
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 25 January 2024
Sec. Plant Biotechnology
Volume 14 - 2023 | https://doi.org/10.3389/fpls.2023.1324401
Tomatoes are susceptible to damage from cold temperatures in all stages of growth. Therefore, it is important to identify genetic resources and genes that can enhance tomato’s ability to tolerate cold. In this study, a population of 223 tomato accessions was used to identify the sensitivity or tolerance of plants to cold stress. Transcriptome analysis of these accessions revealed that SUS3, a member of the sucrose synthase gene family, was induced by cold stress. We further investigated the role of SUS3 in cold stress by overexpression (OE) and RNA interference (RNAi). Compared with the wild type, SUS3-OE lines accumulated less MDA and electrolyte leakage and more proline and soluble sugar, maintained higher activities of SOD and CAT, reduced superoxide radicals, and suffered less membrane damage under cold. Thus, our findings indicate that SUS3 plays a crucial role in the response to cold stress. This study indicates that SUS3 may serve as a direct target for genetic engineering and improvement projects, which aim to augment the cold tolerance of tomato crops.
Tomato (Solanum lycopersicum) is an economically significant vegetable crop globally, originating from tropical and subtropical regions. It is sensitive to low temperatures, which adversely affects tomato growth, development, and ripening (Foolad and Lin, 2001; Venema et al., 2005). Cold stress has become the primary threat to early spring cultivation of tomatoes in Xinjiang, leading to substantial losses for farmers. Therefore, it is significant and valuable to breed modern cold-tolerant varieties in tomato, and it is essential for us to exploit resistant germplasm resources and to further understand the physiological and biochemical processes that promote cold tolerance.
Previous studies on plant cold acclimatization have shown that enzymes involved in the metabolism of antioxidants, lipids, carbohydrates, proline, and polyamines, as well as cold resistance-related proteins, and other osmotic regulation-related proteins, play an important role in plant resistance to cold stress (Li et al., 2011; Sato et al., 2011; Wang et al., 2011; Fan et al., 2012). Moreover, the overexpression of particular genes in pepper (Airaki et al., 2012), grape (Sawicki et al., 2012), and Arabidopsis (Hannah et al., 2005) can significantly improve the transgenic plants’ ability to tolerate cold, drought, and osmotic stresses.
Sucrose synthase (SUS) was first discovered in wheat germ by Cardini et al. (1955), and later, Geigenberger and Stitt (1993) demonstrated its role in regulating various metabolic pathways involving sucrose in plants. The SUS is a multigene family, with each family consisting of at least two gene members. In some species, more SUS families have been found. The tomato SUS family, for example, consists of six genes, namely, SUS1, SUS3, SUS4, SUS5, SUS6, and SUS7, distributed across five chromosomes (Bieniawska et al., 2007). In addition, grape and sugarcane have five SUS family genes identified (Zhang et al., 2013), and more than six have been found in cotton, Arabidopsis, and rice (Tatsuro et al., 2008; Chen et al., 2012; Zou et al., 2013).
The expression of SUS can be prompted by environmental factors, including low oxygen, low or high temperatures, and drought. In tomato roots under low oxygen stress, the overall capacity for sucrose hydrolysis remains unaltered, suggesting that SUS is adopted by tomato to enhance energy utilization efficiency and conserve ATP under hypoxic conditions. The SUS pathway can compensate, at least to some extent, for the inadequate energy supply caused by hypoxia (Sasaki et al., 2001). In barley, the SUS genes exhibit distinct responses to various abiotic stresses. For instance, hypoxia stress can induce HvSUS1, while drought can induce HvSUS3 (Ricard et al., 1998). HbSUS5 expression was found to be enhanced in the roots and leaves of rubber trees (Hevea brasiliensis) in response to low temperature and drought, indicating a significant role in stress response (Xiao et al., 2014). During the maturation of rice seeds, exposure to high temperatures may lead to the occurrence of chalkiness in brown rice. The overexpression of SUS3 provides resistance to chalkiness in brown rice (Takehara et al., 2018).
A previous study showed that the SUS has been identified in many plants, such as tomatoes, tobacco, grape, rice, and Arabidopsis. However, the SUS gene regulation mechanism and biological function and the function of the SUS3 gene underlying cold tolerance have not been further studied. Our study found that SUS3 was highly expressed in the leaves and was induced under cold stress. Overexpression of SUS3 improved its ability in resisting low temperatures during the seedling stage, indicating that SUS3 plays an important role in tomato low-temperature resistance. Our research provides new insight into the biological function of SUS3 and reveals a new gene that contributes to the cold tolerance of tomato.
Two hundred and twenty-three tomato accessions, consisting of 2 Solanum cheesmaniae (wild), 16 S. pimpinellifolium (PIM), 65 S. lycopersicum var. cerasiforme (CER), 105 S. lycopersicum (BIG), and 35 Guangxi accessions (Supplementary Table S1), were collected by our laboratory at Wuhan National Key Laboratory for Germplasm Innovation and Utilization of Horticultural Crops, China, in spring 2018. Four-week-old seedlings were placed in a constant temperature room at 4°C. After 7 days, the symptoms of cold damage on the plants were observed and the images were recorded. Cold damage was graded to five levels: level 0 indicated no symptoms and no sensitivity to the current low temperature; level 1 was characterized by injury spots on the edge of true leaves; level 2 was characterized by large-area injury spots on true leaves; level 3 was characterized by large-area injury spots on true leaves with leaf curling; and level 4 was characterized by wilting of plants and even necrosis of apical meristem (Wang et al., 1996). Three biological replicates were analyzed, and each replicate consisted of 15 individual plants.
Four-week-old seedlings were subjected to cold stress treatments at 4°C for 48 h. Twenty cold-sensitive and 20 cold-tolerant seedlings were selected. After treatment, the third fully expanded leaf from the top of each seedling was collected respectively, frozen in liquid nitrogen, and then stored at −80°C. The leaves were sent to Beijing Novogene Technology Co., Ltd. (Wuhan Branch) for library construction and sequencing. We used the average FPKM (expected number of fragments per kilobase of transcript sequence per million base pairs sequenced) value as a measure of gene expression.
The cold-tolerant A57 (S. lycopersicum cv. Ailsa Craig) was used to study the expression of SUS3. Four-week-old seedlings were subjected to abiotic stress treatments, namely, ABA, ethylene, drought, cold, and salt stress treatments. The methods corresponding to these abiotic stress treatments were described previously (Song et al., 2016). The soil on the surface of the plants was washed off, and the residual water on the surface was absorbed. The plants were divided into five groups for different treatments. Plants in group 1 were placed in a dry environment at room temperature to induce drought stress. The roots of plants in group 2 were immersed in 400 mM of NaCl solution to induce salt stress; plants in group 3 were placed in a 4°C cold chamber to induce cold stress. The plants in group 4 and group 5 were sprayed with 100 μM of ABA and 200 μM of ethylene solutions for hormone induction treatment, respectively. Then, 0, 1, 3, 6, 12, and 24 h after each treatment, the second leaf from the apical meristem was collected, snap-frozen in liquid nitrogen, and stored at −80°C for RNA extraction.
Total RNA was extracted using the TRIzol reagent (Invitrogen, USA) and further reverse-transcribed into cDNA using a HiScript II 1st Strand cDNA Synthesis Kit (Vazyme, China). The relative transcript levels of specific genes were quantified using real-time PCR (qRT-PCR), which was conducted using a QuantStudio™ 6 Flex System (ABI, USA). The expression level of the actin gene (Solyc11g005330) was used as an internal control. For each biological replicate, the relative expression level of SUS3 was quantified using three technical replicates and calculated using the 2−ΔΔCT method. The related primer sequences can be found in Supplementary Table S2.
Cold-sensitive M82 (S. lycopersicum) was selected for transgenic tomato plants. For overexpression constructs, the complete open reading frame (ORF) of SUS3 was amplified from M82 using SUS3-OE primers (Supplementary Table S2), and these constructs were cloned into pHELLSGATE8 vectors. An RNA interference (RNAi) construct, a 412-bp fragment from the SUS3 gene, was amplified from cDNA that was prepared from M82 using the SUS3-Ri primers (Supplementary Table S2) and cloned into pHGRV using Clonase BP (Invitrogen, USA). M82 was transformed with all of the recombinant plasmids using Agrobacterium (strain GV3101)-mediated transformation. The cotyledons of tomato were used for transformation. Positive transgenic plants were screened via PCR with a CaMV35S promoter primer and a gene-specific primer. After PCR and qRT-PCR analyses, several independent homozygous transgenic lines expressing abnormal levels of SUS3 (OE1-2, OE2-4, OE3-2, Ri1-1, Ri8-3, and Ri17-2) and WT (M82) were selected for further analysis.
Six-week-old SUS3-OE lines, SUS3-RNAi lines, and WT were placed in a 4°C constant temperature chamber for 7 days for phenotype observation and photographing. Three replicates were set for each treatment, with 15 seedlings per replicate. The third fully expanded leaf from the apical meristem was harvested and snap-frozen in liquid nitrogen and then stored in a −80°C freezer for physiological and biochemical parameters.
Malondialdehyde (MDA), relative electrolyte leakage levels, and proline were determined as previously described (Campos et al., 2003). The anthrone colorimetry method (Fukao et al., 2006) was adopted to determine the content of soluble sugar. Three biological replicates were analyzed for each treatment.
The fully expanded leaves (first from the apical meristem) from 6-week-old seedlings of SUS3-OE and WT were used for nitroblue tetrazolium (NBT) staining. Three replicates were set for each treatment, with 15 leaves per replicate. The NBT staining method as previously described (Xia et al., 2009) was adopted.
Extraction of enzyme solution: A 0.2-g sample, which had been stored in a −80°C freezer, was weighed and combined with precooled phosphate buffer (pH 7.8, with 0.1 mM of EDTA) and 1.8 mL of 1% PVP. After mixing, the resulting mixture was subjected to centrifugation at 4°C and 12,000 rpm for 15 min. The supernatant obtained was considered to be the enzyme extract. The levels of superoxide dismutase (SOD), catalase (CAT), ascorbate peroxidase (APX), and peroxidase (POD) were quantified through determination as described in previous studies (Jimenez et al., 1997; Morohashi, 2002; Al-aghabary et al., 2005).
To investigate the localization of SUS3, the full-length CDS of SUS3 was cloned into the dual vector pGDG between XhoI and ApaI from the cDNA of A57. To fuse the target gene with the reporter gene, we introduced two additional base pairs CT after the XhoI restriction site. The target gene was fused with the C-terminus of a green fluorescent protein (GFP) reporter gene and expressed under the control of the 35S promoter (Supplementary Table S2) for tomato protoplasts. After 18 h of incubation, the image of GFP and red fluorescent protein (RFP) fluorescence was captured by confocal laser scanning microscopy (Leica SP8, Germany).
In 223 natural populations, 147 were not sensitive to cold stress and no damage was observed on their leaves, while 76 showed sensitivity to cold stress and showed varying degrees of damage. Therefore, based on the degree of damage, the impact of cold damage can be graded to different levels (Figure 1A). To study the effect of cold stress on whole-genome gene expression patterns, RNA-seq was performed in 20 cold-sensitive and 20 cold-tolerant accessions. There were 475 differentially expressed genes (DEGs), consisting of 176 upregulated genes and 299 downregulated genes (Figure 1B). The Gene Ontology (GO) terms of DEGs included heme binding, oxidoreductase activity, tetrapyrrole binding, and iron ion binding (Figure 1C). The Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways of DEGs included starch and sucrose metabolism, biosynthesis of nucleotide sugars, amino sugar and nucleotide sugar metabolism, plant–pathogen interaction, phenylpropanoid biosynthesis, and MAPK signaling pathway (Figure 1D). We detected the expression of 475 DEGs, and a total of 34 significant expression (padj ≤ 0.01) were identified (Supplementary Table S3). Thirty-four DEGs have a higher expression level by RNA-seq (Figure 1E). In order to verify the accuracy of RNA-Seq, 34 DEGs were selected to quantify the expression with M82 (cold-sensitive) and A57 (cold-tolerant) by qRT-PCR (Supplementary Table S4). The findings were largely consistent with RNA-seq. The expression levels of Solyc07g042550 were the highest (Figure 1F), which indicates that SUS3 is a major candidate for having an important influence on cold tolerance (Supplementary Table S3).
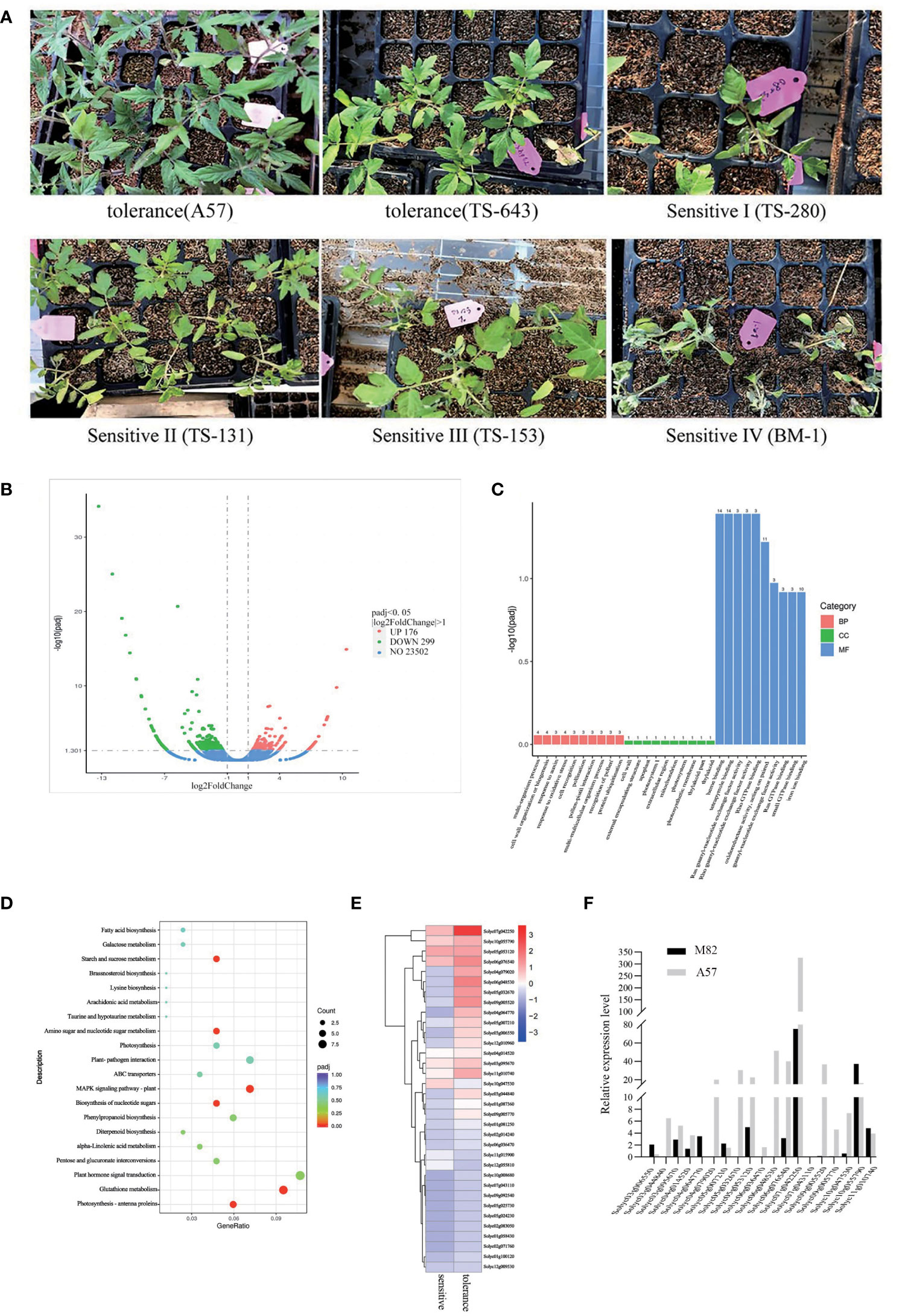
Figure 1 Differentially expressed genes (DEGs) in cold-sensitive and cold-tolerant tomato varieties. (A) Partial cold-sensitive and cold-insensitive tomato varieties. (B) Volcano map of DEGs. (C, D) Significantly enriched GO terms (C) and KEGG pathways (D) of DEGs. (E) Heatmap of gene expression related to sugar metabolism. (F) Gene expression related to sugar metabolism by qRT-PCR.
We analyzed and predicted the structural features and function of SUS3 at the molecular level. The length of the gene is 2,955 bp, which includes an open reading frame (ORF) of 2,418 bp, starting at position 273 with ATG and ending at position 2690 with TAA. It encodes a protein with a length of 806 amino acids and a molecular weight of 92.59 kDa (Supplementary Figure S1A). SUS3 had eight helical segments that had 10–70, 100–250, 270–330, 350–450, 480–500, 550–610, 620–660, and 690–780 amino acid sequences and did not contain a signal peptide, and it was a stable non-transmembrane and hydrophilic protein (Supplementary Figures S1B–D).
In order to verify the protein function of SUS3, the subcellular localization of tomato protoplast confirmed that the GFP was irregularly dotted around the cells (Figure 2A). SUS3 was specifically expressed in the cytoplasm and cell membrane. This result suggests that the SUS3 is mainly located in the cytoplasm and cell membrane. To further investigate the evolutionary relationship of SUS3 and its families, 24 family genes from strawberry (FaSUS1-1~FaSUS1-10, FaSUS2-1~FaSUS2-7, FaSUS3-1~FaSUS3-7), 6 genes from Arabidopsis (AtSUS1~AtSUS6), 13 genes from soybean (GmSUS1-1~GmSUS1-4, GmSUS2-1~GmSUS2-3, GmSUS3-1~GmSUS3-6), 7 genes (OsSUS1~OsSUS7) from rice, 5 genes (ZmSUS1-1~ZmSUS2-1, ZmSUS2-1~ZmSUS2-2, ZSUSm3) from maize, and 22 genes (NtSUS1-1~NtSUS1-13, NtSUS2-1~NtSUS2-4, NtSUS3-3, NtSUS3-5) from tobacco were analyzed. The results showed that the SUS family genes and tobacco SUS genes had high homology, implying that the SUS families and tobacco had similarities in biological functions and growth regulatory mechanisms. Meanwhile, SUS3 has the farthest evolutionary relationship with the other five SUS family genes, and it is speculated that it may exercise different biological functions (Figure 2B).
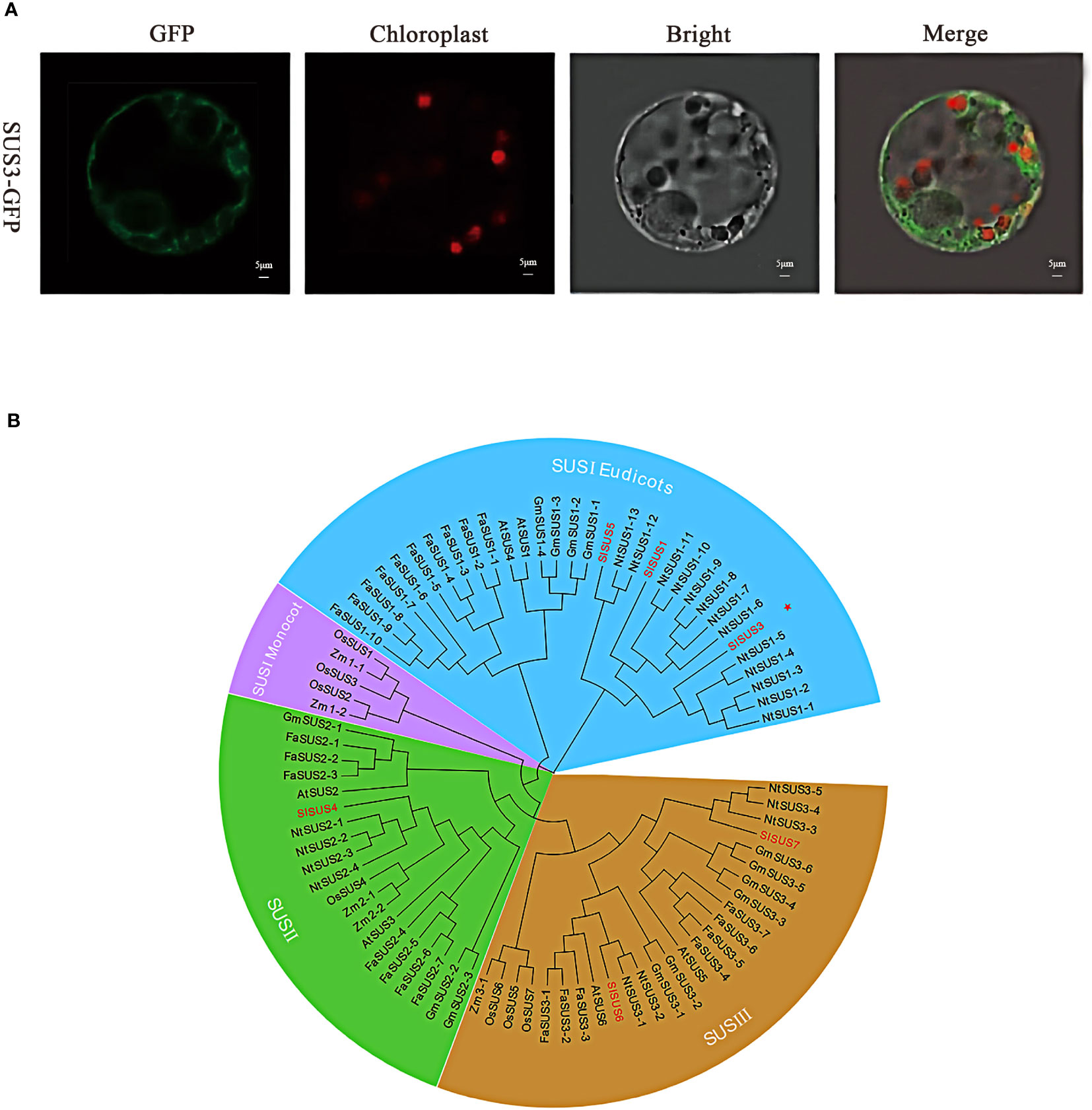
Figure 2 Molecular characterization of SUS3. (A) Subcellular localization of the SUS3 protein in protoplasts. (B) Agrobacterium strains carrying the GFP-SUS3 were digested into protoplasts in tobacco leaves. Bright-field, green fluorescent, red fluorescent, and merged images are shown.
To test whether SUS3 might contribute to responses to other abiotic stresses, we examined the expression pattern of SUS3 in AC plants after subjecting them to ABA, ethylene, drought, cold, and salt stresses (Figure 3). A mild induction of SUS3 expression was detected in response to ABA, ethylene, and salt stresses, and the SUS3 was strongly induced by drought and cold treatment. In the course of 24 h under these treatments, the expression of SUS3 showed an initial increase followed by a decrease. More specifically, the expression level of SUS3 continued to increase significantly within 6–12 h and reached the maximum.
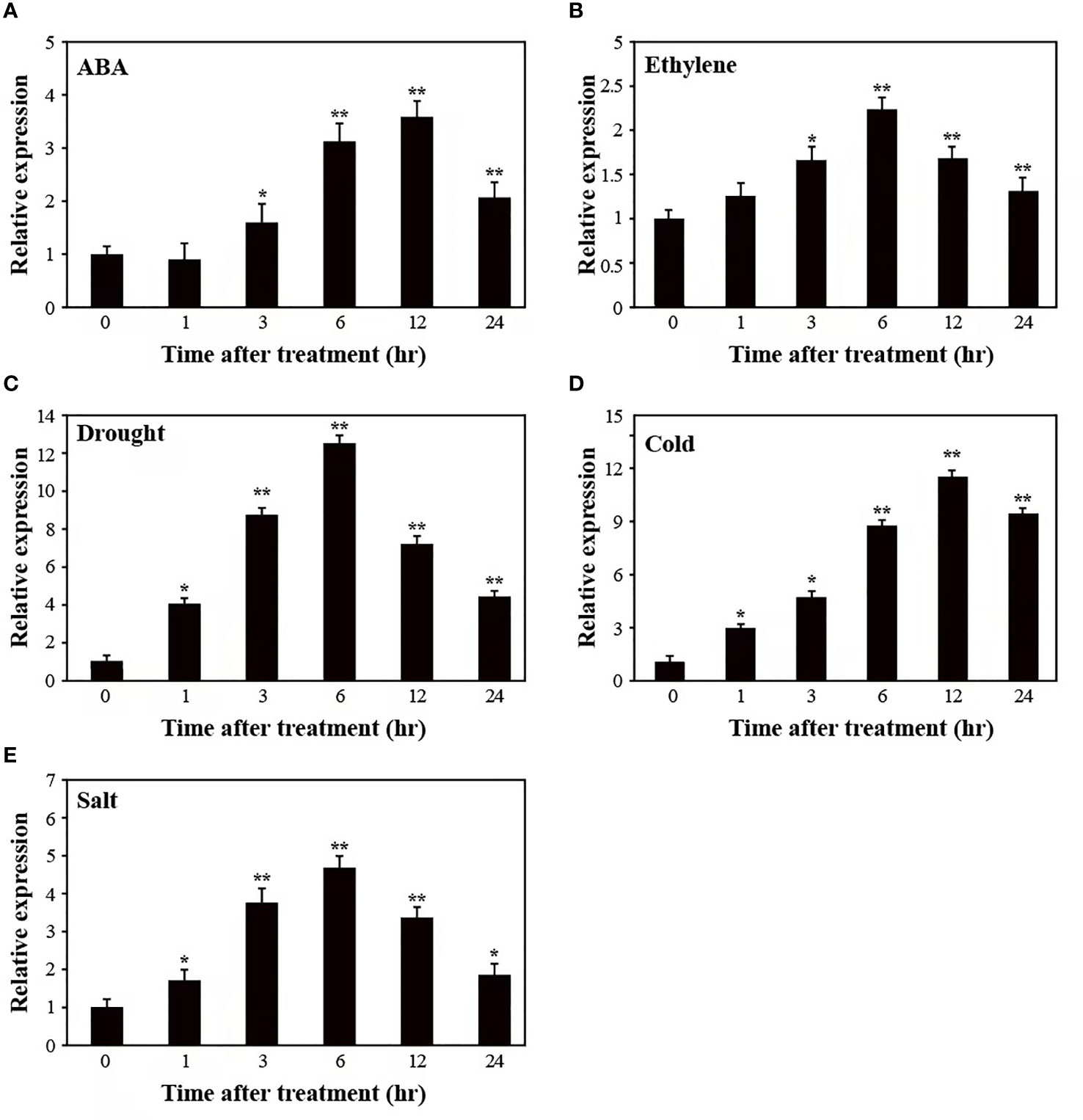
Figure 3 Relative expression pattern of SUS3 under abiotic stress and hormone induction. (A–E) Expression pattern of SUS3 in ABA, ethylene, drought, cold, and salt. Four-week-old seedlings were stressed with ABA (100 μM), ethylene (200 μM), drought (dehydration), cold (4°C), and salt (400 mM of NaCl) treatments for 0, 1, 3, 6, 12, and 24 h, respectively. The relative expression levels were determined using qRT-PCR. The expression levels of the appropriate controls were set to a value of 1. Data are presented as mean ± standard deviation of three biological replicates. Asterisks indicate statistically significant differences between transgenic lines and wild type. *, p < 0.05; **, p < 0.01.
To investigate the biological functions of SUS3, we transformed M82 with the 35S:SUS3 construct. Three independent OE lines (OE1-2, OE2-4, OE3-2) and three RNAi lines (Ri1-1, Ri8-3, Ri17-2) were chosen for further characterization (Figure 4C). We tested the cold tolerance of the SUS3 transgenic seedlings. After cold stress for 7 days, the leaves of the wild type showed wilting, and the stems became shriveled and bent and were unable to uphold upright. In contrast, the SUS3-OE lines in their leaves were capable of preserving regular upward growth (Figure 4A). However, the phenotype of the SUS3-RNAi lines was similar to that of WT under cold stress and did not demonstrate more wilting (Figure 4B). Therefore, it suggests that the loss of SUS3-RNAi lines might not significantly affect cold tolerance.
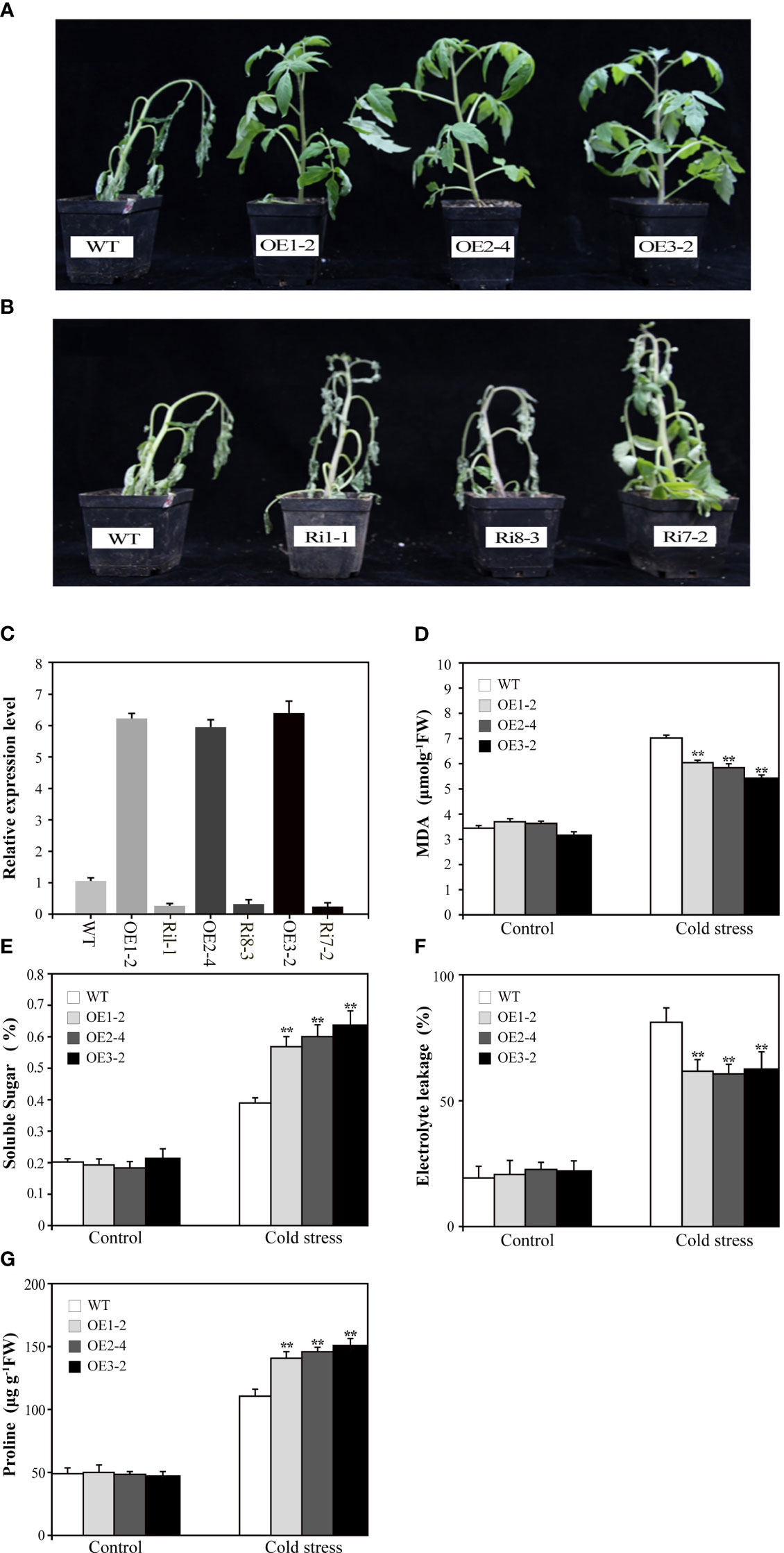
Figure 4 Overexpression of SUS3 improves the cold tolerance in tomato. (A, B) Phenotype of transgenic seedlings expressing abnormal levels of SUS3 and WT seedlings under cold stress conditions. The SUS3-OE lines (OE1-2, OE2-4, and OE3-2), SUS3-RNAi lines (Ri1-1, Ri8-3, and R7-2), and WT seedlings were grown at 4°C for 7 days. (C) Relative expression levels of SUS3 in transgenic lines and WT. Comparison of MDA content (D), soluble sugar (E), relative electrolyte leakage (F), and proline (G) in SUS3-OE lines and WT under cold stress and optimal conditions. Data are presented as mean ± standard deviation of three biological replicates. Asterisks indicate statistically significant differences between transgenic lines and wild type. *, p < 0.05; **, p < 0.01.
To evaluate cold tolerance, we analyzed the MDA, relative electrolyte leakage, and proline and soluble sugar contents between transgenic lines and WT plants under cold stress. Under optimal growth conditions, there is no difference between transgenic lines and WT in the above indexes. After 7 days of cold treatment at 4°C, MDA and relative electrolyte leakage increased in SUS3-OE lines and WT, but the levels were significantly lower in the SUS3-OE lines than in WT (Figures 4D, F). These results indicate that SUS3 can alleviate the damage to cellular membranes that occurs during cold stress. Under cold stress, the contents of both proline and soluble sugar were significantly higher. Moreover, the increase of sugar in leaves of the SUS3-OE lines was significantly higher than in WT.
Various abiotic stresses often result in the excessive accumulation of reactive oxygen species (ROS) in plants, causing substantial damage to multiple cellular structures (Gill and Tuteja, 2010). Therefore, we assessed the levels of superoxide radicals (O2−.) in SUS3-OE lines and WT. After treatment at 4°C for 7 days, the accumulation of O2−. increased in both SUS3-OE lines and WT. However, the accumulation in SUS3-OE lines was significantly lower in comparison to the WT (Figure 5A). This indicates that overexpression of SUS3 has the ability to prevent the accumulation of excessive O2−. free radicals, thereby reducing the damage to plants and enhancing their resistance to cold damage.
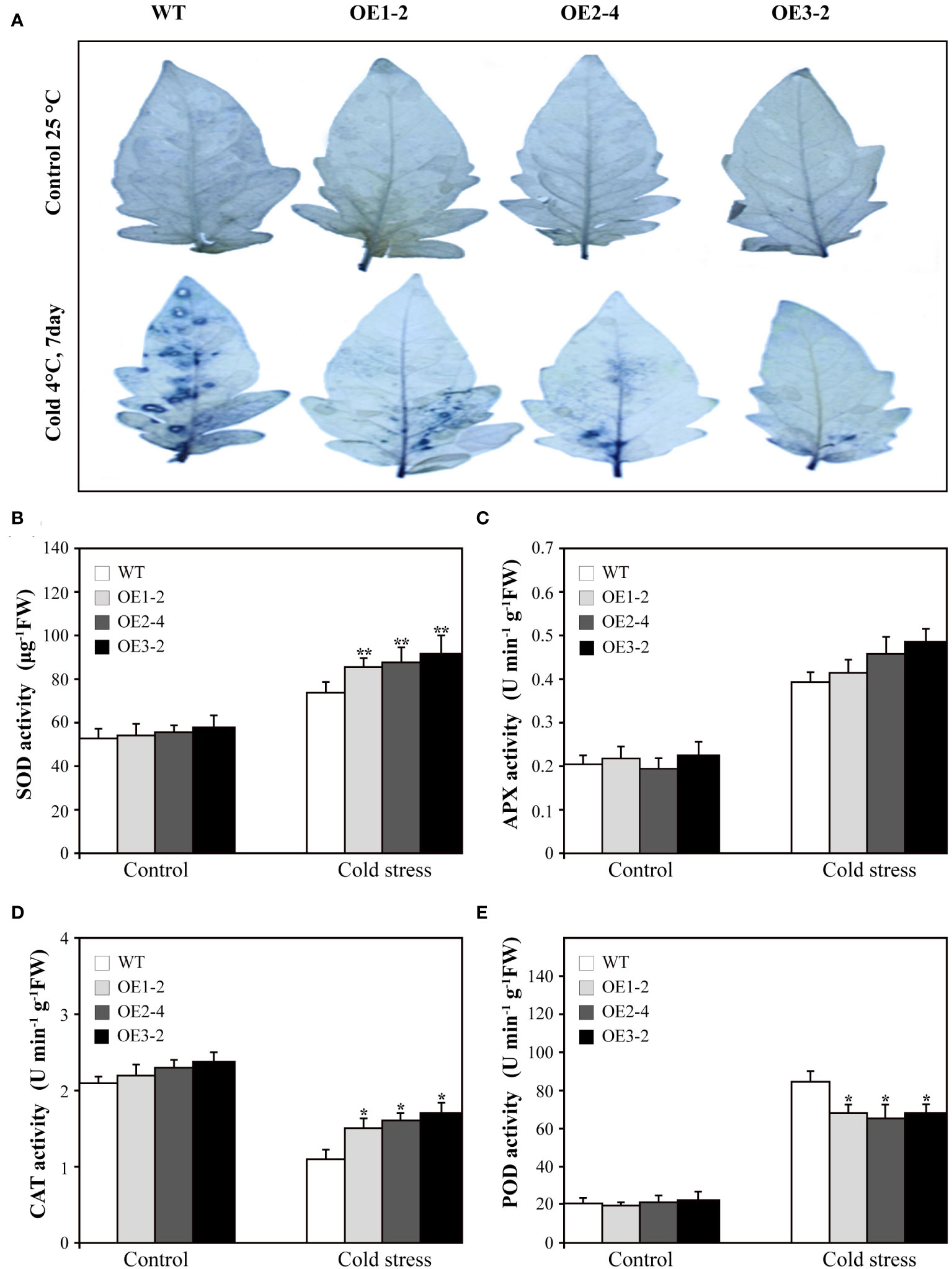
Figure 5 Superoxide radicals(O2−.) accumulation and related enzyme activity of SUS3-OE lines and WT under cold stress and optimal conditions. (A) NBT staining used to detect the accumulation of superoxide radicals. The SUS3-OE lines (OE1-2, OE2-4, and OE3-2) and WT under cold stress and optimal conditions were incubated at 25°C in the dark for 2 h. Comparison of SOD content (B), APX (C), CAT (D), and POD (E) in SUS3-OE lines and WT grown under cold stress and optimal conditions. Data are presented as mean ± standard deviation of three biological replicates. Asterisks indicate statistically significant differences between transgenic lines and wild type. *, p < 0.05; **, p < 0.01.
In order to determine whether overexpression of SUS3 can reduce the damage of plant cells under cold stress, we measured the activities of four key antioxidant enzymes (SOD, CAT, APX, and POD). The results showed that under optimal growth conditions, the activities of the four enzymes did not show significant differences between SUS3-OE lines and WT. However, after 7 days of cold treatment at 4°C, the activity of SOD and CAT was increased in SUS3-OE lines and WT, but the enzyme activity in SUS3-OE lines was significantly higher than in WT (Figures 5B, D). Under cold stress, APX activity in both SUS3-OE lines and WT also increased, but there was no significant difference (Figure 5C). The activity of POD increased in both SUS3-OE lines and WT, but the activity increased more significantly in the WT than in SUS3-OE lines (Figure 5E). Compared with the WT, overexpression of SUS3 increased the activities of SOD and CAT and decreased the activities of POD under cold stress. There was an accumulation of fewer ROS in SUS3-OE lines, thus reducing the oxidative stress injury. This may lead to less ROS accumulation in SUS3-OE lines, thus reducing oxidative stress injury.
In recent years, transcriptomic, proteomic, and metabolomic research tools have been utilized increasingly in plants, which has been noted as a significant trend in botanical studies. Proteomic analyses of tomato fruit during cryopreservation show significant changes in biological processes including defense responses, photosynthesis, and protein degradation, such as heat stress proteins (HSPs), late embryonic developmental enriched proteins (LEAs), ATPases, and 26S proteasome subunits (Page et al., 2010; Sanchez-Bel et al., 2012). Currently, there is no report on the transcriptomics of low-temperature response in tomato seedlings, only reports on low-temperature storage in fruits, and the mechanisms of low-temperature response in seedlings and fruits are considerably different, which to some extent limits the candidate cloning and functional identification of cold-tolerance-related genes in tomato. We used the low-temperature-sensitive and cold-tolerant materials for RNA-seq to compare the expression difference after cold stress. A total of 34 genes were found to be significantly differentially expressed. SUS3 (Solyc07g042550) was upregulated at a considerably higher level in cold-tolerant accessions than cold-sensitive accessions. Additionally, qRT-PCR analyses demonstrated that the expression of this gene was significantly higher in A57, a wild tomato, than in M82, a cultivated tomato. Through this, we confirmed that tomato SUS3 is closely related to cold stress. Previous research indicated that plants are more likely to utilize SUS to promote sucrose catabolism and generate reducing sugars to maintain growth under adverse conditions (Fenando et al., 2000; Bologa et al., 2003). It has been reported that sugar beet, wheat, and cabbage SUS genes are highly expressed under conditions of low temperature (Marana et al., 1990; Hesse and Willmitzer, 1996; Sasaki et al., 2001). It is possible that SUS regulates intracellular soluble sugar content and osmotic pressure to enhance cold tolerance (Crespi et al., 1991). Previous studies have also demonstrated that SUS1 and SUS3 are cold-responsive genes, while SUS1 and SUS3 are respectively induced in response to hypoxia and drought in barley (Barrero-Sicilia et al., 2011). The study examined the effects of SUS3 on cold tolerance and identified a range of tomato accessions that are cold-tolerant or cold-sensitive. This provides a theoretical foundation and enriches the available germplasm for breeding.
Bioinformatics analyses showed that SUS genes are a family. The tomato SUS family has six genes, and they are distributed across five chromosomes. The amino acid number for SUS varies from 805 AA to 891 AA, while the molecular weight ranges between 91.63 kDa and 100.75 kDa. The protein isoelectric point can range from 5.87 to 8.42, and the gene includes 11 to 15 exons (Bieniawska et al., 2007). SUS3 is 2,955 bp long, with an open reading frame of 2,418 bp. SUS3 does not contain a signal peptide and is a stable, hydrophilic, and non-transmembrane protein. Similar results were observed from the analysis of potatoes (Xu et al., 2012) and cotton (Lu et al., 2005). To further investigate the evolutionary relationships of the SUS family, genes can be classified into four major categories: SUSI eudicots, SUSI monocots, SUSII, and SUSIII. SUS1, SUS3, and SUS4 genes are categorized as SUSI eudicots, while the SUS4 gene falls into the SUSII category, and SUS6 and SUS7 belong to the SUSIII category. Notably, no SUS members of the monocotyledon category were found, which is consistent with previous classification results (Van Bel et al., 2017). A phylogenetic tree showed that the significant homology between the SUS in tomato and NtSUS in tobacco implies the similarity in biological functions and regulation mechanisms.
The evaluation of SUS3 showed that it is responsive to cold, drought, salinity, ethylene, and ABA treatments. This gene could play a key role in abiotic stress perception and response. Consequently, our results suggested that SUS3 is crucial in regulating abiotic stress and hormone response. To further evaluate the biological function of tomato SUS3, we obtained transgenic lines in tomato and analyzed their performance under cold stress. SUS3-OE lines showed less leaf wilting and better growth than wild-type plants, indicating that overexpressing SUS3 can improve the tolerance of tomatoes to cold stress. However, the phenotype of the SUS3-RANi lines was similar to that of WT under cold stress and did not demonstrate more leaf wilting. Therefore, it is speculated that SUS3 is not the sole participant in the stress resistance pathway, and other members may have a similar role (Barratt et al., 2009; Baroja et al., 2012).
When plants are subjected to cold stress, cell membrane damage occurs, which can be reflected by relative electrical conductivity and MDA levels (Baud and Graham, 2006; Nguyen et al., 2016). Under cold stress, although the relative electrical conductivity and MDA content in SUS3-OE lines increased due to cold damage, they were significantly lower than in WT. This indicates that SUS3 functions to decrease adversity-induced peroxidation of the plasma membrane. Proline and soluble sugars contribute to maintaining cellular osmotic potential and membrane integrity (Morsy et al., 2007; Szabados and Savoure, 2010). Its accumulation also enhances plant tolerance to abiotic stresses (Cheng et al., 2007; Zhang and Huang, 2010). Overexpressing SUS3 increased proline levels in response to low-temperature stress. Therefore, the SUS3 gene improves cold tolerance in tomato plants. Interestingly, we discovered that the accumulation of O2−. in SUS3-OE lines was significantly lower than that in wild-type plants when subjected to cold stress. SOD and CAT can scavenge ROS, while POD is involved in both ROS generation and ROS scavenging (Cosio and Dunand, 2009). Furthermore, the activities of SOD and CAT related to ROS were significantly higher in the SUS3-OE lines compared with WT. These results suggest that SUS3 improves ROS scavenging capacity and reduces the damage of adversity-induced oxidative stress on cell membranes. Our results showed that overexpressing SUS3 can reduce the damage to plant cells caused by cold stress and enhance the cold tolerance of plants.
In summary, we found that SUS3 is a novel gene whose expression was induced by cold and positively regulates cold tolerance by reducing cell membrane damage. In addition, overexpressing SUS3 increased the cold tolerance in tomato. Furthermore, our analyses of a population indicate that SUS3 is a naturally occurring allele that is significantly enriched in cold-tolerant accessions and the SUS3 allele is significantly distributed in cold-sensitive accessions. These data provide evidence that SUS3 might play a role in cold tolerance. Therefore, our study demonstrates that high expression levels of SUS3 can serve as direct targets for both genetic engineering and selection for enhanced cold tolerance in tomato. These findings provide essential information for breeding.
The original contributions presented in the study are publicly available. This data can be found here: https://www.ncbi.nlm.nih.gov/ accession number PRJNA1065726.
SL: Writing – original draft. YW: Writing – original draft. YYL: Writing – review & editing. CL: Writing – review & editing. YEL: Writing – review & editing. WX: Writing – review & editing. ZY: Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by grants from the National Natural Science Foundation of China (32060685,32360750,32302535), the International Cooperation Promotion Plan of Shihezi University (GJHZ202104), the Scientific Research Foundation of Shihezi University (RCZK202336) and Wuhan Major Project of Key Technologies in Biological Breeding and New Variety Cultivation, grant number 2022021302024852.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1324401/full#supplementary-material
Supplementary Figure 1 | Bioinformatics characteristics of SUS3. (A) Nucleotide and amino acid sequence. (B) the transmembrane amino acid segment. (C) signal peptide (c- score: Original cut score, s- score: Signal score, y- score: General cut score). (D) hydrophilicity/hydrophobicity analyses.
Airaki, M., Leterrier, M., Mateos, R. M., Valderrama, R., Chaki, M., Barroso, J. B., et al. (2012). Metabolism of reactive oxygen species and reactive nitrogen species in pepper (Capsicum annuum L.) plants under low temperature stress. Plant Cell Environ. 35, 281–295. doi: 10.1111/j.1365-3040.2011.02310.x
Al-aghabary, K., Zhu, Z., Shi, Q. (2005). Influence of Silicon Supply on chlorophyll content, chlorophyll fluorescence, and antioxidative enzyme activities in tomato plants under salt stress. J. Plant Nutr. 27, 2101–2115 126. doi: 10.1081/PLN-200034641
Baroja, E., Munoz, F. J., Li, J. (2012). Sucrose synthase activity in the SUS1/SUS2 SUS3/SUS4 Arabidopsis mutant is sufficient to support normal cell lose and starch production. Proc. Natl. Acad. Sci. USA 109, 321–326. doi: 10.1073/pnas.1117099109
Barratt, D. H., Derbyshire, P., Findlay, K., Pike, M., Wellner, N., Lunn, J., et al. (2009). Normal growth of Arabidopsis requires cytosolic invertase but not sucrose synthase. Proc. Natl. Acad. Sci. USA 106 (31), 13124–13129. doi: 10.1073/pnas.0900689106
Barrero-Sicilia, C., Hernando-Amado, S., González-Melendi, P., Carbonero, P. (2011). Structure, expression profile and subcellular localisation of four different sucrose synthase genes from barley. Planta 234 (2), 391–403. doi: 10.1007/s00425-011-1408-x
Baud, S., Graham, I. A. (2006). A spatiotemporal analysis of enzymatic activities associated with carbon metabolism in wild-type and mutant embryos of Arabidopsis using in situhtis tochemistry. Plant J. 46 (1), 155–169. doi: 10.1111/j.1365-313X.2006.02682.x
Bieniawska, Z., PalBarratt, D. H., Garlick, A. P. (2007). Analysis of the sucrose synthase gene-family in Arabidopsis. Plant J.: Cell Mol. Biol. 49 (5), 810–828. doi: 10.1111/j.1365-313X.2006.03011.x
Bologa, K. L., Fernie, A. R., Leisse, A., Loureiro, M., Geigenberger, P. (2003). A bypass of sucrose synthase leads to low internal oxygen and mpaired metabolic performance in growing potato tubers. Plant Physiol. 132, 2058–2072. doi: 10.1104/pp.103.022236
Campos, P. S., Quartin, V., Ramalho, J. C., Nunes, M. A. (2003). Electrolyte leakage and lipide gradation account for cold sensitivity in leaves of coffeasp. Plant Physiol. 160, 283–292. doi: 10.1078/0176-1617-00833
Cardini, C. E., LeloirL, F., Chiriboga, J. (1955). The bio-synthesis of sucrose. J. Biol. Chem. 214, 149–155. doi: 10.1016/S0021-9258(18)70953-8
Chen, A., He, S., Li, F., Li, Z., Ding, M., Liu, Q. (2012). Analyses of the sucrose synthase gene family in cotton: structure, phylogeny and expression patterns. BMC Plant Biol. 12, 1471–2229. doi: 10.1186/1471-2229-12-85
Cheng, C., Yun, K. Y., Ressom, H. W., Mohanty, B., Bajic, V. B., Jia, Y., et al. (2007). An early response regulatory cluster induced by low temperature and hydrogen peroxide in seedlings of chilling-tolerant japonica rice. BMC Genomics 8, 175. doi: 10.1186/1471-2164-8-175
Cosio, C., Dunand, C. (2009). Specific functions of individual class III peroxidase genes. J. Exp. Bot. 60, 391–408. doi: 10.1093/jxb/ern318
Crespi, M. D., Zabaleta, E. J., Pontis, H. G., Salerno, G. L. (1991). Sucrose synthase expression during cold acclimation in wheat. Plant Physiol. 96 (3), 887–891. doi: 10.1104/pp.96.3.887
Fan, W., Zhang, M., Zhang, H., Zhang, P. (2012). Improved tolerance to various abiotic stresses in transgenic sweet potato (Ipomoea batatas) expressing spinach betaine aldehyde dehydrogenase. PloS One 7, e37344. doi: 10.1371/journal.pone.0037344
Fenando, E. P., Boero, C., Gallardo, M., Gonzalez, J. A. (2000). Effect of Na Cl on germination, growth, and soluble sugar content in Chenopodium quinoa Willd. Seeds. Botanical Bull. Academia Sin. (Taipei) 41 (1), 27–34. doi: 10.7016/BBAS.200001.0027
Foolad, M., Lin, G. (2001). Heritability of Early Blight Resistance in a Lycopersicon Esculentum x Lycopersicon Hirsutum Cross Estimated by Correlation between Parent and Progeny. Plant Breeding 120 (2), 173–177.
Fukao, T., Xu, K., Ronald, P. C., Bailey-Serres, J. (2006). A variable cluster of ethylene response factor-like genes regulates metabolic and developmental acclimation responses to submergence in rice. Plant Cell 18, 2021–2034.
Geigenberger, P., Stitt, M. (1993). Sucrose synthase catalyses a readily reversible reaction in vivo in developing potato tubers and other plant tissue. Planta 189, 329–39. doi: 10.1007/BF00194429
Gill, S. S., Tuteja, N. (2010). Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol. Biochem. 48, 909–930. doi: 10.1016/j.plaphy.2010.08.016
Hannah, M. A., Heyer, A. G., Hincha, D. K. (2005). A global survey of gene regulation during cold acclimation in Arabidopsis thaliana. PloS Genet. 1, e26. doi: 10.1371/journal.pgen.0010026
Hesse, H., Willmitzer, L. (1996). Expression analysis of a sucrose synthase gene from sugar beet (Betavlgaris L.). Plant Mol. Biol. 30 (5), 863–872. doi: 10.1007/BF00020799
Jimenez, A., Hernandez, J. A., Del Rio, L. A., Sevilla, F. (1997). Evidence for the presence of the ascorbate-glutathione cycle in mitochondria and peroxisomes of pea leaves. Plant Physiol. 114, 275–284. doi: 10.1104/pp.114.1.275
Li, J., Liu, L., Bai, Y., Finkers, R., Wang, F., Du, Y., et al. (2011). Identification and mapping of quantitative resistance to late blight (Phytophthora infestans) in Solanum habrochaites LA1777. Euphytica 179, 427–438. doi: 10.1007/s10681-010-0340-7
Lu, H., Shen, F., Liu, L., Weifang, S. (2005). Molecular structure characteristics and function prediction analysis of cotton sucrose synthetase (SuSy). Northwest J. Bot. 25 (7), 1372–1376.
Marana, C., Garcia-Olmedo, F., Carbonero, P. (1990). Difffferential expression of two types of sucrose synthase-encoding genes in wheat in response to anaerobiosis, cold shock and light. Gene 88, 167–172. doi: 10.1016/0378-1119(90)90028-P
Morohashi, Y. (2002). Peroxidase activity develops in the micropylar endosperm of tomato seeds prior to radicle protrusion. J. Exp. Bot. 53, 1643–1650. doi: 10.1093/jxb/erf012
Morsy, M. R., Jouve, L., Hausman, J. F., Hoffmann, L., Stewart, J. M. (2007). Alteration of oxidative and carbohydrate metabolism under abiotic stress in two rice (Oryza sativa L.) genotypes contrasting in chilling tolerance. J. Plant Physiol. 164, 157–167. doi: 10.1016/j.jplph.2005.12.004
Nguyen, Q. A., Luan, S., Wi, S. G., Bae, H., Lee, D. S., Bae, H. J. (2016). Pronounced phenotypic changes in transgenic tobacco plants over expressing sucrose synthase may reveal a novel sugar signaling pathway. Front. Plant Sci. 6, 1216. doi: 10.3389/fpls.2015.01216
Page, D., Gouble, B., Valot, B., Bouchet, J. P., Callot, C., Kretzschmar, A., et al. (2010). Protective proteins are differentially expressed in tomato genotypes differing for their tolerance to low-temperature storage. Planta 232, 483–500. doi: 10.1007/s00425-010-1184-z
Ricard, B., Van Toai, T., Chourey, P., Saglio, P. (1998). Evidence for the critical role of sucrose synthase for anoxic tolerance of maize roots using a double mutant. Plant Physiol. 116 (4), 1323–1331. doi: 10.1104/pp.116.4.1323
Sanchez-Bel, P., Egea, I., Sanchez-Ballesta, M. T., Sevillano, L., Del Carmen Bolarin, M., Flores, F. B. (2012). Proteome changes in tomato fruits prior to visible symptoms of chilling injury are linked to defensive mechanisms, uncoupling of photosynthetic processes and protein degradation machinery. Plant Cell Physiol. 53, 470–484. doi: 10.1093/pcp/pcr191
Sasaki, H., Ichimura, K., Imada, S., Yamaki. (2001). Sucrose synthase and sucrose phosphate synthase, but not acid invertase, are regulated by cold acclimation and deacclimation in cabbage seedling. Plant Physiol. 158, 847–852. doi: 10.1078/0176-1617-00391
Sato, Y., Masuta, Y., Saito, K., Murayama, S., Ozawa, K. (2011). Enhanced chilling tolerance at the booting stage in rice by transgenic overexpression of the ascorbate peroxidase gene OsAPXa. Plant Cell Rep. 30, 399–406. doi: 10.1007/s00299-010-0985-7
Sawicki, M., Jeanson, E., Celiz, V., Clément, C., Jacquard, C., Vaillant-Gaveau, N. (2012). Adaptation of grapevine flowers to cold involves different mechanisms depending on stress intensity. PLOS ONE 7 (10), e46976. doi: 10.1371/journal.pone.0046976
Song, J., Yali, X., Shoaib, M., Chuying, Y., Lulu, S., Hanxia, L. (2016). An ATL78-like RING-H2 finger protein confers abiotic stress tolerance through interacting with RAV2 and CSN5B in tomato. Front. Plant Sci. 07, e50785. doi: 10.3389/fpls.2016.01305
Szabados, L., Savoure, A. (2010). Proline: a multifunctional amino acid. Trends Plant Sci. 15, 89–97. doi: 10.1016/j.tplants.2009.11.009
Takehara, K., Murata, K., Yamaguchi, T., Yamaguch, K., Chaya, G., Kido, S., et al. (2018). Thermo responsive allele of sucrose synthase 3 provides high temperature tolerance during the ripening stage in rice (Oryza sativa L). Breed. Sci. 68, 336–342. doi: 10.1270/jsbbs.18007
Tatsuro, H., Grahamn, S., Tomio, T. (2008). An expression analysis profile for the entire sucrose synthase gene family in rice. Plant Sci. 174 (5), 534–543. doi: 10.1016/j.plantsci.2008.02.009
Van Bel, M., Diels, T., Vancaester, E., Kreft, L., Botzki, A., Van De Peer, Y., et al. (2017). PLAZA 4.0: an integrative resource for functional, evolutionary and comparative plant genomics. Nucleic Acids Res. 46, D1190–D1196. doi: 10.1093/nar/gkx1002
Venema, J. H., Linger, P., van Heusden, A. W., Hasselt, P. R., Brüggemann, W. (2005). The inheritance of chilling tolerance in tomato (Lycopersicon spp). Plant Biol. 7, 118–130. doi: 10.1055/s-2005-837495
Wang, X., Li, S., Dong, H., Gao, Z., Dai, S. (1996). Effect of cold stress on several traits during seedling and flowering stages. J. Horticulture 4, 349–354.
Wang, J., Sun, P. P., Chen, C. L., Wang, Y., Fu, X. Z., Liu, J. H. (2011). An arginine decarboxylase gene PtADC from Poncirus trifoliata confers abiotic stress tolerance and promotes primary root growth in Arabidopsis. J. Exp. Bot. 62, 2899–2914. doi: 10.1093/jxb/erq463
Xia, X. J., Wang, Y. J., Zhou, Y. H., Tao, Y., Mao, W. H., Shi, K., et al. (2009). Reactive oxygen species are involved in brassinosteroid-induced stress tolerance in cucumber. Plant Physiol. 150, 801–814. doi: 10.1104/pp.109.138230
Xiao, X., Tang, C., Fang, Y., Yang, M., Zhou, B. H., Qi, J., et al. (2014). Structure and expression profile of the sucrose synthase gene family in the rubber tree: indicative of roles in stress response and sucrose utilization in the laticifers. FEBS J. 281 (1), 291–305. doi: 10.1111/febs.12595
Xu, S., Brill, E., Llewellyn, D. J., Furbank, R. T., Ruan, Y. L. (2012). Overexpression of a potato sucrose synthase gene in cotton accelerates leafexpansion reduces seed abortion and enhances fiber production. Mol. Plant 5, 430–441. doi: 10.1093/mp/ssr090
Zhang, J., Arro, J., Chen, Y., Ming, R. (2013). Haplotype analysis of sucrose synthase gene family in three Saccharum species. BMC Genomics 14, 314–326. doi: 10.1186/1471-2164-14-314
Zhang, Z., Huang, R. (2010). Enhanced tolerance to freezing in tobacco and tomato overexpressing transcription factor TERF2/LeERF2 is modulated by ethylene biosynthesis. Plant Mol. ecular Biol. 73 (3), 241–249. doi: 10.1007/s11103-010-9609-4
Keywords: tomato, sucrose synthase, SUS3, cold stress, transcriptome
Citation: Li S, Wang Y, Liu Y, Liu C, Xu W, Lu Y and Ye Z (2024) Sucrose synthase gene SUS3 could enhance cold tolerance in tomato. Front. Plant Sci. 14:1324401. doi: 10.3389/fpls.2023.1324401
Received: 19 October 2023; Accepted: 26 December 2023;
Published: 25 January 2024.
Edited by:
Shiming Liu, Chinese Academy of Agricultural Sciences, ChinaReviewed by:
Luis Morales-Quintana, Autonomous University of Chile, ChileCopyright © 2024 Li, Wang, Liu, Liu, Xu, Lu and Ye. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhibiao Ye, emJ5ZUBtYWlsLmh6YXUuZWR1LmNu; Wei Xu, eHV3ZWkwNDEyQHNoenUuZWR1LmNu; Yongen Lu, bHV5b25nZW5AbWFpbC5oemF1LmVkdS5jbg==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.