
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 05 December 2023
Sec. Plant Bioinformatics
Volume 14 - 2023 | https://doi.org/10.3389/fpls.2023.1297228
This article is part of the Research TopicCrop Improvement by Omics and BioinformaticsView all 17 articles
Osmotic stress poses a threat to the production and quality of crops. Whirly transcription factors have been investigated to enhance stress tolerance. In this study, a total of 18 Whirly genes were identified from six Triticeae species, which were classified into Whirly1 and Whirly2. The exon–intron structure, conserved motif, chromosomal location, collinearity, and regulatory network of Whirly genes were also analyzed. Real-time PCR results indicated that TaWHY1 genes exhibited higher expression levels in leaf sheaths and leaves during the seedling stage, while TaWHY2 genes were predominantly expressed in roots. Under PEG stress, the expression levels of TaWHY1-7A, TaWHY2-6A, TaWHY2-6B, and TaWHY2-6D were increased, TaWHY1-7D was reduced, and TaWHY1-4A had no significant change. All TaWHY genes were significantly up-regulated in response to NaCl stress treatment. In addition, TaWHY1-7A and TaWHY1-7D mainly enhanced the tolerance to oxidative stress in yeast cells. TaWHY2s mainly improved NaCl stress tolerance and were sensitive to oxidative stress in yeast cells. All TaWHYs slightly improved the yeast tolerance to d-sorbitol stress. The heterologous expression of TaWHY1-7D greatly improved drought and salt tolerance in transgenic Arabidopsis. In conclusion, these results provide the foundation for further functional study of Whirly genes aimed at improving osmotic stress tolerance in wheat.
Wheat (Triticum aestivum L.) is one of the most important staple crops worldwide and a major source of calories for the expanding world population. As a sessile organism, wheat has to suffer from a variety of adverse conditions during the growth and development stages, such as drought and salinization, which contribute to a great reduction in the overall wheat yield and quality (Gupta et al., 2020). Therefore, mining stress-resistant genes and developing improved varieties are the most important strategies to improve wheat yield and quality.
Whirly (WHY) proteins are plant-specific transcription factors binding to single-stranded DNA (ssDNA) to modulate growth and defense responses and located in the chloroplasts, mitochondria, and nucleus (Desveaux et al., 2005; Krupinska et al., 2022; Taylor et al., 2022). Whirly domain consists of four structural topologies, which are characterized by two antiparallel four-stranded β-sheets stabilized by a C-terminal helix-loop-helix motif (Desveaux et al., 2005; Cappadocia et al., 2013; Taylor et al., 2022). Due to the structural similarity with “whirligig,” Whirly transcription factors are named Whirly (Desveaux et al., 2005). The conserved “KGKAAL” motif in the Whirly domains exists extensively in higher plants, which participate in binding to ssDNA and hexamerization of the tetramers forming hollow sphere structures of 12 nm in diameter (Desveaux et al., 2002; Cappadocia et al., 2012). Additionally, Whirly proteins contain a conserved cysteine residue, which might play a vital role in the formation of disulfide bridges between two Whirly proteins (Foyer et al., 2014).
Whirly was initially identified as p24/PBF-2 protein that binds to the elicitor response element (ERE) on the promoter of the pathogen response gene PR-10a in potato (Desveaux et al., 2000). In Arabidopsis, AtWHY1 is targeted to chloroplasts and nucleus (Krause et al., 2005; Ren et al., 2017), which plays a crucial role in regulating telomere length homeostasis (Yoo et al., 2007), maintaining the stability of plastid genome (Marechal et al., 2009), modulating reactive oxygen species (ROS) homeostasis, controlling leaf senescence (Lin et al., 2019), and responding to salicylic acid (SA)-dependent disease resistance (Desveaux et al., 2004). AtWHY1 protein represses the expression of WRKY53 and delays leaf senescence in Arabidopsis (Miao et al., 2013). AtWHY2 is located in the mitochondria and nucleus (Krause et al., 2005; Golin et al., 2020). Overexpression of AtWHY2 leads to mitochondrial dysfunction, early accumulation of senescence-related transcripts (Marechal et al., 2008; Golin et al., 2020), slower growth of pollen tubes, elevation of mtDNA content, and ROS levels in pollen (Cai et al., 2015). AtWHY3 is targeted to chloroplasts, mitochondria, and nucleus in compensation for the lack or mutation of AtWHY1 and AtWHY2 (Krause et al., 2005; Golin et al., 2020). In tomato (Solanum lycopersicum), SlWHY1 and SlWHY2 can be induced by drought and salt stresses (Akbudak and Filiz, 2019). Overexpression of SlWHY1 enhances heat and cold stress tolerance and reduces ROS levels in tomato (Zhuang et al., 2020a; Zhuang et al., 2020b), and SlWHY2 can maintain mitochondrial function under drought stress through interacting with SlRECA2 in tomato (Meng et al., 2020). MeWHY1/2/3 can interact with MeCIPK23 to activate abscisic acid (ABA) biosynthesis and regulate drought resistance in cassava (Manihot esculenta) (Yan et al., 2020). In barley (Hordeum vulgare L.), overexpression of HvWHY1 delays drought-induced leaf senescence (Manh et al., 2023).
Whirly genes have been identified in various plant species, such as Arabidopsis, strawberry, tomato, cassava, and barley (Desveaux et al., 2005; Janack et al., 2016; Yan et al., 2020; Hu and Shu, 2021), however, a comprehensive genome-wide analysis of Whirly genes in Triticeae species has not been investigated. In this study, a genome-wide analysis of Whirly genes was performed in Triticeae species including Triticum aestivum, Triticum urartu, Triticum dicoccoides, Aegilops tauschii, Hordeum vulgare, and Secale cereale to characterize their sequences, gene structures, evolutionary relationships, expression patterns, and stress tolerance under osmotic stress. These results will provide a valuable foundation for further functional investigations of Whirly genes in response to osmotic stress.
Bread wheat cv. Chinese Spring preserved in our laboratory was used in this study, and the sterilized bread wheat seeds were soaked with ddH2O in dark and 4°C condition overnight, then cultured on filter paper wetted with ddH2O in a culture room at 25/18°C with 16-h light/8-h dark for 1 week. Next, 7-day-old bread wheat seedlings with uniform leaf size and root length were selected for subsequent experiments. For drought and salt stress treatments, 7-day-old bread wheat seedlings were cultured under 20% PEG6000 (w/v) and 300 mM NaCl treatments, respectively. In each treatment, the root, leaf sheath, and leaf tissues were collected at 0 h, 1 h, and 6 h, then frozen in liquid nitrogen and stored at −80°C for further investigation.
The protein sequences of Triticum aestivum (Chinese Spring, IWGSC.52), Triticum urartu (Tu 2.0), Triticum dicoccoides (WEWSeq_v1.0), Aegilops tauschii (Aet_v4.0), Hordeum vulgare (IBSC_v2), Brachypodium distachyon (IBI_v3.0), Oryza sativa (Japonica, IRGSP 1.0), Zea mays (B73 RefGen_v4), Solanum lycopersicum (SL3.0), and Arabidopsis thaliana (TAIR10) were downloaded from EnsemblPlants database (http://plants.ensembl.org/index.html). The protein sequence data of Secale cereale (Weining v1) was acquired from the China National Center for Bioinformation (CNCB-NGDC Members and Partners, 2022). To identify candidate Whirly protein sequences, the Hidden Markov Model (HMM) profile of the typical Whirly transcription factor domain (PF08536) (Mistry et al., 2021) was used as a query to search against the protein sequences of these 11 plant species with TBtools software (Chen et al., 2020a). Next, the Pfam (https://www.ebi.ac.uk/interpro/) (Mistry et al., 2021) and SMART (Simple Modular Architecture Research Tool, http://smart.embl.de/) (Letunic et al., 2021) online services were used to further confirm the putative Whirly proteins. The protein length, molecular weight, isoelectric point (pI), and grand average of hydropathy (GRAVY) of the Whirly proteins were analyzed by WheatOmics 1.0 (Ma et al., 2021).
Multiple sequence alignment of Whirly amino acid sequences was performed with ClustalW using the default options in MEGA 11 (Tamura et al., 2021) and visualized by ESPript 3.0 (Gouet et al., 2003). The phylogenetic tree was constructed by using the neighbor-joining (NJ) method with 1,000 bootstrap replicates in MEGA 11 software (Tamura et al., 2021) and visualized by Evolview service (Subramanian et al., 2019).
The exon–intron structures of Whirly genes were analyzed based on TGT (Triticeae-Gene Tribe) (Chen et al., 2020b). The conserved motifs and domains of Whirly family proteins were annotated using the MEME program (Bailey et al., 2009) and SMART website (Letunic et al., 2021) and visualized by TBtools (Chen et al., 2020a). The Swiss-Model program was used to predict the three-dimensional (3D) structure of Whirly proteins (Waterhouse et al., 2018).
The chromosome localization, micro-collinearity, and paralogous/orthologous gene pairs of Whirly genes were identified by using Triticeae-Gene Tribe (TGT) (Chen et al., 2020b). The gene duplication events were determined by Multiple Collinear Scanning Toolkits (MCScanX) (Wang et al., 2012). TBtools was used to calculate the nonsynonymous rate (Ka), synonymous rate (Ks), and the nonsynonymous and synonymous substitution ratio (Ka/Ks) values of the paralogous gene pair with the Nei–Gojobori (NG) method (Chen et al., 2020a).
The upstream transcription factors and downstream target genes of TaWHYs were predicted by using the wheat integrative gene regulatory network (wGRN) (Chen et al., 2023). Protein–protein interactions (PPIs) were analyzed using the STRING database (Von Mering et al., 2003) and WheatOmics 1.0 (Ma et al., 2021).
To investigate the gene expression patterns in bread wheat under drought stress, bread wheat cv. Chinese Spring was planted in a growth chamber under a photoperiod of 16 h/8 h (light/dark). For drought stress, the seedlings were subjected to water-deficit condition during the seedling stage. The leaf tissues were harvested after 0 days, 2 days, 6 days, and 10 days of treatment, and the total RNA of all the collected samples was extracted. A Nanodrop2000 spectrophotometer was used to determine the quantity and quality of the RNA. A total of 12 bread wheat samples (three biological replicates were conducted for each treatment) were sequenced at Majorbio Bio-Pharm Technology Co. Ltd. (Shanghai, China), and paired-end sequencing was performed with an Illumina Novaseq 6000. The transcriptome data have been submitted to NCBI (BioProject ID: PRJNA1003680).
The transcriptome data of different bread wheat tissues (root and shoot) were obtained from NCBI SRA (DRX002485, DRX002486, DRX002487, DRX002491, DRX002492, and DRX002493). The transcriptome data SRX9781249, SRX9781250, SRX9781251, SRX9781252, SRX9781253, SRX9781254, SRX9781255, SRX9781256, SRX9781257, SRX9781258, SRX9781259, and SRX9781260 were used to analyze the gene expression profiles under NaCl stress in leaves during bread wheat seedling stage.
Real-time PCR was performed to detect the expression pattern of Whirly genes according to a previous study (Liu et al., 2022). The total RNA was isolated using RNApure Plant Kit (CWBIO), and the first-strand cDNA was synthesized from 1 μg of total RNA using Prescript III RT ProMix (CISTRO). The real-time PCR was performed using gene-specific primers (Supplementary Table S1) with 2× Ultra SYBR Green qPCR Mix (CISTRO), and the TaActin gene was selected as a reference control. The real-time PCR cycling parameters were 95°C for 30 s, followed by 45 cycles at 95°C for 5 s and 60°C for 30 s, with a melting curve analysis. All reactions were performed on three technical and biological replicates. The relative expression levels of target genes were calculated using the 2−△△CT method (Livak and Schmittgen, 2001).
The coding sequences (CDS) of Whirly genes were cloned into a pGADT7 vector using the ClonExpress II One Step Cloning Kit (Vazyme, Nanjing), then transformed into Saccharomyces cerevisiae (S. cerevisiae) BY4741 or its stress-sensitive mutant BY4741 (Δhog1). The primers are shown in Supplementary Table S1. For osmotic and oxidative stress, the yeast cells Δhog1 carrying the recombinant vector pGADT7-TaWHY2-6A/TaWHY2-6B/TaWHY2-6D/TaWHY1-7A/TaWHY1-7D were cultured in YPD liquid medium (1% yeast extract, 2% peptone, and 2% glucose) at 30°C until density reached an OD600 of 1.0, then serially diluted (10−1, 10−2, 10−3, 10−4) with ddH2O. The cells were spotted onto YPD medium plates (1% yeast extract, 2% peptone, 2% glucose, and 2% agar) containing 1.2 M d-sorbitol, 0.4 M NaCl, or 4.0 mM H2O2 and cultured at 30°C for 3–5 days. The wild-type yeast cells BY4741 and stress-sensitive mutant Δhog1 carrying the empty vector pGADT7 were used as positive and negative controls, respectively.
The coding sequences of TaWHY1-7D were cloned into the pCAMBIA3301-GFP vector, then transformed into Agrobacterium tumefaciens EHA105, and generated 35S:TaWHY1-7D transgenic Arabidopsis lines via the floral dip method. The primers are shown in Supplementary Table S1. The transgenic Arabidopsis lines were selected via spraying 0.5‰ Basta solution. For drought and salt tolerance assays, WT and 35S:TaWHY1-7D transgenic Arabidopsis were treated with drought (water-deficit) and 500 Mm NaCl conditions.
A total of 29 Whirly genes were identified from the protein sequences of 11 plant species via a hidden Markov model (HMM) search. In total, 24 Whirly genes were identified from nine monocotyledon species, comprising six Triticeae species (T. aestivum (6), T. urartu (2), T. dicoccoides (4), Ae. tauschii (2), H. vulgare (2), and S. cereal (2)) and three other monocotyledon species (B. distachyon (2), O. sativa (2), and Z. mays (2)), while five Whirly genes were identified in two dicotyledon species, including S. lycopersicum (2) and A. thaliana (3) (Figure 1A; Supplementary Table S2). To further confirm the reliability of the identified Whirly genes, the expression of Whirly genes was analyzed in T. urartu, T. dicoccoides, S. cereale, H. vulgare, and T. aestivum based on previous published transcriptomic data (Supplementary Table S3). The length of the identified 29 Whirly proteins varied from 223 (HvWHY2-6H) to 286 (ZmWHY1) amino acid residues, with the molecular weights ranging from 24.24 to 31.71 kDa. The pI values ranged from 8.84 (TdWHY1-4A) to 10.81 (SlWHY2), with the calculated grand average of hydrophilic index (GRAVY) varying from −0.207 (AtWHY1) to −0.459 (TaWHY1-4A), suggesting that these 29 Whirly genes encoded highly hydrophilic proteins (Supplementary Table S2).
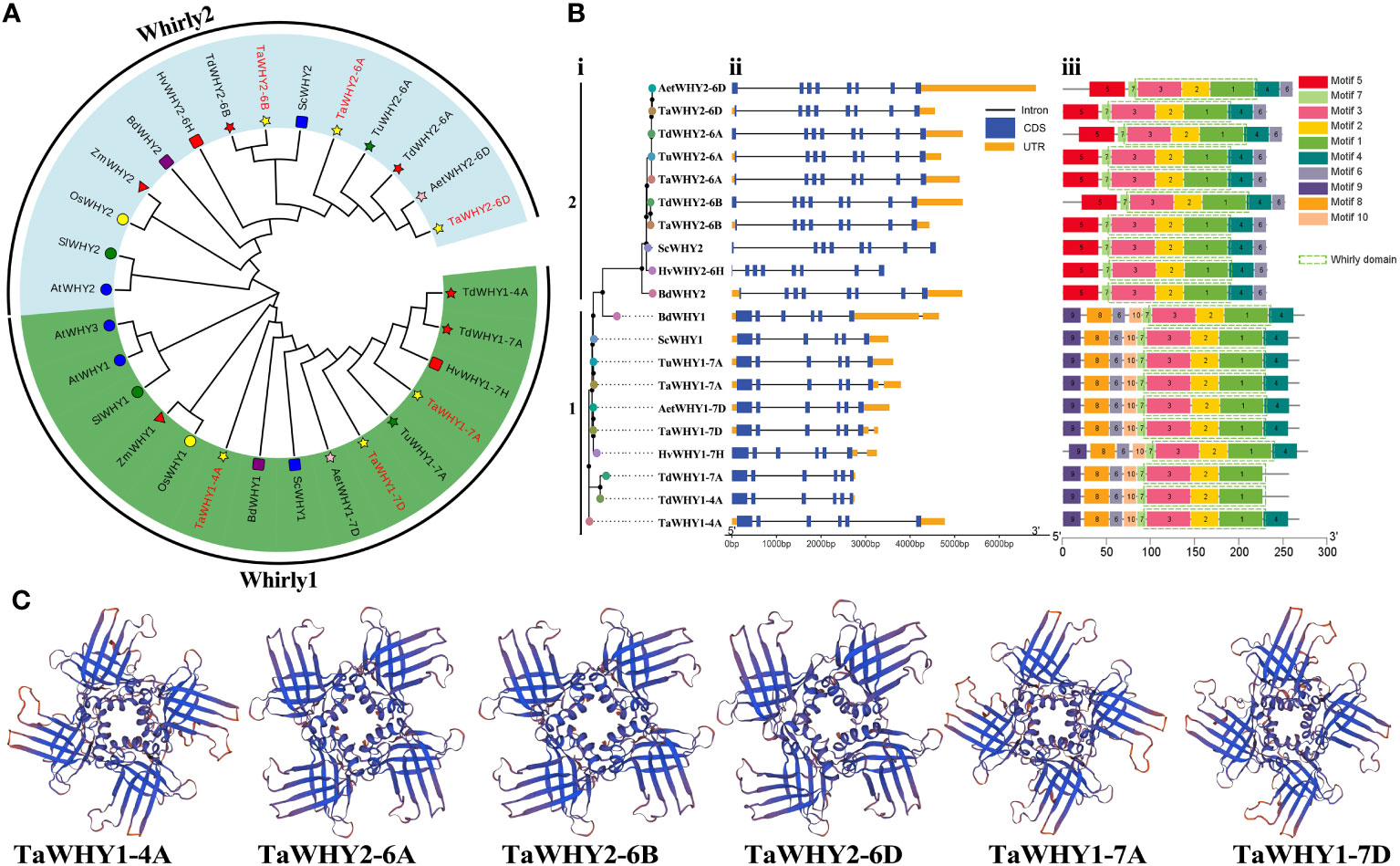
Figure 1 The neighbor-joining (NJ) phylogenetic tree (A), gene structures (B), and 3D structures (C) of Whirly proteins. (A) The tree was constructed using Whirly protein sequences from T. aestivum (Ta), T. urartu (Tu), T. dicoccoides (Td), Ae. tauschii (Aet), H. vulgare (Hv) and S. cereal (Sc), B. distachyon (Bd), O. sativa (Os) and Z. mays (Zm), S. lycopersicum (Sl), and A. thaliana (At) with bootstrap values of 1,000 replicates. Different groups of Whirly proteins are marked by different colors. (B) Phylogenetic classification (i), exon–intron structure (ii), and conserved domain (iii) analyses of Whirly genes in Triticeae species. (C) The Swiss Model program was used to predict the three-dimensional (3D) structure of the Whirly proteins.
To elucidate the evolutionary relationship of Whirly genes, a phylogenetic tree was constructed using these 29 Whirly proteins (Figure 1A). According to the results, Whirly genes were classified into two categories, named group 1 (Whirly1) and group 2 (Whirly2). Bread wheat T. aestivum (AABBDD, hexaploid) has undergone two rounds of natural hybridization events (Levy and Feldman, 2022). Thus, the number of gene family members in T. aestivum (AABBDD) is approximately 1.5- and 3-fold than that in T. dicoccoides (AABB, tetraploid) and other diploid Triticeae species, respectively. Consistently, three Whirly1 or Whirly2 genes were found in T. aestivum, while T. dicoccoides and other diploid Triticeae included two and one Whirly1 or Whirly2 gene, respectively (Figure 1A; Supplementary Table S2).
To investigate the functional divergence of Whirly genes, the exon–intron structures, conserved motifs, and 3D structures of Whirly genes were analyzed in Triticeae species (Figure 1; Supplementary Figure S1). The results revealed that Whirly1 and Whirly2 genes contained six and eight exons in the Triticeae species, respectively. The conserved motif analysis showed that all Whirly proteins contained the Whirly transcription factor domain (PF08536), which consisted of motifs 1, 2, 3, and 7. These also confirmed the reliability of the identified Whirly gene family members. Motif 3 contained the conserved “KGKAAL” sequence, which participated in binding to ssDNA (Supplementary Figure S1) (Desveaux et al., 2002; Cappadocia et al., 2012). Almost all Whirly proteins contained motif 4, except for TdWHY1-4A and TdWHY1-7A, which lacked a portion of the amino acid sequences of motif 4 (Supplementary Figure S1). Motifs 8, 9, and 10 were present in group 1 members, while they were absent in group 2 members. Motif 5 was unique to group 2 members. In addition, all TaWHY proteins contained two anti-parallel four-stranded β-sheets that extend like blades from an α-helical core (Figure 1C), which were consistent with its “whirligig” structure (Desveaux et al., 2005).
The distribution of Whirly genes on the chromosome in six Triticeae species (T. aestivum, T. urartu, T. dicoccoides, Ae. tauschii, H. vulgare, and S. cereal), three other monocotyledon species (B. distachyon, O. sativa, and Z. mays), and two dicotyledon species (S. lycopersicum and A. thaliana) are shown in Supplementary Table S2. In T. aestivum (AABBDD, hexaploid), Whirly1 genes were distributed on chromosomes 4A (TaWHY1-4A), 7A (TaWHY1-7A), and 7D (TaWHY1-7D). Whirly2 genes had three copies in its subgenomes A, B, and D, i.e., TaWHY2-6A, TaWHY2-6B, and TaWHY2-6D were distributed on chromosomes 6A, 6B, and 6D, respectively (Figure 2). Similarly, TdWHY1-4A, TdWHY2-6A, TdWHY2-6B, and TdWHY1-7A were located on chromosomes 4A, 6A, 6B, and 7A in T. dicoccoides (AABB, tetraploid), respectively. AetWHY2-6D and AetWHY1-7D were distributed on chromosomes 6D and 7D in Ae. tauschii (DD, diploid), respectively. TuWHY2-6A and TuWHY1-7A were located on chromosomes 6A and 7A in T. urartu (AA, diploid), respectively. HvWHY2-6H and HvWHY1-7H were located on chromosomes 6H and 7H in H. vulgare (HH, diploid), respectively. In S. cereale (RR, diploid), ScWHY1 and ScWHY2 were distributed on chromosomes 1R and 6R, respectively. Interestingly, the orthologous genes of TaWHY1-4A were not distributed on chromosomes 4A in T. urartu and 4H in H. vulgare, whereas TdWHY1-4A existed on chromosome 4A of T. dicoccoides (Supplementary Table S2). This result suggested that the expansion events of Whirly genes occurred through hybridization and polyploidization.
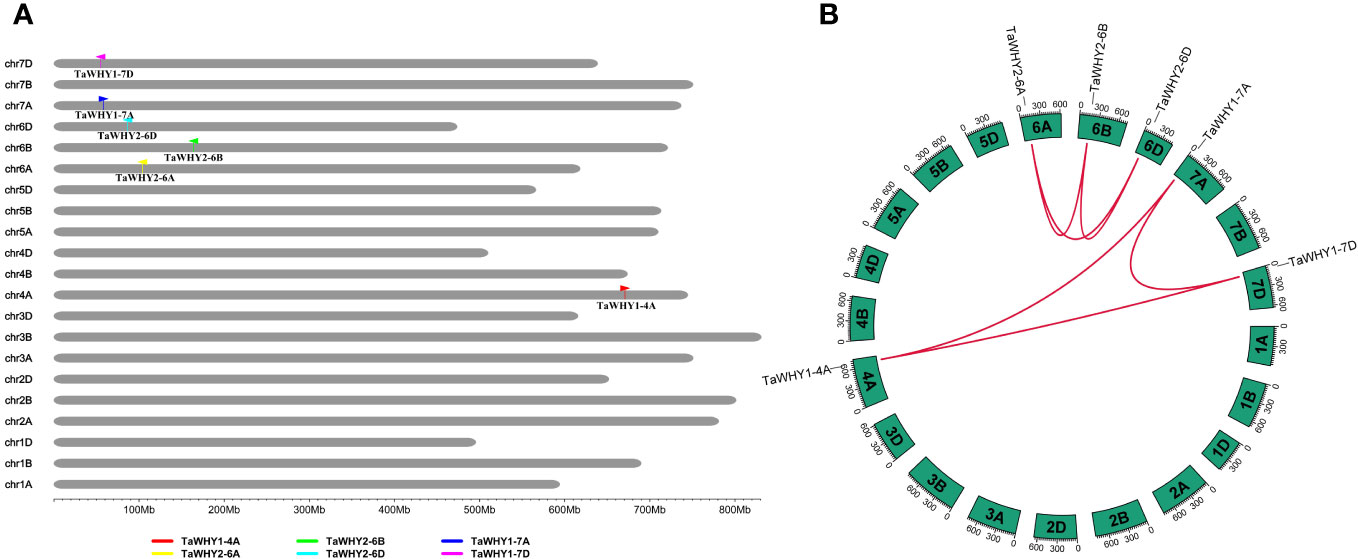
Figure 2 Chromosomal localizations (A) and syntenic relationships (B) among TaWHY genes in T. aestivum. (B) Red lines in the highlight indicate the syntenic TaWHY gene pairs.
To further investigate the evolutionary process of TaWHYs, gene duplication, and micro-collinearity analyses of the Whirly genes were performed (Figure 3; Supplementary Table S4). A total of six paralogous gene pairs of TaWHYs (TaWHY1-4A/TaWHY1-7A/TaWHY1-7D, and TaWHY2-6A/TaWHY2-6B/TaWHY2-6D) were identified in bread wheat genome and expanded by whole-genome duplication (WGD) or segmental duplication events (Figure 2B; Supplementary Table S4). The Ka/Ks values of paralogous gene pairs were all less than 1, indicating that TaWHY genes underwent purifying selection to avoid functional divergence (Supplementary Table S4). Micro-collinearity analysis contributes to the investigation of the inheritance and variation of specific genes in local regions and detecting the origin of specific genes during the hybridization and polyploidization process (Chen et al., 2020b). To explore the origin of Whirly genes in Triticeae species, TaWHY1-4A, TaWHY2-6A, and TaWHY1-7A were used as query genes to analyze the micro-collinearity by TGT (Figure 3). The homologous genes of TaWHY2-6A were detected in the collinearity regions of T. urartu, Ae. tauschii, subgenomes A and B of T. dicoccoides, and subgenomes B and D of T. aestivum, suggesting that the Whirly2 genes and their adjacent genes in the collinearity regions were relatively conserved during evolutionary processes in Triticeae species. However, no homologous genes of TaWHY1-4A and TaWHY1-7A were found in the collinearity regions of subgenome B of T. dicoccoides, and subgenome B of T. aestivum. In addition, the homologous genes of TaWHY1-4A were present in the collinearity regions on chromosome 7A of T. urartu, and 7D of Ae. tauschii, and 7D of T. aestivum, but absent on chromosome 4 of T. urartu, suggesting that TaWHY1-4A and TdWHY1-4A might originate from TuWHY1-7A or AetWHY1-7D.
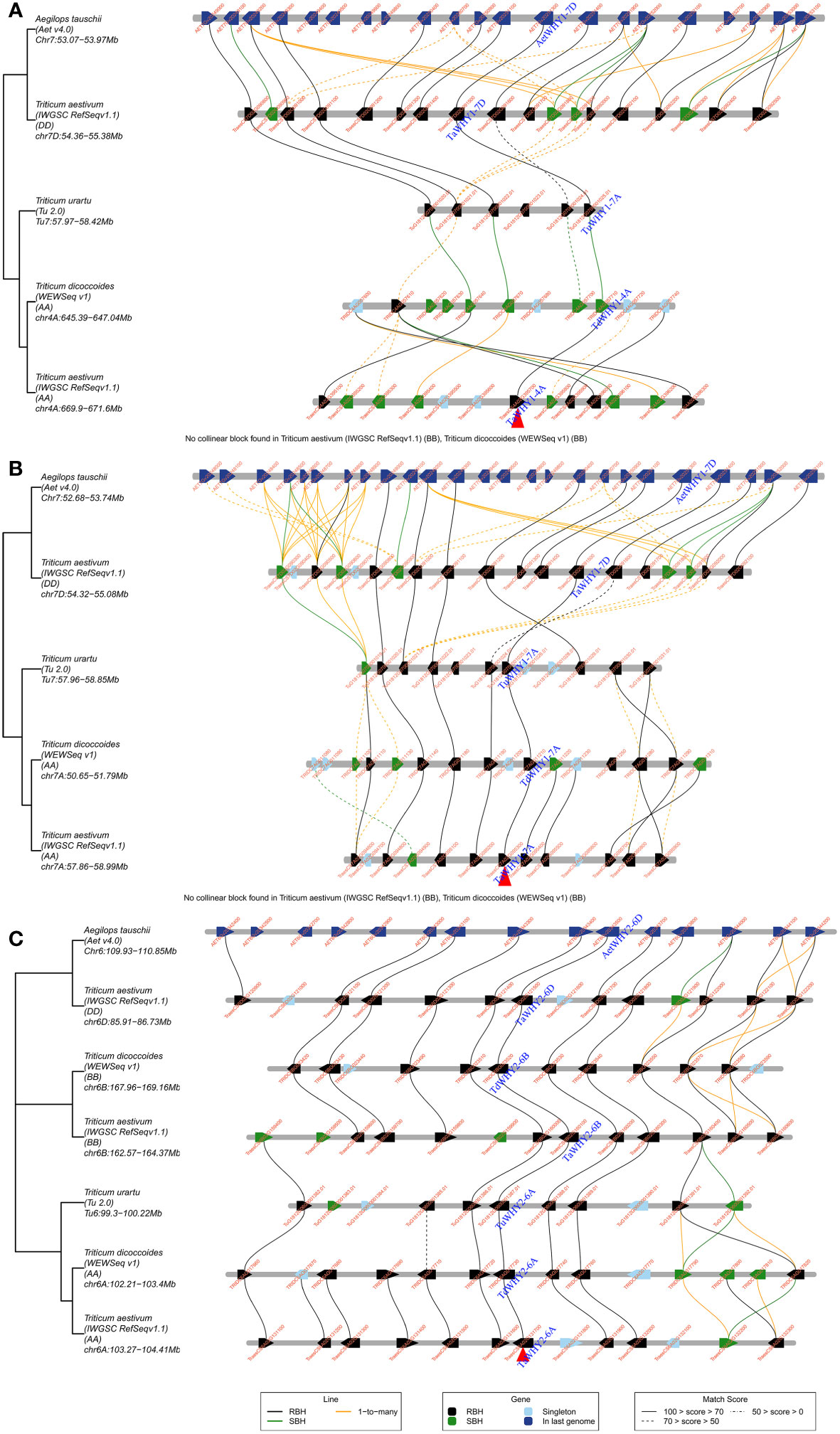
Figure 3 Micro-collinearity analysis of the Whirly gene in Triticeae species. TaWHY1-4A (A), TaWHY1-7A (B), and TaWHY2-6A (C) were used as the query gene, respectively.
To insight into the biological function of TaWHY genes, the transcriptome data and real-time PCR were used to determine the expression patterns of six TaWHY genes in different tissues (leaves, leaf sheaths, and roots during bread wheat seedling stage) and in response to osmotic (drought and salt) stress (Figures 4, 5). The analysis of the transcriptome data revealed that the TaWHY1 genes (TaWHY1-4A, TaWHY1-7A, and TaWHY1-7D) exhibited the highest expression levels in leaves, and the TaWHY2 genes (TaWHY2-6A, TaWHY2-6B, and TaWHY2-6D) showed the highest expression levels in roots (Figure 4A). Consistently, the real-time PCR results showed that TaWHY1 genes (TaWHY1-4A, TaWHY1-7A, and TaWHY1-7D) were highest expressed in leaf sheaths, followed by leaves, and roots during the bread wheat seedling stage. TaWHY2 genes (TaWHY2-6A, TaWHY2-6B, and TaWHY2-6D) exhibited the highest expression level in roots, followed by leaf sheaths, and finally in leaves (Figure 4B).
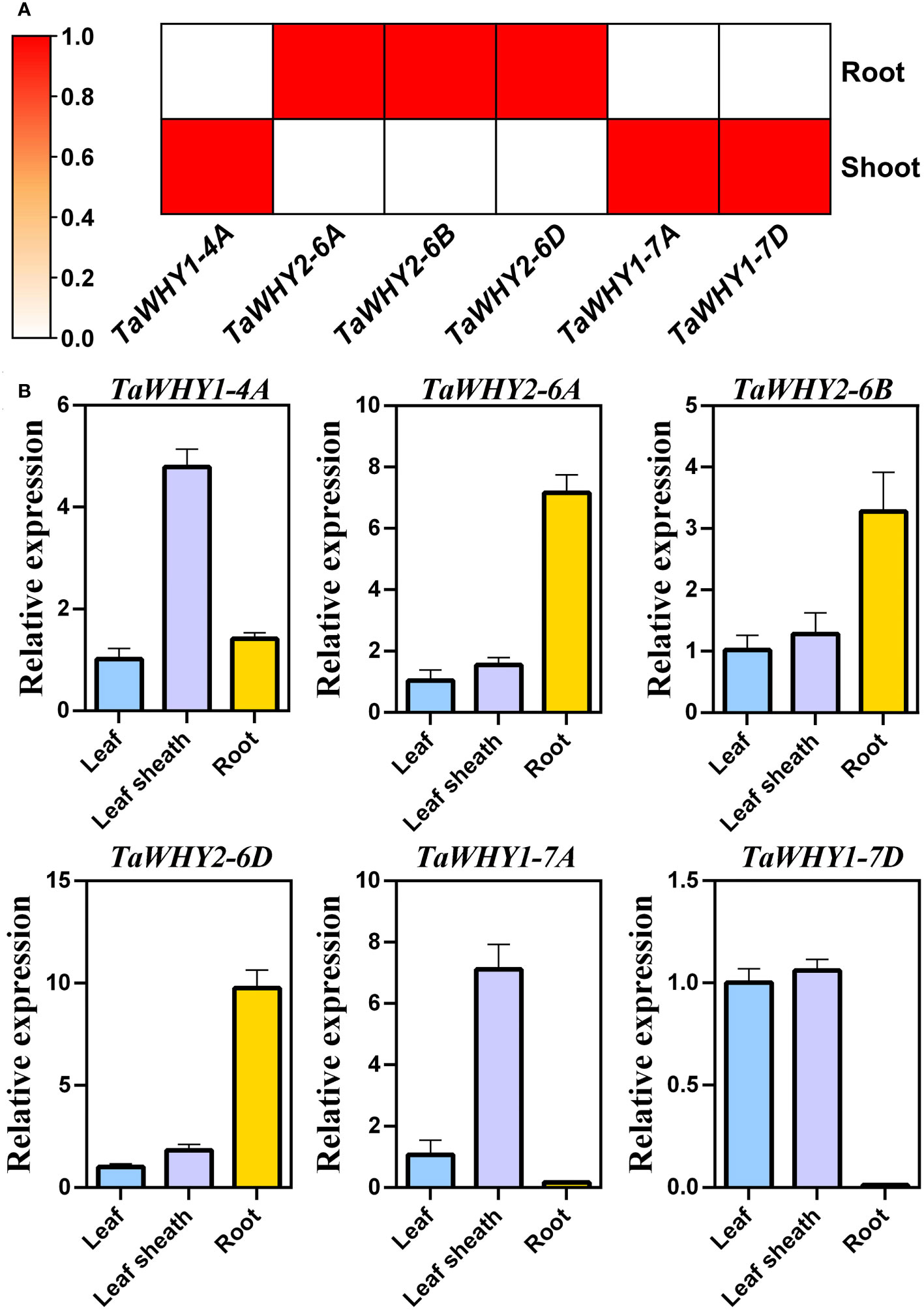
Figure 4 Expression pattern analysis of TaWHYs in different tissues. (A) The expression levels of TaWHY genes in root and shoot were determined through RNA-seq analysis. Fragments per kilobase of exon per million mapped fragments (FPKM) values were used to measure the expression levels of genes. (B) The expression levels of TaWHY genes in the root, leaf sheath, and leaf during the bread wheat seedling stage were determined by real-time PCR. The expression level of the bread wheat actin gene was used as the reference control to standardize the RNA samples for each reaction. Data represent the mean ± SD of three replicates.
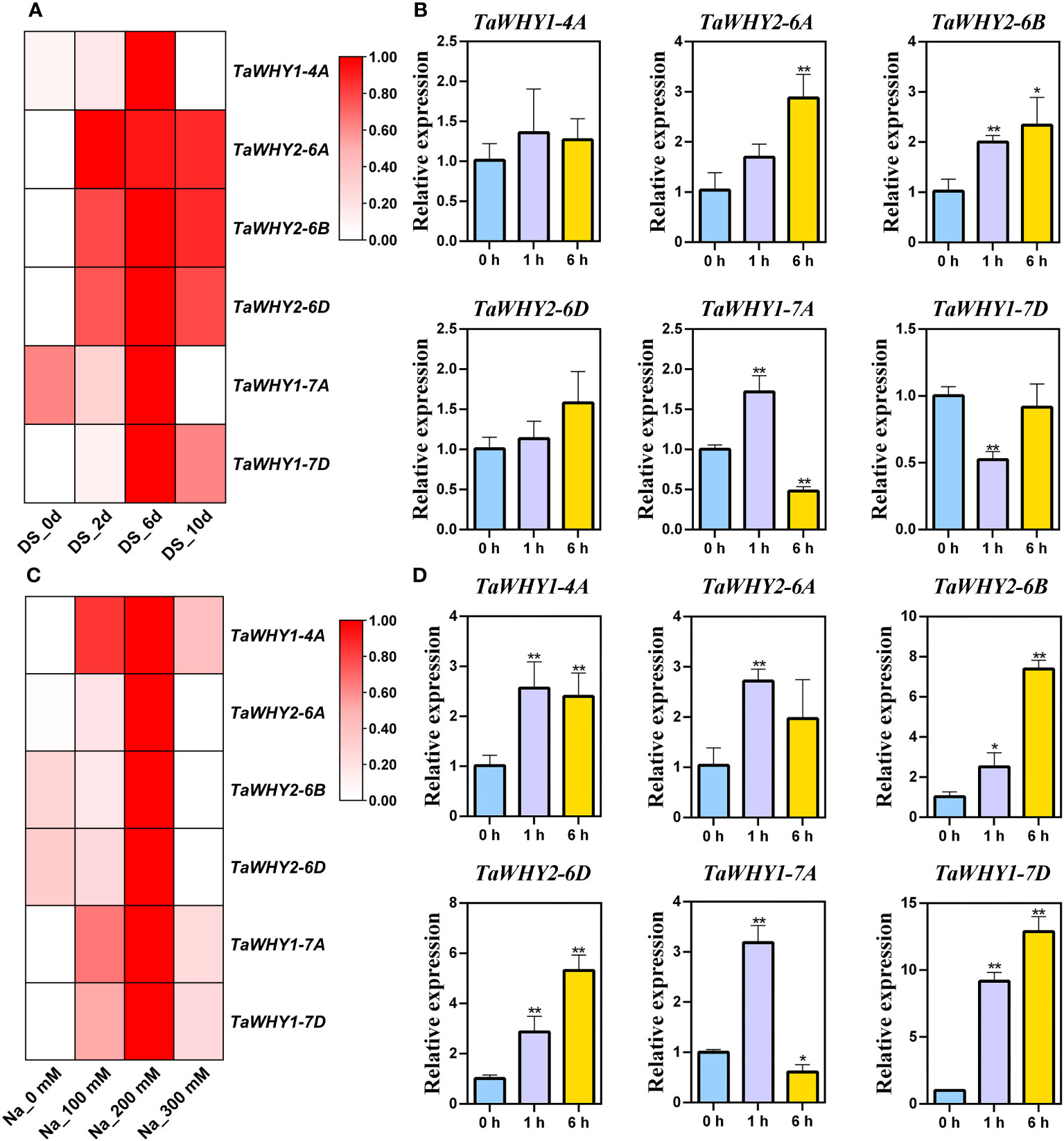
Figure 5 Expression patterns of TaWHY genes in response to osmotic stress. (A) RNA-seq analysis of the expression profiles of TaWHY genes in response to drought stress for 0 days, 2 days, 6 days, and 10 days, respectively. Fragments per kilobase of exon per million mapped fragments (FPKM) values were used to measure the expression levels of genes. (B) The expression profiles of TaWHY genes in bread wheat seedling leaves at 0 h, 1 h, and 6 h after PEG stress treatment. (C) RNA-seq analysis of the expression profiles of TaWHY genes in response to 0 mM, 100 mM, 200 mM, and 300 mM NaCl treatment. FPKM values were used to measure the expression levels of genes. (D) The expression profiles of TaWHY genes in bread wheat seedling leaves at 0 h, 1 h, and 6 h after NaCl stress treatment. The expression level of the bread wheat actin gene was used as the reference control to standardize the RNA samples for each reaction. Data represent the mean ± SD of three replicates. The asterisk indicates significant differences compared with 0 h (control, as 1) based on Student’s t-test (*p < 0.05; **p < 0.01).
After drought stress treatment, RNA-seq analysis revealed that the TaWHY1 genes exhibited the highest expression levels after 6 days of drought treatment, and the expression of TaWHY2 genes increased with the progression of drought stress duration (Figure 5A). The real-time PCR results demonstrated the expression of TaWHY1-7A was up-regulated under PEG stress, peaking at 1 h with 1.6-fold compared with the control, TaWHY1-7D was down-regulated, and TaWHY1-4A was not significantly changed. The expression of TaWHY2-6A, TaWHY2-6B, and TaWHY2-6D (group 2) was gradually up-regulated and reached the highest expression level at 6 h under PEG stress with approximately 2.9-, 2.3-, and 1.6-fold compared with the control, respectively (Figure 5B).
After NaCl treatment, the expression levels of TaWHY genes were significantly up-regulated (Figures 5C, D). The real-time PCR results revealed that the expression levels of TaWHY1-4A, TaWHY2-6A, TaWHY2-6B, TaWHY2-6D, TaWHY1-7A, and TaWHY1-7D were all increased, peaking at 1 h, 1 h, 6 h, 6 h, 1 h, and 6 h with approximately 2.6-, 2.7-, 7.4-, 2.9-, 3.2-, and 12.9-fold compared with the control, respectively (Figure 5D). Therefore, we speculated that TaWHYs might play an important role under osmotic stress.
Transcription factors can interact with different cis-elements in the promoter region of target genes, exerting diverse functions in plant growth, development, and stress response (Strader et al., 2022). To determine the functions of TaWHY genes, upstream transcription factors and downstream target genes of TaWHYs were predicted by using the wheat integrative gene regulatory network (wGRN) (Figure 6; Supplementary Table S5) (Chen et al., 2023). Then, 22, 28, 33, 44, 195, and 187 transcription factors were predicted to regulate the expression of TaWHY1-4A, TaWHY1-7A, TaWHY1-7D, TaWHY2-6A, TaWHY2-6B, and TaWHY2-6D, respectively (Supplementary Table S5). We also conducted an analysis of the expression patterns for the top 30 potential upstream transcription factors and downstream target genes associated with TaWHYs. Under drought stress, the expression patterns of the cytokinin-responsiveGATA transcription factor 1-like gene (TraesCS6B03G0575900) were most similar to TaWHY1-4A. Additionally, the most similar expression patterns were observed in the transcription factor GLK2 (TraesCS3D03G0362600) and TCP family transcription factor TCP5 (TraesCS3A03G0743200, TraesCS3B03G0849100) with TaWHY1-7A. The MYB transcription factor (TraesCS6B03G0466300) and the cytokinin-responsive GATA transcription factor 1-like gene (TraesCS6B03G0575900) exhibited the most similar expression patterns to TaWHY1-7D. Furthermore, the nuclear transcription factor Y subunit C-4-like (TraesCS6A03G0382200) showed the most similar expression patterns to TaWHY2-6A. The transcription factor bHLH49-like gene (TraesCS4D03G0108100) demonstrated the most similar expression patterns to TaWHY2-6B and TaWHY2-6D (Figure 6A; Supplementary Figure S2A). Under salt stress, transcription factors LSD1 (TraesCS1A03G0706000 and TraesCS1B03G0806900) and GLK2 (TraesCS3A03G0376200) exhibited the most similar expression patterns to TaWHY1-4A. The transcription factors GLK2 (TraesCS3A03G0376200), LSD1 (TraesCS1A03G0706000), GATA transcription factor 17-like (TraesCS6A03G0279700), RAP2-9-like (TraesCS7B03G0076700), and Zinc finger CCCH domain-containing protein 44-like (TraesCS7A03G0973900) displayed the most similar expression patterns to TaWHY1-7A, TaWHY1-7D, TaWHY2-6A, TaWHY2-6B, and TaWHY2-6D, respectively (Figure 6A; Supplementary Figure S2B). These transcription factors are highly likely to regulate the expression of TaWHY genes under drought and salt stress.
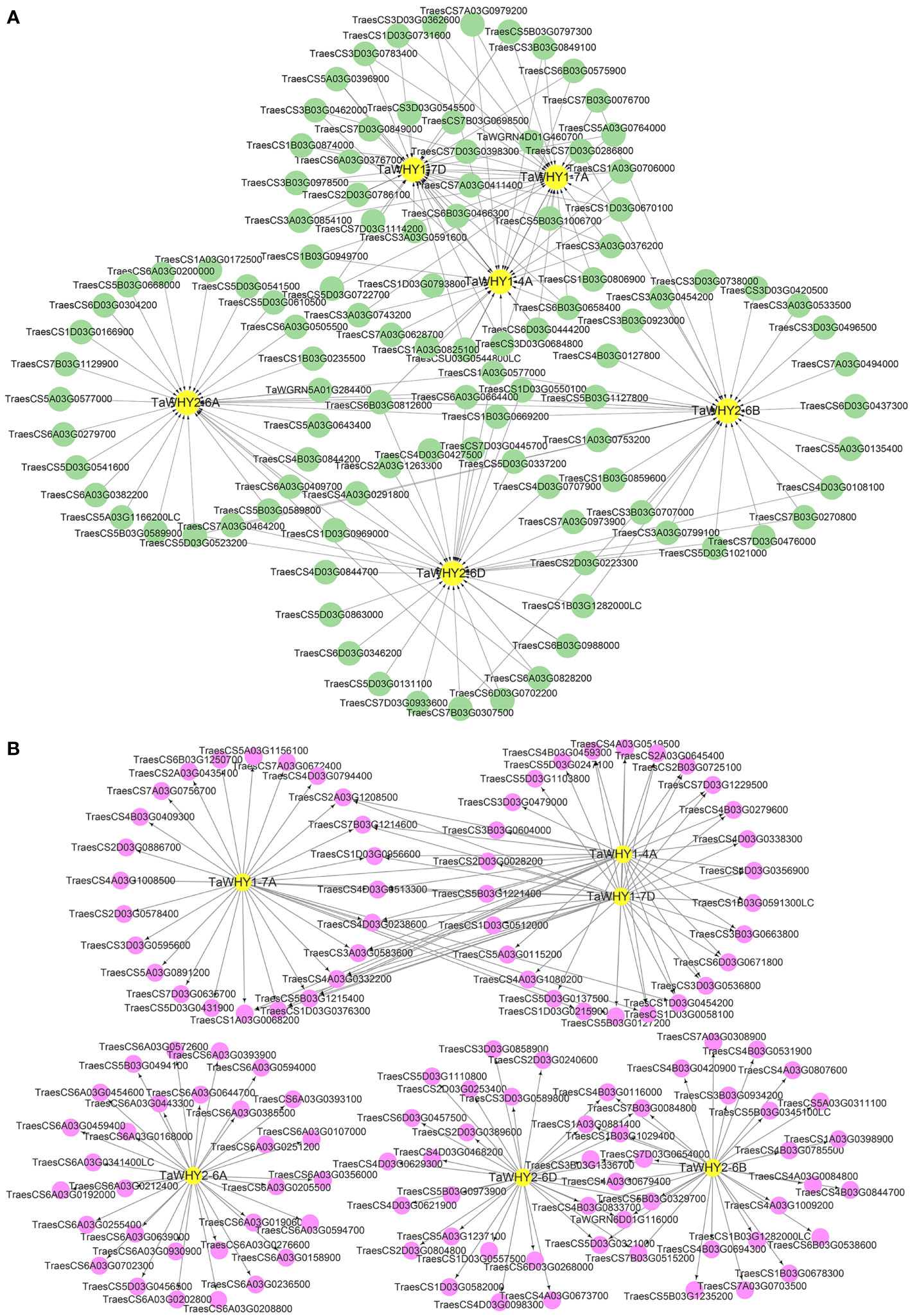
Figure 6 The upstream transcription factor (A) and downstream target gene (B) analyses of TaWHY genes.
TaWHYs, as transcription factors, also regulate downstream target genes in response to osmotic stress. The result suggested that TaWHY1-4A, TaWHY1-7A, TaWHY1-7D, TaWHY2-6A, TaWHY2-6B, and TaWHY2-6D might bind to the promoter of 1,345, 1,181, 1,404, 999, 3,413, and 3,662 downstream target genes, respectively (Supplementary Table S6). Under drought stress, the similar expression patterns were observed in fructokinase-like 1 (TraesCS3A03G0869600), protein fluorescent in blue light (TraesCS5D03G0431900), 2-carboxy-1,4-naphthoquinone phytyltransferase (TraesCS4A03G1008500), 50S ribosomal protein (TraesCS4A03G0332200 and TraesCS6B03G1250700), and starch synthase (TraesCS4D03G0513300) with TaWHY1-7A. The gene of glutamyl-tRNA (Gln) amidotransferase (TraesCS2A03G0645400), CDK5RAP1-like protein (TraesCS4D03G0338300), chaperone protein dnaJ 6-like (TraesCS6A03G0385500), OSB (TraesCS3B03G1336700), and flap endonuclease (TraesCS1B03G1029400) exhibited the most similar expression patterns to TaWHY1-4A, TaWHY1-7D, TaWHY2-6A, TaWHY2-6B, and TaWHY2-6D, respectively (Figure 6B; Supplementary Figure S3A). Similarly, the downstream target genes of the most similar expression patterns with TaWHYs under salt stress were also detected, i.e., TaWHY1-4A with transcription termination/antitermination protein NusG-like (TraesCS5B03G1215400), TaWHY1-7A with protein fluorescent in blue light (TraesCS5D03G0431900), TaWHY1-7D with superoxide dismutase (TraesCS4A03G1080200) and transcription termination/antitermination protein NusG-like (TraesCS5B03G1215400), TaWHY2-6A with eukaryotic translation initiation factor 3 subunit F-like (TraesCS6A03G0205500), TaWHY2-6B with HSP20-like chaperones superfamily protein (TraesCS7D03G0654000) and eukaryotic translation initiation factor 3 subunit K (TraesCS4B03G0785500), and TaWHY2-6D with HSP20-like chaperones superfamily protein (TraesCS7D03G0654000), DNA polymerase delta small subunit-like (TraesCS4B03G0833700) and flap endonuclease 1-A-like (TraesCS1B03G1029400 and TraesCS1A03G0881400) (Figure 6B; Supplementary Figure S3B). The GO enrichment analysis result showed the downstream target genes of TaWHY1s mainly participated in translation, glutaminyl-tRNAGln biosynthesis, protoporphyrinogen IX biosynthetic process, and heme biosynthetic process (Supplementary Figure S4). TaWHY2s might take part in mRNA splicing, RNA binding, and DNA replication (Supplementary Figure S4). It was worth noting that TaWHY1-7D and TaWHY2-6D were predicted to respond to hydrogen peroxide (H2O2) and oxidative stress (Supplementary Figure S4), suggesting TaWHY1-7D and TaWHY2-6D might respond to osmotic stress via regulating ROS homeostasis.
The protein–protein interactions (PPIs) analysis suggested that TaWHY1-4A, TaWHY1-7A, and TaWHY1-7D could interact with 16, 37, and 36 proteins, respectively. TaWHY2-6A, TaWHY2-6B, and TaWHY2-6D interact with 102 proteins (Supplementary Table S7). We identified the interacting proteins with similar expression patterns to TaWHYs under drought stress (Supplementary Figure S5), i.e., TaWHY1-4A was found to interact with fructokinase-like 2 (TraesCS2A02G013600). TaWHY1-7A showed interactions with glutamate-rich WD repeat-containing protein (TraesCS4B02G157000), fructokinase-like 2 (TraesCS2A02G013600), and serine/arginine-rich splicing factor SR34A (TraesCS4D02G168700). TaWHY1-7D demonstrated an interaction with fructokinase-like 2 (TraesCS2A02G013600). Additionally, TaWHY2-6A interacted with DnaJ protein homolog (TraesCS5B02G374900), while TaWHY2-6B and TaWHY2-6D showed interactions with methionine aminopeptidase 1B (TraesCS2B02G448000) and protein OSB2 (TraesCS3B02G536700) (Figure 7; Supplementary Figure S5A). After NaCl treatment, TaWHY1 (TaWHY1-4A, TaWHY1-7A, and TaWHY1-7D) showed the most similar expression patterns with interacting protein single-stranded DNA-binding protein (TraesCS3A02G231400). TaWHY2 (TaWHY2-6A, TaWHY2-6B, and TaWHY2-6D) demonstrated the most similar expression patterns with glutamate-rich WD repeat-containing protein (TraesCS5B02G137200), actin-related protein (TraesCS5B02G422700), chaperone protein dnaJ A6 (TraesCS6B02G274600), and methionine aminopeptidase 1B (TraesCS2D02G231000) (Figure 7; Supplementary Figure S5B). These results suggested the regulatory mechanism of TaWHY genes to avoid or defend against osmotic stress.
To further investigate the function of TaWHY genes under osmotic (d-sorbitol and NaCl) and oxidative (H2O2) stresses, TaWHY2-6A, TaWHY2-6B, TaWHY2-6D, TaWHY1-7A, and TaWHY1-7D were cloned into the pGADT7 vector, and then transformed into the yeast cells BY4741 or stress-sensitive yeast mutant BY4741 (Δhog1) to confirm the ability to improve stress resistance in yeast cells (Figure 8). The results suggested that the growth of the BY4741 or Δhog1 yeast cells carrying these TaWHY genes was not obviously different compared with the control (pGADT7 empty vector) under normal growth conditions. After d-sorbitol treatment, Δhog1 yeast cells overexpressing TaWHYs slightly enhanced their tolerance to d-sorbitol stress in comparison to the negative control. The Δhog1 yeast overexpressing TaWHY2-6A, TaWHY2-6B, and TaWHY2-6D obviously improved the resistance to NaCl stress, but the colonies of Δhog1 with TaWHY1-7A and TaWHY1-7D were slightly increased compared with the negative control under NaCl stress.
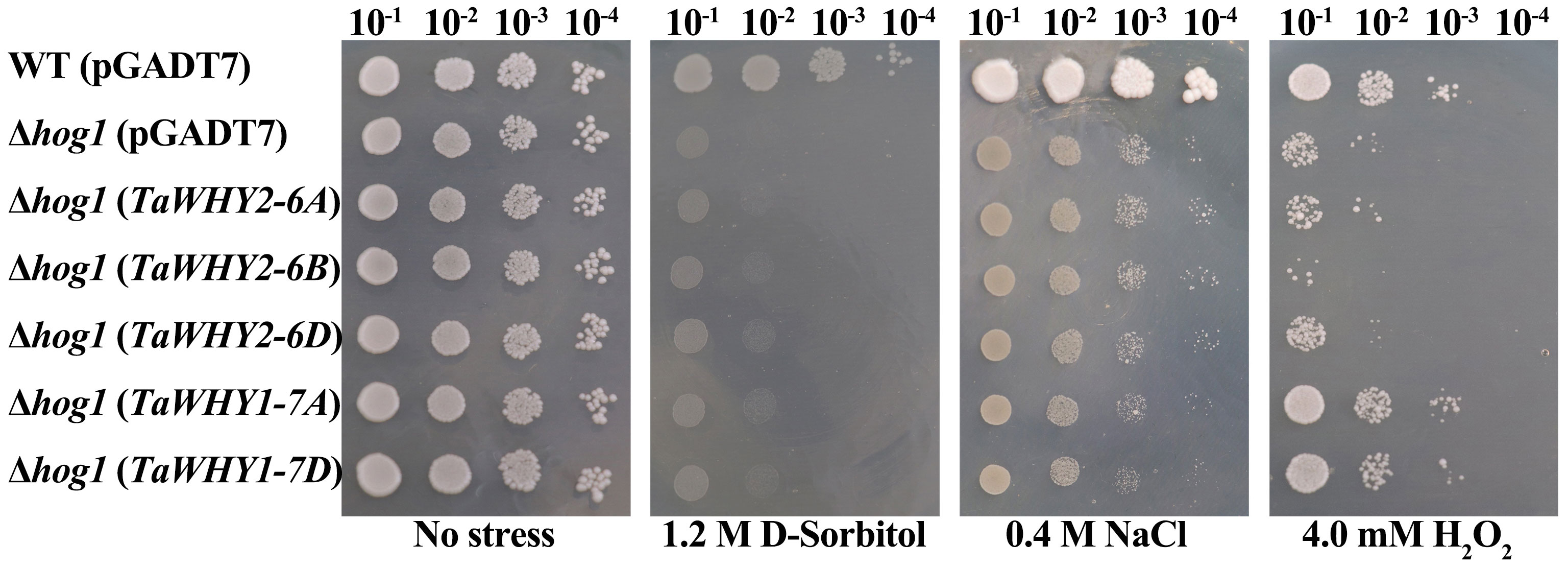
Figure 8 The ability of the tolerance in response to 1.2 M d-sorbitol, 0.4 M NaCl, and 4.0 mM H2O2 stresses in recombinant yeast cells. For osmotic and oxidative stresses, the yeast cells Δhog1 carrying the recombinant vector pGADT7-TaWHY2-6A/TaWHY2-6B/TaWHY2-6D/TaWHY1-7A/TaWHY1-7D were spotted onto YPD medium plates containing 1.2 M d-sorbitol, 0.4 M NaCl, or 4.0 mM H2O2 with serially diluted (10−1, 10−2, 10−3, 10−4) and cultured at 30°C for 3–5 days. The wild-type yeast cells BY4741 and the stress-sensitive mutant Δhog1 carrying the empty vector pGADT7 were used as positive and negative controls, respectively.
Adverse environmental conditions induce ROS production; ROS accumulation can cause oxidative damage to membranes, proteins, and RNA and DNA molecules and even lead to the oxidative destruction of the cell in a process termed oxidative stress; thereby, ROS scavenging is essential for plants to avoid or defend against adverse stress (Choudhury et al., 2017). To determine whether TaWHYs enhanced stress tolerance by scavenging ROS in yeast cells, Δhog1 yeast cells carrying pGADT7-TaWHYs or pGADT7 were grown on YPD medium containing 4.0 mM H2O2, suggesting TaWHY1-7A and TaWHY1-7D strongly enhanced the oxidative stress tolerance in yeast, but the colonies of Δhog1 overexpressing TaWHY2-6A, TaWHY2-6B, and TaWHY2-6D were reduced compared with control. These results indicated that the TaWHY1 and TaWHY2 genes performed diverse functions. TaWHY1 mainly enhanced the tolerance to oxidative stresses; TaWHY2 mainly improved NaCl stress tolerance and was sensitive to oxygen stress; and TaWHY1 and TaWHY2 genes slightly improved the tolerance to d-sorbitol stress.
In order to further confirm the potential role of TaWHY1-7D in response to drought and salt stresses, we generated 35S:TaWHY1-7D transgenic Arabidopsis lines. Three independent transgenic lines (OE4, OE8, and OE10) and wild-type (WT) were chosen for the functional analysis of TaWHY1-7D in response to drought and salt stresses (Figure 9; Supplementary Figure S6). The results showed that there were no obvious phenotypic differences between transgenic and WT plants under normal conditions. After an 8-day drought treatment, the wild-type (WT) plants exhibited wilting and subsequent yellowing. In contrast, the transgenic Arabidopsis overexpressing TaWHY1-7D remained predominantly green. After NaCl treatment for 8 days, both WT and transgenic Arabidopsis lines exhibited growth inhibition compared with CK. The growth inhibition was more severe in WT plants compared to transgenic Arabidopsis. Thus, the heterologous expression of TaWHY1-7D greatly improved drought and salt tolerance in transgenic Arabidopsis.
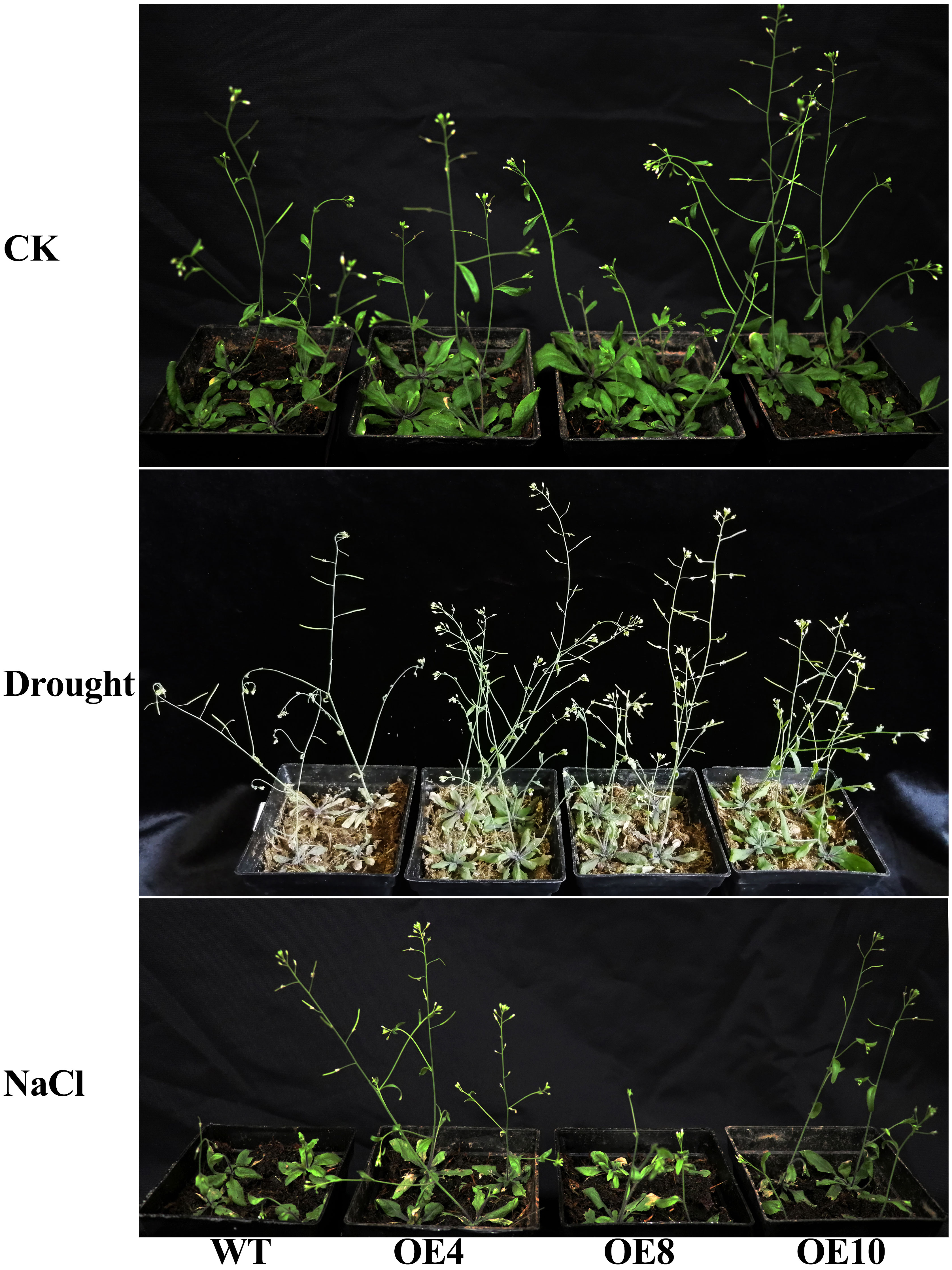
Figure 9 The phenotype of the 35S: TaWHY1-7D transgenic Arabidopsis under drought and NaCl stress. Three independent 35S:TaWHY1-7D transgenic Arabidopsis lines (OE4, OE8, and OE10) and wild type (WT) were chosen for functional analysis of TaWHY1-7D under normal conditions (CK), drought (water-deficit), and salt (NaCl) stress treatments.
Whirly genes have been identified in diverse plant species (Desveaux et al., 2005; Janack et al., 2016; Yan et al., 2020; Hu and Shu, 2021). Most plant species have two kinds of Whirly proteins, Whirly1 and Whirly2, whereas Arabidopsis and cassava have three Whirly proteins (Cappadocia et al., 2013; Yan et al., 2020). As a heterologous hexaploid species composed of three subgenomes A, B, and D, bread wheat (AABBDD) has undergone two rounds of natural hybridization events (Levy and Feldman, 2022). Therefore, bread wheat has six Whirly genes belonging to Whirly1 and Whirly2, and other Triticeae species, including T. urartu (AA, diploid), T. dicoccoides (AABB, tetraploid), Ae. tauschii (DD, diploid), H. vulgare (HH, diploid), and S. cereal (RR, diploid), have two, four, two, two, and two Whirly genes, respectively (Figure 1A; Supplementary Table S2). There was a positive correlation between the number of Whirly genes and that of subgenomes in Triticeae species.
The paralogous Whirly gene pairs TaWHY1-4A/TaWHY1-7A/TaWHY1-7D and TaWHY2-6A/TaWHY2-6B/TaWHY2-6D were identified in T. aestivum genome, which all expanded by WGD or segmental duplication events (Figure 2B; Supplementary Table S4). Interestingly, the paralogous genes of TaWHY1-7A and TaWHY1-7D were found on chromosome 4A instead of chromosome 7B in T. aestivum (Figure 2B). To investigate the origin of TaWHY1-4A, a micro-collinear analysis of TaWHY1-4A was performed. The results showed that the homologous gene of TaWHY1-4A did not exist on subgenome B in other related Triticeae species, but there was homologous gene of TuWHY1-7A on chromosome 7A of T. urartu and AetWHY1-7D on chromosome 7D of Ae. tauschii (Figure 3). Similar events also occurred in the SHMT gene family of T. aestivum (Hu et al., 2022). Therefore, we speculated that the expansion events of Whirly1 genes occurred through hybridization and polyploidization, and TaWHY1-4A and TdWHY1-4A might have originated from TuWHY1-7A or AetWHY1-7D (Figure 3). However, this speculation still needs further research.
Whirly proteins are plant-specific transcription factors that regulate plant development and stress resistance in plants (Krupinska et al., 2022; Taylor et al., 2022). Previous studies mainly focused on the function of Whirly genes under abiotic stress and biotic stresses, such as drought (Yan et al., 2020), salt (Akbudak and Filiz, 2019), chilling (Zhuang et al., 2020b) or light stresses (Swida-Barteczka et al., 2018). Previous studies indicated that AtWHY1 located in chloroplasts and nucleus (Krause et al., 2005; Ren et al., 2017) could repress the expression of WRKY53 and delay leaf senescence in Arabidopsis (Miao et al., 2013), whereas AtWHY2 was located in the mitochondria and nucleus (Krause et al., 2005; Golin et al., 2020). These were consistent with the higher expression of TaWHY1 genes (TaWHY1-4A, TaWHY1-7A, and TaWHY1-7D) in leaf sheaths and leaves and higher expression of TaWHY2 genes (TaWHY2-6A, TaWHY2-6B, and TaWHY2-6D) in roots (Figure 4).
Recently, Whirly genes were reported to improve osmotic stress resistance in plants, such as MeWHYs, which could interact with MeCIPK23 to activate ABA biosynthesis and regulate drought resistance in cassava (Yan et al., 2020). In this study, TaWHY1-7A and three TaWHY2 genes were up-regulated under PEG stress, TaWHY1-7D was down-regulated, and TaWHY1-4A was not significantly changed (Figure 5), suggesting that functional differentiation of Whirly genes occurred. All TaWHYs were up-regulated under NaCl stress (Figure 5) and improved the resistance of NaCl stress in yeast, respectively (Figure 8). The heterologous expression of TaWHY1-7D greatly improved drought and salt tolerance in transgenic Arabidopsis (Figure 9). In addition, Whirly genes have been reported to regulate ROS homeostasis (Lin et al., 2019), and our results also showed that TaWHY1-7A and TaWHY1-7D strongly enhanced the oxidative stress tolerance in yeast cells (Figure 8). ROS scavenging also might be an important reason for the improvement of stress resistance in TaWHY1 genes. However, the growth of Δhog1 overexpressing TaWHY2-6A, TaWHY2-6B, and TaWHY2-6D was inhibited under oxidative stress; these were consistent with a previous study that found that overexpression of AtWHY2 caused the accumulation of ROS in the plant (Cai et al., 2015). The ROS accumulation might cause cellular stress, thus activating the alternative pathway to reduce ROS levels and eliminate the stress (Cai et al., 2015). GO enrichment analysis also showed that TaWHY1-7D and TaWHY2-6D regulated downstream target genes to respond to H2O2 and oxidative stress (Supplementary Figure S4). Based on the above research, we speculate that the Whirly genes may play a vital role in plant resistance to osmotic stress. These results provide useful information for further functional studies of Whirly genes and lay a foundation to improve wheat yield and quality via molecular breeding under osmotic stress.
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
HL: Conceptualization, Data curation, Formal Analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing. XW: Writing – review & editing. WY: Writing – review & editing. WL: Writing – review & editing. YW: Resources, Writing – review & editing. QW: Resources, Writing – review & editing. YZ: Resources, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the National Natural Science Foundation of China (Grant No. 32272159).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1297228/full#supplementary-material
Supplementary Figure 1 | Multiple sequence alignment of the conserved domains of Whirly genes in Triticeae species. Moitf1-10 and DNA binding domain were marked. The conserved cysteine was marked by red triangle.
Supplementary Figure 2 | The FPKM values of upstream transcription factors of TaWHYs under drought (A) and salt (B) stress.
Supplementary Figure 3 | The FPKM values of downstream target genes of TaWHYs under drought (A) and salt (B) stress.
Supplementary Figure 4 | GO enrichment analysis on the downstream target genes of TaWHY genes.
Supplementary Figure 5 | The FPKM values of interacting protein of TaWHYs under drought (A) and salt (B) stress.
Supplementary Figure 6 | The PCR detection (A) and screening (B) of 35S: TaWHY1-7D transgenic Arabidopsis.
Akbudak, M. A., Filiz, E. (2019). Whirly (Why) transcription factors in tomato (Solanum lycopersicum L.): genome-wide identification and transcriptional profiling under drought and salt stresses. Mol. Biol. Rep. 46, 4139–4150. doi: 10.1007/s11033-019-04863-y
Bailey, T. L., Boden, M., Buske, F. A., Frith, M., Grant, C. E., Clementi, L., et al. (2009). MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 37, W202–W208. doi: 10.1093/nar/gkp335
Cai, Q., Guo, L., Shen, Z. R., Wang, D. Y., Zhang, Q., Sodmergen (2015). Elevation of pollen mitochondrial DNA copy number by WHIRLY2: altered respiration and pollen tube growth in arabidopsis. Plant Physiol. 169, 660–673. doi: 10.1104/pp.15.00437
Cappadocia, L., Parent, J. S., Sygusch, J., Brisson, N. (2013). A family portrait: structural comparison of the Whirly proteins from Arabidopsis thaliana and Solanum tuberosum. Acta Crystallogr. Sect F Struct. Biol. Cryst Commun. 69, 1207–1211. doi: 10.1107/S1744309113028698
Cappadocia, L., Parent, J. S., Zampini, E., Lepage, E., Sygusch, J., Brisson, N. (2012). A conserved lysine residue of plant Whirly proteins is necessary for higher order protein assembly and protection against DNA damage. Nucleic Acids Res. 40, 258–269. doi: 10.1093/nar/gkr740
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020a). TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chen, Y., Guo, Y., Guan, P., Wang, Y., Wang, X., Wang, Z., et al. (2023). A wheat integrative regulatory network from large-scale complementary functional datasets enables trait-associated gene discovery for crop improvement. Mol. Plant 16, 393–414. doi: 10.1016/j.molp.2022.12.019
Chen, Y., Song, W., Xie, X., Wang, Z., Guan, P., Peng, H., et al. (2020b). A collinearity-incorporating homology inference strategy for connecting emerging assemblies in the triticeae tribe as a pilot practice in the plant pangenomic era. Mol. Plant 13, 1694–1708. doi: 10.1016/j.molp.2020.09.019
Choudhury, F. K., Rivero, R. M., Blumwald, E., Mittler, R. (2017). Reactive oxygen species, abiotic stress and stress combination. Plant J. 90, 856–867. doi: 10.1111/tpj.13299
CNCB-NGDC Members and Partners (2022). Database resources of the national genomics data center, China national center for bioinformation in 2022. Nucleic Acids Res. 50, D27–D38. doi: 10.1093/nar/gkab951
Desveaux, D., Allard, J., Brisson, N., Sygusch, J. (2002). A new family of plant transcription factors displays a novel ssDNA-binding surface. Nat. Struct. Biol. 9, 512–517. doi: 10.1038/nsb814
Desveaux, D., Despres, C., Joyeux, A., Subramaniam, R., Brisson, N. (2000). PBF-2 is a novel single-stranded DNA binding factor implicated in PR-10a gene activation in potato. Plant Cell 12, 1477–1489. doi: 10.1105/tpc.12.8.1477
Desveaux, D., Marechal, A., Brisson, N. (2005). Whirly transcription factors: defense gene regulation and beyond. Trends Plant Sci. 10, 95–102. doi: 10.1016/j.tplants.2004.12.008
Desveaux, D., Subramaniam, R., Despres, C., Mess, J. N., Levesque, C., Fobert, P. R., et al. (2004). A "Whirly" transcription factor is required for salicylic acid-dependent disease resistance in Arabidopsis. Dev. Cell 6, 229–240. doi: 10.1016/S1534-5807(04)00028-0
Foyer, C. H., Karpinska, B., Krupinska, K. (2014). The functions of WHIRLY1 and REDOX-RESPONSIVE TRANSCRIPTION FACTOR 1 in cross tolerance responses in plants: a hypothesis. Philos. Trans. R Soc. Lond B Biol. Sci. 369, 20130226. doi: 10.1098/rstb.2013.0226
Golin, S., Negroni, Y. L., Bennewitz, B., Klosgen, R. B., Mulisch, M., La Rocca, N., et al. (2020). WHIRLY2 plays a key role in mitochondria morphology, dynamics, and functionality in Arabidopsis thaliana. Plant Direct 4, e00229. doi: 10.1002/pld3.229
Gouet, P., Robert, X., Courcelle, E. (2003). ESPript/ENDscript: Extracting and rendering sequence and 3D information from atomic structures of proteins. Nucleic Acids Res. 31, 3320–3323. doi: 10.1093/nar/gkg556
Gupta, P. K., Balyan, H. S., Sharma, S., Kumar, R. (2020). Genetics of yield, abiotic stress tolerance and biofortification in wheat (Triticum aestivum L.). Theor. Appl. Genet. 133, 1569–1602. doi: 10.1007/s00122-020-03583-3
Hu, Y., Shu, B. (2021). Identifying strawberry Whirly family transcription factors and their expressions in response to crown rot. Notulae Botanicae Horti Agrobotanici Cluj-Napoca 49, 12323. doi: 10.15835/nbha49212323
Hu, P., Song, P., Xu, J., Wei, Q., Tao, Y., Ren, Y., et al. (2022). Genome-wide analysis of serine hydroxymethyltransferase genes in triticeae species reveals that taSHMT3A-1 regulates fusarium head blight resistance in wheat. Front. Plant Sci. 13, 847087. doi: 10.3389/fpls.2022.847087
Janack, B., Sosoi, P., Krupinska, K., Humbeck, K. (2016). Knockdown of WHIRLY1 affects drought stress-induced leaf senescence and histone modifications of the senescence-associated gene HvS40. Plants-Basel 5, 37. doi: 10.3390/plants5030037
Krause, K., Kilbienski, I., Mulisch, M., Rodiger, A., Schafer, A., Krupinska, K. (2005). DNA-binding proteins of the Whirly family in Arabidopsis thaliana are targeted to the organelles. FEBS Lett. 579, 3707–3712. doi: 10.1016/j.febslet.2005.05.059
Krupinska, K., Desel, C., Frank, S., Hensel, G. (2022). WHIRLIES are multifunctional DNA-binding proteins with impact on plant development and stress resistance. Front. Plant Sci. 13, 880423. doi: 10.3389/fpls.2022.880423
Letunic, I., Khedkar, S., Bork, P. (2021). SMART: recent updates, new developments and status in 2020. Nucleic Acids Res. 49, D458–D460. doi: 10.1093/nar/gkaa937
Levy, A. A., Feldman, M. (2022). Evolution and origin of bread wheat. Plant Cell 34, 2549–2567. doi: 10.1093/plcell/koac130
Lin, W., Huang, D., Shi, X., Deng, B., Ren, Y., Lin, W., et al. (2019). H2O2 as a feedback signal on dual-located WHIRLY1 associates with leaf senescence in Arabidopsis. Cells 8, 1585. doi: 10.3390/cells8121585
Liu, H., Yang, W., Zhao, X., Kang, G., Li, N., Xu, H. (2022). Genome-wide analysis and functional characterization of CHYR gene family associated with abiotic stress tolerance in bread wheat (Triticum aestivum L.). BMC Plant Biol. 22, 204. doi: 10.1186/s12870-022-03589-7
Livak, K. J., Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2–ΔΔCT Method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Ma, S., Wang, M., Wu, J., Guo, W., Chen, Y., Li, G., et al. (2021). WheatOmics: A platform combining multiple omics data to accelerate functional genomics studies in wheat. Mol. Plant 14, 1965–1968. doi: 10.1016/j.molp.2021.10.006
Manh, M. B., Ost, C., Peiter, E., Hause, B., Krupinska, K., Humbeck, K. (2023). WHIRLY1 acts upstream of ABA-related reprogramming of drought-induced gene expression in barley and affects stress-related histone modifications. Int. J. Mol. Sci. 24, 6326. doi: 10.3390/ijms24076326
Marechal, A., Parent, J. S., Sabar, M., Veronneau-Lafortune, F., Abou-Rached, C., Brisson, N. (2008). Overexpression of mtDNA-associated AtWhy2 compromises mitochondrial function. BMC Plant Biol. 8, 42. doi: 10.1186/1471-2229-8-42
Marechal, A., Parent, J. S., Veronneau-Lafortune, F., Joyeux, A., Lang, B. F., Brisson, N. (2009). Whirly proteins maintain plastid genome stability in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 106, 14693–14698. doi: 10.1073/pnas.0901710106
Meng, C., Yang, M., Wang, Y., Chen, C., Sui, N., Meng, Q., et al. (2020). SlWHY2 interacts with SlRECA2 to maintain mitochondrial function under drought stress in tomato. Plant Sci. 301, 110674. doi: 10.1016/j.plantsci.2020.110674
Miao, Y., Jiang, J., Ren, Y., Zhao, Z. (2013). The single-stranded DNA-binding protein WHIRLY1 represses WRKY53 expression and delays leaf senescence in a developmental stage-dependent manner in Arabidopsis. Plant Physiol. 163, 746–756. doi: 10.1104/pp.113.223412
Mistry, J., Chuguransky, S., Williams, L., Qureshi, M., Salazar, G. A., Sonnhammer, E. L. L., et al. (2021). Pfam: The protein families database in 2021. Nucleic Acids Res. 49, D412–D419. doi: 10.1093/nar/gkaa913
Ren, Y., Li, Y., Jiang, Y., Wu, B., Miao, Y. (2017). Phosphorylation of WHIRLY1 by CIPK14 shifts its localization and dual functions in Arabidopsis. Mol. Plant 10, 749–763. doi: 10.1016/j.molp.2017.03.011
Strader, L., Weijers, D., Wagner, D. (2022). Plant transcription factors - being in the right place with the right company. Curr. Opin. Plant Biol. 65, 102136. doi: 10.1016/j.pbi.2021.102136
Subramanian, B., Gao, S., Lercher, M. J., Hu, S., Chen, W. H. (2019). Evolview v3: a webserver for visualization, annotation, and management of phylogenetic trees. Nucleic Acids Res. 47, W270–W275. doi: 10.1093/nar/gkz357
Swida-Barteczka, A., Krieger-Liszkay, A., Bilger, W., Voigt, U., Hensel, G., Szweykowska-Kulinska, Z., et al. (2018). The plastid-nucleus located DNA/RNA binding protein WHIRLY1 regulates microRNA-levels during stress in barley (Hordeum vulgare L.). RNA Biol. 15, 886–891. doi: 10.1080/15476286.2018.1481695
Tamura, K., Stecher, G., Kumar, S. (2021). MEGA11: molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 38, 3022–3027. doi: 10.1093/molbev/msab120
Taylor, R. E., West, C. E., Foyer, C. H. (2022). WHIRLY protein functions in plants. Food Energy Secur. 00, e379. doi: 10.1002/fes3.379
Von Mering, C., Huynen, M., Jaeggi, D., Schmidt, S., Bork, P., Snel, B. (2003). STRING: a database of predicted functional associations between proteins. Nucleic Acids Res. 31, 258–261. doi: 10.1093/nar/gkg034
Wang, Y., Tang, H., Debarry, J. D., Tan, X., Li, J., Wang, X., et al. (2012). MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 40, e49. doi: 10.1093/nar/gkr1293
Waterhouse, A., Bertoni, M., Bienert, S., Studer, G., Tauriello, G., Gumienny, R., et al. (2018). SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 46, W296–W303. doi: 10.1093/nar/gky427
Yan, Y., Liu, W., Wei, Y., Shi, H. (2020). MeCIPK23 interacts with Whirly transcription factors to activate abscisic acid biosynthesis and regulate drought resistance in cassava. Plant Biotechnol. J. 18, 1504–1506. doi: 10.1111/pbi.13321
Yoo, H. H., Kwon, C., Lee, M. M., Chung, I. K. (2007). Single-stranded DNA binding factor AtWHY1 modulates telomere length homeostasis in Arabidopsis. Plant J. 49, 442–451. doi: 10.1111/j.1365-313X.2006.02974.x
Zhuang, K., Gao, Y., Liu, Z., Diao, P., Sui, N., Meng, Q., et al. (2020a). WHIRLY1 regulates HSP21.5A expression to promote thermotolerance in tomato. Plant Cell Physiol. 61, 169–177. doi: 10.1093/pcp/pcz189
Keywords: Triticeae species, wheat, Whirly gene, gene expression, osmotic stress
Citation: Liu H, Wang X, Yang W, Liu W, Wang Y, Wang Q and Zhao Y (2023) Identification of Whirly transcription factors in Triticeae species and functional analysis of TaWHY1-7D in response to osmotic stress. Front. Plant Sci. 14:1297228. doi: 10.3389/fpls.2023.1297228
Received: 19 September 2023; Accepted: 20 November 2023;
Published: 05 December 2023.
Edited by:
Xueqiang Wang, Zhejiang University, ChinaReviewed by:
Mehdi Mansouri, Shahid Bahonar University of Kerman, IranCopyright © 2023 Liu, Wang, Yang, Liu, Wang, Wang and Zhao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yanfang Wang, d3lmODYzQDEyNi5jb20=; Qin Wang, d2FuZ3FpbkBiem1jLmVkdS5jbg==; Yanhong Zhao, enloYm9iQDE2My5jb20=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.