
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 06 February 2024
Sec. Plant Pathogen Interactions
Volume 15 - 2024 | https://doi.org/10.3389/fpls.2024.1334958
This article is part of the Research TopicStudy on Plant Differentiation between Beneficial and Pathogenic MicroorganismsView all 9 articles
Introduction: Asexual reproduction of Gastrodia elata Bl. f. glauca S. chow (GeB) produces degeneration with increasing number of GeB. Therefore, we analyzed the microorganisms of GeB seedlings and surrounding soil by Illumina Miseq high-throughput sequencing technology.
Methods: In this study, Illumina Miseq high-throughput sequencing technology was applied to analyze the types and quantities of GeB seedlings and surrounding soil microorganisms in the first to third generations of asexual reproduction, isolated and identified the dominant strains of GeB in the first to third generations and screened the antagonistic bacteria of its pathogenic fungi, and evaluated the effects of beneficial bacteria on the production performance of seedlings planted with GeB.
Results: With an increase in the number of asexual reproductive generations, the number of pathogenic fungi and bacteria in GeB seedlings and the surrounding soil increased, and the number of beneficial fungi and bacteria decreased. Pseudomonas sp., Agrobacterium rhizomes, and Herbaspirillum hiltneri were isolated and identified in the first generation, and Trichoderma harzianum, Penicillium viridiatum, Fusarium oxysporum, and Novosphingobium sp. Were isolated and identified in the third generation. Antagonistic strains of the three pathogenic bacterial strains were screened. In conclusion, beneficial bacteria significantly improved the production performance of asexual reproductive seedlings planted with GeB.
Discussion: In conclusion, our findings suggested that the microorganisms of GeB seedlings and the surrounding soil change as the number of generations of GeB reproduction increases, disrupts the microecological balance of surrounding soil and endophytic microbiomes.This study provides a theoretical basis for the degradation of asexual reproduction in GeB.
The whole family of Orchidaceae has a rich species diversity (more than 28,000 species in the whole family), and the family covers primitive, partial, and complete fungi heterotrophic species (Baasanmunkh et al., 2021). Gastrodia elata Bl. f. glauca S. chow (GeB) is a non-toxic perennial herbaceous heterotrophic plant with a sweet flavor, smooth texture, and various benefits (Hong et al., 2018; Liu et al., 2018; Gong et al., 2021; Shan et al., 2021; Shan et al., 2021; Bae et al., 2022). GeB has no roots and no green leaves, so it cannot directly absorb water and inorganic salts from the soil for the growth of its plants, nor it can photosynthesize to produce organic nutrients for itself. However, GeB relies on symbiotic fungi to provide the nutrients that it needs for growth, so GeB is a typical fungi heterotrophic parasitic herbaceous plant, which relies on assimilation of some fungi hyphae that invade its body to obtain nutrients. There are at least two kinds of fungi in the life of GeB to provide nutrients for it: one kind of fungi is called Mycena, which provides nutrients for the seed when it germinates, so that it grows “mizuna”, and the other kind of fungi is called Armillariella mellea, which provides nutrients for GeB.
The demand for GeB has been increasing. However, wild GeB is scarce. Thus, artificial cultivation has been used to fulfill this demand, solving the problem of GeB supply and demand (Wang J. et al., 2023). The GeB life cycle includes the entire growth process of seeds, protocorms, mizuna, seedlings, mature tubers, and seeds (Li et al., 2020b). Seedlings are derived from asexual and sexual reproduction (Su et al., 2023). The sexually reproducing seeds of GeB have strong resistance and high yield but are often limited by seasonality and site (Yu et al., 2023), a long reproductive cycle (Hong et al., 2002), and many production links (Liu et al., 2022). Asexually reproduced GeB seedlings have short growth cycles and fewer seasonal and site-specific limitations (Park et al., 2012) than sexually reproduced GeB seedlings do. However, asexually reproduced seedlings are prone to degradation, resulting in a low yield of GeB (Han et al., 2020). Thus, clarifying the causes and solutions for the degradation of asexually reproductive seedlings is crucial for the production of GeB.
GeB seedlings undergo morphological changes (Park et al., 2012), and yield declines (de Meeûs et al., 2007) after asexual reproduction and passage. However, the underlying mechanism remains unclear. Endophytes in medicinal plants play an important role in host growth and quality (Naranjo et al., 2023) by providing plants with the nutrients that they need (Guan et al., 2022), promoting plant growth (Liu et al., 2020). Rhizosphere fungi and endophytes of the Orchidaceae family provide nutrients to plants (Barnes et al., 2016), enhance plant stress and disease resistance (Huang et al.,2022), and promote seed germination (Zhang et al., 2021; Jiang et al., 2022). Much valuable work has been conducted on other medicinal plant diseases, but studies on GeB seedling degradation from the perspective of microbial community diversity have not been reported.
The aim of this study was to clarify the causes of the degradation of asexual reproduction in GeB seedlings and to find ways to relieve this degradation. GeB seedlings were used as research objects. First, massive amplicon sequencing (Wang Z. et al., 2023; Chen et al., 2019; Clemmensen et al., 2022) was used to analyze the types and quantities of the first to third generations of asexual reproductive GeB seedlings and surrounding soil microorganisms, and the effects of propagation generations on GeB seedlings and their soil microbial diversity were clarified. Second, the isolation and identification of dominant bacteria were conducted, and the microbial community structure was characterized. The types of dominant beneficial fungi and bacteria, pathogenic fungi, and bacteria were clarified, and the antagonistic bacteria of the pathogenic fungi were screened through confrontation experiments. Finally, the effects of beneficial fungi and bacteria on the production of GeB seedlings were evaluated. Thus, this study provides a theoretical basis for exploring the causes and regulation of asexual reproduction and degradation in GeB.
GeB seedlings and surrounding soil of different propagation generations of GeB in Changbai Mountain were collected from Dabeishan Village, Longquan Town, Jingyu County, Baishan City, Jilin Province (126°30′E–127°16′E, 42°06′N–42°48′N). The study area has an average elevation of 775 m. It has a monsoon climatic zone and a cold-temperate humid climatic zone in the northeastern mountainous areas. In order to avoid the influence of other factors such as climate on changes in the microbial community, three GeB seedlings with reproductive generations in the same planting area from Baishan Jingzhen Tianma Development Co. (Figure 1) were selected for the experiments to ensure that the ground was level. In addition, a thermometer was used for measurement: the temperature was regulated by the density of the shade net, which was maintained at 20°C ± 3°C. In addition, the moisture content of the soil is measured by a hygrometer: the humidity is maintained at 50% ± 5% by artificial watering. This ensures that the study is less exposed to environmental impacts. Five sampling points were selected for the soil, and tubers of the first (first generation of sexual propagation), second (first generation of asexual propagation), and third (second generation of asexual propagation) generations of GeB seedlings by pooled samples were collected using the serpentine sampling method (S sampling method). The S sampling method is as follows: at the determined sampling point, first, use a small soil shovel to remove the surface layer of about 3 cm of soil and, then, tilt down a piece of soil.
Healthy GeB seedlings were selected, and 18 GeB seedlings from different generations were used for microbial community analysis. Soil samples were collected separately from each of the six GeB seedlings and placed in sterile polyethylene bags in the surrounding soil. For every six GeB seedlings, the tubers were cut into small piece; each sample was mixed evenly to prepare a GeB seedling sample. Three replicates were set for each propagation generation, and the seedling and soil samples were replicated.
The soil surrounding the first generation of GeB seedlings was labeled Y, with three replicates (Y1, Y2, and Y3). The soil surrounding the second generation of GeB seedlings was labeled as T, with three replicates (T1, T2, and T3). The soil surrounding the third generation of GeB seedlings was labeled W, with three replicates (W1, W2, and W3).
The first generation of GeB seedlings was labeled A, with three replicates (A1, A2, and A3). The second generation of GeB seedlings was labeled B, with three replicates (B1, B2, and B3). The third generation of GeB seedlings was labeled C, with three replicates (C1, C2, and C3).
DNA extraction and massive sequencing of microbiome amplicons from GeB seedlings and their surrounding soil microorganisms were performed by Shanghai Personal Biotechnology Co., Ltd. In brief, the DNA of each sample was extracted using the OMEGA Soil DNA Kit (Omega Bio-Tek, Norcross, GA, USA), using the manufacturer’s instructions. The quantity and quality of the extracted DNAs were measured using a NanoDrop NC2000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA) and agarose gel electrophoresis, respectively. The DNA was stored at 20°C until the PCR step. For bacteria, the part region of the 16S rRNA gene was amplified using the primer pair 338F/806R (V3–V4 region) for the surrounding soils and 799F/1193R (V5–V7 region) for the GeB seedlings (Table 1). For fungi, the nuclear ribosomal internal transcribed spacer 1 (ITS1) region was amplified using the primer pair ITS5/ITS2 for the surrounding soils and ITS1F/ITS2 for the GeB seedlings (Table 1). Sequencing was performed on an Illumina NovaSeq platform (Illumina, San Diego, CA, USA) to generate 250-bp paired-end reads. All raw data were uploaded to the Sequence Read Archive (SRA), numbers PRJNA1049783, PRJNA1049914, PRJNA1049820, and PRJNA1049958.
Microbiome bioinformatics was performed using QIIME2 2019.4, with slight modification, according to the official tutorials (https://docs.qiime2.org/2019.4/tutorials/). In brief, raw sequence data were demultiplexed using the Demux plugin, followed by primer cutting using the Cutadapt plugin. Sequences were then quality-filtered, denoised, and merged, and the chimera was removed using the DADA2 plugin. Non-singleton amplicon sequence variants (ASVs) were aligned with Multiple Alignment using Fast Fourier Transform (MAFFT) and used to construct a phylogenetic tree using fasttree2. Observed species, Shannon, and Simpson indices as well as other indices were calculated. The alpha diversity index was plotted as a sparse curve according to the Chao1 index, and species annotations in the samples were counted using the amplicon sequence operational taxonomic unit (OTU) sequence. Taxonomy was assigned to ASVs by using the classify-sklearn naive Bayes taxonomy classifier in the Silva database (v138). ITS gene selection was performed using the UNITE database (Release 8.0, https://unite.ut.ee/). 16S rRNA gene, select Greengenes database (Rlease 13.8, http://greengenes.2ndgenome.com/). A Venn diagram was generated to visualize the shared and unique ASVs among samples or groups by using R package “VennDiagram,” based on the occurrence of ASVs across samples regardless of their relative abundance (Zaura et al., 2009). Taxa abundances at the ASV level were statistically compared among samples or groups by using MetagenomeSeq and visualized as Manhattan plots (Zgadzaj et al., 2016). The number of taxonomic units contained in each of the seven taxonomic levels of the resultant kingdom, phylum, class, order, family, genus, and species was analyzed by clustering fungi and bacteria and using R software to produce heat maps. Heat map analysis was performed using Genescloud tools, a free online platform for data analysis. A heat map was plotted using heat map tools on the Genescloud platform (https://www.genescloud.cn). The tool was developed using the pheatmap package (V1.0.8), which was slightly modified to improve the layout style. The data were normalized to z-scores. The package uses popular clustering distances and methods implemented in the dist and hclust functions in R. unweighted pair-group method with arithmetic (UPGMA) cluster analysis uses Genescloud tools, a free online platform for data analysis, and Euclidean distances.
For the isolation of soil microorganisms around the periphery of GeB seedlings, first to third generations of the peripheral soil of GeB seedlings were passed through a 2-mm–pore size sieve. Next, 1 g of it was weighed and placed in 10 mL of sterilized water. The mixture was allowed to stand for 30 min and then thoroughly mixed. It was diluted 103-, 104-, and 105-fold, respectively; placed in a 10-mL centrifuge tube; and stored in the refrigerator at 4°C for later use.
The isolation of microorganisms of GeB seedlings was performed as follows: the surface of the first to third generations of GeB seedlings was disinfected with 75% alcohol after washing with water, the internal tissues of the GeB seedlings were cut into 1 cm × 1 cm tissue blocks with a sterilization knife, and evenly placed on Potato Dextrose Agar (PDA) plates; the culture was continued after inverting the petri dish after 4 h at 28°C.
Twenty microliters of each dilution of soil was added to a petri dish containing PDA and incubated at 28°C for 4 h. Each dish was inverted, and incubation was continued. After mycelial growth at the edge of the tissue, the edge of the colony was sampled using apical purification and added to fresh PDA. Four to five rounds of purification were conducted, and, then, a single-culture isolate was obtained.
Cut GeB seedlings were placed on PDA and incubated. The aforementioned apical purification method and procedures were used to obtain a single-culture isolate.
The cetyltrimethylammonium bromide method was used to extract DNA from the purified and cultured bacterial suspensions by using bacterial and fungi DNA extraction kits (Solarbio). The extracted DNA was purified and stored at −20°C until required. PCR amplification of the ITS region of the fungi and the 16S rRNA gene of the bacterium was performed using universal primers (Table 2). The amplified products were detected using 1% agarose gel electrophoresis, and specific bands were detected using a gel imaging system. The PCR products were stored at 4°C and sent to Jilin Comate Bioscience Co. for sequencing. The PCR reaction system comprised 12.5 μL of Taq MasterMix for PAGE, 1.0 μL each of upstream and downstream primers (10 μmol/L), 1.0μL of DNA template, and 9.5μL of deionized water. The PCR reaction conditions involved 30 cycles of pre-denaturation at 94°C for 5 min, denaturation at 94°C for 30 s, annealing at 58°C/54°C for 30 s, and extension at 72°C for 90 s, followed by a final extension at 72°C for 10 min.
The sequenced base sequences were submitted to the National Center for Biotechnology Information (NCBI) database for a BLAST homology comparison to identify the strains with the highest similarity to each bacterium and determine their biological taxonomic status.
The next aim was to prove the disease-inducing effect of three pathogenic bacteria of GeB seedlings. GeB seedlings of the same origin were rinsed with tap water to remove surface soil and other dirt and then wiped with 75% alcohol. By using the spray inoculation method, 10 μL of 103-fold diluted Trichoderma harzianum, Penicillium viridicatum, and Fusarium oxysporum suspension that contained the spores of the fungi on the surface of GeB seedlings was applied. Each treatment was repeated thrice and finally placed into a sterilized self-sealing bag and incubated at 28°C in the incubator. The same amount of sterile water was sprayed on the surface of healthy GeB seedlings alone as a control. Changes in the surfaces of the GeB seedlings were monitored every 3 days. The curative effects of the two pathogens on GeB seedlings were compared after 2 weeks.
The beneficial bacteria Pseudomonas sp. and Novosphingobium sp. were isolated and identified as described in Section 2.7. The beneficial bacteria Paenibacillus polymyxa (deposited at the China General Microbiological Culture Collection Center, CGMCC: No. 7250) and Germinating strain [Germinating strain D (JQ364945.1) and Germinating strain S (JQ364945.1)] were isolated and preserved in our laboratory and were selected and identified 2.7 isolated and identified single colonies of the GeB pathogens Trichoderma harzianum, Penicillium viridicatum, and Fusarium oxysporum. These were incubated at 28°C overnight in PDB medium; these absorbed 200 μL of spore suspension containing bacteria and fungi and were coated in PDA medium for 3~5 days; and a piece of the PDA was taken from the plate, where the bacteria and fungi grew and the mushrooms cake, and was seeded onto the side of the PDA petri dish. Forty-five PDA mediums were collected, and each PDA medium was treated with one pathogen and one probiotic for confrontation culture. Three replicates were set for each treatment, and the colony confrontation growth of two pathogenic bacteria on GeB seedlings after 1 weeks was compared.
Fungi and bacterial suspensions were prepared as described in Section 2.9. The planting location was the same as that described in Section 2.1, and the continuous cropping farmland planted with GeB in the previous year was selected.
The effect of probiotics on the planting of asexual GeB seedlings was investigated. A method for cultivating GeB in a fungi bag was adopted. Four holes were punched into both sides of each fungi bag filled with Armillariella mellea, and 2 mL of beneficial bacteria liquid was inoculated into each well. Different bags were inoculated with different beneficial bacteria. Sterile water was used as a control, and three replicates were used for each treatment. One asexually propagated GeB seedlings was placed in each well, and each bag contains eight GeB seedlings and weighed a total of 116 g. Plants were sown in May and harvested in October. The production performance (weight, quantity, and infection rate) and pharmacodynamic properties of GeB were determined. The determination of pharmacodynamic components was based on a method in Wu et al. (Wu et al., 2022). The effects of beneficial bacteria on planting sexually propagated GeB seedlings were also investigated. Except for the eight sexually propagated GeB seedlings in each bag, the total of 63.6 g differed, and the other methods were the same as those aforementioned.
As shown in Supplementary Table A1, the percentage of effective sequences for the fungi assay in samples Y, T, and W exceeded 92%, 86%, and 90%, respectively. The percentages of effective sequences for the bacterial assay exceeded 89%, 92%, and 84%, respectively. In samples A, B, and C, the proportion of effective sequences for the fungi assay exceeded 88%, 82%, and 88%, respectively, and the proportion of effective sequences for the bacterial assay exceeded 94%, 93%, and 93%, respectively. Chimeric sequences accounted for less than 10% of the sequences. The high percentage of valid sequences indicated that the sequences obtained from this high-throughput sequencing were suitable for subsequent examination of the interactions between the microbial community structure in GeB seedlings and their surrounding soil.
The sparse curves of fungi and bacteria in GeB seedlings and their surrounding soil from different generations of propagation were further analyzed (Figure 2). The curves tended to flatten when the number of fungi sequences was greater than 30,000, and the number of bacterial sequences was greater than 25,000. The findings indicate that the depth of sequencing could reflect the species information in the samples. The results of species annotation are presented in Table 3. A total of 111 phyla, 104 classes, 247 orders, 130 families, and 480 genera were annotated for fungi. A total of 46 phyla, 372 classes, 1021 orders, 3806 families, and 12,327 genera were annotated for the bacteria in the surrounding soil samples with different propagation generations. A total of 10 phyla, 15 classes, 53 orders, 25 families, and 153 genera were annotated for fungi, and 121 phyla, 444 classes, 194 orders, 1,580 families, and 2,983 genera were annotated for bacteria in GeB seedlings samples with different propagation generations.
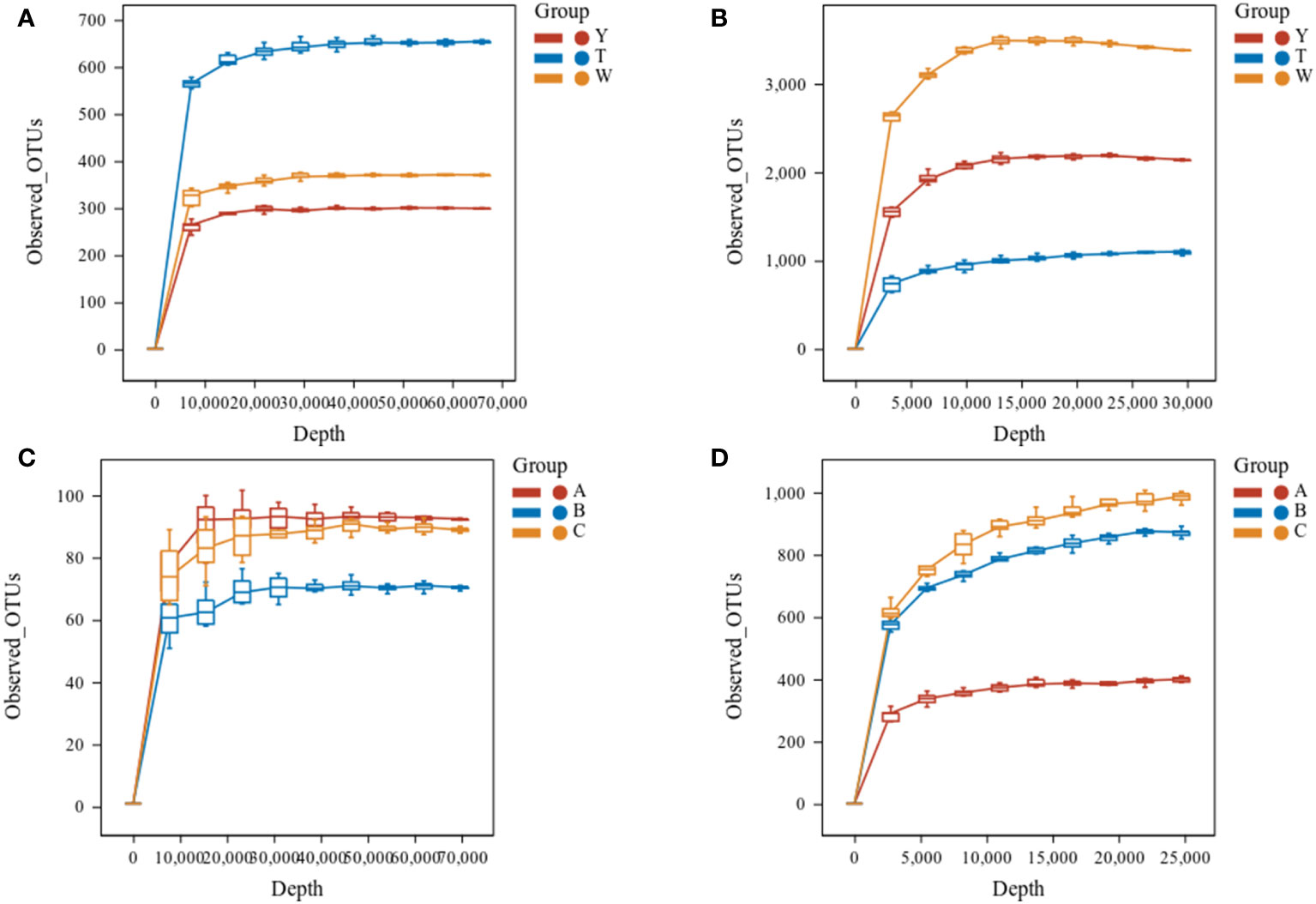
Figure 2 Sparse curves of soil alpha diversity index between the first and third generations of seedlings and their periphery. In a box-and-whisker plot, the meaning of each symbol is as follows: the upper and lower end lines of the box, the upper and lower quartiles [interquartile range (IQR)]; the median line, the median; upper and lower edges, maximum and minimum (extreme values within 1.5× IQR range). (A) Rarefaction curves of soil fungi. (B) Rarefaction curves of soil bacteria. (C) Sparse curve of tuber fungi. (D) Sparse curve of tuber bacteria.
The Venn diagram visualizes the differences and overlaps in the OTUs of fungi and bacterial communities among the samples. The more shared the OTUs, the greater the similarity between the two communities.
The number of species shared by the fungi communities in the surrounding soils of the nine groups of GeB seedlings from different propagation generations was 91. The number of endemic OTUs in each group was 431 (Y), 1,174 (T), and 536 (W). The endemic OTUs ranked as T>W>Y. Shared OTUs comprised 4.2% of the total OTUs. The number of species shared by the bacterial communities was 62. The number of endemic OTUs in each group was 4193 (Y), 1768 (T), and 7525 (W). The endemic OTUs were ranked W>Y>T. Shared bacterial OTUs accounted for 0.4% of the total OTUs (Figures 3A, B).
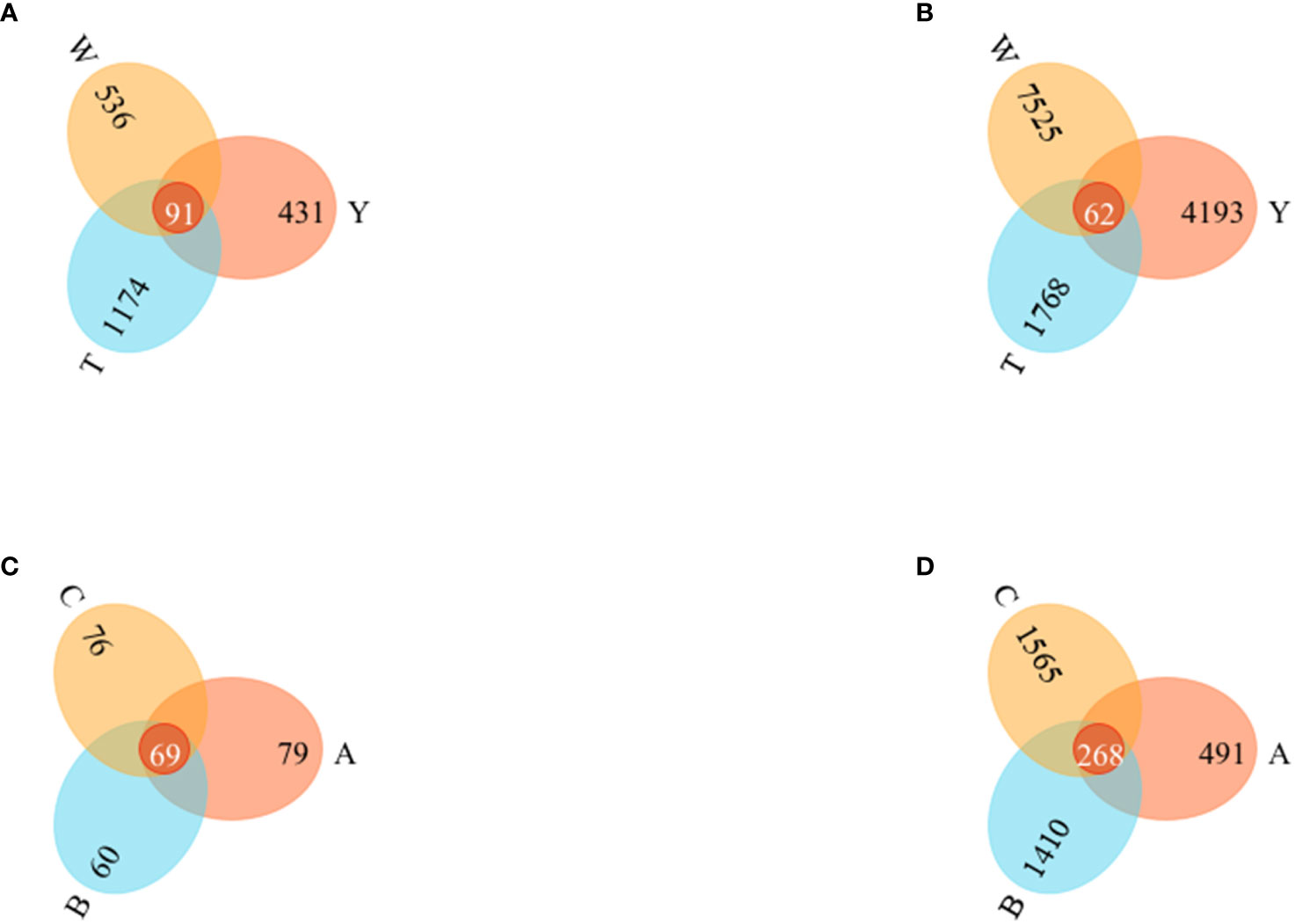
Figure 3 Operational Taxonomic Units (OTUs) Venn diagrams of seedlings from the first to third generations and their surrounding soil. (A) OTUs Venn diagram of soil fungi. (B) OTUs Venn diagram of soil bacteria. (C) OTUS Venn diagram of tuber fungi. (D) OTUs Venn diagram of tuber bacteria.
The number of species common to the fungi community in the nine groups of GeB seedlings from different reproductive generations was 69. The number of unique OTUs in each group was 79 (A), 60 (B), and 76 (C). The unique OTUs were ranked A>C>B. Shared OTUs comprised 4.2% of the total OTUs. The number of species common among the bacterial communities was 268. The number of unique OTUs in each group was 491 (A), 1,410 (B), and 1,565 (C). Unique OTUs were ranked C>B>A. The shared OTUs comprised 0.4% of the total OTUs.
The collective findings indicate that the structure of bacteria and fungi changed significantly after different generations of breeding, but the similarity of microbial and fungi communities was greater in GeB seedlings than in the other groups (Figures 3C, D).
The microbial communities in the surrounding soil of the GeB seedlings and GeB seedlings samples were analyzed for alpha diversity by using a statistical t-test. Chao1 and the Observed species indices were used to indicate richness, with higher values representing higher species richness in the microbial community. The Pielou’s evenness index (Pieloue) was used to represent community evenness, with higher values indicating higher species evenness in the microbial community. Shannon and Simpson indices were used to represent community diversity, with higher values indicating greater species diversity in the microbial community.
In the periphyton soil fungi community of GeB seedlings in sample Y, Chao1, Observed species, Pieloue, Shannon, and Simpson indices were lower than that of samples W and T, indicating that uniformity and richness were lowest, and community diversity was less in the reproduction generation. In the bacterial community of sample W, Chao1, Observed species, Pieloue, Shannon, and Simpson indices were significantly higher than that of samples Y and T (P < 0.05), indicating the highest homogeneity and abundance, richer community diversity in reproduction generation three was also observed (Tables 4A, B).
In the fungi community of GeB seedlings of sample C, Chao1, Observed species, Pieloue, Shannon, and Simpson indices were higher than that of samples A and B, indicating that the uniformity, richness, and community diversity were richest in the propagation of the three generations. In the bacterial community of sample C, Chao1, Observed species, and Shannon indices were higher than that of samples A and B, indicating that the richness, diversity, and community diversity were richest in the propagation of the three generations. However, no significant differences were observed (Tables 4C, D).
In general, the fungi and bacterial community diversity of the third generation of GeB seedlings and their surrounding soils was richer than those of the second and first generations.
At the genus level, the composition of fungi and bacterial communities varied widely different samples (GeB seedlings and their surrounding soil). The sequencing results showed that the dominant fungi genera in the soil around GeB seedlings were Pseudogymnoascus and Fusarium (Supplementary Figure A1A). The dominant bacterial genera in the soil around the GeB seedlings were Escherichia-Shigella and Rhodanobacter (Supplementary Figure A1B). The dominant fungi genera in the GeB seedlings were Alternaria and Penicillium (Supplementary Figure A1C). The dominant bacterial genera in the GeB seedlings were Vibrio and Caulobacter (Supplementary Figure A1D).
A histogram of the top 10 genus level of microorganisms in the periphyton soil of GeB seedlings and the surrounding soil was plotted. As shown in Figure 4, among the surrounding soil fungi, the genera that increased with continued generations (Figure 4A-1) were Fusarium, Humicola, Ilyonectria, Mortierella, unidentified and unclassified fungi, Chaetomium, Trichoderma, Thelonectria, and Ramophialophora. The genera that decreased with continued generation (Figure 4A-2) were Pseudocosmospora, Trichoderma, Anguillospora, Chaetosphaeria, Clonostachys, Mortierella, Mariannaea, Lecanicillium, Ascomycota, and Exophiala.
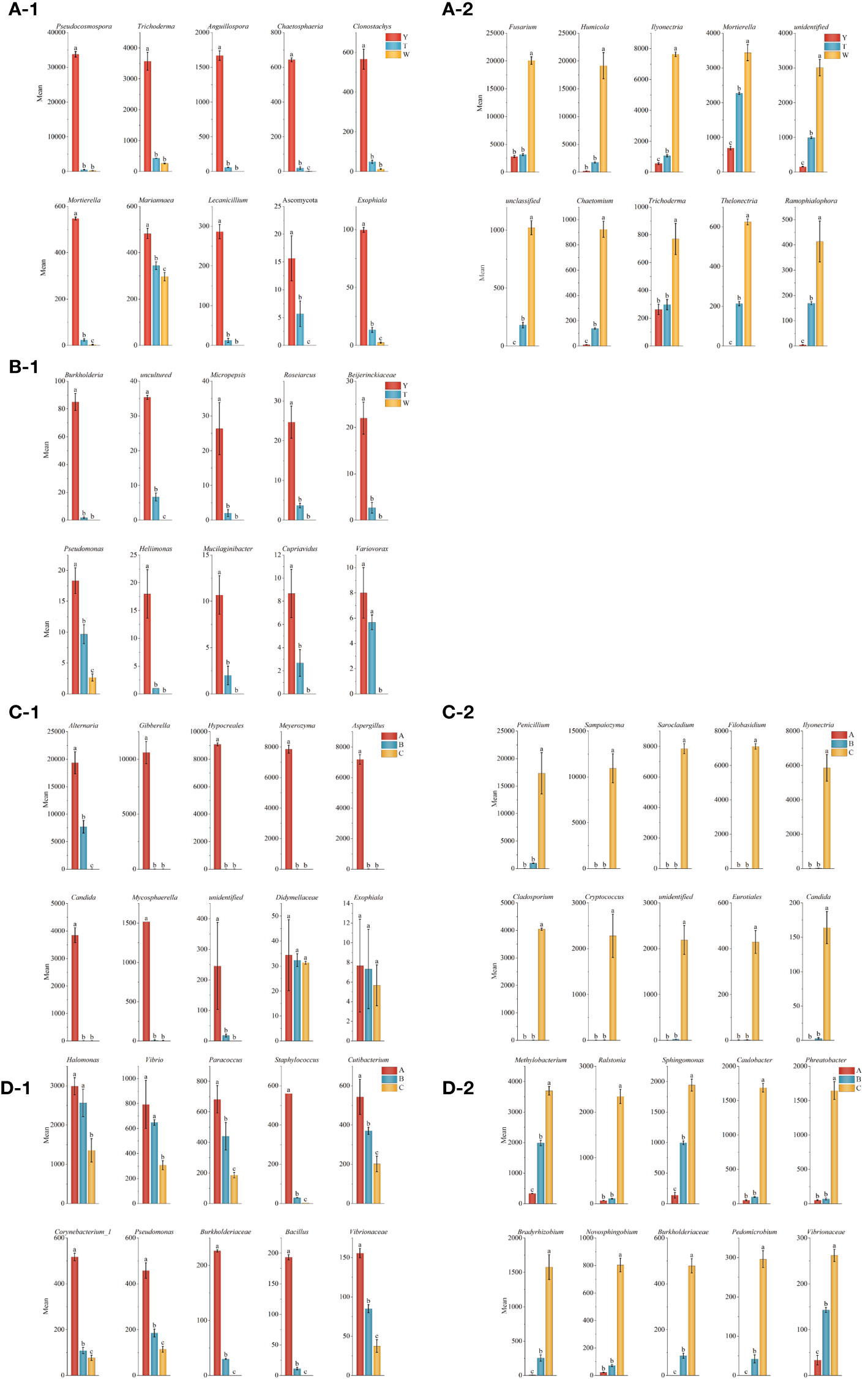
Figure 4 Analysis of dominant strains in the first to third generations of seedlings and their surrounding soil. At the genus level, the top 10 fungi and bacteria in the seedlings of seedlings with three reproductive generations had an increasing and decreasing trend. (A-1) Fungi in the soil around the seedlings decreasing with the number of generations. (A-2) Fungi in the soil around the seedlings increasing with the number of generations. (B-1) Bacteria in the soil around the seedlings decreasing with the number of generations. (C-1) Fungi in seedlings decreasing with the number of generations. (C-2) Fungi in seedlings increasing with the number of generations. (D-1) Bacteria in seedlings decreasing with the number of generations. (D-2) Bacteria in seedlings increasing with the number of generations.
Among the surrounding soil bacteria, 91 genera increased in number with continued generations (Supplementary Table A2). The genera that decreased with continued generations (Figure 4B-1) were Burkholderia, uncultured, Micropepsis, Roseiarcus, Beijerinckiaceae, Pseudomonas, Heliimonas, Mucilaginibacter, Cupriavidus, and Variovorax.
Among the GeB seedlings of fungi, genera that increased with the number of generations were Penicillium, Sampaiozyma, Sarocladium, Filobasidium, Ilyonectria, Cladosporium, Cryptococcus, unidentified Eurotiales, and Candida (Figure 4C-1). Genera that decreased with continued generations were Alternaria, Gibberella, Hypocreales, Meyerozyma, Aspergillus, Candida, Mycosphaerella, unidentified Didymellaceae, and Exophiala (Figure 4C-2).
Among the GeB seedlings of bacteria, genera that increased with continued generations were Methylobacterium, Ralstonia, Sphingomonas, Caulobacter, Phreatobacter, Bradyrhizobium, Novosphingobium, Burkholderiaceae, Pedomicrobium, and Vibrionaceae (Figure 4D-1). Genera that decreased in number with continued generations were Halomonas, Vibrio, Paracoccus, Staphylococcus, Cutibacterium, Corynebacterium_1, Pseudomonas, Burkholderiaceae, Bacillus, and Vibrionaceae (Figure 4D-2).
Because GeB seedlings undergo degradation during asexual reproduction, a reasonable inference is that pathogenic bacteria are present in genera that increase with the number of generations, and beneficial bacteria are present in genera that decrease with the number of generations. The presence of Trichoderma within the periphyton soil of asparagus was significant (P < 0.05) in all three generations of asexually propagated GeB seedlings. Notably, although Trichoderma was present in increasing and decreasing genera, there was a significant difference with the increasing genera, including T. hamatum antagonizing phytopathogenic bacteria (Poveda et al., 2023), which can cause soft rot of Gastrodia (Poveda et al., 2023), and the decreasing genera, including T. atroviride, a plant probiotic that can promote plant health and nutrition (Contreras-Cornejo et al., 2018), increase insect resistance of maize leaves, and inhibit the production of wilt disease in maize crops (Coninck et al., 2020).
In this study, Fusarium increased with the number of generations and was speculated to be a pathogenic bacterium. In addition, Fusarium can cause black rot and soft rot of GeB (Li et al., 2022), which is consistent with the literature. In GeB seedlings, Penicillium raperi increased with increasing generations, and Liu et al. (Liu et al., 2019) found that GeB disease was caused by P. oxalicum. Regarding the Novosphingobium in this study, its abundance increased with increasing algebra. Studies have shown that Novosphingobium can transfer ginsenosides (Kim et al., 2013). The topics of whether the bacteria are beneficial bacterium and antagonistic to other pathogenic bacteria require further research.
The aforementioned findings were also obtained in the immature tuber seedling perimeter soil and GeB seedlings within the common genera Exophiala, Ilyonectria, and Pseudomonas, showing a gradual decrease in Pseudomonas and Exophiala and a gradual increase in Ilyonectria.
Trichoderma, Fusarium, Ilyonectria, Penicillium, Sphingomonas, and Pseudomonas were also correlated (Table 5). With the increasing number of reproductive generations, there was a significant negative correlation between Trichoderma, Fusarium, Ilyonectria, and Penicillium (P < 0.05). Pseudomonas showed a significant positive correlation (P < 0.01). Sphingomonas was significant negatively correlated with Pseudomonas (P < 0.05).
The analysis of species occupying the top 10 differences in species composition was conducted (Figure 5). As shown in the species composition heat map, the genera with significant differences in fungi abundance at the genus level of the soil samples were Apiotrichum, Anguillospora, Pseudogymnoascus, Trichoderma, Humicola, Fusarium, Ilyonectria, Cosmospora, Mortierella, and Nectria. The genera with significant differences in bacterial abundance were Kurthia, Escherichia-Shigella, Enterococcus, Subgroup_6, Bradyrhizobium, Rhodanobacter, Mucilaginibacter, Flavobacterium, Allorhizobium-Neorhizobium-Pararhizobium-Rhizobium, and Sphingomonas.
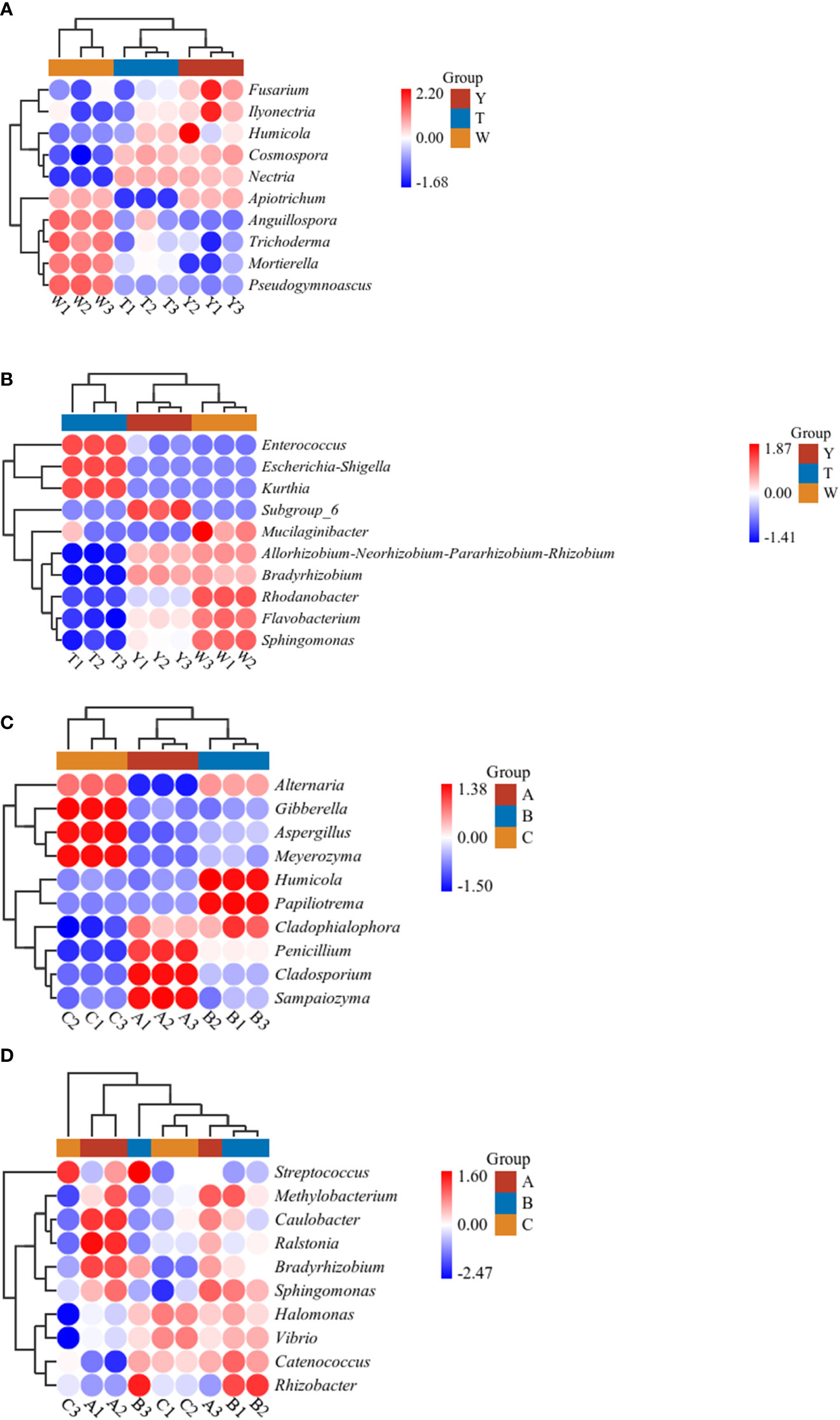
Figure 5 Heat map analysis of soil microbial communities in and around seedlings from the first to third generations. Heat maps are drawn using the heat map tool in the GenesCloud platform (https://www.genescloud.cn). Data were normalized by z-scores, and Euclidean distances were subjected to UPGMA cluster analysis. (A) Heat map analysis of soil fungi. (B) Heat map analysis of soil bacteria. (C) Heat map analysis of tuber fungi. (D) Heat map analysis of tuber bacteria.
At the genus level of the tuber samples, the genera with significant differences in fungi abundance were Gibberella, Sampaiozyma, Penicillium, Cladosporium, Humicola, Alternaria, Meyerozyma, Papiliotrema, Aspergillus, and Cladophialophora. The genera showing significant differences in bacterial abundance were Methylobacterium, Sphingomonas, Bradyrhizobium, Caulobacter, Ralstonia, Rhizobacter, Streptococcus, Catenococcus, Vibrio, and Halomonas. The heat map results showed that different generations of propagation affected the microbial diversity of GeB and the surrounding soil.
Three fungi strains and one bacterial strain were isolated and identified from the surrounding soil of the third generations of propagated GeB. The strains are shown in Figure 6A with base peak maps (Supplementary Figures A2A–D) and sequences (Supplementary Figures A3A–D). The sequence length of this strain was more than 99% homological to the registered genes in the NCBI database. More than 96% coverage of the same base (MT422092.1, MK583349.1, OP881816.1, and KM252978.1, respectively) identifies the fungi as T. harzianum (OR518627.1), P. viridicatum (OR518628.1), and F. oxysporum (OR511432.1) and the bacteria as Novosphingobium (OR529195.1).
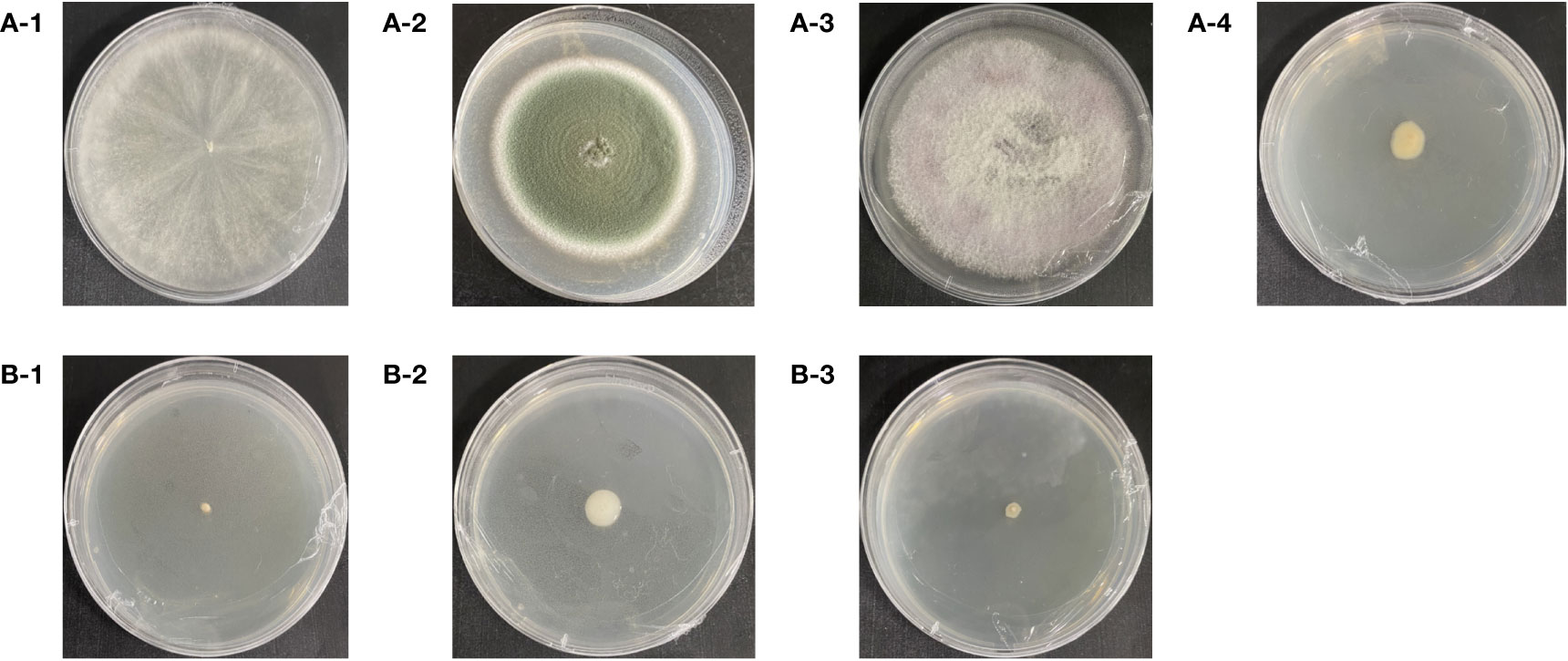
Figure 6 Isolation of soil microorganisms from the first to third generations of seedlings and their surroundings. (A-1) Trichoderma harzianum. (A-2) Penicillium viridicatum. (A-3) Fusarium oxysporum. (A-4) Novosphingobium. (B-1) Pseudomonas sp. (B-2) Agrobacterium rhizomes. (B-3) Herbaspirillum hiltneri.
Three bacteria strains were isolated and identified from the surrounding soil the first generation of propagated GeB. The resulting strains are shown in Figure 6, with the base peak maps (Supplementary Figures A2E–G) and sequences (Supplementary Figures A3E–G). The sequence length of this strain was more than 99% homological to the registered genes in the NCBI database. More than 99% coverage of the same base (AP021904.1, EU420078.1, and KU305712.1, respectively), identifies the bacteria as Pseudomonas sp. (OR501395.1), Agrobacterium rhizomes (OR501394.1), and Herbaspirillum hiltneri (OR588778.1).
The pathogenicity of three pathogenic fungi (P. viridicatum, T. harzianum, and F. oxysporum) in GeB seedlings is shown in Figure 7. When sterile water was sprayed as a control, lesions occurred on the surface of GeB seedlings within 14 days. After spraying the isolated and identified pure P. viridicatum, many brown plaques were formed in a relatively short period of time. After 14 days, a white fluffy mold appeared on the surface of the GeB seedlings, and the white hyphae in the middle of the white hyphae appeared in the later stage, and the GeB seedlings wilted. These results are consistent with the characteristics of pure P. viridicatum infection, further proving that the strain was pure P. viridicatum. After spraying the isolated and identified pure T. harzianum, a small amount of white hyphae formed on the surface of the GeB seedlings within a relatively short period. The edges of the white hyphae gradually changed from white to yellow and finally to green. After 14 days, the GeB seedlings became moist and soft, consistent with the characteristics of T. harzianum, which further proved that the strain was T. harzianum. After spraying F. oxysporum, a small number of white hyphae appeared on the surface of the GeB seedlings. After 14 days, the surface of the white mycelia turned brown with a small amount of sticky substance, the white fluffy mycelia disappeared, and the surface of the GeB seedlings was soft and turned red. These characteristics are consistent with those of F. oxysporum infection, further confirming that the strain was F. oxysporum.
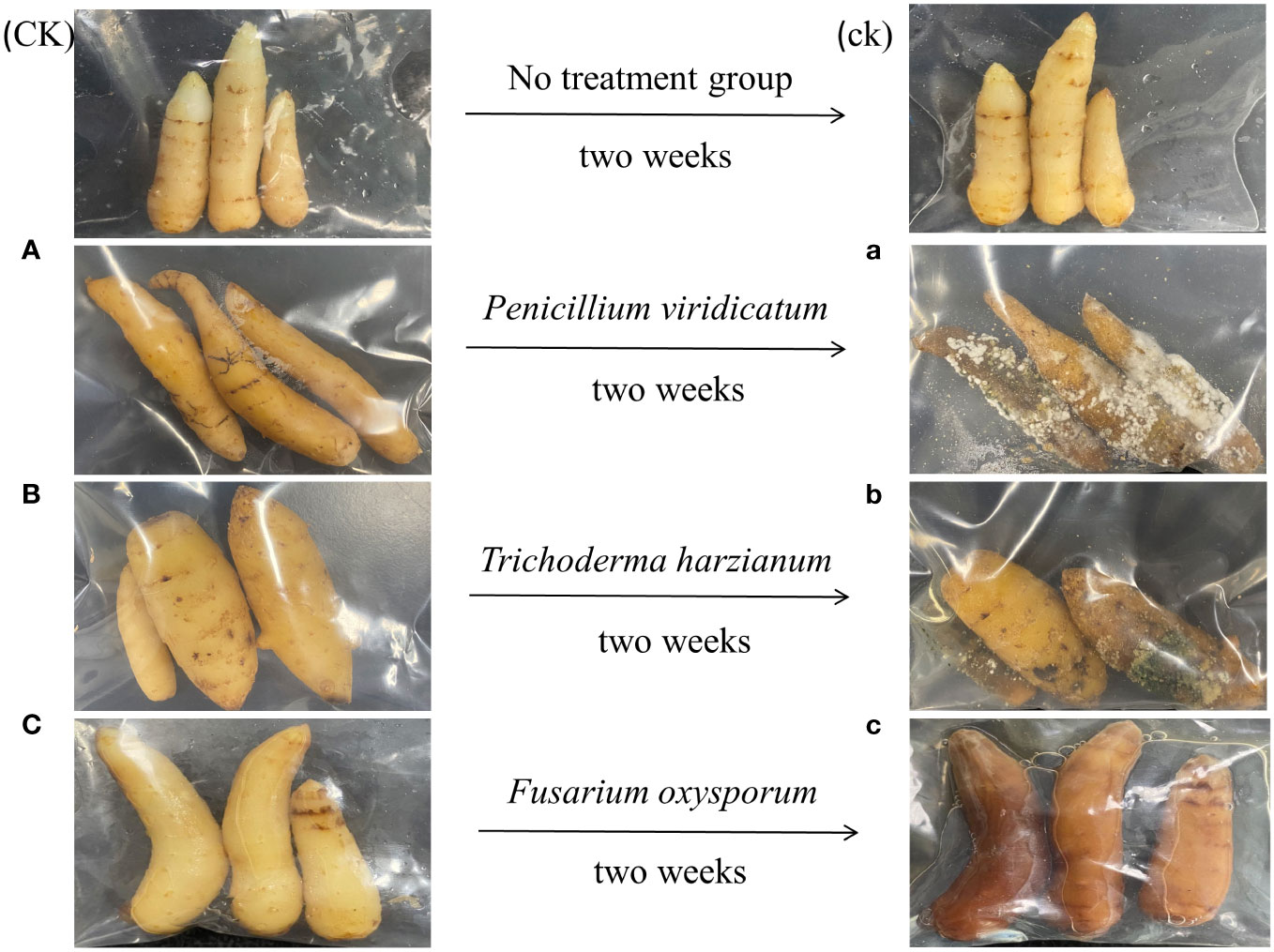
Figure 7 Pathogenic fungi back-grafting verification for isolation and identification of seedlings from the first to third generations and their surrounding soil. On the left is the tuber infected before treatment, and on the right is the tuber 2 weeks after spraying the pathogen. CK, control group sprayed with sterile water; ck, control group after spraying sterile water for 2 weeks. (A) Seedlings that have not been sprayed with Penicillium viridicatum; (a) immature tuber seedlings 2 weeks after spraying Penicillium viridicatum. (B) Seedlings that have not been sprayed with Trichoderma harzianum; (b) seedlings 2 weeks after spraying Trichoderma harzianum. (C) Seedlings that have not been sprayed with Fusarium oxysporum; (c) seedlings 2 weeks after spraying Fusarium oxysporum.
In general, compared with the control group, the GeB seedlings of GeB seedlings in the experimental group had different degrees of decay after spraying the three pathogenic bacteria, but the decay and pathogenicity of P. viridicatum were higher than those of the other two pathogens.
The results of the confrontation assays of three pathogenic fungi (P. viridicatum, T. harzianum, and F. oxysporum) and five strains of beneficial bacteria (Pseudomonas sp., Novosphingobium sp., Paenibacillus polymyxa, Germinating strain D, and Germinating strain S) are shown in Figure 8. There was an antagonistic relationship between the isolated and identified pure P. viridicatum and Germinating bacteria D (Figure 8A). However, there was no obvious antagonistic effect on Pseudomonas sp., Novosphingobium sp., and Paenibacillus polymyxa. Germinating bacteria S. T. harzianum has an antagonistic effect against Paenibacillus polymyxa and Germinating bacteria S (Figure 8B). However, there was no obvious antagonistic effect on Pseudomonas sp. and Novosphingobium sp. Germinating bacteria D. F. oxysporum has an antagonistic effect with Paenibacillus polymyxa and Germinating bacteria S (Figure 8C). However, there was no obvious antagonistic effect with Pseudomonas sp., Novosphingobium sp., and Germinating bacteria D.
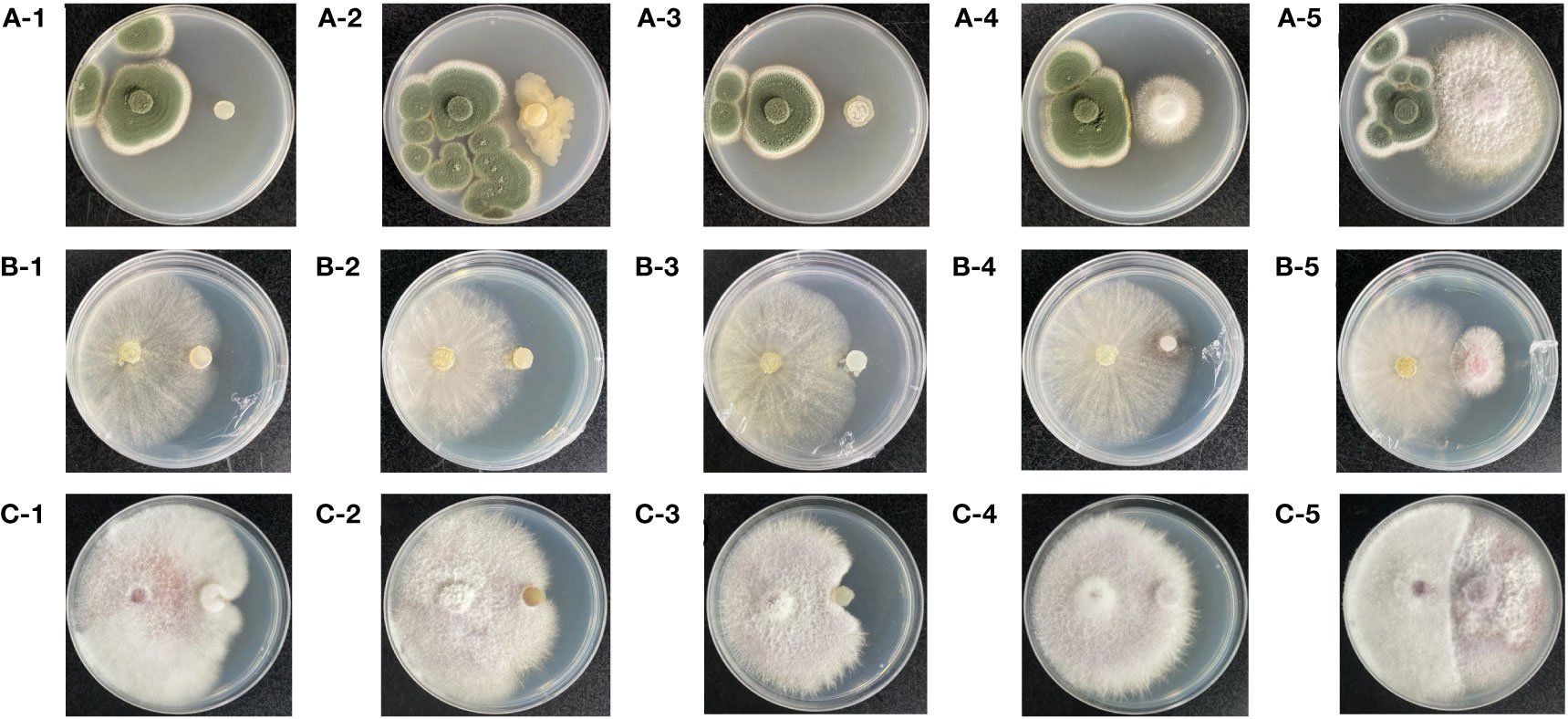
Figure 8 The first to third generations of seedlings and their surrounding soil pathogens and beneficial bacteria were cultured. Observe the growth of colonies and the presence of inhibitory boundaries after 1 week. Pathogenic fungi on the left and beneficial fungi and bacteria on the right. (A-1) Penicillium viridicatum and Pseudomonas sp. (A-2) Penicillium viridicatum and Novosphingobium. (A-3) Penicillium viridicatum and Paenibacillus polymyxa. (A-4) Penicillium viridicatum and Germinating strain D. (A-5) Penicillium viridicatum and Germinating strain S. (B-1) Trichoderma harzianum and Pseudomonas sp. (B-2) Trichoderma harzianum and Novosphingobium. (B-3) Trichoderma harzianum and Paenibacillus polymyxa. (B-4) Trichoderma harzianum and Germinating strain D. (B-5) Trichoderma harzianum and Germinating strain S. (C-1) Fusarium oxysporum and Pseudomonas sp. (C-2) Fusarium oxysporum and Novosphingobium. (C-3) Fusarium oxysporum and Paenibacillus polymyxa. (C-4) Fusarium oxysporum and Germinating strain D. (C-5) Fusarium oxysporum and Germinating strain S.
Treatment of the first generation of GeB seedlings with five probiotic fungi and bacteria with first generation of GeB seedlings had a significant effect on the agronomic traits and infection rate of GeB (P < 0.05; Supplementary Table A3A). Novosphingobium sp. and Pseudomonas sp. significantly increased the weight of GeB (P < 0.05). However, there was no significant difference in the weight of GeB between Paenibacillus polymyxa, Germinating strain D, and Germinating strain S (P > 0.05). Novosphingobium sp. and Pseudomonas sp. significantly increased the number of GeB (P < 0.05). There was no significant difference in the number of GeB between Paenibacillus polymyxa, Germinating strain D, and Germinating strain S compared with that in the control group (P > 0.05). Germinating strain S increased the GeB infection rate significantly (P < 0.05), and there was no significant difference in the infection rate between the other strains and the control group (P > 0.05). Five strains of probiotic fungi-treated and bacteria-treated first generation of GeB seedlings had significant effects on the planting of the six medicinal components of GeB (Supplementary Figure A4; Supplementary Table A4A). The order of gastrodin content from high to low was Germinating strain S > Germinating strain D > Paenibacillus polymyxa > Novosphingobium sp. > CK > Pseudomonas sp. The order of 4-Hydroxybenzyl alcohol content from high to low was Paenibacillus polymyxa > CK > Germinating strain S > Pseudomonas sp. > Germinating strain D > Novosphingobium sp. The order of Parishin E content from high to low is Germinating strain S > Germinating strain D > CK > Novosphingobium sp. > Paenibacillus polymyxa > Pseudomonas sp. The order of Parishin B content from high to low was Germinating strain S > Germinating strain D > CK > Pseudomonas sp. > Novosphingobium sp. > Paenibacillus polymyxa. The order of Parishin C content from high to low was Germinating strain S > Germinating strain D > Paenibacillus polymyxa > Pseudomonas sp. > CK > Novosphingobium sp. The order of Parishin A content from high to low was Germinating strain S > Germinating strain D > Pseudomonas sp. > Novosphingobium sp. > Paenibacillus polymyxa > CK.
Treatment of the third generation of GeB seedlings with five probiotic fungi and bacteria with the third generation of GeB seedlings had a significant effect on the agronomic traits and infection rate of GeB (P < 0.05; Supplementary Table A3B). Pseudomonas sp., Novosphingobium sp., and Germinating strain S significantly increased the weight of GeB (P < 0.05). However, there was no significant difference in the weight of GeB between Germinating strain D and Paenibacillus polymyxa compared with that of CK (P > 0.05). There was no significant difference in the number of GeB between the strains and the CK (P > 0.05). Pseudomonas sp. significantly reduced the GeB infection rate (P < 0.05), and there was no significant difference in the infection rate between other strains and the CK (P > 0.05). Five strains of probiotic fungi-treated and bacteria-treated third generation of GeB seedlings had significant effects on the planting of six medicinal components of GeB (Supplementary Figure A5; Supplementary Table A4B). The order of gastrodin content from high to low was Germinating strain S > Pseudomonas sp. > Germinating strain D > Paenibacillus polymyxa > Novosphingobium sp. > CK. The order of 4-Hydroxybenzyl alcohol content from high to low was Germinating strain D > Germinating strain S > Novosphingobium sp. > Pseudomonas sp. > CK > Paenibacillus polymyxa. The order of Parishin E content from high to low was Pseudomonas sp. > Germinating strain S > Germinating strain D > Paenibacillus polymyxa > Novosphingobium sp. > CK. The order of Parishin B content from high to low was Germinating strain S > Pseudomonas sp. > Germinating strain D > Novosphingobium sp. > Paenibacillus polymyxa > CK. The order of Parishin C content from high to low was Germinating strain S > Pseudomonas sp. > Germinating strain D > Novosphingobium sp. > Paenibacillus polymyxa > CK. The order of Parishin A content from high to low was Germinating strain S > Pseudomonas sp. > Germinating strain D > Paenibacillus polymyxa > Novosphingobium sp. > CK.
The asexual propagation of GeB seedlings was seriously degraded (Xu and Guo, 2000). However, the causes of the degradation of multi-generation asexually reproduced GeB seedlings have not yet been clarified. More than 99% of orchids are partially dependent on symbiotic mycorrhizae, and there is another type of orchid that needs to rely entirely on mycorrhizal nutrition for its entire life cycle (Suetsugu et al., 2017). GeB relies entirely on mycorrhizal nutrition heterotrophic plant of the genus Orchidaceae, which is completely dependent on Mycena and Armillariella mellea for its entire life cycle. The degradation of GeB seedlings reproduced asexually mainly manifested as severe diseases, and the degradation of Orchidaceae has not been reported.
This study is the first to investigate the causes of degradation during the multi-generational propagation of GeB from a microbial perspective. Because GeB has problems such as reduced yield and variety degradation with the increase of generations, it relies on fungi to provide nutrients during the growth of GeB. Therefore, we speculate that the changes in microorganisms are causing their degeneration. The results showed that the bacterial and fungal composition of GeB seedlings and the surrounding soil changed significantly with different propagation generations, which was consistent with our assumptions (Supplementary Figure A1). The alpha diversity analysis demonstrated that the community diversity of fungi and bacteria in the second and third generations of GeB seedlings and the surrounding soil was richer than that of the first generation. In addition, Sections 3.4 and 3.7 showed that there were more pathogenic fungi and bacteria in the third generation of GeB seedlings and the surrounding soil than that in the first and second generations. The increase in the species and number of pathogenic fungi and bacteria may have led to the generational degradation of GeB.
Trichoderma was found in the asexual reproduction GeB seedlings and the surrounding soil, which increased and decreased with the number of reproductive generations, but did not belong to the same species. The increasing strains are T. hamatum causing GeB soft rot (López López et al., 2022; Han M. et al., 2017); the reduced strains were T. atroviride, which increased the insect resistance of maize leaves and inhibited the production of wilt disease in maize crops (Coninck et al., 2020), which was speculated to be a probiotic of GeB seedlings. At the same time, it has been reported that Fusarium (Li et al., 2022), Ilyonectria (Qiao et al., 2019), and Penicillium (Liu et al., 2019) are all pathogenic bacteria of GeB. In this study, these three strains increased with the increase of generations, which verified that they were pathogenic bacteria of GeB, which was consistent with previous studies. Pseudomonas (Adhikari et al., 2001; Kuklinsky-Sobral et al., 2004; Preston et al., 2004) can effectively control phytohormones and the solubilization of minerals, and it is speculated to be a probiotic of GeB. In this study, it decreased with the increase of reproductive generations, and it was the dominant bacterium in the first generation of healthy GeB, which was verified to be a probiotic for GeB seedlings. Sphingomonas (Li et al., 2019) can symbiosis with plants and can play a role in antagonizing plant pathogenic fungi and promoting plant growth, which increases with the increase of reproductive generations (Mazoyon et al., 2023). However, further in-depth research is required to determine whether this is probiotic. In this study, it was concluded that the decrease of beneficial fungi and the increase of pathogenic fungi were the reasons for the degradation of asexual reproduction of GeB seedlings, and it was speculated that the decrease of beneficial fungi may be the reason for the inhibition of pathogenic fungi.
The medicinal plant endophytes that can reduce and prevent the harmful effects of a wide range of pathogens are (Pageni et al., 2014), for example, Cellulomonas marina (Yan et al., 2020), Amycolatopsis (Narsing Rao et al., 2020), Paracoccus endophyticus sp. (Zhang et al., 2019), Pseudomonas sp. (Li et al., 2018), and Sphingomonas mesophila sp. (Li et al., 2019). Pseudomonas and Novosphingobium were isolated in this study. In addition, three fungi strains (T. harzianum, P. viridicatum, and F. oxysporum) and two bacterial strains (A. rhizomes and H. hiltneri) were obtained. These microbes may be valuable in overcoming the degradation of multi-generational asexually propagated GeB seedlings. Amplicon sequencing showed that Penicillium was relatively abundant at the genus level in GeB seedlings. However, Penicillium viridicatum was isolated from the soil around the GeB seedlings. Therefore, the analysis may have used Penicillium sp. as a common strain of GeB seedlings and perisoil microorganisms. In addition, backlash experiments proved that Penicillium viridicatum can cause serious diseases in GeB seedlings and have speculated that Penicillium sp. is an important genus for GeB degeneration. This phenomenon proves that soil microorganisms and plant growth are inseparable (de-Bashan et al., 2020).
According to a review of the literature, there have been no report on the antagonistic bacteria and fungi of the pathogenic fungi of vegetatively propagated GeB seedlings; thus, determining the antagonistic bacteria and fungi of three multi-generation vegetatively propagated GeB seedlings would fill a gap in the literature. In this study, five beneficial bacteria were selected for pathogenic fungi antagonism.
The results showed that the GeB seedlings pathogen P. viridicatum was antagonized by Germinating strain D and P. polymyxa, Germinating strain S antagonized by T. harzianum, and P. polymyxa and Germinating strain S antagonized by F. oxysporum (Figure 8). Therefore, we speculated that supplementation with Pseudomonas sp., Novosphingobium sp., Germinating strain S, Germinating strain D, and P. polymyxa reduces the degradation of asexual GeB seedlings and increases the yield of GeB.
A possible reason for the degradation of asexual reproduction by G. elata Bl. f. glauca S. chow (GeB) on Changbai Mountain was a combined effect of increased pathogenic bacteria, such as P. viridicatum, and decreased beneficial bacteria, such as Pseudomonas. Three strains of P. viridicatum and other pathogenic bacteria were isolated and identified from third generations of asexually propagated GeB seedlings and the soil. Four strains of Pseudomonas and other beneficial bacteria were isolated and identified from first generation of asexually propagated GeB and the surrounding soil. The antagonist of the asexual reproductive degradation pathogen P.viridiatum is Emergence Bacterium D. The antagonists of the asexual reproduction degradation pathogens T. harzianum and F. oxysporum are P. polymyxa and Emergence Bacterium S. In conclusion, beneficial fungi and bacteria reduce the degradation of the asexual reproduction of GeB seedlings.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: GenBank OR511432, OR501395, OR501394, OR518628, OR518627, OR529195, OR588778.
XW: Data curation, Investigation, Methodology, Writing – original draft, Writing – review & editing. YG: Funding acquisition, Supervision, Writing – review & editing. PZ: Investigation, Writing – review & editing. GZ: Investigation, Writing – review & editing. XY: Investigation, Writing – review & editing. QL: Investigation, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study was also supported by the Science and Technology Development Program of Jilin Province (Grant No. 20210401093YY, 20220401110YY), and this study was supported by the National Key Research and Development Programme (Grant No. 2022YFF1300503-04).
We would like to thank the reviewers for their insightful comments and advice on this manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2024.1334958/full#supplementary-material
Adhikari, T. B., Joseph, C. M., Yang, G., Phillips, D. A., Nelson, L. M. (2001). Evaluation of bacteria isolated from rice for plant growth promotion and biological control of seedling disease of rice. Canadian J Microbiol (10), 916–24.
Baasanmunkh, S., Oyuntsetseg, B., Efimov, P., Tsegmed, Z., Vandandorj, S., Oyundelger, K., et al. (2021). Orchids of Mongolia: taxonomy, species richness and conservation status. Diversity (7), 302–302. doi: 10.3390/D13070302
Bae, E.-K., An, C., Kang, M.-J., Lee, S.-A., Lee, S. J., Kim, K.-T., et al. (2022). Chromosome-level genome assembly of the fully mycoheterotrophic orchid Gastrodia elata. G3 Genes|Genomes|Genetics (3). doi: 10.1093/G3JOURNAL/JKAB433
Barnes, E. C., Jumpathong, J., Lumyong, S., Hertweck, C. (2016). Daldionin, an unprecedented binaphthyl derivative, and diverse polyketide congeners from a fungi orchid endophyte. Chem. (Weinheim an der Bergstrasse Germany) (13), 4551-4555. doi: 10.1002/chem.201504005
Chen, Y., Gao, Y., Song, L., Xing, X. (2019). Mycorrhizal fungi community composition in seven orchid species inhabiting Song Mountain, Beijing, China. Science China. Life Sci 62.6 (2019), 838-847. doi: 10.1007/s11427-018-9471-x
Clemmensen, K. E., Ihrmark, K., Durling, M. B., Lindahl, B D. (2023). Sample preparation for fungi community analysis by high-throughput sequencing of barcode amplicons. Methods Mol. Biol. (Clifton N.J.), 37-64. doi: 10.1007/978-1-0716-2871-3_3
Coninck, E., Scauflaire, J., Gollier, M., Legrève, A. (2020). Trichoderma atroviride as a promising biocontrol agent in seed coating for reducing Fusarium damping-off on maize. J. Appl. Microbiol (3), 637-65. doi: 10.1111/jam.14641
Contreras-Cornejo, H. A., Macías-Rodríguez, L., del-Val, E., Larsen, J. (2018). The root endophytic fungus Trichoderma atroviride induces foliar herbivory resistance in maize plants. Appl. Soil Ecol, 45-53. doi: 10.1016/j.apsoil.2017.10.004
de-Bashan, L. E., Nannipieri, P., Antoun, H., Lindermann, R. G. (2020). Application of beneficial microorganisms and their effects on soil, plants, and the environment: the scientific legacy of Professor Yoav Bashan. Biol. Fertil. Soils 4), 1–4. doi: 10.1007/s00374-020-01466-9
de Meeûs, T., Prugnolle, F., Agnew, P. (2007). Asexual reproduction: genetics and evolutionary aspects. Cellular and molecular life sciences. Cell Mol. Life Sci. 64 (11),1355-72. doi: 10.1007/s00018-007-6515-2
Gong, X., Cheng, J., Zhang, K., Wang, Y., Li, S., Luo, Y. (2021). Transcriptome sequencing reveals Gastrodia elata Blume could increase the cell viability of eNPCs under hypoxic condition by improving DNA damage repair ability. J. ethnopharmacol, 114646-114646. doi: 10.1016/J.JEP.2021.114646
Guan, Z., Lin, D., Chen, D., Guo, Y., Lu, Y., Han, Q., et al. (2022). Soil microbial communities response to different fertilization regimes in young Catalpa bungei plantation. Front. Microbiol 13, 948875-948875. doi: 10.3389/FMICB.2022.948875
Han, M., Choi, M. N., Lee, H. R., Park, E. J.. (2017). First Report of Soft Rot Associated With Trichoderma hamatum in Gastrodia elata. Plant Disease 6, 1048-1048. doi: 10.1094/pdis-11-16-1630-pdn
Han, X.-J., Cheng, M.-E., Yuan, Y., Peng, H.-S. (2020). Evolution of commercial specification and experimental identification terms of Gastrodiae Rhizoma. Zhongguo Zhong yao za zhi = Zhongguo zhongyao zazhi = China J. Chin. materia Med (11), 2702-2707. doi: 10.19540/j.cnki.cjcmm.20200329.102
Hong, S. M., An, S. M., Jeong, B. N., Kang, M. H. (2018). Comparative Studies of the in vitro Antioxidant Properties of 80% Ethanol or Water Extracts of Formica rufa L. Gastrodia elata Blume, Cnidium officinale Makino and Their’s Mixture. Korean J. Food Cookery Sci (1), 27-33. doi: 10.9724/kfcs.2018.34.1.27
Hong, I. P., Kim, H. K., Park, J. S., Kim, G. P., Lee, M. W., Guo, S. X. (2002). Physiological characteristics of symbiotic fungi associated with the seed germination of gastrodia elata. Mycobiology (1), 22-26. doi: 10.4489/MYCO.2002.30.1.022
Jiang, Y., Hu, X., Yuan, Y., Guo, X., Chase, M. W., Song, G., et al. (2022). The Gastrodia menghaiensis (Orchidaceae) genome provides new insights of orchid mycorrhizal interactions. BMC Plant Biol (1), 179-179. doi: 10.1186/S12870-022-03573-1
Kim, J.-K., He, D., Liu, Q.-M., Im, W.-T. (2013). Novosphingobium ginsenosidimutans sp. nov., with the ability to convert ginsenoside. J. Microbiol. Biotechnol (4), 444-50. doi: 10.4014/jmb.1212.12053
Kuklinsky-Sobral, J., Araújo, W. L., Mendes, R., Geraldi, I. O., Pizzirani-Kleiner, A. A., Azevedo, J. L. (2004). Isolation and characterization of soybean-associated bacteria and their potential for plant growth promotion. Environmental microbiology (12), 1244-51. doi: 10.1111/J.1462-2920.2004.00658.X.
Li, Y. Y., Guo, S. X., Lee, Y. I. (2020b). Ultrastructural changes during the symbiotic seed germination of Gastrodia elata with fungi, with emphasis on the fungi colonization region. Bot. Stud (1), 4-4. doi: 10.1186/S40529-019-0280-Z
Li, Y.-Q., Narsing, R. M. P., Zhang, H., Wen-Jun, L. (2019). Description of Sphingomonas mesophila sp. nov., isolated from Gastrodia elata Blume. Int. J. system. evolution. Microbiol (4), 1030-1034. doi: 10.1099/ijsem.0.003263
Li, C., Zhang, M., Li, J., Huang, M., Shao, X., Yang, Z. (2022). Fusarium redolens causes black rot disease in Gastrodia elata grown in China. Crop Prot, 155. doi: 10.1016/J.CROPRO.2022.105933
Li, X., Zou, X., Zhang, K., Shi, D. (2018). Draft Genome Sequence of Pseudomonas sp. LS-2, Isolated from a Diseased Gastrodia elata Plant. Microbiol. res. announce (20). doi: 10.1128/MRA.01342-18
Li, Y. Q., Zhang, H., Xiao, M., Dong, Z. Y., Zhang, J. Y., Narsing Rao, M. P., et al. (2020). Cellulomonas endophytica sp. nov., isolated from Gastrodia elata Blume. International Journal of Systematic and Evolutionary Microbiology (5), 3091-3095. doi: 10.1099/ijsem.0.004133
Liu, Y., Gao, J., Peng, M., Si, G. (2018). A review on central nervous system effects of gastrodin. Front. Pharmacol, 20. doi: 10.3389/fphar.2018.00024
Liu, T., Hua, Z., Han, P., Zhao, Y., Zhou, J., Jin, Y., et al. (2022). Mycorrhizosphere Bacteria, Rahnella sp. HPDA25, Promotes the Growth of Armillaria gallica and Its Parasitic Host Gastrodia elata. Front. Microbiol 13, 842893-842893. doi: 10.3389/FMICB.2022.842893
Liu, H., Wu, M., Liu, J., Gao, Y., Ren, A. (2020). Endophyte infection influences arbuscular mycorrhizal fungi communities in rhizosphere soils of host as opposed to non-host grass. J. Soil Sci (2), 995-1009. doi: 10.1111/ejss.12996
Liu, J. T., Zang, P., Gao, Y. G., Zhao, Y., He, Z. M., Zhu, H. Y., et al. (2019). 1st Report of Penicillium oxalicum Causing Blue Mold on Gastrodia elata in Jilin Province, China. Plant Dis (9), 2469-2469. doi: 10.1094/pdis-10-18-1893-pdn
López López, M. E., Del Toro Sánchez Carmen, L., Gutiérrez Lomelí, M., Ochoa Ascencio, S., Aguilar López, J. A., Robles García, M. A., et al. (2022). Isolation and Characterization of Trichoderma spp. for Antagonistic Activity against Avocado (Persea americana Mill) Fruit Pathogens. Horticulturae (8), 714-714. doi: 10.3390/HORTICULTURAE8080714
Mazoyon, C, Firmin, S, Bensaddek, L, Pecourt, A, Chabot, A, Faucon, MP, et al. (2023). Optimizing Crop Production with Bacterial Inputs: Insights into Chemical Dialogue between Sphingomonas sediminicola and Pisum sativum. Microorganisms (7). doi: 10.3390/MICROORGANISMS11071847
Naranjo, H. D., Rat, A., De Zutter, N., De Ridder, E., Lebbe, L., Audenaert, K., et al. (2023). Uncovering genomic features and biosynthetic gene clusters in endophytic bacteria from roots of the medicinal plant alkanna tinctoria tausch as a strategy to identify novel biocontrol bacteria. Microbiol. Spectr (4), e0074723-e0074723. doi: 10.1128/SPECTRUM.00747-23
Huang, M., Gao, D., Lin, L., Wang, S., Xing, S. (2022). Spatiotemporal dynamics and functional characteristics of the composition of the main fungal taxa in the root microhabitat of Calanthe sieboldii (Orchidaceae). BMC Plant Biology (1). doi: 10.1186/S12870-022-03940-Y
Pageni, B. B., Lupwayi, N. Z., Akter, Z., Gan, Y. (2014). Plant growth-promoting and phytopathogen-antagonistic properties of bacterial endophytes from potato (Solanum tuberosum L.) cropping systems. Can. J. Plant Sci (5), 835-844. doi: 10.4141/cjps2013-356
Park, E.-J., Lee, W. Y., Ahn, J. K. (2012). In vitro propagation of myco-heterotrophic Gastrodia elata. Horticulture. Environ. Biotechnol (5), 415-420. doi: 10.1007/s13580-012-0046-y
Poveda, J., Rodríguez, V. M., Abilleira, R., Velasco, P. (2023). Trichoderma hamatum can act as an inter-plant communicator of foliar pathogen infections by colonizing the roots of nearby plants: A new inter-plant “wired communication”. Plant Sci, 111664-11166. doi: 10.1016/J.PLANTSCI.2023.111664
Qiao, M., Tian, W. G., Feng, B., Yu, Z. F., Peng, Z. X. (2019). First Report of Soft Rot Associated with Ilyonectria robusta in Gastrodia elata. Plant Disease (10), 2691-2691. doi: 10.1094/pdis-03-19-0578-pdn
Preston Gail, M. (2004). Plant perceptions of plant growth-promoting Pseudomonas..Philosophical transactions of the Royal Society of London. Series B, Biological sciences (1446), 907-18. doi: 10.1094/pdis-03-19-0578-pdn
Shan, T., Yin, M., Wu, J., Yu, H., Liu, M., Xu, R., et al. (2021). Comparative transcriptome analysis of tubers, stems, and flowers of Gastrodia elata Blume reveals potential genes involved in the biosynthesis of phenolics. Fitoterapia(prepublish), 104988-. doi: 10.1016/J.FITOTE.2021.104988
Su, Z., Yang, Y., Chen, S., Tang, Z., Xu, H. (2023). The processing methods, phytochemistry and pharmacology of Gastrodia elata Bl.: A comprehensive review. J. ethnopharmacol, 116467-116467. doi: 10.1016/J.JEP.2023.116467
Suetsugu, K., Yamato, M., Miura, C., Yamaguchi, K., Takahashi, K., Ida, Y., et al. (2017). Comparison of green and albino individuals of the partially mycoheterotrophic orchid Epipactis helleborine on molecular identities of mycorrhizal fungi, nutritional modes and gene expression in mycorrhizal roots. Mol. Ecol. 6), 1652–1669. doi: 10.1111/mec.14021
Wang, J. (2023). The life of Xuan Zhou, founding father of the Gastrodia elata cultivation and industry in China. Plant Diversity (3), 241-242. doi: 10.1016/J.PLD.2023.03.010
Wang, Z. (2023). High-throughput sequencing technology and its application in epigenetics studies. SHS Web Conferences, 158. doi: 10.1051/SHSCONF/202315801005
Wu, Z., Gao, R., Hong, L., Liao, X., Tang, X., Wang, X., et al. (2022). How steaming and drying processes affect the active compounds and antioxidant types of Gastrodia elata Bl. f. glauca S. chow. Food Res. Int. 157 (Undefined), 111277. doi: 10.1016/j.foodres.2022.111277
Xu, J., Guo, S. (2000). Retrospect on the research of the cultivation of Gastrodia elata Bl, a rare traditional Chinese medicine. Chin. Med. J (8), 686-92.
Yu, E, Liu, Q, Gao, Y, Li, Y, Zang, P, Zhao, Y, et al (2023). An exploration of mechanism of high quality and yield of Gastrodia elata Bl. f. glauca by the isolation, identification, and evaluation of Mycena. Frontiers in Microbiology 13, 1220670-1220670. doi: 10.3389/FMICB.2023.1220670
Zaura, E., Keijser, B. J., Huse, S. M., Crielaard, W. (2009). Defining the healthy “core microbiome” of oral microbial communities. BMC Microbiol. 9, 259. doi: 10.1186/1471-2180-9-259
Zgadzaj, R., Garrido-Oter, R., Jensen, D. B., Koprivova, A., Schulze-Lefert, P., Radutoiu, S. (2016). Root nodule symbiosis in Lotus japonicus drives the establishment of distinctive rhizosphere, root, and nodule bacterial communities. Proc. Natl. Acad. Sci. U S A. 113 (49), E7996–E8005. doi: 10.1073/pnas.1616564113
Zhang, J., Cook, J., Nearing, J. T., Zhang, J., Raudonis, R., Glick, B. R., et al. (2021). Harnessing the plant microbiome to promote the growth of agricultural crops. Microbiol. Res. (prepublish), 126690-. doi: 10.1016/J.MICRES.2020.126690
Keywords: Gastrodia elata Bl. f. glauca S. Chow, Changbai Mountain, asexual reproduction degradation, microbial diversity, isolation and characterization, antagonistic strains
Citation: Wang X, Gao Y, Zang P, Zhang G, Yang X and Liu Q (2024) Decrease in beneficial bacteria and increase in harmful bacteria in Gastrodia seedlings and their surrounding soil are mainly responsible for degradation of Gastrodia asexual propagation. Front. Plant Sci. 15:1334958. doi: 10.3389/fpls.2024.1334958
Received: 08 November 2023; Accepted: 10 January 2024;
Published: 06 February 2024.
Edited by:
Domingo Martinez-Soto, Center for Scientific Research and Higher Education in Ensenada (CICESE), MexicoReviewed by:
Luis Fernando Garcia-Ortega, National Polytechnic Institute of Mexico (CINVESTAV), MexicoCopyright © 2024 Wang, Gao, Zang, Zhang, Yang and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yugang Gao, amxueWR4Z3lnQDE2My5jb20=; Qun Liu, bGl1cXVuMTAyNTI2NTIwOEBzaW5hLmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.