
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 14 February 2024
Sec. Plant Abiotic Stress
Volume 15 - 2024 | https://doi.org/10.3389/fpls.2024.1283912
This article is part of the Research Topic Plant Responses to Salt Stress View all 12 articles
Suaeda dendroides, a succulent euhalophyte of the Chenopodiaceae family, intermittently spread around northern Xinjiang, China, has the ability to grow and develop in saline and alkali environments. The objective of this study was therefore to investigate the underlying molecular mechanisms of S. dendroides response to high salt conditions. 27 sequencing libraries prepared from low salt (200 mM NaCl) and high salt (800 mM NaCl) treated plants at 5 different stages were sequenced using Illumina Hiseq 2000. A total of 133,107 unigenes were obtained, of which 4,758 were DEGs. The number of DEGs in the high salt group (3,189) was more than the low salt treatment group (733) compared with the control. GO and KEGG analysis of the DEGs at different time points of the high salt treatment group showed that the genes related to cell wall biosynthesis and modification, plant hormone signal transduction, ion homeostasis, organic osmolyte accumulation, and reactive oxygen species (ROS) detoxification were significantly expressed, which indicated that these could be the main mechanisms of S. dendroides acclimate to high salt stress. The study provides a new perspective for understanding the molecular mechanisms of halophytes adapting to high salinity. It also provides a basis for future investigations of key salt-responsive genes in S. dendroides.
Soil salinization has emerged as one of the principal abiotic stresses that threaten plant growth and development and restrict the sustainable development of modern agriculture (Lesk et al., 2016; Kopecka et al., 2023). The adverse effects of excessive soil salinity on plants mainly include osmotic and ion stress. Osmotic stress reduces water absorption of plants, resulting in water leakage. A high concentration of salt ions affects the plasma membrane and changes its permeability, which leads to excessive salt ion absorption by plants and excludes the absorption of other nutrient elements. Imbalanced ion absorption causes nutritional imbalance, inhibits growth, and produces single-salt toxicity, causing metabolic disorders (Yang and Guo, 2018; Zhao et al., 2020; Zhang et al., 2022). Understanding the physiological, biochemical, and molecular mechanisms of plant adaptation to salt stress and exploring the genes related to salt tolerance in nature is beneficial for germplasm innovation and cultivating plant varieties with improved salt tolerance. Arabidopsis is widely used in gene functional research and is a model plant for studying molecular mechanisms under various biotic and abiotic stresses. Much effort has been made to breed salt-tolerance crops for agricultural demand, however, the salt tolerance of these crops is limited in their systems. In contrast, halophytes, naturally grown under high salinity conditions, have evolved a series of strategies to adapt to extreme saline environments (Flowers et al., 2015; Calone et al., 2021; Rahman et al., 2021). Learning the mechanisms of salt resistance in halophytic plants and mining key salt-responsive genes are of great significance for cultivating crops with improved salt tolerance.
Halophytes are a kind of plant species that grow and complete their life cycle in saline environments where the salt concentration is greater than 200 mM NaCl, which accounts for about 1% of the world flora (Flowers and Colmer, 2008). Euhalophytes, accumulate excessive Na+ in their succulent leaves and stems and compartmentalize into vacuoles, enabling them to cope with high salt stress (Flowers et al., 2015). The genus Suaeda is a typical euhalophyte, an annual or perennial herb or sub-herb of Chenopodiaceae. It is usually distributed in coastal areas, intertidal zones, deserts, inland salt lakes, and various saline and alkali environments around the world (Wang and Song, 2019; Wang et al., 2022). The suaeda plants grow well with 200 mM NaCl and complete their life cycle in environments where salt and drought co-exist (Song et al., 2009; Guo et al., 2017; Guo et al., 2020). Moreover, the potential of several high salt responsive genes like transporters (NHX1 and HKT1), ion channel (AKT1, TPCA1, SLAC1, aquaporins), antioxidant (APX, CAT1, GST) and osmotic (CMO, BADH and P5CS) have been identified and cloned (Mishra and Tanna, 2017), and some of these genes were explored for developing stress tolerance in the glycophyte (Lv et al., 2008; Wu et al., 2008; Shao et al., 2014; Himabindu et al., 2016; He et al., 2017; Hao et al., 2020). In addition, studies have increasingly found that halophytes can be economical and environmentally friendly candidate species for phytoremediation; excessive salt and heavy metals can be removed from salt and contaminated soils through various strategies to support plant growth (Liang and Shi, 2021; Wang L. et al., 2021). The above research shows that halophytes have important ecological and economic values. A comprehensive study on salt tolerance mechanism of halophytes is crucial for excavating key genes and cultivating stress-resistant varieties, as well as phytoremediation, which is conducive to promoting the sustainable development of agricultural economy in saline-alkali areas.
Suaeda plants can adapt to high concentration saline soil and act as ideal model plants for investigating complicated physiological and molecular mechanisms of salt tolerance. To explore the molecular mechanism of S. dendroides adapting to salt stress conditions, two groups of de novo assembled data were generated from mRNA sequencing of shoot samples from low salt and high salt treated seedlings. Bioinformatics analyses were adopted to compare changes at mRNA levels. These data would contribute to elucidating the molecular mechanisms of S. dendroides acclimate to high salt stress. Furthermore, the transcript information of S. dendroides under high salt treatment was obtained comprehensively, which provided a basis for studying gene expression levels, discovering new transcripts, and exploring the functions of key salt-tolerant genes.
The seeds of S. dendroides were collected from saline soil in Fuhai, Xinjiang Uygur Autonomous Region, China. S. dendroides seeds were sown in plastic pots (12×12 cm), filled with mixed matrix (sand:vermiculite:nutritive soil = 10:1:2, w/w/w) and irrigated with tap water, placed in a plant culture room, maintained a day/night thermoperiod of 25/20°C, a photoperiod of 16 h light/8 h dark, relative humidity of (50 ± 10)%. 10 seedlings with uniform growth were maintained per pot after germination and irrigated with tap water once a week. After one month of incubation, the seedlings were irrigated with 0, 200, 400, 600, 800, and 1,000 mM NaCl solutions. 150 mL of the NaCl solution was applied in salt treatments, while the control pots were irrigated with the same volume of tap water. Throughout the experimental process, water was replenished daily through weighing methods to ensure that the water content of each pot was maintained at 50%.
According to the growth and phenotype of the S. dendroides seedlings under different concentrations of salt stress, the physiological and biochemical parameters were determined after treatment for 10 days. Each variable was determined in three biological replicates.
The shoot samples were taken for biomass and ion content analyses. The fresh weight (FW) of aerial tissues was measured immediately after harvest. The dry weight (DW) was determined by putting the FW samples in an oven at 105 °C for 10 min, then drying for 48 h at 85°C until the mass was constant. Relative water content (RWC) was calculated using the formula: RWC = (FW-DW)/FW×100%. In order to know the salt absorption of S. dendroides plants, Na content in the seedlings under salt treatment was detected. Briefly, 100 mg dried powder from the shoot of each treatment sample was weighed and digested with 5 mL HNO3 overnight and incubated at 80°C for 1~2 h, 120°C for 1~2 h, then 160°C for 4 h, cooling to room temperature, after filtering the extracts, Na and K content was determined by inductively coupled plasma atomic emission spectroscopy (ICP-AES/OES/MS).
Fresh leaf samples were taken for physiological and biochemical parameters analyses, such as chlorophyll content, ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco) activity, malondialdehyde (MDA) content, proline content, soluble sugar content. All the above variables were assayed as the instructions of the reagent kit (Solarbio, China), and the absorbance was measured using a UV2300 ultraviolet visible spectrophotometer (Tianmei, China).
Based on our preliminary physical investigations, 200 mM NaCl is the optimal salt concentration, and 800 mM NaCl is the highest salt concentration for the growth of S. dendroides seedlings (Figure 1). Consequently, the shoot samples were harvested at 0 (before the treatment, marked as C), 1, 5, 10, and 15 days after 200 (marked as L1, L5, L10, and L15) and 800 mM NaCl treatments (marked as H1, H5, H10, and H15) for RNA- sequencing and QPCR. Total RNA extraction from 27 shoot tissues of S. dendroides was according to the manufacturer’s procedure of the EASYspin Plus Plant RNA Kit (Aidlab, China). Qubit® RNA Assay Kit in Qubit® 2.0 Fluorometer (Life Technologies, USA) was used to detect RNA concentration, and RNA Nano 6000 Assay Kit of Agilent Bioanalyzer 2100 system (Life Technologies, USA) was used to evaluate RNA integrity. NEBNext® Ultra™ RNA Library Prep Kit of Illumina® (NEB, USA) was used to generate sequencing libraries following the manufacturer’s recommendations, and library quality was evaluated by the Agilent Bioanalyzer 2100 system. The libraries were sequenced on an Illumina Hiseq platform, and paired-end reads were generated. After filtering adaptor sequences, duplicated sequences, low-quality bases, and undetermined bases, the high-quality clean reads were assembled using Trinity software (Grabherr et al., 2011). Function annotation of the assembled unigenes was carried out through NCBI non-redundant protein sequences (Nr), NCBI non-redundant nucleotide sequences (Nt), Protein family (Pfam), Clusters of Orthologous Groups of proteins (KOG/COG), A manually annotated and reviewed protein sequence database (Swiss-Prot), KEGG Ortholog database (KO) and Gene Ontology (GO) databases.
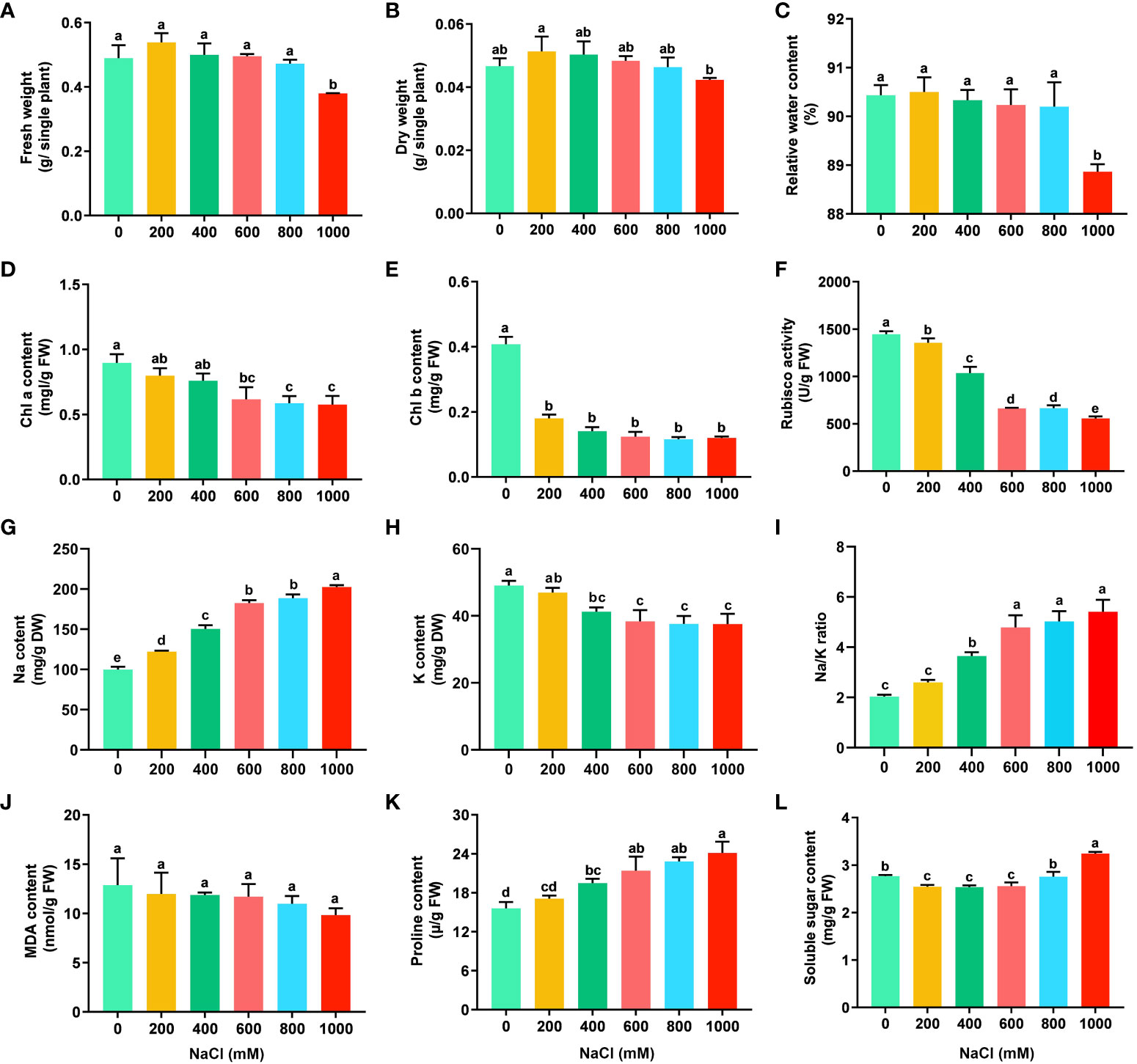
Figure 1 Physiological variables of S. dendroides under salt stress. (A) Fresh weight, (B) Dry weight, (C) Relative water content, (D) Chl a content. (E) Chl b content, (F) Rubisco activity, (G) Na content, (H) K content, (I) Na/K ratio, (J) MDA content, (K) Proline content, (L) Soluble sugar content. X-axis means the concentration of NaCl (mM). Statistical analysis was conducted using SPSS 20.0 software, the significant difference between different treatments was obtained through Tukey’s test of one-way ANOVA analysis. Values are presented as the mean ± the standard error (SE) with three biological replicates. The different letters above the bars indicate the least significant differences between treatments at p < 0.05.
The transcription level of the assembled unigenes was evaluated according to FPKM to identify DEGs between L1 vs C, L5 vs C, L10 vs C, L15 vc C, H1 vs C, H5 vs C, H10 vs C, H15 vs C, H1vs L1, H5 vs L5, H10 vs L10, and H15 vs L15 compares. Differential expression analysis of two conditions/groups was carried out by DESeq2 software (Love et al., 2014). The negative binomial Wald test was used as a statistical test, and then the Benjamini-Hochberg correction was used to obtain the false discovery rate (FDR) (Benjamini and Hochberg, 1995; Love et al., 2014). FDR < 0.05 and |log2FC| > 1 were set as the thresholds for screening of significantly differential expressed genes. VENNY 2.1 (https://bioinfogp.cnb.csic.es/tools/venny/), a free online platform, was used to draw Venn diagrams. TBtools v1.120 was used to construct the heatmaps for gene expression. GO and KEGG enrichment analysis of DEGs were performed using GOseq R packages based on Wallenius non-central hyper-geometric distribution (Young et al., 2010) and KOBAS software (Mao et al., 2005). The sequences of the target DEGs were aligned to the protein sequence of the reference species contained in the STRING database (http://string-db.org/) by using blasx, and an interaction network was constructed by using the protein-protein interaction (PPI) relationship of the reference species. The PPI network of the DEGs was visualized and edited by Cytoscape software v3.9.1 (Shannon et al., 2003).
10 putative DEGs involved in salt conditions were randomly selected for validation of differential expression using QPCR. The primers for the selected genes were designed according to the sequences assembled by S. dendroides RNA sequencing and are listed in Supplementary Table S1. 18S gene of Anabasis aphylla was used as the reference for quantitative expression analysis. 1 ug RNA of 27 samples was reverse transcribed into cDNA with oligo (dT) primers, and the cDNA libraries were used for QPCR. A total of 20µL fluorescent quantitative reaction system contains 10 µL of 2 × PerfectStart® Green qPCR SuperMix (TransGen, China), 2 µL of cDNA template, 0.8 µL of 10 mM primers and 0.4 μL of 50 × ROX Reference Dye. QPCR was performed on an ABI 7500 platform, and the settings were as follows: step 1 95°C, 30 s; step 2 95°C, 5 s, 60°C, 34 s, 40 cycles, followed by the melting curve analysis. Each DEG was analyzed in three biological samples, and each biological sample was repeated three times. 2-ΔΔCT method was used to calculate the relative expression levels of the target genes (Livak and Schmittgen, 2001).
Statistical analysis was conducted using SPSS 20.0 software, the significant differences were analyzed by one-way ANOVA and Tukey’s test (p < 0.05). The images were visualized using GraphPad Prism software (v. 8.3.0.538), and the data was shown as the mean ± standard deviation (SD) value of at least three biological replicates.
The physiological and biochemical features of S. dendroides seedlings under different salt concentrations were performed to estimate the optimal and highest salt concentration for their growth. The seedlings of S. dendroides did not show symptoms of salinity injury after being treated with 200, 400, and 600 mM NaCl treatment compared with the control. The seedlings showed wilting symptoms under 800 and 1,000 mM NaCl treatment and began to recover after 5 days and recovered after 10 days, then gradually returned to normal growth. Under 1000mM NaCl stress, although some seedlings died, most seedlings survived, and the wilting symptoms slowly recovered (Supplementary Figure S1). The fresh weight, dry weight, and relative water content of seedlings showed the highest under 200 mM NaCl treatment, but the difference was not significant compared with the control. However, the fresh weight, dry weight, and relative water content of seedlings treated with 1000 mM NaCl were significantly lower than those of other salt concentrations (Figures 1A-C). The results indicate that a low concentration of NaCl could promote the growth of seedlings. The content of Chl a and Chl b, and Rubisco activity decreased obviously with the increase of NaCl concentration (Figures 1D-F), respectively. Na and K content in the shoots of S. dendroides were determined, and the results showed that Na content was significantly increased with the increase of NaCl concentration. When the NaCl concentration reached 400 mM, the accumulation of Na was 1.5 times that of the control (Figure 1G). The content of K decreased obviously with the increase of salt concentration, which was opposite to Na content (Figure 1H). The trend of Na/K ratio was consistent with that of Na accumulation. When the salt concentration increased to 600 mM, the change of Na/K ratio was moderate and kept a relatively stable level (Figure 1I). The results indicated that S. dendroides could maintain Na/K balance under high salt stress to avoid ion toxicity.
The physiological parameters, such as MDA, proline, and soluble sugar, are usually used to evaluate salt tolerance of plants. In S. dendroides, MDA content decreased gradually with the increase of NaCl concentration, but the difference was not significant (Figure 1J), indicating that salt in this concentration range did not cause the cell membrane lipid peroxidation of S. dendroides. The content of free proline in the seedlings of S. dendroides was significantly increased with the increase of NaCl concentrations (Figure 1K). The soluble sugar content decreased significantly under 200, 400 and 600 mM NaCl treatment, but it was significantly higher than the control when the salt concentration increased to 1000 mM (Figure 1L).
RNA-seq analyses on the S. dendroides with water (C), 200 mM NaCl (L), and 800 mM NaCl (H) treatments at different time points (1, 5, 10, and 15 days) were performed. In total, 27 cDNA libraries were generated and sequenced. A total of 211.13 Gb clean data were obtained, and each sample library yielded a mean size of 7.8 Gb clean data, a Q30 percentage higher than 93.65%, and a GC percentage of 42% (Supplementary Table S2). Pearson’s coefficient of pairwise biological replicates was greater than 0.85, indicating that the consistency among the replicates was high (Supplementary Figure S2). The data shown above demonstrate that the transcriptome sequencing is of high quality. Since S. dendroides has no reference genome, Trinity software was used to do the de novo assembly and get the reference sequence for subsequent analysis. 133,107 unigenes were obtained from 27 libraries. The size distribution was shown in Supplementary Figure S3, the sequence of 38,197 unigenes was longer than 1,000 bp, and 12,286 unigenes was longer than 2000 bp, indicating that some candidate genes have obtained nearly full-length sequence. The mean sequence length of all unigenes was 918 bp, and the N50 length was 1,240 bp (Supplementary Table S3).
133,107 unigene sequences of S. dendroides were aligned based on similarities to the NR, NT, Pfam, Swiss-Prot, KEGG, GO, and KOG databases. The annotation results were shown in Supplementary Table S4, all 133,107 (100%) unigene sequences were annotated at least in one database, and 6,443 (4.84%) aligned to all seven databases. A total of 58,336 (43.82%), 42,308 (31.78%), and 42,637 (32.03%) unigenes had significant BLAST matches in NR, NT, and Pfam databases, respectively. Moreover, 42,637 (32.03%) unigenes were annotated in at least one term of three GO categories (Supplementary Figure S4), 19,118 (14.36%) unigenes were annotated in the KEGG database and classified into 130 KEGG pathways (Supplementary Figure S5). In addition, 11,489 (8.63%) unigenes were annotated in the KOG database and divided into 25 functional categories (Supplementary Figure S6). By comparing the annotation results with the NR database, the distribution of species on the unigene comparison was statistically analyzed. The result showed that S. dendroides transcriptome sequences were well matched with the species of Chenopodium quinoa (29.8%), Beta vulgaris (27.8%), and Spinacia oleracea (19.0%) belonging to Amaranthaceae (Supplementary Figure S7).
DEGs under salt-stressed conditions were defined according to threshold FDR < 0.05 and |log2 fold change| > 1 among different salt-treated samples. A total of 733, 3189, and 992 DEGs were detected between L vs C, H vs C, and H vs L pairwise comparisons, respectively. DEGs were analyzed at each time interval after NaCl treatment (Figure 2A). With the extension of salt treatment time, the number of DEGs increased to 444 and 1,796 after 10 days, which was the peak of salt-responsive genes under both NaCl concentrations, indicating that this period was the key time for S. dendroides to adapt to the salt environment. Down-regulated DEG numbers were more than up-regulated genes among all the treatments. 130 of the 3,189 DEGs were detected among different times of 800 mM NaCl treatment. In addition, only 4 DEGs were detected among 733 DEGs at different times of 200 mM NaCl treatment (Figure 2B). In addition, the expression profiles of DEGs under each treatment were visualized in the heat map (Figure 2C). The expression patterns of most DEGs of samples H1, H5, and H10 were similar and clustered in one class, while the samples H15, C, L1, L5, L10, and L15 were clustered in another class. The results showed that the expression pattern of DEGs under 200 mM NaCl treatment was similar to the control, while the DEGs under 800 mM NaCl treatment were contrary to the control. These results indicated that the euhalophyte S. dendroides gradually adapted to the high salt stress environment, in which the 10 days was a critical time for S. dendroides to cope with the high salt stress environment. Consequently, the following analysis focuses on the DEGs of 800 mM NaCl treatment to get a comprehensive understanding of the adaptive mechanisms of S. dendroides encountering high salt stress.
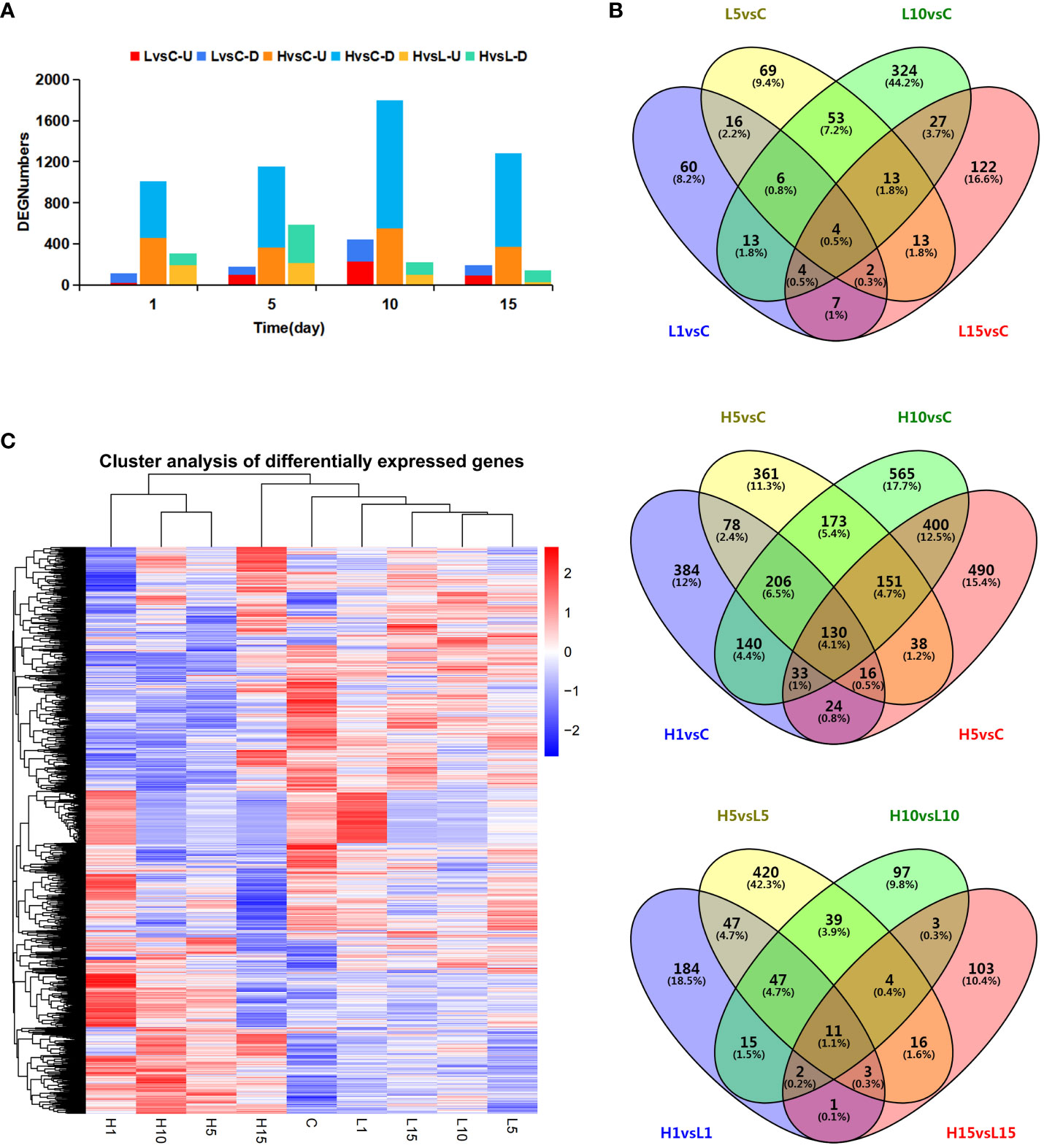
Figure 2 The numbers of all DEGs at different salt-stressed time points of two NaCl concentrations. (A) Numbers of DEGs in pairwise comparisons at different salt-stressed time points of two NaCl concentrations, (B) Venn diagram showed the number of DEGs in different treatment, (C) Heatmap diagrams showed the cluster analysis of DEGs among the treatments. Number of DEGs (FDR< 0.05 and |log 2 Fold Change| > 1) under 200mM (L) and 800mM (H) NaCl stress for 1, 5,10 and 15 day as compared to control samples.
GO and KEGG enrichment analyses were carried out on DEGs at different time points with high salt treatment. GO enrichment analysis showed that the DEGs of H1 vs C, H5 vs C, H10 vs C, and H15 vs C comparisons were significantly enriched in 45, 14, 41, and 39 processes (FDR < 0.05), respectively (Supplementary Table S5). The top 30 Go items of different salt time points were shown in Figures 3A-D, the catalytic activity item was notably enriched at each salt time point, and the DEG numbers were the highest. Besides, carbohydrate metabolic, cell metabolic, methionine metabolic, protein phosphorylation and protein kinase were the most enriched items at each salt treatment. KEGG enrichment results showed that the DEGs of H1 vs C, H5 vs C, H10 vs C, and H15 vs C comparisons were significantly enriched (p-value < 0.05) in 11, 9, 13 and 16 pathways, respectively (Supplementary Table S6). Among these, phenylpropane metabolism, plant hormone signal transduction, cysteine and methionine metabolism, glycerophospholipid metabolism, starch and sucrose metabolism, and ribosome pathway at different salt treatment time points were significantly enriched (Figures 4A-D; Supplementary Table S6). In addition, cell wall-related metabolic pathways were also found in this study, such as the galactose metabolic pathway closely related to pectin, the pentose glucuronic acid conversion, and the flavonoid biosynthesis metabolism closely related to lignin, the phenylalanine metabolic pathway, and keratin and wax biosynthesis. Moreover, the key protein-protein interaction (PPI) network of high NaCl treatment for 10 days was predicted. The result showed that DEGs related to cell wall synthesis and modification, ABA signaling, ethylene biosynthesis, and organic osmolyte accumulation under high salt treatment, which is consistent with GO and KEGG enrichment results. The above results indicate that cell wall synthesis and modification, ABA signaling, ethylene biosynthesis and signal, organic osmolyte accumulation, and ROS homeostasis might be the main adaptation mechanisms of S. dendroides suffering from extreme salt stress.
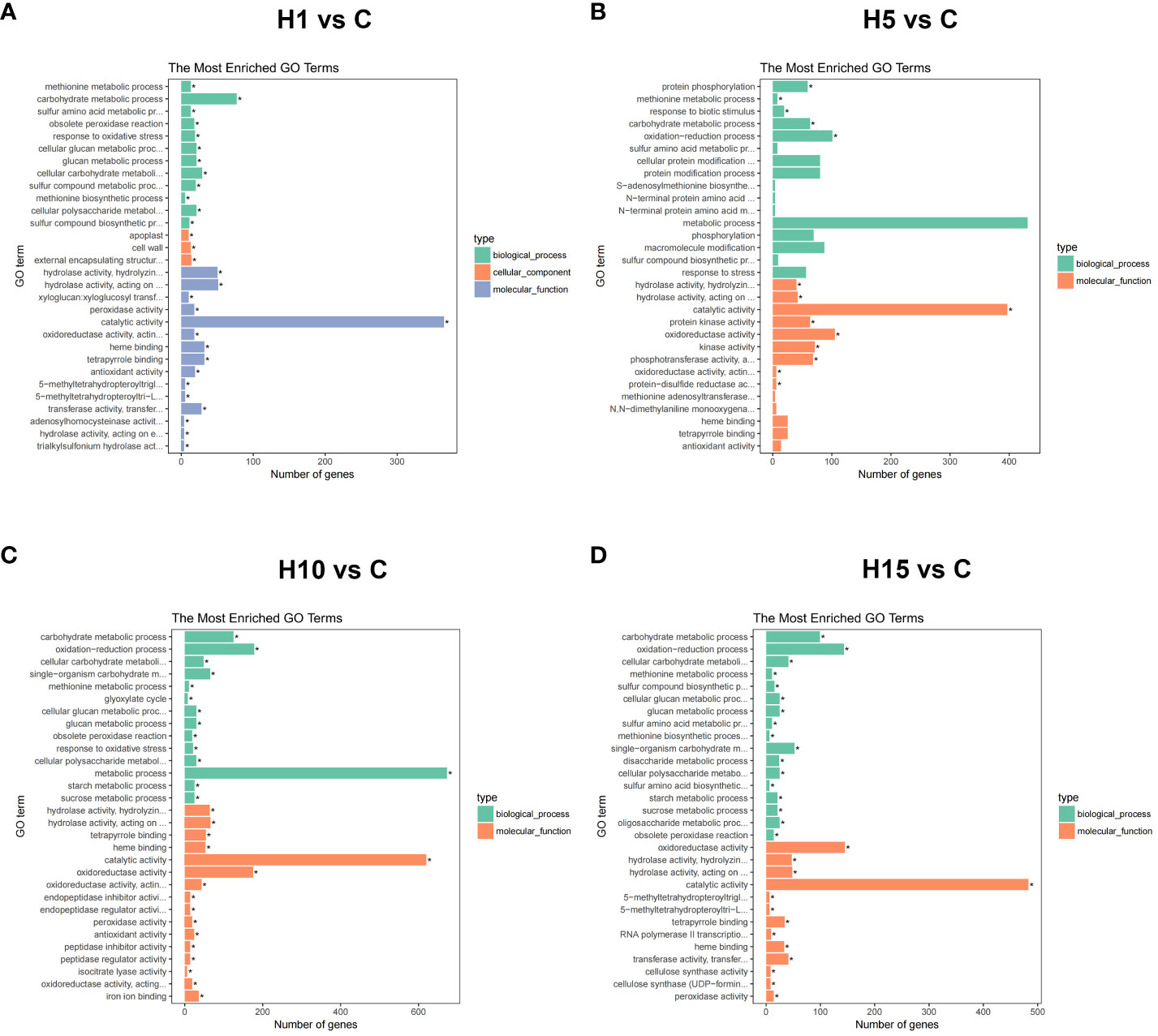
Figure 3 GO enrichment analysis of DEGs in (A) H1 vs C, (B) H5 vs C, (C) H10 vs C, (D) H15 vs C comparisons of high salt stress. Top 30 GO enrichment terms of differently expressed genes significant p-values (p < 0.05). GO terms are mainly divided into three classes: cellular component, molecular function, and biological process. X-axis means the number of DEGs. Y-axis represents GO terms.
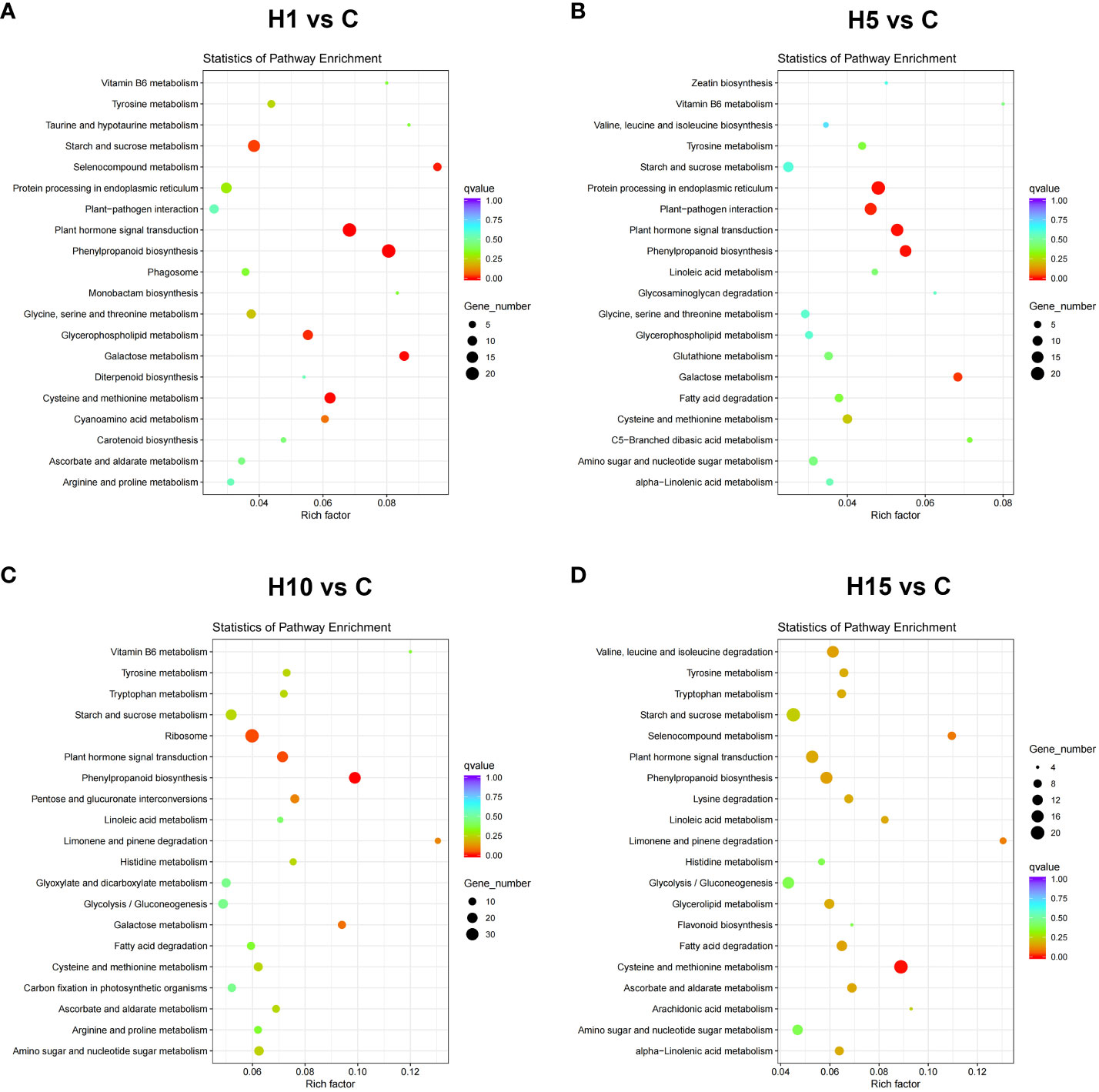
Figure 4 KEGG pathway enrichment analysis of DEGs in (A) H1 vs C, (B) H5 vs C, (C) H10 vs C, (D) H15 vs C comparisons of high salt stress. Top 20 GO enrichment terms of differently expressed genes significant p-values (p < 0.05). X-axis represents Rich factor. Y-axis represents the KEGG pathway.
According to GO, KEGG enrichment, and PPI results, the DEGs involved in cell wall metabolism of S. dendroides under high saline conditions were analyzed. 13 DEGs involved in cellulose synthesis were induced by 800 mM NaCl stress (Figure 5A; Supplementary Table S7). Among these, 4 predicted cellulose synthase A (CESA) genes, including 1 CESA1 and 3 CESA3 genes, which produced cellulose in the primary cell wall (PCW), were up-regulated and significantly increased after 10 days of treatment. Moreover, 3 predicted CESA4, 2 xylem-specific CESA7, and 2 CESA8 genes involved in the biosynthesis of the secondary cell wall (SCW) were up-regulated and dramatically increased after 10 and 15 days of 800 mM NaCl treatment. In addition, 2 COBRA-like (COBLs) genes related to cell wall biosynthesis were up-regulated under 800 mM NaCl treatment and prominently increased after 15 days of the treatment. Moreover, several genes involved in hemicellulose synthesis were detected under high salt stress. 6 glycosyltransferase (GT) family genes involved in the biosynthesis of xylan backbone were identified under high salt treatment (Figure 5A; Supplementary Table S7). Such as glucuronoxylan glucuronosyltransferase (IRX7), beta-1,4-xylosyltransferase (IRX9 and IRX14), galacturonosyltransferase-like 1 (GATL1), UDP-glucuronate:xylan alpha-glucuronosyltransferase 1 (GUX1/PGSIP1 and GUX2/PGSIP3) related to the secondary cell wall biosynthesis, were all significantly increased after 10 days of 800 mM NaCl stress. Meanwhile, 2 non-glycosyltransferase genes, IRREGULAR XYLEM 1 (IRX15) and IRREGULAR XYLEM 1 like (IRX15L), related to xylan backbone synthesis were up-regulated under high salt treatment. 2 cellulose synthase-like protein D5 (CLSD5) related to the biosynthesis of hemicellulose polysaccharides were detected to be significantly expressed under 800 mM NaCl stress. In addition, 6 laccases (LAC) and 30 peroxidases (PER) genes related to the polymerization of lignin monolignol were also found under high salt stress (Figures 5A, 6C; Supplementary Table S7).
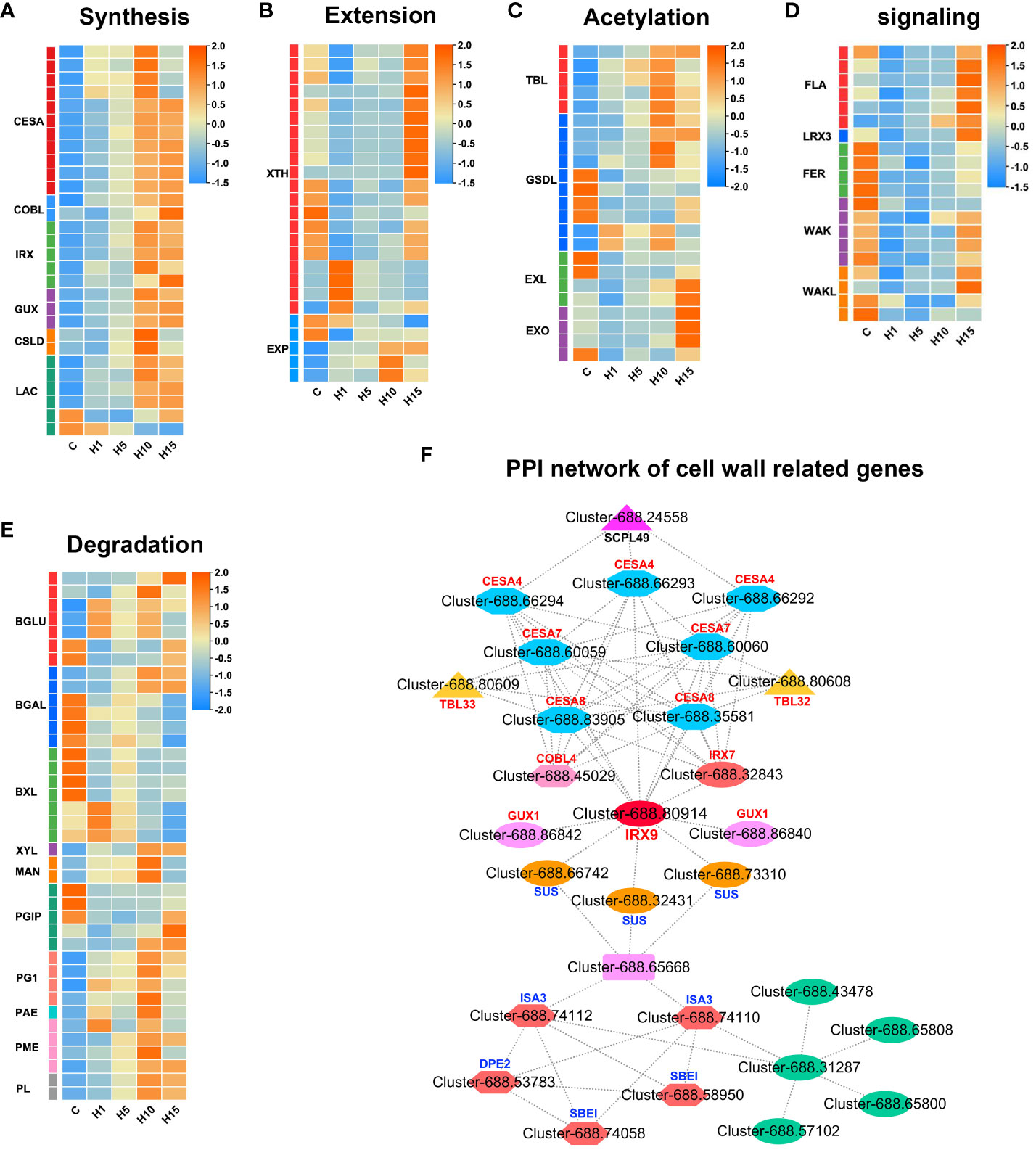
Figure 5 Expression profiles of DEGs related to cell wall (A) synthesis, (B) extension, (C) acetylation, (D) proteins and signaling, (E) degradation, (F) PPI network of cell wall related DEGs in S. dendroides after 10 days of high salt stress. The column names C, H1, H5, H10, and H15 in each heat map are the samples treated with 800 mM NaCl after 0, 1, 5, 10, and 15 days. The expression level is represented by the mean value of FPKM (n=3), and the color scale in the upper right represents the normalized FPKM value.
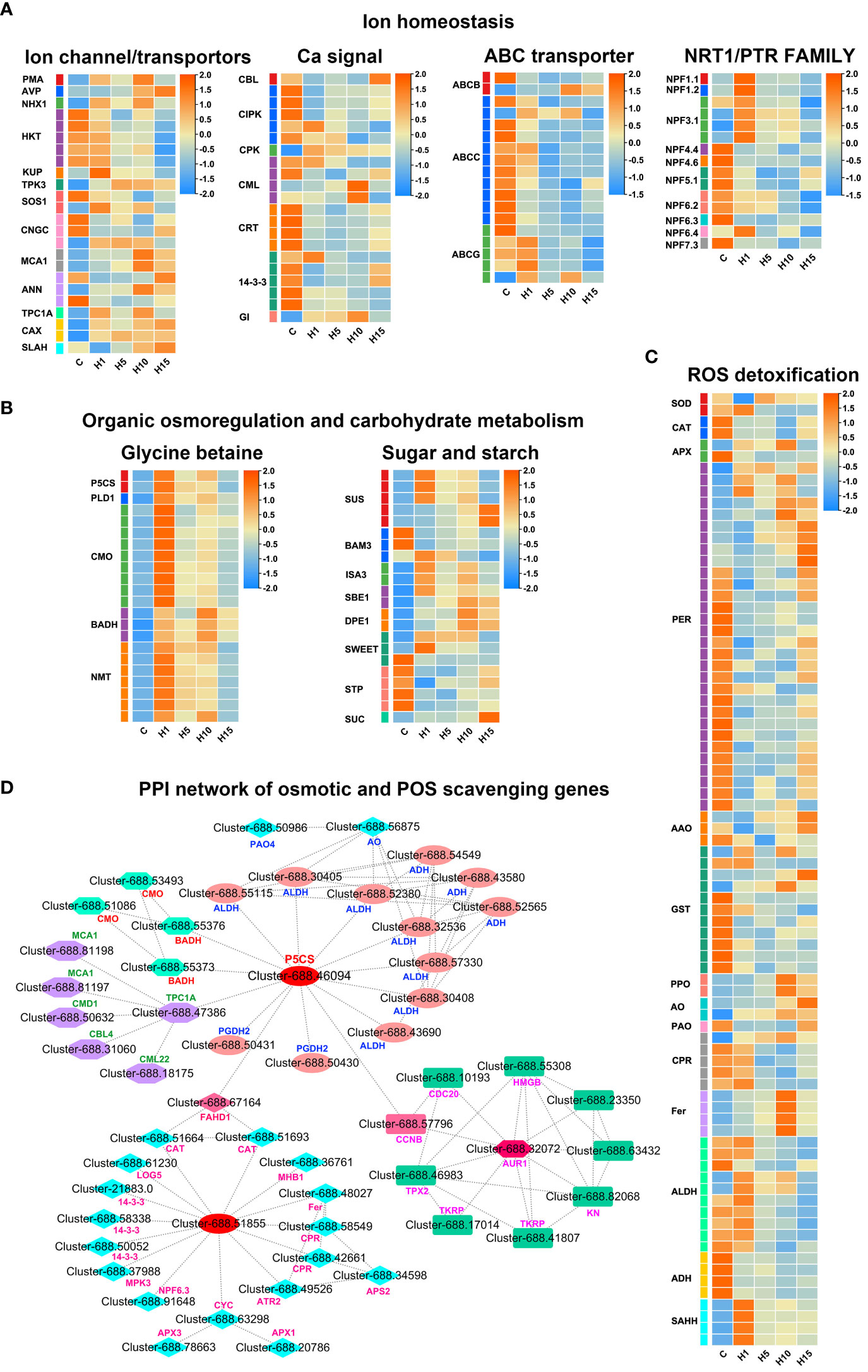
Figure 6 Expression profiles of DEGs related to (A) Ion homeostasis, (B) Organic osmoregulation and carbohydrate metabolism, (C) ROS detoxification, (D) PPI network of osmotic and ROS scavenging related DEGs in S. dendroides after 10 days of high salt stress. The column names C, H1, H5, H10, and H15 in each heat map are the samples treated with 800 mM NaCl after 0, 1, 5, 10, and 15 days. The expression level is represented by the mean value of FPKM (n=3), and the color scale in the upper right represents the normalized FPKM value.
Interestingly, multiple DEGs involved in cell wall modification and remodeling were identified under 800 mM NaCl stress. 20 xyloglucan endotransglucosylase/hydrolase (XTH/XET) and 5 expansins/expansins-like (EXP/EXL) genes that participated in cell wall loosening were identified (Figure 5B; Supplementary Table S7). Several glycosidase encoding DEGs involved in hydrolyzing hemicellulose polysaccharides were obtained, such as 7 β-galactosidase (BGLU), 6 β-galactosidase (BGAL), and 7 β-D-xylosidases (BXL) genes were differentially expressed under 800 mM NaCl stress (Figure 5E; Supplementary Table S7). XYL involved in xyloglucan degradation was down-regulated under high salt treatment. Additionally, pectin metabolic enzymes (Figure 5E; Supplementary Table S7), including pectin acetylesterase (PAE), pectin methyl esterase (PME), polygalacturonase (PG), polygalacturonase inhibitor (PGIP), and pectate lyase (PL) encoding genes involved in cell wall remodeling were obtained under high salt treatment. 4 PMEs, 2 PLs, 4 PG1s, and 1 PAE were up-regulated under 800 mM NaCl treatment (Figure 5E; Supplementary Table S7). 5 trichome birefringence-like (TBL) genes involved in xylan acetylation were up-regulated, 6 GDSL esterase/lipase (GSDL) encoding genes that affecting xylan deacetylation were up-regulated under high salt stress, especially after 10 days, 6 EXORDIUM/EXORDIUM-like (EXO/EXL) genes involved in the regulation of secondary cell wall thickening and lignification were significantly increased after 15 days of high salt stress (Figure 5C; Supplementary Table S7). Moreover, several cell wall genes and receptor-like kinase genes were differentially expressed under high salt treatment (Figure 5D; Supplementary Table S7). 6 DEGs encoding FASCICLIN-like arabinogalactan protein (FLA) were notably increased after 15 days of high salt treatment. 1 leucine-rich repeat extensin-like protein (LRX3) decreased at the early stage of high salt treatment and increased after 15 days. 4 FERONIA (FER) genes of Catharanthus roseus RLK1-like kinase (CrRLK1L) family were down-regulated under high salt stress. 9 Wall-associated receptor kinase (WAK/WAKL) encoding genes were decreased at the early stage of high salt treatment and increased to varying degrees after 15 days of 800 mM NaCl treatment.
Several DEGs associated with ion transport were obtained under 800 mM NaCl treatment in the study (Figure 6A; Supplementary Table 7). A sodium/hydrogen exchanger protein NHX1 involved in vacuolar Na+ transport was significantly up-regulated under 800 mM NaCl treatment. Similarly, a two-pore potassium channel protein TPK3 associated with vacuolar K+ transport, 2 vacuolar Ca2+/H+ exchanger (CAX) genes were significantly up-regulated. DEGs of plasma membrane ATPase (PMA) and vacuolar H+-pyrophosphatases (AVP) were also up-regulated. The results indicated that the up-regulated NHX, PMA, and AVP might be crucial for Na+ sequestration into the vacuolar of S. dendroides. 5 high-affinity K+ transporter (HKT1) encoding genes involved in Na+ transport were obtained, the expression level of 3 genes was significantly increased after 1 day, and 2 were increased significantly after 15 days of high salt treatment. Potassium transporter (KUP) was significantly up-regulated after 1 day of 800 mM NaCl treatment. 3 cyclic nucleotide-gated ion channel (CNGC) genes were obtained, CNGC4 was up-regulated, and 2 CNGC17 genes were down-regulated under high salt treatment. 2 Ca2+ influx channel proteins mid1-complementing activity 1 (MCA1) were up-regulated, especially after 10 and 15 days of 800 mM NaCl treatment. The increasing cytoplasmic Ca2+ are decoded by a series of Ca2+ sensors or binding proteins, such as calcineurin B-like protein (CBL), Ca2+- dependent protein kinases (CPK), Calmodulin (CaM), calmodulin-like proteins (CML), calcium-dependent protein kinase (CIPK) and calreticulin (CRT). 21 DEGs that participated in Ca2+ signaling pathway were obtained, as shown in Figure 6A, CBL4, CIPKs, CMLs, and CRTs were induced by high salt stress. Several nitrate transporter 1/peptide transporter, NRT1/PTR family (NPF) encoding genes related to NO3−/Cl- transport were obtained (Figure 6A). Among these, 8 NPF genes were down-regulated, and 7 were significantly up-regulated after 1 day of high salt stress. Besides, 18 ABC transporters were differently expressed under high salt concentration treatment, in which 3 ABCC22 genes were notably increased after 1 day, 3 genes were up-regulated at all the treatment times, while the remaining 12 genes were down-regulated at all the treatments.
The accumulation of organic adjustment substances in succulent leaves is another important reason for S. dendroides adapting to high salt environment. In this study, 44 DEGs positively correlated with the synthesis of organic adjustment substances proline, betaine, and soluble sugar were identified (Figure 6B). Among these, 2 delta-1-pyrroline-5-carboxylate synthase (P5CS), a key enzyme gene involved in proline synthesis, were significantly up-regulated under 800 mM NaCl treatment. 9 choline monooxygenase (CMO) and 3 betaine aldehyde dehydrogenase (BADH), key enzyme genes participated in betaine synthesis, were also significantly up-regulated. Moreover, 7 phosphoethanolamine N-methyltransferase (NMT) genes were notably up-regulated after 1 day of high salt treatment. Likewise, 5 sucrose synthase (SUS) encoding genes and 2 4-alpha-glucanotransferase (DPE1) involved in soluble carbohydrate accumulation were significantly up-regulated under 800 mM NaCl treatment. 1 sucrose transport protein SUC and 2 bidirectional sugar transporters (SWEET) were significantly increased under high salt stress. These results showed that the up-regulated DEGs of organic osmolyte synthesis under high salt treatment, including proline, betaine, and soluble sugar, play vital roles in osmoregulation of S. dendroides. Antioxidant enzymes and non-enzymatic compounds were crucial for detoxification of ROS under stress conditions. In S. dendroides, the genes of ascorbate peroxidase (APX), superoxide dismutase (SOD), catalase (CAT), PER, glutathione S-transferase (GST), and L-ascorbate oxidase (AAO) were significantly enriched and differentially expressed, to avoiding oxidative stress induced by high salinity (Figure 6C). Besides, polyphenol oxidase (PPO), primary amine oxidase (AO), and ferritin (Fer) encoding genes were up-regulated at all time points after 800 mM NaCl treatment. 10 aldehyde dehydrogenase (ALDH) encoding genes and 4 alcohol dehydrogenase-like (ADH)encoding genes were differently expressed (Figure 6C).
GO and KEGG enrichment results showed that the DEGs under high salt stress were significantly enriched in plant hormone signal transduction pathways like ABA and ethylene signal transduction. Several DEGs that participated in ABA signal transduction were differentially expressed under high salt treatment (Figure 7A). 3 9-cis-epoxycarotenoid dioxygenase (NCED) encoding genes, involved in ABA synthesis, were rapidly up-regulated after 1 day of high salt treatment. 3 genes encoding ABA receptor PYLs were detected, PYR1 and PYL4 were rapidly down-regulated under 800 mM NaCl treatment. 9 protein phosphatase 2C (PP2C) encoding genes were prominently up-regulated after 1 day of 800 mM NaCl treatment. 1 SNF1-related protein kinases (SnRK2) gene was down-regulated under 800 mM NaCl treatment. 2 ABA-responsive element binding factors (ABF2) were increased dramatically after 1 day and declined after 15 days of 800 mM NaCl treatment. 27 DEGs enriched in cysteine and methionine metabolic pathway were mainly associated with ethylene biosynthesis (Figure 7B), such as 5-methyltetrahydropteryltriglutamate homocysteine methyltransferase (METE), S-adenosylmethionine synthase (SAMS), 1-aminocyclopropane-1-carboxylic acid synthase (ACS), 1-aminocyclopropane-1-carboxylic oxidase (ACO) and other key enzymes in ethylene biosynthesis. 5 METE genes and 3 SAMS genes were up-regulated under 800 mM NaCl treatment. 9 ACO and ACO homologs genes were down-regulated.
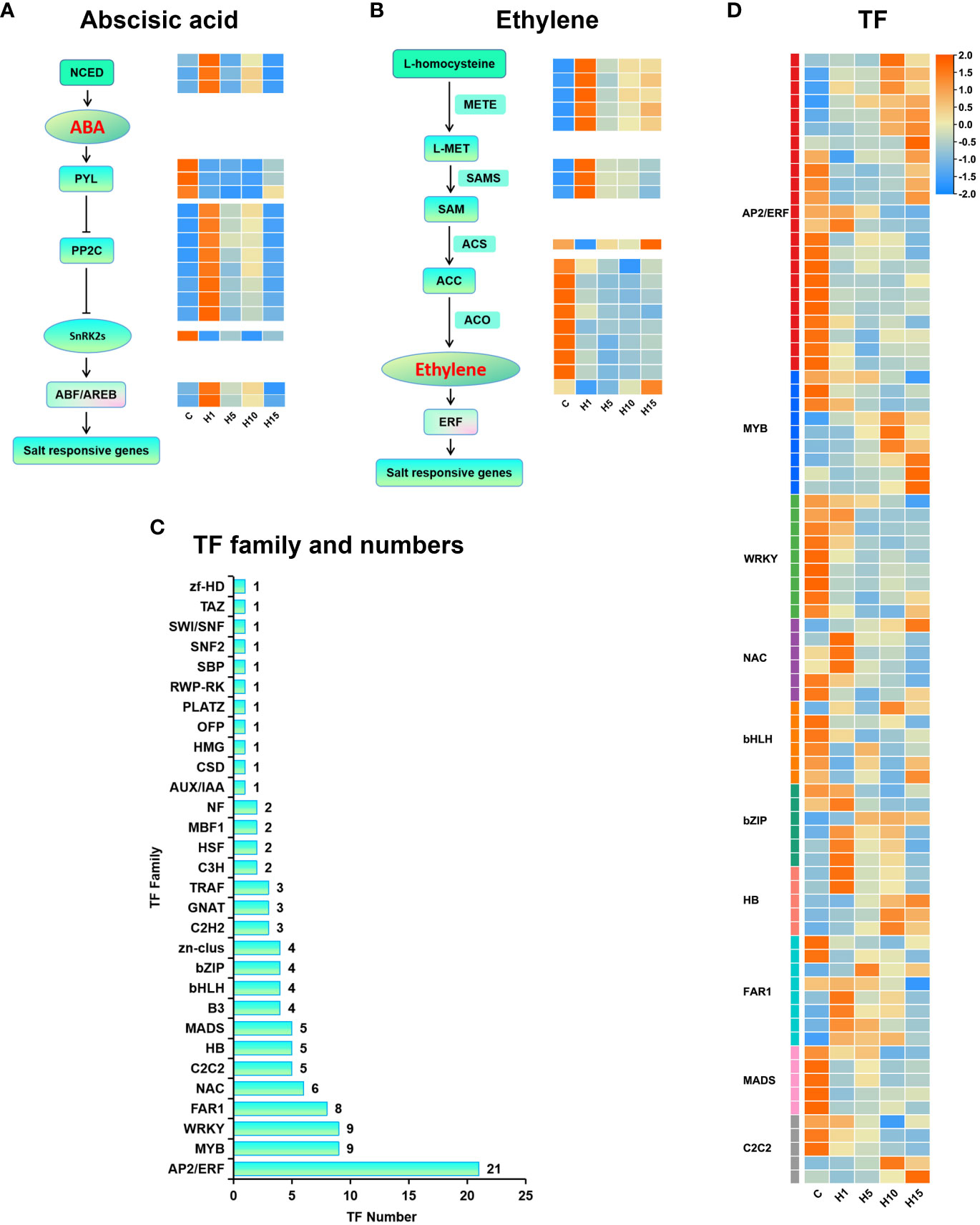
Figure 7 Expression profiles of the plant hormone signal transduction genes and TFs of S. dendroides in response to high salt treatment. (A) ABA synthesis and signal transduction genes, (B) ETH synthesis and signal transduction genes, (C) The numbers of different transcription factor families, (D) Expression profiles of TFs. The column names C, H1, H5, H10, and H15 in each heat map are the samples treated with 800 mM NaCl after 0, 1, 5, 10, and 15 days. The expression level is represented by the mean value of FPKM (n=3), and the color scale in the upper right represents the normalized FPKM value.
Transcription factors (TFs) play a key role in regulating plant resistance to abiotic stress. In this study, 1,864 TF members were obtained in the assembly transcriptome sequence of S. dendroides, and 120 members were differently expressed under high salt treatment. In addition, 26, 36, 64, and 63 TFs were differently expressed in H1 vs C, H5 vs C, H10 vs C, and H15 vs C comparisons, respectively (Supplementary Table S8). The AP2/ERF family is the most abundant TF family in each comparison, followed by WRKY, MYB, FAR1, NAC, MADS, HB, bHLH, and bZIP (Figure 7C), suggesting that these TFs might be the main regulatory factors of S. dendroides plants under high salt treatment. The expression profiles of these major TFs under high salt treatment are displayed in Figure 7D.
In order to verify RNA-seq data, 10 genes involved in different important biological processes were selected for QPCR verification analysis. These 10 genes were involved in ABA signal transduction (Cluster-12993.0 encoding PP2C and Cluster-688.5991 encoding Abscisic acid receptor PYL4), peroxidase (Cluster-25515.0 encoding peroxidase 44-like), Glycerophospholipid metabolism (Cluster-688.49983 encoding phosphoethanolamine N-methyltransferase), transcription factors (Cluster-688.16078 encoding AP2/ERF-ERF and Cluster-688.2897 encoding C2H2), (Cluster-688.57964 encoding BURP domain-containing protein 5-like), and three genes, Cluster-688.64534, Cluster-688.79365, and Cluster-688.79222, highly expressed at all the treatments without any annotation information. The results of QPCR analysis are shown in Figure 8. Moreover, the expression correlation between RNA-seq data and QPCR was calculated, and the correlation coefficient was 0.8853 (Supplementary Figure S8), suggesting that the reliability of RNA-seq data was high and the DEGs obtained from RNA-seq data should be reliable.
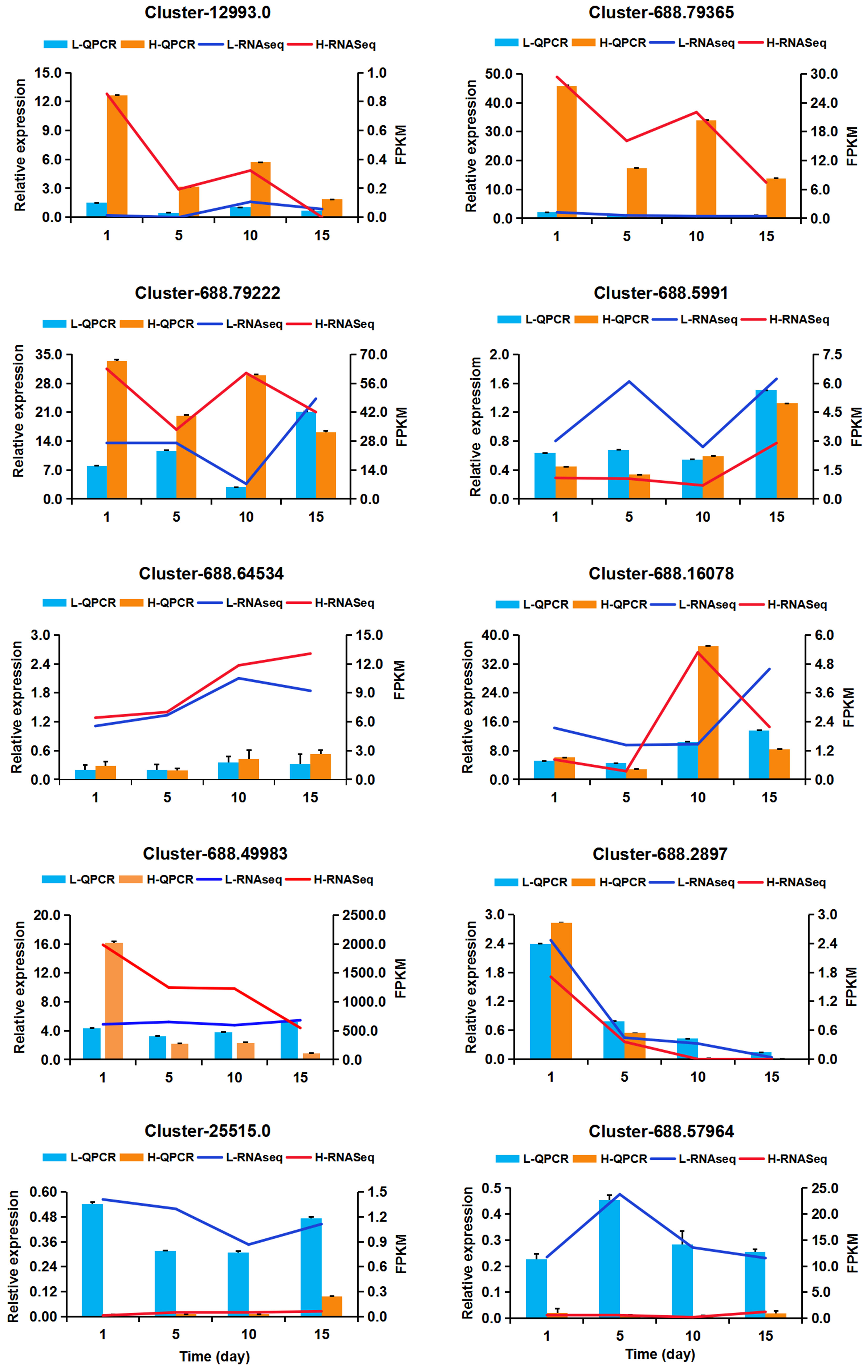
Figure 8 Quantitative real-time PCR validation of 10 DEGs. X-axis means NaCl treatment times. The left Y-axis represents the relative expression level of genes and displayed in bar charts, while the right Y-axis represents the PPKM value and showed by fold lines. Values are presented as the mean ± the standard error (SE) with three biological replicates.
Soil salinization has become a global ecological problem, especially in arid and semi-arid areas, which affects the establishment, development, and growth of plants and eventually leads to crop yield reduction (Lesk et al., 2016; Kopecka et al., 2023). Halophytes grow well in saline soil with a concentration of about 200 mM NaCl or above and survive in extremely harsh environments (Flowers and Colmer, 2008), which provides valuable resources for us to study the complex physiological and molecular mechanisms of plants adapting to abiotic stress. In this study, the adaptive mechanisms of S. dendroides associated with high salt stress were analyzed at the physiological and molecular levels. The fresh weight, dry weight, and relative water content of seedlings showed the highest under 200 mM NaCl treatment (Figures 1A-C), indicating that a low concentration of NaCl could promote the growth of seedlings. Like many succulent euhalophyte species, for example, Suaeda salsa grows optimally at 200 mM NaCl, and the optimal growth is accompanied by an increase in succulence and other morphological changes (Song et al., 2009). Under 800 and 1000mM NaCl stress, the seedlings showed wilting symptoms, and some seedlings even died under 1000mM NaCl. However, the wilting symptoms of the seedlings began to recover after 5 days of the treatment, recovered after 10 days and gradually returned to normal growth. After 10 days of NaCl treatment, the content of Na and K in the shoots of S. dendroides showed that the change of Na/K ratio was moderate and kept a relatively stable level when the salt concentration increased to 600 mM (Figures 1G-I). These indicate that S. dendroides could maintain Na/K balance under high salt stress to avoid ion toxicity. Furthermore, high salt stress can induce the accumulation of proline and soluble sugar, providing osmoprotection and energy for S. dendroides plants to adapt to stress environments. The sequencing result showed that the DEG number was the highest after 10 days of high salt treatment (Figure 2A), indicating that this period was a crucial time for S. dendroides to adapt to the saline environment. According to the expression level of key genes at different treatment times of high salt treatment, our study indicates that cell wall remodeling, ion homeostasis and compartmentalization, osmotic adjustment, and plant hormone signal transduction regulate the adaptation mechanism of S. dendroides suffering from high salt stress (Figure 9).
The plant cell wall is a highly dynamic and complex network, mainly composed of different polysaccharides, lignin, and various structural proteins, which is a barrier to protect plants from the environment (Vaahtera et al., 2019). The cell wall integrity (CWI) maintenance system plays a crucial role in stress sensing and response for plants (Vaahtera et al., 2019; Rui and Dinneny, 2020). At present, a range of plasma membrane-localized receptor-like kinases and cell wall glycoproteins involved in CWI perception and maintenance have been identified. FER belongs to CrRLK1L family and is regarded as a cell wall sensor, which is necessary to activate Ca2+ influx and maintain CWI under salt stress (Feng et al., 2018). Other protein kinase genes, such as WAK and LRX, are thought to bind to cell wall components and play an important role in regulating cell wall integrity (Decreux and Messiaen, 2005; Herger et al., 2019). In our study, 4 FER, 1 LRX3, 5 WAK, and 4 WAKL genes were differently expressed under 800mM NaCl stress (Figure 5D). Several proteins, enzymes, and ions are related to the cell wall modifications in response to abiotic stresses. Generally, salt stress may directly trigger the activation of pectin methyl esterase, which leads to the decrease of pectin methylation degree, thus affecting the characteristics of cell wall (Gigli-Bisceglia et al., 2022). Acetylation is another important pathway of pectin modification (de Souza et al., 2014). Homogalacturonan (HG) is the most abundant pectic polymer, and its remodeling is regulated by special enzymes, such as PME, PAE, PG, or PLL (Senechal et al., 2014). In S. dendroides, some cell wall modification related genes were identified, 4 PME, 5 PL, and 4 PG1 genes were up-regulated under 800 mM NaCl stress (Figure 5E). The result showed that the enzymes involved in pectin modification might be important for S. dendroides to adapt to high salt stress. Additionally, cell wall-loosening protein EXPs and XTHs play important roles in coping with high salt stress. AtXTH30 has a negative effect on salt tolerance (Yan et al., 2019). AtXTH19 and AtXTH23 were induced by brassinosteroids, and the double mutant (atxth19atxth23) was sensitive to salt stress (Xu et al., 2020). EXPs were induced by salt stress, and overexpression of EXP gene was helpful to promote plant salt tolerance (Chen et al., 2017; Jadamba et al., 2020). 20 XTHs and 3 EXPAs in S. dendroides were obtained and differently expressed under 800 mM NaCl stress (Figure 5B). Thus, cell wall remodeling is particularly important for S. dendroides to adapt to a high concentration of Na+ condition.
SCW is thickened cell walls, mainly consisting of cellulose, hemicellulose, lignin, a minor amount of structural proteins and enzymes, which play an essential role in maintaining plant morphology, providing rigid support and ensuring material transportation, and participating in plant biotic and abiotic stress response as a protective barrier (Cosgrove, 2005; Hamann, 2012). In this study, cellulose synthase genes CESA4, CESA7, and CESA8, related to the SCW synthesis, increased prominently after 10 and 15 days of high salt stress (Figure 5A). PPI analysis showed that the expression of COBL4 was positively correlated with the expression of cellulose synthesis genes CESA4, CESA7, and CESA8 related to SCW (Figure 5F). COBL4 encodes a COBRA-like protein that has typical structural characteristics of a glycosylphosphatidylinositol-anchored protein, like sorghum SbBC1, which is homologous to OsBC1 and AtCOBL4, involved in the biosynthesis of cellulose in SCW and affects the mechanical strength of sorghum plants (Li et al., 2019). The expression level of CESA4/7/8 and COBL4 increased significantly under 800 mM NaCl treatment, indicating that high salt stress can promote SCW cellulose synthesis. Xylan is the main component of hemicellulose polysaccharide. Genetic and biochemical analyses revealed that the synthesis of SCW xylan backbone was mediated by IRX10/IRX10L of the GT47 family, IRX9/IRX9L and IRX14/IRX14L of the GT43 family, and GUXs of the GT8 family (Zhong et al., 2019). As the result showed in Figure 5A, the DEGs involved in xylan backbone synthesis, such as IRX9, IRX14, IRX15, and IRX15L, were up-regulated and significantly increased after 10 and 15 days of 800 mM NaCl stress. Moreover, the expression level of the gene IRX7 involved in xylan reduction terminal synthesis and the genes GUX1 and GUX2 involved in side chain synthesis increased significantly after 10 and 15 days of high salt stress. In addition, 5 TBL encoding DEGs were significantly up-regulated after 15 days of 800mM NaCl stress. TBL32 and TBL33 are putative acetyltransferases, which participate in acetyl substitutions of 2-O-GlcA-substituted xylosyl residues, playing important roles in xylan acetylation and normal deposition of the SCW (Yuan et al., 2016). Furthermore, PPI network analysis of DEGs at different treatment times under high salt stress showed that the hub gene IRX9 (Cluster-688.80914) was positively correlated with GUX1, IRX7, CSEA4, CESA7, CESA8 and SUS3. These indicated that high salt stress could promote the synthesis of xylan in SCW of S. dendroides.
However, the coordinated expression of genes involved in cellulose, xylan, and lignin biosynthesis in SCW is mediated by the NAC-MYB transcriptional network (Nakano et al., 2015). A set of closely related NAC domain TFs acts as the primary switch of plant SCW biosynthesis, activating downstream TFs to regulate the entire network of SCW biosynthesis (Mitsuda et al., 2005; Mitsuda et al., 2007). Most of the R2R3-MYB subfamily TFs function as the second layer-master switches of SCW biosynthesis, such as MYB26/103 and MYB46/83 (Xiao et al., 2021). Moreover, the genes involved in ABA synthesis and signal transduction are related to SCW thickening and lignification in Arabidopsis (Endler and Persson, 2011; Wang L. et al., 2020). AtNST1, a NAC domain family TF that can activate the downstream genes related to SCW biosynthesis. Studies have shown that AtNST1 is a key phosphorylation substrate of SnRK2, which acts as the key positive regulator of ABA signal transduction, which regulates SCW formation and lignin deposition in the stem fiber region of Arabidopsis thaliana (Liu et al., 2021). In this study, the expression level of gene NST1 was significantly increased after 15 days of 800 mM NaCl stress (Figure 7D, Supplementary Table S7). MYB4, MYB86, and MYB26/103 were significantly up-regulated after 15 days of high salt stress, and MYB46/83 was notably up-regulated after 10 days (Figure 7D). Therefore, the genes involved in SCW biosynthesis and thickening may be extremely important for S. dendroides adapting to high salt environment. In brief, multiple genes related to cell wall biosynthesis, modification, sensing, and CWI maintenance of S. dendroides were significantly expressed under high salt stress. The results indicated that cell wall remodeling and SCW thickening were the key drivers for S. dendroides plants adapting to high salt stress.
Succulence is another mechanism for euhalophyte plants to adapt to high salt environments. A large amount of inorganic ions and organic osmotic adjustment substances are accumulated in succulent leaves and stems to maintain osmotic balance (Flowers and Colmer, 2015; Rahman et al., 2021). In this study, a series of genes related to inorganic ion transport, including SOS1, HKT1, NHX1, TPK3, PMA, and AVP, were prominently expressed under 800 mM NaCl treatment (Figure 6A, Supplementary Table S7). The plasma membrane Na+/H+ antiporter AtSOS1 is located at the root tips, excluding Na+ from the root, thus reducing Na+absorption (Shi et al., 2000). Na+ influx into cells is mediated mainly by non-selective cation channels and the sodium transporter HKT1. It has been proved that AtHKT1 can unload Na+ from xylem vessels to parenchyma cells and/or control the retrieval of Na+ from xylem to reduce the amount of Na+ transported to shoots (Sunarpi et al., 2005). In addition, SOS1 and HKT1 mediate opposite Na+ fluxes, and coordinate the regulation of Na+ transport and ion homeostasis under salt stress (Wang W.-Y. et al., 2020). Tonoplast located Na+(K+)/H+ antiporter NHX1 has been proven to separate excessive cytoplasmic Na+(K+) into vacuoles, which was motivated by the proton motive force produced by vacuolar H+- ATPase (VHA) and H+-PPase (Yang et al., 2010). The KT/HAK/KUP family genes act as K+/H+ symporter, and most of them are important for K+ absorption in plant roots (Wang Y. et al., 2021). Tonoplast localized AtTPK1 functions in K+ transport across the tonoplast and plays important roles in many physiological processes (Gobert et al., 2007). As a representative euhalophyte, S. dendroides could accumulate a large amount of Na+ in succulent leaves, and maintain the stability of K+ by promoting the ability of K+ transportation under high salt treatment, indicating that S. dendroides has the ability to modulate Na+/K+ balance. Moreover, it has been found that the concentration of cytosolic Ca2+ ([Ca2+]cyt) increases with numerous environmental conditions. Constant [Ca2+]cyt is maintained by Ca2+ influx through Ca2+-permeable ion channels or transporters located in the plasma membrane or tonoplast, and other membranes. MCA1 and MCA2 are responsible for Ca2+ influx in Arabidopsis cells. Over-expression of MCA1 driven by a strong promoter can promote the absorption of Ca2+ in roots and increase cytoplasmic Ca2+ concentration upon hypoosmotic shock (Yamanaka et al., 2010). Two-pore calcium channel protein TPC1 located in the tonoplast acts as a non-selective cation channel, which is co-regulated by voltage and Ca2+ to produce a slow vacuolar current (Ye et al., 2021). besides, CNGC, located mostly in the plasma membrane, enables the absorption of Na+, K+, and Ca2+. AtCNGC10 is involved in K+ and Na+ absorption and long-distance transportation, and AtCNGC17 mediated the influx of osmotically active K+ and the second messenger Ca2+, or both (Jarratt-Barnham et al., 2021). In S. dendroides, 3 CNGC genes were significantly expressed, CNGC4 was significantly increased, while 2 CNGC17 genes decreased at all the time points of 800 mM NaCl treatment (Figure 7A, Supplementary Table S7). Furthermore, cytosolic Ca2+ binds to calcium sensor protein SOS3/CBL4, and subsequently interacts with protein kinase SOS2/CIPK24 to form a complex activator that activates SOS1, a Na+/H+ antiporter, resulting in Na+ efflux from the cytosol. The salt overly sensitive (SOS) pathway comprises CBL4/SOS3, CIPK24/SOS2, and SOS1, which has been considered the key mechanism of Na+ homeostasis and salt tolerance in Arabidopsis (Zhu, 2000). Apart from interacting with SOS1 at the plasma membrane, CIPK24 is also reported to regulate the activity of several tonoplast located transporters by interacting with them, such as Ca2+/H+ antiporter (Cheng et al., 2004), vacuolar V-ATPase (Batelli et al., 2007), and Na+/H+ exchanger (Qiu et al., 2004; Huertas et al., 2012). In this study, CIPK9, the closest SOS2 homolog in S. dendroides, functions in a similar manner to SOS2 in the context of the SOS3-CIPK9-SOS1 complex to mediate salt tolerance in halophytes S. dendroides (Figure 9). These indicate that S. dendroides has an efficient Na+ transport and compartmentalization mechanism to maintain ion homeostasis under high salt environment.
In addition to Na+/K+ homeostasis, higher plants need to adjust the balance of various nutrients in saline environments. Several Arabidopsis NPF genes were discovered to play a crucial role in nitrate uptake and transport (Corratge-Faillie and Lacombe, 2017), such as NRT1.1/AtNPF6.3/CHL1 involved in nitrate uptake. NPF7.3/NRT1.5 plays important roles in K+ translocation from root to shoot, and it also participates in coordination of K+/NO3- distribution in plants. NPF2.5 was expressed in the plasma membrane of root cortical cells, functioned in Cl− efflux from the root, and was up-regulated by NaCl. In the present study, the DEGs involved in the absorption and transportation of some important nutrients were significantly induced by high salt stress. Notably, 15 NPF encoding genes were identified and differently expressed under high salt treatment, in which 7 NPFs, including 4 NPF 3.1 and NPF 1.1/1.2/6.4, were significantly increased after 1 day of high salt stress, while 8 NPFs were down-regulated (Figure 6A, Supplementary Table S7). In addition, NPF4.6/AIT1/NRT1.2 functions as an ABA importer at the site of ABA biosynthesis, plays an important role in regulating the stomatal aperture in inflorescence stems (Kanno et al., 2012). Moreover, it has been found that several members of the ABCG subfamily functioned in ABA transport (Borghi et al., 2015), such as ABCG25, which functions as an ABA exporter from the vascular tissue, and ABCG40 is an ABA uptake transporter in guard cells. A total of 18 ABC family genes were differentially expressed under 800 mM NaCl treatment, including 2 ABCBs, 11 ABCCs, and 5 ABCGs, among which the genes of the ABCC subfamily and the ABCG subfamily were more active under high salt stress (Figure 6A, Supplementary Table S7). Therefore, S. dendroides could promote the absorption and transport of various nutrient elements by increasing the transcription lever of transporters/channels.
PPI analysis showed that P5CS interacted with the DEGs encoding ALDH and BADH, genes involved in Ca2+ signaling and antioxidants (Figure 6C). The ALDH superfamily comprises a variety of enzymes involved in endogenous and exogenous aldehyde metabolism (Tola et al., 2021). P5CS, a rate-limiting enzyme in proline biosynthesis, is a member of ALDH18 family, and its expression is significantly up-regulated in dehydration reactions (Yoshiba et al., 1997). Studies have shown that the accumulation of proline under salt stress is related to the up regulation of P5CS (Mansour and Ali, 2017). Proline plays an essential role in protecting plant growth from unfavorable environmental conditions by osmotic adjustment, working as a molecular chaperone, and protecting the integrity of proteins and enzymes (Ozturk et al., 2021). Nevertheless, phytohormone ABA is a crucial component in integrating multiple signals, controlling downstream stress responses, and influencing proline synthesis and accumulation by regulating the expression of P5CS and P5CR in plants (Verslues and Bray, 2006). The interaction of ABA and the Ca2+ signaling pathway can also regulate proline accumulation in plants under stress conditions. In this study, several DEGs encoding TPC1A, MCA1, CBL4, and CML22 involved in Ca2+ signaling pathway were directly or indirectly related to P5CS (Figure 6C). In addition, proline can also stabilize the antioxidant system and protect the integrity of cell membranes through osmotic adjustment, thereby reducing the influence of ROS and overcoming or repairing stress injury (Banu et al., 2009). In S. dendroides, the major antioxidant enzymes, including SOD, CAT, APX, CPR, and Fer, obviously expressed under high salt stress, may interact with the genes involved in osmoregulation (Figure 6C). BADH, also known as ALDH10 family genes, has been widely studied for its role in stress responses and the production of the osmoprotectant GB (Tola et al., 2021). In plants, PEAMT is the rate-limiting enzyme for synthesizing choline, the precursor of betaine (Nuccio et al., 2000), while CMO and BADH are the key enzymes involved in betaine biosynthesis. It was found that the tolerance of gene CMO and BADH over-expressed plants was enhanced (Shirasawa et al., 2006; Wu et al., 2008). In our study, several key genes involved in GB synthesis were significantly up-regulated under 800 mM NaCl stress, like CMO, BADH, and PEAMT encoding genes were continuously expressed under 800 mM NaCl treatment for 1, 5, 10, and 15 days (Figure 6B). The result above suggests that S. dendroides could synthesize and accumulate a large number of osmoprotectants to protect the plants from high salt stress by regulating the expression patterns of key genes involved in the biosynthesis of compatible solutes.
This study identified potential genes showing a possible adaptation mechanism of halophytic S. dendroides encountering a high salt condition, mainly involved in cell wall metabolism, inorganic ion transport, and organic osmolyte synthesis (Figure 9). A large number of DEGs encoding cell wall sensing, modification, and secondary wall thickening were prominently increased under 800 mM NaCl treatment, which might be important for cell wall repair and integrity maintenance under high salt concentrations. Moreover, the DEGs encoding Na+, K+, Ca2+, and Cl-/NO3- transporters/channels were significantly increased under 800 mM NaCl treatment, which might be beneficial to promote the absorption and transport of nutrient elements under high salt stress. Notably, some genes involved in Na+/K+ transport (such as SOS1, HKT1, NHX, and TPK3) might play important roles in Na+ transporting and sequestering, maintaining ion homeostasis under high saline conditions. Moreover, several genes involved in the synthesis of organic regulators were significantly up-regulated under 800 mM NaCl treatment, suggesting that S. dendroides possesses an effective mechanism to accumulate more osmoprotectants to enhance salt tolerance. Our results contribute a new perspective for understanding the molecular mechanisms of halophytes adapting to high salinity and provide a basis for genetic improvement of salt tolerance in important plants by using excellent genes from halophytic S. dendroides.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: SRA, and the accession numbers PRJNA1018260.
PM: Conceptualization, Methodology, Supervision, Writing – original draft, Writing – review & editing, Formal Analysis, Investigation. JL: Methodology, Writing – review & editing. GS: Methodology, Conceptualization, Project administration, Writing – review & editing, Funding acquisition, Investigation, Resources, Supervision. JZ: Conceptualization, Methodology, Project administration, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This research was funded by the Key Research and Development Task and Special Project of Xinjiang Uygur Autonomous Region (Grant No. 2022B02052-2), the Applied Basic Research Program of Xinjiang Production and Construction Corps (Grant No. 2016AG005), and the Project of Innovation Team Building in Key Areas of Xinjiang Production and Construction Corps (Grant No. 2019CB008).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2024.1283912/full#supplementary-material
Banu, M. N. A., Hoque, M. A., Watanabe-Sugimoto, M., Matsuoka, K., Nakamura, Y., Shimoishi, Y., et al. (2009). Proline and glycinebetaine induce antioxidant defense gene expression and suppress cell death in cultured tobacco cells under salt stress. J. Plant Physiol. 166, 146e156. doi: 10.1016/j.jplph.2008.03.002
Batelli, G., Verslues, P. E., Agius, F., Qiu, Q. S., Fujii, H., Pan, S. Q., et al. (2007). SOS2 promotes salt tolerance in part by interacting with the vacuolar H+-ATPase and upregulating its transport activity. Mol. Cell. Biol. 27, 7781–7790. doi: 10.1128/MCB.00430-07
Benjamini, Y., Hochberg, Y. (1995). Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc B. 57, 289–300. doi: 10.1111/j.2517-6161.1995.tb02031.x
Borghi, L., Kang, J., Ko, D., Lee, Y., Martinoia, E. (2015). The role of ABCG-type ABC transporters in phytohormone transport. Biochem. Soc Trans. 43, 924–930. doi: 10.1042/BST20150106
Calone, R., Bregaglio, S., Sanoubar, R., Noli, E., Lambertini, C., Barbanti, L. (2021). Physiological adaptation to water salinity in six wild halophytes suitable for mediterranean agriculture. Plants 10, 309. doi: 10.3390/plants10020309
Chen, Y.-H., Han, Y.-Y., Kong, X.-Z., Kang, H.-H., Ren, Y.-Q., Wang, W. (2017). Ectopic expression of wheat expansin gene TaEXPA2 improved the salt tolerance of transgenic tobacco by regulating Na+/K+ and antioxidant competence. Physiol. Plant 159, 161–177. doi: 10.1111/ppl.12492
Cheng, N. H., Pittman, J. K., Zhu, J.-K., Hirschi, K. D. (2004). The protein kinase SOS2 activates the Arabidopsis H+/Ca2+antiporter CAX1 to integrate calcium transport and salt tolerance. J. Biol. Chem. 279, 2922–2926. doi: 10.1074/jbc.M309084200
Corratge-Faillie, C., Lacombe, B. (2017). Substrate (un)specificity of arabidopsis NRT1/PTR FAMILY (NPF) proteins. J. Exp. Bot. 68, 3107–3013. doi: 10.1093/jxb/erw499
Cosgrove, D. J. (2005). Growth of the plant cell wall. Nat. Rev. Mol. Cell Biol. 6, 850–861. doi: 10.1038/nrm1746
Decreux, A., Messiaen, J. (2005). Wall-associated kinase WAK1 interacts with cell wall pectins in a calcium-induced conformation. Plant Cell Physiol. 46, 268–278. doi: 10.1093/pcp/pci026
de Souza, A., Hull, P. A., Gille, S., Pauly, M. (2014). Identification and functional characterization of the distinct plant pectin esterases PAE8 and PAE9 and their deletion mutants. Planta 240, 1123–1138. doi: 10.1007/s00425-014-2139-6
Endler, A., Persson, S. (2011). Cellulose synthases and synthesis in arabidopsis. Mol. Plant 4, 199–211. doi: 10.1093/mp/ssq079
Feng, W., Kita, D., Peaucelle, A., Cartwright, H. N., Doan, V., Duan, Q.-H., et al. (2018). The FERONIA receptor kinase maintains cell-wall integrity during salt stress through Ca2+ Signaling. Curr. Biol. 28, 666–675.e5. doi: 10.1016/j.cub.2018.01.023
Flowers, T. J., Colmer, T. D. (2008). Salinity tolerance in halophytes. New Phytol. 179, 945–963. doi: 10.1111/j.1469-8137.2008.02531.x
Flowers, T. J., Colmer, T. D. (2015). Plant salt tolerance: adaptations in halophytes. Ann. Bot. 115, 327–331. doi: 10.1093/aob/mcu267
Flowers, T. J., Munns, R., Colmer, T. D. (2015). Sodium chloride toxicity and the cellular basis of salt tolerance in halophytes. Ann. Bot. 115, 419–431. doi: 10.1093/aob/mcu217
Gigli-Bisceglia, N., van Zelm, E., Huo, W.-Y., Lamers, J., Testerink, C. (2022). Arabidopsis root responses to salinity depend on pectin modification and cell wall sensing. Development 149, dev200363. doi: 10.1242/dev.200363
Gobert, A., Isayenkov, S., Voelker, C., Czempinski, K., Maathuis, F. J. M. (2007). The two-pore channel TPK1 gene encodes the vacuolar K+ conductance and plays a role in K+ homeostasis. Proc. Natl. Acad. Sci. U.S.A. 104, 10726–10731. doi: 10.1073/pnas.0702595104
Grabherr, M. G., Haas, B. J., Yassour, M., Levin, J. Z., Thompson, D. A., Amit, I., et al. (2011). Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 29, 644–652. doi: 10.1038/nbt.1883
Guo, J.-R., Du, M., Tian, H.-Y., Wang, B.-S. (2020). Exposure to high salinity during seed development markedly enhances seedling emergence and fitness of the progeny of the extreme halophyte Suaeda salsa. Front. Plant Sci. 11. doi: 10.3389/fpls.2020.01291
Guo, J.-R., Li, Y.-D., Han, G.-L., Song, J., Wang, B.-S. (2017). NaCl markedly improved the reproductive capacity of the euhalophyte Suaeda salsa. Funct. Plant Biol. 45, 350–361. doi: 10.1071/FP17181
Hamann, T. (2012). Plant cell wall integrity maintenance as an essential component of biotic stress response mechanisms. Front. Plant Sci. 3. doi: 10.3389/fpls.2012.00077
Hao, X.-Y., Li, J.-P., Gao, S.-Q., Tuerxun, Z., Chang, X.-C., Hu, W.-R., et al. (2020). SsPsaH, a H subunit of the photosystem I reaction center of Suaeda salsa, confers the capacity of osmotic adjustment in tobacco. Genes Genom. 42, 1455–1465. doi: 10.1007/s13258-020-00970-4
He, R., Yu, G.-H., Han, X.-R., Han, J., Li, W., Wang, B., et al. (2017). ThPP1 gene, encodes an inorganic pyrophosphatase in Thellungiella halophila, enhanced the tolerance of the transgenic rice to alkali stress. Plant Cell Rep. 36, 1929–1942. doi: 10.1007/s00299-017-2208-y
Herger, A., Dunser, K., Kleine-Vehn, J., Ringli, C. (2019). Leucine-rich repeat extensin proteins and their role in cell wall sensing. Curr. Biol. 29, R851–R858. doi: 10.1016/j.cub.2019.07.039
Himabindu, Y., Chakradhar, T., Reddy, M. C., Kanygin, A., Redding, K. E., Chandrasekhar, T. (2016). Salt-tolerant genes from halophytes are potential key players of salt tolerance in glycophytes. Environ. Exp. Bot. 124, 39–63. doi: 10.1016/j.envexpbot.2015.11.010
Huertas, R., Olias, R., Eljakaoui, Z., Galvez, F. J., Li, J., De Morales, P. A., et al. (2012). Overexpression of SlSOS2 (SlCIPK24) confers salt tolerance to transgenic tomato. Plant Cell Environ. 35, 1467–1482. doi: 10.1111/j.1365-3040.2012.02504.x
Jadamba, C., Kang, K., Paek, N. C., Lee, S. I., Yoo, S. C. (2020). Overexpression of rice expansin 7 (Osexpa7) confers enhanced tolerance to salt stress in rice. Int. J. Mol. Sci. 21, 454. doi: 10.3390/ijms21020454
Jarratt-Barnham, E., Wang, L.-M., Ning, Y.-Z., Davies, J. M. (2021). The complex story of plant cyclic nucleotide-gated channels. Int. J. Mol. Sci. 22, 874. doi: 10.3390/ijms22020874
Kanno, Y., Hanada, A., Chiba, Y., Ichikawa, T., Nakazawa, M., Matsui, M., et al. (2012). Identification of an abscisic acid transporter by functional screening using receptor complex as a sensor. Proc. Natl. Acad. Sci. U.S.A. 109, 9653–9658. doi: 10.1073/pnas.1203567109
Kopecka, R., Kameniarova, M., Cerny, M., Brzobohaty, B., Novak, J. (2023). Abiotic stress in crop production. Int. J. Mol. Sci. 24, 6603. doi: 10.3390/ijms24076603
Lesk, C., Rowhani, P., Ramankutty, N. (2016). Influence of extreme weather disasters on global crop production. Nature 529, 84–87. doi: 10.1038/nature16467
Li, P., Liu, Y.-R., Tan, W.-Q., Chen, J., Zhu, M.-J., Lv, Y., et al. (2019). Brittle culm 1 encodes a cobra-like protein involved in secondary cell wall cellulose biosynthesis in sorghum. Plant Cell Physiol. 60, 788–801. doi: 10.1093/pcp/pcy246
Liang, J.-P., Shi, W.-J. (2021). Cotton/halophytes intercropping decreases salt accumulation and improves soil physicochemical properties and crop productivity in saline-alkali soils under mulched drip irrigation: A three-year field experiment. Field Crop Res. 262, 108027. doi: 10.1016/j.fcr.2020.108027
Liu, C., Yu, H., Rao, X. L., Li, L. G., Dixon, R. A. (2021). Abscisic acid regulates secondary cell-wall formation and lignin deposition in Arabidopsis thaliana through phosphorylation of NST1. Proc. Natl. Acad. Sci. U.S.A. 118, e2010911118. doi: 10.1073/pnas.2010911118
Livak, K. J., Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Love, M. I., Huber, W., Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550. doi: 10.1186/s13059-014-0550-8
Lv, S.-L., Zhang, K.-W., Gao, Q., Lian, L.-J., Song, Y.-J., Zhang, J.-R. (2008). Overexpression of an H+-PPase gene from Thellungiella halophila in cotton enhances salt tolerance and improves growth and photosynthetic performance. Plant Cell Physiol. 49, 1150–1164. doi: 10.1093/pcp/pcn090
Mansour, M. M. F., Ali, E. F. (2017). Evaluation of proline functions in saline conditions. Phytochemistry 140, 52–68. doi: 10.1016/J.Phytochem.2017.04.016
Mao, X.-Z., Cai, T., Olyarchuk, J. G., Wei, L.-P. (2005). Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics 21, 3787–3793. doi: 10.1093/bioinformatics/bti430
Mishra, A., Tanna, B. (2017). Halophytes: potential resources for salt stress tolerance genes and promoters. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.00829
Mitsuda, N., Iwase, A., Yamamoto, H., Yoshida, M., Seki, M., Shinozaki, K., et al. (2007). NAC transcription factors, NST1 and NST3, are key regulators of the formation of secondary walls in woody tissues of Arabidopsis. Plant Cell 19, 270–280. doi: 10.1105/tpc.106.047043
Mitsuda, N., Seki, M., Shinozaki, K., Ohme-Takagi, M. (2005). The NAC transcription factors NST1 and NST2 of Arabidopsis regulate secondary wall thickenings and are required for anther dehiscence. Plant Cell 17, 2993–3006. doi: 10.1105/tpc.105.036004
Nakano, Y., Yamaguchi, M., Endo, H., Rejab, N. A., Ohtani, M. (2015). NAC-MYB-based transcriptional regulation of secondary cell wall biosynthesis in land plants. Front. Plant Sci. 6. doi: 10.3389/fpls.2015.00288
Nuccio, M. L., McNeil, S. D., Ziemak, M. J., Hanson, A. D., Jain, R. K., Selvaraj, G. (2000). Choline import into chloroplasts limits glycine betaine synthesis in tobacco: analysis of plants engineered with a chloroplastic or a cytosolic pathway. Metab. Eng. 2, 300–311. doi: 10.1006/mben.2000.0158
Ozturk, M., Unal, B. T., Garcia-Caparros, P., Khursheed, A., Gul, A., Hasanuzzaman, M. (2021). Osmoregulation and its actions during the drought stress in plants. Physiol. Plant 172, 1321–1335. doi: 10.1111/ppl.13297
Qiu, Q.-S., Guo, Y., Quintero, F. J., Pardo, J. M., Schumaker, K. S., Zhu, J.-K. (2004). Regulation of vacuolar Na+/H+ exchange in Arabidopsis thaliana by the SOS pathway. J. Biol. Chem. 279, 207–215. doi: 10.1074/jbc.M307982200
Rahman, M. M., Mostofa, M. G., Keya, S. S., Siddiqui, M. N., Ansary, M. M. U., Das, A. K., et al. (2021). Adaptive mechanisms of halophytes and their potential in improving salinity tolerance in Plants. Int. J. Mol. Sci. 22, 10733. doi: 10.3390/ijms221910733
Rui, Y., Dinneny, J. R. (2020). A wall with integrity: surveillance and maintenance of the plant cell wall under stress. New Phytol. 225, 1428–1439. doi: 10.1111/nph.16166
Senechal, F., Wattier, C., Rusterucci, C., Pelloux, J. (2014). Homogalacturonan-modifying enzymes: structure, expression, and roles in plants. J. Exp. Bot. 65, 5125–5160. doi: 10.1093/jxb/eru272
Shannon, P., Markiel., A., Ozier, O., Baliga, N. S., Wang, J. T., Ramage, D., et al. (2003). Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504. doi: 10.1101/gr.1239303
Shao, Q., Han, N., Ding, T.-L., Zhou, F., Wang, B.-S. (2014). SsHKT1;1 is a potassium transporter of the C3 halophyte Suaeda salsa that is involved in salt tolerance. Funct. Plant Biol. 41, 790–802. doi: 10.1071/FP13265
Shi, H.-Z., Ishitani, M., Kim, C., Zhu, J.-K. (2000). The Arabidopsis thaliana salt tolerance gene SOS1 encodes a putative Na+/H+ antiporter. Proc. Natl. Acad. Sci. U.S.A. 97, 6896–6901. doi: 10.1073/pnas.120170197
Shirasawa, K., Takabe, T., Takabe, T., Kishitani, S. (2006). Accumulation of glycinebetaine in rice plants that overexpress choline monooxygenase from spinach and evaluation of their tolerance to abiotic stress. Ann. Bot. 98, 565–571. doi: 10.1093/aob/mcl126
Song, J., Chen, M., Feng, G., Jia, Y.-H., Wang, B.-S., Zhang, F.-S. (2009). Effect of salinity on growth, ion accumulation and the roles of ions in osmotic adjustment of two populations of Suaeda salsa. Plant Soil 314, 133–141. doi: 10.1007/s11104-008-9712-3
Sunarpi, Horie, T., Motoda, J., Kubo, M., Yang, H., Yoda, K., et al. (2005). Enhanced salt tolerance mediated by AtHKT1 transporter-induced Na+unloading from xylem vessels to xylem parenchyma cells. Plant J. 44, 928–938. doi: 10.1111/j.1365-313X.2005.02595.x
Tola, A. J., Jaballi, A., Germain, H., Missihoun, T. D. (2021). Recent development on plant aldehyde dehydrogenase enzymes and their functions in plant development and stress signaling. Genes 12, 51. doi: 10.3390/genes12010051
Vaahtera, L., Schulz, J., Hamann, T. (2019). Cell wall integrity maintenance during plant development and interaction with the environment. Nat. Plants 5, 924–932. doi: 10.1038/s41477-019-0502-0
Verslues, P. E., Bray, E. A. (2006). Role of abscisic acid (ABA) and Arabidopsis thaliana ABA-insensitive loci in low water potential-induced ABA and proline accumulation. J. Exp. Bot. 57, 201e212. doi: 10.1093/jxb/erj026
Wang, Y., Chen, Y.-F., Wu, W.-H. (2021). Potassium and phosphorus transport and signaling in plants. J. Integr. Plant Biol. 63, 34–52. doi: 10.1111/jipb.13053
Wang, L., Hart, B. E., Khan, G. A., Cruz, E. R., Persson, S., Wallace, I. S. (2020). Associations between phytohormones and cellulose biosynthesis in land plants. Ann. Bot. 126, 807–824. doi: 10.1093/aob/mcaa121
Wang, W.-Y., Liu, Y.-Q., Duan, H.-R., Yin, X.-X., Cui, Y.-N., Chai, W.-W., et al. (2020). SsHKT1;1 is coordinated with SsSOS1 and SsNHX1 to regulate Na+ homeostasis in Suaeda salsa under saline conditions. Plant Soil 449, 117–131. doi: 10.1007/s11104-020-04463-x
Wang, X.-Y., Shao, X.-T., Zhang, W.-J., Sun, T., Ding, Y.-L., Lin, Z., et al. (2022). Genus Suaeda: advances in phytology, chemistry, pharmacology and clinical application, (1895-2021). Pharmacol. Res. 179, 106203. doi: 10.1016/j.phrs.2022.106203
Wang, F.-L., Song, N.-N. (2019). Salinity-induced alterations in plant growth, antioxidant enzyme activities, and lead transportation and accumulation in suaeda salsa: implications for phytoremediation. Ecotoxicology 28, 520–527. doi: 10.1007/s10646-019-02048-8
Wang, L., Wang, X., Jiang, L., Zhang, K., Tanveer, M., Tian, C.-Y., et al. (2021). Reclamation of saline soil by planting annual euhalophyte Suaeda salsa with drip irrigation: A three-year field experiment in arid northwestern China. Ecol. Eng. 159, 106090. doi: 10.1016/j.ecoleng.2020.106090
Wu, W., Su, Q., Xia, X.-Y., Wang, Y., Luan, Y.-S., An, L.-J. (2008). The Suaeda liaotungensis kitag betaine aldehyde dehydrogenase gene improves salt tolerance of transgenic maize mediated with minimum linear length of DNA fragment. Euphytica 159, 17–25. doi: 10.1007/s10681-007-9451-1
Xiao, R.-X., Zhang, C., Guo, X.-R., Li, H., Lu, H. (2021). MYB Transcription factors and its regulation in secondary cell wall formation and lignin biosynthesis during xylem development. Int. J. Mol. Sci. 22, 3560. doi: 10.3390/ijms22073560
Xu, P.-P., Fang, S., Chen, H.-Y., Cai, W.-M. (2020). The brassinosteroid-responsive xyloglucan endotransglucosylase/hydrolase 19 (XTH19) and XTH23 genes are involved in lateral root development under salt stress in Arabidopsis. Plant J. 104, 59–75. doi: 10.1111/tpj.14905
Yamanaka, T., Nakagawa, Y., Mori, K., Nakano, M., Imamura, T., Kataoka, H., et al. (2010). MCA1 and MCA2 that mediate Ca2+ uptake have distinct and overlapping roles in Arabidopsis. Plant Physiol. 152, 1284–1296. doi: 10.1104/pp.109.147371
Yan, J.-W., Huang, Y., He, H., Han, T., Di, P.-C., Sechet, J., et al. (2019). Xyloglucan endotransglucosylase hydrolase 30 negatively affects salt tolerance in Arabidopsis. J. Exp. Bot. 70, 5495–5506. doi: 10.1093/jxb/erz311
Yang, Y.-Q., Guo, Y. (2018). Elucidating the molecular mechanisms mediating plant salt-stress responses. New Phytol. 217, 523–539. doi: 10.1111/nph.14920
Yang, M.-F., Song, J., Wang, B.-S. (2010). Organ-specific responses of vacuolar H+-ATPase in the shoots and roots of C-3 halophyte Suaeda salsa to NaCl. J. Integr. Plant Biol. 52, 308–314. doi: 10.1111/j.1744-7909.2010.00895.x
Ye, F., Xu, L.-Y., Li, X.-X., Zeng, W.-Z., Gan, N.-H., Zhao, C., et al. (2021). Voltage-gating and cytosolic Ca2+ activation mechanisms of Arabidopsis two-pore channel AtTPC1. Proc. Natl. Acad. Sci. U.S.A. 118, e2113946118. doi: 10.1073/pnas.2113946118
Yoshiba, Y., Kiyosue, T., Nakashima, K., Yamaguchi-Shinozaki, K., Shinozaki, K. (1997). Regulation of levels of proline as an osmolyte in plants under water stress. Plant Cell Physiol. 38, 1095–1102. doi: 10.1093/oxfordjournals.pcp.a029093
Young, M. D., Wakefield, M. J., Smyth, G. K., Oshlack, A. (2010). Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Bio. 11, R14. doi: 10.1186/gb-2010-11-2-r14
Yuan, Y.-X., Teng, Q., Zhong, R.-Q., Haghighat, M., Richardson, E. A., Ye, Z.-H. (2016). Mutations of Arabidopsis TBL32 and TBL33 affect xylan acetylation and secondary wall deposition. PloS One 11, e0146460. doi: 10.1371/journal.pone.0146460
Zhang, H.-M., Zhu, J.-H., Gong, Z.-Z., Zhu, J.-K. (2022). Abiotic stress responses in plants. Nat. Rev. Genet. 23, 104–119. doi: 10.1038/s41576-021-00413-0
Zhao, C.-Z., Zhang, H., Song, C.-P., Zhu, J.-K., Shabala, S. (2020). Mechanisms of plant responses and adaptation to soil salinity. Innov. 1, 100017. doi: 10.1016/j.xinn.2020.100017
Zhong, R.-Q., Cui, D.-T., Ye, Z.-H. (2019). Secondary cell wall biosynthesis. New Phytol. 221, 1703–1723. doi: 10.1111/nph.15537
Keywords: halophyte, Suaeda dendroides, salt stress, transcriptome, adaptive mechanism
Citation: Ma P, Li J, Sun G and Zhu J (2024) Comparative transcriptome analysis reveals the adaptive mechanisms of halophyte Suaeda dendroides encountering high saline environment. Front. Plant Sci. 15:1283912. doi: 10.3389/fpls.2024.1283912
Received: 27 August 2023; Accepted: 30 January 2024;
Published: 14 February 2024.
Edited by:
Zulfiqar Ali Sahito, Zhejiang University, ChinaReviewed by:
Dhananjaya Pratap Singh, Indian Institute of Vegetable Research (ICAR), IndiaCopyright © 2024 Ma, Li, Sun and Zhu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Guoqing Sun, c3VuZ3VvcWluZzAyQGNhYXMuY24=; Jianbo Zhu, empic2h6QDEyNi5jb20=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.