- 1School of Food Industrial Sciences, Miyagi University, Sendai, Japan
- 2Graduate School of Fisheries Sciences, Hokkaido University, Hakodate, Japan
Introduction: To withstand high temperatures that would be lethal to a plant in the naïve state, land plants must establish heat stress memory. The acquisition of heat stress tolerance via heat stress memory in algae has only been observed in the red alga ‘Bangia’ sp. ESS1.
Methods: In this study, we further evaluated the intrinsic ability of this alga to establish heat stress memory by monitoring hydrogen peroxide (H2O2) production and examining the relationship between heat stress memory and the expression of genes encoding nitrogen transporters, since heat stress generally reduces nitrogen absorption. Next, genes encoding nitrogen transporters were selected from our unpublished transcriptome data of ‘Bangia’ sp. ESS1.
Results: We observed a reduction in H2O2 content when heat stress memory was established in the alga. In addition, six ammonium transporter genes, a single-copy nitrate transporter gene and two urea transporter genes were identified. Two of these nitrogen transporter genes were induced by heat stress but not by heat stress memory, two genes showed heat stress memory–dependent expression, and one gene was induced by both treatments. Heat stress memory therefore differentially regulated the expression of the nitrogen transporter genes by reducing heat stress–inducible gene expression and inducing heat stress memory–dependent gene expression.
Discussion: These findings point to the functional diversity of nitrogen transporter genes, which play different roles under various heat stress conditions. The characteristic effects of heat stress memory on the expression of individual nitrogen transporter genes might represent an indispensable strategy for reducing the threshold of sensitivity to recurrent high-temperature conditions and for maintaining nitrogen absorption under such conditions in ‘Bangia’ sp. ESS1.
1 Introduction
Acclimation to changes in environmental conditions is crucial for the survival of plants, as plants cannot move away from abiotic and biotic stresses. Recent studies have demonstrated that plants “memorize” the experience of exposure to non-lethal environmental changes, allowing them to acquire tolerance to subsequent exposure to environmental changes that would normally be lethal (Sharma et al., 2022; Kambona et al., 2023). Indeed, plants withstand extremely high temperatures, which are lethal to plants in the native state, following growth at optimal temperatures for several days after exposure to non-lethal high temperatures (Yamaguchi, 2021a; Yamaguchi et al., 2021b; Ramakrishnan et al., 2022; Charng et al., 2023). The establishment of stress memory is therefore a critical strategy for survival under recurrent environmental changes.
Members of Bangiales, an order of multicellular red algae with a filamentous or foliose shape (Sutherland et al., 2011; Mikami and Takahashi, 2023), grow abundantly in intertidal regions, where temperatures often fluctuate widely (Karsten and West, 2000; Chen et al., 2022). Recent transcriptome analyses indicated that foliose Bangiales algae respond to heat stress via the heat-inducible expression of genes encoding heat shock proteins (HSPs) (Kim et al., 2011; Sun et al., 2015; Wang et al., 2018; Jin et al., 2020; Chang et al., 2021; Yu et al., 2021; Gao et al., 2022; Wi et al., 2023). In the foliose red alga Pyropia yezoensis, membrane fluidization at high temperatures triggers the heat stress–inducible expression of HSP70 and multiprotein bridging factor 1 (MBF1) genes, whereas heat stress–inducible expression of high temperature response 2 (HTR2) and HTR2-like (HTR2L) genes occurs independently of membrane fluidization (Khoa and Mikami, 2022; Mikami and Khoa, 2023). Thus, Bangiales algae perceive and respond to high temperature via heat stress–inducible gene expression, and the responses are regulated by multiple intracellular signal transduction pathways (Khoa and Mikami, 2022). However, little is known about heat stress tolerance and memory in foliose Bangiales.
By contrast, the existence of heat stress memory has been confirmed in filamentous Bangiales. For example, the intrinsic ability to acquire heat stress tolerance by establishing heat stress memory was observed in ‘Bangia’ sp. ESS1 of the ‘Bangia’ 2 group (Kishimoto et al., 2019; Li et al., 2019a; Khoa et al., 2021). However, Bangia atropurpurea acquires heat stress tolerance but is unable to remember heat stress, and ‘Bangia’ sp. ESS2 of the ‘Bangia’ 3 group cannot acquire heat stress tolerance or establish stress memory (Khoa et al., 2021). These findings indicate that physiological responses to heat stress vary among ‘Bangia’ species, although it is unknown whether the presence of stress memory depends on phylogenetic classification (genus Bangia and ‘Bangia’ groups 1, 2, and 3; Sutherland et al., 2011; Mikami and Takahashi, 2023) or the characteristics of individual species.
High temperature usually inhibits the growth of macroalgae (Endo et al., 2020; Fernández et al., 2020; Wu et al., 2022). In addition, high temperature reduces nitrogen uptake and nitrogen content in these algae (Gerard, 1997; Hay et al., 2010; Gouvêa et al., 2017). Thus, growth retardation and reduced nitrogen accumulation are tightly correlated, which is consistent with the finding that nitrogen is an indispensable macronutrient for plant growth in most terrestrial and aquatic ecosystems (Edward and Richard, 2001; Witte, 2011). Indeed, nitrogen fertilization promotes growth under heat stress conditions in brown, green, and red algae (Wang et al., 2014; Gouvêa et al., 2017; Fernández et al., 2020; Wu et al., 2022; Wang et al., 2023). Moreover, high temperature stimulates nitrogen accumulation in meristems of the brown alga Eisenia bicyclis and the green alga Ulva prolifera (Endo et al., 2020; Sato et al., 2021). These findings suggest that the effects of high-temperature stress on nitrogen uptake differ among phyla, genera, and/or species of algae, but how heat stress influences the uptake and accumulation of nitrogen sources in algae remains to be elucidated.
Nitrogen sources comprise organic forms such as urea, amino acids, free peptides, and proteins, as well as the inorganic forms nitrate (NO3−), nitrite (NO2−), and ammonium (NH4+), all of which are major nitrogen sources in soil and seawater (Okumoto and Versaw, 2017). The uptake of these nitrogen sources into cells is mediated by transporters that differentially recognize inorganic or organic nitrogen sources (Gazzarrini et al., 1999; Ludewig et al., 2007, Pinton et al., 2016). For example, the influx of extracellular NH4+ into cells is mediated by ammonium transporters (AMTs; D’Apuzzo et al., 2004), whereas urea uptake into cells occurs via urea transporters (DUR3s; Wang et al., 2008a). In addition, NO3− is imported into cells by nitrate transporters (NRTs), which form a large family with many members and distinct functions in plants (Bai et al., 2013). Thus, AMT, DUR3, and NRT are critical factors in the influx of nitrogen sources.
Despite our increasing knowledge about the presence of nitrogen transporters in terrestrial plants and algae, it is unclear whether there is a relationship between nitrogen uptake and the acquisition of stress tolerance under high-temperature conditions in Bangiales. Because the physiology of the heat stress response in ‘Bangia’ species has been well studied (Notoya and Iijima, 2003; Wang et al., 2008b; Mikami and Kishimoto, 2018; Kishimoto et al., 2019; Li et al., 2019a; Khoa et al., 2021), perhaps ‘Bangia’ sp. ESS1, which memorizes heat stress to acquire heat stress tolerance (Kishimoto et al., 2019; Khoa et al., 2021), could serve as a model system for investigating the expression profiles of nitrogen transporter genes under heat stress in red algae.
In this study, the intrinsic ability of ‘Bangia’ sp. ESS1 to establish heat stress memory was confirmed by monitoring production of hydrogen peroxide (H2O2). Then, the ‘Bangia’ sp. ESS1 genes encoding nitrogen transporters such as AMT, NRT, and DUR3 were identified and their expression profiles were analyzed during the acquisition of heat stress tolerance and the establishment of heat stress memory. Our findings reveal novel aspects of the relationship between heat stress memory and nitrogen uptake under heat stress conditions in Bangiales.
2 Materials and methods
2.1 Algal materials and culture conditions for maintenance
Filamentous gametophytic thalli of ‘Bangia’ sp. ESS1 were harvested in Esashi, Hokkaido, Japan on May 17, 2010 (Hirata et al., 2011) and phylogenetically classified as a member of ‘Bangia’ group 2 (Li et al., 2019a). The alga was maintained clonally as an experimental line in our laboratory in sterilized artificial seawater as described by Li et al. (2019b) under 60–70 μmol photons m−2 s−1 light with a short-day photoperiod (10 h light/14 h dark) at 15°C and aerated with air filtered through a 0.22-μm filter (Whatman, Maidstone, UK). The culture medium was changed weekly.
2.2 Stress treatment of algal materials
Samples (0.05 g fresh weight) of thalli cultured under aeration at 15°C were incubated without agitation in dishes (Azunoru dish, 90 mm diameter × 20 mm height; As One Co., Ltd., Osaka, Japan) containing 50 ml of seawater at 15°C for 7 days to adapt to changes in culture conditions. We previously demonstrated that ‘Bangia’ sp. ESS1 acquires heat stress tolerance by establishing heat stress memory, which is mostly maintained for 2 days after recovery from a 7-day non-lethal stress (Kishimoto et al., 2019). Thus, the samples were then subjected to one of six different stress treatments (Figure 1A): (1) 28°C for 7 days (priming, P); (2) 32°C for 1 day (lethal high temperature-1, LHT-1); (3) 32°C for 6 days (lethal high temperature-6, LHT-6); (4) 28°C for 7 days and 15°C for 2 days (recovery, R); (5) 28°C for 7 days, 15°C for 2 days, and 32°C for 1 day (triggering-1, T-1); and (6) 28°C for 7 days, 15°C for 2 days, and 32°C for 6 days (triggering-6, T-6). The control condition was incubation at 15°C for 7 days. Samples from all stress treatments from three repeated experiments (three samples per treatment) were harvested, frozen in liquid nitrogen, and stored at −80°C prior to H2O2 quantification and gene expression analysis.
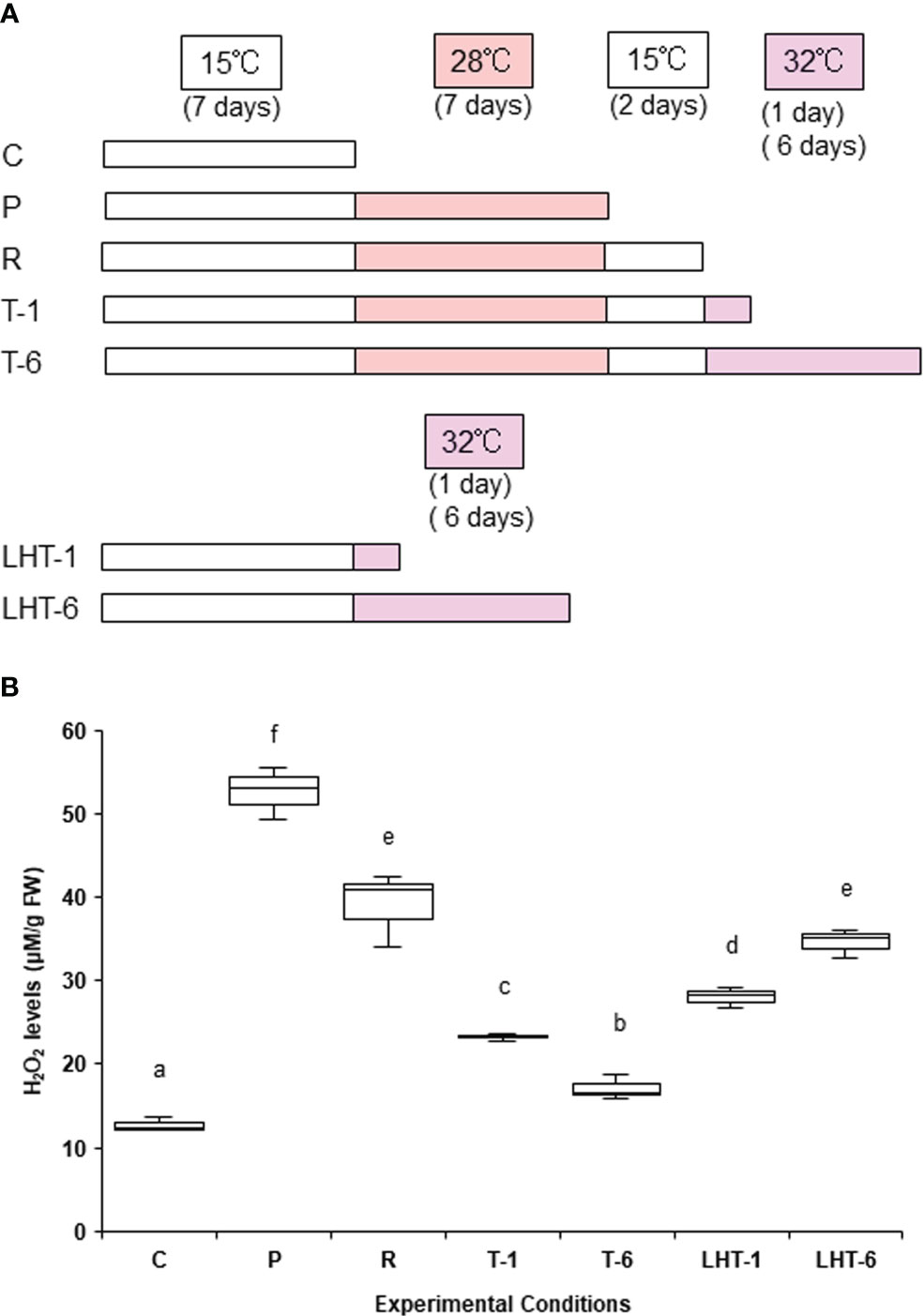
Figure 1 Effects of heat stress memory on hydrogen peroxide (H2O2) production in ‘Bangia’ sp. ESS1. (A) Schematic representation of the experimental conditions, including priming, recovery, and triggering, used to assess the biological significance of heat stress memory in this study. Seven conditions were employed: control, growth at 15°C (C); priming, incubation at 28°C for 7 days after growth at 15°C (P); recovery from 28°C treatment by incubation at 15°C for 2 days (R); triggering, 32°C treatment for 1 or 6 days (T-1 and T-6, respectively); and direct transfer to a lethal high temperature 32°C for 1 or 6 days (LHT-1 and LHT-6, respectively). (B) Quantitative analysis of H2O2 production under various heat stress conditions. H2O2 contents in algae treated with the experimental conditions indicated in (A) were quantified; mean values ± SD per 0.2 g sample fresh weight were calculated from three independent experiments. Different letters denote statistically significant differences (p < 0.05), as determined by one-way ANOVA.
2.3 Quantitative analysis of hydrogen peroxide production
The H2O2 contents of samples treated with the various stress conditions described above were measured as described by Kumar et al. (2011) with slight modifications. Initially, 200-mg (fresh weight) algal samples were extracted with 400 μl of 50 mM Na-acetate buffer (pH 6.5) (1:2, w/v). A 100-μl aliquot of each extract was incubated in a reaction mixture consisting of 50 mM Na-acetate buffer, 1 mM 4-aminoantipyrine, 1 mM 2,4-dichlorophenol, 50 mM MnCl2, and 0.2 mM NADH for 24 h. Finally, the oxidation of aminoantipyrine was measured as the absorbance of the reaction mixture at 510 nm, which was compared to a previously prepared standard curve to determine the H2O2 concentration in each sample.
2.4 Identification of nitrogen transporter genes
Unigenes annotated as putative nitrogen transporter genes were selected from our unpublished transcriptome data from ‘Bangia’ sp. ESS1, and their identity was confirmed by comparing their predicted amino acid sequences with those of known nitrogen transporters by a BLAST search (https://blast.ncbi.nlm.nih.gov/Blast.cgi) after identifying full-length open reading frames (ORFs) with the ORF finder (https://www.ncbi.nlm.nih.gov/orffinder/). Complete mRNA sequences from these genes have been deposited at DDBJ/EMBL/GenBank; their accession numbers are listed next to the species names in the phylogenetic trees.
2.5 Phylogenetic analyses of nitrogen transporters in plants and algae
To examine the evolutionary relationships of the ‘Bangia’ sp. ESS1 nitrogen transporters with those from other plants and algae, neighbor-joining phylogenetic trees were constructed with MEGA 7 software (https://www.megasoftware.net) using ClustalW to align the sequences of orthologs from other organisms. The amino acid sequences of AMTs, NRTs, and DUR3s used for the phylogenetic analysis were obtained from the GenBank, genome, and EST databases. The accession numbers and IDs of these sequences are indicated next to the species names in the phylogenetic trees.
2.6 Total RNA extraction and cDNA synthesis
Total RNA was extracted using a FavorPrep Plant Total RNA Mini Kit (FAVORGEN, Ping Tung, Taiwan) and treated with DNase I to remove any genomic DNA contamination using a TURBO DNA-free kit (Thermo Fisher Scientific, Waltham, MA, USA). Total RNA samples (300 ng) with A260/A280 ratios ranging from 1.9 to 2.1 were used to synthesize first-strand complementary DNA (cDNA) with a PrimeScript 1st strand cDNA Synthesis Kit (Takara Bio, Kusatsu, Japan). The thermal cycling parameters consisted of an initial denaturation step at 98°C for 30 s, followed by 30 cycles of 98°C for 10 s, 60°C for 30 s, and 72°C for 20 s, and a final extension step at 72°C for 5 min.
2.7 Reverse-transcription PCR for quantitative gene expression analysis
Primers for quantitative reverse-transcription PCR (qRT-PCR) were designed using Primer Premier 5 (http://www.premierbiosoft.com) as shown in Supplementary Table S1. To confirm the sizes of the amplified products and the suitability of the primers, a mixture of three cDNA samples was used with all primer sets for PCR with Phusion high-fidelity DNA polymerase and GC buffer (New England BioLabs, MA, USA) according to the manufacturer’s instructions. Primer sets that amplified DNA bands of the expected sizes, as checked by agarose gel electrophoresis, were employed for qPCR. qPCR was performed in a total volume of 20 μl containing 10 μl of 2× SYBR Premix Ex Taq GC, 0.4 μl of ROX Reference Dye, 2 μl of cDNA template, and 0.4 μl (10 μM) of each primer using a SYBR Premix Ex Taq GC kit (Takara Bio, Kusatsu, Japan). The thermal cycling parameters for the AriaMX (3000P) real-time PCR system (Agilent Technologies, CA, USA) consisted of 95°C for 3 min and 40 cycles of 95°C for 5 s and 60°C for 20 s. A dissociation curve was generated to confirm the specificity of amplification at 95°C for 1 min, 55°C for 30 s, and 95°C for 30 s.
2.8 Statistical analysis
Values are means ± SD from triplicate experiments. One-way ANOVA followed by a Tukey-Kramer test was used for multiple comparisons, and significant differences were determined using a cutoff value of p < 0.05.
3 Results
3.1 Heat stress memory reduces hydrogen peroxide production
Since one of the earliest physiological responses to heat stress in a variety of organisms is the production of reactive oxygen species (ROS) (Sachdev et al., 2021; Fortunato et al., 2023), we investigated the heat responses of ‘Bangia’ sp. ESS1 by monitoring its heat stress–dependent production of hydrogen peroxide (H2O2). When ‘Bangia’ sp. ESS1 was exposed to a non-lethal high temperature (28°) for 7 days (priming [P] in Figure 1A), H2O2 highly accumulated (Figure 1B), indicating that exposure to heat stress promotes H2O2 production in this alga as in other organisms.
We then quantified H2O2 contents during the establishment of heat stress memory to explore the relationship between heat stress memory and H2O2 production. When ‘Bangia’ sp. ESS1 was subjected to a 7-day heat stress at 28°C followed by recovery at 15°C for 2 days (recovery [R] in Figure 1A), H2O2 was still elevated at the end of the treatment (Figure 1B), suggesting that the physiological status induced by heat stress was maintained under the non-stressful temperature 15°C. Since stress memory enables survival under naturally lethal high temperatures (Kishimoto et al., 2019), we examined the effects of 32°C treatments on the H2O2 contents of algal samples. When ‘Bangia’ sp. ESS1 was incubated at 32°C for 1 day after a 28°C–15°C treatment (T-1 in Figure 1A), the H2O2 contents decreased, and the decrease was even greater after 6 days at 32°C (T-6 in Figures 1A, B). In contrast, the direct transfer of the alga from 15°C to 32°C for 1 or 6 days (LHT-1 and LHT-6 in Figure 1A) increased H2O2 contents (Figure 1B). These findings indicate that the establishment of heat stress memory decreases the heat stress–dependent production of H2O2 in ‘Bangia’ sp. ESS1.
3.2 Nitrogen transporter genes in ‘Bangia’ sp. ESS1
Based on the functional annotation in our unpublished ‘Bangia’ sp. ESS1 transcriptome data, we identified six unigenes (CL2278, CL2683, CL232, CL2570, CL337, and Unigene24217) as candidate AMT (BE1AMT) genes, one unigene (Unigene22285) as a candidate NRT (BE1NRT) gene, and two unigenes (CL2421 and Unigene31059) as candidate DUR3 (BE1DUR3) genes in ‘Bangia’ sp. ESS1. To explore what type(s) of nitrogen transporters these unigenes encode, we performed phylogenetic analysis using the amino acid sequences of AMTs, NRTs, and DUR3s from ‘Bangia’ sp. ESS1, other algae, and various terrestrial plants.
As shown in Figure 2, five of the six candidate ‘Bangia’ sp. ESS1 AMTs were placed in the plant AMT1 subfamily clade. Although AMTs in terrestrial plants are encoded by a multigene family comprising the AMT1 and AMT2 subfamilies (Couturier et al., 2007; Wittgenstein et al., 2014), no AMT2 subfamily members were identified in ‘Bangia’ sp. ESS1, which is consistent with our previous finding in P. yezoensis (Li et al., 2019b). Accordingly, these unigenes were designated BE1AMT1.1 (CL2278), BE1AMT1.3 (CL232), BE1AMT1.4 (CL2570), BE1AMT1.5 (CL2683), and BE1AMT1.7 (CL337). The names of these unigenes were derived from their AMT1 orthologs in P. yezoensis (Kakinuma et al., 2017; Li et al., 2019b), except for BE1AMT1.7, whose ortholog was not found in P. yezoensis. No orthologs of PyAMT1.2 or PyAMT1.6 were found in ‘Bangia’ sp. ESS1. Four BE1AMT1 family members diverged into four clades containing counterparts from P. yezoensis and Porphyra umbilicalis (Figure 2). For instance, the clade containing BE1AMT1 also contains PyAMT1, the clade containing BE1AMT1.3 and BE1AMT 1.4 contains PyAMT1.3 and PyAMT1.4, and the clade containing BE1AMT1.5 contains PyAMT1.5. It is notable that five BE1AMT1s commonly contained amino acid residues conserved in AMT signature and ammonium ion binding and transport (Supplementary Figure 1), suggesting functionality of these transporter proteins as AMT. These findings point to the conservation of functional diversity among AMT1 genes in Bangiales.
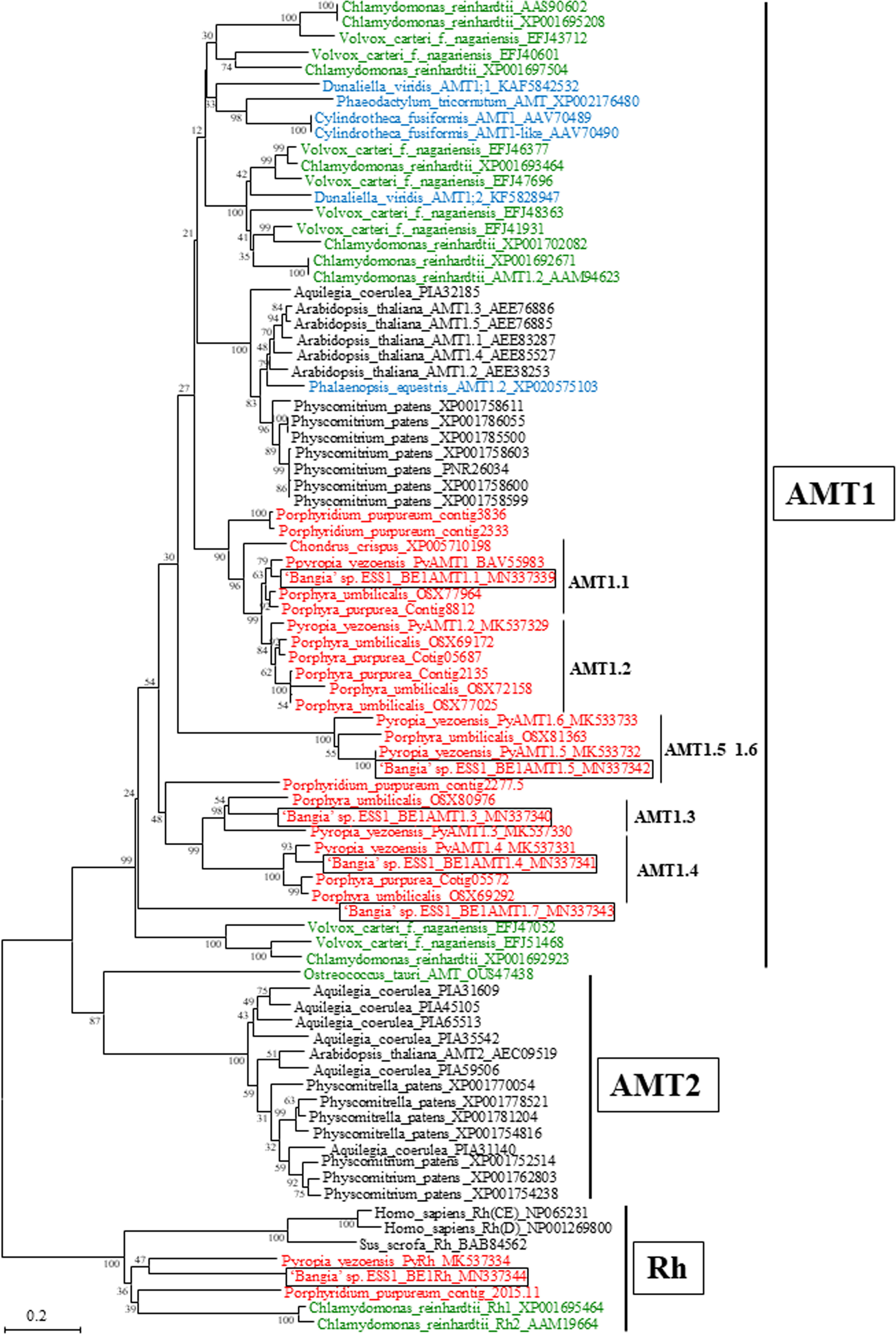
Figure 2 Neighbor-joining-based phylogenetic tree of AMTs from ‘Bangia’ sp. ESS1 compared with those of terrestrial plants and other algae. Boxes indicate BE1AMTs and BE1Rh from ‘Bangia’ sp. ESS1. AMTs from Rhodophyta, Chlorophyta, and Ochrophyta are highlighted by red, green, and blue font, respectively. The bootstrap values from 1000 replicates are indicated at the nodes of the tree. The DDBJ/EMBL/GenBank accession numbers of the AMTs and Rhs used in the phylogenetic analysis are shown next to the species names. Bar, 0.2 substitutions per site.
Unigene24217 falls into the clade containing PyRh (Li et al., 2019b), which is phylogenetically divergent from both AMT1 and AMT2 (Figure 2). Indeed, a product of Unigene24217 shares 30.65% identity with PyRh from P. yezoensis but only 17.39% identity with BE1AMT1. Thus, Unigene24217 was designated BE1Rh (low homology between BE1AMTs and BE1Rh is represented in Supplementary Figure 1).
Unigene22285 formed a multicellular red algal clade with NTR2s from Chondrus crispus, Gracilariopsis chorda, Po. umbilicalis, and P. yezoensis, all of which contain a single NRT2 gene (Figure 3). Thus, Unigene22285 was designated BE1NRT2 that contains NRT2 consensus motif AGWGNLG (Supplementary Figure 2). In land plants, three different gene families of nitrate transporters have been identified, NRT1, NRT2, and NRT3 (Plett et al., 2010), all of which symport NO3− and protons, with low or high affinity for NO3− (Pinton et al., 2016). In addition, the genome of the unicellular green alga Chlamydomonas reinhardtii contains six NRT2 genes (Higuera et al., 2016), and the unicellular red alga Porphyridium purpureum contains two NRT2 genes (Figure 3). Therefore, the presence of a single-copy NRT2 gene (Figure 2; Kakinuma et al., 2008; Brawley et al., 2017; Li et al., 2019b) is unique to multicellular Bangiales.
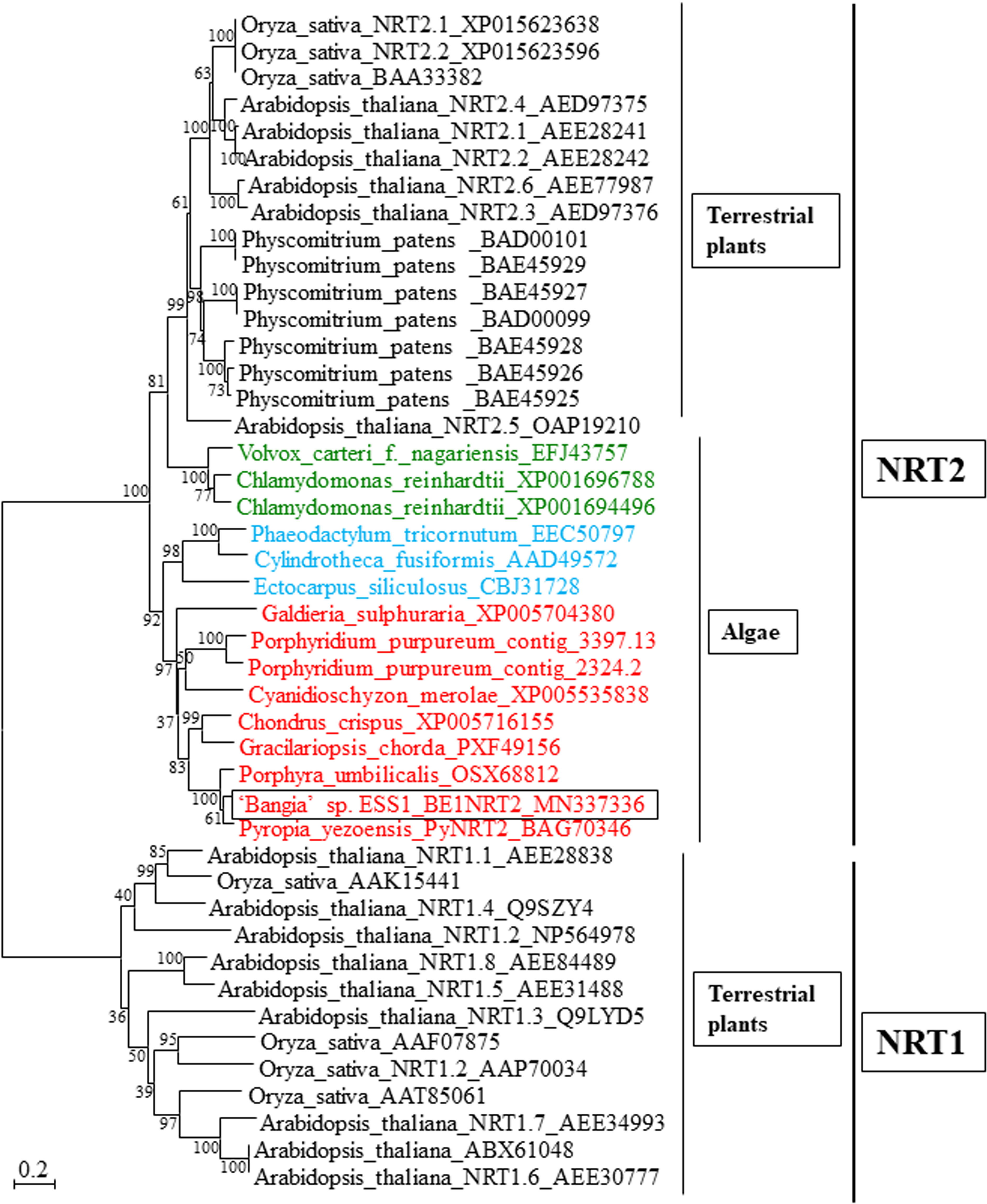
Figure 3 Neighbor-joining-based phylogenetic tree of NRTs from terrestrial plants and algae. The box indicates BE1NRT2 from ‘Bangia’ sp. ESS1. NRT2s from Rhodophyta, Chlorophyta, and Ochrophyta are highlighted by red, green, and blue font, respectively. The bootstrap values from 1000 replicate are indicated at the nodes of the tree. The DDBJ/EMBL/GenBank accession numbers of the NRTs used in the phylogenetic analysis are shown next to the species names. Bar, 0.2 substitutions per site.
There are two red algal clades (I and II) of DUR3s (Figure 4), each of which contains one or two DUR3 family members from other red algae such as P. yezoensis, Po. umbilicalis, C. crispus, and G. chorda (Kakinuma et al., 2008; Collén et al., 2013; Kakinuma et al., 2016; Brawley et al., 2017; Kakinuma et al., 2017; Lee et al., 2018). Since CL2421 was included in clade I with PyDUR3.1 and Unigene31059 belongs to clade II with PyDUR3.2 and PyDUR3.3 (Figure 4), CL2421 and Unigene31059 were designated BE1DUR3.1 and BE1DUR3.2, respectively. These findings are inconsistent with the previous finding that only a single gene encoding DUR3 is present in the genomes of vascular plants (Kojima et al., 2007; Wang et al., 2012; Liu et al., 2014; Pinton et al., 2016). Amino acid sequence of BE1DUR3s represented high homology to those of DUR3s from P. yezoensis and Po. umbilicalis (Supplementary Figure 3), suggesting functionality of two BE1DUR3s as urea transporters.
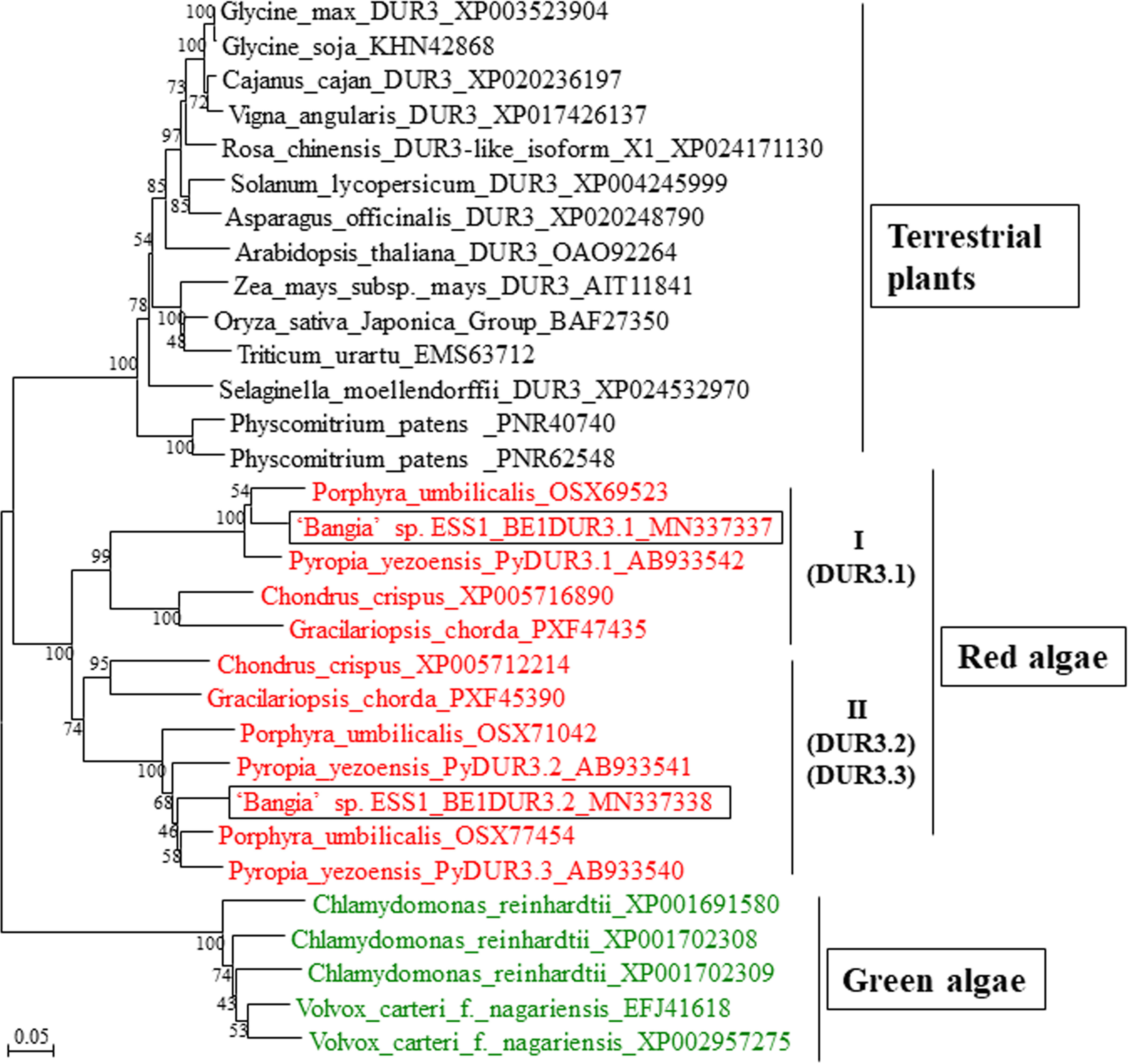
Figure 4 Neighbor-joining-based phylogenetic tree of DUR3s from ‘Bangia’ sp. ESS1 compared with those of terrestrial plants and other algae. Boxes indicate BE1DUR3.1 and BE1DUR3.2 from ‘Bangia’ sp. ESS1. DUR3s from Rhodophyta and Chlorophyta are highlighted by red and green font, respectively. The bootstrap values from 1000 replicates are indicated at the nodes of the tree. The DDBJ/EMBL/GenBank accession numbers of the DUR3s used in the phylogenetic analysis are shown next to the species names. Bar, 0.05 substitutions per site.
3.3 Effects of heat stress memory on the expression of ammonium transporter genes
As shown in Figure 5, the expression of BE1AMT1.3, BE1AMT1.5, and BE1AMT1.7 was highly induced by 7 days of 28°C treatment, although the expression levels of BE1AMT1.1 and BE1AMT1.4 were unchanged. In addition, the recovery treatment (7 day-28°C to 2 day-15°C) reduced the expression levels of all these genes except for BE1AMT1.1, whose expression was induced 5-fold by this treatment. The expression profiles of these genes in the triggering experiments (recovery treatment plus 32°C for 1 or 6 days) varied among genes. For example, no response to the lethal temperature was observed for BE1AMT1.1, whereas the expression of BE1AMT1.4 and BE1AMT1.7 increased by 2 and 8 times, respectively, following 1 day at 32°C and subsequently decreased after 6 days at this temperature. In addition, under the same conditions, BE1AMT1.5 expression decreased, whereas BE1AMT1.3 expression was maintained.
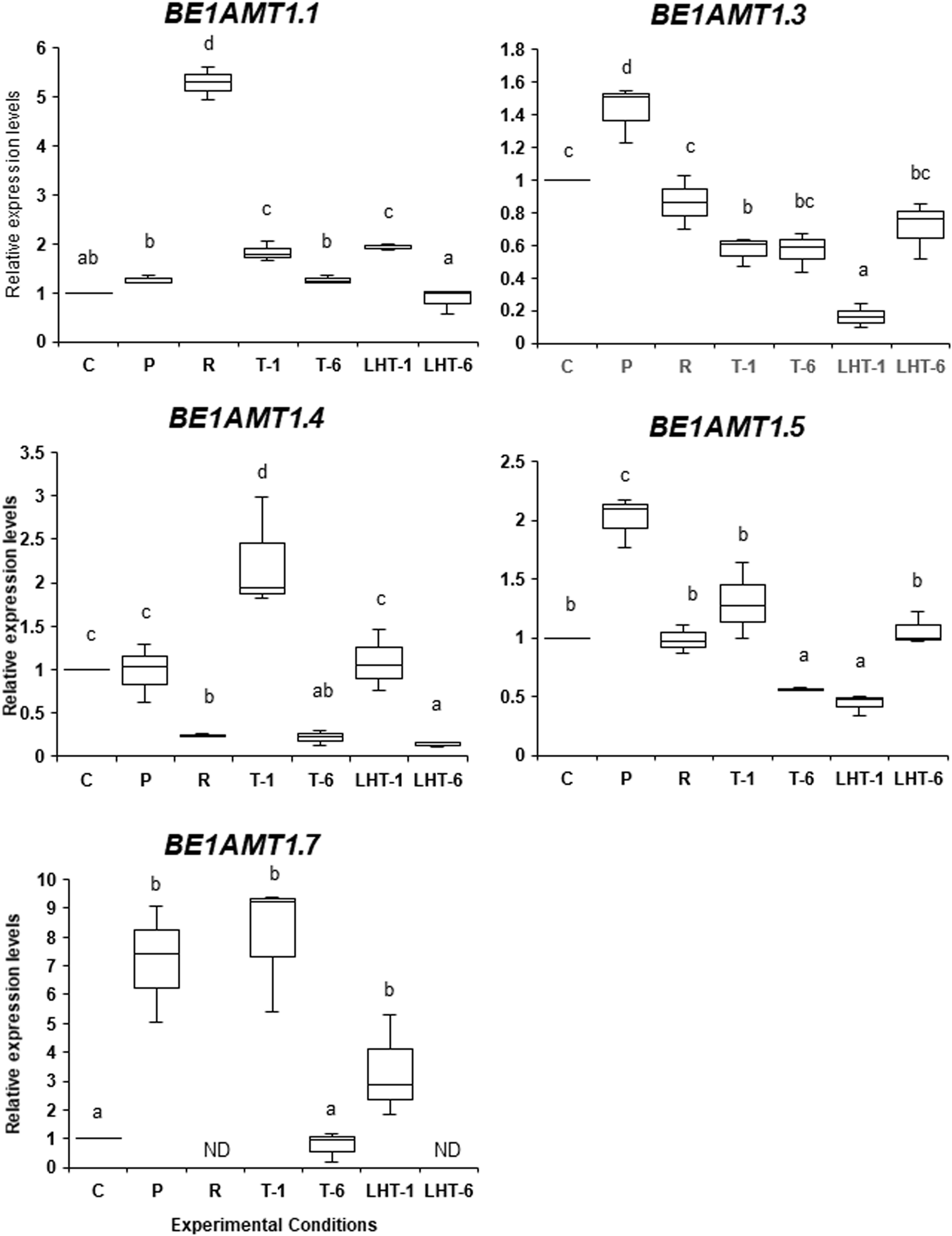
Figure 5 Differences in the expression patterns of the five BE1AMT1 genes under various heat stress conditions. The expression levels of BE1AMT1.1, BE1AMT1.3, BE1AMT1.4, BE1AMT1.5, and BE1AMT1.7 in algal samples treated with the experimental conditions indicated in Figure 1A were measured by qRT-PCR. Relative mRNA levels, which were normalized to the expression of the Actin gene as the reference (Li et al., 2019a), are mean fold changes compared to control (C) samples, with error bars representing the standard deviations of triplicate experiments (n = 3), each with triple technical replicates for qRT-PCR. ND, not detected. Different letters denote statistically significant differences (p < 0.05), as determined by one-way ANOVA.
When ‘Bangia’ sp. ESS1 was incubated at 32°C for 1 or 6 days with no pretreatment (LH-1 and LH-2 in Figure 1), the expression patterns of the BE1AMT1 genes did not strongly vary (Figure 5). For instance, BE1AMT1.1 and BE1AMT1.7 were induced 2-fold by 1 day of 32°C treatment, whereas 6 days of this treatment completely inhibited their expression. BE1AMT1.3 and BE1AMT1.5 expression decreased following 1 day of 32°C treatment and recovered in response to 6 days of treatment. BE1AMT1.4 expression did not significantly change in response to 1 day of heat treatment and decreased in response to 6 days of heat treatment. A comparison of the gene expression profiles between the triggering treatments and the direct transfer to lethal temperature indicated that stress memory influences the expression of BE1AMT1.3, BE1AMT1.4, and BE1AMT1.5, whereas heat stress alone induces the expression of BE1AMT1.7.
3.4 Effects of heat stress memory on nitrate transporter gene expression
As shown in Figure 6, BE1NRT2 expression slightly increased in response to non-lethal heat stress but strongly decreased after direct transfer to lethal high temperature for 1 or 6 days. Expression of BE1NRT2 was normal after 6 days of lethal heat in the triggering treatment, but the triggering treatment caused a more extreme decrease in expression after 1 day at 32°C when compared to direct transfer to the lethal temperature. Although this additional reduction in BE1NRT2 expression by triggering was unexpected, we suggest that this gene is not actively involved in heat stress memory–dependent responses.
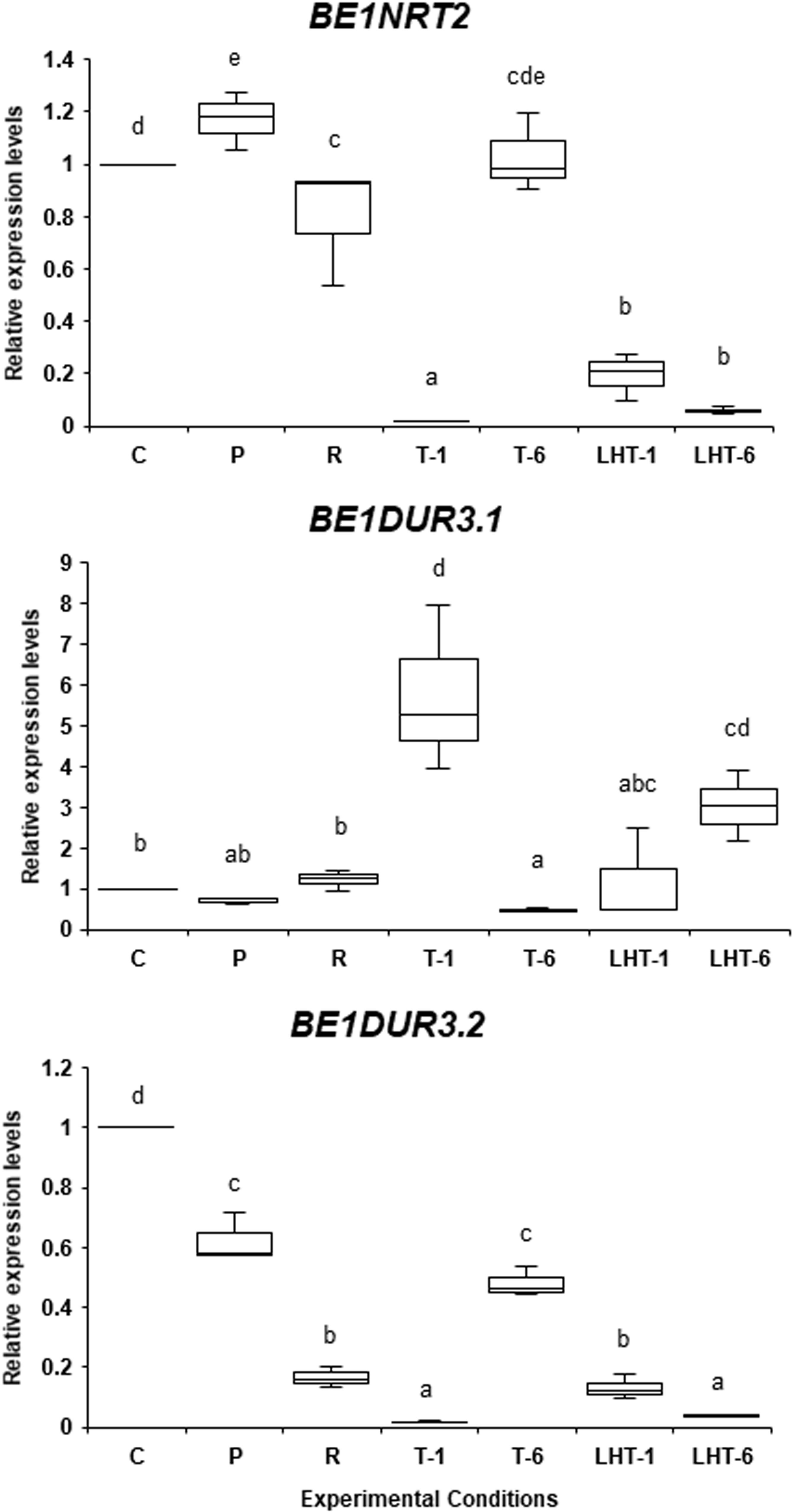
Figure 6 Differences in the expression patterns of BE1NRT2 and BE1DUR3 genes under various heat stress conditions. The expression levels of BE1NRT2, BE1DUR3.1, and BE1DUR3.2 in algal samples treated with the experimental conditions indicated in Figure 1A were examined by qRT-PCR. Relative mRNA levels, which were normalized to the expression of the Actin gene as the reference (Li et al., 2019a), are mean fold changes compared to control (C) samples, with error bars representing the standard deviations of triplicate experiments (n = 3), each with triple technical replicates for qRT-PCR. Different letters denote statistically significant differences (p < 0.05), as determined by one-way ANOVA.
3.5 Effects of heat stress memory on urea transporter gene expression
The expression profile of BE1DUR3.1 was similar to that of BE1AMT1.4: BE1DUR3.1 expression was induced 5-fold by a 1-day exposure to 32°C after the triggering treatment and highly decreased in response to a 6-day exposure to 32°C after the triggering treatment (Figure 6). Although direct exposure to 32°C for 6 days slightly increased the expression of BE1DUR3.1 (Figure 6), BE1AMT4 expression did not increase under the same conditions (Figure 5). These findings indicate that BE1DUR1.1 expression is dependent on heat stress memory. Finally, all treatments reduced the expression of BE1DUR3.2 (Figure 6), suggesting that this gene is not involved in the heat stress response in ‘Bangia’ sp. ESS1.
4 Discussion
Heat stress activates various signal transduction pathways to induce cellular responses to high temperature, including the acquisition of heat stress tolerance and the establishment of heat stress memory (Yamaguchi, 2021a; Yamaguchi, 2021b; Ramakrishnan et al., 2022; Sharma et al., 2022; Charng et al., 2023; Kambona et al., 2023). Although many studies have shed light on these physiological responses in terrestrial plants, our knowledge about tolerance to and memory of heat stress in algae is limited. To date, the presence of an intrinsic ability to establish heat stress memory in algae has only been demonstrated in the red alga ‘Bangia’ sp. ESS1 (Kishimoto et al., 2019; Khoa et al., 2021). Since heat stress generally reduces nitrogen uptake and growth in algae (Gerard, 1997; Hay et al., 2010; Gouvêa et al., 2017; Endo et al., 2020; Fernández et al., 2020; Wu et al., 2022), it is important to address whether the establishment of heat stress memory modulates the uptake of nitrogen sources under heat stress conditions in algae. Therefore, in this study, we identified ‘Bangia’ sp. ESS1 genes encoding the nitrogen transporters BE1AMT1s, BE1NRT2, and BE1DUR3s and examined the effects of heat stress memory on their expression.
We first evaluated the intrinsic ability to establish heat stress memory in ‘Bangia’ sp. ESS1 by monitoring the accumulation of H2O2 under various heat stress conditions. Although a 7-day incubation at 28°C prompted the production of H2O2, heat stress memory reduced the amount of H2O2 produced at 32°C. These results indicate that heat stress memory reduces the sensitivity of the red alga to lethal high temperatures; this results in a reduction in H2O2 content at 32°C by lowering the threshold to initiate the response to heat stress. Heat stress increased H2O2 contents in the red alga Pyropia tenera, a response that was attenuated in a P. tenera mutant with increased heat stress tolerance (Lee and Choi, 2019). These findings support the tight relationship between reduced H2O2 accumulation and the acquisition of heat stress tolerance with low sensitivity to heat stress, although the contribution of heat stress memory to the acquisition of heat stress tolerance in P. tenera has yet to be analyzed.
In addition, the heat stress memory–dependent inhibition of H2O2 accumulation at the lethal temperature suggests that less H2O2 is available to act as a second messenger to activate heat stress signal transduction pathways (Sachdev et al., 2021; Fortunato et al., 2023), which may be related to the modulation of nitrogen transporter gene expression by heat stress memory. Based on this idea, we propose that heat stress memory–dependent inhibition of H2O2 production reduces the expression and activation of ROS scavengers, such as superoxide oxidase (SOD), catalase (CAT), and ascorbate peroxidase (APX) (Shah and Nahakpam, 2012; Hasanuzzaman et al., 2020; Tiwari et al., 2022). If so, this may underlie the reduced sensitivity of algae to heat stress at lethal temperature in response to heat stress memory; however, this notion remains to be confirmed.
This is the first report of nitrogen transporter genes in the genus Bangia. We identified five paralogs of BE1MAT1 that were phylogenetically divided into five subclades, each of which contained the corresponding orthologs from P. yezoensis and Po. umbilicalis. These findings suggest that an ancient Bangiales AMT gene might have diversified into five genes prior to the separation of Pyropia, Porphyra, and Bangia. In addition, we identified two paralogs, BE1DUR3.1 and BE1DUR3.2, belonging to different red algal DUR3 subclades. Since these subclades contained orthologs from G. chorda, C. crispus, Po. umbilicalis, and P. yezoensis, we suggest that the ancestral DUR3 gene might have been present prior to the separation of Bangiophyceae and Florideophyceae. Moreover, in contrast to AMT1s and DUR3s, multicellular Bangiales contain only one NRT2 gene, although multiple NRT2 genes have been identified in unicellular red algae and terrestrial plants (Figure 3). These observations suggest that the ancestral NRT2 gene did not undergo duplication in ancient multicellular algae, but rather diversified in terrestrial plants after their colonization of land.
It is plausible that the differences in these genes between algae and terrestrial plants are due to the differences in their living environments. Since seawater contains high levels of nitrate ion, it appears that the functional diversity of the NRT2 gene is not required in marine algae. The situation is different in soil, where the production of nitrate ions completely depends on the assimilation of nitrogen gas by bacteria to enable the absorption of nitrate ions by plants (Bekele and Yilma, 2021; Guo et al., 2023). Fresh water also contains lower levels of nitrate ions than seawater. This appears to be responsible for the duplication of the NRT2 gene in freshwater algae such as Porphyridium purpureum and C. reinhardtii, which likely contributed to the efficient absorption of low concentrations of nitrogen sources by these algae. By contrast, the requirement for ammonium ions and urea is usually acute in seawater; thus, the multiplication of AMT1 and DUR3 genes might have been necessary for the effective absorption of these molecules by marine algae. Whether there is a relationship between nitrogen transporter gene amplification and the availability of different nitrogen sources in the environment should be verified.
We also addressed whether heat stress and heat stress memory affect the expression levels of nitrogen transporter genes in ‘Bangia’ sp. ESS1. We detected the heat stress–inducible expression of BE1AMT1.3, BE1AMT1.5, and BE1AMT1.7, suggesting that these genes contribute to the early phase of the heat stress response. Of these genes, the expression levels of BE1AMT1.3 and BE1AMT1.5 were reduced if exposure to the lethal temperature came after priming and recovery, pointing to the reduced sensitivity to heat stress–dependent expression due to heat stress memory.
Although this expression pattern appears to be consistent with the production of H2O2, as shown in Figure 1, the effects of heat stress memory on gene expression profiles were complex. For instance, triggering-dependent expression was observed for BE1AMT1.4 and BE1DUR3.1, whose expression was not induced by priming or direct transfer to the lethal temperature, suggesting that heat stress memory increases the sensitivity of these genes to heat stress. A similar pattern was observed for BE1AMT1.7, although priming also induced its expression. In addition, BE1AMT1.1 showed recovery-dependent and priming- and triggering-independent expression, and triggering highly reduced BE1NRT2 and BE1DUR3.2 expression to the levels observed in response to direct transfer to the lethal temperature. These results suggest that the latter two genes are not involved in nitrogen transport under heat stress conditions.
In conclusion, our results indicate that reducing the sensitivity to heat stress by heat stress memory influences mRNA expression in a gene-specific manner. The priming-inducible expression of BE1AMT1.4 and BE1AMT1.5 and triggering-dependent expression of BE1AMT1.4 and BE1DUR3.1 strongly point to functional diversity in the timing of nitrogen absorption during various phases of heat stress. Therefore, we propose that the main effects of heat stress memory are to reduce priming-dependent gene expression by triggering and to induce the trigger-dependent expression of priming-independent genes. These effects appear to underlie the reduced sensitivity of ‘Bangia’ sp. ESS1 to heat stress via the memorization of stress, with differential contributions of nitrogen transporters to the maintenance of nitrogen absorption under heat stress conditions.
We previously demonstrated that ‘Bangia’ sp. ESS1 maintains increased levels of various saturated and monounsaturated fatty acids and decreased levels of various polyunsaturated fatty acids as a physiological basis for heat stress memory (Kishimoto et al., 2019). Perhaps the physical state of the membrane, which depends on fatty acid composition, modulates the activities of membrane-bound nitrogen transporters, which might control the expression patterns of the genes encoding them via a feedback mechanism. Thus, both the activities of nitrogen transporters in membranes in various physical states and the mechanisms regulating nitrogen transporter gene expression under heat stress conditions must be elucidated. Notably, the capability to establish heat stress memory varies among species in the genus Bangia (Khoa et al., 2021). To understand why heat stress response strategies vary among species, comparative analyses of heat stress–dependent H2O2 production and the expression profiles of nitrogen transporter genes should be performed using ‘Bangia’ sp. ESS2, which lacks heat stress memory. These approaches could further our understanding of the relationship between heat stress memory and growth regulation for the survival of Bangiales under unfavorable high-temperature conditions.
5 Conclusion
In this study, we addressed the biological significance of heat stress memory in the red alga ‘Bangia’ sp. ESS1. By comparing changes in the quantities of H2O2 and transcripts from genes encoding nitrogen transporters such as BE1AMT1s, BE1NRT2, and BE1DUR3s, we identified priming- and triggering-dependent sets of nitrogen transporter genes whose expression is repressed and promoted, respectively, by heat stress memory. These different expression profiles promote heat stress tolerance and protect the organism by maintaining nitrogen absorption at high temperatures. Our study provides unique insights into the strategies of heat stress responses and adaptation via the establishment of heat stress memory in aquatic photosynthetic organisms.
Data availability statement
The amino acid sequence data presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article. The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Author contributions
NS: Data curation, Formal analysis, Investigation, Methodology, Validation, Visualization, Writing – review & editing. HK: Data curation, Formal analysis, Investigation, Methodology, Validation, Visualization, Writing – review & editing. KM: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported in part by the designated research fund from Miyagi University.
Acknowledgments
We thank Chengze Li for her technical assistance and Hiroyuki Mizuta, Hokkaido University, for permitting Ho Viet Khoa to study at Miyagi University. Ho Viet Khoa was supported by the Ministry of Education, Culture, Sports, Science, and Technology of Japan.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2024.1331496/full#supplementary-material
References
Bai, H., Euring, D., Volmer, K., Janz, D., Polle, A. (2013). The nitrate transporter (NRT) gene family in poplar. PloS One 8, e72126. doi: 10.1371/journal.pone.0072126
Bekele, M., Yilma, G. (2021). Nitrogen fixation using symbiotic and non-symbioticmicrobes: A review article. Biochem. Mol. Biol. 6, 92–98. doi: 10.11648/j.bmb.20210604.12
Brawley, S. H., Blouin, N. A., Ficko-Blean, E., Wheeler, G. L., Lohr, M., Goodson, H. V., et al. (2017). Insights into the red algae and eukaryotic evolution from the genome of Porphyra umbilicalis (Bangiophyceae, Rhodophyta). Proc. Natl. Acad. Sci. U.S.A. 114, E6361–E6370. doi: 10.1073/pnas.1703088114
Chang, J., Shi, J., Lin, J., Ji, D., Xu, Y., Chen, C., et al. (2021). Molecular mechanism underlying Pyropia haitanensis PhHsp22-mediated increase in the high-temperature tolerance of Chlamydomonas reinhardtii. J. Appl. Phycol. 33, 1137–1148. doi: 10.1007/s10811-020-02351-6
Charng, Y. Y., Mitra, S., Yu, S. J. (2023). Maintenance of abiotic stress memory in plants: Lessons learned from heat acclimation. Plant Cell 35, 187–200. doi: 10.1093/plcell/koac313
Chen, H., Chu, J. S., Chen, J., Luo, Q., Wang, H., Lu, R., et al. (2022). Insights into the ancient adaptation to intertidal environments by red algae based on a genomic and multiomics investigation of Neoporphyra haitanensis. Mol. Biol. Evol. 39, msab315. doi: 10.1093/molbev/msab315
Collén, J., Porcel, B., Carré, W., Ball, S. G., Chaparro, C., Tonon, T., et al. (2013). Genome structure and metabolic features in the red seaweed Chondrus crispus shed light on evolution of the Archaeplastida. Proc. Natl. Acad. Sci. U.S.A. 110, 5247–5252. doi: 10.1073/pnas.1221259110
Couturier, J., Montanini, B., Martin, F., Brun, A., Blaudez, D., Chalot, M. (2007). The expanded family of ammonium transporters in the perennial poplar plant. New Phytol. 174, 137–150. doi: 10.1111/j.1469-8137.2007.01992.x
D’Apuzzo, E., Rogato, A., Simon-Rosin, U., El Alaoui, H., Barbulova, A., Betti, M., et al. (2004). Characterization of three functional high-affinity ammonium transporters in Lotus japonicus with differential transcriptional regulation and spatial expression. Plant Physiol. 134, 1763–1774. doi: 10.1104/pp.103.034322
Edward, H., Richard, M. A. (2001). Nutrients mobilized from leaves ofthaliana during leaf senescence. Arabidopsis J. Plant Physiol. 158, 1317–1323. doi: 10.1078/0176-1617-00608
Endo, H., Inomata, E., Gao, X., Kinoshita, J., Sato, Y., Agatsuma, Y. (2020). Heat stress promotes nitrogen accumulation in meristems via apical blade erosion in a brown macroalga with intercalary growth. Front. Mar. Sci. 7. doi: 10.3389/fmars.2020.575721
Fernández, P. A., Gaitán-Espitia, J. D., Leal, P. P., Schmid, M., Revill, A. T., Hurd, C. L. (2020). Nitrogen sufficiency enhances thermal tolerance in habitat-forming kelp: implications for acclimation under thermal stress. Sci. Rep. 10, 3186. doi: 10.1038/s41598-020-60104-4
Fortunato, S., Lasorella, C., Dipierro, N., Vita, F., de Pinto, M. C. (2023). Redox signaling in plant heat Stress response. Antioxidants 12, 605. doi: 10.3390/antiox12030605
Gao, T., Mo, Z., Tang, L., Yu, X., Du, G., Mao, Y. (2022). Heat shock protein 20 gene superfamilies in red algae: Evolutionary and functional diversities. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.817852
Gazzarrini, S., Lejay, L., Gojon, A., Ninnemann, O., Frommer, W. B., von Wirén, N. (1999). Three functional transporters for constitutive, diurnally regulated, and starvation-induced uptake of ammonium into Arabidopsis roots. Plant Cell 11, 937–948. doi: 10.1105/tpc.11.5.937
Gerard, V. A. (1997). The role of nitrogen nutrition in high-temperature tolerance of the kelp, Laminaria saccharina (Chromophyta). J. Phycol. 33, 800–810. doi: 10.1111/j.0022-3646.1997.00800.x
Gouvêa, L. P., Schubert, N., Martins, C. D. L., Sissini, M., Ramlov, F., Rodrigues, E. R. D. O., et al. (2017). Interactive effects of marine heatwaves and eutrophication on the ecophysiology of a widespread and ecologically important macroalga. Limnol. Oceanogr. 62, 2056–2075. doi: 10.1002/lno.10551
Guo, K., Yang, J., Yu, N., Luo, L., Wang, E. (2023). Biological nitrogen fixation in cereal crops: Progress, strategies, and perspectives. Plant Commun. 4, 100499. doi: 10.1016/j.xplc.2022.100499
Hasanuzzaman, M., Bhuyan, M. H. M. B., Zulfiqar, F., Raza, A., Mohsin, S. M., Mahmud, J. A., et al. (2020). Reactive oxygen species and antioxidant defense in plants under abiotic stress: Revisiting the crucial role of a universal defense regulator. Antioxidants 9, 681. doi: 10.3390/antiox9080681
Hay, K. B., Millers, K. A., Poore, A. G., Lovelock, C. E. (2010). The use of near infrared reflectance spectrometry for characterization of brown algal tissue. J. Phycol. 46, 937–946. doi: 10.1111/j.1529-8817.2010.00890.x
Higuera, J. J., Calatrava, V., González, Z., Mariscal, V., Siverio, J. M., Fernández, E., et al. (2016). NRT2.4 and NRT2.5 are two half-size transporters from the Chlamydomonas NRT2 Family. Agronomy 6, 20. doi: 10.3390/agronomy601002
Hirata, R., Takahashi, M., Saga, N., Mikami, K. (2011). Transient gene expression system established in Porphyra yezoensis is widely applicable in Bangiophycean algae. Mar. Biotechnol. 13, 1038–1047. doi: 10.1007/s10126-011-9367-6
Jin, J., Yang, L., Fan, D., Liu, X., Hao, Q. (2020). Comparative transcriptome analysis uncovers different heat stress responses in heat-resistant and heat-sensitive jujube cultivars. PloS One 15, e0235763. doi: 10.1371/journal.pone.0235763
Kakinuma, M., Coury, D. A., Nakamoto, C., Sakaguchi, K., Amano, H. (2008). Molecular analysis of physiological responses to changes in nitrogen in a marine macroalga, Porphyra yezoensis (Rhodophyta). Cell Biol. Toxicol. 24, 629–639. doi: 10.1007/s10565-007-9053-7
Kakinuma, M., Nakamoto, C., Kishi, K., Coury, D. A., Amano, H. (2017). Isolation and functional characterization of an ammonium transporter gene, PyAMT1, related to nitrogen assimilation in the marine macroalga Pyropia yezoensis (Rhodophyta). Mar. Environ. Res. 128, 76–87. doi: 10.1016/j.marenvres.2016.08.007
Kakinuma, M., Suzuki, K., Iwata, S., Coury, D. A., Iwade, S., Mikami, K. (2016). Isolation and characterization of a new DUR3-like gene, PyDUR3.3, from the marine macroalga (Rhodophyta). Fish. Sci. 82, 171–184. doi: 10.1007/s12562-015-0947-7
Kambona, C. M., Koua, P. A., Léon, J., Ballvora, A. (2023). Stress memory and its regulation in plants experiencing recurrent drought conditions. Theor. Appl. Genet. 136, 26. doi: 10.1007/s00122-023-04313-1
Karsten, U., West, J. A. (2000). Living in the intertidal zone - seasonal effects on heterosides and sun-screen compounds in the red alga Bangia atropurpurea (Bangiales). J. Exp. Mar. Biol. Ecol. 254, 221–234. doi: 10.1016/s0022-0981(00)00280-x
Khoa, H. V., Kumari, P., Uchida, H., Murakami, A., Shimada, S., Mikami, K. (2021). Heat-stress responses differ among species from different ‘Bangia’ clades of Bangiales (Rhodophyta). Plants 22, 1733. doi: 10.3390/plants10081733
Khoa, H. V., Mikami, K. (2022). Membrane fluidization-dependent and -independent pathways are involved in heat stress-inducible gene expression in the marine red alga Neopyropia yezoensis. Cells 11, 1486. doi: 10.3390/cells11091486
Kim, E., Park, H. S., Jung, Y., Choi, D. W., Jeong, W. J., Park, H. S. (2011). Identification of the high-temperature response genes from Porphyra seriata (Rhodophyta) expression sequence tags and enhancement of heat tolerance of Chlamydomonas (Chlorophyta) by expression of the Porphyra HTR2 gene. J. Phycol. 47, 821–828. doi: 10.1111/j.1529-8817.2011.01008.x
Kishimoto, I., Ariga, I., Itabashi, Y., Mikami, K. (2019). Heat-stress memory is responsible for acquired thermotolerance in Bangia fuscopurpurea. J. Phycol. 55, 971–975. doi: 10.1111/jpy.12895
Kojima, S., Bohner, A., Gassert, B., Yuan, L., von Wiren, N. (2007). AtDUR3 represents the major transporter for high-affinity urea transport across the plasma membrane of nitrogen-deficient Arabidopsis roots. Plant J. 52, 30–40. doi: 10.1111/j.1365-313X.2007.03223.x
Kumar, M., Trivedi, N., Reddy, C. R. K., Jha, B. (2011). Toxic effects of imidazolium ionic liquids on the green seaweed Ulva lactuca: oxidative stress and DNA damage. Chem. Res. Toxicol. 24, 1882–1890. doi: 10.1021/tx200228c
Lee, H.-J., Choi, J. (2019). Enhancing temperature tolerance of Pyropia tenera (Bangiales) by inducing mutation. Phycologia 58, 496–503. doi: 10.1080/00318884.2019.1623547
Lee, J., Yang, E. C., Graf, L., Yang, J. H., Qiu, H., Zelzion, U., et al. (2018). Analysis of the draft genome of the red seaweed Gracilariopsis chorda provides insights into genome size evolution in Rhodophyta. Mol. Biol. Evol. 35, 1869–1886. doi: 10.1093/molbev/msy081
Li, C., Ariga, I., Mikami, K. (2019b). Difference in nitrogen starvation-inducible expression patterns among phylogenetically diverse ammonium transporter genes in the red seaweed Pyropia yezoensis. Am. J. Plant Sci. 10, 1325–1349. doi: 10.4236/ajps.2019.108096
Li, C., Irie, R., Shimada, S., Mikami, K. (2019a). Requirement of different normalization genes for quantitative gene expression analysis under abiotic stress conditions in ‘Bangia’ sp. ESS1. J. Aquat. Res. Mar. Sci. 2019, 194–205. doi: 10.29199/2639-4618/ARMS.202037
Liu, G. W., Sun, A. L., Li, D. Q., Athman, A., Gilliham, M., Liu, L. H. (2014). Molecular identification and functional analysis of a maize (Zea mays) DUR3 homolog that transports urea with high affinity. Planta 241, 861–874. doi: 10.1007/s00425-014-2219-7
Ludewig, U., Neuhäuser, B., Dynowski, M. (2007). Molecular mechanisms of ammonium transport and accumulation in plants. FEBS Lett. 581, 2301–2308. doi: 10.1016/j.febslet.2007.03.034
Mikami, K., Khoa, H. V. (2023). Membrane fluidization governs the coordinated heat-inducible expression of nucleus- and plastid genome-encoded heat shock protein 70 genes in the marine red alga Neopyropia yezoensis. Plants 12, 2070. doi: 10.3390/plants12112070
Mikami, K., Kishimoto, I. (2018). Temperature promoting the asexual life cycle program in Bangia fuscopurpurea (Bangiales, Rhodophyta) from Esashi in the Hokkaido Island, Japan. Algal Resour. 11, 25–32. doi: 10.20804/jsap.11.1_2_25
Mikami, K., Takahashi, M. (2023). Life cycle and reproduction dynamics of Bangiales in response to environmental stresses. Semin. Cell Dev. Biol. 134, 14–26. doi: 10.1016/j.semcdb.2022.04.004
Notoya, M., Iijima, N. (2003). Life history and sexuality of archeospore and apogamy of atropurpurea (Roth) Lyngbye (Bangiales, Rhodophyta) from Fukaura and Enoshima. Jpn. Fish. Sci. 69, 799–805. doi: 10.1046/j.1444-2906.2003.00689.x
Okumoto, S., Versaw, W. (2017). Genetically encoded sensors for monitoring the transport and concentration of nitrogen-containing and phosphorus-containing molecules in plants. Curr. Opin. Plant Biol. 39, 129–135. doi: 10.1016/j.pbi.2017.07.004
Pinton, R., Tomasi, N., Zanin, L. (2016). Molecular and physiological interactions of urea and nitrate uptake in plants. Plant Signal Behav. 11, e1076603. doi: 10.1080/15592324.2015.1076603
Plett, D., Toubia, J., Garnett, T., Tester, M., Kaiser, B. N., Baumann, U. (2010). Dichotomy in the NRT gene families of dicots and grass species. PloS One 5, e15289. doi: 10.1371/journal.pone.0015289
Ramakrishnan, M., Zhang, Z., Mullasseri, S., Kalendar, R., Ahmad, Z., Sharma, A., et al. (2022). Epigenetic stress memory: A new approach to study cold and heat stress responses in plants. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.1075279
Sachdev, S., Ansari, S. A., Ansari, M. I., Fujita, M., Hasanuzzaman, M. (2021). Abiotic stress and reactive oxygen species: Generation, signaling, and defense mechanisms. Antioxidants 10, 277. doi: 10.3390/antiox10020277
Sato, Y., Kinoshita, Y., Mogamiya, M., Inomata, E., Hoshino, M., Hiraoka, M. (2021). Different growth and sporulation responses to temperature gradient among obligate apomictic strains of Ulva prolifera. Plants 10, 2256. doi: 10.3390/plants10112256
Shah, K., Nahakpam, S. (2012). Heat exposure alters the expression of SOD, POD, APX and CAT isozymes and mitigates low cadmium toxicity in seedlings of sensitive and tolerant rice cultivars. Plant Physiol. Biochem. 57, 106–113. doi: 10.1016/j.plaphy.2012.05.007
Sharma, M., Kumar, P., Verma, V., Sharma, R., Bhargava, B., Irfan, M. (2022). Understanding plant stress memory response for abiotic stress resilience: Molecular insights and prospects. Plant Physiol. Biochem. 179, 10–24. doi: 10.1016/j.plaphy.2022.03.004
Sun, P., Mao, Y., Li, G., Cao, M., Kong, F., Wang, L., et al. (2015). Comparative transcriptome profiling of Pyropia yezoensis (Ueda) M.S. Hwang & H.G. Choi in response to temperature stresses. BMC Genom. 16, 463. doi: 10.1186/s12864-015-1586-1
Sutherland, J., Lindstrom, S., Nelson, W., Brodie, J., Lynch, M., Hwang, M. S., et al. (2011). A new look at an ancient order: Generic revision of the Bangiales. J. Phycol. 47, 1131–1151. doi: 10.1111/j.1529-8817.2011.01052.x
Tiwari, M., Kumar, R., Min, D., Jagadish, S. V. K. (2022). Genetic and molecular mechanisms underlying root architecture and function under heat stress-A hidden story. Plant Cell Environ. 45, 771–788. doi: 10.1111/pce.14266
Wang, W., Köhler, B., Cao, F., Liu, L. (2008a). Molecular and physiological aspects of urea transport in higher plants. Plant Sci. 175, 467–477. doi: 10.1016/j.plantsci.2008.05.018
Wang, W., Köhler, B., Cao, F., Liu, G., Gong, Y., Sheng, S., et al. (2012). Rice DUR3 mediates high-affinity urea transport and plays an effective role in improvement of urea acquisition and utilization when expressed in Arabidopsis. New Phytol. 193, 432–444. doi: 10.1111/j.1469-8137.2011.03929.x
Wang, Q., Lan, L., Li, H., Gong, Q., Gao, X. (2023). Effects of nitrogen source and concentration on the growth and biochemical composition of the red seaweed Grateloupia turuturu (Halymeniaceae, Rhodophyta). Sustainability 15, 4210. doi: 10.3390/su15054210
Wang, C., Lei, A., Zhou, K., Hu, Z., Hao, W., Yang, J. (2014). Growth and nitrogen uptake characteristics reveal outbreak mechanism of the opportunistic macroalga Gracilaria tenuistipitata. PloS One 9, e108980. doi: 10.1371/journal.pone.0108980
Wang, W., Teng, F., Lin, Y., Ji, D., Xu, Y., Chen, C., et al. (2018). Transcriptomic study to understand thermal adaptation in a high temperature-tolerant strain of Pyropia haitanensis. PloS One 13, e0195842. doi: 10.1371/journal.pone.0195842
Wang, W.-J., Zhu, J.-Y., Xu, P., Xu, J. R., Lin, X. Z., Huang, C. K., et al. (2008b). Characterization of the life history of Bangia fuscopurpurea (Bangiaceae, Rhodophyta) in connection with its cultivation in China. Aquaculture 278, 101–109. doi: 10.1016/j.aquaculture.2008.01.008
Wi, J., Park, E. J., Hwang, M. S., Choi, D. W. (2023). A subfamily of the small heat shock proteins of the marine red alga Neopyropia yezoensis localizes in the chloroplast. Cell Stress Chaperones. 28, 835–840. doi: 10.1007/s12192-023-01375-4
Witte, C.-P. (2011). Urea metabolism in plants. Plant Sci. 180, 431–438. doi: 10.1016/j.plantsci.2010.11.010
Wittgenstein, N. J. V., Le, C. H., Hawkins, B. J., Ehlting, J. (2014). Evolutionary classification of ammonium, nitrate, and peptide transporters in land plants. BMC Evol. Biol. 14, 1–17. doi: 10.1186/1471-2148-14-11
Wu, H., Li, X., Liu, Y., Wang, C., Ji, C., Xu, J. (2022). Increased temperature and nitrogen enrichment inhibit the growth of the golden tide blooming macroalgae Sargassum horneri in the Yellow Sea, China. J. Mar. Sci. Eng. 210, 1692. doi: 10.3390/jmse10111692
Yamaguchi, N. (2021a). Heat memory in plants: histone modifications, nucleosome positioning and miRNA accumulation alter heat memory gene expression. Genes Genet. Syst. 96, 229–235. doi: 10.1266/ggs.21-00040
Yamaguchi, N., Matsubara, S., Yoshimizu, K., Seki, M., Hamada, K., Kamitani, M., et al. (2021b). H3K27me3 demethylases alter HSP22 and HSP17.6C expression in recurring heat in Arabidopsis. Nat. Commun. 12, 3480. doi: 10.1038/s41467-021-23766-w
Keywords: heat stress, stress memory, hydrogen peroxide, nitrogen transporter, gene expression, red alga
Citation: Sato N, Khoa HV and Mikami K (2024) Heat stress memory differentially regulates the expression of nitrogen transporter genes in the filamentous red alga ‘Bangia’ sp. ESS1. Front. Plant Sci. 15:1331496. doi: 10.3389/fpls.2024.1331496
Received: 01 November 2023; Accepted: 22 January 2024;
Published: 05 February 2024.
Edited by:
Dung Tien Le, Bayer Crop Science, VietnamReviewed by:
Yoshinori Utsumi, RIKEN Center for Sustainable Resource Science (CSRS), JapanHa Chu, Vietnam National University, Vietnam
Copyright © 2024 Sato, Khoa and Mikami. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Koji Mikami, bWlrYW1pa0BteXUuYWMuanA=
†These authors have contributed equally to this work
‡Present address: Ho Viet Khoa, Training Department, Ca Mau Community College, Ca Mau, Vietnam
 Natsumi Sato1†
Natsumi Sato1† Ho Viet Khoa
Ho Viet Khoa Koji Mikami
Koji Mikami