- 1College of Agronomy, Shenyang Agricultural University, Shenyang, China
- 2Liaoning Agricultural Vocational and Technical College, Yingkou, China
Introduction: Trehalose is vital for plant metabolism, growth, and stress resilience, relying on Trehalose-6-phosphate synthase (TPS) and Trehalose-6-phosphate phosphatase (TPP) genes. Research on these genes in cultivated peanuts (Arachis hypogaea) is limited.
Methods: This study employed bioinformatics to identify and analyze AhTPS and AhTPP genes in cultivated peanuts, with subsequent experimental validation of AhTPS9’s role in cold tolerance.
Results: In the cultivated peanut genome, a total of 16 AhTPS and 17 AhTPP genes were identified. AhTPS and AhTPP genes were observed in phylogenetic analysis, closely related to wild diploid peanuts, respectively. The evolutionary patterns of AhTPS and AhTPP genes were predominantly characterized by gene segmental duplication events and robust purifying selection. A variety of hormone-responsive and stress-related cis-elements were unveiled in our analysis of cis-regulatory elements. Distinct expression patterns of AhTPS and AhTPP genes across different peanut tissues, developmental stages, and treatments were revealed, suggesting potential roles in growth, development, and stress responses. Under low-temperature stress, qPCR results showcased upregulation in AhTPS genes (AhTPS2-5, AhTPS9-12, AhTPS14, AhTPS15) and AhTPP genes (AhTPP1, AhTPP6, AhTPP11, AhTPP13). Furthermore, AhTPS9, exhibiting the most significant expression difference under cold stress, was obviously induced by cold stress in cultivated peanut, and AhTPS9-overexpression improved the cold tolerance of Arabidopsis by protect the photosynthetic system of plants, and regulates sugar-related metabolites and genes.
Discussion: This comprehensive study lays the groundwork for understanding the roles of AhTPS and AhTPP gene families in trehalose regulation within cultivated peanuts and provides valuable insights into the mechanisms related to cold stress tolerance.
1 Introduction
Trehalose, a symmetrical non-reducing disaccharide formed by two glucose molecules through α, α-1,1-glycosidic bonds, exhibits distinctive physicochemical properties setting it apart from analogous sugars (Paul et al., 2008). It holds a pivotal role in maintaining cellular integrity across various organisms, particularly in plant growth and development (O’Hara et al., 2013). Under adverse conditions like high temperatures, freezing, and osmotic stress, cells synthesize trehalose as a defense mechanism, aiding in osmotic regulation, membrane preservation, and signal transduction (Fernandez et al., 2010; Kosar et al., 2019; Hassan et al., 2023). The precursor of trehalose is trehalose 6-phosphate (Tre6P), which has a dual role in plant metabolism and development (Figueroa and Lunn, 2016). It regulates sucrose production in source leaves to balance the demand for sucrose in growing sink organs. Tre6P also functions as a signal of sucrose availability, influencing developmental decisions such as flowering, embryogenesis, and lateral branching, thereby linking sucrose supply to sink organ growth (Paul et al., 2018). For example, Tre6P modulates plant respiration and metabolism by inhibiting SnRK1 activity, which functions as a key regulator of energy and nutrient responses in plants (Emanuelle et al., 2016). Trehalose synthesis, its versatile functions, and its presence in diverse organisms underline its significance in biological systems. This dynamic sugar, balancing energy and protective roles, holds promise for various applications in plant biotechnology and stress management.
Trehalose synthesis mainly involves two indispensable enzymes: trehalose 6-phosphate (Tre6P) synthase (TPS) and Tre6P phosphatase (TPP) (Smeekens, 2015). In plants, the trehalose biosynthesis pathway involves a sequential enzymatic process: Initially, trehalose-6-phosphate synthase (TPS) facilitates the condensation of uridine diphosphate glucose (UDPG) and glucose 6-phosphate (G6P) to yield an intermediary compound trehalose 6-phosphate (Tre6P). Subsequently, this intermediate is subjected to dephosphorylation by trehalose-6-phosphate phosphatase (TPP), resulting in the synthesis of trehalose (Fichtner and Lunn, 2021). TPS and TPP are key enzymes in the trehalose metabolism pathway. TPS and TPP genes are found in unicellular chlorophyte algae, streptophyte algae, and across all major groups of terrestrial plants, suggesting their presence from the early stages of the green plant lineage. Initially identified in Arabidopsis (AtTPS1, AtTPPA, and AtTPPB), these genes were subsequently recognized as part of extensive gene families after the sequencing of the Arabidopsis genome (Kaul et al., 2000). The Arabidopsis species possesses a total of 11 TPS genes (AtTPS1–AtTPS11) and 10 TPP genes (AtTPPA–AtTPPJ) (Aubourg et al., 2002; Vandesteene et al., 2012). The AtTPS proteins contain TPS and TPP domains, which form two distinct clades in phylogenetic analyses: class I (AtTPS1–AtTPS4) and class II (AtTPS5–AtTPS11) (Leyman et al., 2001). The TPS genes have been extensively studied in the model plant Arabidopsis, as well as in other plant species. These genes play a crucial role in serving as a conduit for sucrose signaling, triggering the production of trehalose during stress responses. The research conducted so far has illuminated the significance of TPS genes, particularly in terms of their capacity to enhance biotic and abiotic stress tolerance when overexpressed or properly expressed in transgenic plants (Iordachescu and Imai, 2008; Vishal et al., 2019; Fichtner et al., 2020). Furthermore, within plants, a substantial family of compact proteins housing the conserved only TPP domain is prevalent. In contrast, the functional roles of TPPs in the context of trehalose production have unveiled an array of distinct expression patterns across various tissues, underscoring their potential functional diversity (Vandesteene et al., 2012; Van Houtte et al., 2013; Du et al., 2022). It’s worth noting, however, that there does exist a degree of overlap in their expression patterns, suggesting a partial redundancy in their functions. Interestingly, through phylogenetic analysis, it has been possible to cluster each TPP and its counterparts from different species into distinct groups (Ma et al., 2007; Li et al., 2008). The induction of TPP expression comes about through varied hormonal and abiotic stress treatments, and the corresponding TPPs have been found to actively participate in plant stress responses. AtTPPF, for instance, assumes a proactive role in safeguarding cells against oxidative damage caused by reactive oxygen species during drought stress by augmenting soluble sugar levels (Lin et al., 2019). On the other hand, lines overexpressing AtTPPD display heightened salt tolerance due to increased sensitivity to redox shifts in two cysteine residues of TPPD. This sensitivity prompts accelerated activity under salt stress, thereby leading to an accumulation of trehalose (Krasensky et al., 2014). The overexpression of OsTPS1 and OsTPP1 within rice plants has been shown to bolster trehalose levels, ultimately enhancing plant survival under low temperature stress (Ge et al., 2008). Moreover, the latter protein, OsTPP1, contributes to the regulation of rice seed germination by engaging in ABA signaling (Wang et al., 2021). Both TPS and TPP gene families have independently expanded in various plant divisions, contributing to the diversity of stress response functions in plants.
Cultivated peanut (Arachis hypogaea L.) is an important oil and economic crop, which is widely planted in the semiarid tropical and subtropical regions (Krishna et al., 2015; Zhuang et al., 2019). Peanut plants are susceptible to various abiotic stresses during their growth, such as temperature, drought, salinity, and metal toxicit (Paul et al., 2001; Shi and Cai, 2009; Puppala et al., 2023). However, among various abiotic stresses, cold stress significantly impairs the growth, production, and quality of many crop plants (Kidokoro et al., 2022; Raza et al., 2023). In recent years, China has experienced an increased frequency of cold damage events, particularly impacting early-sown spring peanuts. These events have a common detrimental effect on various stages of peanut cultivation, including seed germination, growth, development, flowering, and overall yield (Kakani et al., 2002; Chen et al., 2014; Zhang et al., 2019; Xue et al., 2023; Zhang et al., 2023). Identifying key genes that can confer cold stress tolerance is crucial for enhancing crop productivity in regions with low temperatures (Bhat et al., 2022). These genes can be utilized in biotechnological programs to generate improved varieties, which is an urgent requirement in peanut production. Although the TPS and TPP genes has been identified to participate in growth, development, and response to various stress in multiple plant species, there have been no comprehensive analyses of the TPS and TPP gene families in peanut, and previous studies have not yet characterized the role of TPS and TPP genes in responding to environmental stresses. As a tetraploid crop, cultivated peanut contains A and B subgenomes that evolved from two diploid wild peanut varients (Arachis duranensis (AA) and Arachis ipaensis (BB). The sequencing of the whole genome of cultivated and wild peanut has been completed and uploaded, providing an opportunity for the analysis of the TPS and TPP gene familes in the context of cold stress in peanut (Bertioli et al., 2016; Bertioli et al., 2019; Chen et al., 2019; Zhuang et al., 2019).
In this study, we identified 16 TPS and 17 TPP genes in cultivated peanut, and comprehensively analyzed phylogenetic relationships, the chromosomal distributions, gene duplication, motif compositions, gene structures, and cis-acting elements of TPS and TPP genes using bioinformatics methods. Further transcriptome-based tissue-specific expression analysis of AhTPS and AhTPP genes was conducted, and their expression patterns in response to low temperature were assessed using fluorescence quantitative PCR (qPCR) methods. Among these, key genes were functionally characterized in transgenic Arabidopsis to reveal its crucial role in cold tolerance. Our findings lay the groundwork for further investigation into the functions of the AhTPS and AhTPP families in cultivated peanut. Additionally, this research will facilitate their utilization in the genetic improvement of crops.
2 Materials and methods
2.1 Plant materials and treatment
The cold-stress variety Nonghua5 provided by the Peanut Research Institute, Shenyang Agriculture University, Shenyang, China, was used for cultivation and cold treatment. The peanuts underwent sowing, cultivation, and cold treatment following the protocol outlined in Zhang et al. (2023), with minor adjustments. To ensure sterility, the seeds were surface treated using a 3% sodium hypochlorite solution, rinsed thoroughly with distilled water, and placed in darkness for germination. Germinated seeds were then planted in circular plastic pots filled with a mixture of vermiculite and nutrient soil in a 2:1 ratio. The plants were grown in a climate chamber with a 16-hour light (28°C)/8-hour dark (23°C) cycle, a photosynthetic photon flux density of 400 μmol m−2 s−1, and a relative humidity of 70%. To induce cold stress, the temperature in the climate chamber was lowered to 4°C while maintaining other growth conditions. The second leaves were collected at 0, 6, 12, 24, and 48 hours after each treatment, with three biological replicates. The collected leaves were immediately frozen in liquid nitrogen and stored at -80°C.
2.2 Identification of the TPS and TPP family genes in cultivated peanut and wild peanut
To identify genes encoding TPS and TPP proteins of tetraploid cultivated peanuts (Arachis hypogaea) and diploid wild peanuts (Arachis duranensis and Arachis ipaensis) in Peanut genome database (PeanutBase-BLAST, https://www.peanutbase.org/pb_sequenceserver), the sequences of 11 AtTPS and 10 AtTPP proteins were utilized, with an E-value threshold of < 10-5. The identified TPS and TPP gene sequences underwent a conserved domain search (http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi), and manually removing peanut protein sequences lacking TPS or TPP domains. SMART (http://smart.embl.de/) and Pfam (http://pfam.xfam.org/) were employed to confirm the conserved domains of TPS and TPP using the remaining protein sequences. The ExPASy database (https://web.expasy.org/protparam/) was utilized to predict the physicochemical properties of AhTPSs and AhTPPs, including molecular weight, isoelectric point (pI), aliphatic index, and total average hydrophilicity (GRAVY).
2.3 Phylogenetic analysis of AhTPS and AhTPP genes
The phylogenetic tree was constructed using protein sequences from TPS and TPP genes in cultivated and wild diploid peanuts (Arachis hypogaea, Arachis duranensis, and Arachis ipaensis), as well as Arabidopsis, soybean, rice, and tomato. The protein sequences of TPP and TPS from Arachis hypogaea, Arachis duranensis, and Arachis ipaensis were obtained from the peanut genome database (https://www.peanutbase.org/). Similarly, the protein sequences of TPP and TPS from Arabidopsis and rice were downloaded from NCBI (https://www.ncbi.nlm.nih.gov/), while those from soybean and tomato were obtained from the soybean genome database (https://soybase.org/). Multiple sequence alignments were performed using the MUSCLE method, and the phylogenetic tree was constructed using the neighbor-joining (NJ) method with 1000 bootstrap replications, employing MEGA11 software (Tamura et al., 2021). The resulting tree was visualized using EvolView (https://evolgenius.info//evolview).
2.4 Chromosomal location, gene duplication, and synteny analysis
The chromosomal distribution of AhTPSs and AhTPPs was determined by MG2C online software. Gene duplication patterns and collinearity relationships of the AhTPS and AhTPP gene families between tetraploid cultivated and diploid wild peanuts were identified and analyzed using MCScanx of TBtools with default parameters. TBtools was subsequently used to calculate Ka and Ks, with a Ka/Ks to explore the evolutionary dynamics and selection pressure (Chen et al., 2020).
2.5 Analysis of gene structures and motifs
Based on the peanut genome annotation (GFF file), gene exon-intron structures were obtained. TBtools was utilized to construct protein motif and gene structure maps for members of the peanut TPS and TPP gene families. The MEME program (http://meme-suite.org/tools/meme) was employed to assay the conserved motifs of each protein. The parameters used to identify conserved motifs in the protein sequences were set as follows: a maximum of 20 motifs and other optional default parameters.
2.6 Cis-acting element prediction in the upstream region of TPPs and TPSs
In order to explore potential cis-acting regulatory elements within the promoter regions of AhTPP and AhTPS genes, the upstream sequence of 2000 bp preceding the coding region was extracted from the peanut genome sequence. Subsequently, this sequence was subjected to cis-regulatory element prediction using the PlantCARE database (https://bioinformatics.psb.ugent.be/webtools/plantcare/html/). The predicted cis-regulatory elements were categorized based on their regulatory functions, and their distribution within the promoter regions of AhTPP and AhTPS was visualized as a heatmap.
2.7 Expression profile analysis of AhTPSs and AhTPPs in different tissues and under different environmental treatments
The transcriptome-based data of different tissues and under different environmental treatments (different hormonal, low temperature and drought treatments) were retrieved from the PeanutBase database (https://www.peanutbase.org/) and Peanut Genome Resource (http://peanutgr.fafu.edu.cn/index.php) (Clevenger et al., 2016; Zhuang et al., 2019). TBtools was used to generate an expression heatmap of the reads per kilobase per million mapped reads (RPKM) data. The transcriptome data were normalized by log2 (1 +RPKM).
2.8 RNA extraction and quantitative real-time PCR
Total RNA of plant tissues was isolated using the Plant Total RNA Extraction Kit (Tiangen Biotech, Beijing, China) according to the manufacturer’s instructions. Then, cDNA was synthesized by PrimeScriptTM RT Kit (TaKaRa, Japan). AhActin was used as the reference gene, and the specific primers are designed using Primer-BLAST (https://www.ncbi.nlm.nih.gov/tools/primer-blast/). The gene expression analysis was carried out using the SYBR Premix Ex TaqII kit (TliRNaseH Plus) from TaKaRa, Japan, and fluorescence quantitative reactions were detected using ABI7500 from Applied Biosystems, United States. The relative expression analysis was calculated using the 2−ΔΔCT approach.
2.9 Subcelluar localization analysis of AhTPS9 protein
The specific primers (AhTPS9-F/R) were utilized to amplify the complete cDNA of AhTPS9 from tobacco leaves. The coding sequence data for AhTPS9 from PeanutBase (https://www.peanutbase.org/) were used as a reference. The resulting PCR products were connected to the pBWA (V) HS-GLosgfp vector, generating the pBWA (V) HS- AhTPS9-GFP vector which incorporates the green fluorescent protein (GFP) reporter gene. After confirmation through sequencing, the positive clones were transferred into Agrobacterium tumefaciens (EHA105) using electrotransformation. Young seedlings (30 days old) were carefully chosen, and injections were performed into the lower epidermis of leaves. Subsequently, these seedlings were cultured under low light conditions for 2 days. Observation and imaging took place using a laser confocal microscope (Nikon C2-ER, Tokyo, Japan), with the corresponding empty vector serving as a control.
2.10 The AhTPS9 function analysis under cold stress conditions
Gene-specific primers for AhTPS9 were designed to amplify the cDNA sequence in a cold-tolerance genotype Nonghua5 by Primer Premier 6.0. PCR amplification of the coding sequence of AhTPS9 was performed using the TransTaq DNA Polymerase High Fidelity Amplification Kit. The PCR reaction program consisted of an initial denaturation at 94°C for 3 minutes, followed by 30 cycles of denaturation at 94°C for 30 seconds, annealing at 58-60°C for 30 seconds, and extension at 72°C for 1 minute. The final extension was carried out at 72°C for 10 minutes. PCR products were analyzed on a 1% agarose gel, purified using a universal DNA purification recovery kit, and then ligated into the pBWA (V) BS cloning vector. The ligated DNA was transformed into Escherichia coli Top10 competent cells, positive clones were selected, and their identity was confirmed through sequencing. The recombinant plasmid pBWA (V) BS-AhTPS9 was transformed into Agrobacterium tumefaciens EHA105, which was subsequently introduced into wild-type (WT) Arabidopsis using the floral dip method as described by Clough and Bent (1998). Following antibiotic-based screening and PCR verification of the transgenic seedlings, we successfully generated the homozygous transgenic lines in the T2 generation. Subsequently, homozygous T3 progeny was examined and selected for further experimental procedures.
The T3 seeds, previously subjected to a 4°C vernalization treatment on MS medium, were incubated in a growth chamber at 22°C for 14 days. Subsequently, they underwent a 5-day cold treatment at 4°C, followed by phenotypic observations. Columbia wild-type Arabidopsis plants (WT) were subjected to the same cold treatment conditions as controls. Leaf samples were collected at 0h (CK) and 24h after the cold treatment to assess changes in the expression levels of low-temperature-responsive genes in both overexpressing plants and wild-type plants. At 72h after the low-temperature treatment, leaf samples were collected to analyze related metabolites and physiological indicators. All samples are stored in liquid nitrogen for further use.
2.11 Physiological indicator and metabolism measurements
To evaluate the physiological indicators in transgenic and wild plants under cold conditions, the contents of proline and MDA, soluble sugar and starch content were measured using the commercial kits according to the manufacturer’s instructions (Solarbio, Beijing, China). To determine chlorophyll content, leaves of wild-type and transgenic Arabidopsis plants, with and without cold stress treatment, were homogenized in 80% acetone. The homogenized leaves were refrigerated at 4°C for 3 days until they reached near-bleached status. Optical density (OD) values at 645nm and 663nm were then measured, and chlorophyll content was calculated using the formula: (8.02*OD663 + 20.21*OD645) *M/1000/N, where M represents the volume of 80% acetone added, and N is the fresh weight of the aerial plant parts. “The net photosynthetic rate, Fv/Fm, and electrolytic leakage photosynthetic indices were measured using the Li-6400 portable photosynthetic apparatus. Fv/Fm measurements were conducted using the Dual Pam 100, with Arabidopsis plants darkened for a minimum of 30 minutes prior to measurement. The electrolytic leakage experiment method was referenced from Bajji et al. (2002). The measurement of sugar metabolism products, including trehalose, Tre6p, and sucrose content, was conducted using high-performance liquid chromatography-mass spectrometry (HPLC-MS), following the method outlined in Lin et al. (2023).
2.12 Statistical analysis
Three replicates were used for all experiments. Statistically significant data were analyzed using one-way analysis of variance (ANOVA) with the least significant difference (LSD) method. The R package ANOVA (https://statsandr.com/blog/anova-in-r/) was employed for this analysis. The calculated values were presented as means ± standard deviation (SD).
3 Result
3.1 Genome-wide identification and characteristics of AhTPS and AhTPP genes in wild and cultivated peanut
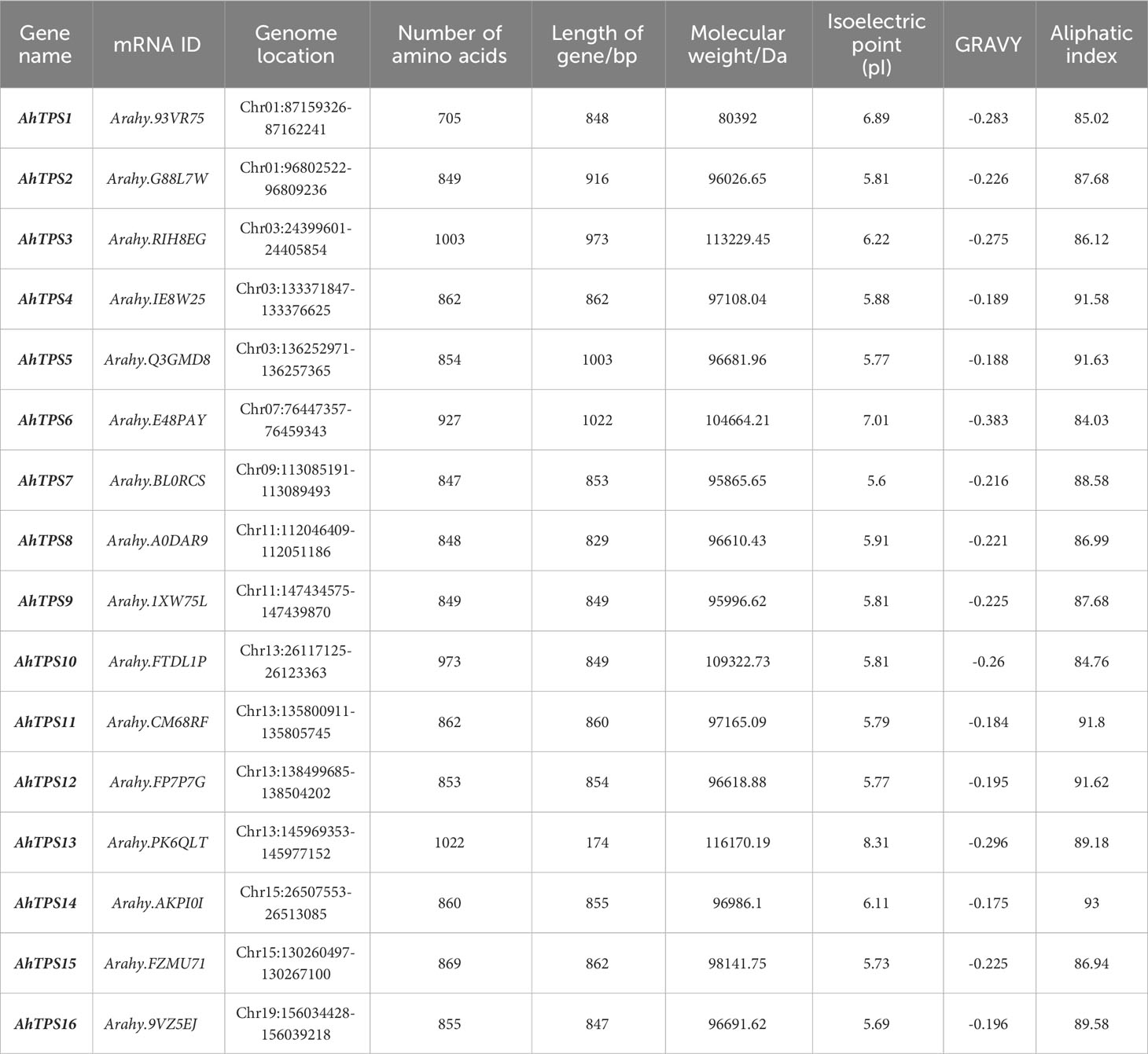
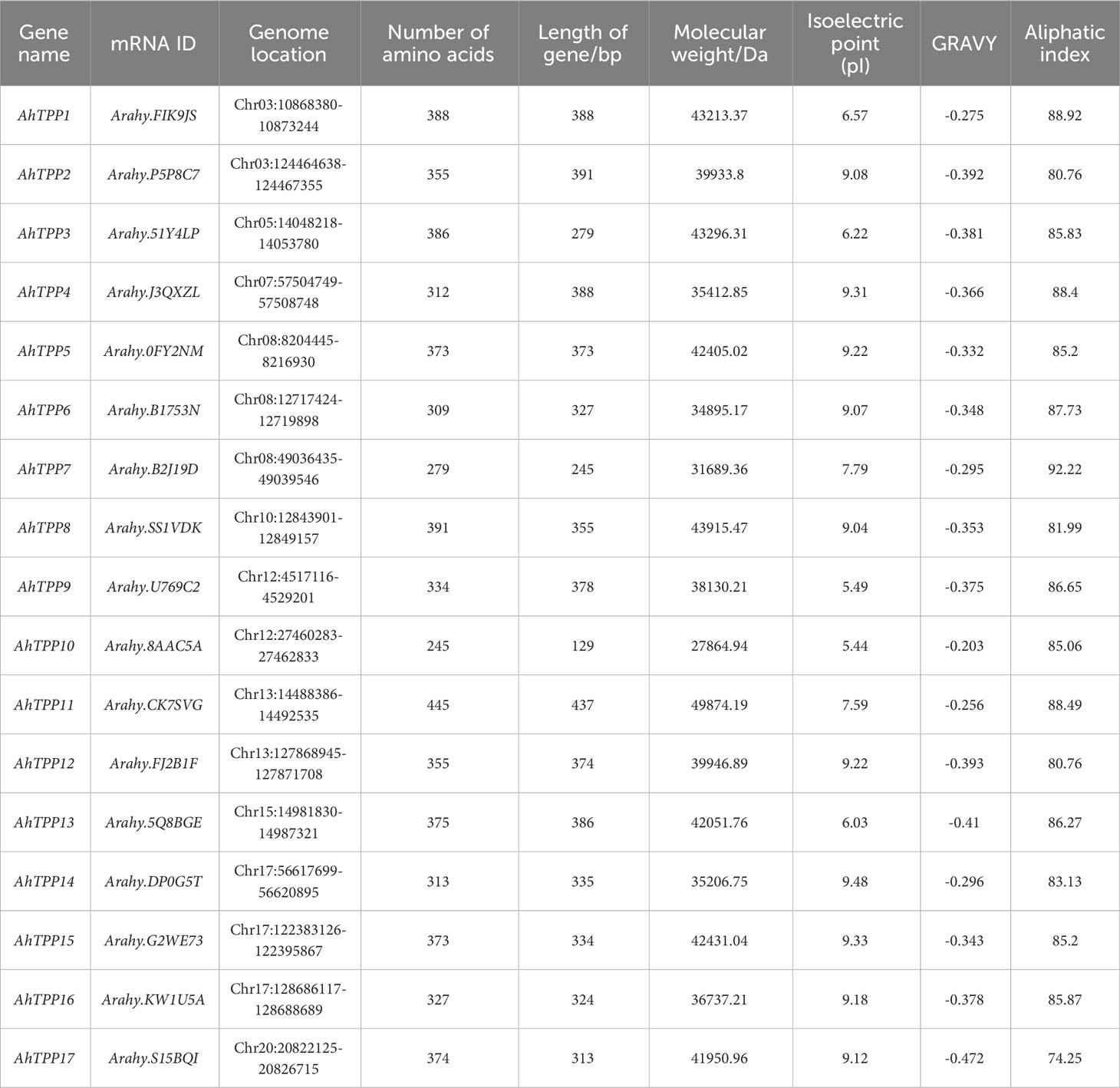
Through BLAST searches and confirmation of conservative domains, a total of 16 TPS and 17 TPP genes were identified in the cultivated peanut genome (Arachis hypogaea). Based on their chromosomal locations, these TPS genes were designated as AhTPS1-16, while the TPP genes were named AhTPP1-17. All 16 TPS genes contained both TPS and TPP conserved domains, whereas the 17 TPP genes each contained a single typical TPP domain (Supplementary Figures 1, 2). Additionally, 19 TPS genes and 15 TPP genes were identified in the wild diploid peanut genomes Arachis duranensis and Arachis ipaensis. In the A. duranensis genome, 9 TPS genes and 7 TPP genes were identified and named AdTPS1 – AdTPS9 and AdTPP1 – AdTPP7, respectively. Meanwhile, A. ipaensis harbored 10 TPS and 8 TPP genes designated as AiTPS1 – AiTPS10 and AiTPP1 – AiTPP8 (Tables 1, 2).
Physicochemical property analysis revealed that the gene length of AhTPSs ranged from 174 to 1022 bp, protein length from 705 to 1022 amino acids, molecular weight from 80.4 kDa to 116.17 kDa. The aliphatic index of AhTPSs ranged from 84.03 to 91.58. Isoelectric points of AhTPS1-AhTPS5, AhTPS7-AhTPS12, and AhTPS14-AhTPS16 fell within the range of 5.01 to 6.89, indicating an acidic nature due to an abundance of acidic amino acids. In contrast, AhTPS6 and AhTPS13 were enriched in basic amino acids. Gene length for AhTPPs ranged from 129 to 437 bp, protein length from 245 to 445 amino acids, and molecular weight from 27864.94 Da to 49874.19 Da. The aliphatic index of AhTPPs ranged from 74.25 to 92.22. Theoretical isoelectric point analysis indicated that AhTPP1, AhTPP3, AhTPP9, AhTPP10 and AhTPP13 had isoelectric points between 5.01 and 6.57, suggesting an acidic nature due to acidic amino acids. Conversely, AhTPP2, AhTPP4-AhTPP8, AhTPP11-AhTPP12, and AhTPS14-AhTPS17 were rich in basic amino acids. The GRAVY (Grand Average of Hydropathy) values for all 16 TPS and 17 TPP proteins were below 0, indicating their hydrophilic nature.
3.2 Phylogenetic analysis of AhTPS and AhTPP gene families
To investigate the evolutionary relationships of the AhTPS and AhTPP gene families, phylogenetic trees were constructed using TPS and TPP family genes from Arabidopsis, soybean, rice, tomato, wild diploid peanut, and cultivated peanut, respectively. Across these species, a total of 87 TPS genes and 79 TPP genes were used to construct systematic phylogenetic trees. In these species, a total of 87 TPS genes and 79 TPP genes were employed to build systematic phylogenetic trees. Based on the phylogenetic relationship of rice and Arabidopsis (Figure 1A), The TPS gene family is categorized into two subfamilies, namely Class I and Class II. Within Class I, there are two subgroups, while Class II is divided into three subgroups. AtTPS1-4 belong to Class II, while AtTPS5-11 belong to Class I. The gene counts of TPS within different taxa, including cultivated peanut, two wild diploid peanuts (Ad/Ai), rice, soybean, Arabidopsis, and tomato, are as follows: Class I-1 (2, 2/2, 1, 5, 1, 2), Class I-2 (0, 0/0, 0, 0, 3, 1), Class II-1 (2, 1/1, 3, 2, 1, 1), Class II-2 (6, 3/4, 2, 7, 3, 2), and Class II-3 (6, 3/3, 5, 6, 3, 5). Except for Class I-2, each group contains at least one gene across the seven species. Comparative analysis suggests a closer genetic relationship between TPS genes in the Arachis genus and GmTPS genes in soybean. Furthermore, AhTPS6 in Class I-1 clusters alongside AtTPS1 and OsTPS1, implying potential functional similarity among genes within the same group.
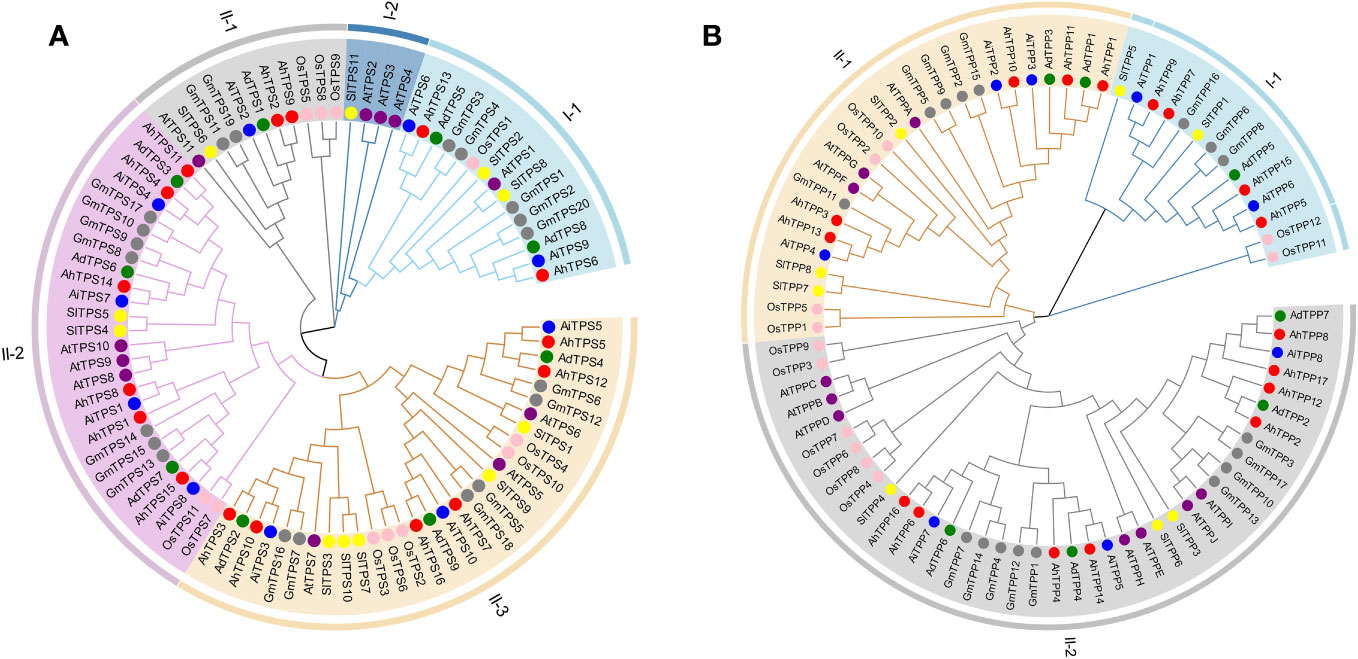
Figure 1 Phylogenetic analysis of TPS (A) and TPP (B) gene family among cultivated peanut (Arachis hypogaea, Ah), wild diploid peanut (Arachis duranensis, Ad and Arachis ipaensis, Ai), Arabidopsis thaliana (At), soybean (Glycine max, Gm), rice (Oryza sativa, Os) and tomato (Solanum lycopersicum, Sl). The phylogenetic tree was constructed using MEGA11 based on the Neighbor-joining method with 1000 bootstrap replicates.
The TPP gene family is divided into two categories (Figure 1B). Class I TPP genes consist of four cultivated peanut genes (AhTPP5, AhTPP7, AhTPP9, and AhTPP15), three wild peanut genes (AdTPP5, AiTPP1, and AiTPP6), three soybean genes (GmTPP6, GmTPP8, and GmTPP16), two rice genes (OsTPP11 and OsTPP12), and two tomato genes (SlTPP1 and SlTPP5); the remaining thirteen TPP genes belong to Class II. Class II TPP genes are divided into two subgroups. AtTPPA, AtTPPG, and AtTPPF belong to Class II-1, while the rest of the AtTPP genes belong to Class II-2. Gene counts within groups for cultivated peanut, wild peanut (Ad/Ai), rice, soybean, Arabidopsis, and tomato are as follows: Class I-1 (4, 1/2, 2, 3, 0, and 2), Class II-1 (5, 2/3, 4, 5, 3, and 3), and Class II-2 (8, 4/3, 6, 9, 7, and 3). The phylogenetic relationships between AhTPPs, GmTPPs, and AtTPPs are closer. AhTPP1, AhTPP3, AhTPP10, AhTPP11, along with AtTPPA, AtTPPG, and AtTPPF, cluster in Class II-1. AhTPP6 and AhTPP16, along with AhTPP4, AhTPP14, AhTPP2, AhTPP12, AhTPP17, and AhTPP8, cluster in Class II-2. AtTPPB, AtTPPC, AtTPPD, AtTPPE, AtTPPH, AtTPPI, and AtTPPJ also cluster in Class II-2. Altogether, TPS and TPP genes exhibit close relationships within subgenomes of cultivated peanuts and wild diploid peanuts, and share a higher degree of similarity with leguminous crop, soybean.
3.3 Chromosomal location, gene duplication, and synteny analysis of AhTPS and AhTPP genes
To study the chromosomal locations of AhTPS and AhTPP genes in the genome of cultivated peanut, the distribution of genes on chromosomes is depicted based on the genome annotation file (GFF) of cultivated peanut (Figure 2). We observed an uneven distribution of the AhTPS and AhTPP genes on the chromosomes of cultivated peanut. The results indicate that peanuts have 20 chromosomes, with 16 AhTPS genes distributed across Chrs01, 03, 07, 09, 11, 13, 15, and 19. Seventeen AhTPP genes are distributed across chromosomes 03, 05, 07, 08, 10, 12, 13, 15, 17, and 20. Most of these genes are located near the ends of the chromosomes. TPS and TPP genes were not identified on certain chromosomes, such as Chr02, Chr04, Chr06, Chr14, Chr16, and Chr18. Based on the gene locations on chromosomes, it was found that some TPS and TPP orthologous genes from the A subgenome (Chr01-10) and the B subgenome (Chr11-20) exhibit consistent positions, such as AhTPS1 and AhTPS8, AhTPS2 and AhTPS9, AhTPP1 and AhTPP11. It suggests their retention during the evolution from wild diploid peanuts to cultivated tetraploid peanut. Conversely, certain genes show distribution differences between the A and B subgenomes. For instance, TPP genes on Chr02 have been lost during the evolutionary process, while Chr12 retains AhTPP9 and AhTPP10.
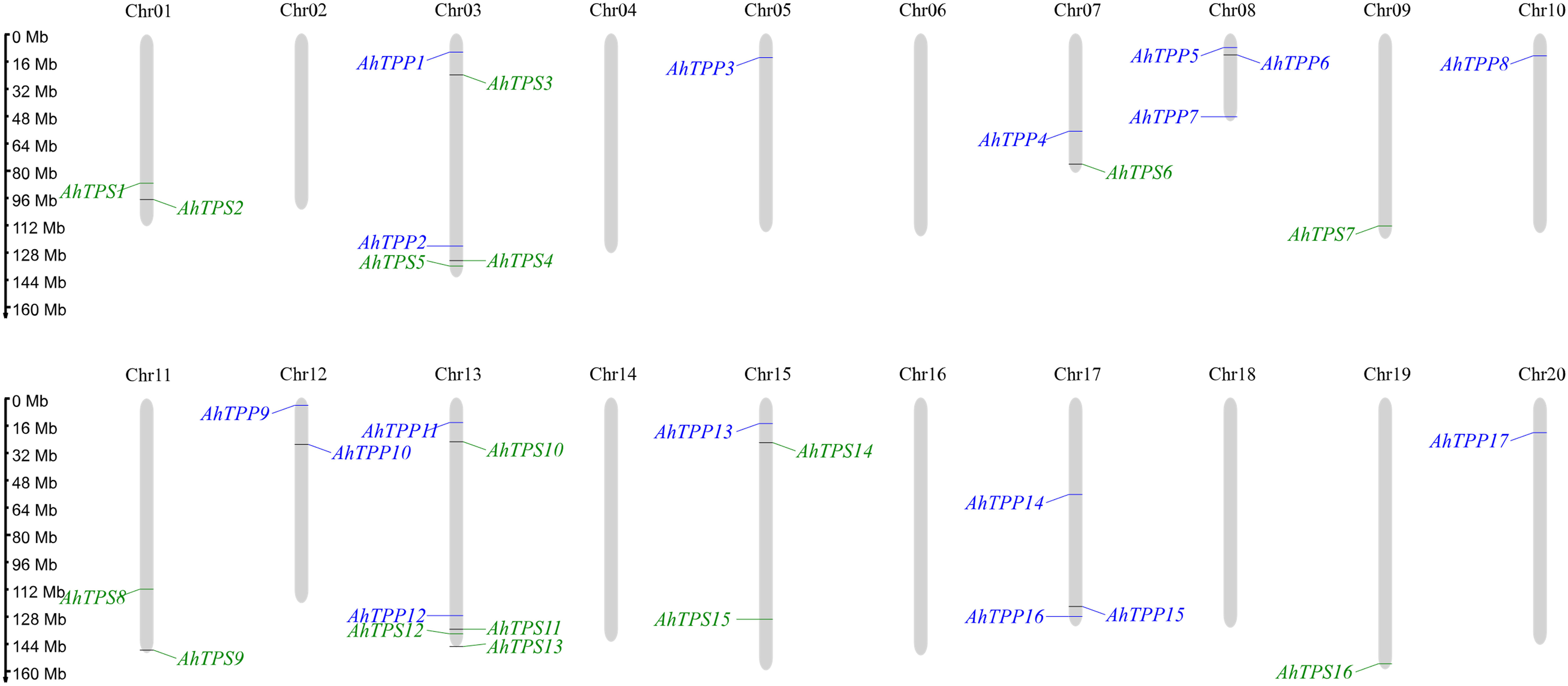
Figure 2 Chromosomal location analysis of AhTPS and AhTPP genes. AhTPSs (green) and AhTPPs (blue) are marked on chromosomes. The scale bar on the left indicates the length of A. hypogaea chromosomes (Mb).
To study evolutionary relationships and reveal the homology relationship of TPS and TPP genes in different Arachis species, two diploid wild peanut, Arachis duranensis and Arachis ipaensis, were selected for collinear analysis the cultivated peanut Arachis hypogaea (Figure 3). According to collinearity analysis, 22 and 21 collinear gene pairs were detected between AhTPS genes of A. duranensis and A. hypogaea, and A. ipaensis and A. hypogaea, respectively (Figure 3A, Supplementary Table 1). Between AhTPP genes of A. duranensis and A. hypogaea, as well as A. ipaensis and A. hypogaea, 19 and 14 collinear gene pairs were respectively identified (Figure 3D, Supplementary Table 2). All TPS and TPP members in cultivated peanuts possess corresponding segments in both wild species, suggesting the genetic conservation of these two family members during the formation of tetraploid peanuts from their wild counterparts. Gene duplication (segmental and tandem duplication) is a major force behind genome evolution. So, Arachis TPS and TPP gene duplication events were evaluated (Supplementary Tables 3, 4). The AhTPS gene family exhibited 12 pairs of segment duplications (Figure 3B), while the AhTPP gene family showed 16 pairs of segment duplications (Figure 3E). No tandem duplications were identified in either the AhTPS or AhTPP gene families. Further collinearity analysis was conducted between the gene families in the diploid wild peanut genomes. Among the TPS gene families of the two wild species, 13 pairs of orthologous genes were identified (Figure 3C), and for the TPP gene families, 6 pairs of orthologous genes were detected (Figure 3F). Some gene pairs, such as AdTPS3-AdTPS6, AdTPS3-AdTPS7, and AiTPS1-AiTPS8 in the TPS gene family (Supplementary Table 3), as well as AdTPP4-AdTPP6 in the TPP gene family (Supplementary Table 4), suggested that these segment duplications had already were present during the evolution of wild diploid peanuts. Discrepancies between the collinearity maps of wild diploid and cultivated peanuts may be attributed to the emergence of new segment duplications or transpositions within the TPS and TPP gene families after the derivation of tetraploid cultivated peanuts from diploid wild peanuts. The ratio of nonsynonymous substitution rate (Ka) to synonymous substitution rate (Ks) provides insights into the evolutionary process and selection pressure. We calculated the Ka/Ks ratio among the Arachis species. For both AhTPS and AhTPP gene members, except for a few genes for which Ka/Ks ratios could not be calculated, most of the calculated Ka/Ks ratios were less than 1, indicating that AhTPS and AhTPP genes were subject to strong purifying selection pressure during evolution (Supplementary Tables 5, 6).
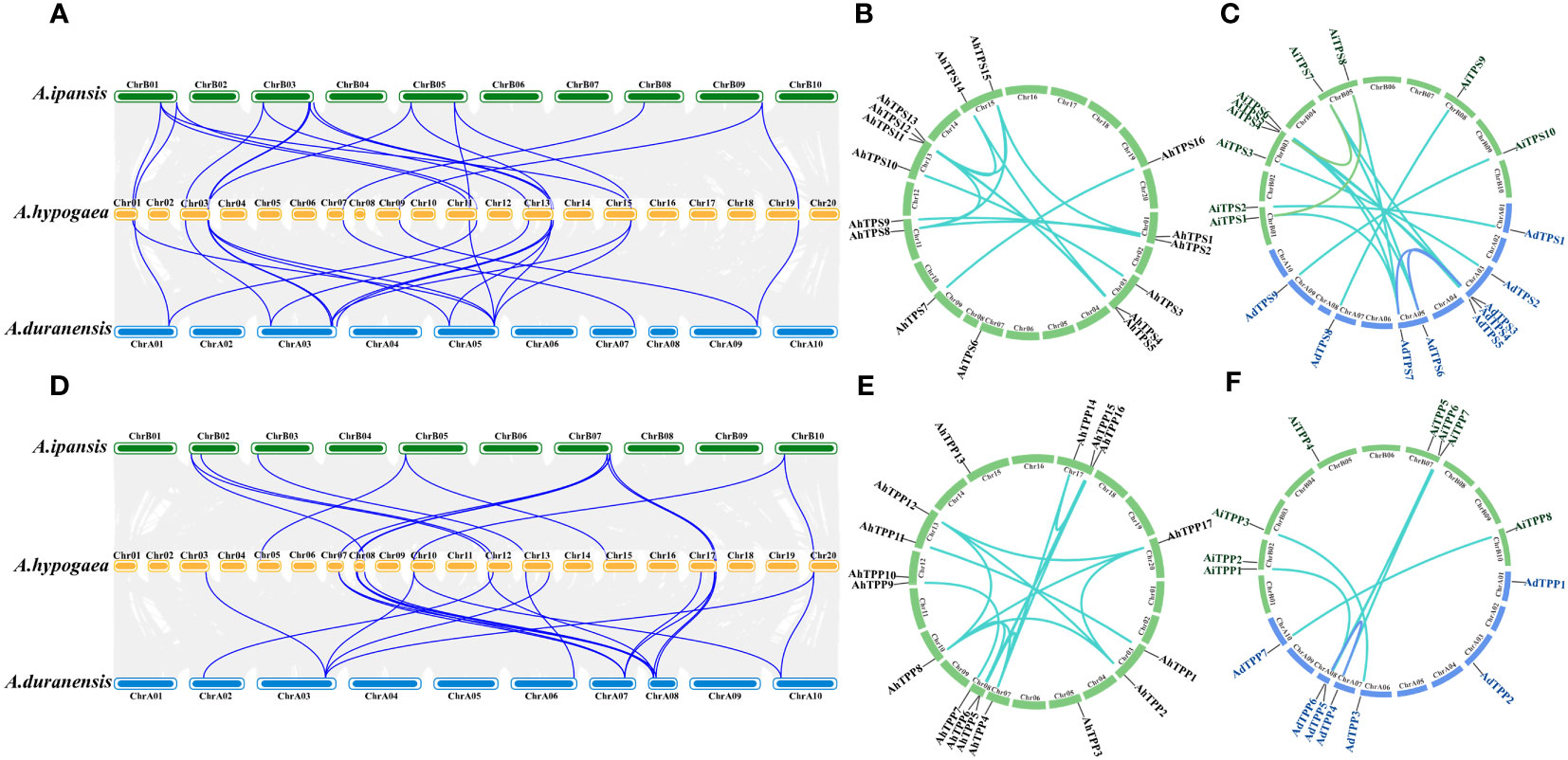
Figure 3 Collinearity analysis of the TPSs (A–C) and TPPs (D–F) in three Arachis species. (A) The syntenic relationship of the AhTPSs between A. ipaensis, A. hypogaea and A. duranensis genomes. (B) The collinearity relationship of AhTPSs within the cultivated peanut (A. hypogaea) genome. (C) The collinearity relationship of AhTPSs between wild diploild peanut A. ipaensis and A. duranensis genomes. (D) The syntenic relationship of the AhTPPSs between A. ipaensis, A. hypogaea and A. duranensis genomes. (E) The collinearity relationship of AhTPPs within the cultivated peanut (A. hypogaea) genome. (F) The collinearity relationship of AhTPPs between wild diploild peanut A. ipaensis and A. duranensis genomes.
3.4 Gene structure and motif composition of AhTPS and AhTPP genes
Gene structure analysis played a crucial role in elucidating the connection between gene family evolution and functional divergence. To unveil the inherent structural attributes of the AhTPS and AhTPP families, an in-depth investigation of gene structures and conserved motifs was conducted (Figure 4). Based on the phylogenetic analysis (Figure 1; Figures 4A,E), Both AhTPS and AhTPP members were divided into 2 groups (I and II), respectively, and a total of 20 conserved motifs are identified (Figures 4B,D). In the AhTPS gene family, Class I genes possess a greater number of introns (19 and 17) compared to Class II genes (4, 5, or 6), indicating a more complex gene structure (Figure 4C). Genes with close phylogenetic relationships often exhibit higher structural similarity. Among the Class I AhTPS genes, AhTPS3 and AhTPS10 have 6 introns each, while AhTPS2 and AhTPS9 have 4 introns each. Concerning the AhTPP gene family, Class I AhTPP genes, AhTPP5 and AhTPP15, have the highest number of introns at 14, whereas AhTPP7 possesses only 7 introns (Figure 4G). In Class I AhTPP genes, AhTPP11 has 14 introns, AhTPP8 and AhTPP17 have 11 introns, AhTPP2 and AhTPP12 contain 12 introns, AhTPP6 and AhTPP16 have 10 introns, and AhTPP8 and AhTPP17 possess 11 introns. Subsequently, we employed the online tool MEME to identify conserved motifs within AhTPS and AhTPP proteins. All AhTPS proteins contain motifs 1, 2, 3, 4, 5, 7, and 15. Except for AhTPS1, all AhTPS proteins contain motifs 6 and 13, and motifs 18 are present in all AhTPS proteins except AhTPS13. Most AhTPS proteins encompass motifs 8, 9, 10, 11, 12, 16, 17, 19, and 20. The fewest motifs (11) are found in AhTPS13. Generally, AhTPP proteins exhibit fewer motifs compared to AhTPS proteins, with AhTPP11 having the lowest count of motifs (13) (Figures 4F,H). All AhTPP proteins share motif 1, while motifs 13 and 20 are exclusively present in AhTPP3 and AhTPP13, motif 17 only in AhTPP4 and AhTPP14, and motif 18 only in AhTPP5 and AhTPP10. Identified conserved domains are listed according to their conservation levels within the AhTPS (Figure 4D) and AhTPP (Figure 4H) gene families. The results reveal that the most conserved motifs in AhTPS are generally located at the N-terminal, whereas the conserved motifs in the AhTPP family are predominantly situated at the C-terminal. This could be attributed to the positioning of TPS domains at the N-terminal and TPP domains at the C-terminal.
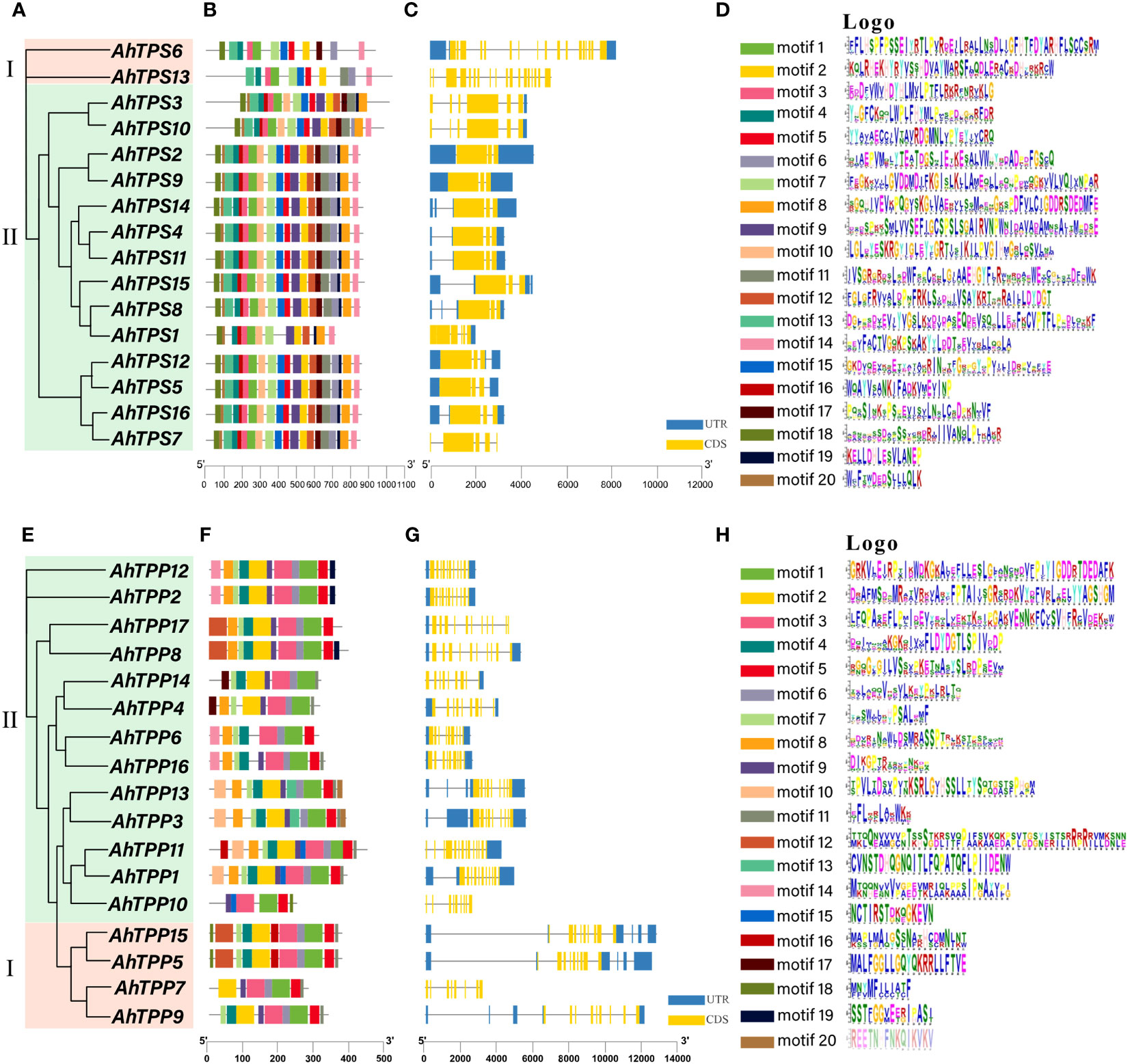
Figure 4 The phylogenetic trees, motifs, and gene intron/exon structures of 16 AhTPS genes and 17 AhTPP genes in cultivated peanut. (A, E) Phylogenetic tree based on AhTPS and AhTPP protein sequences using Neigbour-joining Tree method. (B, F) The motif compositions of AhTPS and AhTPP proteins. specific motifs were indicated using different colors. (C, G) The exon and intron distribution of AhTPS and AhTPP genes. Exons and intron regions are represented by yellow rectangles and grey lines, respectively. (D, H) Sequence logos of 20 motifs in cultivated AhTPS and AhTPP proteins, respectively.
3.5 Cis-element analysis in the upstream region of AhTPS and AhTPP genes
Cis-elements are necessary for gene expression and are widely involved in the regulation of plant growth and development and stress response. To further investigate the potential regulatory mechanisms of the AhTPS and AhTPP genes under stress conditions, we predicted and summarized the cis-regulatory elements within the first 2000 bp segments upstream of AhTPSs and AhTPPs, respectively (Supplementary Tables 7, 8). Cluster analysis of cis-regulatory elements in promoter regions reveals that orthologous genes from A and B subgenomes of cultivated peanut exhibit similar cis-element distributions, both in AhTPS and AhTPP genes (Figure 5). In the AhTPS genes, a total of 61 cis-regulatory elements were identified, including hormone-responsive elements such as abscisic acid (ABA), auxin (IAA), gibberellin (GA), salicylic acid (SA), and methyl jasmonate (MeJA) response elements, as well as stress-related elements like light response, anaerobic response, low temperature, wound, and MYB elements. Almost all AhTPS genes contain a significant number of light-responsive elements. However, considerable variation exists among different genes with respect to other hormones and stress responses. For instance, AhTPS2, AhTPS5, AhTPS9, and AhTPS12 contain numerous ABA-responsive elements, while AhTPS6 and AhTPS13 have a higher abundance of MeJA-responsive elements. In AhTPP genes, a total of 54 cis-regulatory elements were identified, including elements responsive to hormones such as ABA, MeJA, GA, IAA, SA, and stress-related elements like light, anaerobic conditions, and low temperature. Similarly, the most abundant category of cis-elements in AhTPP genes is related to light response. These findings indicate that both AhTPS and AhTPP genes are involved in distinct hormone regulatory pathways and responses to environmental stresses in cultivated peanut.
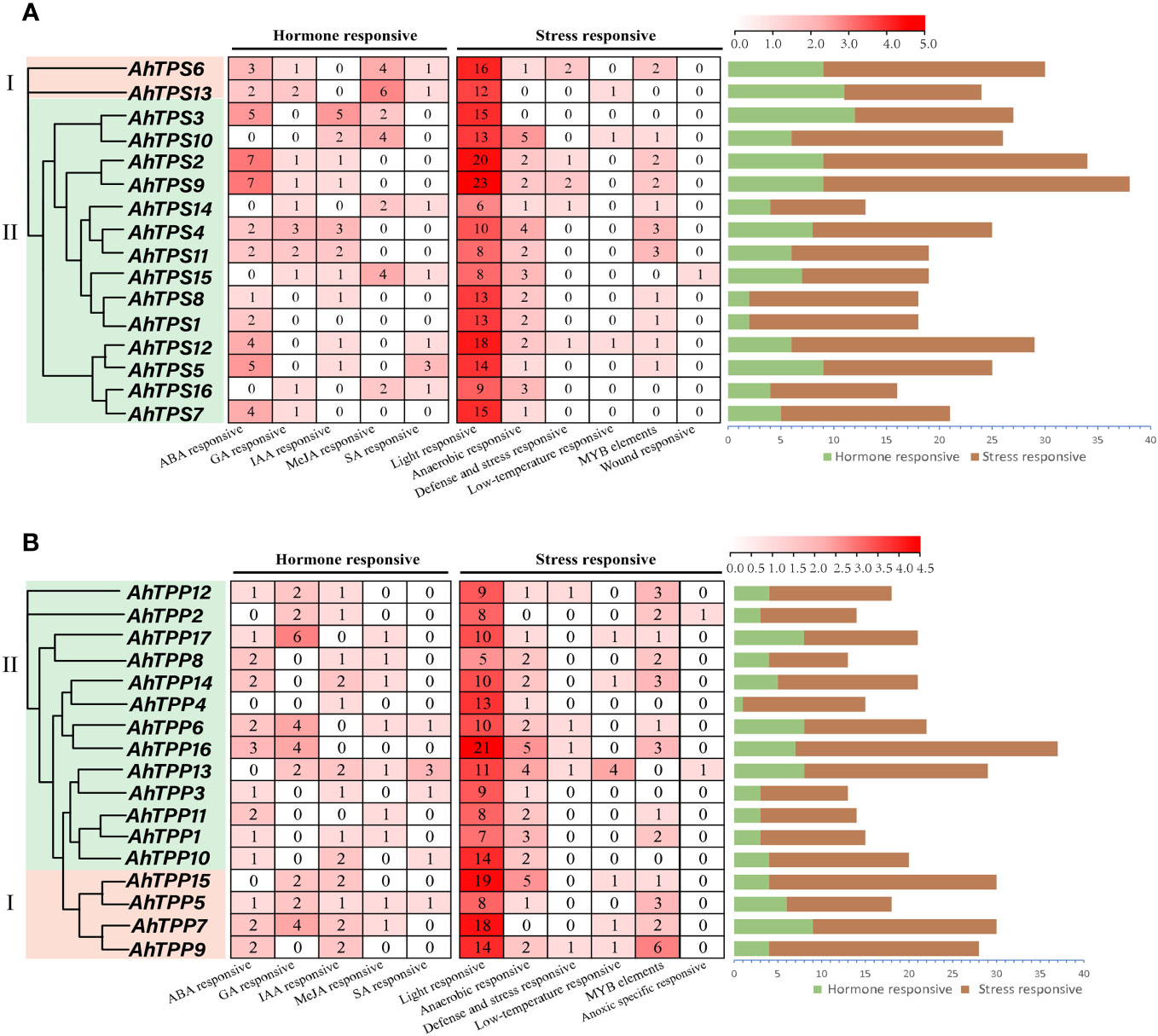
Figure 5 Cis-regulatory elements in the promoters of AhTPS (A) and AhTPP (B) genes in peanut. Various cis-regulatory elements are displayed in different colored boxes.
3.6 Expression analysis of AhTPS and AhTPPs in various tissues and under different environmental treatments
The specific expression patterns of genes can indicate their potential roles in growth and development. We utilized publicly available RNA-seq data related to peanut growth and development from the peanut genome database to investigate the expression profiles of 16 AhTPS and 17 AhTPP genes across various tissues (Clevenger et al., 2016; Zhuang et al., 2019). Furthermore, we conducted a detailed analysis of the expression pattern changes of AhTPSs and AhTPPs in leaves under different hormonal, low temperature, and drought treatments using a publicly available transcriptome dataset (Figure 6). The results suggested that there were differences in the expression levels of the AhTPS genes in many tissue types across multiple developmental stages.
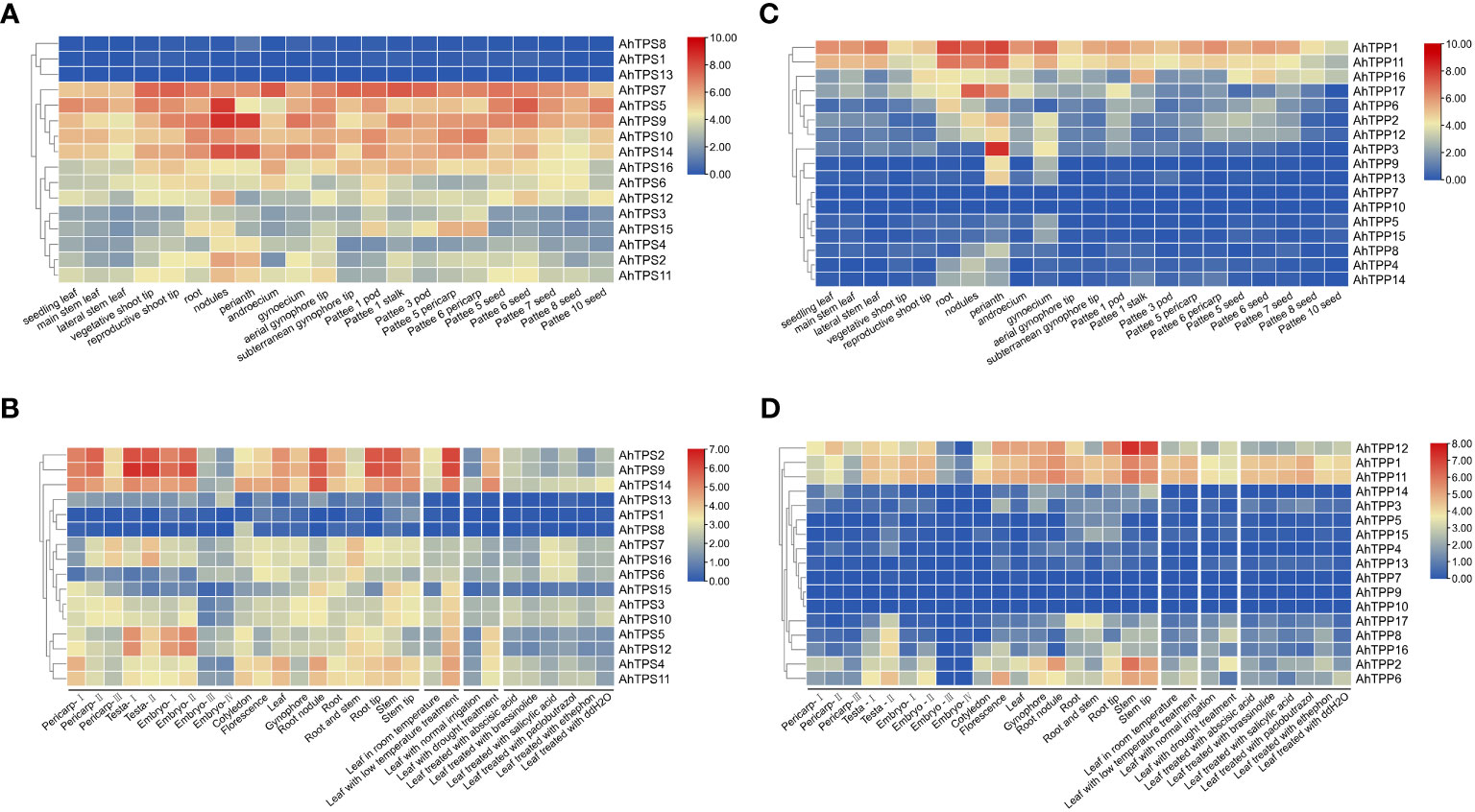
Figure 6 Heat map of AhTPS and AhTPP genes in different tissues, and under hormones and abiotic stresses. The color scale represents Log2 expression values (1+RPKM). (A) Expression profiles of AhTPSs in 22 different tissues (Clevenger et al., 2016). (B) Transcriptome expression of AhTPSs under different tissue, hormones and stress conditions (Zhuang et al., 2019). (C) Expression profiles of AhTPPs in 22 different tissues (Clevenger et al., 2016). (D) Transcriptome expression of AhTPPs under different tissues, hormones and stress conditions (Zhuang et al., 2019).
In both sets of transcriptomic data, the expression levels of AhTPS1, AhTPS8, and AhTPS13 are consistently low across various tissues and treatments, indicating a potential loss of gene function during evolution. Conversely, other AhTPS genes exhibit differential expression across different developmental stages and tissues. Certain genes, such as AhTPS9 and AhTPS14 (Figures 6A,B), maintain consistently high expression levels across nearly all developmental stages and tissues. Some genes, like AhTPS5 and AhTPS12, are predominantly expressed in seed coats and early embryos, while AhTPS2, AhTPS9, and AhTPS14 show significantly higher expression levels in roots and root nodules compared to other AhTPS genes. Moreover, the expression levels of almost all AhTPS genes decline during late embryonic development stages (Embryo-III and Embryo-IV) (Figure 6B). These findings suggest diverse functional roles of different TPS genes in various tissues and developmental stages. Under drought and low-temperature treatments, AhTPS genes exhibit a notable increase in expression, particularly AhTPS2, AhTPS9, and AhTPS14, suggesting their potential importance in peanut’s response to adverse conditions. Regarding hormone treatments, the expression levels of AhTPS6, AhTPS7, and AhTPS16 significantly increase in response to salicylic acid and paclobutrazol treatment, while other AhTPS genes do not show significant expression level changes in response to hormone treatments.
As for AhTPP genes, closely related homologs AhTPP1 and AhTPP11 share similar expression patterns and significantly higher expression levels across various tissues compared to other AhTPP genes (Figures 6C, D). On the other hand, AhTPP7, AhTPP9, AhTPP10, and AhTPP13 consistently exhibit low expression levels across both transcriptomic datasets. AhTPP gene expression also varies in different developmental stages and tissues. For instance, AhTPP3 show higher expression levels in floral organs (Figure 6C), while AhTPP12 and AhTPP17 display elevated expression levels in root tissues. Despite some changes in expression levels in response to stress and hormone treatments, AhTPP genes do not exhibit the same magnitude of expression differences as observed in AhTPS genes (Figure 6D).
3.7 Real-time expression of AhTPSs and AhTPPs under cold treatment
To further explore the expression patterns of the AhTPS and AhTPP genes under low-temperature conditions, we employed qPCR to assess the changes in expression levels of these genes at different time points in the leaves (Figure 7). Primers used for this experiment are included in Supplementary Table 9. Consistent with the expression patterns from transcriptomic data, no expression levels were detected for AhTPS1, AhTPS8, and AhTPS13 within the AhTPS gene family, as well as AhTPP4, AhTPP7, AhTPP9, and AhTPP10 within the AhTPP gene family at any time point under low-temperature treatment (Figure 7A). However, the remaining genes were all detectable. Within the AhTPS gene family, AhTPS2-5, AhTPS9-12, AhTPS14, and AhTPS15 exhibited varying degrees of upregulation in their expression levels under low-temperature treatment. Except for AhTPS3, which reached its highest expression level at 48 hours post-treatment, the other AhTPS genes reached their expression peaks at 12 or 24 hours, indicating a relatively similar expression pattern for these genes under low-temperature treatment. On the other hand, AhTPS6, AhTPS7, and AhTPS16 showed insignificant differences in expression levels under low-temperature treatment, suggesting that these genes might not respond to cold induction. Compared to the AhTPS genes, most of the AhTPP genes did not exhibit substantial differences in expression levels under low-temperature treatment. AhTPP1, AhTPP6, AhTPP11, and AhTPP13 showed elevated expression levels under low-temperature treatment, with the highest expression level observed at 48 hours post-treatment (Figure 7B). Among these, the most significant alteration in expression levels under low-temperature treatment was observed for AhTPP6. However, the expression levels of other genes did not display significant changes.
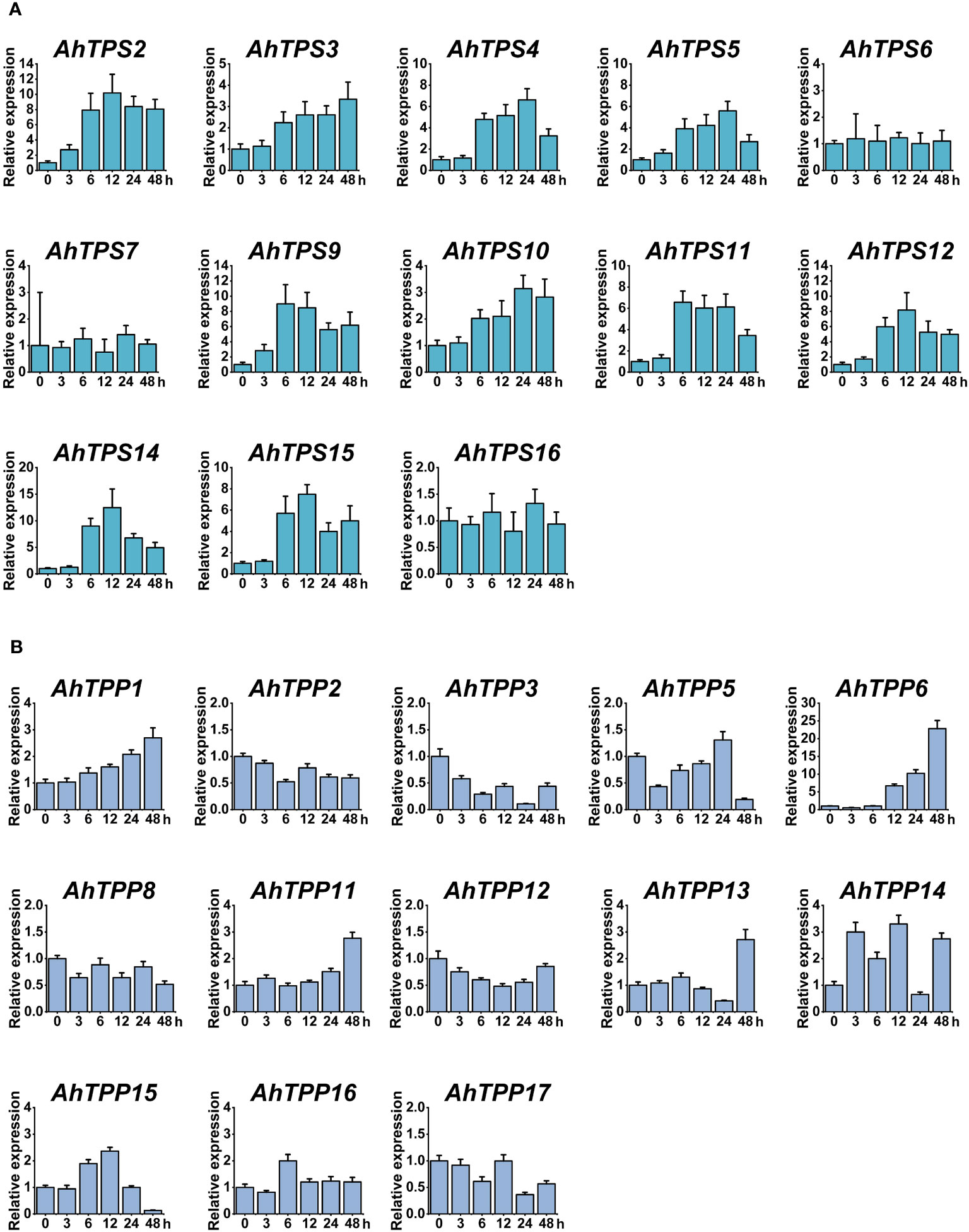
Figure 7 Expression patterns of AhTPS (A) and AhTPP (B) genes in leaf tissues under low-temperature stress. Data represents the mean ± standard deviation of three biological replicates each with three technical replicates. Relative transcript levels of the selected genes were calculated using the 2-ΔΔCt method, with Actin gene in cultivated peanut as the internal reference.
3.8 Subcellular localization of AhTPS9
In order to further elucidate the protein function and expression patterns of the AhTPS gene in peanut, and in conjunction with findings derived from transcriptomic analysis and qPCR validation, the AhTPS9, displaying the most pronounced differential expression under low-temperature conditions, was specifically chosen for subcellular localization analysis. The subcellular distribution of the AhTPS9 protein was investigated using the Agrobacterium-mediated transient expression technique, with green fluorescent protein (GFP) tagging, in the epidermal cells of tobacco leaves. Microscopic imaging of fluorescence revealed that, in leaves transformed with the control vector, GFP exhibited a uniform distribution across the epidermal cells (Figure 8). Furthermore, subsequent to the transient expression of AhTPS9-GFP fusion proteins in tobacco epidermal cells, the GFP signals were observed within both the cytoplasm and the nucleus. These results collectively indicate the dual localization of AhTPS9 within both the nucleus and cytoplasm of tobacco leaves.
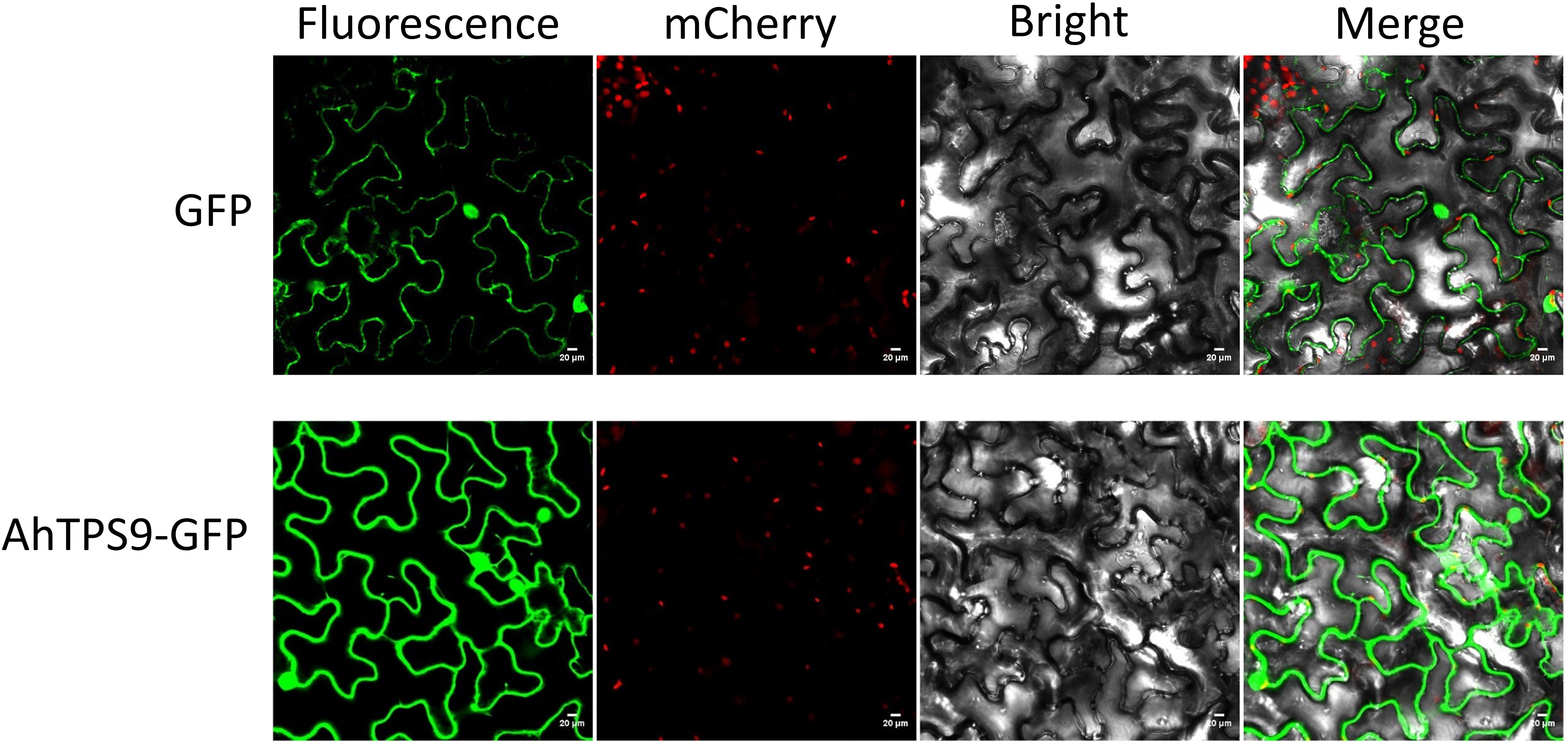
Figure 8 The subcellular localization of AhTPS9 in Nicotiana benthamiana leaf epidermal cells is depicted utilizing a confocal fluorescence microscope. Scale bars represent 20 μm.
3.9 Overexpression of AhTPS9 confers cold stress in transgenic Arabidopsis
We conducted an analysis of AhTPS9 function under cold stress conditions in transgenic Arabidopsis plants. Two transgenic Arabidopsis lines, AhTPS9-OE2 and AhTPS9-OE6, overexpressing AhTPS9, were generated through the construction of AhTPS9 overexpression vectors and genetic transformation of Arabidopsis. Since AhTPS9 is induced by low-temperature stress, both wild-type (WT) and various AhTPS9 overexpression (OE) seedlings were subjected to cold treatment. No observable phenotypic differences under normal conditions (22°C). However, under cold treatment, wild-type Arabidopsis plants exhibited a cold-sensitive phenotype compared to the two AhTPS9 overexpression lines (Figure 9). Physiological indicators indicated lower levels of proline and MDA in the AhTPS9 overexpression lines under low-temperature treatment, suggesting that AhTPS9 can mitigate cold-induced damage in plants.
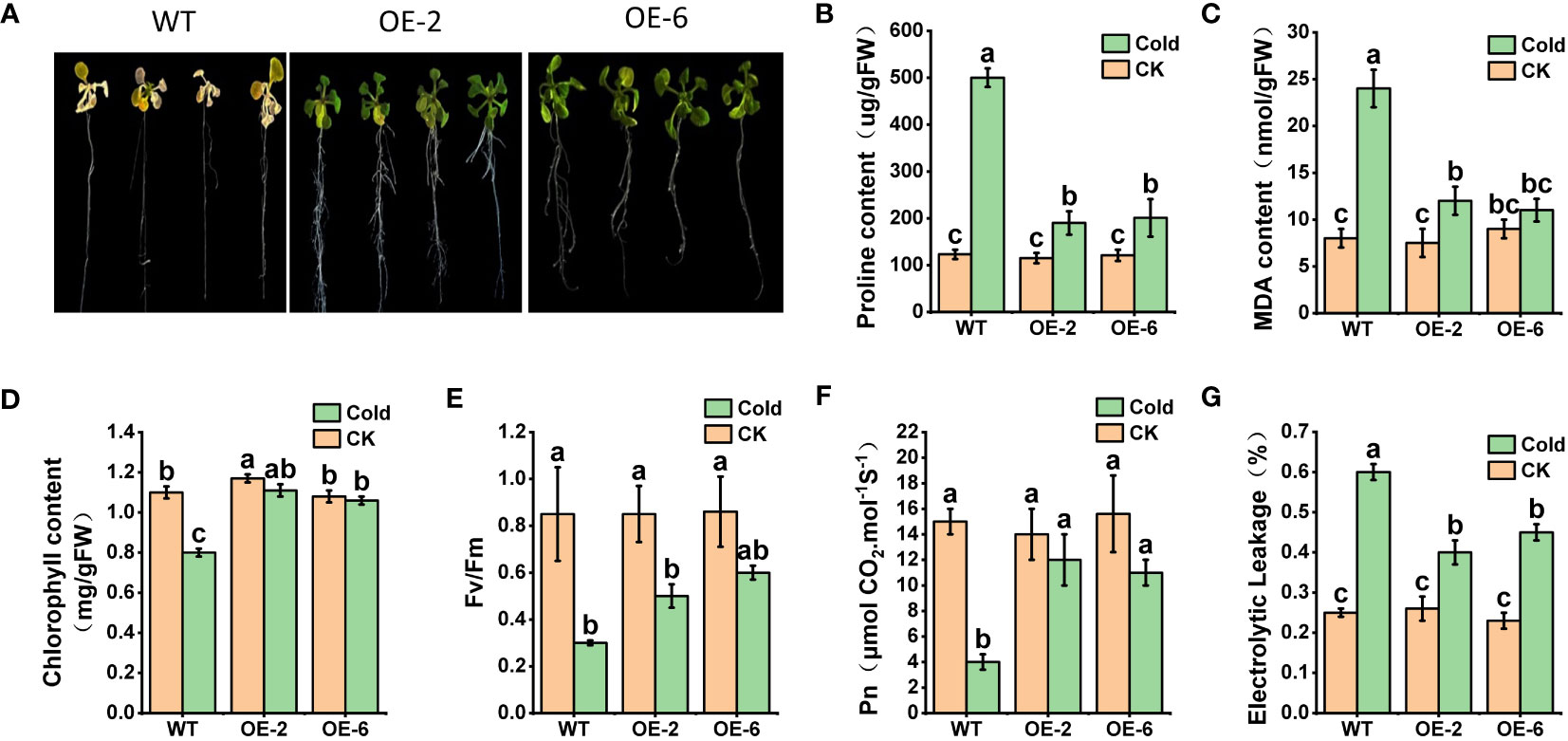
Figure 9 Phenotypes of Arabidopsis overexpressing AhTPS9, OE2 and OE6, along with the wild-type (WT) control, following a 72-hour cold treatment. Arabidopsis plants were grown on MS medium for 14 days and subsequently exposed to a cold environment at 4°C for 72 hours (A). The endogenous levels of stress-responsive parameters, including Proline (B) and MDA (C), were measured. Additionally, photosynthesis-related parameters under cold conditions were assessed, including chlorophyll content (D), Fv/Fm (E), net photosynthetic rate (Pn) (F), and electrical leakage (G). The data presented are expressed as the means ± standard errors (SEs) derived from three independent biological replicates. Statistically significant distinctions among the lines are denoted by different lowercase letters (a–c) based on Duncan’s test (p-value < 0.05).
The measurement of key indicators related to the photosynthetic system revealed that, under normal temperature conditions, there were no significant differences between wild-type Arabidopsis (WT) and the overexpressing plants (AhTPS9-OE2 and AhTPS9-OE6) in terms of Chlorophyll content, Fv/Fm (maximum quantum yield of photosystem II), Pn (photosynthesis rate), and Electrolytic Leakage. However, under low-temperature treatment, the wild-type plants exhibited a significant reduction in Chlorophyll content, Pn, and Fv/Fm, while showing an increase in electrolytic leakage compared to AhTPS9 overexpressing plants (AhTPS9-OE2 and AhTPS9-OE6). This indicates that the overexpression of AhTPS9 has the ability to alleviate the damage caused by low temperatures to the photosynthetic system in Arabidopsis thaliana. Further analysis of sugar metabolites and gene expression levels in the trehalose biosynthesis pathway revealed that, under low-temperature conditions, AhTPS9 expression was significantly higher in the overexpression lines than in the wild type. Additionally, downstream AtTPPI gene expression levels were also elevated in the AhTPS9-OE2 and AhTPS9-OE6 compared to the wild type (Figure 10). Moreover, the overexpression lines exhibited higher levels of Tre6p and trehalose but lower levels of sucrose compared to the wild type. In contrast, the overexpressing plants of AhTPS9 exhibited significantly higher levels of soluble sugars and starch content compared to the wild-type plants. The expression levels of key genes related to sucrose and Tre6P, such as AtSnRK1, were also examined. It was observed that, under normal temperature conditions, the expression of AtSnRK1 was higher in the WT plants than in AhTPS9 overexpressing plants. However, under cold stress conditions, the expression levels of AtSnRK1 significantly decreased. These findings suggest that AhTPS9 may regulate sugar metabolism pathways to alleviate the damage caused by low-temperature stress. Overall, our study demonstrates that AhTPS9 plays a crucial role in enhancing plant tolerance to cold stress, potentially by modulating sugar metabolism pathways.
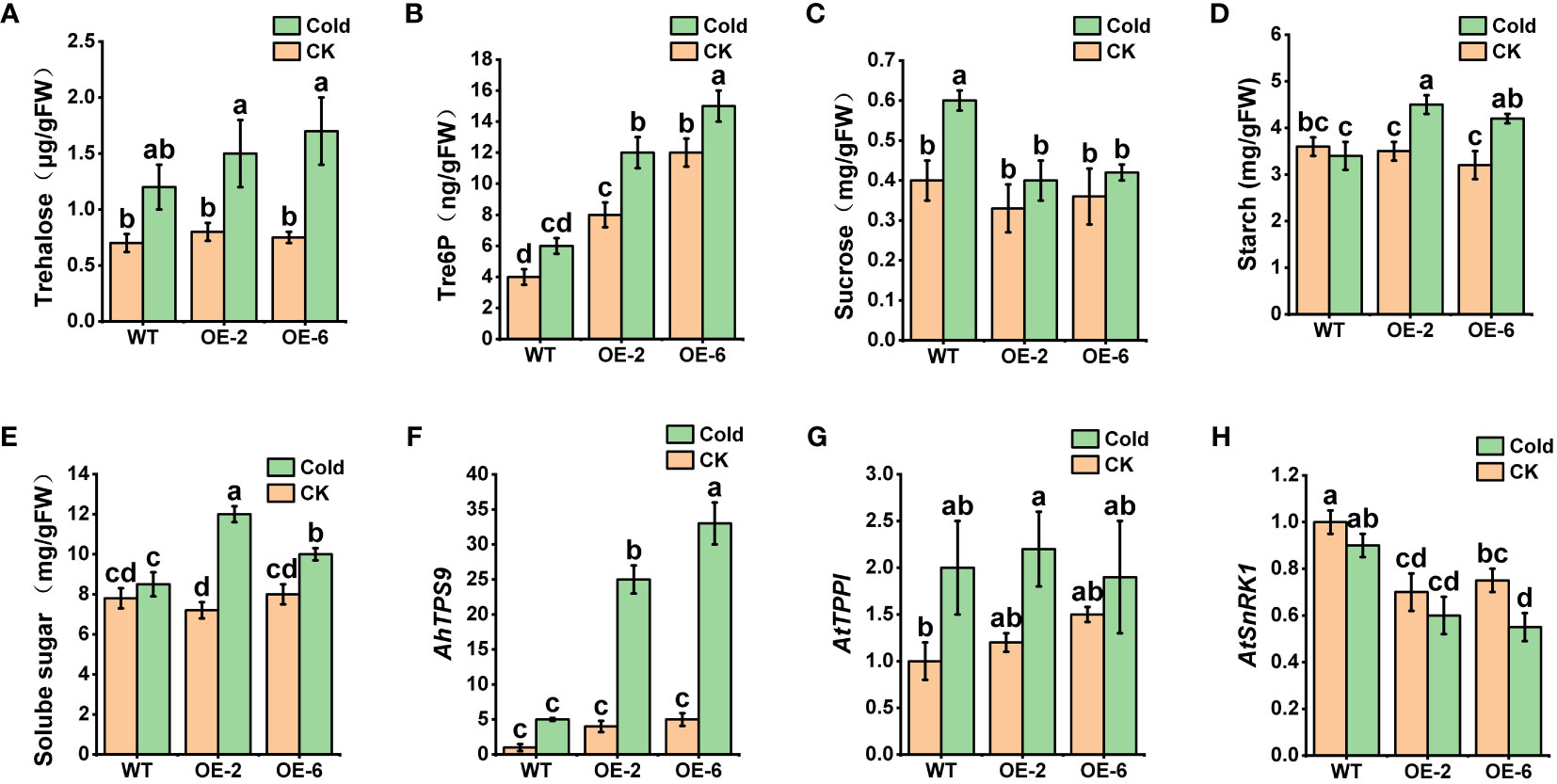
Figure 10 Changes in sugar metabolism and related genes in AhTPS9 overexpressing transgenic plants, including (A) Trehalose content, (B) Tre6P content, (C) sucrose content, (D) starch content, (E) soluble sugar content, as well as the relative expression levels of AhTPS9, AtTPPI, and AtSnRK1 (F–H). The data are presented as means ± standard errors (SEs) derived from three independent biological replicates. Statistically significant differences among the lines are indicated by different lowercase letters (a–c) based on Duncan’s test (p-value < 0.05).
4 Discussion
In response to low-temperature stress, plants rely on molecular mechanisms, with the trehalose biosynthesis pathway and its associated genes playing a crucial role. In plants, trehalose plays a role in regulating the response of plants to various environmental stresses (Paul et al., 2008). Recent research has suggested that the low levels of trehalose in plants may be involved in regulating their response to environmental stress in conjunction with its precursor, Tre6P (Dijken et al., 2004; Schluepmann et al., 2004; O’Hara et al., 2013). This implies that trehalose within plants may serve as an important signaling molecule in mediating the perception and regulation of both biotic and abiotic stresses. Therefore, investigating the trehalose signaling pathway in plants will contribute to the refinement of the plant sugar signaling network and uncovering the mechanisms by which sugars participate in regulating plant growth and development. Key genes in this context include TPS and TPP genes (Eastmond and Graham, 2003). These gene families regulate the synthesis and metabolism of trehalose, serving as vital components in the plant’s response to low temperatures (Kandror et al., 2002; Iordachescu and Imai, 2008). Numerous studies have been conducted on various plant species, such as Arabidopsis, rice, and common bean, revealing the expression and functions of TPS and TPP genes in cold stress responses (Fernandez et al., 2010; Liu et al., 2019; Lin et al., 2023; Liu et al., 2023). They contribute to the maintenance of membrane stability, protein protection, and ultimately enhance the plant’s cold resistance (Elbein et al., 2003). Therefore, in this study, we identified the TPS and TPP genes involved in the synthesis pathway of trehalose in peanuts and explored their roles in the response to low temperatures.
In this study, a total of 16 AhTPS and 17 AhTPP genes were identified in the cultivated peanut genome, while in the genome of wild diploid peanut A. duranensis, there were 9 AdTPS genes and 7 AdTPP genes, and in the genome of A. ipaensis, there were 10 AiTPS genes and 8 AiTPP genes (Figure 1, Table 1). Most of the TPS and TPP gene family members in wild diploid peanuts showed orthologous relationships with genes in the A and B subgenomes of cultivated peanuts, with differences in their numbers possibly arising from new duplication events between the two subgenomes of cultivated peanuts. Although cultivated peanuts have a larger number of AhTPS and AhTPP genes compared to other species such as Arabidopsis, rice, soybean, tomato, watermelon and cucumber (Vogel et al., 2001; Zang et al., 2011; Dan et al., 2021; Yuan et al., 2021; Lin et al., 2023), this may be attributed to the fact that cultivated peanuts, being tetraploids derived from two wild diploid peanuts, A. duranensis and A. ipaensis, have larger genomes (Bertioli et al., 2016; Bertioli et al., 2019; Zhuang et al., 2019). The AhTPS and AhTPP genes have been classified into two main groups based on other crops such as Arabidopsis, rice, and soybean (Figure 6). The AhTPS and AhTPP gene sequences are relatively conserved, with TPS or TPP orthologous genes within the Arachis species clustering together. However, the similarity between non-orthologous genes within the Arachis species is lower than the relationship between the Arachis species and the leguminous crop soybean. This implies that AhTPS genes and AhTPP genes exhibit a higher degree of conservation across species, while the similarity among family members is lower than that between species. Different AhTPS and AhTPP genes may undergo functional differentiation, and various AhTPS and AhTPP members may share similar functions with soybean. Gene duplication plays a crucial role in evolutionary processes, including chromosomal rearrangements, the diversification of gene functions, and the enlargement of gene families (Cannon et al., 2004; Huang and Rieseberg, 2020; Mérot et al., 2020). Hence, the duplication events of AhTPS and AhTPP gene members during the evolutionary process were identified, and the results indicate that both gene families, AhTPS and AhTPP, exhibit similarities in their evolutionary history. This is manifested by the absence of tandem duplications in both gene families, with evolution primarily relying on segment duplications, both in wild diploid peanuts and cultivated tetraploid peanuts (Figure 3). Therefore, the combination of phylogenetic and collinearity analysis results demonstrates that the AhTPS and AhTPP genes maintain a relatively conserved orthologous relationship among species, and the evolutionary process within the peanut species predominantly involves segment duplication.
The analysis of promoter cis-acting elements and gene expression patterns reveals that members of the AhTPS and AhTPP gene families play crucial roles in the growth, development, hormonal regulation, and stress responses of cultivated peanuts (Figures 5–7). The regulation pathways of trehalose in plant growth, development, and stress responses are intricate. Trehalose plays a vital role in safeguarding bioactive compounds and cellular components, such as proteins, nucleic acids, and biological membranes, from harsh environmental conditions like high salinity, drought, extreme temperatures, freezing, and oxidative stress (Nunes et al., 2013; O’Hara et al., 2013). Its most crucial role lies in modulating the synthesis and metabolism of carbohydrates in response to various biological processes and stressors (Oszvald et al., 2018; Hassan et al., 2023). In this study, both the AhTPS and AhTPP gene families were found to contain a significant number of light-responsive elements (Figure 5), suggesting their potential importance in photosynthesis and energy-related pathways. Combining data from various transcriptome databases and qPCR validation (Bertioli et al., 2019; Zhuang et al., 2019), some of these genes exhibited expression throughout the entire growth process of peanuts and participated in different stress and hormone regulatory mechanisms. Notably, the AhTPS family members, AhTPS2, AhTPS9, and AhTPS14, as well as the AhTPP family members, AhTPP1 and AhTPP11, were identified to be involved in such processes. Additionally, AhTPS2 and AhTPS9, along with AhTPP1 and AhTPP11, represented two pairs of orthologous genes originating from the A and B subgenomes, as depicted in Figure 2. These orthologous genes have been retained during the evolutionary process of wild diploid peanut species (Figure 1), indicating their potential significance within the Arachis genus.
To our knowledge, the manipulation of the Arachis hypogaea AhTPS genes in plants has not previously been reported. Combining bioinformatics and gene expression analysis, we observed that the expression levels of AhTPS2 and AhTPS9 were significantly higher than others (Figure 6, Figure 7). These genes exhibited similar gene structures and regulatory elements and showed similar expression levels in different tissues, hormonal treatments, and stress conditions. This suggests that they may have similar functions in the cultivated peanut genome, potentially indicating functional redundancy. To further explore the role of AhTPS genes under cold stress, AhTPS9 was selected for heterologous transformation into Arabidopsis plants to validate its function. Regulation of TPS genes has been shown to enhance abiotic stress tolerance in plants (Pilon-Smits et al., 1998; Garg et al., 2002; Chary et al., 2008; Fernandez et al., 2010; Li et al., 2011; Wang et al., 2016).For instance, constitutive expression of yeast ScTPS1 in potatoes improved drought tolerance but led to pleiotropic growth abnormalities, such as dwarfism, chlorotic leaves, and aberrant root development (Yeo et al., 2000). Transgenic tomato expressing ScTPS1 exhibited improved drought and salt tolerance but also showed other phenotypic changes (Cortina and Culiáñez-Macià, 2005). In tobacco, ScTPS1 expression increased drought tolerance without growth aberrations (Karim et al., 2007). Similarly, overexpression of OsTPS1 in rice improved tolerance to cold, salinity, and drought without visible phenotypic changes (Li et al., 2011). Overexpression of Triticum aestivum TaTPS11 in Arabidopsis enhanced cold tolerance without adverse phenotypes, suggesting its potential value in wheat cold-tolerance breeding (Liu et al., 2019). In this study, in Arabidopsis plants overexpressing AhTPS9, compared to wild-type plants, no significant phenotypic changes were observed during germination and growth processes. However, under cold stress, the AhTPS9-overexpressing plants exhibited improved tolerance, showing reduced levels of proline and MDA, and without significant wilting. This suggests that AhTPS9, similar to OsTPS1 and TaTPS11, can alleviate stress without affecting key phenotypic changes. AhTPS9 can mitigate the damage to the photosynthetic system caused by low temperatures, including chlorophyll content, Pn, and Fv/Fm, which were significantly higher in the overexpressing plants compared to the wild type. TPS genes and their product, Tre6P, have been reported to enhance photosynthesis under stress conditions (Paul et al., 2001). In maize, Tre6P has been shown to regulate photosynthesis and assimilate distribution in reproductive tissues (Oszvald et al., 2018). In tomatoes, co-expression of TPS and TPP enhances photosynthesis under drought and salt stress without affecting plant growth (Lyu et al., 2013). AhTPS9 contains abundant photosynthesis-related elements (Figure 5) and is regulated by multiple hormones (Figure 6). Therefore, the specific regulatory mechanism of AhTPS9 in protecting photosynthesis under low-temperature conditions requires further research.
Sugar metabolism plays a crucial role in plant responses to low-temperature stress (Nägele and Heyer, 2013; Kovi et al., 2016; Hassan et al., 2023). As a soluble sugar, trehalose is present at extremely low levels, making it unable to provide the necessary osmotic stress protection independently. However, the intermediate product Tre6P plays a critical role in sucrose regulation. Tre6P acts as both a signal for sucrose levels and a negative feedback regulatory factor, contributing to the maintenance of sucrose levels within an appropriate range (Figueroa and Lunn, 2016). Tre6P can interact with Sucrose Non-Fermenting 1-Related Kinase 1 (SnRK1) to regulate sucrose levels antagonistically (Baena-Gonzalez and Lunn, 2020). In this study, we also examined key genes related to trehalose metabolism and their metabolic products in transgenic Arabidopsis and its wild type. The results showed that AhTPS9-overexpressing plants exhibited significantly higher levels under low-temperature conditions, leading to the accumulation of Tre6P and a decrease in sucrose levels, which is consistent with the model of Tre6P-SnRK1 regulation of sucrose levels proposed by previous studies (Baena-Gonzalez and Lunn, 2020). AtTPP1 can enhance Arabidopsis cold tolerance by accumulating soluble sugars and jasmonic acid. In this study, the expression level of this gene increased under low-temperature treatment, but it was not affected by AhTPS9. However, it resulted in higher levels of trehalose, suggesting that the upregulation of this gene’s expression may be regulated by CBF genes (Lin et al., 2023). Overexpression of AtTPS11 in Arabidopsis under low-temperature conditions significantly increased starch content (Singh et al., 2011). As an orthologous gene (Figure 1), AhTPS9 similarly increased starch content in overexpressing plants under low-temperature conditions. However, soluble sugar accumulation in overexpressing plants was significantly higher than in the wild type under low-temperature conditions. Therefore, there are complex interactions between sugar metabolism-related genes in the trehalose pathway and cold tolerance, which require further detailed investigation.
5 Conclusion
Collectively, the current study identified 16 AhTPS and 17 AhTPP genes in the peanut genome through bioinformatics analysis. Phylogenetic analysis revealed two distinct subgroups closely related to wild diploid peanuts. Evolutionary patterns suggested gene segmental duplication events and robust purifying selection. Based on expression pattern analysis, AhTPS9 exhibits the most significant differential expression under cold stress. Functional validation revealed that Arabidopsis plants overexpressing AhTPS9 demonstrate enhanced cold tolerance by improving the photosynthetic system and regulating related products and genes involved in sugar metabolism.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Author contributions
CZ: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Writing – original draft, Writing – review & editing. ZH: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Writing – original draft, Writing – review & editing. YL: Data curation, Formal analysis, Investigation, Writing – review & editing. ZL: Data curation, Formal analysis, Investigation, Writing – review & editing. XW: Data curation, Formal analysis, Project administration, Supervision, Validation, Visualization, Writing – review & editing. CJ: Data curation, Formal analysis, Project administration, Supervision, Validation, Visualization, Writing – review & editing. SK: Data curation, Formal analysis, Project administration, Supervision, Validation, Visualization, Writing – review & editing. XL: Data curation, Formal analysis, Project administration, Supervision, Validation, Visualization, Writing – review & editing. SZ: Data curation, Formal analysis, Project administration, Supervision, Validation, Visualization, Writing – review & editing. JW: Data curation, Formal analysis, Project administration, Supervision, Validation, Visualization, Writing – review & editing. HZ: Data curation, Formal analysis, Project administration, Supervision, Validation, Visualization, Writing – review & editing. XZ: Data curation, Formal analysis, Project administration, Supervision, Validation, Visualization, Writing – review & editing. HY: Funding acquisition, Resources, Supervision, Validation, Visualization, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study was supported by the earmarked fund for CARS-13, Liaoning Provincial Natural Science Foundation (2023-MS-214), Liaoning Revitalization Talents Program (XLYC1902002), and Science and Technology Program of Shenyang (No. 21–110-3–17), (No. 23-410-2-08).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1343402/full#supplementary-material
References
Aubourg, S., Lecharny, A., Bohlmann, J. (2002). Genomic analysis of the terpenoid synthase (AtTPS) gene family of Arabidopsis thaliana. Mol. Genet. Genomics 267, 730–745. doi: 10.1007/s00438-002-0709-y
Baena-Gonzalez, E., Lunn, J. E. (2020). SnRK1 and trehalose 6-phosphate–two ancient pathways converge to regulate plant metabolism and growth. Curr. Opin. Plant Biol. 55, 52–59. doi: 10.1016/j.pbi.2020.01.010
Bajji, M., Kinet, J.-M., Lutts, S. (2002). The use of the electrolyte leakage method for assessing cell membrane stability as a water stress tolerance test in durum wheat. Plant Growth Regul. 36, 61–70. doi: 10.1023/A:1014732714549
Bertioli, D. J., Cannon, S. B., Froenicke, L., Huang, G., Farmer, A. D., Cannon, E. K., et al. (2016). The genome sequences of Arachis duranensis and Arachis ipaensis, the diploid ancestors of cultivated peanut. Nat. Genet. 48 (4), 438–446. doi: 10.1038/ng.3517
Bertioli, D. J., Jenkins, J., Clevenger, J., Dudchenko, O., Gao, D., Seijo, G., et al. (2019). The genome sequence of segmental allotetraploid peanut Arachis hypogaea. Nat. Genet. 51 (5), 877–884. doi: 10.1038/s41588-019-0405-z
Bhat, K. A., Mahajan, R., Pakhtoon, M. M., Urwat, U., Bashir, Z., Shah, A. A., et al. (2022). Low temperature stress tolerance: An insight into the omics approaches for legume crops. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.888710
Cannon, S. B., Mitra, A., Baumgarten, A., Young, N. D., May, G. (2004). The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 4 (1), 1–21. doi: 10.1186/1471-2229-4-10
Chary, S. N., Hicks, G. R., Choi, Y. G., Carter, D., Raikhel, N. V. (2008). Trehalose-6-phosphate synthase/phosphatase regulates cell shape and plant architecture in Arabidopsis. Plant Physiol. 146 (1), 97–107. doi: 10.1104/pp.107.107441
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13 (8), 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chen, N., Yang, Q., Hu, D., Pan, L., Chi, X., Chen, M., et al. (2014). Gene expression profiling and identification of resistance genes to low temperature in leaves of peanut (Arachis hypogaea L.). Sci. Hortic. 169, 214–225. doi: 10.1016/j.scienta.2014.01.043
Chen, X., Lu, Q., Liu, H., Zhang, J., Hong, Y., Lan, H., et al. (2019). Sequencing of cultivated peanut, Arachis hypogaea, yields insights into genome evolution and oil improvement. Mol. Plant 12 (7), 920–934. doi: 10.1016/j.molp.2019.03.005
Clevenger, J., Chu, Y., Scheffler, B., Ozias-Akins, P. (2016). A developmental transcriptome map for allotetraploid Arachis hypogaea. Front. Plant Sci. 7. doi: 10.3389/fpls.2016.01446
Clough, S. J., Bent, A. F. (1998). Floral dip: a simplified method for Agrobacterium‐mediated transformation of Arabidopsis thaliana. The plant journal, 16 (6), 753–743. doi: 10.1046/j.1365-313x.1998.00343.x
Cortina, C., Culiáñez-Macià, F. A. (2005). Tomato abiotic stress enhanced tolerance by trehalose biosynthesis. Plant Sci. 169 (1), 75–82. doi: 10.1016/j.plantsci.2005.02.026
Dan, Y., Niu, Y., Wang, C., Yan, M., Liao, W. (2021). Genome-wide identification and expression analysis of the trehalose-6-phosphate synthase (TPS) gene family in cucumber (Cucumis sativus L.). PeerJ 9, e11398. doi: 10.7717/peerj.11398
Dijken, A., Schluepmann, H., Smeekens, S. C. (2004). Arabidopsis trehalose-6-phosphate synthase 1 is essential for normal vegetative growth and transition to flowering. Plant Physiol. 135 (2), 969–977. doi: 10.1104/pp.104.039743
Du, L., Li, S., Ding, L., Cheng, X., Kang, Z., Mao, H. (2022). Genome-wide analysis of trehalose-6-phosphate phosphatases (TPP) gene family in wheat indicates their roles in plant development and stress response. BMC Plant Biol. 22 (1), 120. doi: 10.1186/s12870-022-03504-0
Eastmond, P. J., Graham, I. A. (2003). Trehalose metabolism: a regulatory role for trehalose-6-phosphate? Curr. Opin. Plant Biol. 6 (3), 231–235. doi: 10.1016/S1369-5266(03)00037-2
Elbein, A. D., Pan, Y., Pastuszak, I., Carroll, D. (2003). New insights on trehalose: a multifunctional molecule. Glycobiology 13 (4), 17R–27R. doi: 10.1093/glycob/cwg047
Emanuelle, S., Doblin, M. S., Stapleton, D. I., Bacic, A., Gooley, P. R. (2016). Molecular insights into the enigmatic metabolic regulator, SnRK1. Trends Plant Sci. 21 (4), 341–353. doi: 10.1016/j.tplants.2015.11.001
Fernandez, O., Béthencourt, L., Quero, A., Sangwan, R. S., Clément, C. (2010). Trehalose and plant stress responses: friend or foe? Trends Plant Sci. 15 (7), 409–417. doi: 10.1016/j.tplants.2010.04.004
Fichtner, F., Lunn, J. E. (2021). The role of trehalose 6-phosphate (Tre6P) in plant metabolism and development. Annu. Rev. Plant Biol. 72, 737–760. doi: 10.1146/annurev-arplant-050718-095929
Fichtner, F., Olas, J. J., Feil, R., Watanabe, M., Krause, U., Hoefgen, R., et al. (2020). Functional features of TREHALOSE-6-PHOSPHATE SYNTHASE1, an essential enzyme in Arabidopsis. Plant Cell 32 (6), 1949–1972. doi: 10.1105/tpc.19.00837
Figueroa, C. M., Lunn, J. E. (2016). A tale of two sugars: trehalose 6-phosphate and sucrose. Plant Physiol. 172 (1), 7–27. doi: 10.1104/pp.16.00417
Garg, A. K., Kim, J.-K., Owens, T. G., Ranwala, A. P., Choi, Y. D., Kochian, L. V., et al. (2002). Trehalose accumulation in rice plants confers high tolerance levels to different abiotic stresses. Proc. Natl. Acad. Sci. 99 (25), 15898–15903. doi: 10.1073/pnas.252637799
Ge, L. F., Chao, D. Y., Shi, M., Zhu, M. Z., Gao, J.-P., Lin, H. X. (2008). Overexpression of the trehalose-6-phosphate phosphatase gene OsTPP1 confers stress tolerance in rice and results in the activation of stress responsive genes. Planta 228, 191–201. doi: 10.1007/s00425-008-0729-x
Hassan, M. U., Nawaz, M., Shah, A. N., Raza, A., Barbanti, L., Skalicky, M., et al. (2023). Trehalose: a key player in plant growth regulation and tolerance to abiotic stresses. J. Plant Growth Regul. 42 (8), 4935–4957. doi: 10.1007/s00344-022-10851-7
Huang, K., Rieseberg, L. H. (2020). Frequency, origins, and evolutionary role of chromosomal inversions in plants. Front. Plant Sci. 11. doi: 10.3389/fpls.2020.00296
Iordachescu, M., Imai, R. (2008). Trehalose biosynthesis in response to abiotic stresses. J. Integr. Plant Biol. 50 (10), 1223–1229. doi: 10.1111/j.1744-7909.2008.00736.x
Kakani, V., Prasad, P., Craufurd, P., Wheeler, T. (2002). Response of in vitro pollen germination and pollen tube growth of groundnut (Arachis hypogaea L.) genotypes to temperature. Plant. Cell Environ. 25 (12), 1651–1661. doi: 10.1046/j.1365-3040.2002.00943.x
Kandror, O., DeLeon, A., Goldberg, A. L. (2002). Trehalose synthesis is induced upon exposure of Escherichia coli to cold and is essential for viability at low temperatures. Proc. Natl. Acad. Sci. 99 (15), 9727–9732. doi: 10.1073/pnas.142314099
Karim, S., Aronsson, H., Ericson, H., Pirhonen, M., Leyman, B., Welin, B., et al. (2007). Improved drought tolerance without undesired side effects in transgenic plants producing trehalose. Plant Mol. Biol. 64, 371–386. doi: 10.1007/s11103-007-9159-6
Kaul, S., Koo, H. L., Jenkins, J., Rizzo, M., Rooney, T. (2000). Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. nature 408 (6814), 796–815. doi: 10.1038/35048692
Kidokoro, S., Shinozaki, K., Yamaguchi-Shinozaki, K. (2022). Transcriptional regulatory network of plant cold-stress responses. Trends Plant Sci 27, 922–935. doi: 10.1016/j.tplants.2022.01.008
Kosar, F., Akram, N. A., Sadiq, M., Al-Qurainy, F., Ashraf, M. (2019). Trehalose: a key organic osmolyte effectively involved in plant abiotic stress tolerance. J. Plant Growth Regul. 38, 606–618. doi: 10.1007/s00344-018-9876-x
Kovi, M. R., Ergon, Å., Rognli, O. A. (2016). Freezing tolerance revisited—effects of variable temperatures on gene regulation in temperate grasses and legumes. Curr. Opin. Plant Biol. 33, 140–146. doi: 10.1016/j.pbi.2016.07.006
Krasensky, J., Broyart, C., Rabanal, F. A., Jonak, C. (2014). The redox-sensitive chloroplast trehalose-6-phosphate phosphatase AtTPPD regulates salt stress tolerance. Antioxid. Redox Signaling 21 (9), 1289–1304. doi: 10.1089/ars.2013.5693
Krishna, G., Singh, B. K., Kim, E. K., Morya, V. K., Ramteke, P. W. (2015). Progress in genetic engineering of peanut (Arachis hypogaea L.)—A review. Plant Biotechnol. J. 13 (2), 147–162. doi: 10.1111/pbi.12339
Leyman, B., Van Dijck, P., Thevelein, J. M. (2001). An unexpected plethora of trehalose biosynthesis genes in Arabidopsis thaliana. Trends Plant Sci. 6 (11), 510–513. doi: 10.1016/S1360-1385(01)02125-2
Li, P., Ma, S., Bohnert, H. J. (2008). Coexpression characteristics of trehalose-6-phosphate phosphatase subfamily genes reveal different functions in a network context. Physiol. Plant. 133 (3), 544–556. doi: 10.1111/j.1399-3054.2008.01101.x
Li, H. W., Zang, B. S., Deng, X. W., Wang, X. P. (2011). Overexpression of the trehalose-6-phosphate synthase gene OsTPS1 enhances abiotic stress tolerance in rice. Planta 234, 1007–1018. doi: 10.1007/s00425-011-1458-0
Lin, Q., Wang, J., Gong, J., Zhang, Z., Wang, S., Sun, J., et al. (2023). The Arabidopsis thaliana trehalose-6-phosphate phosphatase gene AtTPPI improve chilling tolerance through accumulating soluble sugar and JA. Environ. Exp. Bot. 205, 105117. doi: 10.1016/j.envexpbot.2022.105117
Lin, Q., Yang, J., Wang, Q., Zhu, H., Chen, Z., Dao, Y., et al. (2019). Overexpression of the trehalose-6-phosphate phosphatase family gene AtTPPF improves the drought tolerance of Arabidopsis thaliana. BMC Plant Biol. 19 (1), 1–15. doi: 10.1186/s12870-019-1986-5
Liu, X., Fu, L., Qin, P., Sun, Y., Liu, J., Wang, X. (2019). Overexpression of the wheat trehalose 6-phosphate synthase 11 gene enhances cold tolerance in Arabidopsis thaliana. Gene 710, 210–217. doi: 10.1016/j.gene.2019.06.006
Liu, W., Zhang, H.-H., Long, Z.-X., Chi, X.-N., Wang, Y.-P. (2023). Identification, evolutionary relationship analysis of the trehalose-6-phosphate synthase (TPS) gene family in common bean (Phaseolus vulgaris) and their expression in response to cold stress. J. Plant Growth Regul. 1-18. doi: 10.1007/s00344-023-11087-9
Lyu, J. I., Min, S. R., Lee, J. H., Lim, Y. H., Kim, J.-K., Bae, C.-H., et al. (2013). Overexpression of a trehalose-6-phosphate synthase/phosphatase fusion gene enhances tolerance and photosynthesis during drought and salt stress without growth aberrations in tomato. Plant Cell. Tissue Organ Cult. (PCTOC). 112, 257–262. doi: 10.1007/s11240-012-0225-7
Ma, S., Gong, Q., Bohnert, H. J. (2007). An Arabidopsis gene network based on the graphical Gaussian model. Genome Res. 17 (11), 1614–1625. doi: 10.1101/gr.6911207
Mérot, C., Oomen, R. A., Tigano, A., Wellenreuther, M. (2020). A roadmap for understanding the evolutionary significance of structural genomic variation. Trends Ecol. Evol. 35 (7), 561–572. doi: 10.1016/j.tree.2020.03.002
Nägele, T., Heyer, A. G. (2013). Approximating subcellular organisation of carbohydrate metabolism during cold acclimation in different natural accessions of Arabidopsis thaliana. New Phytol. 198 (3), 777–787. doi: 10.1111/nph.12201
Nunes, C., Schluepmann, H., Delatte, T. L., Wingler, A., Silva, A. B., Fevereiro, P. S., et al. (2013). Regulation of growth by the trehalose pathway: relationship to temperature and sucrose. Plant Signaling Behav. 8 (12), e26626. doi: 10.4161/psb.26626
O’Hara, L. E., Paul, M. J., Wingler, A. (2013). How do sugars regulate plant growth and development? New insight into the role of trehalose-6-phosphate. Mol. Plant 6 (2), 261–274. doi: 10.1093/mp/sss120
Oszvald, M., Primavesi, L. F., Griffiths, C. A., Cohn, J., Basu, S. S., Nuccio, M. L., et al. (2018). Trehalose 6-phosphate regulates photosynthesis and assimilate partitioning in reproductive tissue. Plant Physiol. 176 (4), 2623–2638. doi: 10.1104/pp.17.01673
Paul, M. J., Gonzalez-Uriarte, A., Griffiths, C. A., Hassani-Pak, K. (2018). The role of trehalose 6-phosphate in crop yield and resilience. Plant Physiol. 177 (1), 12–23. doi: 10.1104/pp.17.01634
Paul, M., Pellny, T., Goddijn, O. (2001). Enhancing photosynthesis with sugar signals. Trends Plant Sci. 6 (5), 197–200. doi: 10.1016/S1360-1385(01)01920-3
Paul, M. J., Primavesi, L. F., Jhurreea, D., Zhang, Y. (2008). Trehalose metabolism and signaling. Annu. Rev. Plant Biol. 59, 417–441. doi: 10.1146/annurev.arplant.59.032607.092945
Pilon-Smits, E. A., Terry, N., Sears, T., Kim, H., Zayed, A., Hwang, S., et al. (1998). Trehalose-producing transgenic tobacco plants show improved growth performance under drought stress. J. Plant Physiol. 152 (4-5), 525–532. doi: 10.1016/S0176-1617(98)80273-3
Puppala, N., Nayak, S. N., Sanz-Saez, A., Chen, C., Devi, M. J., Nivedita, N., et al. (2023). Sustaining yield and nutritional quality of peanuts in harsh environments: Physiological and molecular basis of drought and heat stress tolerance. Front. Genet. 14. doi: 10.3389/fgene.2023.1121462
Raza, A., Charagh, S., Najafi-Kakavand, S., Abbas, S., Shoaib, Y., Anwar, S., et al. (2023). Role of phytohormones in regulating cold stress tolerance: physiological and molecular approaches for developing cold-smart crop plants. Plant Stress 100152. doi: 10.1016/j.stress.2023.100152
Schluepmann, H., Van Dijken, A., Aghdasi, M., Wobbes, B., Paul, M., Smeekens, S. (2004). Trehalose mediated growth inhibition of Arabidopsis seedlings is due to trehalose-6-phosphate accumulation. Plant Physiol. 135 (2), 879–890. doi: 10.1104/pp.104.039503
Shi, G., Cai, Q. (2009). Leaf plasticity in peanut (Arachis hypogaea L.) in response to heavy metal stress. Environ. Exp. Bot. 67 (1), 112–117. doi: 10.1016/j.envexpbot.2009.02.009
Singh, V., Louis, J., Ayre, B. G., Reese, J. C., Shah, J. (2011). TREHALOSE PHOSPHATE SYNTHASE11-dependent trehalose metabolism promotes Arabidopsis thaliana defense against the phloem-feeding insect Myzus persicae. Plant J. 67 (1), 94–104. doi: 10.1111/j.1365-313X.2011.04583.x
Smeekens, S. (2015). From leaf to kernel: trehalose-6-phosphate signaling moves carbon in the field. Plant Physiol. 169 (2), 912–913. doi: 10.1104/pp.15.01177
Tamura, K., Stecher, G., Kumar, S. (2021). MEGA11: molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 38 (7), 3022–3027. doi: 10.1093/molbev/msab120
Vandesteene, L., López-Galvis, L., Vanneste, K., Feil, R., Maere, S., Lammens, W., et al. (2012). Expansive evolution of the trehalose-6-phosphate phosphatase gene family in. Arabidopsis. Plant Physiol. 160 (2), 884–896. doi: 10.1104/pp.112.201400
Van Houtte, H., López-Galvis, L., Vandesteene, L., Beeckman, T., Van Dijck, P. (2013). Redundant and non-redundant roles of the trehalose-6-phosphate phosphatases in leaf growth, root hair specification and energy-responses in Arabidopsis. Plant Signaling Behav. 8 (3), e23209. doi: 10.4161/psb.23209
Vishal, B., Krishnamurthy, P., Ramamoorthy, R., Kumar, P. P. (2019). Os TPS 8 controls yield-related traits and confers salt stress tolerance in rice by enhancing suberin deposition. New Phytol. 221 (3), 1369–1386. doi: 10.1111/nph.15464
Vogel, G., Fiehn, O., Jean-Richard-dit-Bressel, L., Boller, T., Wiemken, A., Aeschbacher, R. A., et al. (2001). Trehalose metabolism in Arabidopsis: occurrence of trehalose and molecular cloning and characterization of trehalose-6-phosphate synthase homologues. J. Exp. Bot. 52 (362), 1817–1826. doi: 10.1093/jexbot/52.362.1817
Wang, G., Li, X., Ye, N., Huang, M., Feng, L., Li, H., et al. (2021). OsTPP1 regulates seed germination through the crosstalk with abscisic acid in rice. New Phytol. 230 (5), 1925–1939. doi: 10.1111/nph.17300
Wang, C. L., Zhang, S. C., Qi, S. D., Zheng, C. C., Wu, C. A. (2016). Delayed germination of Arabidopsis seeds under chilling stress by overexpressing an abiotic stress inducible. GhTPS11. Gene 575 (2), 206–212. doi: 10.1016/j.gene.2015.08.056
Xue, Y., Wu, F., Chen, R., Wang, X., Inkabanga, A. T., Huang, L., et al. (2023). Genome-wide analysis of fatty acid desaturase genes in chia (Salvia hispanica) reveals their crucial roles in cold response and seed oil formation. Plant Physiol. Biochem. 199, 107737. doi: 10.1016/j.plaphy.2023.107737
Yeo, E. T., Kwon, H. B., Han, S. E., Lee, J. T., Ryu, J. C., Byu, M. (2000). Genetic engineering of drought resistant potato plants by introduction of the trehalose-6-phosphate synthase (TPS1) gene from Saccharomyces cerevisiae. Molecules. Cells 10 (3), 263–268. doi: 10.1007/s11240-004-8124-1
Yuan, G., Liu, J., An, G., Li, W., Si, W., Sun, D., et al. (2021). Genome-wide identification and characterization of the trehalose-6-phosphate synthetase (TPS) gene family in watermelon (Citrullus lanatus) and their transcriptional responses to salt stress. Int. J. Mol. Sci. 23 (1), 276. doi: 10.3390/ijms23010276
Zang, B., Li, H., Li, W., Deng, X. W., Wang, X. (2011). Analysis of trehalose-6-phosphate synthase (TPS) gene family suggests the formation of TPS complexes in rice. Plant Mol. Biol. 76, 507–522. doi: 10.1007/s11103-011-9781-1
Zhang, H., Dong, J., Zhao, X., Zhang, Y., Ren, J., Xing, L., et al. (2019). Research progress in membrane lipid metabolism and molecular mechanism in peanut cold tolerance. Front. Plant Sci. 10. doi: 10.3389/fpls.2019.00838
Zhang, H., Yu, Y., Wang, S., Yang, J., Ai, X., Zhang, N., et al. (2023). Genome-wide characterization of phospholipase D family genes in allotetraploid peanut and its diploid progenitors revealed their crucial roles in growth and abiotic stress responses. Front. Plant Sci. 14. doi: 10.3389/fpls.2023.1102200
Keywords: TPS genes, TPP genes, peanut (Arachis hypogaea), cold stress, trehalose
Citation: Zhong C, He Z, Liu Y, Li Z, Wang X, Jiang C, Kang S, Liu X, Zhao S, Wang J, Zhang H, Zhao X and Yu H (2024) Genome-wide identification of TPS and TPP genes in cultivated peanut (Arachis hypogaea) and functional characterization of AhTPS9 in response to cold stress. Front. Plant Sci. 14:1343402. doi: 10.3389/fpls.2023.1343402
Received: 23 November 2023; Accepted: 29 December 2023;
Published: 19 January 2024.
Edited by:
Guowei Li, Shandong Academy of Agricultural Sciences, ChinaReviewed by:
Chinedu Charles Nwafor, University of Nebraska-Lincoln, United StatesYanhao Xu, Hubei Academy of Agricultural Sciences, China
Muhammad Irfan, University of Sargodha, Pakistan
Copyright © 2024 Zhong, He, Liu, Li, Wang, Jiang, Kang, Liu, Zhao, Wang, Zhang, Zhao and Yu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Haiqiu Yu, eXVoYWlxaXVAc3lhdS5lZHUuY24=
†These authors have contributed equally to this work
 Chao Zhong
Chao Zhong Zehua He1†
Zehua He1† Yu Liu
Yu Liu Chunji Jiang
Chunji Jiang Xibo Liu
Xibo Liu Jing Wang
Jing Wang He Zhang
He Zhang Xinhua Zhao
Xinhua Zhao