- 1Key Laboratory of Digital Dry Land Crops of Zhejiang Province, Zhejiang Academy of Agricultural Sciences, Hangzhou, China
- 2Zhejiang Academy of Agricultural Sciences, National Barley Improvement Center, Hangzhou, China
- 3College of Advanced Agricultural Sciences, Zhejiang Agricultural and Forestry University, Hangzhou, China
The chloroplast is an important cellular organelle and metabolic hub, which is not only responsible for plant photosynthesis but is also involved in the de novo biosynthesis of pigments, fatty acids, and hormone metabolisms. Several genes that are responsible for rice leaf color variations have been reported to be directly or indirectly involved in chlorophyll biosynthesis and chloroplast development, whereas a few genes have been functionally confirmed to be responsible for leaf color changes in barley at the molecular level. In this study, we obtained a yellow leaf and dwarf ygl8 mutant from the progeny of Morex (a variety of barley) seeds treated with EMS. We performed bulked-segregant analysis (BSA) and RNA-seq analysis and targeted a UMP kinase encoding gene, YGL8, which generated a splicing site change between exon 5 and 6 of YGL8 due to a G to A single-nucleotide transition in the 5th exon/intron junction in the ygl8 mutant. The splicing site change between exon 5 and 6 of YGL8 had no effects on chloroplast subcellular localization but resulted in an additional loop in the UMP kinase domain, which might disturb the access of the substrates. On one hand, the splicing site change between exon 5 and 6 of YGL8 downregulated the transcriptional expression of chloroplast-encoded genes and chlorophyll-biosynthesis-related genes in a temperature-dependent manner in the ygl8 mutant. On the other hand, the downregulation of bioactive GA-biosynthesis-related GA20ox genes and cell-wall-cellulose-biosynthesis-related CesA genes was also observed in the ygl8 mutant, which led to a reduction in plant height. Our study will facilitate the understanding of the regulation of leaf color and plant height in barley.
Introduction
Photosynthesis, the important process of converting sunlight into chemical energy, provides the basic biochemical substances, such as carbohydrates for plant growth and development, and takes place in the chloroplasts (Nelson and Junge, 2015; Stirbet et al., 2020). Chloroplasts are semi-autonomous organelles and the metabolic centers involved in the synthesis of pigments, such as chlorophyll a and b, nucleotides, amino acids, fatty acids, phytohormones, and many metabolites (Nelson and Junge, 2015; Wang and Grimm, 2015; Daniell et al., 2016; Stirbet et al., 2020). During the course of evolution, most of the chloroplast-encoded genes were transferred into the cell nucleus, and the remaining genes in the chloroplast genome mainly encode proteins for photosynthesis and prokaryotic transcription and translation, including photosynthetic apparatus (Nelson and Junge, 2015), RNA processing factor (matK) (Zoschke et al., 2010; Barthet et al., 2020), RNA polymerase subunits, and ribosomal proteins (Dobrogojski et al., 2020). Thus, the coordination of nuclear-encoded and chloroplast-encoded gene expression is important for chloroplast and plant development (Dobrogojski et al., 2020), and the abnormal development of chloroplasts could decrease photosynthetic efficiency and pigment content, subsequently leading to plant growth retardation and chlorosis (Hein et al., 2009; Zhu et al., 2016; Li et al., 2019; Li et al., 2021).
In rice, more than 100 loci and 40 genes involved in leaf color mutations have been cloned, and the causal genes are mainly directly or indirectly related to chlorophyll biosynthesis and chloroplast development (Chen et al., 2013; Zhou et al., 2013a; Zhou et al., 2013b; Wang et al., 2016; Zhu et al., 2016; Zhou et al., 2017; Chen et al., 2018a; Dong et al., 2019). Although a large number of mutants with full or partial chlorophyll deficiency have been described in barley (Gálová et al., 2000; Prina et al., 2003; Franckowiak and Lundqvist, 2012; Qin et al., 2019), few genes were functionally confirmed at the molecular level due to the relatively late publication of a chromosome-level reference genome (Mascher et al., 2017). The nuclear-encoded PORB (protochlorophyllide oxidoreductase), encoding a vital enzyme in the chlorophyll biosynthesis pathway, was truncated by 60Co-γ ray treatment in the chlorophyll-deficient barley NYB mutant (Liu et al., 2008). The disruption of nuclear-encoded HvCAO, which encodes a chlorophyllide oxygenase and is essential for chlorophyll b biosynthesis, resulted in no chlorophyll b formation and a light green leaf phenotype in a barley fch2 mutant (Mueller et al., 2012). The temperature dependent green-revertible albino leaf of a barley whs18 mutant was caused by a premature stop codon mutation of nuclear-encoded HvFLN1 (fructokinase-1-like), which phosphorylates fructose in the carbohydrate metabolism (Qin et al., 2015). Mutations of nuclear-encoded chloroplast-localized HvCMF7 and HvCMF3, members of the CCT (CONSTANS, CONSTANS-like, and TIMING OF CAB1) motif protein family, caused leaf variegation and chlorophyll-deficient phenotypes, respectively, due to retarded chloroplast development that disrupted chloroplast ribosome formation (Li et al., 2019; Li et al., 2021).
Pyrimidine nucleotides are important building blocks of nucleic acids and also function as cofactors for the biosynthesis of a variety of carbohydrates, such as cellulose, hemicellulose, pectin, and secondary metabolites (Kafer et al., 2004). The UMP kinase is a ubiquitous key enzyme for transferring the phosphate group from ATP to UMP to generate UDP, which is subsequently converted to UTP, CTP, and TTP for RNA and DNA synthesis (Charlier et al., 2018; Witte and Herde, 2020). The malfunction of rice YLG8 (yellow-green leaf 8), which encodes a chloroplast-localized UMP kinase, significantly decreased leaf UDP and UTP content and the transcription and translation of chloroplast-encoded genes, resulting in abnormal chloroplast development, which reduced the chlorophyll content and decreased plant height (Zhu et al., 2016; Chen et al., 2018a; Dong et al., 2019). Additionally, mutation of the Arabidopsis chloroplast-localized UMP kinase encoding Dpt1 (defect in psaA/B transcript accumulation 1) significantly decreased the transcript levels of chloroplast-encoded photosystem I subunits psaA/B and impaired chloroplast development, and the mutant failed to grow photoautotrophically and exhibited pale-yellowish leaves and slower plant growth in MS medium supplemented with sucrose (Hein et al., 2009).
Plant height is regulated by a complex and coordinated network involving gibberellic acid (GA) and cellulose biosynthesis (McFarlane et al., 2014; Wang and Wang, 2022). GA, an important plant hormone, plays a vital role in many processes, such as stem or internode elongation (Gupta and Chakrabarty, 2013). Although more than 130 GAs have been discovered, only four types function as bioactive GAs in flowering plants (He et al., 2020). GA20ox, a 2-oxoglutarate-dependent dioxygenase (2-ODD), plays an essential role in bioactive GA biosynthesis (Hedden, 2020; He et al., 2020). The disruption of GA20ox genes could result in insufficient bioactive GAs and thus decrease the stem or internode length, ultimately affecting plant height (Sasaki et al., 2002; Paciorek et al., 2022). Cellulose, composed of hydrogen-bonded β-1,4 glucans, is the key component of the cell wall, and its biosynthesis mainly relies on the complexes of cellulose synthase (CesA), belonging to the glycosyltranferase-2 (GT-2) superfamily, and CesA-associated proteins (McFarlane et al., 2014). The expression of CesA genes could be activated by a GA-mediated signaling pathway (Huang et al., 2015), and the malfunction of CesA genes could decrease the plant cellulose content and affect cell wall characteristics, as well as cell size and plant morphologies, such as organ size and plant height (Burn et al., 2002; Wang et al., 2012; Chantreau et al., 2015).
To date, numerous proteins involved in chlorophyll biosynthesis, photosystems, and chloroplast development have been reported to be involved in the regulations of rice leaf color. However, our knowledge of the regulation network of barley leaf color variations is still limited. In the present study, we isolated a yellow leaf mutant, ygl8, from the progeny of barley variety Morex seeds treated by EMS and found that the splicing site change between exon 5 and 6 of the chloroplast-localized UMP kinase encoding the HvGYL8 gene, due to a G to A single-nucleotide transition in the 5th exon/intron junction, caused yellow leaf and a dwarf phenotype in the ygl8 mutant in a temperature-dependent manner by downregulating chloroplast-encoded genes and the GA20ox and CesA genes, respectively.
Materials and methods
Plant materials and temperature treatments
Barley (Hordeum vulgare cv. Morex) seeds were treated with 0.03 mol/L EMS for 16 h, and the yellow leaf phenotype mutant was identified in M2 individuals. The mutant was self-pollinated for another five generations to obtain a relatively stably inherited yellow leaf mutant named ylg8. Morex was crossed with the ygl8 mutant, and F1 plants were self-pollinated to generate F2 populations for genetic analysis. The greenhouse and field experiments were carried out for hydroponic experiments and BSA and RNA-seq experiments at the Zhejiang Academy of Agricultural Sciences (Hangzhou, Zhejiang Province in China), respectively.
For hydroponic experiments, the barley seeds were sterilized with 10% commercial NaClO, rinsed with tap water, and then germinated in BSM solution (0.5 mM KCl + 0.1 mM CaCl2) for 2 days. Afterward, BSM was changed to 1/5 Hoagland solution with a photoperiod of 14/10 h, light intensity of 200 ± 25 μmol·m−2·s−1, relative humidity of 60%, and constant temperature of 24°C for 8 days (Cai et al., 2022). For different temperature treatments, the seedlings were grown in incubators with 14/10 h light/dark at a constant air temperature of 10°C, 24°C, or 30°C.
Measurement of chlorophyll a and b and carotenoid contents
Equal fresh weights (approximately 50 mg) of the newly expanded leaves were cut and immersed in 10 ml extract solution (ethanol: acetone: water = 4.5: 4.5: 1) for 16 h in the dark. The homogenates were centrifuged at 3,000 × g for 5 min to remove residual plant debris. The supernatants of these samples were analyzed, and the chlorophyll a and b and carotenoid contents were measured using a spectrophotometer at 663, 645, and 470 nm, and determined using the Lichtenthaler method (Chen et al., 2013).
DNA and RNA template preparation, library construction, and sequencing
The newest fully expanded leaves of Morex and the ygl8 mutant at the four-leaf age were collected 1 month after planting in the field and quickly frozen in liquid nitrogen. The total DNA was extracted following the modified hexadecyl trimethylammonium bromide (CTAB) method (Murray and Thompson, 1980). For whole-genome sequencing, the bulked DNA for MutMap analysis was prepared by mixing DNA from 30 ygl8 individuals in an equal ratio, and the parent DNA for MutMap analysis was prepared by extracting DNA from one Morex sample. A paired-end sequencing library was constructed with an insert size of approximately 350 bp and sequenced on the Illumina Hiseq X Ten platform.
For RNA-seq, the newest fully expanded leaves of Morex and the ygl8 mutant were collected at the stem elongation stage in the field. Total RNA was isolated using an RNAprep Pure Plant Kit (TIANGEN). RNA-seq library construction was carried out following a method described previously (Shen et al., 2014) and sequenced on the Illumina Hiseq X Ten platform. All sequencing was carried out at Biozeron Biotech, Shanghai.
MutMap and RNA-Seq analysis
For MutMap analysis, more than 30 depth re-sequencing raw data of bulked pool and parent Morex ygl8 were filtered using fastp (Chen et al., 2018b) (Version 0.12.4) with the default parameters. The high-quality cleaned reads were aligned to the barley reference genome (Mascher et al., 2021) with BWA (Li and Durbin, 2009) (Version 0.7.17-r1188) using the default parameters. SAMtools (Li et al., 2009) (Version 1.14) was used to convert the mapping results into the sorted and duplication-marked BAM format. SNP detection was performed by BCFtools (Danecek and McCarthy, 2017) (Version 1.14) using the bcftools mpileup command with the parameters ‘–min-MQ 40 –min-BQ 18 –adjust-MQ 50’ and the bcftools call command with the default parameters. The resulting vcf files were filtered using the bcftools filter command with the parameters ‘INFO/MQ>=40 && %QUAL>20’, and the SNP information was extracted using VCFtools (Danecek et al., 2011) (Version 0.1.17). The average SNP-index of the bulked pool of ygl8 and the parent Morex was estimated via the 2 M sliding window with a 10-kb walking step using Mutplot (Sugihara et al., 2022) (Version 2.3.3) with the parameters ‘–N-bulk 30 –window 2000 –step 10 –min-depth 14 –max-depth 90 –N-rep 10000 –min-SNPindex 0.3’. The genome-wide SNP-index plot was obtained using a custom script written in R (version 4.3.0). The SNP variants were functionally annotated using ANNOVAR (Wang et al., 2010) (Version 2019-10-24).
For RNA-seq analysis, the RNA-seq raw sequencing data of leaf tissues of Morex and the ygl8 mutant were filtered using fastp (Chen et al., 2018b) (Version 0.12.4) with the default parameters. The high-quality cleaned reads were aligned to the reference genome consisting of the barley reference genome (Mascher et al., 2021) and chloroplast reference genome (Saski et al., 2007) using HISAT2 (Kim et al., 2019). Following alignments, the raw counts for each gene were derived using featureCounts implemented in the R package Rsubread (Liao et al., 2019) and normalized into the number of transcripts per kilobase of exon sequence in a gene per million mapped reads (TPM) using the TMM method (Robinson and Oshlack, 2010). The read alignments of the RNA-seq data to candidate genes were shown in a sashimi plot implemented in the IGV genome browser. The differentially expressed genes in leaf tissues between Morex and the ygl8 mutant were detected using the DESeq2 method (Love et al., 2014), and the significantly differentially expressed genes were identified with the standard of |log2FC| >1 and an adjusted P < 0.05. The GO and KEGG enrichment analysis were performed using the R package clusterProfiler (Yu et al., 2012) (Version 4.8.2), and the enrichment results were shown using the R package ggplot2 (Wickham, 2016) (Version 3.4.2).
Identification of homologous GA20ox and CesA in barley
The reference barley protein sequences were downloaded from Phytozome (https://phytozome-next.jgi.doe.gov/). Then, Arabidopsis GA20ox (AT4G25420) and CesA (AT4G32410) were used as query proteins to carry out a BLASTP search in the barley protein sequences with an E-value < 1e-10 as the threshold using DIAMOND (Buchfink et al., 2021) (Version 2.0.11) to identify the barley homologous GA20ox and CesA proteins. Furthermore, the hidden Markov models (HMMs) of the zf-UDP (PF14569.9) and Cellulose_synt (PF03552.17) domains, downloaded from Pfam 35.0 (http://pfam-legacy.xfam.org/), were used to identify the barley homologous CesA proteins using HMMER (http://hmmer.org/) with a “trusted cutoff and E-value < 0.01” as the threshold. The HMMs of the DIOX_N (PF14226.9) and 2OG-FeII_Oxy (PF03171.23) domains were used to identify the barley homologous GA20ox proteins. A total of seven homologous CesA and 24 GA20ox were identified in barley.
Construction of vectors
Primers were designed according to the open reading frames of the barley YGL8 and OsYSS1 (Zhou et al., 2017) genes. The corresponding sequences were amplified from the cDNA of barley (Morex or ygl8) or rice and cloned into the pMD18T vector (Takara). The clones were selected by PCR and sequenced for confirmation. For the subcellular localization of YGL8 and ygl8 in tobacco leaves, seamless cloning was used to introduce the YGL8 or ygl8 genes, fused to the GFP gene amplified from the pH7WGF2.0 vector, into the pCAMBIA1300 vector, after the 35S promoter amplified from the pH7WGF2.0 vector in advance, to obtain YGL8-GFP or ygl8-GFP. Additionally, seamless cloning was used to introduce the OsYSS1 gene, fused to the mCherry gene amplified from pSAT4a-mCherry-N1, into a modified pCAMBIA1300 vector to obtain OsYSS1-mCherry. The primer information is listed in Supplementary Table 1.
Subcellular localization of YGL8 in tobacco leaves
The subcellular localization vectors of YGL8 were transiently expressed in tobacco (Nicotiana benthamiana) leaves by Agrobacterium-mediated infiltration. OsYSS1-meCherry was used as a chloroplast marker. The Agrobacterium strain C58C1 harboring p19 was used to prevent the onset of post-transcriptional gene silencing (PTGS) in the infiltrated leaves. Infiltrated tobacco plants were grown for another 3 days for GFP and mCherry imaging using a Zeiss LSM710NLO confocal laser-scanning microscope. The excitation/emission wavelengths were 488 nm for GFP and 561 nm for mCherry.
Protein structure prediction and visualization
The protein structures of YGL8 and ygl8 were predicted using SWISS-MODEL (https://swissmodel.expasy.org/) with the default parameters. The resulting pdb files of the corresponding proteins were compared and shown using PyMOL (DeLano, 2002).
Quantitative real-time PCR
Total RNA was extracted from the newest fully expanded leaves of Morex and the ygl8 mutant at 2 weeks in the hydroponic experiment or stem elongation stage in the field experiment using an RNAprep Pure Plant Kit (Tiangen Co. Ltd., China). First-strand cDNAs were synthesized using a Fasting RT Kit with gDNase (Tiangen Co. Ltd., China). qRT-PCR was performed using the Talent qPCR PreMix (Tiangen Co. Ltd., China), and the barley actin gene was used as the endogenous control. Reactions containing SYBR premix were carried out in final volumes of 20 ul containing 0.3 umol of the appropriate primers and 2 * PCR master mix. The 2-△△t method was used to calculate the relative levels of gene expression. The primers used for qRT-PCR are listed in Supplementary Table 1.
Results
Phenotype of the ygl8 mutant
We isolated a yellow leaf mutant, ygl8, from the progeny of barley variety Morex seeds treated with EMS, and the newly emerged leaves of the ygl8 mutant were yellowish throughout the vegetative growth period (Figures 1A, B). To investigate whether the pigment contents changed in the yellow leaf of the ygl8 mutant, we detected the pigment contents of the newest fully expanded leaves of Morex and the ylg8 mutant grown in hydroponic solution for 3 weeks and observed significantly decreased pigment contents in the ygl8 mutant compared with Morex (Figure 1D). The chlorophyll a and b and carotenoid contents of the ygl8 mutant decreased 15%, 18%, and 29%, respectively, compared with those of Morex (Figure 1D). Additionally, we observed that the plant height of the ygl8 mutant decreased to 83% of that of Morex at the mature stage in the field (Figures 1C, E). These results suggest that pigment synthesis in the leaves of the ygl8 mutant was dramatically inhibited.
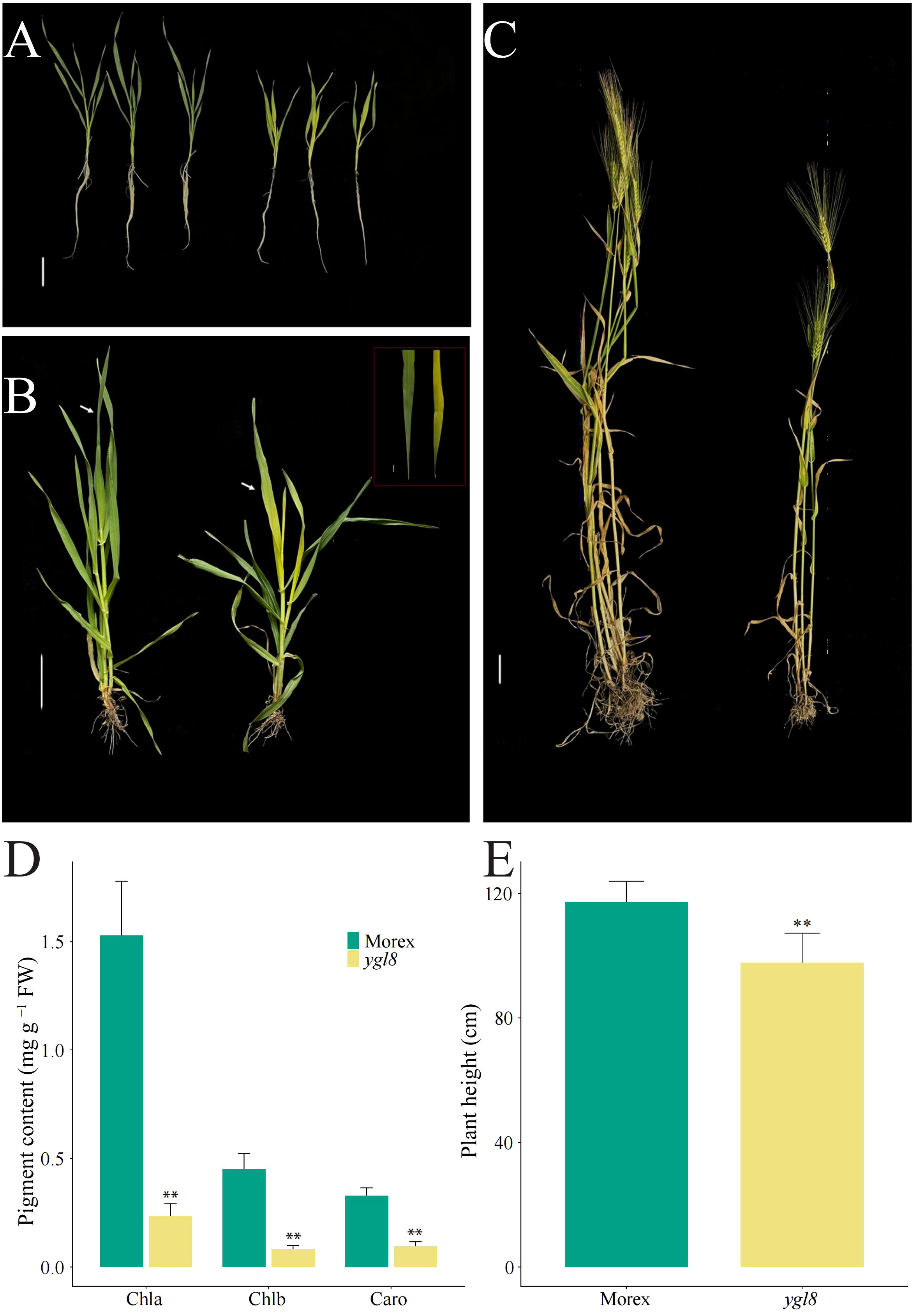
Figure 1 The yellow leaf ygl8 mutant shows significantly decreased pigment content and plant height. The phenotype of Morex (on the left) and the ygl8 mutant (on the right) at the 21-day seedling stage in the greenhouse (A), and at the stem elongation stage (B) and mature stage in the field (C). The white arrows indicate the newly merged leaves. (D, E) The pigment content (D) and plant heights (E) of Morex and the ygl8 mutant at the 21-day seeding stage in the greenhouse and at the mature stage in the field. Chla, Chlb, and Caro represent chlorophyll a, chlorophyll b and carotenoid, respectively. Scale bars: 5 cm. Data are mean + SD (n = 6) for (D, E). Significant differences between Morex and ygl8 mutant are indicated (**P < 0.01; Student’s t test).
Identification of the gene associated with the ygl8 mutant
For the rapid identification of the gene responsible for the yellow leaf phenotype in the ygl8 mutant, the bulked DNA pool, consisting of 30 randomly selected ygl8 individuals with a yellowish leaf phenotype, and Morex were subjected to whole-genome sequencing for SNP detection. In total, we obtained 1,397,745 M and 1,002,230 M clean reads for the Morex and ylg8 bulked pools, respectively (Supplementary Table 2). These reads were aligned to the Morex reference genome (Mascher et al., 2021) and 953,254 SNPs were detected.
For each SNP position, the value of the SNP-index was derived, and the ΔSNP-index values (the difference of the SNP-index of Morex and the bulked ygl8 pool) were plotted against each SNP position on the seven chromosomes (Supplementary Figure 1). Furthermore, 4,312 SNP positions with an ΔSNP-index = 1 were identified on seven chromosomes and functionally annotated according to genetic variant types. Then, we observed 130 SNP positions with an ΔSNP-index = 1 in genic regions, including 2 kb upstream and downstream, and the related 129 genes were focused on as candidate genes for the yellow leaf phenotype in the ygl8 mutant (Supplementary Table 3). Moreover, we compared gene expression in the leaf tissues of Morex and the ygl8 mutant at the stem elongation stage in the field using RNA-seq and found that 59 out of the 129 candidate genes were expressed in the leaf tissues (Supplementary Table 3).
To alleviate the effects of large variations of single ΔSNP-index values, the sliding window average values of the ΔSNP-index were calculated, and we further narrowed down the candidate genomic region to the chromosome 3H: 604,680,000-622,110,000, comprehensively considering the genetic variant types, gene functional annotations, and gene expression (Figure 2A; Supplementary Table 3). There were seven genes with an ΔSNP-index =1 in this region but only four were expressed in our RNA-seq data (Figure 2B). Based on the genetic variant types and functional annotations, we finally focused on HORVU.MOREX.r3.3HG0325830 (named YGL8), which encoded one UMP kinase and contained one G to A single-nucleotide transition in the 5th exon/intron junction in the ygl8 mutant; the transition might lead to a splicing site change between exon 5 and 6 of YGL8 in the ygl8 mutant. Notably, the mutations in the Arabidopsis and rice orthologous YGL8 genes were previously reported to affect chloroplast development, and thus resulted in a low chlorophyll content and yellowish leaf phenotype (Hein et al., 2009; Zhu et al., 2016; Chen et al., 2018a; Dong et al., 2019). To confirm the splicing site change between exon 5 and 6 of the YGL8 gene in the ygl8 mutant, we investigated the read alignments around the YGL8 gene using the RNA-seq data of Morex and the ygl8 mutant and observed the existence of the splicing site change between exon 5 and 6 of YGL8 in the ygl8 mutant (Figure 2C). Furthermore, Sanger sequencing revealed that the splicing site change between exon 5 and 6 of YGL8 resulted in a 30-bp insertion in the 5th exon in the ygl8 mutant (Figure 2C).
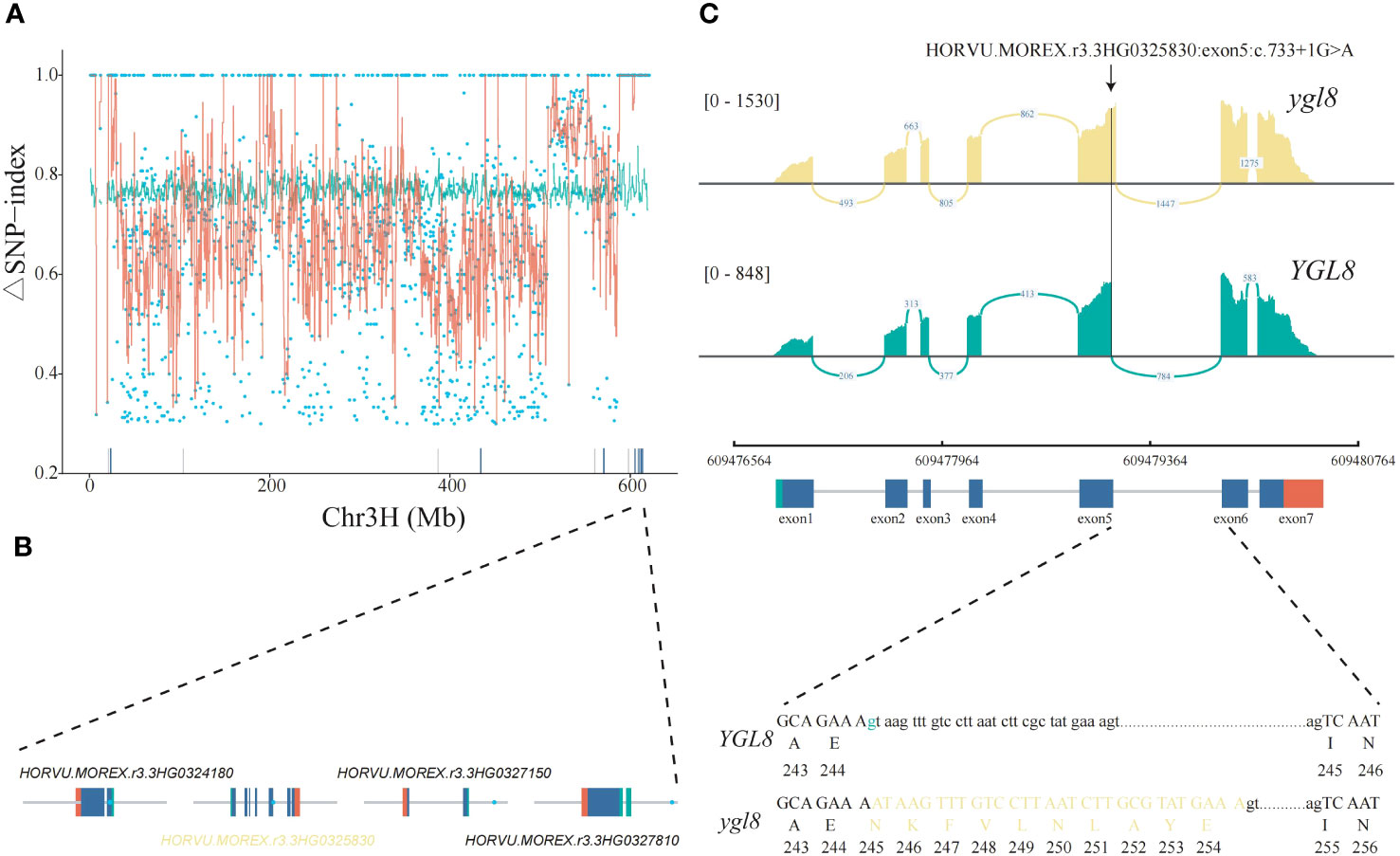
Figure 2 Identification of the candidate gene responsible for the yellow leaf phenotype of the ygl8 mutant by Mutmap and RNA-seq. (A) ΔSNP-index plot of the barley chromosome 3H generated by Mutmap analysis. The red lines correspond to the sliding window average values of the ΔSNP-index of 2-Mb intervals with a 10-Kb increment. The blue points represent SNPs identified between Morex and the ygl8 mutant. The green lines show the 99% confidence limit of the ΔSNP-index value under the null hypothesis. The blue and gray bars above the x axis demonstrate the expressed and unexpressed genes in the leaf RNA-seq data, respectively. (B) The gene structures and mutation positions of the four candidate genes expressed in leaves. The blue points represent the positions with an ΔSNP-index = 1. The green, blue, and red boxes indicate the 5′ UTR, CDS, and 3′ UTR, respectively. (C) The splicing site change between exon 5 and 6 of HORVU.MOREX.r3.3HG0325830 in the ylg8 mutant due to a G to A single-nucleotide transition at the 5th exon/intron junction. The SNP location impacting the splicing site change between exon 5 and 6 of YGL8 in the ygl8 mutant is highlighted with a black arrow. The read alignments are shown in a sashimi plot implemented in the IGV genome browser. Green marks the G to A transition. The exon and intron sequences are indicated in upper and lower cases, respectively. Yellow denotes the added sequences in the exon of the ygl8 gene for the splicing site change between exon 5 and 6.
To investigate whether the yellow leaf phenotype of the ygl8 mutant was controlled by a single gene, we crossed Morex and the ygl8 mutant to obtain F1 progeny, and the F1 plants were self-pollinated to generate F2 progeny. All F1 progeny exhibited the normal green leaf phenotype, and the leaf phenotype of the F2 plants segregated in a 3:1 ratio (150 green: 40 yellow), which indicated that the yellow leaf phenotype of the ygl8 mutant was conferred by a single recessive mutation. These results reveal that the yellow leaf phenotype of the ygl8 mutant might be the result of the splicing site change between exon 5 and 6 of YGL8 due to a G to A single-nucleotide transition in the 5th exon/intron junction.
The chloroplast subcellular localization of YGL8 is not affected by the splicing site change between exon 5 and 6
To comprehensively understand the transcriptional expression patterns of YGL8, the RNA-seq data from different developmental tissues of Morex were derived (Mascher et al., 2017), and the analysis demonstrated that YGL8 was constitutively expressed, with a relatively higher expression level in photosynthetic tissues (Figure 3A). To determine the influences of the splicing site change between exon 5 and 6 on the transcriptional level of YGL8, we examined the transcripts of YGL8 in the 3-week-old leaf tissues of Morex and the ygl8 mutant grown in hydroponic solution, and no significant transcriptional changes were observed (Figure 3B).
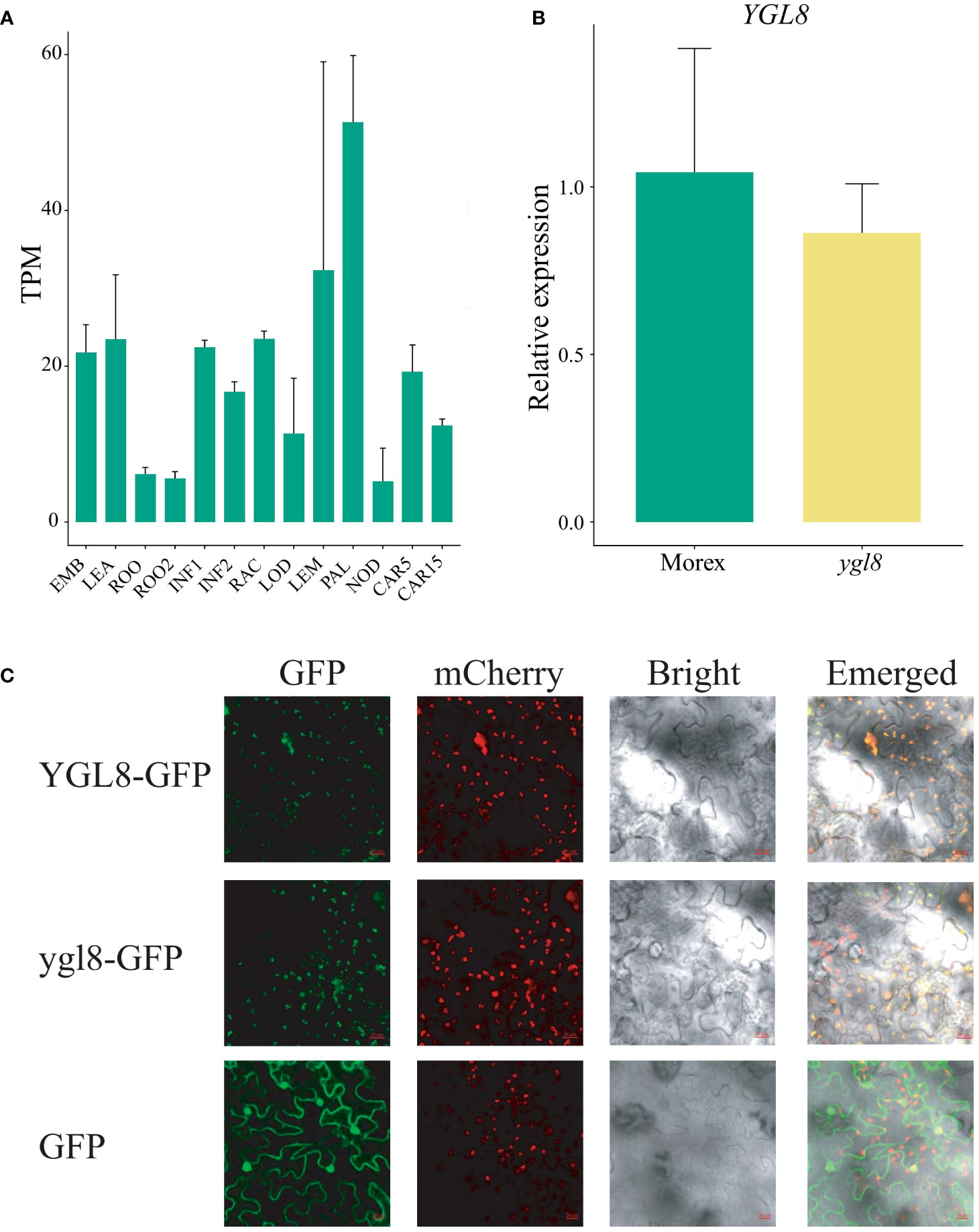
Figure 3 The splicing site change between exon 5 and 6 of YGL8 does not affect the gene transcript level and chloroplast subcellular localization. (A) The YGL8 gene transcript level in the published different developmental tissues (Mascher et al., 2017). EMB, embryo 4 days after planting (dap); LEA, leaf 17 dap; ROO, 17 dap root; ROO2, root 28 dap; INF1, inflorescences 30 dap; INF2, inflorescences 50 dap; RAC, rachis 35 dap inflorescence; LOD, lodicule 42 dap; LEM, lemma 42 dap; PAL, palea 42 dap; NOD, the third stem internode (42 dap); CAR5, caryopsis 5 days post-anthesis (dpa); CAR15, caryopsis 15 dpa. Data are means + SD (n=3). (B) Transcript level of YGL8 in the leaves of Morex and the ygl8 mutant grown in hydroponic culture for 3 weeks. (C) The subcellular localization of YGL8 and ygl8 in tobacco leaves. The GFP were fused to the C terminus of YGL8 (YGL8-GFP) and ylg8 (ylg8-GFP). OsYSS1-mCherry was used as a chloroplast protein marker. The 35S::GFP vector (GFP) was used as a control. The fluorescence signals in tobacco leaves were detected with a confocal laser-scanning microscope. Scale bars: 20 um.
To investigate whether the splicing site change between exon 5 and 6 of YGL8 affected the subcellular localization, the YGL8 or ygl8 fused with GFP (YGL8-GFP or ygl8-GFP) were transiently expressed in tobacco leaves. We observed strong GFP signals of YGL8 and ygl8 in the chloroplast, which coincided with the chloroplast marker signals of OsYSS1-mCherry (Zhou et al., 2017) (Figure 3C). These results demonstrate that the splicing site change between exon 5 and 6 of YGL8 has no effect on the gene transcript and chloroplast subcellular localization.
The splicing site change between exon 5 and 6 of YGL8 affects protein structure
We compared the YGL8 orthologous proteins of different species and observed high similarity among them, especially in the UMP kinase domain (Figure 4A). Given that the splicing site change between exon 5 and 6 of YGL8 was located in the UMP kinase domain (Figure 4A), we assumed that the mutation might affect the protein structure of the conserved UMP kinase domain. To verify this hypothesis, we predicted and compared the protein structures of YGL8 with ygl8 and found that the splicing site change between exon 5 and 6 of YGL8 resulted in an additional loop in the UMP kinase domain structure in the ygl8 protein (Figure 4B). Furthermore, the ten key amino acids, previously reported to be important for interactions with the substrates UMP or UTP (Hein et al., 2009), were examined, and we only observed slight conformational changes for five of the ten amino acids in the predicted ylg8 protein structure (Figure 4C; Supplementary Figure 3). These results suggest that the newly generated loop in ygl8 protein did not significantly affect the UPM domain structure, whereas the additional loop might disturb the access of the substrates to the kinase domain, thus resulting in a full or partial malfunction of the YGL8 gene.
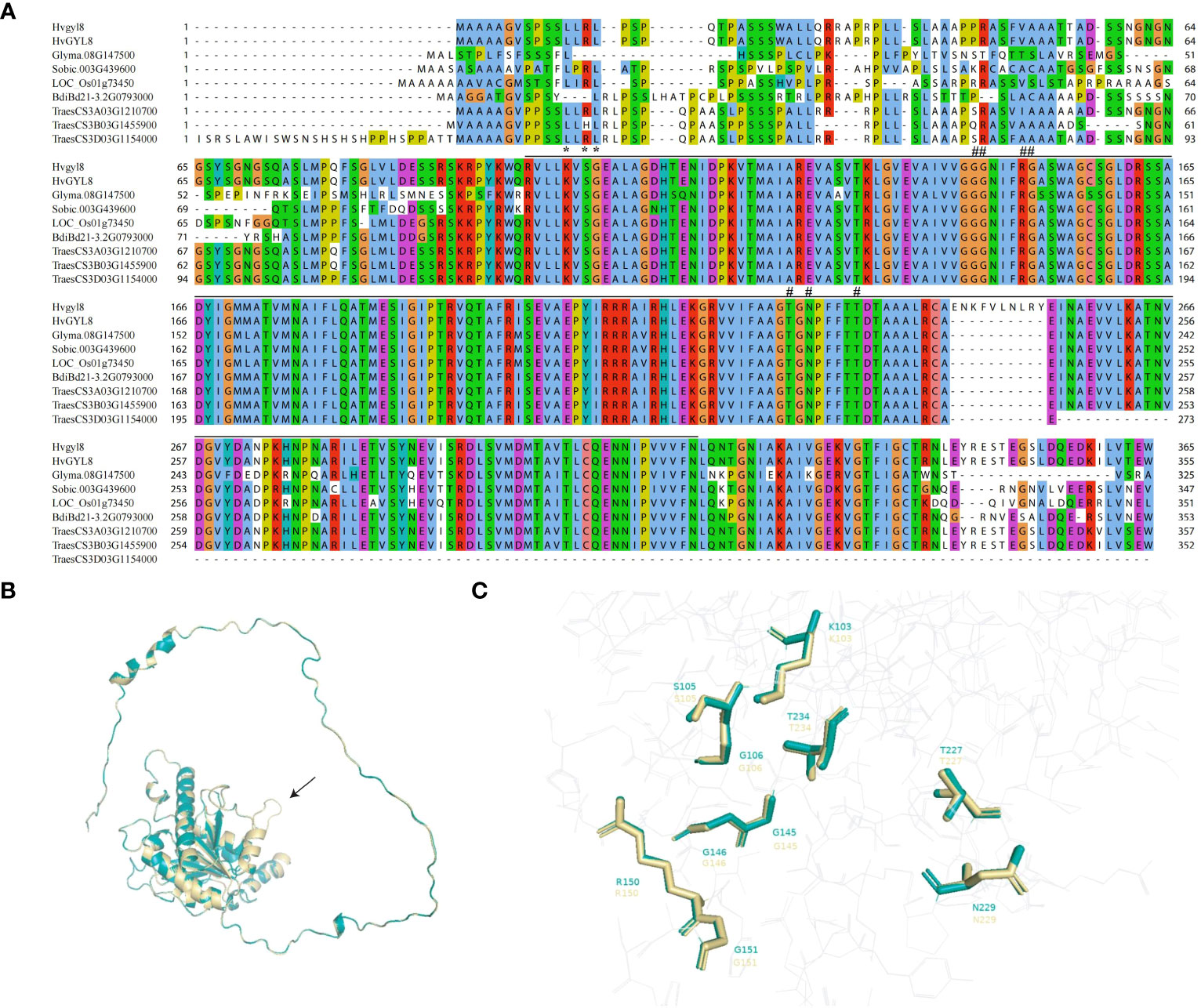
Figure 4 Amino acid alignments of orthologous YGL8 proteins of different plant species and protein structure comparison of barley YGL8 and ygl8. (A) The amino acid sequence alignments of barley YGL8 and ygl8 and the orthologs of soybean, Sorghum, rice, Brachypodium distachyon, and wheat. The black lines indicate the conserved UMP kinase domain. # indicates residues known to interact with UMP. # indicates residues known to interact with either UDP or UTP (Hein et al., 2009). (B) The predicted three-dimensional protein structure comparison of barley YGL8 and ygl8. The black arrow indicates the loop composed of the added 10 amino acids due to the splicing site change between exon 5 and 6 of YGL8. (C) Enlarged image of conformations of the essential amino acids, previously reported to be responsible for substrate binding. The numbers indicate the amino acid positions in the corresponding proteins. The model was constructed according to SWISS-MODEL (https://swissmodel.expasy.org/). The structures are shown using PyMOL. For (B, C), green and yellow represent the barley YGL8 and ygl8 proteins, respectively.
Enrichment analysis of differentially expressed genes in the leaf tissues of the ygl8 mutant
To clarify which genes are affected in terms of transcription levels due to the splicing site change between exon 5 and 6 of YGL8, we investigated the transcriptomes of the leaf tissues of Morex and the ygl8 mutant. Compared with Morex, we observed 2,401 significantly differentially expressed nuclear-encoded genes, including 817 upregulated genes and 1,584 downregulated genes (Figure 5A; Supplementary Table 4). The most significantly enriched GO terms included response to karrikin, chlorophyll biosynthetic process and saccharide biosynthetic process, and response to abiotic stresses such as cold stress and light intensity (Figure 5D; Supplementary Table 6). The most significantly enriched KEGG pathways included photosynthesis, carbon fixation and metabolism, plant hormone signal transduction and MARK signaling pathway, and linoleic acid metabolism (Figure 5E; Supplementary Table 7). These results suggest that photosynthesis and plant signaling transduction are affected by the splicing site change between exon 5 and 6 of the YGL8 gene in the ygl8 mutant.
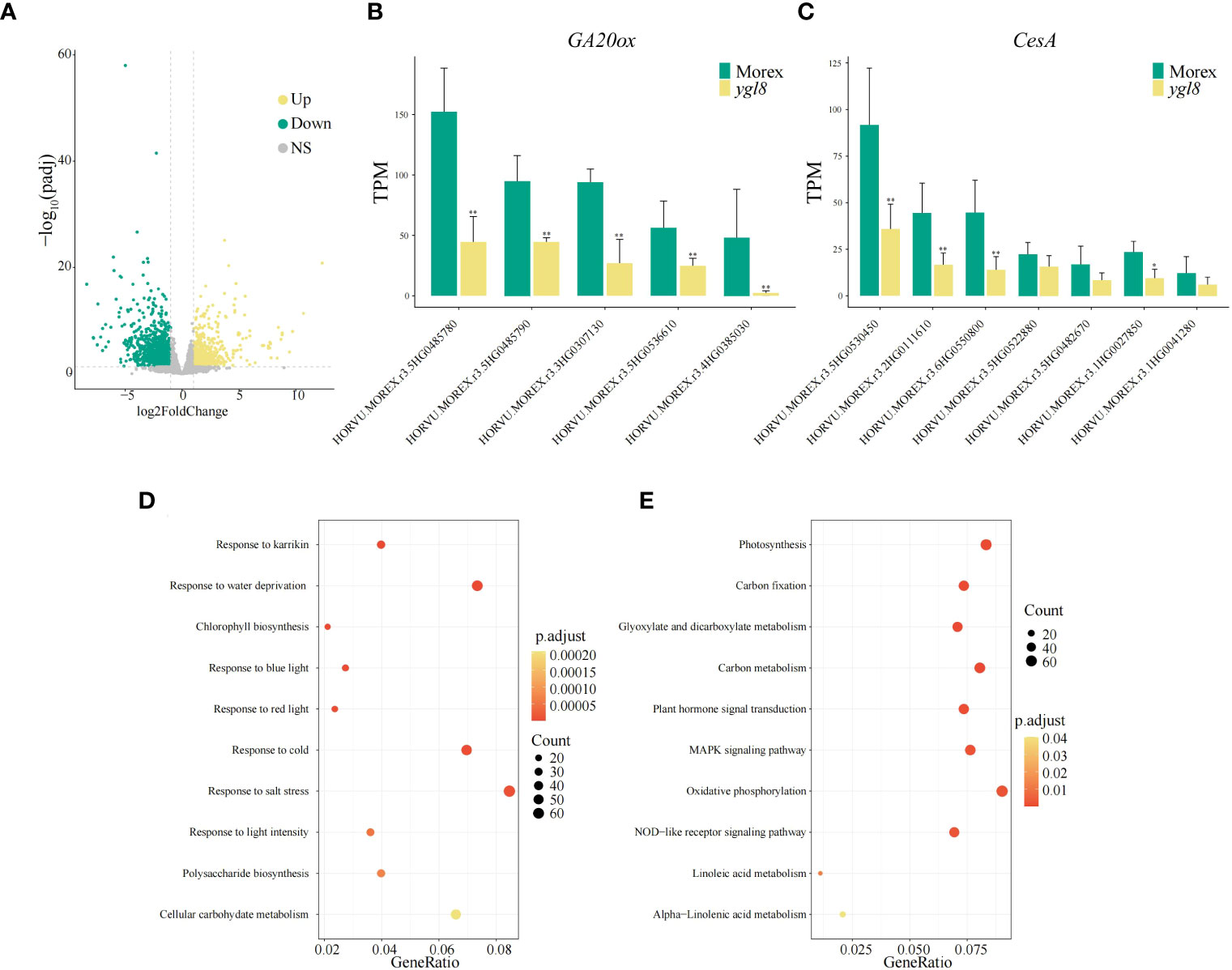
Figure 5 Transcriptome analysis of the leaf tissues of Morex and the ygl8 mutant. (A) Volcano map of differentially expressed genes (DEGs) between Morex and the ygl8 mutant. Up, Down and NS indicate upregulation, downregulation, and no significant changes of gene expression in the ygl8 mutant compared with Morex. (B, C) Transcript levels of differentially expressed GA20ox (B) and CesA (C) homologous genes. Data are mean + SD (n=3) for (B, C). Significant differences between Morex and the ygl8 mutant are indicated (*P.adj < 0.05, **P.adj < 0.01). (D) Top 10 significantly enriched GO terms of DEGs. (E) Top 10 significantly enriched KEGG terms of DEGs. The terms are sorted by p.adjust values in (D, E).
The GA20ox and CesA genes are downregulated in the ygl8 mutant
Bioactive GAs are vital endogenous plant growth regulators, which affect stem and internode elongation partially by influencing the cell wall characteristics via regulating CesA genes (Gupta and Chakrabarty, 2013; Huang et al., 2015). Among the differentially expressed genes in the RNA-seq data from Morex and the ygl8 mutant, we observed that five homologous GA20ox genes and all seven homologous CesA genes were significantly downregulated in the ygl8 mutant (Figures 5B, C; Supplementary Table 8). Furthermore, we examined the gene expression in all 24 homologous GA20ox genes in the leaf and internode tissues from a previous published paper (Mascher et al., 2017) and found that the differentially expressed GA20ox genes in our RNA-seq data had the highest expression levels of all the homologous members in the internode tissues (Supplementary Figure 4; Supplementary Table 9). These results demonstrate that the splicing site change between exon 5 and 6 of YGL8 downregulates the expression of the GA20ox and CesA genes in the ygl8 mutant and thus leads to the dwarf phenotype.
YGL8 is temperature sensitive and affects the expression of chloroplast-encoded and chlorophyll synthetic genes
Many genes responsible for leaf color variations have been reported to be temperature sensitive (Chen et al., 2013; Shi et al., 2015; Liu et al., 2020). To address whether the yellow leaf phenotype of the ygl8 mutant was dependent on the temperature conditions, we grew Morex and the ylg8 mutant in a hydroponic solution under different temperature conditions. Although a more yellowish leaf of the ygl8 mutant was observed at a low temperature (10°C) than at a normal culture temperature (24°C), the newly emerged leaf of the ygl8 mutant was green at a high temperature (30°C) (Figure 6A), which was consistent with the pigment content of the ygl8 mutant (Figure 6B). Furthermore, we examined the transcript levels of the GA20ox and CesA genes, which showed differential expression in our RNA-seq data, in the leaf tissues of Morex and the ygl8 mutant under different temperature conditions. The downregulation of CesA genes in the ygl8 mutant was only observed at a low temperature and no significant transcript changes were observed at normal or high temperatures (Figure 6D). Interestingly, we did not observe obvious transcript changes of GA20ox genes under different temperature conditions in the ygl8 mutant compared with Morex (Figure 6C), inconsistent with our RNA-Seq data (Figure 5B), which might be caused by different developmental sampling stages. Additionally, different conclusions about affected genes have been obtained in the rice ygl8 mutant for different sampling timings (Zhu et al., 2016; Chen et al., 2018a; Dong et al., 2019).
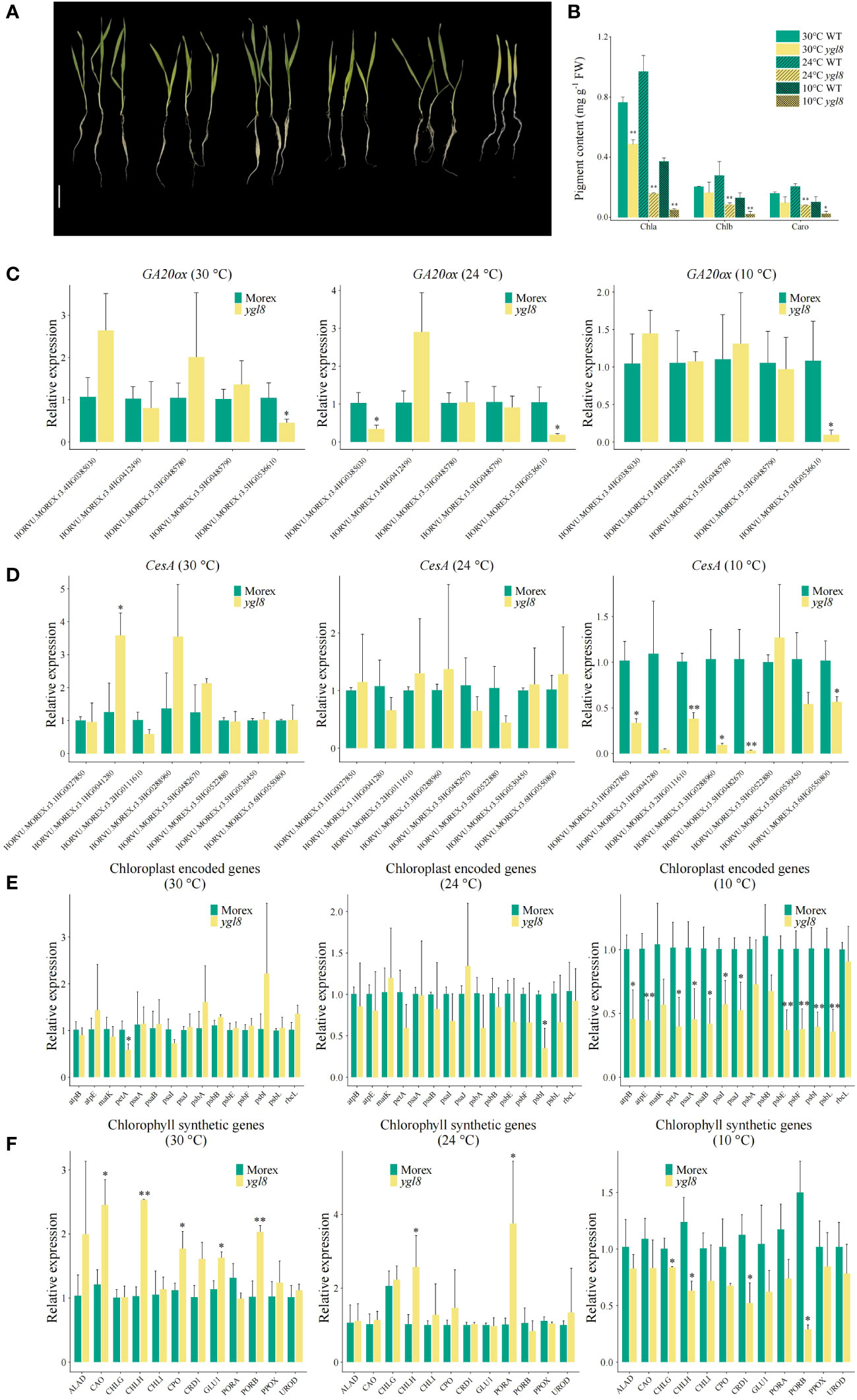
Figure 6 The effects of the splicing site change between exon 5 and 6 of YGL8 are dependent on temperature. (A) The phenotypes of Morex and the ygl8 mutant grown in different temperatures for 2 weeks. Scale bar: 5 cm. (B) The pigment contents of the leaves of Morex and the ygl8 mutant under different temperatures. (C–F) The transcript levels of GA20ox (C), CesA (D), chloroplast-encoded (E), and chlorophyll-biosynthesis-related genes (F) in the leaf tissues of Morex and the ygl8 mutant at different temperatures. Data are mean + SD (n=3). Significant differences between Morex and the ygl8 mutant are indicated (*P < 0.05, **P < 0.01; Student’s t test).
Given that the disruption of chloroplast-localized UMP kinase affected chloroplast-encoded gene expression and resulted in abnormal chloroplast development, subsequently resulting in a chlorophyll-deficient leaf phenotype in Arabidopsis and rice (Hein et al., 2009; Zhu et al., 2016; Chen et al., 2018a; Dong et al., 2019), we examined the chloroplast-encoded and chlorophyll synthetic genes in the ygl8 mutant under different temperature conditions. We found that the chloroplast thylakoid membrane major component encoding genes, including photosystem complex subunit encoding genes (psaA, psaB, psaI, psaJ, psbE, psbF, psbI, and psbL) (Nelson and Junge, 2015), ATP synthase subunit encoding genes (atpB and atpE) (Dobrogojski et al., 2020), and the cytochrome b6/f subunit encoding gene petA (Dobrogojski et al., 2020), showed similar expression patterns in the ygl8 mutant to those in Morex, with downregulation at low temperatures and no significant changes at normal or high temperatures (Figure 6E). Similarly, several chlorophyll synthetic genes were only downregulated in the ygl8 mutant compared with Morex at low temperatures (Figure 6F). These results suggest that the yellow leaf phenotype of the ygl8 mutant is temperature dependent.
Discussion
The chloroplast, as the metabolic hub of a cell, is also the site of hormone metabolism, such as GA and jasmonic acid, and the de novo biosynthesis of pigments such as chlorophyll a and b and abnormal development of chloroplasts could influence the expression of photosynthesis-associated nuclear genes (PhANGs) and pigment production (de Souza et al., 2017; Jan et al., 2022). In this study, we identified a yellow leaf and dwarf ygl8 mutant and found that these phenotypes were caused by a splicing site change between exon 5 and 6 of a UMP kinase encoding YGL8, due to a G to A single-nucleotide transition in the 5th exon/intron junction. The splicing site change between exon 5 and 6 of YGL8 had no effect on the chloroplast subcellular localization (Figures 3B, C) but generated an additional loop in the UMP kinase domain (Figure 4B), which might affect the enzyme function by disturbing the access of substrates and subsequently decreasing chloroplast UDP and UTP contents and impairing DNA and RNA synthesis, consistent with the downregulation of chloroplast-encoded gene expression (Figure 6E). Our RNA-seq results and hydroponic experiments demonstrated that the yellow leaf and plant dwarfism of the ygl8 mutant is caused by the downregulation of chloroplast-encoded genes and GA20ox and CesA genes, respectively (Figures 5B, C, 6C–E), and the effects of the splicing site change between exon 5 and 6 of YGL8 on the phenotype and gene expression are temperature dependent (Figure 6; Supplementary Figure 5).
A few yellow leaf barley mutants have been reported previously (Qin et al., 2015; Wang et al., 2017; Qin et al., 2019; Gajecka et al., 2021). One thermo-inducible chlorophyll-deficient mutant, named V-V-Y, was previously identified from gamma-radiated barley variety Vlamingh seeds, and the vvy mutant showed green leaf at a normal temperature but turned yellowish at temperatures above 30°C (Wang et al., 2017), which was different from the temperature-dependent manner of the ygl8 mutant in this study (Figure 6). In addition, the gene for the vvy mutant was mapped to the long arm of chromosome 4H (Wang et al., 2017); however, the YGL8 gene identified in this study is located on chromosome 3H (Figures 2A, B; Supplementary Table 3). whs18, a natural barley mutant, showed abnormal chloroplast development and stage green-revertible albino under the field condition, and the temperature under the ice-point during growth was necessary for leaf color variations (Qin et al., 2015), which was also different from the temperature-dependent manner of the ygl8 mutant in this study given the observation of a yellow leaf phenotype under constant 24°C or 10°C conditions (Figures 1A, 6A). Mutation of the plastid- and nucleus-localized HvCMF3 gene, a member of the CCT motif protein family, led to a yellowish/light green leaf color due to reduced ribosome accumulation and impaired grana organization and chloroplast development (Li et al., 2021). Another yellow leaf color mutant, ygl, obtained from the progeny of barley cultivar Edamai 934 seeds treated with EMS, showed impaired chloroplast ultrastructure, yellow leaf, and a decreased plant height phenotype. The QTL controlling leaf color phenotype of the ygl mutant was initially mapped in a 12.7 cM interval on the end of chromosome 3H (50.7082–52.0299 cM), but no specific gene responsible for the phenotype was confirmed using the strategy of BSA based on SSR markers and a contig level Morex reference genome (Qin et al., 2019). In this study, we carried out BSA based on high-throughput sequencing and the updated chromosome level Morex reference genome (Mascher et al., 2021) and targeted the YGL8 gene on the end of chromosome 3H (604,680,000–622,110,000 bp). Considering the similar phenotypes and chromosome positions, there is a possibility that YGL8 is the causal gene for the phenotype of the previously reported ygl mutant; however, this needs further confirmation.
The chloroplast can act as an environment sensor and communicate with the nucleus through retrograde signaling pathways to regulate nuclear gene expression in response to developmental cues and stress that affect photosynthesis and growth (Nott et al., 2006; de Souza et al., 2017; Jan et al., 2022). Four distinct retrograde signaling pathways have been recognized in higher vascular plants based on the sources of the signals: the accumulation of tetrapyrrole intermediates such as Mg-protoporphyrin IX, the inhibition of plastid gene expression, changes in plastid redox status, and the production of ROS (Nott et al., 2006; de Souza et al., 2017; Jan et al., 2022). The disruption in retrograde signaling pathways could result in the sustained expression of PhANGs, such as the light-harvesting chlorophyll a/b-binding (LHC) protein encoding gene LHCB1, even under stress conditions, such as photooxidative stress and ABA treatment (Martinez et al., 2003; Koussevitzky et al., 2007; de Souza et al., 2017). Based on the overproduction of ROS, such as H2O2, in leaf tissues in the rice orthologous YGL8 mutant, we expected the transcript levels of LHC genes to be reduced in the ygl8 mutant compared with Morex. Interestingly, we observed the upregulation of many LHC genes in leaf tissues of the ygl8 mutant compared with Morex in our RNA-seq data (Supplementary Figure 5A; Supplementary Table 10), and a similar expression pattern was also observed at low temperatures in hydroponic experiments (Supplementary Figure 5B), demonstrating that the retrograde signaling transduction was disrupted in the ygl8 mutant. Chlorophyll and heme share a common precursor of tetrapyrrole biosynthesis, and at the branchpoint, Mg2+ and Fe2+ are inserted into protoporphyrin IX by Mg-chelatase and ferrochelatase (FC) to subsequently produce chlorophyll and heme, respectively (Jan et al., 2022). The overaccumulation of heme could feedback inhibit the tetrapyrrole biosynthesis pathway (de Souza et al., 2017; Jan et al., 2022) and thus reduce the production of retrograde signaling Mg-protoporphyrin IX, resulting in the sustained expression of the LHCB gene independent of chloroplast status (Martinez et al., 2003). Based on the upregulation of FC1 in the ygl8 mutant at low temperatures in the hydroponic experiment (Supplementary Figure 5B), we speculated that cold-stress-induced ROS decreased LHC gene expression in Morex, whereas the accumulation of heme via increasing FC1 gene expression in the ygl8 mutant partially alleviated the downregulation of the LHC genes, eventually resulting in an upregulation of LHC in the ygl8 mutant compared with Morex (Supplementary Figure 5B). However, other complex mechanisms might be involved in the regulation of LHC gene expression in the ygl8 mutant. In summary, this study facilitates our understanding of the barley leaf color and plant height regulation mechanism, and the ygl8 mutant is a valuable tool for investigating the retrograde signaling pathway in barley in the future.
Conclusion
In this study, we identified a yellow leaf and dwarf ygl8 mutant from the progeny of Morex seeds treated with EMS, and the phenotype of the ygl8 mutant was caused by a splicing site change between exon 5 and 6 of YGL8, which encodes a chloroplast-localized UMP kinase, due to a G to A single-nucleotide transition in the 5th exon/intron junction. The splicing site change between exon 5 and 6 of YGL8 resulted in the insertion of ten amino acids in the UMP kinase domain and might impair enzyme function by disturbing the access of the substrates. The yellow leaf phenotype of the ygl8 mutant was caused by the downregulation of chloroplast-encoded genes and chlorophyll-biosynthesis-related genes in a temperature-dependent manner. For the decreased plant height phenotype of the ygl8 mutant, the downregulation of GA20ox genes, which are responsible for bioactive GA biosynthesis, and CesA genes, which are responsible for cell wall cellulose biosynthesis, relative to Morex was observed. Our study will facilitate the understanding of leaf color and plant height regulation in barley.
Data availability statement
The data presented in the study are deposited in the NCBI repository, accession number PRJNA1027084.
Author contributions
XX: Data curation, Investigation, Writing – original draft. LL: Investigation, Writing – review & editing. KC: Investigation, Writing – review & editing. XS: Investigation, Writing – review & editing. WY: Data curation, Formal analysis, Software, Writing – original draft, Writing – review & editing. JW: Conceptualization, Formal analysis, Funding acquisition, Resources, Supervision, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study was supported by the National Natural Science Foundation of China (32101642), Natural Science Foundation of Zhejiang Province (LQ21C130006), Major Program on Agricultural New Variety Breeding (2021C02064-3-2), and China Agriculture Research System of MOF and MARA (CARS-05-01A-06).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer GZ declared a past collaboration with the author KC to the handling editor.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1327246/full#supplementary-material
References
Barthet, M. M., Pierpont, C. L., Tavernier, E. K. (2020). Unraveling the role of the enigmatic MatK maturase in chloroplast group IIA intron excision. Plant Direct 4, e00208. doi: 10.1002/pld3.208
Buchfink, B., Reuter, K., Drost, H. G. (2021). Sensitive protein alignments at tree-of-life scale using DIAMOND. Nat. Methods 18, 366–368. doi: 10.1038/s41592-021-01101-x
Burn, J. E., Hocart, C. H., Birch, R. J., Cork, A. C., Williamson, R. E. (2002). Functional analysis of the cellulose synthase genes CesA1, CesA2, and CesA3 in Arabidopsis. Plant Physiol. 129, 797–807. doi: 10.1104/pp.010931
Cai, K., Kuang, L., Yue, W., Xie, S., Xia, X., Zhang, G., et al. (2022). Calmodulin and calmodulin-like gene family in barley: Identification, characterization and expression analyses. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.964888
Chantreau, M., Chabbert, B., Billiard, S., Hawkins, S., Neutelings, G. (2015). Functional analyses of cellulose synthase genes in flax (Linum usitatissimum) by virus-induced gene silencing. Plant Biotechnol. J. 13, 1312–1324. doi: 10.1111/pbi.12350
Charlier, D., Nguyen Le Minh, P., Roovers, M. (2018). Regulation of carbamoylphosphate synthesis in Escherichia coli: an amazing metabolite at the crossroad of arginine and pyrimidine biosynthesis. Amino Acids 50, 1647–1661. doi: 10.1007/s00726-018-2654-z
Chen, F., Dong, G., Ma, X., Wang, F., Zhang, Y., Xiong, E., et al. (2018a). UMP kinase activity is involved in proper chloroplast development in rice. Photosynth Res. 137, 53–67. doi: 10.1007/s11120-017-0477-5
Chen, H., Cheng, Z., Ma, X., Wu, H., Liu, Y., Zhou, K., et al. (2013). A knockdown mutation of YELLOW-GREEN LEAF2 blocks chlorophyll biosynthesis in rice. Plant Cell Rep. 32, 1855–1867. doi: 10.1007/s00299-013-1498-y
Chen, S., Zhou, Y., Chen, Y., Gu, J. (2018b). Fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, i884–i890. doi: 10.1093/bioinformatics/bty560
Danecek, P., Auton, A., Abecasis, G., Albers, C. A., Banks, E., DePristo, M. A., et al. (2011). The variant call format and VCFtools. Bioinformatics 27, 2156–2158. doi: 10.1093/bioinformatics/btr330
Danecek, P., McCarthy, S. A. (2017). BCFtools/csq: haplotype-aware variant consequences. Bioinformatics 33, 2037–2039. doi: 10.1093/bioinformatics/btx100
Daniell, H., Lin, C. S., Yu, M., Chang, W. J. (2016). Chloroplast genomes: diversity, evolution, and applications in genetic engineering. Genome Biol. 17, 134. doi: 10.1186/s13059-016-1004-2
DeLano, W. L. (2002). Pymol: An open-source molecular graphics tool. CCP4 Newsl. Protein Crystallogr. 40, 82–92.
de Souza, A., Wang, J. Z., Dehesh, K. (2017). Retrograde signals: integrators of interorganellar communication and orchestrators of plant development. Annu. Rev. Plant Biol. 68, 85–108. doi: 10.1146/annurev-arplant-042916-041007
Dobrogojski, J., Adamiec, M., Luciński, R. (2020). The chloroplast genome: a review. Acta Physiol. Plant. 42, 98. doi: 10.1007/s11738-020-03089-x
Dong, Q., Zhang, Y. X., Zhou, Q., Liu, Q. E., Chen, D. B., Wang, H., et al. (2019). UMP kinase regulates chloroplast development and cold response in rice. Int. J. Mol. Sci. 20 (9), 2107. doi: 10.3390/ijms20092107
Franckowiak, J. D., Lundqvist, U. (2012). Descriptions of barley genetic stocks for 2012. Barley Genet. Newsl 42, 792.
Gajecka, M., Marzec, M., Chmielewska, B., Jelonek, J., Zbieszczyk, J., Szarejko, I. (2021). Changes in plastid biogenesis leading to the formation of albino regenerants in barley microspore culture. BMC Plant Biol. 21, 22. doi: 10.1186/s12870-020-02755-z
Gálová, E., Böhmová, B., Ševčovičová, A. (2000). Analysis of some barley chlorophyll mutants and their response to temperature stress. Photosynthetica 38, 29–35. doi: 10.1023/A:1026735605804
Gupta, R., Chakrabarty, S. K. (2013). Gibberellic acid in plant: still a mystery unresolved. Plant Signal Behav. 8. doi: 10.4161/psb.25504
He, J., Xin, P., Ma, X., Chu, J., Wang, G. (2020). Gibberellin metabolism in flowering plants: an update and perspectives. Front. Plant Sci. 11, 532. doi: 10.3389/fpls.2020.00532
Hedden, P. (2020). The current status of research on gibberellin biosynthesis. Plant Cell Physiol. 61, 1832–1849. doi: 10.1093/pcp/pcaa092
Hein, P., Stockel, J., Bennewitz, S., Oelmuller, R. (2009). A protein related to prokaryotic UMP kinases is involved in psaA/B transcript accumulation in Arabidopsis. Plant Mol. Biol. 69, 517–528. doi: 10.1007/s11103-008-9433-2
Huang, D., Wang, S., Zhang, B., Shang-Guan, K., Shi, Y., Zhang, D., et al. (2015). A gibberellin-mediated DELLA-NAC signaling cascade regulates cellulose synthesis in rice. Plant Cell 27, 1681–1696. doi: 10.1105/tpc.15.00015
Jan, M., Liu, Z., Rochaix, J. D., Sun, X. (2022). Retrograde and anterograde signaling in the crosstalk between chloroplast and nucleus. Front. Plant Sci. 13, 980237. doi: 10.3389/fpls.2022.980237
Kafer, C., Zhou, L., Santoso, D., Guirgis, A., Weers, B., Park, S., et al. (2004). Regulation of pyrimidine metabolism in plants. Front. Biosci. 9, 1611–1625.
Kim, D., Paggi, J. M., Park, C., Bennett, C., Salzberg, S. L. (2019). Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 37, 907–915. doi: 10.1038/s41587-019-0201-4
Koussevitzky, S., Nott, A., Mockler, T. C., Hong, F., Sachetto-Martins, G., Surpin, M., et al. (2007). Signals from chloroplasts converge to regulate nuclear gene expression. Science 316, 715–719. doi: 10.1126/science.1140516
Li, H., Durbin, R. (2009). Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760. doi: 10.1093/bioinformatics/btp324
Li, H., Handsaker, B., Wysoker, A., Fennell, T., Ruan, J., Homer, N., et al. (2009). The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079. doi: 10.1093/bioinformatics/btp352
Li, M., Hensel, G., Mascher, M., Melzer, M., Budhagatapalli, N., Rutten, T., et al. (2019). Leaf variegation and impaired chloroplast development caused by a truncated CCT domain gene in albostrians barley. Plant Cell 31, 1430–1445. doi: 10.1105/tpc.19.00132
Li, M., Hensel, G., Melzer, M., Junker, A., Tschiersch, H., Ruwe, H., et al. (2021). Mutation of the ALBOSTRIANS ohnologous gene hvCMF3 impairs chloroplast development and thylakoid architecture in barley. Front. Plant Sci. 12, 732608. doi: 10.3389/fpls.2021.732608
Liao, Y., Smyth, G. K., Shi, W. (2019). The R package Rsubread is easier, faster, cheaper and better for alignment and quantification of RNA sequencing reads. Nucleic Acids Res. 47, e47. doi: 10.1093/nar/gkz114
Liu, X., Huang, Q., Yang, Y., Tang, J., Zhao, Y., Zhang, J. (2020). Characterization and map-based cloning of the novel rice yellow leaf mutant yl3. J. Plant Biol. 64, 35–44. doi: 10.1007/s12374-020-09275-1
Liu, Z.-L., Yuan, S., Liu, W.-J., Du, J.-B., Tian, W.-J., Luo, M.-H., et al. (2008). Mutation mechanism of chlorophyll-less barley mutant NYB. Photosynthetica 46, 73–78. doi: 10.1007/s11099-008-0013-0
Love, M. I., Huber, W., Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550. doi: 10.1186/s13059-014-0550-8
Martinez, L. O., Jacquet, S., Esteve, J. P., Rolland, C., Cabezon, E., Champagne, E., et al. (2003). Ectopic beta-chain of ATP synthase is an apolipoprotein A-I receptor in hepatic HDL endocytosis. Nat 421, 75–79. doi: 10.1038/nature01250
Mascher, M., Gundlach, H., Himmelbach, A., Beier, S., Twardziok, S. O., Wicker, T., et al. (2017). A chromosome conformation capture ordered sequence of the barley genome. Nat 544, 427–433. doi: 10.1038/nature22043
Mascher, M., Wicker, T., Jenkins, J., Plott, C., Lux, T., Koh, C. S., et al. (2021). Long-read sequence assembly: a technical evaluation in barley. Plant Cell 33, 1888–1906. doi: 10.1093/plcell/koab077
McFarlane, H. E., Doring, A., Persson, S. (2014). The cell biology of cellulose synthesis. Annu. Rev. Plant Biol. 65, 69–94. doi: 10.1146/annurev-arplant-050213-040240
Mueller, A. H., Dockter, C., Gough, S. P., Lundqvist, U., von Wettstein, D., Hansson, M. (2012). Characterization of mutations in barley fch2 encoding chlorophyllide a oxygenase. Plant Cell Physiol. 53, 1232–1246. doi: 10.1093/pcp/pcs062
Murray, M., Thompson, W. (1980). Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 8, 4321–4326. doi: 10.1093/nar/8.19.4321
Nelson, N., Junge, W. (2015). Structure and energy transfer in photosystems of oxygenic photosynthesis. Annu. Rev. Biochem. 84, 659–683. doi: 10.1146/annurev-biochem-092914-041942
Nott, A., Jung, H. S., Koussevitzky, S., Chory, J. (2006). Plastid-to-nucleus retrograde signaling. Annu. Rev. Plant Biol. 57, 739–759. doi: 10.1146/annurev.arplant.57.032905.105310
Paciorek, T., Chiapelli, B. J., Wang, J. Y., Paciorek, M., Yang, H., Sant, A., et al. (2022). Targeted suppression of gibberellin biosynthetic genes ZmGA20ox3 and ZmGA20ox5 produces a short stature maize ideotype. Plant Biotechnol. J. 20, 1140–1153. doi: 10.1111/pbi.13797
Prina, A. R., Arias, M. C., Lainez, V., Landau, A., Maldonado, S. (2003). A cytoplasmically inherited mutant controlling early chloroplast development in barley seedlings. Theor. Appl. Genet. 107, 1410–1418. doi: 10.1007/s00122-003-1391-0
Qin, D., Dong, J., Xu, F., Guo, G., Ge, S., Xu, Q., et al. (2015). Characterization and fine mapping of a novel barley Stage Green-Revertible Albino Gene (HvSGRA) by Bulked Segregant Analysis based on SSR assay and Specific Length Amplified Fragment Sequencing. BMC Genom. 16, 838. doi: 10.1186/s12864-015-2015-1
Qin, D., Li, M., Xu, F., Xu, Q., Ge, S., Dong, J. (2019). Analysis of agronomic characters and preliminary mapping of regulatory genes of a barley yellow-green leaf mutant ygl. J. Triticeae Crop 39, 653–658.
Robinson, M. D., Oshlack, A. (2010). A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 11, 1–9. doi: 10.1186/gb-2010-11-3-r25
Sasaki, A., Ashikari, M., Ueguchi-Tanaka, M., Itoh, H., Nishimura, A., Swapan, D., et al. (2002). Green revolution: a mutant gibberellin-synthesis gene in rice. Nat 416, 701–702. doi: 10.1038/416701a
Saski, C., Lee, S. B., Fjellheim, S., Guda, C., Jansen, R. K., Luo, H., et al. (2007). Complete chloroplast genome sequences of Hordeum vulgare, Sorghum bicolor and Agrostis stolonifera, and comparative analyses with other grass genomes. Theor. Appl. Genet. 115, 571–590. doi: 10.1007/s00122-007-0567-4
Shen, Y., Zhou, Z., Wang, Z., Li, W., Fang, C., Wu, M., et al. (2014). Global dissection of alternative splicing in paleopolyploid soybean. Plant Cell 26, 996–1008. doi: 10.1105/tpc.114.122739
Shi, J., Wang, Y., Guo, S., Ma, L., Wang, Z., Zhu, X., et al. (2015). Molecular mapping and candidate gene analysis of a yellow-green leaf 6 (ygl6) mutant in rice. Crop Sci. 55, 669–680. doi: 10.2135/cropsci2014.07.0483
Stirbet, A., Lazar, D., Guo, Y., Govindjee, G. (2020). Photosynthesis: basics, history and modelling. Ann. Bot. 126, 511–537. doi: 10.1093/aob/mcz171
Sugihara, Y., Young, L., Yaegashi, H., Natsume, S., Shea, D. J., Takagi, H., et al. (2022). High-performance pipeline for mutMap and QTL-seq. PeerJ 10, e13170. doi: 10.7717/peerj.13170
Wang, P., Grimm, B. (2015). Organization of chlorophyll biosynthesis and insertion of chlorophyll into the chlorophyll-binding proteins in chloroplasts. Photosynth Res. 126, 189–202. doi: 10.1007/s11120-015-0154-5
Wang, K., Li, M., Hakonarson, H. (2010). ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 38, e164. doi: 10.1093/nar/gkq603
Wang, S., Wang, Y. (2022). Harnessing hormone gibberellin knowledge for plant height regulation. Plant Cell Rep. 41, 1945–1953. doi: 10.1007/s00299-022-02904-8
Wang, R., Yang, F., Zhang, X. Q., Wu, D., Tan, C., Westcott, S., et al. (2017). Characterization of a thermo-inducible chlorophyll-deficient mutant in barley. Front. Plant Sci. 8, 1936. doi: 10.3389/fpls.2017.01936
Wang, D., Yuan, S., Yin, L., Zhao, J., Guo, B., Lan, J., et al. (2012). A missense mutation in the transmembrane domain of CESA9 affects cell wall biosynthesis and plant growth in rice. Plant Sci. 196, 117–124. doi: 10.1016/j.plantsci.2012.08.002
Wang, Y., Zhang, J., Shi, X., Peng, Y., Li, P., Lin, D., et al. (2016). Temperature-sensitive albino gene TCD5, encoding a monooxygenase, affects chloroplast development at low temperatures. J. Exp. Bot. 67, 5187–5202. doi: 10.1093/jxb/erw287
Witte, C. P., Herde, M. (2020). Nucleotide metabolism in plants. Plant Physiol. 182, 63–78. doi: 10.1104/pp.19.00955
Yu, G., Wang, L. G., Han, Y., He, Q. Y. (2012). clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 16, 284–287. doi: 10.1089/omi.2011.0118
Zhou, Y., Gong, Z., Yang, Z., Yuan, Y., Zhu, J., Wang, M., et al. (2013b). Mutation of the light-induced yellow leaf 1 gene, which encodes a geranylgeranyl reductase, affects chlorophyll biosynthesis and light sensitivity in rice. PloS One 8, e75299. doi: 10.1371/journal.pone.0075299
Zhou, K., Ren, Y., Lv, J., Wang, Y., Liu, F., Zhou, F., et al. (2013a). Young Leaf Chlorosis 1, a chloroplast-localized gene required for chlorophyll and lutein accumulation during early leaf development in rice. Planta 237, 279–292. doi: 10.1007/s00425-012-1756-1
Zhou, K., Ren, Y., Zhou, F., Wang, Y., Zhang, L., Lyu, J., et al. (2017). Young Seedling Stripe1 encodes a chloroplast nucleoid-associated protein required for chloroplast development in rice seedlings. Planta 245, 45–60. doi: 10.1007/s00425-016-2590-7
Zhu, X., Guo, S., Wang, Z., Du, Q., Xing, Y., Zhang, T., et al. (2016). Map-based cloning and functional analysis of YGL8, which controls leaf colour in rice (Oryza sativa). BMC Plant Biol. 16, 134. doi: 10.1186/s12870-016-0821-5
Keywords: UMP kinase, chloroplast development, chlorophyll biosynthesis, gibberellic acid, cellulose synthase, retrograde signaling pathways
Citation: Xia X, Liu L, Cai K, Song X, Yue W and Wang J (2023) A splicing site change between exon 5 and 6 of the nuclear-encoded chloroplast-localized HvYGL8 gene results in reduced chlorophyll content and plant height in barley. Front. Plant Sci. 14:1327246. doi: 10.3389/fpls.2023.1327246
Received: 25 October 2023; Accepted: 30 November 2023;
Published: 12 December 2023.
Edited by:
Jianlong Xu, Chinese Academy of Agricultural Sciences, ChinaCopyright © 2023 Xia, Liu, Cai, Song, Yue and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Junmei Wang, MjF3YW5nam1Ac2luYS5jb20=; Wenhao Yue, eXVld2hAemFhcy5hYy5jbg==
†These authors have contributed equally to this work
 Xue Xia
Xue Xia Lei Liu
Lei Liu Kangfeng Cai
Kangfeng Cai Xiujuan Song1,2,3
Xiujuan Song1,2,3 Wenhao Yue
Wenhao Yue Junmei Wang
Junmei Wang