
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 05 December 2023
Sec. Plant Biotechnology
Volume 14 - 2023 | https://doi.org/10.3389/fpls.2023.1300522
Introduction: WRKY TFs (WRKY transcription factors) contribute to the synthesis of secondary metabolites in plants. Betalains are natural pigments that do not coexist with anthocyanins within the same plant. Amaranthus tricolor (‘Suxian No.1’) is an important leaf vegetable rich in betalains. However, the WRKY family members in amaranth and their roles in betalain synthesis and metabolism are still unclear.
Methods: To elucidate the molecular characteristics of the amaranth WRKY gene family and its role in betalain synthesis, WRKY gene family members were screened and identified using amaranth transcriptome data, and their physicochemical properties, conserved domains, phylogenetic relationships, and conserved motifs were analyzed using bioinformatics methods.
Results: In total, 72 WRKY family members were identified from the amaranth transcriptome. Three WRKY genes involved in betalain synthesis were screened in the phylogenetic analysis of WRKY TFs. RT-qPCR showed that the expression levels of these three genes in red amaranth ‘Suxian No.1’ were higher than those in green amaranth ‘Suxian No.2’ and also showed that the expression level of AtrWRKY42 gene short-spliced transcript AtrWRKY42-2 in Amaranth ‘Suxian No.1’ was higher than that of the complete sequence AtrWRKY42-1, so the short-spliced transcript AtrWRKY42-2 was mainly expressed in ‘Suxian No.2’ amaranth. Moreover, the total expression levels of AtrWRKY42-1 and AtrWRKY42-2 were down-regulated after GA3 treatment, so AtrWRKY42-2 was identified as a candidate gene. Therefore, the short splice variant AtrWRKY42-2 cDNA sequence, gDNA sequence, and promoter sequence of AtrWRKY42 were cloned, and the PRI 101-AN-AtrWRKY42-2-EGFP vector was constructed to evaluate subcellular localization, revealing that AtrWRKY42-2 is located in the nucleus. The overexpression vector pRI 101-AN-AtrWRKY42-2-EGFP and VIGS (virus-induced gene silencing) vector pTRV2-AtrWRKY42-2 were transferred into leaves of ‘Suxian No.1’ by an Agrobacterium-mediated method. The results showed that AtrWRKY42-2 overexpression could promote the expression of AtrCYP76AD1 and increase betalain synthesis. A yeast one-hybrid assay demonstrated that AtrWRKY42-2 could bind to the AtrCYP76AD1 promoter to regulate betalain synthesis.
Discussion: This study lays a foundation for further exploring the function of AtrWRKY42-2 in betalain metabolism.
Amaranth (Amaranthus tricolor L.) is an annual plant with beneficial health effects. It is cultivated in many countries. It is native to India (Parveen et al., 2018) and distributed in southern Asia (Wang et al., 2019), Central Asia, Japan, and other places. Amaranth is rich in betalains, characteristic pigments in Amaranthaceae (Chang et al., 2021), and betalain structure in the species differs from that of betalain in beet. Amaranth is an important resource for betalain extraction and research (Zheng et al., 2016).
Betalains exist in only 15 families in Caryophyllales and the fungus Amanita muscaria (Clement and Mabry, 1996; Polturak et al., 2018). The stability of betalains from different sources varies in the temperature range of 50–90°C; accordingly, botanists use the thermal stability of betacyanins as a chemical classification index (Martins et al., 2017; Carreón-Hidalgo et al., 2022). Betalains are classified as betacyanins and betaxanthins. There are four main types of betacyanins: amaranthin, betanin, gomphrenin, and decarboxybetanin (Strack et al., 2003; Cai et al., 2006). Amaranthin is biosynthesized from betanin (Gins et al., 2002) and has only been detected in the genus Amaranthus. The betalain synthesis pathway in plants has been characterized. Cytochrome P450 enzyme CYP76AD catalyzes tyrosine to l-DOPA (Polturak et al., 2016), Subsequently, l-DOPA is catalyzed to an open cyclo-DOPA by 4,5-dopa dioxygenase extradiol (or DODA enzyme) and then spontaneously forms betalamic acid, which is the core skeleton of all betalain compounds and can spontaneously combine with amino acids or amines to produce betaxanthin (Polturak and Aharoni, 2018; Sheehan et al., 2020). At the same time, dopamine is oxidized by the cytochrome P450 enzyme CYP76AD1 to form cyclo-DOPA, and cyclo-DOPA spontaneously condenses to form beta-glucoside ligand, which then generates betacyanin under the action of beta-glucoside ligand-5-O-glucosyl transferase (Hatlestad et al., 2012; Hatlestad et al., 2015; Alfonso et al., 2019). MYB transcription factors regulate the synthesis of betalain via DODA and CYP76AD1 (Xie et al., 2016; Peng et al., 2019). In addition, WRKY TFs in pitaya participate in the regulation of CYP76AD1 and thus regulate betalain synthesis (Cheng et al., 2017; Zhang et al., 2021).
WRKYs are a plant-specific transcription factor family, named for the conserved WRKYGQK7 amino acid sequence. This family has been identified in Arabidopsis (Birkenbihl et al., 2018), sorghum (Baillo et al., 2020), maize (Hu et al., 2021), pitaya (Chen et al., 2022), Beta vulgaris (Cheng et al., 2017; Wu et al., 2019; Zhang et al., 2021), banana (Jia et al., 2022), and other taxa. WRKY proteins could specifically bind to the TTGAC sequence (also known as W-box) to regulate gene transcription, and their expression is mainly induced by pathogenic bacteria, injury, and SA (salicylic acid) (Li et al., 2020). WRKY proteins are involved in plant development, metabolic regulation (Zhang et al., 2018; Su et al., 2022), plant stress responses (Li et al., 2020), and aging . WRKY TFs regulate betalain metabolism in pitaya. In particular, HmoWRKY40, HpWRKY44, HpWRKY3, and HpWRKY18 play important roles in the color transformation and betalain accumulation of pitaya (Cheng, 2018; Zhang, 2020).
We have identified genes involved in betalain metabolism in amaranth, such as AtrCYP76AD1, AtrDODA, AtrDOPA5-GT, AtrB5-GT, and AtrB6-GT, by transcriptomics analyses (Zheng et al., 2016). However, the specific WRKY TFs in amaranth involved in betalain synthesis and the underlying regulatory mechanisms have not been determined. In this study, we identified WRKY TFs and demonstrated that AtrWRKY42-2 interacts with the promoter of AtrCYP76AD1, thereby regulating the betalain synthesis in amaranth. These findings provide new insight into betalain synthesis.
Seeds of ‘Suxian No.1’ and ‘Suxian No.2’ were provided by Suzhou Academy of Agricultural Sciences. ‘Suxian No.1’ was dark red and rich in betalains. ‘Suxian No.2’ was light green and did not contain betalains (Figure 1). The leaves (functional leaves) of ‘Suxian No.1’ from potted seedlings at about one month were used for transient transformation and virus-induced gene silencing (VIGS). All samples were frozen in liquid nitrogen immediately and stored at −80°C for further analyses. Briefly, 1 mg/L GA3 and 2 mg/L paclobutrazol (PP333) were added into MS medium supplemented with sucrose (30 g/L) and agar (7 g/L) (Pan et al., 2018; Zhao et al., 2019; Xiao et al., 2021). The sterilized seeds of ‘Suxian No.1’ and ‘Suxian No.2’ were inoculated in the medium. Medium without hormones was used as a control. Each treatment was evaluated with 15 bottles and three biological replicates. After 7 days of culture, the hypocotyl at the amaranth seedling stage was collected.

Figure 1 Phenotypic differences between typical potted seedlings of ‘Suxian No.1’ (left) and ‘Suxian No.2’ (right).
Transcriptome data for amaranth were obtained from our constructed database (SRA:SRX7816306–SRX7816311) (Liu et al., 2019). Gene screening was performed using hidden Markov models (HMM). The WRKY domain was downloaded from Pfam (PF03106) (http://pfam.xfam.org/, accessed on October 19, 2021) and was employed to identify all possible WRKY genes in amaranth using HMMER (Chen et al., 2015) with an e-value threshold of -5. Each candidate WRKY gene was further confirmed using SMART (http://smart.embl-heidelberg.de/) and the Conserved Domain Database (CDD, http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi). Physical parameters, including the molecular mass and theoretical isoelectric point (pI) of the deduced WRKY proteins were investigated using ExPASy (http://web.expasy.org/protparam/).
Conserved motifs of AtrWRKY proteins were identified using MEME (Multiple Expectation Maximization for Motif Elicitation) [http://meme-suite.org/tools/meme (accessed on October 29, 2022)].
Amino acid sequence logos of WRKY were generated using the WebLogo platform (https://weblogo.berkeley.edu/logo.cgi). DNAMAN7 (company: Lynnon Corporation; Version 7.0.2.176) was used to compare the amino acid sequences of WRKY homologies in different species.
Full-length amino acid sequences for a phylogenetic analysis were obtained from pitaya (Cheng et al., 2017; Chen et al., 2022) and Beta vulgaris (Wu et al., 2019). A phylogenetic tree was constructed by the neighbor-joining method (NJ) using MEGA11 (Version 11.0.8) with 1000 boot-strap replicates. Evolview (https://www.evolgenius.info/evolview/#/treeview) was used to visualize the phylogenetic tree.
Total RNA was isolated from samples using MolPure Plant Plus RNA Kit (Yeasen, China) according to the manufacturer's instructions. First-strand cDNA was synthesized from 1 μg of total RNA using Recombinant M-MLV reverse transcriptase (TransGen Biotech, Beijing, China). Quantitative real time-PCR (qRT-PCR) was performed in optical 96-well plates using the Roche LightCycler 480 instrument (Roche, Sweden). The reactions were carried out in a 20 μL volume containing 10 μL of SYBR Premix Ex Taq, 0.8 μL of gene-specific primers, 2 μL f diluted cDNA, and 6.4 μL of ddH2O. The PCR conditions were as follows: 30 s at 95°C, 45 cycles of 10 s at 95°C and 20 s at 59°C, followed by 12 s at 72°C. Three biological repeats were performed for each material. SAND (Xiao et al., 2021) was used as the internal reference gene. Excel 2016 was used for data analyses, SPSS 20 was used to evaluate the homogeneity of variances, and the 2−ΔΔCt method was used for quantitative analyses of gene expression. The primer pairs used for the qRT-PCR analysis of WRKY genes are listed in Supplementary Table S1.
The cDNA and gDNA sequences of AtrWRKY42-2 were cloned from ‘Suxian No.1’ and ‘Suxian No.2’ samples. The genomic sequence and its corresponding coding sequences (CDS) were compared using GSDS (Gene Structure Display Server) (http://gsds.cbi.pku.edu.cn/).
The full-length coding sequence of AtrWRKY42-2 was inserted into the pRI 101-AN-EGFP vector (primers are listed in Supplementary Table S1). Agrobacterium strain GV3101 cells carrying pRI 101-AN-AtrWRKY42-2-EGFP and pRI101-AN-EGFP were infiltrated into onion inner epidermis cells. Transient expression of EGFP was recorded using a laser confocal microscope (Olympus model: FV1200 4 laser) at 405 nm. Onion epidermis cells were stained with DAPI to evaluate nuclear localization, and the fluorescence signal was visualized by laser confocal microscopy at 473 nm.
The promoter of AtrWRKY42-2 was cloned from total genomic DNA of ‘Suxian No.1’ using the TaKaRa Genome Walking Kit (see Supplemental Table S1 for specific primers). Promoter elements and conserved binding domains of AtrWRKY42-2 were analyzed using PlantCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/, accessed on November 10, 2022).
pRI 101-AN-AtrWRKY42-2-EGFP and pRI 101-AN-EGFP were transformed into the Agrobacterium tumefaciens strain GV3101. The bacterial cells were resuspended to an OD600 of 0.8–1.0 using MMA buffer (10 mM MES, 10 mM MgCl2, 100 μM acetosyringone) at a 1:1 ratio. Subsequently, bacterial cells were infiltrated into amaranth leaves. Then these seedlings were cultured in the dark at 25°C for 1–2 days. These seedlings were then transferred to a culture room set to 16 h/8 h (light/dark) and 25°C. Plant phenotypes were observed. The transiently transformed plants were cultured for 1 week (including dark treatment for 2 days), and the 7th to 8th true leaves were collected to detect gene expression levels and determine the betalain content.
The gene fragment with the AtrWRKY42-2 conserved domain was cloned into the pTRV2 vector. pTRV1, pTRV2, and pTRV2-AtrWRKY42-2 were transformed into Agrobacterium tumefaciens strain GV3101. The bacterial cells were resuspended to an OD600 of 0.6~0.8 using an MMA buffer. pTRV2 (negative control) and pTRV2- AtrWRKY42-2 were separately infiltrated into amaranth leaves with pTRV1 in a ratio of 1:1. The culture conditions were the same as those described above. The leaves were collected for the detection of gene expression level and the determination of betalain contents.
Betalain contents were determined as described by Hua et al. (Hua et al., 2016) The absorption peaks of betacyanin and betaxanthin at 538 nm and 465 nm were determined by ultraviolet-visible spectrum spectrophotometer (UV-900, Shanghai Yuan Analysis Instrument Co., Ltd.). The betacyanin and betaxanthin contents were calculated according to a molar extinction coefficient of betaxanthin of 5.66 × 104. All determinations were performed in triplicate.
The promoter of AtrCYP76AD1 (see Supplementary Text S2 for the sequence) was ligated to the pHis II yeast reporter vector to form a pPromoter-pHis II recombinant bait vector. AtrWRKY42-2 was ligated into the expression vector pGADT7 to construct the pGADT7-AD recombinant vector and the pGADT7-AtrWRKY42-2 vector plasmid was used to transform the positively verified AtrCYP76AD1- pHis II (primers are listed in Supplementary Table S1). The bait yeast strain Y187 was cultured at 30°C for 3 days. Two deficient media, SD/-Leu-Trp and SD/-Leu-Trp-His-Ade, were used to detect the interaction between AtrWRKY42-2 and the AtrCYP76AD1 promoter based on yeast growth.
A total of 72 putative WRKY genes were screened by a systematic analysis of the transcriptome of A. tricolor. These WRKY genes were termed AtrWRKY1 to AtrWRKY72. The full-length CDS of the AtrWRKY genes ranged from 818 bp (AtrWRKY12 and AtrWRKY31) to 5459 bp (AtrWRKY15) with deduced proteins of 178–1093 amino acids. Furthermore, the computed molecular weights of these WRKY proteins ranged from 19517.74 to 124261.7 Da. The theoretical pI of the deduced AtrWRKY proteins ranged from 5.11 to 9.96. The detailed sequences and annotation are listed in Supplementary Table S3.
A total of 15 conserved motifs were detected from AtrWRKYs using the MEME tool. Each motif contained 15–50 amino acids, and each sequence included 2–12 motifs. Motifs 1 and 3 formed the main structure of the WRKY domain (WRKYGQK), and Motifs 2, 10, and 13 contained a zinc-finger motif. The most frequent motifs of AtrWRKYs were Motif 1, Motif 5, and Motif 2, which showed high amino acid sequence conservation and represent the AtrWRKY domain (Figure 2). AtrWRKY6, AtrWRKY72, AtrWRKY10, and AtrWRKY43 all had Motif 14. However, Motif 9 was only identified in AtrWRKY63.
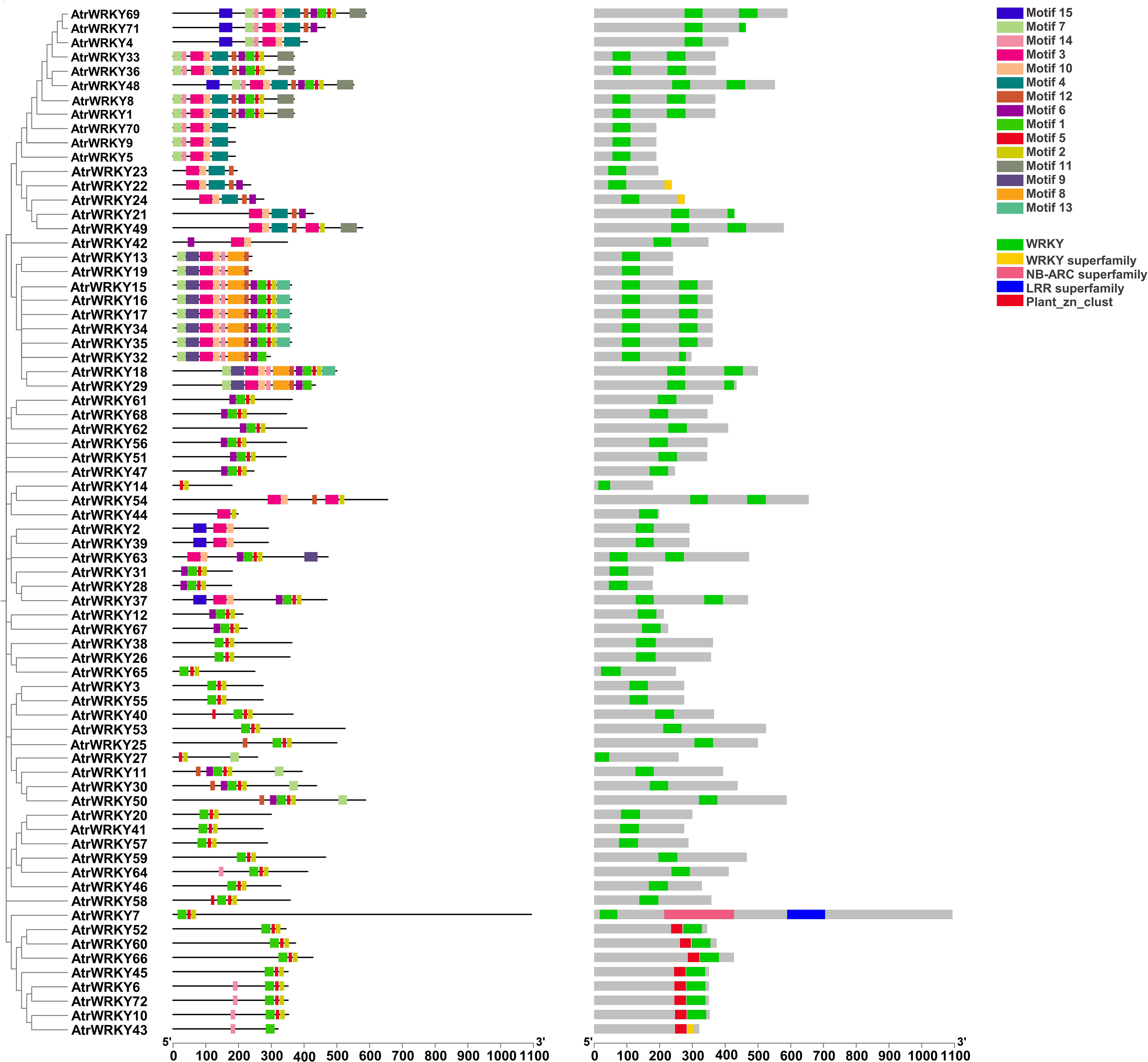
Figure 2 Analysis of conserved motifs and conserved domains of AtrWRKYs. Motif1–15 represents the conserved sequence of AtrWRKYs members. WRKY and WRKY superfamily represent the conservative domain of AtrWRKYs members.LRR superfamily; NB-ARC superfamily represents the LRR and NB-ARC domains; Plant_zn_clust represent plant zinc cluster.(AtrWRKYs: Amaranth WRKY transcription factor family).
At the amino acid sequence level, the family motifs were well conserved (Figures 3A, B). To evaluate the evolutionary history of the WRKY gene family in A. tricolor and to uncover their classification and function in betalain synthesis, 72 WRKY proteins from A. tricolor, Beta vulgaris (betalain), and ‘Guan Huahong’pitaya (betalain) were used to construct a phylogenetic tree using the NJ method implemented in MEGA11. Among the selected AtrWRKY7, AtrWRKY40, and AtrWRKY42-2 proteins, WRKY homologs in the same group/subgroup as those in sugar beet and pitaya may have similar functions. AtrWRKY7 and AtrWRKY40 were homologous to AtWRKY25 and HmoWRKY40, respectively. In particular, AtrWRKY42-2 was homologous to HpWRKY44, AtWRKY44, and BvWRKY44.We predicted that AtrWRKY7, AtrWRKY40, and AtrWRKY42-2 might be involved in betalain synthesis (Figure 3C).
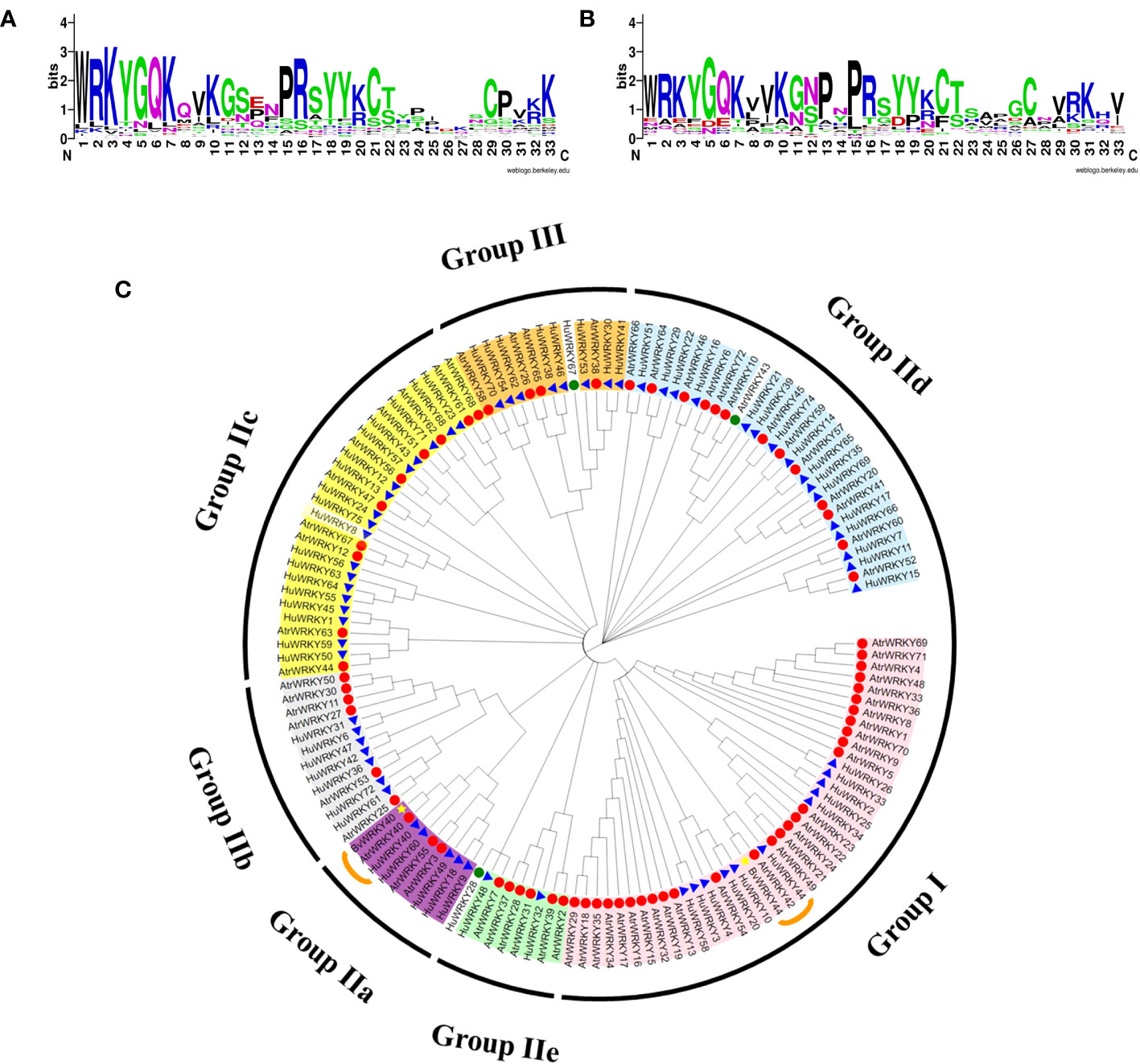
Figure 3 Conserved amino acid sequence analysis and WRKY phylogenetic tree analysis. (A) Comparison of the conserved domain (WRKY-a) of AtrWRKY family members; (B) Comparison of the WRKY-b conserved domain (The higher the bit value in the log map of the domain, the more conserved the corresponding amino acid; WRKY-a: upstream domain of the gene sequence; WRKY-b: downstream domain of gene sequence); (C) WRKY phylogenetic tree (Red circle represents amaranth, blue triangle represents pitaya, and yellow star represents sugar beet. The green circle represents the WRKY members classified in group IV).
An un-rooted phylogenetic tree was constructed based on the alignments of the WRKY motif and zinc-finger motif from amaranth using the NJ method (Figures 2, 3C). The AtrWRKYs were divided into four groups (Groups I–IV). Group I contained two WRKY domains and C2H2 type zinc finger protein domains, including 19 AtrWRKYs. Group II contained one WRKY domain and C2H2 type zinc finger protein domain, including 48 AtrWRKYs. Group III only contained one zinc finger motif of type C2HC and one protein domain (Huang et al., 2012; Wang et al., 2014), including four AtrWRKYs (AtrWRKY65, AtrWRKY38, AtrWRKY58, and AtrWRKY26). AtrWRKY43 belonged to Group IV, which contained one WRKY domain and an incomplete C2H2 type zinc finger protein domain (Eulgem et al., 2000).
As shown in Figure 4, the complete transcript sequence was named AtrWRKY42-1 (CL2645.Contig1_All), and short splice transcript was one of the transcripts of the AtrWRKY42 gene, named AtrWRKY42-2 (CL2645.Contig2_All). AtrWRKY42-2 had only one conserved domain, which was one fewer domain than the complete protein-coding gene, and it could be confirmed as a member of Group II according to the WRKY classification criteria.

Figure 4 Homologous gene protein difference. WRKYGQK: WRKY domain; Zinc finger structure: C-x4-C-x23-H-x-H; Red box is the betalain synthesis-related WRKY specific amino acid sequence. “Atr” stands for Amaranth, “At” stands for Arabidopsis, “Bv” stands for B. vulgaris, and “Hp” stands pitaya.
A sequence alignment of WRKY44 homologues in beet, pitaya, and Arabidopsis (see Supplementary Text S4 for sequence details) showed that the other four proteins were clearly different from the AtWRKY44 protein motif (Figure 4). We further analyzed the AtrWRKY42-2 conserved domains. WRKY proteins involved in betalain synthesis from pitaya, B. vulgaris, Arabidopsis, and amaranth revealed that AtrWRKY42-2 had the same domain and zinc finger structure as those of the WRKY44 cluster. Except for AtWRKY44, the other three genes had special amino acid sequence fragments (Figure 4).
To further identify betalain-related WRKY genes from A. tricolor, the expression patterns of AtrWRKY7\40\42 genes in the leaves of ‘Suxian No.1’ and ‘Suxian No.2’ were determined by RNA-seq. We have previously found that GA3, 2,4-D, and 6-BA promote the growth of amaranth, the accumulation of betacene in amaranth is inhibited by GA3 and 2,4-D, and PP333 promotes the synthesis of betacene (Liu et al., 2018; Pan et al., 2018). Therefore, we determined the expression patterns of AtrWRKY7\40\42 genes under treatment with 1 mg/L GA3 and 2 mg/L paclobutrazol to verify the expression of the putative WRKY TFs. As shown in Figure 4, the expression levels of AtrWRKY7\40\42 were altered in the ‘Suxian No.1’ leaves. In particular, AtrWRKY40 was upregulated under GA3 treatment, AtrWRKY42-2 was downregulated, and AtrWRKY7 expression did not differ between treatment and control groups. AtrWRKY40 was upregulated under paclobutrazol treatment, AtrWRKY7 was downregulated, and AtrWRKY42 was upregulated (Figure 5). Based on these results, we selected AtrWRKY42 as a candidate gene for further analyses. However, the total expression levels of AtrWRKY42-1 and AtrWRKY42-2 in ‘Suxian No.1’ and’Suxian No.2’ were always higher than the expression levels of AtrWRKY42-1, so the short splicing transcript AtrWRKY42-2 expressed mainly in the plants of the two varieties.
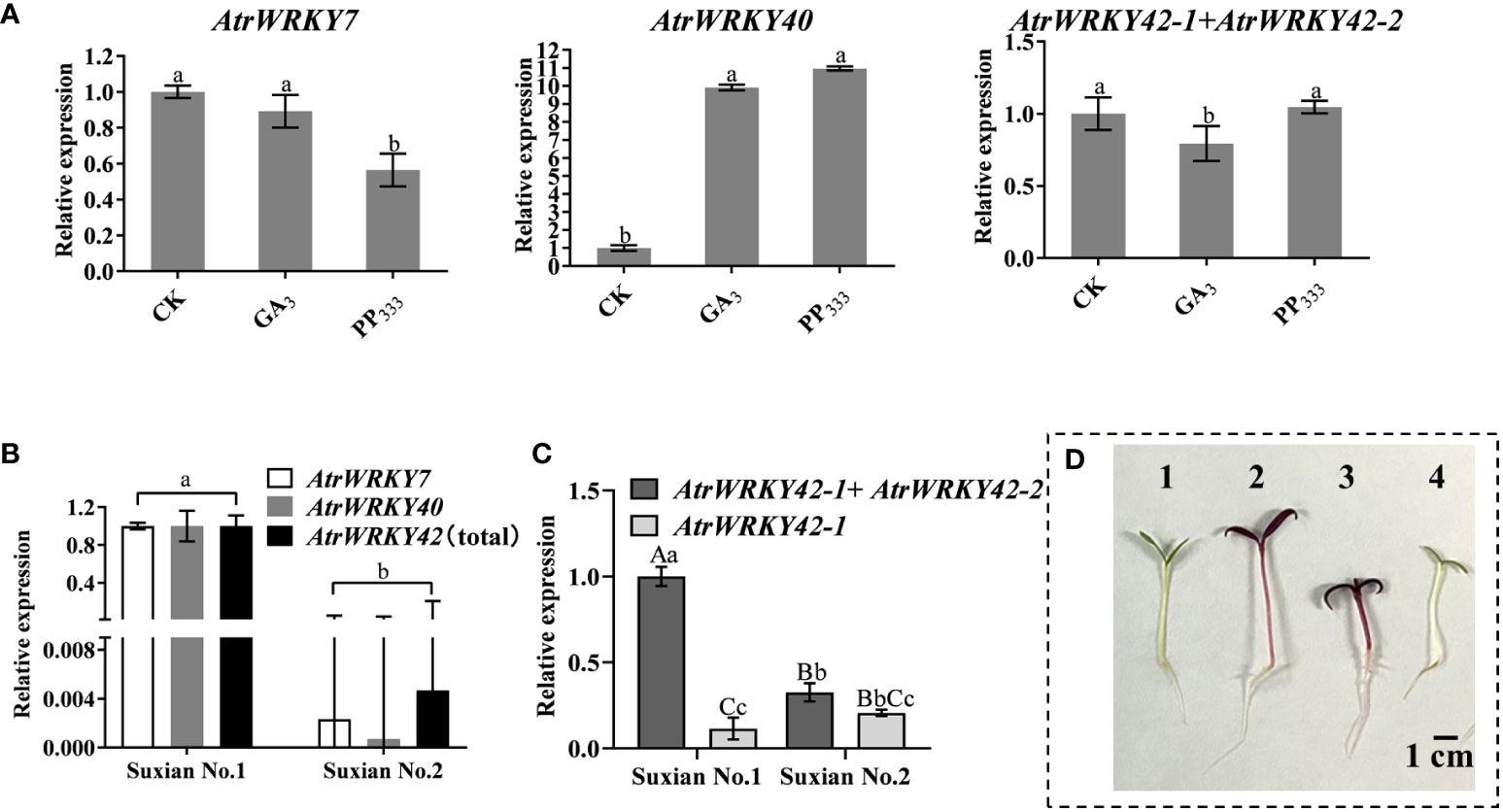
Figure 5 Quantitative analysis of AtrWRKY7, AtrWRKY40, and AtrWRKY42 expression in different varieties and after hormone treatment. (A) Quantitative expression analysis under different hormone treatments; (B) Quantitative expression in different under different varieties; (C). The relative expression levels of AtrWRKY42-1 and AtrWRKY42-2 in two varieties; (D) Plant phenotypes 1–3 indicate the seedlings of ‘Suxian No.1’ treated with GA3, sterile water, and PP333, respectively, and 4 indicates seedlings of ‘Suxian No.2’ treated with sterile water. (CK: ‘Suxian No.1’ blank control; GA3: Optimal concentration of 1 mg/L; PP333: Optimal concentration of 2 mg/L; AtrWRKY42(total): It represents the total expression of AtrWRKY42-1 and AtrWRKY42-2; Unprocessed ‘Suxian No.1’ material (CK) was used for calibration (set as 1) in three independent experiments (n = 3), and the error bar represents twice the standard error. The difference was statistically significant when compared with levels in CK by Student’s t-tests; a, b indicate a significant difference at p < 0.05; Aa, Bb and Cc indicate a significant difference at p < 0.01.
Complementary DNA and genomic DNA of AtrWRKY42-2 were cloned from both ‘Suxian No.1’ and ‘Suxian No.2’. There was no difference in the cDNA sequence of AtrWRKY42-2 between ‘Suxian No.1’ and ‘Suxian No.2’, encoding 348 amino acids, with an isoelectric point (pI) of 7.58 (see Supplementary Table S3), and the gDNA sequences were identical (Figures 6A, B).
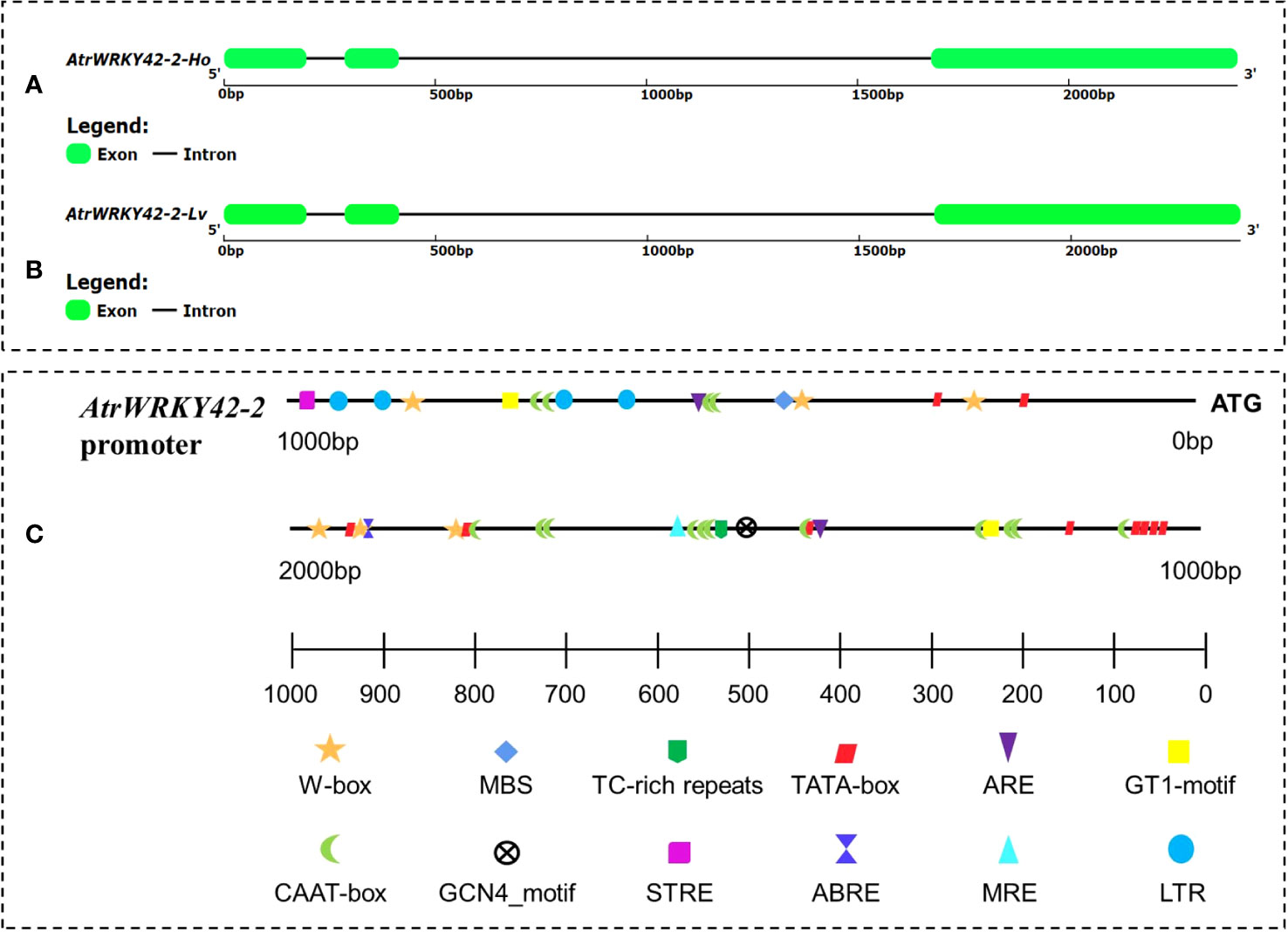
Figure 6 AtrWRKY42-2 gene structure and AtrWRKY42-2 cis-regulatory elements. (A) Gene structure of AtrWRKY42-2 in ‘Suxian No.1’ variety; (B) Gene structure of AtrWRKY42-2 in ‘Suxian No.2’ variety; (C) Distribution of cis-acting elements of the AtrWRKY42-2 promoter.
A 1891 bp promoter region (see Supplementary Text S2 for the sequence) was successfully amplified. In this region, a transcription start site (TSS), a TATA-box 1000 bp upstream of the TSS (Figure 6C), and a large number of other cis-acting elements (Supplementary Table S5) were predicted based on a PlantCARE database analysis.
The full-length CDS of AtrWRKY42-2 was fused with the EGFP gene to analyze its subcellular localization. Ater the transient expression of constructs in the inner epidermis of onion, green fluorescence of AtrWRKY42-2 was detected exclusively in the nuclei (as evaluated by 4’,6-diamidino-2-phenylindole (DAPI) staining); however, in the positive control, the EGFP signal was observed around the cytoplasm, cell membrane, and nuclei (Figure 7).
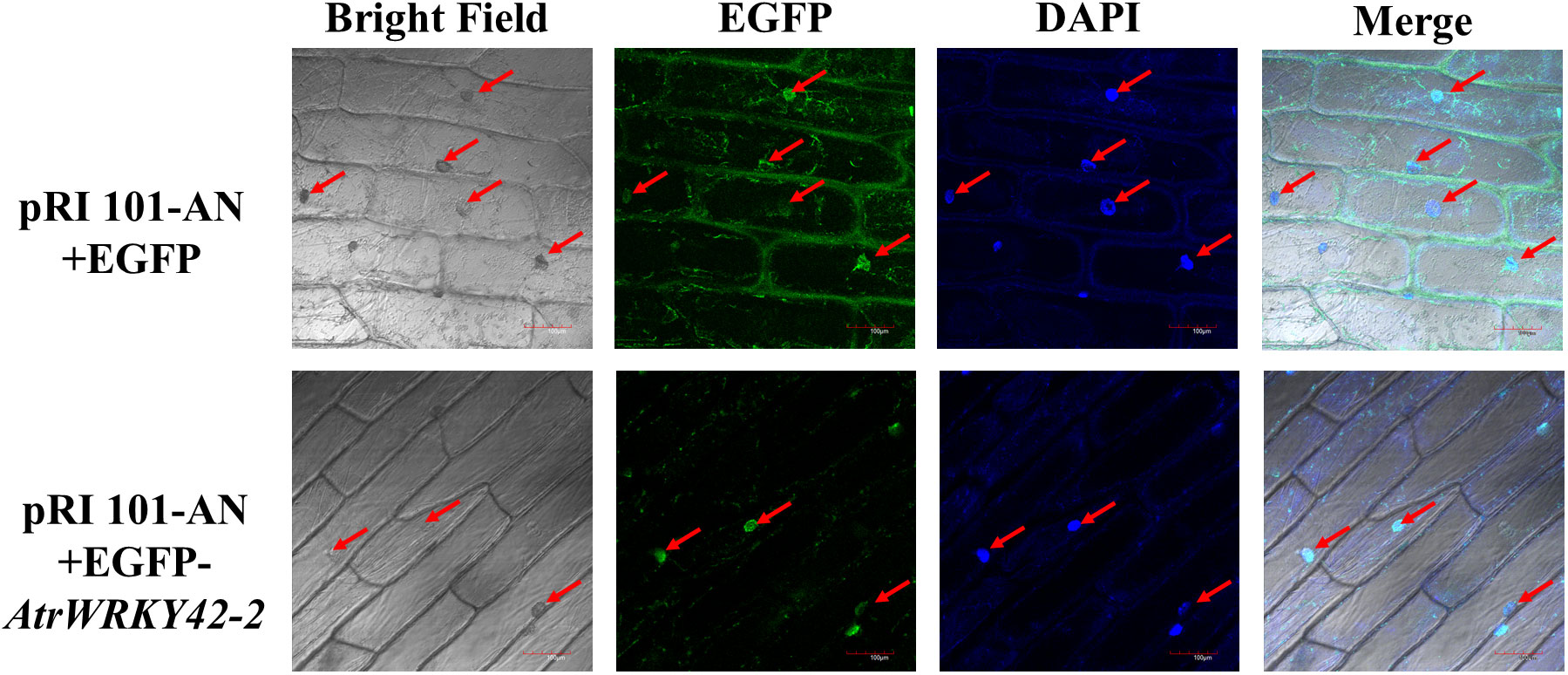
Figure 7 Subcellular localization of pRI 101-AN and AtrWRKY42-2 proteins in onion. Red arrows indicate the location of EGFP and DAPI fluorescence signals in cells. Bars=100μm.
As shown in Figure 8A, when compared with expression in the control and empty plasmid group, the overexpression of AtrWRKY42-2 was visualized as dark red in the scales of ‘Suxian No.1’ amaranth. There were no significant differences in the betaxanthin content among transgenic plants, blank control group, and positive control group. However, the betacyanin content in transgenic plants was significantly higher (P < 0.05) than that in the control (Figure 8B). Results from the RT-qPCR analyses showed that AtrWRKY42-2, CYP76AD1, TyDC, MYB1, B5-GT, and B6-GT (Download ID and primer sequences are shown in Supplementary Table S1), key genes involved in betalain metabolism, were upregulated in plants overexpressing AtrWRKY42-2 (Figure 8C). These findings were consistent with the leaf phenotype and betacyanin content.
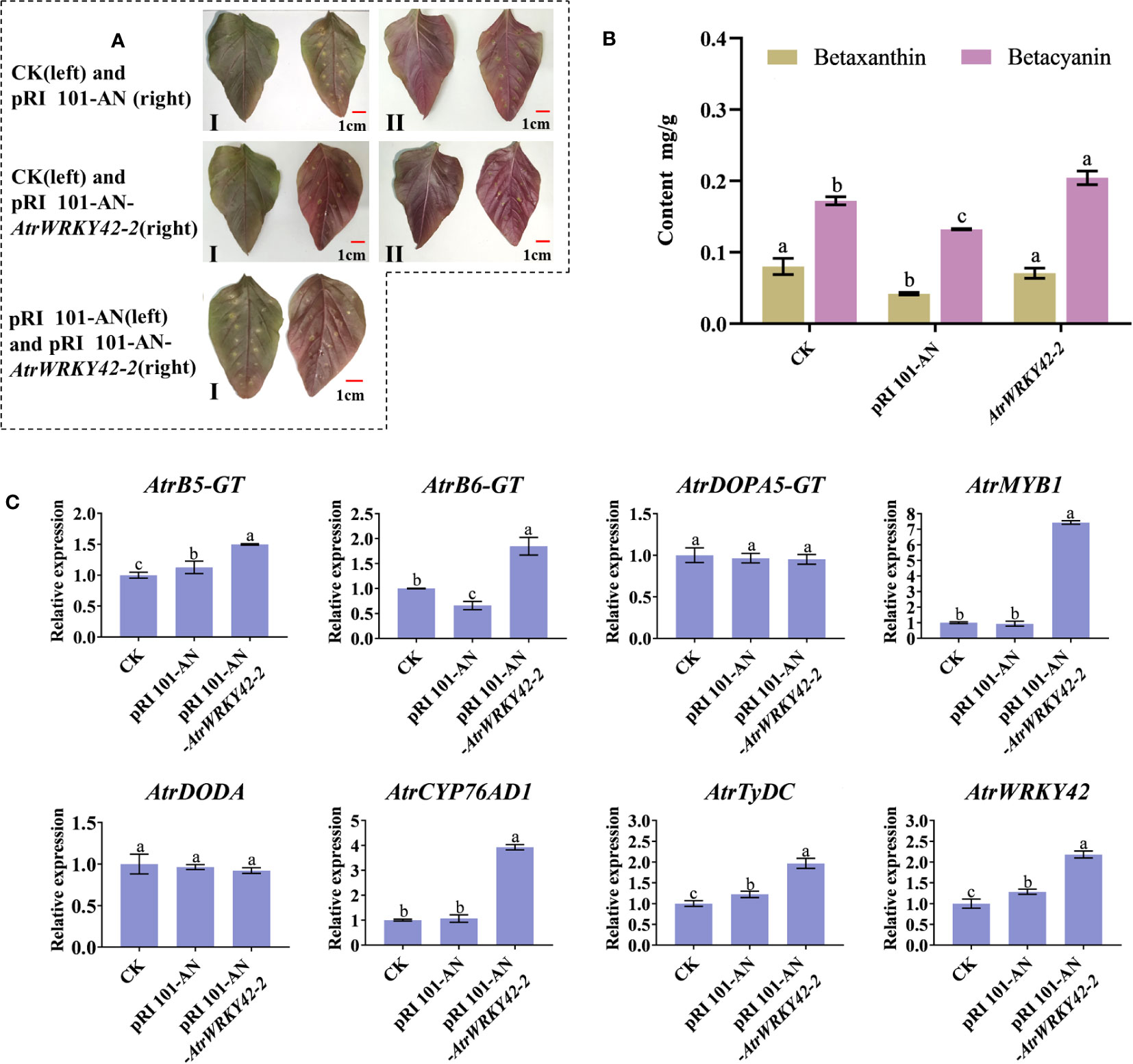
Figure 8 Agrobacterium-mediated transient transformation of the lower epidermis of amaranth leaves, revealing that the overexpression of AtrWRKY42-2 promotes the synthesis of betalain in amaranth. (A) Plant leaves after transient transformation for 7 days (I: Blade face; II: Back of the blade) (B) Betalain content in leaves in plants with different transient transformations (C) Relative expression of betalain synthesis-related genes in leaves of plants with different transient transformations. (a, b and c indicate significant differences at p < 0.05; Bars: 1 cm).
The silencing of AtrWRKY42-2 resulted in new green leaves (although not completely green) after 2–3 weeks of transformation (Figure 9A). There was no significant difference in the betaxanthin content between plants with gene silencing and the control; however, the betacyanin content in leaves was significantly lower in plants with gene silencing than in the control (Figure 9B). The RT-qPCR analysis showed that the expression levels of key genes in betalain synthesis were significantly decreased by gene silencing (Figure 9C). These results indicate that AtrWRKY42-2 plays an important role in the betacyanin biosynthetic pathway of amaranth, and silencing AtrWRKY42-2 inhibited the synthesis of betacyanin.
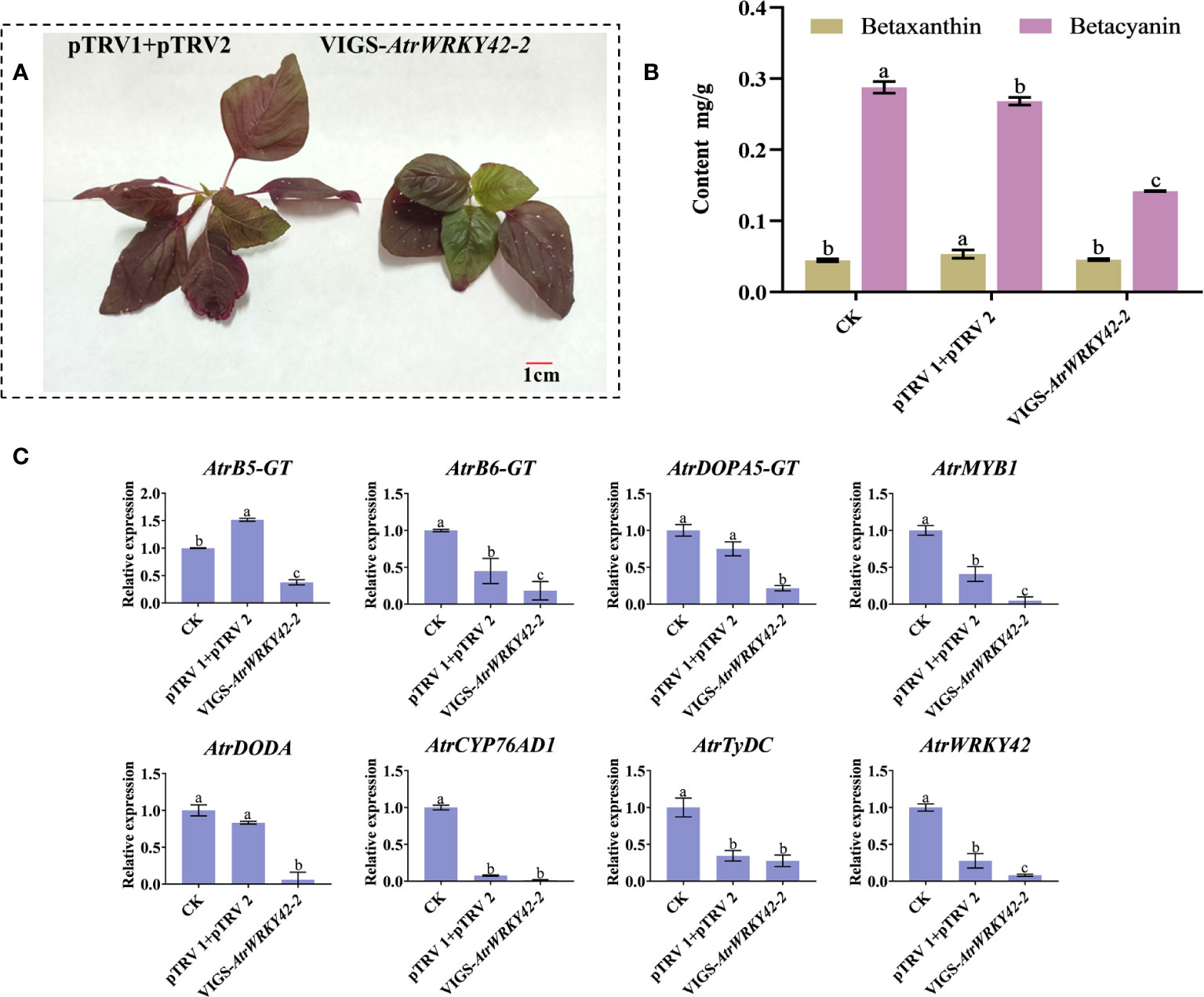
Figure 9 Silencing of AtrWRKY42-2 inhibits betalain production. (A) Positive control plants (left) and VIGS transgenic positive plants (right) Phenotype. (B) Betalain content in leaves of positive control plants and VIGS transgenic plants. (C) Relative expression levels of key genes related to betalain synthesis in transgenic plants with gene silencing. Three biological replicates were performed for each sample (a, b and c indicate significant differences at p < 0.01; Bar = 1 cm).
The promoter of AtrCYP76AD1 was cloned from ‘Suxian No.1’ amaranth (unpublished) (see Supplementary Text S2 for the sequence). A typical W-box motif was identified in the AtrCYP76AD1 promoter, which is a cognate binding site for WRKY TFs, suggesting that WRKY TFs are involved in the regulation of AtrCYP76AD1. Therefore, the interaction between AtrWRKY42-2 and the promoter of AtrCYP76AD1 was evaluated using a yeast one-hybrid assay. Yeast cells carrying only pHis II-AtrCYP76AD1-Pro could not grow on SD/-Leu-Trp-His-Ade medium (Figure 10). Yeast cells containing pHis II-AtrCYP76AD1-Pro+pGADT7-AtrWRKY42-2 were grown on SD/-Leu-Trp-His-Ade medium (Figure 10). These results suggest that AtrWRKY42-2 binds directly to the promoter of AtrCYP76AD1 and may be involved in the biosynthesis of amaranth betalains.
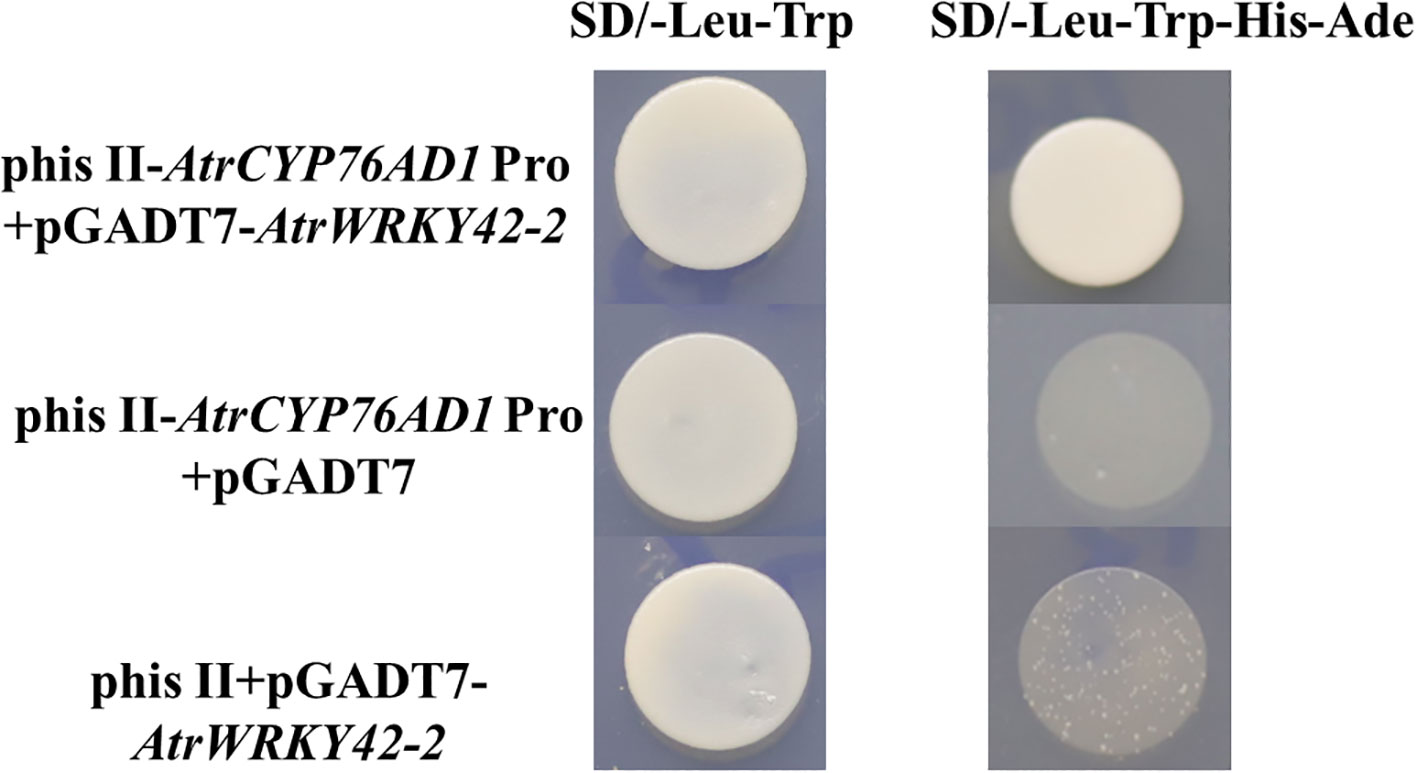
Figure 10 AtrWRKY42-2 and AtrCYP76AD1 promoter yeast one-hybrid assay. The experiment was repeated independently three times. SD/-Leu-Trp and SD/-Leu-Trp-His-Ade indicate yeast single defect medium.
The WRKY TFs are a universal gene family in higher plants, controlling plant growth and development under normal and stress conditions (Jiang et al., 2017; Gangola and Ramadoss, 2020; Li et al., 2020; Jia et al., 2022). However, WRKY gene family members in A. tricolor have not been identified and characterized. We screened 72 AtrWRKY genes from the A. tricolor transcriptome database, which was more than 70 genes reported in pitaya (Chen et al., 2022) and 2 genes in Beta vulgaris (Wu et al., 2019). AtrWRKYs were divided into four subfamilies according to their domain and zinc finger structure, consistent with the previous classification of WRKY TFs (Eulgem et al., 2000; Huang et al., 2012; Wang et al., 2014).
Betalains are a unique lineage in anthocyanin evolution (Alfonso et al., 2019; Li et al., 2019). However, they cannot coexist within the same plant. Betalains have only been detected in Caryophyllales species, with the exception of Caryophyllaceae and Molluginaceae species, as well as some higher fungi. The WRKY TFs has important roles in higher plants, including roles in plant growth and development and secondary metabolite synthesis under the normal and stress conditions (Peng et al., 2019; Hu et al., 2021; Chen et al., 2022). Recent studies have also shown that WRKY TFs are involved in betalain metabolism in pitaya (Gangola and Ramadoss, 2020; Hu et al., 2021; Chen et al., 2022). In pitaya, HmoWRKY40 and HpWRKY44 promote the synthesis of betalains (Cheng et al., 2017; Zhang et al., 2021), while HmoWRKY42 inhibits the synthesis of betalains (Chen et al., 2022). A phylogenetic analysis revealed that AtrWRKY42-2 was homologous to HmoWRKY44 and BvWRKY44. However, the amino acid sequences of BvWRKY44, AtrWRKY42-2, and HpWRKY44 in betalain-producing differed from AtWRKY44 in Arabidopsis (Figure 3C). Some studies have found that AtWRKY44 only regulates the biosynthesis of phenylpropyl and indole alkaloids (Johnson et al., 2002; Mao et al., 2011), no one studied the role of AtWRKY44 in betalain synthesis, so we speculated that amino acid substitutions are involved in the regulation of betalain synthesis. Therefore, we speculated that the amino acid substitutions are involved in the regulation of betalain synthesis. Further studies are needed to verify whether the transfer of AtWRKY44 into A. thaliana mediated by A. tumefaciens regulates anthocyanin synthesis and increases the anthocyanin content, similar to the other three WRKY homologues.
Betalain accumulation could make the amaranth leaves red (Barba-Espin et al., 2018; Fan et al., 2020), which is determined by environmental conditions and transcription factors (Erika et al., 2018; Preczenhak et al., 2019; Guerrero-Rubio et al., 2020; Sakuta et al., 2021; Sokolova et al., 2022). Our previous studies have indicated that various environmental factors and hormones (GA3 and 2,4-D) influence betalain biosynthesis (Xiao et al., 2021). The predicted cis-acting elements of AtrWRKY42-2 contain TC-rich repeats, STREs, and so on based on a PlantCARE database analysis (Supplementary Table S5). Therefore, we speculated that these factors affect betalain synthesis via WRKY TFs. We found that the function of AtrWRKY42-2 in amaranth is similar to those of HpWRKY44 and HmoWRKY40 in pitaya (Cheng et al., 2017; Zhang et al., 2021), and all of these loci promote the accumulation of betalains. When the WRKY domain and zinc finger structure of AtrWRKY42-2 were silenced by VIGS technology, betacyanin synthesis was inhibited. In contrast, betacyanin accumulation in amaranth was promoted by AtrWRKY42-2 overexpression. In analyses of the relative expression of AtrWRKY42-2 and key genes involved in betalain synthesis, we observed that the gene expression levels were associated with the betacyanin content. These results indicated that AtrWRKY42-2 promotes betacyanin synthesis. However, there was no difference in the betaxanthin content between transgenic plant and the control. These results content showed that AtrWRKY42-2 was not involved in betaxanthin synthesis. Perhaps it was synthesis by other CYP76AD members and transcription factors. Previous studies have shown that the synthesis of betacyanin mainly depends on the action of CYP76AD1 to synthesize cyclo-DOPA, and maintaining the ratio of CYP76AD1 and CYP76ADβ (CYP76AD5, 6, 15) can regulate the synthesis ratio of betacyanin and betaxanthin. (Sunnadeniya et al., 2016). However, the regulation of AtrWRKY42-2 in this study only led to the differential expression of key genes involved in the synthesis of betacyanin such as CYP76AD1, and it did not affect the expression of CYP76ADβ subfamily members (CYP76AD5, 6, 15) to exercise hydroxylation. Further functional studies of AtrWRKY7 and AtrWRKY40 are in progress.
WRKY TFs recognize and bind to the TTGAC(C/T) W-box in the promoter regions of target genes (Chen et al., 2019). WRKY TFs can bind not only upstream promoters (Brand et al., 2013; Madhunita and Ralf, 2014) but also to upstream promoters of other genes regulated by WRKY TFs (Cheng et al., 2017; Li et al., 2020; Zhang et al., 2021; Chen et al., 2022; Zheng et al., 2022). The AtrWRKY42-2 promoter sequence contained six W-box elements that specifically bind WRKY TFs. These results indicated that AtrWRKY42-2 binds to itself or other WRKY TFs. In addition, the AtrCYP76AD1 promoter sequence contains a W-box element of WRKY TFs. AtrWRKY42-2 could activate AtrCYP76AD1, a key gene involved in betalain synthesis, by binding to its promoter responsible for betalain biosynthesis in amaranth. HpWRKY44 and HmoWRKY40 could activate HmoCYP76AD1 expression by binding to its promoter responsible for betalain biosynthesis of pitaya (Cheng et al., 2017; Zhang et al., 2021). A yeast one-hybrid assay showed that AtrWRKY42-2 interacts with the AtrCYP76AD1 promoter (Figure 10), providing evidence that AtrWRKY42-2 activates the transcription of AtrCYP76AD1, involved in amaranth betalain biosynthesis (Figure 11). AtrWRKY42-2 transcript factor could regulate downstream gene transcription by binding W-box elements of target gene AtrCYP76AD1 promoter, and then participate in betalain synthesis. The expression level of the key genes in the betalain synthesis pathway was up-regulated or down-regulated in accordance with overexpression or silencing expression of AtrWRKY42-2 (Figures 8–11).
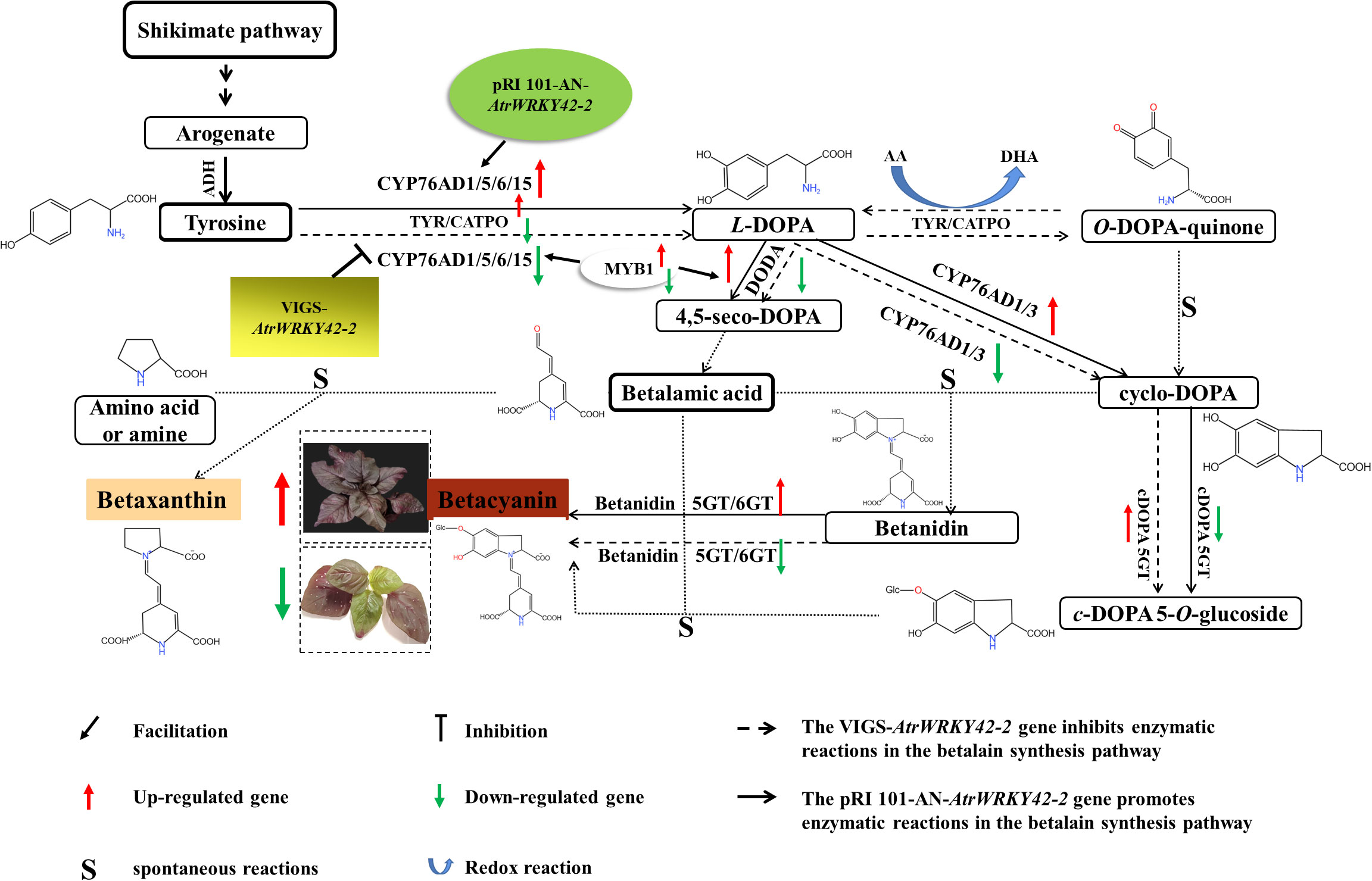
Figure 11 AtrWRKY42-2 is involved in the regulation of betalain synthesis in ‘Suxian No.1’. In this study, Agrobacterium-mediated transient transformation of AtrWRKY42-2 overexpression could promote the betalain synthesis and increase the content of betalain in ‘Suxian No.1’ amaranth. Moreover, the Agrobacteria-mediated virus-induced gene silencing system of AtrWRKY42-2 could inhibit the synthesis of betalain in ‘Suxian No.1’ and reduce the content of betalain. The green and red arrows indicate down-regulation and up-regulation of key genes in the betalain synthesis pathway (TyDC, CYP76AD1, DODA, DOPA5-GT, B5-GT, B6-GT), respectively. AA, ascorbic acid; DHA, dehydroascorbate; GT, glucosyl transferase.
In summary, WRKY family members were screened using HMM in the amaranth transcriptome. A total of 72 WRKY family members were obtained, and these were divided into four groups according to the domain and zinc finger structure. These WRKY TFs were located in the nuclei, with the exception of AtrWRKY50. There were no significant differences in the expression levels of AtrWRKY7\40\42 in the leaves of ‘Suxian No.1’, which may be explained by the complex factors involved in gene expression regulation, such that changes in any single factor do not cause substantial changes in expression. The expression level of AtrWRKY42-2 in ‘Suamaranth 1’ was significantly higher than that of AtrWRKY42-1. A transient overexpression experiment and VIGS assay showed that AtrWRKY42-2 promoted betalain synthesis. A yeast one-hybrid assay demonstrated that AtrWRKY42-2 could bind to the AtrCYP76AD1 promoter to regulate betalain synthesis.
We declare that this manuscript is original, has not been published before, and is not currently being considered for publication elsewhere.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
RY: Writing – original draft, Writing – review & editing. TH: Validation. WS: Conceptualization. ZA: Methodology. ZL: Supervision. SL: Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the Natural Science Foundation of Fujian Province (2023J01449), Innovation Foundation of Fujian Agriculture and Forestry University(KFb22024XA), Rural Revitalization Social Service Team of Fujian Agriculture and Forestry University(11899170125).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1300522/full#supplementary-material
CK, Untreated blank control group; GA3, Gibberellin A3; P333, Paclobutrazol; BLAST, basic local alignment search tool; bp, base pair; CDD, conserved domains database; cDNA, complementary DNA; CDS, coding sequence; CYP, cytochrome P450; DODA:4,5-dopa dioxygenase extradiol; DOPA, 4,5-dihydroxy-phenylalanine; EGFP, Enhanced Green Fluorescent Protein; HMM, Hidden Markov Model; MYB, myb proto-oncogene protein; NCBI, National Center for Biotechnology Information; pI, theoretical isoelectric point; RT-qPCR, reverse transcription quantitative real-time polymerase chain reaction; VIGS, virus-induced gene silencing; WRKY, wrky protein; gDNA, genomic DNA; AtrWRKYs, Amaranth WRKY transcription factor family; TyDC:Tyrosine dehydrogenase; B5-GT, betanidin 5 glucosyltransferase; B6-GT, betanidin 6 glucosyltransferase; PCR, polymerase chain reaction; AD, pGADT7 recombinant plasmid; gDNA, Genomic DNA; cDNA, Transcriptome DNA; DAPI, DAPI staining kit; NJ, neighbor-ending method.
Alfonso, T., Tao, F., Hester, S., Nathanael, W. -H., Samuel, P. B. L. -N., Rui, G., et al. (2019). The evolution of betalain biosynthesis in Caryophyllales. New Phytol. 224, 71–85. doi: 10.1111/nph.15980
Baillo, E. H., Hanif, M. S., Guo, Y., Zhang, Z., Algam, S. A. (2020). Genome-wide Identification of WRKY transcription factor family members in sorghum (Sorghum bicolor (L.) moench). PloS One 15 (8), e0236651. doi: 10.1371/journal.pone.0236651
Barba-Espin, G., Glied-Olsen, S., Dzhanfezova, T., Joernsgaard, B., Lütken, H., Müller, R. (2018). Preharvest application of ethephon and postharvest UV-B radiation improve quality traits of beetroot (Beta vulgaris L. ssp. vulgaris) as source of colourant. BMC Plant Biol. 18, 316. doi: 10.1186/s12870-018-1556-2
Birkenbihl, R. P., Kracher, B., Ross, A., Kramer, K., Finkemeier, I., Somssich, I. E. (2018). Principles and characteristics of the Arabidopsis WRKY regulatory network during early MAMP-triggered immunity. Plant J. Cell Mol. Biol. 96 (3), 487-502. doi: 10.1111/tpj.14043
Brand, L. H., Fischer, N. M., Harter, K., Kohlbacher, O., Wanke, D. (2013). Elucidating the evolutionary conserved DNA-binding specificities of WRKY transcription factors by molecular dynamics and in vitro binding assays. Nucleic Acids Res. 41 (21), 9764-9778. doi: 10.1093/nar/gkt732
Cai, Y., Xing, J., Isun, M., Corke, H. (2006). Rapid identification of betacyanins from Amaranthus tricolor, Gomphrena globosa, and Hylocereus polyrhizus by matrix-assisted laser desorption/ionization quadrupole ion trap time-of-flight mass spectrometry (MALDI-QIT-TOF MS). J. Agric. Food Chem. 54 (18), 6520-6526. doi: 10.1021/JF0609983
Carreón-Hidalgo, J. P., Román-Guerrero, A., Navarro-Ocaña, A., Gómez-Linton, D. R., Franco-Vásquez, D. C., Franco-Vásquez, A. M., et al. (2022). Chemical characterization of yellow-orange and purple varieties of Opuntia ficus-indica fruits and thermal stability of their betalains. J. Food Sci. 88, 161-174. doi: 10.1111/1750-3841.16421
Chang, Y., Chiu, Y., Tsao, N., Chou, Y., Tan, C., Chiang, Y., et al. (2021). Author Correction: Elucidation of the core betalain biosynthesis pathway in Amaranthus tricolor. Sci. Rep. 11, 15634. doi: 10.1038/S41598-021-94665-9
Chen, C., Xie, F., Kamran, S., Hua, Q., Chen, J., Zhang, Z., et al. (2022). Genome-wide identification of WRKY gene family in pitaya reveals the involvement of hmoWRKY42 in betalain biosynthesis. Int. J. Mol. Sci. 23 (18), 10568. doi: 10.3390/IJMS231810568
Chen, X., Li, C., Wang, H., Guo, Z. (2019). WRKY transcription factors: evolution, binding, and action. Phytopathol. Res. 1, 13. doi: 10.1186/s42483-019-0022-x
Chen, Z., Guo, X., Chen, Z., Chen, W., Liu, D., Zheng, Y., et al. (2015). Genome-wide characterization of developmental stage- and tissue-specific transcription factors in wheat. BMC Genomics 16, 125. doi: 10.1186/s12864-015-1313-y
Cheng, M. (2018). Molecular mechanism of wrky transcription factor regulating betalain biosynthesis and sugar metabolism in pitaya fruit. doctorate. Dissertation, (South China Agricultural University). https://kns.cnki.net/KCMS/detail/detail.aspx?dbname=CMFD202002&filename=1018770423.nh
Cheng, M., Huang, Z., Hua, Q., Shan, W., Kuang, J., Lu, W., et al. (2017). The WRKY transcription factor HpWRKY44 regulates CytP450-like1 expression in red pitaya fruit (Hylocereus polyrhizus). Horticulture Res. 4, 17039. doi: 10.1038/hortres.2017.39
Clement, J. S., Mabry, T. J. (1996). Pigment evolution in the caryophyllales: a systematic overview*. Botanica Acta 109 (5), 360–367. doi: 10.1111/j.1438-8677.1996.tb00584.x
Erika, O. H., Jorge, W. C., Jacobo-Velázquez, D. A. (2018). Effects of UVB light, wounding stress, and storage time on the accumulation of betalains, phenolic compounds, and ascorbic acid in red prickly pear (opuntia ficus-indica cv. rojo vigor). Food Bioprocess Technol. 11 (12), 2265-2274. doi: 10.1007/s11947-018-2183-5
Eulgem, T., Rushton, P. J., Robatzek, S., Somssich, I. E. (2000). The WRKY superfamily of plant transcription factors. Trends Plant Sci. 5 (5), 199–206. doi: 10.1016/S1360-1385(00)01600-9
Fan, R., Sun, Q., Zeng, J., Zhang, X. (2020). Contribution of anthocyanin pathways to fruit flesh coloration in pitayas. BMC Plant Biol. 20, 361. doi: 10.1186/s12870-020-02566-2
Gangola, M. P., Ramadoss, B. R. (2020). WRKY transcription factors for biotic and abiotic stress tolerance in plants. Transcription Factors Abiotic Stress Tolerance Plants, 15-28. doi: 10.1016/B978-0-12-819334-1.00002-2
Gins, M., Gins, V. K., Kononkov, P. F. (2002). Change in the biochemical composition of amaranth leaves during selection for increased amaranthine content. Appl. Biochem. Microbiol. 38 (5), 474-479. doi: 10.1023/A:1019980821313
Guerrero-Rubio, M. A., Hernández-García, S., Escribano, J., Jiménez-Atiénzar, M., Cabanes, J., García-Carmona, F., et al. (2020). Betalain health-promoting effects after ingestion in Caenorhabditis elegans are mediated by DAF-16/FOXO and SKN-1/Nrf2 transcription factors. Food Chem. 330, 127228. doi: 10.1016/j.foodchem.2020.127228
Hatlestad, G. J., Akhavan, N. A., Sunnadeniya, R. M., Elam, L., Cargile, S., Hembd, A., et al. (2015). The beet Y locus encodes an anthocyanin MYB-like protein that activates the betalain red pigment pathway. Nat. Genet. 47, 92-96. doi: 10.1038/ng.3163
Hatlestad, G. J., Sunnadeniya, R. M., Akhavan, N. A., Gonzalez, A., Goldman, I. L., Mcgrath, J. M., et al. (2012). The beet R locus encodes a new cytochrome P450 required for red betalain production. Nat. Genet. 44 (7), 92-96. doi: 10.1038/ng.2297
Hu, W., Ren, Q., Chen, Y., Xu, G., Qian, Y. (2021). Genome-wide identification and analysis of WRKY gene family in maize provide insights into regulatory network in response to abiotic stresses. BMC Plant Biol. 21, 427. doi: 10.1186/S12870-021-03206-Z
Hua, Q., Chen, C., Chen, Z., Chen, P., Ma, Y., Wu, J., et al. (2016). Transcriptomic analysis reveals key genes related to betalain biosynthesis in pulp coloration of Hylocereus polyrhizus. Front. Plant Sci. 6, 1179. doi: 10.3389/fpls.2015.01179
Huang, S., Gao, Y., Liu, J., Peng, X., Niu, X., Fei, Z., et al. (2012). Genome-wide analysis of WRKY transcription factors in Solanum lycopersicum. Mol. Genet. Genomics: MGG 287 (6), 495-513. doi: 10.1007/s00438-012-0696-6
Jia, C., Wang, Z., Wang, J., Miao, H., Zhang, J., Xu, B., et al. (2022). Genome-wide analysis of the banana WRKY transcription factor gene family closely related to fruit ripening and stress. Plants 11 (5), 662. doi: 10.3390/PLANTS11050662
Jiang, J., Ma, S., Ye, N., Jiang, M., Cao, J., Zhang, J. (2017). WRKY transcription factors in plant responses to stresses. J. Integr. Plant Biol. 59 (2), 86–101. doi: 10.1111/jipb.12513
Johnson, C. S., Kolevski, B., Smyth, D. R. (2002). TRANSPARENT TESTA GLABRA2, a trichome and seed coat development gene of Arabidopsis, encodes a WRKY transcription factor. Plant Cell 14 (6), 1359–1375. doi: 10.1105/tpc.001404
Li, G., Meng, X., Zhu, M., Li, Z. (2019). Research progress of betalain in response to adverse stresses and evolutionary relationship compared with anthocyanin. Molecules 24 (17), 3078. doi: 10.3390/molecules24173078
Li, W., Pang, S., Lu, Z., Jin, B. (2020). Function and mechanism of WRKY transcription factors in abiotic stress responses of plants. Plants 9 (11), 1515. doi: 10.3390/plants9111515
Liu, S., Guo, G., Wang, W. X., Zhao, C., Peng, L., Zhang, Z., et al. (2018). Effects of exogenous auxin substances and 6-BA on seed germination and seedling growth of Amaranth. Seed 37 (12), 92–96. doi: 10.16590/j.cnki.1001-4705.2018.12.092
Liu, S., Zheng, X., Pan, J., Peng, L., Cheng, C., Wang, X., et al. (2019). RNA-sequencing analysis reveals betalains metabolism in the leaf of Amaranthus tricolor L. PloS One 14 (4), e0216001. doi: 10.1371/journal.pone.0216001
Madhunita, B., Ralf, O. (2014). WRKY transcription factors: Jack of many trades in plants. Plant Signaling Behav. 9 (2), e27700. doi: 10.4161/psb.27700
Mao, G., Meng, X., Liu, Y., Zheng, Z., Chen, Z., Zhang, S. (2011). Phosphorylation of a WRKY transcription factor by two pathogen-responsive MAPKs drives phytoalexin biosynthesis in Arabidopsis. Plant Cell 23 (4), 1639–1653. doi: 10.2307/41433414
Martins, N., Roriz, C. L., Morales, P., Barros, L., Ferreira, I. C. F. R. (2017). Coloring attributes of betalains: a key emphasis on stability and future applications. Food Funct. 8 (4), 1357-1372. doi: 10.1039/c7fo00144d
Pan, J., Peng, L., Zhao, C., Wang, W. X., Zhang, Z., Chen, Y., et al. (2018). Cloning of plant amaARF6 and its response to exogenous hormones in Amaranthus seedlings. J. Northeast Agric. Univ. 49 (5), 24–32. doi: 10.19720/j.cnki.issn.1005-9369.2018.05.004
Parveen, M., Ray, S., Chatterjee, N. C. (2018). Detection and diversity pattern of amaranth Cultivars originated in diverse region of Indian subcontinent. Int. J. Pharma Bio Sci. 9 (3). doi: 10.22376/ijpbs.2018.9.3.b66-71
Peng, L., Wang, Y., Sun, X., Wang, X., Liu, S. (2019). Expression and functional analysis of AmMYB2 related to betalain metabolism of Amaranthus tricolor. Acta Hortic. Sin. 46 (3), 473-485. doi: 10.16420/j.issn.0513-353x.2018-0572
Polturak, G., Aharoni, A. (2018). “La Vie En Rose”: Biosynthesis, sources, and applications of betalain pigments. Mol. Plant 11 (1), 7-22. doi: 10.1016/j.molp.2017.10.008
Polturak, G., Breitel, D., Grossman, N., Sarrion-Perdigones, A., Weithorn, E., Pliner, M., et al. (2016). Elucidation of the first committed step in betalain biosynthesis enables the heterologous engineering of betalain pigments in plants. New Phytol. 210, 269-283. doi: 10.1111/nph.13796
Polturak, G., Heinig, U., Grossman, N., Battat, M., Leshkowitz, D., Malitsky, S., et al. (2018). Transcriptome and metabolic profiling provides insights into betalain biosynthesis and evolution in mirabilis jalapa. Mol. Plant 11, 189-204. doi: 10.1016/j.molp.2017.12.002
Preczenhak, A. P., Orsi, B., Lima, G. P. P., Tezotto-Uliana, J. V., Minatel, I. O., Kluge, R. A. (2019). Cysteine enhances the content of betalains and polyphenols in fresh-cut red beet. Food Chem. 286, 600-607. doi: 10.1016/j.foodchem.2019.02.040
Sakuta, M., Tanaka, A., Iwase, K., Miyasaka, M., Ichiki, S., Hatai, M., et al. (2021). Anthocyanin synthesis potential in betalain-producing Caryophyllales plants. J. Plant Res. 134 (6), 1335-1349. doi: 10.1007/S10265-021-01341-0
Sheehan, H., Feng, T., Walker-Hale, N., Lopez-Nieves, S., Pucker, B., Guo, R., et al. (2020). Evolution of l-DOPA 4,5-dioxygenase activity allows for recurrent specialisation to betalain pigmentation in Caryophyllales. New Phytol. 227 (3), 914-929. doi: 10.1111/nph.16089
Sokolova, D. V., Shvachko, N. A., Mikhailova, A. S., Popov, V. S. (2022). Betalain content and morphological characteristics of table beet accessions: their interplay with abiotic factors. Agronomy 12 (5), 1033. doi: 10.3390/AGRONOMY12051033
Strack, D., Vogt, T., Schliemann, W. (2003). Recent advances in betalain research. Phytochemistry 62 (3), 247-269. doi: 10.1002/chin.200319225
Su, M., Zuo, W., Wang, Y., Liu, W., Zhang, Z., Wang, N., et al. (2022). The WKRY transcription factor MdWRKY75 regulates anthocyanins accumulation in apples (Malus domestica). Funct. Plant Biol. FPB 49 (9), 799-809. doi: 10.1071/FP21146
Sunnadeniya, R., Bean, A., Brown, M., Akhavan, N., Hatlestad, G., Gonzalez, A., et al. (2016). Tyrosine hydroxylation in betalain pigment biosynthesis is performed by cytochrome P450 enzymes in beets (Beta vulgaris). PloS One 11 (2), e0149417. doi: 10.1371/journal.pone.0149417
Wang, K., Liu, Y., Song, Z., Wang, D., Qiu, W., et al. (2019). Chelator complexes enhanced Amaranthus hypochondriacus L. phytoremediation efficiency in Cd-contaminated soils. Chemosphere 237, 124480. doi: 10.1016/j.chemosphere.2019.124480
Wang, L., Zhu, W., Fang, L., Sun, X., Su, L., Liang, Z., et al. (2014). Genome-wide identification of WRKY family genes and their response to cold stress in Vitis vinifera. BMC Plant Biol. 14, 103. doi: 10.1186/1471-2229-14-103
Wu, G., Li, Z., Cao, H., Wang, J. (2019). Genome-wide identification and expression analysis of the WRKY genes in sugar beet ( Beta vulgaris L.) under alkaline stress. PeerJ 7, e7817. doi: 10.7717/peerj.7817
Xiao, F., Zheng, Y., Chen, J., Zhao, C., Chen, H., Wang, L., et al. (2021). Selection and validation of reference genes in all-red Amaranth (;Amaranthus tricolor;L.) seedlings under different culture conditions. J. Hortic. Sci. Biotechnol. 96 (5), 604-613. doi: 10.1080/14620316.2021.1879686
Xie, L., Liu, S., Bai, B. Y., Liu, Y., Lin, M., Cai, S., et al. (2016). Cloning and expression analysis of betalain-related transcription factor gene amMYB1 in amaranthus tricolor L. Acta Botanica Boreali-Occidentalia Sin. 36 (6), 1080-1090. doi: 10.1007/s10529-015-2021-z
Zhang, L. (2020). Transcription factors related to betalain biosynthesis in pitaya screening and functional analyses of MYB and WRKY. Master. Thesis, (South China Agricultural University).
Zhang, L., Chen, C., Xie, F., Hua, Q., Zhang, Z., Zhang, R., et al. (2021). A novel WRKY transcription factor HmoWRKY40 associated with betalain biosynthesis in pitaya (Hylocereus monacanthus) through regulating HmoCYP76AD1. Int. J. Mol. Sci. 22 (4), 2171. doi: 10.3390/IJMS22042171
Zhang, M., Chen, Y., Nie, L., Jin, X., Liao, W., Zhao, S., et al. (2018). Transcriptome-wide identification and screening of WRKY factors involved in the regulation of taxol biosynthesis in Taxus chinensis. Sci. Rep. 8, 5197. doi: 10.1038/s41598-018-23558-1
Zhao, C., Fang, X., Pan, J., Peng, L., Lai, Z., Liu, S. (2019). Cloning and expression analysis of atGAI gene in amaranthus tricolor L. Acta Botanica Boreali-Occidentalia Sin. 39 (2), 199-209. doi: 10.7606/j.issn.1000-4025.2019.02.0199
Zheng, H., Hang, Y., Wang, X., Li, X., Hu, T., Tian, Y., et al. (2022). Research progress of WRKY transcription factor family in medicinal plants. Plant Physiol. J. 58 (6), 1055–1067. doi: 10.13592/j.cnki.ppj.300030
Keywords: Amaranthus tricolor L., betalain, AtrWRKY42-2 gene, relative expression level of WRKY TFs, functional analysis
Citation: Yang R, Huang T, Song W, An Z, Lai Z and Liu S (2023) Identification of WRKY gene family members in amaranth based on a transcriptome database and functional analysis of AtrWRKY42-2 in betalain metabolism. Front. Plant Sci. 14:1300522. doi: 10.3389/fpls.2023.1300522
Received: 23 September 2023; Accepted: 16 November 2023;
Published: 05 December 2023.
Edited by:
Clive Lo, The University of Hong Kong, Hong Kong SAR, ChinaReviewed by:
Jianjun Chen, University of Florida, United StatesCopyright © 2023 Yang, Huang, Song, An, Lai and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shengcai Liu, MTIxNTY5ODkwMEBxcS5jb20=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.