- 1University of Chinese Academy of Sciences (UCAS), Beijing, China
- 2CAS Center for Excellence in Molecular Plant Sciences, Institute of Plant Physiology and Ecology (SIPPE), Chinese Academy of Sciences (CAS), Shanghai, China
- 3Department of Computer Science and Bioinformatics, Khushal Khan Khattak University, Karak, Khyber-Pakhtunkhwa, Pakistan
The dramatic decrease in atmospheric CO2 concentration during Oligocene was proposed as directly linked to C4 evolution. However, it remains unclear how the decreased CO2 concentration directly facilitate C4 evolution, besides its role as a selection pressure. We conducted a systematic transcriptomics and metabolomics analysis under short-term low CO2 condition and found that Arabidopsis grown under this condition showed 1) increased expression of most genes encoding C4-related enzymes and transporters; 2) increased expression of genes involved in photorespiration and pathways related to carbon skeleton generation for ammonium refixation; 3) increased expression of genes directly involved in ammonium refixation. Furthermore, we found that in vitro treatment of leaves with NH4+ induced a similar pattern of changes in C4 related genes and genes involved in ammonium refixation. These data support the view that Arabidopsis grown under short-term low CO2 conditions rewired its metabolism to supply carbon skeleton for ammonium recycling, during which process the expression of C4 genes were up-regulated as a result of a hitchhiking process. This study provides new insights into the adaptation of the C3 model plant Arabidopsis under low CO2 conditions and suggests that low CO2 can facilitate the evolution of C4 photosynthesis beyond the commonly assumed role of being a selection pressure.
Introduction
Although C4 plants only have 8000 species, but it contributes to 23% of the earth’s terrestrial primary productivity (Still et al., 2003; Sage et al., 2007). With over 60 distinct evolutionary origins (Sage et al., 2011; Sage et al, 2018), C4 photosynthesis represents an excellent example of convergent evolution, which attracts attentions among biologists to study the evolutionary history of C4 photosynthesis (Sage, 2001; Sage et al., 2011). According to the present models of C4 evolution (Sage, 2011; Sage, 2012), relocation of photorespiratory CO2 release to the bundle sheath cell (BSC) from the mesophyll cell (MC) is an important early step during the evolution of the C4 photosynthesis (Vogan et al., 2007; Sage, 2012). This step results in C2 photosynthesis, which is characterized by a photorespiratory CO2 pump can elevate the concentration of CO2 in BSC by ~ threefold (Keerberg et al., 2014). Immunogold experiments show that glycine decarboxylase (GDC) is expressed exclusively in the BSC of some intermediates, such as Moricandia arvensis (Rawsthorne et al., 1988; Schluter et al., 2017) and Heliotropium (Vogan et al., 2007). Due to the enrichment of CO2, Ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco) works more efficiently in the BSC, leading to an overall increase in biomass production under condition which supports higher rates of photorespiration (Heckmann et al., 2013; Mallmann et al., 2014), such as low CO2, drought and high temperature (Wingler et al., 1999; Carmo-Silva et al., 2008; Li Y. et al., 2014; Sage et al., 2018).
Accompanied with the release of CO2 by GDC during photorespiration, ammonium (NH4+) is also released (Bauwe et al., 2010), which needs to be refixed since it is toxic (Krogmann et al., 1959). Photorespiratory ammonium can be refixed and integrated into amino acids via the chloroplastic isoform Glutamine synthetase 2 (GS2) and ferredoxin-dependent GOGAT (Fd-GOGAT) (Lam et al., 1996). This process forms the photorespiratory ammonium cycle (Keys, 2006). Photorespiratory CO2 pump results in ammonium imbalance between MC and BSC, which needs to be solved through metabolic shuttles (Nagatani et al., 1971; Lancien et al., 2000; Liu and von Wiren, 2017). Mallman (Mallmann et al., 2014) showed that there are multiple solutions to the ammonium imbalance, with one option, an alanine-pyruvate shuttle, providing a bridge toward C4 photosynthesis. Therefore, ammonium refixation induced by high rate of photorespiration was proposed a bridge toward C4 evolution. However, it raises a question regarding how these metabolic shuttles which recycle ammonium emerge during C4 evolution. Considering that these metabolic shuttles for ammonium recycling require a rather large scale rewiring of the plant’s primary metabolism, quickly evolving such a mechanism when ammonium imbalance in C2 plants appeared would be rather difficult. Importantly, C4 photosynthesis mostly emerged around 30 million years ago and originated independently more than 60 times (Sage et al., 2011), there seems little likelihood for such complex metabolic pathways and its associated regulatory mechanisms to evolve multiple times in such a short time.
All genes involved in C4 photosynthesis are present in C3 ancestral plants and play important house-keeping roles (Aubry et al., 2011); similarly, all the genes involved in the ammonium recycling already exist in gymnosperm species (Valderrama-Martín et al., 2022). It however remains to be tested whether there are regulatory mechanisms coordinating the expression of these genes under conditions favoring photorespiration in typical C3 plants.
The 30-million-year time span subsequent to the Oligocene CO2 decrease during which numerous C4 plants originated suggests that historically low CO2 levels most likely facilitated C4 evolution (Ehleringer et al., 1991; Ehleringer et al, 1997; Christin et al., 2008). Low CO2 condition enhances photorespiration (Li Y. et al., 2014). However, there is little study so far on how this low CO2 directly contributes to the evolutionary formation of the C4 pathway genetically. In this study, we attempt to answer this question through combined transcriptomics and metabolomics analysis. Given that under short-term low CO2 treatment, changes in enzymatic sequences would not be expected, this study focuses on test whether low CO2 altered expressions of genes related to C4 photosynthesis and how. Our results suggest that low CO2 condition indeed induced metabolic rewiring to increase ammonia refixation in the C3 model plant Arabidopsis thaliana, though the signaling pathways through which low CO2 induced observed changed gene expression remains unknown. Since many reactions involved in the ammonia refixation overlap with those for C4 photosynthesis, the mechanisms responsible for the up-regulation of genes related to ammonia refixation inevitably facilitated the up-regulation of genes related to C4 photosynthesis. Therefore, the up-regulation of C4 photosynthesis represents an excellent example of hitchhiking process for the emergence of an important biological process.
Materials and methods
Plant material and growth conditions
Arabidopsis used in this study is the Columbia (Col-0) ecotype. Seeds were surface-sterilized by incubation with 10% (v/v) bleach diluted with ethanol, washed three times with sterile distilled water, and then sown in plastic Petri dishes (diameter 90 mm, depth 20 mm) containing 1/2 MS medium, solidified with 0.7% (w/v) agar and supplemented with 1.5% (w/v) sucrose (pH 5.8). The seeds were placed in the dark at 4°C for three days and germinated for six days in incubators with a photosynthetic photon flux density (PPFD) of 100 μmol m-2s-1 (10 h light/14 h dark cycle) at 22°C. After this, seedlings were transferred to Pindstrup soil and grown in a Percival incubator (Nihonika, Japan) in which CO2 gas was accurately and stably controlled.
Low CO2 treatment
The CO2 condition of 100 ppm (low CO2, LC) and 400 ppm (normal CO2, NC) were applied in two separate Percival incubators (Nihonika, Japan) and maintained throughout this study. Arabidopsis were grown for 14 days under a PPFD of 100 μmol m-2s-1 (10 h light/14 h dark cycle) at 22°C and relative humidity of 65% (Li Y. et al., 2014). After, half of the plants were transferred to low CO2 condition for six days. New fully expanded leaves in three-week-old plants were sampled at 11 am in the morning for transcriptomic and metabolomics analysis, the measurement of chlorophyll content and free ammonium levels under both low CO2 and normal CO2 levels.
Ammonium treatment
To study the effect of ammonium ions on gene expression, seeds were surface-sterilized and sown in a control check (CK) plastic Petri dish (diameter 90 mm, depth 20 mm) in a growth chamber for 10 days at a PPFD of 100 μmol m-2s-1 (10 h light/14 h dark cycle) at 22°C. The composition of the CK was described in (Li et al., 2013), which is composed of 2 mM KH2PO4, 5 mM NaNO3, 2 mM MgSO4, 1 mM CaCl2, 0.1 mM Fe-EDTA, 50 μM H3BO3, 12 μM MnSO4, 1 μM ZnCl2, 1 μM CuSO4, 0.2 μM Na2MoO4, 0.5 gl-1 MES, 1% sucrose, and 0.8% agarose (pH 5.7, adjusted with 1 M NaOH). Half of the seedlings were transferred to the new CK petri dish and the remaining seedlings were transferred to new CK plates which were additionally supplied with 15 mM or 30 mM (NH4)2SO4. Nine seedlings were put on one Petri dish and considered as one biological replicate during sampling. After three days of treatments, leaves were harvested and immediately put into liquid nitrogen and stored at –80°C until RNA extraction. We also put seedlings on the CK plates which were additionally supplied with 30 mM or 60 mM NH4Cl, 15 mM or 30 mM K2SO4, 30 mM or 60 mM KNO3, and 120 mM mannitol to test the effect of other ions (Li et al., 2019).
Measurement of free ammonium content in a leaf
Ammonium content was determined with the colorimetric method based on the phenol hypochlorite assay (Solórzano, 1969; Sarasketa et al., 2014). Briefly, leaves were collected, weighed, and snap-frozen in liquid nitrogen, and then ground with 5 mm zirconia beads. 1 mL H2O was added to the frozen powder. The mixtures were incubated at 80°C for 10 min and then centrifuged at 4000 g and 4°C for 20 min. 100 µL of supernatant was mixed with 200 μL of 0.33 M sodium phenolate, 100 µL of 0.02% sodium nitroprusside; then 200 µL of 2% sodium hypochlorite were added to the mixture. The mixture was incubated at room temperature for 30 min followed by reading absorbance at a wavelength of 630 nm in a spectrophotometer.
Gene expression analysis
For gene expression profiling, we took illuminated new fully expanded leaf samples. Four biological replicates were harvested and frozen in liquid nitrogen immediately and stored at –80°C before RNA extraction. For RNA extraction, the PureLink RNA Mini Kit (Life Technologies Corporation, USA) was used to extract RNA following the manufacturer’s protocol, which includes the removal of gDNA from RNA. The quality of purified RNA was assessed using an Agilent 2100 Bioanalyzer (Agilent, USA) and the RNA samples with RNA Integrity Number (RIN) higher than 7 were used for RNA library construction. RNA libraries were prepared based on standard Illumina (Illumina, Inc., USA) protocols and sequenced using the Illumina X Ten platform in paired-end 150 bp mode. The quality of RNA-seq data (fastq files) was assessed by the FastQC software (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/). RNA-seq analysis was performed by the STAR software (Dobin et al., 2013) with the Arabidopsis reference genome as well as a gene transfer format (GTF) file (downloaded from EnsemblPlants http://plants.ensembl.org/). After generation of the genome index, RNA-seq reads were aligned by STAR with the ‘–quantMode GeneCounts’ option to count reads per gene. Differentially expressed genes (DEGs) were determined by the R package ‘DESeq2’ (Love et al., 2014) which use the Benjamini-Hochberg (BH) adjustment for multiple testing problem with the read counts reported by STAR (Dobin et al., 2013). DEGs between treatment and control were analyzed. Only genes with the adjusted P-value <0.05 (Padj) were considered as DEGs in each treatment.
Functional analysis of differentially expressed genes
Gene transcript abundances and metabolite abundances were visualized applying R package ‘heatmap’ (https://cran.r-project.org/web/packages/pheatmap/index.html). Permutation of Pearson correlations was conducted used the R package Permutation test. Volcano plot and Venn diagrams were plotted applying online tool Bioinformatics (https://www.bioinformatics.com.cn). To identify functional categories of differentially expressed genes, Gene Oncology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses were performed using the Database for Annotation, Visualization and Integrated Discovery (DAVID) (https://david.ncifcrf.gov/). Data were analyzed using the two-tailed Student’s t-test. The term significant is used here for differences or correlations confirmed at * P < 0.05 or better. Details are provided in the figure legends.
Liquid chromatography/mass spectrometry and metabolomics analysis
To enable the sampling of leaves grown under low CO2, we modified the glass door of a Percival incubator to ensure the stability of CO2 condition during leaf sampling. Specifically, we cut two spherical openings in the middle of the glass door and installed two plastic gloves used for sampling.
The Liquid Chromatography/Mass Spectrometry (LC-MS/MS) experiments were performed following (Wang et al., 2014; Arrivault et al., 2019). Fully expanded leaves under light were sampled. For each sample, a leaf area of 1.7 cm2 was sampled and frozen in liquid nitrogen instantaneously. The same leaf position was sampled to measure the fresh weight to calculate specific leaf weight. All leaf samples were cut in situ and immediately transferred into a pre-frozen 2 mL EP tube, then stored in liquid nitrogen for metabolite extraction. After grinding, each sample was fully dissolved with 800 μL extraction buffer (methanol: chloroform = 7:3 (v/v), -20 °C pre-cooling) and incubated under -20°C for 3 hours. Then 560 μL distilled water (ddH2O) was added and mixed with each sample, and 800 μL supernatant was extracted after centrifugation (×2200g, 10min, 4°C). After that, 800 μL buffer (methanol: ddH2O = 1: 1(v/v), 4 °C pre-cooling) was mixed with the sample for another extraction. For each sample, 1.6 mL supernatant in total was obtained by filtering the extraction buffer with a 0.2 μM nylon filter. Among them, 1 mL was used for MS/MS analysis, and 20 μL was used for the QC sample. All extraction operations were performed on ice.
Luna NH2 column (3μm, 100mm*2mm, Phenomenex co. Ltd, USA) was used in the liquid chromatography. The Liquid Chromatography gradient was set with eluent A, which has 10 mM Ammonium acetate and 5% (v/v) acetonitrile solution, with the pH adjusted to 9.5 using ammonium water and eluent B (acetonitrile): 0-1 min, 15% A; 1-8 min, 15-70% A; 8-20 min, 70-95% A; 20-22 min, 95% A; 22-25 min, 15% A. During the mass spectrometry analysis, QTRAP 6500+ (AB Sciex, co. Ltd, USA) was used in the MRM model with all parameters used following (Wang et al., 2014; Arrivault et al., 2019). The condition of all metabolites in samples were calculated based on the “condition-peak area” curve of standard samples and converted to nmol·gFW-1 with specific leaf weight (Arrivault et al., 2019).
Real-time RT–qPCR
The RT–qPCR analysis was conducted as described previously (Chen et al., 2020). The RNA was extracted with the same procedure as described in Gene expression analysis. For reverse transcription, 0.2 μg RNA was reverse transcribed with TranScript One-Step gDNA Removal and cDNA Synthesis SuperMix (TransGen Biotech, China), then SYBR Green Real-Time PCR Master Mix (Yeasen, China) was used for qPCR on the CFX 96 system (Bio-Rad) following the manufacturer’s protocol. The ACTIN 8 (AT1G49240) gene was used as a reference for mRNA normalization. The comparative cycle threshold (Ct) method was used to evaluate the relative gene expression levels. The primers used for the expression analysis are listed in Supplementary Table S1.
Measurement of chlorophyll content
Leaf discs (0.8 cm2) were taken from fully expanded leaves, and chlorophyll was extracted with 80% acetone at 4°C for 24 h in darkness, then the supernatant was used for the absorbance measurement at 652 nm in a spectrophotometer (Arnon, 1949). The total chlorophyll content was calculated with the following formula:
Results
The genes related in primary metabolism were induced under low CO2 condition
To study the effect of low CO2 condition on C3 plants, we examined the physiological, transcriptomic, and metabolomic changes of Arabidopsis under two CO2 conditions: low CO2 at a concentration of 100 ppm (LC) and normal CO2 at a concentration of 400 ppm (NC). Plants grown under low CO2 conditions had lower biomass and decreased chlorophyll than those grown under normal CO2 (Supplementary Figures S1A, B).
The differentially expressed genes (DEGs) were identified between low CO2 and normal CO2 treatments (Supplementary Dataset S1). 2976 genes were upregulated and 2892 genes were downregulated (Supplementary Figure S2A). Gene Ontology (GO) enrichment analysis shows that hexose catabolic process, sugar-phosphatase activity, carbohydrate phosphatase activity, amyloplast, carotene biosynthetic and abscisic acid metabolic process were significantly enriched in upregulated genes (Supplementary Figure S2B; Supplementary Dataset S2), whereas sulfur compound and jasmonic acid-mediated signaling pathway were enriched in the downregulated genes (Supplementary Dataset S2).
In addition, the significantly upregulated genes were mainly participated in metabolic process, including biosynthesis of secondary metabolites, glycolysis/gluconeogenesis, starch and sucrose and carbon metabolism by KEGG analysis (Supplementary Figure S2C; Supplementary Dataset S2). These results show that low CO2 induced changes to gene expression involved in primary metabolism.
Most C4 related genes are upregulated under low CO2 condition in Arabidopsis
Notably, under low CO2, 23 of 26 genes associated with C4 metabolism were significantly increased (Figure 1; Supplementary Dataset S1), including cytoplasmic carbonic anhydrase (CA2, AT5G14740; CA4, AT1G70410) and chloroplast-localized carbonic anhydrase (CA5, AT4G33580). The gene expression of phosphoenolpyruvate carboxylase (PEPC, AT2G42600, AT1G53310), PEPC kinase (PPCK1, AT1G08650), chloroplast/mitochondrial NAD-dependent malate dehydrogenase (pNAD-MDH, AT3G47520; mMDH1, AT1G53240), pyruvate orthophosphate dikinase regulatory protein (PPDK-RP, AT4G21210), alanine aminotransferase (AlaAT1, AT1G17290), and aspartate aminotransferase (AspAT, AT4G31990, AT5G19550, AT2G22250) were also upregulated (Figure 1; Supplementary Dataset S1). In addition, the genes encoding C4 related transporters were upregulated significantly, which includes phosphoenolpyruvate/phosphate translocator (PPT1, AT3G01550; PPT2, AT5G33320), dicarboxylate carriers (DIC, AT4G24570; DIC2, AT2G22500), inorganic pyrophosphatase 2 (PPA2, AT2G18230), plasma membrane protein (PIP1, AT3G61430), and mesophyll envelope protein (MEP2, AT5G23890; MEP3/4, AT5G12470), dicarboxylate transporters (DIT1, AT5G12860; DIT2.1, AT5G64290). Many of these genes, such as PEPC, PPDK and NADP-ME, show increased gene expression under different stress conditions, such as high temperature, drought, high light, and saline conditions (Doubnerova and Ryslava, 2011). Overall, these results show low CO2 condition can cause up-regulation of genes involved in C4 photosynthesis.
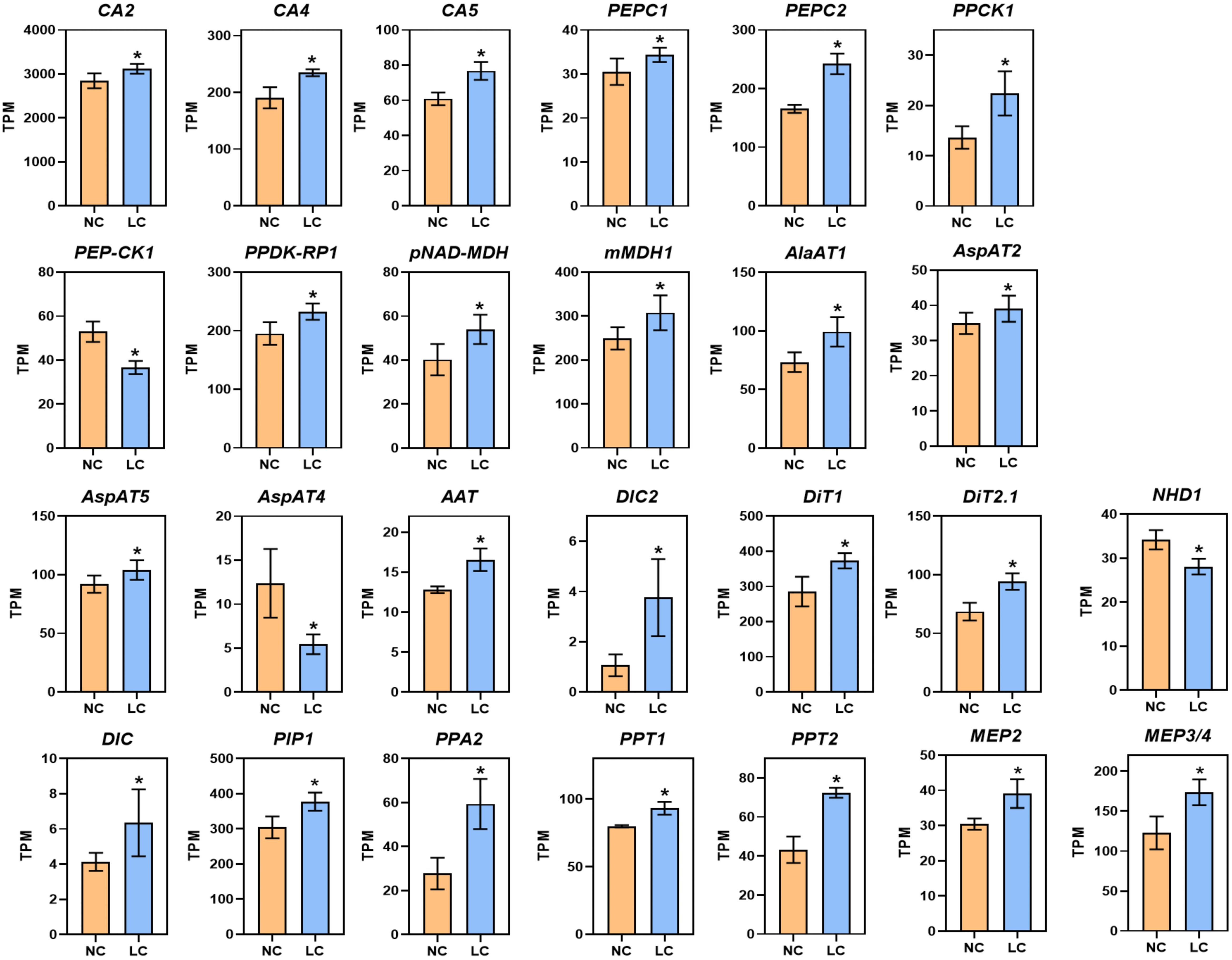
Figure 1 Low CO2 induction of C4 related genes in Arabidopsis. RNA-seq data are plotted as transcript per million (TPM). Gene expression is shown from samples collected from NC (normal CO2, 400 ppm) and LC (low CO2, 100 ppm). CA2, Carbonic Anhydrase 2; CA4, Carbonic Anhydrase 4; CA5, Carbonic Anhydrase 5; PEPC1, Phosphoenolpyruvate Carboxylase 1; PEPC2, Phosphoenolpyruvate Carboxylase 2; PPCK1, PEPC Kinase1; PEP-CK1, Phosphoenolpyruvate Carboxykinase 1; RPDK-RP1, PPDK Regulatory Protein 1; pNAD-MDH, chloroplast NAD-Dependent Malate Dehydrogenase; mMDH1, mitochondrial Malate Dehydrogenase; AlaAT1, Alanine Aminotransferase1; AspAT4, Aspartate Aminotransferase 4; AspAT5, Aspartate Aminotransferase 5; AspAT2, Aspartate Aminotransferase 2; AAT, Aspartate Aminotransferase; PIP1, Plasma Membrane Protein; DIC2, Dicarboxylate Carriers 2; DiT2.1, Dicarboxylate Transport 2.1; DiT1, Dicarboxylate Transporter 1; DIC, Dicarboxylate Carriers; PPA2, Inorganic Pyrophosphatase 2; PPT1, Phosphoenolpyruvate Translocator 1; PPT2, Phosphoenolpyruvate Translocator 2; NHD1, Na+/H+ antiporter; MEP2, Mesophyll Envelope Protein 2; MEP3/4, Mesophyll Envelope Protein 3/4. Data are shown as mean ± s.d (replications n=4). *, adjusted P-value <0.05 which was determined by DESeq2.
We validated 12 genes that are involved in primary metabolism through RT-qPCR. The results of the RT-qPCR were consistent with the RNA-seq analysis as shown in Supplementary Figure S3A, with a Pearson correlation coefficient of 0.75 (Supplementary Figure S3B).
Photorespiratory and ammonium refixation pathway are enhanced under low CO2 condition
Photorespiration was enhanced under low CO2 condition (Li Y. et al., 2014). Photorespiratory ammonium refixation are highly integrated with photorespiratory metabolism (Keys et al., 1978). Therefore, we investigated the expression of genes related to photorespiration and ammonium refixation pathways. The expression of a number of photorespiratory genes, including glycolate oxidase (GOX2, AT3G14415), glutamate: glyoxylate aminotransferase (GGT2, AT1G70580), alanine: glyoxylate aminotransferase (AGT1, AT2G13360), glycine decarboxylase P-protein (GLDP1, AT4G33010; GLDP2, AT2G26080), and mitochondrial transporter A BOUT DE SOUFFLE (Bou, AT5G46800) (Eisenhut et al., 2013), were upregulated (Figure 2A). The gene coding for 2-phosphoglycolate phosphatase (PGLP1, AT5G36700) showed decreased expression (Figure 2A). Concurrent metabolomics analysis shows that the shift to low CO2 condition led to higher intracellular pools of 2-phosphoglycolate (2-PG), glycolate, glycine, and glycerate, which are intermediates in the photorespiratory pathway (Figure 2A). The increased metabolite levels corroborate with the upregulation of photorespiratory gene expression. In addition, the NH4+ levels in leaves were increased by one-fold compared to normal CO2 treatment (Figure 2B).
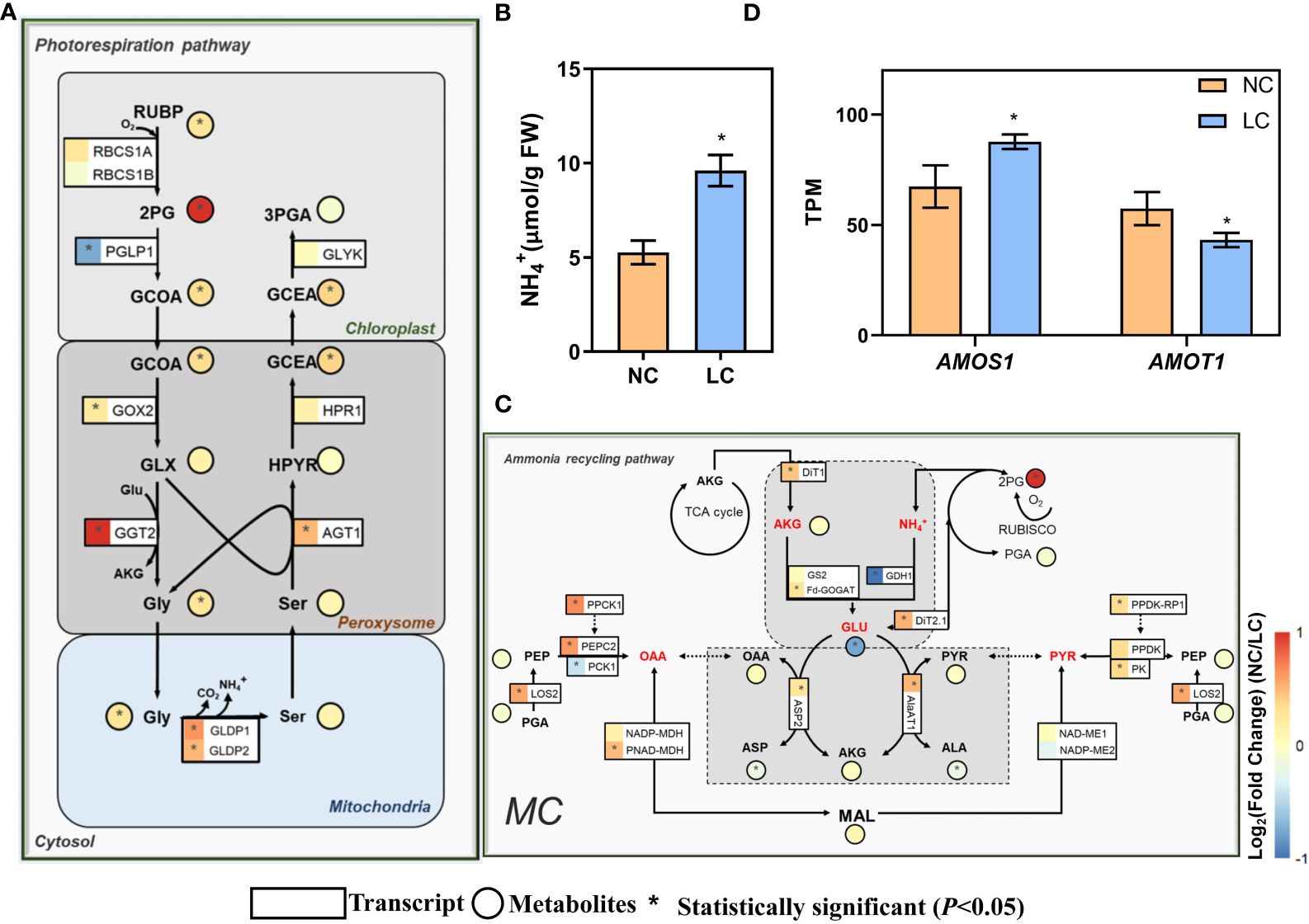
Figure 2 Changes of the gene expression and metabolites concentration on photorespiratory and ammonium recycling pathways under low CO2. (A) Photorespiratory pathway. (B) NH4+ content in the leaf (replications n=4). (C) Ammonium recycling pathway. (D) The changes of expression of ammonium-sensitive genes. AMOS1 (AT5G35220, Ammonium-overly-sensitive 1), AMOT1 (AT3G20770, Ammonium tolerance 1). The relative changes of gene expression and metabolites are shown in log2Fold change in heatmap, which use the same heatmap scale. For gene expression (A, C, D) differentially expressed genes (DEGs) were determined by the R package ‘DESeq2’ with the adjusted P-value <0.05 (replications n=4). For metabolites (A, C) and free NH4+ content in the leaf (B) which were determined by two-sided Student’s t-test (replications n=5) and * indicates P<0.05. Data are shown as mean ± s.d (B, D). The symbols of genes in these metabolic pathways are from the Arabidopsis Information Resource (TAIR) Database (http://www.arabidopsis.org/), and also can get from Supplementary Dataset S1.
For ammonium refixation pathway, the gene coding Fd-GOGAT (AT5G04140) also showed significantly higher expression level under low CO2 condition (Figure 2C). DIT1 and DIT2.1 work in concert in photorespiratory carbon/nitrogen metabolism; the lack of either DiT2.1 or DiT1 led to growth impairment under photorespiratory condition (Kinoshita et al., 2011). In our data, both DiT2.1 and DitT1 showed greater expression levels under low CO2 (Figure 2C). Moreover, the gene expression of AspAT and AlaAT, which are involved in the transamination, showed significantly increase (Figure 2C). The concentration of 2-oxoglutarate (2-OG) showed no significant change (Figure 2C), whereas the concentration of glutamate was reduced by half (Figure 2C). The concentration of oxaloacetate (OAA) and pyruvate were not changed under low CO2 condition, while those of aspartate and alanine were slightly decreased (Figure 2C).
Under low CO2 condition, the expression of the gene AMOS1 (Ammonium-overly sensitive 1) (Li et al., 2012), encoding a plastid metalloprotease that confers sensitivity to ammonium, significantly increased as depicted in Figure 2D. Conversely, the expression of AMOT1 (Ammonium tolerance 1) (Li et al., 2019), which is crucial for mitigating the effects of ammonium toxicity and confers greater tolerance to NH4+ in mutants than the wild type, was markedly reduced (Figure 2D). All these data suggest that Arabidopsis under low CO2 was subjected to ammonium stress under low CO2 condition.
Low CO2 condition reprogramming primary carbon metabolism
The KEGG enrichment analysis of DEGs showed that low CO2 affects genes related to primary carbon metabolism (Supplementary Dataset S2). Among the affected genes, a vast majority of those involved in the tricarboxylic acid cycle (TCA) were markedly upregulated, specifically aconitase (ACO1, AT4G35830), isocitrate dehydrogenase 1 (IDH1, AT4G35260), 2-oxoglutarate dehydrogenase (OGDC, AT3G55410), succinate dehydrogenase (SDH, AT5G66760), and mMDH1 (Figure 3A; Supplementary Dataset S1). However, we observed a significant reduction of the succinate and fumarate in comparison to normal CO2 condition (Figure 3A).
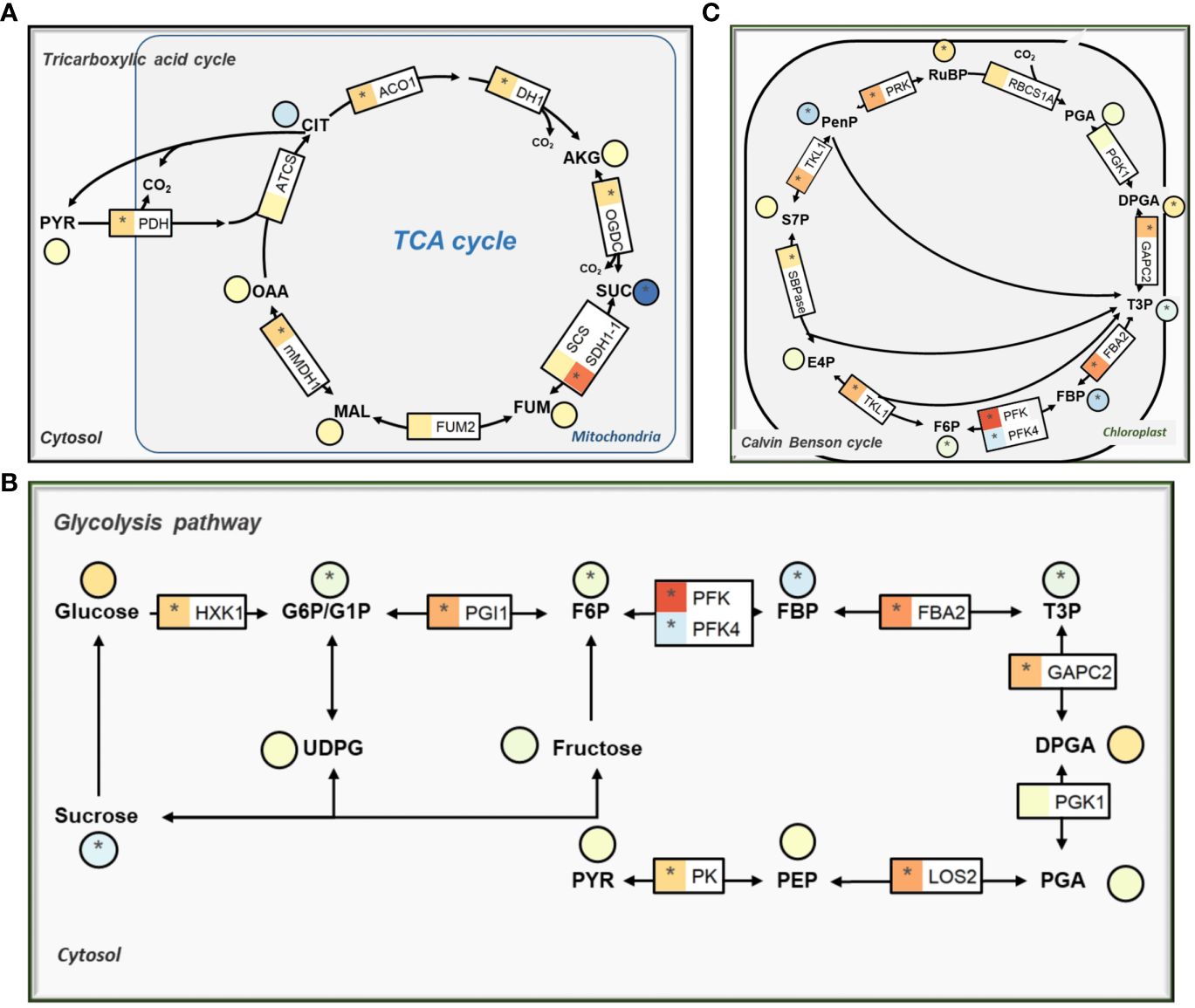
Figure 3 The effect of low CO2 on gene expression and metabolites condition in carbon metabolism pathways. (A) Tricarboxylic acid cycle (TCA cycle). (B) Glycolysis pathway. (C) Calvin-Benson-Bassham (CBB) cycle. The details are same as Figure 2.
The expression levels of genes associated with the glycolysis pathway showed similar trends of changes as those observed among the TCA cycle genes. Notably, most of these genes demonstrated significant upregulation while metabolites like sucrose, glucose-6 phosphate (G6P), fructose-6-phosphate (F6P), fructose-1:6-Bisphosphate (FBP), and triose phosphate (T3P) were significantly reduced under low CO2 treatment (Figure 3B). We observed increased expression levels of enolase (LOS2, AT2G36530), pyruvate kinase family protein (AT5G08570; AT2G36580), and pyruvate dehydrogenase (PDH-E1 BETA, AT1G30120; PDH-E1_ALPHA, AT1G01090). Among the genes related to the Calvin-Benson-Bassham (CBB) cycle, most of the genes showed upregulation in terms of expression, and the majority of the metabolites were affected (Figure 3C). While pentose phosphate (PenP), FBP, and T3P metabolites decreased significantly, other metabolites mostly showed an increase in their levels under the low CO2 condition (Figure 3C).
The 2-PG showed the strongest increase compared to other photorespiratory metabolites (Figure 2), which is in well agreement with the transcriptional repression of PGLP1 (Figure 2). Earlier work found that 2-PG inhibits CBB cycle enzymes TPI and SBPase in Arabidopsis after treatment with low CO2 (Flugel et al., 2017). Such an inhibition may decrease further increase of 2-PG and hence avoid potential greater damage to the cell under low CO2.
These results show that under low CO2 condition Arabidopsis changed expression of primary carbon metabolism and adjusted their metabolite to boost its adaptability to low CO2 condition.
NH4+ treatment increased the expression of genes involved in ammonium refixation
In our results, the leaf NH4+ content increased (Figure 2B) and the ammonium refixation was induced (Figure 2C) following low CO2 treatment. Plants treated with 30 mM NH4+ increased the leaf NH4+ content by at least one-fold, and the NH4+ content increased by at least three-fold with the 60mM NH4+ treatment (Figure 4A). RT-qPCR analysis revealed that NH4+ treatment upregulated the expression of genes involved in ammonium refixation, specifically GS2 and Fd-GOGAT as shown in Figure 4B. Interestingly, this increase was not observed when plants were treated with NH4CI, K2SO4, KNO3, or Mannitol (Li et al., 2019) (Figure 4C).
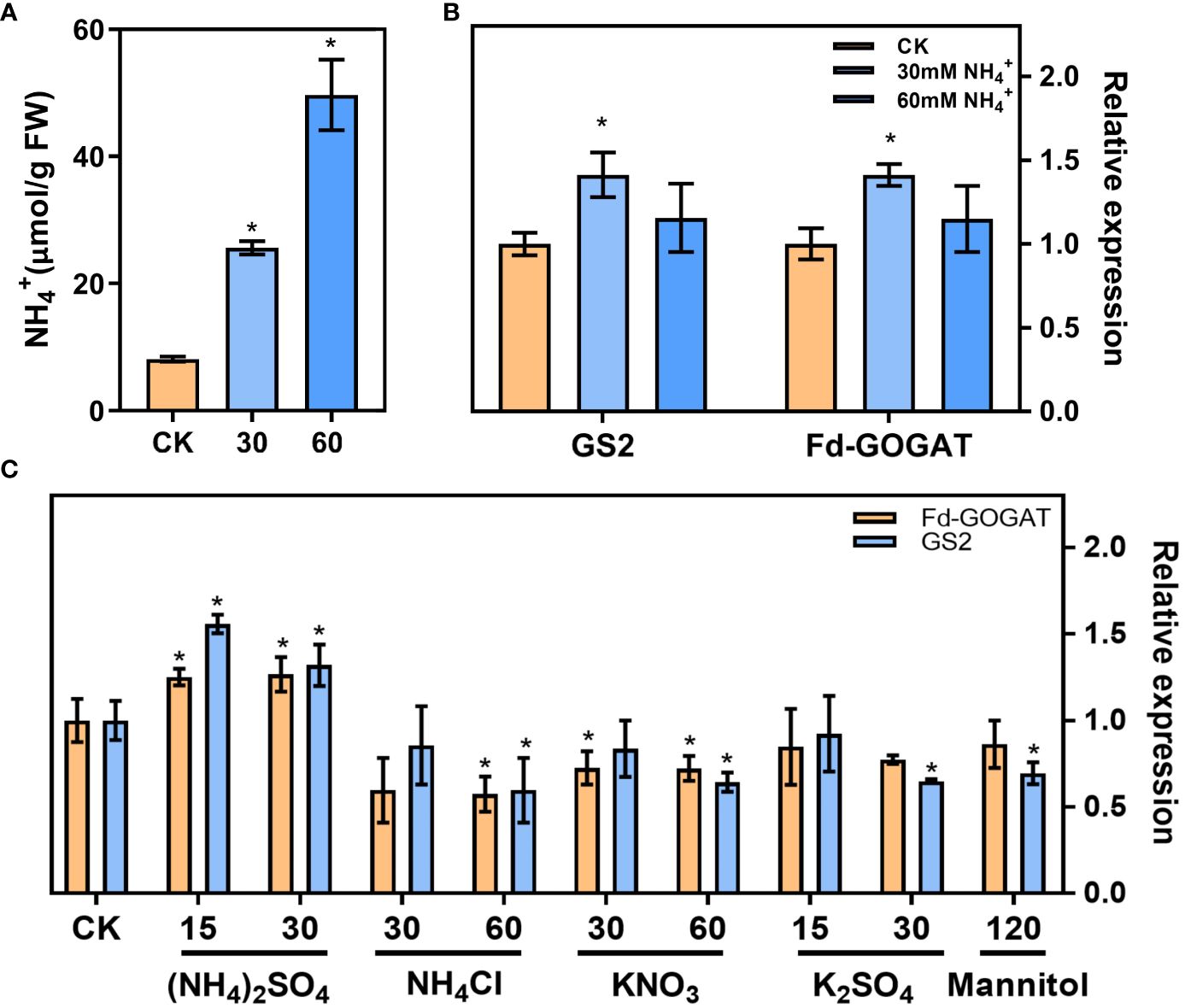
Figure 4 In vitro NH4+ treatment increased the expression of genes involved in ammonium refixation. (A) NH4+ content in the seedlings (Control check, CK; 30 mM NH4+, 30; 60 mM NH4+, 60) (replications n=4). (B) The gene expression of the Glutamine synthetase 2 (GS2) and ferredoxin-dependent GOGAT (Fd-GOGAT) were analyzed using RT–qPCR (replications n=4). (C) The effects of different ion on the gene expression of GS2 and Fd-GOGAT. Data are shown as mean ± s.d, * p<0.05, which were determined by two-sided Student’s t-test.
Comparison of transcriptomics data between NH4+ treatment and low CO2 treatment
After NH4+ treatments, a total of 3920 genes were identified as differentially expressed with 1910 genes upregulated and 2010 genes downregulated (Supplementary Figure S4A). The GO terms for upregulated genes were enriched in stress, nitrogen component metabolism, carbohydrate biosynthetic process and oxoacid metabolism (Supplementary Figure S4B), which is in line with earlier finding that excessive NH4+ induced ammonium refixation related genes, such as glutamate dehydrogenase (Patterson et al., 2010; Daniela et al, 2016), and GS/GOGAT pathway (Liu and von Wiren, 2017). The upregulated genes were also significantly enriched in carbon metabolism and 2-oxocarboxylic acid metabolism (Supplementary Dataset S2), with latter term including pyruvate, oxaloacetate and 2-OG which can enter the TCA cycle to generate carbon skeleton for ammonium refixation.
We compared the transcriptomic changes induced by both NH4+ and low CO2 treatments. We found that DEGs (P<0.05) under both treatments showed a higher Pearson correlation coefficient than those with no significant changes to either treatment (Figure 5A) (P<0.001), which suggest that the treatment of NH4+ and low CO2 caused similar changes. The Venn diagram shows that 525 genes were upregulated under both treatments (Figure 5B; Supplementary Dataset S3), while 464 genes were downregulated under both treatments (Supplementary Figure S4C). The jointly upregulated genes are mostly enriched in glycolysis, starch and sucrose metabolism, carbon metabolism, accounting for 36% of those differentially expressed upregulated genes (Figure 5C). The jointly upregulated genes were also found to be enriched in biological processes related to light stimulation, water deprivation, cold, abscisic acid, and the glycolytic process (Figure 5D). These results suggest that multiple genes involved in primary metabolism were simultaneously affected by both NH4+ and low CO2 treatments.
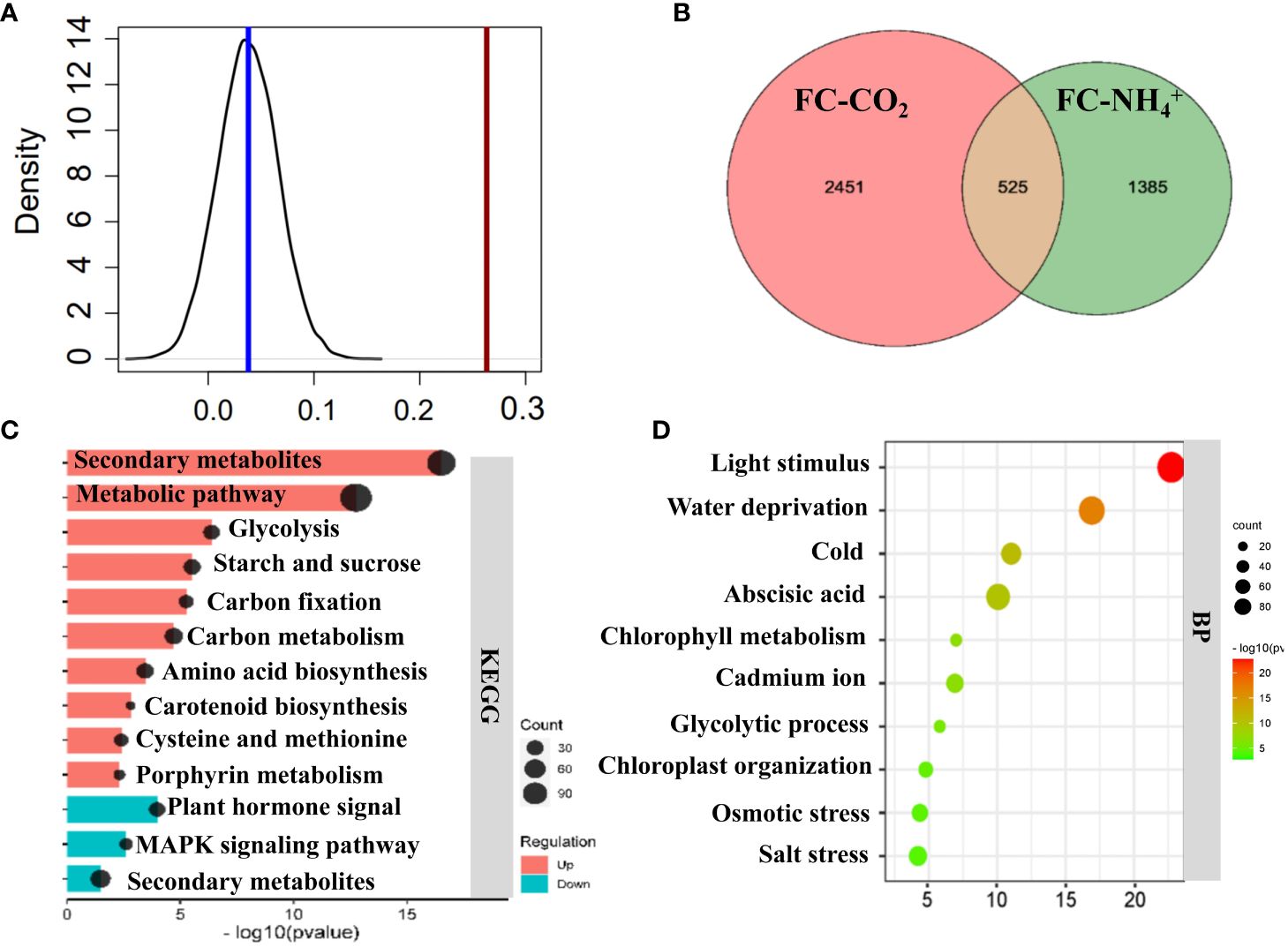
Figure 5 Comparison of transcriptomic profiling between NH4+ and low CO2 treatments. (A) Pearson correlation analysis of transcriptomics data between NH4+ and low CO2 treatment. The black line represents the distribution of Pearson correlations of randomly selected genes (1632) for 1000 times, and the red line represents the Pearson correlation of differentially expressed 1632 genes. A cutoff value for adjusted P-value < 0.05 were used to identify differentially expressed genes. (B) Venn diagrams. The diagram shows the number of genes which are jointly significantly upregulated (adjusted P-value < 0.05) under low CO2 treatment (FC-CO2) and NH4+ treatment (FC-NH4+). Detailed gene information can be found in Supplementary Dataset S3. (C) KEGG enrichment analysis of significantly upregulated genes. (D) Gene ontology (GO) Biological process (BP) enrichment analysis of significantly upregulated genes. The size of the circles represents the enriched gene number, with a larger circle indicating a higher number of genes in the corresponding metabolic pathway category. The horizontal axis represents the P-value. Top 10 significantly enriched results are shown here (C, D). Remaining enriched results can be found in Supplementary Dataset S3. Fisher’s exact text P<0.05.
Influence of NH4+ treatment on the gene expression in primary metabolism
Transcriptomic data from plants treated with NH4+ showed an increase in expression levels of the GS/GOGAT pathway and NADH-dependent glutamate dehydrogenase (GDH), as well as upregulation of nitrate reductase genes (NIA1, AT1G77760; NIA2, AT1G37130) (Masclaux-Daubresse et al., 2010) (Figure 6A), suggesting that NH4+ treatment enhances ammonium refixation metabolism. Additionally, RNA-seq data showed that almost all genes related to the photorespiratory pathway were induced, along with the expressions of genes related to the TCA cycle, Glycolysis pathway, and the CBB cycle (Figure 6A). The transcriptomic results in primary metabolism induced by NH4+ treatment were similar to those induced by low CO2 treatment (Supplementary Figure S5).
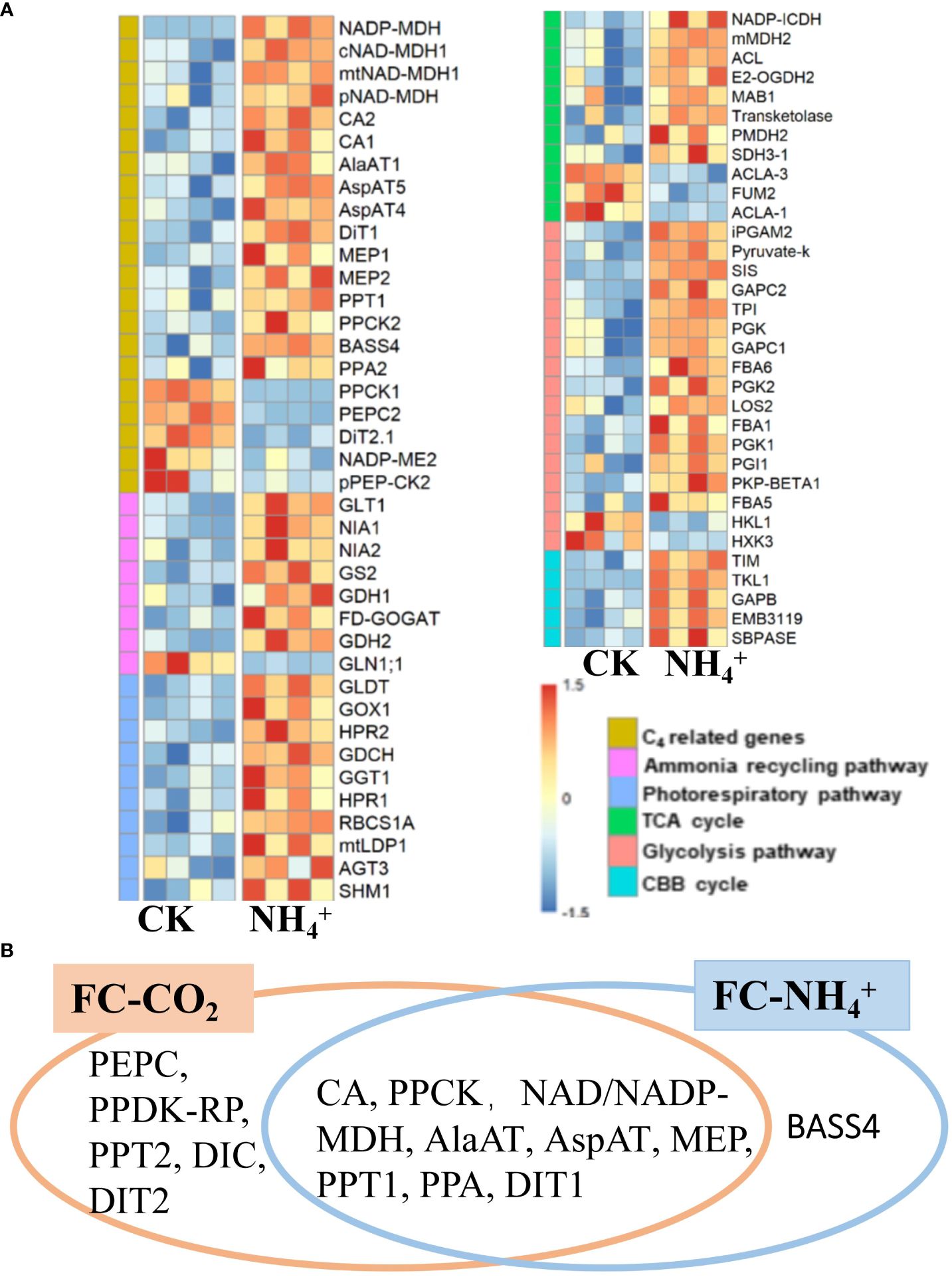
Figure 6 (A) Heatmap of differentially expressed genes of different metabolic pathways under NH4+ treatment (replications n=4). Genes from six pathways were shown as labeled in different colors indicated in the “Pathways” legend. (B) Comparative analysis of C4 related enzymes under low CO2 treatment (FC-CO2) and NH4+ treatment (FC-NH4+). The fisher’s exact text P<0.05.
Further analysis suggested that NH4+ treatment induced the expression of most C4 related genes, including CA (CA1, AT3G01500; cytoplasmic CA2, AT5G14740), NADP-dependent malate dehydrogenase (NADP-MDH, AT5G58330), cytosolic malate dehydrogenase (cMDH1, AT1G04410), NAD-MDH1 (AT1G53240, AT3G47520), and phosphoenolpyruvate carboxylase kinase (PPCK2, AT3G04530) (Figure 6A; Supplementary Dataset S1). Furthermore, some aminotransferases, such as AspAT4, AspAT5 (AT4G31990), and AlaAT1 were up-regulated by NH4+ treatment (Figure 6A; Supplementary Dataset S1). Several transporter genes, including DiT1, PPT1, Sodium Bile acid symporter (BASS4, AT3G56160), PPA2, NHD1, and MEP, were also upregulated by NH4+ treatment (Figure 6A; Supplementary Datasets S1). However, the expression levels of PEPC, PPCK1, and pPEP-CK2 were downregulated (Figure 6A). In addition, we also found that CA, NAD/NADP-MDH, PPCK, AlaAT, AspAT, MEP, PPT1, PPA, and DIT1 were significantly induced by both treatments (Figure 6B). Furthermore, low CO2 treatment induced the upregulation of most C4 related genes, almost including those induced by NH4+ treatment, except for BASS4 (Figure 6B) (Fisher’s exact text P<0.05). Therefore, NH4+ accumulation may be a mechanism that upregulates C4 genes under condition of low CO2 (Figure 6B).
Discussion
The gene expression involved in the primary metabolism is adjusted to increase the capacity of ammonium refixation under low CO2 condition
Arabidopsis grown under low CO2 condition showed a significant decrease in growth (Li Y. et al., 2014) and chlorophyll content compared to the control (Supplementary Figures S1A, B). The stunted growth of Arabidopsis under low CO2 condition can be attributed to the decreased photosynthetic CO2 fixation. The observed decrease in chlorophyll concentration may be related to nitrogen deficiency, as chlorophyll concentration depends on nitrogen content (Ercoli et al., 1993). The high photorespiration under low CO2 leads to an excessive amount of ammonium release in the leaves, which might result in decreased nitrogen content and chlorophyll concentration. Carbon starvation also reduces the capacity of plants to assimilate nitrogen, leading to significant reductions in leaf nitrogen and Rubisco capacity (Dippery et al., 1995; Sage and Coleman, 2001), disrupted C/N ratio (Tcherkez et al., 2017) and induction of autophagic responses (McLoughlin et al., 2020). In our study, carbon starvation resulted in decreased sucrose levels in leaves (Figure 3C). Interestingly, the expression of genes related to amino acid biosynthesis, carbohydrate biosynthesis, carotenoid biosynthesis etc. were increased (Figure 5C). This might be a compensatory effect, which has also been observed in studies of carbon-starved maize (McLoughlin et al., 2020).
The upregulation of PEPC and PEPE-K genes in response to low CO2 has also been observed in our earlier study (Li Y. et al., 2014). In this new study, we found that the expression of almost all core C4 cycle enzymes and transporters were upregulated under low CO2 treatment (Figure 1). Interestingly, a similar upregulation of C4 like pathway genes for carbon fixation was observed in Nannochloropsis oceanica under low CO2 condition (Wei et al., 2019). These data suggest a role of low CO2 condition in the strengthening of C4 gene expression during the Oligocene period when C4 emerged genes during evolution.
Low CO2 condition, like other stresses such as drought and high temperature (Carmo-Silva et al., 2008; Peterhansel and Maurino, 2011), increases photorespiration (Figure 2A) and promotes NH4+ production (Figure 2B) (Frantz et al., 1982; Alencar et al., 2019). Photorespiration is the primary source of NH4+ in higher plants (Leegood et al., 1995; Kumagai et al., 2011). Arabidopsis, being an NH4+ sensitive plant (Cruz et al., 2006), showed stunted plant growth and leaf chlorosis under low CO2 (Supplementary Figures S1A, B) (Li et al., 2012; Li Y. et al., 2014; Esteban et al., 2016). Taken together, these results suggest that Arabidopsis experiences ammonium stress under low CO2 condition.
Previous studies have shown that excess ammonium is toxic (Hachiya et al., 2012; Li B. et al., 2014; Esteban et al., 2016) and needs to be refixed by upregulation of several key enzymes (Cruz et al., 2006; Liu and von Wiren, 2017; Hachiya et al., 2021). Indeed, the GS/GOGAT pathway which is involved in ammonium refixation was enhanced under low CO2 condition (Figure 2C). The TCA cycle under illumination helps provide the carbon skeleton, in particular 2-OG, for ammonium refixation (Nunes-Nesi et al., 2010; Sweetlove et al., 2010; Tcherkez et al., 2017). The gene expression of IDH1 and ACO1, which are used for the generation of 2-OG, were also upregulated in the TCA cycle (Figure 3A). Although the concentration of 2-OG remained unchanged (Arrivault et al., 2009), a significant reduction in the concentrations of fumarate and succinate were observed (Figure 3A), implying that there is an increased flux of 2-OG used for ammonium refixation, rather than decarboxylation in the TCA cycle. Consistent with this, studies have also shown that supplementing with succinate and 2-OG can increase ammonium recirculation and alleviate ammonium toxicity (Yuan et al., 2007; Hachiya et al., 2012).
Previous calculations suggest that the stored leaf citrate content available at the start of light period is insufficient to support the amount of 2-OG required for glutamate production (Stitt et al., 2002). The 2-OG can be generated by reactions catalyzed by aminotransferases. In this aspect, PEPC might play a crucial role during the synthesis of additional 2-OG (Turpin, 1994). Potatoes with overexpressed PEPC display increased 2-OG levels in the leaves (Rademacher et al, 2002); PEPC catalyzes the formation of OAA, which is used in transamination to produce 2-OG. In line with this, the genes encoding PEPC, PEPC-K, AspAT, and MDH are found upregulated under low CO2 condition (Figure 7). In addition, CAs, which are involved in optimizing cytosolic PEPC activity and ensure normal growth under low CO2 condition (DiMario et al., 2016; Weerasooriya et al., 2022), increased significantly under low CO2 condition (Figure 1). Similarly, the expression of PPDK-RP and AlaAT were upregulated, which can also be used to produce more pyruvate and carbon skeletons (Figure 7). Besides, the related transporters, such as Dit1, DIT2.1, showed increased expression as well, which might help meet the increased demand of metabolite transport (Gowik et al., 2011) (Figure 7). All these show that under low CO2 condition, the primary metabolism can be reprogrammed to generate the required carbon skeletons for ammonium refixation.
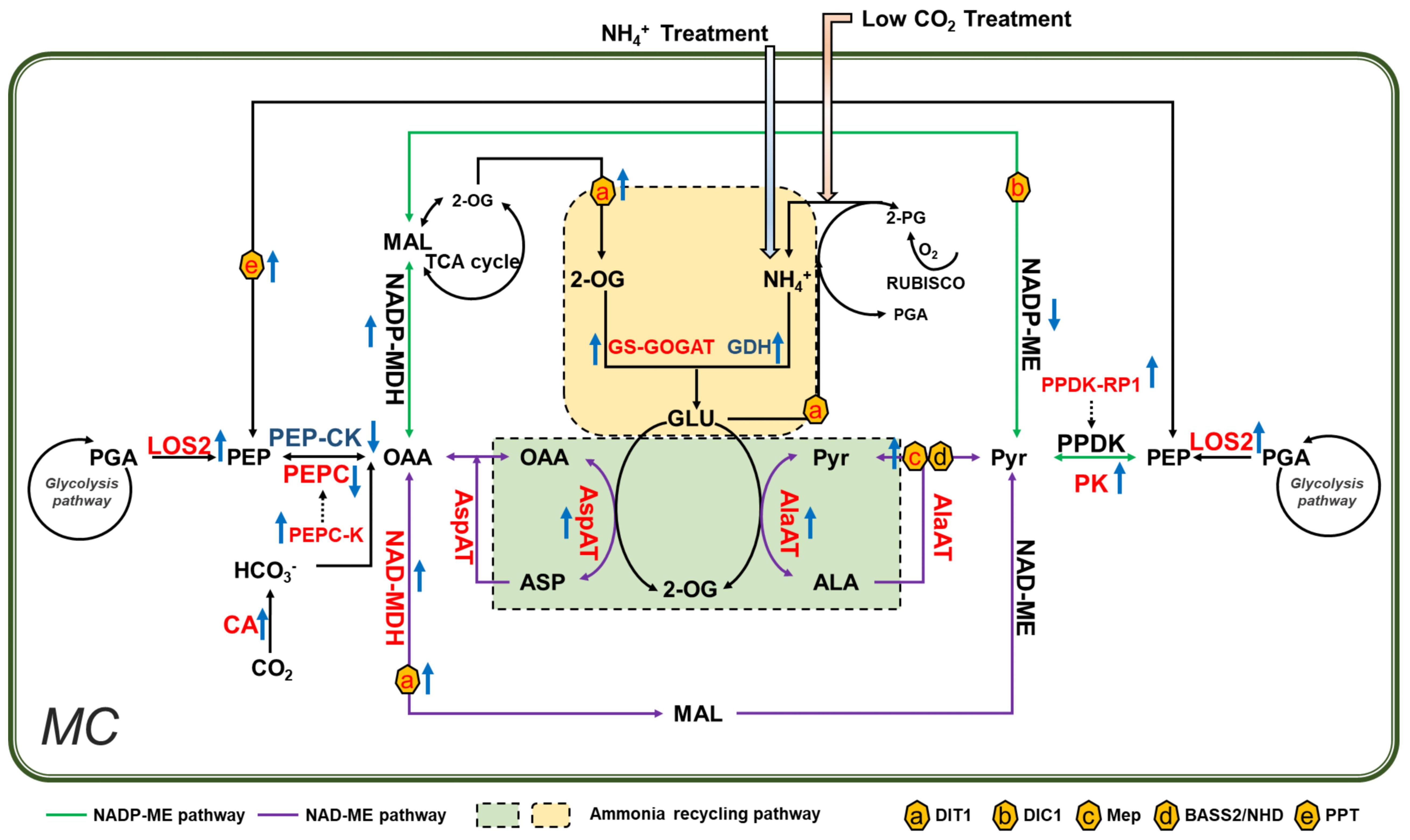
Figure 7 Ammonium accumulation and refixation is a primary mechanism inducing C4 related gene expression under low CO2 condition in Arabidopsis. Low CO2 treatment caused ammonium toxicity in Arabidopsis. Recirculation of ammonium requires reprogramming of primary metabolism reactions that involved in virtually all core C4 genes. In vitro NH4+ treatment induces a similar pattern of changes in C4 related genes and genes involved in primary metabolism. Therefore, we proposed that increased ammonium is a primary mechanism to induce the expression of C4 related gene under low CO2 in Arabidopsis. The Enzymes labeled in different colors were indicated as red/blue: upregulated/downregulated under low CO2. Rising/descending arrows beside the enzymes indicated upregulated/downregulated in NH4+ treatment. NADP-ME and NAD-ME pathways were represented in blue and purple lines respectively. Ammonium recycling pathways were shown in green and yellow rectangles; membrane transporters were indicated in hexagons.
NH4+ accumulation is a possible signal inducing change in the expression of C4 related genes under low CO2 in Arabidopsis
How could low CO2 induce the observed changes in transcriptomics? Here we discuss three possibilities. First, the observed change in transcriptomics might be directly caused by the ammonium accumulation. Indeed, the DEGs identified under low CO2 and NH4+ treatments show correlations (Figure 5A). Specially, under both low CO2 and NH4+ treatments, TCA cycle and related aminotransferases which provide 2-OG for ammonium refixation were significantly upregulated (Figure 6A; Supplementary Figure S4). Furthermore, the C4 related genes were induced by both low CO2 and NH4+ treatments (Test of independence using Fisher test, P<0.05) (Figure 6B). All these suggest that NH4+ accumulation can be a strong candidate signal inducing the expression of C4 related genes under low CO2 in Arabidopsis. However, the detailed mechanism of how ammonium induced these diverse responses is still unknown. Transcriptomic analysis reveals that among the genes activated in response to NH4+, 90% can be regulated by AMOS1 (Li et al., 2012). Interestingly, the transcription level of AMOS1 was increased under low CO2 (Figure 2D), suggesting AMOS1 might be as a potential candidate to induce C4 related gene expression.
Secondly, low CO2 causes decreased photosynthesis, which resulted in decreased biomass production (Supplementary Figure S1A) and decreased concentrations of certain metabolites, such as sucrose (Figure 3B). In general, decreased concentration of carbohydrate enhances expression of genes for CO2 fixation for both C3 and C4 plants and also genes related to mobilization of photosynthesis reserve, and export process (Sheen, 1990; Cheng et al., 1992; Koch, 1996; Ruan, 2014). Given this, decreased sugar concentration might also be a potential mechanism underlining the observed reconfiguration of primary metabolism under low CO2 condition.
Finally, according to (You et al., 2020), ABA and PEPC play crucial roles in enabling Arabidopsis to adapt to low CO2 condition. Intriguingly, there was a notable increase in the expression of ABA related metabolic processes under low CO2 treatment (Supplementary Dataset S2) which implies that ABA signaling might have been involve in the observed changed in the transcriptomic under low CO2 condition as well.
Implication of the existence of regulatory mechanisms inducing ammonium refixation for C4 evolution
One major question to answer in the field of C4 photosynthesis evolution is the detailed process through which rewiring of primary metabolism from C3 to C2 photosynthesis occurred. C2 photosynthesis, which is considered as an intermediate stage during C4 evolution (Bauwe et al., 2010; Heckmann et al., 2013), has been proposed to use ammonium recycling to resolve the issue of ammonium misbalance between BSC and MC (Mallmann et al., 2014). A number of putative pathways to recycle NH4+ were proposed by (Mallmann et al., 2014). These pathways usually use many, though not all, enzymes of the C4 cycle. In all these proposed pathways, ammonium is refixed by GS/GOGAT pathway with carbon skeleton provided by a partial C4 cycle. Several experimental results support this hypothesis. For example, in C3–C4 intermediate Flaveria species, the transcripts GS/GOGAT are upregulated, as also the case for a number of aminotransferases (Mallmann et al., 2014). The question now needs to answer is that, considering that C4 photosynthesis emerged from C3 photosynthesis in a relatively short geological time scale, i.e., only within about ~20 million years (Christin et al., 2008), how could C3 plants rapidly gain such complex regulatory mechanisms regulating ammonium recycling and providing carbon skeleton when the atmospheric CO2 drops at Oligocene?
Our results here show that the regulatory mechanism inducing ammonium recycling pathway pre-existed in C3 plants. Under low CO2 condition or other conditions where the photorespiratory flux and correspondingly the ammonium release flux are high, an innate regulatory mechanism to induce expression of genes involved in ammonium recycling can be activated. This is sensible since conditions causing physiological low CO2 condition, e.g. drought, high temperature, etc. which causes stomatal closure (Ehleringer et al, 1991; Wilkinson and Davies, 2002; Sage et al., 2018), widely exist, mechanisms to enable capturing the released ammonium will confer competitive advantage for plants. This innate ammonium recycling mechanism can be used to solve the challenge of ammonium misbalance in C2 species when this is under demand after the re-allocation of GDC from MC to BSC (Mallmann et al., 2014). This finding is in line with many previous studies which demonstrate that C4 evolution recruited many pre-existing parts and regulatory mechanisms (Aubry et al., 2011; Reyna-Llorens and Hibberd, 2017; Dickinson et al., 2020), which together underlie the parallel independent evolution of C4 photosynthesis.
Summary and perspective
Low CO2 is well recognized to be closely related to the evolution of C4 photosynthesis. Historically, this is mainly discussed from the perspective of low CO2 acting as a selection pressure. This study and also some earlier study show that leaves under low CO2 can also induce changes in the expression of many key C4 genes. Furthermore, here we show that low CO2 induces large scale rewiring of the primary metabolism to increase the capacity of leaves to assimilate ammonium, with ammonium accumulation being a possible candidate signal causing the observed changes. These innate mechanisms which induce the up-regulation of genes involved in ammonium refixation can be used to rebalance the ammonia residue after the GDC is reallocated from MC to BSC. Therefore, we propose that the mechanism responsible for the induction of genes involved in ammonium refixation represents a major regulatory mechanism, which facilitated the evolution of C4 photosynthesis.
Here we list some potential caveats of this study and should be the focus of future studies. First, we point out that during the evolution of C4 photosynthesis, there is a shift in the spatial expression patterns of critical genes involved in C4 photosynthesis (Reeves et al., 2017; Taniguchi et al., 2021). Whether low CO2 might have influenced the spatial expression pattern remains to be studied. Second, in this study, when the CO2 concentration decreased, the CO2:O2 ratio also decreased. Here we do not differentiate whether the observed responses is caused by the decreased CO2 level, or decreased CO2:O2 ratio. Thirdly, in this study, we show that the ammonium might be a potential signal inducing the observed changes in gene expression. However, under both low CO2 and high ammonium concentration, there is an increased carbon assimilation. The possibility of carbon deficiency as a signal for the observed changes also needs to be studied later. Fourthly, the signaling pathway underlies the observe changes in the gene expression needs to be elucidated as well. Elucidation of the detailed molecular mechanism will not only help understand how convergent evolution occurred for C4 photosynthesis, but also help guide future efforts of evolution guided engineering C4 prototype.
Data availability statement
The RNA-seq datasets have been submitted to the NCBI Sequence Read Archive (SRA) under accession number PRJNA842829 (https://www.ncbi.nlm.nih.gov/sra/?term=PRJNA842829).
Author contributions
X-GZ: Funding acquisition, Resources, Supervision, Writing – review & editing. FM: Conceptualization, Investigation, Writing – original draft, Writing – review & editing. YW: Writing – review & editing. NH: Writing – review & editing. M-JL: Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. The work was financially supported by National Key Research and Development Program of China (2020YFA0907600), Strategic Priority Research Program of the Chinese Academy of Sciences (#XDB27020105), Ministry of Science and Technology of China (#XDB37020104), and the National Natural Science Foundation of China (NSFC) general program (#31870214).
Acknowledgments
We thank Dr. Qingqiu Gong, Dr. Qingfeng Song, Yongyao Zhao, Xiaoxiang Ni, Yuhui Huang, Dr. Faming Chen, Dr. Qiming Tang, Dr. Genyun Chen, Jianzhao Yang for technical support and inspiring discussion during the project.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2024.1322261/full#supplementary-material
Abbreviations
2-PG, 2-phosphoglycolate; 2-OG, 2-oxoglutarate; ALA, Alanine; HPYR, Hydroxypyruvate; MAL, Malate; OAA, Oxaloacetate; PenP, Pentose phosphate; PGA, 3-Phosphoglycerate; PYR, Pyruvate; RuBP, Ribulose 1,5-bisphosphate; S7P, Sedoeheptulose-7-phosphate; Ser, Serine; SUC, Succinate; T3P, Triosephosphate; E4P, Erythrose-4-phosphate; F6P, Fructose-6-phosphate; FBP, Fructose bisphosphatase; GCEA, Glycerate; GCOA, Glycolate; GLU, Glutamate; GLX, Glyoxylate; Gly, Glycine; ASP, Aspartate; DPGA, 1,3-Bisphosphoglycerate; G6P/G1P, Glucose 6-phosphate/Glucose 1-phosphate; UDPG, uridine diphosphate glucose.
References
Alencar, V., Lobo, A. K. M., Carvalho, F. E. L., Silveira, J. A. G. (2019). High ammonium supply impairs photosynthetic efficiency in rice exposed to excess light. Photosynth Res. 140, 321–335. doi: 10.1007/s11120-019-00614-z
Arnon, D. I. (1949). Copper enzymes in isolated chloroplasts. Polyphenoloxidase in beta vulgaris. Plant Physiol. 24, 1–15. doi: 10.1104/pp.24.1.1
Arrivault, S., Alexandre Moraes, T., Obata, T., Medeiros, D. B., Fernie, A. R., Boulouis, A., et al. (2019). Metabolite profiles reveal interspecific variation in operation of the Calvin-Benson cycle in both C4 and C3 plants. J. Exp. Bot. 70, 1843–1858. doi: 10.1093/jxb/erz051
Arrivault, S., Guenther, M., Ivakov, A., Feil, R., Vosloh, D., van Dongen, J. T., et al. (2009). Use of reverse-phase liquid chromatography, linked to tandem mass spectrometry, to profile the Calvin cycle and other metabolic intermediates in Arabidopsis rosettes at different carbon dioxide concentrations. Plant J. 59, 826–839. doi: 10.1111/j.1365-313X.2009.03902.x
Aubry, S., Brown, N. J., Hibberd, J. M. (2011). The role of proteins in C(3) plants prior to their recruitment into the C(4) pathway. J. Exp. Bot. 62, 3049–3059. doi: 10.1093/jxb/err012
Bauwe, H., Hagemann, M., Fernie, A. R. (2010). Photorespiration: players, partners and origin. Trends Plant Sci. 15, 330–336. doi: 10.1016/j.tplants.2010.03.006
Carmo-Silva, A. E., Powers, S. J., Keys, A. J., Arrabaca, M. C., Parry, M. A. (2008). Photorespiration in C4 grasses remains slow under drought conditions. Plant Cell Environ. 31, 925–940. doi: 10.1111/j.1365-3040.2008.01805.x
Chen, J. H., Chen, S. T., He, N. Y., Wang, Q. L., Zhao, Y., Gao, W., et al. (2020). Nuclear-encoded synthesis of the D1 subunit of photosystem II increases photosynthetic efficiency and crop yield. Nat. Plants 6, 570–580. doi: 10.1038/s41477-020-0629-z
Cheng, C. L., Acedo, G. N., Cristinsin, M., Conkling, M. A. (1992). Sucrose mimics the light induction of Arabidopsis nitrate reductase gene transcription. Proc. Natl. Acad. Sci. U.S.A. 89, 1861–1864. doi: 10.1073/pnas.89.5.1861
Christin, P. A., Besnard, G., Samaritani, E., Duvall, M. R., Hodkinson, T. R., Savolainen, V., et al. (2008). Oligocene CO2 decline promoted C4 photosynthesis in grasses. Curr. Biol. 18, 37–43. doi: 10.1016/j.cub.2007.11.058
Cruz, C., Bio, A. F., Dominguez-Valdivia, M. D., Aparicio-Tejo, P. M., Lamsfus, C., Martins-Loucao, M. A. (2006). How does glutamine synthetase activity determine plant tolerance to ammonium? Planta 223, 1068–1080. doi: 10.1007/s00425-005-0155-2
Daniela, R., Clément, C., Marjorie, P., Medici, A., Jaeyoon Kim, G., Scalia, D., et al. (2016). Combinatorial interaction network of transcriptomic and phenotypic responses to nitrogen and hormones in the Arabidopsis thaliana root. Sci. Signal. 9, rs13–rs13. doi: 10.1126/scisignal.aaf2768
Dickinson, P. J., Knerova, J., Szecowka, M., Stevenson, S. R., Burgess, S. J., Mulvey, H., et al. (2020). A bipartite transcription factor module controlling expression in the bundle sheath of Arabidopsis thaliana. Nat. Plants 6, 1468–1479. doi: 10.1038/s41477-020-00805-w
DiMario, R. J., Quebedeaux, J. C., Longstreth, D. J., Dassanayake, M., Hartman, M. M., Moroney, J. V. (2016). The Cytoplasmic Carbonic Anhydrases betaCA2 and betaCA4 Are Required for Optimal Plant Growth at Low CO2. Plant Physiol. 171, 280–293. doi: 10.1104/pp.15.01990
Dippery, J. K., Tissue, D. T., Thomas, R. B., Strain, B. R. (1995). Effects of low and elevated CO(2) on C(3) and C(4) annuals : I. Growth and biomass allocation. Oecologia 101, 13–20. doi: 10.1007/BF00328894
Dobin, A., Davis, C. A., Schlesinger, F., Drenkow, J., Zaleski, C., Jha, S., et al. (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. doi: 10.1093/bioinformatics/bts635
Doubnerova, V., Ryslava, H. (2011). What can enzymes of C(4) photosynthesis do for C(3) plants under stress? Plant Sci. 180, 575–583. doi: 10.1016/j.plantsci.2010.12.005
Ehleringer, J. R., Cerling, T. E., Helliker, B. R. (1997). C4 photosynthesis, atmospheric CO2, and climate. Oecologia 112, 285–299. doi: 10.1007/s004420050311
Ehleringer, J. R., Sage, R. F., Flanagan, L. B., Pearcy, R. W. (1991). Climate change and the evolution of C(4) photosynthesis. Trends Ecol. Evol. 6 (3), 95–99. doi: 10.1016/0169-5347(91)90183-X
Ercoli, L., Mariotti, M., Masoni, A., Massantini, F. (1993). Relationship between nitrogen and chlorophyll content and spectral properties in maize leaves. Eur. J. Agron. 2, 113–117. doi: 10.1016/S1161-0301(14)80141-X
Esteban, R., Ariz, I., Cruz, C., Moran, J. F. (2016). Review: Mechanisms of ammonium toxicity and the quest for tolerance. Plant Sci. 248, 92–101. doi: 10.1016/j.plantsci.2016.04.008
Flugel, F., Timm, S., Arrivault, S., Florian, A., Stitt, M., Fernie, A. R., et al. (2017). The photorespiratory metabolite 2-phosphoglycolate regulates photosynthesis and starch accumulation in arabidopsis. Plant Cell 29, 2537–2551. doi: 10.1105/tpc.17.00256
Frantz, T. A., Peterson, D. M., Durbin, R. D. (1982). Sources of ammonium in oat leaves treated with tabtoxin or methionine sulfoximine. Plant Physiol. 69, 345–348. doi: 10.1104/pp.69.2.345
Gowik, U., Brautigam, A., Weber, K. L., Weber, A. P., Westhoff, P. (2011). Evolution of C4 photosynthesis in the genus Flaveria: how many and which genes does it take to make C4? Plant Cell 23, 2087–2105. doi: 10.1105/tpc.111.086264
Hachiya, T., Inaba, J., Wakazaki, M., Sato, M., Toyooka, K., Miyagi, A., et al. (2021). Excessive ammonium assimilation by plastidic glutamine synthetase causes ammonium toxicity in Arabidopsis thaliana. Nat. Commun. 12, 4944. doi: 10.1038/s41467-021-25238-7
Hachiya, T., Watanabe, C. K., Fujimoto, M., Ishikawa, T., Takahara, K., Kawai-Yamada, M., et al. (2012). Nitrate addition alleviates ammonium toxicity without lessening ammonium accumulation, organic acid depletion and inorganic cation depletion in arabidopsis thaliana shoots. Plant Cell Physiol. 53, 577–591. doi: 10.1093/pcp/pcs012
Heckmann, D., Schulze, S., Denton, A., Gowik, U., Westhoff, P., Weber, A. P., et al. (2013). Predicting C4 photosynthesis evolution: modular, individually adaptive steps on a Mount Fuji fitness landscape. Cell 153, 1579–1588. doi: 10.1016/j.cell.2013.04.058
Keerberg, O., Parnik, T., Ivanova, H., Bassuner, B., Bauwe, H. (2014). C2 photosynthesis generates about 3-fold elevated leaf CO2 levels in the C3-C4 intermediate species Flaveria pubescens. J. Exp. Bot. 65, 3649–3656. doi: 10.1093/jxb/eru239
Keys, A. J. (2006). The re-assimilation of ammonia produced by photorespiration and the nitrogen economy of C3 higher plants. Photosynth Res. 87, 165–175. doi: 10.1007/s11120-005-9024-x
Keys, A. J., Bird, I. F., Cornelius, M. J., Lea, P. J., Wallsgrove, R. M., Miflin, B. J. (1978). Photorespiratory nitrogen cycle. Nature 275, 741–743. doi: 10.1038/275741a0
Kinoshita, H., Nagasaki, J., Yoshikawa, N., Yamamoto, A., Takito, S., Kawasaki, M., et al. (2011). The chloroplastic 2-oxoglutarate/malate transporter has dual function as the malate valve and in carbon/nitrogen metabolism. Plant J. 65, 15–26. doi: 10.1111/j.1365-313X.2010.04397.x
Koch, K. E. (1996). Carbohydrate-modulated gene expression in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 47, 509–540. doi: 10.1146/annurev.arplant.47.1.509
Krogmann, D. W., Jagendorf, A. T., Avron, M. (1959). Uncouplers of spinach chloroplast photosynthetic phosphorylation. Plant Physiol. 34, 272–277. doi: 10.1104/pp.34.3.272
Kumagai, E., Araki, T., Hamaoka, N., Ueno, O. (2011). Ammonia emission from rice leaves in relation to photorespiration and genotypic differences in glutamine synthetase activity. Ann. Bot. 108, 1381–1386. doi: 10.1093/aob/mcr245
Lam, H. M., Coschigano, K. T., Oliveira, I. C., Melo-Oliveira, R., Coruzzi, G. M. (1996). The molecular-genetics of nitrogen assimilation into amino acids in higher plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 47, 569–593. doi: 10.1146/annurev.arplant.47.1.569
Lancien, M., Gadal, P., Hodges, M. (2000). Enzyme redundancy and the importance of 2-oxoglutarate in higher plant ammonium assimilation. Plant Physiol. 123, 817–824. doi: 10.1104/pp.123.3.817
Leegood, R. C., Lea, P. J., Adcock, M. D., Häusler, R. E. (1995). The regulation and control of photorespiration. J. Exp. Bot. 46, 1397–1414. doi: 10.1093/jxb/46.special_issue.1397
Li, G., Li, B., Dong, G., Feng, X., Kronzucker, H. J., Shi, W. (2013). Ammonium-induced shoot ethylene production is associated with the inhibition of lateral root formation in Arabidopsis. J. Exp. Bot. 64, 1413–1425. doi: 10.1093/jxb/ert019
Li, B., Li, G., Kronzucker, H. J., Baluska, F., Shi, W. (2014). Ammonium stress in Arabidopsis: signaling, genetic loci, and physiological targets. Trends Plant Sci. 19, 107–114. doi: 10.1016/j.tplants.2013.09.004
Li, B., Li, Q., Xiong, L., Kronzucker, H. J., Kramer, U., Shi, W. (2012). Arabidopsis plastid AMOS1/EGY1 integrates abscisic acid signaling to regulate global gene expression response to ammonium stress. Plant Physiol. 160, 2040–2051. doi: 10.1104/pp.112.206508
Li, Y., Xu, J., Haq, N. U., Zhang, H., Zhu, X. G. (2014). Was low CO2 a driving force of C4 evolution: Arabidopsis responses to long-term low CO2 stress. J. Exp. Bot. 65, 3657–3667. doi: 10.1093/jxb/eru193
Li, G., Zhang, L., Wang, M., Di, D., Kronzucker, H. J., Shi, W. (2019). The Arabidopsis AMOT1/EIN3 gene plays an important role in the amelioration of ammonium toxicity. J. Exp. Bot. 70, 1375–1388. doi: 10.1093/jxb/ery457
Liu, Y., von Wiren, N. (2017). Ammonium as a signal for physiological and morphological responses in plants. J. Exp. Bot. 68, 2581–2592. doi: 10.1093/jxb/erx086
Love, M. I., Huber, W., Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550. doi: 10.1186/s13059-014-0550-8
Mallmann, J., Heckmann, D., Brautigam, A., Lercher, M. J., Weber, A. P., Westhoff, P., et al. (2014). The role of photorespiration during the evolution of C4 photosynthesis in the genus Flaveria. Elife 3, e02478. doi: 10.7554/eLife.02478.021
Masclaux-Daubresse, C., Daniel-Vedele, F., Dechorgnat, J., Chardon, F., Gaufichon, L., Suzuki, A. (2010). Nitrogen uptake, assimilation and remobilization in plants: challenges for sustainable and productive agriculture. Ann. Bot. 105, 1141–1157. doi: 10.1093/aob/mcq028
McLoughlin, F., Marshall, R. S., Ding, X., Chatt, E. C., Kirkpatrick, L. D., Augustine, R. C., et al. (2020). Autophagy plays prominent roles in amino acid, nucleotide, and carbohydrate metabolism during fixed-carbon starvation in maize. Plant Cell 32, 2699–2724. doi: 10.1105/tpc.20.00226
Nagatani, H., Shimizu, M., Valentine, R. C. (1971). The mechanism of ammonia assimilation in nitrogen fixing Bacteria. Arch. Mikrobiol 79, 164–175. doi: 10.1007/BF00424923
Nunes-Nesi, A., Fernie, A. R., Stitt, M. (2010). Metabolic and signaling aspects underpinning the regulation of plant carbon nitrogen interactions. Mol. Plant 3, 973–996. doi: 10.1093/mp/ssq049
Patterson, K., Cakmak, T., Cooper, A., Lager, I., Rasmusson, A. G., Escobar, M. A. (2010). Distinct signalling pathways and transcriptome response signatures differentiate ammonium- and nitrate-supplied plants. Plant Cell Environ. 33, 1486–1501. doi: 10.1111/j.1365-3040.2010.02158.x
Peterhansel, C., Maurino, V. G. (2011). Photorespiration redesigned. Plant Physiol. 155, 49–55. doi: 10.1104/pp.110.165019
Rademacher, T., Häusler, R. E., Hirsch, H.-J., Zhang, L., Lipka, V., Weier, D., et al. (2002). An engineered phosphoenolpyruvate carboxylase redirects carbon and nitrogen flow in transgenic potato plants. Plant J. 32 (1), 25–39. doi: 10.1046/j.1365-313x.2002.01397.x
Rawsthorne, S., Hylton, C. M., Smith, A. M., Woolhouse, H. W. (1988). Photorespiratory metabolism and immunogold localization of photorespiratory enzymes in leaves of C-3 and C-3-C-4 intermediate species of moricandia. Planta 173, 298–308. doi: 10.1007/BF00401016
Reeves, G., Grange-Guermente, M. J., Hibberd, J. M. (2017). Regulatory gateways for cell-specific gene expression in C4 leaves with Kranz anatomy. J. Exp. Bot. 68, 107–116. doi: 10.1093/jxb/erw438
Reyna-Llorens, I., Hibberd, J. M. (2017). Recruitment of pre-existing networks during the evolution of C4 photosynthesis. Philos. Trans. R Soc. Lond B Biol. Sci. 372 (1730), 20160386. doi: 10.1098/rstb.2016.0386
Ruan, Y. L. (2014). Sucrose metabolism: gateway to diverse carbon use and sugar signaling. Annu. Rev. Plant Biol. 65, 33–67. doi: 10.1146/annurev-arplant-050213-040251
Sage, R. F. (2001). Environmental and evolutionary preconditionsfor the origin and diversification of the C4PhotosyntheticSyndrome. Plant Biol. 3, 202–213. doi: 10.1055/s-2001-15206
Sage, A. S. R. R. F. (2011). Photosynthesis and related CO2 concentrating mechanisms. 81–108. doi: 10.1007/978-90-481-9407-0
Sage, R. F. S. (2012). Photorespiration and the evolution of C4 photosynthesis. L.Kocacinar F.Annu. Rev. Plant Biol. 63, 19–47doi: 10.1146/annurev-arplant-042811-105511
Sage, R. F., Christin, P. A., Edwards, E. J. (2011). The C(4) plant lineages of planet Earth. J. Exp. Bot. 62, 3155–3169. doi: 10.1093/jxb/err048
Sage, R. F., Coleman, J. R. (2001). Effects of low atmospheric CO(2) on plants: more than a thing of the past. Trends Plant Sci. 6, 18–24. doi: 10.1016/S1360-1385(00)01813-6
Sage, R. F., Monson, R. K., Ehleringer, J. R., Adachi, S., Pearcy, R. W. (2018). Some like it hot: the physiological ecology of C4 plant evolution. Oecologia 187, 941–966. doi: 10.1007/s00442-018-4191-6
Sage, R. F., Sage, T. L., Pearcy, R. W., Borsch, T. (2007). The taxonomic distribution of C4 photosynthesis in Amaranthaceae sensu stricto. Am. J. Bot. 94, 1992–2003. doi: 10.3732/ajb.94.12.1992
Sarasketa, A., Gonzalez-Moro, M. B., Gonzalez-Murua, C., Marino, D. (2014). Exploring ammonium tolerance in a large panel of Arabidopsis thaliana natural accessions. J. Exp. Bot. 65, 6023–6033. doi: 10.1093/jxb/eru342
Schluter, U., Brautigam, A., Gowik, U., Melzer, M., Christin, P. A., Kurz, S., et al. (2017). Photosynthesis in C3-C4 intermediate Moricandia species. J. Exp. Bot. 68, 191–206. doi: 10.1093/jxb/erw391
Sheen, J. (1990). Metabolic repression of transcription in higher plants. Plant Cell 2, 1027–1038. doi: 10.1105/tpc.2.10.1027
Solórzano, L. (1969). Determination OF AMMONIA IN NATURAL WATERS BY THE PHENOLHYPOCHLORITE METHOD 1 1 this research was fully supported by U.S. Atomic energy commission contract no. ATS (11-1) GEN 10, P.A. 20. Limnol. Oceanogr. 14, 799–801. doi: 10.4319/lo.1969.14.5.0799
Still, C. J., Berry, J. A., Collatz, G. J., DeFries, R. S. (2003). Global distribution of C3and C4vegetation: Carbon cycle implications. Global Biogeochem. Cycles 17, 6–1-6-14. doi: 10.1029/2001GB001807
Stitt, M., Muller, C., Matt, P., Gibon, Y., Carillo, P., Morcuende, R., et al. (2002). Steps towards an integrated view of nitrogen metabolism. J. Exp. Bot. 53, 959–970. doi: 10.1093/jexbot/53.370.959
Sweetlove, L. J., Beard, K. F., Nunes-Nesi, A., Fernie, A. R., Ratcliffe, R. G. (2010). Not just a circle: flux modes in the plant TCA cycle. Trends Plant Sci. 15, 462–470. doi: 10.1016/j.tplants.2010.05.006
Taniguchi, Y. Y., Gowik, U., Kinoshita, Y., Kishizaki, R., Ono, N., Yokota, A., et al. (2021). Dynamic changes of genome sizes and gradual gain of cell-specific distribution of C(4) enzymes during C(4) evolution in genus Flaveria. Plant Genome 14, e20095. doi: 10.1002/tpg2.20095
Tcherkez, G., Gauthier, P., Buckley, T. N., Busch, F. A., Barbour, M. M., Bruhn, D., et al. (2017). Leaf day respiration: low CO2 flux but high significance for metabolism and carbon balance. New Phytol. 216, 986–1001. doi: 10.1111/nph.14816
Turpin, C. (1994). Integrat ion of carbon and ni trogen me tabol ism in plant and algal cells. 45, 577–607. doi: 10.1146/annurev.arplant.45.1.577
Valderrama-Martín, J. M., Ortigosa, F., Ávila, C., Cánovas, F. M., Hirel, B., Cantón, F. R., et al. (2022). A revised view on the evolution of glutamine synthetase isoenzymes in plants. Plant J. 110, 946–960. doi: 10.1111/tpj.15712
Vogan, P. J., Frohlich, M. W., Sage, R. F. (2007). The functional significance of C3-C4 intermediate traits in Heliotropium L. (Boraginaceae): gas exchange perspectives. Plant Cell Environ. 30, 1337–1345. doi: 10.1111/j.1365-3040.2007.01706.x
Wang, L., Czedik-Eysenberg, A., Mertz, R. A., Si, Y., Tohge, T., Nunes-Nesi, A., et al. (2014). Comparative analyses of C(4) and C(3) photosynthesis in developing leaves of maize and rice. Nat. Biotechnol. 32, 1158–1165. doi: 10.1038/nbt.3019
Weerasooriya, H. N., DiMario, R. J., Rosati, V. C., Rai, A. K., LaPlace, L. M., Filloon, V. D., et al. (2022). Arabidopsis plastid carbonic anhydrase betaCA5 is important for normal plant growth. Plant Physiol. 190, 2173–2186. doi: 10.1093/plphys/kiac451
Wei, L., El Hajjami, M., Shen, C., You, W., Lu, Y., Li, J., et al. (2019). Transcriptomic and proteomic responses to very low CO2 suggest multiple carbon concentrating mechanisms in Nannochloropsis oceanica. Biotechnol. Biofuels 12, 168. doi: 10.1186/s13068-019-1506-8
Wilkinson, S., Davies, W. J. (2002). ABA-based chemical signalling: the co-ordination of responses to stress in plants. Plant Cell Environ. 25 (2), 195–210. doi: 10.1046/j.0016-8025.2001.00824.x
Wingler, A., Quick, W. P., Bungard, R. A., Bailey, K. J., Lea, P. J., Leegood, R. C. (1999). The role of photorespiration during drought stress: an analysis utilizing barley mutants with reduced activities of photorespiratory enzymes. Plant Cell Environ. 22, 361–373. doi: 10.1046/j.1365-3040.1999.00410.x
You, L., Zhang, J., Li, L., Xiao, C., Feng, X., Chen, S., et al. (2020). Involvement of abscisic acid, ABI5, and PPC2 in plant acclimation to low CO2. J. Exp. Bot. 71 (14), 4093–4108. doi: 10.1093/jxb/eraa148
Keywords: low CO2, photorespiration, ammonium refixation, regulatory preconditioning, C4 photosynthesis
Citation: Miao F, Wang Y, Haq NUI, Lyu M-JA and Zhu X-G (2024) Rewiring of primary metabolism for ammonium recycling under short-term low CO2 treatment – its implication for C4 evolution. Front. Plant Sci. 15:1322261. doi: 10.3389/fpls.2024.1322261
Received: 16 October 2023; Accepted: 04 July 2024;
Published: 01 August 2024.
Edited by:
Michael Moustakas, Aristotle University of Thessaloniki, GreeceCopyright © 2024 Miao, Wang, Haq, Lyu and Zhu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xin-Guang Zhu, emh1eGdAY2VtcHMuYWMuY24=
 Fenfen Miao1,2
Fenfen Miao1,2 Noor UI Haq
Noor UI Haq Ming-Ju Amy Lyu
Ming-Ju Amy Lyu Xin-Guang Zhu
Xin-Guang Zhu