- 1College of Life Sciences, Key Laboratory of Xinjiang Phytomedicine Resource and Utilization of Ministry of Education, Shihezi University, Shihezi, China
- 2College of Life Sciences, Shaanxi Normal University, Xi’an, China
- 3Geosystems Research Institute, Mississippi State University, Starkville, MS, United States
Introduction: As one of the traditional Chinese medicinal herbs that were most generally used, licorice attracts lots of interest due to its therapeutic potential. Authentic response regulators (ARRs) are key factors in cytokinin signal transduction and crucial for plant growth and stress response processes. Nevertheless, the characteristics and functions of the licorice ARR genes are still unknown.
Results: In present study, a systematic genome-wide identification and expression analysis of the licorice ARR gene family were conducted and 51 ARR members were identified. Collinearity analysis revealed the significant roles of segmental duplications in the expansion of licorice ARR genes. The cis-acting elements associated with development, stress and phytohormone responses were identified, implying their pivotal roles in diverse regulatory processes. RNA-seq and qRT-PCR results suggested that A-type, but not B-type ARRs were induced by zeatin. Additionally, ARRs participated in diverse abiotic stresses and phytohormones responses. Yeast one-hybrid assay demonstrated that GuARR1, GuARR2, GuARR11, GuARR12, GuARR10-1, GuARR10-2 and GuARR14 were able to bind to the promoter of GuARR8-3, and GuARR1, GuARR12 bound to the GuARR8-1 promoter. GuARR1, GuARR2, GuARR11 and GuARR10-2 bound to the GuARR6-2 promoter as well as GuARR12 and GuARR10-2 bound to the GuARR6-1 promoter.
Discussion: Collectively, these findings provide a basis for future ARR genes function investigations, shedding light on the potential medicinal properties and agricultural applications of licorice.
1 Introduction
Cytokinins, initially discovered for their ability in stimulating cell division and modulating root and shoot initiation in culture and have been reported to participate in heterogeneous important biological processes afterward, are essential plant hormones for plant growth, development, and various stress responses (Miller et al., 1955; Miller et al., 1956; Hwang et al., 2002; Heyl and Schmülling, 2003; Kakimoto, 2003; Mizuno, 2004; Ferreira and Kieber, 2005; Aloni et al., 2006). A multi-step phosphorelay system were involved in the cytokinin signal transduction pathway, which comprises three main components: receptor protein histidine kinases (HKs), phosphoric acid group transfer proteins histidine-containing phosphotransfer proteins (HPs), and response regulator (RRs) (Hwang and Sheen, 2001; Müller and Sheen, 2007). In Arabidopsis, serving as transmembrane cytokinin receptors, the HKs (AHK2, AHK3 and AHK4/CYTOKININ RESPONSE 1 (CRE1)/WOODEN LEG (WOL)) can be autophosphorylated on the histidine (His or H) residue and transmit cytokinin signal to HPs (AHP1~AHP5) (Hwang and Sheen, 2001; Inoue et al., 2001; Hutchison et al., 2006; Kim et al., 2012). The phosphorylation group is finally transmitted to the aspartate (Asp or D) residue of ARRs from AHPs in the nucleus, modulating transcription of the downstream genes directly or indirectly (To et al., 2004; Mason et al., 2005; Hill et al., 2013). And among them, the ARRs are central regulators (Jeon and Kim, 2013).
The ARR family members are generally subdivided into A- and B-type ARRs and they all possess a N-terminal receiver domain approximately 120 amino acids harbouring a phosphorylatable invariant Asp residue which are highly conserved (Imamura et al., 1999; Imamura et al., 2003; Schaller et al., 2007; To et al., 2007; Hwang et al., 2012). The A-type ARRs consists of 10 members, including ARR3, ARR4, ARR5, ARR6, ARR7, ARR8, ARR9, ARR15, ARR16, and ARR17, which include a short C-terminal extension apart from the N-terminal receiver domain (To et al., 2007). Acting mainly as the negative-feedback regulation factors in cytokinin signaling, the transcription expression of the A-type ARR genes can be quickly activated by cytokinin (Kieber and Schaller, 2018). The B-type ARRs comprises 11 members, including ARR1, ARR2, ARR10, ARR11, ARR12, ARR13, ARR14, ARR18, ARR19, ARR20, and ARR21, which hold the role of positive regulators in cytokinin signaling and transcriptional activators with potential nuclear localization controlling the transcription of amounts of targets, including A-type ARRs (Mason et al., 2005). Besides the receiver domain, each of them contains a long C-terminal region embodying a Myb-like DNA-binding domain, also named B-motifs or GARP domain (Hosoda et al., 2002). This domain is essential for their role in activating target genes and interacting with other regulators (Rubio et al., 2001; Safi et al., 2017). B-type ARRs are further divided into 3 subfamilies in Arabidopsis. The ARR members (ARR1/2/10/11/12/14/18) in subfamily 1 are associated with cytokinin signal transduction (Mason et al., 2005; Yokoyama et al., 2007). Except for ARR21 in subfamily 2, which can function similarly with subfamily 1 members in cytokinin signal transduction, another member in subfamilies 2 (ARR13) and two subfamily 3 members (ARR19/20) showed no pronounced effects on cytokinin sensitivity (Hill et al., 2013). Additionally, ARR23 was identified as a potential pseudogene B-type ARRs containing the phosphorylated Asp site but a partial B motif (Schaller et al., 2008). ARR22 and ARR24 pertain to C-type ARRs, containing short C-terminal extension and perfect receiver domains with conserved D residuals, whose sequences are distinct from A- and B-type ARRs and are not induced by cytokinin (Kiba et al., 2004; Schaller et al., 2008).
The functions of ARRs have been explored. ARR4 can modulate the circadian light input in Arabidopsis through interacting with PHYTOCHROME B (phyB), acting redundantly with its closest relative ARR3 (Salomé et al., 2006). ARR7 and ARR15 inhibit shoot development and cell multiplication, involving maintenance and regeneration of the meristem (Buechel et al., 2010). ARR4 and ARR8 contribute significantly to the formation of shoot callus (Osakabe et al., 2002). ARR3 and ARR10 participate in regulating the root growth via the multistep phosphorelay pathway, in which Ethylene Response 1 (ETR1) interacts with AHPs and then phosphorylated ARR10 (Zdarska et al., 2019). ARR6, ARR7, and ARR15 play redundant roles in negatively regulating the seed germination (Huang et al., 2017). ARR16 is invovled in the cytokinin signaling pathway, the light as well as jasmonic acid signaling pathways, promoting the growth of hypocotyl in blue light (Srivastava et al., 2019). ARR16/17, together with CLE9/10 and SPEECHLESS (SPCH), regulate the epidermal cell-type composition, producing appropriate numbers and proportions of epidermal cells and stomata for the maintenance of leaf performance in fluctuating environments (Vatén et al., 2018). The phenotype of the arr1arr10arr12 triple mutant showed that ARR1/10/12 together regulate a wide array of downstream processes, including seed size, shoot cell division, light sensitivity and concentration of anthocyanin (Argyros et al., 2008). ARR10 directly targets WUSCHEL (WUS), resulting in enhanced shooting in tissue culture (Zubo et al., 2017). ARR1 interacts with the ARABIDOPSIS TRITHORAX-RELATED 2 (ATXR2) protein to bind to the promoter regions of ARR5 and ARR7, preventing premature WUS activation and transiently repressing cytokinin signaling, thus ensuring the proper cell fate transition (Lee et al., 2021). Mutant phenotypes of rice B-type RRs rr21/22/23 indicate that they participate in regulating sensitivity for cytokinin, growth of leaf and root, development of flower, fertilization, shape of trichome and inflorescence architecture (Worthen et al., 2019). Besides, ARRs are also reported to take part in plant stress response. The arr1/10/12 triple mutant displays enhanced adaptation to drought, which is achieved through the induced anthocyanin biosynthesis, abscisic acid sensitivity, improved cell membrane integrity, and reduced stomatal aperture (Nguyen et al., 2016). ARR1 can mediate the cold-inducible A-type ARRs (ARR5/6/7/15) expression, acting as a cold response positive regulator (Jeon and Kim, 2013). ARR2 is able to bind the defense marker gene PATHOGENESIS-RELATED 1 (PR1), provoking transcription of cytokinin-dependent defense gene, which promotes Arabidopsis resistance to Pseudomonas syringae and the binding was induced by the interaction between a salicylic acid response factor TGACG SEQUENCE-SPECIFIC BINDING PROTEIN (TGA) and ARR2 (Choi et al., 2010). Wheat ARR genes are related to dehydration stress, and different expression patterns were observed for the A- and B-type ARRs in response to cytokinin treatment (Gahlaut et al., 2014). OsARR-B5/7/9/10/16/22/23 showed a response to alkaline salt stress (Rehman et al., 2022). Overexpression of soybean RR2a in Arabidopsis significantly improves alkali stress tolerance (Chen et al., 2018).
Recently, ARR family genes in a lot of species have been examined at length, such as in Arabidopsis, rice, tobacco, tomato, Brassica napus (B. napus), and peach (Hwang et al., 2012; Zeng et al., 2017; Wang et al., 2020; Lv et al., 2021; Jiang et al., 2022; Rehman et al., 2022). However, the ARRs in licorice have not been studied. Licorice, one kind of the traditional Chinese medicinal herbs used commonly with a history of at least 2,000 years, contains a variety of triterpenoids and flavonoids for medicinal purposes (Tang et al., 2015). Licorice has been reported to have abundant pharmacological properties, including anticancer (Zhang et al., 2021), antidiabetic (Yang et al., 2020), anti-inflammatory (Wang et al., 2019), immunoregulatory activities (Aipire et al., 2017), and antiviral (Sun et al., 2019) effects. In addition, it is widely used as a health food and natural sweetener. Due to its medicinal and industrial properties, licorice has become a valuable and attractive trade item, with the export quota bidding reaching 5,200 tons in China in 2022 (http://www.mofcom.gov.cn). In Chinese pharmacopoeia, Glycyrrhiza uralensis Fisch. (G. uralensis), Glycyrrhiza glabra L. (G. glabra), and Glycyrrhiza inflata Batalin (G. inflata) are defined as licorice (National Pharmacopoeia Committee, 2010). The long and complex evolutionary history, as well as the complex genome of licorice, make the analysis and study of gene function more challenging. Therefore, a detailed analysis of ARRs will help to understand how they participate in plant development and stress responses through cytokinin signaling. In this study, the genome-wide identification, expression analysis, and function prediction of the ARR gene family in G. inflata, G. glabra and G. uralensis were performed. A total of 51 ARRs were identified in the three licorice species, and their evolution and structure were analyzed. This study is valuable for further research on the function of ARR gene in licorice and for the cultivation of excellent licorice varieties.
2 Materials and methods
2.1 Identification and the physicochemical properties of licorice ARRs
The G. inflata, G. uralensis and G. glabra whole genome database were assembled by Wuhan Benagen Technology Co., Ltd. The conserved receiver domain (PF00072) and Myb-like DNA-binding domain (PF00249) Hidden Markov Model (HMM) files were downloaded from Pfam (http://pfam.xfam.org/) and used to characterize the ARRs using HMMER software (Potter et al., 2018; El-Gebali et al., 2019). To identify the ARRs in these species, 24 Arabidopsis ARR protein sequences obtained from TAIR database (https://www.arabidopsis.org/) were used as queries to search against the licorice protein databases via the BLASTP program with the e-value 1e-5. The results from both HMMER and BLASTP hits were compared to remove redundancies. Additionally, the NCBI-CDD web server (http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi) and SMART database (http://smart.embl-heidelberg.de) were employed to confirm the licorice ARRs. To learn more about the physicochemical properties of ARRs, ExPASy (http://web.expasy.org/compute_pi/) was used to determine the grand average of hydropathicity (GRAVY), molecular weight, and theoretical isoelectric point (pI) of the identified ARRs (Gasteiger et al., 2003). Furthermore, the subcellular localization of ARRs was predicted applying Cell-PLoc 2.0 (http://www.csbio.sjtu.edu.cn/bioinf/Cell-PLoc-2/).
2.2 Multiple sequence alignment and phylogenetic analysis
A multiple alignment of ARRs protein sequences from licorice and Arabidopsis was performed using ClustalX software with the default settings. Based on the alignment results, an unrooted phylogenetic tree was built through the Neighbor-Joining (NJ) method in MEGA7.0 following the parameters: p-distance, pairwise deletion and bootstrap values (1000 replicates) (Kumar et al., 2016). And the phylogenetic tree was visualized by Evolview v3.
2.3 Gene structure, conserved domain and the conserved motif analysis of the licorice ARRs
The conserved motifs of licorice ARR proteins in G. glabra, G. inflata and G. uralensis were analyzed through MEME (https://meme-suite.org/meme/) (Bailey et al., 2015). To gain more information about the gene structure characteristics, the GFF annotation information of G. glabra, G. inflata and G. uralensis genome was used to map the intron-exon distributions using TBtools (Chen et al., 2020). The licorice ARR protein sequences were submitted to the Pfam database for conserved domain analysis, and then visualized by TBtools.
2.4 Chromosomal location and duplication analysis of licorice ARRs
The chromosomal location information of licorice ARRs was extracted from the G. glabra, G. uralensis and G. inflata genome annotation files, and then visualized through MapChart. Multiple Collinearity Scan toolkit (MCScanX) was used to identify duplication events in each licorice genomes. The synteny of ARRs among three licorices were analyzed using MCScanX and visualized through the Dual Systeny Plotter function in TBtools. The KaKs_Calculator 2.0 was used to determine nonsynonymous (Ka) and synonymous (Ks) substitutions and Ka/Ks ratio between each gene pair (Wang et al., 2010).
2.5 Analysis of cis-acting elements in promoters
The 2 kb upstream sequences of ARRs were extracted by the Gtf/Gff3 Sequences Extractor function in TBtools and submitted to PlantCARE databases for the prediction of cis-acting regulatory elements (Lescot et al., 2002).
2.6 Plant materials and stress and hormone treatments
The G. uralensis seeds dormancy were broken by treating them with 98% concentrated H2SO4 for 50 minutes, followed by the wash with sterilized distilled water for three times. The treated seeds were then cultured in the mixture of soil and vermiculite (2:1, v:v) in the automatic climate chamber with the culture temperature of 28°C during the day and 25°C at night and a relative humidity of 50-55%. The photoperiod was 16 h of light and 8 h of darkness. The 60-day-old seedlings were transferred into Hoagland solution medium added 150 mM NaCl and 10% polyethylene glycol 6000 (PEG6000) for stress treatment. The roots of G. uralensis were collected after 0 h (the control group), 2 h, 6 h, and 12 h treatment. For hormone treatment, 60-day-old seedlings were treated with Hoagland solution medium containing 50 mM ABA, 100 µM methyl jasmonate (MeJA), 100 µM gibberellin (GA), and 100 µM auxin (IAA) for 0 h (the control group), 2 h, 6 h, and 12 h, respectively. In addition, 60-day-old seedlings were treated with Hoagland solution medium containing 0 (the control group), 15, 30, 50, 100 μmol/L zeatin for 6 h. Three biological replicates containting the roots of 15 seedlings in each group were pooled then quickly frozen in liquid nitrogen and stored at -80°C for further use.
2.7 RNA sequencing and quantitative real-time PCR analysis
Leaves and roots samples of G. uralensis treated with 0, 15, 30, 50, 100 μmol/L zeatin for 6 h were used to performed the RNA_seq, which were conducted by Shanghai Majorbio Bio-pharm Biotechnology Co., Ltd. (Shanghai, China) on the Illumina Novaseq 6000 platform (Illumina, San Diego, CA). After trimmed and cleaned by software SeqPrep (https://github.com/jstjohn/SeqPrep) and Sickle (https://github.com/najoshi/sickle), the cleaned data were obtained and then used for de novo assembly using software Trinity (http://trinityrnaseq.sourceforge.net/). After that, the clean data were compared with the downloaded G. uralensis genome through the TopHat2 (http://ccb.jhu.edu/software/tophat/index.shtml) and HISAT2 (http://ccb.jhu.edu/software/hisat2/index.shtml) softwares. The expressiones of GuARRs were normalized as Transcripts Per Million (TPM) values. Total RNA from each sample was extracted with the RNAprep Pure Plant kit (TIANGEN BIOTECH, Beijing, China) and first strand cDNAs were synthesized using the EasyScript One-step gDNA Removal and cDNA Synthesis SuperMix (Vazyme, Nanjing, China). The primers for qRT-PCR were designed through Primer Premier 5.0 and synthesized by TSINKE Biotech (Beijing, China), which were listed in Supplementary Table S1. Guactin (NCBI accession number: EU190972.1) was used as the internal control. The qRT-PCR analysis were performed using the 2×ChamQ SYBR qPCR Master Mix (Vazyme, Nanjing, China) on the Bio-RAD CFX96 Real-Time system (Hercules, CA, USA). The expression level was calculated using 2−ΔΔCt method.
2.8 Yeast one hybrid assays
The 2,000 bp fragments from A-type licorice ARRs promoters were cloned individually into the pLaczi vector. The B-type licorice ARRs coding sequences were cloned into pJG-45 vector. Co-transformed into EGY48 yeast strain, the respective combinations were then selected on SD/-Trp-Ura dropout media. Positive transformants were transferred to the selection medium containing X-gal (5-bromo-4-chloro-3-indolyl-b-D-galactopyranoside) to develop a blue color.
3 Results
3.1 Identification and physicochemical properties of the ARR family genes in licorice
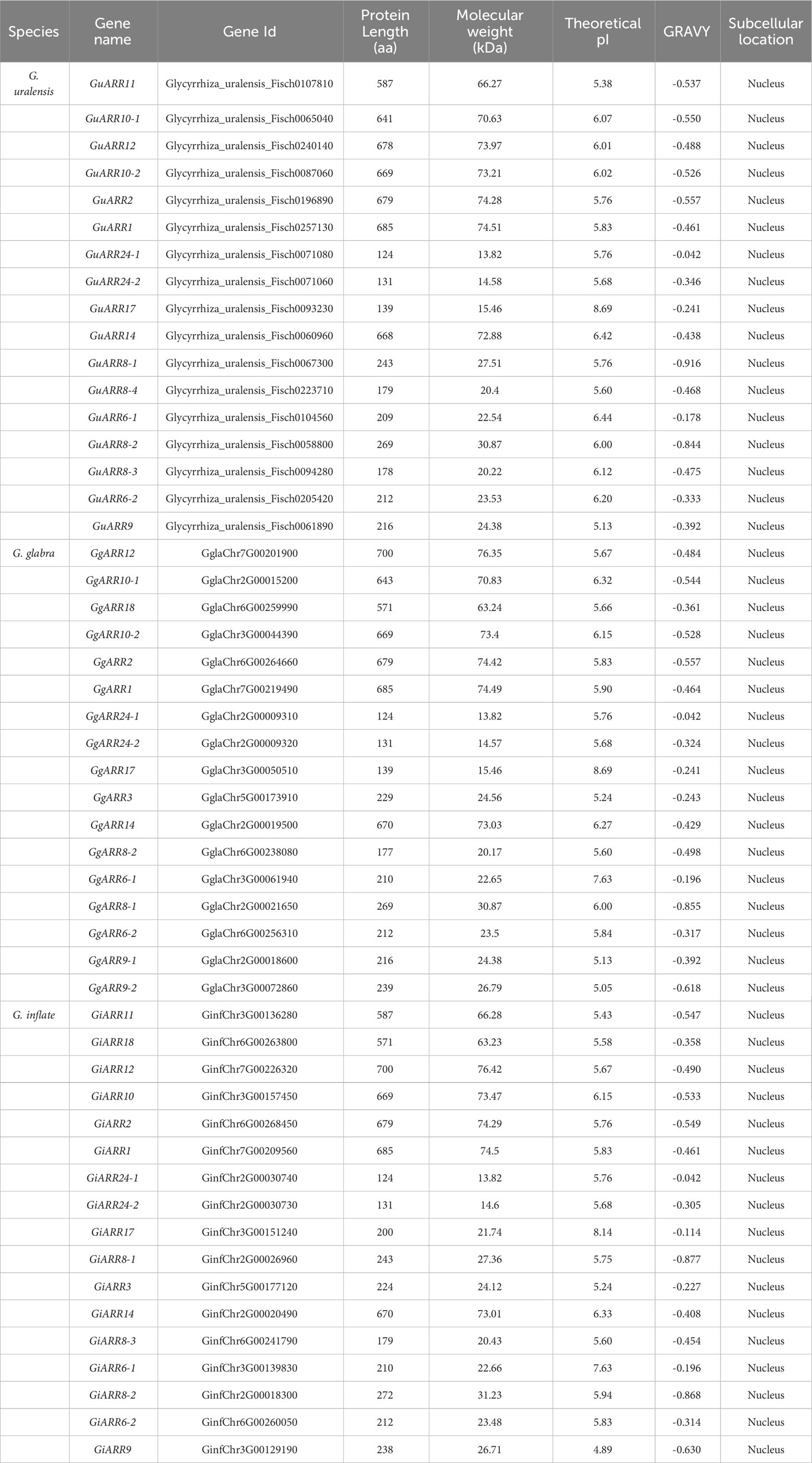
The licorice ARRs were initially identified by searching for conserved response regulator domains. Subsequently, a BLASTP search applying Arabidopsis ARRs sequences as the queries was performed to identify ARRs in the licorice protein database. The results from HMMER and BLASTP were combined to obtain putative licorice ARRs. After removing redundant sequences and sequences lacking complete domains, a total of 51 licorice ARRs were obtained, including 17 G. glabra ARRs (GgARRs), 17 G. uralensis ARRs (GuARRs), and 17 G. inflata ARRs (GiARRs). Supplementary Table S2 presented their amino acid sequences. Among the identified ARRs, 7 GuARRs, 7 GgARRs, and 7 GiARRs contained both the response regulator domain and the Myb-like DNA-binding domain, classifying them as B-type ARRs. The amino acid length of these ARRs ranged from 124 to 700 aa. Their theoretical molecular weights ranged from 13.82 to 76.42 kDa, with an average of 42.53 kDa. Their theoretical isoelectric point (pI) ranged from 4.89 to 8.69 and the GRAVY from -0.916 to -0.042. Subcellular localization analysis predicted their nucleus locations (Table 1, Supplementary Table S3).
3.2 Phylogenetic analysis of ARRs in licorice
To elucidate the evolutionary relationships of ARRs in G. inflata, G. uralensis, G. glabra, and Arabidopsis, we constructed an unrooted Neighbor-Joining (NJ) phylogenetic tree applying sequences of 24 AtARRs, 17 GuARRs, 17 GgARRs, and 17 GiARRs (Figure 1). The 75 ARRs were categorized into three groups, consistenting with the classification of A-, B-, and C-type ARRs in Arabidopsis. Group I comprised 34 A-type ARRs, including 10 AtARRs (AtARR3, AtARR4, AtARR5, AtARR6, AtARR7, AtARR8, AtARR9, AtARR15, AtARR16, AtARR17), 8 GgARRs (GgARR6-1, GgARR6-2, GgARR3, GgARR8-1, GgARR8-2, GgARR9-1, GgARR9-2, GgARR17), 8 GuARRs (GuARR6-1, GuARR6-2, GuARR17, GuARR8-1, GuARR8-2, GuARR8-3, GuARR8-4, GuARR9), and 8 GiARRs (GiARR6-1, GiARR6-2, GiARR3, GiARR17, GiARR8-1, GiARR8-2, GiARR8-3, GiARR9). Thirty-three B-type ARRs were included in groups II, which contained 12 AtARRs (AtARR19, AtARR14, AtARR20, AtARR18, AtARR11, AtARR2, AtARR1, AtARR10, AtARR12, AtARR13, AtARR21, AtARR23), 7 GgARRs (GgARR14, GgARR18, GgARR2, GgARR1, GgARR10-1, GgARR10-2, GgARR12), 7 GuARRs (GuARR14, GuARR11, GuARR2, GuARR1, GuARR10-1, GuARR10-2, GuARR12), and 7 GiARRs (GiARR14, GiARR18, GiARR11, GiARR2, GiARR1, GiARR12, GiARR10). Group III consisted of 2 AtARRs (AtARR22, AtARR24), 2 GgARRs (GgARR24-1, GgARR24-2), 2 GuARRs (GuARR24-1, GuARR24-2), and 2 GiARRs (GiARR24-1, GiARR24-2), which were regarded as C-type ARRs.
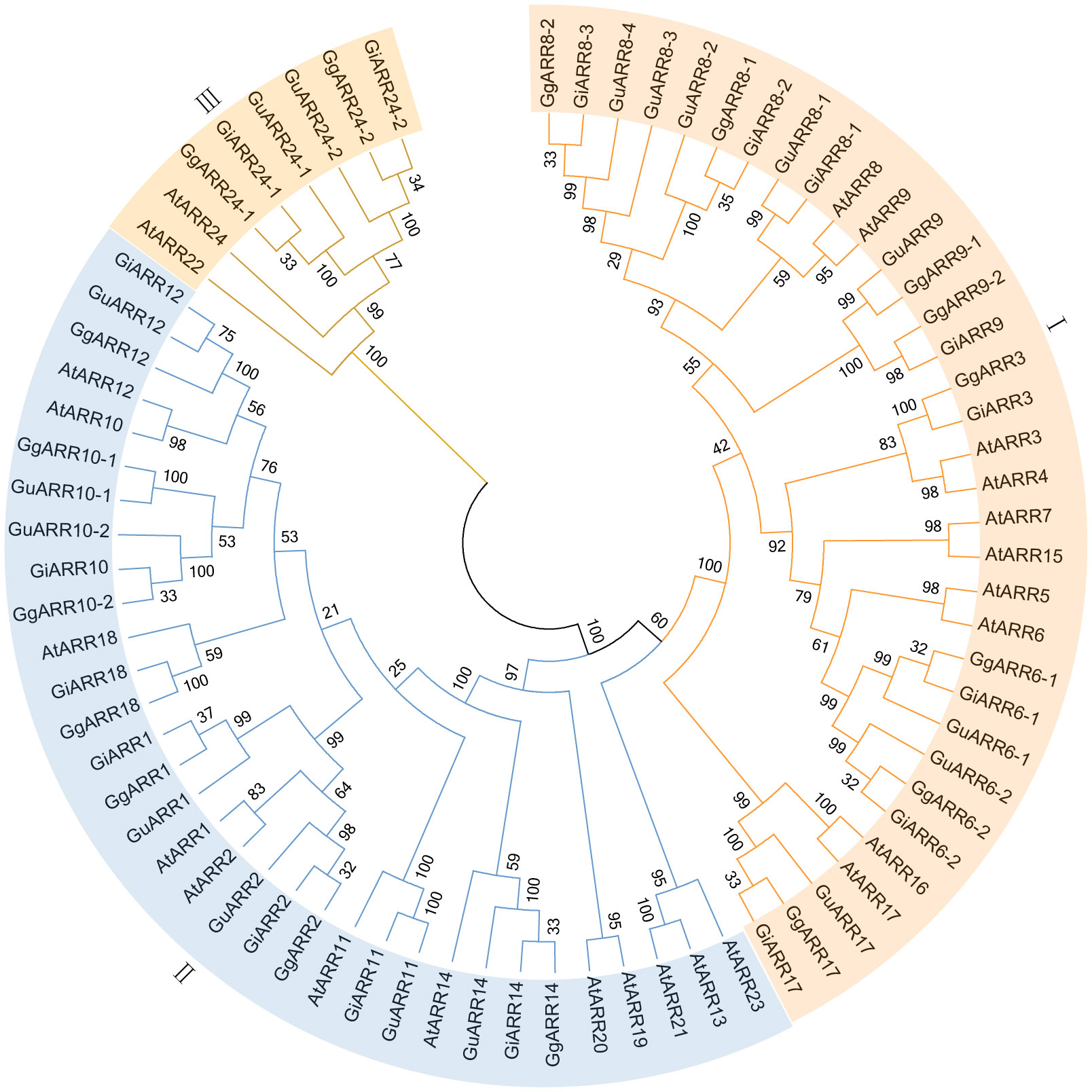
Figure 1 The neighbor-joining phylogenetic tree of ARRs from G. inflata, G. uralensis, G. glabra and A. thaliana using MEGA 7.0 based on their protein sequences. The three kinds of background colors indicated the three groups of the ARRs proteins.
3.3 Sequence alignment of ARRs in licorice
To further investigate the features of the three types of licorices ARRs, the multiple sequence alignments were conducted. The results revealed that all licorice ARRs contained the typical conserved receiver domains containing approximately 120 amino acids, and three conserved phosphorylated amino acid residues existed in which, namely the centeral D1 site, the N-terminal D2 site, and the C-terminal lysine (K) site. A short insertion was exhibited in the A-type licorice ARRs receiver domains (Figure 2). These features of licorice ARRs sequences provided strong support for their classification based on the phylogenetic results.
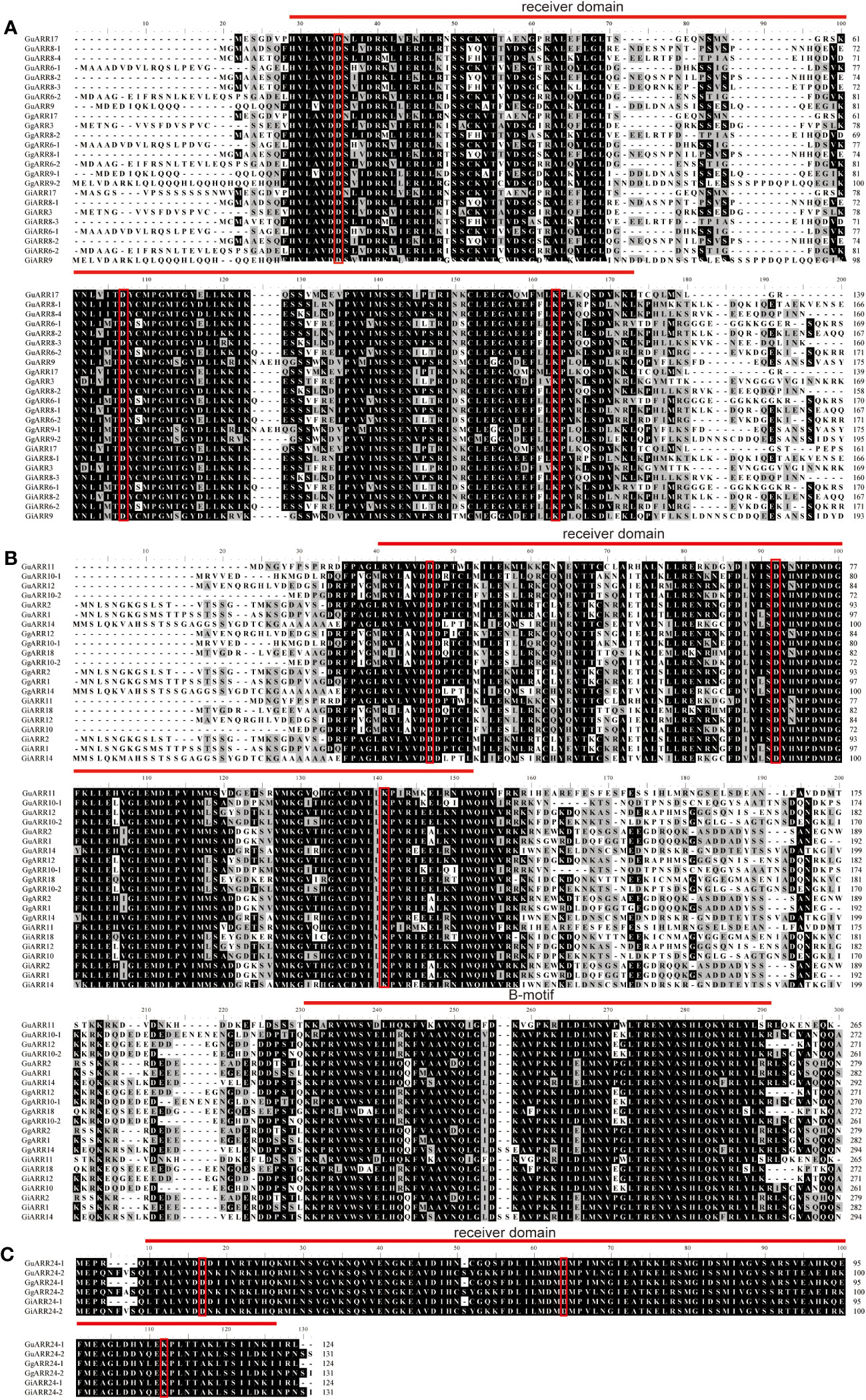
Figure 2 Multiple alignment of the three species of licorice ARRs proteins sequences. (A) Multiple alignment of A-type licorice ARRs proteins sequences. (B) Multiple alignment of B-type licorice ARRs proteins sequences. (C) Multiple alignment of C-type licorice ARRs proteins sequences. The conserved D-D-K residues were highlighted with red rectangles. The receiver domain and B-motif were highlighted with red lines.
3.4 Conserved motif, gene structure and structural domain analysis of ARRs
The MEME analysis identified 8 motifs with a length of 29 to 50 amino acids (Figure 3B, Table 2). These motif sequences were further searched in Pfam for functional identification. Presented in all ARR proteins, motifs 1, 2, and 3 together formed the response regulator receiver domain (Figures 3B, E). Motifs 4 and 5 constituted the Myb-like DNA-binding domain, which were exclusively found in B-type ARRs (Figures 3B, E). The remaining motifs were designated as unknown motifs.
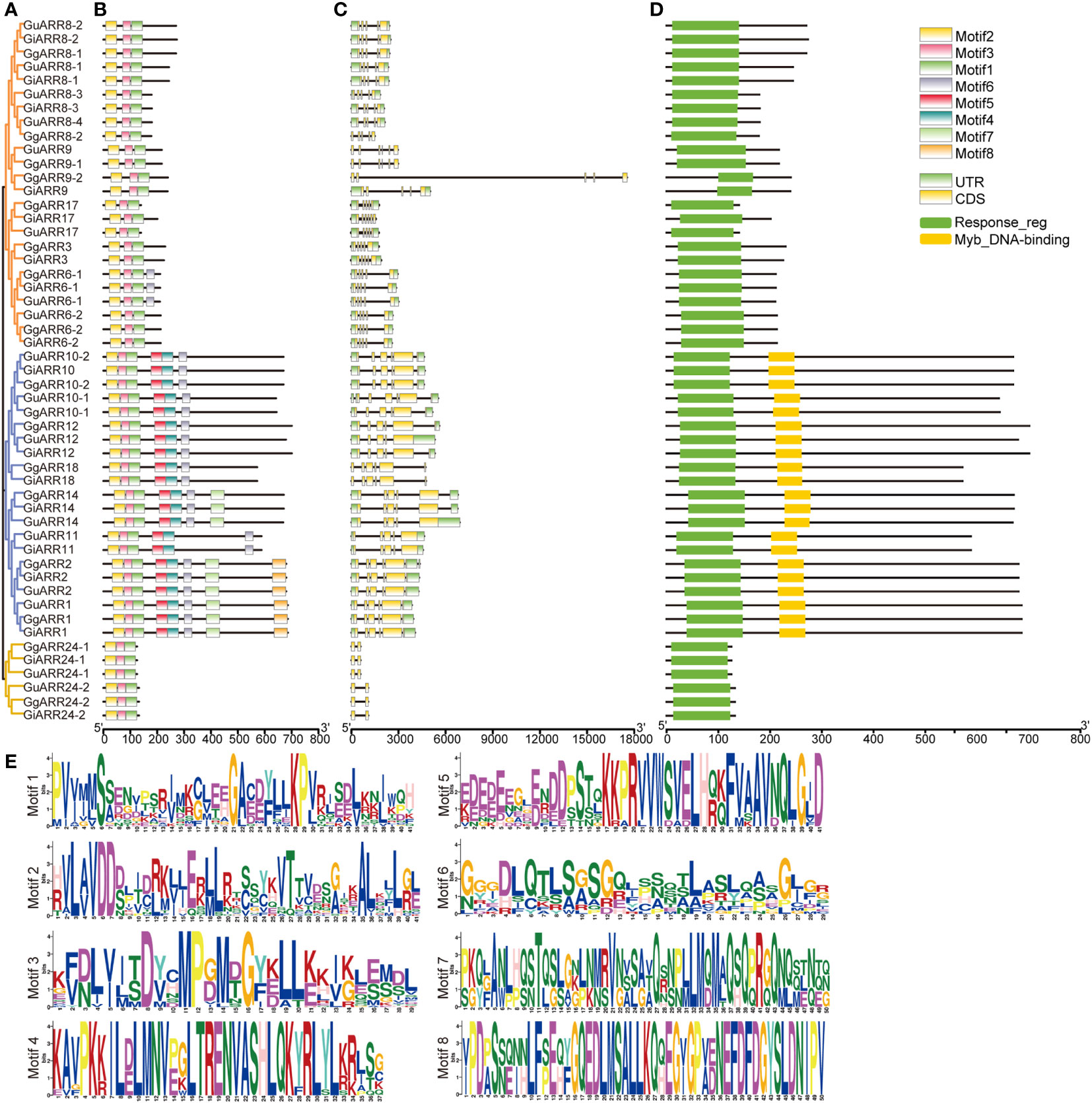
Figure 3 Phylogenetic tree, motifs composition, exon/intron structures and domains of ARRs in licorice. (A) Phylogenetic tree of licorice ARRs constructed with 1000 bootstrap replicates. (B) Conserved motifs of licorice ARRs proteins. A total of 8 motifs were shown. (C) Exon/intron structures of licorice ARRs. Black lines represented introns, yellow rectangles represented exons and green rectangles represented UTRs. (D) Conserved functional domains of licorice ARRs. Yellow rectangles represented Myb-like DNA-binding domains and green rectangles represented the conserved receiver domains. (E) Sequences logos of identified motifs.
A phylogenetic tree of the 51 licorice ARRs was built based on their full-length protein sequences by the NJ method to characterize their evolutionary relationships (Figure 3A). The results showed that these 51 ARRs were divided into three classes, which was consistent with the phylogenetic tree of Arabidopsis ARRs (Figure 1). Gene structures of the licorice ARRs were further investigated, and the numbers of their exon ranged from 2 to 6 (Figure 3C). Most of the A-type ARRs contained 5 exons, except for GuARR9, GgARR9-1, and GiARR17, which each contained 6 exons. Among the 21 licorice B-type ARRs, six members (GuARR12, GgARR14, GuARR14, GiARR14, GuARR11, and GiARR11) contained 5 exons, whereas the remaining B-type ARRs contained six exons. All six C-type ARRs contained two exons. In general, licorice ARRs within the same evolutionary tree branch exhibited similar structures on the exons number and length. To further understand the structure characteristics of the licorice ARR family protein sequences, the protein domains of the three types of licorice ARRs were analyzed using Pfam website and visualized with TBtools. The results displayed that all licorice ARR sequences contained the conserved receiver domain, but the Myb-like DNA-binding domain was present exclusively in B-type ARRs (Figure 3D).
3.5 Chromosome distribution and collinearity analysis of the ARR gene family in licorice
The chromosomal distributions of GgARRs, GuARRs, and GiARRs were visualized according to their genomic positions (Figure 4, Supplementary Table S3). In G. uralensis, 17 GuARRs were mapped on 4 chromosomes unevenly. Specifically, chr2 included the most number of GuARRs, with 7 members (GuARR8-1/2, GuARR14, GuARR9, GuARR10-1, GuARR24-1/2), and chr3 contained 5 GuARRs (GuARR10-2, GuARR17, GuARR8-3, GuARR6-1, GuARR11). In contrast, chr6 and chr7 had fewer GuARR genes, with three (GuARR2, GuARR6-2, GuARR8-4) and two (GuARR1, GuARR12) GuARRs, respectively. The ARRs distributions in G. glabra and G. inflata were similar, with genes located on chr2, 3, 5, 6, and 7. For the GgARRs, 6 members (GgARR8-1, GgARR14, GgARR9-1, GgARR10-1 and GgARR24-1/2) were located on chr2. Chr3 and chr6 contained the same number of ARRs, with GgARR10-2, GgARR17, GgARR6-1, GgARR9-2 located on chr3 and GgARR2, GgARR6-2, GgARR8-2, GgARR18 on chr6. Chr7 comprised GgARR1 and GgARR12. Only GgARR3 was found on chr5. In the case of GiARRs, chr2 and chr3 had the same number of GiARRs. GiARR8-1/2, GiARR14, GiARR24-1/2 were located on chr2, and GiARR6-1, GiARR9/10/11/17 on chr3. GiARR2, GiARR6-2, GiARR8-3, GiARR18 were located on chr6. As it was for GgARRs, chr7 of G. inflata contained GiARR1 and GiARR12, and chr5 contained GiARR3. To understand the expansion mechanism of the ARRs, segmental and tandem duplications events within the licorice genomes were examined. The results showed that the licorice ARR gene family had no tandem duplication gene pairs. However, there were 5, 3, and 6 segmental duplication gene pairs in G. inflata, G. uralensis, and G. glabra genomes, respectively. Among these duplication relationships, GiARR18 was paired with GiARR12 and GiARR10, GiARR6-2 was paired with GiARR6-1 and GiARR3, GgARR18 was paired with GgARR12, GgARR10-1, and GgARR10-2, respectively, while the others were one-to-one paired. GgARR3 was paired with GgARR6-2, GgARR9-1 was paired with GgARR9-2, GgARR10-1 was paired with GgARR10-2. As for the GuARRs, GuARR6-1 was paired with GuARR6-2, GuARR8-3 was paired with GuARR8-4, GuARR10-1 was paired with GuARR10-2 (Figure 5). These results indicated that several licorice ARR genes were possibly generated through gene duplication, and the main driving force for licorice ARR gene evolution was segmental duplication. Additionally, comparative syntenic maps were constructed among the three licorices. The results revealed that 13 GuARRs had a syntenic relationship with 15 GgARRs, accompanying 23 orthologous pairs. Furthermore, there were 21 orthologous pairs between 15 GuARRs and 15 GiARRs, while 15 GgARRs and 13 GiARRs had 25 pairs of syntenic relationships (Figure 6; Supplementary Table S4). These findings suggested the significant roles of these genes in the ARR genes evolution. More than 88% of the 69 gene pairs had a ratio of nonsynonymous substitution rate (Ka) to synonymous substitution rate (Ks) below 1 (Supplementary Table S4), indicating purifying selection during evolution of these genes, and they have relatively conserved functions.
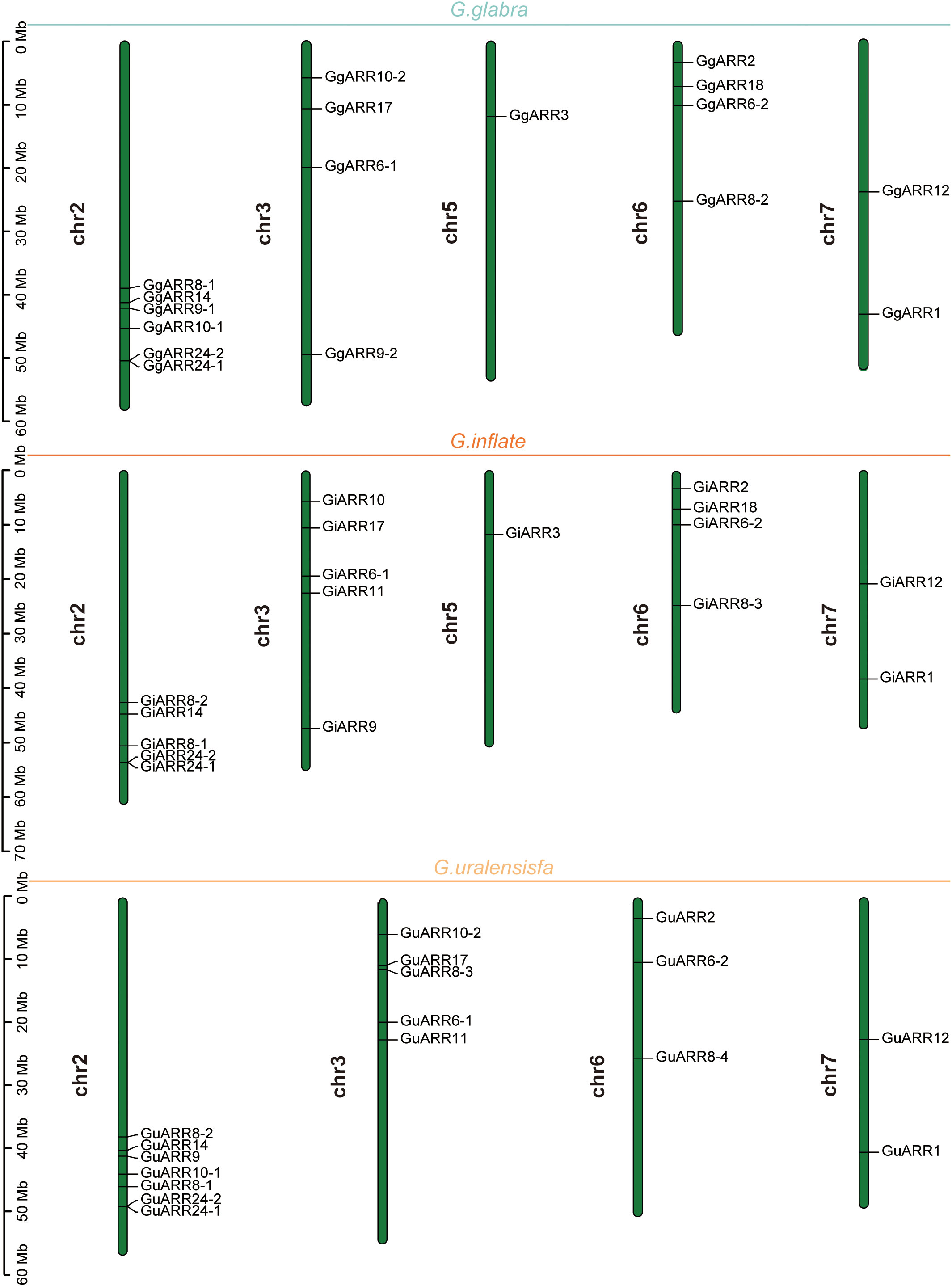
Figure 4 The chromosomal locations of the 51 ARR genes from G. inflata, G. uralensis and G. glabra. The vertical bar on the left side represented the length of chromosome. Mb, megabase.
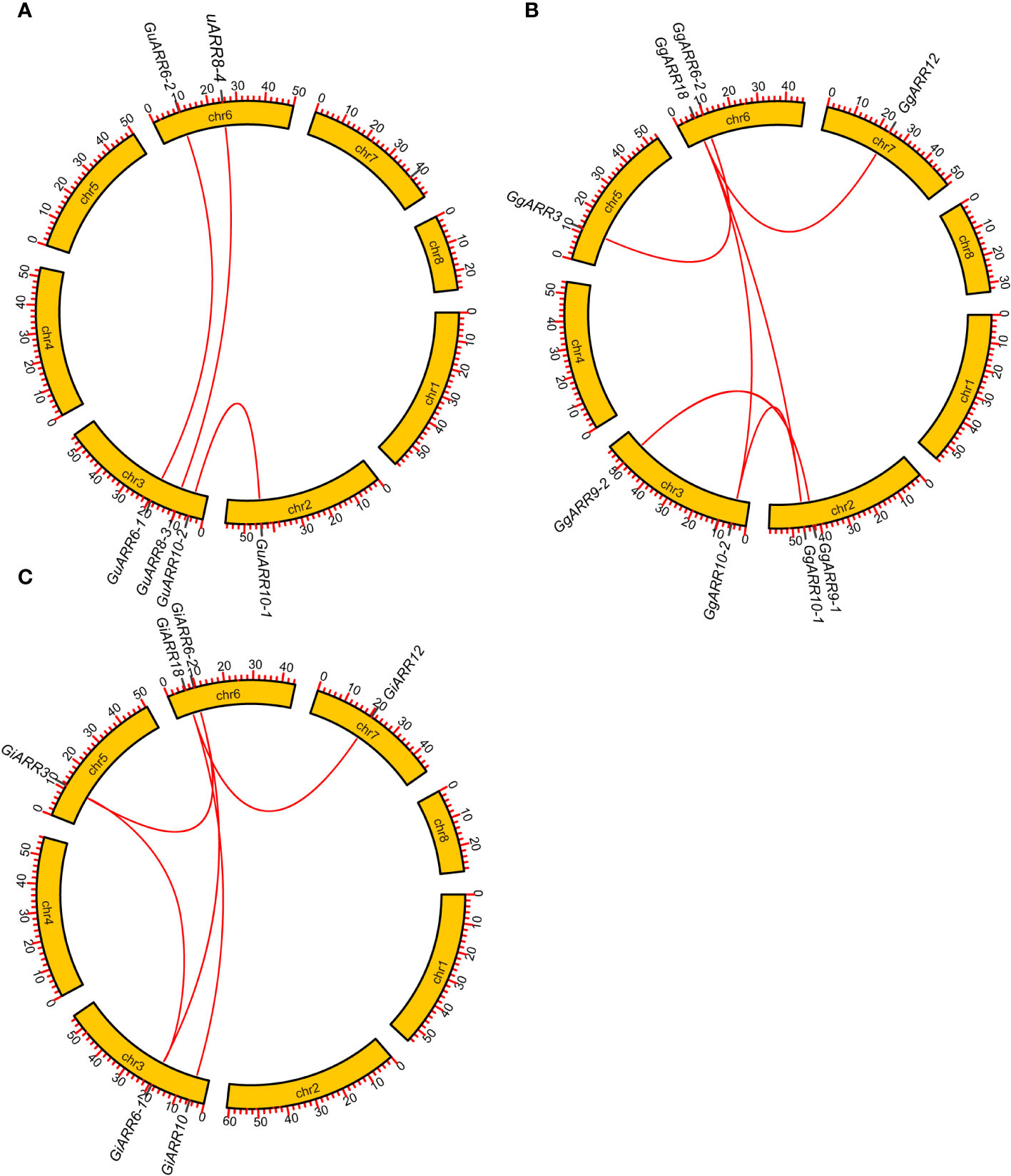
Figure 5 Gene duplication relationship among ARRs of G. inflata, G. uralensis and G. glabra. (A) Gene duplication relationship among GuARRs. (B) Gene duplication relationship among GgARRs. (C) Gene duplication relationship among GiARRs. The scale on the circle was in Megabases.
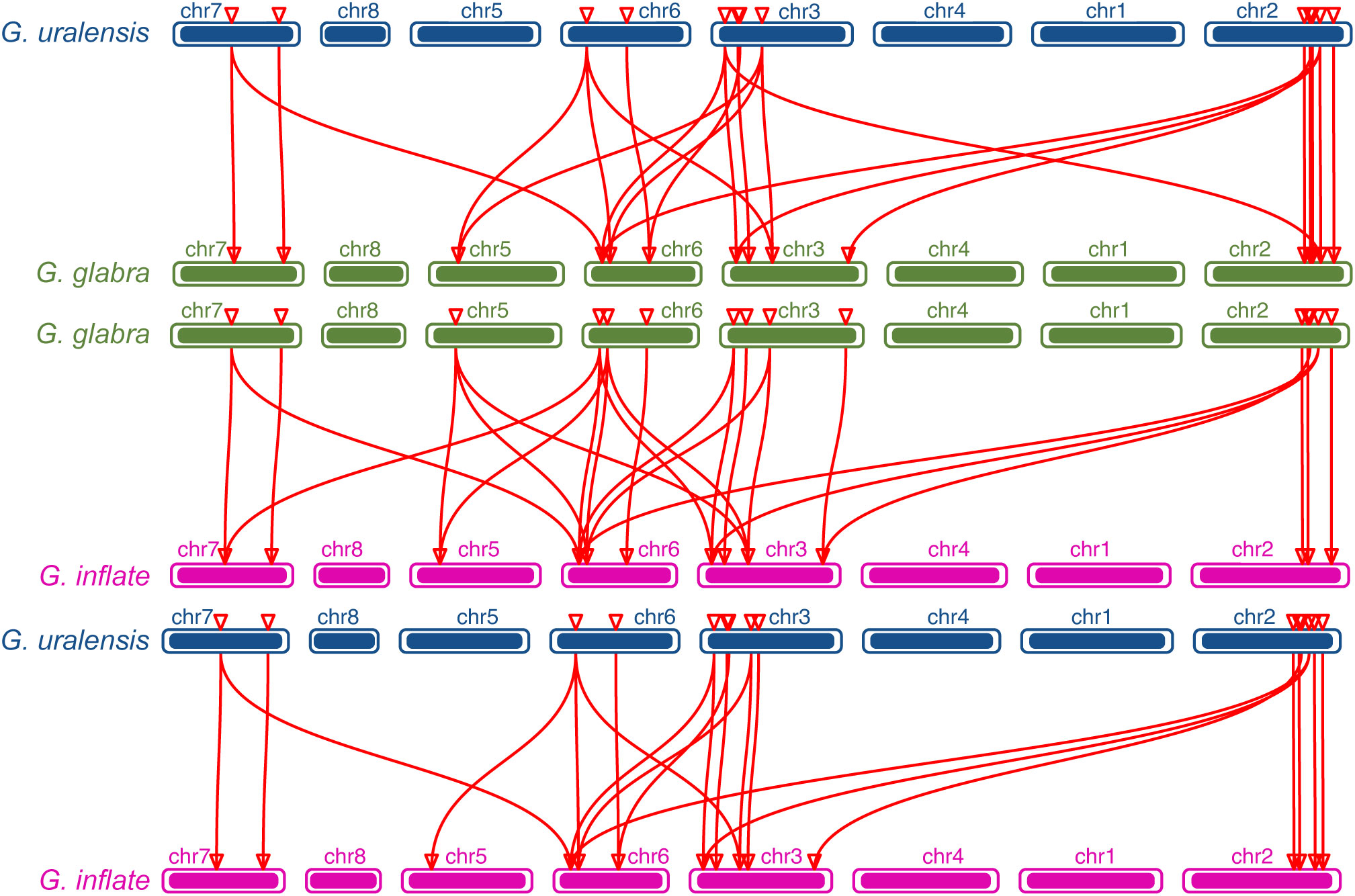
Figure 6 The collinearity relationship between GuARRs and GgARRs, GgARRs and GiARRs, GuARRs and GiARRs.
3.6 Cis-acting regulatory elements in the promoter of ARR genes
To grasp the potential function and regulatory mechanisms of licorice ARR genes, we analyzed their promoter sequences to identify potential cis-acting elements using the PlantCARE database. The analysis showed the existence of 6908 cis-acting elements, and 4904 among them were involved in promoter binding sites, including 285 AT~TATA box (4.13%), 1904 CAAT-box (27.56%), and 2715 TATA-box (39.30%) (Supplementary Table S5). Additionally, a quantity of light-responsive elements were also found in the licorice ARR promoters, including 160 Box 4, 84 G-Box, 45 GT1-motif, 43 GATA-motif, 34 TCT-motif, 31 TCCC-motif, 25 AE-box, 17 GA-motif, 15 I-box, 13 MRE, 11 ATCT-motif, 10 LAMP-element, 10 AT1-motif, 8 chs-CMA1a, 6 ATC-motif, 4 ACE and 4 chs-CMA2a, taking up 7.53% of the total cis-acting elements (Supplementary Table S5). Furthermore, there were 73 plant growth and development related cis-acting elements detected in the licorice ARR genes promoters, in which the circadian (involved in circadian control) was the most with 23, followed by 15 RY-element (seed-specific regulation related), 12 O2-site (zein metabolism regulation related), 11 CAT-box (meristem expression related), 7 MBSI (related to flavonoid biosynthetic gene regulation) and 5 GCN4 motif (involved in endosperm expression) (Figure 7). A total of 398 hormone-related cis-acting elements were existed, in which 18 TGA-element and 3 AuxRR-core were IAA responsive elements, 63 ABRE, 17 ABRE3a and 17 ABRE4 were ABA responsive elements, 32 TCA-element were salicylic acid responsive elements, 40 CGTCA-motif and 40 TGACG-motif were MeJA responsive elements, 12 GARE-motif, 29 P-box and 24 TATC-box were gibberellin responsive elements, and 103 ERE were ethylene response elements. Regarding the 1013 stress-related cis-acting elements, the analysis revealed the presence of 207 MYB, 207 Myc, 119 STRE, 107 ARE, 57 MYB-like sequence, 57 W box, 49 WRE3, 40 as-1, 37 WUN-motif, 36 Myb-binding site, 30 TC-rich repeats, 29 LTR, 26 MBS and 12 MYB recognition site. Overall, the distribution and composition of these diverse cis-acting elements may play a crucial role in shaping the expression of the ARR family genes in response to various stimuli, including light, hormones, abiotic stresses, and licorice plant development.

Figure 7 The numbers of growth and development, phytohormone and stress response related cis-acting elements were shown in licorice ARRs promoter regions. The phylogenetic tree of licorice ARRs was on the left and the classification of ARRs was on the right.
3.7 Expression pattern of GuARRsunder abiotic stress and phytohormone treatments
According to the RNA_seq data, the expression profiles of GuARRs under different concentrations of zeatin treatment were analyzed (Figures 8A, B). The expression results showed that GuARR8-1/2/3, GuARR6-1/2, GuARR1 and GuARR9 were increased in the aboveground part under zeatin treatment, whereas GuARR8-4 was decreased (Figure 8A). In the root, the expressions of GuARR8-1/2/3, GuARR6-1/2, GuARR9, and GuARR17 were significantly induced by zeatin treatment, and GuARR8-4 was up-regulated under 15 μmol/L zeatin treatment (Figure 8B). And the C-type GuARRs had pretty low expression in both aboveground part and root as the TPM values were zero. To validate the RNA_seq data, qRT-PCR was conducted to explore the expressions of A-type and B-type GuARRs in the aboveground part and root under zeatin treatment (Figures 8C, D). QRT-PCR showed that most of the GuARRs showed similar expression patterns with the data from RNA_seq. In roots, A-type GuARRs (GuARR6-1/2, GuARR8-1/2/3/4, GuARR9, GuARR17) showed significant induction by zeatin treatment. GuARR6-1 and GuARR6-2 were induced with 6.53-folds and 16.98-folds after treatment of 50 μmol/L zeatin for 6 h, respectively. The expression of GuARR8-1 and GuARR8-2 were increased with 11.85-folds and 26.06-folds after treatment of 100 μmol/L zeatin, respectively. Expressions of GuARR8-3, GuARR8-4 and GuARR9 were up to 8.10-folds, 2.92-folds and 10.62-folds after treatment of 50 μmol/L zeatin, respectively. In aboveground part, the expression of GuARR17 was up to 9.83-folds after 15 μmol/L zeatin treatment (Figure 8C). Expression of GuARR9 was up to 14.82-folds after 30 μmol/L zeatin treatment. GuARR17, GuARR6-1, GuARR8-1 and GuARR8-2 were induced with 3.10-folds, 7.64-folds, 2.90-folds and 4.12-folds after treatment of 100 μmol/L zeatin, respectively. The expression of GuARR6-2, GuARR8-3 and GuARR8-4 were increased with 4.71-folds, 4.18-folds and 2.98-folds after treatment of 15 μmol/L zeatin, respectively (Figure 8D). These results were consistent with their cytokinin responsive characristic. However, some discrepancies were observed for B-type GuARRs, with GuARR2, GuARR10-1, GuARR11 in roots and GuARR1, GuARR2, GuARR10-2, GuARR12 in aboveground part showing different responses in the qRT-PCR analysis, which should be further verified in the future.
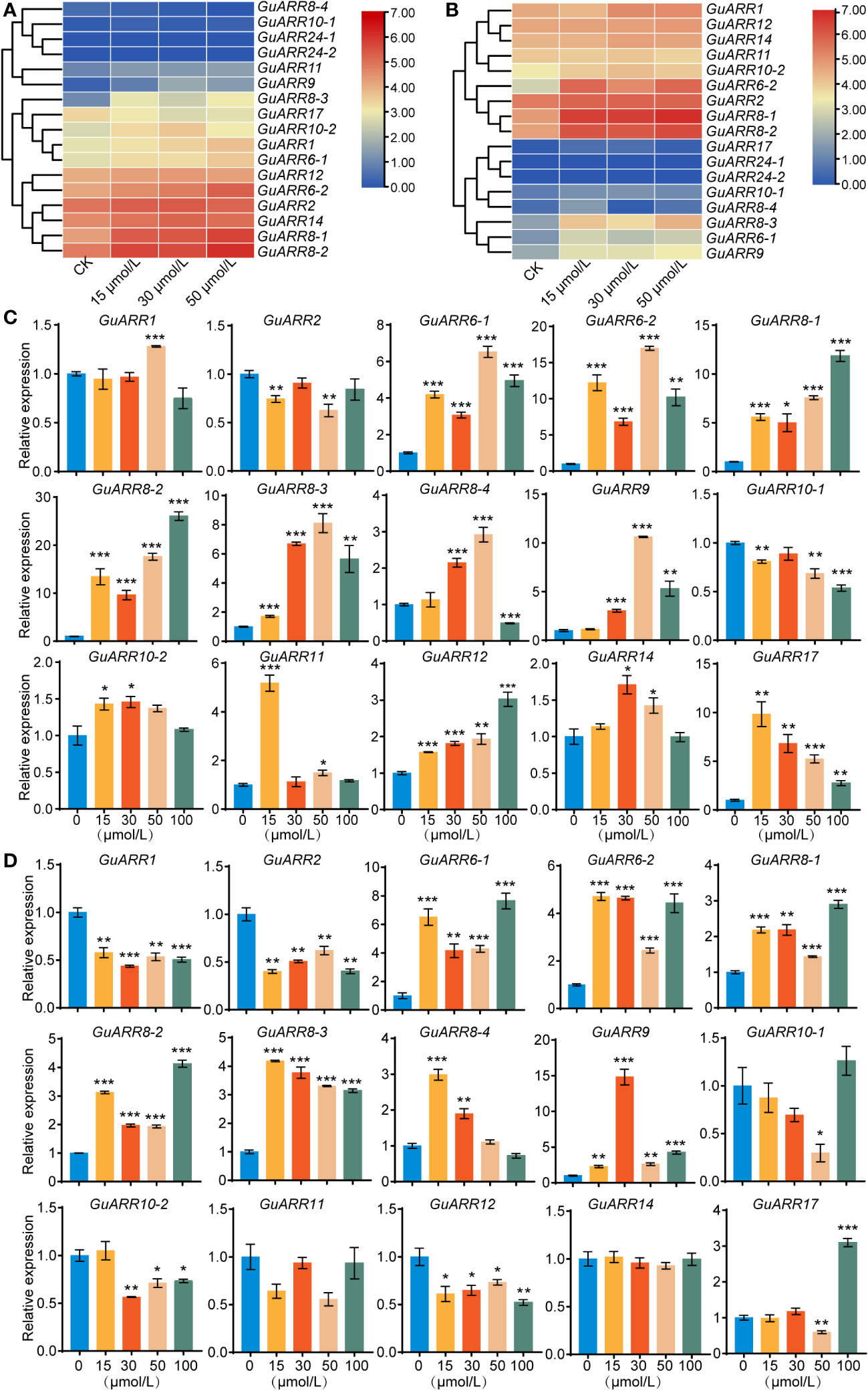
Figure 8 The heatmaps of the expression profiles of GuARRs under various concentrations of zeatin treatments. (A) Heatmap of GuARRs expression profiles in aboveground part. (B) Heatmap of GuARRs expression profiles in roots. The selected GuARRs expression levels in the licorice roots (C) and aboveground part (D) under various concentrations of zeatin treatments via qRT-PCR. Student’s t-test was used to assess significant differences. Significance levels: *, p < 0.05; **, p < 0.01,***, p < 0.001.
Additionally, the expression levels of GuARRs were measured via qRT-PCR under treatments of PEG and NaCl (Figure 9). The results indicated that GuARR2, GuARR10-1, and GuARR12 were induced under PEG stress, whereas the expressions of GuARR1, GuARR6-1, GuARR8-1/2/3/4, GuARR9, GuARR10-2, GuARR11, and GuARR14 were decreased under PEG treatment. Under NaCl treatment, GuARR6-1/2, GuARR8-1/2/3, GuARR10-2, GuARR17, and GuARR11 were up-regulated at 2 h and peaked at 6 h, then their expressions decreased at 12 h. GuARR1, GuARR8-4, and GuARR14 had the highest expression levels after NaCl treatment for 6 h. GuARR12 and GuARR9 were up-regulated at 6 h and 12 h, respectively, but GuARR10-1 was reduced by NaCl treatment.
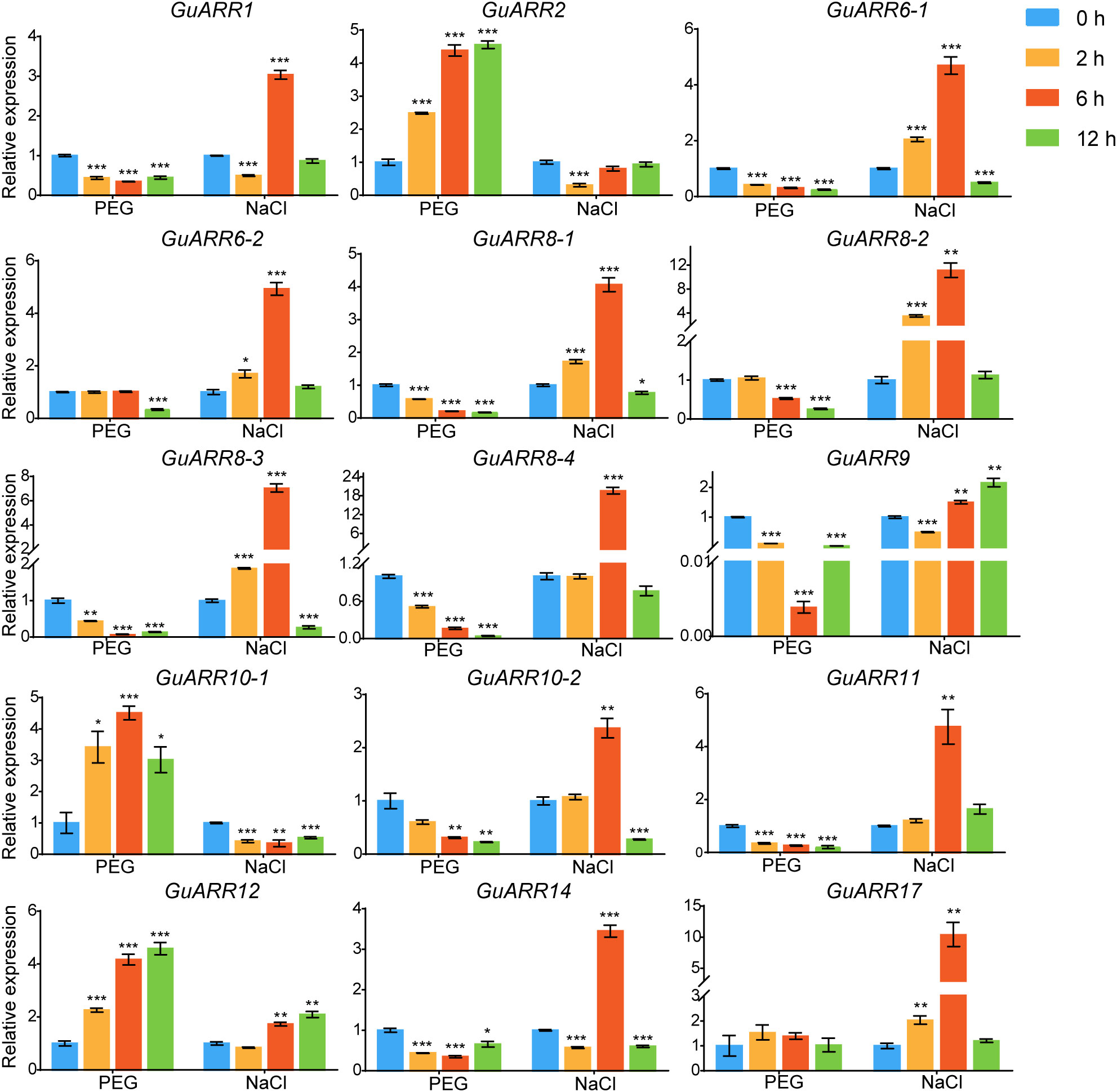
Figure 9 Expression profiles of GuARRs in response to PEG and NaCl treatments based on qRT-PCR analysis. Samples were collected at 0 h, 2 h, 6 h and 12 h after PEG (10%) and NaCl (150 mM) treatment. Student’s t-test was used to assess significant differences. Significance levels: *, p < 0.05; **, p < 0.01,***, p < 0.001.
Furthermore, the expression profiles of A-type and B-type GuARRs response to four kinds of phytohormones were explored using qRT-PCR (Figure 10). Under IAA treatment, GuARR2, GuARR12 and GuARR17 were induced at 6 h and 12 h, while GuARR6-2 peaked at 6 h. Other GuARRs, including GuARR1, GuARR6-1, GuARR8-1/2/3/4, GuARR9, GuARR10-2, GuARR11, and GuARR14, were down-regulated after IAA treatment. The expression of GuARR10-1 was not influenced obviously by IAA. Under GA treatment, GuARR1, GuARR6-1, GuARR8-1/2/3/4, GuARR9, GuARR10-2, and GuARR17 were reduced, whereas GuARR14 was induced significantly at 6 h. GuARR12 and GuARR6-2 were up-regulated at 12 h, and the expression of GuARR11 gradually increased with the extension of treatment time. GuARR2 and GuARR10-1 did not displayed obvious change in their expressions. Most of the GuARR genes, including GuARR1, GuARR6-1/2, GuARR8-1/2/3/4, GuARR9, GuARR10-1/2, GuARR11, and GuARR14, were reduced by ABA treatment, but GuARR2, GuARR12 and GuARR17 were significantly induced. GuARR12 was markedly up-regulated under MeJA treatment, peaking at 6 h. GuARR1 and GuARR2 were up-regulated at 2 h and continued to increase at 6 h, then decreased under the sustained MeJA treatment. GuARR6-2 and GuARR17 had the highest expression level after 6 h of MeJA treatment. GuARR10-1 was dramatically induced under 2 h treatment of MeJA. Other GuARRs, including GuARR6-1, GuARR8-1/2/3/4, GuARR9, GuARR10-2, GuARR11, and GuARR14, were down-regulated under MeJA treatment.
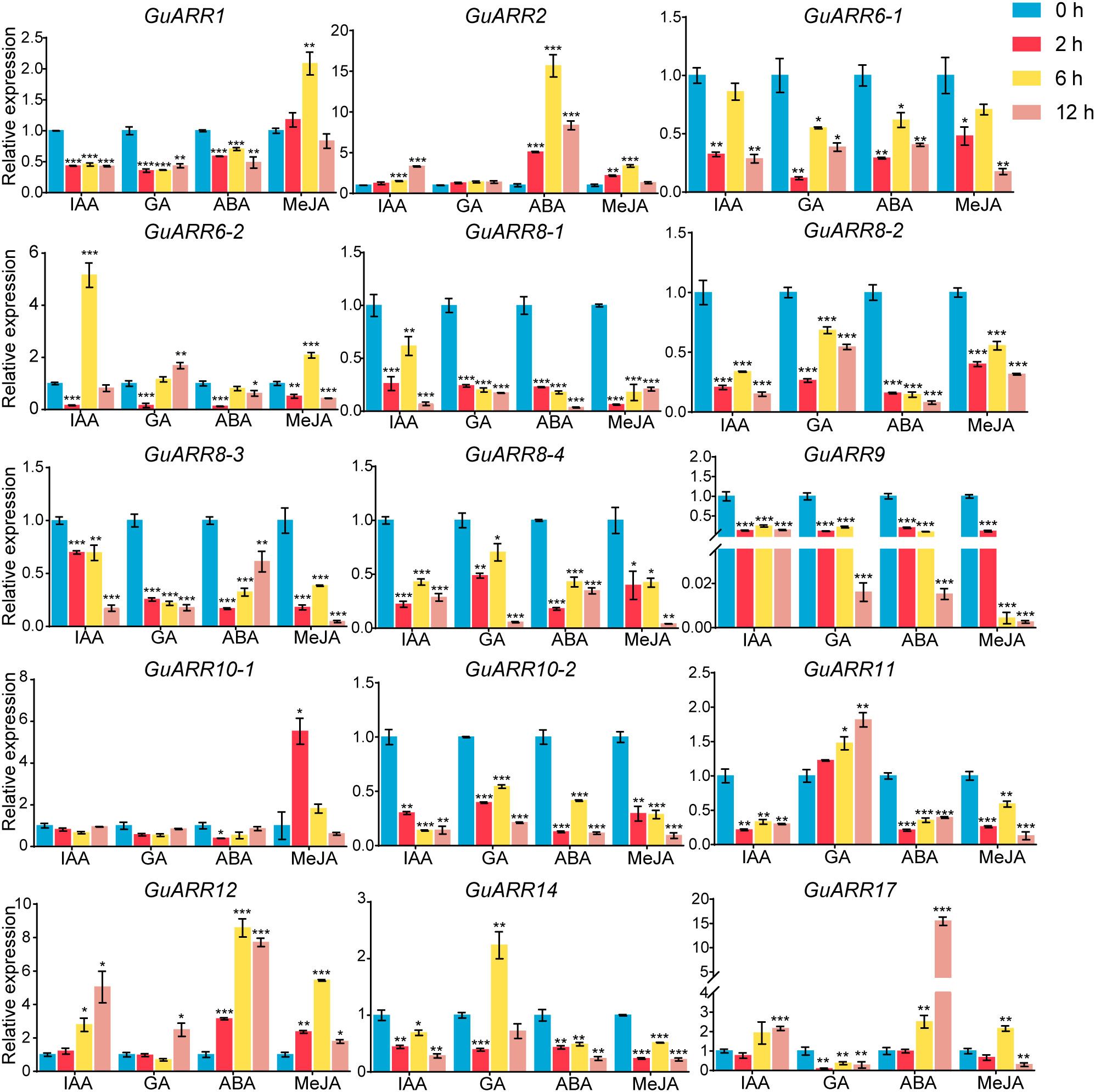
Figure 10 Expression profiles of GuARRs in response to IAA, GA, ABA and MeJA treatments based on qRT-PCR analysis. Samples were collected at 0 h, 2 h, 6 h and 12 h after the four kinds of hormone treatment. Student’s t-test was used to assess significant differences. Significance levels: *, p < 0.05; **, p < 0.01,***, p < 0.001.
3.8 The B-type GuARRs bind to the promoters of A-type GuARRs
To reveal the regulatory effect of B-type ARRs on A-type ARR genes, the binding between B-type licorice ARRs and A-type licorice ARRs promoters was examined using the yeast one-hybrid assay. The coding sequences of seven B-type GuARRs, namely GuARR11, GuARR2, GuARR1, GuARR10-1, GuARR10-2, GuARR12 and GuARR14 were inserted into the pJG45 vector as prey. Additionally, the promoters of eight GuARRs (GuARR17, GuARR8-1/2/3/4, GuARR6-1/2, GuARR9), whose expressions were induced under zeatin treatment, were cloned into the pLacZi vector as bait. And the lacZ reporter activity was firmly activated when GuARR8-3 was concurrently transformed into the yeast cells with GuARR1, GuARR2, GuARR11, GuARR12, GuARR10-1, GuARR10-2, GuARR14 and GuARR8-1 with GuARR1, GuARR12. In addition, the lacZ reporter activity was activated when GuARR6-1 was concurrently transformed with GuARR12, GuARR10-2 as well as GuARR6-2 with GuARR1, GuARR2, GuARR11, GuARR10-2 (Figure 11). These findings provide evidence of the binding between B-type licorice ARRs and A-type licorice ARRs promoters, supporting the regulatory effect of B-type licorice ARRs on A-type licorice ARRs.
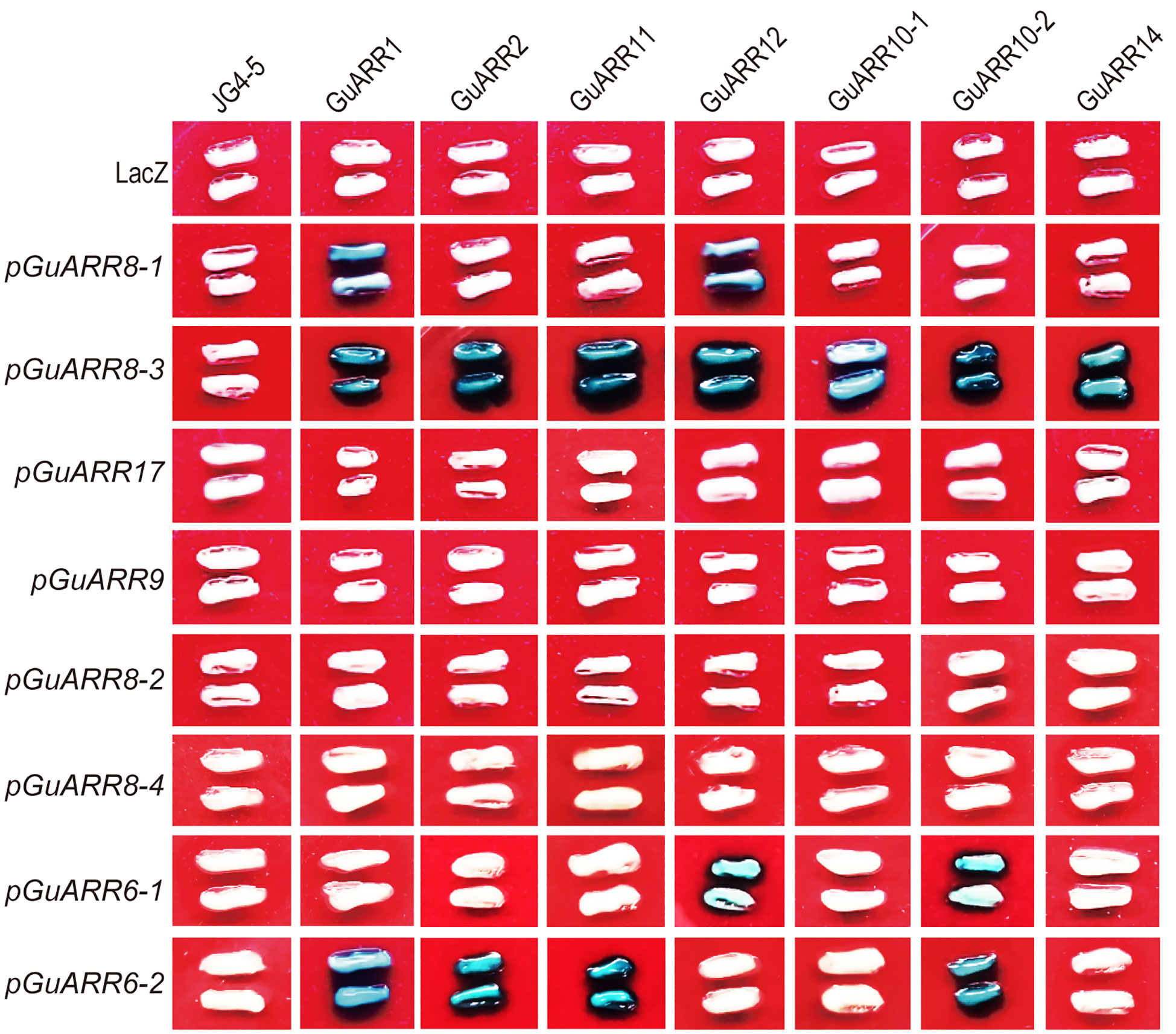
Figure 11 Systematic yeast one-hybrid assay showing the binding of B-type GuARRs to A-type GuARRs promoters. Blue yeast indicates the existence of the interaction.
4 Discussion
Cytokinins are a kind of important plant hormones involved in the regulation of plant growth, development, and plant responses to stresses (Kieber and Schaller, 2018). Authentic response regulators (ARRs) are essential regulators of the widely used His-Asp phosphorelay system present in the cytokinin signaling (Hwang et al., 2012). With the development of bioinformatics, the ARR family gene members in some plants, including Arabidopsis, rice, tobacco, maize, and wild soybean have been studied (Chu et al., 2011; Hwang et al., 2012; Chen et al., 2018; Lv et al., 2021; Rehman et al., 2022). The B-type ARR families have also been analyzed in tomato, B. napus, and peach (Zeng et al., 2017; Wang et al., 2020; Jiang et al., 2022). However, the ARR family in licorice has not been examined. In this study, 17, 17, and 17 ARRs were identified and characterized from G. glabra, G. uralensis and G. inflata respectively, based on the presence of two conserved domains. The physicochemical properties, phylogenetic tree, chromosome locations, gene structures, structural domains, motif distribution, duplication events, cis-acting elements, and expression profiles after treatment with different hormones and abiotic stresses of these ARRs were analyzed, providing a basis for further research on ARR genes.
There were 35, 23, and 12 B-type ARRs identified in B. napus, peach, and tomato, respectively. Arabidopsis, rice, tobacco, maize, and wild soybean contained 21, 28, 59, 28, and 56 ARRs, respectively. The fewer ARRs in licorice than in other species may be a result of homologous gene loss during chromosome replication processes, which was similar in the Jatropha curcas (Geng et al., 2022). The length of amino acid sequences (124 to 700 aa), theoretical isoelectric point (4.89 to 8.69), and GRAVY varied significantly, suggesting the licorice ARRs were highly complex (Table 1, Supplementary Table S3), and comparable results were also obtained in some other species (Wang et al., 2020; Lv et al., 2021). The GRAVY of these ARRs was all negative (-0.916 to -0.042), indicating their hydrophily. Subcellular localization results exhibited that the licorice ARRs were all located in the nucleus, being similar as that in B. napus (Jiang et al., 2022), which may indicate that ARRs mainly function in nucleus and can interact with other transcription factors to modulate various biological processes. All 51 licorice ARRs had the conserved receiver domains harboring the D-D-K sites, which were also found in ARRs of other species (Lv et al., 2021; Geng et al., 2022). And the 21 B-type ARRs also contained a Myb-like DNA-binding domain, which plays a critical role in interacting with other regulators in cytokinin signaling pathways (Rubio et al., 2001; Safi et al., 2017). The locations of licorice ARR genes on chromosomes from G. glabra and G. inflata were similar, as they were all located on chr2, 3, 5, 6, 7, with chr2 and chr3 containing the most number of genes, and chr5 having the fewest number of 1 gene. Chr6 contained 4 genes, and chr7 contained 2 genes. Chr2 of G. uralensis also had the most number of 6 genes, and chr7 of G. uralensis had 2 genes (Figure 4). The results of chromosomal locations indicated the similarity of the 3 kinds of licorice species.
According to the phylogenetic tree, the 51 licorice ARRs were subdivided into three groups, with A-, B-, and C-type ARRs included in the three groups, respectively, which was consistent with the Arabidopsis ARRs (Kiba et al., 2004). Most of the A-type ARRs had 5 exons with 4 introns and the 3’ and 5’ UTR. Most B-type ARRs contained 6 exons with 5 introns and the 3’ and 5’ UTR, while the C-type ARRs contained only 2 exons without UTR regions (Figure 3D). The exons of B-type ARRs were longer than that of the other types of ARRs. Recently, introns have been proved to play an essential part in regulating gene expression (Monteuuis et al., 2019). The varied intron number among different types of ARRs might contribute to licorice ARRs evolution and function diversity, which were also found in other species (Jiang et al., 2022). Interestingly, GgARR9-2 is much longer than other members in the entire family, reaching nearly 1.8 kb with a pretty long intron sequence, which might contribute to the biological function of this gene. A similar phenomenon was found in NtARR27 (Lv et al., 2021). Moreover, the gene structure, conserved motifs numbers and distribution of these ARRs were similar in the same group but differed among different groups, thus licorice ARRs in the same group may possess similar biological functions. Gene duplication events act as a significant factor in expansions and realignments of genomes (Rehman et al., 2022). Plenty of transcription factor families have been reported to experience gene duplication events, like bHLH, NAC, and R2R3-MYB families (He et al., 2022; Liang et al., 2023; Yang et al., 2023). We found 3, 5, and 6 pairs of segmental duplication events in GuARRs, GiARRs, and GgARRs, respectively (Figure 5), while there were no tandem duplications identified, indicating that segmental duplications were the main driving force for the expansion of licorice ARR genes. Additionally, comparative syntenic maps among the three licorices were constructed. The results showed that 13 GuARRs had syntenic relationships with 15 GgARRs, 15 GuARRs and 15 GiARRs had syntenic relationships, while 15 GgARRs and 13 GiARRs had syntenic relationships (Figure 6). Most of these gene pairs bore a ratio of Ka/Ks below 1 (Supplementary Table S4), indicating that they underwent purifying selection in evolution with relatively conserved functions. The cis-acting elements in licorice ARRs promoter regions were analyzed and the presence of important cis-acting elements were found, showing that licorice ARRs may function in the regulation of plant growth, stress, and hormone responses (Figure 7; Supplementary Table S5). Except GgARR3, all the licorice A-type ARRs promoters contained the MYB or MYB recognition site cis-acting elements, indicating that these A-type licorice ARRs may interact with the B-type licorice ARRs. Promoters of three A-type ARRs (GgARR6-2, GuARR6-2, and GiARR6-2) contained a MYB binding site related to the process of flavonoid biosynthesis, which may bind to the Myb-like DNA-binding domain in B-type ARRs, thereby regulating flavonoid biosynthesis (Mason et al., 2005).
Strongly linked to their biological roles, the expression of genes exhibit diverse profiles under specific environmental conditions. Zeatin is a natural plant cytokinin present in higher plants. According to the RNA_seq data, most of the GuARRs exhibited similar expression patterns in the aboveground part and root under zeatin treatment, which may indicate that the GuARRs had similar regulatory mechanism in aboveground part and root. The expressions of GuARR8-1/2/3, GuARR6-1/2, GuARR9 in the aboveground part and all the A-type GuARRs in the root were significantly induced by zeatin treatment, corresponding to their characristic of cytokinin response, thus indicating that A-type GuARRs may be involved in the cytokinin signaling (Kieber and Schaller, 2018). The transcription levels of A-type GuARRs under zeatin treatment explored by the qRT-PCR method generally coincided that in the RNA_seq data, while this was less so for the B-type GuARRs. This result was similar to the discovery of a past research in Jatropha curcas (Geng et al., 2022). This may be due to the fact that B-type ARRs expressions were not significantly influenced by cytokinin. Compared with B-type ARRs, A-type ARRs were more susceptible to induction by cytokinin in Arabidopsis and wheat (Gahlaut et al., 2014; Nguyen et al., 2016). The differences between the expression profiles of A- and B-type GuARRs might suggest the diverse functions of A- and B-type GuARRs.
Roots directly perceive the environmental stress underground. Therefore, root samples from licorice treated with PEG, NaCl, and various phytohormones were used for gene expression analyses response to abiotic stresses and phytohormones using the qRT-PCR method. Plenty of studies have shown that ARR genes are involved in responses to abiotic stresses. For example, ARR5 acts as a positive regulation factor in drought tolerance, and overexpression of ARR5 in plants exhibited more tolerant to drought and hypersensitive to ABA (Huang et al., 2018). ARR1 and ARR12 are involved in promoting the sodium to accumulate in the shoots through modulating the transmittion of sodium ions to the shoot from roots in Arabidopsis (Mason et al., 2010). ARR1/10/12 negatively regulates plant drought stress response, and their triple mutants showed the phenotypes of more tolerance to drought than the wild type (Nguyen et al., 2016). In this study, GuARR2, GuARR10-1, and GuARR12 were significantly induced by drought treatment. Thirteen out of 15 detected GuARRs showed increased trends under NaCl treatment, and 11 out of them, including GuARR6-1/2, GuARR8-1/2/3/4, GuARR10-2, GuARR17, GuARR11, GuARR1, and GuARR14, had the highest expression levels after 6 h of NaCl treatment, and then their expressions decreased at 12 h. GuARR12 and GuARR9 were up-regulated at 6 h and 12 h (Figure 9). This result indicated that these GuARRs can be induced by short-term NaCl stress at the proper concentration but reduced by continuous stress, which might be due to the long-term stress causing a certain degree of damage to plants. Overall, these results indicated that the GuARR genes play underlying roles in drought and NaCl stress responses. Phytohormones, being involved in various signal transduction, exert meaningful roles in plant growth and development. Previous reportes indicated that tomato B-type SlARR genes and CcRR genes exhibited different expressions patterns in response to different phytohormones (Wang et al., 2020; Tao et al., 2023). In addition, BuARRs in B. napus were found to respond to ABA treatment (Jiang et al., 2022). To better understand the roles of GuARRs in phytohormone response, the expression levels of GuARRs under four kinds of phytohormone treatments were measured using the qRT-PCR method, the results illustrated that GuARRs exhibited heterogeneous expression patterns under these phytohormone treatments. Interestingly, GuARR2, GuARR12 and GuARR17 showed up-regulated trends under IAA treatment. GuARR11 and GuARR12 showed increased trends under GA treatment (Figure 10). GuARR2 and GuARR12 were markedly induced by ABA and MeJA treatment and GuARR1 was up-regulated after MeJA treatment for 6 h. It was reported that the SlARR-B1/2/12 in tomato reached the highest expression after treating with MeJA for 6 h (Wang et al., 2020). CcRR1/2/12a were up-regulated upon MeJA treatment and CcRR2 was induced by ABA (Tao et al., 2023). These analyses indicated the essential function of these GuARRs in licorice phytohormone responses. GuARRR12 showed noticeable inductions under all four kinds of phytohormones, indicating a vital roles in the response to phytohormones.
The B-type ARRs possess a Myb-like DNA-binding domain besides the N-terminal receiver domain, which are transcriptional activators in the nucleus and can interact with a set of targets, including the A-type ARRs (Mason et al., 2005). In cytokinin signaling, A-type ARRs serve mainly as negative-feedback regulation factors (Kieber and Schaller, 2018). In this study, all of the A-type GuARR genes had the MYB or MYB recognition site cis-acting elements and were induced by zeatin. All eight A-type GuARRs and seven B-type GuARRs were chosen to examine the binding of B-type GuARRs to A-type GuARRs promoters using the yeast one-hybrid assay. The results indicated that GuARR1, GuARR2, GuARR11, GuARR12, GuARR10-1, GuARR10-2, GuARR14 can bind to the promoter of GuARR8-3, and GuARR1, GuARR12 can bind to the GuARR8-1 promoter. GuARR1, GuARR2, GuARR11 and GuARR10-2 can bind to the GuARR6-2 promoter and GuARR12 and GuARR10-2 can bind to the GuARR6-1 promoter and may regulate their transcription levels (Figure 11). This study provide valuable information for further function research and complex regulatory mechanism of licorice ARR family genes and the further development of licorice functional genomics research.
5 Conclusions
In this study, a total of 51 ARRs were identified in G. uralensis, G. inflata, and G. glabra and were subclassified into three types, A-, B-, and C-type ARRs. A comprehensive analysis of these ARRs, including their evolutionary relationships, physicochemical characteristics, gene structures, chromosomal distributions and cis-acting elements was conducted. Notably, several stress and hormone response cis-elements were found in these licorice ARR genes promoters, showing their latent involvement in stress and hormone signaling pathways. Segmental duplication events were revealed to play a significant role in the licorice ARR genes expansion. These events likely favour to the diversity and complexity of the licorice ARR gene family. Furthermore, the temporospatial expression patterns of GuARRs response to abiotic stress and phytohormones were further characterized by the way of RNA-seq and qRT-PCR analysis, thus suggesting the wide involvement of GuARRs in responses to PEG, NaCl stress, and various phytohormones. Additionally, A- and B-type GuARRs function differently in cytokinin signaling, emphasizing the importance of understanding the distinct roles of different types of ARRs in plant development and stress responses. Lastly, the yeast one-hybrid assay provided evidence that GuARR1, GuARR2, GuARR11, GuARR12, GuARR10-1, GuARR10-2 and GuARR14 binds to the promoter of GuARR8-3, and GuARR1, GuARR12 are able to bind to the promoter of GuARR8-1. GuARR1, GuARR2, GuARR11 and GuARR10-2 can bind to the GuARR6-2 promoter and GuARR12 and GuARR10-2 can bind to the GuARR6-1 promoter. Overall, these findings shed an important light on the function and regulatory mechanisms of ARR genes in Chinese licorice. Further research is required to fully elucidate their functions and roles in plant growth, development, and stress responses. The insights gained from this study will be devoted to a deeper understanding of cytokinin signaling and its impact on plant physiology, which could have significant implications for agricultural and biotechnological applications.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
Author contributions
YS: Formal analysis, Investigation, Writing – original draft. GD: Formal analysis, Investigation, Writing – original draft, Writing – review & editing. HS: Formal analysis, Writing – review & editing. ZL: Writing – review & editing. HL: Writing – review & editing, Funding acquisition, Project administration, Supervision. GX: Funding acquisition, Project administration, Supervision, Writing – review & editing, Conceptualization.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study was supported by Science and Technology Project of Bingtuan (2023AB052), International Science and Technology Cooperation Project of Bingtuan (2020BC002), Science and Technology Project of Huyanghe city (QS2023008), Science and Technology Project of Shihezi University (CXPY202006).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1309802/full#supplementary-material
References
Aipire, A., Li, J., Yuan, P., He, J., Hu, Y., Liu, L., et al. (2017). Glycyrrhiza uralensis water extract enhances dendritic cell maturation and antitumor efficacy of HPV dendritic cell-based vaccine. Sci. Rep. 7, 43796. doi: 10.1038/srep43796
Aloni, R., Aloni, E., Langhans, M., Ullrich, C. I. (2006). Role of cytokinin and auxin in shaping root architecture: regulating vascular differentiation, lateral root initiation, root apical dominance and root gravitropism. Ann. Bot. 97, 883–893. doi: 10.1093/aob/mcl027
Argyros, R. D., Mathews, D. E., Chiang, Y. H., Palmer, C. M., Thibault, D. M., Etheridge, N., et al. (2008). Type B response regulators of Arabidopsis play key roles in cytokinin signaling and plant development. Plant Cell 20, 2102–2116. doi: 10.1105/tpc.108.059584
Bailey, T. L., Johnson, J., Grant, C. E., Noble, W. S. (2015). The MEME suite. Nucleic Acids Res. 43, W39–W49. doi: 10.1093/nar/gkv416
Buechel, S., Leibfried, A., To, J. P., Zhao, Z., Andersen, S. U., Kieber, J. J., et al. (2010). Role of A-type ARABIDOPSIS RESPONSE REGULATORS in meristem maintenance and regeneration. Eur. J. Cell Biol. 89, 279–284. doi: 10.1016/j.ejcb.2009.11.016
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chen, C., Liu, A., Ren, H., Yu, Y., Duanmu, H., Duan, X., et al. (2018). Genome-wide analysis of Glycine soja Response Regulator GsRR genes under alkali and salt stresses. Front. Plant Sci. 9. doi: 10.3389/fpls.2018.01306
Choi, J., Huh, S. U., Kojima, M., Sakakibara, H., Paek, K. H., Hwang, I. (2010). The cytokinin-activated transcription factor ARR2 promotes plant immunity via TGA3/NPR1-dependent salicylic acid signaling in Arabidopsis. Dev. Cell 19, 284–295. doi: 10.1016/j.devcel.2010.07.011
Chu, Z. X., Ma, Q., Lin, Y. X., Tang, X. L., Zhou, Y. Q., Zhu, S. W., et al. (2011). Genome-wide identification, classification, and analysis of two-component signal system genes in maize. Genet. Mol. Res. 10, 3316–3330. doi: 10.4238/2011.December.8.3
El-Gebali, S., Mistry, J., Bateman, A., Eddy, S. R., Luciani, A., Potter, S. C., et al. (2019). The Pfam protein families database in 2019. Nucleic Acids Res. 47, D427–D432. doi: 10.1093/nar/gky995
Ferreira, F. J., Kieber, J. J. (2005). Cytokinin signaling. Curr. Opin. Plant Biol. 8, 518–525. doi: 10.1016/j.pbi.2005.07.013
Gahlaut, V., Mathur, S., Dhariwal, R., Khurana, J. P., Tyagi, A. K., Balyan, H. S., et al. (2014). A multi-step phosphorelay two-component system impacts on tolerance against dehydration stress in common wheat. Funct. Integr. Genomics 14, 707–716. doi: 10.1007/s10142-014-0398-8
Gasteiger, E., Gattiker, A., Hoogland, C., Ivanyi, I., Appel, R. D., Bairoch, A. (2003). ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 31, 3784–3788. doi: 10.1093/nar/gkg563
Geng, X., Zhang, C., Wei, L., Lin, K., Xu, Z. F. (2022). Genome-wide identification and expression analysis of cytokinin response regulator (RR) genes in the woody plant Jatropha curcas and functional analysis of JcRR12 in Arabidopsis. Int. J. Mol. Sci. 23, 11388. doi: 10.3390/ijms231911388
He, F., Zhang, L., Zhao, G., Kang, J., Long, R., Li, M., et al. (2022). Genome-wide identification and expression analysis of the NAC gene family in alfalfa revealed its potential roles in response to multiple abiotic stresses. Int. J. Mol. Sci. 23, 10015. doi: 10.3390/ijms231710015
Heyl, A., Schmülling, T. (2003). Cytokinin signal perception and transduction. Curr. Opin. Plant Biol. 6, 480–488. doi: 10.1016/S1369-5266(03)00087-6
Hill, K., Mathews, D. E., Kim, H. J., Street, I. H., Wildes, S. L., Chiang, Y. H., et al. (2013). Functional characterization of type-B response regulators in the Arabidopsis cytokinin response. Plant Physiol. 162, 212–224. doi: 10.1104/pp.112.208736
Hosoda, K., Imamura, A., Katoh, E., Hatta, T., Tachiki, M., Yamada, H., et al. (2002). Molecular structure of the GARP family of plant Myb-related DNA binding motifs of the Arabidopsis response regulators. Plant Cell 14, 2015–2029. doi: 10.1105/tpc.002733
Huang, X., Hou, L., Meng, J., You, H., Li, Z., Gong, Z., et al. (2018). The antagonistic action of abscisic acid and cytokinin signaling mediates drought stress response in Arabidopsis. Mol. Plant 11, 970–982. doi: 10.1016/j.molp.2018.05.001
Huang, X., Zhang, X., Gong, Z., Yang, S., Shi, Y. (2017). ABI4 represses the expression of type-A ARRs to inhibit seed germination in Arabidopsis. Plant J. 89, 354–365. doi: 10.1111/tpj.13389
Hutchison, C. E., Li, J., Argueso, C., Gonzalez, M., Lee, E., Lewis, M. W., et al. (2006). The Arabidopsis histidine phosphotransfer proteins are redundant positive regulators of cytokinin signaling. Plant Cell 18, 3073–3087. doi: 10.1105/tpc.106.045674
Hwang, I., Chen, H. C., Sheen, J. (2002). Two-component signal transduction pathways in Arabidopsis. Plant Physiol. 129, 500–515. doi: 10.1104/pp.005504
Hwang, I., Sheen, J. (2001). Two-component circuitry in Arabidopsis cytokinin signal transduction. Nature 413, 383–389. doi: 10.1038/35096500
Hwang, I., Sheen, J., Müller, B. (2012). Cytokinin signaling networks. Annu. Rev. Plant Biol. 63, 353–380. doi: 10.1146/annurev-arplant-042811-105503
Imamura, A., Hanaki, N., Nakamura, A., Suzuki, T., Taniguchi, M., Kiba, T., et al. (1999). Compilation and characterization of Arabidopsis thaliana response regulators implicated in His-Asp phosphorelay signal transduction. Plant Cell Physiol. 40, 733–742. doi: 10.1093/oxfordjournals.pcp.a029600
Imamura, A., Kiba, T., Tajima, Y., Yamashino, T., Mizuno, T. (2003). In vivo and in vitro characterization of the ARR11 response regulator implicated in the His-to-Asp phosphorelay signal transduction in Arabidopsis thaliana. Plant Cell Physiol. 44, 122–131. doi: 10.1093/pcp/pcg014
Inoue, T., Higuchi, M., Hashimoto, Y., Seki, M., Kobayashi, M., Kato, T., et al. (2001). Identification of CRE1 as a cytokinin receptor from Arabidopsis. Nature 409, 1060–1063. doi: 10.1038/35059117
Jeon, J., Kim, J. (2013). Arabidopsis response regulator1 and Arabidopsis histidine phosphotransfer protein2 (AHP2), AHP3, and AHP5 function in cold signaling. Plant Physiol. 161, 408–424. doi: 10.1104/pp.112.207621
Jiang, J. J., Li, N., Chen, W. J., Wang, Y., Rong, H., Xie, T., et al. (2022). Genome-wide analysis of the type-B authentic response regulator gene family in Brassica napus. Genes (Basel) 13, 1449. doi: 10.3390/genes13081449
Kakimoto, T. (2003). Perception and signal transduction of cytokinins. Annu. Rev. Plant Biol. 54, 605–627. doi: 10.1146/annurev.arplant.54.031902.134802
Kiba, T., Aoki, K., Sakakibara, H., Mizuno, T. (2004). Arabidopsis response regulator, ARR22, ectopic expression of which results in phenotypes similar to the wol cytokinin-receptor mutant. Plant Cell Physiol. 45, 1063–1077. doi: 10.1093/pcp/pch128
Kieber, J. J., Schaller, G. E. (2018). Cytokinin signaling in plant development. Development 145, dev149344. doi: 10.1242/dev.149344
Kim, K., Ryu, H., Cho, Y. H., Scacchi, E., Sabatini, S., Hwang, I. (2012). Cytokinin-facilitated proteolysis of ARABIDOPSIS RESPONSE REGULATOR 2 attenuates signaling output in two-component circuitry. Plant J. 69, 934–945. doi: 10.1111/j.1365-313X.2011.04843.x
Kumar, S., Stecher, G., Tamura, K. (2016). MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Lee, K., Park, O. S., Go, J. Y., Yu, J., Han, J. H., Kim, J., et al. (2021). Arabidopsis ATXR2 represses de novo shoot organogenesis in the transition from callus to shoot formation. Cell Rep. 37, 109980. doi: 10.1016/j.celrep.2021.109980
Lescot, M., Déhais, P., Thijs, G., Marchal, K., Moreau, Y., Van De Peer, Y., et al. (2002). PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 30, 325–327. doi: 10.1093/nar/30.1.325
Liang, J., Fang, Y., An, C., Yao, Y., Wang, X., Zhang, W., et al. (2023). Genome-wide identification and expression analysis of the bHLH gene family in passion fruit (Passiflora edulis) and its response to abiotic stress. Int. J. Biol. Macromol. 225, 389–403. doi: 10.1016/j.ijbiomac.2022.11.076
Lv, J., Dai, C. B., Wang, W. F., Sun, Y. H. (2021). Genome-wide identification of the ARRs gene family in tobacco (Nicotiana tabacum). Genes Genomics 43, 601–612. doi: 10.1007/s13258-021-01039-6
Mason, M. G., Jha, D., Salt, D. E., Tester, M., Hill, K., Kieber, J. J., et al. (2010). Type-B response regulators ARR1 and ARR12 regulate expression of AtHKT1;1 and accumulation of sodium in Arabidopsis shoots. Plant J. 64, 753–763. doi: 10.1111/j.1365-313X.2010.04366.x
Mason, M. G., Mathews, D. E., Argyros, D. A., Maxwell, B. B., Kieber, J. J., Alonso, J. M., et al. (2005). Multiple type-B response regulators mediate cytokinin signal transduction in Arabidopsis. Plant Cell 17, 3007–3018. doi: 10.1105/tpc.105.035451
Miller, C. O., Skoog, F., Okumura, F. S., Von Saltza, M. H., Strong, F. M. (1956). Isolation, structure and synthesis of kinetin, a substance promoting cell division. J. Am. Chem. Soc 78, 1375–1380. doi: 10.1021/ja01588a032
Miller, C. O., Skoog, F., Von Saltza, M. H., Strong, F. M. (1955). Kinetin, a cell division factor from deoxyribonucleic acid. J. Am. Chem. Soc 77, 1392. doi: 10.1021/ja01610a105
Mizuno, T. (2004). Plant response regulators implicated in signal transduction and circadian rhythm. Curr. Opin. Plant Biol. 7, 499–505. doi: 10.1016/j.pbi.2004.07.015
Monteuuis, G., Wong, J. J. L., Bailey, C. G., Schmitz, U., Rasko, J. E. J. (2019). The changing paradigm of intron retention: regulation, ramifications and recipes. Nucleic Acids Res. 47, 11497–11513. doi: 10.1093/nar/gkz1068
Müller, B., Sheen, J. (2007). Advances in cytokinin signaling. Science 318, 68–69. doi: 10.1126/science.1145461
National Pharmacopoeia Committee (2010). Pharmacopoeia of People’s Republic of China (Beijing: Chemical Industry Press).
Nguyen, K. H., Ha, C. V., Nishiyama, R., Watanabe, Y., Leyva-González, M. A., Fujita, Y., et al. (2016). Arabidopsis type B cytokinin response regulators ARR1, ARR10, and ARR12 negatively regulate plant responses to drought. Proc. Natl. Acad. Sci. U. S. A. 113, 3090–3095. doi: 10.1073/pnas.1600399113
Osakabe, Y., Miyata, S., Urao, T., Seki, M., Shinozaki, K., Yamaguchi-Shinozaki, K. (2002). Overexpression of Arabidopsis response regulators, ARR4/ATRR1/IBC7 and ARR8/ATRR3, alters cytokinin responses differentially in the shoot and in callus formation. Biochem. Biophys. Res. Commun. 293, 806–815. doi: 10.1016/s0006-291x(02)00286-3
Potter, S. C., Luciani, A., Eddy, S. R., Park, Y., Lopez, R., Finn, R. D. (2018). HMMER web server: 2018 update. Nucleic Acids Res. 46, W200–W204. doi: 10.1093/nar/gky448
Rehman, O. U., Uzair, M., Chao, H., Fiaz, S., Khan, M. R., Chen, M. (2022). Role of the type-B authentic response regulator gene family in fragrant rice under alkaline salt stress. Physiol. Plant 174, e13696. doi: 10.1111/ppl.13696
Rubio, V., Linhares, F., Solano, R., Martín, A. C., Iglesias, J., Leyva, A., et al. (2001). A conserved MYB transcription factor involved in phosphate starvation signaling both in vascular plants and in unicellular algae. Genes Dev. 15, 2122–2133. doi: 10.1101/gad.204401
Safi, A., Medici, A., Szponarski, W., Ruffel, S., Lacombe, B., Krouk, G. (2017). The world according to GARP transcription factors. Curr. Opin. Plant Biol. 39, 159–167. doi: 10.1016/j.pbi.2017.07.006
Salomé, P. A., To, J. P., Kieber, J. J., Mcclung, C. R. (2006). Arabidopsis response regulators ARR3 and ARR4 play cytokinin-independent roles in the control of circadian period. Plant Cell 18, 55–69. doi: 10.1105/tpc.105.037994
Schaller, G. E., Doi, K., Hwang, I., Kieber, J. J., Khurana, J. P., Kurata, N., et al. (2007). Nomenclature for two-component signaling elements of rice. Plant Physiol. 143, 555–557. doi: 10.1104/pp.106.093666
Schaller, G. E., Kieber, J. J., Shiu, S. H. (2008). Two-component signaling elements and histidyl-aspartyl phosphorelays. Arabidopsis Book 6, e0112. doi: 10.1199/tab.0112
Srivastava, A. K., Dutta, S., Chattopadhyay, S. (2019). MYC2 regulates ARR16, a component of cytokinin signaling pathways, in Arabidopsis seedling development. Plant Direct 3, e00177. doi: 10.1002/pld3.177
Sun, Z. G., Zhao, T. T., Lu, N., Yang, Y. A., Zhu, H. L. (2019). Research progress of glycyrrhizic acid on antiviral activity. Mini Rev. Med. Chem. 19, 826–832. doi: 10.2174/1389557519666190119111125
Tang, Z. H., Li, T., Tong, Y. G., Chen, X. J., Chen, X. P., Wang, Y. T., et al. (2015). A systematic review of the anticancer properties of compounds isolated from licorice (Gancao). Planta Med. 81, 1670–1687. doi: 10.1055/s-0035-1558227
Tao, S. C., Zhao, L., Mei, J. Q., Abbas, F., Xie, X. T., Yang, Y., et al. (2023). Genome wide identifcation and expression analysis of response regulators family genes in Chinese hickory (Carya cathayensis) suggests their potential roles during grafting. J. Plant Growth Regul. 42, 5099–5115. doi: 10.1007/s00344-022-10898-6
To, J. P., Deruère, J., Maxwell, B. B., Morris, V. F., Hutchison, C. E., Ferreira, F. J., et al. (2007). Cytokinin regulates type-A Arabidopsis Response Regulator activity and protein stability via two-component phosphorelay. Plant Cell 19, 3901–3914. doi: 10.1105/tpc.107.052662
To, J. P., Haberer, G., Ferreira, F. J., Deruère, J., Mason, M. G., Schaller, G. E., et al. (2004). Type-A Arabidopsis response regulators are partially redundant negative regulators of cytokinin signaling. Plant Cell 16, 658–671. doi: 10.1105/tpc.018978
Vatén, A., Soyars, C. L., Tarr, P. T., Nimchuk, Z. L., Bergmann, D. C. (2018). Modulation of asymmetric division diversity through cytokinin and SPEECHLESS regulatory interactions in the Arabidopsis stomatal lineage. Dev. Cell 47, 53–66.e55. doi: 10.1016/j.devcel.2018.08.007
Wang, J., Xia, J., Song, Q., Liao, X., Gao, Y., Zheng, F., et al. (2020). Genome-wide identification, genomic organization and expression profiles of SlARR-B gene family in tomato. J. Appl. Genet. 61, 391–404. doi: 10.1007/s13353-020-00565-5
Wang, L., Zhang, K., Han, S., Zhang, L., Bai, H., Bao, F., et al. (2019). Constituents isolated from the leaves of Glycyrrhiza uralansis and their anti-inflammatory activities on LPS-induced RAW264.7 cells. Molecules 24, 1923. doi: 10.3390/molecules24101923
Wang, D., Zhang, Y., Zhang, Z., Zhu, J., Yu, J. (2010). KaKs_Calculator 2.0: a toolkit incorporating gamma-series methods and sliding window strategies. Genomics Proteomics Bioinf. 8, 77–80. doi: 10.1016/s1672-0229(10)60008-3
Worthen, J. M., Yamburenko, M. V., Lim, J., Nimchuk, Z. L., Kieber, J. J., Schaller, G. E. (2019). Type-B response regulators of rice play key roles in growth, development and cytokinin signaling. Development 146, dev174870. doi: 10.1242/dev.174870
Yang, L., Jiang, Y., Zhang, Z., Hou, J., Tian, S., Liu, Y. (2020). The anti-diabetic activity of licorice, a widely used Chinese herb. J. Ethnopharmacol. 263, 113216. doi: 10.1016/j.jep.2020.113216
Yang, Y., Zhu, J., Wang, H., Guo, D., Wang, Y., Mei, W., et al. (2023). Systematic investigation of the R2R3-MYB gene family in Aquilaria sinensis reveals a transcriptional repressor AsMYB054 involved in 2-(2-phenylethyl) chromone biosynthesis. Int. J. Biol. Macromol. 244, 125302. doi: 10.1016/j.ijbiomac.2023.125302
Yokoyama, A., Yamashino, T., Amano, Y., Tajima, Y., Imamura, A., Sakakibara, H., et al. (2007). Type-B ARR transcription factors, ARR10 and ARR12, are implicated in cytokinin-mediated regulation of protoxylem differentiation in roots of Arabidopsis thaliana. Plant Cell Physiol. 48, 84–96. doi: 10.1093/pcp/pcl040
Zdarska, M., Cuyacot, A. R., Tarr, P. T., Yamoune, A., Szmitkowska, A., Hrdinová, V., et al. (2019). ETR1 integrates response to ethylene and cytokinins into a single multistep phosphorelay pathway to control root growth. Mol. Plant 12, 1338–1352. doi: 10.1016/j.molp.2019.05.012
Zeng, J., Zhu, X., Haider, M. S., Wang, X., Zhang, C., Wang, C. (2017). Genome-wide identification and analysis of the type-B authentic response regulator gene family in peach (Prunus persica). Cytogenet. Genome Res. 151, 41–49. doi: 10.1159/000458170
Zhang, Z., Yang, L., Hou, J., Tian, S., Liu, Y. (2021). Molecular mechanisms underlying the anticancer activities of licorice flavonoids. J. Ethnopharmacol. 267, 113635. doi: 10.1016/j.jep.2020.113635
Zubo, Y. O., Blakley, I. C., Yamburenko, M. V., Worthen, J. M., Street, I. H., Franco-Zorrilla, J. M., et al. (2017). Cytokinin induces genome-wide binding of the type-B response regulator ARR10 to regulate growth and development in Arabidopsis. Proc. Natl. Acad. Sci. U. S. A. 114, E5995–E6004. doi: 10.1073/pnas.1620749114
Keywords: ARR gene family, licorice, evolutionary analyses, expression patterns, stress responses, hormone
Citation: Shi Y, Ding G, Shen H, Li Z, Li H and Xiao G (2024) Genome-wide identification and expression profiles analysis of the authentic response regulator gene family in licorice. Front. Plant Sci. 14:1309802. doi: 10.3389/fpls.2023.1309802
Received: 08 October 2023; Accepted: 18 December 2023;
Published: 10 January 2024.
Edited by:
Ertugrul Filiz, Duzce University, TürkiyeReviewed by:
Muhammad Uzair, National Institute for Genomics and Advanced Biotechnology (NIGAB), PakistanXue Yang, Anhui University of Science and Technology, China
Copyright © 2024 Shi, Ding, Shen, Li, Li and Xiao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hongbin Li, bGloYkBzaHp1LmVkdS5jbg==; Guanghui Xiao, Z3VhbmdodWl4QHNubnUuZWR1LmNu
†These authors have contributed equally to this work
 Yanping Shi
Yanping Shi Guohua Ding
Guohua Ding Haitao Shen
Haitao Shen Zihan Li
Zihan Li Hongbin Li
Hongbin Li Guanghui Xiao
Guanghui Xiao