
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 12 March 2024
Sec. Plant Symbiotic Interactions
Volume 15 - 2024 | https://doi.org/10.3389/fpls.2024.1332745
 Devashish Pathak
Devashish Pathak Archna Suman*
Archna Suman* Pushpendra Sharma
Pushpendra Sharma Krishnan Aswini
Krishnan Aswini Venkadasamy Govindasamy
Venkadasamy Govindasamy Shrikant Gond
Shrikant Gond Rana Anshika
Rana AnshikaCommunity-forming traits (CFts) play an important role in the effective colonization of plant-growth-promoting bacterial communities that influence host plants positively by modulating their adaptive functions. In this study, by considering plant-growth-promoting traits (PGPts) and community-forming traits (CFts), three communities were constructed, viz., SM1 (PGPts), SM2 (CFts), and SM3 (PGPts+CFts). Each category isolates were picked up on the basis of their catabolic diversity of different carbon sources. Results revealed a distinctive pattern in the colonization of the communities possessed with CF traits. It was observed that the community with CFts colonized inside the plant in groups or in large aggregations, whereas the community with only PGPts colonized as separate individual and small colonies inside the plant root and leaf. The effect of SM3 in the microcosm experiment was more significant than the uninoculated control by 22.12%, 27.19%, and 9.11% improvement in germination percentage, chlorophyll content, and plant biomass, respectively. The significant difference shown by the microbial community SM3 clearly demonstrates the integrated effect of CFts and PGPts on effective colonization vis-à-vis positive influence on the host plant. Further detailed characterization of the interaction will take this technology ahead in sustainable agriculture.
In the face of global challenges such as climate change, use of high concentration chemicals, and increasing food demand, sustainable agriculture practices have gained paramount importance. One such sustainable approach is the utilization of plant-growth-promoting bacteria (PGPB), which exhibit characteristics like nutrient solubilization, phytohormone production, and antagonism against pathogens (Javed et al., 2021; Sharf et al., 2021). Selecting the most effective isolates that can colonize effectively in the plant rhizosphere is the crucial step in designing the bioinoculants for integrated nutrient management approach (Oo et al., 2020) to enhance crop productivity while reducing the reliance on chemical fertilizers and pesticides (Cao et al., 2023).
PGPB is a diverse group of soil-dwelling and endophytic bacteria that inhabit the rhizosphere and endosphere of plants (Thakur et al., 2023), where they form intricate interactions with their host plants and contributes to their growth and overall well-being (Shoaib et al., 2020). Harnessing the full potential of PGPB involves not only identifying beneficial bacterial isolates but also their interactions within the complex microbial communities formed in the rhizosphere (Bulgarelli et al., 2013; Trivedi et al., 2020). The intricate communities are characterized by a myriad of interactions (Backer et al., 2018). Successful microbial communities are developed by the individual microbial partners possessing certain community-forming traits (CFts) such as biofilm formation, exopolysaccharide (EPS) production, and others like motility and PHB production. These traits contribute to the establishment of stable and cooperative environments that enhance the survival and functionality of individual microbes in various ecological niches (Wu et al., 2019; Karygianni et al., 2020; Hung et al., 2023). The role of CFts were least understood in plant growth and development.
Besides all these activities of microbial isolates, there is more to reveal about the impact of key dominant genera, which play a major role in community formation (Aswini et al., 2023; Yuan et al., 2023). The presence of key dominant genera in a community may mimic as synthetic natural community (Großkopf and Soyer, 2014; Shayanthan et al., 2022). The concept of synthetic communities (syncoms) has gained prominence as a tool for deciphering the synergistic effects of different microbial species on plant health (Liu et al., 2022; Hao et al., 2023).
Various studies conducted in the past discussed about the plant growth promoters and their effect over various plant species (Bhat et al., 2023; Carreiras et al., 2023). There is a gap to see the integrated effect of the CFts over PGP bacterial activity and their cumulative effect on plant growth and development as well as the practical construction and evaluation of synthetic communities (SMs). This study investigates how these CFts contribute to the establishment of beneficial interactions within the rhizosphere and their subsequent impact on wheat plant growth. We hypothesized that the cumulative effect of CFts and PGP bacterial community works in a better way than only PGP bacterial community.
By assembling PGP bacteria with known CFts, this study aimed to elucidate how these synthetic communities influence wheat plant growth in a controlled microcosm environment by shedding light on these complex interactions. We aimed to contribute valuable insights into sustainable agricultural practices that can mitigate the challenges of an ever-changing world and pave the way for increased crop yields and food security.
Two sources for bacterial isolation were taken: one was from the culture bank of our lab, and another was done by isolation process from wheat rhizospheric soil and endosphere. A total of 187 cultures of wheat and maize was taken from the culture bank of our lab (Marag and Suman, 2018; SaiPrasad et al., 2021), Division of Microbiology, ICAR–IARI, New Delhi, India. Working stock and slants were made in nutrient broth and nutrient agar and kept at 4°C for further work. Root and rhizospheric soil of the wheat variety HD2967 were taken from the field of the Division of Genetics, ICAR–IARI, New Delhi (28° 36′ 50″ N:77° 12′ 32″ E), Nashik (20° 04′ 59″ N:74° 07′ 00″ E), Samastipur (25° 51′ 39″ N:85° 46′ 56″ E), Indore (22° 43′ 31″ N:75° 51′ 55″ E), and Shimla (31° 06′ 12″ N:77° 10′ 20″ E), and HD2833 from Coimbatore (11° 22′ 12″ N:06° 48′ 00″ E) and were collected on the given coordinates mentioned. Samples were taken aseptically to the laboratory and stored at 4°C until further analysis.
Bacteria were isolated from wheat endosphere and rhizosphere of varieties HD2967 and HD2833. Isolation was done by using nutrient agar and tryptic soy agar. First, rhizospheric soil was separated carefully from root samples using sterile brush under aseptic conditions. One gram of soil was serially diluted in 10 ml of saline water (0.8%, w/v), and fourth to fifth dilutions were used to isolate the rhizospheric bacteria. For endospheric bacterial isolation, root samples were surface sterilized with 1% NaOCl for 1 min followed by 30 s of ethanol treatment followed by three times washing with sterile water (Aswini et al., 2023). One gram of root sample was macerated in sterile mortar and pestle, and similar dilutions were used for isolation of endophytic bacteria. The plates were incubated at 28 ± 2°C for 3 days and observed regularly. The isolates were selected based on their morphology, which were purified (Abd El Gawad et al., 2010; Taye et al., 2021) and preserved in 30% glycerol at −20°C till further study.
CFts are the traits which require bacterial quorum sensing mechanism to show their optimality, e.g., biofilm formation and EPS production. Some other traits are also there, which are essential in a community to survive, viz., motility and PHB production. In a community, a proper division of carbon source or nutrients is also essential, which requires different metabolic diversity pattern among isolates (Wei et al., 2020; Dal Bello et al., 2021).
All bacterial isolates were checked for the ability to form biofilm on a microtiter plate (O'Toole, 2011). Biofilm production was done with the crystal violet staining method, where bacterial isolates were first grown separately to reach OD = 1, then reinoculated into the ELISA 96-well plate with biofilm assay medium, i.e., M63 minimal medium [KH2PO4 (22 mM), K2HPO4 (40 mM), and (NH4)2SO4 (15 mM)] supplemented with MgSO4 (1 mM), casamino acid (0.5%), and glucose (0.2%). Freshly prepared bacterial culture and medium were mixed in 1:100 (V/V) ratios in three replicates. Optical density (OD) of grown bacterial culture was taken with respect to the OD of the biofilm or adhered cell to the surface of the wall for 24, 48, and 72 h of growth. Biofilm OD was taken by staining the adhered cell with crystal violet followed by solubilization of the stained cell with 30% acetic acid solution.
EPS production was quantified with Congo red binding assay (Quintieri et al., 2020). Prior to the activity, equal cell density was maintained of each isolate and grown to the OD = 1 in Luria–Bertani (LB) broth. Each isolate was reinoculated in M63 broth for 72 h. After the end of the incubation time, the broth was centrifuged at 10,397 rpm for 5 min, and the pellets of the culture were again suspended in 1 ml of 40 µg ml−1 of Congo red solution. Suspended cultures were incubated for 90 min with vigorous shaking for 10 min at each interval of 15 min for three times. The samples were again centrifuged at 9,600 rpm for 5 min, and OD of the supernatant was taken at 490 nm. The final OD revealed the amount of EPS produced, which was the OD of the blank subtracted from the OD of the inoculated. The amount of EPS production was obtained by matching the OD with the standard curve of different concentrations of Congo red.
Motility of the isolates was performed in the nutrient semi-solid agar media supplemented with 1% TTC. The freshly grown cultures were stabbed straight into the test tube within the medium and were incubated at 35 ± 2°C for 24 h (Kelly and Fulton, 1953). As cells start to replicate and move, they turned TTC into the red colored compound triphenylformazan, and the extent of motility was observed with the expanded colored area inside the tube.
Polyhydroxy butyrate (PHB) is an important compound produced by bacterial isolates, which was used to observe in an abundance of sugar content with minimum competition. PHB is stored within the bacterial cell for use in stressful conditions. The PHB producers were identified with the help of Sudan black dye (60 mg of dye in 200 ml of 70% ethanol). The freshly grown cultures in 1% glucose supplemented nutrient broth for 48 h at 35± 2°C were spotted over the nutrient agar plate (also supplemented with 1% glucose) incubated at 35 ± 2°C for 24 h to allow proper growth. Sudan black dye was poured over the culture plate to cover it all. After 1 min, the extra stain was drained out of the plate and was washed off with the help of 96% ethanol (Williamson and Wilkinson, 1958).
HiCarbo™ Kit (KB 009, HiCarbohydrate™ kit) and HiAssorted™ Biochemical Test Kit (KB002) of HiMedia was used to determine the utilization of various carbon sources as carbohydrates and carboxyl type, along with this some amino acids and enzymatic activities (viz., esculin hydrolyses, urease, β-galactosidase activity, nitrate reductase, and phenylalanine deaminase) by isolates. The kit consists of three strips A, B, and C with all biochemical compounds. A freshly prepared (with 0.5 OD at 620 nm) 50-µl culture of all bacterial isolates was inoculated over the substrate bound inside each well of the strips and incubated at 35± 2°C for 24 h. The change in color showed the positive test for the particular kind of substrate (Kumar et al., 2021).
A set of 294 selected wheat and maize rhizospheric and endospheric microbials was screened for the fixation of nitrogen and solubilization of phosphorus, potassium, zinc, and calcium carbonate. Nitrogen fixation was done with the stab culture in semisolid JNF (Jensen’s nitrogen-free) medium followed by ARA estimation by gas chromatography [GC-2014, Shimadzu, Japan with a flame ionization detector (FID)]. Phosphorus, potassium, and zinc solubilization was observed over the plate of Pikovskaya media (with 0.5% tricalcium phosphate), Aleksandrov media (containing 0.2% of potassium aluminosilicate), and nutrient agar fortified with 0.1% insoluble zinc compound (ZnO), respectively, as described in Verma et al. (2015). Calcite solubilization was observed over the agar plate containing (g 100 ml−1) CaCO3 (0.5), dextrose (1.0), [NH4]2SO4 (0.05), KCl (0.02), MgSO4.7H2O (0.1), yeast extract (0.5), and agar (1.5) with pH 7 (Rana et al., 2015). The plates with respective isolates showing solubilization zones for macro- and micro-nutrients were considered as positive for respective traits. For the quantification of phosphorus, Pikovskaya media (50 ml) were inoculated with each freshly grown positive isolate and incubated at 28 ± 2°C at 180 rpm; the supernatant centrifuged at 8,000 rpm was collected to estimate phosphorus colorimetrically by molybdenum-blue assay (Liu et al., 2014).
All purified isolates were screened for various phytohormones, viz., gibberellic acid (GA3) and indole acetic acid (IAA). GA3 estimation was done as described by Brown and Burlingham (1968). Isolates were inoculated in 20 ml of LB broth and kept overnight for incubation at 30 ± 2°C. After incubation for 7 days, the culture broth was centrifuged at 8,000 rpm for 10 min; 15 ml of supernatant was mixed vigorously with 2 ml of zinc acetate solution followed by mixing of 2 ml of potassium ferrocyanide followed by centrifugation at 8,000 rpm for 10 min and mixing of 5 ml HCl (30%); and the final solution was incubated for 45 min. After the period of incubation, the OD was observed at 254 nm. Five milliliters of 5% HCl was used as blank. The concentration of GA in each culture medium was determined by comparison with the standard GA curve. Indole 3-acetic acids (IAA) estimation was done by the growing of culture in nutrient broth fortified with 1% tryptophan and incubation for 15 days at 28 ± 2°C. The uninoculated tube was maintained as control. The culture broth was centrifuged at 8,000 rpm for 10 min, 2 ml of the supernatant obtained was mixed with 4 ml of Salkowski reagent (50 ml, 35% perchloric acid; 1 ml 0.5 FeCl3) and two drops of concentrated orthophosphoric acid; the mixture was incubated in dark condition for 30 min for color development. The developed pink color depicts IAA production qualitatively; for quantitative estimation, OD was taken at 530 nm by using a spectrophotometer (Halo DB-20S). The concentration of IAA was estimated with the help of a standard curve of 10–100 μg ml−1 following Bric et al. (1991).
ACC-deaminase activity and reduction in ACC was described by Glick (1995) and Li et al. (2011). Accordingly, freshly grown isolates were centrifuged in nutrient broth at 8,000 rpm for 10 min, and the obtained cell pellet was washed with DF medium as described in Glick (1995). The washed pellet was again suspended in DF-ACC medium with a final ACC concentration of 3.0 mmol l−1 for 24 h incubation. The incubated culture was again centrifuged with 0.1 ml supernatant and diluted 10 times with DF medium. The final volume was mixed with double volume of ninhydrine reagent (50 mg ninhydrin + 1.5 mg ascorbic acid + 30 ml ethylene glycol, equal volume of 1 mol l−1 citrate buffer was mixed before use (pH 6.0). The DF medium was considered as blank. Deep purple color to light purple color reveals the extent of ACC deaminase activity, which was measured at 570 nm. The result was compared with the standard curve of ACC in DF medium for quantified value.
The biocontrol activity of microbes involves the traits to suppress or control the growth of harmful or pathogenic microbes through the production of siderophores, hydrogen cyanide (HCN), and ammonia (NH3). The selected isolates were screened for these traits and antagonism test against pathogenic fungi. Siderophore qualitative estimation was done with the change in the color of CAS agar plate from blue to yellow. The quantification of the siderophore was done for qualitatively selected isolates from the plate assay. The selected isolates were inoculated in LB broth. When the cells attained a cfu count of 108 (approximate), the broth was centrifuged, and pellets were discarded. Then, 0.5 ml of the supernatant was mixed with 0.5 ml of CAS reagent prepared as mentioned by Schwyn and Neilands (1987). After 20 min, observations were taken in the form of OD at 630 nm with the help of a spectrophotometer. The reading was quantified in terms of percentage siderophore unit (psu) by Shelley (1994). Ammonia production was identified with the help of Nessler’s reagent, which was added (1 ml) in the well-grown culture broth (10 ml); change in the color from yellow to brown or brickish red confirmed the production of NH3 (Cappuccino and Sherman, 1992). HCN quantitative estimation was done by colorimetric analysis of the changed picric-acid-soaked filter paper. In this regard, the change in the color of alkaline picric-acid-soaked filter paper was eluted in double-distilled water (ddH2O), and OD was taken at 625 nm (Rijavec and Lapanje, 2016). Different concentrations of potassium ferrocyanide compound were used to prepare standard curve for actual cyanide content, which was used to calibrate the HCN produced by bacterial isolates.
Plate assay was carried out for antagonism against pathogenic fungi. All selected isolates were tested for the antagonistic ability against various pathogens., e.g., Tilletia, Bipolaris, Puccinia, and Fusarium over the PDA plates to observe the positive and negative interaction with the pathogenic fungi by bacterial isolates compared with the control plate of the respective fungus (Sharma et al., 2023).
All isolates were selected based on their ability to show better CFts and PGPts. The isolation of genomic DNA was done using the Quick-DNA™ Fungal/Bacterial DNA Miniprep Kit and subjected to a gel run to check the quality of purified DNA. A partial sequence of the 16S rRNA gene was amplified with forward pA (F)—(5′AGAGTTTGATCCTGGCTCAG-3′) and reverse pH (R)—(5′AAGGAGGTGATCCAGCCGCA-3′) as the universal primer using a PCR thermocycler (HiMedia Prima-96). PCR products were delivered to HiMedia for Sanger sequencing and purification. The obtained raw forward and reverse sequences were further processed for contig formation using BioEdit. Misaligned reads in between the contigs, which were used for identification, was checked with the help of Decipher 3.0. To identify, NCBI-BLAST of the final contigs were done and matched with the data of microorganisms having the highest percentage of similarity (>97%) in the NCBI database.
The test of compatibility among isolates was checked over the plate by cross-streaking method. All isolates were cross-streaked one by one so that every isolate could cross another at once with everyone. The isolates that grown together at junction were considered compatible and were taken for further grouping (Pugsley and Oudega, 1987). Along with this, growth patterns of all individual isolates were correlated with the mixed culture (called interaction analysis, not shown here), which revealed about the interaction between the individual isolates.
Each isolate with positive compatibility was grown separately in a 50-ml flask containing 10 ml nutrient broth. Each isolate was allowed to grow up to 1 OD. The selection of microbes for each community was based on a bonitur scale for CFts, PGPts, and CFts+PGPts. The isolates with maximum bonitur value for plant-growth-promoting traits were grouped and termed “PGPts” or SM1. Similarly, the isolates having high bonitur value for community-forming traits were grouped and termed “CFts” or SM2. In addition, the compatible isolates having both the traits (PGPts and CFts) were grouped and termed “CFts+PGPts” or SM3. The final volume (100 ml) of each community was prepared by mixing 10 ml of each isolate, where per milliliter of culture broth contains approximately 1×108 cells.
Surface-sterilized wheat seeds with 1% NaOCl for 60 s followed by ethanol (70%) for 30 s followed by three times wash with sterile double-distilled water (ddH2O) were used to check the colonization potential of the isolates. Each group of the community, which was previously grown separately to reach 1 OD, was allowed to inoculate the prepared seeds for 1.5 h. Inoculated or treated seeds were pressed gently over the soft agar plate (0.8% agar) (five seeds/plate) inside the laminar air flow, to stick over the plates, and were covered with the upper lid. The triplicates of uninoculated (control) and inoculated plates were incubated for 5−7 days in an upright condition at 20°C till the seedling stage. Prior to the visualization, the plates with proper seedlings were flooded with TTC stain, containing TTC (1.5 g l−1) and malic acid (625 mg l−1) in potassium phosphate (0.05 M) buffer (pH 7.0) (Thomas and Sekhar, 2014) for 10 h. Visualization of the leaf and root tissue section was done under the bright field microscope (De winter) after proper washing of the sample with sterile ddH2O.
To evaluate the effect of microbial communities, a pot experiment was taken in the glass house of the Division of Microbiology, ICAR-IARI, New Delhi. The weather condition was 15°C–20°C with 6 h of bright sunny day. The pot (20 cm diameter and 20 cm height) used was presanitized with 1% NaOCl followed by washing with sterile distilled water. The pot was filled with 3 kg of sterile (3 days of sterilization at 120°C 103.42 kPa for 1 h) rooting medium (soil: vermiculite: sand, 3:1:1 ratio). The required fertilizer dose (RDF for wheat 120:60:60 NPK kg ha−1) was mixed with the soil according to the weight of soil in treatments before sowing. Split nitrogen dose application was maintained before and after the germination of wheat seedlings. Overnight grown culture with similar OD was mixed in equal volume according to their communities (CFts, PGPts, and PGPts+CFts). Sterile seeds were treated with each community separately for 2–3 h followed by drying in laminar air flow. The pot experiment was set up in a completely randomized design (CRD) in triplicate having the following treatments: T1–100% RDF, T2–50% RDF + PGP, T3–50% RDF + CFts, T4–50% RDF + PGP + CFts, and T5–50% RDF as negative control. Under this experiment plant biometric data like plant height (cm), root length (cm), and chlorophyll content (nmol cm -1) were taken at 60 DAS and 90 DAS. Along with this, germination percentage and plant dry weight were taken at initiation and harvesting stage, respectively.
Analysis of plant biometric data was done over the OPSTAT for ANOVA followed by Duncan’s post hoc analysis of microcosm experiments, whereas for the visualization of the metabolic pattern of different carbon sources utilization, construction of constellation tree was done for each isolate by hierarchical cluster method in JMP software (v17.0).
Among six wheat samples from different regions of the country, the range of bacterial load in rhizospheric soil was 5.4×105 to 8.7×105 cfu g−1 in which rhizospheric the soil sample from Shimla had the highest count followed by that from Delhi. The range of root endophytic bacterial count was between 2.4×105 and 5.1×105 cfu g−1, in which Shimla had highest count followed by Coimbatore (Table 1). A total of 107 morphotypes were selected in which 53 were rhizospheric soil isolates and 54 were root endophytic isolates (Supplementary Table S1).
General CFts, which include EPS, biofilm production, PHB production, and motility, were considered in this experiment (Figure 1). The range of EPS production is from 2.281 ± 0.021 µg ml−1 to 28.72 ± 0.630 µg ml−1 with mean average production of 14.88 µg ml−1. Among all isolates, the highest producer of the EPS is DWE-1 followed by NWP-12. Whereas in biofilm production, the range of the biofilm formation was found from 0.003 OD to 3.88 OD, among 294 isolates, seven showed the highest production of biofilm, which was >3.8 OD. Among all isolates, there were five levels of motility shown, 0%, 25%, 75%, and 100% based on the coverage area of the test tube, in that 29 isolates were showing maximum motility, which covered 100% of the area of the tube in 24 h. In PHB production, a total of 11 isolates retained the stain of Sudan black dye in their colony and had not wash off from 95% ethanol. Among all 294 isolates, the top 17 cultures were screened based on their bonitur scale and were further kept for community purposes (Table 2; Supplementary Table S2).
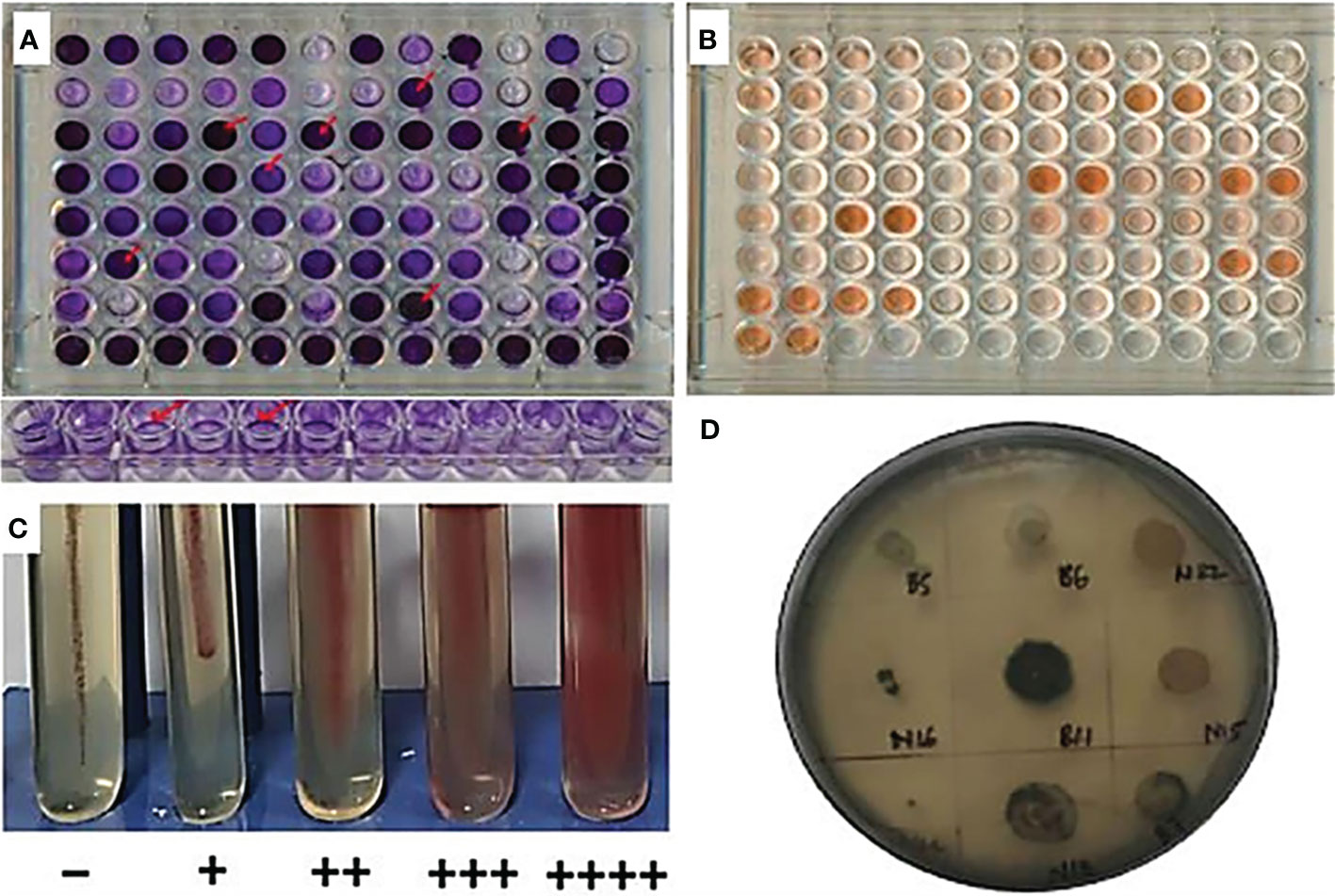
Figure 1 Community forming traits. (A) Crystal violet-stained biofilm in microtiter plates; (B) Congo red dye method for EPS; (C) motility potential of bacterial isolates in nutrient agar with 1% TTC; and (D) Sudan Black dye stained PHB-positive isolate.
Among all 294 isolates, K-solubilization was seen in 58 isolates, and the range of index was from 0.75 to 3.04, in which NWR-15 was showing the highest solubilization index followed by NWR-14. P-solubilization ability ranged between 4.2 ppm and 44.61 ppm, where NWPZ-60 showed the highest solubilization of phosphorus, i.e., 44.61 ppm followed by PHM5–21 (37.45 ppm). Zinc solubilization was seen in 32 isolates with range of 0.678–2.764 of PC4–29. Coming to the Ca complex, a total of 63 isolates out of 294 were showing solubilization, in which 13 isolates were having maximum solubilization potential of Ca complex (Table 3).
In phytohormone production, the evaluation of isolates was done for GA and IAA. The production of GA and IAA was observed by the change in the color of the centrifuged culture broth with respective reagents (Figure 2). In the quantitative estimation of GA production test, out of 294 bacterial strains, 73 bacterial strains showed positive result in the range of 2.40–17.73 ppm. Among all 73 positive isolates, N14 was considered as the highest producer of GA. IAA production was shown by 77 isolates with minimum to maximum range of 0.5–53.73 ppm. Among all, PHM5-68 showed maximum production of IAA, i.e., 53.73 ppm, whereas ACC deaminase activity was observed from 0.05 ppm to 1.58 ppm, in which DWE-2 showed the maximum production among all (Table 3; Supplementary Table S2).
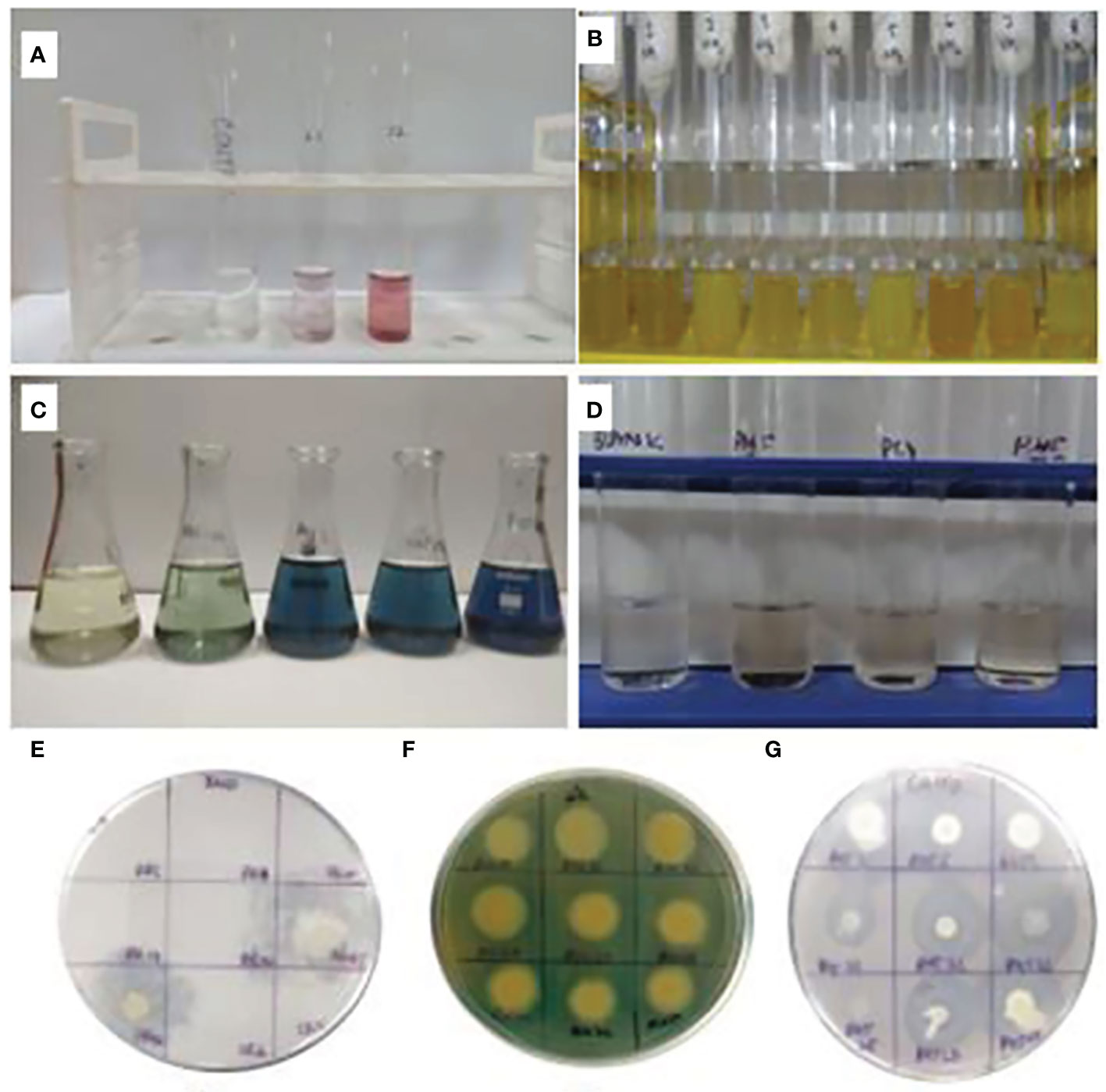
Figure 2 Plant-growth-promoting traits. (A) IAA production; (B) ammonia production; (C) phosphorus estimation; (D) GA activity; (E) Zn solubilization; (F) siderophore production; (G) CaCO3 solubilization.
There are various traits for biocontrol mechanisms that control the growth of harmful or pathogenic microbes. In this study, HCN production, siderophore production, and ammonia production were taken as three important ways responsible for the biocontrol activity. HCN quantification was done to estimate the produced HCN from bacterial isolates. Among all 294 isolates, 87 isolates showed positive behavior for HCN production with a range of 0.58–63.2 ppm. The most potential isolate for HCN production among all isolates was PHM5-9 followed by TWR-9. Ammonia production was seen for 54 isolates from 294 isolates, in which 11 isolates showed higher production (++++) than others. In siderophore quantification, the range of siderophore production was from 8.04 ppm to 61.72 ppm produced from 68 isolates, where PZ-27 showed the highest production of siderophore at 72.06 ppm followed by PC4-29 (61.72 ppm) (Table 3).
Another biocontrol activity was assessed, i.e., antagonism against pathogenic fungi. The antagonistic activity of the isolates was observed against Bipolaris sorokiniana, Fusarium graminearum, Puccinia striiformis, and Tilletia indica. The bacterial isolates showing antagonistic activity did not allow any kind of pathogenic fungi to grow and degenerated the fungal mycelium. Among all 294 bacterial isolates, 58 isolates showed positive activity against the growth of pathogenic fungi. Among the 58 isolated, 16 (PC4-29, NH-3, NH-4, 5, NWP-9, NWP-11, NWP-12, NWP-14, NEP-22, PZ-23, PZ-24, SHZ-36, CZ-44, CZ-45, CZ-46, and DWE-1) showed better inhibition irrespective of the pathogenic fungi (Table 3; Supplementary Figure S3).
Among all 294 isolates, 187 isolates of culture bank and 107 were isolated. All isolates were first identified with CFts and then for PGPts. Out of 294 isolates, 136 isolates showed positive behavior of CFts, and another 67 isolates were positive for PGPts. Out of the two groups when matched together, 131 isolates showed both characteristics. To choose the top 10 isolates for all three groups, bonitur scaling was applied, which gave the ranking of each isolate according to their efficient multi-variate characteristics. The final three groups of isolates were identified, e.g., CFts (Table 2), PGPts (Table 3), and CFts+PGPts (Table 4), based on their peculiar multi-variate traits.
The selected isolates were ensured for their diverse metabolic pattern for the utilization of different carbon sources. The grouping or community was made considering the diverse carbon utilization. In the cladogram of PGP community, results of top 16 bacterial isolates were selected based on their carbon utilization pattern (Figure 3A). The bacterial isolates that form the same clade, e.g., PHM5-22 and NEP-56, PC4-59 and SWE-12, IWE-7 and BWE-6, PHM5-73 and PHM5-9, BWR-15 and PHM5-59, and IWE-6 and PHM5-74, shared similar behavior in the carbon utilization pattern. These bacterial isolates may hinder each other’s growth or may compete for the similar food source. Among top CFt isolates, five clades (PHM5-35 and NWP-12, NWE-11 and SHZ-30, NWE-2 and DWE-3, PHM5-36 and PC4-44, and PHM5-27 and PHM5-34) were formed (Figure 3B). In the formation of PGPts+CFts group, the isolates were again checked for further carbon utilization to ensure the least competition while forming any community, in that eight clades (PHM5-9 and DWE-1, NWR-14 and DWE-2, PC4-29 and PHM5-29, NWP-9 and PHM5-72, IWE-7 and PHM5-73, IWE-6 and PHM5-71, PC4-59 and PHM5-74, and NWE-9 and NWR-15) were formed, which shared the similar carbon utilization pattern (Figure 3C).
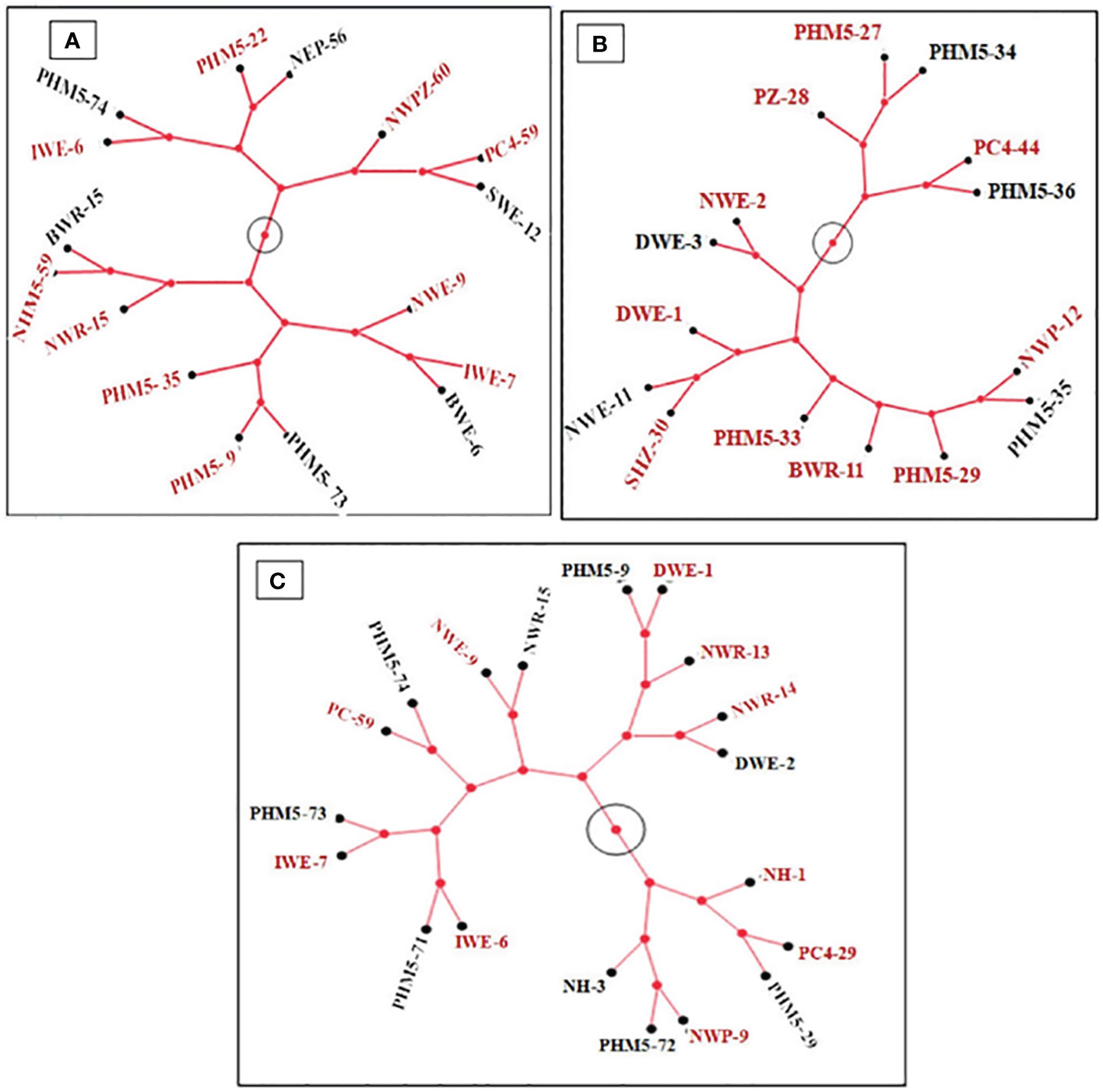
Figure 3 Constellation tree pattern of metabolic diversity of selected isolates for community formation: (A) SM1 (PGPt); (B) SM2 (CFt); (C) SM3 (PGPt+CFt). Isolate numbers in red font in a clad are selected for community development.
The bacterial isolates selected for PGPt communities were streaked across to check the compatibility over the plate. All the bacterial isolates were compatible with each other and were selected for community formation, considering the fact to take one isolate based on the higher ranking of bonitur scale from each clade of metabolic diversity tree, to minimize the competition. Each community was made up of equal number of isolates, i.e., 10. In PGPts, community consists of IWE-6, NWE-9, IWE-7, PC4-59, NWR-15, NWPZ-60, PHM5-9, PHM5-35, PHM5-22, and PHM5-59 isolates, named as SM1. CFts were constructed with DWE-1, NWP-12, BWR-11, PZ-28, SHZ-30, PHM5-27, NWE-2, PHM5-33, PHM5-29, and PC4-44, which was named as SM2, whereas PGPts+CFts was constituted with PC4-29, NWP-9, NWR-14, NH-1, NWR-13, IWE-6, new-9, IWE-7, DWE-1, and PC4-59 named as SM3. The selection of one isolate from each clade was based upon the superiority in the bonitur scale among the two. The final three communities constructed, e.g., SM1 (PGPts), SM2 (CFts), and SM3 (CFts+PGPts), were based on their peculiar multi-variate traits ready for treatments (Table 5).
In all three communities, each of the 10 selected isolates (except the already identified), PCR products were outsourced for Sangers sequencing of the V3–V4 region to HiMedia for identification. The results of the matched contigs were compared on NCBI database with more than 97% of similarity. All the selected isolates of the SM1 community were identified as Enterobacter cloacae subsp. dissolvens strain LMG 2623, Stenotrophomonas pavanii strain ICB89, Bacillus stercoris JCM30051, Klebsiella varicola strain F2R9, Pseudomonas laurentiana strain GSL-010, Bacillus subtilis DSM10, Bacillus haynesii strain NRRL B-41327, Acinetobacter pittii DSM 21653 strain ATCC 19004, Klebsiella variicola strain JCM1662, and Acinetobacter calcoaceticus strain NCCB22016, respectively, for NWE-9, IWE-6, IWE-7, PC4-59, NWR-15, NWPZ-60, PHM5-9, PHM5-35, PHM5-22, and PHM5-59 isolates with more 97% similarity (Table 5; Supplementary Figure S2). The isolates in the SM2 community were identified as B. amyloliquefaciens NBRC15535, B. aerius strain 24K, B. proteolyticus strain MCCC 1A00365, B. velezensis strain CBMB 205, B. velezensis strain FZB42, B. altitudinis strain 41KF2b, B. stercoris JCM30051, B. amyloliquefaciens MPA 1034, Kosakonia cowanii JCM 10956, and Brevibacillus reuszeri strain NBRC 15719, respectively, for DWE-1, NWP-12, BWR-11, PZ-28, SHZ-30, PHM5-27, NWE-2, PHM5-33, PHM5-29, and PC4-44 (Table 5; Supplementary Figure S2). The isolates for SM3 community were identified as B. amyloliquefaciens BCRC 11601, Pantoea agglomerans strain NCTC 9381, B. amyloliquefaciens strain MPA1034, B. altitudinis strain 41KF2b, S. pavanii strain LMG25348, S. pavanii strain ICB89, Enterobacter cloacae subsp. dissolvens strain LMG 2623, B. stercoris JCM30051, B. amyloliquefaciens NBRC15535, and Klebsiella variicola strain F2R9, respectively, for PC4-29, NWP-9, NWR-14, NH-1, NWR-13, IWE-6, NWE-9, IWE-7, DWE-1, and PC4-59 (Supplementary Figure S2; Table 5).
The trans-section view of root and leaf samples was observed under the bright field microscope. In root samples, the microbial group at 18 h of inoculation was colonized more over the root tip and inside the secondary root hairs. It was like they were entering into the root endosphere for all three communities, but the bacterial colonies were absent for the leaf. At 48 h of inoculation, the root cortex region was fully colonized with the microbial communities, and some colonization was also observed inside the leaf. At 72 h of incubation, the colonization was observed more for the root and leaf endosphere compared with the colonization at 48 h. Microbial colonization was observed inside the cell and in intercellular spaces. There were some differences in colonization behavior observed between the three communities. The colonization of the SM2 and SM3 communities was higher, and they were in large aggregates compared with the scattered single cells of the SM1 community (Supplementary Figure S1).
All prepared communities were evaluated by the pot experiments with wheat plants (Figure 4). Plant biometric assays like germination percentage, root length (60 DAS and 90 DAS), shoot length (60 DAS and 90 DAS), plant dry weight (at 90 DAS), and chlorophyll content (60 and 90 DAS) were evaluated under the glass house or semi-control condition. All communities SM1, SM2, and SM3 were compared with the negative control and 100% RDF-treated pots. In germination percentage, T5 (100% RDF) and T4 (SM3 + 50% RDF) showed the highest germination percentage and insignificantly differed with each other. The treatment receiving SM3 + 50% RDF (T4) showed 28% higher germination compared with the control (T1) (Table 6). The chlorophyll content of the plant at 60 DAS and 90 DAS was evaluated with the help of SPAD. The chlorophyll content was significantly higher in T4 than in other treatments T1, T2, and T3 by 37.35%, 21.5%, and 25.14%, respectively, at 60 DAS (Table 6). Plant dry matter was calculated at 90 DAS, which when compared to the negative control with biological treatments, dry weight was 9.11% higher in T4 followed by T2 and T3 by 4.8% and 3.12%, respectively (Table 6). Among the microbial treatments, all were significantly different from each other with higher plant biomass in T4, which was 4.3% and 6% higher than in T2 and T3, respectively. Among all treatments, the T4 (SM3) result was better than the rest of the microbial treatments and negative control irrespective of the plant traits, which showed that there is some cumulative effect of CFts over PGPts.
Wheat, being one of the most pivotal and staple crops, is cultivated across India, covering approximately 29.8 million hectares. Over the past decade, there has been a notable surge in wheat production, with the country’s output escalating from 94.57 million metric tons in the 2012–2013 period to 112.18 million tons in 2022–2023 (Srivastava et al., 2023). Its demand is rising in countries undergoing urbanization and industrialization. In addition to serving as a significant source of starch and energy, wheat also provides substantial quantities of various essential elements crucial for health, including protein, essential vitamins (particularly B vitamins), dietary fiber, and beneficial phytochemicals (Shewry and Hey, 2015). Consequently, it draws the attention of researchers from various domains to enhance both the productivity and quality of grains. Utilizing traditional biofertilizers is a viable approach for the sustainable growth of wheat. However, it comes with certain drawbacks. For instance, employing a single-strain fertilizer may have limited effectiveness in diverse environments due to factors such as competition and predation in exotic surroundings and challenges in achieving proper colonization (Somasegaran and Bohlool, 1990; Bradáčová et al., 2019). These issues can be addressed by employing a consortium, but a significant challenge lies in ensuring compatibility among the members of the consortium and their persistence in different environments. Therefore, concerning microbiology, it is important to advance research in developing wheat endophytic and rhizospheric bacterial communities, which is certain in the colonization of wheat plant and mimics the natural community that also contributes to crop growth and development.
This experiment utilized endophytes and rhizospheric isolates from wheat and maize to develop bacterial communities. Community construction was based on CFts and their PGPts. It was observed that, in addition to the PGPts, various CFts were also present in bacterial isolates, which are essential for community integrity and functioning (Wong et al., 2021). The significant CFts include biofilm synthesis, exopolysaccharide production, polyhydroxybutyrate production, and, to some extent, motility. Motility is a factor that enables beneficial microbes to be attracted towards root exudates and joins the community based on their compatibility, indirectly contributing to community formation. Motility also facilitates the occupation of nutrient-rich niches in the vicinity of the plant, enriching the space with beneficial microbes (Palma et al., 2022). In CFts, several mentioned traits were observed in which the range of EPS was found from 11 to 28.72 µg ml−1, which satisfied with the findings of Mancuso Nichols et al. (2004) and Chambi et al. (2021). Comparing the OD of biofilm formation, the findings of Simoes et al. (2007) and Thomen et al. (2017) were comparable to our observations where biofilm ranges between 0.1 and 4 OD at 540 nm. Motility was measured in the percentage coverage of the test tube by motile cells in a stab culture; a similar experiment was done by Murinda et al. (2002), which matched with our observation. PHB-producing bacteria preserve the extra carbohydrate present in the niche as intracellular reservoir to tackle with the adverse condition. This study covered qualitative analysis of PHB production with Sudan black dye. Black-stained colonies were considered as positive isolates for PHB production, which was matched with the findings of Robert and Iyer (2018).
Combining isolates with potential CFts and PGPts represents a novel approach towards developing plant probiotics. Various scientists have conducted extensive research on the plant-growth-promoting traits of endophytes and rhizospheric bacteria, such as nutrient solubilization, phytohormone production, and antifungal activity (Eid et al., 2021; Sharma et al., 2023). In our study, the plant growth parameters were examined both qualitatively and quantitatively, where nitrogen and phosphorus results came in the range of 4–9.16 nmol C2H4 mg protein−1 h−1 and 6.78–44.61 ppm, respectively, which aligned with the findings of other workers (Gull et al., 2004; Widdig et al., 2019) in the previous studies. The range of results for phytohormone, viz., IAA and GA3 analysis, was 4.43–45.53 ppm and 3.73–12.40 ppm, which was related with the findings of Karadeniz et al. (2006) and Restu et al. (2019), respectively. Siderophore analysis was done with the use of CAS reagent and supernatant of culture broth. The range of total siderophore production was 9.0–61.72 psu. Previous analysis of siderophore production was carried by various scientists (Murakami et al., 2021; Newman, 2023), which satisfied our results.
Including the general screening traits (e.g., PGPts and CFts), microbial communities were developed in this experiment to mimic naturally occurring communities. Previously, in the laboratory, strategies for creating synthetic microbial communities based on functional and/or abundance approaches have been devised (Suman et al., 2022). To validate this, a wet lab synthesis of microbial communities was conducted using key species from heat-sensitive varieties of wheat. These communities were developed and validated in both controlled and field experiments (Aswini et al., 2023). While in this study, the key genera were identified from the selected isolates, their dominance was also confirmed with the dominant genera identified from the metagenome (BioProject-PRJNA944920, PRJNA 954583, and PRJNA 1065154) submitted by the lab in NCBI database. From these projects, the abundant genera found concerning wheat endophytes were Bacillus, Stenotrophomonas, Pseudomonas, Acinetobacter, Enterobacter, Clostridium, and Xanthomonas. The previous experiment, carried out by Tkalec et al. (2022), supported our findings, where they observed abundance of Bacillus species in their samples. The bacterial genera Clostridium and Xanthomonas were excluded from community formation due to their pathogenic nature for plants and animals. The isolates of key dominant species were integrated into the communities based on their multifunctional traits, placing them in their respective community categories (SM1, SM2, and SM3).
After grouping all isolates according to their functional traits, they were exposed to different carbon sources and other compounds to assess their resource requirements. Previous studies by various other scientists (Uroz et al., 2007; Tzamali et al., 2011; Kumar et al., 2021) have shown that in a community, there is a proper distribution of food sources (Passari et al., 2016). In this study, isolates in a clade (which showed same carbon source utilizing pattern) possessing higher bonitur scale for PGPts and CFts were selected for the development of communities. Furthermore, they were checked for compatibility through cross-inoculation on agar plates, which aligned with the findings of Murillo-Roos et al. (2022). All 46 screened bacterial isolates were identified using Sanger sequencing of the V3–V4 region of 16S rDNA. The results indicated that Bacillus was the predominant genus, followed by Enterobactor, and others including Pantoea, Microbacterium, Stenotrophomonas, and Pseudomonas. This result is consistent with the findings of Suman et al. (2020); Alaylar et al. (2021) and Sharma et al. (2023) where most of the rhizospheric and endophytic culturable microbes were dominated by the Bacilli group.
The formed communities were inoculated onto wheat seedlings grown on water agar plates to observe the colonization behavior of communities. Bacterial colonization was assessed using TTC staining, and in our experiments, colonization ability was evaluated at three time intervals: 24 h, 48 h, and 72 h. Initially, colonization was observed at the tip of the secondary root, followed by colonization inside the root and leaf. The observations regarding the colonization of bacterial communities in root and leaf at different periods were consistent with the findings of various workers (Remus et al., 2000; Timmusk et al., 2005; Thomas and Sekhar, 2014). The new behavior of the bacterial community was observed, e.g., formation of aggregation in the community possessed community-forming traits and more single isolated bacterial colony was found in the community, which had only PGPts. The aggregation of the community inside the plant system defines the strength of integrity among individual isolates; being together inside the plant system may impact better than the single isolated bacterial strain. There are various such findings that observed that a bacterial community and consortium showed better results than the application of a single isolated strain (Bradáčová et al., 2019; Santoyo et al., 2021).
In the microcosm experiment, the evaluation of formed communities (PGPt, CFt, and PGPt+CFt) was conducted in sterile soil to assess various plant growth attributes. In our experiments, the germination percentage was found to increase with the application of PGPt+CFt community and did not significantly differ from the treatment with a full dose of fertilizer application. This finding aligns with similar research conducted by Pacheco et al. (2021) and Hussain et al. (2021). The observation of root length was highest in the T1 treatment, followed by T4, at both stages of plant growth. The root growth in T4 was higher compared to the other treatments and the control. Similar results were observed in studies by Khan et al. (2022) and Sukeerthi et al. (2020). Regarding shoot length, the pot with a full dose of fertilizer application showed the highest growth compared to the other treatments, followed by the T4 treatment. In both stages of plant growth, the T4 treatment exhibited better growth than the other two microbial treatments, showing a 54% and 34% increase over the control, which was consistent with the findings of Marag and Suman (2018). In terms of chlorophyll content, treatment T4 consistently showed the highest levels, regardless of the date of observation and treatments. However, the chlorophyll content did not significantly increase at later stages of plant growth compared to the other traits. This finding is consistent with an experiment conducted by Reynolds et al. (2000), where they observed a plateau in the increment of chlorophyll content in wheat plants at the terminal and anthesis times. This supports our result, indicating that at a certain stage, the chlorophyll content of the plant does not increase further. Additionally, a higher chlorophyll content was found in the microbial treatment, which is also consistent with the findings of Khalilzadeh et al. (2018). Plant dry matter was also influenced by the application of microbial treatments, with T4 being only 8% lower than the full dose of fertilizer application and 9.1% higher than the control. This result was aligned with the findings of Bilal et al. (2017) and de Matos Nascimento et al. (2020) in their experiments. A higher plant biomass and other growth attributes were found in the treatments, where the community was showing both the traits of PGP and CF; these findings bound us to consider the effect of community-forming traits, which increases the efficacy of plant-growth-promoting traits of various isolates.
Community-forming traits play a crucial role in facilitating effective rhizosphere colonization and recruitment by plants as endophytes. The integration of efficient PGP bacteria with additional isolates possessing community-forming traits has shown great potential as a probiotic inoculant for enhancing chlorophyll content and plant biomass and further the yield increment. Further exploration of the interactive mechanisms in field trials with the selected communities will open up new strategic avenues for developing innovative bio-inoculants to promote sustainable agriculture.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
DP: Data curation, Formal analysis, Investigation, Methodology, Validation, Visualization, Writing – original draft. AS: Conceptualization, Data curation, Funding acquisition, Investigation, Resources, Supervision, Validation, Writing – original draft, Writing – review & editing. VG: Methodology, Funding acquisition, Resources, Validation, Writing – review & editing. PS: Methodology, Formal analysis, Software, Visualization, Writing – original draft. KA: Investigation, Methodology, Resources, Software, Writing – review & editing. SG: Funding acquisition, Investigation, Methodology, Resources, Supervision, Writing – original draft. RA: Investigation, Methodology, Writing – original draft.
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
The authors thank DST-Science and Engineering Research Board (DST-SERB) researcher project CRG/2019/002146 for funding. ICAR-IARI, Ph.D. fellowship given to author DP as financial assistance is duly acknowledged.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2024.1332745/full#supplementary-material
Abd El Gawad, I. A., Abd El Fatah, A. M., Al Rubayyi, K. A. (2010). Identification and characterization of dominant lactic acid bacteria isolated from traditional rayeb milk in Egypt. J. Am. Sci. 6, 728–735. doi: 10.7537/marsjas061010.85
Alaylar, B., Güllüce, M., Karadayi, M., Koç, T. Y., Karadayi, G. (2021). Rhizospheric PGPR strains of wheat, barley and trefoil grown in Ağrı province. Türk Doğave Fen Dergisi 10, 182–190. doi: 10.46810/tdfd.937841
Aswini, K., Suman, A., Sharma, P., Singh, P. K., Gond, S., Pathak, D. (2023). Seed endophytic bacterial profiling from wheat varieties of contrasting heat sensitivity. Front. Plant Sci. 14. doi: 10.3389/fpls.2023.1101818
Backer, R., Rokem, J. S., Ilangumaran, G., Lamont, J., Praslickova, D., Ricci, E., et al. (2018). Plant growth-promoting rhizobacteria: context, mechanisms of action, and roadmap to commercialization of biostimulants for sustainable agriculture. Front. Plant Sci. 9, 1473. doi: 10.3389/fpls.2018.01473
Bhat, M. A., Mishra, A. K., Jan, S., Bhat, M. A., Kamal, M. A., Rahman, S., et al. (2023). Plant growth promoting rhizobacteria in plant health: A perspective study of the underground interaction. Plants 12, 629. doi: 10.3390/plants12030629
Bilal, M., Ayub, M., Tariq, M., Tahir, M., Nadeem, M. A. (2017). Dry matter yield and forage quality traits of oat (Avena sativa L.) under integrative use of microbial and synthetic source of nitrogen. J. Saudi Soc Agric. Sci. 16, 236–241. doi: 10.1016/j.jssas.2015.08.002
Bradáčová, K., Florea, A. S., Bar-Tal, A., Minz, D., Yermiyahu, U., Shawahna, R., et al. (2019). Microbial consortia versus single-strain inoculants: an advantage in PGPM-assisted tomato production. Agronomy 9, 105. doi: 10.3390/agronomy9020105
Bric, J. M., Bostock, R. M., Silverstone, S. E. (1991). Rapid in situ assay for indole acetic acid production by bacteria immobilized on a nitrocellulose membrane. Appl. Environ. Microbiol. 57, 535–538. doi: 10.1128/aem.57.2.535-538.1991
Brown, M. E., Burlingham, S. K. (1968). Production of plant growth substances by Azotobacter chroococcum. Microbiology 53, 135–144. doi: 10.1099/00221287-53-1-135
Bulgarelli, D., Schlaeppi, K., Spaepen, S., Van Themaat, E. V. L., Schulze-Lefert, P. (2013). Structure and functions of the bacterial microbiota of plants. Annu. Rev. Plant Biol. 64, 807–838. doi: 10.1146/annurev-arplant-050312-120106
Cao, M., Narayanan, M., Shi, X., Chen, X., Li, Z., Ma, Y. (2023). Optimistic contributions of plant growth-promoting bacteria for sustainable agriculture and climate stress alleviation. Environ. Res. 217, 114924. doi: 10.1016/j.envres.2022.114924
Carreiras, J., Cruz-Silva, A., Fonseca, B., Carvalho, R. C., Cunha, J. P., Proença Pereira, J., et al. (2023). Improving grapevine heat stress resilience with marine plant growth-promoting rhizobacteria consortia. Microorganisms 11, 856. doi: 10.3390/microorganisms11040856
Chambi, D., Romero-Soto, L., Villca, R., Orozco-Gutiérrez, F., Vega-Baudrit, J., QuillaGuamán, J., et al. (2021). Exopolysaccharides production by cultivating a bacterial isolate from the hypersaline environment of Salar de Uyuni (Bolivia) in pretreatment liquids of steam-exploded quinoa stalks and enzymatic hydrolysates of curupaú sawdust. J. Ferment. 7, 33. doi: 10.3390/fermentation7010033
Dal Bello, M., Lee, H., Goyal, A., Gore, J. (2021). Resource–diversity relationships in bacterial communities reflect the network structure of microbial metabolism. Nat. Ecol. Evol. 5, 1424–1434. doi: 10.1038/s41559-021-01535-8
de Matos Nascimento, A., Maciel, A. M., Silva, J. B. G., Mendonça, H. V., de Paula, V. R., Otenio, M. H. (2020). Biofertilizer application on corn (Zea mays) increases the productivity and quality of the crop without causing environmental damage. Water Air Soil pollut. 23, 1–10. doi: 10.1007/s11270-020-04778-6
Eid, A. M., Fouda, A., Abdel-Rahman, M. A., Salem, S. S., Elsaied, A., Oelmüller, R., et al. (2021). Harnessing bacterial endophytes for promotion of plant growth and biotechnological applications: an overview. Plants 10, 935. doi: 10.3390/plants10050935
Glick, B. R. (1995). The enhancement of plant growth by free-living bacteria. Can. J. Microbiol. 41, 109–117. doi: 10.1139/m95-015
Großkopf, T., Soyer, O. S. (2014). Synthetic microbial communities. Curr. Opin. Microbiol. 18, 72–77. doi: 10.1016/j.mib.2014.02.002
Gull, M., Hafeez, F. Y., Saleem, M., Malik, K. A. (2004). Phosphorus uptake and growth promotion of chickpea by co-inoculation of mineral phosphate solubilising bacteria and a mixed rhizobial culture. Aust. J. Exp. Agric. 44, 623–628. doi: 10.1071/EA02218
Hao, X., Wang, X., Chen, C., Liu, R., Yin, Y., Yao, J., et al. (2023). Synthetic bacterial communities reshape microbial communities and enhance nutrient supply in desertified land of Northwest China. Appl. Soil Ecol. 189, 104972. doi: 10.1016/j.apsoil.2023.104972
Hung, C. M., Chen, C. W., Huang, C. P., Sheu, D. S., Dong, C. D. (2023). Microbial community structure and potential function associated with poly-3-hydroxybutyrate biopolymer-boosted activation of peroxymonosulfate for waste-activated sludge decontamination. Bioresour. Technol. 369, 128450. doi: 10.1016/j.biortech.2022.128450
Hussain, I., Khan, A., Akbarm, H. (2021). Maize growth in response to beneficial microbes. Humic acid and farmyard manure application. Sarhad J. Agric. 37, 1426–1435. doi: 10.17582/journal.sja/2021/37.4.1426.1435
Javed, S., Javaid, A., Hanif, U., Bahadur, S., Sultana, S., Shuaib, M., et al. (2021). Effect of necrotrophic fungus and PGPR on the comparative histochemistry of Vigna radiata by using multiple microscopic techniques. Microsc. Res. Tech. 84, 2737–2748. doi: 10.1002/jemt.23836
Karadeniz, N., Topcuoğlu, Ş.F., İnan, A. (2006). Auxin, gibberellin, cytokinin and abscisic acid production in some bacteria. World J. Microbiol. Biotechnol. 22, 1061–1064. doi: 10.1007/s11274-005-4561-1
Karygianni, L., Ren, Z., Koo, H., Thurnheer, T. (2020). Biofilm matrixome: extracellular components in structured microbial communities. Trends Microbiol. 28, 668–681. doi: 10.1016/j.tim.2020.03.016
Kelly, A. T., Fulton, M. (1953). Use of triphenyl tetrazolium in motility test medium. Am. J. Clin. Pathol. 23, 512–512. doi: 10.1093/ajcp/23.5_ts.512
Khalilzadeh, R., Seyed Sharifi, R., Jalilian, J. (2018). Growth, physiological status, and yield of salt-stressed wheat (Triticum aestivum L.) plants affected by biofertilizer and cycocel applications. Arid. Land Res. Manage. 32, 71–90. doi: 10.1080/15324982.2017.1378282
Khan, M. Y., Nadeem, S. M., Sohaib, M., Waqas, M. R., Alotaibi, F., Ali, L., et al. (2022). Potential of plant growth promoting bacterial consortium for improving the growth and yield of wheat under saline conditions. Front. Microbiol. 13. doi: 10.3389/fmicb.2022.958522
Kumar, A., Jha, M. N., Singh, D., Pathak, D., Rajawat, M. V. S. (2021). Prospecting catabolic diversity of microbial strains for developing microbial consortia and their synergistic effect on lentil (Lens esculenta) growth, yield and iron biofortification. Arch. Microbiol. 203, 4913–4928. doi: 10.1007/s00203-021-02446-9
Li, Z., Chang, S., Lin, L., Li, Y., An, Q. (2011). A colorimetric assay of 1-aminocyclopropane-1-carboxylate (ACC) based on ninhydrin reaction for rapid screening of bacteria containing ACC deaminase. Lett. Appl. Microbiol. 53, 178–185. doi: 10.1111/lam.2011.53.issue-2
Liu, F. P., Liu, H. Q., Zhou, H. L., Dong, Z. G., Bai, X. H., Bai, P., et al. (2014). Isolation and characterization of phosphate-solubilizing bacteria from betel nut (Areca catechu) and their effects on plant growth and phosphorus mobilization in tropical soils. Biol. Fert. Soils 50, 927–937. doi: 10.1007/s00374-014-0913-z
Liu, H., Qiu, Z., Ye, J., Verma, J. P., Li, J., Singh, B. K. (2022). Effective colonisation by a bacterial synthetic community promotes plant growth and alters soil microbial community. J. Sustain. Agric. Env. 1, 30–42. doi: 10.1002/sae2.12008
Mancuso Nichols, C. A., Garon, S., Bowman, J. P., Raguénès, G., Guezennec, J. (2004). Production of exopolysaccharides by Antarctic marine bacterial isolates. Appl. Microbiol. 96, 1057–1066. doi: 10.1111/j.1365-2672.2004.02216.x
Marag, P. S., Suman, A. (2018). Growth stage and tissue specific colonization of endophytic bacteria having plant growth promoting traits in hybrid and composite maize (Zea mays L.). Microbiol. Res. 214, 101–113. doi: 10.1016/j.micres.2018.05.016
Murakami, C., Tanaka, A. R., Sato, Y., Kimura, Y., Morimoto, K. (2021). Easy detection of siderophore production in diluted growth media using an improved CAS reagent. J. Microbiol. Methods 189, 106310. doi: 10.1016/j.mimet.2021.106310
Murillo-Roos, M., Abdullah, H. S. M., Debbar, M., Ueberschaar, N., Agler, M. T. (2022). Cross-feeding niches among commensal leaf bacteria are shaped by the interaction of strain-level diversity and resource availability. ISME J. 16, 2280–2289. doi: 10.1038/s41396-022-01271-2
Murinda, S. E., Nguyen, L. T., Ivey, S. J., Almeida, R. A., Oliver, S. P. (2002). Novel single-tube agar-based test system for motility enhancement and immunocapture of Escherichia coli O157: H7 by H7 flagellar antigen-specific antibodies. J. Clin. Microbiol. 40, 4685–4690. doi: 10.1128/JCM.40.12.4685-4690.2002
Newman, B. (2023). Bacterial siderophore production on lignin-derived aromatic compounds. M.Sc. Thesis. University of British Columbia. doi: 10.14288/1.0431391
O'Toole, G. A. (2011). Microtiter dish biofilm formation assay. JoVE 47), e2437. doi: 10.3791/2437-v
Oo, K. T., Win, T. T., Khai, A. A., Fu, P. (2020). Isolation, screening and molecular characterization of multifunctional plant growth promoting rhizobacteria for a sustainable agriculture. Am. J. Plant Sci. 11, 773–792. doi: 10.4236/ajps.2020.116055
Pacheco, A. R., Osborne, M. L., Segre, D. (2021). Non-additive microbial community responses to environmental complexity. Nat. Commun. 12, 2365. doi: 10.1038/s41467-021-22426-3
Palma, V., Gutiérrez, M. S., Vargas, O., Parthasarathy, R., Navarrete, P. (2022). Methods to evaluate bacterial motility and its role in bacterial–host interactions. Microorganisms 10, 563. doi: 10.3390/microorganisms10030563
Passari, A. K., Mishra, V. K., Leo, V. V., Gupta, V. K., Singh, B. P. (2016). Phytohormone production endowed with antagonistic potential and plant growth promoting abilities of culturable endophytic bacteria isolated from Clerodendrum colebrookianum Walp. Microbiol. Res. 193, 57–73. doi: 10.1016/j.micres.2016.09.006
Pugsley, A. P., Oudega, B. (1987). Methods for studying colicins and their plasmids, a practical approach (Oxford, UK: IRL Press), 105–161. doi: 10.1007/BF00330613
Quintieri, L., Fanelli, F., Zuhlke, D., Caputo, L., Logrieco, A. F., Albrecht, D., et al. (2020). Biofilm and pathogenesis-related proteins in the foodborne P. fluorescens ITEM 17298 with distinctive phenotypes during cold storage. Front. Microbiol. 11. doi: 10.3389/fmicb.2020.00991
Rana, G., Mandal, T., Mandal, N. K., Sakha, D., Meikap, B. C. (2015). Calcite solubilization by bacteria: A novel method of environment pollution control. Geomicrobiol. J. 32, 846–852. doi: 10.1080/01490451.2015.1010755
Remus, R., Ruppel, S., Jacob, H. J., Hecht-Buchholz, C., Merbach, W. (2000). Colonization behaviour of two enterobacterial strains on cereals. Biol. Fert. Soils. 30, 550–557. doi: 10.1007/s003740050035
Restu, M., Bachtiar, B., Larekeng, S. H. (2019). “Gibberellin and IAA production by rhizobacteria from various private forest,” in IOP conference series. Earth and environmental science, Vol. 270. 1 (Bristol: IOP Publishing). doi: 10.1088/1755-1315/270/1/012018
Reynolds, M. P., Gutierrez-Rodríguez, M., Larque-Saavedra, A. (2000). Photosynthesis of wheat in a warm, irrigated environment: I: genetic diversity and crop productivity. Field Crops Res. 66, 37–50. doi: 10.1016/S0378-4290(99)00077-5
Rijavec, T., Lapanje, A. (2016). Hydrogen cyanide in the rhizosphere: not suppressing plant pathogens, but rather regulating availability of phosphate. Front. Microbiol. 7. doi: 10.3389/fmicb.2016.01785
Robert, R., Iyer, P. R. (2018). Isolation and optimization of PHB (poly-β-hydroxybutyrate) based biodegradable plastics from Chlorella vulgaris. J. bioremediat. biodegrad. 09, 2–5. doi: 10.4172/2155-6199.1000433
SaiPrasad, J., Suman, A., Ramakrishnan, B., Aswini, K. (2021). Diversity and functionality of seed vectored bacterial endophytes in different cultivars of Triticum aestivum L. across ecological boundaries. Res. Square. doi: 10.21203/rs.3.rs-989999/v1
Santoyo, G., Guzmán-Guzmán, P., Parra-Cota, F. I., Santos-Villalobos, S. D. L., Orozco-Mosqueda, M. D. C., Glick, B. R. (2021). Plant growth stimulation by microbial consortia. Agronomy 11, 219. doi: 10.3390/agronomy11020219
Schwyn, B., Neilands, J. B. (1987). Universal chemical assay for the detection and determination of siderophores. Anal. Biochem. 160, 47–56. doi: 10.1016/0003-2697(87)90612-9
Sharf, W., Javaid, A., Shoaib, A., Khan, I. H. (2021). Induction of resistance in chili against Sclerotium rolfsii by plant-growth-promoting rhizobacteria Anagallis arvensis. Egypt. J. Biol. Pest Control. 31, 1–11. doi: 10.1186/s41938-021-00364-y
Sharma, P., Aswini, K., Sai Prasad, J., Kumar, N., Pathak, D., Gond, S., et al. (2023). Characterization of actinobacteria from wheat seeds for plant growth promoting traits and protection against fungal pathogens. J. Basic Microbiol. 63, 439–453. doi: 10.1002/jobm.202200259
Shayanthan, A., Ordoñez, P. A. C., Oresnik, I. J. (2022). The role of synthetic microbial communities (syncoms) in sustainable agriculture. Front. Agron. 4. doi: 10.3389/fagro.2022.896307
Shelley, M. P. (1994). Detection, isolation and characterization of Siderophores. Methods Enzymol. 94, 329–344. doi: 10.1016/0076-6879(94)35151-1
Shewry, P. R., Hey, S. J. (2015). The contribution of wheat to human diet and health. Food Energy Secur. 4, 178–202. doi: 10.1002/fes3.64
Shoaib, A., Ali, H., Javaid, A., Awan, Z. A. (2020). Contending charcoal rot disease of mungbean by employing biocontrol Ochrobactrum ciceri and zinc. Physiol. Mol. Biol. Plants. 26, 1385–1397. doi: 10.1007/s12298-020-00817-y
Simoes, L. C., Simoes, M., Vieira, M. J. (2007). Biofilm interactions between distinct bacterial genera isolated from drinking water. Appl. Environ. Microbiol. 73, 6192–6200. doi: 10.1128/AEM.00837-07
Somasegaran, P., Bohlool, B. B. (1990). Single strain versus multistrain inoculation: Effect of soil mineral N availability on rhizobial strain effectiveness and competition for nodulation on chick-pea, soybean, and dry bean. Appl. Environ. Microbiol. 56, 3298–3303. doi: 10.1128/aem.56.11.3298-3303.1990
Srivastava, A. B., Singh, K. K., Supriya, S. K., Mishra, H., Ahmad, R. (2023). Production and export dynamics of wheat in India. Int. J. Appl. Math. 8, 206–209. doi: 10.22271/maths.2023.v8.i3Sc.1023
Sukeerthi, D., Nachu, N. S., Ashwin, R., Bagyaraj, D. J. (2020). Microbial consortium promotes growth of zinnia and balsam seedlings raised in protrays. J. Floric. Landsc. 6, 4–8. doi: 10.25081/jfcls.2020.v6.6063
Suman, A., Govindasamy, V., Ramakrishnan, B., Aswini, K., SaiPrasad, J., Sharma, P., et al. (2022). Microbial community and function-based synthetic bioinoculants: A perspective for sustainable agriculture. Front. Microbiol. 12. doi: 10.3389/fmicb.2021.805498
Suman, A., Shukla, L., Marag, P. S., Verma, P., Gond, S., Prasad, J. S. (2020). Potential use of plant colonizing Pantoea as generic plant growth promoting bacteria for cereal crops. J. Environ. Biol. 41, 987–994. doi: 10.22438/jeb/41/5/MRN-1250
Taye, Y., Degu, T., Fesseha, H., Mathewos, M. (2021). Isolation and identification of lactic acid bacteria from cow milk and milk products. Sci. World J. e4697445. doi: 10.1155/2021/4697445
Thakur, R., Srivastava, S., Yadav, S. (2023). Multitrait Pseudomonas sp. isolated from the rhizosphere of Bergenia ciliata acts as a growth-promoting bioinoculant for plants. Front. Sustain. Food Syst. 7. doi: 10.3389/fsufs.2023.1097587
Thomas, P., Sekhar, A. C. (2014). Live cell imaging reveals extensive intracellular cytoplasmic colonization of banana by normally non-cultivable endophytic bacteria. AoB Plants 6, 2. doi: 10.1093/aobpla/plu002
Thomen, P., Robert, J., Monmeyran, A., Bitbol, A. F., Douarche, C., Henry, N. (2017). Bacterial biofilm under flow: first a physical struggle to stay, then a matter of breathing. PloS One 12, e0175197. doi: 10.1371/journal.pone.0175197
Timmusk, S., Grantcharova, N., Wagner, E. G. H. (2005). Paenibacillus polymyxa invades plant roots and forms biofilms. Appl. Environ. Microbiol. 71, 7292–7300. doi: 10.1128/AEM.71.11.7292-7300.2005
Tkalec, V., Mahnic, A., Gselman, P., Rupnik, M. (2022). Analysis of seed-associated bacteria and fungi on staple crops using the cultivation and metagenomic approaches. Folia Microbiol. 67, 351–361. doi: 10.1007/s12223-022-00958-5
Trivedi, P., Leach, J. E., Tringe, S. G., Sa, T., Singh, B. K. (2020). Plant–microbiome interactions: From community assembly to plant health. Nat. Rev. Microbiol. 18, 607–621. doi: 10.1038/s41579-020-0412-1
Tzamali, E., Poirazi, P., Tollis, I. G., Reczko, M. (2011). A computational exploration of bacterial metabolic diversity identifying metabolic interactions and growth-efficient strain communities. BMC Syst. Biol. 5, 1–15. doi: 10.1186/1752-0509-5-167
Uroz, S., Calvaruso, C., Turpault, M. P., Pierrat, J. C., Mustin, C., Frey-Klett, P. (2007). Effect of the mycorrhizosphere on the genotypic and metabolic diversity of the bacterial communities involved in mineral weathering in a forest soil. Appl. Environ. Microbiol. 73, 3019–3027. doi: 10.1128/AEM.00121-07
Verma, P., Yadav, A. N., Khannam, K. S., Panjiar, N., Kumar, S., Saxena, A. K., et al. (2015). Assessment of genetic diversity and plant growth promoting attributes of psychrotolerant bacteria allied with wheat (Triticum aestivum) from the northern hills zone of India. Ann. Microbiol. 65, 1885–1899. doi: 10.1007/s13213-014-1027-4
Wei, Y. F., Wang, A. L., Liao, S. A. (2020). Effect of different carbon sources on microbial community structure and composition of ex-situ biofloc formation. Aquaculture 515, 734492. doi: 10.1016/j.aquaculture.2019.734492
Widdig, M., Schleuss, P. M., Weig, A. R., Guhr, A., Biederman, L. A., Borer, E. T., et al. (2019). Nitrogen and phosphorus additions alter the abundance of phosphorus-solubilizing bacteria and phosphatase activity in grassland soils. Front. Environ. Sci. 7. doi: 10.3389/fenvs.2019.00185
Williamson, D. H., Wilkinson, J. F. (1958). The isolation and estimation of the poly-β-hydroxy-butyrate inclusions of Bacillus species. Microbiology 19, 198–209. doi: 10.1099/00221287-19-1-198
Wong, G. C., Antani, J. D., Lele, P. P., Chen, J., Nan, B., Kühn, M. J., et al. (2021). Roadmap on emerging concepts in the physical biology of bacterial biofilms: from surface sensing to community formation. Phys. Biol. 18, 051501. doi: 10.1088/1478-3975/abdc0e
Wu, Y., Peng, C., Xnxin, J., Xueke, N., Dandan, J., Noha, M. A., et al. (2019). Soil biofilm formation enhances microbial community diversity and metabolic activity. Environ. Int. 132, 105116. doi: 10.1016/j.envint.2019.105116
Keywords: biofilm, colonization, microbial community, microcosm, plant growth promoting traits, plant microbe interaction
Citation: Pathak D, Suman A, Sharma P, Aswini K, Govindasamy V, Gond S and Anshika R (2024) Community-forming traits play role in effective colonization of plant-growth-promoting bacteria and improved plant growth. Front. Plant Sci. 15:1332745. doi: 10.3389/fpls.2024.1332745
Received: 09 November 2023; Accepted: 21 February 2024;
Published: 12 March 2024.
Edited by:
Andrea Genre, University of Turin, ItalyReviewed by:
Saurabh Yadav, Hemwati Nandan Bahuguna Garhwal University, IndiaCopyright © 2024 Pathak, Suman, Sharma, Aswini, Govindasamy, Gond and Anshika. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Archna Suman, YXJjaHN1bWFuQHlhaG9vLmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.