
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 12 February 2024
Sec. Plant Pathogen Interactions
Volume 15 - 2024 | https://doi.org/10.3389/fpls.2024.1334189
This article is part of the Research TopicInsights In Plant-Pathogen Interactions: 2023View all 19 articles
 Wenqing Yan1,2
Wenqing Yan1,2 Yunxia Ni1,2
Yunxia Ni1,2 Hui Zhao1,2
Hui Zhao1,2 Xintao Liu2
Xintao Liu2 Min Jia2
Min Jia2 Xinbei Zhao2
Xinbei Zhao2 Yongdong Li2
Yongdong Li2 Hongmei Miao1,3*
Hongmei Miao1,3* Hongyan Liu1,2,3*
Hongyan Liu1,2,3* Haiyang Zhang1,3*
Haiyang Zhang1,3*Leucine-rich repeat receptor-like kinases (LRR-RLKs) can participate in the regulation of plant growth and development, immunity and signal transduction. Sesamum indicum, one of the most important oil crops, has a significant role in promoting human health. In this study, 175 SiLRR-RLK genes were identified in S. indicum, and they were subdivided into 12 subfamilies by phylogenetic analysis. Gene duplication analysis showed that the expansion of the SiLRR-RLK family members in the sesame was mainly due to segmental duplication. Moreover, the gene expansion of subfamilies IV and III contributed to the perception of stimuli under M. phaseolina stress in the sesame. The collinearity analysis with other plant species revealed that the duplication of SiLRR-RLK genes occurred after the differentiation of dicotyledons and monocotyledons. The expression profile analysis and functional annotation of SiLRR-RLK genes indicated that they play a vital role in biotic stress. Furthermore, the protein−protein interaction and coexpression networks suggested that SiLRR-RLKs contributed to sesame resistance to Macrophomina phaseolina by acting alone or as a polymer with other SiLRR-RLKs. In conclusion, the comprehensive analysis of the SiLRR-RLK gene family provided a framework for further functional studies on SiLRR-RLK genes.
Receptor-like protein kinases (RLKs) represent numerous transmembrane kinases that sense stimulation at the cell surface and mediate cell signal transduction by phosphorylation in response to the environment (Trenker and Jura, 2020). Many duplication events of RLKs exist in terrestrial plants (Lehti-Shiu et al., 2009), in which RLKs involved in the stress response show duplications, while those involved in growth and development do not (Shiu et al., 2004), suggesting that duplication events of RLKs are important for terrestrial plants to respond to ever-changing environments (Lehti-Shiu et al., 2012). LRR-RLKs represent the largest family in RLKs, which consist of three protein domains: an LRR domain sensing signal outside the cell, a single-channel transmembrane domain anchoring proteins within the membrane, and a kinase domain involved in signal transduction by autophosphorylation and subsequent specific substrate phosphorylation (Liu et al., 2017).
LRR-RLKs can widely regulate plant development and stress responses by participating in brassinosteroid (BR) and abscisic acid (ABA) signaling pathways. BRI1 (BR insensitive 1), a key LRR-RLK in the BR pathway, could regulate stem elongation, vascular differentiation, seed size, fertility, flowering time and senescence by BR signaling in Arabidopsis by forming the BRI1/BAK1 (BRI1-associated receptor kinase 1) complex (Li et al., 2002; Nam and Li, 2002; Wang et al., 2005). In addition, barley bri1 mutant have multiple effects on disease resistance and plant developmental regulation (Goddard et al., 2014). SERK2 (Somatic embryogenesis receptor kinase 2), another component of the BR pathway, can mediate salt tolerance in rice via BR signaling (Dong et al., 2020). Moreover, OsSERK2 confers rice immunity to Xanthomonas oryzae pv. oryzae by activating the resistance genes XA21 and XA3 (Chen et al., 2014). BAK1 plays an important role in ABA signaling in guard cells. The bak1 mutants exhibited more water loss than the wild type and showed ABA insensitivity in stomatal closure. Additionally, ABA can facilitate the formation of the BAK1/OST1 (Open stomatal 1) complex that mediates ABA-induced stomatal closure in guard cells near the plasma membrane (Shang et al., 2016). Likewise, KIN7 (Kinase 7) is essential in ABA signaling in stomatal closure. Phosphorylation and activation of TPK1 (Tonoplast K+ channel) by the KIN7 is indispensable for ABA- and CO2-mediated stomatal closure (Isner et al., 2018). In addition, LRR-RLKs have been shown to be involved in plant immunity via other phytohormone pathways. PSKR1 (Phytosulfokine receptor 1), an antagonistic regulator between biotrophic and necrotrophic pathogens in plant defense, can mediate plant resistance to pathogens by suppressing salicylic acid-dependent defense while enhancing jasmonic acid-dependent defense (Mosher et al., 2013). However, OsPSKR1 is involved in rice resistance to Pseudomonas syringae DC3000 in rice by activating the expression of PR genes involved in the salicylic acid signaling pathway (Yang et al., 2019). Furthermore, PEPR1 (Pep1 receptor 1) and PEPR2 are involved in plant immunity due to ROS (Reactive oxygen species) production and ethylene signaling (Ma et al., 2016).
The interaction and regulation between LRR-RLK members is intricate during development and stress. For instance, the CLV1 (CLAVATA 1)-CLV2-CRN (CORYNE) trimer is essential in plant stem cell regulation (Bleckmann et al., 2010; Zhu et al., 2010), and the BAK1-TMM (TOO MANY MOUTHS) complex is involved in plant immunity (Jordá et al., 2016). The formation of some LRR-RLK complexes depends on ligand stimulation, for instance, flg22 and elf18 can stimulate FLS2 (Flagellin sensitive 2) and EFR (Elongation factor-Tu receptor) to form dimers with BAK1 and then plant defense is initiated (Roux et al., 2011). Similarly, ligands SCFE1 (Sclerotium culture filtrate ELICITOR1) or NLP20 (Peptide motif) stimulate the formation of the BAK1-SOBIR1 (Suppressor of BIR1-1)-RLP23 complex (Gao et al., 2009), which plays an important role in plant resistance to pathogens. Moreover, there have been fewer studies on the interactions of LRR-RLKs in other plants, with only a few having been confirmed in tomato (Peng and Kaloshian, 2014), tobacco (Franco-Orozco et al., 2017), rice (Chen et al., 2014), Medicago truncatula (Crook et al., 2016) and wheat (Singh et al., 2016).
The large number, the great diversity of structure and function and the intricate interaction networks of LRR-RLKs present a challenge in understanding the functions and mechanisms of LRR-RLK genes in complex signal transduction pathways in plants. Furthermore, the complementary functions between LRR-RLKs indicate the importance of systematic analysis using bioinformatics tools to understand the roles of LRR-RLKs in plants. Recently, the LRR-RLK gene family has been reported in Arabidopsis (Shiu and Bleecker, 2001), soybean (Zhou et al., 2016), wheat (Shumayla et al., 2016), cotton (Sun et al., 2018), rice (Sun and Wang, 2011) and maize (Song et al., 2015). Additionally, the potential roles of LRR-RLKs in response to stresses have been well-studied in Thinopyrum elongatum (Mishra et al., 2021). Sesame charcoal rot caused by M. phaseolina is one of the most serious fungal diseases in sesame production, and threatens the yield and quality of sesame. Although LRR-RLKs are crucial in plant immunity, there is still a lack of systematic studies of the LRR-RLKs in sesame. It is of great practical significance to study LRR-RLK gene family in sesame and their functions related to biotic stresses. In this study, the LRR-RLK gene family in sesame was comprehensively analyzed by phylogeny, structural evolution and expression profile analysis. The potential functions of the sesame LRR-RLK homologous to Arabidopsis were predicted by protein−protein interaction (PPI) and coexpression networks. Our studies tend to gain insight into the functions of the sesame LRR-RLK family and provide new insights into their roles in regulation under M. phaseolina stress at the transcriptome level.
Sesame genome and proteome sequences were provided by Henan Sesame Research Center, Henan Academy of Agricultural Sciences (Zhang et al., 2013; Miao et al., 2023). HMM (Hidden Markov Model) profiles of LRRs (PF00560, PF07723, PF07725, PF12799, PF13306, PF13516, PF13855, PF14580 and PF01816) and Pkinase/Pkinase_Tyr (PF00069 and PF07714) were used for identification of putative LRR-RLKs in S. indicum through HMMER 3.1 (Finn et al., 2011) (E-value < 1 × 10−10). LRR-RLKs identified from Arabidopsis (Shiu and Bleecker, 2001), soybean (Zhou et al., 2016), wheat (Shumayla et al., 2016), cotton (Sun et al., 2018), rice (Sun and Wang, 2011) and maize (Song et al., 2015) were used to run BLASTP with the sesame proteome (E-value < 1 × 10−5). The sum putative LRR-RLKs of the HMMER search result and BLASTP result were used for subsequent analysis. The InterPro database (https://www.ebi.ac.uk/interpro/) was used to confirm the presence of the kinase domain, LRR domain and transmembrane domain in LRR-RLKs in sesame. Sequences that met the above conditions were regarded as LRR-RLKs.
The theoretical isoelectric points (pI) and molecular weights (MW) of SiLRR-RLKs were predicted with Expasy (https://www.expasy.org/). CELLO (http://cello.life.nctu.edu.tw/) was used to predict the subcellular localization while SignalP-5.0 (https://services.healthtech.dtu.dk/service.php?SignalP-5.0) was used for signal peptide prediction. A conserved domain analysis of the sesame SiLRR-RLK family members was performed using the InterPro database. The MEME online server (http://meme-suite.org/) was used to search for conserved motifs. The conserved domains and gene structure of SiLRR-RLKs were visualized by TBtools (Chen et al., 2020). GO annotation was performed on PANNZER 2 (http://ekhidna2.biocenter.helsinki.fi/sanspanz/).
A multiple sequence alignment was performed using ClustalW with the default parameters method based on the aa sequences of the SiLRR-RLK proteins. MEGA 7 software (Kumar et al., 2016) was used to construct a phylogenetic tree of LRR-RLK using the neighbor Joining (NJ) method, and the bootstrap value was set to 1,000. Then the phylogenetic tree was visualized and edited on the iTOL website (https://itol.embl.de/).
The sesame genome file (In fna format) and the genome annotation file (In gff3 format) were used to visualize the chromosome localization with TBtools (Chen et al., 2020). The MCScanX (Wang et al., 2012) program was used to determine collinear orthologous gene duplications (Tandem and segmental duplications) among the sesame LRR-RLK gene family and syntenic LRR-RLK genes between sesame and other plant species. The genome files and annotation files of Solanum tuberosum, Glycine max, Solanum lycopersicum, M. truncatula, A. thaliana, Vitis vinifera, Gossypium hirsutum, Hordeum vulgare, Zea mays, Triticum aestivum, Oryza sativa, Musa acuminata, Setaria italica and Sorghum bicolor were downloaded from the Phytozome database (Goodstein et al., 2012).
RNA-seq data PRJNA892254 was used for in silico expression analysis of diverse sesame tissues. Flower tissues of variety S. indicum var. ‘Zhengzhi No.13’ that showed consistent growth were sampled, and the locations were marked. The capsules at the markers were sampled along with all other tissues (Roots, stems, leaves, capsules and seeds) two weeks later. For in silico expression analysis of sesame seed development, RNA-seq data of variety S. indicum var. ‘Wanzhi No.2’ during seed development comprising 7 days after flowering (7 DAF, S1), 14 DAF (S2), 21 DAF (S3) and full maturity (28 DAF, S4) were used (PRJNA739094) (Zhang et al., 2021). For in silico expression analysis, the RNA-seq data of the disease-resistant variety S. indicum var. ‘Zhengzhi No.13’ infected with M. phaseolina and root tissues were concomitantly collected at 0 h, 12 h, 24 h, 36 h and 48 h post inoculation (PRJNA706471). The data above were downloaded from the SRA database. The reads were filtered, and trimmed using fastp (Chen et al., 2018), then clean reads were mapped to the sesame genome with HISAT2 (version:2.0.4) (Pertea et al., 2015; Pertea et al., 2016). Finally, the FPKM value of each gene was calculated by trimmed mean of M values method (Robinson and Oshlack, 2010).
For in vitro expression analysis of sesame leaves under phytohormone treatment, variety S. indicum var. ‘Zhengzhi No.13’ sesame plants were treated by spraying with 1 mM ABA, SA and MeJA when they grew to 4 pairs of true leaves period. Leaf tissues before treatment and treated post 1h, 3h, 6h, 12h, 24h, 36h and 48h were taken for RNA extraction, reverse transcription and qPCR. The primers of selected 6 SiLRR-RLK genes are listed in Supplementary Table S1, UBQ5 gene was used as a reference gene. There were three replicates for each treatment. The samples were stored at -80°C.
The STRING database (https://string-db.org/) was used to analyze the interaction of sesame LRR-RLK proteins based on orthologs in Arabidopsis with a confidence parameter set at a 0.85 threshold.
Based on a comprehensive search of LRR-RLK genes by HMM profiles and BlastP, 175 LRR-RLK proteins were identified in the sesame genome. The identified LRR-RLK members were given names with the prefix ‘Si’ indicating S. indicum. Phylogenetic analysis of LRR-RLK protein sequences in S. indicum and A. thaliana was carried out (Figure 1). The LRR-RLK of S. indicum was divided into 12 subfamilies together with those of A. thaliana. Group IV had the most members (36), followed by 34 members in group III and 29 members in group IX (Table 1). These three groups comprised 56.57% of SiLRR-RLKs (Figure 1). Group I and V comprising only one member were the smallest subfamily (Table 1). The details about the SiLRR-RLK family, including their accession numbers and characteristics, were given in Supplementary Table S2.
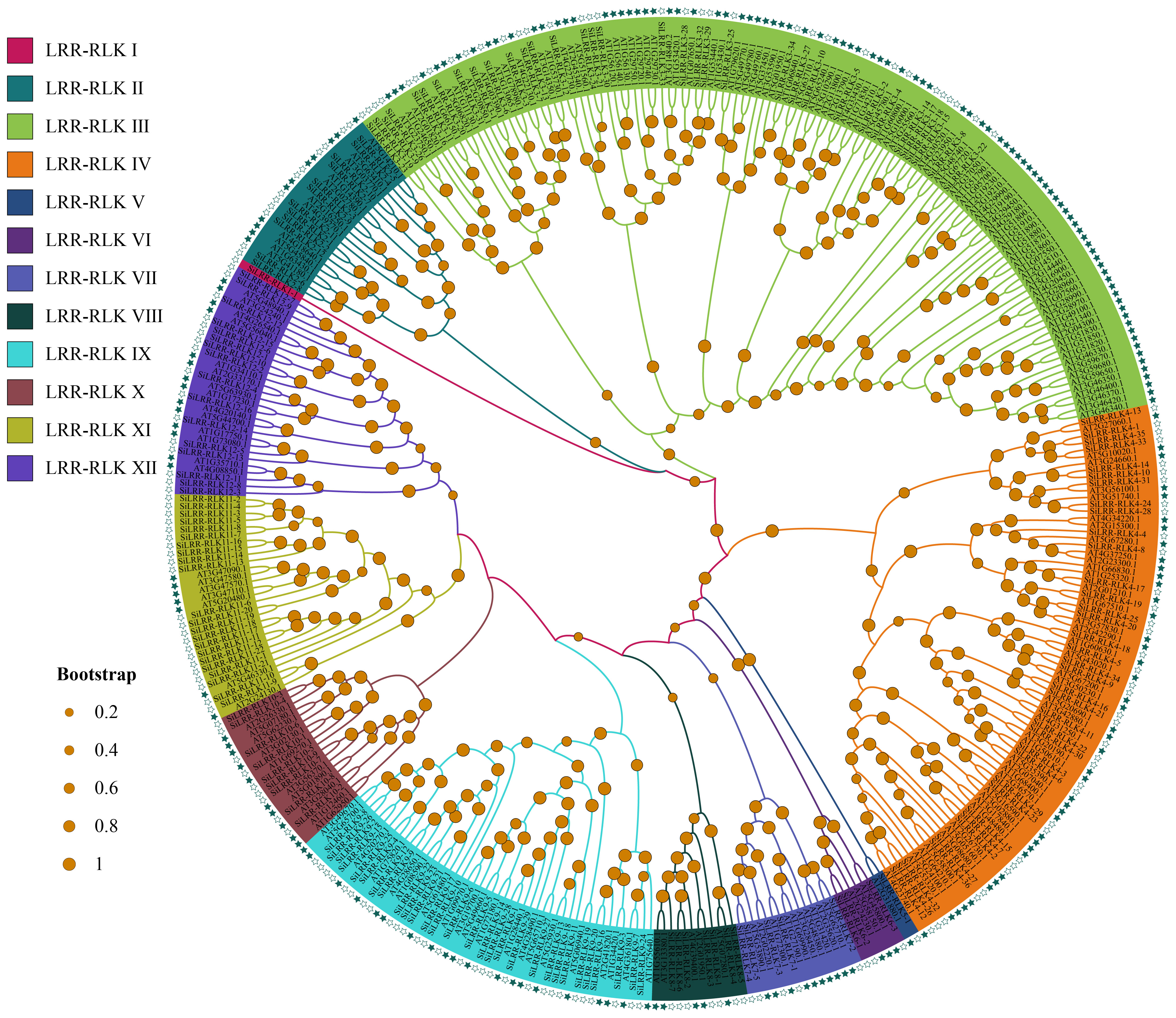
Figure 1 Phylogenetic analysis of the LRR-RLK proteins in S. indicum and A. thaliana. Green hollow pentacles represent LRR-RLKs in S. indicum while green solid pentacles represent those in A. thaliana.
The physicochemical properties of SiLRR-RLKs enabled us to gain insight on their functions. The amino acid (aa) length of SiLRR-RLKs ranged from 466 to 1304 aa. Their isoelectric points (pI) were between 5.14 and 9.56, and their molecular weights ranged from 50.38 to 141.43 kDa (Supplementary Table S2). The summarized information regarding each subfamily was listed in Table 1.
The conserved domains of proteins are closely related to their functions. Based on subfamily classification, conserved motif of SiLRR-RLKs were performed. Results showed that arrangement of motifs in same subfamily were similar (Figure 2). In addition, we identified the conserved domains of SiLRR-RLK proteins and found that they all contain both LRR and kinase domains (Figure 2), illustrating the accuracy of the SiLRR-RLK gene family. LRR-RLKs play a vital role in perceiving signals. Accordingly, a total of 78.29% of SiLRR-RLKs comprised signal peptides in our study (Supplementary Table S2). LRRNT_2 (PF08263) and LRR_8 (PF13855) constituted the major recognition domains in SiLRR-RLKs, which were found in 86.29% and 87.43% of SiLRR-RLKs, respectively. Furthermore, the brassinosteroid receptor island (PF20141) and Malectin (PF11721) domains were found in subfamilies III and VIII, implicating their additional roles in the recognition of BR and other plant hormones. Analysis of conversed motif and domain showed that kinase domains in LRR-RLKs C-terminal were more conserved, illustrating their potential roles in signal transduction. The structural compositions of the SiLRR-RLK genes were also further analyzed (Figure 2). The majority of SiLRR-RLK genes were composed of multiple exons, while only 16 SiLRR-RLKs were intron−less. SiLRR-RLK10-3, SiLRR-RLK10-4 and SiLRR-RLK10-6 have the most 27 exons. The exon−intron arrangement of LRR-RLK genes was conserved in same subfamilies while it varied in subfamilies III and X.

Figure 2 Conserved motifs, conserved domains and gene structures of 12 LRR-RLK subfamilies in S. indicum.
To further explore the potential functions of SiLRR-RLKs, GO (Gene Ontology) annotation was carried out. The results showed that SiLRR-RLKs were mainly involved in phosphorylation and defense response to fungus in terms of biological processes and functioned in kinase activity and ATP binding in molecular function (Figure 3A). All SiLRR-RLKs were predicted to localize to the membrane or plasma membrane. Consistently, all SiLRR-RLKs showed the characteristics of a high aliphatic index and low hydrophilicity (Supplementary Table S2), which further supported the idea that they were located on the plasma membrane. KEGG (Kyoto Encyclopedia of Genes and Genomes) enrichment of SiLRR-RLKs illustrated that SiLRR-RLKs were mainly enriched in thiamine metabolism, MAPK signaling pathway, plant hormone signal transduction and plant−pathogen interaction (Figure 3B). All these results suggest that SiLRR-RLK proteins are essential for signaling recognition and transduction in the stress resistance, growth and development of sesame.
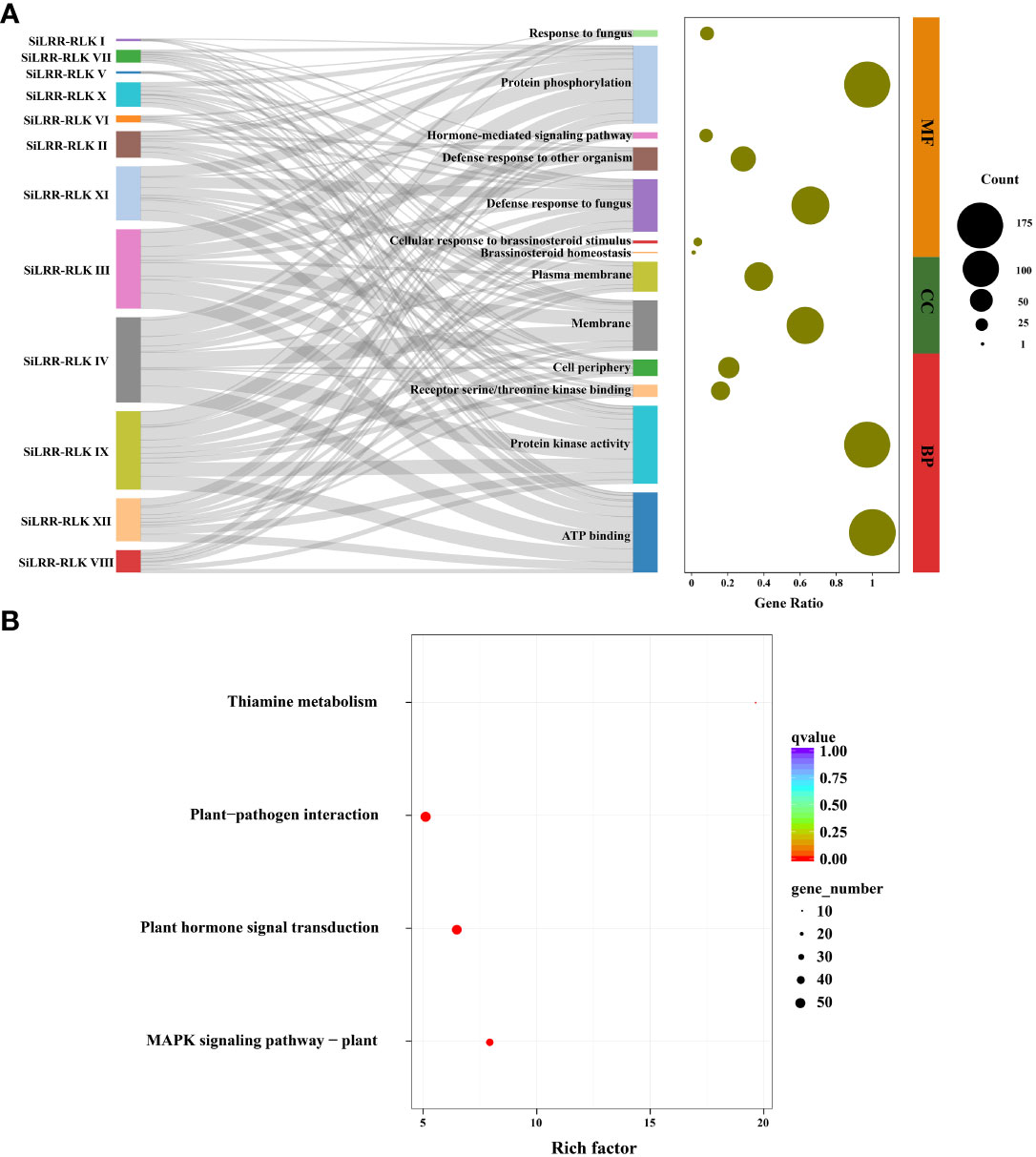
Figure 3 GO annotation and KEGG enrichment of SiLRR-RLK proteins. (A) GO annotation of 12 SiLRR-RLK protein subfamilies. MF, Molecular Function; CC, Cellular Component; BP, Biological Process. (B) KEGG enrichment of SiLRR-RLK proteins.
Gene promoters in plants can regulate the expression of genes to respond to different biotic or abiotic stresses and different growing environments, hence, assessment of cis-elements and transcription factor binding sites in promoters is crucial for understanding transcriptional regulation and gene function (Biłas et al., 2016). The upstream sequences (~2000 bp) of the promoter were obtained to confirm the expression features of SiLRR-RLKs. The cis-elements of the SiLRR-RLK promoters were explored using the PlantCARE database (Lescot et al., 2002). The detailed effects of these motifs (cis-elements) are presented in Supplementary Table S3. SiLRR-RLK promoters contained many cis-elements in response to stresses, illustrating their potential roles in plant responses to adverse environments. Cis-elements of SiLRR-RLK promoters include light-responsive elements, phytohormone-responsive elements, stress-responsive elements and growth and development elements (Figure 4). The most abundant element in SiLRR-RLK promoters was the Box4 (Light) element, followed by the MYC (Drought) and STRE (Stress) elements (Figure 4; Supplementary Table S3), illustrating that SiLRR-RLKs not only had an important role in the light response but were also crucial in the response to both biotic and abiotic stresses. In addition, SiLRR-RLKs possessed the most AAGAA-motif (Seed specific expression) and ERE (Ethylene) elements in terms of growth development and hormone response.
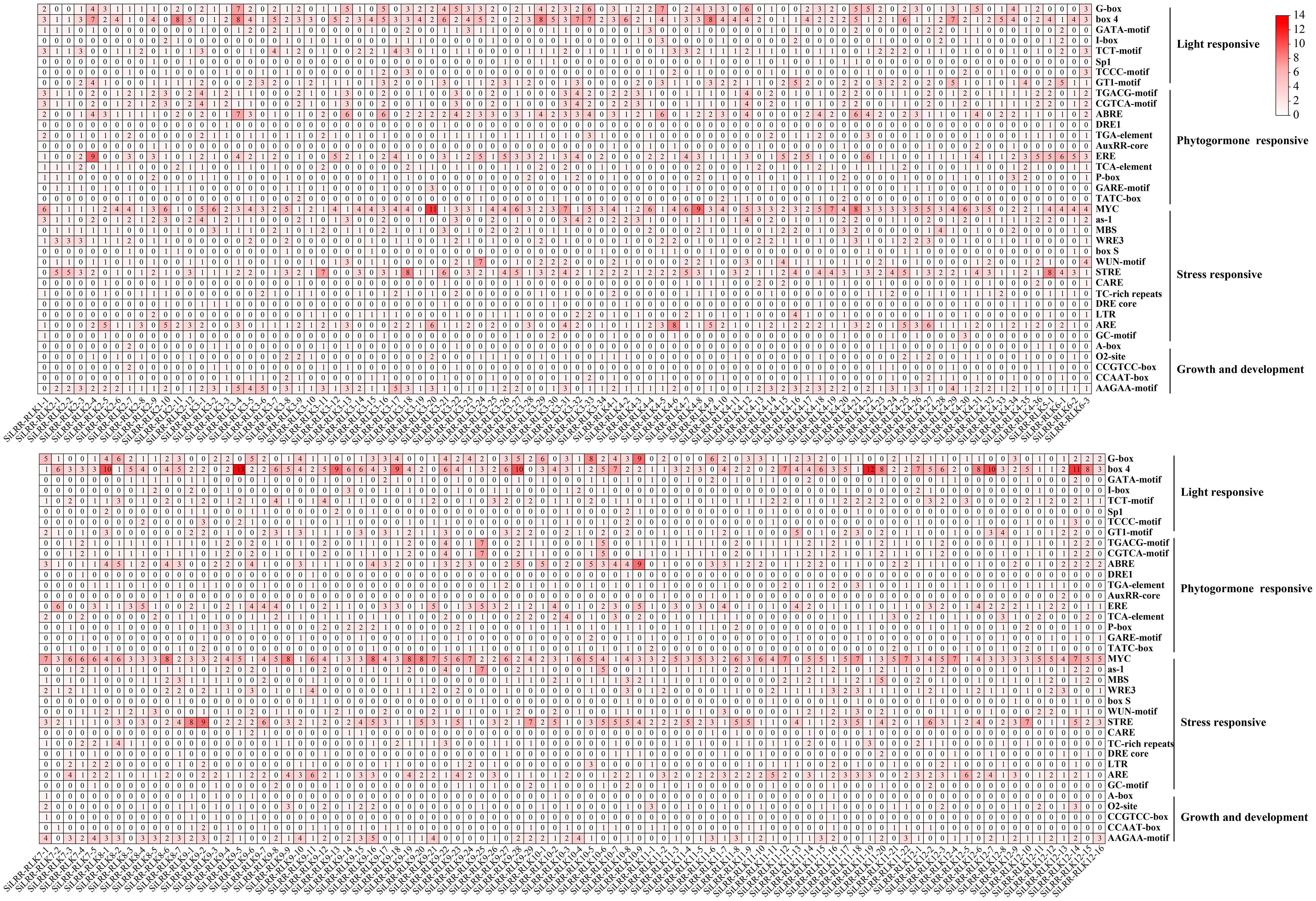
Figure 4 The distribution of cis-elements in SiLRR-RLK promoters. The gradient colors represent the number of cis-elements in SiLRR-RLK promoters.
Transcription factors (TFs) play key roles in many biological processes by regulating the expression of target genes. To investigate the possible regulatory relationship between TFs and SiLRR-RLK genes, the TF binding site prediction on PlantTFDB (Jin et al., 2017) was used. The results showed that the SiLRR-RLK genes could be regulated by 37 TF families (Supplementary Figure S1, Supplementary Table S4). C2H2, MIKC_MADS, MYB, AP2 and Dof were the TFs that can regulate most SiLRR-RLK genes. These TF families are involved in almost every aspect of plant development, hormone signaling, plant defense and stress response, suggesting that SiLRR-RLKs are extensively involved in the growth and stress defense of sesame.
We found that 173 SiLRR-RLKs were unevenly distributed on 13 sesame chromosomes (Chr) while SiLRR-RLK3-34 and SiLRR-RLK4-36 were distributed on an unanchored scaffold (Figure 5A). Twenty-five SiLRR-RLK genes were mapped to Chr 2, followed by 15 SiLRR-RLK genes on Chr 4 and Chr 6. In contrast, minimum SiLRR-RLK genes (9) was found on Chr 5, Chr 7 and Chr 10 (Figure 5B). A total of 48 SiLRR-RLK genes formed 21 gene clusters. Chr 2 and Chr 4 both had maximum gene clusters with four. There are 2-4 SiLRR-RLK members within the gene clusters, most of them contain 2 SiLRR-RLK genes. The tandem duplication SiLRR-RLK genes were identified in each cluster with a threshold of 70% sequence similarity between two aa sequences of SiLRR-RLKs. Of the 21 gene clusters, 12 SiLRR-RLK genes from 7 clusters were considered to be tandem duplicated gene pairs (Figure 5A). In addition, segmental duplication of SiLRR-RLK genes was further analyzed within the sesame genome. 38 segmentally duplicated SiLRR-RLK pairs made by 70 SiLRR-RLK genes were predicted within the sesame genome (Figure 5C). A total of 46.29% of LRR-RLK genes underwent tandem or segmental duplication events, implying that gene duplication events were momentous in the expansion of the SiLRR-RLK gene family.
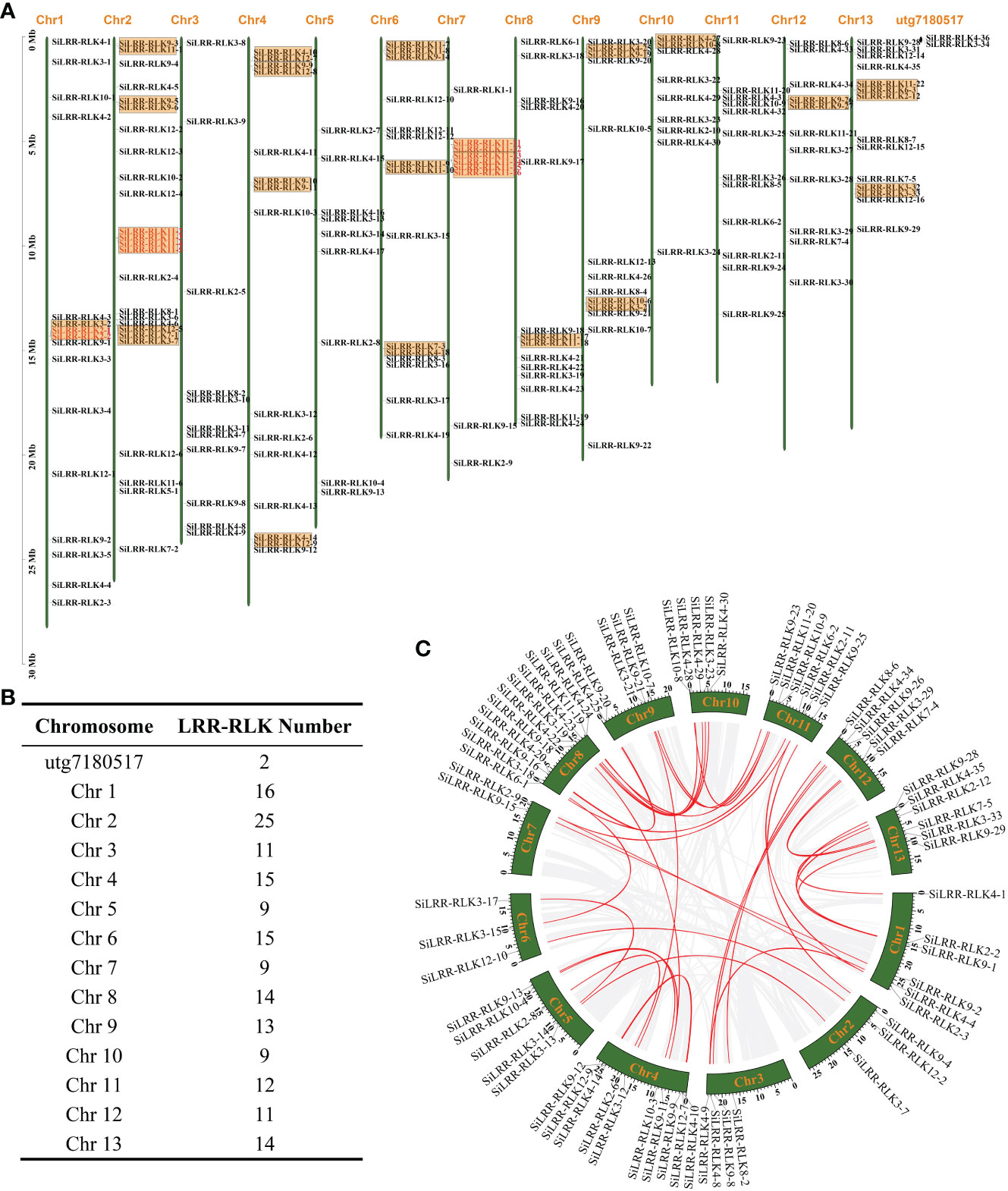
Figure 5 Chromosomal arrangement and gene duplication of SiLRR-RLK genes. (A) 173 SiLRR-RLK genes were mapped to 13 sesame chromosomes while 2 SiLRR-RLK genes mapped to unanchored scaffolds. The orange box indicates a gene cluster and the red names represent tandem duplication genes. (B) Number of SiLRR-RLK genes on each sesame chromosome. (C) The segmental duplication gene pairs of SiLRR-RLK genes. The gray lines indicate all the segmental duplicated gene pairs while red lines highlight the SiLRR-RLK pairs within the sesame genome.
To infer the syntenic relationship of LRR-RLK genes in several plants, seven dicotyledons (G. max, S. lycopersicum, S. tuberosum, G. hirsutum, V. vinifera, A. thaliana and M. truncatula) (Figure 6A) and seven monocotyledons (O. sativa, H. vulgare, Z. mays, T. aestivum, S. italica, M. acuminata and S. bicolor) (Figure 6B) were used for evolution analysis with S. indicum. The LRR-RLK genes are homologous to genes in the dicotyledonous reference plants, and the number of homologous LRR-RLK genes is 120 (G. max), 132 (S. lycopersicum), 131 (S. tuberosum), 114 (G. hirsutum), 113 (V. vinifera), 98 (A. thaliana) and 114 (M. truncatula) (Supplementary Table S5). Nonetheless, only 36 (O. sativa), 16 (H. vulgare), 14 (Z. mays), 20 (T. aestivum), 38 (S. italica), 16 (M. acuminata) and 41 (S. bicolor) homologous LRR-RLK genes existed in monocotyledons (Supplementary Table S5). More homologous LRR-RLK genes were found in dicotyledons than in monocotyledons. In addition, SiLRR-RLK4-27 and SiLRR-RLK10-9 were homologous with all 14 species, suggesting that they are crucial in the evolution of the LRR-RLK gene family. Notably, SiLRR-RLK10-9 also underwent segmental replication events in sesame (Figure 5C).
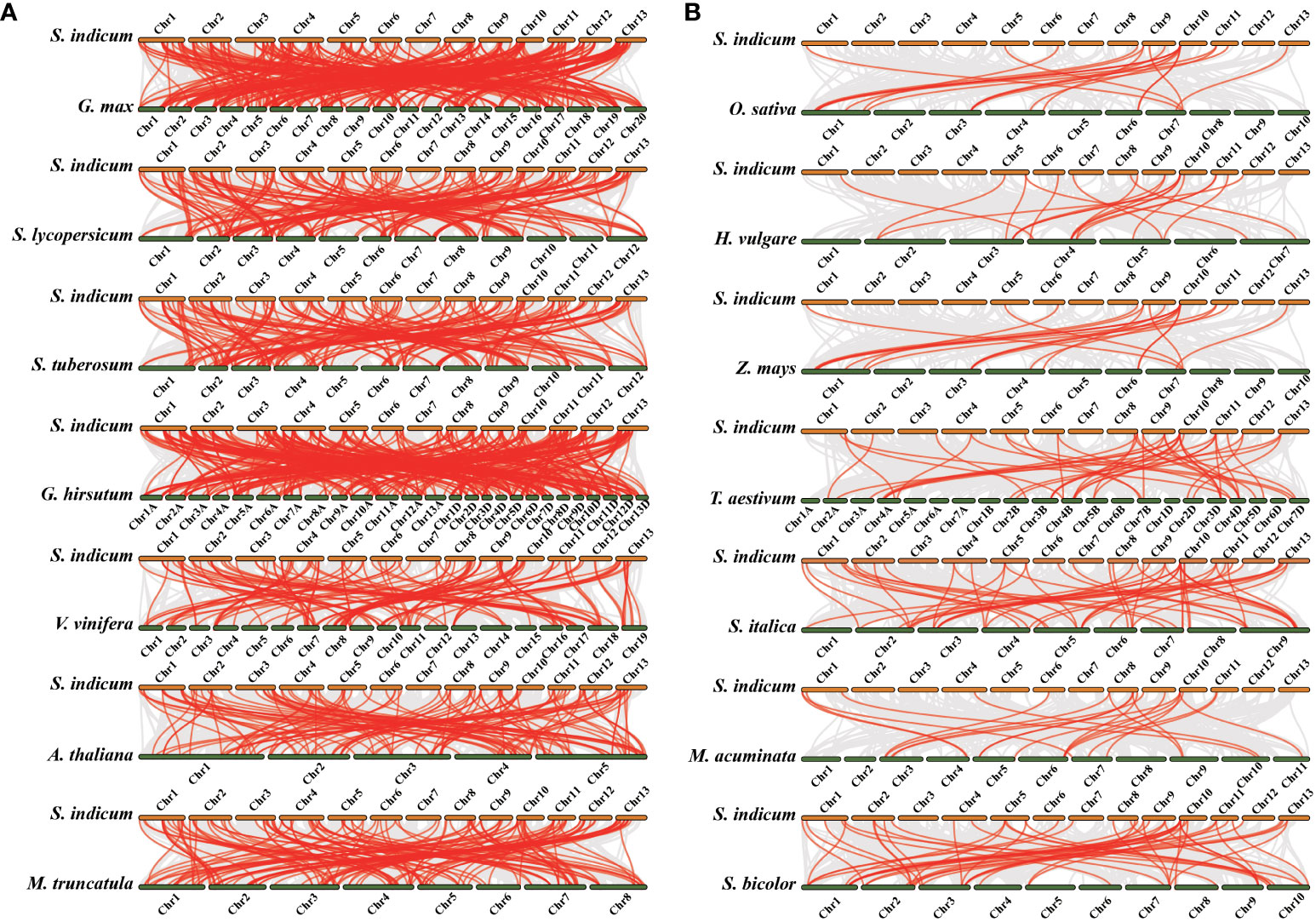
Figure 6 Synteny analysis of LRR-RLK genes between S. indicum and other plant species. The gray lines indicate all the syntenic gene pairs while red lines highlight the SiLRR-RLK pairs between S. indicum and other plant species. (A) Synteny analysis of LRR-RLK genes between S. indicum and dicotyledonous plants. (B) Synteny analysis of LRR-RLK genes between S. indicum and monocotyledonous plants.
To further investigate the evolutionary relationship of LRR-RLK in dicotyledons, phylogenetic analysis of LRR-RLK proteins in dicotyledons was performed (Supplementary Figure S2). The results showed that SiLRR-RLK tended to gather with the LRR-RLKs of A. thaliana and S. tuberosum. We also used the MEME website to search for 10 conserved motifs of all LRR-RLK proteins. We found that the LRR-RLKs in S. indicum shared the most similar motif compositions with A. thaliana and S. tuberosum in the same branch, suggesting that SiLRR-RLKs were more closely related to those of A. thaliana and S. tuberosum.
To gain a broader understanding of the functions of SiLRR-RLKs, we analyzed the divergence in spatial expression among SiLRR-RLK genes. Most SiLRR-RLKs exhibited different expression patterns in different tissues (Figure 7A). Some SiLRR-RLK genes were expressed tissue-characteristically. For instance, SiLRR-RLK8-5 and SiLRR-RLK12-9 were found to be expressed only in seeds, SiLRR-RLK3-13 and SiLRR-RLK4-6 were found to be expressed only in flowers, SiLRR-RLK11-5 and SiLRR-RLK11-18 were found to be expressed only in roots (SiLRR-RLK genes with FPKM values less than 0.1 were not considered expressed) (Supplementary Table S6). The gene expression patterns provided a preliminary clue to its function. 86, 73, 46, 40, 53 and 65 LRR-RLK genes were highly expressed (SiLRR-RLK genes with FPKM value more than 10 were considered as expressed highly) in roots, stems, leaves, flowers, capsules and seeds, respectively (Figure 7B). Of note, 14 SiLRR-RLKs (SiLRR-RLK3-30, SiLRR-RLK3-16, SiLRR-RLK3-10, SiLRR-RLK3-5, SiLRR-RLK10-1, SiLRR-RLK3-22, SiLRR-RLK8-6, SiLRR-RLK6-3, SiLRR-RLK8-2, SiLRR-RLK3-26, SiLRR-RLK3-7, SiLRR-RLK9-8, SiLRR-RLK9-14 and SiLRR-RLK3-1) exhibited constitutively high expression across different tissues, indicating their important roles in the growth and development of sesame (Figure 7B). For example, AtTMK1 (Transmembrane kinase 1), a homolog of SiLRR-RLK3-16, can active GTPase in auxin sensing (Cao et al., 2019). TMK1-mediated auxin signaling regulates membrane-associated clathrin in Arabidopsis roots (Wang et al., 2022), suggesting the importance of SiLRR-RLK3-16 in sensing auxin.
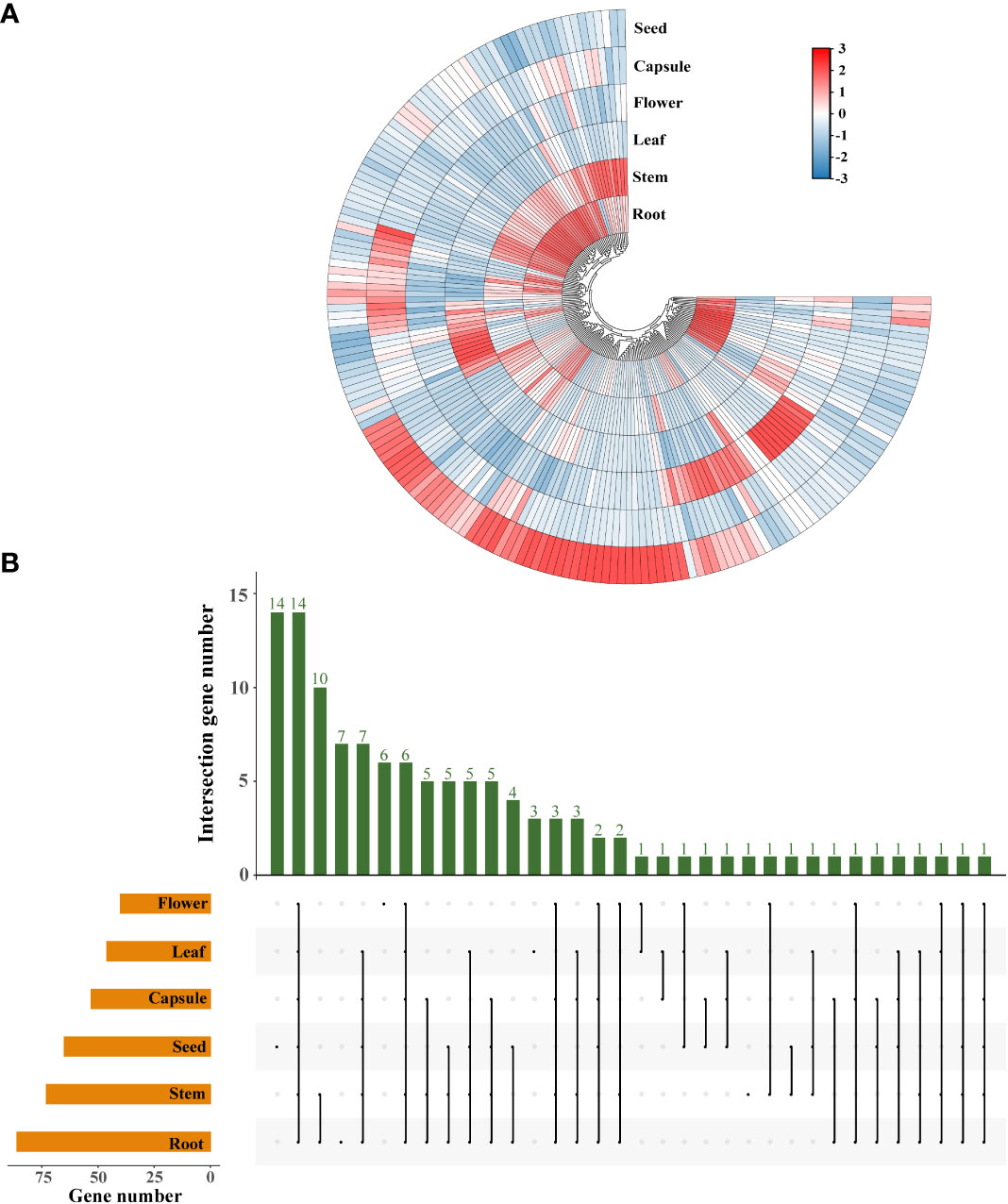
Figure 7 Expression profiles of SiLRR-RLK genes in different tissues. (A) Tissue-specific expression of SiLRR-RLK genes. The color scale shows the range of normalized FPKM values. (B) Number of SiLRR-RLK genes highly expressed in each tissue. Orange bars show the SiLRR-RLK genes highly expressed in each tissue while green bars indicate SiLRR-RLK genes highly expressed across diverse tissues.
The AAGAA motif (seed-specific expression) was found to be the most abundant cis-element in the promoter of SiLRR-RLK in terms of growth and development. Therefore, the expression pattern of SiLRR-RLK in seed development was determined based on PRJNA739094 (Zhang et al., 2021) (Supplementary Figure S3, Supplementary Table S7). The results showed that most SiLRR-RLK genes had a higher expression level in the early stage (S1 and S2) of seed development and then decreased in the later stage (S3 and S4), suggesting potential effects of SiLRR-RLK genes in early seed development. Notably, SiLRR-RLK3-4, SiLRR-RLK3-6, SiLRR-RLK3-17 and SiLRR-RLK9-16 possessed the most AAGAA motifs with five (Figure 4). Among these, SiLRR-RLK3-6 expressed at a low level in all stages. SiLRR-RLK3-4 were induced at early stages S1, S2 and S3. Likewise, SiLRR-RLK3-17 and SiLRR-RLK9-16 were highly expressed during early seed development S1 and S2, implying that SiLRR-RLK3-4, SiLRR-RLK3-17 and SiLRR-RLK9-16 might contribute to sesame seed development. Furthermore, we found several SiLRR-RLKs (SiLRR-RLK3-1, SiLRR-RLK3-7, SiLRR-RLK6-3 and SiLRR-RLK10-4) that were highly expressed during whole seed development (FPKM>10), which may also function in sesame seed development.
LRR-RLK genes were functioned in plant hormone signaling pathways. Therefore, we selected SiLRR-RLK5-1 (Homolog of SOBIR1), SiLRR-RLK9-6 (Homolog of RLK7), SiLRR-RLK3-5 (Homolog of BAK1), SiLRR-RLK8-6 (Homolog of BRI1), SiLRR-RLK12-13 (Homolog of PEPR1) and SiLRR-RLK7-5 (Homolog of PSKR1) genes to investigate their expression patterns under SA, ABA and MeJA treatments (Figure 8).
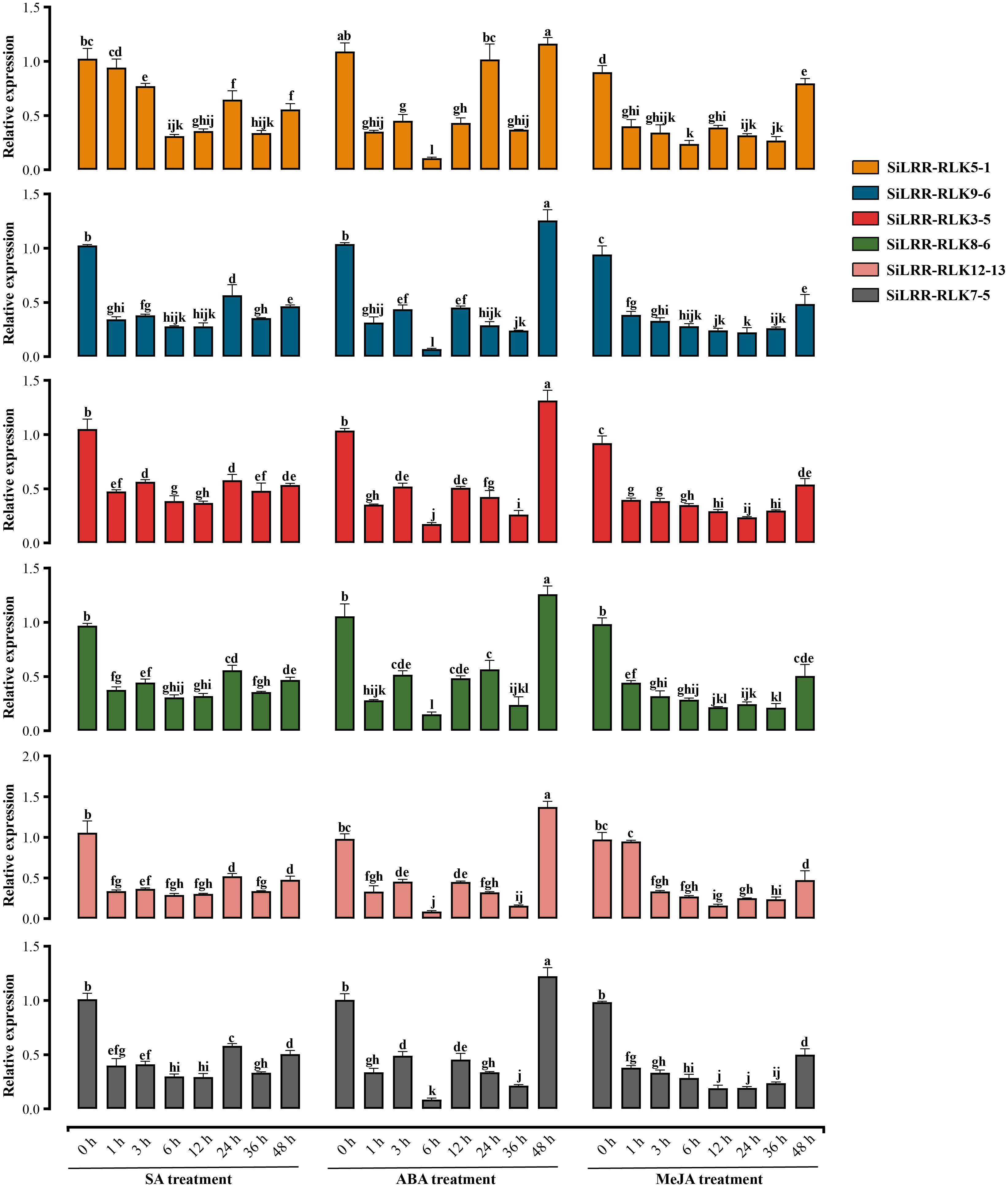
Figure 8 Relative expression level of SiLRR-RLK5-1 (Homolog of SOBIR1), SiLRR-RLK9-6 (Homolog of RLK7), SiLRR-RLK3-5 (Homolog of BAK1), SiLRR-RLK8-6 (Homolog of BRI1), SiLRR-RLK12-13 (Homolog of PEPR1) and SiLRR-RLK7-5 (Homolog of PSKR1) genes at 0 h, 1 h, 3 h, 6 h, 12 h, 24 h, 36 h and 48 h post treated by SA, ABA and MeJA.
The SiLRR-RLK9-6, SiLRR-RLK3-5, SiLRR-RLK8-6, SiLRR-RLK12-13 and SiLRR-RLK7-5 genes were down-regulated significantly at 1 h post SA treatment and remained suppressing within 48 h post SA treatment. However, SiLRR-RLK5-1 was significantly down-regulated expression at 3 h post SA treatment. It is suggested that these genes play a negative role in the early stage (48 h) in SA signaling pathway. Under ABA treatment, SiLRR-RLK5-1, SiLRR-RLK9-6, SiLRR-RLK3-5, SiLRR-RLK8-6, SiLRR-RLK12-13 and SiLRR-RLK7-5 genes were significantly down-regulated at 1h and decreased to the lowest level at 6 h, followed by a significant up-regulation of expression at 48 h (SiLRR-RLK3-5 restored its expression level at 48 h). Under MeJA treatment, SiLRR-RLK5-1, SiLRR-RLK9-6, SiLRR-RLK3-5, SiLRR-RLK8-6, SiLRR-RLK12-13 and SiLRR-RLK7-5 genes were significantly down-regulated, followed by partial restoration of their expression levels at 48 h. Notably, the expression trends of these six genes were similar under phytohormone treatment, suggesting that they may form dimers or polymers in phytohormone signaling pathway and synergistically regulate the downstream pathways.
LRR-RLKs act as cell surface receptors and play a crucial role in signal sensing and transduction. To unravel the function of SiLRR-RLKs in response to pathogen M. phaseolina, the expression patterns of SiLRR-RLK genes under stress were investigated (Figure 9; Supplementary Table S8). Considering that there are many LRR-RLK genes in sesame, we divided them into six clusters based on their expression patterns (FPKM>0) (Supplementary Figure S4). The results showed that the SiLRR-RLK genes in Cluster 5 and Cluster 6 were decreased after inoculation with M. phaseolina, they may mediate the susceptibility in plant immunity (Figure 9; Supplementary Figures S4E, S4F). However, most SiLRR-RLKs were induced at different times in Cluster 1, Cluster 2, Cluster 3 and Cluster 4 under M. phaseolina stress, which further confirmed the widely known disease resistance of LRR-RLKs (Figure 9; Supplementary Figures S4A-D). For instance, SiLRR-RLK11-10 in Cluster 1 were upregulated 4.68-fold at 12 hours after infection while SiLRR-RLK12-6 and SiLRR-RLK9-22 in Cluster 2 were upregulated 4.89- and 10.48-fold at 48 hours after infection, respectively. SiLRR-RLK genes in Cluster 3 were upregulated 2-3 times overall during M. phaseolina treatment. Moreover, the expression of SiLRR-RLK genes in Cluster 4 were continuously induced by M. phaseolina (Supplementary Figures S4A-D, Supplementary Table S8).

Figure 9 Expression patterns of SiLRR-RLK genes under M. phaseolina stress (0 - 48 h). The box plot on the left represents the expression trend of SiLRR-RLK genes in the same cluster, the heatmap in the middle indicates the expression patterns of SiLRR-RLK genes, and the annotation on the right explains the expression trend of SiLRR-RLK genes in each cluster.
The duplicated SiLRR-RLK genes identified were analyzed synchronously with their expression patterns to identify the genes designated for novel functions. The expression patterns of 7 pairs of tandemly duplicated SiLRR-RLK genes and 38 pairs of segmentally duplicated SiLRR-RLK genes during M. phaseolina stress were analyzed in a heatmap (Figure 10). We found that most tandemly and segmentally duplicated SiLRR-RLK genes exhibited antagonistic expression profiles under M. phaseolina stress, suggesting a function of redundancy between SiLRR-RLK genes during sesame disease resistance (Figure 10). There were only a few exceptions, the LRR-RLK11-11:LRR-RLK11-12 gene pair showed a similar expression profile under M. phaseolina stress, that is, they were both induced at post infection.
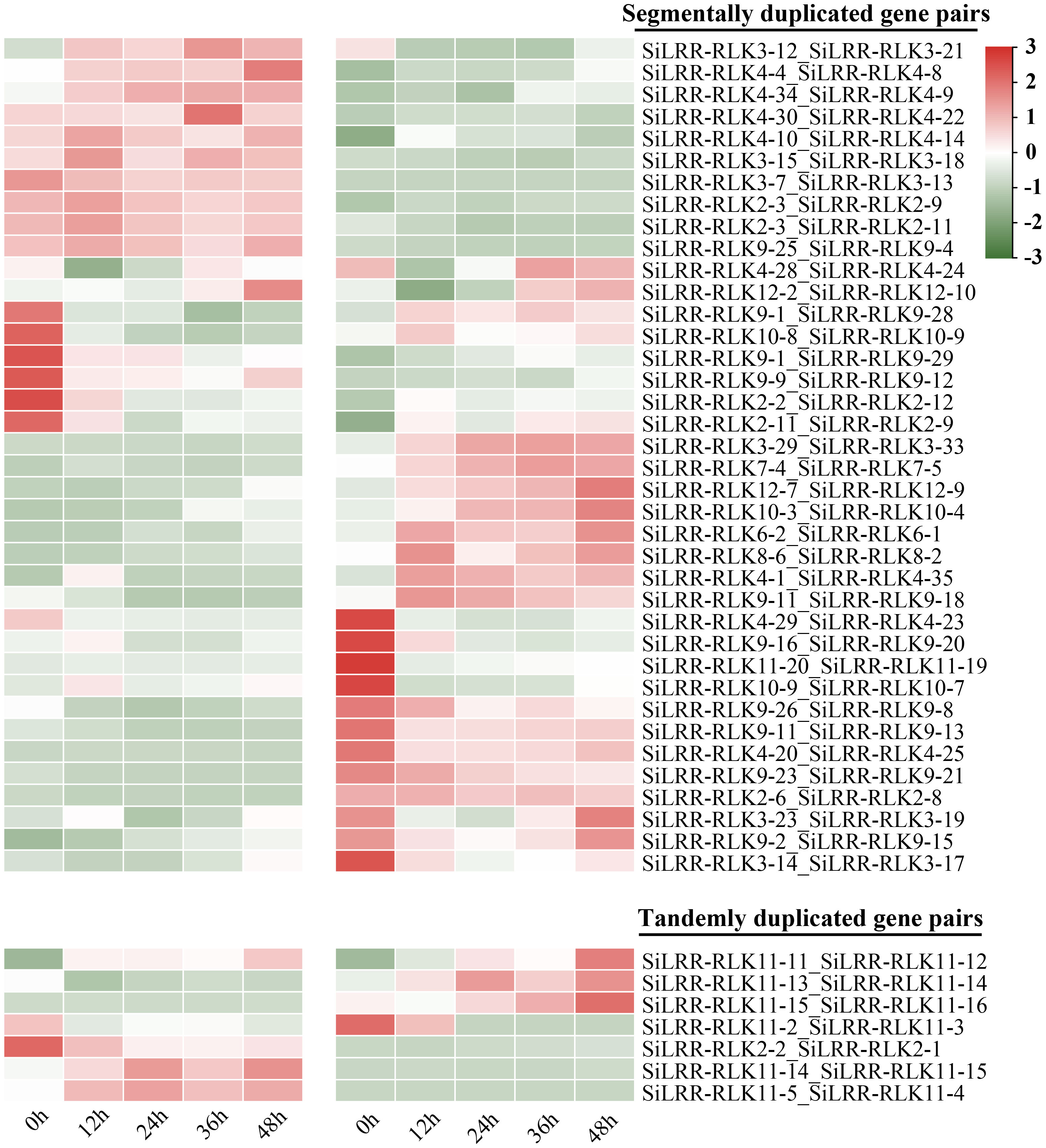
Figure 10 Expression patterns of tandemly and segmentally duplicated SiLRR-RLK gene pairs during M. phaseolina infection (0 - 48 h). “_” represents two SiLRR-RLKs are duplicated gene pairs.
According to well-studied investigations, there are many complex interactions within the LRR-RLK gene family. To further gain insight into the functions of the SiLRR-RLK proteins, we constructed a PPI network by STRING database (https://STRING-db.org/) based on the well-studied LRR-RLKs in Arabidopsis. As shown in Figure 11, the SiLRR-RLK members showed interactions with some other members.
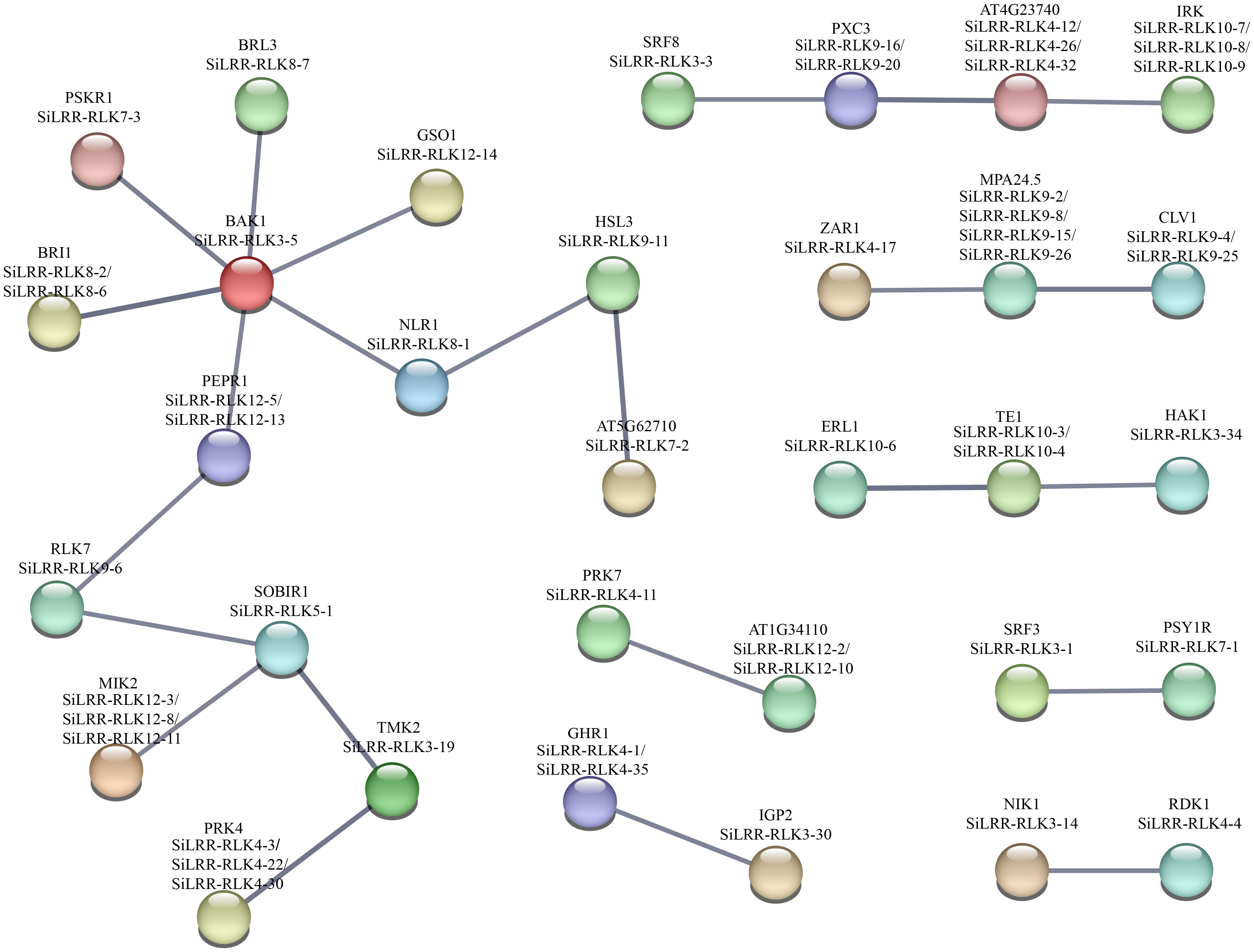
Figure 11 A protein−protein interaction network for SiLRR-RLKs based on their orthologs in Arabidopsis. SiLRR-RLK proteins are shown in brackets with Arabidopsis orthologs.
Obviously, SiLRR-RLK3-5, the homolog of AtBAK1, is the hub functional gene in the PPI network. Studies have showed that AtBAK1, acting as a coreceptor with other proteins, can form complexes (Dimer, trimer or tetramer) such as SOBIR1/BAK1, BAK1/BIR1, ER/BAK1/TMM, BIK1/BAK1/ERL1/ERL2, FLS2/BAK/BIK1 and FLS2/BIK1/RBOHD, and these complexes are all important in relaying signals to downstream components in plant immunity system (Gao et al., 2009; Lu et al., 2010; Wang et al., 2011; Li et al., 2014; Lin et al., 2014; Jordá et al., 2016). In addition, other AtLRR-RLK genes in the network have also been proven to be involved in plant biotic stress, such as SOBIR1, a homolog of SiLRR-RLK5-1, which was reported to form a complex with BAK1 for immunity against the fungi Phytophthora infestans and Sclerotinia sclerotiorum (Gao et al., 2009; Liu et al., 2016). SiLRR-RLK4-17 is homologous to ZAR1, which is a calcium-permeable channel triggering plant immune signaling (Bi et al., 2021). Notably, SiLRR-RLK8-6, a homolog of AtBRI1, was highly expressed in all tissues. BRI1 acts as a BR receptor and is extensively involved in plant growth, development and stresses (Wang et al., 2001). Recent studies have shown that BRI1 can form a heterodimer with SAUR15, which activates the plasma membrane H+-ATPase to promote Arabidopsis organogenesis (Li et al., 2022). In addition, BRI1 is another LRR-RLK that can bind to BAK1, and the BRI1/BAK1 complex regulates stem elongation, vascular differentiation, seed size, fertility, flowering time and senescence by BR signaling in A. thaliana (Li et al., 2002; Nam and Li, 2002; Wang et al., 2005). Notably, BRI1 in cereals has been shown to contribute to disease resistance (Goddard et al., 2014) and drought tolerance (Feng et al., 2015) in plants, indicating that SiLRR-RLKs have complex biological functions by participating in the crosstalk between plant growth and development and stress.
Based on the sequence structure, functional annotation, expression patterns and PPI prediction of SiLRR-RLKs, we can conclude that SiLRR-RLKs are crucial for plant immunity. In addition, previous studies have shown that SiLRR-RLKs are vital components in response to M. phaseolina stress (Yan et al., 2021). To further understand the relationship between SiLRR-RLK genes and sesame disease resistance, the expression patterns of SiLRR-RLK genes under M. phaseolina stress were used for correlation analysis. SiLRR-RLK gene pairs with a Pearson correlation coefficient greater than 0.95 or less than -0.95 suggested a correlation between the two SiLRR-RLK genes. The coexpression network of SiLRR-RLK genes was constructed according to the relationship between SiLRR-RLK genes (Figure 12). In a coexpression network, most genes interact with only a few other genes, while a few interact with a large number of other genes, which are the core genes in this gene network. Core SiLRR-RLK genes in the coexpression network might be vital in sesame resistance to M. phaseolina. Additionally, two core gene sets in the network attracted our attention (Figures 11, 12). One is the BAK1/PEPR1/RLK7/SOBIR1/MIK2 signaling pathway, whose function in plant immunity has been well elucidated in other plants. The other is the SRF8/PXC3/IRK signaling pathway. Notably, there exist a correlation between SiLRR-RLK10-8 and SiLRR-RLK9-20, as well as SiLRR-RLK12-3, SiLRR-RLK12-13, SiLRR-RLK12-8 and SiLRR-RLK9-6 (Figures 11, 12). Combined with coexpression analysis and the PPI network, the core genes, SiLRR-RLK12-3/SiLRR-RLK12-13/SiLRR-RLK12-8/SiLRR-RLK9-6 and SiLRR-RLK10-8/SiLRR-RLK9-20, may be the core components of disease resistance to M. phaseolina in sesame.
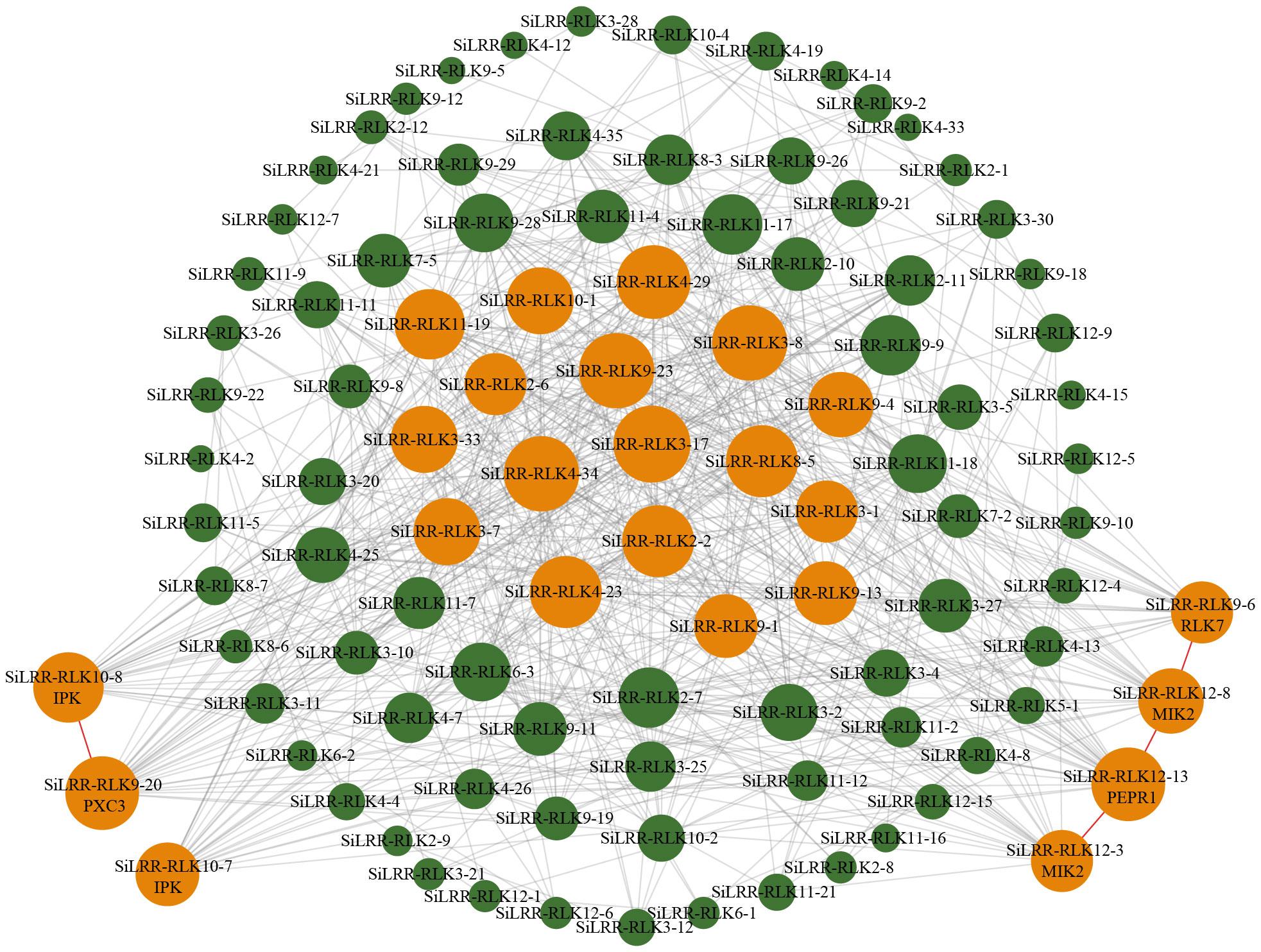
Figure 12 Coexpression network of SiLRR-RLK genes in response to M. phaseolina. The larger nodes represent core SiLRR-RLKs in the network, while the smaller nodes represent noncore SiLRR-RLKs. The size of the node circle is positively correlated with the number of SiLRR-RLKs it interacts. The orange nodes indicate core SiLRR-RLKs that may form multimers to defend against M. phaseolina.
Recently, the LRR-RLK gene family has been identified in many plant species, and the number of LRR-RLK family members varies greatly. The proportion of SiLRR-RLK genes was consistent with Liu et al. (Liu et al., 2017), which showed a 0.67-1.39% proportion in angiosperm species. In higher plants, the number of identified LRR-RLK genes ranged from 180 (C. sativus) to 589 (T. elongatum) (Soltabayeva et al., 2022). A recent study has identified 14 classes resistance (R) genes, including LRR-RLK subclass, in the sesame genome (Miao et al., 2023). In this study, 175 SiLRR-RLK genes were identified from the sesame genome, accounting for 0.73% of the sesame genome and 14.61% of sesame R genes. Although there is a lack of information on the function of LRR-RLK genes in sesame, the evolutionary diversity and function of SiLRR-RLK genes can be inferred from phylogenetic analysis, protein structure, gene structure and expression profiles. The phylogenetic tree revealed that SiLRR-RLKs can be divided into 12 subfamilies. SiLRR-RLKs occurred in almost every major branch together with the Arabidopsis LRR-RLK subfamily (Figure 1), indicating that all Arabidopsis LRR-RLK subfamilies share a common ancestor with sesame. Additionally, the collinearity analysis showed that the homologous LRR-RLK genes existed much more in dicotyledons than monocotyledons (Figure 6). It is implied that the duplication of the LRR-RLK gene probably occurred after the differentiation of dicotyledons and monocotyledons, which has been consistent with a previous investigation(Miao et al., 2023).
Segmental and tandem replication are important drivers of the expansion of gene families, especially in the evolution of plant LRR-RLK gene families (Lehti-Shiu et al., 2009; Lehti-Shiu and Shiu, 2012). A total of 420 (71.31%) LRR-RLK genes with replication events were detected in T. ponticum, involving 191 segmentally duplicated SiLRR-RLK pairs and 145 tandemly duplicated SiLRR-RLK pairs (Mishra et al., 2021). It has been found that 73.3% and 20.3% of LRR-RLK genes in soybean were involve in segmental duplication and tandem duplication (Zhou et al., 2016). Similarly, in the present study, 81 (46.29%) SiLRR-RLK genes involved in 38 segmental pairs and 7 tandem pairs were perceived (Figure 5). Therefore, it is inferred that the expansion of the LRR-RLK gene family is mainly caused by gene segmental duplication. Interestingly, SiLRR-RLK10-9 was found to have collinearity with other 14 species (Figure 6), and SiLRR-RLK10-9 also underwent segmental replication events in sesame, suggesting its key contributions to the expansion of the SiLRR-RLK gene family.
The SiLRR-RLK IV and III subfamilies, representing the two largest subfamilies, exhibited duplication events. SiLRR-RLKs in the IV and III subfamilies were also syntenic with the LRR-RLKs in the other 14 species (Supplementary Table S5). 36 and 34 LRR-RLK members from sesame were found to form the SiLRR-RLK IV and III subfamilies, respectively, based on the phylogenetic tree (Figure 1). Furthermore, there were 16 genes and 12 genes underwent segmental duplication in IV and III subfamilies, respectively (Figures 1, 5C). Under M. phaseolina stress, 13 and 16 SiLRR-RLK genes in IV and III subfamilies were induced significantly (Figure 10, Supplementary Table S8). Notably, proteins from subfamilies IV and III were both assigned to the GO terms phosphorylation and kinase activity (Figure 3A). These results point to the idea that the duplicated events within the SiLRR-RLK IV and III subfamilies during evolution may contribute to the perception of M. phaseolina stress signals in sesame.
The LRR-RLK genes are crucial in recognition and signal transduction in biotic and abiotic stresses, as shown by the fact that their promoters possess many phytohormone and stress responsive cis-elements (Figure 4), hence, the ever-changing environment may also lead to the replication and expansion of the SiLRR-RLK gene family. On the other hand, evidence that there are the ubiquitous redundant functions of SiLRR-RLK genes has indicated that the diversity of SiLRR-RLK genes may also be the result of random genomic drift (Eyüboglu et al., 2007; Albrecht et al., 2008). After duplication, duplicated genes usually accumulate mutations and lead to a functional diversification of LRR-RLK proteins. Arabidopsis LRR-RLKs are primarily involved in regulating plant growth and development and stress responses (Biotic and abiotic stresses) or both (Li and Tax, 2013). In this investigation, KEGG enrichment of SiLRR-RLKs revealed the pathways involved in resistance to M. phaseolina, such as MAPK signaling pathway, plant hormone signal transduction and plant−pathogen interaction pathway (Figure 3B). SiLRR-RLKs may sense extracellular signals, act as early warning genes, and then regulate the early stress response in plants (Lin et al., 2017). In this study, SiLRR-RLKs were shown to play a role in plant−pathogen interactions by transmitting and amplifying signals downstream through protein phosphorylation and kinase activity (Figure 3).
In general, the expression patterns of genes represent their potential functions. Therefore, the expression profiles of SiLRR-RLKs under M. phaseolina stress may help us to gain insight on the function of SiLRR-RLKs. In our research, several genes in response to fungal infection were identified. Many SiLRR-RLKs act as pattern recognition receptors to initiate the PTI (Pathogen-associated molecular pattern-triggered immunity). SiLRR-RLK3-5, homologous to AtBAK1, can be continuously induced during M. phaseolina infection (Supplementary Table S8), implicating its positive function during plant immunity, which is supported by the central position of SiLRR-RLK3-5 in the PPI network (Figure 11). In Arabidopsis, the BAK1/BRI1 complex regulates the cell death process to participate in the immune response by the BR signaling pathway (Li et al., 2002). Likewise, in tobacco, NbBRI1 participates in brassinosteroid-regulated immune responses by regulating the concentrations of H2O2 and NO (Deng et al., 2016). In sesame, SiLRR-RLK8-6, which is homologous to BRI1, were induced during M. phaseolina stress (Supplementary Table S8). Their expression patterns were similar to those of SiLRR-RLK3-5, illustrating that SiLRR-RLK8-6 may form a complex with SiLRR-RLK3-5 and coregulate plant immunity through the BR pathway.
SiLRR-RLK3-14 is homologs of NIK1 (NSP-interacting kinase 1), and the abundance of their transcripts was downregulated at early stage (Supplementary Table S8), which coincides with the fact that NIK1 acts as a negative regulator in plant immunity (Li et al., 2019). BRL3 (BR insensitive1-like 3) in Arabidopsis not only senses FLG22 and regulates ROS homeostasis (Tunc-Ozdemir and Jones, 2017) but also increases permeating agents such as proline in plants, which can improve plant drought tolerance without a penalty in growth (Fàbregas et al., 2018). In sesame, SiLRR-RLK8-7, as a homolog of BRL3, exhibited a upregulation trend during M. phaseolina infection, suggesting that SiLRR-RLK8-7 may play a crucial role in disease resistance (Supplementary Table S8).
The previous results of functional annotation, KEGG enrichment, promoter analysis, expression pattern and PPI network of SiLRR-RLKs all showed that SiLRR-RLKs are crucial in plant immunity. Thus, a coexpression network of SiLRR-RLKs under M. phaseolina stress was constructed. Interestingly, two coexpression immune modules were inferred in the core gene set based on PPI and coexpression network (Figures 11, 12). They are the SiLRR-RLK12-3/SiLRR-RLK12-13/SiLRR-RLK12-8/SiLRR-RLK9-6 module, which is homologous to the Arabidopsis BAK1/PEPR1/RLK7/SOBIR1/MIK2 complex, and SiLRR-RLK10-8/SiLRR-RLK9-20 module, which is homologous to the Arabidopsis SRF8/PXC3/IRK complex. In Arabidopsis, the function of the SOBIR1/BAK1 complex has been well studied, and exogenous NLP20 treatment stimulates the formation of the BAK1/SOBIR1/RLP23 complex and initiates immunity (Gao et al., 2009). Furthermore, the SOBIR1/BAK1 complex could enhance the resistance of Arabidopsis to the fungi P. infestans and S. sculrotiorum (Liu et al., 2016). In addition to BAK1, the function of SOBIR1 in other species has also been studied in detail. In tomato, the homolog of SOBIR1 interacts directly with the disease resistance genes CF-4 and VE1, which mediated the resistance to the fungi Cladosporium fulvum and Verticillium dahliae (Liebrand et al., 2013). SOBIR1 in cotton was reported to interact with bHLH171, phosphorylate bHLH171 and confer resistance to the fungus V. dahliae (Zhou et al., 2019). In tobacco, SOBIR1 can fine-tune ROS production involved in the immune response to the fungus Cladosporium fulvum (Huang et al., 2021). These results suggest that the SiLRR-RLK12-3/SiLRR-RLK12-13/SiLRR-RLK12-8/SiLRR-RLK9-6 module might be important in resistance to the fungus M. phaseolina (Figures 11, 12). They might mediate resistance to the fungus M. phaseolina independently or form dimers or polymers with each other to mediate the immune response to M. phaseolina jointly, which needs further study. However, studies on the immune function of the SRF8/PXC3/IRK (Inflorescence and root apices receptor kinase) module are not clear. In Arabidopsis, PXC3 (Phloem intercalated with xylem-correlated 3) has been shown to interact with BAK1 to regulate vascular development (Xu et al., 2021), but whether it has a function in plant immunity is unknown. Arabidopsis SRF3 (Strubbelig receptor kinase 3) could coordinate immune responses, growth and development in plants (Platre et al., 2022), but the function of SRF8 is uncharted. Therefore, the role of the SiLRR-RLK10-8/SiLRR-RLK9-20 module resistance to M. phaseolina in sesame is not clear, and further experiments are needed to solve this issue.
Whole genome identification and comprehensive analysis of the SiLRR-RLK gene family were carried out in this study. Phylogenetic, structural, evolutionary and expression profile analyses of SiLRR-RLKs revealed the complexity and diversity of the LRR-RLK gene family in sesame and its potential roles under M. phaseolina stress. Furthermore, we found several SiLRR-RLK genes that contributed to resistance to M. phaseolina. Altogether, the results provided a framework for further functional study of SiLRR-RLK genes.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
WY: Data curation, Formal analysis, Investigation, Visualization, Writing – original draft. YN: Data curation, Resources, Writing – review & editing. HZ: Data curation, Resources, Writing – review & editing. XL: Data curation, Resources, Writing – review & editing. MJ: Data curation, Resources, Writing – review & editing. XZ: Resources, Writing – review & editing. YL: Resources, Writing – review & editing. HM: Conceptualization, Funding acquisition, Supervision, Writing – review & editing. HL: Conceptualization, Funding acquisition, Supervision, Writing – review & editing. HZ: Conceptualization, Funding acquisition, Supervision, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. The work was supported by The Key Research Project of the Shennong Laboratory (SN01-2022-04), the Key Project of Science and Technology of Henan Province (201300110600), China Agriculture Research System of MOF and MARA (CARS-14), and Key Research and Development Project of Henan Province (221111520400).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2024.1334189/full#supplementary-material
Supplementary Figure 1 | Statistics of SiLRR-RLK genes regulated by TFs (Genes with TF binding sites were considered to be regulated by TFs).
Supplementary Figure 2 | Phylogenetic analysis and motif compositions of LRR-RLK proteins in six dicotyledons. Green hollow pentacles highlight SiLRR-RLK proteins.
Supplementary Figure 3 | Expression profiles of SiLRR-RLKs during seed development (S1, S2, S3 and S4).
Supplementary Figure 4 | Expression profiles of SiLRR-RLKs of Cluster 1 (A), Cluster 2 (B), Cluster 3 (C), Cluster 4 (D), Cluster 5 (E) and Cluster 6 (F) during M. phaseolina infection (0 - 48 h).
Albrecht, C., Russinova, E., Kemmerling, B., Kwaaitaal, M., De Vries, S. C. (2008). Arabidopsis SOMATIC EMBRYOGENESIS RECEPTOR KINASE proteins serve brassinosteroid-dependent and -independent signaling pathways. Plant Physiol. 148, 611–619. doi: 10.1104/pp.108.123216
Bi, G., Su, M., Li, N., Liang, Y., Dang, S., Xu, J., et al. (2021). The ZAR1 resistosome is a calcium-permeable channel triggering plant immune signaling. Cell 184, 3528–3541.e3512. doi: 10.1016/j.cell.2021.05.003
Biłas, R., Szafran, K., Hnatuszko-Konka, K., Kononowicz, A. K. (2016). Cis-regulatory elements used to control gene expression in plants. Plant Cell 127, 269–287. doi: 10.1007/s11240-016-1057-7
Bleckmann, A., Weidtkamp-Peters, S., Seidel, C. A., Simon, R. (2010). Stem cell signaling in Arabidopsis requires CRN to localize CLV2 to the plasma membrane. Plant Physiol. 152, 166–176. doi: 10.1104/pp.109.149930
Cao, M., Chen, R., Li, P., Yu, Y., Zheng, R., Ge, D., et al. (2019). TMK1-mediated auxin signalling regulates differential growth of the apical hook. Nature 568, 240–243. doi: 10.1038/s41586-019-1069-7
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chen, S., Zhou, Y., Chen, Y., Gu, J. (2018). fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, i884–i890. doi: 10.1093/bioinformatics/bty560
Chen, X., Zuo, S., Schwessinger, B., Chern, M., Canlas, P. E., Ruan, D., et al. (2014). An XA21-associated kinase (OsSERK2) regulates immunity mediated by the XA21 and XA3 immune receptors. Mol. Plant 7, 874–892. doi: 10.1093/mp/ssu003
Crook, A. D., Schnabel, E. L., Frugoli, J. A. (2016). The systemic nodule number regulation kinase SUNN in Medicago truncatula interacts with MtCLV2 and MtCRN. Plant J. 88, 108–119. doi: 10.1111/tpj.13234
Deng, X. G., Zhu, T., Zou, L. J., Han, X. Y., Zhou, X., Xi, D. H., et al. (2016). Orchestration of hydrogen peroxide and nitric oxide in brassinosteroid-mediated systemic virus resistance in Nicotiana benthamiana. Plant J. 85, 478–493. doi: 10.1111/tpj.13120
Dong, N., Yin, W., Liu, D., Zhang, X., Yu, Z., Huang, W., et al. (2020). Regulation of brassinosteroid signaling and salt resistance by SERK2 and potential utilization for crop improvement in rice. Front. Plant Sci. 11, 621859. doi: 10.3389/fpls.2020.621859
Eyüboglu, B., Pfister, K., Haberer, G., Chevalier, D., Fuchs, A., Mayer, K. F., et al. (2007). Molecular characterisation of the STRUBBELIG-RECEPTOR FAMILY of genes encoding putative leucine-rich repeat receptor-like kinases in Arabidopsis thaliana. BMC Plant Biol. 7, 16. doi: 10.1186/1471-2229-7-16
Fàbregas, N., Lozano-Elena, F., Blasco-Escámez, D., Tohge, T., Martínez-Andújar, C., Albacete, A., et al. (2018). Overexpression of the vascular brassinosteroid receptor BRL3 confers drought resistance without penalizing plant growth. Nat. Commun. 9, 4680. doi: 10.1038/s41467-018-06861-3
Feng, Y., Yin, Y., Fei, S. (2015). Down-regulation of BdBRI1, a putative brassinosteroid receptor gene produces a dwarf phenotype with enhanced drought tolerance in Brachypodium distachyon. Plant Sci. 234, 163–173. doi: 10.1016/j.plantsci.2015.02.015
Finn, R. D., Clements, J., Eddy, S. R. (2011). HMMER web server: interactive sequence similarity searching. Nucleic Acids Res. 39, W29–W37. doi: 10.1093/nar/gkr367
Franco-Orozco, B., Berepiki, A., Ruiz, O., Gamble, L., Griffe, L. L., Wang, S., et al. (2017). A new proteinaceous pathogen-associated molecular pattern (PAMP) identified in Ascomycete fungi induces cell death in Solanaceae. New Phytol. 214, 1657–1672. doi: 10.1111/nph.14542
Gao, M., Wang, X., Wang, D., Xu, F., Ding, X., Zhang, Z., et al. (2009). Regulation of cell death and innate immunity by two receptor-like kinases in Arabidopsis. Cell Host Microbe 6, 34–44. doi: 10.1016/j.chom.2009.05.019
Goddard, R., Peraldi, A., Ridout, C., Nicholson, P. (2014). Enhanced disease resistance caused by BRI1 mutation is conserved between Brachypodium distachyon and barley (Hordeum vulgare). Mol. Plant Microbe Interact. 27, 1095–1106. doi: 10.1094/MPMI-03-14-0069-R
Goodstein, D. M., Shu, S., Howson, R., Neupane, R., Hayes, R. D., Fazo, J., et al. (2012). Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 40, D1178–D1186. doi: 10.1093/nar/gkr944
Huang, W. R. H., Schol, C., Villanueva, S. L., Heidstra, R., Joosten, M. (2021). Knocking out SOBIR1 in Nicotiana benthamiana abolishes functionality of transgenic receptor-like protein Cf-4. Plant Physiol. 185, 290–294. doi: 10.1093/plphys/kiaa047
Isner, J. C., Begum, A., Nuehse, T., Hetherington, A. M., Maathuis, F. J. M. (2018). KIN7 kinase regulates the vacuolar TPK1 K(+) channel during stomatal closure. Curr. Biol. 28, 466–472.e464. doi: 10.1016/j.cub.2017.12.046
Jin, J., Tian, F., Yang, D. C., Meng, Y. Q., Kong, L., Luo, J., et al. (2017). PlantTFDB 4.0: toward a central hub for transcription factors and regulatory interactions in plants. Nucleic Acids Res. 45, D1040–d1045. doi: 10.1093/nar/gkw982
Jordá, L., Sopeña-Torres, S., Escudero, V., Nuñez-Corcuera, B., Delgado-Cerezo, M., Torii, K. U., et al. (2016). ERECTA and BAK1 receptor like kinases interact to regulate immune responses in Arabidopsis. Front. Plant Sci. 7, 897. doi: 10.3389/fpls.2016.00897
Kumar, S., Stecher, G., Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Lehti-Shiu, M. D., Shiu, S. H. (2012). Diversity, classification and function of the plant protein kinase superfamily. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 367, 2619–2639. doi: 10.1098/rstb.2012.0003
Lehti-Shiu, M. D., Zou, C., Hanada, K., Shiu, S. H. (2009). Evolutionary history and stress regulation of plant receptor-like kinase/pelle genes. Plant Physiol. 150, 12–26. doi: 10.1104/pp.108.134353
Lehti-Shiu, M. D., Zou, C., Shiu, S.-H. (2012). “"Origin, diversity, expansion history, and functional evolution of the plant receptor-like kinase/pelle family,",” in Receptor-like Kinases in Plants: From Development to Defense. Eds. Tax, F., Kemmerling, B. (Berlin, Heidelberg: Springer Berlin Heidelberg), 1–22.
Lescot, M., Déhais, P., Thijs, G., Marchal, K., Moreau, Y., Van De Peer, Y., et al. (2002). PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 30, 325–327. doi: 10.1093/nar/30.1.325
Li, B., Ferreira, M. A., Huang, M., Camargos, L. F., Yu, X., Teixeira, R. M., et al. (2019). The receptor-like kinase NIK1 targets FLS2/BAK1 immune complex and inversely modulates antiviral and antibacterial immunity. Nat. Commun. 10, 4996. doi: 10.1038/s41467-019-12847-6
Li, L., Li, M., Yu, L., Zhou, Z., Liang, X., Liu, Z., et al. (2014). The FLS2-associated kinase BIK1 directly phosphorylates the NADPH oxidase RbohD to control plant immunity. Cell Host Microbe 15, 329–338. doi: 10.1016/j.chom.2014.02.009
Li, M., Liu, C., Hepworth, S. R., Ma, C., Li, H., Li, J., et al. (2022). SAUR15 interaction with BRI1 activates plasma membrane H+-ATPase to promote organ development of Arabidopsis. Plant Physiol. 189, 2454–2466. doi: 10.1093/plphys/kiac194
Li, J., Tax, F. E. (2013). Receptor-like kinases: key regulators of plant development and defense. J. Integr. Plant Biol. 55, 1184–1187. doi: 10.1111/jipb.12129
Li, J., Wen, J., Lease, K. A., Doke, J. T., Tax, F. E., Walker, J. C. (2002). BAK1, an Arabidopsis LRR receptor-like protein kinase, interacts with BRI1 and modulates brassinosteroid signaling. Cell 110, 213–222. doi: 10.1016/S0092-8674(02)00812-7
Liebrand, T. W., Van Den Berg, G. C., Zhang, Z., Smit, P., Cordewener, J. H., America, A. H., et al. (2013). Receptor-like kinase SOBIR1/EVR interacts with receptor-like proteins in plant immunity against fungal infection. Proc. Natl. Acad. Sci. U.S.A. 110, 10010–10015. doi: 10.1073/pnas.1220015110
Lin, C. W., Huang, L. Y., Huang, C. L., Wang, Y. C., Lai, P. H., Wang, H. V., et al. (2017). Common stress transcriptome analysis reveals functional and genomic architecture differences between early and delayed response genes. Plant Cell Physiol. 58, 546–559. doi: 10.1093/pcp/pcx002
Lin, W., Li, B., Lu, D., Chen, S., Zhu, N., He, P., et al. (2014). Tyrosine phosphorylation of protein kinase complex BAK1/BIK1 mediates Arabidopsis innate immunity. Proc. Natl. Acad. Sci. U.S.A. 111, 3632–3637. doi: 10.1073/pnas.1318817111
Liu, P. L., Du, L., Huang, Y., Gao, S. M., Yu, M. (2017). Origin and diversification of leucine-rich repeat receptor-like protein kinase (LRR-RLK) genes in plants. BMC Evol. Biol. 17, 47. doi: 10.1186/s12862-017-0891-5
Liu, Y., Huang, X., Li, M., He, P., Zhang, Y. (2016). Loss-of-function of Arabidopsis receptor-like kinase BIR1 activates cell death and defense responses mediated by BAK1 and SOBIR1. New Phytol. 212, 637–645. doi: 10.1111/nph.14072
Lu, D., Wu, S., Gao, X., Zhang, Y., Shan, L., He, P. (2010). A receptor-like cytoplasmic kinase, BIK1, associates with a flagellin receptor complex to initiate plant innate immunity. Proc. Natl. Acad. Sci. U.S.A. 107, 496–501. doi: 10.1073/pnas.0909705107
Ma, X., Xu, G., He, P., Shan, L. (2016). SERKing coreceptors for receptors. Trends Plant Sci. 21, 1017–1033. doi: 10.1016/j.tplants.2016.08.014
Miao, H., Wang, L., Qu, L., Liu, H., Sun, Y., Le, M., et al. (2023). Genomic evolution and insights into agronomic trait innovations of Sesamum Species. Plant Commun. 5, 100729. doi: 10.1016/j.xplc.2023.100729
Mishra, D., Suri, G. S., Kaur, G., Tiwari, M. (2021). Comprehensive analysis of structural, functional, and evolutionary dynamics of Leucine Rich Repeats-RLKs in Thinopyrum elongatum. Int. J. Biol. Macromol. 183, 513–527. doi: 10.1016/j.ijbiomac.2021.04.137
Mosher, S., Seybold, H., Rodriguez, P., Stahl, M., Davies, K. A., Dayaratne, S., et al. (2013). The tyrosine-sulfated peptide receptors PSKR1 and PSY1R modify the immunity of Arabidopsis to biotrophic and necrotrophic pathogens in an antagonistic manner. Plant J. 73, 469–482. doi: 10.1111/tpj.12050
Nam, K. H., Li, J. (2002). BRI1/BAK1, a receptor kinase pair mediating brassinosteroid signaling. Cell 110, 203–212. doi: 10.1016/S0092-8674(02)00814-0
Peng, H. C., Kaloshian, I. (2014). The tomato leucine-rich repeat receptor-like kinases SlSERK3A and SlSERK3B have overlapping functions in bacterial and nematode innate immunity. PloS One 9, e93302. doi: 10.1371/journal.pone.0093302
Pertea, M., Kim, D., Pertea, G. M., Leek, J. T., Salzberg, S. L. (2016). Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 11, 1650–1667. doi: 10.1038/nprot.2016.095
Pertea, M., Pertea, G. M., Antonescu, C. M., Chang, T. C., Mendell, J. T., Salzberg, S. L. (2015). StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 33, 290–295. doi: 10.1038/nbt.3122
Platre, M. P., Satbhai, S. B., Brent, L., Gleason, M. F., Cao, M., Grison, M., et al. (2022). The receptor kinase SRF3 coordinates iron-level and flagellin dependent defense and growth responses in plants. Nat. Commun. 13, 4445. doi: 10.1038/s41467-022-32167-6
Robinson, M. D., Oshlack, A. (2010). A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 11, R25. doi: 10.1186/gb-2010-11-3-r25
Roux, M., Schwessinger, B., Albrecht, C., Chinchilla, D., Jones, A., Holton, N., et al. (2011). The Arabidopsis leucine-rich repeat receptor-like kinases BAK1/SERK3 and BKK1/SERK4 are required for innate immunity to hemibiotrophic and biotrophic pathogens. Plant Cell 23, 2440–2455. doi: 10.1105/tpc.111.084301
Shang, Y., Dai, C., Lee, M. M., Kwak, J. M., Nam, K. H. (2016). BRI1-associated receptor kinase 1 regulates guard cell ABA signaling mediated by open stomata 1 in Arabidopsis. Mol. Plant 9, 447–460. doi: 10.1016/j.molp.2015.12.014
Shiu, S. H., Bleecker, A. B. (2001). Receptor-like kinases from Arabidopsis form a monophyletic gene family related to animal receptor kinases. Proc. Natl. Acad. Sci. U.S.A. 98, 10763–10768. doi: 10.1073/pnas.181141598
Shiu, S. H., Karlowski, W. M., Pan, R., Tzeng, Y. H., Mayer, K. F., Li, W. H. (2004). Comparative analysis of the receptor-like kinase family in Arabidopsis and rice. Plant Cell 16, 1220–1234. doi: 10.1105/tpc.020834
Shumayla, Sharma, S., Kumar, R., Mendu, V., Singh, K., Upadhyay, S. K. (2016). Genomic dissection and expression profiling revealed functional divergence in Triticum aestivum leucine rich repeat receptor like kinases (TaLRRKs). Front. Plant Sci. 7, 1374. doi: 10.3389/fpls.2016.01374
Singh, A., Breja, P., Khurana, J. P., Khurana, P. (2016). Wheat brassinosteroid-insensitive1 (TaBRI1) interacts with members of TaSERK gene family and cause early flowering and seed yield enhancement in Arabidopsis. PloS One 11, e0153273. doi: 10.1371/journal.pone.0153273
Soltabayeva, A., Dauletova, N., Serik, S., Sandybek, M., Omondi, J. O., Kurmanbayeva, A., et al. (2022). Receptor-like kinases (LRR-RLKs) in response of plants to biotic and abiotic stresses. Plants (Basel) 11, 2660. doi: 10.3390/plants11192660
Song, W., Wang, B., Li, X., Wei, J., Chen, L., Zhang, D., et al. (2015). Identification of immune related LRR-containing genes in maize (Zea mays L.) by genome-wide sequence analysis. Int. J. Genomics 2015, 231358. doi: 10.1155/2015/231358
Sun, X., Wang, G. L. (2011). Genome-wide identification, characterization and phylogenetic analysis of the rice LRR-kinases. PloS One 6, e16079. doi: 10.1371/journal.pone.0016079
Sun, R., Wang, S., Ma, D., Liu, C. (2018). Genome-wide analysis of LRR-RLK gene family in four Gossypium species and expression analysis during cotton development and stress responses. Genes (Basel) 9, 592. doi: 10.3390/genes9120592
Trenker, R., Jura, N. (2020). Receptor tyrosine kinase activation: From the ligand perspective. Curr. Opin. Cell Biol. 63, 174–185. doi: 10.1016/j.ceb.2020.01.016
Tunc-Ozdemir, M., Jones, A. M. (2017). BRL3 and AtRGS1 cooperate to fine tune growth inhibition and ROS activation. PloS One 12, e0177400. doi: 10.1371/journal.pone.0177400
Wang, X., Li, X., Meisenhelder, J., Hunter, T., Yoshida, S., Asami, T., et al. (2005). Autoregulation and homodimerization are involved in the activation of the plant steroid receptor BRI1. Dev. Cell 8, 855–865. doi: 10.1016/j.devcel.2005.05.001
Wang, Z., Meng, P., Zhang, X., Ren, D., Yang, S. (2011). BON1 interacts with the protein kinases BIR1 and BAK1 in modulation of temperature-dependent plant growth and cell death in Arabidopsis. Plant J. 67, 1081–1093. doi: 10.1111/j.1365-313X.2011.04659.x
Wang, Z. Y., Seto, H., Fujioka, S., Yoshida, S., Chory, J. (2001). BRI1 is a critical component of a plasma-membrane receptor for plant steroids. Nature 410, 380–383. doi: 10.1038/35066597
Wang, Y., Tang, H., Debarry, J. D., Tan, X., Li, J., Wang, X., et al. (2012). MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 40, e49. doi: 10.1093/nar/gkr1293
Wang, Y., Yan, X., Xu, M., Qi, W., Shi, C., Li, X., et al. (2022). TMK1-mediated auxin signal regulates membrane-associated clathrin in Arabidopsis roots. J. Integr. Plant Biol. 65, 82–99. doi: 10.1111/jipb.13366
Xu, K., Jourquin, J., Njo, M. F., Nguyen, L., Beeckman, T., Fernandez, A. I. (2021). The phloem intercalated with xylem-correlated 3 receptor-like kinase constitutively interacts with brassinosteroid insensitive 1-associated receptor kinase 1 and is involved in vascular development in Arabidopsis. Front. Plant Sci. 12, 706633. doi: 10.3389/fpls.2021.706633
Yan, W., Ni, Y., Liu, X., Zhao, H., Chen, Y., Jia, M., et al. (2021). The mechanism of sesame resistance against Macrophomina phaseolina was revealed via a comparison of transcriptomes of resistant and susceptible sesame genotypes. BMC Plant Biol. 21, 159. doi: 10.1186/s12870-021-02927-5
Yang, W., Zhang, B., Qi, G., Shang, L., Liu, H., Ding, X., et al. (2019). Identification of the phytosulfokine receptor 1 (OsPSKR1) confers resistance to bacterial leaf streak in rice. Planta 250, 1603–1612. doi: 10.1007/s00425-019-03238-8
Zhang, H., Miao, H., Wang, L., Qu, L., Liu, H., Wang, Q., et al. (2013). Genome sequencing of the important oilseed crop Sesamum indicum L. Genome Biol. 14, 401. doi: 10.1186/gb-2013-14-1-401
Zhang, Y. P., Zhang, Y. Y., Thakur, K., Zhang, F., Hu, F., Zhang, J. G., et al. (2021). Integration of miRNAs, degradome, and transcriptome omics uncovers a complex regulatory network and provides insights into lipid and fatty acid synthesis during sesame seed development. Front. Plant Sci. 12, 709197. doi: 10.3389/fpls.2021.709197
Zhou, F., Guo, Y., Qiu, L. J. (2016). Genome-wide identification and evolutionary analysis of leucine-rich repeat receptor-like protein kinase genes in soybean. BMC Plant Biol. 16, 58. doi: 10.1186/s12870-016-0744-1
Zhou, Y., Sun, L., Wassan, G. M., He, X., Shaban, M., Zhang, L., et al. (2019). GbSOBIR1 confers Verticillium wilt resistance by phosphorylating the transcriptional factor GbbHLH171 in Gossypium barbadense. Plant Biotechnol. J. 17, 152–163. doi: 10.1111/pbi.12954
Keywords: Sesamum indicum, LRR-RLK, evolution, expression profiles, Macrophomina phaseolina
Citation: Yan W, Ni Y, Zhao H, Liu X, Jia M, Zhao X, Li Y, Miao H, Liu H and Zhang H (2024) Comprehensive analysis of sesame LRR-RLKs: structure, evolution and dynamic expression profiles under Macrophomina phaseolina stress. Front. Plant Sci. 15:1334189. doi: 10.3389/fpls.2024.1334189
Received: 06 November 2023; Accepted: 26 January 2024;
Published: 12 February 2024.
Edited by:
Choong-Min Ryu, Korea Research Institute of Bioscience and Biotechnology (KRIBB), Republic of KoreaReviewed by:
Neelakantan Arumugam, Pondicherry University, IndiaCopyright © 2024 Yan, Ni, Zhao, Liu, Jia, Zhao, Li, Miao, Liu and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hongmei Miao, bWlhb2hvbmdtZWljaGluYUAxNjMuY29t; Hongyan Liu, bGl1aHkxMjE5QDE2My5jb20=; Haiyang Zhang, emhhbmdoYWl5YW5nQHp6dS5lZHUuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.