- Department of Genetics and Biochemistry, Clemson University, Clemson, SC, United States
The unique evolutionary adaptation of legumes for nitrogen-fixing symbiosis leading to nodulation is tightly regulated by the host plant. The autoregulation of nodulation (AON) pathway negatively regulates the number of nodules formed in response to the carbon/nitrogen metabolic status of the shoot and root by long-distance signaling to and from the shoot and root. Central to AON signaling in the shoots of Medicago truncatula is SUNN, a leucine-rich repeat receptor-like kinase with high sequence similarity with CLAVATA1 (CLV1), part of a class of receptors in Arabidopsis involved in regulating stem cell populations in the root and shoot. This class of receptors in Arabidopsis includes the BARELY ANY MERISTEM family, which, like CLV1, binds to CLE peptides and interacts with CLV1 to regulate meristem development. M. truncatula contains five members of the BAM family, but only MtBAM1 and MtBAM2 are highly expressed in the nodules 48 hours after inoculation. Plants carry mutations in individual MtBAMs, and several double BAM mutant combinations all displayed wild-type nodule number phenotypes. However, Mtbam2 suppressed the sunn-5 hypernodulation phenotype and partially rescued the short root length phenotype of sunn-5 when present in a sunn-5 background. Grafting determined that bam2 suppresses supernodulation from the roots, regardless of the SUNN status of the root. Overexpression of MtBAM2 in wild-type plants increases nodule numbers, while overexpression of MtBAM2 in some sunn mutants rescues the hypernodulation phenotype, but not the hypernodulation phenotypes of AON mutant rdn1-2 or crn. Relative expression measurements of the nodule transcription factor MtWOX5 downstream of the putative bam2 sunn-5 complex revealed disruption of meristem signaling; while both bam2 and bam2 sunn-5 influence MtWOX5 expression, the expression changes are in different directions. We propose a genetic model wherein the specific root interactions of BAM2/SUNN are critical for signaling in nodule meristem cell homeostasis in M. truncatula.
1 Introduction
Legumes tightly control nitrogen-fixing symbioses leading to nodulation. The autoregulation of nodulation (AON) pathway negatively regulates the number of nodules formed in response to the metabolic status of the shoot (carbon) and root (nitrogen) (reviewed in Ferguson et al., 2019; Chaulagain & Frugoli, 2021). Genetic analysis has identified multiple genes that when mutated cause plants to hypernodulate, evidence of a network of regulation in AON from both the root and the shoot (reviewed in Roy et al., 2020).
In AON, very early after rhizobial infection, expression of a subset of genes encoding the CLAVATA3/Embryo Surrounding Region (CLE) peptides, MtCLE12 and MtCLE13 in Medicago truncatula, is induced (Mortier et al., 2010). In Lotus japonicus, a similar increase upon infection is observed in the LjCLE-root signaling (RS)1, LjCLE-RS2, and LjCLE-RS3 peptide-encoding genes (Okamoto et al., 2009; Nishida et al., 2016; Hastwell et al., 2017). These CLE peptides are 12–13 amino acids in length and are signaling peptides derived from the C-terminal region of pre-proproteins (Araya et al., 2016; Yamaguchi et al., 2016). In Arabidopsis thaliana, CLE function is associated with regulating cell proliferation and differentiation during development, especially in the shoot and root apical meristems. In M. truncatula, the MtCLE12 peptide has been shown genetically to be modified by an enzyme from the hydroxyproline O-arabinosyltransferase (HPAT) family encoded by the M. truncatula ROOT DETERMINED NODULATION1 (RDN1) gene (Kassaw et al., 2017); the mutation of RDN1 produces a supernodulation phenotype (Schnabel et al., 2011). After nodulation-suppressing CLE peptides are processed, they travel through the xylem to the shoot where they are perceived by a homo- or heterodimeric receptor complex, likely in the parenchyma cells of the vasculature (Kassaw et al., 2017).
Central to this receptor complex is a shoot-acting leucine-rich repeat (LRR) RLK, known as SUPER NUMERARY NODULES (SUNN) in M. truncatula, HYPERNODULATION ABBERANT ROOT FORMATION (HAR1) in L. japonicus, NODULE AUTOREGULATION RECEPTOR KINASE (NARK) in Glycine max, and SYMBIOSIS29 (SYM29) in Pisum sativum (Krusell et al., 2002; Searle et al., 2003; Schnabel et al., 2005). Mutations in the MtSUNN gene produce a hypernodulation phenotype and altered root development (Schnabel et al., 2005). A shoot-controlled increase in nodulation is also observed in the MtCORYNE Tnt1 insertion mutant (crn) of M. truncatula (Crook et al., 2016), as well as the klv mutant (Oka-Kira et al., 2005) and the clv2 mutant (Krusell et al., 2011) in L. japonicus. Bimolecular fluorescence analysis showed that SUNN forms heteromomers with homologous CLV1-interacting proteins CLAVATA2 (CLV2), CORYNE (CRN), and KLAVIER (KLV) (Oka-Kira et al., 2005; Miyazawa et al., 2010; Krusell et al., 2011; Crook et al., 2016). Genetic studies of a hypernodulating mutant in L. japonicus identified a root-responsive gene TOO MUCH LOVE (TML) that regulates nodule numbers in the root from shoot-derived signaling (Magori et al., 2009; Takahara et al., 2013). Knockdown of two TML genes, MtTML1 and MtTML2, showed increased nodule number, suggesting a role for these genes in AON (Gautrat et al., 2019).
Leucine-rich repeat receptor-like kinases (LRR-RLKs) are important in meristem development in most land plants. The similarity between symbiotic AON receptors and their Arabidopsis meristem regulating counterparts like RPK2 (Kinoshita et al., 2010) suggests that like the shoot apical meristem (SAM) and root apical meristem (RAM), LRR-RLK complexes are involved in AON signaling to nodule meristems (Krusell et al., 2002; Crook et al., 2016). Some (but not all) legume CLV2 and RPK2-related mutants show defects in the SAM, but the sunn Atclv1-related mutants do not have SAM defects, indicating overlapping receptor complexes controlling SAM activity and nodule numbers in legumes (Krusell et al., 2002; Schnabel et al., 2005).
Since similar molecules regulate shoot, root, and nodule meristems, we wondered if BAMs, which have not yet been linked to AON in any legume, could be involved in the regulation of the nodule meristem. The Arabidopsis BAMs bind to a wider range of CLE peptides and show more diverse expression patterns compared to CLV1 (reviewed in Yamaguchi et al., 2016). We reasoned that studying BAM expression in M. truncatula could expand our understanding of plant receptor interacting partners’ expression levels and location effects during signal transduction pathways. Since AtCLV1 forms a complex with AtBAMs, MtSUNN might form a complex with MtBAMs to control nodule numbers. Mutational and overexpression experiments of the M. truncatula BAM gene family reported below support a model in which the genetic interaction of BAM2 and SUNN provides a signal to limit nodulation.
2 Materials and methods
2.1 Phylogenetic analysis
The tree was constructed with MEGA X using the maximum likelihood method (Kumar et al., 2018) with bootstrap replicates n = 1,000. The analysis involved 21 amino acid sequences summarized in Supplementary Table 1 for a total of 1,107 positions in the final dataset, with AtCLV1 used to root the tree.
2.2 Mutant line screening
The Tnt-1 mutant lines were isolated from populations of insertion lines obtained from the Nobel Foundation Medicago Mutant Database now located at https://medicagomutant.dasnr.okstate.edu/mutant/index.php described in Tadege et al. (2008). The following Tnt-1 mutant pools were used in this study: Mtbam1 (NF2153), Mtbam2 (NF7126), Mtbam3 (NF2071), Mtbam4 (NF2835), Mtbam5 (NF2488), and sunn-5 (NF2262). Initial PCR screening from pools of Tnt1 insertions in M. truncatula BAMs was carried out to identify plants carrying the insertion in a BAM gene using PCR from genomic DNA. Two primer sets for each gene were designed specifically for capturing the wild-type allele and another specific to flanking regions of the Tnt1 insert and Tnt1 forward primer (see Supplementary Table 2). Plants identified as carrying an insertion were selfed, and the next generation was screened using PCR to obtain single homozygous mutants for all five BAMs from the Tnt-1 pools. Each homozygous mutant was backcrossed to the parental R108 ecotype and reisolated, following the inheritance of the insertion via PCR.
2.3 PCR to identify single and double mutants and sunn-5 structure
A leaf press was made by pressing a leaflet of each plant to a Plant Card (Whatman™, GE Healthcare UK Limited, Amersham, UK) according to the manufacturer’s instructions for long-term storage. A 1.2-mm-diameter piece of Plant Card was excised and washed with Whatman™ FTA Purification Reagent (GE Healthcare UK Limited, UK) followed by TE-1 buffer according to the manufacturer’s instructions and directly used in PCR. The 10-μL PCR used to identify single and double mutants contained 2 μL of gDNA (equivalent to 100 ng RNA), 1 mm each primer, 0.2 mm dNTPs, 1× colorless GoTaq buffer, and 1 U GoTaq (Promega, Madison, WI, USA) with cycling conditions of 95°C for 2 min followed by 40 cycles of 95°C for 5 sec, 55°C for 10 sec, and 72°C for 20 sec. Sequencing of sunn-5 was performed using PCR products amplified from cDNA using pairs of gene-specific primers for sequences JF7092, JF7093, and JF7094 and a Tnt-1 specific primer for JF7091 as referred to in Supplementary Figure 1. PCR conditions were 95°C for 2 min followed by 35 cycles of 95°C for 30 sec, 55°C for 30 sec, and 72°C for 4 min. Sequencing was performed by Arizona Life Sciences.
2.4 Plant crossing
Two pairs of fine-tip forceps (HL-14 #5, Buy in Coins Promotion) and straight-edge scalpels (scalpel blade handle 9303 #3 and scalpel blade 9311 #11, both from Microscopes America, Cumming, GA, USA) were used for keel petal incision and the removal of anthers from the unopened female flower bud and artificial cross-pollination (Veerappan et al., 2014). The mature pods from the successful cross-pollinations were wrapped using micro-perforated polythene sheets (MP1120160T, Prism Pak, Berwick PA, USA) to dry on the plant before harvesting at desiccation. The following crosses were successfully performed and the F2 was screened for double mutants: bam2 × bam3, bam2 × bam4, bam5 × bam4, bam2 × sunn-5, bam3 × sunn-5, and bam3 × sunn-5.
2.5 Plant growth conditions and materials
The identified homozygous bam mutants used for making genetic crosses were grown in a greenhouse, with supplemental lighting to create a 14:10 light:dark (L:D) cycle, a nightly minimum of 18°C, and a daily maximum between 21°C and 27°C. For nodulation screening, the following seeds were used: M. truncatula wild-type Jemalong A17; the AON-defective mutants rdn1-2 (Schnabel et al., 2011), sunn-1, and sunn-4 (Schnabel et al., 2005); and M. truncatula wild-type R108 and Tnt1 mutants bam1-5 (this work), crn (Crook et al., 2016), and cra2 (Huault et al., 2014) in the R108 genetic background (Garmier et al., 2017). Seeds were removed from the pods and scarified in sulfuric acid for 8 min, rinsed five times in dH2O, imbibed in dH2O for 2–3 hours at room temperature, then placed at 4°C for 48–72 hours in the dark to synchronize germination, and allowed to germinate for 1 day at room temperature. This procedure as well as the growth in an aeroponic apparatus is described in Cai et al. (2023). Inoculation of all whole plants screened for nodulation phenotypes was carried out using Sinorhizobium meliloti RM41 (Luyten and Vanderleyden, 2000) for the R108 ecotype with 6 mL each of an OD600= 0.2 culture, and data were collected 14 days post inoculation (dpi). The apparatus was subjected to 14:10 L:D conditions and maintained at room temperature. Nodule number comparisons of wild-type R108 plants and hypernodulating sunn-5 plants to individual single bams plants, bam double-mutant plants, and bam sunn-5 plants were carried out by visual count of individual plant roots using head-mounted magnification glasses.
Transgenic hairy root plants made with Agrobacterium rhizogenes containing the gene constructs described below or the empty vector were grown in TY (selection 25 μg/mL kanamycin) and used for hairy root transformation as described in Limpens et al. (2004). Inoculated seedlings were grown on plates containing modified HMF (Huo et al., 2006). The plates were incubated flat in the growth chamber at 20°C for 3 days to facilitate transformation and then moved to 25°C to facilitate root growth for 2–3 weeks. Transformed roots expressing the DsRED fluorescence marker were identified on the Olympus SZH-10 stereoscopic system (excitation 540 nm; emission detected at 570–625 nm), and untransformed roots were removed. The plants with transformed roots were then grown in perlite for 2 weeks before being used for the nodulation assay described in Kassaw et al. (2017) by inoculation using Sinorhizobium medicae strain ABS7 (Leong et al., 1985; Bekki et al., 1987) for A17 plants or S. meliloti RM41 as described in Kassaw and Frugoli (2012). Nodules were counted at 14 dpi.
Grafting was performed following the protocol of Kassaw and Frugoli (2012), and grafted plants were allowed to grow in the growth chamber at 20°C with a 16-hour photoperiod for 3 days to facilitate transformation and then moved to 25°C with a 16-hour photoperiod to facilitate root growth for 2–3 weeks. Plants were transferred to pots filled with washed, autoclaved perlite and maintained on a light bench with (14:10 L:D) cycle and temperatures between 21°C and 24°C. Plants were watered daily with a 100-fold dilution of water-soluble 20:10:20 (N:P: K) Peat-Lite Special fertilizer (Scotts Company, Marysville, OH, USA) for 5 days. After an additional 4 days of nutrient starvation induced by watering with water alone, the plants were used for inoculation experiments using S. medicae Rm41, and nodule number was assessed at 14 dpi. The survival rate for all grafted plants was approximately 40%–50% except for sunn-5 shoot-to-sunn-5 root plants, which was approximately 10%–20%. The reduced survival of sunn self-grafts is common in both our laboratory and others due to disrupted auxin homeostasis in sunn mutants (van Noorden et al., 2006).
2.5.1 Creation of BAM overexpression constructs
The MtBAM1, MtBAM2, and MtBAM3 sequences were amplified from M. truncatula R108 cDNA using overlap PCR with the primers listed in Supplementary Table 2. Using overlap extension PCR allowed for the cloning of large fragments by fusing two gene halves together to generate large transcripts for cloning (Simionatto et al., 2009). Each transcript was amplified using the following PCRs (10 μL) that contained 2 μL of cDNA (equivalent to 100 ng RNA), 1 mm of each primer, 0.2 mm dNTPs, 1× colorless GoTaq buffer, and 1 U GoTaq (Promega, Madison, WI, USA) with cycling conditions of 95°C for 2 min followed by 40 cycles of 95°C for 5 sec, 55°C for 10 sec, and 72°C for 20 sec. PCR products were gel extracted and purified using the Zymoclean gel DNA recovery kit (Zymo Research, Irvine, CA, USA). The PCR products were then digested with KpnI and SpeI restriction enzymes and ligated into the KpnI and SpeI sites of the pCDsRed/35S vector (Karimi et al., 2002) under the control of the 35S promoter using Escherichia coli DH5α cells. The construct was confirmed by sequencing and transferred to A. rhizogenes strain ArQUA1 (Quandt and Hynes, 1993) by electroporation for use in hairy root transformation.
2.6 A. rhizogenes-mediated plant transformation
Seedlings were transformed as described by Limpens et al. (2004). The hypocotyls of 5-day-old seedlings were cut and transformed by lightly scraping on the surface of Luria–Bertani plates densely grown with A. rhizogenes strain ArQUA1 (Quandt and Hynes, 1993) containing the appropriate binary vector and antibiotic selections at 30°C for 48 hours. After 5 days of cocultivation with the A. rhizogenes in the growth chamber at 23°C at 16:8 L:D, the seedlings were transferred to a nutrient-rich hairy root emergence medium (Limpens et al., 2004) containing 300 µg/mL cefotaxime (Phytotechnology Laboratories, St. Lenexa, KS, USA) sandwiched between two half-round Whatman filter papers grown under the same growth conditions. Five days later, the top filter papers were removed from the plates, and the seedlings were allowed to grow for an additional 5 days on the same emergence medium placed vertically in the same growth chamber. Transformed roots expressing the DsRED fluorescence marker were identified on Olympus SZH-10 stereoscopic system (excitation 540 nm; emission detected at 570–625 nm), untransformed roots were removed as described in Kassaw and Frugoli (2012), and plants were transferred to perlite. For acclimation to perlite, plants were watered for 5 days with a 100-fold dilution of water-soluble 20:10:20 Peat-Lite Special fertilizer (Scotts). Fertilization was then withdrawn, and the plants were hydrated with water alone for an additional 5 days to induce the nitrogen deficiency required for nodulation. Plants were then inoculated with S. meliloti RM41 or S. medicae ABS7 according to ecotype, with 6 mL each of an OD600= 0.2 culture, and data were collected at 14 dpi.
2.7 Quantitative real-time PCR
For RNA extraction, the nodule-forming zone described in Schnabel et al. (2023a) was harvested from 10 plants for each genotype (A17, sunn-1, and sunn-4, rdn2-1, cra2, R108, bam2, bam2 sunn-5, and sunn-5) at multiple time points (0 hours, i.e., prior to inoculation, 12 hours post inoculation (hpi), and 48 hpi) and stored at −80°C. RNA was isolated from nodulating roots using the E.Z.N.A. Plant RNA Kit (Omega Bio-Tek, GA, USA) according to the manufacturer’s instructions. Each RNA sample was digested with RNase-free DNase (Promega) treatment for 40 min to remove genomic DNA contamination. The iScript cDNA Synthesis Kit (Bio-Rad, Hercules, CA, USA) was used to synthesize cDNA from 1 μg of RNA in a 20-μL reaction. The cDNAs were then diluted to 40 μL. All experiments were performed using iTaq™ Universal SYBR® Green Supermix (Bio-Rad, CA, USA) and the Applied Biosystems QuantStudio 3 Real-Time PCR System. Each reaction was performed in triplicate, and results were averaged for a single biological replicate. The total reaction volume was 12.5 μL (5 μL of master mix including 0.5 μL of each primer [0.5 μM final concentration] and 2.0 μL of cDNA). Cycle threshold values were obtained using the QuantStudio 3 software, and expression was determined relative to the internal reference PI4K (see below). Intron spanning primers unique in the M. truncatula genome based on National Center for Biotechnology Information Primer-BLAST analysis were used (Supplementary Table 2). Relative expression levels of genes were assayed using the Pfaffl method (Pfaffl, 2001) relative to previously validated housekeeping reference gene phosphatidylinositol 3- and 4-kinase belonging to ubiquitin family (PI4K; Medtr3g091400 in MtV4.0) (Kakar et al., 2008). Data from the three biological replicates were used to estimate the mean and standard error.
2.8 Statistical analysis
Statistical difference analysis for pairwise mean comparison was calculated by the Tukey–Kramer honestly significant difference (HSD) using JMP Pro 13.1 (https://www.jmp.com/en_us/home.html). For qPCR, absolute quantification was used to compare the quantification cycle (Cq) values of test samples to those of standards (PIK internal reference) of known quantity plotted on a standard curve. The quantity was normalized to a unit amount of nucleic acid (i.e., concentrations). The statistical analysis tests for the probability that the relative expression RE ≠ 1 for data normalized to control.
3 Results
3.1 Phylogenetic and structural analysis of BAM proteins
A phylogenetic analysis by neighbor-joining was performed between the protein sequences of legume and Arabidopsis BAMs, with CLV1 and the related symbiotic kinases used to root the tree (Figure 1). There was not a one-to-one relationship between MtBAMs and AtBAMs; rather, they shared similarities with BAMs from other legume BAM-Like LRR kinases and Arabidopsis. For all the legumes, however, there were two BAM proteins with similarities to AtBAM1 and AtBAM2, including MtBAM1 and MtBAM2. The MtBAM1 and MtBAM2 proteins have 80% sequence similarity between them, while MtBAM1 has 81% similarity to AtBAM1, the highest shared similarity among all comparisons. MtBAM3, MtBAM4, and MtBAM5 were closer phylogenetically to the AtBAM3 protein with a 55%–56% similarity for each. The branching structure suggests that the five BAM genes in legumes arise from duplications of a BAM3 ortholog (Figure 1). To further pursue the idea that these duplicate genes might be specific for nodulation, the expression levels of each MtBAM in individual root tissues were examined during nodule development. Tissue level data from a Laser Capture Microdissection experiment (Schnabel et al., 2023b) and root segment transcriptome data from an unfixed roots experiment (Schnabel et al., 2023a) were examined, following nodule development at 0, 12, 24, 48, and 72 hpi (Figure 2). MtBAM1 and MtBAM2 were expressed in nodule meristems at 48 and 72 hpi at higher levels than the other BAM family members. MtBAM1 and MtBAM2 expression at 12 and 24 hpi was higher in the inner cortical cells at the xylem pole, the cells from which nodules arise (Libbenga and Harkes, 1973). In addition, MtBAM1 and MtBAM2 had higher expression in nodule meristems at 48 and 72 hpi compared to MtBAM3, MtBAM4, and MtBAM5. MtBAM3, MtBAM4, and MtBAM5; all displayed similarly low levels of expression throughout nodule formation, with no clear signature in an individual tissue in this experiment (Figure 2). The expression data suggest that MtBAM1 and MtBAM2 are most likely to be involved in early nodule formation.
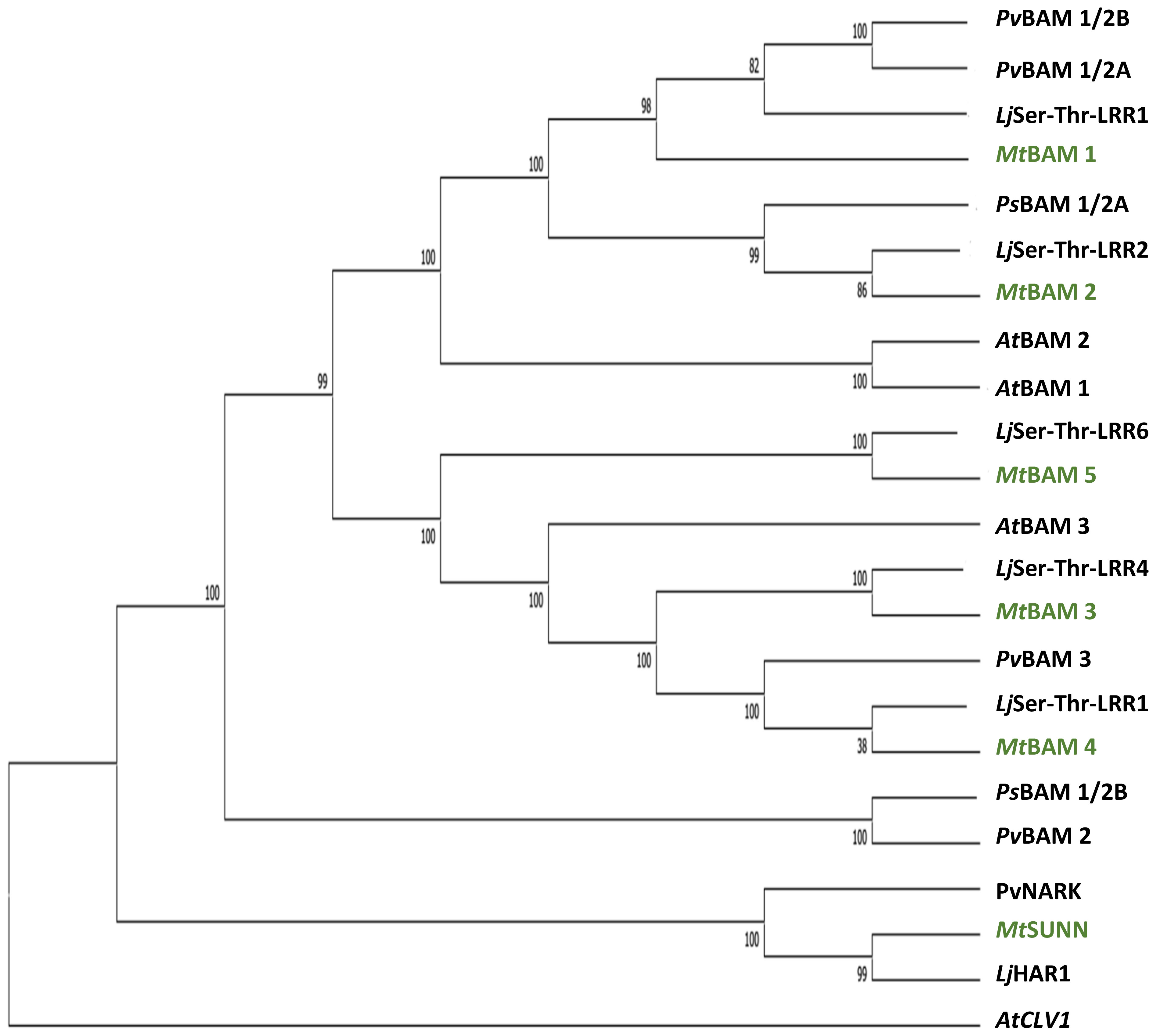
Figure 1 Relatedness of BAM receptor kinases between legumes and Arabidopsis. Phylogenetic tree created using the maximum likelihood method and rooted with AtCLV1, with branches supported by at least 80% of the bootstrap replicates (n = 1,000). Medicago truncatula BAM proteins are highlighted in green. Accession numbers for genes in tree are in Supplementary Table 1.
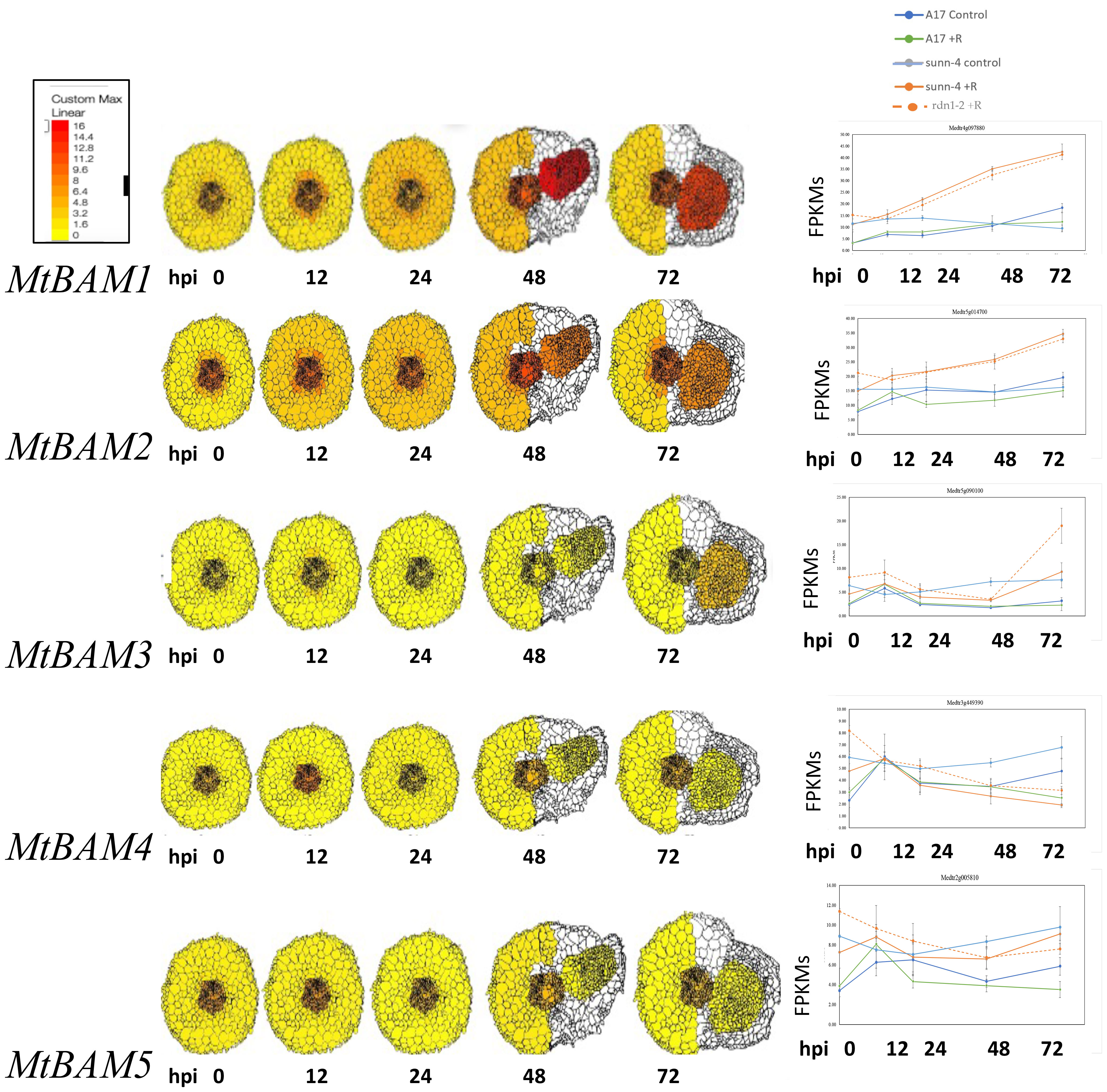
Figure 2 MtBAM expression in individual tissues and whole root segments during nodule formation. The tissue-specific expression patterns on the left for each MtBAM were created using ePlant and the data in Schnabel et al. (2023b) and a fixed maximum for comparison between genes across a time course at 0 to 72 hpi. The whole root expression traces for each MtBAM on the right are from the same time course and conditions, taken from the data in Schnabel et al. (2023a). Expression (FPKMs) of BAMs from 0 to 72 hpi are displayed as blue = A17 control, green = A17 + rhizobia, light blue = sunn-4 control, orange = sunn-4 + rhizobia, and orange dotted = rdn1-2 + rhizobia.
3.2 Root length and nodule number phenotypes of MtBAM mutants are wild type
With the expression data in hand, we generated single homozygous mutants for the BAM genes in M. truncatula using PCR to follow the segregation of Tnt1 insertions in lines isolated from pools of plants from the Nobel Foundation Medicago Mutant Database identified as containing an insertion in an MtBAM gene. The progeny of plants determined to be homozygous for a Tnt-1 insertion in each BAM were tested for nodule number on an aeroponic chamber (see Materials and Methods). All the single MtBAM mutants had Tnt1 insertions in approximately the same place in the LRR domains, and wild-type nodule numbers (Supplementary Figure 2A). Since other nodule regulatory mutants also have root length defects (Schnabel et al., 2005; Schnabel et al., 2010; Schnabel et al., 2011), single mutant plants were grown in the absence of rhizobia, and root length was measured at the equivalent development point to 14 dpi. None of the single MtBAM mutants displayed a root length phenotype different from the wild-type plants (Supplementary Figure 2B). Similar to the observation of no meristem defects in single bam mutants in Arabidopsis (DeYoung et al., 2006), no nodule number or root length phenotype was observed in the M. truncatula single BAM mutants. We also generated double mutants of Mtbam2 with Mtbam3 and Mtbam4, as well as an Mtbam4;Mtbam5 double mutant, to determine if an effect is observable with the loss of more than one BAM, as is the situation with Arabidopsis mutants (DeYoung and Clark, 2008), but these also had no effect on nodule number or root length (Supplementary Figure 3). Interestingly, we were unable to obtain seeds from any cross of the Mtbam1 mutant to any other Mtbam mutant, despite multiple attempts. Examining the ePlant database (Waese et al., 2017), transcriptomic data for MtBAM1 show high levels of expression in tissues such as pods, flowers, stems, and developing seeds (Benedito et al., 2008). When attempting to make crosses with bam1, the stamen looked frail and opaque with few pollen granules visible (data not shown), and this may explain the lack of success generating bam1 bamx double mutants.
3.3 Mtbam2 suppresses the sunn-5 supernodulation phenotype in the double mutant
Since Arabidopsis BAMs were originally isolated by the effects observed in a clv1 mutant background (DeYoung et al., 2006), we reasoned that mutations in MtBAM genes might only have noticeable effects on nodule number in a sunn mutant background. We created a set of bam/sunn double mutants by crossing and isolating F2 progeny using PCR confirmation of the phenotype: bam2 sunn-5, bam3 sunn-5, and bam5 sunn-5. We observed a suppressive effect on nodule number in the bam2 sunn-5 double mutant (Figure 3A). We applied a slightly different observation to the root length phenotype; the sunn-5 short root phenotype was partially rescued by bam2 or bam3, but not bam5 (Figure 3B). Because of the suppressive effect of bam2 on both phenotypes of sunn-5 plants, we investigated the mechanism further.
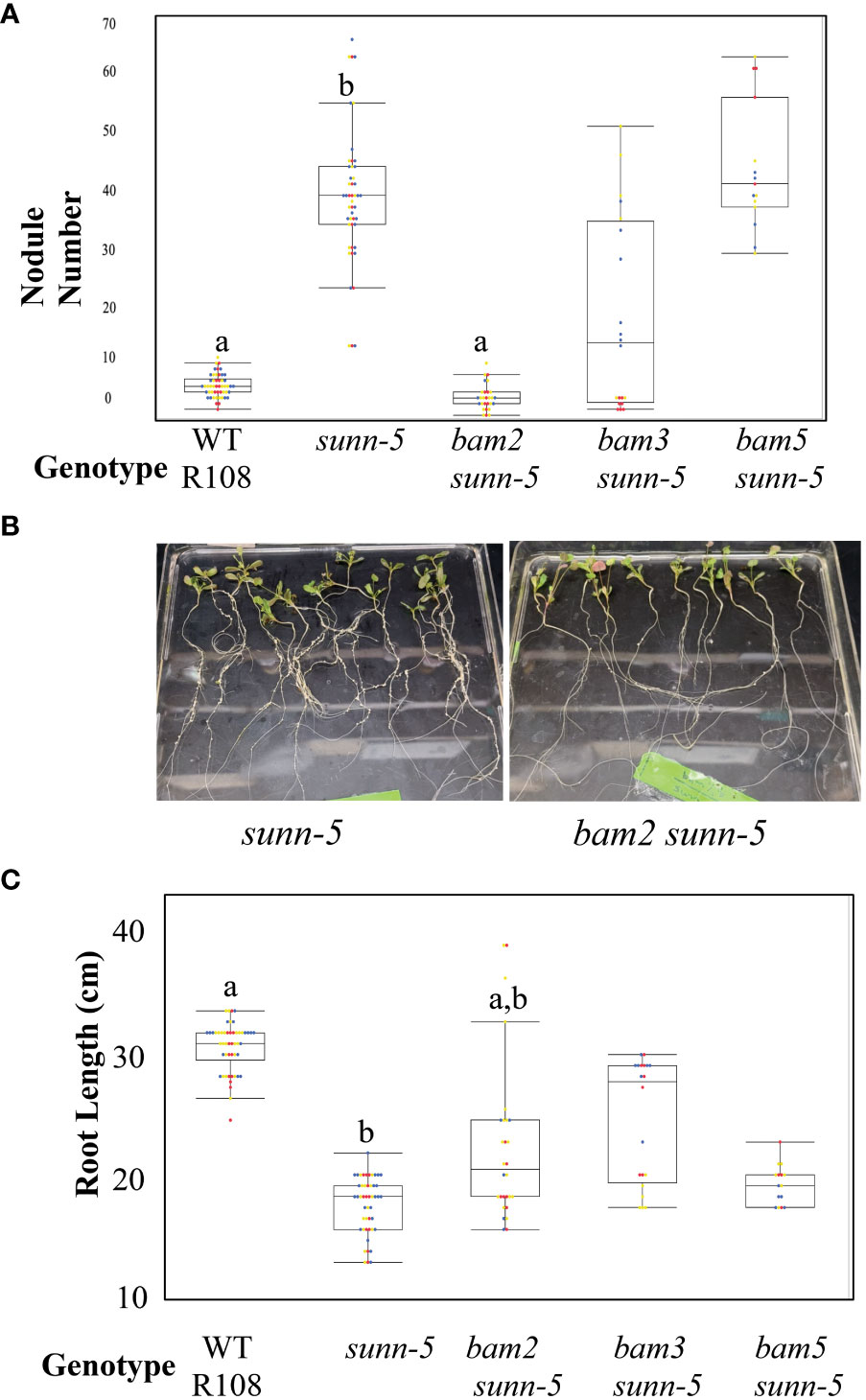
Figure 3 bam2 suppresses nodule number in sunn-5 mutants, while bam2 and bam3 suppress root length defects. (A) Nodule number per plant at 14 dpi. Letters denote significance compared to wild-type (B) photos of sunn-5 and bam2 sunn-5 plants. (C) Root length in wild-type plants compared to sunn-5 and bam double mutants at 14 days without rhizobia. Letters denote significance compared to wild type. Dots represent individual plants. Each dot color is associated with independent biological replicates: rep1 = red, rep2 = yellow, and rep3 = blue. Box plot mean is indicated with a line, while whiskers are ± standard error, pairwise comparison Tukey–Kramer honestly significant difference (HSD), p-value a:b > 0.001. p-value a:a,b > 0.001.
3.4 Grafting to determine localization (shoot or root) of bam2 suppression of sunn-5 phenotype
The sunn mutation has been shown to increase nodule number when a sunn mutant shoot is grafted onto wild-type shoots (Penmetsa et al., 2003). The same grafting experiment was employed to determine the location of the suppression observed in the bam2 sunn5 double mutant. Since AON is a long-distance root to shoot to root regulatory pathway, a set of grafting experiments was designed to determine the location of the effect (Figure 4). Comparing the nodule number of plants created with a bam2 sunn-5 shoot grafted onto a sunn-5 root to that of a sunn-5 shoot on a bam2 sunn-5 root (Figure 4A), suppression of the sunn-5 hypernodulation phenotype occurred when a sunn-5 shoot genotype was grafted onto the bam2 sunn-5 root genotype, indicating that the suppression is root derived (Figure 4A). Since the root is a double mutant, two possibilities arise—either sunn-5 is required for the suppression, or bam2 alone in the roots suppresses the nodule number phenotype of a sunn-5 shoot. To resolve this, a second grafting experiment was designed (Figure 4B) in which the sunn-5 shoot genotype was grafted onto a bam2 sunn-5 root genotype and compared to a sunn-5 shoot genotype grafted onto a bam2 root genotype. Suppression of the sunn-5 hypernodulation phenotype occurred in both graft combinations, suggesting that bam2 can suppress nodule numbers from the roots, with or without sunn-5 being present (Figure 4B).
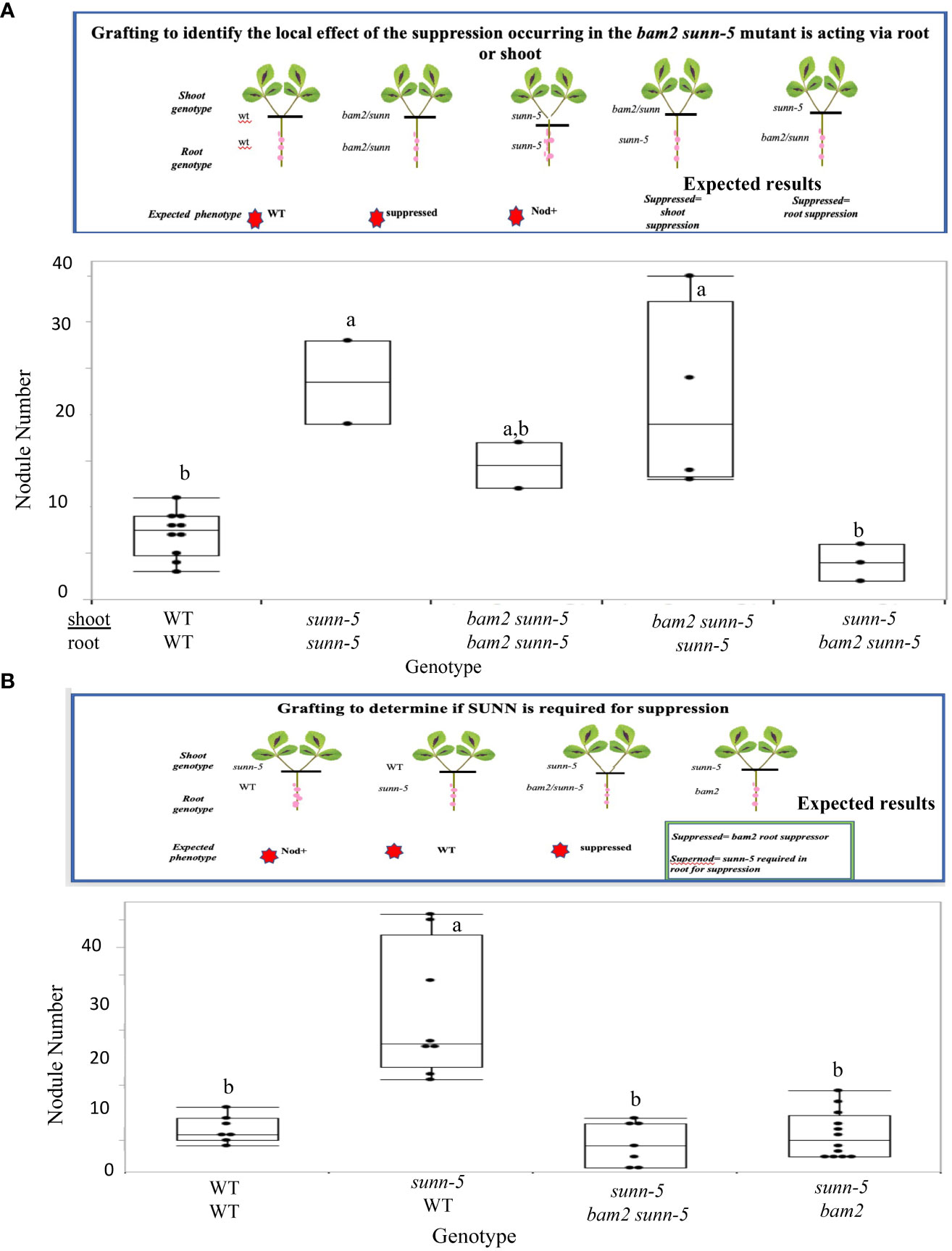
Figure 4 Suppression of the sunn-5 phenotype by bam2 is root derived and occurs regardless of sunn-5 root genotype. (A) Nodule numbers formed on grafted plants to test root/shoot origin of suppression. Gene on top indicates shoot genotype; bottom indicates roots. Results are combined from two individual experiments. Box plot mean is indicated with a line (where more than three data points available), while whiskers are ± standard error, pairwise comparison Tukey–Kramer honestly significant difference (HSD). Letters indicate significance comparisons. p-value a:a,b > 0.01. p-value a:b > 0.001. (B) Nodule numbers formed on grafted plants to determine if SUNN is required. Experimental details are the same as in (A).
3.5 Overexpression of BAM2 increases nodulation in wild-type roots
In addition to phenotyping bam mutants, we observed nodule numbers in transgenic hairy roots overexpressing individual MtBAM genes driven by the CMV 35S promoter compared to control plants carrying empty vector constructs (Figure 5). Overexpression of BAM1 and BAM3 did not alter nodule numbers in wild-type plants of either ecotype, but overexpression of BAM2 in wild-type transgenic hairy roots caused a significant increase in nodule number in both A17 (the ecotype used for genome sequencing) and R108 (an ecotype used for tissue culture-based transformation and the background of the BAM mutant lines). This confirms the specific involvement of BAM2 in nodule number and that the effect is not ecotype specific.
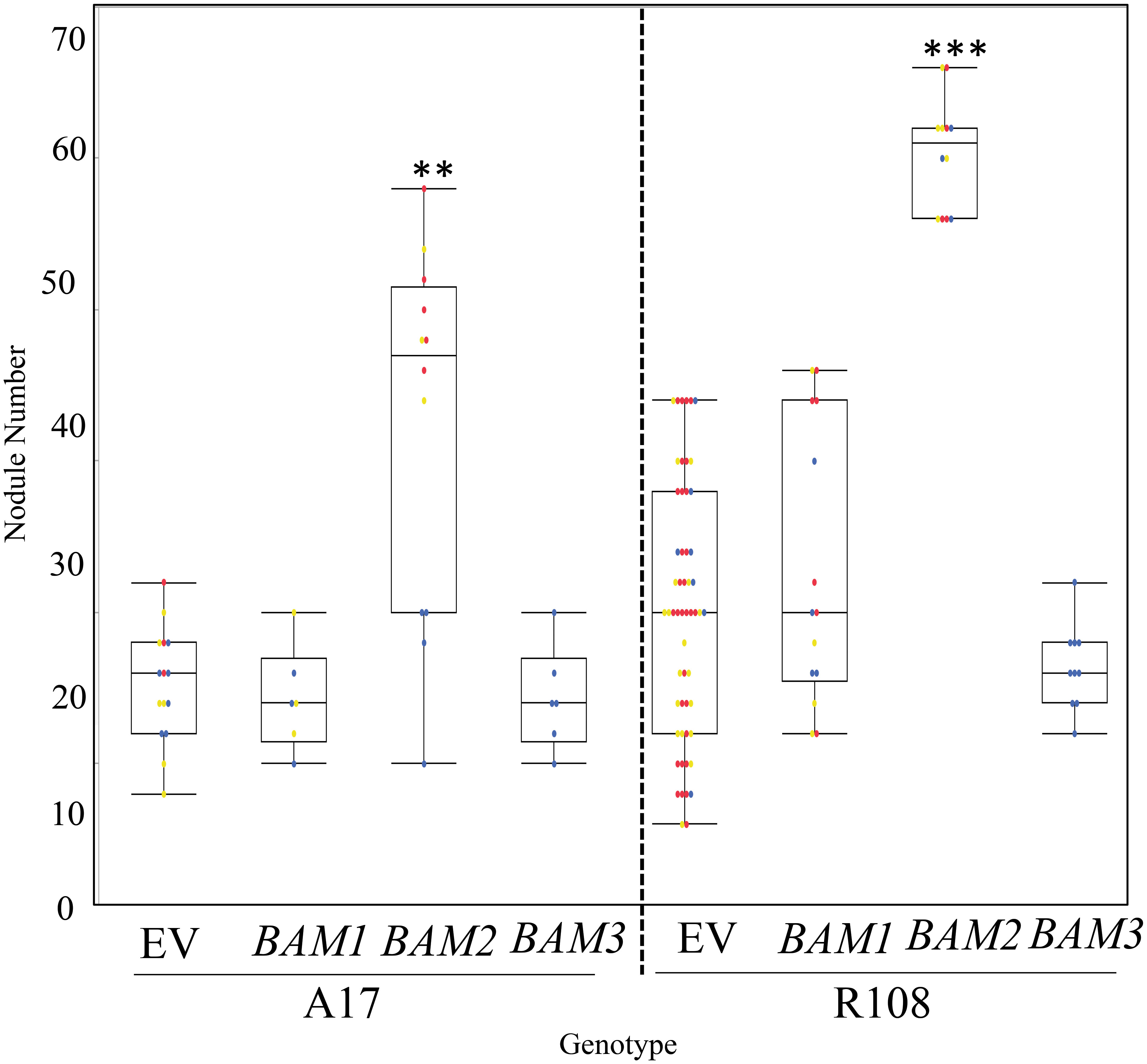
Figure 5 Overexpression of MtBAM2 in wild-type A17 and R108 ecotypes increases nodule number. Nodule number in plants overexpressing BAM1, BAM2, or BAM3 under the 35S promoter (see Materials and Methods) at 14 dpi. A17 ecotype plants are inoculated with Sinorhizobium medicae ABS7, and R108 ecotype plants with Sinorhizobium meliloti RM41. EV = empty vector control. Dots represent individual plants. Each dot color is associated with independent biological replicates: rep1 = red, rep2 = yellow, and rep3 = blue. Box plot mean is indicated with a line, while whiskers are ± standard error, pairwise comparison Tukey–Kramer honestly significant difference (HSD), p-value > 0.01. (**). p-value > 0.001. (***) denotes significance compared to EV control. Pictures of representative plants are in Supplementary Figure 4.
3.6 The effect of overexpression of BAM2 in a sunn mutant background is allele specific
Suppression of nodule number in sunn-5 could arise from disruption of signaling downstream of SUNN, or it could arise from disruption of a signaling step at which BAM and SUNN interact (Meneely, 2020). We tested an allelic series of sunn mutants for indications of interaction between the proteins. The sunn-5 mutant in the R108 background has been used in previous work (Crook et al., 2016; Nowak et al., 2019) and contains a Tnt1 insertion (~2 kb in size) located 406 bp upstream of the ATG start codon in SUNN gene (El-Heba et al., 2015). Using PCR on cDNA (Supplementary Figure 1), we determined that this promoter insertion also removed 584 bases past the start site, resulting in a potential new start in the leucine-rich repeats, as well as a deletion of the end of the repeats, the transmembrane domain and into the kinase domain (Figure 6A); the ability to isolate RNA indicates that a truncated SUNN message is made. Additional sunn mutants exist in the A17 ecotype; the sunn-1 allele in A17 produces a full transcript with only a change in an amino acid in the kinase domain, while the sunn-4 allele contains a premature stop codon immediately after the signal sequence and is considered a null allele (Schnabel et al., 2005). The two alleles vary in nodule number; sunn-1 plants have three to five times the nodules of wild-type plants, while sunn-4 plants have a 10-fold increase in nodule number (Schnabel et al., 2010). A17 is difficult to cross with R108 due to multiple rounds of R108 selection for regeneration and differences in the genomes, which make genetic crosses problematic. Adding to the difficulties, the R108 ecotype nodulates best with a different strain of rhizobia (Hoffmann et al., 1997). Therefore, rather than making the mutants to test suppression, we chose to overexpress BAM2 in the different alleles of sunn in their respective ecotypes, inoculated with the rhizobia best for each ecotype. In contrast to wild-type plants in which overexpression of BAM2 increased nodule number, overexpression of BAM2 in a sunn-5 background suppressed nodule numbers to a wild-type level (Figure 6B). Overexpression of BAM2 also suppresses the hypernodulation phenotype of a sunn-1 mutant, which produces a full-length sunn message with a single amino acid change (Schnabel et al., 2005) but did not suppress hypernodulation in sunn-4 null allele (Figure 6B). When BAM1 and BAM3 were overexpressed in the sunn-4 and sunn-5 mutants, no alteration to the hypernodulating sunn phenotype was observed for either mutant (Figure 6B), again confirming the effect is also specific to the BAM2 gene. The sunn-1 mutant appeared to increase nodule number in response to BAM1 overexpression, but this was not statistically significant because of the distribution spread of nodule number in the empty vector control.
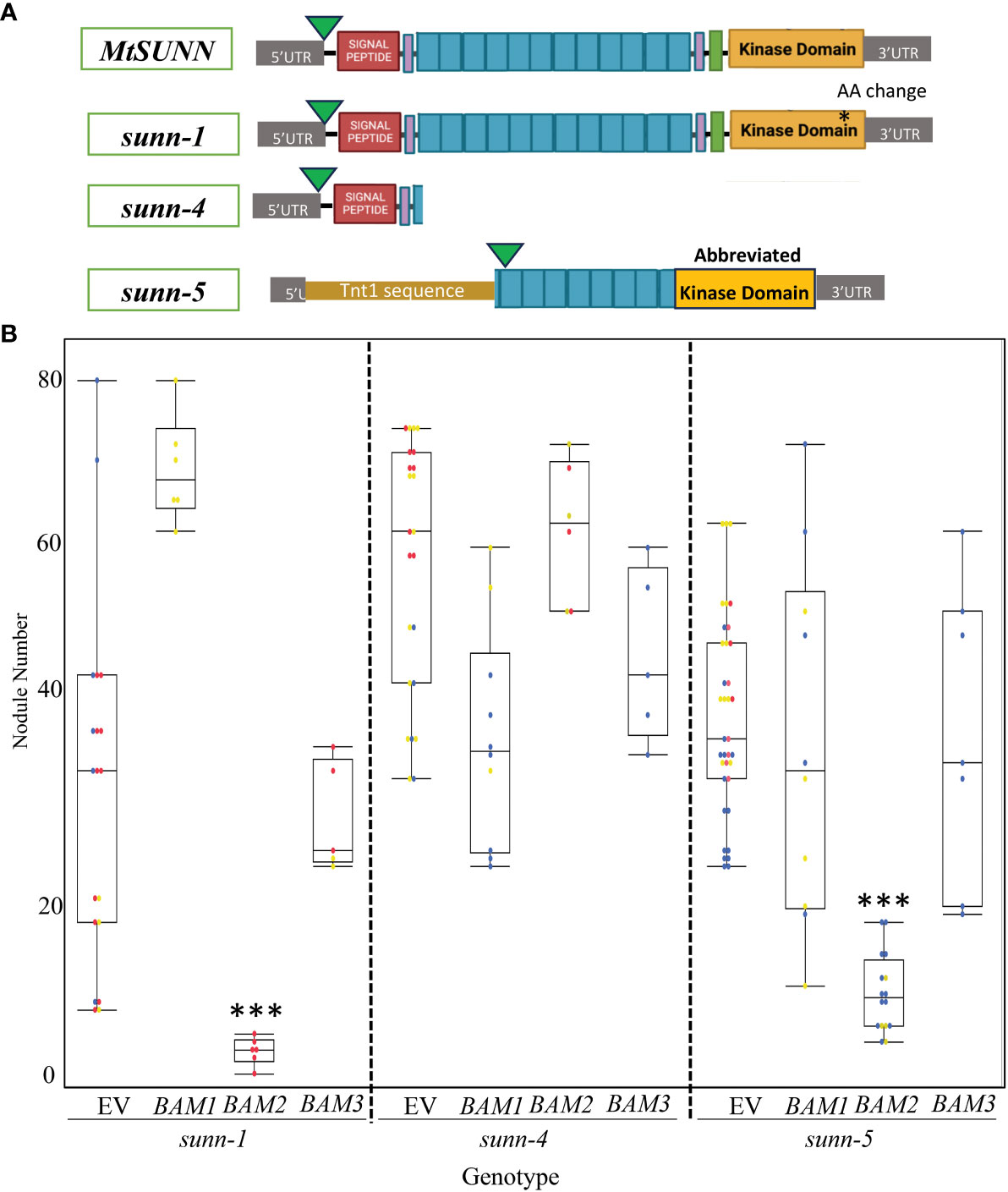
Figure 6 Results of overexpression of MtBAM1, MtBAM2, and MtBAM3 in sunn mutants are allele specific. (A) Diagram of SUNN protein and effects of sunn alleles. Green arrow indicates Met used for start, purple boxes are paired Cys residues, turquoise boxes represent leucine-rich repeats (not to scale; there are 22), and green box is transmembrane domain. (B) Nodulation on plants overexpressing indicated BAM genes. EV = empty vector control. Dots represent individual plants. Each dot color is associated with independent biological replicates: rep1 = red, rep2 = yellow, and rep3 = blue. Box plot mean is indicated with a line, while whiskers are ± standard error, pairwise comparison Tukey–Kramer honestly significant difference (HSD). (***) denotes significance compared to EV, p-value > 0.001. Pictures of representative plants are in Supplementary Figure 4.
3.7 Overexpression of BAMs in other AON mutants
To determine if BAM2 overexpression can suppress nodulation in other hypernodulators, we repeated the previous experiment with three mutants in different portions of the AON pathway. RDN1 functions in the root early in the AON pathway upstream of SUNN, modifying the ligand for the SUNN receptor (Kassaw et al., 2017). The pseudokinase CRN displays an increase in nodule number when mutated and acts at the same point as SUNN in the AON pathway, forming heteromultimers with SUNN (Crook et al., 2016). In contrast, the CRA2 receptor kinase mutants signal in a different part of nodule number regulation, separate from SUNN, conveying information on nitrogen status to the AON pathway through signaling with CEP1 peptide (Laffont et al., 2019). The cra2 mutant does not make nodules because the signal for nitrogen needs is not sent to the roots. None of the BAMs affected the hypernodulation phenotype of rdn1-2 mutants when overexpressed; however, BAM2 overexpression suppressed the hypernodulation phenotype of crn mutants (Figure 7). Interestingly, overexpression of all BAMs tested resulted in the death of all cra2 plants before nodulation could occur, while their empty vector controls remained alive (Figure 7), suggesting that there may be some effect unrelated to nodulation of excess BAM2 in these plants.
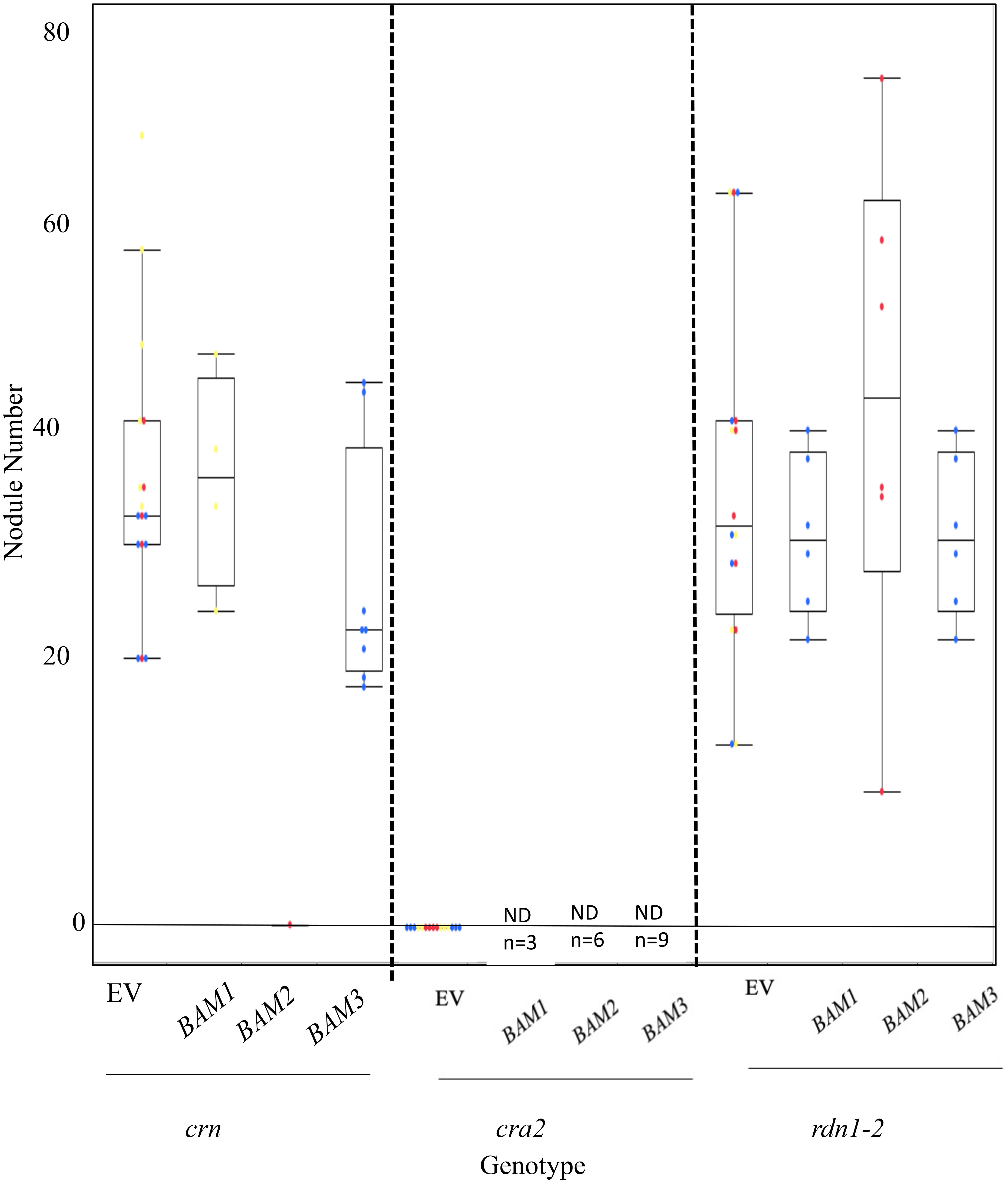
Figure 7 Results of overexpression of MtBAM1, MtBAM2, and MtBAM3 in autoregulation of nodulation (AON) mutants crn and rdn1-2 and cra2 mutants. No significant nodule number phenotype was observed from the overexpression of MtBAM1, MtBAM2, or MtBAM3 in any of the AON mutants. However, MtBAM2 overexpression in crn resulted in almost all plants dying, with only one plant surviving the screening stage. No nodules formed on the sole surviving crn:pMtBAM2 plant. All cra2 plants overexpressing a BAM gene died before screening. EV = empty vector control. Dots represent individual plants. Each dot color is associated with independent biological replicates: rep1 = red, rep2 = yellow, and rep3 = blue. Box plot mean is indicated with a line, while whiskers are ± standard error.
3.8 Expression of genes downstream of BAM/SUNN
The MtWOX5 transcription factor is downstream of SUNN signaling. Since MtWOX5 has been implicated in nodule development in M. truncatula (Osipova et al., 2012), in this work, we examined expression levels in several mutant combinations. We examined the relative expression of MtWOX5 during nodulation in both bam2 and bam2 sunn-5 plants by utilizing a time-course experiment using root segments to assess the relative expression of each marker gene at 0, 12, and 48 hpi. Previous work in our lab measured absolute expression for each gene in wild-type and sunn-4 root segments, and we also used the ePlant resource to confirm the tissue-level expression of these genes. We examined the expression of MtWOX5 at each time point in wild-type, bam2, and bam2 sunn-5 plants using quantitative real-time PCR (Figure 8).
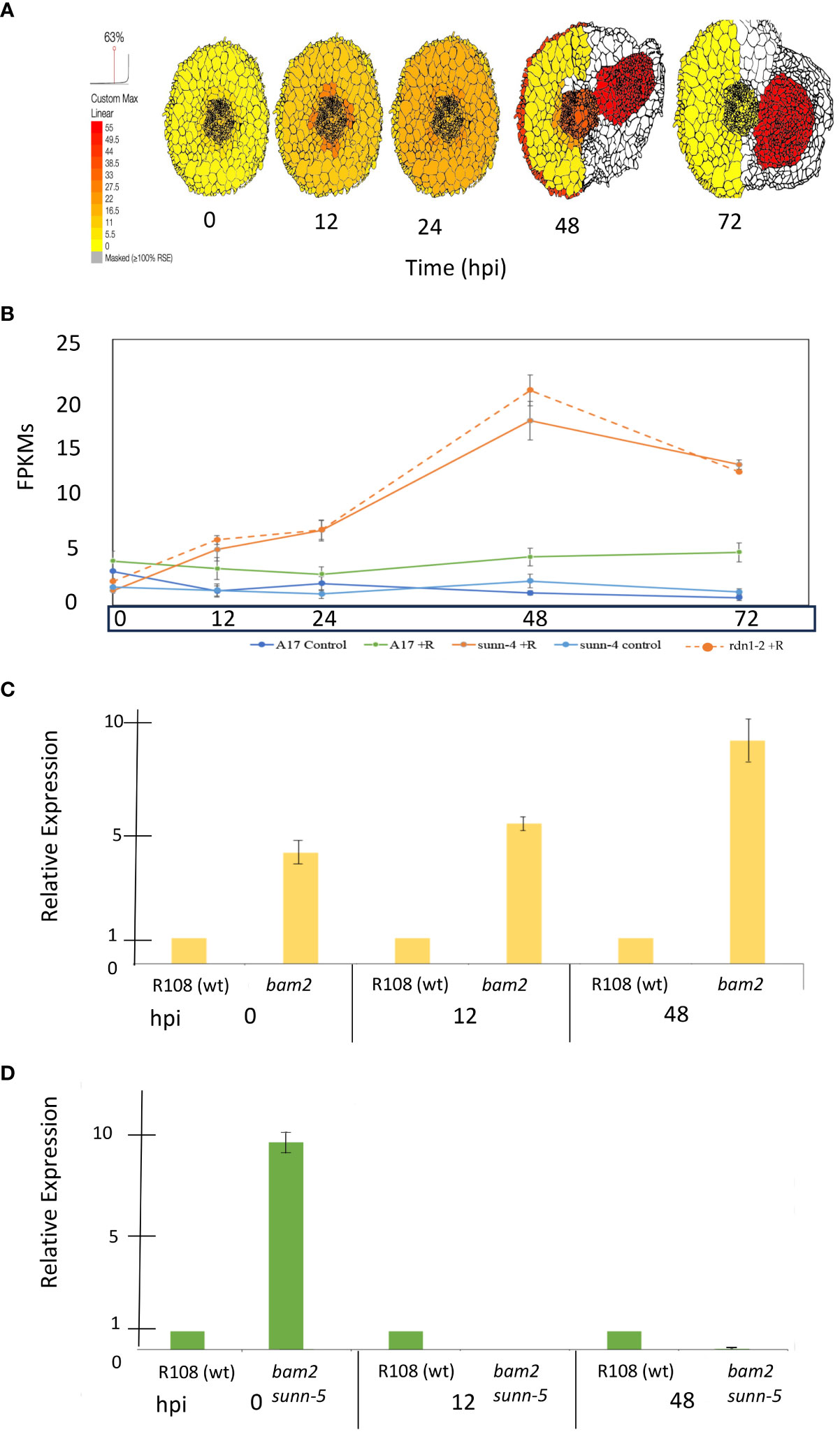
Figure 8 Expression of MtWOX5 during nodule formation. (A) MtWOX5 tissue-specific expression created using ePlant and the data in Schnabel et al. (2023b). (B) Whole root expression traces for WOX5 from the same time course and conditions were taken from the data in Schnabel et al. (2023a). Expression (FPKMs) of WOX5 from 0 to 72 hpi is displayed as blue = A17 control, green = A17 + rhizobia, light blue = sunn-4 control, orange = sunn-4 + rhizobia, and orange dotted = rdn1-2 + rhizobia. (C) Expression of MtWOX5 in bam2 mutants and (D) bam2 sunn-5 mutants. For each time point, relative expression was calculated by fold change compared to the wild-type control at that time point (see Materials and Methods). Data represent three biological and three technical replicates. Error bars represent ± standard error. Note that WOX5 expression was undetectable in the bam2 sunn-5 mutant at 12 hpi and barely detectable at 48 hpi.
MtWOX5 expression is the strongest in developing nodules (Figure 8A) and may reflect the expression of the nodule meristem since the LCM resource colors the entire tissue. MtWOX5 maintains a steady low-level expression throughout nodule development but is slightly higher in the inoculated roots compared to the non-inoculated controls (Figure 8B). In sunn-4 roots, MtWOX5 expression increases at 12 hpi and continues to increase to peak expression in this time course at 48 hpi (Figure 8B). In contrast, in a bam2 background, WOX5 expression as measured using real-time quantitative RT-PCR is greater at 0, 12, and 48 hpi compared to the wild-type control (Figure 8C). However, in the double-mutant bam2 sunn-5 root segments, the highest level of relative expression of MtWOX5 was observed at 0 hpi, with expression less than that of the wild-type control at 12 and 48 hpi in the double mutant (Figure 8D). Expression of the master regulator MtWOX5 decreases after inoculation in the bam2 sunn-5 mutant versus in the single bam2 background in which MtWOX5 increases and stays up after inoculation.
4 Discussion and conclusions
Phylogenetic analysis (Figure 1) suggests the expansion in the BAM gene family between Arabidopsis with three BAM genes, and M. truncatula with five suggests that if the expansion of the gene family resulted in new genes used for nodule meristems, MtBAM4 and MtBAM5 would be the best candidates based on their position with additional legume BAMs in a clade away from most of the rest of the BAMs. However, neither BAM4 nor BAM5 was expressed in nodules (Figure 2), while MtBAM1–3 were expressed in nodules.
Since we did not identify nodule number or root length phenotype in any plants carrying a mutation in a single bam (Supplementary Figure 2) or any of the plants carrying two bam mutations (Supplementary Figure 3), mutation analysis of BAM genes alone did not identify a BAM specific to the regulation of nodule number. The lack of an observable effect from single bam mutants was not unexpected, as single Atbams showed no phenotype (DeYoung et al., 2006). While we did not investigate further, MtBAM1 could be important to multiple developmental systems, as we were unable to generate any double mutants with this line. The discovery that adding a bam2 mutation to a sunn mutation suppressed nodule number to wild-type levels (Figure 3) indicates BAM2 is involved in nodule development, but visualization of this role is only possible when SUNN is disrupted. This effect is specific to bam2 and not observed when sunn-5 is combined with bam3 or bam5. We were unable to test the mutations of bam4 or bam1, as a double mutant with sunn-5 could have a suppressive effect; however, bam4 is not expressed in nodulating roots, making suppression unlikely. Since overexpression of BAM2, but not BAM1 or BAM3, increases nodule number in wild-type plants (Figure 5), multiple lines of evidence support a specific role for BAM2 in nodule number regulation.
The data support the action of BAM2 in nodulation as root specific rather than systemic. While SUNN signals from shoot to root to control nodule number (Penmetsa et al., 2003) and is central to AON (reviewed in Roy et al., 2020), SUNN is expressed throughout the plant in the vasculature (Schnabel et al., 2012), and the function of SUNN in the root has not yet been determined. The localization of the suppressor effect of bam2 in the root (Figure 4A) is particularly interesting, as the SUNN protein is a systemic negative regulator of nodule number; sunn-5 mutants hypernodulate because they cannot send a wild-type regulation signal to the roots to control nodule number. Nevertheless, the mutation in bam2 in the roots of a sunn-5 mutant allows the regulation to proceed, suggesting that the lack of bam2 compensates for the lack of the SUNN signal from the shoots (Figure 4B). Further support of a local rather than systemic effect is the grafting experiment in which a bam2 mutant in the shoot but not the root does not suppress the sunn hypernodulation phenotype (Figure 4A). All sunn mutants have normal nodulation if mutant roots are grafted to wild-type shoots (Penmetsa et al., 2003; Schnabel et al., 2010, Figure 4B), suggesting that whatever the function of SUNN in the root, a mutation of sunn in the root is not causal to hypernodulation. However, the addition of a bam2 mutation in the root along with a sunn mutation changes the response to the shooting signal, supporting action locally in the root versus systemic action.
In Arabidopsis, the differentiation of stem cells in the root apical meristem and cell division in the root is controlled by CLE peptides (Willoughby and Nimchuk, 2021). Recent studies have shown that the SUNN ortholog CLV1 signals through a BAM protein complex to control CLE-mediated signaling of the root apical meristem (Wang et al., 2022) and root phloem development (Hu et al., 2022). Nodule primordia use many of the same regulatory genes, as lateral root primordia (Schiessl et al., 2019) contain a nodule meristem and develop vasculature at the time points used to measure gene expression in this work (48–72 hpi in our growth systems), making the involvement of a BAM quite likely.
In our system, both the sunn-5 and bam2 mutants make transcripts detectable by PCR of cDNA, but sunn-5 is not likely to produce protein due to the lack of regulatory sequences around the possible alternate start in sunn-5 (Figure 6), while the disruption of the coding sequence in all of the bam Tnt1 insertion mutants occurs from insertion of the Tnt-1 sequence in the middle of the LRR region, leading to a truncated protein (Supplementary Figure 1). In Arabidopsis, truncated BAM proteins lacking a kinase domain interfere with meristem homeostasis in a dominant negative manner because of the LRR repeats interacting with other proteins (Wang et al., 2018), but this is not observed in our system, suggesting that the insert in the LRR disrupts any dominant negative effect from the lack of a kinase domain.
The expression of BAM2 in wild-type plant roots before nodulation (0 hpi in Figure 2, right panel) is approximately half the expression of BAM2 in sunn-4 mutant roots and rdn1-2 hypernodulation mutant roots before nodulation begins. BAM2 expression rises in roots of all inoculated plants over the 72-hpi time course, but the rise is larger in the hypernodulation mutants sunn-4 and rdn1-2, and there is no reason not to expect the same pattern in sunn-1 and sunn-5 mutants given that the nodule number phenotypes of these mutants are similar to rdn1-2. If increased BAM2 expression is observed in plant roots that hypernodulate, it is logical that overexpression of BAM2 in wild-type roots increases nodule number. More interesting is the observation that adding more BAM2 to sunn mutant roots already expressing higher levels of BAM2 than wild-type decreases nodule number for sunn-1 and sunn-5 mutant roots but not sunn-4 roots. The decrease in the sunn-1 and sunn-5 roots could be explained if the level of BAM2 from the overexpression construct rises high enough to trigger RNA interference, acting like a bam2 mutation, but the lack of suppression in sunn-4 roots is perplexing. Because the expression of BAM2 in these roots was not measured, it is not possible to rule out the explanation that the construct was not correctly expressed. However, the effect was seen in six plants tested in two independent experiments, giving confidence in the result. Since the transformations are performed in hairy roots in which auxin homeostasis is perturbed, it is possible that the extreme hypernodulation phenotype in the null allele is affected more by the hairy root environment than expression of BAM genes, given that the range of nodule numbers in Figure 6 is below the 125 average nodules per plant in sunn-4 mutants on an aeroponic system in all conditions including the empty vector (Schnabel et al., 2010), adding difficulties to interpretation of the sunn-4 results.
It is important to note that overexpression of wild-type BAM2 did not suppress (or enhance) hypernodulation in the crn and rdn1-2 AON mutant roots, even though the rdn1-2 mutant has high BAM2 expression compared to wild type, so the effect of the mutation is not on the AON pathway itself: the suppression seems to be specific to plant roots carrying mutations in SUNN, with sunn-4 as an exception. Relative expression measurements of MtWOX5 add some insight into how the downstream nodulation signaling may be disrupted in bam2 and bam2 sunn-5 mutants. MtWOX5 expression in the nodule apical meristem (NAM) (Osipova et al., 2011; Roux et al., 2014) provides molecular support for the derivation of nodule development programs from lateral root developmental programs (Hirsch and Larue, 1997; Mathesius et al., 2000; Bright et al., 2005; Desbrosses and Stougaard, 2011). WOX5 gene is downstream from the CLE/CLV1/BAM signaling complex in Arabidopsis (Willoughby & Nimchuk, 2021), but other receptor kinases are involved in connecting CLE/CLV1/BAM signaling to WOX5 expression (Wang et al., 2022). The expression of MtWOX5 is increased compared to the wild type at 0 hpi bam2 mutants and bam2 sunn-5 mutants but is the same as the wild type in sunn-4 and rdn1-2 hypernodulation mutants (Figure 8). In contrast to the steady higher expression observed in sunn-4, rdn1-2, and bam2 mutants at 12 and 48 hpi, the bam2 sunn5 mutant decreases MtWOX5 expression below wild-type levels. Rather than eliminating nodule development, the lower expression is correlated with only a reduction of nodule number in sunn-5 mutants. Likewise, even though bam2 mutants have increased MtWOX5 expression, the number of nodules is not affected, suggesting that other factors outside of MtWOX5 are also involved in nodule number regulation.
In the event of genetic or environmental disruption, plants can initiate a genetic buffering mechanism called “active compensation” in which genes change their behavior to compensate for the disruption (Rodriguez-Leal et al., 2019). For example, in Arabidopsis, the weak clv1 phenotype is genetically buffered by the paralogous BAM receptors through active compensation (Diss et al., 2014; Nimchuk et al., 2015). The loss of CLV1 resulted in an increased expression of BAMs and a change in their expression domains, allowing for the compensation of CLV1 loss (Nimchuk et al., 2015). In Solanum lycopersicum, the loss of SlCLV3, the tomato CLV3 ortholog, triggers an active compensation mechanism, by which the upregulation of SlCLE9 buffers stem cell homeostasis in tomato (Rodriguez-Leal et al., 2019). Considering the related distance between Arabidopsis and tomato, genetic buffering of stem cells reflects a determining feature of indeterminate meristem development, including nodule meristems (Rodriguez-Leal et al., 2019). A similar effect may be happening with BAM2 and SUNN.
We speculate that the loss of, or disruption to, the CLE/SUNN/BAM2 signaling in the roots alters signaling, affecting the nodule-specific transcription factor MtWOX5. We propose a genetic model, wherein the specific root interactions of BAM2/SUNN are critical for signaling in nodule meristem cell homeostasis in M. truncatula. Except for the perplexing sunn-4 overexpression results, an anomaly to pursue in future studies, our data support the involvement of CLE/CLV1/BAM signaling influencing nodule number in M. truncatula.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Author contributions
JT: Data curation, Formal analysis, Investigation, Methodology, Writing – original draft, Writing – review & editing. JF: Conceptualization, Formal analysis, Funding acquisition, Methodology, Resources, Supervision, Visualization, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by NSF 1733470 to JF.
Acknowledgments
We thank Elise Schnabel for the identification of the BAM mutant lines in the Tnt1 pools and the Genetics and Biochemistry Teaching Lab for the use of their real-time PCR machine.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1334190/full#supplementary-material
References
Araya, T., von Wirén, N., Takahashi, H. (2016). CLE peptide signaling and nitrogen interactions in plant root development. Plant Mol. Biol. 91 (6), 607–615. doi: 10.1007/s11103-016-0472-9
Bekki, A., Trinchant, J.-C., Rigaud, J. (1987). Nitrogen fixation (C2H2 reduction) by Medicago nodules and bacteroids under sodium chloride stress. Physiologia Plantarum 71 (1), 61–67. doi: 10.1111/j.1399-3054.1987.tb04617.x
Benedito, V. A., Torres-Jerez, I., Murray, J. D., Andriankaja, A., Allen, S., Kakar, K., et al. (2008). A gene expression atlas of the model legume. Medicago truncatula. Plant J. 55 (3), 504–513. doi: 10.1111/j.1365-313X.2008.03519.x
Bright, L. J., Liang, Y., Mitchell, D. M., Harris, J. M. (2005). The LATD gene of Medicago truncatula is required for both nodule and root development. Mol. Plant-Microbe Interact. 18 (6), 521–532. doi: 10.1094/MPMI-18-0521
Cai, J., Veerappan, V., Arildsen, K., Sullivan, C., Piechowicz, M., Frugoli, J., et al. (2023). A modified aeroponic system for growing small-seeded legumes and other plants to study root systems. Plant Methods 19 (1), 21. doi: 10.1186/s13007-023-01000-6
Chaulagain, D., Frugoli, J. (2021). The regulation of nodule number in legumes is a balance of three signal transduction pathways. Int. J. Mol. Sci. 22 (3), 1117. doi: 10.3390/ijms22031117
Crook, A. D., Schnabel, E. L., Frugoli, J. A. (2016). The systemic nodule number regulation kinase SUNN in Medicago truncatula interacts with MtCLV2 and MtCRN. Plant J. 88 (1), 108–119. doi: 10.1111/tpj.13234
Desbrosses, G. J., Stougaard, J. (2011). Root nodulation: a paradigm for how plant-microbe symbiosis influences host developmental pathways. Cell Host Microbe 10 (4), 348–358. doi: 10.1016/j.chom.2011.09.005
DeYoung, B. J., Bickle, K. L., Schrage, K. J., Muskett, P., Patel, K., Clark, S. E. (2006). The CLAVATA1-related BAM1, BAM2 and BAM3 receptor kinase-like proteins are required for meristem function in Arabidopsis. Plant J. 45 (1), 1–16. doi: 10.1111/j.1365-313X.2005.02592.x
DeYoung, B. J., Clark, S. E. (2008). BAM receptors regulate stem cell specification and organ development through complex interactions with CLAVATA signaling. Genetics 180 (2), 895. doi: 10.1534/genetics.108.091108
Diss, G., Ascencio, D., DeLuna, A., Landry, C. R. (2014). Molecular mechanisms of paralogous compensation and the robustness of cellular networks. J. Exp. Zoology Part B: Mol. Dev. Evol. 322 (7), 488–499. doi: 10.1002/jez.b.22555
El-Heba, G. A. A., Hafez, A., Elsahly, N., Hussien, G. M. (2015). supn, a novel supernodulation mutant in Medicago truncatula. Plant Gene 4, 100–108. doi: 10.1016/j.plgene.2015.07.002
Ferguson, B. J., Mens, C., Hastwell, A. H., Zhang, M., Su, H., Jones, C. H., et al. (2019). Legume nodulation: The host controls the party. Plant Cell Environ. 42 (1), 41–51. doi: 10.1111/pce.13348
Garmier, M., Gentzbittel, L., Wen, J., Mysore, K. S., Ratet, P. (2017). Medicago truncatula: genetic and genomic resources. Curr. Protoc. Plant Biol. 2 (4), 318–349. doi: 10.1002/cppb.20058
Gautrat, P., Mortier, V., Laffont, C., De Keyser, A., Fromentin, J., Frugier, F., et al. (2019). Unraveling new molecular players involved in the autoregulation of nodulation in Medicago truncatula. J. Exp. Bot. 70 (4), 1407–1417. doi: 10.1093/jxb/ery465
Hastwell, A. H., de Bang, T. C., Gresshoff, P. M., Ferguson, B. J. (2017). CLE peptide-encoding gene families in Medicago truncatula and Lotus japonicus, compared with those of soybean, common bean and Arabidopsis. Sci. Rep. 7 (1), 1–13. doi: 10.1038/s41598-017-09296-w
Hirsch, A. M., Larue, T. A. (1997). Is the Legume Nodule a Modified Root or Stem or an Organ sui generis? Crit. Rev. Plant Sci. 16 (4), 361–392. doi: 10.1080/07352689709701954
Hoffmann, B., Trinh, T. H., Leung, J., Kondorosi, A., Kondorosi, E. (1997). A new Medicago truncatula line with superior in vitro regeneration, transformation, and symbiotic properties isolated through cell culture selection. Mol. Plant-Microbe Interact. 10 (3), 307–315. doi: 10.1094/MPMI.1997.10.3.307
Hu, C., Zhu, Y., Cui, Y., Zeng, L., Li, S., Meng, F., et al. (2022). A CLE–BAM–CIK signalling module controls root protophloem differentiation in Arabidopsis. New Phytol. 233 (1), 282–296. doi: 10.1111/nph.17791
Huault, E., Laffont, C., Wen, J., Mysore, K. S., Ratet, P., Duc, G., et al. (2014). Local and systemic regulation of plant root system architecture and symbiotic nodulation by a receptor-like kinase. PloS Genet. 10 (12), e1004891. doi: 10.1371/journal.pgen.1004891
Huo, X., Schnabel, E., Hughes, K., Frugoli, J. (2006). RNAi phenotypes and the localization of a protein::GUS fusion imply a role for Medicago truncatula PIN genes in nodulation. J. Plant Growth Regul. 25 (2), 156–165. doi: 10.1007/s00344-005-0106-y
Kakar, K., Wandrey, M., Czechowski, T., Gaertner, T., Scheible, W.-R., Stitt, M., et al. (2008). A community resource for high-throughput quantitative RT-PCR analysis of transcription factor gene expression in. Medicago truncatula. Plant Methods 4 (1), 1–12. doi: 10.1186/1746-4811-4-18
Karimi, M., Inzé, D., Depicker, A. (2002). GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 7 (5), 193–195. doi: 10.1016/s1360-1385(02)02251-3
Kassaw, T. K., Frugoli, J. A. (2012). Simple and efficient methods to generate split roots and grafted plants useful for long-distance signaling studies in Medicago truncatula and other small plants. Plant Methods 8 (1), 38. doi: 10.1186/1746-4811-8-38
Kassaw, T., Nowak, S., Schnabel, E., Frugoli, J. (2017). Root determined nodulation1 is required for M. truncatula CLE12, but not CLE13, peptide signaling through the SUNN receptor kinase. Plant Physiol. 174 (4), 2445–2456. doi: 10.1104/pp.17.00278
Kinoshita, A., Betsuyaku, S., Osakabe, Y., Mizuno, S., Nagawa, S., Stahl, Y., et al. (2010). RPK2 is an essential receptor-like kinase that transmits the CLV3 signal in Arabidopsis. Development 137 (22), 3911–3920. doi: 10.1242/dev.048199
Krusell, L., Madsen, L. H., Sato, S., Aubert, G., Genua, A., Szczyglowski, K., et al. (2002). Shoot control of root development and nodulation is mediated by a receptor-like kinase. Nature 420 (6914), 422–426. doi: 10.1038/nature01207
Krusell, L., Sato, N., Fukuhara, I., Koch, B. E. V., Grossmann, C., Okamoto, S., et al. (2011). The Clavata2 genes of pea and Lotus japonicus affect autoregulation of nodulation. Plant J. 65 (6), 861–871. doi: 10.1111/j.1365-313X.2010.04474.x
Kumar, S., Stecher, G., Li, M., Knyaz, C., Tamura, K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35 (6), 1547–1549. doi: 10.1093/molbev/msy096
Laffont, C., Huault, E., Gautrat, P., Endre, G., Kalo, P., Bourion, V., et al. (2019). Independent regulation of symbiotic nodulation by the SUNN negative and CRA2 positive systemic pathways. Plant Physiol. 180(1) 559 LP –, 570. doi: 10.1104/pp.18.01588
Leong, S. A., Williams, P. H., Ditta, G. S. (1985). Analysis of the 5′ regulatory region of the gene for δ-aminolevulinic acid synthetase of. Rhizobium meliloti. Nucleic Acids Res. 13 (16), 5965–5976. doi: 10.1093/nar/13.16.5965
Libbenga, K. R., Harkes, P. A. A. (1973). Initial proliferation of cortical cells in the formation of root nodules in Pisum sativum L. Planta 114, 17–28. doi: 10.1007/BF00390281
Limpens, E., Ramos, J., Franken, C., Raz, V., Compaan, B., Franssen, H., et al. (2004). RNA interference in Agrobacterium rhizogenes-transformed roots of Arabidopsis and Medicago truncatula. J. Exp. Bot. 55 (399), 983–992. doi: 10.1093/jxb/erh122
Luyten, E., Vanderleyden, J. (2000). Survey of genes identified in Sinorhizobium meliloti spp., necessary for the development of an efficient symbiosis. Eur. J. Soil Biol. 36 (1), 1–26. doi: 10.1016/S1164-5563(00)00134-5
Magori, S., Oka-Kira, E., Shibata, S., Umehara, Y., Kouchi, H., Hase, Y., et al. (2009). TOO MUCH LOVE, a root regulator associated with the long-distance control of nodulation in Lotus japonicus. Mol. Plant-Microbe Interact. 22 (3), 259–268. doi: 10.1094/MPMI-22-3-0259
Mathesius, U., Weinman, J. J., Rolfe, B. G., Djordjevic, M. A. (2000). Rhizobia can induce nodules in white clover by “hijacking” mature cortical cells activated during lateral root development. Mol. Plant-Microbe Interact. 13 (2), 170–182. doi: 10.1094/MPMI.2000.13.2.170
Meneely, P. (2020). Genetic analysis: genes, genomes, and networks in eukaryotes (USA: Oxford University Press).
Miyazawa, H., Oka-Kira, E., Sato, N., Takahashi, H., Wu, G.-J., Sato, S., et al. (2010). The receptor-like kinase KLAVIER mediates systemic regulation of nodulation and non-symbiotic shoot development in Lotus japonicus. Development 137, 24, 4317 LP–4325. doi: 10.1242/dev.058891
Mortier, V., Den Herder, G., Whitford, R., Van de Velde, W., Rombauts, S., D’haeseleer, K., et al. (2010). CLE peptides control Medicago truncatula nodulation locally and systemically. Plant Physiol. 153 (1), 222 LP–237. doi: 10.1104/pp.110.153718
Nimchuk, Z. L., Zhou, Y., Tarr, P. T., Peterson, B. A., Meyerowitz, E. M. (2015). Plant stem cell maintenance by transcriptional cross-regulation of related receptor kinases. Development 142 (6), 1043 LP–1049. doi: 10.1242/dev.119677
Nishida, H., Handa, Y., Tanaka, S., Suzaki, T., Kawaguchi, M. (2016). Expression of the CLE-RS3 gene suppresses root nodulation in. Lotus japonicus. J. Plant Res. 129 (5), 909–919. doi: 10.1007/s10265-016-0842-z
Nowak, S., Schnabel, E., Frugoli, J. (2019). The Medicago truncatula CLAVATA3-LIKE CLE12/13 signaling peptides regulate nodule number depending on the CORYNE but not the COMPACT ROOT ARCHITECTURE2 receptor. Plant Signaling Behav. 14 (6), 1598730. doi: 10.1080/15592324.2019.1598730
Oka-Kira, E., Tateno, K., Miura, K., Haga, T., Hayashi, M., Harada, K., et al. (2005). klavier (klv), a novel hypernodulation mutant of Lotus japonicus affected in vascular tissue organization and floral induction. Plant J. 44 (3), 505–515. doi: 10.1111/j.1365-313X.2005.02543.x
Okamoto, S., Ohnishi, E., Sato, S., Takahashi, H., Nakazono, M., Tabata, S., et al. (2009). Nod factor/nitrate-induced CLE genes that drive HAR1-mediated systemic regulation of nodulation. Plant Cell Physiol. 50 (1), 67–77. doi: 10.1093/pcp/pcn194
Osipova, M. A., Dolgikh, E. A., Lutova, L. A. (2011). Peculiarities of meristem-specific WOX5 gene expression during nodule organogenesis in legumes. Russian J. Dev. Biol. 42 (4), 226. doi: 10.1134/S1062360411010085
Osipova, M. A., Mortier, V., Demchenko, K. N., Tsyganov, V. E., Tikhonovich, I. A., Lutova, L. A., et al. (2012). WUSCHEL-RELATED HOMEOBOX5 gene expression and interaction of CLE peptides with components of the systemic control add two pieces to the puzzle of autoregulation of nodulation. Plant Physiol. 158 (3), 1329–1341. doi: 10.1104/pp.111.188078
Penmetsa, R. V., Frugoli, J. A., Smith, L. S., Long, S. R., Cook, D. R. (2003). Dual genetic pathways controlling nodule number in Medicago truncatula. Plant Physiol. 131 (3), 998 LP–1008. doi: 10.1104/pp.015677
Pfaffl, M. W. (2001). A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29 (9), e45. doi: 10.1093/nar/29.9.e45
Quandt, J., Hynes, M. F. (1993). Versatile suicide vectors which allow direct selection for gene replacement in gram-negative bacteria. Gene 127 (1), 15–21. doi: 10.1016/0378-1119(93)90611-6
Rodriguez-Leal, D., Xu, C., Kwon, C.-T., Soyars, C., Demesa-Arevalo, E., Man, J., et al. (2019). Evolution of buffering in a genetic circuit controlling plant stem cell proliferation. Nat. Genet. 51 (5), 786–792. doi: 10.1038/s41588-019-0389-8
Roux, B., Rodde, N., Jardinaud, M., Timmers, T., Sauviac, L., Cottret, L., et al. (2014). An integrated analysis of plant and bacterial gene expression in symbiotic root nodules using laser-capture microdissection coupled to RNA sequencing. Plant J. 77 (6), 817–837. doi: 10.1111/tpj.12442
Roy, S., Liu, W., Nandety, R. S., Crook, A., Mysore, K. S., Pislariu, C. I., et al. (2020). Celebrating 20 years of genetic discoveries in legume nodulation and symbiotic nitrogen fixation. Plant Cell 32 (1), 15 LP–15 41. doi: 10.1105/tpc.19.00279
Schiessl, K., Lilley, J. L., Lee, T., Tamvakis, I., Kohlen, W., Bailey, P. C., et al. (2019). NODULE INCEPTION recruits the lateral root developmental program for symbiotic nodule organogenesis in Medicago truncatula. Curr. Biol. 29 (21), 3657–3668. doi: 10.1016/j.cub.2019.09.005
Schnabel, E. L., Chavan, S. A., Gao, Y., Poehlman, W. L., Feltus, F. A., Frugoli, J. A. (2023a). A Medicago truncatula autoregulation of nodulation mutant transcriptome analysis reveals disruption of the SUNN pathway causes constitutive expression changes in some genes, but overall response to Rhizobia resembles wild-type, including induction of TML1 and TML2. Curr. Issues Mol. Biol. 45 (6), 4612–4631. doi: 10.3390/cimb45060293
Schnabel, E., Journet, E.-P., de Carvalho-Niebel, F., Duc, G., Frugoli, J. (2005). The Medicago truncatula SUNN gene encodes a CLV1-like leucine-rich repeat receptor kinase that regulates nodule number and root length. Plant Mol. Biol. 58 (6), 809–822. doi: 10.1007/s11103-005-8102-y
Schnabel, E., Karve, A., Kassaw, T., Mukherjee, A., Zhou, X., Hall, T., et al. (2012). The M. truncatula SUNN gene is expressed in vascular tissue, similarly to RDN1, consistent with the role of these nodulation regulation genes in long distance signaling. Plant Signaling Behav. 7, 1–3. doi: 10.4161/psb.7.1.18491
Schnabel, E. L., Kassaw, T. K., Smith, L. S., Marsh, J. F., Oldroyd, G. E., Long, S. R., et al. (2011). The ROOT DETERMINED NODULATION gene regulates nodule number in roots of Medicago truncatula and defines a highly conserved, uncharacterized plant gene family. Plant Physiol. 157 (1), 328–340. doi: 10.1104/pp.111.178756
Schnabel, E., Mukherjee, A., Smith, L., Kassaw, T., Long, S., Frugoli, J. (2010). The lss supernodulation mutant of Medicago truncatula reduces expression of the SUNN gene. Plant Physiol. 154 (3), 1390–1402. doi: 10.1104/pp.110.164889
Schnabel, E., Thomas, J., El-Hawaz, R., Gao, Y., Poehlman, W., Chavan, S., et al. (2023b). Laser capture microdissection transcriptome reveals spatiotemporal tissue gene expression patterns of M. truncatula roots responding to rhizobia. Mol. Plant-Microbe Interact 36 (12), 805–820. doi: 10.1094/MPMI-03-23-0029-R
Searle, I. R., Men, A. E., Laniya, T. S., Buzas, D. M., Iturbe-Ormaetxe, I., Carroll, B. J., et al. (2003). Long-distance signaling in nodulation directed by a CLAVATA1-like receptor kinase. Science 299 (5603), 109–112. doi: 10.1126/science.107793
Simionatto, S., Marchioro, S. B., Galli, V., Luerce, T. D., Hartwig, D. D., Moreira, Â.N., et al. (2009). Efficient site-directed mutagenesis using an overlap extension-PCR method for expressing Mycoplasma hyopneumoniae genes in Escherichia coli. J. Microbiological Methods 79 (1), 101–105. doi: 10.1016/j.mimet.2009.08.016
Tadege, M., Wen, J., He, J., Tu, H., Kwak, Y., Eschstruth, A., et al. (2008). Large-scale insertional mutagenesis using the Tnt1 retrotransposon in the model legume Medicago truncatula. Plant J. 54 (2), 335–347. doi: 10.1111/j.1365-313X.2008.03418.x
Takahara, M., Magori, S., Soyano, T., Okamoto, S., Yoshida, C., Yano, K., et al. (2013). TOO MUCH LOVE, a novel kelch repeat-containing F-box protein, functions in the long-distance regulation of the legume–Rhizobium symbiosis. Plant Cell Physiol. 54 (4), 433–447. doi: 10.1093/pcp/pct022
van Noorden, G. E., Ross, J. J., Reid, J. B., Rolfe, B. G., Mathesius, U. (2006). Defective long-distance auxin transport regulation in the Medicago truncatula super numeric nodules mutant. Plant Physiol. 140 (4), 1494–1506. doi: 10.1104/pp.105.075879
Veerappan, V., Kadel, K., Alexis, N., Scott, A., Kryvoruchko, I., Sinharoy, S., et al. (2014). Keel petal incision: a simple and efficient method for genetic crossing in Medicago truncatula. Plant Methods 10 (1), 11. doi: 10.1186/1746-4811-10-11
Waese, J., Fan, J., Pasha, A., Yu, H., Fucile, G., Shi, R., et al. (2017). ePlant: visualizing and exploring multiple levels of data for hypothesis generation in plant biology. Plant Cell 29 (8), 1806–1821. doi: 10.1105/tpc.17.00073
Wang, W., Hu, C., Li, X., Zhu, Y., Tao, L., Cui, Y., et al. (2022). Receptor-like cytoplasmic kinases PBL34/35/36 are required for CLE peptide-mediated signaling to maintain shoot apical meristem and root apical meristem homeostasis in Arabidopsis. Plant Cell. 34, 4, 1289–1307. doi: 10.1093/plcell/koab315
Wang, C., Yang, H., Chen, L., Yang, S., Hua, D., Wang, J. (2018). Truncated BAM receptors interfere the apical meristematic activity in a dominant negative manner when ectopically expressed in Arabidopsis. Plant Sci. 269, 20–31. doi: 10.1016/j.plantsci.2018.01.003
Willoughby, A. C., Nimchuk, Z. L. (2021). WOX going on: CLE peptides in plant development. Curr. Opin. Plant Biol. 163, 102056. doi: 10.1016/j.pbi.2021.102056
Keywords: nodulation, meristems, autoregulation of nodulation, M. truncatula, BAM, SUNN
Citation: Thomas J and Frugoli J (2024) Mutation of BAM2 rescues the sunn hypernodulation phenotype in Medicago truncatula, suggesting that a signaling pathway like CLV1/BAM in Arabidopsis affects nodule number. Front. Plant Sci. 14:1334190. doi: 10.3389/fpls.2023.1334190
Received: 06 November 2023; Accepted: 18 December 2023;
Published: 11 January 2024.
Edited by:
Senjuti Sinharoy, National Institute of Plant Genome Research (NIPGR), IndiaReviewed by:
Anindya Kundu, National Institute of Agricultural Botany (NIAB), United KingdomOswaldo Valdes-Lopez, National Autonomous University of Mexico, Mexico
Copyright © 2024 Thomas and Frugoli. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Julia Frugoli, amZydWdvbEBjbGVtc29uLmVkdQ==
 Jacklyn Thomas
Jacklyn Thomas Julia Frugoli
Julia Frugoli