- 1Zhengzhou Fruit Research Institute, Chinese Academy of Agricultural Sciences,Henan Key Laboratory of Fruit and Cucurbit Biology, Zhengzhou Henan, China
- 2Key Lab of Plant Pathology of Hubei Province, College of Plant Science and Technology, Huazhong Agricultural University, Wuhan, China
- 3Zhongyuan Research Center, Chinese Academy of Agricultural Sciences, Xinxiang, Henan, China
- 4Institute of Plant Protection, Xinjiang Academy of Agricultural Sciences, Xinjiang, China
Introduction: Ethylene (ET) is involved in plant responses to viral infection. However, its molecular mechanisms and regulatory network remain largely unknown.
Methods and results: In the present study, we report that cucumber green mottle mosaic virus (CGMMV) in watermelon (Citrullus lanatus) triggers ET production by inducing the expression of ClACO5, a key gene of the ET biosynthesis pathway through transcriptome data analysis and gene function validation. The knock-down of ClACO5 expression through virus-induced gene silencing in watermelon and overexpressing ClACO5 in transgenic Nicotiana benthamiana indicated that ClACO5 positively regulates CGMMV resistance and ET biosynthesis. The salicylic acid-responsive transcription factor gene ClWRKY70 shares a similar expression pattern with ClACO5. We demonstrate that ClWRKY70 directly binds to the W-box cis-element in the ClACO5 promoter and enhances its transcription. In addition, ClWRKY70 enhances plant responses to CGMMV infection by regulating ClACO5 expression in watermelon.
Discussion: Our results demonstrate that the ClWRKY70-ClACO5 module positively regulates resistance to CGMMV infection in watermelon, shedding new light on the molecular basis of ET accumulation in watermelon in response to CGMMV infection.
Introduction
Plants constantly face challenges from various biotic stressors, including viruses, that affect plant growth and development and cause crop losses, posing serious threats to food security (Calil and Fontes, 2017). Upon infections, the physiological and biochemical changes in the host plant are triggered in virus-host interactions. These alterations might lead to plant exhibition of disease symptoms such as mottling, a mosaic, wrinkling of leaves, plant dwarfing, and fruit decay (Mishra et al., 2020). To counter defense against viruses, plants have evolved complex, delicate defense systems to restrict viral infection, including gene silencing, regulation of metabolism, and phytohormone-mediated defense (Incarbone and Dunoyer, 2013). Among them, the accumulation of plant hormones such as salicylic acid (SA), jasmonate (JA), ethylene (ET), and abscisic acid can increase plant resistance to viral infection (Zhu et al., 2014a; Casteel et al., 2015; Alazem and Lin, 2017; Zhang et al., 2017).
In plant development, ET participates in a series of physiological processes, such as seed germination and plant senescence, as well as defense against necrotrophic pathogens (Pieterse et al., 2012; Zhao et al., 2017). While in terms of virus defense, ET could be both a positive and a negative factor. For example, the rice dwarf virus (RDV) increases ET production by interacting with OsSAMS1, an essential component of the ET biosynthesis, resulting in an increase in the enzyme’s activity, thereby increasing susceptibility to RDV (Zhao et al., 2017). Among ET pathway mutants in Arabidopsis, ethylene insensitive 2 (ein2) and ethylene response 1 (etr1) show improved resistance to cauliflower mosaic virus (CaMV) infection, and 1-aminocyclopropane-1-carboxylate synthase (acs6) and ethylene-responsive transcription factor 104 (erf106) show enhanced resistance to tobacco mosaic virus (TMV-cg) infection in crucifers (Love et al., 2007; Chen et al., 2013). By contrast, manipulation of JA and ET signaling in tobacco carrying the resistance gene N conveyed systemic resistance to chilli veinal mottle virus (Zhu et al., 2014a).
ET is synthesized via a two-step reaction: first, ACC synthase (ACS) converts S-adenosylmethionine (SAM) into 1-aminocyclopropane-1-carboxylic acid (ACC), which is then oxidized by ACC oxidase (ACO) to produce ET (Yang and Hoffman, 2003). ACO is a member of the DOXC subclass of the plant 2-oxoglutarate-dependent dioxygenase superfamily (Kawai et al., 2014). ACO genes belong to multigene families in most plants (Booker and DeLong, 2015; Houben and Van de Poel, 2019). Only a few ACOs have been proven to play a role in ET biosynthesis and plant responses to pathogen infection (Kim et al., 1998; Yim et al., 2014; Zeilmaker et al., 2015; Houben and Van de Poel, 2019). ACO genes are under tight regulation and show tissue-specific expression and localization patterns (Blume and Grierson, 1997; Van de Poel et al., 2012; Pattyn et al., 2021). However, only a few transcription factors (TFs) that modulate ACO expression have thus far been identified (Houben and Van de Poel, 2019).
The host gene expressions are altered at transcription to posttranslational level in plant immunity responses at any stage of development. Environmental and/or endogenous cues might activate TFs that bind to the regulatory regions of their target genes, thereby enhancing or inhibiting their transcription (Fan et al., 2014; Chen et al., 2017; Chen and Penfield, 2018; Chen et al., 2018; Zhou et al., 2018; Liu et al., 2021). One of the largest plant TF families, WRKY TFs, is crucial for plant immunity (Kalde et al., 2003; Zhou et al., 2018). WRKY TFs include at least one conserved WRKY domain that binds to W-box motifs in their target genes’ promoters (Hu et al., 2013). For example, WRKY72 increases the susceptibility of rice (Oryza sativa) to bacterial blight by binding to WRKY-binding motifs in the promoter of AOS1 and inhibiting its expression, reducing JA levels (Hou et al., 2019). WRKY8 directly binds to the W-box cis-element in their promoters, regulating the expression of ACS6, ABI4, and ERF104, thus participating in the defense response against TMV-cg (Chen et al., 2013). WRKY70 functions in SA-dependent defense and as a repressor of JA-regulated genes and enhances host defense responses against pathogen infection by activating the expression of many defense-related genes, including PR1, PR2, PR5, and SARD1 (Li et al., 2004). Notably, a link between WRKY70, ACO, and ET accumulation has not been identified, although they are individually associated with plant responses to pathogens.
As an important horticultural crop globally, watermelon (Citrullus lanatus) is vulnerable to a variety of viral diseases. For example, the cucumber green mottle mosaic virus (CGMMV), a species in the genus Tobamovirus, seriously affects watermelon yields and quality (Dombrovsky et al., 2017). No commercial CGMMV-resistant cultivars are currently available. Moreover, little is known about CGMMV resistance genes in watermelons (Cai et al., 2023). Identifying CGMMV-responsive genes might allow for the exploration of the molecular mechanisms underlying CGMMV resistance in watermelon plants, which benefits the molecular breeding of watermelons.
We previously performed transcriptome sequencing to explore the global transcriptome of watermelon leaves systemically infected with CGMMV and found a few differentially expressed genes (DEGs) related to various metabolic pathways (Liu et al., 2023a). In this study, we identified ClACO5, an ACO gene of watermelon, and evaluated its role in ET synthesis and CGMMV resistance. Watermelon WRKY70 (ClWRKY70) promotes its transcription by directly binding to the ClACO5 promoter. In addition, ClWRKY70 expression is regulated by SA, and ClWRKY70 positively regulates plant defense against CGMMV. Hence, we identified a ClWRKY70-ClACO5 regulatory module that governs CGMMV-induced ClACO5 upregulation and ET biosynthesis. This finding reveals the molecular basis of ET accumulation in the response of watermelon to CGMMV infection.
Results
CGMMV induces ET accumulation, and ET treatment enhances CGMMV resistance in watermelon
We previously showed that biosynthesis of the ET precursor ACC is inhibited by CGMMV infection in a susceptible watermelon variety, enhancing its susceptibility to viral infection (Liu et al., 2023a). To explore the role of the ET pathway in plant defense against CGMMV infection, we measured the concentrations of ACC and ET in mock- and CGMMV-infected plants of a susceptible watermelon variety (Zhengkang No. 2) at 10 days post-inoculation (dpi) (Figures 1A, B). At this stage, we observed typical symptoms, including mottle and mosaic patterns on leaves, in CGMMV-infected watermelons. In addition, CGMMV RNAs accumulated to high levels in these plants (Liu et al., 2023a). ACC levels in CGMMV-infected plants were about half of those in mock-inoculated plants (Figure 1A), whereas ET levels were approximately twice as high in CGMMV-infected as in mock-inoculated plants (Figure 1B).
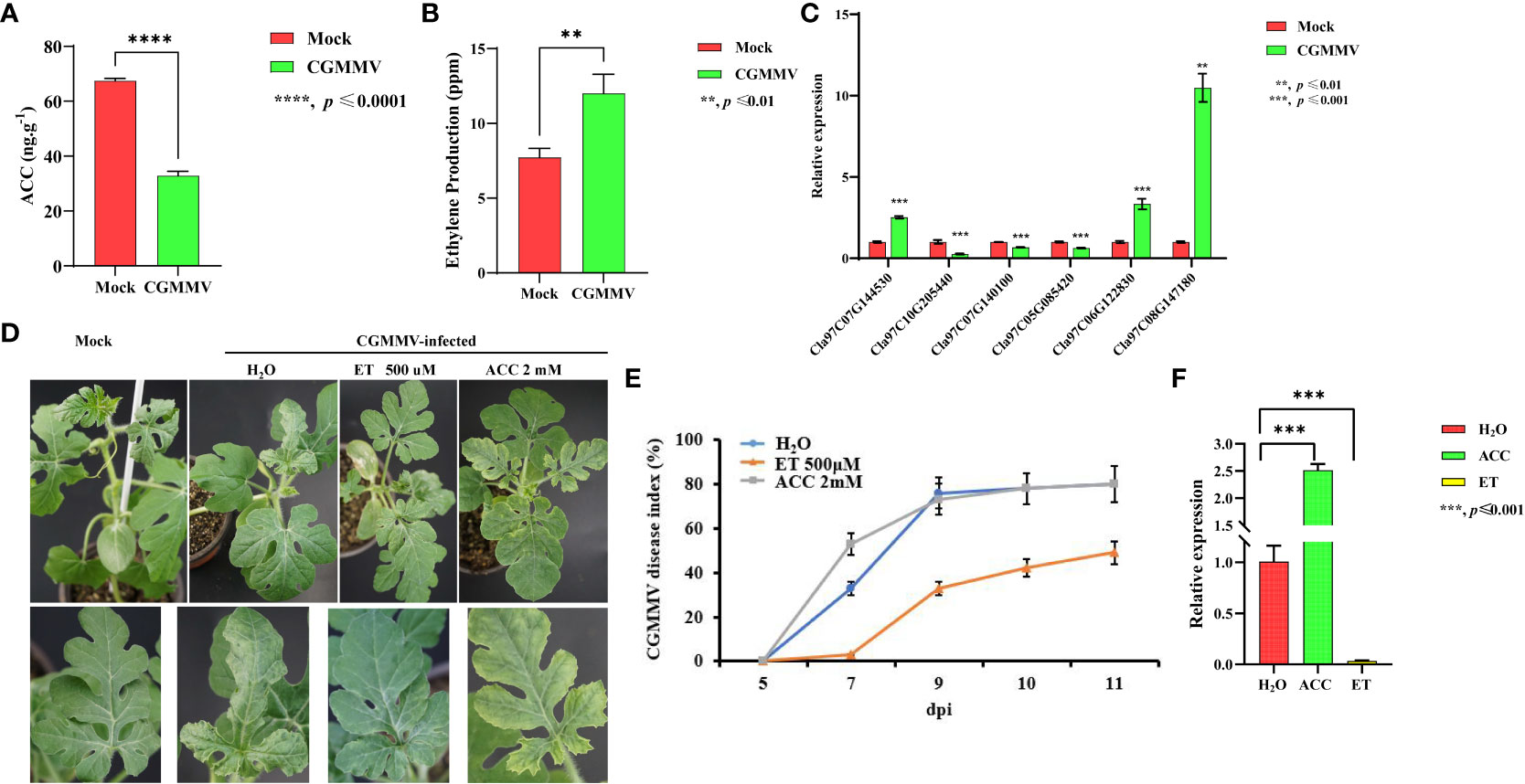
Figure 1 CGMMV induces ET accumulation, and exogenous ET enhances the resistance of watermelon plants to CGMMV. (A, B) ACC (A) and ET (B) levels in mock- and CGMMV-treated watermelon leaves. (C) The transcript levels of ET pathway genes in mock-inoculated or CGMMV-infected watermelon leaves at 10 dpi by RT-qPCR. (D) Phenotypes of 7-day-old seedlings treated with water, ET (500 μM), or ACC (2 mM) before CGMMV treatment. (E) CGMMV disease indices of mock-inoculated or CGMMV-infected watermelon plants at different time points. (F) RT-qPCR of CGMMV CP accumulation in water-, ET (500 μM)-, or ACC (2 mM)-treated watermelon plants. Plants were collected at 10 dpi. Values are means ± SD (n = 3). **p ≤ 0.01, ***p ≤ 0.001 (Student’s t-test).
We wondered whether ACC and ET play opposite roles in antiviral defense in watermelons. To address this question, we sprayed watermelon leaves with ACC (2 mM), ET (500 µM), or H2O (as a control) 72 h before inoculation of CGMMV. At 10 dpi, ACC-treated plants were more susceptible to CGMMV infection, displaying more severe disease symptoms, higher disease indices, and higher levels of viral accumulation than H2O-treated plants (Figures 1D-F). However, ET-treated plants showed that resistance to CGMMV was enhanced, with the exhibition of milder disease symptoms, lower disease indices, and lower levels of virus accumulation, compared to H2O-treated plants (Figures 1D-F). These results undoubtedly suggest that ACC increases the susceptibility of watermelon to CGMMV infection, whereas ET enhances its resistance to this virus.
Identification and analysis of ClACO5
To identify the genes of watermelon responsible for ET accumulation upon CGMMV infection, we analyzed the gene expression profiles of mock- and CGMMV-infected watermelon seedlings by RNA sequencing (RNA-Seq) (Liu et al., 2023a). The expression levels of >1,200 genes significantly altered upon CGMMV infection (Liu et al., 2023a). Gene Ontology (GO) analysis showed that these DEGs were enriched for biological processes in the ET response category (Supplementary Figure S1A). In addition, the expression levels of approximately two-thirds of the 30 ET-related genes were altered in CGMMV-infected watermelons, including genes encoding ET synthases (ACO genes), ET receptors (ETR genes), and ET-responsive TFs (ERF genes) (Supplementary Figure S1B). Specifically, the expression of most ERF, ACO, and ETR genes was inhibited (Figure 1C and Supplementary Figure S1B), whereas the expression of only one ACO (Cla97C07G144530) and two ERF genes (Cla97C06G122830 and Cla97C08G147180) was induced by CGMMV infection. Quantitative reverse-transcription PCR (RT-qPCR) further confirmed the results of RNA-Seq data (Figure 1C). All those results suggest that these ET-related genes might contribute to antiviral defense responses in watermelons.
Three DEGs annotated as ACO genes displayed marked changes in expression (Figure 1C), with only one (Cla97C07G144530) being upregulated upon CGMMV infection. We, therefore, explored whether this gene might be related to the decrease of ACC content and increase of ET content upon CGMMV infection. Sequence analysis showed that the cDNA amplified from the contig included a 1,038-bp open reading frame encoding a protein of 345 amino acids with a predicted molecular mass of 39.7 kDa and an isoelectric point of 6.07. BLAST analysis and sequence alignment of Cla97C07G144530 with its homologs revealed that its predicted amino acid sequence shares 87.54%, and 65.31% amino acid identity with CsACO5 (1-aminocyclopropane-1-carboxylate oxidase 5, XP_004137205) in cucumber (Cucumis sativus) and AtDMR6 (AT5G24530.1) in Arabidopsis thaliana (Supplementary Figure S2), respectively. We named the Cla97C07G144530 gene as ClACO5.
ClACO5 localizes to the cytoplasm and nucleus
Bioinformatics prediction revealed that ClACO5 might be localized to the cytoplasm (Supplementary Figure S3). To confirm this prediction, 35S-ClACO5-GFP or 35S-GFP was co-transformed with a nuclear marker protein H2B (35S-H2B-mCherry) or a membrane marker protein OsMCA1 (35S-OsMCA1-mCherry) into Nicotiana benthamiana leaves. The GFP signals in leaf cells expressing 35S-ClACO5-GFP or 35S-GFP completely overlapped with the signals of mCherry (Figure 2), suggesting that ClACO5 was localized in the cytoplasm and nucleus of N. benthamiana.
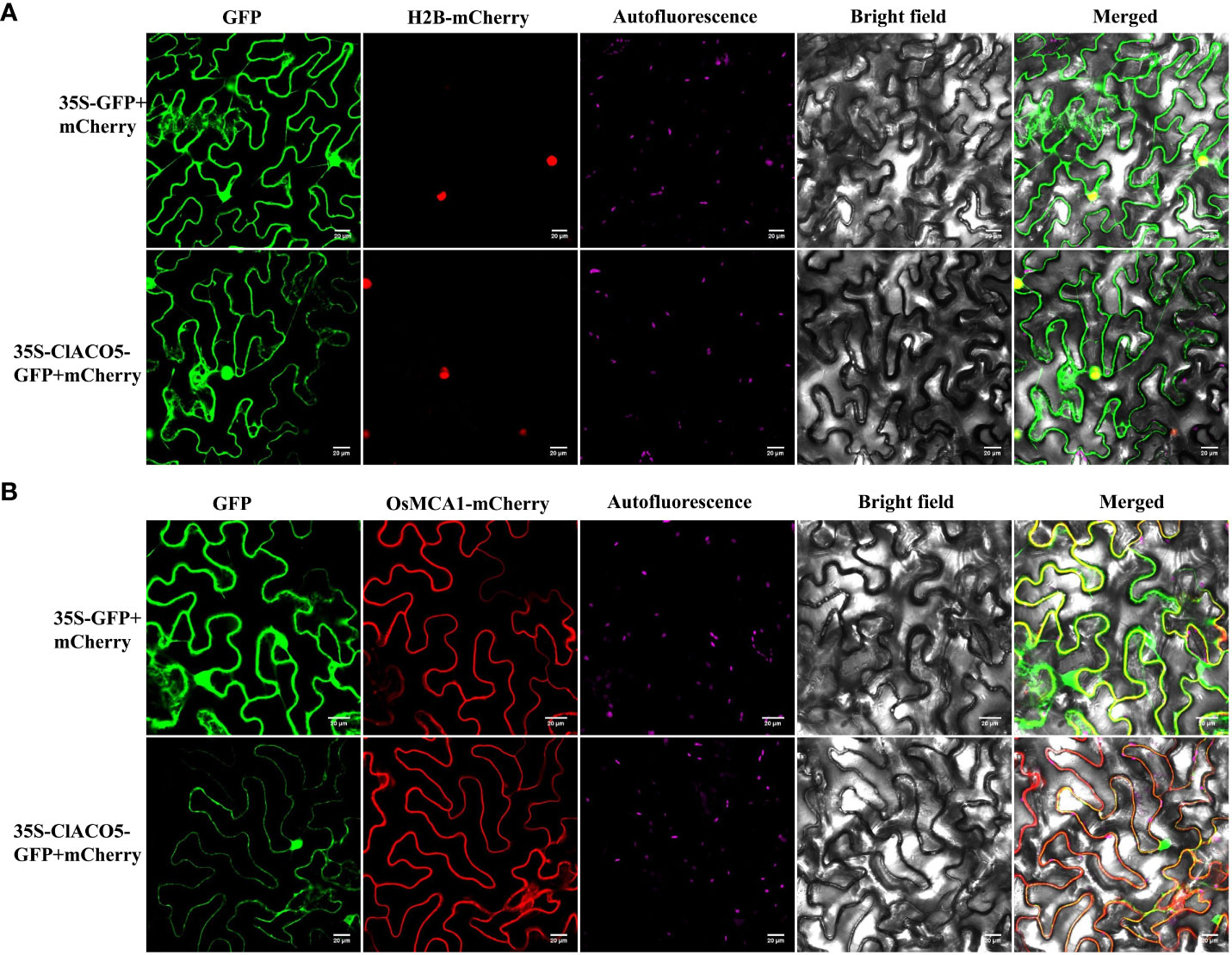
Figure 2 Subcellular localization of ClACO5 protein. Control vector (35S-GFP) or ClACO5-GFP fusion protein (35S-ClACO5-GFP) were co-transformed with a nuclear marker protein H2B (A) and membrane marker protein OsMCA1 (B) fused to mCherry (red fluorescent protein, 35S-H2B-mCherry or 35S- OsMCA1-mCherry) in tobacco (Nicotiana benthamiana) leaves. Bars, 20 μm.
ClACO5 positively contributes to CGMMV resistance
We generated ClACO5-knockdown watermelon plants (pV190-ClACO5) via agroinfiltration by our previously constructed pV190-based virus-induced gene silencing (VIGS) system (Figure 3A). ClACO5 expression levels of silenced plants were only 23% of that in control plants at 20 dpi (Figure 3B), as well as demonstrated less ET accumulation than controls (Figure 3C). To examine the impact of ClACO5 downregulation on CGMMV accumulation, we used systemically infected leaves from control (phytoene desaturase [PDS]-silenced) and ClACO5-silenced watermelon plants to measure CGMMV RNA levels. The accumulation of CGMMV gRNA in ClACO5-knockdown plants increased by 230% and 570% compared to that in PDS-knockdown plants, as revealed by RT-qPCR and RNA gel blot analysis, respectively (Figures 3D, E). These findings manifest that ClACO5 increases ET levels and enhances defense against CGMMV in watermelon plants.
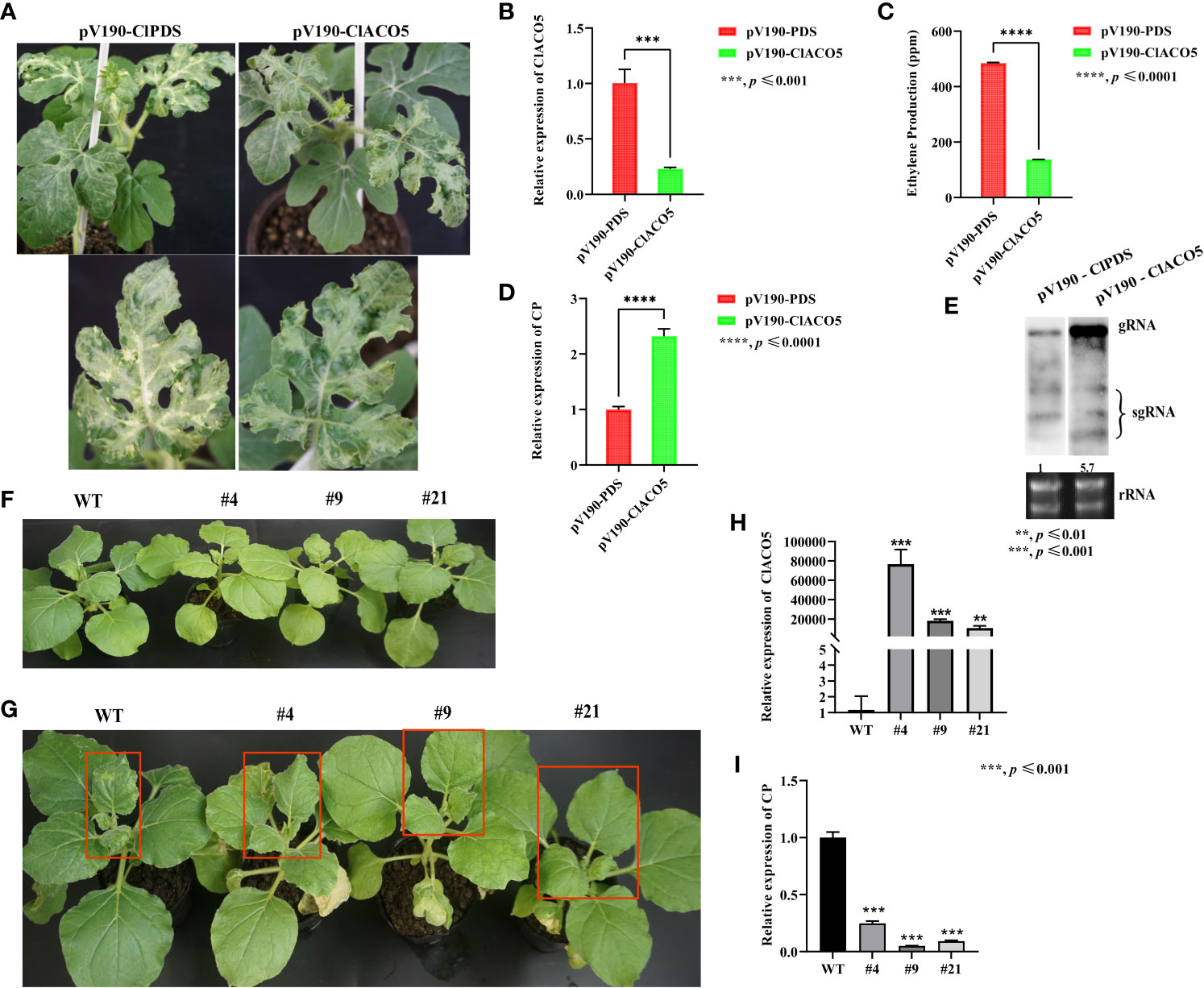
Figure 3 Knockdown and overexpression of ClACO5 enhance CGMMV susceptibility and resistance in watermelon plants and transgenic N. benthamiana. (A) Phenotype of pV190-ClPDS and pV190-ClACO5 in watermelons. (B) The transcript levels of ClACO5 in pV190-ClPDS- and pV190-ClACO5-treated watermelon leaves at 20 dpi by RT-qPCR. (C) Ethylene production of pV190-ClPDS- and pV190-ClACO5-treated watermelon leaves at 20 dpi. (D, E) CGMMV RNA accumulations in the pV190-ClPDS- and pV190-ClACO5-treated watermelon leaves at 20 dpi by RT-qPCR (D) and Northern blotting (E). (F) Phenotype of WT and transgenic N. benthamiana plants under normal growth conditions. (G) Phenotype of WT and transgenic N. benthamiana plants under CGMMV infection conditions. (H) RT-qPCR analysis of the transcript levels of ClACO5 in WT and transgenic tobacco plants. (I) RT-qPCR analysis of CGMMV CP accumulations in transgenic and WT tobacco plants. Watermelon and N. benthamiana plants were all collected at 20 dpi and 10 dpi, respectively. Values are means ± SD (n = 3). **p ≤ 0.01, ***p ≤ 0.001, and ****p ≤ 0.0001 (Student’s t-test). Image of northern blotting was obtained by splicing, as these two samples and other samples were hybridized on the same membrane simultaneously.
To further confirm that ClACO5 functions in plant defense against CGMMV infection, we exogenously overexpressed ClACO5 in N. benthamiana (a CGMMV-susceptible species) and selected three transgenic lines to assess CGMMV resistance. Under normal growth conditions, the wild-type (WT) and transgenic tobacco plants did not exhibit phenotypic differences (Figure 3F), and ClACO5 transcript abundance was significantly higher in the transgenic plants (Figure 3H). However, upon CGMMV infection, these plants showed milder CGMMV disease symptoms, including milder mottle and mosaic symptoms in systemically infected leaves, than the WT plants (Figure 3G). In addition, transgenic N. benthamiana plants accumulated significantly less CGMMV than the WT plants (Figure 3I). Collectively, these results show that knocking down ClACO5 significantly enhanced CGMMV susceptibility, whereas overexpressing this gene enhanced resistance to this virus, indicating that ClACO5 plays a positive role in ET biosynthesis.
ClWRKY70, a transcriptional activator, is induced by CGMMV infection
To identify TFs that regulate ClACO5 expression, we conducted a manual search of the NewPLACE database for the cis-elements in the ClACO5 promoter sequence (~2 kb) involved in responses to plant pathogens. Several W-boxes and MYB motifs in the ClACO5 promoter were identified, which are recognized by WRKY and MYB transcription factors, respectively. An abscisic-acid-response element (ABRE), an SA-responsive element, and a methyljasmonate (MeJA)-responsive element were also found in the ClACO5 promoter (Supplementary Table S1 and Supplementary Figure S4). Analysis of the RNA-Seq data found that CGMMV also induced ClWRKY70 (Cla97C10G206240) expression and that this gene shared similar expression patterns with ClACO5 (Figure 4A). Next, we examined the subcellular localization of ClWRKY70 by transiently co-expressed 35S-ClWRKY6-GFP or 35S-GFP with the nuclear marker gene H2B fused to mCherry in N. benthamiana leaves. The GFP signal from the 35S-GFP vector was ubiquitously distributed throughout the cell. By contrast, the GFP signal from the 35S-ClWRKY70-GFP vector was exclusively localized to the nucleus and co-localized with mCherry (Figure 4C). To examine the transcription-activating activity of ClWRKY70, a transactivation reporter assay was performed. Yeast strain AH109 containing pGBKT7-ClWRKY70 grew well on SD/–Trp or SD/–Trp/–Ade/–His medium, and displayed GAL4 activity on SD/–Trp/–Ade/–His medium supplemented with X-α-Gal, suggesting that ClWRKY70 is a transcriptional activator (Figure 4B).
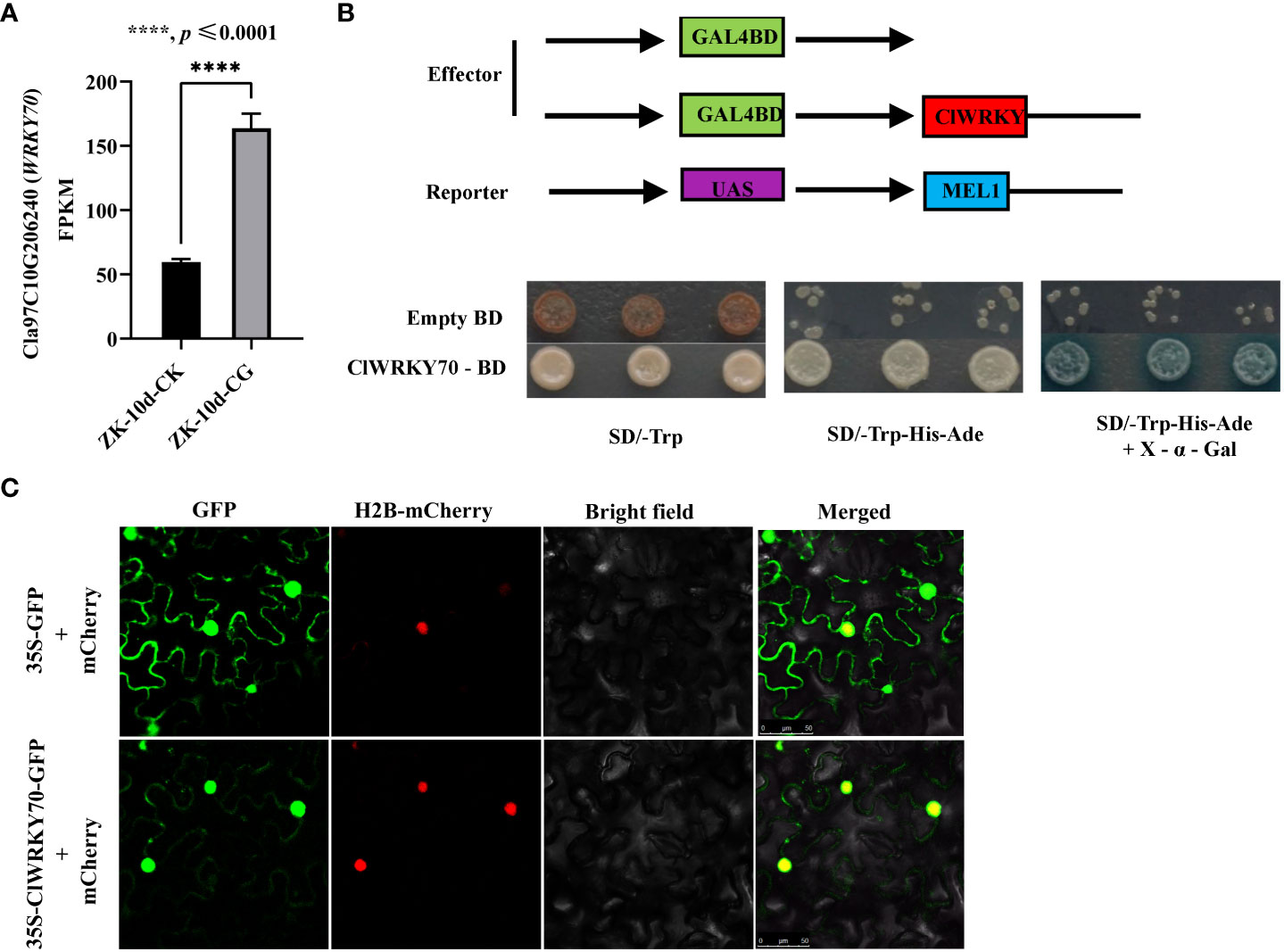
Figure 4 ClWRKY70 expression under CGMMV infection and transcriptional activity assays. (A) ClWRKY70 expression level was assayed after CGMMV infection. (B) Transactivation assay of ClWRKY70 in yeast. (C) Subcellular localization of ClWRKY70 in tobacco. 35S-ClWRKY6-GFP or 35S-GFP was co-transformed with 35S-H2B-mCherry (carried nuclear marker protein H2B) into tobacco leaves. The images of epidermal cells were taken under green (for GFP) and red (for mCherry) fluorescence and bright fields by confocal microscopy. Bars, 50 μm. ZK-10d-CK, ZK-10d-CG, leaves of ‘ZK’ plants inoculated with CGMMV or 0.01 M phosphate butter, were harvested at 10 dpi.
ClWRKY70 binds to the ClACO5 promoter
CGMMV infection increased the abundance of ClWRKY70 and ClACO5 transcripts; these genes play vital roles in plant responses to pathogen infection. We also identified several WRKY-binding motifs in the ClACO5 promoter (Supplementary Table S1). These observations prompted us to examine whether ClWRKY70 interacts with ClACO5 in response to CGMMV infection. A yeast one-hybrid assay (Y1H) confirmed that ClWRKY70 could bind to the W-box element in the ClACO5 promoter (Figure 5B). We also conducted a GUS transactivation experiment in N. benthamiana leaves to further validate this interaction. Leaves co-transformed with pCNF-ClWRKY70, and ClACO5pro-GUS exhibited significantly higher GUS activities than leaves infiltrated with the empty pCNF effector and ClACO5pro-GUS, as shown by the deeper blue color staining image and higher expression level of the GUS gene (Figure 5D).
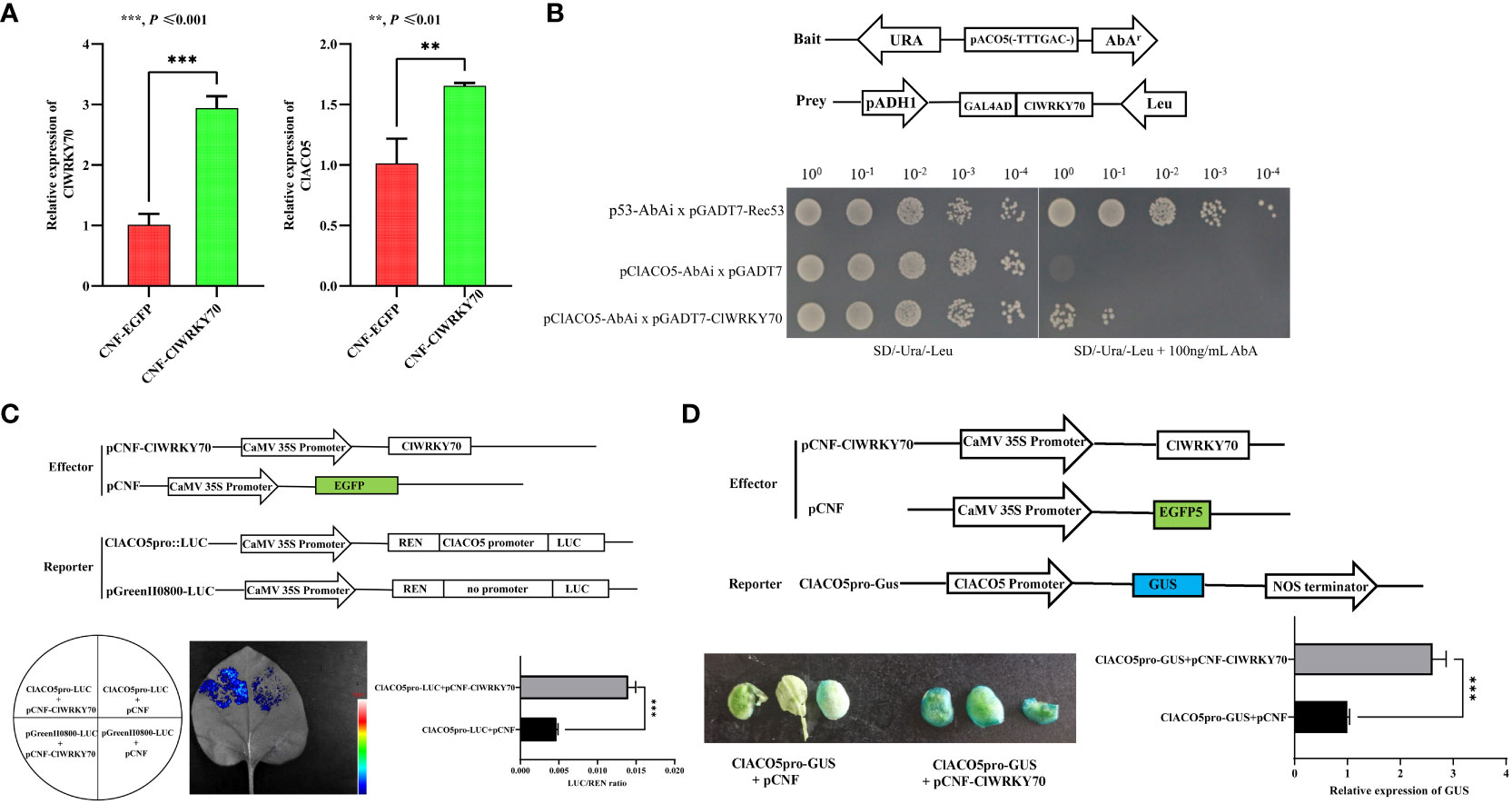
Figure 5 ClWRKY70 activates the transcription of ClACO5. (A) The transcript level of ClWRKY70 and ClACO5 after transient overexpression of ClWRKY70 in watermelon leaves. (B) Yeast one-hybrid (Y1H) analysis using pGADT7-ClWRKY70 as prey, pClACO5-AbAi as bait, and pGADT7-Rec53 and p53-AbAi as positive controls. (C) Luciferase activity analysis showed that higher LUC/REN ratio in the pCNF-ClWRKY70 effect vector than with the pCNF effect vector. The pCNF-ClWRKY70/pCNF effector and ClACO5pro-LUC/pGreenII800-LUC reporter were co-infiltrated into tobacco leaves. (D) GUS activity analysis using ClACO5pro-GUS as reporters and pCNF-ClWRKY70 as an effector. GUS staining images and GUS gene expression levels were visualized and measured.
To further determine the interaction between ClWRKY70 and the ClACO5 promoter, we carried out a dual-luciferase reporter assay. The binding of ClWRKY70 to the ClACO5 promoter led to a nearly three-fold increase in the relative LUC/REN ratio vs. the control (Figure 5C). To verify the role of ClWRKY70 in regulating the expression of ClACO5 in watermelon, we transiently transformed the leaves of 7-day-old seedlings with the pCNF-ClWRKY70 construct or empty vector (pCNF-EGFP). ClWRKY70 transcript levels were significantly higher in leaves transformed with the pCNF-ClWRKY70 construct vs. the empty vector control, and the transcript levels of ClACO5 increased nearly two-fold in leaves transformed with pCNF-ClWRKY70 vs. the pCNF-EGFP control (Figure 5A). Collectively, our results implied that ClWRKY70 functions as a transcriptional activator of ClACO5 and promotes its transcription by binding to the ClACO5 promoter.
ClWRKY70 enhances systemic acquired resistance to CGMMV infection
Since ClWRKY70 participated in plant responses to Acidovorax citrulli infection (Zhang et al., 2020), and ClWRKY70 is an upstream regulator of ClACO5, we reasoned that ClWRKY70 might promote systemic acquired resistance to CGMMV infection. Next, we generated ClWRKY70-overexpressing transgenic N. benthamiana plants and selected three independent lines that had significantly higher ClWRKY70 expression levels than the WT plants (Figure 6B). Under normal growth conditions, the WT and transgenic plants did not display phenotypic differences (Figure 6A). However, after CGMMV infection, transgenic plants exhibited milder mosaic and mottle symptoms in systemically infected leaves compared to WT plants (Figure 6C). In addition, WT plants had higher CGMMV CP transcript levels than transgenic plants (Figure 6D). The aforementioned results suggest that the heterologous expression of ClWRKY70 enhanced CGMMV resistance.
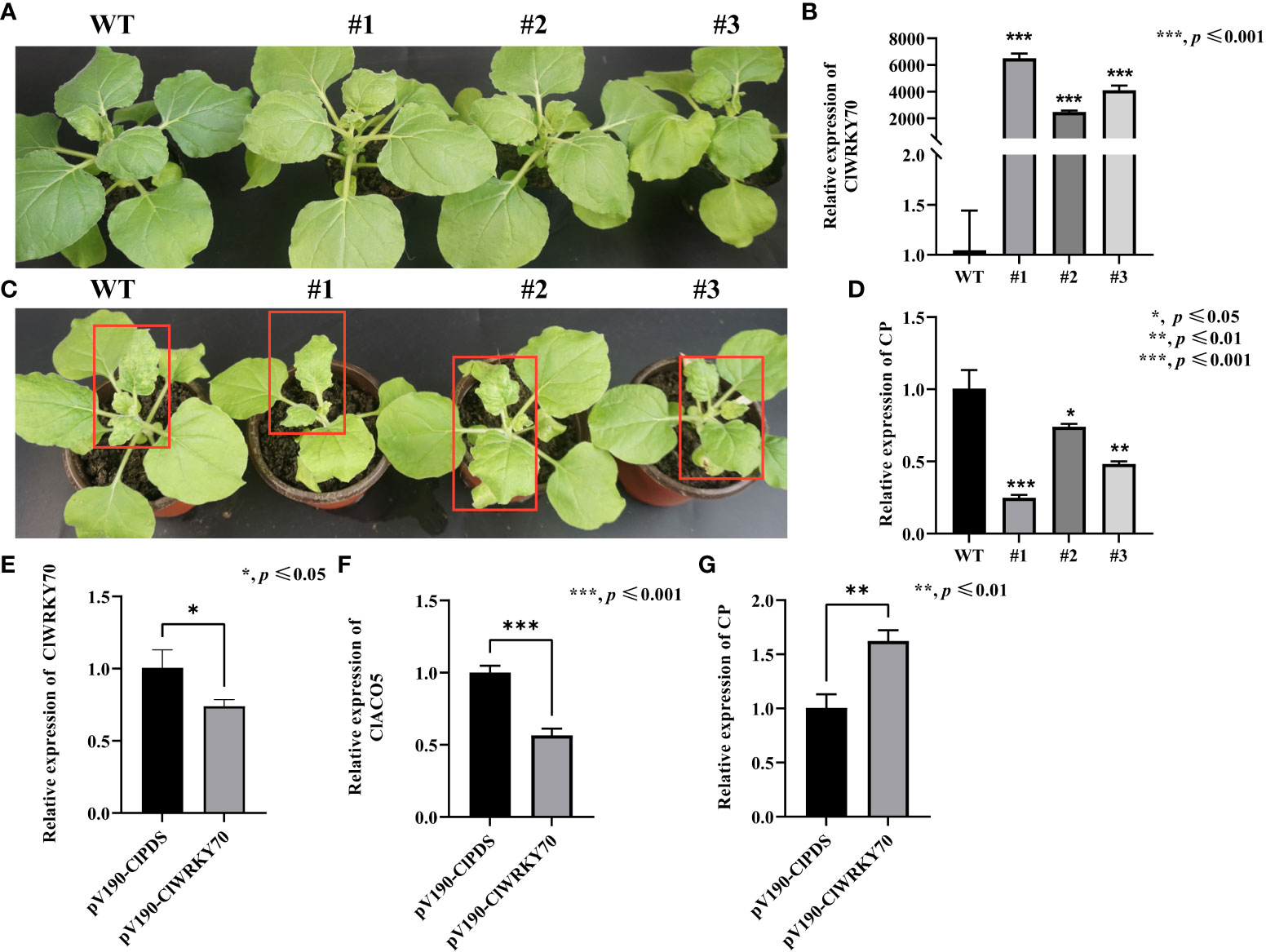
Figure 6 Knockdown and overexpression of ClWRKY70 enhance CGMMV susceptibility and resistance in watermelon plants and transgenic N. benthamiana. (A) Phenotype of WT and transgenic N. benthamiana plants under normal growth conditions. (B) The transcript levels of ClWRKY70 in transgenic and WT N. benthamiana plants by RT-qPCR analysis. (C) Phenotype of transgenic and WT N. benthamiana plants under CGMMV infection conditions. (D) CGMMV CP accumulations in transgenic and WT N. benthamiana plants by RT-qPCR analysis. (E-G) The transcript levels of ClWRKY70 (E), ClACO5 (F), and CGMMV RNA accumulations (G) in pV190-ClPDS- and pV190-ClWRKY70-treated watermelon leaves at 20 dpi by RT-qPCR analysis. Watermelon and N. benthamiana plants were all collected at 20 dpi and 10 dpi, respectively. Values are means ± SD (n = 3). *p ≤ 0.05, **p ≤ 0.01, and ***p ≤ 0.001 (Student’s t-test).
To further probe the function of ClWRKY70 in regulating the host’s susceptibility to CGMMV, we used the pV190 VIGS system to silence ClWRKY70 in watermelon plants. Knockdown of ClWRKY70 resulted in the simultaneous downregulation of ClACO5 (Figures 6E, F). Finally, we analyzed CGMMV accumulation in ClWRKY70-knockdown and control (PDS-silenced) watermelon plants by RT-qPCR, finding that it was 62% higher in the ClWRKY70-knockdown plants (Figure 6G). This indicated that the knockdown of ClWRKY70 increased the susceptibility of watermelons to CGMMV infection.
Discussion
ET production and ClACO5 expression are induced in response to CGMMV infection
Plants have developed complex systems to defend themselves against viral attacks, in which phytohormones play essential roles in facilitating defense signaling in plants (Mishra et al., 2020). ET plays a dual role in plant-virus interactions; it promotes resistance in some interactions and contributes to disease progression in others (Chen et al., 2013; Zhu et al., 2014b; Zhao et al., 2017). For example, the rice dwarf virus (RDV) increases ET production by interacting with OsSAMS1, an essential component of the ET biosynthesis, resulting in an increase in the enzyme’s activity, thereby increasing susceptibility to RDV (Zhao et al., 2017). Southern rice black-streaked dwarf virus (SRBSDV) encoded P6 protein subcellular localization is changed during different stages of SRBSDV infection differentially altering ethylene signaling to support SRBSDV infection and transmission (Zhao et al., 2022). In the current study, we observed a marked increase in ET production in watermelon plants after CGMMV infection. When exogenous ET was applied prior to the mechanical inoculation of CGMMV, watermelon leaves had milder disease symptoms, lower disease indices, and reduced virus accumulation compared with mock plants (Figure 1). These results imply that ET accumulation in watermelon plants might promote resistance to CGMMV infection. The opposite roles of ET in plant responses to viral infection might be related to different virus-host combinations as well as crosstalk between ET and other phytohormones.
ACS and ACO are major enzymes that catalyze the two key steps of ET biosynthesis (Kim et al., 2003). Eight ACS and eight ACO genes of watermelon plants were previously identified (Zhou et al., 2016). In the current research, we found the expressions of three ClACO genes (Cla97C07G140100, Cla97C07G144530, and Cla97C10G205440) were altered by CGMMV infection by analyzing the transcript levels of these genes under CGMMV infection. Among them, only Cla97C07G144530 (ClACO5) was upregulated. Whereas the transcript levels of all ClACS genes were unchanged (Supplementary Figure S1). Thus, ClACO genes play more important roles than ClACS genes in the response of watermelon plants to CGMMV infection.
The expression pattern of ClACO5 is largely consistent with the changes in ET levels in watermelon leaves during CGMMV infection (Figure 3C), and ClACO5 shares high sequence similarity with CuACO5 and AtDMR6 (Supplementary Figure S2). Based on these results, we speculate that ClACO5 may be vital for ET production during CGMMV infection in watermelon leaves. AtDMR6 encodes a putative 2OG-Fe(II) oxygenase associated with defense responses but required for susceptibility to downy mildew (Zeilmaker et al., 2015); it also participates in plant responses to bacteria and oomycetes, flavonoid biosynthesis, and SA catabolism (Falcone Ferreyra et al., 2015; Zhang et al., 2017), but it is unclear whether this gene participates in plant responses to viruses. In the present study, the knockdown of ClACO5 in watermelons conferred enhanced susceptibility to CGMMV infection, whereas heterologous overexpression of this gene in N. benthamiana enhanced resistance to CGMMV infection (Figure 3). Taken together, we conclude that ClACO5 functions in plant responses to CGMMV infection and enhances CGMMV resistance.
ClWRKY70 directly activates the expression of ClACO5
WRKY TFs regulate gene expression by binding directly to the W-box in their target genes’ promoters (Rushton et al., 2010). WRKY70s in some species, such as Arabidopsis, wheat (Triticum aestivum), and tomato (Solanum lycopersicum), play vital roles in plant responses to pathogen infection (Atamian et al., 2012; Wang et al., 2017; Zhou et al., 2018; Liu et al., 2021). However, far less is known about their roles in defense against viral pathogens as compared to fungal or bacterial pathogens. In the present study, we report that ClWRKY70 is induced by CGMMV infection. We observed a marked difference between ClWRKY70-overexpressing transgenic N. benthamiana and WT plants in leaves systemically infected with CGMMV, suggesting that ClWRKY70 might inhibit CGMMV accumulation.
WRKY70 regulates the expression of numerous defense-related genes (Machens et al., 2014), including SARD1, PR1, PR2, and PR5 (Li et al., 2004). WRKY70 may also inhibit ET- and JA-responsive gene expression (Li et al., 2004; Li et al., 2006; Li et al., 2019), but whether it regulates the expression of genes related to phytohormone biosynthesis remains unresolved. In the current study, we found that both ClWRKY70 and ClACO5 were consistently upregulated after CGMMV infection (Figures 1C, 4A). ClWRKY70 encodes a TF that positively regulates watermelon resistance to CGMMV (Supplementary Figures 4, 6). We also showed that the ClACO5 promoter contains a WRKY-binding site. ClWRKY70 directly bound to the ClACO5 promoter and enhanced its activity in N. benthamiana leaves and upregulated this gene in watermelon leaves (Supplementary Figure S4 and Figure 5), implying that ClACO5 is a target gene of ClWRKY70. Based on these results, we propose that ClWRKY70 functions as a major positive regulator of ET biosynthesis by activating ClACO5 expression in watermelon leaves and contributes to the resistance of watermelon plants to CGMMV infection.
SA can promote ClWRKY70 expression (Zhang et al., 2020), and we previously determined that CGMMV infection can induce SA accumulation and that treatment with exogenous SA significantly enhances the CGMMV resistance of watermelons (Liu et al., 2023a). Based on these and the current findings, we propose that SA affects ET biosynthesis by inducing the expression of ClWRKY70 and therefore activation of ClACO5 enhanced resistance of watermelon plants to CGMMV infection. In addition, we previously showed that SA-induced flavonoid biosynthesis plays a vital role in watermelon development and CGMMV resistance (Liu et al., 2023a), whereas AtDMR6, which is highly homologous to ClACO5, has flavone synthase activity (Falcone Ferreyra et al., 2015). Therefore, it would be worth investigating whether SA-induced flavonoid biosynthesis also involves the regulation of ClACO5 expression by ClWRKY70 in watermelon leaves defense against CGMMV infection and whether SA, ET, and flavonoids form a complex transcriptional regulatory network during the defense response of watermelons against CGMMV. Furthermore, we previously found that ET could induce ClAGO5 expression, thus speculating that ethylene may enhance watermelon defense against CGMMV via activating RNA silencing pathway (Liu et al., 2023b).
Taken together, our results demonstrate that ClACO5, localized to the nucleus and cytoplasm, and involved in ET biosynthesis, enhances CGMMV resistance. We propose that CGMMV infection increases SA biosynthesis in watermelon plants, resulting in the activation of downstream signaling pathways. ClWRKY70, involved in SA signaling, regulates ClACO5 expression by interacting with the W-box element in its promoter, thereby increasing ET levels (Figure 7). This model unveils the molecular mechanisms and transcriptional regulatory network of ET accumulation in response to CGMMV infection.
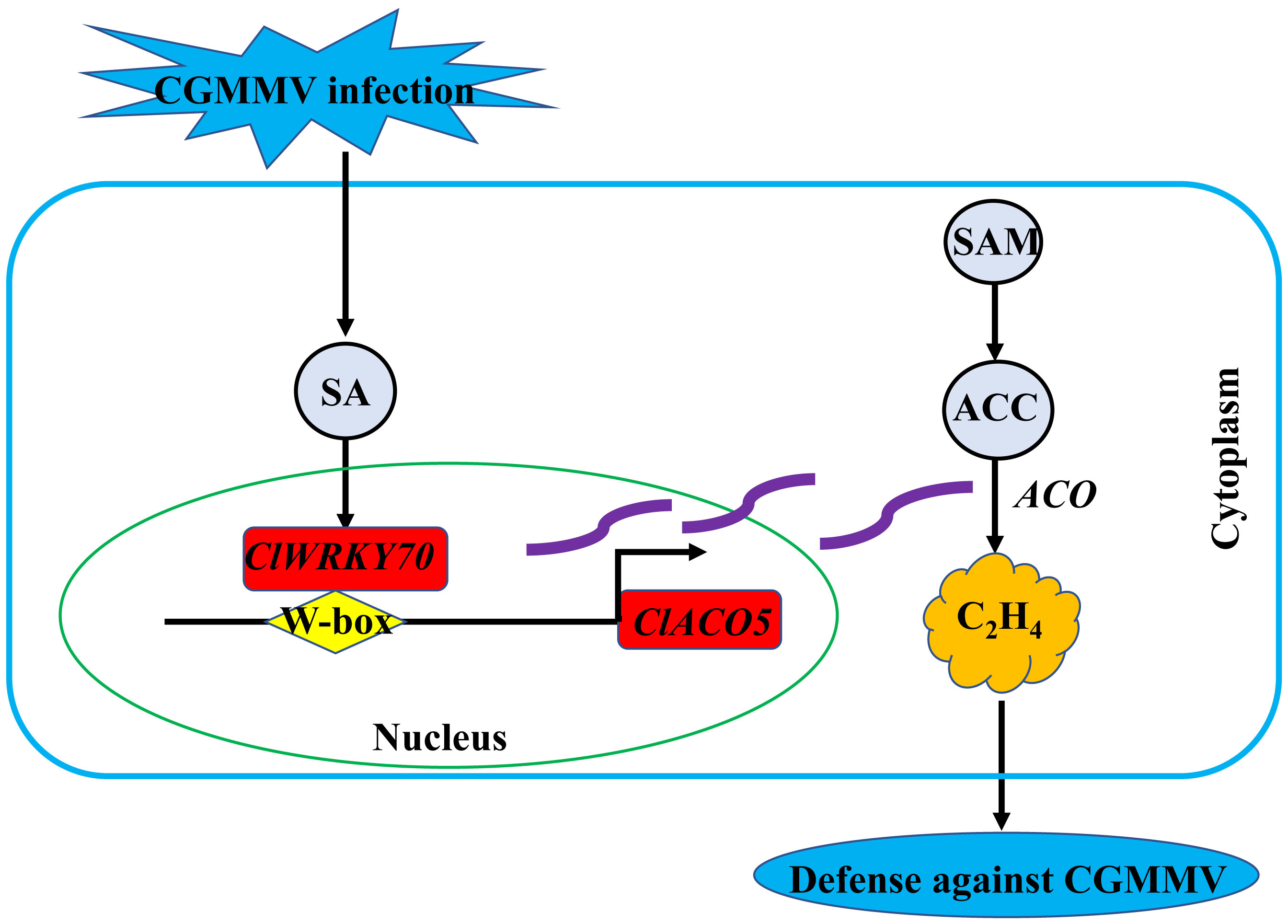
Figure 7 A proposed working model illustrating the regulatory role of SA responsive ClWRKY70-ClACO5 module in ethylene accumulation under CGMMV infection conditions. CGMMV infection increases endogenous SA, which subsequently triggers the SA signaling transduction pathway. ClWRKY70, involved in SA signaling, positively regulates ClACO5 by directly binding to the W-box element and activating the promoter, resulting in the upregulation of ClACO5, which is then integrated into the metabolic pathway for promoting ethylene biosynthesis. The ClACO5 proteins are shown using the purple wavy lines.
Materials and methods
Plant materials and treatments
The susceptible watermelon variety Zhengkang No. 2 and tobacco (N. benthamiana) were used in this research work. To analyze the effects of exogenous ET and ACC on plant responses to CGMMV, 7-day-old watermelon seedlings were sprayed with water, 500 μM ET, or 2 mM ACC for 3 days before CGMMV inoculation. Three biological replicates of leaves were collected 10 days after CGMMV treatment and used for RNA extraction.
Measuring ET production
ET production was measured according to a previous method (Kawano and Shimokawa, 1994). Approximately 5 g of leaf tissue was placed into a glass bottle. The bottle was sealed and incubated at room temperature for at least 5 hours. A 1ml sample of gas was removed from the bottle and analyzed by gas chromatography (Shimadzu GC-2010).
Detection of ACC
Phytohormones ACC contents were detected by MetWare (http://www.metware.cn/) based on the AB Sciex QTRAP 6500 LC-MS/MS platform.
Sequence analysis of ClACO5
Multiple alignment of the putative protein sequences encoded by ClACO5 and its homologs was performed using ClustalW software in conjunction with MEGA 7.0; then, the molecular weight and theoretical isoelectric point of the deduced ClACO5 protein was predicted using ExPASy (http://www.expasy.org), an online tool. The NewPLACE (https://www.dna.affrc.go.jp/PLACE/) database was used to identify the cis-elements in the ClACO5 promoter.
RNA extraction and RT-qPCR analysis
Total RNAs from watermelon leaf tissues were extracted using the method listed in the kit manual (Tiangen, Beijing, China). After the first-strand cDNA was synthesized, five-fold diluted cDNA products were subjected to qPCR using SYBR Green Master Mix (Vazyme, Nanjing, China) on a Roche Real-Time PCR system. We analyzed gene expression levels by the 2–△△CT method (Livak and Schmittgen, 2001). The primers used for RT-qPCR are included in Supplementary Table S2.
Subcellular localization of ClACO5 and ClWRKY70
The ClACO5 and ClWRKY70 coding sequences (CDSs) without the stop codons were amplified via 2×Fast Pfu Master Mix (novoprotein, Suzhou, China) and cloned into the pCNF-GFP vector. The recombinant 35S-ClACO5-GFP, 35S-ClWRKY70-GFP, and 35S-GFP (control) plasmids were transferred into the A.tumefaciens strain GV3101. N. benthamiana leaves were infiltrated with the bacterial suspensions, together with a plasmid expressing nuclear or membrane markers, 35S-H2B-mCherry or 35S-OsMCA1-mCherry (OD600~0.6). We observed the subcellular localizations of 35S-H2B-mCherry, 35S-OsMCA1-mCherry, 35S-ClACO5-GFP, 35S-ClWRKY70-GFP, and 35S-GFP under a confocal laser-scanning microscope.
Transcriptional activation assay
The ClWRKY70 CDS was cloned into the pGBKT7 vector. Yeast strain AH109 cells were transformed with the resulting construct and plated on SD/–Trp, SD/–His/–Trp/–Ade, or SD/–Trp/–Ade/–His supplemented with X-α-Gal. The pGBKT7 + pGADT7-T vector was used as a negative control.
Yeast one-hybrid assay
The ClWRKY70 CDS was ligated into the pGADT7 vector. The 789-bp promoter region containing a W-box cis-element was amplified with primers AD-ClWRKY70-F/R. Then, the amplified fragment was ligated into the pAbAi vector to generate the bait construct. An interaction assay of the ClWRKY70 and ClACO5 promotor fragments was conducted using a Matchmaker™ Gold Yeast One-Hybrid Library Screening System Kit (Clontech, San Francisco, USA).
GUS and dual luciferase reporter assays
The PCR-amplified 789-bp fragment of the ClACO5 promoter was cloned into pCAMBIA3301 and pGreenII 0800-LUC to generate reporter constructs. The ClWRKY70 CDS was cloned into pCNF to produce the effector construct. The reporter and effector constructs were transferred into A. tumefaciens GV3101 with the pSoup helper vector and A. tumefaciens strain GV3101, respectively, which were then used to coinfect 4-week-old tobacco leaves. At 48-72 h after infiltration, LUC and REN activities and GUS activity were measured in the leaves of these plants.
Transient expression in watermelon leaves
The expression construct pCNF-ClWRKY70 was transferred into A. tumefaciens GV3101; the empty vector pCNF was used as a negative control. Watermelon leaves were infiltrated with these cultures, collected 3 days after infiltration, and used to analyze the expression level of ClWRKY70 and ClACO5.
VIGS
A 300-bp fragment of ClACO5 or ClWRKY70 was amplified with gene-specific primers pV190-ClWRKY70-F/R or pV190-ClACO5-F/R (Supplementary Table S2), and cloned into pV190. Then, according to the previous description, all recombinant plasmid were separately transformed and injected into watermelon leaves (Liu et al., 2020). At 20 dpi, systemically infected leaves were collected for qPCR and RNA gel blot analyses.
Stable heterologous expression of ClACO5 or ClWRKY70 in N. benthamiana
The ClACO5 or ClWRKY70 CDS was amplified and ligated into the expression vector pBWA(V)HS-ccdB-GFP (Biorun, Wuhan, China). The A. tumefaciens strain GV3101 harboring the ClACO5 or ClWRKY70 ORF recombinant plasmids was transformed into N. benthamiana. We selected three positive T2 transgenic lines for further experiments.
Evaluation of the CGMMV resistance of ClACO5- or ClWRKY70-overexpressing
N. benthamiana plants
N. benthamiana plants from the WT line and three overexpressed ClACO5 transgenic lines (OE#4, OE#9, and OE#21) or three overexpressed ClWRKY70 transgenic lines (OE#1, OE#2, and OE#3) were inoculated with CGMMV. At 10 dpi, the plants were observed, and leaves were collected for measurement of target gene expression and CGMMV accumulation.
RNA gel blot analysis
The leaves of watermelon plants from the VIGS experiment were collected, and total RNA was extracted and used to analyze CGMMV accumulation with a DIG Northern Starter kit (Roche, Mannheim, Germany). The detailed method refers to a previously described method (Liu et al., 2017).
Statistical analysis
The inoculations of CGMMV were repeated at least twice with at least three replicates for each line and each time point. GraphPad Prism 9.0 was used to process all data and analyze statistical differences using Student’s t-tests at significance levels of p ≤ 0.05 (*), p ≤ 0.01 (**), p ≤ 0.001 (***), and p ≤ 0.0001 (****).
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
Author contributions
ML: Conceptualization, Data curation, Formal Analysis, Investigation, Methodology, Software, Validation, Writing – original draft. BK: Methodology, Writing – review & editing. HW: Methodology, Project administration, Writing – review & editing. BP: Project administration, Writing – review & editing. LL: Methodology, Writing – review & editing. NH: Resources, Supervision, Writing – review & editing. QG: Funding acquisition, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was funded by the National Natural Science Foundation of China (Grant No. 31572147), the Agricultural Science and Technology Innovation Program (CAAS-ASTIP-2022-ZFRI-09), the China Agriculture Research System of MOF and MARA (CARS-25), and the Science and Technology Major Project of Xinjiang Uygur Autonomous Region, China (2023A02009).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1332037/full#supplementary-material
References
Alazem, M., Lin, N. S. (2017). Antiviral roles of abscisic acid in plants. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.01760
Atamian, H. S., Eulgem, T., Kaloshian, I. (2012). SlWRKY70 is required for Mi-1-mediated resistance to aphids and nematodes in tomato. Planta 235, 299–309. doi: 10.1007/s00425-011-1509-6
Blume, B., Grierson, D. (1997). Expression of ACC oxidase promoter-GUS fusions in tomato and Nicotiana plumbaginifolia regulated by developmental and environmental stimuli. Plant J. 12, 731–746. doi: 10.1046/j.1365-313x.1997.12040731.x
Booker, M. A., DeLong, A. (2015). Producing the ethylene signal: regulation and diversification of ethylene biosynthetic enzymes. Plant Physiol. 169, 42–50. doi: 10.1104/pp.15.00672
Cai, L., Liu, J., Wang, S., Gong, Z., Yang, S., Xu, F., et al. (2023). The coiled-coil protein gene WPRb confers recessive resistance to Cucumber green mottle mosaic virus. Plant Physiol. 191, 369–381. doi: 10.1093/plphys/kiac466
Calil, I. P., Fontes, E. P. B. (2017). Plant immunity against viruses: antiviral immune receptors in focus. Ann. Bot. 119, 711–723. doi: 10.1093/aob/mcw200
Casteel, C. L., Alwis, M. D., Bak, A., Dong, H., Whitham, S. A., Jander, G. (2015). Disruption of ethylene responses by turnip mosaic virus mediates suppression of plant defense against the green peach aphid vector. Plant Physiol. 169, 209–218. doi: 10.1104/pp.15.00332
Chen, J., Nolan, T. M., Ye, H, Zhang, M., Tong, H., Xin, P., et al. (2017). Arabidopsis WRKY46, WRKY54, and WRKY70 transcription factors are involved in brassinosteroid-regulated plant growth and drought responses. Plant Cell 29, 1425–1439. doi: 10.1105/tpc.17.00364
Chen, L., Zhao, Y., Xu, S., Zhang, Z., Xu, Y., Zhang, J., et al. (2018). OsMADS57 together with OsTB1 coordinates transcription of its target OsWRKY94 and D14 to switch its organogenesis to defense for cold adaptation in rice. New Phytol. 218, 219–231. doi: 10.1111/nph.14977
Chen, M., Penfield, S. (2018). Feedback regulation of COOLAIR expression controls seed dormancy and flowering time. Science 360, 1014–1017. doi: 10.1126/science.aar7361
Chen, L., Zhang, L., Li, D., Wang, F., Yu, D. (2013). WRKY8 transcription factor functions in the TMV-cg defense response by mediating both abscisic acid and ethylene signaling in Arabidopsis. Proc. Natl. Acad. Sci. United States America 110, E1963–E1971. doi: 10.1073/pnas.1221347110
Dombrovsky, A., Tran-Nguyen, L. T. T., Jones, R. A. C. (2017). Cucumber green mottle mosaic virus: Rapidly increasing global distribution, etiology, epidemiology, and management. Annu. Rev. Phytopathol. 55, 231–256. doi: 10.1146/annurev-phyto-080516-035349
Falcone Ferreyra, M. L., Emiliani, J., Jose Rodriguez, E., Alina Campos-Bermudez, V., Grotewold, E., Casati, P., et al. (2015). The identification of Maize and Arabidopsis Type I flavone synthases links flavones with hormones and biotic interactions. Plant Physiol. 169, 1090–1107. doi: 10.1104/pp.15.00515
Fan, M., Bai, M. Y., Kim, J. G., Wang, T., Oh, E., Chen, L., et al. (2014). The bHLH transcription factor HBI1 mediates the trade-off between growth and pathogen-associated molecular pattern-triggered immunity in Arabidopsis. Plant Cell 26, 828–841. doi: 10.1105/tpc.113.121111
Hou, Y., Wang, Y., Tang, L., Tong, X., Wang, L., Liu, L., et al. (2019). SAPK10-mediated phosphorylation on WRKY72 releases its suppression on jasmonic acid biosynthesis and bacterial blight resistance. iScience 16, 499–510. doi: 10.1016/j.isci.2019.06.009
Houben, M., Van de Poel, B. (2019). 1-Aminocyclopropane-1-Carboxylic Acid Oxidase (ACO): The enzyme that makes the plant hormone ethylene. Frontier Plant Sci. 10. doi: 10.3389/fpls.2019.00695
Hu, Y., Chen, L., Wang, H., Zhang, L., Wang, F., Yu, D. (2013). Arabidopsis transcription factor WRKY8 functions antagonistically with its interacting partner VQ9 to modulate salinity stress tolerance. Plant J. 74, 730–745. doi: 10.1111/tpj.12159
Incarbone, M., Dunoyer, P. (2013). RNA silencing and its suppression: novel insights from in planta analyses. Trends Plant Sci. 18, 382–392. doi: 10.1016/j.tplants.2013.04.001
Kalde, M., Barth, M., Somssich, I. E., Lippok, B. (2003). Members of the Arabidopsis WRKY group III transcription factors are part of different plant defense signaling pathways. Mol. Plant-Microbe Interact. 16, 295–305. doi: 10.1094/mpmi.2003.16.4.295
Kawai, Y., Ono, E., Mizutani, M. (2014). Evolution and diversity of the 2-oxoglutarate-dependent dioxygenase superfamily in plants. Plant J. 78, 328–343. doi: 10.1111/tpj.12479
Kawano, T., Shimokawa, K. (1994). A simple method for vacuum extraction and quantitative determination of internal ethylene of excised apple tissue. J. Japanese Soc. Hortic. Sci. 63, 453–459. doi: 10.2503/jjshs.63.453
Kim, Y. S., Choi, D., Lee, M. M., Lee, S. H., Kim, W. T. (1998). Biotic and abiotic stress-related expression of 1-aminocyclopropane-1-carboxylate oxidase gene family in Nicotiana glutinosa L. Plant Cell Physiol. 39, 565–573. doi: 10.1093/oxfordjournals.pcp.a029406
Kim, C. Y., Liu, Y., Thorne, E. T., Yang, H., Fukushige, H., Gassmann, W., et al. (2003). Activation of a stress-responsive mitogen-activated protein kinase cascade induces the biosynthesis of ethylene in plants. Plant Cell 15, 2707–2718. doi: 10.1105/tpc.011411
Li, J., Brader, G., Kariola, T., Palva, E. T. (2006). WRKY70 modulates the selection of signaling pathways in plant defense. Plant J. 46, 477–491. doi: 10.1111/j.1365-313X.2006.02712.x
Li, J., Brader, G., Palva, E. T. (2004). The WRKY70 transcription factor: a node of convergence for jasmonate-mediated and salicylate-mediated signals in plant defense. Plant Cell 16, 319–331. doi: 10.1105/tpc.016980
Li, N., Han, X., Feng, D., Yuan, D., Huang, L.-J. (2019). Signaling crosstalk between salicylic acid and ethylene/jasmonate in plant defense: do we understand what they are whispering? Int. J. Mol. Sci. 20, 671. doi: 10.3390/ijms20030671
Liu, L., Peng, B., Zhang, Z., Wu, Y., Miras, M., Aranda, M. A., et al. (2017). Exploring different mutations at a single amino acid position of cucumber green mottle mosaic virus replicase to attain stable symptom attenuation. Phytopathology 107, 1080–1086. doi: 10.1094/phyto-03-17-0107-r
Liu, M., Liang, Z., Aranda, M. A., Hong, N., Liu, L., Kang, B., et al. (2020). A cucumber green mottle mosaic virus vector for virus-induced gene silencing in cucurbit plants. Plant Methods 16, 9. doi: 10.1186/s13007-020-0560-3
Liu, H., Liu, B., Lou, S., Bi, H., Tang, H., Tong, S., et al. (2021). CHYR1 ubiquitinates the phosphorylated WRKY70 for degradation to balance immunity in Arabidopsis thaliana. New Phytol. 230, 1095–1109. doi: 10.1111/nph.17231
Liu, M., Kang, B., Wu, H., Aranda, M. A., Peng, B., Liu, L., et al. (2023a). Transcriptomic and metabolic profiling of watermelon uncovers the role of salicylic acid and flavonoids in the resistance to cucumber green mottle mosaic virus. J. Exp. Bot. 74, 5218–5235. doi: 10.1093/jxb/erad197
Liu, M., Wu, H., Hong, N., Kang, B., Peng, B., Liu, L., et al. (2023b). Argonaute 1 and 5 proteins play crucial roles in the defence against cucumber green mottle mosaic virus in watermelon. Mol. Plant Pathol. 24, 961–972. doi: 10.1111/mpp.13344
Livak, K. J., Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Love, A. J., Laval, V., Geri, C., Laird, J., Tomos, A. D., Hooks, M. A., et al. (2007). Components of Arabidopsis defense- and ethylene-signaling pathways regulate susceptibility to Cauliflower mosaic virus by restricting long-distance movement. Mol. Plant-Microbe Interact. 20, 659–670. doi: 10.1094/mpmi-20-6-0659
Machens, F., Becker, M., Umrath, F., Hehl, R. (2014). Identification of a novel type of WRKY transcription factor binding site in elicitor-responsive cis-sequences from Arabidopsis thaliana. Plant Mol. Biol. 84, 371–385. doi: 10.1007/s11103-013-0136-y
Mishra, J., Srivastava, R., Trivedi, P. K., Verma, P. C. (2020). Effect of virus infection on the secondary metabolite production and phytohormone biosynthesis in plants. 3 Biotech. 10, 547. doi: 10.1007/s13205-020-02541-6
Pattyn, J., Vaughan-Hirsch, J., Van de Poel, B. (2021). The regulation of ethylene biosynthesis: a complex multilevel control circuitry. New Phytol. 229, 770–782. doi: 10.1111/nph.16873
Pieterse, C. M. J., van der Does, D., Zamioudis, C., Leon-Reyes, A., Van Wees, S. C. M. (2012). Hormonal modulation of plant immunity. Annual review of cell and developmental biology 28, 489–521. doi: 10.1146/annurev-cellbio-092910-154055
Rushton, P. J., Somssich, I. E., Ringler, P., Shen, Q. J. (2010). WRKY transcription factors. Trends Plant Sci. 15, 247–258. doi: 10.1016/j.tplants.2010.02.006
Van de Poel, B., Bulens, I., Markoula, A., Hertog, M. L., Dreesen, R., Wirtz, M., et al. (2012). Targeted systems biology profiling of tomato fruit reveals coordination of the Yang cycle and a distinct regulation of ethylene biosynthesis during postclimacteric ripening. Plant Physiol. 160, 1498–1514. doi: 10.1104/pp.112.206086
Wang, J., Tao, F., An, F., Zou, Y., Tian, W., Chen, X., et al. (2017). Wheat transcription factor TaWRKY70 is positively involved in high-temperature seedling plant resistance to Puccinia striiformis f. sp tritici. Mol. Plant Pathol. 18, 649–661. doi: 10.1111/mpp.12425
Yang, S. F., Hoffman, N. E. (2003). Ethylene biosynthesis and its regulation in higher plants. Annual.Review .Plant Physiol. 35, 155–189. doi: 10.1146/annurev.pp.35.060184.001103
Yim, W. J., Kim, K. Y., Lee, Y. W., Sundaram, S. P., Lee, Y., Sa, T. M. (2014). Real time expression of ACC oxidase and PR-protein genes mediated by Methylobacterium spp. in tomato plants challenged with Xanthomonas campestris pv. vesicatoria. J. Plant Physiol. 171, 1064–1075. doi: 10.1016/j.jplph.2014.03.009
Zeilmaker, T., Ludwig, N. R., Elberse, J., Seidl, M. F., Berke, L., Van Doorn, A., et al. (2015). DOWNY MILDEW RESISTANT 6 and DMR6-LIKE OXYGENASE 1 are partially redundant but distinct suppressors of immunity in Arabidopsis. Plant J. 81, 210–222. doi: 10.1111/tpj.12719
Zhang, Y., Zhao, L., Zhao, J., Li, Y., Wang, J., Guo, R., et al. (2017). S5H/DMR6 encodes a salicylic acid 5-hydroxylase that fine-tunes salicylic acid homeostasis. Plant Physiol. 175, 1082–1093. doi: 10.1104/pp.17.00695
Zhang, X., Yang, Y., Zhao, M., Yang, L., Jiang, J., Walcott, R., et al. (2020). Acidovorax citrulli Type III effector AopP suppresses plant immunity by targeting the watermelon transcription factor WRKY6. Frontier Plant Sci. 11. doi: 10.3389/fpls.2020.579218
Zhang, L., Zhang, F., Melotto, M., Yao, J., He, S. Y. (2017). Jasmonate signaling and manipulation by pathogens and insects. J. Exp. Bot. 68, 1371–1385. doi: 10.1093/jxb/erw478
Zhao, Y., Cao, X., Zhong, W., Zhou, S., Li, Z., An, H., et al. (2022). A viral protein orchestrates rice ethylene signaling to coordinate viral infection and insect vector-mediated transmission. Mol. Plant 15, 689–705. doi: 10.1016/j.molp.2022.01.006
Zhao, S., Hong, W., Wu, J., Wang, Y., Ji, S., Zhu, S., et al. (2017). A viral protein promotes host SAMS1 activity and ethylene production for the benefit of virus infection. Elife 6, e27529. doi: 10.7554/eLife.27529
Zhou, M., Guo, S., Zhang, J., Zhang, H., Li, C., Tang, X., et al. (2016). Comparative dynamics of ethylene production and expression of the ACS and ACO genes in normal-ripening and non-ripening watermelon fruits. Acta Physiologiae Plantarum 38, 228. doi: 10.1007/s11738-016-2248-x
Zhou, M., Lu, Y., Bethke, G., Harrison, B. T., Hatsugai, N., Katagiri, F., et al. (2018). WRKY70 prevents axenic activation of plant immunity by direct repression of SARD1. New Phytol. 217, 700–712. doi: 10.1111/nph.14846
Zhu, F., Xi, D., Yuan, S., Xu, F., Zhang, D., Lin, H.. (2014a). Salicylic acid and jasmonic acid are essential for systemic resistance against tobacco mosaic virus in Nicotiana benthamiana. Mol. Plant-Microbe Interact. 27, 567–577. doi: 10.1094/mpmi-11-13-0349-R
Zhu, F., Xi, D., Deng, X., Peng, X., Tang, H., Chen, Y., et al. (2014b). The Chilli veinal mottle virus regulates expression of the tobacco mosaic virus resistance gene n and jasmonic acid/ethylene signaling is essential for systemic resistance against chilli veinal mottle virus in tobacco. Plant Mol. Biol. Rep. 32, 382–394. doi: 10.1007/s11105-013-0654-4
Keywords: ethylene, ClACO5, cucumber green mottle mosaic virus, ClWRKY70 regulation, watermelon
Citation: Liu M, Kang B, Wu H, Peng B, Liu L, Hong N and Gu Q (2024) Ethylene enhances resistance to cucumber green mottle mosaic virus via the ClWRKY70-ClACO5 module in watermelon plants. Front. Plant Sci. 14:1332037. doi: 10.3389/fpls.2023.1332037
Received: 02 November 2023; Accepted: 19 December 2023;
Published: 11 January 2024.
Edited by:
Maite F. S. Vaslin, Federal University of Rio de Janeiro, BrazilReviewed by:
Ragunathan Devendran, Martin Luther University of Halle-Wittenberg, GermanyTong Zhang, South China Agricultural University, China
Copyright © 2024 Liu, Kang, Wu, Peng, Liu, Hong and Gu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qinsheng Gu, Z3VxaW5zaGVuZ0BjYWFzLmNu; Huijie Wu, d3VodWlqaWVAY2Fhcy5jbg==
 Mei Liu
Mei Liu Baoshan Kang
Baoshan Kang Huijie Wu
Huijie Wu Bin Peng
Bin Peng Liming Liu
Liming Liu Ni Hong
Ni Hong Qinsheng Gu
Qinsheng Gu