
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Plant Sci. , 15 December 2023
Sec. Plant Metabolism and Chemodiversity
Volume 14 - 2023 | https://doi.org/10.3389/fpls.2023.1330586
This article is part of the Research Topic Proanthocyanidins and Isoflavonoids View all 12 articles
 Changfu Li
Changfu Li Yansheng Zhang*
Yansheng Zhang*The pathway for forming isoflavonoid skeletal structure is primarily restricted to the Leguminosae family. Subsequent decorations on the compound backbone by tailoring enzymes would change their biological and medicinal properties. Pueraria lobata is a leguminous plant, and as a traditional Chinese medicine its roots have been ascribed a number of pharmacological activities. Glycosylation and methylation are the main modifying processes in isoflavonoid metabolism in P. lobata roots, resulting in the accumulation of unique glycosylated and methylated end isoflavonoid compounds. For instance, daidzein 8-C-glucoside (i.e., puerarin) and puerarin derivatives are produced only by the Pueraria genus. Puerarin has been established as a clinical drug for curing cardiovascular diseases. To better understand the characteristic isoflavonoid metabolism in P. lobata, this review attempts to summarize the research progress made with understanding the main glycosylation and methylation of isoflavonoids in P. lobata and their biosynthetic enzymes.
Among the 26 Pueraria species listed in the plant database (www.theplantlist.org), only three species, Pueraria lobata (Willd.) ohwi (Figure 1A), Pueraria thomsonii Benth, and Pueraria peduncularis Benth, have been included into Chinese Pharmacopoeia (Wang et al., 2020). The dried root of P. lobata (Figure 1B), also called Ge-Gen in China, is used as traditional herb medicine mainly for treating cardiovascular diseases, vascular hypertension, and diabetes (Ehrman et al., 2007; Wong et al., 2011). Modern pharmacological studies have revealed hepatoprotective(Sun et al., 2019), anti-inflammatory (Xi C. et al., 2023), anti-boneloss (Yang et al., 2017), and anti-cancer effects (Ahmad et al., 2020) of P. lobata extracts.
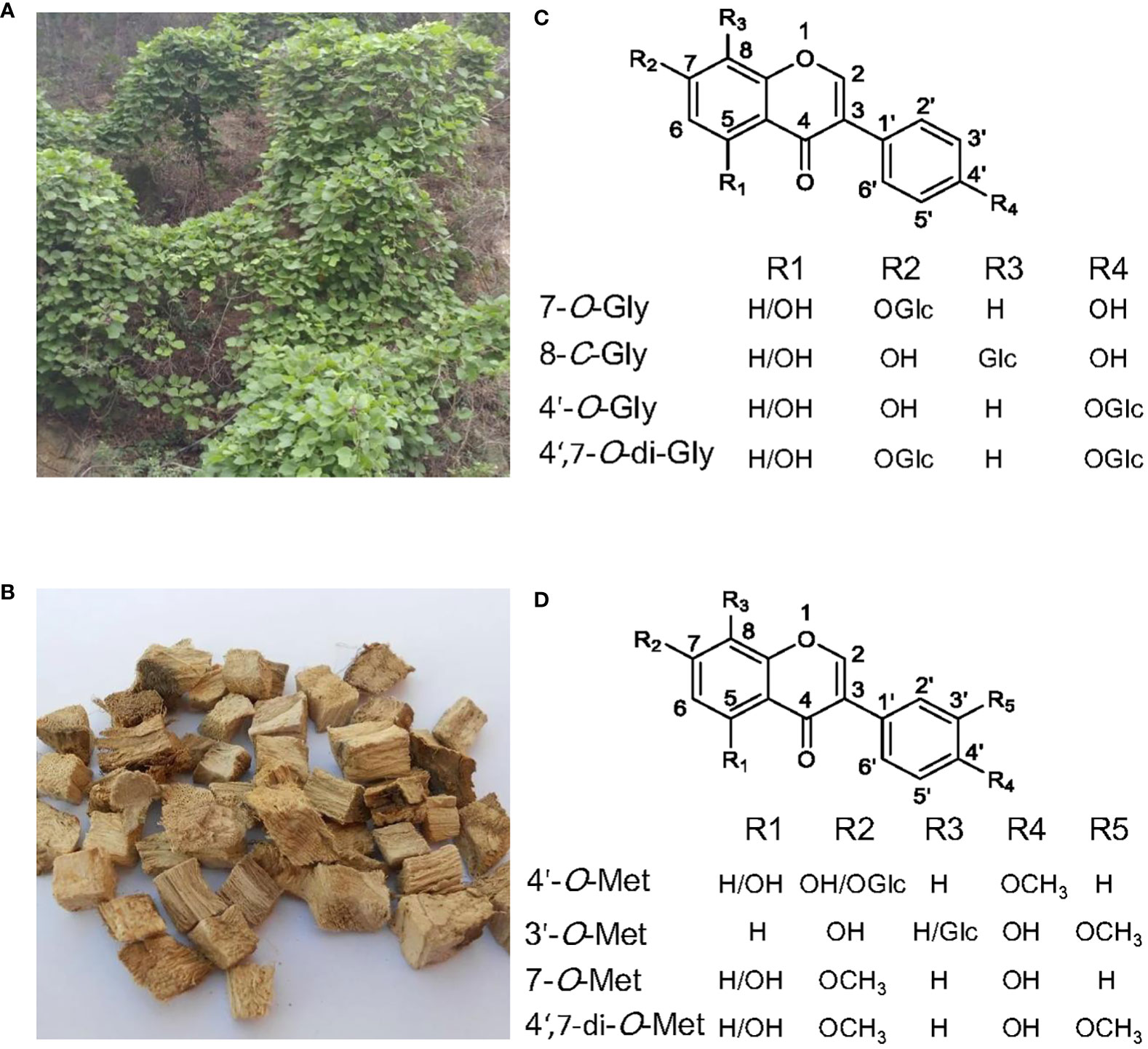
Figure 1 The medicinal plant Pueraria lobata and the main glycosylated and methylated isoflavonoids accumulated in it. (A) P. lobata plant. (B) The dried roots of P. lobata used in traditional Chinese medicine. (C) Structures of the main isoflavone glucosides. (D) Structures of the main O-methylated isoflavonoids.
Isoflavonoid compounds are considered the main bioactive components of P. lobata (Rong et al., 1998). A significant example is puerarin (i.e., daidzein 8-C-glucoside), an isoflavone that has been used as a prescribed drug in clinical practice for the treatment of cardiovascular diseases (Zhou et al., 2021). The biosynthetic pathway for the formation of isoflavonoid backbone is predominantly conserved in legumes (Falcone Ferreyra et al., 2012). Subsequent modifications on the isoflavonoid skeleton, such as glycosylation and methylation, result in the difference in isoflavonoid composition between different leguminous species. For instance, puerarin and its glycosylated and/or methylated derivatives (e.g. 3’-methoxy puerarin, 6’’-O-xylosylpuerarin and puerarin 4’-O-glucoside) are produced only by the species within the Pueraria genus (Wang et al., 2020), therefore conferring their unique medicinal value for human. Phytochemical studies revealed glycosylation and methylation as the two major modifications in isoflavonoid metabolism in P. lobata (Wang et al., 2020). As a consequence, pharmacological activities of P. lobata have focused mainly on the specifically glycosylated or/and methylated isoflavonoids, such as puerarin (Wang et al., 2022) and 3’-methoxy puerarin (Zhao et al., 2007). There is intense interest in identifying the enzymes responsible for the glycosylation and methylation reactions for isoflavonoid metabolism in P. lobata. This review considers the recent progress in understanding the biochemistry of the glycosylation and methylation for isoflavonoid metabolism in P. lobata.
Isoflavonoid glycosides from Pueraria are mainly the C- and O-glycosides (Wang et al., 2020). The glycol-conjugation towards Pueraria isoflavonoids occurs primarily at the positions of O-7, C-8, and O-4’ (Wang et al., 2020) (Figure 1C). Mono-glycosylation at C-8 or O-7 seems to be prevalent in P. lobata, as the most abundant isoflavone glycosides in P. lobata include the 7-O-glucosides of genistein and daidzein, and the 8-C-glucosides of daidzein (i.e., puerarin) (Ohshima et al., 1988; Rong et al., 1998; Wang et al., 2016). P. lobata root also accumulates the 4’-O-glucosides of puerarin, genistein, and daidzein (Ohshima et al., 1988; Li et al., 2010; Wang et al., 2016), and daidzein 4’,7-O-diglucosides (Keung et al., 1996). Glycosylation reaction is enzymatically driven by uridine diphosphate (UDP)-sugar glycosyltransferases (UGTs), and the UGTs involved in plant secondary metabolism usually belong to the family 1 UGTs, which possess the signature PSPG (plant secondary product glycosyltransferase consensus sequence) motif at their C-terminal (Vogt and Jones, 2000).
The common sites for methylation of P. lobata isoflavonoids are O-4’, O-3’, and O-7 (Figure 1D). The majority of methylated isoflavonoids in P. lobata are 4’-O-methylated isoflavones represented by formononetin (4’-O-methyldaidzein) and biochanin A (5-hydroxy formononetin), while the 3’- and 7-O-methylated isoflavones are produced in much less amounts (Rong et al., 1998). The 4’- and 7-O-methylated isoflavones are also produced in other leguminous plant species, including Medicago truncatula, Glycyrrhiza echinata, Medicago sativa, and Lotus japonicus (He et al., 1998; Akashi et al., 2003; Deavours et al., 2006). Interestingly, the occurrence of 3’-O-methylated isoflavones seems to be restricted to the Pueraria genus. The O-methylation reaction is catalyzed by an OMT which transfers a methyl from the donor SAM (S-adenosyl-L-methionine) to a hydroxyl moiety of an acceptor.
The 8-C-glycosylation is required for puerarin formation. There is great interest in understanding the biochemical process for the formation of the 8-C-glycosyl group in puerarin (Inoue and Fujita, 1977; Wang et al., 2017; Bao et al., 2022). Some data remain contradictory, particularly regarding the step at which the 8-C-glucosyl group is introduced (Figure 2). Early labeling studies had proposed an upstream intermediate isoliquiritigenin at the chalcone stage, but not daidzein at the isoflavone stage, as an acceptor for the 8-C-glycosylation (Inoue and Fujita, 1977). However, an enzyme assay using the Pueraria root crude protein provided an implication that the C-glucosyl unit in puerarin might be introduced at the isoflavanone stage (He et al., 2011). This assumption is prone to being considered because that a number of flavone C-GTs recognize 2-hydroxyflavanone intermediates as their natural substrates (Brazier-Hicks et al., 2009; Nagatomo et al., 2014; Hirade et al., 2015). Nonetheless, Xi et al. revealed that there are no orthologs of 2-hydroxyflavanone C-GTs in P. lobata (Xi H.T. et al., 2023), indicating that if the C-glycosylation for puerarin biosynthesis occurs at the isoflavanone stage, it is probably catalyzed by a phylogenetically distinct UGT. A very recent labeling study provided evidence that both isoliquiritigenin and daidzein could be incorporated into puerarin in vivo (Adolfo et al., 2022), suggesting that the 8-C-glycosylation can happen at either the chalcone or isoflavone stage, or simultaneously at both levels.
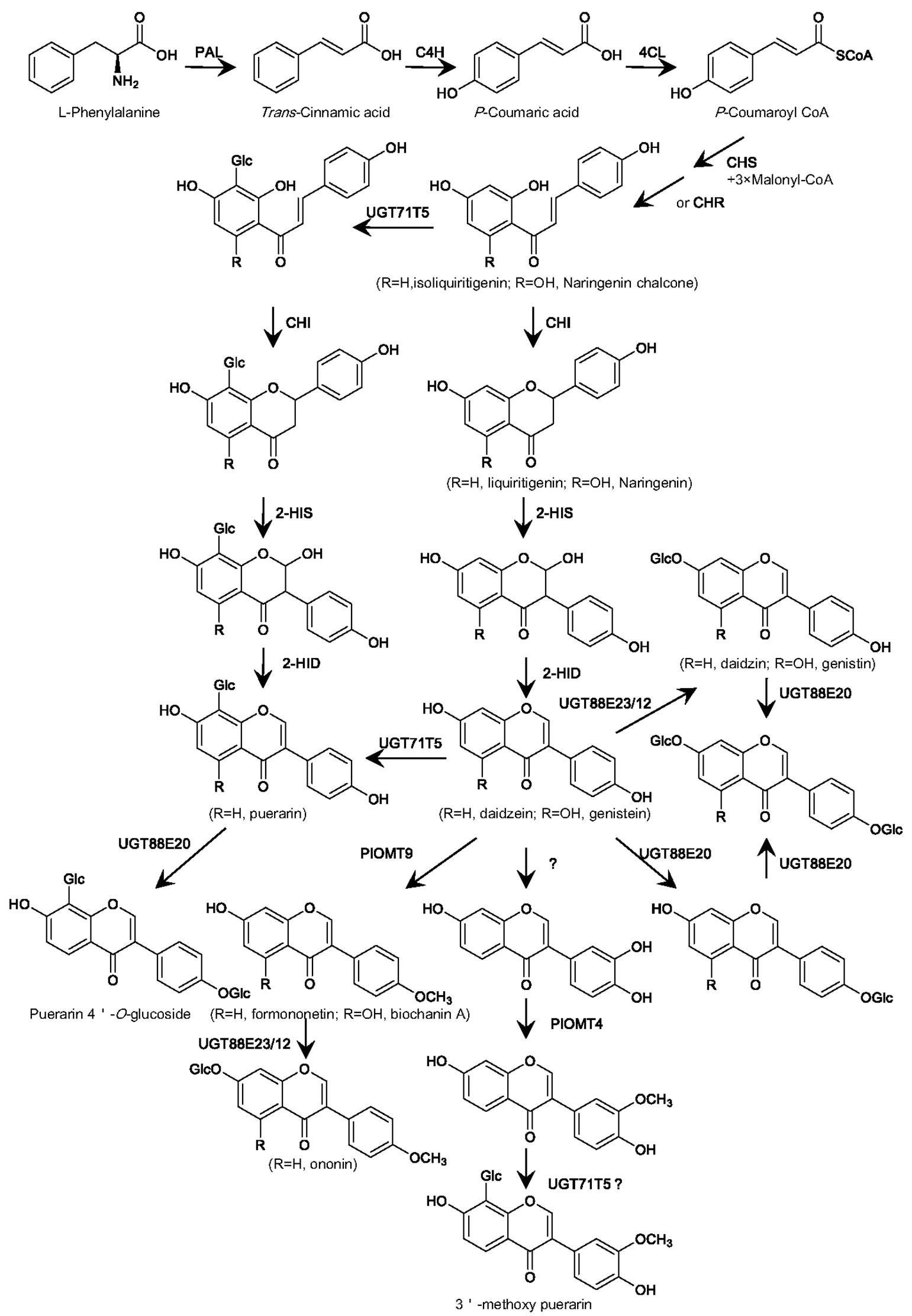
Figure 2 Proposed biosynthesis in P. lobata of the main glycosylated and methylated isoflavonoids based on the properties of the P. lobata UGTs and OMTs characterized so far. PAL, phenylalanine ammonialyase; C4H, cinnamate 4-hydroxylase; 4CL, 4-coumarate CoA ligase; CHS, chalcone synthase; CHR, chalcone reductase; CHI, chalcone isomerase; 2-HIS, 2-hydroxyisoflavanone synthase; 2-HID, 2-hydroxyisoflavanone dehydratase; UGT, UDP-glucosyltransferase; OMT, O-methyltransferase.
For the first time, a P. lobata C-GT (namely PlUGT43), which directly transfers a glucose group to the C-8 position of daidzein leading to puerarin, was molecularly cloned from the root of P. lobata by Wang et al. (Wang et al., 2017). Through the in vitro assays, PlUGT43 was found to have no or negligible activity with the chalcone intermediate isoliquiritigenin (Wang et al., 2017). However, its isoform (officially named UGT71T5), which shares 99.72% sequence identity with PlUGT43, was recently reported to be capable of catalyzing the C-glycosylation activity against both daidzein and isoliquiritigenin (Adolfo et al., 2022). Incubation of the recombinant PlUGT43 or UGT71T5 with 2-hydroxyisoflavanone did not generate a product matching the 2-hydroxyisoflavanone C-glycoside (Wang et al., 2017; Adolfo et al., 2022), indicating that they had no C-glycosylation activity with 2-hydroxyisoflavanone. The RNAi-mediated down-regulation of UGT71T5 caused a strong reduction in the levels of puerarin in P. lobata hairy roots (Adolfo et al., 2022), confirming that PlUGT43 (UGT71T5) functions as a C-GT at least partially for puerarin biosynthesis in P. lobata. Interestingly, when the 2-HIS (2-hydroxyisoflavanone synthase; see its place in the pathway in Figure 2), which is the entry enzyme catalyzing the formation of isoflavonoid backbone (Steele et al., 1999; Jung et al., 2000), was down-regulated in P. lobata hairy roots, the potential C-glycosides of isoliquiritigenin and/or liquiritigenin significantly accumulated, when compared to that in the control roots (Adolfo et al., 2022). This data clearly supports that introduction of the C-glucosyl group can take place at the chalcone stage and then the 2-HIS would be able to accommodate the C-glycosides as substrates (Figure 2). Recently, another variant of PlUGT43, designated PlCGT, was identified from P. lobata by Ye et al. (Bao et al., 2022). PlCGT shows 97.01% amino acid identity with PlUGT43, and this variant essentially catalyzes the same C-glycosylation activity as PlUGT43 (Bao et al., 2022). An ortholog of PlUGT43, named PtUGT8, was also isolated from P. thomsonii species (Duan et al., 2022). Despite exhibiting a high sequence identity (96.52%) to PlUGT43, PtUGT8 was shown as primarily having a 7-O-glucosylation activity toward isoflavones whereas not catalyzing the 8-C-glycosylation reaction as does by PlUGT43 (Duan et al., 2022). In view of a previous finding that O- and C-GT can be easily shifted by changing only a few amino acids (Gutmann and Nidetzky, 2012), a subtle difference in the active sites of PtUGT8 and PlUGT43 may plausibly account for this discrepancy.
Taken together, combination of the published data from the in vitro assays using the recombinant UGTs, the in vivo labeling experiments, and the transgenic studies of P. lobata hairy roots strongly supports that during puerarin biosynthesis, the C-glucosylation reaction takes place most likely at either the chalcone or isoflavone stage, or both.
The 7-O-glycosylation is common for isoflavonoid metabolism in leguminous plant species. In the 1980s, a relatively pure protein bearing the isoflavone 7-O-glucosylation activity was first purified from Cicer arietinum L (Koster and Barz, 1981). Later, genes encoding isoflavone 7-O-glucosyltransferases were isolated from the cell suspension cultures of Glycyrrhiza echinata (Nagashima et al., 2004), the roots (Noguchi et al., 2007) and seeds (Dhaubhadel et al., 2008) of Glycine max.
A total of six P. lobata UGTs, named PlUGT1(official UGT designation UGT88E12), PlUGT13(UGT88H1), PlUGT4(UGT72Y3), PlUGT15(UGT88E23), PlUGT57(UGT84F7) and UGT88A40, had been characterized by in vitro assays as having an isoflavone 7-O-glucosylation activity on daidzein and genistein (Li et al., 2014; Wang et al., 2019; Adolfo et al., 2022). Analysis of the substrate conversion rate and enzymatic kinetic parameter revealed that both PlUGT1 and PlUGT15 are highly specific for isoflavones with no or little activity against other acceptors, including chalcones, flavanones, flavones, and flavonols (Li et al., 2014; Wang et al., 2019). The PlUGT13, PlUGT4, or UGT88A40 can accept relatively broad substrates and glycosylates substrates at different positions (Li et al., 2014; Adolfo et al., 2022). Although PlUGT57 also shows a strict substrate preference for isoflavone aglycones, its catalytic efficiency (Kcat/Km; 2.10 × 103 M-1s-1) toward daidzein, when UDP-glucose is used as a sugar donor, is about 20-fold lower than UGT88E12 (3.79 × 104 M-1s-1), and 80-fold lower than UGT88E23(1.75 × 105 M-1s-1) (Wang et al., 2019). Phylogenetic analysis (Li et al., 2014; Wang et al., 2019) revealed that PlUGT1 and PlUGT15 showed a close relationship with a G. max 7-O-UGT GmIF7GT (UGT88E3) that shows a substrate preference for isoflavones (Noguchi et al., 2007). Therefore, members of the UGT88E family are believed to truly contribute to the 7-O-glycosylation in isoflavonoid metabolism in P. lobata.
The presence of 4’-O-glucosides of daidzein, genistein, and puerarin (Ohshima et al., 1988; Fang et al., 2006), and 4’,7-O-diglucoside of daidzein (Zhang et al., 2013) in P. lobata tissues suggests the occurrence of UGTs specific for 4’-O-glycosylating these compounds.
One full-length cDNA encoding PlUGT2 (officially assigned as UGT88E20) was identified and cloned from P. lobata using an RNA-sequencing approach (Wang et al., 2016). Tissue-specific expression analysis indicated that the transcript of PlUGT2 was higher in roots relative to stems and leaves. Phylogenetic analysis (Wang et al., 2016) showed that PlUGT2 was grouped into the same clade with GmUGT1 and GmUGT7 from Glycine max, which are the flavone 4’-O-UGTs (Funaki et al., 2015). The purified protein of PlUGT2 could catalyze either O-4’- or O-7-glucosylation of genistein, daidzein, liquiritigenin, and naringenin, yielding their mono-4’-O- or 7-O-glucosides. Interestingly, PlUGT2 consecutively glycosylates these mono-glucosides to di-glucosides with both O-4’ and O-7 being glucosylated (Wang et al., 2016). In comparison with the mono-glycosylation, the di-glycosylation activity catalyzed by PlUGT2 is much lower, consistent with the fact that the 4’,7-O-di-glucosides are produced at extremely low levels in P. lobata tissues(Zhang et al., 2013). PlUGT2 is the first 4’-O-glucosyltransferase identified from P. lobata. Recently, Adolfo et al. (Adolfo et al., 2022) reported another P. lobata UGT, named UGT73C42, which catalyzes either 4’- or 7-O-glucosylation of various polyphenolic compounds, including chalcones, flavones, and isoflavones.
PlUGT2 could also catalyze 4’- or 7-O-glucosylation of puerarin (Wang et al., 2016), which is currently a clinical drug for curing cardiovascular diseases (Zhou et al., 2021). Although puerarin is currently a prescribed drug, its low water solubility is still a serious drawback in clinical applications (Wang et al., 2012; Chen et al., 2021). Glycosylation is an efficient way to increase water solubility(Liu et al., 2016), thus, the identification of PlUGT2 would provide such an opportunity.
Many isoflavonoids are O-methylated with the methoxy residue improving their biological activities by increasing liposolubility (Wen et al., 2017). P. lobata accumulates 3’-methoxy-derivatives of isoflavones, including 3’-methoxydaidzein, 3’-methoxydaidzin, 3’-methoxypuerarin, and 3’-methoxyformononetin (Rong et al., 1998; Li et al., 2016b; Wang et al., 2020). Relative to the puerarin itself, its derivative 3’-methoxypuerarin exhibited better protective effects on cerebral ischemic-reperfusion injury in rats (Zhao et al., 2007).
One OMT, designated PlOMT4, was cloned (Li et al., 2016b) from P. lobata based on the P. lobata transcriptome database (Wang et al., 2015). Tissue-specific expression analysis revealed that PlOMT4 was expressed most highly in roots, and its transcript was up-regulated by MeJA (Li et al., 2016b). PlOMT4 was found to have the activity of methylating 3’-hydroxy daidzein to form 3’-methoxy-daidzein (Li et al., 2016b). PlOMT4 has no activity with the isoflavonoid substrates with free hydroxyl groups at either C7 or C4’ (Li et al., 2016b), suggesting the methylation activity of PlOMT4 is region-specific. In addition, PlOMT4 is inactive with 3’-hydroxy puerarin (Li et al., 2016b), indicating that the 8-C-glucosylation of 3’-hydroxy daidzein prevents methylation at the 3’-position, and thereby the 3’-methylation should take place prior to the 8-C-glycosylation during 3’-methoxy-puerarin biosynthesis. PlOMT4 seems to be the only isoflavone specific 3’-O-methyltransferase so far identified from plant species.
In the early 1970s, scientists began a search for the isoflavone 4’-O-methyltransferase (I4’OMT) from plants. At a protein level, a methyltransferase, which catalyses the 4’-O-methylation of the isoflavone daidzein, was purified from Cicer arietinum L (Wengenmayer et al., 1974), indicating that the 4’-O-methylation for biosynthesis of 4’-O-methylated isoflavonoids can take place at the isoflavone stage. However, in alfalfa (Medicago sativa L.) seedlings, radiolabeled daidzein is not incorporated into 4’-O-methylated isoflavonoids (Dewick and Martin, 1979). Paradoxically, biosynthesis of the 4’-O-methylated isoflavonoids in alfalfa suspension cells strongly correlates with the isoflavone 7-O-methyltransferase (I7OMT) activity (Edwards and Dixon, 1991; He et al., 1998), and over-expression of the I7OMT led to enhanced levels of 4’-O-methylated isoflavonoids in the elicited alfalfa leaves(He and Dixon, 2000). In the elicited alfalfa leaves, the operationally soluble I7OMT re-locates to the endoplasmic reticulum where the 2-HIS naturally resides, leading to an interesting hypothesis that the association with other isoflavonoid pathway enzymes may change the region-specificity of I7OMT from the 7- to 4’-position in vivo (Liu and Dixon, 2001). On the other hand, an enzyme assay with the Glycyrrhiza echinata cell-free extract demonstrated that the 4’-O-methylation occurs at the level of 2,7,4’-trihydroxyisoflavanone (Akashi et al., 2000). This is supported by the molecular cloning and characterization of cDNAs encoding the 2,7, 4’-tri-hydroxyisoflavanone 4’-O-methyltransferase (HI4’OMT) from Glycyrrhiza echinata (Akashi et al., 2003), and Medicago truncatula (Deavours et al., 2006). The isoflavone daidzein could not be converted by HI4’OMT, and it only recognizes 2-trihydroxy-isoflavanone as the direct methyl acceptor (Akashi et al., 2003), suggesting that HI4’OMT catalyzes the 4’-O-methylation reaction only at the isoflavanone stage. Therefore, the history leading to the finding of isoflavonoid 4’-O-methyltransferases demonstrates two alternative pathways likely involved: one is the simplest 4’-O-methylation occurring at the isoflavone stage, and the other is the reaction performed at the level of 2-hydroxyisoflavanones. From P. lobata, Li et al. identified a novel isoflavone 4’-O-methyltransferase (designated PlOMT9) that is capable of directly 4’-O-methylating isoflavones (Li et al., 2016a). Because that PlOMT9 shows the highest degree of amino acid identity with the isoflavone 7-O-methyltransferases (I7OMTs), PlOMT9 was initially presumed as an I7OMT. However, yeast cells expressing PlOMT9 efficiently performed the 4’-O-methylation of daidzein, genistein, prunetin, and isoformononetin (Li et al., 2016a), demonstrating that PlOMT9 functions actually as a I4’OMT. The I4’OMT activity catalyzed by PlOMT9 was further confirmed by in vitro assays using the purified recombinant PlOMT9 (Li et al., 2016a). Moreover, the recombinant PlOMT9 was not active with 2,7,4’-trihydroxy-isoflavanone, which is the natural substrate of HI4’OMT (Akashi et al., 2003). In addition to the main I4’OMT activity, PlOMT9 retains an extremely low 7-O-methylation activity, such as O-methylating daidzein at C7 position to yield trace amounts of isoformononetin. Over-expression of PlOMT9 in Glycine max hairy roots increased the levels of formononetin and ononin (formononetin 7-O-glucoside) by 111.2% and 940.9%, respectively, in comparison with the controls. P. lobata contains a HI4’OMT-like enzyme (Li et al., 2016a), which shares 73% amino acid identity with the HI4’OMT from G. echinata (Akashi et al., 2003), but it is inactive either with 2,7,4’-trihydroxy-isoflavanone or the isoflavone daidzein (Li et al., 2016a).
In summary, utilizing the transcriptomic analysis, in combination with in vitro biochemical analysis of recombinant protein, provides a strong basis for understanding the biosynthetic mechanism of glycosylation and methylation of isoflavonoids in P. lobata (Figure 2). For the O-glycosylation of isoflavonoids in P. lobata, either 7-O- or 4’-O-glycosyltransferase protein is from members of the UGT88E subgroup. For the C-glycosylation of isoflavonoids, PlUGT43 (official designated UGT71T5) is the only isoflavone C-GT identified from plants so far. For the O-methylation of isoflavonoids in P. lobata, both 3’- and 4’-O-methyltransferases perform the methylation reactions at the isoflavone stage, directly utilizing isoflavones as the best acceptors.
Of particular value among the isoflavonoids are puerarin and its derivatives, which are produced exclusively in Pueraria species. Puerarin has been established as a clinical drug to deal with cardiovascular diseases (Wang et al., 2022). By expressing the PlUGT43, in combination with other pathway genes, the production of puerarin directly from glucose could be achieved in yeast at a concentration of 72.8 mg/L (Liu et al., 2021). The poor water solubility of puerarin is still a challenge in narrowing its treatment window in clinical usage (Liu et al., 2016). Considering that glycosylation is the most effective way to increase water solubility of small molecules (Li et al., 2004), the PlUGT2, which is capable of glucosylating puerarin, would provide an excellent template for further designing novel enzymes to increase the water solubility of puerarin.
CL: Writing – original draft. YZ: Funding acquisition, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the National Natural Science Foundation of China (32270416; 31870275; 31170284), and the National Key R&D Program of China (2018YFC1706200).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Adolfo, L. M., Burks, D., Rao, X. L., Alvarez-Hernandez, A., Dixon, R. A. (2022). Evaluation of pathways to the-glycosyl isoflavone puerarin in roots of kudzu (Pueraria montana lobata). Plant Direct 6 (9), e442. doi: 10.1002/pld3.442
Ahmad, B., Khan, S., Liu, Y., Xue, M. Z., Nabi, G., Kumar, S., et al. (2020). Molecular mechanisms of anticancer activities of Puerarin. Cancer Manage. Res. 12, 79–90. doi: 10.2147/Cmar.S233567
Akashi, T., Sawada, Y., Aoki, T., Ayabe, S. (2000). New scheme of the biosynthesis of formononetin involving 2,7,4’-trihydroxyisoflavanone but not daidzein as the methyl acceptor. Biosci. Biotechnol. Biochem. 64 (10), 2276–2279. doi: 10.1271/bbb.64.2276
Akashi, T., Sawada, Y., Shimada, N., Sakurai, N., Aoki, T., Ayabe, S. (2003). cDNA cloning and biochemical characterization of S-adenosyl-L-methionine: 2,7,4’-trihydroxyisoflavanone 4’-O-methyltransferase, a critical enzyme of the legume isoflavonoid phytoalexin pathway. Plant Cell Physiol. 44 (2), 103–112. doi: 10.1093/pcp/pcg034
Bao, Y. O., Zhang, M., Qiao, X., Ye, M. (2022). Functional characterization of a C-glycosyltransferase from Pueraria lobata with dual-substrate selectivity. Chem. Commun. 58 (88), 12337–12340. doi: 10.1039/d2cc04279g
Brazier-Hicks, M., Evans, K. M., Gershater, M. C., Puschmann, H., Steel, P. G., Edwards, R. (2009). The C-glycosylation of flavonoids in cereals. J. Biol. Chem. 284 (27), 17926–17934. doi: 10.1074/jbc.M109.009258
Chen, H., Pang, Z., Qiao, Q., Xia, Y., Wei, Y., Gao, Y., et al. (2021). Puerarin-na chelate hydrate simultaneously improves dissolution and mechanical behavior. Mol. Pharm. 18 (7), 2507–2520. doi: 10.1021/acs.molpharmaceut.1c00005
Deavours, B. E., Liu, C. J., Naoumkina, M. A., Tang, Y., Farag, M. A., Sumner, L. W., et al. (2006). Functional analysis of members of the isoflavone and isoflavanone O-methyltransferase enzyme families from the model legume Medicago truncatula. Plant Mol. Biol. 62 (4-5), 715–733. doi: 10.1007/s11103-006-9050-x
Dewick, P. M., Martin, M. (1979). Biosynthesis of pterocarpan, isoflavan and coumestan metabolites of Medicago sativa: Chalcone,isoflavone and isoflavanone precursors. Phytochemistry 18, 597–602. doi: 10.1016/S0031-9422(00)84267-3
Dhaubhadel, S., Farhangkhoee, M., Chapman, R. (2008). Identification and characterization of isoflavonoid specific glycosyltransferase and malonyltransferase from soybean seeds. J. Exp. Bot. 59 (4), 981–994. doi: 10.1093/jxb/ern046
Duan, H. Y., Wang, J., Zha, L. P., Peng, H. S., Zhao, Y. P., Yuan, Y., et al. (2022). Molecular cloning and functional characterization of an isoflavone glucosyltransferase from. Chin. J. Nat. Med. 20 (2), 133–138. doi: 10.1016/S1875-5364(21)60105-X
Edwards, R., Dixon, R. A. (1991). Isoflavone O-methyltransferase activities in elicitor-treated cell suspension cultures of Medicago sativa. Phytochemistry 30, 2597–2606. doi: 10.1016/0031-9422(91)85107-B
Ehrman, T. M., Barlow, D. J., Hylands, P. J. (2007). Phytochemical informatics of traditional Chinese medicine and therapeutic relevance. J. Chem. Inf. Model. 47 (6), 2316–2334. doi: 10.1021/ci700155t
Falcone Ferreyra, M. L., Rius, S. P., Casati, P. (2012). Flavonoids: biosynthesis, biological functions, and biotechnological applications. Front. Plant Sci. 3. doi: 10.3389/fpls.2012.00222
Fang, C. B., Wan, X. C., Tan, H. R., Jiang, C. J. (2006). Separation and determination of isoflavonoids in several kudzu samples by high-performance capillary electrophoresis (HPCE). Annali Di Chimica 96 (1-2), 117–124. doi: 10.1002/adic.200690002
Funaki, A., Waki, T., Noguchi, A., Kawai, Y., Yamashita, S., Takahashi, S., et al. (2015). Identification of a highly specific isoflavone 7-O-glucosyltransferase in the soybean (Glycine max (L.) Merr.). Plant Cell Physiol. 56 (8), 1512–1520. doi: 10.1093/pcp/pcv072
Gutmann, A., Nidetzky, B. (2012). Switching between O- and C-Glycosyltransferase through exchange of active-site motifs. Angewandte Chemie-International Edition 51 (51), 12879–12883. doi: 10.1002/anie.201206141
He, X. Z., Blount, J. W., Ge, S. J., Tang, Y. H., Dixon, R. A. (2011). A genomic approach to isoflavone biosynthesis in kudzu (Pueraria lobata). Planta 233 (4), 843–855. doi: 10.1007/s00425-010-1344-1
He, X. Z., Dixon, R. A. (2000). Genetic manipulation of isoflavone 7-O-methyltransferase enhances biosynthesis of 4’-O-methylated isoflavonoid phytoalexins and disease resistance in alfalfa. Plant Cell 12 (9), 1689–1702. doi: 10.1105/tpc.12.9.1689
He, X. Z., Reddy, J. T., Dixon, R. A. (1998). Stress responses in alfalfa (Medicago sativa L). XXII. cDNA cloning and characterization of an elicitor-inducible isoflavone 7-O-methyltransferase. Plant Mol. Biol. 36 (1), 43–54. doi: 10.1023/a:1005938121453
Hirade, Y., Kotoku, N., Terasaka, K., Saijo-Hamano, Y., Fukumoto, A., Mizukami, H. (2015). Identification and functional analysis of 2-hydroxyflavanone C-glucosyltransferase in soybean (Glycine max). FEBS Lett. 589 (15), 1778–1786. doi: 10.1016/j.febslet.2015.05.010
Inoue, T., Fujita, M. (1977). Biosynthesis of puerarin in Pueraria root. Chem. Pharm. Bull. 25, 3226–3231. doi: 10.1248/cpb.25.3226
Jung, W., Yu, O., Lau, S. M. C., O’Keefe, D. P., Odell, J., Fader, G., et al. (2000). Identification and expression of isoflavone synthase, the key enzyme for biosynthesis of isoflavones in legumes. Nat. Biotechnol. 18 (2), 208–212. doi: 10.1038/72671
Keung, W. M., Lazo, O., Kunze, L., Vallee, B. L. (1996). Potentiation of the bioavailability of daidzin by an extract of Radix puerariae. Proc. Natl. Acad. Sci. U.S.A. 93 (9), 4284–4288. doi: 10.1073/pnas.93.9.4284
Koster, J., Barz, W. (1981). UDP-glucose:isoflavone 7-O-glucosyltransferase from roots of chick pea (Cicer arietinum L.). Arch. Biochem. Biophys. 212 (1), 98–104. doi: 10.1016/0003-9861(81)90347-7
Li, D., Park, S. H., Shim, J. H., Lee, H. S., Tang, S. Y., Park, C. S., et al. (2004). In vitro enzymatic modification of puerarin to puerarin glycosides by maltogenic amylase. Carbohydr. Res. 339 (17), 2789–2797. doi: 10.1016/j.carres.2004.09.017
Li, G., Zhang, Q., Wang, Y. (2010). Chemical constituents from roots of Pueraria lobata. Zhongguo Zhong Yao Za Zhi 35 (23), 3156–3160. doi: 10.4268/cjcmm20102314
Li, J., Li, C. F., Gou, J. B., Wang, X., Fan, R. Y., Zhang, Y. S. (2016a). An alternative pathway for formononetin biosynthesis in. Front. Plant Sci. 7. doi: 10.3389/Fpls.2016.00861
Li, J., Li, C. F., Gou, J. B., Zhang, Y. S. (2016b). Molecular cloning and functional characterization of a novel isoflavone 3’-O-methyltransferase from. Front. Plant Sci. 7. doi: 10.3389/Fpls.2016.00793
Li, J., Li, Z. B., Li, C. F., Gou, J. B., Zhang, Y. S. (2014). Molecular cloning and characterization of an isoflavone 7-O-glucosyltransferase from. Plant Cell Rep. 33 (7), 1173–1185. doi: 10.1007/s00299-014-1606-7
Liu, C. J., Dixon, R. A. (2001). Elicitor-induced association of isoflavone O-methyltransferase with endomembranes prevents the formation and 7-O-methylation of daidzein during isoflavonoid phytoalexin biosynthesis. Plant Cell 13 (12), 2643–2658. doi: 10.1105/tpc.13.12.2643
Liu, G., Liu, Z., Yuan, S. (2016). Recent advances in methods of puerarin biotransformation. Mini Rev. Med. Chem. 16 (17), 1392–1402. doi: 10.2174/1389557516666160505114456
Liu, Q. L., Liu, Y., Li, G., Savolainen, O., Chen, Y., Nielsen, J. (2021). De novo biosynthesis of bioactive isoflavonoids by engineered yeast cell factories. Nat. Commun. 12 (1), 6085. doi: 10.1038/S41467-021-26361-1
Nagashima, S., Inagaki, R., Kubo, A., Hirotani, M., Yoshikawa, T. (2004). cDNA cloning and expression of isoflavonoid-specific glucosyltransferase from cell-suspension cultures. Planta 218 (3), 456–459. doi: 10.1007/s00425-003-1118-0
Nagatomo, Y., Usui, S., Ito, T., Kato, A., Shimosaka, M., Taguchi, G. (2014). Purification, molecular cloning and functional characterization of flavonoid C-glucosyltransferases from Fagopyrum esculentum M. (buckwheat) cotyledon. Plant J. 80 (3), 437–448. doi: 10.1111/tpj.12645
Noguchi, A., Saito, A., Homma, Y., Nakao, M., Sasaki, N., Nishino, T., et al. (2007). A UDP-glucose:isoflavone 7-O-glucosyltransferase from the roots of soybean (Glycine max) seedlings. J. Biol. Chem. 282 (32), 23581–23590. doi: 10.1074/jbc.M702651200
Ohshima, Y., Okuyama, T., Takahashi, K., Takizawa, T., Shibata, S. (1988). Isolation and high performance liquid chromatography (HPLC) of isoflavonoids from the Pueraria root. Planta Med. 54 (3), 250–254. doi: 10.1055/s-2006-962420
Rong, H., Stevens, J. F., Deinzer, M. L., Cooman, L. D., Keukeleire, D. D. (1998). Identification of isoflavones in the roots of Pueraria lobata. Planta Med. 64 (7), 620–627. doi: 10.1055/s-2006-957534
Steele, C. L., Gijzen, M., Qutob, D., Dixon, R. A. (1999). Molecular characterization of the enzyme catalyzing the aryl migration reaction of isoflavonoid biosynthesis in soybean. Arch. Biochem. Biophys. 367 (1), 146–150. doi: 10.1006/abbi.1999.1238
Sun, Y. J., Zhang, H. M., Cheng, M., Cao, S. J., Qiao, M., Zhang, B. L., et al. (2019). New hepatoprotective isoflavone glucosides from Pueraria lobata (Willd.) Ohwi. Nat. Prod. Res. 33 (24), 3485–3492. doi: 10.1080/14786419.2018.1484461
Vogt, T., Jones, P. (2000). Glycosyltransferases in plant natural product synthesis: characterization of a supergene family. Trends Plant Sci. 5 (9), 380–386. doi: 10.1016/s1360-1385(00)01720-9
Wang, D., Bu, T., Li, Y. Q., He, Y. Y., Yang, F., Zou, L. (2022). Pharmacological activity, pharmacokinetics, and clinical research progress of puerarin. Antioxidants 11 (11), 2121. doi: 10.3390/Antiox11112121
Wang, X., Fan, R., Li, J., Li, C., Zhang, Y. (2016). Molecular cloning and functional characterization of a novel (Iso)flavone 4’,7-O-diglucoside glucosyltransferase from Pueraria lobata. Front. Plant Sci. 7. doi: 10.3389/fpls.2016.00387
Wang, X., Li, S. T., Li, J., Li, C. F., Zhang, Y. S. (2015). De novo transcriptome sequencing in Pueraria lobata to identify putative genes involved in isoflavones biosynthesis. Plant Cell Rep. 34 (5), 733–743. doi: 10.1007/s00299-014-1733-1
Wang, X., Li, C. F., Zhou, C., Li, J., Zhang, Y. S. (2017). Molecular characterization of the C-glucosylation for puerarin biosynthesis in Pueraria lobata. Plant J. 90 (3), 535–546. doi: 10.1111/tpj.13510
Wang, X., Li, C. F., Zhou, Z. L., Zhang, Y. S. (2019). Identification of three (Iso)flavonoid glucosyltransferases from Pueraria lobata. Front. Plant Sci. 10. doi: 10.3389/Fpls.2019.00028
Wang, Y., Ma, Y., Ma, Y., Du, Y., Liu, Z., Zhang, D., et al. (2012). Formulation and pharmacokinetics evaluation of puerarin nanocrystals for intravenous delivery. J. Nanosci. Nanotechnol. 12 (8), 6176–6184. doi: 10.1166/jnn.2012.6436
Wang, S. G., Zhang, S. M., Wang, S. P., Gao, P., Dai, L. (2020). A comprehensive review on Pueraria: Insights on its chemistry and medicinal value. Biomed. Pharmacother. 131, 110734. doi: 10.1016/J.Biopha.2020.110734
Wen, L. R., Jiang, Y. M., Yang, J. L., Zhao, Y. P., Tian, M. M., Yang, B. (2017). Structure, bioactivity, and synthesis of methylated flavonoids. Ann. New York Acad. Sci. 1398 (1), 120–129. doi: 10.1111/nyas.13350
Wengenmayer, H., Ebel, J., Grisebach, H. (1974). Purification and properties of a S-adenosylmethionine: isoflavone 4’-O-methyltransferase from cell suspension cultures of Cicer arietinum L. Eur. J. Biochem. 50 (1), 135–143. doi: 10.1111/j.1432-1033.1974.tb03881.x
Wong, K. H., Li, G. Q., Li, K. M., Razmovski-Naumovski, V., Chan, K. (2011). Kudzu root: Traditional uses and potential medicinal benefits in diabetes and cardiovascular diseases. J. Ethnopharmacol. 134 (3), 584–607. doi: 10.1016/j.jep.2011.02.001
Xi, C., Zhang, M. Y., Li, B. T., Meng, X. W., Xu, S. C., Du, H., et al. (2023). Metabolomics of the anti-inflammatory effect of Pueraria lobata and Pueraria lobata var. Thomsonii in rats. J. Ethnopharmacol. 306, 116144. doi: 10.1016/J.Jep.2023.116144
Xi, H. T., Zhu, Y. R., Sun, W. W., Tang, N., Xu, Z. Q., Shang, X. H., et al. (2023). Comparative transcriptome analysis of provides candidate genes involved in puerarin biosynthesis and its regulation. Biomolecules 13 (1), 170. doi: 10.3390/Biom13010170
Yang, X., Yang, Y., Zhou, S., Gong, X., Dai, Q., Zhang, P., et al. (2017). Puerarin stimulates osteogenic differentiation and bone formation through the ERK1/2 and p38-MAPK signaling pathways. Curr. Mol. Med. 17 (7), 488–496. doi: 10.2174/1566524018666171219101142
Zhang, Z., Lam, T. N., Zuo, Z. (2013). Radix Puerariae: an overview of its chemistry, pharmacology, pharmacokinetics, and clinicpal use. J. Clin. Pharmacol. 53 (8), 787–811. doi: 10.1002/jcph.96
Zhao, T. F., Han, J., Chen, Y. Q., Wan, H. T., Bie, X. D. (2007). The mechanism of 3-methoxy puerarin on decreasing the cerebral ischemia-reperfusion injury in rats. Asia Pacific J. Clin. Nutr. 16, 302–304.
Keywords: glycosylation, methylation, isoflavonoid, pueraria, biosynthesis
Citation: Li C and Zhang Y (2023) Glycosylation and methylation in the biosynthesis of isoflavonoids in Pueraria lobata. Front. Plant Sci. 14:1330586. doi: 10.3389/fpls.2023.1330586
Received: 31 October 2023; Accepted: 01 December 2023;
Published: 15 December 2023.
Edited by:
Deyu Xie, North Carolina State University, United StatesReviewed by:
Mehran Dastmalchi, McGill University, CanadaCopyright © 2023 Li and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yansheng Zhang, emhhbmd5czFAc2h1LmVkdS5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.