- 1School of Life Sciences, University of Hawaiʻi, Honolulu, HI, United States
- 2Department of Tropical Plant and Soil Sciences, University of Hawaiʻi, Honolulu, HI, United States
Introduction: Sida fallax (Malvaceae) is the most widespread and variable taxon of Malvaceae in the Hawaiian Islands, growing with a diversity of morphological forms in different habitats including Midway Atoll, Nihoa, and all the main islands. Morphological variation exists within and among populations. The study aimed to investigate the genetic variation within and among populations from various habitats and geographic locations throughout the Hawaiian range of S. fallax.
Methods: A total of 124 samples, with up to five samples per population where possible, were collected from 26 populations across six of the main Hawaiian Islands (Kauaʻi, Oʻahu, Maui, Molokaʻi, Lānaʻi, and Hawaiʻi) and Nihoa in the Northwestern Hawaiian Islands. The sampling strategy encompassed collecting populations from different habitats and geographic locations, including coastal and mountain ecotypes, with many intermediate morphological forms. Multiplexed ISSR genotyping by sequencing (MIG-seq) was used to detect single nucleotide polymorphisms (SNP) and genetic differences among individuals and populations were evaluated using PCO analyses.
Results: The relationship of FST with the geographical distance between the populations was assessed using the Mantel test. The results showed that populations on a single island were more closely related to each other and to populations on islands within their respective groups than they were to populations on other islands.
Discussion: The overall genetic relationships among islands were, to a large extent, predictive based on island position within the chain and, to a lesser extent, within island topography.
1 Introduction
Oceanic archipelagos have long been considered natural laboratories for studying evolution because of their role in speciation and adaptive radiation (Darwin, 1859; Lack, 1947; Carlquist, 1974; Schluter, 2000). The remote archipelago of the Hawaiian Islands (ca. 4000 km from the nearest continental landmass) supports a variety of geographically isolated habitats resulting from the volcanic nature of the islands and ranging from mesic mountain to dry coastal zones. In addition, on the youngest island, lava flows continuously destroy old habitats and create new ones (Sohmer and Gustafson, 1987; Westerbergh and Saura, 1994; Wagner et al., 1999). Furthermore, some plant characteristics can facilitate adaptive radiation. For example, Takayama et al. (2018) suggests that herbaceous perennial herbs with leaky self-incompatible breeding systems, good intra-island dispersal capabilities, and flexible structural and physiological systems are more likely to undergo adaptive radiation in island environments. Importantly, the progenitors of adaptively radiated groups in islands are those that have already been successful in adaptations to different environments in source areas and have undergone eco-geographic speciation. In addition, diversification could be facilitated by ancestral genetic variation and hybridization (Choi et al., 2021). Consequently, the flora of the Hawaiian Islands exhibits some notable cases of adaptive radiation (Wagner and Funk, 1995; Ziegler, 2002; Keeley and Funk, 2011). In many cases, adaptive radiation is accompanied by speciation. Examples include the silversword alliance (Asteraceae; Baldwin and Sanderson, 1998; Carlquist et al., 2003; Landis et al., 2018), Hawaiian Bidens (Asteraceae; Knope et al., 2012, 2020) lobeliads (Campanulaceae; Givnish et al., 2009), mints (Lamiaceae; Lindqvist et al., 2003), Cyrtandra (Gesneriaceae; Johnson et al., 2019) and Schiedea (Caryophyllaceae; Sakai et al., 2006). In these examples, the evolution of widely divergent growth forms and morphologies were derived from a single colonization and are found in a wide range of habitats (Wagner and Funk, 1995; Price and Wagner, 2004). In other cases, adaptive radiation was not followed by abundant speciation and species have highly variable intraspecific morphological diversity. One example is Metrosideros polymorpha Gaud., the dominant tree species in the Hawaiian Islands (Percy et al., 2008).
Sida fallax (Malvaceae) is widely distributed throughout the Pacific region. It is the most widespread and variable taxon of Malvaceae in the Hawaiian Islands, growing with a diversity of morphological forms and in different habitats of Midway Atoll, Nihoa, and all the main islands. Morphological variation exists both within and among populations, particularly in plant stature, leaf size and shape, degree of pubescence, and inflorescence characters. There are numerous forms that were named for this species in the Hawaiian language recognizing this diversity in plant growth and habitat (Neal, 1948; Handy and Handy, 1991; Krauss, 1993). Ecologically, it is found in a variety of habitats from dry and mesic coastal zones to upland forest habitats, with an elevation range from 0 to 2,000 m (Gagné and Cuddihy, 1990).
There are two ecological forms of Sida fallax with many intermediates in the Hawaiian Islands (Figure 1) (Wagner et al., 1999; Yorkston and Daehler, 2006). A low elevation ecotype occurs along beaches and in dry, coastal shrublands. These are low sprawling shrubs that are typically less than 50 cm tall with small (up to 4-cm long), densely pubescent ovate leaves. This form can take the extreme form of a prostrate subshrub in coastal communities. Sida fallax from other Pacific locations exhibit this form only. In contrast, the mountain ecotype is an erect shrub that typically reaches 1–1.5 m in height (in some cases exceeding 2 m) with large (commonly 4 to 8 cm long), glabrous or nearly glabrous leaves. These forms are found in upland communities and mesic forest sites, usually away from coastal influences (Wagner et al., 1999; Yorkston and Daehler, 2006). The many intermediate morphological types between these extreme forms, many of which were recognized by early Hawaiians (see above), occur in a variety of habitats throughout the Hawaiian Islands.

Figure 1 Ecotype variation within Sida fallax. Beach ecotype (A) habit, (B) flower and smaller densely pubescent, ovate leaves; intermediate/inland ecotype (C) habit, (D) flower buds and mildly pubescent, acute leaves; mountain ecotype (E) habit, (F) flower and larger glabrous, acute leaves.
Several studies have been carried out on the different ecotypes of S. fallax. Stephens (2000) compared the ecophysiology of beach and mountain ecotypes and found that the beach ecotype had a greater tolerance of drought than the mountain ecotype. Experimental crosses carried out by Yorkston and Daehler (2006) between the beach and mountain forms demonstrated a lack of fertility barriers between them. Triplett et al. (2024) observed that coastal leaves of S. fallax exhibited a greater photosynthetic advantage from amphistomy compared to nearby montane leaves in more closed forest environments. Furthermore, a phylogenetic study of all ecotypes of S. fallax from the Hawaiian Islands and areas throughout the Pacific supported it as a single species with very little variation among the nuclear and chloroplast gene regions examined (Pejhanmehr, 2022; Pejhanmehr et al., 2022). Examples of species ecotypes that are genetically different are known (Korecký et al., 2021).The objective of the present study was to investigate the genetic variation within and among populations from the various habitats and geographic locations throughout the Hawaiian range of S. fallax. The various ecological and morphological forms of S. fallax discussed above occur on many of the islands. However, the expectation is that genetic relations will mirror the dispersion among islands. Parallel evolution is occurring with morphologically different forms on a single island being more closely related to each other than they are to morphologically similar forms on different islands.
2 Materials and methods
2.1 Sampling and DNA extraction
When possible, up to five samples were selected from collections representing 26 distinct populations of Sida fallax throughout the Hawaiian Islands. For some populations, fewer samples were available. Collections represented widely separated populations from the coastal strand to montane zones and habitats ecologically intermediate between these two extremes. Similarly, collections within populations were made from widely spaced individuals. Ecotypes were assigned based on proximity to coastal communities and elevation, and were identified as either “beach” or “mountain/inland”. Populations were from six of the main Hawaiian Islands (Kauaʻi, Oʻahu, Maui, Molokaʻi, Lānaʻi, and Hawaiʻi) and Nihoa in the Northwestern Hawaiian Islands (Figure 2; Table 1). Vouchers were deposited in the Joseph F. Rock (HAW) herbarium of the University of Hawaiʻi at Mānoa. Total genomic DNA was extracted using the CTAB method of Doyle and Doyle (1987) with some modifications (Morden et al., 1996) and deposited in the Hawaiian Plant DNA Library (HPDL) at the University of Hawaiʻi at Mānoa.
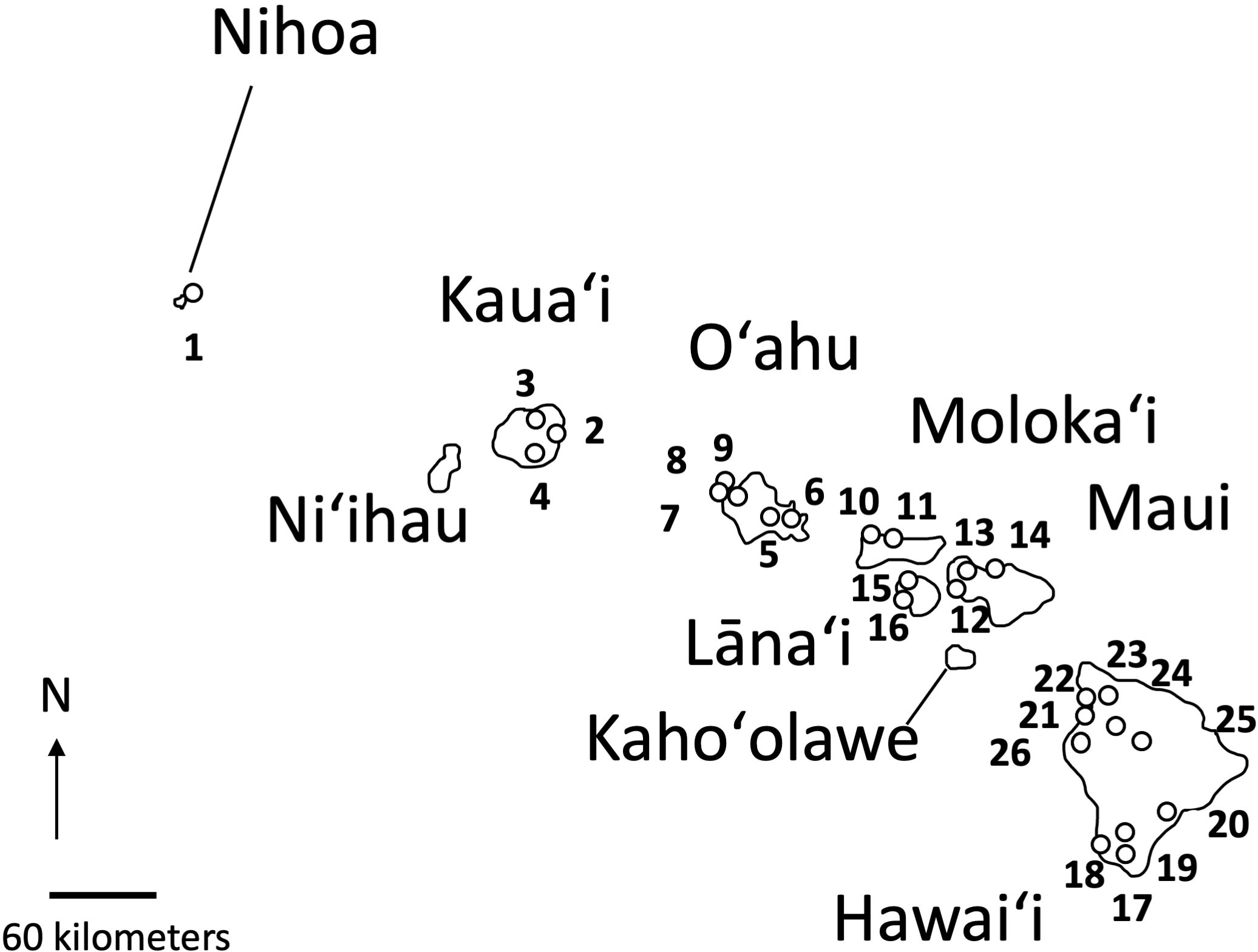
Figure 2 Location of the 26 populations of Sida fallax on Nihoa and the main Hawaiian Islands. Numbers indicate the populations identified in Table 1.
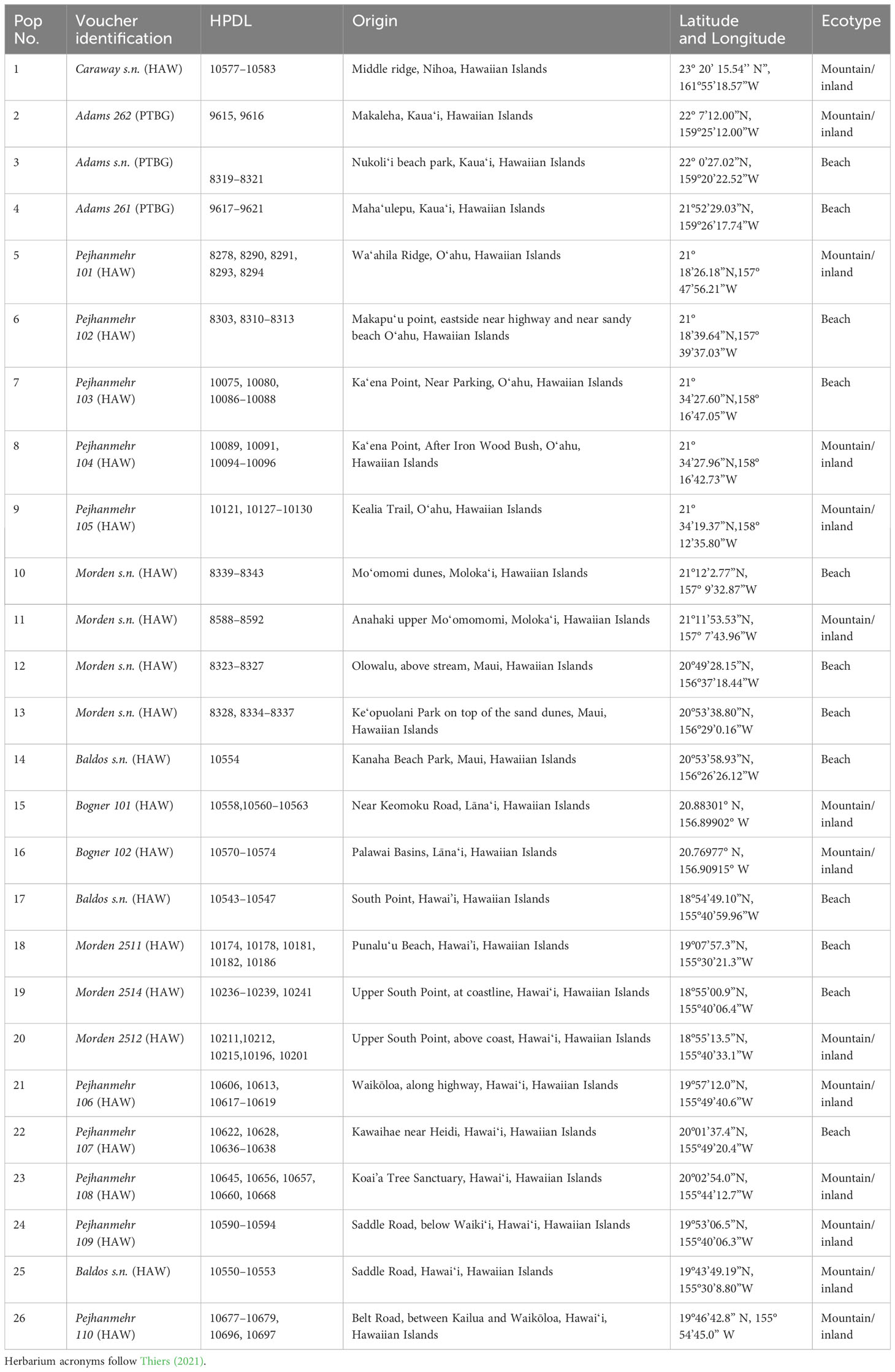
Table 1 Taxa used for Sida fallax population genetic analyses with population number, voucher identification, Hawaiian Plant DNA Library accession number (HPDL), collection locality, and ecotype.
2.2 MIG-seq library construction and sequencing
In total, 124 samples from 26 populations (Figure 2; Table 1) were selected for Multiplexed ISSR genotyping by sequencing (MIG-seq) to detect single nucleotide polymorphisms (SNP) (Suyama and Matsuki, 2015). MIG-seq library preparation and primer selection were performed according to Suyama and Matsuki (2015). After measurement of the final concentration of the libraries, approximately 10 pM of the libraries were used for sequencing on an Illumina MiSeq Sequencer (Illumina, San Diego, California), using a MiSeq Reagent Kit v3 (150 cycle, Illumina).
2.3 Quality control and SNP detection
Adapter and anchor sequence trimming of raw data were performed according to Suyama and Matsuki (2015), using FASTQ Trimmer (Galaxy Version 1.1.5) (Blankenberg et al., 2010; Afgan et al., 2018). High-quality reads were assessed using the Filter by Quality application (Galaxy Version 1.0.2+galaxy0) (Gordon, 2010; Afgan et al., 2018) with the criterion of q = 30 and p = 40 (q: quality cut-off value, p: percent of bases in a sequence that must have quality equal to or higher than q). Cutadapt (Galaxy Version 3.4+galaxy0) (Martin, 2011; Afgan et al., 2018) was used to remove reads from extremely short library entries (Maximum error rate = 0.01). Filtered reads were assembled using de novo map pipelines (ustacks, cstacks, sstacks, gstacks) in Stacks2: de novo map (Galaxy Version 2.4+galaxy1) (Catchen et al., 2011, Catchen et al., 2013; Afgan et al., 2018) with the following settings: Mismatches allowed between loci when processing a single individual = 2, Mismatches allowed between loci when building the catalog = 1, minimum percentage of individuals in a population required to process a locus for that population = 0, and minimum number of populations a locus must be present in to process a locus = 1.
2.4 Population genetic analysis
The assembled reads (gstackes) were used as input for population genetic analysis using Stacks2: populations (Galaxy Version 2.4+galaxy1) (Catchen et al., 2011, Catchen et al., 2013; Afgan et al., 2018) with the following settings: Minimum percentage of individuals in a population required to process a locus for that population = 0, Minimum number of populations a locus must be present in to process a locus = 1, minimum percentage of individuals across populations required to process a locus = 0, Fraction of samples possessing both loci to prune remaining samples from analysis = 1. Robustness of results was examined by choosing different numbers for Minimum percentage of individuals in a population required to process a locus for that population, and minimum percentage of individuals across populations required to process a locus, i.e., 0 and 0.7, 0.5 and 0, 0.7 and 0, 0 and 0.5, 0.4 and 0, 0.3 and 0.1, 0.45 and 0, 0.3 and 0, 0.1 and 0, and 0.2 and 0. The outputs of Stacks2: populations were used as input data to generate covariance values from population allele frequencies using GenoDive v2.0b27 (Meirmans and Van Tienderen, 2004) to investigate relationships of individuals within and among populations. Genetic similarity indices were calculated using both Gower (1971) and Nei and Li (1979) similarity coefficients for populations using MVSP Plus ver. 3.1 (Kovach, 2007). Relationships within and among populations were estimated from the similarity matrixes using principal coordinate analysis (PCO) with MVSP Plus ver. 3.1 (Kovach, 2007) using Gower similarity (Gower, 1971). The outputs of Stacks2: populations that included the SNP sites of all the samples was converted to VCF format and used to reconstruct a neighbor joining tree in VCF2PopTree (Supplementary Figure 1) (Subramanian et al., 2019). The relationship of FST with the geographical distance between the populations was assessed using the function mantel.test in ade4 library of the R package ape v5.3 (Dray and Dufour, 2007; R Core Team, 2021) with 9,999 permutations. Finally, a scatter plot was created using ggplot2 function in R (Wickham, 2016) to visualize the relationship between FST values and the distances between paired populations and ecotypes.
3 Results
3.1 Numbers of reads, SNPs, and polymorphic loci
The number of reads per sample ranged from 1,789 to 216,730 in the raw data (from 1,539 to 210,631 after quality filtering) and the removal rate varied from 0 to 11% for 124 individuals. The forward and reverse reads were treated independently because they do not overlap and could not be combined. The results of both reads were compared and provided near-identical outcomes, and only the reverse reads are presented here. Stacks verified 57,475 loci composed of 3,838,616 (bp) sites. Twenty-five sites that were pseudo-variant (PCR error) loci and detected only in one sample were filtered (Suyama and Matsuki, 2015), and 12,796 variant sites (or SNP) remained. The mean genotyped sites per locus was 66.79 bp (stderr 0.01).
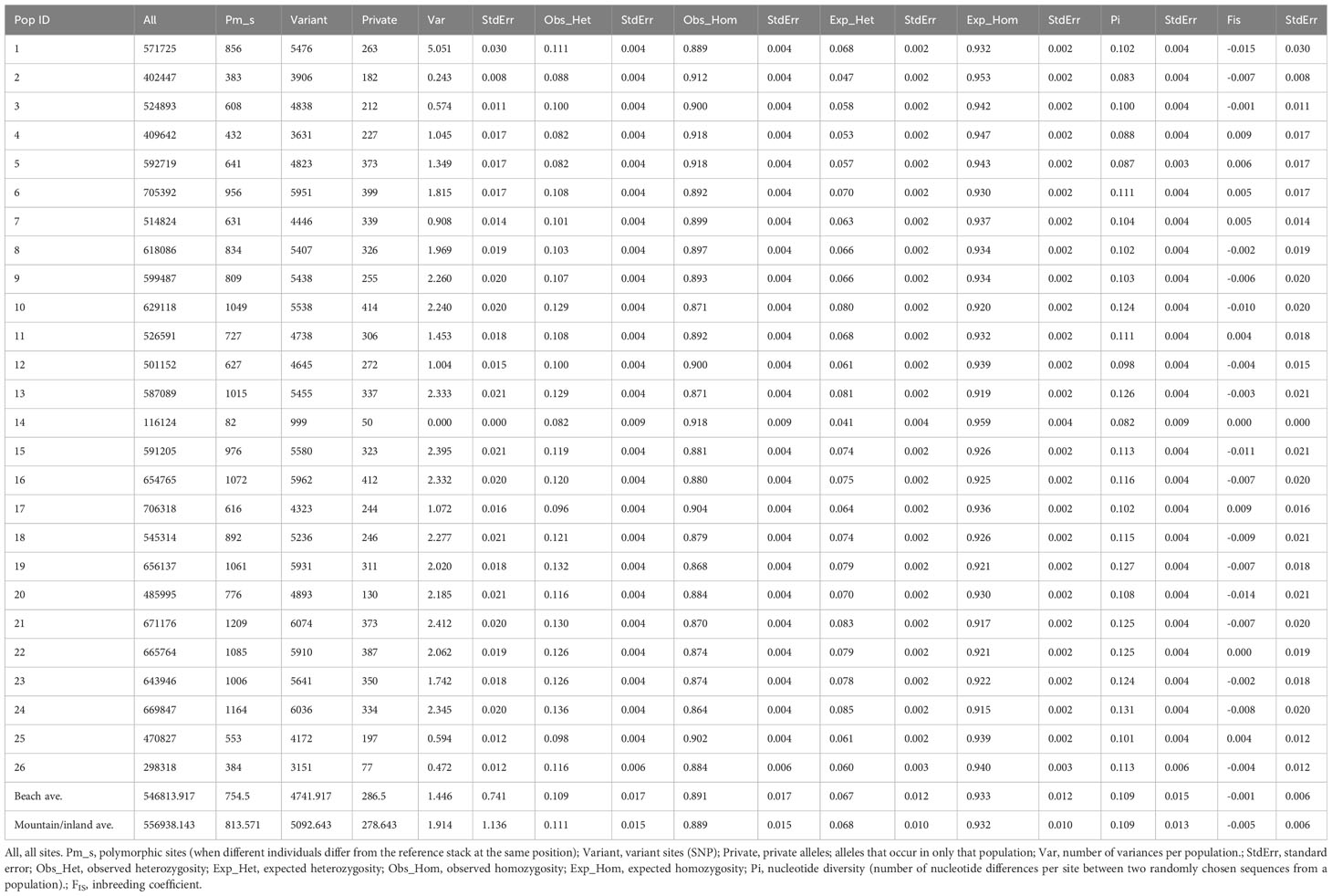
The population diversity statistics are summarized in Table 2. Polymorphic sites per population ranged from 82 to 1209 and variant sites (SNP) from 999 to 6074. Private alleles (alleles occurring in only a single population) ranged from 50 to 414 among populations. Observed heterozygosity ranged from 0.082 to 0.136 and nucleotide diversity (Pi) from 0.082 to 0.131. Among all population diversity statistics, the smallest values belonged to population 14 from Maui Beach where only one individual was found. The largest values for each statistic were mostly from Hawaiʻi Island (populations 17 and 24, mountain/inland habitats) and Lānaʻi (population 21, beach habitat). The inbreeding coefficient (FIS) ranged from -0.015 on Nihoa (population 1, mountain/inland habitat of Nihoa) to 0.009 on both Kauaʻi (population 4, beach habitat) and Hawaiʻi (population 17, beach habitat).The average value of observed heterozygosity comparison between beach (0.109) and mountain/inland (0.111) habits were similar.
3.2 Population differentiation and genetic distance
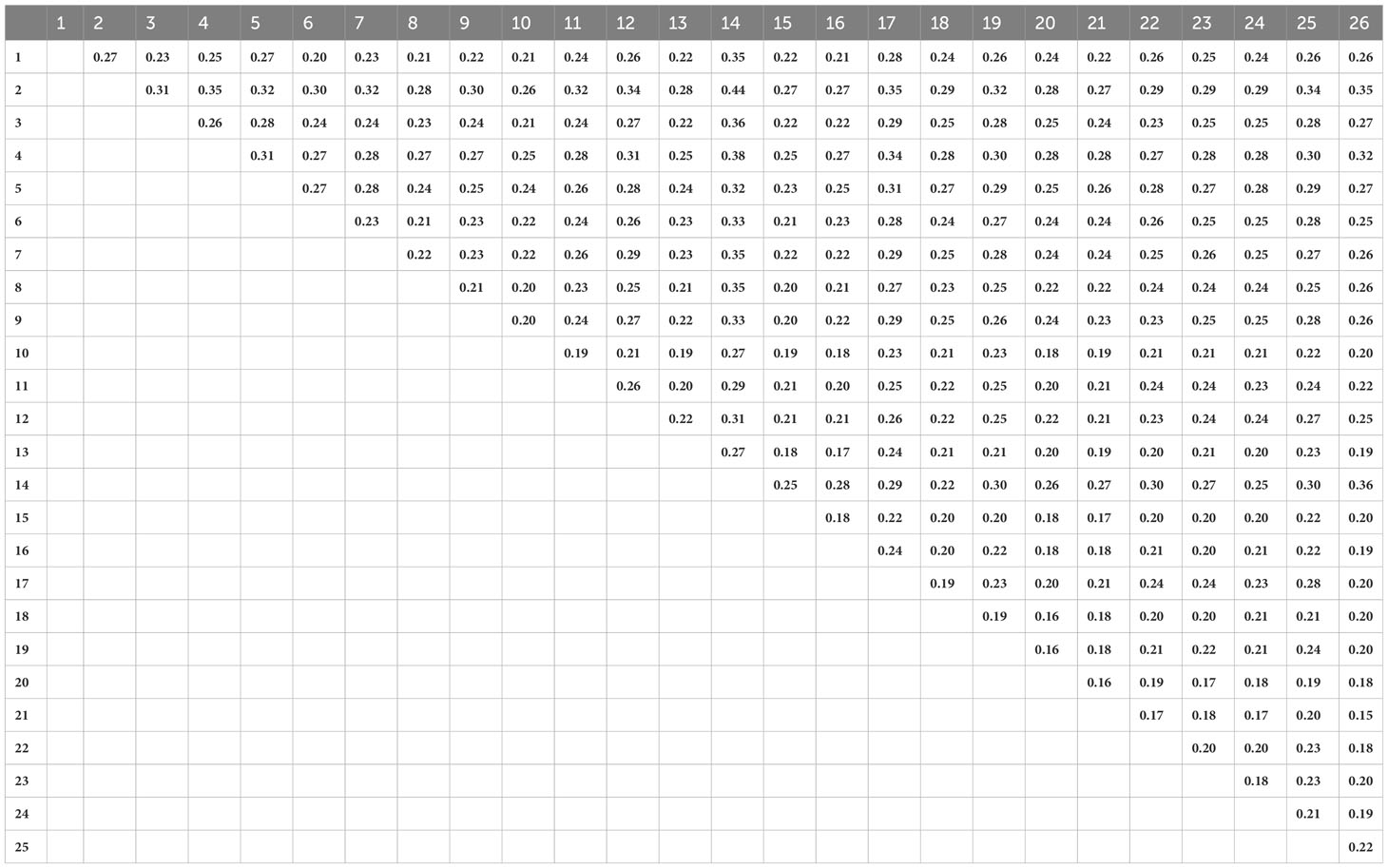
The Mantel test identified a significant positive correlation between genetic and geographic distances among S. fallax populations (r=0.295, p = 0.0209). Populations that were more closely associated geographically were also more closely related genetically. Overall, most FST values were approximately 0.20 with very low genetic variance among the 26 populations (Table 3). However, populations that were located on the same island and in similar habitats had smaller FST values, and populations on different islands and in different habitats had larger FST values. The smallest pairwise FST was between two populations from the mountain/inland habitat of Hawaiʻi Island (populations 21 and 26) that were located close to each other (FST = 0.15). In contrast, populations from different habitats and islands showed the largest FST, for example population 2 from a mountain/inland habitat on Kauaʻi and population 14 from a beach habitat on Maui (FST = 0.44). The scatter plot also indicates a significant positive correlation between genetic and geographic distances among S. fallax populations (Supplementary Figure 2). Additionally, it shows that beach-to-beach population pairs have a higher average FST than mountain/inland paired populations.
3.3 Genetic Assignment
Genetic differences among individuals and populations from across the range of habitat and locations of S. fallax in the Hawaiian Islands were evaluated using PCO analyses. Three main islands groupings were evident: 1) Oʻahu, Kauaʻi and Nihoa; 2) Maui, Molokaʻi, and Lānaʻi (collectively referred to as Maui Nui); 3) Hawaiʻi Island. Populations from each island grouping intersect at the center of the graph (Figures 3, 4). The phylogenetic analysis indicated the same island grouping as well (Supplementary Figure 1).
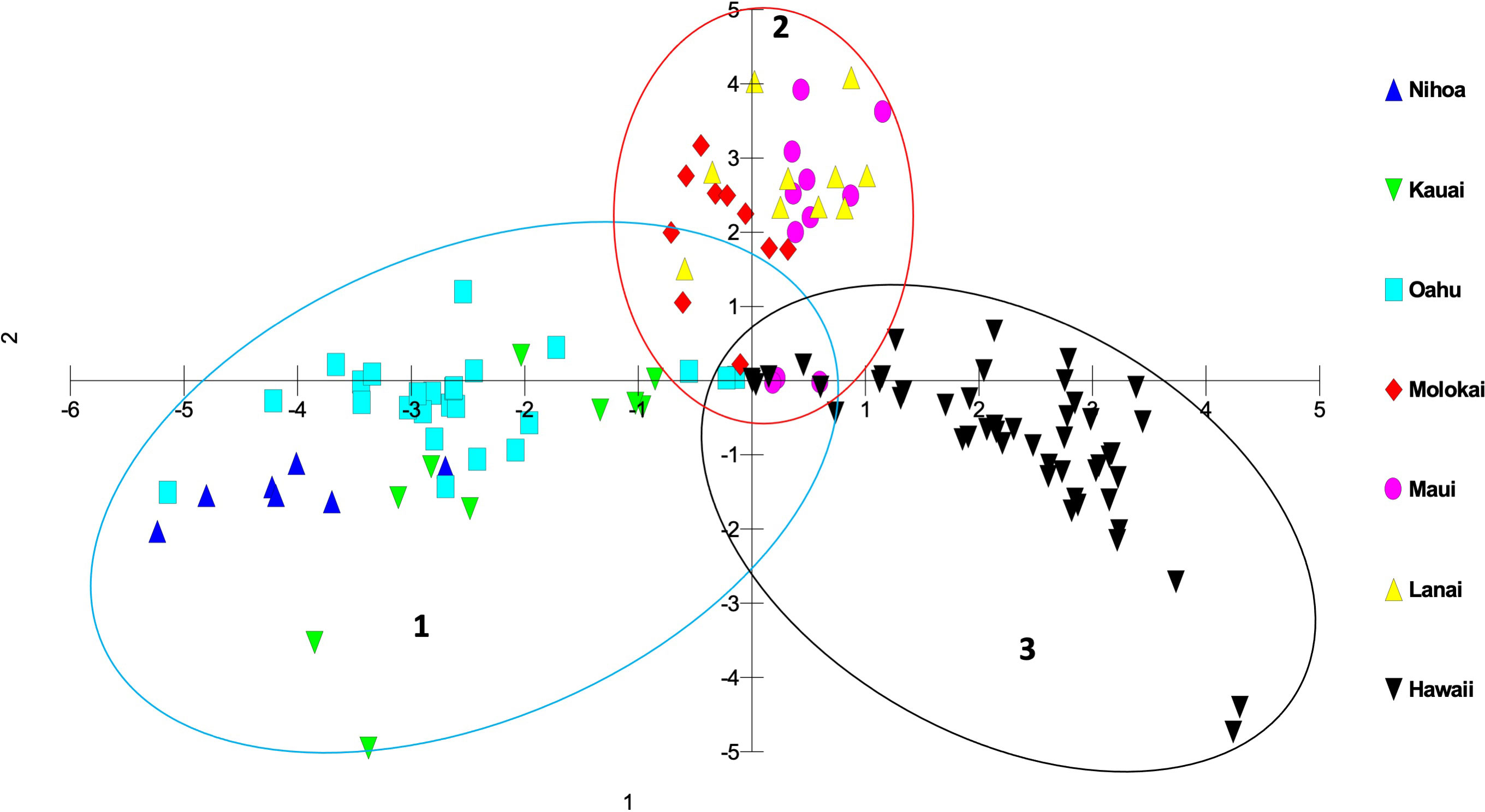
Figure 3 Principal coordinate analysis using MIG-seq data identifying individuals from each island in the Hawaiian Islands using a matrix of covariance values calculated from population allele frequencies. The three groups of S. fallax populations include 1) Nihoa, Oʻahu and Kauaʻi; 2) Maui, Molokaʻi, and Lānaʻi (Maui Nui); 3) Hawaiʻi.
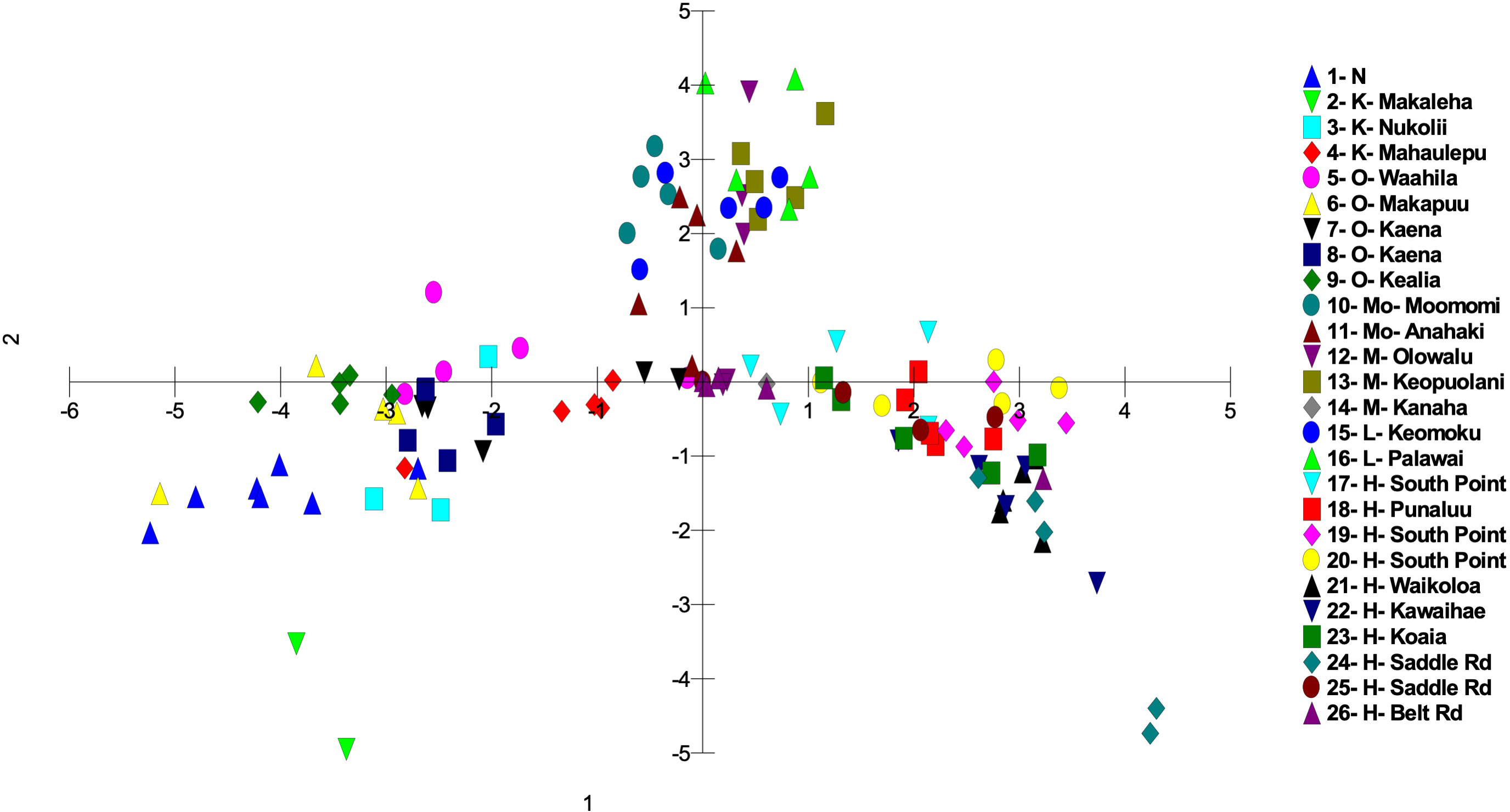
Figure 4 Principal coordinate analysis using MIG-seq data identifying the 26 S. fallax populations in across the Hawaiian Islands using a matrix of covariance values calculated from population allele frequencies. Island abbreviations: N, Nihoa; K, Kauaʻi; O, Oʻahu; M, Maui; Mo, Molokaʻi; L, Lānaʻi; H, Hawaiʻi (Table 1). Population numbers correspond to those in Figure 2.
There was a trend of coastal/beach populations occurring more predominantly near the zone of intersection (ZOI) of the island groups and the mountain/inland or most isolated populations being more distant from this zone of intersection (Figure 5). Within the Oʻahu-Kauaʻi-Nihoa group, populations near the ZOI include the coastal Kaʻena Point, Oʻahu (population 7), and Mahaʻulepu, Kauaʻi (Population 4) populations. One individual from Waʻahila (a mountain population; population 5) was located within the ZOI, but the remaining individuals from this population were not. Other populations outside the ZOI included those from Makaleha and Nukoliʻi from Kauaʻi; and Kealia, inland Kaʻena, and Makapuʻu from Oʻahu. Plants from Nihoa (the island being well separated from the main Hawaiian Islands) were well differentiated from the ZOI (Figure 2). The habitat of Nihoa is largely mountain basaltic slopes with no beach habitat.
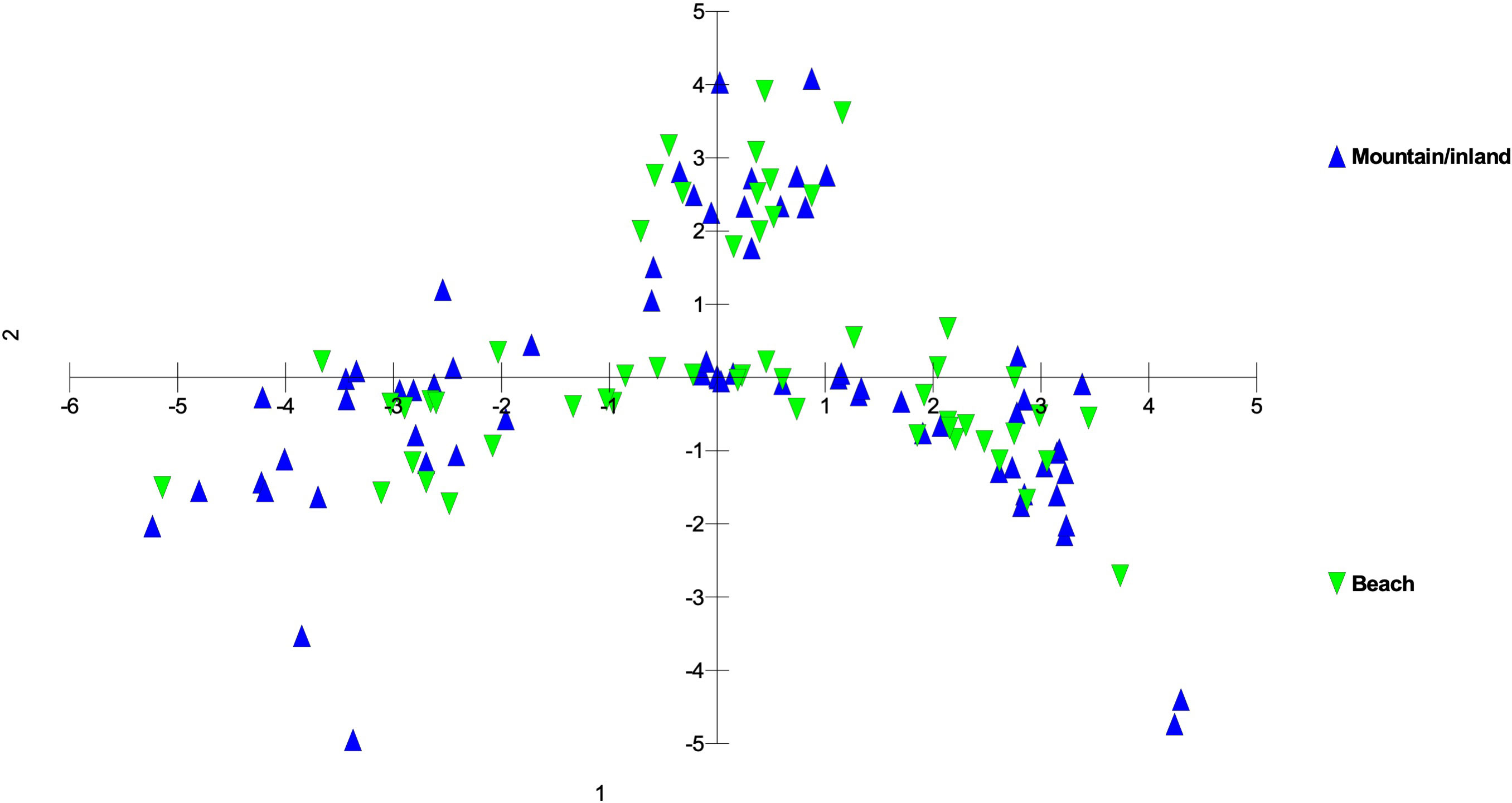
Figure 5 Principal coordinate analysis using MIG-seq data of S. fallax populations in Hawaiian Islands distinguished by habitat - mountain or inland vs. beach ecotypes - based on the same analysis presented in Figure 3. Nihoa plants, although near coastal were considered inland as there is no beach habitat on that island.
The second island group consisted of populations from Maui Nui. All Maui Nui populations were collected from beaches or inland regions close to the beach. A few individuals from Molokaʻi (population 11) were within the ZOI (Figure 4), but most fell outside it. Maui Nui plants form a cohesive group that does not exhibit extensive diversification and does not extend as far from the ZOI as do the other two groups.
The third island group consisted of plants from Hawaiʻi Island. As the largest island, more populations (10) were investigated, throughout the island, than on the other islands. Populations extend from the ZOI, with plants from South Point and Belt Road (populations 17 and 26, respectively) relatively close to the ZOI to populations well separated from the ZOI, notably those from Saddle Road (population 24). There was broad overlap in coastal/beach and mountain/inland populations within the diversity of the Hawaiʻi Island group.
4 Discussion
It is common in the analysis of population variation among species that are distributed throughout the Hawaiian Islands that individuals and populations within an island show a closer genetic relationship to other populations on that island than to populations from other islands. This has been demonstrated repeatedly using other methods for a wide variety of genera including Hibiscus (Malvaceae; Huppman, 2013), Erythrina (Fabaceae; Grave, 2018), and Sesbania (Fabaceae; Cole and Morden, 2021). In this study of Sida fallax, the population organization reflected a Nihoa-Kauaʻi-Oʻahu group, a Maui Nui group, and a Hawaiʻi Island group. Populations within each of these three groups broadly overlapped genetically and were distinct from those in the other two groups, suggesting that each group has retained intra-group connectivity. On closer inspection of the variation in the previous studies noted above it is evident that, although the island populations are distinct, they also are consistent with these same three groups of islands found for Sida fallax. A major distinction in this study is that the three groups are connected (the ZOI) suggesting gene flow still exists among them. This is probably a reflection of Sida fallax having many coastal populations and fruits with indehiscent mericarps capable of long-distance dispersal via ocean currents. These factors allow it to maintain genetic cohesion among populations throughout the Hawaiian Islands and even across the Pacific Ocean (Pejhanmehr, 2022; Pejhanmehr et al., 2022) as has been found in other groups (Baum et al., 2004; Takayama et al., 2008; Areces-Berazain and Ackerman, 2016; Areces-Berazain and Ackerman, 2017).
Within each of the three island groups, populations were largely intermixed genetically and there was no island-by-island separation. There was no clear genetic delineation between beach and mountain/inland ecotypes. However, there was some evidence of more coastal or beach populations being closer to the zone of intersection and inland populations more distant from it, suggesting diversification within each of these groups. However, there was also a large degree of overlap suggesting continuing introgression among the forms of Sida fallax as well as across the habitats. As found by Yorkston and Daehler (2006), there are no fertility barriers among populations and forms of the species. Sida fallax are pollinated by native insects such as yellow-faced bees, Hylaeus spp., and non-native insects such as honeybees, Apis mellifera, and other generalist Hymenoptera, butterflies, and moths (Yorkston and Daehler, 2006; Shay et al., 2016; Shay and Drake, 2018; Aslan et al., 2019). Fruits of other Sida spp. were demonstrated to disperse readily by air or water when mature (Raju and Rani, 2016).
Some populations fell far from the zone of intersection. These included the Nihoa, Makaleha (Kauaʻi), and Saddle Road (Hawaiʻi) mountain ecotype populations, along with some individuals from beach ecotype populations. The mountain ecotypes were more ecologically isolated from other parts of islands so they may have some unique genetic differences. Moreover, the beach ecotypes could receive seeds from other parts of the Pacific via ocean currents and accumulate some unique genetic diversity. This suggests that there is ongoing population diversification, but gene flow among populations is keeping them from fully segregating from others within their groups.
Overall, populations on a single island were more closely related to each other and to populations on islands within their respective groups than they were to populations on other islands. Due to the higher probability of long-distance seed dispersal via ocean currents for beach ecotype populations, they exhibit somewhat higher genetic diversity among themselves rather than mountain ecotype populations that are more isolated (Supplementary Figure 2). The overall genetic relationships among islands were to a large extent predictive based on island position within the chain, and, to a lesser extent, within island topography.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, PRJNA979219.
Ethics statement
Written informed consent was obtained from the individual(s) for the publication of any identifiable images or data included in this article.
Author contributions
MP: Conceptualization, Data curation, Investigation, Writing – original draft, Writing – review & editing. MK: Conceptualization, Formal analysis, Writing – review & editing. MY: Formal analysis, Methodology, Writing – review & editing. CM: Conceptualization, Funding acquisition, Investigation, Project administration, Resources, Supervision, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. Financial support was gratefully provided by UHM School of Life Sciences and Pacific Cooperative Studies Unit (PCSU), and Harold and Elizabeth St. John Scholarship (to MP) from UH Mānoa.
Acknowledgments
We express our gratitude to the following people for helping with this research project: Tom Ranker, Alison Sherwood, and Robert Cowie for all their helpful comments and support, Emiko Oguri for MIG-seq library preparation and sequencing, and Jesse Adams for collecting samples from Kauaʻi island. We thank the following organizations: UH School of Life Sciences, Harold and Elizabeth St. John Scholarship, and Pacific Cooperative Studies Unit (PCSU) for financial support for this project. The content of this manuscript has previously appeared online as PhD dissertation of Mersedeh Pejhanmehr (Pejhanmehr, 2022). This is publication SoLS # 199 from the School of Life Sciences, University of Hawai’i at Mānoa.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2024.1304078/full#supplementary-material
Supplementary Figure 1 | Neighbor Joining tree using MIG-seq data for 124 individuals in 26 populations of Sida fallax across the Hawaiian Islands. Island abbreviations: N, Nihoa; K, Kauaʻi; O, Oʻahu; M, Maui; Mo, Molokaʻi; L, Lānaʻi; H, Hawaiʻi. Population numbers correspond to those in Table 1. Blue color: mountain/inland populations; Green color: beach populations.
Supplementary Figure 2 | Scatter plot showing FST values versus distances between paired populations. Mtn: mountain.
References
Afgan, E., Baker, D., Batut, B., Van Den Beek, M., Bouvier, D., Ech, M., et al. (2018). The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2018 update. Nucleic Acids Res. 46, W537–W544. doi: 10.1093/nar/gky379
Areces-Berazain, F., Ackerman, J. D. (2016). Phylogenetics, delimitation and historical biogeography of the pantropical tree genus Thespesia (Malvaceae, Gossypieae). Bot. J. Linn. Soc. 181, 171–198. doi: 10.1111/boj.12414
Areces-Berazain, F., Ackerman, J. D. (2017). Diversification and fruit evolution in eumalvoids (Malvaceae). Bot. J. Linn. Soc 184, 401–417. doi: 10.1093/botlinnean/box035
Aslan, C. E., Shiels, A. B., Haines, W., Liang, C. T. (2019). Non-native insects dominate daytime pollination in a high-elevation Hawaiian dryland ecosystem. Am. J. Bot. 106, 313–324. doi: 10.1002/ajb2.1233
Baldwin, B. G., Sanderson, M. J. (1998). Age and rate of diversification of the Hawaiian silversword alliance (Compositae). Proc. Natl. Acad. Sci. U.S.A. 95, 9402–9406. doi: 10.1073/pnas.95.16.9402
Baum, D. A., Smith, S. D. W., Yen, A., Alverson, W. S., Nyffeler, R., Whitlock, B. A., et al. (2004). Phylogenetic relationships of Malvatheca (Bombacoideae and Malvoideae; Malvaceae sensu lato) as inferred from plastid DNA sequences. Am. J. Bot. 91, 1863–1871. doi: 10.3732/ajb.91.11.1863
Blankenberg, D., Gordon, A., Von Kuster, G., Coraor, N., Taylor, J., Nekrutenko, A., et al. (2010). Manipulation of FASTQ data with galaxy. Bioinformatics 26, 1783–1785. doi: 10.1093/bioinformatics/btq281
Carlquist, S. (1974). Island biology (New York: Columbia University Press). doi: 10.5962/bhl.title.63768
Carlquist, S., Baldwin, B. G., Carr, G. D. (2003). Tarweeds and silverswords: evolution of the Madiinae (Asteraceae) (Saint Louis: Missouri Botanical Garden Press).
Catchen, J. M., Amores, A., Hohenlohe, P., Cresko, W., Postlethwait, J. H. (2011). Stacks: building and genotyping loci de novo from short-read sequences. G3 Genes Genomes Genet. 1, 171–182. doi: 10.1534/g3.111.000240
Catchen, J., Hohenlohe, P. A., Bassham, S., Amores, A., Cresko, W. A. (2013). Stacks: an analysis tool set for population genomics. Mol. Ecol. 22, 3124–3140. doi: 10.1111/mec.12354
Choi, J. Y., Dai, X., Alam, O., Peng, J. Z., Rughani, P., Hickey, S., et al. (2021). Ancestral polymorphisms shape the adaptive radiation of Metrosideros across the Hawaiian Islands. Proc. Natl. Acad. Sci. U.S.A. 118, 1–10. doi: 10.1073/pnas.2023801118
Cole, D. M., Morden, C. W. (2021). Population divergence and evolution of the Hawaiian endemic Sesbania tomentosa (Fabaceae). Pacific Sci. 75, 447–467. doi: 10.2984/75.4.1
Doyle, J., Doyle, J. (1987). A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull. 19, 11–15.
Dray, S., Dufour, A. B. (2007). The ade4 package: Implementing the duality diagram for ecologists. J. Stat. Software 22, 1–20. doi: 10.18637/jss.v022.i04
Gagné, W. C., Cuddihy, W. L. (1990). “Vegetation,” in Manual of the flowering plants of Hawai’i. Eds. Wagner, W. L., Herbst, D. R., Sohmer, S. H. (University of Hawai’i Press, Honolulu, HI).
Givnish, T. J., Millam, K. C., Mast, A. R., Paterson, T. B., Theim, T. J., Hipp, A. L., et al. (2009). Origin, adaptive radiation and diversification of the Hawaiian lobeliads (Asterales: Campanulaceae). Proc. R. Soc. B. Biol. Sci. 276, 407–416. doi: 10.1098/rspb.2008.1204
Gordon, A. (2010) FASTQ/A short-reads pre-processing tools. Available at: http://hannonlab.cshl.edu/fastx_toolkit/.
Gower, J. C. (1971). A general coefficient of similarity and some of its properties. Biometrics 27, 857–871. doi: 10.2307/2528823
Grave, E. F. (2018). Reproductive ecology and population genetics of Hawaiian wiliwili, Erythrina sandwicensis (Fabaceae) (Honolulu: University of Hawai‘i at Mānoa).
Handy, E. S. C., Handy, E. G. (1991). Native planters in old Hawaii – their life, lore, and environment (Honolulu: Bishop Museum Press).
Huppman, E. R. H. (2013). Analysis of relationships among endemic Hawaiian Hibiscus (Honolulu: University of Hawai‘i at Mānoa).
Johnson, M. A., Pillon, Y., Sakishima, T., Price, D. K., Stacy, E. A. (2019). Multiple colonizations, hybridization and uneven diversification in Cyrtandra (Gesneriaceae) lineages on Hawai’i Island. J. Biogeogr. 46, 1178–1196. doi: 10.1111/jbi.13567
Keeley, S. C., Funk, V. A. (2011). “Origin and evolution of Hawaiian endemics: new patterns revealed by molecular phylogenetic studies,” The biology of island floras. Ed. Bramwell, D. (Cambridge: Cambridge University Press).
Knope, M. L., Funk, V. A., Johnson, M. A., Wagner, W. L., Datlof, E. M., Johnson, G., et al. (2020). Dispersal and adaptive radiation of Bidens (Compositae) across the remote archipelagoes of Polynesia. J. Syst. Evol. 58, 805–822. doi: 10.1111/jse.12704
Knope, M. L., Morden, C. W., Funk, V. A., Fukami, T. (2012). Area and the rapid radiation of Hawaiian Bidens (Asteraceae). J. Biogeogr. 39, 1206–1216. doi: 10.1111/j.1365-2699.2012.02687.x
Korecký, J., Čepl, J., Stejskal, J., Faltinová, Z., Dvořák, J., Lstibůrek, M., et al. (2021). Genetic diversity of Norway spruce ecotypes assessed by GBS-derived SNPs. Sci. Rep. 11, 1–12. doi: 10.1038/s41598-021-02545-z
Kovach (2007). Kovach computing services 1987-99. Multi variate statistical package, V. 3.0 (Pentraeth, Wales: Kovach Computing Services).
Landis, M. J., Freyman, W. A., Baldwin, B. G. (2018). Retracing the Hawaiian silversword radiation despite phylogenetic, biogeographic, and paleogeographic uncertainty. Evol. (N. Y). 72, 2343–2359. doi: 10.1111/evo.13594
Lindqvist, C., Motley, T. J., Jeffery, J. J., Albert, V. A. (2003). Cladogenesis and reticulation in the Hawaiian endemic mints (Lamiaceae). Cladistics 19, 480–495. doi: 10.1111/j.1096-0031.2003.tb00384.x
Martin, M. (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 17, 10. doi: 10.14806/ej.17.1
Meirmans, P. G., Van Tienderen, P. H. (2004). GENOTYPE and GENODIVE: Two programs for the analysis of genetic diversity of asexual organisms. Mol. Ecol. Notes 4, 792–794. doi: 10.1111/j.1471-8286.2004.00770.x
Morden, C. W., Caraway, V., Motley, T. J. (1996). Development of a DNA library for native Hawaiian plants. Pacific Sci. 50, 324–335. Available at: http://hdl.handle.net/10125/2904
Nei, M., Li, W.-H. (1979). Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc. Natl. Acad. Sci. U.S.A. 76, 5269–5273. doi: 10.1073/pnas.76.10.5269
Pejhanmehr, M. (2022). Phylogenetic affinities among Sida species and allied genera (Malvaceae: Malveae), and examination of Sida fallax within the Hawaiian Islands and throughout the Pacific (Honolulu: University of Hawai‘i at Mānoa).
Pejhanmehr, M., Yorkston, M., Morden, C. W. (2022). Low genetic diversity in the highly morphologically diverse Sida fallax walp. (Malvaceae) throughout the pacific. Pacific Sci. 76, 361–376. doi: 10.2984/76.4.2
Percy, D. M., Garver, A. M., Wagner, W. L., James, H. F., Cunningham, C. W., Miller, S. E., et al. (2008). Progressive island colonization and ancient origin of Hawaiian Metrosideros (Myrtaceae). Proc. R. Soc B. 275, 1479–1490. doi: 10.1098/rspb.2008.0191
Price, J. P., Wagner, W. L. (2004). Speciation in Hawaiian angiosperm lineages: cause, consequence, and mode. Evol. (N. Y). 58, 2185–2200. doi: 10.1111/j.0014-3820.2004.tb01597.x
Raju, A. J. S., Rani, D. S. (2016). Pollination ecology of Sida acuta, S. cordata and S. cordifolia (Malvaceae). Phytol. Balc. 22, 363–376.
R Core Team. (2021). R: A language and environment for statistical computing. R foundation for statistical computing. Available at: https://www.R-project.org/.
Sakai, A. K., Weller, S. G., Wagner, W. L., Nepokroeff, M., Culley, T. M. (2006). Adaptive radiation and evolution of breeding systems in Schiedea (Caryophyllaceae), an endemic Hawaiian genus. Ann. Missouri Bot. Gard. 93, 49–63. doi: 10.3417/0026-6493(2006)93[49:ARAEOB]2.0.CO;2
Schluter, D. (2000). The ecology of adaptive radiation (Oxford: Oxford University Press). doi: 10.1093/oso/9780198505235.001.0001
Shay, K. R., Drake, D. R. (2018). Pollination biology of the Hawaiian coastal vine Jacquemontia sandwicensis (Convolvulaceae). Pacific Sci. 72, 485–499. doi: 10.2984/72.4.8
Shay, K., Drake, D. R., Taylor, A. D., Sahli, H. F., Euaparadorn, M., Akamine, M., et al. (2016). Alien insects dominate the plant-pollinator network of a Hawaiian coastal ecosystem. Pacific Sci. 70, 409–429. doi: 10.2984/70.4.2
Sohmer, S. H., Gustafson, R. (1987). Plants and flowers of Hawai’i (Honolulu: University of Hawaii Press).
Stephens, M. L. (2000). The comparative ecophysiology of mountain and coastal populations of Sida fallax Walp. (Malvaceae) in Hawaii (Honolulu: University of Hawai‘i at Mānoa).
Subramanian, S., Ramasamy, U., Chen, D. (2019). VCF2PopTree: a client-sidesoftware to construct population phylogeny from genome-wide SNPs. PeerJ 7, e8213. doi: 10.7717/peerj.8213
Suyama, Y., Matsuki, Y. (2015). MIG-seq: An effective PCR-based method for genome-wide single-nucleotide polymorphism genotyping using the next-generation sequencing platform. Sci. Rep. 5, 1–12. doi: 10.1038/srep16963
Takayama, K., Crawford, D. J., López-Sepúlveda, P., Greimler, J., Stuessy, T. F. (2018). Factors driving adaptive radiation in plants of oceanic islands: a case study from the Juan Fernández Archipelago. J. Plant Res. 131, 469–485. doi: 10.1007/s10265-018-1023-z
Takayama, K., Tateishi, Y., Murata, J., Kajita, T. (2008). Gene flow and population subdivision in a pantropical plant with sea-drifted seeds Hibiscus tiliaceus and its allied species: evidence from microsatellite analyses. Mol. Ecol. 17, 2730–2742. doi: 10.1111/j.1365-294X.2008.03799.x
Thiers, B. M. (2021). Index Herbariorum. Available at: https://sweetgum.nybg.org/science/ih/.
Triplett, G., Buckley, T. N., Muir, C. D. (2024). Amphistomy increases leaf photosynthesis more in coastal than montane plants of Hawaiian ‘ilima (Sida fallax). Am. J. Bot. 111 (2), e16284. doi: 10.1002/ajb2.16284
Wagner, W. L., Funk, V. A. (1995). Hawaiian biogeography: evolution on a hot spot archipelago (Washington, D.C: Smithsonian Institution Press).
Wagner, W. L., Herbst, D. R., Sohmer, S. H. (1999). Manual of the Flowering Plants of Hawaiʻi, revised edition (Honolulu, Hawaii, USA: University of Hawaiʻi Press).
Westerbergh, A., Saura, A. (1994). Genetic differentiation in endemic Silene (Caryophyllaceae) on the Hawaiian Islands. Am. J. Bot. 81, 1487–1493. doi: 10.1002/j.1537-2197.1994.tb15634.x
Wickham, H. (2016). ggplot2: elegant graphics for data analysis (New York: Springer-Verlag). doi: 10.1007/978-3-319-24277-4
Yorkston, M., Daehler, C. C. (2006). Interfertility between Hawaiian ecotypes of Sida fallax (Malvaceae) and evidence of a hybrid disadvantage. Int. J. Plant Sci. 167, 221–230. doi: 10.1086/499615
Keywords: Sida fallax, population genetics, Hawaiian Islands, MIG-seq, ecotypes
Citation: Pejhanmehr M, Kantar MB, Yorkston M and Morden CW (2024) Population genetics of Sida fallax Walp. (Malvaceae) in the Hawaiian Islands. Front. Plant Sci. 15:1304078. doi: 10.3389/fpls.2024.1304078
Received: 28 September 2023; Accepted: 12 February 2024;
Published: 01 March 2024.
Edited by:
Shu-Miaw Chaw, Academia Sinica, TaiwanReviewed by:
Rob Smissen, Manaaki Whenua Landcare Research, New ZealandManosh Kumar Biswas, University of Leicester, United Kingdom
Copyright © 2024 Pejhanmehr, Kantar, Yorkston and Morden. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Clifford W. Morden, Y21vcmRlbkBoYXdhaWkuZWR1
†Present address: Mersedeh Pejhanmehr, Department of Plant Biology, Carnegie Institution for Science, Stanford, CA, United States
 Mersedeh Pejhanmehr
Mersedeh Pejhanmehr Michael Benjamin Kantar
Michael Benjamin Kantar Mitsuko Yorkston1
Mitsuko Yorkston1 Clifford W. Morden
Clifford W. Morden