagent development Co., LTD., Wuhan, China). VIGS-mediated silencing of ZjHXK5 and ZjHXK6 gene expression in jujube fruits according to the previous methods (Tian et al., 2014).
Transient and stable transformation
The plasmids confirmed by sequencing were named pK7WG2D-ZjHXK5-GFP, pK7WG2D-ZjHXK6-GFP and pTRV2-ZjHXK5, pTRV2-ZjHXK6, and transformed into Agrobacterium strains GV3101 and EHA105. Agrobacterium strains GV3101 carrying pK7WG2D-ZjHXK5-GFP and pK7WG2D-ZjHXK6-GFP were separately used for injection into one-month-old tobacco leaves (Sparkes et al., 2006) and cv ‘Jing39’ ripening jujube fruits with similar size. After injection, dark treat for 1 d, followed by normal light treatment for 2 d. Agrobacterium strains GV3101 carrying pTRV2-ZjHXK5 and pTRV2-ZjHXK6, were separately used for injection into cv ‘Dongzao’ ripening jujube fruits with similar size. After injection, dark treat for 1 d, followed by normal light treatment for 4 d (Tian et al., 2014). Agrobacterium EHA105 carrying pK7WG2D-ZjHXK5-GFP and pK7WG2D-ZjHXK6-GFP were used for infection of Micro-Tom tomato (Sun et al., 2015) and ‘Jing39’ jujube callus, respectively. Fruits and callus were collected from T2 generation positive lines.
Determination of the sugar component content
Sugar component extraction was performed following the method described by previous methods (Bartolozzi et al., 1997). Previous studies have indicated that the major sugar components in jujube fruits include fructose, glucose, and sucrose (Hubbard et al., 1991; Li et al., 2007; Rashwan et al., 2020), accounting for over 95% of the total sugar content. Therefore, in this study, the total content of these three sugar components was considered as the total sugar content.
Expression analysis by qRT-PCR
Following previous method, the ZjUBQ was used as the internal reference gene for qRT-PCR analysis (Zhang et al., 2015). A reaction mixture composed of 10 μL as follows: 0.5 μL of cDNA template, 0.2 μL for each forward and reverse primer, 5 μL of PowerUp TM SYBR TM Green Master Mix, and 4.1 μL of ddH2O. The 2^-ΔΔCT method was used for calculate the relative gene expression levels (Livak and Schmittgen, 2001).
Data analysis
The data was statistically analyzed by GraphPad Prism (Mitteer and Greer, 2022). Duncan’s method was used to detect differences at P ≤ 0.05. The heat map of expression was drawn with TBtools-II (Chen et al., 2023).
Results
Identification and characterization of HXK gene family members in jujube
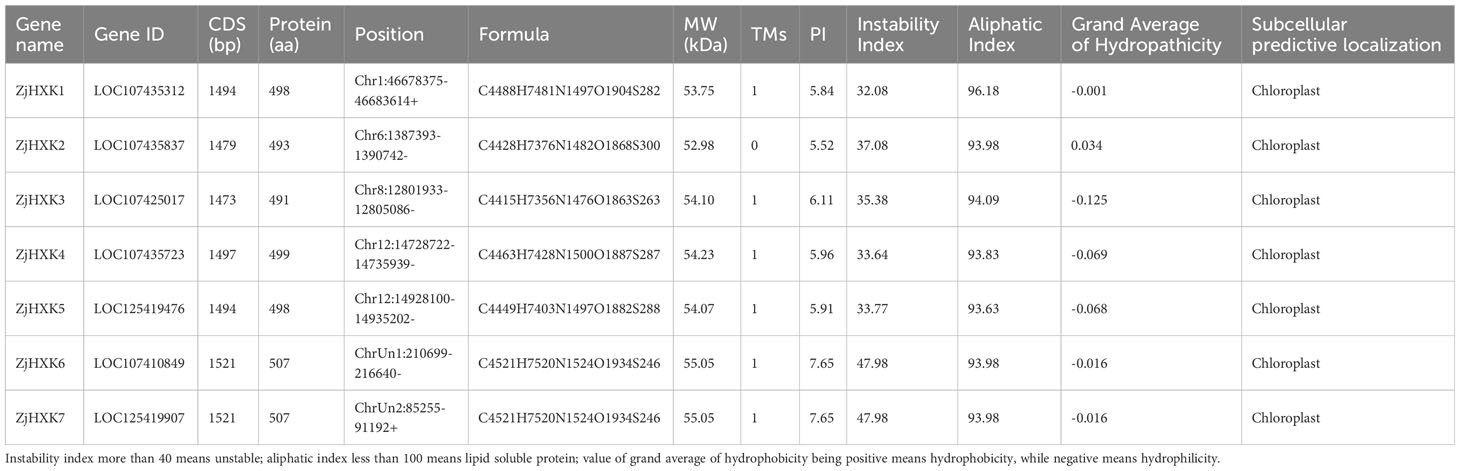
A total of seven ZjHXK candidate sequences were confirmed, all of them contain conserved domains PF00349.24 and PF03727.19 (Supplementary Figure 1). It’s worth noting that these candidate sequences were distributed non-uniformly across chromosomes 1, 6, 8, 12 and an unidentified chromosome, and they were named as ZjHXK1to ZjHXK7 based on their chromosomal locations. The physicochemical properties of ZjHXK (Table 1) showed that coding sequence (CDS) lengths ranged from 1473 to 1521bp. The molecular weights of ZjHXK proteins varied from 52.98 to 55.05 kDa, and the isoelectric points (pI) ranged from 5.52 to 7.65. Notably, ZjHXK2 protein was predicted located extracellularly and not a transmembrane protein. All but ZjHXK6 and ZjHXK7 were stable proteins. All ZjHXK proteins were lipophilic protein. except ZjHXK2, other ZjHXK was hydrophilic protein. Subcellular prediction analysis revealed that all ZjHXK were localized within the chloroplast.
Protein signaling peptides, glycosylation and phosphorylation sites analysis
The signal peptide analysis of ZjHXK proteins revealed that all ZjHXK protein can consider as non-secretory proteins (Supplementary Figure 2). Phosphorylation site analysis found that ZjHXK proteins had many multiple potential phosphorylation sites (Supplementary Figure 3). such as ZjHXK6 and ZjHXK7 exhibited the highest number of phosphorylation sites, including 29 Serine (Ser), 15 Threonine (Thr), and 3 Tyrosine (Tyr) phosphorylation sites. Glycosylation sites analysis of ZjHXK proteins (Supplementary Figure 4) indicated that ZjHXK1 had a single typical N-glycosylation site located at amino acid position 229. In contrast, ZjHXK2 lacks N-glycosylation sites.
Prediction of secondary and tertiary structure of proteins
The secondary structure predictions of ZjHXK proteins indicate that the ZjHXK protein were composed of α-helices, random coils, extended strands, and β-turns. Among them, the α-helices was the main component, ranging from 40.97% to 47.25% (Supplementary Figure 5). Each ZjHXK protein contains two distinct tertiary structures, with significant differences between them (Figure 1).
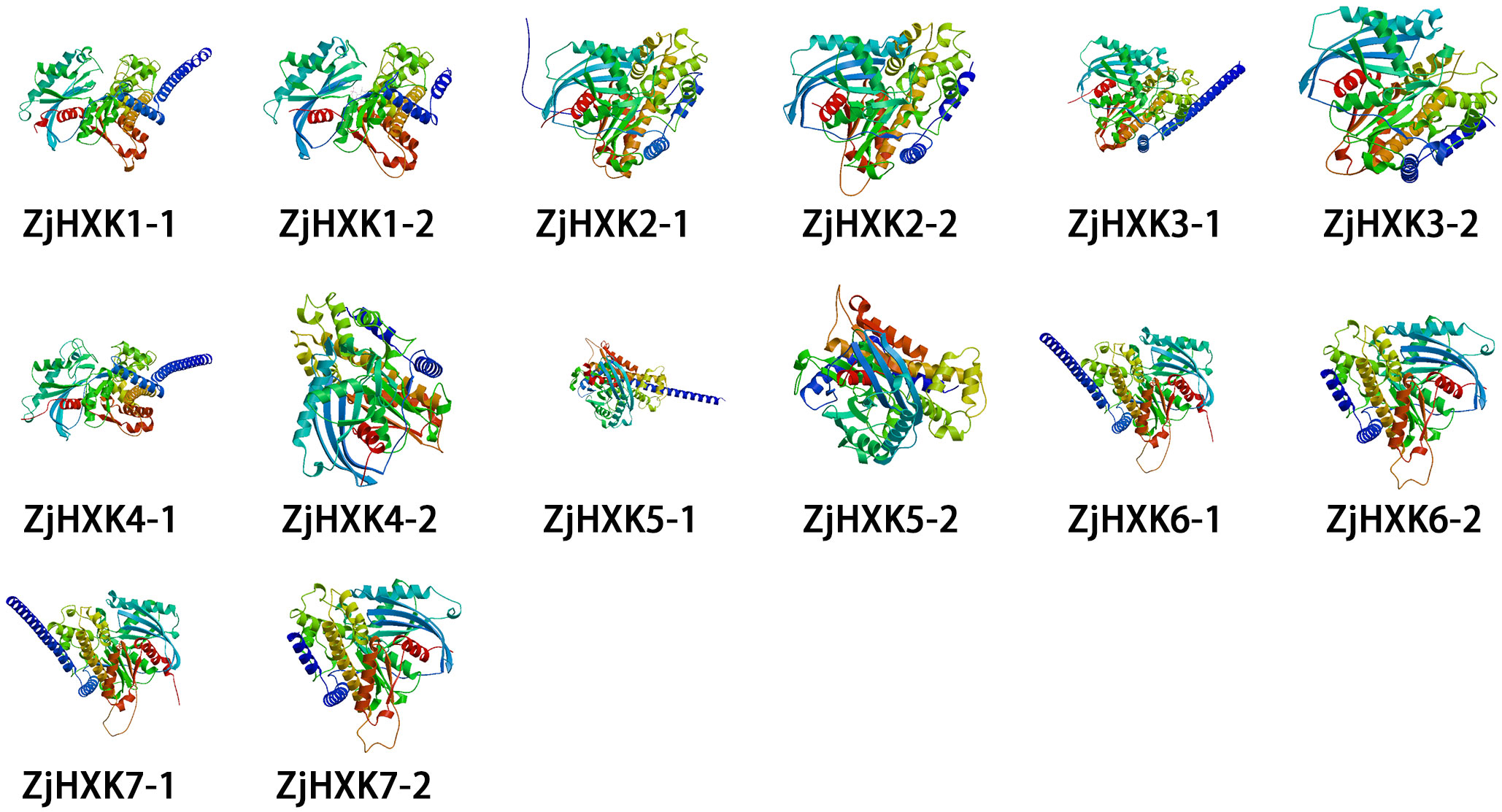
Figure 1 Tertiary structure analysis. There may be multiple tertiary structure models for the same protein sequence.
Phylogenetic analysis
The protein sequences of Arabidopsis HXK (AtHXK), apple HXK (MdHXK), peach HXK (PpHXK), pear HXK (PbHXK), and sweet orange HXK (CsHXK) were used to construct a neighbor-joining phylogenetic tree (Figure 2). These genes were classified into groups A, B and C, the group B and groups C further divided into two subgroups. Among them, ZjHXK members were distributed in all five subgroups, with ZjHXK3 in group A, ZjHXK6 and ZjHXK7 in subgroup B1, ZjHXK2 in subgroup B2, ZjHXK1 in subgroup C1, ZjHXK4 and ZjHXK5 in subgroup C2. Group A genes were mostly alkaline proteins, subgroup B1 protein were non-transmembrane proteins, subgroup B2 proteins have the highest proportion of extended chains, subgroup C1 contains more cis-acting elements related to stress response, and C2 subgroup proteins have the highest proportion of α-helices.
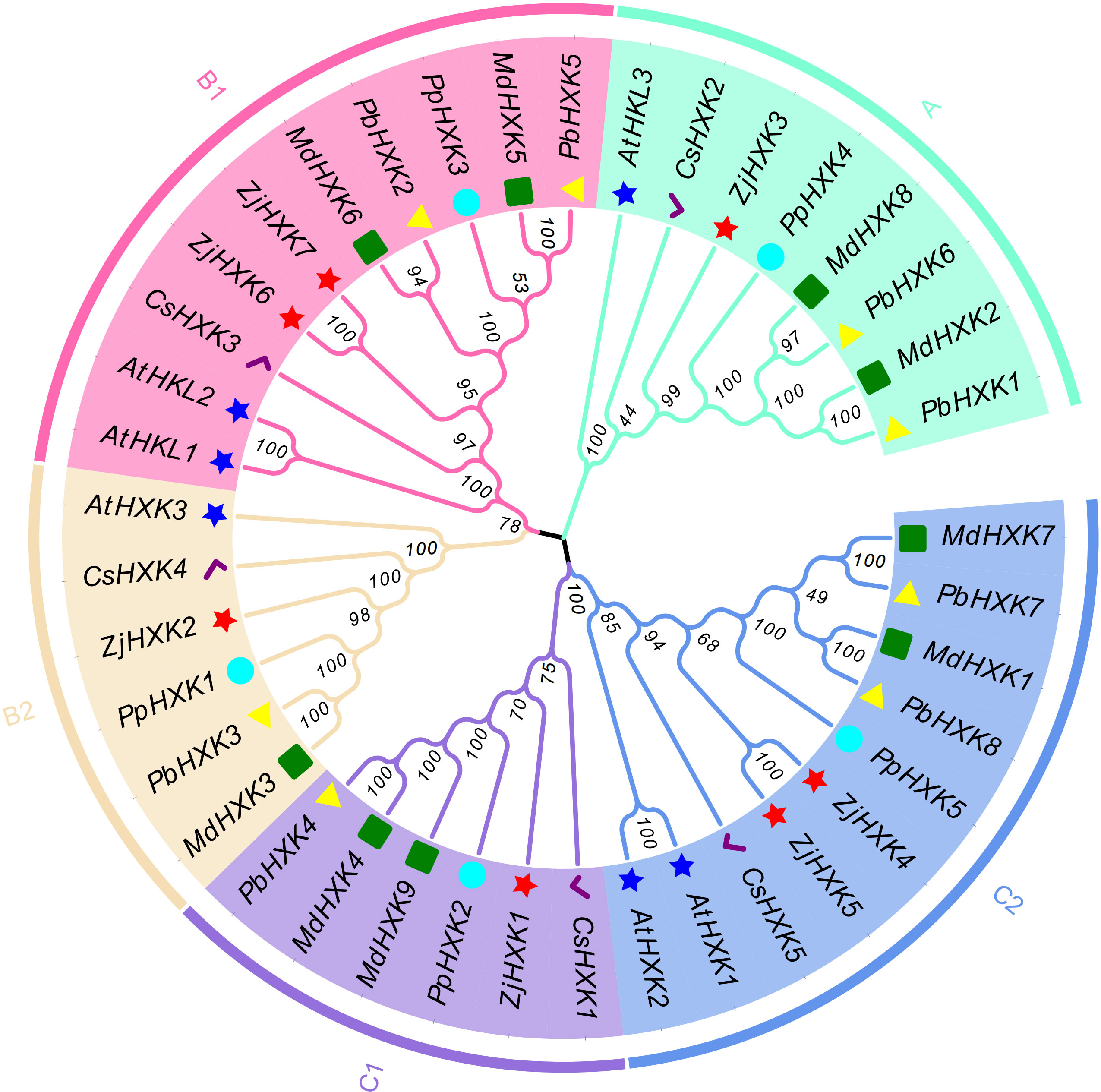
Figure 2 Phylogenetic analysis of HXK proteins from Jujube and other plants. At, Arabidopsis thaliana; Md, Malus domestica; Pp, Prunus persica; Pb, Pyrus bretschneideri; Cs, Citrus sinensis. The red star, the yellow triangle, the blue star, the green rect, the cyan circle and the purple tick were represented respectively Ziziphus jujuba, Pyrus bretschneideri, Arabidopsis thaliana, Malus domestica, Prunus persica and Citrus sinensis. A, B and C represent the three groups, respectively.
Gene structure and conserved motifs of ZjHXK genes
The gene structure analysis (Figure 3A) showed that each of the ZjHXK genes contained nine exons separated by eight introns. Among these, ZjHXK3 exhibits the shortest gene length, while longest for ZjHXK4. The MEME of ZjHXK proteins found that 10 putative conserved motifs were identified in all ZjHXK proteins except ZjHXK2 and ZjHXK3. In addition, the ZjHXK2 gene lacked motif 7, and the ZjHXK3 gene lacked motif 9 (Figure 3B). The lengths of the conserved motifs ranged from 28 to 50 aa (Figure 3C).
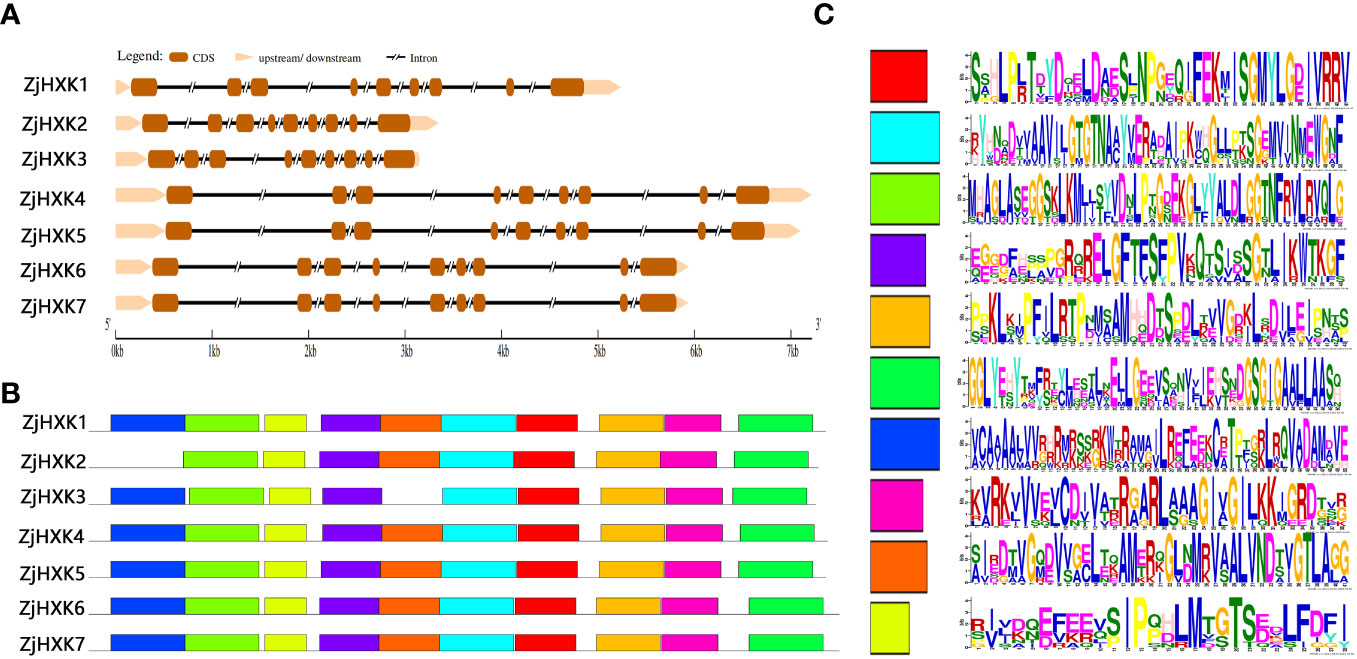
Figure 3 The gene structure and conserved motifs of ZjHXK genes in jujube. The gene structure of ZjHXK genes (A), in which brown lines represent exons and black lines represent introns. The conserved motifs of ZjHXK proteins (B), in which the conserved motifs are indicated by colored boxes. Sequences of the 10 conserved motifs in the ZjHXK proteins (C).
Promoter cis-element analysis
Promoter cis-acting regulatory elements results showed the there were many cis-acting regulatory elements involved in hormones, stress, and light in the promoter regions (Figure 4). Among these elements, there were relatively more cis-acting regulatory elements associated with hormone response in ZjHXK promoter regions, while the number of cis-acting regulatory elements related to stress response was relatively lower. It is worth noting that the MeJA-responsiveness cis-acting elements, linked to hormone response, were the most abundant. In addition, ZjHXK2 displayed the highest number of ABRE and G-Box cis-acting elements, whereas ZjHXK4 and ZjHXK5 had only one cis-acting element related to light response but the highest number of cis-acting elements associated with stress response.
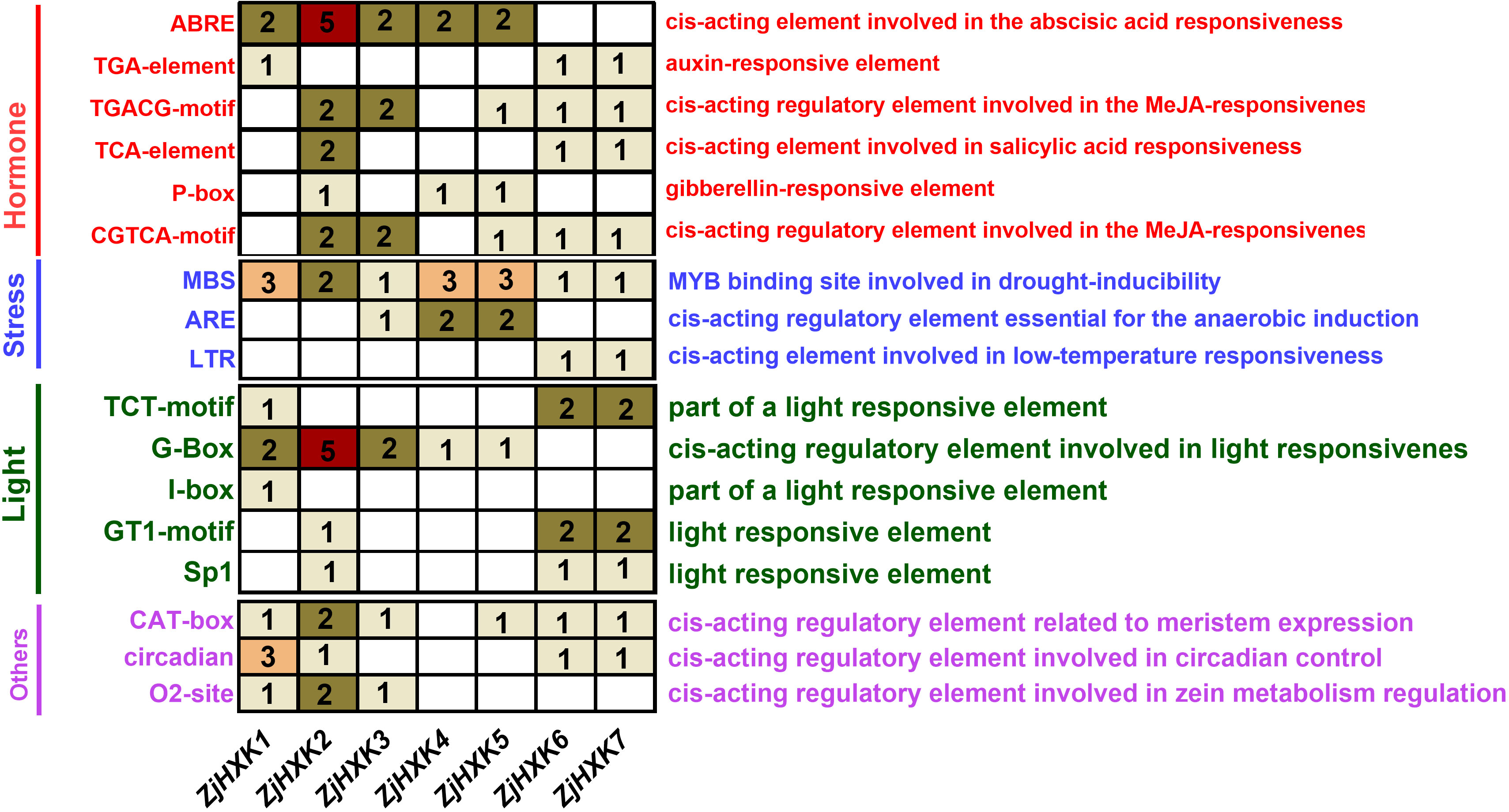
Figure 4 Promoter cis-element analysis of HXK gene family. The figure does not show all cis-acting elements, but only the cis-acting elements that are prevalent on ZjHXK. The cis-acting elements related to hormones were shown in red, those related to stress were shown in blue, and those related to light were shown in green. The others are shown in purple. The number in the box represents the number of corresponding cis-acting elements in the corresponding promoter sequence.
Intragenomic and intergenomic collinearity analysis
Through intragenomic collinearity analysis, two pairs of collinear gene pairs were identified, namely, ZjHXK1 and ZjHXK5, and ZjHXK6 and ZjHXK7 (Figure 5A). Through intergenomic collinearity analysis, genetic differences and gene duplications of HXK genes between jujube and Arabidopsis were explored. Collinearity was found between four ZjHXK gene family members and three Arabidopsis HXK genes (Figure 5B). Specifically, ZjHXK4 (chr 12), ZjHXK5 (chr 12) and ZjHXK1 (chr 1), exhibit collinearity with AtHXK2 (chr 2) and AtHXK1 (chr 4). ZjHXK3 (chr 8) shared collinearity with AtHKL3 (chr 4). Additionally, during the evolutionary process in different species, duplicated genes may have undergone changes or been lost.
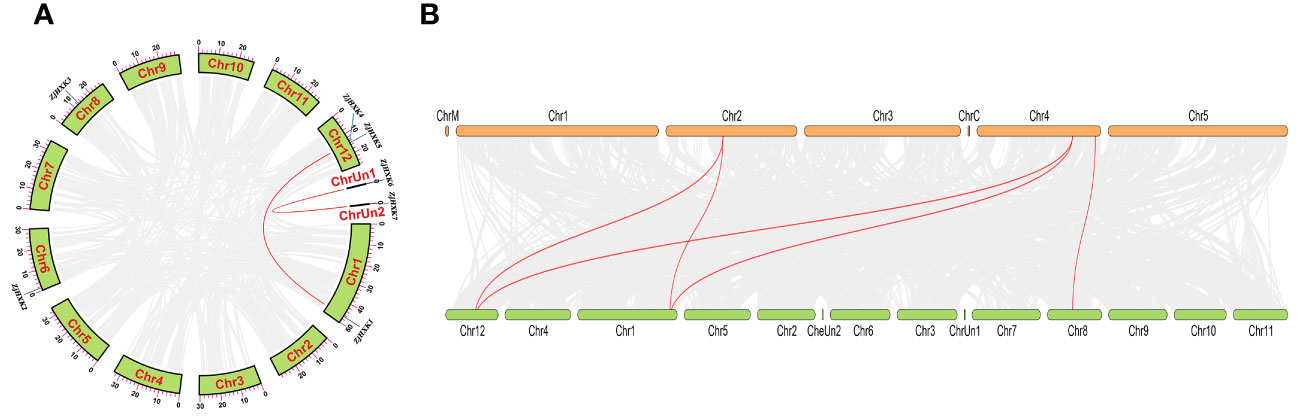
Figure 5 Collinearity analysis of the HXK gene families in Ziziphus jujuba (A), between Ziziphus jujuba and Arabidopsis (B). The red lines connect two genes which exist multicollinearity. In a, the squares around the circles represent 12 chromosomes of Ziziphus jujuba. Among them, Ziziphus jujuba has two sequence that has not been assembled into chromosomes. In b, the green boxes represent chromosomes of Ziziphus jujuba. The orange boxes represent the chromosomes of Arabidopsis.
Sugar composition and ZjHXK gene expression patterns in different developmental stages of fruit
We determined the sugar composition of cv ‘Fucuimi’ and cv ‘Jing39’ fruit at different developmental stages using gas chromatography. The results showed that the trends in sugar composition changes in cv ‘Fucuimi’ and cv ‘Jing39’ fruit during fruit growth were similarly. However, throughout the entire developmental process, cv ‘Fucuimi’ exhibited significantly higher sugar composition levels compared to cv ‘Jing39’ (Figure 6A).
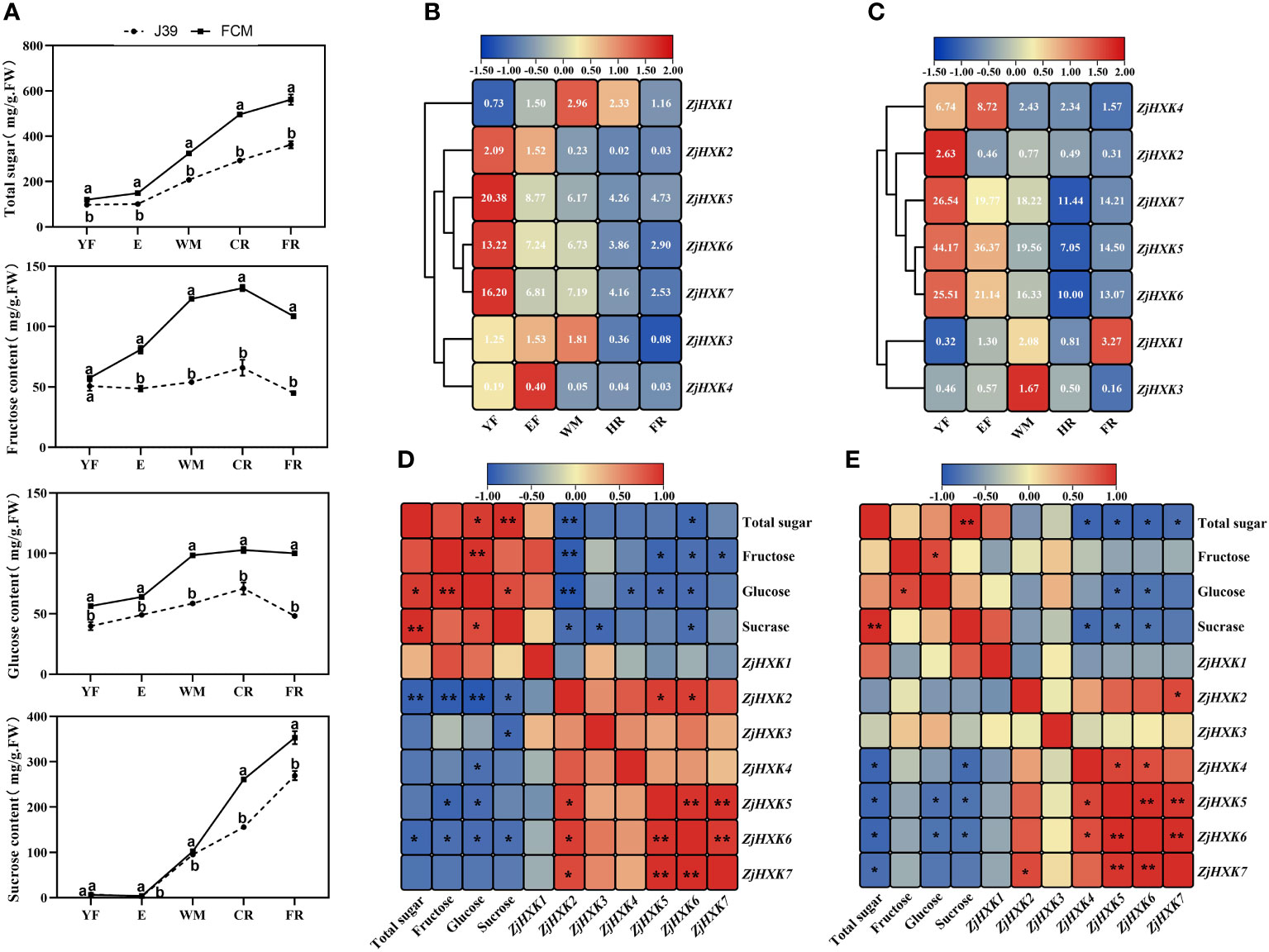
Figure 6 Sugar content of ‘Fucuimi’ and ‘Jing39’ (A), ZjHXK gene family expression levels of ‘Fucuimi’ (B) and ‘Jing39’ (C) during fruit development, correlation analysis of ‘Fucuimi’ (D) and ‘Jing39’ (E) between the expression and sugar component. In a, the same lowercase letters indicate no significant difference at the 0.05 level between the two varieties in the same period. YF, E, WM, CR and FR represented fruits at young fruit stage, expanding stage, white mature stage, half-red stage and ripening stage, respectively. The numbers in b & c were FPKM values, the bluer the color, the lower the expression, and the redder the expression. d and e are based on Pearson coefficients, with blue to red indicating negative to positive correlations. *, p < 0.05; **, p < 0.01.
Some ZjHXK exhibit similar expression patterns during fruit development of cv ‘Fucuimi’ and cv ‘Jing39’ (Figures 6B, C). Among them, the expression level of ZjHXK3 increases as the fruit grows until maturity stage (WM). In addition, the expression levels of ZjHXK2, ZjHXK4, ZjHXK5, ZjHXK6, and ZjHXK7 decrease as the fruit matures. Notably, ZjHXK5, ZjHXK6, and ZjHXK7 consistently exhibit higher expression levels throughout the whole fruit development period. Correlation analysis between sugar content and the expression levels of ZjHXK genes in cv ‘Fucuimi’ and cv ‘Jing39’ fruit suggested that ZjHXK5 and ZjHXK6 were significantly negatively correlated with total sugar, fructose, glucose, and sucrose content (Figures 6D, E).
In addition, we also analyzed the expression levels of ZjHXK in six different plant tissues (root, leaf, flower, stem, branch, and fruit) (Supplementary Figure 6). We found that ZjHXK2 was almost not expressed in leaves and fruit but exhibits higher expression in branches. ZjHXK3 maintains low expression levels throughout the development period. ZjHXK4 and ZjHXK5 have higher expression levels in leaves and flowers, while ZjHXK6 and ZjHXK7 exhibit higher expression in roots and stems.
Transient transformation of ZjHXK5 and ZjHXK6
To validate the roles of ZjHXK5 and ZjHXK6 in sugar metabolism, we conducted transient transformation in heterologous tobacco leaves and endogenous jujube fruit (Supplementary Figure 7). The total sugar, fructose, glucose, and sucrose content in tobacco leaves and jujube fruit were significant decrease after overexpressed ZjHXK5 and ZjHXK6 (Figures 7A, B). However, through VIGS-mediated silencing of ZjHXK5 and ZjHXK6 gene expression, we found an increase in sugar components in jujube fruits, with a significant elevation in both total sugar and sucrose content (Figure 7C).

Figure 7 Sugar composition analysis of ZjHXK5 and ZjHXK6 in transiently transformed tobacco leaves (A) and jujube fruits (B), as well as VIGS-Mediated silencing of jujube fruit gene expression (C). * and ** represent significant differences between treatment and control at 0.05 and 0.01 levels, respectively.
Stable genetic transformation of ZjHXK5 and ZjHXK6
Through genetic transformation, after stable overexpression of ZjHXK5 and ZjHXK6 (Supplementary Figure 8), the contents of fructose, glucose and sucrose in tomato fruit and jujube callus were also significantly decreased (Figures 8A–D). In addition, we noted that the growth rates of calli decreased, and root length, plant height, leaflet length, and stem thickness of tomato were significantly lower than the control.
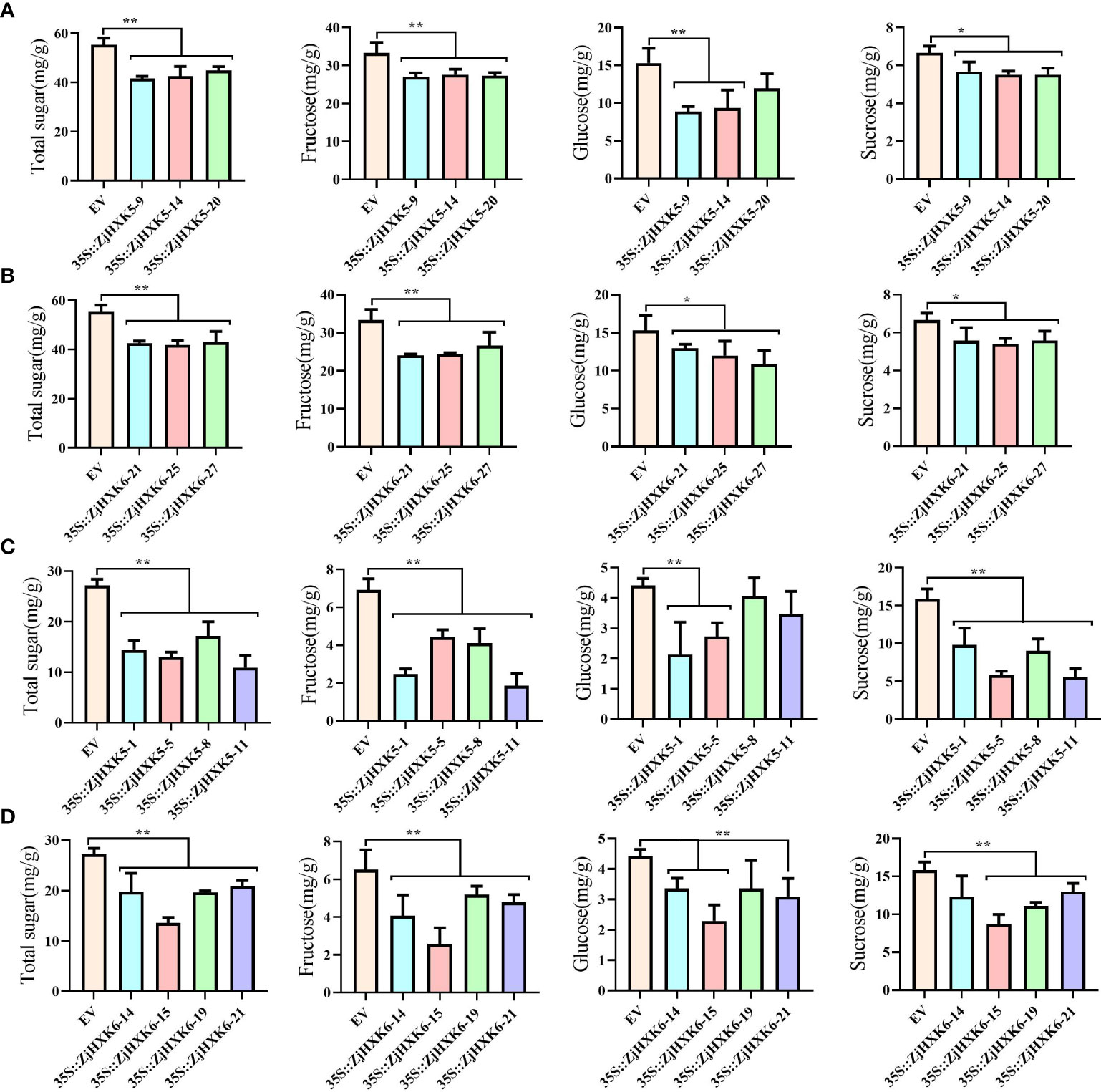
Figure 8 Stable genetic transformation of 35S::ZjHXK5 and 35S::ZjHXK6 in Micro-Tom (A, B) and ‘Jing39’ callus (C, D). * and ** represent significant differences between treatment and control at 0.05 and 0.01 levels, respectively.
Effect of exogenous MeJA on sugar component content in jujube
The promoter regions of ZjHXK5 and ZjHXK6 genes contain many cis-regulatory elements responsive to MeJA. Therefore, we designed MeJA treatments with different gradients in jujube calli. The expression of ZjHXK5 and ZjHXK6 was significantly down-regulated after treatment with different concentrations of MeJA (Figure 9A). Similarly, T1, T2 and T3 treatments significantly increased the total sugar content, and T1 and T2 treatments also had significant effects on the sugar composition (Figure 9B). In addition, we observed that as the MeJA treatment concentration increased, the callus browning rate increased, while the growth rate decreased, suggesting that T1 or T2 may the optimal treatment concentrations.

Figure 9 Changes in gene expression levels (A) and sugar composition (B) of ZjHXK5 and ZjHXK6 in ‘Jing39’ callus tissues treated with different concentrations of MeJA. T1, T2, T3, and T4 correspond to MeJA treatments at concentrations of 0.05, 0.10, 0.20, and 0.30 mmol/L, respectively. * and ** represent significant differences between treatment and control at 0.05 and 0.01 levels, respectively.
Discussion
HXK gene not only plays a crucial role in various biological processes by catalyzing glucose phosphorylation, but also participates in intracellular signaling, thereby effect plant growth and development (Jang et al., 1997; Granot, 2007). To date, members of the HXK gene family have been identified in numerous species, including six in Arabidopsis (Karve et al., 2008), ten in rice (Cho et al., 2006), nine in maize (Zhang et al., 2016), seven in cassava, and ten in apple (Zhu et al., 2021). In this study, seven ZjHXK genes was identified at gene-wide level. Phosphorylation events are involved in various cellular processes that affect the subcellular localization and stability of target proteins (Johnson, 2009). In this study, a greater number of phosphorylation sites were predicted in ZjHXK proteins, suggesting their susceptibility to post-translational modification events.
The tertiary structure, which involves further twisting and folding on the basis of secondary structure, contributes to a better understanding of gene functionality. Notably, in previous research, the tertiary structure analysis of HXK members was neglected (Huang et al., 2020; Liao et al., 2022). In this study, all ZjHXK proteins possess two distinct tertiary structures with substantial differences between them. This suggests that different HXK tertiary structures may execute distinct functions in various biological processes. This underscores the adaptability and multifunctionality of proteins. It is important to note that the existence of these two tertiary structures may potentially lead to divergent enzymatic functions.
Previous research has shown that the characteristics of gene intron/exon sequences are crucial for understanding gene function and evolutionary relationships (Liao et al., 2020). In this study, it was found that the number of introns and coding sequences (CDS) in ZjHXK gene family members was consistent, further indicating the high conservation in the evolution of ZjHXK genes. The results of conserved motif analysis support this result, as many HXK family members share the same conserved motifs. This is one of the main reasons why some genes cluster together in phylogenetic trees. According to the phylogenetic analysis, HXK is classified into subfamilies A, B1, B2, and C2, corresponding to subfamilies V, IV, I, and II in Arabidopsis, which was consistent with previous studies (Dai et al., 2002; Kim et al., 2013; Geng et al., 2017). The jujube genome has undergone frequent interchromosomal fusions and segmental duplications, but a recent whole-genome duplication has not occurred (Liu et al., 2014). Multiple collinearity analysis of HXK suggested that HXK had originated through duplication events among its own family members. Intraspecific genome collinearity analysis of HXK revealed two pairs of collinear gene pairs, ZjHXK1 and ZjHXK5, and ZjHXK6 and ZjHXK7, indicating homologous genes on different chromosomes during the course of evolution. These genes further confirmed the high conservation of the HXK gene family. Inter-specific genome collinearity showed that four ZjHXK gene family members exhibited collinearity with three Arabidopsis HXK genes, suggesting some similarity between jujube and Arabidopsis HXK genes. It is also possible that duplicate genes may have been altered or lost during the evolutionary process in different species (Yang et al., 2023).
Promoter cis-regulatory element analysis revealed that all members of the ZjHXK family possess multiple hormone-responsive elements, light-responsive elements, and stress-responsive elements. However, there were significant differences in the types and number of cis-regulatory elements in the same orientation among these family members. These findings suggest that multiple homologous genes have gradually evolved during the process of plant development, thereby avoiding situations in which mutations in a single gene result in the loss of function and slow down or halt growth (Magadum et al., 2013). In addition, ZjHXK genes were also involved in plant growth and development. Further analysis of cis-elements highly related to sugar components indicated that ZjHXK promoters were predominantly associated with hormone-regulatory elements, with ABRE elements being the most common. Overexpression of AtHXK in Arabidopsis led to increased sensitivity to cytokinin in cell division, implying that ZjHXK1 in jujube may regulate plant hormone sensitivity.
As is widely recognized, hexokinase (HXK) is known to participate in the regulation of fruit sugar content by catalyzing the phosphorylation of hexose, thereby influencing sugar metabolism and accumulation. Its activity and expression levels are likely to play a pivotal role in determining fruit sugar content and sweetness. Previous studies have indicated that jujube is a fruit with relatively high sugar content (Bartolozzi et al., 1997; Sparkes et al., 2006; Sun et al., 2015). By assessing the gene expression levels and sugar component changes in high and low sugar jujube fruits at various developmental stages, we can gain a more comprehensive understanding of the mechanisms underlying sugar accumulation. Gene expression profile analysis revealed that the majority of ZjHXK genes exhibit a downregulation in expression levels as the fruit matures, while the trend in sugar component content is opposite to this. Correlation analysis suggests that ZjHXK5 and ZjHXK6 genes may play a significant role in the high-sugar accumulation phenotype of jujube, consistent with functional studies in pear (Zhao et al., 2019) and apple (Zhu et al., 2021). One of the most interesting results is that ZjHXK genes were expressed in multiple tissues, but their expression levels vary significantly, indicating that the ZjHXK genes perform distinct functions in different organs.
Due to the difficulty of stable genetic transformation in jujube, even if transgenic plants are obtained, it will take 3-5 years to fruit. Therefore, most studies of gene function rely on instantaneous transformation of callus or stable expression in model plants. Some fruit trees are also identified gene function by this way (Anjanappa and Gruissem, 2021). In this study, ZjHXK5 and ZjHXK6 were been confirmed that can negatively regulate sugar metabolism, consistent with previous studies, such as transgenic tomato leaves expressing AtHXK1 exhibited a decrease in photosynthetic efficiency, accelerated senescence, and varying degrees of reduction in young fruit weight, starch content, and soluble sugar content in mature fruits (Dai et al., 1999). It can be inferred that ZjHXK5 and ZjHXK6 accelerate the phosphorylation of hexoses, resulting in reduction of sugar composition, and thus regulate plant growth and development. In addition, a large number of studies have shown that the expression of HXK genes in plants is influenced by certain exogenous hormones (Hitoshi, 2006; Pimenta Lange and Lange, 2006; Mishra et al., 2009). Both the ZjHXK5 and ZjHXK6 promoters contain MeJA-responsive elements, including TGACG-motif and CGTCA-motif. Upon subjecting jujube callus tissues to MeJA treatment, it was observed that the expression of ZjHXK5 and ZjHXK6 genes was significantly downregulated. Simultaneously, sugar components exhibited a significant increase under MeJA treatment at concentrations of 0.05 mmol/L and 0.10 mmol/L. This suggests that ZjHXK5 and ZjHXK6 were downregulated under MeJA induction and can effectively regulate sugar accumulation at suitable concentrations. MeJA treatment before fruit picking can not only increase the soluble solids and total sugar content of the fruit (Li et al., 2023), but MeJA treatment after fruit picking can also increase the antioxidant capacity and phenolic content of the fruit, thereby extending the shelf life of the fruit (Wang et al., 2021). Although the present study does not provide a detailed explanation of the underlying mechanisms, it holds valuable reference value for a deeper understanding of the molecular mechanisms governing sugar composition regulation in jujube fruits. Furthermore, it offers a new avenue of investigation into the HXK gene family.
Conclusions
In this study, we have provided a comprehensive understanding of the identification of the HXK gene family in jujube, as well as the identified of key genes related sugar metabolism. Seven ZjHXK genes were identified in the jujube genome. All ZjHXK genes exhibit similar gene structures, conserved motifs, and subcellular localization. Many common and unique cis-elements were identified within the ZjHXK promoter regions. Additionally, a phylogenetic tree of HXK genes was constructed, classifying them into five subgroups. Through the correlation analysis of ZjHXK expression levels and sugar component content, we found that ZjHXK5 and ZjHXK 6 were the key genes regulating sugar. The sugar content was decreased after overexpression in jujube calli, tobacco leaf and tomato fruit. Transient silencing of ZjHXK5 and ZjHXK6 genes results in a significant increase in sucrose and total sugar content. Furthermore, ZjHXK5 and ZjHXK6 were downregulated under MeJA treatment, which significantly enhanced sugar component content. In summary, our study lays the foundation for further exploration of the molecular mechanisms of sugar metabolism in jujube.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
Author contributions
PT: Writing – original draft. GL: Writing – review & editing. DL: Resources, Writing – original draft. XZ: Visualization, Writing – original draft. WZ: Data curation, Writing – original draft. QX: Writing – review & editing. CW: Supervision, Writing – review & editing. JW: Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This project was supported by the National Natural Science Foundation of China (32260729) and Supported by the Earmarked Fund of Xinjiang Jujube Industrial Technology System (XJCYTX-01).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2024.1335120/full#supplementary-material
References
Anjanappa, R. B., Gruissem, W. (2021). Current progress and challenges in crop genetic transformation. J. Plant Physiol. 261, 153411. doi: 10.1016/j.jplph.2021.153411
Bailey, T. L., Williams, N., Misleh, C., Li, W. W. (2006). MEME: discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 34, W369–W373. doi: 10.1093/nar/gkl198
Bartolozzi, F., Bertazza, G., Bassi, D., Cristoferi, G. (1997). Simultaneous determination of soluble sugars and organic acids as their trimethylsilyl derivatives in apricot fruits by gas-liquid chromatography. J. Chromatogr. A 758, 99–107. doi: 10.1016/S0021-9673(96)00709-1
Chen, C., Wu, Y., Li, J., Wang, X., Zeng, Z., Xu, J., et al. (2023). TBtools-II: A “one for all, all for one” bioinformatics platform for biological big-data mining. Mol. Plant S1674-2052 (23), 00281–00282. doi: 10.1016/j.molp.2023.09.010
Chen, J., Tsim, K. W. K. (2020). A review of edible jujube, the ziziphus jujuba fruit: A heath food supplement for anemia prevalence. Front. Pharmacol. 11. doi: 10.3389/fphar.2020.593655
Cho, J., Ryoo, N., Ko, S., Lee, S., Lee, J., Jung, K., et al. (2006). Structure, expression, and functional analysis of the hexokinase gene family in rice (Oryza sativa L.). Planta 224, 598–611. doi: 10.1007/s00425-006-0251-y
Dai, N., Kandel-Kfir, M., Petreikov, M., Hanael, R., Levin, I., Ricard, B., et al. (2002). The tomato hexokinase LeHXK1 cloning, mapping, expression pattern and phylogenetic relationships. Plant Sci. 163, 581–590. doi: 10.1016/S0168-9452(02)00166-8
Dai, N., Schaffer, A., Petreikov, M., Shahak, Y., Giller, Y., Ratner, K., et al. (1999). Overexpression of Arabidopsis hexokinase in tomato plants inhibits growth, reduces photosynthesis, and induces rapid senescence. Plant Cell 11, 1253–1266. doi: 10.1105/tpc.11.7.1253
Geng, M., Yao, Y., Wang, Y., Wu, X., Sun, C., Li, R., et al. (2017). Structure, expression, and functional analysis of the hexokinase gene family in cassava. Int. J. Mol. Sci. 18, 1041. doi: 10.3390/ijms18051041
Granot, D. (2007). Role of tomato hexose kinases. Funct. Plant biology: FPB 34, 6, 564–6, 570. doi: 10.1071/FP06207
Granot, D., Kelly, G., Stein, O., David, S. R. (2014). Substantial roles of hexokinase and fructokinase in the effects of sugars on plant physiology and development. J. Exp. Bot. 65, 809–819. doi: 10.1093/jxb/ert400
Hitoshi, S. (2006). CYTOKININS: Activity, biosynthesis, and translocation. Annu. Rev. Plant Biol. 57, 431–449. doi: 10.1146/annurev.arplant.57.032905.105231
Hu, B., Jin, J., Guo, A. Y., Zhang, H., Luo, J., Gao, G. (2015). GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics 31, 1296–1297. doi: 10.1093/bioinformatics/btu817
Huang, T., Luo, X., Wei, M., Shan, Z., Zhu, Y., Yang, Y., et al. (2020). Molecular cloning and expression analysis of sucrose phosphate synthase genes in cassava (Manihot esculenta Crantz). Sci. Rep. 10, 20707. doi: 10.1038/s41598-020-77669-9
Hubbard, N. L., Pharr, D. M., Huber, S. C. (1991). Sucrose phosphate synthase and other sucrose metabolizing enzymes in fruits of various species. Physiologia Plantarum 82, 191–196. doi: 10.1111/j.1399-3054.1991.tb00080.x
Jang, J. C., León, P., Zhou, L., Sheen, J. (1997). Hexokinase as a sugar sensor in higher plants. Plant Cell 9, 5–19. doi: 10.1105/tpc.9.1.5
Jang, J. C., Sheen, J. (1994). Sugar sensing in higher plants. Plant Cell 6 (11), 1665–1679. doi: 10.1016/S1360-1385(97)89545-3
Ji, Q., Wang, R., Chen, K., Xie, Z., Li, S., Wang, D., et al. (2023). Comparative transcriptome profiling analysis provides insight into the mechanisms for sugar change in Chinese jujube (Ziziphus jujuba Mill.) under rain-proof cultivation. Plant Genome 16, e20341. doi: 10.1002/tpg2.20341
Johnson, L. N. (2009). The regulation of protein phosphorylation. Biochem. Soc. Trans. 37, 627–641. doi: 10.1042/bst0370627
Karni, L., Aloni, B. (2002). Fructokinase and Hexokinase from Pollen Grains of Bell Pepper (Capsicum annuum L.): Possible Role in Pollen Germination under Conditions of High Temperature and CO2 Enrichment. Ann. Bot. 90, 607–612. doi: 10.1093/aob/mcf234
Karve, R., Lauria, M., Virnig, A., Xia, X., Rauh, B. L., Moore, B. (2010). Evolutionary lineages and functional diversification of plant hexokinases. Mol. Plant 3, 334–346. doi: 10.1093/mp/ssq003
Karve, A., Rauh, B. L., Xia, X., Kandasamy, M., Meagher, R. B., Sheen, J., et al. (2008). Expression and evolutionary features of the hexokinase gene family in Arabidopsis. Planta 228, 3, 411–425. doi: 10.1007/s00425-008-0746-9
Kim, Y. M., Heinzel, N., Giese, J. O., Koeber, J., Melzer, M., Rutten, T., et al. (2013). A dual role of tobacco hexokinase 1 in primary metabolism and sugar sensing. Plant Cell Environ. 36, 1311–1327. doi: 10.1111/pce.12060
Li, J., Fan, L., Ding, S., Ding, X. (2007). Nutritional composition of five cultivars of chinese jujube. Food Chem. 103, 454–460. doi: 10.1016/j.foodchem.2006.08.016
Li, B., Zhang, X., Han, C., Duan, R., Yang, J., Xue, H. (2023). Effects of methyl jasmonate on fruit coloration and quality improvement in pears (Pyrus bretschneideri). Agronomy 13, 2409. doi: 10.3390/agronomy13092409
Liao, G., Li, Y., Wang, H., Liu, Q., Zhong, M., Jia, D., et al. (2022). Genome-wide identification and expression profiling analysis of sucrose synthase (SUS) and sucrose phosphate synthase (SPS) genes family in Actinidia chinensis and A. eriantha. BMC Plant Biol. 22, 215. doi: 10.1186/s12870-022-03603-y
Liao, G., Liu, Q., Li, Y., Zhong, M., Huang, C., Jia, D., et al. (2020). Identification and expression profiling analysis of ascorbate peroxidase gene family in Actinidia chinensis (Hongyang). J. Plant Res. 133, 715–726. doi: 10.1007/s10265-020-01206-y
Liao, G., Xu, Q., Allan, A. C., Xu, X. (2023). L-Ascorbic acid metabolism and regulation in fruit crops. Plant Physiol. 192, 1684–1695. doi: 10.1093/plphys/kiad241
Liu, M., Wang, J., Wang, L., Liu, P., Zhao, J., Zhao, Z., et al. (2020). The historical and current research progress on jujube-a superfruit for the future. Horticulture Res. 7, 119. doi: 10.1038/s41438-020-00346-5
Liu, M., Zhao, J., Cai, Q., Liu, G., Wang, J., Zhao, Z., et al. (2014). The complex jujube genome provides insights into fruit tree biology. Nat. Commun. 5, 5315. doi: 10.1038/ncomms6315
Livak, K. J., Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Magadum, S., Banerjee, U., Murugan, P., Gangapur, D., Ravikesavan, R. (2013). Gene duplication as a major force in evolution. J. Genet. 92, 155–161. doi: 10.1007/s12041-013-0212-8
Mishra, B. S., Singh, M., Aggrawal, P., Laxmi, A. (2009). Glucose and auxin signaling interaction in controlling Arabidopsis thaliana seedlings root growth and development. PloS One 4, e4502. doi: 10.1371/journal.pone.0004502
Mitteer, D. R., Greer, B. D. (2022). Using graphpad prism’s heat maps for efficient, fine-grained analyses of single-case data. Behav. Anal. Pract. 15, 505–514. doi: 10.1007/s40617-021-00664-7
Moore, B., Zhou, L., Rolland, F., Hall, Q., Cheng, W. H., Liu, Y. X., et al. (2003). Role of the Arabidopsis glucose sensor HXK1 in nutrient, light, and hormonal signaling. Science 300, 332–336. doi: 10.1126/science.1080585
Pimenta Lange, M. J., Lange, T. (2006). Gibberellin biosynthesis and the regulation of plant development. Plant Biol. 8, 281–290. doi: 10.1055/s-2006-923882
Polit, J. T., Ciereszko, I. (2009). In situ activities of hexokinase and fructokinase in relation to phosphorylation status of root meristem cells of Vicia faba during reactivation from sugar starvation. Physiologia Plantarum 135 (4), 342–350. doi: 10.1111/j.1399-3054.2008.01201.x
Rashwan, A., Karim, N., Shishir, M. R. I., Bao, T., Lu, Y., Chen, W. (2020). Jujube fruit: A potential nutritious fruit for the development of functional food products. J. Funct. Foods 75 (9), 104205. doi: 10.1016/j.jff.2020.104205
Rolland, F., Baena-Gonzalez, E., Sheen, J. (2006). Sugar sensing and signaling in plants: conserved and novel mechanisms. Annu. Rev. Plant Biol. 57, 675–709. doi: 10.1146/annurev.arplant.57.032905.105441
Roth, M. S., Westcott, D. J., Iwai, M., Niyogi, K. K. (2019). Hexokinase is necessary for glucose-mediated photosynthesis repression and lipid accumulation in a green alga. Commun. Biol. 2, 347. doi: 10.1038/s42003-019-0577-1
Saddhe, A. A., Manuka, R., Penna, S. (2021). Plant sugars: Homeostasis and transport under abiotic stress in plants. Physiologia Plantarum 171, 739–755. doi: 10.1111/ppl.13283
Sami, F., Yusuf, M., Faizan, M., Faraz, A., Hayat, S. (2016). Role of sugars under abiotic stress. Plant Physiol. Biochem. 109, 54–61. doi: 10.1016/j.plaphy.2016.09.005
Sparkes, I. A., Runions, J., Kearns, A., Hawes, C. (2006). Rapid, transient expression of fluorescent fusion proteins in tobacco plants and generation of stably transformed plants. Nat. Protoc. 1, 2019–2025. doi: 10.1038/nprot.2006.286
Sun, S., Kang, X., Xing, X., Xu, X., Cheng, J., Zheng, S., et al. (2015). Agrobacterium-mediated transformation of tomato (Lycopersicon esculentum L. cv. Hezuo 908) with improved efficiency. Biotechnol. Biotechnol. Equip. 29, 861–868. doi: 10.1080/13102818.2015.1056753
Swartzberg, D., Hanael, R., Granot, D. (2011). Relationship between hexokinase and cytokinin in the regulation of leaf senescence and seed germination. Plant Biol. 13, 439–444. doi: 10.1111/j.1438-8677.2010.00376.x
Tian, J., Pei, H., Zhang, S., Chen, J., Chen, W., Yang, R. (2014). TRV–GFP: a modified tobacco rattle virus vector for efficient and visualizable analysis of gene function. J. Exp. Bot. 65, 311–322. doi: 10.1093/jxb/ert381
Veramendi, J., Fernie, A. R., Leisse, A., Willmitzer, L., Trethewey, R. N. (2002). Potato hexokinase 2 complements transgenic Arabidopsis plants deficient in hexokinase 1 but does not play a key role in tuber carbohydrate metabolism. Plant Mol. Biol. 49, 491–501. doi: 10.1023/a:1015528014562
Wang, S. Y., Shi, X. C., Liu, F. Q., Laborda, P. (2021). Effects of exogenous methyl jasmonate on quality and preservation of postharvest fruits: A review. Food Chem. 353, 129482. doi: 10.1016/j.foodchem.2021.129482
Wasaya, A., Rehman, I., Mohi Ud Din, A., Hayder Bin Khalid, M., Ahmad Yasir, T., Mansoor Javaid, M., et al. (2023). Foliar application of putrescine alleviates terminal drought stress by modulating water status, membrane stability, and yield- related traits in wheat (Triticum aestivum L.). Front. Plant Sci. 14. doi: 10.3389/fpls.2022.1000877
Xiao, W., Sheen, J., Jang, J. C. (2000). The role of hexokinase in plant sugar signal transduction and growth and development. Plant Mol. Biol. 44 (4), 451–461. doi: 10.1023/a:1026501430422
Xu, W., Wei, Y., Wang, X., Han, P., Chen, Y., Xu, F., et al. (2021). Molecular cloning and expression analysis of hexokinase genes in peach fruit under postharvest disease stress. Postharvest Biol. Technol. 172, 111377. doi: 10.1016/j.postharvbio.2020.111377
Yang, C., Zhao, X., Luo, Z., Wang, L., Liu, M. (2023). Genome-wide identification and expression profile analysis of SWEET genes in Chinese jujube. PeerJ 11, e14704. doi: 10.7717/peerj.14704
Zhang, C., Huang, J., Li, X. (2015). Identification of appropriate reference genes for RT-qPCR analysis in Ziziphus jujuba Mill. Scientia Hortic. 197, 166–169. doi: 10.1016/j.scienta.2015.09.026
Zhang, Z., Li, X., Zhang, C., Zou, H., Wu, Z. (2016). Isolation, structural analysis, and expression characteristics of the maize nuclear factor Y gene families. Biochem. Biophys. Res. Commun. 478, 752–758. doi: 10.1016/j.bbrc.2016.08.020
Zhao, B., Qi, K., Yi, X., Chen, G., Liu, X., Qi, X., et al. (2019). Identification of hexokinase family members in pear (Pyrus × bretschneideri) and functional exploration of PbHXK1 in modulating sugar content and plant growth. Gene 711, 143932. doi: 10.1016/j.gene.2019.06.022
Keywords: bioinformatics analysis, hexokinase, jujube, methyl jasmonate, sugar metabolism
Citation: Tong P, Liao G, Lu D, Zhou X, Zhang W, Xu Q, Wu C and Wang J (2024) ZjHXK5 and ZjHXK6 negatively regulate the sugar metabolism of Ziziphus jujuba Mill.. Front. Plant Sci. 15:1335120. doi: 10.3389/fpls.2024.1335120
Received: 08 November 2023; Accepted: 22 January 2024;
Published: 12 February 2024.
Edited by:
Jihong Hu, Northwest A&F University, ChinaReviewed by:
Xiaoxu Li, Chinese Academy of Agricultural Sciences (CAAS), ChinaMingjun Li, Northwest A&F University, China
Copyright © 2024 Tong, Liao, Lu, Zhou, Zhang, Xu, Wu and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jiangbo Wang, d2pib2tAMTI2LmNvbQ==; Cuiyun Wu, d2N5YnlAMTYzLmNvbQ==
†ORCID: Cuiyun Wu, orcid.org/0000-0002-0178-3524
Jiangbo Wang, orcid.org/0000-0001-5315-3658
 Panpan Tong1,2
Panpan Tong1,2 Guanglian Liao
Guanglian Liao Xiaofeng Zhou
Xiaofeng Zhou Qiang Xu
Qiang Xu Cuiyun Wu
Cuiyun Wu Jiangbo Wang
Jiangbo Wang