- 1National Engineering Research Center for Agriculture in Northern Mountainous Areas, Agricultural Technology Innovation Center in Mountainous Areas of Hebei Province, Hebei Agricultural University, Baoding, Hebei, China
- 2College of Life Sciences, Hebei Agricultural University, Baoding, China
Introduction: Walnut blight is a serious bacterial disease that affects the yield and quality of walnuts. Pantoea agglomerans is one of the main causative agents of walnut blight. However, there have been few studies on the response of walnuts to P. agglomerans infection.
Methods: In this study, the soluble sugar, photosynthesis, antioxidant enzyme activities, and secondary metabolites were measured, and the transcriptomic analysis was performed to determine the response of walnut tissue cultures to P. agglomerans infection.
Results: After pathogen inoculation, the soluble sugar content decreased, and photosynthesis was inhibited. Antioxidant enzyme (superoxide dismutase and peroxidase) activities and secondary metabolites (phenol and flavonoid) contents increased, especially in the early stages of inoculation. Transcriptomic analysis revealed that the phenylpropanoid biosynthesis pathway is induced after infection, and pathogen infection promotes ABA and ethylene signal transduction and inhibits auxin signaling. In addition, SA and JA-related gene expression was altered after inoculation with P. agglomerans, and the FLS- and calcium-mediated disease resistance signaling pathways were activated. Furthermore, our results suggested an involvement of the R-protein RPM-mediated disease resistance pathway in the response of walnuts to bacterial infections.
Discussion: Our findings indicated that phenylpropanoid biosynthesis, hormone signal transduction, and plant-pathogen interaction have key roles in pathogenic inoculation, which provide insights into the molecular mechanisms in the response of walnuts to P. agglomerans infection.
1 Introduction
Walnuts (Juglans regia L.) are one of the four most popular nuts in the world, with high nutritional value and health benefits in their seed kernel (Zhang et al., 2020; Ji et al., 2021). In addition, walnut is an important economic tree, which brings economic benefits and ecological value to the mountainous people and environment in China. Walnut blight is a serious bacterial disease for walnut production worldwide and causes severe damage to the green tissue of the walnut tree, such as bud, leaves, fruits and so on (Mikulic-Petkovsek et al., 2011; Gašić et al., 2022).
Xanthomonas arboricola pv. juglandis (Xaj) was initially identified as the causative agent of walnut blight (Vauterin et al., 1995). It causes brown apical necrosis in walnut fruit and premature fruit drop, leading to significant yield losses (Moragrega et al., 2011). Studies have indicated that Xaj may cause primary infection in young walnut fruits, and Fusarium spp. and Alternaria spp. may participate in the induction of brown apical necrosis, causing secondary infection (Moragrega and Özaktan, 2010). Pantoea agglomerans is the causative agent of walnut blight, which causes internal brown apical necrosis in walnut (Yang et al., 2011). P. agglomerans belongs to the Enterobacteriaceae family (Cheng et al., 2013; Dutkiewicz et al., 2016) and can infect various hosts. Recently, it has been reported that P. agglomerans can cause soft rot disease in cabbage (Guo et al., 2020), leaf shot hole disease in peaches (Zang et al., 2021), necrotic symptoms and dieback in pistachios (Zamorano et al., 2022), blight disease in melons (She et al., 2021), and necrotic disease in mangoes (Gutierrez-Barranquero et al., 2019), jujuba (She et al., 2019), and plums (Li et al., 2020).
The susceptibility of walnuts to bacterial blight depends on the environmental conditions, leafing date, cultivar, soil characteristics, and disease history (Woeste et al., 1992; Olson et al., 1997; Radix et al., 1998; Frutos and Ortega, 2012; Moragrega and Llorente, 2023). The occurrence of walnut blight is more serious in early leafing cultivars and wet years (Mikulic-Petkovsek et al., 2011). Radix et al. (1998) suggested that soil could influence the susceptibility of walnuts to blight by changing the polyphenol content in walnut tissues. Walnut species vary significantly in their resistance to walnut blight, although no species is immune to walnut blight so far (Soltani and Aliabadi, 2010; Frutos and Ortega, 2012). In general, Juglans sigllata accessions are more resistant to walnut blight than J. regia accessions in the field (Shengke, 1987). Belisario and Zoina (1995) reported that J. regia is the walnut species most sensitive to walnut blight. However, there are differences in resistance among walnut genotypes within the same species (Bandi et al., 2015). Jiang et al. (2019b) evaluated the resistance of 18 walnut genotypes, including J. sigllata, J. regia and their hybrids, against Xaj and found that the ‘Qingxiang’ (J. regia) variety displayed a relatively strong resistance to blight.
When pathogens infect plants, they produce reactive oxygen species (ROS) (del Río, 2015). To eliminate the damage caused by ROS in plant cells, plants have evolved a complex network of antioxidant systems, including superoxide dismutase (SOD), peroxidase (POD), carotenoids, ascorbate, etc (Foyer et al., 1994; Li et al., 2015). Moreover, phenolics are involved in the defense against pathogens. Differences in the phenolic content of healthy fruits may indicate differences in inherent disease resistance among cultivars. The phenolic content of resistant apple cultivars was higher than that of susceptible cultivars (Petkovsek et al., 2009). However, Mikulic-Petkovsek et al. (2011) showed that the Seifersdorfer cultivar, with a high phenolic content in its fruit, showed extremely high susceptibility to Xaj, and the Fernette cultivar, with a low phenolic content, was quite resistant to Xaj. Solar et al. (2006) found that seasonal variations in phenolic compounds in annual shoots have a greater influence than phenolic content on the susceptibility of the shoots to walnut blight pathogens. Sagawa et al. (2020) performed proteome analysis using Chandler walnut (J. regia) fruits inoculated with Xaj and found that the infection enhanced oxidative stress and degradation processes, and biosynthetic metabolism was inhibited in the fruits. In addition, phytohormone-associated defense proteins changed after infection.
Although walnut blight has been reported for many years (Yang et al., 2011; Lamichhane, 2014; Higuera et al., 2015; Chakraborty et al., 2016), previous studies have mostly focused on the isolation of the causative agents and changes in secondary metabolites in infected tissues. The response mechanism of walnuts during host–pathogen interactions is not fully understood. Moreover, most studies have focused on the bacterial blight caused by Xaj, and even fewer studies have focused on the response to P. agglomerans. However, P. agglomerans has been reported as a biocontroller of postharvest fruit diseases to control Penicillium digitatum in mandarins, Penicillium italicum in oranges, and Rhizopus stolonifer in pears and apples (Nunes et al., 2001; Soto et al., 2018). Therefore, it is important to understand the pathogenic potential of P. agglomerans to avoid its undesired or dangerous effects on plants and humans when used as a biocontroller. This study aimed at analyzing the physiological responses and transcriptome data from tissue cultures of ‘Qingxiang’ walnuts inoculated with P. agglomerans to further understand the plant-pathogen interactions.
2 Materials and methods
2.1 Plant materials and treatments
Tissue cultures of ‘Qingxiang’ walnuts were used in this study. The walnuts were cultured on Driver and Kuniyuki Walnut (DKW) medium with 1.5 mg/L 6-benzylaminopurine (6-BA) and 0.02 mg/L indole butyric acid (IBA) at 23 ± 2 °C with a 16-h light/8-h dark photoperiod. Tissue cultures were sub-cultured every 25 days. DKW medium contained 1.416 g/L NH4NO3, 0.265 g/L KH2PO4, 1.559 g/L K2SO4, 0.74 g/L MgSO4·7H2O, 0.149 g/L CaCl2·2H2O, 1.968 g/L Ca(NO3)2·4H2O, 33.5 mg/L MnSO4·H2O, 0.39 mg/L Na2MoO4·2H2O, 17 mg/L Zn(NO3)2·6H2O, 4.8 mg/L H3BO3, 2.5 mg/L CuSO4·5H2O, 33.8 mg/L FeSO4·7H2O, 45.4 mg/L Na2EDTA·2H2O, 0.1 g/L inositol, 2 mg/L glycine, 1 mg/L nicotinic acid, and 2 mg/L VB1.
P. agglomerans, a walnut blight pathogen, was isolated from infected walnut tissue. Walnut tissue cultures were inoculated with P. agglomerans by stem wound immersion. Samples were collected at 0 (C0), 1 (T1), 8 (T2), and 15 (T3) days post-inoculation (dpi) for physiological data determination and transcriptome analysis. Sixty seedlings were inoculated and three independent replicates were performed.
2.2 Measure of soluble sugar content
The soluble sugar content was determined using the Anthrone colorimetric method (Luo and Huang, 2011). The samples were ground, mixed with 80% alcohol, and boiled for 20 min. The supernatant was collected. Next, 80% alcohol was added to the residue, and the mixture was boiled for 20 min. The two supernatants were then combined. The extract was added to Anthrone reaction, and the mixture was boiled for 10 min. After cooling, absorbance was measured at 620 nm in a spectrophotometer.
2.3 Determination of Fv/Fm
The maximum quantum yield of PSII (Fv/Fm) was measured using chlorophyll fluorescence imaging systems (FluorCam 7). The seedlings were kept in the dark for 20 minutes before measurement.
2.4 Determination of total phenolic content
The total phenolic content was determined using the Folin–Ciocalteu assay (Li et al., 2012). The sample was ground in liquid nitrogen, mixed with 95% ethanol, and centrifuged to obtain the supernatant. The supernatant was added to the Folin–Ciocalteu reaction, allowed to stand for 3 min, and then added to Na2CO3. The mixture was incubated for 30 min and the absorbance was measured at 760 nm in a spectrophotometer.
2.5 Determination of flavonoid contents
The aluminum chloride assay was used to determine the flavonoid content according to Ayele et al. (2022). The samples were ground in liquid nitrogen, mixed with 70% ethanol, and sonicated for 30 min to obtain the crude extract. Crude extracts were added to 5% NaNO2 and incubated for 5 min. Then, 10% AlCl3 was added, and the mixture was incubated for 1 min. NaOH solution was added, and the mixture was incubated for 10 min. Absorbance was measured at 510 nm in a spectrophotometer.
2.6 Transcriptome analysis
Total RNA was extracted from seedlings in the C0, T1, T2, and T3 groups. Transcriptome data were sequenced using the Illumina HiSeq platform (Bolger et al., 2014). Clean reads were assembled and aligned to the walnut reference genome V1.0 (http://xhhuanglab.cn/data/juglans.html) using HISAT2 (Kim et al., 2019). The value of fragments per kilobase of exon per million mapped reads (FPKM) was applied to measure the expression level of genes. Differentially expressed genes (DEGs) were identified using FC≥2 and FDR < 0.01 (Love et al., 2014). The functions of the DEGs were annotated using the GO (The Gene Ontology Consortium 2018) and KEGG databases (https://www.genome.jp/kegg) (Kanehisa et al., 2007).
2.7 Validation of RNA-seq by quantitative reverse-transcription PCR
Ten DEGs in the transcriptome were selected, and their expression levels were assessed by qRT-PCR using SYBR Premix Ex Taq (Takara), following the manufacturer’s instructions. The reaction system consisted of 10 μL of SYBR, 1 μL of forward primer (10 μmol/L), 1 μL of reverse primer (10 μmol/L), 1 μL of cDNA, and 7 μL of ddH2O. JrACT2 (XM_018972062.1) gene was used as the loading control. Relative expression level was calculated using 2-ΔΔCT method. All PCR reactions were performed in triplicate. The primers used in this study are listed in Table S1.
3 Results
3.1 Phenotypic analysis of walnut tissue cultures under pathogen treatment
To test the effect of the walnut blight pathogen on ‘Qingxiang’ walnuts, we analyzed the phenotypic traits of walnut tissue cultures treated with P. agglomerans. Compared with 0 dpi, there were no obvious symptoms in tissue cultures at 1 dpi. At 8 dpi, black necrotic spots appeared on the lower leaves of walnut tissue cultures. At 15 dpi, walnut tissue cultures showed more serious symptoms, with complete necrosis of the lower leaves and black stems, and moreover, the upper leaf margin was also necrotic (Figure 1A). We determined the soluble sugar content of walnut tissue cultures at different time points. The results showed that the soluble sugar content decreased significantly with inoculation treatment, and the highest content was detected at 0 dpi. The decrease from 0 to 1 dpi was drastic, whereas the decrease from 1 to 15 dpi was moderate (Figure 1B). We further determined Fv/Fm in walnut tissue cultures and found no significant difference in Fv/Fm between 0–8 dpi and a significant decreased in Fv/Fm at 15 dpi (Figure 1C). These results indicate that walnut blight pathogens affect plant growth and photosynthesis, and plants consume a large amount of soluble sugar in response to the early stages of pathogen infection.
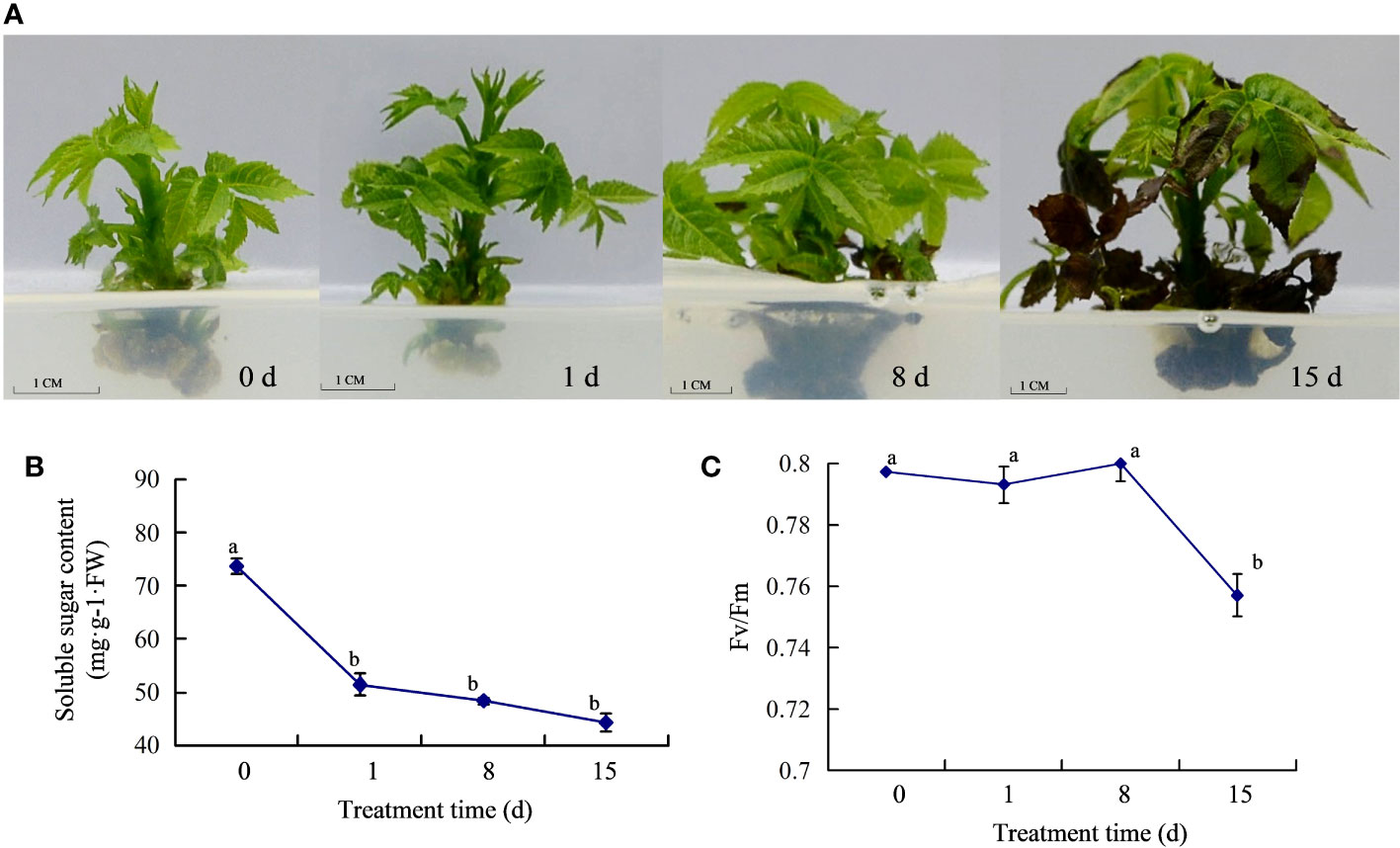
Figure 1 Response of walnut tissue cultures to walnut blight pathogen. (A), Phenotypes of the walnuts inoculated with pathogen; (B), Determination of soluble sugar content; (C), Determination of Fv/Fm.
3.2 Physiological responses of walnut tissue cultures under pathogen treatment
We measured the activity of antioxidant enzymes in walnut tissue cultures following pathogen treatment. The results showed that after infection, SOD activity increased gradually, with no significant change in activity at 0–8 dpi, and the highest activity at 15 dpi (Figure 2A). However, POD activity first increased and then decreased, reaching its highest value at 1 dpi, which was approximately six times the value at 0 dpi (Figure 2B). Previous studies showed that phenols and flavonoids play vital roles in disease resistance. Therefore, we determined the total phenol and flavonoid content in the infected tissues. The results showed that both total phenol and flavonoid contents increased sharply in the early stage of inoculation, reached a maximum at 1 dpi, and then decreased (Figures 2C, D), but the contents were still higher than those at 0 dpi, indicating that secondary metabolites may play a major role in the early stage of infection.
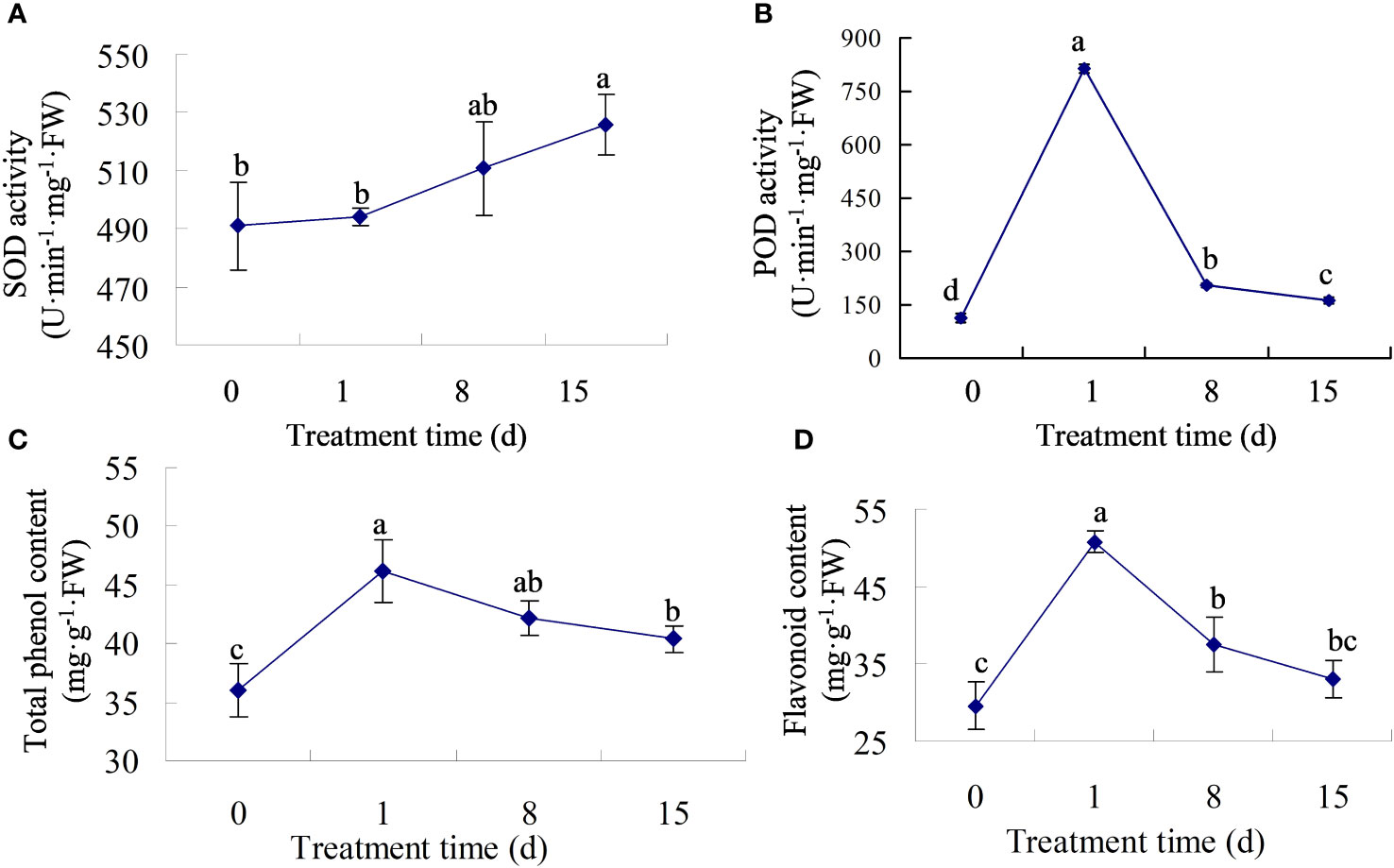
Figure 2 The physiological traits of walnut tissue cultures under walnut blight pathogen treatment. (A, B), Determination of SOD and POD activity. (C, D), Determination of total phenol and flavonoid contents.
3.3 RNA-sequencing results
To detect comparative gene expression changes in walnut tissue cultures in response to walnut blight pathogens, transcriptome analysis was performed on seedlings inoculated with P. agglomerans at 0 (C0), 1 (T1), 8 (T2), and 15 dpi (T3) using a high-throughput sequencing platform. The quality of the transcriptome data of the 12 samples was assessed. Detailed sequencing and assembly data are presented in Table S2. A total of 292,966,049 clean reads were obtained from mapping of the walnut genome sequence. The GC content was between 45.77–46.68%, and the Q30 bases were distributed between 94.01–95.01%. Principal component analysis (PCA) showed that sample C0-3 was clustered with T2 (Figure S1), so this sample was removed in the subsequent analysis. There were 2,337 DEGs in C0vsT1 with 962 up-regulated and 1,375 down-regulated genes (Table S3). In C0vsT2, there were 1,524 DEGs with 970 upregulated and 554 downregulated genes (Table S4). However, in comparison C0vsT3, 7,086 DEGs were found, with 3,742 upregulated and 3,344 downregulated genes (Table S5), suggesting that the gene changes were most dramatic in walnut tissue cultures at 15 dpi.
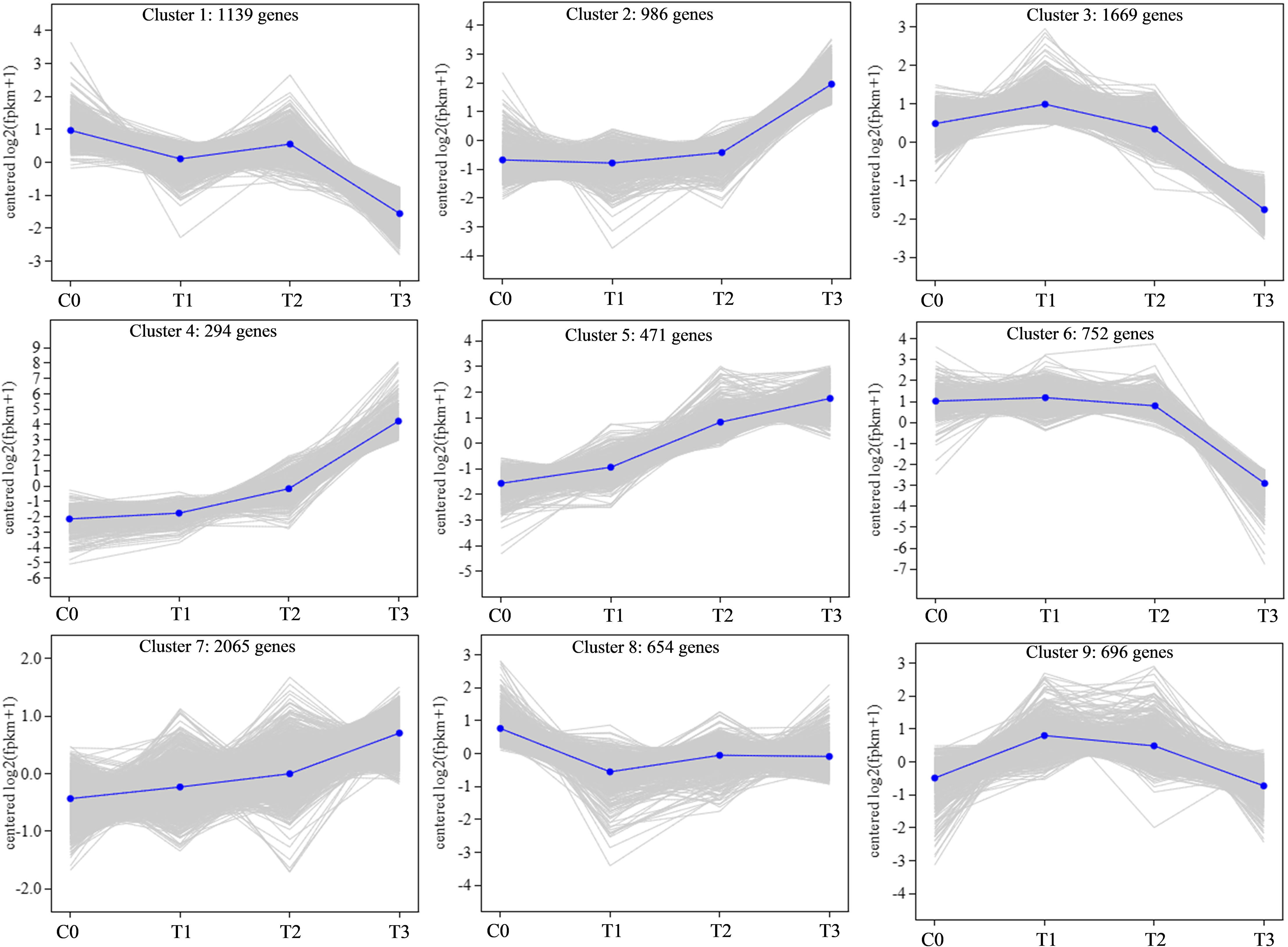
3.4 Co-expression patterns of DEGs
Co-expression patterns were analyzed to detect gene expression trends in walnut tissue cultures during walnut blight pathogen infection. Nine statistically significant profiles were identified (Figure 3; Table S6). Overall, the expression of genes in clusters 2, 4, 5, and 7 was upregulated after infection, particularly from T2 to T3. Gene expression in clusters 1, 3, and 6 was downregulated after infection, particularly from T2 to T3. Gene expression in cluster 8 first decreased and then increased, and gene expression in cluster 9 first increased and then decreased. In addition, in the early stage of pathogen treatment (C0-T1), the genes in cluster 8 changed significantly and were mainly enriched in the circadian rhythm–plant, porphyrin and chlorophyll metabolism, peroxisome, and inositol phosphate metabolism categories (Figure 4; Table S7), and the genes were downregulated. Genes in clusters 4 and 5 changed mainly in the middle stage (T1-T2), which was mainly enriched in phenylpropanoid biosynthesis, diterpenoid biosynthesis, sesquiterpenoid and triterpenoid biosynthesis, MAPK signaling pathway-plant, and plant-pathogen interaction categories, and these genes were upregulated. Dramatically upregulated genes in the later stage (T2-T3) were mainly in clusters 2 and 4, which were enriched in phenylpropanoid biosynthesis, MAPK signaling pathway-plant, and amino acid metabolism categories, and the downregulated genes in the later stage were mainly in clusters 1, 3, and 6, which were mainly enriched in photosynthesis, carbon metabolism, biosynthesis of amino acids, phenylpropanoid biosynthesis, plant hormone signal transduction, and other categories. We analyzed the DEGs in clusters 1-6 in which the DEGs were significantly downregulated or upregulated after pathogen inoculation.
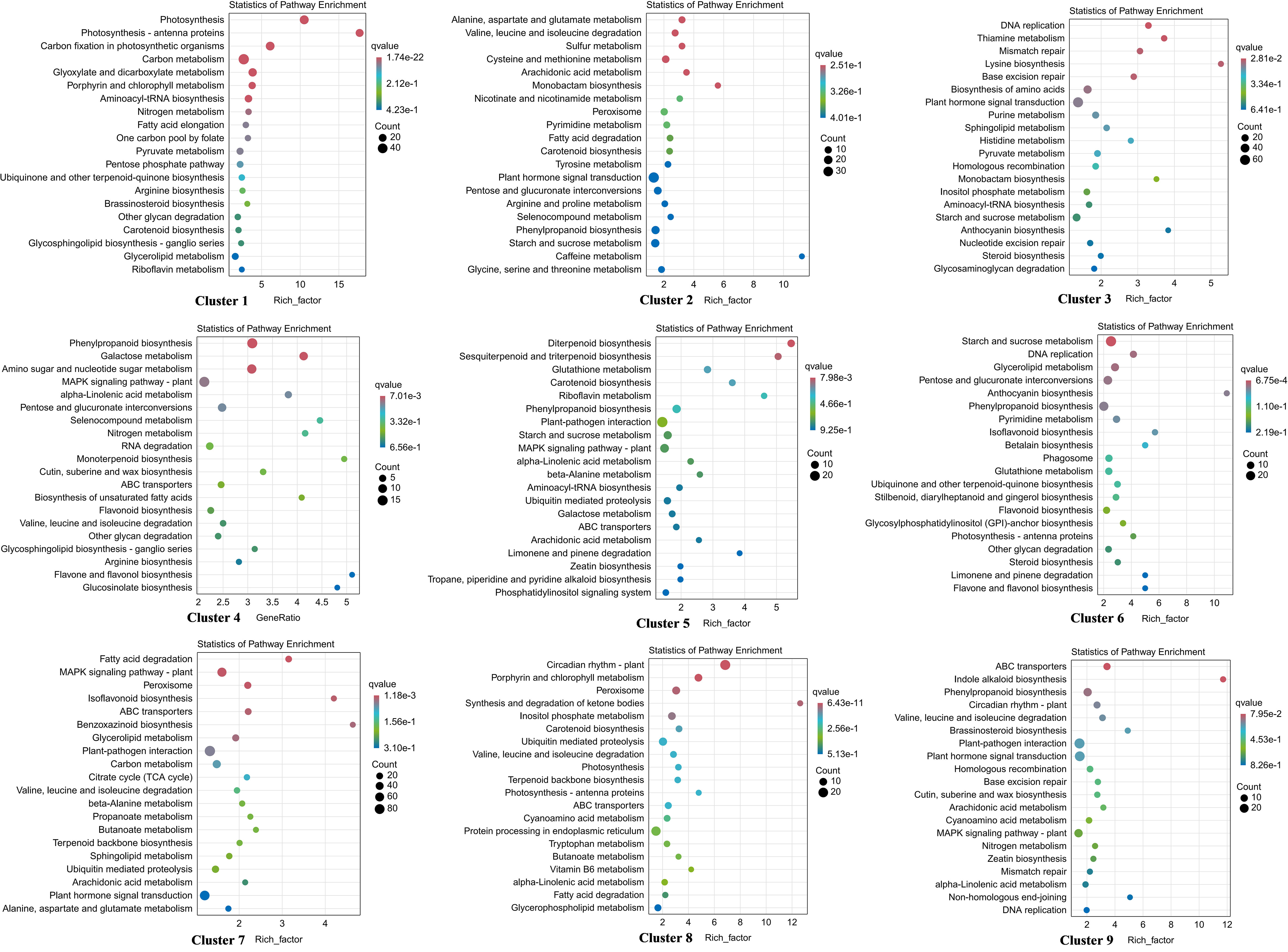
Figure 4 KEGG enrichment of DEGs in 9 profiles. The larger the bubble is, the more the number of DEGs involved. The redder the color of bubbles is, the more significant the enrichment is.
3.5 DEGs involved in plant hormone signal transduction
To analyze the response of hormone signals to P. agglomerans infection in walnut tissue cultures, we analyzed the DEGs involved in hormone signal transduction (Table S8). First, we analyzed the expression of auxin-related genes. Many auxin response gene expression changed after inoculation, including the early response genes Aux/IAA, SAUR, GH3 and the auxin response factor gene ARF. Twenty-five Aux/IAA were detected, and only two genes, AUX1 (JreChr14G10665) and IAA (JreChr02G10754), were upregulated after inoculation, with the highest expression levels at T3. The expression of the remaining Aux/IAA was gradually downregulated or upregulated initially and then downregulated after inoculation, and the expression level was the lowest at T3 (Figure 5A). Three GH genes, including GH3.1 (JreChr05G10405 and JreChr05G10425), GH3.6 (JreChr12G11319) and 7 SAUR genes SAUR32 (JreChr05G11465 and JreChr05G11464), SAUR36 (JreChr11G11531 and JreChr14G11097), SAUR71 (JreChr06G10870), SAUR6B (JreChr08G12047 and JreChr15G11383), were upregulated, and all the other SAUR and GH genes were downregulated (Figures 5B, C). In addition, the expression of seven ARF genes was downregulated (Figure 5D). Ethylene, ABA, SA, and JA are involved in plant stress responses. Therefore, we analyzed the gene expression of these hormone signaling pathways. SA is involved in disease resistance by inducing the expression of pathogenesis-related (PR) genes. There were four PR1 genes; the expression of JreChr05G10873 and JreChr05G10921 was induced, while the other two genes were repressed (Figure 5E). 1-aminocyclopropane-1-carboxylate synthase (ACS) catalyses the synthesis of ethylene precursor ACC. Two ACS genes (JreChr01G13882 and JreChr11G11292) were upregulated at T2 and T3, and four ethylene-responsive transcription factor genes, ERF1B and ERFC3 were upregulated (Figure 5F). For ABA signal transduction, ten protein phosphatase 2C genes were detected. Two genes, PP2C.50 (JreChr16G10051) and PP2C.53 (JreChr15G11806), were highly expressed at C0 and T1, and then decreased following inoculation, whereas the other PP2C genes were upregulated, especially at T3 (Figure 5G). During JA signal transduction, jasmonic acid-amino synthetase genes, JAR (JreChr02G10297 and JreChr01G10944), were highly expressed at C0-T2 and down-regulated at T3. In addition, we obtained nine TIFY genes; the expression of TIFY6B showed a downward trend, and TIFY10b was first upregulated and then decreased. Expression of the other five TIFY genes was gradually upregulated after inoculation (Figure 5H).
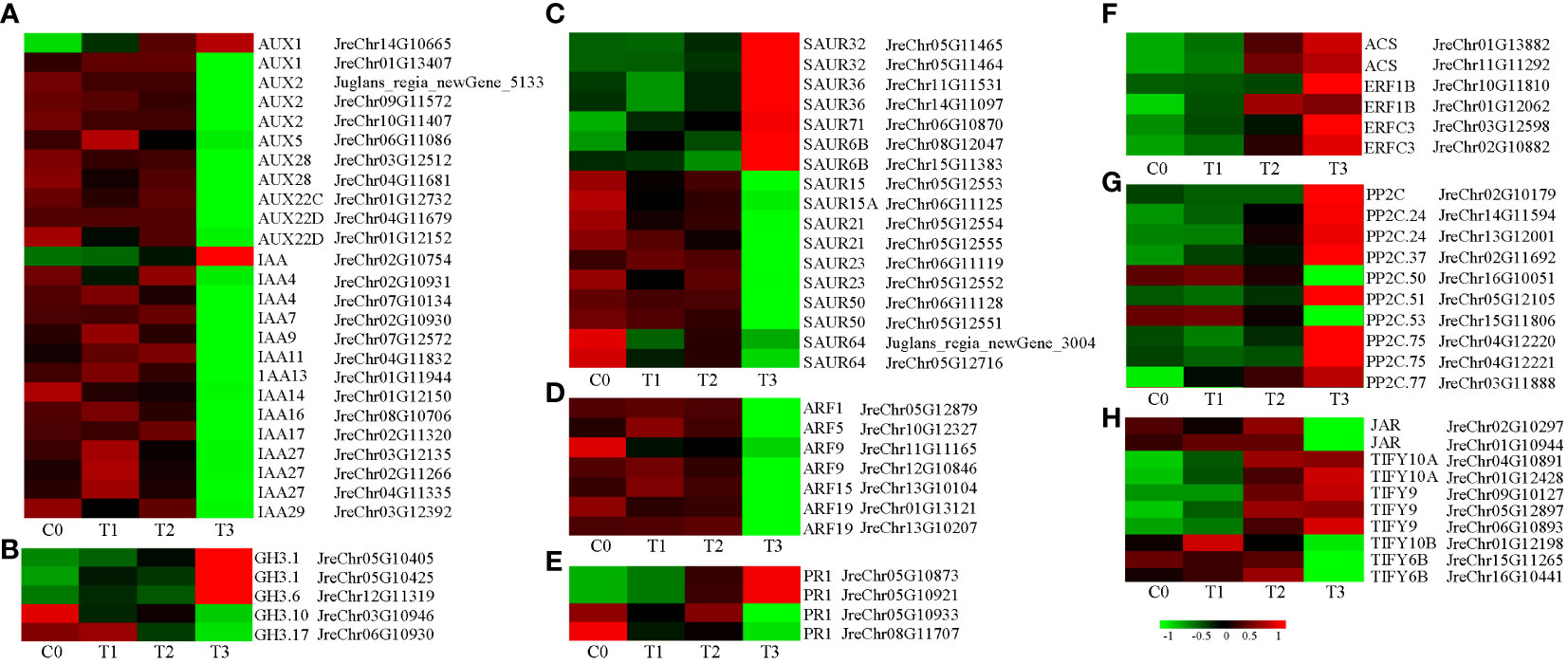
Figure 5 Expression of genes involved in hormone signal transduction after pathogen inoculation. (A–D), Expression of auxin-related genes AUX/IAA, GH3, SAUR, and ARF. (E), Expression of SA-related genes. (F), Expression of ethylene-related genes. (G), Expression of ABA-related genes. (H), Expression of JA-related genes. C0, T1, T2, and T3 refer to the 0, 1, 8, and 15 days after pathogen inoculation, respectively. Red and green indicate up- and down-regulation, respectively. Values are shown as log2(fpkm+1), which obtained through gene co-expression analysis.
3.6 DEGs involved in phenylpropanoid biosynthesis
Phenylpropane biosynthesis is an important secondary metabolic pathway in plants that plays an important role in the stress response. There are 65 DEGs involved in the phenylpropanoid biosynthesis pathway in clusters 1–6 (Figure 6; Table S9). The expression of CES (JreChr02G11615 and JreChr04G11602), C3’H (JreChr09G12012), REF1 (JreChr04G11390), F6H (JreChr13G10907), and CCoAOMT (JreChr05G12881) genes were upregulated by pathogen induction, and the expression levels were highest at T3. UGT72E (JreChr01G11984 and JreChr01G11970) was downregulated after infection, with the highest expression at C0. There were 16 POX genes; six genes (JreChr03G10690, JreChr04G10636, JreChr03G11462, JreChr15G11746, JreChr14G11423, and JreChr01G13501) had higher expression levels at C0–T2, whereas the remaining 10 genes had low expression at the early stage, and their expression increased gradually after infection. Six CAD genes (JreChr09G10469, JreChr10G10465, JreChr09G10465, Juglans_regia_newGene_3795, JreChr06G11577, and JreChr05G10125) were upregulated, with the highest expression at T3, and two CAD genes (JreChr08G12114 and JreChr03G10217) were downregulated. However, the expression of most bglX, CCR, HCT, COMT, and TOGT1 genes was higher at C0–T2, and the expression levels were the lowest at T3, indicating that genes involved in the phenylpropanoid pathway were greatly affected by P. agglomerans, especially at T3.
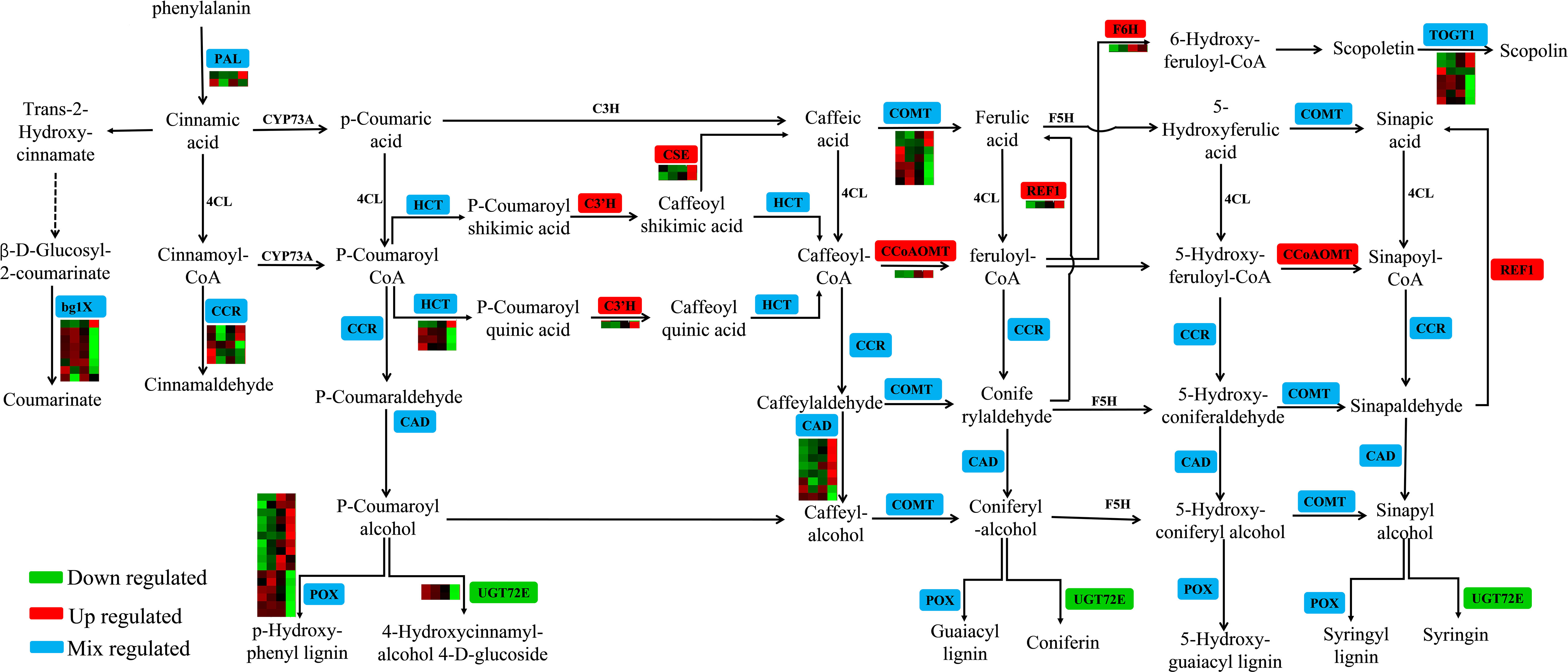
Figure 6 Expression of genes involved in phenylpropanoid biosynthesis. For the heatmap, each row represents a gene, and four columns represent C0, T1, T2, and T3, respectively.
3.7 DEGs involved in plant-pathogen interaction
Many genes involved in plant-pathogen interactions were differentially expressed (Table S10). The receptor-like protein kinase FLS2 recognizes the flagellin protein flg22 to activate pathogen-associated molecular patterns (PAMP)-triggered immunity (PTI). FLS2 interacts with BAK1 to activate the downstream MAPK signaling pathway (Kourelis and van der Hoorn, 2018; Zhou and Zhang, 2020). After infection, eight FLS2 genes were detected and their expression first increased and then decreased except for JreChr05G11743, JreChr03G12298, and JreChr16G10740. The expression of BAK1 genes was similar to FLS2. MAPKKK18, MAPKK5, and MAPKK9 were upregulated, whereas MAPKK1 and MAPKK6 were downregulated after inoculation (Figure 7A). R proteins recognize different pathogen effectors that activate effector-triggered immunity (ETI). There are three types of R genes: PTO, RPM1, and RPS2. Three PTO genes (JreChr04G12080, JreChr09G10406, and JreChr03G13221) were upregulated at T1 and then decreased. The other six PTO genes were down-regulated after inoculation (Figure 7B). Except for JreChr03G10302 and JreChr04G12542, the RMP1 genes were upregulated in infected tissues (Figure 7B). In addition, P. agglomerans infection changed the calcium signaling pathway. Cyclic nucleotide-gated channels (CNGC) are Ca2+ channels responsible for cytoplasmic Ca2+ signaling in plants. CNGC1 (JreChr05G12110) and CNGC15b (Juglans_regia_newGene_5511) were downregulated, the other CNGC genes expression first increased and then decreased, with the highest expression observed at T1. CPK showed the highest expression at T1. Among calcium-binding protein (CML) genes, CML3 was downregulated, CML30 and CML41 (JreChr02G10892) were highly expressed at C0 and T2. CML1, CML8, CML41 (JreChr01G11675), CML45, and CML47 were upregulated in infected tissues, with the highest expression at T3 (Figure 7C). Furthermore, we analyzed the expression of transcription factors involved in plant-pathogen interactions and found that two types of transcription factor genes were differentially expressed. Seven bHLH137 genes were downregulated, with the lowest expression observed in T3. Except for three WRKY22 genes (JreChr15G12139, JreChr15G12133, and JreChr08G10036), the other WRKY genes were all upregulated after inoculation, with the highest expression at T3, which showed an opposite trend to bHLH137, indicating that these WRKY genes may have opposite functions to bHLH137 in response to pathogen infection (Figure 7D).
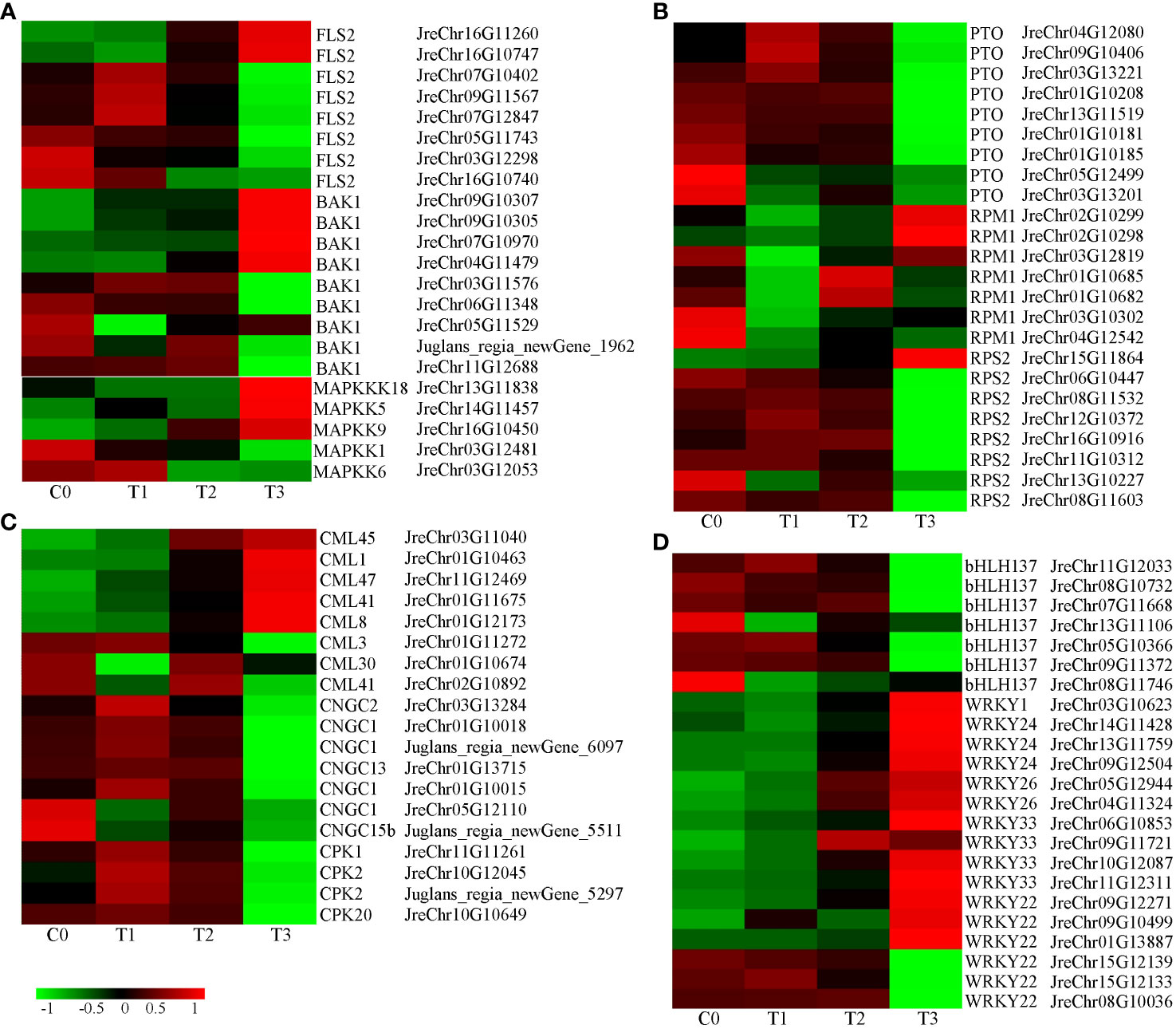
Figure 7 Expression of genes involved in plant-pathogen interaction. (A), Expression of genes involved in flag22-FLS2 mediated disease resistance pathway. (B), Expression of R genes. (C), Expression of calcium signaling-related genes. (D), Expression of transcription factor genes bHLH and WRKY.
3.8 qRT-PCR validation of DGEs
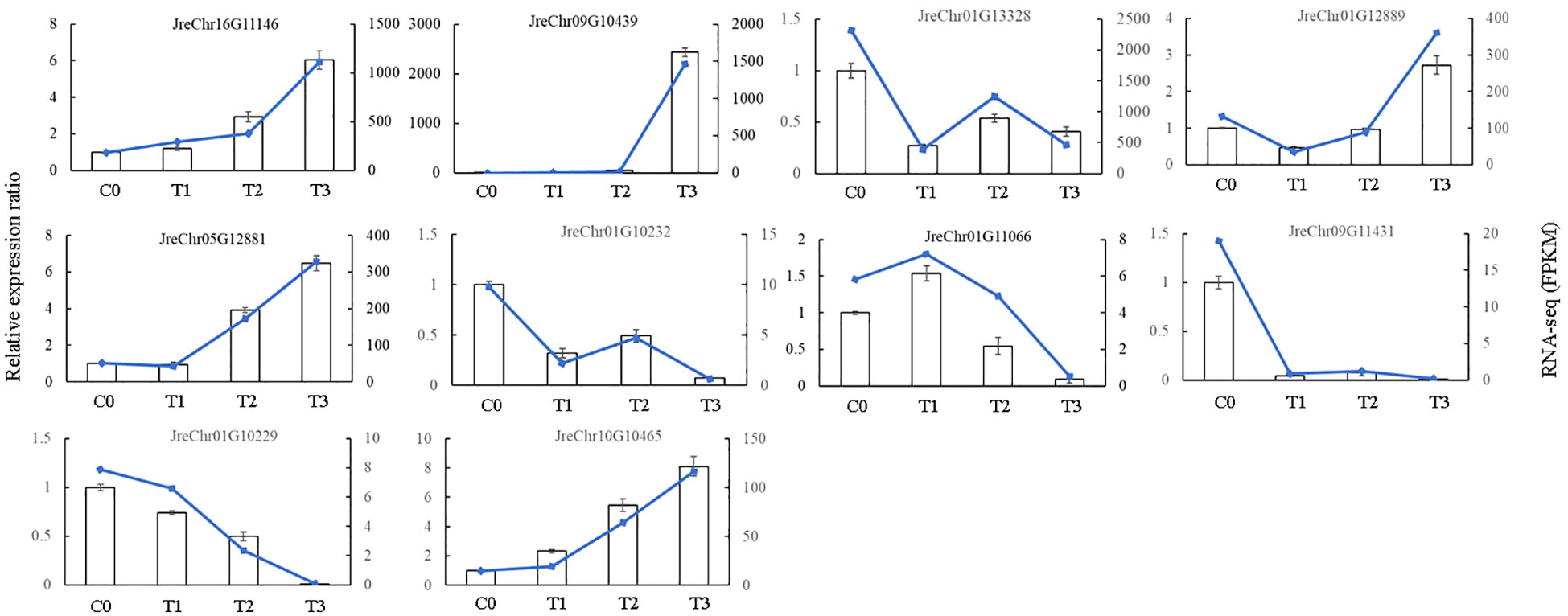
To verify the accuracy of our RNA-seq data, we randomly selected ten DEGs in the phenylpropanoid pathway for PCR validation. The results showed that the trend in gene expression obtained by qRT-PCR was consistent with that obtained by RNA-seq (Figure 8), indicating that the RNA-seq data were reliable.
4 Discussion
Walnut blight is a severe disease in walnuts that can cause serious economic losses. Currently, copper-based biocides are commonly used to control walnut blight. However, these biocides can easily cause bacteria to develop resistance and pollute the environment (Jiang et al., 2019a). Therefore, it is necessary to explore the plant-pathogen interactions in walnuts infected with bacterial blight. In this study, transcriptome data were collected to understand the response mechanisms of walnuts to P. agglomerans infection.
When attacked by pathogens, ROS rapidly accumulate in plant cells. To avoid excessive ROS damage to cells, plants have evolved antioxidant systems, including SOD, POD, ascorbate peroxidase (APX), catalase (CAT), etc (De Gara et al., 2003). In maize inoculated with Fusarium verticillioides and Piriformospora indica, the SOD, CAT, and glutathione S-transferase (GST) activities in the roots significantly increased (Kumar et al., 2009). In Cucumber plants, the activity of antioxidant enzymes such as SOD, POD, and CAT was significantly increased after Cucumber mosaic cucumovirus inoculation (Sofy et al., 2020). Similar results were found in our study; the POD activity increased after P. agglomerans infection, especially in the early stage (Figure 2B), and moreover, SOD activity increased, although the increase was slower (Figure 2A).
More researches proved that the secondary metabolites such as flavonoids and phenols played important roles in plant defense reactions and plant–pathogen interaction (Sofy et al., 2020). Mikulic-Petkovsek et al. (2011) found that the cumulative content of polyphenols in fruits was weakly correlated with walnut blight susceptibility, and the total content of the analyzed phenolic compounds increased after infection with Xaj, especially in the walnut cultivars Cisco and Zdole. Petkovšek et al. (2008) found that all the phenolic substances detected, including chlorogenic acid, caffeic acid, p-coumaric acid, rutin, catechin, epicatechin, and quercetin-3-O-rhamnoside, were higher in infected leaves than in healthy leaves. Similar results were found in walnuts, in which the total phenolic content and total analyzed phenolic content of leaves infected with Xanthomonas campestris pv. Juglandis was higher than that in healthy leaves (Medic et al., 2022). After infection with V. inaequalis, the ability to accumulate flavanols differs between resistant and susceptible apple cultivars (Treutter and Feucht, 1990). However, Sierotzki and Gessler (1993) showed that preformed flavan-3-ols and disease resistance were not positively correlated in Malus x domestica and Venturia. In our study, the total phenol and flavonoid content was accumulated rapidly in the early stages of pathogen inoculation (Figures 2C, D), although there were no obvious symptoms at this time (Figure 1). In addition, transcriptome data analysis showed that the expression levels of many genes involved in the phenylpropane pathway were higher in the early stages of infection (Figure 6), suggesting that secondary metabolite pathways may be involved in the response of walnuts to bacterial blight, especially in the early stages. In addition to secondary metabolites and antioxidant enzymes, the soluble sugar content and Fv/Fm changed after pathogen infection and showed a downward trend (Figures 1B, C), indicating that P. agglomerans infection altered energy metabolism and photosynthesis and then inhibited walnut tissue culture growth.
Plant hormones, such as SA, ethylene, and JA, play important roles in disease resistance and crosstalk. Abdelrahman et al. (2017) found that SA signaling-related genes PR1 and JA biosynthesis and signaling-related genes PLDα1 (phospholipase D alpha 1), OPR (12-oxophytodienoate reductase), and JIP23 (jasmonate-induced protein 23 KD) were upregulated in resistant wild Asparagus kiusianus stems relative to susceptible A. officinalis after P. asparagi infection. Sagawa et al. (2020) found that the PR proteins PR1, PR2, PR3, and PR5 significantly accumulated in walnut fruits with Xaj infection, and moreover, the ethylene biosynthesis proteins 1-aminocyclopropane-1-carboxylate oxidase (ACC) and S-adenosylmethionine synthase (SAM) were increased. Kong et al. (2020) showed that auxin signaling antagonizes SA signaling by regulating Pseudomonas syringae pv. tomato DC3000 infection through Arabidopsis lateral roots. Zhao and Li (2021) suggested that SA, JA, ethylene, ABA, auxin, GA, cytokinins, and brassinosteroids (BRs) act together to regulate plant-virus interactions. In our study, the expression of most auxin-related genes was higher in the early stage of pathogen infection and decreased in the later stage, while the expression of ethylene- and ABA-related genes showed the opposite trend (Figure 5), indicating that P. agglomerans infection inhibited auxin signaling and promoted ethylene and ABA signaling, whereas SA and JA signaling may play roles throughout the process of plant-pathogen interaction. Whether these hormones have synergistic or antagonistic effects on the response of walnuts to pathogenic infections requires further investigation.
Plants have evolved a two-tiered immune system that includes the PTI and ETI. Flg22 is a 22-amino acid peptide located at the N-terminus of flagellin, which is recognized by a variety of plants and induces PTI. After P. agglomerans inoculation, flg22-FLS2 mediated signaling changed significantly, and more FLS2 and BAK1 genes first increased and then decreased (Figure 7A), indicating that PT1 was induced in walnut tissues by P. agglomerans treatment. Calcium is a secondary messenger that plays an important role in the activation of the plant immune system. Our results showed that the expression of calcium signaling pathway genes first increased or decreased (Figure 7C), indicating that pathogen inoculation activated calcium signaling pathways. However, the expression of R-protein genes PTO and RPS was mainly suppressed at T3, and RPM was upregulated at T2 or T3 (Figure 7B), indicating that the ETI pathway may be involved in the response of walnuts to pathogen infection. Further studies are needed to determine which R-proteins play roles in the recognition of blight pathogen in walnut.
5 Conclusion
Our results indicated that P. agglomerans infection affects energy metabolism and photosynthesis, induces the phenylpropanoid biosynthesis pathway, and promotes the accumulation of phenols and flavonoids in walnut tissue cultures. In addition, bacterial infection inhibits auxin signaling and promotes ABA and ethylene signal transduction. FLS- and calcium-mediated disease resistance pathways are activated, and the R-protein gene RPM may be involved in the response of walnuts to pathogen infection. These results provide insights into the molecular mechanisms in the response of walnuts to P. agglomerans infection.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Files, further inquiries can be directed to the corresponding author/s.
Author contributions
X-HA: Formal analysis, Writing – original draft, Writing – review & editing. NW: Formal analysis, Writing – original draft. HW: Funding acquisition, Project administration, Writing – review & editing. YL: Formal analysis, Validation, Writing – review & editing. X-YS: Formal analysis, Validation, Writing – review & editing. SZ: Data curation, Formal analysis, Writing – review & editing. YT: Project administration, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the National Key Research and Development Program (2022YFD1600402), the Key Research and Development Program of Hebei Province (21326352D), and the Natural Science Foundation of Hebei Province (C2023204269), and the Science and Technology Project of Hebei Education Department (QN2021075).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1294643/full#supplementary-material
Supplementary Figure 1 | The PCA analysis of sequencing samples.
Supplementary Table 1 | The primers used for qRT-PCR.
Supplementary Table 2 | The quality statistic of the sequencing data.
Supplementary Table 3 | DEGs in the C0vsT1.
Supplementary Table 4 | DEGs in the C0vsT2.
Supplementary Table 5 | DEGs in the C0vsT3.
Supplementary Table 6 | DEGs in each cluster.
Supplementary Table 7 | The KEGG analysis of DEGs in each cluster.
Supplementary Table 8 | DEGs involved in hormone signal transduction.
Supplementary Table 9 | DEGs involved in phenylpropanoid biosynthesis.
Supplementary Table 10 | DEGs involved in plant-pathogen interaction.
References
Abdelrahman, M., Suzumura, N., Mitoma, M., Matsuo, S., Ikeuchi, T., Mori, M., et al. (2017). Comparative de novo transcriptome profiles in Asparagus officinalis and A. kiusianus during the early stage of Phomopsis asparagi infection. Sci. Rep. 7, 2608. doi: 10.1038/s41598-017-02566-7
Ayele, D. T., Akele, M. L., Melese, A. T. (2022). Analysis of total phenolic contents, flavonoids, antioxidant and antibacterial activities of Croton macrostachyus root extracts. BMC Chem. 16, 30. doi: 10.1186/s13065-022-00822-0
Bandi, A., Hevesi, M., Szani, Z., TÓTh, M. (2015). Assessment of bacterial blight tolerance of persian walnut based on immature nut test. Notulae Scientia Biologicae 7, 253–257. doi: 10.15835/nsb729620
Belisario, A., Zoina, A. (1995). Occurrence of Persian (English) walnut blight and its control in the nursery. Eur. J. For. Pathol. 25, 224–231. doi: 10.1111/j.1439-0329.1995.tb01006.x
Bolger, A. M., Lohse, M., Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Chakraborty, S., Britton, M., Martínez-García, P. J., Dandekar, A. M. (2016). Deep RNA-Seq profile reveals biodiversity, plant–microbe interactions and a large family of NBS-LRR resistance genes in walnut (Juglans regia) tissues. AMB Express 6, 1–13. doi: 10.1186/s13568-016-0182-3
Cheng, A., Liu, C. Y., Tsai, H. Y., Hsu, M. S., Yang, C. J., Huang, Y. T., et al. (2013). Bacteremia caused by Pantoea agglomerans at a medical center in Taiwa –2010. J. Microbiology Immunol. Infection 46, 187–194. doi: 10.1016/j.jmii.2012.05.005
De Gara, L., de Pinto, M. C., Tommasi, F. (2003). The antioxidant systems vis-à-vis reactive oxygen species during plant–pathogen interaction. Plant Physiol. Biochem. 41, 863–870. doi: 10.1016/S0981-9428(03)00135-9
del Río, L. A. (2015). ROS and RNS in plant physiology: an overview. J. Exp. Bot. 66, 2827–2837. doi: 10.1093/jxb/erv099
Dutkiewicz, J., Mackiewicz, B., Lemieszek, M. K., Golec, M., Milanowski, J. (2016). Pantoea agglomerans: a mysterious bacterium of evil and good. Part IV. Beneficial effects. Ann. Agric. Environ. Med. 23, 206–222. doi: 10.5604/12321966.1203879
Foyer, C. H., Descourvieres, P., Kunert, K. J. (1994). Protection against oxygen radicals: an important defence mechanism studied in transgenic plants. Plant Cell Environ. 17, 507–523. doi: 10.1111/j.1365-3040.1994.tb00146.x
Frutos, D., Ortega, G. (2012). Search for juglans regia genotypes resistant/tolerant to Xanthomonas abboricola pv. juglandis in the framework of cos action 873. J. OF Plant Pathol. 94, S1.37–S1.46. doi: 10.4454/jpp.v94i1sup.007
Gašić, K., Zlatković, N., Kuzmanović, N. (2022). Polyphasic study of phytopathogenic bacterial strains associated with deep bark canker of walnut in Serbia revealed a new species, Brenneria izbisi sp. nov. Front. Plant Sci., 13, 1055186. doi: 10.3389/fpls.2022.1055186
Guo, M., Liu, Y., Liu, S. N., Qu, Q. Z., Cui, T. F., Zhang, Y. W. (2020). First report of bacterial soft rot caused by Pantoea agglomerans on chinese cabbage in China. Plant Dis. 104, 277. doi: 10.1094/PDIS-04-19-0746-PDN
Gutierrez-Barranquero, J. A., Cazorla, F. M., Antonio Tores, J., de Vicente, A. (2019). Pantoea agglomerans as a new etiological agent of a bacterial necrotic disease of mango trees. Phytopathology 109, 17–26. doi: 10.1094/PHYTO-06-18-0186-R
Higuera, G., González-Escalona, N., Véliz, C., Vera, F., Romero, J. (2015). Draft genome sequences of four Xanthomonas arboricola pv. juglandis strains associated with walnut blight in Chile. Genome Announcements 3, e01160-15. doi: 10.1128/genomeA.01160-15
Ji, F., Ma, Q., Zhang, W., Liu, J., Feng, Y., Zhao, P., et al. (2021). A genome variation map provides insights into the genetics of walnut adaptation and agronomic traits. Genome Biol. 22, 1–22. doi: 10.1186/s13059-021-02517-6
Jiang, S., Han, S., He, D., Cao, G., Fang, K., Xiao, X., et al. (2019a). The accumulation of phenolic compounds and increased activities of related enzymes contribute to early defense against walnut blight. Physiol. Mol. Plant Pathol. 108, 101433. doi: 10.1016/j.pmpp.2019.101433
Jiang, S., Han, S., He, D., Cao, G., Zhang, F., Wan, X. (2019b). Evaluating walnut (Juglans spp.) for resistance to walnut blight and comparisons between artificial inoculation assays and field studies. Australas. Plant Pathol. 48, 221–231. doi: 10.1007/s13313-019-0621-0
Kanehisa, M., Araki, M., Goto, S., Hattori, M., Hirakawa, M., Itoh, M., et al. (2007). KEGG for linking genomes to life and the environment. Nucleic Acids Res 36, D480–D484. doi: 10.1093/nar/gkm882
Kim, D., Paggi, J. M., Park, C., Bennett, C., Salzberg, S. L. (2019). Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 37, 907–915. doi: 10.1038/s41587-019-0201-4
Kong, X., Zhang, C., Zheng, H., Sun, M., Zhang, F., Zhang, M., et al. (2020). Antagonistic interaction between auxin and SA signaling pathways regulates bacterial infection through lateral root in Arabidopsis. Cell Rep. 32, 108060. doi: 10.1016/j.celrep.2020.108060
Kourelis, J., van der Hoorn, R. A. L. (2018). Defended to the nines: 25 years of resistance gene cloning identifies nine mechanisms for R protein function. Plant Cell 30, 285–299. doi: 10.1105/tpc.17.00579
Kumar, M., Yadav, V., Tuteja, N., Johri, A. K. (2009). Antioxidant enzyme activities in maize plants colonized with Piriformospora indica. Microbiology 155, 780–790. doi: 10.1099/mic.0.019869-0
Lamichhane, J. R. (2014). Xanthomonas arboricola diseases of stone fruit, almond, and walnut tees: progress toward understanding and management. Plant Dis. 98, 1600–1610. doi: 10.1094/PDIS-08-14-0831-FE
Li, C., Jia, Y., Tian, Y., Zhou, L., Sun, W., Deng, J., et al. (2020). First report of necrotic disease caused by Pantoea agglomerans on Plum (Prunus salicina) in China. Plant Dis. 104, 1248–1248. doi: 10.1094/PDIS-10-19-2131-PDN
Li, X., Lin, J., Gao, Y., Han, W., Chen, D. (2012). Antioxidant activity and mechanism of Rhizoma Cimicifugae. Chem. Cent. J. 6, 140. doi: 10.1186/1752-153X-6-140
Li, S., Tan, H. Y., Wang, N., Zhang, Z. J., Lao, L., Wong, C. W., et al. (2015). The role of oxidative stress and antioxidants in liver diseases. Int. J. Mol. Sci. 16, 26087–26124. doi: 10.3390/ijms161125942
Love, M. I., Huber, W., Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 1–21. doi: 10.1186/s13059-014-0550-8
Luo, X., Huang, Q. (2011). Relationships between leaf and stem soluble sugar content and tuberous root starch accumulation in cassava. J. Agric. Sci. 3, 64. doi: 10.5539/jas.v3n2p64
Medic, A., Jakopic, J., Hudina, M., Solar, A., Veberic, R. (2022). Identification and quantification of major phenolic constituents in Juglans regia L. leaves: healthy vs. infected leaves with Xanthomonas campestris pv. juglandis using HPLC-MS/MS. J. King Saud Univ. - Sci. 34, 101890. doi: 10.1016/j.jksus.2022.101890
Mikulic-Petkovsek, M., Slatnar, A., Veberic, R., Stampar, F., Solar, A. (2011). Phenolic response in green walnut husk after the infection with bacteria Xanthomonas arboricola pv. juglandis. Physiol. Mol. Plant Pathol. 76, 159–165. doi: 10.1016/j.pmpp.2011.09.006
Moragrega, C., Llorente, I. (2023). Effects of leaf wetness duration, temperature, and host phenological stage on infection of walnut by Xanthomonas arboricola pv. juglandis. Plants 12, 2800. doi: 10.3390/plants12152800
Moragrega, C., Matias, J., Aletà, N., Montesinos, E., Rovira, M. (2011). Apical necrosis and premature drop of Persian (English) walnut fruit caused by Xanthomonas arboricola pv. juglandis. Plant Dis. 95, 1565–1570. doi: 10.1094/PDIS-03-11-0259
Moragrega, C., Özaktan, H. (2010). Apical necrosis of persian (English) walnut (Juglans regia): an update. J. Plant Pathol. 92, S67–S71. doi: jstor.org/stable/41998757
Nunes, C., Usall, J., Teixidó, N., Viñas, I. (2001). Biological control of postharvest pear diseases using a bacterium, Pantoea agglomerans CPA-2. Int. J. Food Microbiol. 70, 53–61. doi: 10.1016/S0168-1605(01)00523-2
Olson, W. H., Buchner, R., Adaskaveg, J. E., Lindow, S. E. (1997). Walnut blight control in California. III International Walnut Congress 442, 361–366. doi: 10.17660/ActaHortic.1997.442.57
Petkovšek, M. M., Stampar, F., Veberic, R. (2008). Increased phenolic content in apple leaves infected with the apple scab pathogen. J. Plant Pathol. 90, 49–55. doi: jstor.org/stable/41998458
Petkovsek, M. M., Stampar, F., Veberic, R. (2009). Seasonal changes in phenolic compounds in the leaves of scab-resistant and susceptible apple cultivars. Can. J. Plant Sci. 89, 745–753. doi: 10.4141/CJPS08202
Radix, P., Bastien, C., Jay-Allemand, C., Charlot, G., Seigle-Murandi, F. (1998). The influence of soil nature on polyphenols in walnut tissues. A possible explanation of differences in the expression of walnut blight. Agronomie 18, 627–637. doi: 10.1051/agro:19981002
Sagawa, H. D. C., de A. B. Assis, R., Zaini, P. A., Wilmarth, P. A., Phinney, B. S., Moreira, L. M., et al. (2020). Proteome analysis of walnut bacterial blight disease. Int. J. Mol. Sci. 21, 7453. doi: 10.3390/ijms21207453
She, X. M., Yu, L., Lan, G. B., Tang, Y. F., Deng, M. G., Li, Z. G., et al. (2019). First report of necrotic disease caused by Pantoea agglomerans on Ziziphus jujuba in China. Plant Dis. 103, 1405. doi: 10.1094/PDIS-01-19-0025-PDN
She, X., Yu, L., Lan, G., Tang, Y., Deng, M., Li, Z., et al. (2021). Pantoea agglomerans causing blight disease on pepino melon (Solanum muricatum) in China. Crop Prot. 139, 105385. doi: 10.1016/j.cropro.2020.105385
Shengke, X. (1987). Gene resources of Juglans and genetic improvement of Juglans regia in China. Sci. Silvae Sin. 3, 342–350.
Sierotzki, H., Gessler, C. (1993). Flavan-3-ol content and the resistance of Malus × domestica to Venturia inaequalis (Cke.) Wint. Physiol. Mol. Plant Pathol. 42, 291–297. doi: 10.1006/pmpp.1993.1027
Sofy, A. R., Dawoud, R. A., Sofy, M. R., Mohamed, H. I., Hmed, A. A., El-Dougdoug, N. K. (2020). Improving regulation of enzymatic and non-enzymatic antioxidants and stress-related gene stimulation in Cucumber mosaic cucumovirus-infected Cucumber plants treated with glycine betaine, chitosan and combination. Molecules 25, 2341. doi: 10.3390/molecules25102341
Solar, A., Colarič, M., Usenik, V., Stampar, F. (2006). Seasonal variations of selected flavonoids, phenolic acids and quinones in annual shoots of common walnut (Juglans regia L.). Plant Sci. 170, 453–461. doi: 10.1016/j.plantsci.2005.09.012
Soltani, J., Aliabadi, A. A. (2010). Genetic resistance to bacterial blight disease in Persian walnut. Eur. J. Plant Pathol. 128, 65–70. doi: 10.1007/s10658-010-9629-6
Soto, F., Tramón, C., Aqueveque, P., de Bruijn, J. (2018). Microorganismos antagonistas que inhiben el desarrollo de patógenos en post-cosecha de limones (Citrus limon L.). Chilean J. Agric. Anim. Sci. 34, 173–184. doi: 10.4067/S0719-38902018005000406
Treutter, D., Feucht, W. (1990). The pattern of flavan-3-ols in relation to scab resistance of apple cultivars. J. Hortic. Sci. 65, 511–517. doi: 10.1080/00221589.1990.11516087
Vauterin, L., Hoste, B., Kersters, K., Swings, J. (1995). Reclassification of xanthomonas. Int. J. Systematic Evolutionary Microbiol. 45, 472–489. doi: 10.1099/00207713-45-3-472
Woeste, K. E., McGranahan, G. H., Schroth, M. N. (1992). Variation among persian walnuts in response to inoculation with Xanthomonas campestris pv. juglandis. J. Am. Soc. Hortic. Sci. 117, 527–531. doi: 10.21273/JASHS.117.3.527
Yang, K. Q., Qu, W. W., Liu, X., Liu, H. X., Hou, L. Q. (2011). First report of Pantoea agglomerans causing brown apical necrosis of walnut in China. Plant Dis. 95, 773–773. doi: 10.1094/PDIS-01-11-0060
Zamorano, A., Zuñiga, T., Córdova, P., Higuera, G., Bertaccini, A., Fiore, N. (2022). Pantoea agglomerans-induced dieback in Pistachio in Chile. Horticulturae 8, 1052. doi: 10.3390/horticulturae8111052
Zang, R., Song, L. L., Xu, C., Yin, X. L., Qiao, Z. X., Geng, Y. H., et al. (2021). Clarification of the etiology of peach bacteria leaf shot hole caused by Pantoea spp. in Henan province, China, based on morphological characteristics and multi-locus sequences analysis. Eur. J. Plant Pathol. 161, 253–271. doi: 10.1007/s10658-021-02319-3
Zhang, J., Zhang, W., Ji, F., Qiu, J., Song, X., Bu, D., et al. (2020). A high-quality walnut genome assembly reveals extensive gene expression divergences after whole-genome duplication. Plant Biotechnol. J. 18, 1848–1850. doi: 10.1111/pbi.13350
Zhao, S., Li, Y. (2021). Current understanding of the interplays between host hormones and plant viral infections. PloS Pathog. 17, e1009242. doi: 10.1371/journal.ppat.1009242
Keywords: walnut blight, physiological response, transcriptomic analysis, phenylpropanoid biosynthesis, hormone signal
Citation: An XH, Wang N, Wang H, Li Y, Si XY, Zhao S and Tian Y (2023) Physiological and transcriptomic analyses of response of walnuts (Juglans regia) to Pantoea agglomerans infection. Front. Plant Sci. 14:1294643. doi: 10.3389/fpls.2023.1294643
Received: 15 September 2023; Accepted: 21 November 2023;
Published: 05 December 2023.
Edited by:
Yue Ma, Shenyang Agricultural University, ChinaCopyright © 2023 An, Wang, Wang, Li, Si, Zhao and Tian. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hongxia Wang, MTUwOTc3OTMyMDNAMTYzLmNvbQ==; Yi Tian, NDIwNzQwNzI1QHFxLmNvbQ==
† These authors have contributed equally to this work
 Xiu-Hong An1†
Xiu-Hong An1† Hongxia Wang
Hongxia Wang Shugang Zhao
Shugang Zhao Yi Tian
Yi Tian