- 1Department of Plant & Microbial Biology, University of California, Berkeley, Berkeley, CA, United States
- 2Plant Stress and Germplasm Development Unit, Cropping Systems Research Laboratory, U.S. Department of Agriculture-Agricultural Research Service, Lubbock, TX, United States
- 3Plant Gene Expression Center, U.S. Department of Agriculture-Agricultural Research Service, Albany, CA, United States
Introduction: A fundamental developmental switch for plants is transition from vegetative to floral growth, which integrates external and internal signals. INDETERMINATE1 (Id1) family proteins are zinc finger transcription factors that activate flowering in grasses regardless of photoperiod. Mutations in maize Id1 and rice Id1 (RID1) cause very late flowering. RID1 promotes expression of the flowering activator genes Early Heading Date1 (Ehd1) and Heading date 1 (Hd1), a rice homolog of CONSTANS (CO).
Methods and results: Mapping of two recessive late flowering mutants from a pedigreed sorghum EMS mutant library identified two distinct mutations in the Sorghum bicolor Id1 (SbId1) homolog, mutant alleles named sbid1-1 and sbid1-2. The weaker sbid1-1 allele caused a 35 day delay in reaching boot stage in the field, but its effect was limited to 6 days under greenhouse conditions. The strong sbid1-2 allele delayed boot stage by more than 60 days in the field and under greenhouse conditions. When sbid1-1 and sbid1-2 were combined, the delayed flowering phenotype remained and resembled that of sbid1-2, confirming late flowering was due to loss of SbId1 function. Evaluation of major flowering time regulatory gene expression in sbid1-2 showed that SbId1 is needed for expression of floral activators, like SbCO and SbCN8, and repressors, like SbPRR37 and SbGhd7.
Discussion: These results demonstrate a conserved role for SbId1 in promotion of flowering in sorghum, where it appears to be critical to allow expression of most major flowering regulatory genes.
Introduction
Plant flowering behavior is determined by transcriptional and posttranscriptional signaling networks that promote flowering under inductive photoperiods and also repress flowering under non-inductive photoperiods. A conserved integration point for flowering signals is the CONSTANS (CO) - FLOWERING LOCUS T (FT) regulatory module (Song et al., 2015), named for genes first discovered in Arabidopsis thaliana. CO encodes a transcription factor with B-box domains and a signature CO, CO-LIKE and TIMING OF CAB1 (CCT) domain that is a member of a widely conserved family in plants (Putterill et al., 1995; Griffiths et al., 2003). Arabidopsis FT protein is part of the larger plant PEBP-related family conserved throughout flowering plants containing FT-like florigen-related proteins (Danilevskaya et al., 2008; Turck et al., 2008). Leaf expressed FT-like proteins in Arabidopsis, rice, tomato, and cucurbits serve as molecular florigens that stimulate floral development at the shoot apical meristem (Lifschitz et al., 2006; Jaeger and Wigge, 2007; Lin et al., 2007; Tamaki et al., 2007; Notaguchi et al., 2008). The primary role of CO is regulation of FT expression. Whether CO protein activates or represses its FT target genes varies among plants. In Arabidopsis, CO activates FT expression under floral inductive long-day (LD) photoperiods to promote flowering (Samach and Coupland, 2000; Valverde et al., 2004; Song et al., 2012). The rice CO ortholog Heading date 1 (Hd1) upregulates expression of the FT ortholog Heading date 3a (Hd3a) in floral inductive SD photoperiods but is a repressor in LD (Yano et al., 2000; Kojima et al., 2002). Hd1 potentially adopts its repressive function by participating in a co-repressor complex with the LD-expressed flowering repressor rice PSEUDORESPONSE REGULATOR 37 (OsPRR37) (Koo et al., 2013). In this role, Hd1 also represses the expression of rice Early heading date 1 (Ehd1). Ehd1 encodes a B-type response regulator that promotes Hd3a expression under SD separate from Hd1 (Doi et al., 2004; Itoh et al., 2010; Zhao et al., 2015). CONSTANS OF Zea mays1 (CONZ1) is an ortholog of rice Hd1 (Miller et al., 2008) proposed to regulate the maize florigen-related gene Zea mays CENTRORADIALIS 8 (ZCN8). ZCN8 is a presumed florigen, because silencing ZCN8 expression delays flowering in maize (Meng et al., 2011) and transgenic phloem-specific expression of ZCN8 in Arabidopsis produces early flowering in an ft mutant background (Lazakis et al., 2011).
Sorghum CONSTANS (SbCO) acts upstream to promote the expression of SbEhd1 and sorghum CENTRORADIALIS 8 (SbCN8), the co-linear ortholog of maize ZCN8, and SbCN12, a PEBP-family gene orthologous between maize and sorghum (Murphy et al., 2011; Yang et al., 2014b). Both SbCN8 and SbCN12 possess florigen activity when overexpressed in Arabidopsis (Wolabu et al., 2016). Collectively, SbCN8 and SbCN12 are regulated by SbCO and SbEhd1 (Murphy et al., 2014; Yang et al., 2014b), consistent with this set of genes acting as the CO-FT module in sorghum.
Additional upstream regulators of SbCO and SbEhd1 are the repressors sorghum PRR37 (SbPRR37) (Murphy et al., 2011) and sorghum Grain number, plant height, and heading date (SbGhd7) (Murphy et al., 2014). SbPRR37 encodes a type of transcriptional repressor originally discovered as core circadian clock genes in Arabidopsis (Farre and Liu, 2013), but SbPRR37 has no contribution to sorghum circadian clock function (Murphy et al., 2011). Differentially functional SbPRR37 alleles underlie the flowering time-associated Maturity locus Ma1, which has the largest impact on sorghum flowering time (Quinby, 1945; Quinby, 1967). As is typical of PRR37 proteins, SbPRR37 contains a CCT domain like Hd1/CO, but it belongs to a distinct protein subfamily (Murakami et al., 2003; Murphy et al., 2011). SbGhd7 is a homolog of rice Ghd7 discovered as a quantitative trait locus contributing to natural variation in heading date (Xue et al., 2008). Ghd7 proteins also contain a CCT domain but do not contain the B-box domains found in Hd1/CO (Murphy et al., 2014). Genetic variation at SbGhd7 corresponds to sorghum Maturity locus Ma6 (Rooney and Aydin, 1999; Murphy et al., 2014). SbPRR37 and SbGhd7 inhibit flowering under LD conditions, by repressing the expression of flowering activators, primarily SbEhd1 but also SbCO, to ultimately repress the expression of florigen-related genes like SbFT, SbCN8, and SbCN12 (Murphy et al., 2011; Murphy et al., 2014). Under LD, SbPRR37 is also proposed to interact with SbCO as part of a repressor complex that turns SbCO into an inhibitor of florigen gene expression (Yang et al., 2014b).
Genes in the INDETERMINATE1 (Id1) family act as activators of flowering in grasses. The maize id1 mutant is very late flowering (Singleton, 1946; Colasanti et al., 1998). Mutations in the rice Id1 homolog, RID1, cause a never-flowering phenotype (Wu et al., 2008). RID1 contributes to stimulation of Ehd1 expression to promote flowering in LD and SD photoperiods (Matsubara et al., 2008; Park et al., 2008). Id1 proteins belong to a family of transcription factors characterized by a unique arrangement of four Cys2His2-type zinc finger motifs (Colasanti et al., 2006). Maize Id1 protein binds to DNA in vitro at an 11-bp DNA sequence motif (Kozaki et al., 2004). RID1 has been demonstrated to bind promoter regions of rice Hd3a and RFT1 in vitro (Deng et al., 2017). Also, ChIP-seq analysis with RID1 demonstrated it binds upstream of several flowering time genes, including rice Hd1 (Zhang et al., 2022b). Comparison of chromatin modifications in the maize id1 mutant to the wild type indicated that Id1 influences the chromatin state around florigen genes ZCN8 and ZCN7, which is apparent as changes in histone acetylation (Mascheretti et al., 2015). Similarly, RID1 is involved in regulating chromatin accessibility and histone methylation at the promoters of Hd3a and RTF1 (Zhang et al., 2022a).
To identify genes that promote flowering in sorghum, late-flowering mutants were identified in a pedigreed sorghum EMS mutant library in the BTx623 genetic background (Jiao et al., 2016). Two independent late-flowering mutants discovered in this population each substantially delayed the timing of boot stage, an early visual indicator of flowering. The causal mutation in each line was identified by whole-genome resequencing and with preexisting sequence knowledge of the EMS mutations present in the parental population. In each case, the causal mutation was in the sorghum Id1 (SbId1) homolog. The sbid1-1 allele is a non-synonymous mutation that changes a conserved amino acid in the second zinc finger domain of SbId1. The sbid1-2 allele is a nonsense mutation that results in a predicted SbId1 protein approximately half its normal size. Plants carrying the weaker sbid1-1 eventually flowered in the field, but plants with the sbid1-2 allele did not. The flowering delay in sbid1-1 was reduced when plants were grown under greenhouse conditions to nearly that of wild-type plants. When sbid1-1 and sbid1-2 were combined, the flowering phenotype was that of sbid1-2. Evaluation of major flowering time regulatory gene expression in the sbid1-2 background showed that Id1 is needed for expression of floral activators, like SbCO and SbCN8, and repressors, like SbPRR37 and SbGhd7. These results demonstrate a conserved role for SbId1 in promotion of flowering in sorghum, where it appears to be critical to allow expression of most major regulatory genes.
Methods
Plant lines and environmental conditions
All sorghum lines were the BTx623 genetic background carrying the ms8 allele (Xin et al., 2017). The M2-1299 and M2-0483 lines were from a collection of 256 whole-genome-sequenced M4 EMS-mutagenized sorghum lines described by Jiao et al. (2016). Plants were screened for the sbid1-1 and sbid1-2 mutations in SbId1 by Derived Cleaved Amplified Polymorphic Sequences PCR with the primers in Supplementary Table 3 together with restriction enzymes Hpy188I and HpyCH4V, respectively, from New England Biolabs (neb.com). The Hyp188I restriction enzyme cleaves the sbid1-1 mutant-derived PCR fragment, but not the wild-type PCR fragment, and the HpyCH4V restriction enzyme does not cleave the sbid1-2 mutant-derived PCR fragment but cleaves the wild-type PCR fragment.
Greenhouse flowering time trials were under LD conditions of 16-h days and 8-h nights. Natural sunlight was supplemented with LED lighting from either LumiGrow Pro325 (lumigrow.com) or DayBreak LED GrowLuXx (daybreakled.com) fixtures. Daytime temperature was set to 26°C, and nighttime temperature was set to 20°C. Seedlings were sown in 4-in. peat pots filled with SuperSoil from The Scotts Company (scotts.com) and transplanted to 13-L pots filled with corn soil (composed of aged wood fines, green waste compost, fir bark, grape compost, rice hulls, chicken manure, red lava, and sandy loam mixed by American Soil and Stone (Richmond, CA)) when seedlings reached the three-to-four-leaf stage (10 to 15 days old). Greenhouse plants were watered twice daily and received 20–20–20 N–P–K fertilizer once a week. These trials were conducted over the same season as field trials, beginning in late May and finishing in late September. The exceptions were experiments allowing sbid1-2 or sbid1-2/id1-1 lines to flower and set seed that extended into December and January.
Field-grown plants for flowering time trials were grown at the University of California, Davis Vegetable Crops facility in Davis, CA, during the summers of 2019–2023. Plants were started by seed sown directly to soil in rows that were watered and fertilized by drip irrigation. Field season began in late May and finished late September or early October.
Plants for analysis of leaf gene expression were grown to the four-leaf stage (approximately 2 weeks old) in peat pots under standard greenhouse conditions, and then pots were transferred to Percival Scientific model PGC-36C9 growth chambers (percival-scientific.com) set to a light:dark cycle of 16-h light: 8-h dark. The light period was 26°C, and the dark period was 22°C. Light was provided by white LEDs (wavelength range 400–700 nm) at a total fluence rate of 360 µM photons/m2 s. Plants were watered by subirrigation twice a week, once with water and once with a water solution containing 134 ppm of Peters Professional 20–20–20 fertilizer (ICL, icl-sf.com).
BSA-seq mapping of sbid1-1 and sbid1-2 alleles
Genomic DNA was prepared from leaf tissue taken from late-flowering mutant plants with the Qiagen DNeasy Plant Mini kit according to the manufacturer’s recommendations (qiagen.com). The genomic DNA was precipitated with 1/10 volumes of 3 M sodium acetate and 2.5 volumes of 100% ethanol to remove contaminating salts, the DNA pellet dissolved in Ambion nuclease-free water (thermofisher.com), and the DNA concentration determined with the Qubit dsDNA Quantification Assay Kit (thermofisher.com).
A genomic DNA pool for deep sequencing was made by combining 500 ng of genomic DNA from 20 mutant individuals. Sequencing library construction and deep sequencing were performed by Novogene (novogene.com). Briefly, DNA was fragmented by sonication, poly-A tailed, ligated to adapters for Illumina sequencing, and PCR amplified with Illumina primers P5 and P7 index oligos. PCR fragments were purified with the AMPure XP system (beckman.com) and the library size distribution checked by Agilent 2100 Bioanalyzer (agilent.com) and quantified by qPCR. Libraries were 150-bp pair-end sequenced on an Illumina NovaSeq 6000 machine to greater than 10X genome coverage. The Illumina sequencing FASTQ files were deposited at the NCBI Sequence Read Archive (SRA) and are available at accession numbers SRR25730657 (https://www.ncbi.nlm.nih.gov/sra/SRR25730657) and SRR25668575 (https://www.ncbi.nlm.nih.gov/sra/SRR25668575) (Xin and Harmon, 2023). The BSA-seq workflow was run online on the cloud-based analysis platform SciApps (sciapps.org) with default settings according to the protocol described by Wang et al. (2021) using Sorghum bicolor genome assembly v3.0.1 and annotation v3.0 from Phytozome v13 (phytozome-next.jgi.doe.gov/).
Analysis of flowering time gene expression by qPCR
Plants at the leaf 5 stage (approximately 21 days old) were sampled by cutting across the leaf 5 ligule with a razor blade. All the tissue extending above the ligule was collected, the midvein removed from mature leaves with a razor blade, wrapped in aluminum foil, flash frozen by immersion in liquid nitrogen, and stored at −80°C. Samples were taken at 4-h intervals from plants under a regular day–night cycle or an opposite day–night cycle, so that each sampling time collected two time points separated by 12 h. Samples at ZT0 were collected after lights in the growth chamber turned on and samples at ZT16 were collected after lights turned off. Two individual plants were collected per time point in two independent experiments, to generate a total of four biological replicates for each genotype. White LED headlamps covered with two layers of Roscolux 89 (Moss Green; us.rosco.com) filter were used to aid tissue collection under a green safelight.
Tissue samples were hand-ground under liquid nitrogen with a mortar and pestle. Total RNA was isolated from approximately 100 mg of tissue with the Qiagen RNeasy Plant Mini Kit and on-column DNase I digestion with the Qiagen RNase-Free DNase Set according to the manufacturer’s recommendations (qiagen.com). Total RNA was precipitated with 1/10 volumes of 3 M sodium acetate and 2.5 volumes of 100% ethanol to remove contaminating salts and the RNA pellet dissolved in Ambion nuclease-free water (thermofisher.com). RNA concentration was determined with a NanoDrop spectrophotometer (nanodrop.com). First-strand cDNA was synthesized from 2 µg of total RNA with Thermo Fisher Scientific Maxima Reverse Transcriptase and oligo(dT)18 according to the manufacturer’s recommendations (thermofisher.com). The final products were diluted with four volumes of Milli-Q water (EMD Millipore, Hayward, CA), and this served as a template for qPCR with the primers listed in Supplementary Table 3.
Two technical replicate qPCR reactions were composed and performed as described previously (Bendix et al., 2013). Cq values were calculated with the regression function for each primer set in Bio-Rad CFX Manager Software (Bio-Rad, Hercules, CA). Relative transcript level was calculated as 2^(Cqnormalizer − Cqexperimental), where Cqnormalizer is the geometric mean of the Cq values for the normalizer 1 and normalizer 2 primer sets (Supplementary Table 3). Relative expression level was relative transcript level normalized to the average relative transcript level for all timepoints, genotypes, and replicates.
Identification of normalizer transcripts for qPCR
The normalizer transcripts for qPCR analysis were selected based on constant temporal and uniform leaf expression characteristics in public RNA-seq datasets. First, transcripts were identified that displayed moderate expression that was constant across the 24 samples in a 72-h diurnal time series made from leaves of the BTx623 inbred line (Lai et al., 2020). Transcripts with an average FPKM value between 25 and 500 were selected as a moderately expressed set. Transcripts from this moderately expressed set were selected for constant diurnal expression based on a JTK_CYCLE (Hughes et al., 2010) adjusted p-value of 1.0, a percent coefficient of variation <8, and a ratio of highest/lowest expression <1.4. The percent coefficient of variation was calculated as %CV = ((standard deviation/average FPKM) × 100). This constant diurnal expression set contained 27 genes. Second, the genes from the constant diurnal expressed set were filtered for uniform expression in RNA-seq libraries from 197 leaf samples from the BTx623 and RTx430 inbred lines made as part of the 2016 EPICON field trial (Varoquaux et al., 2019). Uniform expression was determined based on a percent coefficient of variation <30 and a ratio of highest/lowest expression <7 for normalized counts for all leaf samples. The primers in Supplementary Table 3 targeting transcripts from genes Sobic.008G153500 and Sobic.010G068400 were found to exhibit 95%–105% efficiency and uniform Cq values with cDNA from a range of tissues and times of day.
Results
Identification of the sbid1-1 allele, an EMS mutation with a conditional late-flowering phenotype
Screening of a pedigreed sorghum EMS mutant library (Jiao et al., 2016) for visual growth and developmental phenotypes identified a late-flowering individual from the 15M2-1299 line. This plant was crossed to the wild-type male sterile 8 (ms8) line (Xin et al., 2017) to generate material for mapping the causal mutation. The flowering behavior of the selfed mutant and the F1 ms8/mutant progeny was then evaluated in the field at Davis, California. Flowering was scored as the number of days from sowing to boot stage, an early visual indication of flowering. Plants were considered at boot stage when the collar of the flag leaf appeared in the whorl. The F1 progeny and wild-type control plants both flowered after an average of 51 (± 4.3 standard deviation, SD) and 52 (± 4.1 SD) days, respectively, whereas mutant progeny reached boot stage after an average of 95 (± 7.9 SD) days (Supplementary Figure 1A). Next season, the F2 progeny were grown out to confirm the late-flowering phenotype and to collect samples for mapping the position of the mutation. Out of 72 F2 plants, 20 individuals were late flowering, reaching boot stage more than 40 days later than their siblings (Supplementary Figure 1B). The 20 late-flowering individuals from this trial were used as the mapping population to identify the EMS mutation responsible for the later-flowering phenotype.
Genome resequencing and bulk segregant analysis (BSA) mapping associated the late-flowering phenotype with an EMS-derived lesion in a sorghum Id1 (SbId1) homolog. A genomic DNA pool from the 20 F2 late-flowering individuals was deep sequenced, and these data were used in the BSA-seq mapping pipeline (Wang et al., 2021). This approach takes advantage of the existing database of positional information and mutation effect predictions for EMS-derived small nucleotide polymorphisms (SNPs) in the mutant library to identify SNPs linked with the mutant phenotype and determine candidate non-synonymous, deleterious mutations. A 45.5-Mb region on chromosome 1 had SNPs linked to the flowering phenotype (Supplementary Figure 2A). Within this region were 10 EMS-derived non-synonymous mutations within coding regions of genes and above a linking probability of 5. Five of these mutations were predicted to be deleterious to gene function (Supplementary Table 1). Of these potentially deleterious mutations, an obvious candidate was the C to T transition within SbId1 (Sobic.001G242900) that resulted in the substitution of a conserved proline at position 155 with a serine (P155S) within the second zinc finger domain (Supplementary Figures 2, 3, and Supplementary Table 1). This potential SbId1 mutant allele was named sbid1-1.
In subsequent field trials, the sbid1-1 mutation cosegregated in backcross 1 (BC1) F2 populations with a late-flowering phenotype like that of the original mutant line. Consistent with a recessive allele, plants homozygous for sbid1-1 reached boot stage at an average of 104 (± 8.0 SD) days, which was later than plants genotyping as heterozygous for this mutation and wild type that reached boot stage at an average of 66 days (± 7.0 SD) (Figures 1A, B). In the same field trials, two different plantings of a homozygous mutant BC1F3 line required an average of 92 (± 6.6 SD) days to begin booting when all wild-type plants achieved boot stage within an average of 66 (± 5.4 SD) days (Figure 1C). Also, 17 out of 123 plants did not boot upon termination of the trial at 113 days after sowing. These results indicate that the mutation corresponding to the sbid1-1 allele is associated with a significant delay in flowering.
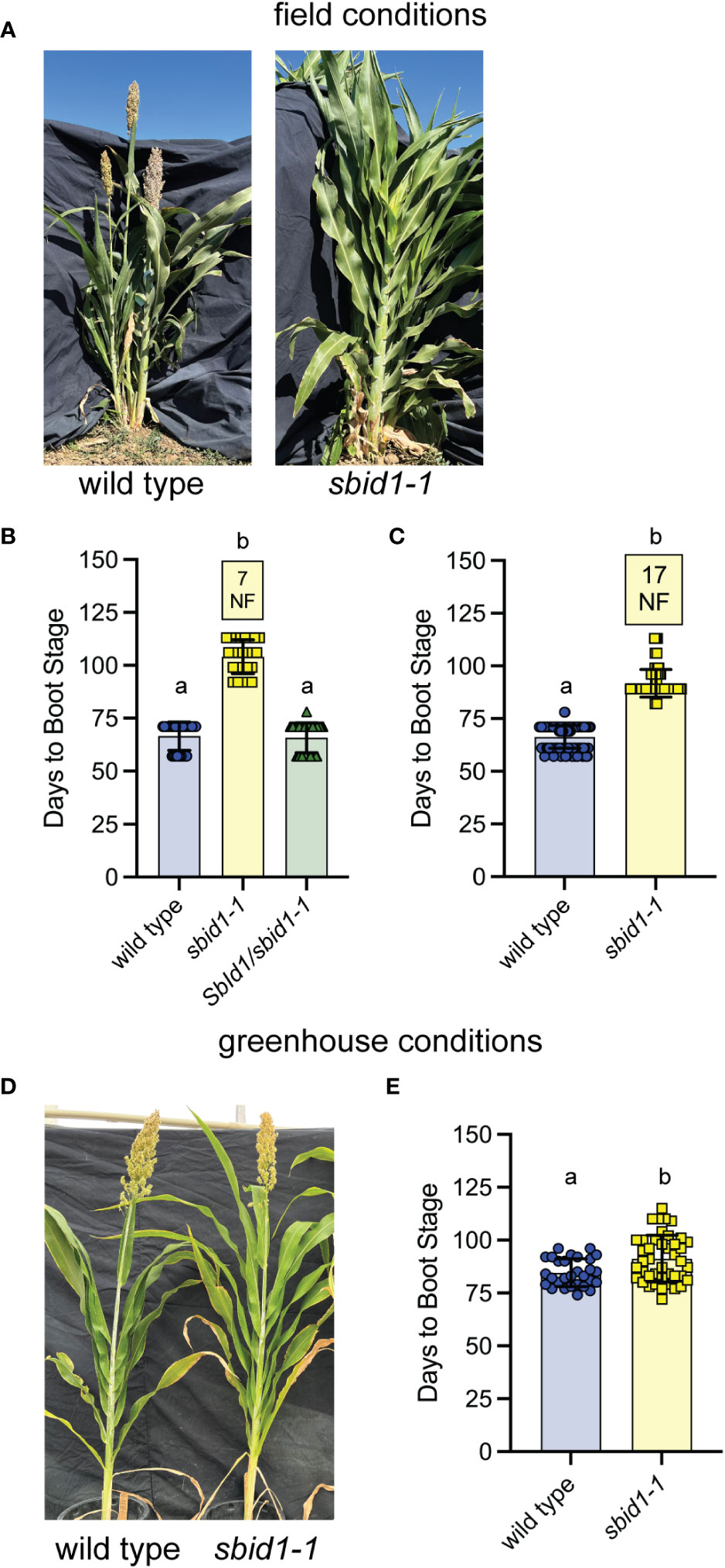
Figure 1 Homozygous sbid1-1 plants have a more pronounced flowering time delay under field conditions. (A) Representative wild-type (BTx623 inbred) and sbid1-1 plants at 109 days from sowing in the field. (B, C) Days to boot stage from sowing in field conditions at Davis, CA. Data are combined from two field trials. (B) Wild type (n = 28), sbid1-1 (n = 25), and SbId1/sbid1-1 heterozygotes (n = 60) from selfed sbid1-1 heterozygous parent. (C) Wild-type (n = 150) and sbid1-1 plants (n = 166). “NF” in yellow-colored boxes indicates the number of plants that had not flowered in at least 113 days after sowing. (D) Representative wild-type (BTx623 inbred) and sbid1-1 plants grown under greenhouse conditions at 107 days from sowing. (E) Days to boot stage from sowing for wild-type (n = 29) and sbid1-1 (n = 42) plants under greenhouse conditions. Means sharing a common letter are not significantly different by unpaired two-tailed t-test at the p <0.05 level of significance. Error bars are ± standard deviation.
Flowering time trials in the greenhouse revealed that the effect of the sbid1-1 allele is weaker under these conditions. Across three independent greenhouse trials starting in May to June of 2021–2023, sbid1-1 reached boot stage at an average of 91 (± 11 SD) days, which was 6 days later than the wild-type average of 85 (± 6.5 SD) days (Figures 1D, E), instead of the >30-day delay apparent in the field trials. These results show that the flowering delay caused by sbid1-1 flowering was greatest in the field, and this effect was almost fully mitigated by unknown aspects of growth under greenhouse conditions.
Identification of the EMS-derived sbid1-2 allele that essentially blocks flowering
Plants carrying the late-flowering mutation in the 15M2-0483 EMS line were discovered in an F2 population from a backcross of an unrelated mutant (with normal flowering behavior) to the ms8 BTx623 line. Tissue samples were taken from 20 late-flowering individuals in this population to capture their genetic information, since the extreme late-flowering phenotype of these plants could have potentially interfered with recovery of seeds. Genomic DNA was prepared from these plants for mapping the position of the causal mutation with the BSA-seq pipeline. Mapping based on sequence information from this genomic DNA pool identified SNPs from regions on chromosomes 1, 4, 7, and 10 as potentially linked to the late-flowering phenotype (Supplementary Figure 2B). BSA-seq called three non-synonymous mutations as linked at a linking probability of 5 with the only one predicted to be deleterious in a gene of unknown function (Supplementary Table 2). However, just under the significance threshold was a C to T transition in SbId1 that produced a nonsense mutation replacing the glutamine at position 199 with a stop codon (Q199*) in the third zinc finger domain (Supplementary Figure 2B). This mutation truncates the 428 amino acid ID1 protein to approximately half its usual length (Supplementary Figure 3). This putative SbId1 mutant allele was named sbid1-2.
To confirm that the sbid1-2 mutation was linked to the flowering phenotype, 15M2-0483 F2 populations were grown out in the field and greenhouse, and plants were screened by PCR for the sbid1-2 mutation and were scored for days to reach boot stage. Out of 39 plants in the field, the four plants genotyping as homozygous for sbid1-2 did not boot after 140 days, whereas all plants genotyping as heterozygous or wild type at this position in Id1 reached boot stage within 71 days. This result indicates that sbid1-2 is a recessive allele. In two subsequent field trials that began 17 days apart in Davis, CA, none of the 113 sbid1-2 mutant plants reached boot stage after 113 days from sowing, but all 142 wild-type plants reached this stage with a population average of 62 (± 3.2 SD) days (Figures 2A, B).
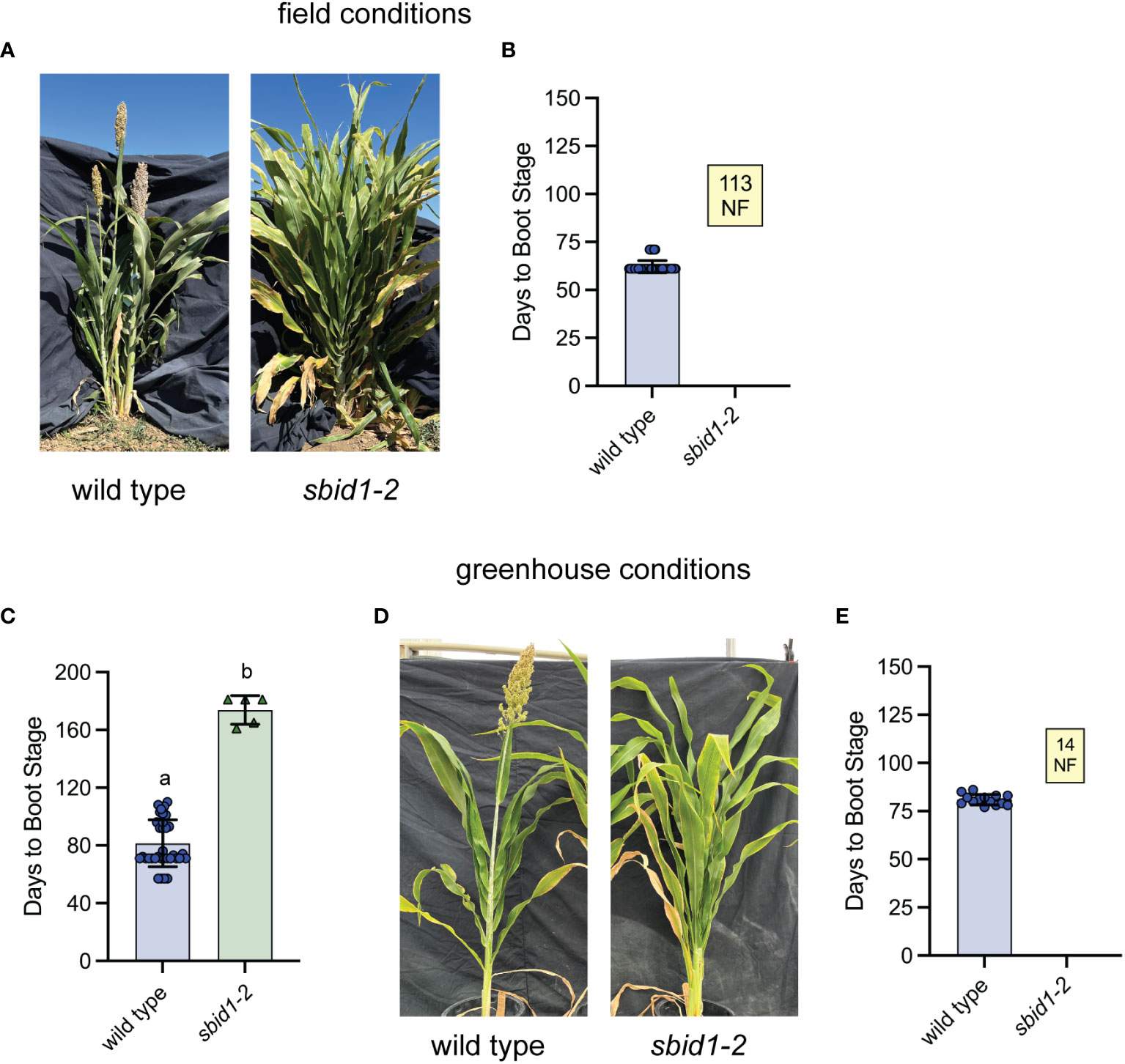
Figure 2 Homozygous sbid1-2 plants do not achieve boot stage under standard field or greenhouse growth conditions. (A) Representative wild-type (BTx623 inbred) and sbid1-2 plants at 109 days from sowing in the field. (B) Days to boot stage from sowing for wild type (n = 142) and sbid1-2 (n = 113) in separate two field trials in Davis, CA. (C, E) Days to boot stage from sowing under greenhouse conditions. (C) Wild type (n = 36) and sbid1-2 (n = 5) in a trial where sbid1-1 plants were allowed to reach boot stage and set seed. (D) Representative wild-type (BTx623 inbred) and sbid1-2 plants grown under greenhouse conditions at 107 days from sowing. (E) Wild-type (n = 14) and sbid1-2 (n = 14) plants. “NF” in the yellow-colored box indicates the number of plants that had not flowered in at least 120 days after sowing. Means sharing a common letter are not significantly different by unpaired two-tailed t-test at p < 0.05 level of significance. Error bars are ± standard deviation.
The flowering delay associated with the sbid1-2 allele was also profound under greenhouse conditions. In the first greenhouse trial, the four sbid1-2 homozygous plants recovered did not boot after terminating the experiment at 150 days, whereas the average days to boot stage for eight heterozygous plants was 90 (± 4 SD) days, only 7 days later than the wild-type average of 83 (± 7 SD) days. In the next greenhouse trial, five homozygous sbid1-2 plants were maintained as long as was required to reach boot stage and set seed. The average booting time for these mutant plants was 173 (± 10 SD) days with the earliest being 161 days and the latest 181 days (Figure 2C). The average boot time for the wild type in this trial was 81 (± 16 SD) days. The progeny of these homozygous sbid1-2 mutant plants in the greenhouse did not achieve boot stage after more than 110 days, whereas the wild type required an average of 81 (± 2.7 SD) days (Figures 2D, E). These results demonstrate that plants homozygous for this C to T transition in SbId1 have a profound flowering delay representing effectively a block in flowering for the length of typical field trials, but sbid1-2 plants eventually flower when maintained for approximately 170 days in the greenhouse.
Combining the sbid1-1 and sbid1-2 allele demonstrates non-complementation, confirming flowering requires SbId1 activity
Complementation tests were performed to confirm that the sbid1-1 and sbid1-2 mutations were responsible for delaying flowering in each mutant background. F1 plants with the sbid1-1 and sbid1-2 alleles combined were generated by pollinating a male sterile sbid1-2 heterozygote with pollen from an sbid1-1 homozygous plant. F2 populations were made by allowing F1 plants to self in the greenhouse.
In the first greenhouse trial, F1 plants genotyping as sbid1-2/sbid1-1 took an average of 181 (± 12.3 SD) days to reach boot stage, with a range of 167 to 203 days (Figure 3A). In contrast, F1 progeny genotyping as SbId1/sbid1-1 reached boot stage at an average of 95 (± 4.9 SD) days, comparable with the average of 93 (± 12.7 SD) days for wild-type plants. The sbid1-2/sbid1-1 behavior was like homozygous sbid1-2 plants started at the same time and grown under the same conditions (Figures 2C, E). sbid1-2/sbid1-1 F2 plants in a second greenhouse trial behaved similarly. The group of plants genotyping as either SbId1/id1-1, sbid1-2/SbId1, and wild type all reached boot stage at approximately 68 days, but none of the sbid1-2/sbid1-1 plants did so after 100 days when the experiment was terminated (Figure 3B).
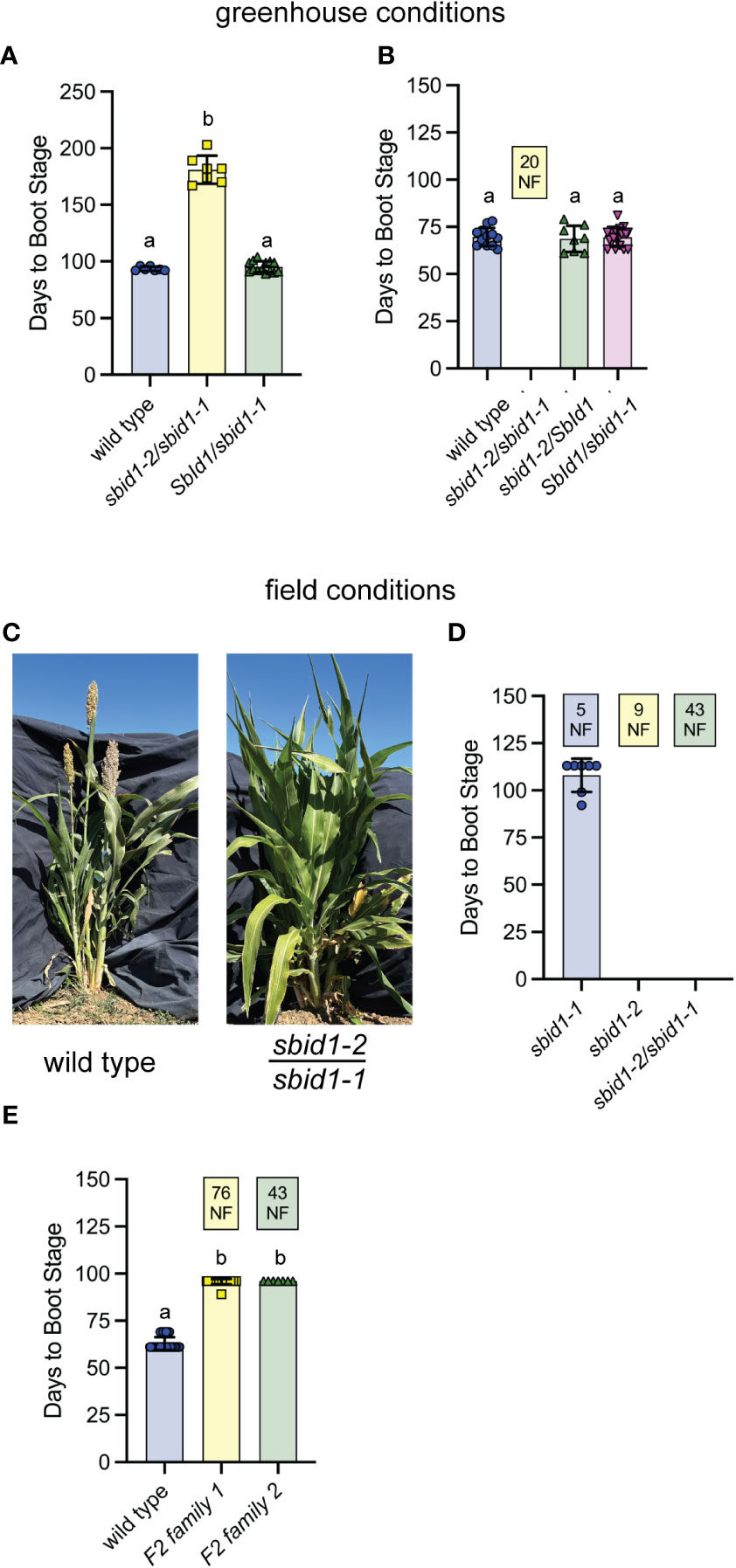
Figure 3 Non-complementation by sbid1-1 and sbid1-2 alleles confirms SbId1 is needed to promote flowering. (A, B) Days to boot stage under greenhouse conditions in 2022 (A) and 2023 (B). (A) Wild-type (BTx623) (n = 7), sbid1-2/sbid1-1 (n = 7), and SbId1/sbid1-1 (n = 14) from pollination of a male sterile (ms8) sbid1-2 heterozygous panicle with sbid1-1 homozygous pollen. All plants were allowed to reach boot stage. (B) Wild-type and sbid1-2/sbid1-1 (n = 20), sbid1-2/SbId1 (n = 8), and SbId1/sbid1-1 (n = 18) from the pollination of a male sterile (ms8) sbid1-2 heterozygous panicle with sbid1-1 heterozygous pollen. “NF” in the yellow-colored box indicates the number of plants that had not flowered in at least 120 days after sowing. (C) Representative wild-type (BTx623 inbred) and sbid1-2/sbid1-1 heterozygous plants at 109 days from sowing in the field. (D, E) Days to boot stage in the field at Davis, CA. (D) sbid1-2/sbid1-1 (n = 43), sbid1-1 (n = 12), and sbid1-2 (n = 9) from selfed sbid1-2/sbid1-1 plants. “NF” in colored boxes indicates the number of plants for that genotype that had not flowered in at least 113 days after sowing. The genotype of all plants in (A, D) was determined with PCR. (E) Wild type (n = 67) and individuals from either a pool of selfed sbid1-2/sbid1-1 plants (F2 family 1; n = 100) and one selfed sbid1-2/sbid1-1 plant (F2 family 2; n = 50). These plants were not tested for genotype at SbId1. “NF” in the colored box indicates the number of plants that had not flowered in at least 96 days after sowing. Means sharing a common letter are not significantly different by unpaired two-tailed t-test at p <0.05 level of significance. Error bars are ± standard deviation.
Comparable results were also obtained in field trials with PCR genotyped F2 plants. Out of a total of 64 progeny from two different self-fertilized sbid1-1/sbid1-2 F1 plants, 7 of the 12 sbid1-1/sbid1-1 flowered within the 113 days of the trial, but none of the 9 sbid1-2/sbid1-2 or the 43 sbid1-2/sbid1-1 plants had flowered (Figures 3C, D). Two additional F2 families from two different selfed sbid1-2/sbid1-1 parents were grown out in the same field, but these plants were not genotyped at the SbId1 locus. One family of 100 had 24 plants achieve boot stage after 96 days, whereas the second family of 50 had seven plants at boot stage after 96 days (Figure 3E). A wild-type population of 89 plants sown at the same time reached boot stage at an average of 63 (± 3.4 SD) days (Figure 3E). These results show that the mutations present in SbId1 in the sbid1-1 and sbid1-2 alleles were responsible for delayed flowering. Furthermore, the severity of the sbid1-2/sbid1-1 phenotype more closely matched the strong sbid1-2 allele than the weaker sbid1-1 allele, which was most evident under greenhouse conditions.
The sbid1-2 mutant allele significantly reduces expression of flowering time genes
To gain insight into the origin of late flowering caused by loss of SbId1 function, the effect of the sbid1-2 allele on the expression level and daily accumulation pattern, or waveform, of major flowering time regulatory genes was examined in the leaves of plants at the leaf 5 stage (approximately 21 days old), at 4-h intervals over 24 h under LD conditions (i.e., 16-h days and 8-h nights). Evaluation of SbId1 transcript levels in wild type and sbid1-2 showed that levels were lower in the mutant at each timepoint, but the waveform was similar between mutant and wild type (Figure 4A). Since rice RID1 contributes to upregulation of the floral activator Ehd1 under LD photoperiods (Matsubara et al., 2008; Park et al., 2008), levels of the SbEhd1 transcript were determined for these time courses. No difference in SbEhd1 levels was apparent between sbid1-2 and wild type at any of the time points (Figure 4B). Also, the waveform for SbEhd1 accumulation in id1-2 matched the wild type, which was comparable with the SbEhd1 waveform described previously for LD conditions (Murphy et al., 2011). In contrast, floral activator SbCO transcript accumulation in sbid1-2 was low at all time points and lacked the wild-type waveform (Murphy et al., 2011) that rose beginning at 8 h after dawn (zeitgeber time (ZT) 8), peaked at the beginning of the dark period at ZT16, and extended to 4 h after dawn at ZT4 (Figure 4C). Expression of florigen-encoding genes SbCN8 and SbCN12 was also lower in sbid1-2 than in the wild type, with a notable lack of peak expression at ZT4 (Figures 4D, E). These findings were consistent with a diminishment of SbCN8 and SbCN12 upregulation by SbCO in sbid1-2 and, consequently, a block in promotion of the transition to flowering by these florigen genes.
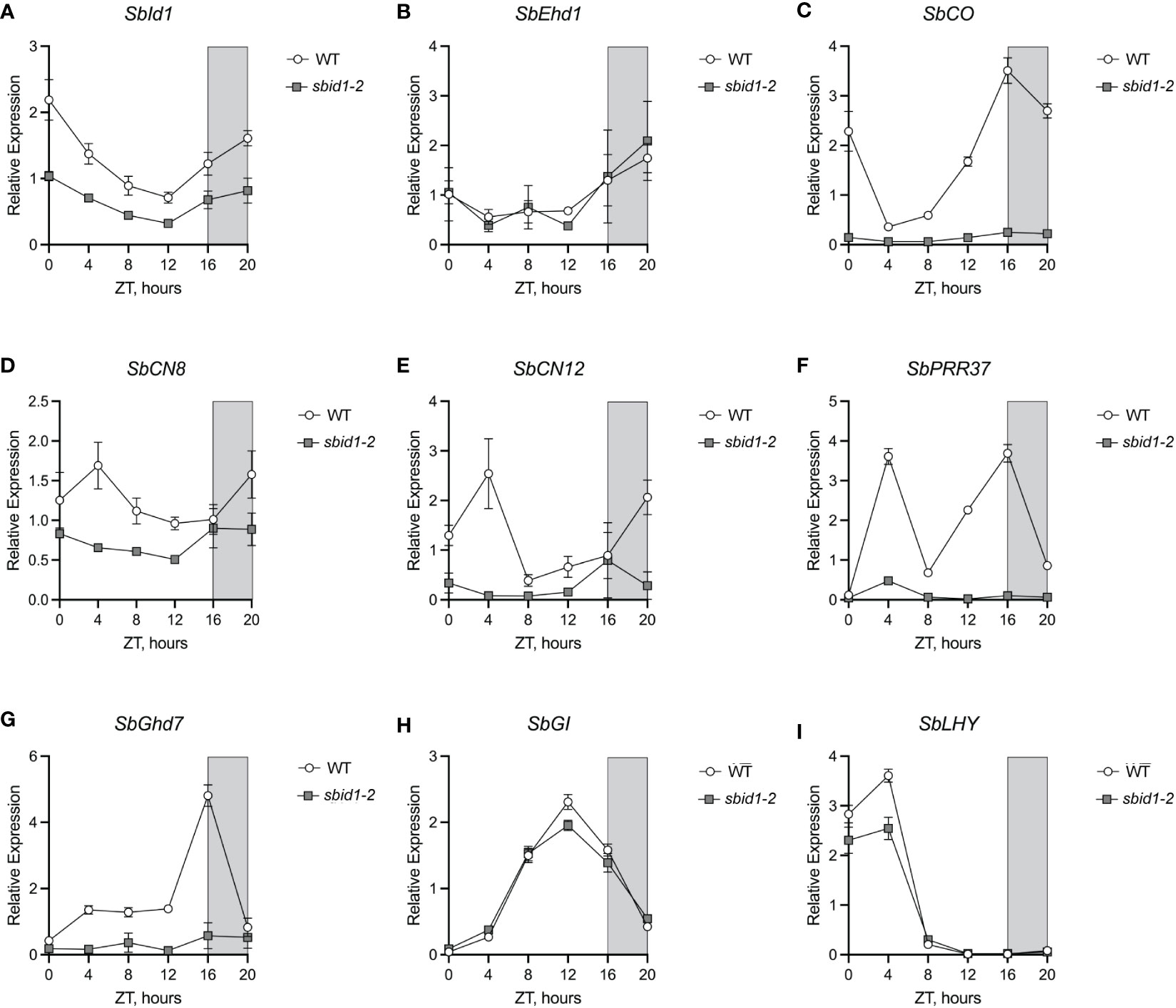
Figure 4 Loss of SbId1 activity interferes with expression of major flower time activators and repressors. Relative expression of (A) SbId1, (B) SbEhd1, (C) SbCO, (D) SbCN8, (E) SbCN12, (F) SbPRR37, (G) SbGhd7, (H) SbGI, and (I) SbLHY in wild-type (white circles) or sbid1-2 (gray squares) plants under LD conditions (16-h light and 8-h darkness). Zeitgeber time (ZT) in hours and ZT0 corresponded to lights on at dawn and ZT16 corresponded to lights off at night (gray shading). Points are the mean of four individual plants from two independent experiments. Error bars are ± standard error of the mean.
Although the BTx623 genetic background has the inactive sbprr37-3 (ma1) and sbghd7-1 (ma6) alleles (Murphy et al., 2011; Murphy et al., 2014), the expression of SbPRR37 and SbGhd7 was compared between sbid1-2 and the wild type to determine if SbId1 contributes to transcriptional regulation of these major floral repressors. Both SbPRR37 and SbGhd7 transcripts were low at all time points in sbid1-2 (Figures 4F, G). SbPRR37 expression in sbid1-2 lacked the major peaks at ZT4 in the morning and at ZT16 at the beginning of night apparent in the wild type (Figure 4F) and described previously for SbPRR37 expression under LD (Murphy et al., 2011). Similarly, the primary SbGhd7 peak at ZT16 in the wild type (Murphy et al., 2014) did not occur in sbid1-2 (Figure 4G). These results show that SbId1 contributes to activation of these floral repressor genes. The alteration in gene expression in sbid1-2 was not due to a change in overall rhythmic gene expression, since expression waveforms for SbGI, a flowering regulator and circadian clock gene (Abdul-Awal et al., 2020), and SbLHY, a presumed circadian clock gene (Lai et al., 2020), were similar between sbid1-2 and the wild type, except for reduced peak levels for both transcripts (Figures 4H, I). Thus, Id1 is needed for expression of the major regulatory genes for flowering, including floral activators and repressors.
Discussion
Two mutants with significant delays in flowering time discovered in a pedigreed sorghum EMS mutant library were identified as novel sbid1 mutant alleles. Plants with the weaker sbid1-1 non-synonymous mutation had booting delayed by more than 30 days in the field but behaved similar to the wild type under greenhouse conditions. The condition-sensitive nature of sbid1-1 makes it a potentially useful tool to dissect the activity of sbid1 in the future. The strong sbid1-2 allele, a nonsense mutation that encodes for a truncated SbId1 protein, required approximately 170 days to reach boot stage, effectively blocking flowering under typical field conditions. Plants carrying sbid1-1 and sbid1-2 together had the strong flowering phenotype of sbid1-2, confirming that the phenotype arises from loss of SbId1 function. Evaluation of expression for major flowering time regulatory genes in the sbid1-2 background showed that SbId1 is needed for expression of floral activators, like SbCO and its target genes SbCN8 and SbCN12. Flowering time repressors SbPRR37 and SbGhd7 also had significantly reduced expression in the mutant background.
The strong flowering time delay caused by sbid1-1 and sbid1-2 mirrors the effect of id1 and rid1 mutants on flowering in maize (Singleton, 1946; Colasanti et al., 1998) and rice (Matsubara et al., 2008; Wu et al., 2008), respectively. However, analysis of the gene expression profile in sbid1-2 highlights potential differences in or previously unknown aspects of the contribution of SbId1 to the flowering time regulatory system. There was no indication that SbId1 influences SbEhd1 expression, unlike the case of RID1 in rice (Matsubara et al., 2008; Park et al., 2008; Wu et al., 2008). Loss of RID1 activity eliminates a peak in Ehd1 expression that occurs at midday, which reduces Ehd1-promoted florigen gene expression (Matsubara et al., 2008; Park et al., 2008; Wu et al., 2008). Any impact of sbid1-2 on SbEhd1 expression may have been obscured by the lack of midday SbEhd1 upregulation in the wild-type plants evaluated here. Regardless, a lack of SbCO, SbCN8, and SbCN12 upregulation in sbid1-2 indicates SbId1 protein promotes flowering through the SbCO-SbCN8/SbCN12 module. In this role, the SbId1 protein appears to be required to upregulate SbCO expression under LD conditions to promote SbCN8 and SbCN12 expression.
The fact that these sbid1 alleles caused substantial flowering time delays in a genetic background lacking the activity of SbPRR37 and SbGhd7 demonstrates that Id1 activity does not depend on these major flowering repressors. The BTx623 inbred carries the sbprr37-3 allele of ma1, which encodes a Lys162Asp substitution in SbPRR37 (Murphy et al., 2011), and the sbghd7-1 allele of ma6, which is a 5-bp insertion in the SbGhd7 coding sequence (Murphy et al., 2014). SbPRR37 and SbGhd7 delay flowering under LD conditions by repressing the expression of SbEhd1 and inhibiting SbCO activation of florigen genes (Murphy et al., 2011; Murphy et al., 2014; Yang et al., 2014b).
Nevertheless, gene expression analysis indicated that SbId1 is needed for expression of these repressor genes. SbPRR37 and SbGhd7 transcript levels were below basal levels in sbid1-1, with expression patterns lacking the peaks at midday (ZT8) and the transition from day to night (ZT16). Light signals mediated by phytochromes A and C, which represent maturity loci Ma3 (Childs et al., 1997) and Ma5 (Yang et al., 2014a), promote expression of SbPRR37and SbGhd7 (Yang et al., 2014a). Therefore, it is conceivable that SbId1 plays a role in light-promoted upregulation of SbPRR37 and SbGhd7. Similarly, rice Ghd7 may be upregulated by RID1 (Wu et al., 2008), indicating that Id1 regulation of these major repressors is likely highly conserved.
These results demonstrate that SbId1 potentially contributes to gene expression in all the major arms of flowering time regulation in sorghum to promote (Wu et al., 2008) upregulation of both activators like SbCO and SbCN8/SbCN12, as well as repressors SbPRR37 and SbGhd7. Since RID1 and maize Id1 impact histone modifications to increase chromatin accessibility at the promoters of their respective florigen genes (Mascheretti et al., 2015; Zhang et al., 2022a), it is tempting to speculate that a general mechanism of Id1 protein action is establishment of active chromatin states at promoters of major regulatory genes to activate the photoperiod regulatory network. This model suggests that a conserved role for Id1 proteins is to establish the capacity for flowering time regulation, serving as a master regulator as proposed for RID1 in rice (Wu et al., 2008). In this role, SbId1 is needed for expression of both activators and repressors, not solely as an activator of downstream activators. Important tests of this idea are to evaluate the role of SbId1 in genetic backgrounds like bioenergy sorghum accessions with intact systems for photoperiodic flowering time control, and to identify what changes, if any, mutations in SbId1, such as the alleles described here, have on the epigenetic states across the sorghum genome, most importantly at flowering time genes.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: NCBI BioProject, PRJNA1006376, https://www.ncbi.nlm.nih.gov/bioproject/PRJNA1006376 NCBI SRA, SRR25730657, https://www.ncbi.nlm.nih.gov/sra/SRR25730657 NCBI SRA, SRR25668575, https://www.ncbi.nlm.nih.gov/sra/SRR25668575.
Author contributions
SD: Formal analysis, Investigation, Writing – review & editing. JC: Investigation, Writing – review & editing. ZX: Investigation, Writing – review & editing, Formal analysis. FH: Conceptualization, Formal analysis, Funding acquisition, Investigation, Methodology, Supervision, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by USDA-ARS (project number 2030-21000-049-00D) to FH.
Acknowledgments
Thank you to Lia Poasa, Shawna Kelley, and Emilio Corona from the PGEC greenhouse, as well as Israel Herrera and Luis Loza at UC Davis Vegetable Crops, for excellent care of plants. Thank you to the Sponsored Projects for Undergraduate Research program at UC Berkeley for support of SD.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1304822/full#supplementary-material
Supplementary Figure 1 | The mutation from the M2-1299 line significantly increases the number of days to reach boot stage.
Supplementary Figure 2 | BSAseq results for mapping of late flowering alleles from M2-1299 and M2-0483.
Supplementary Figure 3 | Alignment of Id1 homologs from C4 and C3 grasses.
References
Abdul-Awal, S. M., Chen, J., Xin, Z., Harmon, F. G. (2020). A sorghum gigantea mutant attenuates florigen gene expression and delays flowering time. Plant Direct. 4 (11), e00281. doi: 10.1002/pld3.281
Bendix, C., Mendoza, J. M., Stanley, D. N., Meeley, R., Harmon, F. G. (2013). The circadian clock-associated gene gigantea1 affects maize developmental transitions. Plant Cell Environ. 36 (7), 1379–1390. doi: 10.1111/pce.12067
Childs, K. L., Miller, F. R., Cordonnier-Pratt, M. M., Pratt, L. H., Morgan, P. W., Mullet, J. E. (1997). The sorghum photoperiod sensitivity gene, Ma3, encodes a phytochrome B. Plant Physiol. 113 (2), 611–619. doi: 10.1104/pp.113.2.611
Colasanti, J., Tremblay, R., Wong, A. Y. M., Coneva, V., Kozaki, A., Mable, B. K., et al. (2006). The maize INDETERMINATE1 flowering time regulator defines a highly conserved zinc finger protein family in higher plants. BMC Genomics 7 (1), 158. doi: 10.1186/1471-2164-7-158
Colasanti, J., Yuan, Z., Sundaresan, V. (1998). The indeterminate gene encodes a zinc finger protein and regulates a leaf-generated signal required for the transition to flowering in maize. Cell 93 (4), 593–603. doi: 10.1016/S0092-8674(00)81188-5
Danilevskaya, O. N., Meng, X., Hou, Z., Ananiev, E. V., Simmons, C. R. (2008). A genomic and expression compendium of the expanded PEBP gene family from maize. Plant Physiol. 146 (1), 250–264. doi: 10.1104/pp.107.109538
Deng, L., Li, L., Zhang, S., Shen, J., Li, S., Hu, S., et al. (2017). Suppressor of rid1 (SID1) shares common targets with RID1 on florigen genes to initiate floral transition in rice. PloS Genet. 13 (2), e1006642. doi: 10.1371/journal.pgen.1006642
Doi, K., Izawa, T., Fuse, T., Yamanouchi, U., Kubo, T., Shimatani, Z., et al. (2004). Ehd1, a B-type response regulator in rice, confers short-day promotion of flowering and controls FT-like gene expression independently of Hd1. Genes Dev. 18 (8), 926–936. doi: 10.1101/gad.1189604
Farre, E. M., Liu, T. (2013). The PRR family of transcriptional regulators reflects the complexity and evolution of plant circadian clocks. Curr. Opin. Plant Biol. 16 (5), 621–629. doi: 10.1016/j.pbi.2013.06.015
Griffiths, S., Dunford, R. P., Coupland, G., Laurie, D. A. (2003). The evolution of CONSTANS-like gene families in barley, rice, and arabidopsis. Plant Physiol. 131, 1855–1867. doi: 10.1104/pp.102.016188
Hughes, M. E., Hogenesch, J. B., Kornacker, K. (2010). JTK_CYCLE: an efficient nonparametric algorithm for detecting rhythmic components in genome-scale data sets. J. Biol. Rhythms. 25 (5), 372–380. doi: 10.1177/0748730410379711
Itoh, H., Nonoue, Y., Yano, M., Izawa, T. (2010). A pair of floral regulators sets critical day length for Hd3a florigen expression in rice. Nat. Genet. 42 (7), 635–638. doi: 10.1038/ng.606
Jaeger, K. E., Wigge, P. A. (2007). FT protein acts as a long-range signal in Arabidopsis. Curr. Biol. 17 (12), 1050–1054. doi: 10.1016/j.cub.2007.05.008
Jiao, Y., Burke, J., Chopra, R., Burow, G., Chen, J., Wang, B., et al. (2016). A sorghum mutant resource as an efficient platform for gene discovery in grasses. Plant Cell. 28 (7), 1551–1562. doi: 10.1105/tpc.16.00373
Kojima, S., Takahashi, Y., Kobayashi, Y., Monna, L., Sasaki, T., Araki, T., et al. (2002). Hd3a, a rice ortholog of the Arabidopsis FT Gene, promotes transition to flowering downstream of Hd1 under short-day conditions. Plant Cell Physiol. 43 (10), 1096–1105. doi: 10.1093/pcp/pcf156
Koo, B. H., Yoo, S. C., Park, J. W., Kwon, C. T., Lee, B. D., An, G., et al. (2013). Natural variation in OsPRR37 regulates heading date and contributes to rice cultivation at a wide range of latitudes. Mol. Plant 6 (6), 1877–1888. doi: 10.1093/mp/sst088
Kozaki, A., Hake, S., Colasanti, J. (2004). The maize ID1 flowering time regulator is a zinc finger protein with novel DNA binding properties. Nucleic Acids Res. 32 (5), 1710–1720. doi: 10.1093/nar/gkh337
Lai, X., Bendix, C., Yan, L., Zhang, Y., Schnable, J. C., Harmon, F. G. (2020). Interspecific analysis of diurnal gene regulation in panicoid grasses identifies known and novel regulatory motifs. BMC Genomics 21 (1), 428. doi: 10.1186/s12864-020-06824-3
Lazakis, C. M., Coneva, V., Colasanti, J. (2011). ZCN8 encodes a potential orthologue of Arabidopsis FT florigen that integrates both endogenous and photoperiod flowering signals in maize. J. Exp. Botany. 62 (14), 4833–4842. doi: 10.1093/jxb/err129
Lifschitz, E., Eviatar, T., Rozman, A., Shalit, A., Goldshmidt, A., Amsellem, Z., et al. (2006). The tomato FT ortholog triggers systemic signals that regulate growth and flowering and substitute for diverse environmental stimuli. Proc. Natl. Acad. Sci. U S A. 103 (16), 6398–6403. doi: 10.1073/pnas.0601620103
Lin, M-KMK, Belanger, H., Lee, Y-JYJ, Varkonyi-Gasic, E., Taoka, KIK-I, Miura, E., et al. (2007). FLOWERING LOCUS T protein may act as the long-distance florigenic signal in the cucurbits. Plant Cell. 19 (5), 1488–1506. doi: 10.1105/tpc.107.051920
Mascheretti, I., Turner, K., Brivio, R. S., Hand, A., Colasanti, J., Rossi, V. (2015). Florigen-encoding genes of day-neutral and photoperiod-sensitive maize are regulated by different chromatin modifications at the floral transition. Plant Physiol. 168 (4), 1351–1363. doi: 10.1104/pp.15.00535
Matsubara, K., Yamanouchi, U., Wang, Z.-X., Minobe, Y., Izawa, T., Yano, M. (2008). Ehd2, a rice ortholog of the maize INDETERMINATE1 gene, promotes flowering by up-regulating ehd1. Plant Physiol. 148 (3), 1425–1435. doi: 10.1104/pp.108.125542
Meng, X., Muszynski, M. G., Danilevskaya, O. N. (2011). The FT-likeZCN8 gene functions as a floral activator and is involved in photoperiod sensitivity in maize. Plant Cell. 23 (3), 942–960. doi: 10.1105/tpc.110.081406
Miller, T. A., Muslin, E. H., Dorweiler, J. E. (2008). A maize CONSTANS-like gene, conz1, exhibits distinct diurnal expression patterns in varied photoperiods. Planta 227 (6), 1377–1388. doi: 10.1007/s00425-008-0709-1
Murakami, M., Ashikari, M., Miura, K., Yamashino, T., Mizuno, T. (2003). The evolutionarily conserved OsPRR quintet: rice pseudo-response regulators implicated in circadian rhythm. Plant Cell Physiol. 44 (11), 1229–1236. doi: 10.1093/pcp/pcg135
Murphy, R. L., Klein, R. R., Morishige, D. T., Brady, J. A., Rooney, W. L., Miller, F. R., et al. (2011). Coincident light and clock regulation of pseudoresponse regulator protein 37 (PRR37) controls photoperiodic flowering in sorghum. Proc. Natl. Acad. Sci. U S A. 108 (39), 16469–16474. doi: 10.1073/pnas.1106212108
Murphy, R. L., Morishige, D. T., Ja, B., WL, R., Yang, S., Klein, P. E., et al. (2014). Ghd7 (Ma6) represses sorghum flowering in long days: alleles enhance biomass accumulation and grain production. Plant Genome. 7 (2), 1–10. doi: 10.3835/plantgenome2013.11.0040
Notaguchi, M., Abe, M., Kimura, T., Daimon, Y., Kobayashi, T., Yamaguchi, A., et al. (2008). Long-distance, graft-transmissible action of arabidopsis FLOWERING LOCUS T protein to promote flowering. Plant Cell Physiol. 49 (11), 1645–1658. doi: 10.1093/pcp/pcn154
Park, S. J., Kim, S. L., Lee, S., Je, B. I., Piao, H. L., Park, S. H., et al. (2008). Rice Indeterminate 1 (OsId1) is necessary for the expression of Ehd1 (Early heading date 1) regardless of photoperiod. Plant J. 56 (6), 1018–1029. doi: 10.1111/j.1365-313X.2008.03667.x
Putterill, J., Robson, F., Lee, K., Simon, R., Coupland, G. (1995). The CONSTANS gene of Arabidopsis promotes flowering and encodes a protein showing similarities to zinc finger transcription factors. Cell 80 (6), 847–857. doi: 10.1016/0092-8674(95)90288-0
Quinby, J. R., Karper, R. E. (1945). The Inheritance of Three Genes That Influence Time of Floral Initiation and Maturity Date in Milo1. Agron. J. 37, 916–936. doi: 10.2134/agronj1945.00021962003700110006x
Quinby, J. R. (1974). The Maturity Genes of Sorghum. In: Advances in Agronomy. Norman, AG., editor. (Academic Press) 19, 267–305. doi: 10.1016/S0065-2113(08)60737-3
Rooney, W. L., Aydin, S. (1999). Genetic control of a photoperiod-sensitive response in Sorghum bicolor (L.) Moench. Crop Sci. 39 (2), 397–400. doi: 10.2135/cropsci1999.0011183X0039000200016x
Samach, A., Coupland, G. (2000). Time measurement and the control of flowering in plants. Bioessays 22 (1), 38–47. doi: 10.1002/(SICI)1521-1878(200001)22:1<38::AID-BIES8>3.0.CO;2-L
Singleton, W. R. (1946). Inheritance of indeterminate growth in maize. J. Heredity. 37 (2), 61–64. doi: 10.1093/oxfordjournals.jhered.a105582
Song, Y. H., Shim, J. S., Kinmonth-Schultz, H. A., Imaizumi, T. (2015). Photoperiodic flowering: time measurement mechanisms in leaves. Annu. Rev. Plant Biol. 66 (1), 441–464. doi: 10.1146/annurev-arplant-043014-115555
Song, Y. H., Smith, R. W., To, B. J., Millar, A. J., Imaizumi, T. (2012). FKF1 conveys timing information for CONSTANS stabilization in photoperiodic flowering. Science 336 (6084), 1045–1049. doi: 10.1126/science.1219644
Tamaki, S., Matsuo, S., Wong, H. L., Yokoi, S., Shimamoto, K. (2007). Hd3a protein is a mobile flowering signal in rice. Science 316 (5827), 1033–1036. doi: 10.1126/science.1141753
Turck, F., Fornara, F., Coupland, G. (2008). Regulation and identity of florigen: FLOWERING LOCUS T moves center stage. Annu. Rev. Plant Biol. 59 (1), 573–594. doi: 10.1146/annurev.arplant.59.032607.092755
Valverde, F., Mouradov, A., Soppe, W., Ravenscroft, D., Samach, A., Coupland, G. (2004). Photoreceptor regulation of CONSTANS protein in photoperiodic flowering. Science 303 (5660), 1003–1006. doi: 10.1126/science.1091761
Varoquaux, N., Cole, B., Gao, C., Pierroz, G., Baker, C. R., Patel, D., et al. (2019). Transcriptomic analysis of field-droughted sorghum from seedling to maturity reveals biotic and metabolic responses. Proc. Natl. Acad. Sci. U.S.A 116, 27124–27132. doi: 10.1073/pnas.1907500116
Wang, L., Lu, Z., Regulski, M., Jiao, Y., Chen, J., Ware, D., et al. (2021). BSAseq: an interactive and integrated web-based workflow for identification of causal mutations in bulked F2 populations. Bioinformatics 37 (3), 382–387. doi: 10.1093/bioinformatics/btaa709
Wolabu, T. W., Zhang, F., Niu, L., Kalve, S., Bhatnagar-Mathur, P., Muszynski, M. G., et al. (2016). Three FLOWERING LOCUS T-like genes function as potential florigens and mediate photoperiod response in sorghum. New Phytologist. 210 (3), 946–959. doi: 10.1111/nph.13834
Wu, C., You, C., Li, C., Long, T., Chen, G., Byrne, M. E., et al. (2008). RID1, encoding a Cys2/His2-type zinc finger transcription factor, acts as a master switch from vegetative to floral development in rice. Proc. Natl. Acad. Sci. U S A. 105 (35), 12915–12920. doi: 10.1073/pnas.0806019105
Xin, Z., Harmon, F. G. (2023). Genome sequencing for bulk segregant analysis mapping of sorghum late flowering mutants. NCBI BioProject. Available at: https://www.ncbi.nlm.nih.gov/bioproject/PRJNA1006376.
Xin, Z., Huang, J., Smith, A. R., Chen, J., Burke, J., Sattler, S. E., et al. (2017). Morphological characterization of a new and easily recognizable nuclear male sterile mutant of sorghum (Sorghum bicolor). PloS One 12 (1), e0165195. doi: 10.1371/journal.pone.0165195
Xue, W., Xing, Y., Weng, X., Zhao, Y., Tang, W., Wang, L., et al. (2008). Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice. Nat. Genet. 40 (6), 761–767. doi: 10.1038/ng.143
Yang, S., Murphy, R. L., Morishige, D. T., Klein, P. E., Rooney, W. L., Mullet, J. E. (2014a). Sorghum phytochrome B inhibits flowering in long days by activating expression of SbPRR37 and SbGHD7, repressors of SbEHD1, SbCN8 and SbCN12. PloS One 9 (8), e105352. doi: 10.1371/journal.pone.0105352
Yang, S., Weers, B. D., Morishige, D. T., Mullet, J. E. (2014b). CONSTANS is a photoperiod regulated activator of flowering in sorghum. BMC Plant Biol. 14 (1), 148. doi: 10.1186/1471-2229-14-148
Yano, M., Katayose, Y., Ashikari, M., Yamanouchi, U., Monna, L., Fuse, T., et al. (2000). Hd1, a major photoperiod sensitivity quantitative trait locus in rice, is closely related to the Arabidopsis flowering time gene CONSTANS. Plant Cell. 12 (12), 2473–2484. doi: 10.1105/tpc.12.12.2473
Zhang, S., Deng, L., Cheng, R., Hu, J., Wu, C.-Y. (2022a). RID1 sets rice heading date by balancing its binding with SLR1 and SDG722. J. Integr. Plant Biol. 64 (1), 149–165. doi: 10.1111/jipb.13196
Zhang, S., Deng, L., Zhao, L., Wu, C. (2022b). Genome-wide binding analysis of transcription factor Rice Indeterminate 1 reveals a complex network controlling rice floral transition. J. Integr. Plant Biol. 64 (9), 1690–1705. doi: 10.1111/jipb.13325
Zhao, J., Chen, H., Ren, D., Tang, H., Qiu, R., Feng, J., et al. (2015). Genetic interactions between diverged alleles of Early heading date 1 (Ehd1) and Heading date 3a (Hd3a)/ RICE FLOWERING LOCUS T1 (RFT1) control differential heading and contribute to regional adaptation in rice (Oryza sativa). New Phytologist. 208 (3), 936–948. doi: 10.1111/nph.13503
Keywords: EMS mutagenesis, bulk segregant analysis, whole-genome resequencing, flowering time, gene expression, photoperiodic flowering, Sorghum bicolor
Citation: De Riseis S, Chen J, Xin Z and Harmon FG (2023) Sorghum bicolor INDETERMINATE1 is a conserved primary regulator of flowering. Front. Plant Sci. 14:1304822. doi: 10.3389/fpls.2023.1304822
Received: 30 September 2023; Accepted: 14 November 2023;
Published: 13 December 2023.
Edited by:
Tesfamichael Kebrom, Prairie View A&M University, United StatesReviewed by:
John Mullet, Texas A&M University, United StatesXiaoyu Weng, The University of Texas at Austin, United States
Copyright © 2023 De Riseis, Chen, Xin and Harmon. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Frank G. Harmon, ZnJhbmsuaGFybW9uQHVzZGEuZ292; Zmhhcm1vbkBiZXJrZWxleS5lZHU=
†ORCID: Samuel De Riseis, orcid.org/0000-0001-7668-3096
Junping Chen, orcid.org/0000-0001-9060-8017
Zhanguo Xin, orcid.org/0000-0003-1471-7785
Frank G. Harmon, orcid.org/0000-0001-7017-5373
 Samuel De Riseis
Samuel De Riseis Junping Chen2†
Junping Chen2† Frank G. Harmon
Frank G. Harmon