
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 31 January 2024
Sec. Plant Abiotic Stress
Volume 15 - 2024 | https://doi.org/10.3389/fpls.2024.1285879
This article is part of the Research TopicDeciphering Mechanisms of Plant Adaptation and Resistance Under Cold Temperature StressView all 8 articles
Tetrastigma hemsleyanum is a valuable herb widely used in Chinese traditional and modern medicine. Winter cold severely limits the artificial cultivation of this plant, but the physiological and molecular mechanisms upon exposure to cold stress in T. hemsleyanum are unclear. T. hemsleyanum plants with different geographical origins exhibit large differences in response to cold stress. In this research study, using T. hemsleyanum ecotypes that exhibit frost tolerance (FR) and frost sensitivity (FS), we analyzed the response of cottage seedlings to a simulated frost treatment; plant hormones were induced with both short (2 h) and long (9 h) frost treatments, which were used to construct the full-length transcriptome and obtained 76,750 transcripts with all transcripts mapped to 28,805 genes, and 27,215 genes, respectively, annotated to databases. Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis showed enrichment in plant hormone signaling pathways. Further analysis shows that differently expressed genes (DEGs) concentrated on calcium signaling, ABA biosynthesis and signal transduction, and ethylene in response to cold stress. We also found that endogenous ABA and ethylene content were increased after cold treatment, and exogenous ABA and ethylene significantly improved cold tolerance in both ecotypes. Our results elucidated the pivotal role of ABA and ethylene in response to cold stress in T. hemsleyanum and identified key genes.
Tetrastigma hemsleyanum Diels et Gilg is an herbaceous climber that is widely distributed in tropical and subtropical regions, mainly in provinces of south and southwest China (Wang et al., 2015). In China, T. hemsleyanum is used as traditional medicine “San ye qing,” a broad-spectrum antibiotic material against fever and inflammation (Li, 1998). It has also exhibited antioxidant, antivirus, antitumor, immunomodulatory, and hypoglycemic effects in modern pharmacological research studies (Ji et al., 2021). Because of its slow growth, it usually takes 3–5 years to meet the requirements of commercial medicinal materials, so it is a precious perennial medicinal resource. Due to overexploitation, wild resources have been on the verge of extinction in recent years (Peng et al., 2019) and the supply of medicinal herbs is largely dependent on cultivation. For synthesized utilization of land and imitating wild conditions to improve the quality, artificial T. hemsleyanum is usually planted in the mountains, causing the herb to suffer from winter cold frequently.
Cold stress is a major environmental factor that limits agricultural production (Einset et al., 2007). Low temperatures limit CO2 fixation coupled with an overreduction in the electron transport chain, leading to photosynthesis suppression and dramatic increases in reactive oxygen species (ROS) (Maleki and Ghorbanpour, 2018). In response to abiotic stresses, plants have evolved highly complex adaptive mechanisms. Once the receptor is triggered by cold stimulation, a signal is spread by transduction pathways, and induced gene-expression changes subsequently cause physiological changes, including an increase in antioxidant levels, induction of influx of cellular calcium ions, alteration in membrane lipid composition, adjustment of hormone levels, and changes in electrolyte leakage and soluble proteins.
Low temperatures, especially winter minimum temperatures, are one of the primary forces determining the geographic boundaries of many plant species (Pither, 2003), but physiological tolerance to cold also varies among populations within a species (Armstrong et al., 2020). T. hemsleyanum plants with different geographical origins contained enormous genetic variability (Peng et al., 2015), which caused different morphological characteristics and environmental adaptability. It provides convenience for ascertaining the cold response and regulatory networks of cold tolerance in T. hemsleyanum.
Short-read RNA-Seq (mainly using Illumina technology) has been used for over a decade; however, Illumina sequencers are appropriate only for short-read-length sequencing because of information loss caused by fragments during sample preparation and spliced after sequencing (Cui et al., 2020). Nanopore sequencing technology is a new approach that can sequence single long DNA and RNA molecules (Deamer et al., 2016; Wang Y. H. et al., 2021). Since the first nanopore sequencer, MinION, was provided by Oxford Nanopore Technologies (ONT), it has been used extensively to assemble the initial reference genomes of many non-model organisms (Scheunert et al., 2020), but only several researchers in transcriptomics have explored the molecular mechanisms in plants to date, such as in Arabidopsis (Cui et al., 2020; Parker et al., 2020; Qin et al., 2022), rose (Li et al., 2021), peach (Liu et al., 2021), quinoa (Zheng et al., 2022), and potatoes (Xiong et al., 2022).
In this research study, we monitored the phenotypes of T. hemsleyanum seedlings at their preferred temperature, 2 h and 9 h frost stress, and sequenced their full-length transcripts using the ONT method. As a result, we identified numerous genes that may take part in the cold stress response. These genes are involved in hormone and signal transduction pathways, especially the biosynthesis and signal transduction of ABA and ethylene. Endogenous ABA and ethylene contents were increased after cold treatment, and exogenous ABA and ethylene significantly improved cold tolerance. These results indicated the pivotal role of ABA and ethylene in the cold stress response of T. hemsleyanum. This study provides insight into the cold stress response pathway in T. hemsleyanum and thus provides a basis for improving the resistance of this cash crop to cope with increasingly frequent cold waves in the major grain-producing area south of the Yangtze River in China.
T. hemsleyanum collected from different geographical areas exhibits various cold tolerances. When exposed to −3°C, the leaves of sensitive T. hemsleyanum ecotype seedlings exhibited severe damage; most sensitive ecotype seedlings died after exposure to −4°C for 12 h (Figure 1). It is difficult to take typical tissue for further analysis. Therefore, in this study, cottage seedlings of T. hemsleyanum ecotypes that exhibited frost tolerance (FR) and frost sensitivity (FS) were selected and exposed to a simulated frost treatment (−2°C) for analysis of physiological indicators and gene expression. In contrast, a −4°C simulated frost treatment was performed to evaluate the cold tolerance survival rate.
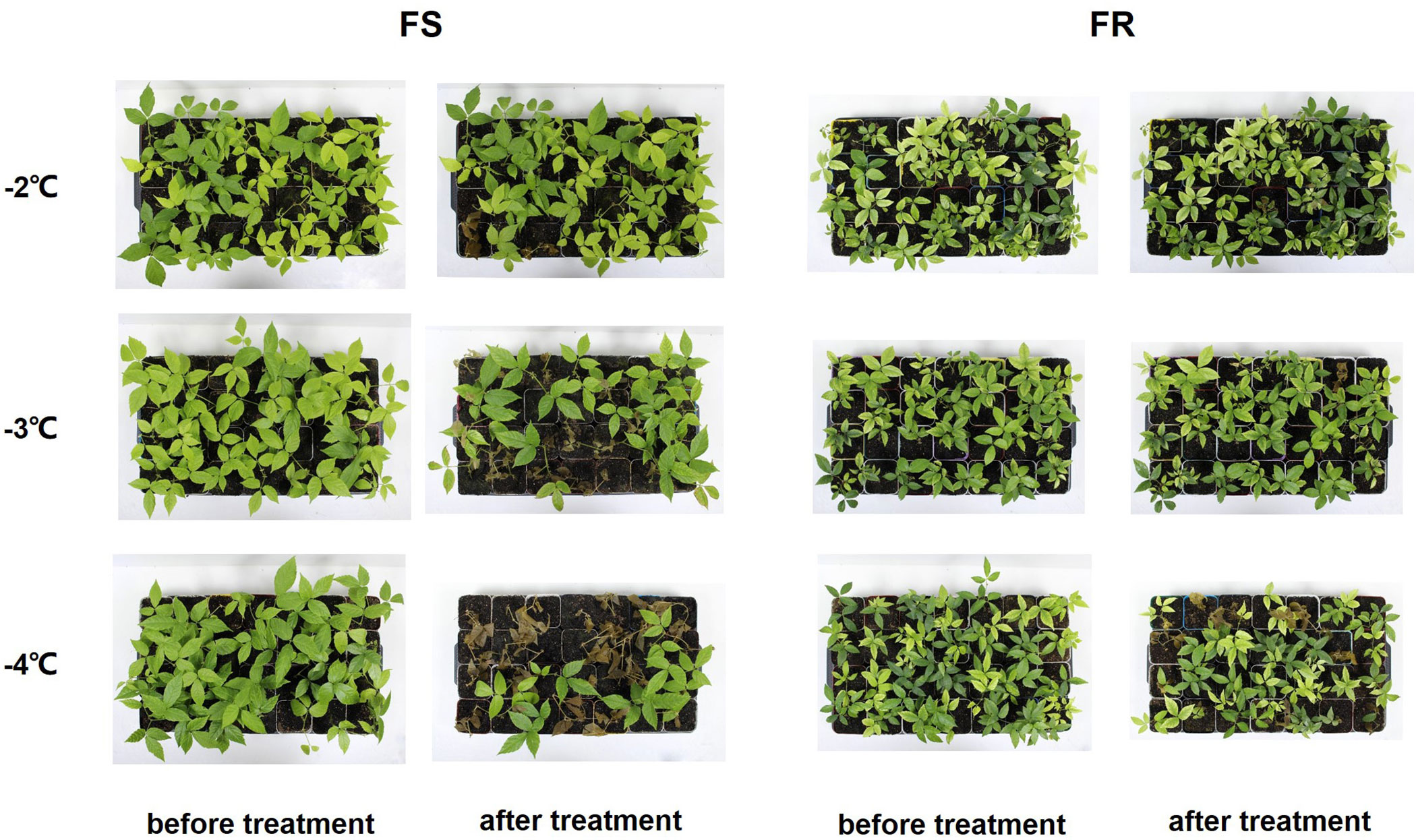
Figure 1 Cold injury in different T. hemsleyanum cottage ecotypes. Representative images of 2-month-old cottage seedlings of FR and FS grown under control (CK) and exposed to a simulated frost treatment for 12 h (T) were exhibited to demonstrate the difference in frost tolerance between the two ecotypes.
To investigate the molecular mechanisms underlying the frost response, transcriptome sequencing was performed on the leaves of T. hemsleyanum ecotype seedling growth at normal temperature (25°C, CK), exposed to frost conditions (−2°C) for a short time (2 h) and a long time (9 h).
A total of 18 libraries from six samples (three biological replicates per sample) were obtained. The clean data output from each sample was at least 6 GB, and 89%~92% of clean reads, namely, 5.40–8.95 million, were full-length reads. After mapping to the T. hemsleyanum genome, 4.83~7.94 million reads were mapped, and mapped rates in all libraries exceeded 97% (Supplementary Table 1).
A total of 76,750 transcripts were obtained, and 53,190 transcripts were newly found. A total of 28,805 transcripts were mapped to existing genes, and 4,121 were new genes (Figure 2A). A total of 27,215 genes were annotated in databases, and the percentage that was annotated was 94.5% (Figure 2A).
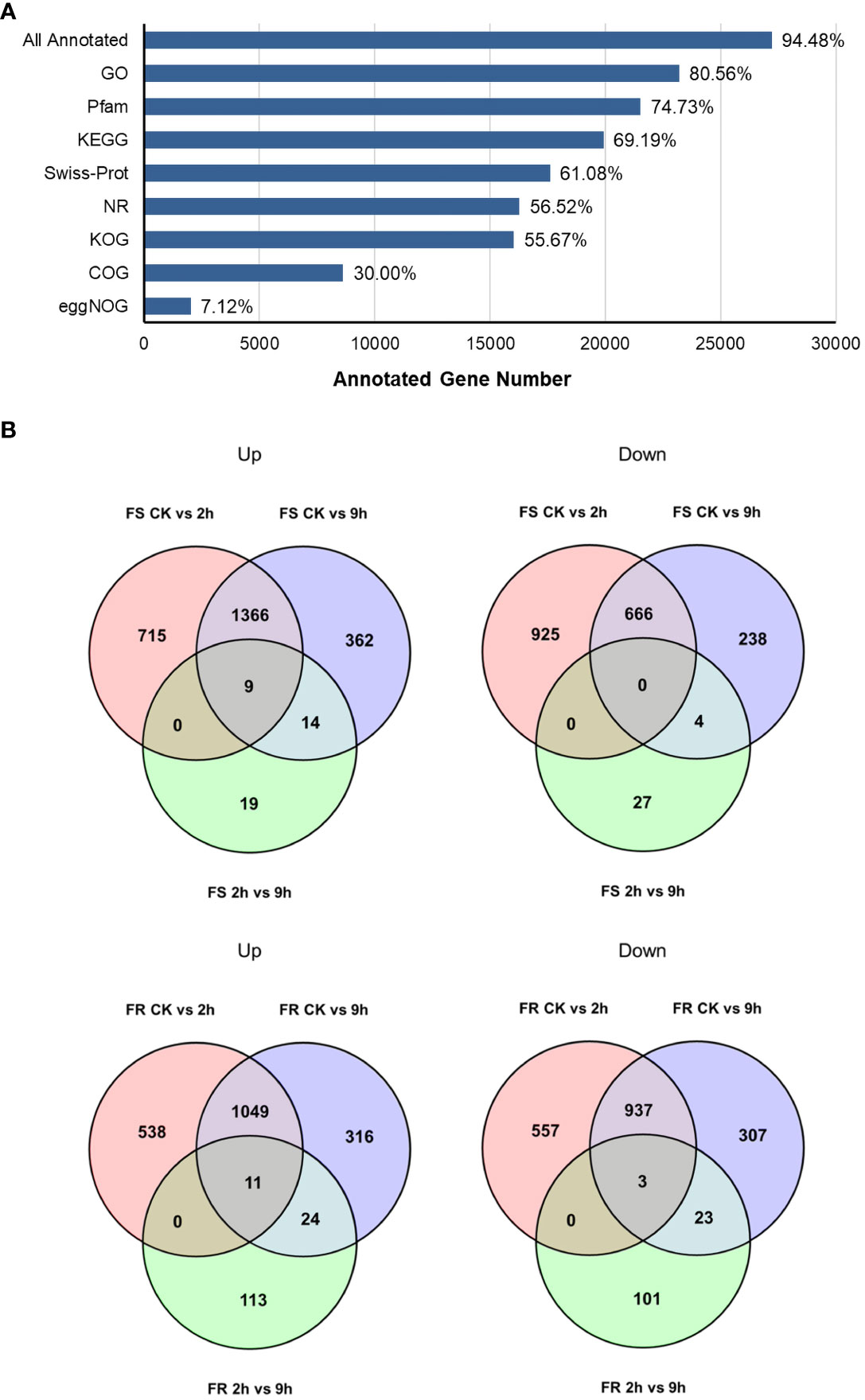
Figure 2 DEGs in T. hemsleyanum in response to cold stress. (A) Functional annotations of all genes in the indicated databases. (B) The number of DEGs induced and reduced by cold.
To further elucidate the changes regulated by cold stress in two T. hemsleyanum ecotypes with different cold tolerances, we analyzed the differently expressed genes (DEGs) in FS and FR after cold treatment for 2 h and 9 h relative to CK, respectively, and DEGs in cold treatment for 9 h relative to 2 h. DEGs were identified based on a false discovery rate (FDR) ≤0.05 and fold change ≥2. A total of 6,086 DEGs were detected in all the 18 cDNA libraries: 3,683 genes were differentially expressed after 2 h and 3,095 after 9 h of cold stress exposure in FS, and 2,659 genes were differentially expressed after 2 h and 2,670 after 9 h of cold stress exposure in FR (Figure 2B). Generally speaking, the 9-h-simulated frost treatment resulted in more DEGs, but most of these were already present after 2 h, with only 73 DEGs in FS and 275 DEGs in FR differentially expressed after 9 h of cold treatment relative to 2 h, which may suggest a rapid response of transcriptomic changes, which already happened a short time after applying cold stress. There were more DEGs in FS than in FR compared with their respective controls, indicating that some adaptive traits already exist in the more cold-tolerant FR at moderate temperature conditions, resulting in FR being less affected and the sensitive variety FS being more affected by cold stress.
KOBAS 2.0 software was used for the Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis of DEGs in 2-h and 9-h treatment samples relative to controls in both ecotypes. DEGs were annotated to 134 KEGG reference pathways (Figure 3A), 26 KEGG pathways induced by cold were predominantly significantly enriched in at least one dataset, and 58 KEGG pathways suppressed by cold were predominantly significantly enriched in at least one dataset (Supplementary Figure 1).
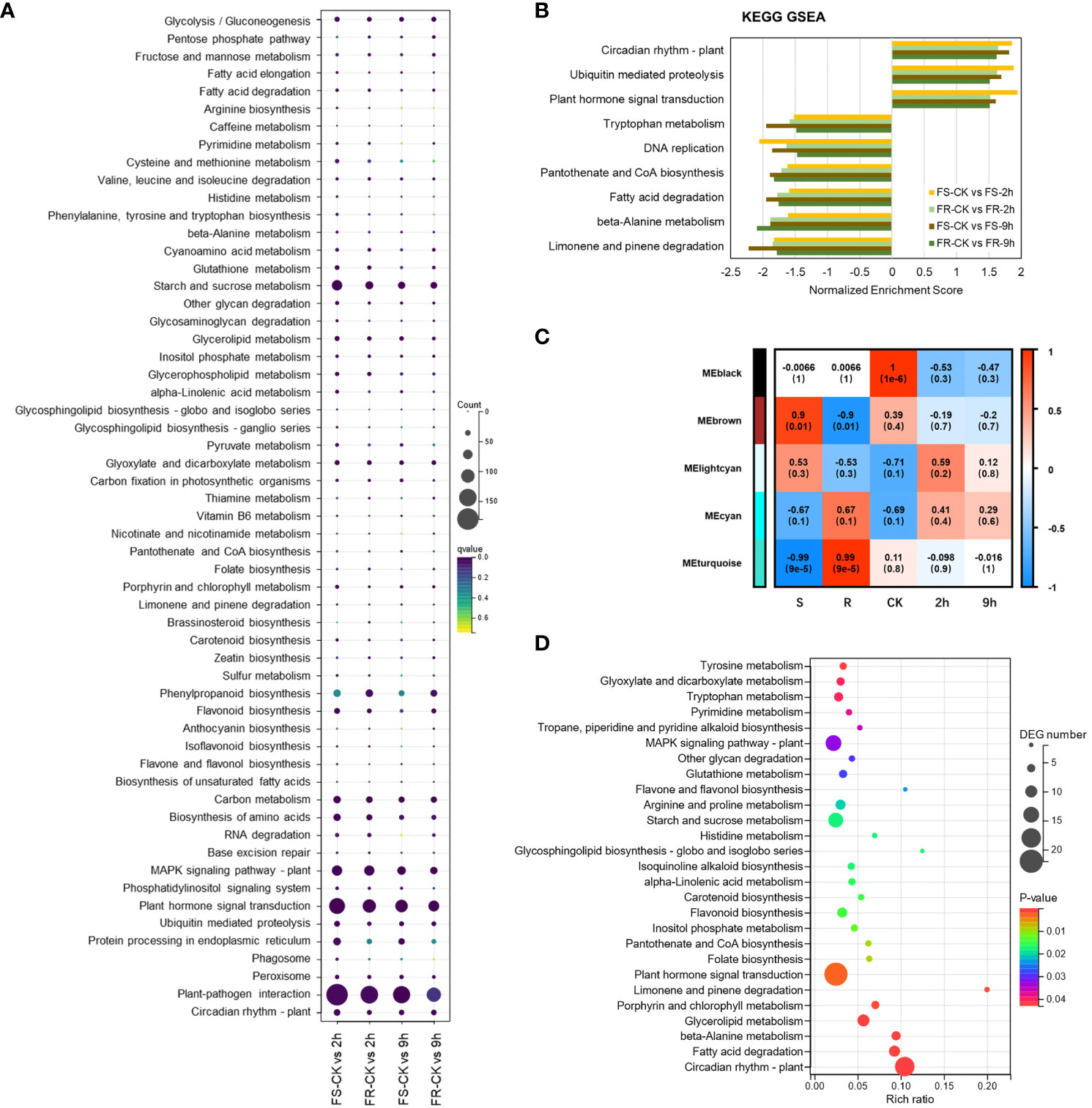
Figure 3 KEGG clustering analysis of DEGs. (A) KEGG pathway enrichment analysis based on DEGs. (B) KEGG pathway enrichment analysis by GSEA. (C) Relationships between module eigengenes and external traits in WGCNA. Each row represents a consensus module, and each column represents a specific trait. The numbers in each grid represent the correlation coefficients between module eigengenes and corresponding traits, with P values indicated in parentheses below. (D) KEGG pathway enrichment analysis based on DEGs in the black module (P < 0.05).
DEGs participating in inositol phosphate metabolism (ko00562), glyoxylate and dicarboxylate metabolism (ko00630), MAPK signaling pathway (ko04016), plant hormone signal transduction (ko04075), circadian rhythm (ko04712), and ubiquitin-mediated proteolysis (ko04120) were predominantly upregulated, and fatty acid degradation (ko00071), starch and sucrose metabolism (ko00500), other glycan degradation (ko00511), limonene and pinene degradation (ko00903), base excision repair (ko03410), beta-alanine metabolism (ko00410), pantothenate and CoA biosynthesis (ko00770), and peroxisome biosynthesis (ko04146) were predominantly downregulated after 2 h and 9 h of cold stress in both ecotypes.
To further elucidate the cold-induced transcriptomic variations, gene set enrichment analyses (GSEAs) were performed (Figure 3B). Plant hormone signal transduction (ko04075), circadian rhythm (ko04712), and ubiquitin-mediated proteolysis (ko04120) pathways were also induced. At the same time, fatty acid degradation (ko00071), limonene and pinene degradation (ko00903), beta-alanine metabolism (ko00410), pantothenate and CoA biosynthesis (ko00770), DNA replication (ko03030), and tryptophan metabolism (ko00380) were suppressed significantly in both ecotypes after the 2-h and 9-h treatments. It indicated that these pathways might be central mechanisms in response to low-temperature stress.
To better understand the correlation between these DEGs and the variation in the cold stress response, we performed a weighted correlation network analysis (WGCNA). Co-expression modules were generated by linking to the traits, including ecotypes and cold treatments. A total of five modules were generated (Figure 3C), and consistent with the pairwise comparison, modules colored black showed a significant correlation with normal atmospheric temperature, DEGs in black module obviously enrichment in plant hormone signal transduction (ko04075) (Figure 3D), but no modules correlating with the 2-h or 9-h cold treatments were found.
Changes in membrane fluidity are widely considered to be involved in temperature sensing. Both cold and heat alter cellular membrane fluidity, which can affect the structure and/or activity of membrane-localized proteins such as Ca2+ channels, thereby triggering Ca2+ influx, a crucial process for inducing temperature-responsive gene expression.
In this study, we found six genes encoding calmodulin proteins, and four were induced by cold (Figure 4). There were two upregulated calmodulin proteins highly expressed; 1,621 counts of The01G014180 were detected in samples at normal temperature in FS, significantly upregulated by 6.9-fold (11,123 counts) in the 2-h cold treatment and by 4.5-fold (7,294 counts) in the 9-h cold treatment. A total of 209 counts of ONT.15400 were detected in CK, upregulated by 20.7-fold (43,293 counts) in the 2-h cold treatment and 31.2-fold (6,542 counts) in the 9-h cold treatment in FS. This may cause OXI1 and three out of four RbohD genes to become upregulated in response to cold stress, especially OXI1 (The14G001130) upregulated 6.5-fold after 2 h and 17.8-fold after 9 h of cold treatments in FS. These may be key genes in the response to cold stress.
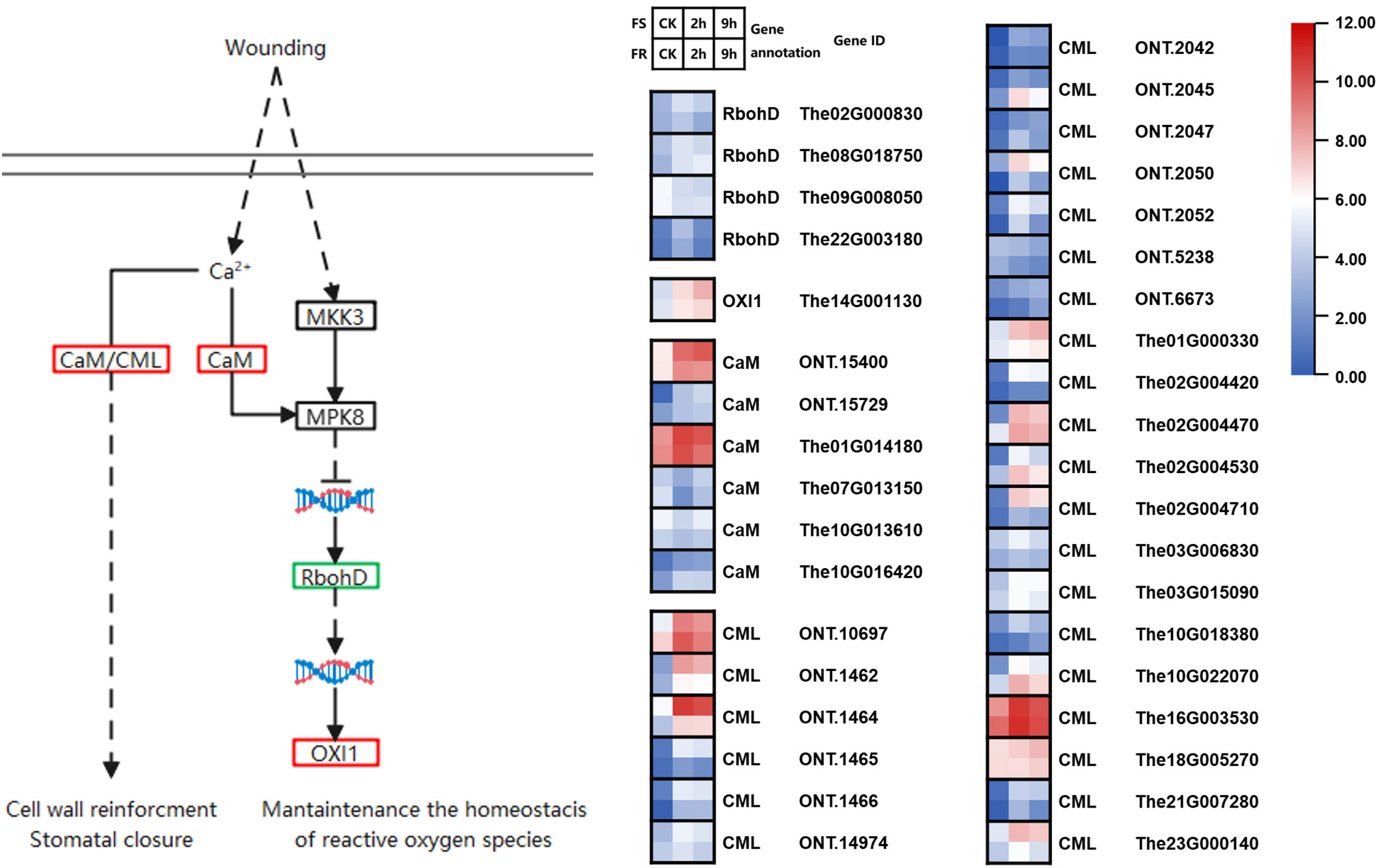
Figure 4 Cold-responsive DEGs involved in temperature sensing and calcium signal pathways. Each colored cell in the heatmap represents log2 of the DEG CPM (mean of three biological replicates); red represents induced genes, and blue represents repressed genes. A schematic diagram of signaling pathway genes; red boxes represent all annotated genes that were upregulated, blue boxes represent all annotated genes that were downregulated, green boxes represent both up and downregulated genes found in this location, and black boxes represent no DEGs found in this location.
These key genes were also induced in FR but slightly less than that in FS. A total of 2,034 counts of The01G014180 were detected in CK samples, but it was upregulated by 4.3-fold (8,741 counts) in the 2-h cold treatment and by 1.86-fold (3,780 counts) in the 9-h cold treatment in FR. A total of 227 counts of The01G014180 were detected in CK samples, upregulated by 8.73-fold (1,985 counts) in the 2-h cold treatment and by 7.36-fold (1,673 counts) in the 9-h cold treatment in FR. The expression of ONT.15400 was 2.18- and 3.91-fold in FS compared with FR after 2-h and 9-h cold treatments. The same expression pattern also happened in OXI1 (The14G001130).
In addition, 26 genes encoding calcium-binding protein (CML) were found, and 25 were induced by cold exposure. Most expressed lower, but three highly upregulated CML expression. Among them, ONT.10697 and The16G003530 were expressed more in FR than in FS at normal temperature and after cold stress, whereas ONT.1464 was expressed more in FS. CML was found to regulate stress response through the nitrogen monoxide (NO) signaling pathway; these three CMLs may be key genes as well.
Abscisic acid (ABA) is a vital phytohormone that plays a key role in plant stress responses. In this research study, DEGs were significantly clustered in plant hormone signal transduction, whereas many DEGs in ABA biosynthesis were upregulated; we speculated that ABA might be related to the T. hemsleyanum cold response.
There were 11 DEGs annotated to six KEGG catalogs in the ABA synthesis pathway, including protein lutein deficient 5 (LUT5, ONT.690), β-carotene 3-hydroxylase (crtZ, The13G012840), zeaxanthin epoxidase (ZEP, ONT.20306, The02G023090, and The05G017100), 9-cis-epoxycarotenoid dioxygenase (NCED, The08G020520, The15G003070, and The15G003170), ABA DEFICIENT 2 (ABA2, The01G012390, and The20G009070), and abscisic-aldehyde oxidase (AAO3, ONT.11866). Most DEG levels were low or downregulated by cold, including three NCEDs, which is the committing step in ABA synthesis; for instance, around 179 counts of The15G003070 (NCED) were detected in FS and 668 in FR at normal temperature, which was 3.7-fold that in FS, it was downregulated by 13.8-fold after 2 h and 7.2-fold after 9 h of cold stress in FS, and it was downregulated by 21.7-fold after 2 h and 20.2-fold after 9 h of cold stress in FR. However, the crtZ gene (The13G012840) and one ZEP gene (The02G023090) were highly expressed and upregulated by cold (Figure 5A).
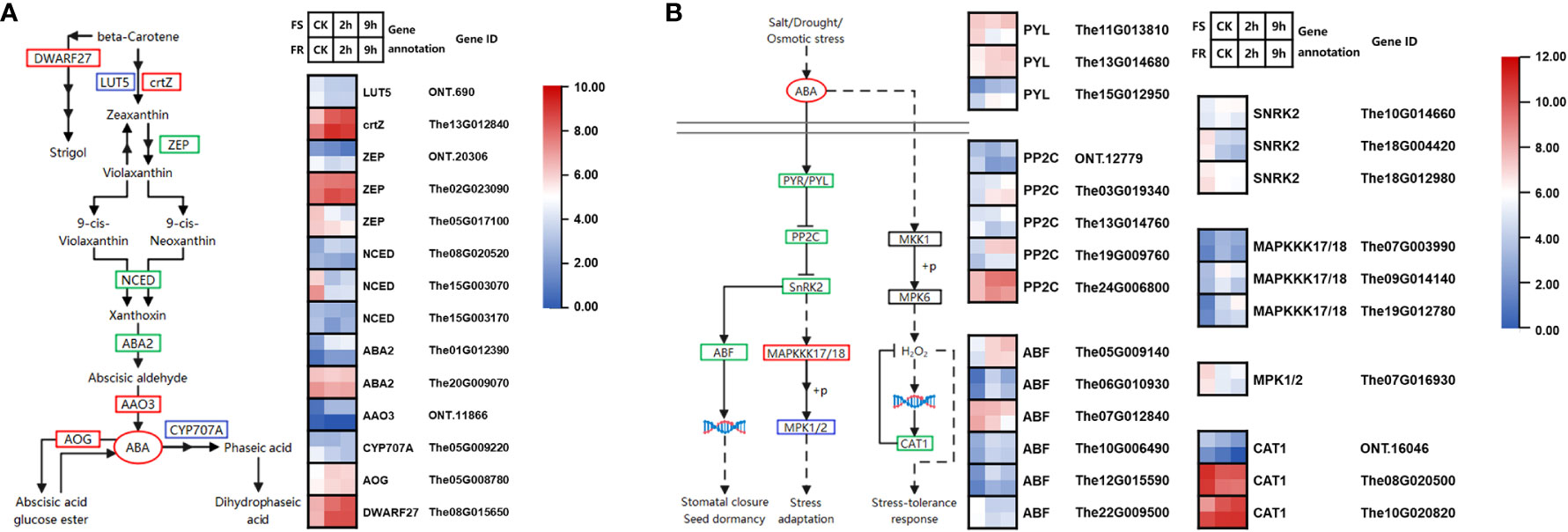
Figure 5 Cold-responsive DEGs regulate ABA biosynthesis and signaling pathways. (A) Cold-responsive DEGs in ABA biosynthesis. (B) Cold-responsive DEGs in the ABA signaling pathway. Each colored cell in the heatmap represents log2 of the DEG CPM (mean of three biological replicates); red represents induced genes, and blue represents repressed genes. A schematic diagram of signaling pathway genes; red boxes represent all annotated genes that were upregulated, blue boxes represent all annotated genes that were downregulated, green boxes represent both up and downregulated genes found in this location, and black boxes represent no DEGs found in this location.
Around 1,000 counts of The02G023090 (ZEP) were detected in both ecotypes at normal temperatures. It was upregulated by 2.5-fold after 2 h and 2.0-fold after 9 h of cold stress in FR but not significantly upregulated in FS. As for The13G012840 (crtZ), 1,130 counts were detected in FR at normal temperature, which was 4.7-fold that in FS. The expression of The13G012840 was upregulated to 5,212 at 2 h and 3,977 at 9 h after cold treatments in FR, whereas it was also upregulated to 2,011 at 2 h and 2,752 at 9 h after cold treatments in FS. The13G012840 and The02G023090 may be key genes leading to the difference in cold tolerance between FR and FS.
CYP707A gene in the downstream ABA synthesis pathway transforms ABA to inactive metabolites such as phasic acid and dihydrophaseic acid; the CYP707A gene (The05G009220) was suppressed by cold in FR, whereas abscisate beta-glucosyltransferase (AOG), which controls ABA glycosylation for storage, was upregulated in both FS and FR, so that available ABA in plants increased.
In addition, the expression of DWARF27 (The08G015650), which controls the biosynthesis of strigol, was significantly upregulated and highly expressed. Strigol promotes the growth and nutrient uptake efficiency of inter- and endosymbiotic microorganisms, which may enhance the cold tolerance of plants in another way.
We found 17 DEGs annotated to four KEGG catalogs in the ABA signaling pathways (Figure 5B). Protein phosphatase 2C (PP2Cs) negatively regulated downstream genes, and ABA binds to PYRABACTIN RESISTANCE 1 (PYR1), which in turn binds to and inhibits PP2Cs. Two DEGs encoding PYR/PYL (The13G014680 and The15G012950) and three DEGs encoding PP2C (The03G019340, The19G009760, and The24G006800) were upregulated by cold, especially The24G006800 which was highly expressed and expressed more in FS, which may be the most critical DEG in the ABA signaling pathway.
There were two SUCROSE NONFERMENTING 1-RELATED PROTEIN KINASE 2 genes (SnRK2, The18G004420, and The18G012980) downregulated by cold, but finally, four ABA-RESPONSIVE ELEMENT BINDING PROTEIN genes (ABF, The05G009140, The06G010930, The10G006490, and The12G015590) were upregulated. In addition, mitogen-activated protein kinase kinase kinase 17 (MAPKKK17) was induced by SnRK2 and positively regulated stress adaption; in this research study, three MAPKKK17 (The07G003990, The09G014140, and The19G012780) were also upregulated by cold.
ABA can also regulate stress tolerance response through the MAPK pathway. There were two DEGs encoding CAT that were highly expressed and regulated by cold; the expression of The08G020500 was suppressed, whereas The10G020820 was induced by 4.4-fold at 2 h and 6.87 at 9 h in FS, whereas its expression was 5.7-fold in FR compared with FS; after cold stress, it reached more than 10,000 counts.
The precursor of ethylene is produced from the methionine salvage pathway. Peroxides produced by cellular damage during stress were sensed. The signal was transduced to upregulate ethylene biosynthesis through the MAPK pathway. Two DEGs encoding mitogen-activated protein kinase (MPK3, ONT.16948, and The19G006960) were upregulated by cold; the expression of The19G006960 was 3.7-fold higher in FR than in FS, and it was upregulated by 9.5-fold at 2 h and 6.4-fold at 9 h after cold treatment in FS. In contrast, it was upregulated by 2.8-fold at 2 h and 1.5-fold at 9 h after cold treatment in FR.
We found 19 DEGs annotated to 10 KEGG catalogs in the methionine salvage pathway; six KEGG catalogues were upregulated, namely, tyrosine aminotransferase (TAT), S-adenosylmethionine decarboxylase (speD), 1,2-dihydroxy-3-keto-5-methylthiopentene dioxygenase (mtnD), adenosylhomocysteinase (AHCY), cystathionine gamma-synthase (DNMT3B), and S-adenosylmethionine synthetase (metK). Among these, two metK genes (The0G010940, The17G003520) and two speD genes (The03G011170, The23G007700) were highly expressed and significantly induced, which may be key genes in promoting the methionine cycle (Figure 6A).
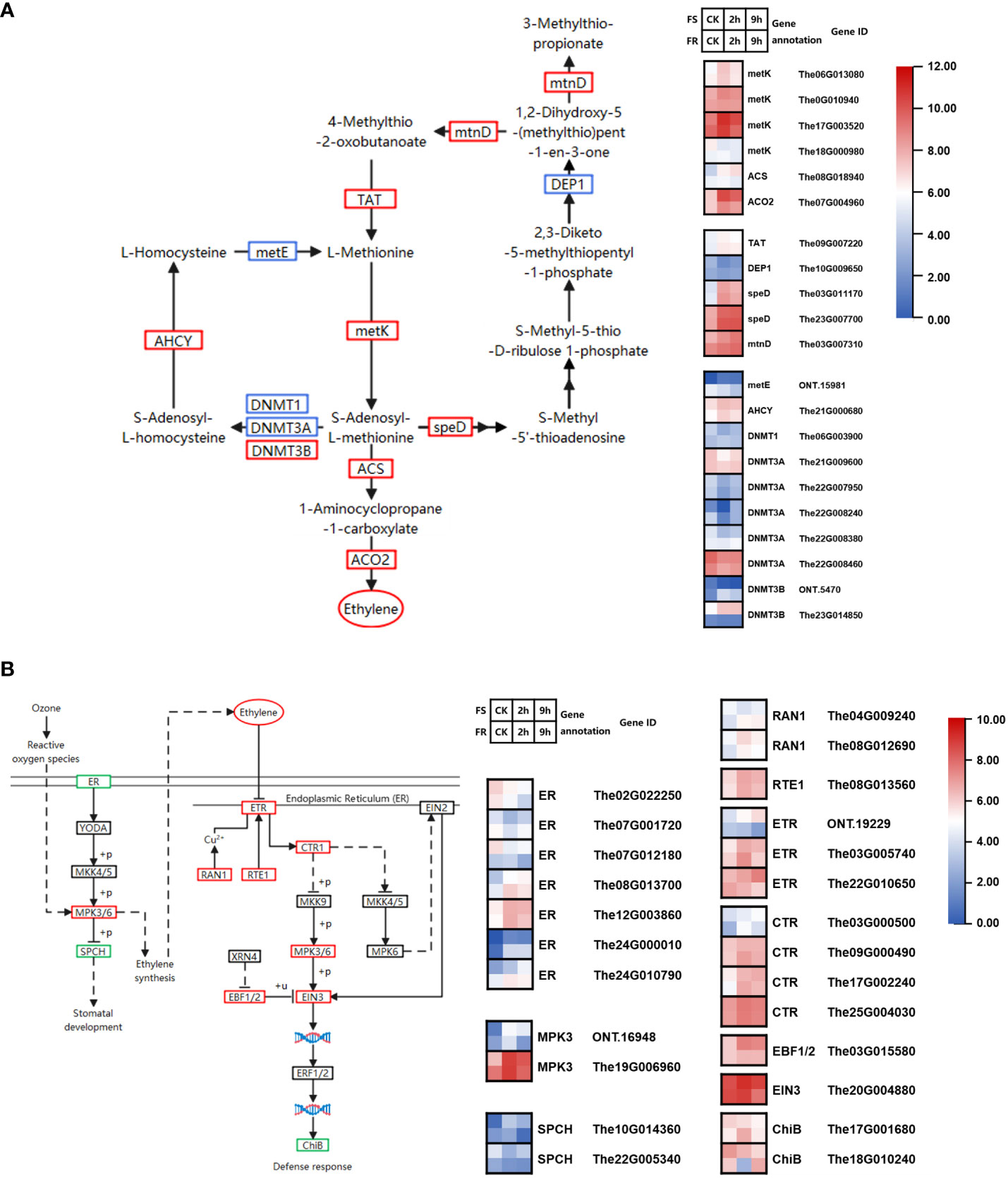
Figure 6 Cold-responsive DEGs regulate ethylene biosynthesis and signaling pathways. (A) Cold-responsive DEGs in ethylene biosynthesis. (B) Cold-responsive DEGs in the ethylene signaling pathway. Each colored cell in the heatmap represents log2 of the DEG CPM count values (mean of three biological replicates); red represents induced genes, and blue represents repressed genes. A schematic diagram of signaling pathway genes; red boxes represent all annotated genes that were upregulated, blue boxes represent all annotated genes that were downregulated, green boxes represent both up- and downregulated genes found in this location, and black boxes represent no DEGs found in this location.
1-Aminocyclopropane-1-carboxylate synthase (ACS) and aminocyclopropanecarboxylate oxidase (ACO2) catalyze SAM to produce ethylene. The08G018940 encoding ACS and The07G004960 encoding ACO2 were upregulated, especially the expression of The07G004960 which was induced by 13.1-fold at 2 h and 8.2-fold at 9 h after cold treatment in FS and 3.9-fold at 2 h and 2.5-fold at 9 h after cold treatment in FR.
There were nine DEGs annotated to four KEGG catalogs in the ethylene synthesis pathway, including ETR (ONT.19229, The03G005740 and The22G010650), CTR1 (The03G000500, The09G000490, The17G002240, and The25G004030), EIN3 (The20G004880), and EBF (The03G015580). All DEGs were upregulated by cold stress. Among them, EIN3 was highly expressed and significantly upregulated by cold stress (Figure 6B).
To verify the reliability and accuracy of RNA-seq data, quantitative real-time PCR (qRT-PCR) was used to detect the relative expression levels of 19 important and typical genes participating in temperature sensing, ABA, and ethylene biosynthesis and signaling pathways. Most of these genes were significantly and substantially induced by the 2-h and 9-h cold treatments (Figures 7A–C). The expression pattern detected with qRT-PCR was highly consistent with that detected using transcriptomics (R2 = 0.8873) (Figure 7D); it revealed the reliability of these RNA-seq data.
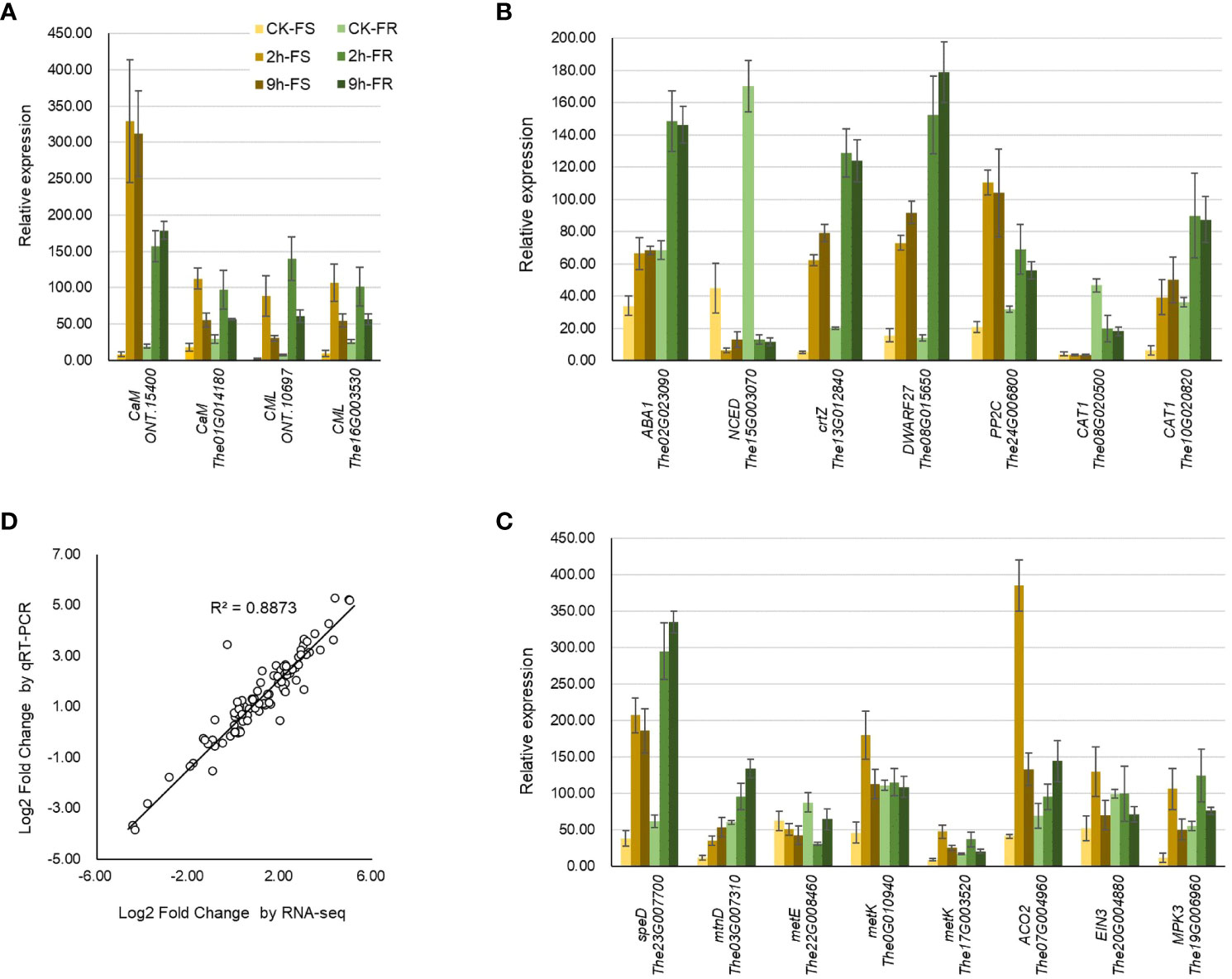
Figure 7 qRT-PCR analysis of different expressed genes involved in cold response. Relative expression of the key DEGs involved in (A) temperature sensing, biosynthesis, and signaling of (B) ABA and (C) ethylene. (D) Expression pattern validation by analyzing the linear dependence relation between the log2 fold change in key DEGs obtained from RNA-seq and qRT-PCR. The gene expression was determined by fold change relative to the reference gene MDH. All data were collected from three biological replicates and three technical replicates for each sample.
Since we found that the expressions of key genes related to ABA and ethylene were induced in response to cold stress, the ABA and ethylene contents in T. hemsleyanum after simulated frost treatments were examined. Endogenous ABA and ethylene were both induced by chilling stress and increased during −2°C chilling (Figures 8A, B). There was no significant difference between ABA in FS and FR, whereas the ethylene level was higher in FS than in FR during a 12-h treatment. After 12 h of chilling, the ABA content increased by 52.6% in FS and 57.0% in FR whereas the ethylene increased more strongly, by 107.7% in FS and 225.0% in FR. The results also revealed the positive role of ABA and ethylene in response to chilling at the metabolic level.
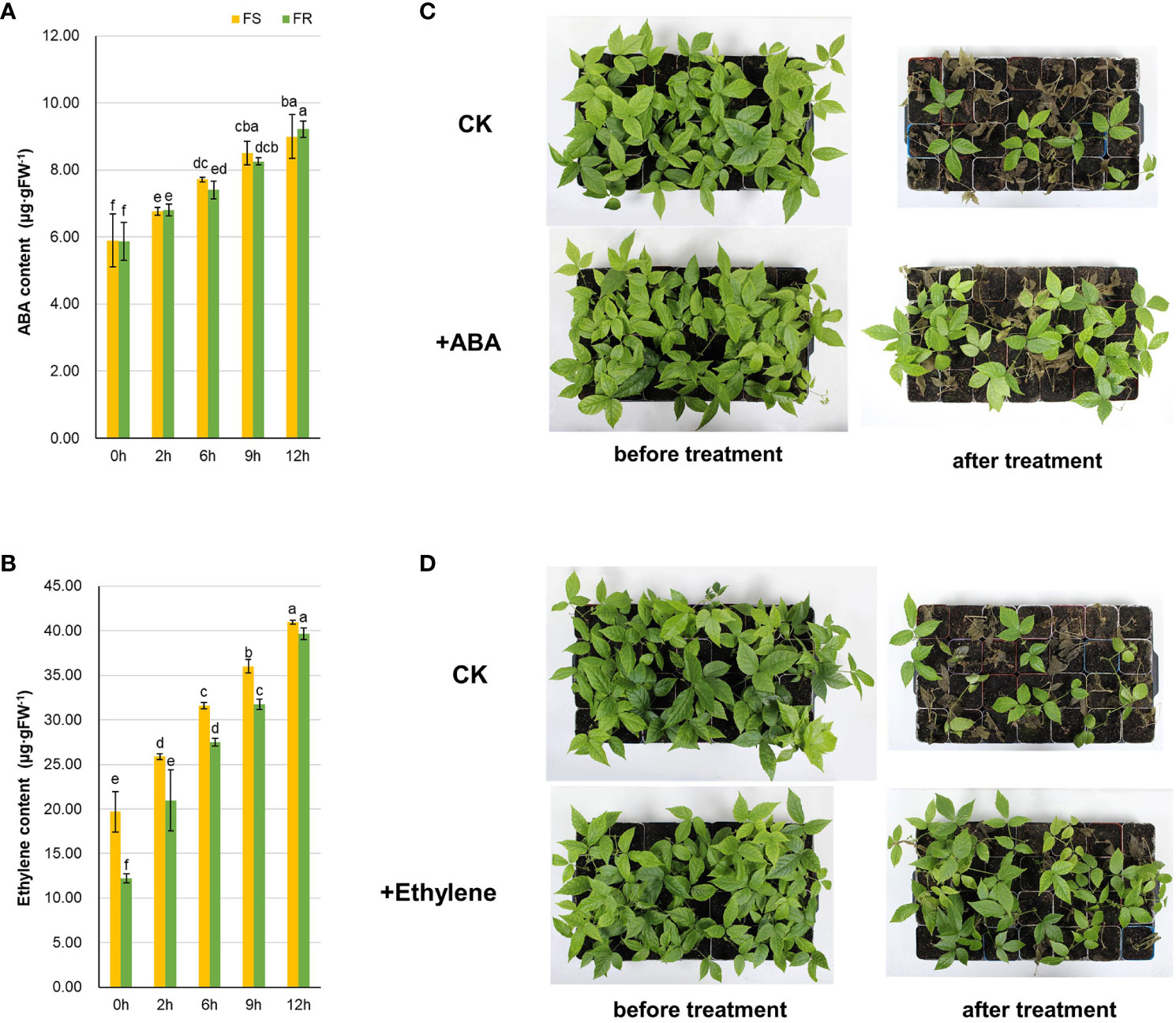
Figure 8 Effects of ABA and ethylene on cold tolerance in T. hemsleyanum. The variation in endogenous ABA (A) and ethylene (B) in FS and FR after chilling. Effect of exogenous ABA (C) and ethylene (D) on cold stress response after chilling. The content of plant hormones was detected after −2°C at different times, and data are the mean ± SD of three biological replicates. The letters indicate significant differences (multiple comparisons by Duncan, P < 0.05). The effect of exogenous phytohormones was tested in FS seedlings, and the phenotypes were recorded after −4°C treatment for 12 h.
In order to verify it further, exogenous phytohormones were sprayed on the FS seedlings, and −4°C chilling was applied. The chilling injury was obviously alleviated by ABA and ethylene, especially ethylene (Figures 8C, D). The survival rate was increased by 10.7% with ABA and 25.0% with ethylene.
Tetrastigma hemsleyanum is an herb that is widely used in Chinese traditional and modern medicine. As a thermophilous plant, winter cold severely limits the artificial cultivation of this plant. Several studies have been carried out aimed at the cold response in T. hemsleyanum, but the mechanisms of cold sensitivity in this plant remain unclear. In this study, the transcriptomes in T. hemsleyanum seedlings of the cold-resistant variety FR and the cold-sensitive variety FS exposed to −2°C for 2 h and 9 h were compared and analyzed to decipher the T. hemsleyanum cold stress response.
A total of 76,750 transcripts and 28,805 genes were obtained, 4,121 of which were new genes; furthermore, the sequence was obtained using the latest ONT with exceptional read length, further enriching the existing T. hemsleyanum gene bank.
Calcium is a secondary messenger used in many plant signaling processes. Cell membrane solidification induced by cold can affect the structure and/or activity of membrane-localized proteins, thereby triggering Ca2+ influx, a crucial process for inducing temperature-responsive gene expression (Knight et al., 1996; Ding et al., 2020). Plants possessed several major systems to sense and conduct Ca2+ signaling, including CaM (calmodulin)/CMLs (CaM-like proteins), CCaMK (Ca2+- and Ca2+/CaM-dependent protein kinase), CDPKs (Ca2+-dependent protein kinases), and CBLs (calcineurin B-like proteins), CIPKs (CBL-interacting protein kinases) (Galon et al., 2010). In this research study, we found six genes encoding calmodulin proteins; four were induced by cold, and two were highly expressed, which may cause OXI1 and three out of four RbohD genes to be upregulated in response to cold stress. In addition, we found 26 genes encoding CMLs, 25 induced by cold, and three were highly expressed. The results demonstrated that calcium signaling was regulated in response to the cold stress in T. hemsleyanum.
Except for the genes controlled by Ca2+, RbohD and OXI1 are also regulated by ROS and ROS-generating stimuli independently, playing important roles in the plant immune response to pathogens. Specifically, RbohD regulates oxidative burst during perception of pathogen-associated molecular patterns (PAMPs) by pattern recognition receptors (PRRs), OXI1 kinase is required for activation of mitogen-activated protein kinases (MAPKs), and an essential part of the signal transduction pathway links oxidative burst signals to diverse downstream responses (Rentel et al., 2004; Kadota et al., 2015). In this research study, two out of three cold-induced RbohD genes were expressed more in FS, and OXI1 was dramatically induced, and the expression in FS was 2.61-fold that in FR, which might cause more serious injury in FS after cold stress.
ABA is a vital phytohormone that regulates many essential physiological and biochemical processes, and it has a key role in stress resistance during plant growth and development (Huang et al., 2017). Stress rapidly triggers ABA production (Yamaguchi-Shinozaki and Shinozaki, 2006; Li et al., 2021), which causes ABA levels to increase and cold tolerance to be enhanced, such as in Arabidopsis and wheat (Choi et al., 2000; Hou et al., 2010). ABA is perceived by pyrabactin resistance (PYR)/PYR1-like (PYL)/regulatory components of ABA receptor (RCAR) receptors, which inactivate PP2C, resulting in activating the protein kinase SnRK2; SnRK2s activate many proteins via protein phosphorylation regulating stomatal closure (Lv et al., 2018). Group A PP2Cs interacted physically with SnRK2s in various combinations and efficiently inactivated ABA-activated SnRK2s via dephosphorylation of multiple Ser/Thr residues in the activation loop. This step was suppressed by the RCAR/PYR ABA receptors in response to ABA in Arabidopsis (Singh et al., 2015). The center stage in the ABA signaling pathway of PP2Cs was also verified in rice (Xue et al., 2008). In this research study, most DEGs in the ABA synthesis pathway were expressed at low levels or downregulated by cold, but genes encoding crtZ and ZEP were significantly induced by cold, which might be vital genes causing ABA to increase during short-term cold response in T. hemsleyanum. Some DEGs encoding PYL, PP2C, SnRK2, and ABF detected in this research study were also upregulated by cold, which may regulate numerous target genes in ABA-dependent pathways and ABA functional responses.
Ethylene is a crucial plant growth regulator that mediates cold stress responses in a species-dependent manner (Wang Y. C. et al., 2021). For example, ethylene positively affects the cold tolerance of tomato (Lycopersicon esculentum) (Ciardi et al., 1997). By contrast, ethylene levels are negatively correlated with the cold tolerance of Medicago truncatellid (Zhao et al., 2014). Low temperatures promote ethylene release in grapevines, and exogenous ACC (1-aminocyclopropane-1-carboxylat, the precursor of ethylene) increased the grapevine cold tolerance (Sun et al., 2016); ethylene positively regulated grapevine cold tolerance by modulating AP2/ERF and WRKY transcription factors, and the response of ABA and IAA during cold stress may be regulated by ETH signaling (Hou et al., 2023). T. hemsleyanum is also classified as a member of Vitisl. In this research study, DEGs encoding MPK3, which promoted ethylene synthesis, were upregulated, DEGs annotated to 12 KEGG catalogs in the ethylene synthesis pathway, and 10 KEGG catalogs were upregulated, including vital genes ACS and ACO2. DEGs annotated to four KEGG catalogs in the ethylene synthesis pathway were all upregulated by cold stress. It demonstrated that ethylene might be the major cold-responsive phytohormone within the 9 h stress treatment of T. hemsleyanum.
In addition, phytohormones can also regulate target genes through the MAPK signaling pathway. MAPK signaling is regulated by posttranslational modifications such as phosphorylation and ubiquitination, which might be the reason that we found no DEGs annotated to KEGG catalogs in some intermediate loci of transduction pathways, but DEGs encoding CAT were drastically changed. Posttranslational regulation might also happen in other nodes of phytohormone signaling pathways.
The 9-cis epoxycarotenoid dioxygenase (NCED) is considered to be a key rate-limiting enzyme in ABA biosynthesis (González-Silva et al., 2011); previous research has shown that cold stress caused significant upregulation of NCEDs at 24 h in T. hemsleyanum (Peng et al., 2021). However, in this research study, two out of three NCEDs were suppressed by cold, and one out of three NCEDs were only significantly induced in FS. All NCED genes were expressed at low levels. It indicated that the 9-h cold treatment is not sufficient to activate some cold response methods; it is necessary to spend more time to observe long-term transcriptional regulations.
Many vital DEGs we found, such as ONT.15400 (CaM), The10G020820 (CAT), and The19G006960 (MPK3), were expressed more in FR leaves at normal temperature, which might help form the cold tolerance. Some inherent traits that cannot be found in the transcriptome, such as the composition of the membrane, proline, and soluble sugar construction, might have reduced the cold injury before the cold response in the tissue of cold tolerance ecotypes, which may be the reason why more DEGs and more dramatic expression changes were found in the seedlings of sensitive ecotypes.
The change in expression level changed the endogenous ABA content, and exogenous ABA improved the cold tolerance of FS, consistent with previous research, which found endogenous ABA content at 0°C in other T. hemsleyanum ecotypes (Peng et al., 2021). Furthermore, we found a positive role of ethylene in response to cold stress, and the effect of ethylene might be stronger than ABA within 12 h.
Two Tetrastigma hemsleyanum (Sanyeqing) ecotypes grown in the plant garden of Hangzhou Academy of Agricultural Sciences (Hangzhou, Zhejiang Province, China) were involved in this research study. FS has large leaves and stout semi-cylindrical stalks; FR has medium-sized leaves and medium-thickness, cylindrical stalks.
Stem segments with one node and one bud were clipped and planted in peat. After 2 months of growth, cutting seedlings that had built root systems and sprouted five to six nodes were selected.
Selected seedlings were exposed to a simulated frost treatment (−2°C) for 0 h, 2 h, and 9 h. Three samples were performed for each cultivar, and each sample contained 12 plants. Leaves at the third and fourth nodes from the tip were collected and stored at −80°C for further analysis.
RNA samples were prepared using an RNA simple Total RNA Kit (DP411, TIANGEN), RNA integrality was tested by agarose gel (LabChip GX, Agient2100), and the RNA concentration was detected using a NanoDrop 2000 (Thermo Fisher Scientific).
Library preparation was performed according to the standard protocol provided by ONT. A mass of 1 μg total RNA was prepared for cDNA libraries using the cDNA-PCR Sequencing Kit (SQK-LSK110+EXP-PCB096) protocol provided by ONT. The template-switching activity of reverse transcriptases enriches full-length cDNAs and adds defined PCR adapters directly to both ends of the first-strand cDNA, followed by cDNA PCR for 14 cycles with LongAmp Tag (NEB). The PCR products were then subjected to ONT adaptor ligation using T4 DNA ligase (NEB). Agencourt XP beads were used for DNA purification according to the ONT protocol. The final cDNA libraries were added to the PromethION Flow Cell (R9 Version, FLO-PRO002, Nanopore) and run on the PromethION platform at Biomarker Technology Company (Beijing, China).
Raw reads were first filtered with a minimum average read quality score = 6 and a minimum read length = 350 bp. Ribosomal RNA was discarded after mapping to the rRNA database. Next, full-length, non-chimeric (FLNC) transcripts were determined by searching for primers at both ends of the reads. Clusters of FLNC transcripts were obtained after mapping to the reference genome with mimimap2 (2.16), and consensus isoforms were obtained after polishing within each cluster by pinfish (v0.1.0).
Consensus sequences were mapped to the reference genome using minimap2 (2.16). Mapped reads were further collapsed by the cDNA_Cupcake (5.8) package with min-coverage = 85% and min-identity = 90%. A 5′ difference was not considered when collapsing redundant transcripts. The coding sequence (CDS) was predicted using TransDecoder (v3.0.0).
Quantification of gene/transcript expression levels and differential expression analysis
Full-length reads were mapped to the reference transcriptome sequence. Reads with a match quality above 5 were used to quantify further. Expression levels were estimated by reads per gene/transcript per 10,000 mapped reads. For samples with biological replicates, differential expression analysis of two conditions/groups was performed using the DESeq2 R package (1.6.3) (Love et al., 2014). DESeq2 provides statistical routines for determining differential expression in digital gene expression data using a model based on the negative binomial distribution. The resulting P values were adjusted using Benjamini and Hochberg’s approach for controlling the FDR. Genes with an FDR <0.05 and fold change ≥2 found by DESeq2 were assigned as differentially expressed. Heatmaps were generated by TBtools (https://github.com/CJ-Chen/TBtools).
Gene Ontology (GO) enrichment analysis was implemented using the GOseq R (2.18.0) packages based on Wallenius non-central hyper-geometric distribution (GO database released on 2020-06-01) (Young et al., 2010). KOBAS (Mckenna et al., 2010) software was used to test the statistical enrichment of DEGs in KEGG pathways (KEGG version is 20191220). The obtained novel transcript sequences were also aligned to the NR (202009) (Deng et al., 2006), Swissprot (202005) (Apweiler et al., 2004), COG (COG2014) (Tatusov et al., 2000), KOG (KOG2003) (Koonin et al., 2004), and Pfam (Pfam33.1) (Kanehisa et al., 2004) databases. GSEA was performed using online tools in BMKCloud (www.biocloud.net).
WGCNA with all 18 libraries acquired from ONT RNA-seq was performed using the WGCNA R package in BMKCloud (www.biocloud.net) (Langfelder and Horvath, 2008). Co-expression network modules were identified based on counts per million mapped reads (CPM) ≥10, variation of CPM ≥0.5; the minimum module size was 10, the minimum height for merging modules was 0.3, and the soft threshold power was picked by the program automatically. The central hub genes were defined as those with module eigengene-based connectivity (kME) >0.7 within an assigned module, and modules were correlated with ecotypes and treatments to identify co-expression.
Plant tissue was the same batch as that used for transcriptome sequencing. RNA samples were prepared using the RNeasy Plant Mini Kit (Qiagen), and DNA was eliminated using an RNase-Free DNase Set (Qiagen). cDNAs were synthesized using SuperScript™ III First-Strand Synthesis SuperMix for qRT-PCR (Invitrogen). The samples were amplified in Power SYBR® Green PCR Master Mix (Applied Biosystems) and detected using a CFX384 Real-Time PCR System (Bio-Rad). Three samples were performed for each cultivar, and three replicates were performed for each sample. The MDH (malic dehydrogenase) gene was selected as an internal reference gene. The information on DEGs whose expression have been detected is offered in Supplementary Table 2. The primers were designed using Beacon Designer 7.8 and Primer Premier 6.0 (Supplementary Table 3).
Plant samples used for phytohormone measurement were prepared as described in 2.1. A mass of 0.1g tissue from each biological replicate was crushed in 1 mL of PBS solution (0.01 mol·L−1, pH = 7.2–7.4). The measurement was performed according to the standard protocol of ELISA kits provided by mlbio Biotechnology (https://www.mlbio.cn/).
Plants used for evaluation of the effect of exogenous phytohormones were cultivated as described in 4.1. A total of 28 seedlings were included for each treatment; 100 mL of 500 μmol/L ABA or 60 mg/L ethylene was sprayed on seedlings and cultivated for 3 days and then sprayed with exogenous phytohormones and cultivated for 3 days again. Treated seedlings were exposed to a −4°C simulated frost treatment for 12 h.
The original data of ONT transcriptome presented in the study are deposited in the database Genome Sequence Archive (GSA) of CNCB-NGDC, accession number SRR14575715-SRR14575720 under bioproject accession PRJCA016284. Other data presented in this study can be found in Supplementary Material.
LQ: Conceptualization, Funding acquisition, Methodology, Project administration, Resources, Supervision, Writing – original draft, Writing – review & editing. SY: Data curation, Investigation, Visualization, Writing – original draft, Writing – review & editing. NL: Investigation, Writing – review & editing. EY: Writing – review & editing. JY: Funding acquisition, Project administration, Resources, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This research was supported by Zhejiang Provincial Natural Science Foundation of China under Grant of Public Welfare Technology Application (LGN21C130002) and Hangzhou Academy of Agricultural Sciences under Grant of Technological Innovation and Demonstration Extension (2022HNCT07).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2024.1285879/full#supplementary-material
Apweiler, R., Bairoch, A., Wu, C. H., Barker, W. C., Boeckmann, B., Ferro, S., et al. (2004). UniProt: the universal protein knowledgebase. Nucleic Acids Res. 32, D115–D119. doi: 10.1093/nar/gkh131
Armstrong, J. J., Takebayashi, N., Wolf, D. E. (2020). Cold tolerance in the genus Arabidopsis. Am. J. Bot. 107 (3), 489–497. doi: 10.1002/ajb2.1442
Choi, H. I., Hong, J. H., Ha, J. O., Kang, J. Y., Kim, S. Y. (2000). ABFs, a family of ABA-responsive element binding factors. J. Biol. Chem. 275, 1723–1730. doi: 10.1074/jbc.275.3.1723
Ciardi, J. A., Deikman, J., Orzolek, M. D. (1997). Increased ethylene synthesis enhances chilling tolerance in tomato. Physiol. Plant 101, 333–340. doi: 10.1111/j.1399-3054.1997.tb01005.x
Cui, J. W., Shen, N., Lu, Z. G., Xu, G. L., Wang, Y. Y., Jin, B. (2020). Analysis and comprehensive comparison of PacBio and nanopore-based RNA sequencing of the Arabidopsis transcriptome. Plant Methods 16, 85. doi: 10.1186/s13007-020-00629-x
Deamer, D., Akeson, M., Branton, D. (2016). Three decades of nanopore sequencing. Nat. Biotechnol. 34, 518–524. doi: 10.1038/nbt.3423
Deng, Y. Y., Li, J. Q., Wu, S. F., Zhu, Y. P., Chen, Y., Fuchu, H. E. (2006). Integrated NR database in protein annotation system and its localization. Comput. Eng. 32, 71–74. doi: 10.1109/INFOCOM.2006.241
Ding, Y. L., Shi, Y. T., Yang, S. H. (2020). Molecular regulation of plant responses to environmental temperatures. Mol. Plant 13, 544–564. doi: 10.1016/j.molp.2020.02.004
Einset, J., Winge, P., Bones, A. (2007). ROS signaling pathways in chilling stress. Plant Signal Behav. 2, 365–367. doi: 10.4161/psb.2.5.4461
Galon, Y., Finkler, A., Fromm, H. (2010). Calcium-regulated transcription in plants. Mol. Plant 3, 653–669. doi: 10.1093/mp/ssq019
González-Silva, N., López-Lara, I. M., Reyes-Lamothe, R., Taylor, A. M., Sumpton, D., Thomas-Oates, J., et al. (2011). The dioxygenase-encoding olsD gene from Burkholderia cenocepacia causes the hydroxylation of the amide-linked fatty acyl moiety of ornithine-containing membrane lipids. Biochemistry 50, 6396–6408. doi: 10.1021/bi200706v
Hou, Y. D., Guo, Z. F., Yi, Y., Li, H. N., Li, H. G., Chen, L. J., et al. (2010). Effects of cold acclimation and exogenous pytohormone abscisic acid treatment on physiological indicators of winterness wheat. J. Plant Sci. 5, 125–136. doi: 10.3923/jps.2010.125.136
Hou, Y. J., Wong, D. C. J., Li, Q. Y., Zhou, H. M., Zhu, Z. F., Gong, L. Z., et al. (2023). Dissecting the effect of ethylene in the transcriptional regulation of chilling treatment in grapevine leaves. Plant Physiol. Biochem. 196, 1084–1097. doi: 10.1016/j.plaphy.2023.03.015
Huang, X. B., Shi, H. Y., Hu, Z. R., Liu, A., Amombo, E., Chen, L., et al. (2017). ABA is involved in regulation of cold stress response in Bermudagrass. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.01613
Ji, T., Ji, W. W., Wang, J., Chen, H. J., Peng, X., Cheng, K. J., et al. (2021). A comprehensive review on traditional uses, chemical compositions, pharmacology properties and toxicology of Tetrastigma hemsleyanum. J. Ethnopharmacol. 264, 113247. doi: 10.1016/j.jep.2020.113247
Kadota, Y., Shirasu, K., Zipfel, C. (2015). Regulation of the NADPH oxidase RBOHD during plant immunity. Plant Cell Physiol. 56, 1472–1480. doi: 10.1093/pcp/pcv063
Kanehisa, M., Goto, S., Kawashima, S., Okuno, Y., Hattori, M. (2004). The KEGG resource for deciphering the genome. Nucleic Acids Res. 32 (Database issue), D277–D280. doi: 10.1093/nar/gkh063
Knight, H., Trewavas, A. J., Knight, M. R. (1996). Cold calcium signaling in Arabidopsis involves two cellular pools and a change in calcium signature after acclimation. Plant Cell 8, 489–503. doi: 10.1105/tpc.8.3.489
Koonin, E. V., Fedorova, N. D., Jackson, J. D., Jacobs, A. R., Krylov, D. M., Makarova, K. S., et al. (2004). A comprehensive evolutionary classification of proteins encoded in complete eukaryotic genomes. Genome Biol. 5 (2), R7. doi: 10.1186/gb-2004-5-2-r7
Langfelder, P., Horvath, S. (2008). WGCNA: an R package for weighted correlation network analysis. BMC Bioinform. 9, 559. doi: 10.1186/1471-2105-9-559
Li, W., Fu, L. F., Geng, Z. W., Zhao, X. J., Liu, Q. H., Jiang, X. Q. (2021). Physiological characteristic changes and full-length transcriptome of rose (Rosa chinensis) roots and leaves in response to drought stress. Plant Cell Physiol. 61, 2153–2166. doi: 10.1093/pcp/pcaa137
Liu, J. J., Bao, Y., Zhong, Y. M., Wang, Q., Liu, H. F. (2021). Genome-wide association study and transcriptome of olecranon-type traits in peach (Prunus persica L.) germplasm. BMC Genomics 22, 702. doi: 10.1186/s12864-021-08017-y
Love, M. I., Huber, W., Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550. doi: 10.1186/s13059-014-0550-8
Lv, X. Z., Li, H. Z., Chen, X. X., Xiang, X., Guo, Z. X., Yu, J. Q., et al. (2018). The role of calcium-dependent protein kinase in hydrogen peroxide, nitric oxide and ABA-dependent cold acclimation. J. Exp. Bot. 69, 4127–4139. doi: 10.1093/jxb/ery212
Maleki, M., Ghorbanpour, M. (2018). “Cold tolerance in plants: molecular machinery deciphered,” in Biochemical, Physiological and Molecular Avenues for Combating Abiotic Stress Tolerance in Plants. Eds. Maleki, M., Ghorbanpour, M. (London, United Kingdom: Academic Press), 57–71. doi: 10.1016/B978-0-12-813066-7.00004-8
Mckenna, A., Hanna, M., Banks, E., Sivachenko, A., Cibulskis, K., Kernytsky, A., et al. (2010). The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303. doi: 10.1101/gr.107524.110
Parker, M. T., Knop, K., Sherwood, A. V., Schurch, N. J., Mackinnon, K., Gould, P. D., et al. (2020). Nanopore direct RNA sequencing maps the complexity of Arabidopsis mRNA processing and m6A modification. eLife 9, e49658. doi: 10.7554/eLife.49658
Peng, X., Ji, Q. Y., Fan, S., Zhang, Y. J., Zhang, J. J. (2015). Genetic diversity in populations of the endangered medicinal plant Tetrastigma hemsleyanum revealed by ISSR and SRAP markers: implications for conservation. Genet. Resour. Crop Evol. 62, 1069–1078. doi: 10.1007/s10722-014-0210-6
Peng, X., Li, M. J., Wu, H., Chen, H. J., Zhang, Z. Y. (2021). Co-regulation role of endogenous hormones and transcriptomics profiling under cold stress in Tetrastigma hemsleyanum. J. Plant Growth Reg. 40, 1992–2006. doi: 10.1007/s00344-020-10246-6
Peng, X., Wu, H., Chen, H., Zhang, Y., Qiu, D., Zhang, Z. (2019). Transcriptome profiling reveals candidate flavonol-related genes of Tetrastigma hemsleyanum under cold stress. BMC Genomics 20, 687. doi: 10.1186/s12864-019-6045-y
Pither, J. (2003). Climate tolerance and interspecific variation in geographic range size. Proc. R. Soc. B 270, 475–481. doi: 10.1098/rspb.2002.2275
Qin, H., Ou, L., Gao, J., Chen, L. X., Wang, J. W., Hao, P., et al. (2022). DENA: training an authentic neural network model using nanopore sequencing data of Arabidopsis transcripts for detection and quantification of N6-methyladenosine on RNA. Genome Biol. 23, 25. doi: 10.1186/s13059-021-02598-3
Rentel, M. C., Lecourieux, D., Ouaked, F., Usher, S. L., Petersen, L., Okamoto, H., et al. (2004). OXI1 kinase is necessary for oxidative burst-mediated signalling in Arabidopsis. Nature 427, 858–861. doi: 10.1038/nature02353
Scheunert, A., Dorfner, M., Lingl, T., Oberprieler, C. (2020). Can we use it? On the utility of de novo and reference-based assembly of nanopore data for plant plastome sequencing. PloS One 15, e0226234. doi: 10.1371/journal.pone.0226234
Singh, A., Jha, S. K., Bagri, J., Pandey, G. K. (2015). ABA inducible rice protein phosphatase 2C confers ABA insensitivity and abiotic stress tolerance in Arabidopsis. PloS One 10, e0125168. doi: 10.1371/journal.pone.0125168
Sun, X. M., Zhao, T. T., Gan, S. H., Ren, X. D., Fang, L. C., Karungo, S. K., et al. (2016). Ethylene positively regulates cold tolerance in grapevine by modulating the expression of ethylene RESPONSE FACTOR 057. Sci. Rep. 6, 24066. doi: 10.1038/srep24066
Tatusov, R. L., Galperin, M. Y., Natale, D. A., Koonin, E. V. (2000). The COG database: a tool for genome scale analysis of protein functions and evolution. Nucleic Acids Res. 28, 33–36. doi: 10.1093/nar/28.1.33
Wang, Y. C., Jiang, H. Y., Mao, Z. L., Liu, W. J., Jiang, S. H., Xu, H. F., et al. (2021). Ethylene increases the cold tolerance of apple via the MdERF1B–MdCIbHLH1 regulatory module. Plant J. 106, 379–393. doi: 10.1111/tpj.15170
Wang, Y. H., Jiang, W. M., Comes, H. P., Hu, F. S., Qiu, Y. X., Fu, C. X. (2015). Molecular phylogeography and ecological niche modelling of a widespread herbaceous climber, Tetrastigma hemsleyanum (Vitaceae): Insights into Plio-Pleistocene range dynamics of evergreen forest in subtropical China. New Phytol. 206, 852–867. doi: 10.1111/nph.13261
Wang, Y. H., Zhao, Y., Bollas, A., Wang, Y., Au, K. F. (2021). Nanopore sequencing technology, bioinformatics and applications. Nat. Biotechnol. 39, 1348–1365. doi: 10.1038/s41587-021-01108-x
Xiong, J., Tang, X. H., Wei, M. Z., Yu, W. J. (2022). Comparative full-length transcriptome analysis by Oxford nanopore technologies reveals genes involved in anthocyanin accumulation in storage roots of sweet potatoes (Ipomoea batatas L.). PeerJ 10, e13688. doi: 10.7717/peerj.13688
Xue, T., Wang, D., Zhang, S., Ehlting, J., Ni, F., Jakab, S., et al. (2008). Genome-wide and expression analysis of protein phosphatase 2C in rice and Arabidopsis. BMC Genomics 9, 550. doi: 10.1186/1471-2164-9-550
Yamaguchi-Shinozaki, K., Shinozaki, K. (2006). Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu. Rev. Plant Biol. 57, 781–803. doi: 10.1146/annurev.arplant.57.032905.105444
Young, M. D., Wakeeld, M. J., Smyth, G. K., Oshlack, A. (2010). Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol. 11, R14. doi: 10.1186/gb-2010-11-2-r14
Zhao, M. G., Liu, W. J., Xia, X. Z., Wang, T. Z., Zhang, W. H. (2014). Cold acclimation-induced freezing tolerance of Medicago truncatula seedlings is negatively regulated by ethylene. Physiol. Plant 152, 115–129. doi: 10.1111/ppl.12161
Keywords: Tetrastigma hemsleyanum, full-length transcriptome, cold stress, ABA, ethylene
Citation: Qian L, Yin S, Lu N, Yue E and Yan J (2024) Full-length transcriptome reveals the pivotal role of ABA and ethylene in the cold stress response of Tetrastigma hemsleyanum. Front. Plant Sci. 15:1285879. doi: 10.3389/fpls.2024.1285879
Received: 31 August 2023; Accepted: 10 January 2024;
Published: 31 January 2024.
Edited by:
Karl J. Niklas, Cornell University, United StatesReviewed by:
Tianyuan Zhang, Huazhong Agricultural University, ChinaCopyright © 2024 Qian, Yin, Lu, Yue and Yan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jianli Yan, eWFuamlhbmxpMDBAZ21haWwuY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.