
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 23 January 2024
Sec. Plant Metabolism and Chemodiversity
Volume 15 - 2024 | https://doi.org/10.3389/fpls.2024.1307489
This article is part of the Research Topic Proanthocyanidins and Isoflavonoids View all 12 articles
 Emily J. Clayton1,2
Emily J. Clayton1,2 Nishat S. Islam1
Nishat S. Islam1 Kelsey Pannunzio1,2
Kelsey Pannunzio1,2 Kuflom Kuflu1
Kuflom Kuflu1 Ramtin Sirjani1,2
Ramtin Sirjani1,2 Susanne E. Kohalmi2
Susanne E. Kohalmi2 Sangeeta Dhaubhadel1,2*
Sangeeta Dhaubhadel1,2*Soybean (Glycine max) produces a class of phenylalanine (Phe) derived specialized metabolites, isoflavonoids. Isoflavonoids are unique to legumes and are involved in defense responses in planta, and they are also necessary for nodule formation with nitrogen-fixing bacteria. Since Phe is a precursor of isoflavonoids, it stands to reason that the synthesis of Phe is coordinated with isoflavonoid production. Two putative AROGENATE DEHYDRATASE (ADT) isoforms were previously co-purified with the soybean isoflavonoid metabolon anchor ISOFLAVONE SYNTHASE2 (GmIFS2), however the GmADT family had not been characterized. Here, we present the identification of the nine member GmADT family. We determined that the GmADTs share sequences required for enzymatic activity and allosteric regulation with other characterized plant ADTs. Furthermore, the GmADTs are differentially expressed, and multiple members have dual substrate specificity, also acting as PREPHENATE DEHYDRATASES. All GmADT isoforms were detected in the stromules of chloroplasts, and they all interact with GmIFS2 in the cytosol. In addition, GmADT12A interacts with multiple other isoflavonoid metabolon members. These data substantiate the involvement of GmADT isoforms in the isoflavonoid metabolon.
Soybean (Glycine max [L.] Merr) is an important grain legume grown worldwide. Soybean seeds are high in oil and protein content, making them a nutrient-rich food source for both livestock and humans (Hill and Breidenbach, 1974; Messina, 2010). As one of the most cultivated oilseed crops, agricultural waste from soybean harvest has potential applications as a biofuel source (Yong and Wu, 2022). Soybeans also produce a legume unique class of specialized metabolites called isoflavonoids. Isoflavonoids are important signaling molecules, as they are essential for interaction between legumes and nitrogen-fixing bacteria (Phillips and Kapulnik, 1995; Subramanian et al., 2006; Hassan and Mathesius, 2012). Isoflavonoid glyceollins act as phytoalexins and provide defense against abiotic and biotic stress, such as resistance against the soil-borne pathogen Phytophthora sojae (Subramanian et al., 2006; Lozovaya et al., 2007), that cause massive crop losses (Bradley et al., 2021; Chandra et al., 2022). Furthermore, there is some evidence suggesting that isoflavonoids such as genistein and glyceollin have health benefits (Lamartiniere, 2000; Dixon and Ferreria, 2002; Sarkar and Li, 2003; Cederroth and Nef, 2009; Messina, 2010; Bhat et al., 2021). As such, isoflavonoids have been the target of traditional breeding and metabolic engineering to improve cultivar resistance to biotic and abiotic stress (Scott et al., 2021; Yousefi-Taemeh et al., 2021).
The biosynthesis of isoflavonoids is a complex process that is derived from the phenylpropanoid metabolism (Winkel, 2001), downstream from the synthesis of the aromatic amino acid phenylalanine (Phe). As shown in Figure 1, the first committed step of (iso)flavonoid biosynthesis is the action of CHALCONE SYNTHASE (CHS) to synthesize a chalcone scaffold from which all (iso)flavonoids are built (Winkel, 2006; Dastmalchi and Dhaubhadel, 2014). The key branch point of isoflavonoid synthesis from the flavonoid biosynthetic pathway is catalyzed by the cytochrome P450 enzyme ISOFLAVONE SYNTHASE (IFS) where naringenin and liquiritigenin are converted to isoflavones genistein and daidzein, respectively (Winkel, 2006; Dastmalchi and Dhaubhadel, 2014; García-Calderón et al., 2020). Many of the enzymes involved in (iso)flavonoid biosynthesis have been shown to form protein-protein interactions (Winkel, 2004; Jorgensen et al., 2005; Dastmalchi et al., 2016). The phenylpropanoid pathway was first proposed to form a multienzyme metabolon by Helen Stafford (1974) and further confirmed by Hrazdina and Wagner (1985), as an explanation for the efficiency of substrate channeling in specialized metabolite biosynthesis. Further evidence demonstrated that the metabolon includes enzymes involved in flavonoid synthesis, such as CHS, CHALCONE ISOMERASE (CHI), and FLAVONOL SYNTHASE (FLS) (Winkel, 2004; Nakayama et al., 2019). In soybean, this metabolon also includes key isoflavonoid biosynthetic enzymes CHALCONE REDUCTASE (CHR) and IFS (Figure 2) (Dastmalchi et al., 2016). The isoflavonoid biosynthetic metabolon is associated with the cytosolic surface of the endoplasmic reticulum (ER), anchored by the two ER membrane cytochrome P450 monooxygenase enzymes CINNAMATE 4-HYDROXYLASE (C4H) and IFS (Jorgensen et al., 2005; Dastmalchi et al., 2016). To date, the flavonoid metabolon has been identified in a wide range of plant species (Owens et al., 2008; Crosby et al., 2011; Waki et al., 2016; Nakayama et al., 2019), conferring a variety of advantages such as metabolic channeling of substrates, or the sequestering of toxic intermediates (Moller, 2010; Pareek et al., 2021; Zhang and Fernie, 2021).
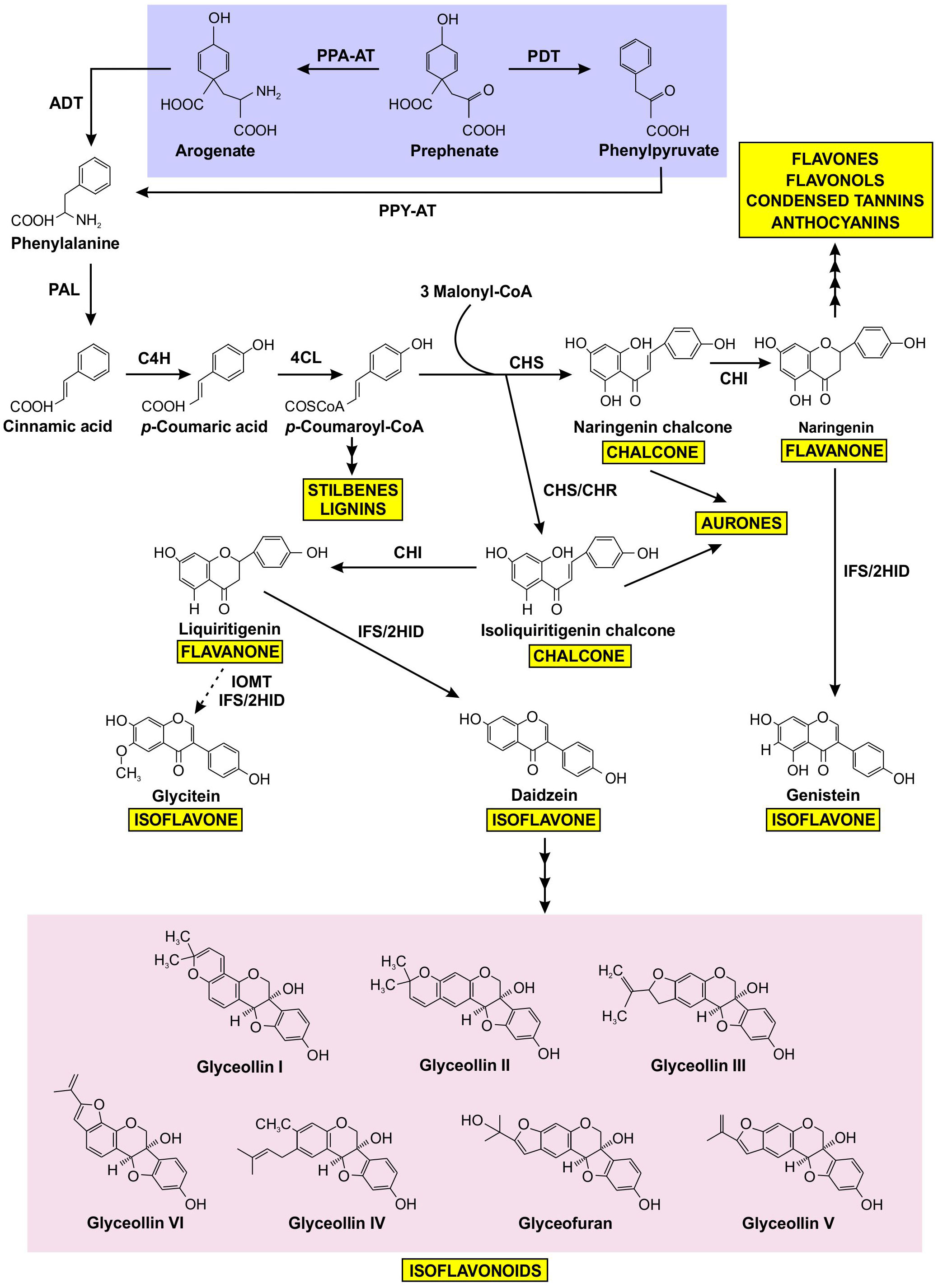
Figure 1 Prephenate from the shikimate pathway uses two different mechanisms to produce phenylalanine, which is subsequently channeled into the production of a variety of phenylpropanoids, including isoflavonoids. Phenylalanine produced by the arogenate and phenylpyruvate branches are highlighted in purple, isoflavonoids are highlighted in pink, and other phenylpropanoid end products are highlighted in yellow. The dotted arrow represents speculative steps, and multiple arrows indicate two or more steps in the pathway. PPA-AT, prephenate-aminotransferase; PDT, prephenate dehydratase; PPY-AT, phenylpyruvate aminotransferase; PPA-AT, prephenate aminotransferase; ADT, arogenate dehydratase; PAL, phenylalanine ammonia lyase; C4H, cinnamate 4-hydroxylase; 4CL, 4–coumarate: CoA ligase; CHS, chalcone synthase; CHR, chalcone reductase; CHI, chalcone isomerase; IFS, isoflavone synthase; 2HID, 2-hydroxyisoflavanone dehydratase; I2’H, isoflavone 2’-hydroxylase; IOMT, isoflavone O–methyltransferase.
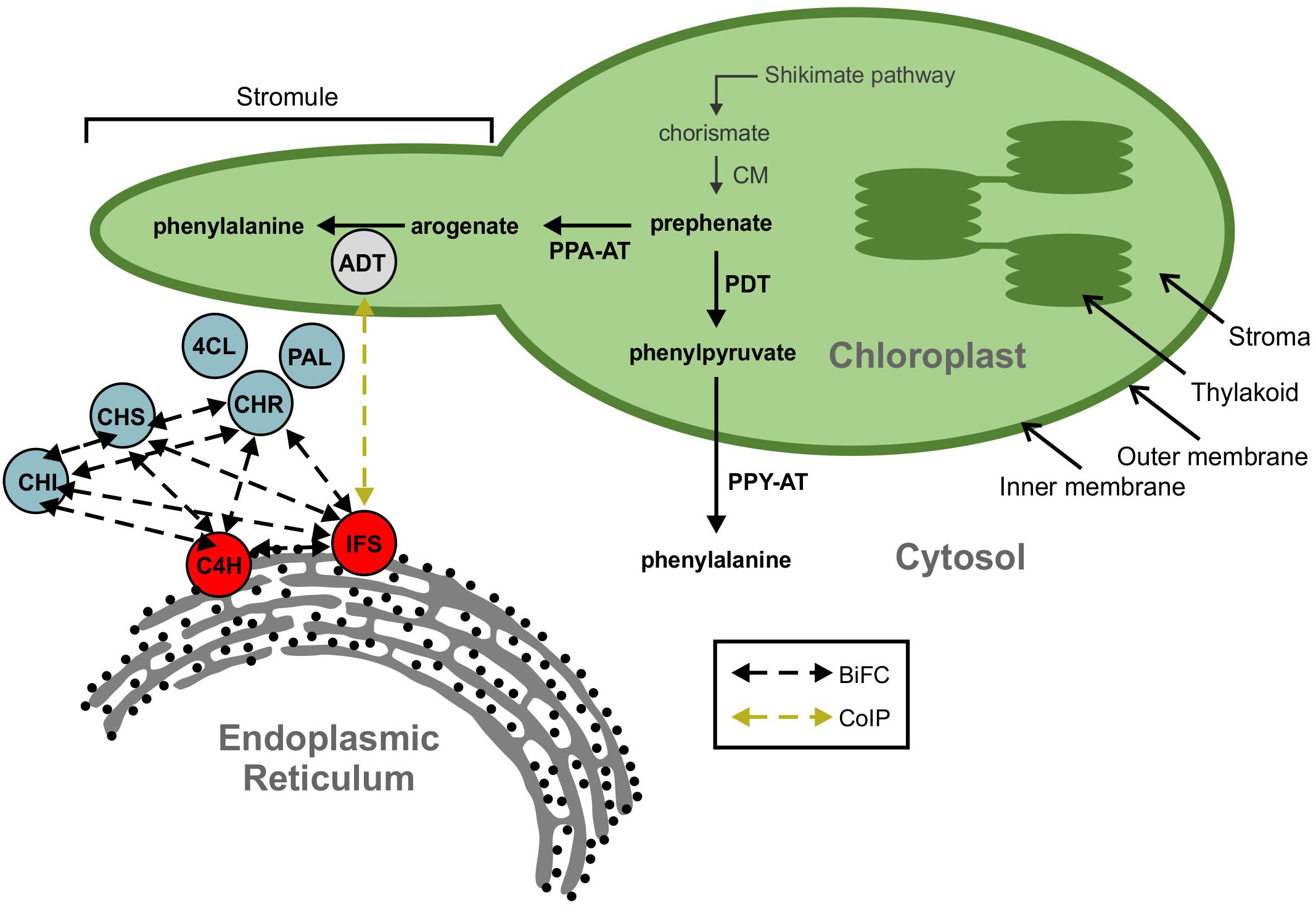
Figure 2 A model showing ADT interaction with the isoflavonoid metabolon. The isoflavonoid metabolon is made up of cytoplasm-localized enzymes (blue) anchored to the cytosolic face of the ER membrane through interaction with cytochrome P450 enzymes, IFS and C4H (red). Protein-protein interactions between multiple members of the isoflavonoid metabolon have been determined by BiFC (black dashed arrows). An interaction between IFS and an ADT (grey) was also detected via Co-IP (yellow dashed arrow). ADT enzymes are involved in phenylalanine biosynthesis and are localized to the chloroplast along with the shikimate pathway and other aromatic amino acid biosynthetic machinery. Subcellular compartments are indicated by grey fonts. ADT, arogenate dehydratase; PAL, phenylalanine ammonia lyase; 4CL, 4-coumarate-CoA ligase; C4H, cinnamate 4-hydroxylase; CHI, chalcone isomerase; CHR, chalcone reductase; CHS, chalcone synthase; CM, chorismite mutase; IFS, isoflavone synthase; PDT, prephenate dehydratase; PPA-AT, prephenate aminotransferase; PPY-A, phenylpyruvate aminotransferase.
When characterizing the soybean isoflavonoid metabolon, we detected an interaction between GmIFS2 and two putative AROGENATE DEHYDRATASES (ADTs), Glyma.12G181800.1 and Glyma.13G319000.1 (Dastmalchi et al., 2016). ADTs catalyze the final step in Phe biosynthesis via the arogenate pathway (Maeda and Dudareva, 2012) in which prephenate is converted to arogenate by a PREPHENATE AMINOTRANSFERASE (PPA-AT; (Graindorge et al., 2010; Maeda et al., 2011), followed by a decarboxylation/dehydration to Phe, catalyzed by an ADT (Jung et al., 1986; Ehlting et al., 2005; Cho et al., 2007). The interaction between ADT and IFS observed in our previous work was an unexpected finding (Dastmalchi et al., 2016), as ADTs have been shown to localize within the chloroplasts in other plant species, consistent with the chloroplastic localization of shikimate and arogenate pathway enzymes (Cho et al., 2007; Rippert et al., 2009; Maeda et al., 2010; El-Azaz et al., 2016). However, a second Phe biosynthetic route, the prephenate pathway, had been described where prephenate is first decarboxylated/dehydrated into phenylpyruvate by a PREPHENATE DEHYDRATASE (PDT; (Cotton and Gibson, 1965), which is then transaminated into Phe by the action of a PHENYLPYRUVATE AMINOTRANSFERASE (PPY-AT; (Cotton and Gibson, 1965; Fazel et al., 1980). While prephenate pathway is predominantly used by microbes, there was evidence for existence of the prephenate pathway in plants. PPY-ATs had been identified in several plant species (Watanabe et al., 2002; Kaminaga et al., 2006; Warpeha et al., 2006), and PDT activity had been reported in Petunia hybrida, Arabidopsis, Oryza sativa and Pinus pinaster, though the dual ADT/PDT enzymes have a preference for arogenate over prephenate (Cho et al., 2007; Yamada et al., 2008; Maeda et al., 2010; El-Azaz et al., 2016; El-Azaz et al., 2022). Recently, cytosolic Phe biosynthesis via the prephenate pathway was reported in Petunia x hybrida (Yoo et al., 2013; Qian et al., 2019) and is expected to act in parallel with the arogenate pathway. As the isoflavonoid metabolon is anchored to the cytosolic face of the ER, the two GmADTs identified as part of the isoflavonoid metabolon (Dastmalchi et al., 2016) could be part the cytosolic Phe synthesis route, and may have PDT activity. As most plant genomes encode at least two, if not more ADT genes, there are likely many more than two GmADT isoforms present in soybean as it is a paleopolyploid. These isoforms may also interact with the isoflavonoid metabolon. However, the complete GmADT family has not been characterized.
Here we identified additional ADT family members in soybean and describe their gene structure, phylogeny, tissue-specific gene expression and subcellular localization. We demonstrate that some members of the GmADT family contain PDT activity in yeast complementation analysis. Despite the fact that all GmADTs were detected in the chloroplast, we confirmed their ability to interact with GmIFS2 and multiple other isoflavonoid metabolon enzymes in planta. Together, these data suggest that members of the GmADT family are associating with the isoflavonoid metabolon, and those with PDT activity could be supplying Phe to the metabolon through a cytosolic prephenate pathway.
Nicotiana benthamiana was grown on PRO-MIX® BX MYCORRHIZAETM soil (Rivière-du-Loup, Canada) in a growth room set to 16 h light at 24°C and 8 h dark at 20°C with 60% relative humidity and a light intensity of 80-100 μmol m-2s-1.
Soybean cultivar Williams 82 seeds were planted in sterile pots containing PRO-MIX® BX MYCORRHIZAE™ soil (Premier Tech Home and Garden, Rivière-du-Loup, QC) and maintained in a growth room under a 16 h light and 8 h dark cycle at 24°C with 60-70% humidity and a light intensity of 250 μmol m-2s-1. The plants were watered with a fertilizer solution containing nitrogen-phosphorus-potassium (20-8-20). At the flowering stage of soybean plants, stem, leaf, root, and flower tissue were harvested, frozen in liquid nitrogen, and stored at -80°C.
Candidate soybean ADTs (GmADTs) were identified by mining the soybean genome in Phytozome 13 (https://phytozome-next.jgi.doe.gov/). Two previously identified soybean ADTs Glyma.13G319000.1 and Glyma.12G181800.1 (GmADT13A and GmADT12A, Table 1) (Dastmalchi et al., 2016) were used in BLAST searches against the soybean genome database (Glycine max Wm82.a4.v1). Each unique gene identified from the two initial input sequences was used in BLAST search again to look for all possible GmADTs. Multiple sequence alignments were performed using protein sequences of ADTs in Clustal Omega (Sievers et al., 2011), and visualized in boxshade using pyBoxshade (https://github.com/mdbaron42/pyBoxshade). TargetP was used for subcellular localization and cleavage site prediction (Emanuelsson et al., 2007). The gene and transcript data (in gff3 format) for GmADTs were retrieved from Phytozome 13 for Wm82.a4.v1 genome assembly and the gene structure model was generated using TBtools (Chen et al., 2020).
For phylogenetic analysis, the predicted transit peptide sequences were removed from plant ADTs and putative GmADTs according to Cho et al. (2007). The chorismate mutase domain of the E. coli P-protein was removed according to Zhang et al. (1998). The mature protein sequences were aligned using ClustalW and the tree was constructed with the bootstrap set to 1,000 replicates using MEGAX software (Kumar et al., 2018).
Soybean RNA-seq data was retrieved from Phytozome 13 database with expression values in FPKM (Wang et al., 2019). A heatmap was generated using log2-transformed normalized transcript abundance values using TBtools (Chen et al., 2020). Gene cluster in the Newick tree was generated in MEGAX and imported into the heatmap.
Total RNA was extracted from soybean tissues (50-70 mg) using the RNeasy plant Mini kit (Qiagen). An on column DNase I (Promega) treatment was performed prior to RNA elution from each sample. Total RNA (1 μg) was used to synthesize cDNA using oligo dT primers and SuperScript IV First Strand Synthesis System (Thermofisher) as per manufacturer’s instructions.
The coding regions of GmADTs were amplified using RT-PCR with gene-specific primers (Supplementary Table 1) and cloned into pDONR-Zeo (Invitrogen) using BP clonase® (Invitrogen), followed by transformation into E. coli DH5α via electroporation. The recombinant entry clones were confirmed by sequencing and then recombined with the destination vectors pEarleyGate101 (pEG101) for subcellular localization (Earley et al., 2006) and pEarleyGate201-YN and pEarleyGate202-YC for in planta protein-protein interaction assays (Lu et al., 2010) in an LR recombination reaction (Invitrogen). The expression clones were transformed into Agrobacterium tumefaciens GV3101. GmIFS2 (Glyma.13G173500) in pEarleyGate201-YN and pEarleyGate202-YC were obtained from Dastmalchi et al. (2016).
For the PDT assay, each of the GmADTs with a 6×His-C-terminal fusion was recombined into the destination vector pAG423GAL-ccdB-ECFP (Addgene plasmid # 14173; http://n2t.net/addgene:14173; RRID : Addgene_14173) using Gateway technology as described above and transformed into Saccharomyces cerevisiae pha2 (Ackerman et al., 1992) using Frozen-EZ Yeast Transformation II™ kit (Zymo Research). Transformants were screened on minimal synthetic dextrose (SD)/-His plates.
For subcellular localization, A. tumefaciens harboring pEG101 containing GmADTs were transformed into N. benthamiana leaves by infiltration as described by Sparkes et al. (2006). For protein-protein interaction by BiFC, fusions containing YN and YC fragments of YFP were co-infiltrated into N. benthamiana leaves in a 1:1 (v/v) mixture as described before. The protein expression was visualized 48 h post-infiltration by using the Olympus FV1000 confocal microscope under a 60× water immersion objective lens. For YFP visualization, the excitation wavelength was set to 514 nm and emission was collected at 520-550 nm. For chloroplast-visualization, the natural auto-fluorescence produced by chlorophyll was harnessed by exciting the chlorophyll at 600 nm and emission was collected at 640-700 nm.
For the pha2 complementation assay (Bross et al., 2011), yeast cultures were grown in appropriately supplemented liquid raffinose media overnight at 30°C with shaking. Each culture diluted in double distilled water to a final density of 5 x 104 cells/mL, and 10 μL of cells were spotted on appropriate selection media. SD plates prepared with different carbon sources (glucose, galactose, and raffinose) were used. The GAL1 promoter in the destination vectors is expressed or repressed by galactose or glucose, respectively, whereas raffinose has no influence on the regulation of the promoter (St John and Davis, 1981; Lohr et al., 1995). Plates of each carbon source were made either with a -histidine (-His), or a -histidine-phenylalanine (-His-Phe) dropout powder. The lack of His selects for the presence of the ADT expression vector, while the lack of Phe selects for PDT activity when ADT proteins are induced. Negative (untransformed pha2 and empty destination vector) and positive (WT AtADT2) controls were spotted on every plate, and all complementation tests were repeated at least 3 times. Images were taken with a digital camera (Canon EOS 70D) six and thirteen days after spotting, and a single representative image of each construct is shown.
Yeast cultures were grown overnight in glucose media, washed twice with double distilled water, then grown overnight to an OD600 of 0.6-0.8 in glucose media as a negative control, or in galactose media to induce ADT expression. Total protein was extracted using the yeast alkaline lysis method (Kushnirov, 2000). Protein extracts were then size separated on an SDS-PAGE gel (6% stacking gel, 12% separating gel). Following electrophoresis, the resolved proteins were visualized with Coomassie Brilliant Blue. Gels were incubated at room temperature for 30 minutes with shaking, and then destained for 2-3 hrs to remove background stain before an image was taken.
GmADT-6×His and CHR14-6xHis fusion proteins were detected using Western blot analysis with a monoclonal mouse anti-His primary antibody (1:1200, Sigma, SAB1305538). A goat anti-mouse HRP conjugate secondary antibody (BioRad, 1706516) was used in all cases, and HRP activity was visualized using the Clarity ECL kit (BioRad, 1705061).
To identify all the members of the GmADT gene family, we used Glyma.12G181800 (GmADT12A) and Glyma.13G319000 (GmADT13A) sequences as queries in a BLAST search in the G. max Wm82.a4.v1 genome database. These two searches identified 10 GmADTs. Each of these GmADTs were used separately as a query sequence in the BLAST search in the soybean genome database. This process was repeated until no new ADT was discovered (Table 1). The following nomenclature was developed: GmADTs were numbered according to the chromosome on which they are encoded and a letter (A, B, etc) was added if more than one GmADT was located on the same chromosome.
The 10 identified ADT loci in the soybean genome are distributed across five different chromosomes, with chromosome 12 containing four GmADTs (GmADT12A, GmADT12B, GmADT12C and GmADT12D) while chromosomes 11 (GmADT11A, GmADT11B) and 13 (GmADT13A, GmADT13B) each contain two GmADTs. Chromosomes 9 and 17 carry only one GmADT each (GmADT9 and GmADT17, Table 1). The putative GmADT genes encode proteins with a calculated molecular mass ranging from 23.1 to 46.9 kDa (Table 1).
A multiple sequence alignment of previously characterized ADTs from Arabidopsis thaliana (AtADTs), Petunia x hybrida (PhADTs) and Pinus pinaster (PpADTs) and the deduced amino acid sequences of GmADTs revealed that, similar to other plant ADTs, GmADTs also contain a putative N-terminal transit peptide, an internal catalytic domain, and a C-terminal ACT domain (Figure 3). As expected the transit peptide regions are highly variable while the catalytic and ACT domains of the GmADT, AtADT, PhADT and PpADT proteins exhibit substantial levels of sequence conservation. Comparing GmADT sequences, GmADT9 is the most varied member of the family and shares only 49.5-63.3% sequence identity with other isoforms at the amino acid level. A pairwise percentage identity of full-length GmADTs sequences at the amino acid and nucleotide levels varied from 49.5 to 96.2% and 50.6 to 94.8%, respectively (Table 2). Most GmADTs except GmADT12C and GmADT13B contain the conserved TRF triad (Figure 3, green box) in the catalytic domain that is critical for substrate binding and prephenate/arogenate catalysis (Zhang et al., 2000; Hsu et al., 2004; Tan et al., 2008). Instead of the TRF triad, GmADT12C contains an SRY sequence instead. In addition, key ligand binding motifs (ESRP and GALV) in the ACT domain (Pohnert et al., 1999; Tan et al., 2008) are also shared by the GmADTs and other ADTs. However, the C-terminal region of the catalytic domain and the entire ACT domain from GmADT13B, and the C-terminus of the ACT domain from GmADT12D are missing. As such, GmADT12D contains the GALV but lacks the ESRP regulatory motif due to C-terminal truncation. Soybean cultivar Williams 82 whole genome sequence has been reassembled multiple times, however, this discrepancy was observed consistently including in the most recent release Glycine max var. Williams 82-ISU-01 (https://phytozome-next.jgi.doe.gov/info/GmaxWm82ISU_01_v2_1). As GmADT13B lacks the entirety of the ACT domain, it is likely not allosterically regulated if it is a functional protein.
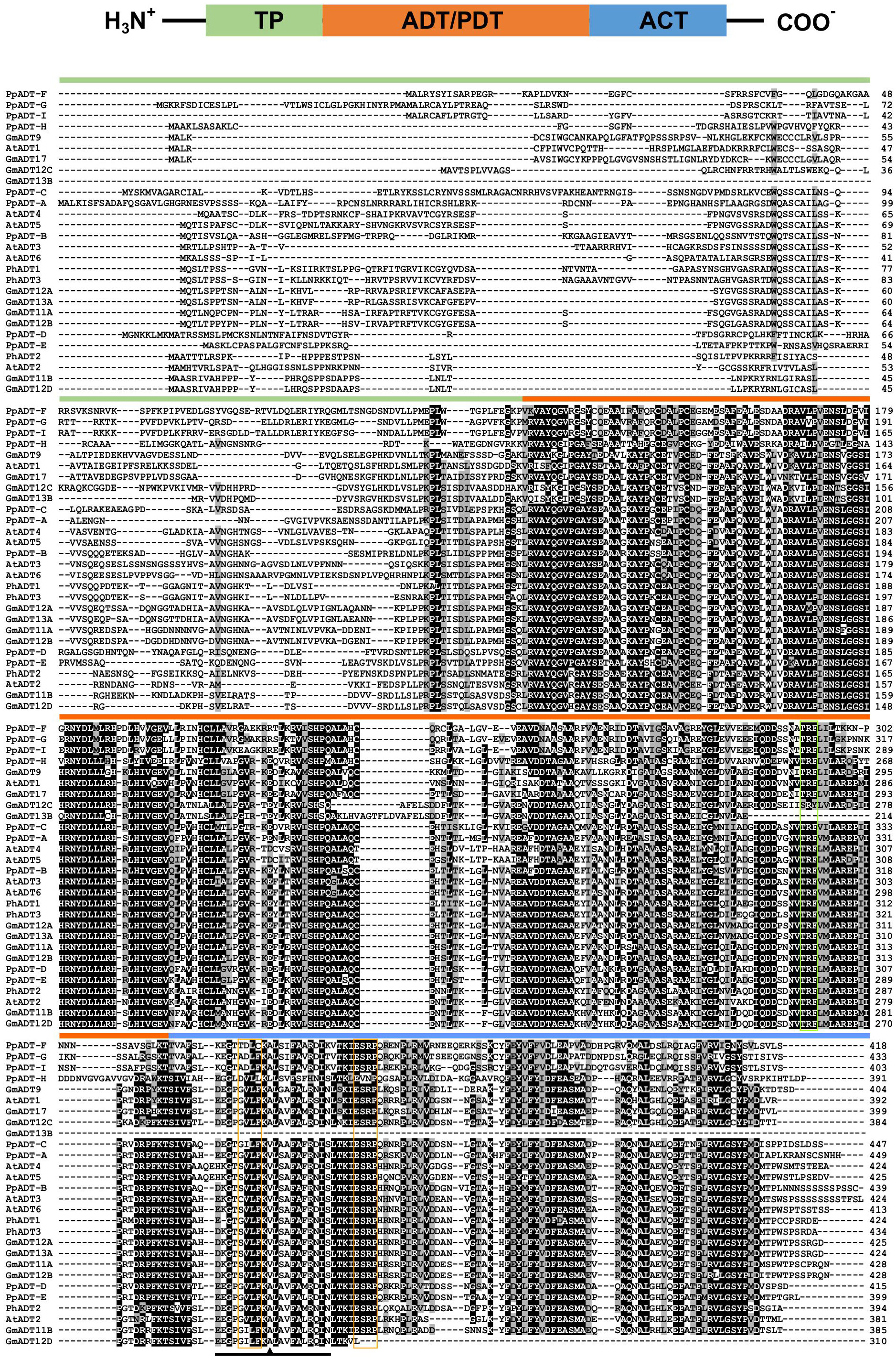
Figure 3 Protein domains and multiple sequence alignment of GmADTs and AtADTs. A schematic diagram (top) of predicted protein domains of ADTs include transit peptide (TP), catalytic ADT or PDT domain and regulatory ACT domain (modified from Cho et al., 2007). The amino acid sequences of the GmADTs, Arabidopsis ADTs (AtADT), Petunia hybrida (PhADTs) and Pinus pinaster (PpADTs) were aligned using ClustalO. Black shading indicates identical residues, grey shading indicates similar amino acid residues and dashed lines indicate gaps. Colored bars above the sequences indicate predicted protein domains color coded as shown on top. Green boxe indicate TRF motif. Conserved A (A314 in PpADT) residue conferring PDT activity is shown by the arrow head within the PAC domain (black line). Yellow boxes indicate GALV and ESRP domains.
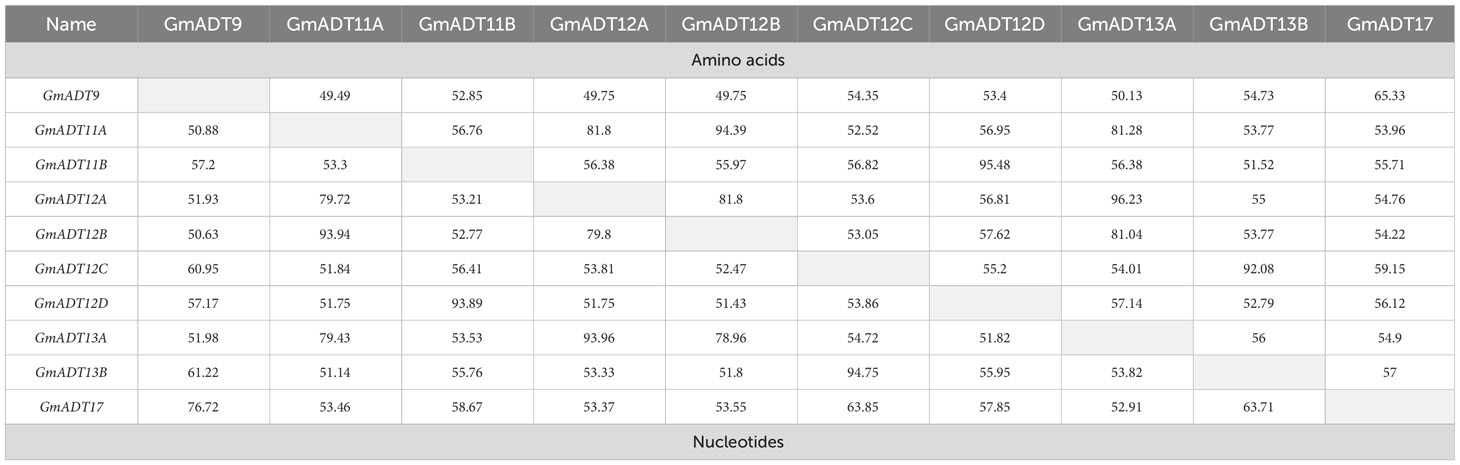
Table 2 Pairwise coding region and amino acid sequence comparisons of the soybean GmADT gene family.
Originaly ADTs were thought to be monofunctional enzymes like bacterial PDTs. However it has been demonstrated that some ADT isozymes can also act as PDTs (Cho et al., 2007; Maeda et al., 2010; Bross et al., 2011; El-Azaz et al., 2016; Qian et al., 2019). ADT and PDT enzymes catalyze the same biochemical reaction (decarboxylation/dehydration) using very similar substrates, and many key catalytic and regulatory residues are conserved between them (Cho et al., 2007; Tan et al., 2008). Previous work to determine the sequences responsible for this dual substrate recognition in Pinus pinaster ADTs (PpADTs) identified a 22 amino acid region at the N-terminus of the ACT domain, termed the PDT activity conferring domain, or PAC domain (El-Azaz et al., 2016). Additionally, a critical alanine residue (Ala314 in PpADT-G) was to be sufficient to define prephenate substrate recognition in the PpADTs. Five GmADTs contain this alanine residue within the PAC domain: GmADT9, GmADT11B, GmADT12C, GmADT12D and GmADT17 (Figure 3), suggesting these isoforms are ADT/PDTs.
An analysis of GmADT gene structure indicated that four GmADTs (GmADT11A, GmADT12A, GmADT12B and GmADT13A) contained no introns in their open reading frame while six contained multiple introns (Figure 4A). GmADT9, GmADT11B, GmADT12C and GmADT17 contained 10 introns of varying sizes ranging from 81 to 1444 nucleotides while GmADT12D and GmADT13B contained eight and five introns, respectively. Among all the GmADT family members, only GmADT11B and GmADT12D contained introns in its 3`UTR.
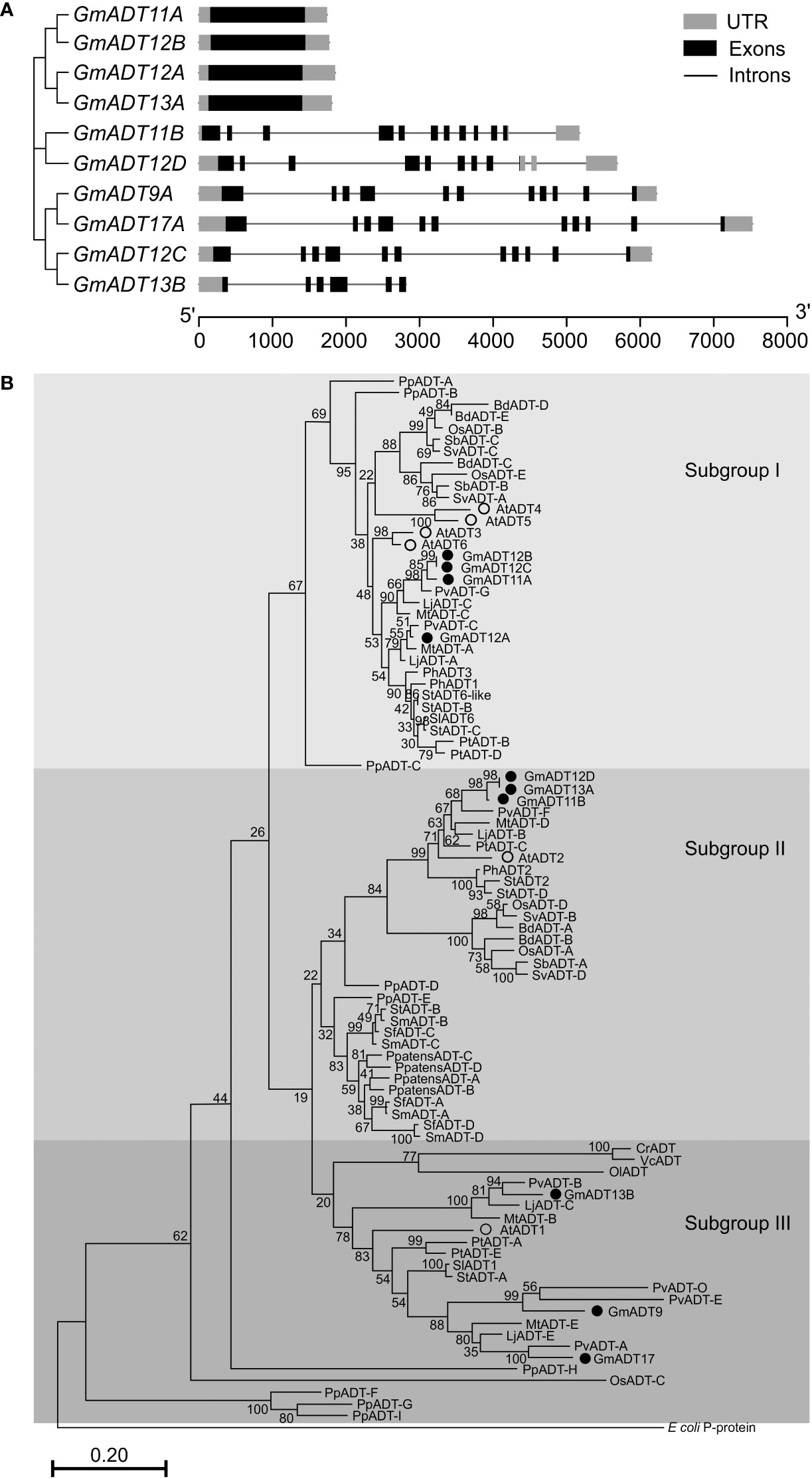
Figure 4 Gene structure and phylogenetic analysis of GmADTs. (A) Genomic structure of GmADTs were drawn to scale using gene models of GmADTs, retrieved from Phytozome 13 for soybean (G. max Wm82.a4.v1) genome and the gene cluster was imported from a neighbor-joining tree of GmADTs. (B) Mature protein sequences of GmADTs, AtADTs, PpADTs and PhADTs were identified using TargetP, aligned using ClustalΟ and a phylogenetic tree was constructed by neighbor-joining method using MEGAX. Bootstrap values (1000 replicates) are shown as percentages next to branch points. The PDT domain of the E. coli P-protein was included as an outgroup. At, Arabidopsis thaliana; Pp, Pinus pinaster; Ph, Petunia hybrida. Accession numbers: AtADT1, AT1G11790.1; AtADT2, AT3G07630.1; AtADT3, AT2G27820.1; AtADT4, AT3G44720.1; AtADT5, AT5G22630.1; AtADT6, AT1G08250.1; PhADT1, ACY79502.1; PhADT2, ACY79503.1; PhADT3, ACY79504.1; PpADTA, APA32582.1; PpADTB, APA32583.1; PpADTC, APA32584.1; PpADTD, APA32585.1; PpADTE, APA32586.1; PpADTF, APA32587.1; PpADTG, APA32588.1; PpADTH, APA32589.1; PpADTI, APA32590.1; E. coli, WP_115444483.1.
To illustrate the evolutionary relationship among soybean ADTs and other characterized ADT proteins, a phylogenetic analysis was performed using the predicted amino acid sequences of their mature proteins. As shown in Figure 4B, similar to AtADTs (Cho et al., 2007), soybean ADTs form three distinct subgroups. GmADT9, GmADT13B, and GmADT17 grouped in ‘subgroup III’ with AtADT1. Three GmADTs, GmADT11B, GmADT12D, and GmADT13A grouped into ‘subgroup II’ with AtADT2 and PhADT2 while four GmADTs, GmADT11A, GmADT12A, GmADT12B and GmADT12C, grouped in ‘Subgroup I’ with AtADT3, AtADT4, AtADT5 and AtADT6. Similar to other angiosperms, at least one GmADT isoform is present in each subgroup. In addition, GmADTs in each subgroup are clustered with ADT sequences with other legume species (Medicago truncatula, Lotus japonicus, and Phaseolus vulgaris). As observed for most of the gene families in soybean, GmADTs mostly group in pairs, consistent with its recent whole genome duplication (Schmutz et al., 2010; Yuan and Song, 2023). Subgroups II and III contain other plant ADTs that have been shown to act as PDTs (eg. AtADT1, AtADT2, PpADT-G, PhADT2).
To determine the mRNA expression patterns of the GmADT gene family members in soybean tissues, we utilized the transcriptome dataset available in the public domain as a resource (Wang et al., 2019). As shown in Figure 5, the dataset obtained from Phytozome 13 database consisted of transcript accumulation in root, nodules, stem, leaf, flowers (open and unopen) and seed tissues collected during the development from early to mature seeds. The maximum fragments per kilobase of transcript per million mapped reads (FPKM) values of GmADTs varied from 0.5 (GmADT9 in early seed development) to 36.1 (GmADT12C in flowers). While the majority of GmADTs were expressed in most of the tissue analyzed, each gene family member displayed a unique tissue-specific expression pattern. GmADT12A and GmADT13A transcripts accumulated to highest levels in root tissues while the transcripts of only GmADT12D and GmADT17 were found in nodules. Transcript accumulation for GmADT9, GmADT11B, GmADT12A, GmADT12D and GmADT17 was higher during early seed development whereas expression of GmADT13B was detected in leaf, flower and seed tissue at mid-maturation stage. Despite lacking the transit peptide and the ACT domain, the predicted GmADT13B transcript levels were detectable, thus potentially producing only a truncated protein (Figure 3; Table 1). Therefore, it is likely that GmADT13B is non-functional as ADT/PDT and was excluded from further characterization in this study.
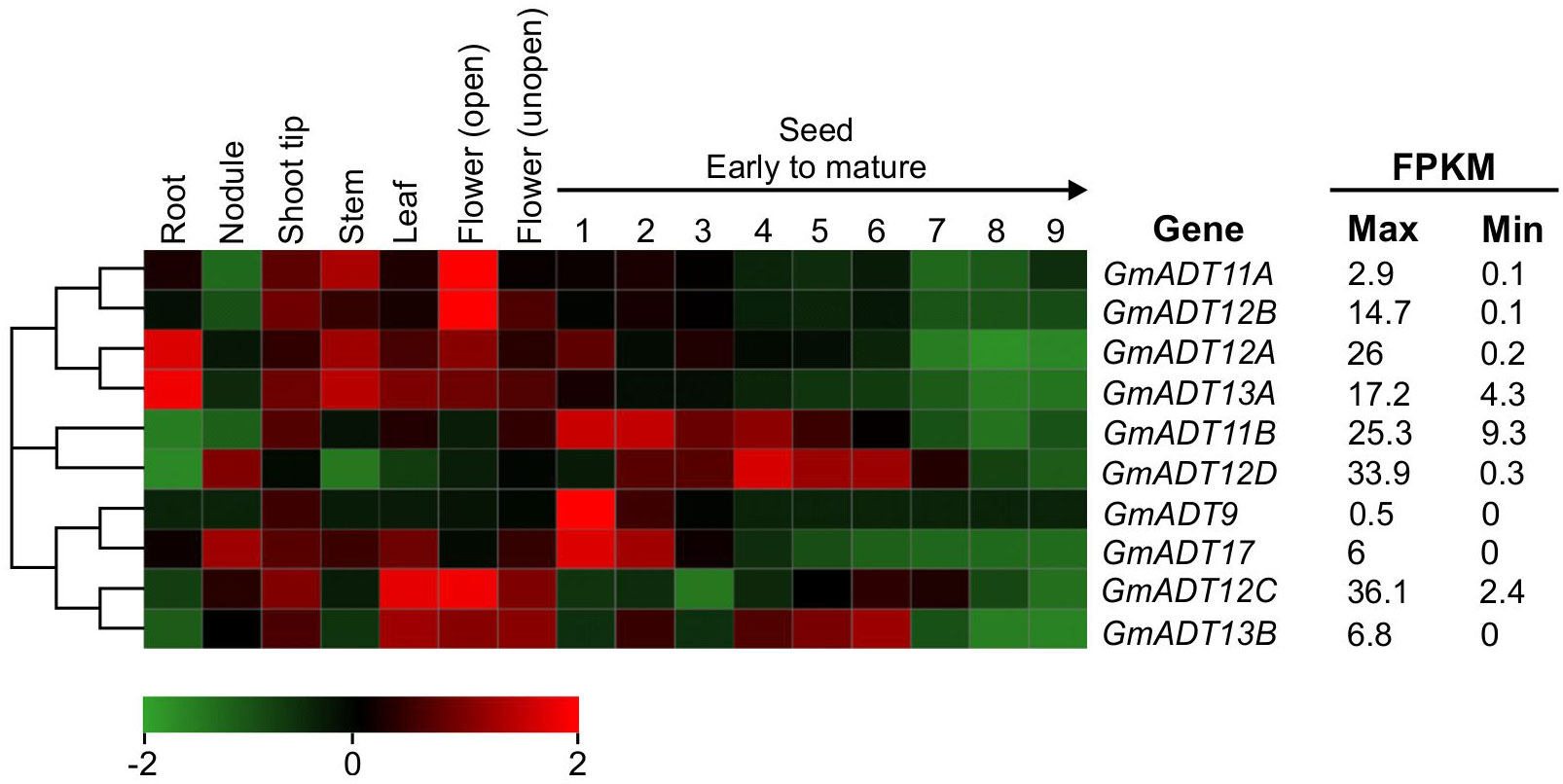
Figure 5 mRNA expression analysis of GmADTs. The transcriptome data of GmADT across different tissues were retrieved from Phytozome 13 database (Wang et al., 2019) for heatmap generation. The maximum (max) and minimum (min) expression values for each gene are shown and the gene cluster was imported from neighbor-joining tree generated with GmADT protein sequences in MEGAX. The black arrow on top of 1 to 9 indicates seed developmental stages from early (1) to mature (9). The color scale below the heatmap indicates transcript abundance values in log2 transformed across each row. Red and green indicating high and low levels of transcript abundance, respectively. FPKM, fragments per kilobase of transcript per million.
Plant ADTs were previously reported to localize to the chloroplasts in multiple species including Arabidopsis, petunia and pine (Rippert et al., 2009; Maeda et al., 2010; El-Azaz et al., 2016; Bross et al., 2017). To confirm if the GmADTs also are chloroplast localized, we investigated the subcellular localization of the nine GmADTs that were initially annotated in Phytozome as full length ADTs (as predicted truncated protein, GmADT13B was excluded from further analysis). GmADTs were translationally fused to YFP, transiently expressed in N. benthamiana leaves, and visualized by confocal microscopy. The red auto-fluorescence generated by chlorophyll was used as a chloroplast marker. As shown in Figure 6, all nine GmADTs showed chloroplastic localization, specifically a stromule sub-plastidial localization. Stromules resemble tail-like or bead-like protrusions from the chloroplast body (Arimura et al., 2001; Pyke and Howells, 2002; Kwok and Hanson, 2004b; Hanson and Hines, 2018). This stromule sub-plastidial localization of the GmADTs is consistent with the reports for other plant ADTs (Maeda et al., 2010; El-Azaz et al., 2016; Bross et al., 2017). Furthermore, GmADT11B displayed additional thin, elongated sub-plastidial localization at the poles of the chloroplasts (Figure 6, inset), distinct from the other GmADTs but similar to those observed for AtADT2, an AtADT likely involved in chloroplast division (Abolhassani Rad, 2017).
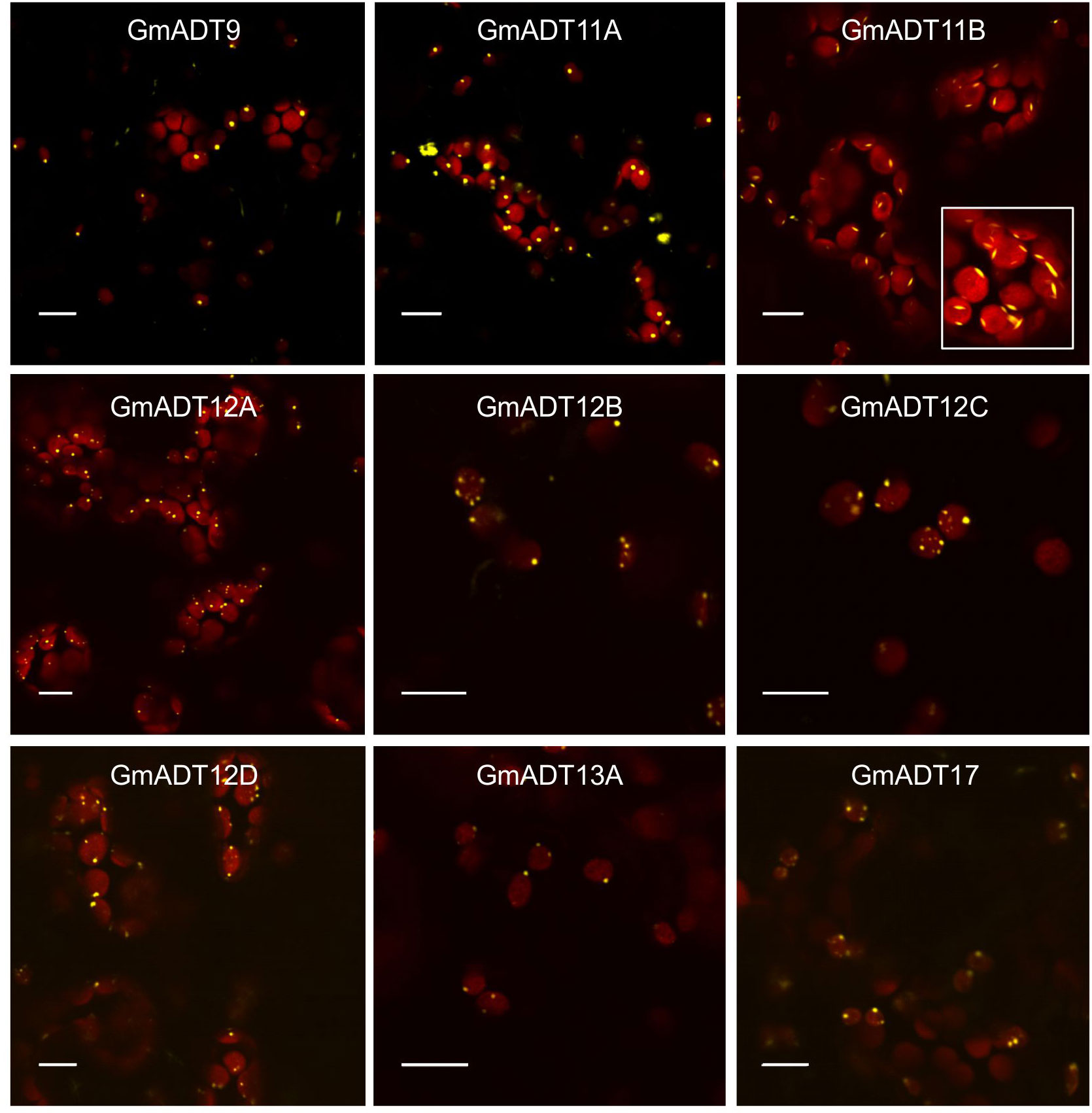
Figure 6 Subcellular localization of the GmADTs. A translational fusions of GmADT-YFP were transiently expressed in N. benthamiana leaf and visualized by confocal microscopy. Confirmation of localization was performed through co-localization of the GmADT-YFP fusion with the chloroplast autofluorescence (in red). Scale bars represent 10 µM.
To determine if the GmADT and GmIFS2 interaction detected via Co-IP is actually occurring in-planta, we conducted Bimolecular Fluorescence Complementation (BiFC) assay to assess protein-protein interactions. In our BiFC system we used N. benthamiana, which synthesizes flavonoids but not isoflavonoids. However, we are introducing the key branch enzyme leading to isoflavonoid synthesis, GmISF2. The flavonoid synthesis machinery upstream of IFS2 is the same in both legumes and non legumes, allowing us to use the N. benthamiana transient expression system to assess GmADT interactions with GmIFS2. As we were expressing soybean genes in a heterologous system, all contructs were expessed using a 35S promoter to ensure comparable expression levels (Odell et al., 1985; Lu et al., 2010; Tsugama et al., 2013; Amack and Antunes, 2020; Liang et al., 2023).
Each GmADT was translationally fused to the C-terminal half of YFP (GmADT-YC) and GmIFS2 to the N-terminal half of YFP (GmIFS-YN). GmADT-YC and GmIFS2-YN constructs were co-expressed in N. benthamiana leaves and the protein-protein interaction was monitored using confocal microscopy (Figure 7). Co-expression of GmADT and GmIFS2 constructs were also performed in reciprocal combination (GmADT-YN and GmIFS2-YC (Supplementary Figure 1). As shown in Figure 7, the detection of YFP fluorescence indicating either close proximity or direct interaction between each of the GmADT family members and GmIFS2 was confirmed. The observed reticulate pattern of fluorescence indicated that the interaction was occurring at the ER surface where GmIFS2 is localized. We also detected an interaction between GmIFS2 and an Arabidopsis ADT, AtADT5. Similar results were obtained for the reciprocal combinations (Supplementary Figure 1). As negative controls, we also tested GmIFS2 with proteins unrelated to isoflavonoid synthesis and specialized metabolism: a protoanthocyanidin transporter protein from common bean (Figure 7, PvMATE8) and an Arabidopsis seed storage protein CRUCIFERIN1 (Figure 7, AtCRA1). No signal was detected when co-expressing GmIFS2 with PvMATE8 or AtCRA1 (Figure 7).
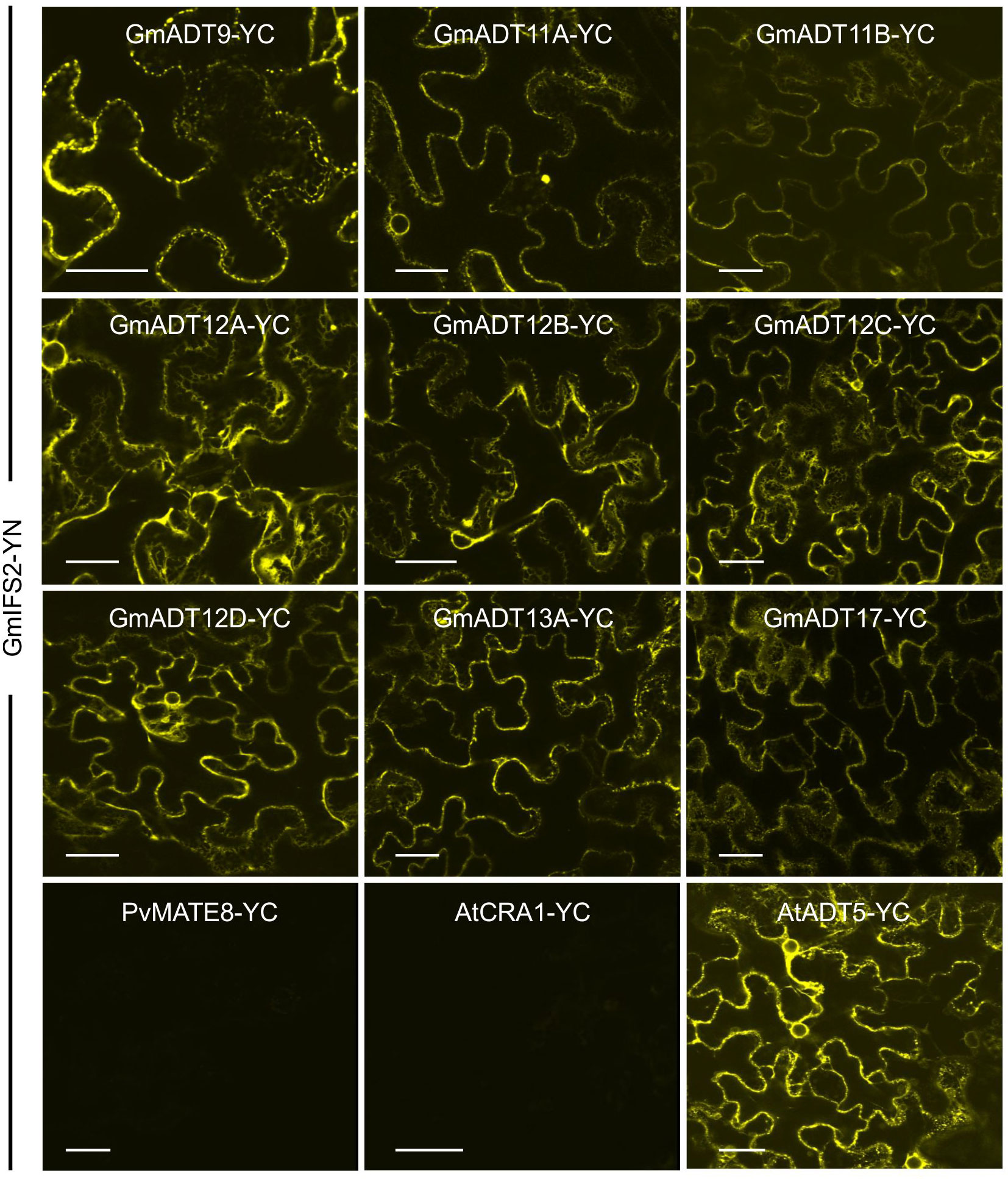
Figure 7 GmADTs and GmIFS2 interact in planta at the ER. Bi-directional interaction between GmADT isoforms and GmIFS2 by co-expression of translational fusions with N (YN)- or C (YC)- terminal fragments of YFP in N. benthamiana as assayed by BiFC. Proximity of the proteins results in a YFP fluorescence and localization of interaction as monitored by confocal microscopy. The interaction between chloroplast-localized GmADT and ER-localized GmIFS2 appear to be localized to the ER. As representative negative controls, GmMATE8-YC or AtCRA1-YC co-infiltrated with GmIFS2 (YN) is shown. Scale bars represent 30 µM.
To investigate if GmADTs interact with the soluble enzymes of the (iso)flavonoid pathways, interaction between GmADT12A and GmCHS8, GmCHR14, GmCHI2 were also assayed by BiFC (Figure 8). GmADT12A was chosen as a representative ADT based on its putative involvement in the isoflavonoid metabolon (Dastmalchi et al., 2016). We also evaluated the interaction between GmADT12A and two other isoflavonoid ER anchors, GmC4H2 and GmIFS1 (Dastmalchi et al., 2016; Khatri et al., 2023). The interaction of GmADT12A with GmC4H2 and GmIFS1 was restricted to the surface of the ER (Figures 8I,II). Interestingly, the interaction of GmADT12A with soluble nucleo-cytoplasmic-localized proteins GmCHS8, GmCHR14 and GmCHI2 was detected in both the nucleus and cytosol (Figures 8III-V). No interaction was observed between GmADT12A and GmCYP1, AtCRA1 or PvMATE8 (Figures 8VI–VIII). No discrepancies were observed when testing the reciprocal combinations (Supplementary Figure 2).
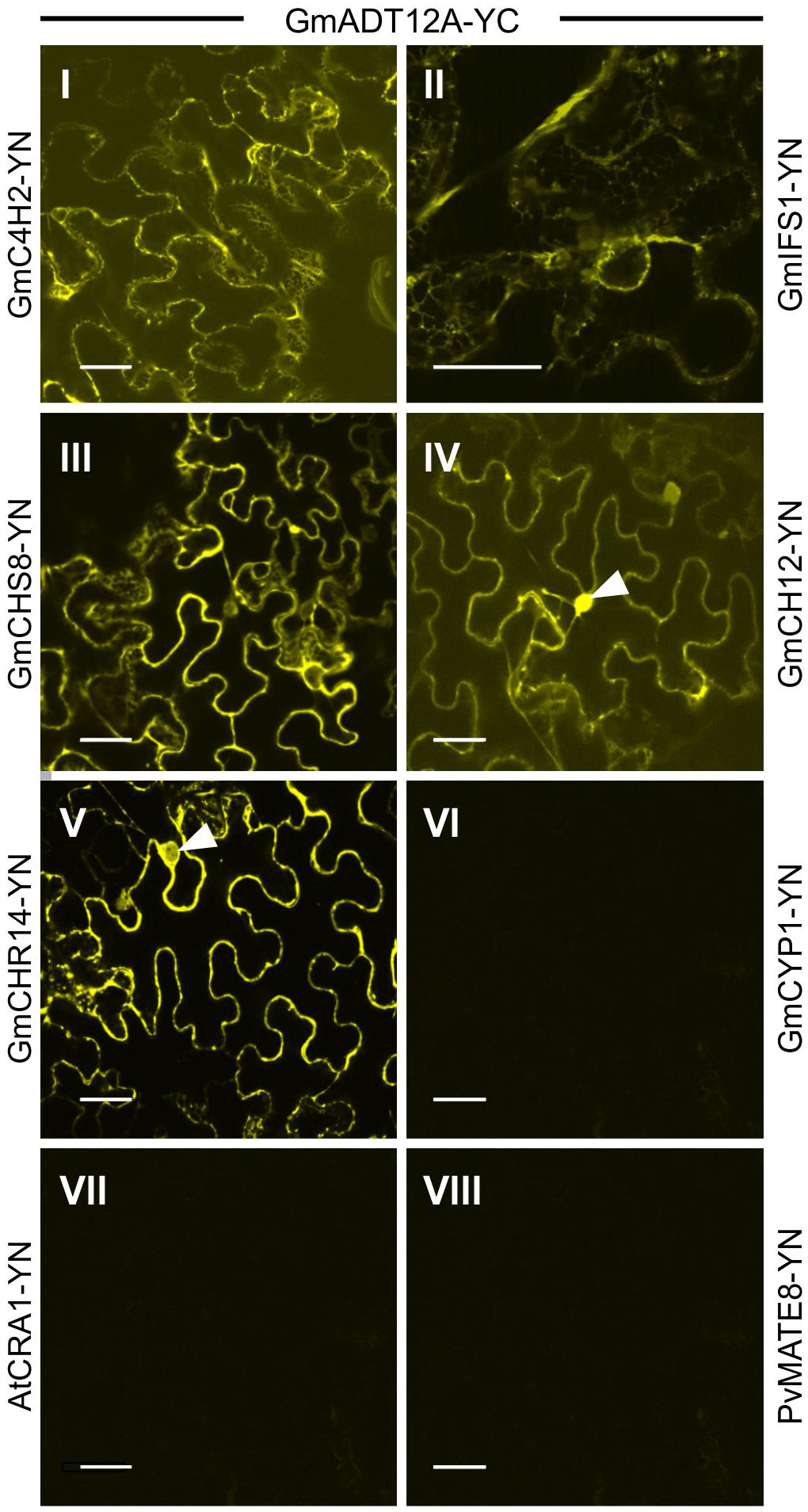
Figure 8 Interaction of GmADT12A with soluble and ER-localized isoflavonoid biosynthetic pathway enzymes by co-expression of translational fusions with N (YN)- or C (YC)- terminal fragments of YFP in N. benthamiana as assayed by BiFC. Proximity of the proteins results in a YFP fluorescence and localization of interaction as monitored by confocal microscopy. (I) and (II) The interaction between chloroplast localized GmADT12A and ER localized GmC4H2 and GmIFS1 appear to be localized to the ER. (III), (IV) and (V) GmADT12A and soluble enzymes GmCHS8, GmCHR14 and GmCHI2 appear to be localized to the cytoplasm and nucleus. White arrow heads point to nuclei. (VI), (VII) and (VIII) GmCYP1 is a nucleo-cytosolic protein (Mainali et al., 2017). No interaction was observed between GmADT12A and GmCYP1 or AtAtCRA1 or PvMATE8 (negative control). Scale bars represent 30 µM.
Sequence analysis suggested that some members of the GmADT family may have PDT activity, specifically GmADT9, GmADT11B, GmADT12C, GmADT12D and GmADT17 (Figure 3). To assess PDT activity of the GmADTs, the pha2 yeast complementation assay (Bross et al., 2011) was performed. The S. cerevisiae pha2 strain lacks the endogenous PDT protein, and can be complemented by a GmADT with PDT activity. Further, the pha2 assay is inducible by galactose. Thus, only in the prescence of galactose GmADTs are expressed, and any GmADT with PDT activity will synthesize Phe and cells will grow without Phe supplementation.
The untransformed pha2 strain and pha2 transformed with an empty vector were used as negative controls, and they do not complement the strain (Figure 9, pha2 and empty vector). As a positive control, AtADT2 was used as it was previously shown to have PDT activity (Cho et al., 2007; Bross et al., 2011). As expected, no growth was observed when cells were grown with glucose or raffinose as carbon sources. When induced with galactose, GmADT9, GmADT11A, GmADT11B, GmADT12A and GmADT17 complemented the pha2 strain, indicating these GmADT isoforms have PDT activity (Figure 9).

Figure 9 pha2 complementation analysis of GmADTs. GmADTs were expressed in the yeast pha2 strain and spotted onto SD media supplemented with either glucose (Glu), raffinose (Raf) or galactose (Gal). Media with each carbon source were also prepared either with or without phenylalanine (Phe). S. cerevisiae pha2 strains transformed with GmADT were diluted to a final density of 5 x 104 cells/mL and then equal volume of each was plated. Shown are representative growth spots, with images taken after 13 days of incubation. As negative controls, untransformed pha2 strain and the pha2 strain transformed with the empty expression vector were included. AtADT2 was used as a positive control. GmADT9, GmADT11A, GmADT11B, GmADT12A and GmADT17 complemented the pha2 strain, demonstrating these GmADT enzymes possess PDT activity.
Next, Western blots were performed to confirm that all 6×His-tagged GmADTs were expressed in the pha2 yeast strain (Supplementary Figure 3). Total soluble protein was extracted (Kushnirov, 2000), and equal volumes of extract were analyzed by SDS-PAGE. Coomassie staining was performed to ensure protein was successfully extracted (Supplementary Figure 3), and purified 6×His-GmCHR14 was used as a positive control. Signals from GmADT9, GmADT11A, GmADT11B, GmADT12A and GmADT13A were observed, however the remaining GmADTs (GmADT12B, GmADT12C, GmADT12D, and GmADT17) were not detected (Supplementary Figure 3). While GmADT17 appeared to be expressed sufficiently to complement the pha2 strain, we cannot definitively conclude that GmADT12B, GmADT12C, and GmADT12D lack PDT activity, as they do not appear to be expressed in yeast.
Plant specialized metabolism is a tightly regulated network of overlapping pathways with many participating enzymes, all being controlled temporally during development, in response to external stimuli, and spatially in different plant tissues and subcellular organelles. Many of the enzymes involved in these biosynthetic pathways are encoded by multi-gene families (Boudet, 2007), and can direct carbon flux toward specific metabolic pathways (Cannon et al., 2004; Corea et al., 2012b). In the present study, we identified nine members of the GmADT family, examined their tissue-specific expression, and subcellular localization. We determined that five of the nine GmADTs possess PDT activity in the in vivo pha2 complementation assay. We further showed that GmADTs interact with GmIFS2 in vivo. In particular, GmADT12A exhibits PDT activity, is expressed in root tissue and interacts with GmIFS2, substantiating its involvement in the isoflavonoid metabolon (Dastmalchi et al., 2016).
There have been multiple whole genome duplications (WGDs) in the evolutionary history of soybean. Aside from the ancient WGD shared by all angiosperms (Young and Bharti, 2012), soybean has undergone two more recent WGDs: one just prior to radiation of legumes (~58-59 million years ago), and one Glycine-specific WGD that ocurred ~13-14 million years ago (Shoemaker et al., 2006; Schmutz et al., 2010; Yuan and Song, 2023). As a result, soybean has many duplicated genes, including the GmADT family members. There is often a period of gene loss following WGDs leading to the pseudogenization and loss of the redundant genes, but neofunctionalization of duplicates can also occur (Lynch and Force, 2000; Young and Bharti, 2012). Our data demonstrate that most of the GmADTs we identified encode functional proteins, indicating the GmADT isoforms are not fully redundant. The GmADTs are expressed in all soybean tissues, however they have unique temporal and spatial expression patterns, suggesting they are differentially regulated, and could differentially contribute to phenylpropanoid and (iso)flavonoid biosynthesis. GmADT9 and GmADT17 have diverged the most in sequence, sharing only ~50-63% identity with the other GmADTs at the nucleotide level and protein levels. Generally, GmADT isoforms that share high sequence identity, such as GmADT11A and GmADT12B, had similar expression profiles reflecting their shared evolutionary history. Though GmADT11B and GmADT12D share 93% nucleotide sequence identity, their expression patterns differ in the nodules, stem and throughout seed development, suggesting differences in regulation of gene expression and perhaps different contributions to specialized metabolism between these closely related isoforms. ADT isoforms from Arabidopsis have also been shown to be differentially expressed and contribute uniquely to metabolism (Corea et al., 2012b; Chen et al., 2016; Muhammad et al., 2023). For example, GmADT11A, GmADT12A, GmADT12B, and GmADT13A exhibited high mRNA expression in stems (Figure 5), and these GmADTs group with AtADT4 and AtADT5 in subgroup I (Figure 4). Of the six isoforms of the AtADT family, AtADT4 and AtADT5 contributed greatly to lignin production in stem tissue (Corea et al., 2012a) suggesting a role in lignin production for GmADT11A, GmADT12A, GmADT12B, and GmADT13A. El-Azaz and colleagues (2018) demonstrated that AtADT2 expression was crucial for normal seed development in Arabidopsis. Further, the adt2 seed phenotype could be rescued by both ADT and PDT activity, by expressing either AtADT3 or PHA2, the PDT from S. cerevisiae (El-Azaz et al., 2018). GmADT11B and GmADT12D are the highest expressed soybean isoforms in seeds that also group with AtADT2 in subgroup II, indicating they may also have an important role in seed development. GmADT12D and GmADT17 are the most highly expressed in the nodules (Figure 5), suggesting these isoforms play an important role in the nodulation process. Taken together, the soybean GmADT have unique expression patterns that allows the isoforms to differentially contribute to Phe biosynthesis and ultimately specialized metabolism.
A consquence of WGDs is the alteration entire genetic networks, which can provide short-term advantages to biotic and abiotic stress tolerance (Van de Peer et al., 2017). In plants, many genes involved in stress responses and development are also present as large gene families (Cannon et al., 2004; Pan et al., 2022). In addition, the legume-specific WGD is suggested to have had a profound effect on nodulation and symbiosis (Young et al., 2011). Further, duplications in the flowering plant lineage has lead to a clade of allosterically deregulated ADT enzymes (referred to as type-II), and at least one member of this clade is present in most angiosperm genomes (El-Azaz et al., 2022). Type-I ADTs are tightly allosterically regulated by Phe, and include PpADT-G, AtADT1, GmADT9, GmADT11B and GmADT17. Type-II ADTs are much more loosely regulated, and include PpADT-C, AtADT4, and the remaining GmADT isoforms. These type-II isoforms are though to contribute to the accumulation of large quantities of Phe in the cell, which can potentially be shuttled toward specialized metabolite synthesis (El-Azaz et al., 2022; Muhammad et al., 2023). As isoflavonoids are a diverse class of compounds important for establishing symbioses with Rhizobia, a large family of differentially regulated and loosely allosterically regulated ADTs may allow more flexible control over (iso)flavonoid synthesis, in addition to some degree of functional redundancy conferred by large gene families (Kafri et al., 2009; El-Azaz et al., 2022; Muhammad et al., 2023).
Most ADT enzymes characterized to date are localized to the chloroplast, specifically to stromules (Rippert et al., 2009; Maeda et al., 2010; El-Azaz et al., 2016; Bross et al., 2017). However, some ADT isozymes localize to the nucleus or cytosol, for example, AtADT5 and AtADT6, respectively (Bross et al., 2017). We determined that the GmADTs were all localized to stromules. Since all nine GmADTs have an N-terminal chloroplast transit peptide, this suggests all GmADTs participate in phenylalanine biosynthesis via the well described chloroplast localized arogenate pathway. This is not surprising, as most plants described to date have a family of plastidic ADTs to convert arogenate to Phe (Jung et al., 1986; Ehlting et al., 2005; Cho et al., 2007; Rippert et al., 2009; Maeda and Dudareva, 2012; El-Azaz et al., 2016), and the GmADTs are similar in sequence to other characterized plant ADTs.
However, we observed interactions with GmIFS2 for all nine GmADTs that occur at the ER. In addition, GmADT12A also interacts both GmC4H2 and GmIFS1 at the ER membrane, GmCHS8 in the cytosol, and with GmCHI2 and GmCHR14 in the cytosol and the nucleus (Figure 8). The interaction of GmADT12A with GmCHI2 and GmCHR14 in the nucleus adds to the growing body of evidence that has detected (iso)flavonoid synthesis enzymes in the nucleus. Nucleo-cytoplasmic localizations have previously been reported for both GmCHI2 and GmCHR14 (Dastmalchi and Dhaubhadel, 2015; Sepiol et al., 2017). In addition, nuclear localization of CHI and CHS enzymes have also been reported in Arabidopsis and grapevine (Saslowsky et al., 2005; Wang et al., 2016; Watkinson et al., 2018; Wang et al., 2019). Localization of the (iso)flavonoid biosynthetic machinery in nucleus would allow direct deposition of these compounds in the nucleus (Saslowsky et al., 2005), however the role of (iso)flavonoids in the nucleus is not entirely clear. As flavonoids can bind DNA and have been shown to bind histones, regulation of gene expression by binding DNA associated proteins or DNA directly have been suggested as possible roles for these compounds in the nucleus (Hodek et al., 2002; Ramadass et al., 2003). Alternatively, the GmADTs and other (iso)flavonoid synthesis enzymes in the nucleus could have a secondary role there in regulating gene expression as retrograde signals (Isemer et al., 2012).
It is unlikely that the detected interaction is due to overpression of both GmIFS2 and the GmADTs, as no interaction was detected with PvMATE8 or AtCRA1 using the same detection system, indicating the interaction is specific to the GmADTs and GmIFS2. Furthermore, the same promoter was used for both detection of subcellular localization and protein-protein interactions, indicating the difference in GmADT localization is not soley due to overexpression using the 35S promoter. In addition, it has been shown that subcellular localization patterns detected in tobacco with a 35S promoter are indicative of what happens in the native organism. For example, AtADT5 nuclear localization was first determined in tobacco and was then confirmed when expressed under its native promoter in Arabidopsis (Bross et al., 2017), indicating the alternate nuclear localization was not an articact of expression in tobacco.
The current data substantiate our previous investigation of the isoflavonoid metabolon that was performed using the soybean hairy roots expressing GmIFS2-YFP, and GmADT12A and GmADT13A were pulled down in Co-IP analysis (Dastmalchi et al., 2016). Among the members of GmADT family, root-specific expression was only observed for GmADT12A and GmADT13A (Figure 5) which is consistent with only these two isoforms being identified by the Co-IP analysis. As the GmADTs are differentially expressed, isoflavonoid metabolon composition may vary depending on the GmADT isoform expressed in a given tissue. It has also been suggested that there could be more than one metabolon dedicated to the synthesis of different metabolites, given the availability of enzyme isoforms and their substrate preferences (Ehlting et al., 1999; Winkel, 2004; Li et al., 2015). One also needs to consider that in addition to tissue-specific expression of isoforms, metabolons are inherently transient and dynamic in nature. Consequently, there could be a high turnover of GmADT isoforms participating in the isoflavonoid metabolon, providing flexibility in metabolon assembly, substrate sequestering, metabolic channeling potential, and stress response (Winkel, 2002; Moller, 2010; Hawes et al., 2015).
Most plant ADT families have least one isoform with predicted or demonstrated PDT activity, suggesting that both the prephenate and arogenate pathways are active in plants. It is thought that the Phe is predominantly synthesized via the arogenate pathway in the plastids. However, cytosolic PhPPY-AT and PhCM isoforms have been characterized (Yoo et al., 2013; Qian et al., 2019), and a cytosolic prephenate pathway has been described in Petunia hybrida (Qian et al., 2019). The possible advantages associated with the prephenate pathway, or the degree to which the prephenate pathway contributes to Phe synthesis is still unclear.
We determined that five GmADT isoforms had detectable PDT activity using the pha2 complementation test: GmADT9, GmADT11A, GmADT11B, GmADT12A and GmADT17. As GmADT9 and GmADT17 fall within subgroup I, and GmADT11B in subgroup II in the ADT/PDT phylogenetic tree (Figure 4B), this result is not surprising. These three GmADT isoforms contain the critical Ala314 residue in the PAC domain that confers PDT activity to PpADT-B and PpADT-G (El-Azaz et al., 2016). Interestingly, GmADT11A and GmADT12A were also found to have PDT activity, but neither isoform contains the critical Ala314 residue. In fact, GmADT11A and GmADT12A both cluster within subgroup III, with isoforms from Arabidopsis and P. pinaster that do not have PDT activity. While some key resiudes for PDT activity have been identified in the PAC domain, it was noted that mutations in this domain had a variety of outcomes on PDT and ADT activity (El-Azaz et al., 2016; El-Azaz et al., 2022). It is possible that other unidentified amino acid residues within the PAC domain are important in defining PDT activity in soybean and other plant species. Further, by sequence alone, GmADT12C and GmADT12D were expected to act as PDTs. However, these two GmADT isoforms were among the GmADT proteins not well expressed in the pha2 strain. As such, they could still have PDT activity if assessed using another method. We expect all the GmADTs to act as ADTs, given the sequence similarity to other plant ADTs, though we did not characterize ADT activity here. Determining the kinetics of the GmADTs with both arogenate and prephenate as a substrate should also be investigated in future, as it has been demonstrated that the Arabidopsis and Petunia ADT/PDTs prefer arogenate over prephenate as a substrate (Cho et al., 2007; Maeda et al., 2010). The proportion of phenylalanine synthesized via either pathway could also be tissue-specific, depending on whether the ADT isoform expressed in a given tissue has PDT activity.
As the cytoplasmic isoflavonoid metabolon assembles on the cytosolic face of the ER, we initially hypothesized that participating GmADTs are cytosolic, but no cytosolic GmADT isoforms were observed in this study when expressed alone. This begs the question, how can plastid-localized enzymes interact with a cytosolic metabolon? One possibility is that some of the GmADTs contain alternative transcriptional start sites, as seen for PhADT3 (Qian et al., 2019), resulting in ADTs lacking a transit peptide that localize in the cytosoplasm (Figure 10A). Having transcript variants, with or without transit peptides, could allow differential contribution in response to various stress conditions or signaling cascades like nodulation induction. Alternatively, the full-length GmADTs could also associate with the metabolon in the cytosol prior to their transit to the chloroplast.
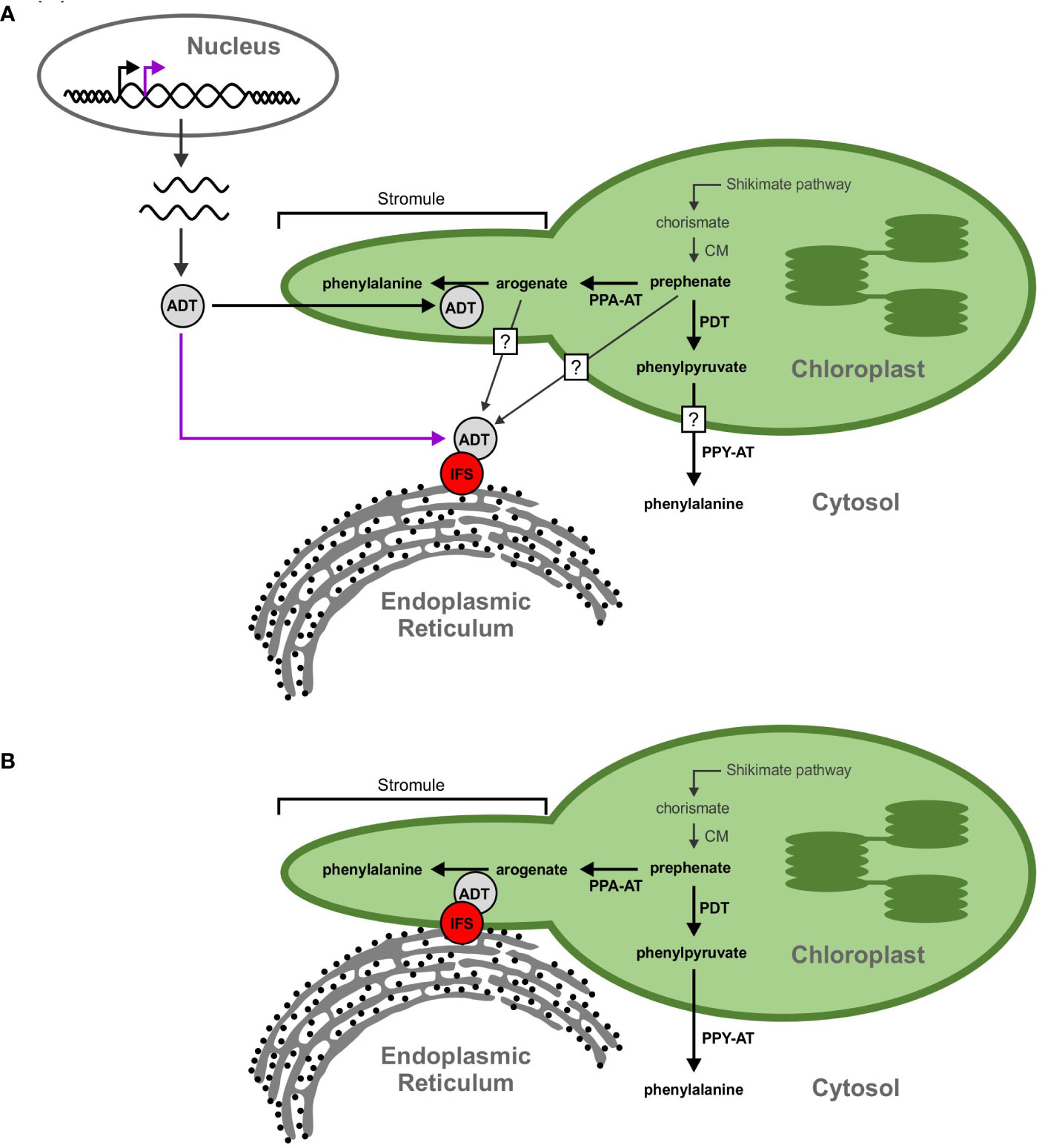
Figure 10 Proposed models for the GmADT-GmIFS2 interaction. (A) Full length ADT proteins contain an N-terminal transit peptide that directs them to the chloroplast (black arrow). However, alternate transcriptional start sites could result in a protein that lacks the transit peptide and remains in the cytosol (purple arrow). The cytosolic ADT protein would be available to interact with IFS and participate in the isoflavonoid metabolon. Substrates (prephenate, arogenate, phenylpyruvate, etc) could be transported out of the chloroplast, or they could passively diffuse across the chloroplast membrane, facilitated by the larger surface area of stromules. Prephenate could also be synthesized directly in the cytosol by cytosolic isoforms of CM. (B) Transorganelle continuity is another model allowing to explain the interaction between ADT and IFS proteins. The ER is a dynamic organelle that has been shown to interact with other organelles, namely the mitochondria and nucleus. Interactions between the ER and chloroplast membranes could bring the isoflavonoid metabolon and ADTs in close proximity.
Another explanation for the involvement of GmADTs in the isoflavonoid metabolon is trans-organelle continuity (Figure 10B). The ER is a dynamic organelle (Allen and Brown, 1988; Bayer et al., 2017; Kriechbaumer and Brandizzi, 2020), and there is evidence that the ER can associate with various organelles in the cell, including the chloroplast (Schattat et al., 2012; Phillips and Voeltz, 2016; Barton et al., 2018). It has been demonstrated in mammalian and plant cells that the ER forms contacts with peroxisomes, mitochondria, the plasma membrane and plasmodesmata (Hawes et al., 2015; Bayer et al., 2017; Perico and Sparkes, 2018; Li et al., 2022). The ER has also been shown to be involved in nuclear envelope formation and reshaping during cell division, and may facilitate protein localization to the inner nuclear membrane (Anderson and Hetzer, 2007; Blackstone, 2017; Ungricht and Kutay, 2017).
Multiple contact sites have been demonstrated between the ER membrane and chloroplasts (Griffing, 2011; Block and Jouhet, 2015), and it has even been suggested that the ER and chloroplast membranes can become contiguous (Whatley et al., 1991). ER morphology changes have also been shown to correlate with the growth and retraction of stromules (Schattat et al., 2011). Stromules have been reported to expedite the transport and/or diffusion of small molecules and proteinsbetween the plastid and surrounding cellular compartments, and have been shown to be involved in various signaling and defense responses (Gray et al., 2001; Hanson and Hines, 2018; Perico and Sparkes, 2018), as well as plastid-to-plastid contact sites, and possible contacts with other organelles (Kwok and Hanson, 2004a; Erickson et al., 2017). It has also been demonstrated that ER lumen localized enzymes have access to tocopherol substrates at the inner plastid membrane (Mehrshahi et al., 2013), making trans-orgnalle continuity a compelling argument for GmADT access to the (iso)flavonoid metabolon. Stromule localization of GmADT proteins places them in a dynamic sub-plastidial environment, facilitating interactions with the ER, and subsequent association with the (iso)flavonoid metabolon. GmADT involvement in the (iso)flavonoid metablon would directly link flux toward Phe biosynthesis to the synthesis of specialized metabolites.
Organelle interactions offer a variety of advantages. Substrates do not need to be exported from organelles and diffused across the cytosol. Required substrates and intermediates are sequestered, limiting byproduct formation and increasing pathway efficiency (Moller, 2010; Zhang and Fernie, 2021). Regulating organelle contacts could also provide another avenue to regulate flux through pathways, in addition to transcriptional and translational regulation of enzymatic components.
We have identified nine GmADT family members and demonstrated their interaction with GmIFS2 and other (iso)flavonoid metabolon enzymes using an N. benthamiana BiFC expression system. While we previously reported the interaction of GmADT12A and GmADT13A with GmIFS2 in soybean roots, confirmation of the interaction between GmIFS2 and the other GmADTs in soybean tissue is still required. In addition, we have demonstrated that at least five of the nine GmADTs have PDT activity by yeast complementation analysis in the pha2 strain. However, there are still questions surrounding the GmADT family members and their involvement in (iso)flavonoid synthesis. In this work, we did not assess the ADT activity of the soybean ADTs. While they appear to be homologous to other plant ADTs, and share many of the sequences defining ADT activity and regulation, quantifying the ADT activity of the GmADT family is still necessary. Further, the factors influencing flux between the arogenate pathway and the prephenate pathway, and how GmADTs contribute phenylalanine to the (iso)flavonoid metabolon are yet unknown. Finally, determining the extent to which tissue-specific expression influences the (iso)flavonoid metabolon and ADT isoform involvement would provide insights into the differential regulation and diverging functions of gene family members.
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
EC: Data curation, Investigation, Formal analysis, Methodology, Writing – original draft. NI: Data curation, Methodology, Software, Validation, Writing – review & editing. KP: Writing – review & editing, Investigation. KK: Investigation, Writing – review & editing, Validation. RS: Investigation, Writing – review & editing. SK: Writing – review & editing, Supervision. SD: Supervision, Writing – review & editing, Conceptualization, Data curation, Funding acquisition, Investigation, Project administration, Resources, Validation, Visualization.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This research was supported by Agriculture and Agri-Food Canada’s Genomics Research and Development Initiative Abase grants (J-001826) and a Natural Sciences and Engineering Research Council of Canada’s Discovery Grant (385922-2011, 044661-2018 RGPIN) to SD.
The authors thank Ling Chen and Alex Molnar for technical assistance.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2024.1307489/full#supplementary-material
Supplementary Figure 1 | GmADT and GmIFS2 interact in planta at the ER. Reciprocal combination of GmADT isoforms and GmIFS2 (as shown in )and their interaction by co-expression of translational fusions with N (YN)- or C (YC)- terminal fragments of YFP in N. benthamiana as assayed by BiFC.
Supplementary Figure 2 | GmADT12A-YN and isoflavonoid biosynthetic enzymes-YC interaction (reciprocal combination of ).
Supplementary Figure 3 | Western blot detection of GmADTs in S. cerevisiae pha2. (A) GmADTs- his were expressed in the yeast pha2 strain and detected by western blotting analysis using anti-his antibody. (B) Coomassie stained gel of total protein under non-inducing (glucose) condition. (C) Coomassie stained gel of total protein under inducing (galactose) condition.
Abolhassani Rad, S. (2017). The mystery of nuclear localization of AROGENATE DEHYDRATASE 5 from Arabidopsis thaliana (London, ON: The University of Western Ontario).
Ackerman, S. H., Martin, J., Tzagoloff, A. (1992). Characterization of ATP11 and detection of the encoded protein in mitochondria of Saccharomyces cerevisiae. J. Biol. Chem. 267, 7386–7394. doi: 10.1016/S0021-9258(18)42529-X
Allen, N. S., Brown, D. T. (1988). Dynamics of the endoplasmic reticulum in living onion epidermal cells in relation to microtubules, microfilaments, and intracellular particle movement. Cell Motil. 10, 153–163. doi: 10.1002/cm.970100120
Amack, S. C., Antunes, M. S. (2020). CaMV35S promoter – A plant biology and biotechnology workhorse in the era of synthetic biology. Curr. Plant Biol. 24, 100179. doi: 10.1016/j.cpb.2020.100179
Anderson, D., Hetzer, M. (2007). Nuclear envelope formation by chromatin-mediated reorganization of the endoplasmic reticulum. Nat. Cell Biol. 9, 1160–1166. doi: 10.1038/ncb1636
Arimura, S., Hirai, A., Tsutsumi, N. (2001). Numerous and highly developed tubular projections from plastids observed in Tobacco epidermal cells. Plant Sci.: an Int. J. Exp. Plant Biol. 160, 449–454. doi: 10.1016/s0168-9452(00)00405-2
Barton, K. A., Wozny, M. R., Mathur, N., Jaipargas, E. A., Mathur, J. (2018). Chloroplast behaviour and interactions with other organelles in Arabidopsis thaliana pavement cells. J. Cell Sci. 131, jcs202275. doi: 10.1242/jcs.202275
Bayer, E. M., Sparkes, I. A., Vanneste, S., Rosado, A. (2017). From shaping organelles to signalling platforms: the emerging functions of plant ER–PM contact sites. Curr. Opin. Plant Biol. 40, 89–96. doi: 10.1016/j.pbi.2017.08.006
Bhat, S. S., Prasad, S. K., Shivamallu, C., Prasad, K. S., Syed, A., Reddy, P., et al. (2021). Genistein: A potent anti-breast cancer agent. Curr. Issues Mol. Biol. 43, 1502–1517. doi: 10.3390/cimb43030106
Blackstone, C. (2017). Protein targeting: ER leads the way to the inner nuclear envelope. Curr. Biol. 27, 1284–1286. doi: 10.1016/j.cub.2017.10.037
Block, M. A., Jouhet, J. (2015). Lipid trafficking at endoplasmic reticulum-chloroplast membrane contact sites. Curr. Opin. Cell Biol. 35, 21–29. doi: 10.1016/j.ceb.2015.03.004
Boudet, A. M. (2007). Evolution and current status of research in phenolic compounds. Phytochemistry 68, 2722–2735. doi: 10.1016/j.phytochem.2007.06.012
Bradley, C. A., Allen, T. W., Sisson, A. J., Bergstrom, G. C., Bissonnette, K. M., Bond, J., et al. (2021). Soybean yield loss estimates due to diseases in the United States and Ontario, Canada, from 2015 to 2019. Plant Health Prog. 22, 483–495. doi: 10.1094/PHP-01-21-0013-RS
Bross, C. D., Corea, O. R., Kaldis, A., Menassa, R., Bernards, M. A., Kohalmi, S. E. (2011). Complementation of the pha2 yeast mutant suggests functional differences for arogenate dehydratases from Arabidopsis thaliana. Plant Physiol. Biochem. 49, 882–890. doi: 10.1016/j.plaphy.2011.02.010
Bross, C. D., Howes, T. R., Abolhassani Rad, S., Kljakic, O., Kohalmi, S. E. (2017). Subcellular localization of Arabidopsis arogenate dehydratases suggests novel and non-enzymatic roles. J. Exp. Bot. 68, 1425–1440. doi: 10.1093/jxb/erx024
Cannon, S. B., Mitra, A., Baumgarten, A., Young, N. D., May, G. (2004). The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 4, 10. doi: 10.1186/1471-2229-4-10
Cederroth, C. R., Nef, S. (2009). Soy, phytoestrogens and metabolism: A review. Mol. Cell Endocrinol. 304, 30–42. doi: 10.1016/j.mce.2009.02.027
Chandra, S., Choudhary, M., Bagaria, P. K., Nataraj, V., Kumawat, G., Choudhary, J. R., et al. (2022). Progress and prospectus in genetics and genomics of Phytophthora root and stem rot resistance in soybean (Glycine max L.). Front. Genet. Nov 14, 13. doi: 10.3389/fgene.2022.939182
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chen, Q., Man, C., Li, D., Tan, H., Xie, Y., Huang, J. (2016). Arogenate dehydratase isoforms differentially regulate anthocyanin biosynthesis in Arabidopsis thaliana. Mol. Plant 9, 1609–1619. doi: 10.1016/j.molp.2016.09.010
Cho, M.-H., Corea, O. R. A., Yang, H., Bedgar, D. L., Laskar, D. D., Anterola, A. M., et al. (2007). Phenylalanine biosynthesis in Arabidopsis thaliana. Identification and characterization of arogenate dehydratases. J. Biol. Chem. 282, 30827–30835. doi: 10.1074/jbc.M702662200
Corea, O. R., Bedgar, D. L., Davin, L. B., Lewis, N. G. (2012a). The arogenate dehydratase gene family: towards understanding differential regulation of carbon flux through phenylalanine into primary versus secondary metabolic pathways. Phytochemistry 82, 22–37. doi: 10.1016/j.phytochem.2012.05.026
Corea, O. R. A., Ki, C., Cardenas, C. L., Kim, S. J., Brewer, S. E., Patten, A. M., et al. (2012b). Arogenate dehydratase isoenzymes profoundly and differentially modulate carbon flux into lignins. J. Biol. Chem. 287, 11446–11459. doi: 10.1074/jbc.M111.322164
Cotton, R. G. H., Gibson, F. (1965). Biosynthesis of phenylalanine and tyrosine - enzymes converting chorismic acid into prephenic acid and their relationships to prephenate dehydratase and prephenate dehydrogenase. Biochim. Biophys. Acta 100, 76–88. doi: 10.1016/0304-4165(65)90429-0
Crosby, K. C., Pietraszewska-Bogiel, A., Gadella, T. W. J., Winkel, B. S. J. (2011). Forster resonance energy transfer demonstrates a flavonoid metabolon in living plant cells that displays competitive interactions between enzymes. FEBS Lett. 585, 2193–2198. doi: 10.1016/j.febslet.2011.05.066
Dastmalchi, M., Bernards, M. A., Dhaubhadel, S. (2016). Twin anchors of the soybean isoflavonoid metabolon: evidence for tethering of the complex to the endoplasmic reticulum by IFS and C4H. Plant J. 85, 689–706. doi: 10.1111/tpj.13137
Dastmalchi, M., Dhaubhadel, S. (2014). “Soybean seed isoflavonoids: biosynthesis and regulation,” in Phytochemicals – Biosynthesis, Function and Application (Cham: Springer International Publishing), 1–21.
Dastmalchi, M., Dhaubhadel, S. (2015). Soybean chalcone isomerase: evolution of the fold, and the differential expression and localization of the gene family. Planta 241, 507–523. doi: 10.1007/s00425-014-2200-5
Dixon, R. A., Ferreria, D. (2002). Genistein. Phytochemistry 60, 205–211. doi: 10.1016/S0031-9422(02)00116-4
Earley, K. W., Haag, J. R., Pontes, O., Opper, K., Juehne, T., Song, K., et al. (2006). Gateway-compatible vectors for plant functional genomics and proteomics. Plant J. 45, 616–629. doi: 10.1111/j.1365-313X.2005.02617.x
Ehlting, J., Büttner, D., Wang, Q., Douglas, C. J., Somssich, I. E., Kombrink, E. (1999). Three 4-coumarate:coenzyme A ligases in Arabidopsis thaliana represent two evolutionarily divergent classes in angiosperms. Plant J.: Cell Mol. Biol. 19, 9–20. doi: 10.1046/j.1365-313X.1999.00491.x
Ehlting, J., Mattheus, N., Aeschliman, D. S., Li, E. Y., Hamberger, B., Cullis, I. F., et al. (2005). Global transcript profiling of primary stems from Arabidopsis thaliana identifies candidate genes for missing links in lignin biosynthesis and transcriptional regulators of fiber differentiation. Plant J. 42, 618–640. doi: 10.1111/j.1365-313X.2005.02403.x
El-Azaz, J., Canovas, F. M., Avila, C., de la Torre, F. (2018). The arogenate dehydratase ADT2 is essential for seed development in Arabidopsis. Plant Cell Physiol. 59, 2409–2420. doi: 10.1093/pcp/pcy200
El-Azaz, J., Cánovas, F. M., Barcelona, B., Ávila, C., de la Torre, F. (2022). Deregulation of phenylalanine biosynthesis evolved with the emergence of vascular plants. Plant Physiol. 188, 134–150. doi: 10.1093/plphys/kiab454
El-Azaz, J., de la Torre, F., Ávila, C., Cánovas, F. M. (2016). Identification of a small protein domain present in all plant lineages that confers high prephenate dehydratase activity. Plant J. 87, 215–229. doi: 10.1111/tpj.13195
Emanuelsson, O., Brunak, S., von Heijne, G., Nielsen, H. (2007). Locating proteins in the cell using TargetP, SignalP and related tools. Nat. Prot. 2, 953–971. doi: 10.1038/nprot.2007.131
Erickson, J. L., Kantek, M., Schattat, M. H. (2017). Plastid-nucleus distance alters the behavior of stromules. Front. Plant Sci. 8, 1135. doi: 10.3389/fpls.2017.01135
Fazel, A. M., Bowen, J. R., Jensen, R. A. (1980). Arogenate (pretyrosine) is an obligatory intermediate of L-tyrosine biosynthesis - confirmation in a microbial mutant. P Natl. Acad. Sci. U.S.A. 77, 1270–1273. doi: 10.1073/pnas.77.3.1270
García-Calderón, M., Pérez-Delgado, C. M., Palove-Balang, P., Betti, M., Márquez, A. J. (2020). Flavonoids and isoflavonoids biosynthesis in the model legume Lotus japonicus; Connections to nitrogen metabolism and photorespiration. Plants 9, 774. doi: 10.3390/plants9060774
Graindorge, M., Giustini, C., Jacomin, A. C., Kraut, A., Curien, G., Matringe, M. (2010). Identification of a plant gene encoding glutamate/aspartate-prephenate aminotransferase: The last homeless enzyme of aromatic amino acids biosynthesis. FEBS Lett. 584, 4357–4360. doi: 10.1016/j.febslet.2010.09.037
Gray, J. C., Sullivan, J. A., Hibberd, J. M., Hansen, M. R. (2001). Stromules: Mobile protrusions and interconnections between plastids. Plant Biol. 3, 223–233. doi: 10.1055/s-2001-15204
Griffing, L. R. (2011). Laser stimulation of the chloroplast/endoplasmic reticulum nexus in tobacco transiently produces protein aggregates (boluses) within the endoplasmic reticulum and stimulates local er remodeling. Mol. Plant 4, 886–895. doi: 10.1093/mp/ssr072
Hanson, M. R., Hines, K. M. (2018). Stromules: probing formation and function. Plant Physiol. 176, 128–137. doi: 10.1104/pp.17.01287
Hassan, S., Mathesius, U. (2012). The role of flavonoids in root-rhizosphere signalling: opportunities and challenges for improving plant-microbe interactions. J. Exp. Bot. 63, 3429–3444. doi: 10.1093/jxb/err430
Hawes, C., Kiviniemi, P., Kriechbaumer, V. (2015). The endoplasmic reticulum: A dynamic and well-connected organelle. J. Integr. Plant Biol. 57, 50–62. doi: 10.1111/jipb.12297
Hill, J. E., Breidenbach, R. W. (1974). Proteins of soybean seeds.1. Isolation and characterization of major components. Plant Physiol. 53, 742–746. doi: 10.1104/pp.53.5.742
Hodek, P., Trefil, P., Stiborová, M. (2002). Flavonoids-potent and versatile biologically active compounds interacting with cytochromes P450. Chemico Biol. Interact. 139, 1–21. doi: 10.1016/S0009-2797(01)00285-X
Hrazdina, G., Wagner, G. J. (1985). Metabolic pathways as enzyme complexes: Evidence for the synthesis of phenylpropanoids and flavonoids on membrane associated enzyme complexes. Arch. Biochem. Biophys. 237, 88–100. doi: 10.1016/0003-9861(85)90257-7
Hsu, S. K., Lin, L. L., Lo, H. H., Hsu, W. H. (2004). Mutational analysis of feedback inhibition and catalytic sites of prephenate dehydratase from Corynebacterium glutamicum. Arch. Microbiol. 181, 237–244. doi: 10.1007/s00203-004-0649-5
Isemer, R., Mulisch, M., Schäfer, A., Kirchner, S., Koop, H. U., Krupinska, K. (2012). Recombinant Whirly1 translocates from transplastomic chloroplasts to the nucleus. FEBS Lett. 586, 85–88. doi: 10.1016/j.febslet.2011.11.029
Jorgensen, K., Rasmussen, A. V., Morant, M., Nielsen, A. H., Bjarnholt, N., Zagrobelny, M., et al. (2005). Metabolon formation and metabolic channeling in the biosynthesis of plant natural products. Curr. Opin. Plant Biol. 8, 280–291. doi: 10.1016/j.pbi.2005.03.014
Jung, E., Zamir, L. O., Jensen, R. A. (1986). Chloroplasts of higher-plants synthesize L-phenylalanine via L-arogenate. Proc. Natl. Acad. Sci. U.S.A. 83, 7231–7235. doi: 10.1073/pnas.83.19.7231
Kafri, R., Springer, M., Pilpel, Y. (2009). Genetic redundancy: new tricks for old genes. Cell 136, 389–392. doi: 10.1016/j.cell.2009.01.027
Kaminaga, Y., Schnepp, J., Peel, G., Kish, C. M., Ben-Nissan, G., Weiss, D., et al. (2006). Plant phenylacetaldehyde synthase is a bifunctional homotetrameric enzyme that catalyzes phenylalanine decarboxylation and oxidation. J. Biol. Chem. 281, 23357–23366. doi: 10.1074/jbc.M602708200
Khatri, P., Chen, L., Rajcan, I., Dhaubhadel, S. (2023). Functional characterization of Cinnamate 4-hydroxylase gene family in soybean (Glycine max). PloS One 18, e0285698. doi: 10.1371/journal.pone.0285698
Kriechbaumer, V., Brandizzi, F. (2020). The plant endoplasmic reticulum: an organized chaos of tubules and sheets with multiple functions. J. Microsc. 280, 122–133. doi: 10.1111/jmi.12909
Kumar, S., Stecher, G., Li, M., Knyaz, C., Tamura, K. (2018). MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Kushnirov, V. V. (2000). Rapid and reliable protein extraction from yeast. Yeast 16, 857–860. doi: 10.1002/1097-0061(20000630)16:9<857::AID-YEA561>3.0.CO;2-B
Kwok, E. Y., Hanson, M. R. (2004a). Plastids and stromules interact with the nucleus and cell membrane in vascular plants. Plant Cell Rep. 23, 188–195. doi: 10.1007/s00299-004-0824-9
Kwok, E. Y., Hanson, M. R. (2004b). Stromules and the dynamic nature of plastid morphology. J. Microsc. 214, 124–137. doi: 10.1111/j.0022-2720.2004.01317.x
Lamartiniere, C. A. (2000). Protection against breast cancer with genistein: a component of soy. Am. J. Clin. Nutr. 71, 1705S–1707S. doi: 10.1093/ajcn/71.6.1705S
Li, C., Duckney, P., Zhang, T., Fu, Y., Li, X., Kroon, J., et al. (2022). TraB family proteins are components of ER-mitochondrial contact sites and regulate ER-mitochondrial interactions and mitophagy. Nat. Commun. 13, 5658. doi: 10.1038/s41467-022-33402-w
Li, Y., Kim, J. I., Pysh, L., Chapple, C. (2015). Four Isoforms of Arabidopsis 4-coumarate-CoA ligase have overlapping yet distinct roles in phenylpropanoid metabolism. Plant Physiol. 169, 2409–2421. doi: 10.1104/pp.15.00838
Liang, B., Cao, J., Wang, R., Fan, C., Wang, W., Hu, X., et al. (2023). ZmCIPK32 positively regulates germination of stressed seeds via gibberellin signal. Plant Physiol. Biochem. 199, 107716. doi: 10.1016/j.plaphy.2023.107716
Lohr, D., Venkov, P., Zlatanova, J. (1995). Transcriptional regulation in the yeast GAL gene family: a complex genetic network. FASEB J. 9, 777–787. doi: 10.1096/fasebj.9.9.7601342
Lozovaya, V. V., Lygin, A. V., Zernova, O. V., Ulanov, A. V., Li, S., Hartman, G. L., et al. (2007). Modification of phenolic metabolism in soybean hairy roots through down regulation of chalcone synthase or isoflavone synthase. Planta 225, 665–679. doi: 10.1007/s00425-006-0368-z
Lu, Q., Tang, X., Tian, G., Wang, F., Liu, K., Nguyen, V., et al. (2010). Arabidopsis homolog of the yeast TREX-2 mRNA export complex: components and anchoring nucleoporin. Plant J. 61, 259–270. doi: 10.1111/j.1365-313X.2009.04048.x
Lynch, M., Force, A. (2000). The probability of duplicate gene preservation by subfunctionalization. Genetics 154, 459–473. doi: 10.1093/genetics/154.1.459
Maeda, H., Dudareva, N. (2012). The shikimate pathway and aromatic amino Acid biosynthesis in plants. Annu. Rev. Plant Biol. 63, 73–105. doi: 10.1146/annurev-arplant-042811-105439
Maeda, H., Shasany, A. K., Schnepp, J., Orlova, I., Taguchi, G., Cooper, B. R., et al. (2010). RNAi suppression of Arogenate Dehydratase1 reveals that phenylalanine is synthesized predominantly via the arogenate pathway in petunia petals. Plant Cell. 22, 832–849. doi: 10.1105/tpc.109.073247
Maeda, H., Yoo, H. J., Dudareva, N. (2011). Prephenate aminotransferase directs plant phenylalanine biosynthesis via arogenate. Nat. Chem. Biol. 7, 19–21. doi: 10.1038/nchembio.485
Mainali, H. R., Vadivel, A. K. A., Li, X., Gijzen, M., Dhaubhadel, S. (2017). Soybean cyclophilin GmCYP1 interacts with an isoflavonoid regulator GmMYB176. Sci. Rep. 7, 39550. doi: 10.1038/srep39550
Mehrshahi, P., Stefano, G., Andaloro, J. M., Brandizzi, F., Froehlich, J. E., DellaPenna, D. (2013). Transorganellar complementation redefines the biochemical continuity of endoplasmic reticulum and chloroplasts. Proc. Natl. Acad. Sci. 110, 12126–12131. doi: 10.1073/pnas.1306331110
Messina, M. (2010). Insights gained from 20 years of soy research. J. Nutr. 140, 2289–2295. doi: 10.3945/jn.110.124107
Muhammad, D., Alameldin, H. F., Oh, S., Montgomery, B. L., Warpeha, K. M. (2023). Arogenate dehydratases: unique roles in light-directed development during the seed-to-seedling transition in Arabidopsis thaliana. Front. Plant Sci. 14. doi: 10.3389/fpls.2023.1220732
Nakayama, T., Takahashi, S., Waki, T. (2019). Formation of flavonoid metabolons: functional significance of protein-protein interactions and impact on flavonoid chemodiversity. Front. Plant Sci. 10. doi: 10.3389/fpls.2019.00821
Odell, J. T., Nagy, F., Chua, N.-H. (1985). Identification of DNA sequences required for activity of the cauliflower mosaic virus 35S promoter. Nature 313, 810–812. doi: 10.1038/313810a0
Owens, D. K., Alerding, A. B., Crosby, K. C., Bandara, A. B., Westwood, J. H., Winkel, B. S. J. (2008). Functional analysis of a predicted flavonol synthase gene family in Arabidopsis. Plant Physiol. 147, 1046–1061. doi: 10.1104/pp.108.117457
Pan, L., Luo, Y., Wang, J., Li, X., Tang, B., Yang, H., et al. (2022). Evolution and functional diversification of catalase genes in the green lineage. BMC Genomics 23, 411. doi: 10.1186/s12864-022-08621-6
Pareek, V., Sha, Z., He, J., Wingreen, N. S., Benkovic, S. J. (2021). Metabolic channeling: predictions, deductions, and evidence. Mol. Cell. 81, 3775–3785. doi: 10.1016/j.molcel.2021.08.030
Perico, C., Sparkes, I. (2018). Plant organelle dynamics: cytoskeletal control and membrane contact sites. Nat. Rev. Mol. Cell Biol. 220, 381–394. doi: 10.1111/nph.15365
Phillips, D. A., Kapulnik, Y. (1995). Plant isoflavonoids, pathogens and symbionts. Trends Microbiol. 3, 58–64. doi: 10.1016/S0966-842X(00)88876-9
Phillips, M. J., Voeltz, G. K. (2016). Structure and function of ER membrane contact sites with other organelles. Nat. Rev. Mol. Cell Biol. 17, 69–82. doi: 10.1038/nrm.2015.8
Pohnert, G., Zhang, S., Husain, A., Wilson, D. B., Ganem, B. (1999). Regulation of phenylalanine biosynthesis. Studies on the mechanism of phenylalanine binding and feedback inhibition in the Escherichia coli P-protein. Biochemistry-Us 38, 12212–12217. doi: 10.1021/bi991134w
Pyke, K. A., Howells, C. A. (2002). Plastid and stromule morphogenesis in tomato. Ann. Bot. 90, 559–566. doi: 10.1093/aob/mcf235
Qian, Y., Lynch, J. H., Guo, L., Rhodes, D., Morgan, J. A., Dudareva, N. (2019). Completion of the cytosolic post-chorismate phenylalanine biosynthetic pathway in plants. Nat. Commun. 10, 15. doi: 10.1038/s41467-018-07969-2
Ramadass, P., Meerarani, P., Toborek, M., Robertson, L. W., Hennig, B. (2003). Dietary flavonoids modulate PCB-induced oxidative stress, CYP1A1 induction, and AhR-DNA binding activity in vascular endothelial cells. Toxicol. Sci. 76, 212–219. doi: 10.1093/toxsci/kfg227
Rippert, P., Puyaubert, J., Grisollet, D., Derrier, L., Matringe, M. (2009). Tyrosine and phenylalanine are synthesized within the plastids in Arabidopsis. Plant Physiol. 149, 1251–1260. doi: 10.1104/pp.108.130070
Sarkar, F. H., Li, Y. W. (2003). Soy isoflavones and cancer prevention. Cancer Invest. 21, 744–757. doi: 10.1081/CNV-120023773
Saslowsky, D. E., Warek, U., Winkel, B. S. (2005). Nuclear localization of flavonoid enzymes in Arabidopsis. J. Biol. Chem. 280, 23735–23740. doi: 10.1074/jbc.M413506200
Schattat, M., Barton, K., Baudisch, B., Klösgen, R. B., Mathur, J. (2011). Plastid stromule branching coincides with contiguous endoplasmic reticulum dynamics. Plant Physiol. 155, 1667–1677. doi: 10.1104/pp.110.170480
Schattat, M. H., Griffiths, S., Mathur, N., Barton, K., Wozny, M. R., Dunn, N., et al. (2012). Differential coloring reveals that plastids do not form networks for exchanging macromolecules. Plant Cell. 24, 1465–1477. doi: 10.1105/tpc.111.095398
Schmutz, J., Cannon, S. B., Schlueter, J., Ma, J., Mitros, T., Nelson, W., et al. (2010). Genome sequence of the palaeopolyploid soybean. Nature 463, 178–183. doi: 10.1038/nature08670
Scott, I. M., McDowell, T., Renaud, J. B., Krolikowski, S. W., Chen, L., Dhaubhadel, S. (2021). Soybean (Glycine max L Merr) host-plant defenses and resistance to the two-spotted spider mite (Tetranychus urticae Koch). PloS One 16, e0258198. doi: 10.1371/journal.pone.0258198
Sepiol, C. J., Yu, J., Dhaubhadel, S. (2017). Genome-wide identification of chalcone reductase gene family in soybean: insight into root-specific GmCHRs and Phytophthora sojae resistance. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.02073
Shoemaker, R. C., Schlueter, J., Doyle, J. J. (2006). Paleopolyploidy and gene duplication in soybean and other legumes. Curr. Opin. Plant Biol. 9, 104–109. doi: 10.1016/j.pbi.2006.01.007
Sievers, F., Wilm, A., Dineen, D., Gibson, T. J., Karplus, K., Li, W., et al. (2011). Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7, 539–545. doi: 10.1038/msb.2011.75
Sparkes, I. A., Runions, J., Kearns, A., Hawes, C. (2006). Rapid, transient expression of fluorescent fusion proteins in tobacco plants and generation of stably transformed plants. Nat. Prot. 1, 2019–2025. doi: 10.1038/nprot.2006.286
Stafford, H. A. (1974). “Possible multienzyme complexes regulating the formation of C6-C3 phenolic compounds and lignins in higher plants,” in Recent Advances in Phytochemistry (New York: Elsevier), 53–79.
St John, T. P., Davis, R. W. (1981). The organization and transcription of the galactose gene cluster of Saccharomyces. J. Mol. Biol. 152, 285–315. doi: 10.1016/0022-2836(81)90244-8
Subramanian, S., Stacey, G., Yu, O. (2006). Endogenous isoflavones are essential for the establishment of symbiosis between soybean and Bradyrhizobium japonicum. Plant J. 48, 261–273. doi: 10.1111/j.1365-313X.2006.02874.x
Tan, K. M., Li, H., Zhang, R. G., Gu, M. Y., Clancy, S. T., Joachimiak, A. (2008). Structures of open (R) and close (T) states of prephenate dehydratase (PDT) - Implication of allosteric regulation by L-phenylalanine. J. Struct. Biol. 162, 94–107. doi: 10.1016/j.jsb.2007.11.009
Tsugama, D., Liu, S., Takano, T. (2013). A bZIP protein, VIP1, interacts with Arabidopsis heterotrimeric G protein β subunit, AGB1. Plant Physiol. Biochem. 71, 240–246. doi: 10.1016/j.plaphy.2013.07.024
Ungricht, R., Kutay, U. (2017). Mechanisms and functions of nuclear envelope remodelling. Nat. Rev. Mol. Cell Biol. 18, 229–245. doi: 10.1038/nrm.2016.153
Van de Peer, Y., Mizrachi, E., Marchal, K. (2017). The evolutionary significance of polyploidy. Nat. Rev. Genet. 18, 411–424. doi: 10.1038/nrg.2017.26
Waki, T., Yoo, D., Fujino, N., Mameda, R., Denessiouk, K., Yamashita, S., et al. (2016). Identification of protein-protein interactions of isoflavonoid biosynthetic enzymes with 2-hydroxyisoflavanone synthase in soybean (Glycine max (L.) Merr.). Biochem. Bioph. Res. Co. 469, 546–551. doi: 10.1016/j.bbrc.2015.12.038
Wang, J., Hossain, M. S., Lyu, Z., Schmutz, J., Stacey, G., Xu, D., et al. (2019). SoyCSN: Soybean context-specific network analysis and prediction based on tissue-specific transcriptome data. Plant Direct. 3, e00167. doi: 10.1002/pld3.167
Wang, H., Wang, W., Zhan, J., Yan, A., Sun, L., Zhang, G., et al. (2016). The accumulation and localization of chalcone synthase in grapevine (Vitis vinifera L.). Plant Physiol. Biochem. 106, 165–176. doi: 10.1016/j.plaphy.2016.04.042
Warpeha, K. M., Lateef, S. S., Lapik, Y., Anderson, M., Lee, B. S., Kaufman, L. S. (2006). G-protein-coupled receptor 1, G-protein Galpha-subunit 1, and prephenate dehydratase 1 are required for blue light-induced production of phenylalanine in etiolated Arabidopsis. Plant Physiol. 140, 844–855. doi: 10.1104/pp.105.071282
Watanabe, S., Hayashi, K., Yagi, K., Asai, T., Mactavish, H., Picone, J., et al. (2002). Biogenesis of 2-Phenylethanol in Rose Flowers: Incorporation of [2H8]L-Phenylalanine into 2-Phenylethanol and its β-D-Glucopyranoside during the Flower Opening of Rosa `Hoh-Jun’ and Rosa damascena Mill. Biosci. Biotechnol. Biochem. 66, 943–947. doi: 10.1271/bbb.66.943
Watkinson, J. I., Bowerman, P. A., Crosby, K. C., Hildreth, S. B., Helm, R. F., Winkel, B. S. J. (2018). Identification of MOS9 as an interaction partner for chalcone synthase in the nucleus. PeerJ 6, e5598. doi: 10.7717/peerj.5598
Whatley, J. M., McLean, B., Juniper, B. E. (1991). Continuity of chloroplast and endoplasmic reticulum membranes in Phaseolus vulgaris. New Phytol. 117, 209–217. doi: 10.1111/j.1469-8137.1991.tb04901.x
Winkel, B. S. J. (2001). It takes a garden. How work on diverse plant species has contributed to an understanding of flavonoid metabolism. Plant Physiol. 127, 1399–1404. doi: 10.1104/pp.010675
Winkel, B. S. J. (2002). Biosynthesis of flavonoids and effects of stress. Curr. Opin. Plant Biol. 5, 218–223. doi: 10.1016/S1369-5266(02)00256-X
Winkel, B. S. J. (2004). Metabolic channeling in plants. Annu. Rev. Plant Biol. 55, 85–107. doi: 10.1146/annurev.arplant.55.031903.141714
Winkel, B. S. J. (2006). “The biosynthesis of flavonoids,” in The Science of Flavonoids (New York, NY: Springer New York), 71–95.
Yamada, T., Matsuda, F., Kasai, K., Fukuoka, S., Kitamura, K., Tozawa, Y., et al. (2008). Mutation of a rice gene encoding a phenylalanine biosynthetic enzyme results in accumulation of phenylalanine and tryptophan. Plant Cell. 20, 1316–1329. doi: 10.1105/tpc.107.057455
Yong, K. J., Wu, T. Y. (2022). Second-generation bioenergy from oilseed crop residues: Recent technologies, techno-economic assessments and policies. Energy Conversion Manag. 267, 115869. doi: 10.1016/j.enconman.2022.115869
Yoo, H., Widhalm, J. R., Qian, Y., Maeda, H., Cooper, B. R., Jannasch, A. S., et al. (2013). An alternative pathway contributes to phenylalanine biosynthesis in plants via a cytosolic tyrosine:phenylpyruvate aminotransferase. Nat. Commun. 4, 2833. doi: 10.1038/ncomms3833
Young, N. D., Bharti, A. K. (2012). Genome-enabled insights into legume biology. Annu. Rev. Plant Biol. 63, 283–305. doi: 10.1146/annurev-arplant-042110-103754
Young, N. D., Debellé, F., Oldroyd, G. E. D., Geurts, R., Cannon, S. B., Udvardi, M. K., et al. (2011). The Medicago genome provides insight into the evolution of rhizobial symbioses. Nature 480, 520–524. doi: 10.1038/nature10625
Yousefi-Taemeh, M., Lin, J., Ifa, D. R., Parrott, W., Kovinich, N. (2021). Metabolomics differences of Glycine max QTLs resistant to soybean looper. Metabolites 11, 710. doi: 10.3390/metabo11100710
Yuan, J., Song, Q. (2023). Polyploidy and diploidization in soybean. Mol. Breed. 43, 51. doi: 10.1007/s11032-023-01396-y
Zhang, Y., Fernie, A. R. (2021). Metabolons, enzyme–enzyme assemblies that mediate substrate channeling, and their roles in plant metabolism. Plant Commun. 2, 100081. doi: 10.1016/j.xplc.2020.100081
Zhang, S., Pohnert, G., Kongsaeree, P., Wilson, D. B., Clardy, J., Ganem, B. (1998). Chorismate mutase-prephenate dehydratase from Escherichia coli. Study of catalytic and regulatory domains using genetically engineered proteins. J. Biol. Chem. 273, 6248–6253. doi: 10.1074/jbc.273.11.6248
Keywords: soybean, arogenate dehydratase, isoflavone synthase, phenylalanine, isoflavonoid, metabolon, specialized metabolites
Citation: Clayton EJ, Islam NS, Pannunzio K, Kuflu K, Sirjani R, Kohalmi SE and Dhaubhadel S (2024) Soybean AROGENATE DEHYDRATASES (GmADTs): involvement in the cytosolic isoflavonoid metabolon or trans-organelle continuity? Front. Plant Sci. 15:1307489. doi: 10.3389/fpls.2024.1307489
Received: 04 October 2023; Accepted: 03 January 2024;
Published: 23 January 2024.
Edited by:
Deyu Xie, North Carolina State University, United StatesReviewed by:
Xianzhi He, North Carolina State University, United StatesCopyright © 2024 Susanne E. Kohalmi and His Majesty the King in Right of Canada, as represented by the Minister of Agriculture and Agri-Food Canada for the contribution of Emily J. Clayton, Nishat S. Islam, Kelsey Pannunzio, Kuflom Kuflu, Ramtin Sirjani and Sangeeta Dhaubhadel. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sangeeta Dhaubhadel, sangeeta.dhaubhadel@agr.gc.ca
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.