
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 22 November 2023
Sec. Plant Abiotic Stress
Volume 14 - 2023 | https://doi.org/10.3389/fpls.2023.1308721
This article is part of the Research Topic Surviving and Thriving: How Crops Perceive and Respond to Temperature Stress View all 13 articles
TIFY [TIF(F/Y)XG] proteins are a plant particular transcription factor family that regulates plant stress responses. Therefore, to fill this gap, we investigated CaTIFY genes in pepper. Gene structure and conserved motifs of the pepper TIFY gene family were systematically analyzed using sequence alignment analysis, Cis-acting element analysis, transcriptomic data, and RT-qPCR analysis, and their expression patterns were further analyzed using Virus-Induced Gene Silencing (VIGS) and cold stress reactive oxygen species (ROS) response. We identified 16 CaTIFY genes in pepper, which were dispersed among seven subgroups (JAZI, JAZII, JAZIII, PPD, TIFY, and ZIM/ZML). Several CaTIFY members had stress-related harmonic-responsive elements, and four (CaTIFY7, CaTIFY10b, CaTIFY1b, and CaTIFY6b) had low-temperature-responsive elements. Transcriptomic data and RT-qPCR analysis revealed that the TIFY genes in pepper displayed different expression patterns in the roots, stems, leaves, flower fruits, and seeds. In particular, CaTIFY7 was highly expressed in young leaves, and CaTIFY10b was highly expressed in roots. CaTIFYs participated in the regulation of several different abiotic stresses and CaTIFY7 and CaTIFY10b were significantly induced by cold stress. Additionally, Virus-Induced Gene Silencing (targeting CaTIFY7 and CaTIFY10b) resulted in plants that were sensitive to cold stress. Conversely, overexpression of CaTIFY7 and CaTIFY10b enhanced plant cold tolerance by promoting the expression of genes related to cold stress and the ROS response. CaTIFY7 and CaTIFY10b interacted with themselves and CaTIFY7 also interacted with CaTIFY10b in the yeast two-hybrid (Y2H) system. Our data provide a basis for further analysis of the role of pepper TIFY genes in cold-stress responses in the future.
The TIFY gene family encodes a transcription factor unique to plants characterized by a highly conserved TIFY domain, which plays an important regulatory role in plant growth, development, and stress tolerance. The TIFY domain is composed of 36 amino acids and a TIF[F/Y] XG core region (Vanholme et al., 2007). Many studies have shown that the TIFY family of proteins can be divided into four subfamilies: TIFY, JAZ, PPD, and ZML (Bai et al., 2011). The first TIFY gene was identified in Arabidopsis thaliana (Nishii et al., 2000). Subsequently, the TIFY gene family was identified in rice (Ye et al., 2009), maize (Heidari et al., 2021), cassava (Zheng et al., 2022), tomato (Heidari et al., 2021), and other species (Huang et al., 2016; Sirhindi et al., 2016; Xia et al., 2017; He et al., 2020; Du et al., 2022).
The TIFY protein is a key regulator of the jasmonic acid signaling pathway, participates in defense and stress responses during plant development, and plays a major role in responses to biological and abiotic stresses (Thireault et al., 2015; Ebel et al., 2018). There are few reports on the response of the TIFY family to biotic stress, mainly focusing on the abiotic stress response of the TIFY family. Multiple TIFY genes have been shown to respond to drought and salt stress in arabidopsis, apples, grapes, rice, soybeans, and corn (Ye et al., 2009; Zhang et al., 2012; Li et al., 2015; Zhou et al., 2015; Meng et al., 2019; Zhao et al., 2020). In rice, the overexpression of OsTIFY11a significantly improves its tolerance to salt and dehydration stress (Ye et al., 2009). In addition, OsJAZ1 interacts with OsbHLH148, a transcription regulator in the jasmonic acid signaling pathway, resulting in enhanced drought tolerance in rice (Seo et al., 2011). Among solanaceous plants, only the TIFY genes of tomato have been identified, and transcriptome data show that most SlJAZ genes respond to salt, osmotic stress, or JA-ABA treatments (Chini et al., 2017).
With the rapid development of transcriptome sequencing, an increasing number of TIFY genes have been found to respond to low-temperature stress (Ye et al., 2009; Zhang et al., 2012; Zhang et al., 2015; Huang et al., 2016; He et al., 2020). Most BdTIFY genes are responsive to one or more abiotic stressors, including drought, salinity, low temperature, and heat (Zhang et al., 2015). In grapes, VvJAZ4, 5, 9, 11, and VvZML1 respond to cold stress and are regulated by drought or salinity (Zhang et al., 2012). In Brassica napus, recent research has found that TIFY11b, TIFY10a, TIFY3, TIFY5a/b, and TIFY6 are induced by cold acclimation and overexpression of BnaJAZ7⁃A3/BnaJAZ7⁃C3 enhances cold injury stress (He et al., 2020). In rice, some OsTIFY genes are induced by cold but not induced or even repressed by ABA, which suggests OsTIFY participated in the cold-stress response via ABA-independent signaling pathways (Ye et al., 2009). Studies on Arabidopsis have found that the interaction between JAZ1/JAZ4 and C-repeat binding factor (CBF) inducers inhibits the function of transcriptional ICE1, thereby weakening the expression of CBFs (Huang et al., 2012; Hu et al., 2013). Furthermore, the overexpression of JAZ1 or JAZ4 inhibits the response of Arabidopsis to freezing stress (Hu et al., 2013). However, few studies have revealed the function of the TIFY protein in plant low-temperature responses.
Pepper (Capsicum annum L.) is a versatile vegetable belonging to the Solanaceae family and is an economically important crop worldwide. The average yield loss caused by low temperatures is enormous (Chinnusamy et al., 2007; Legris et al., 2017). Although there have been many advances in understanding the cold resistance mechanisms of crops, there are still few reports on the low-temperature tolerance of pepper. Transcription factors also play an important role in the response of pepper to low-temperature stress; however, only a small number of transcription factor families, including MADS, NAC, and bHLH, have been shown to participate in the low-temperature regulation process in pepper (Guo et al., 2015; Chen et al., 2019; Hou et al., 2020; Zhang et al., 2020; Wang et al., 2022; Zhang et al., 2023). So far, the TIFY protein in pepper has not been identified, and its role in the cold-stress response has not been studied. Therefore, to fill this gap, we investigated CaTIFY genes in pepper.
In this study, we identified 16 genes encoding TIFY proteins in the pepper genome. Chromosomal position, phylogenetics, gene structure, conserved domains, and promoter cis-element analyses were carried out to provide genome-wide identification and investigation of CaTIFY genes. Furthermore, the expression of all CaTIFY genes was investigated in various tissues and organs at different developmental stages and under various abiotic stresses, using published transcriptome data. We also examined their expression profiles after cold treatments and in different tissues by RT-qPCR assays and found CaTIFY7 and CaTIFY10b were induced by cold stress in leaves significantly. Gene function analysis of CaTIFY7 and CaTIFY10b showed that they positively regulated cold-stress tolerance in pepper plants. Further analysis demonstrated that the downregulation or overexpression of CaTIFY7 and CaTIFY10b influenced the expression levels of cold-stress and reactive oxygen species (ROS)-related genes. Further, the Y2H assay proved CaTIFY7 and CaTIFY10b interacted with themselves and CaTIFY7 also interacted with CaTIFY10b. By performing this study, we have laid a foundation for further research on the function and molecular mechanism of the TIFY gene family in the pepper stress response and provide a basis for improving plant stress resistance in the future.
TIFY domain (PF06200) hidden Markov file was downloaded from Pfam database (http://pfam.xfam.org/). HMMER software was used to screen for possible TIFY proteins in the pepper genome and candidate members were submitted to Pfam and SMART (https://smart.embl.de/) online programs to verify the composition of conservative structural domains for further screening.
The CDS and genome sequence of the pepper TIFY gene family were submitted to the GSDS (http://gsds.gao-lab.org/) online website for visual analysis of the intron–exon substructure of the TIFY gene family.
TBtools was used to extract the TIFY protein sequence of pepper and MEGA software was used to perform multiple sequence alignments on the TIFY protein sequence of pepper. The functional domain of the TIFY protein in chili pepper was analyzed based on conserved domain sequence characteristics.
First, we used MEGA5 software to conduct multiple sequence alignments of Arabidopsis, rice, tomato, and identified pepper TIFY protein sequences. Alignment results were used to build a phylogenetic tree based on the maximum parsimony method, and the bootstrap value was set to 1000. Next, the online program Itol (https://itol.embl.de/) was used to further refine the evolutionary tree.
We obtained the genome sequences and GFF files of peppers from the PepperHub database (http://122.205.95.132/index.php) (Liu et al., 2017). TBtools software was used to extract the chromosome length and location information of TIFY genes in the pepper genome from the GFF files and to perform visual analysis.
Conserved motifs in CaTIFYs were predicted using MEME-Suite (https://meme-suite.org/meme/tools/meme) and visualized using TBtools. Specific parameters were set as follows: ‘Any number of repetitions’ was selected as the site distribution, and up to ten motifs were retrieved.
First, 2000 bp upstream DNA sequences of the pepper TIFY gene CDS were extracted from the pepper genome sequence using the TBtools software. We then uploaded these sequences to the PlantCARE database (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) to obtain cis-acting element information on the promoter of CaTIFY genes in pepper. Subsequently, the cis-acting elements related to stress and hormones were classified. TB tools software was used to visualize the analysis of CaTIFY gene promoters in pepper.
The identified BvTIFY protein sequence was uploaded to the STRING database (https://cn.string-db.org/) for prediction of the biological functions of homologous CaTIFY and other proteins and their potential interactions.
RNA-seq data of CaTIFY family members in different tissues and organs (roots, shoots, young leaves, mature leaves, old leaves, flowers, fruits, and seeds) under several abiotic stresses (salt, drought, H2O2, heat, and cold) were obtained from the PepperHub Database. HEMI software was used to visualize the expression levels of CaTIFYs.
Peppers (Capsicum annuum L.) were cultivated under a 25°C temperature and 16/8-h day/night photoperiod. Pepper plants of uniform size were selected and exposed to cold stress (4°C) at the 6-8 true leaf stage in growth chambers. Leaves and roots were sampled at 1, 3, 6, 12, and 24 h after cold treatment.
TRIzol and chloroform reagents were used to extract total RNA from pepper leaves and roots. A HiScript II 1st Strand cDNA Synthesis Kit (+ gDNA wiper) (Vazyme, Nanjing, China) was used for reverse transcription. ChamQ Universal SYBR® qPCR Master Mix (Vazyme) and CFX384 Real-Time PCR Detection System (Bio-Rad, Hercules, CA, USA) were used to perform RT-qPCR. The quantitative results were analyzed by the 2−ΔΔCt method and the CaUBI3 gene (Wan et al., 2011) was used as the internal control for normalizing gene expression. Each experiment was performed in triplicate. The primers used in the qPCR analysis are listed in Supplementary Table 1.
The silence-specific 312 bp fragment of CaTIFY7 was selected and inserted into the pTRV2 vector between the KpnI and XhoI sites to form the pTRV2:CaTIFY7 recombinant plasmid. The pTRV2:CaTIFY7, pTRV2, and pTRV1 were individually transformed into Agrobacterium GV3101 cells. Suspensions of pTRV2:CaTIFY7 and pTRV2 were individually mixed with equal volumes of pTRV1 and injected into the cotyledons of two true-leaf-stage pepper seedlings as described previously (Wang et al., 2013). The same experimental method was used for CaTIFY10b and specific 331 bp fragments of CaTIFY10b were used to build a carrier.
The full-length CaTIFY7 cDNA (Capana01g003720) and CaTIFY10b cDNA (Capana07g000750) were obtained from the Pepper Genome Database. PCR-amplified products of CaTIFY7 and CaTIFY10b were cloned into empty vectors to generate 35S:CaTIFY7 and 35S:CaTIFY10b. The empty vectors 35S:CaTIFY7 and 35S:CaTIFY10b were individually transformed into Agrobacterium GV3101. The suspensions were infiltrated into the leaves of pepper plants at the eight-leaf stage as described previously (Cai et al., 2021). After 48 h, half of the infiltrated plants were moved to 4 °C treatment. All infiltrated leaves were collected for further use 24 h after cold stress.
To assess cold-stress biochemical indices, 3,3’-diaminobenzidine (DAB) and nitro blue tetrazolium staining (Coolaber, Beijing, China) were used to stain the transformed leaves as per the manufacturer’s instructions.
To determine the relationship between CaTIFY7 and CaTIFY10b, we performed a Y2H assay using the MatchmakerTM Two-Hybrid System (Clontech, USA). The CDS of CaTIFY7 and CaTIFY10b were amplified with specific primers and cloned into the pGBKT7-BD vector between the EcoRI and PstI sites and the pGADT7-AD vector between the EcoRI and BamHI sites. The fusion pGBKT7-BD and pGADT7-AD constructs were co-transformed into the Y2H Gold strain and screened for SD/-Leu/-Trp. Then multiple monoclonal clones were mixed and spotted onto SD/-Leu/-Trp/-His/-Ade media at 30°C for 3-5 d. The fusion of pGBKT7-53 and pGADT7-T was used as a positive control.
To identify TIFY members in pepper, the TIFY domain (PF06200) was used to screen for possible TIFY proteins in the pepper genome. 16 TIFY genes were identified in the pepper genome and named CaTIFYs, according to their homologous TIFY genes in Arabidopsis (Figure 1). The numbers of amino acids in CaTIFYs ranged from 135 to 411 and their molecular weights ranged from 14.61 to 45.7 KDa. The theoretical PIs of the CaTIFYs ranged from 4.65 to 10.68 (Table 1). In addition, the subcellular location prediction analysis showed that all CaTIFYs were located in the nucleus.
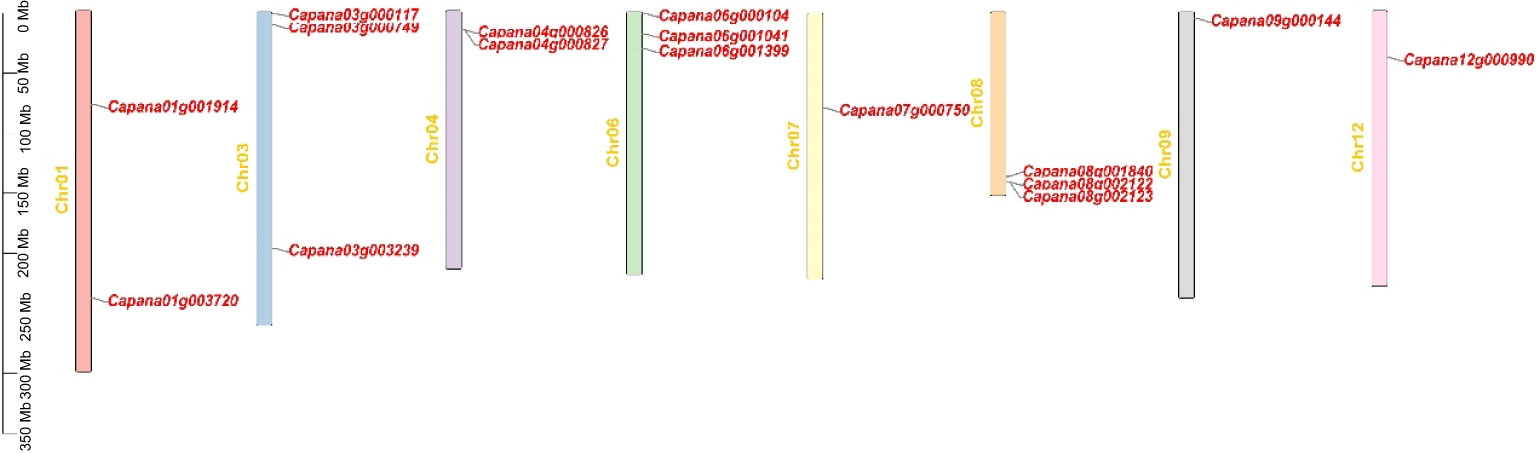
Figure 1 Distribution of the TIFY genes on chromosomes of pepper. The scale on the right is in million bases (Mb).
Multiple sequence comparisons revealed that all members of the TIFY gene family contained TIFY and Jaz domains (Figure 2A). The TIFY domain of the CaTIFY proteins was more conserved than the Jaz domain. CaTIFY3, CaTIFY9, and CaTIFY10d contained almost no Jaz domains (Figure 2A). In addition, conserved motifs were also identified. As shown in Figure 2B, a total of 10 motifs were predicted in CaTIFY proteins using the MEME website and TBtools software. The numbers of CaTIFY motifs ranged from one (CaTIFY8) to five (CaTIFY10b/c). There were four motifs in the seven CaTIFY (CaTIFY1b, CaTIFY1c, CaTIFY2, CaTIFY4a, CaTIFY4b, CaTIFY10a, and CaTIFY10d) proteins and their proportions exceeded 30% of the total proteins. Almost all proteins had motif1 and motif2, except for CaTIFY8, which only had motif1. Proteins in the same clade (for example CaTIFY10a, CaTIFY10b, CaTIFY10c, and CaTIFY10d) contained similar conserved motifs. However, CaTIFY1a had only two motifs, unlike CaTIFY1b and CaTIFY1c which had four motifs.
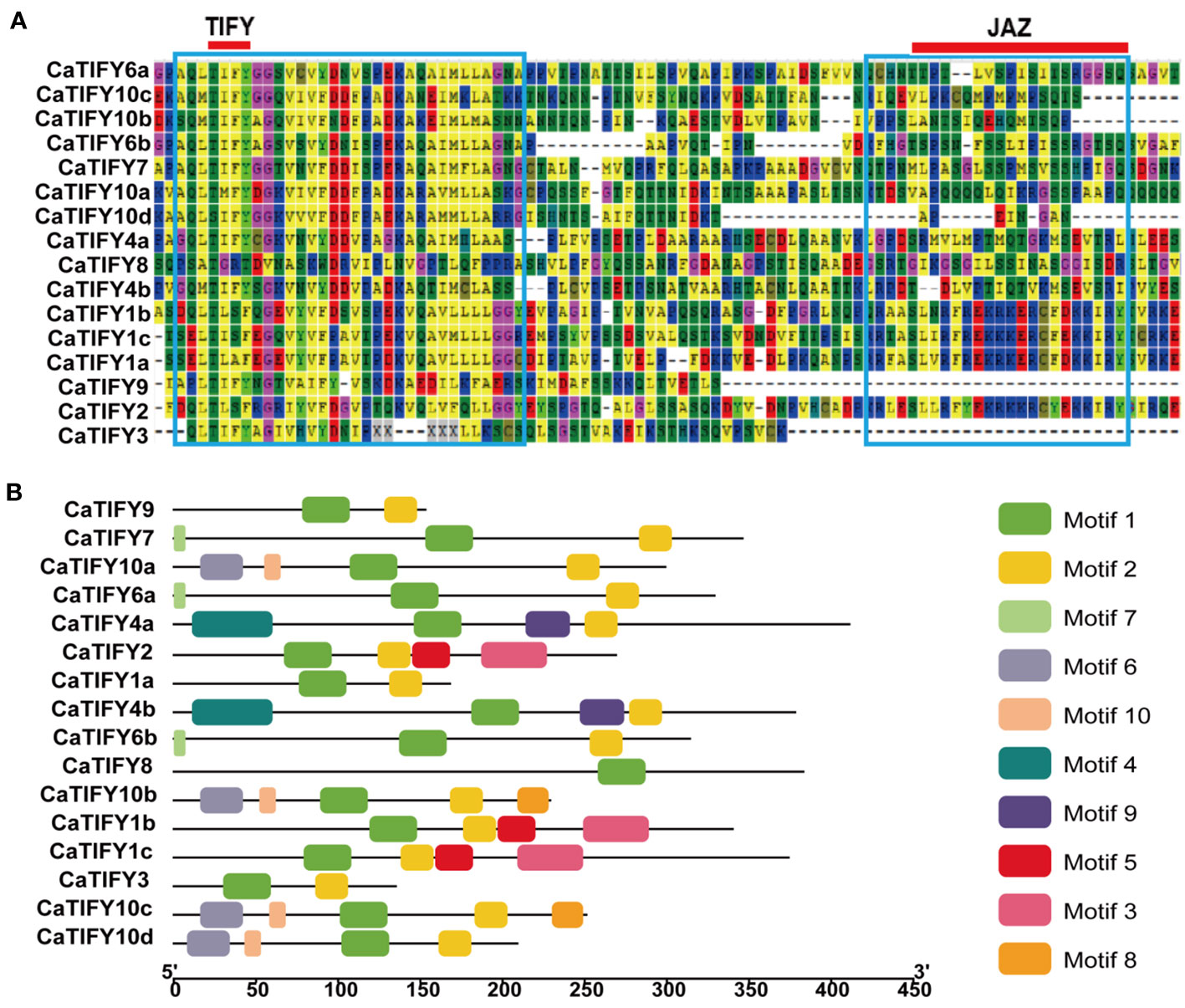
Figure 2 Multiple sequences alignment of CaTIFYs and conserved motif distributions of CaTIFYs proteins in pepper. (A) The blue boxes indicate conserved amino acids. The red line indicate the core amino acids of TIFY and Jas domain. (B) Distribution of the conserved motifs in pepper TIFY proteins. Ten conserved motifs are marked with different colored boxes. The scale bar indicates 50 aa.
To investigate the phylogenetic relationships among the CaTIFYs, Pepper CaTIFY, rice OsTIFY, Arabidopsis AtTIFY, and tomato SlTIFY sequences were used to construct a phylogenetic tree (Figure 3). Phylogenetic analysis showed that CaTIFY proteins could be divided into 11 groups belonging to the JAZ, PPD, TIFY, and ZIM/ZML groups. In pepper, no TIFY gene belonged to JAZ7/8 (the AtTIFY5 homologous gene). Among the CaTIFYs, TIFY10 had the highest number of homologous genes. Rice has 11 homologous genes of TIFY10/11 and there were four homologous genes in pepper. Notably, although tomatoes and peppers are Solanaceae plants, there are only four TIFY homologous genes in tomatoes.
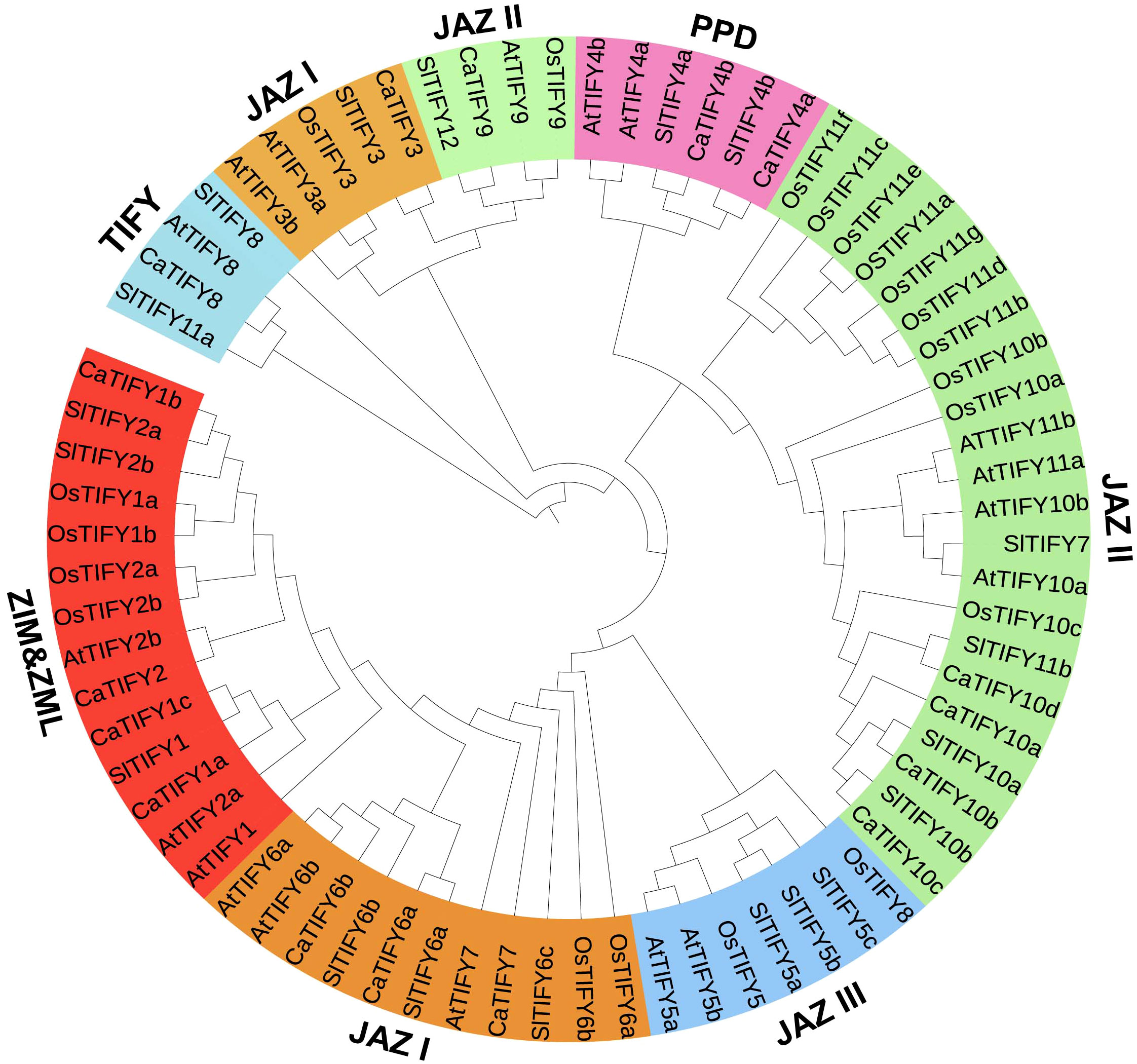
Figure 3 Evolutionary relationship of the TIFY proteins across different plant species. 58 TIFY proteins come from Arabidopsis (18), rice (20), tomato(19), and pepper(16).The resulting 9 groups are classified in four subfamilies, which belong to JAZ, PPD, TIFY and ZIM/ZML.
To understand the structural diversity of TIFY genes, we further analyzed the structure of the CaTIFY gene family. The intron–exon substructures of CaTIFY genes are visualized in Supplementary Figure 1. According to the evolutionary relationships of the CaTIFY protein, there were significant differences in sequence length and structure among genes, indicating that large sequence differences existed among the different CaTIFY genes (Supplementary Figure 1). The number of CaTIFY introns ranged between 1 and 9, of which CaTIFY1c contained nine introns and CaTIFY9 had only one. All other CaTIFY intron numbers ranged from 2 to 8. There were no strong correlations between phylogeny and exons/introns. Genes with high homology did not have similar gene structures and the length of introns varied greatly, indicating significant gene differentiation in the TIFY family in pepper.
Cis-acting elements can affect and predict gene expression in response to stress or hormones. The 2000 bp region upstream of the start codon of each pepper TIFY gene was extracted from the Pepper Genome Database. PlantCARE software was used to analyze the cis-acting elements related to stress or hormones. As shown in Figure 4, 81.2% of the members of the TIFY gene family in peppers had anaerobic induction elements. Of the five hormones, 25% of the CaTIFY members had auxin-responsive elements and 56.2% of the CaTIFY members had ABA-, MeJA-, and gibberellin-responsive elements. Fifty percent of CaTIFY members had a salicylic acid-responsive element. In addition, four CaTIFY members (CaTIFY7, CaTIFY10b, CaTIFY1b and CaTIFY6b) had a low-temperature-responsive element, four CaTIFY members (CaTIFY7, CaTIFY2, CaTIFY8 and CaTIFY10b) had drought induction elements, and four CaTIFY members had defense- and stress-responsive elements (Figure 4).
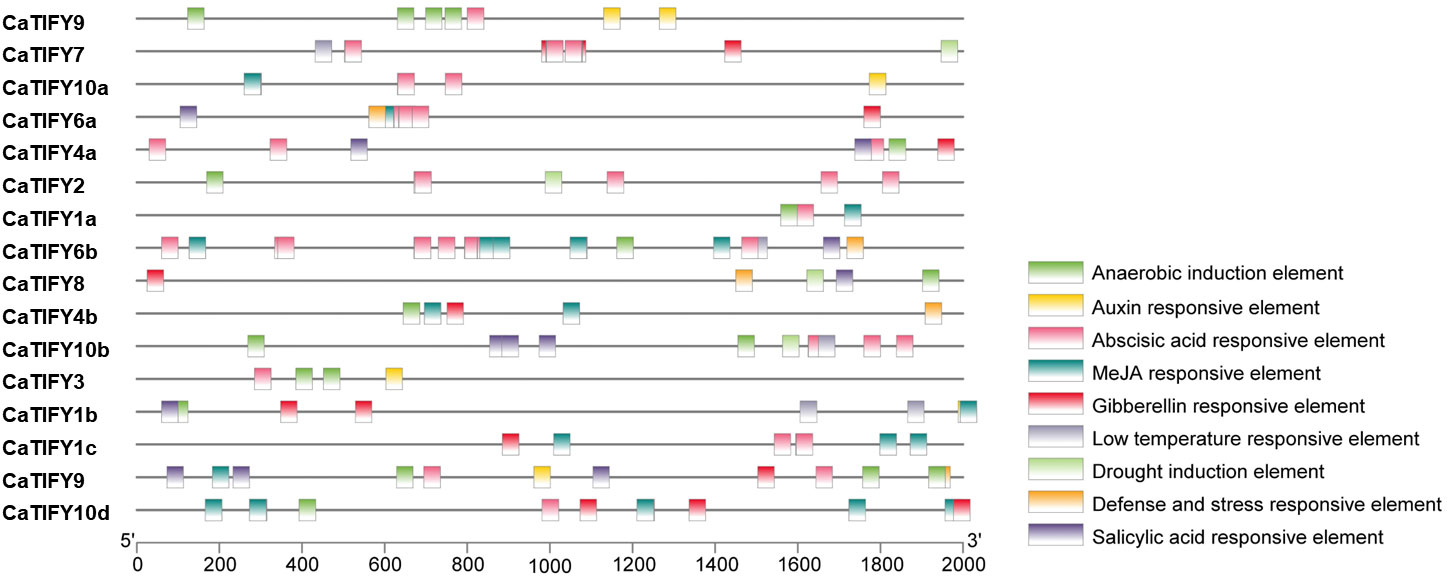
Figure 4 Promoter cis-element analysis of CaTIFY genes. Nine cis-acting elements related to abiotic stress are detected in each species, in which they are represented by distinct colored boxes.
To understand the expression patterns of CaTIFY genes in different pepper tissues, the relative expression levels of CaTIFY family genes in eight tissues (roots, shoots, young leaves, mature leaves, old leaves, flowers, fruits, and seeds) were determined based on published RNA-seq data. As is shown in Supplementary Figure 3A, a heat map was generated to illustrate the expression profiles. The relative expression levels of the 16 CaTIFY genes differed among the eight tissues (roots, shoots, young leaves, mature leaves, old leaves, flowers, fruits, and seeds). The expression levels of CaTIFY1c, CaTIFY2, CaTIFY3, and CaTIFY6b were very low in all tissues. CaTIFY7 was highly expressed in young leaves, mature leaves, old leaves, flowers, and fruits but had very low transcription levels in roots, shoots, and seeds (Supplementary Figure 3A). Significantly, the expression level of CaTIFY10b in the roots was the highest among all CaTIFY genes, but its expression level in other tissues was relatively low, especially in the leaves and seeds. Additionally, CaTIFY10c showed a tissue-specific expression pattern similar to that of CaTIFY10b. However, its overall expression level was lower than that of CaTIFY10b (Supplementary Figure 3A). Interestingly, although CaTIFY10d is homologous to CaTIFY10a/b/c, its tissue-specific expression patterns were completely different. CaTIFY10d was hardly expressed in roots and seeds but had relatively high expression levels in flowers. Subsequently, we validated the tissue expression patterns of these CaTIFY genes in pepper leaves using RT-qPCR (Supplementary Figures 2A–P). Consistent with the transcriptome data, CaTIFY7 was highly expressed in young leaves and flowers and CaTIFY10b was highly expressed in roots (Supplementary Figures 2J, N). Overall, most CaTIFY genes had the highest expression levels in leaves. The exceptions were CaTIFY1b and CaTIFY6a, which showed the highest expression levels in roots, and CaTIFY6b, which showed the highest expression level in flowers (Supplementary Figure 2).
To characterize the abiotic-responsive CaTIFY genes, the Fragments Per Kilobase of exon model per million mapped fragments (FPKM) values of each gene in leaves and roots treated with NaCl, mannitol, H2O2, heat, or cold were determined (Supplementary Figures 3B–F). Under salt stress, the transcript levels of TIFY4a, TIFY6b, and TIFY8 in leaves were significantly repressed, whereas the expression levels of TIFY6b and TIFY8 in the roots were induced by NaCl treatment. TIFY10b and TIFY10c were strongly induced by salt stress, especially in roots (Supplementary Figure 3B). Mannitol treatment induces drought stress responses in plants. As shown in Supplementary Figure 3C, TIFY7, TIFY10b, and TIFY10c were induced by drought stress in the leaves and roots of pepper. TIFY6a and TIFY6b were strongly induced by drought stress in roots, whereas TIFY6b was strongly repressed by drought stress in leaves. H2O2 is one of the most abundant reactive oxygen species (ROS) in cells, and its accumulation causes oxidative damage to cells. It is a key signaling molecule involved in plant growth, development, and stress resistance. In the leaves, TIFY6a, TIFY7, TIFY10a, TIFY10b, and TIFY10c were induced by H2O2 whereas TIFY6b was reduced by H2O2. TIFY6a, TIFY6b, TIFY7, TIFY8, TIFY10a, TIFY10b and TIFY10c were induced by H2O2 in roots while the expression level of TIFY1a was reduced by H2O2 in roots (Supplementary Figure 3D).
Temperature is an important factor that restricts plant growth, and either too high or too low a temperature can cause stress on plant growth. Under high-temperature stress, TIFY6a and TIFY7 were strongly repressed, whereas TIFY1a and TIFY10b were induced by heat stress (Supplementary Figure 3E). Additionally, TIFY6a, TIFY6b, TIFY10b, and TIFY10c were induced by heat stress (Supplementary Figure 3E). As expected, cold stress also affected the expression of some TIFY genes in pepper plants. As shown in Supplementary Figure 3F, TIFY10a, TIFY10b, and TIFY10c were strongly induced by cold stress, whereas they were not induced by chilling in either roots or leaves. Surprisingly, TIFY6a and TIFY6b were strongly induced by cold stress in the roots but were repressed by cold stress in the leaves. The expression levels of TIFY1a in the roots and leaves reduced under cold treatment. The responses of the other genes to cold stress were not significant.
To examine the changes in CaTIFY gene expression of pepper in response to cold stress, the pepper seedlings were subjected to 4 °C treatment. We determined the abundance of transcripts of sixteen CaTIFY genes under cold stress using RT-qPCR to confirm the results of the RNA-seq data. As shown in Figure 5, CaTIFY7 and CaTIFY10b were significantly induced by cold stress in leaves, whereas CaTIFY1b, CaTIFY1c, CaTIFY2, and CaTIFY4b were significantly reduced by cold treatment (Figures 5B–D, G, J, N). Additionally, the expression of CaTIFY3, CaTIFY6a and CaTIFY10d tended to be induced after cold stress treatment (Figures 5E, H, P). However, the constitutive expression levels of CaTIFY3, CaTIFY6a and CaTIFY10d were much lower than CaTIFY7 and CaTIFY10b in leaf and under low-temperature treatment as shown in published transcriptome data (Supplementary Figures 3A, F). In summary, all of the results identified CaTIFY7 and CaTIFY10b as candidates for further studies on the potential important roles of CaTIFY genes in enhancing low-temperature stress tolerance in pepper leaves.
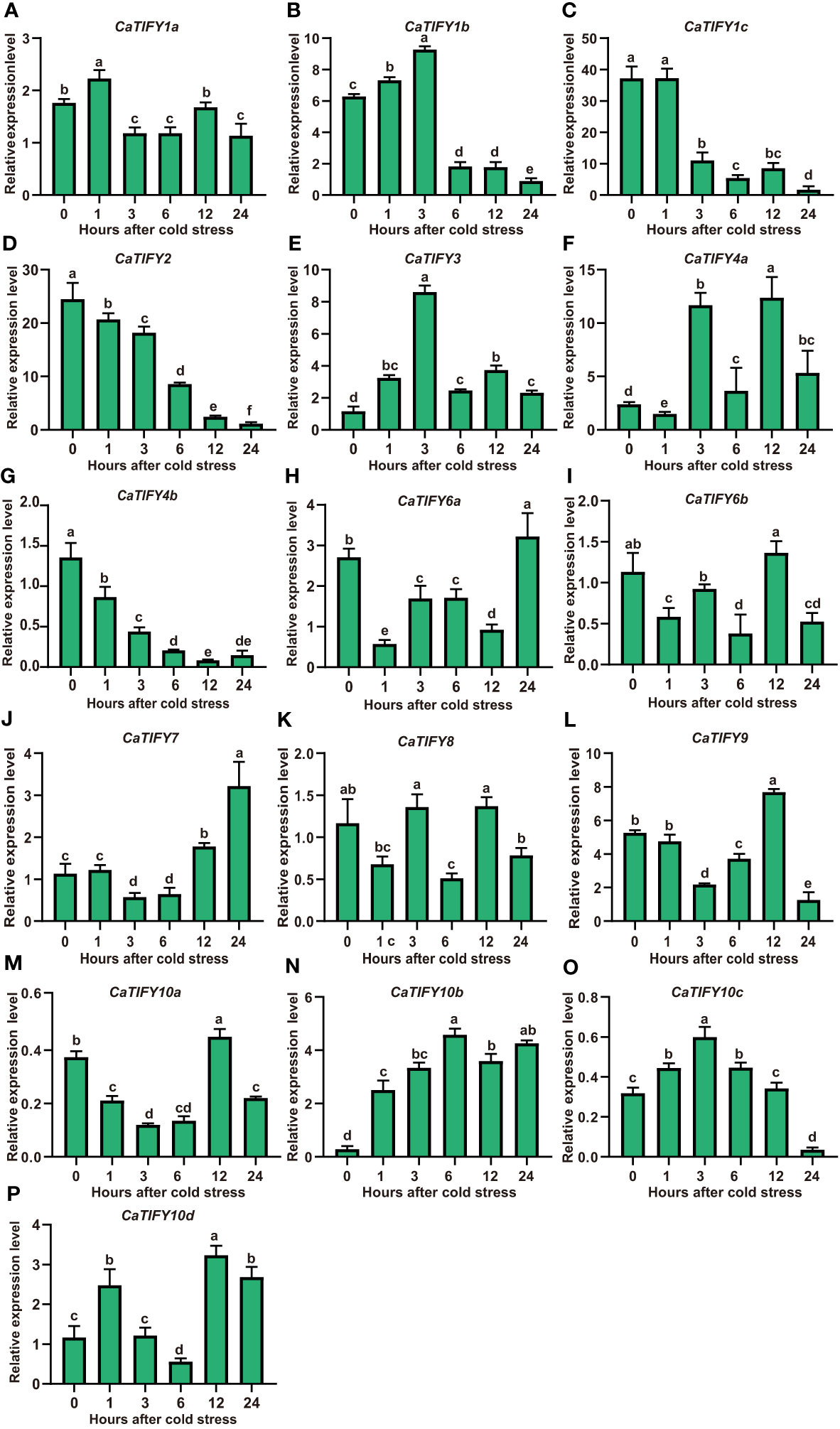
Figure 5 RT-qPCR analysis of expression patterns of CaTIFY genes in pepper leaf under cold stress. (A–P) Expression levels of sixteen CaTIFY genes were measured using RT-qPCR at different times under 4 °C cold stress. RT-qPCR data were normalized using CaUBI3 as the reference gene and were displayed relative to 0 h. Different letters indicate significant differences between samples according to the Student-Newman-Keuls test (P <0.05).
To confirm the role of CaTIFY7 in pepper under cold stress, Virus-Induced Gene Silencing (VIGS) was used to knock down the gene. Silencing CaTIFY7 significantly increased the sensitivity of pepper to cold stress. As shown in Figure 6A, pepper plants transformed with TRV2:00 and TRV2:CaTIFY7-silenced both showed normal, undifferentiated growth under normal conditions. However, the growth of pepper plants transformed with an empty vector was significantly better than those with CaTIFY7 under 4 °C cold stress treatment. RT-qPCR analysis revealed that the CaTIFY7 silencing efficiency was high and showed a 68.6% or 80.6% reduction compared to the control group under normal or cold stress, respectively (Figure 6B). CaTIFY7 knockdown led to significant downregulation of cold-induced genes (CaERD15, CaCBF1a, CaCBF1b, CaCOR47-like, and CaKIN) and ROS-related genes (CaPOD, CaSOD, CaCAT2, and CaAPX1) under cold stress (Figures 6C–K). CaTIFY7 overexpression markedly increased the reactive oxygen species content and expression levels of cold-induced genes and ROS-related genes under cold stress (Figures 7A–L). Preliminary results indicated that CaTIFY7 plays a positive role in regulating pepper tolerance to cold stress.
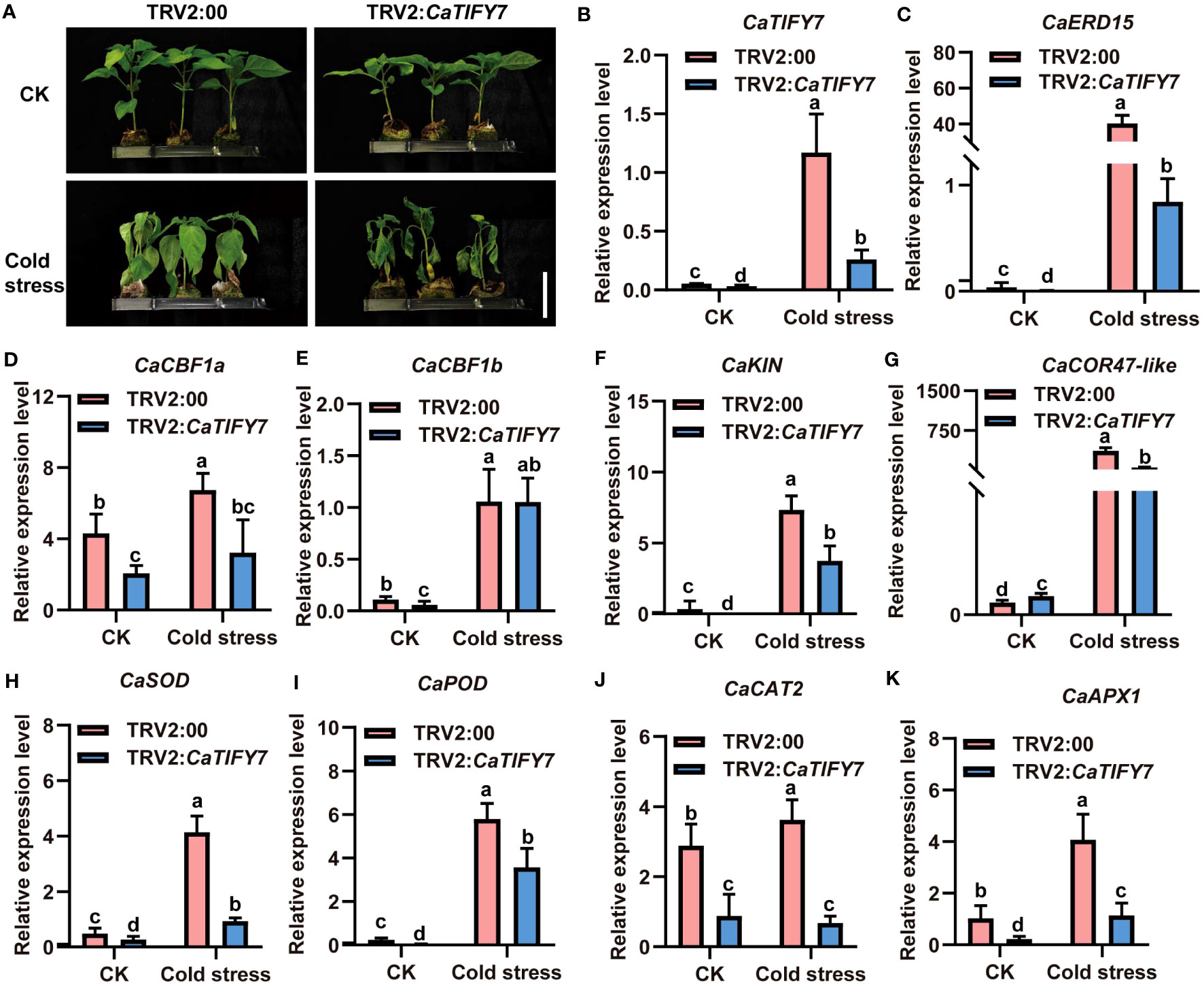
Figure 6 Knock down of CaTIFY7 in pepper weaken plant tolerance significantly under cold stress. (A) The phenotype of pepper plants with reduced expression CaTIFY7 under 4 °C cold stress for 48 (h) Bar=4 cm. (B) The silencing efficiency of CaTIFY7 under normal or cold stress. (C-K) The expression level of CaERD15, CaCBF1a, CaCBF1b, CaKIN, CaCOR47-like, CaSOD, CaPOD, CaCAT2 and CaAPX1 were analyzed in transgenic pepper plants with TRV2:CaTIFY7 under normal or cold stress. CaUBI3 were used as the reference gene. All the values are the averages ± SD from three independent experiments. Different letters indicate significant differences between samples according to the Student-Newman-Keuls test (P <0.05).
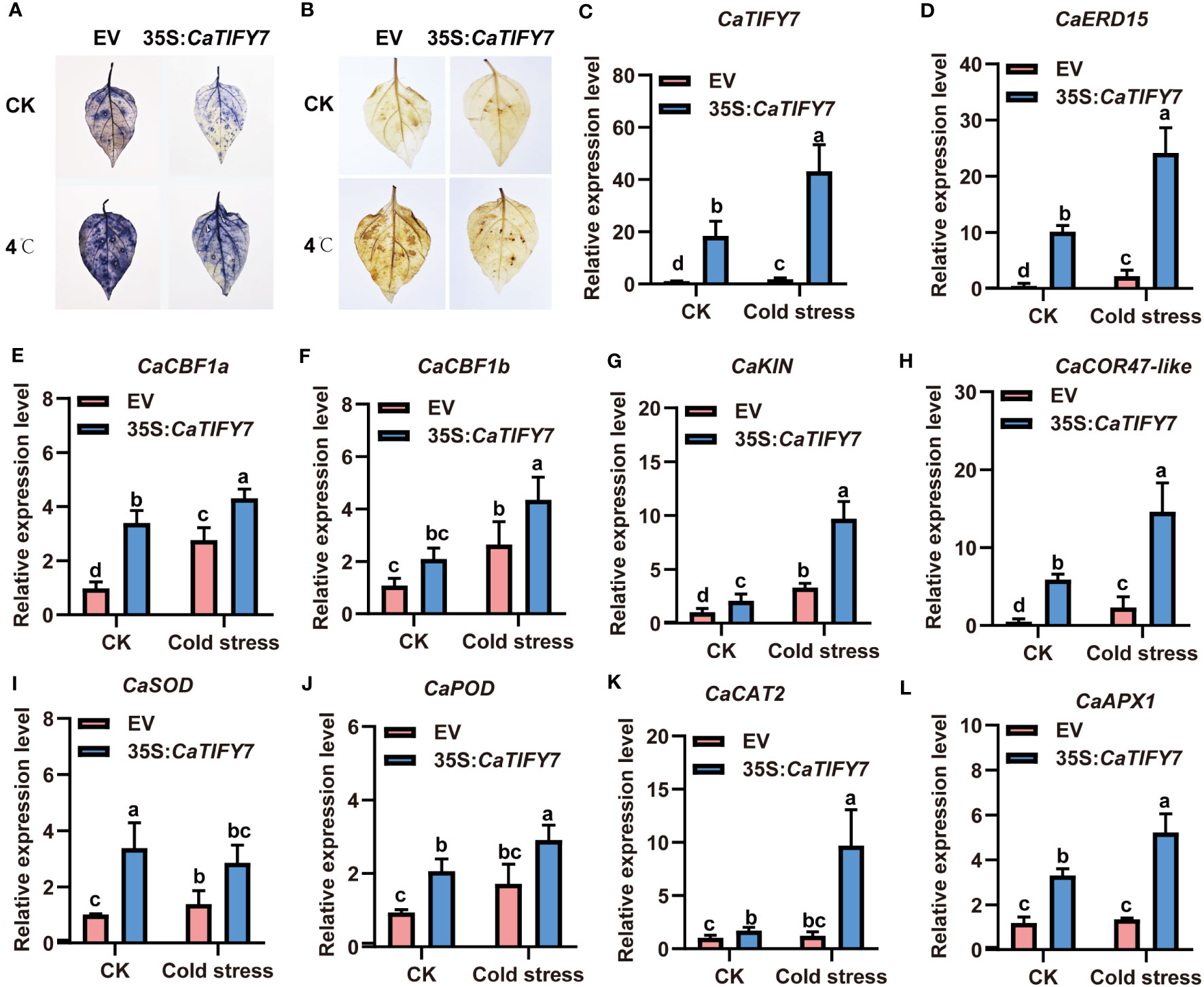
Figure 7 Overexpression of CaTIFY7 enhances cold stress tolerant in pepper. (A) O2- detecting with NBT after cold stress. (B) H2O2 staining with DAB under normal or cold stress. (C) The expression level of CaTIFY7 were analyzed in transgenic pepper plants having 35S:CaTIFY7 under normal or cold stress. (D-L) The transcription level of CaERD15, CaCBF1a, CaCBF1b, CaKIN, CaCOR47-like, CaSOD, CaPOD, CaCAT2 and CaAPX1 were detected in transgenic pepper plants with 35S:CaTIFY7 after cold stress treatment. The gene expression levels were normalized to those of CaUBI3. The relative transcript level was determined and normalized using the reference level and averaged over the three technical replicates. All the values are the averages ± SD from three independent experiments. Different letters indicate significant differences between samples according to the Student-Newman-Keuls test (P <0.05).
To further confirm the function of CaTIFY10b in cold-stress response, transgenic pepper leaves containing the 35S:CaTIFY10b vector were generated to explore the role of CaTIFY10b in cold tolerance. After 24 h, the CaTIFY10b-silenced plants showed a more severe degree of withering than the control plants (Figure 8A). The RT-qPCR results showed that CaTIFY10b expression was successfully reduced in TRV2:CaTIFY10b-silenced plants (Figure 8B). Subsequently, the expression of cold-induced and ROS-related genes in CaTIFY10b-silenced pepper leaves was analyzed and the knockdown of CaTIFY10b significantly suppressed the expression of cold-induced and ROS-related genes under cold stress (Figures 8C–K). As expected, overexpression of CaTIFY10b also increased the reactive oxygen content in plant leaves and significantly increased the expression of these genes under cold stress in pepper (Figures 9A–L). These results showed that CaTIFY10b positively regulated cold tolerance in pepper by promoting the expression of stress-related genes.
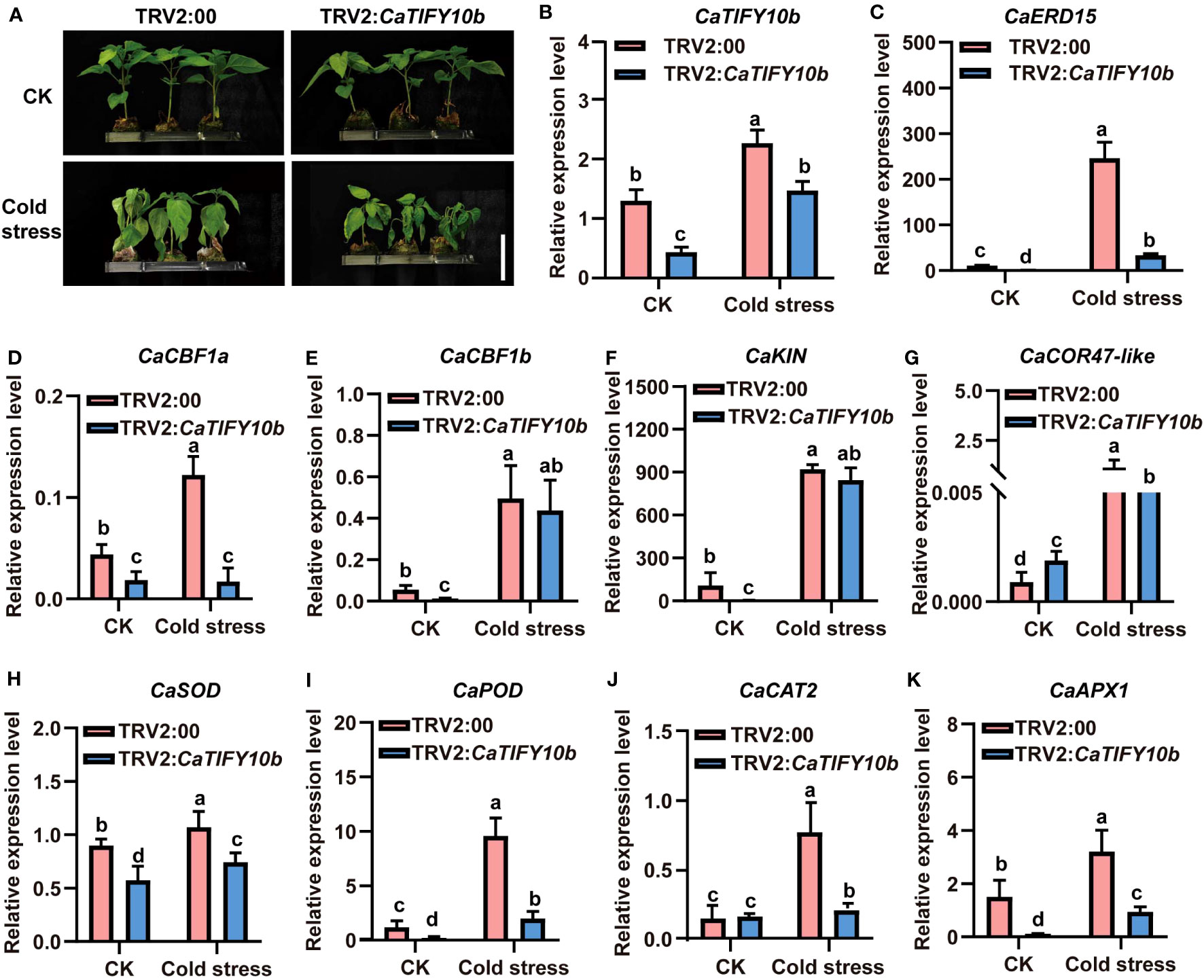
Figure 8 Knock down of CaTIFY10b in pepper seedlings improves sensitiveness under cold stress. (A) The phenotype of pepper plants with suppression of CaTIFY10b under cold stress for 48 (h) Bar=4 cm. (B) The expression level of CaTIFY10b were analyzed in transgenic pepper plants under normal or cold stress. (C-K) The transcription level of CaERD15, CaCBF1a, CaCBF1b, CaKIN, CaCOR47-like, CaSOD, CaPOD, CaCAT2 and CaAPX1 were detected in transgenic pepper plants with TRV2:CaTIFY7 under normal or cold stress. The gene expression levels were normalized to those of CaUBI3. All the values are the averages ± SD from three independent experiments. Different letters indicate significant differences between samples according to the Student-Newman-Keuls test (P <0.05).
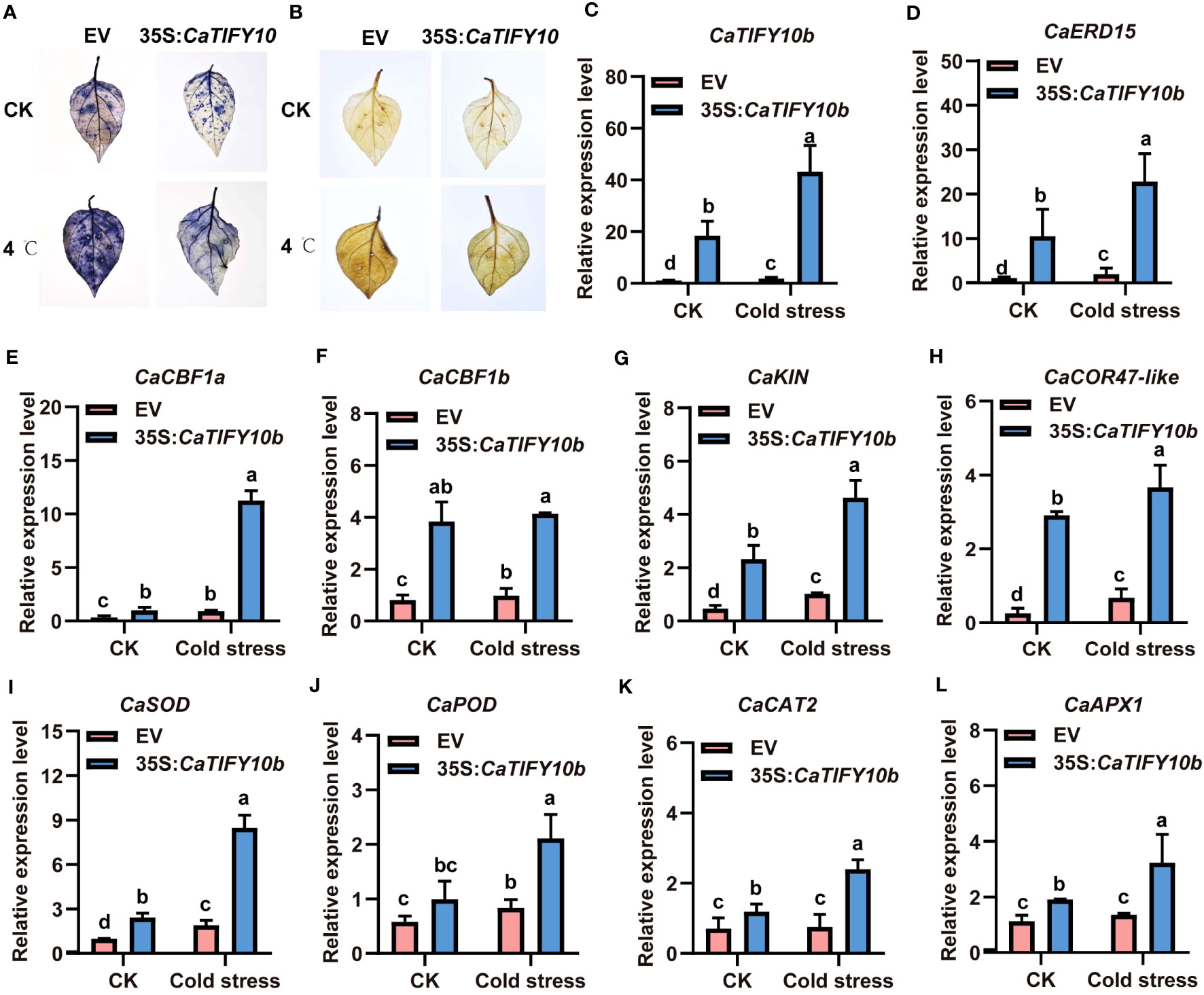
Figure 9 Overexpression of CaTIFY10b positively regulated cold stress response in pepper. (A) O2- detecting with NBT under normal or cold stress. (B) H2O2 staining with DAB after cold stress. (C) The transcription level of CaTIFY10b were analyzed in transgenic pepper plants having 35S:CaTIFY10b under normal or cold stress. (D-L) The expression of cold stress and ROS-related genes were detected in transgenic pepper plants with 35S:CaTIFY10b after cold stress treatment. CaUBI3 were used as the reference gene. All the values are the averages ± SD from three independent experiments. Different letters indicate significant differences between samples according to the Student-Newman-Keuls test (P <0.05).
Because CaTIFY7 and CaTIFY10b are both involved in cold-stress reactions, it would be interesting to explore whether they can form heterodimers in response to cold-stress signals. The interaction network of the 16 CaTIFY proteins was predicted using the STRING database, and the results are shown in Supplementary Figure 4. Surprisingly, the results indicated that CaTIFY7 and CaTIFY10b may have a conserved interaction relationship in the model plant. To confirm this relationship, we performed a yeast two-hybrid analysis to verify their interaction. As shown in Figure 10, CaTIFY7 interacted with CaTIFY10b in the Y2H assay (Figure 10). In addition, CaTIFY7 and CaTIFY10b interacted with each other (Figure 10). These results showed that CaTIFY7 and CaTIFY10b may form heterodimers or homodimers to regulate the cold-stress response in pepper.
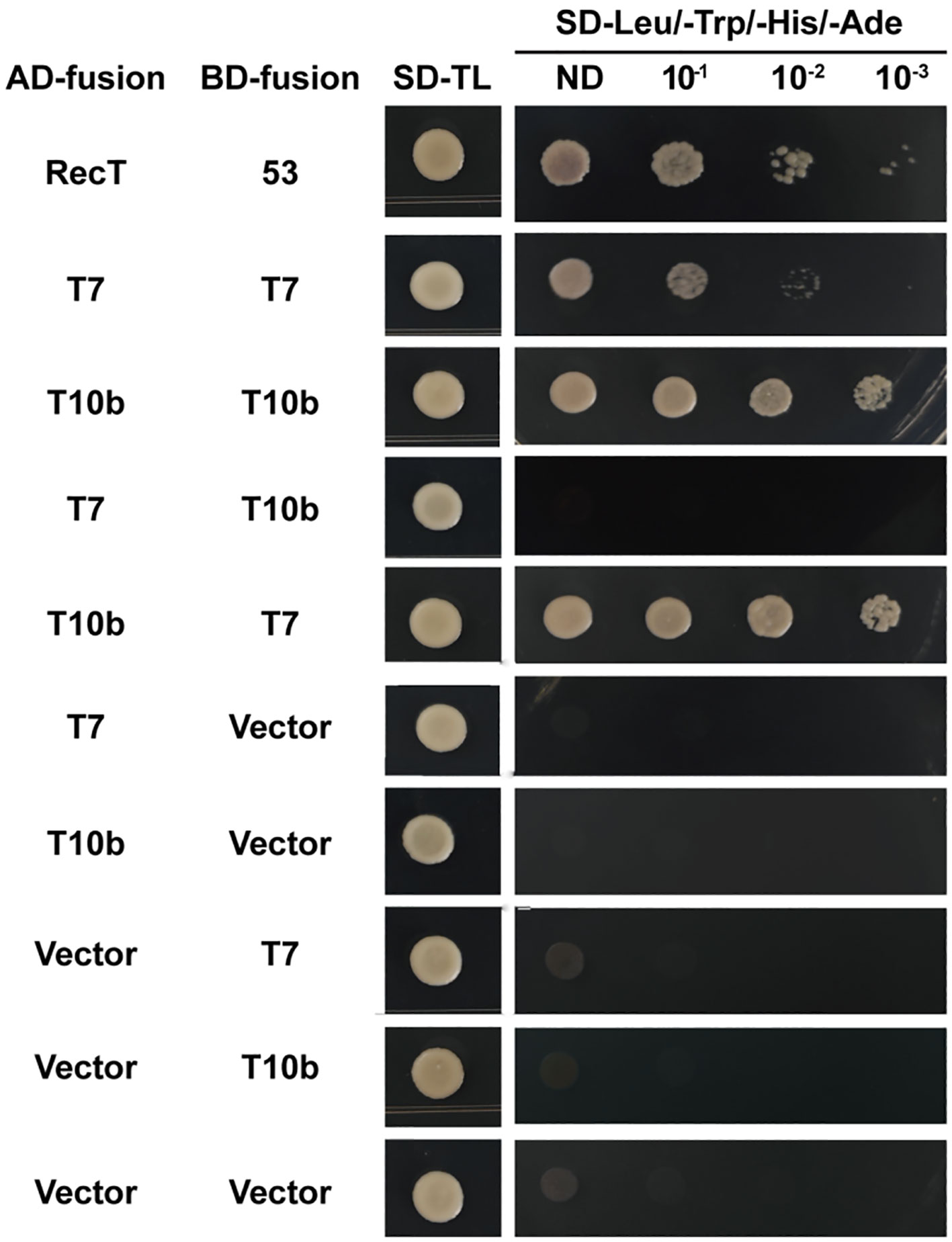
Figure 10 Y2H assay between CaTIFY7 and CaTIFY10b. CaTIFY7 interacts with itself or CaTIFY10b and CaTIFY10b interacts with itself or CaTIFY7 in the Y2H assay. Protein interactions were analyzed by the growth of yeast in the medium without leucine, tryptophan, histidine and adenine medium.
TIFY proteins are defined as TIFY transcription factors because of their specific binding to consensus sequences. TIFY proteins are important regulators of abiotic and biotic stress responses in plants. Several TIFY proteins have been identified in various plant species. However, the TIFY gene family in Solanaceae crops, including pepper, has not previously been identified or characterized. We identified sixteen TIFY genes in pepper and the TIFY gene family members all contained the conservative TIFY⁃motif and JAZ domains (Figures 1; 2). The lack of a homologous gene for Arabidopsis AtTIFY5 among the TIFY members in pepper suggests that this homologous gene may have undergone selective loss in pepper. All CaTIFY genes were distributed on chromosomes 1, 3, 4, 6, 7, 8, 9, and 12 and only CaTIFY1b/c formed a homologous gene cluster on chromosome 8 (Figure 3).
Many elements were related to hormones and stress in the CaTIFYs promoter of peppers, but there were significant differences among the different CaTIFY genes (Figure 4). Most members of the TIFY gene family in pepper had anaerobic induction elements, and more than half of the TIFY genes responded to ABA, MeJA, salicylic acid, and gibberellin. In addition, some genes contained different stress response elements, suggesting that CaTIFYs may be involved in the regulation of different hormone signal transduction pathways, thereby responding to environmental stress.
The TIFY family is widely distributed in plants and its members play important roles in regulating the development of stems, leaves, and flowers in plants (Zhu et al., 2011; Hakata et al., 2012; He et al., 2015; Baekelandt et al., 2018). To better understand the differential expression of CaTIFYs genes in peppers, we analyzed the gene expression of CaTIFYs in different tissues. RNA-seq data showed that the expression levels of the majority of pepper TIFY did not show tissue-specific expression patterns (Supplementary Figures 2, 3). The expression levels of CaTIFY7 were high in most tissues, suggesting that they have an important role in pepper development. Notably, CaTIFY10 was most highly expressed in the roots, suggesting that it has a key role in the root development and growth of pepper.
Next, we analyzed the gene expression patterns of this family under different stress conditions (Supplementary Figure 3). Salt stress induced expression of TIFY6b, TIFY8, TIFY10b, and TIFY10c in roots, but repressed the transcription levels of TIFY6b and TIFY4a in leaves, suggesting that TIFY6b expression is regulated by different mechanisms in different tissues. The significant induction of TIFY10b and TIFY10c expression suggests that they are likely to participate in salt stress responses, which is consistent with the function of their homologous genes (GmTIFY10e and GmTIFY10g) in the positive regulation of salt tolerance in soybeans (Liu et al., 2022). Under drought stress, the induced expression of TIFY10a, TIFY10b, and TIFY10c in leaves and roots indicates that TIFY10 might play an important role in the drought response of pepper. Based on the discovery that multiple members of the TIFY family enhance the drought resistance of different plants (Ye et al., 2009; Zhao et al., 2016; Zhang et al., 2023), it can be predicted that CaTIFY10a/b/c may also regulate drought resistance in pepper.
Among all TIFY genes, the responses of TIFY6a, TIFY6b, TIFY7 and TIFY10b to high temperatures were the most significant (Supplementary Figure 3). The induced expression of TIFY6a, TIFY6b, and TIFY10b, and the inhibited expression of TIFY7, suggest that TIFY6a, TIFY6b, and TIFY10b might play a role in enhancing high-temperature resistance in pepper while the function of TIFY10b might be the opposite. However, there are almost no reports on the function of TIFY in responses of plants to high temperatures and this is a direction worth studying further. Similarly, the function of TIFY during the low-temperature response is unclear. In pepper, we found that several TIFY (TIFY1a, TIFY4a, TIFY6a, TIFY6b, TIFY10a, TIFY10b, and TIFY10c) genes responded to cold stress, suggesting that they may be important candidate genes for the cold response in pepper.
To cope with low-temperature stress, plants have evolved a “cold adaptation” mechanism. Many transcription factors play an important role in the regulation of cold-stress adaptation in plants (Chinnusamy et al., 2010; Ding et al., 2019). It has long been known that the TIFY gene responds to cold stress in rice, rape, tomato, bamboo, and grain sorghum (Ye et al., 2009; Huang et al., 2016; Chini et al., 2017; He et al., 2020; Du et al., 2022). However, there have been few reports on the function of TIFY in plant responses to low temperatures. Our results confirmed that pepper CaTIFY7 and CaTIFY10b were positive regulators of plant responses under cold stress, where they regulated the expression levels of cold-induced and ROS-related genes (Figures 6–9). We also demonstrated that CaTIFY7 and CaTIFY10b play important roles in pepper plants under salt stress.
To further analyze the regulatory mechanism of CaTIFY7 and CaTIFY10b in terms of protein interactions, we analyzed the CaTIFY protein interaction network using the STRING database (Supplementary Figure 4). Many CaTIFYs, except for CaTIFY1b, CaTIFY1c, CaTIFY1a, and CaTIFY2, interacted with each other to form complexes, indicating that most CaTIFYs formed heterodimers that may affect protein function. Some members of the CaTIFY family formed a complex regulatory network with COI1 protein, MYC2 transcription factor, and Ninja and bHLH13 proteins. It’s known that COI1/JAZs/MYC2 acts as the core jasmonic acid-signaling module in the plant jasmonic acid signaling pathway (Chini et al., 2009; Chung and Howe, 2009), which means that these CaTIFYs have a conserved function in sensing JA signals and regulating JA levels in pepper. However, CaTIFY1a/b/c and CaTIFY2 are independent of this signaling pathway, suggesting that these genes may exert regulatory functions through unique mechanisms. Interestingly, CaTIFY7 interacted with CaTIFY10b, as demonstrated by the Y2H assay (Figure 10), suggesting that CaTIFY7 and CaTIFY10b act as heterodimers or homodimers to regulate the cold-stress response in pepper.
We identified 16 CaTIFY genes in pepper, of which CaTIFY7 and CaTIFY10b were significantly upregulated by cold stress. Silencing CaTIFY7 and CaTIFY10b significantly repressed the cold tolerance of transgenic pepper plants and the expression of cold-induced and ROS-related genes. Conversely, overexpression of CaTIFY7 and CaTIFY10b enhanced cold tolerance by promoting the expression of cold-induced and ROS-related genes. Further, we showed that CaTIFY7 and CaTIFY10b interacted with each other and that CaTIFY7 interacted with CaTIFY10b. This study provides a basis for further research on how TIFY family members regulate cold tolerance in plants.
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
XW: Data curation, Formal Analysis, Investigation, Methodology, Software, Writing – original draft, Writing – review & editing. NL: Writing – original draft, Conceptualization, Data curation, Formal Analysis, Writing – review & editing. TZ: Writing – original draft, Investigation, Project administration. KX: Conceptualization, Data curation, Writing – original draft, Formal Analysis. SG: Conceptualization, Data curation, Project administration, Writing – original draft. YY: Conceptualization, Project administration, Resources, Writing – original draft. MY: Funding acquisition, Supervision, Validation, Writing – review & editing. FW: Funding acquisition, Supervision, Validation, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This research was funded by National High Technology Research and Development Program of China (2021YFD1600300-4-2), Key Research and Development Program of Hubei Province (2022BBA0066, 2022BBA0060), Key Research and Development Program of Hubei Province (2023BBB044), Major Project of Hubei Hongshan Laboratory (2022hszd009), Postdoctoral Innovation Practice Positions of Hubei Province, Youth Fund of Hubei Academy of Agricultural Sciences (2024NKYJJ06), High-quality Seed Industry Development Program of Hubei Province and China Agriculture Research System (CARS-23-G28).
We thank Professor Feng Li (Huazhong Agricultural University) for his gift of pTRV1-C2b and pTRV2-C2b vectors. We would like to thank Editage (www.editage.cn) for English language editing.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1308721/full#supplementary-material
Baekelandt, A., Pauwels, L., Wang, Z., Li, N., De Milde, L., Natran, A., et al. (2018). Arabidopsis leaf flatness is regulated by PPD2 and NINJA through repression of CYCLIN D3 genes. Plant Physiol. 178 (1), 217–232. doi: 10.1104/pp.18.00327
Bai, Y., Meng, Y., Huang, D., Qi, Y., Chen, M. (2011). Origin and evolutionary analysis of the plant-specific TIFY transcription factor family. Genomics. 98, 128–136. doi: 10.1016/j.ygeno.2011.05.002
Cai, W., Yang, S., Wu, R., Cao, J., Shen, L., Guan, D., et al. (2021). Pepper NAC-type transcription factor NAC2c balances the trade-off between growth and defense responses. Plant Physiol. 186 (4), 2169–2189. doi: 10.1093/plphys/kiab190
Chen, R., Ma, J., Luo, D., Hou, X., Ma, F., Zhang, Y., et al. (2019). CaMADS, a MADS-box transcription factor from pepper, plays an important role in the response to cold, salt, and osmotic stress. Plant Sci. 280, 164–174. doi: 10.1016/j.plantsci.2018.11.020
Chini, A., Ben-Romdhane, W., Hassairi, A., Aboul-Soud, M. A. M. (2017). Identification of TIFY/JAZ family genes in Solanum lycopersicum and their regulation in response to abiotic stresses. PloS One 12 (6), e0177381. doi: 10.1371/journal.pone.0177381
Chini, A., Boter, M., Solano, R. (2009). Plant oxylipins: COI1/JAZs/MYC2 as the core jasmonic acid-signalling module. FEBS J. 276 (17), 4682–4692. doi: 10.1111/j.1742-4658.2009.07194.x
Chinnusamy, V., Zhu, J. K., Sunkar, R. (2010). Gene regulation during cold stress acclimation in plants. Methods Mol. Biol. 639, 39–55. doi: 10.1007/978-1-60761-702-0_3
Chinnusamy, V., Zhu, J., Zhu, J. K. (2007). Cold stress regulation of gene expression in plants. Trends Plant Sci. 12, 444–451. doi: 10.1016/j.tplants.2007.07.002
Chung, H. S., Howe, G. A. (2009). A critical role for the TIFY motif in repression of jasmonate signaling by a stabilized splice variant of the JASMONATE ZIM-domain protein JAZ10 in Arabidopsis. Plant Cell. 21 (1), 131–145. doi: 10.1105/tpc.108.064097
Ding, Y., Shi, Y., Yang, S. (2019). Advances and challenges in uncovering cold tolerance regulatory mechanisms in plants. New Phytol. 222, 1690–1704. doi: 10.1111/nph.15696
Du, Q., Fang, Y., Jiang, J., Chen, M., Li, X., Xie, X. (2022). Genome-wide identification and characterization of the JAZ gene family and its expression patterns under various abiotic stresses in Sorghum bicolor. J. Integr. Agric. 21 (12), 3540–3555. doi: 10.1016/j.jia.2022.08.104
Ebel, C., BenFeki, A., Hanin, M., Solano, R., Chini, A. (2018). Characterization of wheat (Triticum aestivum) TIFY family and role of Triticum durum TdTIFY11a in salt stress tolerance. PloS One 13 (7), e0200566. doi: 10.1371/journal.pone.0200566
Guo, W. L., Wang, S. B., Chen, R. G., Chen, B. H., Du, X. H., Yin, Y. X., et al. (2015). Characterization and expression profile of CaNAC2 pepper gene. Front. Plant Sci. 6. doi: 10.3389/fpls.2015.00755
Hakata, M., Kuroda, M., Ohsumi, A., Hirose, T., Nakamura, H., Muramatsu, M., et al. (2012). Overexpression of a rice TIFY gene increases grain size through enhanced accumulation of carbohydrates in the stem. Biosci. Biotechnol. Biochem. 76 (11), 2129–2134. doi: 10.1271/bbb.120545
He, X., Kang, Y., Li, W., Liu, W., Xie, P., Liao, L., et al. (2020). Genome-wide identification and functional analysis of the TIFY gene family in the response to multiple stresses in Brassica napus L. BMC Genomics 21, 736. doi: 10.1186/s12864-020-07128-2
He, D. H., Lei, Z. P., Tang, B. S., Xing, H. Y., Zhao, J. X., Jing, Y. L. (2015). Identification and analysis of the TIFY gene family in Gossypium raimondii. Genet. Mol. Res. 14 (3), 10119–10138. doi: 10.4238/2015
Heidari, P., Faraji, S., Ahmadizadeh, M., Ahmar, S., Mora-Poblete, F. (2021). New insights into structure and function of TIFY genes in Zea mays and Solanum lycopersicum: A Genome-Wide Comprehensive Analysis. Front. Genet. 12. doi: 10.3389/fgene.2021.657970
Hou, X. M., Zhang, H. F., Liu, S. Y., Wang, X. K., Zhang, Y. M., Meng, Y. C., et al. (2020). The NAC transcription factor CaNAC064 is a regulator of cold stress tolerance in peppers. Plant Sci. 291, 110346. doi: 10.1016/j.plantsci.2019.110346
Hu, Y., Jiang, L., Wang, F., Yu, D. (2013). Jasmonate regulates the inducer of CBF expression-C-repeat binding factor/DRE binding factor1 cascade and freezing tolerance in Arabidopsis. Plant Cell. 25 (8), 2907–2924. doi: 10.1105/tpc.113.112631
Huang, Z., Jin, S. H., Guo, H. D., Zhong, X. J., He, J., Li, X., et al. (2016). Genome-wide identification and characterization of TIFY family genes in Moso Bamboo (Phyllostachys edulis) and expression profiling analysis under dehydration and cold stresses. PeerJ. 4, e2620. doi: 10.7717/peerj.2620
Huang, G. T., Ma, S. L., Bai, L. P., Zhang, L., Ma, H., Jia, P., et al. (2012). Signal transduction during cold, salt, and drought stresses in plants. Mol. Biol. Rep. 39 (2), 969–987. doi: 10.1007/s11033-011-0823-1
Legris, M., Nieto, C., Sellaro, R., Prat, S., Casal, J. J. (2017). Perception and signalling of light and temperature cues in plants. Plant J. 90 (4), 683–697. doi: 10.1111/tpj.13467
Li, X., Yin, X., Wang, H., Li, J., Guo, C., Gao, H., et al. (2015). Genome-wide identification and analysis of the apple (Malus×domestica Borkh.) TIFY gene family. Tree Genet. Genomes. 11 (1), 808. doi: 10.1007/s11295-014-0808-z
Liu, F., Yu, H., Deng, Y., Zheng, J., Liu, M., Ou, L., et al. (2017). PepperHub, an informatics hub for the chili pepper research community. Mol. Plant 10, 1129–1132. doi: 10.1016/j.molp.2017.03.005
Liu, Y. L., Zheng, L., Jin, L. G., Liu, Y. X., Kong, Y. N., Wang, Y. X., et al. (2022). Genome-wide analysis of the soybean TIFY family and identification of GmTIFY10e and GmTIFY10g response to salt stress. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.845314
Meng, L., Zhang, T., Geng, S., Scott, P. B., Li, H., Chen, S. (2019). Comparative proteomics and metabolomics of JAZ7-mediated drought tolerance in Arabidopsis. J. Proteomics. 196, 81–91. doi: 10.1016/j.jprot.2019.02.001
Nishii, A., Takemura, M., Fujita, H., Shikata, M., Yokota, A., Kohchi, T. (2000). Characterization of a novel gene encoding a putative single zinc-finger protein, ZIM, expressed during the reproductive phase in Arabidopsis thaliana. Biosci. Biotechnol. Biochem. 64, 1402–1409. doi: 10.1271/bbb.64.1402
Seo, J. S., Joo, J., Kim, M. J., Kim, Y. K., Nahm, B. H., Song, S. I., et al. (2011). OsbHLH148, a basic helix-loop-helix protein, interacts with OsJAZ proteins in a jasmonate signaling pathway leading to drought tolerance in rice. Plant J. 65 (6), 907–992. doi: 10.1111/j.1365-313X.2010.04477.x
Sirhindi, G., Sharma, P., Arya, P., Goel, P., Kumar, G., Acharya, V., et al. (2016). Genome-wide characterization and expression profiling of TIFY gene family in pigeonpea (Cajanus cajan (L.) Millsp.) under copper stress. J. Plant Biochem. Biotechnol. 25, 301–310. doi: 10.1007/s13562-015-0342-6
Thireault, C., Shyu, C., Yoshida, Y., St Aubin, B., Campos, M. L., Howe, G. A. (2015). Repression of jasmonate signaling by a non-TIFY JAZ protein in Arabidopsis. Plant J. 82 (4), 669–667. doi: 10.1111/tpj.12841
Vanholme, B., Grunewald, W., Bateman, A., Kohchi, T., Gheysen, G. (2007). The TIFY family previously known as ZIM. Trends Plant Sci. 12, 239–244. doi: 10.1016/j.tplants.2007.04.004
Wan, H., Yuan, W., Ruan, M., Ye, Q., Wang, R., Li, Z., et al. (2011). Identification of reference genes for reverse transcription quantitative real-time PCR normalization in pepper (Capsicum annuum L.). Biochem. Biophys. Res. Commun. 416 (1-2), 24–30. doi: 10.1016/j.bbrc.2011.10.105
Wang, J. E., Liu, K. K., Li, D. W., Zhang, Y. L., Zhao, Q., He, Y. M., et al. (2013). A novel peroxidase CanPOD gene of pepper is involved in defense responses to phytophtora capsici infection as well as abiotic stress tolerance. Int. J. Mol. Sci. 14 (2), 3158–3177. doi: 10.3390/ijms14023158
Wang, Z., Zhang, Y., Hu, H., Chen, L., Zhang, H., Chen, R. (2022). CabHLH79 acts upstream of CaNAC035 to regulate cold stress in pepper. Int. J. Mol. Sci. 23 (5), 2537. doi: 10.3390/ijms2305253
Xia, W., Yu, H., Cao, P., Luo, J., Wang, N. (2017). Identification of TIFY family genes and analysis of their expression profiles in response to phytohormone treatments and Melampsora larici-populina infection in poplar. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.00493
Ye, H., Du, H., Tang, N., Li, X., Xiong, L. (2009). Identification and expression profiling analysis of TIFY family genes involved in stress and phytohormone responses in rice. Plant Mol. Biol. 71 (3), 291–305. doi: 10.1007/s11103-009-9524-8
Zhang, Y., Gao, M., Singer, S. D., Fei, Z., Wang, H., Wang, X. (2012). Genome-wide identification and analysis of the TIFY gene family in grape. PloS One 7 (9), e44465. doi: 10.1371/journal.pone.0044465
Zhang, H., Guo, J. Z., Chen, X., Zhou, Y., Pei, Y., Chen, L., et al. (2023). Transcription factor CabHLH035 promotes cold resistance and homeostasis of reactive oxygen species in pepper. Hortic. Plant J. doi: 10.1016/j.hpj.2023.03.007
Zhang, H., Ma, F., Wang, X., Liu, S., Saeed, U. H., Hou, X., et al. (2020). Molecular and functional characterization of CaNAC035, an NAC transcription factor from pepper (Capsicum annuum L.). Front. Plant Sci. 11. doi: 10.3389/fpls.2020.00014
Zhang, L., You, J., Chan, Z. (2015). Identification and characterization of TIFY family genes in Brachypodium distachyon. J. Plant Res. 128 (6), 995–1005. doi: 10.1007/s10265-015-0755-2
Zhao, C., Pan, X., Yu, Y., Zhu, Y., Kong, F., Sun, X., et al. (2020). Overexpression of a TIFY family gene, GsJAZ2, exhibits enhanced tolerance to alkaline stress in soybean. Mol. Breeding. 40 (3), 255–265. doi: 10.1007/s11032-020-01113-z
Zhao, G., Song, Y., Wang, C., Butt, H. I., Wang, Q., Zhang, C., et al. (2016). Genome-wide identification and functional analysis of the TIFY gene family in response to drought in cotton. Mol. Genet. Genom. 291 (6), 2173–2187. doi: 10.1007/s00438-016-1248-2
Zheng, L., Wan, Q., Wang, H., Guo, C., Niu, X., Zhang, X., et al. (2022). Genome-wide identification and expression of TIFY family in cassava (Manihot esculenta Crantz). Front. Plant Sci. 13. doi: 10.3389/fpls.2022.1017840
Zhou, X., Yan, S., Sun, C., Li, S., Li, J., Xu, M., et al. (2015). A Maize Jasmonate Zim-Domainprotein, ZmJAZ14, associates with the JA, ABA, and GA signalingpathways in transgenic Arabidopsis. PloS One 10 (3), e0121824. doi: 10.1371/journal.pone.0121824
Keywords: pepper (Capsicum annuum L.), CaTIFY, genome-wide analysis, cold stress, gene expression
Citation: Wang X, Li N, Zan T, Xu K, Gao S, Yin Y, Yao M and Wang F (2023) Genome-wide analysis of the TIFY family and function of CaTIFY7 and CaTIFY10b under cold stress in pepper (Capsicum annuum L.). Front. Plant Sci. 14:1308721. doi: 10.3389/fpls.2023.1308721
Received: 07 October 2023; Accepted: 06 November 2023;
Published: 22 November 2023.
Edited by:
Zemin Wang, Gansu Agricultural University, ChinaReviewed by:
Wang Aimin, Jiangsu Normal University, ChinaCopyright © 2023 Wang, Li, Zan, Xu, Gao, Yin, Yao and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Fei Wang, MjA5NjY5NTRAcXEuY29t; Minghua Yao, eWFvbWhfMjAwOEAxMjYuY29t
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.