
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 19 January 2024
Sec. Plant Physiology
Volume 14 - 2023 | https://doi.org/10.3389/fpls.2023.1305299
This article is part of the Research TopicAquaporins: Dynamic Role and Regulation, Volume IIView all 4 articles
Background: Oat (Avena sativa L.) belongs to the early maturity grass subfamily of the Gramineae subfamily oats (Avena) and has excellent characteristics, such as tolerance to barrenness, salt, cold, and drought. Aquaporin (AQP) proteins belong to the major intrinsic protein (MIP) superfamily, are widely involved in plant growth and development, and play an important role in abiotic stress responses. To date, previous studies have not identified or analyzed the AsAQP gene family system, and functional studies of oat AQP genes in response to drought, cold, and salt stress have not been performed.
Methods: In this study, AQP genes (AsAQP) were identified from the oat genome, and various bioinformatics data on the AQP gene family, gene structure, gene replication, promoters and regulatory networks were analyzed. Quantitative real-time PCR technology was used to verify the expression patterns of the AQP gene family in different oat tissues under different abiotic stresses.
Results: In this study, a total of 45 AQP genes (AsAQP) were identified from the oat reference genome. According to a phylogenetic analysis, 45 AsAQP were divided into 4 subfamilies (PIP, SIP, NIP, and TIP). Among the 45 AsAQP, 23 proteins had interactions, and among these, 5AG0000633.1 had the largest number of interacting proteins. The 20 AsAQP genes were expressed in all tissues, and their expression varied greatly among different tissues and organs. All 20 AsAQP genes responded to salt, drought and cold stress. The NIP subfamily 6Ag0000836.1 gene was significantly upregulated under different abiotic stresses and could be further verified as a key candidate gene.
Conclusion: The findings of this study provide a comprehensive list of members and their sequence characteristics of the AsAQP protein family, laying a solid theoretical foundation for further functional analysis of AsAQP in oats. This research also offers valuable reference for the creation of stress-tolerant oat varieties through genetic engineering techniques.
Oat (Avena sativa L.) belongs to the early-maturity grass subfamily of the Gramineae subfamily oats (Avena) and is an annual grain feeding crop of the genus A. sativa in the family of Gramineae. According to the seed type, oats can be divided into skin oats and naked oats. Oats exhibits barrenness, resistance to salinity, cold and drought, and a high grass yield, is rich in nutrients, has high palatability and other excellent characteristics, and is widely distributed in 42 countries and regions around the globe (Zhou et al., 1999; Wu et al., 2019; Ye et al., 2023; Zhang et al., 2023). A. sativa also plays an important role in promoting the development of grassland animal husbandry and livestock feeding in winter and spring and provides an ecological pasture for soil salinization and desertification control (Gang and Zhang, 2020). The cultivation of A. sativa is important for restoring degraded natural pastures and improving the utilization of saline soils and can also play a unique role in meeting the challenges of food security and promoting grassland animal husbandry and ecological environmental protection (Nan et al., 2017; Shi et al., 2023).
Aquaporins (AQPs) are a class of small molecular intrinsic membrane proteins with major intrinsic protein domains (MIPs) located on the cell membrane. Their primary function is to form “pores” on the cell membrane, allowing for the controlled movement of water in and out of cells, forming narrow channels that facilitate single-file transport. This enables signal transduction within plant cells and regulates the dynamic balance of water, promoting rapid transmembrane water transport (Wu et al., 2015; Afzal et al., 2016; Singh et al., 2020). The AQP family was first discovered in humans and then widely isolated in animals, plants, fungi, and bacteria (Hu et al., 2015; Feng et al., 2018; Zhang et al., 2022). Since the isolation of the first plant water channel protein AtTIP1 in Arabidopsis thaliana (Zhang and Du, 2014), an increasing number of AQP have been found in plants, such as Zea mays L., Oryza sativa Linn. and Lycopersicon esculentum Miller (Chaumont et al., 2001; Sakurai et al., 2005; Li et al., 2018). Based on structural features, plant AQP proteins can be categorized into seven subfamilies: plasma membrane intrinsic proteins (PIPs), vesicular membrane intrinsic proteins (tonoplast intrinsic proteins, TIPs), Nod26-like membrane intrinsic proteins (NIPs), small basic intrinsic proteins (SIPs), X intrinsic proteins (XIPs), GlpF-like intrinsic proteins (GIPs),.and hybrid intrinsic proteins (HIPs) (Bienert and Chaumont, 2011; Abascal et al., 2014; Kapilan et al., 2018). GIPs and HIPs are currently found only in mosses, ferns and algae (Danielson and Johanson, 2008; Anderberg et al., 2012; Maurel et al., 2015). Seven subfamilies have different functions and subcellular localizations. Specifically, PIPs, XIPs, SIPs, and some NIPs are usually localized in plasma membranes, and TIPs are mostly located in vesicular membranes (Takano et al., 2006; Maeshima and Ishikawa, 2008; Bienert et al., 2011). NIPs can also localize to the membrane of the ER (Diehn et al., 2015). All AQP have a conserved hourglass structure, which comprises six transmembrane (TM) helices (TM1-TM6) formed by six α-helix bundles and five loops (Loops A-E) (TÖrnroth-Horsefield et al., 2006). Plant AQP can not only mediate the transport of water between plant tissues but also act as an ion-selective permeable membranes to effectively mediate the transmembrane transport of other small molecules, nutrient elements, and metal ions, among others, in plants and effectively improve the stress tolerance of plant by affecting the operation of small-molecule solutes such as ions, gases, and heavy metals in the plant body (Yuan, 2022). The overexpression of AtTIP5;1 in A. thaliana enhances the tolerance to boron toxicity (Pang et al., 2010); Mutations in AtNIP3;1 or AtNIP7;1 improve the tolerance to arsenate toxicity (Xu et al., 2015; Lindsay and Maathuis, 2016); and AtPIP1;2 can mediate CO2 transport (Boudichevskaia et al., 2015).
Abiotic stresses such as salinity, drought, flooding, and high- and low- temperature stresses exert inhibitory effects on plant growth, and plants can only survive under adversity conditions, especially in widespread saline and arid environments, by resisting the damage caused by water deficit through the formation of various proteins, such as AQP to evolve specific physiological mechanisms adapted to adversity stresses (Hu et al., 2012; Zhang et al., 2013). Many studies have demonstrated that AQP proteins are not only widely involved in plant growth and development but can also respond to multiple abiotic stresses. GmPIP1:6 could improve the salt resistance of Glycine max (Li et al., 2015). In barley leaves, the expression of the HvPIP1;6 gene increased under salt stress conditions, promoting leaf growth (Fricke et al., 2006). The overexpression of rice OsPIP1-1 or OSPIP2-2 in A. thaliana significantly improves the drought tolerance of the plant (Guo et al., 2006), and the overexpression of OsPIP2 in rice can improve the survival rate of rice under low-temperature stress (Li et al., 2008). Soybean GmTIP1-1 enhances soybean resistance to abiotic stress through its interaction with GmSNARE proteins involved in abiotic stress regulation and stress-responsive protein GmF-box (Li et al., 2019). The ectopic overexpression of TsTIP1;2 in A. thaliana resulted in a significant enhancement of plant tolerance to drought, salt, and oxidative stresses (Wang et al., 2014). Overexpression of the PvPIP2;9 gene in Panicum virgatum resulted in increased plant height, leaf length, aboveground biomass, cellulose content, and protein content under drought stress (Zhang et al., 2020).
To date, no study has identified or analyzed the AsAQP gene family system, and functional studies of A. sativa AQP genes in response to drought, cold and salt stress have not been conducted. In this study, we performed a gene-wide identification of the AsAQP family using bioinformatics methods and analyzed the phylogenetic relationship among AsAQP, their gene structure, and their chromosome distribution through a covariance analysis and an analysis of the protein interaction network of AsAQP. A quantitative real-time PCR (qRT-PCR) analysis was performed to examine the gene expression patterns of AsAQP in different tissues and their responses to cold, drought and salt stress. An overall expression analysis of A. sativa revealed the role of the AQP members in the different biological processes of A. sativa and these findings lay the foundation for further research on the biological functions of the family members and provides a theoretical basis for a functional analysis of the AQP proteins under abiotic stresses.
Whole genome files of A. sativa (GCA_022788535) were downloaded from the Ensembl Plants website (https://plants.ensembl.org/index.html, accessed on 19 April 2023). The sequences of A. thaliana AQP family proteins were downloaded from the TAIR database (https://www.arabidopsis.org/, accessed on 10 May 2023), a hidden Markov model (HMM) of AQP was downloaded from the Pfam database (https://pfam.xfam.org/, accessed on 12 May 2023), and the sequences of 35 A. thaliana AQP proteins were used as templates for comparison with the A. sativa genome using TBtools software, with an e-value of 10. The identified genes were screened, duplicates and shorter sequences were removed, and the candidate genes obtained from the preliminary identification were validated against the candidate AQP protein sequences through the online website NCBI Conserved Domain Search online website (https://www.ncbi.nlm.nih.gov/structure/cdd/wrpsb.cgi, accessed on 14 May 2023). The conserved structural domains of the candidate AQP protein sequences were validated, and the validation results were visualized using the software TBtools to eliminate redundant repeats and incomplete sequences that did not contain the MIP structural domains with the aim of obtaining all candidate genes of the A. sativa AQP gene family.
The AQP protein sequences of A. sativa were compared with those of wheat and A. thaliana using the ClustalW algorithm, and the phylogenetic tree was constructed with MEGA 7.0 software using the neighboring method. Based on the results of phylogenetic analysis, AsAQP were categorized into four classes: PIP, TIP, NIP and SIP.
The gene structure of the A. sativa AQP family members was analyzed using the online tool MEME (http://meme-suite.org/tools/meme) and the A. sativa genome annotation file, and their conserved motifs were analyzed based on a predicted value of 10. TBtools was used to visualize and map the obtained gene structure and the distribution of introns and exons, and the conserved motif information was visualized and mapped.
Chromosomal positions of A. sativa AQP family genes were mapped using TBtools software, and 2000 bp upstream of the start codon was extracted as the promoter sequence, and cis-acting element prediction of the promoter was performed using Plantcare.
To further analyze the A. sativa AQP family genes and to investigate whether the A. sativa AQP genes contain tandem duplications or segmental duplications, the duplication events of the AQP genes were analyzed using MCScanX software. To investigate the replication relationship between the A. sativa AQP gene family and other species, the covariance of A. thaliana, T. aestivum, and Z. mays with the A. sativa AQP gene were analyzed using the One Step MCScanX module of TBtools software. Chromosome distribution maps of the covariance of A. sativa AQP were plotted using Advanced Circos of TBtools software.
The AsAQP protein sequences were uploaded using the online software STRING (https://string-db.org/, accessed on 20 July 2023). Using the A. thaliana database sequence as reference, the AsAQP interacting proteins were predicted according to the known interactions between A. thaliana AQP proteins. Optimizing protein-protein interaction networks with Cytoscape 3.7.2 (Doncheva et al., 2019).
The oat variety selected in this experiment was Avena sativa cv. Mengyan No.1. Seeds with full grains were selected, cultivated in a petri dish until spouting, and then transplanted into a seedling cup at a vermiculite- to- nutrient soil ratio of 1:1, with 9 seeds per cup, and cultured in a greenhouse (day/night 25°C/18°C, 65%~75% relative humidity). The light intensity was 200-250 μmol·m-2s-2 and the seedlings were thinned once they grew to two-leaf monophase. After 14 days, each cup were subjected to stress. The plants in the cold treatment group were incubated in an incubator at 4°C, whereas those in the control group were cultured normally in a long-day incubator. The seedlings in the cold treatment group were sampled after 6 h, 12 h, 24 h and 48 h of treatment, and the untreated seedlings were also sampled as controls. In this study, 20% PEG6000 was used to simulate drought stress, and samples were collected at 6 time points (0 h, 2 h, 4 h, 8 h, 12 h and 24 h) during PEG6000 treatment. Moreover, 150 mM NaCl was used to simulate salt stress, and samples were collected at 6 time points (0 h, 1 h, 3 h, 6 h, 12 h and 24 h) during this treatment. Different tissue parts of A. sativa growing to the waxy ripening stage were sampled. Three biological replicates, each of which consisted of three single plants, were included in this study, and the samples (all of the germinated seedlings) were immediately frozen in liquid nitrogen and stored at -80°C until used for RNA extraction.
The FastPure Plant Total RNA Isolation Kit (TIANGEN, Beijing, China) was used for the isolation of total RNA. The RNA quality was assessed using the NanoDrop™ One/OneC microvolume UV-Vis spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA) and checked using RNAse-free 1% (w/v) agarose gel electrophoresis. cDNA was then synthesized using a TransScript II One-Step gDNA Removal and cDNA Synthesis SuperMix kit (Vazyme, Nanjing, China). The qRT-PCR primers were designed based on the full-length coding sequences (CDS) of 20 AsAQP genes, with A. sativa AsActin (KP257585.1) gene used as the reference gene (Xiang et al., 2020) (Table S1). The expression levels of 20 AsAQP genes in different tissues and organs and under drought, salt and cold stress were measured by qRT-PCR, which was performed using a Light Cycle96 with TB Green® Premix Ex Taq™ II (TAKARA, Beijing, China).The thermal cycle program was as follows: an initial step at 95°C for 3 min, followed by 40 cycles of 95°C for 15 s and 60°C for 30 s. Three biological replicates of each response were included in the experiment, and the data were analyzed using the 2-ΔΔCT method. The expression heatmaps of 20 genes in different tissues and organs and under different abiotic stresses were established by using TBtools software. Detailed qpcr data for different abiotic stress treatments in oats are available in (Table S2).
Characterization of the structural domain integrity of the AQP family genes identified by the hidden Markov model resulted in the identification of 45 family members, which is a higher number than the found in A. thaliana, which is consistent with the fact that the genome of A. sativa is larger than that of A. thaliana. The amino acid number of A. sativa AQP family members ranged from 176 to 324, the molecular weight of the proteins ranged from 18.84 kD to 34.93 kD, and the isoelectric point size was in the range of 5.33 to 11.12. Most of the A. sativa AQP members had 5-6 transmembrane structural domains, but AVESA.00001b.r3. 6Cg0003382.4 contained only three transmembrane structural domains, probably because the other transmembrane structural domains were lost during evolution (Table 1).
To further analyze the evolutionary relationship of AQP family genes and to mine the homologous evolutionary relationship between A. sativa and other species, this study constructed a phylogenetic tree used 25 protein sequences from T. aestivum 30 protein sequences from A. thaliana and 45 protein sequences from A. sativa. The A. sativa AQP proteins co-clustered into four subfamilies; 12 members of the TIP subfamily, 19 members of the PIP subfamily, 10 members of the NIP subfamily and 4 members of the SIP subfamily. The TIP, PIP, and NIP subfamilies had similar numbers of members whereas the SIP subfamily had substantially fewer members than the others, which was consistent with the distribution of members among the subfamilies of the A. thaliana AQP family (Figure 1).
To analyze the sequence conservation of AQP gene families, the conserved motifs of 45 AQP family members were identified by MEME, and a total of 10 motifs were selected and outputted. The results showed greater similarity in the types and numbers of conserved motifs among members within the same subfamily and the, frequencies of Motif 2, Motif 3, Motif 4, Motif 6 and Motif 8 were higher. Motif 2, Motif 4, Motif 6 and Motif 8 appeared more frequently, and 45 and 44 family members contained Motif 2 and Motif 4, respectively. In addition, high consistency of motifs was observed among the same subfamily, but some unique motifs were found among the different subfamilies, e.g., all members of the PIP subfamily had Motif 5, and Motif 5 was not observed in members of the other subfamilies. All five members of the NIP subfamily (1Dg0000683.1, 1Ag0000703.1, 1Cg0001352.1, 3Cg0000766.1 and 6Dg0000172.1) contain the unique Motif 10 (Figure 2). The position and type of motifs in the unified subfamily were found to be highly conserved, whereas specific motifs may exist in different subfamilies, which may reflect of the functional conservatism among subfamilies.
To obtain a more in-depth understanding of the function of the A. sativa AQP gene family, the cis-acting elements located 2000 bp upstream of the promoters of the 45 members were analyzed (Figure 3). The identified cis-acting elements can be broadly categorized into light-response-related elements, abiotic stress-related elements, phytohormone-responsive elements, and growth-development-responsive elements, which suggests that the AQP gene family may play a role in the growth and development of A. sativa and in coping with a variety of environmental changes. Moreover, 44 family members contained light-responsive elements, and the AQP gene family also contained various phytohormone-responsive elements, such as the growth hormone-responsive element AuxRE, the salicylic acid-responsive element TCA-element, and the gibberellin-responsive element GARE-motif, etc. The AQP gene family was also identified as the most effective in the development of A. sativa. Several elements related to the responses to abiotic stresses, including the anaerobic response element ARE, and the low-temperature response element LTR were identified. In addition, several elements related to growth and development were also found.
Based on the genomic information of A. sativa, the distribution of 45 A. sativa AQP genes on 21 chromosomes was analyzed (Figure 4). The results showed that these genes were unevenly distributed on 20 chromosomes and chromosome 4C did not contain an AQP gene. More than six genes were located on chromosome 6C. Moreover, the analysis revealed the presence of four genes on chromosomes 1A and 6A, three genes on chromosomes 1D, 2D, 4D, and 6D, two genes on chromosomes 2A, 3C, 7C, and 7D, and only one gene on chromosomes 1C, 2C, 3A, 3D, 4A, 5A, 5C, 5D, and 7A. AVESA.00001b.r3.3Ag0000886.1 is not shown because it was not found to be localized on a chromosome and could therefore not be plotted in the figure.
The phenomenon of gene duplication is widespread and plays an integral role in plant evolution. To analyze the evolutionary process among A. sativa AQP family genes, genome-wide duplication events among AQP family members were identified and analyzed and MCScan X software was used to draw Circos plots This analysis showed that a total of 24 genes formed genome-wide duplication pairs, that the 24 pairs of homologous genes were segmentally duplicated and that the duplication pairs resulted in many homologues of AsAQP genes among chromosomes, which increases the likelihood of evolution (Figure 5). These findings suggests that gene duplication events play an important role in the expansion of AsAQP genes.
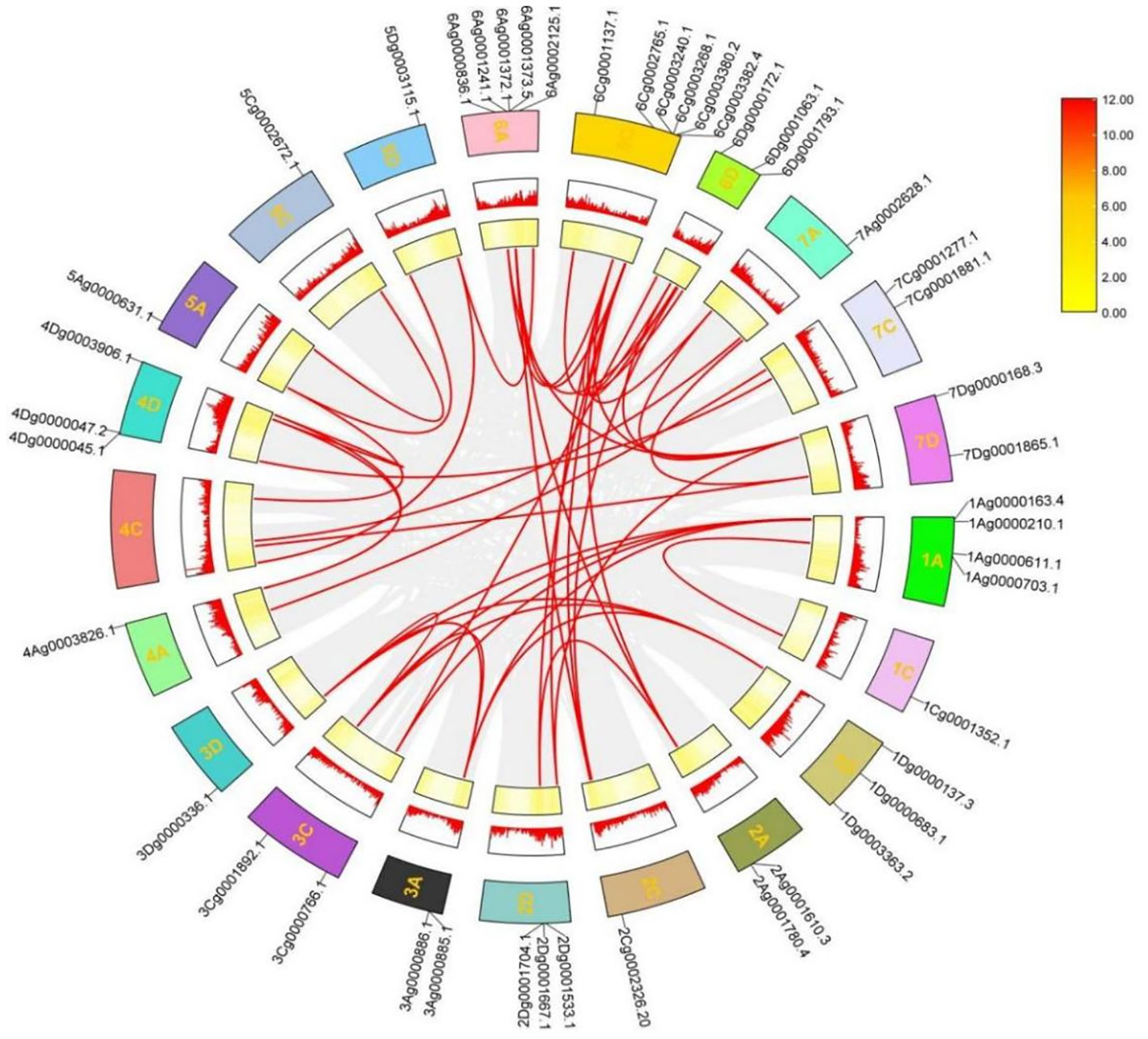
Figure 5 Chromosomal distribution and intragroup covariance analysis of the AsAQP genes. The gray section represents the collinearity of all genes within A. sativa; the red line represents the collinearity within the AQP gene family.
To further understand the possible evolutionary events of the AQP gene family in different crops, covariance analysis was performed on A. sativa simultaneously with A.thaliana and T. aestivum (Figure 6). The results revealed the existence of many collinear genes existed in A. sativa, A. thaliana and T. aestivum. Moreover, 4225 collinearity pairs were found between A. sativa and A. thaliana, which accounted for 4.59% of the total number of genes in A. sativa. In addition, 21 co-collinearity pairs were detected between A. sativa and A. thaliana, and 44,515 collinearity pairs were observed between A. sativa and Z. mays, which accounted for 43.01% of the total number of genes in A. sativa. Additionally, 35 collinearity pairs were found between A. sativa and Z. mays, and 45, 200 collinearity pairs were detected between A. sativa and T. aestivum, which accounted for 48.50% of the total number of genes in A. sativa. Seventy-six homologous gene pairs were also detected in A. sativa and T.aestivum.
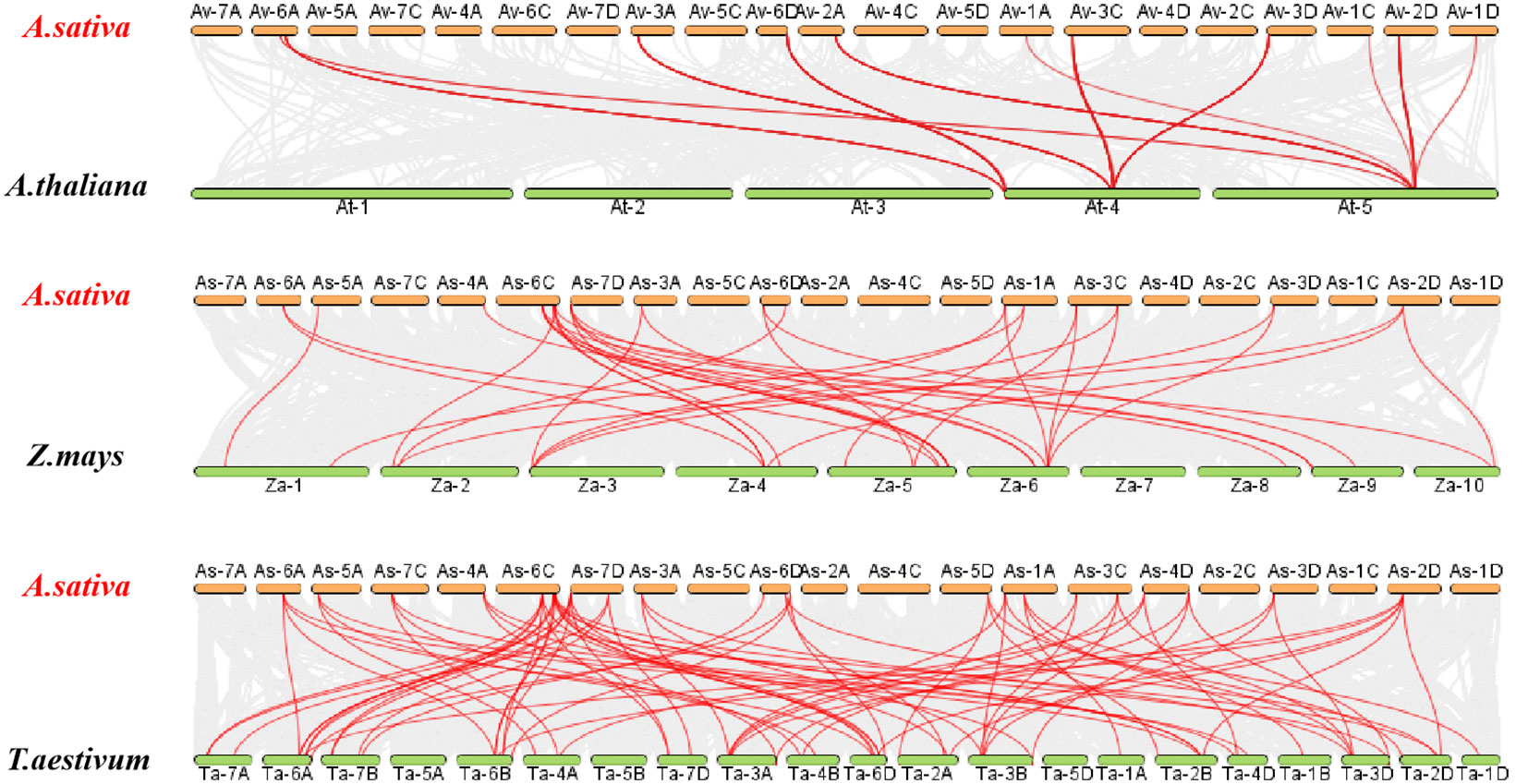
Figure 6 Analysis of AQP gene collinearity between A. sativa and three representative plants. The grey lines in the background indicate blocks of collinearity within A. sativa and the indicated plants, whereas the red lines indicate homozygous AQP gene pairs.
To better understand the function of the A. sativa AQP gene family, 45 A. sativa AQP proteins were uploaded to the STRING online database and compared with the A. thaliana protein database as a reference. Construction of the interaction network of A. sativa AQP proteins (Figure 7) revealed 23 nodes (representing AsAQP proteins) and 102 edges (representing interactions between proteins) in the regulatory network. This network indicated that 23 of the 45 AsAQP had interaction relationships, and among these, 5Ag0000633.1 had interaction relationships with 16 AsAQP proteins, which was the largest number of interacting proteins. This finding suggests that 5Ag0000633.1 may be at the core of the AQP gene family.
Twenty genes were randomly selected from the four subfamilies of A. sativa AQP proteins (TIP, PIP, NIP and SIP), which contained different protein domains and cis-elements, and their tissue-specific expression was studied. The expression heatmap of A. sativa AQP family genes in different tissues was drawn (Figure 8A). The results showed that 20 genes of different AsAQP subfamilies showed differential expressed in different organs of A. sativa, and the expression levels of most TIP subfamily genes, such as 7Dg0001865.1, 1Dg0003363.2, and 3Dg0000336, were higher in the stem nodes and spikes than in the stems and leaves. The expression of 1Ag0000611.1 was higher in spike and leaf. The expression of PIP subfamily genes was higher in stem nodes than in other tissues. Most of the NIP subfamily genes with the exception of 7Dg0000168.3 and 6Ag0000836.1 exhibited higher expression in spikes. Among the SIP subfamily genes, 4Dg0000047.3 and 5Ag0000631.1 were expressed at higher levels in stems, spikes and stem nodes. The expression levels of most genes were higher in the spikes and stems of A. sativa but lower in leaves and stems, which may be closely related to their functions and participation in A. sativa growth and development at different stages.
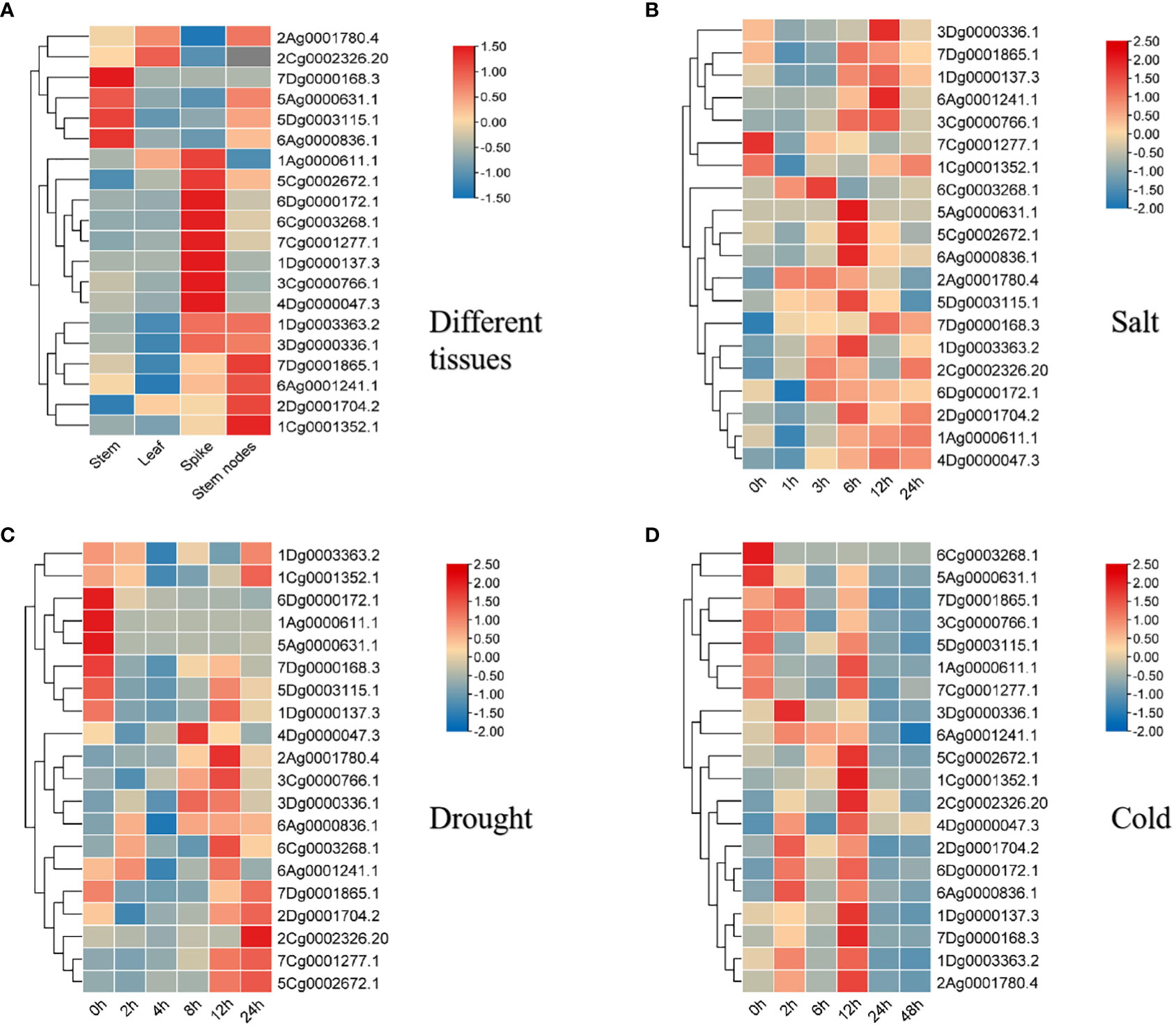
Figure 8 Expression heatmap of the A. sativa AQP gene family. (A) Tissue expression analysis of AsAQP genes in A. sativa (B) Expression of AsAQP genes under salt stress conditions. (C) Expression of AsAQP genes under drought stress conditions. (D) Expression of AsAQP genes under cold stress conditions.
To understand the response of AsAQP gene to abiotic stress, we further analyzed the expression patterns of 20 members of the AQP gene family under three abiotic stresses (salt, cold and drought). The results presented in Figure 8B show that 20 AsAQP genes exhibit varying degrees of response to the three stress treatments, and 6Cg0003268.1 was significantly upregulated after 3 h of NaCl treatment. 2Dg0001741.2, 1Dg0003363.2, 5Dg000311.1, 6Ag0000836.1, 5Cg0002672.1, and 5Ag0000631.1 were significantly upregulated and peaked after 6 h of NaCl treatment. In addition, several AQP genes were upregulated after 12 h of NaCl treatment, indicating that some AQP genes involved in the response of A. sativa to salt stress. A cluster analysis showed that AsAQP genes located in the same evolutionary branch exhibit certain similarities in response to NaCl treatment. For example, 7Dg0001865.1 and 1Dg0000137.3 showed similar expression patterns after NaCl treatment, and their expression levels were upregulated after the same treatment duration. However, 2Ag0001780.4 and 5Dg0003115.1, which are located in the same evolutionary branch, were exceptions: the expression of 2Ag0001780.4 was significantly upregulated after 1 h of NaCl treatment and then decreased, whereas the expression of 5Dg0003115.1 was significantly upregulated after 6 h of NaCl treatment. These results indicate that the 2Ag0001780.4 and 5Dg00031151.1 genes may play a significant role in the response to NaCl stress but play different roles at different times, and their functions may differ, suggesting that these genes may respond to NaCl stress through interaction. The results presented in Figure 8C showed that the expression levels of 7Dg0001865.1, 7Cg0001277.1, 2Cg0002326.20, 2Dg0001704.2, 7Dg0001865.1 and 1Cg0001352.1 were upregulated and peaked after PEG treatment for 24 h. The expression levels of 4Dg0000047.3, 2Ag0001788.4 and 2Cg0002326.20 were significantly upregulated after PEG treatment for different durations, suggesting that some AQP genes were related to the response of A. sativa to drought stress. As shown in Figure 8D, the expression levels of 3Dg0000336.1, 1Cg0001352.1, 6Ag0000836.1, 4Dg0000047.3 and 2Cg0002326.20 were upregulated after low temperature treatment at 4°C for different durations, indicating that the expression of some AQP genes was induced by low-temperature stress. Among these genes, 1Cg0001352.1 was the most highly induced gene. 6Cg0003268.1 was inhibited under low-temperature stress, and the expression levels of the remaining 19 AsAQP genes were not inhibited with increases in the stress duration, moreover, the expression levels of most genes peaked after 12 h under low-temperature stress.
With the rapid development of genome sequencing technology, an increasing number of plant genomes have been successfully sequenced, which has facilitated the identification of a wide range of plant genomes (Rose et al., 2021). The sequencing of the A. sativa genome has been completed, and its genomic data are online, which facilitates studies of the A. sativa gene family and gene evolution (Peng et al., 2022). Water is the basis of plant life, and water molecules are able to enter the cell from the environment through the cell membrane and participate in life activities. Seventy percent of transmembrane transport is accomplished through specific membrane proteins known as AQP proteins, and the AQP genes involved in the regulation of plant growth and development are abundant, diverse, and widely distributed in the plant cells (Wang and Cui, 2016). Plant AQP proteins are intrinsic membrane proteins belonging to the MIP superfamily that play important roles in water transport across membranes, cellular osmoregulation, seed germination, lateral root production, carbon fixation, nutrient uptake and stress resistance (Chen et al., 2023). AQP are widely distributed in plant genomes, and genome-wide analyses of AQP in a variety of higher plants have been reported. However, studies on analyses of the functions of the A. sativa AQP family have not been conducted. Therefore, a genome-wide analysis of AQP in A. sativa is of great importance.
In this study, 45 AsAQP family members were isolated and identified from the A. sativa genome. This number of its gene family members is greater than that the numbers found in A. thaliana, O. sativa, and Z. mays (Chaumont et al., 2001; Johanson et al., 2001; Sakurai et al., 2005), which demonstrates the diversity of these genes among different species, and it is hypothesized that gene duplication and functional differentiation of AQP genes occured during the process of plant evolution (Dassanayake et al., 2011). Based on these findings combined with the evolutionary tree of the AQP protein family, all AsAQP can be divided into four subfamilies (PIP;TIP;NIP and SIP), which is consistent with the AQP delineation categories of AQP that have been previously studied by researchers (Mahnaz and Parviz, 2023). Due to the highly compartmentalized nature of plant cells, the subcellular localization of aquaporins (AQP) exhibits diversity. AQP are present in all cellular and subcellular compartments of plants (Zhang et al., 2017). The AsAQP family consists of four subfamilies with different subcellular localizations, including the endoplasmic reticulum, vacuoles, and cytoplasm. The different subfamily members are localized to different positions in the cell, and this localization is likely closely associated with the molecular functions of the proteins. However, this study found that the A. sativa PIP subfamily member 2Ag0001780.4 is localized to the endoplasmic reticulum, while the other PIP subfamily members are localized to the cytoplasm. In A. thaliana, the NIP subfamily member NIP5;1 is localized to the plasma membrane, while another subfamily member, NIP2;1, is localized to the endoplasmic reticulum (Mizutani et al., 2006; Takano et al., 2006). This is similar to the results of this study, where members of the same subfamily exhibit different subcellular localizations. In addition, the study also found diverse localizations among the TIP subfamily members. In A. sativa, seven TIP subfamily members, such as 4Ag0003826.1, 4Dg0003906.1, 1Ag0000611.1, 3Cg0001892.1, 3Ag0000885.1, 1Ag0000210.1 and 1Dg0003363.2, are localized to the cytoplasm, while five members, such as 3Dg0000336.1, 3Ag0000886.1, 7Dg0001865.1, 6Cg0003268.1 and 2Dg0001667.1, are localized to vacuoles. The subcellular localization of ScTIP1;1 in the grass species Stipa capillacea Keng is on the Vacuoles, and its gene expression can be induced by salt, drought, and low temperature stress (Dong et al., 2016). Similarly, all PeTIPs in bamboo are localized on the Vacuoles, and RNA-seq data and qPCR results confirm their involvement in response to low temperature, drought, and strong light stress (Zhu et al., 2021). Based on this, it can be speculated that members of the AsTIPs subfamily in A. sativa, such as 3Dg0000336.1, 3Ag0000886.1, 7Dg0001865.1, 6Cg0003268.1, and 2Dg0001667.1, play different roles in combating external stressors, thereby regulating plant tolerance to abiotic stresses such as drought, salt stress, and cold.
In the present protein interaction regulatory network of the AsAQP gene family, NIP family members exhibit strong protein interactions with other AQP subfamilies, especially the SIP and PIP subfamilies. The NIP subfamily member 1Ag0000703.1 also interacts with a boron transporter (BOR4), which is speculated to be involved in the transport of boric acid and other substances. Boric acid is an essential trace element in pollen tube development, fertilization and other reproductive processes (Masataka et al., 2014), TIP subfamily members, some members of the PIP family and a palmitoyl transferase protein (PAT24) interact with palmitoyl transferase proteins involved in protein palmitoylation, regulation of cellulose biosynthesis processes, and regulation of secondary cell wall biogenesis (Chen et al., 2021). TIP, NIP, and PIP family members also interact with NAC091, which belongs to the NAC family of transcription factors, a superfamily of plant-specific transcription factors. A. thaliana NAC091 is a transcription factor that interacts with turnip crinkle virus (TCV) capsid protein and is involved in regulation of the A. thaliana defense response to TCV (Tao et al., 2005; Teresa et al., 2014). NAC091 is also upregulated by NaCl, low temperature (4°C) and ABA, among other stresses, and can also transfer ER stress signals from the plasma membrane to the nucleus; thus, this transcription factor plays an important role in alleviating ER stress and promoting cell survival (Jian, 2020). It can be inferred that some members of the A. sativa AQP family interact with the transcription factor NAC091 to play an important role in the regulation of abiotic stress in forage plants.
In an environment with an abiotic stress, such as salt, drought and low temperature, dry air, ion stress and temperature changes in the surrounding environment will cause water absorption difficulties in plants. The signal molecules generated by water loss enable plants to maintain their water balance by changing the expression of AQP (Sutka et al., 2011). An analyze of the expression patterns of AsAQP family genes under low temperature, salt and drought stress indicated that most AsAQP genes may be involved in the abiotic stress response. Low temperature stress upregulates the expression of 15 AsAQP family genes, which suggests that these genes may play a positive regulatory role in the molecular regulatory pathways that enhance the low-temperature adaptability of A. sativa, and these genes may be important candidate genes involved in the response of A. sativa to cold stress. GhTIP1;1 in cotton also exhibits an obvious positive regulatory effect, and upregulation of this gene in A. thaliana significantly increases the expression level of cold-response genes (Gongmin et al., 2022). Moreover, OsPIP2;7 overexpression in rice improves the survival rate of rice under low-temperature stress and affects the expression levels of other AQP genes in rice (Li et al., 2008). Salt stress significantly upregulated the expressions of 1Dg0003363.2, 2Ag0001780.4 and 6Ag0000836.1, which predicted that these three genes could positively regulate the salt tolerance of A. sativa, indicating that the positive regulatory function of AQP gene under salt and cold stress is relatively common. Drought stress inhibited the expression of 11 AsAQP family genes, which suggests that these genes may play a negative regulatory role in the molecular pathways that enhance drought adaptability in A. sativa. The AQP gene expression patterns of A. thaliana were substantially different under drought stress; for example, AtPIP2;1 and AtPIP2;2 expression was down-regulated under drought stress (Jin, 2015), but AtPIP1;3 transcription was also detected; and AtPIPs expression was up-regulated in a few groups (Alexandersson et al., 2010). The analysis of OsPIP1 expression in rice under drought stress revealed that the expression of these two genes was up-regulated, whereas the expression of all OsPIP2s genes were down-regulated (Liu et al., 2006). These results indicate that the AQP gene expression patterns in plants under drought stress are complex. Similar to the above-described results, this study found that AsAQP genes in A. sativa under drought stress mainly exhibit two expression patterns: an “up-down-regulated” expression pattern, and a “down-up-regulated” expression pattern. These results indicate that the expression of AQP gene expression in plants under drought stress is regulated by a complex signaling network, and the regulatory mechanism of AQP genes in plant drought resistance needs further study. 6Cg0003268.1 and 3Dg0000336.1 belong to the NIP subfamily types and show similar expression patterns under drought and salt stress, which further indicates that the evolutionarily similar genes exhibit functional redundancy to a certain extent. Moreover, the same type of gene also shows functional differentiation, and low-temperature treatment strongly inhibited the expression of 6Cg0003268.1 but only slightly change the expression level of 1Ag0000611.1. The expression of 6Ag0000836.1 was significantly up-regulated under salt, drought and cold stress, which suggests that this gene is an important candidate gene involved in the biotic stress response. A. sativa is an important and high-quality forage that is sensitive to low temperature, drought and high salt (Song et al., 2013; Kamal et al., 2022). Further study of AsAQP gene functions in the abiotic stress response is of great significance for the germplasm conservation and molecular assisted breeding of A. sativa.
In this study, the AQP gene family in the whole genome of A. sativa was analyzed, and a total of 45 AQP genes were identified through analyses of the evolutionary relationships, gene structure, gene duplication, promoter and regulatory network of the AQP gene family based on bioinformatic data. According to the A. thaliana AQP subfamily classification, the AQP gene family is divided into 4 subfamilies, and the PIP and SIP subfamilies have the highest (19) and fewest (4) members. AQP genes from the same evolutionary branch were found to have similar genes and structures. Among the 45 AsAQP, 23 proteins had interaction relationships, and 5Ag0000631.1 had the largest number of interacting proteins. An expression analyses revealed that these 20 genes were expressed in all tissues, and the expression of these genes showed marked differences among different tissues and organs, indicating that these AsAQP genes have different functions in the growth and development of A. sativa. The expression analysis of 20 AsAQP genes under cold, drought and salt stress showed that the expression of 6Ag0000836.1 gene of the NIP subfamily was significantly upregulated under different abiotic stresses, and this gene could be further verified as a key candidate gene involved in the responses to stress. The results of this study provide the complete members of the AsAQP protein family and their sequence characteristics and can serve as a reference and theoretical basis for further analyses of the structural characteristics of AsAQP and improving the stress resistance of A. sativa.
The oat data presented in the study are deposited in the Ensembl Plants website repository, accession number GCA_022788535”; Arabidopsis thaliana data presented in the study are deposited in the TAIR database repository.
XZ: Conceptualization, Data curation, Formal Analysis, Visualization, Writing- original draft, Writing-review & editing. DY: Writing-original draft. LM: Investigation,Writing-review & editing. XW: Validation, Visualization, Writing-review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by grants from the China Agriculture Research System of MOF and MARA(CARS-34), the National Crop Germplasm Resources Center(NCGRC-63), the Agricultural Science and Technology Innovation Program (ASTIP-IAS10).
We are grateful to Qingxue Jiang and Zhipeng Zhang for their contributions in the sampling investigation for this study.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1305299/full#supplementary-material
Abascal, F., Irisarri, I., Zardoya, R. (2014). Diversity and evolution of membrane intrinsic proteins. Biochim. Biophys. Acta 1840 (5), 1468–1148. doi: 10.1016/j.bbagen.2013.12.001
Afzal, Z., Howton, C. T., Sun, Y., Shahid Mukhtar, M. (2016). The roles of aquaporins in plant stress responses. J. Dev. Biol. 4 (1), 9. doi: 10.3390/jdb4010009
Alexandersson, E., Danielson Jonas, A. H., Moparthi, V.K., Fontes, M., Kjellbom, Johanson, P. U. (2010). Johan RådeTranscriptional regulation of aquaporins in accessions of Arabidopsis in response to drought stress. Plant J. 61 (4), 650–660. doi: 10.1111/j.1365-313X.2009.04087.x
Anderberg, H. I., Kjellbom, P., Johanson, U. (2012). Annotation of Selaginella moellendorffii major intrinsic proteins and the evolution of the protein family in terrestrial plants. Front. Plant Sci. 3. doi: 10.3389/fpls.2012.00033
Bienert, G. P., Chaumont, F. (2011). Plant aquaporins: roles in water homeostasis, nutrition, and signaling processes. Transporters. Pumps. Plant Signaling 7, 3–36. doi: 10.1007/978-3-642-14369-4_1
Bienert, G. P., Manuela, D. B., Jahn, T. P., Boutry, M., François, C. (2011). Solanaceae XIPs are plasma membrane aquaporins that facilitate the transport of many uncharged substrates. Plant J. 66 (2), 306–317. doi: 10.1111/J.1365-313X.2011.04496.X
Boudichevskaia, A., Heckwolf, M., Kaldenhoff, R. (2015). T-DNA insertion in aquaporin gene AtPIP1;2 generates transcription profiles reminiscent of a low CO2 response. Plant Cell Environ. 38 (11), 2286–2298. doi: 10.1111/pce.12547
Chaumont, F., Barrieu, F., Wojcik, E., Chrispeels, M. J., Jung, R. (2001). Aquaporins constitute a large and highly divergentprotein family in maize. Plant Physiol. 125 (3), 1206–1215. doi: 10.1104/PP.125.3.1206
Chen, D. Q., Hao, F. H., Mu, H. Q., Ahsan, N., Stacey, G. (2021). S-acylation of P2K1 mediates extracellular ATP-induced immune signaling in Arabidopsis. Nat. Commun. 12, 2750. doi: 10.1038/S41467-021-22854-1
Chen, Y., You, C. H., Chen, Y. L., Sun, T. T., Zhao, Z. N., Huang, T. C., et al. (2023). Research progress on structure, function and regulation of plant NIP aquaporins. Chin. Sci. Bull. 68 (14), 1799–1813. doi: 10.1360/TB-2022-1110
Danielson, J. A., Johanson, U. (2008). Unexpected complexity of the aquaporin gene family in the moss Physcomitrella patens. BMC Plant Biol. 8, 45. doi: 10.1186/1471-2229-8-45
Dassanayake, M., Oh, D. H., Haas, J. S., et al. (2011). The genome of the extremophile crucifer Thellungiella parvula. Nat. Genet. 43, 913–918. doi: 10.1038/ng.889
Diehn, A. T., EBernhardt, B., et al. (2015). Genome-wide identification of aquaporin encoding genes in Brassica oleracea and their phylogenetic sequence comparison to Brassica crops and Arabidopsis. Front. Plant Sci. 6. doi: 10.3389/fpls.2015.00166
Doncheva, N. T., Morris, J. H., Gorodkin, J., Jensen, L. J. (2019). Cytoscape StringApp: Network analysis and visualization of proteomics Data. J. Proteome Res. 18 (2), 623–632. doi: 10.1021/acs.jproteome.8b00702
Dong, C., Yang, Y., Sun, X., Li, W., Yang, S. H., Huang, R., et al. (2016). Cloning of the ScTIP1;1 gene in Miscanthus sinensis and its response to abiotic stress. J. Plant Sci. 34 (01), 99–108. doi: 10.11913/PSJ.2095-0837.2016.10099
Feng, Z. J., Xu, S. C., Liu, N., Zhang, G.W., Hu, Q.Z., Xu, Z.S., et al. (2018). Identification of the AQPs members involved in abiotic stress re-sponses from Arabidopsis. Gene 646, 64–73. doi: 10.1016/j.gene.2017.12.048
Fricke, W., Akhiyarova, G., Wei, W., Alexandersson, E., Miller, A., Kjellbom, P.O., et al. (2006). The short-term growthresponse to salt of the developing barley leaf. J. Exp. Bot. 57 (5), 1079–1095. doi: 10.1093/jxb/erj095
Gang, Y. H., Zhang, H. B. (2020). Status and role of oat in sustainable development of grassland animal husbandry in Qinghai Province. Qinghai. Pratacult. 29 (01), 54–57. doi: 10.3969/j.issn.1008-1445.2020.01.013
Gongmin, C., Mengdi, W., Zhang, L. Y., Wei, H.L., Wang, H.T., Lu, J.H., et al. (2022). Overexpression of a cotton aquaporin gene ghTIP1;1-like confers cold tolerance in transgenic Arabidopsis. Int. J. Mol. Sci. 23 (3), 1361. doi: 10.3390/IJMS23031361
Guo, L., Wang, Z. Y., Lin, H., Cui, W.E., Chen, J., Liu, M.H., et al. (2006). Expression and functional analysis of the rice plasmamembrane intrinsic protein gene family. Cell Res. 16 (3), 277–286. doi: 10.1038/sj.cr.7310035
Hu, W., Hou, X., Huang, C., Yan, Y., Tie, W.W., Ding, Z.H., et al. (2015). Genome-wide identification and expression analyses of aquaporin gene family during development and abiotic stress in banana. Int. J. Mol. Sci. 16 (8), 19728–19751. doi: 10.3390/ijms160819728
Hu, W., Yuan, Q., Wang, Y., Cai, R., Deng, X., Wang, J., et al. (2012). Overexpression of a wheat aquaporin gene,TaAQPs8,enhances salt stress tolerance in transgenic tobacco. Plant Cell Physiol. 53 (12), 2127–2141. doi: 10.1093/pcp/pcs154
Jian, Y. (2020). Cloning of AtNAC091 membrane-binding transcription factor and its biological function under stress (China: Guizhou normal university).
Jin, M. (2015). The role of PIP aquaporins in response to various environmental scenarios in Arabidopsis thaliana (China: Inner Mongolia University).
Johanson, U., Karlsson, M., Johansson, I., Gustavsson, S., Sjö̈vall, S., Fraysse, L., et al. (2001). The complete set of genes encoding major intrinsic proteins in Arabidopsis provides a framework for a new nomenclature for major intrinsic proteins in plants. Plant Physiol. 126 (4), 1358–1369. doi: 10.1104/pp.126.4.1358
Kamal, N., Tsardakas, R. N., Bentzer, J., Gundlach, H., Haberer, G., Juhász, A., et al. (2022). The mosaic oat genome gives insights into a uniquely healthy cereal crop. Nature 606 (7912), 113–119. doi: 10.1038/s41586-022-04732-y
Kapilan, R., Vaziri, M., Zwiazek, J. J. (2018). Regulation of aquaporins in plants under stress. Biol. Res. 51 (1), 4. doi: 10.1186/s40659-018-0152-0
Li, J., Ban, L. P., Wen, H. Y., Wang, Z., Dzyubenko, N., Chapurin, V., et al. (2015). An aquaporin protein is associated with drought stress tolerance. Biochem. Biophys.Res. Commun. 459 (2), 208–213. doi: 10.1016/j.bbrc.2015.02.052
Li, F., He, X. H., Li, Y. K., Yi, Y. (2018). Bioinformatic analysisof tomato aquaporin gene family. Genomics Appl. Biol. 37 (09), 3950–3957. doi: 10.13417/j.gab.037.003950
Li, S., Huang, Y., Xuan, H., Huang, L., Zhao, J.M., Wang, H.T., et al. (2019). Cloning and functional study of soybean GmTIP1-1 gene. J. Nanjing. Agric. University. 42 (5), 793–801. doi: 10.7685/jnau.201901024
Li, G. W., Zhang, M. H., Cai, W. M., Sun, W.N., Su, W.A. (2008). Characterization of OsPIP2;7, a water channel protein in rice. Plant Cell Physiol. 49 (12), 1851–1858. doi: 10.1093/pcp/pcn166
Lindsay, E. R., Maathuis, F. J. (2016). Arabidopsis thaliana NIP7;1 is involved in tissue arsenic distribution and tolerance in response to arsenate. FEBS Lett. 590 (6), 779–786. doi: 10.1002/1873-3468.12103
Liu, H. Y., Sun, W. N., Su, W. A., Tang, Z. C. (2006). Co-regulation of water channels and potassium channels in rice. Physiol. Plant 128 (1), 58–69. doi: 10.1111/j.1399-3054.2006.00709.x
Maeshima, M., Ishikawa, F. (2008). ER membrane aquaporins in plants. Pflug. Arch. Eur. J. Phy. 456 (4), 709–716. doi: 10.1007/S00424-007-0363-7
Mahnaz, Y., Parviz, H. (2023). Genome-wide analysis of aquaporin gene family in Triticum turgidum and its expression profile in response to salt stress. Genes 14 (1), 202. doi: 10.3390/GENES14010202
Masataka, U., Sheliang, W., Takehero, K., Shuji, S., Katsushi, Y., Toru, F., et al. (2014). Identification and characterizationofan Arabidopsis mutant with altered localization of NIP5;1, a plasma membrane boric acid channel,reveals the requirement ford-Galactoseinendomembrane organization. Plant Cell Physiol. 55 (4), 704–714. doi: 10.1093/pcp/pct191
Maurel, C., Boursiac, Y., Luu, D. T., Santoni, V., Shahzad, Z., Verdoucp, L., et al. (2015). Aquaporins in plants. Physiol. Rev. 95 (4), 1321–1358. doi: 10.1152/physrev.00008.2015
Mizutani, M., Watanabe, S., Nakagawa, T., Maeshima, M. (2006). Aquaporin NIP2;1 is mainly localized to the ER membrane and shows root-specific accumulation in Arabidopsis thaliana. Plant Cell Physiol. 47 (10), 1420–1426. doi: 10.1093/pcp/pcl004.54
Nan, M., Zhao, G. Q., Chai, J. K. (2017). Analysis and comprehensive evaluation of genetic diversity of germplasm phenotypic traits of forage oat in semiarid region of loess plateau. Acta Agrestia. Sin. 25 (06), 1197–1205. doi: 10.11733/j.issn.1007-0435.2017.06.006
Pang, Y., Li, L., Ren, F., Lu, P., Wei, P., Cai, J., et al. (2010). Overexpression of the tonoplast aquaporin AtTIP5;1 conferred tolerance to boron toxicity in Arabidopsis. Genet. Genomics 37 (06), 389–397. doi: 10.1016/S1673-8527(09)60057-6
Peng, Y. Y., Yang, H. H., Guo, L. C., Deng, C., Wang, C.L., Wang, Y.B., et al. (2022). Reference genome assemblies reveal the origin and evolution of allohexaploid oat. Nat. Genet. 54, 1248–1258. doi: 10.1038/S41588-022-01127-7
Rose, A.M., Scott, H., B, P. F., VanBuren, R. (2021). Representation and participation across 20 years of plant genome sequencing. Nat. Plants. 7 (12), 1571–1578. doi: 10.1038/S41477-021-01031-8
Sakurai, J., Ishikawa, F., Yamaguchi, T., Uemura, M., Maeshima, M. (2005). Identification of 33 rice aquaporin genes and analysis of their expression and function. Plant Cell Physiol. 46 (9), 1568–1577. doi: 10.1093/pcp/pci172
Shi, Q., Cai, T., Liu, Y., Du, J., Wudabala, M., Ma, L., et al. (2023). Analysis of the current development status of the forage and feed industry in Inner Mongolia and suggestions for countermeasures. Anim. Husb. Feed. Sci. 44 (02), 103–112. doi: 10.12160/j.issn.1672-5190.2023.02.016
Singh, K. R., Deshmukh, R., Muthamilarasan, M., Rani, R., Prased, M. (2020). Versatile roles of aquaporin in physiological processes and stress tolerance in plants. Plant Physiol. Biochem. 149, 178–189. doi: 10.1016/j.plaphy.2020.02.009
Song, L., Alicia, H., Nobuhiro, S., Shulaev, E., Luo, X.Z., Cenariu, D., et al. (2013). Linking genes of unknown function with abiotic stress responses by high-throughput phenotype screening. Physiol. Plant 148 (3), 322–333. doi: 10.1111/ppl.12013
Sutka, M., Li, G., Boudet, J., Boursiac, Y., Doumas, P., Maurel, C. (2011). Natural variation of root hydraulics in Arabidopsis grown in normal and salt-stressed conditions. Plant Physiol. 155 (3), 1264–1276. doi: 10.1104/pp.110.163113
Takano, J., Wada, M., Ludewig, U., Schaaf, G., Wirén, Fujiwara, T. (2006). The Arabidopsis major intrinsic protein NIP5;1 is essential for efficient boron uptake and plant development under boron limitation. Plant Cell. 18 (6), 1498–1509. doi: 10.1105/TPC.106.041640
Tao, R., Feng, Q., T. Jack, M. (2005). The nuclear localization of the Arabidopsis transcription factor TIP is blocked by its interaction with the coat protein of Turnip crinkle virus. Virology 331 (2), 316–324. doi: 10.1016/j.virol.2004.10.039
Teresa, D., Feng, Q., Paul, T., Morris, T.J. (2014). Turnip crinkle virus coat protein inhibits the basal immune response to virus invasion in Arabidopsis by binding to the NAC transcription factor TIP. Virology 449, 207–214. doi: 10.1016/j.virol.2013.11.018
TÖrnroth-Horsefield, S., Wang, Y., Hedfalk, K., Johanson, U., Karlsson, M., Tajkhorshid, E., et al. (2006). Structural mechanism of plant aquaporin gating. Nature 439 (7077), 688–694. doi: 10.1038/nature04316
Wang, L. L., Chen, A. P., Zhong, N. Q., Liu, N., Wu, X. M., Wang, F., et al. (2014). The Thellungiella salsuginea tonoplast aquaporin TsTIP1;2 functions in protection against multiple abiotic stresses. Plant Cell Physiol. 55 (1), 148–161. doi: 10.1093/pcp/pct166
Wang, W. C., Cui, K. H. (2016). Abiotic stress on the research progress of plant hole protein expression regulation. J. Plant Physiol. 52 (04), 423–430. doi: 10.13592/j.cnki.ppj.2016.0043
Wu, X., Du, C. X., Yang, B. B., Fan, H.F. (2015). Research review on plant aquaporins. J. Zhejiang. A. F. University. 32 (05), 789–796. doi: 10.11833/j.issn.2095-0756.2015.05.020
Wu, X. M., Chen, Z. J., Chi, H. W., Jia, H. L., Yang, R. D., Shi, Y. H, et al (2019). Comparison of agronomic traits and quality of 25 forage Oat cultivars. Pratac. Sci 40 (07), 1902–1912. doi: 10.11829/j.issn.1001-0629.2022-1005
Xiang, D., Man, L., Wang, Q., Wang, X.D., Zhang, W.W., Liu, P., et al. (2020). Cloning of the oat protein kinase gene AsSnRK2.10 and analysis of its response to abiotic stress. J. Agric. Biotechnol. 28 (11), 1923–1935. doi: 10.3969/j.issn.1674-7968.2020.11.003
Xu, W., Dai, W., Yan, H., Li, S., Shen, H., Chen, Y., et al. (2015). Arabidopsis NIP3;1 Plays an important role in arsenic uptake and root to shoot translocation under arsenite stress conditions. Mol. Plant 8 (5), 722–733. doi: 10.1016/j.molp.2015.01.005
Ye, X. L., Gan, Z., Wan, Y., Xiang, D.B., Wu, X.Y., Wu, Q., et al. (2023). Research progress and prospect of breeding for forage oat. Acta Pratacult. Sin. 32 (02), 160–177. doi: 10.11686/cyxb2022263
Yuan, Z. Y. (2022). Genome-wide identification and expression analysis of water channel protein gene family in sweet potato. Mol. Plant Breed. 20 (05), 1452–1461. doi: 10.13271/j.mpb.020.001452
Zhang, L., Du, X. G. (2014). Recent advances in plant aquaporins. Plant Sci. J. 32 (3), 304–314. doi: 10.3724/SP.J.1142.2014.30304
Zhang, Y. X., Fei, S. H., Xu, Y.m., He, Y., Zhu, Z. J., Liu, Y. Y. (2022). The structure, function and expression analysis of the nodulin 26-like intrinsic protein subfamily of plant aquaporins in tomato. Sci. Rep. 12 (01), 9180. doi: 10.1038/S41598-022-13195-0
Zhang, J., Gao, W. B., Yan, L., Zhang, Z.W., Zhou, H.T., Wu.B., et al. (2023). Identification and evaluation of salt-alkali tolerance of Oat germplasm resources and screening of salt-alkali tolerance germplasm. Acta Agron. Sin. 49 (06), 1551–1561. doi: 10.3724/SP.J.1006.2023.21032
Zhang, J., Li, D., Zou, D., Luo, F., Wang, X., Zheng, Y., et al. (2013). A cotton gene encoding a plasma membrane aquaporin is involved in seedling development and in response todrought stress. Acta Biochim. Biophys. Sin. 45 (2), 104–114. doi: 10.1093/abbs/gms096
Zhang, L., Peng, D., Zhang, J., Zhang, F. C. (2017). Function and regulatory mechanisms of aquaporins in plant stress resistance. Mol. Plant Breed. 15 (11), 4441–4450. doi: 10.13271/j.mpb.015.004441
Zhang, J., Wen, W. W., Li, H., Lu, Q. Y., Xu, B., Huang, B. R. (2020). Overexpression of an aquaporin gene PvPIP2;9 improved biomass yield,protein content,drought tolerance and water use efficiency in switchgrass (Panicum virgatum L.). GCB. Bioenergy. 12 (11), 979–991. doi: 10.1111/gcbb.12751
Zhou, X., Jelle, V., Murph, Y., Zhang, F. C. (1999). Progenitor germplasm of domesticated hexaploid oat. Crop Sci. 39 (04), 1208–1214. doi: 10.2135/cropsci1999.0011183X003900040042x
Keywords: Avena sativa L., aquaporin, gene family, genome-wide, abiotic stress
Citation: Zhou X, Yi D, Ma L and Wang X (2024) Genome-wide analysis and expression of the aquaporin gene family in Avena sativa L.. Front. Plant Sci. 14:1305299. doi: 10.3389/fpls.2023.1305299
Received: 01 October 2023; Accepted: 19 December 2023;
Published: 19 January 2024.
Edited by:
Rupesh Deshmukh, Central University of Haryana, IndiaReviewed by:
Madhab Kumar Sen, Czech University of Life Sciences Prague, CzechiaCopyright © 2024 Zhou, Yi, Ma and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lin Ma, bWFsaW5AY2Fhcy5jbg==; Xuemin Wang, d2FuZ3h1ZW1pbkBjYWFzLmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.