- 1Ministry of Education Key Laboratory for Transboundary Ecosecurity of Southwest China, Yunnan Key Laboratory of Plant Reproductive Adaptation and Evolutionary Ecology and Centre for Invasion Biology, Institute of Biodiversity, School of Ecology and Environmental Science, Yunnan University, Kunming, China
- 2Ministry of Education Key Laboratory for Biodiversity Science and Ecological Engineering, Institute of Biodiversity Science, Fudan University, Shanghai, China
Seedling emergence is an essential event in the life cycle of plants. Most invasive plants have an advantage in population colonization over native congeners. However, differential seedling emergence between invasive plants and native congeners, especially their mechanisms, have rarely been explored. In this study, we show that the seedlings of invasive Phytolacca americana emerge faster compared to native P. acinosa. Genome-wide transcriptomes of initially germinated seeds versus seedlings at 4 days after germination (DAG) suggested that differentially expressed genes (DEGs) in the photosynthesis-antenna proteins pathway were up-regulated in both P. americana and P. acinosa, while DEGs in starch and sucrose metabolism were significantly down-regulated in P. americana. Gene expression analysis indicated that photosynthesis-related DEGs reached their highest level at 3 DAG in P. americana, while they peaked at 4 DAG in P. acinosa. We also identified one β-amylase gene in P. americana (PameAMYB) that showed the highest expression at 1 DAG, and two β-amylase genes in P. acinosa that expressed lower than PameAMYB at 0 and 1 DAG. Enzymatic activity of β-amylases also suggested that P. americana had the highest activity at 1 DAG, which was earlier than P. acinosa (at 4 DAG). Soluble sugars, the main source of energy for seedling emergence, were showed higher in P. americana than in P. acinosa, and reached the highest at 4 DAG that positively affected by photosynthesis. These results indicate that the rapid seedling emergence of invasive P. americana benefited from the high soluble sugar content produced by starch metabolism and photosynthesis. Altogether, this work contributes to our fundamental knowledge on physiological and molecular mechanisms for plant invasion success.
Introduction
Seedling emergence occurs after seed germination, when the shoot and root of the seed are formed (Csontos, 2003), and is an essential prerequisite for vegetative growth, population colonization, and dispersal of plants (Verdú and Traveset, 2005; Leger et al., 2019; Larson et al., 2020). However, emerging seedlings are vulnerable to many extrinsic factors, such as water, pests, and maternal effects (Lamichhane et al., 2018; Chen et al., 2020). Seed-to-seedling transition is also affected by intrinsic factors, including plant genotypes (Graebner et al., 2012; Souza and Fagundes, 2014; Yu et al., 2020). Therefore, short time intervals between seed germination and seedling emergence are usually helpful for successful establishment of seedlings.
Seedling emergence is highly dependent on plant physiological status. Typically, soluble sugars are the main source of energy for seedling emergence (Penfield et al., 2005). Hydrolytic enzymes, including α-amylase, β-amylase, and α-glucosidase, are the primary enzymes that break down starch into soluble sugars (Zeeman et al., 2010). In Triticum aestivum, post-germination seedling growth is positively regulated by starch degradation that is catalyzed by α-amylase and α-glucosidase (Sun et al., 2020). The β-amylase activity in Cyclobalanopsis gilva was found to be higher in germinated seeds compared to non-germinated seeds (Zaynab et al., 2018). Therefore, high hydrolytic enzyme activity in germinated seeds is crucial for rapid seedling emergence.
Invasive plants cause serious ecological consequences and pose great threats to the economy and human health (Rands et al., 2010; Vila et al., 2011; Rai and Singh, 2020). Most invasive plants grow more rapidly than their native congeners, such as Copaifera langsdorffii versus C. oblongifolia, Spartina densiflora versus S. maritime, as well as Phytolacca americana versus P. acinosa (Fagundes et al., 2020; Infante-Izquierdo et al., 2020; Liu et al., 2022). Functional traits of plants, like specific leaf area and photosynthetic rate, have been an active point of research on the rapidity of plant growth (Feng et al., 2009; Liu et al., 2022). However, the mechanisms that underlie the physiological and molecular differences of plant growth between invasive and native congeners are still poorly understood, especially during the transition from seeds to seedlings.
P. americana is an herbaceous perennial plant native to North America that occupies a wide range of habitats in China, and is known to be poisonous to mammals (Xu et al., 2012). Meanwhile, P. acinosa is the congener of P. americana that is native to China. In this study, we compared the differences in seedling emergence of invasive P. americana and native P. acinosa. Firstly, the number of emerged seedlings for the two species was recorded in the 10 days after seed germination (DAG). Subsequently, genome-wide transcriptomes were sequenced for emerged seedlings at 0 and 4 DAG for both species. According to the differential metabolic pathways between P. americana and P. acinosa, photosynthesis-related differentially expressed genes (DEGs) and differentially expressed β-amylase genes were further investigated. Additionally, gene expression and enzymatic activity of β-amylase, and total soluble sugar content were determined to investigate the effect of photosynthesis and starch metabolism on seedling emergence. These results would help illustrate the physiological and molecular mechanisms of differential seedling emergence between the two congeners, and further reveal the invasion mechanisms of P. americana.
Materials and methods
Plant materials
Seeds of P. americana (24°49′ N, 102°52′ E) and P. acinosa (25°26′ N, 104°19′ E) were collected from the field in August 2020 in Yunnan, China. Mature racemes, that grew under adequate sunlight in the canopy, were randomly collected from each plant species. Seeds were obtained after the raceme flesh were removed (Liu D. et al., 2020). All seeds were stored in the laboratory at the Institute of Biodiversity, School of Ecology and Environmental Science, Yunnan University. The 1,000-seed weight of P. americana and P. acinosa was recorded as 7.55 ± 0.08 g and 7.56 ± 0.03 g, respectively. For germination, all seeds were soaked in 98% H2SO4 for 15 min to break dormancy. Subsequently, seeds were separately placed in Petri dishes (d = 9 cm) with 20 mL of 1% agar medium (1g agar and 100 mL ddH2O) and cultured in a growth chamber (Yiheng, Shanghai, China) with a 14 h light: 10 h dark cycle at 27°C and 25°C, respectively.
Germination rate, seedling emergence, and seedling biomass measurement
To compare the difference in time intervals between seed germination and seedling emergence, we first calculated seed germination rate of P. americana and P. acinosa. Six seeds of one species were placed in a Petri dish with agar medium and cultured as described above. Ten replicates were performed. The seeds were considered to have germinated when approximately 1 mm of the radicle protruded from the seed coat. At day 3, the radicle of the seed emerged, and the number of germinated seeds was recorded. The germination rate was determined as the number of germinated seeds at day 3 versus the total number of tested seeds.
The seeds that germinated at day 3 were employed for seedling emergence tests and treated as 0 DAG. Twenty initially germinated seeds of P. americana and P. acinosa were separately transferred into another 1% agar medium and cultured as described above. Three replicates were conducted. Seedlings with spread cotyledons were used for seedling emergence analysis (Larson et al., 2020). The number of emerged seedlings was recorded daily for 10 days. At 10 DAG, the fresh weight of the seedlings was measured using an analytical balance (BSA223S, OLABO, Shandong, China).
RNA isolation, Illumina sequencing, and data processing
Seedlings at 4 DAG of each species were individually collected for transcriptome analysis, and 0 DAG seedlings were used as controls (Supplementary Figure S1). Eight germinated seeds were used for one replicate, and three biological replicates were conducted. Total RNA was extracted using the RNAprep Pure Plant Kit (Tiangen Biotech Co., Ltd, Beijing, China) following the manufacturer’s instructions. RNA quality and purity were determined using a NanoPhotometer N60 (Implen, Germany) and an Agilent Bioanalyzer 2100 system (Agilent Technologies, CA, USA). A sample of 1 μg RNA was used, and cDNA libraries were constructed separately, as previously described (Liu D. et al., 2020). Transcripts at the genome-wide transcriptional level were separately determined using pair-end (150 bp) RNA sequencing (RNA-Seq) on an Illumina NovaSeq 6000 platform at Biomarker Technology Co. (Beijing, China).
Clean reads were obtained by removing adaptor, reads with Q30 (probability of incorrect base call = 1/1000) of less than 85%, and reads containing more than 10% unknown bases per read from raw reads. After trimming, transcripts were assembled by pooling clean reads for all individual plant treatments using Trinity (v2.5.1), with min_kmer_cov set to 2 (Grabherr et al., 2011). To quantify gene expression, clean reads were mapped back to the transcriptome dataset, and read numbers were obtained using RSEM (v1.2.19; Li and Dewey, 2011). The mapped reads were normalized according to the fragment per kilobase of exon model per million mapped reads (FPKM; Mortazavi et al., 2008). Transcripts with a P-adjusted (P-adj) value of < 0.05 between two groups (each group with three biological replicates) and absolute log2 (0 DAG verse 4 DAG) > 1 were assigned as DEGs (Anders and Huber, 2010; Wu et al., 2021). Gene ontology (GO) enrichment analysis of the DEGs was performed using the topGO R package (v2.28.0) based on the Kolmogorov-Smirnov test (Alexa and Rahnehfuhrer, 2016). KOBAS2.0 software was used to test the statistical enrichment of DEGs in the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways (Xie et al., 2011). Raw data were deposited in the Sequence Read Archive of the National Center for Biotechnology Information (accession number: PRJNA992375).
Candidate gene selection
We selected DEGs from KEGG pathways with P < 0.05 between the two developmental stages, and DEGs with FPKM value > 10 and log2foldchange (0 DAG versus 4 DAG) > 2 or < -2 in each pathway were filtered and then analyzed in R (4.1.3, R Core Development Team, 2020) using the GOplot package.
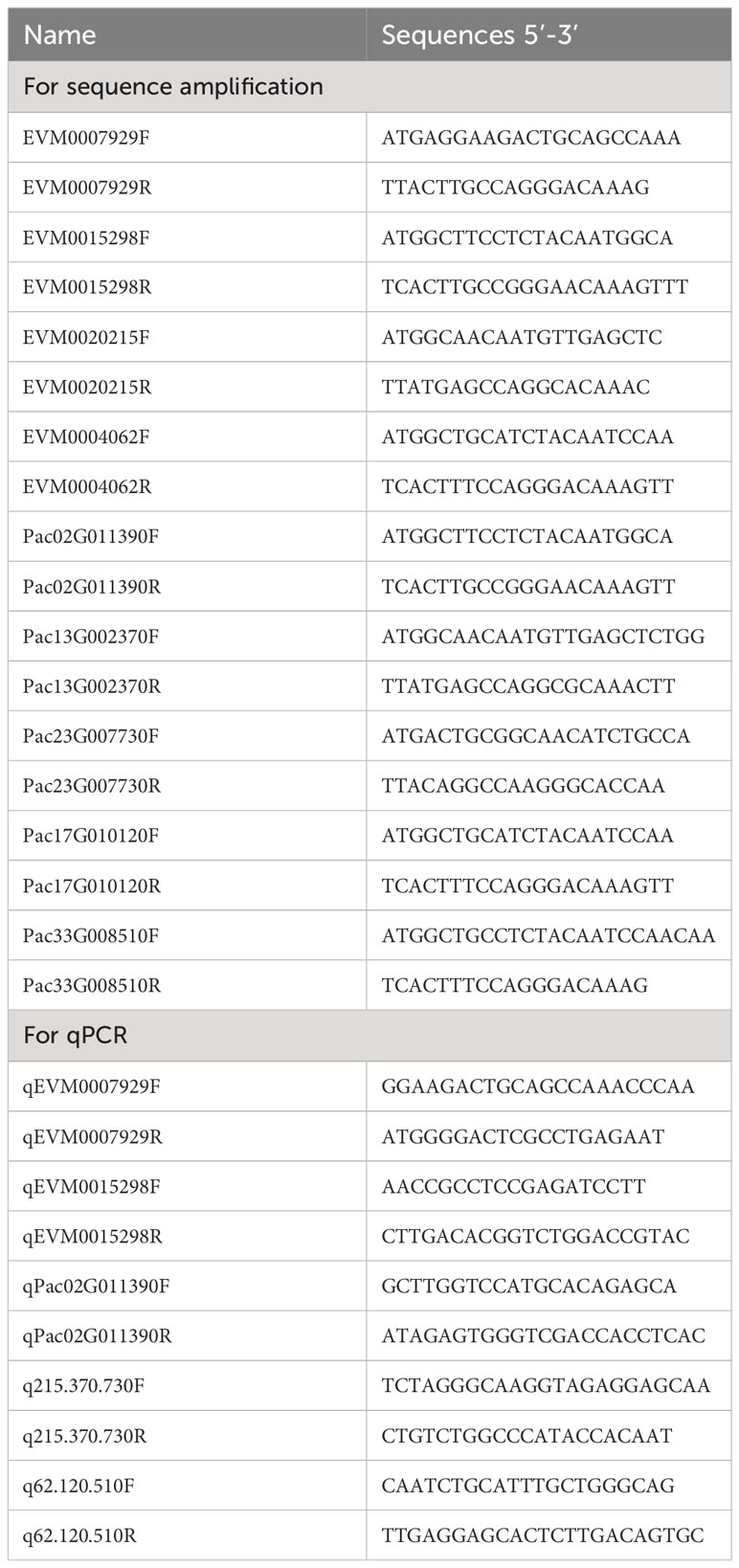
We found that DEGs enriched in the photosynthesis-antenna proteins pathway were up-regulated in both P. americana and P. acinosa. To better analyze the phylogenetic relationship among these DEGs, we constructed a phylogenetic tree according to their deduced amino acid sequences with MEGA 11.0 using the Neighbor-Joining method with 1000 bootstrap replicates (Hall and Notes, 2013). FPKM values for the DEGs were also presented using iTOL (https://itol.embl.de/). To further analyze gene expression, some homologous DEGs between the two species were randomly selected. These genes were amplified with the primers listed in Table 1 and cloned into a sequencing vector BluntSimple (TransGen, China) for full sequencing. Quantitative polymerase chain reaction (qPCR) was then performed to determine the gene expression of the seedlings at 0, 1, 2, 3, and 4 DAG.
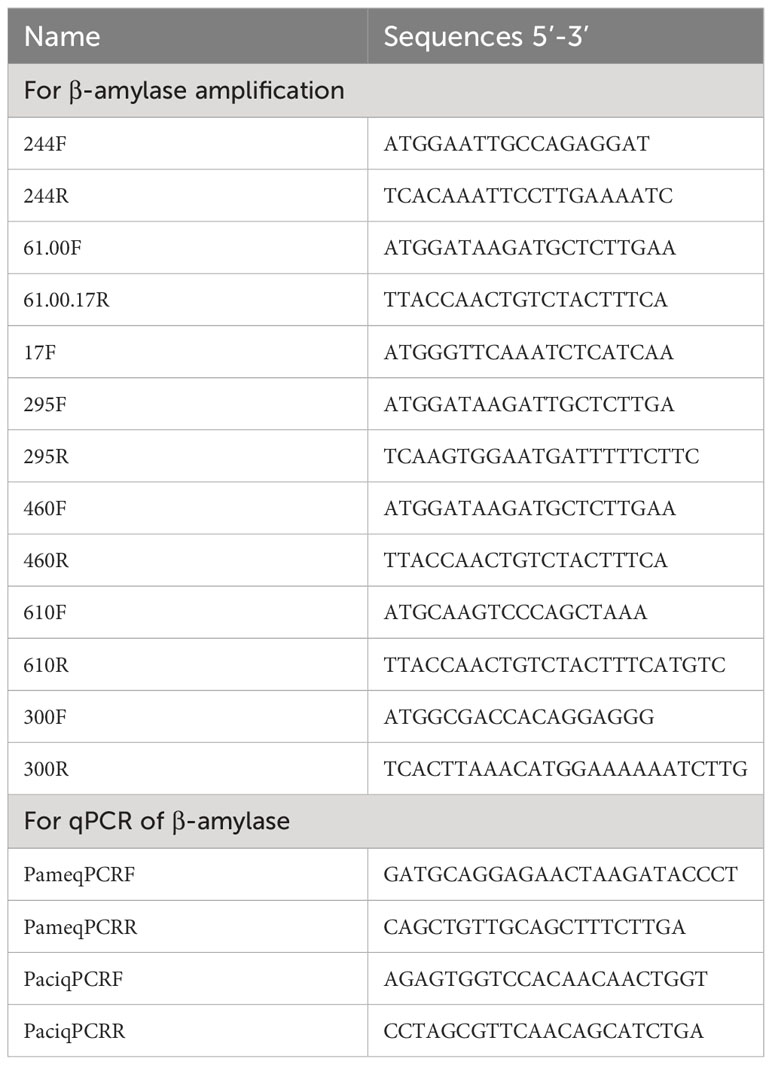
We also found that β-amylase genes in starch and sucrose metabolism were significantly down-regulated compared with other DEGs in the selected KEGG pathways in P. americana. An additional phylogenetic tree was constructed based on the deduced amino acid sequences for the β-amylase DEGs in P. americana and P. acinosa together with β-amylases with reviewed functions in UniProt (Hall and Notes, 2013). According to the reference genomes of P. americana and P. acinosa, candidate β-amylase genes were amplified with the primers listed in Table 2, and then sequenced. Substrate-binding and protein active sites of the deduced amino acid sequences from the amplified sequences were verified according to the homologous β-amylase in Ipomoea batatas (Yoshida and Nakamura, 1991) using SnapGene (4.2.4, GSL, Biotech, USA). qPCR was also conducted to analyze the β-amylase gene expression of the seedlings at 0, 1, 2, 3, and 4 DAG.
cDNA preparation and qPCR analysis
Emerged seedlings at 0, 1, 2, 3, and 4 DAG of the two species were separately collected and stored at -80°C after flash freezing in liquid nitrogen. Total RNAs were extracted as described above, and cDNA was subsequently prepared using 1.0 μg RNA using the FastQuant RT kit (Tiangen Biotech Co., Ltd, Beijing, China) according to the manufacturer’s instructions.
SnapGene was used to design the primers to quantify target gene expression (Tables 1, 2). qRCR analysis was conducted using SuperReal PreMix Plus kits (SYBR Green; Tiangen Biotech Co., Ltd., Beijing, China) and an ABI QuantStudio 7 Flex PCR system (Applied Biosystems, Carlsbad, CA, USA). Samples were detected separately in 20 μL reaction mixture volumes with an initial thermal profile of 15 min at 95°C, followed by 40 cycles of 10 s at 95°C and 32 s at 60°C. Three technical replicates were performed for each sample, and three biological samples were used.
For qPCR analysis, we used the absolute quantitative method where the copy number of each gene was determined according to a standard curve. The standard curve was generated using a 10-fold serial dilution of standard plasmid ranging from 1 × 106 to 1 × 102 copies/μL target fragments (Liu et al., 2016).
β-amylase activity and soluble sugar content in P. americana and P. acinosa
Emerged seedlings collected at 0, 1, 2, 3, and 4 DAG of the two species were also used to determine β-amylase activity and soluble sugar content. The β-amylase activity and soluble sugar content of each sample were separately determined following the instructions of β-amylase kits (Grace Biotechnology Co., Ltd. Suzhou, China) and soluble sugar content kits (Grace Biotechnology Co., Ltd. Suzhou, China) using a microplate reader (SuperMax 3100, Flash, China).
To validate the role photosynthesis played in the production of soluble sugars during seedling emergence, we cultured initial germinating seeds of the two species in the dark, and then collected emerged seedlings at 0, 1, 2, 3, and 4 DAG of the two species. Soluble sugar content of each sample was determined as described above, and three replicates were performed for each treatment.
Data analysis
Data were analyzed using R. Student’s t-tests were used to compare germination rates, the number of emerged seedlings on the same day, and the fresh weight of the 10 days old seedlings between P. americana and P. acinosa. Likewise, expression levels of target genes, β-amylase activity, and soluble sugar content of the samples collected at the same time were also compared using t-tests. The number of emerged seedlings, expression level of target genes, β-amylase activity, and soluble sugar content of the same species at different times were analyzed using one-way analysis of variance (one-way ANOVA) and pair-wise post hoc analyses by least significant difference (LSD) in the agricolae package. Statistical significance was set at P < 0.05.
Results
Germination rate, seedling emergence, and seedling fresh weight for P. americana and P. acinosa
After being treated with H2SO4 to break seed dormancy, we found P. americana and P. acinosa seeds began to germinate at day 3, and their germination rates were not significantly different (t-test, P = 0.1152; Figure 1A). These germinated seeds were then used for the following experiments.
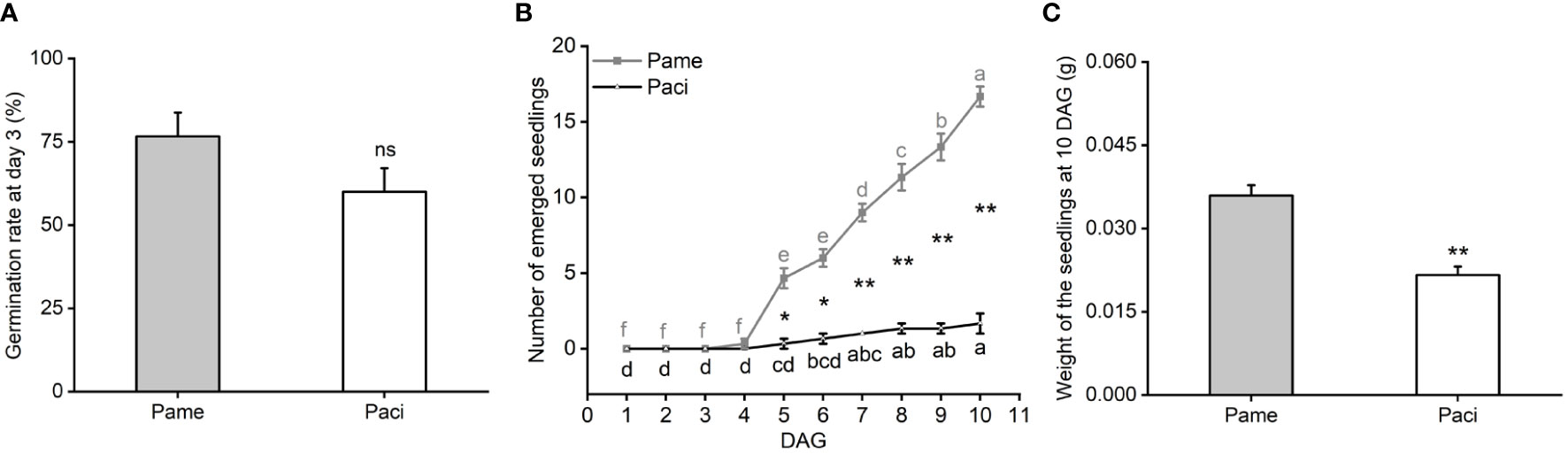
Figure 1 Germination rate at day 3 after seed dormancy breaking treatment (A), number of emerged seedlings with spread cotyledons (B) and weight of the 10 DAG seedlings (C) of Phytolacca americana (Pame) and Phytolacca acinosa (Paci). DAG: day(s) after germination. Values represent the mean ± standard error. Different letters in the same shade as the line indicate that the means differ significantly among the tested time points under one-way ANOVA followed by least significant difference (LSD) test. Asterisks indicate a significant difference between the two species at the same time point under Student’s t-test. *P < 0.05, **P < 0.01, ns P > 0.05.
After culturing the initially germinated seeds for 10 days, we found that P. americana seedlings began to emerge at 4 DAG (one-way ANOVA, PPame < 0.001), while P. acinosa began to emerge at 5 DAG (one-way ANOVA, PPaci < 0.001). At 10 DAG, approximately 80% of the tested P. americana seeds developed into seedlings, while less than 10% of the P. acinosa seedlings developed. Additionally, the number of emerged P. americana seedlings was significantly higher than that of P. acinosa (t-test; P5 DAG = 0.004; P6 DAG = 0.001; P7 DAG < 0.001; P8 DAG < 0.001; P9 DAG < 0.001; P10 DAG < 0.001; Figure 1B). At 10 DAG, the fresh weight of P. americana seedlings was also significantly higher than that of P. acinosa seedlings (t-test, P = 0.004; Figure 1C). Together, these results indicate that the time intervals between seed germination and seedling emergence of invasive P. americana were shorter than that of native P. acinosa.
Genome-wide transcriptome data analysis
RNA-Seq data of the tested samples were evaluated using Pearson’s correlation coefficient (r) and showed high consistency (r > 0.94) within groups (Supplementary Figure S2). The average FPKM of each group was then used for DEG analysis. In total, 7,806 and 11,340 DEGs were identified in the 0 vs 4 DAG seedlings of P. americana and P. acinosa, respectively. Of these, 4,341 and 3,465 genes in P. americana, and 6,779 and 4,561 genes in P. acinosa were up- and down-regulated, respectively (Figure 2A). In both P. americana and P. acinosa, 5,705 DEGs showed more than 50% similarity and were treated as the intersection of the Venn diagram (Figure 2B; Supplementary Material). We also found 2,101 DEGs only in the 0 vs 4 DAG seedlings for P. americana, and 6,511 DEGs only in the 0 vs 4 DAG seedlings for P. acinosa.
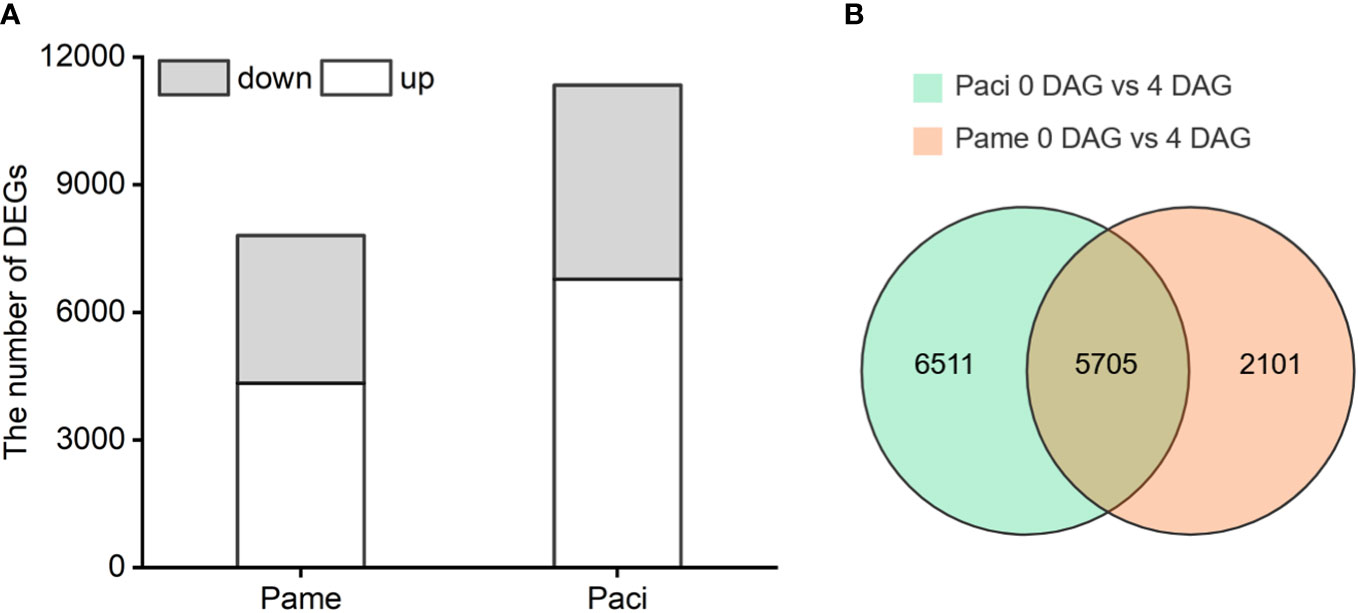
Figure 2 Differentially expressed genes (DEGs) of the 0 vs 4 DAG seedlings of Phytolacca americana (Pame) and Phytolacca acinosa (Paci) (A), and homologous DEGs between 0 vs 4 DAG seedlings of P. americana and P. acinosa (B). DAG: day(s) after germination. Grey represents the number of down-regulated DEGs, and white represents the number of up-regulated DEGs.
According to the annotations on the GO database, P. americana seedling DEGs were highly enriched in the metabolic and cellular processes in the biological process category, cell and cell part in the cellular component category, as well as binding and catalytic activity in the molecular function category. The same trend was also observed in P. acinosa seedlings. However, the number of DEGs in P. acinosa was higher than that in P. americana (Supplementary Figure S3).
KEGG pathway enrichment analysis suggested that 26 pathways were differentially enriched by the DEGs in P. americana, while 19 pathways were enriched in P. acinosa (Supplementary Figure S4). Among these KEGG pathways, 12 pathways were only found in P. americana seedlings, while only five were in P. acinosa seedlings. Furthermore, energy metabolism pathways, such as butanonate metabolism, pentose phosphate pathway, and starch and sucrose metabolism, were only found in P. americana. According to the FPKM values, DEGs for the photosynthesis-antenna proteins pathway in both species were highly up-regulated. However, some DEGs for starch and sucrose metabolism were significantly down-regulated in P. americana (Supplementary Figures S5, S6).
DEGs in photosynthesis-antenna proteins pathway
In total, we filtered 23 photosynthesis-antenna proteins pathway DEGs in the 0 vs 4 DAG seedlings for P. americana and 37 DEGs in P. acinosa seedlings. The phylogenetic tree suggested that photosynthesis-antenna proteins pathway DEGs between P. americana and P. acinosa were highly similar. Even though these DEGs were significantly up-regulated, many DEGs in P. acinosa seedlings were more highly expressed compared to their homogeneous genes in P. americana (Figure 3).
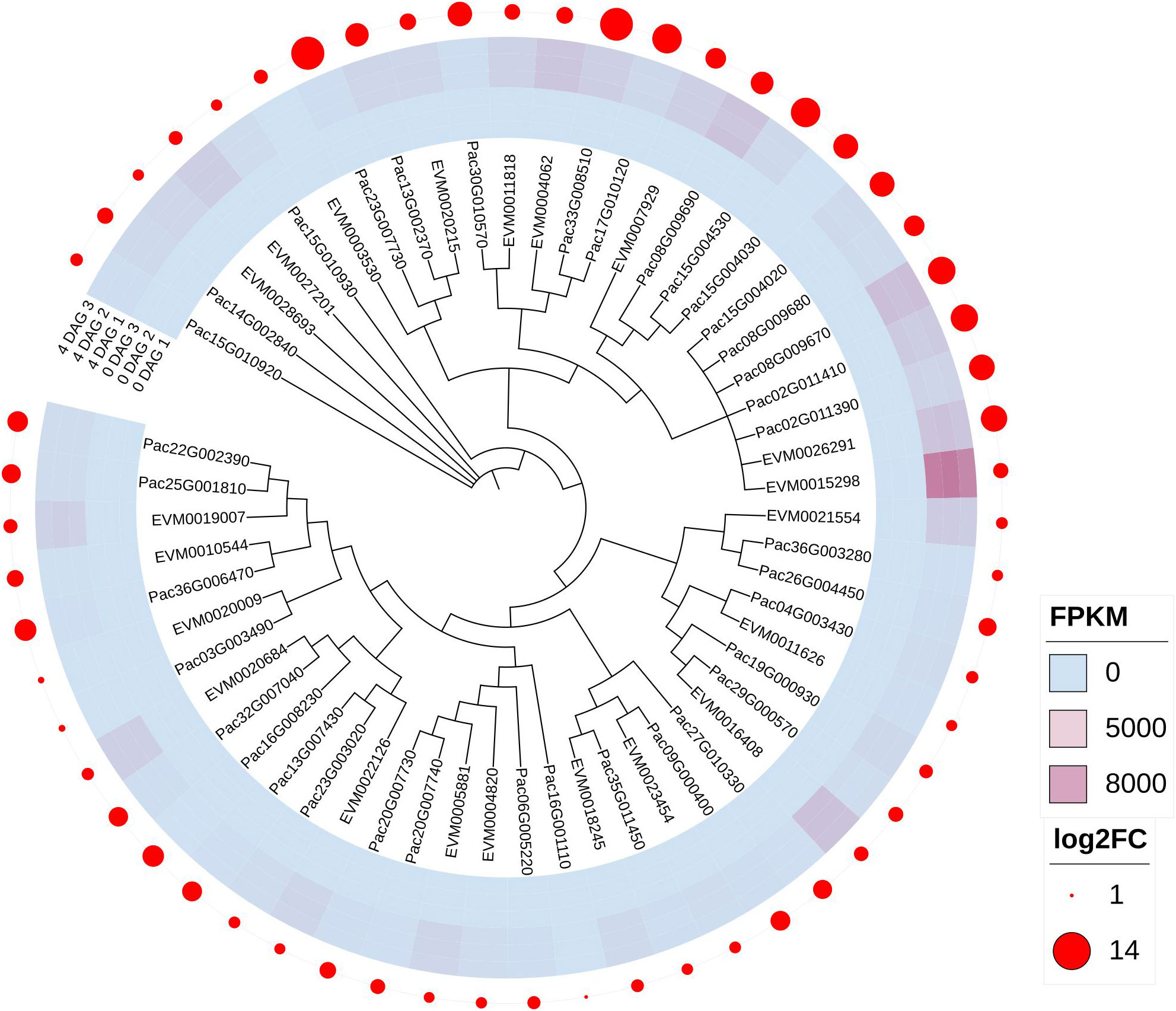
Figure 3 Differentially expressed genes (DEGs) enriched in the photosynthesis-antenna proteins pathway between Phytolacca americana and Phytolacca acinosa. DAG: day(s) after germination. FPKM represent the fragment per kilobase of exon model per million mapped reads value of each DEG, and log2FC is the log2foldchange (0 DAG versus 4 DAG) of each DEG. EVM is the DEG in P. americana, and Pac is the DEG in P. acinosa.
Additionally, we compared the gene expression among seedlings collected at 0, 1, 2, 3, and 4 DAG, and found that the tested DEGs in P. americana had the highest expression at 3 DAG, while in P. acinosa they showed the highest expression at 4 DAG (Figure 4).
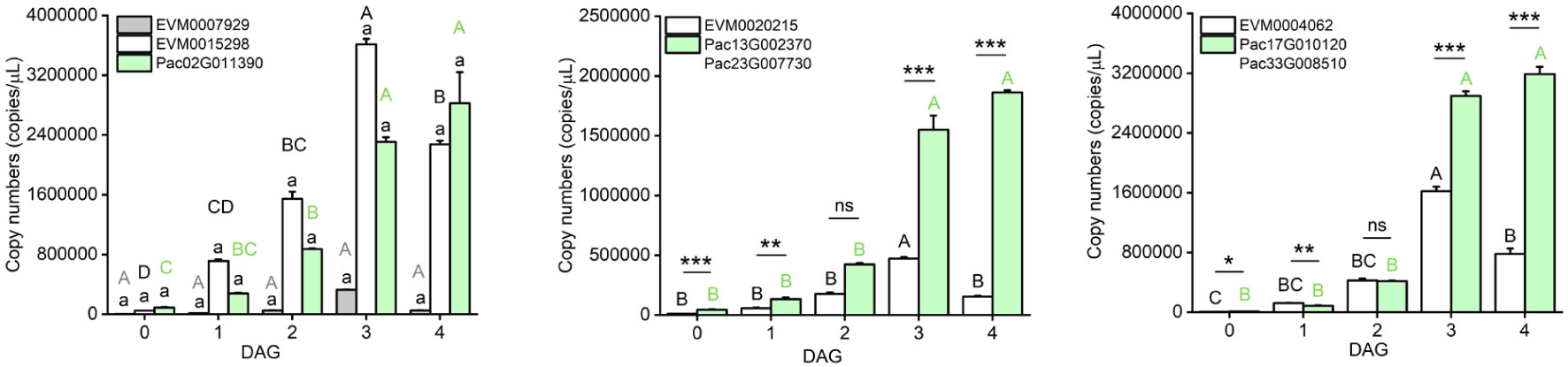
Figure 4 Expression level of homologous fragments of genes involved in photosynthesis-antenna proteins pathway between Phytolacca americana and Phytolacca acinosa. DAG: day(s) after germination. Values represent the mean ± standard error. Different letters in the same color as the column indicate that the means differ significantly among the tested time points under one-way ANOVA followed by least significant difference (LSD) test. Asterisks indicate a significant difference between the two species at the same time point under Student’s t-test. *P < 0.05, **P < 0.01, ***P < 0.001, ns P > 0.05.
Candidate genes of β-amylase
We then selected and compared DEGs involved in the synthesis of 21 enzymes for starch and sucrose metabolism. Genes for β-glucosidase and β-amylase in P. americana were differentially expressed during the first four days after germination (Figure 5). The phylogenetic tree of β-amylase in P. americana and P. acinosa together with other plants reported in UniProt were constructed and showed that five DEGs for β-amylases in P. americana and three DEGs in P. acinosa were clustered with β-amylase in I. batatas (IbatAMYB; Figure 6A).
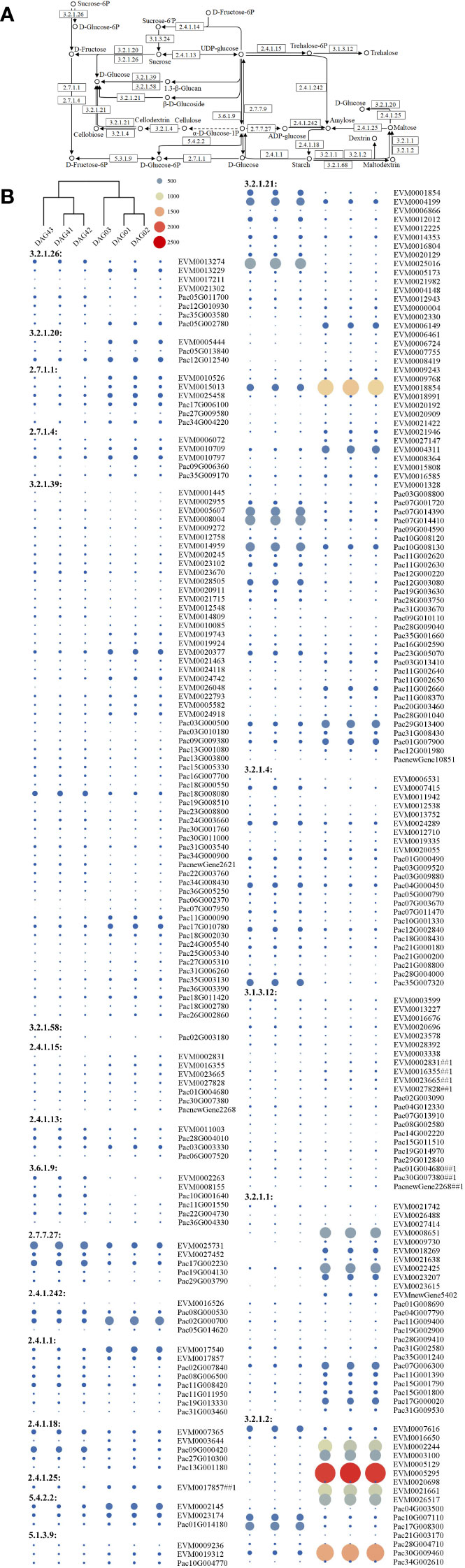
Figure 5 Starch and sucrose metabolism pathway and the related differentially expressed genes (DEGs) in Phytolacca americana and Phytolacca acinosa. (A) Starch and sucrose metabolism pathway that DEGs were detected in P. americana and P. acinosa. (B) Heatmap of the DEGs based on the fragment per kilobase of exon model per million mapped reads values. DAG: day(s) after germination. ## indicates the sequence is related to more than one enzyme. DAG01, DAG02, and DAG03 are the three replicates of 0 DAG seedlings, while DAG41, DAG42, and DAG43 are the three replicates of 4 DAG seedlings.
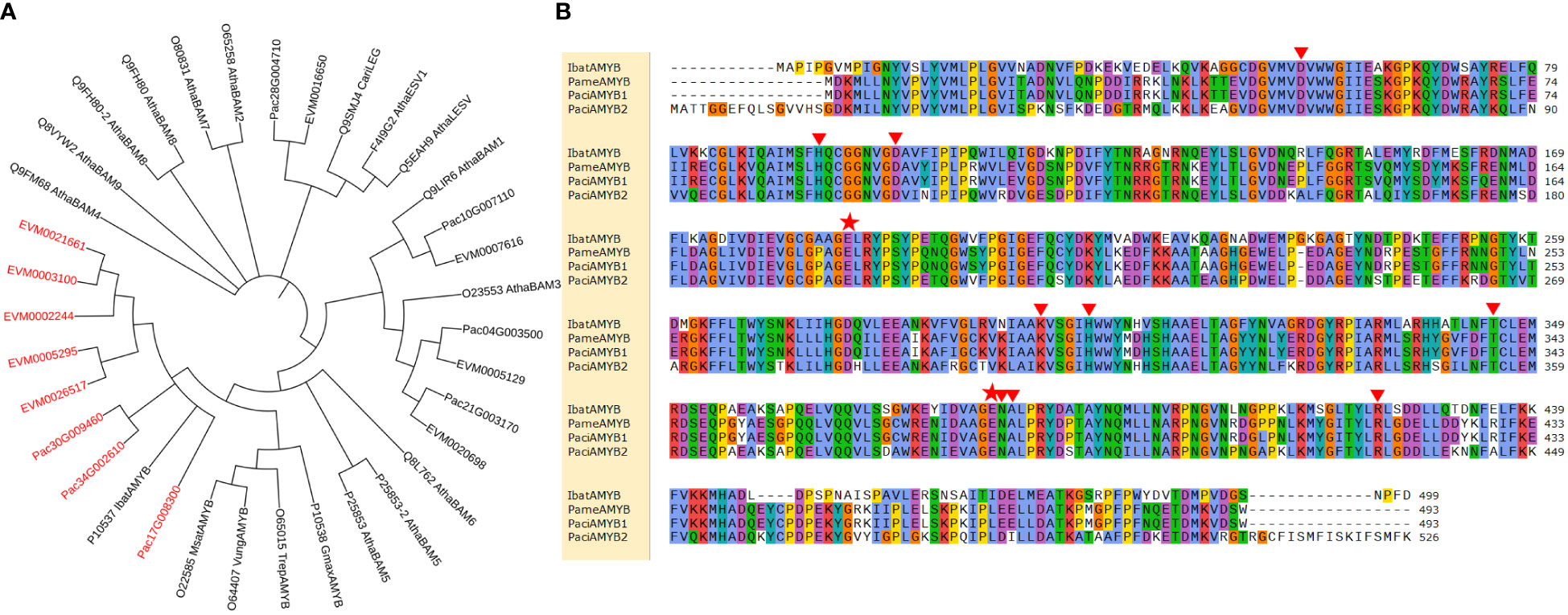
Figure 6 β-amylase in Phytolacca americana and Phytolacca acinosa. (A) Phylogenetic tree of β-amylase in P. americana and P. acinosa together with other reported β-amylases in Uniprot. (B) Sequence alignment of β-amylase in P. americana (PameAMYB), P. acinosa (PaciAMYB1 and PaciAMYB2), and Ipomoea batatas (IbatAMYB). EVM is the DEG in P. americana, and Pac is the DEG in P. acinosa. Red font represents the target sequences. Red triangles indicate the substrate binding sites of the proteins, and red pentangles show the active sites of the proteins.
Further PCR amplification and sequence alignment revealed that four of the five β-amylase genes in P. americana were amplified and showed the consensus sequence (named PameAMYB). However, the remaining transcript could not be amplified. Additionally, two of the three transcripts for β-amylase in P. acinosa were confirmed to be the same and were named PaciAMYB1 and PaciAMYB2. Further, sequence alignment of IbatAMYB, PameAMYB, PaciAMYB1 and PaciAMYB2 suggested that all the binding and active sites were present among the deduced amino acid sequences (Figure 6B). Together, these results suggest the presence of functional β-amylase in both P. americana and P. acinosa.
Gene expression and enzymatic activity of β-amylase
At 0 DAG, β-amylase gene expression was high for both species. PameAMYB expression was higher in P. americana at 1 DAG, but then sharply decreased (one-way ANOVA, PPame < 0.001; Figure 7A). In comparison, PaciAMYB1 decreased from 0 DAG (one-way ANOVA, PPaci < 0.001). Although the gene expression level of PameAMYB was lower at 2, 3, and 4 DAG compared to PaciAMYB1, PameAMYB was highly expressed at 0 and 1 DAG (t-test, P0 DAG = 0.018, P1 DAG < 0.001, P2 DAG = 0.001, P3 DAG < 0.001, P4 DAG < 0.001).
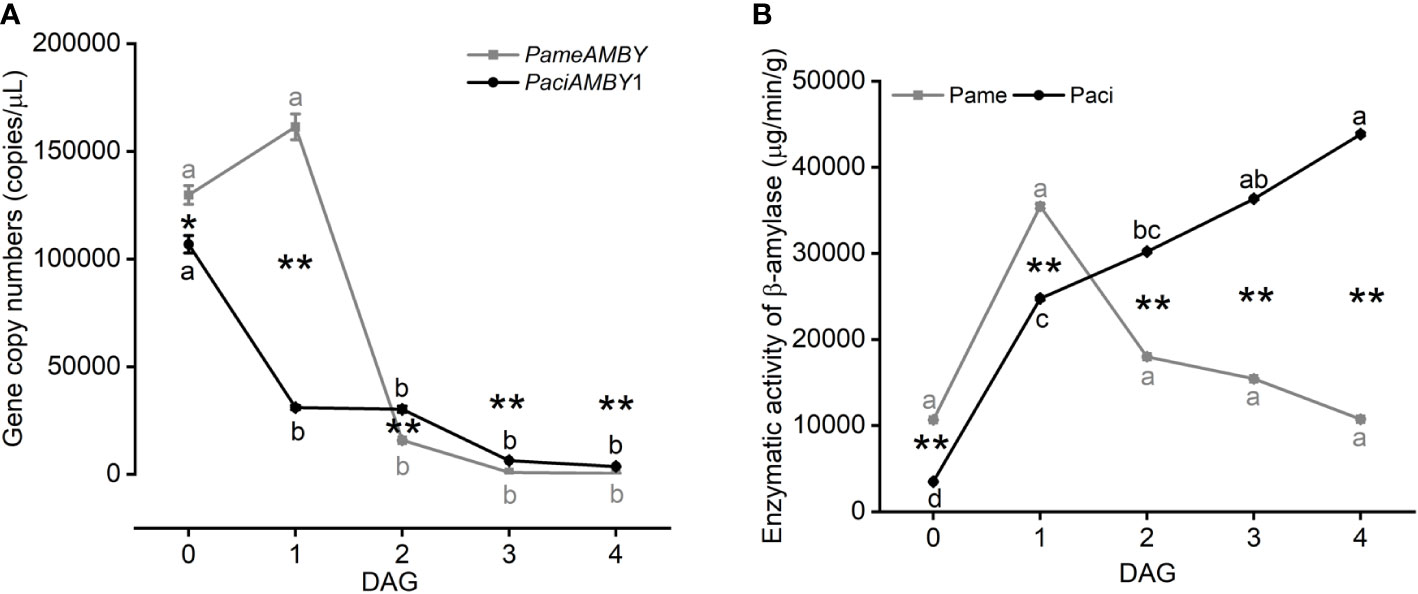
Figure 7 Gene expression of PameAMBY and PaciAMBY1 (A), and enzymatic activity of β-amylase (B) among samples of Phytolacca americana (Pame) and Phytolacca acinosa (Paci). DAG: day(s) after germination. Values represent the mean ± standard error. Different letters in the same shade as the line indicate significantly different means of the species under one-way ANOVA followed by least significant difference (LSD) test. Asterisks indicate a significant difference between the two species at the same time point under Student’s t-test. *P < 0.05, **P < 0.01.
The enzymatic activity measurements suggested that β-amylase activity was the highest at 1 DAG in P. americana and decreased at 4 DAG to a level similar to 0 DAG. However, β-amylase activity was not significantly different during those four days (one-way ANOVA, PPame = 0.265; Figure 7B). In P. acinosa, the β-amylase activity gradually increased (one-way ANOVA, PPaci < 0.001) and began to be higher than P. americana at 2 DAG (t-test, P0 DAG < 0.001, P1 DAG < 0.001, P2 DAG < 0.001, P3 DAG < 0.001, P4 DAG < 0.001), and this trend could be related to the expression of PaciAMBY2, since it was up-regulated at 0 vs 4 DAG in P. acinosa (log2FC of PaciAMBY2 = 4.75, Supplementary Material).
Total soluble sugars determination
Total soluble sugar content in P. americana increased in the first two days after germination, and reached its highest level at 4 DAG (one-way ANOVA, PPame < 0.001; Figure 8A). However, total soluble sugars in P. acinosa increased from 0 to 3 DAG, but then decreased (one-way ANOVA, PPaci < 0.001). At 0 DAG, the soluble sugar content was not significantly different between the two species (t-test, P = 0.748), and was higher at 1 (t-test, P = 0.045) and 2 (t-test, P = 0.009) DAG in P. americana compared to P. acinosa, but then decreased at 3 DAG (t-test, P = 0.149). However, it was markedly higher in P. americana than in P. acinosa at 4 DAG (t-test, P = 0.001).
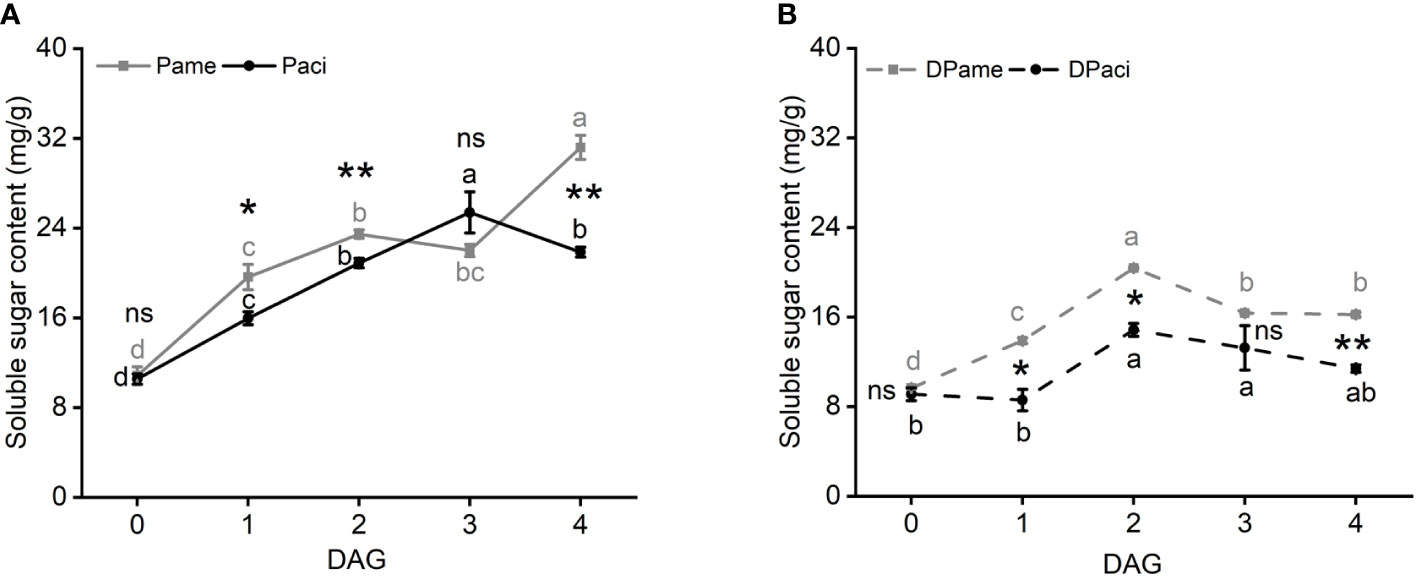
Figure 8 Total soluble sugar content among samples of Phytolacca americana (Pame) and Phytolacca acinosa (Paci) cultured in light/dark cycle (A) or in the dark (B). Pame: P. americana seedlings are cultured in the light/dark cycle; Paci: P. acinosa seedlings are cultured in the light/dark cycle. DPame: P. americana seedlings are cultured in the dark; DPaci: P. acinosa seedlings are cultured in the dark. Values represent the mean ± standard error. Different letters in the same shade as the line indicate significantly different means of the species under one-way ANOVA followed by least significant difference (LSD) test. Asterisks indicate a significant difference between the two species at the same time point under Student’s t-test. *P < 0.05, **P < 0.01, ns P > 0.05.
When the initially germinated seeds were cultured in the dark, the two species showed a similar variation in total soluble sugar content (one-way ANOVA; PPame < 0.001; PPaci = 0.035), even though the sugar content in P. americana was higher than that in P. acinosa (t-test, P0 DAG = 0.443, P1 DAG = 0.034, P2 DAG = 0.011, P3 DAG = 0.259, P4 DAG = 0.006). Moreover, soluble sugar content in P. americana did not significantly increase at 4 DAG (Figure 8B).
The germinated seeds cultured in the light-dark cycle also showed higher total soluble sugar content compared to those cultured in the dark (t-test, P. americana: P0 DAG = 0.325, P1 DAG = 0.030, P2 DAG = 0.007, P3 DAG = 0.004, P4 DAG = 0.002; P. acinosa: P0 DAG = 0.146, P1 DAG = 0.006, P2 DAG = 0.003, P3 DAG = 0.022, P4 DAG = 0.001). These results suggest that photosynthesis is vital for the formation of soluble sugars in P. americana at 4 DAG.
Discussion
Invasive P. americana shows faster seedling emergence compared to native P. acinosa
It has been reported that invasive plants show faster seedling emergences compared to native ones, such as Bromus tectorum (Griffith et al., 2014), Sorghum halepense (Reichmann et al., 2016), and Spartina densiflora (Infante-Izquierdo et al., 2020). In our study, even though P. americana and P. acinosa seeds began to germinate at the same time and showed similar germination rates, the invasive P. americana seedlings emerged more rapidly than native P. acinosa (Figures 1A, B). Typically, emerged seedlings use resources for growth and, therefore, gain competitive ability. Under resource competition, early seedling emergence could lead to disproportional resource utilization and create fecundity advantages (Dyer et al., 2000). We found that P. americana showed seedling emergence one day earlier than P. acinosa, which suggest that invasive P. americana could utilize resources earlier than native P. acinosa. Ross and Harper (1972) reported that 95% of plant biomass variation could be accounted for by time since emergence. Here, the biomass of P. americana seedlings at 10 DAG was significantly higher than P. acinosa seedlings (Figure 1C). This is also similar to the previous data that P. americana had a higher total biomass and exhibited a higher reproductive capacity compared to P. acinosa (Liu et al., 2022). Additionally, to avoid the effect of seed batch on seed germination and seedling emergence, we compared the data among seeds collected in 2019, 2021 to 2023, and found similar data among one particular species during the years (Supplementary Figures S7, S8).
Our transcriptome data also showed that many genes were differentially expressed at 0 vs 4 DAG in both P. americana and P. acinosa seedlings. Total DEGs in P. acinosa were higher than in P. americana (Figure 2), which may be related to the ploidy level of these plants, since P. americana is diploid and P. acinosa is autotetraploid. Similar results were found in autotetraploid rice and its diploid donor (Wang et al., 2022). GO analysis of the 0 vs 4 DAG seedlings of P. americana and P. acinosa also suggest that the two species undergo similar development (Figure 3). However, the KEGG pathway enrichment analysis suggests that the differential KEGG pathways between the two congeners are not entirely consistent (Supplementary Figure S3). More pathways are activated in invasive P. americana than in native P. acinosa, especially pathways related to energy metabolism. Studies found that genes in slowly and rapidly developing Panicum virgatum are differentially expressed in pathways related to diterpenoid biosynthesis, thiamine metabolism, and circadian rhythm (Zhang et al., 2019). The inconsistent results might be caused by the varying developmental stages of tested samples.
Photosynthesis and β-amylase play important roles in P. americana’s rapid seedling emergence
High photosynthetic rates in the aerial parts of invasive Mikania micrantha contribute to its rapid growth (Liu et al., 2020). Additionally, the rapid formation of the photosynthetic system promotes post-germination growth of Pennisetum glaucum (Wu et al., 2021), and is highly dependent on cotyledon photosynthesis (Zhang et al., 2008). In our study, photosynthesis related pathways are activated in the emerged seedlings from both P. americana and P. acinosa, and the induced genes involved in these photosynthesis-antenna proteins pathways share high similarities in the two species (Figure 3). However, photosynthesis-related genes show the highest expression at 3 DAG in P. americana, but are the highest at 4 DAG in P. acinosa (Figure 4), which may facilitate the seedling emergence of P. americana.
In addition, amylase genes are positively correlated with invasion success of Dendroctonus valens (Liu F. et al., 2020). In our study, β-amylase genes in starch and sucrose metabolism are differentially expressed at 0 vs 4 DAG seedlings of P. americana (Figure 5). Although β-amylase plays a minor role in storage starch degradation in wheat (Kaneko et al., 2000; Sun et al., 2020), β-amylase activity in P. americana is the highest at 1 DAG, which is in accordance with its gene expression level of PameAMYB, and continuous substrate catalysis leads to the accumulation of soluble sugars, and a soluble sugar content decrease at 3 DAG with increasing consumption during growth (Figures 7, 8). However, soluble sugars could also be formed by the photosynthetic system in the following days, as evidenced by the low soluble sugar content in emerged P. americana seedlings grown in the dark (Figure 8). Therefore, our data indicate that high soluble sugars produced by starch metabolism and photosynthesis contributes to P. americana rapid seedling emergence. Still, the role of PameAMYB in rapid seedling emergence of invasive P. americana should be further tested using genome editing in the future.
In conclusion, our results show that invasive P. americana seedlings emerge faster than native P. acinosa. KEGG pathway analysis of 0 vs 4 DAG seedlings suggested that DEGs in photosynthesis-antenna proteins pathway were up-regulated in both P. americana and P. acinosa, while DEGs in starch and sucrose metabolism were significantly down-regulated in P. americana. Gene expression analysis indicated that photosynthesis-related DEGs in P. americana seedlings reached the highest level at 3 DAG, while P. acinosa seedlings showed their peak at 4 DAG. We also identified one β-amylase gene in P. americana (PameAMYB) that showed the highest expression at 1 DAG, and two β-amylase genes in P. acinosa that PaciAMYB1 expressed the highest at 0 DAG, while PaciAMYB2 at 4 DAG. However, at 0 DAG, the expression of PaciAMYB1 was lower than PameAMYB. Enzymatic activity of β-amylases determination suggested that P. americana had the highest β-amylases activity at 1 DAG, which was earlier than in P. acinosa (at 4 DAG). Soluble sugars, the main source of energy for seedling emergence, were showed higher in P. americana than in P. acinosa, and reached the highest at 4 DAG. Moreover, soluble sugar content was significantly decreased at 4 DAG in P. americana when cultured the germinated seeds in the dark. These results suggest that high soluble sugar content in P. americana produced by starch metabolism and photosynthesis benefits the rapid seedling emergence, enhancing our knowledge on physiological mechanisms of invasion success.
Data availability statement
The original data presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Author contributions
DL: Conceptualization, Funding acquisition, Investigation, Software, Validation, Writing – original draft, Writing – review & editing. ML: Data curation, Investigation, Writing – original draft. RJ: Funding acquisition, Writing – review & editing. BL: Funding acquisition, Writing – review & editing. YW: Funding acquisition, Supervision, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the National Natural Science Foundation of China (32101413 and U2102218) and National Key Research and Development Program (2022YFC2601100).
Acknowledgments
We would like to thank Dr. John Measey at Yunnan University, China, and Dr. Kelly Barr at the University of California, Los Angeles, and Editage (www.editage.com) for English language editing. We are also grateful to Yue Zhou in Yunnan University for her help during the experiments.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2024.1255698/full#supplementary-material
References
Alexa, A., Rahnehfuhrer, J. (2016). topGO: Enrichment analysis for Gene Ontology. R package version. 2.28.0 (Cranio: Bioconductor). doi: 10.18129/B9.bioc.topGO
Anders, S., Huber, W. (2010). Differential expression analysis for sequence count data. Genome Biol. 11, R106. doi: 10.1186/gb-2010-11-10-r106
Chen, F., Zhou, W., Yin, H., Luo, X., Chen, W., Liu, X., et al. (2020). Shading of the mother plant during seed development promotes subsequent seed germination in soybean. J. Exp. Bot. 71, 2072–2084. doi: 10.1093/jxb/erz553
Csontos, P. (2003). Weed ecology in natural and agricultural systems. Agric. Ecosyst. Environ. 4, 243–244. doi: 10.1016/j.agee.2004.03.002
Dyer, A. R., Fenech, A., Rice, K. J. (2000). Accelerated seedling emergence in interspecific competitive neighborhoods. Ecol. Lett. 3, 523–529. doi: 10.1111/j.1461-0248.2000.00187.x
Fagundes, M., Cuevas-Reyes, P., Araujo, W. S., Faria, M. L., Valerio, H. M., Pimenta, M. A., et al. (2020). Influence of light availability and seed mass on germinability and initial growth of two congeneric species of Fabaceae. Acta Botanica Mexicana 127, e1638. doi: 10.21829/abm127.2020.1638
Feng, Y., Lei, Y., Wang, R., Zheng, Y. (2009). Evolutionary tradeoffs for nitrogen allocation to photosynthesis versus cell walls in an invasive plant. PNAS 106, 1853–1856. doi: 10.1073/pnas.0808434106
Grabherr, M. G., Haas, B. J., Yassour, M., Levin, J. Z., Thompson, D. Z., Amit, I., et al. (2011). Full length transcriptome assembly from RNA Seq data without a reference genome. Nat. Biotechnol. 29, 644–U130. doi: 10.1038/nbt.1883
Graebner, R. C., Callaway, R. M., Montesinos, D. (2012). Invasive species grows faster, compete better, and shows greater evolution toward increased seed size and growth than exotic non-native congeners. Plant Ecol. 213, 545–553. doi: 10.1007/s11258-012-0020-x
Griffith, A. B., Andonian, K., Weiss, C. P., Loiket, M. E. (2014). Variation in phenotypic plasticity for native and invasive populations of Bromus tectorum. Biol. Invasions 16, 2627–2638. doi: 10.1007/s10530-014-0692-3
Hall, B. J., Notes, A. (2013). Building phylogenetic trees from molecular data with MEGA. Mol. Biol. Evol. 30 (5), 1229–1235. doi: 10.1093/molbev/mst012
Infante-Izquierdo, M. D., Polo-Ávila, A., Sanjosé, I., Castillo, J. M., Nieva, F. J. J., Grewell, B. J. (2020). Effects of heavy metal pollution on germination and early seedling growth in native and invasive Spartina cordgrasses. Mar. pollut. Bull. 158, 111367. doi: 10.1016/j.marpolbul.2020.111376
Kaneko, T., Kihara, M., Ito, K., Takeda, K. (2000). Molecular and chemical analysis of β-amylase-less mutant barley in Tibet. Plant Breed. 119, 383–387. doi: 10.1046/j.1439-0523.2000.00502.x
Lamichhane, J. R., Debaeke, P., Steinberg, C., You, M., J.Barbetti, M., Aubertot, J.-N. (2018). Abiotic and biotic factors affecting crop seed germination and seedling emergence: a conceptual framework. Plant Soil. 432, 1–28. doi: 10.1007/s11104-018-3780-9
Larson, J. E., Anacker, B. L., Wanous, S., Funk, J. L. (2020). Ecological strategies begin at germination: traits, plasticity and survival in the first 4 days of plant life. Funct. Ecol. 34, 968–979. doi: 10.1111/1365-2435.13543
Leger, E. A., Atwater, D. Z., James, J. J. (2019). Seed and seedling traits have strong impacts on establishment of a perennial bunchgrass in invaded semi-arid systems. J. Appl. Ecol. 56, 1343–1354. doi: 10.1111/1365-2664.13367
Li, B., Dewey, N. C. (2011). RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinf. 12, 323. doi: 10.1186/1471-2105-12-323
Liu, D., Chen, L., Chen, C., An, X., Zhang, Y., Wang, Y., et al. (2020). Full-length transcriptome analysis of Phytolacca americana and its congener P. icosandra and gene expression normalization in three Phytolaccaceae species. BMC Plant Biol. 20, 396. doi: 10.1186/s12870-020-02608-9
Liu, D., Chen, L., Chen, C., Zhou, Y., Xiao, F., Wang, Y., et al. (2022). Effect of plant VOCs and light intensity on growth and reproduction performance of an invasive and a native Phytolacca species in China. Ecol. Evol. 12, e8522. doi: 10.1002/ece3.8522
Liu, D., Li, P., Han, Y., Lei, W., Hou, M. (2016). Low temperature storage of southern rice black-streaked dwarf virus-infected rice plants cannot sustain virus transmission by the vector. J. Econ. Entomol. 109, 57–61. doi: 10.1093/jee/tov277
Liu, F., Wickham, J. D., Cao, Q., Lu, M., Sun, J. (2020). An invasive beetle-fungus complex is maintained by fungal nutritional-compensation mediated by bacterial volatiles. ISME J. 14, 2829–2842. doi: 10.1038/s41396-020-00740-w
Liu, B., Yan, J., Li, W., Yin, L., Li, P., Yu, H. (2020). Mikania micrantha genome provides insights into the molecular mechanism of rapid growth. Nat. Commun. 11, 340. doi: 10.1038/s41467-019-13926-4
Mortazavi, A., Williams, B. A., McCue, K., Schaeffer, L., Wold, B. (2008). Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 5, 621–628. doi: 10.1038/nmeth.1226
Penfield, S., Graham, S., Graham, I. A. (2005). Storage reserve mobilization in germinating oilseeds: Arabidopsis as a model system. Biochem. Soc. Trans. 33, 380–383. doi: 10.1042/BST0330380
R Core Development Team (2020). R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. Available at: https://www.R-project.org/
Rai, P. K., Singh, J. S. (2020). Invasive alien plant species: their impact on environment, ecosystem services and human health. Ecol.indic. 111, 106020. doi: 10.1016/j.ecolind.2019.106020
Rands, M. R. W., Adams, W. M., Bennun, L., Butchart, S. H. M., Clements, A., Coomes, D., et al. (2010). Biodiversity conservation: challenges beyond 2010. Science 329, 1298–1303. doi: 10.1126/science.1189138
Reichmann, L. J., Schwinning, S., Polley, H. W., Fay, P. A. (2016). Traits of an invasive grass conferring an early growth advantage over native grasses. J. Plant Ecol. 9, 672–681. doi: 10.1093/jpe/rtw014
Ross, M. A., Harper, J. L. (1972). Occupation of biological space during seeding establishment. J. Ecol. 60, 77–88. doi: 10.2307/2258041
Souza, M. L., Fagundes, M. (2014). Seed size as key factor in germination and seedling development of Copaifera langsdorffii (Fabaceae). Am. J. Plant Sci. 5, 48412. doi: 10.4236/ajps.2014.517270
Sun, M., Tuan, P. H., Izydorczyk, M. S., Ayele, B. T. (2020). Ethylene regulates post-germination seedling growth in wheat through spatial and temporal modulation of ABA/GA balance. J. Exp. Bot. 71, 1985–2004. doi: 10.1093/jxb/erz566
Verdú, M., Traveset, A. (2005). Early emergence enhances plant fitness: a phylogenetically controlled meta-analysis. Ecology 86, 1385–1394. doi: 10.1890/04-1647
Vila, M., Espinar, J. L., Hejda, M., Hulme, P. E., Jarosik, V., Maron, J. L., et al. (2011). Ecological impacts of invasive alien plants: a meta-analysis of their effects on species, communities and ecosystems. Ecol. Lett. 14, 702–708. doi: 10.1111/j.1461-0248.2011.01628.x
Wang, N. N., Wang, S. Y., Qi, F., Wang, Y. K., Lin, Y. J., Zhou, Y. M., et al. (2022). Autotetraploidization gives rise to differential gene expression in response to saline stress in rice. Plants-Basel. 11, 3114. doi: 10.3390/plants11223114
Wu, B., Sun, M., Zhang, H., Yang, D., Lin, C., Khan, I., et al. (2021). Transcriptome analysis revealed the regulation of gibberellin and the establishment of photosynthetic system promote rapid seed germination and early growth of seedling in pearl millet. Biotechnol. Biofuels. 14, 94. doi: 10.1186/s13068-021-01946-6
Xie, C., Mao, X., Huang, J., Ding, Y., Wu, J., Dong, S., et al. (2011). KOBAS 2.0: a web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 39, W316–W322. doi: 10.1093/nar/gkr483
Xu, H., Qiang, S., Genovesi, P., Ding, H., Wu, J., Meng, L., et al. (2012). An inventory of invasive alien species in China. NeoBiota 15, 1–26. doi: 10.3897/neobiota.15.3575
Yoshida, N., Nakamura, K. (1991). Molecular cloning and expression in Escherichia coli of cDNA encoding the subunit of sweet potato β-amylase. J. Biochem. 110, 196–121. doi: 10.1093/oxfordjournals.jbchem.a123556
Yu, F., Wan, W., Lv, M. J., Zhang, J. L., Meng, L. S. (2020). Molecular mechanism underlying the effect of the intraspecific alternation of seed size on plant drought tolerance. J. Agric. Food Chem. 68, 703–711. doi: 10.1021/acs.jafc.9b06491
Zaynab, M., Pan, D., Fatima, M., Chen, S., Chen, W. (2018). Transcriptomic approach to address low germination rate in Cyclobalnopsis gilva seeds. S. Afr. J. Bot. 119, 286–294. doi: 10.1016/j.sajb.2018.09.024
Zeeman, S. C., Kossmann, J., Smith, A. M. (2010). Starch: its metabolism, evolution, and biotechnological modification in plants. Annu. Rev. Plant Biol. 61, 209–234. doi: 10.1146/annurev-arplant-042809-112301
Zhang, J. L., Siemann, E., Tian, B., Huang, W., Ding, J. Q. (2019). Differences in seed properties and germination between native and introduced populations of Triadica sebifera. J. Plant Ecol. 13, 70–77. doi: 10.1093/jpe/rtz048
Keywords: rapid seedling emergence, invasive Phytolacca americana, β-amylase, photosynthesis, soluble sugar
Citation: Liu D, Liu M, Ju R, Li B and Wang Y (2024) Rapid seedling emergence of invasive Phytolacca americana is related to higher soluble sugars produced by starch metabolism and photosynthesis compared to native P. acinosa. Front. Plant Sci. 15:1255698. doi: 10.3389/fpls.2024.1255698
Received: 27 July 2023; Accepted: 03 January 2024;
Published: 26 January 2024.
Edited by:
Aleš Kladnik, University of Ljubljana, SloveniaReviewed by:
Weiqiang Li, Chinese Academy of Sciences (CAS), ChinaSzymon Kubala, Polish Academy of Sciences, Poland
Copyright © 2024 Liu, Liu, Ju, Li and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Danfeng Liu, ZGZsQHludS5lZHUuY24=; Yi Wang, eWl3YW5nQHludS5lZHUuY24=
 Danfeng Liu
Danfeng Liu Maoye Liu1
Maoye Liu1 Ruiting Ju
Ruiting Ju Yi Wang
Yi Wang