- Key Laboratory of Biology and Genetic Improvement of Horticultural Crops (South China), Ministry of Agriculture and Rural Affairs/Guangdong Vegetable Engineering and Technology Research Center/College of Horticulture, South China Agricultural University, Guangzhou, China
Eggplant (Solanum melongena) is an economically important crop and rich in various nutrients, among which rutin that has positive effects on human health is found in eggplant. Glycosylation mediated by UDP-glycosyltransferases (UGTs) is a key step in rutin biosynthesis. However, the UGT gene has not been reported in eggplant to date. Herein, 195 putative UGT genes were identified in eggplant by genome-wide analysis, and they were divided into 17 subgroups (Group A-P and Group R) according to the phylogenetic evolutionary tree. The members of Groups A, B, D, E and L were related to flavonol biosynthesis, and rutin was the typical flavonol. The expression profile showed that the transcriptional levels of SmUGT genes in Clusters 7-10 were closely related to those of rutin biosynthetic pathway genes. Notably, SmUGT89B2 was classified into Cluster 7 and Group B; its expression was consistent with rutin accumulation in different tissues and different leaf stages of eggplant. SmUGT89B2 was located in the nucleus and cell membrane. Virus-induced gene silencing (VIGS) and transient overexpression assays showed that SmUGT89B2 can promote rutin accumulation in eggplant. These findings provide new insights into the UGT genes in eggplant, indicating that SmUGT89B2 is likely to encode the final enzyme in rutin biosynthesis.
Introduction
As an important horticultural crop, eggplant (Solanum melongena L.) is the third most cultivated Solanaceae species in the world, next to potato (Solanum tuberosum L.) and tomato (Solanum lycopersicum L.) (Barchi et al., 2021). Eggplant is planted on 1.85 million hectares with a worldwide production of 56.30 million tons, which is mainly cultivated in India and China, and consumed worldwide (FAO, see www.fao.org) (Martínez-Ispizua et al., 2021; Wang et al., 2022). Flavonoids are one of the most valued compounds in eggplant fruit: anthocyanins are the highest, followed by flavonol (Martínez-Ispizua et al., 2021). Currently, all kinds of flavonols have been identified in eggplant fruit, including quercetin, rutin, myricetin, quercetin-3-diglucoside, kaempferol-3,7-diglucoside, kaempferol-diglucoside and quercetin-3-rhamnoside (Singh et al., 2017; Sulli et al., 2021). Flavonols are plant-specialized metabolites with important functions in regulate plant development, signaling, and stress response (Daryanavard et al., 2023).
Rutin is a typical flavonol and plays a key role in defense responses against abiotic and biotic stresses, such as resistance to low temperatures, adaptation to environmental change, and tolerance to fungal and viral pathogens (Suzuki et al., 2005; Yang et al., 2022). Rutin has wide range of pharmacological activities and high antioxidant capacity. It is widely used in clinical medicine, e.g., for anticarcinogenic, antiviral and anti-inflammation treatment and for the protection of nerves and the cardiovascular system (Glasser et al., 2002; Jimenez-Aliaga et al., 2011; Luca et al., 2020). Rutin is the diglycoside form of quercetin, which is the main flavonol in vegetables and fruits. It is the most popular dietary flavonoid compound in the human diet (Hosseinzadeh and Nassiri-Asl, 2014; Tobar-Delgado et al., 2023). Although the biosynthetic pathway of flavonol is clear, the enzyme in the final two glycosylation steps of quercetin conversion into rutin remains elusive (Figure S1A) (Vogt, 2010; Zhang et al., 2017; Shen et al., 2022). UGT encodes the final enzyme in the rutin biosynthetic pathway and determines the pharmaceutical functions of rutin (Jing et al., 2016).Glycosylation is one of the most important modifications. Glycosylation catalyzed by glycosyltransferase is an important modification process in plants and is one of the final steps involved in secondary metabolite synthesis (Caputi et al., 2012). Glycosyltransferase catalyzes the transfer of glycosyl from activated donor molecules onto various small molecule receptors, leading to the production of di-saccharides, poly-glycosides, and diversified glycosides of noncarbohydrate moieties (Caputi et al., 2012; Wilson and Tian, 2019). The plant secondary product glycosyltransferase (PSPG) box is the binding site of the sugar donor. It consists of a conserved sequence of 44 amino acid residues in the UGT of plants near the C-terminus (Gachon et al., 2005; Offen et al., 2006). The UDP-glycosyltransferase superfamily is a large family that possesses redundant functions and complex structures and presents distinct but overlapping substrate specificities (Mackenzie et al., 1997). Members of the UGT superfamily are named using the acknowledged convention (Figure S1B). Numbers from 1 to 50 are utilized for animals, 51 to 70 for yeasts and amoebae, 71 to 100 for plants and 101 to 200 for bacteria (Figure S1B) (Ross et al., 2001). With the development of molecular biology, an increasing number of UGT genes have been cloned and characterized, which are enrich of rutin biosynthesis. Rutin is formed by the two-step glycosylation of quercetin. For example, FaGT6 was proven to be responsible for the first step, which catalyzed quercetin to 3-O-glucosides (Griesser et al., 2008). UGT genes with similar functions in the first step of glycosylation have been reported in both soybean (Kovinich et al., 2010) and grape (Mizohata et al., 2013). GmUGT79A6 was identified as rhamnose-glycosyltransferase involved in the second step of rutin glycosylation (Rojas et al., 2014). In addition, FeUGT79A8 (Koja et al., 2018) and FtUGT73BE5 (Yin et al., 2020) were reported to promote rutin accumulation in buckwheat. Many UGT genes involved in key secondary metabolite biosynthesis have been successfully identified and investigated in different plant species, including Arabidopsis (Paquette et al., 2003), pomegranate (Wilson et al., 2019), buckwheat (Yin et al., 2020), soybean (Yin et al., 2017), and tea (Cui et al., 2016; Dai et al., 2022). However, the genes of rutin biosynthesis are still unknown in eggplant.
In this study, the pangenome of eggplant was used to identify the UGT family and obtain potential UGT genes that might be related to rutin biosynthesis. A total of 195 UGT candidate genes were identified in eggplant, and their gene structure and evolutionary relationship were analyzed. Five UGT candidate genes were identified with rutin biosynthesis via expression pattern and correlation analysis. Furthermore, SmUGT89B2 was proven to participate in rutin biosynthesis in eggplant by using VIGS and transient overexpression assays.
Material and methods
Identification of UGTs in the eggplant genome and their chromosomal locations
UGT families were retrieved from the eggplant genome V4.0 downloaded from the Solanaceae genomics database (https://solgenomics.net/) by using the following two methods: 44 amino acid sequences of PSPG-box were used as query sequences to blast the eggplant, and the Hidden Markov Model (HMM) profile of PF00201. The members of UGT families needed to meet the requirement of the two methods. The NCBI Conserved Domain Search Service (Batch CD-Search) and SMART were used to filter redundant sequences. The full-length amino acid sequences, molecular weight (MW), theoretical isoelectric point (PI) and instability index of the UGT proteins were predicted by the ExPASy server. Based on the gff3 file of eggplant, the distribution of UGT genes on chromosomes was retrieved and visualized by using TBtools (Chen et al., 2020).
Sequence alignment and phylogenetic analysis
The full-length amino acid sequences of all SmUGTs were aligned by Clustal X2 software with default values used for all parameters. The phylogenetic evolutionary tree was constructed using the maximum likelihood method through MEGA X software with 2000 bootstrap replications (Kumar et al., 2018). The 122 UGT protein sequences of Arabidopsis used in the tree were downloaded from the site (http://www.p450.kvl.dk/index).
Gene structure and conserved motif analysis
Based on the relationship of the coding sequence and its corresponding genomic DNA sequence from the gff3 file, the gene structure of SmUGTs was visualized by using TBtools. Conserved motifs or domains were searched by MEME (https://meme-suite.org/meme/tools/meme)(Bailey et al., 2015), with parameters as follows: number of motif predictions, 20; minimum motif width, 6; maximum motif width, 50 (Bailey et al., 2009), and then visualized by using TBtools.
RNA-seq data analysis
The transcriptome data at different developmental stages of eggplant fruit were obtained from previous eggplant genome research (Barchi et al., 2019). A heatmap reflecting the expression patterns of SmUGTs at different developmental stages of eggplant fruit was constructed by TBtools.
Plant materials
The E801 inbred line used in the study was provided by our lab. Eggplant seeds were sown in hole trays when the seedlings grew to the three-leaf stage and transplanted to the experimental farm (South China Agricultural University, Qilin). The different tissues of eggplant, including the roots, stems, leaves, flowers and commercial fruits, were collected, immediately frozen in liquid nitrogen and then stored at -80°C for RNA extraction and determination of rutin content. The different developmental stages of eggplant fruits at 5 DPA, 12 DPA, 15 DPA, 22 DPA and 30 DPA were collected, and immediately frozen in liquid nitrogen and then stored at -80°C for RNA extraction and determination of rutin content (Figure S2).
Total RNA extraction and quantitative real-time PCR
Total RNA was extracted from eggplant tissues by an Eastep Super Total RNA Extraction Kit (Promega, WI, USA) according to Liu et al. (Liu et al., 2023). First-strand cDNA was synthesized by using a cDNA Kit PLUS (B0003) (EZBioscience, Roseville, MN, USA) according to the manufacturer’s instructions. All primer sequences are listed in Table S2, including reference genes. The relative expression of UGT genes was calculated as described by Song et al. (Song et al., 2020). The qPCRs using different tissues were performed with biological triplicates.
Determination of rutin content
The rutin content was detected according to Dong et al. (Dong et al., 2020). Fresh tissues (0.1 g) were ground into a fine powder with liquid nitrogen, extracted with 2 mL ethanol (60%) and centrifuged for 10 min at 12000 rpm. The supernatant was transferred to a new 2 mL centrifuge tube and placed in a metal bath at 80 °C for 2 h. After heating, 0.4 mL of supernatant was mixed with 0.4 mL of AlCl3 (0.1 mol/L) and 0.6 mL of CH3COOK (1 mol/L) solution and then added to 0.6 mL of ethanol (60%). The mixture was shaken and allowed to stand for 30 minutes. The absorbance of the solution was measured using a microplate reader at 420 nm. Determination of rutin content was performed in triplicate.
Subcellular localization analysis
To confirm the Subcellular localization of SmUGT89B2, the coding DNA sequence (CDS) of SmUGT89B2 was cloned from eggplant. Its coding sequence without a terminator codon was cloned and inserted into a vector fused with EGFP carrying the CaMV 35S promoter to generate the 35S: SmUGT89B2-EGFP construct by using one-step cloning (C112, Vazyme, China) according to the manufacturer’s instructions. The recombinant plasmid was transformed into competent cells of Agrobacterium strain GV3101. Agrobacterium tumefaciens GV3101 containing the corresponding constructs was incubated with YEP medium at 28°C. The bacterial cells were collected and modulated 0.4-0.6 at 600 nm using MS medium (10 mM 2-(4-morpholino)-ethane sulfonic acid, 10 mM MgCl2, and 200 μM acetosyringone, pH = 5.6) and then placed in darkness at 28°C for 2 h. Agrobacterium tumefaciens GV3101 containing the 35S:SmUGT89B2-EGFP construct and empty vector were mixed with the NLS-DsRed construct in equal proportions (1:1). The mixed solution was infiltrated into the abaxial side of an Nicotiana benthamiana leaf. Three days later, the GFP signal of leaves that were inoculated with Agrobacterium tumefaciens was observed by using fluorescence microscopy (Leica, Germany) (Song et al., 2023). The primers used are listed in Table S2.
Virus-induced gene silencing assays
The full-length CDS of SmUGT89B2 was predicted to be a hypothetically optimal fragment (300 bp) by using the Solanaceae database (http://vigs.solgenomics.net/). The 300 bp fragment was amplified and ligated to the pTRV2 vector. pTRV2-SmUGT89B2, pTRV1 and pTRV2 were transformed into competent cells of Agrobacterium strain GV3101. Agrobacterium tumefaciens GV3101 containing pTRV2-SmUGT89B2 and pTRV1 were mixed in equal proportions, and the bacterial solution was infiltrated into the abaxial side of 30-day-old seedlings of eggplant with a 1 mL syringe. The eggplant plants were grown in a growth chamber under growth conditions as follows: 60–70% humidity, 22-25°C, 16 h/8 h (light/dark cycle). The primers used are listed in Table S2.
Transient overexpression analysis
Transient overexpression analysis was performed according to a previous method (Wang et al., 2022). The full-length coding sequence of SmUGT89B2 was cloned and inserted into the PC1300 vector. The recombinant vector was transformed into competent cells of Agrobacterium strain GV3101. The bacterial solution containing the corresponding constructs was infiltrated into the abaxial side of eggplant (E801) leaves at the five-leaf stage, and the plants were grown in a growth chamber. After 5 days, the leaves of eggplant were collected, frozen in liquid nitrogen and stored at −80°C for RNA extraction and determination of rutin content. The primers used are listed in Table S2.
Results
Identification and chromosomal localization of eggplant UGT genes
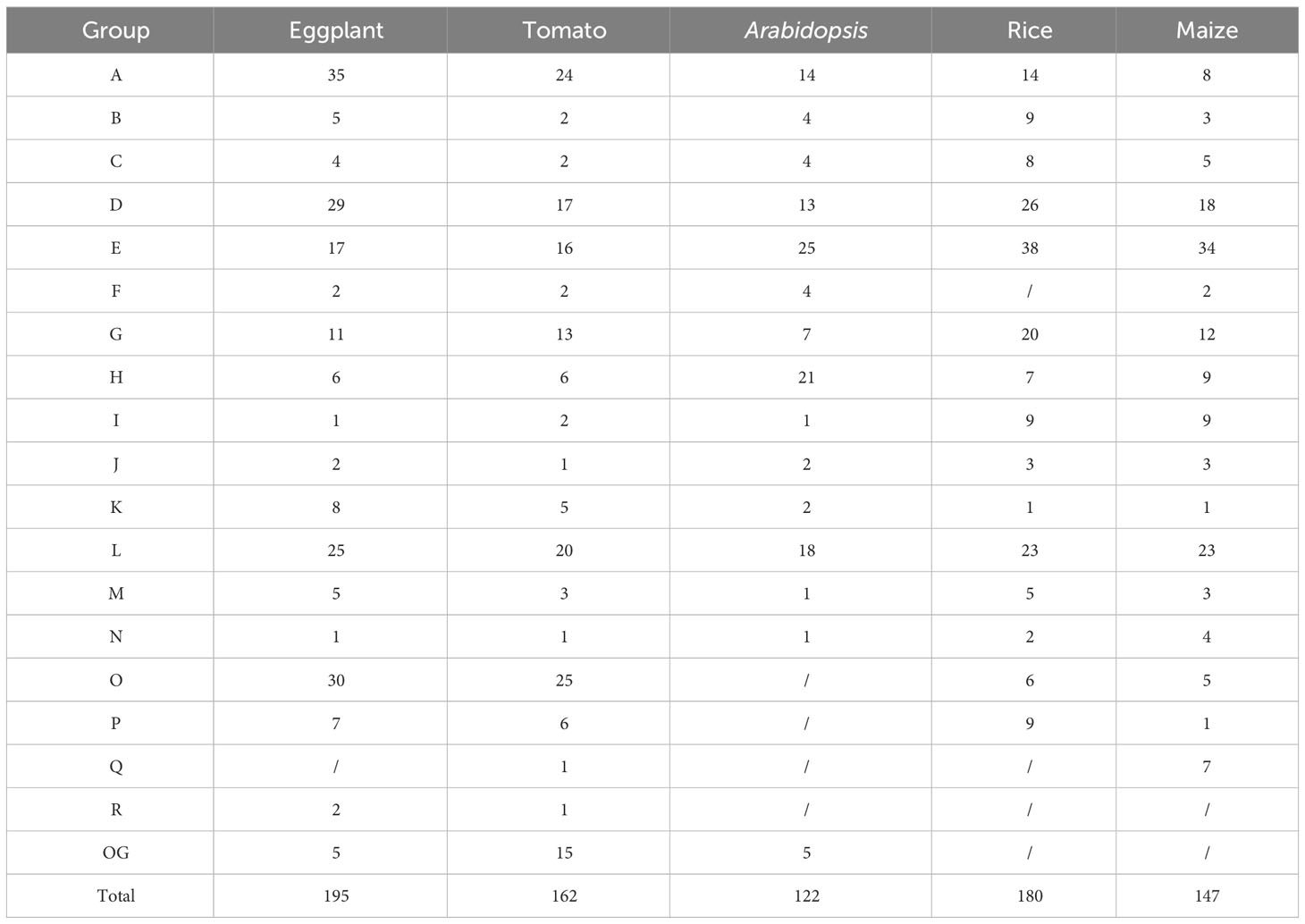
Based on the V4 version of the eggplant genome, the HMM model was used for searching UGT genes. After redundancy was removed by SMART and CDD-Search, a total of 195 UGT genes were identified. All candidate UGT genes contained a conserved UDPGT domain, which encoded amino acid sequences ranging from 154 aa to 1197 aa in length. The molecular weight (MW) of these proteins ranged from 17.02 kDa to 134.07 kDa, and the scope of the theoretical pI was 4.81 to 9.23. Most of the proteins were stable, and only fifty-seven (29% of the total) of SmUGT proteins were unstable (Table S1). Analysis of their chromosome location distribution showed that all candidate UGT genes were mapped on chromosomes 1-12, except for SMEL4_00g012200, which was not assembled to any chromosomes due to sequencing and assembly technology limitations. Among these chromosomes, chromosome 11 contained the largest number of SmUGT members (36). Chromosome 1, chromosome 5, and chromosome 10 contained 17, 23, and 26 members, respectively. Chromosome 12 (7) included the fewest number of SmUGT members (7) (Figure 1).
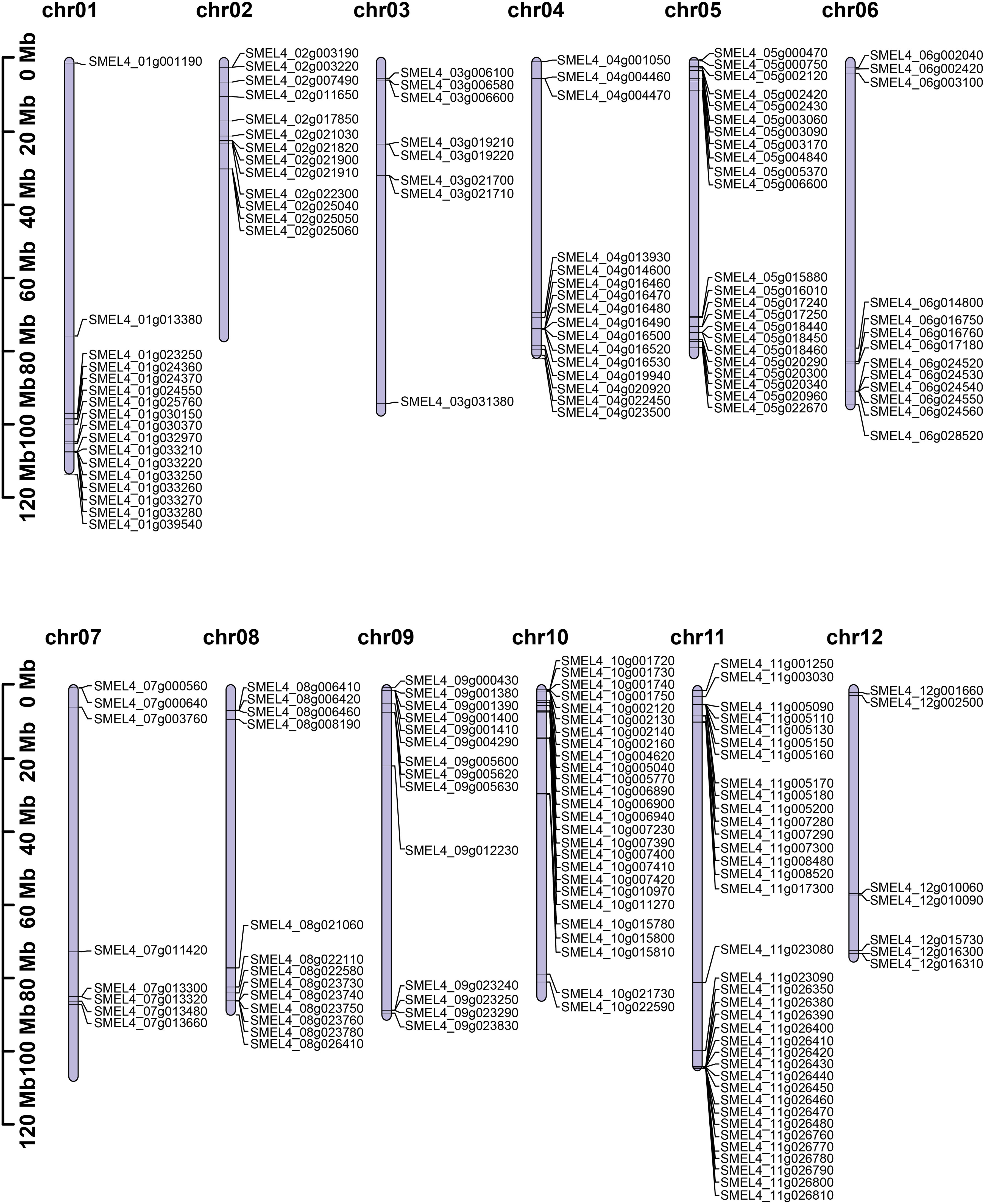
Figure 1 The distribution of chromosome locations. The cylinder represents the chromosomes, and the numbers above the column represent the number of chromosomes.
Phylogenetic analysis of eggplant UGT family proteins
To clarify the phylogenetic relationship of the UGT family in eggplant, a phylogenetic tree was constructed by the maximum likelihood method with all of the identified SmUGT protein sequences and those in other plants, including 122 Arabidopsis UGTs, two UGT genes of Group P in rice (Li et al., 2014) and four UGT genes of Groups Q and O in maize (Li et al., 2014; Huang et al., 2015). These UGT proteins from rice and maize were conducive to the classification of the SmUGT phylogenetic group. According to the phylogenetic tree, SmUGTS were clustered into 17 phylogenetic groups (A-P, R) and an outgroup (Figure 2A). As reported in Arabidopsis, AtUGT84B1 from Group L was involved in glycosylation of IAA (Mateo-Bonmati et al., 2021). Both AtUGT79B1 and AtUGT91A1 from Group A were proven to mediate anthocyanin modification (Ono et al., 2010). AtUGT89C1, AtUGT73C6 and AtUGT78D1, which belonged to Groups B, D and E, respectively, were related to glycosylation of flavonol (Jones et al., 2003; Yonekura-Sakakibara et al., 2007). It was likely that members of Groups A, B, D, E and L were related to phytohormone regulation and secondary metabolite biosynthesis.
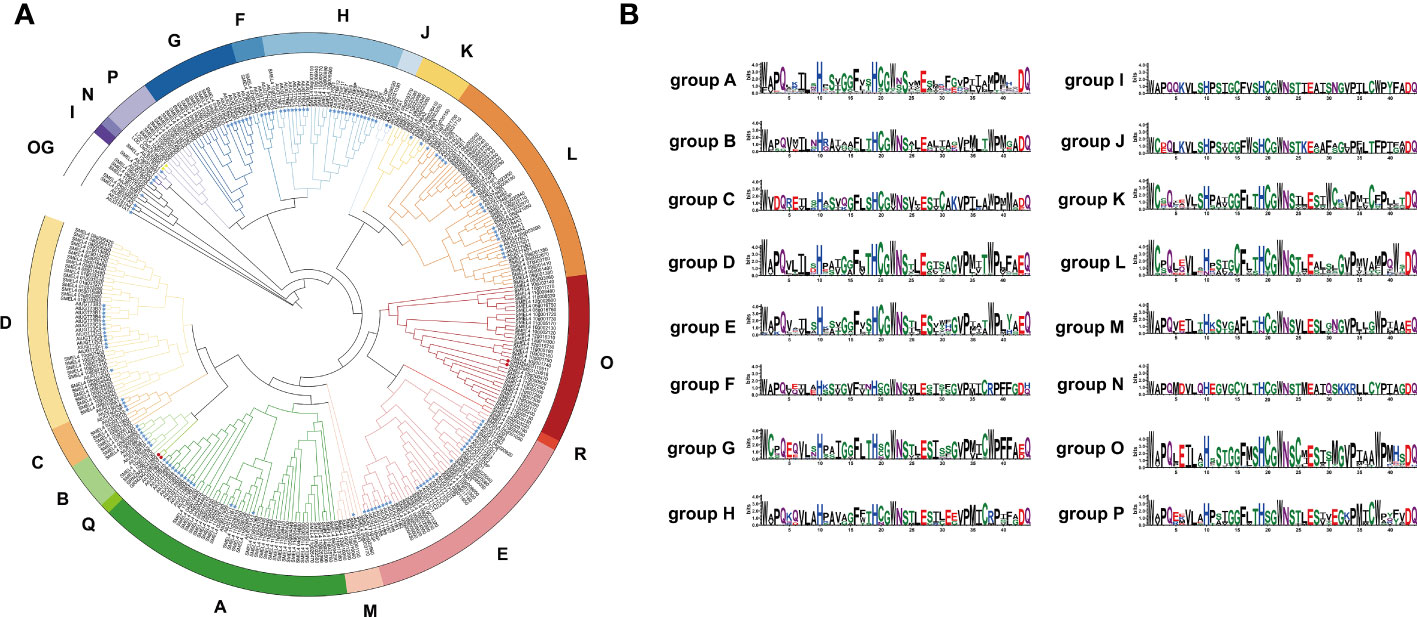
Figure 2 (A) Phylogenetic tree of the UGT family members. The tree consisted of all the identified UGT genes from eggplant, two UGT genes from rice (Li et al., 2014), four UGT genes from maize (Li et al., 2014; Huang et al., 2015) and all Arabidopsis UGT genes (Wilson and Tian, 2019). The tree was constructed using the maximum likelihood method. Different colored branches represent different groups. The blue star represents Arabidopsis. The yellow triangle represents rice, and the red circle represents maize. (B) Logos of the PSPG motif of phylogenetic Groups A - P generated by WebLogo (Crooks et al., 2004). The height of the amino acid indicates the frequency observed in multiple alignment.
To further understand the distribution of the UGTs across the phylogenetic range covered in different plant species, the number of UGT proteins distributed in phylogenetic groups is summarized in Table 1. Notably, some significant differences were observed in these phylogenetic groups. Compared with tomato, the members of eggplant UGTs were absent in Group Q, whereas the members of Solanaceae UGTs were present in Groups O, P and R compared with Arabidopsis, especially the members of Group R only found in Solanaceae. Group A contained the largest numbers of SmUGT members (35), followed by Groups O (30) and D (29). Groups F, I, J and N included a small number of UGT members in these plants. Compared with that of other species, the members of Group O in Solanaceae species had significant expansion, as well as tomato. The results indicated that Group O might play a vital role in Solanaceae plants.
The PSPG-box was located in the UDPGT conserved domain. The PSPG box was distinct in different phylogenetic groups, in which highly conserved residues were observed at positions 1 (W), 4 (Q), 8 (L), 10 (H), 19-24 (HC/SGWNS), 27 (E), and 44 (Q) (Figure 2B). Some amino acids could interact directly with UDP-sugar donors to form hydrogen bonds, which were related to the evolution and function of enzymes. The amino acid residue Q (44) has been reported to determine the sugar donor specificity of UGTs, which affects the catalytic efficiency of glucosyl transfer activity (Ono et al., 2010; Caputi et al., 2012).
Gene structure and conserved motif analysis of SmUGTs
To investigate the structural features of SmUGT proteins, conserved amino acid sequences and gene structure were analyzed. Except for the outgroup, there were 89 members without introns (46%). The number of intron regions ranged from 1 to 7 in the rest of the members (Figure 3B). SMEL4_05g000470 (from Group O) was one of the most complex genes and had 7 introns. All members of Groups G, H, I, J, K, N, and P had introns. All SmUGT amino acid sequences were submitted to MEME, and a total of 20 conserved motifs were obtained (Table S3). These motifs possessed 11-50 residues and were named motif 1-motif 20. Motif 1, located in the PSPG box, existed in all SmUGT genes and served as a specific motif of the UGT family. In general, the same group has a similar motif distribution, whereas different groups possess a specific distribution of motifs. As shown in Figure 3A, motif 14 was mainly distributed in Groups O and A, while motif 16 and motif 18 were primarily present in Groups D and M. Motif 13 and motif 19 were found only in Group O.
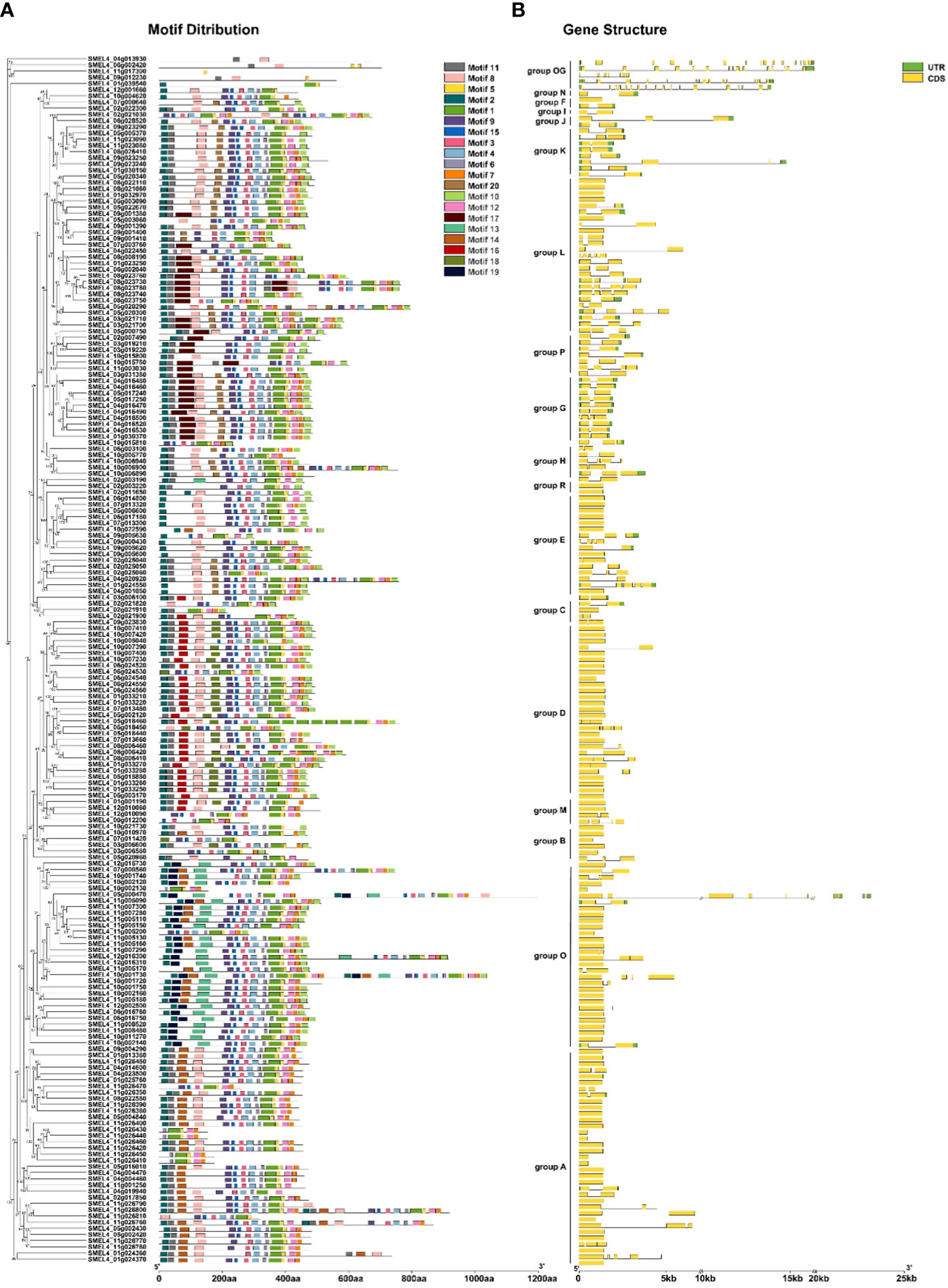
Figure 3 Conserved motifs and gene structure of SmUGTs. The phylogenetic tree was constructed by all UGT gene sequences using the maximum likelihood method, and the conserved motif (A) and gene structure (B) were placed orderly on the right side of the phylogenetic tree. Different colored boxes represent different motifs, and the box length represents the motif length in the motif distribution. The yellow box indicates exons; the green box indicates UTRs; and the black line indicates introns.
Expression analysis of SmUGTs in different developmental stages of eggplant fruit
Expression patterns can predict the biological functions of genes to a certain extent. To investigate the underlying functions of candidate UGT genes during rutin biosynthesis processes, transcriptome data of eggplant at different developmental stages from a public database were downloaded (Barchi et al., 2019). The expression patterns of candidate SmUGTs and rutin biosynthesis genes in different developmental stages of eggplant fruit were analyzed. The rutin biosynthesis genes were highly expressed in stage 3 (Figure 4A). A total of 23 SmUGTs (12%) were expressed at a level that could not be detected. Based on the similar expression trend, all SmUGTs in different developmental fruit stages were hierarchically clustered and divided into 10 clusters (Figure 4B). The expression patterns of Clusters 7, 8, 9 and 10 were similar to the expression patterns of rutin biosynthesis genes, and their expression levels gradually increased with fruit development. Therefore, the members of Clusters 7–10 served as novel candidates for the regulation of rutin biosynthesis.
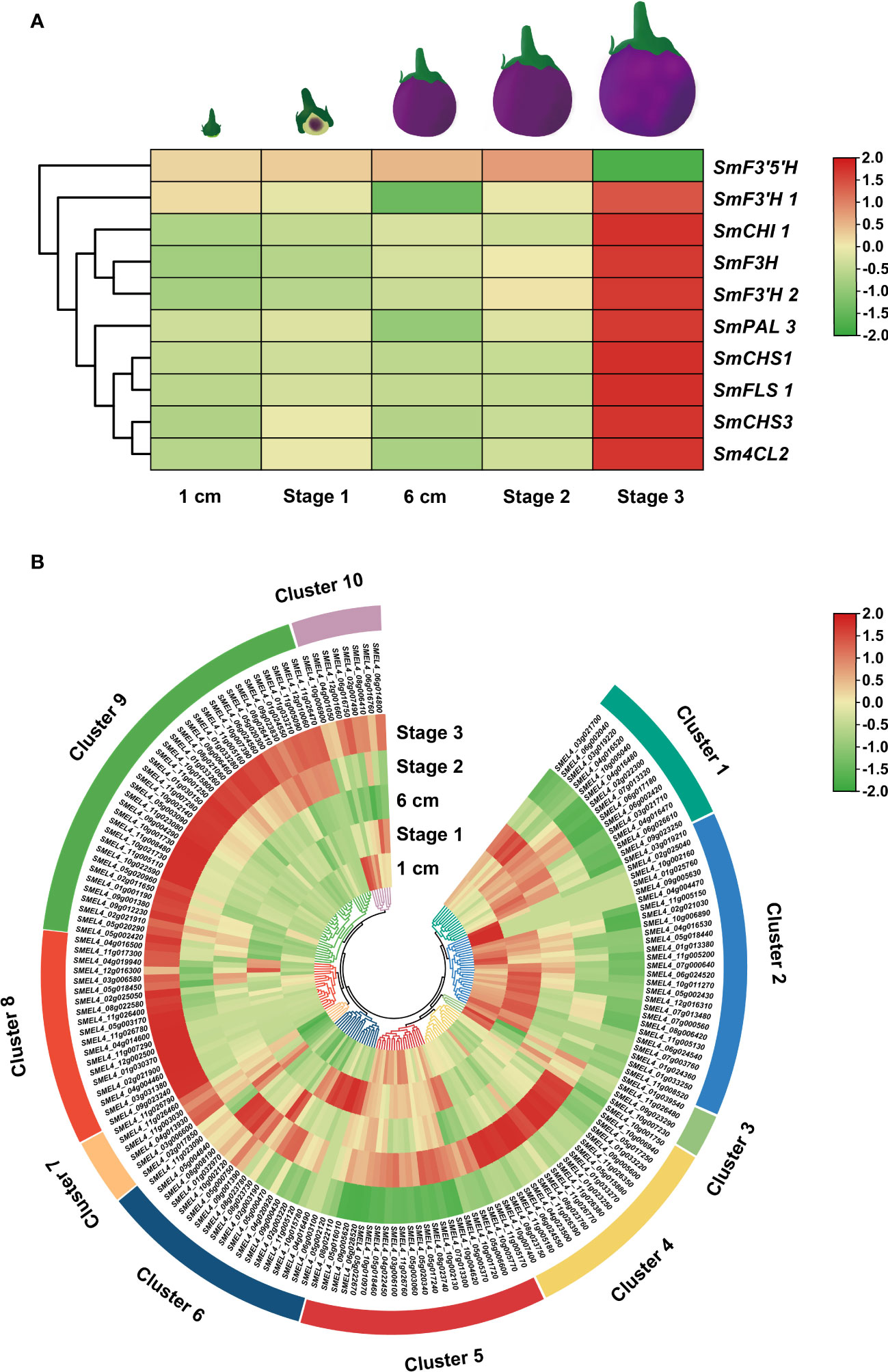
Figure 4 Expression patterns of rutin biosynthesis (A) and UGT (B) genes. Fruit 1 cm represents 3-4 days after flowering (DAF); fruit stage 1 represents 8-14 DAF; fruit 6 cm represents 18-21 DAF; fruit stage 2 represents approximately 38 DAF; fruit stage 3 represents 55-60 DAF.
Expression patterns of SmUGTs in different tissues and different developmental stages of fruits and leaves in eggplant
To further identify SmUGT genes involved in rutin biosynthesis in eggplant, five SmUGT genes that belonged to Groups A-F and L with a higher expression level were selected from candidate Clusters 7 to 10 to perform qRT−PCR assays. These five genes were renamed by the UGT Nomenclature Committee, called SmUGT71BA3, SmUGT72B88, SmUGT73A69, SmUGT75L62 and SmUGT89B2, respectively (Table S2). Rutin accumulated in all tissues of eggplant, and the highest content was found in leaves, followed by flowers and pericarp. Meanwhile, the highest content was found at 5 DPA, and with the development of fruit, the rutin content gradually decreased (Figure 5A). The expression of SmUGT72B88, SmUGT73A69 and SmSmUGT89B2 was the highest in leaves, followed by flowers and fruits, which was consistent with the accumulation of rutin in different tissues of eggplant (Figure 5B). However, SmUGT71BA3 was highly expressed in roots and fruits compared with that in other tissues. The expression of SmUGT75L62 was highest in pericarp, whereas it was transcribed at a low level in stem that was not detectable. The expression pattern of SmUGT71BA3 was inversely represented in pericarp and pulp, showing a significant decrease in pericarp and a marked increase in pulp from 5 DPA to 30 DPA. In the other four genes, including SmUGT72B88, SmUGT73A69, SmUGT75L62 and SmUGT89B2, their own expression levels were similar in the pericarp and pulp. The expression levels of SmUGT73A69 and SmUGT75L62 exhibited a growing tendency with the process of fruit development, while SmUGT72B88 and SmUGT89B2 were observed to have higher expression at 5 DPA and then showed a decrease at 12 DPA. Their expression levels were not significantly different from 12 DPA to 30 DPA, except for that of SmUGT89B2, which increased at 30 DPA (Figure 5B).
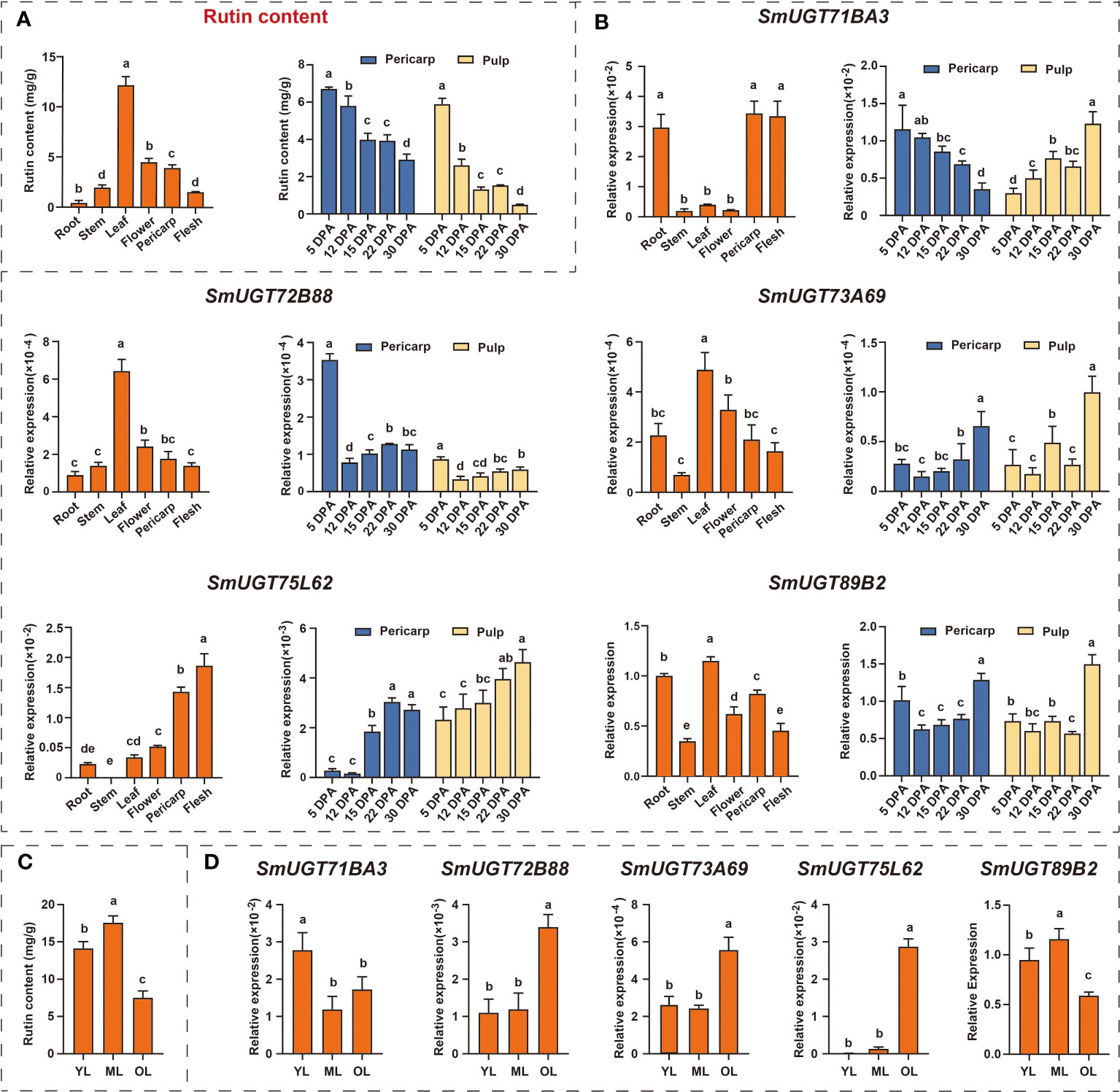
Figure 5 The expression patterns of five SmUGT genes in different tissues and different developmental stages of leaves and fruit in eggplant (E801). (A) Rutin content in different eggplant tissues (roots, stems, leaves, flowers and pericarp and pulp). (B) Expression pattern of five SmUGT genes in different eggplant tissues (roots, stems, leaves, flowers, pericarp and flesh). Data are the mean ± SE (n = 3). Different lowercase letters in the figures represent significantly different values (P < 0.05, Tukey’s test). (C) The rutin content at different developmental stages of leaves in E801. (D) Expression of five SmUGT genes at different developmental stages of leaves. Different lowercase letters in the figures represent significantly different values (P < 0.05, Tukey’s test).
To further explore the expression patterns of these five genes, leaves at different developmental stages were used for verification. During the course of leaf senescence, the rutin content gradually increased from young leaves to mature leaves and then decreased, with a peak at mature leaves (Figure 5C). As shown in Figure 5D, the transcription levels of SmUGT72B88, SmUGT73A69 and SmUGT75L62 increased with the process of leaf senescence. The expression patterns of SmUGT89B2 maintained good agreement with the accumulation of rutin in different developing leaves.
Sequence analysis and subcellular location of SmUGT89B2
To determine whether SmUGT89B2 is involved in rutin biosynthesis in eggplant, the amino acid sequence of SmUGT89B2 was analyzed. The full-length CDS of SmUGT89B2 was cloned from the E801 inbred line. Its protein encoded 483 amino acids. The molecular weight (MW) of SmUGT89B2 was 53.83 kDa, and theoretical PI was 6.44. It is an unstable protein, and the instability index was 42.04. The aliphatic index (95.51) was predicted that SmUGT89B2 was a hydrophilic protein (Table S1). Several UGT proteins that have been reported to be involved in flavonol glycosylation were obtained (Carbonell-Bejerano et al., 2014; Yin et al., 2017; Koja et al., 2018; Yin et al., 2020), and some UGT proteins from other Solanaceae species that shared the highest sequence identity with SmUGT89B2 were retrieved from public databases (NCBI, https://www.ncbi.nlm.nih.gov/). All of them were used to construct an unrooted phylogenetic tree based on their multiple sequence alignment. As shown in the phylogenetic tree, the UGT genes involved in flavonol glycosylation in the different species had a distant evolutionary relationship with SmUGT89B2. The conserved PSPG-box motif existed in all UGT amino acid sequences, in which some amino acids were highly conserved (Figure 6A) (Caputi et al., 2012). To clarify the subcellular localization of SmUGT89B2, its sequence was fused in frame with the EGFP gene to generate the 35S:SmUGT89B2-EGFP construct (Figure 6B). The DsRed gene served as the nuclear marker by fusion with a nuclear localization signal (NLS) (Sun et al., 2020). The coexpression of either 35S: EGFP or 35S:SmUGT89B2-EGFP with DsRed in epidermal cells of Nicotiana benthamiana leaves indicated that SmUGT89B2 was localized in the nucleus and cell membrane, as well as in the cytoplasm (Figure 6B). It was likely that SmUGT89B2 functioned in the whole cell.
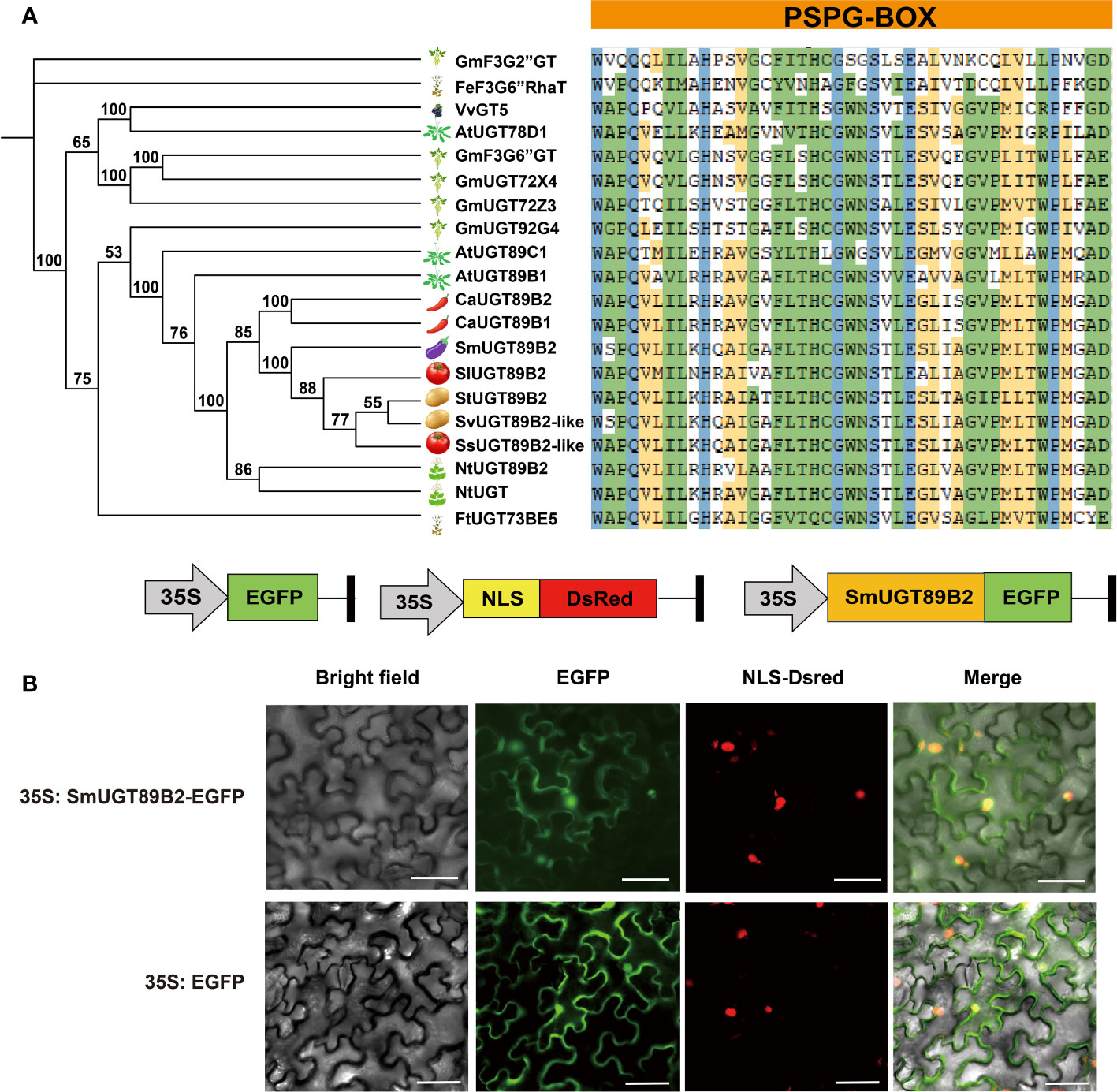
Figure 6 Sequence analysis and subcellular localization of SmUGT89B2. (A) Multiple alignment of PSPG-box motif (right) and a phylogenetic tree (left) of SmUGT89B2 with homologous genes from other plant species. The sequences of other plant species were obtained from NCBI and soybean database. A phylogenetic tree was built via MEGA X using the maximum likelihood method with 2000 bootstrap replicates. (B) Subcellular localization of SmUGT89B2. EGFP represents enhanced GFP. NLS-DsRed served as a nuclear marker, and fluorescence was visualized using an inverted fluorescence microscope. Scale bar = 50 μm.
SmUGT89B can positively promote rutin accumulation in eggplant
To determine whether SmUGT89B2 was associated with rutin accumulation in eggplant, the transcriptional level of SmUGT89B2 was silenced in the E801 inbred line through virus-induced gene silencing (VIGS). Three silenced eggplant lines that possessed the highest silencing efficiency were selected to determine SmUGT89B2 expression and the rutin content. As shown in Figure 7B, the expression level of SmUGT89B2 was significantly decreased in silenced lines, reducing by approximately 60% of that in the control plants. Simultaneously, the rutin content in the silenced leaves decreased by approximately 40% of that in the control leaves. To further confirm whether SmUGT89B2 plays a role in rutin biosynthesis in eggplant, a transiently overexpressed assay was performed in eggplant seedlings at the five-leaf stage. The SmUGT89B2 transcription levels increased 8.8-28-fold in the overexpressed leaf compared with those in the control, consistent with the 1.2-1.7-fold increase in rutin content in overexpressed leaf (Figure 7D). These results suggested that SmUGT89B2 played an important role in rutin biosynthesis in eggplant.
Discussion
Glycosyltransferase is a supergene family, and plant UGTs are classified into the first family (GT1) of the glycosyltransferase (GT) family. Glycosylated metabolites generated by UDP-dependent glycosyltransferases play important roles in plant interactions with the environment as well as human and animal nutrition (Wilson and Tian, 2019). The UGT family has been identified in various plants, including Arabidopsis (123) (Li et al., 2001), cotton (274) (Xiao et al., 2019), maize (147) (Li et al., 2014), peach (168) (Wu et al., 2017), buckwheat (106) (Yin et al., 2020), soybean (212) (Yin et al., 2017), tea (178) (Cui et al., 2016), pomelo (145) (Wu et al., 2020), and Brassica rapa (147) (Yu et al., 2017). The numbers of UGT genes present significant differences between various plants. In this study, a total of 195 UGT genes identified from the eggplant genome were divided into 17 groups by constructing a phylogenetic tree with other plant UGTs, containing 14 highly conserved phylogenetic groups (A-N) and three new phylogenetic groups (O, P, and R) (Figure 2A). Group O contained 30 UGT members in eggplant, while it contained approximately the same amount of UGT members in tomato (25) (Wilson and Tian, 2019) and tabacoo (40) (Yang et al., 2023). In contrast, the number of UGT members was dramatically reduced in rice (6), maize (5) and other dicotyledon plants (Wilson and Tian, 2019). Thus, the members of Group O were significantly expanded in Solanaceae plants and seem to be unique to Solanaceae species. UGT709G2 from Group P shows a substrate preference for UDP-Glc and catalyzes myricetin 3-O-rhamnoside into myricetin 3-O-glucosyl rhamnoside in montbretia (Irmisch et al., 2018). The members of Group R were recently discovered and first identified in buckwheat. FeUGT708C1 from Group R could recognize and catalyze the C-glycosylation of trihydroxy acetophenone-like compounds (Nagatomo et al., 2014). It appears that the members of new phylogenetic groups are related to the glycosylation of secondary compounds in different plants. Group R was absent in all Brassicales but present in Solanaceae species. which was probably responsible for the distant relationship between them. According to the phylogenetic evolutionary tree, five SmUGT genes with two Arabidopsis genes (AtUGT80A2 and AtUGT80B1) were classified into a separate out group (OG) (Figure 2A), in which the PSPG-box was less conserved. Members of the OG group have been proposed to modify sterols and lipids in other plants (Paquette et al., 2003). It is likely that the five genes may be involved in the formation of sterols and lipids in eggplant.
The members defined in the same phylogenetic subgroup generally possess similar functions (Soler et al., 2015; Song et al., 2020). Some UGT genes belonging to Groups B, D, E and L have been identified. AtUGT73C6 (Group D) can catalyze the transfer of glucose from UDP-glucose to kaempferol-3-O-rhamnoside and quercetin-3-O-rhamnoside (Jones et al., 2003). FaGT6, which belongs to the UGT71 family, catalyzes quercetin into 3-O-glucoside (Griesser et al., 2008). UGT75 family members are mostly flavonoid-5-O-glycosyltransferases, which are mainly involved in the synthesis of plant anthocyanins (Noguchi et al., 2009; He et al., 2015). Therefore, the members from Groups B, D, E and L might possess the underlying function of participating in flavonol biosynthesis.
Based on the sequence analysis of a multigene family, its gene expression compared with that in the expression profiles of known genes and complementation of the metabolite pattern in combination with reverse genetics are conducive to understanding the underlying function of a single member from the superfamily (Offen et al., 2006). The expression patterns of SmUGT members from Clusters 7 to 10 exhibited good agreement with the transcription levels of rutin biosynthetic genes in eggplant (Figure 4B). Thus, the members of Clusters 7 to 10 acted as novel candidates for rutin biosynthesis. SmUGT89B2 found in Cluster 7 was classified into Group B, as well as AtUGT89C1. AtUGT89C1 has been proposed to be involved in flavonol biosynthesis in Arabidopsis (Yonekura-Sakakibara et al., 2007). It seems that SmUGT89B2 may be involved in flavonol biosynthesis in eggplant. The other four UGT genes observed in RNA-Seq data with high RPKM values, including SmUGT71BA3 and SmUGT72B88 (Group E), SmUGT73A69 from Group D and SmUGT75L62 from Group L, which were found in Cluster 9, were selected for qRT−PCR assays, as well as SmUGT89B2 (Figure 4B).
The expression pattern of these five genes was unlike the rutin accumulation pattern at different developmental stages of fruit in eggplant (Figure 5A). The rutin content was the highest at the young fruit stage. It seems that plants can activate self-protection processes via the production of secondary metabolites against undesirable circumstances, such as UV irradiation, cold stress, and desiccation stress (Suzuki et al., 2005; Yang et al., 2012). This defense reaction was previously reported in anthocyanin accumulation (Naing and Kim, 2021). A large number of secondary metabolites are needed to protect plants, especially in tender tissues. The expression of SmUGT89B2 was similar to the accumulation pattern of rutin in eggplant (Figure 5). Therefore, SmUGT89B2 served as a novel candidate gene for rutin biosynthesis in this study. The subcellular localization results showed that SmUGT89B2 was localized in the nucleus and cell membrane, as well as in the cytoplasm (Figure 6B). Most UGT genes have multiple subcellular localizations to control where specific pathway products accumulate (Yi et al., 2020; Daryanavard et al., 2023). Transient overexpression and silencing expression of SmUGT89B2 in eggplant indicated that SmUGT89B2 could affect rutin accumulation (Figure 7). It was likely that SmUGT89B2 encoded the final enzyme in the rutin biosynthetic pathway. Although the functions of UGT89B2 involved in rutin biosynthesis in eggplants in vivo have been identified in this study, the enzyme activity and biochemical characteristics of UGT89B2 should be further identified. Which step of SmUGT89B2 involved in two glycosylation steps of quercetin transform to rutin also need to be verified by catalytic experiments. But the exploration of this catalytic experiment is certainly the most difficult because UGT glycosyltransferase possesses substrate diversity. In the future, enzymatic tests of the SmUGT89B2 will be the key works for our studies.
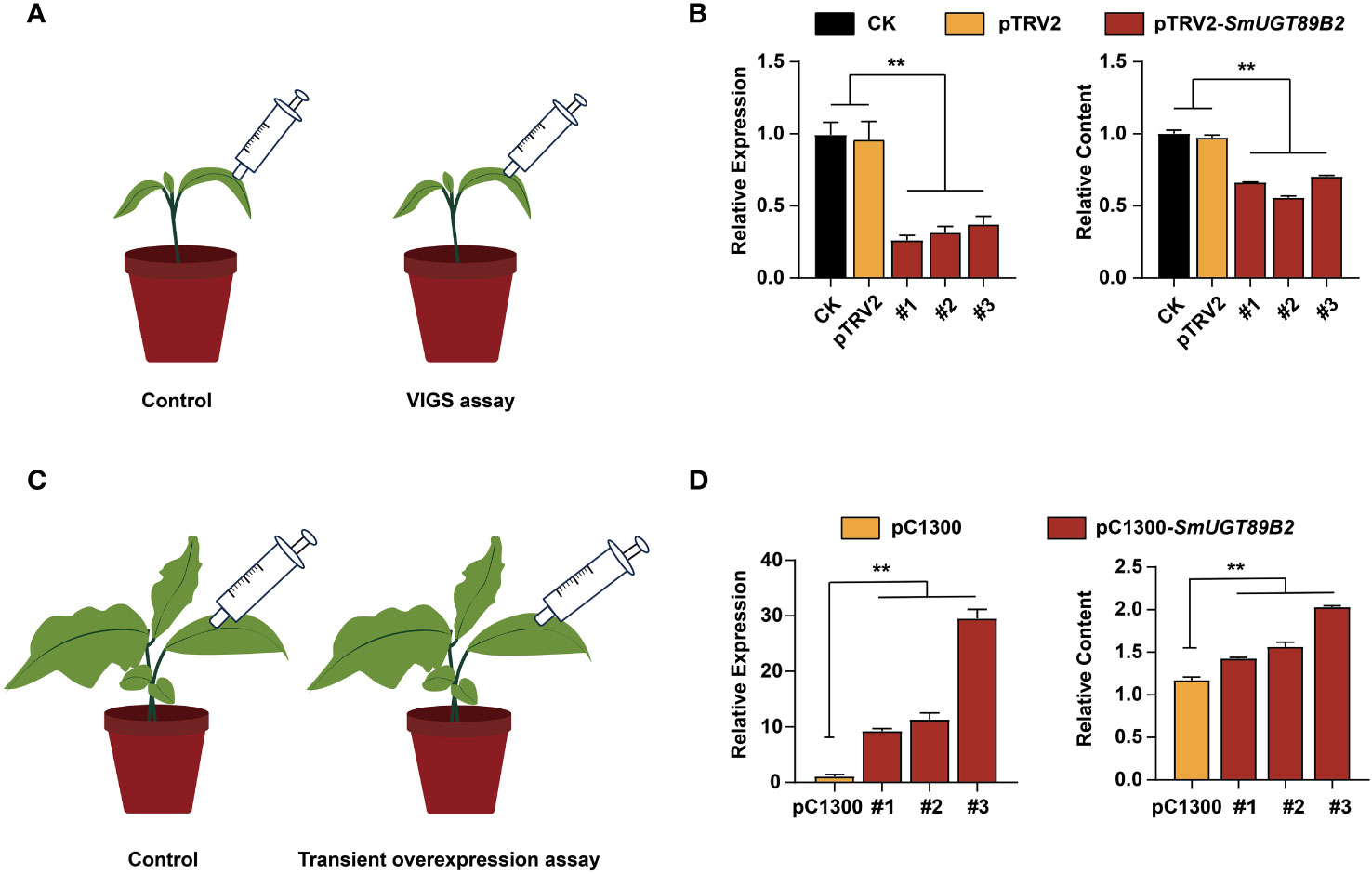
Figure 7 Silencing and transient overexpression of SmUGT89B2 in eggplant. (A) Schematic diagram of the SmUGT89B VIGS assay. (B) The expression of SmUGT89B and relative rutin content in leaves of SmUGT89B-silenced eggplants. CK were injected with water. pTRV2 was injected with Agrobacterium harboring an empty vector. Data are the mean ± SE (n = 3). (C) Schematic diagram of the transient overexpression assay for SmUGT89B2. (D) The expression of SmUGT89B and relative rutin content in leaves of SmUGT89B-overexpressed eggplants. pC1300 were the plants whose leaves were injected with Agrobacterium harboring an empty vector. mean ± SE (n = 3). Student’s t test was used to indicate significant differences compared to those in the empty vector control (**P < 0.01).
Conclusion
Overall, all UGT genes from the eggplant genome through the HMM model and PSPG-box motif were searched in the study. A total of 195 SmUGT genes were identified, which were divided into 17 phylogenetic subgroups according to the classification of Arabidopsis, maize and rice. The conserved motifs and gene structures of candidate UGT genes were analyzed. According to the expression profile analysis, Clusters 7-10 served as candidate clusters, in which the five genes SmUGT71BA3, SmUGT72B88, SmUGT73A69, SmUGT75L62 and SmUGT89B2 were selected as candidate genes. However, among these selected genes, SmUGT89B had a higher transcription level in eggplant, and its expression pattern was similar to the accumulation trend in eggplant by qRT−PCR assay. VIGS and transient overexpression demonstrated that SmUGT89B2 could positively promote rutin biosynthesis in eggplant.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author contributions
YG: Data curation, Formal analysis, Writing – original draft, Visualization, Writing – review & editing. BY: Writing – original draft. RL: Formal analysis, Visualization, Writing – original draft. BS: Data curation, Validation, Writing – original draft. YL: Data curation, Validation, Writing – original draft, Formal analysis. YZ: Writing – original draft. ZQ: Data curation, Project administration, Writing – original draft. SY: Project administration, Writing – original draft, Writing – review & editing. BC: Funding acquisition, Project administration, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study was supported by the Key R&D Projects in Guangdong Province(2022B0202080003), Seed Industry Revitalisation Project of Guangdong (2022-NPY-00-026), and Vegetable Industry System Innovation Team Project of Guangdong (2022KJ110).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1310080/full#supplementary-material
References
Bailey, T. L., Boden, M., Buske, F. A., Frith, M., Grant, C. E., Clementi, L., et al. (2009). Meme suite: tools for motif discovery and searching. Nucleic. Acids Res. 37, W202–W208. doi: 10.1093/nar/gkp335
Bailey, T. L., Johnson, J., Grant, C. E., Noble, W. S. (2015). The meme suite. Nucleic. Acids Res. 43 (W1), W39–W49. doi: 10.1093/nar/gkv416
Barchi, L., Pietrella, M., Venturini, L., Minio, A., Toppino, L., Acquadro, A., et al. (2019). A chromosome-anchored eggplant genome sequence reveals key events in Solanaceae evolution. Sci. Rep. 9 (1), 11769. doi: 10.1038/s41598-019-47985-w
Barchi, L., Rabanus-Wallace, M. T., Prohens, J., Toppino, L., Padmarasu, S., Portis, E., et al. (2021). Improved genome assembly and pan-genome provide key insights into eggplant domestication and breeding. Plant. J. 107 (2), 579–596. doi: 10.1111/tpj.15313
Caputi, L., Malnoy, M., Goremykin, V., Nikiforova, S., Martens, S. (2012). A genome-wide phylogenetic reconstruction of family 1 UDP-glycosyltransferases revealed the expansion of the family during the adaptation of plants to life on land. Plant J. 69 (6), 1030–1042. doi: 10.1111/j.1365-313X.2011.04853.x
Carbonell-Bejerano, P., Diago, M. P., Martinez-Abaigar, J., Martinez-Zapater, J. M., Tardaguila, J., Nunez-Olivera, E. (2014). Solar ultraviolet radiation is necessary to enhance grapevine fruit ripening transcriptional and phenolic responses. BMC Plant Biol. 14, 183. doi: 10.1186/1471-2229-14-183
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). Tbtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13 (8), 1194–1202. doi: 10.1016/j.molp.2020.06.009
Crooks, G. E., Hon, G., Chandonia, J. M., Brenner, S. E. (2004). Weblogo: a sequence logo generator. Genome Res. 14 (6), 1188–1190. doi: 10.1101/gr.849004
Cui, L., Yao, S., Dai, X., Yin, Q., Liu, Y., Jiang, X., et al. (2016). Identification of UDP-glycosyltransferases involved in the biosynthesis of astringent taste compounds in tea (Camellia sinensi). J. Exp. Bot. 67 (8), 2285–2297. doi: 10.1093/jxb/erw053
Dai, X., Shi, X., Yang, C., Zhao, X., Zhuang, J., Liu, Y., et al. (2022). Two UDP-glycosyltransferases catalyze the biosynthesis of bitter flavonoid 7-O-neohesperidoside through sequential glycosylation in tea plants. J. Agric. Food. Chem. 70 (7), 2354–2365. doi: 10.1021/acs.jafc.1c07342
Daryanavard, H., Postiglione, A. E., Muhlemann, J. K., Muday, G. K. (2023). Flavonols modulate plant development, signaling, and stress responses. Curr. Opin. Plant Biol. 72, 2350. doi: 10.1016/j.pbi.2023.102350
Dong, R., Yu, B., Yan, S., Qiu, Z., Lei, J., Chen, C., et al. (2020). Analysis of vitamin P content and inheritance models in eggplant. Hortic. Plant J. 6 (4), 240–246. doi: 10.1016/j.hpj.2020.05.005
Gachon, C. M. M., Langlois-Meurinne, M., Saindrenan, P. (2005). Plant secondary metabolism glycosyltransferases: the emerging functional analysis. Trends Plant Sci. 10 (11), 542–549. doi: 10.1016/j.tplants.2005.09.007
Glasser, G., Graefe, E. U., Struck, F., Veit, M., Gebhardt, R. (2002). Comparison of antioxidative capacities and inhibitory effects on cholesterol biosynthesis of quercetin and potential metabolites. Phytomedicine 9 (1), 33–40. doi: 10.1078/0944-7113-00080
Griesser, M., Vitzthum, F., Fink, B., Bellido, M. L., Raasch, C., Munoz-Blanco, J., et al. (2008). Multi-substrate flavonol O-glucosyltransferases from strawberry (Fragaria×ananassa) achene and receptacle. J. Exp. Bot. 59 (10), 2611–2625. doi: 10.1093/jxb/ern117
He, F., Chen, W., Yu, K., Ji, X., Duan, C., Reeves, M. J., et al. (2015). Molecular and biochemical characterization of the UDP-glucose: anthocyanin 5-O-glucosyltransferase from Vitis amurensis. Phytochemistry 117, 363–372. doi: 10.1016/j.phytochem.2015.06.023
Hosseinzadeh, H., Nassiri-Asl, M. (2014). Review of the protective effects of rutin on the metabolic function as an important dietary flavonoid. J. Endocrinol. Invest. 37 (9), 783–788. doi: 10.1007/s40618-014-0096-3
Huang, J., Pang, C., Fan, S., Song, M., Yu, J., Wei, H., et al. (2015). Genome-wide analysis of the family 1 glycosyltransferases in cotton. Mol. Genet. Genomics 290 (5), 1805–1818. doi: 10.1007/s00438-015-1040-8
Irmisch, S., Jo, S., Roach, C. R., Jancsik, S., Yuen, M. M. S., Madilao, L. L., et al. (2018). Discovery of UDP-glycosyltransferases and BAHD-acyltransferases involved in the biosynthesis of the antidiabetic plant metabolite montbretin a. Plant Cell. 30 (8), 1864–1886. doi: 10.1105/tpc.18.00406
Jimenez-Aliaga, K., Bermejo-Bescos, P., Benedi, J., Martin-Aragon, S. (2011). Quercetin and rutin exhibit antiamyloidogenic and fibril-disaggregating effects in vitro and potent antioxidant activity in APPswe cells. Life Sci. 89 (25-26), 939–945. doi: 10.1016/j.lfs.2011.09.023
Jing, R., Li, H. Q., Hu, C. L., Jiang, Y. P., Qin, L. P., Zheng, C. J. (2016). Phytochemical and pharmacological profiles of three Fagopyrum buckwheats. Int. J. Mol. Sci. 17 (4), 589. doi: 10.3390/ijms17040589
Jones, P., Messner, B., Nakajima, J. I., Schaffner, A. R., Saito, K. (2003). UGT73C6 and UGT78D1, glycosyltransferases involved in flavonol glycoside biosynthesis in arabidopsis thaliana. J. Biol. Chem. 278 (45), 43910–43918. doi: 10.1074/jbc.M303523200
Koja, E., Ohata, S., Maruyama, Y., Suzuki, H., Shimosaka, M., Taguchi, G. (2018). Identification and characterization of a rhamnosyltransferase involved in rutin biosynthesis in Fagopyrum esculentum (common buckwheat). Biosci. Biotechnol. Biochem. 82 (10), 1790–1802. doi: 10.1080/09168451.2018.1491286
Kovinich, N., Saleem, A., Arnason, J. T., Miki, B. (2010). Functional characterization of a UDP-glucose: flavonoid 3-O-glucosyltransferase from the seed coat of black soybean (Glycine max (L.) Merr.). Phytochemistry 71 (11-12), 1253–1263. doi: 10.1016/j.phytochem.2010.05.009
Kumar, S., Stecher, G., Li, M., Knyaz, C., Tamura, K. (2018). Mega x: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35 (6), 1547–1549. doi: 10.1093/molbev/msy096
Li, Y., Baldauf, S., Lim, E., Bowles, D. J. (2001). Phylogenetic analysis of the UDP-glycosyltransferase multigene family of Arabidopsis thaliana. J. Biol. Chem. 276 (6), 4338–4343. doi: 10.1074/jbc.M007447200
Li, Y., Li, P., Wang, Y., Dong, R., Yu, H., Hou, B. (2014). Genome-wide identification and phylogenetic analysis of family-1 UDP glycosyltransferases in maize (Zea mays). Planta 239 (6), 1265–1279. doi: 10.1007/s00425-014-2050-1
Liu, R., Shu, B., Wang, Y., Yu, B., Wang, Y., Gan, Y., et al. (2023). Transcriptome analysis reveals key genes involved in the eggplant response to high-temperature stress. Environ. Exp. Bot. 211, 105369. doi: 10.1016/j.envexpbot.2023.105369
Luca, S. V., Macovei, I., Bujor, A., Miron, A., Skalicka-Woźniak, K., Aprotosoaie, A. C., et al. (2020). Bioactivity of dietary polyphenols: the role of metabolites. Crit. Rev. Food. Sci. Nutr. 60 (4), 626–659. doi: 10.1080/10408398.2018.1546669
Mackenzie, P. I., Owens, I. S., Burchell, B., Bock, K. W., Bairoch, A., Belanger, A., et al. (1997). The UDP glycosyltransferase gene superfamily: recommended nomenclature update based on evolutionary divergence. Pharmacogenetics 7 (4), 255–269. doi: 10.1097/00008571-199708000-00001
Martínez-Ispizua, E., Calatayud, Á., Marsal, J. I., Mateos-Fernández, R., Díez, M. J., Soler, S., et al. (2021). Phenotyping local eggplant varieties: commitment to biodiversity and nutritional quality preservation. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.696272
Mateo-Bonmati, E., Casanova-Saez, R., Simura, J., Ljung, K. (2021). Broadening the roles of UDP-glycosyltransferases in auxin homeostasis and plant development. New Phytol. 232 (2), 642–654. doi: 10.1111/nph.17633
Mizohata, E., Okuda, T., Hatanaka, S., Nakayama, T., Horikawa, M., Nakayama, T., et al. (2013). Crystallization and preliminary X-ray crystallographic analysis of UDP-glucuronic acid: flavonol-3-O-glucuronosyltransferase (Vvgt5) from the grapevine Vitis vinifera. Acta Crystallogr. F. 69 (Pt 1), 65–68. doi: 10.1107/S1744309112045095
Nagatomo, Y., Usui, S., Ito, T., Kato, A., Shimosaka, M., Taguchi, G. (2014). Purification, molecular cloning and functional characterization of flavonoid C-glucosyltransferases from Fagopyrum esculentum M. (Buckwheat) cotyledon. Plant J. 80 (3), 437–448. doi: 10.1111/tpj.12645
Naing, A. H., Kim, C. K. (2021). Abiotic stress-induced anthocyanins in plants: their role in tolerance to abiotic stresses. Physiol. Plant 172 (3), 1711–1723. doi: 10.1111/ppl.13373
Noguchi, A., Horikawa, M., Fukui, Y., Fukuchi-Mizutani, M., Iuchi-Okada, A., Ishiguro, M., et al. (2009). Local differentiation of sugar donor specificity of flavonoid glycosyltransferase in lamiales. Plant Cell. 21 (5), 1556–1572. doi: 10.1105/tpc.108.063826
Offen, W., Martinez-Fleites, C., Yang, M., Kiat-Lim, E., Davis, B. G., Tarling, C. A., et al. (2006). Structure of a flavonoid glucosyltransferase reveals the basis for plant natural product modification. EMBO. J. 25 (6), 1396–1405. doi: 10.1038/sj.emboj.7600970
Ono, E., Homma, Y., Horikawa, M., Kunikane-Doi, S., Imai, H., Takahashi, S., et al. (2010). Functional differentiation of the glycosyltransferases that contribute to the chemical diversity of bioactive flavonol glycosides in grapevines (Vitis vinifera). Plant Cell. 22 (8), 2856–2871. doi: 10.1105/tpc.110.074625
Paquette, S., Moller, B. L., Bak, S. (2003). On the origin of family 1 plant glycosyltransferases. Phytochemistry 62 (3), 399–413. doi: 10.1016/s0031-9422(02)00558-7
Rojas, R. F., Rodriguez, T. O., Murai, Y., Iwashina, T., Sugawara, S., Suzuki, M., et al. (2014). Linkage mapping, molecular cloning and functional analysis of soybean gene Fg2 encoding flavonol 3-O-glucoside (1 → 6) rhamnosyltransferase. Plant Mol. Biol. 84 (3), 287–300. doi: 10.1007/s11103-013-0133-1
Ross, J., Li, Y., Lim, E., Bowles, D. J. (2001). Higher plant glycosyltransferases. Genome Biol. 2 (2), REVIEWS3004. doi: 10.1186/gb-2001-2-2-reviews3004
Shen, N., Wang, T., Gan, Q., Liu, S., Wang, L., Jin, B. (2022). Plant flavonoids: classification, distribution, biosynthesis, and antioxidant activity. Food Chem. 383, 132531. doi: 10.1016/j.foodchem.2022.132531
Singh, A. P., Wang, Y., Olson, R. M., Luthria, D., Banuelos, G. S., Pasakdee, S., et al. (2017). LC-MS-MS analysis and the antioxidant activity of flavonoids from eggplant skins grown in organic and conventional environments. Food Nutr. Sci. 08 (09), 873–888. doi: 10.4236/fns.2017.89063
Soler, M., Camargo, E. L., Carocha, V., Cassan-Wang, H., San, C. H., Savelli, B., et al. (2015). The eucalyptus grandis R2R3-MYB transcription factor family: evidence for woody growth-related evolution and function. New Phytol. 206 (4), 1364–1377. doi: 10.1111/nph.13039
Song, J., Chen, C., Zhang, S., Wang, J., Huang, Z., Chen, M., et al. (2020). Systematic analysis of the capsicum ERF transcription factor family: identification of regulatory factors involved in the regulation of species-specific metabolites. BMC Genomics 21 (1), 573. doi: 10.1186/s12864-020-06983-3
Song, J., Sun, B., Chen, C., Ning, Z., Zhang, S., Cai, Y., et al. (2023). An R-R-type MYB transcription factor promotes non-climacteric pepper fruit carotenoid pigment biosynthesis. Plant J. 115 (3), 724–741. doi: 10.1111/tpj.16257
Sulli, M., Barchi, L., Toppino, L., Diretto, G., Sala, T., Lanteri, S., et al. (2021). An eggplant recombinant inbred population allows the discovery of metabolic QTLs controlling fruit nutritional quality. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.638195
Sun, B., Zhou, X., Chen, C., Chen, C., Chen, K., Chen, M., et al. (2020). Coexpression network analysis reveals an MYB transcriptional activator involved in capsaicinoid biosynthesis in hot peppers. Hortic. Res. 7 (1), 162. doi: 10.1038/s41438-020-00381-2
Suzuki, T., Honda, Y., Mukasa, Y. (2005). Effects of UV-B radiation, cold and desiccation stress on rutin concentration and rutin glucosidase activity in tartary buckwheat (Fagopyrum tataricum) leaves. Plant Sci. 168 (5), 1303–1307. doi: 10.1016/j.plantsci.2005.01.007
Tobar-Delgado, E., Mejía-España, D., Osorio-Mora, O., Serna-Cock, L. (2023). Rutin: family farming products’ extraction sources, industrial applications and current trends in biological activity protection. Molecules 28 (15), 5864. doi: 10.3390/molecules28155864
Wang, Y., Li, C., Yan, S., Yu, B., Gan, Y., Liu, R., et al. (2022). Genome-wide analysis and characterization of eggplant F-box gene superfamily: gene evolution and expression analysis under stress. Int. J. Mol. Sci. 23 (24), 16049. doi: 10.3390/ijms232416049
Wilson, A. E., Tian, L. (2019). Phylogenomic analysis of UDP-dependent glycosyltransferases provides insights into the evolutionary landscape of glycosylation in plant metabolism. Plant J. 100 (6), 1273–1288. doi: 10.1111/tpj.14514
Wilson, A. E., Wu, S., Tian, L. (2019). PgUGT95B2 preferentially metabolizes flavones / flavonols and has evolved independently from flavone / flavonol UGTs identified in Arabidopsis thaliana. Phytochemistry 157, 184–193. doi: 10.1016/j.phytochem.2018.10.025
Wu, B., Gao, L., Gao, J., Xu, Y., Liu, H., Cao, X., et al. (2017). Genome-wide identification, expression patterns, and functional analysis of UDP glycosyltransferase family in peach (Prunus persica L. Batsch). Front. Plant Sci. 8. doi: 10.3389/fpls.2017.00389
Wu, B., Liu, X., Xu, K., Zhang, B. (2020). Genome-wide characterization, evolution and expression profiling of UDP-glycosyltransferase family in pomelo (Citrus grandis) fruit. BMC Plant Biol. 20 (1), 459. doi: 10.1186/s12870-020-02655-2
Xiao, X., Lu, Q., Liu, R., Gong, J., Gong, W., Liu, A., et al. (2019). Genome-wide characterization of the UDP-glycosyltransferase gene family in upland cotton. 3 Biotech. 9 (12), 453. doi: 10.1007/s13205-019-1984-1
Yang, C., Fang, X., Wu, X., Mao, Y., Wang, L., Chen, X. (2012). Transcriptional regulation of plant secondary metabolism f. J. Integr. Plant Biol. 54 (10), 703–712. doi: 10.1111/j.1744-7909.2012.01161.x
Yang, L., Kang, Y., Liu, J., Li, N., Sun, H., Ao, T., et al. (2022). Foliar spray with rutin improves cadmium remediation efficiency excellently by enhancing antioxidation and phytochelatin detoxification of Amaranthus hypochondriacus. Int. J. Phytoremediat. 24 (10), 1060–1070. doi: 10.1080/15226514.2021.1999902
Yang, Q., Zhang, Y., Qu, X., Wu, F., Li, X., Ren, M., et al. (2023). Genome-wide analysis of UDP-glycosyltransferases family and identification of UDP genes involved in abiotic stress and flavonol biosynthesis in Nicotiana tabacum. BMC Plant Biol. 23 (1), 204. doi: 10.1186/s12870-023-04208-9
Yi, S., Kuang, T., Miao, Y., Xu, Y., Wang, Z., Dong, L., et al. (2020). Discovery and characterization of four glycosyltransferases involved in anthraquinone glycoside biosynthesis in Rubia yunnanensis. Org. Chem. Front. 7 (17), 2442–2448. doi: 10.1039/D0QO00579G
Yin, Q., Han, X., Han, Z., Chen, Q., Shi, Y., Gao, H., et al. (2020). Genome-wide analyses reveals a glucosyltransferase involved in rutin and emodin glucoside biosynthesis in tartary buckwheat. Food Chem. 318, 126478. doi: 10.1016/j.foodchem.2020.126478
Yin, Q., Shen, G., Di, S., Fan, C., Chang, Z., Pang, Y. (2017). Genome-wide identification and functional characterization of UDP-glucosyltransferase genes involved in flavonoid biosynthesis in glycine max. Plant Cell Physiol. 58 (9), 1558–1572. doi: 10.1093/pcp/pcx081
Yonekura-Sakakibara, K., Tohge, T., Niida, R., Saito, K. (2007). Identification of a flavonol 7-O-rhamnosyltransferase gene determining flavonoid pattern in arabidopsis by transcriptome coexpression analysis and reverse genetics. J. Biol. Chem. 282 (20), 14932–14941. doi: 10.1074/jbc.M611498200
Yu, J., Hu, F., Dossa, K., Wang, Z., Ke, T. (2017). Genome-wide analysis of UDP-glycosyltransferase super family in Brassica rapa and Brassica oleracea reveals its evolutionary history and functional characterization. BMC Genomics 18 (1), 474. doi: 10.1186/s12864-017-3844-x
Keywords: eggplant, UGT superfamily, rutin, biosynthesis, functional identification
Citation: Gan Y, Yu B, Liu R, Shu B, Liang Y, Zhao Y, Qiu Z, Yan S and Cao B (2023) Systematic analysis of the UDP-glucosyltransferase family: discovery of a member involved in rutin biosynthesis in Solanum melongena. Front. Plant Sci. 14:1310080. doi: 10.3389/fpls.2023.1310080
Received: 09 October 2023; Accepted: 21 November 2023;
Published: 22 December 2023.
Edited by:
Xiang Pu, Sichuan Agricultural University, ChinaReviewed by:
Tian Xie, Chinese Academy of Sciences (CAS), ChinaQinggang Yin, China Academy of Chinese Medical Sciences, China
Copyright © 2023 Gan, Yu, Liu, Shu, Liang, Zhao, Qiu, Yan and Cao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shuangshuang Yan, c3N5YW5Ac2NhdS5lZHUuY24=; Bihao Cao, Y2FvYmgwMUBzY2F1LmVkdS5jbg==
†These authors have contributed equally to this work
 Yuwei Gan
Yuwei Gan Bingwei Yu†
Bingwei Yu† Yonggui Liang
Yonggui Liang Zhengkun Qiu
Zhengkun Qiu Shuangshuang Yan
Shuangshuang Yan Bihao Cao
Bihao Cao